“The field of bacterial viruses is a fine playground for serious children who ask ambitious questions.” Max Delbruck
The first bacterial intron, a self-splicing group I intron, was found to interrupt the thymidylate synthase (td) gene of the Escherichia coli phage T4 (11). The second and third bacterial group I introns were found to interrupt the aerobic (nrdB) and anaerobic (nrdD [initially named sunY]) ribonucleotide reductases of phage T4 (29, 90), and another group I intron was soon discovered in the DNA polymerase gene of SPO1, a Bacillus phage (25). From this (admittedly) small sampling of phage genomes, one might have naively expected that group I introns would be abundant in phage or bacterial genomes, especially since subsequent laboratory experiments demonstrated that group I introns could propagate themselves (by a process called homing) throughout populations of intron-minus alleles with near 100% efficiency (5, 68). That a similar homing phenomenon had also been previously demonstrated for a group I intron in the large rRNA gene of yeast mitochondria (34) gave additional support to the notion that group I introns should be able to spread efficiently throughout populations. However, this expected outcome has never been realized in natural phage populations; some phage populations harbor many introns, while other related phage populations are strangely lacking in any introns whatsoever (Table 1). Why do group I introns have an unusual distribution in phage and bacterial genomes, and what potential barriers might exist to prevent their spread?
TABLE 1.
Distribution of group I introns in bacteria and bacteriophages
| Bacterium or bacteriophage | Species | Gene or enzyme | Endonucleasea | Splicingb | Reference(s) |
|---|---|---|---|---|---|
| Bacteria | |||||
| Gram-negative | A. tumefaciens | tRNACCUArg | None | Yes | 69 |
| Azoarcus sp. strain BH72 | tRNACAUIle | None | Yes | 69 | |
| Chlamydial | S. negevensis ZT | 23S rRNA | LAGLIDAG | No | 21 |
| Cyanobacterial | Many | tRNAUAALeu | None | Yes | 40, 64, 74, 87 |
| Many | tRNAfMet | Nonec | Yes | 6, 64 | |
| Bacteriophages | |||||
| Gram-negative bacteria | E. coli/T4, T6, RB3, LZ2, Tu1A | Thymidylate synthase (td) | I-TevI (GIY-YIG) | Yes | 11, 29, 67 |
| E. coli/T4 | Anaerobic ribonucleotide reductase (nrdD) | I-TevII (GIY-YIG) | Yes | 29, 67 | |
| E. coli/T4, RB3 | Aerobic ribonucleotide reductase (nrdB) | Fragment | Yes | 67 | |
| Gram-positive bacteria | B. subtilis/β22 | td | Fragment | Yes | 1 |
| B. subtilis/SPO1 | DNA polymerase | I-HmuI (H-N-H) | Yes | 25, 26, 27 | |
| B. subtilis/SP82 | DNA polymerase | I-HmuII (H-N-H) | Yes | 26, 27 | |
| B. subtilis/φe | DNA polymerase | I-HmuIII (H-N-H) | Yes | 26 | |
| B. subtilis/SPβ prophage | Ribonucleotide reductase (nrdF) | yosQ (H-N-H) | Yes | 45 | |
| S. thermophilus/Sb3 | Lysin | H-N-H | Yes | 24 | |
| L. lactis/r1t | ORF40 | H-N-H | Yes | 61 | |
| Lactococcus delbrueckii/LL-H | Terminase | ORF168 (H-N-H) | Yes | 54 | |
| Staphylococcus aureus/Twort | ORF142 introns 1, 2, 3 | None | Yes | 44 |
When known, endonuclease names are given (e.g., I-TevI). “Fragment” refers to a truncated ORF, and “None” means no ORF was present in the intron. Endonucleases are also identified by which family they belong to (GIY-YIG, H-N-H, His-Cys Box, or LAGLIDAG).
For the majority of group I introns, evidence for splicing is based on in vitro assays. Only a few introns have been shown to be mobile in vivo.
One tRNAfMet gene, that of Synechocystis sp. strain 6803, contains a novel endonuclease, I-SspI (R. Bonocora and D. A. Shub, submitted for publication).
A similar question might be asked of group II introns in bacteria. Much was made of the initial finding of group II introns in bacteria, as their discovery added fuel to the debate concerning the evolutionary origins of eukaryotic spliceosomal introns (22, 70) which have both structural and functional similarities to group II introns (53, 79, 84). Yet, the number of group II introns in bacteria is small, many of which are inferred only from database matches to reverse transcriptases or maturases encoded within known introns (13, 41, 73, 89), and only two have been shown to splice or be mobile in vivo (48, 49, 55, 80) (Table 2). While group II intron homing is mechanistically distinct from group I intron homing, the principle is similar; group II introns home from intron-containing to intronless alleles. Many elegant biochemical and genetic experiments have unraveled the complexities of bacterial group II intron homing (14, 48, 49), and based on these results, there seems no a priori reason why group II introns should not be able to spread efficiently through populations of intron-minus alleles. The paucity of bacterial group II introns becomes even more perplexing given the recent demonstration of group II intron transposition to novel chromosomal sites (15). That group II introns are abundant in mitochondrial and chloroplast genomes (52) and present in bacterial genomes but at lower levels (and absent in phages) only adds to the mystery surrounding the lack of group II introns in bacteria, as mitochondria and chloroplasts are typically prokaryotic in genome organization and ultimately are derived from two distinct bacterial lineages. Do similar barriers that prevent group I introns from spreading throughout bacterial populations also apply to preventing the spread of group II introns in bacteria?
TABLE 2.
Distribution of group II introns in bacteria
| Bacterium | Speciesa | Intron/insertion siteb | ORF | Splicingc | Reference(s) |
|---|---|---|---|---|---|
| Gram-negative | E. coli ECOR 43, 17, 21, 31, 67 | IntA/H-repeat element | Yes | ND | 23 |
| E. coli ECOR 44, 47 | IntB/H-repeat element | Yes | ND | 23 | |
| E. coli ECOR 9, 52, 60, 17, 20, 23, 24 | IntC/plasmid-related ORF | Yes | ND | 23 | |
| E. coli ECOR 9, 38 | IntD/IS3411 | Yes | ND | 23 | |
| E. coli | ND/pR471a inferred—retron? | Yes | ND | 41 | |
| Azotobacter vinelandii UWR | AvUWRx.1/ND | ?? | ND | 22 | |
| Sphingomonas aromaticivorans F199 | ND/ND | Yes | ND | 73 | |
| Pseudomonas alcaligenes NCIB 9867 | Xln6/pRP4 | Yes | Inferred | 89 | |
| Cyanobacterial | Calothrix sp. strain PCC7601 | Cal.x1/ND | Yes | Yes | 22 |
| Calothrix sp. strain PCC7101 | Cal.x2/ssDNA binding proteind | Yes | ND | 22 | |
| Gram-positive | L. lactis ML3 | Ll.LtrB/relaxase gene, pRS01 | Yes | Yes | 55, 56 |
| L. lactis 712 | IntL/congjugation gene mobA | Yes | Yes | 80 | |
| S. meliloti GR4 | RmInt1/ISRm2011-2, pRmeGR4b | Yes | Yes | 48 | |
| Streptococcus pnemoniae | ND/ND | Yes | ND | 13 |
Some group II introns were identified from various members of the E. coli ECOR collection, as listed by their strain number.
Insertion sites of some group II introns were not identified, indicated by ND.
There is no evidence for splicing for the majority of putatively identified group II introns, indicated by ND.
ssDNA, single-stranded DNA.
WHAT IS INTRON HOMING?
Homing is the process by which group I and II introns spread through a population of homologous (or cognate) intronless alleles (17) (Fig. 1). Homing is completely dependent on and initiated by intron-encoded proteins, and for group I and group II introns, completion of the homing process relies on host-encoded proteins (for a review, see reference 42). Group I intron homing is initiated by a DNA endonuclease (usually called a homing endonuclease) encoded within the intron itself. Four families of endonucleases were initially identified on the basis of conserved amino acid motifs and structural similarities; they are the GIY-YIG (9, 51), LAGLIDAG (31, 50), His-Cys box (60), and H-N-H (28, 83) families (for reviews, see references 3 and 4). However, structural similarities in active-site architecture have recently been found in representative H-N-H and His-Cys box endonuclease crystal structures, suggesting that these two families could possibly be merged into one (39). Homing endonucleases of all families recognize lengthy DNA sequences (14 to 40 bp) that span the analogous intron insertion site in the intronless allele (4) and mostly introduce a double-strand break (DSB). Since the recognition sequence of the intron-encoded endonuclease is interrupted by the intron, intron-containing alleles are immune to cleavage by their own endonucleases.
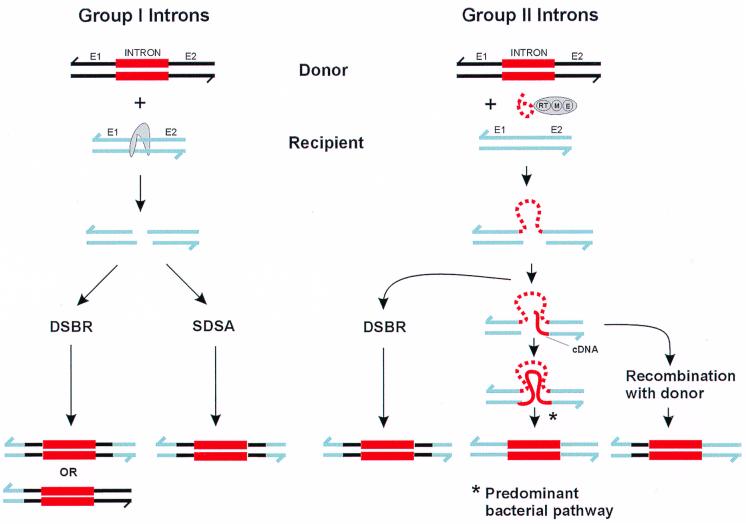
FIG. 1.
Schematic of group I and II intron homing pathways. Solid black lines represent upstream (E1) and downstream (E2) exons of donor (intron-containing) alleles, and blue-gray lines indicate recipient (intronless) alleles. Group I or II introns are depicted by red boxes. Broken red lines indicate intron RNA, while solid red lines indicate cDNA synthesized by the reverse transcriptase (RT) function of group II intron proteins. The gray tooth-shaped structure represents a group I intron endonuclease. Group II intron proteins possess two functions in addition to RT, namely, maturase (M) and endonuclease (E) activities.
In group I intron homing, host- and phage-encoded DNA repair proteins use the intron-containing allele as a template to repair the DSB in the intronless allele, thus copying the intron and its endonuclease gene into the intron-minus allele (Fig. 1). In the E. coli bacteriophage T4, two repair pathways, DSB repair (DSBR) and synthesis-dependent strand annealing (SDSA) are implicated in repair of the DSB during intron acquisition (57). These repair pathways differ in that resolution of the Holliday junction in DSBR results in both crossover and noncrossover products, whereas no Holliday junctions are found and no crossover products are generated in the SDSA pathway.
For group II introns, remarkably, the intron RNA forms a complex with the intron-encoded protein to initiate homing by recognizing a specific DNA sequence (Fig. 1). The intron RNA is an essential catalytic component of this process (49, 76, 93). Because group II intron homing can proceed through a reverse-transcribed copy of the intron RNA, the process has been termed retrohoming to distinguish it from endonuclease-mediated homing of group I introns, which proceeds strictly through DNA intermediates (16). And, unlike group I intron homing where the intron-encoded endonuclease is sufficient to promote homing into intron-minus alleles, group II intron homing relies on two functions of the intron-encoded protein in addition to the endonuclease function; maturase, which promotes intron splicing, and reverse transcriptase, which generates intermediates of the retrohoming process (reviewed in reference 43). Whereas in bacteria, group II intron proteins are encoded by self-contained open reading frames (ORFs) with independent promoters (91), in yeast mitochondria, they are produced as fusion proteins with the upstream exon (75). Subsequent processing of this polyprotein yields a product that initially functions as a maturase to promote stabilization of intron structures necessary for splicing. Splicing of the intron from the pre-mRNA releases an intron lariat that forms a RNP with the intron-encoded protein (43). This complex binds to the target sequence in the intron-minus alleles where the intron RNA cleaves the sense strand of the DNA at the intron insertion site by a reverse splicing reaction, and the intron becomes covalently attached to the intron-minus allele. Cleavage of the antisense strand is carried out by the endonuclease function of the intron-encoded protein, and the 3′ end serves as the primer for cDNA synthesis by the reverse transcriptase function of the intron-encoded protein using the integrated intron as the template (92).
It is at this point in the homing process where bacterial and yeast mitochondrial group II homing can potentially differ (Fig. 1). Retrohoming of the Lactococcus lactis Ll.LtrB intron proceeds independent of homologous recombination (14), whereas the yeast introns require recombination (20). In the bacterial pathway, the intron RNA undergoes a reverse splicing reaction and serves as a template for complete cDNA synthesis (14). In contrast, and depending on the yeast mitochondrial group II intron, cDNA synthesis may not proceed to the end of the intron, and completion of the retrohoming process is thus dependent on the cDNA invading the intron donor allele and on homologous recombination between the donor and recipient alleles (20). Recombination results in coconversion of flanking exon sequences, a result not observed in the bacterial retrohoming pathway.
WHERE ARE GROUP I AND II INTRONS FOUND?
Although group I and II introns are found in a diverse phylogenetic sampling of bacteria and phages, both exhibit an unusual genomic distribution (Tables 1 and 2). For group I introns, three interesting themes emerge. First, group I introns do not interrupt protein-coding genes in bacterial chromosomes, they interrupt only tRNA genes (40, 64, 65, 74, 87; information found at the Comparative RNA website of The Institute for Cellular and Molecular Biology, The University of Texas at Austin, Austin [http://www.rna.icmb.utexas.edu]). There are 74 known bacterial chromosomal group I introns, and 73 interrupt tRNA genes (information found at the Comparative RNA website). However, most of these introns are very similar, being isolated from closely related species or different strains of the same species of bacterium. In total, four different tRNAs are interrupted by introns, tRNAUAALeu, tRNACCUArg, tRNAfMet, and tRNACAUIle. There is only one example of an interrupted bacterial rRNA gene, that of the 23S ribosomal DNA (rDNA) of Simkania negevensis ZT (21). Although the interrupting sequence can be folded into a secondary structure very similar to group I introns of the IB4 subgroup, typical of organellar rRNA introns and atypical of bacterial tRNA introns, there is no evidence for splicing in vivo as assayed by reverse transcription-PCR. The single example of a chromosomally encoded group I intron in a protein-coding gene is in the ribonucleotide reductase gene (nrdF) of SPβ, an integrated prophage of Bacillus subtilis (45).
Second, the converse is true for phage-encoded group I introns; these introns are always found interrupting protein-coding genes, never structural RNA genes (some phages carry tRNA genes). Furthermore, most group I phage introns interrupt highly conserved protein-coding genes, many of which are involved in some aspect of DNA metabolism. This discovery prompted speculation that the function of phage introns might be the regulation of DNA metabolism, thus accounting for their retention in size-constrained phage genomes (25, 81; see below).
Third, homing endonucleases are typically encoded within phage group I introns, not in chromosomal group I introns, with two exceptions. The first exception is the tRNAfMet intron of Synechocystis that contains an ORF that potentially codes for a small basic protein not similar to any known endonuclease (6), and the second is a LAGLIDAG endonuclease ORF within the S. negevensis ZT 23S rDNA insertion (21). However, in spite of the tendency for phage introns to encode endonucleases, not all phage introns do, and some appear to have remnants of ORFs, evidenced by the phage T4 nrdB intron (19) and the Bacillus phage β22 thymidylate synthase intron (1). Perhaps the best example of endonuclease-lacking phage introns is found in the Staphylococcus phage Twort; three group I introns interrupt a late-transcribed gene (ORF142) of unknown function, and none of them code for endonucleases (44). Generally then, phage introns that interrupt highly conserved protein-coding genes code for homing endonucleases, whereas bacterial chromosome-encoded group I introns interrupt tRNA genes and do not code for endonucleases.
Although there are fewer known examples of group II introns than of group I introns in bacteria, one trend is immediately clear: most group II introns are inserted in, or associated with, other mobile genetic elements (23, 32, 48, 56, 59, 80) (Table 2). The best studied bacterial group II intron, the Ll.LtrB intron of L. lactis ML3, is inserted in the conjugative relaxase gene essential for transfer of the plasmid pRS01 (56). The same intron, but designated IntL, was discovered independently in a chromosomally encoded gene, mobA, required for conjugation in L. lactis 712 (80). Another group II intron, RmInt1, is inserted within an insertion sequence of the IS630-Tc1/IS3 retroposon superfamily that is found on a plasmid, pRmeGR4b, of Sinorhizobium meliloti (48). Four group II introns have been identified in isolates of the E. coli ECOR collection, and all of them are associated with some form of mobile DNA (23, 36). Two introns from distinct ECOR isolates are inserted at different positions within the mobile Hinc repeat of Rhs elements. A third E. coli intron, found in two different ECOR strains, is inserted within a sequence that is very similar to IS3411, and the fourth intron is inserted in an ORF of unknown function but is related to an ORF encoded by T-DNA of Agrobacterium tumefaciens.
DISTRIBUTION OF GROUP I INTRONS IN PHAGE POPULATIONS
It is perhaps ironic that self-splicing group I introns were discovered as a property of T-even phages of gram-negative bacteria, for gram-negative bacterial phages are where group I introns are least abundant (18). Apart from the td, nrdD, and nrdB introns in T-even phages, no other group I introns have been discovered in any gram-negative bacterial phage. Conversely, group I introns are more abundant in phages of gram-positive bacteria (1, 24, 25, 26, 45, 54, 61), exemplified by the Staphylococcus phage Twort, which possesses at least five introns (44). While it is tempting to generalize about group I intron distribution in phages, it is worth noting that only two detailed studies have been performed; one concerned with distribution of the three T4 introns in related T-even phages (18) and the other concerned with the distribution of a lysin intron in Streptococcus thermophilus phages (24).
Phage T4 is unusual with respect to other T-even phages in that it possesses three group I introns. Two of the three, the td and nrdD introns, are mobile, while the third intron, nrdB, is not (68). T-even phage RB3 also harbors a nrdB intron very similar to the T4 intron, but the RB3 intron is almost twice as long because it possesses a complete ORF coding for a functional endonuclease (which T4 nrdB does not [19]). The T4 nrdB intron thus appears to carry a deletion of the ORF relative to the RB3 nrdB intron, but the endonuclease was presumably intact and functional at some point. Other T-even phages are also surprisingly bereft of introns; for instance, phage T6 has only one intron, td, and T2 has no introns (67). (One T2 strain, T2W, harbors both the td and nrdD introns. However, experimental evidence suggests that these introns were inherited during laboratory propagation of T2 and T4.)
In an effort to better understand the distribution of introns in natural T-even phage populations, Eddy screened 32 phages, a number of which were novel isolates of mammalian fecal matter from the Denver Zoo or from clarified sewage samples of city water treatment plants (18). The results are surprising. Using a combination of GTP labeling of RNA, primer extension sequencing, and PCR fingerprinting, Eddy was able to detect a total of eight intron occurrences in all phages examined. In phage isolates from which PCR products were obtained, the nrdD and nrdB introns appear to be very rare, present in only 1 of 30 and 2 of 31 phages, respectively. However, the td intron is more widespread, present in 5 of 22 phages from which a thymidylate synthase gene could be amplified. Also, Eddy found that every phage isolate except one could infect at least four E. coli strains differing in cell surface receptors. Thus, it seems unlikely that the paucity of introns in T-even phages is due to lack of opportunity of intron-carrying phages to infect similar host cell populations as intronless phages.
The second detailed study of group I intron distribution reaches a somewhat different conclusion than that of the above survey (24). S. thermophilus is an economically important bacterium for the dairy industry, and as such, its phages have been subject to much study. Five independent phage isolates have been completely sequenced, allowing a detailed PCR survey of related phages for the presence of introns. Foley and coworkers initially found a self-splicing group I intron, complete with an H-N-H endonuclease ORF, interrupting the lysin gene of phage Sf1 (24). When they expanded their PCR survey to encompass 61 phages isolated from geographically and ecologically distinct locales, they found that 31 of the phages contained the intron, four of which have deletions within the H-N-H ORF. Thus, it appears that intron spread in this phage population has been much more successful than intron spread in populations of T-even phages.
T-EVEN INTRONS: RECENT ARRIVALS OR LONG-TERM RESIDENTS?
Of the distribution of introns in T-even phages, Eddy (18) commented that “Although variable intron occurrence has been said to imply that the introns are spread through the population by virtue of their mobility, it actually seems somewhat puzzling that these highly infectious introns do not occur in most or all T-even isolates.” What then accounts for the sporadic and limited distribution of introns in the T-even phage family? One argument is that the introns have only recently arrived in T-even phages and have not yet had sufficient time or opportunity to spread throughout other T-even phages. However, we present the following points supporting an alternative view that the T4 introns have been present for a significant period of time in the T-even population (2).
First, expression of the three T4 intron-encoded ORFs is tightly regulated (30). All three intron-containing genes are expressed as early or middle transcripts from promoters upstream of the respective genes, but translation of the intron endonuclease gene from the pre-mRNA is prevented by the presence of secondary structures that occlude ribosome binding sites. As intron endonucleases are expressed from late promoters found within the introns, these RNA transcripts do not include secondary structures to occlude the endonuclease gene ribosome binding site, thus facilitating translation of the endonuclease. This mode of regulation must be the result of specific adaptation in T4 or in another T-even-like phage, since the highly unusual T4 late promoter sequences are present within the introns themselves. Second, the intron-encoded endonucleases I-TevI, I-TevII, and I-TevIII appear to have adapted to the T-even environment as codon usage is optimized for T4 (82). Third, both I-TevI and I-TevII, the nrdD intron endonuclease, make predominantly minor groove contacts with their DNA recognition sequences (7, 47). This mode of DNA contact is likely an adaptation to modified T-even phage DNA, which in T4 contains bulky α- and β-glucosylated hydroxymethyl cytosine residues in the major groove (46). These adaptations are counter to the introns and endonucleases being recent arrivals in the T-even phage population.
WHY ARE PHAGE INTRONS IN THE GENES THAT THEY ARE IN?
The startling discovery that phage T4 group I introns were inserted within genes of nucleotide metabolism (thymidylate synthase [td], aerobic [nrdB], and anaerobic [nrdD] ribonucleotide reductases) led to the proposal that the function of these introns was to regulate levels of nucleotide precursors for DNA synthesis through regulation of splicing (81). This proposal was strengthened by the discovery of another group I intron in a gene involved in DNA metabolism, the DNA polymerase of Bacillus phage SPO1 (25). Thus, a rationale was provided for the retention of seemingly optional introns in size-constrained genomes subject to pressures of genome streamlining for rapid replication. However, this presumed function for the T4 introns began to lose attractiveness with the discovery that close relatives of T4 lacked introns in these genes (67), with the (unpublished) experimental observation that T4 introns could be deleted without detectable detriment to the phage (18), and with the finding that some phage introns do not interrupt genes of nucleotide metabolism (44, 54, 61).
Why then do phage introns tend to be inserted within highly conserved genes involved in some aspect of DNA metabolism? One possible reason relates to the relatively high specificity of group I intron-encoded endonucleases. In general, homing endonucleases recognize relatively long stretches of DNA (14 to 40 bp) and, depending on the individual endonuclease, tolerate various numbers of nucleotide substitutions within their respective recognition sequences (35). Yet, homing endonucleases in phage populations are presented with a limited number of potential recognition sequences because phages have relatively small genomes. Small genome size also constrains the phage with respect to number of genes it can ultimately encode, so phages tend to carry genes coding essential functions: proteins involved in replication, transcription, and morphogenesis. Thus, for phage-encoded homing endonucleases, one possible strategy to maximize spread to related phages comes from using conserved nucleotide sequences, which are likely to lie within genes that function in replication and transcription, as recognition sites.
Furthermore, a specific prediction would be that the recognition sites of homing endonucleases are nucleotide sequences corresponding to important functional domains within those proteins, as these sequences are more likely to be conserved between phages. For instance, the recognition sequence of I-TevI, the phage T4 thymidylate synthase intron endonuclease, encompasses 37 bp (7, 8) that includes codons for the highly conserved Arg218, Ser219, Asp221, and Asn229 (E. coli numbering), all of which are functionally critical (10). Thymidylate synthase catalyzes the reductive methylation of dUMP by 5,10-methylene-5,6,7,8-tetrahydrofolate to produce dTMP and 7,8-dihydrofolate. In the crystal structure of the E. coli enzyme complexed with dUMP, Arg218 and Ser219 form one hydrogen bond each with the phosphate of dUMP. Ser219 and Asp221 are thought to be important for the enzyme's reduced affinity for ribose nucleotides because these residues would sterically hinder binding of nucleotides containing a 2′-OH group. In addition, Asp221 and Asn229 each form hydrogen bonds with the pyrimidine ring of dUMP. That other homing endonucleases also have recognition sequences that encompass functionally critical residues can be easily verified as additional structures and mutational data become available.
Intron insertion into essential phage genes also imposes negative selection on the phage to remove the intron, since inexact deletion of an intron from an essential gene may potentially affect codons corresponding to amino acid residues critical for function. Ironically, precise deletion of the intron from an individual phage within a population would provide positive selection for retention of the endonuclease ORF, because the homing site would be regenerated and become a substrate for endonucleases of other phage within the same population. Of course, no hypothesis is bulletproof, and there are already examples of endonuclease-containing introns interrupting genes not involved in replication or transcription (the lysin gene of S. thermophilus phages [24] and the terminase gene of Lactobacillus phage LL-H [54]). It is possible, however, that these endonucleases are also targeting conserved nucleotide sequences within their respective genes.
HOW DO BACTERIAL INTRONS GET AROUND?
Intron homing is an efficient but confining process that results in the spread and potential fixation of a group I or II intron in a population of intronless alleles. Nevertheless, homing does not promote intron dissemination to nonallelic sites. Intron transposition, however, can do this.
Intron transposition to nonallelic sites has been inferred from well-documented studies for fungal mitochondrial group II introns (58, 77). Group II intron transposition in bacteria was also inferred from studies on the distribution and insertion sites of an intron in strains of S. meliloti (48). Recently, Cousineau and coworkers experimentally demonstrated retrotransposition for the first time, showing movement of the L. lactis Ll.LtrB group II intron to novel sites in the chromosome (15). Unlike retrohoming, retrotransposition can occur independently of the endonuclease function of the intron-encoded protein but is dependent on host recombinase functions. Furthermore, retrotransposition events resulted in introns with various degrees of splicing function, as judged from primer extension analysis of L. lactis RNA isolated from strains harboring novel intron insertions. Given these results, it is curious and surprising that group II introns are not more widely distributed throughout bacterial or phage genomes.
What evidence, if any, exists for group I intron transposition in bacteria? In vitro and in vivo experimental evidence suggests that reverse splicing of group I intron RNA into nonallelic RNA is one possible mechanism of intron transposition (85). Because as little as 4 to 6 bp of sequence complementarity is required between the intron and target RNA to initiate a reverse splicing event, many potential sites exist within any population of RNA molecules. Indeed, Roman and Woodson (71) have demonstrated reverse splicing in vivo of the Tetrahymena large-subunit (LSU) RNA group I intron into E. coli 23S RNA at 11 novel sites. The abundance of structural RNA transcripts (rRNAs and tRNAs) has led to the suggestion that such RNAs would be preferential targets for reverse splicing and transposition, providing a possible explanation for the prevalence of group I introns in bacterial tRNA genes (3). Nevertheless, the limiting step in reverse-splicing-mediated group I intron transposition must surely be reverse transcription of the intron and target RNA and subsequent recombination, given that the requisite reverse transcriptase enzyme activity is not prevalent in many bacterial cells.
The relaxed sequence specificity of some intron-encoded endonucleases may also provide a means by which group I introns could invade new alleles (7, 8), as DSBs might result from the recognition and cleavage of degenerate versions of homing sites by intron endonucleases. Whether the same pathways that repair DSBs in allelic homing events could repair DSBs generated at nonallelic sites is determined by the amount of homology between flanking exon sequences. Detailed studies on the homology requirements for T4 td intron homing suggest that efficient homing is supported by exon homologies of 50 bp or greater. However, homing events were still observed with limiting exon homology (10 bp) or when homology is confined to only one exon in T4 phages carrying a mutation in the 3′-5′ exonuclease DexA, which functions to degrade 3′ tails of DSBs (33, 66). Illegitimate recombination may be involved in the repair of DSBs with limiting exon homology and thus provide a potential pathway for group I intron invasion at nonallelic sites.
HOW EFFICIENT IS INTRON TRANSPOSITION?
One factor that must influence intron dissemination in bacteria is the frequency and efficiency of group I and II intron transposition versus the frequency and efficiency of homing. Homing is an extremely efficient process, driven in part by the high affinity of group I intron endonucleases or group II intron RNP complexes for their respective recognition sequences, resulting in near 100% conversion of recipient intron-minus alleles to intron-plus alleles. In contrast, transposition is an infrequent process due, in part, to the reduced affinity of intron-encoded proteins for variants of their recognition sequences and to heterology between intron-containing and intronless alleles. Furthermore, for phage-encoded endonucleases, whose expression is temporally restricted (30) and of low abundance, opportunities for transposition are probably rare.
Intron transposition, whether mediated by reverse splicing or by an endonuclease, places the intron in a nonnative environment, with potential effects on intron splicing. For instance, although the Tetrahymena LSU RNA group I intron can reverse splice into novel sites in E. coli 23S RNA, the forward-splicing efficiency is 10-fold lower than from its natural site (72). Likewise, group II intron transposition to novel locations in L. lactis resulted in forward-splicing rates lower than those observed from the intron's natural splice junction (15). These results suggest that RNA sequence and structure can potentially influence sites at which transposition events result in splicing-competent introns.
WHAT MIGHT SELECT AGAINST INTRON INSERTION INTO CHROMOSOMAL GENES?
Without exception, group I introns found in bacterial chromosomes are never inserted in protein-coding genes (Table 1). Why is this so? One possibility immediately comes to mind, which has been suggested by others (11, 62, 78, 81, 86). In bacteria, unlike eukaryotic nuclei, transcription (hence splicing) and translation are coupled. For introns that encode endonucleases and thus exceed 1,000 bp in length, the ribosome is positioned over the 5′ splice site of the intron RNA before RNA polymerase has reached the 3′ splice site. Movement of the ribosome into the intron may prevent formation of helices that stabilize the catalytic core of the intron and potentially delay splicing of the intron (63). Group I introns inserted in phage protein-coding genes have overcome this problem by the presence of a stop codon almost immediately after the start of the intron sequence at the 5′ splice boundary (or no further than structural element P2) (11, 82). Presumably, this stop codon acts to release the ribosome from the RNA, thus freeing the intron RNA to fold into a catalytically active structure. Ironically, the ribosome may also aid in the folding of the intron core by preventing spurious secondary or tertiary interactions between sequences in the 5′ exon and intron RNA (78). Introns inserted in tRNA or rRNA genes do not face this problem of coupled transcription and translation, which may account for the prevalence of group I introns in bacterial tRNA genes.
Nikolcheva and Woodson have shown that group I introns are potentially toxic to bacteria because of possible inhibition of ribosome formation by the intron RNA (62). Using a heterologous system in which a splicing-defective Tetrahymena rRNA group I intron was inserted into the analogous position of the E. coli 23S RNA, they demonstrated that unspliced 23S RNA accumulated in 50S ribosomes, and these complexes failed to assemble complete 70S ribosomes. In addition, using intron constructs competent for splicing, spliced intron RNA remained noncovalently attached to the 50S ribosome particle, probably through interaction of the intron internal guide sequence with the splice junction in 23S RNA. This association was correlated with slow growth of bacterial strains harboring the group I intron. Thus, there may be strong selection against group I intron insertion into rRNA genes in bacteria.
DOES NONSPECIFIC ENDONUCLEASE ACTIVITY INFLUENCE INTRON DISTRIBUTION?
Ironically, the very same relaxed sequence specificity of group I intron endonucleases that affords them the ability to bind and cleave variants of their recognition sequences may limit genomic retention because of the potentially toxic effects of extraneous DSBs in the host chromosome. One of the more curious observations concerning bacterial group I introns is that, with one experimentally demonstrated exception, endonuclease-containing introns are not found in bacterial chromosomes (Table 1). Yet, the increased genome size of bacteria relative to that of phages could be of benefit only to endonuclease-containing introns, since the number of potential targets, and thus opportunities for transposition, is increased. DSBs generated at ectopic sites on the bacterial chromosome by endonucleases binding to and cleaving degenerate versions of their recognition sequences must be repaired for the cell to survive, and repair is initiated by homoglous sequences on daughter chromosomes. However, laboratory wild-type strains of E. coli lack the proper enzyme activities to efficiently repair DSBs (12). Furthermore, when grown in minimal medium, potential repair of any DSB in E. coli is further hindered by the lack of multiple chromosomes (38). Perhaps it is not surprising then that endonuclease-containing introns have been most successful in colonizing smaller, multicopy genomes, such as those found in phages, mitochondria, or plastids.
AMBITIOUS QUESTIONS, SOME ANSWERS, MORE QUESTIONS
One problem in attempting to provide reasons for the current genomic and phylogenetic distribution of group I and II introns in bacteria is that there is no expectation as to what intron distribution should be. Efficient homing of group I and II introns in laboratory experiments does not necessarily imply efficient homing in natural populations. Little is known about the dynamics of natural populations of phages (or bacteria) or the frequency of coinfection of the same host cell, factors that surely affect opportunities for intron homing and/or transposition. Studies of intron dynamics at the population level, such as those which highlight surprising differences in group I intron distribution of gram-negative and gram-positive bacterial phages, only emphasize difficulties in applying lessons learned about intron homing from laboratory experiments to natural phage populations. Our view of intron distribution in bacteria is also influenced by laboratory experiments that examine homing events over a limited number of phage or bacterial generations and then the experiments are discarded. Homing events that occur in natural populations must become fixed in bacteria or phages, yet nothing is known about the selective pressures that influence this process.
It is probably correct to state that there is no single reason that can adequately explain the current distribution of group I and II introns in bacteria. Undoubtedly, the number of bona fide group I and II introns in bacteria and phages will increase as more and more prokaryotic genomes are sequenced, as RNA folding and search algorithms become more sophisticated, and as already putatively identified introns are confirmed experimentally. Yet basic questions and observations concerning intron distribution in natural bacterial and phage populations will still apply, and one obvious and exciting future avenue of research is to experimentally address some of the factors that influence intron dissemination in bacteria and their phages.
ACKNOWLEDGMENTS
We thank Archana Belle, Rick Bonocora, Benoit Cousineau, Vicky Derbyshire, Cheryl Eifert, Michelle Gilson, Markus Landthaler, and Niles Lehman for discussion and comments on the manuscript.
D.R.E. was supported by a postdoctoral fellowship from the Medical Research Council of Canada, M.B. was supported by NIH grants GM39422 and GM44844, and D.A.S. was supported by NIH grant GM37746.
REFERENCES
- 1.Bechhofer D H, Hue K K, Shub D A. An intron in the thymidylate synthase gene of Bacillus bacteriophage β22: evidence for independent evolution of a gene, its group I intron, and the intron open reading frame. Proc Natl Acad Sci USA. 1994;91:11669–11673. doi: 10.1073/pnas.91.24.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belfort M. Bacteriophage introns: parasites within parasites? Trends Genet. 1989;5:209–213. doi: 10.1016/0168-9525(89)90083-8. [DOI] [PubMed] [Google Scholar]
- 3.Belfort M, Perlman P S. Mechanisms of intron mobility. J Biol Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 4.Belfort M, Roberts R J. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell-Pedersen D, Quirk S, Clyman J, Belfort M. Intron mobility in phage T4 is dependent upon a distinctive class of endonucleases and independent of DNA sequences encoding the intron core: mechanistic and evolutionary implications. Nucleic Acids Res. 1990;18:3763–3770. doi: 10.1093/nar/18.13.3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biniszkiewicz D, Cesnaviciene E, Shub D A. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryk M, Quirk S M, Mueller J E, Loizos N, Lawrence C, Belfort M. The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J. 1993;12:2141–2149. doi: 10.1002/j.1460-2075.1993.tb05862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryk M, Belisle M, Mueller J E, Belfort M. Selection of a remote cleavage site by I-TevI, the td intron-encoded endonuclease. J Mol Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 9.Burger G, Werner S. The mitochondrial URF1 gene in Neurospora crassa has an intron that contains a novel type of URF. J Mol Biol. 1985;186:231–242. doi: 10.1016/0022-2836(85)90100-7. [DOI] [PubMed] [Google Scholar]
- 10.Carreras C W, Santi D V. The catalytic mechanism and structure of thymidylate synthase. Annu Rev Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 11.Chu F K, Maley G F, Maley F, Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc Natl Acad Sci USA. 1984;81:3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clyman J, Belfort M. trans and cis requirements for intron mobility in a prokaryotic system. Genes Dev. 1992;6:1269–1279. doi: 10.1101/gad.6.7.1269. [DOI] [PubMed] [Google Scholar]
- 13.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 14.Cousineau B, Smith D, Lawrence-Cavanagh S, Mueller J E, Yang J, Mills D, Manias D, Dunny G, Lambowitz A M, Befort M. Retrohoming of a bacterial group II intron: mobility via complete reverse splicing, independent of homologous DNA recombination. Cell. 1998;94:451–462. doi: 10.1016/s0092-8674(00)81586-x. [DOI] [PubMed] [Google Scholar]
- 15.Cousineau B, Lawrence S, Smith D, Belfort M. Retrotransposition of a bacterial group II intron. Nature. 2000;404:1018–1021. doi: 10.1038/35010029. [DOI] [PubMed] [Google Scholar]
- 16.Curcio M J, Belfort M. Retrohoming: cDNA-mediated mobility of group II introns requires a catalytic RNA. Cell. 1996;84:9–12. doi: 10.1016/s0092-8674(00)80987-3. [DOI] [PubMed] [Google Scholar]
- 17.Dujon B, Belfort M, Butow R A, Jacq C, Lemieux C, Perlman P S, Vogt V M. Mobile introns: definition of terms and recommended nomenclature. Gene. 1989;82:115–118. doi: 10.1016/0378-1119(89)90035-8. [DOI] [PubMed] [Google Scholar]
- 18.Eddy S R. Introns in the T-even bacteriophages. Ph.D. thesis. Boulder: University of Colorado; 1992. [Google Scholar]
- 19.Eddy S R, Gold L. The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev. 1991;5:1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- 20.Eskes R, Yang J, Lambowitz A M, Perlman P S. Mobility of yeast mitochondrial group II introns: engineering a new site specificity and retrohoming via full reverse splicing. Cell. 1997;88:865–874. doi: 10.1016/s0092-8674(00)81932-7. [DOI] [PubMed] [Google Scholar]
- 21.Everett K D, Kahane S, Bush R M, Friedman M G. An unspliced group I intron in the 23S rRNA links Chlamydiales, chloroplasts, and mitochondria. J Bacteriol. 1999;181:4734–4740. doi: 10.1128/jb.181.16.4734-4740.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferat J-L, Michel F. Group II self-splicing introns in bacteria. Nature. 1993;364:358–361. doi: 10.1038/364358a0. [DOI] [PubMed] [Google Scholar]
- 23.Ferat J-L, Le Gouar M, Michel F. Multiple group II self-splicing introns in mobile DNA from Escherichia coli. C R Acad Sci. 1994;317:141–148. [PubMed] [Google Scholar]
- 24.Foley S, Bruttin A, Brussow H. Widespread distribution of a group I intron and its three deletion derivatives in the lysin gene of Streptococcus thermophilus bacteriophages. J Virol. 1999;74:611–618. doi: 10.1128/jvi.74.2.611-618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodrich-Blair H, Scarlato V, Gott J M, Xu M-Q, Shub D A. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell. 1990;63:417–424. doi: 10.1016/0092-8674(90)90174-d. [DOI] [PubMed] [Google Scholar]
- 26.Goodrich-Blair H, Shub D A. The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res. 1994;22:3715–3721. doi: 10.1093/nar/22.18.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodrich-Blair H, Shub D A. Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell. 1996;84:211–221. doi: 10.1016/s0092-8674(00)80976-9. [DOI] [PubMed] [Google Scholar]
- 28.Gorbalenya A E. Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci. 1994;3:1117–1120. doi: 10.1002/pro.5560030716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gott J M, Shub D A, Belfort M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labelling of RNA in vitro. Cell. 1986;47:81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- 30.Gott J M, Zeeh A, Bell-Pederson D, Ehrenman K, Belfort M, Shub D A. Genes within genes: independent expression of phage T4 intron open reading frames and the genes in which they reside. Genes Dev. 1988;2:1791–1799. doi: 10.1101/gad.2.12b.1791. [DOI] [PubMed] [Google Scholar]
- 31.Hensgens L A, Bonen L, de Haan M, van der Horst G, Grivell L A. Two intron sequences in yeast mitochondrial COX1 gene: homology among URF-containing introns and strain-dependent variation in flanking exons. Cell. 1983;32:379–389. doi: 10.1016/0092-8674(83)90457-9. [DOI] [PubMed] [Google Scholar]
- 32.Huang C C, Narita M, Yamagata T, Itoh Y, Endo G. Structure analysis of a class II transposon encoding the mercury resistance of the gram-positive bacterium Bacillus megaterium MB1, a strain isolated from Minimata Bay, Japan. Gene. 1999;234:361–369. doi: 10.1016/s0378-1119(99)00184-5. [DOI] [PubMed] [Google Scholar]
- 33.Huang Y-J, Parker M M, Belfort M. Role of exonucleolytic degradation in group I intron homing in phage T4. Genetics. 1999;153:1501–1512. doi: 10.1093/genetics/153.4.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 35.Jurica M S, Stoddard B L. Homing endonucleases: structure, function, and evolution. Cell Mol Life Sci. 1999;55:1304–1326. doi: 10.1007/s000180050372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoop V, Brennicke A. Evidence for a group II intron in Escherichia coli inserted into a highly conserved reading frame associated with mobile DNA sequences. Nucleic Acids Res. 1994;22:1167–1171. doi: 10.1093/nar/22.7.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowalski J C, Belfort M, Stapleton M A, Holpert M, Dansereau J T, Pietrokovski S, Baxter S M, Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krasin F, Hutchinson F. Repair of DNA double-strand breaks in Escherichia coli, which requires recA function and the presence of a duplicate genome. J Mol Biol. 1977;116:81–98. doi: 10.1016/0022-2836(77)90120-6. [DOI] [PubMed] [Google Scholar]
- 39.Kuhlmann U C, Moore G R, James R, Kleanthous C, Hemmings A M. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- 40.Kuhsel M G, Strickland R, Palmer J D. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 41.Kulaeva O I, Koonin E V, Wootton J C, Levine A S, Woodgate R. Unusual insertion element polymorphisms in the promoter and terminator regions of the mucAB-like genes of R471a and R466b. Mutat Res. 1998;397:247–262. doi: 10.1016/s0027-5107(97)00222-4. [DOI] [PubMed] [Google Scholar]
- 42.Lambowitz A M, Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 43.Lambowitz A M, Caprara M G, Zimmerly S, Perlman P S. Group I and II ribozymes as RNPs: clues to the past and guides to the future. In: Gesteland R F, Cech T R, Atkins J F, editors. The RNA world. 2nd ed. (monograph 37). Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 451–485. [Google Scholar]
- 44.Landthaler M, Shub D A. Unexpected abundance of self-splicing group I introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc Natl Acad Sci USA. 1999;96:7005–7010. doi: 10.1073/pnas.96.12.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lazarevic V, Soldo B, Dusterhoft A, Hilbert H, Mauel C, Karamata D. Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc Natl Acad Sci USA. 1998;95:1692–1697. doi: 10.1073/pnas.95.4.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lehman I R, Pratt G A. On the structure of the glycosylated hydroxymethylcytosine nucleotides of T2, T4 and T6. J Biol Chem. 1960;235:3254–3259. [PubMed] [Google Scholar]
- 47.Loizos N, Silva G H, Belfort M. Intron-encoded endonuclease I-TevII binds across the minor groove and induces two distinct conformational changes in its DNA substrate. J Mol Biol. 1996;255:412–424. doi: 10.1006/jmbi.1996.0034. [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Abarca F, Zekri S, Toro N. Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol Microbiol. 1998;28:1295–1306. doi: 10.1046/j.1365-2958.1998.00894.x. [DOI] [PubMed] [Google Scholar]
- 49.Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny G M, Belfort M, Lambowitz A M. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michel F, Cummings D J. Analysis of class I introns in a mitochondrial plasmid associated with senescence of Podospora anserina reveals extraordinary resemblance to the Tetrahymena ribosomal intron. Curr Genet. 1985;10:69–79. doi: 10.1007/BF00418495. [DOI] [PubMed] [Google Scholar]
- 51.Michel F, Dujon B. Genetic exchange between bacteriophage T4 and filamentous fungi? Cell. 1986;46:323. doi: 10.1016/0092-8674(86)90651-3. [DOI] [PubMed] [Google Scholar]
- 52.Michel F, Umesono K, Ozeki H. Comparative and functional anatomy of group II catalytic introns—a review. Gene. 1989;82:5–30. doi: 10.1016/0378-1119(89)90026-7. [DOI] [PubMed] [Google Scholar]
- 53.Michel F, Ferat J L. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 54.Mikkonen M, Alatossava T. A group I intron in the terminase gene of Lactobacillus delbrueckii subsp. lactis phage LL-H. Microbiology. 1995;141:2183–2190. doi: 10.1099/13500872-141-9-2183. [DOI] [PubMed] [Google Scholar]
- 55.Mills D A, Manias D A, McKay L L, Dunny G M. Homing of a group II intron from Lactococcus lactis subsp. lactis ML3. J Bacteriol. 1997;179:6107–6111. doi: 10.1128/jb.179.19.6107-6111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mills D A, McKay L L, Dunny G M. Splicing of a group II intron involved in the conjugative transfer of pRS01 in lactococci. J Bacteriol. 1996;178:3531–3538. doi: 10.1128/jb.178.12.3531-3538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mueller J E, Clyman J, Huang Y J, Parker M M, Belfort M. Intron mobililty in phage T4 occurs in the context of recombination-dependent DNA replication by way of multiple pathways. Genes Dev. 1996;10:351–364. doi: 10.1101/gad.10.3.351. [DOI] [PubMed] [Google Scholar]
- 58.Mueller M W, Allmaier M, Eskes R, Schweyen R J. Transposition of group II intron aI1 in yeast and invasion of mitochondrial genes at new locations. Nature. 1993;366:174–176. doi: 10.1038/366174a0. [DOI] [PubMed] [Google Scholar]
- 59.Mullany P, Pallen M, Wilks M, Stephen J R, Tabaqchali S. A group II intron in a conjugative transposon from the gram-positive bacterium, Clostridium difficile. Gene. 1996;174:145–150. doi: 10.1016/0378-1119(96)00511-2. [DOI] [PubMed] [Google Scholar]
- 60.Muscarella D E, Ellison E L, Ruoff B M, Vogt V M. Characterization of I-PpoI, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Mol Cell Biol. 1990;10:3386–3396. doi: 10.1128/mcb.10.7.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nauta A. Molecular characterization and exploitation of the temperate Lactococcus lactis bacteriophage rIt. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1997. [Google Scholar]
- 62.Nikolcheva T, Woodson S A. Association of a group I intron with its splice junction in 50S ribosomes: implications for intron toxicity. RNA. 1997;3:1016–1027. [PMC free article] [PubMed] [Google Scholar]
- 63.Ohman-Heden M, Ahgren-Stalhandske A, Hahne S, Sjoberg B M. Translation across the 5′-splice site interferes with autocatalytic splicing. Mol Microbiol. 1993;7:975–982. doi: 10.1111/j.1365-2958.1993.tb01189.x. [DOI] [PubMed] [Google Scholar]
- 64.Paquin B, Kathe S D, Nierzwicki-Bauer S A, Shub D A. Origin and evolution of group I introns in cyanobacterial tRNA genes. J Bacteriol. 1997;179:6798–6806. doi: 10.1128/jb.179.21.6798-6806.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paquin B, Heinfling A, Shub D A. Sporadic distribution of tRNACCUArg introns among α-purple bacteria: evidence for horizontal transmission and transposition of a group I intron. J Bacteriol. 1999;181:1049–1053. doi: 10.1128/jb.181.3.1049-1053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parker M M, Belisle M, Belfort M. Intron homing with limited exon homology: illegitimate double-strand-break repair in intron acquisition by phage T4. Genetics. 1999;153:1513–1523. doi: 10.1093/genetics/153.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quirk S M, Bell-Pedersen D, Tomaschewski J, Ruger W, Belfort M. The inconsistent distribution of introns in the T-even phages indicates recent genetic exchanges. Nucleic Acids Res. 1988;17:301–315. doi: 10.1093/nar/17.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quirk S M, Bell-Pedersen D, Belfort M. Intron mobility in the T-even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell. 1989;56:455–465. doi: 10.1016/0092-8674(89)90248-1. [DOI] [PubMed] [Google Scholar]
- 69.Reinhold-Hurek B, Shub D A. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- 70.Roger A J, Doolittle W F. Why introns-in-pieces? Nature. 1993;364:289–290. doi: 10.1038/364289a0. [DOI] [PubMed] [Google Scholar]
- 71.Roman J, Woodson S A. Integration of the Tetrahymena group I intron into bacterial rRNA by reverse splicing in vivo. Proc Natl Acad Sci USA. 1998;95:2134–2139. doi: 10.1073/pnas.95.5.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roman J, Rubin M H, Woodson S A. Sequence specificity of in vivo reverse splicing of the Tetrahymena group I intron. RNA. 1999;5:1–13. doi: 10.1017/s1355838299981244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romine M F, Stillwell L C, Wong K-K, Thurston S J, Sisk E C, Sensen C, Gaasterland T, Fredrickson J K, Saffer J D. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rudi K, Jakobsen K S. Cyanobacterial tRNALeuUAA group I introns have a polyphyletic origin. FEMS Microbiol Lett. 1997;156:293–298. doi: 10.1016/s0378-1097(97)00446-1. [DOI] [PubMed] [Google Scholar]
- 75.Saldanha R, Mohr G, Belfort M, Lambowitz A M. Group I and II introns. FASEB J. 1993;7:15–24. doi: 10.1096/fasebj.7.1.8422962. [DOI] [PubMed] [Google Scholar]
- 76.Saldanha R, Chen B, Wank H, Matsuura M, Edwards J, Lambowitz A M. RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry. 1999;38:9069–9083. doi: 10.1021/bi982799l. [DOI] [PubMed] [Google Scholar]
- 77.Sellem C H, Lecellier G, Belcour L. Transposition of a group II intron. Nature. 1993;366:176–178. doi: 10.1038/366176a0. [DOI] [PubMed] [Google Scholar]
- 78.Semrad K, Schroeder R. A ribosomal function is necessary for efficient splicing of the T4 phage thymidylate synthase intron in vivo. Genes Dev. 1998;12:1327–1337. doi: 10.1101/gad.12.9.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sharp P A. Five easy pieces. Science. 1991;254:663. doi: 10.1126/science.1948046. [DOI] [PubMed] [Google Scholar]
- 80.Shearman C, Godon J-J, Gasson M. Splicing of a group II intron in a functional transfer gene of Lactococcus lactis. Mol Microbiol. 1996;21:45–53. doi: 10.1046/j.1365-2958.1996.00610.x. [DOI] [PubMed] [Google Scholar]
- 81.Shub D A, Xu M-Q, Gott J M, Zeeh A, Wilson L D. A family of autocatalytic group I introns in bacteriophage T4. Cold Spring Harb Symp Quant Biol. 1987;52:193–200. doi: 10.1101/sqb.1987.052.01.024. [DOI] [PubMed] [Google Scholar]
- 82.Shub D A, Gott J M, Xu M-Q, Lang F B, Michel F, Tomaschewski J, Pedersen-Lane J, Belfort M. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc Natl Acad Sci USA. 1988;85:1151–1155. doi: 10.1073/pnas.85.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shub D A, Goodrich-Blair H, Eddy S R. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 84.Weiner A M. mRNA splicing and autocatalytic introns: distant cousins or the products of chemical determinism? Cell. 1993;72:161–164. doi: 10.1016/0092-8674(93)90654-9. [DOI] [PubMed] [Google Scholar]
- 85.Woodson S A, Cech T R. Reverse self-splicing of the Tetrahymena group I intron: implication for the directionality of splicing and for intron transposition. Cell. 1989;57:335–345. doi: 10.1016/0092-8674(89)90971-9. [DOI] [PubMed] [Google Scholar]
- 86.Woodson S A. Ironing out the kinks: splicing and translation in bacteria. Genes Dev. 1998;12:1243–1247. doi: 10.1101/gad.12.9.1243. [DOI] [PubMed] [Google Scholar]
- 87.Xu M Q, Kathe S D, Goodrich-Blair H, Nierzwicki-Bauer S A, Shub D A. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990;250:1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- 88.Yang J, Zimmerly S, Perlman P S, Lambowitz A M. Efficient integration of an intron RNA into double-stranded DNA by reverse splicing. Nature. 1996;381:332–335. doi: 10.1038/381332a0. [DOI] [PubMed] [Google Scholar]
- 89.Yeo C C, Tham J M, Yap M W, Poh C L. Group II intron from Pseudomonas alcaligenes NCIB 9867 (P25X): entrapment in plasmid RP4 and sequence analysis. Microbiology. 1997;143:2833–2840. doi: 10.1099/00221287-143-8-2833. [DOI] [PubMed] [Google Scholar]
- 90.Young P, Ohman M, Xu M Q, Shub D A, Sjoberg B-M. Intron-containing T4 bacteriophage gene sunY encodes an anaerobic ribonucleotide reductase. J Biol Chem. 1994;269:20229–20232. [PubMed] [Google Scholar]
- 91.Zhou L, Manias D A, Dunny G L. Regulation of intron function: efficient splicing in vivo of a bacterial group II intron requires a functional promoter within the intron. Mol Microbiol. 2000;37:639–652. doi: 10.1046/j.1365-2958.2000.02033.x. [DOI] [PubMed] [Google Scholar]
- 92.Zimmerly S, Guo H, Perlman P S, Lambowitz A M. Group II intron mobility occurs by target DNA-primed reverse transcription. Cell. 1995;82:1243–1247. doi: 10.1016/0092-8674(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 93.Zimmerly S, Guo H, Eskes R, Yang J, Perlman P S, Lambowitz A M. A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell. 1995;83:529–583. doi: 10.1016/0092-8674(95)90092-6. [DOI] [PubMed] [Google Scholar]