Abstract
Judged by migration of its lipopolysaccharide (LPS) in gel electrophoresis, the O antigen of Rhizobium etli mutant strain CE166 was apparently of normal size. However, its LPS sugar composition and staining of the LPS bands after electrophoresis indicated that the proportion of its LPS molecules that possessed O antigen was only 40% of the wild-type value. Its LPS also differed from the wild type by lacking quinovosamine (2-amino-2,6-dideoxyglucose). Both of these defects were due to a single genetic locus carrying a Tn5 insertion. The deficiency in O-antigen amount, but not the absence of quinovosamine, was suppressed by transferring into this strain recombinant plasmids that shared a 7.8-kb stretch of the R. etli CE3 lps genetic region α, even though this suppressing DNA did not carry the genetic region mutated in strain CE166. Strain CE166 gave rise to pseudonodules on legume host Phaseolus vulgaris, whereas the mutant suppressed by DNA from lps region α elicited nitrogen-fixing nodules. However, the nodules in the latter case developed slowly and were widely dispersed. Two other R. etli mutants that had one-half or less of the normal amount of O antigen also gave rise to pseudonodules on P. vulgaris. The latter strains were mutated in lps region α and could be restored to normal LPS content and normal symbiosis by complementation with wild-type DNA from this region. Hence, the symbiotic role of LPS requires near-normal abundance of O antigen and may require a structural feature conferred by quinovosamine.
Rhizobium etli bacteria induce bean plants to form root nodules in which they fix nitrogen. As nodules develop, the bacteria penetrate to interior plant cell layers by means of an infection thread, in which a linear bacterial colony is surrounded by a tubular wall and plant plasma membrane that separates the bacteria from plant cytoplasm (24).
Mutants of R. etli that lack the O-antigen portion of the lipopolysaccharide (LPS) are defective in infection (6, 10, 30, 35). Fewer nodules are elicited by such mutants per region of susceptible root, and in the nodules that are formed, very few bacteria can be recovered. Microscopic examination reveals that the infection threads in these nodules are distended and cease development within the root hair or the subjacent cell layer (35). Closely resembling the development of nodules elicited by mutants that do not infect at all (46), the resulting nodule structure lacks most features of a normal mature nodule and has been described as a pseudonodule (27, 35). O-antigen-deficient mutants of Rhizobium leguminosarum and Bradyrhizobium japonicum also exhibit defects in infection and cause aberrant development of nodules on their respective host legumes (4, 12, 22, 23, 37, 38, 39, 44).
Although these studies demonstrate that the presence of O antigen is important in the symbiosis, the specific structural features that are required are still not established (30). However, there is evidence that addresses the question of how much O antigen each bacterium must possess in order to be symbiotically proficient. In R. etli mutant strain CE166 the proportion of LPS molecules that possess O antigen is less than normal (10). Strain CE166 is as defective in symbiosis as mutants that lack O antigen completely. Based on these properties of strain CE166, it has been argued that R. etli must have more O antigen than this strain in order to infect bean plants proficiently. However, work to be presented shows that the LPS of strain CE166 also lacks quinovosamine (QuiN) (2-amino-2,6-dideoxyglucose), which is contained in wild-type O-antigen-containing LPS as a single residue (6, 16, 17). Therefore, by itself, the phenotype of strain CE166 allows the possibility that either of these LPS features—the relative content of O-antigen-containing LPS or the presence of QuiN—is crucial in the symbiosis.
In previous work it was reported that the O-antigen deficiency and the symbiotic defects of this mutant fortuitously were suppressed by the introduction of lps DNA that does not contain the genetic locus that is mutated in CE166 (10). The biochemical and genetic basis for this suppression was examined further in the work described below. In particular, the suppressed strain was examined for QuiN content in its LPS. The symbiotic phenotype resulting from this genetic suppression was scrutinized more completely as well.
Two other mutants that synthesize less O-antigen-containing LPS than normal were isolated. They exhibited the same symbiotic phenotype as CE166.
The results strengthen the argument that the relative amount of O antigen is an important symbiotic determinant. At the same time, some of the findings are consistent with previous suggestions (12, 20) that specific LPS residues may be important for infection and/or nodule development. In addition, consideration of the proposed structure of the LPS and the absence of QuiN in CE166 raise intriguing questions about the fidelity of complex polysaccharide biosynthesis.
MATERIALS AND METHODS
Growth of bacteria.
Bacteria were grown to stationary phase in TY broth—a medium composed of 0.5% tryptone, 0.3% yeast extract, and 10 mM calcium chloride (3, 41)—at 30°C in Fernbach flasks (for LPS chemical analysis) or glass tubes (for plant inoculation) shaken at 200 rpm.
Isolation of mutants CE394 and CE395.
Wild-type strain CE3 (Table 1) was subjected to transposon mutagenesis with Tn5 suicide plasmid pSUP2021 (10, 42). Kanamycin-resistant colonies were screened for binding of monoclonal antibody JIM28 under conditions in which the LPS I of strain CE3 changes so that the antibody no longer binds (32). The details of the screening procedure will be described in a forthcoming report (V. J. Neumann, D. M. Duelli, and K. D. Noel, unpublished data). Certain properties of these strains have been reported previously (31).
TABLE 1.
Strains and plasmids used in this study
| Straina or plasmidb | Relevant characteristic(s) | Reference(s) or source |
|---|---|---|
| Strains | ||
| CE3 | R. etli wild-type LPS and symbiosis | 33 |
| CE166 | lps-166::Tn5, QuiN− LPS, low I/IIc | 10 |
| CE166α | CE166 carrying pCOS109.11 | This work |
| CE374 | lps-233::Tn5 | 32, 45 |
| CE394 | lps-394::Tn5, altered LPS and I/II | This work |
| CE394α | CE394 carrying pCOS109.11 | This work |
| CE395 | lps-395::Tn5, altered LPS and I/II | This work |
| CE395α | CE395 carrying pCOS109.11 | This work |
| CE408d | mTn5SSgusA20 in CE3 | This work |
| CE433d | mTn5SSgusA20 in CE109 (lacks LPS I) | This work |
| CE434d | mTn5SSgusA20 in CE166 | This work |
| CE436d | mTn5SSgusA20 in CE166α | This work |
| CE451 | lps-166::Tn5 in CE3 by homogenotization | This work |
| Plasmids | ||
| pCOS109.11 | R. etli CE3 lps region α in 31-kb insert | 10, 29 |
| pDEL2 | 22-kb DNA from pCOS109.11 insert | 11 |
| pDEL2-3 | 14-kb DNA from pDEL2 insert | 11 |
| pDEL14 | 16-kb DNA from pCOS109.11 insert | 11 |
| pDEL21 | 9-kb DNA from pCOS109.11 insert | 11 |
| pEQ166 | pJQ200 carrying the 11-kb lps-166::Tn5 EcoRI fragment | This work |
| pJQ200 | Vector for homogenotization; Gm sacB | 40 |
Strain CE3 is a streptomycin-resistant derivative of R. etli nodule isolate CFN42 (33). All other strains were derived from CE3.
All R. etli DNA inserts, except in pEQ166, were carried in vector pLAFR1 (18).
I/II, LPS I/LPS II ratio. The apparent ratios of LPS I/LPS II of the strains decrease in the following order: CE3 and CE374 (equal ratios), CE395, CE166, CE394.
mTn5SSgusA20 was inserted at unknown locations after introduction into the respective parent strains on plasmid pCAM120 (47). These strains were used to compare extents of infection. Their LPS SDS-PAGE and symbiotic phenotypes were the same as those of respective parent strains (CE3, CE109, CE166, and CE166α) (data not shown).
Construction of strain CE451.
The 11-kb EcoRI fragment of strain CE166 that contains the Tn5 insertion was cloned into the EcoRI site of pBluescript and subcloned into plasmid pJQ200 (40) by ApaI and XbaI digestion of vector and source DNA. This construct (pEQ166) (Table 1) was transferred into strain CE3, and substitution of the lps-166 mutant locus for the corresponding wild-type locus was selected by plating on TY agar containing kanamycin (30 mg/ml) and 5% sucrose (40). Strain CE451 was one of the colonies arising on this plate.
SDS-polyacrylamide gel electrophoresis (PAGE).
Sodium dodecyl sulfate (SDS) extracts of washed cells from fully grown cultures were subjected to electrophoresis in 18% polyacrylamide, and the gels were stained by the periodate-silver procedure as described previously (10). Relative amounts of LPS in different bands were assessed by analysis of the silver staining with an Ambis optical imaging system (32).
LPS chemical analysis.
LPS was extracted by the hot phenol-water method (6) and purified by either Sepharose 4B chromatography (8, 10) or by polymyxin-agarose affinity chromatography (17). Neutral sugars and aminosugars were quantified by gas chromatography on SP2330 columns (Supelco) as alditol acetate derivatives after hydrolysis in 2 M trifluoroacetic acid at 121°C for 2 h (1), whereas the content of acidic sugars was determined by gas chromatography on DB-1 (J & W Scientific) and SPB-1 (Supelco) columns as trimethylsilyl (TMS) methyl glycoside derivatives after methanolysis in methanolic 1 M HCl (17). The main GC peak contributed by glucuronic acid (GlcA) during TMS methyl glycoside formation coincides with a peak contributed by C14 hydroxy fatty acids; therefore, some samples were first treated at 100°C with 1% acetic acid to remove lipid A before subjecting the soluble material to derivatization and GC analysis. High-performance anion exchange chromatography (HPAEC) on a Carbo Pac PA-1 column (Dionex), eluted with a sodium acetate gradient in 100 mM sodium hydroxide, was carried out after treatment of the purified LPS with 1% acetic acid at 100°C as previously specified (7).
Complementation or suppression of mutants with cloned wild-type DNA.
Cloned DNA in Escherichia coli was transferred to R. etli recipients by conjugation as described previously (10, 11, 13), using plasmid pRK2013 or its derivative pRK600 (15) to mobilize the plasmids listed in Table 1.
Nodulation tests.
R. etli strains were tested for nodulation of Phaseolus vulgaris L. cv. Midnight Black Turtle Soup in growth pouches as previously described (35). Nitrogenase activity was measured by the acetylene reduction catalyzed by intact nodulated roots (19, 34), after which the nodules were stripped off the roots, counted, and weighed. Bacterial CFU were determined from crushed, surface-sterilized nodules (35).
Infection by strains labelled with gusA (Table 1) was determined after making longitudinal hand sections from 1-cm-long segments of nodulated roots with fresh portions of double-edged razor blades. The slices were placed immediately in 1 ml of β-glucuronidase (GUS) staining solution (50 mM sodium phosphate [pH 7.0], 1 mM EDTA, 0.1% Sarkosyl, 0.1% Triton X-100, 1 mM ferricyanide, 1 mM ferrocyanide, and 500 μg of 5-bromo-4-chloro-3-indonyl-β-glucuronic acid [X-GlcA] [from Biosynth] per ml) in a loosely capped 13-by-100-mm glass tube and incubated for 30 min to 24 h at 30°C with shaking in the dark (47). Staining was arrested by removing the staining solution and replacing it with the same solution without X-GlcA or ferricyanide or ferrocyanide. Sharply defined blue infection traces were observed in wet mounts of the slices by light microscopy.
RESULTS
O-antigen structure and content of CE166.
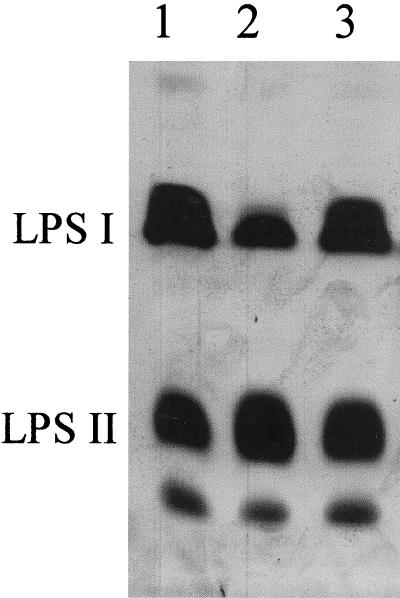
The SDS-PAGE gel in Fig. 1 compares the LPS profiles of strain CE166 and wild-type strain CE3. The slower-migrating bands labeled as LPS I carry the polysaccharide portion designated as O antigen, whereas faster-migrating bands labeled as LPS II do not (6). The relative deficit of LPS I in CE166 is apparent. From image analysis of the silver staining of similar gels, the mutant appeared to harbor 34% as much LPS I as the wild-type strain (relative to the same amount of LPS II). However, the migration in SDS-PAGE suggested that the mutant LPS I was approximately the same size as the wild-type LPS I.
FIG. 1.
SDS-PAGE comparison of relative amounts of LPS I and LPS II in strains CE166, CE166α, and wild-type CE3. SDS extracts of washed cell pellets were loaded to give roughly equal amounts of total LPS. The band that migrates faster than LPS II comigrates with purified lipid A. Lanes: 1, CE3; 2, CE166; 3, CE166α.
The sugar compositions of the wild-type and the mutant LPSs are given in Table 2. The contents of the O-antigen sugars fucose, O-methyl-6-deoxyhexoses, and glucuronic acid relative to core sugars galactose and galacturonic acid indicated that the mutant had about 40% as much O-antigen as the wild type. The mutant LPS was devoid of QuiN, which in the wild type is present as a single residue per LPS I molecule (16, 17). The gas chromatograms after either alditol acetate or TMS methylglycoside derivatization failed to show any signal at the elution time of this sugar. In addition, mass spectral scans for the signature ion of 2-aminosugars (e.g., m/z 173 from the TMS methylglycoside) detected only glucosamine. To have escaped detection, the mutant must have less than 1% of the wild-type content of QuiN.
TABLE 2.
Sugar compositions of purified LPSs
| Sugarb | Molar ratioa found in strain:
|
|||
|---|---|---|---|---|
| CE3 | CE166 | CE166α | CE395 | |
| QuiN | 0.6 | ND | ND | 0.4 |
| 2OMe6Dhx | 0.9 | 0.4 | 0.8 | ND |
| 3OMe6Dhx | 2.9 | 1.3 | 2.5 | 1.6 |
| Fuc | 3.6 | 1.4 | 2.6 | 2.2 |
| GlcA | 3.0 | 1.7 | 2.4 | 1.9 |
| Man | 1.9 | 1.4 | 1.7 | 1.5 |
| Gal | 1.0 | 1.0 | 1.0 | 1.0 |
| GalA | 2.9 | 2.8 | 2.9 | 3.0 |
| Kdo | 2.1 | 1.9 | 2.2 | — |
| GlcN | 0.8 | 0.9 | 0.9 | 0.9 |
Entries are molar ratios of the sugars normalized to 1.0 galactose. They are the mean values from five parallel analyses of CE3, CE166, and CE166α LPS and three additional parallel analyses of CE3 and CE166 LPS in two laboratories over the course of 10 years. In the data for these three strains, the average standard deviation of the mole ratios before normalization was 21% of the mean value. The CE395 data are mean values from two determinations. ND, not detected (<0.01); —, obscured by TMS methyl glycoside peaks due to residual EDTA in the CE395 LPS after Sepharose 4B chromatography.
The sugars in boldface type are found in the portion of the R. etli CE3 designated as the O antigen (16, 17). Abbreviations: 2OMe6Dhx, 2-O-methyl-6-deoxyhexose; 3OMe6Dhx, 3-O-methyl-6-deoxy hexose; Fuc, fucose; Man, mannose; Gal, galactose; GalA, galacturonic acid; GlcN, glucosamine. Migration with methylated standards indicates that 2OMe6Dhx and 3OMe6Dhx are 2-O-methyl-fucose and 3-O-methyl-6-deoxytalose, respectively, in strain CE3 (16) (Fig. 4).
In other respects the LPS of the mutant was similar to the wild-type LPS. The relative proportions of galacturonic acid, galactose, and mannose (Table 2) suggested that the inner core region of the LPS (17) remains intact in CE166. This inference was supported by HPAEC of oligosaccharides released by mild acid cleavage at 3-deoxy-d-manno-2-octulonic acid (Kdo) residues. After hydrolysis with 1% acetic acid at 100°C, the characteristic HPAEC peaks that arise from wild-type LPS core structure (7) were found in the hydrolysate of CE166 LPS also (data not shown).
The LPS I of CE166 also resembled the wild-type LPS I antigenically. It reacted with the four monoclonal antibodies (JIM26, JIM27, JIM28, and JIM29) known to recognize the LPS I of R. etli CE3 but not the LPS I of other Rhizobium strains that have been tested (3, 45). Also like the wild type (32, 45), mutant CE166 altered the LPS structure during growth in the presence of bean exudates or low pH, such that antibodies JIM28 and JIM29 no longer bound (data not shown).
Linkage of two LPS defects to same genetic locus.
Strain CE451 was selected as a recombinant in which the 11-kb EcoRI fragment of CE166 carrying the Tn5 insertion had replaced the corresponding wild-type DNA of strain CE3. This strain exhibited the sugar composition of strain CE166, specifically the absence of QuiN and the deficiency in O-antigen sugars. On SDS-PAGE strain CE451 exhibited the LPS I deficiency of CE166, having about one-third the LPS I content of wild-type CE3 relative to LPS II (data not shown).
Suppression by DNA from R. etli lps region α.
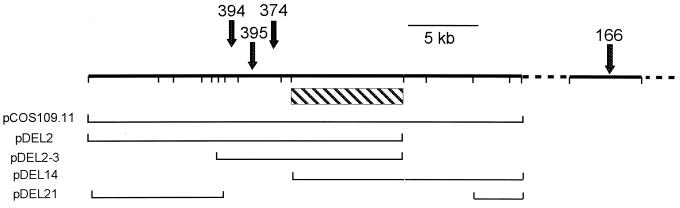
Recombinant plasmid pCOS109.11 carries 30 kb of R. etli DNA (Fig. 2). The genes within this DNA that affect LPS are known collectively as lps region α (29). In previous work (10) this plasmid was transferred into mutant CE166 to create strain CE166α (Table 1). In strain CE166α the abnormal ratio of LPS I and LPS II of strain CE166 is suppressed (Fig. 1). By image analysis of LPS staining on SDS PAGE (e.g., Fig. 1), the LPS I content of strain CE166α was at least 85% of the wild-type value relative to LPS II. This conclusion also was supported by the contents of O-antigen sugars, which averaged about 80% of the wild-type values (Table 2). Although the mean values of the molar ratios of O-antigen sugars in CE166α LPS were uniformly lower than those of the wild type (Table 2), the differences were only marginally statistically significant at the 90% confidence level for Fuc and Man contents, and not significant even at that level for differences in the O-methyl-deoxyhexoses. On the other hand, the differences between CE166 and CE166α in contents of O-antigen sugars were statistically significant at the 99% confidence level in a t test.
FIG. 2.
The R. etli CE3 lps α and lps-166 genetic regions. The lps α locus is a contiguous stretch of DNA shown on the left. The dotted line indicates that lps-166 is separated from the lps α locus by an undetermined distance along the chromosome. Ticks on the upper line indicate the positions of EcoRI sites. The relative locations of the Tn5 insertions of four mutants in this study are indicated by arrows above this line. The lines below indicate stretches of DNA from lps α that are contained in the specified plasmids. The lps α DNA responsible for the suppression of lps-166 is located in the region indicated by the striped box.
Although the amount of LPS I was obviously boosted in CE166α, the LPS of this strain clearly retained one deficiency of CE166 LPS. It lacked QuiN (Table 2).
Using SDS-PAGE as the assay, shorter portions of this lps genetic region (11) were tested for suppression of CE166. Plasmids pDEL2, pDEL2-3, and pDEL14 (Fig. 2) also suppressed the LPS I/LPS II deficiency, whereas pDEL21 did not (data not shown). Therefore, the suppression appears to require one or more genes in the 7.8-kb region common to both pDEL2-3 and pDEL14 (Fig. 2).
Other R. etli mutants deficient in LPS I amount.
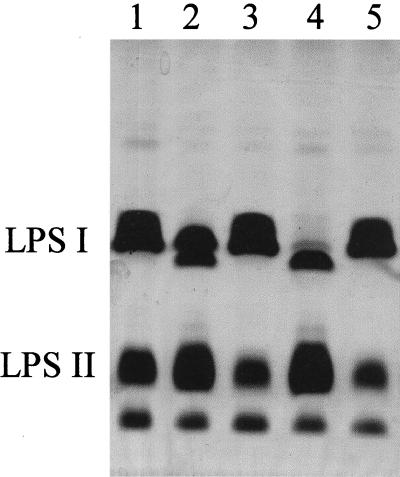
Like strain CE166, mutant strains CE394 and CE395 (Table 1) also made LPS I in lower amounts than the wild type (Fig. 3). The purified LPS from CE395, the strain with the higher LPS I content, had the normal content of QuiN (Table 2). Aside from having lower ratios of all O-antigen sugars, it specifically lacked the 2-O-methyl-6-deoxyhexose found in the wild type (Table 2).
FIG. 3.
SDS-PAGE of SDS extracts of washed cell pellets of strains showing variation in LPS I content and the same strains after complementation with lps region α. Lanes: 1, wild-type CE3; 2, CE395; 3, CE395α; 4, CE394; 5, CE394α.
The Tn5 insertion mutations in strains CE394 and CE395 were located in EcoRI fragments within the lps α region (Fig. 2), as shown by DNA-DNA hybridization. Southern blots of EcoRI digests of the DNA from strains CE394, CE395, and CE3 (wild type) were probed with labeled pCOS109.11. A wild-type 0.93-kb band was replaced with a 6.5-kb band in CE394, and an 8.5-kb band was present in CE395 instead of the 2.9-kb wild-type band. These size increases are consistent with mutation by insertion of the 5.8-kb transposon. In this type of analysis, CE166 DNA gave the same hybridization pattern with pCOS109.11 as wild-type DNA.
Plasmid pCOS109.11 restored the normal LPS I/LPS II ratio when transferred into these mutants (CE394α and CE395α [Fig. 3]). As expected from the location of the insertion mutations, plasmids pDEL2 and pDEL2-3 (Fig. 2) also restored the normal LPS I amount, whereas plasmids pDEL14 and pDEL21 did not (data not shown). The genetic region in which these mutations map and which complements these mutants, therefore, is distinct from the locus that suppresses CE166.
Symbiotic phenotypes.
Mutant CE166 gave rise to the pseudonodulation typical of certain auxotrophs and mutants that lack LPS I (10, 27); the (pseudo)nodules developed in a slow, stunted fashion, were highly dispersed along the root, and never exhibited nitrogen-fixing activity (Table 3). Infections were examined by using a derivative of strain CE166 carrying gusA (strain CE434 [Table 1]) and viewing the staining of GUS activity in hand sections of (pseudo)nodules by light microscopy. The results supported previous observations (10) that infections by CE166 stopped within the infected root hairs or at most penetrated one to two cell layers beyond. Infections by a gusA-containing derivative of mutant CE109 (strain CE433, Table 1), which lacks O antigen completely (6), gave results indistinguishable from those of CE166.
TABLE 3.
Symbiotic performance of complemented and suppressed strains
| Time and inoculant | AR/planta | Nodules/plantc
|
Specific ARb | |
|---|---|---|---|---|
| No. | wt | |||
| 14 d.a.i. | ||||
| CE3 | 100 | 60 | 45 | 100 |
| CE166α | 1 | 12 | 7 | 10 |
| CE166αNd | 2 | 11 | 6 | 16 |
| CE374 | 2 | 16 | 10 | 11 |
| CE394α | 42 | 45 | 29 | 67 |
| CE395α | 36 | 52 | 32 | 53 |
| 20 d.a.i. | ||||
| CE3 | 100 | 63 | 78 | 100 |
| CE166e | <0.1 | 25 | 2 | <0.1 |
| CE166α | 29 | 17 | 39 | 59 |
| 26 d.a.i. | ||||
| CE3 | 100 | 68 | 120 | 100 |
| CE166e | <0.1 | 60 | —f | <0.1 |
| CE166α | 25 | 18 | 71 | 42 |
| CE374 | 92 | 31 | 88 | 123 |
| CE395α | 110 | 48 | 123 | 112 |
| CE394α | 123 | 50 | 125 | 116 |
AR (acetylene reduction) normalized to values exhibited by strain CE3 (which is set to 100).
AR per total nodule fresh weight, expressed as percent of the wild-type value.
Number and total fresh weight (mg) of the nodules per plant.
An isolate from nodules of CE166α.
Representative of results with CE394 and CE395.
—, small pseudonodules (not weighed).
On the other hand, strain CE166α gave rise to true nodule development, although many fewer nodules appeared and their development lagged at least 4 days behind the development incited by the wild type. For instance, at 14 days after inoculation (d.a.i.), these nodules were smaller and much less active than those incited by the wild type (Table 3). However, although never as many nodules developed, at 20 d.a.i. the nodules were larger and nearly as active as those of the wild type (Table 3). Whereas nodules inoculated by CE166 averaged 104 CFU/nodule, those inoculated with either CE3 or CE166α averaged 2 × 107 CFU/nodule at this time. At 26 d.a.i. the nodules induced by the suppressed mutant averaged twice the mass of the wild-type nodules, although the specific nitrogenase activity was still less than wild type (Table 3). Similarly, infections by CE166α (viewed by GUS staining of gusA-containing derivative CE346 [Table 1]) were greatly behind those of the wild type at 16 d.a.i. but were comparable to those of the wild type by 26 d.a.i. Bacterial isolates from the eventually well-developed nodules incited by CE166α gave the same delay in nodule development as CE166α (CE166αN of Table 3). CE166 transconjugants carrying pDEL2-3 or pDEL14 gave results similar to those of CE166α (data not shown).
Mutants CE394 and CE395 exhibited the same symbiotic block as CE166. However, when strains CE394 and CE395 were complemented with wild-type alleles carried on plasmids pCOS109.11, pDEL2, or pDEL2-3, their symbiotic properties were similar to those of the wild type and clearly superior to CE166 suppressed by carrying these same plasmids (Table 3).
In summary, the tested strains fell into three groups according to development of the symbiosis. Group I (strains CE3, CE394α, and CE395α) had normal or near-normal proficiency. Group II consisted of strains CE166α and CE374, the latter being a previously documented strain that appears to have very subtle alterations in LPS I structure (32). These strains elicited markedly fewer nodules, whose development was obviously slower (at least 4 days behind that of the wild type) but eventually had near-normal specific nitrogen-fixing activity and greater masses than normal. Group III (strains CE166, CE394, and CE395) elicited sparsely distributed pseudonodules that contained very low numbers of bacteria and lacked nitrogen-fixing activity.
DISCUSSION
Structural alterations of CE166 LPS.
Clearly, there are at least two alterations: the absence of QuiN and the reduced amount of O antigen in the LPS population. In other respects the structure of strain CE166 LPS I appears to be similar to that of the wild type, as revealed by antigenic reactions, sugar ratios, and SDS-PAGE migration.
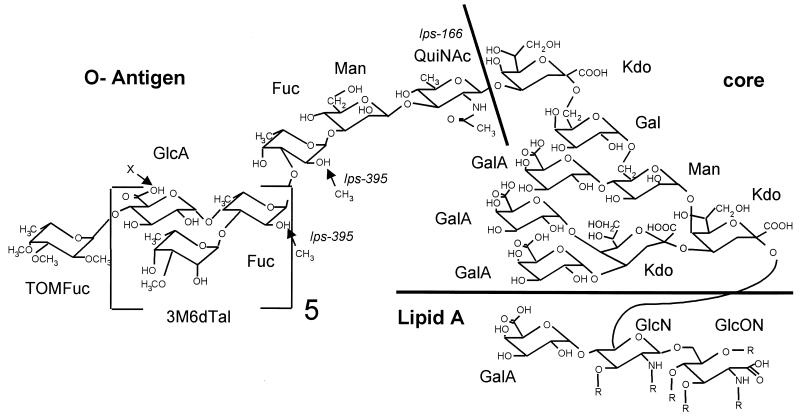
The absence of QuiN is intriguing. Structural analysis indicates that QuiN (as its N-acetyl derivative) is present as a 1,3-linked residue in the wild-type LPS structure at the junction between the core oligosaccharide and O-antigen repeating unit (Fig. 4) (16, 17), such that its elimination should truncate the LPS at that point. One possibility is that it is substituted by another residue to allow for LPS I production in strain CE166. The sugar compositions of CE166 and CE166α LPS do not reveal an obvious candidate for this substituted residue, however. In particular, glucosamine does not appear to be substituted for QuiN (Table 2).
FIG. 4.
Proposed structure of LPS I of R. etli CE3 (16, 17). Features affected in mutants CE166 and CE395 are noted. Designation as O antigen is arbitrarily assigned to all contiguous residues released with the Kdo most distal to lipid A after mild hydrolysis that cleaves at Kdo residues (16, 23). Chemical analysis indicates that Fuc residues are only partially O-methylated at the positions shown (16). R, the various hydroxyfatty acyl groups that are ester- and amide-linked at the positions indicated in the lipid A portion; TOMFuc, tri-O-methylated Fuc; 3M6dTal, 3-O-methyl-6-deoxytalose; QuiNAc, N-acetylquinovosamine; GlcON, 2-aminogluconate. All other abbreviations are defined in Table 2.
Genetic and biochemical basis of the effect of lps α DNA on CE166.
Genetic analysis indicates that the effect of pCOS109.11 is one of extragenic suppression. The cloned EcoRI fragment of CE166 that contains the Tn5 insertion is not of a size expected of an insertion in any of the EcoRI fragments of pCOS109.11, nor does it hybridize with the DNA of pCOS109.11 in Southern blots (9, 10). Likewise, the pattern of EcoRI bands revealed by hybridization with pCOS109.11 as a probe is the same in both CE166 and CE3.
That the Tn5 insertion is the cause of both mutant phenotypes is supported by the results of transferring the Tn5- mutated locus into the wild-type genetic background, by low-resolution chromosomal linkage analysis in previous work (9, 10), and by homogenotization with the Tn5-mutated EcoRI fragment of CE166 in this study. Kmr transconjugants had the SDS-PAGE and symbiotic phenotypes of CE166. Importantly, analysis of transconjugant strain CE451 shows that the Tn5-mutated locus confers both the LPS I deficiency and the absence of QuiN, whereas the DNA within pCOS109.11 (lps region α) suppresses only the LPS I deficiency.
Very recent single-pass nucleotide sequencing outward from the Tn5 insertion of CE166 indicates that it is situated in an open reading frame specifying a protein of 325 amino acids that is homologous to a family of enzymes involved in deoxysugar synthesis (K. D. Noel, unpublished data). The highest matches are to two proteins (WbfT and WbpV, with 50 and 47% identity, respectively) believed to be involved in the second step of the synthesis of UDP-QuiNAc from UDP-GlcNAc (2).
The DNA responsible for the suppression apparently lies within the 7.8-kb EcoRI fragment common to plasmids pDEL2-3 and pDEL14. This DNA exists already in CE166, presumably with wild-type functionality if the mutant phenotypes are conferred by the Tn5 insertion mapping elsewhere. Therefore, it is likely that the suppression is due to the multicopy dosage of a gene or genes in this 7.8-kb EcoRI fragment.
There are several reasonable hypotheses for the biochemical mechanism of suppression, with the basic premise being that the gene encodes a transferase, O-antigen exporter, or LPS ligase with lax specificity or that it specifies a regulatory factor, and that multiple genetic copies lead to higher concentration of the encoded protein. Variations of this idea are worth pursuing as a means of producing wild-type amounts of other polysaccharides having an altered or absent residue, so that the biological importance of the residue itself can be tested. A case in point is the 2-O-methylation of fucose that is absent from mutant CE395 and whose absence probably is the cause of the lower O-antigen synthesis or attachment to lipid A.
Implications regarding requirements of LPS structure for symbiosis.
Comparisons of the phenotypes of strains CE166, CE166α, and CE3 indicate that LPS I must be more abundant than LPS II in order that a strain be successful in the symbiosis between R. etli and bean roots. The structure conferred by a particular residue, QuiN, may also be important, but the argument based on the data of this study for this LPS I feature is equivocal.
Restoration of near-normal LPS I/LPS II ratio in CE166 harboring pCOS109.11 (and its derivatives pDEL2-3 and pDEL14) was accompanied by sufficiently restored symbiotic proficiency that CE166α infections yielded well-developed nodules with nitrogen-fixing activity. An obvious inference is that relative abundance of LPS I is crucial in symbiosis. This conclusion is supported also by the phenotypes of strains CE394 and CE395. In fact, there are no exceptions to this idea among the approximately 30 R. etli mutants altered in LPS structure. Strain CE395 has the most LPS I of any mutant obviously deficient in LPS I; this mutant and all strains carrying less LPS I fail at an early stage in infection. It should be noted that all extant strains having a lower LPS I/LPS II ratio exhibit additional abnormalities in LPS structure, e.g., the abnormal LPS I banding of CE394 and CE395 (Fig. 3), the methylated sugar missing from strain CE395, and the absence of QuiN from mutant CE166. Nevertheless, the only obvious feature common to CE166, CE394, and CE395 is deficiency in the relative amount of LPS I.
Although near restoration of O-antigen amount in CE166α overcomes the absolute block in infection suffered by mutant 166, nodule development by CE166α is slower and sparser than normal. The difference in symbiosis between CE166α and wild type might be due to either of two deficiencies in LPS structure in strain CE166α. According to SDS-PAGE, strain CE166α consistently had a lower LPS I/LPS II ratio than the wild type, and its content of O-antigen sugars was lower (although without high statistical confidence). Hence, that its symbiotic phenotype was intermediate between those of CE166 and the wild type could be because LPS I content in CE166α is not restored completely to wild-type levels. On the other hand, the LPS of strain CE166α also differed from wild-type LPS in lacking QuiN. It may be that this portion of the LPS structure is specifically involved in promoting nodule development (perhaps by promoting an interaction required in infection thread development).
Another possibility is that CE166α suffers problems caused by multiple copies of plasmid pCOS109.11 or its possible instability (36). However, mutants CE394 and CE395 complemented with pCOS109.11 behave in symbiosis much more like wild type than CE166α (Table 3); therefore, this possibility seems less likely.
Comparison with mutant CE374 and LPS mutants of other species.
The LPS and symbiotic phenotypes of CE166α are reminiscent of the phenotypes of R. etli mutant strain CE374, whose properties also have suggested that specific LPS structure plays a role in the frequency of nodule formation and in the rate of infection. Strain CE374 appears to have wild-type amounts of LPS I, but its LPS I displays subtle antigenic differences and minor differences in SDS-PAGE banding compared with the wild type (32, 45). Although the symbiotic deficiencies of strain CE374 are very similar to those of CE166α, strain CE374 is not deficient in QuiN. In fact, analysis of sugar composition has not revealed an obvious difference between the LPSs of mutant CE374 and the wild type. The difference may be in acid-labile “decorations” of LPS I residues.
Examples from other species of LPS mutants that elicit delayed and/or slow nodule development include Sinorhizobium meliloti mutant Rm6963 on alfalfa (25, 28) and Bradyrhizobium elkanii mutant SM1 on soybean (26, 43). In each case, more than one sugar appears to be missing from the LPS, but substantial (undetermined) structure remains. Soybean presents an interesting parallel with bean (P. vulgaris) in regard to LPS and symbiosis. Both plants form determinate nodules, and soybean forms severely stunted pseudonodules when inoculated with B. japonicum mutants that lack O antigen entirely (44). Conclusions about how closely the case of B. elkanii SM1 parallels those of R. etli CE166α and CE374, however, await more definitive information about the LPS structures of all three mutants.
Possible biological roles of these LPS I structural features.
Some of the inferences from this work can be summed up in the following hypotheses. Insufficient O antigen leads to complete blockage of infection soon after infection starts (perhaps due to plant toxins that abundant O antigen can prevent from penetrating to targets on the rhizobial cell [14]). This role may be relatively independent of particular structure. However, certain other aspects of the infection process are greatly facilitated by specific structural features of the LPS, possibly including a feature contributed by the QuiN residue. Changes in these LPS features slow infection and, thereby, nodule development and also lead to lower incidence of nodules per infectible region of the root.
That subtle alterations in LPS structure slow infection suggests, as one hypothesis, that infection requires structurally specific interactions between the bacterial LPS and plant receptors on the plasma membrane. Analogous interactions with animal cell receptors have been proposed as part of the activity of LPS in certain bacterial pathogens (21). In developing nodules these interactions might direct the targeting of machinery for membrane growth to the sites at which the bacteria impinge directly on the plant plasma membrane, including the site at which the infection thread is initiated (5) and, later, the tip of the infection thread (30). Similar suggestions have been made previously (24, 44). The trifoliin A protein of clover roots has been proposed as a receptor of the LPS of R. leguminosarum bv. trifolii strains (12, 20). Interestingly, trifoliin A binds at much higher affinity to LPS containing QuiN and the binding is inhibited by QuiN (20). It would be of interest to investigate whether there are LPS-binding proteins from bean that interact preferentially with R. etli CE3 LPS containing QuiN and whether such proteins are associated with infection threads.
ACKNOWLEDGMENTS
K.D.N. acknowledges the technical assistance of Valerie Neumann, Tina Thorp, Kevin Barleben, and Jodie Box.
This work was supported by grants DE-FG02-98ER20307 from the U.S. Department of Energy (to K.D.N.) and GM39583 from the National Institutes of Health (to R.W.C.) as well as DOE grant DE-FG09-93ER20097 to the CCRC.
REFERENCES
- 1.Albersheim P, Nevins D J, English P D, Karr A. A method for the analysis of sugars in plant cell-wall polysaccharides by gas-liquid chromatography. Carbohydr Res. 1967;5:340–345. [Google Scholar]
- 2.Belanger M, Burrows L L, Lam J S. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology. 1999;145:3505–3521. doi: 10.1099/00221287-145-12-3505. [DOI] [PubMed] [Google Scholar]
- 3.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 4.Brink B A, Miller J, Carlson R W, Noel K D. Expression of Rhizobium leguminosarum CFN42 genes for lipopolysaccharide in strains derived from different R. leguminosarum soil isolates. J Bacteriol. 1990;172:548–555. doi: 10.1128/jb.172.2.548-555.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan D A, Torrey J G. The structural basis for infection of root hairs of Trifolium repens by Rhizobium. Can J Bot. 1981;59:1647–1664. [Google Scholar]
- 6.Carlson R W, Kalembasa S, Turowski D, Pachori P, Noel K D. Characterization of the lipopolysaccharide from a Rhizobium phaseoli mutant that is defective in infection thread development. J Bacteriol. 1987;169:4923–4928. doi: 10.1128/jb.169.11.4923-4928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson R W, Reuhs B, Chen T, Bhat U R, Noel K D. Lipopolysaccharide core structures in Rhizobium etli and mutants deficient in O-antigen. J Biol Chem. 1995;270:11783–11788. doi: 10.1074/jbc.270.20.11783. [DOI] [PubMed] [Google Scholar]
- 8.Carlson R W, Sanders R E, Napoli C, Albersheim P. Host-symbiont interactions. III. Purification and characterization of Rhizobium lipopolysaccharides. Plant Physiol. 1978;62:912–917. doi: 10.1104/pp.62.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cava J R. Genetic analysis of lipopolysaccharide and purine biosynthesis and their requirement for nodulation in Rhizobium leguminosarum strain CFN42. Ph.D. dissertation. Milwaukee, Wis: Marquette University; 1988. [Google Scholar]
- 10.Cava J R, Elias P M, Turowski D A, Noel K D. Rhizobium leguminosarum CFN42 genetic regions encoding lipopolysaccharide structures essential for complete nodule development on bean plants. J Bacteriol. 1989;171:8–15. doi: 10.1128/jb.171.1.8-15.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cava J R, Tao H, Noel K D. Mapping of complemention groups within a Rhizobium leguminosarum CFN42 chromosomal region required for lipopolysaccharide synthesis. Mol Gen Genet. 1990;221:125–128. [Google Scholar]
- 12.Dazzo F B, Truchet G L, Hollingsworth R L, Hrabak E M, Pankratz H S, Philip-Hollingsworth S, Salzwedel J L, Chapman K, Appenzeller L, Sqartini A, Gerhold D, Orgambide G. Rhizobium lipopolysaccharide modulates infection thread development in white clover root hairs. J Bacteriol. 1991;173:5371–5384. doi: 10.1128/jb.173.17.5371-5384.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ditta G, Stanfield S, Corbin D, Helinski D R. Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA. 1980;77:7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenschenk L, Diebold R, Perez-Lesher J, Peterson A C, Peters A K, Noel K D. Inhibition of Rhizobium etli polysaccharide mutants by Phaseolus vulgaris root compounds. Appl Environ Microbiol. 1994;60:3315–3322. doi: 10.1128/aem.60.9.3315-3322.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finan T M, Kunkel B, De Vos G F, Signer E R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986;167:66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg L S, Bhat U R, Carlson R W. Structural characterization of the O-antigenic polysaccharide of the lipopolysaccharide from Rhizobium etli strain CE3. A unique O-acetylated glycan of discrete size, containing 3-O-methyl-6-deoxy-l-talose and 2,3,4-tri-O-methyl-l-fucose. J Biol Chem. 2000;275:18851–18863. doi: 10.1074/jbc.M001090200. [DOI] [PubMed] [Google Scholar]
- 17.Forsberg L S, Carlson R W. The structures of the lipopolysaccharides from Rhizobium etli strains CE358 and CE359. The complete structure of the core region of R. etli lipopolysaccharides. J Biol Chem. 1998;273:2747–2757. doi: 10.1074/jbc.273.5.2747. [DOI] [PubMed] [Google Scholar]
- 18.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 19.Hardy, R. W. F., R. D. Holsten, E. K. Jackson, and R. C. Burns. The acetylene-ethylene assay for nitrogen fixation: laboratory and field evaluations. Plant Physiol. 43:1185–1207. [DOI] [PMC free article] [PubMed]
- 20.Hrabak E M, Urbano M R, Dazzo F B. Growth-phase-dependent immunodeterminants of Rhizobium trifolii lipopolysaccharide which bind trifoliin A, a white clover lectin. J Bacteriol. 1981;148:697–711. doi: 10.1128/jb.148.2.697-711.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacque M, Paradis S E. Adhesin-receptor interactions in Pasteurellaceae. FEMS Microbiol Rev. 1998;22:45–59. doi: 10.1016/s0168-6445(98)00007-2. [DOI] [PubMed] [Google Scholar]
- 22.Kannenberg E L, Rathbun E A, Brewin N J. Molecular dissection of structure and function in the lipopolysaccharide of Rhizobium leguminosarum strain 3841 using monoclonal antibodies and genetic analysis. Mol Microbiol. 1992;6:2477–2487. doi: 10.1111/j.1365-2958.1992.tb01424.x. [DOI] [PubMed] [Google Scholar]
- 23.Kannenberg E L, Reuhs B L, Forsberg L S, Carlson R W. Lipopolysaccharides and K-antigens from bacteria belonging to the Rhizobiaceae family: their structures, biosynthesis, and functions. In: Spaink H, Kondorosi A, Hooykaas P, editors. The Rhizobiaceae: molecular biology of model plant-associated bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1998. pp. 119–154. [Google Scholar]
- 24.Kijne J W. The Rhizobium infection process. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman & Hall; 1991. pp. 349–398. [Google Scholar]
- 25.Lagares A, Caetano-Anolles G, Niehaus K, Lorenzen J, Ljunggren H D, Puhler A, Favelukes G. A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J Bacteriol. 1992;174:5941–5952. doi: 10.1128/jb.174.18.5941-5952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maier R J, Brill W J. Involvement of Rhizobium japonicum O antigen in soybean nodulation. J Bacteriol. 1978;133:1294–1299. doi: 10.1128/jb.133.3.1295-1299.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newman J D, Schultz B W, Noel K D. Dissection of nodule development by supplementation of Rhizobium leguminosarum biovar phaseoli purine auxotrophs with 4-aminoimidazole-5-carboxamide riboside. Plant Physiol. 1992;99:401–408. doi: 10.1104/pp.99.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niehaus K, Lagares A, Puhler A. A Sinorhizobium meliloti lipopolysaccharide mutant induces effective nodules on the host plant Medicago sativa (alfalfa) but fails to establish a symbiosis with Medicago truncatula. Mol Plant-Microbe Interact. 1998;11:906–914. [Google Scholar]
- 29.Noel K D. Rhizobial polysaccharides required in symbioses with legumes. In: Verma D P S, editor. Molecular signals in plant-microbe communications. Boca Raton, Fla: CRC Press; 1992. pp. 341–357. [Google Scholar]
- 30.Noel K D, Duelli D M. Rhizobium lipopolysaccharide and its role in symbiosis. In: Triplett E W, editor. Prokaryotic nitrogen fixation: a model system for analysis of a biological process. Wymondham, United Kingdom: Horizon Scientific Press; 2000. pp. 415–431. [Google Scholar]
- 31.Noel K D, Duelli D M, Neumann V J. Changes in Rhizobium lipopolysaccharide structure induced by host compounds. In: Legocki A, Bothe H, Puhler A, editors. Biological fixation of nitrogen for ecology and sustainable agriculture. Berlin, Germany: Springer-Verlag; 1997. pp. 107–110. [Google Scholar]
- 32.Noel K D, Duelli D M, Tao H, Brewin N J. Antigenic change in the lipopolysaccharide of Rhizobium etli CFN42 induced by exudates of Phaseolus vulgaris. Mol Plant-Microbe Interact. 1996;9:180–186. [Google Scholar]
- 33.Noel K D, Sanchez A, Fernandez L, Leemans J, Cevallos M A. Rhizobium phaseoli symbiotic mutants with transposon Tn5 insertions. J Bacteriol. 1984;158:148–155. doi: 10.1128/jb.158.1.148-155.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noel K D, Stacey G, Tandon S R, Silver L E, Brill W J. Rhizobium japonicum mutants defective in symbiotic nitrogen fixation. J Bacteriol. 1982;152:485–494. doi: 10.1128/jb.152.1.485-494.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noel K D, Vandenbosch K A, Kulpaca B. Mutations in Rhizobium phaseoli that lead to arrested development of infection threads. J Bacteriol. 1986;168:1392–1401. doi: 10.1128/jb.168.3.1392-1401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Connell M, Noel T C, Yeung E C, Hynes M, Hynes M F. Decreased symbiotic effectiveness of Rhizobium leguminosarum strains carrying plasmid RP4. FEMS Microbiol Lett. 1998;161:275–283. doi: 10.1111/j.1574-6968.1998.tb12958.x. [DOI] [PubMed] [Google Scholar]
- 37.Perotto S, Brewin N J, Kannenberg E L. Cytological evidence for a host defense response that reduces cell and tissue invasion in pea nodules by lipopolysaccharide-defective mutants of Rhizobium leguminosarum strain 3841. Mol Plant-Microbe Interact. 1994;7:99–112. [Google Scholar]
- 38.Poole P S, Schofield N A, Reid C J, Drew E M, Walshaw D L. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology. 1994;140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- 39.Priefer U B. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar viciae VF39. J Bacteriol. 1989;171:6161–6168. doi: 10.1128/jb.171.11.6161-6168.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection of gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 41.Sherwood M T. Inhibition of Rhizobium trifolii by yeast extracts or glycine is prevented by calcium. J Gen Microbiol. 1970;71:351–358. [Google Scholar]
- 42.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 43.Stacey G, Paau A S, Noel K D, Maier R J, Silver L E, Brill W J. Mutants of Rhizobium japonicum defective in nodulation. Arch Microbiol. 1982;132:219–224. [Google Scholar]
- 44.Stacey G, So J-S, Roth L E, Lakshmi B, Carlson R W. A lipopolysaccharide mutant of Bradyrhizobium japonicum that uncouples plant from bacterial differentiation. Mol Plant-Microbe Interact. 1991;4:332–340. doi: 10.1094/mpmi-4-332. [DOI] [PubMed] [Google Scholar]
- 45.Tao H, Brewin N J, Noel K D. Rhizobium leguminosarum CFN42 lipopolysaccharide antigenic changes induced by environmental conditions. J Bacteriol. 1992;174:2222–2229. doi: 10.1128/jb.174.7.2222-2229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.VandenBosch K A, Noel K D, Kaneko Y, Newcomb E H. Nodule initiation elicited by noninfective mutants of Rhizobium phaseoli. J Bacteriol. 1985;162:950–959. doi: 10.1128/jb.162.3.950-959.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other Gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]