Abstract
Oxazolidinone antibiotics inhibit bacterial protein synthesis by interacting with the large ribosomal subunit. The structure and exact location of the oxazolidinone binding site remain obscure, as does the manner in which these drugs inhibit translation. To investigate the drug-ribosome interaction, we selected Escherichia coli oxazolidinone-resistant mutants, which contained a randomly mutagenized plasmid-borne rRNA operon. The same mutation, G2032 to A, was identified in the 23S rRNA genes of several independent resistant isolates. Engineering of this mutation by site-directed mutagenesis in the wild-type rRNA operon produced an oxazolidinone resistance phenotype, establishing that the G2032A substitution was the determinant of resistance. Engineered U and C substitutions at G2032, as well as a G2447-to-U mutation, also conferred resistance to oxazolidinone. All the characterized resistance mutations were clustered in the vicinity of the central loop of domain V of 23S rRNA, suggesting that this rRNA region plays a major role in the interaction of the drug with the ribosome. Although the central loop of domain V is an essential integral component of the ribosomal peptidyl transferase, oxazolidinones do not inhibit peptide bond formation, and thus these drugs presumably interfere with another activity associated with the peptidyl transferase center.
During the course of evolution, a disproportionately large number of natural antibiotics have been selected to act upon the ribosome. In the majority of cases, these drugs bind to ribosomes by interacting directly with rRNA (8). Due to the presence of multiple copies of rRNA genes in most species, it is difficult for a sensitive organism to develop resistance by mutating the antibiotic binding site, which is probably one of the main reasons why the ribosome has been repeatedly selected as an antibiotic target.
Conditions created by the extensive and sometimes uncontrolled use of natural and synthetic antibiotics for antimicrobial therapy have promoted the selection and rapid spread of resistant pathogens that exhibit high tolerance to many drugs, including those which are targeted against the ribosome. Although the occurrence of antibiotic resistance mutations in rRNA genes is fairly rare in comparison with other types of resistance, a number of such cases have been reported, especially in those pathogens which contain only one or two copies of rRNA operons in their chromosome (B. Vester and S. Douthwaite, submitted for publication).
The rapidly growing incidence of drug resistance in pathogenic bacteria urges the development of new antibiotics. Several new drugs targeted against the ribosome are currently being developed, including the oxazolidinones (3, 18). After first being identified as prospective antimicrobial agents in 1987 (32), oxazolidinones were abandoned for some time due to their high toxicity. Later on, new derivatives with superior pharmacological properties were found (3, 16), and recently one of the oxazolidinone antibiotics, linezolid (Fig. 1A), has been approved for clinical use. Linezolid shows excellent activity against gram-positive bacteria, including those resistant to other ribosome-targeted drugs (36).
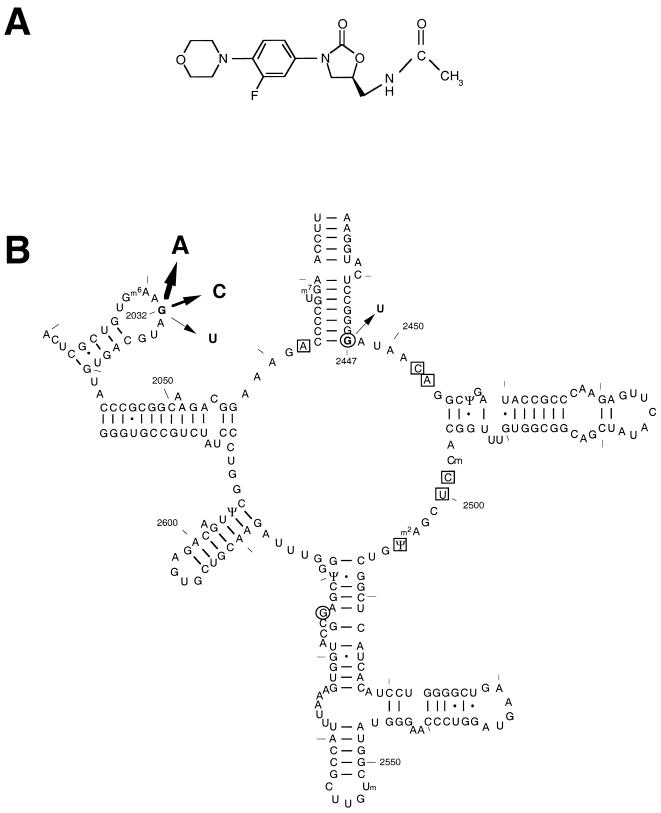
FIG. 1.
(A) Chemical structure of linezolid, an oxazolidinone. (B) Location of linezolid resistance mutations in E. coli 23S rRNA. The secondary structure of the segment of E. coli 23S rRNA encompassing the central loop of domain V and the neighboring regions is shown (17, 25). E. coli linezolid resistance mutations described in this paper are shown by arrows, the thickness of which is proportional to the level of linezolid resistance that each mutation confers. Also marked are the positions of nucleotide substitutions that confer linezolid resistance in H. halobium (boxed) and in S. aureus and E. faecalis (circled) (19; Swaney et al., 38th ICAAC).
Oxazolidinones inhibit protein synthesis in Bacteria both in vivo and in cell-free systems (9, 15, 30). Several hypotheses regarding the mode of oxazolidinone action have been proposed; these include hampering mRNA and/or tRNA binding, inhibition of formation of the first peptide bond, and interference with translocation (5, 9, 14, 15, 21, 30, 33). However, a convincing description of the mechanism of the drug action still remains elusive. Binding studies have revealed interaction of the drug with the large ribosomal subunit (20). Discovery of the mutations G2447U and G2576U (Escherichia coli numeration) in the 23S rRNA of laboratory-generated oxazolidinone-resistant Staphylococcus aureus and Enterococcus faecium strains have underscored the importance of rRNA for drug binding and/or action (S. M. Swaney, D. L. Shinabarger, R. D. Schaadt, J. H. Bock, J. L. Slightom, and G. E. Zurenko, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-104, 1998; G. E. Zurenko, W. M. Todd, B. Hafkin, B. Meyers, C. Kauffman, J. Bock, J. Slightom, and D. Shinabarger, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-848, 1999) (see Fig. 1B). This notion was strongly supported by analysis of oxazolidinone-resistant mutants of the archaeon Halobacterium halobium (19). All of more than 20 analyzed spontaneous Linr H. halobium mutants contained mutations in 23S rRNA. The mutations were clustered within the central loop of domain V, arguably the most functionally important rRNA region in the ribosome. Thus, resistance mutations in gram-positive bacteria and in archaea clearly implicate the central region of domain V of 23S rRNA as a primary component of the oxazolidinone binding site on the ribosome.
The results of these in vivo experiments are, however, in apparent conflict with the recent in vitro studies claiming that oxazolidinones protect A2114 and A2119 in E. coli 23S rRNA, as well as A864 in 16S rRNA, from chemical modification (21). Furthermore, in this study, a photochemically active oxazolidinone derivative was cross-linked to the 23S rRNA in the E. coli ribosome at positions U2113, U2118, and C2153. The seeming discrepancy between these data and those from the mutational analysis could result either from differences in the approaches used or from idiosyncrasies in the drug interaction with E. coli ribosomes.
In order to clarify the location of the drug binding site on the bacterial ribosomes, we characterized oxazolidinone-resistant mutations in E. coli rRNA. Isolation of Linr mutants containing the G2032A mutation in 23S rRNA, and the resistance conferred by other substitutions at position 2032 as well as at position 2447, strongly argued in favor of the central region of domain V being the main site of oxazolidinone interaction with the ribosome.
MATERIALS AND METHODS
Strains, plasmids, materials, and enzymes.
E. coli strains JM109 (35) [endA1 recA1 gyrA96 thi hsdR17(rK−mK+) relA1 supE44 Δ(lac-proAB) (F′ traD36 proAB lacIqZΔM15)] and HN818 (K-12 acrAB::Tn903) (provided by H. Nikaido, University of California, Berkeley) were used for propagation of the wild-type and mutant plasmids and for drug sensitivity testing. The E. coli mutator strain XL1-Red (Stratagene) (endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac mutD5 mutS mutT Tn10) was used for mutagenesis of plasmid pKK3535. Cells were grown at 37°C in Luria-Bertani (LB) medium (29) containing antibiotics when necessary.
Plasmids pKK3535 (4), pNK, and pLK35 (12) (Table 1) are pBR322 derivatives containing the entire rrnB operon of E. coli under the control of P1 and P2 (pKK3535 and pNK) or the lambda PL (pLK35) promoter. The G2447A mutation was introduced into pKK3535 using the Quick-Change site-directed mutagenesis kit (Stratagene) in combination with two oligonucleotide primers, CTCCGGGTATAACAGG and CCTGTTATACCCGGAG (the mutagenizing position is underlined). Mutagenesis at 23S rRNA position 2032 of the pKK3535 plasmid was performed in an M13 bacteriophage construct (11). This construct contained the 2.7-kb SalI/BamHI fragment of pKK3535 (4), which comprises the 3′ end of the rrnB operon, including the 3′ half of 23S rRNA. Two cloning steps were necessary to place the 2032 mutations back into their original context in pKK3535. First, the mutagenized 2.7-kb fragments were inserted into the SalI/BamHI sites of pSK41 (12). The 7.1-kb BglII/SacI fragments were then excised from the new pSK41 constructs and ligated with the 3.9-kb BamHI/SacI fragment of pKK3535. Plasmid pNK is functionally identical to pKK3535 and differs structurally only in that a 903-bp NaeI/BamHI fragment has been removed from the inactivated tetracycline resistance gene of the vector. With the exception of the single base substitutions at position 2032 of the 23S rRNA gene, plasmids pNK2032A, pNK2032C, and pNK2032T are identical to the wild-type pNK.
TABLE 1.
Plasmids used in this study
| Plasmid designation | Relevant characteristics | Source or reference |
|---|---|---|
| pKK3535 | pBR322 carrying a 7,514-bp fragment of E. coli chromosome spanning the entire rrnB operon with its natural promoters | 4 |
| pKK2032A | pKK3535 containing a G2032A mutation in the 23S rRNA gene | 11 |
| pKK2447U | pKK3535 containing a G2447U mutation in the 23S rRNA gene | This work |
| pLK35 | pKK3535 derivative containing the rrnB operon expressed under the control of the lambda PL promoter | 12 |
| pLK2032A | pLK35 containing a G2032A mutation in the 23S rRNA gene | 11 |
| pNK | pKK3535 derivative; has the same 7,514-bp insert of rrnB with its natural promoters; from the pBR322 portion, a 903-bp fragment spanning positions 10961 to 11864 has been deleted | This work |
| pNK2032A | pNK containing a G2032A mutation in the 23S rRNA gene | This work |
| pNK2032U | pNK containing a G2032U mutation in the 23S rRNA gene | This work |
| pNK2032C | pNK containing a G2032C mutation in the 23S rRNA gene | This work |
Linezolid was from Pharmacia Corporation. Restriction enzymes were from MBI Fermentas, and chemicals were from Fisher Scientific. DNA sequencing was performed with the Sequenase kit from U.S. Biochemicals using a set of primers complementary to the E. coli 23S rRNA (22).
Isolation of Linr mutants in JM109 cells.
Plasmid pKK3535, which contains the entire rrnB operon of E. coli (4), was randomly mutagenized by being passed through the E. coli mutator strain XL1-Red (Stratagene), as recommended by the supplier. E. coli strains JM109 and HN818 were transformed with the mutagenized pKK3535 and then plated onto LB agar plates containing 100 μg of ampicillin/ml and either 4 mM linezolid (for strain JM109) or 8 μM linezolid (for strain HN818). After 24 h of incubation, individual colonies were picked and grown overnight in 3 ml of LB medium containing 100 μg of ampicillin/ml. Plasmids were then isolated and used to transform fresh competent cells. Plasmids conferring a Linr phenotype upon retransformation were used for sequence analysis in which the 3′ half of the 23S rRNA, including domains IV, V, and VI, was sequenced from a set of specific primers.
Testing antibiotic sensitivity.
Antibiotic sensitivity was determined in liquid cultures and on plates. The overnight cultures of E. coli HN818 cells transformed with the wild-type or mutant plasmids grown in LB medium containing 100 μg of ampicillin/ml were diluted 500-fold (to a final concentration that gave an A650 of 0.01) into fresh medium containing 100 μg of ampicillin/ml and varying linezolid concentrations and were grown overnight. Optical densities (expressed as A650) of cell cultures were determined and plotted. Antibiotic sensitivity testing on agar plates was performed by diluting exponentially growing cultures and spotting 20-μl drops onto the surfaces of agar plates (27). Plates were photographed after incubation for 12 to 24 h at 37°C.
Quantification of mutant rRNA expression.
E. coli HN818 cells transformed with wild-type or mutant pNK plasmids were grown in liquid cultures in the presence of 50 μg of ampicillin/ml. RNA was extracted from exponentially growing cells or from ribosomes using TRIzol reagent (GIBCO BRL). The ratio of mutant to wild-type 23S rRNA was determined by a primer extension technique (31) using an oligonucleotide primer (TCTTGCCGCGGGTACACTGC) complementary to the 23S rRNA segment 2035 to 2054.
Peptidyl transferase assay.
The effect of linezolid on peptidyl transferase activity was tested under “fragment reaction” conditions (23). The reaction was performed in 50 μl of a buffer containing 20 mM Tris-HCl (pH 8.0), 20 mM MgCl2, 400 mM KCl, 0.4 mM puromycin, 10 pmol of E. coli ribosomes, and 1 pmol (65,000 cpm) of [35S]formyl-methionyl tRNA. Linezolid or carbomycin was added to 1.25 or 0.4 mM, respectively, and the reaction was initiated by adding 25 μl of cold methanol. Tubes were incubated for 30 min on ice, and the reaction was stopped by adding 10 μl of 10 M NaOH. After incubation for 20 min at 37°C, 200 μl of 1 M KH2PO4 was added and the product of the peptidyl transferase reaction, [35S]formyl methionyl puromycin, was extracted with 1 ml of ethyl acetate. A 0.7-ml volume of the ethyl acetate phase was mixed with 8 ml of scintillation cocktail and counted.
RESULTS
Isolation of Linr mutants.
Wild-type E. coli is intrinsically tolerant to oxazolidinone antibiotics due to efflux of the drug (J. M. Buysse, W. F. Demyan, D. S. Dunyak, D. Stapert, J. C. Hamel, and C. W. Ford, Abstr. 36th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-42, 1996). However, we found that, at high concentrations (2 mM), linezolid suppresses growth of the E. coli laboratory strain JM109. Therefore, this strain was used in the initial experiments on the selection of Linr mutations. Because of the presence in E. coli of seven rRNA (rrn) operon copies (26), spontaneous mutations arising in one operon copy usually remain undetected, as they are masked by the excess of wild-type rRNA. Therefore, in order to identify rRNA mutations conferring resistance to linezolid, mutant rRNA genes were introduced into cells on a multicopy plasmid (31). The pKK3535 plasmid containing the entire rrnB operon of E. coli (4) was randomly mutagenized by being passed through an E. coli mutator strain, and the resulting library of mutant plasmids was then transferred to a nonmutator strain, JM109. Linr clones were selected on agar plates containing 4 mM linezolid. Several colonies appeared on the plate after 24 h of incubation. However, only 2 out of 12 clones tested maintained a Linr phenotype after restreaking. The Linr phenotype of these two clones was cotransferable with the plasmid, indicating that the resistance determinant resided in the plasmid. Sequencing of a portion of the plasmid-borne 23S rRNA gene revealed the presence of the G2032A mutation in the 23S rRNA of both Linr clones (Fig. 1B).
While these experiments were under way, E. coli strain HN818 became available to us; in this strain the gene for the AcrAB transporter, involved in drug efflux, has been disrupted. This strain is significantly more sensitive to linezolid than wild-type E. coli; the linezolid MIC for this strain is only 20 μM in liquid culture and 8 μM on agar plates. Selection of Linr mutants was repeated in strain HN818 using the same pKK3535 mutant plasmid library that was used in the original selection experiments with strain JM109. In this case, resistant mutants were selected on agar plates containing 8 μM linezolid. Mutant pKK3535 plasmids conferring the transferable Linr phenotype were isolated from three randomly picked clones. Sequence analyses of the plasmid-borne 23S rRNA gene also showed the presence of the G2032A mutation in all three of these Linr clones. The mutation was absent in several randomly picked Lins clones. The independent occurrence of the same G2032A mutations in all of the Linr clones isolated in two selection experiments, using different E. coli strains, established a clear correlation between the appearance of this mutation and a Linr phenotype.
Testing the effect of the G2032A mutation engineered in the 23S rRNA gene on linezolid resistance.
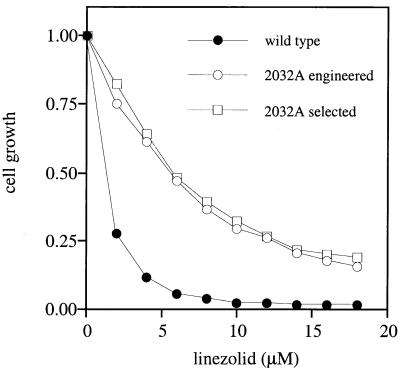
Finding the G2032A mutation in plasmids isolated from all the Linr clones did not exclude the potential presence of other mutations in these plasmids which theoretically could contribute to resistance. Therefore, in order to test whether the single G2032A mutation was sufficient to confer linezolid resistance, this mutation was engineered in the wild-type pKK3535 plasmid by site-directed mutagenesis (11). The resulting plasmid, pKK2032A, was introduced into the Lins strain HN818, and the level of linezolid tolerance was compared with that of cells harboring either the wild-type pKK3535 plasmid or the plasmid from one of the previously selected Linr clones (Fig. 2). The plasmids isolated by selection and by engineering of the G2032A mutation both conferred the same level of linezolid resistance. Thus, the G2032A point mutation is sufficient to render E. coli cells resistant to linezolid.
FIG. 2.
Linezolid sensitivities of E. coli cells transformed with wild-type pKK3535, pKK3535 containing the engineered G2032A mutation (2032A engineered), or a mutant pKK3535 plasmid isolated from one of the selected linezolid-resistant clones (2032 selected). Cell cultures were grown in the presence of 100 μg of ampicillin/ml and varying concentrations of linezolid, as described in Materials and Methods. Optical densities (A650) of cultures were determined after 5 h of growth and normalized relative to the optical densities of cultures grown in the absence of linezolid.
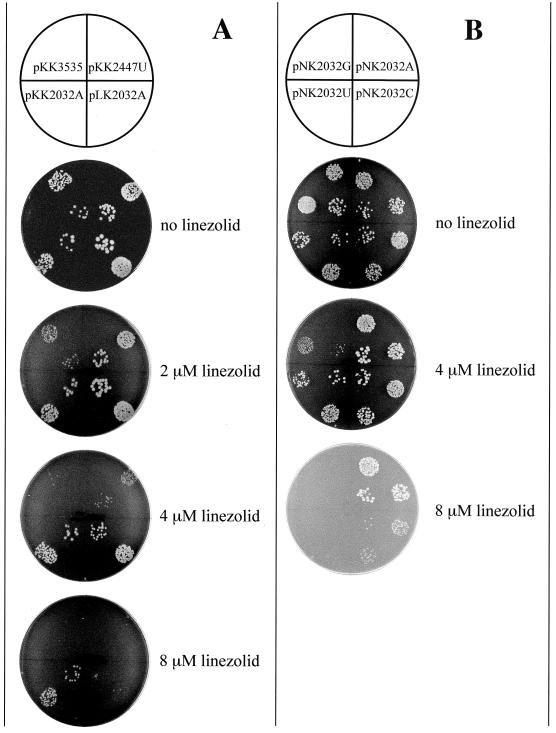
In E. coli cells containing plasmid-borne mutated rRNA genes, mutant ribosomes coexist with chromosome-encoded wild-type ribosomes. Thus the extent of antibiotic resistance is expected to correlate with the fraction of mutant ribosomes present in the cell. To verify that, we compared the linezolid resistance of cells transformed with different plasmids carrying the G2032A mutation in the 23S rRNA gene. In the pKK3535 plasmid, expression of the rRNA operon is controlled by native P1 and P2 promoters. In pLK35, the rRNA operon is transcribed under the control of the lambda PL promoter, which is weaker than the P1–P2 promoter system (12) (Table 1). Accordingly, more than 50% of the ribosomes in pKK3535-transformed cells contain plasmid-encoded rRNA (reference 1 and our unpublished results), whereas mutant ribosomes account for only 25% of the ribosomal population in pLK35-transformed cells (28). As expected, the linezolid resistance of cells expressing mutant (G2032A) 23S rRNA from plasmid pLK35 was diminished compared to that of cells expressing mutant 23S rRNA from plasmid pKK3535 (Fig. 3A). This result established the codominant nature of the G2032A Linr mutation.
FIG. 3.
Spot test analysis of the linezolid resistance of E. coli cells transformed with various plasmid constructs. Aliquots (20 μl) of exponential cultures containing ca. 20 and 200 cells were spotted onto LB agar plates containing 100 μg of ampicillin/ml and varying concentrations of linezolid. Plates were photographed after 24 h of incubation. (A) Cells were transformed with either the wild-type plasmid pKK3535, one of its mutant versions containing G2032A or G2447U (pKK2032A or pKK2447U), or the pLK35 plasmid containing the G2032A mutation (pLK2032A). (B) Cells were transformed with the wild-type plasmid pNK or with one of its mutant versions containing G2032A, G2032U, or G2032C.
Testing other substitutions at position 2032 for Linr.
In order to compare the effects of different mutations at position G2032 on cell sensitivity to linezolid, each of the three possible nucleotide substitutions was engineered at this position in the 23S rRNA gene of the pNK plasmid. Like plasmid pKK3535, plasmid pNK contains the entire wild-type rrnB operon, but pNK is altered in the vector portion to give a more favorable restriction site configuration that facilitates cloning (see Materials and Methods). The level of expression of mutant rRNA from the pNK-derived plasmids is the same as that from pKK3535 (data not shown). The wild-type plasmid (pNK) and its three mutant versions (pNK2032A, pNK2032U, and pNK2032C) were introduced into HN818 cells, and the level of expression of mutant 23S rRNA was verified using the primer extension technique (31). Mutant 23S rRNA accounted for 73, 61, and 74% of the total cellular 23S rRNA in the A2032, U2032, and C2032 mutants, respectively. There was no significant difference in the ratio of wild-type to mutant 23S rRNA in the fractions of individual subunits, 70S ribosomes, or polysomes (data not shown), showing that the mutations at position G2032 did not interfere significantly with the ribosome function. The linezolid resistance of transformants was tested on agar plates (Fig. 3B) and in liquid cultures. Linezolid resistance increased in the order G2032 (wild type)→U2032→C2032→A2032. This result was consistent with MICs determined in liquid cultures: 20 μM (for the wild type), 40 μM (for the U mutant), 60 μM (for the C mutant), and 80 μM (for the A mutant). Since mutant rRNAs are expressed at comparable levels in all the mutants, the observed difference in linezolid resistance caused by A, U, or C substitutions at position G2032 is most probably explained by differential effects of the mutations on drug binding and/or action.
In addition to substitutions at position 2032 of the 23S rRNA gene, another mutation, G2447U, was engineered in plasmid pKK3535. This mutation was previously found in a laboratory-selected Linr strain of S. aureus (Swaney et al., 38th ICAAC). As can be seen in Fig. 3, the resistance of E. coli cells expressing the G2447U mutation was comparable to the low linezolid resistance conferred by the G2032U mutation.
Effect of linezolid on peptidyl transferase reaction catalyzed by the large ribosomal subunit.
Position G2032 is located in the immediate vicinity of the central loop of domain V of 23S rRNA. This region is known to constitute an essential component of the ribosomal peptidyl transferase center. Therefore, we examined the effect of linezolid on the activity of ribosomal peptidyl transferase. The peptidyl transferase assay was performed under “fragment reaction” conditions (in the presence of 33% methanol) (23), using formyl-methionyl-tRNA as a donor substrate and puromycin as an acceptor (34). Even at a very high concentration (1.25 mM), linezolid did not affect the reaction, suggesting that peptidyl transferase activity is not the primary target of this drug (data not shown).
DISCUSSION
In this paper we present evidence that mutations in E. coli 23S rRNA confer resistance to an oxazolidinone antibiotic, linezolid. Our findings provide new evidence that rRNA plays an important role in the binding and/or action of oxazolidinones, and they reinforce our previous conclusion that oxazolidinones interact with the large ribosomal subunit in the vicinity of its peptidyl transferase center.
All five Linr mutants isolated in two independent experiments contained the same G2032A mutation. Though this result suggests that G2032A is the “best” Linr mutation, it does not exclude a possibility that other mutations can also confer linezolid resistance; indeed, two other substitutions engineered at position 2032, as well as the G2447U mutation, rendered cells resistant to low concentrations of the drug. All of the previously characterized Linr mutations found in the rRNA genes of S. aureus, Enterococcus faecalis, and H. halobium were localized within or in close proximity to the central loop of domain V of 23S rRNA, indicating that this rRNA region is important for drug binding (19; Swaney et al. 38th ICAAC). Cross-linking of G2032 to A2054 and C2055 (10) shows that in the ribosome tertiary structure the 2032 loop is in an immediate proximity to the central loop. Thus, the RNA structure encompassing this hairpin, the central loop of domain V, and several adjacent RNA elements emerge as the primary binding site of oxazolidinone antibiotics. Although we cannot completely exclude the possibility that mutations in the central region of domain V induce an allosteric conformation change that affects drug binding to a distant rRNA site, this scenario seems unlikely, given a broad spectrum of known Linr mutations clustered in or near the central loop of domain V (see Fig. 1B). In addition, oxazolidinones can compete for binding to the ribosome with chloramphenicol and lincomycin, which are known to interact with the central loop of domain V (20). The mutations at position 2032, which were shown in this paper to confer linezolid resistance, render ribosomes resistant to lincosamides and chloramphenicol as well (6, 11). Thus, our mutational data provide a structural basis for the previous suggestion that the binding sites of chloramphenicol and lincomycin overlap with the binding site of linezolid (20).
Analysis of oxazolidinone resistance mutations in E. coli (this paper) and other organisms (19; Swaney et al., 38th ICAAC) clearly points to the central loop of domain V and several adjacent hairpins as the primary binding site of interaction of oxazolidinones with the ribosome. This conclusion, however, seems to contradict the results of in vitro experiments in which oxazolidinones were footprinted and cross-linked to the E. coli 23S rRNA segment 2110 to 2155 (21), since in the ribosome tertiary structure, the 2110-to-2155 region appears to be too far from the central region of domain V to allow simultaneous interaction of an oxazolidinone molecule with these two rRNA regions (2, 24). The incongruity between the mutational and cross-linking data may potentially be explained by structural differences between linezolid and the compounds used by Matassova et al. (the latter lacked the fluorine atom, while the morpholine moiety was replaced by pyridyl or azido-benzyl groups). Alternatively, the discrepancy between genetic and biochemical data may result from the intrinsic difference between the in vivo and in vitro approaches. This consideration is aggravated by the fact that oxazolidinones are notorious for their high nonspecific binding to the ribosome in vitro (20). Indeed, we found that under artificial in vitro conditions, an azido derivative of linezolid cross-links nonspecifically to rRNA and ribosomal proteins, while performance of cross-linking under more natural conditions limits drug cross-linking exclusively to the central loop of domain V (unpublished data). Therefore, we favor the view that the primary site of oxazolidinone action is located in the vicinity of the central loop of domain V of 23S rRNA.
The central region of domain V of 23S rRNA represents one of the most conserved rRNA segments (13). Therefore, it is not surprising that the oxazolidinone-binding site is apparently conserved between the evolutionarily distant domains of Archaea and Bacteria. This is illustrated by the clustering of oxazolidinone resistance mutations in the vicinity of the central loop of domain V both in bacteria (S. aureus, E. faecalis, E. coli) and in an archaeon (H. halobium) (19; Swaney et al., 38th ICAAC). Nevertheless, the spectrum of resistance mutations is clearly organism specific. One conceivable reason for this specificity is that nucleotide changes that are permissible in one organism may be deleterious in others. It is also possible, however, that the precise orientation of the drug on the ribosomes of different species may vary to some extent.
The mode of action of oxazolidinones remains largely unknown. The central loop of domain V and the neighboring rRNA segments constitute an integral part of the ribosomal peptidyl transferase center. A number of drugs which interact with the central region of 23S rRNA inhibit catalysis of peptide bond formation (7). Among them are chloramphenicol and lincomycin, whose binding sites overlap with that of oxazolidinones (20). In spite of that, neither linezolid nor several of the other tested oxazolidinone derivatives inhibited the peptidyl transferase activity of the E. coli ribosome. Similarly, no inhibition of peptidyl transferase activity by oxazolidinones was observed in several other systems (15, 19, 30). Therefore, activity of the tested oxazolidinones appears to be targeted against a ribosomal function other than catalysis of peptide bond formation but probably indirectly related to the activity of ribosomal peptidyl transferase.
ACKNOWLEDGMENTS
This work was supported by research grants from the National Institutes of Health (GM53762) and Pharmacia Corporation to A.S.M. and by grants from the Danish Natural Sciences Research Council and Danish Medical Research Council to S.D.
We thank H. Nikaido for making strain HN818 available to us.
REFERENCES
- 1.Aagaard C, Rosendahl G, Dam M, Powers T, Douthwaite S. Specific structural probing of plasmid-coded ribosomal RNAs from Escherichia coli. Biochimie. 1991;73:1439–1444. doi: 10.1016/0300-9084(91)90176-2. [DOI] [PubMed] [Google Scholar]
- 2.Ban N, Nissen P, Hansen J, Capel M, Moore P B, Steitz T A. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 3.Brickner S J, Hutchinson D K, Barbachyn M R, Manninen P R, Ulanowicz D A, Garmon S A, Grega K C, Hendges S K, Toops D S, Ford C W, Zurenko G E. Synthesis and antibacterial activity of U-100592 and U-100766, two oxazolidinone antibacterial agents for the potential treatment of multidrug-resistant gram-positive bacterial infections. J Med Chem. 1996;39:673–679. doi: 10.1021/jm9509556. [DOI] [PubMed] [Google Scholar]
- 4.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 5.Burghardt H, Schimz K L, Müller M. On the target of a novel class of antibiotics, oxazolidinones, active against multidrug-resistant Gram-positive bacteria. FEBS Lett. 1998;425:40–44. doi: 10.1016/s0014-5793(98)00194-x. [DOI] [PubMed] [Google Scholar]
- 6.Cseplö A, Etzold T, Schell J, Schreier P H. Point mutations in the 23S rRNA genes of four lincomycin resistant Nicotiana plumbaginifolia mutants could provide new selectable markers for chloroplast transformation. Mol Gen Genet. 1988;214:295–299. doi: 10.1007/BF00337724. [DOI] [PubMed] [Google Scholar]
- 7.Cundliffe E. Antibiotic inhibitors of ribosome function. In: Gale E F, Cundliffe E, Reynolds P E, Richmond M H, Waring M J, editors. The molecular basis of antibiotic action. London, United Kingdom: John Wiley & Sons; 1981. pp. 402–545. [Google Scholar]
- 8.Cundliffe E. Recognition sites for antibiotics within rRNA. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 479–490. [Google Scholar]
- 9.Daly J S, Eliopoulos G M, Reiszner E, Moellering R C., Jr Activity and mechanism of action of DuP 105 and DuP 721, new oxazolidinone compounds. J Antimicrob Chemother. 1988;21:721–730. doi: 10.1093/jac/21.6.721. [DOI] [PubMed] [Google Scholar]
- 10.Doring T, Greuer B, Brimacombe R. The three-dimensional folding of ribosomal RNA; localization of a series of intra-RNA cross-links in 23S RNA induced by treatment of Escherichia coli 50S ribosomal subunits with bis-(2-chloroethyl)-methylamine. Nucleic Acids Res. 1991;19:3517–3524. doi: 10.1093/nar/19.13.3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douthwaite S. Functional interactions within 23S rRNA involving the peptidyl transferase center. J Bacteriol. 1992;174:1333–1338. doi: 10.1128/jb.174.4.1333-1338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douthwaite S, Powers T, Lee J Y, Noller H F. Defining the structural requirements for a helix in 23S ribosomal RNA that confers erythromycin resistance. J Mol Biol. 1989;209:655–665. doi: 10.1016/0022-2836(89)93000-3. [DOI] [PubMed] [Google Scholar]
- 13.Egebjerg J, Larsen N, Garrett R A. Structural map of 23S rRNA. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The ribosome: structure, function, and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 168–179. [Google Scholar]
- 14.Eustice D C, Feldman P A, Slee A M. The mechanism of action of DuP 721, a new antibacterial agent: effects on macromolecular synthesis. Biochem Biophys Res Commun. 1988;150:965–971. doi: 10.1016/0006-291x(88)90723-1. [DOI] [PubMed] [Google Scholar]
- 15.Eustice D C, Feldman P A, Zajac I, Slee A M. Mechanism of action of DuP 721: inhibition of an early event during initiation of protein synthesis. Antimicrob Agents Chemother. 1988;32:1218–1222. doi: 10.1128/aac.32.8.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ford C W, Hamel J C, Wilson D M, Moerman J K, Stapert D, Yancey R J, Jr, Hutchinson D K, Barbachyn M R, Brickner S J. In vivo activities of U-100592 and U-100766, novel oxazolidinone antimicrobial agents, against experimental bacterial infections. Antimicrob Agents Chemother. 1996;40:1508–1513. doi: 10.1128/aac.40.6.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutell R R, Larsen N, Woese C R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones R N, Barrett M S. Antimicrobial activity of SCH 27899, oligosaccharide member of the everninomicin class with a wide gram-positive spectrum. J Clin Microb Infect. 1995;1:35–43. doi: 10.1111/j.1469-0691.1995.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 19.Kloss P, Xiong L, Shinabarger D L, Mankin A S. Resistance mutations in 23S rRNA identify the site of action of the protein synthesis inhibitor linezolid in the ribosomal peptidyl transferase center. J Mol Biol. 1999;294:93–101. doi: 10.1006/jmbi.1999.3247. [DOI] [PubMed] [Google Scholar]
- 20.Lin A H, Murray R W, Vidmar T J, Marotti K R. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob Agents Chemother. 1997;41:2127–2131. doi: 10.1128/aac.41.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matassova N B, Rodnina M V, Endermann R, Kroll H P, Pleiss U, Wild H, Wintermeyer W. Ribosomal RNA is the target for oxazolidinones, a novel class of translational inhibitors. RNA. 1999;5:939–946. doi: 10.1017/s1355838299990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moazed D, Noller H F. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci USA. 1991;88:3725–3728. doi: 10.1073/pnas.88.9.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Monro R E, Marcker K A. Ribosome-catalysed reaction of puromycin with a formylmethionine-containing oligonucleotide. J Mol Biol. 1967;25:347–350. doi: 10.1016/0022-2836(67)90146-5. [DOI] [PubMed] [Google Scholar]
- 24.Mueller F, Sommer I, Baranov P, Matadeen R, Stoldt M, Wohnert J, Gorlach M, Van Heel M, Brimacombe R. The 3D arrangement of the 23S and 5S rRNA in the Escherichia coli 50S ribosomal subunit based on a cryo-electron microscopic reconstruction at 7.5 Å resolution. J Mol Biol. 2000;298:35–59. doi: 10.1006/jmbi.2000.3635. [DOI] [PubMed] [Google Scholar]
- 25.Noller H F, Kop J, Wheaton V, Brosius J, Gutell R R, Kopylov A M, Dohme F, Herr W, Stahl D A, Gupta R, Woese C R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981;9:6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nomura M, Post L E. Organization of ribosomal genes and regulation of their expression in Escherichia coli. In: Chambliss G, Craven G R, Davies J, Davis K, Kahan L, Nomura M, editors. Ribosomes: structure, function, and genetics. Baltimore, Md: University Park Press; 1980. pp. 671–691. [Google Scholar]
- 27.Powers T, Noller H F. Dominant lethal mutations in a conserved loop in 16S rRNA. Proc Natl Acad Sci USA. 1990;87:1042–1046. doi: 10.1073/pnas.87.3.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosendahl G, Hansen L H, Douthwaite S. Pseudoknot in domain II of 23S rRNA is essential for ribosome function. J Mol Biol. 1995;249:59–68. doi: 10.1006/jmbi.1995.0280. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Shinabarger D L, Marotti K R, Murray R W, Lin A H, Melchior E P, Swaney S M, Dunyak D S, Demyan W F, Buysse J M. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob Agents Chemother. 1997;41:2132–2136. doi: 10.1128/aac.41.10.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sigmund C D, Ettayebi M, Borden A, Morgan E A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 32.Slee A M, Wuonola M A, McRipley R J, Zajac I, Zawada M J, Bartholomew P T, Gregory W A, Forbes M. Oxazolidinones, a new class of synthetic antibacterial agents: in vitro and in vivo activities of DuP 105 and DuP 721. Antimicrob Agents Chemother. 1987;31:1791–1797. doi: 10.1128/aac.31.11.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swaney S M, Aoki H, Ganoza M C, Shinabarger D L. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother. 1998;42:3251–3255. doi: 10.1128/aac.42.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan G T, DeBlasio A, Mankin A S. Mutations in the peptidyl transferase center of 23S rRNA reveal the site of action of sparsomycin, a universal inhibitor of translation. J Mol Biol. 1996;261:222–230. doi: 10.1006/jmbi.1996.0454. [DOI] [PubMed] [Google Scholar]
- 35.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 36.Zurenko G E, Yagi B H, Schaadt R D, Allison J W, Kilburn J O, Glickman S E, Hutchinson D K, Barbachyn M R, Brickner S J. In vitro activities of U-100592 and U-100766, novel oxazolidinone antibacterial agents. Antimicrob Agents Chemother. 1996;40:839–845. doi: 10.1128/aac.40.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]