Abstract

This paper delves into the polymorphism of 2-[3-(trifluoromethyl)anilino]benzoic acid, commonly referred to as flufenamic acid (FA), a pharmaceutical agent employed in treating inflammatory conditions. The central focus of the study is on a newly unearthed solvatomorphic structure of FA in methanol (FAM), and a thorough comparison is conducted with the commercially available standard structure. Employing a comprehensive approach, including X-ray crystallography, Hirshfeld surface analysis, density functional theory (DFT), molecular docking, and molecular dynamics (MD) simulations, the research aims to unravel the structural and functional implications of solvatomorphism. The X-ray crystal structure analysis brings to light notable differences between the standard FA and solvatomorphic FAM, showcasing variations in intermolecular interactions and crystal packing. Key features such as hydrogen bonding, π···π stacking, and C–H···π interactions are identified as influential factors shaping the stability and conformation of the compounds. Hirshfeld surface analysis further quantifies the nature and contribution of intermolecular interactions, providing a comprehensive perspective on molecular stability. Density functional theory offers valuable electronic structure insights, highlighting disparities in frontier molecular orbitals between FA and FAM. Molecular docking studies against prostaglandin D2 11-ketoreductase explore potential drug interactions, unveiling distinct binding modes and hydrogen bonding patterns that shed light on how the solvatomorphic structure may impact drug–target interactions. In-depth molecular dynamics simulations over 100 ns investigate the stability of the protein–ligand complex, with root mean square deviation and root mean square fluctuation analyses revealing minimal deviations and affirming the stability of FAM within the active site of the target protein.
Introduction
2-[3-(Trifluoromethyl)anilino]benzoic acid or flufenamic acid, an active pharmaceutical ingredient used to treat conditions like rheumatoid arthritis, osteoarthritis, and various inflammatory ailments, is indeed a standout example in the world of polymorphism. There have been reports of nine different forms of this compound.1−12 The initial crystal structure, later referred to as form III, was documented in 1973,13 and almost a decade later, form I was identified.14 Interestingly, these two forms are enantiotropically related. The extensive polymorphism exhibited by flufenamic acid has led to the exploration of various strategies to control its polymorphic behavior, including the utilization of soluble additives and the induction of heteronucleation through polymers.15
The duality of polymorphism in solid-state materials is evident. The influence of crystal structure on physicochemical properties can give rise to novel and potentially beneficial functions in metastable polymorphs.16−18 On the flip side, the unforeseen discovery of a new crystal form for a commercial compound can incur substantial costs.19,20 These aspects carry notable importance in the pharmaceutical sector, where the efficacy and regulatory adherence of active pharmaceutical ingredients (APIs) frequently hinge on a particular polymorphic form.21,22 Hence, the identification of novel polymorphic forms, understanding the factors influencing their occurrence, and characterizing polymorphic phase transitions are crucial endeavors.
Polymorphism holds crucial significance in industries such as pharmaceuticals, pigments, and electronics, given its inherent presence in discovery and development processes. Nearly all substances possess the capacity to adopt two or more solid phases, allowing for the manipulation of a compound’s physical and chemical characteristics through various polymorphic forms. Jean-Paul Garnier, CEO of GlaxoSmithKline, underscores the relevance of polymorphism by noting that approximately 50% of drug candidates entering clinical trials face efficacy and safety concerns, while an additional 40% encounter challenges related to patents, solubility, and drug interactions, emphasizing its impact on the success of pharmaceutical endeavors.23,24 This statement emphasizes the essential requirement to regulate and manipulate the desired physicochemical properties throughout the drug development process. Structural conformations or polymorphism can either diminish or improve the properties of drug candidates, making them pivotal considerations in pharmaceutical research and development.
In the realm of scientific research, in silico methods have become indispensable, offering a diverse array of computational tools applicable across scientific domains spanning drug discovery, materials science, and beyond. These methods harness the capabilities of algorithms and simulations to scrutinize and predict molecular interactions, thereby providing valuable insights into the behavior of molecules under varied conditions.25,26 Of particular significance in recent times is the burgeoning interest in computer-aided drug discovery (CADD) approaches, driven by their potential to address the scale, time, and cost challenges inherent in conventional experimental methodologies. CADD encompasses a series of computational steps, including the identification of potential drug targets, the virtual screening of extensive chemical libraries for promising drug candidates, the subsequent optimization of candidate compounds, and in silico assessments of their potential toxicity. Following these computational processes, selected candidate compounds undergo confirmation through in vitro/in vivo experiments. This strategy reduces the number of chemical compounds requiring experimental evaluation, concurrently increasing the success rate by eliminating inefficient and toxic candidates from consideration.27 CADD has proven successful in bringing novel drug compounds to market across diverse diseases, exemplified by its application in developing drugs inhibiting human immunodeficiency virus (HIV)-1 (e.g., atazanavir, saquinavir, indinavir, and ritonavir), anticancer drugs (e.g., raltitrexed), and antibiotics (e.g., norfloxacin).28−31 The integration of machine learning techniques with CADD approaches has further enhanced their accuracy and efficiency.32 This evolution in drug discovery is underscored by the advent of advanced computational methodologies such as molecular modeling, cheminformatics, bioinformatics, artificial intelligence, and machine learning. These theoretical disciplines have become integral components of modern approaches to drug discovery, collectively contributing to a more comprehensive and efficient exploration of potential therapeutic compounds.33
In this context, here, we compare the newly discovered solvatomorphic structure of flufenamic acid with the commercially available standard structure to investigate drug interactions through in silico studies. The study includes detailed structural analysis, Hirshfeld and NCI validations, and molecular docking and dynamic simulation studies of the new solvatomorphic structure of flufenamic acid in comparison with the standard structure. This comprehensive study provides valuable insights into the polymorphism of flufenamic acid and its solvatomorphic form. The findings contribute to a better understanding of how solvatomorphism influences the physicochemical properties of pharmaceutical compounds, with potential implications for drug development and manufacturing processes, particularly in the treatment of inflammatory conditions.
Materials and Methods
The compound 2-[3-(trifluoromethyl)anilino]benzoic acid was purchased from Sigma-Aldrich and further used for crystal growth. The compound was dissolved with ethanol and methanol separately and kept for crystal growth under slow evaporation.
Single-Crystal X-ray Structure Determination
The X-ray crystal structure analysis methodology described involves several key steps, including data collection, data processing, structure determination, refinement, and validation. Here’s an elaborate explanation of each step:
Data collection: Single crystal selection: Defect-free single crystals of the material (FAM crystals) grown in both ethanol and methanol solvents are selected for analysis.
Polarizing microscope inspection: The selected crystals are inspected under a polarizing microscope to ensure that they are of high quality and suitable for X-ray data collection.
X-ray data collection: Instrumentation: X-ray intensity data are collected using a Rigaku XtaLAB Mini X-ray diffractometer.
Experimental setup: The experiment is conducted at a temperature of 293 K. Goniometer settings: The goniometer settings include fixing χ at 54° and φ ranging from 0 to 360° with a scan width of 0.5°. Exposure parameters: The exposure time is set to 3 s, and the sample-to-detector distance is 50.0 mm. X-ray source: The X-ray diffractometer operates at 50 kV and 12 mA with MoKα radiation of wavelength λ = 0.71073 Å.
Data processing: Software: The collected X-ray intensity data are processed using Crystal-clear-SM Expert-2 software.34,35 Direct methods: Direct approaches are adopted to determine the initial structure results.
Structure determination: Software tools: SHELXS and SHELXL36,37 software programs are used for structure determination. OLEX2: These programs are implemented in the OLEX2 software suite.38 Full-Matrix Least-Squares vs F2: Full-matrix least-squares refinement against F2 is employed to correct for the initial atomic coordinates.
Refinement: Software tools: Refinement of the crystal structure is carried out by using SHELXL software. Full-Matrix Least-Squares Refinement: The refinement process involves optimizing the atomic coordinates and other parameters to fit the experimental X-ray diffraction data.
Geometric restraints: PLATON39 is used for crystal geometry exploration, and restraints are applied to maintain realistic bond lengths and angles.
Software tools: PLATON and MERCURY 4.2.040 are used for crystal geometry validation and visualizing the molecular structure and analyzing the crystal packing.
The X-ray crystal structure analysis methodology involves selecting high-quality single crystals, conducting X-ray data collection under controlled conditions, processing the data using specialized software, determining the initial structure through direct methods, refining the structure through least-squares refinement, and validating the results using crystallographic tools.
Hirshfeld Surface Analysis
Hirshfeld surface (HS) analysis, along with accompanying two-dimensional fingerprint plots, offers qualitative insights into diverse intermolecular interactions. These analyses were performed for FA and FAM molecules utilizing CrystalExplorer software,41 based on the CIF data. The HS analysis involves mapping with a normalized contact distance (dnorm), which is defined in terms of the nearest internal (di) and external distance (de). This analytical approach permits the visualization and quantification of the complex’s intermolecular interactions, represented through a spectrum of colors ranging from red to blue.42
Density Functional Theory
Utilizing the Gaussian 09 program,43 the optimization of CPPQC’s structural geometry and subsequent energy calculations were conducted through density functional theory (DFT), employing the 6-311++G(d,p) basis set. The exchange component of the DFT computations employed a three-parameter hybrid function combined with the Lee–Yang–Parr (LYP) correlation function.44 Following geometry optimization, diverse properties were assessed. The Khon–Sham frontier molecular orbitals (FAOs) were computed, and the optimized geometry was used to determine the reduced density gradient (RDG). For evaluating the energy gap of the frontier molecular orbitals and associated global reactive parameters, Koopman’s approximation was employed,45 and Koopmans’ theorem proves beneficial as it obviates the need to compute the distinct energies of the original molecule and its ion separately for determining the ionization energy and electron affinity. Visualization of the frontier molecular orbitals (FAOs) was achieved using GaussView 6.0.46 Analysis of the reduced density gradient (RDG) was executed utilizing Multiwfn 3.847 software, and the outcomes were visualized with Visual Molecular Dynamics (VMD) software.48
DFT is employed to investigate the electronic structure and energetics of the molecules. It is a quantum mechanical approach that provides accurate predictions of molecular properties, such as HOMO–LUMO energies, molecular orbitals, and global reactivity parameters. In the context of this study, DFT is crucial for understanding how the choice of solvent (ethanol and methanol) affects the electronic structure of flufenamic acid and its solvatomorphic form (FAM). DFT calculations allow researchers to predict the stability of different molecular conformations and analyze the impact of solvents on the molecular structure, including changes in bond distances, angles, and intermolecular interactions.
Molecular Docking Studies
The docking process plays a crucial role in examining the interactions between an inhibitor and its target molecule. To investigate the binding strength of a solvatomorphic structure found in methanol solvent with the prostaglandin D2 11-ketoreductase protein, we utilized MGL tools 1.5.649,50 and AutoDock Vina.51 We began by obtaining the three- dimensional X-ray crystallographic structure of the target protein (PDB ID: 1S2C) from the Protein Data Bank (www.rcsb.org). Subsequently, we extracted the ligands bound to the protein using Biovia Discovery Studio Visualizer.52 Further, we conducted molecular docking studies, during which we placed a novel quinoline derivative into the active site of the 1S2C protein using AutoDock tools. This process yielded a negative binding affinity value expressed in kilocalories per mole for the docked complex. Following this, we visualized the resulting protein–ligand complex and analyzed the interactions between the ligand and the binding sites using Biovia Discovery Studio Visualizer.53
Molecular docking is used to predict the binding modes and interactions between small molecules (ligands) and target proteins. It helps identify potential binding sites, understand the nature of interactions, and assess binding affinity. In this study, molecular docking is applied to investigate how flufenamic acid and its solvatomorphic form interact with prostaglandin D2 11-ketoreductase (AKR1C3), a relevant target in the context of anti-inflammatory drugs.
Molecular Dynamic (MD) Simulation
Molecular dynamic (MD) simulation studies were conducted to assess the stability of the protein–ligand complex interaction, utilizing the academic version of Desmond modules within the Schrodinger 2020-2 suite.53 The MD simulation, guided by the more favorable docking results, began by immersing the complex into a cubic box filled with TIP3P water molecules to ensure proper solvation. Complex preparation and evaluation were carried out using the OPLS3 force field. To achieve a neutral system, a sophisticated algorithm was initially employed followed by the Marlyna–Tobias–Klein method. Subsequently, the relaxed system underwent a 100 ns simulation, maintaining a constant pressure of 1 bar and employing a Nose–Hoover thermostat set to 300 K under NPT ensembles. Stability assessment was conducted through the examination of root mean square deviation (RMSD), root mean square fluctuations (RMSF), and the hydrogen bond fingerprint profile, providing insights into the potential energy stability of the protein–ligand complex.
They provide information about the stability, flexibility, and conformational changes in molecular complexes. In this study, molecular dynamics simulations are used to explore the stability of the protein–ligand complex formed between AKR1C3 and flufenamic acid (FA) or its solvatomorphic form (FAM) over a 100 ns simulation period. RMSD and RMSF analyses help assess the stability of the ligands within the binding pocket of the protein, providing valuable insights into the long-term behavior of the molecular system.
Results and Discussion
Crystal Growth
The compound was separately dissolved in ethanol and methanol, and slow evaporation was employed to facilitate crystal growth. The X-ray structural analysis revealed that the ethanol solution produced the typical structure of flufenamic acid (FA), whereas the methanol solution yielded a novel solvatomorphic structure of flufenamic acid (FAM), resulting in the formation of rod-shaped white crystals within a week.
X-ray Crystal Structure Description
The spatial arrangement of atoms in the crystal lattice was determined by using single-crystal X-ray diffraction. FA and FAM structures are crystallized in a monoclinic crystal system with different space group C2/c and P21/n, respectively, comprising octamers and tetramers in each unit cell (Z = 8 and Z = 4). The ORTEP representation of FA and FAM drawn at 50% probability is depicted in Figure 1, which gives insights about the electron density distribution around each atoms. The structural refinement parameters are listed in Table 1.
Figure 1.
ORTEP of the FA (a) and FAM (b) molecules with thermal ellipsoids drawn at 50% probability.
Table 1. Crystal Data and Structure Refinement Parameters of FA and FAM Compounds.
| parameter | FA | FAM |
|---|---|---|
| empirical formula | C14H10F3NO2 | C15H14F3NO3 |
| formula weight | 281.23 | 313.28 |
| crystal system | monoclinic | monoclinic |
| space group | C2/c | P21/n |
| temperature | 293 K | 293 K |
| wavelength | 0.71073 Å | 0.71075 Å |
| a | 40.04(4) Å | 19.329(16) Å |
| b | 5.17(4) Å | 10.302(9) Å |
| c | 12.35(2) Å | 19.470(16) Å |
| β | 92.29° | 157.342° |
| volume | 2555(20) Å3 | 1494(2) Å3 |
| Z | 8 | 4 |
| density | 1.462 mg m–3 | 1.393 mg m–3 |
| μ | 0.127 mm–1 | 0.121 mm–1 |
| F000 | 1152 | 648 |
| final [I > 2σ(I)] | R1 = 0.0864, wR2 = 0.1533 | R1 = 0.0935, wR2 = 0.2800 |
| R indices (all data) | R1 = 0.2051, wR2 = 0.1989 | R1 = 0.1334, wR2 = 0.3197 |
| reflections collected | 6845 | 8199 |
| independent reflections | 2900 | 3370 |
| index ranges | –39 ≤ h ≤ 51 | –25 ≤ h ≤ 4 |
| –6 ≤ k ≤ 6 | 0 ≤ k ≤ 13 | |
| –15 ≤ l ≤ 15 | –4 ≤ l ≤ 25 | |
| θ range for data collection | 3.06 to 27.48° | 3.21 to 27.47° |
| largest diff. peak and hole | 0.233 and −0.226 e Å–3 | 0.876 and −0.762 e Å–3 |
The FA compound exhibits significant nonplanar characteristics, with both benzene rings lying in entirely distinct planes, resulting in an interplanar angle of 43.67°. Conversely, when the molecule includes methanol as a solvent (FAM), the inter planar angle increases to 49.25°. This elevation of interplanar angle by 5.58° in the FAM molecule can be attributed to the presence of methanol solvent (MeOH). Additionally, in the FA molecule, the position of the O2 atom in benzoic acid is nearly coplanar with the surrounding atoms, while in the FAM molecule, it deviates by approximately 0.235 Å from this coplanar arrangement.
An association between FA and MeOH in the FAM molecule is established via a hydrogen bond interaction C15–H15C···O2 measuring a hydrogen-acceptor distance of 1.832 Å with a dihedral angle of 172.4° between them. Both the compounds showcase a variety of interactions including intramolecular, intermolecular, π···π stackings, and C–H···π interactions.
The crystal structure of FA demonstrates the existence of three intermolecular hydrogen bond interactions, which results in two five-membered S(5) and one six-membered S(6) planar ring formation (Figure 2a), whereas the FAM compound displays two intermolecular hydrogen bonds, which lead to the formation of S(5) and S(6) planar rings (Figure 2b).
Figure 2.
Various intramolecular interactions lead to the formation of S(5) and S(6) synthons in FA (a) and FAM (b).
As a consequence of classical O2–H2···O1 hydrogen bond (symmetry code: 3/2 – x, 1/2 – y, −z) interactions between adjacent FA molecules connect head-to-head leads to the establishment of supramolecular R22(8) ring synthon and these pair of molecules interconnected by C14···C11 short contacts, which indeed results in the formation 1D molecular linear chains along the b-axis (Figure 3). Additional hydrogen bond interactions were observed between neighboring molecules, resulting in the emergence of R33(14) ring motif (Figure 4). Further, detailed structure analysis showcased that the existence of π···π stacking interactions occur specifically between the centroids of trifluorotoulene (Cg1) and benzoic acid (Cg2) moieties. Remarkably, the intriguing aspect is that the distance between the centroids of trifluorotoulene in adjacent molecules matches precisely with the distance between the centroids of benzoic acid (Cg1···Cg1 = Cg2···Cg2 = 5.170 Å).
Figure 3.
R22(8) ring motif bridges molecular chains in FA.
Figure 4.
R22(8) and R33(14) supramolecular synthon along the b-axis plane established in FA molecules.
In addition to the aforementioned interactions, the molecule exhibits a dual set of interactions involving lone pairs, specifically C–H···π and C–O···π interactions with the centroids of trifluorotoluene and benzoic acid within the FA molecule, respectively. These intriguing interactions are defined by a C9–H9···Cg1 distance of 2.873 Å (symmetry code: x, −1 + y, z) and a C14–O1···Cg2 distance of 3.78 Å (symmetry code: x, 1 + y, z), respectively. It is noteworthy that such interactions with lone pairs are absent in the FAM molecule. Moreover, the compound’s crystal growth along the crystallographic b-axis is significantly impacted by the existence of π···π stacking interactions, as depicted in Figure 5b. These stacking interactions occur between the centroids of trifluorotoluene and the benzoic acid moieties. The combined effect of these π···π stacking interactions, along with C9–H9···Cg1 and C14–O1···Cg2 interactions, actively contributes to the development of 1D linear molecular arrays along the crystallographic b-axis, as illustrated in Figure 5a,b. Importantly, these molecular arrays play a crucial role in enhancing the structural stability of the compound.
Figure 5.
C–H···π, C–O···π (a), and π···π stacking interactions (b) between FA molecules resulting in the crystal growth along the crystallographic b-axis.
Structural observation of the FAM molecule unveiled the engagement of the methanol molecule in five hydrogen bond interactions: C15–H15c···O2, C15–H15a···O1, C15–H15b···O2, O3–H3a···O1, and O3···H2–C2 (D-A (Å): 2.787, 2.795, 2.787, 3.268 and A-D (Å): 2.678, respectively) as a donor in four interactions and as an acceptor in the remaining one interaction, respectively (Figure 6a). The neighboring molecules are linked through these interactions of methanol solvents via nonclassical hydrogen bonds leading to the formation of dimeric supramolecular architecture of R44(12). Such dimers are further linked through the R24(9) ring motif, which is a result of O3–H3a···O1, C15–H15b···O2, and C15–H15c···O2 hydrogen bond interactions, which build the molecule along the crystallographic b-axis (Figure 6b).
Figure 6.
Interaction environment of a methanol molecule (a) and R44(12) and R24(9) supramolecular architecture formation (b) in the crystal structure of FAM.
Further analysis of the FAM molecule showed a variety of π···π stacking interactions. These are facilitated by the involvement of a methanol solvent, which acts as a mediator between adjacent molecules. These interactions are particularly evident in the arrangement of centroids, where distinct centroid distances between Cg1 and Cg2 have been observed (Cg1 is the centroid of trifluorotoulene, and Cg2 is the centroid of benzoic acid). These variations in centroid distances, as outlined in Table 2 and visually represented in the corresponding Figure 7, can be attributed to the presence of methanol solvent.
Table 2. Potential Hydrogen Bonds and Short Ring Interactionsa.
|
Potential hydrogen bonds | |||||
|---|---|---|---|---|---|
| donor-H···acceptor | D-H (Å) | H···A (Å) | D···A (Å) | D-H···A (°) | symmetry |
| FA | |||||
| N1—H1···O1 | 0.86 | 2.14 | 2.70(2) | 122 | |
| O2—H2···O1 | 0.82 | 1.85 | 2.66(2) | 174 | 3/2 – x, 1/2 – y, −z |
| C12–H12···O2 | 0.93 | 2.38 | 2.73(2) | 102 | |
| FAM | |||||
| N1–H1···O1 | 0.861(10) | 2.079(14) | 2.734(13) | 132.4(10) | |
| O3–H15A···O1 | 0.82(6) | 2.48(8) | 3.268(6) | 160(18) | 3/2 – x, −1/2 + y, 1/2 – z |
| C15–H15A···O1 | 0.959(15) | 1.928(16) | 2.795(5) | 149.2(19) | 1 – x, 1 – y, −z |
| C15–H15B···O2 | 0.97(4) | 1.83(4) | 2.787(8) | 174(4) | –1/2 + x, 1/2 – y, –1/2 + z |
| C15–H15C···O2 | 0.961(8) | 1.832(6) | 2.787(6) | 172.4(19) | |
| Short ring and C–H···Cg interactions | ||||||
|---|---|---|---|---|---|---|
| Cg(I) | Cg(J) | Cg(I)-Cg(J) (Å) | α (°) | β (°) | γ (°) | symmetry |
| FA | ||||||
| Cg1 | Cg1 | 5.17(4) | 0.0(2) | 36.4 | 36.4 | x, −1 + y, z |
| Cg1 | Cg1 | 5.17(4) | 0.0(2) | 36.4 | 36.4 | x, 1 + y, z |
| Cg1 | Cg2 | 4.79(4) | 43.3(2) | 28.7 | 65.6 | x, 1 + y, z |
| Cg2 | Cg1 | 5.25(4) | 72.1(2) | 29.0 | 84.3 | x, −y, 1/2 + z |
| Cg2 | Cg2 | 5.17(4) | 0.03(18) | 50.0 | 50.0 | x, −1 + y, z |
| Cg2 | Cg2 | 5.17(4) | 0.03(18) | 50.0 | 50.0 | x, 1 + y, z |
| C9–H9 | Cg1 | 2.87 | 9.86 | x, −1 + y, z | ||
| FAM | ||||||
| Cg1 | Cg1 | 5.584(11) | 0.0(3) | 52.0 | 52.0 | 1 – x, 1 – y, 1 – z |
| Cg1 | Cg1 | 5.708(6) | 76.0(3) | 23.2 | 89.8 | –1/2 + x, 1/2 – y, −1/2 + z |
| Cg1 | Cg2 | 5.470(7) | 46.6(3) | 35.4 | 82.0 | –1 + x, y, −1 + z |
| Cg1 | Cg2 | 5.277(8) | 60.1(3) | 42.4 | 37.7 | –1/2 + x, 1/2 – y, −1/2 + z |
| Cg2 | Cg1 | 5.277(8) | 60.1(3) | 37.7 | 42.4 | 1/2 + x, 1/2 – y, 1/2 + z |
| Cg2 | Cg2 | 5.090(5) | 31.1(2) | 36.6 | 61.7 | –1/2 + x, 1/2 – y, –1/2 + z |
Cg(I) and Cg(J): centroids of the rings, Cg(I)-Cg(J): centroid distance between ring I and ring J, α: dihedral angle between mean planes I and J, β:angle between the centroid vector Cg(I)···Cg(J), the normal to the plane (I), γ: angle between the centroid vector Cg(I)···Cg(J), the normal to the plane (J), and slippage: distance between Cg(I) and perpendicular projection of Cg(J) on ring I (Ang).
Figure 7.

π···π stacking interactions between FAM molecules mediated through MeOH solvent.
The study revealed conformational changes in FAM compared with the standard FA, primarily influenced by the choice of solvents (ethanol and methanol). In FAM, the interplanar angle increased by 5.58°, attributed to the presence of methanol solvent. Notably, the position of O2 in benzoic acid exhibited a deviation of approximately 0.235 Å from the coplanar arrangement in FAM. These structural alterations impact intermolecular interactions. FAM showcased distinctive hydrogen bond interactions with methanol, forming supramolecular architectures. Additionally, FAM displayed varied π···π stacking interactions, facilitated by methanol, influencing centroid distances. These conformational changes, particularly in hydrogen bonding and stacking patterns, can significantly affect the molecular properties and stability of FAM, offering insights into its potential pharmaceutical applications.
Hirshfield Surface Analysis
Hirshfeld surface analysis (HSA) emerges as the most effective method for analyzing the distinct intermolecular interactions within the cystalline environment. The presence of methanol in the FAM conformer that affects the distinct nature and individual contribution of each intermolecular interaction in the FA conformer can be unveiled by the utilization of HSA. This analytical approach becomes crucial in comprehending the structural stability of the molecule, as it reveals the structural stability of molecules through interactions such as C–H···π, C–O···π, and π···π stacking. HSA enriches the understanding and examination of contributions arising from interactions between distinct pairs of atoms, which can be facilitated through the 2D fingerprint plot analysis. Using CIF as an input, the HS of FA and FAM molecules have been superimposed across dnorm, shape index, and curvedness surfaces (Figure 8), employing transparency to facilitate the visual representation of various molecular components that significantly contributed to the stability of compound.
Figure 8.
dnorm (a), shape index (b), and curvedness (c) mapped on the Hirshfeld surface of the FA and FAM molecules.
The investigation of 2D fingerprint plots unveiled the quantitative distribution of molecular contacts, with each contributing to the overall composition of the Hirshfeld surface. Within these interactions, H···H contacts demonstrated the most significant contribution in both the FA and FAM molecules, accounting for 26.8 and 30.7% of their respective Hirshfeld surfaces (Figure 9). However, in the FA molecule, C–H interactions emerged as the second-highest contributor at 21.2%, whereas in the FAM molecule, it assumed the third position. In contrast, F–H interactions took the second spot, contributing 22.8% to the overall interactions, while C–H interactions ranked third in terms of contribution. Additionally, the increase in the O–H contribution in the FAM molecule is observed, which is attributed to the presence of MeOH in it. Other interactions also made notable contributions to the HS.
Figure 9.
2D fingerprint plots of the FA (top) and FAM (bottom) compounds visually illustrate the distinct contributions of each interaction to the overall Hirshfeld surface.
From the dnorm surface, we can clearly observe only two dark red spots near the H2 and O1 atoms of the FA molecule, which confirm the presence of a strong hydrogen bond, which is involved in the formation of the R22(6) synthon. The number of red spots is greater in FAM molecules and is observed near the O1 and O2 atoms of FA and the hydrogen atoms of methanol solvent, highlighting the hydrogen bond interactions (Table 2), which results in R44(12) supramolecular architecture and verifies the increase in the O–H contribution as observed in FPs.
Similarly, the shape index on the Hirshfeld surface reveals red triangular areas that represent concave stacking interactions, and blue triangular regions depict the convex ring atoms of the molecule. The surface’s curvedness, gauged by the root-mean-square curvature, distinguishes areas with low curvature, indicating flatness, from those with high curvature, denoting sharp curvatures. The presence of flat regions on the surface signifies the occurrence of π···π stacking interactions.
HSA provides a detailed, visually accessible representation of intermolecular interactions within the crystal lattice of both flufenamic acid (FA) and its methanol solvatomorph (FAM). By mapping regions of the crystal where significant interactions occur, it offers insights into the nature and spatial distribution of these interactions. The analysis helps elucidate the influence of the methanol solvent on FAM’s structure. By comparing HSA results between FA and FAM, researchers can pinpoint variations in the distribution and strength of intermolecular interactions, particularly focusing on the role of methanol in forming hydrogen bonds and stabilizing the crystal structure. 2D fingerprint plots derived from HSA provide a quantitative breakdown of molecular contacts, revealing the contribution of various interactions to the overall stability of the compounds. This allows for a systematic evaluation of the role of specific interactions, such as C–H···π, C–O···π, and π···π stacking, in maintaining structural integrity. Hirshfeld surface features: The analysis of distinct features on the Hirshfeld surface, such as the presence of red spots near specific atoms, aids in the identification of strong hydrogen bonds and short contacts. This information is crucial for understanding the formation of supramolecular architectures and molecular chains within the crystal lattice.
Frontier Molecular Orbitals (FAOs)
The electronic structures of the molecules were further investigated using quantum computational techniques (DFT). The molecular ground state was optimized in the spin state to determine the configuration with the lowest energy. This optimization was conducted by using the B3LYP functional in conjugation with the 6-311G(d,p) basis set.
Table 3 comprises the values of HOMO (highest occupied molecular orbital) and LUMO (lowest unoccupied molecular orbital) energies, the energy gap between them, and the related global reactive parameters encompassing the global electrophilicity index (ω), global hardness (η), chemical potential (μ), and global softness (s) obtained using the values of HOMO and LUMO energy differences. The energy gap reflects the chemical stability of the molecule. Figure 10 displays the 3D orbitals, where green and red colors depict the positive and negative phases of the molecular orbital wave function, respectively. Chemical potential signifies the propensity of electrons to escape from a stable system; a negative value denotes stability. Hardness indicates the resistance of a chemical system’s cloud to deformation under minor perturbations during chemical processes. Molecules with larger HOMO–LUMO gaps are characterized as harder and less reactive, while those with smaller gaps are considered softer and more reactive. The HOMO of the FA and FAM molecules is predominantly localized over the benzoic acid, amine, and carbon atoms of trifluorobenzene ring. Meanwhile, LUMO energies of the both the molecules are mainly concentrated over the amine and benzoic acid atoms. The energy gap of the both the molecules varies by 0.154 eV. The molecule’s global hardness reflects its intermolecular stability. Electrophilicity relates to the capacity of an electrophile to acquire additional electronic charge and its resistance to electron transfer, there by influencing stability.
Table 3. Quantum Chemical Parameters of FA and FAM Obtained by DFT Methods.
| parameters | FA | FAM |
|---|---|---|
| EHOMO (eV) | –6.2056 | –6.1675 |
| ELUMO (eV) | –1.9489 | –2.0648 |
| energy gap (ΔE) (eV) | 4.2567 | 4.1027 |
| ionization potential (IP) (eV) | 6.2056 | 6.1675 |
| electron affinity (EA) (eV) | 1.9489 | 2.0648 |
| electronegativity (χ) (eV) | 4.0772 | 4.1161 |
| global hardness (η) (eV) | 2.1283 | 2.0513 |
| softness (σ) (eV–1) | 0.4698 | 0.4875 |
| chemical potential(μ) (eV) | –4.0772 | –4.1161 |
| electrophilicity index (ω) (eV) | 3.9053 | 4.1296 |
Figure 10.

Frontier molecular orbital and its energy gap.
QTAIM Analysis
Bader’s AIM tool (QTAIM and NCI) is used to delve deeper into the intermolecular interactions. This technique is frequently used to identify and categorize intramolecular associations in molecular systems. It works by emphasizing regions of minimized electron density between atoms, where the RDG reduces to zero. NCI plots further characterize molecular environments, classifying them into stabilizing (blue) and destabilizing (red) regions. The combination of NCI with RDG analysis allows for the comprehensive exploration of the intricate network of noncovalent interactions within a molecule.
Figure 11a–c present the 3D visualization of NCI for FA and FAM with an isosurface value of 0.68. The intramolecular and intermolecular interactions, detailed in Table 2, are validated through brownish-green-colored disc-shaped surfaces in both molecules. Notably, Figure 11 illustrates strong intramolecular hydrogen bonding between the oxygen of the acid group and the hydrogen of the amine group (O1···H1–N1) in both FA and FAM, with a slight difference in bond distance (0.059 Å) depicted as blue-colored iso-surface discs. The red-colored discs within the trifluoro and benzoic acid rings signify repulsion or steric effects within the molecular rings. The RDG plot, derived from RDG against the product of electron density and the sign of Hessian eigenvalue (λ2) denoted as sign(λ2)ρ (Figure 11b), evaluates the strength of weak interactions, indicating a repulsive nature in deep red (0.011 to 0.05 atomic units), strong attractive forces in deep blue dots (negative values from −0.02 to −0.05 atomic units), and manifestation of van der Waals effects in the region between −0.02 to 0.01 of sign(λ2)ρ, represented by green dots.
Figure 11.
Noncovalent intramolecular interactions are visualized using isosurfaces with a threshold value set at 0.65 (a). The 2D RDG scatter graph provides a graphical representation of the properties and features of these interactions (b). Atoms-in-molecules topology analysis to evaluate the intermolecular O2–H2···O1 bond in FA and the C15–H15c···O2, C15–H15a···O1, and O3–H3a···O1 bonds in FAM H-bond binding energy at their BCPs (c).
Intermolecular H-Bond Binding Energy
Intermolecular H-bond binding energy was assessed by predicting the H-bond binding energy (BE) based on the electron density at bond critical points (BCPs) corresponding to H-bonds in the FA and FAM molecule. This analysis, depicted in Figure 11c and detailed in Table 4, was conducted using Bader’s theory implemented in the Multiwfn package with optimized parameters. Bader’s theory identifies interatomic interactions through BCPs of the (3,-1) type and the presence of bond paths between atoms. The summarized data in Table 4 reveal that the ρ(r) values are below 0.1 a.u. and ∇2ρ(r) at their BCPs are positive, confirming the fulfillment of intermolecular noncovalent interaction criteria for the O2–H2···O1 bond in FA and the C15–H15c···O2, C15–H15a···O1, and O3–H3a···O1 bonds in FAM. H-bond binding energies (Table 4) indicate their noncovalent nature, signifying the presence of strong hydrogen bonds.
Table 4. B3LYP-6-311(d,p) Calculated QTAIM Topological Descriptors at the BCP of the Hydrogen Bond and Other Hydrogen Bond Indices for the FA and FAM Molecules.
| interactions | BCPρ(a.u.) | ρBCP(a.u.) | V(r)(a.u.) | G(r)(a.u.) | EHB(kJ/mol) |
|---|---|---|---|---|---|
| O2–H2···O1 (FA) | 107 | 0.0852 | –0.0935 | 0.0616 | –122.742 |
| C15–H15c···O2 (FAM) | 74 | 0.0107 | –0.0075 | 0.0010 | –9.8456 |
| C15–H15a···O1 (FAM) | 64 | 0.00571 | –0.0039 | 0.0057 | –5.119 |
| O3–H3a···O1 (FAM) | 69 | 0.0438 | –0.0517 | 0.0398 | –67.869 |
Molecular Docking Analysis
Molecular docking analysis for FA and FAM with prostaglandin D2 11-ketoreductase (AKR1C3) is carried out using the X-ray crystal structures of AKR1C3 with bound indomethacin and flufenamic acid (PDB ID: 1S2C). Prostaglandin D2 11-ketoreductase (AKR1C3) is a significant enzyme involved in the intricate web of pharmacological interactions. It plays a role in prostaglandin production and is targeted by certain nonsteroidal anti-inflammatory drugs (NSAIDs). Recent crystal structures reveal how NSAIDs such as indomethacin and flufenamic acid interact with AKR1C3. These findings highlight AKR1C3 as a COX-independent target for NSAIDs, potentially leading to new cancer therapies with reduced side effects. The AKR1C3′s multifaceted role connects it to both COX-dependent and COX-independent pathways, making it a fascinating player in the world of pharmacology and potential cancer treatments. X-ray crystal structures of AKR1C3 complexed with nonsteroidal anti-inflammatory drugs (NSAIDs) indomethacin and flufenamic acid are reported at resolutions of 1.8 and 1.7 Å, respectively. In the indomethacin complex, a single molecule is located in the active site, while flufenamic acid binds to both the active site and the beta-hairpin loop at the opposite end of the central beta-barrel. Two additional crystal structures at resolutions of 1.20 and 2.1 Å reveal acetate occupancy in the active site, filling the proposed oxyanion hole. These structural insights highlight AKR1C3 as a COX-independent target for NSAIDs, offering a foundation for the development of new cancer therapies with potentially reduced COX-dependent side effects.
Cartoon representation of docking poses of FA (color by carbon atoms: black) and FMA (color by carbon atoms: pink) in the active site of targeted protein is depicted in Figure 12. The structure of AKR1C3 where FA bound within the active site was determined at a high resolution of 1.8 Å.54,55 The most crucial binding interaction between FA and protein, responsible for the inhibitory effect, involves hydrogen bonding between the carboxylic acid atom of the drug and the oxyanion site residues Tyr-55 and His-117. Additionally, the other oxygen atom in this group forms a hydrogen bond with an adjacent water molecule, which is part of an integral and conserved water network, observed in other enzyme structures. The CF3 group of the drug occupies the SP1 active site pocket and forms hydrogen bonds with a water molecule and the hydroxyl group of Tyr-216. The two aromatic rings of the drug molecule are securely held in place by van der Waals interactions, particularly through face-to-face and edge-stacking interactions with aromatic protein side chains. The benzoic acid ring primarily interacts with the side chains of Tyr-24, Trp-227, and Phe-306, while the phenylamine ring is surrounded by the side chains of Trp-86, Asn-167, and Phe-311 (Figure 13 and Table 5).
Figure 12.
Cartoon representation of docking poses of FA (color by carbon atoms: black) and FMA (color by carbon atoms: pink) in the active site of targeted protein.
Figure 13.
2D (left) and 3D (right) representations of protein–ligand interactions of FA.
Table 5. Interactions of FA and FAM with the Targeted Protein.
| protein (amino acids) | ligand | interaction type | bond distance (Å) |
|---|---|---|---|
| FA | |||
| A:TYR 55 | oxygen | conventional hydrogen bond | 2.54 |
| A:TYR 216 | hydrogen | conventional hydrogen bond | 2.93 |
| A:ASN 167 | hydrogen | halogen (fluorine) | 3.35 |
| A:ASN 167 | hydrogen | halogen (fluorine) | 3.14 |
| A:ASN 167 | π ring of trifluoromethyl | π-donor hydrogen bond | 3.98 |
| A:TYR24 | oxygen | π-anion | 4.65 |
| A:TYR24 | π ring of benzoic acid | π-anion | 5.16 |
| A:PHE 311 | π ring of trifluoro-methyl aniline | π-π stacked | 4.92 |
| A:PHE 306 | π ring of trifluoro-methyl aniline | π-π stacked | 5.97 |
| A:PHE 306 | π ring of benzoic acid | π-π stacked | 5.02 |
| A:TRP 227 | π ring of benzoic acid | alkyl | 4.51 |
| A:LEU 54 | π ring of benzoic acid | π-alkyl | 5.17 |
| FAM | |||
| A:LEU 268 | van der Waals | ||
| A:HIS 117 | π ring of trifluoro-methyl aniline | attractive charge | 4.55 |
| A:TYR 55 | oxygen | conventional hydrogen bond | 2.18 |
| A:TYR 23 | oxygen | conventional hydrogen bond | 2.44 |
| A:TYR 24 | oxygen | conventional hydrogen bond | 1.97 |
| A:LYS 84 | oxygen | π-cation | 4.13 |
| A:GLN 222 | π ring of benzoic acid | π-donor hydrogen bond | 3.01 |
| A:TYR 55 | π ring of trifluoro-methyl aniline | π-π stacked | 5.45 |
| A:TYR:24 | π ring of benzoic acid | π-π T-shaped | 5.95 |
| A:TYR 24 | π ring of trifluoro-methyl aniline | π-π T-shaped | 5.30 |
| A:TYR 24 | carbon | alkyl | 4.16 |
| A:TRP 227 | carbon | alkyl | 4.35 |
| A:LEU 54 | carbon | alkyl | 4.89 |
| A:LYS 270 | π ring of benzoic acid | π-alkyl | 5.31 |
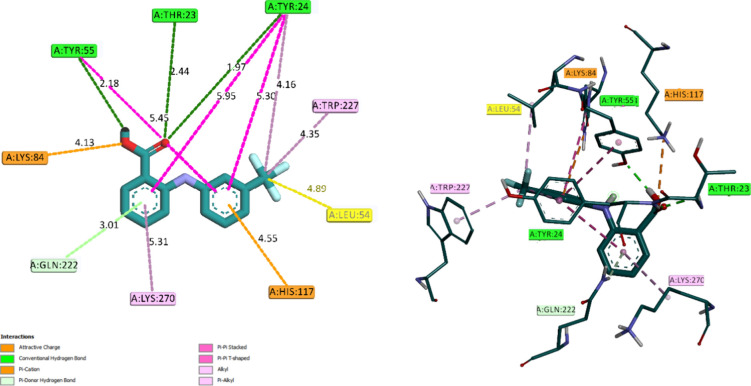
The interaction mode of the novel compound FAM was analyzed by using molecular docking. As depicted in the Figure 13, FAM binds within the active site of AKR1C3 with various interactions. The carboxylate group of FAM occupies the acetate-binding site, forming hydrogen bonds between oxygen atoms O1 and O2 with Thr23 and Tyr55, respectively (Figure 15). O1 of FAM also forms an additional hydrogen bond with Tyr24, acting as a double acceptor. The two aromatic rings of the FAM molecule are anchored in place through van der Waals interactions with aromatic residues, including His117, Tyr55, Tyr24, Gln222, and Lys270, as indicated in detail in Table 5 and Figure 14.
Figure 15.
RMSD plot of protein (a), ligand showing the minimal deviations (b), and RMSF graph (c) representing dynamic behavior of active site amino acids during the simulation period.
Figure 14.
2D (left) and 3D (right) representations of protein–ligand interactions of FAM.
Molecular Dynamics Simulation Study
A molecular dynamics (MD) simulation lasting 100 ns was conducted to gain deeper insights into the binding mode and stability of the protein–ligand complex. The primary goal was to analyze the dynamic behavior and interactions occurring between the target protein and the FAM molecule. Several metrics were employed to evaluate the complex throughout the simulation period. The root-mean-square deviation (RMSD) was used to assess the stability of the binding mode, offering insights into how the complex structure fluctuated over time. Additionally, the protein–ligand interactions were thoroughly examined to understand the nature and strength of the intermolecular contacts during the simulation. This analysis yielded valuable insights into the dynamic behavior of the complex and its potential impact on binding stability. Figures 15 and 16 visually illustrate the results of these analyses, providing a comprehensive understanding of the complex’s behavior throughout the MD simulation.
Figure 16.
Protein–ligand interactions showcased in the histogram plot (a) and 2D view (b).
Root-Mean-Square Deviation (RMSD)
Figure 15a,b presents the visual representations of the RMSD plot for the ligand and protein, respectively, during the simulation. The RMSD plot offers insights into the deviations in the structures of the ligand and protein over the simulation period. In the ligand-RMSD plot, it is observed that the FAM molecule exhibited fluctuations of up to 1.2 Å at 22 and 75 ns. However, for the rest of the simulation, the ligand maintained a minimal deviation of 0.8 Å, indicating the stability of the FAM molecule within the protein’s binding pocket. In compare, the protein achieved initial stability within the first 10 ns and remained relatively stable throughout the entire simulation period, with deviations ranging from 2.4 to 3.6 Å (within a 2 Å range). This suggests that the protein structure maintains a relatively consistent conformation and experiences limited deviations during the simulation.
Root-Mean-Square Fluctuation (RMSF)
Figure 15c depicts the magnitude of fluctuation and dynamic behavior of individual amino acids within the protein structure during the simulation. This comprehensive analysis over the simulation duration aimed to enhance our understanding of the protein’s flexibility. We calculated the average root-mean-square fluctuation (RMSF) for each amino acid in the 1S2C protein while considering the presence of the FAM molecule. Active site residues showed very least fluctuations [TYR 24 (0.75 Å), TYR 55 (1.14 Å), TYR 23 (0.61 Å), LEU 54 (1.17 Å), LYS 84 (0.049 Å), HIS 117 (0.58 Å), GLN 222 (1.19 Å), TRP 227 (1.), and LYS 84 (0.62 Å)] during the simulation period. The resulting RMSF plot indicated that the binding process with the receptor remained stable, with minimal impact on the protein’s flexibility throughout the simulation period.
Protein–Ligand Interactions
Hydrogen bonds play a crucial role in drug discovery, influencing drug specificity and stability in protein–ligand interactions. In our simulations (as shown in Figures 16), across the AKR family, there are four amino acids, Asp50, Tyr55, Lys84, and His117, that are highly conserved. It has been suggested that these four amino acids work together to form a catalytic tetrad, playing a crucial role in catalyzing the oxidation of alcohols or the reduction of ketones through a push–pull mechanism.40,41 FAM formed vital hydrogen bonds with ASP50 and LYS84 residues for 99 and 49% of the simulation time, respectively. TYR24 also contributed by forming bonds with FAM’s trifluoroaniline ring for 55% of the simulation. Notably, FAM’s amine group intermittently interacted with LYS84 through the oxygen atom of the carboxylic group (69% of the simulation), indicating its role in complex stabilization.
Exploration of the Specific Amino Acid Residues Involved in Interactions with Both FA and FAM and Comparing the Binding Affinities and Docking Scores
The molecular docking analysis revealed distinct binding modes for flufenamic acid (FA) and its solvatomorphic derivative (FAM) with prostaglandin D2 11-ketoreductase (AKR1C3). The key interactions between the ligands and the protein are summarized in Table 5, and the figures provide a visual representation of the docking poses and interactions.
Docking score for FA: −8.9 kcal/mol and for FAM: −9.5 kcal/mol
FA Binding Interactions
Conventional Hydrogen Bond with TYR 55 (2.54 Å) and TYR 216 (2.93 Å).
Halogen (Fluorine) Hydrogen Bond with ASN 167 (3.35 3.14 Å).
π-Donor Hydrogen Bond with ASN 167 (3.98 Å).
π-Anion with TYR 24 (4.65 Å) and a π ring of benzoic acid (5.16 Å).
π-π was stacked with PHE 311, PHE 306, and TRP 227.
Alkyl interacts with TRP 227, LEU 54, and π-Alkyl with LEU 54.
FAM Binding Interactions
van der Waals interaction with LEU 268.
Attractive charge with HIS 117 (π ring of trifluoro-methyl aniline, 4.55 Å).
Conventional Hydrogen Bond with TYR 55 (2.18 Å), TYR 23 (2.44 Å), and TYR 24 (1.97 Å).
π-Cation with LYS 84 (4.13 Å).
π-Donor Hydrogen Bond with GLN 222 (3.01 Å).
π-π Stacked with TYR 55, TYR 24, and the π ring of trifluoro-methyl aniline.
π-π T-shaped with TYR 55, TYR 24, and a π ring of benzoic acid.
Alkyl interactions with TYR 24, TRP 227, LEU 54, and LYS 270.
Molecular Dynamics Simulation
The RMSD plot indicates the stability of FAM within the protein’s binding pocket with minimal deviation. The protein structure maintains a relatively consistent conformation with a limited number of deviations during the simulation. The RMSF analysis shows the least fluctuations in active site residues, indicating stable binding with minimal impact on protein flexibility.
Implications
FAM exhibits a more negative docking score, suggesting stronger binding affinity than FA. Specific interactions with key amino acid residues contribute to the stability of the protein–ligand complex. Further exploration of these interactions and comparative analysis may provide insights into the potential differences in drug efficacy or selectivity. Overall, the study provides valuable information about the binding modes, stability, and potential implications for the drug efficacy of FA and its solvatomorphic derivative FAM with AKR1C3.
Key Differences between FAM and FA and the Potential Implications for Drug Development
Utilizing single-crystal X-ray diffraction, we explored the spatial arrangement of atoms in FA and a novel solvatomorphic structure named FAM, obtained from ethanol and methanol solutions, respectively. The structures exhibited monoclinic crystal systems with different space groups (C2/c and P21/n), comprising octamers and tetramers in each unit cell (Z = 8 and Z = 4). Notably, FAM revealed an increased interplanar angle due to the presence of methanol solvent presence.
FA demonstrated three intermolecular hydrogen bonds forming S(5) and S(6) planar rings, resulting in 1D molecular linear chains along the b-axis. In contrast, FAM displayed two intermolecular hydrogen bonds, yielding one S(5) and S(6) planar ring. π···π stacking interactions between trifluorotoluene and benzoic acid moieties played a significant role in both compounds, contributing to the development of 1D linear molecular arrays along the b-axis. Intriguingly, FAM showcased additional interactions involving lone pairs, which were absent in FA.
HSA revealed distinct intermolecular interactions within the crystalline environment. Notably, H···H contacts contributed significantly to both FA and FAM surfaces. FAM exhibited increased the level of the O–H contribution, attributed to methanol presence. The analysis facilitated a deeper understanding of the molecular stability through C–H···π, C–O···π, and π···π stacking interactions.
Quantum computational techniques provided insights into FA and FAM electronic structures. The HOMO and LUMO energies, along with the energy gap, highlighted the variations. FAOs were primarily localized over the benzoic acid, amine, and carbon atoms of the trifluorobenzene ring.
NCI analysis validated intramolecular associations, revealing strong intramolecular hydrogen bonding. RDG analysis further characterized weak interactions, highlighting regions of repulsion, attraction, and van der Waals effects.
Docking studies with prostaglandin D2 11-ketoreductase (AKR1C3) showcased crucial hydrogen bonding interactions between FA/FAM and the protein. FAM’s carboxylate group formed hydrogen bonds with Thr23 and Tyr55, occupying the acetate-binding site.
MD simulations revealed stable ligand-protein interactions, as indicated by the RMSD and RMSF plots. Notably, the FAM maintained a minimal deviation within the binding pocket, affirming its stability. Protein–ligand interactions, especially hydrogen bonds, played a pivotal role in the complex stability.
This study provides a comprehensive understanding of FA polymorphism, particularly the solvatomorphic structure of FAM. The unique interactions observed in FAM, including those with methanol solvent, present intriguing prospects for drug development. The variations in intermolecular interactions, electronic structures, and binding modes between FA and FAM underscore the potential impact of solvatomorphism on the pharmaceutical properties. These findings contribute valuable insights that could influence drug development strategies, ultimately enhancing therapeutic agents targeting inflammatory conditions.
Conclusions
In conclusion, this research has provided a comprehensive exploration of flufenamic acid (FA) polymorphism, with a specific focus on a newly identified solvatomorphic structure named FAM in a methanol solvent. The central objective of this study was to investigate whether solvatomorphism induces conformational changes that might influence the interactions of the drug compared to those of the standard FA. The X-ray crystal structure analysis of flufenamic acid (FA) and its solvatomorph (FAM) revealed distinct structural features influenced by the choice of solvent. FA exhibited significant nonplanar characteristics, while FAM, formed in methanol, displayed a novel solvatomorphic structure with altered interplanar angles. Hydrogen bonding interactions, including a notable C–H···O bond between FA and methanol in FAM, played a crucial role in crystal formation. FA demonstrated 1D linear molecular chains and π···π stacking interactions, contributing to structural stability. In contrast, FAM exhibited a dimeric supramolecular architecture facilitated by methanol-mediated hydrogen bonds. Hirshfeld surface analysis highlighted the impact of methanol on intermolecular interactions in the FAM, providing a quantitative distribution of molecular contacts. Noncovalent interactions, analyzed through frontier molecular orbitals and NCI plots, demonstrated the stability of FA and FAM, with varying contributions from hydrogen bonding, van der Waals forces, and steric effects. Molecular docking studies with prostaglandin D2 11-ketoreductase (AKR1C3) revealed the binding modes of FA and FAM, emphasizing critical hydrogen bonding interactions. Molecular dynamics simulations confirmed the stability of the protein–ligand complex with minimal deviations in RMSD and RMSF. The study sheds light on the influence of solvent choice on the crystal structures of FA and FAM, providing valuable insights into their molecular interactions, stability, and potential pharmaceutical applications. The comprehensive analysis combines experimental and computational approaches to unravel the intricate details of these compounds, contributing to a broader understanding of crystal engineering and drug design.
Acknowledgments
Author J.R.R. and H.A.A. extend their appreciation and acknowledge Researchers Supporting Project number (RSP2024R54) for all funding from King Saud University, Riyadh, Saudi Arabia.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c07520.
The supplementary crystallographic data for this paper can be obtained free of charge via https://www.ccdc.cam.ac.uk/structures/, or by e-mailingdata_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: + 44(0)1223–336033”. (CIF)
The authors declare no competing financial interest.
Supplementary Material
References
- Kuhnert-Brandstatter M.; Borka L.; Friedrich-Sander G. Zur ¨ Polymorphie von Arzneimitteln: Flufenaminsaure und BL 191. Arch. Pharm. 1974, 307, 845–853. 10.1002/ardp.19743071107. [DOI] [PubMed] [Google Scholar]
- Krc J. Jr. Crystallographic properties of flufenamic acid. Microscope 1977, 25, 31–45. [Google Scholar]
- Galdecki Z.; Glowka M. L.; Gorkiewicz Z. Polymorphism of 2-(3trifluoromethylphenylamino)benzoic acid. Acta Polym. Pharm. 1978, 35, 77–79. [PubMed] [Google Scholar]
- Burger A.; Ramberger R. Thermodynamische Beziehungen zwischen polymorphen Modifikationen: Flufenaminsaure und Mefe-naminsaure. Microchim. Acta 1980, 73, 17–28. 10.1007/BF01197228. [DOI] [Google Scholar]
- Lee E. H.; Boerrigter S. X. M.; Rumondor A. C. F.; Chamarthy S. P.; Byrn S. R. Formation and solid-state characterization of a salt-induced metastable polymorph of flufenamic acid. Cryst. Growth Des. 2008, 8, 91–97. 10.1021/cg7008607. [DOI] [Google Scholar]
- Surov A. O.; Terekhova I. V.; Bauer-Brandl A.; Perlovich G. L. Thermodynamic and structural aspects of some fenamate molecular crystals. Cryst. Growth Des. 2009, 9, 3265–3272. 10.1021/cg900002q. [DOI] [Google Scholar]
- Espinell J. R. H.; López-Mejías V.; Stelzer T. Revealing Polymorphic Phase Transformations in Polymer-Based Hot Melt Extrusion Processes. Cryst. Growth Des. 2018, 18, 1995–2002. 10.1021/acs.cgd.7b01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nartowski K. P.; Malhotra D.; Hawarden L. E.; Sibik J.; Iuga D.; Zeitler J. A.; Fábián L.; Khimyak Y. Z. 19 F NMR Spectroscopy as a Highly Sensitive Method for the Direct Monitoring of Confined Crystallization within Nanoporous Materials. Angew. Chem. 2016, 128, 9050–9054. 10.1002/ange.201602936. [DOI] [PubMed] [Google Scholar]
- Case D. H.; Srirambhatla V. K.; Guo R.; Watson R. E.; Price L. S.; Polyzois H.; Cockcroft J. K.; Florence A. J.; Tocher D. A.; Price S. L. Successful computationally directed templating of metastable pharmaceutical polymorphs. Cryst. Growth Des. 2018, 18, 5322–5331. 10.1021/acs.cgd.8b00765. [DOI] [Google Scholar]
- Cookman J.; Hamilton V.; Price L. S.; Hall S. R.; Bangert U. Visualising early-stage liquid phase organic crystal growth via liquid cell electron microscopy. Nanoscale 2020, 12, 4636–4644. 10.1039/C9NR08126G. [DOI] [PubMed] [Google Scholar]
- Sanabria Ortiz K.; Hernandez Espinell J. R.; Ortiz Torres D.; Lopez-Mejías V.; Stelzer T. Polymorphism in solid dispersions. ´. Cryst. Growth Des. 2020, 20, 713–722. 10.1021/acs.cgd.9b01138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler A. R.; Ragbirsingh R.; McMonagle C. J.; Waddell P. G.; Heaps S. E.; Steed J. W.; Thaw P.; Hall M. J.; Probert M. R. Encapsulated nanodroplet crystallization of organic-soluble small molecules. Chem. 2020, 6, 1755–1765. 10.1016/j.chempr.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J. F. 3′-Trifluoromethyldiphenylamine-2-carboxylic acid, C14H10F3NO2 flufenamic acid. Cryst. Struct. Commun. 1973, 2, 459–461. [Google Scholar]
- Krishna Murthy H.; Bhat T.; Vijayan M. Structure of a new crystal form of 2-{[3-(trifluoromethyl) phenyl] amino} benzoic acid (flufenamic acid). Acta Crystallogr., Sect. B: Struct. Crystallogr. Cryst. Chem. 1982, 38, 315–317. 10.1107/S0567740882002763. [DOI] [Google Scholar]
- Lopez-Mejías V.; Kampf J. W.; Matzger A. J. Nonamorphism ´ in Flufenamic Acid and a New Record for a Polymorphic Compound with Solved Structures. J. Am. Chem. Soc. 2012, 134, 9872–9875. 10.1021/ja302601f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentili D.; Gazzano M.; Melucci M.; Jones D.; Cavallini M. Polymorphism as an additional functionality of materials for technological applications at surfaces and interfaces. Chem. Soc. Rev. 2019, 48, 2502–2517. 10.1039/C8CS00283E. [DOI] [PubMed] [Google Scholar]
- Chung H.; Diao Y. Polymorphism as an emerging design strategy for high-performance organic electronics. J. Mater. Chem. C 2016, 4, 3915–3933. 10.1039/C5TC04390E. [DOI] [Google Scholar]
- Di Martino P.; Guyot-Hermann A. M.; Conflant P.; Drache M.; Guyot J. C. A new pure paracetamol for direct compression: The orthorhombic form. Int. J. Pharm. 1996, 128, 1–8. 10.1016/0378-5173(95)04127-3. [DOI] [Google Scholar]
- Rietveld I. B.; Ceolin R. Rotigotine: unexpected polymorphism ´ with predictable overall monotropic behavior. J. Pharm. Sci. 2015, 104, 4117–4122. 10.1002/jps.24626. [DOI] [PubMed] [Google Scholar]
- 30. Chen Z.; Li Y.; Chen E.; Hall D. L.; Darke P. L.; Culberson C.; Shafer J. A.; Kuo L. C. Crystal structure at 1.9-A resolution of human immunodeficiency virus (HIV) II protease complexed with L-735,524, an orally bioavailable inhibitor of the HIV proteases. J. Biol. Chem. 1994, 269, 26344–26348. 10.1016/S0021-9258(18)47199-2. [DOI] [PubMed] [Google Scholar]
- Chung H.; Diao Y. Polymorphism as an emerging design strategy for high-performance organic electronics. J. Mater. Chem. C 2016, 4, 3915–3933. 10.1039/C5TC04390E. [DOI] [Google Scholar]
- Lee A. Y.; Erdemir D.; Myerson A. S. Crystal polymorphism in chemical process development. Annu. Rev. Chem. Biomol. Eng. 2011, 2, 259–280. 10.1146/annurev-chembioeng-061010-114224. [DOI] [PubMed] [Google Scholar]
- Elliott J.; Hancock B. Pharmaceutical materials science: an active new frontier in materials research. MRS Bull. 2006, 31, 869–873. 10.1557/mrs2006.205. [DOI] [Google Scholar]
- Gardner C. R.; Walsh C. T.; Almarsson Ö. Drugs as materials: valuing physical form in drug discovery. Nat. Rev. Drug Discovery 2004, 3 (11), 926–934. 10.1038/nrd1550. [DOI] [PubMed] [Google Scholar]
- Dow Jones Newswires. (2003). ″GlaxoSmithKline on Track to Launch 11 Drugs by Dec. 2003.″ New York, NY, USA. 25. Shaker B.; Ahmad S.; Lee J.; Jung C.; Na D., 2021. In silico methods and tools for drug discovery. Comput. Biol. Med., 137, p.104851. 10.1016/j.compbiomed.2021.104851 [DOI] [PubMed] [Google Scholar]
- Oselusi S. O.; Dube P.; Odugbemi A. I.; Akinyede K. A.; Ilori T. L.; Egieyeh E.; Sibuyi N. R.; Meyer M.; Madiehe A. M.; Wyckoff G. J.; Egieyeh S. A. The role and potential of computer-aided drug discovery strategies in the discovery of novel antimicrobials. Computers in biology and medicine 2024, 169, 107927 10.1016/j.compbiomed.2024.107927. [DOI] [PubMed] [Google Scholar]
- Segall M. D.; Barber C. Addressing toxicity risk when designing and selecting compounds in early drug discovery. Drug Discovery Today 2014, 19, 688–693. 10.1016/j.drudis.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Robinson B. S.; Riccardi K. A.; Gong Y. F.; Guo Q.; Stock D. A.; Blair W. S.; Terry B. J.; Deminie C. A.; Djang F.; Colonno R. J.; Lin P. F. BMS-232632, a Highly Potent Human Immunodeficiency Virus Protease Inhibitor That Can Be Used in Combination with Other Available Antiretroviral Agents. Antimicrob. Agents Chemother. 2000, 44, 2093–2099. 10.1128/AAC.44.8.2093-2099.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krohn A.; Redshaw S.; Ritchie J. C.; Graves B. J.; Hatada M. H. Novel binding mode of highly potent HIV-proteinase inhibitors incorporating the (R)- hydroxyethylamine isostere. J. Med. Chem. 1991, 34, 3340–3342. 10.1021/jm00115a028. [DOI] [PubMed] [Google Scholar]
- Kempf D. J.; Marsh K. C.; Denissen J. F.; McDonald E.; Vasavanonda S.; Flentge C. A.; Green B. E.; Fino L.; Park C. H.; Kong X.-P. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Nat. Acad. Sci. 1995, 92, 2484–2488. 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson A. C. The process of structure-based drug design. Chem. Biol. 2003, 10, 787–797. 10.1016/j.chembiol.2003.09.002. [DOI] [PubMed] [Google Scholar]
- Vamathevan J.; Clark D.; Czodrowski P.; Dunham I.; Ferran E.; Lee G.; Li B.; Madabhushi A.; Shah P.; Spitzer M.; Zhao S. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discovery 2019, 18, 463–477. 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto-Martínez F. D.; López-López E.; Juárez-Mercado K. E.; Medina-Franco J. L. Computational drug design methods—current and future perspectives. Silico Drug Des. 2019, 19. 10.1016/B978-0-12-816125-8.00002-X. [DOI] [Google Scholar]
- Rigaku Corporation and its Global Subsidiaries, 2011, Expert 2.0 r15. Software for Data Collection and Processing.
- Pflugrath J. W. The finer things in X-ray diffraction data collection. Acta Crystallographica Section D: Biological Crystallography 1999, 55 (10), 1718–1725. 10.1107/S090744499900935X. [DOI] [PubMed] [Google Scholar]
- Sheldrick G. M. Phase annealing in SHELX-90: direct methods for larger structures. Acta Crystallogr. A 1990, 46 (6), 467–473. 10.1107/S0108767390000277. [DOI] [Google Scholar]
- Sheldrick G. M. Crystal structure refinement with SHELXL. Acta Crystallogr. C 2015, 71 (1), 3–8. 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolomanov O. V.; Bourhis L. J.; Gildea R. J.; Howard J. A.; Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009, 42 (2), 339–341. 10.1107/S0021889808042726. [DOI] [Google Scholar]
- Spek A. L. PLATON, an integrated tool for the analysis of the results of a single crystal structure determination. Acta Crystallogr. A 1990, 46 (s1), c34–c34. [Google Scholar]
- 21. Macrae C. F.; Sovago I.; Cottrell S. J.; Galek P. T. A.; McCabe P.; Pidcock E.; Platings M.; Shields G. P.; Stevens J. S.; Towler M.; Wood P. A. Mercury 4.0: from visualization to analysis, design and prediction. J. Appl. Crystallogr. 2020, 226–235. 10.1107/S1600576719014092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman P. R.; Turner M. J.; McKinnon J. J.; Wolff S. K.; Grimwood D. J.; Jayatilaka D.; Spackman M. A. CrystalExplorer: A program for Hirshfeld surface analysis, visualization and quantitative analysis of molecular crystals. J. Appl. Crystallogr. 2021, 54 (3), 1006–1011. 10.1107/S1600576721002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman M. A.; McKinnon J. J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4 (66), 378–392. 10.1039/B203191B. [DOI] [Google Scholar]
- Frisch M. J.; Trucks G.; Schlegel H. B.; Scuseria G. E.; Robb M. A.; Cheeseman J. R.; Scalmani G.; Barone V.; Mennucci B.; Petersson G. A.; Nakatsuji H.; Caricato M.; Li X.; Hratchian H. P.; Izmaylov A. F.; Bloino J.; Zheng G.; Sonnenberg J. L.; Hada M.; Ehara M.; Toyota K.; Fukuda R.; Hasegawa J.; Ishida M.; Nakajima T.; Honda Y.; Kitao O.; Nakai H.; Vreven T.; Montgomery J.; Peralta J. E.; Ogliaro F.; Bearpark M.; Heyd J. J.; Brothers E.; Kudin K. N.; Staroverov V. N.; Kobayashi R.; Normand J.; Raghavachari K.; Rendell A. P.; Burant J. C.; Iyengar S. S.; Tomasi J.; Cossi M.; Rega N.; Millam J. M.; Klene M.; Knox J. E.; Cross J. B.; Bakken V.; Adamo C.; Jaramillo J.; Gomperts R.; Stratmann R. E.; Yazyev O.; Austin A. J.; Cammi R.; Pomelli C. S.; Ochterski J.W.; Martin R. L.; Morokuma K.; Zakrzewski V. G.; Voth G. A.; Salvador P.; Dannenberg J. J.; Dapprich S.; Daniels A. D.; Farkas F. J. B.; Ortiz J. V.; Cioslowski J.; Fox D. J.. Gaussian 09, revision a. 01, (2009) Gaussian Inc..
- Gill P. M.; Johnson B. G.; Pople J. A.; Frisch M. J. The performance of the Becke—Lee—Yang—Parr (B—LYP) density functional theory with various basis sets. Chem. Phys. Lett. 1992, 197 (4–5), 499–505. 10.1016/0009-2614(92)85807-M. [DOI] [Google Scholar]
- Manne R.; Åberg T. Koopmans’ theorem for inner-shell ionization. Chem. Phys. Lett. 1970, 7, 282–284. 10.1016/0009-2614(70)80309-8. [DOI] [Google Scholar]
- Dennington R.; Keith T.; Millam J.. GaussView, version 6. Semichem Inc.: Shawnee Mission, KS. (2009). [Google Scholar]
- Lu T.; Chen F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. Journal of Molecular Graphics. 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Morris G. M.; Huey R.; Lindstrom W.; Sanner M. F.; Belew R. K.; Goodsell D. S.; Olson A. J. Autodock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 16, 2785–2791. 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt J.; Santos-Martins D.; Tillack A. F.; Forli S. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61 (8), 3891–3898. 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O.; Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31 (2), 455–461. 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biovia Dassault Systemes; Discovery studio visualizer, Discovery studio visualizer 17.20, San Diego, CA, USA, 936. San Diego, 2017. [Google Scholar]
- Schrödinger D.Release 2023–2: Desmond Molecular Dynamics System; D. E. Shaw Research: New York, NY, 2023. Maestro-Desmond InteroperabilityTools, Schr̈odinger: New York, NY, (2023). [Google Scholar]
- Lovering A. L.; Ride J. P.; Bunce C. M.; Desmond J. C.; Cummings S. M.; White S. A. Crystal structures of prostaglandin D(2) 11-ketoreductase (AKR1C3) in complex with the non-steroidal anti-inflammatory drugs flufenamic acid and indomethacin. Cancer Res. 2004, 64 (5), 1802–1810. 10.1158/0008-5472.CAN-03-2847. [DOI] [PubMed] [Google Scholar]
- Lovering A. L.; Ride J. P.; Bunce C. M.; Desmond J. C.; Cummings S. M.; White S. A. Crystal structures of prostaglandin D2 11-ketoreductase (AKR1C3) in complex with the nonsteroidal anti-inflammatory drugs flufenamic acid and indomethacin. Cancer research 2004, 64 (5), 1802–1810. 10.1158/0008-5472.CAN-03-2847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.