ABSTRACT
The utilization of ATP within cells plays a fundamental role in cellular processes that are essential for the regulation of host–pathogen dynamics and the subsequent immune response. This study focuses on ATP-binding proteins to dissect the complex interplay between Staphylococcus aureus and human cells, particularly macrophages (THP-1) and keratinocytes (HaCaT), during an intracellular infection. A snapshot of the various protein activity and function is provided using a desthiobiotin-ATP probe, which targets ATP-interacting proteins. In S. aureus, we observe enrichment in pathways required for nutrient acquisition, biosynthesis and metabolism of amino acids, and energy metabolism when located inside human cells. Additionally, the direct profiling of the protein activity revealed specific adaptations of S. aureus to the keratinocytes and macrophages. Mapping the differentially activated proteins to biochemical pathways in the human cells with intracellular bacteria revealed cell-type-specific adaptations to bacterial challenges where THP-1 cells prioritized immune defenses, autophagic cell death, and inflammation. In contrast, HaCaT cells emphasized barrier integrity and immune activation. We also observe bacterial modulation of host processes and metabolic shifts. These findings offer valuable insights into the dynamics of S. aureus–host cell interactions, shedding light on modulating host immune responses to S. aureus, which could involve developing immunomodulatory therapies.
IMPORTANCE
This study uses a chemoproteomic approach to target active ATP-interacting proteins and examines the dynamic proteomic interactions between Staphylococcus aureus and human cell lines THP-1 and HaCaT. It uncovers the distinct responses of macrophages and keratinocytes during bacterial infection. S. aureus demonstrated a tailored response to the intracellular environment of each cell type and adaptation during exposure to professional and non-professional phagocytes. It also highlights strategies employed by S. aureus to persist within host cells. This study offers significant insights into the human cell response to S. aureus infection, illuminating the complex proteomic shifts that underlie the defense mechanisms of macrophages and keratinocytes. Notably, the study underscores the nuanced interplay between the host’s metabolic reprogramming and immune strategy, suggesting potential therapeutic targets for enhancing host defense and inhibiting bacterial survival. The findings enhance our understanding of host–pathogen interactions and can inform the development of targeted therapies against S. aureus infections.
KEYWORDS: Staphylococcus aureus, HaCaT cells, THP-1 cells, host–pathogen interactions, activity-based protein profiling (ABPP), ATP-interacting proteins, host immune response, bacterial metabolism
INTRODUCTION
In the complex interface of host–pathogen dynamics, Staphylococcus aureus infections across varied anatomical sites elicit a cascade of cellular responses. S. aureus causes conditions from minor skin irritations to life-threatening sepsis, altering its metabolism and virulence to thrive within host environments like macrophages and keratinocytes (1, 2). It counters host defenses by interrupting pathways like NF-κB and neutralizing reactive oxygen species (ROS) to protect itself from oxidative bursts and antimicrobial peptides (AMPs). In keratinocytes, S. aureus alters surface proteins to evade recognition and secretes proteases that cleave AMPs, rendering them ineffective (3–6). S. aureus expertly exploits host mechanisms, manipulating ATP-binding proteins to alter inflammatory responses, potentially exacerbating immune reactions, causing tissue damage, and promoting its spread (7).
Keratinocytes, which guard the epidermal barrier, often serve as the first line of defense against microbial intruders. Meanwhile, professional macrophages patrol the systemic environment, carrying out specialized immunological functions, including phagocytosis and initiation of the adaptive immune response to eliminate pathogens (8). Bacterial infections trigger complex interactions between various immune cells and non-immune cells, utilizing distinct yet complementary mechanisms to recognize and eliminate pathogens. Professional phagocytes, notably macrophages, are pivotal in phagocytosis, antigen presentation, and directing immune responses whereas non-professional phagocytes, like keratinocytes, represent non-immune cells that are crucial in the defense against pathogens (9). Macrophages utilize pattern recognition receptors (PRRs) such as Toll-like receptors and Nod-like receptors to identify bacterial pathogen-associated molecular patterns and initiate phagocytosis (10, 11). Inside these phagosomes, enzymes like NADPH oxidase generate ROS, complemented by lysosomal acidification and proteases to eradicate bacteria (9, 12). Keratinocytes rely mainly on PRRs to produce AMPs and cytokines and chemokines to recruit immune cells. AMPs, particularly LL-37, can directly disrupt bacterial membranes (10, 13, 14). Upon PRR activation, both cell types trigger the NF-κB and inflammasome pathways, leading to an inflammatory response and the release of cytokines such as IL-1β and IL-18, highlighting their collaborative yet distinct contributions to immune defense (15, 16).
Macrophages detect pathogens and respond by reprogramming their metabolic pathways, known as the “immunometabolic shift.” This shift facilitates various immune functions and pivots on the availability and utilization of ATP, a crucial energy storage molecule in living organisms (17, 18). ATP also plays a critical role in the signaling processes orchestrating these metabolic adjustments in both bacteria and host cells during infection. These metabolic switches are closely associated with specific ATP-interacting proteins that coordinate essential biochemical reactions (18, 19). Given their importance in cellular functions, these ATP-interacting proteins have been considered key targets for antimicrobial development.
Unveiling the active fraction of ATP-interacting proteome during S. aureus infection in human cells through activity-based protein profiling can revolutionize our understanding of host–pathogen interactions. This chemoproteomic technique utilizes activity-based probes to capture and investigate the functional state of enzymes, transcending the limitations of transcriptomics and conventional proteomics by focusing on enzymatic activity rather than mere presence. The targeted enrichment involved in this chemoproteomic approach is also beneficial for identifying low-abundance proteins and functional characterization of hypothetical proteins (20). Nucleotide acyl phosphate probes are utilized for selective, covalent labeling and isolation of active kinases, ATPases, and other ATP-binding proteins that are pivotal in human cellular processes, including both healthy and cancerous cells, as well as in microbial systems. This technique has facilitated the mapping of signal transduction pathways and provided insights into the regulatory mechanisms governing cell proliferation, apoptosis, and energy metabolism (21–24).
In this study, we utilized a desthiobiotin-ATP probe for the targeted profiling of ATP-interacting proteins in S. aureus and human cells during intracellular infection. Specifically, we used HaCaT cells, an immortalized human keratinocyte cell line, and THP-1 cells, a human monocytic cell line (THP1), to explore the biosynthetic and metabolic pathways adopted by bacteria during infection. Furthermore, we examined the differential immune responses elicited by these keratinocytes (non-professional phagocytes) and monocytes (professional phagocytes). This comparative approach to understanding the host–pathogen interplay during S. aureus infection provides profound insights into the pathogen’s pathogenesis and elucidates the complex molecular mechanisms that regulate these interactions.
MATERIALS AND METHODS
Bacterial strains and growth conditions
The S. aureus strain used is the USA300_JE2, a derivative of a methicillin-resistant S. aureus variant USA300_FPR3757. All strains used in this study are detailed in Table S1. S. aureus transposon mutants used in this study were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus for distribution through BEI Resources, NIAID, NIH, as part of the following reagent: Nebraska Transposon Mutant Library Screening Array, NR-48501. For the infection of the human cell lines, the S. aureus strain USA300_JE2 was cultured in tryptic soy broth (TSB) at 37°C at 200 rpm. The overnight cultures were subcultured in fresh TSB and grown at 37°C at 200 rpm to an optical density at 600 nm (OD600) of 1.0. Bacterial pellets were washed twice with ice-cold phosphate-buffered saline (PBS) and resuspended in Roswell Park Memorial Institute (RPMI) or Dulbecco’s modified Eagle’s medium (DMEM) depending on the experimental setup.
Complementation of transposon mutants
The primers for cloning the genes for complementation are detailed in Table S2. The open reading frames of the genes were amplified from S. aureus USA300_JE2 genomic DNA by PCR, using Q5 Hot Start High-Fidelity DNA Polymerase (New England Biolabs). The resulting PCR products were then ligated into the EcoRI/KpnI sites of the pCM29-sgfp vector using the ClonExpress II One-Step Cloning Kit (Vazyme). These constructs were transformed into Escherichia coli DH5α for screening and selection of the correct inserts by PCR. To improve the efficiency of transformation and overcome the restriction barrier, the recombinant plasmids were transformed into and extracted from E. coli IM01B. The resulting constructs were subsequently electroporated into the corresponding S. aureus transposon mutants, and the transformants were verified by PCR with plasmid-specific primers.
Cell culture
HaCaT cells (Cytion) were cultured in DMEM with 10% fetal bovine serum (FBS) (Sigma) in T75 plates. THP-1 cells (TIB-202, ATCC) were cultured in RPMI 1640 media with 10% FBS (Sigma) in T75 plates. All cells were grown to ~80% confluency at 37°C with 5% CO2. THP-1 monocyte differentiation was initiated by incubating the cells with 10 ng/mL phorbol 12-myristate 13-acetate (Sigma-Aldrich) for 2 days at 37°C in a 5% CO2 environment. Following this 2-day differentiation period, the culture medium was aspirated, and the cells were washed three times with PBS. Subsequently, the cells were further incubated for 1 day in fresh RPMI medium supplemented with 10% FBS.
Infection of human cell lines for protein profiling
THP-1 macrophages and HaCaT cell lines were infected with the S. aureus USA300_JE2 at a multiplicity of infection (MOI) of 10 or 100, depending on the experimental setup. Two parameters were considered for the choice of MOI: to ensure sufficient bacterial load for detecting intracellular bacterial proteins and to avoid significant cytotoxic effects on human cell lines. An MOI of 10 was sufficient for general infection assays, while an MOI of 100 was effective for protein profiling, allowing adequate colonization and protein accumulation without significant cytotoxicity to HaCaT or THP-1 cell lines. The human cells were incubated with bacteria for 1 hour at 37°C in the respective media, RPMI 1640 media for THP-1 and DMEM for HaCaT. The non-internalized bacteria were removed by washing the host cells three times with PBS, followed by incubation with fresh complete media of RPMI 1640 or DMEM supplemented with 100 µg/mL gentamicin for 1 hour at 37°C. The media were removed, the cells were washed three times with ice-cold PBS, and the cell culture plates were placed on ice for protein extraction. As a negative control, the THP-1 cells, HaCaT cells, and bacteria were incubated separately in the DMEM and RPMI media.
Profiling of human and S. aureus ATP-interacting proteins using desthiobiotin-ATP probe
Three sets of samples were prepared for each cell type to investigate the interaction with S. aureus. For the first set, THP-1 cells and HaCaT cells were infected with S. aureus in RPMI 1640 and DMEM media, respectively. The second set comprised THP-1 and HaCaT cells cultured alone in their respective growth media. The third set consisted of S. aureus cultured independently under identical conditions to those of the THP-1 and HaCaT cells (Fig. 1A). Following infection, cells were scraped off the plates into ice-cold lysis buffer (25 mM Tris HCl, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 5% glycerol) with protease and phosphatase inhibitors (Pierce Protease and Phosphatase Inhibitor Mini Tablets, EDTA-free, Thermo Fisher). The cell suspension was transferred into 2-mL screw-cap tubes containing 100 µL of 0.1-mm glass beads and lysed with bead-beating. The cell lysates were centrifuged for 20 min at 4°C and 14,000 rpm. The supernatant (total lysate) was transferred to a new tube. The lysis buffer was exchanged for the reaction buffer (4 M urea + lysis buffer) using the Zeba Spin Desalting Columns (Thermo Fisher). Following buffer exchange, the lysate protein concentration was measured using the Qubit protein assay kit, and the protein concentration was adjusted to 2 mg/mL using the reaction buffer. Each sample was mixed with 10 µL of 1 M MgCl2 and incubated for 1 min at room temperature. The ActivX desthiobiotin-ATP probe (Thermo Fisher) was reconstituted in ultrapure water to 20 µM, and 10 µL was added to each sample followed by incubation for 10 min at room temperature. Following labeling, 50 mL of 50% high-capacity streptavidin agarose resin slurry was added to each sample and incubated for 1 hour at room temperature with constant mixing on a rotator. The samples were centrifuged at 100 × g for 1 min to pellet the resin. The resins were washed two times with 500 mL of the reaction buffer, and the bound protein was eluted by adding 2× Laemmli reducing sample buffer and boiled for 5 min.
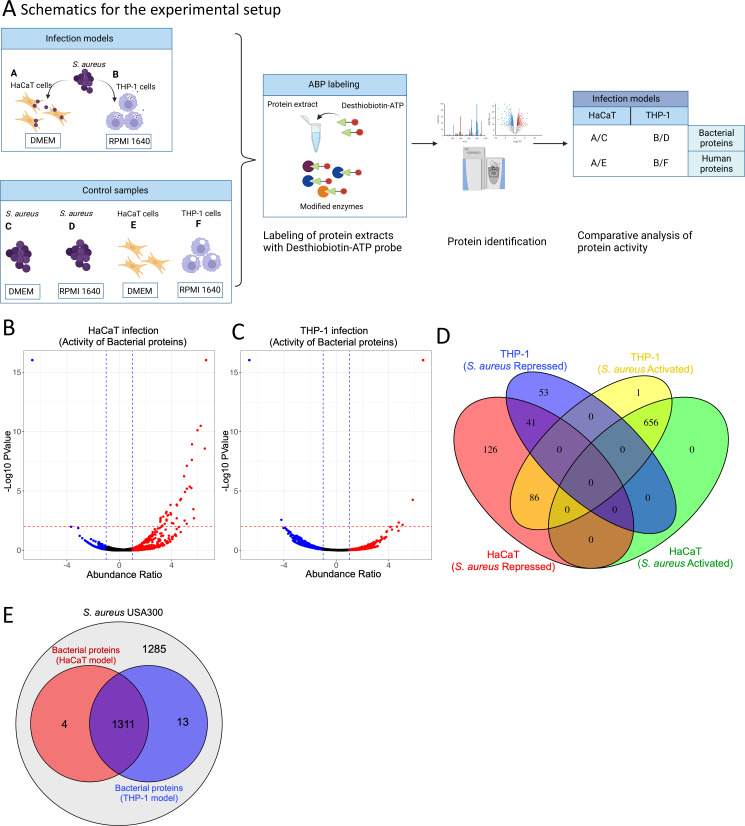
Fig 1.
(A) Schematic representation of the experimental setup including infection, ATP-interacting protein profiling, and data analysis. The diagram gives an overview of the S. aureus infection of THP-1 and HaCaT cells and the control cultures of the human cells and bacteria grown in isolation. Probe labeling of the protein extracts from the samples was performed using the desthiobiotin-ATP probe and the modified proteins identified using mass spectrometry. After infection, the differentially activated proteins in the S. aureus and host cells were identified by comparing them with the control samples, which are bacteria and human cell lines grown in isolation. (B and C) Activated and repressed proteins in S. aureus following infection in the HaCaT and THP-1 cells and chemoproteomic enrichment, respectively. The abundance ratio is expressed as a log2 value of the relative activity of the proteins. (D) Differentially activated and repressed S. aureus protein distribution following infection in the HaCaT and THP-1 cells. (E) The general protein profile derived from the ATP probe in the S. aureus cell following infection in the THP-1 and HaCaT cells and their overlap with the total proteome in the S. aureus USA300 genome using the hidden Markov model (HMM) profile derived from the UniProt database.
Mass spectrometry and data analysis
Protein samples were initially reduced with dithiothreitol and subsequently alkylated using iodoacetamide. Trypsin digestion was performed at a concentration of 6 ng/µL (Promega, #V511A). Post-digestion, peptides were purified with an Omix C18 tip (A57003100, Agilent), and the resulting eluates were dried via evaporation. For Liquid chromatography-mass spectrometry (LC-MS) analysis, the dried samples were reconstituted in 15 µL of 0.1% formic acid, and a quantity of 0.5 µg of peptides per sample was injected. The peptide mixtures, prepared in 0.1% formic acid, were loaded onto an EASY-nLC1200 system (Thermo Fisher Scientific) coupled with an EASY-Spray column (C18, 2 µm, 100 Å, 50 µm × 50 cm). A gradient of 5%–80% acetonitrile in 0.1% formic acid was applied to fractionate the peptides over a 60-min period at a flow rate of 300 nL/min. The fractionated peptides were then subjected to analysis on an Orbitrap Exploris 480 mass spectrometer (Thermo Scientific). Data acquisition was set to the data-dependent mode, employing a Top40 method. For protein identification, the acquired data were searched against the S. aureus USA300_FPR3757 and human proteome databases using Proteome Discoverer 3.0 software. The search parameters included a peptide mass tolerance of 10 ppm and a fragment mass tolerance of 0.02 Da. A stringent false detection rate (FDR) threshold of 5% was applied for peptide identification. To reinforce the reliability of the results, only peptides with at least two unique identifications were considered, and each sample was analyzed in triplicate to ensure reproducibility. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE (25) partner repository with the data set identifier PXD049003.
Functional annotation and pathway analysis
To functionally annotate proteins differentially regulated in S. aureus and human cell lines (HaCaT and THP-1) post-infection, we integrated EggNOG for orthologous group classification, KEGG pathway for metabolic and signaling roles, and gene ontology (GO) annotations. EggNOG provided insights into conserved domains and functions based on evolutionary history, while KEGG mapping highlighted their involvement in specific pathways. GO annotations were used to categorize proteins into biological processes (objectives the protein contributes to), molecular functions (biochemical activities), and cellular components (protein’s cellular location).
Classification of ATP-interacting proteins in S. aureus
Protein sequences of S. aureus were retrieved from the UniProt database. A comprehensive set of known ATP-binding motifs was compiled from literature and existing protein databases. These motifs were used to construct a HMM profile using the HMMER software suite to detect ATP-binding domains. The complete proteome of S. aureus USA300 FPR3757 genome and the total proteome from the HaCaT and THP-1 infection models were screened against the constructed HMM profile. This was aimed at identifying potential ATP-interacting proteins by matching the amino acid sequences of the proteins to the ATP-binding HMM profile.
Galleria infection assay
Larvae of Galleria melonella were obtained from Reptilutstyr AS (Norway). For infection experiments, S. aureus strains were grown overnight in TSB, washed twice in PBS, and suspended in PBS to a final concentration of 1.0 × 105 CFU/mL. Larvae of approximately equal weight were inoculated with 20 µL of this bacterial suspension, resulting in an infection dose of 1 × 103 CFU/larva. Bacteria were injected into the hemocoel of the larvae between the last pair of legs using a 30-G syringe microapplicator (0.30 mm [30 G] × 8 mm, BD Micro-Fine Demi). As a control, larvae were mock-inoculated with 20 µL of a PBS solution. Survival of the larvae was observed every 3 hours at 37°C. Larvae were regarded dead when they were not moving upon repeated physical stimulation.
Time-course human cell line infection assay
For the time-course infection assay, THP-1 and HaCaT cells were inoculated with the S. aureus strains at a MOI of 10. The THP-1 and HaCaT cells were cultured for 1 hour with the bacteria in RPMI 1640 and DMEM, respectively, without antibiotics. To eliminate non-internalized bacteria, the cells were washed thrice with PBS and then incubated in fresh complete media containing 50 µg/mL gentamicin. At designated time points, the media were discarded, the cells were washed with PBS and lysed with 0.01% Triton X-100 in PBS, and the lysates were serially diluted and plated on Mueller–Hinton agar. After 24 hours of incubation at 37°C, the bacterial colonies were counted.
Time-course human cell line cytotoxicity assay
THP-1 and HaCaT cells were infected with the S. aureus strains at a MOI of 10 for 1 hour within RPMI 1640 and DMEM, respectively. Subsequently, cells were washed thrice with PBS to eliminate non-internalized bacteria. They were cultured in fresh RPMI 1640 or DMEM media, each supplemented with 50 µg/mL gentamicin, throughout the time-course assay. Post-incubation, the media were aspirated, cells were washed thrice with PBS, and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide reagent was diluted in the respective cell culture media and added to the cells, followed by incubation at 37°C for 2 hours. The wells were washed twice with PBS, and dimethyl sulfoxide (DMSO) was added to solubilize the formazan crystals. Alongside these samples, control assays with uninfected cells were conducted to ensure assay validity (Fig. S1). The absorbance at 520 nm was measured using a BioTek Synergy H1 plate reader.
Statistical analysis
Normality and homogeneity of variance were assessed using Shapiro–Wilk and Levene’s tests, respectively. For data that were normally distributed with homogenous variances, a one-way analysis of variance (ANOVA) followed by Dunnett’s post hoc test was used for comparisons against the wild type (WT). Data that were not normally distributed were analyzed using the Kruskal–Wallis test followed by Dunn’s post hoc test for multiple comparisons.
RESULT AND DISCUSSION
Profiling of ATP-interacting proteins in S. aureus
Employing a desthiobiotin-ATP probe facilitated the targeted investigation of proteins exhibiting altered activity profiles during S. aureus infection in human keratinocyte (HaCaT) and monocyte (THP-1) cell lines. Proteomic samples from these cells, both independently cultured and infected with S. aureus, were labeled with the probe for enrichment and subsequent analysis. By comparing the proteomic data from the infected state to that of cells grown under controlled media conditions, we delineated the activity profile of both bacterial and human proteins (from HaCaT and THP-1 cells) that were activated or repressed during infection (Fig. 1A). To enhance the selectivity of our chemoproteomic analysis using the desthiobiotin-ATP probe, which exhibits potential cross-reactivity with various metabolic enzymes and chaperones, we implemented a stringent exclusion criterion. Proteins exhibiting consistency with the predetermined threshold cut-offs in all biological replicates were retained. This selective approach yielded a set of proteins with consistent activity across the experimental conditions, facilitating accurate comparative analysis across our experimental and control data sets. The combined data sets (Table S3) show the bacterial proteins resulting from the verification of activity-dependent protein interactions in our study.
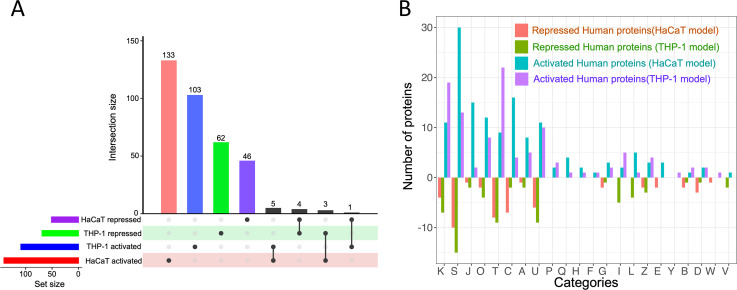
S. aureus exhibited distinct protein activation profiles when infecting the HaCaT and THP-1 cells (Fig. 1B through D). While a significant number of proteins (656) were activated in both HaCaT and THP-1 cells, only 41 proteins were commonly repressed across both cell models. Interestingly, 86 proteins activated in THP-1 cells corresponded with those repressed in HaCaT cells. In contrast, the THP-1 model showed repression of 53 bacterial proteins, whereas in the HaCaT cell, 126 S. aureus proteins were repressed, indicating unique bacterial responses to different host cell environments (Fig. 1D). Furthermore, approximately half of the proteins from S. aureus USA300 were identified in the infection models using the ATP probe (Fig. 1E).
To categorize the differentially activated protein sets identified using the ATP probe, we developed an HMM using data from the UniProt database, focusing on well-known bacterial protein domains that interact with ATP or nucleotides. Subsequently, we organized the proteins from each model into their respective functional or structural categories. Among the identified proteins, a substantial group comprised proteins with nucleotide-binding domains, with the majority showing no overlap with proteins possessing well-defined ATP-interacting domains (Fig. S2). The overlapping classification of ATP-interacting proteins can be attributed to certain proteins containing multiple domains. Furthermore, enriched proteins that did not match any of the defined ATP- or nucleotide-binding domains were classified as “Unassigned,” which mainly encompassed hypothetical proteins, proteins with poorly annotated domains, and metabolic enzymes. The molecular basis for the interaction of these proteins with the ATP probe is unclear, and it cannot be ruled out that the enrichment of some of these targets is the result of non-specific interactions, in which case the observed differences between different biological conditions represent differences in “abundance” rather than “activity” (Fig. S2). It is also possible that some proteins may have been enriched indirectly based on their interaction with other ATP probe-labeled enzymes within molecular complexes. However, nucleotide acyl phosphate probes have been shown previously to reveal novel ATP-binding sites where ATP might act as allosteric regulators of enzyme function (26). Allosteric control of enzyme activity through ATP is common for metabolic enzymes such as phosphofructokinases PfkA (27, 28) and might also be a potential novel function for unassigned proteins in our data set. The specificity of these interactions may be addressed in follow-up studies, e.g., using a competition assay with ATP and other nucleotides (26, 29). The unassigned proteins provide valuable insights into uncharacterized proteins with ATP-interacting functions, potentially shedding light on their roles in metabolism, stress responses, and host–pathogen interactions.
Functional classification of bacterial enzymes
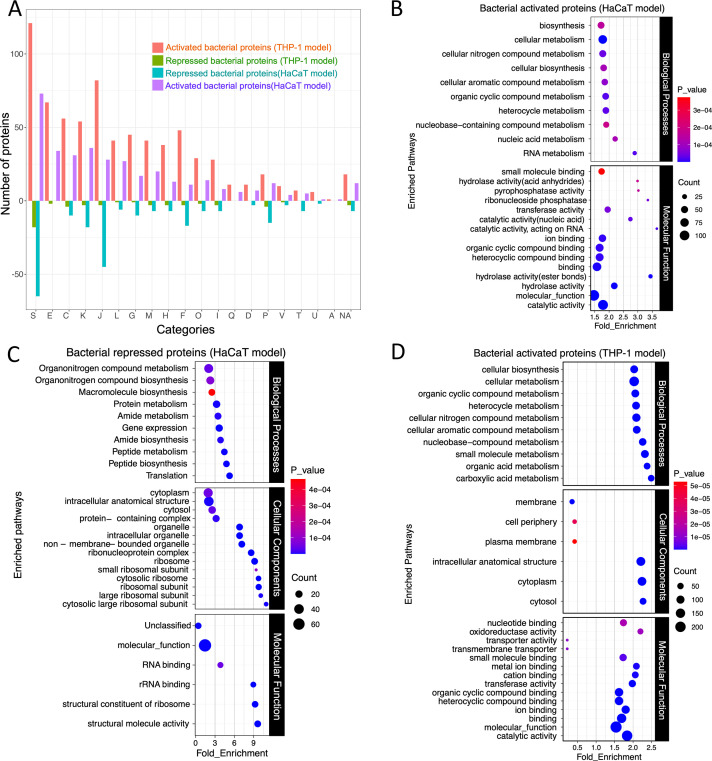
We employed a comprehensive approach combining enrichment analysis, GO, and cluster of orthologous groups (COG) analysis for a broader perspective on protein functions by considering evolutionary relationships, metabolic pathways, and molecular interactions. Based on the COG analysis, the enrichment of proteins in categories includes amino acid transport and metabolism (E), energy production and conversion (C), transcription (K), translation and ribosomal biogenesis (J), replication, recombination, and repair (L), cell wall/membrane biogenesis (M), and carbohydrate transport and metabolism (G) (Fig. 2A). The comparative analysis using the COG categories of the activated and repressed bacterial proteins from the HaCaT and THP-1 infections shows that specific functional categories are more affected by the bacterial presence in one cell type over the other, which indicates a targeted bacterial strategy for adapting to specific host cellular environments. Specifically, it was observed that in the THP-1 model, there is a higher number of activated bacterial proteins in most categories than in the HaCaT model. On the other hand, in the HaCaT model, there are more repressed proteins as compared to the THP-1 model (Fig. 2A), suggesting cell-type-specific interactions and bacterial response strategies, which could also indicate a strategic bacterial adaptation to the intracellular environment of that specific host cell type. For example, the category related to transcription (K) and translation (J) shows a substantial number of repressed proteins in the HaCaT model (Fig. 2A). This repression could reflect S. aureus adaptive response by downregulating proteins involved in transcription and translation to conserve energy and resources or avoiding the host’s immune detection mechanisms.
Fig 2.
(A) COG analysis of the activated and repressed S. aureus proteins following infection in the THP-1 and HaCaT cells and chemoproteomic enrichment. The length of the up and down bars represents the number of activated and repressed proteins, respectively. The bar chart illustrates the differential activation of S. aureus proteins categorized by COG during infection in the THP-1 and HaCaT cells. Each bar represents the degree to which proteins within a specific COG category are either activated or repressed. The x-axis labels, denoted by single-letter codes, correspond to distinct functional COG categories. The GO analysis of the (B) bacterial activated and (C) repressed proteins following infection in the HaCaT cells and (D) the bacterial activated proteins following infection in the THP-1 cells.
Additional GO analysis of the differentially active bacterial proteins from HaCaT and THP-1 infection models revealed enrichment in metabolic pathways utilizing various biomolecules such as carboxylic and organic acids and organic substance biosynthesis (Fig. 2B and D). The distribution of molecular functions in S. aureus proteins after infection in HaCaT and THP-1 indicates their diverse roles, including catalytic activities, transport functions, and binding activities, suggesting involvement in enzymatic reactions, molecule transport, and interactions with ions and small molecules (Fig. 2B through D). During infection in the HaCaT cells, most enriched bacterial processes included proteins involved in catalytic activities, RNA-related functions, phosphatase activity, and acid anhydride hydrolase activity (Fig. 2B). In contrast, in the THP-1 infection, proteins involved in molecular functions such as transporter activity, oxidoreductase activity, nucleotide binding, and metal ion binding were prominent (Fig. 2B through D). These distinctions suggest that in the HaCaT model, the bacteria activate proteins related to enzymatic and RNA-related processes, while in the THP-1 infection, most activated proteins function in transport, redox reactions, and nucleotide interactions.
During infection in the HaCaT cells, the repression of bacterial proteins associated with translation, peptide, and protein metabolism when they are located intracellularly (Fig. 2C) suggests a downregulation of the bacterial protein synthesis machinery, which could conserve energy or divert resources toward other processes necessary for survival or immune evasion. This aligns with the activation of pathways involved in RNA and nucleic acid metabolism (Fig. 2B), indicating an increased focus on genetic regulation and adaptation by the bacterium. Additionally, the downregulation of bacterial molecular pathways like translation, peptide biosynthesis, gene expression, and macromolecule and organonitrogen compound biosynthesis during S. aureus infection in HaCaT cells indicates a shift in bacterial metabolic priorities (Fig. 2C).
Bacterial energy metabolic pathways during intracellular location
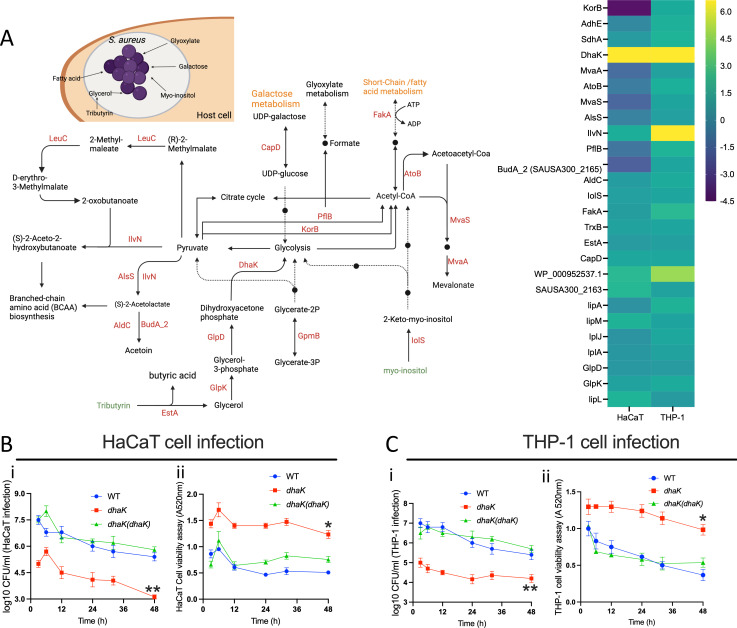
Investigation into bacterial protein-mediated metabolic signatures, prompted by the enrichment of energy production and conversion pathways during infections, revealed their critical role in catabolic pathways typical of intracellular pathogens. These pathways include fatty acid degradation, the tricarboxylic acid cycle, glycolysis, pyruvate metabolism, and formate production (Fig. 3A).
Fig 3.
(A) The metabolic map and heatmap show S. aureus proteins involved in energy metabolism and carbon source utilization after infection in HaCaT and THP-1 cells. The heatmap shows the relative activity of the bacterial proteins. The heatmap scale shows the log2 value for the protein activity. The map renders the pathways derived from the KEGG and EggNOG databases. (B and C) The time-course HaCaT and THP-1 infection and cell viability assays following infection with the S. aureus strains; the WT, transposon mutant of dhaK, and its in-trans complemented strain [dhaK(dhaK)]. The data represent the mean ± SD of three independent experiments. A P-value >0.05 is considered significant (*P < 0.01; **P < 0.001).
While some intracellular bacteria, such as Listeria monocytogenes and Mycobacterium tuberculosis, may prefer glycerol and fatty acids as carbon sources, respectively (30), our observations in S. aureus infecting both HaCaT and THP-1 cells indicate significant activation of DhaK, a dihydroxyacetone kinase involved in glycerol metabolism (31) (Fig. 3A and B). We also observed increased enrichment of EstA/FphF, an esterase that we recently identified in another chemoproteomic study using serine hydrolase-reactive fluorophosphonate probes (32, 33), but this esterase does not possess domains likely to directly interact with ATP. Increased activity of EstA and DhaK, associated with the utilization of tributyrin via glycerol, suggests that glycerol may serve as a significant carbon source for metabolic processes during intracellular infection. In support of this, in L. monocytogenes, a mutant strain lacking DhaK, which is defective in glycerol and dihydroxyacetone utilization, has been shown to exhibit a reduced intracellular replication rate (34, 35). Using our S. aureus strain background, we confirmed that a DhaK transposon mutant displayed a reduced ability to infect/replicate in both THP-1 and HaCaT cells [Fig. 3B(i) and C(i)]. Moreover, this mutation correlates with a decrease in cytotoxicity in these cell lines [Fig. 3B(ii) and C(ii)]. Complementation with DhaK-expressing plasmid rescued the phenotypes in both cell lines (Fig. 3B and C).
Proteins like CapD and IolS, involved in galactose and myo-inositol catabolism, respectively, were activated in both HaCaT and THP-1 infections (Fig. 3A), highlighting the bacterium’s ability to catabolize host-derived molecules to fuel its metabolic pathways (36, 37).
In both HaCaT and THP-1 cells, the activation of proteins for lipoic acid biosynthesis (LipA, LipL, and LipM) and scavenging (LplA, LplJ) was observed, with higher activity noted in THP-1 cells (Fig. 3A and B). This suggests that S. aureus relies on both its innate biosynthesis and external scavenging mechanisms for lipoic acid during infection, which is key for its defense against reactive oxygen and nitrogen species from macrophages, aiding in its evasion of immune clearance (38, 39). Additionally, in both HaCaT and THP-1 cell infection, bacterial protein FakA involved in fatty acid and acetate metabolism (40) showed increased activity, indicating a preference for fatty acid metabolism (Fig. 3A and B).
The enzyme PflB, which converts pyruvate to acetyl-CoA and formate under anaerobic conditions, was uniquely active in the THP-1 model (Fig. 3A and B), while KorB (SAUSA300_1183), also involved in pyruvate to acetyl-CoA conversion, was activated in THP-1 (41, 42). This dual activation of PflB and KorB for pyruvate metabolism could indicate metabolic flexibility or redundancy in the S. aureus approach to energy production, allowing it to adapt to the intracellular environment of the macrophages.
Furthermore, the enzymes AtoB, MvaS, and MvaA (Fig. 3A) play a crucial role in the mevalonate biosynthesis pathway and were found to be activated during infection in THP-1 cells but repressed during HaCaT infection (Fig. 3B). In S. aureus, mevalonate is essential for the primary metabolism of bacteria, and it is the only route for the biosynthesis of isoprenoids, which are crucial for cell wall formation, respiratory energy generation, and oxidative stress protection (43, 44).
Bacterial amino acid metabolism and transport during intracellular location
Intracellular bacteria like S. aureus can de novo synthesize or scavenge amino acids for metabolic energy, cellular integrity, and biosynthesis of vital compounds, including nucleotides and cell wall components (30, 45, 46).
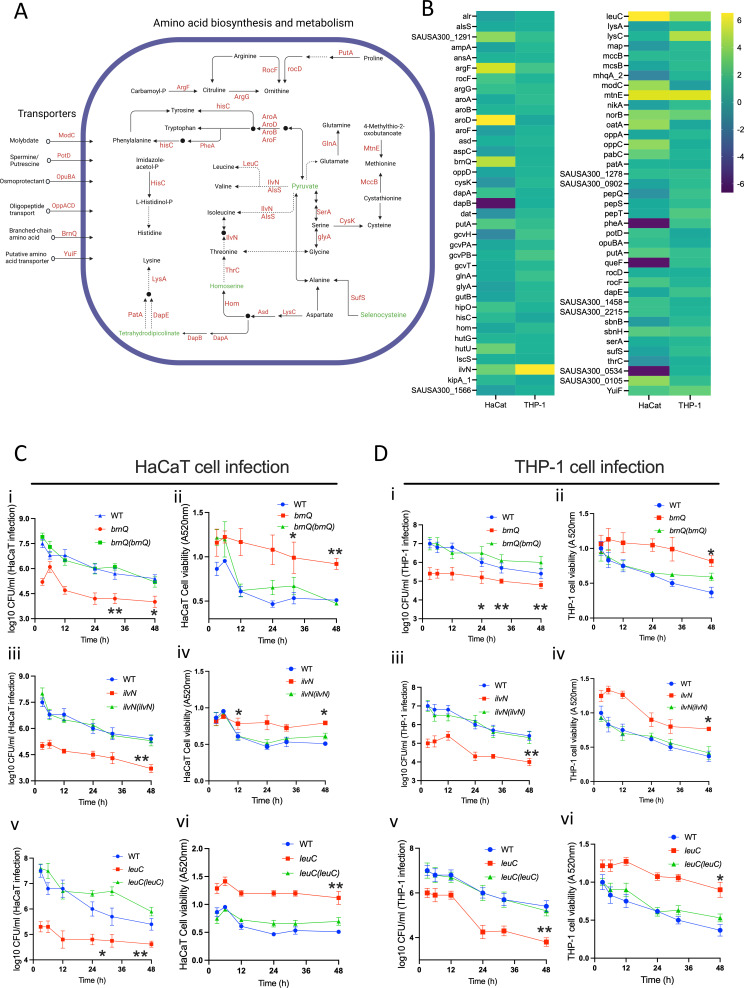
During HaCaT cell infection, we noted an elevated activity of the S. aureus amino acid transport system BrnQ (SAUSA300_1300), higher than during THP-1 cell infections (Fig. 4A and B). BrnQ is essential for the uptake of branched-chain amino acids, including leucine, isoleucine, and valine (47). To evaluate this, we infected HaCaT and THP-1 cells with WT S. aureus, a transposon mutant of brnQ and its complemented strain. The brnQ mutation markedly diminished S. aureus ability to infect both THP-1 and HaCaT cells over prolonged periods. This mutation simultaneously reduced the cytotoxic effects on the cells. The complementation of brnQ in these mutants restored their infective proficiency and cytotoxicity to wild-type levels [Fig. 4C(i and ii) and D(i and ii)], confirming the effect of the mutation.
Fig 4.
(A) A diagrammatic rendering of the amino acid synthesis and transport pathway involving bacterial proteins activated and repressed following infection in the THP-1 and HaCaT cells. (B) The heatmap shows the relative activity (log2 value) of bacterial proteins involved in the amino acid synthesis, metabolism, and transport pathway. (C and D) The time-course HaCaT and THP-1 infection and cell viability assay following infection with the WT S. aureus strains; transposon mutants of brnQ, IlvN, and leuC; and their corresponding complemented strains brnQ(brnQ), ilvN(ilvN), and leuC(leuC). The data represent the mean ± SD of three independent experiments. A P-value >0.05 is considered significant. *P < 0.01; **P < 0.001.
Simultaneously, IlvN and LeuC, involved in the biosynthesis of branched-chain fatty acids, were activated during infection in both HaCaT and THP-1 cells (Fig. 4A and B), indicating a dual strategy of biosynthesis and scavenging by the bacteria in response to intracellular location. The transposon mutants of both ilvN and leuC display reduced infection capabilities and reduced cytotoxicity in both HaCaT and THP-1 cell lines during prolonged infection [Fig. 4C(iii to vi) and D(iii to vi)]. IlvN, in conjunction with other activated proteins like AlsS, BudA, and AldC, plays a vital role in acetoin production (Fig. 3A; Fig. S3). This compound acts as a storage form of surplus carbon and energy and is generated during glucose fermentation. Additionally, it serves as an external electron acceptor during anaerobic conditions (48, 49).
AroD, a key enzyme in the shikimate pathway involved in the biosynthesis of aromatic amino acids (phenylalanine, tyrosine, and tryptophan), was highly enriched during HaCaT cell line infection compared to that in THP-1 cells (Fig. 4A and B). In some other bacteria, the AroD-catalyzed product 3-dehydroshikimate, combined with citrate and spermidine, contributes to synthesizing the siderophore petrobactin, which is, however, not reported for S. aureus (50). Yet, the high activation of PotD, a protein involved in spermidine transport, in bacteria during HaCaT infection may indicate a convergence of these metabolic pathways also in S. aureus (Fig. 4A and B). During both HaCaT and THP-1 infections, the AroA and AroB, which are responsible for downstream chorismate production (51), showed lower levels of activity. On the other hand, PheA, which is involved in phenylalanine biosynthesis, was activated during THP-1 infection but repressed during HaCaT infection. The MtnE and MccB involved in the methionine salvage pathway (52) exhibited similar activation levels during infection in both HaCaT and THP-1 cell lines (Fig. 4A and B).
The proteins GcvH, GcvT, GcvPA, and GcvPB, which are involved in glycine metabolism to lipoyllysine, showed increased activity during THP-1 infection compared to the HaCaT cell infection (Fig. 4B; Fig. S3). Lipoyllysine is essential for intermediary metabolism, including the tricarboxylic acid cycle, and is required for the breakdown of glucose and the generation of acetyl-CoA (53). Given the observed activation of proteins related to lipoic acid biosynthesis and salvage pathways in S. aureus during its infection of human cells (Fig. 3B; Fig. S3), it is likely that lipoic acid-dependent post-translational modifications, particularly those involving the glycine cleavage system, play an essential role in the bacterial survival strategy within the host cells.
Additionally, HutU, which is involved in histidine metabolism and L-glutamate, aspartate, and alanine biosynthesis (Fig. 4B; Fig. S3), and the RocF/Arg, which participates in arginine degradation to urea (54), were activated in the HaCaT infection model (Fig. 4B; Fig. S3).
Bacterial virulence and stress response during infection of keratinocytes and macrophages
Central to bacterial infections is the expression of specific proteins that play pivotal roles in virulence, direct damage to host tissues, orchestration of nutrient acquisition in nutrient-limited conditions, and the subversion of the host’s immune defenses. Here, SbnE, involved in staphyloferrin B production (55), and the major facilitator superfamily transporter NorB showed significant activation in both infection models (Fig. S3). In the HaCaT infection model, S. aureus proteins such as RadA (a DNA repair protein) and SAUSA300_2299 (a multidrug resistance protein A transporter) were highly activated. PflB involved in response to NO stress (56) was activated during THP-1 infection, which aligns with macrophage reactive nitrogen species production (Fig. S3). Furthermore, proteins related to oxidative stress response, like BshB (involved in bacillithiol biosynthesis), LepA (influencing vacuolar modeling for host immune evasion), and RocF (involved in putrescine and urea production necessary for reactive nitrogen species detoxification), were identified. Additionally, proteins such as SAUSA300_2140, IsdB, and PerR, associated with iron-siderophore biosynthesis, were also found to be activated (Fig. S3). Beyond the well-known pathways enriched based on GO, KEGG, and COG categories, various characterized and uncharacterized bacterial proteins mediating stress response, virulence, and survival during infection showed differential activity in both models (Fig. S4).
Bacterial two-component systems activated during infection in HaCaT and THP-1 cells
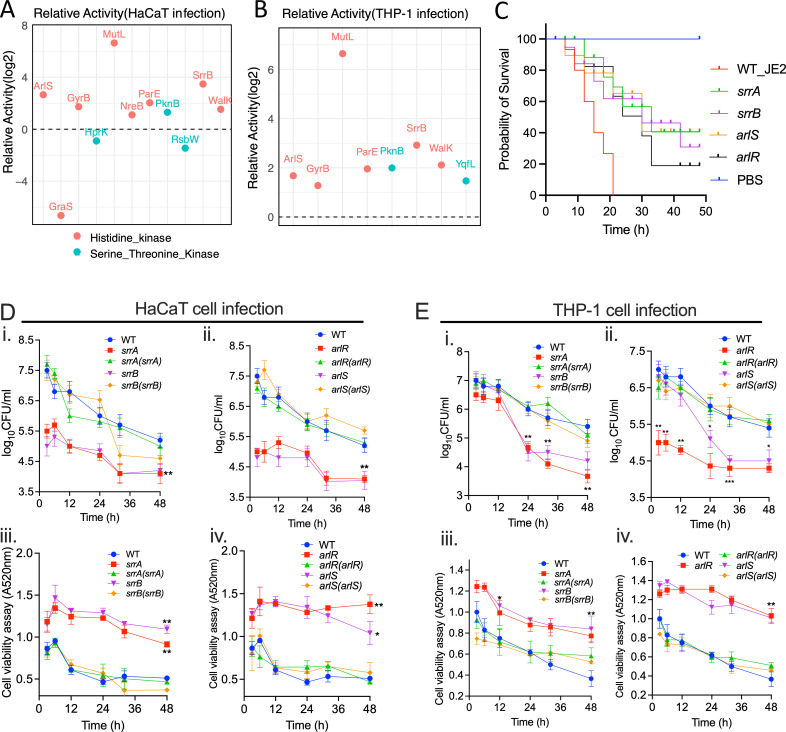
The bacterial two-component systems (TCSs) and other kinases sense the environment, enabling bacteria to adapt accordingly. Profiling the bacterial proteins allowed us to identify a set of histidine kinases including some belonging to the TCS, as well as serine-threonine kinases in S. aureus.
In the HaCaT infection model, the TCS sensor kinases ArlS, NreB, SrrB, and WalK exhibited increased activity, whereas the GraS was repressed (Fig. 5A). Furthermore, during infection in THP-1 cells, the histidine kinases ArlS, SrrB, and WalK were upregulated (Fig. 5B). Notably, the accessory component YycI, which forms a complex WalKR-YycHI to regulate virulence determinants and stress response pathways in S. aureus, showed increased activity in both infection models (Fig. S3). Using the Galleria melonella infection model, we confirmed that transposon mutants deficient in the histidine kinase or response regulators of the SrrAB and ArlSR showed increased survival of infected larvae compared to WT strains (Fig. 5C). To further investigate the role of the TCS in host infection, we infected THP-1 and HaCaT cells with transposon mutants and complements of the TCSs, SrrAB and ArlSR. During the prolonged infection, we observed that the mutants of both sensor kinases and response regulators exhibited reduced infection capacity and cytotoxicity in both cell lines. However, this attenuated virulence was restored to the WT level in their respective complemented strains (Fig. 5D and E). The WalKR, an essential TCS required for S. aureus viability (57), was not included in the virulence analysis due to the unavailability of corresponding mutants. In S. aureus, the WalKR regulates enzymes involved in the synthesis and degradation of the bacterial cell wall, which is essential for maintaining cellular integrity, cell shape, and cell division (57, 58). It has been shown to elicit host inflammatory responses by controlling autolytic activity (59, 60). Additionally, it helps the bacteria adapt to environmental stresses, such as changes in osmotic pressure or exposure to antibiotics that target the cell wall (61–63). The WalKR system can also influence the expression of virulence factors and play a role in biofilm formation (58, 60).
Fig 5.
Relative activity of S. aureus histidine kinases and serine/threonine kinases following infection in (A) HaCaT cells and (B) THP-1 cells. (C) Galleria melonella survival following infection by the wild-type S. aureus USA300_JE2 and the transposon mutants of the TCSs SrrAB and ArlSR. PBS was used as a control. (D and E) The time-course HaCaT and THP-1 infection and cell viability assay following infection with the WT S. aureus strains; transposon mutants of srrA, srrB, arlR, and arlS; and their corresponding complemented strains srrA(srrA), srrB(srrB), arlS(arlR), and arlR(arlR). The data represent the mean ± SD of three independent experiments. A P-value >0.05 is considered significant. *P < 0.01; **P < 0.001.
In both infection models, S. aureus serine-threonine kinase PknB was active, whereas YqfL activation was specific to THP-1 cell infections. Conversely, HprK, a serine kinase, and RsbW, a serine-threonine-protein kinase, were slightly suppressed during HaCaT cell infection (Fig. 5A and B). These kinases are vital for regulating cellular processes; for example, HprK regulates carbon metabolism; PknB is tied to cell growth, division, and cellular quiescence; and RsbW is associated with the stress response (64–67). Also, the GyrB, ParE, and MutL were all activated in bacteria during infection in both cell lines (Fig. 5A and B). ParE and GyrB are both involved in the manipulation of DNA supercoiling and are part of topoisomerase complexes, whereas MutL is involved in the repair of DNA mismatches (68, 69). All three are essential for the maintenance and regulation of the bacterial genome integrity.
Distribution and functional categorization of human proteins in mammalian cells infected with S. aureus
After S. aureus infection, THP-1 and HaCaT cells displayed cell-specific responses in their profile of activated and repressed proteins (Fig. 6A). Functional categorization revealed that a significant number of proteins are associated with categories such as K (transcription), T (signal transduction mechanisms), J (translation, ribosomal structure, and biogenesis), C (energy production and conversion), and O (post-translational modification, protein turnover, and chaperones) (Fig. 6B) reflecting the host’s effort to control the infection by regulating immune responses, metabolic adaptation, and ensuring protein functionality. Interestingly, there were minimal overlaps in the activated and repressed proteins between the two cell lines (Fig. 6A and B), signifying that while there are some common elements in response to S. aureus infection, each cell type also employs unique strategies that are likely tailored to their specific roles in the immune system. The combined data sets (Table S4) show the THP-1 and HaCaT proteins resulting from the verification of activity-dependent protein interactions in our study.
Fig 6.
(A) Distribution of activated and repressed human proteins from HaCaT and THP-1 cells following S. aureus infection. (B) This bar chart illustrates the differential activation of human proteins categorized by COG in THP-1 and HaCaT cells in response to S. aureus infection. Each bar represents the degree to which proteins within a specific COG category are either activated or repressed. The x-axis labels, denoted by single-letter codes, correspond to distinct functional COG categories.
Functional profiling of HaCaT proteins in response to intracellular S. aureus infection
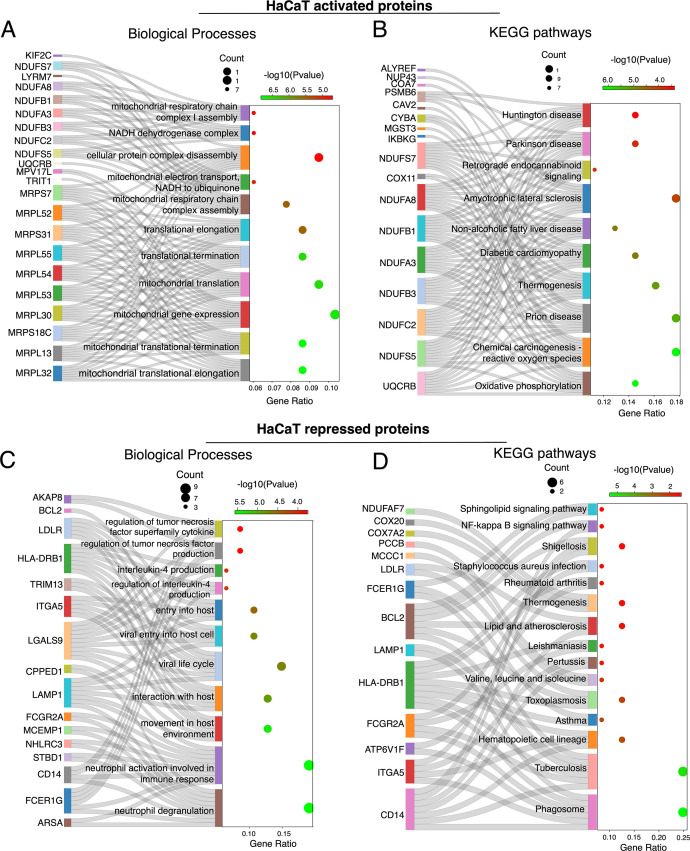
The gene ontology and biochemical pathway analysis of the differentially activated proteins from the HaCaT cell following S. aureus infection profoundly impacted cellular energy metabolism and signaling. Many of the activated proteins in the HaCaT cells were involved in mitochondrial processes, including mitochondrial translation, gene expression, respiratory chain complex assembly, and electron transport (Fig. 7A). This suggests that S. aureus infection in HaCaT cells may lead to an increase in ATP production or indicate mitochondrial stress or damage induced by the bacteria. Also, the enrichment in pathways related to oxidative phosphorylation, thermogenesis, and electron transport to ubiquinone suggests an alteration in energy metabolism and potential oxidative stress in the infected cells (70–72) (Fig. 7A and B). The activation of the chemical carcinogenesis-reactive oxygen species pathway in HaCaT cells during S. aureus infection involved the CYBA, which is involved in ROS production, and MGST3, which protects against oxidative stress (73–75) and IKBKG directly involved in the activation of the NF-κB pathway (Fig. 7B) (15, 76). Various components of the mitochondrial electron chain transport components and NADH dehydrogenase complex (NDUFS7, NDUFA8, NDUFA3, NDUFB1, NDUFC2, and NDUFS5) (Fig. 7A and B) indicate that the cells are potentially trying to eliminate the bacteria through oxidative stress and the energy demands required for immune response (77–80).
Fig 7.
Sankey dot plot showing the flow of activated proteins to their respective (A) biological processes and (B) KEGG pathway enrichment in HaCaT cells following S. aureus infection. (C) Sankey dot plots depicting the repressed proteins and their corresponding biological processes and (D) KEGG pathway enrichment in HaCaT cells following S. aureus infection. The dot size indicates gene count, and color intensity represents the −log10(P-value).
Annotations for cellular components and molecular functions showed a strong influence on mitochondrial aspects, including the mitochondrial inner membrane, mitochondrial ribosome, mitochondrial respiratory chain complexes, and NADH dehydrogenase complex I (Fig. S5A and B), indicating that the infection has a significant impact on energy metabolism and ATP demand. Pathways related to oxidoreductase activity, particularly those affecting NAD(P)H and enzymes like glutathione peroxidase, suggest potential redox imbalance and oxidative stress (74, 75). Additionally, pathways for binding activities such as transferrin receptor and long-chain fatty acid binding indicate shifts in cellular transport and metal ion homeostasis (Fig S5A and B). In general, the activated protein in the HaCaT cells indicates that S. aureus may either disrupt mitochondrial energy production or provoke the cell to enhance mitochondrial function as a protective measure.
Conversely, proteins repressed in HaCaT cells post-infection were significantly enriched in pathways related to immune response alterations, intracellular trafficking, and energy metabolism (Fig. 7C and D). This included the downregulation of neutrophil-related pathways (neutrophil activation involved in immune response and neutrophil degranulation) and cytokines like interleukin-4 and TNF and the repression of proteins involved in the phagosome pathway (Fig. 7C and D) (81, 82). Additionally, the repression in pathways annotated for shigellosis, tuberculosis, and toxoplasmosis and even S. aureus infections (involving BCL2, FCGR2A, HLA-DRB1, FCER1G, CD14, and LAMP1 that have functions in apoptosis, phagocytosis, and inflammatory response) (Fig. 7D) may reflect a deviation of the immune system induced by S. aureus (81–85).
Interestingly, the ATP-interacting protein profiles of the host and bacteria give evidence for their metabolic interdependencies that might be indicative of the competition for resources. Whereas S. aureus upregulates BrnQ branched-chain amino acid (BCAA) transport systems, likely to sequester BCAAs for its growth (Fig. 4A and B), the repression of MCCC1 and PCCB (Fig. 7D), which are crucial for BCAA catabolism, suggests that HaCaT cells try to conserve BCAAs for immune function and tissue repair (86, 87).
Finally, the repression of cellular components related to endocytic vesicles, secretory granules, and lysosomes could impact antigen processing and pathogen defense, while the downregulation of binding activities such as immunoglobulin and ubiquitin binding may affect immune recognition and protein degradation processes (Fig. S5C and D).
Functional profiling of THP-1 proteins in response to intracellular S. aureus infection
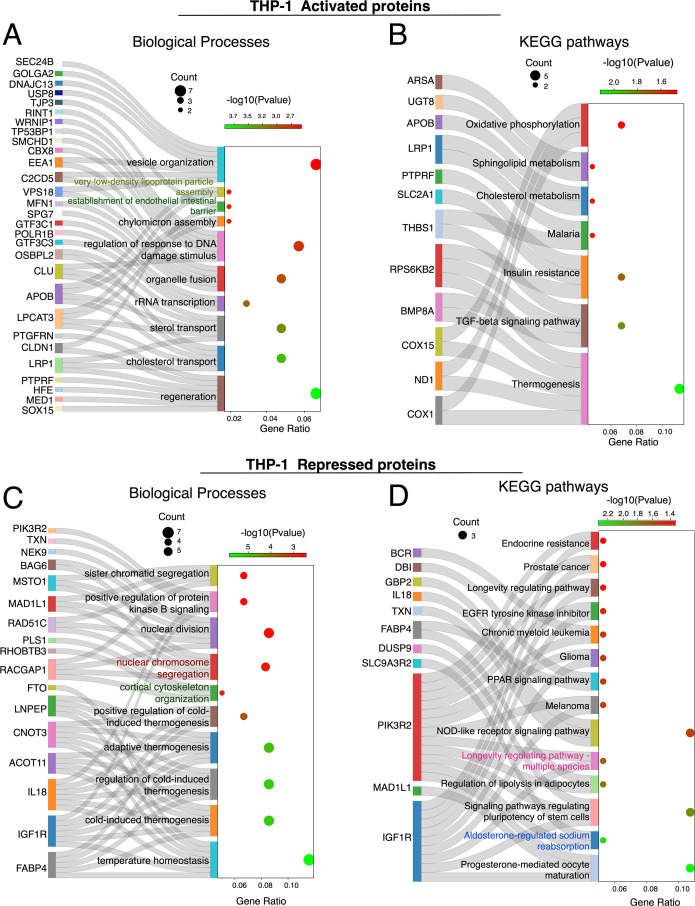
THP-1 cells responded to S. aureus infection through changes in lipid metabolism, DNA integrity, and immune response. The enriched pathways included those related to cholesterol and sterol transport, suggesting active modulation of lipid metabolism during infection (Fig. 8A and B). Cholesterol and sphingolipid metabolism are essential for membrane structure, signaling pathways, and apoptosis (88). Additionally, WRNIP1, TP53BP1, CLU, SMCHD1, CBX8, and RINT1 were chemoproteomically enriched in S. aureus-infected THP1 cells (Fig. 7E). These proteins are involved in responding to DNA damage, chromatin regulation, and double-strand break repair, indicating potential DNA damage due to bacterial invasion or modulation by the host to limit bacterial replication (89–93). Furthermore, proteins like SPG7, MFN1, VPS18, C2CD5, EEA1, USP8, DNAJC13, GOLGA2, and SEC24B were enriched (Fig. 8A), and these are critical for organelle fusion and vesicle organization and play vital roles in cellular processes such as autophagy, apoptosis, lysosomal degradation of pathogens, antigen presentation, and cytokine release, all essential for the immune system’s defense against bacterial infection (94, 95).
Fig 8.
Sankey dot plot illustrating the activated proteins and their related (A) biological processes and (B) KEGG pathway enrichment in THP-1 cells following S. aureus infection. (C) Sankey dot plots of repressed proteins and their biological processes and (D) KEGG pathway in THP-1 cells following S. aureus infection. The dot size indicates gene count, and color intensity represents the −log10(P-value).
Further analysis of activated THP-1 proteins revealed their roles in lipid transport and metabolism involving proteins such as CLU, OSBPL2, and APOB (96–98), cell membrane dynamics (early endosome, coated vesicle, vesicle tethering complex, vesicle lumen, and desmosome), oxidoreductase activities, and heme-copper terminal oxidase activity (Fig. S6A and B), which are associated with mitochondrial function, the electron transport chain, cellular energy metabolism, and responses to oxidative stress (99, 100). Regarding energy production and metabolism, COX1, ND1, and COX15 play roles in energy metabolism and mitochondrial function (101, 102) (Fig. S6A and B).
The repressed proteins in THP-1 cells during S. aureus infection might indicate disruptions in cellular processes including metabolism, cell division, signaling, and intracellular trafficking. While cold-induced or adaptive thermogenesis typically involves converting energy into heat through proteins like ACOT11, FABP4, IL18, IGF1R, CNOT3, and FTO (103–107) (Fig. 8C), this process bypasses ATP synthase, releasing energy as heat rather than ATP (108, 109). However, due to the energy demands of the THP-1 cell response to bacterial infection, processes like thermogenesis, which consume energy without ATP production, are suppressed (108). Thus, during infection, the cell prioritizes energy pathways that support ATP production, crucial for powering immune responses. This metabolic shift underscores the cell’s adaptive ability to ensure survival during infection-induced stress.
The KEGG pathway analysis of the THP-1 repressed proteins revealed enrichment in pathways such as progesterone-mediated oocyte maturation, endocrine resistance, melanoma, glioma, chronic myeloid leukemia, prostate cancer, EGFR tyrosine kinase inhibitor resistance, and longevity regulating pathways, primarily involving proteins PIK3R2 and IGFIR linked to the PI3K/Akt signaling pathway (Fig. 8D) (110). Furthermore, proteins related to vesicles and endosomes, critical for cellular trafficking, material exchange, and DNA replication, were suppressed during bacterial infections (Fig. S6D). This may reflect a bacterial strategy to inhibit cell proliferation and potentially manipulate cell death to their advantage (111). Additionally, proteins facilitating molecular functions, such as GTPase and small GTPase binding, essential for intracellular trafficking, signaling, and cytoskeletal organization, were also repressed (Fig. S6C).
Immune and metabolic shifts in THP-1 and HaCaT cells during bacterial challenge
Protein mapping in host cells exposed to S. aureus reveals distinct, cell type-specific immune adaptations. THP-1 cells activated proteins associated with immune defense, autophagy, and inflammation. In contrast, HaCaT cells activated proteins that support cellular barrier integrity and trigger immune responses, reflecting tailored strategies for bacterial invasion (Fig. 9).
Fig 9.
(A) List of human immune response pathways. Heatmap and pathway mapping of differentially activated human proteins from the HaCaT and THP-1 cells following S. aureus infection. The heatmap scale shows the log2 value for the protein activity.
Upon examining the link between protein activity and pathways in THP-1 cells, we observed a prioritization of immune defense mechanisms, including enhanced pathogen clearance and immune activation, autophagic cell death, and inflammatory responses during pathogen exposure. Proteins such as TSP1, LPCAT3, BMP8A, TP63, PIK3R4, and KS6B2 play a crucial role in autophagy, apoptosis, and ferroptosis facilitating pathogen clearance, remove compromised cells, and limit the spread of infection (71, 112–116). Also, following exposure to pathogens, PERP, an apoptosis-inducing protein (117, 118), showed higher activation levels in THP-1 cells than in HaCaT cells (Fig. 9). Furthermore, proteins like INO1 and PIK3C2A for inositol synthesis, essential in cellular signaling, growth, proliferation, differentiation, survival, and intracellular trafficking, were activated in THP-1 cells upon infection (119, 120). Notably, myo-inositol also plays a role in depolarizing macrophages, enhancing phagocytosis (121), and inducing autophagy independently of the mammalian target of rapamycin (mTOR) signaling pathway (122)
Exposure to pathogens has been found to change the metabolic pathway of host cells. In this case, the repression of SLC2A1, a glucose transporter, in HaCaT cells and its activation in THP-1 cells reflect their metabolic needs for immune response (Fig. 9). SLC2A1 is involved in glycolysis by supporting glucose uptake and promoting anaerobic metabolism via the hypoxia-inducing factor (HIF) signaling pathway (123, 124). Therefore, by limiting glucose uptake, HaCaT cells can restrict the availability of glucose, which is a major carbon source for S. aureus metabolism. This innate defense strategy is an example of “nutritional immunity” (125). Additionally, PECR, involved in fatty acid oxidation, was repressed in THP-1 cells but activated in HaCaT cells (Fig. 9). This leads to a shift in the metabolic reprogramming of immune cells during infection from fatty acid metabolism to glycolysis, known as the Warburg effect (126, 127). Many intracellular pathogens have evolved to take advantage of the Warburg metabolism of the host cell or to push the host cell into a state of increased glycolysis (126, 127). The Warburg metabolism, mediated by the mTOR/HIF-1alpha pathway, is crucial for producing IL-22 and cytokines during intracellular infections (97, 98, 128).
Regarding nutritional immunity, host cells often limit iron availability to inhibit bacterial proliferation. Both LPCAT3 (activated only in THP-1 cells) and HFE (activated in both HaCaT and THP-1 cells) (Fig. 9) proteins are involved in iron regulation mechanisms via the iron-dependent programmed cell death (ferroptosis) pathway (129–131).
HaCaT cells engage in mechanisms that mitigate excessive inflammation and cellular damage, preserving cellular integrity and the skin’s protective barrier. The upregulation of NEMO, PECR, and P2RX4, implicated in NF-κB, IL-17, peroxisome, and calcium signaling pathways (Fig. 8), demonstrates the HaCaT cells’ strategic response to S. aureus. This includes bolstering the immune response via the NF-κB pathway and enhancing antimicrobial and inflammatory actions through IL-17 signaling. NEMO, through NF-κB, is essential for keratinocyte survival, proliferation, and migration in response to various stressors such as cytokines and bacterial and viral products (132–134). In contrast to the THP-1 cells, the proteins PIK3R4, KS6B2, BMP8A, TSP1, LPCAT3, and TP63 were suppressed in HaCaT cells. These proteins are involved in autophagy, Salmonella infection, phagosome, ferroptosis, apoptosis, and the P53 signaling pathway (121). PECR, a peroxisome reductase, is involved in fatty acid synthesis (135), vital in forming the epidermal permeability barrier, and keratinocytes function as building blocks and energy generation, storage, and metabolism (136, 137). The activation of PECR in HaCaT cells (Fig. 9) may suggest an increased synthesis of fatty acids to maintain the keratinocyte layer disrupted by the infection. PNN located in epithelial cell desmosomes regulates cell integrity and adhesion (138). Host factors such as NEAT-1, associated with PNN, trigger macrophage inflammation (139, 140). Silencing NEAT-1 by ALYREF repression reduces ATP and mitochondrial energy production (141). Additionally, PNN overexpression creates a stable adhesive state, impeding cell transition and affecting epithelial tissue structure and function (142). Hence, suppressing PNN can help HaCaT cells maintain the immune barrier and facilitate post-bacterial challenge recovery.
The differential activities of the proteins in both HaCaT and THP-1 cells could result from the bacteria’s modulation of host immune processes as an immune evasion mechanism. For instance, disrupting the tight junction due to the repression of CLD1 and ZO-3 in HaCaT cells (Fig. 9) can have immediate implications for cell integrity and could represent the success of bacteria in compromising barriers and facilitating invasion and progression of infection. S. aureus is known to disrupt tight junctions to facilitate invasion and spread (143, 144). Also, various bacteria have been reported to subvert host defenses to promote inflammation, depending on the bacterial needs and the phase of infection by targeting the NF-κB pathway (129).
Conclusion
This study delves into the interplay between ATP-interacting proteins in S. aureus and human cells, specifically THP-1 macrophages and HaCaT keratinocytes, during an infection. By employing chemical probes designed to target specific proteins selectively, our research unveils distinctive patterns of protein activation within S. aureus and human cells highlighting the strategies and adaptations during host–pathogen interactions. A graphical summary of the pathways and proteins activated during the S. aureus–human cell interaction is illustrated in Fig. S7.
These findings pinpoint potential targets for therapeutic interventions and contribute to a more comprehensive understanding of the dynamics that unfold during S. aureus infections. This knowledge serves as the foundation for developing interventions to disrupt bacterial virulence or fortify the host’s defense mechanisms, thereby aiding in the ongoing battle against antibiotic resistance. Moreover, the insights gained into S. aureus metabolic adjustments during infection hold promise for the creation of novel antimicrobials that target these pathways. Additionally, our research unveils strategies to modulate host immune responses, potentially leading to the development of immunomodulatory therapies that enhance the host’s defense mechanisms or mitigate damage caused by inflammation. The findings of this study open diverse avenues for future research, for a deeper exploration of molecular-level interactions between hosts and pathogens.
ACKNOWLEDGMENTS
This work was funded through a grant from the Centre for New Antibacterial Strategies (CANS) to K.H., M.J., and C.S.L. and a CANS start-up grant to C.S.L. Mass spectrometry-based proteomic analyses were performed by the UiT Proteomics and Metabolomics Core Facility (PRiME). This facility is a member of the National Network of Advanced Proteomics Infrastructure (NAPI), which is funded by the Research Council of Norway INFRASTRUKTUR-program (project number: 295910).
Nebraska transposon mutant library (NTML) was provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution through BEI Resources, NIAID, NIH: NTML Screening Array, NR-48501.
Contributor Information
Stephen Dela Ahator, Email: Stephen.d.ahator@uit.no.
Mona Johannessen, Email: mona.johannessen@uit.no.
Kathryn C. Milligan-McClellan, University of Connecticut, Storrs, Connecticut, USA
DATA AVAILABILITY
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (25) partner repository with the data set identifier PXD049003.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/msystems.00179-24.
Fig. S1 to S7.
Strains.
Primers.
Activity of S. aureus proteins.
Activity of human proteins.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Brown AF, Leech JM, Rogers TR, McLoughlin RM. 2014. Staphylococcus aureus colonization: modulation of host immune response and impact on human vaccine design. Front Immunol 4:507. doi: 10.3389/fimmu.2013.00507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bien J, Sokolova O, Bozko P. 2011. Characterization of virulence factors of Staphylococcus aureus: novel function of known virulence factors that are implicated in activation of airway epithelial proinflammatory response. J Pathog 2011:601905. doi: 10.4061/2011/601905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Foster TJ. 2005. Immune evasion by staphylococci. Nat Rev Microbiol 3:948–958. doi: 10.1038/nrmicro1289 [DOI] [PubMed] [Google Scholar]
- 4. De Jong NWM, Van Kessel KPM, Van Strijp JAG. 2019. Immune evasion by Staphylococcus aureus. Microbiol Spectr 7:2–7. doi: 10.1128/microbiolspec.GPP3-0061-2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wójcik-Bojek U, Różalska B, Sadowska B. 2022. Staphylococcus aureus—a known opponent against host defense mechanisms and vaccine development—do we still have a chance to win? Int J Mol Sci 23:948. doi: 10.3390/ijms23020948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diamond G, Beckloff N, Weinberg A, Kisich KO. 2009. The roles of antimicrobial peptides in innate host defense. Curr Pharm Des 15:2377–2392. doi: 10.2174/138161209788682325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen H, Zhang J, He Y, Lv Z, Liang Z, Chen J, Li P, Liu J, Yang H, Tao A, Liu X. 2022. Exploring the role of Staphylococcus aureus in inflammatory diseases. Toxins (Basel) 14:464. doi: 10.3390/toxins14070464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quaresma JAS. 2019. Organization of the skin immune system and compartmentalized immune responses in infectious diseases. Clin Microbiol Rev 32:10–1128. doi: 10.1128/CMR.00034-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pidwill GR, Gibson JF, Cole J, Renshaw SA, Foster SJ. 2020. The role of macrophages in Staphylococcus aureus infection. Front Immunol 11:620339. doi: 10.3389/fimmu.2020.620339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ryu S, Song PI, Seo CH, Cheong H, Park Y. 2014. Colonization and infection of the skin by S. aureus: immune system evasion and the response to cationic antimicrobial peptides. Int J Mol Sci 15:8753–8772. doi: 10.3390/ijms15058753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Askarian F, Wagner T, Johannessen M, Nizet V. 2018. Staphylococcus aureus modulation of innate immune responses through toll-like (TLR), (NOD)-like (NLR) and C-type lectin (CLR) receptors. FEMS Microbiol Rev 42:656–671. doi: 10.1093/femsre/fuy025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hirayama D, Iida T, Nakase H. 2017. The phagocytic function of macrophage-enforcing innate immunity and tissue homeostasis. Int J Mol Sci 19:92. doi: 10.3390/ijms19010092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lunjani N, Ahearn-Ford S, Dube FS, Hlela C, O’Mahony L. 2021. Mechanisms of microbe-immune system dialogue within the skin. Genes Immun 22:276–288. doi: 10.1038/s41435-021-00133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piipponen M, Li D, Landén NX. 2020. The immune functions of keratinocytes in skin wound healing. Int J Mol Sci 21:8790. doi: 10.3390/ijms21228790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu T, Zhang L, Joo D, Sun S-C. 2017. NF-κB signaling in inflammation. Signal Transduct Target Ther 2:17023. doi: 10.1038/sigtrans.2017.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rahman MM, McFadden G. 2011. Modulation of NF-κB signalling by microbial pathogens. Nat Rev Microbiol 9:291–306. doi: 10.1038/nrmicro2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gauthier T, Chen W. 2022. Modulation of macrophage immunometabolism: a new approach to fight infections. Front Immunol 13:780839. doi: 10.3389/fimmu.2022.780839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Russell DG, Huang L, VanderVen BC. 2019. Immunometabolism at the interface between macrophages and pathogens. Nat Rev Immunol 19:291–304. doi: 10.1038/s41577-019-0124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ganeshan K, Chawla A. 2014. Metabolic regulation of immune responses. Annu Rev Immunol 32:609–634. doi: 10.1146/annurev-immunol-032713-120236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Benns HJ, Wincott CJ, Tate EW, Child MA. 2021. Activity-and reactivity-based proteomics: recent technological advances and applications in drug discovery. Curr Opin Chem Biol 60:20–29. doi: 10.1016/j.cbpa.2020.06.011 [DOI] [PubMed] [Google Scholar]
- 21. Patricelli MP, Nomanbhoy TK, Wu J, Brown H, Zhou D, Zhang J, Jagannathan S, Aban A, Okerberg E, Herring C, Nordin B, Weissig H, Yang Q, Lee J-D, Gray NS, Kozarich JW. 2011. In situ kinase profiling reveals functionally relevant properties of native kinases. Chem Biol 18:699–710. doi: 10.1016/j.chembiol.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wolfe LM, Veeraraghavan U, Idicula-Thomas S, Schürer S, Wennerberg K, Reynolds R, Besra GS, Dobos KM. 2013. A chemical proteomics approach to profiling the ATP-binding proteome of Mycobacterium tuberculosis. Mol Cell Proteomics 12:1644–1660. doi: 10.1074/mcp.M112.025635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffman MA, Fang B, Haura EB, Rix U, Koomen JM. 2018. Comparison of quantitative mass spectrometry platforms for monitoring kinase ATP probe uptake in lung cancer. J Proteome Res 17:63–75. doi: 10.1021/acs.jproteome.7b00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Franks CE, Campbell ST, Purow BW, Harris TE, Hsu K-L. 2017. The ligand binding landscape of diacylglycerol kinases. Cell Chem Biol 24:870–880. doi: 10.1016/j.chembiol.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, Walzer M, Wang S, Brazma A, Vizcaíno JA. 2022. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res 50:D543–D552. doi: 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okerberg ES, Dagbay KB, Green JL, Soni I, Aban A, Nomanbhoy TK, Savinov SN, Hardy JA, Kozarich JW. 2019. Chemoproteomics using nucleotide acyl phosphates reveals an ATP binding site at the dimer interface of procaspase-6. Biochemistry 58:5320–5328. doi: 10.1021/acs.biochem.9b00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lovingshimer MR, Siegele D, Reinhart GD. 2006. Construction of an inducible, pfkA and pfkB deficient strain of Escherichia coli for the expression and purification of phosphofructokinase from bacterial sources. Protein Expr Purif 46:475–482. doi: 10.1016/j.pep.2005.09.015 [DOI] [PubMed] [Google Scholar]
- 28. Guixé V, Rodríguez PH, Babul J. 1998. Ligand-induced conformational transitions in Escherichia coli phosphofructokinase 2: evidence for an allosteric site for MgATP2-. Biochemistry 37:13269–13275. doi: 10.1021/bi980576p [DOI] [PubMed] [Google Scholar]
- 29. Adachi J, Kishida M, Watanabe S, Hashimoto Y, Fukamizu K, Tomonaga T. 2014. Proteome-wide discovery of unknown ATP-binding proteins and kinase inhibitor target proteins using an ATP probe. J Proteome Res 13:5461–5470. doi: 10.1021/pr500845u [DOI] [PubMed] [Google Scholar]
- 30. Eisenreich W, Dandekar T, Heesemann J, Goebel W. 2010. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol 8:401–412. doi: 10.1038/nrmicro2351 [DOI] [PubMed] [Google Scholar]
- 31. Sauvageot N, Ladjouzi R, Benachour A, Rincé A, Deutscher J, Hartke A. 2012. Aerobic glycerol dissimilation via the Enterococcus faecalis DhaK pathway depends on NADH oxidase and a phosphotransfer reaction from PEP to DhaK via EIIADha. Microbiology (Reading) 158:2661–2666. doi: 10.1099/mic.0.061663-0 [DOI] [PubMed] [Google Scholar]
- 32. Fellner M, Lentz CS, Jamieson SA, Brewster JL, Chen L, Bogyo M, Mace PD. 2020. Structural basis for the inhibitor and substrate specificity of the unique Fph Serine hydrolases of Staphylococcus aureus. ACS Infect Dis 6:2771–2782. doi: 10.1021/acsinfecdis.0c00503 [DOI] [PubMed] [Google Scholar]
- 33. Lentz CS, Sheldon JR, Crawford LA, Cooper R, Garland M, Amieva MR, Weerapana E, Skaar EP, Bogyo M. 2018. Identification of a S. aureus virulence factor by activity-based protein profiling (ABPP). Nat Chem Biol 14:609–617. doi: 10.1038/s41589-018-0060-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joseph B, Mertins S, Stoll R, Schär J, Umesha KR, Luo Q, Müller-Altrock S, Goebel W. 2008. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol 190:5412–5430. doi: 10.1128/JB.00259-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joseph B, Przybilla K, Stühler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. 2006. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol 188:556–568. doi: 10.1128/JB.188.2.556-568.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carvalho SM, de Jong A, Kloosterman TG, Kuipers OP, Saraiva LM. 2017. The Staphylococcus aureus α-acetolactate synthase ALS confers resistance to nitrosative stress. Front Microbiol 8:1273. doi: 10.3389/fmicb.2017.01273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Michell RH. 2007. Evolution of the diverse biological roles of inositols. Biochemical Society Symposia 74:223–246. doi: 10.1042/BSS2007c19 [DOI] [PubMed] [Google Scholar]
- 38. O’Riordan M, Moors MA, Portnoy DA. 2003. Listeria intracellular growth and virulence require host-derived lipoic acid. Science 302:462–464. doi: 10.1126/science.1088170 [DOI] [PubMed] [Google Scholar]
- 39. Laczkovich I, Teoh WP, Flury S, Grayczyk JP, Zorzoli A, Alonzo F. 2018. Increased flexibility in the use of exogenous lipoic acid by Staphylococcus aureus. Mol Microbiol 109:150–168. doi: 10.1111/mmi.13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zachary D, Bose JL. 2018. Redirection of metabolism in response to fatty acid kinase in Staphylococcus aureus. J Bacteriol 200:00345–18. doi: 10.1128/JB.00345-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Becker A, Fritz-Wolf K, Kabsch W, Knappe J, Schultz S, Volker Wagner AF. 1999. Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat Struct Biol 6:969–975. doi: 10.1038/13341 [DOI] [PubMed] [Google Scholar]
- 42. Martina L, Manuel L, Diana M, Michael L, Andreas P, Friedrich G. 2011. Pyruvate formate lyase acts as a formate supplier for metabolic processes during anaerobiosis in Staphylococcus aureus. J Bacteriol 193:952–962. doi: 10.1128/JB.01161-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Matsumoto Y, Yasukawa J, Ishii M, Hayashi Y, Miyazaki S, Sekimizu K. 2016. A critical role of mevalonate for peptidoglycan synthesis in Staphylococcus aureus. Sci Rep 6:22894. doi: 10.1038/srep22894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Carl JB, Xiaoyu S, Jianshi T. 2009. The mevalonate pathway of Staphylococcus aureus. J Bacteriol 191:851–861. doi: 10.1128/JB.01357-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Passalacqua KD, Charbonneau M-E, O’riordan MXD. 2016. Bacterial metabolism shapes the host–pathogen interface, p 15–41. In Virulence mechanisms of bacterial pathogens [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alreshidi MM, Dunstan RH, Macdonald MM, Gottfries J, Roberts TK. 2019. The uptake and release of amino acids by Staphylococcus aureus at mid-exponential and stationary phases and their corresponding responses to changes in temperature, pH and osmolality. Front Microbiol 10:3059. doi: 10.3389/fmicb.2019.03059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaiser JC, Sen S, Sinha A, Wilkinson BJ, Heinrichs DE. 2016. The role of two branched‐chain amino acid transporters in Staphylococcus aureus growth, membrane fatty acid composition and virulence. Mol Microbiol 102:850–864. doi: 10.1111/mmi.13495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Xiao Z, Xu P. 2007. Acetoin metabolism in bacteria. Crit Rev Microbiol 33:127–140. doi: 10.1080/10408410701364604 [DOI] [PubMed] [Google Scholar]
- 49. Fuchs S, Pané-Farré J, Kohler C, Hecker M, Engelmann S. 2007. Anaerobic gene expression in Staphylococcus aureus. J Bacteriol 189:4275–4289. doi: 10.1128/JB.00081-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tripathi A, Schofield MM, Chlipala GE, Schultz PJ, Yim I, Newmister SA, Nusca TD, Scaglione JB, Hanna PC, Tamayo-Castillo G, Sherman DH. 2014. Baulamycins A and B, broad-spectrum antibiotics identified as inhibitors of siderophore biosynthesis in Staphylococcus aureus and Bacillus anthracis. J Am Chem Soc 136:1579–1586. doi: 10.1021/ja4115924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Horsburgh MJ, Foster TJ, Barth PT, Coggins JR. 1996. Chorismate synthase from Staphylococcus aureus. Microbiology (Reading) 142 ( Pt 10):2943–2950. doi: 10.1099/13500872-142-10-2943 [DOI] [PubMed] [Google Scholar]
- 52. Sekowska A, Dénervaud V, Ashida H, Michoud K, Haas D, Yokota A, Danchin A. 2004. Bacterial variations on the methionine salvage pathway. BMC Microbiol 4:1–17. doi: 10.1186/1471-2180-4-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cronan JE. 2016. Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiol Mol Biol Rev 80:429–450. doi: 10.1128/MMBR.00073-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Halsey CR, Lei S, Wax JK, Lehman MK, Nuxoll AS, Steinke L, Sadykov M, Powers R, Fey PD. 2017. Amino acid catabolism in Staphylococcus aureus and the function of carbon catabolite repression. mBio 8:10–1128. doi: 10.1128/mBio.01434-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheung J, Beasley FC, Liu S, Lajoie GA, Heinrichs DE. 2009. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Mol Microbiol 74:594–608. doi: 10.1111/j.1365-2958.2009.06880.x [DOI] [PubMed] [Google Scholar]
- 56. Hochgräfe F, Wolf C, Fuchs S, Liebeke M, Lalk M, Engelmann S, Hecker M. 2008. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J Bacteriol 190:4997–5008. doi: 10.1128/JB.01846-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dubrac S, Msadek T. 2004. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol 186:1175–1181. doi: 10.1128/JB.186.4.1175-1181.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dubrac S, Boneca IG, Poupel O, Msadek T. 2007. New insights into the walk/Walr (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol 189:8257–8269. doi: 10.1128/JB.00645-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gajdiss M, Monk IR, Bertsche U, Kienemund J, Funk T, Dietrich A, Hort M, Sib E, Stinear TP, Bierbaum G. 2020. YycH and YycI regulate expression of Staphylococcus aureus autolysins by activation of WalRK phosphorylation. Microorganisms 8:870. doi: 10.3390/microorganisms8060870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Delauné A, Dubrac S, Blanchet C, Poupel O, Mäder U, Hiron A, Leduc A, Fitting C, Nicolas P, Cavaillon J-M, Adib-Conquy M, Msadek T. 2012. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect Immun 80:3438–3453. doi: 10.1128/IAI.00195-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Howden BP, McEvoy CRE, Allen DL, Chua K, Gao W, Harrison PF, Bell J, Coombs G, Bennett-Wood V, Porter JL, Robins-Browne R, Davies JK, Seemann T, Stinear TP. 2011. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog 7:e1002359. doi: 10.1371/journal.ppat.1002359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jansen A, Türck M, Szekat C, Nagel M, Clever I, Bierbaum G. 2007. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus. Int J Med Microbiol 297:205–215. doi: 10.1016/j.ijmm.2007.02.002 [DOI] [PubMed] [Google Scholar]
- 63. Delaune A, Poupel O, Mallet A, Coic Y-M, Msadek T, Dubrac S. 2011. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS One 6:e17054. doi: 10.1371/journal.pone.0017054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mir M, Asong J, Li X, Cardot J, Boons G-J, Husson RN. 2011. The extracytoplasmic domain of the Mycobacterium tuberculosis Ser/Thr kinase PknB binds specific muropeptides and is required for PknB localization. PLoS Pathog 7:e1002182. doi: 10.1371/journal.ppat.1002182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Monedero V, Poncet S, Mijakovic I, Fieulaine S, Dossonnet V, Martin-Verstraete I, Nessler S, Deutscher J. 2001. Mutations lowering the phosphatase activity of HPr kinase/phosphatase switch off carbon metabolism. EMBO J 20:3928–3937. doi: 10.1093/emboj/20.15.3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Alper S, Dufour A, Garsin DA, Duncan L, Losick R. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol 260:165–177. doi: 10.1006/jmbi.1996.0390 [DOI] [PubMed] [Google Scholar]
- 67. Huemer M, Mairpady Shambat S, Hertegonne S, Bergada-Pijuan J, Chang C-C, Pereira S, Gómez-Mejia A, Van Gestel L, Bär J, Vulin C, Pfammatter S, Stinear TP, Monk IR, Dworkin J, Zinkernagel AS. 2023. Serine-threonine phosphoregulation by PknB and Stp contributes to quiescence and antibiotic tolerance in Staphylococcus aureus. Sci Signal 16:eabj8194. doi: 10.1126/scisignal.abj8194 [DOI] [PubMed] [Google Scholar]
- 68. Fujimoto-Nakamura M, Ito H, Oyamada Y, Nishino T, Yamagishi J-I. 2005. Accumulation of mutations in both gyrB and parE genes is associated with high-level resistance to novobiocin in Staphylococcus aureus. Antimicrob Agents Chemother 49:3810–3815. doi: 10.1128/AAC.49.9.3810-3815.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Prunier A-L, Leclercq R. 2005. Role of mutS and mutL genes in hypermutability and recombination in Staphylococcus aureus. J Bacteriol 187:3455–3464. doi: 10.1128/JB.187.10.3455-3464.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dunham-Snary KJ, Surewaard BGJ, Mewburn JD, Bentley RET, Martin AY, Jones O, Al-Qazazi R, Lima PAD, Kubes P, Archer SL. 2022. Mitochondria in human neutrophils mediate killing of Staphylococcus aureus. Redox Biol 49:102225. doi: 10.1016/j.redox.2021.102225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mulcahy ME, O’Brien EC, O’Keeffe KM, Vozza EG, Leddy N, McLoughlin RM. 2020. Manipulation of autophagy and apoptosis facilitates intracellular survival of Staphylococcus aureus in human neutrophils. Front Immunol 11:565545. doi: 10.3389/fimmu.2020.565545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Greenlee-Wacker MC, Rigby KM, Kobayashi SD, Porter AR, DeLeo FR, Nauseef WM. 2014. Phagocytosis of Staphylococcus aureus by human neutrophils prevents macrophage efferocytosis and induces programmed necrosis. J Immunol 192:4709–4717. doi: 10.4049/jimmunol.1302692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Steinmetz-Späh J, Liu J, Singh R, Ekoff M, Boddul S, Tang X, Bergqvist F, Idborg H, Heitel P, Rönnberg E, Merk D, Wermeling F, Haeggström JZ, Nilsson G, Steinhilber D, Larsson K, Korotkova M, Jakobsson P-J. 2022. Biosynthesis of prostaglandin 15dPGJ2-glutathione and 15dPGJ2-cysteine conjugates in macrophages and mast cells via MGST3. J Lipid Res 63:100310. doi: 10.1016/j.jlr.2022.100310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jakobsson P-J, Mancini JA, Riendeau D, Ford-Hutchinson AW. 1997. Identification and characterization of a novel microsomal enzyme with glutathione-dependent transferase and peroxidase activities. J Biol Chem 272:22934–22939. doi: 10.1074/jbc.272.36.22934 [DOI] [PubMed] [Google Scholar]
- 75. Ueno N, Takeya R, Miyano K, Kikuchi H, Sumimoto H. 2005. The NADPH oxidase Nox3 constitutively produces superoxide in a p22phox-dependent manner: its regulation by oxidase organizers and activators. J Biol Chem 280:23328–23339. doi: 10.1074/jbc.M414548200 [DOI] [PubMed] [Google Scholar]
- 76. Solt LA, May MJ. 2008. The IκB kinase complex: Master regulator of NF-κB signaling. Immunol Res 42:3–18. doi: 10.1007/s12026-008-8025-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Padavannil A, Ayala-Hernandez MG, Castellanos-Silva EA, Letts JA. 2021. The mysterious multitude: structural perspective on the accessory subunits of respiratory complex I. Front Mol Biosci 8:798353. doi: 10.3389/fmolb.2021.798353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Lunnon K, Keohane A, Pidsley R, Newhouse S, Riddoch-Contreras J, Thubron EB, Devall M, Soininen H, Kłoszewska I, Mecocci P, Tsolaki M, Vellas B, Schalkwyk L, Dobson R, Malik AN, Powell J, Lovestone S, Hodges A, AddNeuroMed Consortium . 2017. Mitochondrial genes are altered in blood early in Alzheimer's disease. Neurobiol Aging 53:36–47. doi: 10.1016/j.neurobiolaging.2016.12.029 [DOI] [PubMed] [Google Scholar]
- 79. Giachin G, Bouverot R, Acajjaoui S, Pantalone S, Soler-López M. 2016. Dynamics of human mitochondrial complex I assembly: implications for neurodegenerative diseases. Front Mol Biosci 3:43. doi: 10.3389/fmolb.2016.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mckenzie M, Ryan MT. 2010. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB Life 62:497–502. doi: 10.1002/iub.335 [DOI] [PubMed] [Google Scholar]
- 81. Ruvolo PP, Deng X, May WS. 2001. Phosphorylation of Bcl2 and regulation of apoptosis. Leukemia 15:515–522. doi: 10.1038/sj.leu.2402090 [DOI] [PubMed] [Google Scholar]
- 82. Haziot A, Ferrero E, Köntgen F, Hijiya N, Yamamoto S, Silver J, Stewart CL, Goyert SM. 1996. Resistance to endotoxin shock and reduced dissemination of gram-negative bacteria in CD14-deficient mice. Immunity 4:407–414. doi: 10.1016/s1074-7613(00)80254-x [DOI] [PubMed] [Google Scholar]
- 83. Krzewski K, Gil-Krzewska A, Nguyen V, Peruzzi G, Coligan JE. 2013. LAMP1/CD107a is required for efficient perforin delivery to lytic granules and NK-cell cytotoxicity. Blood 121:4672–4683. doi: 10.1182/blood-2012-08-453738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Castro-Dopico T, Dennison TW, Ferdinand JR, Mathews RJ, Fleming A, Clift D, Stewart BJ, Jing C, Strongili K, Labzin LI, Monk EJM, Saeb-Parsy K, Bryant CE, Clare S, Parkes M, Clatworthy MR. 2019. Anti-commensal IgG drives intestinal inflammation and type 17 immunity in ulcerative colitis. Immunity 50:1099–1114. doi: 10.1016/j.immuni.2019.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yamasaki S, Ishikawa E, Sakuma M, Hara H, Ogata K, Saito T. 2008. Mincle is an ITAM-coupled activating receptor that senses damaged cells. Nat Immunol 9:1179–1188. doi: 10.1038/ni.1651 [DOI] [PubMed] [Google Scholar]
- 86. Son SM, Park SJ, Stamatakou E, Vicinanza M, Menzies FM, Rubinsztein DC. 2020. Leucine regulates autophagy via acetylation of the mTORC1 component raptor. Nat Commun 11:3148. doi: 10.1038/s41467-020-16886-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kalousek F, Darigo MD, Rosenberg LE. 1980. Isolation and characterization of propionyl-CoA carboxylase from normal human liver. evidence for a protomeric tetramer of nonidentical subunits. J Biol Chem 255:60–65. [PubMed] [Google Scholar]
- 88. Codini M, Garcia-Gil M, Albi E. 2021. Cholesterol and sphingolipid enriched lipid rafts as therapeutic targets in cancer. Int J Mol Sci 22:726. doi: 10.3390/ijms22020726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kong L-J, Meloni AR, Nevins JR. 2006. The Rb-related p130 protein controls telomere lengthening through an interaction with a Rad50-interacting protein, RINT-1. Mol Cell 22:63–71. doi: 10.1016/j.molcel.2006.02.016 [DOI] [PubMed] [Google Scholar]
- 90. Vandamme J, Völkel P, Rosnoblet C, Le Faou P, Angrand P-O. 2011. Interaction proteomics analysis of polycomb proteins defines distinct PRC1 complexes in mammalian cells. Mol Cell Proteomics 10:M110. doi: 10.1074/mcp.M110.002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gurzau AD, Chen K, Xue S, Dai W, Lucet IS, Ly TTN, Reversade B, Blewitt ME, Murphy JM. 2018. FSHD2-and BAMS-associated mutations confer opposing effects on SMCHD1 function. J Biol Chem 293:9841–9853. doi: 10.1074/jbc.RA118.003104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Chapman JR, Sossick AJ, Boulton SJ, Jackson SP. 2012. BRCA1-associated exclusion of 53BP1 from DNA damage sites underlies temporal control of DNA repair. J Cell Sci 125:3529–3534. doi: 10.1242/jcs.105353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tsurimoto T, Shinozaki A, Yano M, Seki M, Enomoto T. 2005. Human Werner helicase interacting protein 1 (WRNIP1) functions as a novel modulator for DNA polymerase δ. Genes Cells 10:13–22. doi: 10.1111/j.1365-2443.2004.00812.x [DOI] [PubMed] [Google Scholar]
- 94. Buratta S, Tancini B, Sagini K, Delo F, Chiaradia E, Urbanelli L, Emiliani C. 2020. Lysosomal exocytosis, exosome release and secretory autophagy: the autophagic-and endo-lysosomal systems go extracellular. Int J Mol Sci 21:2576. doi: 10.3390/ijms21072576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Cui B, Lin H, Yu J, Yu J, Hu Z. 2019. Autophagy and the immune response, p 595–634. In Autophagy: biology and diseases: basic science [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Wang H, Ma Q, Qi Y, Dong J, Du X, Rae J, Wang J, Wu W-F, Brown AJ, Parton RG, Wu J-W, Yang H. 2019. ORP2 delivers cholesterol to the plasma membrane in exchange for phosphatidylinositol 4, 5-bisphosphate (PI (4, 5) P2). Mol Cell 73:458–473. doi: 10.1016/j.molcel.2018.11.014 [DOI] [PubMed] [Google Scholar]
- 97. Whitfield AJ, Barrett PHR, van Bockxmeer FM, Burnett JR. 2004. Lipid disorders and mutations in the APOB gene. Clin Chem 50:1725–1732. doi: 10.1373/clinchem.2004.038026 [DOI] [PubMed] [Google Scholar]
- 98. Yeh FL, Wang Y, Tom I, Gonzalez LC, Sheng M. 2016. TREM2 binds to apolipoproteins, including APOE and CLU/APOJ, and thereby facilitates uptake of amyloid-beta by microglia. Neuron 91:328–340. doi: 10.1016/j.neuron.2016.06.015 [DOI] [PubMed] [Google Scholar]
- 99. Okamoto M, Shimogishi M, Nakamura A, Suga Y, Sugawara K, Sato M, Nishi R, Fujisawa A, Yamamoto Y, Kashiba M. 2021. Differentiation of THP-1 monocytes to macrophages increased mitochondrial DNA copy number but did not increase expression of mitochondrial respiratory proteins or mitochondrial transcription factor A. Arch Biochem Biophys 710:108988. doi: 10.1016/j.abb.2021.108988 [DOI] [PubMed] [Google Scholar]
- 100. Gao Y, Meyer B, Sokolova L, Zwicker K, Karas M, Brutschy B, Peng G, Michel H. 2012. Heme-copper terminal oxidase using both cytochrome c and ubiquinol as electron donors. Proc Natl Acad Sci USA 109:3275–3280. doi: 10.1073/pnas.1121040109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Antonicka H, Mattman A, Carlson CG, Glerum DM, Hoffbuhr KC, Leary SC, Kennaway NG, Shoubridge EA. 2003. Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway, causing early-onset fatal hypertrophic cardiomyopathy. Am J Hum Genet 72:101–114. doi: 10.1086/345489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ndi M, Marin-Buera L, Salvatori R, Singh AP, Ott M. 2018. Biogenesis of the bc1 complex of the mitochondrial respiratory chain. J Mol Biol 430:3892–3905. doi: 10.1016/j.jmb.2018.04.036 [DOI] [PubMed] [Google Scholar]
- 103. Zhang X, Luo S, Wang M, Cao Q, Zhang Z, Huang Q, Li J, Deng Z, Liu T, Liu C-L, Meppen M, Vromman A, Flavell RA, Hotamışlıgil GS, Liu J, Libby P, Liu Z, Shi G-P. 2022. Differential Il18 signaling via Il18 receptor and Na-Cl co-transporter discriminating thermogenesis and glucose metabolism regulation. Nat Commun 13:7582. doi: 10.1038/s41467-022-35256-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, Glunk V, Sousa IS, Beaudry JL, Puviindran V, Abdennur NA, Liu J, Svensson P-A, Hsu Y-H, Drucker DJ, Mellgren G, Hui C-C, Hauner H, Kellis M. 2015. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med 373:895–907. doi: 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Li X, Morita M, Kikuguchi C, Takahashi A, Suzuki T, Yamamoto T. 2017. Adipocyte‐specific disruption of mouse Cnot3 causes lipodystrophy. FEBS Lett 591:358–368. doi: 10.1002/1873-3468.12550 [DOI] [PubMed] [Google Scholar]
- 106. Shu L, Hoo RLC, Wu X, Pan Y, Lee IPC, Cheong LY, Bornstein SR, Rong X, Guo J, Xu A. 2017. A-FABP mediates adaptive thermogenesis by promoting intracellular activation of thyroid hormones in brown adipocytes. Nat Commun 8:14147. doi: 10.1038/ncomms14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Boucher J, Mori MA, Lee KY, Smyth G, Liew CW, Macotela Y, Rourk M, Bluher M, Russell SJ, Kahn CR. 2012. Impaired thermogenesis and adipose tissue development in mice with fat-specific disruption of insulin and IGF-1 signalling. Nat Commun 3:902. doi: 10.1038/ncomms1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schieber AMP, Ayres JS. 2016. Thermoregulation as a disease tolerance defense strategy. Pathog Dis 74:ftw106. doi: 10.1093/femspd/ftw106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Watanabe M, Yamamoto T, Mori C, Okada N, Yamazaki N, Kajimoto K, Kataoka M, Shinohara Y. 2008. Cold-induced changes in gene expression in brown adipose tissue: implications for the activation of thermogenesis. Biol Pharm Bull 31:775–784. doi: 10.1248/bpb.31.775 [DOI] [PubMed] [Google Scholar]
- 110. Mayer IA, Arteaga CL. 2016. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med 67:11–28. doi: 10.1146/annurev-med-062913-051343 [DOI] [PubMed] [Google Scholar]
- 111. Yáñez-Mó M, Siljander PR-M, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, et al. 2015. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4:27066. doi: 10.3402/jev.v4.27066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Yehualaeshet T, O’Connor R, Green-Johnson J, Mai S, Silverstein R, Murphy-Ullrich JE, Khalil N. 1999. Activation of rat alveolar macrophage derived L-TGFβ-1 by Plasmin requires interaction with TSP-1 and the TSP-1 cell surface receptor, CD36. Am J Pathol 155:841–851. doi: 10.1016/s0002-9440(10)65183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wang Y, Lu H, Wang Z, Li Y, Chen X. 2022. TGF-β1 promotes autophagy and inhibits apoptosis in breast cancer by targeting TP63. Front Oncol 12:865067. doi: 10.3389/fonc.2022.865067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Birgisdottir ÅB, Mouilleron S, Bhujabal Z, Wirth M, Sjøttem E, Evjen G, Zhang W, Lee R, O’Reilly N, Tooze SA, Lamark T, Johansen T. 2019. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy 15:1333–1355. doi: 10.1080/15548627.2019.1581009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Terfve CDA, Wilkes EH, Casado P, Cutillas PR, Saez-Rodriguez J. 2015. Large-scale models of signal propagation in human cells derived from discovery phosphoproteomic data. Nat Commun 6:8033. doi: 10.1038/ncomms9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Tang D, Kroemer G. 2020. Peroxisome: the new player in ferroptosis. Signal Transduct Target Ther 5:273. doi: 10.1038/s41392-020-00404-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Davies L, Gray D, Spiller D, White MRH, Damato B, Grierson I, Paraoan L. 2009. P53 apoptosis mediator PERP: localization, function and caspase activation in uveal melanoma. J Cell Mol Med 13:1995–2007. doi: 10.1111/j.1582-4934.2008.00590.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Attardi LD, Reczek EE, Cosmas C, Demicco EG, McCurrach ME, Lowe SW, Jacks T. 2000. PERP, an apoptosis-associated target of P53, is a novel member of the PMP-22/Gas3 family. Genes Dev 14:704–718. doi: 10.1101/gad.14.6.704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Arcaro A, Zvelebil MJ, Wallasch C, Ullrich A, Waterfield MD, Domin J. 2000. Class II phosphoinositide 3-kinases are downstream targets of activated polypeptide growth factor receptors. Mol Cell Biol 20:3817–3830. doi: 10.1128/MCB.20.11.3817-3830.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Deranieh RM, He Q, Caruso JA, Greenberg ML. 2013. Phosphorylation regulates myo-inositol-3-phosphate synthase: a novel regulatory mechanism of inositol biosynthesis. J Biol Chem 288:26822–26833. doi: 10.1074/jbc.M113.479121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chen X-H, Zhang B-W, Li H, Peng X-X. 2015. Myo-inositol improves the host’s ability to eliminate balofloxacin-resistant Escherichia coli. Sci Rep 5:10720. doi: 10.1038/srep10720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. 2005. Lithium induces autophagy by inhibiting Inositol monophosphatase. J Cell Biol 170:1101–1111. doi: 10.1083/jcb.200504035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Hudson CC, Liu M, Chiang GG, Otterness DM, Loomis DC, Kaper F, Giaccia AJ, Abraham RT. 2002. Regulation of hypoxia-inducible factor 1α expression and function by the mammalian target of rapamycin. Mol Cell Biol 22:7004–7014. doi: 10.1128/MCB.22.20.7004-7014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Düvel K, Yecies JL, Menon S, Raman P, Lipovsky AI, Souza AL, Triantafellow E, Ma Q, Gorski R, Cleaver S, Vander Heiden MG, MacKeigan JP, Finan PM, Clish CB, Murphy LO, Manning BD. 2010. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell 39:171–183. doi: 10.1016/j.molcel.2010.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Pajon C, Fortoul MC, Diaz-Tang G, Marin Meneses E, Kalifa AR, Sevy E, Mariah T, Toscan B, Marcelin M, Hernandez DM, Marzouk MM, Lopatkin AJ, Eldakar OT, Smith RP. 2023. Interactions between metabolism and growth can determine the co-existence of Staphylococcus aureus and Pseudomonas Aeruginosa. Elife 12:e83664. doi: 10.7554/eLife.83664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Escoll P, Buchrieser C. 2018. Metabolic reprogramming of host cells upon bacterial infection: why shift to a Warburg‐Like metabolism? FEBS J 285:2146–2160. doi: 10.1111/febs.14446 [DOI] [PubMed] [Google Scholar]
- 127. Proal AD, VanElzakker MB. 2021. Pathogens hijack host cell metabolism: intracellular infection as a driver of the Warburg effect in cancer and other chronic inflammatory conditions. Immunometabolism 3. doi: 10.20900/immunometab20210003 [DOI] [Google Scholar]
- 128. Oosting M, Kerstholt M, Ter Horst R, Li Y, Deelen P, Smeekens S, Jaeger M, Lachmandas E, Vrijmoeth H, Lupse M, Flonta M, Cramer RA, Kullberg BJ, Kumar V, Xavier R, Wijmenga C, Netea MG, Joosten LAB. 2016. Functional and genomic architecture of Borrelia burgdorferi-induced cytokine responses in humans. Cell Host Microbe 20:822–833. doi: 10.1016/j.chom.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 129. Amaral EP, Costa DL, Namasivayam S, Riteau N, Kamenyeva O, Mittereder L, Mayer-Barber KD, Andrade BB, Sher A. 2019. A major role for ferroptosis in Mycobacterium tuberculosis–induced cell death and tissue necrosis. J Exp Med 216:556–570. doi: 10.1084/jem.20181776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Bagayoko S, Meunier E. 2022. Emerging roles of ferroptosis in infectious diseases. FEBS J 289:7869–7890. doi: 10.1111/febs.16244 [DOI] [PubMed] [Google Scholar]
- 131. Jacolot S, Yang Y, Paitry P, Férec C, Mura C. 2010. Iron metabolism in macrophages from HFE hemochromatosis patients. Mol Genet Metab 101:258–267. doi: 10.1016/j.ymgme.2010.07.010 [DOI] [PubMed] [Google Scholar]
- 132. Takao J, Yudate T, Das A, Shikano S, Bonkobara M, Ariizumi K, Cruz PD. 2003. Expression of NF‐κB in epidermis and the relationship between NF‐κB activation and inhibition of keratinocyte growth. Br J Dermatol 148:680–688. doi: 10.1046/j.1365-2133.2003.05285.x [DOI] [PubMed] [Google Scholar]
- 133. Israël A. 2010. The IKK complex, a central regulator of NF-κB activation. Cold Spring Harb Perspect Biol 2:a000158. doi: 10.1101/cshperspect.a000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Zilberman-Rudenko J, Shawver LM, Wessel AW, Luo Y, Pelletier M, Tsai WL, Lee Y, Vonortas S, Cheng L, Ashwell JD, Orange JS, Siegel RM, Hanson EP. 2016. Recruitment of A20 by the C-terminal domain of NEMO suppresses NF-κB activation and autoinflammatory disease. Proc Natl Acad Sci USA 113:1612–1617. doi: 10.1073/pnas.1518163113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Das AK, Uhler MD, Hajra AK. 2000. Molecular cloning and expression of mammalian peroxisomaltrans-2-enoyl-coenzyme A reductase cDNAs. J Biol Chem 275:24333–24340. doi: 10.1074/jbc.M001168200 [DOI] [PubMed] [Google Scholar]
- 136. Khnykin D, Miner JH, Jahnsen F. 2011. Role of fatty acid transporters in epidermis: implications for health and disease. Dermatoendocrinol 3:53–61. doi: 10.4161/derm.3.2.14816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Schmuth M, Jiang YJ, Dubrac S, Elias PM, Feingold KR. 2008. Thematic review series: skin lipids. peroxisome proliferator-activated receptors and liver X receptors in epidermal biology. J Lipid Res 49:499–509. doi: 10.1194/jlr.R800001-JLR200 [DOI] [PubMed] [Google Scholar]
- 138. Joo J-H, Alpatov R, Munguba GC, Jackson MR, Hunt ME, Sugrue SP. 2005. Reduction of Pnn by RNAi induces loss of cell-cell adhesion between human corneal epithelial cells. Mol Vis 11:133–142. [PubMed] [Google Scholar]
- 139. Yang Y, Li M, Ma Y, Ye W, Si Y, Zheng X, Liu H, Cheng L, Zhang L, Zhang H, Zhang X, Lei Y, Shen L, Zhang F, Ma H. 2022. LncRNA NEAT1 potentiates SREBP2 activity to promote inflammatory macrophage activation and limit hantaan virus propagation. Front Microbiol 13:849020. doi: 10.3389/fmicb.2022.849020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Chen D-D, Hui L-L, Zhang X-C, Chang Q. 2019. NEAT1 contributes to ox‐LDL‐induced inflammation and oxidative stress in macrophages through inhibiting miR‐128. J Cell Biochem 120:2493–2501. doi: 10.1002/jcb.27541 [DOI] [PubMed] [Google Scholar]
- 141. Klec C, Knutsen E, Schwarzenbacher D, Jonas K, Pasculli B, Heitzer E, Rinner B, Krajina K, Prinz F, Gottschalk B, Ulz P, Deutsch A, Prokesch A, Jahn SW, Lellahi SM, Perander M, Barbano R, Graier WF, Parrella P, Calin GA, Pichler M. 2022. ALYREF, a novel factor involved in breast carcinogenesis, acts through transcriptional and post-transcriptional mechanisms selectively regulating the short NEAT1 isoform. Cell Mol Life Sci 79:391. doi: 10.1007/s00018-022-04402-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Shi Y, Tabesh M, Sugrue SP. 2000. Role of cell adhesion–associated protein, pinin (DRS/memA), in corneal epithelial migration. Invest Ophthalmol Vis Sci 41:1337–1345. [PubMed] [Google Scholar]
- 143. Guttman JA, Finlay BB. 2009. Tight junctions as targets of infectious agents. Biochim Biophys Acta 1788:832–841. doi: 10.1016/j.bbamem.2008.10.028 [DOI] [PubMed] [Google Scholar]
- 144. Becker KA, Fahsel B, Kemper H, Mayeres J, Li C, Wilker B, Keitsch S, Soddemann M, Sehl C, Kohnen M, Edwards MJ, Grassmé H, Caldwell CC, Seitz A, Fraunholz M, Gulbins E. 2018. Staphylococcus aureus alpha-toxin disrupts endothelial-cell tight junctions via acid sphingomyelinase and ceramide. Infect Immun 86:10–1128. doi: 10.1128/IAI.00606-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S7.
Strains.
Primers.
Activity of S. aureus proteins.
Activity of human proteins.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (25) partner repository with the data set identifier PXD049003.