Abstract
The periplasmic protein FepB of Escherichia coli is a component of the ferric enterobactin transport system. We overexpressed and purified the binding protein 23-fold from periplasmic extracts by ammonium sulfate precipitation and chromatographic methods, with a yield of 20%, to a final specific activity of 15,500 pmol of ferric enterobactin bound/mg. Periplasmic fluid from cells overexpressing the binding protein adsorbed catecholate ferric siderophores with high affinity: in a gel filtration chromatography assay the Kd of the ferric enterobactin-FepB binding reaction was approximately 135 nM. Intrinsic fluorescence measurements of binding by the purified protein, which were more accurate, showed higher affinity for both ferric enterobactin (Kd = 30 nM) and ferric enantioenterobactin (Kd = 15 nM), the left-handed stereoisomer of the natural E. coli siderophore. Purified FepB also adsorbed the apo-siderophore, enterobactin, with comparable affinity (Kd = 60 nM) but did not bind ferric agrobactin. Polyclonal rabbit antisera and mouse monoclonal antibodies raised against nearly homogeneous preparations of FepB specifically recognized it in solid-phase immunoassays. These sera enabled the measurement of the FepB concentration in vivo when expressed from the chromosome (4,000 copies/cell) or from multicopy plasmids (>100,000 copies/cell). Overexpression of the binding protein did not enhance the overall affinity or rate of ferric enterobactin transport, supporting the conclusion that the rate-limiting step of ferric siderophore uptake through the cell envelope is passage through the outer membrane.
Iron, an essential nutrient for the growth of bacteria, serves as a cofactor for enzymes, as a redox center in electron carriers such as cytochromes and iron-sulfur proteins, and as a global regulator of many cellular biosynthetic and metabolic systems. However, iron is initially inaccessible to bacteria in their natural environments. Within animal fluids and tissues, proteins such as transferrin, lactoferrin, or ferritin sequester iron, while in neutral or basic aqueous environments outside the host, iron rapidly oxidizes and precipitates in ferric hydroxide polymers (28). Microbes respond to iron unavailability by synthesizing and secreting small organic molecules with high affinity for Fe3+ that liberate the metal from its organic or inorganic complexes. These molecules, called siderophores, are usually hydroxamate or catecholate compounds that form hexadentate complexes with iron (29). Although its chelation by siderophores solves the dilemma of iron unavailability, the molecular dimensions of the metal complexes (∼750 Da) create a second problem: ferric siderophores are too large to enter bacterial cells through the general porin channels of the outer membrane. Consequently, bacteria produce high-affinity, energy-dependent cell envelope transport systems that recognize, bind, and transport ferric siderophores into the cytoplasm (for a review, see reference 14). The high specificity and affinity of these iron acquisition systems allow bacteria to proliferate at even very low external iron concentrations.
Escherichia coli and several other species of Enterobacteriaceae secrete the catecholate siderophore enterobactin. The outer membrane (OM) protein FepA binds ferric enterobactin and transports it to the periplasm (23, 36). Efficient ferric enterobactin transport into the cytoplasm, however, requires a soluble periplasmic protein, FepB, as shown by complementation with hybrid lambda phage (35). Evidence exists that FepB binds ferric enterobactin: an LPP-OmpA-FepB fusion protein expressed on the E. coli cell surface adsorbed the ferric siderophore (44). Native FepB functions in the periplasm to facilitate the transfer of ferric enterobactin to a multisubunit inner membrane permease, FepCDG. This final stage of transport into the cell probably requires ATP hydrolysis (42).
When iron is abundant, its ferrous complex with the Fur protein negatively regulates fepB, just as it controls other genes encoding iron transport proteins, such that measurable transcription does not occur (2, 6, 7). The 318-amino-acid pro-FepB contains a cleavable leader sequence, has a calculated molecular mass of 34.3 kDa, and associates with the cytoplasmic membrane (34). The 292-amino-acid mature FepB protein has a calculated molecular mass of 31.6 kDa and exists in the periplasm (34). However, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the periplasmic fraction revealed three distinct FepB bands, with molecular masses of 36.5, 33.5, and 31.5 kDa (33, 34). Although the origin of these isoforms is unknown, previous work eliminated two potential causes: FepB contains no cysteine, ruling out the presence of alternative, disulfide-stabilized forms, and double-labeled experiments with 32P- and 35S-labeled methionine did not demonstrate posttranslational phosphorylation of FepB (6).
Our experiments show that soluble FepB, in crude form in periplasmic extracts or in purified form, avidly binds ferric enterobactin. We generated antibodies to FepB, studied its expression and functional importance in vivo, and quantitatively characterized its binding affinity and specificity by equilibrium methods.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli strains BN1071 (49), KDF541 (49), and BL21(DE3) (Novagen) carry chromosomal fepB+ genes; the latter strain contains chromosomally encoded T7 RNA polymerase under the control of the isopropyl-β-d-thiogalactoside (IPTG)-inducible lacUV5 promoter. DK214 (35) (provided by C. F. Earhart) is fepB. Plasmid pME13-18 (6) (provided by C. F. Earhart) carries the wild-type E. coli fepB gene under its natural promoter. p72 (provided by M. A. McIntosh) carries fepB+ under T7 promoter control, which allowed us to regulate its expression from the plasmid in strain BL21(DE3) by manipulating the concentration of IPTG in the culture medium. pIB3 and pIB51 are multicopy plasmids that also carry fepB+ under the control of its natural promoter. To create them, we PCR amplified fepB and its upstream flanking region from p72 and inserted the product into pUC18 and pHSG398 (48), respectively, using BamHI and SalI sites. Both of these clonings eliminated the small open reading frame (orf1) immediately upstream of fepB and its promoter (33, 34). We sequenced both constructs to verify the integrity of the fepB promoter region and structural gene, using an ALF-Express automated DNA sequencer (Pharmacia). pITS449 is a pUC18 derivative that carries wild-type fepA (31).
Bacteria were grown in Luria-Bertani broth (24) and in some experiments were subcultured at 1% into T (22) or morpholinepropanesulfonic acid (MOPS) (27) minimal medium. The cultures were shaken at 37°C with vigorous aeration to mid-log phase. To maximize FepB expression, we added IPTG to 10−4 M and further incubated the cultures for 1.5 h.
Ferric siderophores.
59Fe-enterobactin (59FeEnt) was prepared by chromatography over Sephadex LH20 in sodium phosphate buffer (26). Ferric complexes of enantioenterobactin and agrobactin were previously described (49).
Osmotic shock fluid.
Periplasmic extracts were prepared by a modified osmotic shock procedure (30). A 50-ml volume of bacterial culture was centrifuged at 10,000 × g for 10 min, and the pellet was washed with 1 ml of 30 mM Tris (pH 8.0) and resuspended in 250 μl of 30 mM Tris (pH 8.0) containing 20% sucrose. After the addition of 2.5 μl of 0.1 M EDTA, the cell suspension was incubated at room temperature for 15 min with occasional swirling. The cells were pelleted by centrifugation and resuspended in 1 ml of cold shock solution (5 ml of water, 2.5 μl of 1 M MgCl2). After 10 min in an ice bath, the cells were pelleted by centrifugation at 5,000 × g for 20 min, and the supernatant, containing the periplasmic fluid, was decanted, not pipetted, into a new tube. The procedure was repeated to maximize the yield.
FepB purification.
BL21(DE3) p72 (13.5 liters) was grown to mid-log phase and induced with 10−4 M IPTG in 900-ml LB aliquots in Fernbach flasks to ensure adequate aeration. All purification procedures were performed at 4°C. The osmotic shock procedure was appropriately scaled to the larger volume of cell suspension, and FepB was precipitated from the periplasmic extracts with a 45 to 80% ammonium sulfate cut. After dialysis against 10 mM Tris (pH 7.4), the protein solution was loaded onto a DE-52 anion-exchange column in the same buffer and eluted with a gradient of 0 to 0.3 M NaCl. FepB eluted at approximately 0.15 M NaCl. As a final step, the pooled DE-52 fractions were chromatographed on Sephacryl S-100 HR in Tris-buffered saline (TBS) (pH 7.4). Protein concentrations were determined by the MicroBCA assay (Pierce, Rockford, Ill.) using bovine serum albumin as a standard.
Chromatographic binding determinations.
Binding of FeEnt to crude wild-type FepB was assayed by column chromatography. Various amounts of 59FeEnt were added to 300 μl of periplasmic fluid. After 10 min on ice, a small amount of glycerol was added, and the sample was chromatographed on a 1.5- by 30-cm column of Sephadex LH20 equilibrated in 50 mM Tris (pH 6.9); 0.55-ml fractions were collected. The column was washed with 50 mM EDTA to remove any residual 59Fe.
Intrinsic fluorescence.
All buffers were filtered to eliminate precipitates. Using an SLM 8000C fluorimeter, upgraded to 8100 capability with automated shutters and polarizers (SLM Instruments, Rochester, N.Y.), the excitation and emission maxima for FepB were 280 and 327 nm, respectively. These settings were used for fluorescence measurements of siderophore binding by FepB. At temperatures from 4 to 25°C, using purified FepB (see Fig. 1, pooled fractions 15 to 21; >90% 33.8-kDa band), the binding-reaction mixtures reached equilibrium in a few seconds (data not shown). With an integration time of 5 s, we recorded fluorescence intensities after the addition of various amounts of siderophores to FepB (44 nM) in TBS (pH 7.4). After subtraction of the emission spectrum of the siderophore itself (in TBS [pH 7.4]), the data were corrected for dilution effects and contaminating fluorescence from impurities in the sodium phosphate buffer. Finally, as a negative control of FeEnt binding, the fluorescence of bovine serum albumin in TBS (pH 7.4), was recorded in its presence and absence. No changes in bovine serum albumin fluorescence occurred, demonstrating the specificity of the binding of catecholate siderophores to FepB.
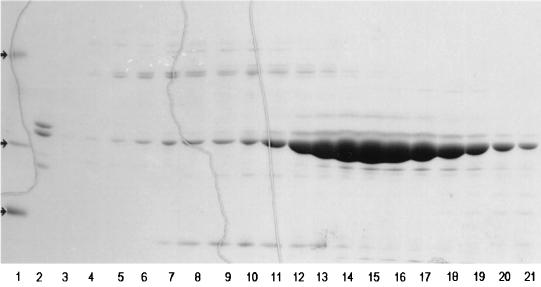
FIG. 1.
SDS-PAGE of FepB chromatography on Sephacryl S100. Periplasmic fluid containing overexpressed FepB was subjected to ammonium sulfate precipitation and anion-exchange (DE-52 column) chromatography. The purest fractions were consolidated, concentrated by ammonium sulfate precipitation, and chromatographed over Sephacryl S-100. Lanes 1, molecular mass markers (arrows), consisting of phosphorylase b (94 kDa), carbonic anhydrase (29 kDa), and lysozyme (14 kDa); 2, starting material; 3 to 21, fractions from S-100. The purest fractions (lanes 15 to 20) were collected, pooled, and used for binding experiments and animal immunizations.
N-terminal sequencing.
SDS-PAGE (1) of purified FepB stained with Coomassie blue revealed two major bands of 33.8 and 31.5 kDa. The N-terminal 15 amino acids of each band were sequenced by sequential Edman degradation (21) at the Protein and Nucleic Acid Shared Facility of the Medical College of Wisconsin.
Antibody generation.
FepB, denatured by boiling in 1% SDS for 10 min, was added to native FepB in a 1:1 molar ratio. For polyclonal antisera, the mixture was emulsified with complete Freund's adjuvant and 100 μg of protein was injected into mice or rabbits. The animals were boosted with the same amount, emulsified in incomplete Freund's adjuvant, weekly for a month, and serum was collected. Monoclonal antibodies were made as previously described (13, 26).
Western immunoblots.
Whole-cell lysates (5 × 108 cells/lane [31]) were solubilized in SDS-PAGE sample buffer by boiling for 5 min and resolved on 12% polyacrylamide gels (1). Electrophoresis, electrotransfer to nitrocellulose paper, antibody staining, and colorimetric development were performed as previously described (26). For quantitation of FepB expression, the nitrocellulose was incubated overnight with rabbit polyclonal anti-FepB sera, incubated with 125I-protein A (8, 20), and subjected to audioradiography.
Colicin susceptibility.
The sensitivity to colicins B and D was determined by limiting dilution on a lawn of the test bacteria.
RESULTS
Purification of FepB.
The binding protein in periplasmic fluid from BL21(DE3)/p72 precipitated over a broad range of ammonium sulfate concentrations, from 45 to 80%. After dialysis of the precipitate in 10 mM Tris (pH 7.4), the protein solution was loaded onto an 80-ml DE-52 anion-exchange column (5.7 by 22 cm). The column was washed with 5 volumes of 10 mM Tris (pH 7.4) and eluted with a linear gradient of NaCl. Fractions containing FeEnt binding activity were pooled and concentrated by ammonium sulfate precipitation, and samples with the highest specific activity were fractionated on a column of Sephacryl S-100 HR (1.3 by 117 cm) in TBS (pH 7.4). The purest fractions were pooled and stored (Fig. 1). This procedure resulted in 23-fold purification of FepB: the 9 mg we obtained from 75 g of wet cell paste had a specific activity of 15,500 pmol of 59FeEnt bound/mg (Table 1), a stoichiometry of approximately 0.5.
TABLE 1.
Purification of FepB
| Purification step | Volume (ml) | Total activity (U)a | Amt of protein (mg/ml) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Periplasmic fluid | 500.0 | 687,000 | 2.0 | 687 | 1 | 100 |
| Ammonium sulfate precipitation | 20 | 321,000 | 7.1 | 2,260 | 3.3 | 47 |
| DE-52 chromatography | 20.5 | 149,000 | 0.9 | 8,075 | 11.8 | 22 |
| Sephacryl S-100 chromatography | 11.2 | 139,000 | 0.8 | 15,513 | 22.6 | 20 |
Activity units are arbitrary, based on the binding of 59FeEnt measured by the column chromatography assay.
Gel filtration of FepB on Sephacryl S-100 HR, with appropriate protein standards, showed a molecular mass of 33.8 kDa. Rf measurements for purified FepB from SDS-PAGE (Fig. 1) also estimated its molecular mass as 33.8 kDa, suggesting that its native form is monomeric. SDS-PAGE also revealed a second major periplasmic protein in BL21/p72, of 31.5 kDa, that reacted with anti-FepB sera in immunoblots. The N-terminal 15 amino acids of the 33.5- and 31.5-kDa protein bands were identical to each other and to those of the deduced primary structure of the mature protein (33, 34). The first amino acid of mature FepB, which corresponds to A27 in pro-FepB, confirmed its signal peptide as residues 1 to 26.
59FeEnt binding by FepB.
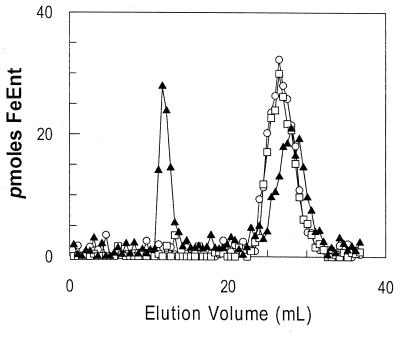
When chromatographed together on the methylated dextran resin Sephadex LH20, the hydrophilic protein FepB eluted first and the smaller, hydrophobic siderophore eluted much later. Adsorption of 59FeEnt to the periplasmic protein resulted in their coelution early in the column profile. In these initial experiments, we monitored FepB elution by measuring its absorbance at 280 nm and 59FeEnt by counting the radioactivity of the fractions. FepB-59FeEnt binding-reactions performed with extracts from BL21(DE3)/p72, which encodes FepB on a high-copy-number plasmid, separated into two peaks of radioactivity on Sephadex LH20 (Fig. 2): the first peak contained FepB and 59FeEnt, and the second contained only 59FeEnt. On the other hand, chromatography of binding-reactions performed with fluids from the fepB strain DK214, DK214 containing the low-copy-number fepB+ plasmid pME13-18, the fepB+ strain BL21(DE3), or the entA fepB+ strain BN1071, all obtained from cells grown in iron-deficient minimal medium, showed only one peak, of free 59FeEnt (Fig. 2). From these results, it was apparent that overexpression of FepB is necessary for detection of 59FeEnt binding in the column assay.
FIG. 2.
Chromatographic measurement of 59FeEnt-FepB binding. 59FeEnt and periplasmic fluid containing overexpressed FepB were mixed and chromatographed over Sephadex LH20. Extracts from DK214 (fepB) (○), BL21(DE3) (chromosomal fepB+) (□), and BL21(DE3)/p72 (high-copy-number, IPTG-inducible fepB+) (▴) were tested. The first peak represents ligand bound to FepB, while the second peak represents free 59FeEnt.
Western blots of column fractions with anti-FepA monoclonal antibody 41 (26) detected a very small amount of FepA in the periplasmic extracts (data not shown). However, the cochromatography of 59FeEnt with protein on the column did not derive from the presence of FepA, because similar FepA contamination occurred in extracts from the fepB strain DK214, which did not measurably bind FeEnt in the assay.
Affinity of purified FepB for catecholate ferric siderophores.
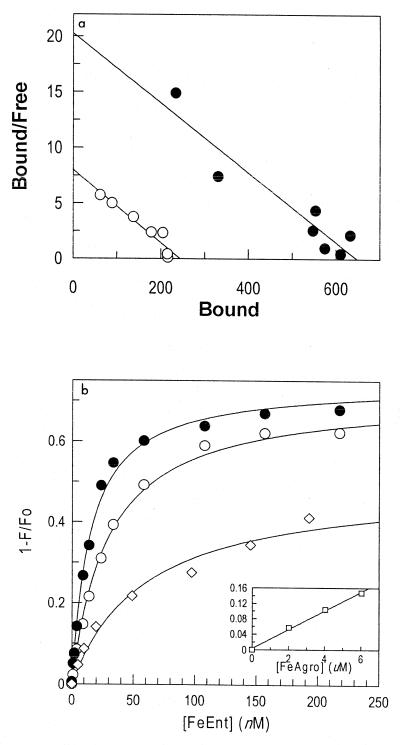
Aliquots of periplasmic fluid containing overexpressed FepB were mixed with different concentrations of 59FeEnt and chromatographed on Sephadex LH20. The counts for the first peak were summed and denoted as bound ligand. The summed counts for the second peak, which contained unbound 59FeEnt, were denoted as free ligand. From these data, we determined the ratio of bound ligand to free ligand at equilibrium (Fig. 3a). Analysis of binding data from crude FepB, in periplasmic extracts, produced an apparent Kd of 124 ± 17 nM; analogous experiments with purified FepB yielded a Kd of 145 ± 29 nM.
FIG. 3.
Binding of catecholate ferric siderophores to FepB. (a) Scatchard (41) analysis of 59FeEnt binding to FepB, measured by the column chromatography assay, using either periplasmic fluid (○) (Kd = 124 nM) or purified FepB (●) (Kd = 145 nM). (b) Fluorescence measurements using excitation and emission wavelengths of 280 and 327 nm, respectively, in the presence of Ent (◊), FeEnt (●), or FeEnEnt (○). FepB fluorescence emissions were quenched when the protein bound the ferric siderophores, and the concentration dependence of the binding (1 − F/F0) was used to estimate the affinity of the interactions. The Kd values of the binding equilibria were 60 nM for Ent, 29 nM for FeEnt, and 15 nM for FeEnEnt.
Intrinsic fluoresence spectroscopy of the four tryphophans in FepB (residues 29, 89, 152, and 209) provided another measure of the affinity of the interaction with FeEnt. Binding of the ferric siderophore did not shift the excitation or emission maxima of purified FepB (Fig. 1, pooled fractions 15 to 21; >90% 33.8-kDa band), suggesting that the tryptophans did not experience any significant change in environment. However, saturation with FeEnt reduced the fluorescence intensity of FepB by approximately 70%. The concentration dependence of this decrease showed a midpoint (Kd) at 29 ± 1.4 nM (Fig. 3b), roughly fivefold lower than that observed in the column assay. The discrepancy between the two measurements probably derives from the much shorter time frame of the spectroscopic method, which increases the accuracy of the bound/free ratio determinations (see Discussion).
Saturation of FepB with ferric enantioenterobactin (FeEnEnt) decreased the fluorescence intensity of the binding protein 75%, slightly more than that observed during the binding of FeEnt. The midpoint of this transition reflected a Kd of 14.5 ± 1 nM. Again, the excitation and emission maxima did not change. Comparable experiments with the apo-siderophore enterobactin showed that it also bound to FepA with high affinity (Kd = 60 nM). However, ferric agrobactin did not specifically adsorb to FepB. Whereas enterobactin, FeEnt, and FeEnEnt engendered a >70% decrease in the intrinsic fluorescence of FepB at saturation (0.2 μM), ferric agrobactin did not manifest saturation binding (Fig. 3b). Its addition to solutions of FepB only marginally changed the fluorescence of the binding protein (<15% at concentrations up to 6 μM), presumably as a result of collisional quenching. Thus, FepB did not recognize ferric agrobactin.
Anti-FepB sera.
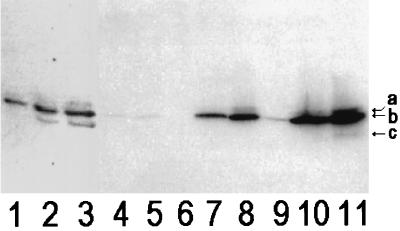
We raised polyclonal antisera against purified FepB in rabbits and monoclonal antibodies to it in mice. Western blots with the rabbit anti-FepB confirmed the absence of the binding protein in the fepB strain DK214 and its presence in the fepB+ (chromosomal or plasmid) strains (Fig. 4). FepB expressed from the chromosome or from low-copy-number plasmids was difficult to visualize in Coomassie blue-stained SDS-PAGE gels.
FIG. 4.
Chromosomal and plasmid-mediated expression of FepB in E. coli strains. The bacterial strains of interest were grown in Luria-Bertani broth, subcultured into MOPS minimal medium, grown to mid-log phase, lysed in SDS-PAGE sample buffer (5 × 108 cells), and subjected to Western immunoblotting with polyclonal rabbit anti-FepB and 125I-protein A. Lanes: 1 to 3, 1, 2.5, and 5 μg of purified FepB, respectively; 4 to 11, lysates from KDF541/pITS449, BN1071, DK214, DK214/pIB3, DK214/pIB51, BL21, BL21/p72, and BL21/p72 plus IPTG, respectively. The positions of the 36.5-kDa (a), 33.8-kDa (b), and 31.5-kDa (c) bands are marked by arrows.
Monoclonal antibodies from the 37 anti-FepB hybridomas that we raised, purified, and subcloned reacted with FepB in an enzyme-linked immunosorbent assay and with denatured FepB in Western blots. Protein A did not react with any of the MAbs; we used the polyclonal rabbit antisera for quantitation of FepB expression.
Effect of FepB overexpression on 59FeEnt uptake.
Using the variety of fepB-containing plasmids, we expressed the binding protein at various levels (Fig. 4) and measured the effects on 59FeEnt uptake (Table 2). Although FepB was necessary for transport of the ferric siderophore, variations in its concentration did not affect either the overall affinity or the rate of 59FeEnt uptake. This was best seen in 59FeEnt uptake experiments by strains expressing FepB from multicopy plasmids (pIB3, pIB51, and p72). These constructions produced FepB at 9- to 35-fold-higher levels than those resulting from chromosomal expression by the natural promoter, with little effect on the rate of 59FeEnt transport (Table 2, Fig. 4). Furthermore, we compared the concentrations of chromosomal fepB in three different strains (KDF541/pITS449, BN1071, and BL21) and plasmid-mediated fepB in two different backgrounds (DK214 and BL21). In Table 2 the 59FeEnt binding data and also the colicin susceptibility measured the affinity and amount of FepA in the outer membrane (from Kd and capacity, and percent killing, respectively), and we observed some variation of this parameter in the different strains. The transport data characterized the overall affinity and rate of the uptake process into the cytoplasm (from Km and Vmax, respectively). The comparison of FepB expression levels and 59FeEnt uptake rates in the different strains demonstrated that variations in the periplasmic concentration of FepB did not significantly change the velocity of 59FeEnt transport. For example, in the isogenic series of BL21, an approximately 10-fold increase in FepB concentration did not alter the Vmax of 59FeEnt uptake. Instead, the transport rate depended on the amount of FepA in the outer membrane, as established by the isogenic pair KDF541/pITS449 and BN1071, which have the same amount of FepB (chromosomal level) but different amounts of FepA. These results support the idea that passage through the outer membrane is the rate-limiting step of 59FeEnt transit through the cell envelope. Furthermore, from the chromosomal expression results, we calculated that each bacterial cell contains approximately 3,800 FepB proteins. The multicopy plasmids produced as many as 135,000 copies per cell (p72 plus IPTG).
TABLE 2.
FeEnt uptake by strains with chromosomeor plasmid-encoded FepB
| Strain | FeEnt
|
Colicin B killing (%)e | Colicin D killing (%)e | |||
|---|---|---|---|---|---|---|
| Binding
|
Transport
|
|||||
| Kda | Capacityb | Kmc | Vmaxd | |||
| KDF541/pITS449 | 0.09 | 111 | 0.3 | 181 | 100 | 100 |
| BN1071 | 0.103 | 27.6 | 0.829 | 120 | 4 | 2.5 |
| DK214 | 0.938 | 11.4 | 0 | 2 | 1 | |
| DK214/pIB3 | 0.584 | 11.6 | 1.45 | 83 | 2 | 2.5 |
| DK214/pIB51 | 0.896 | 27.3 | 1.5 | 179 | 2 | 2.5 |
| BL21 | 1.2 | 77.6 | 0.817 | 127 | 50 | 50 |
| BL21/p72 | 0.924 | 67.3 | 0.957 | 140 | 20 | 25 |
| BL21/p72 + IPTG | 0.538 | 83.2 | 0.711 | 116 | NDf | ND |
Kd (nanomolar) was determined by measurement of 59FeEnt binding to bacteria at 0°C. All kinetic and thermodynamic constants were obtained by analysis of the data with Grafit 4 (Erithacus Software Ltd., London, United Kingdom).
59FeEnt binding capacity (picomoles/109 cells).
Km (nanomolar) was determined for the 59FeEnt uptake reaction into live bacteria at 37°C.
Vmax (picomoles/109 cells/minute) was determined from 59FeEnt uptake measurements.
Colicin B or D killing is expressed relative to that of KDF541/pITS449 (24), which contains chromosomal FepB and multicopy plasmid-encoded FepA (a pUC18 derivative).
ND, not determined.
DISCUSSION
The native structure of the FepB protein may resemble that of other gram-negative bacterial periplasmic binding proteins, including those of the sugar (MalE [43], MglB [50], RbsB [25], and AraF [9]), amino acid (HisJ [32], LivJ [40], LivK [38], and GltP [12]), and iron (3) transport systems: a double-lobed kidney bean that closes around the ligand during binding. However, the FeEnt-FepB system differs from other binding protein-dependent transport systems in several ways. First, FepB and another iron binding periplasmic protein of E. coli, FhuD (4, 17) are synthesized at low levels relative to sugar (MalE [16]) and amino acid (HisJ [45]) binding proteins and the iron binding protein Fbp from Haemophilus influenzae (10). Even when derepressed by iron starvation, chromosomally encoded FepB was not detected in Coomassie blue-stained gels of periplasmic extracts. We needed overexpression to see FepB in SDS-PAGE, and similar findings were reported for FhuD (17, 18).
Second, the affinity of FepB for its ligand is apparently greater than that of other characterized periplasmic binding proteins. The Kd of the FeEnt-FepB interaction lies in the nanomolar range, while those of FhuD (37) and sugar or amino acid binding proteins (MalE [47] and HisJ [51]) are micromolar. It is most relevant to address this difference in light of the similar fluorescence measurements that were recorded for MalE, FhuD, and FepB and in relationship to the nature of the outer membrane components of the three transport systems. The comparison suggests that in specific porin-mediated transport, the affinity of the outer membrane protein (LamB: Kd = 2 μM [15, 46, 47]) for the sugar maltose is comparable to that of the periplasmic protein (MalE: Kd = 1 μM [47]), whereas in the ligand-gated porin systems, the affinity of the outer membrane protein (FepA: Kd = 0.1 nM [31]; FhuA: Kd = 50 to 100 nM [19]) for the ferric siderophore considerably exceeds that of the periplasmic protein (FepB: Kd = 30 nM [this study]; FhuD: Kd = 1 μM [37]). Thus, specific porin and ligand-gated porin transport occur with fundamentally different parameters. The low-affinity nature of maltose uptake means that the carbon source must reach high external concentrations to achieve uptake into the periplasm. In this case, in which solute efflux may occur through the open maltoporin channel, equilibration across the outer membrane creates high periplasmic concentrations of the sugar, which are appropriately matched by the micromolar Kd of the MalE binding reaction. In ferric siderophore transport systems, on the other hand, efflux of the solute apparently does not occur, because of the gating of transport through the TonB- and energy-dependent outer membrane receptor protein. However, ferric siderophore uptake systems accumulate iron from very low external concentrations, and under such conditions the periplasmic concentration of the solute may not reach micromolar levels, hence the need for a binding protein with higher affinity. In both transport systems, the binding proteins function with sufficient affinity to create a periplasmic pool of bound solute; this complex presumably is the substrate for the inner membrane permease system.
Our results also show differences between the ferric catecholate binding protein, FepB, and the ferric hydroxamate binding protein, FhuD. The latter functions in the transport of ferrichrome, aerobactin, and coprogen. All three siderophores protect FhuD from degradation by proteinase K (17) and have measurable affinity for His-tagged FhuD (37). The broad specificity and lower affinity of FhuD may stem from the lack of a deep cleft in its tertiary structure (5). Conversely, FepB exclusively bound ferric enterobactin (natural or enantio); it did not accept the relatively similar catecholate ferric agrobactin. Other experiments suggest that FepB does not recognize ferric vibriobactin, a catecholate siderophore with structural similarity to ferric agrobactin (52).
The existence of multiple electrophoretic forms of FepB represents a third difference from sugar and amino acid binding proteins. Like others (6), we observed three FepB isoforms, of 36.5, 33.8, and 31.5 kDa. However, they were only apparent when FepB was heavily overexpressed (Fig. 4): normal production from the chromosome produced a homogeneous band of 33.8 kDa. The 31.5- and 33.8-kDa polypeptides had identical N termini, refuting the possibility of different signal peptidase cleavage sites. Thus, the 33.8-kDa band is not pro-FepB (predicted from the sequence as 34.3 kDa). Rather, the 36.5-kDa band (6) is probably unprocessed pro-FepB. The 33.8-kDa protein which was the major FepB isoform both in vivo and after purification, may contain a posttranslational modification with lipid (for a review, see reference 39), as is found for other FeEnt binding proteins, CeuE of Campylobacter jejuni and ViuP of Vibrio cholerae (52). FepB does not contain the most common lipid attachment site (Cys in the motif LLAAC [11, 48]); in fact, the protein does not contain cysteine, and so if lipidation does occur, it takes place at a novel site. The 31.5-kDa band probably represents mature, nonposttranslationally modified FepB (predicted to be 31.6 kDa), which appears under overexpression conditions because such high concentrations of the binding protein overload the lipidation system. Unlike FepB, FhuD has only a proform and a mature form (17, 18), and so lipidation is apparently not a requisite feature of binding proteins associated with TonB-dependent transport systems.
The Sephadex LH20 chromatography assay independently demonstrated binding between FeEnt and FepB, but it was of limited value for quantitative affinity determinations, because of the time required to perform the experiment. Assuming a simple equilibrium with adsorption of FeEnt to FepB at the diffusion limit, a Kd of 30 nM predicts a dissociation half-life for FeEnt-FepB of <1 s. Therefore, as the complex traverses the resin, some of the FeEnt will release from it and fractionate away from the bound peak, causing an underestimation of the equilibrium concentration of FeEnt-FepB. This explains the lower estimate of affinity: the Kd of 130 nM that we obtained from the chromatographic method suggests that about 75% of the FeEnt that initially adsorbed to FepB dissociated and separated from the binding protein during chromatography.
ACKNOWLEDGMENTS
We thank Jean Michel Betton (Institut Pasteur, Paris, France) for helpful discussions, Hashimoto Gotoh (National Institute of Genetics, Tokyo, Japan) for the gift of cloning vector pHSG398, and Charles Earhast and Mark McIntosh for bacterial strains and plasmids.
This work was supported by grant GM53836 from the National Institutes of Health and grant MCB9709418 from the National Science Foundation to P.E.K.
REFERENCES
- 1.Ames G F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974;249:634–644. [PubMed] [Google Scholar]
- 2.Bagg A, Neilands J B. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruns C M, Nowalk A J, Arvai A S, McTigue M A, Vaughan K G, Mietzner T A, McRee D E. Structure of Haemophilus influenzae Fe(+3)-binding protein reveals convergent evolution within a superfamily. Nat Struct Biol. 1997;4:919–924. doi: 10.1038/nsb1197-919. [DOI] [PubMed] [Google Scholar]
- 4.Burkhardt R, Braun V. Nucleotide sequence of the fhuC and fhuD genes involved in iron (III) hydroxamate transport: domains in FhuC homologous to ATP-binding proteins. Mol Gen Genet. 1987;209:49–55. doi: 10.1007/BF00329835. [DOI] [PubMed] [Google Scholar]
- 5.Clarke T E, Ku S Y, Dougan D R, Vogel H J, Tari L W. The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat Struct Biol. 2000;7:287–291. doi: 10.1038/74048. [DOI] [PubMed] [Google Scholar]
- 6.Elkins M F, Earhart C F. Nucleotide sequence and regulation of the Escherichia coli gene for ferrienterobactin transport protein FepB. J Bacteriol. 1989;171:5443–5451. doi: 10.1128/jb.171.10.5443-5451.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forsgren A, Sjoquist J. “Protein A” from Staphylococcus aureus. 3. Reaction with rabbit gamma-globulin. J Immunol. 1967;99:19–24. [PubMed] [Google Scholar]
- 9.Gilliland G L, Quiocho F A. Structure of the l-arabinose-binding protein from Escherichia coli at 2.4 A resolution. J Mol Biol. 1981;146:341–362. doi: 10.1016/0022-2836(81)90392-2. [DOI] [PubMed] [Google Scholar]
- 10.Harkness R E, Chong P, Klein M H. Identification of two iron-repressed periplasmic proteins in Haemophilus influenzae. J Bacteriol. 1992;174:2425–2430. doi: 10.1128/jb.174.8.2425-2430.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayashi S, Wu H C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990;22:451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao C D, Sun Y J, Rose J, Wang B C. The crystal structure of glutamine-binding protein from Escherichia coli. J Mol Biol. 1996;262:225–242. doi: 10.1006/jmbi.1996.0509. [DOI] [PubMed] [Google Scholar]
- 13.Klebba P E, Benson S A, Bala S, Abdullah T, Reid J, Singh S P, Nikaido H. Determinants of OmpF porin antigenicity and structure. J Biol Chem. 1990;265:6800–6810. [PubMed] [Google Scholar]
- 14.Klebba P E, Newton S M. Mechanisms of solute transport through outer membrane porins: burning down the house. Curr Opin Microbiol. 1998;1:238–247. doi: 10.1016/s1369-5274(98)80017-9. [DOI] [PubMed] [Google Scholar]
- 15.Klebba P E, Newton S M, Charbit A, Michel V, Perrin D, Hofnung M. Further genetic analysis of the C-terminal external loop region in Escherichia coli maltoporin. Res Microbiol. 1997;148:375–387. doi: 10.1016/S0923-2508(97)83868-5. [DOI] [PubMed] [Google Scholar]
- 16.Koman A, Harayama S, Hazelbauer G L. Relation of chemotactic response to the amount of receptor: evidence for different efficiencies of signal transduction. J Bacteriol. 1979;138:739–747. doi: 10.1128/jb.138.3.739-747.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koster W, Braun V. Iron (III) hydroxamate transport into Escherichia coli. Substrate binding to the periplasmic FhuD protein. J Biol Chem. 1990;265:21407–21410. [PubMed] [Google Scholar]
- 18.Koster W, Braun V. Iron-hydroxamate transport into Escherichia coli K12: localization of FhuD in the periplasm and of FhuB in the cytoplasmic membrane. Mol Gen Genet. 1989;217:233–239. doi: 10.1007/BF02464886. [DOI] [PubMed] [Google Scholar]
- 19.Locher K P, Rosenbusch J P. Oligomeric states and siderophore binding of the ligand-gated FhuA protein that forms channels across Escherichia coli outer membranes. Eur J Biochem. 1997;247:770–775. doi: 10.1111/j.1432-1033.1997.t01-1-00770.x. [DOI] [PubMed] [Google Scholar]
- 20.Marchalonis J J. An enzymic method for the trace iodination of immunoglobulins and other proteins. Biochem J. 1969;113:299–305. doi: 10.1042/bj1130299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987;262:10035–10038. [PubMed] [Google Scholar]
- 22.McIntosh M A, Earhart C F. Coordinate regulation by iron of the synthesis of phenolate compounds and three outer membrane proteins in Escherichia coli. J Bacteriol. 1977;131:331–339. doi: 10.1128/jb.131.1.331-339.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIntosh M A, Earhart C F. Effect of iron of the relative abundance of two large polypeptides of the Escherichia coli outer membrane. Biochem Biophys Res Commun. 1976;70:315–322. doi: 10.1016/0006-291x(76)91144-x. [DOI] [PubMed] [Google Scholar]
- 24.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 25.Mowbray S L, Cole L B. 1.7 Å X-ray structure of the periplasmic ribose receptor from Escherichia coli. J Mol Biol. 1992;225:155–175. doi: 10.1016/0022-2836(92)91033-l. [DOI] [PubMed] [Google Scholar]
- 26.Murphy C K, Kalve V I, Klebba P E. Surface topology of the Escherichia coli K-12 ferric enterobactin receptor. J Bacteriol. 1990;172:2736–2746. doi: 10.1128/jb.172.5.2736-2746.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neilands J B. Siderophores: structure and function of microbial iron transport compounds. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 29.Neilands J B, Erickson T J, Rastetter W H. Stereospecificity of the ferric enterobactin receptor of Escherichia coli K-12. J Biol Chem. 1981;256:3831–3832. [PubMed] [Google Scholar]
- 30.Neu H C, Heppel L A. On the surface localization of enzymes in E. coli. Biochem Biophys Res Commun. 1964;17:215–219. doi: 10.1016/0006-291x(64)90386-9. [DOI] [PubMed] [Google Scholar]
- 31.Newton S M, Igo J D, Scott D C, Klebba P E. Effect of loop deletions on the binding and transport of ferric enterobactin by FepA. Mol Microbiol. 1999;32:1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 32.Oh B H, Kang C H, De Bondt H, Kim S H, Nikaido K, Joshi A K, Ames G F. The bacterial periplasmic histidine-binding protein. structure/function analysis of the ligand-binding site and comparison with related proteins. J Biol Chem. 1994;269:4135–4143. [PubMed] [Google Scholar]
- 33.Ozenberger B A, Nahlik M S, McIntosh M A. Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J Bacteriol. 1987;169:3638–3646. doi: 10.1128/jb.169.8.3638-3646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pierce J R, Earhart C F. Escherichia coli K-12 envelope proteins specifically required for ferrienterobactin uptake. J Bacteriol. 1986;166:930–936. doi: 10.1128/jb.166.3.930-936.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierce J R, Pickett C L, Earhart C F. Two fep genes are required for ferrienterochelin uptake in Escherichia coli K-12. J Bacteriol. 1983;155:330–336. doi: 10.1128/jb.155.1.330-336.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugsley A P, Reeves P. Characterization of group B colicin-resistant mutants of Escherichia coli K-12: colicin resistance and the role of enterochelin. J Bacteriol. 1976;127:218–228. doi: 10.1128/jb.127.1.218-228.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohrbach M R, Braun V, Koster W. Ferrichrome transport in Escherichia coli K-12: altered substrate specificity of mutated periplasmic FhuD and interaction of FhuD with the integral membrane protein FhuB. J Bacteriol. 1995;177:7186–7193. doi: 10.1128/jb.177.24.7186-7193.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sack J S, Trakhanov S D, Tsigannik I H, Quiocho F A. Structure of the l-leucine-binding protein refined at 2.4 A resolution and comparison with the Leu/Ile/Val-binding protein structure. J Mol Biol. 1989;206:193–207. doi: 10.1016/0022-2836(89)90532-9. [DOI] [PubMed] [Google Scholar]
- 39.Sankaran K, Gupta S D, Wu H C. Modification of bacterial lipoproteins. Methods Enzymol. 1995;250:683–697. doi: 10.1016/0076-6879(95)50105-3. [DOI] [PubMed] [Google Scholar]
- 40.Saper M A, Quiocho F A. Leucine, isoleucine, valine-binding protein from Escherichia coli. Structure at 3.0-A resolution and location of the binding site. J Biol Chem. 1983;258:11057–11062. [PubMed] [Google Scholar]
- 41.Scatchard G. The attractions of proteins for small molecules and ions. Ann N Y Acad Sci. 1949;51:660–672. [Google Scholar]
- 42.Shea C M, McIntosh M A. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol Microbiol. 1991;5:1415–1428. doi: 10.1111/j.1365-2958.1991.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 43.Spurlino J C, Lu G Y, Quiocho F A. The 2.3-A resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 44.Stephens D L, Choe M D, Earhart C F. Escherichia coli periplasmic protein FepB binds ferrienterobactin. Microbiology. 1995;141:1647–1654. doi: 10.1099/13500872-141-7-1647. [DOI] [PubMed] [Google Scholar]
- 45.Stern M J, Prossnitz E, Ames G F. Role of the intercistronic region in posttranscriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol Microbiol. 1988;2:141–152. doi: 10.1111/j.1365-2958.1988.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 46.Szmelcman S, Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975;124:112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szmelcman S, Schwartz M, Silhavy T J, Boos W. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild-type and lambda-resistant mutants as measured by fluorescence quenching. Eur J Biochem. 1976;65:13–19. doi: 10.1111/j.1432-1033.1976.tb10383.x. [DOI] [PubMed] [Google Scholar]
- 48.Takeshita S, Sato M, Toba M, Masahashi W, Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61:63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- 49.Thulasiraman P, Newton S M, Xu J, Raymond K N, Mai C, Hall A, Montague M A, Klebba P E. Selectivity of ferric enterobactin binding and cooperativity of transport in gram-negative bacteria. J Bacteriol. 1998;180:6689–6696. doi: 10.1128/jb.180.24.6689-6696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vyas N K, Vyas M N, Quiocho F A. The 3 A resolution structure of a D-galactose-binding protein for transport and chemotaxis in Escherichia coli. Proc Natl Acad Sci USA. 1983;80:1792–1796. doi: 10.1073/pnas.80.7.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wolf A, Shaw E W, Oh B H, De Bondt H, Joshi A K, Ames G F. Structure/function analysis of the periplasmic histidine-binding protein. Mutations decreasing ligand binding alter the properties of the conformational change and of the closed form. J Biol Chem. 1995;270:16097–16106. doi: 10.1074/jbc.270.27.16097. [DOI] [PubMed] [Google Scholar]
- 52.Wyckoff E E, Valle A M, Smith S L, Payne S M. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J Bacteriol. 1999;181:7588–7596. doi: 10.1128/jb.181.24.7588-7596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]