Abstract
Borrelia burgdorferi, the causative agent of Lyme disease, has recently been demonstrated to infect and enhance the invasive properties of breast cancer cells, while also influencing the expression of inflammatory chemokines (CXCL8 and CXCL10). This study investigates the presence of B. burgdorferi in invasive breast cancer tissues using commercially available, FDA-approved breast cancer tissue microarrays consisting of 350 ductal, 32 lobular, and 22 intraductal invasive breast carcinomas, alongside 29 normal breast tissues. Employing fluorescent immunohistochemical staining and high-resolution imaging, the findings revealed that approximately 20% of invasive lobular and ductal carcinomas, followed by 14% of intraductal carcinomas, tested positive for B. burgdorferi, while all normal breast tissues tested negative. PCR analysis further confirmed the presence of B. burgdorferi DNA in breast cancer tissues. Moreover, 25% of B. burgdorferi-positive tissues exhibited expression of both chemokines, CXCL8 and CXCL10, which was not observed in B. burgdorferi-negative tissues. Analysis of available patient data, including age, indicated a correlation between older patients and B. burgdorferi-positive tissues. This study validates the presence of B. burgdorferi in invasive breast cancer tissues and highlights the involvement of key CXCL family members associated with inflammatory processes.
Keywords: Lyme disease, breast cancer, Borrelia burgdorferi infection, inflammation
Introduction
According to the American Cancer Society, infectious agents are associated with 20% of all cancers [1]. In recent years, among infectious agents, bacterial infections have been the center point for the emergence and progression of different types of cancer. Pathogenic bacteria have been shown to promote chronic inflammation and evasion of the immune system leading to a specialized niche tissue microenvironment that can lead to enhanced cell proliferation and increased risk of tumorigenesis [2].
The most well-known instance of bacterial infections leading to cancer development is Helicobacter pylori infection causing gastric cancer [3, 4]. Additionally, the significance of various pathogens in cancer, such as Salmonella typhi in gall bladder cancer, Chlamydia spp. in pulmonary Mucosa-Associated Lymphoid Tissue (MALT) lymphoma, Streptococcus spp. in colorectal carcinoma, and chronic infection and inflammation via Chlamydia spp. and Mycoplasma spp. in lung and ovarian cancer, has been recognized by several studies [5, 6]. Staphylococcus epidermidis and strains of Escherichia coli have also been found in breast cancer; however, the causal relationship has not been firmly established [5, 7].
A recent microbiome study examining various types of breast cancer tissues identified DNA signatures of several human pathogens, including Borrelia burgdorferi, a spirochetal bacterium, the causative agent of Lyme disease [8]. The presence of B. burgdorferi has been associated with poor prognosis. This prompts inquiry into whether this spirochete might play a role in aggressive breast cancers, as prior studies have connected B. burgdorferi infection with malignancies such as cutaneous B-cell lymphoma and non-Hodgkin lymphoma [9–11].
Many pathogens can penetrate host cells and use the host niche environment for colonization. They replicate in the cellular compartments and cross host barriers for dissemination [12]. B. burgdorferi was previously found to be capable of disseminating and colonizing into multiple host organs and tissues which aid in its pathogenicity and overall survival [13]. Intracellular localization of B. burgdorferi has also been reported for different mammalian cells which could help the bacteria evade environmental changes, antibiotic treatment, and the host immune system [14–19]. The pathogenicity of B. burgdorferi was also seen to be able to persist in different tissues through mechanisms that manipulate the innate and adaptive immune response along with host-protein interaction to induce vascular and extracellular matrix changes [20].
A recent in vitro study also demonstrated that B. burgdorferi can invade triple-negative breast cancer cells at greater rates than normal mammary epithelial cells and increase cancer cell invasive potential [19]. Further studies showed a significant change in genome-wide RNA and miRNA expressions upon B. burgdorferi infection in triple-negative breast cancer and normal mammary epithelial cells [21, 22]. The genes affected were further analyzed for their role in physiological processes, including host inflammation and cancer signaling pathways. It was discovered that B. burgdorferi actively alters cellular responses to favor pathways associated with inflammation and tumor progression [23]. One of the major findings from this study was that several members of the C-X-C motif chemokine family, especially CXCL8 and CXCL 10 were highly elevated after B. burgdorferi infection [22]. The CXCL8 protein, also known as interleukin 8 (IL-8), is secreted by cells of the innate immune system and serves as a chemoattractant to recruit neutrophils [23]. CXCL10, on the other hand, exerts its functions through its receptor CXCR3, inducing pleiotropic effects such as the stimulation of NK cells, monocytes, and T-cell migration. It also serves as a key regulatory factor in the context of the ‘cytokine storm’ [23]. The importance of these findings warrants additional investigation into the presence of B. burgdorferi within cancer tissues to explore deeper into the underlying processes.
This study aims to investigate the presence of B. burgdorferi antigen/DNA in commercially available, FDA-approved paraffin-embedded breast cancer tissue arrays. B. burgdorferi localization and morphology were evaluated by IHC staining combined with high-resolution microscopic analysis of these breast cancer tissues. Moreover, previously reported inflammatory markers induced by B. burgdorferi, such as CXCL8 and CXCL10, were analyzed to elucidate potential correlations between the presence of B. burgdorferi and host inflammatory responses.
Materials and methods
Breast cancer tissue arrays
Commercially available FDA-approved breast cancer paraffin-embedded tissue arrays containing breast cancer tissues were purchased from US Biomax, Inc., Rockville, MD (BR2161, BN1021, BR2082, BR2082a, BR041, BR082b) and Novus Biologicals USA (NBP2-42080). Together these tissue arrays contained 404 cases covering all the common types of breast cancer tissues from female patients of various age groups with different pathological diagnoses and grades of tumor. This study also included 29 normal breast cancer tissues as controls. Each breast cancer tissue core was 5 μm in thickness and 1 mm in diameter.
Immunohistochemical (IHC) staining
The breast cancer tissue arrays were deparaffinized on a slide warmer for 15 min and followed with three 5-min xylene washes at room temperature (RT). The tissue array was then rehydrated by immersing in 100, 90, and 70% methanol wash for 5 min each respectively, and placed under slow-running tap water for 25 min to remove the traces of alcohol from the slide. The slides were rinsed with 1X Phosphate Buffered Saline (PBS, Sigma-Aldrich, St. Louis. MO, USA) mixed with 1% bovine serum albumin solution (Sigma-Aldrich) and distilled water for 5 min each. The excess solution around the tissue was gently wiped using Kim wipes. The water bath was equilibrated to 99 °C and slides were immersed into a Coplin jar with preheated mediated antigen retrieval buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 10 min. The Coplin jar was then removed from the water bath and set at RT for 20 min to cool the buffer. This step was followed by placing the slides under slow-running tap water to remove the traces of the sodium citrate buffer for 5 min. Then slides were washed 3x with 1X PBS +1% BSA and distilled water for 5 min each.
The tissue sections were then blocked with a 1:200 dilution of goat serum (Thermo Fisher Scientific, Waltman, MA, USA) in 1X PBS for 1 h in a humidified chamber at RT. Excess solution was gently dabbed on a Kim wipe, and the slides were rinsed with 1X PBS +1% BSA and distilled water for 5 min each. Different sets of tissue array slides were stained for CXCL8 using polyclonal IL8/CXCL8 rabbit antibody (Cat#A2541, ABclonal Technology, Woburn, MA, USA) and for CXCL10 using polyclonal CXCL10 antibody (Cat#DF6417, Affinity Biosciences, Cincinnati, OH, USA). Both antibodies were diluted to 1:200 in 1X PBS before being added to the tissue slides. The slides were incubated at 4 °C overnight in a humidified chamber. The next day the slides were rinsed 3x with 1X PBS +1% BSA and distilled water for 5 min each. The tissue sections were then treated with a 1:200 dilution of the secondary anti-rabbit antibody with a fluorescent red tag (goat anti-rabbit IgG (H+L), DyLight 594 conjugated, (Cat#35569, Invitrogen, Waltman, MA, USA) and incubated for an hour at RT. The excess solution around the tissue was gently wiped and washed as mentioned above. The slides were then treated with B. burgdorferi monoclonal antibody (Cat#MA1-7006, Invitrogen) which was diluted to 1:200 with 1X PBS and added to the slides. The slides were incubated at 4 °C overnight in a humidified chamber. The next day the slides were washed 3x with 1X PBS +1% BSA and distilled water for 5 min each.
The tissue sections were then treated with a 1:200 dilution of the secondary anti-mouse antibody with a fluorescent green tag (goat anti-mouse IgG (H+L), DyLight 488 conjugated, Cat#35502, Invitrogen) and incubated for an hour at RT than washed as mentioned above. The slide sections were then counterstained with 0.1% Sudan black (Sigma) for 20 min at RT. Slides were washed again one last time with 1XPBS and then mounted with Prolong Diamond Antifade Mounting media with DAPI (Thermo Fisher Scientific). Images were taken and processed using a Nikon Eclipse 80i and Leica Thunder widefield fluorescent microscope at 200X and 400X magnifications.
DNA Extraction/PCR
Genomic DNA was extracted from paraffin-embedded tissue arrays using a Qiagen AllPrep DNA/RNA FFPE Kit (Qiagen Cat# 80234, Hilden, Germany). First, the slides were deparaffinized as described above and the tissues on the slides were scraped off using a sterile scalpel and collected in sterile microcentrifuge tubes. The genomic DNA was then isolated using the DNA extraction protocol as suggested by the manufacturer. The DNA samples were quantified using a BioTek (Winooski, VT, USA) Microplate Spectrophotometer.
The DNA samples were tested with standard PCR using primers designed to amplify B. burgdorferi CTP synthase genes using published primers [24]. CTP synthase sequences were amplified in nested reactions. First-round primers were F: 5′-ATTGCAAGTTCTGAGAATA-3′ and R: 5′-CAAACATTACGAGCAAATTC-3′ in a 50 μL reaction with HotStarTaq buffer (Qiagen), 25 pmoles of each primer, MgCl2 adjusted to 2.5 mM, and 1.5 units HotStarTaq (Qiagen, Cat# 203203). Reaction conditions were an initial denaturation at 94 °C for 15 min, followed by 30 cycles of 94 °C/30 s, 48 °C/30 s, 72 °C/30 s, and a final extension at 72 °C/5 min. The nested reaction primers were F: 5′-GATATGGAAAATATTTTATTTATTG-3′ and R: 5′AAACCAAGACAAATTCCAAG-3′ in a 50 µL reaction containing 25 pmoles of each primer, 2.5 mM MgCl2, 1.5 units HotStarTaq. Reaction conditions were an initial denaturation at 94 °C for 15 min, followed by 35 cycles of 94 °C/30 s, 50 °C/30 s, 72 °C/30 s, and a final extension at 72 °C/5 min. The PCR products were analyzed by standard agarose gel-electrophoresis technique and Hi-Lo DNA maker was used to determine the size of the obtained bands (Minnesota Molecules, Minneapolis, MN).
Sequence analysis
The amplified DNA was purified using the QIAGEN PCR purification kit (Qiagen) following the manufacturer's instructions. To confirm the amplified products belonged to Borrelia spp, the purified DNA samples were sent for sequencing in both directions with forward and reverse primers to Eurofins Genomics, Louisville, KY. The sequencing results were matched against the NCBI server using the Basic Local Alignment Search Tool (BLAST, https://blast.ncbi.nlm.nih.gov/Blast.cgi) which confirmed the data obtained through PCR.
Ethics
It is not applicable because this study used FDA-approved, commercially available breast cancer tissue array slides.
Results
The first part of this study was to determine the presence of B. burgdorferi in human breast cancer tissues in FDA-approved, commercially available breast cancer tissue arrays using fluorescent (IHC) staining technique with B. burgdorferi sensu stricto (s.s.) specific monoclonal antibodies.
The tissue array slides included 350 ductal, 32 lobular, 22 intraductal invasive breast carcinomas, along with 29 normal breast tissues. The IHC results demonstrated that approximately 20% of invasive breast ductal carcinoma, 19% lobular, and 14% intraductal invasive breast carcinomas were positive for B. burgdorferi antigen while all the normal control breast tissues were negative (Table 1).
Table 1.
B. burgdorferi-positive breast cancer samples and normal breast tissues according to the pathology of tissue
| Type of Tissue | Percentage of B. burgdorferi sensu stricto positive samples (%) |
| Invasive ductal carcinoma (350) | 20 |
| Invasive lobular carcinoma (32) | 19 |
| Intraductal carcinoma (22) | 14 |
| Normal breast tissues (29) | 0 |
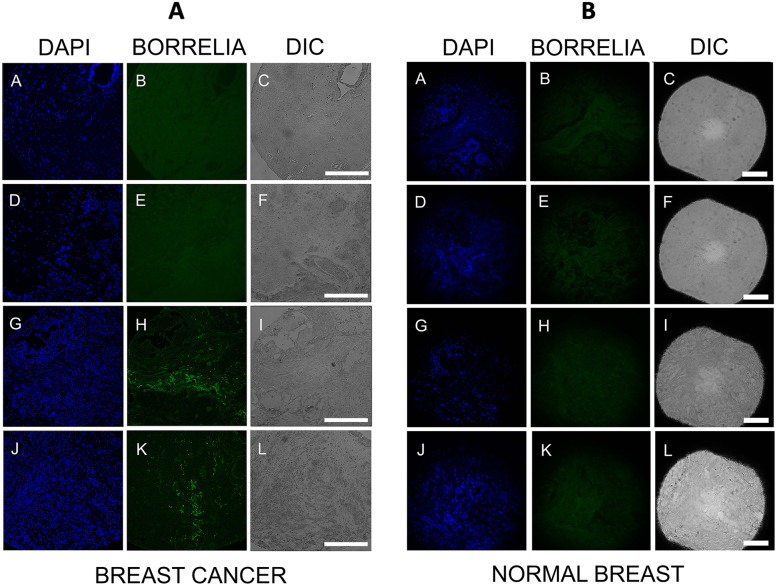
Figure 1A and B provide the representative IHC images for B. burgdorferi-positive and negative breast cancer tissues and normal breast biopsy tissues. B. burgdorferi-positive breast cancer tissues of invasive ductal and lobular carcinomas which had fluorescent green staining are shown in Fig. 1A; Panels H, K respectively. B. burgdorferi-negative breast cancer tissues with no detectable green signal can be seen in Fig. 1A; Panels B, E. There was no B. burgdorferi specific staining in any of the control samples from healthy donors (Fig. 1B; Panels B, E, H, K).
Fig. 1.
Representative images of B. burgdorferi specific IHC staining of breast cancer and normal breast tissue arrays. Fig. 1A, Panels B and E show B. burgdorferi-negative breast cancer tissues and Panels H and K show B. burgdorferi-positive breast cancer tissues (green staining). Panels A, D, G, J show the DAPI stained nucleus of the corresponding tissues. Panels C, F, I, L: Differential interference microscopy (DIC) demonstrating the size and tissue morphology of the corresponding tissues. Fig. 1B: Panels B, E, H, K show B. burgdorferi-negative normal breast tissues and Panels A, D, G, J show the DAPI nuclear stain of the corresponding normal breast tissues. Panels C, F, I, L: Differential interference microscopy (DIC) demonstrating the size and tissue morphology of the corresponding tissues. White scale bar: 100 µm
To visualize the tissue morphology in the different biopsy samples, differential interference microscopy (DIC) was used (Fig. 1A and B; Panels C, F, I, L). DAPI nuclear staining was utilized to detect nuclei of the cells (Fig. 1A and B: Panels A, D, G, J).
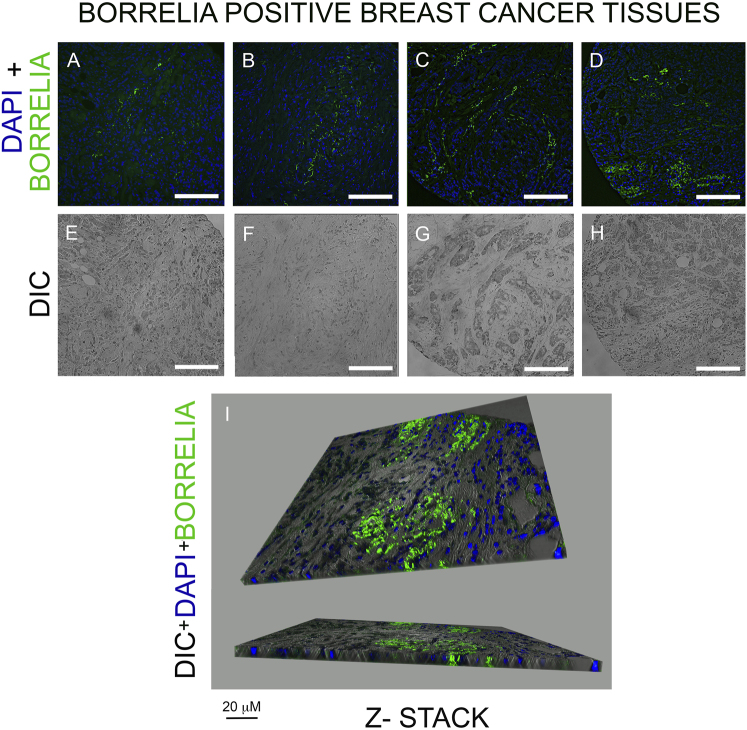
The B. burgdorferi-positive breast cancer tissue sections were further visualized with a Leica Thunder widefield fluorescence microscope in the breast cancer tissues along with their 3D distribution (Fig. 2). The DAPI/Borrelia merged images visualize the presence of the spirochetal form along with some B. burgdorferi aggregates at specific sites of the tissues (Fig. 2: Panels: A–D). The tissue morphology of the different biopsy samples was visualized using DIC (Fig. 2; Panels E, F, G, H).
Fig. 2.
Representative images of B. burgdorferi-specific IHC staining of breast cancer tissue array. Panels A–D: Merged DAPI (blue nuclei staining) and B. burgdorferi (green staining) showing the localization of B. burgdorferi spirochetes and aggregates in breast cancer tissues Panels E–H: Differential interference microscopy (DIC) demonstrating the size and tissue morphology of the corresponding tissues. Panel I: A representative Leica Thunder Z-stack 3D image of B. burgdorferi-positive breast cancer tissue (Panel D). White scale bar: 100 µm
In Fig. 2, Panel I demonstrates a 3D image made by Z-stacks (using Leica Thunder microscopy) of the B. burgdorferi-positive sample (Fig. 2, Panel D) visualizing that the spirochetes aggregated inside the breast cancer tissu.
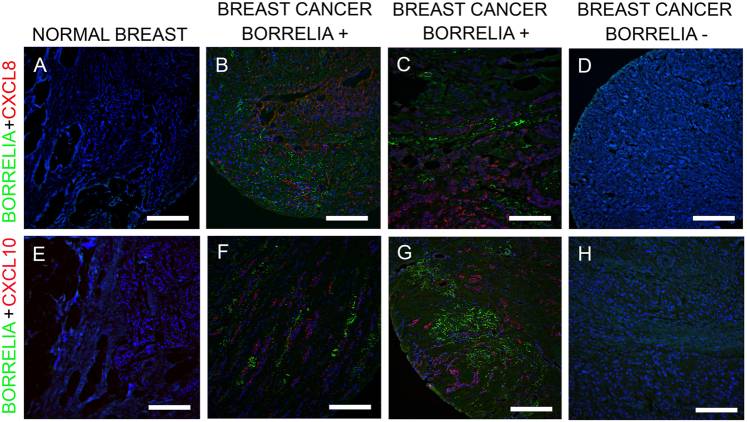
The study further focused on determining if there was any evidence of inflammatory stimuli due to the presence of B. burgdorferi infection, using the previously reported inflammatory markers CXCL8 and CXCL10. The obtained IHC results suggest evidence for inflammatory response localized near the B. burgdorferi spirochetes and aggregates (Fig. 3; Panels B, C, F, and G). On the contrary, the breast cancer tissues were negative for B. burgdorferi (Fig. 3; Panels D and H) and normal breast tissues had no detectable CXCL8 and CXCL10 staining (Fig. 3; Panels A and E).
Fig. 3.
Representative images of CXCL8 and CXCL10 inflammatory markers on B. burgdorferi-positive and negative breast cancer and normal breast tissues. Panels: B and C show B. burgdorferi-positive breast cancer tissues stained Borrelia monoclonal antibody (green) and CXCL8 monoclonal antibody (red). Panels: F and G show B. burgdorferi-positive breast cancer tissues stained Borrelia monoclonal antibody (green) and CXCL10 monoclonal antibody (red). Panels: D and H show no staining on B. burgdorferi-negative breast cancer tissue samples for both inflammatory markers, CXCL8 and CXCL10 respectively. Panels: A and E show negative control; normal breast tissues are negative for B. burgdorferi and both inflammatory markers. White scale bar: 100 µm
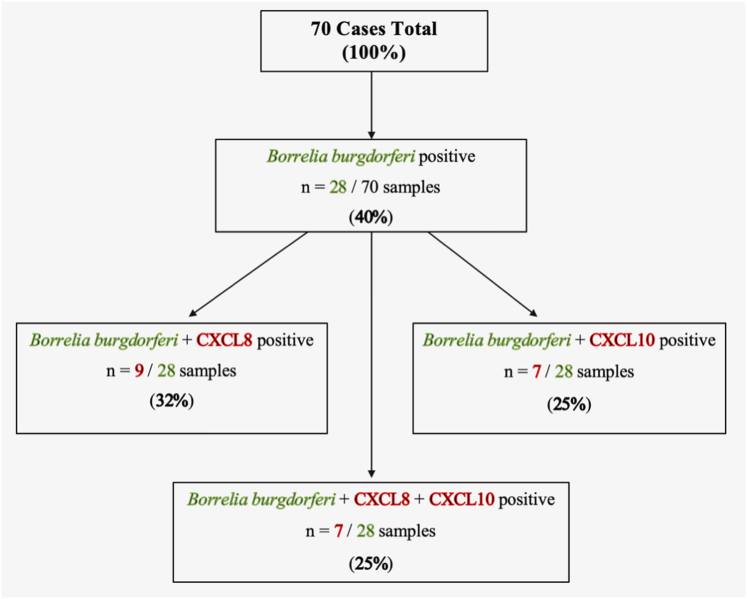
Quantitative analysis of the presence of CXCL8 and CXCL10 on the B. burgdorferi-positive and negative breast cancer samples with 70 cases/marker showed that the tissue samples positive for B. burgdorferi were also positive for both markers in about 25% of the cases. Individually, out of the B. burgdorferi-positive tissues, 32% were positive for the CXCL8 marker and 25% were positive for the CXCL10 marker from their respective tissue samples (Fig. 4). Interestingly, all the inflammatory marker-positive tissue samples were found to be ductal carcinoma except a single lobular carcinoma sample in the CXCL8-positive tissues.
Fig. 4.
A summary diagram of the B. burgdorferi, CXCL8 and CXCL10-positive and negative breast cancer tissues with a total of 70 cases
The B. burgdorferi-positive breast cancer and B. burgdorferi-negative normal breast tissue samples were used to confirm the presence of B. burgdorferi DNA using a nested pyrG-specific PCR reaction as described previously [33]. Due to the small size of the tissue samples, tissues from ten different cases of B. burgdorferi-positive invasive ductal carcinoma were pooled together for DNA extraction. The amplified PCR product was then evaluated using gel electrophoresis and direct sequencing. Figure 5 demonstrates that a positive 603 bp long amplicon was obtained from the extracted breast cancer DNA samples as well as from the positive B. burgdorferi DNA control but not from the ‘No DNA’ negative control samples. The PCR amplified products were further confirmed for the presence of Borrelia spp. by direct sequencing analysis and mapped using the Basic Local Alignment Search Tool (BLAST, Fig. 6). The maximum hits observed were B. burgdorferi s.s. specific strains (Identity: 99%).
Fig. 5.
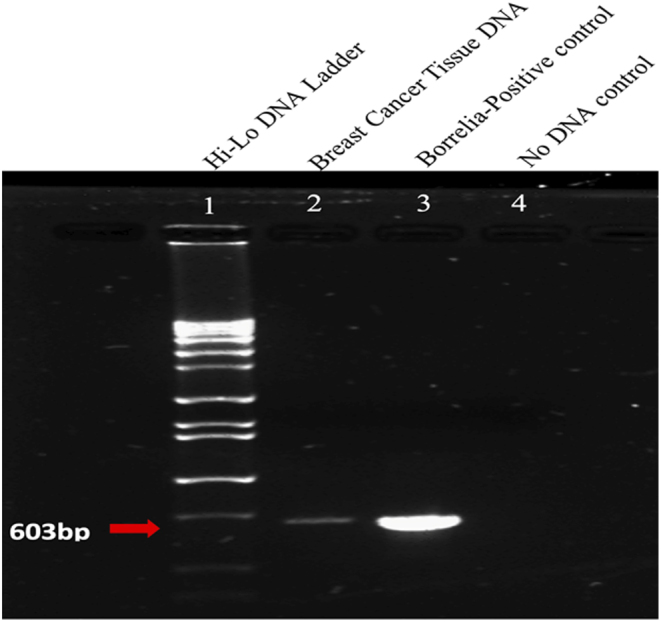
A representative gel electrophoresis image for the B. burgdorferi amplified PCR products using pyrG gene as the target. Lanes 1: Hi-Lo DNA maker, Lane 2: PCR amplified breast cancer tissue DNA, Lane 3: positive control, B. burgdorferi DNA and Lane 4: No DNA control
Fig. 6.
A representative pairwise alignment analysis of the sequencing results of the PCR product of B. burgdorferi-positive breast cancer tissues using the NCBI BLAST tool
This study also aimed to find whether breast cancer progression and metastasis correlate with the presence of B. burgdorferi using available patient clinical data from the commercial tissue arrays. Breast cancer grade and patient age at the time of obtaining the biopsy tissues were considered as primary criteria for the quantitative analysis of B. burgdorferi-positive tissues out of the total number of tissues for each criterion.
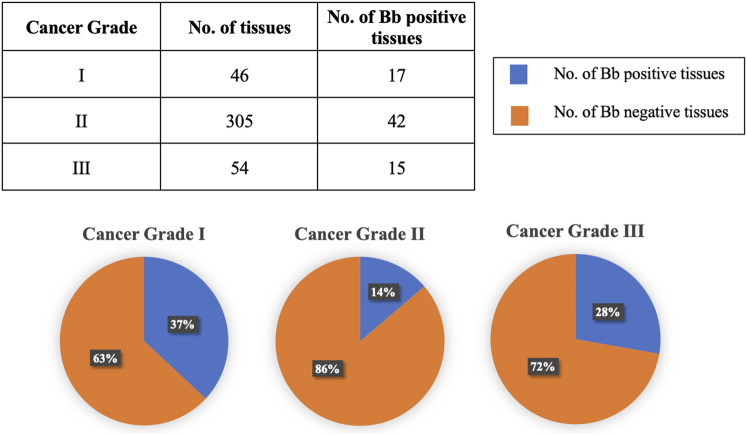
The cancer grade data for patient tissue samples are categorized as follows: Grade I, resembling normal breast cells and exhibiting slow growth; Grade II, displaying characteristics less like normal breast cells and growing faster than usual; Grade III, consisting of abnormal breast cells and typically showing aggressive growth. The analysis considered the correlation between cancer growth rate and the percentage of B. burgdorferi-positive tissue samples, revealing the following distribution: Grade I (37%), Grade II (14%), and Grade III (28%) (see Fig. 7).
Fig. 7.
Represents a quantitative analysis of B. burgdorferi-positive samples categorized by their respective cancer grade. The pie charts illustrate the percentage of B. burgdorferi-positive tissues (depicted in blue) relative to the total number of tissues identified within each grade. Additionally, the table above provides the numerical tissue count for each grade alongside the number of B. burgdorferi-positive samples in each grade
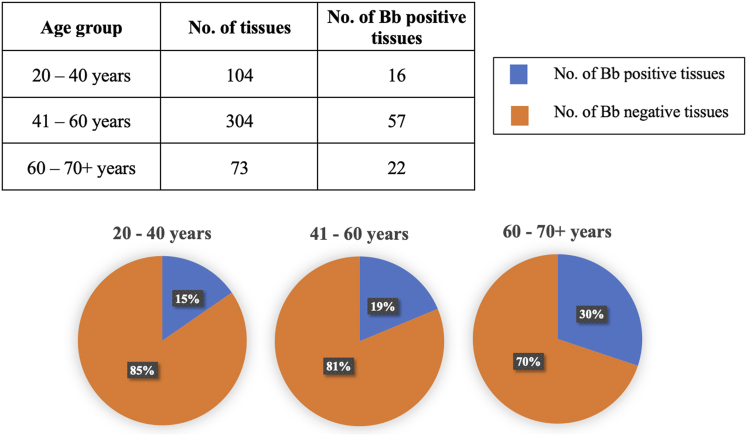
To investigate a possible correlation between patient age and B. burgdorferi-positive breast cancer tissues, patients were divided into three age groups. The percentages of B. burgdorferi-positive samples within each age group were as follows: 20–40 years (15%), 41–60 years (19%), and 61–70+ years (30%) (see Fig. 8).
Fig. 8.
Comparison of B. burgdorferi-positive samples identified across various age groups ranging from 20 to 70+ years. The pie charts illustrate the percentages of B. burgdorferi-positive tissues (depicted in blue) relative to the total number of tissues within each age group. Additionally, the table above provides the numerical tissue count and the number of B. burgdorferi-positive samples in each age group
Discussion
This study investigated the presence of B. burgdorferi in various types of breast cancer tissues, including invasive lobular, ductal, and intraductal carcinoma. Immunohistochemistry staining was conducted on FDA-approved human breast tissue samples to detect B. burgdorferi s.s. antigens. The results revealed B. burgdorferi spirochetes/aggregates in approximately 20% of invasive lobular and ductal carcinoma samples, followed by 14% in intraductal carcinoma samples. The presence of B. burgdorferi in tissue samples identified by immunohistochemistry was further confirmed using nested PCR and direct sequencing analysis to detect Borrelia DNA. Microscopic analysis of the B. burgdorferi-positive tissues revealed individual spirochetes and aggregates distributed deep within the breast cancer tissues. Negative control samples of normal breast tissues showed no evidence of either B. burgdorferi antigens or DNA. Inflammatory marker staining conducted on these breast cancer tissues revealed a pronounced host inflammatory response, with significant staining observed for both CXCL8 and CXCL10 markers in 25% of the B. burgdorferi-positive breast cancer tissue samples.
Quantitative analysis of individual patient clinical data for cancer grade revealed that grades I and III are the most frequently observed grades associated with the detection of B. burgdorferi-positive tissues. Additionally, analysis based on patient age criteria indicated that older patients (60+ years old) were more likely to have a higher incidence of B. burgdorferi-positive breast cancer tissue.
Recently, there has been growing speculation about the influence of bacterial microbiota on cancer development and its progression. For instance, another spiral-shaped bacterium, H. pylori is recognized as a causative agent for gastric cancer and MALT lymphoma [4]. Studies have identified two potential mechanisms by which H. pylori contributes to cancer development [25]. Firstly, H. pylori infection induces chronic inflammation of the gastric mucosa, leading to conditions such as atrophy and intestinal metaplasia. Secondly, it has been demonstrated that H. pylori can produce, release, and modify bacterial virulence factors within the tissue microenvironment, thereby promoting cancer progression [25]. Another bacterial genus implicated in various cancers, including cervical, ovarian, and lung cancers, is Chlamydia [26–28]. Interestingly, previous studies have documented the co-infection of B. burgdorferi with H. pylori and Chlamydia spp. in patients with Lyme disease [29, 30].
Infectious diseases have long been implicated in chronic infection-related inflammation [31]. Chronic inflammation is closely associated with the initiation of cancer [32, 33], with studies indicating that chemokines, along with cytokines, can act as early inflammatory mediators that influence the host response to pathogen exposure [34]. Chemokines belonging to the C-X-C family are known to play a role in leukocyte recruitment and regulate the tumor microenvironment by controlling tumor cell proliferation, metastasis, invasion, angiogenesis, and therapeutic resistance [23]. Numerous studies on Chlamydia trachomatis have demonstrated its capacity to upregulate C-X-C chemokine levels at the site of infection, thereby inducing an intense inflammatory response [35, 36].
B. burgdorferi has long been associated with chronic inflammation in various organs. A classic example is the skin condition characterized by dense lymphocytic infiltration known as acrodermatitis chronica atrophicans (ACA) [37]. Additionally, a study demonstrated the adaptive and resistant nature of B. burgdorferi, where lingering and debilitating symptoms persisted in a Lyme disease patient despite years of antibiotic treatment [38]. This study revealed the presence of B. burgdorferi in multiple organs, accompanied by an infiltrating CD3+ T-lymphocytic response. Numerous other reports have indicated that B. burgdorferi can upregulate inflammatory mediators such as interleukins and CXCL chemokines in host cells [39–41].
Inflammatory markers within the CXCL family were investigated in relation to chemo/cytokine-mediated neuroinflammation in the cerebrospinal fluid of Borrelia-infected patients [40]. The study concluded that abnormal serum antibody indices and elevated levels of CXCL8 and CXCL10 indicated a potential connection to Borrelia-mediated inflammation [40]. A similar upregulation was observed in human brain microvascular endothelial cells (hBMECs) infected with Borrelia bavariensis, wherein CXCL8 and CXCL10 were significantly elevated, correlating with cellular stress and the innate immune response [41].
Gene ontology analysis of these markers revealed signaling cascades associated with cell communication, organization of the extracellular matrix, cellular responses triggered by pattern recognition receptors, and antigen processing. These pathways suggest potential mechanisms exploited by B. burgdorferi to traverse the blood-brain barrier (BBB) [41].
A recent transcriptomic analysis of B. burgdorferi-infected breast cancer cell line (MDA-MB-231) and a normal mammary epithelial cell line (MCF10A) revealed a spectrum of differentially expressed genes modulated by the intracellular invasion of B. burgdorferi [22]. Ontological analysis of these genes demonstrated multiple markers associated with infection, inflammation, and cancer development [22]. This study highlighted a significant trend with an upregulation of chemokines, including members of the C-X-C motif chemokine family (such as CXCL8 and CXCL10), in both cell lines upon B. burgdorferi infection.
The CXCL8 and CXCL10 chemokines have been implicated in stimulating tumor angiogenesis and promoting the migration and invasion of breast and ovarian tumor cells [42–46]. Our study revealed localized CXCL8 and CXCL10 inflammatory responses near the B. burgdorferi spirochetes and aggregates, accounting for 25% of the B. burgdorferi-positive tissues for both markers (32% for CXCL8 alone). Interestingly, this observation was predominantly found in ductal carcinomas. These findings raise the question of whether other inflammatory markers, such as additional members of the CXCL family or IL-6 which were shown to increase after B. burgdorferi infection of triple-negative breast cancer cells [22] could also be involved. Furthermore, it would be also interesting to determine what is the deciding mechanism that stimulates certain inflammatory pathways in breast carcinomas.
In a follow-up study, we aim to explore the connection of B. burgdorferi infection to other inflammatory markers, including other members of the CXCL family, IL-6, C-reactive protein, and tumor necrosis factor-α, all of which have been reported to be highly elevated in breast cancer. Additionally, we will investigate the potential coinfection of B. burgdorferi-positive breast cancer tissues with H. pylori and Chlamydia spp., as coexistence has been demonstrated in B. burgdorferi infected skin tissues.
A significant limitation of this study lies in the restricted availability of individual patient clinical data, which hampers our ability to fully understand the correlation between B. burgdorferi infection, cancer grade, and patient age. For instance, the variability in sample sizes across different cancer grades undermines the support for the hypothesis suggesting that B. burgdorferi infection may contribute to cancer progression. Similarly, the correlation between patient age groups and B. burgdorferi infection lacks sufficient data for a comprehensive analysis, warranting the need for a larger patient dataset to validate the findings of this study. Other factors that could enhance our understanding include assessing the correlation between breast cancer types based on hormone receptor status and TNM staging classification, which unfortunately was unavailable for the majority of patient biopsy samples and therefore not incorporated into our analysis. Although limitations exist, these tissue biopsy microarrays are FDA-approved and commercially available making them reliable and easily reproducible for similar cancer studies.
In summary, this study suggests that pathogenic infections such as B. burgdorferi may play a role in promoting breast cancer and/or contributing to persistent infection by modulating the immune-inflammatory response within the tumor microenvironment. The findings illustrate B. burgdorferi's capability not only to disseminate as spirochetes but also to form aggregates deep within breast cancer tissues. Furthermore, B. burgdorferi was observed to induce high levels of localized inflammatory responses near the infected area via the CXCL8 and CXCL10 chemokines. Consequently, further investigation into inflammatory markers is imperative to elucidate B. burgdorferi's role in cancer progression and immune response manipulation. This research could offer opportunities for developing a diagnostic marker panel to monitor B. burgdorferi infection and cancer progression.
Funding sources
This research received no external funding.
Author contributions
Conceptualization, G.G., J.Y.S., N.J.P, S.T.; methodology, N.J.P., S.T., J.Y.S., G.G.; validation, N.J.P., S.T., G.G., J.Y.S. and J.J.; formal analysis, N.J.P., S.T., G.G., J.Y.S.; investigation, N.J.P., S.T., G.G., J.Y.S. and J.J.; resources, E.S.; data curation, N.J.P., S.T., G.G., and J.Y.S.; writing—original draft preparation, N.J.P. and E.S.; writing—review and editing N.J.P., S.T., J.J., J.M.P., and E.S.; visualization, N.J.P., S.T., J.J., J.M.P., E.S.; supervision, E.S.; project administration, E.S. All authors have read and agreed to the published version of the manuscript.
Conflict of interest
The authors have declared that no competing interests exist.
Data availability statement
The data presented in this study are available at a reasonable request from the corresponding author.
Acknowledgment
This work was supported by the Pink Clover Foundation and National Philanthropic Trust. The supporter had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript. The authors would also like to thank Dr. Joerg Nikolaus at the Yale West Campus Imaging Core, for providing training and guidance for the facility.
References
- 1.American Cancer Society . Cancer facts & figures. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html. Accessed February 11, 2024.
- 2.Dalton-Griffin L, Kellam P. Infectious causes of cancer and their detection. J Biol. 2009;8(7):67. 10.1186/jbiol168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pucułek M, Machlowska J, Wierzbicki R, Baj J, Maciejewski R, Sitarz R. Helicobacter pylori associated factors in the development of gastric cancer with special reference to the early-onset subtype. Oncotarget. 2018;9(57):31146–31162. 10.18632/oncotarget.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elsalem L, Jum’ah AA, Alfaqih MA, Aloudat O. The bacterial microbiota of gastrointestinal cancers: role in cancer pathogenesis and therapeutic perspectives. Clin Exp Gastroenterol. 2020;13:151–185. 10.2147/CEG.S243337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummins J, Tangney M. Bacteria and tumours: causative agents or opportunistic inhabitants? Infect Agent Cancer. 2013;8(1):11. 10.1186/1750-9378-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yusuf K, Sampath V, Umar S. Bacterial infections and cancer: exploring this association and its implications for cancer patients Int J Mol Sci. 2023;24(4):3110–3110. 10.3390/ijms24043110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The microbiota of breast tissue and its association with breast cancer. Appl Environ Microbiol. 2016;6(3):146–153. 10.1556/1886.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee S, Tian T, Wei Z, Shih N, Feldman MD, Peck KN, et al. Distinct microbial signatures associated with different breast cancer types. Front Microbiol. 2018;9:951. 10.3389/fmicb.2018.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garbe C, Stein H, Dienemann D, Orfanos CE. Borrelia burgdorferi-associated cutaneous B cell lymphoma: clinical and immunohistologic characterization of four cases. J Am Acad Dermatol. 1991;24(4):584–590. 10.1016/0190-9622(91)70088-j. [DOI] [PubMed] [Google Scholar]
- 10.Jelić S, Filipović-Ljesković I. Positive serology for Lyme disease borrelias in primary cutaneous B-cell lymphoma: a study in 22 patients; is it a fortuitous finding? Hematol Oncol. 1999;17(3):107–116. . [DOI] [PubMed] [Google Scholar]
- 11.Schöllkopf C, Melbye M, Munksgaard L, Smedby KE, Rostgaard K, Glimelius B, et al. Borrelia infection and risk of non-Hodgkin lymphoma. Blood. 2008;111(12):5524–5529. 10.1182/blood-2007-08-109611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribet D, Cossart P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015;5(3):173–183. 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Hyde JA. Borrelia burgdorferi keeps moving and carries on: a review of borrelial dissemination and invasion. Front Immunol. 2017;8:114. 10.3389/fimmu.2017.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, Sturrock A, Weiss JJ. Intracellular localization of Borrelia burgdorferi within human endothelial cells. Infect Immun. 1991;59:671–678. 10.1128/iai.59.2.671-678.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Georgilis K, Peacocke M, Klempner MS. Fibroblasts protect the Lyme disease spirochete, Borrelia burgdorferi, from ceftriaxone in vitro. J Infect Dis. 1992;166:440–444. 10.1093/infdis/166.2.440. [DOI] [PubMed] [Google Scholar]
- 16.Klempner MS, Noring R, Rogers RA. Invasion of human skin fibroblasts by the Lyme disease spirochete, Borrelia burgdorferi. J Infect Dis. 1993;167:1076–1081. 10.1093/infdis/167.5.1074. [DOI] [PubMed] [Google Scholar]
- 17.Montgomery RR, Malawista SE. Entry of Borrelia burgdorferi into macrophages is end-on and leads to degradation in lysosomes. Infect Immun. 1996;64:2867–2872. 10.1128/iai.64.7.2867-2872.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livengood JA, Gilmore RD Jr. Invasion of human neuronal and glial cells by an infectious strain of Borrelia burgdorferi. Microbes Infect. 2006;8:2832–2840. 10.1016/j.micinf.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Gaur G, Sawant JY, Chavan AS, Khatri VA, Liu Y-H, Zhang M, et al. Effect of invasion of Borrelia burgdorferi in normal and neoplastic mammary epithelial cells. Antibiotics. 2021;10(11):1295. 10.3390/antibiotics10111295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tracy KE, Baumgarth N. Borrelia burgdorferi manipulates innate and adaptive immunity to establish persistence in rodent reservoir hosts. Front Immunol. 2017;8:116. 10.3389/fimmu.2017.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Debbarma A, Mansolf M, Khatri VA, Valentino JA, Sapi E. Effect of Borrelia burgdorferi on the expression of miRNAs in breast cancer and normal mammary epithelial cells. Microorganisms. 2023;11(6):1475. 10.3390/microorganisms11061475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khatri VA, Paul S, Patel NJ, Thippani S, Sawant JY, Durkee KL, et al. Global transcriptomic analysis of breast cancer and normal mammary epithelial cells infected with Borrelia burgdorferi. Eur J Microbiol Immunol. 2023;13(3):63–76. 10.1556/1886.2023.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou C, Gao Y, Ding P, Wu T, Yi G. The role of CXCL family members in different diseases. Cell Death Discov. 2023;9:212. 10.1038/s41420-023-01524-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sapi E, Pabbati N, Datar A, Davies EM, Rattelle A, Kuo BA. Improved culture conditions for the growth and detection of Borrelia from human serum. Int J Med Sci. 2013;10(4):362–376. 10.7150/ijms.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogiatzi P, Cassone M, Luzzi I, Lucchetti C, Otvos L Jr, Giordano A. Helicobacter pylori as a class I carcinogen: physiopathology and management strategies. J Cell Biochem. 2007;102(2):264–273. 10.1002/jcb.21375. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Siddique A, Khan AA, Wang Q, Malik A, Jan AT, et al. Chlamydia trachomatis Infection: their potential implication in the etiology of cervical cancer. J Cancer. 2021;12:4891–4900. 10.7150/jca.58582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arcia Franchini AP, Iskander B, Anwer F, Oliveri F, Fotios K, Panday P, et al. The role of Chlamydia trachomatis in the pathogenesis of cervical cancer. Cureus. 2022;14(1):e21331. 10.7759/cureus.21331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wittman AJ, Jackson LA, Vaughan TL. Chlamydia pneumoniae and lung cancer: epidemiologic evidence. Cancer Epidemiol Biomarkers Prev. 2005 Apr;14(4):773–8. 10.1158/1055-9965.EPI-04-0599. [DOI] [PubMed] [Google Scholar]
- 29.Middelveen MJ, Filush KR, Bandoski C, Kasliwala RS, Melillo A, Stricker RB, et al. Mixed Borrelia burgdorferi and Helicobacter pylori biofilms in Morgellons disease dermatological specimens. Healthcare (Basel). 2019;7(2):70. 10.3390/healthcare7020070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapi E, Gupta K, Wawrzeniak K, Gaur G, Torres J, Filush K, et al. Borrelia and Chlamydia can form mixed biofilms in infected human skin tissues. Eur J Microbiol Immunol. 2019;9(2):46–55. 10.1556/1886.2019.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furman D, Campisi J, Verdin E, Carrera-Bastos P, Targ S, Franceschi C, et al. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25(12):1822–1832. 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao H, Wu L, Yan G, Chen Y, Zhou M, Wu Y, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Sig Transduct Target Ther 2021;6:263. 10.1038/s41392-021-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L, Deng H, Cui H, Feng J, Zuo Z, Deng J, et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget 2017;9(6):7204–7218. 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan C, Latter JL, Amirshahi A, Symonds I, Finnie J, Bowden N, et al. Progesterone activates multiple innate immune pathways in Chlamydia trachomatis-infected endocervical cells. Am J Reprod Immunol. 2013;71(2):165–177. 10.1111/aji.12168. [DOI] [PubMed] [Google Scholar]
- 36.Cheong HC, Cheok YY, Chan YT, Tang TF, Sulaiman S, Looi CY, et al. Chlamydia trachomatis plasmid-encoding Pgp3 protein induces secretion of distinct inflammatory signatures from HeLa cervical epithelial cells. BMC Microbiol. 2023;23(1):58. 10.1186/s12866-023-02802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asbrink E, Hovmark A, Hederstedt B. The spirochetal etiology of acrodermatitis chronica atrophicans Herxheimer. Acta Derm Venereol. 1984;64(6):506–512. [PubMed] [Google Scholar]
- 38.Sapi E, Kasliwala RS, Ismail H, Torres JP, Oldakowski M, Markland S, et al. The long-term persistence of Borrelia burgdorferi antigens and DNA in the tissues of a patient with Lyme Disease. Antibiotics. 2019;8(4):183. 10.3390/antibiotics8040183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Habicht GS, Katona LI, Benach JL. Cytokines and the pathogenesis of neuroborreliosis: Borrelia burgdorferi induces glioma cells to secrete interleukin-6. J Infect Dis. 1991;164(3):568–574. 10.1093/infdis/164.3.568. [DOI] [PubMed] [Google Scholar]
- 40.Liba Z, Nohejlova H, Capek V, Krsek P, Sediva A, Kayserova J. Utility of chemokines CCL2, CXCL8, 10 and 13 and interleukin 6 in the pediatric cohort for the recognition of neuroinflammation and in the context of traditional cerebrospinal fluid neuroinflammatory biomarkers. PLoS One. 2019;14(7):e0219987. 10.1371/journal.pone.0219987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tkáčová Z, Bhide K, Mochnáčová E, Petroušková P, Hruškovicová J, Kulkarni A, et al. Comprehensive mapping of the cell response to Borrelia bavariensis in the brain microvascular endothelial cells in vitro using RNA-Seq. Front Microbiol. 2021;12. 10.3389/fmicb.2021.760627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong X, Liao X, Qiu S, Xu, H, Zhang S, Wang S, et al. CXCL8 in tumor biology and its implications for clinical translation. Front Mol Biosci. 2022;9:723846. 10.3389/fmolb.2022.723846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu X, Wang Q, Du H, Hao H. CXCL8 and the peritoneal metastasis of ovarian and gastric cancer. Front Immunol. 2023;14:1159061. 10.3389/fimmu.2023.1159061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M, Choi HY, Woo JW, Chung YR, Park SY. Role of CXCL10 in the progression of in situ to invasive carcinoma of the breast. Sci Rep. 2021;11(1):18007. 10.1038/s41598-021-97390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jin J, Li Y, Muluh TA, Zhi L, Zhao Q. Identification of CXCL10-relevant tumor microenvironment characterization and clinical outcome in ovarian cancer. Front Genet. 2021;12:678747. 10.3389/fgene.2021.678747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.K Au K, Peterson N, Truesdell P, Reid-Schachter G, Khalaj K, Ren R, et al. CXCL10 alters the tumour immune microenvironment and disease progression in a syngeneic murine model of high-grade serous ovarian cancer. Gynecol Oncol. 2017;145(3):436–445. 10.1016/j.ygyno.2017.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available at a reasonable request from the corresponding author.