Abstract
Bacteroides thetaiotaomicron, a gram-negative obligate anaerobe, utilizes polysaccharides by binding them to its cell surface and allowing cell-associated enzymes to hydrolyze them into digestible fragments. We use the starch utilization system as a model to analyze the initial steps involved in polysaccharide binding and breakdown. In a recent paper, we reported that one of the outer membrane proteins involved, SusG, had starch-degrading activity but was not sufficient for growth on starch. Moreover, SusG alone did not have detectable starch binding activity. Previous studies have shown that starch binding is essential for starch utilization. In this paper, we report that four other outer membrane proteins, SusC through SusF, are responsible for starch binding. Results of 14C-starch binding assays show that SusC and SusD both contribute a significant amount of starch binding. SusE also appears to contribute substantially to starch binding. Using affinity chromatography, we show in vitro that these Sus proteins interact to bind starch. Moreover, protease accessibility of either SusC or SusD greatly increased when one was expressed without the other. This finding supports the hypothesis that SusC and SusD interact in the outer membrane. Evidence from additional protease accessibility studies suggests that SusC, SusE, and SusF are exposed on the cell surface. Our results demonstrate that SusC and SusD act as the major starch binding proteins on the cell surface, with SusE enhancing binding. SusF's role in starch utilization has yet to be determined, although the fact that starch protected it from proteolytic attack suggests that it does bind starch.
Bacteroides thetaiotaomicron, a gram-negative obligate anaerobe, can utilize polysaccharides very efficiently as a source of carbon and energy. Early studies on polysaccharide utilization by B. thetaiotaomicron showed that the enzymes that break down these polysaccharides are cell associated, with most of the enzymatic activity located in the periplasm or cytoplasm. Subsequent studies revealed that binding of the polysaccharide to the cell surface prior to hydrolysis was an important step in polysaccharide utilization (1, 2). This strategy for polysaccharide utilization may allow the bacterium to sequester hydrolysis products more efficiently and may even allow it to affix itself to a polysaccharide-containing particle.
The process of polysaccharide utilization by Bacteroides spp. has been best studied in the case of the starch utilization system of B. thetaiotaomicron. A cluster of eight starch utilization (sus) genes has been identified. One of the genes encodes a regulatory protein (SusR). When cells are grown on maltose or starch, SusR appears to activate the promoters of susA and the susB–G operon. It is not known whether maltose, the presumed inducer of sus gene expression, is bound to SusR or sensed in some other way. Three genes in the cluster encode starch-degrading enzymes (SusA, SusB, and SusG). SusG and SusA are neopullulanases that cleave starch into mono- and disaccharides. SusB is an α-glucosidase that acts on the products of SusA and SusG. SusG has a very low activity compared to SusA. In fact, only when susA was disrupted was it possible to detect SusG activity (12). Despite its low activity, however, SusG is essential for growth on starch. SusG is also the only one of these enzymes that is exposed on the cell surface (12). SusA is a periplasmic enzyme, and SusB has been tentatively localized to the cytoplasm (1, 12).
SusC appears to be a porin that allows uptake of maltodextrins from maltose (G2) to maltoheptaose (G7), because a mutant producing only SusC but not SusD through SusG could grow as well as the wild type on maltodextrins. A mutant lacking SusC could grow on glucose and poorly on maltose or maltotriose but not on the higher maltodextrins. The fact that SusG in combination with SusC, but without SusD through SusF, was not sufficient to allow cells to grow on starch suggested that binding of starch and further processing of it were complex. That is, SusG was not simply degrading starch on the cell surface and releasing the products for uptake through SusC.
Previously, we found that SusG made little contribution to the binding of starch to the bacterial surface (12). Nor was SusG alone able to bind starch to the cell surface. Binding of starch appears to be mediated by one or more of the other outer membrane proteins (OMP) that have no detectable enzymatic activity (SusC, SusD, SusE, and SusF). SusC alone was not sufficient to bind labeled starch to the cell surface, but a combination of SusC and SusD allowed cells to bind about 70% of the starch bound by the wild type. Since a mutation in susC had a polar effect on susD, it was not clear whether SusD alone was sufficient for this starch binding or whether SusC was playing a significant role as well. Presumably, SusE or SusF or both were necessary to account for the full level of starch binding seen with wild-type cells. If SusC, SusD, SusE, and SusF were all involved in binding starch, one or more of them should be exposed on the cell surface. Similarly, if SusC is in fact a porin for oligosaccharides as the results of previous genetic experiments suggested, SusC should be surface exposed. In this paper, we report the analysis of surface accessibility of these starch binding proteins. Furthermore, we provide genetic and biochemical evidence that SusC, SusD, and SusE appear to interact with each other to bind starch.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
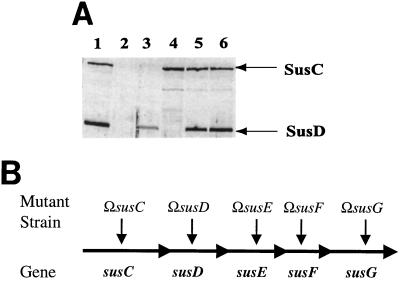
The bacterial strains and plasmids used in this study are listed in Table 1. All Escherichia coli strains used in this study were grown in Luria-Bertani broth or on Luria-Bertani agar at 37°C. B. thetaiotaomicron 5482, transposon-generated derivatives, and single-disruption mutants used in this study have been described previously (10). For clarification purposes, polar disruption mutants of the Sus operon are diagrammed in Fig. 1B.
TABLE 1.
Strains and plasmidsa
| Strain or plasmid | Relevant characteristic(s) | Description or source (reference) |
|---|---|---|
| E. coli | ||
| DH5αMCR | RecA Gns | Hanahan (5) |
| B. thetaiotaomicron | ||
| BT5482 | Wild type; Gnr | Anaerobe Laboratory, Virginia Polytechnic Institute, Blacksburg |
| BT4007 | Wild type; Tetr Gnr G7+ Am+ | B. thetaiotaomicron 5482 (a Rifr strain) with CTn DOT |
| ΩsusC | Tetr Gnr G7− Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron 5482 chromosome in the susC gene (pBT1-SC) (10) |
| ΩsusD | Tetr Gnr G7+ Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron 5482 chromosome in the susD gene (10) |
| ΩsusE | Tetr Gnr G7+ Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron 5482 chromosome in the susE gene (10) |
| ΩsusF | Tetr Gnr G7+ Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron 5482 chromosome in the susF gene (10) |
| ΩsusG | Tetr Gnr G7+ Am− | Bacteroides suicide vector pBT-1 containing a PCR-generated 0.61-kbp KpnI-BamHI fragment inserted in the B. thetaiotaomicron chromosome 5482 in the susG gene (10) |
| Plasmids | ||
| pBT-1 | Knr (Tcr) | RSF1010-based suicide vector used to make insertional disruptions (14) |
| pNLY1::PsusA | Apr Cmr (Cmr) | pACYC-based shuttle vector containing the susA promoter used to express genes in trans (this study) |
| pSDC27 | Apr Cmr (Cmr) | pNLY1::PsusA containing a StuI-PvuII fragment from a B. thetaiotaomicron chromosome cloned downstream of the susA promoter (this study) |
Abbreviations: G7, maltoheptose; Am, amylopectin; Ap, ampicillin; Cm, chloramphenicol; Em, erythromycin; Gn, gentamicin; Tc, tetracycline; CTn, conjugative transposon; CTn DOT, conjugative transposon which contains the tetQ gene. For the plasmids shown, antibiotic resistances not in parentheses are expressed only in E. coli, and antibiotic resistances in parentheses are expressed only in B. thetaiotaomicron.
FIG. 1.
(A) Immunoblot showing SusD expression from a multicopy plasmid. Approximately 50 μg of protein was loaded in each lane. All membrane fractions were obtained from cells grown on defined medium with maltose as the sole carbohydrate source. Lanes: 1, membrane fraction from B. thetaiotaomicron 5482; 2, membrane fraction from B. thetaiotaomicron ΩsusC; 3, membrane fraction from B. thetaiotaomicron ΩsusC(pSDC27); 4, membrane fraction from B. thetaiotaomicron ΩsusD; 5, membrane fraction from B. thetaiotaomicron ΩsusD(pSDC27); 6, membrane fraction from B. thetaiotaomicron ΩsusE. This and all other immunoblots shown in this paper were scanned using an Epson Perfection 636U scanner and incorporated into a figure using both Adobe Photoshop 5.5 and Adobe Illustrator 8.0. (B) The Sus operon, showing polar insertional disruption mutations used in these studies.
Bacteroides strains were grown initially in a prereduced Trypticase-yeast extract-glucose medium. For optimal induction of starch utilization genes, cells were transferred to a defined medium containing maltose (0.3%) as the sole carbohydrate source. To test for growth on starch, we inoculated cells into a defined medium with amylopectin or pullulan (0.3%) as the sole carbohydrate source. In the text, the assertion that a particular mutant could not grow on starch means that there was no increase in turbidity after inoculation into amylose or amylopectin medium after several days of incubation. The following antibiotic concentrations were used in this study: ampicillin, 200 μg/ml; chloramphenicol, 15 μg/ml (E. coli) or 20 μg/ml (B. thetaiotaomicron); erythromycin, 10 μg/ml; gentamicin, 200 μg/ml; and tetracycline, 1 μg/ml.
Chemicals.
14C-starch (Nicotiana tabacum 1) was purchased from DuPont NEN. Amylopectin, pullulan, proteinase K, n-octyl-β-d-glucopyranoside, and phenylmethylsulfonyl fluoride were purchased from Sigma Corp.
DNA methods.
Isolation of plasmids was done using a Wizard Plus DNA purification system (Promega Corp., Madison, Wis.). Dephosphorylation reactions and restriction digests were performed in accordance with the manufacturer's instructions (Bethesda Research Laboratories, Bethesda, Md., or New England Biolabs, Beverly, Mass.). Transformation of E. coli DH5αMCR was done by the method of Lederberg and Cohen (7). Constructs generated in E. coli were transferred to Bacteroides recipients as described by Shoemaker et al. (13).
Membrane preparation.
Membranes were prepared by the ultracentrifugation method of Valentine and Salyers (16). Cells were grown in a defined medium with maltose as the sole carbohydrate source (0.3%) to late log phase (optical density at 650 nm of 0.6 to 0.8). The cells were washed once with 20 mM potassium phosphate buffer (pH 7.2) and resuspended in 5 ml of the same buffer. These cells were disrupted by sonication. After the cell extract was separated from insoluble material by centrifugation, the whole membranes (both inner and outer membranes) were pelleted from the cell extract by ultracentrifugation (200,000 × g for 2.5 h at 4°C). The soluble fraction was collected, and the membrane pellet was washed once with 20 mM potassium phosphate buffer and pelleted again by ultracentrifugation under the same conditions. The membrane pellet was resuspended in 20 mM potassium phosphate buffer, and the membranes were dispersed by sonication.
Expression of susD in trans.
We wished to express susD independently of the other starch-associated OMPs. To achieve this, we cloned the susD gene downstream of the susA promoter on a shuttle vector, pNLY1::PsusA. This vector was used previously to express susG in trans. This construct was designated pSDC27. We introduced pSDC27 into ΩsusC, which does not express any of the starch-associated OMPs, to create the strain ΩsusC(pSDC27). We also introduced this plasmid into ΩsusD, a disruption mutant that produced SusC, but not SusD through SusG, to determine if the resulting strain, ΩsusD(pSDC27), acted similarly to ΩsusE in terms of protein expression and binding characteristics. Using antisera directed against SusC and SusD, we determined by immunoblot analysis whether SusC and SusD were expressed in ΩsusC(pSDC27) and ΩsusD(pSDC27). We also used immunoblot analysis to confirm that the other Sus OMPs were not being produced in these strains. Moreover, we determined that these mutants were not able to grow on starch.
14C-starch binding experiments.
To determine the contribution of various Sus OMPs to binding of starch to intact cells, we measured binding activities of various mutants using a modification of the procedure of Anderson and Salyers (1). Intact cells were used instead of membranes because previous studies showed that membranes isolated from cells bound much less labeled starch than intact cells (1). Cells were grown in a defined medium containing 0.3% maltose to an optical density at 650 nm of 0.5 to 0.6. The cells were pelleted by centrifugation and washed twice with phosphate-buffered saline (PBS) (pH 7.4) to dissociate any loose capsular material. The cells were resuspended in PBS to an optical density at 650 nm of 0.4. Subsequently, the cell suspensions were incubated in a mixture of 14C-starch and unlabeled amylopectin (200 μg/ml in PBS stock) for 5 min under aerobic conditions. Under aerobic conditions, the cells do not internalize or accumulate starch except for that initially bound (1). The 5-min time point is used for convenience, since we have found previously that harvesting cells at earlier or later times makes no difference in the amount of starch bound. Apparently, whatever starch is going to be bound is bound within the first minute, and the amount does not increase even with incubation times of several hours. Under aerobic conditions, there is no evidence for translocation and uptake of the starch molecules. Under anaerobic conditions, uptake can be demonstrated (1). To separate binding from uptake, we use the aerobic conditions.
The cells were harvested by centrifugation for 45 s, and the supernatant fluid was discarded. The cell pellet was washed twice with 500 μl of PBS without disrupting the cell pellet. After the washes, the cell pellet was resuspended in 100 μl of PBS buffer, transferred to 2 ml of scintillation fluid, and counted on a Beckman 600IS scintillation counter. In previous experiments, we had found that starch bound to the cells was bound tightly enough not to be dislodged by washing with buffer. Thus, the binding we are measuring is irreversible, and there is no evidence that this binding involves transport of the starch into the cell.
B. thetaiotaomicron 4007 was used as a wild-type control because it contained the tetQ gene, which was used in the chromosomal insertional disruptions to generate the other mutants tested. We regard binding by the strain ΩsusC as nonspecific binding and subtracted these values from the binding seen in other strains. Values are reported in micrograms of starch bound per milligram of cell protein. These values were obtained by multiplying the total counts per minute by a dilution factor, which was the ratio of labeled starch to total starch in each assay. That number was converted by an empirical constant (based on observed counts per minute per given amount of starch) to disintegrations per minute, which allowed the total micrograms of starch bound to be calculated by using the reported values of 2.2 × 106 disintegrations per min per μg of starch. Experimental values were standardized to the whole-cell protein concentration, which was determined using a Bio-Rad DC protein assay kit, using bovine serum albumin as a standard.
Proteolysis experiments.
Accessibility to proteinase K was used to determine whether SusC, SusD, SusE, or SusF proteins were exposed on the cell surface according to the procedure of Shipman et al. (12). Cells were inoculated with 0.5 to 1.0 ml of an overnight culture of VPI-grown cells to 100 ml of defined media containing 0.3% maltose. These cells were grown to an optical density at 650 nm of 0.6 to 0.8 and harvested by centrifugation at room temperature. The cell pellet was washed twice at room temperature with 100 mM potassium phosphate buffer (pH 7.2) in order to dissociate any loose capsular material from the cells. Subsequently, the cells were resuspended in 9 ml of 100 mM potassium phosphate buffer. Fresh proteinase K (20-mg/ml stock) was added to a final concentration of 2 mg/ml. This high concentration of proteinase K was required to see any degradation of outer membrane proteins. The cells were incubated at 37°C with occasional mixing. Samples of 2 ml each were removed at various intervals. Phenylmethylsulfonyl fluoride was added to a final concentration of 10 mM for each sample to stop proteinase K activity. The cells were harvested by centrifugation and washed with 2 ml of a 100 mM potassium phosphate buffer-phenylmethylsulfonyl fluoride solution. The final cell pellet was resuspended in 100 mM potassium phosphate buffer.
After the cells were disrupted by sonication, the protein concentration was determined using a Bio-Rad DC protein assay kit with bovine serum albumin as a standard. Approximately 100 μg of protein from each sample was resuspended in Laemmli buffer and electrophoresed on a sodium dodecyl sulfate–8% polyacrylamide gel (SDS–8% PAGE). The gel was transferred to a Bio-Rad Trans-Blot nitrocellulose membrane, and SusC, SusD, SusE, and SusF proteins were detected using antisera directed against the corresponding protein. The secondary antibody used was a goat anti-mouse antibody conjugated with horseradish peroxidase supplied in the Bio-Rad Opti-4CN kit, which also supplied the detection substrate. As a control, we repeated the above-described procedure except for the addition of proteinase K to ascertain whether SusC, SusD, SusE, and SusF were stable during the incubation period in the absence of proteinase K. In some cases, long incubation times were used. This raises the question of whether after such long incubations the outer membrane was still intact. To confirm that the outer membrane was still intact, we routinely tested for the presence of a periplasmic marker, SusA (12). In all experiments shown here, SusA was intact and present at the same level at the end of the digestion process as at the beginning (data not shown). The SusA antibody was detected by using a Bio-Rad goat anti-rabbit–horseradish peroxidase Opti-4CN substrate kit. To determine if starch would protect the proteins from cleavage by proteinase K, we followed the procedure as outlined above but incubated wild-type cells with proteinase K and amylopectin (final concentration, 2 mg/ml).
Affinity chromatography.
To determine whether the Sus OMPs were acting as a complex, we tested their binding to an amylose resin mixture (New England Biolabs), which contained amylose covalently bonded to agarose beads. A volume of 10 ml of this resin was packed into a chromatography column. The resin was washed with 5 column volumes of MBP buffer (20 mM Tris, 20 mM NaCl, 20 mM EDTA) and used for the following experiments. After each experiment, the column was regenerated according to the manufacturer's instructions.
Cells were grown in 700 ml of a defined medium supplemented with 0.3% maltose. After reaching an optical density at 650 nm of 0.8 to 1.0, these cells were harvested by centrifugation. The cell pellet was washed once with 20 mM potassium phosphate buffer, and a membrane fraction was obtained according to the procedure of Valentine and Salyers (16). Membranes were resuspended in 20 mM potassium phosphate buffer and dispersed by sonication. The protein concentration was determined using a Bio-Rad DC assay kit with bovine serum albumin as a standard. This suspension was added to 0.1 M KPO4–0.15 M KCl buffer supplemented with 1.5% n-octyl-β-d-glucopyranoside to a concentration of 5 mg of protein per ml of buffer. Previous work has shown that these conditions release all the Sus proteins from the membranes (10). After the membranes were solubilized, remaining unsolubilized material was pelleted by ultracentrifugation, and the solubilized proteins were collected for further purification. In one experiment, membrane proteins from ΩsusC(pSDC27) were incubated with membrane proteins from ΩsusD overnight at 4°C before loading onto the column.
Prior to loading of the proteins, the affinity matrix was washed with 1 column volume of MBP buffer supplemented with 0.75% n-octyl-β-d-glucopyranoside for detergent equilibration with the sample to be loaded. This step was needed to ensure that the protein remained solubilized in the column. Next, the solubilized membrane protein (25 to 30 mg of cell protein) was loaded onto the column, and the column was washed with 5 column volumes of MBP buffer with 0.75% n-octyl-β-d-glucopyranoside at an S/V ratio of 2 column volumes/h. This wash was collected for further analysis. The proteins that remained on the column were eluted with 5 column volumes of the same buffer, to which maltose had been added (final concentration, 100 mM). This maltose eluant was collected for further analysis.
The maltose eluant and wash were concentrated more than 50-fold by tangential flow-filtration (Amicon Centri-prep concentrator MW 10,000). The concentrated proteins were resuspended in Laemmli buffer and electrophoresed on an SDS–8% polyacrylamide gel. The proteins were transferred to a Bio-Rad Trans-Blot nitrocellulose membrane. This nitrocellulose membrane was treated with antisera directed against the appropriate Sus proteins using a goat anti-mouse–horseradish peroxidase Opti-4CN substrate kit.
In one experiment, solubilized membranes from a mutant that produced SusC but not SusD and solubilized membranes from a cell that produced SusD but not SusC were mixed and incubated prior to passage through the column. The purpose of this experiment was to determine whether one of these proteins was helping to fold the other one or whether their cooperation in attaching to the starch column was more likely to be due to interaction that allowed them to stick to the column, whereas they were incapable of doing this alone.
RESULTS
Both SusC and SusD are needed for starch binding by intact cells.
Previous 14C-starch binding assays had shown that a mutant expressing only SusC and SusD (ΩsusE) had approximately 70% of wild-type starch binding activity. A mutant expressing SusC alone (ΩsusD) had very little starch binding activity. It is important to note that this assay measures tight binding of starch to the cell surface and does not involve transport of starch into the cell. During our assays, the cells bound starch almost immediately but did not continue to accumulate it over time (1; present study). Nor do the cells appear to lose starch over time, as would be expected if SusG, the OMP with starch-degrading activity, were degrading and releasing starch from the cells. That is, after several hours of incubation at 37°C, there is no decrease in the amount of labeled starch bound to wild-type cells. This suggests that whatever degradation of starch is carried out by SusG under these conditions is coupled to retention of starch, presumably by other proteins in the starch surface binding complex.
These initial 14C-starch binding assays by Reeves et al. were done at subsaturating concentrations of starch (10). To make sure that findings of this earlier study were not affected by the use of a single, low concentration of starch, we modified this procedure to determine starch binding at saturating concentrations of starch. Using these new conditions, we wished to determine whether both SusC and SusD or SusD alone was required for this binding activity. To determine whether SusD alone was sufficient for binding, we transferred pSDC27, an expression vector containing susD, into a strain expressing none of the starch binding OMPs (ΩsusC). According to an immunoblot of the membrane fraction, this strain produced SusD at a level about two times lower than the wild type (Fig. 1). In contrast, when susC was expressed from the chromosome and susD was expressed from the plasmid, the SusD protein was present at wild-type levels. This suggests a possible stabilizing effect of SusC on the production or assembly of the SusD protein, which we provide evidence for in a later section.
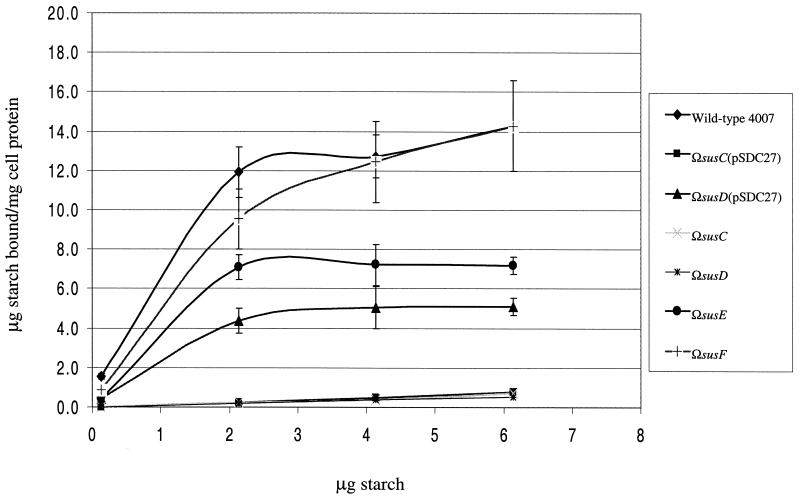
We used the strains expressing SusC and/or SusD to determine whether SusD was sufficient for starch binding. As shown in Fig. 2, the strains expressing SusC only (ΩsusD) or SusD only [ΩsusC(pSDC27)] had no significant starch binding activity. However, strains expressing both SusC and SusD [ΩsusE and ΩsusD(pSDC27)] bound starch at about half of wild-type levels at saturating starch concentrations (Fig. 2). Expression of susD in trans from the plasmid [ΩsusD(pSDC27)] rather than from the chromosome (ΩsusE) affected starch binding very little. The level of binding was somewhat lower when SusD was provided from the plasmid, as expected from the lower level of SusD produced by this strain (Fig. 2). This result confirmed that the clone was complementing successfully the chromosomal disruption of susD, even though the gene was present in multiple copies and was somewhat underproduced on the plasmid. Thus, SusD is not sufficient for starch binding, and both SusD and SusC are needed for significant starch binding by B. thetaiotaomicron. Nevertheless, SusC and SusD are not sufficient for growth, since cells expressing both OMPs but not SusE, SusF, and SusG did not grow at all on starch.
FIG. 2.
Starch binding by B. thetaiotaomicron mutants compared to that of the wild type. For simplicity, error bars are shown for B. thetaiotaomicron 4007, ΩsusF, ΩsusE, and ΩsusD(pSDC27) only. Error bars for other cases are comparable in size.
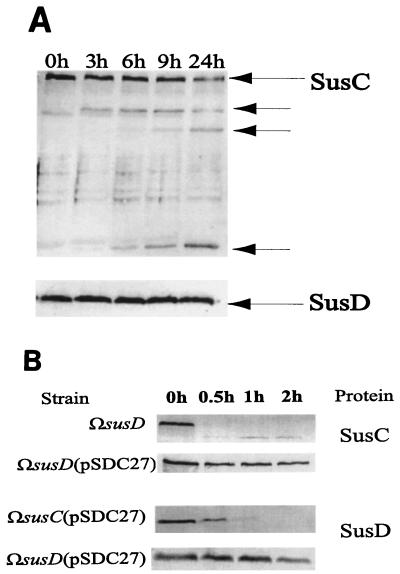
Since SusC and SusD were clearly important for starch binding, we reasoned that one or both of them might be exposed on the cell surface. We tested for surface exposure by determining accessibility to proteinase K of the protein in intact cells. This technique had shown previously that another starch OMP, SusG, was exposed on the cell surface. In all cases, we used SusA as a periplasmic marker to ensure that the outer membrane barrier was not breached by the proteinase K (12). In no case did the concentration of SusA that was detectable in Western blots change even after prolonged incubation (data not shown). Both SusC and SusD were more resistant to proteolytic attack than SusG. We were able to detect breakdown products for SusC after 3 h, and the number and intensity of these products increased at longer incubation times, although SusC was never degraded completely during the course of this experiment (Fig. 3A). This result suggested that SusC was surface exposed. Since, however, SusC was degraded completely by proteinase K within 3 h in membranes from disrupted cells (data not shown), it is probably exposed mostly on the periplasmic side of the outer membrane.
FIG. 3.
(A) Immunoblots showing proteolytic sensitivity of SusC and SusD in wild-type intact cells expressing all the Sus proteins. Approximately 100 μg of protein from cell extracts was loaded in each lane. Cells were treated with proteinase K (final concentration, 2 mg/ml). Degradation products of SusC due to proteinase K are shown by the arrows under the original SusC. No degradation products were observed for SusD. The lanes are labeled according to the time that had elapsed after addition of proteinase K. In all cases, the amount of the periplasmic protein, SusA, was the same at all stages of digestion, and no breakdown products were detected (not shown). (B) Immunoblots showing changes in proteolytic sensitivities of SusC and SusD in various mutants. Approximately 100 μg of protein from cell extracts was loaded in each lane. The protein detected on the immunoblot is shown on the right, with the corresponding mutant strains expressing the protein labeled on the left-hand side. The lanes are labeled according to the time that had elapsed after addition of proteinase K.
By contrast, we did not detect any degradation of SusD during the incubation period. Thus, it appeared initially that SusD was not surface exposed. However, SusD was not degraded even after disrupted membranes from cells were treated with proteinase K. After 24 h, SusD was still present in the sample (data not shown). Thus, SusD is inaccessible to proteinase K in wild-type membranes. SusD itself is not protease resistant, however. We show in an experiment described below that SusD, produced independently of the other Sus OMPs, is protease sensitive in intact cells. Thus, SusD appears to be well protected either by the other starch OMPs or by its configuration in the outer membrane or both.
SusC and SusD interact in the outer membrane.
Since both SusC and SusD were necessary for starch binding, it seemed likely that they interacted with each other in the outer membrane. A first line of evidence for such an interaction came from mutants expressing either SusC (ΩsusD) or SusD [ΩsusC(pSDC27)] alone. The proteins in these mutants showed considerably different protease sensitivities than proteins in a wild-type cell which produced all the starch OMPs. SusC and SusD, when expressed by themselves, were degraded significantly after 30 min of proteolytic attack (Fig. 3B). This was a much faster degradation than that seen in wild-type cells. Addition of amylopectin during treatment with proteinase K did not protect SusC and SusD when they were produced (data not shown). This finding agrees with the 14C-starch binding data, which show that SusC and SusD individually do not bind starch.
In a mutant in which SusC was produced from a chromosomal gene and SusD was produced from the plasmid [ΩsusD(pSDC27)], both proteins were not degraded as readily by proteinase K as was SusC or SusD alone (Fig. 3B). This change in protease accessibility suggests that SusE and SusF are not required for protection from protease attack and that SusC and SusD interact to stabilize each other in the outer membrane. This interaction also seems to be necessary for starch binding, since cells only bind starch when both SusC and SusD are expressed. Another explanation of the results of this experiment is that SusC or SusD acts as a chaperonin to ensure the proper folding of the other protein. In a later section, results are presented that argue against this hypothesis and for an interaction that allows the two proteins to bind starch as a multimer.
SusE and SusF contribute to starch binding.
Since SusC and SusD accounted for only about half of wild-type binding activity, it seemed likely that SusE or SusF or both would be responsible for the rest. When SusE was produced along with SusC and SusD (ΩsusF), the strain bound amounts of starch similar to those bound by the wild type (Fig. 2). Thus, SusE was making some contribution to starch binding. We had seen this same wild-type binding activity in a strain expressing SusC, SusD, SusE, and SusF (ΩsusG) (12). Thus, SusF seemed to play a limited role, if any, in binding, according to this assay.
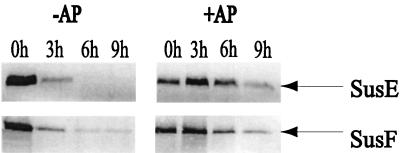
We used the protease accessibility assay described above on wild-type intact cells to determine if SusE and SusF were exposed on the cell surface. We found that both SusE and SusF were accessible to proteinase K. SusE and SusF were degraded significantly after 3 h (Fig. 4). To determine if bound starch was able to protect either of these proteins from protease digestion, we incubated intact cells with potassium phosphate buffer containing amylopectin with proteinase K. The amylopectin inhibited degradation of both SusE and SusF, since both were present in significant amounts after 9 h of incubation with proteinase K (Fig. 4). SusC and SusD were also expressed in these cells, but we saw no differences upon treatment with amylopectin in the protease digestion patterns for these proteins. But since SusC was slightly degraded and SusD was not degraded by proteinase K in the absence of amylopectin, it is difficult to determine the actual effect of the amylopectin treatment for these particular proteins. To ensure that amylopectin itself was not affecting proteinase K activity, we demonstrated that SusG, a protein shown to have very limited binding activity (12), was degraded by proteinase K in the presence of amylopectin (data not shown). These results suggest that SusE and SusF contribute to starch binding, even though the contribution of SusF to binding of starch by intact cells was not detectable.
FIG. 4.
Immunoblots showing proteolytic sensitivity and protection from proteolysis by amylopectin of SusE and SusF in wild-type cells. Approximately 100 μg of protein from cell extracts was loaded in each lane. The immunoblots are labeled above according to whether amylopectin was added to the proteinase K treatment. +AP, addition of amylopectin; −AP, no amylopectin was added. The lanes are labeled according to the time that had elapsed after addition of proteinase K. In all cases, the amount of the periplasmic protein SusA was the same at all stages of digestion, and no breakdown products were detected (not shown).
SusC, SusD, and SusE interact in vitro to bind starch.
As another approach to determine if starch OMPs interacted to bind starch, we characterized these interactions biochemically using an amylose-agarose column. To determine whether the starch binding OMPs would bind to the column, solubilized membrane proteins from B. thetaiotaomicron were loaded onto the column. This experiment was first performed on proteins from wild-type cells to determine the selectivity of this method for the starch-associated OMPs. Proteins that bound to the amylose resin were eluted with a high concentration of maltose. Figure 5A shows that the column bound mainly the starch-associated OMPs, SusC through SusG. A few other faint bands of unknown identity were evident on SDS gels of material eluted from the column, but SusC through SusG were the major proteins. These maltose eluant proteins were confirmed as starch-associated OMPs by immunoblotting, using antisera directed against each protein (data not shown).
FIG. 5.
(A) SDS-PAGE gel showing selectivity of amylose-agarose affinity chromatography for Sus OMPs. Molecular markers are shown to the left of the blot. Each protein labeled on the right was confirmed by immunoblots using antisera directed against the corresponding protein. Lanes: 1, membrane proteins of B. thetaiotaomicron 5482; 2, n-octyl-β-d-glucopyranoside-solubilized membrane proteins; 3, wash fraction of n-octyl-β-d-glucopyranoside-solubilized membrane proteins after loading onto an amylose-agarose column; 4, maltose eluant fraction of n-octyl-β-d-glucopyranoside-solubilized membrane proteins. (B) Immunoblots showing changes between various mutants in amylose binding activity of SusC, SusD, and SusE. The protein is labeled on the right of the blot, with the mutant strains expressing the corresponding protein labeled to the left of the blot. Lanes: 1, membrane fraction of the mutant; 2, n-octyl-β-d-glucopyranoside-solubilized membrane fraction of the mutant; 3, wash fraction of n-octyl-β-d-glucopyranoside-solubilized membrane proteins of the mutant after loading onto amylose-agarose column; 4, maltose eluant of n-octyl-β-d-glucopyranoside-solubilized membrane fraction of mutants. Although not evident in the figure, lane 4 of the immunoblot for mutant ΩsusE did show a low level of SusC. The designation ΩsusD + ΩsusC(pSDC27) indicates that solubilized membrane fractions from a strain producing only SusD and one producing only SusC were incubated together overnight before being loaded on the column.
When solubilized membrane proteins from strains expressing either SusC (ΩsusD) or SusD [ΩsusC(pSDC27)] alone were run through the column, the corresponding protein was present in the wash but not in the maltose elution fraction (Fig. 5B). Thus, when expressed individually, these proteins did not bind the starch column. This finding is consistent with the results of the 14C-starch binding assay. Interestingly, when both proteins were expressed either in the ΩsusD(pSDC27) or ΩsusE strain, SusD was retained on the column. Most of SusC washed through the column, although a very small amount relative to the amount of SusD did appear in the maltose eluant (Fig. 5B). This result suggests that SusC and SusD together interact in such a way as to allow the protein(s) to bind to the starch column, but not as effectively as in the wild type. This may help explain why the mutant expressing just SusC and SusD did not bind starch at wild-type levels. Additionally, when membrane proteins from the ΩsusC(pSDC27) and ΩsusD strains were mixed before loading onto the column, both SusC and SusD were retained by the column (Fig. 5B). This provides evidence that these proteins, when expressed individually, are folding properly and can interact with each other in vitro but are not as effective a complex as the complete suite of binding proteins in wild-type cells.
When SusE was expressed along with SusC and SusD (in the strain ΩsusF), all three proteins were retained on the starch column (Fig. 5B; SusE is not shown). In fact, SusC was now retained on the column, not eluted in the wash. These results provide additional evidence of interactions between SusC, SusD, and SusE. This overall enhancement of binding activity by SusE agrees with the results from the 14C-starch binding assay, where SusE expression enhanced binding by intact cells to nearly wild-type levels.
DISCUSSION
We had shown previously that SusG is responsible for the starch-degrading activity detectable in the outer membrane fraction of B. thetaiotaomicron but did not contribute to starch binding by the cells (12). In this study we showed that SusC, SusD, and SusE together bind as much starch as wild-type cells. Evidence that these proteins form a complex comes from two sources. First, when either SusC or SusD was present in outer membranes without the other, no starch binding occurred. When both were present, cells could bind at 50% of the wild-type level. Moreover, SusC and SusD alone were each much more accessible to exogenously added proteinase K than when both were present. This finding suggests that SusC and SusD stabilize each other in the outer membrane.
A second line of evidence for complex formation was the finding that although SusC and SusD alone did not bind to a starch column, SusC and SusD together allowed SusD and some SusC to be retained on the column. The fact that SusC was not retained as efficiently as SusD could be a reflection of the fact that when these proteins are in a membrane, their interaction is stabilized, whereas in a solubilized form it is weaker. The surprising thing about this finding was that SusD interacted so strongly with the column when so little SusC was retained, whereas SusD alone did not bind the column at all. One possible interpretation is that SusC causes some conformational change in SusD or actually modifies SusD in some way that persists even when SusC is absent. We have seen no evidence for a covalent modification of SusD when SusC is present. That is, migration of SusD in SDS gels appears unchanged. An even more surprising finding was that mixing SusC and SusD from different strains before passing them over the column resulted in more binding of SusC to the column. This result shows that SusC is not acting as a chaperone for SusD or vice versa, because both proteins were properly folded enough to be stably maintained in the cell. Yet it is difficult to understand why mixing them after production and localization would lead to a more effective binding complex. What this does show, however, is that SusC and SusD can associate even in the solubilized form to form a complex that allows them to bind the starch column.
Results from the 14C-starch binding assay, the proteinase K accessibility experiments, and the in vitro column assay support the hypothesis that SusE plays a role in starch binding and may stabilize the SusC-SusD-SusE complex. With SusE present, binding of starch by intact cells was increased. Moreover, starch protected SusE from proteinase K digestion. Finally, in the in vitro column assay, SusE allowed SusC to be retained more efficiently along with SusD on the column. The role of SusF remains a mystery. Results from the starch binding assay suggest that its role in starch binding is minimal. Yet it is clearly exposed on the bacterial surface, and starch protected it from proteinase K digestion. One possible explanation for this result is that SusE, which does enhance binding, might have protected SusF simply by tethering the starch so that nearby SusF was partially protected. If SusF does play a role in starch binding, it appears to be a minor one. An unanswered question is why the cells need SusE and SusF at all, since SusC and SusD are sufficient to bind starch. These proteins do not add to the tightness of binding of starch to the cells, because binding to cells producing only SusC and SusD is just as irreversible as binding to wild-type cells. One possible explanation comes from considering what the binding assay does not measure: translocation of the starch. Under conditions used to measure binding, there is no accumulation of label by the cells after the initial binding step. Thus, the assay presumably measures binding independently of uptake and further utilization of starch. At present, there is no assay for the putative translocation step. Possibly SusE and SusF play a role in this step. Also absent from most of the mutants used in this study was SusG, one of the starch-degrading enzymes. SusE and SusF may play a role in interacting with SusG, which makes no contribution to starch binding.
Results of the protease accessibility experiments show that SusE and SusF are surface exposed. The data for SusC and SusD are less clear-cut. The fact that when SusC and SusD were produced separately in intact cells they were accessible to protease digestion suggests that they are surface exposed. When both are present, however, digestion by exogenous protease was minor in the case of SusC and not detectable in the case of SusD. SusC has many homologues in the B. thetaiotaomicron genome and in the Porphyromonas gingivalis genome (6, 11). The amino acid sequence of SusC, together with the fact that SusC is necessary and sufficient for utilization of intermediate-sized oligomers of glucose (9), suggests that SusC might be a porin. In this role, SusC would have to be surface exposed in order to admit the oligosaccharides to the periplasmic space.
Since SusD is important for binding long-chain starch, it would seem that this protein too should be exposed on the surface. It may be that the interaction of SusC and SusD, further stabilized by membrane components, produces a complex that renders SusD inaccessible to protease attack. This possibility is supported by the observation of protease accessibility in membranes isolated from disrupted cells. SusC in these membrane fragments was rapidly digested by proteinase K, while SusD was stable even after SusC had disappeared. Yet when SusC was not present at all, SusD was degraded completely, even in intact cells. As with the column data, this finding supports the possibility that SusD interacts with SusC.
The starch utilization system of B. thetaiotaomicron has unique features compared to other studied systems. The majority of components are involved in substrate attachment rather than hydrolysis. Other characterized surface-associated multiprotein complexes are composed mainly of enzymes and other proteins that help localize or assemble them to the complex. The complex that is most closely analogous to the starch utilization system is the cellulosome found in cellulolytic clostridial species. The cellulosome is a complex of cellulases and scaffolding proteins which is either secreted into the extracellular medium or embedded in the cell surface (3). For the cellulosome, the cellulose binding domains and catalytic sites are primarily on the same protein. There is one protein, CipC, which may play a role similar to that of the combination of SusC and SusD. It is thought to keep cellulose in a position favorable to enzymatic attack by other proteins (8). Nevertheless, in contrast to the Sus system, the cellulosome utilizes a majority of enzymes rather than noncatalytic binding proteins for its function.
The interactions of the Sus OMPs are very important for binding and presumably for translocation as well. A similar phenomenon can be found in the maltose transport system in the cytoplasmic membrane of E. coli. Three proteins act as a membrane-associated multiprotein complex to transport maltose into the cytoplasm: MalF, MalG, and MalK (4). Through similar protease accessibility experiments, Traxler and Beckwith found that MalF and MalG interact and assemble in the inner membrane to form a functional translocation complex with two copies of MalK (15). Our system differs in several aspects from this one. First, the complex is associated with the outer membrane. Second, it not only translocates but strongly binds and then cleaves a large substrate either before or during translocation into the periplasm. The starch utilization system of B. thetaiotaomicron appears to be the first example of an outer membrane-associated multiprotein complex that separates two functions, substrate binding and hydrolysis, using different proteins for each function.
Although the Sus proteins characterized to date mediate the early steps in starch utilization, there are clearly other proteins that should be part of the utilization process that have still not been identified. These include such proteins as the equivalents of E. coli MalF, MalG, and MalK. There may be redundant systems for maltose utilization in B. thetaiotaomicron, because to date no mutants have been found that fail to use maltose.
ACKNOWLEDGMENTS
We thank Kyu Hong Cho for excellent technical advice on the affinity chromatography experiments as well as many helpful discussions. We also thank James Imlay for critical review of the manuscript and many helpful suggestions throughout this project.
REFERENCES
- 1.Anderson K L, Salyers A A. Biochemical evidence that starch breakdown by Bacteroides thetaiotaomicron involves outer membrane starch-binding sites and periplasmic starch-degrading enzymes. J Bacteriol. 1989;171:3192–3198. doi: 10.1128/jb.171.6.3192-3198.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K L, Salyers A A. Genetic evidence that outer membrane binding of starch is required for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1989;171:3199–3204. doi: 10.1128/jb.171.6.3199-3204.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 4.Davidson A L, Nikaido H. Purification and characterization of the membrane-associated components of the maltose transport system from Escherichia coli. J Biol Chem. 1991;266:8946–8951. [PubMed] [Google Scholar]
- 5.Hanahan D. Studies on the transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 6.Hanley S A, Aduse-Opoku J, Curtis M A. A 55-kilodalton immunodominant antigen of Porphyromonas gingivalis W has arisen via horizontal gene transfer. Infect Immun. 1999;67:1157–1171. doi: 10.1128/iai.67.3.1157-1171.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lederberg E M, Cohen S M. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974;119:1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pages S, Gal L, Belaich A, Gaudin C, Tardif C, Belaich J P. Role of scaffolding protein CipC of Clostridium cellulolyticum in cellulose degradation. Appl Environ Microbiol. 1997;179:2810–2816. doi: 10.1128/jb.179.9.2810-2816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves A R, D'Elia J N, Frias J, Salyers A A. A Bacteroides thetaiotaomicron outer membrane protein that is essential for utilization of maltooligosaccharides and starch. J Bacteriol. 1996;178:823–830. doi: 10.1128/jb.178.3.823-830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves A R, Wang G-R, Salyers A A. Characterization of four outer membrane proteins that play a role in utilization of starch by Bacteroides thetaiotaomicron. J Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salyers A A, Shoemaker N, Cooper A, D'Elia J, Shipman J A. Genetic methods for diverse prokaryotes. 29th ed. Vol. 29. London, England: Academic Press Ltd.; 1999. Genetic methods for Bacteroides species; pp. 229–249. [Google Scholar]
- 12.Shipman J A, Cho K H, Siegel H A, Salyers A A. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoemaker N B, Getty C, Guthrie E P, Salyers A A. Regions in Bacteroides plasmids pBFTM10 and pB8-51 that allow Escherichia coli-Bacteroides shuttle vectors to be mobilized by IncP plasmids and by a conjugative Bacteroides tetracycline resistance element. J Bacteriol. 1986;166:959–965. doi: 10.1128/jb.166.3.959-965.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tancula E, Feldhaus M J, Bedzyk L A, Salyers A A. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J Bacteriol. 1992;174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Traxler B, Beckwith J. Assembly of a hetero-oligomeric membrane protein complex. Proc Natl Acad Sci USA. 1992;89:10852–10856. doi: 10.1073/pnas.89.22.10852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valentine P, Salyers A A. Analysis of proteins associated with growth of Bacteroides ovatus on the branched galactomannan guar gum. Appl Environ Microbiol. 1992;58:1534–1540. doi: 10.1128/aem.58.5.1534-1540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]