Abstract
Background
Only a subset of patients with gastric cancer experience long-term benefits from immune checkpoint inhibitors (ICIs). Currently, there is a deficiency in precise predictive biomarkers for ICI efficacy. The aim of this study was to develop and validate a pathomics-driven ensemble model for predicting the response to ICIs in gastric cancer, using H&E-stained whole slide images (WSI).
Methods
This multicenter study retrospectively collected and analyzed H&E-stained WSIs and clinical data from 584 patients with gastric cancer. An ensemble model, integrating four classifiers: least absolute shrinkage and selection operator, k-nearest neighbors, decision trees, and random forests, was developed and validated using pathomics features, with the objective of predicting the therapeutic efficacy of immune checkpoint inhibition. Model performance was evaluated using metrics including the area under the curve (AUC), sensitivity, and specificity. Additionally, SHAP (SHapley Additive exPlanations) analysis was used to explain the model’s predicted values as the sum of the attribution values for each input feature. Pathogenomics analysis was employed to explain the molecular mechanisms underlying the model’s predictions.
Results
Our pathomics-driven ensemble model effectively stratified the response to ICIs in training cohort (AUC 0.985 (95% CI 0.971 to 0.999)), which was further validated in internal validation cohort (AUC 0.921 (95% CI 0.839 to 0.999)), as well as in external validation cohort 1 (AUC 0.914 (95% CI 0.837 to 0.990)), and external validation cohort 2 (0.927 (95% CI 0.802 to 0.999)). The univariate Cox regression analysis revealed that the prediction signature of pathomics-driven ensemble model was a prognostic factor for progression-free survival in patients with gastric cancer who underwent immunotherapy (p<0.001, HR 0.35 (95% CI 0.24 to 0.50)), and remained an independent predictor after multivariable Cox regression adjusted for clinicopathological variables, (including sex, age, carcinoembryonic antigen, carbohydrate antigen 19-9, therapy regime, line of therapy, differentiation, location and programmed death ligand 1 (PD-L1) expression in all patients (p<0.001, HR 0.34 (95% CI 0.24 to 0.50)). Pathogenomics analysis suggested that the ensemble model is driven by molecular-level immune, cancer, metabolism-related pathways, and was correlated with the immune-related characteristics, including immune score, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data score, and tumor purity.
Conclusions
Our pathomics-driven ensemble model exhibited high accuracy and robustness in predicting the response to ICIs using WSIs. Therefore, it could serve as a novel and valuable tool to facilitate precision immunotherapy.
Keywords: Immune Checkpoint Inhibitor, Pathology, Gastric Cancer
WHAT IS ALREADY KNOWN ON THIS TOPIC
The effectiveness of immune checkpoint inhibition is intricately influenced by the intricate interplay between tumor, microenvironment, and host factors, resulting in the overall treatment outcome. H&E-stained whole slide images contain valuable information about the tumor microenvironment. Previous studies have demonstrated the potential of using pathomics signatures to predict the effectiveness of immunotherapy. The main emphasis of these investigations was on establishing imaging substitutes for distinct molecular indicators, including but not limited to microsatellite instability, Epstein-Barr virus infection status, or programmed death ligand 1(PD-L1) expression. The primary objective, however, was not to explore the potential of pathomics features in augmenting the prediction of immunotherapy efficacy on their own.
WHAT THIS STUDY ADDS
we have developed and validated a pathomics-driven ensemble model for direct prediction of immunotherapy efficacy.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Through rigorous validation with data from multiple centers, the pathomics-driven ensemble model has shown its predictive value beyond traditional risk factors. Overall, the pathomics-driven ensemble model represents a novel and valuable tool for precision immunotherapy in gastric cancer.
Introduction
Gastric cancer (GC) is a commonly diagnosed malignancy and stands as the third most prevalent cause of cancer-associated death globally.1 Immune checkpoint inhibitors (ICIs) targeting programmed death (ligand) 1 (PD-(L)1) emerged as the primary treatment option for patients lacking targeted therapeutic alternatives.2–4 Despite the effectiveness of ICIs in numerous tumor types, patients with GC exhibit heterogeneous responses to ICIs.4 5 The durable response of ICIs continues to be limited to a small subset of patients.3 5 Effective predictive biomarkers play a crucial role in distinguishing patients who have a higher likelihood of benefiting from immunotherapy with ICIs, compared with those who may benefit more from alternative treatment modalities.
In GC, various molecular indicators such as tumor mutation burden (TMB), PD-L1 expression, microsatellite instability (MSI), and Epstein-Barr virus (EBV) infection status, have been proposed to identify susceptibility to PD-(L)1 inhibitors.6–8 However, these features are inadequate in capturing the full variability in outcomes, likely due to the complex nature of the immune response to cancer.8–10 For example, PD-L1 expression is currently the most commonly used indicator for predicting the efficacy of immune therapy in clinical practice, but studies have found that patients with low or no expression of PD-L1 can also benefit from anti-PD-(L)1 treatment, indicating the need for improved predictive value.8 Microsatellite instability-high (MSI-H) and TMB are also markers for immune therapy, but the proportion of MSI-H patients in advanced GC is only about 4%–7%.5 11 Additionally, there are limitations in MSI and TMB testing, such as high sample requirements, lack of standardized criteria, susceptibility to testing methods and analysis techniques, and high costs involved.11 12
The effectiveness of immune checkpoint inhibition is intricately influenced by the intricate interplay between tumor, microenvironment, and host factors, resulting in the overall treatment outcome.13–16 H&E-stained whole slide images (WSI) contain valuable information about the tumor microenvironment. Pathomics analyses hold potential in a range of tasks.17 18 These tasks include quantifying the tissue microenvironment, conducting comprehensive image-based omics analyses, identifying morphological features associated with prognosis, and establishing links between morphology and treatment response.17 18
Previous studies have demonstrated the potential of using pathomics signatures to predict the effectiveness of immunotherapy.19–21 Nevertheless, the main emphasis of these investigations was on establishing imaging substitutes for distinct molecular indicators, including but not limited to MSI, EBV infection status, or PD-L1 expression. The primary objective, however, was not to explore the potential of pathomics features in augmenting the prediction of immunotherapy efficacy on their own.20–22 While these studies emphasize the potential of pathomics in capturing significant biological information, their limitations lie in the fact that these molecular characteristics only act as partial indicators for predicting the efficacy of ICIs. Furthermore, previous studies have overlooked the interpretation of the complex pathomics models.20–22
In this study, we aimed to develop and validate a pathomics-driven ensemble model for direct prediction of immunotherapy efficacy and to interpret the ensemble model. We conducted a multicenter investigation to train and validate an ensemble model based on pathomics features, aiming to predict the therapeutic efficacy of immune checkpoint inhibition in GC. Furthermore, we employed SHAP (SHapley Additive exPlanations) analysis and pathogenomics analysis to interpret the predictions made by ensemble models from different perspectives.
Methods
Study design and participants
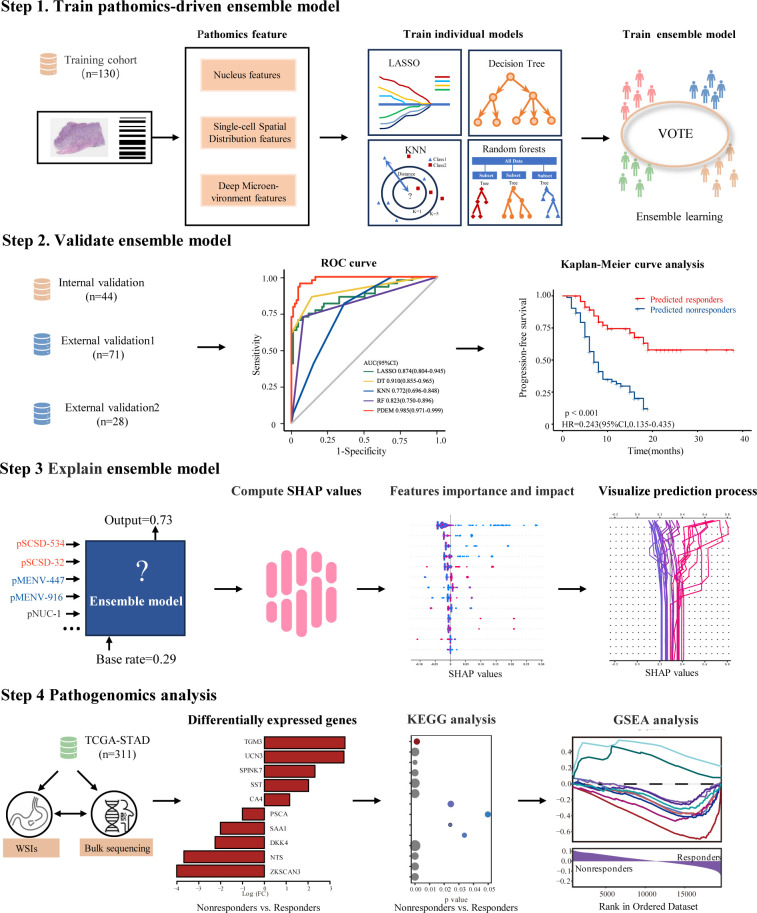
Within this multicenter study, clinical data and whole-slide H&E images from 584 patients were retrospectively gathered and analyzed (figure 1). To predict the response to ICIs, this study retrospectively included patients diagnosed with GC who underwent ICIs at Sun Yat-sen University Cancer Center (n=174), Nanfang Hospital of Southern Medical University (n=71), and Guangdong Provincial Hospital of Chinese Medicine (n=28) from January 2019 to December 2021. The patients from the three centers were divided into four cohorts: the training cohort (n=130) and internal validation cohort (n=44) from Sun Yat-sen University Cancer Center, and the external validation cohort 1 (n=71) and external validation cohort 2 (n=28) from Nanfang Hospital of Southern Medical University and Guangdong Provincial Hospital of Chinese Medicine, respectively. The online supplemental methods contain a comprehensive list of the primary criteria for inclusion and exclusion.
Figure 1.
Study design overview. In this multicenter study, a retrospective collection and analysis of whole-slide H&E images and clinical data were conducted for 584 patients. An ensemble learning model was developed and validated using pathomics features with the objective of predicting the therapeutic efficacy of immune checkpoint inhibition in gastric cancer. Model performance was assessed using metrics such as area under the curve, sensitivity, specificity, positive predictive value, and negative predictive value. Additionally, SHAP (SHapley Additive exPlanations) analysis was employed to explain the model’s predicted values as the sum of the attribution values for each input feature, and pathogenomics analysis was employed to explain the molecular mechanisms underlying the predictions made by the model. GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes; KNN, k-nearest neighbors; LASSO, least absolute shrinkage and selection operator; pMENV, pathomics deep microenvironment features; pNUC, pathomics nucleus features; pSCSD, pathomics single-cell spatial distribution features; ROC curve, receiver operating characteristic curve; TCGA-STAD, The Cancer Genome Atlas-stomach adenocarcinoma.
jitc-2024-008927supp001.pdf (7.4MB, pdf)
Baseline information, such as age, sex, carcinoembryonic antigen (CEA) levels, tumor location, differentiation, carbohydrate antigen 19-9 (CA19-9) levels, PD-L1 expression, therapy regimen, ICI therapy line, and follow-up data was collected. The Combined Positive Score for PD-L1 expression was calculated by dividing the sum of PD-L1-positive tumor cells (showing either partial or complete membrane staining), lymphocytes, and macrophages (with either membrane staining or intracellular staining, or both) by the total number of viable tumor cells, and then multiplying the result by 100.3
After receiving immunotherapy, patients have regular follow-up appointments. Initially, visits are scheduled every 3 months for the first 2 years, then switch to every 6 months thereafter. During each visit, radiologists evaluate treatment response based on imaging studies, like abdominal enhanced CT scans, using the response evaluation criteria in solid tumours (RECIST) (V.1.1) criteria, which categorize the response as progressive disease (PD), stable disease (SD), partial response (PR), or complete response (CR). In this study, patients are categorized as responders (CR/PR) or non-responders (SD/PD) based on their treatment response.15 23–25 Patient survival is measured through both progression-free survival and overall survival. Progression-free survival is the time from starting immunotherapy to tumor progression or death from any cause. Overall survival is the time from starting immunotherapy to death from any cause.
In order to validate and interpret the pathomics-driven model at the molecular level, we assembled an independent cohort consisting of 416 patients with GC from The Cancer Genome Atlas (TCGA) database accessed through the Genomic Data Commons (https://gdc.cancer.gov/). The cohort was selected based on the inclusion of high-quality pathological images. Cases lacking image resolution or exhibiting suboptimal image quality (such as the presence of pen marks or inadequate staining) were excluded from the analysis. Ultimately, this data set included 311 patients with GC for validation (online supplemental figure 1).
Image acquisition and processing
The Aperio ScanScope Scanner system (Leica Biosystems) was used to scan all selected slides with the ×40 objective. The resulting images were then digitized and saved as svs. format files. To manage these files, the Aperio ImageScope software (V.12.4.6) was employed. To ensure adequate image quality, all WSIs were reviewed. WSIs captured at the 40× magnification (0.25 µm/pixel) were analyzed whenever available, while some slides were scanned at 20× and their corresponding images were used. The public clustering-constrained attention multiple instance learning (CLAM) repository was employed to perform automated tissue segmentation for each WSI.26 Subsequently, two expert pathologists meticulously examined and refined the regions of interest (ROIs) using the ImageScope software (online supplemental figure 2).
Extraction of pathomics features from images
To develop a model based on pathomics features, three types of quantitative pathomics features were extracted: pathomics nucleus feature, pathomics deep microenvironment features, and pathomics single-cell spatial distribution features. Three categories encompass information on individual cell morphology, cellular spatial distribution, and the overall microenvironment, providing a holistic insight into the tumor microenvironment (online supplemental figure 2).
First, pathomics tumor nucleus features were extracted. After segmenting tumor nuclei using a HoVer-Net model27 for each ROI, we extracted three categories of pathomics nucleus features, including nuclear intensity, morphology, and texture features, using the “MeasureObjectIntensity”, “MeasureObjectSizeShape”, and “Measure Texture” modules in the CellProfiler platform.28 The extracted features were aggregated by mean, median, SD, 25-quantiles, and 75-quantiles of the values for the ROI in each slide. In total, 525 pathomics nucleus features (pNUC) features were generated for each patient.
Next, deep microenvironment pathomics features were extracted. Image patches of size 256×256 were extracted, without overlap, from all identified tissue regions after segmentation. Subsequently, a pretrained ResNet50 model on ImageNet was used as an encoder to convert each 256×256 patch into a 1024-dimensional feature vector, using spatial average pooling after the third residual block.
Finally, single-cell spatial distribution pathomics features were extracted. A pretrained HoVer-Net model27 on the PanNuke data set29 was employed to segment and classify cells in the ROI as tumor cells, lymphocytes, stromal cells, dead cells, or non-neoplastic epithelial cells. To generate the
red, green, and blue (RGB) image, we calculated the quantity of tumor cells, lymphocytes, and stromal cells per square unit on a 16×16 µm2 grid. The resulting image consisted of density maps for tumor cells, lymphocytes, and stromal cells, represented by the red, green, and blue channels, respectively. Lastly, the same pretrained ResNet50 model used for feature extraction of the tumor microenvironment was applied to the RGB image to capture different cells and their spatial organization patterns.
Pathomics-driven ensemble model development and validation
This study employed an ensemble strategy to predict ICIs response. Specifically, the ensemble approach used several algorithms, including the least absolute shrinkage and selection operator, k-nearest neighbors, decision trees, random forests, and a voting regressor. The voting regressor, which is an ensemble meta-estimator, was fitted with the base regressors using the entire data set. The individual predictions from each base regressor were averaged using the voting regressor to generate the final prediction. For comparison purposes, a range of machine learning methods, such as logistic regression, support vector machine, were employed, using the scikit-learn libraries.
Interpretation of the model
The utilization of SHAP, a game theoretic technique, allows for the interpretation of results produced by various machine learning models.30–32 In order to address the challenge of black-box predictions in machine learning models, we used an explanatory model called SHAP to analyze the data produced by the ensemble model. This allowed us to gain insights into the decision-making process of the algorithm. Essentially, the SHAP explainer model calculates SHAP values to measure the impact of each input feature on the predicted output. These values were then employed to prioritize features and visually depict significant associations. A higher value indicates the significance of the predictor and its substantial influence on the prediction of therapeutic effectiveness in immune checkpoint inhibition. SHAP analysis were implemented using the shap libraries (V.0.42.1).
Molecular validation and interpretation of the model
We used the TCGA-stomach adenocarcinoma (STAD) data set, which comprises publicly accessible genomic/transcriptomic data along with corresponding WSIs, for conducting pathogenomics analysis. The gene expression data and WSIs from 311 patients in the TCGA-STAD cohort were used for analysis.
After processing the WSIs, we computed predicted classifications and performed Gene Set Enrichment Analyses to uncover the molecular pathways linked to the pathomics-driven ensemble model. Genes significantly correlated with the predicted classification were identified using the Spearman’s rank test. Correction for multiple testing was carried out using the Benjamini-Hochberg method. In order to elucidate the biological implications of the pathomics-driven ensemble model, Gene Set Enrichment Analysis33 34 and Kyoto Encyclopedia of Genes and Genomes35 analyses were performed, respectively. The default values were applied to all parameters, and a statistically significant adjusted p value of <0.05 was used. The association between the pathomics-driven ensemble model and immune-related features was additionally examined using gene expression data. The Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) score, immune score, stroma score, and tumor purity were calculated using the estimate package (V.17) in R.36 The ESTIMATE score, immune score, stroma score, and tumor purity were presented based on the distribution of samples.
Statistical analysis
Statistical analyses in this study were conducted using R (V.4.1.0), SPSS (V.26.0, IBM), or Python (V.3.6.5). A t-test was used to compare continuous variables, while a χ2 test or Fisher’s exact test was used to compare categorical variables, as appropriate. Univariate and multivariate Cox regression analyses were employed to investigate the effect of survival clinicopathological variables. The Pearson correlation coefficient was used to evaluate pairwise correlations among pathomics features. The survival times of patients in two groups were estimated using the Kaplan-Meier estimator and compared using the log-rank test. The DeLong test was used to estimate the performance of the model in predicting ICIs response. All statistical analyses were conducted using a two-sided approach, considering a p value<0.05 as indicative of statistical significance.
Results
Patient characteristics
A total of 584 eligible patients were enrolled in the retrospective studies. These comprised of 130 patients in the training cohort, 44 patients in the internal validation cohort, 71 patients in the external validation cohort 1, 28 patients in the external validation cohort 2, and 311 patients in the TCGA-STAD cohort. The training cohort (n=130) and internal validation cohort 1 (n=44) were obtained from Sun Yat-sen University Cancer Center (Guangzhou, China). The external validation cohort 1 (n=71) and external validation cohort 2 (n=28) were obtained from Nanfang Hospital of Southern Medical University (Guangzhou, China) and Guangdong Provincial Hospital of Chinese Medicine (Guangzhou, China), respectively. Comprehensive details regarding demographic and clinical characteristics can be retrieved from table 1. The distribution of treatment lines, ICI regimens, and tumor response evaluations according to RECIST (V.1.1) across different cohorts is presented in online supplemental figure 3.
Table 1.
Patient characteristics in all cohorts
| Training cohort (n=130) |
Internal validation cohort (n=44) |
External validation cohort 1 (n=71) |
External validation cohort 2 (n=28) |
|||||||||
| Responders (n=44) |
Non-responders (n=86) | P value | Responders (n=21) |
Non-responders (n=23) | P value | Responders (n=36) |
Non-responders (n=35) | P value | Responders (n=12) |
Non-responders (n=16) | P value | |
| Age | 0.99 | 0.57 | 0.18 | 0.05 | ||||||||
| Median, IQR (years) | 58 (52–66) | 62 (48–66) | 60 (49–68) | 63 (50–69) | 57 (50–65) | 62 (54–67) | 54 (46–63) | 63 (57–67) | ||||
| Sex | 0.39 | 0.05 | 0.12 | 0.24 | ||||||||
| Male | 28 (64) | 48 (56) | 5 (24) | 11 (48) | 24 (67) | 29 (83) | 6 (50) | 12 (75) | ||||
| Female | 16 (36) | 38 (44) | 16 (76) | 12 (52) | 12 (33) | 6 (17) | 6 (50) | 4 (25) | ||||
| Location | 0.35 | 0.12 | 1.00 | |||||||||
| Cardia | 9 (20) | 24 (28) | 7 (33) | 8 (35) | 1.00 | 9 (25) | 6 (17) | 4 (33) | 4 (25) | |||
| Body | 8 (18) | 17 (20) | 3 (14) | 4 (17) | 8 (22) | 7 (20) | 3 (25) | 5 (31) | ||||
| Antrum | 25 (57) | 36 (42) | 10 (48) | 9 (39) | 19 (53) | 17 (49) | 5 (42) | 7 (44) | ||||
| Whole | 2 (5) | 9 (10) | 1 (5) | 2 (9) | 0 | 5 (14) | 0 | 0 | ||||
| Differentiation | 0.31 | 0.91 | 0.10 | 0.60 | ||||||||
| Well | 2 (5) | 3 (3) | 0 | 0 | 1 (3) | 1 (3) | 0 | 1 (6) | ||||
| Moderate | 9 (20) | 12 (14) | 3 (14) | 3 (13) | 9 (25) | 3 (9) | 0 | 1 (6) | ||||
| Poor or undifferentiation | 33 (75) | 71 (83) | 18 (86) | 20 (87) | 26 (72) | 31 (89) | 12 (100) | 14 (88) | ||||
| CEA | 0.15 | 0.39 | 0.53 | 1.00 | ||||||||
| Elevated | 16 (36) | 21 (24) | 10 (48) | 8 (35) | 7 (19) | 9 (26) | 5 (42) | 7 (44) | ||||
| Normal | 28 (64) | 65 (76) | 11 (52) | 15 (65) | 29 (81) | 26 (74) | 7 (58) | 9 (56) | ||||
| CA19-9 | 0.85 | 0.21 | 0.09 | 0.36 | ||||||||
| Elevated | 14 (32) | 26 (30) | 7 (33) | 12 (52) | 3 (8) | 8 (23) | 1 (8) | 4 (25) | ||||
| Normal | 30 (68) | 60 (70) | 14 (67) | 11 (48) | 33 (92) | 27 (77) | 11 (92) | 12 (75) | ||||
| PD-L1 expression (CPS) | 0.14 | 0.51 | 0.38 | 1.00 | ||||||||
| <1 | 7 (16) | 19 (22) | 3 (14) | 7 (30) | 9 (25) | 9 (26) | 3 (25) | 4 (25) | ||||
| 1–10 | 22 (50) | 51 (59) | 9 (43) | 8 (35) | 23 (64) | 18 (51) | 5 (42) | 7 (44) | ||||
| >10 | 15 (34) | 16 (19) | 9 (43) | 8 (35) | 4 (11) | 8 (23) | 4 (33) | 5 (31) | ||||
| Line of immune checkpoint inhibitor therapy | 0.15 | 0.40 | <0.01 | 1.00 | ||||||||
| 1 | 10 (23) | 30 (35) | 18 (86) | 18 (78) | 31 (86) | 13 (37) | 5 (42) | 7 (44) | ||||
| ≥2 | 34 (77) | 56 (65) | 3 (14) | 5 (22) | 5 (14) | 22 (63) | 7 (58) | 9 (56) | ||||
| Therapy regimen | 0.13 | 0.29 | 1.00 | 0.57 | ||||||||
| Anti-PD-(L)1 immune checkpoint inhibitor monotherapy | 3 (7) | 14 (16) | 18 (86) | 16 (70) | 2 (6) | 2 (6) | 8 (67) | 10 (63) | ||||
| Immune checkpoint inhibitor plus chemotherapy | 41 (93) | 72 (84) | 3 (14) | 7 (30) | 34 (94) | 33 (94) | 4 (33) | 6 (38) | ||||
Data are n (%), or median (IQR).
CA19-9, carbohydrate antigen 19-9; CEA, carcinoembryonic antigen; CPS, Combined Positive Score of PD-L1 expression; PD-L1, programmed death ligand 1.
Out of the participants, 44 (34%) of the 130 in the training cohort, 21 (48%) of the 44 in the internal validation cohort, 36 (51%) of the 71 in the external validation cohort 1, and 12 (43%) of the 28 in the external validation cohort 2 were classified as responders (table 1). No significant differences were observed in the baseline characteristics between responders and non-responders, except for the line of ICI therapy in the external validation cohort 1, which was found to be statistically significant (p<0.05) (table 1).
Pathomics-driven ensemble model development and validation
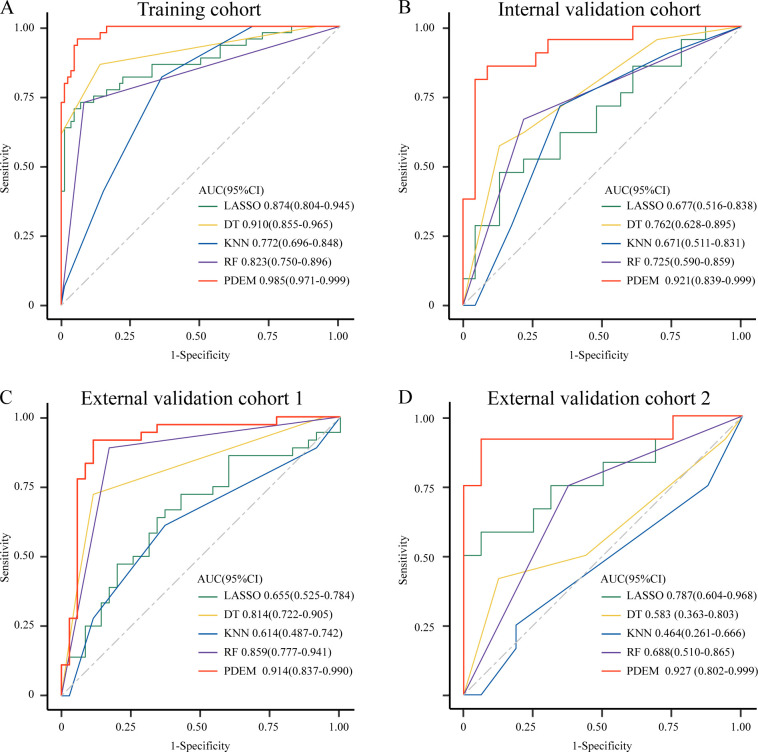
For the purpose of this study, we extracted three types of quantitative pNUC, single-cell spatial distribution features (pSCSD), and deep microenvironment features (pMENV) from baseline H&E-stained slides. Then, we developed a pathomics-driven ensemble model (PDEM) to predict the response of patients with GC after ICIs treatment (figure 1). The ensemble model demonstrated favorable predictive accuracy for response to ICIs in the training cohort, with an area under the curve (AUC) of 0.985 (95% CI 0.971 to 0.999). Moreover, the ensemble model exhibited a remarkable sensitivity of 0.955 (95% CI 0.893 to 0.999) and specificity of 0.942 (95% CI 0.892 to 0.991) in the training cohort, as shown in table 2. Following successful development in the training cohort, the ensemble model exhibited high accuracy in predicting response to ICIs in the internal validation cohort (AUC 0.921 (95% CI 0.839 to 0.999)), as well as in external validation cohorts 1 (AUC 0.914 (95% CI 0.837 to 0.990)) and 2 (AUC 0.927 (95% CI 0.802 to 0.999)). The ensemble model maintained a high sensitivity (95% CI 0.857 to 0.917) and specificity (95% CI 0.78 to 0.938) in the three validation cohorts. The ensemble model demonstrated negative predictive value (NPV) greater than 0.87 in all cohorts, with a positive predictive value (PPV) surpassing 0.89 (table 2) (online supplemental figure 4).
Table 2.
Prediction performance of pathomics-driven ensemble model versus individual prediction models and CPS in multiple cohorts
| Training cohort | Internal validation cohort | External validation cohort 1 | External validation cohort 2 | |
| Pathomics-driven ensemble model | ||||
| AUC | 0.985 (0.971 to 0.999) | 0.921 (0.839 to 0.999) | 0.914 (0.837 to 0.990) | 0.927 (0.802 to 0.999) |
| Specificity | 0.942 (0.892 to 0.991) | 0.913 (0.798 to 0.999) | 0.886 (0.780 to 0.991) | 0.938 (0.819 to 0.999) |
| Sensitivity | 0.955 (0.893 to 0.999) | 0.857 (0.707 to 0.999) | 0.917 (0.826 to 0.999) | 0.917 (0.760 to 0.999) |
| NPV | 0.976 (0.943 to 0.999) | 0.875 (0.743 to 0.999) | 0.912 (0.816 to 0.999) | 0.938 (0.819 to 0.999) |
| PPV | 0.894 (0.805 to 0.982) | 0.900 (0.769 to 0.999) | 0.892 (0.792 to 0.992) | 0.970 (0.760 to 0.999) |
| LASSO | ||||
| AUC | 0.874 (0.804 to 0.945) | 0.677 (0.516 to 0.838) | 0.655 (0.525 to 0.784) | 0.787 (0.604 to 0.968) |
| Specificity | 0.953 (0.909 to 0.998) | 0.870 (0.732 to 0.999) | 0.657 (0.500 to 0.814) | 0.938 (0.819 to 0.999) |
| Sensitivity | 0.705 (0.468 to 0.874) | 0.476 (0.263 to 0.69) | 0.639 (0.482 to 0.796) | 0.583 (0.304 to 0.862) |
| NPV | 0.863 (0.794 to 0.932) | 0.645 (0.477 to 0.814) | 0.639 (0.482 to 0.796) | 0.750 (0.560 to 0.940) |
| PPV | 0.886 (0.78 to 0.991) | 0.769 (0.54 to 0.998) | 0.657 (0.500 to 0.814) | 0.875 (0.646 to 0.999) |
| KNN | ||||
| AUC | 0.772 (0.696 to 0.848) | 0.671 (0.511 to 0.831) | 0.614 (0.487 to 0.742) | 0464 (0.261 to 0.666) |
| Specificity | 0.64 (0.538 to 0.741) | 0.652 (0.458 to 0.847) | 0.629 (0.468 to 0.789) | 0.813 (0.621 to 0.999) |
| Sensitivity | 0.818 (0.704 to 0.932) | 0.714 (0.521 to 0.908) | 0.611 (0.452 to 0.77) | 0.250 (0.005 to 0.495) |
| NPV | 0.873 (0.791 to 0.955) | 0.714 (0.521 to 0.908) | 0.611 (0.452 to 0.77) | 0.591 (0.385 to 0.796) |
| PPV | 0.537 (0.418 to 0.657) | 0.652 (0.458 to 0.847) | 0.629 (0.468 to 0.789) | 0.500 (0.100 to 0.900) |
| Decision tree | ||||
| AUC | 0.910 (0.855 to 0.965) | 0.762 (0.628 to 0.895) | 0.814 (0.722 to 0.905) | 0.583 (0.363 to 0.803) |
| Specificity | 0.86 (0.787 to 0.934) | 0.870 (0.732 to 0.999) | 0.886 (0.780 to 0.991) | 0.875 (0.713 to 0.999) |
| Sensitivity | 0.864 (0.762 to 0.965) | 0.571 (0.360 to 0.783) | 0.722 (0.576 to 0.869) | 0.417 (0.138 to 0.696) |
| NPV | 0.925 (0.867 to 0.983) | 0.690 (0.521 to 0.858) | 0.756 (0.625 to 0.888) | 0.667 (0.465 to 0.868) |
| PPV | 0.760 (0.642 to 0.878) | 0.800 (0.598 to 0.999) | 0.867 (0.745 to 0.988) | 0.714 (0.380 to 0.999) |
| Random forest | ||||
| AUC | 0.823 (0.750 to 0.896) | 0.725 (0.590 to 0.859) | 0.859 (0.777 to 0.941) | 0.688 (0.510 to 0.865) |
| Specificity | 0.919 (0.861 to 0.976) | 0.783 (0.614 to 0.951) | 0.829 (0.704 to 0.953) | 0.625 (0.388 to 0.862) |
| Sensitivity | 0.727 (0.596 to 0.859) | 0.667 (0.465 to 0.868) | 0.889 (0.786 to 0.992) | 0.750 (0.505 to 0.995) |
| NPV | 0.868 (0.799 to 0.938) | 0.72 (0.544 to 0.896) | 0.879 (0.767 to 0.990) | 0.769 (0.540 to 0.998) |
| PPV | 0.821 (0.700 to 0.941) | 0.737 (0.539 to 0.935) | 0.842 (0.726 to 0.958) | 0.600 (0.352 to 0.848) |
| CPS | ||||
| AUC | 0.585 (0.490 to 0.681) | 0.581 (0.420 to 0.741) | 0.459 (0.338 to 0.580) | 0.508 (0.298 to 0.718) |
| Specificity | 0.814 (0.732 to 0.896) | 0.304 (0.116 to 0.492) | 0.257 (0.112 to 0.402) | 0.688 (0.460 to 0.915) |
| Sensitivity | 0.341 (0.201 to 0.481) | 0.857 (0.707 to 0.999) | 0.75 (0.609 to 0.891) | 0.333 (0.067 to 0.6) |
| NPV | 0.707 (0.617 to 0.797) | 0.700 (0.416 to 0.984) | 0.500 (0.269 to 0.731) | 0.579 (0.357 to 0.801) |
| PPV | 0.484 (0.308 to 0.668) | 0.529 (0.362 to 0.697) | 0.509 (0.375 to 0.644) | 0.444 (0.120 to 0.769) |
| SVM | ||||
| AUC | 0.886 (0.822 to 0.950) | 0.553 (0.377 to 0.728) | 0.585 (0.450 to 0.720) | 0.422 (0.189 to 0.655) |
| Specificity | 0.767 (0.678 to 0.857) | 0.565 (0.363 to 0.768) | 0.686 (0.532 to 0.84) | 0.750 (0.538 to 0.962) |
| Sensitivity | 0.932 (0.857 to 0.999) | 0.619 (0.411 to 0.827) | 0.528 (0.365 to 0.691) | 0.333 (0.067 to 0.600) |
| NPV | 0.957 (0.908 to 0.999) | 0.619 (0.411 to 0.827) | 0.586 (0.435 to 0.736) | 0.600 (0.385 to 0.815) |
| PPV | 0.672 (0.554 to 0.790) | 0.565 (0.363 to 0.768) | 0.633 (0.461 to 0.806) | 0.500 (0.154 to 0.846) |
| Logistic regression | ||||
| AUC | 0.641 (0.566 to 0.716) | 0.508 (0.391 to 0.625) | 0.528 (0.490 to 0.566) | 0.563 (0.410 to 0.715) |
| Specificity | 0.942 (0.892 to 0.991) | 0.826 (0.671 to 0.981) | 0.999 (0.999 to 1.000) | 0.875 (0.713 to 0.999) |
| Sensitivity | 0.341 (0.201 to 0.481) | 0.190 (0.023 to 0.358) | 0.056 (0.019 to 0.130) | 0.250 (0.005 to 0.495) |
| NPV | 0.736 (0.654 to 0.819) | 0.528 (0.365 to 0.691) | 0.507 (0.389 to 0.625) | 0.609 (0.409 to 0.808) |
| PPV | 0.750 (0.560 to 0.940) | 0.500 (0.154 to 0.846) | 0.999 (0.999 to 1.000) | 0.600 (0.171 to 0.999) |
Data are mean (95% CI).
AUC, area under the curve; CPS, Combined Positive Score of PD-L1 expression; KNN, k-nearest neighbors; LASSO, least absolute shrinkage and selection operator; NPV, negative predictive value; PD-L1, programmed death-ligand 1; PPV, positive predictive value; SVM, support vector machine.
The four individual predictions from the least absolute shrinkage and selection operator (LASSO), k-nearest neighbors (KNN), decision trees (DT), and random forests (RF) are ultimately fused into the pathomics-driven ensemble model. Compared with individual predictions in the validation cohorts, the ensemble model exhibited superior performance in predicting response to ICIs (figure 2). In the training cohort, the DT model exhibited a slightly lower AUC (0.910 (95% CI 0.855 to 0.965)) compared with the ensemble model. However, in the validation cohorts, the DT model demonstrated significantly lower AUCs, ranging from 0.583 (95% CI 0.363 to 0.803) to 0.814 (95% CI 0.722 to 0.905). The RF model outperformed the other individual predictions with a higher AUC in the internal validation cohort (AUC 0.725 (95% CI 0.590 to 0.859)), as well as in the external validation cohorts 1 and 2 (AUC 0.859 (95% CI 0.777 to 0.941) and 0.688 (95% CI 0.510 to 0.865), respectively). However, its AUC was still lower compared with the PDEM (table 2). We compared three different combinations of ensemble models. The results indicate that there is no significant difference in the AUC of the ensemble models among the three combinations in the training set. However, in the validation sets, there is a difference in the AUC of the ensemble models among the three combinations, with the KNN+LASSO+DT+RF ensemble model demonstrating greater stability and generalization capability (online supplemental table 1).
Figure 2.
Comparison of prediction performance between the pathomics-driven ensemble model and individual prediction models in the training and validation cohorts. The ensemble model integrated LASSO, KNN, decision trees, and random forests model. Receiver operating characteristic curves of predictive performance for immunotherapy effect in patients with gastric cancer among the four individual predictions (LASSO, KNN, decision trees, random forests) and the pathomics-driven ensemble model in the training cohort (A) internal validation cohort (B) external validation cohort 1 (C) and external validation cohort 2 (D). AUC, area under curve; DT, decision trees; KNN, k-nearest neighbors; LASSO, least absolute shrinkage and selection operator; PDEM, the pathomics-driven ensemble model; RF, random forests.
To delve deeper, a comparison was conducted between the PDEM and PD-L-1 expression level (Combined Positive Score (CPS)), as well as other commonly used models like support vector machine (SVM) and logistic regression (LR). The PDEM outperforms CPS and other modeling methods in predicting the efficacy of ICIs (online supplemental figure 5). In contrast to the PDEM, CPS exhibited significantly lower AUCs values ranging from 0.459 (95% CI 0.338 to 0.580) to 0.585 (95% CI 0.490 to 0.681) across all cohorts, accompanied by reduced sensitivity and specificity. Compared with the PDEM, the SVM model exhibited a lower AUC, ranging from 0.422 (95% CI 0.189 to 0.655) to 0.886 (95% CI 0.822 to 0.950), while the LR model demonstrated significantly inferior AUC values, ranging from 0.508 (95% CI 0.391 to 0.625) to 0.641 (0.566 to 0.716), across all cohorts (table 2). The DeLong test was used to examine the receiver operating characteristic curves, which demonstrated that the PDEM exhibited a substantial enhancement in the AUC for predicting the response to ICIs, surpassing both CPS and alternative modeling techniques (online supplemental table 2). Lastly, we compared the prediction performance between the PDEM and models based on individual types of pathomics features in both the training and internal validation cohorts. The results demonstrate that the model integrating the three types of pathological features outperforms the models based on single pathological features in predicting the efficacy of immunotherapy for GC (online supplemental figure 6).
Prognostic value of pathomics-driven ensemble model
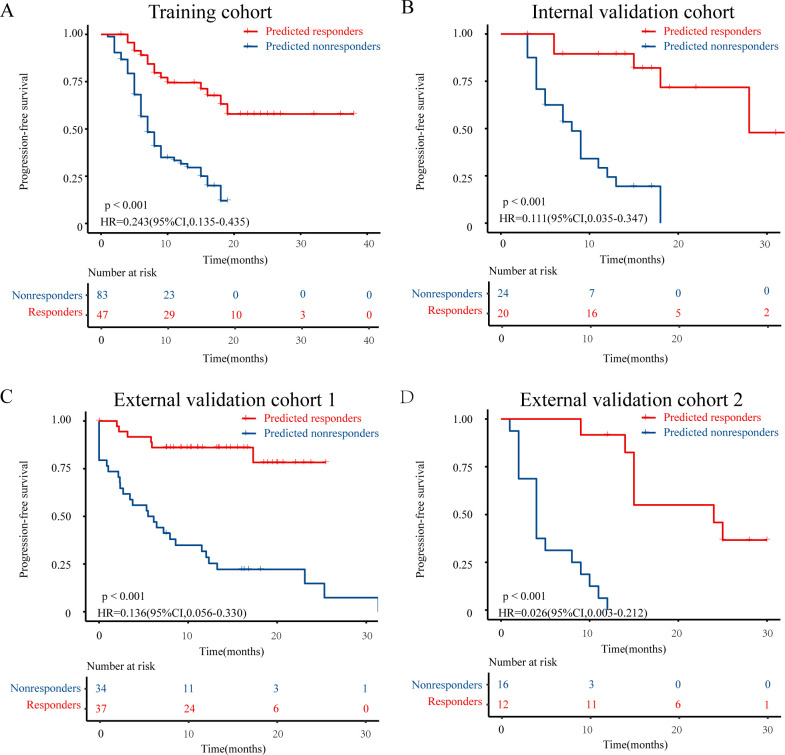
The Kaplan-Meier analysis revealed that the responders predicted by the PDEM had a significantly better progression-free survival (PFS) and overall survival compared with the non-responders (all p<0.05, figure 3, online supplemental figure 7). This finding is consistent with the results observed in patients who actually responded to immune checkpoint inhibition (all p<0.05, online supplemental figures 8 and 9).
Figure 3.
Progression-free survival Kaplan-Meier curve analysis of prediction populations. Patients identified as “predicted responders” by pathomics-driven ensemble model presented favorable progression-free survival than that of patients identified as “predicted nonresponders” in the training cohort (A) internal validation cohort (B) external validation cohort 1 (C) and external validation cohort 2 (D).
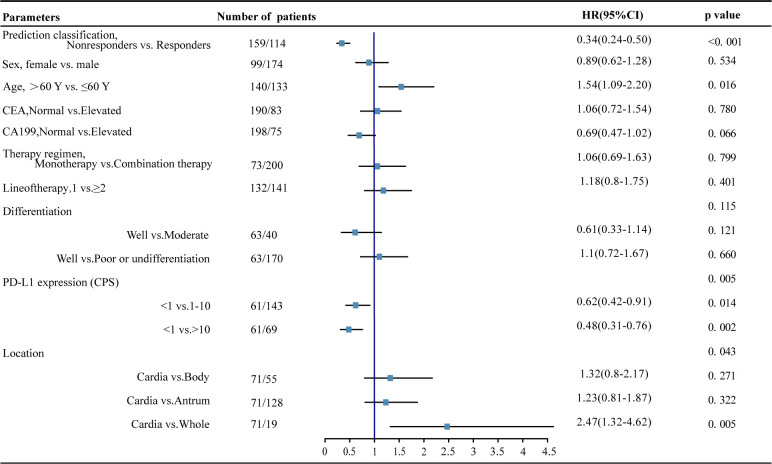
Univariate Cox regression analysis demonstrated that the prediction signature of PDEM was an independent prognostic factor for PFS in patients with GC who underwent immunotherapy (p<0.001, HR 0.35 (95% CI 0.24 to 0.50), online supplemental figure 10). Furthermore, in the multivariable Cox regression analysis adjusting for clinicopathological variables such as sex, age, CEA, CA19-9, therapy regime, line of therapy, differentiation, location, and PD-L1 expression, the prediction signature of the PDEM remained an independent prognostic factor for progression-free survival across all patients (p<0.001, HR 0.34 (95% CI 0.24 to 0.50), figure 4).
Figure 4.
Forest plot for the multivariate cox regression analysis of progression-free survival. CA19-9, carbohydrate antigen 19-9; CEA, carcinoembryonic antigen; CPS, Combined Positive Score of PD-L1 expression;PD-L1, programmed death-ligand 1.
We evaluated the prognostic value of PDEM within each subgroup of patients as defined by clinicopathological variables. our findings revealed significant differences in PFS between patients in two predicted classifications in all subgroups defined by sex, age, CEA, CA19-9, therapy regime, line of therapy, differentiation, location and PD-L1 expression (all p<0.05, online supplemental figure 11). Taken together, these data suggest that the prediction signature of the PDEM is a robust independent prognostic factor in patients with GC who underwent immunotherapy.
Interpretation of pathomics-driven ensemble model
Complex models like ensemble models are not easily understandable. We cannot rely on the original model itself for its interpretation. Instead, it is imperative for us to employ a simplified interpretive framework, termed as an approximation of the primary model’s interpretation. In the interpretation of feature attribution, SHAP functions as an additive method, explaining the model’s predicted values as the cumulative sum of attribution values assigned to each input feature. A significant SHAP value indicates the significance impact of the predictor on predicting the effectiveness of immune checkpoint inhibition therapy.
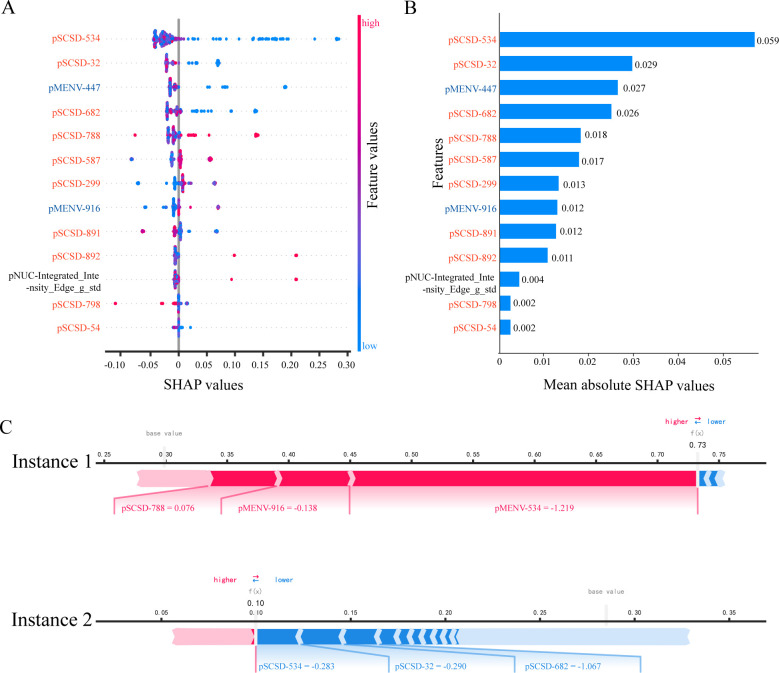
First and foremost, we compute the feature importance and impact using SHAP. We rank the feature importance in descending order and identify 13 pathomics features that strongly influence the prediction of ICIs efficacy. The results are presented through beeswarm summary plot (figure 5A), feature importance plot (figure 5B), and heatmap plot (online supplemental figure 12). Both the feature importance plot and the beeswarm summary plot reveal that pSCSD-534 has a significant impact on the prediction of ICIs efficacy. A smaller value of pSCSD-534 indicates a higher probability of benefiting from ICIs for patients with GC (figure 5A,B, online supplemental figure 12). The mutual correlations among the important features were calculated using Pearson correlation coefficients (online supplemental figure 13).
Figure 5.
Interpretation of pathomics-driven ensemble model by SHAP. (A) Beeswarm summary plot of feature importance from SHAP analysis. The beeswarm plot is designed to display an information-dense summary of how the top features in a data set impact the model’s output. Each observation in the data is represented by a single dot on each feature row. The vertical axis indicates the features, ordered from top to bottom, based on their importance as predictors. The placement of the dot along the feature row is dictated by the corresponding feature’s SHAP value, and the accumulation of dots within each feature row illustrates its density. The feature values determined the color of the dots, with pink representing a direct association with the response to immune checkpoint inhibitors, while blue indicating an inverse association with the response to immune checkpoint inhibitors. (B) Feature importance plot. Passing a matrix of SHAP values to the bar plot function creates a global feature importance plot, where the global importance of each feature is taken to be the mean absolute value for that feature across all the given samples. The model’s predictions of the response to immune checkpoint inhibitors are significantly influenced by predictors exhibiting large mean SHAP values. (C) SHAP force plots. The force plots show how the model arrived at its decision. The ensemble model predicts the probability of response to immune checkpoint inhibitors, with the bolded value indicating the likelihood. Pink represents predictors that are positively associated with response, while blue represents predictors that are negatively associated. Instance 1: the SHAP force plot reveals the identification of a “responder” case that was correctly predicted. Instance 2: the SHAP force plot reveals the identification of a case as “nonresponder” by pathomics-driven ensemble model. pMENV, pathomics deep microenvironment features; pNUC, pathomics nucleus features; pSCSD, pathomics single-cell spatial distribution features; SHAP, SHapley Additive exPlanations.
Furthermore, we visualized the prediction process using SHAP. SHAP provides explanations for individual instance predictions, and SHAP values can be combined for a global interpretation. The force plot (figure 5C) and the decision plot (online supplemental figure 14) effectively illustrate how the model reached its decision. In figure 5C, instance 1, SHAP values explain the predicted probability of ICIs efficacy for patient with GC as follows: the baseline is an average prediction probability of 0.29, the probability of predicting benefit from ICIs is 0.73. Notably, the main influential factors that force the patient’s potential benefit from ICIs are pSCSD-788 value of 0.076, pMENV-916 value of −0.138, and a pMENV-534 value of −1.219.
Molecular validation of pathomics-driven ensemble model
To explain the biological foundations of the PDEM, we conducted an in-depth analysis of pathogenomics. The WSIs were processed to determine the predicted classification, and Gene Set Enrichment Analyses were conducted to identify the molecular pathways linked to the pathomics-based ensemble model.
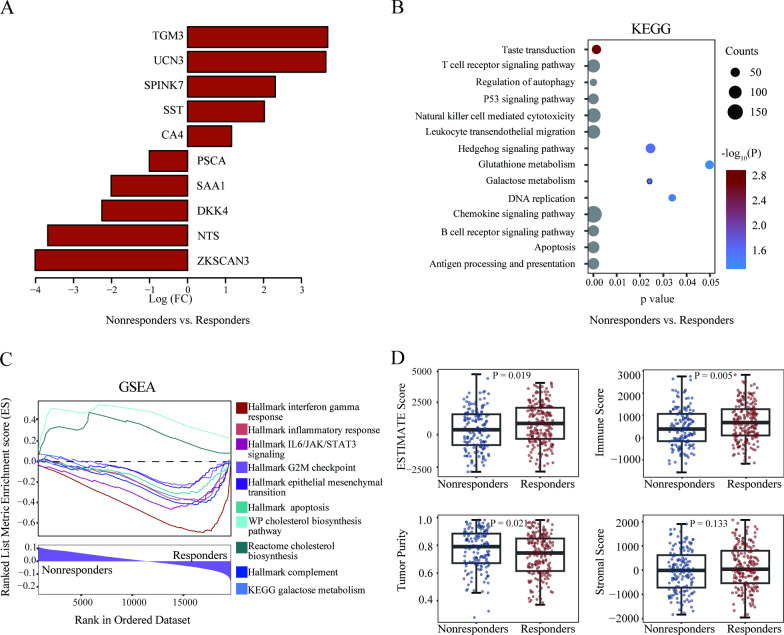
To investigate the transcriptional differences between responders and non-responders identified by the ensemble model, the differentially expressed genes between them were identified (figure 6A). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis indicated that these differentially expressed genes were mainly enriched in categories associated with the immune system, cancer hallmarks, and metabolism-related pathways (figure 6B).
Figure 6.
Pathogenomics analysis of the pathomics-driven ensemble model. (A) The differentially expressed genes between responders and non-responders. (B) Visualization of the top enriched KEGG pathways by gene counts along with p values in responders versus non-responders. (C) Gene Set Enrichment Analysis delineated the molecular pathways significantly associated with the pathomics-driven ensemble model. (D) Associations between the pathomics-driven ensemble model and immune-related characteristics. ESTIMATE score, immune score, stroma score, and tumor purity were presented in responders versus non-responders. ESTIMATE, Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data; GSEA, Gene Set Enrichment Analysis; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Further Gene Set Enrichment Analysis demonstrated that genes highly expressed in the predicted responders showed significant enrichment in immune-related pathways such as complement, interferon gamma response and inflammatory response pathway. Additionally, the predicted responders exhibited enrichment in tumor suppressor pathways, such as the G2M checkpoint and apoptosis pathway. Interestingly, we also observed the enrichment of several metabolism-related pathways. The galactose metabolism pathway was enriched in the predicted responders, whereas the cholesterol biosynthesis pathway was enriched in the predicted non-responders (figure 6C). In general, these results align with the favorable prognosis and response rates observed in the predicted responders, and could indicate potential targets for therapeutic intervention to overcome resistance ICIs in the predicted non-responders.
Subsequently, we investigated the associations between the PDEM and immune-related characteristics. Significant differences were observed in immune score, ESTIMATE score, and tumor purity between responders and non-responders identified by the ensemble model. The responders exhibited higher immune scores and lower ESTIMATE scores and tumor purity (figure 6D). These findings suggest that the ensemble model is driven by molecular-level immune cell infiltration, which holds promise for predicting the efficacy of ICIs.
Discussion
In this study, we developed a PDEM to predict response after immune checkpoint inhibition in patients with GC. Furthermore, we employed SHAP analysis and pathogenomics analysis to interpret the predictions made by ensemble models from diverse perspectives. The crucial role of predicting the potential benefits of ICIs treatment in patients can be effectively accomplished by utilizing the PDEM. The ensemble model demonstrates excellent performance in accurately distinguishing patients who are more likely to benefit from ICIs treatment, as evidenced by favorable AUC, high sensitivity, specificity, PPV, and NPV. Our study offers a dependable and reproducible tool for pretreatment prediction of ICIs response, thereby facilitating the clinical implementation of computer-assisted personalized management for patients diagnosed with GC.
In previous pathomics analyses, the main emphasis has been on establishing imaging surrogates for specific molecular biomarkers, such as MSI, EBV infection status, or PD-L1 expression.20–22 These studies aimed to predict the response to immune checkpoint inhibition by capturing relevant biological information. However, the limited scope of these efforts arises from the fact that these biological characteristics serve as incomplete predictors of ICI response, as they only capture a portion of the intricate and diverse molecular characteristics linked to responsiveness. As a result, the imaging surrogates for these molecular markers are unlikely to surpass the markers themselves, and relying solely on imaging does not contribute to improving prediction accuracy. To overcome this constraint, our goal is to fully exploit the capabilities of imaging through the utilization of an artificial intelligence framework designed for direct outcome prediction.
For the purpose of this study, we extracted three types of quantitative pathomics features: nucleus features, single-cell spatial distribution features, and deep microenvironment features. From a biological standpoint, these three types of pathological features hold distinct biological significance and complement each other. They encompass information on individual cell morphology, cellular spatial distribution, and the overall microenvironment, providing a comprehensive understanding of the tumor microenvironment. We explore three levels of complementary features to fully exploit the pathological image features associated with ICIs.
Another strength of our study is the utilization of pathogenomics analysis to investigate and validate the genetic components of our model. Through this analysis, we identified multiple immune-related pathways and confirmed their significance in the decision-making process of the model. The elucidation of these molecular mechanisms provides valuable insights for clinicians, aiding their understanding of the model and guiding fundamental research on the mechanisms underlying the efficacy of ICIs.
From a clinical perspective, the utilization of the pathomics-based ensemble model holds promise in facilitating personalized treatment decisions for individuals diagnosed with GC. Compared with several molecular predictors, the PDEM demonstrates superior predictive performance. Compared with the CPS, the PDEM shows higher AUC, sensitivity, and specificity. This model can identify potential responders to ICIs among patients with low CPS scores and identify individuals who may not benefit from ICIs within the high CPS score group.
With the increasing prevalence of gastroscopy, pathological examination has become a routine diagnostic procedure for diagnosing GC. Pathological image-based predictive models have a wide range of applications. The PDEM predicts the ICIs response in patients with postoperative GC but also shows potential for selecting candidates who may benefit from neoadjuvant ICIs. Furthermore, compared with conventional histopathological techniques, PDEM has exhibited promising prospects in enhancing the accuracy, repeatability, and efficiency of pathological diagnosis.
Our study has several significant limitations. These include potential bias due to the retrospective, multicenter nature of the study and heterogeneity in data sources. Although our putative model has been validated on two external cohorts, it requires further validation on larger prospective cohorts comprising homogeneous patient populations, treatments, and image modalities. Second, the ensemble model driven by pathomics features demonstrates the potential for predicting the efficacy of neoadjuvant ICIs based on gastric endoscopic pathological slides. However, the model was trained and validated using postoperative pathological slides. It is necessary to further validate the model in a GC cohort undergoing neoadjuvant immunotherapy. Third, efforts should be made to improve the interpretability of the ensemble model. We have conducted pathogenomics analysis pertaining to model predictions. However, we have not explored the correlation between pathomics features and molecular mechanisms. Future investigations aimed at comprehending the underlying mechanisms of these pathomics features and their performance will aid in establishing causal patho-immunogenomic relationships, thereby unraveling the biological intricacies that drive ensemble prediction. Additionally, H&E stained tissue section serves as the foundation of anatomical pathology diagnosis. the heterogeneity inherent in tumors and the limitations in sampling for pathological specimens, primarily due to the potential lack of representativeness in small biopsy specimens, should be taken into consideration.
In conclusion, we have developed and validated a PDEM to predict the effectiveness of immune checkpoint inhibition in patients with GC. Through extensive validation with data from multiple centers, our model has shown its predictive value beyond traditional risk factors. However, further research is needed to confirm these findings and evaluate the clinical applicability of our proposed pathological imaging-based biomarker. Large-scale prospective trials are necessary to refine our results and determine the usefulness of this biomarker in guiding personalized treatment selection for patients with GC.
Acknowledgments
The authors would like to thank The Cancer Genome Atlas for providing the genomic and pathomics data used in this study.
Footnotes
ZH, ZZ and XY contributed equally.
Correction notice: This article has been corrected since it was first published online. The affiliations for author Zhicheng Zhang have been updated to (2) Department of Gastroenterology, The First Hospital of Jilin University, Changchun, Jilin, China and (3) JancsiLab, JancsiTech, Hongkong, China.
Contributors: YJ and GL conceived and designed the study. ZH, XY, XW, JW, TZ, WW, and WX acquired the data. ZZ, Zhe Li, SS, and MTI did the statistical analyses. ZZ, AAG, Zihan Li, and LY developed, trained, and applied the ensemble model. WW, WX, GL, and YJ verified the underlying raw data; all authors had access to the data presented in the manuscript; all authors analyzed and interpreted the data; ZH, TZ, and Zhe Li prepared the first draft of the manuscript; YJ conceived and oversaw the project, interpreted data, reviewed the manuscript, and acts as guarantor; and all authors contributed to manuscript preparation.
Funding: This work was supported by China Postdoctoral Science Foundation (NO:2022M721497), Key-Area Research and Development Program of Guangdong Province (NO: 2021B0101420005), President Foundation of Nanfang Hospital, Southern Medical University (NO: 2023B058), National Natural Science Foundation of China (NO: 82102156), Guangzhou Basic and Applied Basic Research Foundation (NO:2023A04J0453), and Guangdong Provincial Hospital of Chinese Medicie Zhaoyang Talent Project (NO: ZY2022YL27).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the Institutional Review Board of Nanfang Hospital of Southern Medical University (NFEC2023375), Guangdong Provincial Hospital of Chinese Medicine (BF2019-066), and Sun Yat-sen University Cancer Center (SL-B2022-751-02). Informed consent was waived since this was a retrospective study.
References
- 1. Sung H, Ferlay J, Siegel RL, et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clinicians 2021;71:209–49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2. Shitara K, Van Cutsem E, Bang Y-J, et al. Efficacy and safety of pembrolizumab or pembrolizumab plus chemotherapy vs chemotherapy alone for patients with first-line, advanced gastric cancer: the KEYNOTE-062 phase 3 randomized clinical trial. JAMA Oncol 2020;6:1571–80. 10.1001/jamaoncol.2020.3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janjigian YY, Shitara K, Moehler M, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (Checkmate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40. 10.1016/S0140-6736(21)00797-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Salas-Benito D, Pérez-Gracia JL, Ponz-Sarvisé M, et al. Paradigms on immunotherapy combinations with chemotherapy. Cancer Discov 2021;11:1353–67. 10.1158/2159-8290.CD-20-1312 [DOI] [PubMed] [Google Scholar]
- 5. Kono K, Nakajima S, Mimura K. Current status of immune checkpoint inhibitors for gastric cancer. Gastric Cancer 2020;23:565–78. 10.1007/s10120-020-01090-4 [DOI] [PubMed] [Google Scholar]
- 6. Kim ST, Cristescu R, Bass AJ, et al. Comprehensive molecular characterization of clinical responses to PD-1 inhibition in metastatic gastric cancer. Nat Med 2018;24:1449–58. 10.1038/s41591-018-0101-z [DOI] [PubMed] [Google Scholar]
- 7. McGrail DJ, Pilié PG, Rashid NU, et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 2021;32:661–72. 10.1016/j.annonc.2021.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Di Bartolomeo M, Morano F, Raimondi A, et al. Prognostic and predictive value of microsatellite instability, inflammatory reaction and PD-L1 in gastric cancer patients treated with either adjuvant 5-FU/LV or sequential FOLFIRI followed by cisplatin and docetaxel: a translational analysis from the ITACA-S trial. Oncologist 2020;25:e460–8. 10.1634/theoncologist.2019-0471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim JY, Kim WG, Kwon CH, et al. Differences in immune contextures among different molecular subtypes of gastric cancer and their prognostic impact. Gastric Cancer 2019;22:1164–75. 10.1007/s10120-019-00974-4 [DOI] [PubMed] [Google Scholar]
- 10. Ji Z, Peng Z, Gong J, et al. Hyperprogression after immunotherapy in patients with malignant tumors of digestive system. BMC Cancer 2019;19:705. 10.1186/s12885-019-5921-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang J, Xiu J, Farrell A, et al. Mutational analysis of microsatellite-stable gastrointestinal cancer with high tumour mutational burden: a retrospective cohort study. Lancet Oncol 2023;24:151–61. 10.1016/S1470-2045(22)00783-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakamura Y, Kawazoe A, Lordick F, et al. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: an emerging paradigm. Nat Rev Clin Oncol 2021;18:473–87. 10.1038/s41571-021-00492-2 [DOI] [PubMed] [Google Scholar]
- 13. Moutafi M, Rimm DL. Putting the Microenvironment into the immunotherapy companion diagnostic. Clin Cancer Res 2021;27:3812–4. 10.1158/1078-0432.CCR-21-1238 [DOI] [PubMed] [Google Scholar]
- 14. Jiang Y, Zhou K, Sun Z, et al. Non-invasive tumor microenvironment evaluation and treatment response prediction in gastric cancer using deep learning radiomics. Cell Rep Med 2023;4:101146. 10.1016/j.xcrm.2023.101146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang W, Jiang Y, Xiong W, et al. Noninvasive imaging of the tumor immune microenvironment correlates with response to immunotherapy in gastric cancer. Nat Commun 2022;13:5095. 10.1038/s41467-022-32816-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zeng D, Wu J, Luo H, et al. Tumor microenvironment evaluation promotes precise checkpoint Immunotherapy of advanced gastric cancer. J Immunother Cancer 2021;9:e002467. 10.1136/jitc-2021-002467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van der Laak J, Litjens G, Ciompi F. Deep learning in histopathology: the path to the clinic. Nat Med 2021;27:775–84. 10.1038/s41591-021-01343-4 [DOI] [PubMed] [Google Scholar]
- 18. Bera K, Schalper KA, Rimm DL, et al. Artificial intelligence in digital pathology — new tools for diagnosis and precision oncology. Nat Rev Clin Oncol 2019;16:703–15. 10.1038/s41571-019-0252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie J, Luo X, Deng X, et al. Advances in artificial intelligence to predict cancer immunotherapy efficacy. Front Immunol 2023;13:1076883. 10.3389/fimmu.2022.1076883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med 2019;25:1054–6. 10.1038/s41591-019-0462-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hinata M, Ushiku T. Detecting immunotherapy-sensitive subtype in gastric cancer using histologic image-based deep learning. Sci Rep 2021;11:22636. 10.1038/s41598-021-02168-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y, Sun Z, Wan L, et al. Tumor microenvironment characterization for assessment of recurrence and survival outcome in gastric cancer to predict chemotherapy and immunotherapy response. Front Immunol 2022;13:890922. 10.3389/fimmu.2022.890922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Y, Jia K, Sun Y, et al. Predicting response to immunotherapy in gastric cancer via multi-dimensional analyses of the tumour immune microenvironment. Nat Commun 2022;13:4851. 10.1038/s41467-022-32570-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwartz LH, Litière S, de Vries E, et al. RECIST 1.1-update and clarification: from the RECIST committee. Eur J Cancer 2016;62:132–7. 10.1016/j.ejca.2016.03.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schwartz LH, Seymour L, Litière S, et al. RECIST 1.1 - Standardisation and disease-specific adaptations: perspectives from the RECIST working group. Eur J Cancer 2016;62:138–45. 10.1016/j.ejca.2016.03.082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu MY, Williamson DFK, Chen TY, et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat Biomed Eng 2021;5:555–70. 10.1038/s41551-020-00682-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Graham S, Vu QD, Raza SEA, et al. Hover-Net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med Image Anal 2019;58:101563. 10.1016/j.media.2019.101563 [DOI] [PubMed] [Google Scholar]
- 28. Weisbart E, Tromans-Coia C, Diaz-Rohrer B, et al. Cellprofiler Plugins – an easy image analysis platform integration for containers and python tools. J Microsc 2023. 10.1111/jmi.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gamper J, et al. Pannuke dataset extension, insights and baselines. arXiv 2020. [Google Scholar]
- 30. Gómez-Ramírez J, Ávila-Villanueva M, Fernández-Blázquez MÁ. Selecting the most important self-assessed features for predicting conversion to mild cognitive impairment with random Forest and Permutation-based methods. Sci Rep 2020;10:20630. 10.1038/s41598-020-77296-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lundberg S, Lee SI. A unified approach to interpreting model predictions. Nips, 2017. [Google Scholar]
- 32. Shokouhmand A, Aranoff ND, Driggin E, et al. Efficient detection of aortic stenosis using morphological characteristics of cardiomechanical signals and heart rate variability parameters. Sci Rep 2021;11:23817. 10.1038/s41598-021-03441-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mootha VK, Lindgren CM, Eriksson K-F, et al. PGC-1Α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–73. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 35. Kanehisa M, Furumichi M, Sato Y, et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res 2023;51:D587–92. 10.1093/nar/gkac963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2024-008927supp001.pdf (7.4MB, pdf)
Data Availability Statement
Data are available upon reasonable request.