Abstract
Objective. Deep learning reconstruction (DLR) algorithms exhibit object-dependent resolution and noise performance. Thus, traditional geometric CT phantoms cannot fully capture the clinical imaging performance of DLR. This study uses a patient-derived 3D-printed PixelPrint lung phantom to evaluate a commercial DLR algorithm across a wide range of radiation dose levels. Method. The lung phantom used in this study is based on a patient chest CT scan containing ground glass opacities and was fabricated using PixelPrint 3D-printing technology. The phantom was placed inside two different size extension rings to mimic a small- and medium-sized patient and was scanned on a conventional CT scanner at exposures between 0.5 and 20 mGy. Each scan was reconstructed using filtered back projection (FBP), iterative reconstruction, and DLR at five levels of denoising. Image noise, contrast to noise ratio (CNR), root mean squared error, structural similarity index (SSIM), and multi-scale SSIM (MS SSIM) were calculated for each image. Results. DLR demonstrated superior performance compared to FBP and iterative reconstruction for all measured metrics in both phantom sizes, with better performance for more aggressive denoising levels. DLR was estimated to reduce dose by 25%–83% in the small phantom and by 50%–83% in the medium phantom without decreasing image quality for any of the metrics measured in this study. These dose reduction estimates are more conservative compared to the estimates obtained when only considering noise and CNR. Conclusion. DLR has the capability of producing diagnostic image quality at up to 83% lower radiation dose, which can improve the clinical utility and viability of lower dose CT scans. Furthermore, the PixelPrint phantom used in this study offers an improved testing environment with more realistic tissue structures compared to traditional CT phantoms, allowing for structure-based image quality evaluation beyond noise and contrast-based assessments.
Keywords: deep learning reconstruction, CT imaging phantoms, dose reduction, image quality assessment, 3D-printing, 3D-printed phantom
1. Introduction
Over the last few years, there has been substantial interest in the development and clinical use of deep learning reconstruction (DLR) algorithms for improving computed tomography (CT) image quality and reducing radiation dose (Koetzier et al 2023). For decades, filtered back projection (FBP) was the dominant reconstruction algorithm due to its numerical stability and fast computation time (Willemink and Noël 2019). However, at lower doses, FBP image quality drops and image noise increases dramatically (Koetzier et al 2023). With continued interest in dose reduction (Brenner and Hall 2007), especially in pediatric populations (Miglioretti et al 2013, Nagayama et al 2021, Sun et al 2021, Son et al 2022), clinical CT imaging has begun moving away from FBP toward newer solutions such as iterative reconstruction (IR) which preserves image quality at lower doses. Various forms of IR have demonstrated significant potential to minimize noise and thus to reduce dose compared to FBP (Willemink et al 2013). However, limitations in IR including unnatural noise texture (Willemink et al 2013, Philips Healthcare 2024) and extended reconstruction time (Willemink et al 2013, Koetzier et al 2023) have resulted in a push for further innovation in reconstruction solutions.
DLR for CT has emerged as a novel solution for improving image quality and reconstruction time while preserving FBP-like noise textures. These algorithms utilize artificial neural networks such as convolutional neural networks (CNNs) (Kang et al 2017, Chen et al 2017) or generative adversarial networks (GANs) (Wolterink et al 2017) which are trained to produce optimized output images from lower dose input data. DLR frameworks can be broadly categorized as either indirect, where a deep learning network is used alongside FBP or IR, or direct, in which the network directly converts sinogram data to image data without FBP or IR (Koetzier et al 2023). Many different implementations of DLR have been proposed in academic research (Wu et al 2017, Yang et al 2018, Bao et al 2019) as well as introduced clinically by CT vendors (Hsieh et al 2019, Boedeker 2021, Philips Healthcare 2024).
With the rise of commercially available DLR algorithms, there has been an increase in studies evaluating DLR. Multiple patient and phantom studies have demonstrated that DLR can improve image quality at low doses through enhanced lesion detectability and reduced noise (Akagi et al 2019, Nakamura et al 2019, Nagayama et al 2021, Sun et al 2021, Greffier et al 2022a, Miyata et al 2022, Park et al 2022, Greffier et al 2022b, Mikayama et al 2022, Son et al 2022, Greffier et al 2023a). These studies utilize quantitative metrics such as signal-to-noise ratio (SNR), contrast-to-noise ratio (CNR), noise, detectability index (d’), and noise power spectrum (NPS). In addition, qualitative scores for various aspects of subjective image quality have been obtained via reader studies by experienced radiologists. The literature has shown that various implementations of DLR can reduce dose by about 30%–71% compared to hybrid iterative reconstruction (HIR) methods while preserving diagnostic image quality (Koetzier et al 2023).
While there are many promising results regarding DLR performance, there are several limitations to current studies. First, due to the nonlinear nature of DLR, images reconstructed with DLR demonstrate object-dependent resolution and noise (Li et al 2022, Solomon et al 2020, Higaki et al 2020, Greffier et al 2023b). Traditional CT phantoms used in DLR evaluation studies are often composed of simple geometric shapes which are not designed to represent realistic tissue structures (Greffier et al 2022a, 2022b, Mikayama et al 2022). As a result, general image quality metrics such as noise and CNR measured on traditional CT phantoms cannot fully capture the clinical imaging performance of DLR (Samei et al 2019). Second, clinical imaging studies using patient data are often limited by sample size and restricted by radiation dose exposure concerns (Akagi et al 2019, Greffier et al 2023a, Lyu et al 2023), which limit the acceptable dose range as well as the number of times a patient can be scanned. Furthermore, patient scans do not have reliable ground truth images for comparison and thus cannot be used to assess the structural accuracy of a reconstructed image. A clinical scenario in which structural accuracy is important is lung CT imaging with ground glass opacity (GGO) findings. Subtle differences in shape (round versus polygonal, with or without radial growths) and texture (presence or absence of solid densities) in a GGO can lead to differences in image interpretation and clinical decision making (Infante et al 2009). Because of this, the accurate reconstruction of such structures and details is critical to ensuring the highest quality of patient care. Previous studies have investigated the general image quality and detectability of reconstructed structures but have not directly addressed the question of reconstruction accuracy of complex structures and textures such as those found in GGOs. Image reconstruction for clinical scenarios such as this require evaluation beyond what is currently available with phantom and patient studies.
This study proposes to use a patient-derived PixelPrint (Mei et al 2022a, Shapira et al 2023, 2022) phantom as a novel solution to address the current limitations in the evaluation of DLR performance. PixelPrint is a technology which produces 3D-printed patient-based phantoms which demonstrate highly detailed tissue structures, realistic textures, and accurate attenuation profiles. PixelPrint software converts 3D CT images into geometric code (g-code) instructions for fused filament fabrication (FFF) 3D printers by taking advantage of the partial volume effect to produce desired Hounsfield Unit (HU) values (Shapira et al 2022). Previous studies have demonstrated a high degree of HU and geometric similarity between scans of PixelPrint phantoms and their reference patient scans (Mei et al 2022b). Furthermore, reader studies demonstrated that there was no clinically significant difference in image quality assessment between reading a phantom lung image and reading a patient lung image (Shapira et al 2023). Compared to standard geometric CT imaging phantoms, PixelPrint phantoms demonstrate realistic tissue morphology and thus can more fully capture the clinical imaging performance of DLR. Compared to patient data, PixelPrint phantoms allow for more flexibility in radiation dose usage and have more accurate ground truth images with which to assess the structural precision of DLR images.
This study utilized a 3D-printed PixelPrint lung phantom to evaluate the clinical imaging performance of a commercial DLR algorithm, precise Image (PI) (Philips Healthcare, Cleveland, OH, USA) (Philips Healthcare 2024) compared to FBP and IR, with particular focus on the question of structural accuracy of reconstructed anatomy. PI is an example of a direct DLR algorithm and utilizes simulated low dose sinogram data for CNN training (Koetzier et al 2023, Philips Healthcare 2024). The PixelPrint phantom was scanned with a large range of radiation doses to investigate the dose reduction potential of each algorithm. As image quality is affected by patient size (i.e. CT images of large patients tend to have higher noise and reduced image quality compared to smaller patients), two different phantom sizes were included in the performance assessment to examine the generalizability of results to different patient sizes.
2. Methods
2.1. Patient CT scan selection
The institutional review board (IRB) at the University of Pennsylvania approved this retrospective study (IRB Protocol #853697). A single patient chest CT scan containing multiple subsolid GGOs representing metastatic lesions was retrospectively selected as the model for the 3D-printed phantom in this study (figure 1). The image was taken from the Hospital of the University of Pennsylvania PACS system and anonymized. GGO lesions are an example of highly detailed lung structures in which accurate reconstruction of textures and shapes is clinically important. The scan and reconstruction parameters of the patient CT scan are listed in table 1.
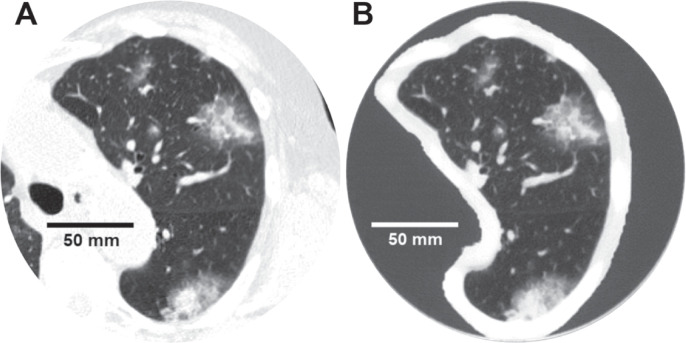
Figure 1.
(A) Image of the left lung from the patient chest CT scan which was used to generate the phantom. (B) CT scan of the printed phantom, scanned at 20 mGy and 120 kVp and reconstructed using PI-Sharp. WL: −500, WW: 1100 HU.
Table 1.
Scan and reconstruction parameters of the patient CT image.
| Scanner model | Philips spectral CT 7500 |
|---|---|
| Scan mode | Helical |
| Tube voltage | 120 kVp |
| Tube current | 173 mA |
| Rotation time | 0.4 s |
| Helical pitch | 1.15 |
| Exposure | 60 mAs |
| CTDIvol | 4.7 mGy |
| Collimation | 128 × 0.625 mm |
| Slice thickness | 1 mm |
| Slice increment | 1 mm |
| Reconstruction filter | YA |
| Reconstructed field of view | 368 × 368 mm2 |
| Matrix size | 512 × 512 pixel2 |
| Pixel spacing | 0.7188 mm |
2.2. Phantom fabrication
The phantom was fabricated using PixelPrint technology (Shapira et al 2022, 2023) to produce a realistic patient-specific lung CT phantom. The entire phantom was 3D-printed as one piece using polylactic acid (PLA) filament on an FFF printer (Lulzbot TAZ Sidekick with M175 v2 tool head, Fargo Additive Manufacturing Equipment 3D, LLC Fargo, ND, USA). The phantom was designed as a 20 cm diameter cylinder containing the segmented left lung positioned in the center of the cylinder. A 4 cm scan length containing a large (4.5 × 3.2 cm2) GGO was selected. The left lung was segmented using an open-source automated U-net lung segmentation model (Hofmanninger et al 2020). A 1 cm border of tissue surrounding the lung including parts of the ribs, thoracic muscles, and mediastinum was also included in the phantom. The regions inside of the segmented lung and border were printed using PixelPrint technology to modulate density and accurately reproduce the HU profiles of the patient image, within the HU range attainable with PLA (−867 to 115 HU) (Mei et al 2022b). Regions of the cylinder outside of the border were printed with a constant infill ratio of 15% (corresponding to ∼ −800 HU).
2.3. Image acquisition and reconstruction
The phantom was scanned with a default high resolution chest imaging protocol on a conventional CT scanner (Incisive CT, Philips Healthcare, Cleveland, OH, USA). Multiple scans were acquired with varying radiation dose levels ranging from 0.5 to 20 mGy. Scans were repeated three times at each dose level and each scan was reconstructed using FBP, an iterative reconstruction algorithm (iDose (Miglioretti et al 2013)) at a single noise level (Level 3), and DLR (PI) at five levels with increasingly aggressive noise reduction (Sharper, Sharp, Standard, Smooth, Smoother) (table 2). Additional scan and reconstructions parameters common to all scans are listed in table 3.
Table 2.
Varying radiation dose levels used for phantom scanning and the different methods used for reconstruction.
| Exposure [mAs] | CTDIvol [mGy] | Reconstruction algorithms |
|---|---|---|
| 250 a | 20 a | FBP, YC Filter a |
| 235 | 19 | |
| 185 | 15 | iDose4 Level 3, YC filter |
| 148 b | 12 b | Precise Image (PI), Lung Definition, Sharper/Sharp/Standard/Smooth/Smoother |
| 111 c | 9 c | |
| 74 | 6 | |
| 49 | 4 | |
| 25 d | 2 d | |
| 12 | 1 | |
| 6 | 0.5 |
Dose and reconstruction parameters used for ground truth image.
Diagnostic reference level for a 29–33 cm water-equivalent diameter patient (Radiology ACo 2018).
Achievable dose level for a 29–33 cm water-equivalent diameter patient (Radiology ACo 2018).
Lung cancer screening level (Kazerooni et al 2014).
Table 3.
CT scan and reconstruction parameters for the phantom scans.
| Scanner model | Philips incisive CT |
|---|---|
| Scan mode | Helical |
| Tube voltage | 120 kVp |
| Rotation time | 0.5 s |
| Helical pitch | 1 |
| Collimation | 64 × 0.625 mm |
| Slice thickness | 1 mm |
| Slice increment | 0.5 mm |
| Reconstructed field of view | 350 × 350 mm2 |
| Matrix size | 512 × 512 pixel2 |
| Pixel spacing | 0.6836 mm |
2.4. Extension rings
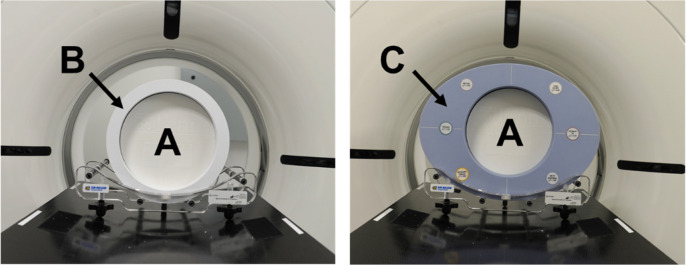
Patient size has a significant impact on the noise and image quality of CT images and, thus, can affect the performance of DLR. To mimic different patient sizes, the lung phantom was placed inside two different size extension rings during scanning (figure 2). A custom 25 × 25 cm2 water-equivalent extension ring with a 20 cm cylindrical bore was 3D printed using PLA filament. The lung phantom was placed in this custom extension ring to represent a small-sized patient (small phantom), resulting in a total water equivalent diameter (D w) of about 19 cm. To represent a medium-sized patient, the lung phantom was placed in the 20 cm bore of a 30 × 40 cm2 multi-energy CT phantom (MECT) (Sun Nuclear, WI, USA) extension ring (medium phantom). The D w of the phantom plus MECT extension ring was about 30 cm. The small and medium phantom D w’s are representative of patient D w’s of an average pediatric (McCollough et al 2022) and adult (Kanal et al 2017) chest, respectively. The scan and reconstruction parameters outlined in tables 2 and 3 were repeated for each phantom size.
Figure 2.
The PixelPrint lung phantom (A) placed inside a 25 × 25 cm2 3D printed extension ring (B) to represent a small-sized patient and placed inside of a 30 × 40 cm2 MECT extension ring (C) to represent a medium-sized patient.
2.5. Image analysis
Image noise and CNR were calculated for each reconstruction and dose combination. The image noise was calculated for a 2 × 2 cm2 region of interest (ROI) across 10 consecutive slices in a homogeneous region of the phantom background lung parenchyma. The CNR was calculated between the GGO lesion and the background lung parenchyma where the GGO ROI was a 2 × 2 cm2 ROI over 14 consecutive slices inside of the GGO lesion. The equations used for noise and CNR calculations were:
where is the standard deviation of HU values in the background lung ROI, is the mean HU in the GGO ROI, and is the mean HU in the background lung ROI.
In addition to these general image quality metrics, the structural accuracy of the reconstructed images was evaluated using the image similarity metrics: root mean squared error (RMSE), structural similarity index measure (SSIM) (Wang et al 2004), and multi-scale SSIM (MS SSIM) (Wang et al 2003), using the highest dose (20 mGy) FBP image as the ground truth image. RMSE provides a direct intensity-based comparison between the reconstructed image and the ground truth, while SSIM takes luminance, contrast, and structural features into consideration. MS SSIM further expands on SSIM by generalizing the SSIM algorithm to incorporate image information at a variety of different resolution scales. The use of several similarity metrics helps ensure that the results are robust to different methods of assessing how closely the reconstructed images match the ground truth. All similarity metrics were measured in a 13.5 × 13.5 cm2 ROI across 50 consecutive slices within the 3D printed phantom. The RMSE and SSIM were calculated for each image using the open source Python package skimage.metrics (van der Walt et al 2014), and the MS SSIM was calculated using the open source python library pytorch-msssim (Pytorch-msssim 2023). All ROIs used for these calculations are shown in figure 3, and the same ROIs were used for each of the reconstructed images. Since the 20 mGy scans were used as the ground truth, they were excluded from the sample for all image metric calculations.
Figure 3.
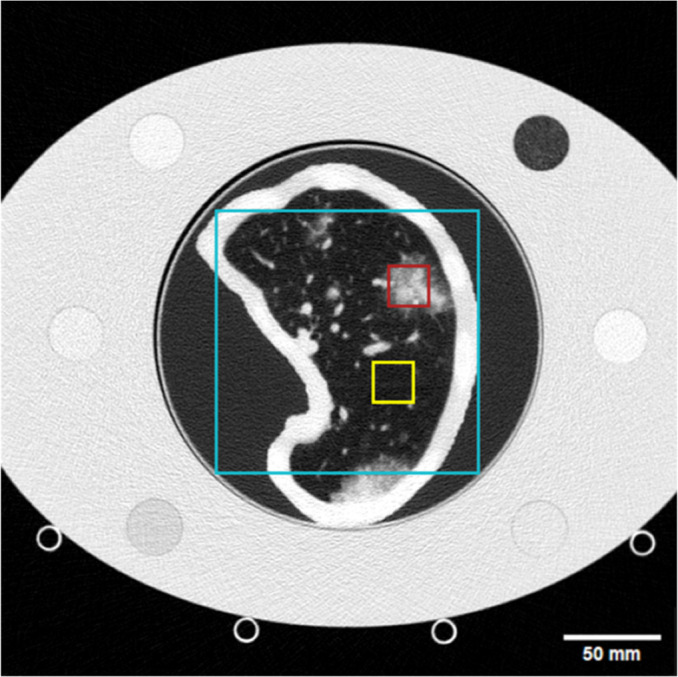
CT scan of the phantom placed inside of the medium size extension ring with marked ROIs used for image quality metric measurements. The yellow box encompasses the background lung ROI used for image noise and CNR calculations, the red box shows the GGO ROI used to calculate CNR, and the cyan box represents the ROI used for RMSE, SSIM, and MS SSIM measurements. WL: −450, WW: 1100 HU.
2.6. Statistical analysis
The performance of each dose and reconstruction combination was evaluated in comparison to the performance of the FBP images of scans taken at 12 mGy, which is the diagnostic reference level for 29–33 cm water equivalent diameter patients (Radiology ACo 2018). A two-sample, one-tailed t-test was performed for each image metric using the open-source python package Scipy statistical functions (Virtanen et al 2020). Effects were considered statistically significant where which after applying the Bonferroni post hoc correction results in The potential dose reduction of each reconstruction algorithm was then determined by finding the lowest dose measured at which there was no statistically significant decrease in image quality from the reference for any measured metric.
3. Results
3.1. Comparison of reconstruction algorithms
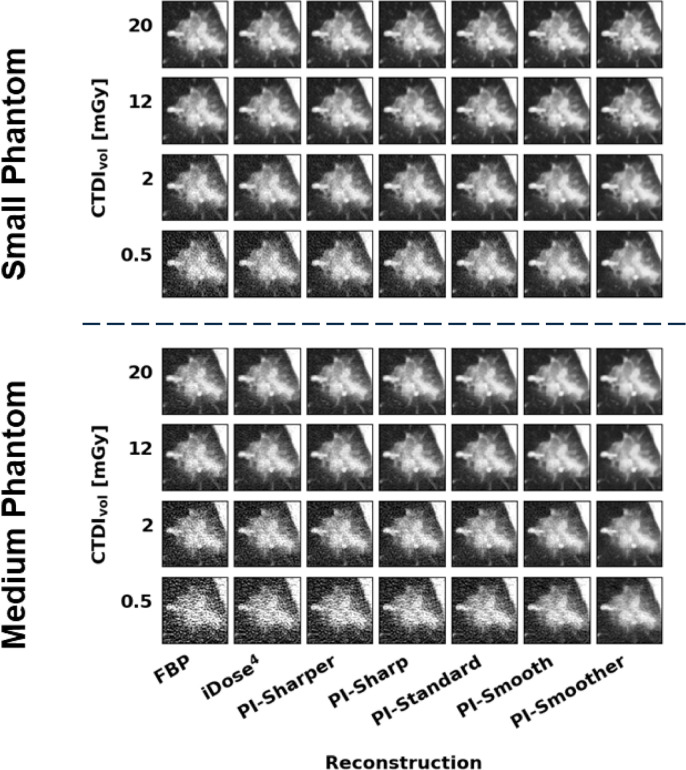
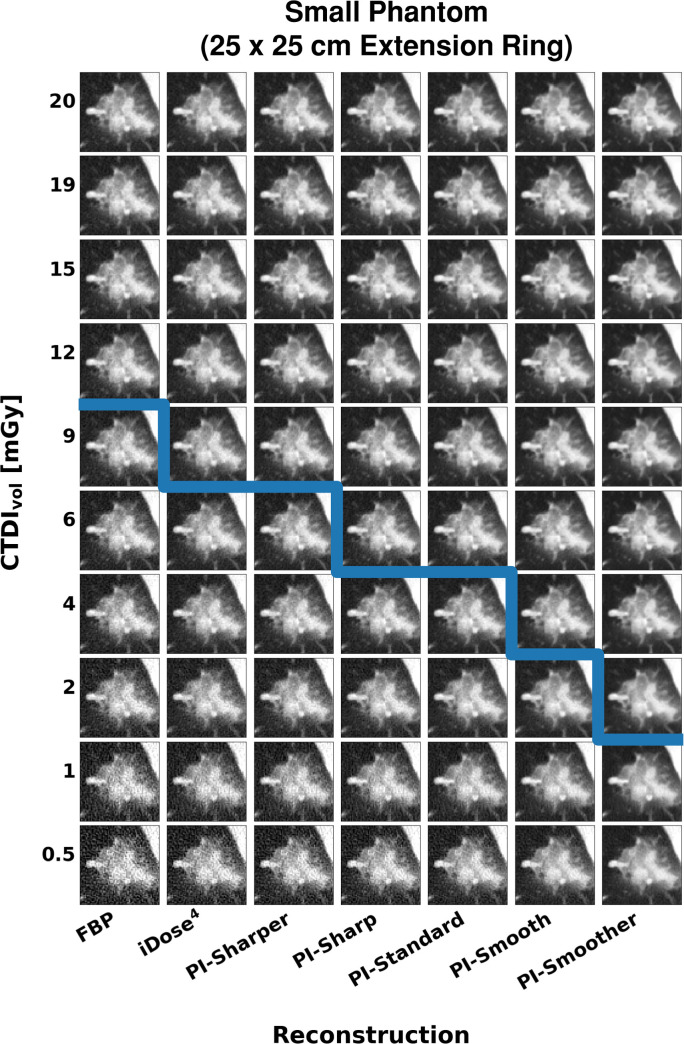
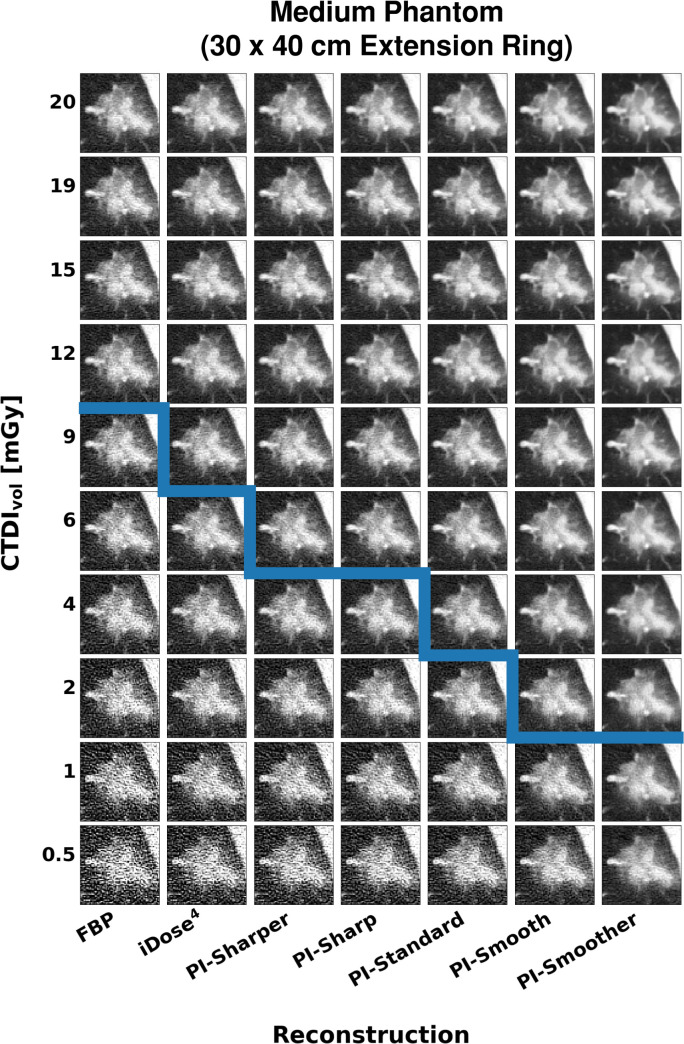
PI demonstrates superior performance compared to both FBP and iterative reconstruction for all measured metrics in both phantom sizes. The image quality of FBP images is noticeably degraded by noise at lower doses while low dose scans reconstructed with iDose (Miglioretti et al 2013) and PI have image quality which more closely resemble the highest dose FBP image. This effect is demonstrated visually in figure 4 and confirmed quantitatively by the measured metrics. The results of each metric are represented in figures 5 and 6, and tables A1–A10 show the t-test statistics.
Figure 4.
Images of the GGO lesion taken from the small phantom (top) and medium phantom (bottom) at several dose and reconstruction combinations. Figures A1 and A2 in the appendix show the GGO lesion images from all the dose levels collected. WL: −500, WW: 1000 HU.
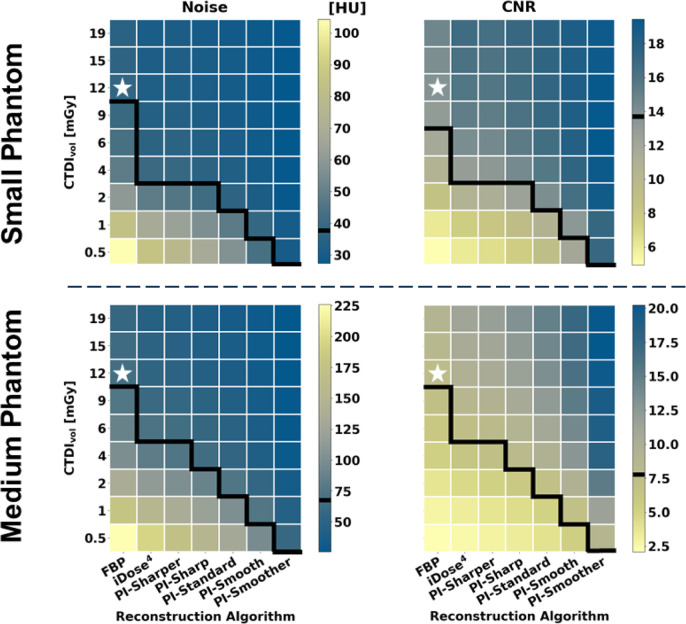
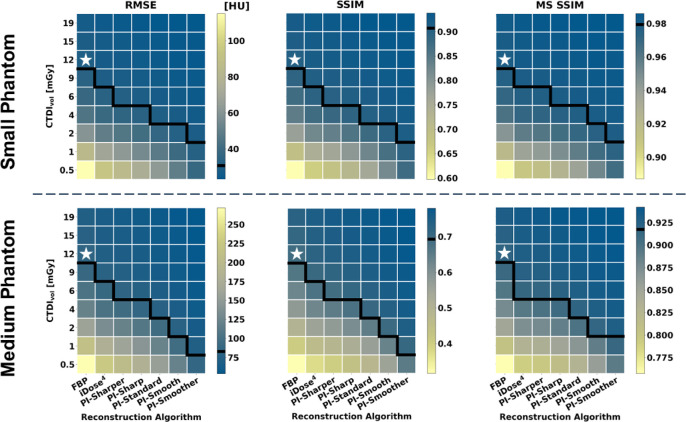
Figure 5.
Heatmaps displaying results of the image metric calculations for Noise (left) and CNR (right) from both the small phantom (top) and medium phantom (bottom). A white star is used to designate the dose and reconstruction combination used as the reference for statistical comparison for each metric. The value for this reference group is indicated on the color bars by a black line. The black lines on the heatmaps separate values that are statistically better than or equivalent to the reference (above the line) from those that are statistically worse than the reference (below the line).
Figure 6.
Heatmaps displaying results of the image metric calculations for RMSE (left), SSIM (middle), and MS SSIM (right) from both the small phantom (top) and medium phantom (bottom). The white stars and black lines in this figure have the same function as in figure 5.
All metrics show that iDose4 is capable of dose reduction compared to FBP and that PI shows further dose reduction compared to iDose4. Furthermore, more aggressive noise reduction, i.e. smoother levels of PI, showed improved performance over less aggressive noise reduction, i.e. sharper levels of PI. When only considering noise and CNR, the different levels of PI achieved dose reduction capabilities between 67%–96% for the small phantom and between 50%–96% for the medium phantom, respectively (figure 5). However, the results of the image similarity metrics RMSE, SSIM, and MSSIM show more conservative dose reduction estimates compared to the estimates obtained from noise and CNR alone. When considering the image similarity metric results, PI demonstrated lower dose reduction capabilities of 25%–83% in the small phantom and 50%–83% in the medium phantom (figure 6). Thus, these image similarity metrics provide additional information about the structural accuracy of the reconstructed images that is not captured by general image quality metrics like noise and CNR.
3.2. Phantom size effects
The image quality of the small phantom reconstructions showed an average of approximately 40% improvement across all metrics compared to the matched doses and reconstructions of the medium phantom. Analysis of noise and CNR suggest that there is a slight increase in dose reduction capabilities of PI in the small phantom (67%–96%) compared to the medium phantom (50%–96%). However, the image similarity metrics show the opposite trend, with slightly lower dose reduction capabilities in the small phantom (25%–83%) compared with the medium phantom (50%–83%). Overall, the results from the two phantom sizes showed similar trends in dose reduction.
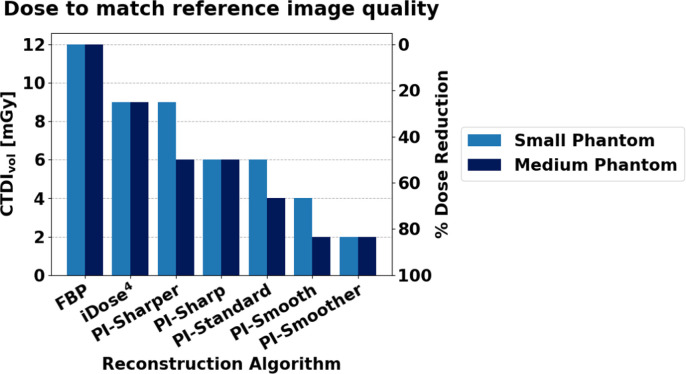
3.3. Summarized potential dose reduction capabilities
The overall dose reduction of each reconstruction algorithm compared to FBP was determined using the minimum dose at which all metrics matched or exceeded the reference performance. These minimum doses are summarized in figure 7 with corresponding dose reduction percentages indicated on the right axis. For both the small phantom and medium phantom, PI demonstrated dose reduction capabilities up to 83% for the highest level of denoising (PI-Smoother).
Figure 7.
The minimum doses (left axis) required to match or exceed all image quality metrics of the reference images for each reconstruction algorithm, along with the corresponding percent dose reduction (right axis).
4. Discussion
This study examined the clinical imaging performance of a DLR algorithm, PI, compared to FBP and IR by utilizing a custom-made patient derived PixelPrint lung phantom. The results show that PI is capable of dose reduction for this clinical scenario between 25% and 83% compared to FBP depending on the denoising level of the algorithm and phantom size. This suggests that in some cases PI can produce diagnostic level image quality even for CT scans acquired at lung cancer screening doses of <3 mGy (Kazerooni et al 2014). This could mean more effective lung cancer screening and/or reduced radiation burden. Furthermore, these dose reductions are achieved with more natural noise textures compared to IR. The unnatural or ‘plastic’ looking textures in IR images are often attributed to a leftward shift of the NPS curve (Ehman et al 2014, Szczykutowicz et al 2022). DLR algorithms have been reported to offer varying degrees of improvement in noise texture depending on the specific algorithm assessed and the denoising level used (Szczykutowicz et al 2022). A study by Greffier et al that looked specifically at the NPS of Standard through Smoother levels of PI showed that the average spatial frequency and peak spatial frequencies of PI were closer to those of FBP compared to iDose4, suggesting more favorable noise textures (Greffier et al 2023b). These results are complementary to the results presented in the current study.
Despite the differences in study design, this study demonstrated similar trends in noise reduction, image quality improvement, and dose reduction capabilities of PI with previous literature evaluating PI. Greffier et al investigated the use of PI for evaluating liver metastases in a patient study (Greffier et al 2023a) which demonstrated that more aggressive levels of denoising for PI (Smooth and Smoother) resulted in better performance in the lowest dose scans, as shown in this study. In a separate study examining the use of PI in chest imaging, Greffier et al reported dose reductions of 58% and 83% compared to iDose4 Level 4 for PI-Smooth and PI-Smoother respectively, based on task-based image quality assessment of simulated ground glass opacifications in a standard, geometric phantom (Greffier et al 2022a). Similarly, our study results show that for a medium-sized phantom, if the results from MS SSIM are excluded, PI-Smooth and PI-Smoother have a 58% and 88% dose reduction potential respectively compared to iDose4 level 3. However, when MS SSIM is included the dose reduction potential becomes more conservative. The correlation between our results and previously published literature help to validate the use of PixelPrint phantoms in the evaluation of DLR.
Studies evaluating other DLR algorithms have also demonstrated reduced noise and improved lesion detectability in DLR compared to IR or FBP (Son et al 2022, Miyata et al 2022, Park et al 2022, Greffier et al 2022b, Koetzier et al 2023). The exact percentage of dose reduction reported was heavily dependent upon many factors including the clinical scenario, the reference dose, reference reconstruction algorithm, DLR algorithm and denoising strength, and specific metrics evaluated. Overall, the results from studies of several other reconstruction algorithms included a wide range of dose reduction estimates between 30% and 85%, which is similar to the results for PI.
Furthermore, this study demonstrates the additional information which evaluations using PixelPrint phantoms can provide compared to patients and standard phantoms. Unlike in patient studies, an evaluation using PixelPrint phantoms allows for the comparison of a wide range of doses beyond the dose range typically acceptable in clinical practice. Thus, ground truth data can be obtained by acquiring a higher than standard radiation dose. Additionally, the dose reduction capability of DLR can be probed more precisely by repeatedly acquiring increasingly lower dose data and comparing the resultant images directly. The ability to acquire images without any patient motion between scans facilitates comparison between images via similarity metrics such as RMSE.
Compared to traditional CT phantoms, the presence of clinically relevant structures and details in PixelPrint phantoms is advantageous because it enables comparison of reconstruction accuracy for complex structures. Reconstruction accuracy can be evaluated by using image similarity metrics such as SSIM to compare a reconstructed image to the selected ground truth. The inclusion of these image similarity metrics resulted in more conservative dose reduction estimates compared to the results obtained using only general image quality metrics such as noise and CNR. This may be because although the DLR algorithm can essentially tune noise levels to almost any desired amount, some information from detailed structures may not be recoverable. As a result, analyzes on non-clinical structures such as those found in traditional CT phantoms cannot adequately capture these algorithms’ diagnostic imaging performance. PixelPrint phantoms can also provide additional information about patient size dependency in DLR dose reduction. A previous patient study using general image quality metrics showed that iDose4 achieved higher dose reduction in smaller patients versus larger patients (Arapakis et al 2014). In the present study, only considering the noise and CNR measurements results in the same trend while the inclusion of image similarity metrics results in a reversal of the trend such that the small phantom size has slightly reduced dose reduction potential. Finally, it has been reported that a possible concern with DLR is that if certain lesions are not well represented in training sets, these lesions may not be reconstructed accurately in DLR images (Nagayama et al 2021). This is not something that can be tested with standard geometric phantoms but can be easily investigated using different PixelPrint phantoms with various lesions and known ground truth images. These findings suggest that the use of PixelPrint phantoms in conjunction with image similarity metrics provides valuable information which is not available from standard phantoms or patient studies alone for determining dose reduction capability.
The present study has a few limitations. First, PI was compared to only one denoising level of iDose4, the default level for lung imaging. To form a more robust understanding of the improvement that PI affords over IR, it would be valuable to compare PI to more denoising levels of iDose (Miglioretti et al 2013). Similarly, the same phantom could be used to evaluate and compare multiple DLR algorithms including both commercial and open-source algorithms. Second, this study only utilized one phantom and thus only one example of patient anatomy. Future studies involving more phantoms from different patients could improve our insights into the behavior of PI in different clinical scenarios and disease states. This could be especially useful in rarer or more unique clinical cases where patient data is limited. Third, this study does not include a reader study with subjective image quality scores. However, there are other existing studies that include a reader study (Greffier et al 2022a, 2023b, Philips Healthcare 2024) and those results show good alignment with the results of the present study. Fourth, the HU range of the phantom used in this study is limited to between −867 and 115 HU for the PLA material used. Other printing materials are being investigated in order to increase the HU range of PixelPrint phantoms (Mei et al 2022a). Finally, the raw projection data corresponding to the patient images used to create the PixelPrint phantom was unavailable, preventing a direct comparison between PI performance on phantom data and its performance on the source patient data.
5. Conclusion
This study demonstrates the dose reduction capabilities of a DLR algorithm, Precise Image, in the context of lung imaging with GGOs. For this clinical scenario, PI has the capability of producing diagnostic image quality at up to 83% lower radiation dose, even surpassing the dose reduction capabilities of iterative reconstruction. These results are consistent with existing literature evaluating DLR. Images reconstructed using PI demonstrate not only improved noise and contrast compared to FBP and iterative reconstruction, but also improved structural accuracy of lung features such as GGO lesions. The use of PI can improve the clinical utility and viability of lower dose CT scans, ultimately improving patient care while reducing radiation exposure.
The PixelPrint phantom used in this study offers an improved testing environment with more realistic tissue structures and attenuation profiles compared to other CT phantoms. This is particularly important for the evaluation of nonlinear reconstruction algorithms such as DLR. Thus, PixelPrint phantoms can elevate the clinical relevance of phantom evaluations of new and emerging CT technologies, which will lead to more rapid translation of these technologies into medical practice.
Acknowledgments
This work was partially funded by the National Institutes of Health through the following Grants: R01EB035092, R01EB030494, R01EB031592, R01HL166236, and R01CA249538. PN has received a hardware grant and research grant funding from Philips Healthcare. SH, AP, and EW are employees of Philips Healthcare. The other authors have no relevant conflicts of interest to disclose.
Author contributions.
Conception: JI, SH, PN, Design: JI, SH, KM, AP, LR, PN, Data acquisition: SH, EW, Analysis: JI, Drafting: JI, Revising: JI, SH, KM, AP, OS, LL, GG, PN.
Appendix.
Figure A1.
Images of the GGO lesion taken from the small phantom at each dose and reconstruction combination. WL: −500, WW: 1000 HU. Images above the blue line match or exceed the diagnostic reference level (12 mGy) image quality for all measured metrics.
Figure A2.
Images of the GGO lesion taken from the medium phantom at each dose and reconstruction combination. WL: −500, WW: 1000 HU. Images above the blue line match or exceed the diagnostic reference level (12 mGy) image quality for all measured metrics.
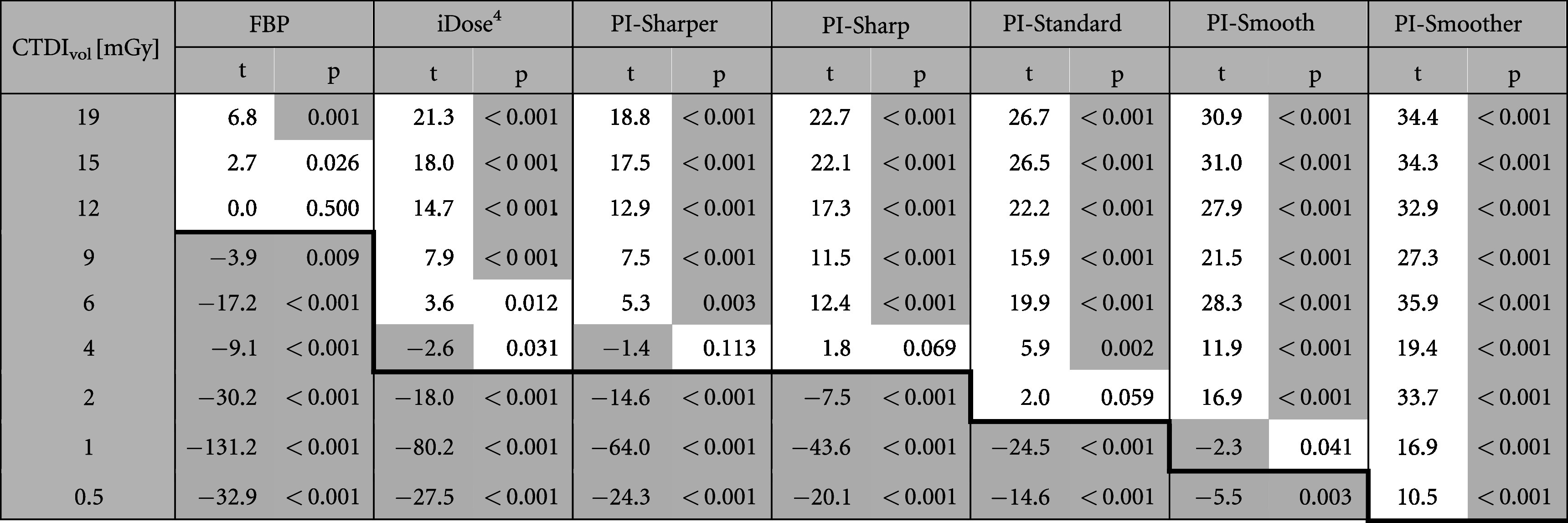
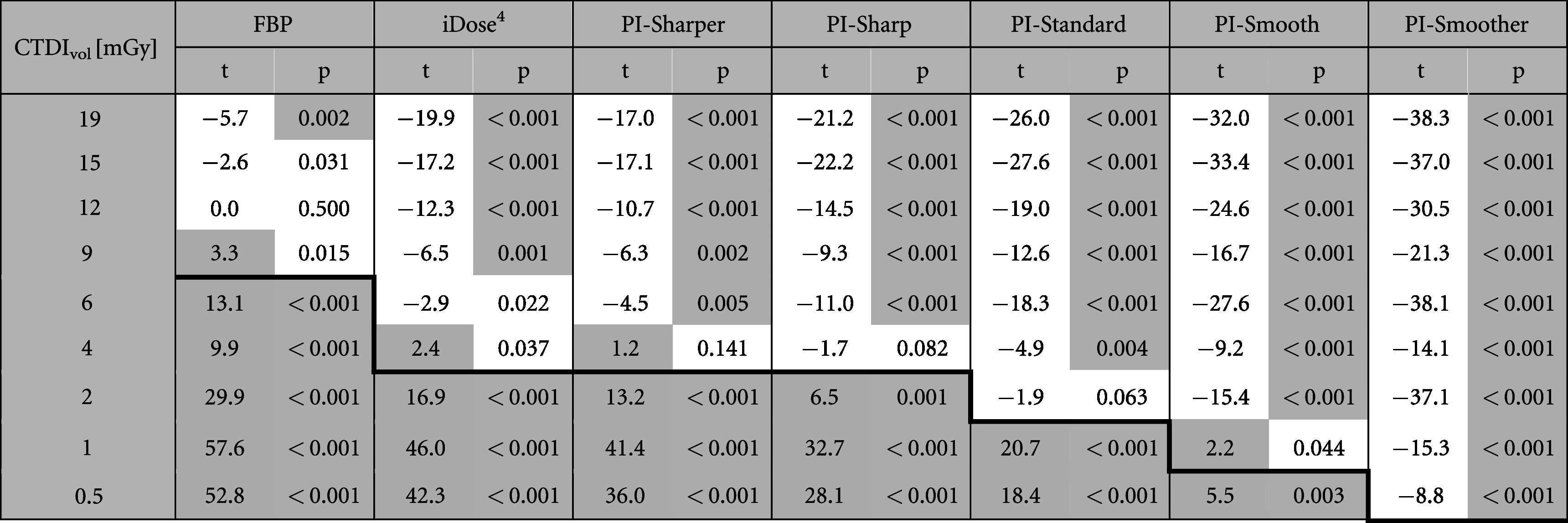
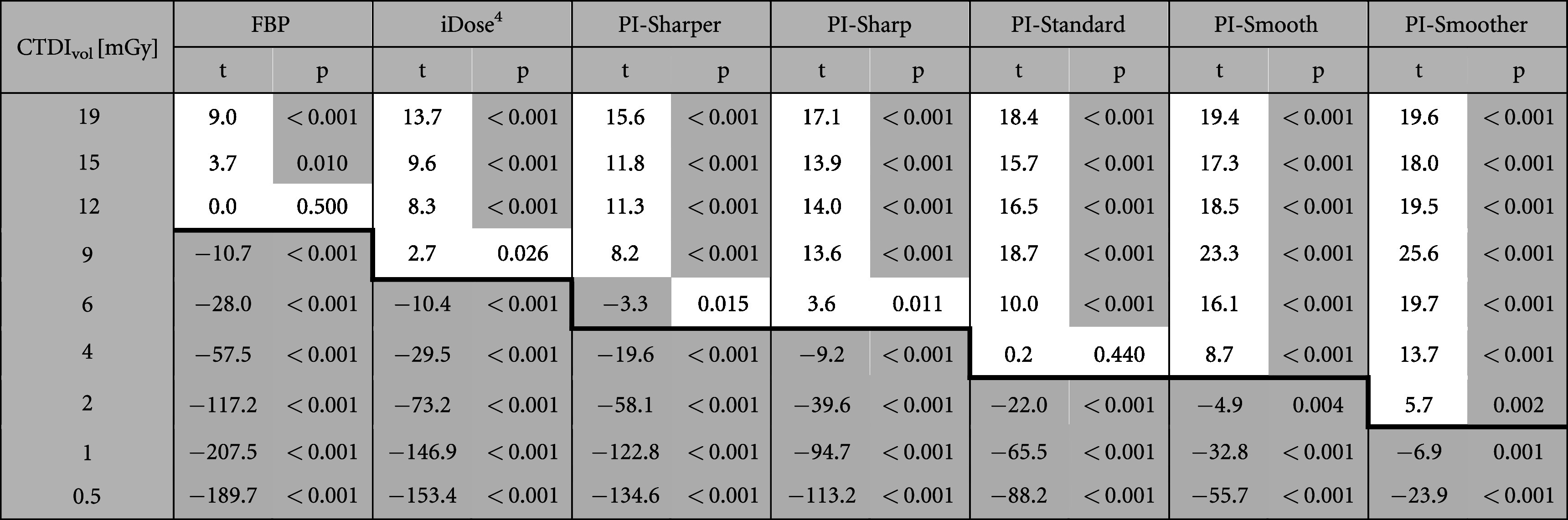
Tables A1–A10. t-values and p-values obtained from the two-sample, one-tailed student’s t-test for each image metric calculated. The cells highlighted with red indicate where the t-value signifies worse performance (t < 0 for Noise and RMSE, t > 0 for CNR, SSIM, and MS SSIM) than the reference (FBP, 12 mGy). The cells highlighted in blue indicate where the p-value suggests a statistically significant result (p < 0.01). Cells above the thick black line have performance that is better than or not statistically different than the reference (t > 0 and/or p > 0.01). Cells below the thick black line have performance that is statistically worse than the reference (t < 0 and p < 0.01).
Table A1.
Noise—small phantom.
|
Table A2.
CNR—small phantom.
|
Table A3.
RMSE—small phantom.
|
Table A4.
SSIM—small phantom.
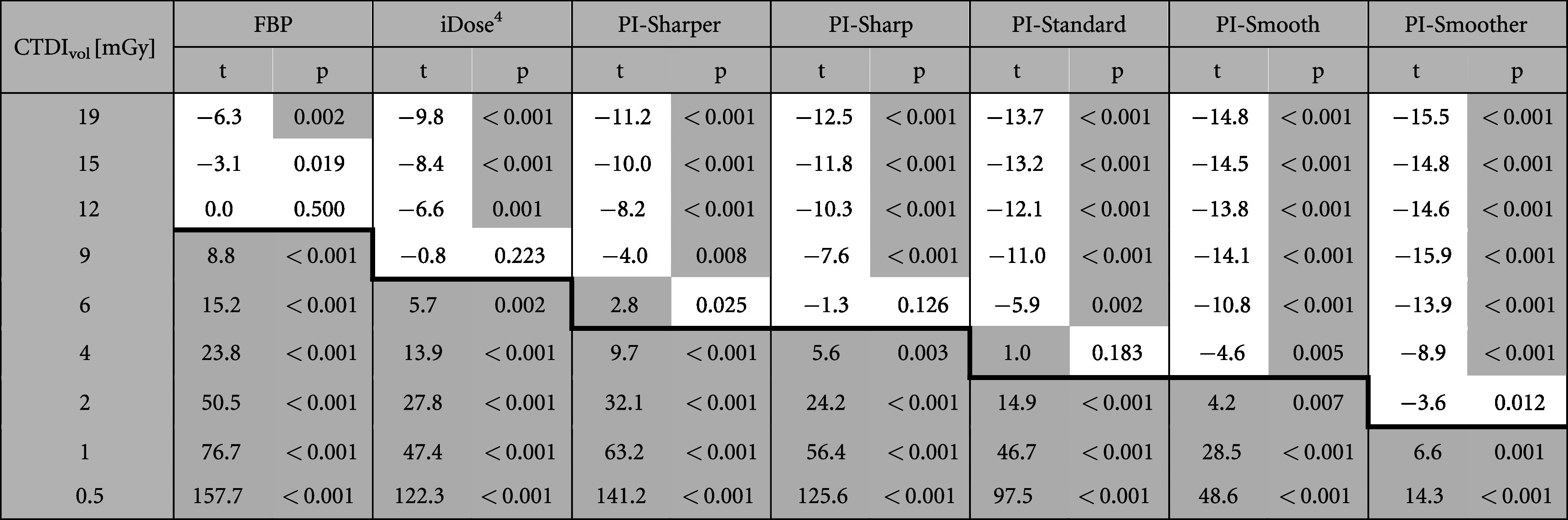
|
Table A5.
MS SSIM—small phantom.
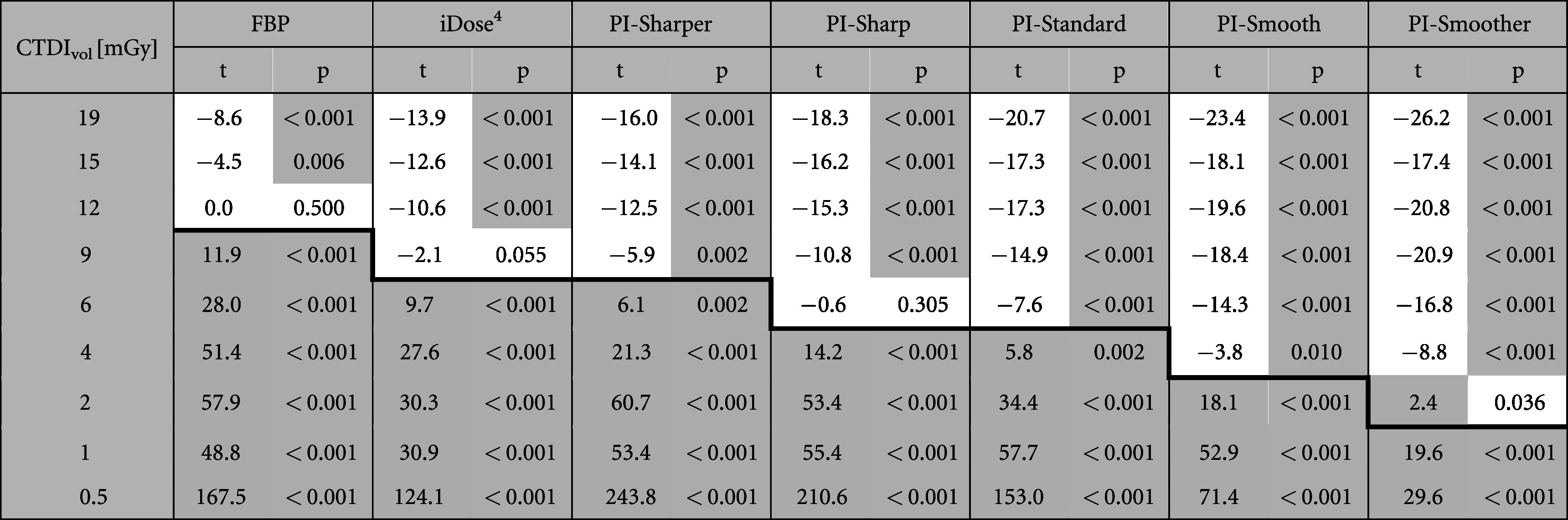
|
Table A6.
Noise— medium phantom.
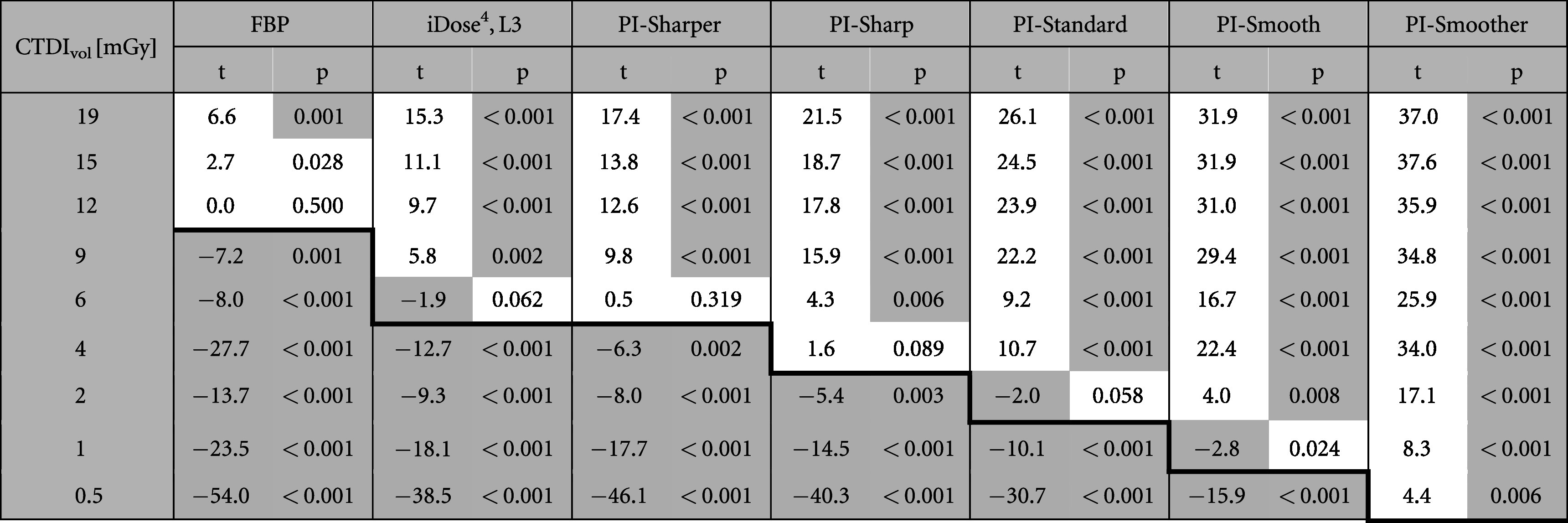
|
Table A7.
CNR—medium phantom.
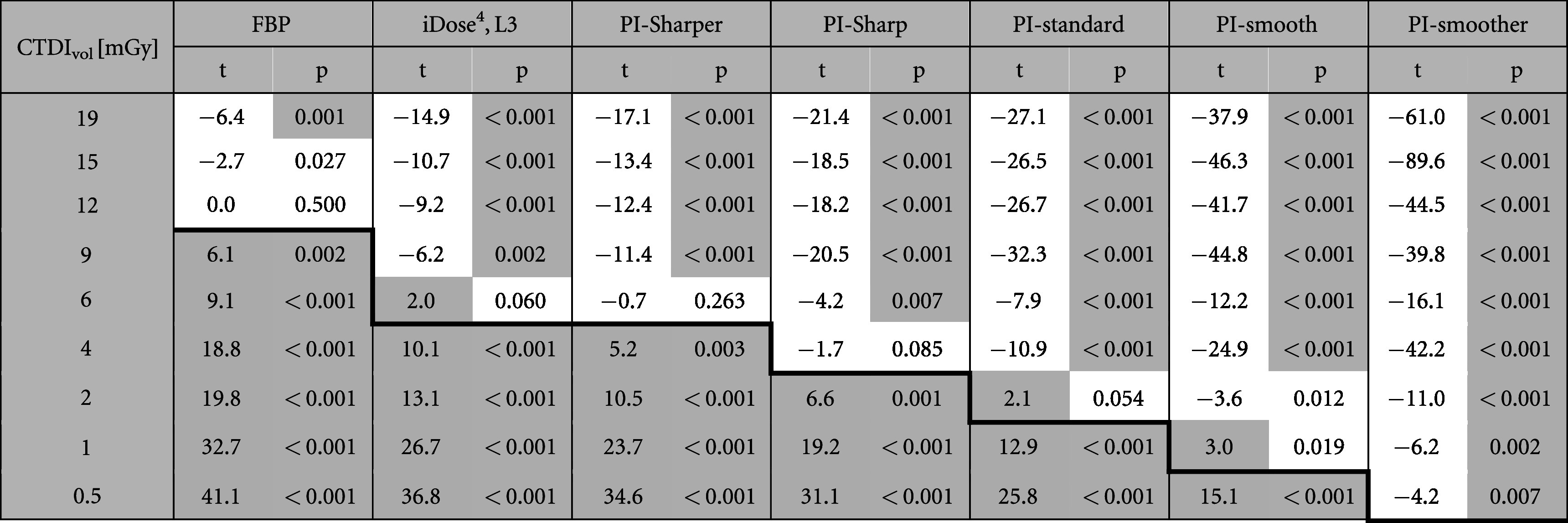
|
Table A8.
RMSE—medium phantom.
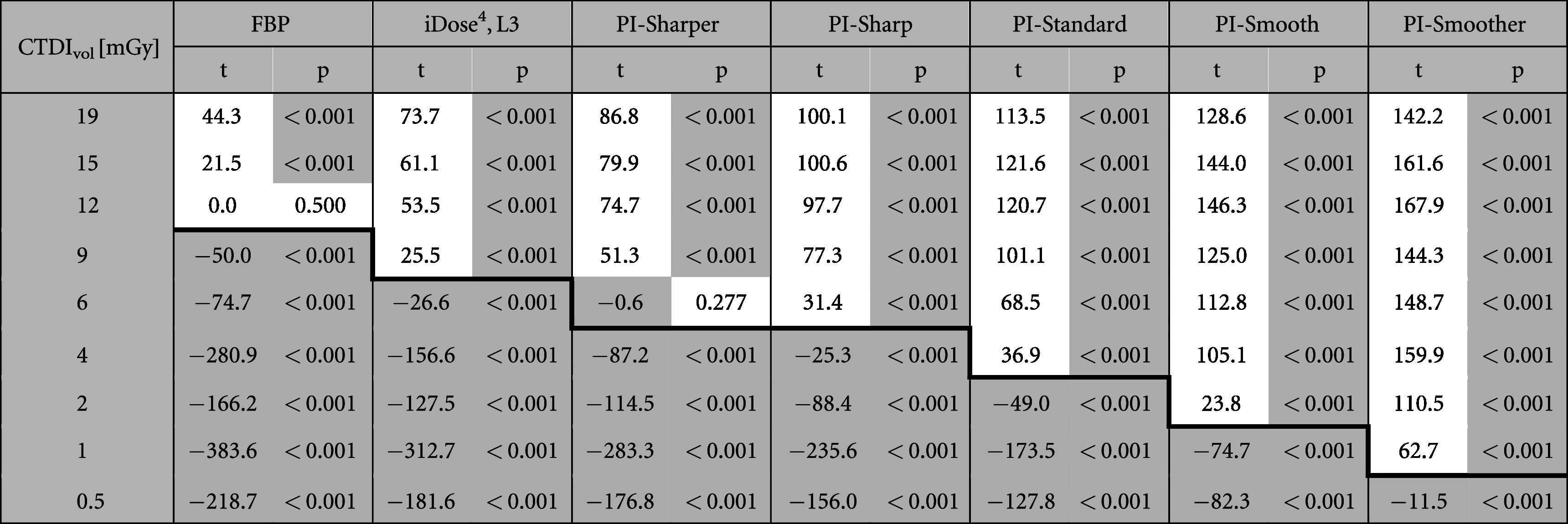
|
Table A9.
SSIM—medium phantom.
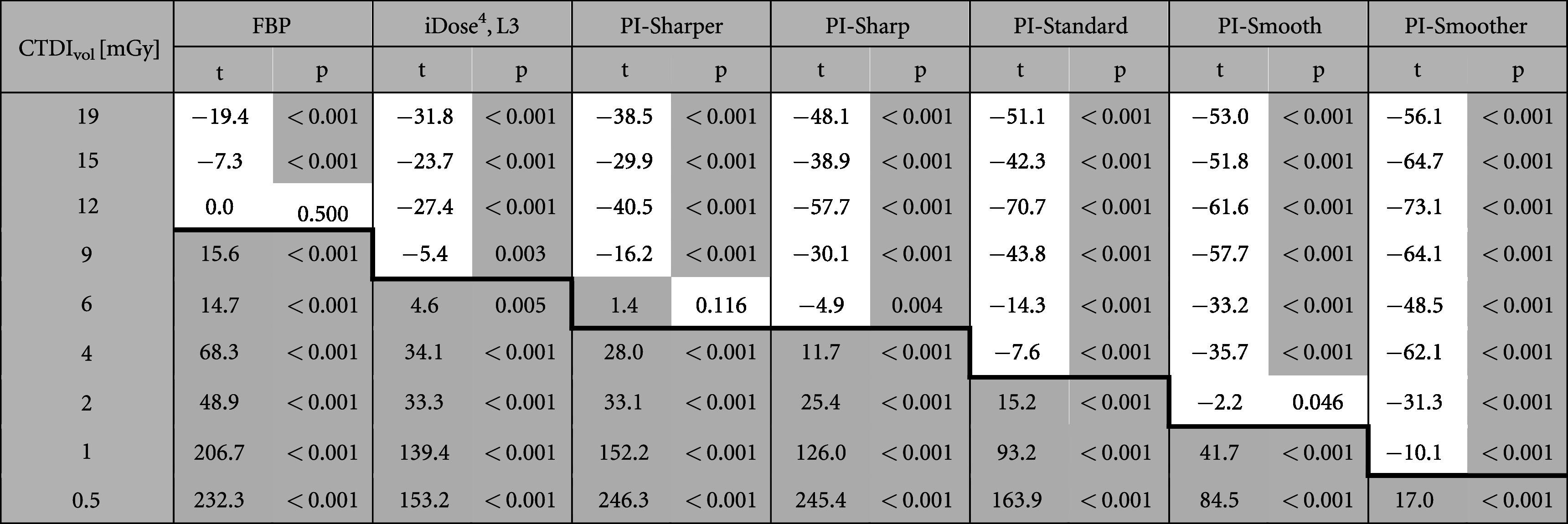
|
Table A10.
MS SSIM—medium phantom.
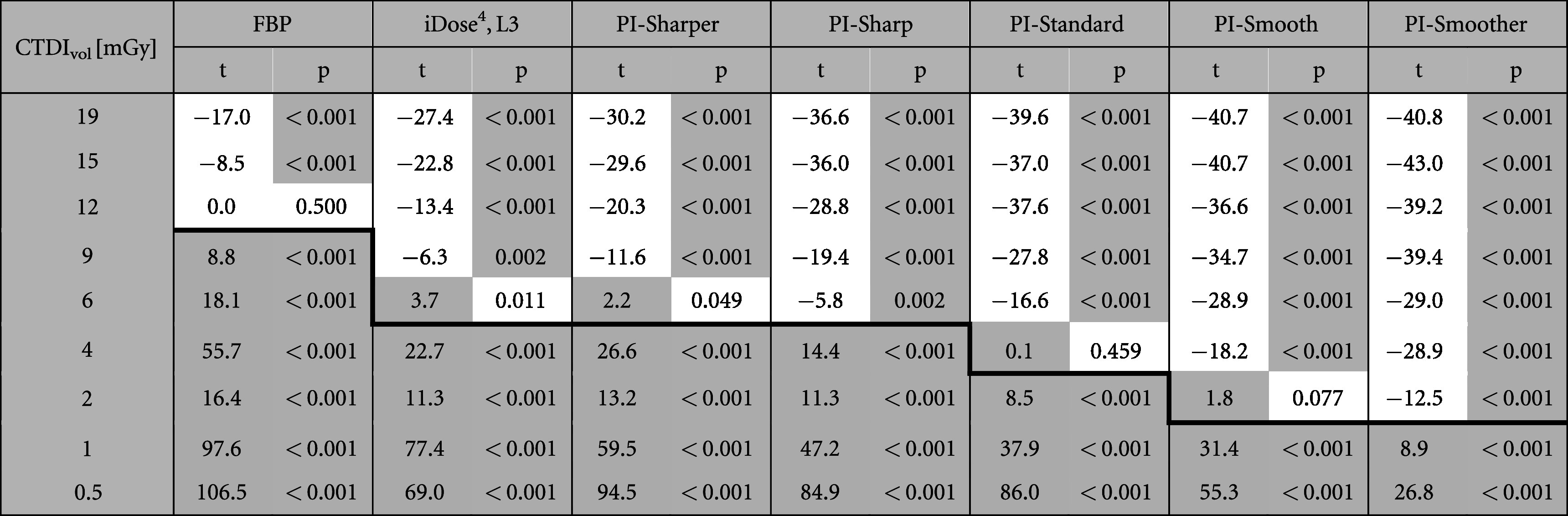
|
Data availability statement
The data cannot be made publicly available upon publication because they are not available in a format that is sufficiently accessible or reusable by other researchers. The data that support the findings of this study are available upon reasonable request from the authors.
References
- Akagi M, et al. Deep learning reconstruction improves image quality of abdominal ultra-high-resolution CT. Eur. Radiol. 2019;29:6163–71. doi: 10.1007/s00330-019-06170-3. [DOI] [PubMed] [Google Scholar]
- Arapakis I, et al. Using ‘iDose4’ iterative reconstruction algorithm in adults’ chest-abdomen-pelvis CT examinations: effect on image quality in relation to patient radiation exposure. Br. J. Radiol. 2014;87:20130613. doi: 10.1259/bjr.20130613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao P, et al. Convolutional sparse coding for compressed sensing CT reconstruction. IEEE Trans. Med. Imaging. 2019;38:2607–19. doi: 10.1109/TMI.2019.2906853. [DOI] [PubMed] [Google Scholar]
- Boedeker K. 2021. AiCE deep learning reconstruction: bringing the power of ultra-high resolution CT to routine imaging Canon Med. Syst. https://global.medical.canon/publication/ct/2019WP_AiCE_Deep_Learning .
- Brenner D J, Hall E J. Computed tomography—an increasing source of radiation exposure. New Engl. J. Med. 2007;357:2277–84. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- Chen H, et al. Low-dose CT with a residual encoder-decoder convolutional neural network. IEEE Trans. Med. Imaging. 2017;36:2524–35. doi: 10.1109/TMI.2017.2715284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehman E C, et al. Methods for clinical evaluation of noise reduction techniques in abdominopelvic CT. Radiographics. 2014;34:849–62. doi: 10.1148/rg.344135128. [DOI] [PubMed] [Google Scholar]
- Greffier J, et al. Impact of an artificial intelligence deep-learning reconstruction algorithm for CT on image quality and potential dose reduction: a phantom study. Med. Phys. 2022a;49:5052–63. doi: 10.1002/mp.15807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greffier J, et al. Comparison of two deep learning image reconstruction algorithms in chest CT images: a task-based image quality assessment on phantom data. Diagn. Intervent. Imaging. 2022b;103:21–30. doi: 10.1016/j.diii.2021.08.001. [DOI] [PubMed] [Google Scholar]
- Greffier J, et al. First results of a new deep learning reconstruction algorithm on image quality and liver metastasis conspicuity for abdominal low-dose CT. Diagnostics. 2023a;13:1182. doi: 10.3390/diagnostics13061182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greffier J, et al. Improved image quality and dose reduction in abdominal CT with deep-learning reconstruction algorithm: a phantom study. Eur. Radiol. 2023b;33:699–710. doi: 10.1007/s00330-022-09003-y. [DOI] [PubMed] [Google Scholar]
- Higaki T, et al. Deep learning reconstruction at CT: phantom study of the image characteristics. Acad. Radiol. 2020;27:82–7. doi: 10.1016/j.acra.2019.09.008. [DOI] [PubMed] [Google Scholar]
- Hofmanninger J, Prayer F, Pan J, Röhrich S, Prosch H, Langs G. Automatic lung segmentation in routine imaging is primarily a data diversity problem, not a methodology problem. Eur. Radiol. Exp. 2020;4:50. doi: 10.1186/s41747-020-00173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Liu E, Nett B, Tang J, Thibault J-B, Sahney S. A New Era of Image Reconstruction: TrueFidelity™. White paper (JB68676XX) GE Healthcare; 2019. [Google Scholar]
- Infante M, et al. Differential diagnosis and management of focal ground-glass opacities. Eur. Resp. J. 2009;33:821–7. doi: 10.1183/09031936.00047908. [DOI] [PubMed] [Google Scholar]
- Kanal K M, Butler P F, Sengupta D, Bhargavan-Chatfield M, Coombs L P, Morin R L. US diagnostic reference levels and achievable doses for 10 adult CT examinations. Radiology. 2017;284:120–33. doi: 10.1148/radiol.2017161911. [DOI] [PubMed] [Google Scholar]
- Kang E, Min J, Ye J C. A deep convolutional neural network using directional wavelets for low-dose x-ray CT reconstruction. Med. Phys. 2017;44:e360–75. doi: 10.1002/mp.12344. [DOI] [PubMed] [Google Scholar]
- Kazerooni E A, et al. ACR-STR practice parameter for the performance and reporting of lung cancer screening thoracic computed tomography (CT): 2014 (Resolution 4) J. Thorac. Imaging. 2014;29:310–6. doi: 10.1097/RTI.0000000000000097. [DOI] [PubMed] [Google Scholar]
- Koetzier L R, et al. Deep learning image reconstruction for CT: technical principles and clinical prospects. Radiology. 2023;306:e221257. doi: 10.1148/radiol.221257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, et al. Local linearity analysis of deep learning CT denoising algorithms. SPIE. 2022;12304:123040T. doi: 10.1117/12.2646371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu P, et al. Deep learning reconstruction CT for liver metastases: low-dose dual-energy versus standard-dose single-energy. Eur. Radiol. 2024;34:28–38. doi: 10.1007/s00330-023-10033-3. [DOI] [PubMed] [Google Scholar]
- McCollough C H, et al. Dependence of water-equivalent diameter and size-specific dose estimates on CT tube potential. Radiology. 2022;303:404–11. doi: 10.1148/radiol.210860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei K, et al. PixelPrint: three-dimensional printing of patient-specific soft tissue and bone phantoms for CT. Seventh International Conference on Image Formation in X-Ray Computed Tomography (ICIFXCT 2022); Baltimore, United States. 2022; Proc. SPIE Int. Soc. Opt. Eng.; 2022a. 123042G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei K, et al. Three-dimensional printing of patient-specific lung phantoms for CT imaging: emulating lung tissue with accurate attenuation profiles and textures. Med. Phys. 2022b;49:825–35. doi: 10.1002/mp.15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miglioretti D L, et al. The use of computed tomography in pediatrics and the associated radiation exposure and estimated cancer risk. JAMA Pediatrics. 2013;167:700–7. doi: 10.1001/jamapediatrics.2013.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikayama R, et al. Deep-learning reconstruction for ultra-low-dose lung CT: volumetric measurement accuracy and reproducibility of artificial ground-glass nodules in a phantom study. Br. J. Radiol. 2022;95:20210915. doi: 10.1259/bjr.20210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, et al. The evaluation of the reduction of radiation dose via deep learning-based reconstruction for cadaveric human lung CT images. Sci. Rep. 2022;12:12422. doi: 10.1038/s41598-022-16798-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagayama Y, et al. Deep learning-based reconstruction for lower-dose pediatric CT: technical principles, image characteristics, and clinical implementations. Radiographics. 2021;41:1936–53. doi: 10.1148/rg.2021210105. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, et al. Deep learning-based CT image reconstruction: initial evaluation targeting hypovascular hepatic metastases. Radiol. Artif. Intell. 2019;1:e180011. doi: 10.1148/ryai.2019180011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Shin J, Min I K, Bae H, Kim Y E, Chung Y E. Image quality and lesion detectability of lower-dose abdominopelvic CT obtained using deep learning image reconstruction. Korean J. Radiol. 2022;23:402–12. doi: 10.3348/kjr.2021.0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa J Pytorch-msssim. Pytorch-msssim. November 17, 2021 ( https://github.com/jorge-pessoa/pytorch-msssim)
- Philips Healthcare 2021 AI for significantly lower dose and improved image quality.
- Radiology ACo 2018. Acr–Aapm–Spr Practice Parameter For Diagnostic Reference Levels And Achievable Doses In Medical X-Ray Imaging.
- Samei E, et al. Performance evaluation of computed tomography systems: summary of AAPM Task Group 233. Med. Phys. 2019;46:e735–56. doi: 10.1002/mp.13763. [DOI] [PubMed] [Google Scholar]
- Shapira N, et al. PixelPrint: three-dimensional printing of realistic patient-specific lung phantoms for CT imaging. SPIE Medical Imaging; San Diego, California, United States: Proc SPIE Int Soc Opt Eng; 2022. 120310N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira N, et al. Three-dimensional printing of patient-specific computed tomography lung phantoms: a reader study. PNAS Nexus. 2023;2:pgad026. doi: 10.1093/pnasnexus/pgad026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon J, Lyu P, Marin D, Samei E. Noise and spatial resolution properties of a commercially available deep learning-based CT reconstruction algorithm. Med. Phys. 2020;47:3961–71. doi: 10.1002/mp.14319. [DOI] [PubMed] [Google Scholar]
- Son W, et al. Comparison of a deep learning-based reconstruction algorithm with filtered back projection and iterative reconstruction algorithms for pediatric abdominopelvic CT. Korean J. Radiol. 2022;23:752–62. doi: 10.3348/kjr.2021.0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, et al. Improving the image quality of pediatric chest CT angiography with low radiation dose and contrast volume using deep learning image reconstruction. Quant. Imaging Med. Surg. 2021;11:3051. doi: 10.21037/qims-20-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczykutowicz T P, Toia G V, Dhanantwari A, Nett B. A review of deep learning CT reconstruction: concepts, limitations, and promise in clinical practice. Curr. Radiol. Rep. 2022;10:101–15. doi: 10.1007/s40134-022-00399-5. [DOI] [Google Scholar]
- van der Walt S, et al. Scikit-image: image processing in Python. Peerj. 2014;2:e453. doi: 10.7717/peerj.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtanen P, et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–72. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Simoncelli E P, Bovik A C. Multiscale structural similarity for image quality assessment. IEEE; The Thrity-Seventh Asilomar Conference on Signals, Systems & Computers; IEEE; 2003. pp. 1398–402. [DOI] [Google Scholar]
- Wang Z, Bovik A C, Sheikh H R, Simoncelli E P. Image quality assessment: from error visibility to structural similarity. IEEE Trans. Image Process. 2004;13:600–12. doi: 10.1109/TIP.2003.819861. [DOI] [PubMed] [Google Scholar]
- Willemink M J, et al. Iterative reconstruction techniques for computed tomography: I. Technical principles. Eur. Radiol. 2013;23:1623–31. doi: 10.1007/s00330-012-2765-y. [DOI] [PubMed] [Google Scholar]
- Willemink M J, Noël P B. The evolution of image reconstruction for CT—from filtered back projection to artificial intelligence. Eur. Radiol. 2019;29:2185–95. doi: 10.1007/s00330-018-5810-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolterink J M, Leiner T, Viergever M A, Išgum I. Generative adversarial networks for noise reduction in low-dose CT. IEEE Trans. Med. Imaging. 2017;36:2536–45. doi: 10.1109/TMI.2017.2708987. [DOI] [PubMed] [Google Scholar]
- Wu D, Kim K, Fakhri G E, Li Q. Iterative low-dose CT reconstruction with priors trained by artificial neural network. IEEE Trans. Med. Imaging. 2017;36:2479–86. doi: 10.1109/TMI.2017.2753138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, et al. Low-dose CT image denoising using a generative adversarial network with wasserstein distance and perceptual loss. IEEE Trans. Med. Imaging. 2018;37:1348–57. doi: 10.1109/TMI.2018.2827462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data cannot be made publicly available upon publication because they are not available in a format that is sufficiently accessible or reusable by other researchers. The data that support the findings of this study are available upon reasonable request from the authors.