Abstract
Purpose
To describe the clinical, electrophysiological and genetic spectrum of inherited retinal diseases associated with variants in the PRPH2 gene.
Methods
A total of 241 patients from 168 families across 15 sites in 9 countries with pathogenic or likely pathogenic variants in PRPH2 were included. Records were reviewed for age at symptom onset, visual acuity, full-field ERG, fundus colour photography, fundus autofluorescence (FAF), and SD-OCT. Images were graded into six phenotypes. Statistical analyses were performed to determine genotype–phenotype correlations.
Results
The median age at symptom onset was 40 years (range, 4–78 years). FAF phenotypes included normal (5%), butterfly pattern dystrophy, or vitelliform macular dystrophy (11%), central areolar choroidal dystrophy (28%), pseudo-Stargardt pattern dystrophy (41%), and retinitis pigmentosa (25%). Symptom onset was earlier in retinitis pigmentosa as compared with pseudo-Stargardt pattern dystrophy (34 vs 44 years; P = 0.004). The median visual acuity was 0.18 logMAR (interquartile range, 0–0.54 logMAR) and 0.18 logMAR (interquartile range 0–0.42 logMAR) in the right and left eyes, respectively. ERG showed a significantly reduced amplitude across all components (P < 0.001) and a peak time delay in the light-adapted 30-Hz flicker and single-flash b-wave (P < 0.001). Twenty-two variants were novel. The central areolar choroidal dystrophy phenotype was associated with 13 missense variants. The remaining variants showed marked phenotypic variability.
Conclusions
We described six distinct FAF phenotypes associated with variants in the PRPH2 gene. One FAF phenotype may have multiple ERG phenotypes, demonstrating a discordance between structure and function. Given the vast spectrum of PRPH2 disease our findings are useful for future clinical trials.
Keywords: pattern dystrophy, peripherinopathy, OCT, fundus autofluorescence, CACD
Inherited retinal diseases (IRDs) are the most common cause of blindness in the working age group.1 Disease-associated variants in the PRPH2 gene were the fourth most common cause of IRD (5.2%) in the United Kingdom.2 To date, 352 disease-causing variants in PRPH2 are listed in the Human Gene Mutation Database with autosomal dominant, and less commonly, recessive and digenic (with retinal outer segment membrane protein 1, ROM1, OMIM*180721) inheritance.3 PRPH2 (OMIM *179605), encodes a photoreceptor-specific tetraspanin, essential for the formation and maintenance of rod and cone outer segments.4 Dominant variants in PRPH2 lead to central areolar choroidal dystrophy (CACD, OMIM #613105), retinitis pigmentosa (RP7, OMIM #608133), pseudo-Stargardt pattern dystrophy (PSPD, OMIM #169150), butterfly pattern dystrophy (BPD), and vitelliform macular dystrophy (VMD, OMIM #608161). Disease-associated variants in the PRPH2 gene have been reported in pedigrees with marked interfamilial and intrafamilial variability and penetrance.4,5
To date, no clear genotype–phenotype correlations in human PRPH2 disease have been firmly established.4,6–9 This may be attributable to the relatively small sample sizes of previous clinical studies, variable penetrance, and no standardized system for phenotype grading using multimodal imaging. Some variability may be due to genetic modifiers in ROM1 and ABCA4.7,8 Poloschek et al.7 found patients who carry the p.Arg172Trp variant in PRPH2 demonstrated an isolated MD, whereas those carrying additional variants in the ROM1 or ABCA4 gene exhibited a more severe phenotype. However, Leroy et al.8 excluded variants in ROM1 as a modifier for an RP phenotype in PRPH2 disease. A recent case series of 19 PRPH2 patients by Bianco et al.9 proposed that missense variants in the D2 loop were associated with a cone–rod dystrophy (CRD). To date, pathogenic missense variants at six codon positions (Arg142, Arg172, Arg195, Ile196, Arg203, and Gly208) have been reported to cause CACD.9–17 In contrast, truncating variants (nonsense and frameshift) have been associated with a range of phenotypes including VMD, BPD, PSPD, and RP.
This study describes the clinical and genetic spectrum of PRPH2-associated IRD, including 22 novel pathogenic variants. Herein we report genotype–phenotype correlations in the largest multicenter case series of 241 patients harbouring 91 unique pathogenic or likely pathogenic PRPH2 variants. Given the marked variability of disease, our findings will be useful for defining cohorts for inclusion in future clinical trials.
Methods
Study Population
A retrospective cohort study of patients carrying a pathogenic or likely pathogenic variant in the PRPH2 gene. Family members carrying the same variant were also included where available. Patients were identified from the Australian Inherited Retinal Diseases Registry and DNA bank with phenotypic data acquired through the Western Australia Retinal Degeneration Study, an extensive database for IRDs that currently includes more than 900 patients referred from 2017 to December 2023 at the Lions Eye Institute (Perth, Australia). Additional data were collected from the Centre for Eye Research Australia (Melbourne, Australia), Save Sight Institute (Sydney, Australia), Royal Brisbane and Women's Hospital (Brisbane, Australia), New Zealand National Eye Centre (Auckland, New Zealand), IRCCS Fondazione Bietti (Rome, Italy), IRCCS San Raffaele Scientific Institute (Milan, Italy), IRD-PT registry18 (Coimbra, Portugal), Amsterdam University Medical Centers (Amsterdam, the Netherlands), National Taiwan University Hospital, (Taipei, Taiwan), Singapore National Eye Centre (Singapore, Singapore), The Hospital for Sick Children (Toronto, Ontario, Canada), Shiley Eye Institute, University of California San Diego (San Diego, CA, USA), and Wills Eye Hospital (Philadelphia, PA, USA).
This study was approved by the institutional review boards of the University of Western Australia (2021/ET000151), Sir Charles Gairdner Hospital (RGS04985, 1998-115), Royal Victorian Eye and Ear Hospital/Centre for Eye Research Australia (19/1443H), New Zealand Ministry of Health (NTX/08/12/123), Auckland District Health Board (A+4290), the Save Sight Institute (2022/PID01932), Sezione IFO/Fondazione Bietti (NEU_01-2014), Vita-Salute San Raffaele University (MIRD2020), University of Coimbra (CE-125/2019), Erasmus Medical Center (NL34152.078.10), National Taiwan University Hospital (IRB 201408082RINC), Singapore ethics review board (2015/2766), University of California San Diego (IRB 120516), Wills Eye Hospital (IRB 2021-85), and the Hospital for Sick Children (REB-1000017804). It adhered to the tenets of the Declaration of Helsinki, and informed consent was obtained from all participants or their legal guardians.
Clinical Data Collection
Data were collected for age at symptom onset, family history, visual acuity (VA), electrophysiological testing, and clinical imaging, including SD-OCT, fundus autofluorescence (FAF), and colour imaging. Age at onset was defined as the year in which the first symptoms were noted. VA was measured on either a Snellen-style or a logMAR-style chart, such as the Early Treatment Diabetic Retinopathy letter chart. Optos ultrawide field FAF imaging (Optos PLC, Dunfermline, UK) was used for phenotype grading where possible. Heidelberg HRA+OCT (Heidelberg Engineering, Heidelberg, Germany) 55° or 30° blue wavelength FAF images were graded when Optos images were not available. All SD-OCT scans were obtained using Heidelberg Spectralis.
FAF grading was performed by two independent examiners (FKC and RCHJ) and included the following phenotypes: normal, VMD, BPD, CACD, PSPD, and RP. The CACD phenotype, as originally described by Hoyng et al.19 and Boon et al.,20 had a well-defined oval region of stippled hyper and hypoautofluorescence in the central macular with or without a radial configuration of hyperautfluorescence at the border and hypoautofluorescent patches within.19 As per Boon et al.20 using color, FAF, and fluorescein angiography, CACD may reach the temporal vascular arcade superiorly and inferiorly and encompass the optic nerve, nasally with no peripapillary sparing. We only included cases with stages 2 to 4 of the Hoyng et al.19 classification system, which was based on color and fluorescein angiography. This was due to the difficulty in distinguishing stage 1 CACD from VMD or BPD. The PSPD phenotype included focal hyperautofluorscent or fleck-like lesions, with or without stippled hypoautofluorescence spreading, centrifugally beyond the arcades resembling Stargardt disease. The RP phenotype was characterized by peripheral hypoautofluorescence with or without a central hyperautofluorscent ring. Any grading disagreements were resolved by a third examiner (CJFB).
Electrophysiology was performed in accordance with the International Clinical Electrophysiology of Vision Society Standards.21–23 Full-field ERG traces were reviewed, and component parameters were extracted and compared with a control cohort at Sir Charles Gairdner Hospital, Perth, Australia. A cone dystrophy (COD) was noted if the light-adapted (LA) responses showed a-wave reduction and flicker delay, whereas the dark-adapted (DA) responses were within the normal range. CRD was defined by a more severe LA than DA response component deficit. A rod–cone dystrophy (RCD) was defined by a more severe DA than LA response component deficit (typically with no signal detectable in the DA 0.01 cd/m2 test). MD was defined by a reduced P50 on the pattern ERG (PERG) or central response density loss on multifocal ERG with the DA and LA response components within the normal range.
Some of these cases have been published previously by Heath Jeffery et al.24 (n = 12), Bianco et al.9 (n = 19), and Antonelli et al.25 (n = 28).
Genetic Data Collection
Variants in the PRPH2 gene were identified using a range of molecular strategies over time. All variants were confirmed with Sanger sequencing or whole exome sequencing. Genetic testing was performed with next-generation sequencing. Pathogenicity of PRPH2 variants was assigned based on the American College of Medical Genetics and Genomics guidelines and associated literature.26,27 Previous disease associations were explored in the Human Gene Mutation Database, Leiden Open Variation Database (http://databases.lovd.nl/shared/genes/PRPH2), and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/). Potential pathogenicity of variants was assessed in silico with different tools, including functional pathogenicity and protein stability predictors using PolyPhen2,28 SIFT,29 REVEL,30 and CADD31 for missense variants, SIFT-Indel32,33 and VEST-434 for in-frame amino acid alterations and frameshift variants, Mutation Taster35 and ENTPRISE-X36 for nonsense and delins, and Splice AI37 for splicing-altering variants.
Statistical Analyses
All analyses were performed in R software version 4.1.3 (The R Core Team, Vienna, Austria) and R Studio version 2022.07.1 (RStudio Team, Boston, MA, USA). Categorical data was expressed as proportions, and continuous data as means with standard deviations or a median and interquartile range (IQR). For genotype–phenotype correlations, genotypes were stratified into exon deletions, missense, in-frame indels, frameshift, nonsense, splicing, and start–loss. Clinical features considered for statistical analysis were age at symptom onset, VA, FAF grading, and ERG parameters. Visual impairment was recorded according to the World Health Organization: mild (VA, <20/40 and ≥20/60), moderate (VA, <20/60 and ≥20/200), severe (VA, <20/200 and ≥20/400), and blindness (VA, <20/400). VA data measured on a Snellen-style chart was converted to logMAR equivalent, and off-chart levels of vision were assigned a logMAR value of 2.0 for counting fingers, 2.3 for hand movements, and 2.6 for light perception. A locally estimated scatterplot smoothing curve was constructed using VA from the better-seeing eye (lowest logMAR value) to illustrate the average evolution of VA with age across the cohort. Interocular symmetry in VA was determined by Bland-Altman analysis and Spearman rho. One-way ANOVA and Kruskal–Wallis testing compared clinical variables across different FAF phenotypes. Mann–Whitney U testing was used to compare ERG parameters in the PRPH2 cohort with controls. Bonferroni correction was applied, where appropriate, for post hoc comparisons and multiple testing. Statistical significance was set at a P value of less than 0.05.
Results
Clinical Presentation and Visual Function
A total of 241 patients from 168 presumably unrelated families with a PRPH2-associated IRD were recruited at a median age of 56 years (range, 7–89 years). Age at symptom onset was available for 189 patients and 20 were asymptomatic at their last review. The median age at symptom onset was 40 years (IQR, 30–50 years; range, 4–78 years) (Table 1). The 20 asymptomatic patients were last reviewed at a median age of 39 years (range, 10–65 years). VA for the right and left eye was available for 241 patients. Median VA was 0.18 logMAR (IQR, 0–0.54 logMAR), or 20/30 Snellen equivalent (IQR, 20/20–20/80 Snellen equivalent) for the right eye, and 0.18 logMAR (IQR, 0–0.42 logMAR) or 20/30 Snellen equivalent (IQR, 20/20–20/60 Snellen equivalent) for the left eye. Although some interocular differences were large (−2.20 to +2.50), the mean VA between right and left eye was not significantly different (paired t test, P = 0.11).
Table 1.
Clinical Characteristics of PRPH2 Patients
| Characteristics | PRPH2 Cohort (n = 241) |
|---|---|
| Age at imaging (y) (n = 241) | |
| Mean ± SD | 54.8 ± 15.5 |
| Median (IQR) | 55.8 (Q1, 44.9; Q3, 66.9) |
| Range | 7–89 |
| Sex (M:F) | 118:123 |
| Age at symptom onset (n = 189) | |
| Median (IQR) | 40 (Q1, 30; Q3, 50) |
| Range | 4–78 |
| Last available VA in the right eye (n = 241) | |
| Median VA, in logMAR (IQR) | 0.18 (Q1, 0; Q3, 0.54) |
| Median VA, in Snellen equivalent (IQR) | 20/30 (Q1, 20/20; Q3, 20/80) |
| Last available VA in the left eye (n = 241) | |
| Median VA, in logMAR (IQR) | 0.18 (Q1, 0; Q3, 0.42) |
| Median VA, in Snellen equivalent (IQR) | 20/30 (Q1, 20/20; Q3, 20/60) |
| FAF, n/total (%) | |
| Normal | 13/241 (5%) |
| BPD/VMD | 27/241 (11%) |
| CACD | 68/241 (28%) |
| PSPD | 98/241 (41%) |
| RP | 35/241 (15%) |
| ERG, no/total (%) | |
| Normal ffERG and PERG | 8/100 (8%) |
| MD | 21/100 (21%) |
| COD | 16/100 (16%) |
| Cone-rod dystrophy | 17/100 (17%) |
| Rod-cone dystrophy | 38/100 (38%) |
ffERG, full-field ERG.
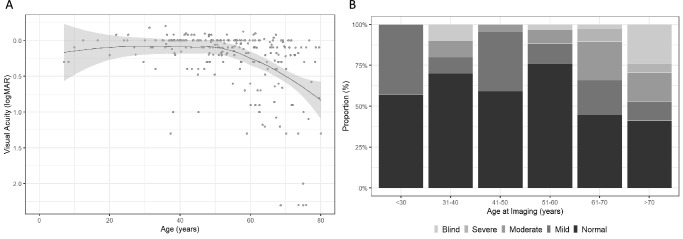
Overall, VA showed stability until the fifth decade with a decline thereafter (Fig. 1). The proportion of patients with normal vision in their better seeing eye also declined with age (Fig. 1). Of 241 patients, 55 (22.8%) showed interocular asymmetry in VA greater than 0.3 logMAR. The 95% limits of agreements were −0.97 to +1.07 logMAR (Supplementary Material S1).
Figure 1.
Locally estimated scatterplot smoothing (LOESS) curve using VA from the better-seeing eye illustrating the average evolution of VA with age across the entire cohort (A). Proportion (%) of PRPH2 patients with normal, mild (VA, <20/40 and ≥20/60), moderate (VA, <20/60 and ≥20/200), severe visual impairment (VA, <20/200 and ≥20/400) and blindness (VA, <20/400) by age at imaging (B).
Fundus Autofluorescence
Of 241 PRPH2 disease-associated variant carriers FAF was normal in 13 (5%), at a median age of 33 years (range, 7–71 years); of these 13 patients, 10 (77%) were asymptomatic (Fig. 2). Twenty-seven patients (11%) demonstrated a BPD (n = 21) or VMD (n = 6) FAF phenotype (Fig. 3), of which 3 (11%) were asymptomatic and the remaining 24 had a median age at symptom onset of 44 years (range, 26–70 years). The CACD FAF phenotype (Fig. 4) was observed in 68 patients (28%), with a median age at symptom onset of 40 years (range, 14–70 years) in 55 patients; 3 patients were asymptomatic (age, 13–55 years). PSPD (Fig. 5) was seen in 98 patients (41%) with a median age at symptom onset of 40 years (range, 7–78 years) in 73 patients; 4 patients were asymptomatic (range, 30–55 years). RP (Fig. 6) was observed in 35 patients (15%) with a median age at symptom onset of 33 years (range, 4–63 years) in 34 patients. Table 2 summarises the clinical features of each FAF phenotype. There were no significant difference in the sex distribution (P = 0.751). A younger age at imaging was observed in the normal FAF group (P < 0.001), which was likely a result of familial screening. Age at symptom onset was lower in the RP group as compared with BPD/VMD (P = 0.048) or PSPD (P = 0.015). VA in the better seeing eye was worse in the CACD group as compared with BPD/VMD (P = 0.035). An age-related decline in VA from the sixth decade was observed across all CACD, PSPD, and RP phenotype groups (Supplementary Material S2).
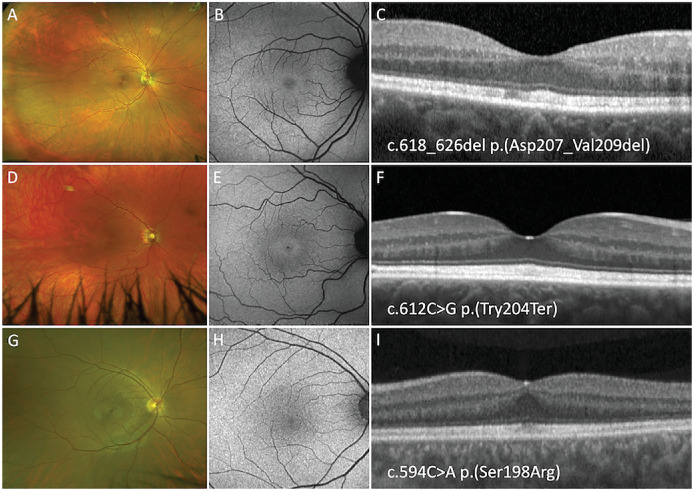
Figure 2.
Optos pseudocolor, FAF, and SD-OCT imaging of a 40-year-old woman (A, B, C), a 39-year-old woman (D, E, F), and a 40 year-old-man (G, H, I) harboring the p.(Asp207_Val209del), p.(Try204Ter), and p.(Ser198Arg) PRPH2 variants respectively. All three patients show normal pseudocolor and FAF imaging with thickening of band 2 on SD-OCT.
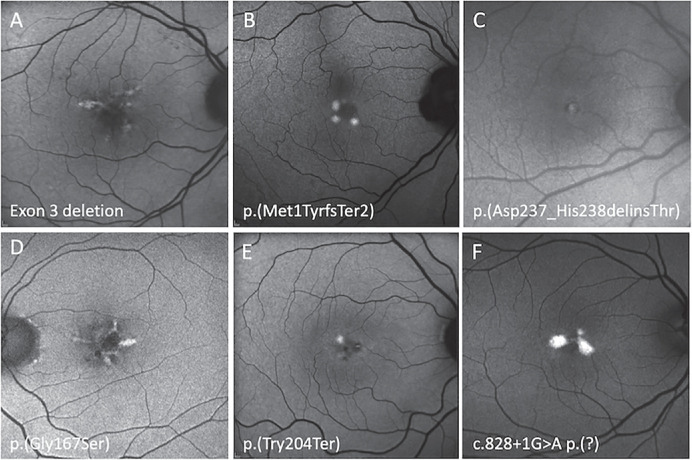
Figure 3.
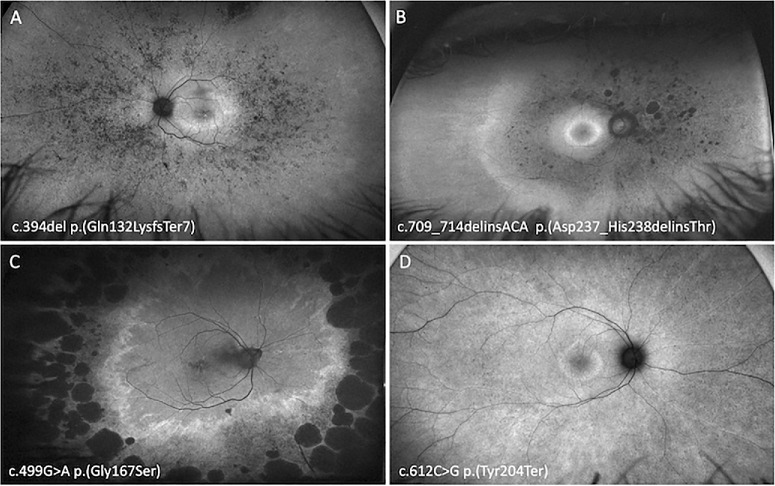
Optos FAF imaging of six PRPH2 patients exhibiting focal central macula hyperautofluorescence defined as a BPD or VMD. An 88-year-old woman with a VA of 20/25 harboring exon 3 deletion (A), a 49-year-old woman with a VA of 20/20 harboring p.(Met1TyrfsTer2) (B), a 47-year-old woman with a VA of 20/16 harboring p.(Asp237_His238delinsThr) (C), a 66-year-old man with a VA of 20/25 harboring p.(Gly167Ser) (D), a 57-year-old woman with a VA of 20/20 harboring p.(Tyr204Ter) (E), and a 66-year-old woman with a VA of 20/25 harboring c.828+1G>A (F).
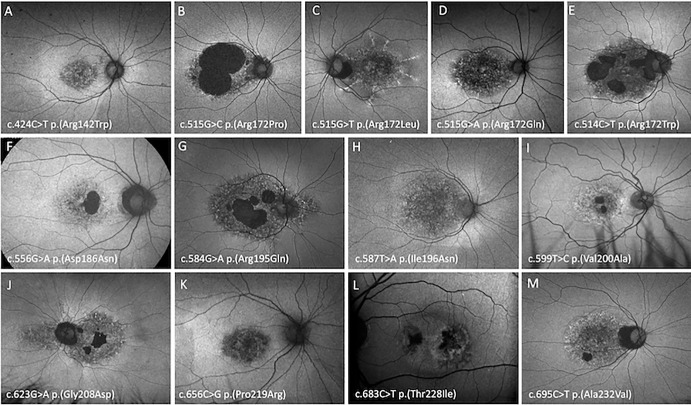
Figure 4.
Optos FAF imaging of PRPH2 patients demonstrating the CACD phenotype. A 53-year-old woman with VA of 20/25 harboring p.(Arg142Trp) (A), a 45-year-old man with a VA of 20/60 harboring p.(Arg172Pro) (B), an 89-year-old woman with a VA of 20/250 harboring p.(Arg172Leu) (C), a 46-year-old woman with a VA of 20/20 harboring p.(Arg172Gln) (D), a 67-year-old woman with a VA of 20/60 harboring p.(Arg172Trp) (E), a 33-year-old man with a VA of 20/20 harboring p.(Asp186Asn) (F), a 53-year-old man with a VA of 20/60 harboring p.(Arg195Gln) (G), a 27-year-old man with a VA of 20/40 harboring p.(Ile196Asn) (H), a 48-year-old man with a VA of counting fingers harboring p.(Val200Ala) (I), a 68-year-old man with a VA of 20/120 harboring p.(Gly208Asp) (J), a 61-year-old woman with a VA of 20/30 harboring p.(Pro219Arg) (K), a 61-year-old man with a VA of 20/15 harboring p.(Thr228Ile), (L) and a 53-year-old woman with a VA of 20/30 harboring p.(Ala232Val) (M).
Figure 5.
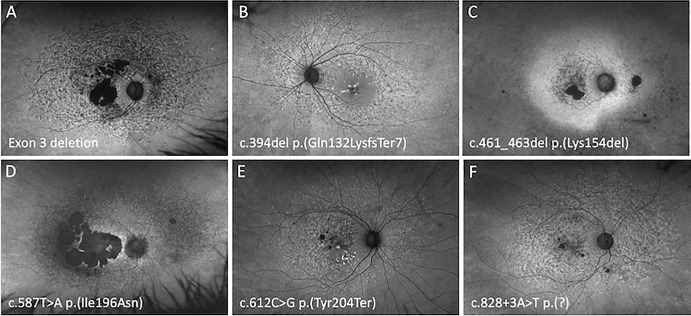
Optos FAF imaging of PRPH2 patients demonstrating the PSPD phenotype. A 66-year-old woman with a VA of 20/200 harboring an exon 3 deletion (A), a 55-year-old woman with a VA of 20/80 harboring p.(Gln132LysfsTer7) (B), a 59-year-old woman with a VA of 20/20 harboring p.(Lys154del) (C), a 61-year-old woman with a VA of 20/40 harboring p.(Ile196Asn) (D), a 51-year-old woman with a VA of 20/20 harboring p.(Tyr204Ter) (E), and a 67-year-old woman with a VA of 20/90 harboring c.828+3A>T (F).
Figure 6.
Optos FAF imaging of PRPH2 patients demonstrating the RP phenotype. A 48-year-old woman with VA of 20/40 harboring p.(Gln132LysfsTer7) (A), a 55-year-old woman with a VA of 20/25 harboring p.(Asp237_His238delinsThr) (B), a 71-year-old woman with a VA of 20/30 harboring p.(Gly167Ser) (C), and a 36-year-old woman with a VA of 20/25 harboring p.(Tyr204Ter) (D).
Table 2.
Clinical Characteristics of Each Phenotype Group
| Phenotype Group | Normal | BPD/VMD | CACD | PSPD | RP | P Value |
|---|---|---|---|---|---|---|
| No. of cases | 13 | 27 | 68 | 98 | 35 | |
| Sex | ||||||
| Female:male | 7:6 | 17:10 | 35:35 | 48:50 | 18:17 | 0.751a |
| Age at imaging | ||||||
| Mean ± SD | 32 ± 17 | 56 ± 13 | 53 ± 16 | 60 ± 12 | 52 ± 15 | |
| < 0.001†,‡ | ||||||
| Median (IQR) | 33 (20–40) | 56 (48–66) | 54 (42–63) | 61 (51–70) | 52 (38–62) | |
| Asymptomatic count/total (%) | 10/13 (77%) | 3/27 (11%) | 3/58 (5%) | 4/77 (5%) | 0/34 (0%) | |
| Age of symptom onset | ||||||
| Mean ± SD | 31 ± 34 | 45 ± 12 | 41 ± 13 | 44 ± 14 | 34 ± 17 | |
| 0.004†,§ | ||||||
| Median (IQR) | 20 (13–45) | 44 (40–51) | 40 (34–50) | 40 (35–53) | 33 (20–48) | |
| Better eye VA (logMAR) | ||||||
| Median (IQR) | 0 (0–0.1) | 0.02 (0–0.1) | 0.2 (0–0.7) | 0.1 (0–0.3) | 0.1 (0–0.2) | 0.002‖,¶ |
| Electrophysiology | ||||||
| Normal* | 2 | 2 | 4 | 0 | 0 | |
| Macular dystrophy | 0 | 3 | 9 | 9 | 0 | |
| Cone dystrophy | 0 | 4 | 7 | 5 | 0 | |
| Cone-rod dystrophy | 0 | 0 | 2 | 15 | 0 | |
| Rod-cone dystrophy | 0 | 0 | 0 | 15 | 23 |
Normal full-field and PERG.
χ2 test.
One-way ANOVA.
The normal group was significantly lower than all other groups (all P < 0.001). The PSPD group was significantly higher than both the CACD (P = 0.012) and RP (P = 0.017) groups.
The RP group was younger than both BPD/VMD (P = 0.048) and PSPD (P = 0.015)
Kruskal–Wallis test.
VA for the BPD/VMD group was significantly lower than the CACD croup (P = 0.035)
Electrophysiology
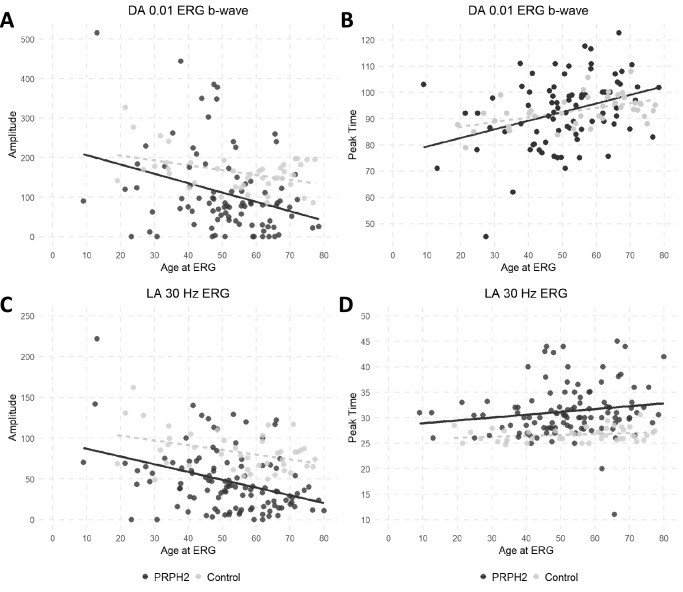
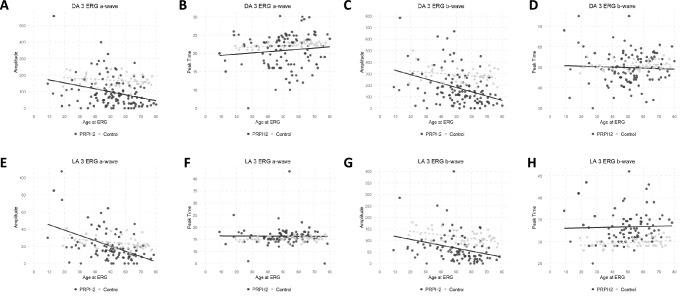
Full-field, pattern, and multifocal ERG (mfERG) data were available for 100 patients and this was performed at a median age of 52 years (range, 9–81 years). Of these patients, 29 (29%) had no generalized cone or rod dysfunction on full-field ERG. Of those remaining, 16 patients (16%) had isolated COD, 17 (17%) had CRD, and 38 (38%) had RCD (Table 2). Of the 29 patients with a normal full-field ERG, 26 had a PERG and 16 had a mfERG. Of those with a PERG, 8 (31%) were subnormal, whereas all 16 mfERG (100%) showed reduced response densities indicating macular dysfunction. Combining our PERG and mfERG data, 21 of 29 without generalized retinal dysfunction had evidence of localized macular dysfunction. Of the 12 with electrooculogram (EOG) and no generalized dysfunction on full-field ERG, 2 (16.7%) had a reduced Arden ratio (<1.7).38 Of the 87 patients with detectable a- and b-waves with the DA3.0 response, 7 (8%) had a reduced b:a ratio (<1.2) in one or both eyes. In contrast, 31 patients (31%) had a reduced b:a ratio (<3.0) with the LA3.0 response in one or both eyes. Thus, this reduced b:a ratio was more frequently observed with the LA3.0 response as compared with DA3.0 (Supplementary Material S3). A cohort of controls (female:male ratio of 20:24) with a mean age of 54 years (range, 19–77 years) were enrolled from one site and tested with the RETIport 3.2 (Roland Consult, Brandenburg, Germany). Comparisons were made between the controls and the entire PRPH2 cohort (Fig. 7). Information regarding the regression lines featured in Figures 7 and 8 is provided in Supplementary Material S3. Further comparisons were performed against a subset of PRPH2 patients who were tested under the same conditions and at the same institution as the controls (Table 3, Supplementary Material S3).
Figure 7.
Scatter plots for electrophysiologic parameters and age. The full-field ERG component amplitudes and peak times in PRPH2 patients (black circles) and unaffected controls (grey circles) are plotted against age. Data are shown for the DA 0.01 ERG (A and B), LA 30-Hz ERG (C and D). Regression lines are shown for the PRPH2-associated retinopathy (solid line) and control (grey broken line) data.
Figure 8.
Scatter plots for electrophysiologic parameters and age. The major full-field ERG component amplitudes and peak times in PRPH2 patients (black circles) and unaffected controls (grey circles) are plotted against age. Data are shown for the DA3.0 ERG a-wave (A, B) and b-wave (C, D), LA 3.0 ERG a-wave (E, F) and b-wave (G, H). Regression lines are shown for the PRPH2-associated retinopathy (solid line) and control (grey broken line) data.
Table 3.
Amplitudes and Peak Times of the Full-Field ERG Components in a Control Group Compared With PRPH2 at a Single Center
| Stimulus | Component | Parameter | PRPH2 n = 25 Median (5th, 95th) | Control n = 44 Median (5th, 95th) | P Value* |
|---|---|---|---|---|---|
| DA 0.01 | b-wave | Amplitude peak time | 81 (0, 349) | 162 (97, 253) | <0.001 |
| 94 (75, 110) | 93 (85, 101) | 1.000 | |||
| DA 3.0 | a-wave | Amplitude peak time | 72 (0, 271) | 163 (127, 235) | <0.001 |
| 22 (13, 26) | 22 (21, 23) | 1.000 | |||
| b-wave | Amplitude peak time | 137 (13, 466) | 280 (211, 372) | <0.001 | |
| 50 (37, 62) | 50 (48, 55) | 1.000 | |||
| LA 30-Hz | flicker | Amplitude peak time | 39 (4, 123) | 81 (53, 126) | <0.001 |
| 30 (26, 42) | 27 (25, 28) | <0.001 | |||
| LA 3.0 | a-wave | Amplitude peak time | 15 (0, 56) | 23 (16, 38) | 0.003 |
| 16 (13, 20) | 15 (14, 17) | 0.214 | |||
| b-wave | Amplitude peak time | 52 (4, 168) | 107 (77, 166) | <0.001 | |
| 33 (29, 41) | 30 (28, 32) | <0.001 |
DA, dark adapted; LA, light adapted.
Amplitudes are in microvolts and peak times are in milliseconds. Statistical significance was established using the Mann–Whitney U test.
Statistically significant after Bonferroni correction.
Our PRPH2 cohort showed a steeper decline in amplitude with age for both the DA0.01 and LA30 Hz when compared with controls (Fig. 7). The peak time was delayed in the LA30 Hz indicating cone system dysfunction. The a-wave in the DA3.0 and LA3.0 showed a greater age-related decline in the PRPH2 group as compared with controls (Fig. 8). A delayed peak time was observed in the LA3.0 b-wave. A subanalysis at one Australian site between the PRPH2 cohort and controls showed a significant difference in all DA and LA amplitude parameters (P < 0.001) with significant delays in the LA30 Hz and LA3.0 b-wave implicit times (P < 0.001) (Table 3). Of those exhibiting a normal or BPD/VMD FAF phenotype, most had a normal ERG or isolated macular dysfunction, whereas those with a PSPD or RP phenotype typically showed generalized rod and cone abnormalities (Table 2). Conversely, those with CACD had a varied electrophysiological phenotype ranging from a normal full-field and PERG (n = 4), MD (n = 9), COD (n = 7), to CRD (n = 2) (Supplementary Material S3).
Genetic Characteristics
Ninety-one unique variants were identified consisting of 46 missense, 5 inframe deletions, 21 frameshift, 12 nonsense, 2 exon deletions, 4 splice-site, and 1 start–loss. All variants were likely pathogenic or pathogenic based on American College of Medical Genetics criteria (Tables 4 and 5, Supplementary Material S4). Among the 46 missense variants at 36 codon positions, 5 amino acid substitutions at 5 codon positions and 1 inframe deletion–insertion were novel (Fig. 9, Supplementary Material S4). Sixteen of the 40 truncating or start–loss variants were also novel.
Table 4.
Missense Variant List With In Silico Prediction and Phenotype
| Codon | cDNA | Amino Acid | Exon | Protein Domain | PolyPhen2 Var | SIFT | REVEL | CADD | Mutation Taster | Grantham Score | Clinical Phenotype | ACMG Class | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13 | c.37C>T | p.Arg13Trp | 1 | NH2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.502 | 25.3 | Deleterious (88|12) | 101 | RP | 5 | Sohocki et al.64; Koyanagi et al.65; Peeters et al.39 |
| 38 | c.112G>A | p.Gly38Arg | 1 | 1st TM | Probably damaging (1.000) | Affect protein function (0.00) | 0.876 | 26.6 | Deleterious (81|19) | 125 | PSPD | 4 | Novel |
| 41 | c.122T>C | p.Leu41Pro | 1 | 1st TM | Probably damaging (0.981) | Affect protein function (0.00) | 0.916 | 27.3 | Deleterious (77|23) | 98 | BPD | 4 | Peeters et al.39; Bianco et al.9 |
| 138 | c.413T>G | p.Met138Arg | 1 | D2 | Probably damaging (0.99) | Affect protein function (0.00) | 0.856 | 27.5 | Deleterious (85|15) | 91 | PSPD | 4 | Bianco et al.9 |
| 141 | c.422A>G | p.Tyr141Cys | 1 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.881 | 29.3 | Deleterious (86|14) | 194 | PSPD | 5 | Sohocki et al.64; Stone et al.67; Sullivan et al.66; Khani et al.68 |
| 142 | c.424C>T | p.Arg142Trp | 1 | D2 | Probably damaging (0.948) | Affect protein function (0.00) | 0.635 | 25 | Deleterious (74|26) | 101 | CACD | 5 | Hoyng et al.10; Klevering et al.69; Boon et al.20 |
| 153 | c.457A>G | p.Lys153Glu | 1 | D2 | Probably damaging (0.999) | Affect protein function (0.00) | 0.864 | 28.3 | Deleterious (89|11) | 56 | PSPD | 5 | Peeters et al.39 |
| c.458A>G | p.Lys153Arg | 1 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.828 | 27.8 | Deleterious (85|15) | 26 | RP | 5 | Jacobson et al.70; Reeves et al.47; Peeters et al.39 | |
| 167 | c.499G>A | p.Gly167Ser | 1 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.953 | 28.8 | Deleterious (84|16) | 56 | N; BPD; VMD; PSPD; RP | 5 | Testa et al.71; Coco-Martin et al.72 |
| 172 | c.514C>T | p.Arg172Trp | 1 | D2 | Probably damaging (0.967) | Affect protein function (0.00) | 0.67 | 26 | Deleterious (80|20) | 101 | CACD | 5 | Wells et al.11; Duncan et al.73; Reeves et al.47 |
| c.515G>A | p.Arg172Gln | 1 | D2 | Benign (0.103) | Affect protein function (0.03) | 0.453 | 26.6 | Deleterious (64|36) | 43 | CACD | 5 | Wells et al.11; Wroblewski et al.12; Payne et al.50 | |
| c.515G>C | p.Arg172Pro | 1 | D2 | Possibly damaging (0.886) | Affect protein function (0.01) | 0.63 | 27.3 | Deleterious (57|43) | 103 | CACD | 4 | LOVD | |
| c.515G>T | p.Arg172Leu | 1 | D2 | Possibly damaging (0.642) | Affect protein function (0.01) | 0.491 | 26.8 | Benign (45|55) | 102 | CACD | 4 | Novel | |
| 178 | c.533A>G | p.Glu178Arg | 1 | D2 | Probably damaging (0.978) | Affect protein function (0.00) | 0.875 | 26.6 | Deleterious (93|7) | 43 | PSPD; RP | 5 | Peeters et al.39 |
| 179 | c.535T>G | p.Trp179Gly | 1 | D2 | Probably damaging (0.999) | Affect protein function (0.00) | 0.909 | 29.9 | Deleterious (90|10) | 184 | RP | 5 | Colombo et al.74 |
| 185 | c.554T>C | p.Leu185Pro | 1 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.915 | 29 | Deleterious (98|2) | 98 | BPD | 5 | Peeters et al.39 |
| 186 | c.556G>A | p.Asp186Asn | 1 | D2 | Benign (0.124) | Affect protein function (0.00) | 0.382 | 23.6 | Deleterious (94|6) | 23 | CACD | 4 | Kohl et al.13; Martin-Merida et al.75; Almoguera et al.76 |
| 195 | c.584G>A | p.Arg195Gln | 2 | D2 | Probably damaging (0.978) | Affect protein function (0.00) | 0.761 | 32 | Deleterious (58|42) | 43 | CACD | 5 | Da Palma et al.17; Alapati et al.51 |
| 196 | c.587T>A | p.Ile196Asn | 2 | D2 | Possibly damaging (0.815) | Affect protein function (0.00) | 0.306 | 29.4 | Deleterious (61|39) | 194 | CACD; PSPD | 5 | Reeves et al.47; Peeters et al.39; Bianco et al.9 |
| 198 | c.594C>A | p.Ser198Arg | 2 | D2 | Probably damaging (0.999) | Affect protein function (0.00) | 0.258 | 26.7 | Deleterious (88|12) | 110 | N; PSPD; RP | 4 | Novel |
| c.594C>G | p.Ser198Arg | 2 | D2 | Probably damaging (0.983) | Affect protein function (0.00) | 0.259 | 26.4 | Deleterious (88|12) | 110 | RP | 5 | Sullivan et al.66; Manes et al.77; Peeters et al.39 | |
| 200 | c.599T>C | p.Val200Ala | 2 | D2 | Benign (0.314) | Affect protein function (0.00) | 0.281 | 28.9 | Deleterious (80|20) | 64 | CACD | 4 | ClinVar, LOVD |
| 208 | c.623G>A | p.Gly208Asp | 2 | D2 | Benign (0.206) | Affect protein function (0.03) | 0.712 | 22.6 | Deleterious (41|59) | 94 | N; CACD | 4 | Reeves et al.47; Bianco et al.9; Trujillo et al.47; Birtel et al.58 |
| 209 | c.625G>A | p.Val209Ile | 2 | D2 | Probably damaging (0.998) | Affect protein function (0.00) | 0.631 | 26.4 | Benign (27|73) | 29 | RP | 5 | Coco-Martin et al.72 |
| c.625G>T | p.Val209Phe | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.927 | 29.1 | Deleterious (90|10) | 50 | PSPD | 5 | Carss et al.78; Peeters et al.39 | |
| 210 | c.628C>T | p.Pro210Ser | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.928 | 28.7 | Deleterious (92|8) | 74 | PSPD | 5 | Peeters et al.39 |
| c.629C>G | p.Pro210Arg | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.89 | 27.5 | Deleterious (95|5) | 103 | BPD | 5 | Boon et al.20; Duncan et al.73; Reeves et al.47 | |
| 211 | c.633C>A | p.Phe211Leu | 2 | D2 | Probably damaging (0.997) | Affect protein function (0.00) | 0.771 | 24.1 | Deleterious (92|8) | 22 | RP | 5 | Ekstrom et al.79; Peeters et al.39 |
| 212 | c.634A>C | p.Ser212Arg | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.957 | 28.7 | Deleterious (82|18) | 110 | PSPD | 5 | ClinVar |
| c.635G>A | p.Ser212Asn | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.797 | 28.5 | Deleterious (70|30) | 46 | PSPD | 4 | Novel | |
| c.636C>G | p.Ser212Arg | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.901 | 32 | Deleterious (96|4) | 110 | N; PSPD | 5 | ClinVar | |
| 213 | c.638G>A | p.Cys213Tyr | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.946 | 31 | Deleterious (100|0) | 194 | PSPD | 5 | Zhang et al.80; Duncan et al.73; Stone et al.67 |
| 214 | c.642C>G | p.Cys214Trp | 2 | D2 | Probably damaging (1.000) | Affect protein function (0.00) | 0.874 | 27.7 | Deleterious (99|1) | 215 | PSPD | 5 | Bianco et al.9 |
| 216 | c.646C>T | p.Pro216Ser | 2 | D2 | Possibly damaging (0.771) | Affect protein function (0.00) | 0.682 | 25.5 | Deleterious (81|19) | 74 | PSPD | 5 | Fishman et al.81; Stone et al.67; Coco-Martin et al.72 |
| 218 | c.653C>G | p.Ser218Trp | 2 | D2 | Probably damaging (0.999) | Affect protein function (0.00) | 0.764 | 27.9 | Deleterious (80|20) | 177 | BPD | 4 | Novel |
| 219 | c.656C>G | p.Pro219Arg | 2 | D2 | Possibly damaging (0.713) | Affect protein function (0.00) | 0.731 | 23.3 | Deleterious (83|17) | 103 | N; CACD | 5 | Payne et al.50; Peeters et al.39 |
| 220 | c.659G>A | p.Arg220Gln | 2 | D2 | Probably damaging 0.987 | Affect protein function (0.00) | 0.724 | 31 | Deleterious (85|15) | 43 | BPD; PSPD | 5 | Jacobson et al.70; Ellingford et al.82; |
| c.659G>C | p.Arg220Pro | 2 | D2 | Probably damaging (0.993) | Affect protein function (0.00) | 0.882 | 31 | Deleterious (89|11) | 103 | PSPD | 5 | Reeves et al.47; Peeters et al.39 | |
| 222 | c.665G>C | p.Cys222Ser | 2 | D2 | Possibly damaging (0.999) | Affect protein function (0.00) | 0.918 | 29.1 | Deleterious (95|5) | 112 | PSPD; RP | 4 | Ahmad et al.83; Liu et al.84; Peeter et al.39 |
| 228 | c.683C>T | p.Thr228Ile | 2 | D2 | Possibly damaging (0.776) | Affect protein function (0.00) | 0.771 | 24.8 | Deleterious (92|8) | 89 | CACD | 4 | Alapati et al.51; Reeves et al.47; Peeters et al.39 |
| 232 | c.695C>T | p.Ala232Val | 2 | D2 | Possibly damaging (0.871) | Affect protein function (0.00) | 0.694 | 26.8 | Deleterious (69|31) | 64 | CACD | 4 | ClinVar |
| 246 | c.738G>C | p.Trp246Cys | 2 | D2 | Possibly damaging (0.986) | Affect protein function (0.00) | 0.956 | 32 | Deleterious (87|13) | 215 | RP | 4 | Manes et al.77; Peeters et al.39 |
| 250 | c.749G>C | p.Cys250Ser | 2 | D2 | Probably Damaging (1.000) | Affect protein function (0.00) | 0.968 | 28.6 | Deleterious (97|3) | 112 | PSPD | 5 | ClinVar |
| 284 | c.850C>G | p.Arg284Gly | 3 | COOH | Probably damaging (0.976) | Affect protein function (0.01) | 0.753 | 26.2 | Benign (41|59) | 125 | BPD | 4 | Bianco et al.9 |
| 289 | c.866C>T | p.Ser289Leu | 3 | COOH | Possibly damaging (0.501) | Affect protein function (0.00) | 0.212 | 24.5 | Deleterious (69|31) | 145 | RP | 5 | de Breuk et al.85; Peeters et al.39 |
| 316 | c.946T>G | p.Trp316Gly | 3 | COOH | Benign (0.031) | Tolerated (0.07) | 0.157 | 22.7 | Deleterious (67|33) | 184 | RP | 4 | Huang et al.86; Peeters et al.39 |
ACMG, American College of Medical Genetics; COOH, carboxyl terminus; D2, D2 loop; NH2, amine terminus; TM, transmembrane.
Table 5.
Truncating and Start–Loss Variant List With Phenotype
| Codon | Nucleotide Change | Protein Consequence | Mutation Type | Exon | Protein Domain | Phenotype | ACMG Class | References |
|---|---|---|---|---|---|---|---|---|
| c.1A>T | p.Met1? | Start loss | 1 | N/A | PSPD | 5 | Peeters et al.39 | |
| c.(828+1_829-1)_(*1_?)del | Exon 3 deletion | Exon deletion | 3 | N/A | BPD; PSPD | 4 | Novel | |
| deletion of exon 1 | Exon 1 deletion | Exon deletion | 1 | N/A | N; PSPD | 5 | Novel | |
| c.582-1G>A | p.(?) | Splicing | intron 1 | N/A | PSPD | 5 | Reeves et al.47; Bianco et al.9 | |
| c.828+1G>A | p.(?) | Splicing | intron 2 | N/A | BPD | 5 | Peeters et al.39 | |
| c.828+2T>C | p.(?) | Splicing | intron 2 | N/A | PSPD | 5 | Peeters et al.39 | |
| c.828+3A>T | p.(?) | Splicing | intron 2 | N/A | PSPD | 5 | Peeters et al.39; Sullivan et al.94 | |
| 46 | c.136C>T | p.Arg46Ter | Nonsense | 1 | D1 | BPD; PSPD; RP | 5 | Bianco et al.9 |
| 97 | c.290G>A | p.Trp97Ter | Nonsense | 1 | C | BPD; VMD; PSPD; RP | 5 | Bianco et al.9 |
| 97 | c.291G>A | p.Trp97Ter | Nonsense | 1 | 3rd TM | N | 4 | Novel |
| 126 | c.372_381delinsAAGCTGA | p.Leu126Ter | Nonsense | 1 | D2 | N | 4 | LOVD |
| 179 | c.536G>A | p.Trp179Ter | Nonsense | 1 | D2 | RP | 4 | LOVD |
| 191 | c.571G>T | p.Glu191Ter | Nonsense | 1 | D2 | PSPD; RP | 5 | Birtel et al.58; Duncker et al.95; Peeters et al.39 |
| 195 | c.583C>T | p.Arg195Ter | Nonsense | 2 | D2 | PSPD | 5 | Peeters et al.39; Alapati et al.51 |
| 197 | c.589A>T | p.Lys197Ter | Nonsense | 2 | D2 | PSPD | 4 | Novel |
| 204 | c.612C>G | p.Tyr204Ter | Nonsense | 2 | D2 | N; BPD; PSPD; RP | 5 | Birtel et al.58; Peeters et al.39 |
| 236 | c.708C>A | p.Tyr236Ter | Nonsense | 2 | D2 | PSPD | 5 | Novel |
| 257 | c.771_772delinsGA | p.Tyr257Ter | Nonsense | 2 | D2 | PSPD | 4 | Bianco et al.9 |
| 300 | c.897dup | p.Glu300Ter | Nonsense | 3 | COOH | BPD | 4 | Novel |
| 1 | c.1_7del | p.Met1TyrfsTer2 | Frameshift | 1 | NH2 | BPD; PSPD; RP | 4 | Novel |
| 12 | c.33del | p.Lys12SerfsTer12 | Frameshift | 1 | 1st TM | BPD | 5 | Novel |
| 23 | c.68del | p.Met23ArgfsTer15 | Frameshift | 1 | 1st TM | VMD; PSPD | 4 | Bianco et al.9 |
| 25 | c.73_74del | p.Trp25ValfsTer19 | Frameshift | 1 | 1st TM | VMD; PSPD | 5 | Kajiwara et al.87 |
| 38 | c.113del | p.Gly38AspfsTer4 | Frameshift | 1 | 1st TM | BPD | 5 | Yang et al.88; Reeves et al.47 |
| 42 | c.115delinsTTGTTCCT | p.Lys42ValfsTer5 | Frameshift | 1 | D1 | PSPD | 4 | Novel |
| 60 | c.172_176dup | p.Val60IlefsTer7 | Frameshift | 1 | 2nd TM | PSPD | 5 | Novel |
| 72 | c.212_213insAT | p.Cys72SerfsTer28 | Frameshift | 1 | C | RP | 4 | Novel |
| 87 | c.259_266del | p.Asp87GlnfsTer87 | Frameshift | 1 | D2 | PSPD | 5 | Reeves et al.47; Carss et al.78 |
| 132 | c.394del | p.Gln132LysfsTer7 | Frameshift | 1 | D2 | VMD; PSPD; RP | 5 | Boon et al.40 |
| 147 | c.440del | p.Pro147LeufsTer6 | Frameshift | 1 | D2 | PSPD | 4 | Novel |
| 148 | c.441del | p.Gly148AlafsTer5 | Frameshift | 1 | D2 | PSPD | 5 | Kajiwara et al.87; Boon et al.40; Birtel et al.58 |
| 156 | c.468_481delinsG | p.Ile156MetfsTer96 | Frameshift | 1 | D2 | PSPD | 4 | Novel |
| 193 | c.578_579del | p.Lys193ArgfsTer24 | Frameshift | 1 | D2 | BPD | 5 | Birtel et al.58 |
| 216 | c.646_649delinsGG | p.Pro216GlyfsTer84 | Frameshift | 2 | COOH | PSPD | 5 | Reeves et al.47 |
| 236 | c.702_706dup | p.Tyr236SerfsTer22 | Frameshift | 2 | D2 | PSPD | 4 | Bianco et al.9 |
| 246 | c.734dup | p.Trp246ValfsTer55 | Frameshift | 3 | COOH | PSPD | 4 | Sodi et al.89 |
| 301 | c.903del | p.Ser301ArgfsTer23 | Frameshift | 3 | COOH | RP | 4 | Novel |
| 304 | c.910_911insG | p.Gln304ArgfsTer88 | Frameshift+Elong | 3 | COOH | RP | 4 | Novel |
| 315 | c.943dup | p.Thr315AsnfsTer77 | Frameshift+Elong | 3 | COOH | RP | 4 | Novel |
| 322 | c.964_965del | p.Ser322CysfsTer69 | Frameshift+Elong | 3 | COOH | PSPD | 4 | Reeves et al.47 |
| 154 | c.461_463del | p.Lys154del | Inframe deletion | 1 | D2 | PSPD | 5 | Weleber et al.90 |
| 169 | c.505_507del | p.Asn169del | Inframe deletion | 1 | D2 | PSPD | 5 | van Lith-Verhoeven et al.91 |
| 207 | c.618_626del | p.Asp207_Val209del | Inframe deletion | 2 | D2 | N; RP | 5 | Kalyanasundaram et al.92 |
| 237 | c.709_714delinsACA | p.Asp237_His238delinsThr | Inframe deletion | 2 | D2 | N: VMD: RP | 4 | Novel |
| 271 | c.811_813del | p.Leu271del | Inframe deletion | 2 | D2 | N; PSPD; RP | 5 | Jin et al.93 |
ACMG, American College of Medical Genetics; COOH, carboxyl terminus; D2, D2 loop; NH2, amine terminus; TM, transmembrane.
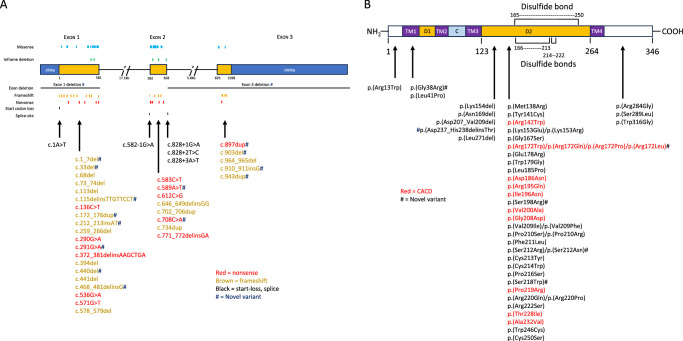
Figure 9.
Position and type of PRPH2 truncating variants (nonsense, frameshift, start-loss, splice site) by exon location (A). Missense and inframe deletion variants and their protein positions within our PRPH2 cohort (B).
Of the 51 variants that altered protein sequence, 45 were localized to the D2 loop, whereas 1 resided in the N-terminal, 2 were in the transmembrane domain 1 (TMD1), and 3 were in the C-terminal (Fig. 9). The most common missense variants were at codon positions Arg172 (n = 35), Gly208 (n = 16), Gly167 (n = 12), and Glu178 (n = 10). There were 33 frameshift or nonsense variants occurring within exon 1 (n = 20), exon 2 (n = 7), and exon 3 (n = 6) that elicited a premature stop codon. The most common truncating variants occurred at positions Trp97Ter (n = 9), Tyr204Ter (n = 8), and Arg46Ter (n = 7) and at nucleotides c.394del p.(Gln132LysfsTer7) (n = 6) and c.259_266del p.(Asp87GlnfsTer87) (n = 6).
There were 71 patients with a CACD FAF phenotype carrying 13 missense variants at 10 codon positions. One of these missense variants p.(Ile196Asn) also manifested PSPD in one case and two cases with p.(Gly208Asp) and p.(Pro219Arg) had a normal FAF. In 85 patients with truncating, splice or start–loss variants, FAF phenotypes included PSPD (58%), BPD/VMD (21%), RP (16%), and normal (5%). The three most common frameshift and nonsense variants showed a similar FAF distribution (PSPD 57.5%, BPD/VMD 22.5%, RP 17.5%, and normal 2.5%). Notably, no truncating variants manifested a CACD phenotype. The three most common missense variants: p.(Tyr141Cys), p.(Gly167Ser) and p.(Glu178Arg) showed a similar FAF phenotype distribution (PSPD 67.7%, RP 22.6%, BPD/VMD 6.5%, and normal 3.2%). In-frame deletions appeared to be associated with a larger proportion of patients with a normal FAF (33% vs. 5%) and a lower frequency of PSPD as compared with truncating or null variants (33% vs. 58%).
Discussion
This large, international, multicenter study describes the clinical and genetic spectrum of 241 patients with 91 unique pathogenic or likely pathogenic PRPH2 variants. Although the severity of PRPH2-associated IRD can vary considerably, VA often remained relatively stable until the fifth decade, with a constant decline thereafter. There was a range of retinal presentations including five distinct FAF phenotypes. Within each FAF phenotype group, there was often a spectrum of electrophysiological changes. Importantly, we observed 13 specific missense variants in the PRPH2 gene that were strongly associated with a CACD phenotype. Conversely truncating variants resulted in variable FAF phenotypes.
In previous PRPH2 cohorts, clinical and electrophysiological terms have often been used without a clear distinction between their structural and functional associations.9,25,39 Some studies classified patients as having a pattern dystrophy based on morphological features, including macular pigmentary changes in combination with functional features such as a normal or minimally abnormal full-field ERG.9 In contrast, we classified patients into normal, BPD, VMD, PSPD, CACD, and RP groups based on their FAF pattern. We showed one FAF phenotype may have multiple ERG phenotypes and vice versa (Table 2). For example, PSPD was associated with MD (21%), COD (11%), CRD (34%), and RCD (34%) ERG phenotypes. Our observation of concurrent cone and rod dysfunction (68%) was like that reported by Boon et al. where one-half of the 17 patients with PSPD had cone and rod dysfunction.40 Conversely, a COD was observed across the BPD, CACD, and PSPD FAF phenotypes groups. We observed a reduced b:a ratio in 8% and 31% of the rod and cone responses, respectively. This may be attributable to additional inner retinal dysfunction as previously described by Ba-Abbad et al.41 The reduced b:a ratio in the DA3.0 response may also represent a predominance of DA cone responses with markedly reduced rods (manifesting as a photopic hill phenomenon in the dark). Thus, future studies need to incorporate the red DA3.0 and DA10.0 responses to look for DA cone responses and the photopic hill phenomenon, respectively.42 This discordance between FAF and ERG phenotyping supports our proposal that clinicians should incorporate both standardized FAF and ERG grading systems. Because three ERG groups have been proposed in ABCA4-associated retinopathy,43 future longitudinal studies should investigate the prognostic value of ERG grading for predicting visual outcomes in PRPH2-asssociated IRD.
Of 91 different PRPH2 variants, there were 21 frameshift, 12 nonsense, 2 exon deletions, 4 splice-site, and 1 start–loss, of which most were expected to result in loss of function or haploinsufficiency. Animal studies44,45 suggest that haploinsufficiency affects rods more than cones, leading to RP. However, clinical case series have shown conflicting genotype–phenotype correlations. In a case series of 40 Japanese patients harboring 17 PRPH2 variants, Oishi et al.46 found no clear genotype–phenotype correlations. Reeves et al.47 found an association of exon 1 variants with CRD, PSPD, and RP, whereas a pattern dystrophy was associated with variants in exon 2. Bianco et al.9 observed that loss-of-function variants were associated with a mild pattern dystrophy and the addition of ABCA4 variants resulted in a more severe phenotype. Peeters et al.39 listed the missense variants exclusively associated with RP or a pattern dystrophy. In contrast with animal studies, we demonstrated a predominance of PSPD in more than one-half of these patients. This result was replicated in a subanalysis of 40 patients carrying the three most common truncating variants. Variable FAF phenotypes were observed in 73 patients with missense variants that were not associated with a CACD phenotype. The similar FAF phenotypic spectrum of these missense variants to truncating variants suggests these specific amino acid substitutions may lead to haploinsufficiency through retention or mislocalization of the PRPH2 protein to the photoreceptor inner segment.48,49 Our observation that none of the 81 patients with truncating variants demonstrated a CACD FAF phenotype warrants further consideration.
CACD was observed in 71 patients harboring 13 missense variants at 10 codon positions. Nearly all of these variants exhibited a CACD phenotype with the exception of p.(Ile196Asn), which also manifested as a PSPD in one patient. Before this paper the CACD phenotype had only been associated with missense variants at five codon positions. The p.(Pro219Arg) and p.(Thr228Ile) variants have been reported with a macular and pattern dystrophy respectively, despite no FAF imaging.50,51 Similarly, p.(Asp186Asn) was reported to manifest a CRD and RP phenotype without supportive FAF imaging.13 Although substitutions at Arg172 to Trp or Gln have been described, p.(Arg172Pro) and p.(Arg172Leu) have not been reported previously to cause CACD. Despite previous reports suggesting truncating PRPH2 variants can cause CACD, we propose CACD only presents with specific missense variants. Antonelli et al.25 described a case of CACD with p.(Trp246ValfsTer55). FAF imaging, however, was atypical, demonstrating central macular hypoautofluorescence, and hyperautofluorescence fleck-like deposits that extended beyond the vascular arcades. Daftarian et al.52 described p.(Gln239Ter) to be associated with CACD. Their FAF imaging, however, was more suggestive of a PSPD. To date, Boon et al.20 have published the largest case series of CACD including 103 patients, most of whom carried the p.(Arg142Trp) variant. Other PRPH2 variants, including p.(Arg172Trp),53–55 p.(Arg172Gln),53,12 p.(Arg195Gln),17,51 p.(Arg195Leu),14,15,56 p.(Asp196Asn),9,39,47 p.(Arg203Pro),16 and p.(Gly208Asp)13,57,58 have also been associated with CACD. Nonpenetrance has been associated with p.(Arg172Trp) up to the age of 24.5 Full penetrance was initially described with p.(Arg142Trp); however, Boon et al.4 later described three families with evidence of nonpenetrance. In our cohort, two cases with normal FAF harbored the p.(Gly208Asp) and p.(Pro219Arg) variants; the former was reported previously to have incomplete penetrance.59 Both variants were also shown to exhibit a CACD phenotype in this study. We found some carriers may remain asymptomatic up to 69 years of age. Within our cohort, nonpenetrance, defined as an absence of macular abnormalities on fundus color or FAF imaging, was seen in nearly 6% of patients (13/241) and observed up to 71 years of age. In contrast, Boon et al.4 proposed nonpenetrance may be up to 21% in CACD-specific variants; however, this estimate may not be generalizable to our cohort. Finally, a knock-in animal model of CACD due to p.(Arg195Leu) demonstrated a progressive decrease in VA and ERG amplitudes in both scotopic and photopic conditions.60 Our ERG data for CACD showed a similar trend toward greater cone than rod involvement as reported previously by Hoyng and Deutman.19 It is intriguing that these animals exhibited evidence of disrupted communication between photoreceptors, bipolar and horizontal cells, supporting the observation of a reduced b:a ratio.
The current study is subject to the known limitations of retrospective clinical data entry, such as missing data and variable protocols, as well as devices used for VA, FAF, and ERG acquisition. A prior study described discordance in VA derived from a Snellen chart compared with a logMAR chart.61 Thus, any observed trend in VA should be interpreted with caution. Future prospective studies using a standardized VA measurement will reduce this bias. An analysis of age-related changes in VA, FAF and ERG was not possible owing to an absence of longitudinal data. Examination of the ERG waveforms for the DA10.0 response would have been useful to elucidate the presence of the photopic hill phenomenon given the reduced b:a ratio we observed in the DA3.0. In addition, we did not include juxtafoveal lesions as part of the CACD spectrum, because these were often difficult to differentiate from VMD and BPD FAF phenotypes. Future prospective studies with structural and functional parameters will enhance our current knowledge of disease progression and provide more reliable and standardized biomarkers for tracking PRPH2 disease progression.62,63
This study found a large variability in VA, FAF, and ERG phenotypes in patients with molecularly confirmed pathogenic or likely pathogenic variants in the PRPH2 gene. We observed significant discordance between FAF and ERG phenotypes. To our knowledge, this work represents the largest PRPH2 cohort with multimodal imaging. We report a novel genotype–phenotype correlation whereby patients harboring 13 missense variants at 10 codon positions predominately manifested a CACD phenotype. Additional studies, including cellular disease models of CACD-specific variants and longitudinal evaluation with prospective data, are still required. The detailed clinical and genetic information provided here will be useful for clinicians to aid their work-up of patients with a PRPH2-associated IRD.
Supplementary Material
Acknowledgments
Disclosure: R.C. Heath Jeffery, None; J.A. Thompson, Retina Australia; J. Lo, None; E.S. Chelva, None; S. Armstrong, None; J.S. Pulido, None; R. Procopio, None; A.L. Vincent, None; L. Bianco, None; M. Battaglia Parodi, None; L. Ziccardi, None; G. Antonelli, None; L. Barbano, None; J.P. Marques, None; S. Geada, None; A. L. Carvalho, None; W. Chao Tang, None; C. Mun Chan, None; C.J.F. Boon, None; J. Hensman, None; T.-C. Chen, None; C.-Y. Lin, None; P.-L. Chen, None; A. Vincent, None; A. Tumber, None; E. Heon, None; J.R. Grigg, NHMRC APP1116360, APP1099165, APP1109056; R.V. Jamieson, NHMRC APP1116360, APP1099165, APP1109056; E.E. Cornish, None; B.M. Nash, None; S. Borooah, Foundation Fighting Blindness Grant CD-GT-0918-0746-SEI and Nixon Visions Foundation; L.N. Ayton, Novartis, Apellis, National Health & Medical Research Council (NHMRC) GNT1195713; A.C. Britten-Jones, None; T.L. Edwards, None; J.B. Ruddle, None; A. Sharma, None; R.G. Porter, None; T.M. Lamey, Retina Australia; T.L. McLaren, Retina Australia; S. McLenachan, None; D. Roshandel, None; F.K. Chen, Future Health Research and Innovation Fund, the McCusker Charitable Foundation, Channel 7 Telethon Trust, Retina Australia, NHMRC GNT1116360, GNT1188694, GNT1054712 and MRF1142962
References
- 1. Heath Jeffery RC, Mukhtar SA, McAllister IL, et al.. Inherited retinal diseases are the most common cause of blindness in the working-age population in Australia. Ophthalmic Genet. 2021; 42(4): 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pontikos N, Arno G, Jurkute N, et al.. Genetic basis of inherited retinal disease in a molecularly characterized cohort of more than 3000 families from the United Kingdom. Ophthalmology. 2020; 127(10): 1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN.. The human gene mutation database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014; 133(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boon CJ, den Hollander AI, Hoyng CB, Cremers FP, Klevering BJ, Keunen JE.. The spectrum of retinal dystrophies caused by mutations in the peripherin/RDS gene. Prog Retina Eye Res. 2008; 27(2): 213–235. [DOI] [PubMed] [Google Scholar]
- 5. Michaelides M, Holder GE, Bradshaw K, Hunt DM, Moore AT.. Cone-rod dystrophy, intrafamilial variability, and incomplete penetrance associated with the R172W mutation in the peripherin/RDS gene. Ophthalmology. 2005; 112(9), 1592–1598. [DOI] [PubMed] [Google Scholar]
- 6. Ikelle L, Makia M, Lewis T, et al.. Comparative study of PRPH2 D2 loop mutants reveals divergent disease mechanism in rods and cones. Cell Mol Life Sci. 2023; 80(8): 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Poloschek CM, Bach M, Lagrèze WA, et al.. ABCA4 and ROM1: implications for modification of the PRPH2-associated macular dystrophy phenotype. Invest Ophthalmol Vis Sci. 2010; 51(8): 4253–4265. [DOI] [PubMed] [Google Scholar]
- 8. Leroy BP, Kailasanathan A, De Laey JJ, Black GC, Manson FD. Intrafamilial phenotypic variability in families with RDS mutations: Exclusion of ROM1 as a genetic modifier for those with retinitis pigmentosa. Br J Ophthalmol. 2007; 91(1): 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bianco L, Arrigo A, Antropoli A, et al.. PRPH2-associated retinopathy: novel variants and genotype-phenotype correlations. Ophthalmol Retina. 2023; 7(5): 450–461. [DOI] [PubMed] [Google Scholar]
- 10. Hoyng CB, Heutink P, Testers L, Pinckers A, Deutman AF, Oostra BA.. Autosomal dominant central areolar choroidal dystrophy caused by a mutation in codon 142 in the peripherin/RDS gene. Am J Ophthalmol. 1996; 121(6): 623–629. [DOI] [PubMed] [Google Scholar]
- 11. Wells J, Wroblewski J, Keen J, et al.. Mutations in the human retinal degeneration slow (RDS) gene can cause either retinitis pigmentosa or macular dystrophy. Nat Genet. 1993; 3(3): 213–218. [DOI] [PubMed] [Google Scholar]
- 12. Wroblewski JJ, Wells JA 3rd, Eckstein A, et al.. Macular dystrophy associated with mutations at codon 172 in the human retinal degeneration slow gene. Ophthalmology. 1994; 101(1): 12–22. [DOI] [PubMed] [Google Scholar]
- 13. Kohl S, Christ-Adler M, Apfelstedt-Sylla E, et al.. RDS/peripherin gene mutations are frequent causes of central retinal dystrophies. J Med Genetics. 1997; 34(8): 620–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yanagihashi S, Nakazawa M, Kurotaki J, Sato M, Miyagawa Y, Ohguro H.. Autosomal dominant central areolar choroidal dystrophy and a novel Arg195Leu mutation in the peripherin/RDS gene. Arch Ophthalmol. 2003; 121(10): 1458–1461. [DOI] [PubMed] [Google Scholar]
- 15. Keilhauer CN, Meigen T, Weber BH.. Clinical findings in a multigeneration family with autosomal dominant central areolar choroidal dystrophy associated with an Arg195Leu mutation in the peripherin/RDS gene. Arch Ophthalmol. 2006; 124(7): 1020–1027. [DOI] [PubMed] [Google Scholar]
- 16. Choi H, Cloutier A, Lally D.. PRPH2-associated macular dystrophy in 4 family members with a novel mutation. Ophthalmic Genet. 2022; 43(2): 235–239. [DOI] [PubMed] [Google Scholar]
- 17. Da Palma MM, Martin D, Salles MV, et al.. Retinal dystrophies and variants in PRPH2. Arq Bras Oftalmol. 2019; 82(2): 158–160. [DOI] [PubMed] [Google Scholar]
- 18. Marques JP, Carvalho AL, Henriques J, et al.. Design, development and deployment of a web-based interoperable registry for inherited retinal dystrophies in Portugal: the IRD-PT. Orphanet J Rare Dis. 2020; 15(1): 304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hoyng CB, Deutman AF.. The development of central areolar choroidal dystrophy. Graefes Arch Clin Exp Ophthalmol. 1996; 234(2): 87–93. [DOI] [PubMed] [Google Scholar]
- 20. Boon CF, Klevering JB, Cremers FPM, et al.. Central areolar choroidal dystrophy. Ophthalmology. 2009; 116(4): 771–782. [DOI] [PubMed] [Google Scholar]
- 21. Robson AG, Frishman LJ, Grigg J, et al.. ISCEV Standard for full-field clinical electroretinography (2022 update). Doc Ophthalmol. 2022; 144: 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bach M, Brigell M, Hawlina M, et al.. ISCEV standard for clinical pattern electroretinography (PERG) - 2012 update. Doc Ophthalmol. 2013; 126: 1–7. [DOI] [PubMed] [Google Scholar]
- 23. Hood D, Bach M, Brigell M, et al.. ISCEV Standard for clinical multifocal electroretinography (2011 edition). Doc Ophthalmol. 2012; 124: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heath Jeffery RC, Thompson JA, Lamey TM, et al.. Longitudinal analysis of functional and structural outcome measures in PRPH2-associated retinal dystrophy. Ophthalmol Retina. 2023; 7(1): 81–91. [DOI] [PubMed] [Google Scholar]
- 25. Antonelli G, Parravano M, Barbano L, et al.. Multimodal study of PRPH2 gene-related retinal phenotypes. Diagnostics (Basel). 2022; 12(8): 1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Richards S, Aziz N, Bale S, et al.. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015; 17(5): 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abou Tayoun AN, Pesaran T, DiStefano MT, et al.. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018; 39(11): 1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adzhubei IA, Schmidt S, Peshkin L, et al.. A method and server for predicting damaging missense mutations. Nat Methods. 2010; 7(4): 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ng PC, Henikoff S.. Predicting deleterious amino acid substitutions. Genome Res. 2001; 11(5): 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ioannidis NM, Rothstein JH, Pejaver V, et al.. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016; 99(4): 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J, A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014; 46(3): 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu J, Ng PC.. SIFT Indel: predictions for the functional effects of amino acid insertions/deletions in proteins. PLoS One. 2013; 8(10): e77940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu J, Ng PC.. Predicting the effects of frameshifting indels. Genome Biol. 2012; 13(2): R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Douville C, Masica DL, Stenson PD, et al.. Assessing the pathogenicity of insertion and deletion variants with the Variant Effect Scoring Tool (VEST-Indel). Hum Mutat. 2016; 37(1): 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwarz JM, Rodelsperger C, Schuelke M, Seelow D.. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Meth. 2010; 7(8): 575–576. [DOI] [PubMed] [Google Scholar]
- 36. Zhou H, Gao M, Skolnick J. ENTPRISE-X: predicting disease-associated frameshift and nonsense mutations. PLoS One. 2018; 13(5): e0196849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jaganathan K, Kyriazopoulou Panagiotopoulou S, et al.. Predicting splicing from primary sequence with deep learning. Cell. 2019; 176(3): 535–548.e24. [DOI] [PubMed] [Google Scholar]
- 38. Constable PA, Bach M, Frishman LJ; for the International Society for Clinical Electrophysiology of Vision; ISCEV standard for clinical electro-oculography (2017 update). Doc Ophthalmol. 2017; 134(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Peeters MHC, Khan M, Rooijakkers AAMB, et al.. PRPH2 mutation update: in silico assessment of 245 reported and 7 novel variants in patients with retinal disease. Hum Mutat. 2021; 42(12): 1521–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Boon CJF, van Schooneveld MJ, den Hollander AI, et al.. Mutations in the peripherin/RDS gene are an important cause of multifocal pattern dystrophy simulating STGD1/fundus flavimaculatus. Br J Ophthalmol. 2007; 91: 1504–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ba-Abbad R, Robson AG, Yap YC, et al.. Prph2 mutations as a cause of electronegative ERG. Retina. 2014; 34(6): 1235–1243. [DOI] [PubMed] [Google Scholar]
- 42. Holder GE, Mahroo O.. Electronegative ERG or pseudo-negative ERG? Doc Ophthalmol. 2022; 145(3): 283–286. [DOI] [PubMed] [Google Scholar]
- 43. Lois N, Holder GE, Bunce C, Fitzke FW, Bird AC.. Phenotypic subtypes of Stargardt macular dystrophy—fundus flavimaculatus. Arch Ophthalmol. 2001; 119(3): 359–369. [DOI] [PubMed] [Google Scholar]
- 44. Stricker HM, Ding X, Quiambao A, et al.. The Cys214–>Ser mutation in peripherin/rds causes a loss-of-function phenotype in transgenic mice. Biochem J. 2005; 388(Pt2): 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cheng T, Peachey NS, Li S, et al.. The effect of peripherin/rds haploinsufficiency on rod and cone photoreceptors. J Neurosci. 1997; 17(21): 8118–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Oishi A, Fujinami K, Mawatari G, et al.. Genetic and phenotypic landscape of PRPH2-associated retinal dystrophy in Japan. Genes (Basel). 2021; 12(11): 1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reeves MJ, Goetz KE, Guan B, et al.. Genotype-phenotype associations in a large PRPH2-related retinopathy cohort. Hum Mutat. 2020; 41(9): 1528–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Salinas RY, Baker SA, Gospe SM, et al.. A single valine residue plays an essential role in peripherin/RDS targeting to photoreceptor outer segments. PLoS One. 2013; 8(1): e54292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loewen CJR, Moritz OL, Tam BM, et al.. The role of subunit assembly in peripherin-2 targeting to rod photoreceptor disk membranes and retinitis pigmentosa. Mol Biol Cell. 2003; 14(8): 3400–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Payne AM, Downes SM, Bessant DA, et al.. Founder effect, seen in the British population, of the 172 peripherin/RDS mutation-and further refinement of genetic positioning of the peripherin/RDS gene. Am J Hum Genet. 1998; 62(1): 192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alapati A, Goetz K, Suk J, et al.. Molecular diagnostic testing by eyeGENE: analysis of patients with hereditary retinal dystrophy phenotypes involving central vision loss. Invest Ophthalmol Vis Sci. 2014; 55(9): 5510–5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Daftarian N, Mirrahimi M, Sabbaghi H, et al.. PRPH2 mutation as the cause of various clinical manifestations in a family affected with inherited retinal dystrophy. Ophthalmic Genet. 2019; 40(5): 436–442. [DOI] [PubMed] [Google Scholar]
- 53. Downes SM, Fitzke FW, Holder GE, et al.. Clinical features of codon 172 RDS macular dystrophy: similar phenotype in 12 families. Arch Ophthalmol. 1999; 117: 1373–1383. [DOI] [PubMed] [Google Scholar]
- 54. Nakazawa M, Wada Y, Tamai M.. Macular dystrophy associated with monogenic Arg172Trp mutation of the peripherin/RDS gene in a Japanese family. Retina. 1995; 15: 518–523. [DOI] [PubMed] [Google Scholar]
- 55. Piguet B, Heon E, Munier FL, et al.. Full characterization of the maculopathy associated with an Arg-172-Trp mutation in the RDS/peripherin gene. Ophthalmic Genet. 1996; 17: 175–186. [DOI] [PubMed] [Google Scholar]
- 56. Albertos-Arranz H, Sanchez-Saez X, Martinez-Gil N, et al.. Phenotypic differences in a PRPH2 mutation in members of the same family assessed with OCT and OCTA. Diagnostics. 2021; 11(5): 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Trujillo MJ, Bueno J, Osorio A, et al.. Three novel RDS-peripherin mutations (689delT, 857del17, G208D) in Spanish families affected with autosomal dominant retinal degenerations. Mutations in brief no. 147. Hum Mutat. 1998; 12(1): 70. [DOI] [PubMed] [Google Scholar]
- 58. Birtel J, Eisenberger T, Gliem M, et al.. Clinical and genetic characteristics of 251 consecutive patients with macular and cone/cone-rod dystrophy. Sci Rep. 2018; 8(1): 4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Soucy M, Kolesnikova M, Kim AH, Tsang SH.. Phenotypic variability in PRPH2 as demonstrated by a family with incomplete penetrance of autosomal dominant cone-rod dystrophy. Doc Ophthalmol. 2023; 146(3): 267–272. [DOI] [PubMed] [Google Scholar]
- 60. Ruiz-Pastor MJ, Sanchez-Saez X, Kutsyr O, et al.. Prph2 knock-in mice recapitulate human central areolar choroidal dystrophy retinal degeneration and exhibit aberrant synaptic remodeling and microglial activation. Cell Death Dis. 2023; 14(11): 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen FK, Agelis L, Peh KK, et al.. Visual acuity fractions derived from a Snellen chart and letter scores on the early treatment diabetic retinopathy study chart. Asia Pac J Ophthalmol. 2014; 3(5): 277–285. [DOI] [PubMed] [Google Scholar]
- 62. Heath Jeffery RC, Lo J, Thompson JA, et al.. Analysis of the outer retinal bands in ABCA4 and PRPH2-associated retinopathy using OCT. Ophthalmol Retina. 2024; 8(2): 174–183. 2023 May [online ahead of print]. [DOI] [PubMed] [Google Scholar]
- 63. Renner AB, Fiebig BS, Weber BHF, et al.. Phenotypic variability and long-term follow-up of patients with known and novel PRPH2/RDS gene mutations. Am J Ophthalmol. 2009; 147(3): 518–530. [DOI] [PubMed] [Google Scholar]
- 64. Sohocki MM, Daiger SP, Bowne SJ, et al.. Prevalence of mutations causing retinitis pigmentosa and other inherited retinopathies. Hum Mutat. 2001; 17(1): 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koyanagi Y, Akiyama M, Nishiguchi KM, et al.. Genetic characteristics of retinitis pigmentosaw in 1204 Japanese patients. J Med Genet. 2019; 56(10)662–670. [DOI] [PubMed] [Google Scholar]
- 66. Sullivan LS, Bowne SJ, Birch DG, et al.. Prevalence of disease-causing mutations in families with autosomal dominant retinitis pigmentosa: a screen of known genes in 200 families. Invest Ophthalmol Vis Sci. 2006; 47(7): 3052–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stone EM, Andorf JL, Whitmore SS, et al.. Clinically focused molecular investigation of 1000 consecutive families with inherited retinal disease. Ophthalmology. 2017; 124(9): 1314–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Khani SC, Karoukis AJ, Young JE, et al.. Late-onset autosomal dominant macular dystrophy with choroidal neovascularisation and nonexudative maculopathy associated with mutation in the RDS gene. Invest Ophthalmol Vis Sci. 2003; 44(8): 3570–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Klevering BJ, van Driel M, van Hogerwou AJM, et al.. Central areolar choroidal dystrophy associated with dominantly inherited drusen. Br J Ophthalmol. 2002; 86(1): 91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jacobson SG, Cideciyan AV, Maguire AM, et al.. Preferential rod and cone photoreceptor abnormalities in heterozygotes with point mutations in the RDS gene. Exp Eye Res. 1996; 63(5): 603–608. [DOI] [PubMed] [Google Scholar]
- 71. Testa F, Marini V, Rossi S, et al.. A novel mutation in the RDS gene in an Italian family with pattern dystrophy. Br J Ophthalmol. 2005; 89(8): 1066–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Coco-Martin RM, Sanchez-Tocino HT, Desco C, et al.. PRPH2-related retinal diseases: broadening the clinical spectrum and describing a new mutation. Genes (Basel). 2020; 11(7): 773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Duncan JL, Talcott KE, Ratnam K, et al.. Cone structure in retinal degeneration associated with mutations in the peripherin/RDS gene. Invest Ophthalmol Vis Sci. 2011; 52(3): 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Colombo L, Maltese PE, Castori M, et al.. Molecular epidemiology in 591 Italian probands with non-syndromic retinitis pigmentosa and Usher syndrome. Invest Ophthalmol Vis Sci. 2021; 62(2): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Martin-Merida I, Aguilera-Garcia D, Fernandez-San JP, et al.. Toward the mutational landscape of autosomal dominant retinitis pigmentosa: a comprehensive analysis of 258 Spanish families. Invest Ophthalmol Vis Sci. 2018; 59(6): 2345–2354. [DOI] [PubMed] [Google Scholar]
- 76. Almoguera B, Li J, Fernandez-San PJ, et al.. Application of whole exome sequencing in six families with an initial diagnosis of autosomal dominant retinitis pigmentosa: lessons learned. PLoS One. 2015:10(7): e0133624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Manes G, Guillaumie T, Vos WL, et al.. High prevalence of PRPH2 in autosomal dominant retinitis pigmentosa in France and characterisation of biochemical and clinical features. Am J Ophthalmol. 2015; 159(2): 302–314. [DOI] [PubMed] [Google Scholar]
- 78. Carss KJ, Arno G, Erwood M, et al.. Comprehensive rare variant analysis via whole-genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017; 100(1): 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ekstrom U, Ponjavic V, Abrahamson M, et al.. Phenotypic expression of autosomal dominant retinitis pigmentosa in a Swedish family expressing a Phe-211-Leu variant of peripherin/RDS. Ophthalmic Genet. 1998; 19(1): 27–37. [DOI] [PubMed] [Google Scholar]
- 80. Zhang K, Garibaldi DC, Li Y, et al.. Butterfly-shaped pattern dystrophy: a genetic, clinical, and histopathological report. Arch Ophthalmol. 2002; 120(4): 485–490. [DOI] [PubMed] [Google Scholar]
- 81. Fishman GA, Stone E, Gilbert LD, et al.. Clinical features of a previously undescribed codon 216 (proline to serine) mutation in the peripherin/retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Ophthalmology. 1994; 101(8): 1409–1421. [DOI] [PubMed] [Google Scholar]
- 82. Ellingford JM, Barton S, Bhaskar S, et al.. Molecular findings from 537 individuals with inherited retinal disease. J Med Genet. 2016; 53(11): 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ahmad OR, Ayyagari R, Zacks DN.. A novel missense mutation in the rds/peripherin gene associated with retinal pattern dystrophy. Retin Cases Brief Rep. 2010; 4(1): 84–85. [DOI] [PubMed] [Google Scholar]
- 84. Liu X, Tao T, Zhao L, et al.. Molecular diagnosis based on comprehensive genetic testing in 800 Chinese families with non-syndromic inherited retinal dystrophies. Clin Exp Ophthalmol. 2021; 49(1): 46–59. [DOI] [PubMed] [Google Scholar]
- 85. de Breuk A, Acar IE, Kersten E, et al.. Development of a genotype assay for age-related macular degeneration: the eye-risk consortium. Ophthalmology. 2021; 128(11): 1604–1617. [DOI] [PubMed] [Google Scholar]
- 86. Huang L, Li S, Xiao X, et al.. Screening for variants in 20 genes in 130 unrelated patients with cone-rod dystrophy. Mol Med Rep. 2013; 7(6): 1779–1785. [DOI] [PubMed] [Google Scholar]
- 87. Kajiwara K, Hahn LB, Mukai S, et al.. Mutations in the human retinal degeneration slow gene in autosomal dominant retinitis pigmentosa. Nature. 1991; 354(6353): 480–483. [DOI] [PubMed] [Google Scholar]
- 88. Yang Z, Lin W, Moshfeghi DM, et al.. A novel mutation in the RDS/Peripherin gene causes adult-onset foveomacular dystrophy. Am J Ophthalmol. 2003; 135(2): 213–218. [DOI] [PubMed] [Google Scholar]
- 89. Sodi A, Mucciolo DP, Giorgio D, et al.. Clinical and molecular findings in patients with pattern dystrophy. Ophthalmic Genet. 2021; 42(5): 577–587. [DOI] [PubMed] [Google Scholar]
- 90. Weleber RG, Carr RE, Murphey WH, et al.. Phenotypic variation including retinitis pigmentosa, pattern dystrophy, and fundus flavimaculatus in a single family with a deletion of codon 153 or 154 of the peripherin/RDS gene. Arch Ophthalmol. 1993; 111(11): 1531–1542. [DOI] [PubMed] [Google Scholar]
- 91. van Lith-Verhoeven JJC, van den Helm B, Deutman AF, et al.. A peculiar autosomal dominant macular dystrophy caused by an asparagine deletion at codon 169 in the peripherin/RDS gene. Arch Ophthalmol. 2003; 121(10): 1452–1457. [DOI] [PubMed] [Google Scholar]
- 92. Kalyanasundaram TS, Black GC, O'Sullivan J, Bishop PN. A novel peripherin/RDS mutation resulting in a retinal dystrophy with phenotypic variation. Eye(Lond). 2009; 23(1): 237–239. [DOI] [PubMed] [Google Scholar]
- 93. Jin Z-B, Mandai M, Yokota T, et al.. Identifying pathogenic genetic background of simplex or multiplex retinitis pigmentosa patients: a large scale mutation screening study. Med Genet. 2008; 45(7): 465–472. [DOI] [PubMed] [Google Scholar]
- 94. Sullivan LS, Guilford SR, Birch DG, Daiger SP.. A novel splice site mutation in the gene for peripherin/RDS causing dominant retinal degeneration. Invest Ophthalmol Vis Sci. 1996; 37: 1145. [Google Scholar]
- 95. Duncker T, Tsang SH, Woods RL, et al.. Quantitative fundus autofluorescence and optical coherence tomography in PRPH2/RDS- and ABCA4-associated disease exhibiting phenotypic overlap. Invest. Ophthalmol. Vis. Sci. 2015; 56(5): 3159–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.