Significance
Tools to track the primary cellular anabolic reductant, NADPH, are essential for understanding organismal health and disease. Here, we demonstrate that the variations in hydrogen stable isotope ratios of lipids are useful indicators of redox metabolism in the yeast Saccharomyces cerevisiae, a model microorganism for industry and studies of eukaryotic disease including cancer. Large differences in lipid hydrogen isotope ratios distinguish respiration and fermentation in yeast, reflecting shifts in the metabolic origins of NADPH. Changes in NADPH demand associated with cellular biochemical composition are also recorded in lipid hydrogen isotope ratios. These results provide promising tools to characterize eukaryotic metabolism, thus helping to elucidate the role of NADPH in health and disease.
Keywords: lipids, yeast, NADPH, fractionation, hydrogen isotope
Abstract
Shifts in the hydrogen stable isotopic composition (2H/1H ratio) of lipids relative to water (lipid/water 2H-fractionation) at natural abundances reflect different sources of the central cellular reductant, NADPH, in bacteria. Here, we demonstrate that lipid/water 2H-fractionation (2εfattyacid/water) can also constrain the relative importance of key NADPH pathways in eukaryotes. We used the metabolically flexible yeast Saccharomyces cerevisiae, a microbial model for respiratory and fermentative metabolism in industry and medicine, to investigate 2εfattyacid/water. In chemostats, fatty acids from glycerol-respiring cells were >550‰ 2H-enriched compared to those from cells aerobically fermenting sugars via overflow metabolism, a hallmark feature in cancer. Faster growth decreased 2H/1H ratios, particularly in glycerol-respiring cells by 200‰. Variations in the activities and kinetic isotope effects among NADP+-reducing enzymes indicate cytosolic NADPH supply as the primary control on 2εfattyacid/water. Contributions of cytosolic isocitrate dehydrogenase (cIDH) to NAPDH production drive large 2H-enrichments with substrate metabolism (cIDH is absent during fermentation but contributes up to 20 percent NAPDH during respiration) and slower growth on glycerol (11 percent more NADPH from cIDH). Shifts in NADPH demand associated with cellular lipid abundance explain smaller 2εfattyacid/water variations (<30‰) with growth rate during fermentation. Consistent with these results, tests of murine liver cells had 2H-enriched lipids from slower-growing, healthy respiring cells relative to fast-growing, fermenting hepatocellular carcinoma. Our findings point to the broad potential of lipid 2H/1H ratios as a passive natural tracker of eukaryotic metabolism with applications to distinguish health and disease, complementing studies that rely on complex isotope-tracer addition methods.
Fractionation of hydrogen stable isotopes at natural abundance [2H/1H ratios typically expressed as δ2H values in permil (‰)] between lipids and growth water (lipid/water 2H-fractionation, 2εlipid/water) varies greatly and is a potential tool to track metabolism in the present and past (1–4). Lipid production is biochemically well characterized and hydrogen has potential for large isotopic fractionations (5). In prokaryotes, lipid/water 2H-fractionation can shift from −200‰ to +300‰ within the same species in response to variations in central metabolism that lead to different enzymatic sources of nicotinamide adenine dinucleotide phosphate (NADPH), the primary anabolic reductant in cells (1, 2).
In eukaryotes, the metabolic basis of lipid/water 2H-fractionations is poorly understood. Similar fractionations observed in photosynthetic eukaryotes compared to photosynthetic prokaryotes are consistent with the same NADPH source (6–13). This suggests that 2H-fractionations in eukaryotes should also record the NADPH source shifts accompanying different modes of heterotrophy. Isotope variability could be enhanced as a result of more complex metabolite trafficking at metabolite branchpoints (5) associated with the cellular compartmentalization in eukaryotes. Conversely, restriction of metabolite transport across intracellular compartment membranes in eukaryotes might mute isotopic signals of NADPH supply. A mechanistic understanding of natural abundance 2H/1H ratios would provide insights into underlying metabolic processes in eukaryotic heterotrophs.
Respiration and fermentation are two common metabolisms in eukaryotic heterotrophs distinguished by large differences in NADPH cycling. They have been extensively studied in the yeast Saccharomyces cerevisiae in diverse contexts, including in medicine where it is considered a microbial model of cancer metabolism (14, 15). S. cerevisiae engages in overflow metabolism when using sugars such as glucose as the growth substrate. This mode, termed the “Crabtree effect,” is analogous to the characteristic “Warburg effect” of cancer cells, in which energy is mainly generated by fermentation rather than normal respiration (15). In both yeast and cancer cells, sugar fermentation produces “overflow” products despite the presence of oxygen, fulfilling a need for replenishment of oxidized redox cofactor NAD+ and continued adenosine triphosphate (ATP) production by substrate-level phosphorylation (16). Importantly, the higher glycolytic and pentose phosphate fluxes enabled by overflow metabolism satisfy the increased demands for biosynthetic precursors and protection against oxidative stress needed for fast cell proliferation, a key challenge of cancer cells (17, 18). Conversely, nonfermentable substrates such as glycerol are respired using central carbon pathways distinct from those in fermentative metabolism in yeast (Fig. 1 and SI Appendix, Fig. S1) (19–21).
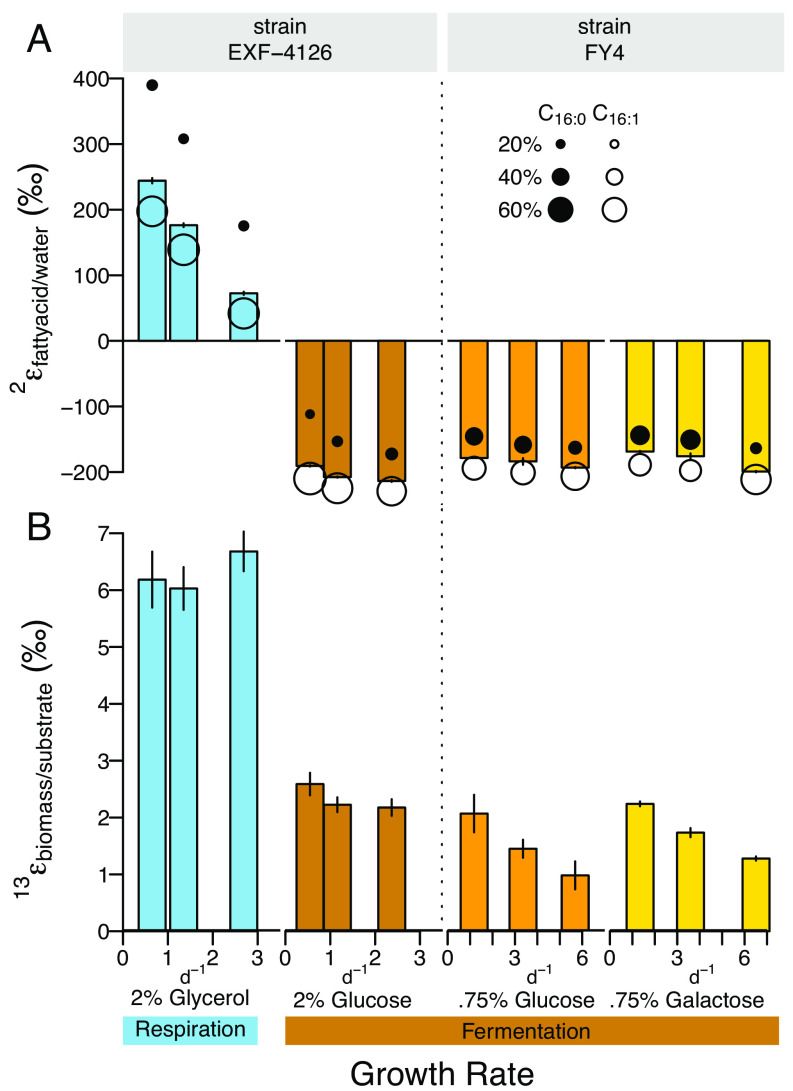
Fig. 1.
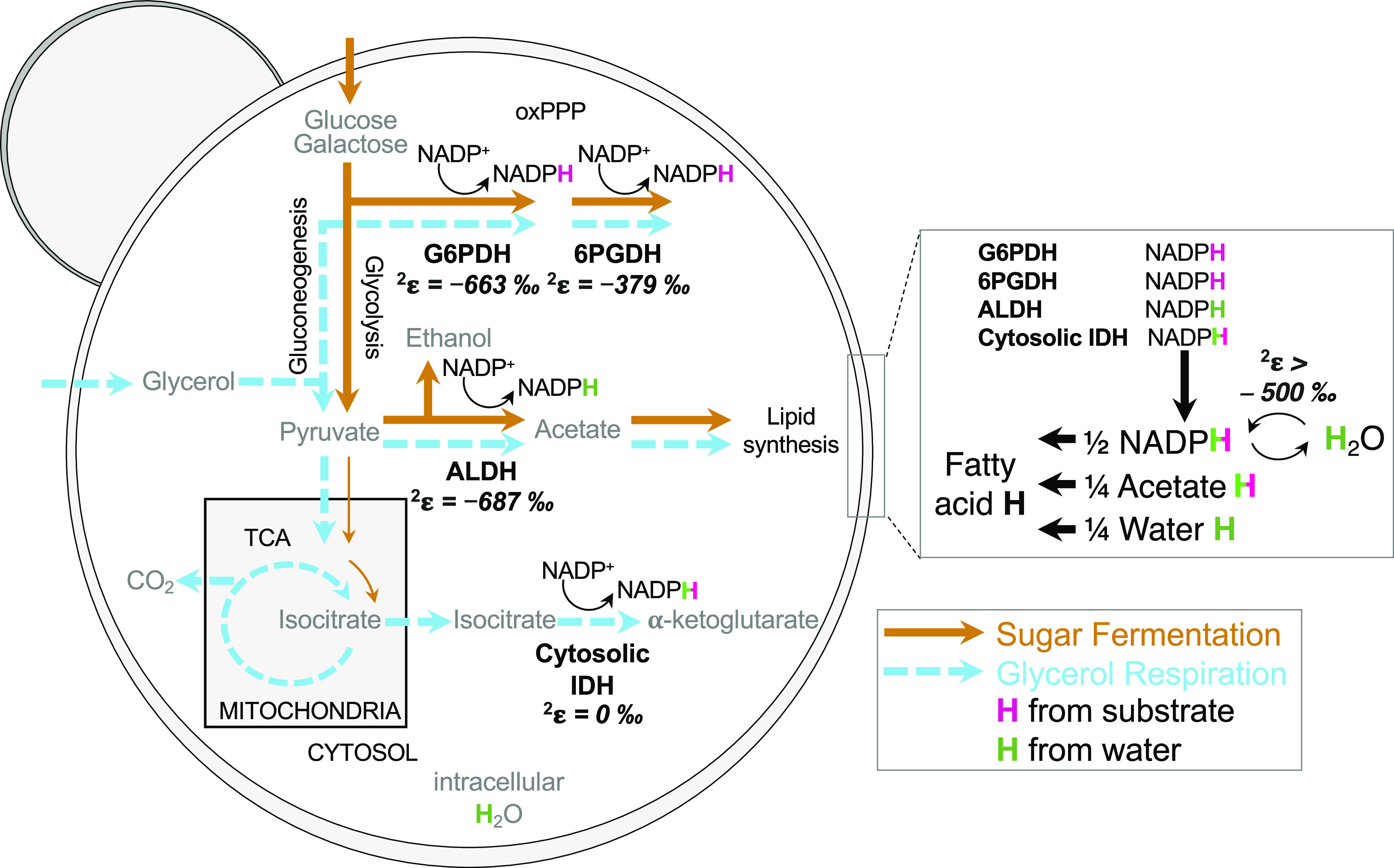
Yeast S. cerevisiae central carbon metabolism pathways, NADPH hydrogen (H) sources, and deuterium kinetic isotope effects of NADPH supply enzymes indicated as enzyme 2H-fractionation factors (2ε). Sugar fermentation (brown arrows) and glycerol-respiration (blue dashed arrows) induce variable activity of NADPH supply enzymes (G6PD, glucose-6-phosphate dehydrogenase; 6PGDH; 6-phosphogluconate dehydrogenase in the oxidative pentose phosphate pathway (oxPPP); ALDH, aldehyde dehydrogenase; IDH, isocitrate dehydrogenase; TCA, tricarboxylic acid cycle; FE, flavin enzymes). H on NADPH originates from carbon substrate (pink H) and water (green H) and is transferred to fatty acids, accounting for half of lipid H, with the remainder derived from acetate (one quarter), and intracellular water (one quarter); some NAD(P)H hydrogen may exchange with water hydrogen by flavin enzymes (24–26). Detailed H tracking and deuterium kinetic isotope effects in SI Appendix, Fig. S1 and Table S1.
The large differences in central carbon metabolism typically correspond to the different NADPH sources used for fermentative and respiratory growth (22, 23). Shifts in NADPH source can be imprinted on the lipid/water 2H-fractionation, as demonstrated in bacteria (1). The largest contributor of carbon-bound lipid (e.g., fatty acid) hydrogen in organisms is NADPH (24), and its isotopic composition has been shown to be influenced by a number of factors. Most important are the unique deuterium kinetic isotope effects (dKIEs) of different NADP+-reducing enzymes (SI Appendix, Table S1) and variable fluxes through these enzymes due to the operation of different central metabolic pathways (1, 2). Additional metabolic signals in NADPH can derive from the isotope composition of substrate sources of NADPH hydrogen (5), and enzyme-catalyzed exchange of hydrogen on NADPH with that in water and with nicotinamide adenine dinucleotide (NADH) (24–29) SI Appendix, SI Discussion 1).
In yeast, four enzymes are sources of cytosolic NADPH used for lipid biosynthesis (Fig. 1) (22, 23). Glucose-6-phosphate dehydrogenase (G6PDH) and 6-phosphogluconate dehydrogenase (6PGDH) are sources in the oxidative pentose phosphate pathway (oxPPP). Cytosolic NADP+-dependent isocitrate dehydrogenase (cIDH) provides a source of NADPH and alpha-ketoglutarate to the cytosol, whereas NADPH from mitochondrial IDH (mIDH) is not available for fatty acid synthesis in the cytosol. The last source of cytosolic NADPH is aldehyde dehydrogenase (ALDH) in pyruvate degradation to acetate during ethanol production and/or lipid biosynthesis (Fig. 1 and SI Appendix, Fig. S1). Three of the four NADPH-generating enzymes (G6PDH, 6PGDH, and ALDH) are active during both glucose fermentation and glycerol-respiration (22, 23). These have normal dKIEs (SI Appendix, Table S1), which strongly favor 1H-containing substrates (30–32), and produce 2H-depleted NADPH. However, cIDH is only expressed during respiratory growth (33), and it has no dKIE (SI Appendix, Table S1) (34, 35), resulting in 2H/1H ratios of produced NADPH equal to that of isocitrate. Yeast lack a transhydrogenase (36) that can directly couple NADH-NADPH redox cycling, thus reducing the number of controls on NADPH 2H/1H ratios (29).
Using S. cerevisiae as a model organism, we found large differences in lipid/water 2H-fractionation between respiring and fermenting cells and variable influences of growth rate. Shifts in lipid/water 2H-fractionation reflect shifts in the hydrogen isotopic composition of NADPH. Those differences primarily depend on NADPH sources through changes in the activity and dKIE of NADPH-generating enzymes and can be slightly modulated by its sinks. Given similarities in yeast and cancer metabolism (14, 15), a test with murine cells revealed a shift in 2H-fractionation between healthy hepatocytes and proliferating hepatoma (37, 38) that is qualitatively consistent with yeast data. Our results demonstrate the potential of lipid/water 2H-fractionation to track NADPH cycling in metabolic processes important for health and disease.
Results and Discussion
We cultured S. cerevisiae under sugar-fermenting and glycerol-respiring conditions at varying growth rates. To compare the effects of metabolism and strain, we used two S. cerevisiae strains: strain EXF-4126, capable of growth by aerobic fermentation on glucose or respiration on glycerol in minimal media (39), and strain FY4, which ferments sugars but does not respire glycerol in minimal media (40). To force cells to use a single metabolism we used nitrogen-limited chemostats (SI Appendix, Fig. S5) to control growth rate and maintain exponential growth, avoiding the diauxic shift that occurs in aerobic batch cultures upon glucose exhaustion when cells switch to ethanol utilization (41). All chemostat cultures had very low bioavailable nitrogen, representative of most natural environmental conditions (42). We measured 2H/1H ratios of fatty acids, growth water, and carbon substrate for all cultures. In cultures with strain EXF-4126, which tested the slowest growth rates, we also monitored key NADP+-reducing enzyme activities, redox cofactor ratios, and biomass composition to elucidate the cellular mechanisms behind lipid δ2H variability. As a preliminary comparison with mammalian cancer, we then measured lipid/water 2H-fractionation for healthy murine hepatocytes and proliferating HEPA1-6 hepatoma cells, a model cell line of murine liver cancer (37, 38).
Yeast Lipid/Water 2H-Fractionation Depends Chiefly on Mode of Energy Metabolism and Secondarily on Growth Rate.
Lipid/water 2H-fractionations were extremely sensitive to carbon substrate (Fig. 2A and SI Appendix, Fig. S2), varying between large 2H-enrichments and depletions in fatty acid 2H/1H ratios relative to water. Fatty acids from glycerol-respiring cells were >550‰ heavier in 2H than from fermenting cells in carbohydrate-fed chemostats, with palmitic acid yielding the most extreme enrichments. These effects are larger than those observed so far in prokaryotes from substrate-induced metabolic shifts (1). Substrate δ2H, which varied by only ~60‰ between glycerol and glucose and ~30‰ between glucose and galactose (SI Appendix, Fig. S3), cannot explain the much larger shifts in 2H/1H ratios relative to water [and neither can variations in water δ2H up to ~100‰ (SI Appendix, Figs. S2 and S3)]. The observed changes in lipid/water 2H-fractionation must thus stem from metabolism-specific processes induced by the class of carbon substrate, an interpretation supported by 2H-fractionation consistency across strain and sugar type (Fig. 2).
Fig. 2.
S. cerevisiae (A) lipid/water 2H-fractionation and (B) biomass/substrate 13C-fractionation for chemostat growth on fermentable sugar and nonfermentable glycerol substrates at different growth rates. Strain EXF-4126 grown with 2% glycerol (blue), 2% glucose (brown); strain FY4 grown with 0.75% glucose (orange), 0.75% galactose (yellow). Panel A bars, abundance weighted average 2εfattyacid/water of n = 3 replicate sampling timepoints; error bars are SD. Palmitic (n-C16:0) and palmitoleic (n-C16:1) acid 2εfattyacid/water indicated by filled and open circles, respectively (circle size, relative abundance of 16 carbon fatty acids). Panel B bars are biomass/substrate 13C-fractionation, mean of 3 to 6 replicate sampling timepoints; error bars are SD of 3 to 6 timepoint replicates. See details in SI Appendix, Figs. S2 and S3 and Datasets S1–S3.
Chemostat experiments enabled direct investigation of the growth rate sensitivity of lipid/water 2H-fractionation. As expected, carbon substrate had the largest effect on lipid/water 2H-fractionation; nonetheless, the effect of growth rate was still significant. In the glycerol-fed chemostat, this fractionation decreased (less 2H-enrichment) linearly as growth rate increased, with a maximum difference of 200‰ for palmitic acid (R2 = 0.99, P << 0.001). The 2H-fractionation in sugar-fed cells was much less sensitive to growth rate (Fig. 2A and SI Appendix, Fig. S9) with 2H-enrichments of <60‰ (e.g., from −110 to −170‰ or less, R2 = 0.74 to 0.97, P << 0.001). The largest differences in 2H-fractionation between glycerol- and glucose-based growth occurred in the slowest (~0.5 d−1) growth rate chemostats.
Cellular Redox Status, Ethanol Production, and Biomass Carbon Isotopes Vary between Respiration and Fermentation.
To identify the metabolic causes of lipid/water 2H-fractionation in yeast (Fig. 2A), we evaluated cellular redox state (SI Appendix, Fig. S4) and central metabolic pathways used for the cycling of energy cofactors during growth on sugars and glycerol (Figs. 2B and 3 and SI Appendix, Figs. S2–S13 and Datasets S1–S4). Ratios of NAD+/NADH and NADP+/NADPH inform the redox balance of catabolism and anabolism, respectively. Higher NAD+/NADH ratios and three times faster increase of NAD+/NADH with growth rate in glycerol-fed chemostats relative to glucose-fed chemostats (SI Appendix, Fig. S4) illustrate the greater oxidative capacity of respiratory processes for NAD+ regeneration compared to overflow metabolism (16). This aligns with ~fivefold higher ethanol concentrations in the glucose- vs. glycerol-fed chemostats (SI Appendix, Fig. S5). More oxidized NAD+/NADH ratios in glycerol-respiration are also consistent with its larger biomass/substrate carbon isotope fractionation (Fig. 2B), which reflects the net effects of all isotopically fractionating steps of substrate metabolism (Fig. 1). Glycerol-fed biomass had a three times larger 13C-fractionation than glucose-fed biomass (Fig. 2B) similar to results in other fungus species (43, 44) but in contrast to other heterotrophs in which biomass δ13C varies minimally (45). These data confirm the respiratory growth on glycerol and the fermentative growth on glucose in chemostats across a range of growth rates.
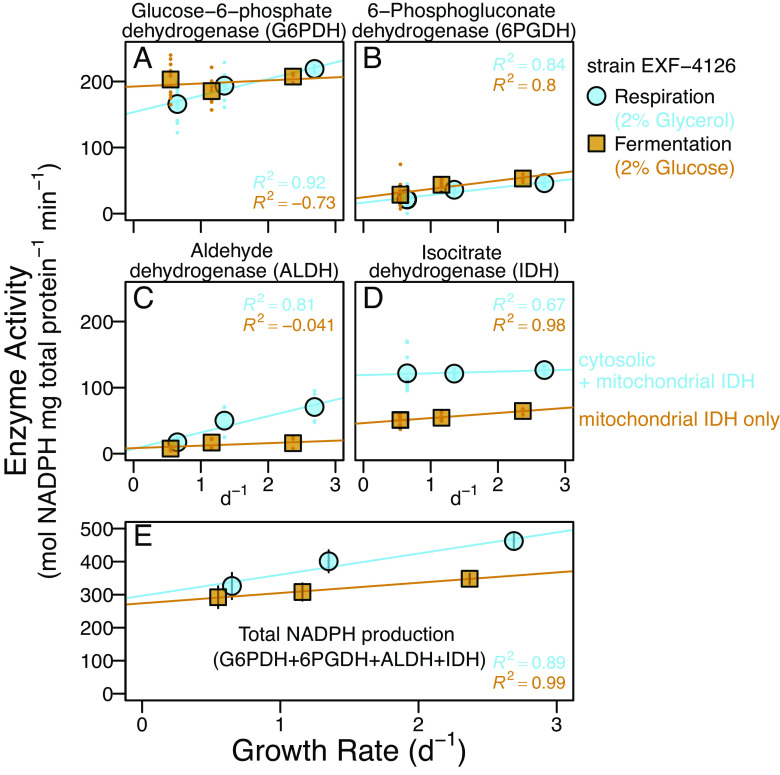
Fig. 3.
NADP+-reducing (NADPH-producing) enzyme activities in glycerol-respiring (blue circles) and glucose-fermenting (brown squares) chemostats of strain EXF-4126. (A–D) Enzyme activities of 4 to 11 individual sample replicates (colored dots) and their averages (circles and squares). Measured IDH activity for glucose-grown cells is assumed to represent mIDH as the cytosolic form is glucose repressed (33). (E) Total NAPDH production rates and 1 SD propagated errors, assuming comparable total protein extract efficiencies across enzyme assays. All linear regressions are based on averages with adjusted R2. See Datasets S1–S4 for details.
Unlike NAD+/NADH ratios, NADP+/NADPH ratios at each growth rate were equivalent for glucose and glycerol-fed cultures (SI Appendix, Fig. S4). These ratios increase with growth rate (SI Appendix, Fig. S4), reflecting the greater use of reductant at faster growth. The nearly identical responses of NADP+/NADPH ratios in glycerol and glucose cultures indicate that the mechanism for lipid/water 2H-fractionation variability does not directly result from redox balance but instead lies in the constellations of NADP+-reducing enzymes that contribute to the NADPH pool during fermentation and respiration (Fig. 1).
NADPH Source Control on Yeast Lipid/Water 2H-Fractionation: Respiration vs. Fermentation.
To quantify NADPH sources from different metabolisms (Fig. 1) and their effects on lipid/water 2H-fractionation, we measured the activities of NADP+-reducing enzymes: G6PDH, 6PGDH, ALDH, and IDH (IDH as both mitochondrial and cytosolic) (Fig. 3). Higher total NAPDH production rates (Fig. 3E) and ~equal NADP+/NADPH ratios (SI Appendix, Fig. S4A) in glycerol-respiring compared to fermenting cells indicate greater anabolic costs of glycerol-based growth.
Since NADPH is the largest contributor of lipid H (Fig. 1 and SI Appendix, Fig. S1), lipid 2H/1H ratios track NADPH 2H/1H ratios. These are set by the relative fluxes of reductant from NADP+-reducing enzymes and their dKIEs which can vary by as much as ~700‰ (Fig. 1 and SI Appendix, Table S1). Hydrogen exchange between NADPH and NADH is absent in yeast as it lacks the necessary transhydrogenase enzyme (23) and substrate δ2H and growth water δ2H had no to minimal effect on lipid 2H/1H ratios. Sizeable shifts in fractionation should correspond with changes in NADPH contribution from enzymatic sources with large differences in dKIEs.
The two oxPPP enzymes, G6PDH and 6PGDH, were important sources of NADPH for glucose fermentation and glycerol-respiration at each growth rate (Fig. 3 A and B and SI Appendix, Fig. S6 A and B). For every condition, G6PDH was the largest source of NADPH (~50 to 70%), while 6PGDH contributions were <15% (SI Appendix, Fig. S6 A and B). Both oxPPP enzymes have large normal dKIEs (Fig. 1 and SI Appendix, Table S1), strongly favoring their 1H-containing substrates (glucose-6-phosphate and 6-phosphogluconate) to donate H to NADP+ (30, 31). However, these enzymes varied the least between substrates (Fig. 3 A and B and SI Appendix, Fig. S7) and thus played a minimal role in shifting 2H-fractionation from negative to positive values (Fig. 2) in response to substrate.
ALDH activity in the respiring chemostat was two to five times higher than in fermenting chemostats (Fig. 3 and SI Appendix, Fig. S7), but its contribution to the total NAPDH flux remained <16% in any culture (SI Appendix, Fig. S6C). Importantly, ALDH also has a normal dKIE (32) (SI Appendix, Table S1) and thus cannot explain 2H-enrichment in fatty acids from glycerol cultures.
IDH activity was two times greater in respiring than in the fermenting cells (Fig. 3D and SI Appendix, Figs. S6D and Fig. S7) and the second most important respiratory NADPH source after G6PDH (SI Appendix, Fig. S6). In glycerol-fed cells, activities reflect mitochondrial plus cytosolic IDH, while those in glucose-fed cells reflect only the mIDH (33). Since NAD(P)(H) pools remain within cellular compartments (36), mIDH-derived NADPH cannot affect cytosolic lipids. Cytosolic IDH provides relatively 2H-enriched NAPDH compared to all other NADPH sources in yeast (Fig. 1) since there is little to no kinetic isotope effect associated with the transfer of isocitrate-H from cIDH to NADP+ (34, 35) (SI Appendix, Table S1). The relatively 2H-enriched NADPH supplied by the high cIDH activity during respiratory growth (Fig. 3 and SI Appendix, Fig. S6) can explain 2H-enrichment in lipids from glycerol-fed chemostats (Fig. 2). This is supported by the very high correlation (Pval < 0.001) of IDH-derived NAPDH with lipid 2H-fractionations (Fig. 4A and SI Appendix, Fig. S9 and Table S2). Our data also suggest a sensitive method to quantify NAPDH sources in yeast based on 2H-fractionation. For example, we find a 1-percentage point increase in the total IDH contribution to NADPH production results in a ~30‰ increase in 2H-fractionation, with an estimated uncertainty of 5‰ based on the error of the slope (SI Appendix, Table S2).
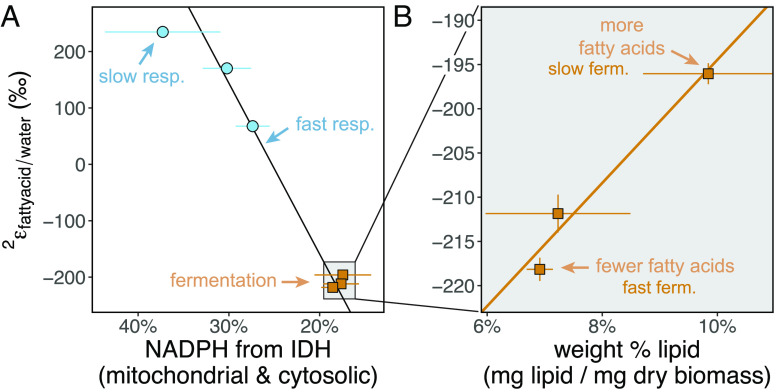
Fig. 4.
Fatty acid/water 2H-fractionations are primarily controlled by (A) relative NADPH supply from IDH and can be modulated by shifts in (B) biomass lipid levels. (A) Yeast 2εfattyacid/water vs. total IDH contributions to NADPH production (%, note inverse x-axis scale) calculated using enzyme activities (Pearson rtotIDH = 0.98, ptotIDH < 0.001, York corr < 0.001, York = 0.08, SI Appendix, Table S2). Total IDH activity in fermentation should solely reflect mIDH (33). (B) Yeast 2εfattyacid/water in glucose-fed yeast biomass vs. the relative abundance of fatty acids (Pearson rlipid% = 0.98, plipid% = 0.12, York corr = 0.12, York = 0.3). Error bars, 1 SD propagated error from enzyme activities (n = 4 to 11 replicates, Fig. 3) and biomass composition measurements (n = 3 replicates). Details in SI Appendix, Fig. S9 and Datasets S1–S4.
The possibility of an inverse isotope effect for NADP+-specific IDH (34, 46, 47) (SI Appendix, Table S1) is intriguing and could help explain the remarkably 2H-enriched fatty acids in slow-growing glycerol-fed cells (Fig. 2). In the absence of transhydrogenase enzymes in yeast (23), NADH cycling does not directly affect lipid δ2H since NAD(P)(H) pools do not cross cellular compartment membranes and are separately regulated (36). But it is possible that the substrates feeding all four enzymes in glycerol-respiring cells are additionally heavier in 2H. This could stem from TCA cycle fractionations that produce 2H-enriched isocitrate (see discussion in ref. 1). Additionally, intracellular water could be isotopically distinct from growth water through water-producing metabolic reactions (48–50), which might directly impact lipid-H during lipid biosynthesis (Fig. 1). Water hydrogen exchange with precursor metabolites to NAPD+-reducing enzymes (e.g., the exchange of 2H in cell water onto gluconeogenic metabolites by fast, reversible sugar isomerization reactions with an inverse dKIE, SI Appendix, Fig. S1) may also have helped increase 2H in glycerol-grown lipids (see SI Appendix, SI Discussion 1 for details). In particular, the existence of isotopically distinct pools of intracellular water is more apparent as metabolism speeds up, but it is difficult to predict whether this water is enriched or depleted relative to medium (50). The decrease in 2H-enrichment with faster glycerol growth (Fig. 2) could reflect a 2H-depleted intracellular water pool formed by gluconeogenesis that directly contributes to lipid biosynthesis, muting the 2H-enrichment effect of NAPDH. However, the very high correlations of 2H-fractionation with IDH activity (Fig. 4A) indicate any water effect is minor. Collectively, yeast data are consistent with previous work on prokaryotes which established NADPH supply via diverse enzymes as a primary control on lipid hydrogen isotope ratios across different metabolisms (1, 2).
NADPH Source Control on Yeast Lipid/Water 2H-Fractionation for Slow and Fast Respiration.
In glycerol-respiring cells, faster growth was associated with large variations in enzyme activities (Fig. 3 and SI Appendix, Fig. S6) and in 2H-fractionation (up to ~200‰ 2H-depletion with faster growth) (Fig. 2). Faster growth in glucose-fermenting cells was associated with less variability in enzyme activity (Fig. 3 and SI Appendix, Fig. S6) and much smaller 2H-depletion (Fig. 2A). This indicates important differences in the metabolic mechanisms used by respiring and fermenting yeast to adjust their growth rate.
Faster growth has larger anabolic requirements for NAPDH and cellular precursors, as shown by higher total NAPDH production rates for both substrates (Fig. 3E). In glycerol chemostats, NAPDH supplied by IDH (~constant across growth rates, Fig. 3D) was supplemented with increasing amounts from oxPPP and ALDH enzymes (Fig. 3 A–C and SI Appendix, Fig. S6) to support nucleotide and lipid biosynthesis (Fig. 1 and SI Appendix, Fig. S1). As a result, the relative contributions of 2H-enriched NAPDH from IDH decreased (Fig. 4A and SI Appendix, Fig. S6) while 2H-depleted NADPH contributions from oxPPP dehydrogenases and ALDH (all with large normal dKIEs, Fig. 1 and SI Appendix, Table S1) increased with faster growth. This dynamic in glycerol-fed cells is reflected accordingly as large changes in 2H-fractionation with faster growth (~200‰ 2H-depletion as the IDH contribution to total NADPH production decreased from ~37 ± 6 percent to 27 ± 2 percent, Fig. 4A and Dataset S2). This sensitivity to IDH is in line with that observed for changes in fractionation depending on growth substrate.
For the glucose-fermenting chemostats at different growth rates, relative changes in NADPH sources are barely detectable compared to those occurring in glycerol-respiration (Figs. 3 and 4A and SI Appendix, Fig. S6). Relative contributions from G6PDH scarcely decrease with growth rate, and contributions from 6PGDH only slightly increase (SI Appendix, Fig. S6). Glucose repression of cIDH (33) indicates the measured IDH activity is mitochondrial. This activity did not vary with growth rate (Fig. 4A and Dataset S2), and did not affect cytosolic lipids (36), as discussed above. The enzyme data are consistent with the relatively minor changes in 2H-fractionation (<30‰ decrease) in response to faster growth by fermentation (Figs. 2 and 4).
It is notable that only 6PGDH but not G6PDH had slightly increased activity at higher growth rate in glucose-fed cultures (Fig. 3 and SI Appendix, Fig. S6), most likely to help fulfill the greater demand for pentose phosphates in addition to NADPH. The small change in oxPPP enzyme activities in glucose-fermenting cells might reflect the glycolytic origins of oxPPP precursors, which are readily available for synthesis of biomass carbon constituents and NADPH under all conditions. In contrast, glycerol-respiring cells, which rely on TCA cycle activity for energy and certain precursors, must operate gluconeogenesis and related upper central metabolic pathways to obtain sufficient anabolic precursors for fast growth.
NADPH Sink Controls on Yeast Lipid/Water 2H-Fractionation in Slow vs. Fast Fermentation.
Given the unremarkable changes in NADPH sourcing within glucose-fermenting cells, we hypothesized that the small lipid 2H-fractionation shifts with growth rate could instead reflect NADPH demand. Variation in lipid isotopes with NADPH consumption has been documented (4, 5). To evaluate this possibility, we analyzed biomass composition focusing on lipids (weight abundance in biomass, Fig. 4B); fatty acid compositions SI Appendix, Fig. S8, elemental stoichiometry (C:N ratios, SI Appendix, Fig. S8), and bulk biomass carbon stable isotope fractionation (Fig. 2B, SI Appendix, Fig. S13, and Dataset S4).
Glucose-fermenting cells had higher fatty acid relative abundances in biomass, greater % lipid unsaturation, and higher biomass C:N ratios than glycerol-respiring cells (SI Appendix, Fig. S8). We identified a significant linear relationship (Pearson Pval < 0.1, York Pval < 0.2) between fractionation and the reciprocal of fatty acid relative abundance in biomass (Fig. 4B, SI Appendix, Fig. S9, and Dataset S3). Correlations with fatty acid weight abundance and lipid unsaturation (% unsaturated lipids) were also notable but less well supported (Pearson Pval = 0.12, York Pval < 0.3, SI Appendix, Fig. S11 and Dataset S3). In glycerol-respiring cells, we found no change in the biomass fatty acid abundances and lipid composition (% unsaturated fatty acids) with growth rate (SI Appendix, Figs. S8 and S9 and Dataset S3). These data point to biomass lipid abundance in sugar-fermenting cultures as the driver for their small shifts in 2H-fractionation with growth rate (Fig. 2A).
Carbon flows at branchpoints leading to different compound classes play a well-established role in the compound-specific carbon isotopic fractionations relative to biomass (5). Following this, we hypothesized changes in NADPH flux between lipids, which are the most 2H-depleted of cellular compounds (5), and other organic compounds caused the small shifts in lipid/water 2H-fractionation in fermenting chemostats (Fig. 4B). We tested this with an isotopic model based on mass balance between lipids and other biomass precursor compounds, their relative NAPDH demands, and their 2H-fractionations (SI Appendix, Fig. S10). Indeed the 2H-depletion of glucose 2εfattyacid/water with faster fermentative growth (Fig. 4B) can be explained by greater expression of the large biosynthetic dKIE of lipid biosynthesis that originates from decreased NAPDH allocation to lipid synthesis vs. other materials (e.g., proteins, nucleic acids), which are less 2H-depleted and require less NADPH (SI Appendix, Fig. S10).
Lipid accumulation in the slowest growing glucose-fermenting cells which contained ~20 percent more palmitoleic acid than in fast growth (SI Appendix, Fig. S8) likely reflected extreme nutrient limitation (51) and dumping of excess NADPH. This is consistent with lower NADP+/NADPH ratios at slower growth (SI Appendix, Fig. S4). Faster input of nitrogen, the limiting nutrient in chemostats (SI Appendix, Fig. S5), enabled faster steady-state growth and the accumulation of more nitrogen-rich protein and RNA (52) and less build-up of carbon-rich lipid stores (53). This dynamic resulted in decreasing biomass C/N ratios at faster growth rates (SI Appendix, Figs. S8D and S13), as expected in glucose-fed cells (54).
Other Metabolic Isotope Signals in Yeast.
The production of unsaturated fatty acids from saturated fatty acids by desaturase enzymes with normal dKIE is expected to yield 2H-depleted unsaturated lipids relative to their saturated counterparts (55). We observed variable 2H-depletions between palmitoleic acid (one unsaturation) to fully saturated palmitic acid (2ε16:1/16:0 between ~ −50‰ and −150‰, SI Appendix, Figs. S11 and S12), especially in glycerol-fed cells (Fig. 2A and SI Appendix, Figs. S11 and S12). These could signify changes in the production/degradation turnover patterns between saturated and unsaturated lipid biosynthesis (SI Appendix, SI Discussion 2).
The 13C-enrichment of biomass from glycerol-respiration relative to that from sugar fermentation is also interesting (Fig. 2) and could reflect multiple metabolic differences. These include a large site-specific 13C isotopic fractionation associated with decarboxylation of substrate (56) and/or minimal 13C-fractionation by glycolysis enzymes (57) relative to the possible 13C preference of gluconeogenesis in certain fungi (58). Finally, it could reflect autophagic or possibly apoptotic yeast biomass recycling (59, 60), in which the release of 13C-depleted CO2 leads to 13C-enrichment of the biomass, which is consistent with enzyme activity, lipid unsaturation, and cell behavior in chemostats (SI Appendix, SI Discussion 3).
Conceptual Model of NADPH Source and Sink Control of Yeast Lipid/Water 2H-Fractionation.
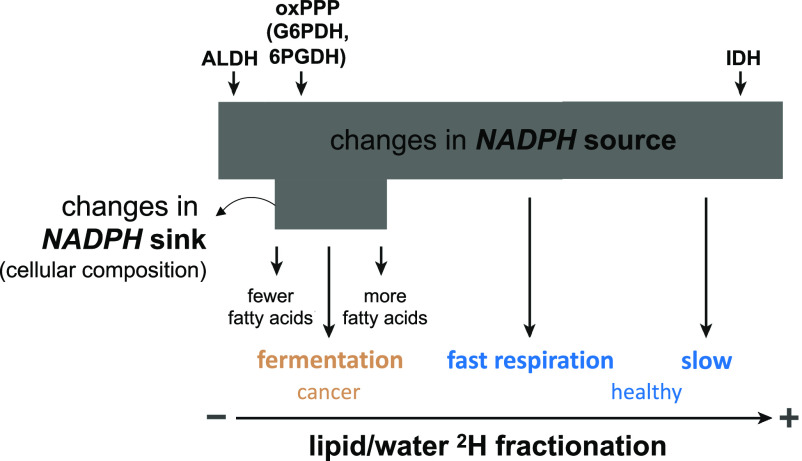
Our data exemplify how variations in NADPH sources and sinks across metabolisms and growth rates act in tandem to constrain the 2H-composition of the NADPH pool and determine lipid/water 2H-fractionation in yeast (Fig. 5). NADPH sources are the principal control on fractionation. The enzymatic sources of NADPH (Fig. 1), which have diverse dKIEs (SI Appendix, Table S1), strongly depend on yeast metabolism (Fig. 1) (22, 23) and can be affected by the NADPH demands of variable growth rates (e.g., NADPH sources shift with the rate of glycerol-fed growth, Fig. 4). This sets the stage for the largest differences in NADPH 2H/1H ratios, which are reflected in lipid 2H-compositions and thus in lipid/water 2H-fractionation (Fig. 5) due to the role of NADPH as a primary hydride donor for fatty acid biosynthesis (Fig. 1). NADPH consumption can act as a secondary and relatively minor control on lipid 2H (Fig. 5) through changes in the proportion of NADPH allocated between lipids and other cellular constituents, which vary in their NADPH requirements and biosynthetic 2H-fractionations (61–63). Additional factors such as antioxidant demand and enzyme-mediated hydrogen atom exchange (SI Appendix, SI Discussion 4) and enzyme substrate H (SI Appendix, SI Discussion 1) are possible tertiary levers on NADPH 2H/1H ratios.
Fig. 5.
Schematic diagram showing the primary importance of NADPH sources and secondary importance of NADPH sinks on the 2H-composition of NAPDH and lipid/water 2H-fractionation (2εlipid/water). The size of the gray box indicates the magnitude of change. Other tertiary sources include exchange of hydrogen atoms on NADPH with that in water and metabolites (SI Appendix, SI Discussions 1 and 4). IDH, isocitrate dehydrogenase; G6PDH, glucose-6-phosphate dehydrogenase, and 6PGDH, 6-phosphogluconate dehydrogenase in the oxPPP.
The data and interpretations presented here may help explain the diversity of growth rate vs. lipid/water 2H-fractionation trends observed so far in other microorganisms (SI Appendix, Fig. S14 and SI Discussion 5). Despite the fundamental differences between prokaryotes and compartmentalized eukaryotes, collectively yeast data are consistent with previous work on prokaryotes which established NADPH supply via diverse enzymes and δ2H of the substrate as the primary levers controlling lipid hydrogen isotope ratios across different metabolisms (1, 2).
Lipid/Water 2H-Fractionation in Healthy and Cancerous Murine Liver Cells.
Studies of disease metabolism often rely on metabolic flux analyses (MFA) that use isotopes introduced into metabolism at tracer levels (>10s of 1,000 per mil) in laboratory settings (64, 65). In contrast, changes in the natural abundance stable isotope composition of biomass and metabolites, traditionally applied to track biogeochemical processes, offer a passive/noninvasive tool to track metabolic processes important for health and disease. Importantly, the breadth of samples supporting such studies could be very large and include both living and nonliving organisms.
In view of metabolic similarities between yeast and cancer cells (15), we hypothesized that healthy cells using respiration would produce 2H-enriched lipids relative to cancer cells reliant on overflow metabolism due to respiration’s larger cytosolic IDH supply of NADPH. In a direct assessment of lipid 2H for disease, we cultured slow-growing, primary hepatocytes (liver cells) and fast proliferating, HEPA1-6 hepatocellular carcinoma cells (37, 38) from mice for 24 h on glucose-supplemented rich medium plates (65). Our pilot measurements of lipid/water 2H-fractionation (Table 1), which reflect both de novo synthesized lipids and those assimilated from diet or medium, are qualitatively consistent with both the growth rate and substrate patterns observed in yeast. Fast-growing cancer (fermenting) cells had lower lipid/water 2H-fractionation compared to slow-growing healthy (respiring) cells (Table 1, see schematic in Fig. 5). NADPH fluxes for de novo lipid synthesis have been determined by MFA in the same healthy murine hepatocytes and in the HepG2 human hepatocarcinoma-lineage under the conditions employed here (65). These results suggest IDH activity as the source of the observed lipid 2H-enrichment in respiring murine hepatocytes, similar to yeast. The sensitivity of hepatocyte lipid/water 2H-fractionation to IDH activity is much lower than in yeast. This likely reflects the large but unconstrained direct contribution of dietary and medium lipids [e.g., ~50 to 90% total lipids in healthy murine hepatocytes (65)] to lipid 2H, which would mute the NADPH source signals present in the de novo synthesized lipid pool. Other factors may also affect the interpretation of the data (SI Appendix, SI Discussion 6). Further study, including additional replicates, cell types, tissues, and species, is needed to assess lipid/water 2H-fractionation as a tool for NADPH flux tracking in animals.
Table 1.
Fatty acid/water 2H-fractionations for healthy and cancerous murine liver cells
| Sample | Metabolism | Growth (qualitative) | 2εC16:0/water (‰) | SD 2εC16:0/water (‰) | 2εC18:0/water (‰) | SD 2εC18:0/water (‰) |
|---|---|---|---|---|---|---|
| Primary hepatocyte | Respiration, healthy | Slow/no growth | −200 | 2.0 | −137 | 2.7 |
| Hepatoma (Hepa1-6) | Fermentation, cancer | Fast growth | −218 | 0.4 | −192 | 1.2 |
NADPH fluxes for de novo lipid synthesis were previously characterized by isotope tracing methods in the same or similar cell types (65). Data derive from n = 1 sample of each cell type and SD are from 3 to 4 analytical measurements.
The normal and aberrant operation of NADPH flows is thought to distinguish health and disease across many of life’s lineages, including cancer (65), viral infections in coccolithophores (66), postviral syndromes (67) and immune dysfunction (68) in humans, and bacterially infected plant crops (69). Our collective data, in conjunction with those of prokaryotes (1, 2), strongly point to the underrecognized potential of 2H-fractionation as a noninvasive screening tool for NADPH source tracking with diverse applications.
Conclusion
Studies of disease metabolism typically rely on complex stable isotope tracer additions into living organisms under specialized settings. The application of natural abundance stable isotope ratios that act as natural process tracers to study disease is underexplored. We have shown that lipid/water 2H-fractionation is sensitive to broad differences in NADPH cycling between respiration and overflow metabolism in yeast and, according to our preliminary data, in healthy and diseased murine liver cells. Our results help establish natural abundance hydrogen isotope ratios of lipids as a screening tool of metabolic change in eukaryotic organisms with potential wide applicability. Natural abundance 2H/1H ratios in lipids complement and provide direction for more complex and resource-intensive isotope tracer and -omics approaches that aim to quantify pools and fluxes of various metabolites. Our results point to diverse opportunities for developing natural abundance isotope tools to detect and characterize disease in eukaryotic organisms, including in humans.
Materials and Methods
Yeast Cell Cultures.
S. cerevisiae strain EXF-4126 (39) was obtained from The Microbial Culture Collection Ex, University of Ljubljana. Strain FY4 was obtained from the Rabinowitz lab at Princeton University. Chemostats consisted of autoclave-sterilized 2 L bioreactors (Chemglass Inc.,) at ~1 L working volumes at 30 °C, 400 rpm, aerated with filtered house air, and equipped with a pH/Temp/ORP InPro sensor (3253i Mettler-Toledo LLC, Columbus, OH), optical O2 sensors for strain EXF-4126 only (6860i Mettler-Toledo LLC, Columbus, OH), and a data-logging scale to continuously monitor conditions. Nitrogen-limited minimal media contained 0.053 g L−1 ammonium sulfate and 1.7 g L−1 yeast nitrogen base without ammonium sulfate and amino acids. Carbon substrates were 2% (20 g L−1) glucose or glycerol (strain EXF-4126) or 0.75% (7.5 g L−1) of glucose or galactose (strain FY4, which does not grow on glycerol). Batch cultures of strain FY4 were also grown in 150 mL baffled polycarbonate flasks, shaken at 250 rpm. See details on media, chemostat setup, operation, sampling, supernatant analyses, and strain integrity in SI Appendix, SI Methods 1.
Murine Liver Cell Cultures.
Primary hepatocytes isolated from wild-type C57BL/6 mice were cultured overnight in high glucose (4.5 g L−1, the same glucose stock used for yeast cell culturing) Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 1% penicillin–streptomycin, 100 nM insulin, 100 nM dexamethasone, and 1% GlutaMAX in collagen-coated six-well plates in a 37 °C, 5% CO2 incubator (65). Murine hepatoma HEPA1-6 cells from the American Type Culture Collection ATCC (CRL-1830) were cultured in DMEM with 10% fetal bovine serum (65). For plate experiments, primary hepatocytes and Hepa1-6 cells were grown for 24 h under identical conditions in the primary hepatocyte medium described above with the additional amendment of 0.584 g L−1 glutamine (SAFC). Plate experiments were terminated at 24 h by scraping cells off plates into collection tubes on dry ice and frozen until analysis. Mouse rearing followed previously described methods (65). Mouse work was approved by the Princeton University Institute Animal Care and Use Committee.
Fatty Acid Extraction and Analysis.
Fatty acids were extracted using 9:1 dichloromethane:methanol after freeze-drying samples. Cells collected on filters (strain FY4 cultures) were extracted by Automated Solvent Extraction (ASE-200, Dionex Corp., Sunnyvale, CA) at 1,500 psi and 100 °C for three 5 min cycles. Biomass from the 2% glycerol and glucose chemostats (strain EXF-4126) was quantitatively extracted via sonication and methylated in a single step (70) from 20 to 30 mg of dried biomass plus a recovery standard. Fatty acids from primary hepatocytes and HEPA1-6 cells were also extracted via sonication and methylated in a single step (70). Lipids were identified and quantified using a Trace 1310 gas chromatograph equipped with a flame ionization detector and an ISQTM mass spectrometer (MS) (Thermo Fisher Scientific, Waltham, MA). Lipids in sufficient abundance for isotope analysis were palmitic (n-C16:0), palmitoleic (n-C16:1), and oleic (n-C18:1) fatty acids. Fatty acid hydrogen isotope ratios were measured using a Trace 1310 gas chromatograph-GC Isolink II coupled to a 253 Plus isotope ratio mass spectrometer (IRMS) via a ConFlo IV (Thermo Fisher Scientific, Waltham, MA) in the Dept. of Geological Sciences, University of Colorado, Boulder. See SI Appendix, SI Methods 2 for detailed instrument settings.
Water and Substrate Hydrogen Isotopic Analysis.
Water samples from fresh media and filtered supernatant were analyzed for δ2Hwater by GasBench-IRMS at the UC Davis Stable Isotope Lab. Biosynthetic 2H-fractionation between media and lipids was calculated as 2εlipid/water = (2α − 1), where 2α = (δ2Hlipid + 1)/(δ2Hwater + 1). Substrate hydrogen isotope composition was determined by both substrate acetylation and equilibration methods (71), yielding average δ2H values of nonexchangeable substrate H of δ2Hglycerol = −45 ± 5‰, δ2Hglucose = 8 ± 10‰, and δ2Hgalactose = 40 ± 20‰. See SI Appendix, SI Methods 3 for details.
Biomass Biochemical Composition and Carbon Isotope Analysis.
Biomass samples for elemental analyses were collected from cultures, stored frozen, or freeze-dried until further analyses (see SI Appendix, SI Methods 4 for details). Total C and N of oven-dried (60 °C, overnight) samples prepared in tin capsules (9 × 10 mm, Elemental Microanalysis and Costech) were analyzed on a Vario ISOTOPE select CHNOS Elemental Analyzer (Elementar, Langenselbold, Germany), with measurements calibrated with an in-house aminocaproic acid standard (ACROS). Gases continued to the ISOPRIME IRMS (Elementar, Langenselbold, Germany) for subsequent isotope analysis and calibrated to certified standards including USGS25, USGS40, USGS65, USGS41, and our in-house aminocaproic acid standard (δ15N = +4.75‰; δ13C = −27.5‰). Biomass/substrate 13C isotope fractionation calculated as 13εbiomass/substrate = (13α − 1), where 13α = (δ13Cbiomass + 1)/(δ13Csubstrate + 1).
Enzyme and NAD(P)(H) Assays.
Enzyme and NAD(P)+/NAD(P)H assays were performed on fresh or frozen (−80 °C, less than 1 wk) cells for strain EXF-4126 only, using 35 to 50 mL samples obtained simultaneously from each batch culture collected from a sterile 50 mL syringe or from chemostat overflow into sterile 50 mL centrifuge tubes on ice (approximately 15 to 30 min collection time depending on growth rate). For the enzyme assays, samples were pelletized (2 °C, 1 min, max rpm), rinsed with 1 mL wash buffer [100 mM Tris HCL, pH = 8, 10 mM MgCl2, and protease inhibitor tabs (5 Complete Mini, EDTA-free tabs per 50 mL buffer)], repelletized, and then either frozen or processed immediately. See SI Appendix, SI Methods 5 for details. The NAD(P)(H) assays followed methods from ref. 72.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Acknowledgments
This work was funded by the Simons Foundation (732763 to A.E.M.), Princeton University Department of Geosciences Harry Hess Postdoctoral Fellowship (A.E.M.), and the High Meadows Environmental Institute Carbon Mitigation Initiative (X.Z.). We thank members of the Wing and Kopf labs (CU), especially N. Chaudhry, J.Colangelo-Lillis, C. Asamoto, and N. Dildar for culturing and analytical aid; S. Haynes, L. Sall, and I. Bystrom (PU) for chemostat help; T. Xiao (Rabinowitz lab, PU) for strain FY4; E. Delgado and R. Doucett (UC Davis) for analyzing water samples; S. Cunningham and K. Graves (MLML) for analyzing nutrient samples; S. Oleynik, J. Lueders-Dumont, K. Luxem, and V. Luu (PU) for EA-IRMS aid; B. Wing, S. Hammer, and Z. Garvin for sampling suggestions; and L. Reji, R. Darnajoux, E. Han, and other members of Ward-Zhang joint group meetings for feedback. We thank F. Morel for careful review of the manuscript and excellent recommendations of two anonymous reviewers to improve manuscript readability. We acknowledge the analytical contributions of the CU Boulder Earth Systems Stable Isotope Lab Core Facility (RRID:SCR_019300) and the Princeton University Geosciences Phillips Fund.
Author contributions
A.E.M. and X.Z. designed research; A.E.M., S.H.K., Z.Z., J.M., D.B.N., and A.L.M. performed research; A.E.M., S.H.K., Z.Z., D.B.N., A.L.M., and X.Z. contributed new reagents/analytic tools; A.E.M., S.H.K., Z.Z., J.M., D.B.N., A.L.M., and X.Z. analyzed data; and A.E.M., S.H.K., D.B.N., and X.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Ashley E. Maloney, Email: ashley.maloney@colorado.edu.
Xinning Zhang, Email: xinningz@princeton.edu.
Data, Materials, and Software Availability
Analysis scripts data have been deposited in GitHub (https://github.com/kopflab/2024_maloney_et_al) (73). All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Zhang X., Gillespie A. L., Sessions A. L., Large D/H variations in bacterial lipids reflect central metabolic pathways. Proc. Natl. Acad. Sci. U.S.A. 106, 12580–12586 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wijker R. S., Sessions A. L., Fuhrer T., Phan M., 2H/1H variation in microbial lipids is controlled by NADPH metabolism. Proc. Natl. Acad. Sci. U.S.A. 116, 1–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heinzelmann S. M., et al. , Impact of metabolism and growth phase on the hydrogen isotopic composition of microbial fatty acids. Front. Microbiol. 6, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osburn M. R., Dawson K. S., Fogel M. L., Sessions A. L., Fractionation of hydrogen isotopes by sulfate- and nitrate-reducing bacteria. Front. Microbiol. 7, 1–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hayes J. M., Fractionation of carbon and hydrogen isotopes in biosynthetic processes. Rev. Mineral. Geochem. 43, 225–227 (2001). [Google Scholar]
- 6.Schouten S., et al. , The effect of temperature, salinity and growth rate on the stable hydrogen isotopic composition of long chain alkenones produced by Emiliania huxleyi and Gephyrocapsa oceanica. Biogeosciences 3, 113–119 (2006). [Google Scholar]
- 7.van der Meer M. T. J., et al. , Large effect of irradiance on hydrogen isotope fractionation of alkenones in Emiliania huxleyi. Geochim. Cosmochim. Acta 160, 16–24 (2015). [Google Scholar]
- 8.Sachs J. P., “Hydrogen isotope signatures in the lipids of phytoplankton” in Treatise on Geochemistry, H. D. Holland, K. K. Turekian, Eds. (Elsevier Ltd., ed. 2, 2014), pp. 79–94. [Google Scholar]
- 9.Ladd S. N., Sachs J. P., Hydrogen isotope response to changing salinity and rainfall in Australian mangroves. Plant Cell Environ. 38, 2674–2687 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Sachs J. P., Maloney A. E., Gregersen J., Effect of light on 2H/1H fractionation in lipids from continuous cultures of the diatom Thalassiosira pseudonana. Geochim. Cosmochim. Acta 209, 204–215 (2017). [Google Scholar]
- 11.Sachs J. P., Kawka O. E., The influence of growth rate on 2H/1H fractionation in continuous cultures of the coccolithophorid Emiliania huxleyi and the diatom Thalassiosira pseudonana. PLoS One 10, e0141643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cormier M., Werner R. A., Leuenberger M. C., Kahmen A., 2H-enrichment of cellulose and n-alkanes in heterotrophic plants. Oecologia 189, 1–9 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Estep M. F., Hoering T. C., Stable hydrogen isotope fractionations during autotrophic and mixotrophic growth of microalgae. Plant Physiol. 67, 474–477 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Botstein D., Fink G. R., Yeast: An experimental organism for 21st century biology. Genetics 189, 695–704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natter K., Kohlwein S. D., Yeast and cancer cells—Common principles in lipid metabolism. Biochim. Biophys. Acta 1831, 314–326 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luengo A., et al. , Increased demand for NAD+ relative to ATP drives aerobic glycolysis. Mol. Cell 81, 691–707.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang P., Du W., Wu M., Regulation of the pentose phosphate pathway in cancer. Protein Cell 5, 592–602 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.S. Y. Lunt, M. G. Vander Heiden, Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 27, 441–464 (2011). [DOI] [PubMed] [Google Scholar]
- 19.Xiberras J., Klein M., Nevoigt E., Glycerol as a substrate for Saccharomyces cerevisiae based bioprocesses—Knowledge gaps regarding the central carbon catabolism of this ‘non-fermentable’ carbon source. Biotechnol. Adv. 37, 107378 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Klein M., et al. , Towards the exploitation of glycerol’s high reducing power in Saccharomyces cerevisiae-based bioprocesses. Metab. Eng. 38, 464–472 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Xiberras J., Klein M., Prosch C., Malubhoy Z., Nevoigt E., Anaplerotic reactions active during growth of Saccharomyces cerevisiae on glycerol. FEMS Yeast Res. 20, foz086 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Minard K. I., McAlister-Henn L., Sources of NADPH in yeast vary with carbon source. J. Biol. Chem. 280, 39890–39896 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Bruinenberg P. M., The NADP(H) redox couple in yeast metabolism. Antonie Van Leeuwenhoek 52, 411–429 (1986). [DOI] [PubMed] [Google Scholar]
- 24.Schmidt H.-L., Werner R. A., Eisenreich W., Systematics of 2H patterns in natural compounds and its importance for the elucidation of biosynthetic pathways. Phytochem. Rev. 2, 61–85 (2003). [Google Scholar]
- 25.Zhang Z., Chen L., Liu L., Su X., Rabinowitz J. D., Chemical basis for deuterium labeling of fat and NADPH. J. Am. Chem. Soc. 139, 14368–14371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito K., Akihiko K., Shigenobu O., Yousuke S., Tamio Y., Incorporation of hydrogen atoms from deuterated water and stereospecifically deuterium-labeled nicotinamide nucleotides into fatty acids with the Escherichia coli fatty acid synthetase system. Biochim. Biophys. Acta 618, 202–213 (1980). [PubMed] [Google Scholar]
- 27.Simon H., Kraus A., “Hydrogen isotope transfer in biological processes” in Isotopes in Organic Chemistry, Buncel E., Lee C., Eds. (Elsevier Science, 1976), pp. 153–229. [Google Scholar]
- 28.Ghisla S., Massey V., Mechanisms of flavoprotein-catalyzed reactions. Eur. J. Biochem. 181, 1–17 (1989). [DOI] [PubMed] [Google Scholar]
- 29.Leavitt W. D., Flynn T. M., Suess M. K., Bradley A. S., Transhydrogenase and growth substrate influence lipid hydrogen isotope ratios in Desulfovibrio alaskensis G20. Front. Microbiol. 7, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermes J. D., Roeske C. A., O’Leary M. H., Cleland W. W., Use of multiple isotope effects to determine enzyme mechanisms and intrinsic isotope effects. Malic enzyme and glucose 6-phosphate dehydrogenase. Biochemistry 21, 5106–5114 (1982). [DOI] [PubMed] [Google Scholar]
- 31.Rendina A. R., Hermes J. D., Cleland W. W., Use of multiple isotope effects to study the mechanism of 6-phosphogluconate dehydrogenase. Biochemistry 23, 6257–6262 (1984). [DOI] [PubMed] [Google Scholar]
- 32.Vedadi M., Meighen E., Critical glutamic acid residues affecting the mechanism and nucleotide specificity of Vibrio harveyi aldehyde dehydrogenase. Eur. J. Biochem. 246, 698–704 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Contreras-Shannon V., Lin A. P., McCammon M. T., McAlister-Henn L., Kinetic properties and metabolic contributions of yeast mitochondrial and cytosolic NADP+-specific isocitrate dehydrogenases. J. Biol. Chem. 280, 4469–4475 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Cook P. F., W. W., Cleland, pH variation of isotope effects in enzyme-catalyzed reactions. 1. Isotope- and pH-dependent steps the same. Biochemistry 20, 1797–1805 (1981). [DOI] [PubMed] [Google Scholar]
- 35.O’Leary M. H., Multiple isotope effects on enzyme-catalyzed reactions. Annu. Rev. Biochem. 58, 377–401 (1989). [DOI] [PubMed] [Google Scholar]
- 36.van Dijken J. P., Scheffers W. A., Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol. Lett. 32, 199–224 (1986). [Google Scholar]
- 37.Darlington G. J., Bernhard H. P., Miller R. A., Ruddle F. H., Expression of liver phenotypes in cultured mouse hepatoma cells. J. Natl. Cancer Inst. 64, 809–819 (1980). [PubMed] [Google Scholar]
- 38.Cassim S., Raymond V. A., Dehbidi-Assadzadeh L., Lapierre P., Bilodeau M., Metabolic reprogramming enables hepatocarcinoma cells to efficiently adapt and survive to a nutrient-restricted microenvironment. Cell Cycle 17, 903–916 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swinnen S., et al. , Re-evaluation of glycerol utilization in Saccharomyces cerevisiae: Characterization of an isolate that grows on glycerol without supporting supplements. Biotechnol. Biofuels 6, 1–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao T., Khan A., Shen Y., Chen L., Rabinowitz J. D., Glucose feeds the tricarboxylic acid cycle via excreted ethanol in fermenting yeast. Nat. Chem. Biol. 18, 1380–1387 (2022). [DOI] [PubMed] [Google Scholar]
- 41.Brauer M. J., Saldanha A. J., Dolinski K., Botstein D., Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell 16, 2503–2517 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Elser J. J., et al. , Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 10, 1135–1142 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Will O. H., Tieszen L. L., Gerlach T., Kellen M., Alteration of carbon isotope ratios by eight Ustilago species on defined media. Bot. Gaz. 150, 152–157 (1989). [Google Scholar]
- 44.Abraham W. R., Hesse C., Pelz O., Ratios of carbon isotopes in microbial lipids as an indicator of substrate usage. Appl. Environ. Microbiol. 64, 4202–4209 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blair N., et al. , Carbon isotopic fractionation in heterotrophic microbial metabolism. Appl. Environ. Microbiol. 50, 996–1001 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramachandran N., Durbano M., Colman R. F., Kinetic isotope effects in the NAD- and NADP-specific isocitrate dehydogneases of pig heart. FEBS Lett. 49, 129–133 (1974). [DOI] [PubMed] [Google Scholar]
- 47.O’Leary M. H., Limburg J. A., Isotope effect studies of the role of the metal ions in isocitrate dehydrogenase. Biochemistry 16, 1129–1135 (1977). [DOI] [PubMed] [Google Scholar]
- 48.Kreuzer H. W., et al. , Detection of metabolic fluxes of O and H atoms into intracellular water in mammalian cells. PLoS One 7, e39685 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kreuzer-Martin H. W., Ehleringer J. R., Hegg E. L., Oxygen isotopes indicate most intracellular water in log-phase Escherichia coli is derived from metabolism. Proc. Natl. Acad. Sci. U.S.A. 102, 17337–17341 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kreuzer-Martin H. W., Lott M. J., Ehleringer J. R., Hegg E. L., Metabolic processes account for the majority of the intracellular water in log-phase Escherichia coli cells as revealed by hydrogen isotopes. Biochemistry 45, 13622–13630 (2006). [DOI] [PubMed] [Google Scholar]
- 51.He Q., et al. , Oleaginicity of the yeast strain Saccharomyces cerevisiae D5A. Biotechnol. Biofuels 258, 1–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lange H. C., Heijnen J. J., Statistical reconciliation of the elemental and molecular biomass composition of Saccharomyces cerevisiae. Biotechnol. Bioeng. 75, 334–344 (2001). [DOI] [PubMed] [Google Scholar]
- 53.Cortassa S., Aon J. C., Aon M. A., Fluxes of carbon, phosphorylation, and redox intermediates during growth of Saccharomyces cerevisiae on different carbon sources. Biotechnol. Bioeng. 47, 193–208 (1995). [DOI] [PubMed] [Google Scholar]
- 54.Larsson C., von Stockar U., Marison I., Gustafssont L., Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J. Bacteriol. 175, 4809–4816 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chikaraishi Y., Suzuki Y., Naraoka H., Hydrogen isotopic fractionations during desaturation and elongation associated with polyunsaturated fatty acid biosynthesis in marine macroalgae. Phytochemistry 65, 2293–2300 (2004). [DOI] [PubMed] [Google Scholar]
- 56.Zhang B. L., Buddrus S., Trierweiler M., Martin G. J., Characterization of glycerol from different origins by 2H- and 13C-NMR studies of site-specific natural isotope fractionation. J. Agric. Food Chem. 46, 1374–1380 (1998). [Google Scholar]
- 57.DeNiro M. J., Epstein S., Mechanism of carbon isotope fractionation associated with lipid synthesis. Science 197, 261–263 (1977). [DOI] [PubMed] [Google Scholar]
- 58.Kohzu A., et al. , Dynamics of 13C natural abundance in wood decomposing fungi and their ecophysiological implications. Soil Biol. Biochem. 37, 1598–1607 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Carmona-Gutierrez D., et al. , Apoptosis in yeast: Triggers, pathways, subroutines. Cell Death Differ. 17, 763–773 (2010). [DOI] [PubMed] [Google Scholar]
- 60.Kourtis N., Tavernarakis N., Autophagy and cell death in model organisms. Cell Death Differ. 16, 21–30 (2009). [DOI] [PubMed] [Google Scholar]
- 61.Xu J. Z., Yang H. K., Zhang W. G., NADPH metabolism: A survey of its theoretical characteristics and manipulation strategies in amino acid biosynthesis. Crit. Rev. Biotechnol. 38, 1061–1076 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Slekar K. H., Kosman D. J., Culotta V. C., The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 271, 28831–28836 (1996). [DOI] [PubMed] [Google Scholar]
- 63.Celton M., et al. , A comparative transcriptomic, fluxomic and metabolomic analysis of the response of Saccharomyces cerevisiae to increases in NADPH oxidation. BMC Genomics 13, 1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Antoniewicz M. R., A guide to 13C metabolic flux analysis for the cancer biologist. Exp. Mol. Med. 50, 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Z., et al. , Serine catabolism generates liver NADPH and supports hepatic lipogenesis. Nat. Metab. 3, 1608–1620 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bidle K. D., Vardi A., A chemical arms race at sea mediates algal host-virus interactions. Curr. Opin. Microbiol. 14, 449–457 (2011). [DOI] [PubMed] [Google Scholar]
- 67.Paul B. D., Lemle M. D., Komaroff A. L., Snyder S. H., Redox imbalance links COVID-19 and myalgic encephalomyelitis/chronic fatigue syndrome. Proc. Natl. Acad. Sci. U.S.A. 118, 1–10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Panday A., Sahoo M. K., Osorio D., Batra S., NADPH oxidases: An overview from structure to innate immunity-associated pathologies. Cell. Mol. Immunol. 12, 5–23 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balmant K. M., et al. , Redox proteomics of tomato in response to Pseudomonas syringae infection. Hortic. Res. 2, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rodríguez-Ruiz J., Belarbi E. H., Sánchez J. L. G., Alonso D. L., Rapid simultaneous lipid extraction and transesterification for fatty acid analyses. Biotechnol. Tech. 12, 689–691 (1998). [Google Scholar]
- 71.Holloway-Phillips M. M., et al. , Species variation in the hydrogen isotope composition of leaf cellulose is mostly driven by isotopic variation in leaf sucrose. Plant Cell Environ. 45, 2636–2651 (2022). [DOI] [PubMed] [Google Scholar]
- 72.Kern S. E., Price-Whelan A., Newman D. K., “Extraction and Measurement of NAD(P)+ and NAD(P)H” in Pseudomonas Methods and Protocols, Methods in Molecular Biology, Filloux A., Ramos J.-L., Eds. (Springer Science+Business Media, 2014), pp. 311–323. [DOI] [PubMed] [Google Scholar]
- 73.Maloney A. E., Kopf S., Zhang X., KopfLab/2024_maloney_et_al. Github. https://github.com/kopflab/2024_maloney_et_al. Deposited 15 April 2024.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Data Availability Statement
Analysis scripts data have been deposited in GitHub (https://github.com/kopflab/2024_maloney_et_al) (73). All study data are included in the article and/or supporting information.