Abstract
Now that the meningococcal genome sequence has been completed, the lack of a suitable method for saturation mutagenesis remains a major obstacle to the unraveling of the pathogenic propensity of Neisseria meningitidis. Here, we demonstrate that in vitro Himar1 mariner transposition on chromosomal or PCR-amplified meningococcal DNA, which is subsequently reintroduced into N. meningitidis by natural transformation, is an extremely efficient mutagenesis method. Southern blot analysis, sequencing the Himar1 insertion point in numerous transposition mutants, and a limited screening of the mutant libraries for clones impaired in maltose catabolism confirmed that Himar1 transposed randomly in N. meningitidis. Taken together, these data demonstrate that Himar1 in vitro transposition can lead to the exhaustive mutagenesis of N. meningitidis, allowing for the first time a genomic-scale mutational analysis of this important human pathogen.
With the completion of the genomic sequences of two different strains, Z2491 (24) and MC58 (38), Neisseria meningitidis entered the postgenomic era (35). The first example of the utility of such a sequence was the in silico identification of several promising vaccine candidates against serogroup B meningococci (28) for which no vaccine is available. It is very likely that further deciphering of its genomic content will lead to a better comprehension of the genetic determinants essential for the meningococcus peculiar properties. However, as already noted for other bacteria whose genomes have been sequenced, almost half (46.3%) of the identified genes in MC58 (38) encode proteins with no predictable function. In addition, some of the functions assigned on the basis of sequence homologies may turn out to be erroneous. Therefore, to convert sequence data into meaningful biological information, two kinds of mutagenesis procedures are required: one allowing the rapid mutagenesis of defined genes for the identification of the linked phenotypes, and one allowing random mutagenesis of the whole genome for the identification of the genes responsible for defined phenotypic traits.
Unfortunately, none of the presently available mutagenesis methods seems usable as such for a genomic-scale mutagenesis of N. meningitidis. Indeed, although facilitated by the fact that the meningococcus is naturally transformable, allelic exchange mutagenesis remains mostly a gene-by-gene analysis method. Unless a high-throughput strategy involving numerous laboratories is envisioned, as for the budding yeast Saccharomyces cerevisiae (42), allelic exchange seems impracticable for a saturation mutagenesis of N. meningitidis. For example, mutagenesis of the 2,230 predicted open reading frames (ORFs) in the genome of Z2491 (24) would require at least 4,460 cloning steps (one step to clone each gene and another one to introduce a resistance cassette within each of them). Moreover, some genomic regions are known to be refractory to cloning in Escherichia coli and would therefore be difficult to mutagenize. Therefore, transposon mutagenesis seems to be the method of choice. However, conventional transposon mutagenesis has been limited in Neisseria mostly due to the lack of suitable neisserial transposons. Moreover, derivatives of well-known E. coli mobile elements, such as Tn5, are not active in Neisseria (39). Consequently, conjugative transposons and shuttle mutagenesis have been used to create mutant libraries in Neisseria. Conjugative transposons like Tn916 (9) are large elements that are self-transferable by conjugation to a variety of bacteria, where they integrate into the chromosome by transposition. When introduced into N. meningitidis, Tn916 (15) and a Tn916-like derivative (21) transposed into different sites, as evidenced by the identification of mutants presenting precise genetic defects (8, 34, 37). Conjugative transposons are, however, not practical for saturation mutagenesis of N. meningitidis for several reasons: (i) the frequency of transposition is low, and thus the number of mutants falls far short of the number statistically required for the mutagenesis of all N. meningitidis genes; (ii) the insertions are not perfectly stable (36); and (iii) transposition presents some site specificity. Some of these drawbacks are absent in shuttle mutagenesis, where cloned neisserial DNA is mutated by transposition within E. coli and subsequently transferred into Neisseria, where it inactivates the corresponding genes via allelic exchange (33). This technique has been refined with the construction of versatile minitransposons that permit the creation of lacZ transcriptional fusions (6), the production of phoA fusions (5, 12) for the identification of exported proteins that may play a role in the pathogenesis (14), and signature tag mutagenesis of N. meningitidis (7). Although it makes possible the creation of large libraries of mutants, which are predicted to be stable due to the use of minitransposons, this method is limited in part by the fact that there are genes refractory to cloning in E. coli which are difficult to mutate.
Because in vitro transposition systems, using mobile elements such as Tn7 (11), Himar1 (2), and Ty1 (30), are particularly suitable for the mutagenesis of naturally competent bacteria, as has been demonstrated in Haemophilus influenzae and Streptococcus pneumoniae, we reasoned that the use of such a system in N. meningitidis would overcome the problems associated with the previous transposition systems. Indeed, in addition to the advantages previously listed for shuttle mutagenesis, in vitro transposition does not require the target DNA to be cloned. DNA is mutated in vitro, using a minitransposon and a purified transposase, and is reintroduced into the bacterium to be mutated, where it inactivates the corresponding genes via allelic exchange. Because Himar1 mariner transposition is random and very efficiently mediated by a single protein that is easily overproduced and purified in E. coli (18), we chose to use this mobile element to develop an in vitro transposon mutagenesis method for N. meningitidis. We thus constructed a minitransposon consisting of Himar1 inverted repeat sequences flanking a gene coding for kanamycin resistance in N. meningitidis and an uptake sequence (10) that is required for the DNA to be taken up during natural transformation. We demonstrated that this Himar1 derivative efficiently transposed on PCR products, allowing the site-specific mutagenesis of defined genes, but also on chromosomal DNA, thereby permitting for the first time the creation of comprehensive mutant libraries. Therefore, in conjunction with the genome sequence, in vitro Himar1 transposition allows a genomic-scale mutational analysis of N. meningitidis.
MATERIALS AND METHODS
Bacterial cultures.
E. coli was routinely grown on liquid or solid Luria-Bertani medium, and kanamycin was used at 20 μg/ml when required.
Because N. meningitidis Z2491, whose genome sequence has been recently completed by the Sanger Centre (24), is not transformable, we opted for Z5463, formerly designated C396, another strain that was isolated from a patient with meningitis in The Gambia in 1983 (1). Z5463 is a naturally transformable serogroup A strain that belongs to the same clonal group as Z2491, thus allowing the use of the genomic sequence of the latter strain. N. meningitidis was grown on GCB agar medium (Difco) containing the Kellog supplements (16), i.e., 4 g of glucose per liter, 200 ng of thiamine per liter, 5 mg of ferric nitrate per liter, and 100 ng of l-glutamine per liter. The plates were incubated for 16 h at 37°C in a moist atmosphere containing 5% CO2.
To assay maltose catabolism by N. meningitidis mutants, individual clones were grown for 6 h at 37°C on 96-well microtiter plates containing GCB liquid medium with 2% of a solution of phenol red (2 g/liter) and supplemented with the Kellog supplements, except that glucose was replaced by maltose. Sugar-degrading bacteria change the color of the medium from red to yellow.
Molecular biology techniques.
Routine molecular biology techniques were carried out as recommended (32). DNA sequences, performed with the Big-Dye primer sequencing kit, were read on an ABI-Prism 310 automated sequencer (PE Applied Biosystems). Southern blot analysis was done as previously described (25).
N. meningitidis chromosomal DNA was prepared as follows from overnight (ON) cultures on GCB agar plates. A loopful of bacteria from one plate was resuspended in 500 μl of lysis buffer (50 mM NaCl, 20 mM EDTA, 50 mM Tris [pH 8], 1% sodium dodecyl sulfate, 100 μg of proteinase K), and the sample was incubated for 5 to 10 min at 42°C. The clear lysate was extracted several times with phenol-chloroform, and the DNA was concentrated by ethanol precipitation.
Construction of vectors.
Plasmid pMM2611 containing the mini-Himar1, consisting of the first and last 100 bp of Himar1, was constructed by PCR-ligation-PCR mutagenesis (3) using pMM26 as a template (19). The primers used for the first half-reaction were T2828r (5′-TGAAAAAGGAAGAGTATGAG-3′) and 76rSma (5′-TACCCGGGAATCATTTGAAGGTTGGTAC-3′). T2380f (5′-TTACATGATCCCCCATGTTG-3′) and 1218f (5′-TCGCTCTTGAAGGGAACTATG-3′) were used for the second half-reaction. The assembly reaction used the ligation products from each half-reaction and primers T2828r and T2380f. The final PCR product was cloned into the EcoRV site of pCDNAII (Invitrogen). The transposon donor vector pSM1, which was used as a donor for all in vitro transposition reactions, was constructed by cloning the aphA-3 gene (41), encoding kanamycin resistance (Kmr) in Neisseria, and a neisserial uptake sequence (10) into the mini-Himar1 present on plasmid pMM2611. The 1.5-kbp kanamycin resistance cassette was amplified using primers Km6 (5′-CGGGATCCGCCGTCTGAACCAGCGAACCATTTGAGG-3′), where the uptake sequence (10) is in bold, and Km7 (5′-ACGCGTCGACGCTTTTTAGACATCTAAATCTAGG-3′). The PCR fragment was made blunt ended with T4 DNA polymerase and cloned into the unique SmaI site of pMM2611.
The ssa1 gene, previously identified by subtractive hybridization (26), was chosen as a target in our site-specific mutagenesis experiments. It was PCR amplified from Z5463 chromosomal DNA using primers oligo R (5′-CGTGCCTGAAATGTCGTTAC-3′) and oligo F (5′-ACCCCAACCTTCCCTACAAA-3′). Where indicated, the 1.5-kbp PCR product was directly cloned into the pCR2.1-TOPO vector using the TOPO TA cloning kit (Invitrogen) as outlined by the manufacturer.
Transformation of N. meningitidis.
An optimized transformation protocol was designed for this study to detect rare transposition events. Bacteria grown ON on GCB plates were resuspended at a final optical density at 550 nm of ca. 5 in transformation buffer, i.e., supplemented GCB liquid medium containing 5 mM MgCl2. Aliquots (200 μl) of the previous bacterial suspension were transferred into the wells of a 24-well tissue culture plate, and approximately 1 μg DNA was added. Transformation mixtures were incubated for 30 min at 37°C with strong shaking. Then 1.8 ml of prewarmed transformation buffer was added, and incubation was continued for 2 h at 37°C. It is important to note that during this outgrowth, no increase of the bacterial population was observed. Transformants were selected on GCB agar plates containing 200 μg of kanamycin per ml. We reproducibly obtained approximately 8 × 105 transformants/μg of chromosomal DNA.
In vitro transposition reactions.
Purification of a hyperactive C9 mutant transposase of Himar1 (17) and in vitro transposition reactions (18) were performed essentially as described previously. However, some parameters known to enhance the rate of transposition in vitro (19), such as incubation time, temperature, and MgCl2 concentration, were optimized in our reactions. Transposition reactions were carried out in 10% glycerol–2 mM dithiothreitol–250 μg of bovine serum albumin per ml–25 mM HEPES [pH 7.9]–100 mM NaCl–10 mM MgCl2. They contained, in a final volume of 20 μl, at least 500 ng of donor and target DNAs and 120 nM transposase. After a 3-h incubation at 30°C, a 10-min exposure at 75°C was done to inactivate the transposase and the reaction products were directly purified using the Qiaex II gel extraction kit (Qiagen). Before the DNA was transformed into N. meningitidis, single-stranded gaps, introduced upon Himar1 transposition (18), were repaired. Gaps in the DNA were first filled, for 30 min at 16°C, with T4 DNA polymerase in a final volume of 20 μl. Reactions were carried out in 1× buffer 2 (NEB) containing 50 ng of bovine serum albumin per μl and 1 mM concentrations of each deoxynucleoside triphosphate. The enzyme was heat inactivated by a 10-min exposure at 75°C. After addition of 2 μl of ligation mix (22 mM ATP, 5 U of T4 DNA ligase), the mixture was further incubated ON at 16°C. Usually half of the repaired transposition products (11 μl) were used to transform N. meningitidis. Clones that had inserted the transposon into the chromosome were selected on kanamycin-containing plates.
Mapping of Himar1 insertion sites.
Because the identification of genomic DNA sequences flanking the transposon in transposon mutants is a long and labor-intensive procedure, especially when numerous clones are to be analyzed, we used a simple and efficient ligation-mediated PCR technique (LMPCR) that was previously used for identifying the flanking sequences in mycobacterial transposon mutants (29). The linkers were formed by annealing LMP1 (5′-TAGCTTATTCCTCAAGGCACGAGC-3′) with LMP2 (5′-GATCGCTCGTGC-3′) or LMP3 (5′-CCGGGCTCGTGC-3′); the underlined sequences correspond to complementary sequences in the primers, whereas sequences complementary to cohesive ends generated by Sau3AI (LMP2) or NgoAIV (LMP3) are in bold. Sau3AI and NgoAIV were chosen because they generate relatively short DNA fragments upon restriction of N. meningitidis chromosomal DNA. The complete genomic sequence of Z2491 (24) contains, on average, one Sau3AI site every 1,001 bp and one NgoAIV site every 1,728 bp. After ligation of the linkers to digested genomic DNA, the insertion sites were amplified with AmpliTaq Gold DNA polymerase (PE Applied Biosystems) using LMP1 and IR1 (5′-CCGGGGACTTATCAGCCAACC-3′), an outward primer internal to the 27-bp inverted repeats of the Himar1 element. PCR products were gel purified using the Qiaex II gel extraction kit and directly sequenced using LMP1 as a primer. When the amplified fragments were too long to be sequenced in extenso, they were cloned using the TOPO TA cloning kit before being sequenced using suitable primers from the kit. Sequences were mapped onto the genome of N. meningitidis Z2491 using the BLASTN (4) and Artemis programs of the Sanger Centre (http://www.sanger.ac.uk/Projects/N_meningitidis). When appropriate, the interrupted ORFs were compared with the National Center for Biotechnology Information database by using the TBLASTN program (4).
RESULTS
Transposition on defined DNA fragments: site-specific mutagenesis.
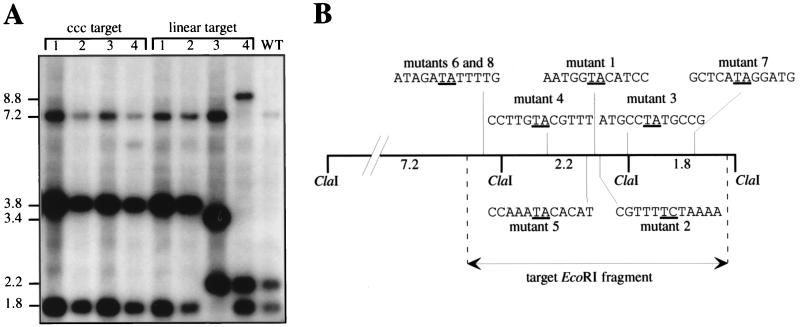
As already mentioned, an important aspect of genomic analysis is the capability to engineer, in the shortest possible time, numerous site-specific mutations in order to identify the phenotypes linked with the proteins encoded by defined genes. To test the feasibility of Himar1 transposition for this purpose in N. meningitidis, we used as a target the neisserial hrtA locus, which presents significantly enhanced transformation and recombination efficiencies in the meningococcus (7), to increase the probability of detecting transposon mutants. The covalently closed circular (CCC) pHC6 plasmid (7), containing the hrtA locus as a 4.5-kbp EcoRI fragment, was thus subjected in vitro to Himar1 mutagenesis. Transforming a single transposition reaction product into N. meningitidis Z5463 typically gave 2,000 Kmr transformants. A total of 20 randomly picked colonies were analyzed by Southern blotting of ClaI-digested chromosomal DNA, using the pHC6 plasmid as a probe. Because there are two ClaI sites 2 kbp apart in the hrtA locus, three hybridizing fragments of 7.2, 2.2, and 1.8 kbp, respectively, were obtained with wild-type Z5463 DNA (Fig. 1A). Because ClaI does not cut in the mini-Himar, we expected transposon mutants to present equally three hybridizing fragments, one of which should present a 1.6-kbp increase in length corresponding to the size of the inserted mini-Himar1. Indeed, all of the analyzed Kmr transformants, four of which are shown in Fig. 1A, presented the same hybridizing pattern, with three ClaI fragments of 7.2, 3.8, and 1.8 kbp. Therefore, the 1.6-kbp increase in length observed for the central ClaI fragment, 2.2 kbp in the wild type, confirmed that the transformants indeed resulted from the transposition of Himar1 into the hrtA locus.
FIG. 1.
Southern blot analysis (A) and mapping of Himar1 insertions into the hrtA locus (B) of representative transposon mutants. (A) The target fragment containing the hrtA locus was either cloned (CCC plasmid) or linear. Genomic DNA of eight mutants was digested with ClaI and probed with the pHC6 plasmid containing the hrtA locus on a 4.5-kbp EcoRI fragment. N. meningitidis Z5463 genomic DNA (WT) was included as a control. Molecular sizes are indicated in kilobase pairs. (B) Distribution of eight insertions on the ClaI restriction map of the hrtA genomic locus. The sequences of the insertions sites are highlighted, and the target dinucleotide that is duplicated upon transposition is underlined. Note that the mutants whose Himar1 insertion sites were sequenced are different from the mutants shown in panel A.
To test the feasibility of using uncloned DNA as a target for Himar1 mutagenesis, the hrtA locus was also mutagenized as a gel-purified 4.5-kbp EcoRI fragment. When a single mutagenesis reaction product was transformed into N. meningitidis Z5463, 200 Kmr transformants were selected. There was thus a 10-fold decrease in the number of transformants compared to the previous experiments, where CCC DNA was used as a target. This might reflect the different conformation of the DNA in the CCC plasmid, to which Himar1 is known to be sensitive (19). Southern blot analysis of eight randomly picked transformants, four of which are shown in Fig. 1A, confirmed that the tested clones were transposition mutants. Indeed, they presented one hybridizing ClaI fragment that was 1.6 kbp longer than the corresponding fragment seen in the wild-type strain. Interestingly, unlike what was observed when CCC DNA was used as the target, transposon insertions were more evenly scattered within the target hrtA locus: five insertions in the central 2.2-kbp ClaI fragment, two in the left 7.2-kbp fragment, and one in the right 1.8-kbp fragment (Fig. 1A). To easily identify sequences flanking the Himar1 after insertional mutagenesis and thus analyze the randomness of the transposition, we adapted LMPCR (29) to Neisseria. After amplifying them by LMPCR, we sequenced and mapped the Himar1 insertion sites in the hrtA locus for eight mutants (Fig. 1B). Except for the insertion sites of mutants 6 and 8, in the left 7.2-kbp ClaI fragment (Fig. 1B), which were identical and thus probably siblings, the mapping revealed that the insertion sites were different, demonstrating that Himar1 displayed no apparent site specificity. As previously noted by others (18, 31), transposition occurred mainly after a TA dinucleotide (in seven of eight mutants) that was consequently duplicated.
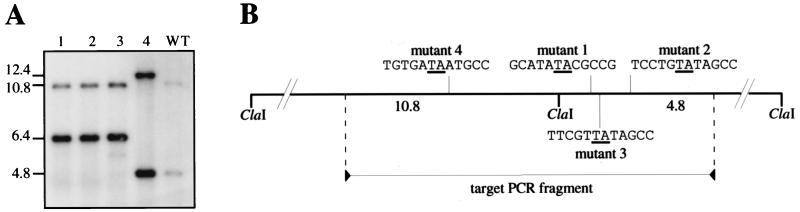
To demonstrate the general validity of Himar1 in vitro transposition for site-specific mutagenesis in N. meningitidis, transposition was performed on a neisserial locus presenting a normal rate of transformation, unlike hrtA. We chose as a target the ssa1 gene, which was previously identified by RDA subtractive hybridization (26). This gene was particularly interesting because it is absent from the commensal N. lactamica and is homologous to a serine protease, a virulence-associated protein from Pseudomonas fluorescens (26). We amplified by PCR part of the ssa1 gene from N. meningitidis on a 1.5-kbp fragment and subjected it to Himar1 in vitro transposition either directly or after cloning. As expected, we obtained fewer Kmr transformants than in the previous experiment with the highly transformable hrtA locus. Typically, we obtained 5 to 10 Kmr transformants per single mutagenesis reaction. Interestingly, as opposed to the previous results with the hrtA locus, the prior cloning of the PCR fragment did not seem to favor Himar1 transposition. Several Kmr transformants were analyzed by Southern blotting, using the ssa1 PCR product as a probe (Fig. 2A). ClaI-digested wild-type Z5463 DNA, included as a control, gave two hybridizing fragments of 4.8 and 10.8 kbp because the target contains a ClaI restriction site (Fig. 2A). As in the previous experiments, all the tested transformants were transposition mutants because they presented one hybridizing fragment with a 1.6-kbp increase in length, corresponding to the size of the inserted mini-Himar1. Of four mutants analyzed, three had insertions in the right 4.8-kbp ClaI fragment whereas one had the mini-Himar1 inserted in the left 10.8-kbp ClaI fragment (Fig. 2A). The insertions sites of Himar1 in the different mutants were amplified by PCR using as primers a sequence in the Himar1 inverted repeat and a sequence in the target gene. The different lengths of the amplified fragments (data not shown) confirmed that the four transposon insertion sites were different, which was evidenced by sequencing and mapping them on the ssa1 genomic locus (Fig. 2B). In all four mutants, transposition occurred after a TA dinucleotide that was consequently duplicated. In conclusion, in vitro Himar1 transposition is of general use for site-specific mutagenesis in N. meningitidis.
FIG. 2.
Southern blot analysis (A) and mapping of the insertions into the ssa1 gene (B) of four representative transposon mutants. (A) Genomic DNA of N. meningitidis Z5463 (WT) and of four mutants (1 to 4) resulting from transposition into the ssa1 gene was digested with ClaI and probed with the 1.5-kbp PCR fragment corresponding to ssa1 that was used as a target for transposition. Molecular sizes are indicated in kilobase pairs. (B) Distribution of the insertions on the ClaI restriction map of the ssa1 genomic locus. The sequences of the insertion sites are highlighted, and the target dinucleotide that is duplicated upon transposition is underlined.
Transposition on chromosomal DNA: random mutagenesis.
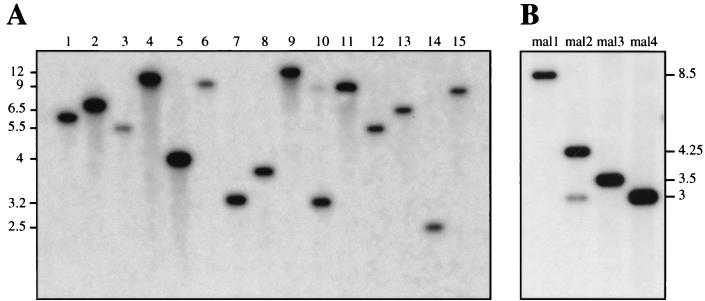
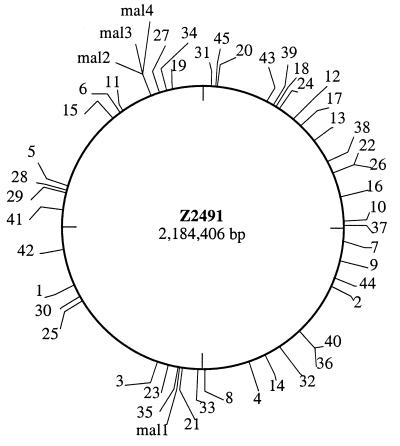
Considering the above results, we expected in vitro Himar1 transposition to be a suitable tool for the creation of comprehensive transposon mutant libraries in N. meningitidis, the lack of which has been an important obstacle to the genetic characterization of its pathogenic propensity. We therefore performed Himar1 mutagenesis on Z5463 chromosomal DNA and reproducibly obtained up to 10,000 Kmr colonies per single reaction. ClaI-digested DNA of more than 50 clones, of which 15 are shown in Fig. 3A, was analyzed by Southern blotting using the aphA-3 cassette as a probe. The hybridization patterns for the tested mutants, singly hybridizing ClaI fragments of different sizes (Fig. 3A), were in agreement with a random distribution of Himar1 insertions on the chromosome. This was confirmed by Southern analysis of EcoRV-digested chromosomal DNA (data not shown). By using LMPCR (29), we amplified the Himar1 insertion sites in 45 mutants. The sequencing of the amplified fragments again confirmed that transposition occurred at random with no apparent site specificity (Table 1). The positioning of the insertion sites on the N. meningitidis Z2491 genomic sequence showed that they were present within every quadrant of the genome (Fig. 4), suggesting that there is no obvious hot spot for Himar1 transposition. The chromosomal locations into which transposition had occurred were readily identified because our raw sequences, except for minor sequencing errors, were almost identical to parts of the complete sequence of Z2491 (24). Because the coding density of Z2491 is 82.9% (24), we expected a similar percentage of the sequenced transposition sites to be within ORFs. Indeed, in agreement with a random distribution of Himar1 in the chromosome, in 35 of 45 mutants (77.8%) transposition occurred within ORFs, 42.9% of which could not be ascribed a potential function (Table 1).
FIG. 3.
Southern blot analysis of random insertions into the chromosome. Genomic DNA of 15 (1 to 15) representative random transposon mutants (A) and four (mal1 to mal4) mutants that present an impaired ability to utilize maltose (B) was digested with ClaI and probed with a PCR product that corresponds to the kanamycin resistance gene present in the minitransposon. Molecular sizes are indicated in kilobase pairs.
TABLE 1.
Sequence and location of random Himar1 insertion sites and potential function of the interrupted genes
| Mutant | Sequence at point of insertiona | Point of insertiona | NmORFb | Potential functionb (organism) |
|---|---|---|---|---|
| 1 | ATCGACAATAATGCTACA | 1489020 | 1582 | Unknown |
| 2 | GTTGGCCATATATTGCCA | 696195 | 708 | N-acetyl-β-glucosaminidase (Vibrio furnissii) |
| 3 | GGACGATATAGAACGTGG | 1202078 | 1274 | Phosphoglyceromutase (Zymomonas mobilis) |
| 4 | AAACCGCGTATTGTACGT | 973969 | —c | |
| 5 | ATCTTATGTATGATAATT | 1743815 | 1799 | Unknown |
| 6 | GCAGTCCGTATAAGTATT | 1963690 | 2024 | Transferrin binding protein 1 (N. meningitidis) |
| 7 | GCCGACCATACTGCCGAA | 581224 | 608 | Carbamoyl phosphate synthase small subunit (N. gonorrhoeae) |
| 8 | CGGCAAGATAGGTGATGT | 1084167 | 1136 | Homoserine O-acetyltransferase (Leptospira meyeri) |
| 9 | GAACGAAATACCCATCGA | 628684 | 643 | Lipopolysaccharide biosynthesis protein (Thermotoga maritima) |
| 10 | CGGCATTATATAGAAACC | 526471 | — | |
| 11 | GGTTGATTGAAAAGGCGG | 1972409 | — | |
| 12 | AAGAAGCGTAGGCTTTGC | 240924 | 257 | 3-Phosphoglycerate kinase (Alcaligenes eutrophus) |
| 13 | TACCCCCATATGGTGTTG | 313623 | — | |
| 14 | TTTTATTGTATCAATACT | 933137 | — | |
| 15 | TAAAAGGATATAGCCTGA | 1941332 | 2006 | Unknown |
| 16 | TACCGGCGTAAGATATTG | 466730 | 478 | Serine protease homologue (Pseudomonas fluorescens) |
| 17 | CGATAATATATTATTCAT | 265542 | — | |
| 18 | CAGATAAATACCCATACC | 190812 | 207 | Unknown |
| 19 | GGTCGATGTATTGGACTC | 2103164 | 2155 | PilG pilus assembly protein (N. meningitidis) |
| 20 | CCACAGTATAAAATTATT | 33411 | 40 | Unknown |
| 21 | GAGGGCACTATTCCAAAT | 1140781 | 1200 | Hia adhesin (Haemophilus influenzae) |
| 22 | ATCACCAATATTTCCGCC | 404824 | 430 | HrpA helicase (E. coli) |
| 23 | ACACGGTATAAGCATCCG | 1174994 | 1248 | Unknown |
| 24 | AACCGAACTATACGCCCA | 200134 | — | |
| 25 | CAGCCTTATATTGTGCGT | 1447005 | 1548 | Unknown |
| 26 | CAAATCATTATCACTGAG | 407012 | 432 | Unknown |
| 27 | TGCACTTGTATTTGCCTT | 2073103 | 2122 | Unknown |
| 28 | ATCATAAGTAACCTTGAT | 1729621 | 1786 | Periplasmic putrescine binding protein (E. coli) |
| 29 | TTCCAATCTAAGCAGAAC | 1723027 | 1778 | Secreted protease (E. coli) |
| 30 | TTGGGGAATACTCATTCT | 1458816 | 1557 | Unknown |
| 31 | CATAGCGGTATGCATAGG | 16031 | 20 | Unknown |
| 32 | GGCGTGCATATTTATCGT | 890614 | 915 | Unknown |
| 33 | CCGAGCTGTACTTGGGCG | 1104221 | 1153 | Succinyl-CoAd synthetase β subunit (E. coli) |
| 34 | TCGTTCCGTATCCCAGTT | 2095153 | 2147 | Lactoylglutathione lyase (N. meningitidis) |
| 35 | AGACGGCATACCGCTCAG | 1150268 | 1206 | Oxidoreductase (Helicobacter pylori) |
| 36 | GTCGTCTTTATGGCGGAC | 829845 | 849 | Unknown |
| 37 | AAAGGTAGTAGTCGTAAC | 538662 | 561 | DNA adenine methylase pseudogene (N. meningitidis) |
| 38 | TTCGATAATAATTCGCAA | 377284 | — | |
| 39 | GTGTATAATATATAACAT | 182083 | — | |
| 40 | TAAGGATGTAGCGTCCGA | 828989 | 848 | UTP-glucose-1-phosphate uridylyltransferase (E. coli) |
| 41 | TATCATTATATTAATATG | 1681423 | — | |
| 42 | AGGAAAACTATCCGCAGA | 1577037 | 1660 | ATP-dependent DNA helicase (E. coli) |
| 43 | CGAGGCCGTAATGGTAGA | 166035 | 184 | Unknown |
| 44 | TCTACCCATAATACGCAT | 675145 | 688 | FhaB (Bordetella pertussis) |
| 45 | CAAACAAATAGGGGACTG | 33147 | 39 | Unknown |
| mal1 | TGGCTTCATACAAATGAC | 1146010 | 1205 | d-Lactate dehydrogenase (E. coli) |
| mal2 | GTTCGTAGAAGTCGAAAT | 2049901 | 2098 | Maltose phosphorylase (Lactobacillus sanfranciscensis) |
| mal3 | GAATCATGTATTTACCCC | 2050315 | 2098 | Maltose phosphorylase (L. sanfranciscensis) |
| mal4 | TTGCGGCATACAGATAGA | 2053125 | 2100 | Maltose/H+ symporter (Daucus carota) |
The point of insertion of the transposon was after the dinucleotide in bold letters that was duplicated upon insertion.
When insertions occurred into one of the 2,230 predicted ORFs from the Z2491 sequence (24), its number (NmORF) and potential function are indicated.
—, insertions occurred in intergenic regions.
CoA, coenzyme A.
FIG. 4.
Distribution of the random Himar1 insertions on the genome map of N. meningitidis Z2491. The insertions sites were localized by comparing their sequences (see Table 1) to the genomic database at the Sanger Centre.
Another classical possibility to confirm that transposition is random is to demonstrate that mutants presenting a defined genetic defect can be found within the mutant libraries. Therefore, we decided to perform a limited screening in search of N. meningitidis mutants that have lost the ability to utilize maltose, mainly for three reasons: (i) maltose utilization can readily be pinpointed in vitro by a simple colorimetric test; (ii) this test has been used for decades in clinical typing to distinguish between the meningococcus (Mal+) and the gonococcus (Mal−); and (iii) maltose utilization usually depends on several genes, which was expected to facilitate the identification of Mal− mutants in a limited screen such as the one we intended to perform. This screening led to the identification of 4 of 480 clones (0.8% of the tested mutants), denoted mal1 to mal4, with an impaired capacity to produce acid during maltose catabolism. Mutants mal3 and mal4 were clearly Mal−, whereas mal1 and mal2 presented a delayed change of color in the colorimetric test but were ultimately Mal+. The Southern blot analysis revealed that each mutant presented a unique hybridization pattern (Fig. 3B), confirming that they resulted from different transposition events. Indeed, the sequences of the Himar1 insertion sites in the four mutants were different (Table 1). As expected, transposition occurred within ORFs that showed similarities to genes encoding proteins involved in sugar transport and catabolism (Table 1). In mal1, Himar1 was inserted within a gene encoding a putative respiratory d-lactate dehydrogenase, a membrane-bound flavoenzyme that feeds electrons into the respiratory chain in E. coli (22). Therefore, the delayed Mal+ phenotype in the mal1 mutant suggests that membrane potential is important in the meningococcus for maltose import or degradation. In mal2, mal3, and mal4, mini-Himar1 insertions occurred within a 6-kbp region (Fig. 5). The mal2 and mal3 mutants presented different insertions into the same ORF, encoding a maltose phosphorylase, an enzyme catalyzing the phosphorolysis of maltose into glucose-1-phosphate. In E. coli (22), phosphoglucomutase converts glucose-1-phosphate into glucose-6-phosphate, which subsequently enters the glycolysis pathway. Interestingly, an ORF homologous to the β-phosphoglucomutase gene from Lactococcus lactis was found after the maltose phosphorylase gene and is probably cotranscribed with it, because the two ORFs are separated by only 12 bp (Fig. 5). However, a polar effect of the inserted transposon on the phosphoglucomutase gene can be excluded because this gene is duplicated elsewhere in the genome. The Mal− phenotype of mal3 suggests that the maltose phosphorylase is essential for the entry of maltose into the glycolysis pathway. On the other hand, the Mal+ phenotype of mal2 is probably explained by the fact that Himar1 transposed near the 3′ end of the gene without entirely disrupting its function (Fig. 5). Mal4 was mutated in an ORF with similarities to a gene from carrot encoding a sucrose/H+ symporter. The Mal− phenotype of mal4 therefore suggests that the interrupted gene may encode a maltose/H+ symporter that could be responsible for the entry of maltose into the cell. However, because the interrupted ORF is probably cotranscribed (Fig. 5) with an ORF that is present 3 bp downstream, homologous to a gene encoding mutarotase from Streptococcus thermophilus catalyzing the interconversion of the α and β anomeric forms of sugars, a polar effect, though unlikely, cannot be ruled out.
FIG. 5.
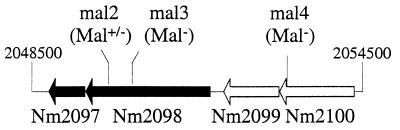
Himar1 insertions in mutants mal2, mal3, and mal4 occur within a 6-kbp region of the N. meningitidis genome. The ORFs, numbered as in the Z2491 sequence (24), encode a β-phosphoglucomutase (Nm2097), a maltose phosphorylase (Nm2098), a mutarotase (Nm2099), and a putative maltose/H+ symporter (Nm2100).
DISCUSSION
We have developed an extremely simple transposon-mediated mutagenesis system, based on in vitro Himar1 transposition (18), that is suitable for mutagenesis of N. meningitidis both in a site-directed (defined genomic DNA fragments) and in a random (chromosomal DNA) fashion. As opposed to previously available mutagenesis methods, in vitro Himar1 transposition presents several particularly desirable features that make it suitable for a genomic-scale mutagenesis of the meningococcus. The mobile element used for mutagenesis is a small minitransposon, approximately 1.6 kbp, that consists only of a kanamycin resistance gene and a neisserial uptake sequence (10) flanked by the inverted repeats of Himar1. Because the transposon lacks the gene encoding the transposase, it can no longer transpose once it is inserted in the chromosome, which is expected to render the mutants perfectly stable. The small size of the mini-Himar1 and the presence of convenient restriction sites will facilitate the possible construction of minitransposons containing either promoterless reporter genes, such as lacZ, phoA, or gfp, or unique short DNA tags. The construction of tagged transposons opens the way for the development of signature-tagged mutagenesis (13), a powerful approach to the identification of virulence genes in vivo. As demonstrated initially in Salmonella enterica serovar Typhimurium (13) and subsequently in numerous pathogenic bacteria (27), signature-tagged mutagenesis could greatly facilitate the screening of mutant libraries in search of N. meningitidis mutants affected in virulence.
We demonstrate that neisserial DNA, whether chromosomal or PCR amplified, is a suitable target for in vitro Himar1 transposition. Therefore, as opposed to allelic exchange and shuttle mutagenesis, there is no need to clone the target DNA before subjecting it to mutagenesis. Consequently, genes that are refractory to cloning in E. coli can nevertheless be mutagenized. Moreover, the use of large PCR fragments, as demonstrated in H. influenzae (18), could allow the simultaneous site-directed mutagenesis of many genes. This could facilitate the mutational analysis of large regions of the chromosome that carry numerous genes which are possibly important for virulence, as, for example, the chromosomal regions found only in the pathogenic Neisseria species that were identified using subtractive hybridization (26). When chromosomal DNA was used as a target, the frequency of transposition was high compared to that for previously available mutagenesis methods, which permitted the creation of mutant libraries of strain Z5463 numbering tens of thousands of mutants. Because N. meningitidis Z2491, which has been sequenced by the Sanger Centre (24), is not transformable, the very closely related strain Z5463 seems a good choice for starting a mutational analysis of N. meningitidis virulence. However, if needed, other N. meningitidis strains can be mutagenized provided that they are transformable, keeping in mind that the number of mutants that can be obtained will be highly dependent on the transformation efficiency. For example, when in vitro Himar1 transposition was performed on a variant of 8013, a serogroup C strain (20) that is less transformable than Z5463, a 20-fold reduction in the number of mutants obtained was observed (data not shown).
Most importantly, from the sequences of 61 insertion sites—8 in hrtA, 4 in ssa1, and 49 random insertions—it can be deduced that Himar1 transposition occurs with no apparent site specificity. All the mutants we tested resulted from independent transposition events, except two mutants with mutations in the hrtA locus that showed the same point of insertion. In this case, it is likely that the two clones were siblings even though it cannot be excluded that this insertion site, nucleotide 290 in the 4,472-bp hrtA locus, is a “warm spot” for Himar1 transposition. As previously reported by others (18, 19, 31), in 95% of the analyzed mutants transposition occurred after TA dinucleotides. This very limited preference is in no way a problem for N. meningitidis random mutagenesis, since TA dinucleotides occur, on average, approximately every 17 bp in the genome. However, even the specificity for TA dinucleotides could be relaxed by replacing Mg2+ with Mn2+ in the transposition reaction mixtures (18). Another demonstration that Himar1 transposition was random came from a limited screen in search of mutants presenting an impaired maltose catabolism. Of the analyzed mutants, 0.8% presented an impaired capacity to produce acid from maltose degradation and were mutated within genes that showed similarities to those encoding proteins involved in sugar transport and catabolism. Our results suggest that maltose import into N. meningitidis may be driven by the flow of protons across the membrane, an unorthodox uptake system for maltose already described in Lactobacillus sanfranciscensis (23). Interestingly, this was also suggested by the recent in silico analysis of the complete sequence of MC58 (38). However, comprehensive mutational and biochemical analyses are of course required to give a broader view of the maltose catabolic pathway in the meningococcus. Moreover, our results allow us to speculate on the reasons why N. gonorrhoeae is naturally Mal−. Indeed, a search of the preliminary genome sequence of the gonococcus at the University of Oklahoma (http://dna1.chem.ou.edu/gono.html) revealed that the maltose phosphorylase and the putative maltose/H+ symporter genes, the genes we identified as specific to maltose catabolism, are both present in N. gonorrhoeae as pseudo-genes. They both contain frameshifts and/or stop codons. Hence, the inability of N. gonorrhoeae to utilize maltose is the result of an evolutionary process during which the gonococcus accumulated mutations in genes of the maltose catabolic pathway and ultimately lost the capacity to utilize maltose.
In conclusion, in vitro Himar1 transposition should allow N. meningitidis mutational analysis on a scale that was previously unfeasible. Hopefully, mutagenesis studies, in conjunction with the genome sequences (24, 38), will lead to a better understanding of the molecular mechanisms of meningococcal meningitis, a disease that remains an important public health burden (40).
ACKNOWLEDGMENTS
We thank U. Vogel and M. Frosch (University of Würzburg) for providing plasmid pHC6. We thank J.-L. Beretti for help in purifying the Himar1 transposase. We are grateful to C. R. Tinsley and J.-M. Reyrat for critical reading of the manuscript. E. Abachin and G. Quesne are acknowledged for automated sequencing. The Sanger Centre and the Gonococcal Genome Sequencing Project (University of Oklahoma) are gratefully acknowledged for making genome sequences publicly available before publication.
This work was supported by grants from the Institut National de la Santé et de la Recherche Médicale, Université Paris V-René Descartes, and Fondation pour la Recherche Médicale.
REFERENCES
- 1.Achtman M, Neibert M, Crowe B A, Strittmatter W, Kusecek B, Weyse E, Walsh M J, Slawig B, Morelli G, Moll A, Blake M. Purification and characterization of eight class 5 outer membrane protein variants from a clone of Neisseria meningitidis serogroup A. J Exp Med. 1988;168:507–525. doi: 10.1084/jem.168.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerley B J, Rubin E J, Camilli A, Lampe D J, Robertson H M, Mekalanos J J. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc Natl Acad Sci USA. 1998;95:8927–8932. doi: 10.1073/pnas.95.15.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. Bio/Technology. 1995;18:746–750. [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schäffer A A, Zang J, Zang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle-Vavra S, Seifert H S. Shuttle mutagenesis: a mini-transposon for producing PhoA fusions with exported proteins in Neisseria gonorrhoeae. Gene. 1995;155:101–106. doi: 10.1016/0378-1119(94)00890-5. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra S, Seifert H S. Shuttle mutagenesis: two mini-transposons for gene mapping and for lacZ transcriptional fusions in Neisseria gonorrhoeae. Gene. 1993;129:51–57. doi: 10.1016/0378-1119(93)90695-y. [DOI] [PubMed] [Google Scholar]
- 7.Claus H, Frosch M, Vogel U. Identification of a hotspot for transformation of Neisseria meningitidis by shuttle mutagenesis using signature-tagged transposons. Mol Gen Genet. 1998;259:363–371. doi: 10.1007/s004380050823. [DOI] [PubMed] [Google Scholar]
- 8.Erwin A L, Stephens D S. Identification and characterization of auxotrophs of Neisseria meningitidis produced by Tn916 mutagenesis. FEMS Microbiol Lett. 1995;127:223–228. doi: 10.1111/j.1574-6968.1995.tb07477.x. [DOI] [PubMed] [Google Scholar]
- 9.Gawron-Burke C, Clewell D B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982;300:281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- 10.Goodman S D, Scocca J J. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1988;85:6982–6986. doi: 10.1073/pnas.85.18.6982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gwinn M L, Stellwagen A E, Craig N L, Tomb J-F, Smith H O. In vitro Tn7 mutagenesis of Haemophilus influenzae Rd and characterization of the role of atpA in transformation. J Bacteriol. 1997;179:7315–7320. doi: 10.1128/jb.179.23.7315-7320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas R, Kahrs A F, Facius D, Allmeier H, Schmitt R, Meyer T F. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130:23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 13.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 14.Kahrs A F, Bihlmaier A, Facius D, Meyer T F. Generalized transposon shuttle-mutagenesis in Neisseria gonorrhoeae: a method for isolating epithelial cell invasion-defective mutants. Mol Microbiol. 1994;12:819–831. doi: 10.1111/j.1365-2958.1994.tb01068.x. [DOI] [PubMed] [Google Scholar]
- 15.Kathariou S, Stephens D S, Spellman P, Morse S A. Transposition of Tn916 to different sites in the chromosome of Neisseria meningitidis: a genetic tool for meningococcal mutagenesis. Mol Microbiol. 1990;4:729–735. doi: 10.1111/j.1365-2958.1990.tb00643.x. [DOI] [PubMed] [Google Scholar]
- 16.Kellog D S, Jr, Peacock W L, Jr, Deacon W E, Brown L, Pirle C I. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lampe D J, Akerley B J, Rubin E J, Mekalanos J J, Robertson H M. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc Natl Acad Sci USA. 1999;96:11428–11433. doi: 10.1073/pnas.96.20.11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lampe D J, Churchill M E A, Robertson H M. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 19.Lampe D J, Grant T E, Robertson H M. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nassif X, Lowy J, Stenberg P, O'Gaora P, Ganji A, So M. Antigenic variation of pilin regulates adhesion of Neisseria meningitidis to human epithelial cells. Mol Microbiol. 1993;8:719–725. doi: 10.1111/j.1365-2958.1993.tb01615.x. [DOI] [PubMed] [Google Scholar]
- 21.Nassif X, Puaoi D, So M. Transposition of Tn1545-Δ3 in the pathogenic neisseriae: a genetic tool for mutagenesis. J Bacteriol. 1991;173:2147–2154. doi: 10.1128/jb.173.7.2147-2154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. [Google Scholar]
- 23.Neubauer H, Glaasker E, Hammes W P, Poolman B, Konings W N. Mechanism of maltose uptake and glucose excretion in Lactobacillus sanfrancisco. J Bacteriol. 1994;176:3007–3012. doi: 10.1128/jb.176.10.3007-3012.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkhill J, Achtman M, James K D, Bentley S D, Churcher C, Klee S R, Morelli G, Basham D, Brown D, Chillingworth T, Davies R M, Davis P, Devlin K, Feltwell T, Hamlin N, Holroyd S, Jagels K, Leather S, Moule S, Mungall K, Quail M A, Rajandream M-A, Rutherford K M, Simmonds M, Skelton J, Whitehead S, Spratt B G, Barrell B G. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 25.Pelicic V, Jackson M, Reyrat J-M, Jacobs W R, Jr, Gicquel B, Guilhot C. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94:10955–10960. doi: 10.1073/pnas.94.20.10955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perrin A, Nassif X, Tinsley C. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect Immun. 1999;67:6119–6129. doi: 10.1128/iai.67.11.6119-6129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perry R D. Signature-tagged mutagenesis and the hunt for virulence factors. Trends Microbiol. 1999;7:385–388. doi: 10.1016/s0966-842x(99)01582-6. [DOI] [PubMed] [Google Scholar]
- 28.Pizza M, Scarlato V, Masignani V, Giuliani M M, Arico B, Comanducci M, Jennings G T, Baldi L, Bartolini E, Capecchi B, Galeotti C L, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood D W, Jeffries A C, Saunders N J, Granoff D M, Venter J C, Moxon E R, Grandi G, Rappuoli R. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- 29.Prod'hom G, Lagier B, Pelicic V, Hance A J, Gicquel B, Guilhot C. A reliable amplification technique for the characterization of genomic DNA sequences flanking insertion sequences. FEMS Microbiol Lett. 1998;158:75–81. doi: 10.1111/j.1574-6968.1998.tb12803.x. [DOI] [PubMed] [Google Scholar]
- 30.Reich K A, Chovan L, Hessler P. Genome scanning in Haemophilus influenzae for identification of essential genes. J Bacteriol. 1999;181:4961–4968. doi: 10.1128/jb.181.16.4961-4968.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubin E J, Akerley B J, Novik V N, Lampe D J, Husson R N, Mekalanos J J. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci USA. 1999;96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Seifert H S, Ajioka R S, Paruchuri D, Heffron F, So M. Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J Bacteriol. 1990;172:40–46. doi: 10.1128/jb.172.1.40-46.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens D S, McAllister C F, Zhou D, Lee F K, Apicella M A. Tn916-generated, lipooligosaccharide mutants of Neisseria meningitidis and Neisseria gonorrhoeae. Infect Immun. 1994;62:2947–2952. doi: 10.1128/iai.62.7.2947-2952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauss E J, Falkow S. Microbial pathogenesis: genomics and beyond. Science. 1997;276:707–712. doi: 10.1126/science.276.5313.707. [DOI] [PubMed] [Google Scholar]
- 36.Swartley J S, McAllister C F, Hajjeh R A, Heinrich D W, Stephens D S. Deletions of Tn916-like transposons are implicated in tetM-mediated resistance in pathogenic Neisseria. Mol Microbiol. 1993;10:299–310. doi: 10.1111/j.1365-2958.1993.tb01956.x. [DOI] [PubMed] [Google Scholar]
- 37.Swartley J S, Stephens D S. Identification of a genetic locus involved in the biosynthesis of N-acetyl-d-mannosamine, a precursor of the (β2→8)-linked polysialic acid capsule of serogroup B Neisseria meningitidis. J Bacteriol. 1994;176:1530–1534. doi: 10.1128/jb.176.5.1530-1534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tettelin H, Saunders N J, Heidelberg J, Jeffries A C, Nelson K E, Eisen J A, Ketchum K A, Hood D W, Peden J F, Dodson R J, Nelson W C, Gwinn M L, DeBoy R, Peterson J D, Hickey E K, Haft D H, Salzberg S L, White O, Fleischmann R D, Dougherty B A, Mason T, Ciecko A, Parksey D S, Blair E, Cittone H, Clark E B, Cotton M D, Utterback T R, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith H O, Fraser C M, Moxon E R, Rappuoli R, Venter J C. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 39.Thomas C E, Carbonetti N H, Sparling P F. Pseudo-transposition of a Tn5 derivative in Neisseria gonorrhoeae. FEMS Microbiol Lett. 1996;145:371–376. doi: 10.1111/j.1574-6968.1996.tb08603.x. [DOI] [PubMed] [Google Scholar]
- 40.Tikhomirov E, Santamaria M, Esteves K. Meningococcal disease: public health burden and control. World Health Stat Q. 1997;50:170–177. [PubMed] [Google Scholar]
- 41.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5"-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 42.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, Chu A M, Connelly C, Davis K, Dietrich F, Dow S W, El Bakkoury M, Foury F, Friend S H, Gentalen E, Giaever G, Hegemann J H, Jones T, Laub M, Liao H, et al. Functional characterization of the Saccharomyces cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]