Significance
An essential part of conserving biodiversity is to find and protect areas with high species richness and endemism (hotspots). We combine 20 y of field surveys, distributional data and new molecular data to analyze diversity patterns in Chinese amphibians, one of the most endangered groups of animals in one of Earth’s most diverse countries. We analyze 521 described species of Chinese amphibians and identify 100 potential cryptic species. We find 10 hotspots of exceptional biodiversity, six of which are new relative to previously known hotspots. These six new hotspots are associated with mountain ranges in southeastern China and are mostly unprotected and highly threatened by human pressure. Protecting these six new hotspots should be a high conservation priority.
Keywords: amphibians, biodiversity, China, cryptic species, hotspots
Abstract
Identifying and protecting hotspots of endemism and species richness is crucial for mitigating the global biodiversity crisis. However, our understanding of spatial diversity patterns is far from complete, which severely limits our ability to conserve biodiversity hotspots. Here, we report a comprehensive analysis of amphibian species diversity in China, one of the most species-rich countries on Earth. Our study combines 20 y of field surveys with new molecular analyses of 521 described species and also identifies 100 potential cryptic species. We identify 10 hotspots of amphibian diversity in China, each with exceptional species richness and endemism and with exceptional phylogenetic diversity and phylogenetic endemism (based on a new time-calibrated, species-level phylogeny for Chinese amphibians). These 10 hotspots encompass 59.6% of China’s described amphibian species, 49.0% of cryptic species, and 55.6% of species endemic to China. Only four of these 10 hotspots correspond to previously recognized biodiversity hotspots. The six new hotspots include the Nanling Mountains and other mountain ranges in South China. Among the 186 species in the six new hotspots, only 9.7% are well covered by protected areas and most (88.2%) are exposed to high human impacts. Five of the six new hotspots are under very high human pressure and are in urgent need of protection. We also find that patterns of richness in cryptic species are significantly related to those in described species but are not identical.
Earth may be undergoing a major biodiversity crisis (1, 2). For example, recent reports have shown catastrophic declines in animal populations since 1970 (3), with similar declines in plants and fungi (4). To help stave off this biodiversity crisis, recent conferences have emphasized the idea of protecting 30% of the planet’s surface area for conservation by 2030 (5–8). Understanding spatial patterns of species richness is essential for targeting locations that will maximize the protection of biodiversity.
One important approach for protecting biodiversity is to target hotspots that harbor exceptional species richness and endemism (9). For example, the 25 global biodiversity hotspots initially defined by Myers et al. (9) harbor 44% of Earth’s vascular plant species and 35% of terrestrial vertebrate species but cover only 1.4% of Earth’s land surface. Thus, these hotspots can protect considerable species richness in a relatively small area. Therefore, there is an urgent need to identify and protect additional regions of high diversity and endemism that can help preserve Earth’s biodiversity.
A substantial proportion of Earth’s plant and animal diversity occurs in China. China is considered one of 17 megadiverse countries (10). China is estimated to harbor 15% of the world’s vertebrate species and 12% of the plant species, in an area covering only ~6% of Earth’s land surface (11, 12). Therefore, conservation in China is crucial for protecting global biodiversity.
China includes four biodiversity hotspots traditionally recognized by Conservation International (https://www.conservation.org/). Three of these span international boundaries, including the Himalayas, Mountains of Central Asia, and Indo-Burma hotspots. By contrast, the Hengduan Mountains hotspot lies almost entirely (>98%) within China (13), and is one of the most diverse temperate hotspots (14). The area encompassed by these four hotspots in China is located largely in the western part of the country, near the Qinghai-Xizang Plateau.
Recent studies have highlighted that conservation priorities in China might be taxonomically and spatially biased (15, 16). In particular, most of the country’s protected areas are located in the western portion (13). These protected areas seem to function moderately well for mammals and birds but might not be suitable for other groups (15). Thus, there is a pressing need to thoroughly evaluate spatial biodiversity patterns in other groups.
Amphibians are crucial for protection of biodiversity in China. Amphibians are the most endangered group of vertebrates globally (17, 18), with 41% of species considered at risk of extinction (19). Unfortunately, amphibians are generally less frequently and intensively studied compared to mammals and birds (20). They are also underrepresented in planning protected areas in China (16). Hu et al. (21) performed an excellent analysis of spatial patterns in phylogenetic and genetic diversity in Chinese land vertebrates. However, they did not focus on amphibians nor on identifying fine-scale areas for conservation (e.g., they suggested all of South China and Southwest China as conservation priorities).
Conservation of amphibians (and many other groups) is also complicated by the presence of many cryptic species (22, 23). These cryptic species might go undetected and unprotected because they are not morphologically distinguishable (24–27). Cryptic species may also have smaller geographic ranges than morphology-based species, making them more vulnerable to extinction (28). However, to our knowledge, no studies have conducted large-scale analyses that integrate morphologically cryptic species in conservation planning.
Here, we present an analysis of China’s amphibian biodiversity. We first describe spatial diversity patterns based on our own field surveys, on published and new distributional and molecular data, and on analyses of potential cryptic species identified with molecular data. We then identify hotspots of amphibian biodiversity in China, and use these results to help inform conservation planning for China’s amphibians and other groups.
Results
Spatial Diversity Patterns and Hotspots.
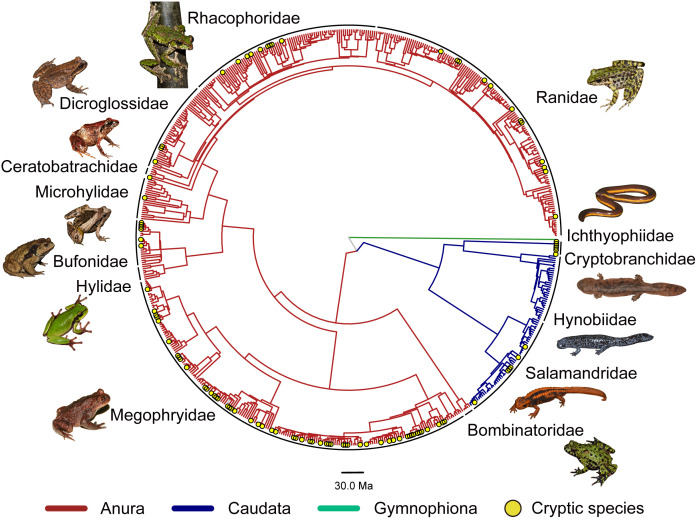
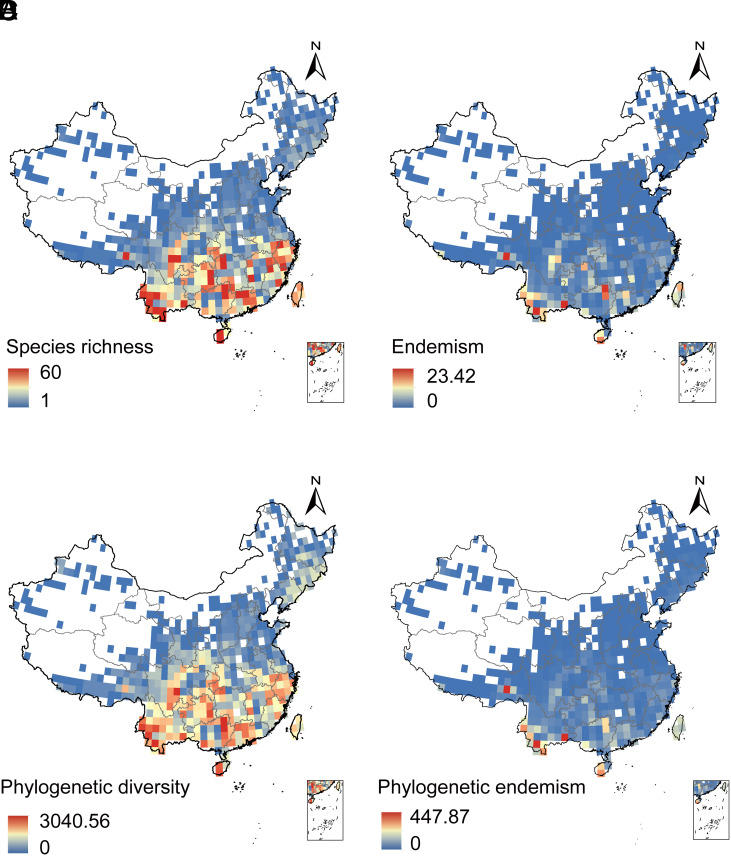
We collected distribution data from 548 described amphibian species in China, accounting for 98.4% (548/557) of all described Chinese amphibian species by the end of 2020 (Dataset S1). Our molecular analyses identified 100 potential cryptic species (Datasets S1 and S2 and SI Appendix, Results). We integrated almost all of these species into a new large-scale, time-calibrated phylogeny that included 621 Chinese amphibian species (Fig. 1), including 521 described species and 100 potential cryptic species. These 100 cryptic species were similar in age to the described species (SI Appendix, Fig. S1 and Results). We estimated the distribution of 647 species (548 described and 99 cryptic) using new field surveys and a summary of published distributional data. We then mapped the distribution of these species among 567 grid cells of 1 × 1 degree size (~111 × 111 km2). Species richness (Fig. 2A) and endemism (Fig. 2B) were then mapped on these grid cells. Based on the new phylogeny (Fig. 1), we also mapped spatial patterns in phylogenetic diversity (Fig. 2C) and phylogenetic endemism (Fig. 2D). We also mapped diversity patterns separately for each major amphibian clade (anurans, caudates, caecilians; SI Appendix, Fig. S2). A UPGMA clustering analysis of these 567 grid cells identified eight large-scale biogeographic regions (SI Appendix, Fig. S3 and Methods and Results).
Fig. 1.
Time-calibrated phylogeny of Chinese amphibians for estimating phylogenetic diversity and phylogenetic endemism. The yellow filled circles indicate the potential cryptic species identified in this study. The phylogeny included 521 described Chinese amphibian species (93.5% of currently described species; Dataset S1) and 100 potential cryptic species. The tree is available in newick format in Dataset S5. Photos courtesy of Chenqi Lu, Peng Guo, Hui Zhao, Yufan Wang, and Bao-Lin Zhang.
Fig. 2.
Diversity patterns in Chinese amphibians. Spatial patterns of (A) species richness, (B) weighted species endemism, (C) phylogenetic diversity, and (D) phylogenetic endemism. The white grid cells lacked recorded amphibian species and were excluded from the analyses, and were mostly high-elevation and arid regions. The gray lines indicate the provincial boundaries of China.
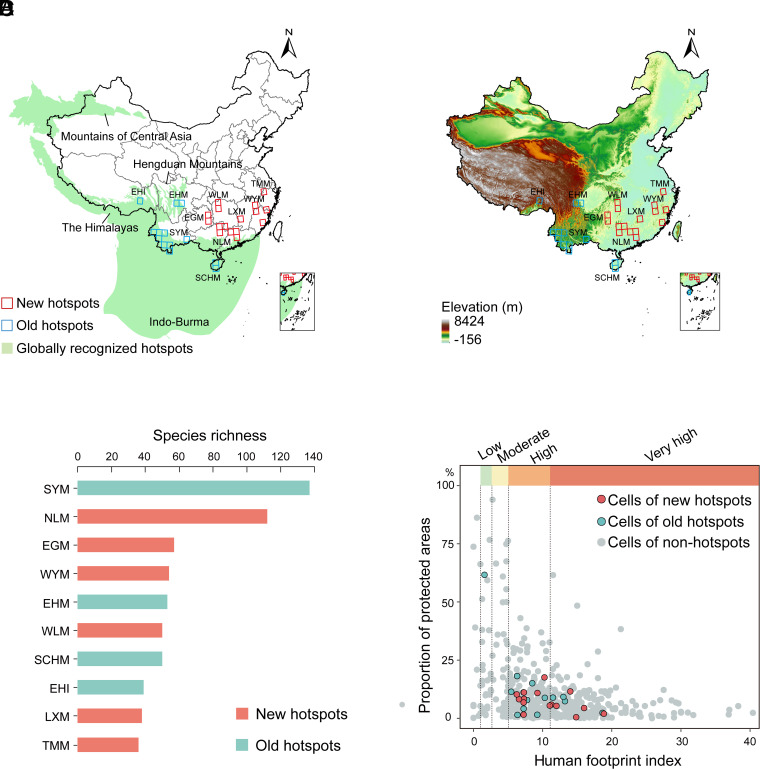
The 567 grid cells were first ranked based on their total species richness (described species plus cryptic species). There were 29 cells in the upper 95th percentile (Fig. 3A) and the cell with the highest richness was in the Nanling Mountains (SI Appendix, Fig. S4). We grouped these 29 cells into 10 geographically distinct biodiversity hotspots (Fig. 3A) considering their affiliated biogeographic regions (SI Appendix, Fig. S3A), mountain ranges (Fig. 3B), and species composition (SI Appendix, Fig. S5). Among these 10 hotspots (each including multiple cells), the Southwest Yunnan Mountains and Nanling Mountains had the highest and second-highest total richness, respectively (Fig. 3C). We also ranked grid cells of these 10 hotspots based on their endemism, phylogenetic diversity, and phylogenetic endemism (SI Appendix, Table S1). Among the 29 high-richness cells, 12 were in the upper 95th percentile across all diversity metrics (SI Appendix, Table S1). These cells were located in the Nanling Mountains, Eastern Hengduan Mountains, Hainan Island, and Southwest Yunnan Mountains (SI Appendix, Table S1). Our results are mostly based on the 95th percentile, but alternative hotspots based on 99th percentile and 90th percentile were also identified (SI Appendix, Fig. S6 and Results). Using the 99th percentile, there were only four cells, which were in the Nanling Mountains, Southwest Yunnan Mountains, and Eastern Hengduan Mountains (SI Appendix, Fig. S6A). Using the upper 90th percentile instead (SI Appendix, Fig. S6B) led to a large increase in the number of high-diversity cells in South China (from 18 to 39) and a smaller increase in western China (from 11 to 16).
Fig. 3.
Hotspots of Chinese amphibian diversity. (A) The 29 highest-diversity grid cells (upper 95th percentile of species richness, square grid cells). We grouped these 29 grid cells into 10 geographically distinct biodiversity hotspots (orange grid cells): Eastern Himalayas (EHI), Eastern Hengduan Mountains (EHM), Southwest Yunnan Mountains (SYM), South-Central Hainan Mountains (SCHM), Nanling Mountains (NLM), Wuling Mountains (WLM), Eastern Guizhou Mountains (EGM), Luoxiao Mountains (LXM), Wuyi Mountains (WYM), and Tianmu Mountains (TMM). Green regions indicate the four previously recognized global biodiversity hotspots in China (Himalayas, Hengduan Mountains, Indo-Burma, and Mountains of Central Asia). Cells within these four globally recognized hotspots are colored blue and referred to as “old hotspots”, whereas those outside are colored red and referred to as “new hotspots”. The gray lines indicate the provincial boundaries of China. (B) Elevation map of China showing all hotspot grid cells. (C) Total species richness of each hotspot. Abbreviations of hotspots as above. (D) Proportion of protected areas and human footprint index of each grid cell, including grid cells of new hotspots (red), old hotspots (blue), and nonhotspots (gray).
Among the 10 hotspots recognized here, four overlapped with three previously recognized hotspots (Fig. 3A), including the Himalayas (Eastern Himalayas here), Hengduan Mountains (Eastern Hengduan Mountains here), and the Indo-Burma hotspot (mountains of Yunnan Province and Hainan Island). The other six hotspots were newly found here (Fig. 3A) and were broadly distributed in southeastern China, and included the Eastern Guizhou, Luoxiao, Nanling, Tianmu, Wuling, and Wuyi mountain ranges. One grid cell in the Nanling Mountains (longitude 113.5 and latitude 23.5), partially overlapped (41.2%) with the Indo-Burma hotspot. We classified it in the new Nanling Mountains hotspot given that this is where the majority of the grid cell is. Further, the Indo-Burma hotspot in this part of China generally corresponds to lowlands, not highlands.
The 10 hotspots identified here represented 58.0% (381/657) of all Chinese amphibian species (59.6% described plus 49.0% cryptic) among only 5% of the grid cells (SI Appendix, Fig. S7). They contained 55.6% of all endemic species and 43.0% of all narrowly endemic species (those restricted to a single grid cell; SI Appendix, Fig. S8). The six new hotspots contained 28.3% of Chinese amphibians (SI Appendix, Fig. S9) including 34.1% of Chinese endemics and 14.5% of narrow endemics. The described and cryptic species both showed high richness in these 10 hotspots (SI Appendix, Fig. S10), and there was a significant relationship (r2 = 0.49; P < 0.0001) between richness of described species and potential cryptic species among all grid cells (SI Appendix, Fig. S11).
The overall patterns for amphibians generally reflect those from frogs (SI Appendix, Fig. S2), which make up most of Chinese amphibian diversity. Salamanders also showed similar patterns (SI Appendix, Fig. S2), with high richness in the newly defined montane hotspots in South China (e.g., Nanling, Tianmu). However, unlike frogs, salamanders had limited richness in grid cells in Hainan and southwestern China (Yunnan Province) and high richness in northeast China (Jilin Province) and northern Taiwan. Caecilians have very limited richness, with only a few species in the southernmost parts of China (SI Appendix, Fig. S2).
We found a strong correlation between species richness and the remaining indices, especially phylogenetic diversity (SI Appendix, Fig. S12). We performed additional analyses based on phylogenetic diversity standardized for richness. These analyses showed high values in the Southwest Yunnan Mountains hotspot (SI Appendix, Fig. S13), but many other grid cells with high values had very low richness (e.g., grid cells with single species in northern China). These were not emphasized as biodiversity hotspots here, since our primary emphasis is on species richness. Average divergence times of species (SI Appendix, Fig. S13) were also largely unrelated to richness.
Conservation Effectiveness and Human Pressure.
We assessed the spatial overlap of each species’ geographic range with protected areas, with conservation threats (human footprint index), and with the hotspots found here (SI Appendix, Figs. S14–S17). By overlaying the distribution range of each amphibian species and a spatial layer of protected areas (SI Appendix, Fig. S14), we estimated that 14.1% of these amphibian species (93/657) were completely included in current protected areas in China (100% covered; SI Appendix, Fig. S15A), whereas 18.9% (124/657) were entirely outside protected areas (not covered; SI Appendix, Fig. S15A). The gap species (0 to 20% covered) accounted for 50.8% of Chinese amphibians (334/657) and 78.0% of the species in the six new hotspots (145/186; SI Appendix, Fig. S15 A and B). Among the 186 species in the six new hotspots, only 9.7% are 100% covered by protected areas (SI Appendix, Fig. S15B). Increasing the scope of protected areas in these six new hotspots could benefit 43.4% of all gap species (145/334) and 13.0% (10/77) of partial gap species (20 to 90% covered; SI Appendix, Fig. S15 A and B).
By overlaying the distribution range of each species and a spatial layer describing human impacts (SI Appendix, Fig. S16 and Methods), we estimated that 77.8% of Chinese species (511/657) occurred at least partly in high human-pressure areas (SI Appendix, Fig. S17A; part of their range overlapped with high-human pressure areas). Among species that occurred largely in high-pressure areas (>50% of their range), 26.1% (43/165) occurred in the new hotspots (SI Appendix, Fig. S17). Among the 186 species in the new hotspots, only 11.8% were completely outside of high-pressure areas (SI Appendix, Fig. S17B; nonthreatened). The remaining 88.2% were at least partly exposed to high human impacts (SI Appendix, Fig. S17B; partially threatened, threatened, and severely threatened).
Overall, these 10 hotspots were poorly protected areas and suffered from high human pressure, except for the Eastern Himalayas (Fig. 3D and SI Appendix, Table S2). Twenty-seven of the 29 hotspot grid cells (from nine hotspots) were classified as facing high human pressure (human footprint index 6 to 11) or very high (>12) human pressure (Fig. 3D and SI Appendix, Table S2). The other two hotspot grid cells experience low and moderate pressure (Eastern Himalayas [EHI] and Southwestern Yunnan Mountains [SYM]), respectively.
Discussion
Based on a comprehensive analysis of Chinese amphibian species, we identified 10 hotspots having exceptional species richness, endemism, and phylogenetic diversity (Fig. 3). These hotspots encompass 58.0% of all Chinese amphibian species and 49.0% of cryptic species (SI Appendix, Fig. S7). Only four of these hotspots correspond with previously known global biodiversity hotspots. The six new hotspots occur in the mountains of southeastern China and harbor 28.3% of Chinese amphibian species (SI Appendix, Figs. S7 and S8).
There is an urgent need for protection of these six new hotspots. South China has recently experienced rapid economic growth and severe loss of protected areas (15, 29, 30). Human pressure is intense in these new hotspots (Fig. 3D). By contrast, the global hotspots in western China (the Himalayas and Hengduan Mountains) have generally been the focus of more conservation efforts (13, 15). Yet, the Nanling Mountains in South China have much higher amphibian richness than the Hengduan Mountains (Fig. 3C). Previous assessments of Chinese biodiversity have highlighted the mismatch between the large protected areas in western and northeastern China and the many biodiverse areas in South China (15, 31). Our study specifically pinpoints the Nanling Mountains and five other mountain ranges in South China as high-priority areas for protection. These six new hotspots harbor many Critically Endangered amphibian species (19), such as the salamanders Hynobius maoershanensis (Hynobiidae) and Paramesotriton labiatus (Salamandridae) and the frog Leptobrachella maoershanensis (Megophryidae). Besides amphibians, they also contain numerous other rare and threatened animal and plant species, such as the viper Protobothrops mangshanensis and the tree Firmiana danxiaensis (32). The Nanling Mountains also have exceptional species richness and phylogenetic diversity for angiosperm plants (33) and possibly many other taxa as well. The endemic species in these mountain ranges might also be especially vulnerable to climate change, given that the limited maximum heights of these ranges reduce the potential for montane species to disperse to cooler, higher elevations.
In summary, we identify six new hotspots of amphibian biodiversity in China. Many of these new hotspots have higher richness and endemism than traditionally recognized global hotspots in China (Fig. 3C and SI Appendix, Fig. S4 and Table S1). They are also relatively unprotected and are under high pressure from human activities (Fig. 3D). Protecting these new hotspots is imperative as Chinese amphibians are more imperiled (77.8% threatened; SI Appendix, Fig. S17) than amphibians globally [41% threatened; (34)]. More broadly, we present possibly the first large-scale analysis of richness patterns in cryptic species. Our results suggest that described species alone offer an incomplete picture of species richness but that spatial richness patterns are broadly correlated between described and cryptic species.
Materials and Methods
Data Collection and Assembly.
Chinese amphibians included 557 described species by the end of 2020 (Dataset S1), belonging to 13 families and 62 genera (35). Among these species, 340 are endemic to China. We also estimated the number of potential cryptic species, using molecular data from 2,306 individuals sampled through our fieldwork by 2020 and three species-delimitation methods (SI Appendix, Methods).
Data on species distributions were assembled from multiple sources, resulting in the most comprehensive dataset on the distribution of Chinese amphibians to date (548 described species, 99 cryptic species). Previous studies had more limited sampling of Chinese amphibians (94 to 401 species), such as Meng et al. (36), Chen et al. (37), and Hu et al. (21) Much of these data were from fieldwork by the authors (from 2001 to 2020), with specimens now deposited in the Kunming Institute of Zoology of the Chinese Academy of Sciences. Specimens were collected throughout China, except for the grasslands of Inner Mongolia, extreme high elevations of the Xizang Plateau, and the desert in Xinjiang (SI Appendix, Fig. S3 A and B). These arid areas generally lack suitable habitat for amphibians (31, 38). For each species, fieldwork was focused on sampling their full geographic range and type localities. Tissue samples and GPS coordinates were taken from each specimen.
We also incorporated additional sources of distribution data. These included the Global Biodiversity Information Facility (GBIF) database, nearly all published national, provincial, and local faunas, and published literature associated with the taxonomy of Chinese amphibians. We cross-checked these locality data against the approximate geographic range determined by our newly generated molecular dataset (SI Appendix, Methods) to ensure that the former occurred within the latter. Species-distribution models were not used to generate range maps given the possible overprediction of species ranges, as found for Neotropical amphibians (39).
A total of 11,745 occurrences records from 647 Chinese amphibian species were collected, including 548 described species and 99 cryptic species. We lacked distribution data for 10 species (nine described and one cryptic) because they lacked precise occurrence records. These 10 species were excluded from mapping analyses.
Mapping Distribution Ranges.
Distribution records were gridded using the R package letsR (40) to calculate and visualize large-scale distribution patterns. To minimize the effect of unequal sampling area, we excluded 44 grid cells with <50% land area following Kreft and Jetz (41) and He et al. (42). Grid cells that appeared to lack amphibians (see white areas in Fig. 2) were excluded (n = 290). These included the high-elevation regions of the Qinghai-Xizang plateau and the desert and grassland areas in northern China. This left 567 grid cells of 1 × 1 degree size (~111 × 111 km2). UPGMA analysis was performed on the included grid cells to delimit large-scale biogeographic regions for Chinese amphibians (SI Appendix, Methods). Standard maps from the Chinese government were used (http://www.sbsm.gov.cn). Species were considered narrowly endemic if they were restricted to one grid cell.
We note that there are also some grid cells with low species richness in southern China. The low richness of these cells may be related to a variety of factors, including human impacts (urbanization and agriculture), lower sampling effort in human-impacted areas, and lower species richness at low elevations (even without human impacts). However, our goal was not explaining why some grid cells have unusually low species richness, but rather on identifying grid cells with exceptionally high richness. In China, these high-richness grid cells correspond to montane regions, which appears to be a general pattern in amphibians (43–45).
Estimation of Diversity Metrics.
We calculated the species richness of each grid cell by summing the number of species in that cell. We estimated the cell’s weighted endemism (46) as the inverse of each species’ distribution range (represented by the number of grid cells that each species occupies) summed across all species present in that grid cell. Endemism, weighted endemism, phylogenetic diversity, and phylogenetic endemism for each cell were calculated using the R package phyloregion (47).
To estimate phylogenetic diversity and phylogenetic endemism, we first estimated a time-calibrated phylogeny that included most Chinese amphibian species (93.5%; 521/557). We used concatenated sequences of 12 nuclear genes (BDNF, C-MYC, CXCR4, H3A, NCX1, POMC, RAG1, RAG2, RHOD, SIA, SLC8A3, and TYR) and 7 mitochondrial genes (12S rRNA, 16S rRNA, COI, CYTB, ND1, ND2, and ND3). These genes were selected based on their use in large-scale phylogenetic studies of amphibians (48–50). Most sequences were obtained from GenBank following protocols detailed in those studies (stopping at the end of 2020). When a species had multiple individuals with data on GenBank, we chose the individual with the most genetic markers. The COI sequences of many cryptic species identified in this study were newly generated (SI Appendix, Methods). GenBank accession numbers are in Dataset S3. The molecular dataset included 521 described Chinese amphibian species and 100 cryptic species and 1,057 non-Chinese species. A total of 36 described Chinese amphibian species were not included because we lacked tissue samples and data were unavailable on GenBank. Detailed methods for estimating the time-calibrated phylogeny are provided in SI Appendix, Methods. The maximum-likelihood tree and time-calibrated phylogeny (both in newick format) are available in Datasets S4 and S5, respectively.
We performed correlation analyses among species richness, species endemism, phylogenetic diversity, and phylogenetic endemism using the R package corrplot (51). To control for the potential confounding effects of richness on phylogenetic diversity, we calculated the standardized effective size of phylogenetic diversity (52). We calculated standardized effective size of phylogenetic diversity SES-PD using the R package phyloregion (47). The SES-PD is calculated by dividing the difference between the observed PD and the mean value of 1,000 randomly generated PDs by the SD of the randomly generated PDs. We also calculated mean divergence times among species in grid cells (SI Appendix, Fig. S13).
Identifying Biodiversity Hotspots.
Many studies have shown that selecting the top 5% of land areas with the highest richness is optimal for identifying biodiversity hotspots (53–56). Accordingly, we ranked all cells based on their total number of species and considered cells in the upper 95th percentile as hotspots (top 29 of 567). We also compared these results to those using the upper 99th and 90th percentiles (SI Appendix, Results and Fig. S6http://www.pnas.org/lookup/doi/10.1073/pnas.2320674121#supplementary-materials). We overlaid the grid cells of hotspots identified here with a global hotspot layer (9, 57) to determine whether these grid cells are located in previously recognized hotspots. Cells that were not within previously recognized hotspots were considered new to this study.
Species Protection and Exposure to Human Pressure.
We assessed each species using two data types: i) the proportion of the geographic range of each species that is within a protected area; and ii) an estimate of human pressure within each species’ geographic range. Following Rodrigues et al. (58), we overlaid the distribution range of each species and the layer of protected areas in ArcGIS 10.2. The distribution range of each species was represented by a minimum-convex polygon generated by multiple GPS coordinates using ArcGIS 10.2. We used the most comprehensive dataset of protected areas in China (15). This dataset consisted of a vector layer in which the minimum distance that can be represented is ~125 m. This level of resolution is standard for large-scale geographic analyses (15, 59). Based on the percentage of each species’ range that fell inside the protected areas, we classified species into four groups (60): i) unprotected: species’ range was completely outside protected areas; ii) gap: maximum of 20% covered by protected areas; iii) partial gap: 21 to 90% covered by protected areas; and iv) well-covered: >90% covered by protected areas.
To quantify human threats to biodiversity (61), we used the human footprint index raster layer (1 km2 resolution) to represent human pressure (62). The human footprint index is calculated based on data from remote sensing and from field survey data for multiple variables, including the extent of built environments, human population density, and the presence of railways and roads (62). A human footprint index of 0 indicates no human pressure, 1 to 2 indicates low pressure, 3 to 5 moderate pressure, 6 to 11 high pressure, and 12 to 50 very high pressure (62). We then overlaid the distribution range of each species and the human pressure layer. Based on the percentage of each species’ range that fell inside the high human pressure areas, we classified species into four groups: i) severely threatened: species’ range was completely inside the very high human pressure areas (100%); ii) threatened: >50% in the very high-pressure areas; iii) partially threatened: ≤50% and >0%, and iv) nonthreatened: completely outside the very high-pressure areas.
Protected Areas and Human Pressure on Hotspots.
To assess the conservation status of hotspots identified here, we estimated the percentage of hotspots covered by protected areas by overlaying the 29 hotspot grid cells and the protected areas in ArcGIS 10.2. To assess average human pressure within each hotspot, we took the average values of the human footprint index across all 1-km2 grids within each 1 × 1 degree grid cell of the 29 hotspots grid cells, using ArcGIS 10.2.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (TXT)
Dataset S05 (TXT)
Dataset S06 (TXT)
Dataset S07 (TXT)
Dataset S08 (XLSX)
Acknowledgments
This work was supported by the Strategic Priority Research Program of Chinese Academy of Sciences (CAS) to Wei Xu and J. Chai (XDB31000000); National Key R&D Program of China (2022YFC2602500) and Yunnan Revitalization Talent Support Program Science & Technology Champion Project to J. Che; National Natural Science Foundation of China (NSFC 32300383), and Yunnan Applied Basic Research Projects (202301AT070431) to Wei Xu; National Natural Science Foundation of China (NSFC 32100371), Science and Technology Basic Resources Investigation Program of China (2021FY100203), Major Science and Technique Programs in Yunnan Province (202102AA310055) to Y.-H.W.; National Natural Science Foundation of China (NSFC 31900323), and Open Project from the State Key Laboratory of Genetic Resources and Evolution (GREKF21-02) to J.-M.C.; Key R&D program of Yunnan Province, China (202303AH310055) and China’s Biodiversity Observation Network (Sino-BON) to W.G.; Program of Yunnan Forestry and Grassland Administration (2022GF258D-10) and Survey of Wildlife Resources in Key Areas of Tibet (ZL202203601) to K.W.; Animal Branch of the Germplasm Bank of Wild Species, CAS (the Large Research Infrastructure Funding) to T.Y. and J. Che; Spring City Plan: the High-level Talent Promotion and Training Project of Kunming (2022SCP001) to Y.-P.Z. We thank the National Forestry and Grassland Administration, local Forestry Department and National Reserve of China for permission to undertake field surveys and for other assistance. We thank the Genome Center of Biodiversity from the Kunming Institute of Zoology (CAS) for support in generating the DNA sequence data. We are grateful to Yun-Yu Wang, Jun-Xiao Yang, Li-Min Ding, Hai-Tao Zhao, Jia-Zhong Wang, Liang Zhang, Li Ding, Jinzhong Fu, and many others for help with field surveys and providing samples. We thank two anonymous reviewers for helpful comments on the manuscript.
Author contributions
Y.-P.Z., J.J.W., and J. Che designed research; Wei Xu, Y.-H.W., W.-W.Z., Y.-P.Z., J.J.W., and J. Che performed research; Wei Xu, Y.-H.W., Weihua Xu, Y.C., Z.-Y.L., X.-Y.L., D.-R.Z., and C. Lu analyzed data; W.-W.Z., H.-M.C., B.-L.Z., J.-M.C., D.-Q.R., H.Z., F.Y., Z.Y., K.J., J.-Q.J., M.H., D.Z., L.-J.W., Y.Z., J.-T.L., J.J., X.-M.Z., C. Li, T.Y., Z.D., G.-G.Z., J. Chai, W.-G.Z., and J. Che collected data; and Wei Xu, Y.-H.W., W.G., K.W., J.J.W., and J. Che wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Although PNAS asks authors to adhere to United Nations naming conventions for maps (https://www.un.org/geospatial/mapsgeo), our policy is to publish maps as provided by the authors.
Contributor Information
John J. Wiens, Email: wiensj@arizona.edu.
Jing Che, Email: chej@mail.kiz.ac.cn.
Data, Materials, and Software Availability
All R codes used in this study have been stored on GitHub at https://github.com/CheLab-KIZ/Paper-codes/tree/main/CJPA202401 (63), along with intermediate analysis files. All other data are included in the manuscript and/or supporting information.
Supporting Information
References
- 1.Barnosky A. D., et al. , Has the Earth’s sixth mass extinction already arrived? Nature 471, 51–57 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Ceballos G., Ehrlich P. R., Raven P. H., Vertebrates on the brink as indicators of biological annihilation and the sixth mass extinction. Proc. Natl. Acad. Sci. U.S.A. 117, 13596–13602 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WWF, “Executive summary” in Living Planet Report 2018. Aiming Higher, Grooten M., Almond R. E. A., Eds. (WWF, Gland, Switzerland, 2018). [Google Scholar]
- 4.Antonelli A., et al. , State of the World’s Plants and Fungi 2020 (Royal Botanic Gardens, Kew, UK, 2020). [Google Scholar]
- 5.Baillie J., Zhang Y. P., Space for nature. Science 361, 1051 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Dinerstein E., et al. , A global deal for nature: Guiding principles, milestones, and targets. Sci. Adv. 5, eaaw2869 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waldron A., et al. , “Protecting 30% of the planet for nature: Costs, benefits and economic implications” in Campaign for Nature (2020). https://pure.iiasa.ac.at/id/eprint/16560/1/Waldron_Report_FINAL_sml.pdf. Accessed 25 October 2023.
- 8.Oewg C. B. D., First Draft of the Post-2020 Global Biodiversity Framework CBD/WG2020/3/3 (CBD, 2021). [Google Scholar]
- 9.Myers N., Mittermeier R. A., Mittermeier C. G., Da Fonseca G. A., Kent J., Biodiversity hotspots for conservation priorities. Nature 403, 853–858 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Mittermeier R. A., “Primate diversity and the tropical forest: Case studies from Brazil and Madagascar and the importance of the megadiversity countries” in Biodiversity, Wilson E. O., Ed. (National Academy Press, 1988). [Google Scholar]
- 11.Jiang Z., Luo Z., Assessing species endangerment status: Progress in research and an example from China. Biodivers. Sci. 20, 612–622 (2012). [Google Scholar]
- 12.Wang L., Jia Y., Zhang X., Qin H., Overview of higher plant diversity in China. Biodivers. Sci. 23, 217–224 (2015). [Google Scholar]
- 13.Mi X., et al. , The global significance of biodiversity science in China: An overview. Natl. Sci. Rev. 8, nwab032 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Körner C., Spehn E. M., Mountain Biodiversity: A Global Assessment (Parthenon, Boca Raton, FL, 2002). [Google Scholar]
- 15.Xu W., et al. , Strengthening protected areas for biodiversity and ecosystem services in China. Proc. Natl. Acad. Sci. U.S.A. 114, 1601–1606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B. V., Pimm S. L., How China expanded its protected areas to conserve biodiversity. Curr. Biol. 30, R1334–R1340 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Wake D. B., Vredenburg V. T., Are we in the midst of the sixth mass extinction? A view from the world of amphibians. Proc. Natl. Acad. Sci. U.S.A. 105, 11466–11473 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alroy J., Current extinction rates of reptiles and amphibians. Proc. Natl. Acad. Sci. U.S.A. 112, 13003–13008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.IUCN, The IUCN Red List of Threatened Species. Version 2021-3 (IUCN, 2021). https://www.IUCNiucnredlist.org. [Google Scholar]
- 20.Bishop P. J., et al. , The amphibian extinction crisis—What will it take to put the action into the amphibian conservation action plan? S.A.P.I.EN.S. 5, 97–111 (2012). [Google Scholar]
- 21.Hu Y., et al. , Spatial patterns and conservation of genetic and phylogenetic diversity of wildlife in China. Sci. Adv. 7, eabd5725 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy R. W., Advances in herpetological research emanating from China. Zool. Res. 37, 4–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Womack M. C., et al. , State of the Amphibia 2020: A review of five years of amphibian research and existing resources. Ichthyol. Herpetol. 110, 638–661 (2021). [Google Scholar]
- 24.Hanken J., Why are there so many new amphibian species when amphibians are declining? Trends Ecol. Evol. 14, 7–8 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Vieites D. R., et al. , Vast underestimation of Madagascar’s biodiversity evidenced by an integrative amphibian inventory. Proc. Natl. Acad. Sci. U.S.A. 106, 8267–8272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K., et al. , The updated checklists of amphibians and reptiles of China. Biodivers. Sci. 28, 189–218 (2020). [Google Scholar]
- 27.Chai J., et al. , Discovery of a wild, genetically pure Chinese giant salamander creates new conservation opportunities. Zool. Res. 43, 469–480 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J., Slik F., Zheng S., Lindenmayer D. B., Undescribed species have higher extinction risk than known species. Conserv. Lett. 15, e12876 (2022). [Google Scholar]
- 29.Chen J., Progress in development and management of nature reserves in China. Report to the Standing Committee of National People’s Congress. www.mep.gov.cn/xxgk/hjyw/201607/t20160701_356571.shtml. Accessed June 2018.
- 30.Ma Z., et al. , Changes in area and number of nature reserves in China. Conserv. Biol. 33, 1066–1075 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H., et al. , Report on Baseline Assessment of China’s Biodiversity (Sciences Press, Beijing, China, 2013). [Google Scholar]
- 32.Huang J. L., Miao S. Y., Deng Y., The core resources and scientific protection values of Guangdong Nanling National Park. Nat. Park 5, 1–7 (2020). [Google Scholar]
- 33.Lu L. M., et al. , Evolutionary history of the angiosperm flora of China. Nature 554, 234–238 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann M., et al. , The impact of conservation on the status of the world’s vertebrates. Science 330, 1503–1509 (2010). [DOI] [PubMed] [Google Scholar]
- 35.AmphibiaChina, The Database of China’s Amphibians (Kunming Institute of Zoology (CAS), Kunming, China, 2020), https://www.amphibiachina.org/. [Google Scholar]
- 36.Meng K., Tao Y., Murphy R. W., Li S. Q., Does plate tectonics determine distributional patterns of extant animals? A case study using the amphibians of China. Acta Zootaxon. Sin. 38, 679–686 (2013). [Google Scholar]
- 37.Chen Y., Zhang J., Jiang J., Nielsen S. E., He F., Assessing the effectiveness of China’s protected areas to conserve current and future amphibian diversity. Divers. Distrib. 23, 146–157 (2016). [Google Scholar]
- 38.Fei L., Hu S., Ye C., Huang Y., Eds., Fauna Sinica, Amphibia. Anura (Science Press, Beijing, China, 2009), vol. 2. [Google Scholar]
- 39.Munguía M., Rahbek C., Rangel T. F., Diniz-Filho J. A. F., Araújo M. B., Equilibrium of global amphibian species distributions with climate. PLoS ONE 7, e34420 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vilela B., Villalobos F., letsR: A new R package for data handling and analysis in macroecology. Methods Ecol. Evol. 6, 1229–1234 (2015). [Google Scholar]
- 41.Kreft H., Jetz W., A framework for delineating biogeographical regions based on species distributions. J. Biogeogr. 37, 2029–2053 (2010). [Google Scholar]
- 42.He J., Kreft H., Gao E., Wang Z., Jiang H., Patterns and drivers of zoogeographical regions of terrestrial vertebrates in China. J. Biogeogr. 44, 1172–1184 (2017). [Google Scholar]
- 43.Smith S. A., Nieto Montes de Oca A., Reeder T. W., Wiens J. J., A phylogenetic perspective on elevational species richness patterns in Middle American treefrogs: Why so few species in lowland tropical rainforests? Evolution 61, 1188–1207 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Hutter C. R., Lambert S. M., Wiens J. J., Rapid diversification and time explain amphibian richness at different scales in the Tropical Andes. Earth’s most biodiverse hotspot. Am. Nat. 190, 828–843 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Rahbek C., et al. , Humboldt’s enigma: What causes global patterns of mountain biodiversity? Science 365, 1108–1113 (2019). [DOI] [PubMed] [Google Scholar]
- 46.Crisp M. D., Laffan S., Linder H. P., Monro A., Endemism in the Australian flora. J. Biogeogr. 28, 183–198 (2001). [Google Scholar]
- 47.Daru B. H., Karunarathne P., Schliep K., phyloregion: R package for biogeographic regionalization and macroecology. Methods Ecol. Evol. 11, 1483–1491 (2020). [Google Scholar]
- 48.Pyron R. A., Wiens J. J., A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61, 543–583 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Pyron R. A., Biogeographic analysis reveals ancient continental vicariance and recent oceanic dispersal in amphibians. Syst. Biol. 63, 779–797 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Jetz W., Pyron R. A., The interplay of past diversification and evolutionary isolation with present imperilment across the amphibian tree of life. Nat. Ecol. Evol. 2, 850–858 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Wei T., Simko V., R package “corrplot”: Visualization of a Correlation Matrix (2021), https://github.com/taiyun/corrplot. Accessed 28 January 2024.
- 52.Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J., Phylogenies and community ecology. Annu. Rev. Ecol. Evol. Syst. 33, 475–505 (2002). [Google Scholar]
- 53.Prendergast J. R., Wood S. N., Lawton J. H., Eversham B. C., Correcting for variation in recording effort in analyses of diversity hotspots. Biodivers. Lett. 1, 39–53 (1993). [Google Scholar]
- 54.Tang Z., Wang Z., Zheng C., Fang J., Biodiversity in China’s mountains. Front. Ecol. Evol. 4, 347–352 (2006). [Google Scholar]
- 55.Huang J., et al. , Diversity hotspots and conservation gaps for the Chinese endemic seed flora. Biol. Conserv. 198, 104–112 (2016). [Google Scholar]
- 56.Xue T., et al. , Prioritizing conservation of biodiversity in an alpine region: Distribution pattern and conservation status of seed plants in the Qinghai-Tibetan Plateau. Glob. Ecol. Conserv. 32, e01885 (2011). [Google Scholar]
- 57.Hoffman M., et al. , Biodiversity Hotspots (version 2016.1) (2016.1). Zenodo (2016). 10.5281/zenodo.3261807. Accessed 8 April 2021. [DOI]
- 58.Rodrigues A. S. L., et al. , Effectiveness of the global protected area network in representing species diversity. Nature 428, 640–643 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Xu W., et al. , Transforming protected area management in China. Trends Ecol. Evol. 34, 762–766 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Peng S., et al. , Preserving the woody plant tree of life in China under future climate and land-cover changes. Proc. R. Soc. B: Biol. Sci. 289, 20221497 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martins J. H., Camanho A. S., Gaspar M. B., A review of the application of driving forces–pressure–state–impact–response framework to fisheries management. Ocean Coast. Manag. 69, 273–281 (2012). [Google Scholar]
- 62.Venter O., et al. , Sixteen years of change in the global terrestrial human footprint and implications for biodiversity conservation. Nat. Commun. 7, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu W., et al. , Source code for “Hidden hotspots of amphibian biodiversity in China”. GitHub. https://github.com/CheLab-KIZ/Paper-codes/tree/main/CJPA202401. Deposited 16 February 2024. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (TXT)
Dataset S05 (TXT)
Dataset S06 (TXT)
Dataset S07 (TXT)
Dataset S08 (XLSX)
Data Availability Statement
All R codes used in this study have been stored on GitHub at https://github.com/CheLab-KIZ/Paper-codes/tree/main/CJPA202401 (63), along with intermediate analysis files. All other data are included in the manuscript and/or supporting information.