Abstract
The α-acetolactate decarboxylase (ALDC) gene, aldB, is the penultimate gene of the leu-ilv-ald operon, which encodes the three branched-chain amino acid (BCAA) biosynthesis genes in Lactococcus lactis. Its product plays a dual role in the cell: (i) it catalyzes the second step of the acetoin pathway, and (ii) it controls the pool of α-acetolactate during leucine and valine synthesis. It can be transcribed from the two promoters present upstream of the leu and ilv genes (P1 and P2) or independently under the control of its own promoter (P3). In this paper we show that the production of ALDC is limited by two mechanisms. First, the strength of P3 decreases greatly during starvation for BCAAs and under other conditions that generally provoke the stringent response. Second, although aldB is actively transcribed from P1 and P2 during BCAA starvation, ALDC is not significantly produced from these transcripts. The aldB ribosome binding site (RBS) appears to be entrapped in a stem-loop, which is itself part of a more complex RNA folding structure. The function of the structure was studied by mutagenesis, using translational fusions with luciferase genes to assess its activity. The presence of the single stem-loop entrapping the aldB RBS was responsible for a 100-fold decrease in the level of aldB translation. The presence of a supplementary secondary structure upstream of the stem-loop led to an additional fivefold decrease of aldB translation. Finally, the translation of the ilvA gene terminating in the latter structure decreased the level of translation of aldB fivefold more, leading to the complete extinction of the reporter gene activity. Since three leucines and one valine are present among the last six amino acids of the ilvA product, we propose that pausing of the ribosomes during translation could modulate the folding of the messenger, as a function of BCAA availability. The purpose of the structure-dependent regulation could be to ensure the minimal production of ALDC required for the control of the acetolactate pool during BCAA synthesis but to avoid its overproduction, which would dissipate acetolactate. Large amounts of ALDC, necessary for operation of the acetoin pathway, could be produced under favorable conditions from the P3 transcripts, which do not contain the secondary structures.
Synthesis of the three branched-chain amino acids (BCAA), leucine, isoleucine, and valine, has been studied in detail in organisms as diverse as bacteria, fungi, and plants (for reviews, see references 6, 22, and 47). A particular feature of BCAA synthesis is that several steps of this pathway are carried out by the same enzymes. Genes encoding the enzymes responsible for BCAA synthesis are often clustered, e.g., the ilvBNC-leuACBD operon of Bacillus subtilis, the ilvBNC operon of Corynebacterium glutamicum, and the ilvGMEDA operon of Escherichia coli. In Lactococcus lactis subsp. lactis, the structural genes for BCAA synthesis are present in a single operon that also contains three additional genes (Fig. 1A) (16, 19). One of these genes, aldB, encodes an α-acetolactate (AL) decarboxylase (ALDC), an enzyme usually involved in the catabolic degradation of AL to acetoin in the 2,3-butanediol pathway.
FIG. 1.
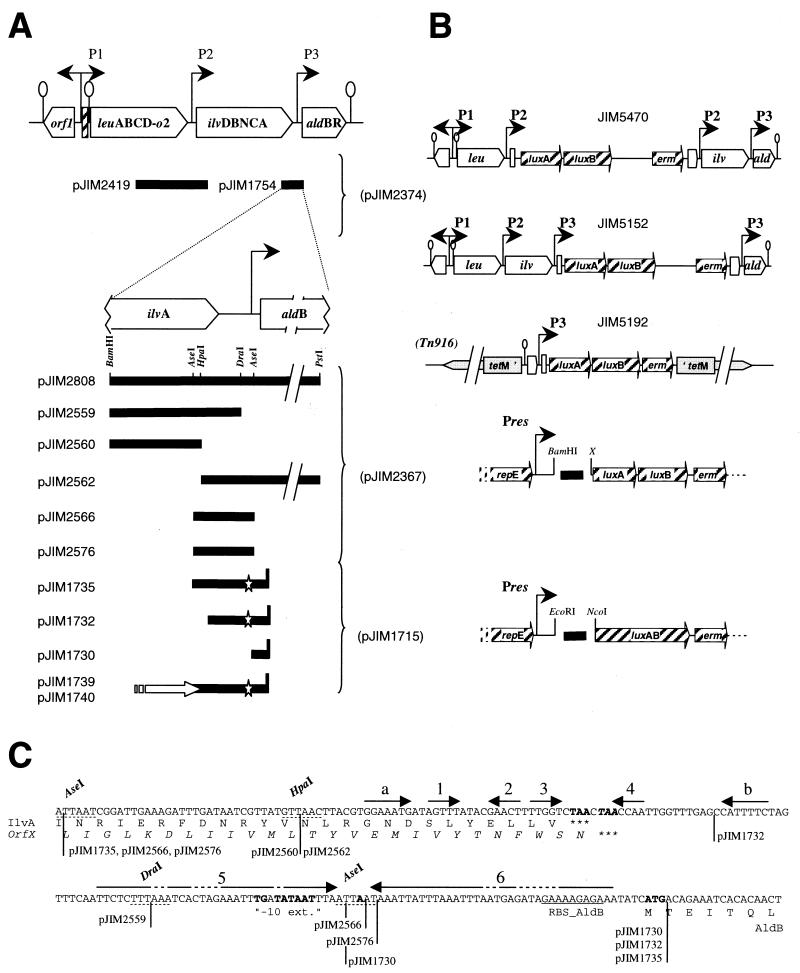
Organization of the L. lactis leu-ilv-ald cluster and schematic representation of the constructions used in this work. (A) Locations of the fragments (horizontal bars) fused with the luciferase genes. The vectors used are indicated in parentheses on the right, and the designations of the final constructions are given on the left. The stars indicate the mutation inactivating P3, the vertical bar indicates the start codon of aldB (pJIM1730 to -1740), and the white arrow indicates the 5′ end of the hisC gene fused or not with ilvA (pJIM1740 and pJIM1739, respectively). (B) Schematic representation of the final constructs after integration in the chromosome for JIM5470, -5152, and -5192 or carried on replicative plasmids. P1, P2, and P3 refer to promoters (represented by arrows). Pres is a constitutive promoter from the vector present downstream of repE, encoding the protein allowing plasmid replication. tetM and erm are the resistance markers from Tn916 and pAMβ1. luxA, luxB, and luxAB are the native lux genes from Vibrio harveyi and the fused version. Lollipops show the terminators. (C) Sequence of the intergenic ilvA-aldB region. The arrows indicate the repeats that could form potential secondary structure of the messengers initiated at P1 or P2 by the pairing of 1-2, 3-4, 5-6, and a-b, as shown in Fig. 4. The sequence in boldface is the −10 extended box (“−10 ext.”) and the transcriptional start of P3. The ORFs translated below the sequence encode IlvA, AldB, and (in italics) the ORF translated in pJIM1739 (OrfX). The vertical lines show the ends of the cloned fragments in the indicated plasmids.
AL is a central metabolite involved in both anabolism (synthesis of leucine and valine) and catabolism (production of acetoin). The control of its partition between the two pathways is thus of particular importance. Previous work has shown that in L. lactis, the product of aldB was responsible for (i) the degradation of AL produced during the catabolism of sugars (18) and (ii) the regulation of the pool of AL in the cell during BCAA metabolism (19). This is important since IlvBN, the first enzyme of the pathway, is not subject to feedback control in L. lactis (3). To exert a role in the metabolic regulation of BCAA biosynthesis, ALDC should be expressed during BCAA starvation. However, to avoid the dissipation of the pool of AL during the biosynthesis, the activity of this enzyme should be tightly controlled. A first level of control, at the enzymatic level, was previously described. L. lactis ALDC is activated allosterically in vitro by leucine (36). The activation of ALDC in vivo by addition of leucine in the medium induces valine starvation (19).
Despite the low activity of ALDC in the absence of leucine, its synthesis at high levels during BCAA synthesis could have a negative effect on BCAA synthesis in the cell. This consideration raises the question of a possible regulation of ALDC synthesis. Indeed, it may be advantageous for the cell to adjust the amount of ALDC in response to different environmental conditions. For instance, ALDC may be required at high levels during carbon limitation or aerobiosis, conditions that induce a shift from lactic homofermentation to mixed acid fermentation, often concomitant with high fluxes to acetoin. In contrast, ALDC may be required at low levels to regulate the AL pool during BCAA biosynthesis (19).
A previous study suggested that the transcriptional level of the specific aldB promoter is almost constitutive, regardless of the presence of BCAA (19). Moreover, aldB is transcribed from two promoters, one situated upstream of the leu-ilv operon and the other within it, since there is no terminator upstream of aldB. This feature has not been found in other microorganisms, where aldB is under the control of a promoter independent from BCAA biosynthesis genes (32, 41). The transcriptional pattern of aldB in L. lactis is thus in apparent contradiction with the previous assumption that the production of ALDC should be limited during BCAA starvation, when the leu-ilv operon is strongly transcribed.
The analysis of the sequence of the intergenic region between ilvA and aldB indicated that the mRNA could fold in a complex structure entrapping the aldB ribosome binding site (RBS) (40). It was proposed that such folding could reduce the translation of aldB produced from the upstream transcripts. Here we show that the translation of aldB from the messengers initiated upstream of its specific promoter is indeed inhibited. In addition we show that the transcription from the specific aldB promoter is tightly regulated by amino acid starvation. This suggests that the conjunction of the two mechanisms regulates ALDC synthesis to adjust optimally its level to the cell requirement in L. lactis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are presented in Table 1 and Fig. 1B and C. L. lactis subsp. lactis NCDO2118 was grown at 30°C in M17 medium (46) or in chemically defined medium (CDM) (referred to as MCD in Fig. 2 and 3) (37), in which the sugar and amino acid compositions were modified as described in Results. The two amino acid analogs arginine hydroxamate and serine hydroxamate were added at 250 μg/ml in CDM without arginine and serine, respectively. E. coli and B. subtilis were grown in Luria-Bertani medium at 37°C. Transformation of L. lactis, E. coli, and B. subtilis was performed by standard procedures (1, 21, 43). When necessary, erythromycin (100 μg/ml for E. coli, 5 μg/ml for L. lactis, and 0.5 μg/ml for B. subtilis), ampicillin (50 μg/ml for E. coli), or tetracycline (10 μg/ml) was added to the medium.
TABLE 1.
Bacterial strains, plasmids, and mutagenic oligonucleotides
| Strain or plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| L. lactis | ||
| NCDO2118 | L. lactis subsp. lactis natural isolate | NCDO |
| JIM4460 | NCDO2118, aldB::tetM | 19 |
| JIM5152 | NCDO2118, Emr, P3::lux on pJIM1754 integrated in the chromosome | This work |
| JIM5192 | NCDO2118, transconjugant with Tn916::P3::luxAB-erm from JIM5244 | This work |
| JIM5470 | NCDO2118, Emr, P2::lux on pJIM2419 integrated in the chromosome | This work |
| Other species | ||
| JIM5027 | B. subtilis containing Tn916 from Enterococcus faecalis | This work |
| JIM5244 | JIM5027, Tn916::P3::luxAB-erm by double crossover of pJIM2842 | This work |
| TG1 | E. coli, supE thi Δ(lac-proAB) hsdD5 F+traD36 proAB lacI ZDM15 | 15 |
| Plasmids | ||
| pVE6004 | Emr thermosensitive derivative of pGKV12 | 30 |
| pVE6007 | Cmr thermosensitive derivative of pGKV12 | 30 |
| pBluescript | Ampr, M13 ori, pBR322 ori | Stratagene |
| pJIM500 | 18.5-kb XbaI fragment of L. lactis chromosome containing the leu-ilv-ald operon in pIL253 | 16 |
| pJIM540 | 1.2-kb BamHI-EcoRI fragment of pJIM500 in pBluescript | 19 |
| pJIM540Δ* | pJIM540Δ (Δ = deleted by HindIII-PstI), amplified by PCR with P3mut3 and P3mut4, cut by NsiI, blunted by T4 polymerase and ligated; P3 contains the G-to-A and T-to-A substitutions | This work |
| pJIM2802 | Emr, contains P3::luxAB upstream of the erm gene in pJIM2367 | 39 |
| Integrative plasmids | ||
| pJIM2374 | Emr, ORI(pWV01ΔrepA), integrative transcriptional fusion vector with the luxAB genes | |
| pJIM1754 | pJIM2374 with 0.564-kb BamHI-PstI fragment containing P3 from pJIM540 | This work |
| pJIM2419 | pJIM2374 with 3.82-kb EcoRI fragment containing P2 from pJIM500 | This work |
| pJIM2842 | pVE6007 containing tetM from Tn916; the 5.2-kb XcaI fragment containing terminator-P3-luxAB-erm cassette from pJIM2802 is inserted in HindII from tetM | This work |
| Plasmids for analysis of P3 promoter | ||
| pJIM2367 | Emr, promoter probe vector with the luxAB genes | 39 |
| pJIM2808 | pJIM2367 containing BamHI-PstI fragment from pJIM540 | This work |
| pJIM2559 | pJIM2367 containing 171-bp BamHI-DraI fragment from pJIM540 | This work |
| pJIM2560 | pJIM2367 containing 261-bp BamHI-HpaI fragment from pJIM540 | This work |
| pJIM2562 | pJIM2367 containing 393-bp HpaI-PstI fragment from pJIM540 | This work |
| pJIM2566 | pJIM2367 containing 148-bp AseI-AseI fragment from pJIM540 | This work |
| pJIM2576 | pJIM2367 carrying a 150-bp PCR fragment obtained from pJIM2566 with P3mut1 and a plasmid oligonucleotide; the fragment contains the wild-type P3 | This work |
| pJIM2589 | Like pJIM2576 with mutation in the P3 −10 extended box (Table 4) | This work |
| pJIM2596 | Like pJIM2576 with mutation in the P3 −10 extended box (Table 4) | This work |
| pJIM2586 | Like pJIM2576 with mutation in the P3 −10 extended box (Table 4) | This work |
| pJIM2591 | Like pJIM2576 with mutation in the P3 −10 extended box (Table 4) | This work |
| pJIM2593 | Like pJIM2576 with mutation in the P3 −10 extended box (Table 4) | This work |
| pJIM2594 | Like pJIM2576 with mutation in the P3 −10 extended box (Table 4) | This work |
| Plasmids for translational study of aldB | ||
| pJIM781 | XhoI-BamHI (PCR processed) PCR fragment containing the RBS and the beginning of L. lactis hisC | C. Delome, personal communication |
| pJIM1299 | Ligated AseI fragment from pJIM2566 and AseI-EcoRI fragment from pJIM540Δ*, amplified by PCR with oligonucleotide aldB1 and an oligonucleotide from the plasmid pJIM540; cloned in BamHI of pBluescript | This work |
| pJIM1711 | BamHI PCR fragment of 113 bp amplified with oligonucleotides aldB1 and aldB2 from pJIM1299 cloned in BamHI of pBluescript | This work |
| pJIM1715 | Emr, transcriptional-translational fusion vector with luxAB fusion gene processed to introduce a NdeI site overlapping the ATG start codon | 39 |
| pJIM1730 | pJIM1715 containing the 50-bp AseI-NdeI fragment from pJIM1299 in SpeI (T4 polymerase blunted)-NdeI | This work |
| pJIM1732 | pJIM1715 containing the BamHI-NdeI PCR fragment from pJIM1711 in BamHI-NdeI | This work |
| pJIM1735 | pJIM1715 containing the BamHI-NdeI fragment from pJIM1299 in BamHI-NdeI | This work |
| pJIM1739 | pJIM1735 containing the XhoI (T4 polymerase blunted)-BamHI fragment from pJIM781 in SmaI-BamHI | This work |
| pJIM1740 | pJIM1739 derivative with BamHI cut and filled by T4 polymerase | This work |
| Mutagenic oligonucleotides | ||
| P3mut1 | ATCGAATTCATTAATTAAATTAT(ACGT)T(AT)AAATTTCTAGTGa | This work |
| P3mut3 | GGGCATGCATTTAAAATAATTTAATTAAT | This work |
| P3mut4 | ACGGATGCATTTCTAGTGATTTAAAGAG | This work |
| aldB1 | CCGGATCCATATGATTTCTCTTTCTATCTC; the ATG from NdeI is the start codon of AldB | This work |
| aldB2 | GGGGATCCATTTTCTAGTTTCAATTCTC | This work |
The rules, boldface, and italics indicate the restriction sites designed in the oligonucleotide, the base changed compared to the wild-type sequence, and the start codon used for the translation of luxAB, respectively.
FIG. 2.
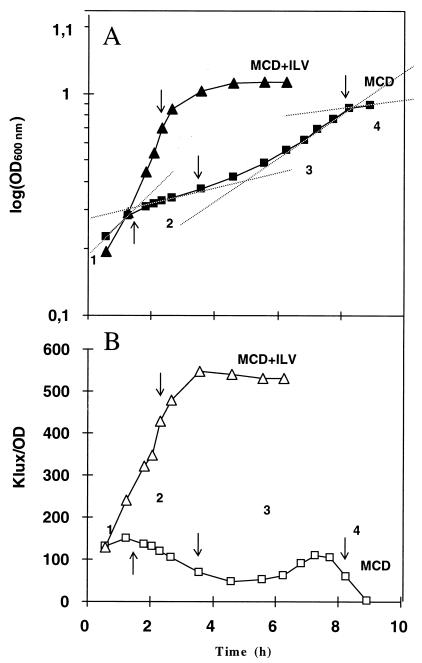
Growth curves (A) and luciferase expression from a P3::lux fusion carried by Tn916 in JIM5192 (B). The cells were grown in CDM with (triangles) or without (squares) BCAA. The arrows mark the transitions between the different phases of growth. In CDM without isoleucine, leucine, and valine (ILV) the phases were as follows: 1, growth after inoculation; 2, adaptation to ILV starvation; 3, exponential growth; and 4, stationary phase. Dashed lines indicate the slopes of the curves in the different phases. Results of a representative experiment is shown, out of three that were carried out.
FIG. 3.
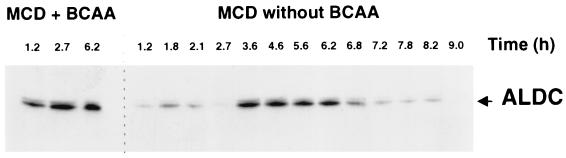
Western blotting with ALDC antibodies, carried out on total cell extracts from JIM5192 grown in CDM with and without BCAA, as indicated. The samples were taken during the experiment presented in Fig. 2, at the times indicated above the lanes.
DNA and RNA manipulations.
Plasmids and chromosomal DNA were prepared as previously described (27, 43, 44). Southern blotting, DNA hybridization, and other molecular techniques were performed as described previously (43). DNA probes were labeled by nick translation using [α-32P]dCTP (Amersham) according to the recommendations of the supplier (Boehringer Mannheim). The sequences of all constructions were verified using a Taq Dye Primer Cycle sequencing kit and 370A sequencer (Applied Biosystems). RNA preparation and Northern hybridizations were performed as described previously (19).
Plasmids construction and mutagenesis.
The relevant steps for construction of plasmids and strains are summarized in Table 1. PCR fragments were usually cloned in pBS and sequenced before their insertion in the final vector. The sites adjacent to the cloned PCR fragments in pBS were often used to facilitate the constructions. Fusion of the pBS derivatives with the appropriate vectors followed by the deletion of the pBS backbone in L. lactis was also frequently used.
Campbell-like integrations were performed as follows. Integrative plasmids derived from pJIM2374 conferring erythromycin resistance were first established at 30°C in L. lactis NCDO2118, which contains the thermosensitive helper plasmid pVE6007, carrying a chloramphenicol resistance gene. In a second step, the strains carrying pJIM2374-derived plasmids integrated in the chromosome were selected on erythromycin at 37°C, a nonpermissive temperature for pVE6007 replication (30). Chromosomal gene replacements were done by a two-step procedure (4). The derivative of Tn916 carrying a P3::lux fusion and the erythromycin marker in tetM was constructed in B. subtilis and then transferred by conjugation to L. lactis, to give JIM5192.
Analysis of ALDC synthesis by Western blotting.
The cells were recovered from the medium by centrifugation and suspended in lysis buffer (Tris [50 mM, pH 8], NaCl [0.1 M], saccharose [10%], lysozyme [1 mg/ml]) for 30 min at 4°C. The suspension was adjusted to an optical density (OD) at 600 nm of 10. Before the sample was loaded on the gel, loading buffer was added and the samples were boiled for 2 minutes. Proteins were subjected to electrophoresis in sodium dodecyl sulfate-polyacrylamide gels (24) and blotted on polyvinylidene difluoride membrane (Millipore). ALDC was revealed by immunochemiluminescence, using anti-ALDC polyclonal antibodies and ECL detection (Amersham). The antibodies were produced in rabbit after three injections of 20 μg of ALDC (36) and included in liposomes.
Luciferase assay.
One milliliter of culture was mixed with 5 μl of nonaldehyde, and the light emission was measured immediately in a Bertold luminometer. Lux activity was monitored during the entire length of the growth to obtain an accurate measure of the transcriptional levels at the different growth stages. Values given in this work are those read on a plot of lux versus OD at an OD of 0.5.
RESULTS
Characterization of P3 elements by directed mutagenesis.
The P3 promoter is specific for ald genes. Its transcriptional start was previously determined (Fig. 1C) (19). An extended −10 promoter box (TGNTATAAT) without a −35 box is present at an appropriate distance from the 5′ end of this transcript. A deletion analysis of the region containing P3 was performed in order to characterize the cis elements necessary for its full activity. First, we subcloned a 0.56-kb BamHI-PstI fragment containing P3 in the promoter probe vector pJIM2367 in front of the lux genes. The plasmid obtained, pJIM2808 (Fig. 1A), produced about 2,000 klx/OD unit when introduced in L. lactis NCDO2118. This strong luciferase activity indicates that the fragment carries the elements necessary for the full activity of P3. The deletion of the region downstream of DraI in pJIM2559 and pJIM2560 (Fig. 1C) led to a loss of lux activity, as expected for the removal of P3. Deletion of the sequence upstream of HpaI in pJIM2562 did not affect the activity of P3. However, the 148-bp AseI fragment that contains the start point of transcription (Fig. 1C, pJIM2566) gave a 50-fold-reduced activity. Finally, the addition of two more bases after the AseI site restored full P3 activity (pJIM2576). This indicates that the P3 promoter overlaps the AseI site.
To better characterize the effect of the TGN motif on P3 activity, we have mutagenized these nucleotides with the help of degenerated oligonucleotides (Table 1). The 150-bp PCR products were inserted in the promoter probe vector pJIM2367. We obtained six different combinations of mutations, with one or two changes from the consensus present in P3 (Table 2). The replacement of the first nucleotide from the −10 box (TATAAT) by C or A decreased the activity of P3 more than 1,000-fold. The G-to-A change in the TGN motif also decreased the activity of P3 1,000-fold, showing that this base is absolutely required for significant activity of this promoter. Lastly, double changes provoked an even stronger decrease of the transcriptional activity. For example, G-to-A combined with T-to-C changes almost completely abolished the activity of the promoter. These experiments establish the importance of the TG part of the promoter.
TABLE 2.
Mutations affecting the activity of P3
| Plasmid | Promotera | Luciferase activityb | Ratioc |
|---|---|---|---|
| pJIM2576 (wild type) | TG A TATAAT | 2,000 | 1 |
| pJIM2589 | TG A cATAAT | 6 | 3.0 × 10−3 |
| pJIM2596 | TG A aATAAT | 0.45 | 2.2 × 10−4 |
| pJIM2586 | Ta A TATAAT | 10 | 5.0 × 10−3 |
| pJIM2591 | Ta A cATAAT | 0.7 | 3.5 × 10−4 |
| pJIM2593 | Ta A gATAAT | 1.4 | 7.0 × 10−4 |
| pJIM2594 | Ta A aATAAT | 0.43 | 2.1 × 10−4 |
The −10 consensus sequence is in uppercase, and the mutation is in lowercase.
Specific luciferase activity in kilolux/OD unit at an OD of 0.5.
The rate was calculated as the ratio of the luciferase activity produced from the promoter to that produced by the wild type.
Regulation of P3 by global response to starvation.
To study the activity of P3 in a chromosomal context without the interference of the two upstream promoters, we introduced the P3::lux fusion in the tetM gene of Tn916 in JIM5192, as shown in Fig. 2. When JIM5192 was cultivated in CDM with BCAA, the luciferase activity increased constantly during growth. In contrast, in the absence of BCAA, the activity fluctuated at low levels. The fluctuations were correlated with the growth rate. Indeed, the growth curve can be divided in four phases. During the first, upon inoculation, growth was fast, possibly due to BCAA remaining in the inoculum. During the second, growth was slow, possibly because of the latency necessary to start the BCAA synthesis. The growth resumed during the third phase and stopped when the culture reached the fourth, stationary phase. The luciferase activity had a tendency to increase during rapid growth phases (1 and 3) and to decrease during the slow growth and stationary phases (2 and 4). This suggests that P3 transcriptional activity is growth rate dependant or is low in cells starved for BCAA (during latency or stationary phase).
To determine the parameters affecting P3 activity, we carried out several experiments with JIM5192: (i) the growth rate was modulated independently of amino acid starvation, and (ii) cells were starved for amino acids other than BCAA. The growth rate was modulated by modifying the sugar in the medium. Glucose gave the highest growth rate (generation time [Tg], 50 min) and lactose gave the lowest (Tg, 150 min), while galactose was intermediate (Tg, 90 min). The luciferase activity was two- to threefold higher in CDM with lactose or galactose than in CDM with glucose during exponential growth. This result rules out the hypothesis that a high growth rate mediates an increase of P3 activity. The effect of starvation for amino acids other than BCAA was tested in two experiments. First, JIM5192 was inoculated in CDM without histidine or methionine. Starvation for these amino acids severely reduces the growth of L. lactis. Under these conditions, the activity of P3 remained low. Second, cells were grown in medium lacking arginine or serine in presence of analogs of these amino acids (arginine or serine hydroxamate, respectively). The addition of these analogs is necessary to provoke starvation for the cognate amino acid in JIM5192, since this strain is a prototroph for both. A marked decrease in P3 activity was observed. Taken together, these results suggest that P3 is controlled by a general response to starvation.
ALDC synthesis correlates with P3, but not P1 and P2, activity.
The production of ALDC was monitored by Western blotting with polyclonal antibodies directed against the L. lactis ALDC (Fig. 3). In the presence of BCAA in the medium, the amount of ALDC was similar during the growth and stationary phases. However, in the absence of BCAA, this amount varied during growth. ALDC was detectable after inoculation and then disappeared during the lag required for adaptation to BCAA starvation. When the growth resumed, ALDC was produced again, and it then decreased to a nondetectable level after the end of the exponential growth phase. The production of ALDC thus seems to occur mainly during a phase in which the activity of P3, measured by luciferase fusion in JIM5192, increased (Fig. 2).
To measure more precisely the level of transcription originating upstream of P3, the luxAB reporter genes were placed downstream of P2 and P3 on the integrative plasmids pJIM2419 and pJIM1754, respectively (Fig. 1). The assay of the luciferase activities from NCDO2118 derivatives carrying these fusions on the chromosome allowed the measurement of the combined transcription level from P1 plus P2 and from P1 plus P2 plus P3, respectively. In the presence of the three BCAA, the transcription level was fivefold lower upstream than downstream of P3 (16 and 82 klx, respectively). When the three BCAA were omitted, the levels of transcription became similar for the two fusions (62 versus 82 klx, respectively). This confirms that the activity of P3 is reduced under partial or full BCAA starvation and that the transcriptional level from the two upstream promoters is high. The fact that ALDC production was concomitant with the transcription from P3 but not from P1 and P2 suggests that, although they are abundant during BCAA starvation, the messengers originating from P1 and P2 do not yield a significant amount of this enzyme.
Inhibition of aldB translation.
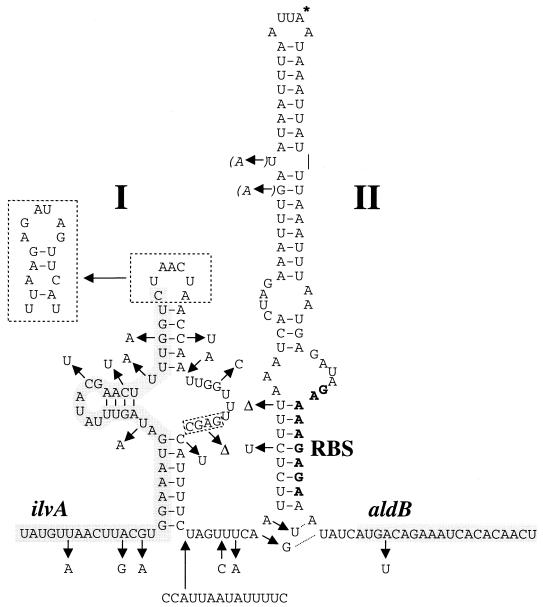
The fact that ALDC is not produced when P1 and P2 messengers are abundant suggests that the production of this protein is inhibited at a posttranscriptional level. The possible messenger folding in the region of the aldB translational start was previously described and was proposed to modulate the translation of ALDC (40). Figure 4 shows a modification of the previously proposed folding, in order to take into account the sequence variability found in other L. lactis strains. Indeed, in closely related strains the important regulatory features, such as DNA motifs and secondary structures, are usually conserved, whereas variations occur at higher frequency in the other noncoding regions (9). In the present case, the previously proposed cloverleaf-like structure is not conserved, since a 10-bp insertion and several mutations that destabilize it were found in L. lactis subsp. cremoris strains. However, this region can produce a secondary structure named stem I in all cases. The end of the coding frame of ilvA, in which the 12 last codons code for four leucine residues and one valine residue, terminates in the top bulge of stem I (Fig. 1C and 4). A second structure (stem II), with a free energy of −12.4 kcal/mol, entraps the RBS and is almost invariable in all L. lactis strains, since only three changes were found in the paired region. None of these changes significantly modifies the stability of the structure, since two are compensatory changes (G-C to A-U) and the third results in a G-U pairing instead of G-C (Fig. 4).
FIG. 4.
Potential secondary structure present upstream of aldB in L. lactis subsp. lactis NCDO2118 and L. lactis subsp. cremoris MG1363. The structure presented is that of NCDO2118, and the changes found for MG1363 are indicated by arrows. Δ indicates deleted bases; arrows directed toward the sequence indicate insertions. The bulge of stem I (boxed by a dashed line) and the spacer between stems I and II are larger in MG1363. Two additional potential pairings at the bottom of stem II for MG1363 are indicated by dashed lines. The two base changes introduced to inactivate P3 are indicated in italics and are placed in parentheses (on the left of stem II). The base corresponding to the transcriptional start of P3 is marked with an asterisk at the top of stem II. The sequences belonging to the ilvA and aldB ORFs are shaded. The sequence of the aldB RBS is in boldface.
Several fragments of this region were subcloned in pJIM1715, a vector allowing measurement of the efficiency of a translational signal cloned upstream of luxAB (39). The DNA fragment used as starting material for these constructions contains two changes that inactivate P3 (see above) (Table 2), since the activity of this promoter would interfere with the intended experiments. The two changes should not alter the stability of stem II significantly, since the first one is a replacement of a G-U pairing by A-U and the second is in a bulge (Fig. 4). The fragments end with an NdeI restriction site introduced at the start codon of aldB that allow production of a translational fusion with the luxAB gene present in the vector pJIM1715 (Fig. 1B and C). The 53-bp fragment, carried by pJIM1730, containing the RBS of ALDC without the complementary region of stem II, showed a strong luciferase activity (Table 3), while the 113-bp fragment present in pJIM1732, containing the entire stem II, displayed 22- to 125-fold-lower luciferase activity. This indicates that the translation of the aldB::lux fusion was probably by the formation of stem II. pJIM1735, pJIM1739, and pJIM1740 carry a 215-bp fragment containing stems I and II. The last two plasmids carry additional inserts of 400 and 404 bp, allowing translation of peptides out of and in frame of ilvA, respectively (Fig. 1C). The 215-bp fragment reduced the translation of luxAB more than fivefold compared to pJIM1732. Interestingly, the translation of the end of ilvA in pJIM1740, but not that of the frameshifted open reading frame (ORF) in pJIM1739, reduced luciferase activity to the level of the background noise (Table 3).
TABLE 3.
Analysis of translation of aldB::lux fusions
| Plasmid | Insert | M17
|
CDM with BCAA
|
CDM without BCAA
|
|||
|---|---|---|---|---|---|---|---|
| Lux activitya | Inhibition factorb | Lux activity | Inhibition factor | Lux activity | Inhibition factor | ||
| pJIM1730 | RBS | 200 | 1 | 250 | 1 | 130 | 1 |
| pJIM1732 | Stem II (anti-RBS/RBS) | 9.3 | 22 | 2.0 | 125 | 1.9 | 68 |
| pJIM1735 | Stems I and II (tRNA-like/anti-RBS/RBS) | 1.6 | 125 | 0.49 | 510 | 0.53 | 245 |
| pJIM1739 | Stems I and II, ′ORF(+1) translated | 1.2 | 167 | 0.60 | 417 | 0.31 | 419 |
| pJIM1740 | Stems I and II, ′ilvA translated | 0.45 | 444 | 0.10c | >2,500 | 0.11c | >1,200 |
In kilolux per OD unit.
Ratio of luciferase activity to that obtained with pJIM1730.
Value at the threshold level.
The effect of the availability of BCAA in the medium on the rate of translation of different constructs was tested by growing the cells in M17 or in CDM with and without BCAA (Table 3). The Lux activity measured in M17 was three- to fourfold higher than that in CDM, except for the strain carrying the control plasmid pJIM1730. The differences observed in CDM with or without BCAA with any of the plasmids were not significant. In the case of pJIM1740, which has all the features of the gene in its chromosomal context, the luciferase value in M17 was measurable and more than fourfold higher than the threshold of detection reached in CDM.
DISCUSSION
The dual role of ALDC in L. lactis, both in the catabolism of pyruvate and in the biosynthesis of BCAA, implies a tight control of its synthesis in order to avoid an excess of its activity during BCAA biosynthesis but ensure sufficient activity for acetoin production (19). The present study identifies different elements involved in the regulation of ALDC synthesis.
Translational control of aldB transcribed from leu-ilv promoters.
The transcripts initiated at P1 and P2 in the absence of isoleucine and leucine terminate downstream of the ald genes. However, no detectable ALDC is produced from these transcripts, as shown by Western blot analysis. This observation suggests that ALDC synthesis is subject to a posttranscriptional control. The activity of a luciferase reporter gene under the control of the aldB RBS decreases up to more than 2,500-fold in the presence of upstream sequences. Messengers containing these sequences might fold into secondary structures that entrap the RBS and thus inhibit the translation of aldB (Fig. 5). Folding of messengers in the RBS region is already known to inhibit translation, for example, in the bacteriophage MS2 coat genes and several other E. coli genes (10, 11). The rate of synthesis of these proteins might be related to the fraction of mRNA molecules in which the RBS is unfolded and would thus depend on the free energy of the local secondary structures. Unexpectedly, the 100-fold inhibition due to stem II is much lower than the 30,000-fold inhibition calculated with the model proposed by de Smit and van Duin (10, 11). The difference between the calculated and the experimental values might be due to (i) a difference in the effect of secondary structures on RBS efficiency particular to L. lactis or (ii) factors interfering with the folding of stem II.
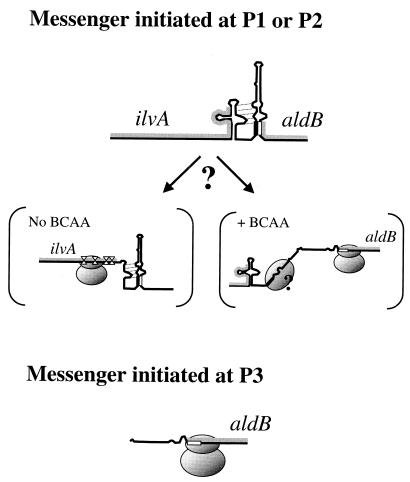
FIG. 5.
Model for the regulation of translation of aldB. The translation of the messengers initiated at P1 and P2 is inhibited by the formation of stem II. The strength of translation inhibition is enhanced by the presence of stem I and the translation of ilvA (upper panel). Messenger initiated at P3 lacks the RBS-entrapping structure, and the ribosome can always load (lower panel). The RBS is represented by an open box, and its complementary sequence in stem II is represented by a filled box; the gray lines represent the ilvA and aldB ORFs on the messengers. In brackets are presented two hypotheses to explain the enhanced inhibition due to stem I and the translation of ilvA. Under BCAA limitation, the ribosome could stall at the leucine and valine codons located at the end of ilvA (diamonds). This would allow the stabilization of stem II, which entraps the aldB RBS, possibly by the formation of a pseudoknot. In the presence of BCAA, a possible activation of aldB translation could result from the destabilization of the structure made by stem II, either by the formation of stem I or by the binding of a factor facilitating the loading of a ribosome. From our experimental data, ilvA translation would increase the stability of stem II more than 20-fold. In the presence of BCAA, the destabilization of stem II would be incomplete and would still lead to a significant decrease of translation efficiency of aldB compared to a messenger initiated at P3.
The first hypothesis is not in agreement with data calculated from previous work on a possible contribution of mRNA secondary structure to translation initiation efficiency in L. lactis (48). In that work, the translation of in-frame lacZ fusions under the control of RBSs potentially trapped in secondary structures was inhibited at a level similar to the theoretical values. The difference between the calculated and the observed values found in the present study might thus be due to cellular factors or flanking sequences that would interfere with the folding of stem II. Factors such as proteins interacting with the mRNA and interfering with the folding around the RBS have been reviewed by McCarthy and Gualerzi (33) and Voorma (49). Interestingly, the presence of stem I caused a significant decrease in the translation of aldB, suggesting that stems I and II might interact.
The stronger translational inhibition of aldB by stem II in the presence of stem I is reminiscent of the translational regulation of the E. coli S10 operon (13). In this operon, a secondary structure is necessary to stabilize the stem-loop that sequesters the RBS of S10. Interestingly, the translation of ilvA, ending in structure I, increased the translation inhibition. This effect is specific to the translation of ilvA, since the translation of the +1-shifted ORF yields no significant effect. We suggest that the additional inhibition might be due to a particular feature of the ilvA coding frame. Indeed, the very end of ilvA encodes a high number of leucine residues, a feature that may induce ribosome stalling during BCAA starvation. Translational regulation by stalling ribosomes that change the secondary structure in the vicinity of RBS has already been described (28, 35). Moreover, the effect of translation of specific codons, including leucine codons, on the folding of mRNA has been extensively documented in the context of the study of transcription attenuation (7, 25). We assume that the rate of translation of ilvA, including possible stalling of the ribosomes at the end of this gene, could modulate the folding of structure I (Fig. 5). The very strong inhibition of translation of the lux reporter gene in CDM did not allow us to confirm this hypothesis. Moreover, the mechanism leading to this regulation might be different from those, already described, that are dependent on a long duration of ribosome stalling during the translation of a specialized peptide. Lastly, we can consider two models to account for the postulated activation of ALDC synthesis by BCAA (Fig. 5): (i) part of stem I could stabilize stem II when ribosomes stall in ilvA, and (ii) stem II could be partially destabilized by an additional factor when stem I is formed or ribosomes are released. The presence of this potential factor would explain the difference between the experimental and theoretical values for the stem II-dependent translational inhibition.
ALDC synthesis from its specific promoter, P3.
The aldB information is carried not only on mRNA initiated at P1 and P2 but also on shorter messengers that start at a position corresponding to the loop of stem II (19). We confirm here the earlier hypothesis proposing that the short messengers are produced from P3, a specific promoter, rather than by the processing of the longer messenger, since (i) a 150-bp fragment containing the potential promoter and the start of transcription allowed full transcriptional activity, (ii) directed mutagenesis of P3 promoter −10 extended consensus bases inactivates the transcriptional activity of this fragment, and (iii) P1 and P2 transcripts are repressed in the presence of BCAA, whereas the intensity of the messengers is maximal in rich medium. P3 does not have a −35 consensus box but instead contains an extended TGN consensus upstream of the −10 box. A single substitution, TGATAATAT to TATATAAT, inactivates P3 to the same extent as the replacement of the first T of the conventional −10 box. The TG motif is recognized by a region of the RNA polymerase ς70 subunit conserved in all bacteria (2). Interestingly, the effect of the G-to-A mutation is much stronger in L. lactis than in E. coli, since this change decreases the activity of P3 200-fold, versus 20- and 50-fold for PRE and galP1, respectively (23). This might reflect the greater importance of this motif, which is very frequent in gram-positive bacteria such as B. subtilis (20), Streptococcus (42), and Lactococcus (12). The complete absence of a −35 box does not impede P3 activity, the level of transcription of which is comparable to those of promoters from several highly expressed glycolytic genes (E. Jamet and P. Renault, data not shown).
Interestingly, the translation of aldB on the P3 messengers is not inhibited by RNA folding, since they start after sequence complementary to the RBS, present in stem II. Initiation of transcription from promoters located downstream of sequences that inhibit translation was shown in enterobacteria for pyrC and pyrD (26, 45), the T4 lysozyme gene (17), and the tnp gene from IS10 (29). The formation of the hairpin in the extended 5′ transcript of galE was interpreted as a biological precaution against the activation of a gene by transcription from unrelated promoters. The presence of a strong promoter in front of aldB could allow the production of large amounts of ALDC, independently of the need of the cell for acetolactate. However, P3 activity sharply decreases during BCAA starvation. Interestingly, P3 activity is also reduced during starvation for other amino acids, such as methionine, and during adaptation phases after change in the carbon source. These observations suggest that P3 is controlled by a general response, such as the stringent control. In E. coli, this global response to starvation is mediated by the accumulation of ppGpp (8). In streptococci, ppGpp was found to accumulate under conditions of amino acid starvation and glucose exhaustion (34). Since a homologue of spoT, the gene responsible for ppGpp synthesis in gram-positive bacteria, is also present in lactococci (38), we propose that P3 is controlled by a similar regulation.
Conclusion.
In this work, we show that ALDC synthesis is inhibited at the translational level when it is transcribed from P1 and P2, the two promoters induced during BCAA starvation. However, P3, a third promoter, allows the synthesis of ALDC independent of this translational control. Surprisingly, the transcription of aldB from this promoter is not strongly activated when pyruvate catabolism is shifted to the acetoin-butanediol pathway as in other bacteria, such as B. subtilis, Klebsiella terrigena, and Oenococcus oeni (5, 14, 41). The transcription from P3 is reduced during lag phases, probably under the control of the stringent response and is active almost constitutively during exponential growth phases. Lastly, ALDC activity is further regulated at a posttranslational level: (i) its enzymatic activity requires the presence of leucine in the cell (19, 36), and (ii) ALDC is rapidly degraded when cells are shifted to BCAA starvation. The multiple controls for the synthesis of ALDC should allow a rapid adjustment of the amount of this enzyme, which is required at different levels in response to the environmental conditions. Indeed, during BCAA synthesis, the pool of AL could be controlled right away by ALDC, to counterbalance the absence of IlvBN retroinhibition by the products of the BCAA pathway (3, 19). During a shift to heterofermentation, the acetoin pathway will be activated without delay, since the expression of AlsS, the catabolic acetolactate synthase and the first enzyme of this pathway, is constitutive (31).
ACKNOWLEDGMENT
This work was partially financed by contract BIO2-CT94-3055 of the European Union.
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barne K A, Bown J A, Busby S J, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson K H, Godon J J, Renault P, Griffin H G, Gasson M J. Effect of ilvBN-encoded alpha-acetolactate synthase expression on diacetyl production in Lactococcus lactis. Appl Microbiol Biotechnol. 1996;45:107–111. [Google Scholar]
- 4.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blomqvist K, Nikkola M, Lehtovaara P, Suihko M L, Airaksinen U, Straby K B, Knowles J K C, Penttila M E. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes. J Bacteriol. 1993;175:1392–1404. doi: 10.1128/jb.175.5.1392-1404.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo J M. Leucine biosynthesis in prokaryotes. In: Herrman K M, Somerville R L, editors. Amino-acid biosynthesis and regulation. Reading, Mass: Addison-Wesley; 1983. pp. 267–284. [Google Scholar]
- 7.Carter P W, Barktus J M, Calvo J M. Transcription attenuation in Salmonella typhimurium: the significance of rare leucine codons in the leu leader. Proc Natl Acad Sci USA. 1986;83:8127–8131. doi: 10.1073/pnas.83.21.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 9.Delorme C, Ehrlich S D, Renault P. Regulation of expression of the Lactococcus lactis histidine operon. J Bacteriol. 1999;181:2026–2037. doi: 10.1128/jb.181.7.2026-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Smit M H, van Duin J. Control of translation by mRNA secondary structure in Escherichia coli. A quantitative analysis of literature data. J Mol Biol. 1994;244:144–150. doi: 10.1006/jmbi.1994.1714. [DOI] [PubMed] [Google Scholar]
- 11.de Smit M H, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci USA. 1990;87:7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vos W M. Gene cloning and expression in lactic acid bacteria. FEMS Microbiol Rev. 1987;46:281–295. [Google Scholar]
- 13.Freedman L P, Zengel J M, Archer R H, Lindahl L. Autogenous control of the S10 ribosomal protein operon of Escherichia coli: genetic dissection of transcriptional and posttranscriptional regulation. Proc Natl Acad Sci USA. 1987;84:6516–6520. doi: 10.1073/pnas.84.18.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garmyn D, Monnet C, Martineau B, Guzzo J, Cavin J F, Divies C. Cloning and sequencing of the gene encoding alpha-acetolactate decarboxylase from Leuconostoc oenos. FEMS Microbiol Lett. 1996;145:445–450. doi: 10.1111/j.1574-6968.1996.tb08614.x. [DOI] [PubMed] [Google Scholar]
- 15.Gilson T J. Ph.D. thesis. Cambridge, England: University of Cambridge; 1984. [Google Scholar]
- 16.Godon J J, Chopin M C, Ehrlich S D. Branched-chain amino acid biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol. 1992;174:6580–6589. doi: 10.1128/jb.174.20.6580-6589.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gott J M, Zeeh A, Bell-Pedersen D, Ehrenman K, Belfort M, Shub D A. Genes within genes: independent expression of phage T4 intron open reading frames and the genes in which they reside. Genes Dev. 1988;12B:1791–1799. doi: 10.1101/gad.2.12b.1791. [DOI] [PubMed] [Google Scholar]
- 18.Goupil N, Corthier G, Ehrlich S D, Renault P. Imbalance of leucine flux in Lactococcus lactis and its use for the isolation of diacetyl-overproducing strains. Appl Environ Microbiol. 1996;62:2636–2640. doi: 10.1128/aem.62.7.2636-2640.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goupil-Feuillerat N, Godon J-J, Cocaign-Bousquet M, Ehrlich S D, Renault P. Dual role of alpha-acetolactate decarboxylase in Lactococcus lactis subsp. lactis. J Bacteriol. 1997;179:6585–6593. doi: 10.1128/jb.179.20.6285-6293.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helmann J D. Compilation and analysis of Bacillus subtilis sigma(A)-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holo H, Nes I F. High-frequency transformation by electroporation of Lactococcus lactis subsp.cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol. 1989;55:3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohlhaw G B. The leucine biosynthetic pathway in yeast: compartmentation, enzyme regulation, gene expression. In: Barak Z, Chipman D M, Schloss J V, editors. Biosynthesis of branched chain amino acids. Weinheim, Germany: VCH; 1990. pp. 33–42. [Google Scholar]
- 23.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Landick R, Turnbough C L, Yanovski C. Transcription attenuation. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 26.Liu J, Turnbough C L., Jr Identification of the Shine-Dalgarno sequence required for expression and translational control of the pyrC gene in Escherichia coli K-12. J Bacteriol. 1994;176:2513–2516. doi: 10.1128/jb.176.9.2513-2516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loureiro Dos Santos A L, Chopin A. Shotgun cloning in Streptococcus lactis. FEMS Microbiol Lett. 1987;42:209–212. [Google Scholar]
- 28.Lovett P S, Rogers E J. Ribosome regulation by the nascent peptide. Microbiol Rev. 1996;60:366. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma C K, Kolesnikow T, Rayner J C, Simons E L, Yim H, Simons R W. Control of translation by mRNA secondary structure: the importance of the kinetics of structure formation. Mol Microbiol. 1994;14:1033–1047. doi: 10.1111/j.1365-2958.1994.tb01337.x. [DOI] [PubMed] [Google Scholar]
- 30.Maguin E, Duwat P, Hege T, Ehrlich S D, Gruss A. New thermosensitive plasmid for gram-positive bacteria. J Bacteriol. 1992;174:5633–5638. doi: 10.1128/jb.174.17.5633-5638.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marugg J D, Goelling D, Stahl U, Ledeboer A M, Toonen M Y, Verhue W M, Verrips C T. Identification and characterization of the alpha-acetolactate synthase gene from Lactococcus lactis subsp. lactis biovar diacetylactis. Appl Environ Microbiol. 1994;60:1390–1394. doi: 10.1128/aem.60.4.1390-1394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayer D, Schlensog V, Bock A. Identification of the transcriptional activator controlling the butanediol fermentation pathway in Klebsiella terrigena. J Bacteriol. 1995;177:5261–5269. doi: 10.1128/jb.177.18.5261-5269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy J E G, Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990;6:78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- 34.Mechold U, Malke H. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J Bacteriol. 1997;179:2658–2667. doi: 10.1128/jb.179.8.2658-2667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narayanan C S, Dubnau D. Demonstration of erythromycin-dependent stalling of ribosomes on the ermC leader transcript. J Biol Chem. 1987;262:1766–1771. [PubMed] [Google Scholar]
- 36.Phalip V, Monnet C, Schmitt P, Renault P, Godon J J, Divies C. Purification and properties of the alpha-acetolactate decarboxylase from Lactococcus lactis subsp. lactis NCDO 2118. FEBS Lett. 1994;351:95–99. doi: 10.1016/0014-5793(94)00820-5. [DOI] [PubMed] [Google Scholar]
- 37.Poolman B, Konings W N. Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol. 1988;170:700–707. doi: 10.1128/jb.170.2.700-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rallu F, Gruss A, Ehrlich S D, Maguin E. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol Microbiol. 2000;35:517–528. doi: 10.1046/j.1365-2958.2000.01711.x. [DOI] [PubMed] [Google Scholar]
- 39.Renault P, Corthier G, Goupil N, Delorme C, Ehrlich S D. Plasmid vectors for Gram-positive bacteria switching from high to low copy number. Gene. 1996;183:175–182. doi: 10.1016/s0378-1119(96)00554-9. [DOI] [PubMed] [Google Scholar]
- 40.Renault P, Godon J-J, Goupil N, Delorme C, Corthier G, Ehrlich S D. Metabolic operons in lactococci. Dev Biol Stand. 1995;85:431–441. [PubMed] [Google Scholar]
- 41.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Simon D, Chopin A. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie. 1988;70:559–566. doi: 10.1016/0300-9084(88)90093-4. [DOI] [PubMed] [Google Scholar]
- 45.Sorensen K I. Conformational heterogeneity in the Salmonella typhimurium pyrC and pyrD leader mRNAs produced in vivo. Nucleic Acids Res. 1994;22:625–631. doi: 10.1093/nar/22.4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terzaghi B, Sandine W E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Umbarger H E. Biosynthesis of the branched-chain amino acids. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 353–367. [Google Scholar]
- 48.Vandeguchte M, Vanderlende T, Kok J, Venema G. A possible contribution of mRNA secondary structure to translation initiation efficiency in Lactococcus lactis. FEMS Microbiol Lett. 1991;81:201–208. doi: 10.1111/j.1574-6968.1991.tb04746.x. [DOI] [PubMed] [Google Scholar]
- 49.Voorma H O. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 759–777. [Google Scholar]