Abstract
Objective:
To explore the therapeutic mechanism of Mori Cortex against osteosarcoma (OS), we conducted bioinformatics prediction followed by in vitro experimental validation.
Methods:
Gene expression data from normal and OS tissues were obtained from the GEO database and underwent differential analysis. Active Mori Cortex components and target genes were extracted from the Traditional Chinese Medicine System Pharmacology database. By intersecting these targets with differentially expressed genes in OS, we identified potential drug action targets. Using the STRING database, a protein-protein interaction network was constructed. Subsequent analyses of these intersected genes, including Gene Ontology enrichment and Kyoto Encyclopedia of Genes and Genomes pathway enrichment, were performed using R software to elucidate biological processes, molecular functions, and cellular components, resulting in the simulation of signaling pathways. Molecular docking assessed the binding capacity of small molecules to signaling pathway targets. In vitro validations were conducted on U-2 OS cells. The CCK8 assay was used to determine drug-induced cytotoxicity in OS cells, and Western Blotting was employed to validate the expression of AKT, extracellular signal-regulated kinases (ERK), Survivin, and Cyclin D1 proteins.
Results:
Through differential gene expression analysis between normal and OS tissues, we identified 12,364 differentially expressed genes. From the TCSMP database, 39 active components and 185 therapeutic targets related to OS were derived. The protein-protein interaction network indicated that AKT1, IL-6, JUN, VEGFA, and CASP3 might be central targets of Mori Cortex for OS. Molecular docking revealed that the active compound Morusin in Mori Cortex exhibits strong binding affinity to AKT and ERK. The CCK8 assay showed that Morusin significantly inhibits the viability of U-2 OS cells. Western Blot demonstrated a reduction in the p-AKT/AKT ratio, the p-ERK/ERK ratio, Survivin, and Cyclin D1.
Conclusion:
Mori Cortex may exert its therapeutic effects on OS through multiple cellular signaling pathways. Morusin, the active component of Mori Cortex, can inhibit cell cycle regulation and promote cell death in OS cells by targeting AKT/ERK pathway.
Keywords: bioinformatics, in vitro experiment, Mori Cortex, Morusin, osteosarcoma
1. Introduction
Osteosarcoma (OS) is the most prevalent primary malignant bone tumor among children and adolescents.[1] Typically, it originates in the long bones, such as the femur, tibia, and humerus.[2] Due to its high proliferative and invasive nature, the bone tissues and nearby structures are rapidly damaged, while also metastasizing to other areas.[3] Currently, the clinical treatment for OS primarily involves chemotherapy and radical surgical resection.[4] However, the long-term survival rate for patients diagnosed with metastatic or recurrent OS has been only 25% to 30% over the past decades.[5] Most patients are young, and the side effects of chemotherapy or surgery can have a profound impact on their quality of life.[6] Thus, developing more effective and less toxic treatments for young OS patients is urgently needed.
With the exploration of traditional Chinese medicine (TCM), increasing evidence suggests that Chinese herbs have beneficial therapeutic effects on cancer patients.[7] Therefore, Chinese medicine treatments as complementary or alternative therapies have been widely accepted.[8] Mori Cortex is derived from the white inner bark of young mulberry roots, and possesses antiinflammatory, antioxidative stress, and immune-boosting properties.[9–11] Morusin is a typical prenylated flavonoid compound found in Mori Cortex.[12] Previous studies have reported that Morusin exerts anticancer activity against melanoma by activating the MAPK/STAT3 signaling pathway.[13] Furthermore, Morusin can induce apoptosis in lung cancer cells by increasing intracellular ROS levels and inhibiting the PI3K/AKT pathway.[14] Hence, the potential anticancer activity of Morusin in OS warrants investigation.
Network pharmacology in bioinformatics is an emerging discipline.[15] It integrates techniques and content from fields such as pharmacology, systems biology, and computational biology, reflecting its systemic and holistic characteristics.[16] Through the methods of network pharmacology, we constructed a multi-level “drug-ingredient-target-disease” interaction network to explore the specific mechanisms of TCM’s regulatory effects on the body and analyze the connections between drugs and diseases.[17] This approach bridges TCM with modern medicine.[18] This study utilized network pharmacology and in vitro experiments to explore and preliminarily validate the potential mechanism of Mori Cortex in treating OS. We provided a solid theoretical foundation for further in-depth research into the pharmacological mechanisms of Mori Cortex in treating OS.
2. Methods
2.1. Screening of active components of Mori Cortex with protein targets of OS
Gene expression profiles for normal and OS tissues were extracted from the GEO database (https://www.ncbi.nlm.nih.gov/gds/). The “limma” and “pheatmap” packages in R software were used for differential gene expression analysis to identify OS protein targets. The Traditional Chinese Medicine System Pharmacology (TCMSP) database (http://tcmspw.com/tcmsp.php) was utilized to search for the chemical components of Mori Cortex. Criteria were set for oral bioavailability ≥30% and drug likeness ≥0.18 to ascertain the active ingredients of Mori Cortex. Subsequently, targets corresponding to these active ingredients were identified. An intersection of the targets from these active ingredients and the differential genes in OS was determined. Using Cytoscape software, a visualization of the “drug-active ingredient-target protein-disease” relationship was constructed.
2.2. Construct a protein-protein interaction network
Construction of protein-protein interaction (PPI) networks is crucial for understanding cellular physiology in both normal and disease states. In this study, the STRING database (http://string-db.org/) was used to conduct PPI network analysis on the intersected gene set, with the species limited to “Homo sapiens” and a confidence score >0.4. We imported the data obtained from the STRING database into Cytoscape 3.6.0 and subsequently analyzed the network’s degree, betweenness centrality, average shortest path, and closeness centrality using the NetworkAnalyzer plugin. Then, we sorted the genes based on their degree values and selected the top 20 genes with scores above the average as core markers.
2.3. Enrichment analysis
Gene Ontology (GO) enrichment analysis on the common targets between Mori Cortex and OS was performed utilizing the “enrichplot” and “ggplot2” packages in R, focusing biological processes, molecular functions, and cellular components. We filtered the enriched pathways based on an adjusted P ≤ .05. The top 5 gene enrichments for biological processes, molecular functions, and cellular components were listed. Similarly, using R software, we performed Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on the common targets of Mori Cortex and OS and annotated the pathways involved for each gene to further investigate the association between these pathways and the disease.
2.4. Molecular docking
To ensure molecular docking accuracy, a systematic protocol including ligand and receptor processing, parameter optimization and control measures was adopted: Ligand processing: The 3D structure of the intended docking target was procured from the PubChem database in mol2 format. Using AutodockTools 1.5.6, the ligand molecule underwent hydrogenation, charging, ligand root identification, rotatable bond specification, and was eventually saved in pdbqt format. Receptor processing: The core 3D structure of the target protein was sourced from the RCSB protein database (www.rcsb.org/). Upon importing into AutodockTools 1.5.6, hydrogen atoms were added, Gasteiger charge was computed, nonpolar hydrogens were bound, and the entity was designated as a receptor before saving as a pdbqt file. Docking parameters: The coordinates and box size essential for Vina molecular docking were determined. Exhaustiveness was set at 15, while default values were retained for the remaining parameters. Control and output: Leveraging Autodockvina 1.1.2, semiflexible docking was conducted. The conformation exhibiting the highest affinity was selected as the ultimate docking conformation.
2.5. Cell culture
Before use, the U-2 OS cells were cryopreserved in liquid nitrogen in a mixture of 10% DMSO and fetal bovine serum. After thawing, U-2 OS cells containing 10% FBS, 2 mM l-glutamine, 100 units/mL of penicillin, and 100 μL Cultured in 90% McCoy’s 5A medium with g/mL streptomycin. These cells were seeded and cultured using the previous method in a 75 cm2 tissue culture flask at a temperature of 37°C in a moist 5% CO2 environment.
2.6. CCK8
We seeded 1 × 104 U-2 OS cells in each well of a 96-well plate. After incubating them in culture media containing 0, 5, 10, 15, and 20 μM of Mori Cortex for 48 hours, the cell viability was assessed using the CCK8 assay. Firstly, the treated cells were mixed with the CCK8 reagent according to the recommended ratio. They were then incubated at 37°C for a specific duration. Finally, the absorbance was measured at a designated wavelength using a microplate reader to determine the cell viability.
2.7. Western Blot
For protein extraction, U-2 OS cells were divided into 2 groups: the control group without added Mori Cortex and the experimental group with Mori Cortex (20 μmol/L). After 48 hours, the culture medium was removed, the cells were then washed 3 times with PBS. Subsequently, a precooled protein lysis buffer containing 10% phosphatase inhibitor was added to the cells. The cells were then scraped off using a cell scraper and collected into a 1.5 mL EP tube. The tube was placed on ice, and the cells were lysed for 20 minutes. After lysis, an appropriate amount of loading buffer was added, and the sample was heated at 95°C for 10 minutes and stored for future use. Proteins were separated by 10% polyacrylamide gel electrophoresis and, when the bromophenol blue was near the bottom of the gel, transferred onto a PVDF membrane (Millipore, Billerica, MA). The membrane was then blocked with 10% skim milk at room temperature for 30 minutes, followed by overnight incubation with the primary antibody at 4°C. The next day, the membrane was washed 3 times with TBST buffer and incubated with a goat antirabbit or antimouse secondary antibody labeled with horseradish peroxidase at 37°C for 1 hour.
3. Results
3.1. Obtaining targets for Mori Cortex therapy in OS
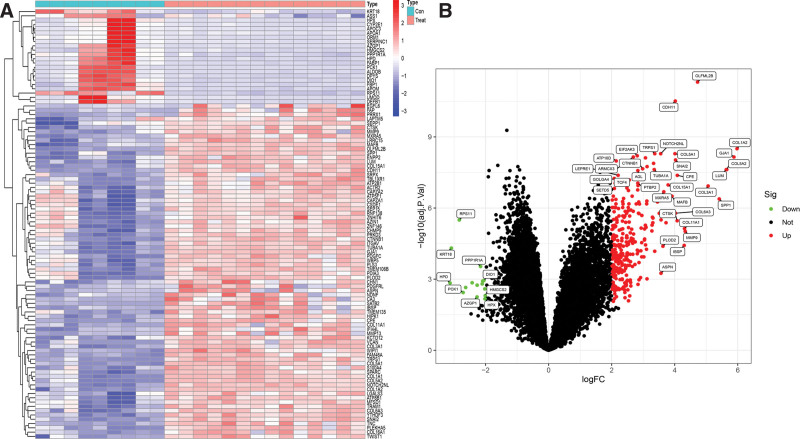
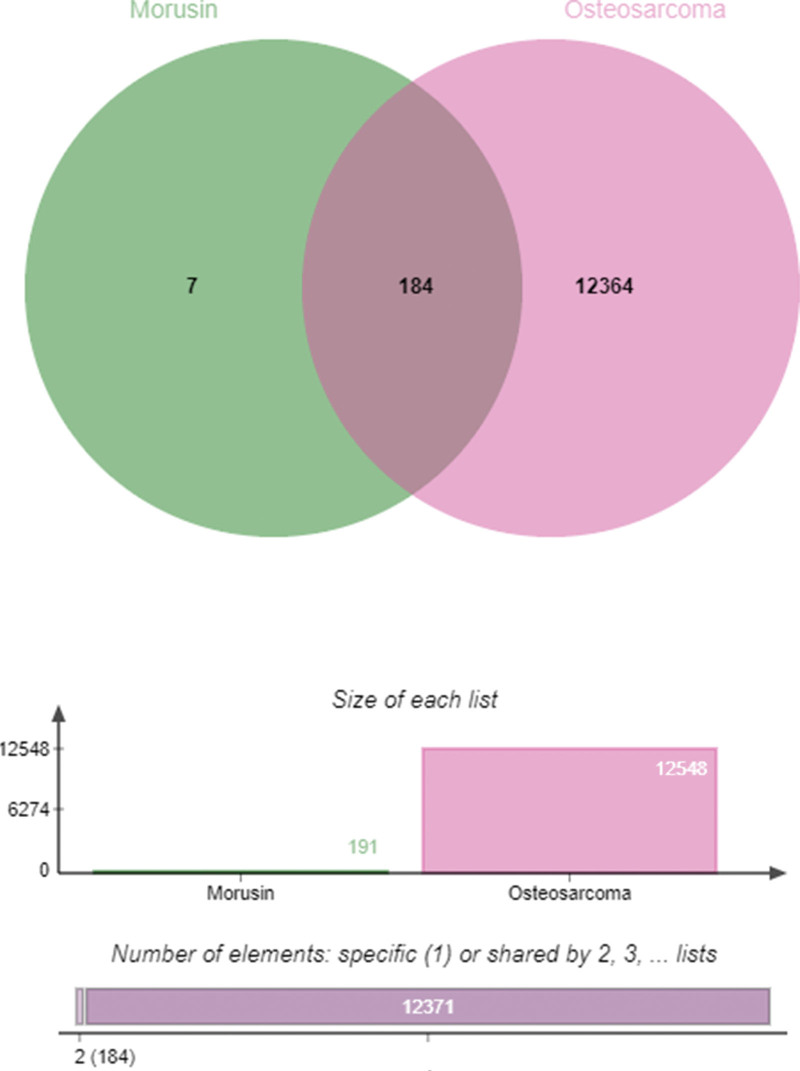
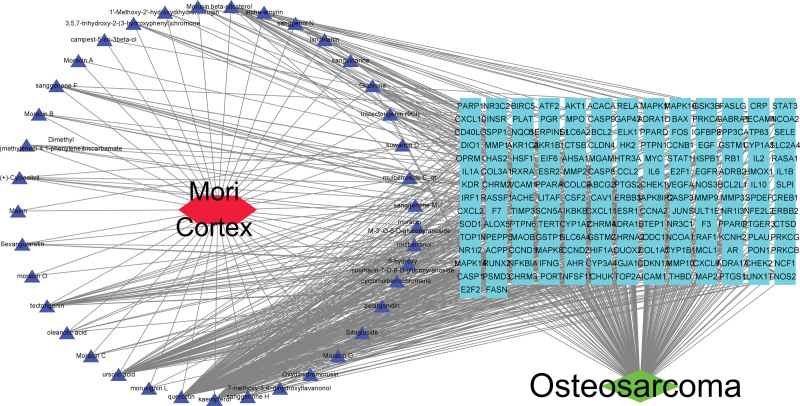
We retrieved gene expression data for normal and OS tissues (GSE16088) from the GEO database. Using R software, we identified 12,364 genes that were differentially expressed between normal and OS tissues, considering them as therapeutic targets for OS (Fig. 1). Utilizing the TCMSP database with criteria set at drug likeness ≥18% and oral bioavailability ≥30%, we pinpointed 40 effective components and 191 targets of Mori Cortex. Subsequently, by intersecting these drug targets with the differentially expressed genes from the tissues, we identified 187 common genes. Venny 2.1 software was utilized to illustrate a Venn diagram (Fig. 2), and Cytoscape 3.7.0 software was employed to map out the Mori Cortex drug-OS target network (Fig. 3).
Figure 1.
Gene expression analysis of normal and OS tissues. (A) Heatmap and (B) volcano plot. OS = osteosarcoma.
Figure 2.
Venn diagram of Mori Cortex and OS. OS = osteosarcoma.
Figure 3.
Network of active ingredients and targets of Mori Cortex.
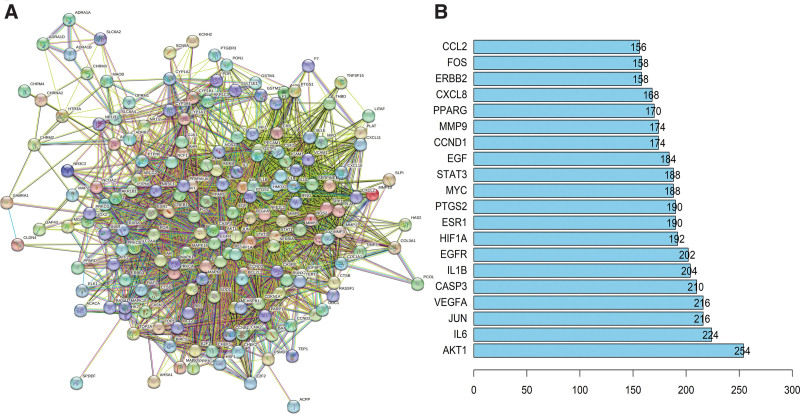
3.2. Protein-protein interaction network and core genes
In this comprehensive study, the potential common target genes of Mori Cortex and OS were imported into the STRING database for analysis. To ensure the data accuracy, the species was specifically set to humans, and default settings were applied for other related parameters. Through this process, we successfully constructed a detailed PPI network diagram, which is presented in Figure 4A. To further refine our analysis, we employed the Cytoscape 3.6.0 software to conduct an exhaustive topological structure analysis of this PPI network. Based on the degree centrality of each gene, we specifically selected genes with scores exceeding 75, considering them as key targets in this study. Subsequently, we listed the top 20 most critical target genes and showcased them in Figure 4B. Notably, the genes AKT1, IL-6, and JUN exhibited outstanding performance in terms of degree centrality, ranking as the top 3 respectively.
Figure 4.
Constructing PPI network and selecting core genes. (A) PPI network diagram of Mori Cortex-OS common targets. (B) Twenty key targets selected by PPI topology analysis based on degree. OS = osteosarcoma, PPI = protein-protein interaction.
3.3. Enrichment analysis
3.3.1. GO enrichment analysis
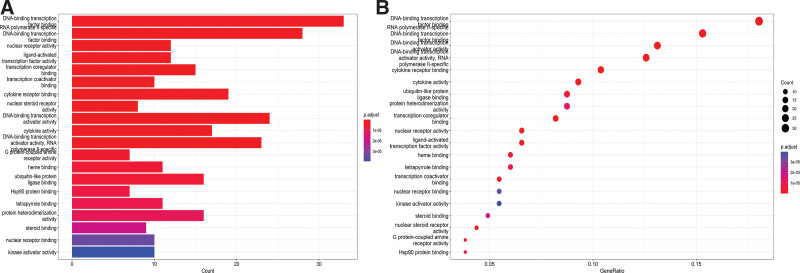
By using the GO database to conduct enrichment analysis on the intersecting genes between Mori Cortex and OS, a total of 169 GO annotations were obtained. Based on the Count value, we found that the core biological processes of Mori Cortex treating OS are mainly related to DNA-binding transcription factor binding, RNA polymerase II-specific DNA-binding transcription factor binding, DNA-binding transcription activator activity, DNA-binding transcription activator activity specific to RNA polymerase II, cytokine receptor binding, receptor ligand activity, cytokine activity, ubiquitin-like protein ligase binding and protein heterodimerization activity (Fig. 5).
Figure 5.
GO enrichment analysis. (A) Barplot and (B) Dotplot. GO = Gene Ontology.
3.3.2. KEGG enrichment analysis
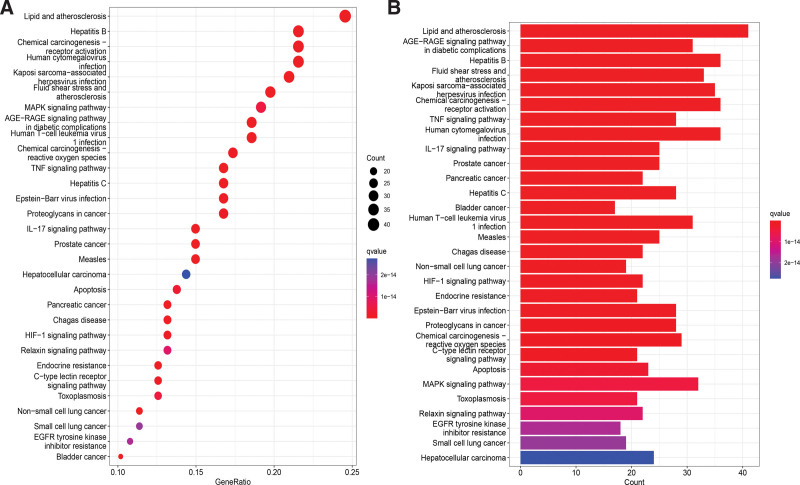
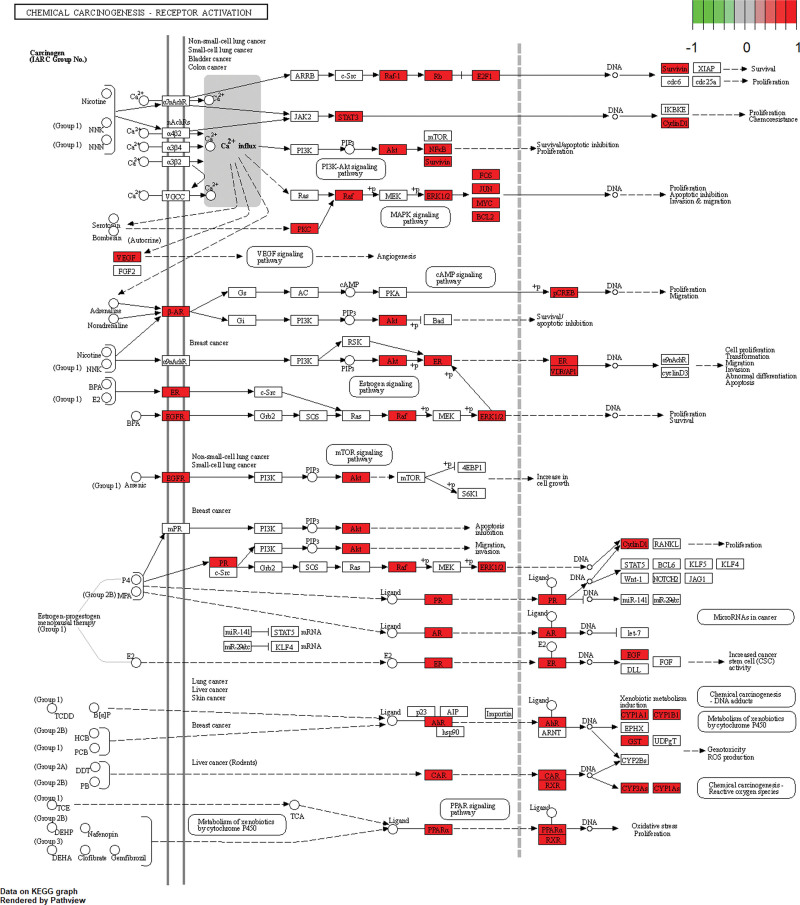
KEGG enrichment identified 183 pathways. Based on the P value, the top 30 KEGG annotations were filtered. The primary pathways involved include Lipid and atherosclerosis, AGE-RAGE signaling pathway in diabetic complications, Hepatitis B, Fluid shear stress and atherosclerosis, Kaposi sarcoma-associated herpesvirus infection, Chemical carcinogenesis - receptor activation, TNF signaling pathway, Human cytomegalovirus infection, IL-17 signaling pathway, and Prostate cancer (Fig. 6). Through the analysis of the simulated signaling pathways derived from KEGG enrichment, we found that Mori Cortex might exert anti-OS effects by inhibiting the AKT/extracellular signal-regulated kinases (ERK) pathway (Fig. 7).
Figure 6.
KEGG enrichment analysis. (A) Dotplot and (B) Barplot. KEGG = Kyoto Encyclopedia of Genes and Genomes.
Figure 7.
KEGG simulated signaling pathway analysis. KEGG = Kyoto Encyclopedia of Genes and Genomes. ERK = extracellular signal-regulated kinases.
3.4. Molecular docking
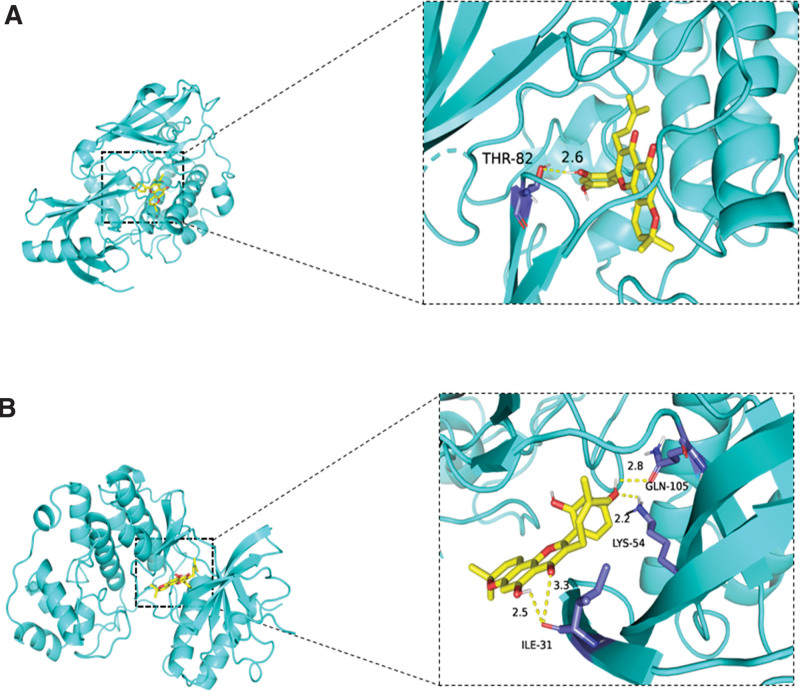
Based on the enrichment analysis results, we found that the key signaling pathways for Mori Cortex in treating OS are AKT and ERK. Therefore, we performed molecular docking of these 2 hub genes with Morusin, a significant active compound in Mori Cortex. When the binding energy is <0 kJ/mol, the small molecule ligand can spontaneously bind to the protein receptor. If the binding energy is <−5.0 kJ/mol or lower, it indicates a better binding capability between the 2. We found that Morusin can bind with AKT and ERK, with binding energies of −10.9 kcal/mol and −7.9 kcal/mol, respectively (Fig. 8).
Figure 8.
Molecular docking. (A) Molecular docking model of Morusin with AKT. (B) Molecular docking model of Morusin with ERK. ERK = extracellular signal-regulated kinases.
3.5. Morusin can inhibit cell cycle regulation and promote cell death in OS cells by targeting AKT/ERK pathway
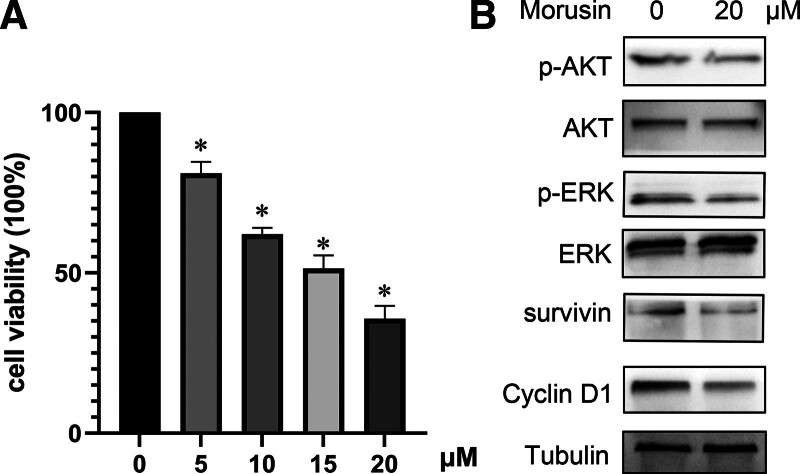
The results from the CCK8 assay indicated that within 48 hours, Morusin can curtail the viability of U-2 OS cells in a dose-dependent manner. As the concentration of the drug rises, there’s a consistent decline in cell survival. Compared to the control group at 0 μmol/L, a higher concentration of Morusin (20 μmol/L) significantly diminished the survival rate of U-2 OS cells (P < .05) (Fig. 9A).
Figure 9.
In vitro experiments. (A) Cell viability assay, U-2 OS cells were treated with 0, 5, 10, 15, and 20 μmol/L Morusin for 48 hours, and cell viability was detected by CCK-8 assay. *P < .05, compared with 0 μmol/L group. (B) Western Blot showed that the expression of p-AKT/AKT, p-ERK/ERK, surviving, and Cyclin D1, U-2 OS cells were treated with 20 μmol/L Morusin for 48 hours. ERK = extracellular signal-regulated kinases, OS = osteosarcoma.
To further validate the anti-OS mechanism of Morusin as determined by KEGG enrichment analysis and molecular docking, we used Western Blotting to assess the expression of pathway proteins in the U-2 OS cell line treated with 0 and 20 μmol/L Morusin for 48 hours. The results revealed that after 48 hours of treatment with Morusin (20 μmol/L), the ratios of p-AKT/AKT and p-ERK/ERK decreased, and the protein levels of Survivin and Cyclin D1 were reduced (P < .05) (Fig. 9B).
4. Discussion
Although we have conducted numerous studies over the past few decades, there has still been no new treatment method that can effectively improve the survival rate of patients with metastatic OS.[19] At present, TCM has become a hot topic in the field of cancer treatment, and its therapeutic effects on various cancers have been widely recognized.[20] Mori Cortex is widely used in TCM due to its pharmacological effects, which include nourishing Yin, soothing the liver, lowering blood pressure, and providing antiinflammatory properties.[21] There are numerous records of Mori Cortex in historical medical texts, and it is frequently prescribed for symptoms like dizziness, tinnitus, vertigo, and hypertension.[22] The active compounds found in Mori Cortex, such as Moracin A and Morusin, endow it with distinct pharmacological characteristics, highlighting its significance in TCM treatments.[23] Therefore, this study explores the anti-OS mechanism of Mori Cortex and its main active component, Morusin, using network pharmacology and in vitro experimental methods.
Through the TCMSP database, we obtained 40 active components of Mori Cortex and 191 therapeutic targets. By intersecting with the differentially expressed genes related to OS obtained from the GEO database analysis, we found that 184 drug targets matched with the OS-related differential genes. Through the Mori Cortex component-OS target network diagram, it was found that the main anti-OS components of Mori Cortex include Moracin A, quercetin, Oxydihydromorusin, Moracin D, Morusin, etc. There is a wealth of literature reporting the antitumor properties of components like quercetin and Moracin A.[24,25] Given Morusin’s strong antioxidant and antiinflammatory capabilities, this study aims to explore the antitumor characteristics of Morusin.[26] The literature suggests that Mori Cortex can serve as a functional food/herbal remedy for the prevention of obesity and its associated metabolic diseases by regulating abnormal adipocyte metabolism involving the AMPK signaling pathway.[21] Meanwhile, Morusin can inhibit the promotion of prostate cancer cell apoptosis and counteract the Warburg effect by suppressing the ROS-mediated FOXM1/c-Myc signaling axis.[12] In the simulated pathway generated based on KEGG enrichment, the AKT and ERK pathways are the key signaling pathways for Mori Cortex’s anti-OS cellular responses. Numerous studies have shown that AKT plays a pivotal role in various cellular processes, such as glucose metabolism, apoptosis, cell proliferation, transcription, and cell migration.[27] It serves as a central node in the cellular signaling pathway, particularly in pathways related to cell survival and insulin signaling.[28] Studies have shown that TAGLN2 is induced in the hypoxic microenvironment of glioblastoma PDX cell lines, and it enhances cellular resistance to standard treatments by binding and inhibiting PTEN, thus activating the PI3K/Akt pathway.[29] Danphenolic acid F inhibits the growth of KRAS-dependent lung cancer cells via the PI3K/AKT signaling pathway.[30] ERK is a key member of the MAPK family.[31] It typically participates in signal transduction from the cell membrane to the nucleus, regulating various cellular processes, including proliferation, differentiation, migration, and survival.[32] Furthermore, MC1R signaling can accelerate G1/S transition through the cAMP-CREB/ATF-1 and ERK-NFκB pathways, promoting the progression of breast cancer.[33] The molecular docking results indicate that Morusin demonstrates a favorable binding affinity to the AKT and ERK proteins. The CCK8 assay confirmed that Morusin effectively suppresses the vitality of U-2 OS cells. Western Blot results revealed that at a concentration of 20 μmol/L, Morusin significantly inhibits the phosphorylation of the AKT and ERK proteins, and concurrently suppresses the expression of Survivin and Cyclin D1. These findings suggest that Morusin might induce apoptosis in OS cells by inhibiting the AKT/ERK signaling pathway and regulating the cell cycle. However, to ascertain the specificity and safety of Morusin, more experimental validations are required.
5. Conclusion
In summary, Mori Cortex exhibits a commendable therapeutic effect against OS, and its primary active component, Morusin, may inhibit cell cycle regulation and promote OS cell apoptosis by suppressing the AKT/ERK pathway. This study introduces a novel perspective on the therapeutic mechanism of TCM Mori Cortex against OS, laying the groundwork and providing insights for further in-depth exploration of its specific mechanism.
Author contributions
Conceptualization: Bo Chen.
Data curation: Yuanhui Wang.
Investigation: Ling Wang, Dongke Xie.
Methodology: Yuanhui Wang.
Software: Ling Wang, Dongke Xie.
Visualization: Ling Wang.
Writing – original draft: Yuanhui Wang.
Writing – review & editing: Bo Chen.
Abbreviations:
- ERK
- extracellular signal-regulated kinases
- GO
- Gene Ontology
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- OS
- osteosarcoma
- PPI
- protein-protein interaction.
This study was conducted through bioinformatics data mining and in vitro cell experiments, thus exempt from institutional review board approval.
The authors have no funding and conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Wang Y, Wang L, Xie D, Chen B. Investigating the molecular mechanism of Mori Cortex against osteosarcoma by bioinformatics analysis and in vitro experimental. Medicine 2024;103:20(e38261).
Contributor Information
Yuanhui Wang, Email: wyl012235@163.com.
Ling Wang, Email: wyl012235@163.com.
Dongke Xie, Email: xiedongke@foxmail.com.
References
- [1].Mair K, Von Werne K, Kalman NS, et al. Multi-institutional experience of proton therapy for osteosarcoma in the Proton Collaborative Group (PCG) prospective registry. Int J Radiat Oncol Biol Phys. 2023;117(2s):e322. [Google Scholar]
- [2].Ye G, Jiao Y, Deng L, et al. Beauvericin suppresses the proliferation and pulmonary metastasis of osteosarcoma by selectively inhibiting TGFBR2 pathway. Int J Biol Sci. 2023;19:4376–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen W, Li Z, Yu N, et al. Bone-targeting exosome nanoparticles activate Keap1/ Nrf2/ GPX4 signaling pathway to induce ferroptosis in osteosarcoma cells. J Nanobiotechnol. 2023;21:355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang P, Song Y, Li H, et al. SIRPA enhances osteosarcoma metastasis by stabilizing SP1 and promoting SLC7A3-mediated arginine uptake. Cancer Lett. 2023;576:216412. [DOI] [PubMed] [Google Scholar]
- [5].Campos F, Téres R, Sebio A, Bettim BB, Martinez-Trufero J. Survival differences of patients with resected extraskeletal osteosarcoma receiving two different (Neo)adjuvant chemotherapy regimens: a systematic review and meta-analysis. Clin Oncol (R Coll Radiol). 2023;35:e720–7. [DOI] [PubMed] [Google Scholar]
- [6].Eichler M, Hentschel L, Singer S, et al. Health related quality of life over time in German sarcoma patients. An analysis of associated factors - results of the PROSa study. Front Endocrinol (Lausanne). 2023;14:1166838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liang Y, Liu M, Cheng Y, Wang X, Wang W. Prevention and treatment of rheumatoid arthritis through traditional Chinese medicine: role of the gut microbiota. Front Immunol. 2023;14:1233994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li Y, Yao L. Traditional Chinese medicine enables the development of small-molecule inhibitors of HSP47, future therapeutic implication in venous thromboembolism. Chin J Nat Med. 2023;21:641–2. [DOI] [PubMed] [Google Scholar]
- [9].You S, Jang M, Kim GH. Mori cortex radicis attenuates high fat diet-induced cognitive impairment via an IRS/Akt signaling pathway. Nutrients. 2020;12:1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].You S, Kim GH. Protective effect of Mori Cortex radicis extract against high glucose-induced oxidative stress in PC12 cells. Biosci Biotechnol Biochem. 2019;83:1893–900. [DOI] [PubMed] [Google Scholar]
- [11].Kim HM, Han SB, Lee KH, et al. Immunomodulating activity of a polysaccharide isolated from Mori Cortex Radicis. Arch Pharm Res. 2000;23:240–2. [DOI] [PubMed] [Google Scholar]
- [12].Koo JI, Sim DY, Lee H-J, et al. Apoptotic and anti-Warburg effect of Morusin via ROS mediated inhibition of FOXM1/c-Myc signaling in prostate cancer cells. Phytother Res. 2023;37:4473–87. [DOI] [PubMed] [Google Scholar]
- [13].Zhao K, Dai Q, Wu J, Wei Z, Duan Y, Chen B. Morusin enhances the antitumor activity of MAPK pathway inhibitors in BRAF-mutant melanoma by inhibiting the feedback activation of STAT3. Eur J Cancer. 2022;165:58–70. [DOI] [PubMed] [Google Scholar]
- [14].Wang J, Liu X, Zheng H, et al. Morusin induces apoptosis and autophagy via JNK, ERK and PI3K/Akt signaling in human lung carcinoma cells. Chem Biol Interact. 2020;331:109279. [DOI] [PubMed] [Google Scholar]
- [15].Fu L, Zhao L, Li F, et al. Pharmacological mechanism of quercetin in the treatment of colorectal cancer by network pharmacology and molecular simulation. J Biomol Struct Dyn. 2023:1–12. [DOI] [PubMed] [Google Scholar]
- [16].Noor F, Asif M, Ashfaq UA, Qasim M, Tahir Ul Qamar M. Machine learning for synergistic network pharmacology: a comprehensive overview. Brief Bioinform. 2023;24:bbad120. [DOI] [PubMed] [Google Scholar]
- [17].Li X, Liu Z, Liao J, Chen Q, Lu X, Fan X. Network pharmacology approaches for research of traditional Chinese medicines. Chin J Nat Med. 2023;21:323–32. [DOI] [PubMed] [Google Scholar]
- [18].Zhao L, Zhang H, Li N, et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J Ethnopharmacol. 2023;309:116306. [DOI] [PubMed] [Google Scholar]
- [19].van Ewijk R, Cleirec M, Herold N, et al.; FOSTER Consortium (Fight OsteoSarcoma Through European Research), Work Package 3 on Recurrent/Refractory Osteosarcoma Trials. A systematic review of recent phase-II trials in refractory or recurrent osteosarcoma: can we inform future trial design? Cancer Treat Rev. 2023;120:102625. [DOI] [PubMed] [Google Scholar]
- [20].Wang X, Hou L, Cui M, Liu J, Wang M, Xie J. The traditional Chinese medicine and non-small cell lung cancer: from a gut microbiome perspective. Front Cell Infect Microbiol. 2023;13:1151557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ma LL, Yuan Y-Y, Zhao M, et al. Mori Cortex extract ameliorates nonalcoholic fatty liver disease (NAFLD) and insulin resistance in high-fat-diet/streptozotocin-induced type 2 diabetes in rats. Chin J Nat Med. 2018;16:411–7. [DOI] [PubMed] [Google Scholar]
- [22].Li L, Wang Y, Liu F, Xu Y, Bao H. Study on the effect of deep eutectic solvent liquid phase microextraction on quality standard, antitussive, and expectorant of Sangbaipi decoction. J Anal Methods Chem. 2021;2021:9999406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lim CY, Kim H, Chung MS. Mori Cortex Radicis extract inhibits human norovirus surrogate in simulated digestive conditions. Food Sci Biotechnol. 2021;30:1243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ostovar S, Pourmadadi M, Zaker MA. Co-biopolymer of chitosan/carboxymethyl cellulose hydrogel improved by zinc oxide and graphene quantum dots nanoparticles as pH-sensitive nanocomposite for quercetin delivery to brain cancer treatment. Int J Biol Macromol. 2023;253(Pt 4):127091. [DOI] [PubMed] [Google Scholar]
- [25].Soung NK, Kim H-M, Asami Y, et al. Mechanism of the natural product moracin-O derived MO-460 and its targeting protein hnRNPA2B1 on HIF-1α inhibition. Exp Mol Med. 2019;51:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cho AR, Park W-Y, Lee H-J, et al. Antitumor effect of morusin via G1 arrest and antiglycolysis by AMPK activation in hepatocellular cancer. Int J Mol Sci. 2021;22:10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Glaviano A, Foo ASC, Lam HY, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer. 2023;22:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang J, Wang C, Hou Y, et al. Molecular mechanisms of Thrombospondin-2 modulates tumor vasculogenic mimicry by PI3K/AKT/mTOR signaling pathway. Biomed Pharmacother. 2023;167:115455. [DOI] [PubMed] [Google Scholar]
- [29].Ye T, Chen R, Zhou Y, et al. Salvianolic acid A (Sal A) suppresses malignant progression of glioma and enhances temozolomide (TMZ) sensitivity via repressing transgelin-2 (TAGLN2) mediated phosphatidylinositol-3-kinase (PI3K)/ protein kinase B (Akt) pathway. Bioengineered. 2022;13:11646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hou X, Zhou C, Liang Z, et al. Salvianolic acid F suppresses KRAS-dependent lung cancer cell growth through the PI3K/AKT signaling pathway. Phytomedicine. 2023;121:155093. [DOI] [PubMed] [Google Scholar]
- [31].Samson SC, Khan AM, Mendoza MC. ERK signaling for cell migration and invasion. Front Mol Biosci. 2022;9:998475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sugiura R, Satoh R, Takasaki T. ERK: a double-edged sword in cancer. ERK-dependent apoptosis as a potential therapeutic strategy for cancer. Cells. 2021;10:2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chelakkot VS, Thomas K, Romigh T, et al. MC1R signaling through the cAMP-CREB/ATF-1 and ERK-NFκB pathways accelerates G1/S transition promoting breast cancer progression. NPJ Precis Oncol. 2023;7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]