Summary:
The avian dorsal telencephalon has two vast territories, the nidopallium and the mesopallium, both of which have been shown to contribute substantially to higher cognitive functions. From their connections, these territories have been proposed as equivalent to mammalian neocortical layers 2 and 3, various neocortical association areas, or the amygdala, but whether these are analogies or homologies by descent is unknown. We investigated the molecular profiles of the mesopallium and the nidopallium with RNAseq. Gene expression experiments established that the mesopallium, but not the nidopallium, shares a transcription factor network with the intratelencephalic class of neocortical neurons, which are found in neocortical layers 2, 3, 5 and 6. Experiments in alligators demonstrated that these neurons are also abundant in the crocodilian cortex and form a large mesopallium-like structure in the dorsal ventricular ridge. Together with previous work, these molecular findings indicate a homology by descent for neuronal cell types of the avian dorsal telencephalon with the major excitatory cell types of mammalian neocortical circuits: the layer 4 input neurons, the deep layer output neurons and the multi-layer intratelencephalic association neurons. These data raise the interesting possibility that avian and primate lineages evolved higher cognitive abilities independently through parallel expansions of homologous cell populations.
Introduction:
Advanced cognitive abilities are scarce in the animal world, despite a vast diversity of animal forms, behaviors, and nervous system anatomies [1]. The most widely known varieties of cognition are found in mammals including elephants, dolphins, and primates [2, 3]. Mammalian intelligence is invariably associated with the presence of a large and complex neocortex, by far the largest structure of the human brain [4, 5].
Less familiar are the cognitive abilities of birds, which were overlooked until the mid-20th century [6]. Birds have large brains relative to body size, and avian brains are packed with neurons at a density greater than that of the primate brain [7]. Large, cell-dense brains may mediate the ability of some birds to perform cognitively demanding tasks with primate-like skill [8]: they manufacture and use tools to solve problems [9], can recognize themselves in a mirror [10], can form abstract concepts [11], and can communicate vocally [12]. Birds evolved their cognitive abilities convergently with primates [13], as reflected in the dramatic differences in their forebrain anatomies. Birds do not have a brain structure that remotely resembles the neocortex in morphology.
Birds are archosaur reptiles, a group including living crocodilians and extinct dinosaurs. Neither birds nor non-avian reptiles possess a six-layered neocortex, the neuroanatomical hallmark of all extant mammals [14]. In place of a laminar structure, the bird dorsal telencephalon (DT) contains territories of clustered neuronal cell bodies, or nuclei [15]. The bird DT is organized into two main divisions, the Wulst and the dorsal ventricular ridge (DVR), which are demarcated by a shallow sulcus called the vallecula. The DVR is further subdivided into the mesopallium, nidopallium, and arcopallium (Figure 1). Whether any of these bird DT territories are homologous in any way to the neocortex is a longstanding problem in evolutionary neuroscience [16], but it is certain that bird intelligence does not require a laminated neocortex. In particular, avian DT neurons lack the layer-spanning apical dendrites that are a defining feature of the cerebral cortex [17].
Figure 1. Connectional anatomy of the avian dorsal telencephalon.
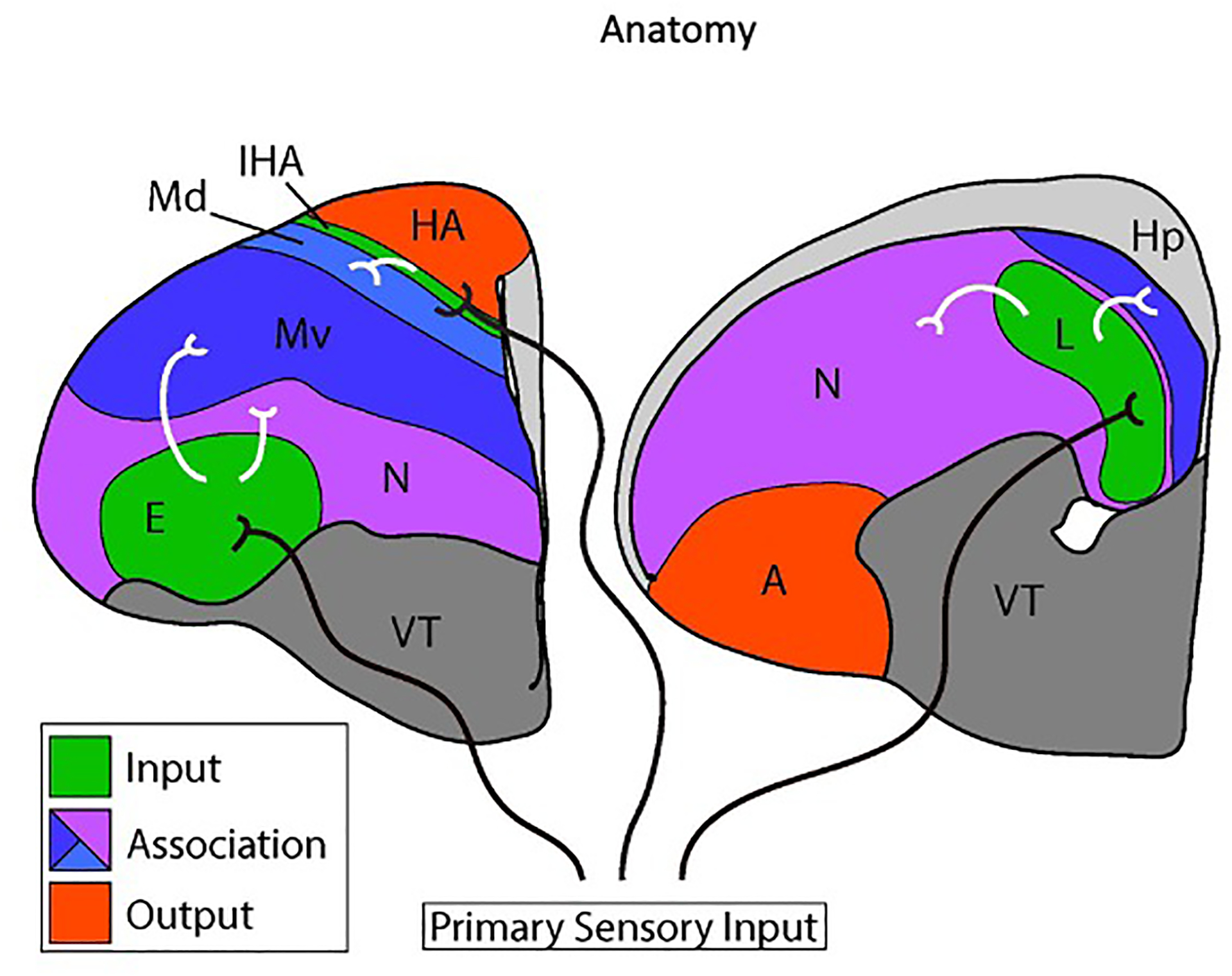
Two coronal cross sections through the chick telencephalon are shown at intermediate (left) and posterior (right) levels. Medial is to the right and dorsal is at the top. Only the left hemisphere is shown. The two main divisions of the avian dorsal telencephalon are the Wulst and the dorsal ventricular ridge (DVR). The Wulst includes the hyperpallium apicale (HA), the interstitial nucleus of the hyperpallium apicale (IHA), and the dorsal mesopallium (Md). The DVR comprises the ventral mesopallium (Mv), the nidopallium (N), and the arcopallium (A). The hippocampus (Hp) is a medial dorsal telencephalon structure likely homologous to the mammalian hippocampus. The ventral telencephalon (VT) is indicated.
Primary input cell populations are shaded in green and output populations are red. Association territories are shaded in blue or purple. Axons originating in the dorsal thalamus (black axons) transmit visual information to the entopallium (E), auditory information to Field L (L), and visual and somatosensory information to the IHA. The Md, Mv, and nidopallium receive secondary sensory information from primary input nuclei, represented by white axons. The association territories interconnect extensively with each other and with output populations (full circuitry not shown).
Bird intelligence may, however, arise from neural circuitry fundamentally similar to that of the neocortex. The bird DT, like the neocortex, receives visual, auditory, and somatosensory input from dorsal thalamic axons (Figure 1, black axons to green nuclei). The targets of ascending sensory information in birds are clearly delimited nuclei that by circuitry resemble the input neurons of neocortical layer 4 (L4). Information output from the bird DT arises from cells in the arcopallium and the HA (Figure 1, red nuclei), which project to brainstem motor centers and are similar to the subcortical projection neurons of neocortical layer 5 (L5). These cross-species similarities in connectivity led Harvey Karten to propose that input and output neurons of mammals and birds are homologous at the cell-type level [18, 19]. Recent studies provided strong support for Karten’s cell-type homology hypothesis by demonstrating input and output cells express conserved molecular markers: avian input cells are enriched for expression of the neocortical L4 genes KCNH5/EAG2 and RORB, while avian output cells express L5 markers including FEZF2, CACNA1H, and SULF2 [17]. Input and output cells, including their defining connections and gene expression profiles, were likely inherited from the last common ancestor of amniotes.
There are two massive and enigmatic territories in the avian DT, the mesopallium and nidopallium, which are neither strictly input nor output in their connectivity. These territories are not the targets of ascending primary sensory information and their axonal projections do not leave the telencephalon (but see below for nidopallium) [20, 21]. Their functions can be described as associational: they contribute to the integration of multiple sensory modalities and perform higher-order information processing. The mesopallium and nidopallium are believed to contribute significantly to avian cognition, learning, and memory [22].
The mesopallium was previously thought to be restricted to the DVR, but recent molecular studies show that the ventral Wulst expresses many genes characteristic of the DVR mesopallium [23]. These studies suggest the mesopallium has a bi-partite organization, with a dorsal mesopallium in the Wulst (Figure 1, Md in light blue) abutting a ventral mesopallium in the DVR (Figure 1, Mv in dark blue). Md and Mv form links between primary input and other DT cell populations. Mv receives axons from two DVR input territories, the visual entopallium and the auditory Field L (Figure 1, white axons) [20]. Mv projects in turn to the arcopallium and nidopallium, especially to the posterior nidopallium [20]. Md receives information from the IHA, the primary Wulst input population, and sends axons to the Wulst output nucleus, the HA (Figure 1, IHA) [24–26]. Both mesopallium divisions project to the major basal ganglia nucleus in the ventral telencephalon, the striatum [27].
The nidopallium is a heterogeneous structure with many connectional similarities to the mesopallium (Figure 1, purple). It lies ventral and posterior to the mesopallium, and just above the striatum. The nidopallium contains both input and associational cell populations. The input nuclei entopallium and Field L are embedded within the nidopallium as subnuclei (Figure 1). The vast bulk of the nidopallium, however, does not receive direct primary sensory input but instead receives secondary information from the entopallium and Field L (Figure 1, white axons) [28, 29]. Its posterior division is heavily interconnected with the mesopallium and projects to the striatum as well as the arcopallium [21, 22].
Conflicting homologies of the mesopallium and nidopallium have been proposed based on differing criteria. Some researchers argue the mesopallium and nidopallium are homologous to the temporal neocortex due to their similar positions in lateral DT [30]. A second view proposes the mesopallium is homologous to neocortical layers 2 and 3 (L2/3) based on similar gene expression [31]. Other researchers note the nidopallium and mesopallium are nuclear territories ventral and lateral to the bulk of the Wulst. They argue the nidopallium and mesopallium are not comparable to the neocortex at all, but are instead homologous to subcortical nuclei in the mammalian piriform lobe such as the amygdala and claustrum [32, 33].
Gene expression profiles, particularly for transcription factors, can provide deep insight into cell-type identities [34]. Conserved transcription factors often indicate that the evolutionary origins of some cell types greatly predate the emergence of the tissue-level structures in which they are found [35]. We characterized with RNA sequencing the molecular features of the chick mesopallium and compared them with those of the nidopallium and the input and output nuclei. Our findings provide a novel solution to the enigma of the mesopallium, namely a cell-type homology with the intratelencephalic neurons of the mammalian neocortex, including callosal projection neurons. Our molecular data, together with previous connectional studies, suggest that intratelencephalic neurons were present in the amniote last common ancestor and now populate highly divergent telencephalic structures in mammals, reptiles, and birds.
Results:
HOLT analysis of the mesopallium transcriptome identifies specific marker genes
To test whether the mammalian neocortex contains homologues of avian DT association territories, we performed an unbiased, forward screen for molecular markers of the chicken mesopallium and nidopallium. Tissue was collected by dissection from seven territories in embryonic day 14 (E14) chicken DT: anterior mesopallium (M); posterior nidopallium (N); the sensory input populations entopallium (E), Field L (L), and basorostralis (Bas); and the output nuclei HA and arcopallium (A) (Figure S1). Purified RNAs from these tissues were submitted for Illumina 100 base pair, paired-end RNA sequencing. Reads were assembled with Trinity software and transcript abundance was estimated with RSEM.
To identify marker genes restricted to the mesopallium and nidopallium, we devised the HOLT (High-to-Low Ordering of Like Transcripts) method (Figure 2A,B). For the mesopallium, we selected transcripts expressed at 2.5 fold greater FPKM (fragments per kilobase of transcript per million mapped reads) values in the mesopallium compared with the arcopallium and the entopallium (Figure 2A). These filters removed broadly expressed neuronal genes and glial and housekeeping genes. We ranked the filtered transcripts in descending order of expression in the mesopallium and selected transcripts for protein-coding genes expressed at 5 FPKM or higher. In trial experiments we had found that 5 FPKM is near the threshold of detection by in situ hybridization (ISH). The cumulative HOLT filters yielded 78 highly expressed transcripts for protein-coding genes enriched in the mesopallium compared with input and output cell populations (Table S1). The 15 most abundant genes are presented in Figure 2C. A parallel differential expression analysis with edgeR returned 18 genes also identified with the HOLT method, but missed several key mesopallium-specific factors (see STAR Methods).
Figure 2. HOLT analysis of the mesopallium transcriptome.
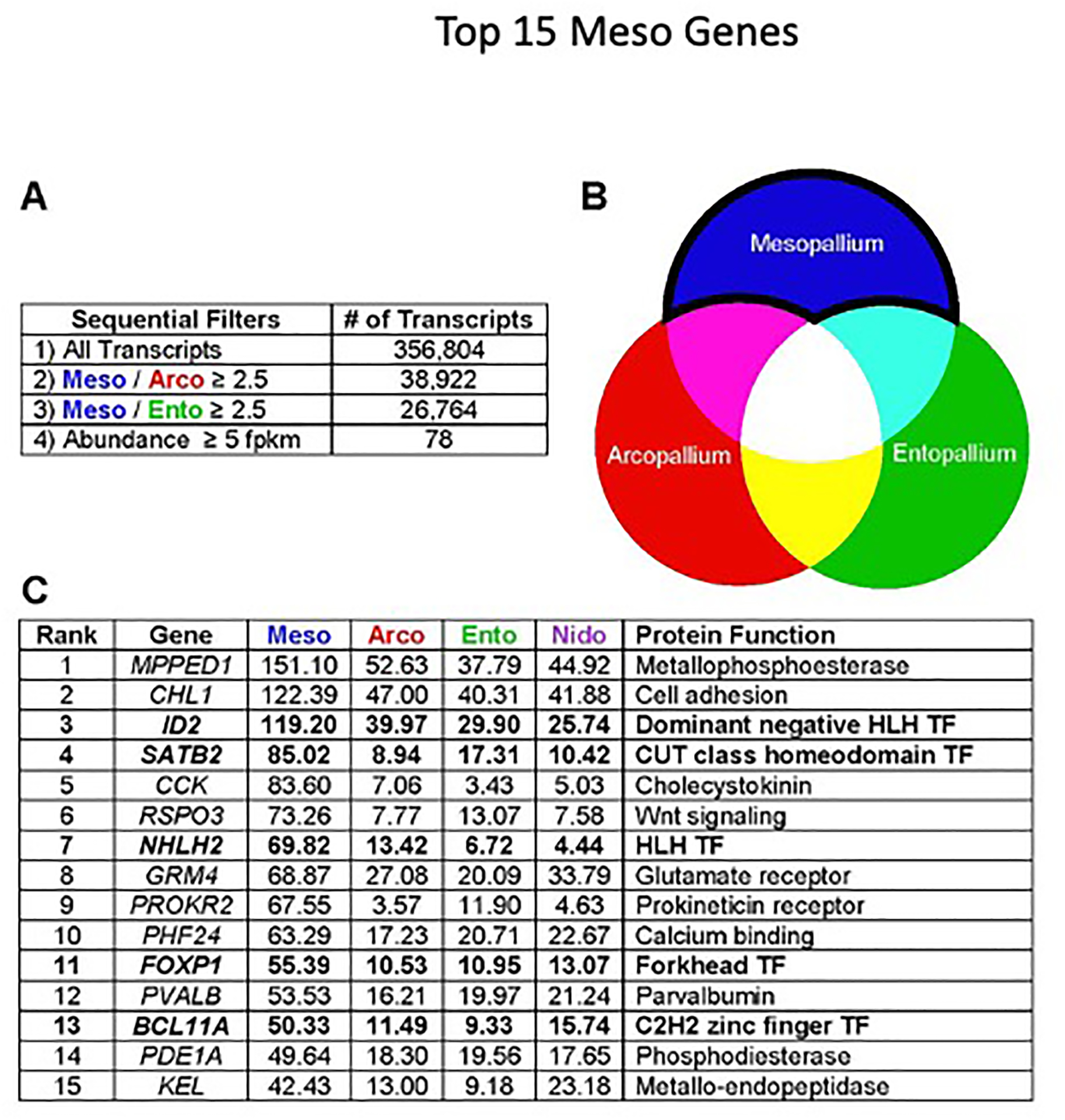
(A) The HOLT pipeline is based on serial pairwise comparisons of transcript abundance. We first selected transcripts enriched in mesopallium compared with arcopallium, and from these, selected transcripts enriched in mesopallium compared with entopallium. We then ordered the mesopallium-selective transcripts by FPKM value.
(B) Visual representation of target transcripts: our HOLT approach is designed to detect transcripts enriched in mesopallium association neurons, but not in output or input neurons.
(C) The 15 most highly expressed mesopallium-enriched genes. Transcription factors are highlighted in bold type. Values indicate expression levels in FPKM. TF, Transcription Factor.
ISH experiments confirmed that candidate genes identified by the HOLT method are expressed in the mesopallium, and often with great specificity (Figure S2A–F). Most marker genes identified two delineable territories: a thick ventral district and a thinner dorsal one. Comparison with published expression patterns for DRD5 (#42, Table S1) identifies these two territories as Mv and Md [15, 36]. In addition, five of the top 78 transcripts were previously identified as mesopallium markers by Jarvis et al. (#8, 11, 17, 21, and 28) [23]. Mv and Md are most clearly demonstrated in staining for NHLH2 transcripts (Figure S2G–N).
These gene expression data establish that the mesopallium is a unitary molecular territory. It has dorsal and ventral divisions that share extensive similarity in gene expression [23, 37]. None of the mesopallium HOLT candidates examined proved to be selective markers of either the Md or Mv. Moreover, Md and Mv have been both shown to form intratelencephalic connections, and to lack primary sensory input and long output projections (Atoji and Wild 2012). These data prompt the hypothesis that the mesopallium, like the input and output nuclei of the avian DT, is dominated by a principal class of excitatory neurons.
Six transcription factors are enriched in dorsal and ventral chicken mesopallium
We anticipated that transcription factors would be more likely than other classes of molecules to show conservation at the cell-type level across species. Such factors may also be functionally required for the developmental specification of mesopallium cell-type identity. Five of the 15 most abundant mesopallium-specific transcripts code for transcription factor molecules (Figure 2C, bolded). One additional mesopallium transcription factor marker, EMX1, was not detected by the HOLT method but was previously identified (Table S1) [38]. ISH experiments confirmed that all six transcription factors are highly enriched in the mesopallium at E14 (data not shown) and postnatal day zero (Figure 3A–F). While each expression pattern is unique, they share important similarities. Each factor is expressed throughout the entire mesopallium across its anteroposterior and mediolateral axes, and each labels Md and Mv domains with clear boundaries. Of these six factors, only NHLH2 was not detected in postnatal mouse neocortex or alligator mesopallium (data not shown).
Figure 3. Expression patterns of mesopallium-enriched transcription factors.
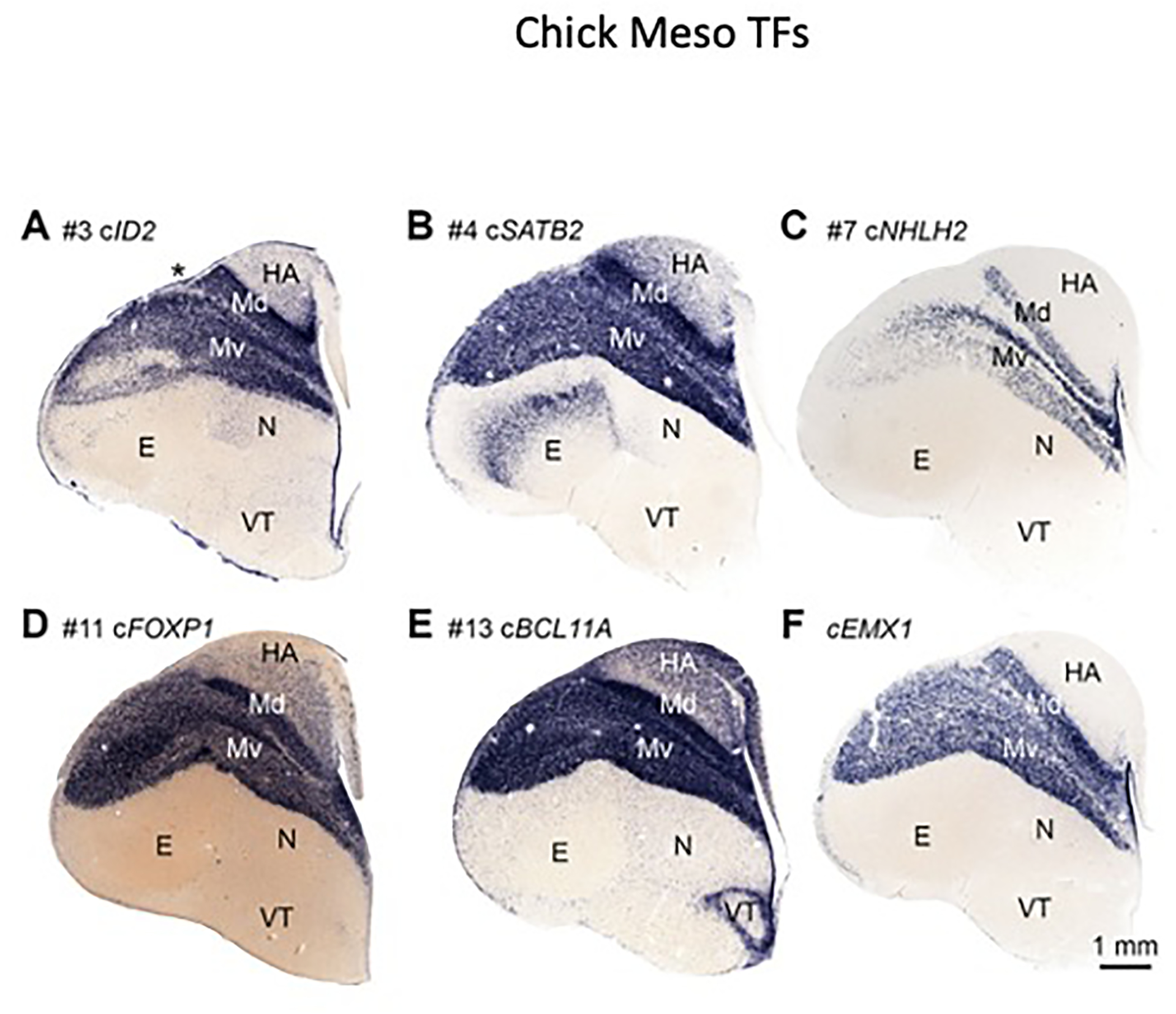
In situ hybridization on coronal sections through postnatal day zero chick telencephalon. Orientation as in Figure 1. Six transcription factors are highly enriched in dorsal (Md) and ventral (Mv) mesopallium:
(A) #3 ID2, (B) #4 SATB2, (C) #7 NHLH2, (D) #11 FOXP1, (E) #13 BCL11A, and (F) EMX1. Numbers refer to ranking in Figure 2C. Asterisk in A indicates the vallecula.
We identified five transcription factors (ID2, SATB2, FOXP1, BCL11A, and EMX1) that together constitute a candidate gene regulatory network for mesopallium cell-type identity. These five factors will be collectively referred to as the MesoTFN (mesopallium transcription factor network).
Mesopallium molecular organization is conserved across birds
MesoTFN expression extends from the Mv into the Wulst, but it is unclear how to relate the Wulst expression to anatomy previously described in the literature due to inconsistent nomenclature. Four layer-like territories, or pseudolayers [26], are traditionally recognized in the Wulst. From medial to lateral, these subdivisions are the HA, IHA, HI, and HD [15]. The classic HD is the lateral-most division of the Wulst and abuts the DVR. Jarvis et al. named the mesopallium-like Wulst territory the Md [23]. However, it remains unclear whether some birds have an HD or HI, or both, that are molecularly distinct from the Md. If the Md extends to the IHA, there is no room for a non-mesopallium territory and the classic HD/HI corresponds to Md. If the Md does not extend to the IHA, and there is a territory between them, this territory may represent a separate HD or HI.
We performed ISH on adjoining sections from E20 chicken telencephalon for the genes FOXP1 and KCNH5. FOXP1 labels the Md, which is almost fully contained within the Wulst as indicated by its location dorsal to the vallecula (Figure S3A, asterisk). KCNH5 labels the IHA a short distance dorsal to the vallecula (Figure S3B). Serial section alignment demonstrated that the FOXP1(+) and KCNH5(+) territories abut in anterior DT, with no intervening FOXP1(−)/KCNH5(−) zone that could correspond to a distinct HD or HI.
The chicken Gallus gallus is a member of the avian superorder Galloanserae. Most extant birds belong to the Neoaves, which diverged from the Galloanserae 90 million years ago [39]. The absence of HI/HD observed in chicken may be an ancestral characteristic, with Neoaves gaining these additional territories later. Alternatively, chickens may have lost HD/HI. To test for these possibilities, we examined FOXP1 and KCNH5 expression in the starling Sturnus vulgaris. Traits present in the chicken and the starling, a member of the Neoaves, are likely to be ancestral to and conserved across birds.
The starling Wulst, as recognized by the position of the vallecula (Figure S3C–F, red asterisk), dominates the anterior DT. The large starling Wulst would be a good candidate to harbor an HD/HI territory between the Md and IHA, if such a territory is present in birds. Starling FOXP1 is expressed in a Wulst domain dorsal to the vallecula as well as a DVR domain (Figure S3C). The starling Md extends to the IHA, as identified by KCNH5 (Figure S3D). At intermediate (Figure S3E) and more posterior (Figure S3F) levels of the telencephalon, starling BCL11A clearly identifies both Md and Mv. The starling does not appear to possess a candidate HD/HI separate from the Md. These data raise the possibility that all Neognath birds (Neoaves and Galloanserae) have a Wulst Md and a DVR Mv, both of which express MesoTFN factors. It will be important, however, to search for a separate HI/HD territory in a bird of prey, such as a diurnal owl, where a prominent HI and HD have been cytoarchitectonically described [25].
MesoTFN factors in mouse neocortex
Expression of the MesoTFN has been previously reported in the mouse neocortex. Mouse Satb2 is expressed in excitatory callosal projection neurons of upper and deep neocortical layers. In the absence of Satb2 function, genetically marked callosal neurons misexpress the output cell determinant Bcl11b/Ctip2 and convert to an output-like cell fate [40]. Bcl11a/Ctip1 is co-expressed with Satb2 in callosal neurons, and promotes specification of callosal identity at the expense of output cell identity [41]. Foxp1 is coexpressed with Satb2, but is excluded from output neurons [42]. Mouse Id2 is also expressed in both upper and deep neocortical layers (Figure S4) [43]. We report here that Emx1 is expressed in postnatal mouse neocortex strongly in L2/3 and at a reduced level in L5 (Figure S4). Thus, five of the six mesopallium-specific transcription factors we identified by HOLT are expressed in the mouse neocortex. None of these factors is specific to any particular neocortical layer, but all of them are expressed in the upper layers L2/3, which are thought to contain exclusively intratelencephalic projection neurons. Satb2, Bcl11a, and Foxp1 are expressed in the deep layers, but they are in intratelencephalic neurons and not Bcl11b-expressing output neurons [40–42].
Chicken MesoTFN factors are co-expressed in excitatory neurons
These connectional and gene expression data suggest that neocortical intratelencephalic cell types are enriched in the avian mesopallium. If correct, then the MesoTFN factors should be co-expressed with the excitatory neuron marker VGLUT2. Chicken VGLUT2 is strongly expressed in both the mesopallium and nidopallium (Figure 4A). Two-color fluorescent in situ hybridization (FISH) experiments established that BCL11A is extensively co-expressed with VGLUT2 within the mesopallium (Figure 4B,C). Scattered BCL11A(+)/VGLUT2(−) cells in the nidopallium likely identify inhibitory interneurons (Figure 4B,C), consistent with BCL11A expression in the ventral telencephalon (Figure 3E). Within the mesopallium, the vast majority of BCL11A(+) cells do not express the inhibitory neuron marker GAD1 (Figure 4D–F). MesoTFN factors may be expressed by a single principal cell type or by multiple mutually exclusive but similarly distributed cell types. In mouse, Satb2 and Bcl11a are co-expressed and are each genetically required for the specification of intratelencephalic neurons [40, 41]; co-expression of these factors in the chick mesopallium would provide the strongest support for a cell-type homology. FISH experiments demonstrated that most, if not all, SATB2-expressing mesopallium neurons also express BCL11A (Figure 4G–I).
Figure 4. Co-expression analysis of mesopallium transcription factors.
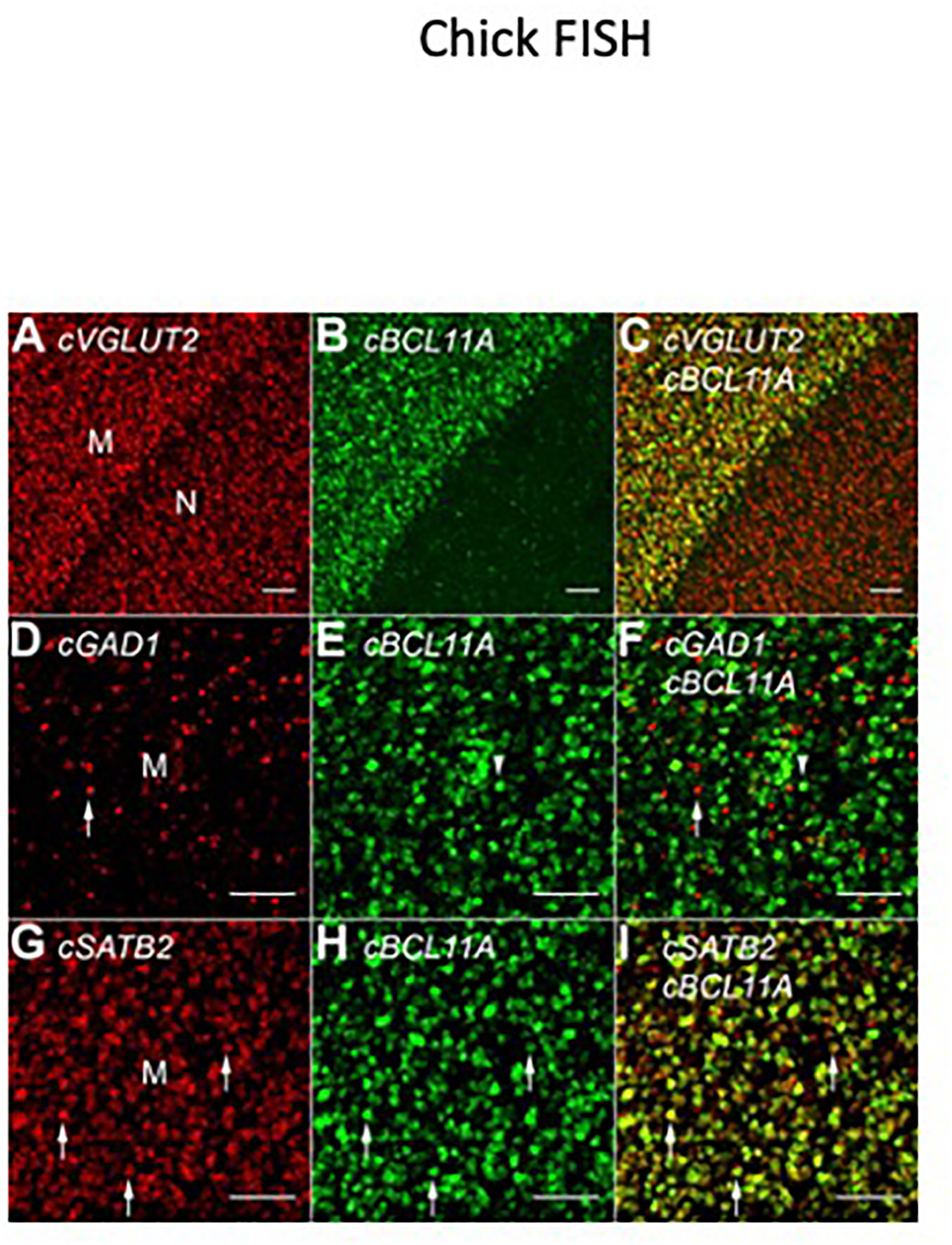
(A) Vesicular glutamate transporter 2 (VGLUT2) is expressed in excitatory neurons of the dorsal telencephalon, including the mesopallium (M) and nidopallium (N). A thin, cell-poor lamina separates these territories.
(B) BCL11A is expressed throughout the mesopallium and in scattered cells in the nidopallium.
(C) An overlay of the fluorescent channels shows that BCL11A co-localizes with VGLUT2 in the mesopallium.
(D–F) Most BCL11A-expressing cells in the mesopallium do not express the inhibitory neuron marker glutamate decarboxylase 1 (GAD1). The arrow and arrowhead indicate a singly labeled interneuron and BCL11A(+) neuron, respectively.
(G–I) The transcription factors SATB2 and BCL11A are extensively co-expressed by presumed excitatory neurons in the mesopallium. Arrows indicate three examples of double labeled cells.
All scale bars are 100 μm.
MesoTFN factors are expressed in the alligator DVR and cerebral cortex
The striking similarities of avian mesopallium neurons and mammalian neocortical intratelencephalic neurons suggest that both sets of cell types are descendents of a common cell population present in the last common ancestor of mammals and birds. This common ancestor gave rise to all extant reptiles (Figure 5A). We therefore tested whether MesoTFN factors are present in the DT of a non-avian reptile.
Figure 5. Molecular identification of intratelencephalic neurons in the alligator dorsal cortex and DVR.
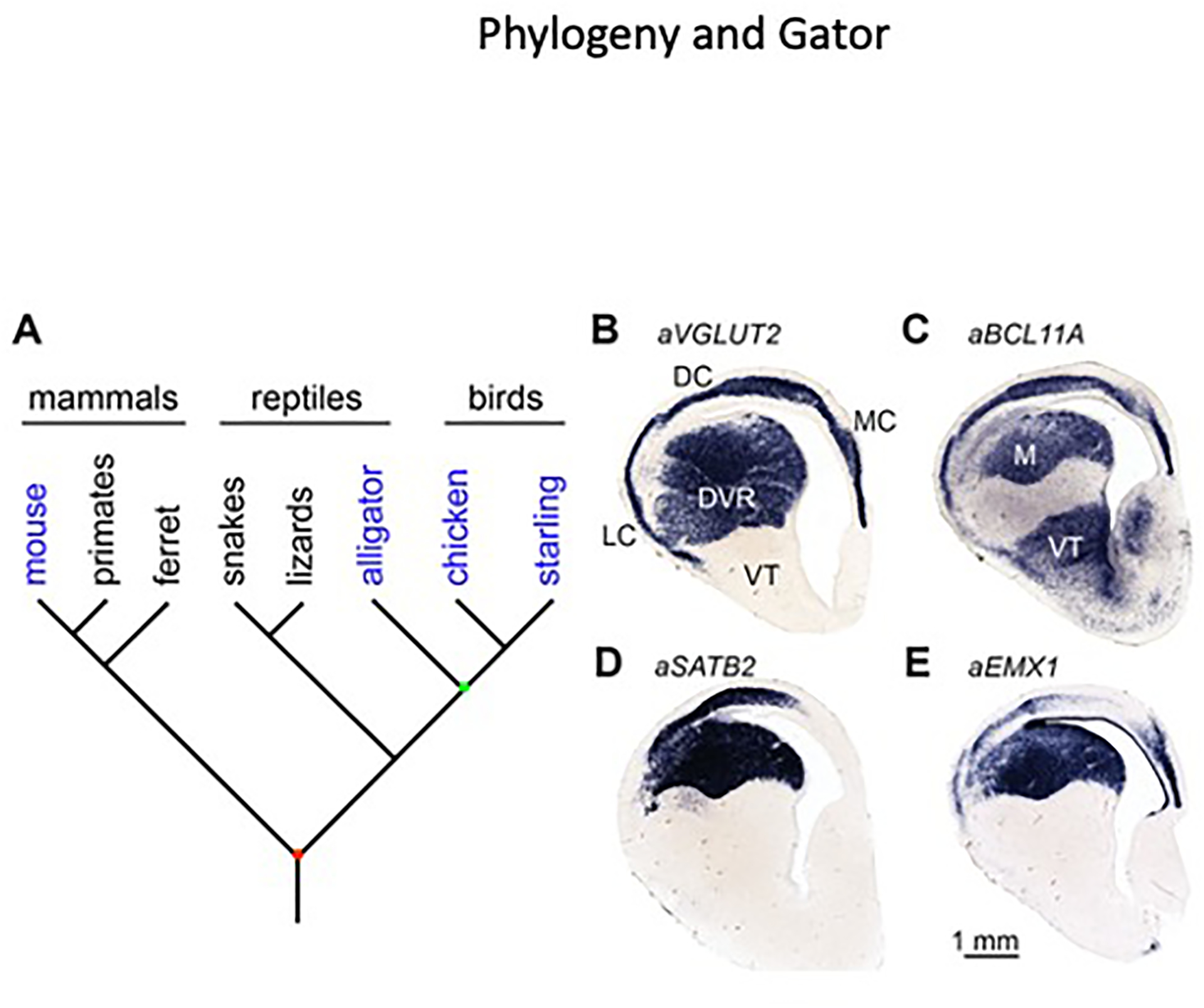
(A) Extant amniotes include mammals, reptiles, and birds. Species examined in this study are shaded in blue. Connectional and molecular data from mammals and birds raise the possibility that intratelencephalic neurons were present in the last common ancestor of amniotes (red node), an animal that lived approximately 320 million years ago. In this case we would expect to find intratelencephalic neurons in a reptile, barring secondary losses. Reptiles are paraphyletic with respect to birds, meaning there is no common ancestor of reptiles that is not also the ancestor to birds. Crocodilians including Alligator mississippiensis are more closely related to birds than are other reptiles. The last common ancestor of crocodilians and birds (green node) lived around 240 million years ago [44].
(B) Alligator VGLUT2 is expressed throughout the dorsal telencephalon, including the DVR and medial (MC), dorsal (DC), and lateral (LC) cortices.
(C–E) The avian mesopallium markers (C) BCL11A, (D) SATB2, and (E) EMX1 identify a crocodilian mesopallium (M) and a dorsal cortex cell population.
The American alligator, Alligator mississippiensis, has a large, avian-like DVR with clear cytoarchitectonic subdivisions (Figure 5B). Crocodilians are the closest living relatives of birds, but these groups diverged more than 240 million years ago [44]. Unlike birds, but similar to all other studied reptiles, the alligator has a three-layered cerebral cortex. The cerebral cortex comprises three mediolateral subdivisions: a medial cortex, widely believed to be homologous with the mammalian hippocampus (Figure 5B, MC) [45], a lateral cortex that may be homologous with the mammalian olfactory cortex (Figure 5B, LC) [46], and a dorsal cortex that by position may be homologous with the avian Wulst and at least part of the mammalian neocortex (Figure 5B, DC). ISH experiments in late-stage alligator embryos established that BCL11A, SATB2, EMX1, ID2, and FOXP1 are expressed in a large territory in the dorsal DVR, which we denote the alligator mesopallium (Figure 5C–E and data not shown). All five MesoTFN factors are also expressed in an alligator dorsal cortex cell population (Figure 5C–E and data not shown). There are no tracing studies of intratelencephalic circuitry in alligators, but these gene expression data strongly suggest that there are two populations of reptile intratelencephalic cells: one in the DVR and the other in the dorsal cortex. This pattern is reminiscent of the avian arrangement with a DVR Mv and a Wulst Md.
Chicken mesopallium arises from a fate-restricted territory in anterior telencephalon
Our data from two distantly related bird species, a reptile, and a mammal provide strong evidence that intratelencephalic neurons were present in the amniote last common ancestor. These neurons are abundant in the upper layers of the mammalian neocortex and are intermingled with the connectionally and molecularly distinct subcerebral output neurons in the deep layers (Figure 6, Mouse). The avian mesopallium, however, appears to be a more homogeneous territory where densely clustered intratelencephalic neurons are spatially segregated from other excitatory cell types (Figure 6, Chicken). This anatomy raises the possibility that chicken mesopallium cells are born in a dedicated DT progenitor territory, one that gives rise only to the mesopallium. This would be a considerable divergence from the model for neocortical neurogenesis, in which all major excitatory cell types are generated across the neocortical primordium.
Figure 6. Distribution of intratelencephalic neurons in mammals, reptiles, and birds.
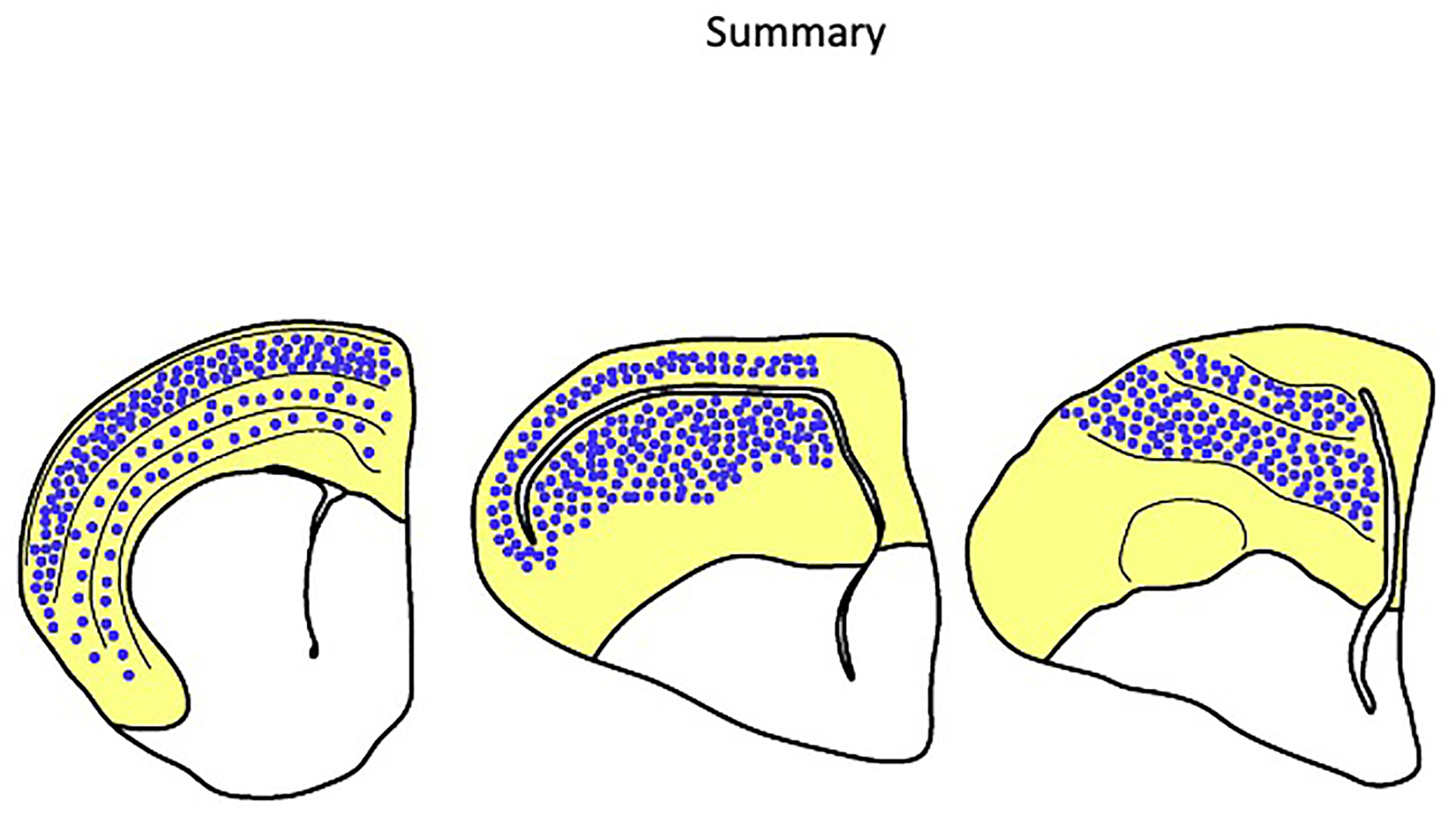
Schematics of the telencephalon of a mouse (left), alligator (middle), and chicken (right). The dorsal telencephalon is shaded in grey. In the mammalian neocortex, the vast majority of excitatory neurons in the upper layers 2 and 3 are intratelencephalic (IT) neurons (blue dots). IT neurons are also interspersed with output neurons in the deep layers. Molecular data suggests IT neurons are present in the alligator cerebral cortex and form a mesopallium in the dorsal DVR. Unlike the arrangement in the neocortex, avian IT neurons form coherent clusters segregated from other major excitatory neuron populations.
Whole mount ISH for SATB2 on Hamburger-Hamilton stage 30 (HH30, E6) chick embryos demonstrates a district of SATB2(+) cells in anterior telencephalon with a sharp posterior boundary (Figure 7A). At E8, SATB2(+) cells remain packed in an anterior territory (Figure 7B,C). Early SATB2 expression is consistent with the local generation of mesopallium cells. Gene expression evidence is, however, insufficient for fate mapping. These results could also be explained by the production of mesopallium precursor cells throughout the DT that coalesce through migration before expressing SATB2. A third possibility is that mesopallium cells are born in a restricted territory that also produces other cell types. To distinguish among these possibilities, we fate-mapped anterior telencephalon with an indelible genetic marker.
Figure 7. A mesopallium-restricted progenitor field in chicken telencephalon.
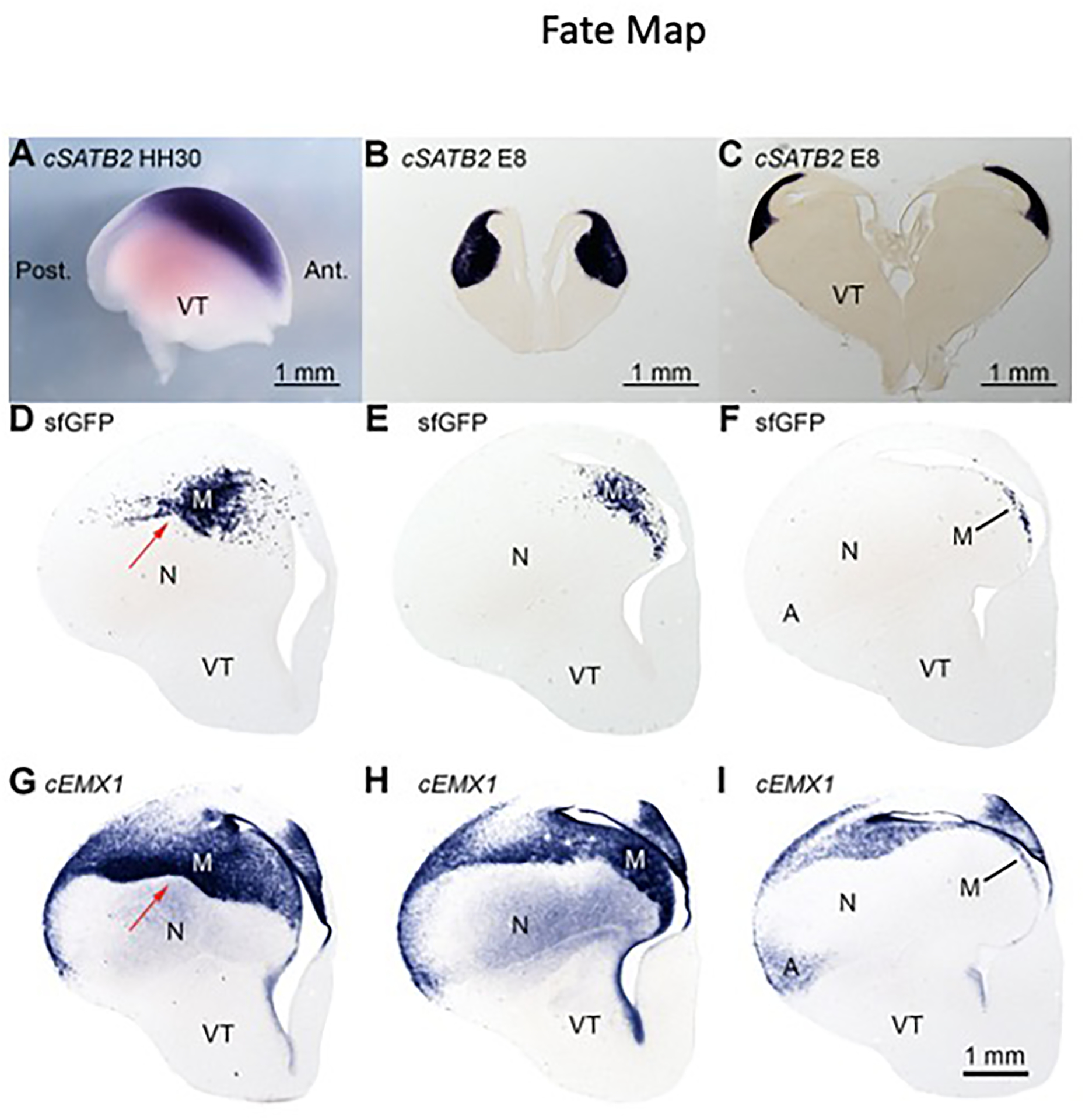
(A) Whole mount in situ hybridization for SATB2 in HH30 chicken. Only the telencephalon is pictured, lateral view with anterior to right. This is the earliest stage at which SATB2 transcript was detected, suggesting that mesopallium neurons are born in a restricted anterior territory. A, Anterior; P, Posterior.
(B) Anterior and (C) intermediate sections from an E8 telencephalon labeled for SATB2 transcripts. Staining indicates that early mesopallium precursors occupy a coherent anterior territory.
(D–F) Three sections from an E14 telencephalon electroporated with PBXW-sfGFP and CDV-hypbase plasmids, labeled with a riboprobe for sfGFP.
(G–I) EMX1-labeled sections adjacent to the above sfGFP sections. Descendants of electroporated cells, identified by sfGFP staining, are confined to the mesopallium and form a sharp ventral boundary with the nidopallium (red arrow).
We co-electroporated PBXW-sfGFP and CDV-hypbase plasmids into the anterior telencephalon of HH23/24 chicken embryos. PBXW-sfGFP encodes the superfolder GFP molecule flanked by PiggyBac transposition sequences. CDV-hypbase encodes the PiggyBac transposase, which inserts sfGFP and its promoter into the genome. We detected the descendants of electroporated progenitors by ISH for the sfGFP transcript at E14 and observed labeling restricted to the mesopallium (Figure 7D–F). At all three levels of telencephalon shown, labeling is confined to the mesopallium as indicated by EMX1 staining on serially adjoining sections (Figure 7G–I). The characteristic contours of the mesopallium are evident by sfGFP-expressing cells distributed along the mesopallium ventral border (Figure 7D) and the crescent-shaped posterior mesopallium (Figure 7E,F). Staining for sfGFP is absent from the nidopallium and arcopallium. This result strongly supports the existence of a fate-restricted territory in anterior DT that gives rise to mesopallium intratelencephalic cells, but not to other cell types, including input and output cells.
DACH2, the only known nidopallium transcription factor marker gene, is weakly expressed in the mouse neocortex
The identification of a MesoTFN raises the question of whether the avian nidopallium possesses a separate TFN and, if so, whether mammalian DT contains a nidopallium homolog. We used the HOLT method to identify nidopallium-specific transcripts (Table S2). This approach recovered the single known nidopallium marker, the transcription factor DACH2 (#32) [47]. We examined expression patterns of 30 of these nidopallium-enriched genes, including the transcriptional regulators MEIS2, LBH, NR2F2, PBX3, SOX2, FEZF2, NR0B1, BCL11B, DLX6, FOXP2, and CUX1 (data not shown). None of these is a pannidopallium marker gene. Even DACH2 is not truly a nidopallium-specific marker because it additionally labels the IHA, the primary input nucleus of the Wulst (Figure S5A–C). Dach2 is currently the only nidopallium-specific transcription factor available for potentially identifying a mammalian nidopallium homolog. At postnatal day six, mouse Dach2 probes labeled weakly a sparse cell population distributed in a laminar pattern across the neocortex (Figure S5D). The Dach2(+) lamina lies just deep to L4 and may correspond to the upper L5 sublayer L5a. No conclusions on any nidopallium homology can be drawn from the expression of this single marker, but Dach2 expression does provide a genetic access point for further characterization of this cell population. Mouse Dach2 was not detected in the basolateral amygdala, which has been proposed as a mammalian homolog to the avian nidopallium [32, 33].
Discussion:
The classic conception of neocortical circuitry is the canonical neocortical column. Sensory information first reaches L4/input neurons, which are linked to L5/output neurons through relays in L2/3. This circuity is believed to form the foundation of neocortex function across neocortical areas and across distantly related mammalian species. A broader view of neocortical cell types recognizes that L2/3 neurons belong to a larger class, the intratelencephalic neurons [48]. These neurons are defined not by their layer, but by their connections. Intratelencephalic (or IT) neurons, as the name implies, send projections only to telencephalic targets including the striatum [49]. Most importantly, IT neurons underlie the intracortical circuity linking higher-order neocortical association areas [50]. Many IT cells are callosal neurons that project to the contralateral cerebral hemisphere through the corpus callosum [51]. Neocortical IT cells in layers 3 and 5 were first formally identified by Charles Wilson, through their projections to the contralateral striatum [52]. Our previous studies provided molecular evidence for neocortical input and output neuron homologs in the avian and turtle telencephalon [17]. IT cell homologs, however, have not to date been recognized outside of mammals, apart from the characterization of striatally projecting avian DT neurons [27, 53].
Archosaur reptiles do not possess a corpus callosum or any comparable DT commissure [54, 55]. However, birds have two vast association territories, the mesopallium and the nidopallium. The avian association territories receive input from other DT cell populations, most notably from the primary sensory input cells found in the entopallium and Field L, and connect broadly throughout the cerebral hemispheres. In particular, the mesopallium and the nidopallium interconnect with one another and send projections to the avian DT output nuclei. Based on connections, the mesopallium and the nidopallium are strong candidate homologs of mammalian neocortical IT neurons. We took advantage of the unique nuclear morphology of the avian DT to test whether the mesopallium or the nidopallium shares molecular properties with any neocortical cell population. Our tissue dissections, HOLT bioinformatics, and ISH confirmation studies proved remarkably successful: we identified six mesopallium-specific transcription factors and numerous non-transcription factor markers (see Figure S2). Five of the transcription factors, the MesoTFN (Satb2, Bcl11a, Foxp1, Id2, and Emx1), are expressed by mouse IT neurons. Satb2 and Bcl11a, which we found were co-expressed in mesopallium neurons, are genetically required for mouse IT neuron specification [40, 41]. We further found that at least two MesoTFN factors (BCL11A and FOXP1) are conserved across birds, and all five are expressed in the alligator dorsal cortex and mesopallium. Future studies, including on snakes and lizards, are likely to uncover further conservation of the MesoTFN across birds and reptiles.
Karten’s proposal of homologous cell types in the amniote DT provided an important conceptual advance for comparative neuroanatomy [18]. Previous generations of anatomists carved vertebrate brains into large territories including the neocortex, amygdala, Wulst, DVR, striatum, and so on. Recent attempts to compare telencephala at the level of these structures bred a dizzying array of conflicting interpretations [6, 56]. Karten recognized that the fundamental unit of brain organization is not a territory or structure, but rather the neuronal cell type defined by its circuit contributions. This indicates neuronal cell types can be reorganized over evolutionary time into new and highly divergent structures without changing their fundamental connectional and molecular identities. Here, we add a third conserved cell type to Karten’s original concept of conserved DT input and output cells, the IT cell. The common ancestor of amniotes, which gave rise to mammals, reptiles, and birds, had input, output, and IT cells. This hypothesis provides a clear resolution to the problem of the mesopallium and substantially pushes back the evolutionary time of origin of a significant neocortical cell type.
Most attempts to identify neocortex homologs in non-mammals conceptualized the neocortex in terms of layers. As a result, previous authors were fixated on the question of whether L2/3 homologs exist in birds and reptiles. This approach led to conflicting conclusions. On one hand, some authors concluded that the upper layers are a mammalian invention [57]: they were added de novo in early mammals to a primitive three-layered reptilian dorsal cortex. A second interpretation is that birds have a homolog of L2/3 in the mesopallium [31]. This idea is not supported by the available gene expression data, as none of the mesopallium-specific transcription factors we examined in mouse were restricted to neocortical L2/3. Rather, L2/3 neurons are a subset of IT neurons, a larger class defined principally by connections and, now, by additional gene expression data. As a technical point, neocortical L4/input neurons are also considered to be a subset of IT neurons [48], and the mesopallium homology we propose is with the non-L4 IT neurons of upper and deep layers (Figure 6).
The proposed IT neuron homology clarifies that cell-type homologies are not structural homologies of neocortical layers and avian nuclei. Homologs need not be confined to a particular layer or nucleus. An important implication of our hypothesis is that reptiles and birds do not have separate homologs of neocortical layers 2 and 3, as suggested previously [23]. Instead, IT neurons diversified into subclasses in upper and deep neocortical layers only after mammals diverged from reptiles. Similarly, the mammalian neocortex is unlikely to harbor separate homologs of the avian Md or Mv. These dorsal and ventral IT subpopulations may have emerged specifically in the reptilian lineage after it split from mammals.
Birds may be the most intelligent vertebrates outside of primates, but the two groups developed their wits independently. This could have been accomplished by the evolutionary elaboration of the same or of different neural circuits and cell types. Mammalian IT neuron populations, and in particular those of the upper layers, are thought to increase in number disproportionately in large-brained mammals. As the thickness of the neocortical sheet expands through phylogeny, especially in the primate lineage relative to rodents, upper neocortical layers come to occupy an increasingly large fraction of the total cortical depth [58]. Similarly, in large-brained birds like parrots and owls, the avian mesopallium and nidopallium are enlarged to a greater extent than are the primary sensory nuclei such as the entopallium [59]. Selective expansion of associative telencephalic cell populations may therefore represent a common mechanism for the evolution of higher cognitive functions in birds and mammals. These independent expansions may have involved homologous associational IT cell types, which would be a striking example of the parallel evolution of the neural bases of cognition.
Establishing the mesopallium as a territory of neocortical IT-homologous neurons highlights the mystery of the avian nidopallium. What is the mammalian nidopallium homolog, if not IT cells? The nidopallium could be homologous to neocortical L4/input cells, but this possibility is unsupported because only primary sensory divisions of the nidopallium (entopallium and Field L) express L4 marker genes [17]. The nidopallium makes associational connections similar to those of the mesopallium, but does not express the key mammalian IT determinants BCL11A and SATB2. We found that the nidopallium marker Dach2 is expressed in mouse neocortical L5a, but this data point should not be over-interpreted. Mouse Dach2 expression may identify a poorly understood input-subtype, output-subtype, or IT-like cell. Alternatively, Dach2 may have been independently recruited into the mammalian DT and the avian DT. Future studies should seek to identify nidopallium homologs in anamniote taxa [60].
Despite extensive conservation at the level of circuitry, the cellular architectures of the mammalian neocortex and the avian DT are dramatically divergent. Neocortical input, output, and IT neurons are distributed across the neocortical surface area and are often intermixed. Birds took a very different evolutionary path by grouping like-classes of cells into spatially segregated nuclei. These anatomical differences are surely accompanied by fundamental differences in the developmental mechanisms of cell-type specification and tissue morphogenesis. The avian DT nuclear architecture suggests that the different cell types arise from spatially segregated, dedicated progenitor pools in the ventricular zone. Our fate mapping data shows that this is the case, at least for avian IT cells. The avian mesopallium offers several advantages over the neocortex for investigating the early development of this fundamental cell class. In this study, we exploited avian DT anatomy, specifically the large size of DT nuclei, to identify novel marker genes and candidate cellular determinants of the mesopallium. The chicken model system for experimental molecular embryology may now facilitate the identification of IT-cell-specifying signals operating during early telencephalic patterning.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Steven Briscoe (stevendb@uchicago.edu).
Experimental model and subject details
All animal procedures were reviewed and approved by the University of Chicago Institutional Animal Care and Use Committee (IACUC).
The sex of the chickens, starlings, mice, and alligators was not determined.
The chicken Gallus gallus
Embryonic and hatchling chickens were raised from fertilized White Leghorn chicken (Gallus gallus) eggs purchased from Sunnyside Hatchery (Beaver Dam, Wisconsin). These eggs were incubated at 38°C with 90% relative humidity in a Model 1502 circulated air incubator (GQF Manufacturing Company, Savannah, Georgia). Eggs were manually rotated 180° three times per day for the first four days of incubation to promote viability. Embryonic day zero (E0) begins at the start of incubation, post-hatching day zero (P0) on the day of hatching.
The mouse Mus musculus
Postnatal CD-1 mice (Mus musculus) were provided by the University of Chicago Transgenic Facility, via the laboratory of E. Grove at the University of Chicago.
The starling Sturnus vulgaris
Adult, wild-caught starlings (Sturnus vulgaris) were provided by the laboratory of D. Margoliash at the University of Chicago.
The alligator Alligator mississippiensis
Fertilized alligator (Alligator mississippiensis) eggs were provided by Ruth Elsey and colleagues at the Rockefeller Wildlife Refuge (Grand Chenier, Louisiana). Alligator eggs were transported by car in maternally provided nesting material in moistened, perforated Styrofoam boxes. Eggs were subsequently transferred to a GQF incubator where they were incubated at 30°C and 90% relative humidity. Embryos were staged according to the criteria of Ferguson [61]. We studied stage 25 animals, which are defined as resembling “a miniature version of a hatchling, with a considerable volume of external yolk and a large umbilical region.” Stage 25 typically corresponds to sixty days after egg laying and one week before hatching.
Method Details
RNA sequencing and analysis
Embryonic day 14 chicken brains were dissected out whole and sliced into 300 μm thick sections with a McIlwain Tissue Chopper (The Mickle Laboratory Engineering Company). Nuclei were dissected from sections in diethyl pyrocarbonate (DEPC, Sigma-Aldrich) treated PBS using forceps and transferred to RNAlater (Life Technologies) where they were stored overnight at 4°C.
RNA was extracted using Trizol Reagent, suspended in nuclease-free water, and submitted to the University of Chicago Functional Genomics Core Facility for Poly A+ selected library preparation and Illumina 100 base pair paired-end directional sequencing. Raw reads were quality trimmed with Trimmomatic [62], digitally normalized, and assembled de novo with Trinity [63, 64]. Trimmed reads were mapped onto assembled transcripts using RSEM and abundance estimates were TMM-normalized (trimmed mean of M-values). Fragments per kilobase of transcript per million mapped reads (FPKM) were used for direct comparisons across samples by controlling for length of transcript (longer transcripts generate more fragments) and for total numbers of fragments per sample.
To identify transcripts enriched in particular nuclei, we devised the differential expression analysis of High-Low Ordering of Like Transcripts (HOLT). HOLT is a series of sequential pairwise comparisons of transcript abundance that progressively filters reads to those enriched in particular samples. For instance, in order to identify transcripts enriched in sample A compared with samples B and C, we performed the following operations (order does not matter and comparison ratios can be varied for stringency): A/B > 2.5 then A/C > 2.5. The filtered transcript list was ordered by abundance in sample A to identify the most highly expressed candidate genes. Transcripts were annotated using BLASTN and BLASTX (NCBI) searches. Trial experiments with a reduced stringency (2 fold enrichment) proved to yield many transcripts not selective for the mesopallium in ISH experiments. In contrast, 3-fold enrichment was too stringent, excluding mesopallium-selective marker genes identified with 2.5 fold filtering.
We compared the list of candidate genes identified with the HOLT Method to those identified with edgeR, an established method of differential expression analysis [65]. We used edgeR to identify genes differentially expressed across all seven collected DT territories with a minimum 2-fold change in expression, a false discovery rate (FDR) of 0.05, and a minimum FPKM of 5. This edgeR analysis identified 21 candidate protein-coding genes as upregulated in the mesopallium compared with the entopallium and the arcopallium. EdgeR recovered NHLH2 and SATB2, but missed the key mesopallium-specific transcription factors ID2, FOXP1, and BCL11A (see Supplemental Table 1). EdgeR did not detect any transcription factors not also detected with the HOLT method. Overall, all but three of the transcripts for protein-coding genes identified by edgeR were also identified by HOLT. We conclude that there is significant overlap in the results obtained with edgeR and the HOLT method. Compared to edgeR, the HOLT method has the distinct advantages of being simple, intuitive, and highly effective for the identification of molecular marker genes.
cDNA preparation
Dissected brain tissue of all species was flash-frozen on dry ice and stored at −80°C, or was used immediately for RNA extraction. Tissue was homogenized with a pestle and RNA was extracted with Trizol Reagent (Invitrogen) following manufacturer instructions. RNA was either stored at −80°C in RNase-free water (Sigma) or used immediately for cDNA synthesis [SuperScript III 1st strand cDNA kit (Fisher Scientific)] following manufacturer instructions. cDNA was diluted in RNase-free water and stored at −20°C.
Molecular cloning
For all species examined, EST sequence was readily available from the National Center for Biotechnology Information (NCBI). Polymerase chain reaction (PCR) primers were designed using deposited sequence and Primer3 online software (bioinfo.ut.ee/primer3-0.4.0/). PCR reaction mixtures comprised standard reagents and concentrations (1x polymerase buffer, 0.75mM MgCl2, 200 μM dNTP, 0.2 μM primers, 1.25 units Taq DNA polymerase per 50 μL solution). PCR reactions were performed using a RoboCycler Gradient 40 (Stratagene). Reaction solutions were initially heated to 94°C for 5 minutes, followed by 40 cycles of 94°C for 1 minute, 50–55°C for 1 minute, and 72°C for 1 minute per kilobase of product length. A final elongation step was performed at 72°C for 10 minutes. PCR product length was estimated by running 5 μl of the 50 μl reaction solution on a 1% agarose gel alongside a DNA ladder. When necessary, target DNA bands were gel-purified using a Zymoclean Gel DNA Recovery Kit (Zymo Research).
PCR products were ligated into the pGEM-T Easy Vector (Promega) for 1–7 days at 4°C, then transfected into DH5α competent cells at 42°C for 30 seconds. Transformed cells were plated on ampicillin-positive LB agar plates with X-Gal (Goldbio) for blue/white selection and incubated overnight at 37°C. Plates were stored at 4°C for 1–4 days to allow for blue-colored colonies to darken. White colonies were picked and miniprepped using PureLink Quick Plasmid DNA Miniprep Kits (Invitrogen). Plasmids were sequenced with T7 and SP6 primers at the on-campus UC Comprehensive Cancer Center DNA Sequencing Facility. cDNA sequences were analyzed with BLAST (NCBI) and annotated with A Plasmid Editor software (ApE, M. Wayne Davis). Plasmids were stored at −20°C.
Brain tissue preparation
Embryonic chicken brains up to E14 and stage 25 alligator brains were collected by decapitation, dissection, and immersion fixation in 4°C PFA (solution of 4% paraformaldehyde in PBS). Late embryonic and hatchling chickens, adult starlings, and postnatal mice were anesthetized with Euthasol deeply and perfused transcardially with PFA prior to tissue collection.
All fixed brain tissue was cryogenically protected by equilibration in 30% sucrose/PBS overnight at 4°C before sectioning. Sucrose-saturated brains were mounted on a freezing sledge microtome (Leica SM2000R) and sections were cut at a thickness of 20–24 microns. Sections were collected in DEPC-PBS, mounted onto Superfrost Plus slides (Fisher Scientific), and dried for 20 minutes at room temperature in a fume hood before further processing for in situ hybridization. Whenever possible, tissue was sliced, mounted, and processed in a single day for maximal in situ hybridization signal.
Single-color section in situ hybridization
RNA probe template preparation.
75 μg of miniprepped plasmid DNA were linearized by overnight restriction digestion, standardly employing SacII or Spe1 enzymes (New England Biolabs). Other enzymes were chosen as needed to avoid internal cDNA cut sites. Digested DNA was treated with 5 μl 10% SDS (Bio-Rad) and 5 μl proteinase K (Roche) for 15 minutes at 37°C, phenol/chloroform (Acros Organics) extracted, and ethanol precipitated. The dried DNA pellet was resuspended in 10 μl of Sigma water.
DIG-conjugated antisense RNA probe synthesis.
RNA transcription reactions comprised 4 μl water, 4 μl 10x bovine serum albumin, 2 μl 10x transcription buffer, 2 μl 10x DIG-labeling nucleotide mix (Roche), 2 μl Protector RNase inhibitor (Roche), 2 μl DTT, 2 μl template DNA, and 2 μl of RNA polymerase T7/SP6 (New England Biolabs). The transcription reaction was carried out overnight at 37°C. The reaction solution was treated with 4 μl of RNase-free DNase (Roche) for 1 hour at 37°C. RNA was purified with two consecutive ethanol/LiCl precipitations and suspended in 100 μl of formamide (Sigma-Aldrich). RNA in formamide was heated at 50°C for 15 minutes to remove secondary structure before being imaged on a 1% agarose gel with ethidium bromide. RNA probes were judged acceptable if one or more bright, tight bands were visible on the gel.
Pre-hybridization tissue processing.
Completely dried slides bearing brain sections were post-fixed in PFA for 15 minutes at room temperature and rinsed three times with DEPC-PBS (PBS treated with diethyl pyrocarbonate). Tissue was then treated with proteinase K in incubation buffer (1.5 μl stock proteinase K per 20 ml 100 mM Tris-HCL pH8.0, 50 mM EDTA pH 8.0) for 30 minutes at 37°C. Digested tissue was fixed in PFA for 15 minutes, rinsed three times with DEPC-PBS, and transferred to 5-slot slide mailers (Evergreen Scientific) containing 15 ml hybridization solution (50% formamide, 5x SSC, 1% SDS, 0.25 grams yeast RNA [Roche], and 0.1 grams heparin sulfate [Alfa aesar] per 500 ml).
Hybridization and antibody binding.
Slides in hybridization solution were heated in a 73°C Isotemp 210 water bath (Fisher Scientific) for 30 minutes. Hybridization solution was then decanted into a 50 ml conical tube (Denville Scientific), rapidly mixed with 95 μl antisense RNA probe reaction, and returned to the slide mailer for an overnight incubation at 73°C. The next day, RNA solution was decanted into a 50 ml conical and stored at −20°C for up to 2 additional uses. Slides were washed three times for one hour each at 73°C in preheated Solution X (50% formamide, 2x SSC, 1% SDS). Following Solution X washes, slides were washed briefly three times in TBST (A 10x TBS stock was first prepared with 250 ml Tris-HCl pH 7.5, 80 g NaCl, 2 g KCl per 1 L. This was diluted and combined with 10 ml Tween for 1 L of 1X TBST). Slides were blocked for 1 hr in 10% lamb serum (Invitrogen) in TBST at room temperature.
Hybridized complexes of antisense probe/target mRNA were detected with anti-DIG Fab fragments conjugated to the colorigenic enzyme alkaline phosphatase (Roche). The antibody-enzyme conjugate was preadsorbed with chick embryo powder/1% lamb serum in TBST, and then diluted to 1:5000 in TBST/1% lamb serum (3 μl of antibody per 15 ml of TBST/1% lamb serum per mailer). Antibody solution was poured into slide mailers and antibody binding was carried out for 2 hours at room temperature on a rocker. Antibody solution was decanted and stored at 4°C for up to 2 additional uses. Unbound antibody was removed by washing slides three times for 15 min in TBST. Slides were briefly washed once in NTMT (100 mM Tris-HCl pH 9.5, 100 mM NaCl, 50 mM MgCl2, 1% Tween).
Color reaction.
Antibody/mRNA was detected using phosphatase histochemistry. 5-bromo-4-chloro-3-indolyl phosphate (BCIP, Denville Scientific) stocks were produced by diluting 50 mg powder per 1 ml 100% dimethyl formamide (DMF, Acros Organics). The formazan nitro blue tetrazolium (NBT, Denville Scientific) stock was produced by mixing 100 mg powder in 1 ml 70% DMF in water. Color reagent stocks were stored at −20°C. Slides were incubated in 4 μl BCIP and 4 μl NBT stock per 1 ml NTMT at room temperature for 1–7 days. For best results, post-hybridization washes through to the color reaction were done in a single day. Color reaction solution was replenished every day for the duration of color development. The length of color reaction depended on several factors including length of probe used and abundance of target mRNA in the tissue. Highly expressed target genes can develop a clear signal in minutes, while low-abundance target genes may require 5 days of incubation to be visible. Following the color reaction, slides were washed twice in Stop TE buffer (10 mM Tris-HCl pH 7.5, 10 mM EDTA pH 8.0) then once more for 2 hours to overnight. Slides were washed overnight in TBST and fixed overnight in 10% Formalin (Fisher Scientific) in PBS. Slides were rinsed in PBS, dried, dehydrated in an ethanol series, soaked in Histoclear (National Diagnostics) and coverslipped using Eukitt mounting medium (Sigma-Aldrich) and microscope cover glass (Fisher Scientific).
Two-color fluorescent in situ hybridization (FISH)
Antisense RNA probe synthesis.
DIG- and DNP-labeled RNA probes were employed for all FISH experiments. RNA probes were synthesized as described above, but with DNP-labeled UTP and other ribonucleotides (Perkin Elmer) substituted for DIG labeling mix as needed.
Pre-hybridization tissue processing.
Slides with brain sections were prepared as described above, but with one additional step. To inactivate endogenous peroxidases, tissue was incubated in 2% H2O2/50% MeOH in DEPC-PBS for 20 minutes at room temperature prior to proteinase K treatment.
Hybridization, antibody binding, and color reaction.
Both probes were hybridized simultaneously overnight at 73°C, but detected by antibodies sequentially. Slides were washed three times for one hour each at 73°C in preheated Solution X. Following Solution X washes, slides were washed briefly three times in TBST. Slides were blocked in 20% DIG Blocking Buffer (Sigma-Aldrich) in TBST for 1 hr at 4°C. Slides were carefully dried with a paper towel, and a box was drawn around the brain slices with a hydrophobic marker. Slides were set horizontally in a dark, moistened chamber for the first antibody incubation. Anti-DNP-HRP (Perkin Elmer) and Anti-DIG-POD (Roche) antibodies were preadsorbed as described above and diluted 1:200 in TBSTB. 200 μl of antibody solution was then pipetted directly upon brain slices and incubated for 2 hrs at room temperature. Slides were wiped of hydrophobic residue, washed three times in TBST for 15 minutes each, and washed twice in TNT (0.1M Tris HCl [pH 7.5], 0.15M NaCl, 0.05% Tween 20) for 5 minutes each.
Slides were again dried, traced with the hydrophobic pen, and placed in the dark moist chamber for the color reaction. The selected tyramide, either Cy3 or Cy5 (Perkin Elmer), was diluted 1:50 in amplification diluent and 150 μl was pipetted atop each slide for a 1 hr color reaction. Slides were washed three times for 15 minutes each in TNT, quenched in 2% H2O2/DEPC-PBS for 20 minutes, and washed two times in TBST for 15 minutes and once more overnight. The second antibody reaction was then carried out, repeating steps through the second color reaction. Following the second color reaction, slides were rinsed in PBS and coverslipped using Fluoromount-G (SouthernBiotech).
Single-color whole mount in situ hybridization
Whole brains were dissected from chick embryos in PBS, then fixed overnight in PFA at 4°C. Tissue was washed in PTW (1% Tween in PBS) two times for five minutes each at room temperature. Tissue was dehydrated in 25%, 50%, 75% and 100% MeOH in PTW for ten minutes each, followed by overnight incubation in 100% MeOH at −20°C. Tissue was rehydrated in 75%, 50%, and 25% MeOH in PTW for five minutes each, then washed in PTW twice for five minutes. Tissue was incubated in 6% hydrogen peroxide in PTW for 60 minutes at room temperature on a rocker, then washed two times in PTW for five minutes each. Tissue was incubated in detergent mix three times for 30 minutes each at room temperature, post-fixed in 1% Tween in PFA for 20 minutes, washed twice in PTW for five minutes each, then stored in glass scintillation vials with 5 ml of hybridization solution at −20°C.
Tissue was prehybridized for 1 hr and hybridized overnight at 73°C with 33 μl of the 100 μl RNA transcription solution, prepared as described above. Tissue was washed in preheated Solution X four times for 30 minutes each. TBST was then added, which causes tissue to settle to the bottom of the vial, and washed three more times in TBST for five minutes each. Tissue was blocked in 10% lamb serum in TBST for two hours at room temperature. Incubation solution was then replaced with five ml 1% lamb serum in TBST. Antibody solution was prepared as described above. One μl of antibody was added to each five ml scintillation vial. Tissue was incubated overnight at 4°C on a rocker.
The brains were washed in TBST three times for five minutes each, five times for 60 minutes each, and overnight at room temperature. Color reaction was performed as described above.
In ovo electroporation
Transgene overexpression was accomplished using in ovo electroporation of embryonic chicken telencephalon [66]. Chicken eggs for electroporation were incubated on their sides and rotated regularly. Eggs were set in the incubator on the evening of receipt and electroporated in the morning 4 days later; electroporated eggs were therefore E3.5 or HH23, an ideal electroporation stage given the size and accessibility of the telencephalon. The day before electroporation, 4 ml of albumin were extracted from the pointed end of the egg with a needle attached to a syringe. The day of electroporation, a small window was cut into the egg directly above the expected location of the embryo. Four drops of Ringer’s solution (9 g NaCl, 0.42 g KCl, and 0.16 g CaCl2 per 1 L water) were deposited on the embryo for hydration.
Borosilicate Glass Capillaries (World Precision Instruments, Inc) were tapered with a Model P-97 Flaming/Brown micropipette puller (Sutter Instrument Co.), and their tips manually cut open to a width of 40 μm. Gene expression plasmids were prepared using an Origene Maxiprep System, phenol/chloroform extraction, ethanol precipitation, and resuspension in 100 μl TE buffer (Origene) for a concentration of approximately 5–15 μg DNA per μl TE. The electroporation mix comprised one or more plasmids at a final concentration of 1–2 μg per plasmid per μl TE with 0.02% Fast Green dye. The electroporation mix was back-filled into the open glass capillary using a PV830 Pneumatic PicoPump (World Precision Instruments).
The filled capillary was inserted into the right telencephalon of the chick embryo and the electroporation mix was injected until green dye filled the lateral ventricle. A negative electrode (made from 0.025 mm tungsten wire) was inserted into the right telencephalon and a positive electrode (0.063 mm platinum wire) was positioned outside but very close to the neuroepithelium. A Model 2100 Isolated Pulse Stimulator (A-M systems) was used to generate an electrical field by delivering three 25 millisecond pulses of 9 V each with an inter-pulse period of 1 second. The appearance of bubbles along both electrodes indicated a successful pulse delivery. For more restricted electroporation sites, 1 pulse was delivered. The electric field generated drives DNA from the ventricle into neural progenitor cells. Both electrodes were subsequently dipped into 5M NaOH and wiped with a Kimwipe (Kimtech Science) to remove any encrusted film. Eggs were sealed with Scotch packing tape and returned to the incubator. Because the electroporation mix always included a fluorescent reporter molecule, embryos could be examined the next morning using a Leica Fluo III fluorescent microscope to screen for successfully electroporated animals. Eggs displaying reported fluorescence were given 4 drops of Ringer’s solution, sealed with Parafilm “M” Laboratory Film (Pechiney Plastic Packaging), and returned to the incubator for another 10 days (to E14).
Image capture and analysis
Images of stained brain sections were captured with a Leica Fluo III microscope or a Zeiss Axioskop 40, a mounted AxioCam HRc color camera (Zeiss), and AxioVision software (version 4.8, Zeiss). Images were cropped and corrected for brightness using Photoshop CS6 (Adobe Systems).
Fluorescent images were captured using a Leica SP5 2 photon microscope at the University of Chicago Integrated Light Microscopy Core. Images were adjusted for contrast using FIJI software [67].
Illustrations were produced using Illustrator CS6 (Adobe Systems).
Supplementary Material
Table S2 (Related to Figures 1 and 2) Top 145 nidopallium coding-transcripts
For the nidopallium HOLT analysis, we selected transcripts enriched in the nidopallium in comparison with the mesopallium by 2.5 fold. This resulted in 97,261 candidate transcripts. From these, we selected coding genes expressed at 5 FPKM or higher in nidopallium, yielding 145 genes. The 145 most abundant nidopallium-specific genes are listed by descending levels of expression. Transcriptional regulator genes are in bold type.
Table S1 (Related to Figure 2) Top 78 mesopallium coding-transcripts
The 78 most abundant mesopallium-specific genes are listed by descending levels of expression. Transcriptional regulator genes are in bold type. EMX1 was not recovered by the HOLT method because it is expressed in a subnucleus of the arcopallium, but it is included at the bottom of the list. Gene names marked with a plus (+) were confirmed as mesopallium-specific with ISH. Those with a minus (−) were expressed in the mesopallium but were not mesopallium-specific. There were no false positives among the genes examined by ISH. Genes without either a plus or minus sign were not examined by ISH. Gene names with (eR) were also detected by a parallel bioinformatic analysis with edgeR (see STAR Methods).
Figure S1 (Related to Figure 2) Chick dorsal telencephalon nuclei collected for RNAseq analysis
In situ hybridization for the conserved input cell marker KCNH5 on E14 chick telencephalon. Five coronal sections from a single cerebral hemisphere are shown in anterior to posterior sequence. The seven indicated cell populations were harvested from embryonic day 14 telencephalon: (A) 1. Hyperpallium apicale (HA), 2. Mesopallium (M), (B) 3. Nucleus basorostralis (Bas), (C) 4. Entopallium (E), (D) 5. Field L (L), (E) 6. Nidopallium (N), and 7. Arcopallium (A). RNA was purified from these cell populations and submitted for sequencing.
Figure S2 (Related to Figures 2 and 3) In situ validation of mesopallium candidate genes identified by HOLT
(A–F) In situ hybridization for candidate mesopallium marker genes identified by the HOLT method. All sections are from embryonic day 14 chick telencephalon. Our approach recovered many highly specific mesopallium marker genes. Gene expression suggests that the mesopallium has a consistent internal molecular identity and is dominated by a single principal neuronal type.
(G–N) An anteroposterior series of sections from a single postnatal day zero chick telencephalon labeled for NHLH2 transcript. The Wulst dorsal mesopallium (Md) and the DVR ventral mesopallium (Mv) are clearly distinguished and can be followed along the anteroposterior axis. (N) Caudal mesopallium, for instance, is continuous with the rostral part of Mv. Asterisk in G-K indicates the vallecula, a shallow sulcus that defines the boundary between the DVR ventrally and the Wulst dorsally.
Figure S3 (Related to Figure 3) Mesopallium molecular organization is conserved across birds
We performed in situ hybridization for the mesopallium marker FOXP1 and the IHA marker KCNH5 on serial sections through embryonic day 20 chick telencephalon. Asterisks in all panels indicate the vallecula.
(A–B) The chick Md, as labeled by FOXP1, extends from the vallecula to the IHA, seen in (B). We conclude that our chicken Md is equivalent to the structure referred to as HD by avian neuroanatomists.
(C–D) Adjacent sections through the anterior telencephalon of an adult starling demonstrate that the FOXP1-expressing mesopallium extends to the (D) KCNH5-expressing IHA.
(E–F) Sections through (E) intermediate and (F) posterior levels of the starling telencephalon demonstrate conserved expression of BCL11A in Md and Mv.
Figure S4 (Related to Figure 3) Emx1 and Id2 are expressed in the postnatal mouse neocortex
Expression patterns of the avian mesopallium transcription factors Satb2, Bcl11a, and Foxp1 in the mouse neocortex were previously reported. (A) Emx1 and (B) Id2 are also expressed in the mouse neocortex at postnatal day six (P6). Both are prominently expressed in L2/3 in addition to the deep layers.
Figure S5 (Related to Figure 1) The nidopallium transcription factor Dach2 is expressed in mouse neocortical layer 5
(A–C) An anteroposterior series through a single embryonic day 20 chick telencephalon labeled for DACH2 transcript. The entire nidopallium (N) is labeled, with sharp boundaries where it meets the mesopallium (M), arcopallium (A), and ventral telencephalon (VT). We did not identify other molecular markers that capture this pattern. Chicken DACH2 is also expressed in the IHA (A).
(D) Mouse Dach2 is expressed in a thin, laminar pattern in the neocortex at postnatal day six. This transcript is present at extremely low levels and was detected by combination in situ hybridization employing two non-overlapping probes. Mouse Dach2 expression was also detected in the olfactory cortex, the cortical amygdala, and the septum (data not shown). Expression was not detected in the basolateral amygdala, which has been proposed as a mammalian homolog of the avian nidopallium [32].
(E) A section adjacent to that in D labeled with a riboprobe for the well-characterized L5/output cell marker Fezf2. Comparison with Dach2 staining indicates that the Dach2 lamina is contained within L5, possibly in an upper part of L5 corresponding to L5a. The arrowheads in D indicate the somatosensory whisker barrels, prominent clusters of L4/input cells in rodent primary somatosensory cortex. They are faintly visible as grey blobs and are located just above the Dach2 stripe, further suggesting that Dach2 labels cells in L5a.
Acknowledgements:
We thank the Grove lab at the University of Chicago for providing mice and sharing reagents, the Margoliash lab at the University of Chicago for providing starlings, and Ruth Elsey and colleagues at the Rockefeller Wildlife Refuge for providing alligator eggs. We thank Vytas Bindokas and the Integrated Light Microscopy Core Facility at University of Chicago for assistance with imaging, and Timothy Sanders for providing overexpression plasmids. We thank Tanya Shpigel for editing expertise. S.D.B. thanks his funding sources: the Molecular and Cellular Biology Training Program (T32 GM007183) and the Developmental Biology Training Program (HD055164).
Data and Software Availability
RNA sequencing reads have been deposited in the Short Read Archive at NCBI under Bioproject number PRJNA416004. Antisense RNA probe sequences are being uploaded to the nucleotide database at NCBI.
References:
- 1.Roth G (2015). Convergent evolution of complex brains and high intelligence. Phil. Trans. R. Soc. B 370, 20150049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates LA, Poole JH, and Byrne RW (2008). Elephant cognition. Curr. Biol 18, R544–546. [DOI] [PubMed] [Google Scholar]
- 3.Marino L (2004). Dolphin cognition. Curr. Biol 14, R910–911. [DOI] [PubMed] [Google Scholar]
- 4.Sousa AMM, Meyer KA, Santpere G, Gulden FO, and Sestan N (2017). Evolution of the human nervous system function, structure, and development. Cell 170, 226–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florio M, Borrell V, and Huttner WB (2017). Human-specific genomic signatures of neocortical expansion. Curr. Opin. Neurobiol 42, 33–44. [DOI] [PubMed] [Google Scholar]
- 6.Striedter GF (2005). Principles of Brain Evolution, (Sunderland, MA: Sinauer Associates; ). [Google Scholar]
- 7.Olkowicz S, Kocourek M, Lucan RK, Portes M, Fitch WT, Herculano-Houzel S, and Nemec P (2016). Birds have primate-like numbers of neurons in the forebrain. Proc. Natl. Acad. Sci. USA 113, 7255–7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balakhonov D, and Rose J (2017). Crows rival monkeys in cognitive capacity. Sci. Rep 7, 8809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weir AA, Chappell J, and Kacelnik A (2002). Shaping of hooks in New Caledonian crows. Science 297, 981. [DOI] [PubMed] [Google Scholar]
- 10.Prior H, Schwarz A, and Gunturkun O (2008). Mirror-induced behavior in the magpie (Pica pica): evidence of self-recognition. PLoS Biol. 6, e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wasserman EA (2016). Conceptualization in pigeons: the evolution of a paradigm. Behav. Processes 123, 4–14. [DOI] [PubMed] [Google Scholar]
- 12.Pepperberg IM (1999). The Alex Studies: Cognitive and Communicative Abilities of Grey Parrots, (Cambridge, MA: Harvard University Press; ). [Google Scholar]
- 13.Emery NJ, and Clayton NS (2004). The mentality of crows: convergent evolution of intelligence in corvids and apes. Science 306, 1903–1907. [DOI] [PubMed] [Google Scholar]
- 14.Ulinski PS (1983). Dorsal Ventricular Ridge: A Treatise on Forebrain Organization in Reptiles and Birds, (New York: John Wiley & Sons; ). [Google Scholar]
- 15.Reiner A, Perkel DJ, Bruce LL, Butler AB, Csillag A, Kuenzel W, Medina L, Paxinos G, Shimizu T, Striedter G, et al. (2004). Revised nomenclature for avian telencephalon and some related brainstem nuclei. J. Comp. Neurol 473, 377–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dugas-Ford J, and Ragsdale CW (2015). Levels of homology and the problem of neocortex. Annu. Rev. Neurosci 38, 351–368. [DOI] [PubMed] [Google Scholar]
- 17.Dugas-Ford J, Rowell JJ, and Ragsdale CW (2012). Cell-type homologies and the origins of the neocortex. Proc. Natl. Acad. Sci. USA 109, 16974–16979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karten HJ (1969). The organization of the avian telencephalon and some speculations on the phylogeny of the amniote telencephalon. Ann. N. Y. Acad. Sci 167, 164–179. [Google Scholar]
- 19.Karten HJ (2015). Vertebrate brains and evolutionary connectomics: on the origins of the mammalian ‘neocortex’. Phil. Trans. R. Soc. B 370, 20150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Atoji Y, and Wild JM (2012). Afferent and efferent projections of the mesopallium in the pigeon (Columba livia). J. Comp. Neurol 520, 717–741. [DOI] [PubMed] [Google Scholar]
- 21.Atoji Y, and Wild JM (2009). Afferent and efferent projections of the central caudal nidopallium in the pigeon (Columba livia). J. Comp. Neurol 517, 350–370. [DOI] [PubMed] [Google Scholar]
- 22.Gunturkun O (2005). The avian ‘prefrontal cortex’ and cognition. Curr. Opin. Neurobiol 15, 686–693. [DOI] [PubMed] [Google Scholar]
- 23.Jarvis ED, Yu J, Rivas MV, Horita H, Feenders G, Whitney O, Jarvis SC, Jarvis ER, Kubikova L, Puck AE, et al. (2013). Global view of the functional molecular organization of the avian cerebrum: mirror images and functional columns. J. Comp. Neurol 521, 3614–3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakamori T, Sato K, Atoji Y, Kanamatsu T, Tanaka K, and Ohki-Hamazaki H (2010). Demonstration of a neural circuit critical for imprinting behavior in chicks. J. Neurosci 30, 4467–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karten HJ, Hodos W, Nauta WJ, and Revzin AM (1973). Neural connections of the “visual wulst” of the avian telencephalon. Experimental studies in the piegon (Columba livia) and owl (Speotyto cunicularia). J. Comp. Neurol 150, 253–278. [DOI] [PubMed] [Google Scholar]
- 26.Medina L, and Reiner A (2000). Do birds possess homologues of mammalian primary visual, somatosensory and motor cortices? Trends Neurosci. 23, 1–12. [DOI] [PubMed] [Google Scholar]
- 27.Veenman CL, Wild JM, and Reiner A (1995). Organization of the avian “corticostriatal” projection system: a retrograde and anterograde pathway tracing study in pigeons. J. Comp. Neurol 354, 87–126. [DOI] [PubMed] [Google Scholar]
- 28.Brauth SE, and McHale CM (1988). Auditory pathways in the budgerigar. II. Intratelencephalic pathways. Brain Behav. Evol 32, 193–207. [DOI] [PubMed] [Google Scholar]
- 29.Krutzfeldt NO, and Wild JM (2004). Definition and connections of the entopallium in the zebra finch (Taeniopygia guttata). J. Comp. Neurol 468, 452–465. [DOI] [PubMed] [Google Scholar]
- 30.Butler AB, Reiner A, and Karten HJ (2011). Evolution of the amniote pallium and the origins of mammalian neocortex. Ann. N. Y. Acad. Sci 1225, 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki IK, Kawasaki T, Gojobori T, and Hirata T (2012). The temporal sequence of the mammalian neocortical neurogenetic program drives mediolateral pattern in the chick pallium. Dev. Cell 22, 863–870. [DOI] [PubMed] [Google Scholar]
- 32.Bruce LL, and Neary TJ (1995). The limbic system of tetrapods: a comparative analysis of cortical and amygdalar populations. Brain Behav. Evol 46, 224–234. [DOI] [PubMed] [Google Scholar]
- 33.Puelles L, Ayad A, Alonso A, Sandoval JE, MartInez-de-la-Torre M, Medina L, and Ferran JL (2016). Selective early expression of the orphan nuclear receptor Nr4a2 identifies the claustrum homolog in the avian mesopallium: impact on sauropsidian/mammalian pallium comparisons. J. Comp. Neurol 524, 665–703. [DOI] [PubMed] [Google Scholar]
- 34.Wagner GP (2014). Homology, Genes, and Evolutionary Innovation, (Princeton, NJ: Princeton University Press; ). [Google Scholar]
- 35.Strausfeld NJ, and Hirth F (2013). Deep homology of arthropod central complex and vertebrate basal ganglia. Science 340, 157–161. [DOI] [PubMed] [Google Scholar]
- 36.Sun Z, and Reiner A (2000). Localization of dopamine D1A and D1B receptor mRNAs in the forebrain and midbrain of the domestic chick. J. Chem. Neuroanat 19, 211–224. [DOI] [PubMed] [Google Scholar]
- 37.Chen CC, Winkler CM, Pfenning AR, and Jarvis ED (2013). Molecular profiling of the developing avian telencephalon: regional timing and brain subdivision continuities. J. Comp. Neurol 521, 3666–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernandez AS, Pieau C, Reperant J, Boncinelli E, and Wassef M (1998). Expression of the Emx-1 and Dlx-1 homeobox genes define three molecularly distinct domains in the telencephalon of mouse, chick, turtle and frog embryos: implications for the evolution of telencephalic subdivisions in amniotes. Development 125, 2099–2111. [DOI] [PubMed] [Google Scholar]
- 39.Jarvis ED, Mirarab S, Aberer AJ, Li B, Houde P, Li C, Ho SY, Faircloth BC, Nabholz B, Howard JT, et al. (2014). Whole-genome analyses resolve early branches in the tree of life of modern birds. Science 346, 1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, and McConnell SK (2008). Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron 57, 364–377. [DOI] [PubMed] [Google Scholar]
- 41.Woodworth MB, Greig LC, Liu KX, Ippolito GC, Tucker HO, and Macklis JD (2016). Ctip1 regulates the balance between specification of distinct projection neuron subtypes in deep cortical layers. Cell. Rep 15, 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hisaoka T, Nakamura Y, Senba E, and Morikawa Y (2010). The forkhead transcription factors, Foxp1 and Foxp2, identify different subpopulations of projection neurons in the mouse cerebral cortex. Neuroscience 166, 551–563. [DOI] [PubMed] [Google Scholar]
- 43.Rubenstein JL, Anderson S, Shi L, Miyashita-Lin E, Bulfone A, and Hevner R (1999). Genetic control of cortical regionalization and connectivity. Cereb. Cortex 9, 524–532. [DOI] [PubMed] [Google Scholar]
- 44.Green RE, Braun EL, Armstrong J, Earl D, Nguyen N, Hickey G, Vandewege MW, St John JA, Capella-Gutierrez S, Castoe TA, et al. (2014). Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346, 1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Striedter GF (2016). Evolution of the hippocampus in reptiles and birds. J. Comp. Neurol 524, 496–517. [DOI] [PubMed] [Google Scholar]
- 46.Scalia F, Halpern M, and Riss W (1969). Olfactory bulb projections in the south american caiman. Brain Behav. Evol 2, 238–262. [Google Scholar]
- 47.Szele FG, Chin HK, Rowlson MA, and Cepko CL (2002). Sox-9 and cDachsund-2 expression in the developing chick telencephalon. Mech. Dev 112, 179–182. [DOI] [PubMed] [Google Scholar]
- 48.Harris KD, and Shepherd GM (2015). The neocortical circuit: themes and variations. Nat. Neurosci 18, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shepherd GM (2013). Corticostriatal connectivity and its role in disease. Nat. Rev. Neurosci 14, 278–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Felleman DJ, and Van Essen DC (1991). Distributed hierarchical processing in the primate cerebral cortex. Cereb. Cortex 1, 1–47. [DOI] [PubMed] [Google Scholar]
- 51.Economo MN, Clack NG, Lavis LD, Gerfen CR, Svoboda K, Myers EW, and Chandrashekar J (2016). A platform for brain-wide imaging and reconstruction of individual neurons. Elife 5, e10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson CJ (1987). Morphology and synaptic connections of crossed corticostriatal neurons in the rat. J. Comp. Neurol 263, 567–580. [DOI] [PubMed] [Google Scholar]
- 53.Reiner A, Stern EA, and Wilson CJ (2001). Physiology and morphology of intratelencephalically projecting corticostriatal-type neurons in pigeons as revealed by intracellular recording and cell filling. Brain Behav. Evol 58, 101–114. [DOI] [PubMed] [Google Scholar]
- 54.Letzner S, Simon A, and Gunturkun O (2016). Connectivity and neurochemistry of the commissura anterior of the pigeon (Columba livia). J. Comp. Neurol 524, 343–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riss W, Halpern M, and Scalia F (1969). The quest for clues to forebrain evolution - The study of reptiles. Brain Behav. Evol 2, 1–50. [Google Scholar]
- 56.Belgard TG, Montiel JF, Wang WZ, Garcia-Moreno F, Margulies EH, Ponting CP, and Molnar Z (2013). Adult pallium transcriptomes surprise in not reflecting predicted homologies across diverse chicken and mouse pallial sectors. Proc. Natl. Acad. Sci. USA 110, 13150–13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheung AF, Pollen AA, Tavare A, DeProto J, and Molnar Z (2007). Comparative aspects of cortical neurogenesis in vertebrates. J. Anat 211, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hutsler JJ, Lee DG, and Porter KK (2005). Comparative analysis of cortical layering and supragranular layer enlargement in rodent carnivore and primate species. Brain Res. 1052, 71–81. [DOI] [PubMed] [Google Scholar]
- 59.Rehkamper G, Frahm HD, and Zilles K (1991). Quantitative development of brain and brain structures in birds (galliformes and passeriformes) compared to that in mammals (insectivores and primates). Brain Behav. Evol 37, 125–143. [DOI] [PubMed] [Google Scholar]
- 60.Suryanarayana SM, Robertson B, Wallén P, and Grillner S (2017). The lamprey pallium provides a blueprint of the mammalian layered cortex. Curr. Biol 27, 1–14. [DOI] [PubMed] [Google Scholar]
- 61.Ferguson MWJ (1985). Reproductive biology and embryology of the crocodilians. In Biology of the Reptilia, Volume 14, Development A, Gans C, Billett F and Maderson PFA, eds. (New York: John Wiley & Sons; ), pp. 329–492. [Google Scholar]
- 62.Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCorrison JM, Venepally P, Singh I, Fouts DE, Lasken RS, and Methe BA (2014). NeatFreq: reference-free data reduction and coverage normalization for De Novo sequence assembly. BMC Bioinformatics 15, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc 8, 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schurch NJ, Schofield P, Gierliński M, Cole C, Sherstnev A, Singh V, Wrobel N, Gharbi K, Simpson GG, Owen-Hughes T, et al. (2016). How many biological replicates are needed in an RNA-seq experiment and which differential expression tool should you use? RNA 22, 839–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Agarwala S, Sanders TA, and Ragsdale CW (2001). Sonic hedgehog control of size and shape in midbrain pattern formation. Science 291, 2147–2150. [DOI] [PubMed] [Google Scholar]
- 67.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2 (Related to Figures 1 and 2) Top 145 nidopallium coding-transcripts
For the nidopallium HOLT analysis, we selected transcripts enriched in the nidopallium in comparison with the mesopallium by 2.5 fold. This resulted in 97,261 candidate transcripts. From these, we selected coding genes expressed at 5 FPKM or higher in nidopallium, yielding 145 genes. The 145 most abundant nidopallium-specific genes are listed by descending levels of expression. Transcriptional regulator genes are in bold type.
Table S1 (Related to Figure 2) Top 78 mesopallium coding-transcripts
The 78 most abundant mesopallium-specific genes are listed by descending levels of expression. Transcriptional regulator genes are in bold type. EMX1 was not recovered by the HOLT method because it is expressed in a subnucleus of the arcopallium, but it is included at the bottom of the list. Gene names marked with a plus (+) were confirmed as mesopallium-specific with ISH. Those with a minus (−) were expressed in the mesopallium but were not mesopallium-specific. There were no false positives among the genes examined by ISH. Genes without either a plus or minus sign were not examined by ISH. Gene names with (eR) were also detected by a parallel bioinformatic analysis with edgeR (see STAR Methods).
Figure S1 (Related to Figure 2) Chick dorsal telencephalon nuclei collected for RNAseq analysis
In situ hybridization for the conserved input cell marker KCNH5 on E14 chick telencephalon. Five coronal sections from a single cerebral hemisphere are shown in anterior to posterior sequence. The seven indicated cell populations were harvested from embryonic day 14 telencephalon: (A) 1. Hyperpallium apicale (HA), 2. Mesopallium (M), (B) 3. Nucleus basorostralis (Bas), (C) 4. Entopallium (E), (D) 5. Field L (L), (E) 6. Nidopallium (N), and 7. Arcopallium (A). RNA was purified from these cell populations and submitted for sequencing.
Figure S2 (Related to Figures 2 and 3) In situ validation of mesopallium candidate genes identified by HOLT
(A–F) In situ hybridization for candidate mesopallium marker genes identified by the HOLT method. All sections are from embryonic day 14 chick telencephalon. Our approach recovered many highly specific mesopallium marker genes. Gene expression suggests that the mesopallium has a consistent internal molecular identity and is dominated by a single principal neuronal type.
(G–N) An anteroposterior series of sections from a single postnatal day zero chick telencephalon labeled for NHLH2 transcript. The Wulst dorsal mesopallium (Md) and the DVR ventral mesopallium (Mv) are clearly distinguished and can be followed along the anteroposterior axis. (N) Caudal mesopallium, for instance, is continuous with the rostral part of Mv. Asterisk in G-K indicates the vallecula, a shallow sulcus that defines the boundary between the DVR ventrally and the Wulst dorsally.
Figure S3 (Related to Figure 3) Mesopallium molecular organization is conserved across birds
We performed in situ hybridization for the mesopallium marker FOXP1 and the IHA marker KCNH5 on serial sections through embryonic day 20 chick telencephalon. Asterisks in all panels indicate the vallecula.
(A–B) The chick Md, as labeled by FOXP1, extends from the vallecula to the IHA, seen in (B). We conclude that our chicken Md is equivalent to the structure referred to as HD by avian neuroanatomists.
(C–D) Adjacent sections through the anterior telencephalon of an adult starling demonstrate that the FOXP1-expressing mesopallium extends to the (D) KCNH5-expressing IHA.
(E–F) Sections through (E) intermediate and (F) posterior levels of the starling telencephalon demonstrate conserved expression of BCL11A in Md and Mv.
Figure S4 (Related to Figure 3) Emx1 and Id2 are expressed in the postnatal mouse neocortex
Expression patterns of the avian mesopallium transcription factors Satb2, Bcl11a, and Foxp1 in the mouse neocortex were previously reported. (A) Emx1 and (B) Id2 are also expressed in the mouse neocortex at postnatal day six (P6). Both are prominently expressed in L2/3 in addition to the deep layers.
Figure S5 (Related to Figure 1) The nidopallium transcription factor Dach2 is expressed in mouse neocortical layer 5
(A–C) An anteroposterior series through a single embryonic day 20 chick telencephalon labeled for DACH2 transcript. The entire nidopallium (N) is labeled, with sharp boundaries where it meets the mesopallium (M), arcopallium (A), and ventral telencephalon (VT). We did not identify other molecular markers that capture this pattern. Chicken DACH2 is also expressed in the IHA (A).
(D) Mouse Dach2 is expressed in a thin, laminar pattern in the neocortex at postnatal day six. This transcript is present at extremely low levels and was detected by combination in situ hybridization employing two non-overlapping probes. Mouse Dach2 expression was also detected in the olfactory cortex, the cortical amygdala, and the septum (data not shown). Expression was not detected in the basolateral amygdala, which has been proposed as a mammalian homolog of the avian nidopallium [32].
(E) A section adjacent to that in D labeled with a riboprobe for the well-characterized L5/output cell marker Fezf2. Comparison with Dach2 staining indicates that the Dach2 lamina is contained within L5, possibly in an upper part of L5 corresponding to L5a. The arrowheads in D indicate the somatosensory whisker barrels, prominent clusters of L4/input cells in rodent primary somatosensory cortex. They are faintly visible as grey blobs and are located just above the Dach2 stripe, further suggesting that Dach2 labels cells in L5a.
Data Availability Statement
RNA sequencing reads have been deposited in the Short Read Archive at NCBI under Bioproject number PRJNA416004. Antisense RNA probe sequences are being uploaded to the nucleotide database at NCBI.


