Abstract
Methylobacterium dichloromethanicum DM4 grows with dichloromethane as the unique carbon and energy source by virtue of a single enzyme, dichloromethane dehalogenase–glutathione S-transferase. A mutant of the dichloromethane-degrading strain M. dichloromethanicum DM4, strain DM4-1445, was obtained by mini-Tn5 transposon mutagenesis that was no longer able to grow with dichloromethane. Dichloromethane dehalogenase activity in this mutant was comparable to that of the wild-type strain. The site of mini-Tn5 insertion in this mutant was located in the polA gene encoding DNA polymerase I, an enzyme with a well-known role in DNA repair. DNA polymerase activity was not detected in cell extracts of the polA mutant. Conjugation of a plasmid containing the intact DNA polymerase I gene into the polA mutant restored growth with dichloromethane, indicating that the polA gene defect was responsible for the observed lack of growth of this mutant with dichloromethane. Viability of the DM4-1445 mutant was strongly reduced upon exposure to both UV light and dichloromethane. The polA′-lacZ transcriptional fusion resulting from mini-Tn5 insertion was constitutively expressed at high levels and induced about twofold after addition of 10 mM dichloromethane. Taken together, these data indicate that DNA polymerase I is essential for growth of M. dichloromethanicum DM4 with dichloromethane and further suggest an important role of the DNA repair machinery in the degradation of halogenated, DNA-alkylating compounds by bacteria.
Dichloromethane (DCM) is an organic solvent produced industrially in large amounts for a wide range of technical applications (Halogen Solvents Industry Alliance [http://www.hsia.org/white_papers/methchlor.htm]). Its low boiling point and high solubility in water make it a frequently encountered environmental contaminant (36, 46). The toxicity of DCM to mammals continues to be investigated intensively (9, 14, 21, 40, 45), but its causes are not yet fully characterized at the molecular level. Many specialized aerobic methylotrophic bacteria have been isolated from soil and groundwater environments contaminated with DCM for their ability to grow with DCM as the sole source of carbon and energy (49). Such bacteria rely on a single enzyme, DCM dehalogenase, for this purpose. DCM dehalogenase, which can make up to 20% of the soluble protein during bacterial growth with DCM, was purified and shown to catalyze the glutathione-dependent transformation of DCM to formaldehyde, used in both biomass and energy production, and to two molecules of hydrochloric acid (31). The corresponding gene dcmA was cloned (33) from Methylobacterium dichloromethanicum DM4 (15) (formerly Methylobacterium sp. strain DM4), Methylophilus sp. strain DM11 (3), and, more recently, from several other DCM-degrading strains (49, 50). Sequence analysis indicates that DCM dehalogenases belong to the glutathione S-transferase (GST) enzyme family (27, 47). DCM dehalogenases were the first bacterial GSTs to be characterized, but it is becoming clear that the genomes of some gram-negative bacteria may contain more than a dozen GST genes (48). In all higher organisms with an aerobic lifestyle, GSTs serve as versatile, relatively nonspecific catalysts for the conjugation and subsequent detoxification of reactive electrophilic compounds (27). In bacteria, however, GST often represent specialized catabolic enzymes with an essential role in the mineralization of toxic chemicals (47, 48).
Although the conversion of DCM to formaldehyde by DCM-degrading bacteria only requires the presence of DCM dehalogenase, growth-supporting GST-mediated dehalogenation of DCM may impose quite drastic requirements upon bacterial metabolism. On the one hand, DCM-converting GSTs from mammals and methylotrophic bacteria have toxic and mutagenic effects in Salmonella enterica serovar Typhimurium (18, 44) and Methylobacterium (18). On the other hand, the massive production of hydrochloric acid by cytosolic DCM dehalogenase during growth with DCM suggests that DCM-degrading methylotrophic bacteria may have evolved efficient systems for the maintenance of intracellular pH and for the excretion of chloride ions. These aspects of bacterial dehalogenation metabolism have been rather neglected until now and, in large part, remain to be explored. In the present work, we have used minitransposon insertion mutagenesis to identify genes associated with DCM metabolism in the DCM-degrading strain M. dichloromethanicum DM4. We report that a mutant of this strain disrupted in the gene encoding DNA polymerase I, an enzyme with a well-known role in DNA repair (16), is no longer able to grow with DCM as the sole carbon source. This suggests an important role for the DNA repair machinery during bacterial mineralization of DCM.
MATERIALS AND METHODS
Materials.
Restriction and DNA modifying enzymes used in cloning were from Fermentas. Oligonucleotides were purchased from Microsynth (Balgach, Switzerland). Escherichia coli DNA polymerase I and Klenow fragment were from New England Biolabs. All other chemicals were analytical grade or better and were purchased from Fluka except where noted.
Bacterial strains, media, and growth conditions.
E. coli strains DH5α (GIBCO/BRL Life Technologies) and XL1-Blue (Stratagene) were used for cloning, and E. coli strains S17-1 (41) and S17-1λpir (38) were used as donor strains in biparental mating experiments. E. coli strains were grown under shaking at 37°C in Luria-Bertani medium (2), with kanamycin (25 mg/liter), ampicillin (100 mg/liter), and tetracycline (25 mg/liter) antibiotics as required. M. dichloromethanicum DM4 wild type (17) and derivatives of the mini-Tn5 insertion mutant strain DM4-1445 were grown at 30°C in liquid minimal medium (MM) on a rotary shaker at 150 rpm in glass flasks with gastight mininert caps (Supelco), with methanol (40 mM) and/or DCM (10 mM) as described (19). Solid media contained (per liter) 15 g of agar and 50 mg of cycloheximide. Bacterial growth in liquid cultures was determined by monitoring optical density at 600 nm (OD600). MM agar plates were incubated in 3-liter gastight glass jars to which 960 μl of methanol (MeOH) (yielding 40 mM final concentration) and/or 380 μl of DCM (10 mM final concentration) was added.
Mini-Tn5 mutagenesis.
Mini-Tn5 transposon mutagenesis (13) of M. dichloromethanicum DM4 was performed by biparental plate conjugation of E. coli S17-1λpir containing plasmid pUT/mini-Tn5lacZ1 (12) with wild-type strain DM4. A mixture of 50 μl of resuspended and 20-fold-concentrated cultures of donor (OD600 = 0.5) and recipient (OD600 = 1.0) strains was spotted on nutrient broth agar (Difco) at 30°C for 24 h. Kanamycin-resistant transconjugants were obtained by spreading the mating mixture on MM agar plates containing kanamycin (5 mg/liter) and incubation for 7 to 10 days in 3-liter gastight jars with 40 mM methanol as the carbon source. Colonies were patched on agar plates of the same medium and screened for impairment of growth with DCM.
DNA isolation and manipulation.
Preparation of total DNA, restriction enzyme digestions, cloning, and Southern blot analysis were performed by standard procedures (2). Southern blot analysis of total DNA from DM4-1445, restricted with several enzymes, was performed with a 795-bp digoxigenin (DIG)-labeled fragment of the kanamycin gene generated by PCR with primers 5′-GAGCATCAAATGAAACTGC-3′ and 5′-CATATTCAACGGGAAACG-3′ using PCR DIG labeling mix (Boehringer Mannheim). The hybridizing 8.5-kb PstI fragment was cloned into pBluescript KSII(+) (Stratagene) from total PstI-digested DNA of the DM4-1445 mutant by selection for kanamycin resistance. Detection of the polA gene by Southern blot analysis was performed with a 530-bp DIG-labeled DNA probe generated by PCR with the primers 5′-GACCGAAGAGACGCAACC-3′ and 5′-CCTGACTCGAAATCGTAGAACC-3′ selected from sequence analysis of the cloned PstI fragment from mutant DM4-1445. The wild-type polA gene was cloned by ligation of a 7.5-kb SmaI/SalI fragment of M. dichloromethanicum DM4 with pBluescript KSII(+) vector cut with SmaI/SalI. A 4.7-kb SmaI/HindIII subfragment containing the entire polA gene was then ligated to broad-host-range cloning vector pVK100 (30) cut with BglII, blunted with T4 DNA polymerase, and then digested with HindIII. The resulting plasmid pME8112 was transformed into E. coli S17-1 and conjugated into mutant strain DM4-1445 by biparental mating as described above.
Sequence analysis.
The 4.7-kb SmaI/HindIII fragment containing the entire polA gene was sequenced by primer walking on both strands with the DyeTerminator kit using an ABI377 automated sequencer (Perkin-Elmer). Sequences were analyzed with the Genetics Computer Group sequence analysis package (version 10), and the assembled sequence was deposited in the EMBL database (accession no. AJ242630). Similarity searches were performed by using gapped BLAST and PSI-BLAST programs (1) for comparisons against public protein and gene databases.
Preparation of cell extracts.
Bacterial cultures (100 ml) were harvested, resuspended in 3 ml of cold extraction buffer (50 mM Tris-HCl [pH 7.5], 2 mM dithiothreitol, 0.5 mM EDTA, 25% [wt/vol] glycerol), and disrupted in a French pressure cell (three times at 55 MPa). Cell debris were removed by centrifugation (30,000 × g at 4°C for 45 min), and the protein concentration was determined in the supernatant using a commercial Bradford reagent (Bio-Rad) and bovine serum albumin (Sigma) as the standard. Cell extracts were either analyzed directly or flash frozen in liquid nitrogen and stored at −80°C.
DCM dehalogenase assay.
Specific activity of DCM dehalogenase was measured in triplicate in cell extracts (2 to 20 μg protein) using a previously described coupled enzyme assay with formaldehyde dehalogenase (51).
DNA polymerase assay.
Nicked DNA was prepared from calf thymus DNA (0.8 mg/ml) with DNase I (0.26 μg/ml) in a solution containing 8 mM Tris-HCl (pH 8), 16 mM NaCl, 4 mM MgCl2, 0.8 mM EDTA, and bovine serum albumin (200 μg/ml) for 10 min at 37°C and isolated by ethanol precipitation after extraction. DNA polymerase activity was detected in cell extracts separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (10% acrylamide) containing nicked calf thymus DNA (125 μg/ml) and fibrinogen (40 μg/ml) using previously described methods (5, 22) with minor modifications. A prestained broad-range protein marker (New England Biolabs) was run to facilitate subsequent analysis. After electrophoresis, gels were washed for 24 h in 10 mM Tris buffer (pH 7.6) and further incubated overnight in the same buffer containing 2 mM MgCl2; a 15 μM concentration (each) of dATP, dGTP, and dTTP; and 100 μCi of [α-32P]dCTP (Amersham). Gels were then washed three times in 5% trichloroacetic acid containing 1% sodium pyrophosphate, dried, and autoradiographed for 48 h.
β-Galactosidase assay.
β-Galactosidase activity was measured in cell extracts by standard methods (37) and expressed as o-nitrophenyl-β-d-galactoside hydrolyzed (in nanomoles/minute/milligram of protein), using a value of 4.5 mM−1 cm−1 for the extinction coefficient at 420 nm (8).
Chloride determination.
Chloride was determined colorimetrically as its ferric thiocyanate complex as described previously (4). Samples of growing cultures were centrifuged at 4°C (13,000 rpm, 10 min, Heraeus biofuge) and cell supernatants were first diluted 1:1 with 30% H2O2 and heated at 80°C for 2 min. Six hundred microliters of this solution was then mixed with 200 μl of 0.25 M ferric ammonium sulfate, dissolved in 9 N nitric acid and then with 200 μl of saturated mercuric thiocyanate in ethanol. The absorbance at 460 nm was determined after 10 min of incubation at room temperature, and the chloride concentration was determined with a standard curve of 0 to 500 μM NaCl.
Viability assessment.
Bacterial survival after various treatments was determined using a spot plating technique. Serially diluted cultures (10- to 109-fold) were spotted (5 μl) in triplicate onto MM agar plates. Plates were then incubated in 3-liter gastight containers containing 40 mM MeOH, 10 mM DCM, or both carbon sources for 7 days at 30°C. Dilutions with spots containing 5 to 30 colonies were counted, and the resulting average was expressed as the ratio of viable cells in treated cultures to those in untreated cultures. In the case of UV exposure, cells were grown in MM with 40 mM MeOH, washed, serially diluted, and spotted on plates as described above. Plates were irradiated for different times with UV light (254 nm) from a germicidal lamp (Stratalinker UV model 1800 cross-linker; Stratagene) at a rate of 5 J m−2 s−1 and then incubated in the dark as described above. For determination of survival after DCM treatment, cultures were grown in MM with 40 mM MeOH and treated with 10 mM DCM at an OD600 of 0.05, and viability of the cultures was determined at different time points after addition of 10 mM DCM as described above.
RESULTS
Isolation of strain DM4-1445, a mutant of M. dichloromethanicum DM4 unable to grow with DCM as the sole carbon source.
Random transposition of the mini-Tn5lacZ1 element into the chromosome of M. dichloromethanicum DM4 was obtained by biparental conjugation with E. coli S17-1λpir (38) harboring plasmid pUT/mini-Tn5lacZ1 (12). Mini-Tn5 mutants of strain DM4 were obtained on solid MM with MeOH as the carbon source by selecting for the kanamycin resistance afforded by the minitransposon element. Screening of about 1,000 kanamycin-resistant clones for the loss of the ability to grow on plates with DCM as the carbon source yielded 10 independent mutants of strain DM4. However, this mutagenesis experiment was not performed to saturation, since no mutant was recovered that featured a mini-Tn5 insertion in the DCM dehalogenase gene itself. One of the mutants obtained, DM4-1445 was also unable to grow with DCM in liquid culture (Table 1) and was therefore investigated in detail.
TABLE 1.
Growth rates of shaken batch cultures of M. dichloromethanicum DM4 and its polA derivatives with different carbon sourcesa
| Strain | Growth rate (h−1) on carbon source
|
|||
|---|---|---|---|---|
| MeOH | DCM | MeOH-DCM | MeOH-CH2O | |
| DM4 wild-type | 0.19 | 0.08 | 0.12 | 0.19 |
| DM4-1445 | 0.17 | —b | 0.08 | 0.17 |
| DM4-1445(pME8112) | 0.17 | 0.08 | 0.11 | 0.19 |
The concentration of MeOH was 40 mM, that of DCM was 10 mM, and that of formaldehyde (CH2O) was 1 mM. Standard errors are <0.01 h−1 in all cases.
—, no growth.
Cloning and sequence analysis of the gene disrupted by minitransposon insertion in mutant DM4-1445.
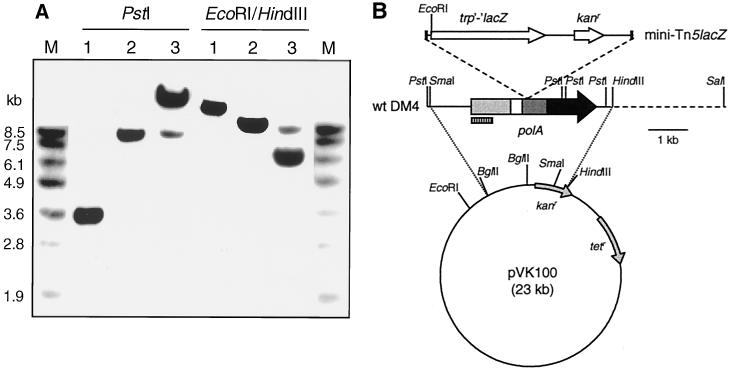
Hybridization with a DIG-labeled probe specific for the kanamycin resistance gene of the mini-Tn5 element confirmed that only one minitransposon insertion had occurred in mutant DM4-1445 (data not shown). The insertion was located on an 8.5-kb PstI DNA fragment (Fig. 1A) which was rescued from total digested DNA by selection for kanamycin resistance. Sequencing of the cloned PstI DNA fragment revealed that the gene disrupted by minitransposon insertion in mutant DM4-1445 displayed strong similarity to the DNA polymerase I gene (polA) of E. coli (28). Hybridization with a DIG-labeled probe to the rescued M. dichloromethanicum DM4 polA gene fragment confirmed that M. dichloromethanicum DM4 only contained one copy of the polA gene (Fig. 1A) and led to the cloning of a 7.5-kb SmaI-SalI fragment from total DNA of wild-type DM4 containing the polA gene in its entirety (Fig. 1B). Sequence analysis of the polA gene from M. dichloromethanicum DM4 showed that the corresponding gene product was most closely related to DNA polymerase I from Rhizobium leguminosarum (24) and E. coli (28) (54 and 42% pairwise identity at the protein level, respectively). Unlike some homologs lacking the sequence region encoding the 3′-5′ exonuclease activity (see, e.g., reference 34), the PolA protein sequence from M. dichloromethanicum DM4 featured the three sequence domains corresponding to the 5′-3′ exonuclease, proofreading 3′-5′ exonuclease, and DNA polymerase activities of DNA polymerase I from E. coli.
FIG. 1.
Southern blot analysis of the polA gene of M. dichloromethanicum DM4. (A) Chromosomal DNA (5 μg) from wild-type M. dichloromethanicum DM4 (lanes 1), mutant M. dichloromethanicum DM4-1445 (lanes 2), and mutant DM4-1445 conjugated with complementing plasmid pME8112 and digested with PstI or with HindIII/EcoRI (lanes 3) was hybridized with a polA-specific DIG-labeled probe after agarose gel electrophoresis. Lanes M, molecular size markers. (B) Corresponding schematic map of cloned DNA fragments with the DM4 polA gene, indicating the point of insertion of the mini-Tn5 element (12) in mutant DM4-1445 (top), and vector pVK100 (30) used for mutant complementation with the wild-type (wt) polA gene. Restriction sites relevant for Southern analysis and the location of the DIG-labeled probe (striped box) are shown. Segments corresponding to 5′-3′ exonuclease, 3′-5′ exonuclease, and DNA polymerase domains of the polA gene product are indicated as light grey, dark grey and black boxes, respectively.
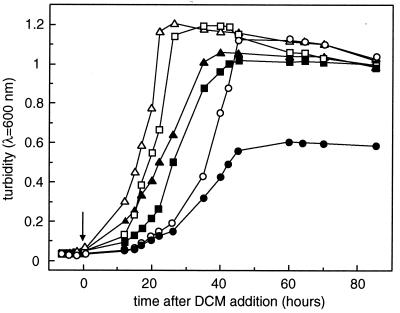
Complementation of the growth defect of DM4-1445 on DCM with the intact polA gene.
The growth rate of wild-type M. dichloromethanicum DM4 is 0.19 h−1 with methanol and 0.08 h−1 with DCM under the conditions used (Table 1; Fig. 2). In addition to its lack of growth with DCM as the sole carbon source, mutant DM4-1445 showed delayed and poorer growth with 40 mM MeOH in the presence of 10 mM DCM (Table 1; Fig. 2). Plasmid pME8112, a derivative of the IncP shuttle plasmid pVK100 (30) containing a SmaI-HindIII fragment with the intact polA gene, restored growth of the mutant with DCM as the unique carbon source (Table 1), and relieved the growth defect observed for growth with mixtures of MeOH and DCM (Table 1; Fig. 2). Some plasmids, such as ColE1 in E. coli (29) and an IncQ plasmid in R. leguminosarum (10), were previously shown to require DNA polymerase I for replication. Both plasmid pME8112 and its parent vector pVK100 were stably replicated in the polA mutant as well as in wild-type strain DM4 (data not shown). The DM4 polA mutant contained about three copies of plasmid pME8112 per cell (Fig. 1A).
FIG. 2.
Effect of DCM on growth of wild-type M. dichloromethanicum DM4 and of the polA mutant. Strains DM4 wild-type (squares), mutant DM4-1445 (circles), and complemented mutant DM4-1445(pME8112) (triangles) were grown at 30°C in minimal medium with 40 mM MeOH as the unique carbon source (open symbols). The growth behavior of cultures additionally treated with 10 mM DCM at an OD600 of 0.05 (arrow) is depicted with filled symbols. Data from one representative experiment are shown.
Biochemical characterization of the growth defect of mutant DM4-1445 with DCM as the carbon source.
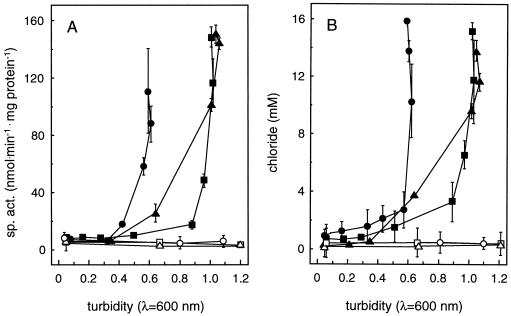
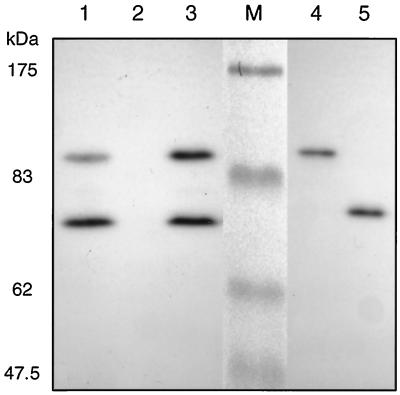
Absence of growth of mutant DM4-1445 with DCM (Table 1) was not due to lack of active DCM dehalogenase: mutant DM4-1445 contained the DCM dehalogenase gene and expressed the corresponding protein, as verified by Southern and Western analysis, respectively (data not shown). In addition, intracellular levels of reduced glutathione in the mutant were similar to that of the wild-type strain (about 200 nmol/mg of protein [data not shown]). Measurements of DCM dehalogenase activity in cell extracts of cells grown with 40 mM MeOH and 10 mM DCM confirmed the normal onset of DCM dehalogenase expression at about 20 h after DCM addition in mutant DM4-1445 (Fig. 3A; compare Fig. 2). Levels of DCM dehalogenase activity in mutant DM4-1445 also appeared normal in vivo, as evidenced by the amount of chloride ions excreted into the growth medium (Fig. 3B). DNA polymerase activity was then investigated in SDS-PAGE gels of cell extracts containing nicked DNA that were incubated with deoxynucleotides including α-32P-labeled dCTP. No DNA polymerase activity could be detected in the resulting autoradiogram with cell extracts of mutant DM4-1445 (Fig. 4). In contrast, extracts of wild-type DM4 and of the mutant strain complemented with plasmid pME8112 containing an intact polA gene showed bands of very similar sizes to DNA polymerase I and Klenow fragment of E. coli run as controls (Fig. 4). These results suggest that the lack of a functional polA gene product is responsible for the observed growth defect of mutant DM4-1445 with DCM as the carbon source.
FIG. 3.
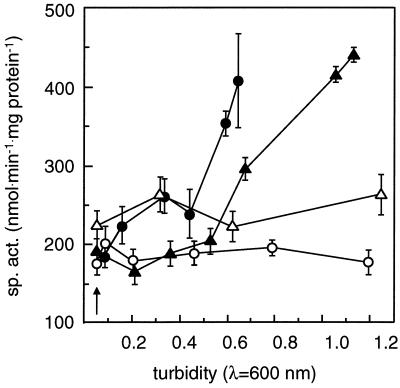
Induction of DCM dehalogenase after addition of DCM. DCM dehalogenase induction during growth is given as a function of culture turbidity. (A) Specific activity in crude extracts harvested at various stages of growth; (B) concentration of chloride ions released in the growth medium (symbols as defined in the legend to Fig. 2). Error bars, standard errors of triplicate measurements.
FIG. 4.
DNA polymerase I activity of wild-type M. dichloromethanicum DM4 and of the polA mutant. Shown is an autoradiogram of cell extract protein (100 μg) of wild-type strain DM4 (lane 1), mutant DM4-1445 (lane 2), and complemented mutant DM4-1445(pME8112) (lane 3), separated by SDS-PAGE in a gel containing nicked DNA, after incubation with dideoxynucleotides and α-32P-labeled dCTP (see Materials and Methods). Lane M, prestained marker from the scanned gel; lanes 4 and 5, E. coli DNA polymerase I and Klenow fragment, respectively (0.02 U [∼0.1 ng] each).
Expression of the polA gene upon DCM addition.
Insertion of the mini-Tn5lacZ1 element in the polA gene resulted in a transcriptional polA′-lacZ fusion in mutant DM4-1445 (Fig. 1B). Expression of the gene fusion was investigated in cell extracts of mutant DM4-1445 and of complemented mutant DM4-1445(pME8112) harvested at various stages of growth with and without addition of DCM (Fig. 5). Only negligible β-galactosidase activity was detected in protein extracts of the wild-type strain (<20 nmol/min/mg of protein [data not shown]). The polA gene was constitutively expressed at a high level during growth with MeOH as the carbon source in both polA mutant and complemented strains, in agreement with the reported estimate of 400 copies of DNA polymerase I for an E. coli cell (16). Addition of DCM resulted in an approximately twofold increase of β-galactosidase activity which accompanied the induction of DCM dehalogenase expression (Fig. 5; compare Fig. 3).
FIG. 5.
Effect of DCM on the expression of the polA′-lacZ fusion in the DM4-1445 polA mutant background. The β-galactosidase specific activity (sp. act.) arising from the polA′-lacZ transcriptional fusion was determined in cell extracts of M. dichloromethanicum DM4-1445 (circles) and DM4-1445(pME8112) (triangles) grown with MeOH as the carbon source with (filled symbols) and without (open symbols) prior treatment with 10 mM DCM at an OD600 of 0.05 and expressed as a function of the optical density of the cultures at the time of cell harvest. Error bars, standard errors of triplicate measurements.
Sensitivity of M. dichloromethanicum DM4 to toxic insults.
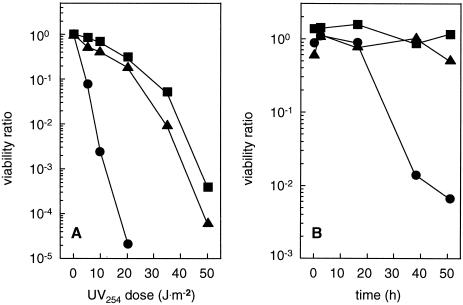
Mutant DM4-1445 showed a markedly lower resistance to the DNA cross-linking agents UV light (Fig. 6A) and mitomycin (data not shown) typical of that observed previously with polA mutants of various bacteria (e.g., see references 22 and 42) (Fig. 6A). In contrast, the degrees of resistance of wild-type and complemented mutant strains to UV and mitomycin were undistinguishable and comparable to those previously reported for E. coli (42), taking the low growth rate of M. dichloromethanicum into account (doubling time of about 3.6 h with MeOH at 30°C [Table 1]). Most notably, the polA mutant from strain DM4 was also sensitive to treatment with DCM (Fig. 6B). The viability of bacteria plated on MM after the addition of 10 mM DCM markedly decreased upon induction of the DCM dehalogenase (after about 20 h) (compare Fig. 2 and 3), while that of wild-type and complemented mutant strains remained unaffected.
FIG. 6.
Effects of UV light and DCM on the viability of wild-type M. dichloromethanicum DM4 and of the polA mutant. Methanol-grown cultures of M. dichloromethanicum DM4 wild-type (squares), DM4-1445 (circles), and DM4-1445(pME8112) (triangles) were exposed to UV light at 254 nm (5 J · m−2 · s−1) (A) or treated with 10 mM DCM at an OD600 of 0.05 (B) for various times before plating on MM with 40 mM MeOH. Bacterial viability is expressed relative to that of untreated cultures.
DISCUSSION
The data reported here show that disruption of the polA gene in M. dichloromethanicum DM4 is responsible for the observed growth defect of mutant DM4-1445 with DCM. This provides a clear indication that repair of DNA damage is important for DCM metabolism by Methylobacterium expressing DCM dehalogenase-GST. DNA polymerase I is a multidomain protein with 5′-3′ exonuclease, 3′-5′ exonuclease, and DNA polymerase activity involved in DNA repair processes (16). The DM4-1445 polA strain may still express the 5′-3′ exonuclease domain of DNA polymerase I in active form, since insertion of the minitransposon element occurred downstream of the section encoding this activity (Fig. 1A). In any case, the disruption of the 3′-5′ exonuclease domain by minitransposon insertion led to loss of DNA polymerase function (Fig. 4) and was sufficient to prevent growth of M. dichloromethanicum DM4 with DCM.
Although the toxic agent in DCM metabolism remains to be characterized in detail, several observations, summarized below, provide suggestive clues as to its identity. Clearly, a role for DCM itself can be excluded under the experimental conditions used: wild-type strain DM4 grows with 10 mM DCM without its viability being detectably affected (49), and DCM is nontoxic to a mutant of M. dichloromethanicum DM4 lacking the DCM dehalogenase gene (49).
Thus, GST-mediated DCM turnover is likely to be responsible for poor growth of the polA mutant in the presence of DCM (Fig. 2). Formaldehyde, the product of DCM conversion by DCM dehalogenase-GST, can give rise to DNA cross-links and is a known cytotoxic agent and mutagen (9, 20, 35). Indeed, mutations in the polA gene are known to be detrimental for viability of E. coli upon formaldehyde exposure (43). Several lines of evidence indicate, however, that formaldehyde is unlikely to be the main toxic agent associated with DCM conversion. First, the growth rate of Methylobacterium with MeOH, which involves formaldehyde as a central metabolic intermediate, was not affected in the DM4-1445 polA mutant (Table 1). Second, the presence of 1 mM formaldehyde did not affect growth and viability of both the wild type and the polA mutant (Table 1 and data not shown [2 mM formaldehyde led to a prolonged lag phase prior to growth in both wild-type and polA mutant strains]). Finally, constitutive expression from plasmids of the DCM-active GST theta 1-1 from rat in the presence of DCM was more toxic and mutagenic to Methylobacterium and S. enterica serovar Typhimurium TA1535 than that of bacterial DCM dehalogenases, despite a lower conversion rate of DCM to formaldehyde by the rat enzyme under the conditions used (18). In addition, formaldehyde requires a proficient nucleotide excision repair machinery to unfold its mutagenic effects (52), but S. enterica serovar Typhimurium TA1535 is excision repair deficient (26).
Inability of the polA mutant to cope with intracellular acid production would constitute another possible explanation for the toxicity of GST-mediated DCM conversion. Expression of the apparently monocistronic polA gene is upregulated twofold upon acidification of the culture medium in E. coli (23), and a twofold increase in β-galactosidase activity arising from the polA transcriptional fusion in the DM4-1445 mutant was also observed after addition of DCM (Fig. 5). The exact nature of the inducer(s) of polA expression during growth of strain DM4 with DCM remain to be determined. However, the possibility that the intracellular pH was lowered to a detrimental level in the mutant can be excluded since DCM dehalogenase, which is inactive below pH 6 (S.V., unpublished data), was as active in the polA mutant as in the wild-type strain (Fig. 3B).
Nevertheless, the finding that DNA polymerase I is essential for DCM metabolism in Methylobacterium suggests that the main toxic agent of GST-mediated conversion of DCM is able to cause extensive DNA damage. In addition, vailable experimental data indicate that this agent is not DCM, formaldehyde, or hydrochloric acid, but rather an intermediate in the reaction catalyzed by DCM dehalogenase. S-Chloromethylglutathione is generally believed to be formed during the reaction of DCM to formaldehyde catalyzed by DCM dehalogenase-GSTs (11, 21) but remains poorly characterized due to its high reactivity: S-fluoromethylglutathione, the less-reactive fluorinated homolog of S-chloromethylglutathione, is hydrolyzed with a half-life of 5.8 min at room temperature in D2O (11). Transient formation of a compound with a 19F-nuclear magnetic resonance signal compatible with S-fluoromethylglutathione was observed with DCM dehalogenase from Methylophilus sp. DM11 incubated with glutathione and chlorofluoromethane (6). Chemically synthesized S-chloromethylglutathione alkylated deoxyguanosine in vitro (11), and the main product of this reaction was characterized as S-[1-N2-deoxyguanosinylmethyl]glutathione (11, 44). Nevertheless, the formation of DNA lesions as a consequence of S-chloromethylglutathione formation during conversion of DCM by DCM dehalogenase-GST remains to be demonstrated. Certainly, an obvious role for DNA polymerase I in DCM metabolism would be in participating in nucleotide excision repair of bulky base adducts formed upon reaction of DNA with S-chloromethylglutathione and in preventing stalling of the UvrABC excision nuclease protein complex (7, 25, 32, 39), which would presumably recognize such lesions. Whether DNA adducts are formed during DCM conversion by DCM dehalogenase is currently being investigated and should contribute to clarifying the role of DNA repair processes in DCM metabolism. More generally, the polA mutant of M. dichloromethanicum DM4 provides a first indication that genes other than that involved in the dehalogenation reaction itself are needed for bacterial growth with halogenated methanes. It is expected that the characterization of other minitransposon mutants showing impaired growth with DCM will shed light on the nature of these accessory requirements.
ACKNOWLEDGMENTS
We gratefully acknowledge Thomas Leisinger for helpful discussions, encouragement, and support.
This research was supported by the Swiss National Research Foundation (grant 3100-50602.97 to S.V.).
REFERENCES
- 1.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley; 2000. [Google Scholar]
- 3.Bader R, Leisinger T. Isolation and characterization of the Methylophilus sp. strain DM11 gene encoding dichloromethane dehalogenase/glutathione S-transferase. J Bacteriol. 1994;176:3466–3473. doi: 10.1128/jb.176.12.3466-3473.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann J G, Sanik J. Determination of trace amounts of chlorine in naphtha. Anal Chem. 1957;29:241–243. [Google Scholar]
- 5.Blank A, Silber J R, Thelen M P, Dekker C A. Detection of enzymatic activities in sodium dodecyl sulfate-polyacrylamide gels: DNA polymerases as model enzymes. Anal Biochem. 1983;135:423–430. doi: 10.1016/0003-2697(83)90705-4. [DOI] [PubMed] [Google Scholar]
- 6.Blocki F A, Logan M S P, Baoli C, Wackett L P. Reaction of rat liver glutathione S-transferases and bacterial dichloromethane dehalogenase with dihalomethanes. J Biol Chem. 1994;269:8826–8830. [PubMed] [Google Scholar]
- 7.Caron P R, Kushner S R, Grossman L. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc Natl Acad Sci USA. 1985;82:4925–4929. doi: 10.1073/pnas.82.15.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casabadan M J, Mastinez-Arias A, Shapira S-K, Chou J. β-galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 9.Casanova M, Bell D A, d' A. Heck H. Dichloromethane metabolism to formaldehyde and reaction of formaldehyde with nucleic acids in hepatocytes of rodents and humans with and without glutathione S-transferase T1 and M1 genes. Fund Appl Toxicol. 1997;37:168–180. [PubMed] [Google Scholar]
- 10.Crank S F, Downie J A. Isolation of a DNA polymerase I (polA) mutant of Rhizobium leguminosarum that has significantly reduced levels of an IncQ-group plasmid. Mol Gen Genet. 1994;243:119–123. doi: 10.1007/BF00283884. [DOI] [PubMed] [Google Scholar]
- 11.Dechert S. Untersuchungen zum Wirkmechanismus der Mutagenität und Tumorigenität von Dichlormethan und seinen Metaboliten. Ph.D. Thesis. Würzburg, Germany: Universität Würzburg; 1995. [Google Scholar]
- 12.De Lorenzo V, Herrero M, Jakubzik Z, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion in Gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 14.Dhillon S, von Burg R. Methylene chloride. J Appl Toxicol. 1995;15:329–335. doi: 10.1002/jat.2550150417. [DOI] [PubMed] [Google Scholar]
- 15.Doronina N V, Trotsenko Y A, Tourova T P, Kuznetzov B B, Leisinger T. Methylopila helvetica sp. nov. and Methylobacterium dichloromethanicum sp. nov.—novel aerobic facultatively methylotrophic bacteria utilizing dichloromethane. Syst Appl Microbiol. 2000;23:210–218. doi: 10.1016/S0723-2020(00)80007-7. [DOI] [PubMed] [Google Scholar]
- 16.Friedberg E C, Walker G C, Siede W. DNA repair and mutagenesis. Washington, D.C.: ASM Press; 1995. [Google Scholar]
- 17.Gälli R, Leisinger T. Plasmid analysis and cloning of the dichloromethane-utilization genes of Methylobacterium sp. DM4. J Gen Microbiol. 1988;134:943–952. doi: 10.1099/00221287-134-4-943. [DOI] [PubMed] [Google Scholar]
- 18.Gisi D, Leisinger T, Vuilleumier S. Enzyme-mediated dichloromethane toxicity and mutagenicity of bacterial and mammalian dichloromethane-active glutathione S-transferases. Arch Toxicol. 1999;73:71–79. doi: 10.1007/s002040050589. [DOI] [PubMed] [Google Scholar]
- 19.Gisi D, Willi L, Traber H, Leisinger T, Vuilleumier S. Effects of bacterial host and dichloromethane dehalogenase on the competitiveness of methylotrophic bacteria growing with dichloromethane. Appl Environ Microbiol. 1998;64:1194–1202. doi: 10.1128/aem.64.4.1194-1202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graves R J, Trueman P, Jones S, Green T. DNA sequence analysis of methylene chloride-induced HPRT mutations in chinese hamster ovary cells: comparison with the mutation spectrum obtained for 1,2-dibromoethane and formaldehyde. Mutagenesis. 1996;11:229–233. doi: 10.1093/mutage/11.3.229. [DOI] [PubMed] [Google Scholar]
- 21.Green T. Methylene chloride induced mouse liver and lung tumours: an overview of the role of mechanistic studies in human safety assessment. Hum Exp Toxicol. 1997;16:3–13. doi: 10.1177/0960327197016001021. [DOI] [PubMed] [Google Scholar]
- 22.Gutman P D, F P, Minton K W. Restoration of the DNA damage resistance of Deinococcus radiodurans DNA polymerase mutants by Escherichia coli DNA polymerase I and Klenow fragment. Mutat Res. 1994;314:87–97. doi: 10.1016/0921-8777(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 23.Hickey E W, Hirshfield I N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl Environ Microbiol. 1990;56:1038–1045. doi: 10.1128/aem.56.4.1038-1045.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y-P, Downie J A, Ito J. Primary structure of the DNA polymerase I gene of an α-Proteobacterium, Rhizobium leguminosarum, and comparison with other family A DNA polymerases. Curr Microbiol. 1999;38:355–359. doi: 10.1007/pl00006816. [DOI] [PubMed] [Google Scholar]
- 25.Husain I, van Houten B, Thomas D C, Abdel-Monem M, Sancar A. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc Natl Acad Sci USA. 1985;82:6774–6778. doi: 10.1073/pnas.82.20.6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inman M A, Butler M A, Connor T H, Matney T S. The effects of excision repair and the plasmid pKM101 on the induction of His+ revertants by chemical agents in Salmonella typhimurium. Teratogen Carcinogen Mutagen. 1983;3:491–501. doi: 10.1002/1520-6866(1990)3:6<491::aid-tcm1770030605>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 27.Josephy P D. Molecular toxicology. Oxford: Oxford University Press; 1997. [Google Scholar]
- 28.Joyce C M, Kelley W S, Grindley N D F. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982;257:1958–1964. [PubMed] [Google Scholar]
- 29.Kingsbury D T, Helinski D R. DNA polymerase as a requirement for the maintenance of the bacterial plasmid colicinogenic factor E1. Biochem Biophys Res Commun. 1970;41:1538–1544. doi: 10.1016/0006-291x(70)90562-0. [DOI] [PubMed] [Google Scholar]
- 30.Knauf V C, Nester E W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 31.Kohler-Staub D, Leisinger T. Dichloromethane dehalogenase of Hyphomicrobium sp. strain DM2. J Bacteriol. 1985;162:676–681. doi: 10.1128/jb.162.2.676-681.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert B, Roques B P, Le Pecq J-B. Induction of an abortive and futile DNA repair process in E. coli by the antitumor DNA bifunctional intercalator, ditercalinium: role in polA in death induction. Nucleic Acids Res. 1988;16:1063–1078. doi: 10.1093/nar/16.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.La Roche S D, Leisinger T. Sequence analysis and expression of the bacterial dichloromethane dehalogenase structural gene, a member of the glutathione S-transferase supergene family. J Bacteriol. 1990;172:164–171. doi: 10.1128/jb.172.1.164-171.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawyer F C, Stoffel S, Saiki R K, Myambo K, Drummond R, Gelfand D H. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from Thermus aquaticus. J Biol Chem. 1989;264:6427–6439. [PubMed] [Google Scholar]
- 35.Ma T-H, Harris M M. Review of the genotoxicity of formaldehyde. Mutat Res. 1988;196:37–59. doi: 10.1016/0165-1110(88)90027-9. [DOI] [PubMed] [Google Scholar]
- 36.Mckay D, Shiu W Y, Ma K C. Volatile organic chemicals. Vol. 3. Boca Raton, Fla: Lewis Publishers; 1993. pp. 400–406. [Google Scholar]
- 37.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 38.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae require toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sancar A. DNA excision repair. Annu Rev Biochem. 1998;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 40.Sherratt P J, Manson M M, Thomson A M, Hissink E A M, Neal G E, van Bladeren P J, Green T, Hayes J D. Increased bioactivation of dihaloalkanes in rat liver due to induction of class theta glutathione S-transferase T1-1. Biochem J. 1998;335:619–630. doi: 10.1042/bj3350619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–790. [Google Scholar]
- 42.Sweet D M, Moseley B E B. The resistance of Micrococcus radiodurans to killing and mutation by agents which damage DNA. Mutat Res. 1976;34:175–186. doi: 10.1016/0027-5107(76)90122-6. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Morita T, Kawazoe Y. Mutagenic characteristics of formaldehyde on bacterial systems. Mutat Res. 1985;156:153–161. doi: 10.1016/0165-1218(85)90058-8. [DOI] [PubMed] [Google Scholar]
- 44.Thier R, Taylor J B, Pemble S E, Humphreys W G, Persmark M, Ketterer B, Guengerich F P. Expression of mammalian glutathione S-transferase 5-5 in Salmonella typhimurium TA1535 leads to base-pair mutations upon exposure to dihalomethanes. Proc Natl Acad Sci USA. 1993;90:8576–8580. doi: 10.1073/pnas.90.18.8576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thier R, Wiebel F A, Hinkel A, Burger A, Brüning T, Morgenroth K, Senge T, Wilhelm M, Schulz T G. Species differences in the glutathione transferase GSTT1-1 activity towards the model substrates methyl chloride and dichloromethane in liver and kidney. Arch Toxicol. 1998;72:622–629. doi: 10.1007/s002040050552. [DOI] [PubMed] [Google Scholar]
- 46.van Agteren M H, Keuning S, Janssen D B. Handbook on biodegradation and biological treatment of hazardous organic compounds. Dordrecht, The Netherlands: Kluwer; 1998. pp. 79–91. [Google Scholar]
- 47.Vuilleumier S. Bacterial glutathione S-transferases: what are they good for? J Bacteriol. 1997;179:1431–1441. doi: 10.1128/jb.179.5.1431-1441.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vuilleumier, S. Bacterial dichloromethane dehalogenases and the detoxification of xenobiotics: dehalogenation through glutathione conjugation and beyond, in press. In: R. E. Hoagland, R. E. Zablotowicz, and J. C. Hall (ed.), Biotransformations in plants and microorganisms. ACS Symposium Series, vol. 777. Oxford University Press, Oxford, United Kingdom.
- 49.Vuilleumier, S. Coping with a halogenated one-carbon diet: aerobic dichloromethane-mineralising bacteria, in press. In S. Agathos and W. Reineke (ed.), Biotechnology for the environment. Focus on biotechnology series, vol. 3. Kluwer Academic Publishers BV, Dordrecht, The Netherlands.
- 50.Vuilleumier S, Gisi D, Stumpp M T, Leisinger T. Bacterial dichloromethane dehalogenases: a particular brand of glutathione S-transferases. Clin Chem Enzymol Commun. 2000;8:367–378. [Google Scholar]
- 51.Vuilleumier S, Leisinger T. Protein engineering studies of dichloromethane dehalogenase/glutathione S-transferase from Methylophilus sp. strain DM11. Ser12 but not Tyr6 is required for enzyme activity. Eur J Biochem. 1996;239:410–417. doi: 10.1111/j.1432-1033.1996.0410u.x. [DOI] [PubMed] [Google Scholar]
- 52.Zijlstra J A. Liquid holding increases mutation induction by formaldehyde and some other cross-linking agents in Escherichia coli K12. Mutat Res. 1989;210:255–261. doi: 10.1016/0027-5107(89)90086-9. [DOI] [PubMed] [Google Scholar]