Abstract
A gene designated thnD, which is required for biodegradation of the organic solvent tetralin by Sphingomonas macrogoltabidus strain TFA, has been identified. Sequence comparison analysis indicated that thnD codes for a carbon-carbon bond serine hydrolase showing highest similarity to hydrolases involved in biodegradation of biphenyl. An insertion mutant defective in ThnD accumulates the ring fission product which results from the extradiol cleavage of the aromatic ring of dihydroxytetralin. The gene product has been purified and characterized. ThnD is an octameric thermostable enzyme with an optimum reaction temperature at 65°C. ThnD efficiently hydrolyzes the ring fission intermediate of the tetralin pathway and also 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid, the ring fission product of the biphenyl meta-cleavage pathway. However, it is not active towards the equivalent intermediates of meta-cleavage pathways of monoaromatic compounds which have small substituents in C-6. When ThnD hydrolyzes the intermediate in the tetralin pathway, it cleaves a C-C bond comprised within the alicyclic ring of tetralin instead of cleaving a linear C-C bond, as all other known hydrolases of meta-cleavage pathways do. The significance of this activity of ThnD for the requirement of other activities to mineralize tetralin is discussed.
Tetralin (1,2,3,4-tetrahydronaphthalene) is a toxic organic solvent since its accumulation in the cell membranes because of its lipophilic character may lead to changes in their structure and function (35, 36) and also because of the formation of toxic hydroperoxides in the cell (12). Tetralin is produced for industrial purposes from naphthalene by catalytic hydrogenation or from anthracene by cracking, and it is widely used as a degreasing agent and solvent for fats, resins, and waxes; as a substitute for turpentine in paints, lacquers, and shoe polishes; and also in the petrochemical industry in connection with coal liquefaction (15).
Tetralin is a bicyclic molecule composed of an aromatic and an alicyclic moiety which share two carbon atoms. In principle, initial transformation of tetralin may involve metabolization of either the aromatic or the alicyclic ring, thus rendering the corresponding alicyclic or aromatic intermediate. However, since pathways known to metabolize aromatic rings are quite different from those acting on alicyclic rings (7, 38), mineralization of tetralin could require the recruitment of two types of metabolic pathways. In spite of this interesting characteristic, very little is known about tetralin utilization by bacteria, and just a few bacterial strains able to grow on tetralin as the only carbon and energy source have been reported (33). By identifying accumulated intermediates, several reports suggest that some bacteria, such as Pseudomonas stutzeri AS39 (31), initially hydroxylate and further oxidize the alicyclic ring, while others, such as Corynebacterium sp. strain C125 (34), initially dioxygenate the aromatic ring which is cleaved in the extradiol position (meta-cleavage pathway). A strain designated TFA, which is able to grow by using tetralin as the only carbon and energy source, was recently isolated and ascribed to the species Sphingomonas macrogoltabidus. Initial characterization of this strain showed that TFA cannot grow by using other aromatic compounds such as naphthalene, biphenyl, or xylenes as carbon and energy sources (22). Physical and genetic analysis of mutants led to the identification of a genomic region involved in tetralin biodegradation which comprises two divergent operons (22). Identification of thnC, coding for an extradiol dioxygenase, and characterization of the reaction catalyzed by its gene product indicated that biodegradation of tetralin by strain TFA also involves an initial metabolization of the aromatic ring by a meta-cleavage catabolic pathway (2). However, a complete biodegradation pathway has not yet been established.
An important step in meta-cleavage pathways is the hydrolysis of the ring fission product. Substrate specificity of hydrolases catalyzing this step may be a key determinant of the selectivity of the pathway with respect to the degradation of various aromatic compounds (13, 14). These enzymes belong to the group of β-ketolases (EC 3.7.1.-), which hydrolyze carbon-carbon bonds of β-diketones. C-C bond hydrolytic cleavage has been considered a relatively rare enzymatic reaction type and has been little studied in spite of its potential in organic synthesis (27). However, characterization of catabolic meta-cleavage pathways of different aromatic compounds in the last years has led to the identification of an increasing number of hydrolases catalyzing C-C bond cleavage in different gram-positive and gram-negative bacterial species. Some of them have been purified and characterized (3, 5, 8, 10, 13, 18, 25, 32).
Hydrolases involved in catabolic meta-cleavage pathways of aromatic compounds are a class of β-ketolases (27) which hydrolyze linear C-C bonds of vinylogous 1,5-diketones formed by the dioxygenative meta-cleavage of activated arenes. This results in production of a vinylpyruvate and a carboxylate. Sequence comparisons of some of these hydrolases indicate that they are members of the α/β-hydrolase fold superfamily of enzymes (8, 26), which contain a catalytic triad with the configuration nucleophile-acid-histidine in order of amino acid sequence. In vinylogous β-ketolases and other hydrolytic enzymes, the catalytic triad serine-aspartate-histidine is well conserved, and mutagenesis studies on XylF, the hydrolase of the TOL pathway, have shown that these residues are critical for catalysis (8).
We report here the sequence analysis of a gene designated thnD, which codes for a hydrolase involved in tetralin biodegradation by S. macrogoltabidus strain TFA; the overproduction, purification, and characterization of ThnD; and the identification of the reaction product. Unlike all other known hydrolases involved in meta-cleavage pathways of aromatic compounds, which cleave linear C-C bonds, ThnD hydrolyzes a C-C bond comprised in the alicyclic ring of the tetralin derivative, thus yielding a single dicarboxylate product.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Escherichia coli DH5α (17) was used for cloning and isolation of DNA for sequencing. NCM631/pIZ227 (16), a strain producing the T7 RNA polymerase, was used to overproduce ThnD. E. coli strains were routinely grown in Luria-Bertani medium. Strain TFA and its mutant derivative K6 (22) were grown in mineral medium (9) with tetralin in the vapor phase and β-hydroxybutyrate (1 g liter−1) as the carbon and energy source.
Plasmids pIZ608, pIZ591, and pIZ578 have been described elsewhere (2, 22). A 1,349-bp StuI-ClaI fragment from pIZ608 was cloned into pBluescript II KS(+) (Stratagene) and commercially sequenced (Boehringer Mannheim). The same StuI-ClaI fragment was cloned into pIZ578 linearized with SmaI and ClaI to construct pIZ670. To construct pIZ671, a 104-bp deletion was generated in pIZ670 by oligonucleotide site-directed mutagenesis, as previously described (23), using the oligonucleotide 5′GAAGGTCGTAGTGAAGC3′. The deletion of 104 bp was designed to join the second codon of thnD to the frame coding for a His10 tag located upstream.
Overexpression, purification, and electrophoretic conditions.
For overexpression of thnD, E. coli NCM631/pIZ227 was transformed with pIZ670 or pIZ671. The resulting transformants were grown in Luria-Bertani liquid medium at 26°C to reach an optical density at 600 nm of 0.7. They were then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) overnight (10 to 12 h). Cells were harvested by centrifugation, frozen in liquid nitrogen, broken with aluminum oxide 90 (Merck), and suspended in 0.01 volume of 20 mM Tris-HCl (pH 8.0) plus 100 mM NaCl. Purification of the His-tagged ThnD was performed by affinity chromatography with a Co2+-bound resin, following the instructions of the TALON metal affinity resin user manual (Clontech Laboratories, Inc.). Imidazole (0.2 M) was used to elute the protein. Sample preparation and sodium dodecyl sulfate-polyacrylamide gel electrophoresis were performed essentially as described (24). Gels were stained with GELCODE Blue stain reagent (Pierce).
Chemicals.
Dihydroxytetralin (DHT) was chemically synthesized (2). Catechol, 3-methylcatechol, 4-methylcatechol, and 2,3-dihydroxybiphenyl were purchased from Aldrich at the highest purity available. The corresponding ring fission products were synthesized biologically by whole cells of NCM631/pIZ227,pIZ591, which overproduce the broad-substrate-specificity extradiol dioxygenase ThnC (2).
Activity assays.
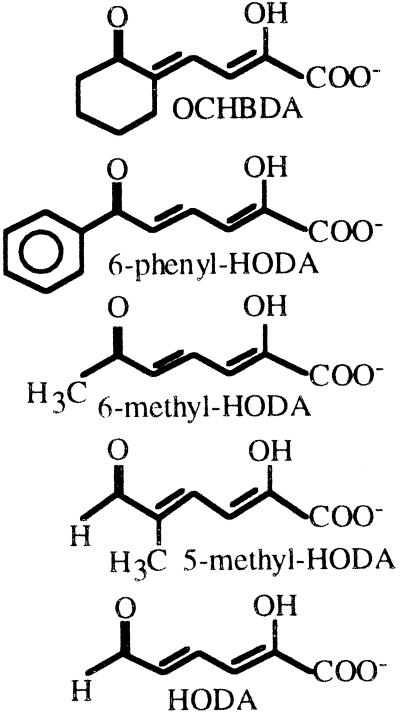
One unit of enzyme activity was defined as the amount of enzyme that converts 1 μmol of substrate per min. Hydrolase activity towards the different ring fission products, which are yellow compounds, was determined in 20 mM Na2HPO4-KH2PO4 buffer (pH 7.2), by measuring substrate consumed at 65°C. The extinction coefficients used for the following substrates were 2-hydroxy-4-(2-oxocyclohexyl)-buta-2,4-dienoic acid (OCHBDA), λmax = 336 nm, ɛ = 12.26 mM−1 cm−1 (2); 2-hydroxy-6-oxohexa-2,4-dienoic acid (HODA), λmax = 375 nm, ɛ = 36 mM−1 cm−1; 2-hydroxy-6-oxohepta-2,4-dienoic acid (6-methyl-HODA), λmax = 388 nm, ɛ = 13.8 mM−1 cm−1; 2-hydroxy-5-methyl-6-oxohexa-2,4-dienoic acid (5-methyl-HODA), λmax = 382 nm, ɛ = 28.1 mM−1 cm−1; and 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoic acid (6-phenyl-HODA), λmax = 434 nm, ɛ = 13.2 mM−1 cm−1 (20). Protein concentration was determined by the method of Bradford (4), with bovine serum albumin as the standard. All assays were quantified using a Beckman DU 640 spectrophotometer equipped with a thermojacketed cuvette holder.
Molecular weight determination.
The relative molecular weight of the native enzyme was determined by gel filtration through a Pharmacia Biotech Sephacryl S-300 HR column (15-ml bed volume) calibrated with horse pancreas ferritine (Mr, 440,000), bovine liver catalase (Mr, 232,000), rabbit muscle aldolase (Mr, 158,000), and ovalbumin (Mr, 45,000) as reference proteins. Samples of 60 μl were loaded to the column, which was equilibrated with buffer containing 50 mM Tris-HCl (pH 8.0) and 100 mM NaCl. Protein elution from the column was in the same buffer at a flow rate of 0.4 ml min−1. Fractions of 225 μl were collected and assayed for hydrolase activity.
Identification of intermediates.
The products from the hydrolase reaction were separated and detected by high-pressure liquid chromatography (HP 1100 series; Hewlett-Packard, Waldbronn, Germany) equipped with a diode array detector, using a reversed-phase column (ODS Hypersil 5 μm, 250 by 2 mm; Hewlett-Packard).
To identify the hydrolase reaction product in the tetralin pathway, induced whole cells of NCM631/pIZ227,pIZ591 were used to produce high amounts of the yellow compound from DHT. Cells were removed by centrifugation, and purified hydrolase was added to the supernatant. When the yellow compound turned over, the supernatant was acidified to pH 2, and the hydrolase product was extracted with Bakerbond spe extraction columns (Mallinckrodt Baker B.V.), following the supplier's instructions. The product was eluted with 1/7.5 sample volume of methanol. The solvent was subsequently changed to dichloromethane in order to just methylate the carboxyl groups. The solution was methylated with diazomethane (6). A sample of the methylated product was concentrated to dryness under a stream of N2. The residue was dissolved in methanol and blown for 30 min with hydrogen using PtO2 as the catalyst or trimethylsilylated with N,O-bis-trimethylsilylfluoroacetamide (28). Afterwards the solution was filtered, concentrated under nitrogen and injected directly in the gas chromatography-mass spectrometry (GC-MS) system. The resulting products were analyzed by GC-MS using a Fisons mass selective detector (MD800; VG Analytical, Manchester, United Kingdom) with a DB-5 MS fused silica column (inner diameter, 30 m by 0.25 mm; 0.25-μm film thickness; J&W Scientific, Folsom, Calif.). The column program temperature used was 150°C (5-min isothermal) to 240°C (final hold, 15 min) at 4°C/min. The injector was 250°C, and the transfer line temperature was 250°C. The carrier gas (helium) flow rate was 1 ml min−1. The end of column was inserted directly into the ion source block. The mass spectra were generated in EI+ (electron ionization positive mode) at 70 eV.
Sequence analysis comparison.
The resulting sequence of 1,346 bp was initially compared to those in the databases using the BLASTp and tBLASTn programs (1). Sequences which showed high similarity to that of strain TFA were aligned using the ClustalX program (37) with default parameters. A distance matrix and a phylogenetic tree were constructed using the neighbor-joining method (30) and visualized with the TreeView program.
Nucleotide sequence accession number.
The nucleotide sequence reported here has been submitted to the DDBJ, EMBL, and GenBank nucleotide sequence databases under accession no. AF204963.
RESULTS AND DISCUSSION
Cloning and sequencing of thnD and sequence analysis of hydrolases.
A collection of nonpolar KIXX insertion mutants of strain TFA unable to grow on tetralin was previously constructed (22). When grown in the presence of β-hydroxybutyrate plus tetralin, mutant K6 produced a yellow pigment with two absorption peaks at 336 nm and 417 nm, which shifted depending on the pH. Absorption spectra of the accumulated compound at different pHs (data not shown) matched those of OCHBDA, the identified product of the ring fission reaction catalyzed by the extradiol dioxygenase ThnC (2). This result suggested that in the mutant K6, the catabolic pathway of tetralin was blocked in the step following meta-cleavage of the aromatic ring. A 1,349-bp DNA fragment encompassing the insertion site in the mutant K6 was subcloned from the plasmid pIZ608 and sequenced.
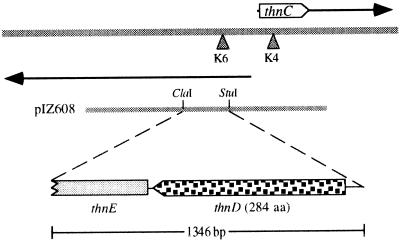
Translation of the nucleotide sequence in all possible reading frames revealed the existence of a complete open reading frame (ORF) of 284 amino acids followed by an ORF for which only a partial sequence was obtained (Fig. 1). Comparison of the deduced amino acid sequence of the complete ORF to those in the databases revealed that it was highly similar to hydrolases of different meta-cleavage pathways of aromatic compounds. Similarity was highest to BphD encoded in the plasmid pNL1 of Sphingomonas aromaticivorans F199 (29) and to EtbD from Rhodococcus sp. strain RHA1 (19), with which it showed 53 and 51% identity, respectively. The incomplete ORF showed high similarity to hydratases of meta-cleavage pathways. Therefore, the identified ORFs were designated thnD and thnE, respectively (Fig. 1). Restriction analysis of the sequence showed that the KIXX cassette was inserted at the second codon of thnD in the mutant K6. Direction of transcription of thnD and thnE is opposite to that of the previously identified gene thnC (2) (Fig. 1), thus confirming the existence of two divergent operons involved in tetralin biodegradation. This was previously inferred by complementation analysis of mutants (22).
FIG. 1.
Representation of the genomic region of strain TFA involved in tetralin biodegradation, showing the divergent thnC and thnD genes. Arrows represent divergent operons. Triangles represent locations of KIXX insertions in mutants unable to grow on tetralin as the only carbon source. aa, amino acids.
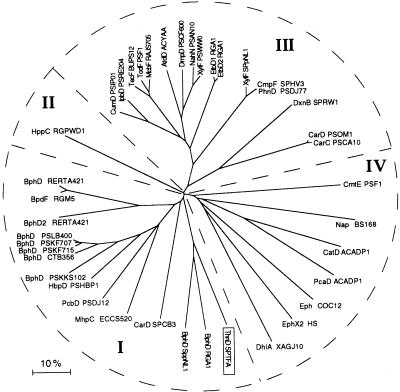
A dendrogram resulting from the comparison of amino acid sequences of hydrolases involved in catabolic meta-cleavage pathways of aromatic compounds and other hydrolases showing significant similarity to ThnD is shown in Fig. 2. The enzymes have been divided in four groups according to their sequence similarity relationships. Except MhpC, all hydrolases of group I are involved in biodegradation of bicyclic molecules such as biphenyls, carbazole, or tetralin. On the other hand, most of the enzymes of group III are involved in biodegradation of monocyclic compounds, though DxnB, CarD PSOM1, and CarC PSCA10, comprising the highly divergent branch of group III, are part of catabolic pathways of bicyclic molecules. Group II is represented by a single hydrolase involved in biodegradation of 3-hydroxyphenylpropionic acid. Except CmtE, which is a C-C bond hydrolase, the heterogeneous group IV is composed of carbon-heteroatom hydrolases such as carboxylesterases (represented by Nap), enol-lactone hydrolases involved in catabolic ortho-cleavage pathways (CatD and PcaD), epoxide hydrolases (Eph), and the haloalkane dehalogenase DhlA. Except DhlA and epoxide hydrolases, all other enzymes in Fig. 2 are serine hydrolases since the catalytic triad serine-aspartate-histidine is conserved.
FIG. 2.
Dendrogram showing the best tree obtained by the neighbor-joining method, from the alignment of 39 sequences showing significant similarity to ThnD. Scale represents distance expressed as percentage of divergence. The ThnD sequence is boxed. GenBank accession numbers for the following other hydrolases are given parenthetically: BphD RGA1 (D78322), BphD SPpNL1 (AF079317), CarD SPCB3 (AF060489), MhpC ECCS520 (Y09555), PcbD PSDJ12 (D44550), HbpD PSHBP1 (U73900), BphD PSKKS102 (M26433), BphD CTB356 (L34338), BphD PSKF715 (M33813), BphD PSKF707 (D85851), BphD PSLB400 (X66123), BphD2 RERTA421 (AB014348), BpdF RGM5 (U44891), BphD RERTA421 (D88016), HppC RGPWD1 (U89712), CumD PSIP01 (D83955), IpbD PSRE204 (AF006691), TecF BUPS12 (U78099), TodF PSF1 (Y18245), McbF RAJS705 (AJ006307), AtdD ACYAA (AB008831), DmpD PSCF600 (X52805), NahN PSAN10 (AF039534), XylF PSpWW0 (M64747), EtbD1 RGA1 (AB004320), EtbD2 RGA1 (AB004321), XylF SPpNL1 (AF079317), CmpF SPHV3 (Z84817), PhnD PSDJ77 (U83881), DxnB SPRW1 (X72850), CarD PSOM1 (AB001723), CarC PSCA10 (D89064), CmtE PSF1 (U24215), Nap BS168 (AB001488), CatD ACADP1 (AF009224), PcaD ACADP1 (L05770), Eph COC12 (AJ224332), EphX2 HS (L05779), DhlA XAGJ10 (M26950).
In general, sequence divergences of hydrolases of meta-cleavage pathways were higher than that of the corresponding extradiol dioxygenases (11). On the other hand, as for extradiol dioxygenases, sequences of hydrolases involved in degradation of bicyclic compounds and those involved in degradation of monoaromatic compounds tend to cluster together in two different groups (I and III in Fig. 2). Therefore, sequence similarity relationships among hydrolases also reflect substrate specificity. However, there are notable exceptions, such as MhpC and HppC; CumD and CmtE; and CarC, CarDPSOM1, and CarDPSCB3. Hydrolases within each set of enzymes hydrolyze the same substrate, but their sequences show very low similarity and cluster in different groups (Fig. 2), which suggests functional convergence of some hydrolases. This and the overall high divergence found among these hydrolases are consistent with the view that members of the α/β-hydrolase fold family do not have to share significant primary structure similarity in spite of having a similar mechanism of reaction (26).
Comparison of ThnD to sequences in the databases did not show similarity to other C-C bond hydrolases cleaving 1,3-diketones, such as fumarylacetoacetate hydrolases or kynureninases (27). Alignment of these sequences together with those in Fig. 2 showed an overall divergence of 80 to 95%. Therefore, C-C bond hydrolases cleaving vinylogous 1,5-diketones are more related in sequence to C-heteroatom hydrolases (group IV) than to C-C bond hydrolases cleaving 1,3-diketones, in spite of the latter being ascribed to the same biochemical group (EC 3.7.1.-).
Overproduction and purification of ThnD.
Involvement of ThnD in degradation of tetralin was shown by the Thn− phenotype of the mutant K6 (22), which bears a nonpolar KIXX insertion in thnD. The sequence of ThnD and the identification of OCHBDA as the intermediate accumulated in the mutant K6 suggested that ThnD catalyzes hydrolysis of the ring fission product of DHT. However, to confirm that thnD codes for a hydrolase catalyzing the step following aromatic ring cleavage, the catalysis reaction had to be characterized. In order to identify, overproduce, and purify the thnD gene product in a single step for subsequent analysis, the E. coli strain NCM631/pIZ227 was transformed with plasmid pIZ670 or pIZ671. pIZ670 drives production of native ThnD, while pIZ671 should drive production of an His10-tagged ThnD, which also contains the peptide signal for the protease factor Xa.
The overproducing strain NCM631/pIZ227 bearing pIZ670 accumulated a protein with an apparent molecular mass of 32 kDa (not shown), consistent with the molecular weight deduced from its coding sequence (32,040). When bearing the plasmid pIZ671, the overproducing strain accumulated to significantly higher amounts a product of 35 kDa, consistent with the predicted molecular weight of the His-tagged protein (34,293). Approximately half of the His-tagged protein was soluble and could be purified by affinity chromatography with a cobalt-bound resin (data not shown). Attempts to remove the N-terminal tail of the purified His-tagged protein with the factor Xa were unsuccessful, thus suggesting that its signal sequence is occluded in the native conformation of this protein. Therefore, the His-tagged form was used for subsequent characterization.
Using both the E. coli crude extract enriched in native ThnD and the purified His-tagged ThnD, a spectrophotometric hydrolase activity assay was set, based on consumption of OCHBDA, which was biologically produced from DHT by ThnC (2).
Biochemical properties of ThnD.
Enzyme activity assays using purified ThnD were run at different temperatures to estimate its optimum reaction temperature. The rate of enzyme-dependent substrate consumption increased with increasing temperature until a maximum was reached at 65°C (not shown). Stability of the hydrolase activity was also tested by incubating His-tagged ThnD at room temperature and at 70°C and measuring activity at different times. Activity of the enzyme was fully stable for 8 h at room temperature, and heat treatment at 70°C for 2 h resulted in 78% of the initial activity (not shown). These results indicate that ThnD is a thermostable hydrolase efficiently working at high temperatures. This is an interesting characteristic of ThnD given the potential interest of this class of enzymes in organic synthesis (27).
Subunit composition of purified His-tagged ThnD was estimated by calibrated gel filtration chromatography. The elution volume of maximum hydrolase activity corresponded to a size of 273 ± 9 kDa (not shown), which indicated that active His-tagged ThnD consists of eight subunits. Oligomeric structure of hydrolases involved in meta-cleavage pathways appears to be variable, since monomers (5), dimers (18, 25), tetramers (13, 32) and octamers (18) have been previously described. Although the octameric structure has been shown for the His-tagged protein, this may be the likely structure of native ThnD since BphD from Rhodococcus sp. strain RHA1, which showed high similarity to ThnD and an optimum reaction temperature at 65°C, is also an octamer (18).
Purified His-tagged ThnD was found to obey the classical Michaelis-Menten kinetic. Assuming an octameric structure, the kinetic parameters calculated for the His-tagged protein at its optimum temperature were as follows: turnover number (kcat), 92.1 s−1; Km, 26.3 μM; and kcat/Km, 35.23 × 10−5 M−1 s−1. Activity assays using crude extracts enriched in native ThnD showed that Km for OCHBDA of native or His-tagged ThnD was the same, thus suggesting that the His tag does not dramatically affect ThnD activity.
The capacity of ThnD to hydrolyze other ring fission products of catabolic meta-cleavage pathways of monocyclic or bicyclic aromatic compounds was also tested. Consistently with its clustering with BphD enzymes (Fig. 2), ThnD also efficiently hydrolyzed 6-phenyl-HODA (Table 1), the ring fission product of 2,3-dihydroxybiphenyl, which has a large substituent at C-6. Kinetic parameters of the reaction using this substrate were similar to those shown using OCHBDA, thus suggesting that ThnD could be proficient in biphenyl meta-cleavage pathways. Benzoate was identified as a reaction product of the hydrolysis of 6-phenyl-HODA (not shown), thus suggesting that the hydrolysis mechanism of ThnD is similar to that previously described for BphD (32) or MhpC (21) hydrolases. ThnD very poorly hydrolyzed 6-methyl-HODA, the ring fission product of 3-methylcatechol, consistent with the view that the size of the substituent at C-6 is important for substrate affinity (32). The aldehydes HODA and 5-methyl-HODA, the ring fission products of catechol and 4-methylcatechol, respectively, were hydrolyzed even less efficiently.
TABLE 1.
Activity of ThnD towards different ring fission products
| Substratea | Relative activity (%) | Km (μM) | kcat (s−1) | kcat/Km (10−5) (M−1 s−1) |
|---|---|---|---|---|
 |
100 | 26.3 | 92.5 | 35.23 |
| 45.7 | 34.8 | 48.8 | 14.03 | |
| 1 | NDb | ND | ND | |
| <1 | ND | ND | ND | |
| <1 | ND | ND | ND | |
The enol tautomer of each substrate, which is the most abundant form in solution, is depicted. The oxo group of OCHBDA may also partially be in the hydroxyl form.
ND, not determined.
Identification of the hydrolysis product in the tetralin pathway.
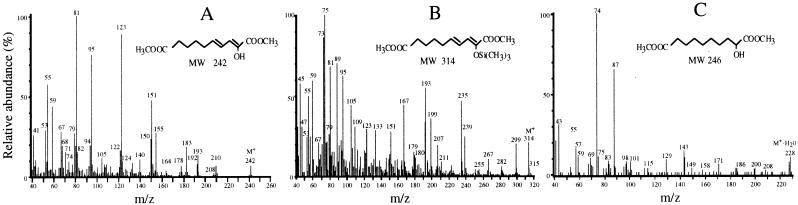
The hydrolysis reaction product(s) of the tetralin pathway was methylated in order to protect the carboxylic groups. GC-MS of the methylated sample showed a major mass peak eluting at 16.75 min. The mass spectrum taken at the top of the peak is shown in Fig. 3A. A weak ion at m/z 242 was thought of as the molecular ion, whose molecular mass matches that of the dimethyl ester of 2-hydroxydeca-2,4-dienedioic acid. A set of even sequential fragments at m/z 210, 150, 122, and 94 shows successive neutral losses of CH3OH, HCOOCH3, CO, and CO, respectively, from the molecular ion and provides evidence of two carboxymethylated groups and an additional C-O bond in the molecule. Another set of odd key fragments at m/z 183, 151, and 123 corresponds to sequential elimination of CH3COO, CH3OH, and CO, respectively. Subsequent ketene (CH2⩵CO) elimination gives the m/z 81 ion as the base peak of the spectrum. Loss of water at m/z 224 was not observed as could be expected if a hydroxyl group were present. This process occurs predominantly by 1-4 elimination through a six-membered intermediate. Absence of this ion may be interpreted as a consequence of the high stability of conjugated double bonds in C-2 to C-4.
FIG. 3.
Mass spectra of the product of hydrolysis modified by methylation (A), methylation and trimethylsilylation (B), or methylation and hydrogenation (C).
The presence of a hydroxyl group was confirmed by analyzing an aliquot of the methylated sample which had been additionally trimethylsilylated. The major peak eluted at 20.36 min and showed a mass spectrum (Fig. 3B) in which the molecular mass of the resulting adduct appeared at m/z 314. The ion at m/z 299 evidenced the loss of one methyl from the trimethylsilyl group, which is typical of trimethylsilylated compounds. The set of key fragment ions at m/z 267 and 235 shows the sequential double elimination of CH3OH from the m/z 299 ion, while m/z 193 represents the loss of CH3OH and the trimethylsilyl group from this ion. The m/z 199 ion represents the cleavage at C-5–C-6 of the molecule containing a double bond in C-4–C-5. Subsequent elimination of trimethylsilyl or methanol from the last ion gives the m/z 167 or 109, respectively. Evidence for the presence of the trimethylsilyl group was provided by the couple of fragments at m/z 73 and m/z 75.
To ascertain the presence of unsaturated double bonds, a second aliquot of the methylated sample was hydrogenated. The mass chromatogram showed a major peak eluting at 19.96 min whose mass spectrum is shown in Fig. 3C. Instead of the molecular ion at expected m/z 246, the highest ion found was at m/z 228, which should be interpreted as a dehydration process suffered by the alcohol prior to being ionized in the ion source, due to a loss of charge stabilization around the conjugated double bonds once they were eliminated from the original compound. The m/z 200 ion is an ethylene expulsion, which is frequently coupled to water elimination. The spectrum of the hydrogenated compound showed a typical fragmentation pattern of methyl esters, with a strong ion at m/z 74 (MacLafferty rearrangement) as base peak of the spectrum and the set of ions at m/z 87, 101, 115, 129, 143, etc., from the linear chain.
Taken together, these data indicate that hydrolysis of the ring fission product in the tetralin pathway results in a single dicarboxylic product with a molecular weight of 214, which contains one hydroxyl group and two double bonds, one of which should be in C-4. Since hydrolysis of ring fission products in other biodegradation pathways involves cleavage of a linear C-C bond, thus releasing two hydrolytic products, it is obvious that the hydrolysis reaction of OCHBDA catalyzed by ThnD results in a significantly different product. However, this is consistent with the hydrolysis of the C-5–C-6 bond of the ring fission product described for other hydrolases. Since C-5 and C-6 of the vinylogous β-diketone in OCHBDA are part of the alicyclic ring (Table 1), hydrolysis of this substrate results in alicyclic ring opening, thus releasing a single linear compound instead of two hydrolytic products.
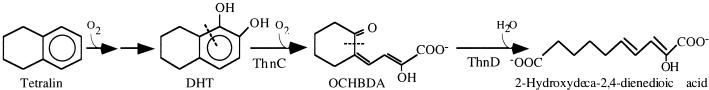
The ability of ThnD to cleave the alicyclic ring of tetralin has strong implications for its catabolism, thus conferring unique characteristics on this pathway. In catabolic pathways of bicyclic molecules such as biphenyl or naphthalene, the two rings are degraded at two distinct stages, each requiring a complete set of enzymes (39). In the tetralin pathway, the aromatic and the alicyclic rings are opened in two subsequent steps by an extradiol dioxygenase and a hydrolase activity (Fig. 4). Further catabolism of the resulting product catalyzed by potential hydratase, aldolase, and aldehyde dehydrogenase activities, which are common activities in degradation pathways of aromatic compounds, would yield pyruvate and the dicarboxylic acid pimelate, which could then enter the general metabolic pathways of the bacteria. Although these activities have not yet been described in the strain TFA, the gene immediately downstream of thnD potentially codes for a hydratase (Fig. 1). In turn, cleavage of the alicyclic ring of tetralin by ThnD makes unnecessary for mineralization of tetralin the use of enzymatic activities involved in degradation of alicyclic rings. Thus, it appears that one set of enzymes typically involved in degradation of one aromatic ring is sufficient to degrade both the aromatic and the alicyclic ring of tetralin.
FIG. 4.
Tetralin biodegradation pathway, showing the subsequent cleavage of both rings.
ACKNOWLEDGMENTS
This work was supported by the Spanish Comisión Interministerial de Ciencia y Tecnología, grant BIO96-0908; by the European Union under the ENVIRONMENT Program, contract EV5V-CT92-0192; and by fellowships of the Spanish Ministerio de Educación to M.J.H. and to E.A.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andújar E, Hernáez M J, Kaschabek S R, Reineke W, Santero E. Identification of an extradiol dioxygenase involved in tetralin biodegradation: gene sequence analysis, purification and characterization of the gene product. J Bacteriol. 2000;182:789–795. doi: 10.1128/jb.182.3.789-795.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayly R C, di Berardino D. Purification and properties of 2-hydroxy-6-oxo-2,4-heptadienoate hydrolase from two strains of Pseudomonas putida. J Bacteriol. 1978;134:30–37. doi: 10.1128/jb.134.1.30-37.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–252. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Bünz P V, Falchetto R, Cook A M. Purification of two isofunctional hydrolases (EC 3.7.1.8) in the degradative pathway for dibenzofuran in Sphingomonas sp. strain RW1. Biodegradation. 1993;4:171–178. doi: 10.1007/BF00695119. [DOI] [PubMed] [Google Scholar]
- 6.Cohen J D. Convenient apparatus for the generation of small amounts of diazomethane. J Chromatogr. 1984;303:193. [Google Scholar]
- 7.Dagley S, Evans W C, Ribbons D W. New pathways in the oxidative metabolism of aromatic compounds by microorganisms. Nature (London) 1960;188:560–566. doi: 10.1038/188560a0. [DOI] [PubMed] [Google Scholar]
- 8.Díaz E, Timmis K N. Identification of functional residues in a 2-hydroxymuconic semialdehyde hydrolase. A new member of the α/β hydrolase-fold family of enzymes which cleaves carbon-carbon bonds. J Biol Chem. 1995;270:6403–6411. doi: 10.1074/jbc.270.11.6403. [DOI] [PubMed] [Google Scholar]
- 9.Dorn E, Hellwig M, Reineke W, Knackmuss H-J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99:61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- 10.Duggleby C J, Williams P. Purification and properties of the 2-hydroxy-6-oxohepta-2,4-dienoate hydrolase (2-hydroxymuconic semialdehyde hydrolase) encoded by the TOL plasmid pWW0 from Pseudomonas putida mt-2. J Gen Microbiol. 1986;132:717–726. [Google Scholar]
- 11.Eltis L, Bolin J. Evolutionary relationships among extradiol dioxygenases. J Bacteriol. 1996;178:5930–5937. doi: 10.1128/jb.178.20.5930-5937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferrante A A, Augliera J, Lewis K, Klibanov A M. Cloning of an organic solvent-resistance gene in Escherichia coli: the unexpected role of alkylhydroperoxide reductase. Proc Natl Acad Sci USA. 1995;92:7617–7621. doi: 10.1073/pnas.92.17.7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa K, Hirose J, Suyama A, Zaiki T, Hayashida S. Gene components responsible for discrete substrate specificity in the metabolism of biphenyl (bph operon) and toluene (tod operon) J Bacteriol. 1993;175:5224–5232. doi: 10.1128/jb.175.16.5224-5232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa K, Tomizuka N, Kamibayashi A. Effect of chlorine substitution on the bacterial metabolism of various polychlorinated biphenyls. Appl Environ Microbiol. 1979;38:301–310. doi: 10.1128/aem.38.2.301-310.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaydos R M. Naphthalene. In: Grayson M, Eckroth D, editors. Kirk-Othmer encyclopedia of chemical technology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1981. pp. 698–719. [Google Scholar]
- 16.Govantes F, Molina-López J A, Santero E. Mechanism of coordinated synthesis of the antagonistic regulatory proteins NifL and NifA of Klebsiella pneumoniae. J Bacteriol. 1996;178:6817–6823. doi: 10.1128/jb.178.23.6817-6823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Hatta T, Shimada T, Yoshihara T, Yamada A, Masai E, Fukuda M, Kiyohara H. Meta-fission product hydrolases from a strong PCB degrader Rhodococcus sp. RHA1. J Ferment Bioeng. 1998;85:174–179. [Google Scholar]
- 19.Hauschild J E, Masai E, Sugiyama K, Hatta T, Kimbara K, Fukuda M, Yano K. Identification of an alternative 2,3-dihydroxybiphenyl 1,2-dioxygenase in Rhodococcus sp. strain RHA1 and cloning of the gene. Appl Environ Microbiol. 1996;62:2940–2946. doi: 10.1128/aem.62.8.2940-2946.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heiss G, Stolz A, Kuhm A E, Müller C, Klein J, Altenbuchner J, Knackmuss H-J. Characterization of a 2,3-dihydroxybiphenyl dioxygenase from the naphthalenesulfonate-degrading bacterium strain BN6. J Bacteriol. 1995;177:5865–5871. doi: 10.1128/jb.177.20.5865-5871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson I M, Bugg T D. Pre-steady-state kinetic analysis of 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid 5,6-hydrolase: kinetic evidence for enol/keto tautomerization. Biochemistry. 1997;36:12252–12258. doi: 10.1021/bi971116j. [DOI] [PubMed] [Google Scholar]
- 22.Hernáez M J, Reineke W, Santero E. Genetic analysis of biodegradation of tetralin by a Sphingomonas strain. Appl Environ Microbiol. 1999;65:1806–1810. doi: 10.1128/aem.65.4.1806-1810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Lam W W, Bugg T D. Purification, characterization, and stereochemical analysis of a C-C hydrolase: 2-hydroxy-6-keto-nona-2,4-diene-1,9-dioic acid 5,6-hydrolase. Biochemistry. 1997;36:12242–12251. doi: 10.1021/bi971115r. [DOI] [PubMed] [Google Scholar]
- 26.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, Sussman J L, Verschueren K H, Goldman A. The alpha/beta hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 27.Pokorny D, Steiner W, Ribbons D M. β-Ketolases—forgotten hydrolytic enzymes? Trends Biotechnol. 1997;15:291–286. [Google Scholar]
- 28.Poole C F. Recent advances in the silylation of organic compounds for gas chromatography. In: Blau K, King G S, editors. Handbook of derivatives for chromatography. London, United Kingdom: Heyden; 1978. pp. 152–200. [Google Scholar]
- 29.Romine M, Stillwell L, Wong K-K, Thurston S, Sisk E, Sensen C, Gaasterland T, Fredrickson J, Saffer J. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber A F, Winkler U K. Transformation of tetralin by whole cells of Pseudomonas stutzeri AS 39. Eur J Appl Microbiol Biotechnol. 1983;18:6–10. [Google Scholar]
- 32.Seah S T, Terracina G, Bolin J T, Riebel P, Snieckus V, Eltis L. Purification and preliminary characterization of a serine hydrolase involved in the microbial degradation of polychlorinated biphenyls. J Biol Chem. 1998;273:22943–22949. doi: 10.1074/jbc.273.36.22943. [DOI] [PubMed] [Google Scholar]
- 33.Sikkema J, de Bont J A M. Isolation and initial characterization of bacteria growing on tetralin. Biodegradation. 1991;2:15–23. [Google Scholar]
- 34.Sikkema J, de Bont J A M. Metabolism of tetralin (1,2,3,4-tetrahydronaphthalene) in Corynebacterium sp. strain C125. Appl Environ Microbiol. 1993;59:567–572. doi: 10.1128/aem.59.2.567-572.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sikkema J, de Bont J A M, Poolman B. Interactions of cyclic hydrocarbons with biological membranes. J Biol Chem. 1994;269:8022–8028. [PubMed] [Google Scholar]
- 36.Sikkema J, Poolman B, Konings W N, de Bont J A M. Effects of the membrane action of tetralin on the functional and structural properties of artificial and bacterial membranes. J Bacteriol. 1992;174:2986–2992. doi: 10.1128/jb.174.9.2986-2992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trudgill P W. Microbial degradation of the alicyclic ring. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker; 1984. pp. 131–180. [Google Scholar]
- 39.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]