Abstract
The ytrABCDEF operon of Bacillus subtilis was deduced to encode a putative ATP-binding cassette (ABC) transport system. YtrB and YtrE could be the ABC subunits, and YtrC and YtrD are highly hydrophobic and could form a channel through the cell membrane, while YtrF could be a periplasmic lipoprotein for substrate binding. Expression of the operon was examined in cells grown in a minimal medium. The results indicate that the expression was induced only early in the stationary phase. The six ytr genes form a single operon, transcribed from a putative ςA-dependent promoter present upstream of ytrA. YtrA, which possesses a helix-turn-helix motif of the GntR family, acts probably as a repressor and regulates its own transcription. Inactivation of the operon led to a decrease in maximum cell yield and less-efficient sporulation, suggesting its involvement in the growth in stationary phase and sporulation. It is known that B. subtilis produces acetoin as an external carbon storage compound and then reuses it later during stationary phase and sporulation. When either the entire ytr operon or its last gene, ytrF, was inactivated, the production of acetoin was not affected, but the reuse of acetoin became less efficient. We suggest that the Ytr transport system plays a role in acetoin utilization during stationary phase and sporulation.
Genome sequencing of Bacillus subtilis revealed some 4,100 protein-coding genes, a quarter of which corresponded to a number of gene families (8). The largest family, of at least 78 members, encodes the ATP-binding cassette (ABC) proteins (16), suggesting that the ABC transport systems play an important role in this organism. Bacterial ABC transport systems, functionally classified as the ABC importers and extruders, are known as multisubunit machinery comprising membrane-associated protein subunits (2). A typical ABC transport system consists of two hydrophobic membrane-spanning domains (MSDs) and two hydrophilic ABC domains localized at the cytoplasmic face of the cell membrane. The ABC proteins are also called traffic ATPases (1) and contain the ABC domains that supply the energy for the active transport by hydrolyzing ATP. In the bacterial ABC importers, the ABC domains and the MSDs are commonly present on separate polypeptides, and in several systems, the ABC domains have been shown to associate tightly with two hydrophobic proteins containing MSDs (7). In addition, all the bacterial ABC import systems include a periplasmic substrate-binding protein that interacts with a coming substrate, binds to it, and presents it to the import complex (2). It is also known that in gram-positive species, all of such substrate-binding proteins are periplasmic lipoproteins that are anchored to the membrane via N-terminal hydrophobic lipid extension (16).
Recently, Quentin and coworkers reported a computerized prediction, aiming to categorize the 78 ABC proteins of B. subtilis into systems in combination with their MSD and substrate-binding protein partners (16). They were able to classify the 78 systems into 11 subfamilies, further split into 6 subfamilies for importers and 5 for extruders. However, over two-thirds of the members of ABC proteins of B. subtilis have not been functionally characterized. Here we report characterization of one of those functionally unknown ABC transport systems, the ytrABCDEF gene cluster, found at 3118.3 kb on the B. subtilis chromosome (8). YtrB and YtrE were predicted to encode ABC proteins with a nucleotide-binding domain (8, 16). YtrB exhibited weak similarity to PotG of Escherichia coli, involved in putrescine import (14), while YtrE showed homology with BacH of Enterococcus faecalis, required for bacteriocin export (23). YtrC and YtrD, which are paralogous, with 47% identity in 333 amino acid residues, are highly hydrophobic and are plausible MSD proteins, since they were predicted to possess nine and eight transmembrane segments, respectively (16). YtrF could be an MSD protein, because of its putative four transmembrane segments (16), but was also predicted to be a periplasmic lipoprotein (22). It was proposed that the above five gene products could be split into two independent export systems, YtrBCD and YtrEF (16). However, organization of the six ytr genes suggests an alternative possibility, that they might form a single operon for an integrated ABC transport system. The first gene product of the putative operon, YtrA, was proposed to be a transcriptional regulator, since it shares significant similarity with N-terminal regions of many bacterial regulators that contain a helix-turn-helix motif and belong to the GntR family (5, 26), such as FarR of E. coli (15), HutC of Klebsiella aerogenes (20), and GntR of B. subtilis (10). It could possibly regulate this operon.
Acetoin is a product of fermentative metabolism in many prokaryotic and eukaryotic microorganisms including Bacillus spp. (12). Strains of B. subtilis grown in media which contain enough fermentative carbon sources, such as glucose or fructose, produce acetoin as an external carbon storage compound and then reuse it as a carbon and energy source later, during stationary phase and sporulation (9). In B. subtilis, the genes encoding the enzymes for acetoin production have been reported to form a single operon, alsSD (17). In addition, it has been proposed that there are at least two systems for acetoin catabolism: one encoded by the acuABC genes and the other by the acoABCLR genes (4, 6). However, the mechanism for the production and utilization of acetoin by B. subtilis is not fully understood, and nothing is known about transport systems specific for acetoin. We report here that the ytrABCDEF genes form a single operon, probably encoding an ABC transport system involved in the reuse of acetoin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains used in this study are listed in Table 1. Plasmid pMUTIN2mcs (lacZ lacI amp erm) is an integration vector for the targeted gene inactivation in B. subtilis (24). It replicates in E. coli but not in B. subtilis and carries an erythromycin resistance gene that is active in B. subtilis when the plasmid DNA is integrated into the chromosome. In addition, it carries a promoterless lacZ reporter derived from E. coli and the spac promoter regulated by the lac repressor encoded by lacI (25), which allows the expression of the genes downstream of the integration site. E. coli cells harboring plasmids were grown on Luria broth (LB) (18) containing ampicillin (50 μg/ml). B. subtilis cells were grown on the following media containing erythromycin (0.3 μg/ml) when needed: tryptose blood agar base (Difco) supplemented with 0.18% glucose (referred to as TBABG), a sporulation medium (DSM) (19), a minimal medium containing 0.4% glucose as the carbon source (MM) (29), and S1 medium (21).
TABLE 1.
Bacterial strains used in this study
| Species and strain | Genotype | Source or referencea |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | Wild-type strain |
| BFS45 | trpC2 Pytr::pMUTIN2mcs | This work (pytrA3B→168) |
| BFS47 | trpC2 ytrF-pMUTIN2mcs | This work (pytrF1→168) |
| BFS53 | trpC2 ΔytrA::pMUTIN2mcs | This work |
| FU349 | trpC2 ytrF::pMUTIN2mcs | This work (pytrFd→168) |
| E. coli C600 | supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21 | 30 |
Arrows indicate transformation from donor DNA to recipient strain.
Construction of B. subtilis mutant strains.
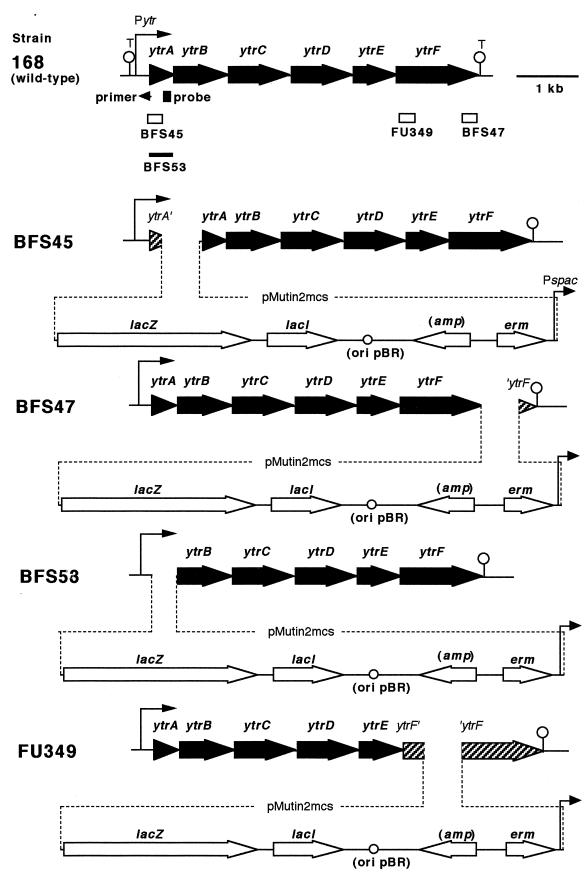
B. subtilis strains BFS45, BFS47, and FU349 were constructed as follows. DNA fragments corresponding to part of the ytrA and ytrF genes (Fig. 1) were amplified by PCR using specific primer pairs and chromosomal DNA of B. subtilis 168 as a template. The specific primer pairs used for the constructions were as follows (restriction sites are underlined): for strain BFS45, ytrAE primer (5′-CCGGAATTCAAAGGTTCAGATCGTATAG-3′) and ytrAB (5′-CGCGGATCCATATATAGGTCCCTCTGC-3′); for BFS47, ytrFH (5′-CCCAAGCTTTTTTAGGCTGTGTGATTG-3′) and ytrFB (5′-CGCGGATCCCGTTCGCATTATAATTCTC-3′); for FU349, ytrFdH (5′-CCCAAGCTTGGCTTCGCGTCTTTATGACG-3′) and ytrFdB (5′-CGCGGATCCGTACGATTGCCTAAAGTAGC-3′). The PCR product for the BFS45 construction was trimmed with EcoRI and BamHI and ligated with pMUTIN2mcs that had been digested with EcoRI and BamHI. For construction of strains BFS47 and FU349, HindIII and BamHI were used instead of EcoRI and BamHI. The ligated DNAs were used to transform E. coli C600 to ampicillin resistance on LB plates. Cloning of the right PCR products into pMUTIN2mcs was confirmed by DNA sequencing. The resulting plasmids, pytrA3B, pytrF1, and pytrFd, were used to transform B. subtilis 168 to erythromycin resistance on TBABG, yielding B. subtilis strains BFS45, BFS47, and FU349, respectively. Correct integration of a single copy of the plasmids into the respective positions by a single-crossover event was confirmed by Southern blot analysis.
FIG. 1.
Genetic organization of the ytr locus of each of B. subtilis strains used in this study. The ytr genes and the pMUTIN2mcs-borne genes are presented as solid and open arrows, respectively. Beneath the genetic organization of strain 168 (wild type), the DNA stretches cloned into pMUTIN2mcs (open boxes) or replaced (thick line) are indicated with the name of the respective mutants. Positions of a probe for the Northern analysis (Fig. 2) and a primer used for extension analysis (Fig. 3) are indicated with a closed box and a small arrowhead, respectively. Positions of the ytr promoter (Pytr) and two putative terminators (T) deduced from the DNA sequence are also indicated. In the diagrams of the four mutant strains, dotted lines indicate integration of the pMUTIN2mcs into the chromosome, and truncated genes are shaded. The IPTG-inducible spac promoter (Pspac), which is regulated by the lac repressor encoded by lacI, drives expression of the downstream genes. The lacZ reporter is expressed under the control of the region upstream of the integration site.
B. subtilis strain BFS53 with a deletion of ytrA was constructed as follows. Two DNA fragments (approximately 1.5 kb) were amplified by PCR using specific primer pairs and chromosomal DNA of B. subtilis 168 as a template; one fragment corresponded to a region upstream of the ytrA gene, and the other corresponded to a downstream one. All the PCR was done using GeneAmp XL PCR kit (Perkin-Elmer). The specific primer pairs used were as follows (restriction sites are underlined): for the upstream fragment, ytrAU1 primer (5′-CATTTTTCGTGCATGCGG-3′) and ytrAU2 (5′-CGCGGATCCTTTCTATACGATCTGAACC-3′); and for the downstream fragment, ytrAD1 (5′-CCCAAGCTTAGGAAATCAGCGCTGATG-3′) and ytrAD2 (5′-ATAGAAAGCGGATTTGCC-3′). The upstream and downstream fragments were trimmed with BamHI and HindIII, respectively, and then ligated with pMUTIN2mcs that had been digested with BamHI and HindIII. Ligated DNA was directly used for transformation of B. subtilis 168 to erythromycin resistance on TBABG. In such transformants, ytrA gene was expected to be replaced by pMUTIN2mcs via a double-crossover event. The replacement was confirmed by PCR analysis using the primer pair of ytrAU1 and ytrAD2 (Fig. 1).
RNA techniques.
Cells of B. subtilis strains were inoculated into MM to an optical density at 600 nm (OD600) of 0.05 and incubated at 37°C with shaking. The cells were harvested 5 h (T−1, 1 h before the beginning of stationary phase), 6 h (T0, point defined as the beginning of stationary phase), 7 h (T1, 1 h after the beginning of stationary phase), and 8 h (T2, 2 h after the beginning of stationary phase) after inoculation, at OD600s of approximately 1.5, 2.8, 4.0, and 4.1, respectively. Total RNA of the cells was extracted by mixing the cells with glass beads, phenol, and cetyltrimethylammonium bromide, and then purified as described previously (3). Alternatively, the cells were inoculated into DSM and grown for 1.5 h (exponential growth), 3 h (transition between exponential growth and stationary phase), 4 h (point defined as the beginning of sporulation), 5 h (1 h after the beginning of sporulation), 7 h (3 h after the beginning of sporulation), and 9 h (5 h after the beginning of sporulation), to give OD600s of approximately 0.4, 1.5, 2.1, 2.3, 2.0, and 2.3, respectively. Total RNA was prepared as described above.
For Northern blot analysis, RNA samples were electrophoresed in a glyoxal gel, transferred to a Hybond-N membrane (Amersham), and hybridized with a labeled probe as described previously (28). To prepare the probe, part of the ytrA coding region (Fig. 1) was amplified by PCR using chromosomal DNA of B. subtilis 168 as a template and a primer pair, ytrAC1 (5′-AGAATGCAAAAACGACACTGG-3′) and ytrAC2 (5′-TCACATCAGCGCTGATTTCC-3′). The PCR product was labeled radioactively by using Bca BEST labeling kit (Takara Shuzo) and [α-32P]dCTP (ICN Biomedicals).
To map a 5′ end of the ytr transcript by primer extension, 50 μg of each RNA was annealed to a primer (5′-CCGATCATCACAAGAACAAG-3′) that had been labeled at its 5′ end by a MEGALABEL kit (Takara Shuzo) and [γ-32P]ATP (Amersham). Primer extension reactions were carried out as described previously (28).
Determination of acetoin concentration in culture media.
B. subtilis cells were cultured at 30°C for 16 h on TBABG plates containing erythromycin when needed. Cells were harvested, washed once with freshly prepared MM, inoculated into MM to an OD600 of 0.05, and grown at 37°C with shaking. When needed, 1 h (T1) and 2 h (T2) after the entry into stationary phase, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and exogenous 10 mM acetoin (Aldrich) were added to the culture, respectively. A portion (1 ml) of the culture was withdrawn at different times during the growth, the cells were removed by centrifugation, and the concentration of acetoin in the supernatant was measured as described previously (4).
RESULTS
Organization of the ytrABCDEF genes.
The ytrABCDEF gene cluster is located at 3118.3 kb on the B. subtilis chromosome (8). As shown in Fig. 1, the six ytr genes are oriented in the same direction. There are only two transcription terminator-like sequences within this region: one located about 300 bp upstream of the ytrA translation start site and the other just downstream of ytrF (11). The intergenic regions between the ytr genes are very short; even the longest one, between ytrC and ytrD, is only 28-bp long. Moreover, translation stop and start regions overlap for ytrA and ytrB, ytrB and ytrC, and ytrE and ytrF. Such overlap can lead to translational coupling (13), suggesting that these genes are cotranscribed. Taken together, the organization of the six ytr genes suggests that they might form an operon.
The ytrABCDEF genes form an operon expressed early in the stationary phase.
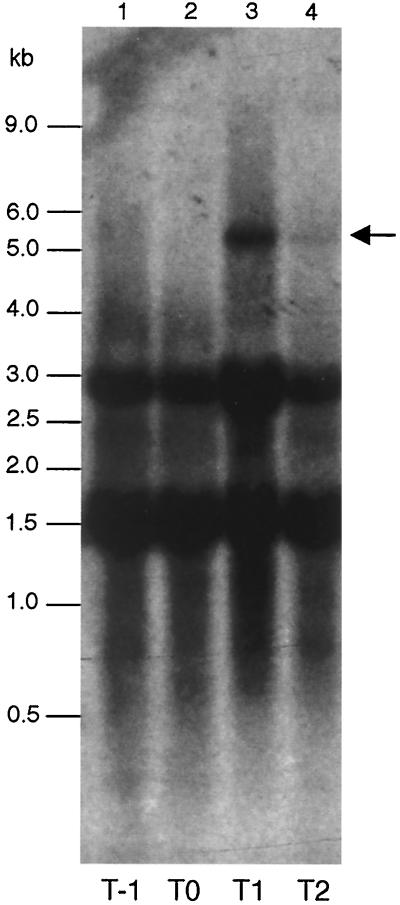
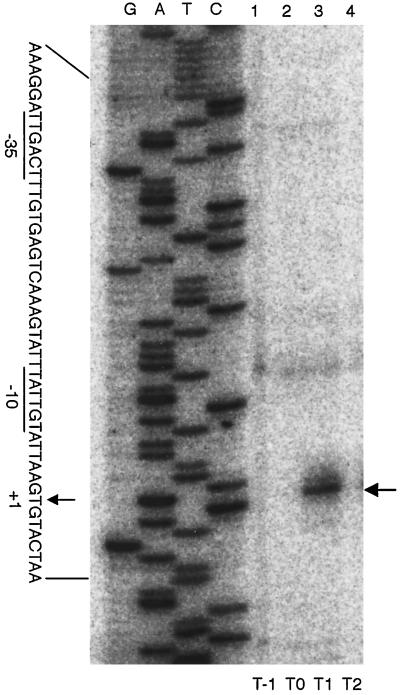
RNA samples prepared from cells of strain 168 grown in MM were subjected to a Northern analysis, using a ytrA specific probe. As shown in Fig. 2, a specific transcript was present 1 h after the entry in stationary phase (T1) but disappeared almost completely 1 h later (T2). The size of the transcript was estimated to be about 5.5-kb, which would just cover all the ytr genes. Interestingly, no such transcript was detected in cells grown in DSM, a nutrient sporulation medium less rich in glucose (19) (data not shown). A 5′ end of the transcript was determined by primer extension analysis (Fig. 3). It mapped 244-bp upstream of the ytrA translation start site, close to a ςA-dependent promoter-like sequence with typical −10 and −35 regions. These results suggest that the ytrABCDEF genes form an operon, which is transcribed early in the stationary phase in the cells grown in MM.
FIG. 2.
Northern analysis of the ytr transcript. Cells of strain 168 were grown in MM, and RNA samples were prepared at various time points; lanes 1, 2, 3, and 4 contained the RNA prepared at T−1, T0, T1, and T2, respectively. The RNA samples were subjected to a Northern analysis targeting the ytr transcript using the probe (Fig. 1). The position of the ytr transcript is indicated with an arrow. The two strong bands at about 1.5 and 3 kb are due to nonspecific hybridization to rRNAs.
FIG. 3.
Mapping of a 5′ end of the ytr transcript. The RNA samples were subjected to a primer extension analysis using the 32P-labeled primer (Fig. 1); lanes 1, 2, 3, and 4, contained the reactions using RNAs prepared at T−1, T0, T1, and T2, respectively. Reverse transcripts were subjected to a gel electrophoresis together with dideoxy sequencing reactions carried out by using the same primer (lanes G, A, T, and C). The position of a major reverse transcript is indicated with an arrow on the right side of the panel. On the left side, the 5′ end position is marked with an arrow (+1, as the transcription start site) in the nucleotide sequence of the noncoding strand of the ytr promoter region, where plausible −10 and −35 regions are underlined.
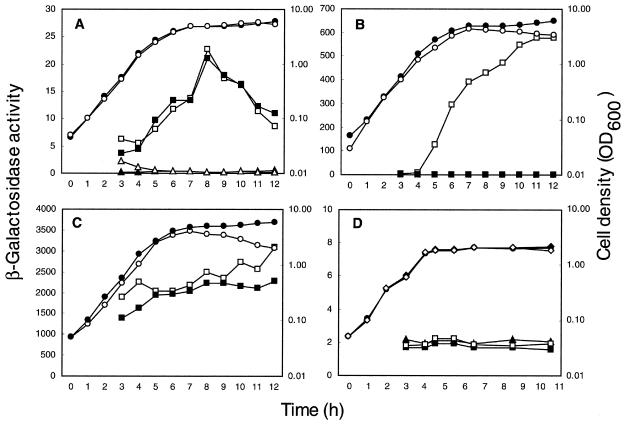
To verify this conclusion, the expression of the ytr operon was monitored with the aid of reporter gene. A strain carrying pMUTIN2mcs in the chromosome was used for this purpose (Fig. 1, strain BFS47), having a lacZ gene immediately downstream of ytrF, the last gene of the putative operon. As shown in Fig. 4A, the β-galactosidase activity in BSF47 cells grown in MM was elevated at the transition between exponential growth and stationary phase to give a peak activity early in the stationary phase and decreased thereafter. In contrast, only a negligible activity was found in cells grown in DSM (Fig. 4D). This expression pattern coincides well with that revealed by the transcription analysis and confirms that the ytr genes form an operon, which is expressed maximally early in the stationary phase in the cells grown in MM.
FIG. 4.
Growth and expression of the lacZ reporter of strains of B. subtilis. Strains BFS47 (ytrF-pMUTIN2mcs) (A), BFS45 (Pytr::pMUTIN2mcs) (B), and BFS53 (ΔytrA::pMUTIN2mcs) (C) were grown in MM (open square, β-galactosidase activity; open circle, cell density) and in MM with 1 mM IPTG (closed square, β-galactosidase activity). Strain 168 (wild type) was also grown in MM (open triangle, β-galactosidase activity; closed circle, cell density) and in MM with 1 mM IPTG (closed triangle, β-galactosidase activity). (D) Strains BFS47 (squares, β-galactosidase activity; diamonds, cell density) and 168 (closed triangle, β-galactosidase activity; closed circle, cell density) were grown in DSM supplemented with (open symbols) and without (closed symbols) 5 mM acetoin; exogenous acetoin was added to the culture 3 h after the inoculation. The experiments were repeated at least three times for each strain and medium with similar results; representative results are shown. β-Galactosidase activity (nanomoles of 2-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per milligram of protein) was determined as described previously (28).
The ytrA gene encodes a negative regulator for the ytr operon.
Sequence analysis indicated that YtrA may be a transcriptional regulator of the GntR family. To investigate the possibility that it regulates the ytr operon, two mutant strains, BFS45 and BFS53, were constructed (Fig. 1). In the first, BFS45, the entire ytr operon including the ytrA gene is under the control of the IPTG-inducible spac promoter (25) and the activity of the natural ytr promoter can be monitored by the expression of lacZ. In the second, BFS53, the ytrA gene is replaced with pMUTIN2mcs and the ytr promoter activity can be monitored as in BFS45, but on a ytrA-null background.
As shown in Fig. 4B when the BFS45 cells were grown in the presence of IPTG, where ytrA was expressed throughout the cell growth, the β-galactosidase activity was essentially nil. On the other hand in the absence of IPTG, where ytrA was repressed, the activity reached high levels upon transition between exponential growth and stationary phase and did not decrease thereafter as observed in the BFS47 cells (Fig. 4A). These results indicate that YtrA negatively regulates the ytr promoter. This conclusion is strengthened by the fact that β-galactosidase activity was constitutively high in the ytrA-null BFS53 cells, both in the presence and the absence of IPTG (Fig. 4C). It should be noted that in the absence of IPTG β-galactosidase activity in the BFS45 cells was not constant but increased regularly, never reaching values as high as those observed in the ytrA null cells. This could be due to a leakage of the spac promoter in the absence of IPTG, allowing some synthesis of the repressor protein in the BFS45 cells.
The ytr operon is involved in acetoin utilization.
Inactivation of the ytr operon, by lack of IPTG in strain BFS45 or by disruption in strain BFS53, led to a slight but reproducible decrease in cell yield at the end of growth in MM (Fig. 4B and C, compare filled and open circles). This observation, together with the fact that the operon is expressed maximally early in the stationary phase (Fig. 2, 3, and 4A), led us to hypothesize that it might be involved in establishment and/or maintenance of stationary growth in a medium containing glucose. Since it is known that B. subtilis cells grown in glucose-rich media such as MM produce acetoin as an external carbon storage compound and then reuse it during stationary phase and sporulation (9), we examined the effect of inactivation of the ytr operon on production and utilization of acetoin.
B. subtilis strains (168 [wild type], BFS45, and BFS47) were grown in MM without IPTG, and the concentration of acetoin in the culture media was measured (Fig. 5A). With the wild-type cells, acetoin appeared early in the stationary phase, accumulated to a concentration of approximately 3 mM at T2, and was almost fully consumed within 3 h. This indicates that the cells could produce and reuse acetoin under these growth conditions. Production and reuse of acetoin by the BFS47 cells, which carry pMUTIN2mcs downstream of ytrF and thus retain the intact ytr operon, were similar to those of the wild-type strain. In contrast, BFS45 cells, which do not express the operon in the absence of IPTG, produced acetoin as the wild-type, but reused it with a substantial delay. All three strains produced and reused acetoin at the same rate in the presence of IPTG, which allowed expression of the ytr operon in BFS45 cells (Fig. 5B). These results indicate that the ytr operon plays a role in acetoin consumption.
FIG. 5.
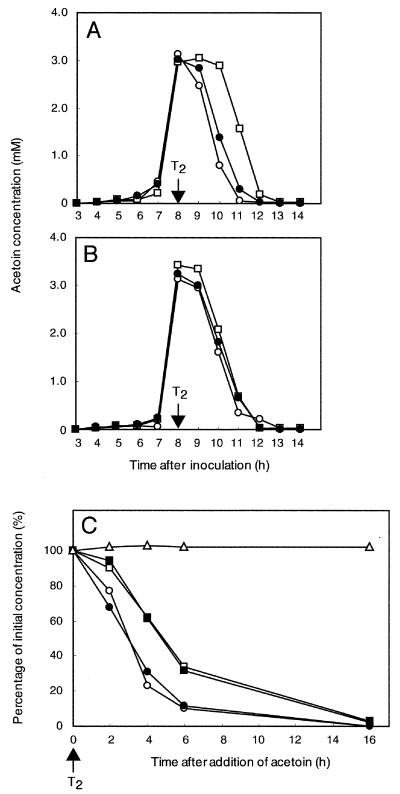
Production and consumption of acetoin by strains of B. subtilis. (A and B) Spontaneous production and consumption of acetoin. Strains of B. subtilis were grown in MM in the absence (A) or presence (B) of 1 mM IPTG added at T1. The concentration of acetoin in the culture medium was determined at various time points after the inoculation as indicated: open circle, strain 168 (wild type); closed circle, strain BFS47 (ytrF-pMUTIN2mcs); open square, strain BFS45 (Pytr::pMUTIN2mcs). (C) Consumption of acetoin added exogenously. Cells of the strains were grown in MM, and at T2 acetoin (10 mM) was added into the cultures exogenously. The concentration of acetoin in the medium was measured at various time points as indicated (100% corresponds to approximately 13 mM), and the percentage of the remaining acetoin is shown: open circles, strain 168 (wild type); closed circles, strain BFS47 (ytrF-pMUTIN2mcs); open squares, strain BFS45 (Pytr::pMUTIN2mcs); closed squares, strain FU349 (ytrF::pMUTIN2mcs); open triangles, cell-free medium. The experiments were repeated at least three times for each strain and medium with similar results; representative results are shown.
To confirm this conclusion, an excess of acetoin (10 mM) was added to the culture at the time of maximal acetoin accumulation (T2) and its concentration was measured thereafter (Fig. 5C). After 6 h less than 10% of the acetoin was present in cultures of the wild-type and BFS47 cells, whereas almost 40% remained in the BFS45 culture. A similar delay in acetoin consumption was observed with another mutant strain, FU349 (Fig. 1), which lacks the ytrF gene (Fig. 5C). Addition of IPTG to the BFS45 culture did not abolish the delay, possibly because the induction of the operon was insufficient to consume the excess acetoin (data not shown).
As mentioned above, acetoin is generally used as a carbon and energy source during stationary phase and sporulation. The utilization during sporulation was tested in a manner similar to that described by Grundy et al. (4) (Table 2). Cells grown in MM until T2 were washed and resuspended in the S1 medium (21) supplemented with or without acetoin as a carbon source and then incubated further until T26; glycine was added to S1 since it was reported that utilization of acetoin under these condition may require it, although the physiological significance of this is unknown (4). The wild-type and strain BFS47 cells, which have the intact ytr operon, grew continuously in MM until T26 to cell densities of about 3 × 108 and sporulated at frequencies of about 60%. In S1 without acetoin, the two strains grew to lower cell densities and sporulated with lower efficiencies. Addition of acetoin to this medium increased slightly but reproducibly both the cell density and sporulation frequency of the two strains. As expected, the strains BFS45 and FU349 cells, which have a defective ytr operon, grew in MM to lower cell densities and also exhibited lower sporulation frequencies than the two ytr-proficient strains. Interestingly, addition of acetoin to S1 did not enhance either the cell density or sporulating ability of these two ytr-deficient strains. The results imply that the ytr mutants, which showed the slower acetoin consumption, do not use acetoin during sporulation as efficiently as the wild-type strains.
TABLE 2.
Sporulation of strains of B. subtilis
| Strain (relevant genotype) | Mediuma | T26 viable cells (CFU ml−1) | T26 spores (CFU ml−1)b | Sporulation (%)c |
|---|---|---|---|---|
| 168 (wild-type) | MM | 3.0 × 108 | 1.9 × 108 | 63 |
| S1d | 5.9 × 107 | 1.0 × 107 | 16 | |
| S1 + acetoin | 1.5 × 108 | 4.1 × 107 | 27 | |
| BSF45 (Pytr::pMUTIN2mcs) | MM | 2.1 × 108 | 3.7 × 107 | 18 |
| S1 | 5.4 × 107 | 3.0 × 106 | 5.5 | |
| S1 + acetoin | 3.0 × 107 | 5.0 × 105 | 1.7 | |
| BFS47 (ytrF-pMUTIN2mcs) | MM | 3.2 × 108 | 2.0 × 108 | 62 |
| S1 | 2.2 × 107 | 2.0 × 106 | 9.1 | |
| S1 + acetoin | 1.1 × 108 | 1.7 × 107 | 15 | |
| FU349 (ytrF::pMUTIN2mcs) | MM | 1.2 × 108 | 2.5 × 107 | 21 |
| S1 | 3.3 × 107 | 3.0 × 106 | 8.9 | |
| S1 + acetoin | 3.1 × 107 | 2.2 × 106 | 7.0 |
Cells were grown until 2 h after the end of exponential growth (T2) in MM at 37°C with shaking, and then the culture was divided into three parts. The cells in the first part were allowed to grow continuously in MM until T26. The cells in the second and the third parts were harvested once, washed and resuspended in S1 medium in the presence and absence of acetoin, respectively, and grown until T26.
Sporulation was quantified by measuring survival after incubation at 70°C for 15 min. The number of CFU was determined by spreading the cells onto TBABG containing erythromycin when needed. Experiments were repeated three times for each strain and medium with similar results; representative results are presented.
Sporulation frequency is relative to the viable cell amount at T26 of each culture of the strains.
S1 medium contained 10 mM glycine and was supplemented with 10 mM acetoin as indicated.
The results above implied a possibility that the ytr operon might be induced by acetoin. To investigate this possibility, we tested the effect of acetoin addition on the expression of the operon by monitoring the lacZ reporter in BFS47 cells. As described above, the β-galactosidase activity in the cells grown in DSM was negligible (Fig. 4D), indicating that the operon was not normally expressed, and production of acetoin under these growth conditions was hardly detected (data not shown). Addition of acetoin (5 mM; higher than the maximal concentration accumulated in MM) to the culture did not lead to higher β-galactosidase activity (Fig. 4D), suggesting that acetoin did not induce the ytr operon.
DISCUSSION
The present work led to several conclusions. (i) The ytrABCDEF genes of B. subtilis form an operon, which is expressed early in the stationary phase in the cells grown in the glucose-rich MM (Fig. 2, 3, and 4A). (ii) The ytr operon is likely transcribed from a promoter, negatively regulated by the expression of ytrA, encoding a protein of the GntR family of repressors (Fig. 3 and 4B and C). (iii) Inactivation of the ytr operon leads to a slower consumption of acetoin in the culture medium during stationary phase (Fig. 5). (iv) This slower consumption of acetoin results in a lower cell density and sporulation frequency (Table 2). (v) Acetoin addition does not induce the ytr operon (Fig. 4D).
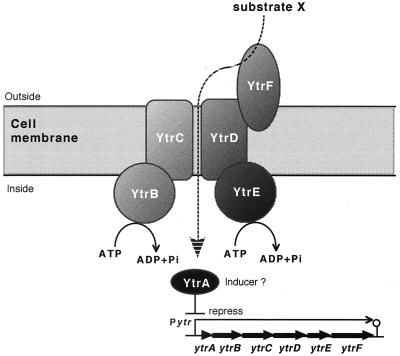
The six ytr genes form an operon transcribed as a 5.5-kb mRNA, mainly in the beginning of the stationary phase in cells grown in MM (Fig. 2 and 4A). The short intergenic regions where translation stop and start sites of the ytr genes overlap, might result in translational coupling, conducive to controlling the synthesis of the gene products with a nearly 1:1 stoichiometry (13). It is plausible that the YtrBCDEF proteins might be the subunits of an integrated ABC transport system, the model of which is depicted in Fig. 6. YtrB and YtrE are situated on the internal side of the cell membrane and function as the ABC proteins to supply the energy for the transport, YtrC and YtrD are the MSD proteins that make a channel through the cell membrane, and YtrF is the substrate-binding protein. In addition, YtrA regulates the expression of the entire operon presumably as a repressor (Figs. 4B and C).
FIG. 6.
Putative functions of the ytr gene products. The ytrABCDEF operon of B. subtilis encodes a putative ABC import system responsible for a substrate, X; YtrB and YtrE are the ABC subunits, the hydrophobic YtrC and YtrD are the MSD proteins that form a channel through the cell membrane, YtrF is a periplasmic lipoprotein for substrate binding, and YtrA may regulate Pytr as a repressor.
The inactivation of the ytr operon led to a less-efficient utilization of acetoin, but acetoin production was not affected (Fig. 5 and Table 2). Consequently, a simple hypothesis is that the Ytr ABC transport system imports acetoin. In this case B. subtilis cells have also another acetoin import system, since the acetoin utilization by the ytr mutants was impaired only partially. A more complex formal alternative is that the system transports a yet-unknown substance required for efficient acetoin catabolism. Since the effect of ytr inactivation is observed in a minimal medium, such a substance would have to be produced and excreted by the B. subtilis cells in parallel with acetoin. We know of no precedent for such a process and consider it unlikely. However, further work is required to rule out this alternative.
YtrA was proposed to be a repressor of the GntR family. Here we show that it regulates negatively the ytr operon. Its binding site might be a nucleotide sequence which overlaps the transcription start site (defined as +1) and shows a partial dyad symmetry: −5TtaAGTGTAcTAaTT-G-AAgTAaTACACTatA+26 (latter half, mismatches, and the center of the symmetry are shown in italic type, lower case letters, and underlined, respectively). Typical members of the GntR family consist of approximately 250 amino acid residues (5). Their N-terminal half forms a DNA binding domain containing a helix-turn-helix motif (5), and the DNA binding is modulated through interaction between their C-terminal domain and the specific effector (27). It is noteworthy that YtrA comprises only 130 amino acid residues, suggesting that its C-terminal domain is too small to accommodate the effector binding. Acetoin seems not to be the effector, since its addition did not induce the reporter gene present in the strain BFS47 (Fig. 4D). Further work is required to identify the effector and to analyze YtrA binding to DNA.
ACKNOWLEDGMENTS
We thank Hironobu Katsumata, Ryosuke Tanaka, Atsushi Umeda, and Shigeru Yamada for their technical assistance; Petar P. Pujic for valuable discussions; Choong-Min Kang for his critical reading of the manuscript; and Valérie Vagner for providing pMUTIN2mcs. This work was supported by grant JSPS-RFTF96L00105 from the Japan Society for the Promotion of Science and grant BIO4-CT95-0278 from the EU.
REFERENCES
- 1.Ames G F-L, Miura C S, Shyamala V. Bacterial periplasmic permeases belong to a family of transport proteins operating from Escherichia coli to human: traffic ATPases. FEMS Microbiol Rev. 1990;6:429–446. doi: 10.1111/j.1574-6968.1990.tb04110.x. [DOI] [PubMed] [Google Scholar]
- 2.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujita Y, Yoshida K, Miwa Y, Yanai N, Nagakawa E, Kasahara Y. Identification and expression of the Bacillus subtilis fructose-1,6-bisphosphatase gene (fbp) J Bacteriol. 1998;180:4309–4313. doi: 10.1128/jb.180.16.4309-4313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grundy F J, Waters D A, Takova T Y, Henkin T M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993;10:259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- 5.Haydon D J, Guest J R. A new family of bacterial regulatory proteins. FEMS Microbiol Lett. 1991;63:291–295. doi: 10.1016/0378-1097(91)90101-f. [DOI] [PubMed] [Google Scholar]
- 6.Huang M, Oppermann-Sanio F B, Steinbüchel A. Biochemical and molecular characterization of the Bacillus subtilis acetoin catabolic pathway. J Bacteriol. 1999;181:3837–3841. doi: 10.1128/jb.181.12.3837-3841.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerpolla R E, Shyamala V K, Klebba P, Ames G F-L. The membrane-bound proteins of periplasmic permeases form a complex. J Biol Chem. 1991;266:9857–9865. [PubMed] [Google Scholar]
- 8.Kunst F, Ogasawara N, Moszer I, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 9.Lopez J, Thoms B. Beziehungen zwischen katabolischer Repression und Sporulation bei Bacillus subtilis. Arch Microbiol. 1976;109:181–186. doi: 10.1007/BF00425133. [DOI] [PubMed] [Google Scholar]
- 10.Miwa Y, Fujita Y. Purification and characterization of a repressor for the Bacillus subtilis gnt operon. J Biol Chem. 1988;263:13252–13257. [PubMed] [Google Scholar]
- 11.Moszer I. The complete genome of Bacillus subtilis: from sequence annotation to data management and analysis. FEBS Lett. 1998;430:28–36. doi: 10.1016/s0014-5793(98)00620-6. [DOI] [PubMed] [Google Scholar]
- 12.Nakano M, Dailly Y P, Zuber P, Clark D P. Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth. J Bacteriol. 1997;179:6749–6755. doi: 10.1128/jb.179.21.6749-6755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oppenheim D S, Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980;95:785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pistocchi R, Kashiwagi K, Miyamoto S, Nukui E, Sadakata Y, Kobayashi H, Igarashi K. Characteristics of the operon for a putrescine transport system that maps at 19 minutes on the Escherichia coli chromosome. J Biol Chem. 1993;268:146–152. [PubMed] [Google Scholar]
- 15.Quail M A, Dempsey C E, Guest J R. Identification of a fatty acyl responsive regulator (FarR) in Escherichia coli. FEBS Lett. 1994;356:183–187. doi: 10.1016/0014-5793(94)01264-4. [DOI] [PubMed] [Google Scholar]
- 16.Quentin Y, Fichant G, Denizot F. Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J Mol Biol. 1999;287:467–484. doi: 10.1006/jmbi.1999.2624. [DOI] [PubMed] [Google Scholar]
- 17.Renna M C, Najimudin N, Winik L R, Zahler S A. Regulation of the Bacillus subtilis alsS, alsD, and alsR genes involved in post-exponential-phase production of acetoin. J Bacteriol. 1993;175:3863–3875. doi: 10.1128/jb.175.12.3863-3875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 19.Schaeffer P, Millet J, Aubert J P. Catabolite repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwacha A, Bender R A. Nucleotide sequence of the gene encoding the repressor for the histidine utilization genes of Klebsiella aerogenes. J Bacteriol. 1990;172:5477–5481. doi: 10.1128/jb.172.9.5477-5481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugae K, Freese E. Requirement for acetate and glycine (or serine) for sporulation without growth of Bacillus subtilis. J Bacteriol. 1970;104:1074–1085. doi: 10.1128/jb.104.3.1074-1085.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tjalsma H, Kontinen V P, Pragai Z, Wu H, Meima R, Venema G, Bron S, Sarvas M, van Dijl J M. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis. Signal peptidase II is required for the efficient secretion of α-amylase, a non-lipoprotein. J Biol Chem. 1999;274:1698–1707. doi: 10.1074/jbc.274.3.1698. [DOI] [PubMed] [Google Scholar]
- 23.Tomita H, Fujimoto S, Tanimoto K, Ike Y. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J Bacteriol. 1997;179:7843–7855. doi: 10.1128/jb.179.24.7843-7855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 25.Yansura D G, Henner D J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci USA. 1984;81:439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida K, Fujita Y, Sarai A. Missense mutations in the Bacillus subtilis gnt repressor that diminish operator binding ability. J Mol Biol. 1993;231:167–174. doi: 10.1006/jmbi.1993.1270. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida K, Ohmori H, Miwa Y, Fujita Y. Bacillus subtilis gnt repressor mutants that diminish gluconate-binding ability. J Bacteriol. 1995;177:4813–4816. doi: 10.1128/jb.177.16.4813-4816.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida K, Aoyama D, Ishio I, Shibayama T, Fujita Y. Organization and transcription of the myo-inositol operon, iol, of Bacillus subtilis. J Bacteriol. 1997;179:4591–4598. doi: 10.1128/jb.179.14.4591-4598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoshida K, Ishio I, Nagakawa I E, Yamamoto Y, Yamamoto M, Fujita Y. Systematic study of gene expression and transcription organization in the gntZ-ywaA region of the Bacillus subtilis genome. Microbiology. 2000;146:573–579. doi: 10.1099/00221287-146-3-573. [DOI] [PubMed] [Google Scholar]
- 30.Young R A, Davis R W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci USA. 1983;80:1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]