Abstract
B cells are important in tuberculosis (TB) immunity, but their role in the human lung is understudied. Here, we characterize B cells from lung tissue and matched blood of patients with TB and found they are decreased in the blood and increased in the lungs, consistent with recruitment to infected tissue, where they are located in granuloma associated lymphoid tissue. Flow cytometry and transcriptomics identify multiple B cell populations in the lung, including those associated with tissue resident memory, germinal centers, antibody secretion, proinflammatory atypical B cells, and regulatory B cells, some of which are expanded in TB disease. Additionally, TB lungs contain high levels of Mtb-reactive antibodies, specifically IgM, which promotes Mtb phagocytosis. Overall, these data reveal the presence of functionally diverse B cell subsets in the lungs of patients with TB and suggest several potential localized roles that may represent a target for interventions to promote immunity or mitigate immunopathology.
Subject terms: Tuberculosis, Respiratory tract diseases
Using flow cytometry and transcriptomic analyses, the authors characterize the functional diversity of B cell subsets in the lungs of patients with tuberculosis.
Introduction
In 2021, Mycobacterium tuberculosis (Mtb) was estimated to infect a quarter of the world’s population, cause tuberculosis (TB) in 10.6 million and kill 1.6 million people1. Mtb is spread when exhaled/coughed aerosolized droplets are inhaled into another individual’s lungs where infection propagates. A characteristic of pulmonary TB is the generation of granuloma2, typically formed from clusters of infected alveolar macrophages surrounded by a cuff of lymphocytes, including T cells, B cells and various innate lymphoid cells. In addition, B cells are frequently observed within lymphocyte aggregates near granuloma, referred to as granuloma-associated lymphoid tissue (GrALT)3. GrALT has been observed in mice4–8, non-human primates (NHP)8–10 and humans8,11,12 and is typically associated with protective immunity. However, the role of B cells in the immune response to TB within the lung remains poorly understood.
In humans, TB leads to a reduction in circulating B cells11,13–17, which recover following successful treatment11,15,18. In mice, B cell knock out or depletion results in greater susceptibility to TB4,6, with adoptive transfer resulting in reversed lung pathology7,19. In non-human primates, B cell depletion increases bacterial burden in lung lesions10. Mechanistically, recent studies implicate B cells in orchestrating the CD4+ T cell response within GrALT, affecting both the TH1 and TH17 protective responses3,20–23. In addition, B cell follicle area correlated with lung IL17 mRNA levels, and B cells from pleural fluid were critical mediators of the IL17 and IL22 responses24–26. Tissue-resident memory B cells reside in the lung and can mature into antibody-secreting plasma cells27. Several studies demonstrate a role for Mtb-specific antibodies in protective immunity to TB in both NHPs and humans22,23,28–32. Furthermore, active and latent TB can be differentiated based on antibody glycosylation profiles29,33 and the resulting differences in the Fc effector functions (binding FcγRIII) including antibody-mediated phagocytosis29.
Together, these studies demonstrate B cell involvement during TB and highlight important canonical and non-canonical functions that contribute to TB immunity. However, data from humans in the lung compartment is lacking. Here, we demonstrate that TB-infected lung tissues are enriched for CD19+ B cells, which are associated with granuloma. Lung B cells were predominantly of a memory phenotype and expressed markers associated with residency and germinal centre homing. In addition, the lung was enriched for antibody-secreting cells (ASC) and unique putative regulatory B cell phenotypes. Using a granuloma biomimetic model, we found that B cells from healthy donors contribute to control of Mtb growth, while this function is inconsistent in patients with TB, suggesting potential impairment of these cells during active disease. Finally, the lungs of patients with TB were enriched for Mtb-specific antibodies relative to control lungs, especially IgM, which enhanced Mtb phagocytosis by primary human cells. These findings highlight the diversity of B cell function in human TB.
Results
Comparison of CD19+ B cells in blood and lung tissue compartments
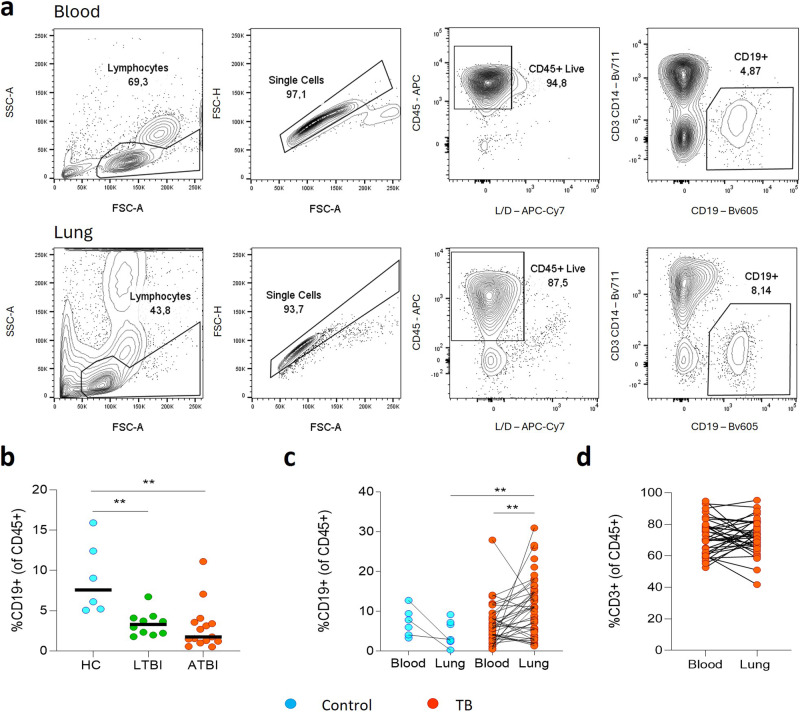
First, we compared the relative frequencies of B cells in the peripheral blood of non-TB control patients, patients with latent TB (LTBI) and active TB (ATBI) (Fig. 1a, b). LTBI was confirmed by a positive Interferon Gamma Release Assay (IGRA) test in participants with no signs, symptoms, or history of TB disease. Non-TB controls were similarly asymptomatic but were IGRA negative16. The frequency of CD19+ B cells was reduced in the blood of LTBI and ATBI compared to healthy controls, consistent with published data11,13–16. We then measured the frequency of B cells in homogenized lung tissue from patients with TB, obtained from individuals enrolled in the AHRI lung resection cohort34, undergoing surgical resection to treat TB sequelae (Fig. 1a, c). Additional samples were analysed from patients with no history of TB, who were undergoing surgical resection of lung cancer and from whom macroscopically uninvolved tissue was obtained. These controls were not IGRA tested, and thus latent TB infection could not be excluded, although TB prevalence is so high in KwaZulu-Natal, most donors are highly likely to have been TB-exposed. The lung tissue of patients with TB was highly enriched for B cells compared to matched blood samples, with a median frequency of approximately 9% of CD45+, reaching as high as 31% in some patients (Fig. 1c), significantly higher than in lung tissue from non-TB controls (median of 3%). In contrast, CD3+ T cells were not significantly enriched in lung tissue from patients with TB compared to matched blood, although these cells were more abundant than B cells (median 72%, Fig. 1d).
Fig. 1. Frequencies of CD19+ B cells differ between blood and lung compartments.
a Representative flow plots identifying CD19+ B cells from peripheral blood mononuclear cells (PBMC) and resected lung tissue. b B cell frequencies in the blood were compared for a set of healthy controls (HC, n = 6), patients with latent TB (LTBI, n = 10) or active TB (ATBI, n = 15), demonstrating reduced circulating B cells in active TB. c B cell frequencies were compared for lung tissue versus matched blood of cancer control patients (in cyan, n = 8) and patients with TB (in red, n = 60). B cell numbers were increased in the lung compared to the blood in patients with TB, and also relative to cancer control lung tissue. d In contrast, comparative frequencies of CD3+ T cells between lung tissue and matched blood of patients with TB showed no significant difference. Statistical analyses were performed using the Mann–Whitney test between unmatched samples and the Wilcoxon test for matched/paired samples. P values are denoted by ** <0.01.
Histological localization of B cells in proximity to granulomatous structures
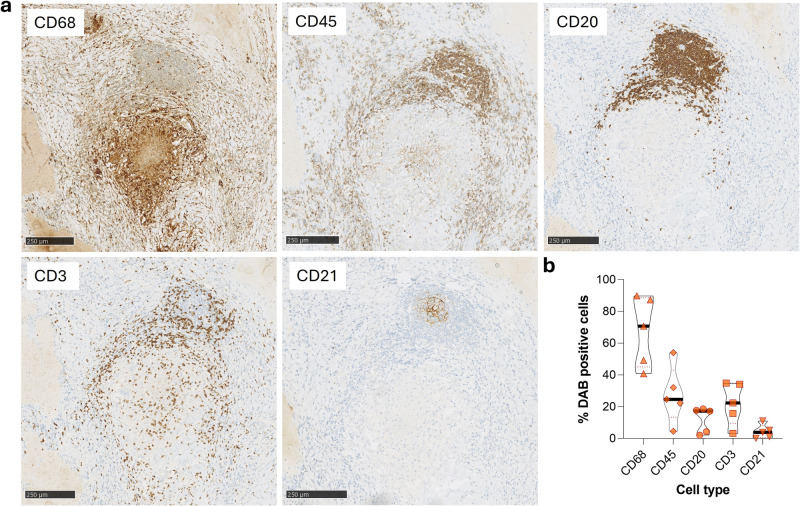
To determine the localization of B cells within lung tissue of patients with TB, we conducted histological assessment of lung sections from participants with distinct granulomatous lesions. The canonical TB lung granuloma has a CD68+ macrophage core surrounded by a cuff of CD45+ lymphocytes (Fig. 2a). This cuff comprises CD3+ T cells with some also present within the granuloma core. CD20+ staining reveals a distinct B cell aggregate in close association with the granuloma, consistent with previously described GrALT8,11,12. B cells also associate with the granuloma cuff, primarily adjacent to the GrALT. The GrALT also contains CD3+ T cells, consistent with previous reports and with lymphoid follicles. In addition, CD21 staining is present within the centre of the GrALT, suggestive of follicular dendritic cells8,35, although mature B cells can express CD21 within germinal centres36. Of interest were the staining patterns of anti-CD20, anti-CD3 and anti-CD21 in particular (Supplementary Fig. 1). CD20 and CD3 staining are characteristic of lymphoid cells whereas CD21 stained cells displaying a more dendrite-like pattern. The CD20+ cells occupy most of the aggregate, whereas the CD3+ cells are more peripheral, and the CD21+ cells seem to occupy the centre of the aggregate. Quantitation of granulomatous lesion containing lungs from 5 different patients with TB show a roughly equivalent frequency of T and B cells associated with lung granuloma, in the region of 20% of nucleated (haematoxylin+) cells (Fig. 2b). As some surface markers are co-expressed by different cells, the total exceeds 100%. Overall, these data suggest that B cells are enriched in the lung during TB infection in humans and make up a significant proportion of granuloma-associated lymphocytes.
Fig. 2. Immunohistochemistry of human lung tissue resection from a patient with TB demonstrates B cell aggregates adjacent to the granuloma.
a Serial sections of lung tissue from patients with TB were stained for macrophages (anti-CD68), leucocytes (anti-CD45), B cells (anti-CD20), T cells (anti-CD3) and dendritic cells (anti-CD21). b The diaminobenzidine (DAB) stained cells were enumerated and expressed as % DAB positive cells of total nucleated (haematoxylin+) cells, per total slide area imaged. The relative frequencies from a total of five different tissue samples from patients with TB are compared.
Prominent memory and antibody-secreting cell (ASC) phenotypes in the lung compartment including CD69+ memory
Next, we examined the phenotype of B cells in matched blood and lung samples from participants with TB and non-TB controls (Fig. 3). First, we examined the B cell maturation state based on the canonical memory marker CD27 and activation marker CD3837 (Fig. 3a). Notably, CD27+CD38- memory B cells were enriched in lung tissue from patients with TB compared to matched blood, which was dominated by CD27-CD38- naive and CD27-CD38+ transitional B cells (Fig. 3b). The same trend was observed in control lung samples, although the enrichment of memory B cells was not significant and memory B cells were significantly more abundant in lungs from patients with TB compared to controls. CD27+CD38hi antibody-secreting cells (ASCs) were present in lung tissue, and were significantly enriched compared to matched blood, but were significantly higher in non-TB control lung tissue. In addition, lung derived memory B cells were highly enriched for the expression of the tissue residence marker CD6927 (Fig. 3c). Finally, from a subset of lung samples, lung draining lymph nodes were studied as an alternative comparator tissue enriched with B cells (Fig. 3d, Supplementary Fig. 2)38. Lung tissue had increased memory subsets compared to matched lymph nodes, and as expected, lymph nodes contained a low prevalence of ASCs38.
Fig. 3. B cell transitional, naive, memory and antibody-secreting cell population frequency demonstrates differences between compartments that are mainly disease-independent.
a Representative flow plots of the canonical transitional (Trans), naive, memory (Mem) and antibody-secreting cell (ASC) B cell phenotypes derived from staining CD19+ B cells with CD27 and CD38 (cancer control in cyan, n = 8; patients with TB in red, n = 60). b The relative frequencies of these four B cell phenotypes were compared between blood and lung tissue compartments. c The frequency of CD69+ memory B cells was assessed (cancer control in cyan, n = 5; patients with TB in red, n = 25). d This analysis was extended to lung draining lymph nodes (LN, n = 17) from the TB cohort for comparison. A representative gating strategy for LN derived B cells is included as Supplementary Fig. 2. Statistical analyses were performed using the Mann–Whitney test between unmatched samples, the Wilcoxon test for matched/paired samples and a three-way Kruskal–Wallis comparison. P values are denoted by * ≤0.05; ** <0.01; *** <0.001 and **** <0.0001.
Transcriptional analysis of lung-derived B cells
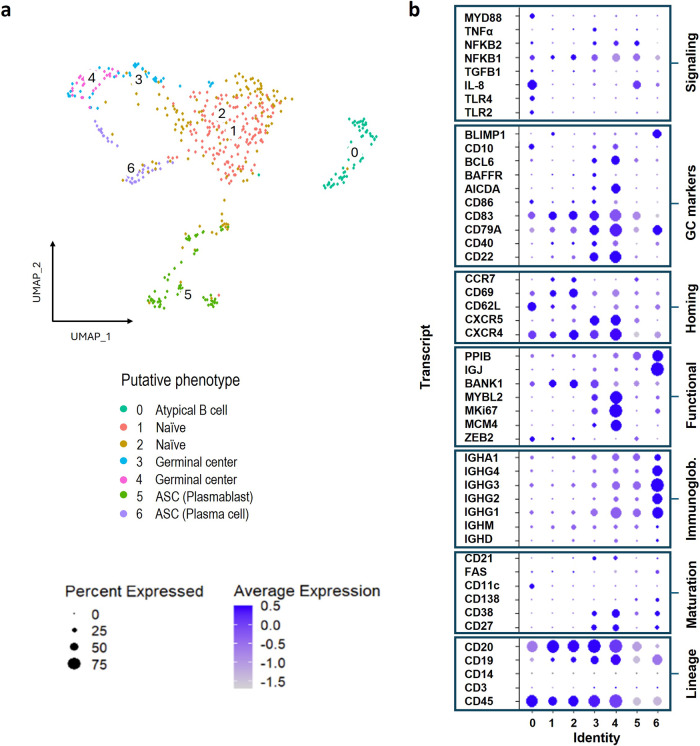
Having observed an enrichment of memory B cell subsets in lung tissue from patients with TB, we interrogated these in a single cell RNA sequence (scRNAseq) analysis of additional lung tissue samples obtained from the same lung cohort. Of a total 20962 single cells recovered from 9 resected lungs from patients with TB, 404 displayed canonical B cell markers including the lineage markers CD19 and CD20. We focused on leucocytes, and sub clustering of this limited dataset identified a total of 7 unique B cell subsets, shown by UMAP projection (Fig. 4a). A curated list of B cell genes was used to assign putative functional phenotypes to the 7 subsets (Fig. 4b). This includes populations expressing genes associated with memory B cells (3 and 4) which expressed CD21, a B cell coreceptor involved in T cell-dependent signalling, in addition to CD27 and CD38, and ASCs (5 and 6), expressing the highest levels of genes associated with immunoglobulin heavy chains, including subclasses IgG 1-4 as well as IgA. These cells also expressed CD138 and BLIMP1, both markers of long-lived plasma cells37,39, and IgJ and PPIB, involved in the production of IgM and IgA antibodies39–42 and protein secretion, respectively43. Consistent with flow cytometry data, lung B cells express the gene encoding CD69, especially subsets 1 and 2, which also express CCR7 and CXCR4. Subsets 3 and 4 lack CCR7 but co-express CXCR4 and CXCR5. All three chemokine receptors are involved in trafficking within the germinal centre (GC)7,39,44, and may thus represent B cell populations associated with GrALT. In line with this, populations 3 and 4 express Bcl6, a key regulator of the GC response45–47 and AICDA, which is expressed during B cell receptor (BCR) affinity maturation48, as well as the GC marker CD1035. Population 3 and 4 also express the highest levels of CD21 and CD22, both essential to B cell GC survival36,49 and CD79a, a marker of mature B cells9. The immunoglobulin and CD79a transcript abundance generally increased from populations 0 to 6, and since both are associated with the B cell surface receptor, this also suggests the local B cell population is maturing9,50. Several genes associated with genome replication (MCM4)45, proliferation (MKi67)42,45 and cell cycle progression (MYBL2)46 are also highly expressed. Population 0, on the other hand, is distinguished by CD11c expression, which is a marker of atypical B cells51–54, and ZEB2, recently identified as a key transcription factor promoting the development of this cell type55. This population also expresses genes encoding TLR2, TLR4 and MyD88, a common adaptor molecule for all TLRs except TLR356. Interestingly, BANK-1 was recently shown to signal together with MyD88 and TLRs to co-ordinate innate immune signalling B cells, including the production of IL-8, and population 0 expresses both BANK-1 and high levels of the IL-8 gene57. Thus, scRNAseq data supports the presence of GC-like B cell populations, ASC-like and atypical/innate-like B cells in lung tissue from patients with TB. The next aim was to identify similarities between these B cell populations putatively identified by sequencing, with those identified by flow cytometry.
Fig. 4. Differential gene expression identifies subclusters amongst B cells isolated from human lung tissue from patients with TB.
scRNA sequencing was used to explore the differentially expressed genes in B cells isolated from human lung tissue from patients with TB (n = 9). a B cells were clustered based on their differential gene expression and visualised by UMAP with a total of seven putative clusters identified (0 to 6). b These B cell clusters were interogated for their expression of specific B cell-associated genes to delineate their likely functions.
Transcriptional heterogeneity of B cells translates to several B cell phenotypes identified by flow cytometry, highlighting compartmental heterogeneity
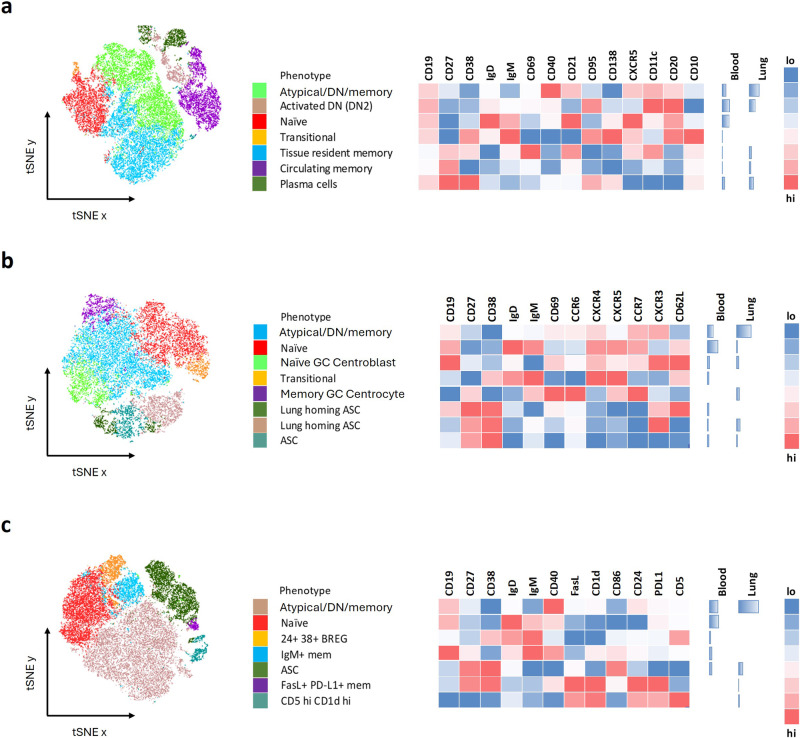
We obtained 13 additional lung samples and analysed lung homogenate and matched blood samples by flow cytometry using three separate antibody panels, each elaborating on a core set of shared markers included in Figs. 1 and 3: CD19, CD27, CD38, IgD and IgM (Fig. 5). We initially analysed these data using an unbiased clustering algorithm to identify unique B cell populations based on the markers in each flow cytometry panel (Fig. 5a–c). Consistent with the initial flow cytometry profiling (Fig. 3), the main phenotypic differences were based on B cell maturation, with the blood containing more transitional and naive B cells, compared to increased memory and ASC frequencies in the lung. A single patient sample was used to generate a representative tSNE plot (Fig. 5), to gain an overview of the number of unique B cell phenotypes associated with the lung and blood compartments. We then interrogated any phenotypes of interest in a total of 13 patients with TB to assess compartmental differences (Fig. 6, Supplementary Fig. 3).
Fig. 5. tSNE plot comparison of B cell phenotypes further refines blood and lung tissue associated compartments.
In order to study the complexity of the B cell phenotypes associated with blood and lung tissue compartments of patients with TB, three separate flow cytometry phenotyping panels were used. a Assesed the level of maturation of the B cells, b the expression of different homing markers and c the relative expression of regulatory markers. The different B cell populations were clustered spatially using tSNE, with a single patient presented here, followed by a key describing the major phenotypes each of which is assigned a different colour. The relative expression of the specific markers associated with each phenotype were plotted alongside in a heatmap. Since the blood and lung samples were concatenated into a single file, the relative frequencies of cells in the blood or lung tissue compartment could be compared as shown alongside the heatmap.
Fig. 6. tSNE-derived unique B cell phenotypes associated with the blood or lung compartment of patients with TB.
a Double negative (DN) B cells (CD27-IgD-) were assessed and the activated (DN2) phenotype (CD21-CD11c+) was enriched in the lungs of patients with TB. b GC homing (CXCR4+CXCR5+) transitional and naive B cells were enriched in the blood compartment. c The blood compartment was enriched for a tranistional regulatory (CD24+CD38+) population. d The lung compartment contained a unique activated B cell memory (CD27+CXCR5+CD62L-CD69+CCR6+CCR7+) population. e Lung-derived ASC populations expressed a CXCR3+CD62L- phenotype relative to the blood-derived counterparts. Other B cell phenotypes associated with regulatory functions were enriched in the lung, including f a CD5+ phenotype (CD5hiCD40-CD1d+) and g a CD27+ regulatory population (CD27+CD24+FasL+PDL1+). A set of 13 samples from patients with TB were used in these analyses. Statistical analyses were performed using the Wilcoxon test for matched/paired samples. P values are denoted by * ≤0.05 and ** <0.01.
Common to all three panels was a prominent B cell phenotype that was CD27mid/lo, CD38-, IgD- and IgM- (Fig. 5a green, b blue, c tan) designated as an atypical/double negative (DN) memory population. The double negative phenotype is defined as being negative for CD27 and IgD51. This was expanded in the lung compared to matched blood. As shown in flow panel 1 (Fig. 5a), two such populations were apparent, both of which express the key atypical B cell marker CD11c, consistent with the scRNAseq data (Fig. 4). The second population resembles an activated double negative (DN2) phenotype, as it expresses the highest levels of CD11c, and is CD20hi and CXCR5-51–54. This activated DN2 phenotype was enriched in the lung when assessed across the 13 patients (Fig. 6a, Supplementary Fig. 3a). Several studies have observed an enrichment of DN2 B cells in the blood under inflammatory conditions51–54 and further enrichment in the lung of patients with TB supports their potential importance at the site of infection. In addition, in the second panel, the putative atypical B cell population (Fig. 5b blue) expresses high levels of CXCR3, which would facilitate homing to inflamed tissue sites via CXCL1027,39,58.
Naive (Fig. 5, red) and transitional (Fig. 5, orange) B cell phenotypes were dominant in the blood, as seen in the initial flow cytometry phenotyping data (Fig. 3), with the transitional phenotype expressing CD10 and CD138 as expected (Fig. 5a)37. Both naive and transitional phenotypes displayed high expression of CXCR5 associated with homing to germinal centres (Figs. 5b and 6b, Supplementary Fig. 3b)7,39,44, and both phenotypes were enriched in circulation (Fig. 6B). The naive GC centroblast-like phenotype (Fig. 5b, lime green) expressed homing markers including CD62L, CXCR4, CXCR5 and CCR7, consistent with population 1 and 2 identified by scRNAseq (Fig. 4). A transitional B cell phenotype, CD24+ CD38+, which is associated with regulatory functions42,59, was enriched in the blood compared to lung (Figs. 5c and 6c, Supplementary Fig. 3c). In contrast, the purple population in Fig. 5b, which expressed a phenotype resembling the putative GC populations 3 and 4 from the scRNAseq data (Fig. 4), being CD27+CXCR5+CD62L-CD69+CCR7+ and CCR6+, was significantly enriched in the lung (Fig. 6d, Supplementary Fig. 3d).
The CD27+CD38hi ASC-like populations observed by scRNAseq (Fig. 4) were also apparent by flow cytometry in the lung, including CD138+ plasma cells (Fig. 5a). ASCs could be separated based on expression of CXCR3 and CD62L (Fig. 5b). CXCR3 facilitates homing to sites of inflammation, and CD62L (L-selectin) is essential for migration of lymphocytes into tissue and is lost as B cells mature into plasmablasts and finally plasma cells60. CXCR3+CD62L+ ASC were significantly enriched in the blood, whereas CXCR3+CD62L- ASC were enriched in the lung (Figs. 5b and 6e and Supplementary Fig. 3e), consistent with antibody-secreting populations (5 and 6) observed by sequencing (Fig. 4). In addition, two phenotypes associated with regulatory functions were significantly associated with the lung environment, one expressing a CD5+CD1d+ phenotype (Figs. 5c and 6f and Supplementary Fig. 3f)26,61 and the other a memory CD27+CD24+ phenotype59 (Fig. 6g and Supplementary Fig. 3g). The second putative regulatory population also expressed PDL-1, the receptor for PD-1, important for regulating the T cell response in TB3,62,63. The importance of antigen-specific memory B cells and the PDL-1/PD-1 axis for localising TFH cells within GrALT and mediating TB control in both mice and macaques was recently demonstrated3. Together these flow cytometry data confirm the presence of diverse B cell phenotypes in lung tissue from patients with TB, including an expanded CD11c expressing atypical B cell population, subsets potentially associated with GC activity, antibody production, and regulation.
B cells impact Mtb growth in a 3D granuloma biomimetic model
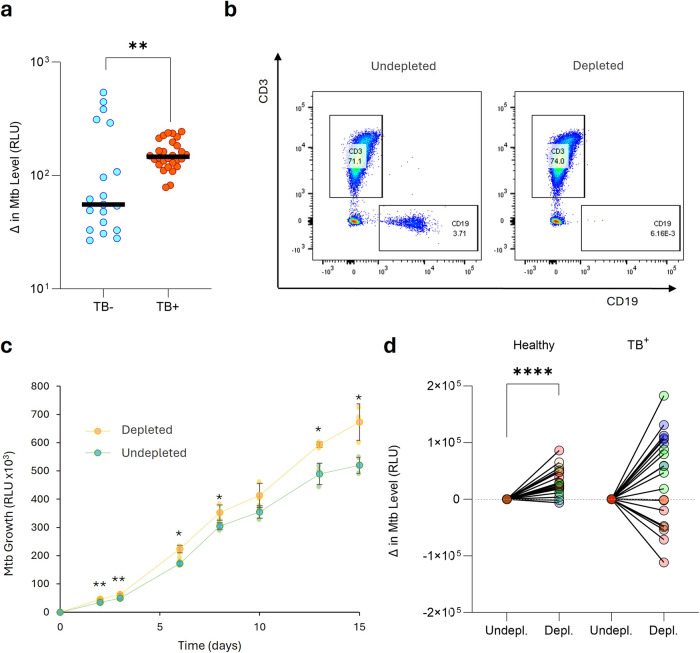
To investigate the potential effect of B cells on Mtb growth, we turned to a 3D biomimetic culture model that resembles the granuloma microenvironment64,65. The model uses peripheral blood mononuclear cells (PBMC) infected with bioluminescent Mtb. These Mtb-infected PBMC are then encapsulated in collagen/alginate microspheres that contain an extracellular matrix scaffold. Each microsphere therefore represents a 3D microenvironment that mimics an individual granuloma. Without B cell depletion, the overall Mtb growth rate was significantly greater in undepleted PBMC from patients with TB compared to control samples, consistent with immune dysregulation during TB disease (Fig. 7a). CD19+ B cells were depleted from PBMC using positive magnetic selection and Mtb growth kinetics measured compared to non-depleted microspheres (Fig. 7b). Using PBMC from four healthy donors, with each condition performed in triplicate, demonstrated that B cell depletion caused a consistent and significant increase in Mtb growth (Fig. 7c, d). Interestingly though, the effect of depletion on PBMC from patients with active TB prior to the initiation of drug therapy was highly variable, with Mtb growth increasing in two of the donors and decreasing in the remaining two (Fig. 7d). Together, these data demonstrate that B cells can regulate Mtb growth, but this effect is variable in active TB suggesting their function can be altered by ongoing disease.
Fig. 7. B cells contribute to host control of Mtb in healthy donors in a 3D biomimetic model, but in patients with TB the effect is highly variable.
a Comparatively, Mtb grew more rapidly in PBMC derived from patients with TB. B cells were depleted from PBMCs, of which four donors were healthy and four had active TB, and compared to undepleted PBMCs. b Representative FACS plots of the depletion process. c B cell depletion leads to significantly higher Mtb growth in a healthy donor PBMC sample, representative of four healthy donors. d Mtb outgrowth data at day 7 from a total of four PBMC donors from each group are summarized, demonstrating the highly variable effect of B cell depletion in patients with TB. Data were normalized to the undepleted control group and compared by Wilcoxon matched-pairs signed rank test. Standard deviation is presented in (c). P values are denoted by * ≤0.05; ** <0.01 and **** <0.0001.
Lung tissue derived antibodies show Mtb specificity and enhance bacterial phagocytosis
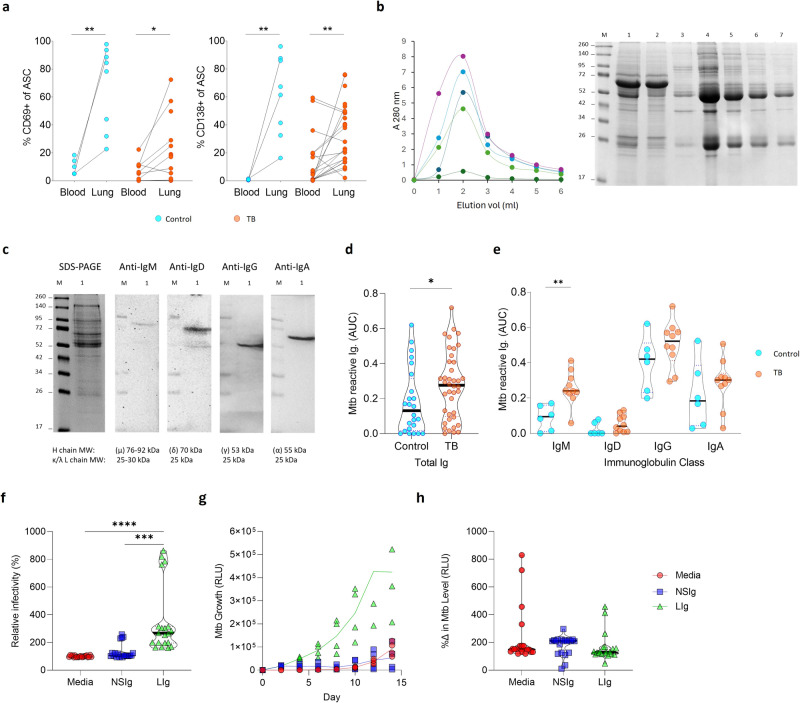
Having observed an expansion of ASC subsets in lung tissue from patients with TB, we compared the relative frequency of ASCs of TB and non-TB patients and found an enrichment in lung tissue in both groups (Fig. 8a). The ASCs expressed the tissue residence marker CD69 and the plasma cell marker CD13827,37. These compartmental differences prompted us to investigate the antibody profiles in lungs from patients with TB. Therefore, we isolated antibodies from lung tissue homogenates using a thiophilic gradient (Fig. 8b)66 and confirmed the presence of IgM, IgD, IgG and IgA (Fig. 8c; IgE was not detected). The purified antibodies were reactive to whole Mtb lysate by ELISA, and significantly enriched in the patients with TB (Fig. 8d). Although individual Mtb-reactive antibody classes showed similar upregulation, only IgM was significantly upregulated in the patients with TB (Fig. 8e). Interestingly in this highly TB endemic population, there was no difference in the reactivity of plasma-derived antibodies from the same TB and non-TB diseased participants (Supplementary Fig. 4). These data indicate the enrichment of TB-specific antibodies in TB diseased human lung tissue, supporting a role at the site of disease.
Fig. 8. Mature CD138+ plasma cells are enriched in the lung tissue, resulting in an upregulated Mtb specific antibody response that enhances Mtb phagocytosis.
a The tissue residence marker CD69 was expressed on antibody-secreting cells (ASC), with the majority displaying a mature plasma cell CD138+ phenotype in both cancer controls (n = 8) and patients with TB (n = 13 to 25). b Antibodies were isolated from homogenised lung tissue supernatants using a thiophilic resin, with example elution profiles from five lungs measured at 280 nm, and then examined on a reducing 12.5% SDS-PAGE gel. The molecular weight marker (lane M), was followed by the initial lung homogenate supernatant sample (lane 1), the unbound sample (lane 2) followed by the eluted fractions 1–5 from the elution profile alongside (lanes 3–7). c Peak eluents (lane 1) were tested for the presence of IgM, IgD, IgG and IgA using class-specific secondary antibodies by western blot. d Mtb-specific antibody binding was investigated in lung samples from cancer controls (n = 6) and patients with TB (n = 10), detecting both total reactivity and e class-specific responses in a direct ELISA-based approach, expressed as area under the curve (AUC). An equal concentration range (100 to 0.8 µg/ml) of isolated antibody was used to standardize the assay and allow for comparison between patients. Antibody frequencies were compared by Mann–Whitney test. f Mtb phagocytosis by PBMCs was compared between untreated Mtb versus Mtb treated with non-specific immunoglobulin (NSIg) or lung immunoglobulin (LIg) from patients with TB. The lung-derived immunoglobulin increased phagocytosis, while non-specific immunoglobulin had no effect. Data are presented as summary data from five immunoglobulin samples from patients with TB, tested with three different PBMC donors, with each experiment done in quadruplicate and infectivity was measured by luminescence. Data were normalized to the media control and the donor matched mean differences were compared by one-way ANOVA. g Representative Mtb growth using immunoglobulin from patients with TB comparing with untreated and non-specific immunoglobulin in the 3D biomimetic model of TB. h Mtb luminescence at day 8 normalized to day 0 initial infectious load shows no overall difference in growth rate over time. P values are denoted by * ≤0.05; ** <0.01; *** <0.001 and **** <0.0001.
Finally, to confirm the specificity of these antibodies and to test their functionality, purified antibodies were studied using the 3D biomimetic Mtb culture model65. Antibodies purified from lung homogenate of patients with TB enhanced the phagocytosis of Mtb, shown by increased uptake of luminescent Mtb compared to isotype controls (Fig. 8f). Although the flow cytometry does not determine if the bacteria are phagocytosed or cell-associated, the cells are extensively washed post-infection, and we have previously shown that Mtb growth in this system is predominantly intracellular64. Subsequent growth of Mtb over the next two weeks was markedly greater when Mtb had been treated with these antibodies (Fig. 8g). However, when the results were normalized to the initial infectious load, no significant change in the growth rate was observed, suggesting the antibodies do not modulate subsequent proliferation, at least within this model system (Fig. 8h). Thus, these data confirm the presence of Mtb specific antibodies in TB affected lung tissue, which enhance the phagocytosis of mycobacteria.
Discussion
Diverse lines of evidence, from human studies and animal models, demonstrate that B cells are involved in the host response to Mtb, but B cells represent a multi-faceted immune subset, and their precise role has remained elusive. The frequency of B cells in circulation is reduced during active TB11,13–17 and infection is known to induce B cell containing tertiary lymphoid follicles in the lung referred to as granuloma-associated lymphoid tissue (GrALT)3,8,11,12. Using matched blood and lung samples, we show that these phenomena overlap, with decreased B cells in the blood reflecting an increase in the lungs of patients with TB. Lung B cell frequency was extremely high in some participants, and, by histology, they were found to be associated with the granuloma in similar quantities to T cells. We next performed a detailed characterization of B cell phenotypes in human TB lung tissue using complementary scRNAseq and flow cytometry approaches and interrogate the potential functional impact in a cellular model. For technical reasons, it was not possible to perform scRNAseq analysis and FACS analysis on the same samples, however, the fact that we observe similar changes at both the transcriptional and protein level supports the phenotypic changes observed by each approach, despite analysis being in different samples.
A central focus of our study was to compare B cells in the lung compartment with those in circulation, which have been the focus of most studies in humans. In contrast to a predominantly naive and transitional phenotype in the blood, lung B cells were mostly of a memory and ASC phenotype and expressed the tissue residence marker CD69, consistent with resident memory cells67. There is limited data regarding human lung derived B cell phenotypes, but our data are supported by Barker et al. and Tan et al., who also observed CD69+ expression68,69. Mechanistically, from mouse models, the recruitment of B cells to the lung requires local antigen encounter27,69,70. The initial seeding of memory B cells in the lung is derived from the mediastinal lymph node69, with lung resident cells expressing CXCR3, CCR6 and CD6927,69. These cells patrol the lung and upon local antigen encounter either form part of induced bronchus-associated lymphoid tissue (iBALT), or GrALT in the case of TB, or differentiate into ASC27,70. We observed memory B cells and ASCs expressing all three markers, supporting their tissue residence. Interestingly, Tan et al. detected a resting memory B cell phenotype and little expression of GC-associated markers Bcl6, Ki67 and AICDA in healthy lungs69. In contrast, we observed two memory B cell populations expressing these transcripts in our scRNAseq data, in addition to a memory B cell phenotype expressing both CCR6 and CCR7 homing markers by flow cytometry. We also detected both transcripts and surface expression of the plasma cell marker CD138. Together, our findings reflect the diseased state of these lungs from patients with TB, suggestive of local antigen encounter by B cells in the lung and their involvement in GrALT, with differentiation into ASCs and hence a tissue-resident B cell response.
Several of our findings point toward the possible function or interaction of B cells with T cells within the GrALT. CCR7 facilitates migration of B cells toward the T cell zone within GC, allowing for B cell help from TFH cells44,71 and a key interaction with TFH cells includes the CD40-CD40L interaction72, all of which were expressed by lung B cells. Linge et al. demonstrate that B cells are an important source of IL6 and that the TH1 and TH17 response23 is affected when IL6 is knocked out of B cells21. Regarding putative regulatory B cells observed in the lung, recent evidence suggests that antigen experienced memory B cells expressing PDL-1 are important in recruiting TFH cells into GC3. Moreover, Zhang et al. show that CD5hiCD1d+ B cells can inhibit IL-17 production by T cells26. These cells were present in lung tissue from patients with TB and have previously been observed in circulation of patients with ATBI and expanded in those with cavitation25,32. The local inflamed milieu of the lung is also reflected by the presence of atypical B cells, which are associated with an inflamed state51,53. The atypical cells expressed TLR2 and 4, of which the former could detect bacterial lipoprotein such as ManLAM for example, resulting in MyD88 signalling and the IL8 response, which could result in the local recruitment of immune cells including T cells73,74. Importantly, CD11c B cells have been associated with many inflammatory conditions in humans including granulomatous lung diseases such as sarcoidosis75. In that study, these were termed age-associated B cells (ABCs), which share the same markers and probably represent the same or very similar B cell populations76. In addition, atypical or ABC are associated with many autoimmune conditions including Lupus and multiple sclerosis, and TB shares clinical similarities with such autoimmune conditions77,78. It is possible, therefore, that the atypical B cells enriched in lungs from patients with TB play a role in the immunopathogenesis of TB, through mechanisms including antibody and cytokine production and cross talk with T cells. Overall, the heterogeneity of B cells observed in lung tissue from patients with TB is consistent with multiple putative roles in TB protection and pathogenesis. We do concede that these data are only descriptive of the array of B cells found in the human lung with TB, and specific functional roles remain to be mechanistically proven.
To investigate the effect of B cells on Mtb in the lung, we used a granuloma biomimetic model and found that depletion of B cells increases Mtb growth. This is consistent with mouse6,7 and NHP10 studies showing an increased pulmonary bacterial burden in the absence of B cells. In addition, Joosten et al. observed a negative correlation between B cell frequency and bacterial growth in humans, using a whole blood mycobacterial growth inhibition assay79. Consequently, all these studies support a protective role for B cells, but notably in our model this control is inconsistent when PBMCs from patients with active TB were used. We believe it is likely to reflect the fact that the patients with TB were, by chance, at very different stages of disease severity. For example, in more severe TB disease we expect higher frequencies of pro-inflammatory atypical B cells, which may contribute to Mtb growth. The depletion of B cells in these cases may reduce Mtb growth, both in the 3D model and by analogy, in the lung. This is supported by the fact that Mtb growth was higher in PBMCs from patients with TB vs controls before B cell depletion (Fig. 7a). However, additional experiments are required to explore this hypothesis further. This suggests that B cells can contribute to control of Mtb growth, but also that the B cell compartment is disrupted during active TB. In a longitudinal study, Moreira-Teixeira et al. observed a decreased B cell signature with increased disease progression in the blood of patients with LTBI, LTBI-progressors and patients with ATBI, consistent with our observation16. Similarly, several studies showed that the proportions of B cell populations and transcriptional signatures are affected by TB11,15,16,80.
Finally, we analysed the antibodies present in TB-affected lung tissue and found significantly increased levels of those specific for Mtb. This was observed for all antibody classes, but only reached significance for IgM. Interestingly, IgM is known to be a potent activator of the complement system, which is a strong biomarker of TB disease81. In addition, IgM memory B cells are associated with induced lymphoid tissue in mucosal barriers82, suggesting a potential link with the GrALT observed in TB affected lungs. IgM memory B cells are emerging as important effectors of both adaptive and innate-like immune responses in mucosal barriers82. Antibodies from the lungs of infected patients enhanced uptake of Mtb, confirming both specificity and consistent with opsonisation increasing phagocytosis of mycobacteria83. However, we did not detect an effect on subsequent Mtb growth in our granuloma biomimetic model. A strong IgM response was associated with protection against TB disease in NHPs vaccinated with BCG via an intravenous route84. In addition, studies in humans have shown enhanced phagocytosis, phagolysosome fusion and macrophage killing using plasma antibodies isolated from TB resistors compared to those with active TB disease29,85. As lung antibodies used in this study were isolated from subjects with advanced TB lung disease, it is therefore perhaps expected that they failed to reduce Mtb growth despite enhanced phagocytosis. However, the fact that TB-specific antibodies are present at the site of disease in humans supports a role in the immune response to Mtb infection in humans.
B cell phenotype and function differs significantly between the lung and circulation, and B cells can regulate the host-pathogen interaction in TB. B cells isolated from human lung tissue displayed unique phenotypic subsets including populations consistent with tissue-resident memory, GC activity, atypical and regulatory B cells, suggesting potentially diverse roles in the immune response to Mtb. In addition, Mtb-specific antibodies are enriched in lung tissue from patients with TB and augment phagocytosis of Mtb. These data show B cells are likely to contribute to the host response to TB in humans at multiple points and highlight that considering tissue-specific phenotypes is likely to be essential to fully understand their overall impact. Future studies will aim to assess the TB specificity of the B cells in the GrALT and focus on spatial transcriptomic approaches to investigate the nature of the B cell follicles within the lung and if they form part of protection or pathology.
Methods
Cohort description
This study made use of a cohort of patients that underwent thoracotomy or lung resection surgeries. This included patients with a history of TB and non-TB-associated pathologies, with or without HIV. Written informed consent was obtained from all enrolled participants and surgeries were performed at either the King Dini-Zulu Hospital Complex or Inkosi Albert Luthuli Hospital in Durban, KwaZulu-Natal, South Africa. Patients were assessed for the extent of lung disease (cavitation and/or bronchiectasis) via HRCT. Candidates for surgery had to pass a fitness test as determined by Karnofsky score, 6-min walk test, spirometry, and arterial blood gas. Patients with massive hemoptysis were further assessed for their general condition, effort tolerance prior to hemoptysis, arterial blood gas measurement, serum albumin level and HRCT imaging. Tissues were collected following surgery to remove the irreversibly damaged lobes or lungs. Resected lung tissue was dissected into smaller tissue samples on site and transferred to RPMI 1640 media containing 10% FBS and kept on ice. The sections selected for analysis represented visibly low-, mid- and severely diseased areas of the lung, which were combined and processed86. Therefore, the isolated cells were representative of the entire lung and were not necessarily only of granulomatous origin. Samples were received for processing within 1 h of dissection and processed immediately in a BSL3 facility.
The cohort consisted largely of a Black African population 82% (90% of males and 74% of females). A total of 76 lung samples were collected, including 17 lung-draining lymph nodes and all samples with matched blood (Table 1). About half of the cohort was HIV positive, with positivity skewing slightly toward females. Most resections were TB-related (76%), with a median age of 41 years, and a relatively similar age distribution amongst males and females. Close to two-thirds of cases were males, and of these 45% were active/previous smokers. Lung cancer resections were dissected, and the healthy tissue margins were retained as the TB-negative control lung samples86.
Table 1.
Descriptive table of the study cohort (n = 76)
| Gender | + Lymph node | HIV +ve | TB | Smokers | Agea | |
|---|---|---|---|---|---|---|
| Male | 47 (62%) | 7 | 20 (43%) | 34 (72%) | 21 (45%) | 42 (35-48) |
| Female | 29 (38%) | 10 | 19 (66%) | 24 (83%) | 1 (3%) | 39 (34,47) |
| Total | 76 | 17 | 39 (51%) | 58 (76%) | 22 (29%) |
The total sample numbers and percentages in each column are highlighted in bold.
aMedian and IQR are indicated.
Histology
A section of lung was cut and transferred to 10% buffered formalin to fix. The sample was then processed in a vacuum filtration processor using a xylene-free method and isopropanol as the main substitute fixative. The tissues were embedded in paraffin wax and then cut into 4 µm sections, baked at 60 °C for 15 min, dewaxed using two xylene changes and rehydrated with descending grades of alcohol to water. These were then Hematoxylin and Eosin (H&E) stained using standard procedures, then dehydrated again in ascending grades of alcohol, cleared in xylene and mounted with a distyrene, plasticizer and xylene solution (DPX). The H&E slides were used to assess specimen quality before proceeding to immunohistochemistry.
Tissue sections (4 µm) of specimens of good quality were mounted on charged slides, and heated at 56 °C for 15 min. Sections were dewaxed in xylene, rinsed in 100% ethanol and one change of SVR (95%), washed under running water for 2 min followed by antigen retrieval via Heat Induced Epitope Retrieval (HIER) in Tris-HCl (pH 6.0) for 30 min. Slides were cooled for 15 min and rinsed under running water for 2 min. Endogenous peroxide activity was blocked using 3% H2O2 for 10 min at room temperature (RT), followed by a wash in PBST and blocking with protein block (Novolink) for 5 min at RT. Slides were incubated with primary antibodies for CD20 (M0755-CD20cy-L26, DAKO), CD68 (ab192847, Abcam), CD45 (M0701-2B11 + PD7/26, DAKO), CD3 (ab16669, Abcam) and CD21 (M0784-1F8, Dako), followed by washing and incubation with the polymer (Novolink) for 30 min at RT. Finally, the slides were washed and stained with diaminobenzidine (DAB) for 5 min, washed and counterstained with hematoxylin for 2 min, washed and blued in 3% ammoniated water for 30 s, washed, dehydrated, and mounted in DPX. Slides were imaged using a Hamamatsu NDP slide scanner (Hamamatsu NanoZoomer RS2, Model C10730-12) and its viewing software (NDP.View2). The red, green, and blue colour balance was kept at 100% whereas gamma correction was maintained between 0.7 and 2. Brightness (60–110%) and contrast (100–180%) settings varied slightly between slides depending on staining quality. Resolution was 230 nm/pixel yielding file sizes of 2–4.4 GB. Contrast, brightness, and intensity of exported images (jpg format) were minimally adjusted using CorelDraw 2020. DAB-stained cells were quantified using QuPath (https://qupath.github.io/) with the operator blinded to the nature of the sample.
Peripheral blood mononuclear cell (PBMC) isolation
Blood samples were collected into EDTA tubes and processed within 4 h of collection. Samples were centrifuged 930 × g for 5 min and 1 ml plasma fractions were aliquoted and stored at −80 °C. The remaining red blood cell pellet was made up to three times its pellet volume with PBS warmed to room temperature. PBMCs were isolated using a Ficoll-Paque (Amersham Biosciences, Little Chalfont, UK) density gradient sedimentation method as per manufacturer’s instructions. Isolated PBMCs were cryopreserved in 10% DMSO in heat inactivated foetal bovine serum (FBS, ThermoFischer Scientific) and stored in liquid nitrogen until use.
Lung mononuclear cell (LMC) isolation
Tissue specimens were mechanically dissociated using scissors followed by GentleMACs (Miltenyi Biotec) homogenization for 15 s in RPMI 1640, 10% FBS, 40 µg collagenase D (Roche) and 40 U/ml DNAse I (SIGMA-Aldrich). Samples were then incubated for 30 min at 37 °C, followed by another 75 s homogenization cycle and passed through 70 µm cell strainer (Corning). Following a 5 min 930 × g centrifugation (Beckman Coulter, Allegra X-12R), the sample pellet was suspended in 5 ml and passed through a 40 µm cell strainer and centrifuged as before. Finally, the pellet was treated with 5 ml red blood cell lysis solution (QIAGEN) for 5 min, made up to 30 ml with PBS and centrifuged again. Cells were counted and split into equal numbers (1–5 million) to stain for flow cytometry. Lung draining lymph nodes were processed similarly without the need for GentleMACs homogenization or collagenase D and DNAse I treatment.
Flow cytometry
Isolated LMCs were processed for flow cytometry on the same day to avoid reduced viability following cryopreservation and thawing. To facilitate running matched blood samples on the same day as stained LMCs, isolated PBMCs were thawed on the day the lung samples were processed. Briefly, cryopreserved PBMCs were thawed, washed, and rested in RPMI 1640 containing 10% FBS for 1 h in a 37 °C, 5% CO2 incubator prior to staining. Between 1–5 million PBMCs or LMCs were stained with the respective B cell surface marker panels (Supplementary Tables 1 to 3) for 20 min at RT in the dark. Samples were washed twice with PBS and suspended in 250 µl 2% PFA-PBS and kept at 4 °C in the dark. Samples were acquired on a BD FACS Aria Fusion III and data analysed using FlowJo version 9.9.6 (Tree Star).
Lung homogenate and plasma antibody isolation
All homogenate supernatants throughout the LMC isolation protocol were reserved for isolation of patient lung-derived immunoglobulins. The homogenate supernatants were centrifuged at 10,000 RCF for 10 min to pellet any remaining cellular debris. These were then filtered through a 0.22 µm syringe filter (Millipore, Merck). This clarified supernatant was then incubated with a Thiophilic resin (Pierce, ThermoFischer) for isolation of immunoglobulins66. For plasma antibodies, 2 ml plasma was loaded onto a thiophilic resin. Each sample type had a designated thiophilic column to avoid cross contamination of purified immunoglobulin pools. Both sample types were incubated with the resin for 1 h at room temperature, after which the bound immunoglobulins were eluted as per the manufacturer’s instructions. The resulting eluted sample absorbance was measured at 280 nm and fractions of interest were run on SDS-PAGE and western blot to assess sample purity, after which they were pooled by sample type. Sterile glycerol was added to 50% (v/v) and the pooled samples were stored at −20 °C.
SDS-PAGE and western blot of isolated immunoglobulins
Isolated immunoglobulin fractions were prepared for analysis on 12.5% reducing SDS-PAGE gels87. Samples were prepared in an equal volume of reducing SDS-PAGE sample buffer containing 10% (v/v) β-mercaptoethanol and boiled for 5 min before loading on duplicate SDS-PAGE gels. Once completed, one gel was stained with Coomassie brilliant blue R250 (SIGMA) as a reference gel, while the second was transferred (Trans-Blot Turbo, Bio-Rad) to nitrocellulose membrane for western blotting. PBS with 0.1% Tween-20 (PBST) was used throughout for western blotting. Membranes were blocked with 5% low-fat milk powder in PBST for 1 h, washed twice with PBST and incubated with immunoglobulin class-specific primary antibodies (all at 1:5000 dilution) for 2 h: Goat anti-human IgD-HRPO (cat. no 2030-05, Southern Biotech), Goat anti-human IgA-HRPO (cat. no 2050-05, Southern Biotech), Donkey anti-human IgG-HRPO (cat. no 709-036-073, Jackson ImmunoResearch), Donkey anti-human IgM-HRPO (cat. no 709-036-098, Jackson ImmunoResearch). Blots were washed again with PBST followed by a final wash and detection with enhanced chemiluminescence substrate (Pierce, ThermoFischer) using a ChemiDocTM imager (Bio-Rad).
ELISA to test Mtb specificity of isolated immunoglobulins
Mycobacterium tuberculosis (Mtb) H37Rv cultures were used to prepare a lysate for ELISAs. The Mtb lysate protein concentration was determined by Pierce 660 nm protein assay (Pierce, ThermoFischer) and used to coat ELISA plates at 10 µg/ml of lysate in PBS, 100 µl/well overnight at 4 °C. Plates were washed three times between each incubation step, using 200 µl/well PBS with 0.1% Tween-20 (PBST). The ELISA plate wells were blocked with 0.5% BSA in PBST for 1 h at 37 °C. This was followed by adding either the plasma or lung isolated immunoglobulins at a starting concentration of 100 µg/ml, 100 µl/well and serially diluting at 1:5, giving a final range from 100 to 0.8 µg/ml. Plates were incubated for 1 h at 37 °C. Finally, the plates were incubated for 1 h at 37 °C with the class-specific secondary antibodies as described for the western blotting protocol and detected using TMB substrate. The final titration curves were compared between samples as Area Under the Curve (AUC) readouts.
Mycobacterium tuberculosis (Mtb) culture
Standard Mtb H37Rv and bioluminescent Mtb H37Rv88 cultures were maintained in Middlebrook 7H9 medium (Difco) supplemented with 10% OADC, 0.2% glycerol, 0.01% Tyloxapol (SIGMA). For the bioluminescent H37Rv Mtb the media was supplemented with 25 µg/ml kanamycin. Cultures were grown to 1 × 108 CFU/ml Mtb (OD = 0.6) at which point they were suitable for infections using a multiplicity of infection (M.O.I.) of 0.1.
Bio-electrospray (BES) 3D culture biomimetic model
A 3D culture biomimetic model, designed to mimic the host-pathogen interaction64,89, was used to assess the impact of B cell depletion or antibody supplementation on the growth of Mtb.
Freshly isolated PBMCs were used for the BES experiments. Cells were infected with Mtb overnight in a T75 flask whereafter they were detached, washed, and mixed with 1.5% sterile alginate (Pronova UP MVG alginate, Nova Matrix) with human collagen (Advanced BioMatrix) to give a final concentration of 5 × 106 cells/ml. The PBMCs were encapsulated using an electrostatic bead generator (Nisco, Zurich, Switzerland) at a 10 ml/h flow rate using a Harvard syringe fitted with a 0.7 mm external diameter nozzle and dropping into a 100 mM CaCl2 ionotropic HBSS gelling bath as described in detail by Tezera et al.64,89. The resulting ~600 µm diameter microspheres containing the Mtb infected PBMCs were washed twice with HBSS with Ca2+/Mg2+ and transferred to RPMI 1640 supplemented with 10% human AB serum, 25 µg/ml kanamycin, 1% ampicillin. The beads were transferred to Eppendorffs and incubated at 37 °C, 5% CO2. The use of a bioluminescent Mtb allowed growth to be monitored using a luminometer (GloMax 20/20 Luminometer, Promega), with readings taken every 2 to 3 days over a 15-day period.
The initial set of BES experiments involved a B cell depletion step using a EasySepTM CD19 positive selection kit II (STEMCELL), with the control PBMCs remaining “untouched”. B cell depletion was performed prior to Mtb infection as described above. To confirm the CD19 depletion efficacy, samples were stained with a basic flow cytometry antibody panel, using L/D, CD45, CD3 and CD19 antibodies as listed in Supplementary Table 1. Cultures were maintained for a total of 15 days post infection and luminescence was measured every 2 to 3 days.
For the second set of BES experiments, PBMCs were left untouched, however, Mtb was treated with the antibodies or control non-specific human IgG (SIGMA, cat. 56834-25mg) for 1 h prior to infection of the PBMCs overnight and a media-only control was included. Relative infectivity was measured immediately, or cultures within microspheres were maintained for 15 days, and luminescence was measured as before.
Seq-Well single-cell RNA sequencing
LMCs were prepared for seq-well as previously described90. Cells were diluted to a single cell suspension of 15,000 cells in 200 µl RPMI, 10% FBS and loaded onto a polymethylsiloxane (PDMS) array pretreated with R10 media for 15 min. Once the cells settled into the microwells, the microarray was washed with PBS (SIGMA) then sealed with a plasma functionalized polycarbonate membrane (Sterlitech). The sealed arrays were incubated at 37 °C for 40 min, followed by 20 min at room temperature in a guanidinium thiocyanate (SIGMA), EDTA (ThermoFischer), 1% β-mercaptoethanol (SIGMA) and sarkosyl (SIGMA) buffer. Capture beads were then allowed to hybridize with released mRNA in a hybridization PBS, NaCl (ThermoFischer), MgCl2 (SIGMA) and polyethylene glycol (PEG) (SIGMA) buffer, with gentle shaking at 60 rpm for 40 min. The beads with the captured mRNA were collected from the wells with three washes of a Tris-HCl (ThermoFischer) buffer containing NaCl and MgCl2 and centrifugation at 2500 × g for 5 min after each wash.
cDNA synthesis was done in a mastermix containing Maxima H Minus Reverse Transcriptase, Maxima buffer, dNTPs, RNAse inhibitor, a template switch oligonucleotide and PEG for 30 min at room temperature, followed by an overnight incubation at 52 °C with end-over-end mixing. The cDNA was digested with exonuclease and complementary DNA denatured from the capture beads with a 5 min incubation in NaOH (SIGMA) and a wash with Tris-HCl, EDTA and Tween-20 (ThermoFischer). PCR amplification was then performed following suspension of the beads in a mastermix containing Klenow Fragment (NEB), dNTPs, PEG and dN-SMRT oligonucleotide and incubating for 45 min at 38 °C. Hereafter the PCR product was cleaned twice with an AMPure XP SPRI (Agencourt) bead cleanup at 0.6 and 0.8 times sample volume ratios. The product quality was assessed by Agilent Tape station hsD5000, confirming an expected 1 kbp amplicon with little or no primer dimers below 200 bp. The DNA libraries were quantified (Qubit High Sensitivity DNA kit) before preparation for Illumina sequencing using the Nextera XT DNA sample preparation kit, with 900 pg of each library added to the tagmentation reaction. The amplified product was cleaned again using the AMPure XP SPRI beads and the final libraries pooled before loading onto the NovaSeq 6000 using a paired-end read structure with a read 1 primer: read 1: 20 bases, read 2: 50 bases, read 1 index: 8 bases.
The sequencing reads were analysed by aligning them to the hg19 genome assembly and processing them according to the Drop-Seq Computation Protocol v2.0 (https://github.com/broadinstitute/Drop-seq). This yields a cell by gene matrix which was then transformed to loge(UMI + 1) using the Seurat R package v3.1.0 (https://satijalab.org/seurat/) and scaled by a factor of 10,000. The overall quality was assessed by the read distribution, the number of transcripts, and genes per cell. A Uniform Manifold Approximation and Projection (UMAP) was used for dimensionality reduction with the min distance set to 0.5 and the neighbours set to 30. To cluster cells with similar transcriptional profiles, an unsupervised clustering algorithm, FindClusters, was used with the resolution set to 0.5. These clusters were then further divided using a differential expression test, FindAllMarkers, in Seurat, setting “test.use” to “wilcox”, Wilcoxon-adjusted p value cutoff <0.01.
Statistics and reproducibility
All analyses, except for Seqwell data, were performed in Prism (v9; GraphPad Software Inc., San Diego, CA, USA). Nonparametric tests were used throughout, with Mann–Whitney and Wilcoxon tests used for unmatched and paired samples, respectively, and a three-way ANOVA, Kruskal–Wallis comparison. P values less than 0.05 were considered statistically significant and denoted by * ≤0.05; ** <0.01; *** <0.001 and **** <0.0001.
Study approval
Human patients undergoing thoracotomy or lung resection surgery for TB or non-TB-associated pathologies gave written informed consent for a blood draw and tissue collection. The study protocol, data collection tools and associated consent forms were approved by the University of KwaZulu-Natal Biomedical Research Ethics Committee (BE 019/13). The CUBS study protocol for blood collection from healthy donors and patients with active TB was also approved (BE 022/13). Healthy blood donor study ethical approval was provided by the National Research Ethics Service Committee South Central—Southampton A, ref 13/SC/0043, Southampton, United Kingdom. Participant compensation was approved by the relevant ethics committees.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
A.L. was supported by the Wellcome Trust (210662/Z/18/Z) and the BMGF (OPP1137006). R.K. and P.E. were supported by a Dorothy Temple Cross grant from the UK Medical Research Foundation (MRF-131-0002-RG-ELKI-C0850). P.E. was supported by the UK Medical Research Council (MR/P023754/1 and MR/W025728/1).
Author contributions
R.K. conducted experimental work, data analysis and writing of the manuscript; P.O. provided initial data, assisted with data analysis and writing; D.M. assisted with data analysis and writing; L.T. conducted some biomimetic model experiments, assisted with data analysis and writing; M.A., I.M., M.C., A.N. and M.M. assisted in sample preparation; M.C. assisted with “R” analyses; F.K., K.K. and R.M. obtained samples and analysed clinical information; K.L. and K.N. assisted with microscopy; A.S. established the lung cohort; A.K.S. assisted with data analysis; P.E. helped with data interpretation, study design, and manuscript preparation; and A.L. is the senior author who designed and implemented this study, analysed the data, and cowrote the manuscript with R.K.
Peer review
Peer review information
Communications Biology thanks Ananya Gupta, Stephen Cose, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Joanna Hester and Dario Ummarino. A peer review file is available.
Data availability
The source data behind the graphs in the paper can be found in Supplementary Data 1. Further inquiries can be directed to the corresponding author. The raw single-cell RNA sequencing data have not been shared publicly due to ethical restrictions, given the at-risk nature of people with HIV, but are available on request through the corresponding authors and their contact to the IRB, Prof. Shenuka Singh (Singhshen@ukzn.ac.za) or the UKZN ethics board (BREC@ukzn.ac.za).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Robert Krause, Email: Robert.krause@ahri.org.
Al Leslie, Email: al.leslie@ahri.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06282-7.
References
- 1.Organization, W. H. Global Tuberculosis Report 2022 (2022).
- 2.O’Garra A, et al. The immune response in tuberculosis. Annu. Rev. Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 3.Swanson RV, et al. Antigen-specific B cells direct T follicular-like helper cells into lymphoid follicles to mediate Mycobacterium tuberculosis control. Nat. Immunol. 2023;24:855–868. doi: 10.1038/s41590-023-01476-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choreno-Parra JA, et al. Mycobacterium tuberculosis HN878 infection induces human-like B-cell follicles in mice. J. Infect. Dis. 2020;221:1636–1646. doi: 10.1093/infdis/jiz663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hertz D, Dibbern J, Eggers L, von Borstel L, Schneider BE. Increased male susceptibility to Mycobacterium tuberculosis infection is associated with smaller B cell follicles in the lungs. Sci. Rep. 2020;10:5142. doi: 10.1038/s41598-020-61503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linge, I., Kondratieva, E. & Apt, A. Prolonged B-lymphocyte-mediated immune and inflammatory responses to tuberculosis infection in the lungs of TB-resistant mice. Int. J. Mol. Sci.24, 1140 (2023). [DOI] [PMC free article] [PubMed]
- 7.Maglione PJ, Xu J, Chan J. B cells moderate inflammatory progression and enhance bacterial containment upon pulmonary challenge with Mycobacterium tuberculosis. J. Immunol. 2007;178:7222–7234. doi: 10.4049/jimmunol.178.11.7222. [DOI] [PubMed] [Google Scholar]
- 8.Slight SR, et al. CXCR5(+) T helper cells mediate protective immunity against tuberculosis. J. Clin. Invest. 2013;123:712–726. doi: 10.1172/JCI65728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esaulova E, et al. The immune landscape in tuberculosis reveals populations linked to disease and latency. Cell Host Microbe. 2021;29:165–178.e168. doi: 10.1016/j.chom.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phuah J, et al. Effects of B cell depletion on early Mycobacterium tuberculosis infection in cynomolgus macaques. Infect. Immun. 2016;84:1301–1311. doi: 10.1128/IAI.00083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joosten SA, et al. Patients with tuberculosis have a dysfunctional circulating B-cell compartment, which normalizes following successful treatment. PLoS Pathog. 2016;12:e1005687. doi: 10.1371/journal.ppat.1005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulrichs T, et al. Human tuberculous granulomas induce peripheral lymphoid follicle-like structures to orchestrate local host defence in the lung. J. Pathol. 2004;204:217–228. doi: 10.1002/path.1628. [DOI] [PubMed] [Google Scholar]
- 13.Corominas M, et al. B-lymphocytes and co-stimulatory molecules in Mycobacterium tuberculosis infection. Int. J. Tuberc. Lung Dis. 2004;8:98–105. [PubMed] [Google Scholar]
- 14.Hernandez J, et al. Low number of peripheral blood B lymphocytes in patients with pulmonary tuberculosis. Immunol. Invest. 2010;39:197–205. doi: 10.3109/08820130903586346. [DOI] [PubMed] [Google Scholar]
- 15.Roy Chowdhury R, et al. A multi-cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature. 2018;560:644–648. doi: 10.1038/s41586-018-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreira-Teixeira L, et al. Mouse transcriptome reveals potential signatures of protection and pathogenesis in human tuberculosis. Nat. Immunol. 2020;21:464–476. doi: 10.1038/s41590-020-0610-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Manna, M. P. et al. Impact of Mycobacterium tuberculosis infection on human B cell compartment and antibody responses. Cells11, 2906 (2022). [DOI] [PMC free article] [PubMed]
- 18.du Plessis WJ, Keyser A, Walzl G, Loxton AG. Phenotypic analysis of peripheral B cell populations during Mycobacterium tuberculosis infection and disease. J. Inflamm. 2016;13:23. doi: 10.1186/s12950-016-0133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosio CM, Gardner D, Elkins KL. Infection of B cell-deficient mice with CDC 1551, a clinical isolate of Mycobacterium tuberculosis: delay in dissemination and development of lung pathology. J. Immunol. 2000;164:6417–6425. doi: 10.4049/jimmunol.164.12.6417. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, et al. B cells promote granulomatous inflammation during chronic Mycobacterium tuberculosis infection in mice. PLoS Pathog. 2023;19:e1011187. doi: 10.1371/journal.ppat.1011187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Linge I, et al. Pleiotropic effect of IL-6 produced by B-lymphocytes during early phases of adaptive immune responses against TB infection. Front Immunol. 2022;13:750068. doi: 10.3389/fimmu.2022.750068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Darrah PA, et al. Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature. 2020;577:95–102. doi: 10.1038/s41586-019-1817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Darrah PA, et al. Airway T cells are a correlate of i.v. Bacille Calmette-Guerin-mediated protection against tuberculosis in rhesus macaques. Cell Host Microbe. 2023;31:962–977.e968. doi: 10.1016/j.chom.2023.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang M, et al. B cell infiltration is associated with the increased IL-17 and IL-22 expression in the lungs of patients with tuberculosis. Cell Immunol. 2011;270:217–223. doi: 10.1016/j.cellimm.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Zhang M, et al. Anti-tuberculosis treatment enhances the production of IL-22 through reducing the frequencies of regulatory B cell. Tuberculosis. 2014;94:238–244. doi: 10.1016/j.tube.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M, et al. CD19(+)CD1d(+)CD5(+) B cell frequencies are increased in patients with tuberculosis and suppress Th17 responses. Cell Immunol. 2012;274:89–97. doi: 10.1016/j.cellimm.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 27.MacLean AJ, et al. Secondary influenza challenge triggers resident memory B cell migration and rapid relocation to boost antibody secretion at infected sites. Immunity. 2022;55:718–733.e718. doi: 10.1016/j.immuni.2022.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simmons JD, et al. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat. Rev. Immunol. 2018;18:575–589. doi: 10.1038/s41577-018-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu LL, et al. A functional role for antibodies in tuberculosis. Cell. 2016;167:433–443.e414. doi: 10.1016/j.cell.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu LL, et al. IFN-gamma-independent immune markers of Mycobacterium tuberculosis exposure. Nat. Med. 2019;25:977–987. doi: 10.1038/s41591-019-0441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawahara JY, Irvine EB, Alter G. A case for antibodies as mechanistic correlates of immunity in tuberculosis. Front Immunol. 2019;10:996. doi: 10.3389/fimmu.2019.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rijnink WF, Ottenhoff THM, Joosten SA. B-Cells and antibodies as contributors to effector immune responses in tuberculosis. Front Immunol. 2021;12:640168. doi: 10.3389/fimmu.2021.640168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grace PS, et al. Antibody subclass and glycosylation shift following effective TB treatment. Front. Immunol. 2021;12:679973. doi: 10.3389/fimmu.2021.679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogongo, P. et al. Tissue-resident-like CD4+ T cells secreting IL-17 control Mycobacterium tuberculosis in the human lung. J. Clin. Invest.131, e142014 (2021). [DOI] [PMC free article] [PubMed]
- 35.Gars E, Butzmann A, Ohgami R, Balakrishna JP, O’Malley DP. The life and death of the germinal center. Ann. Diagn. Pathol. 2020;44:151421. doi: 10.1016/j.anndiagpath.2019.151421. [DOI] [PubMed] [Google Scholar]
- 36.Fischer MB, et al. Dependence of germinal center B cells on expression of CD21/CD35 for survival. Science. 1998;280:582–585. doi: 10.1126/science.280.5363.582. [DOI] [PubMed] [Google Scholar]
- 37.Clavarino G, et al. Novel strategy for phenotypic characterization of human B lymphocytes from precursors to effector cells by flow cytometry. PLoS ONE. 2016;11:e0162209. doi: 10.1371/journal.pone.0162209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Day S, et al. Comparison of blood and lymph node cells after intramuscular injection with HIV envelope immunogens. Front Immunol. 2022;13:991509. doi: 10.3389/fimmu.2022.991509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J., Jr. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102:592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- 40.Calame KL. Plasma cells: finding new light at the end of B cell development. Nat. Immunol. 2001;2:1103–1108. doi: 10.1038/ni1201-1103. [DOI] [PubMed] [Google Scholar]
- 41.Xu, A. Q., Barbosa, R. R. & Calado, D. P. Genetic timestamping of plasma cells in vivo reveals tissue-specific homeostatic population turnover. eLife9, e59850 (2020). [DOI] [PMC free article] [PubMed]
- 42.Lin W, et al. Circulating plasmablasts/plasma cells: a potential biomarker for IgG4-related disease. Arthritis Res. Ther. 2017;19:25. doi: 10.1186/s13075-017-1231-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gothel SF, Marahiel MA. Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell Mol. Life Sci. 1999;55:423–436. doi: 10.1007/s000180050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pereira JP, Kelly LM, Cyster JG. Finding the right niche: B-cell migration in the early phases of T-dependent antibody responses. Int. Immunol. 2010;22:413–419. doi: 10.1093/intimm/dxq047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bunting KL, et al. Multi-tiered reorganization of the genome during B cell affinity maturation anchored by a germinal center-specific locus control region. Immunity. 2016;45:497–512. doi: 10.1016/j.immuni.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lefebvre C, et al. A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol. Syst. Biol. 2010;6:377. doi: 10.1038/msb.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranuncolo SM, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat. Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 48.Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- 49.Meyer SJ, et al. CD22 controls germinal center B cell receptor signaling, which influences plasma cell and memory B cell output. J. Immunol. 2021;207:1018–1032. doi: 10.4049/jimmunol.2100132. [DOI] [PubMed] [Google Scholar]
- 50.Salerno F, et al. An integrated proteome and transcriptome of B cell maturation defines poised activation states of transitional and mature B cells. Nat. Commun. 2023;14:5116. doi: 10.1038/s41467-023-40621-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenks SA, Cashman KS, Woodruff MC, Lee FE, Sanz I. Extrafollicular responses in humans and SLE. Immunol. Rev. 2019;288:136–148. doi: 10.1111/imr.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jenks SA, et al. Distinct effector B cells induced by unregulated toll-like receptor 7 contribute to pathogenic responses in systemic lupus erythematosus. Immunity. 2020;52:203. doi: 10.1016/j.immuni.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Woodruff MC, et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 2020;21:1506–1516. doi: 10.1038/s41590-020-00814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krause, R. et al. HIV skews the SARS-CoV-2 B cell response towards an extrafollicular maturation pathway. eLlife11, e79924 (2022). [DOI] [PMC free article] [PubMed]
- 55.Gao, X. et al. Zeb2 drives the formation of CD11c+ atypical B cells to sustain germinal centers that control persistent infection. Sci. Immunol. 11, eadj4748 (2024). [DOI] [PubMed]
- 56.Kang SM, et al. MyD88 plays an essential role in inducing B cells capable of differentiating into antibody-secreting cells after vaccination. J. Virol. 2011;85:11391–11400. doi: 10.1128/JVI.00080-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Georg I, Diaz-Barreiro A, Morell M, Pey AL, Alarcon-Riquelme ME. BANK1 interacts with TRAF6 and MyD88 in innate immune signaling in B cells. Cell Mol. Immunol. 2020;17:954–965. doi: 10.1038/s41423-019-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muehlinghaus G, et al. Regulation of CXCR3 and CXCR4 expression during terminal differentiation of memory B cells into plasma cells. Blood. 2005;105:3965–3971. doi: 10.1182/blood-2004-08-2992. [DOI] [PubMed] [Google Scholar]
- 59.Hasan MM, et al. CD24(hi)CD38(hi) and CD24(hi)CD27(+) human regulatory B cells display common and distinct functional characteristics. J. Immunol. 2019;203:2110–2120. doi: 10.4049/jimmunol.1900488. [DOI] [PubMed] [Google Scholar]
- 60.Scharer CD, et al. Antibody-secreting cell destiny emerges during the initial stages of B-cell activation. Nat. Commun. 2020;11:3989. doi: 10.1038/s41467-020-17798-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yuan C, et al. Mycobacterium tuberculosis mannose-capped lipoarabinomannan induces IL-10-producing B cells and hinders CD4(+)Th1 immunity. iScience. 2019;11:13–30. doi: 10.1016/j.isci.2018.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed, M., Tezera, L. B., Elkington, P. T. & Leslie, A. J. The paradox of immune checkpoint inhibition re-activating tuberculosis. Eur. Respir. J.60, 2102512 (2022). [DOI] [PMC free article] [PubMed]
- 63.Tezera, L. B. et al. Anti-PD-1 immunotherapy leads to tuberculosis reactivation via dysregulation of TNF-alpha. eLife9, e52668 (2020). [DOI] [PMC free article] [PubMed]
- 64.Tezera, L. B. et al. Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. eLife6, e21283 (2017). [DOI] [PMC free article] [PubMed]
- 65.Reichmann, M. T. et al. Integrated transcriptomic analysis of human tuberculosis granulomas and a biomimetic model identifies therapeutic targets. J. Clin. Invest.131, e148136 (2021). [DOI] [PMC free article] [PubMed]
- 66.Hutchens TW, Porath J. Thiophilic adsorption of immunoglobulins–analysis of conditions optimal for selective immobilization and purification. Anal. Biochem. 1986;159:217–226. doi: 10.1016/0003-2697(86)90331-3. [DOI] [PubMed] [Google Scholar]
- 67.Weisel NM, et al. Comprehensive analyses of B-cell compartments across the human body reveal novel subsets and a gut-resident memory phenotype. Blood. 2020;136:2774–2785. doi: 10.1182/blood.2019002782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barker, K. A. et al. Lung-resident memory B cells protect against bacterial pneumonia. J. Clin. Invest.131, e141810 (2021). [DOI] [PMC free article] [PubMed]
- 69.Tan HX, et al. Lung-resident memory B cells established after pulmonary influenza infection display distinct transcriptional and phenotypic profiles. Sci. Immunol. 2022;7:eabf5314. doi: 10.1126/sciimmunol.abf5314. [DOI] [PubMed] [Google Scholar]
- 70.Allie SR, et al. The establishment of resident memory B cells in the lung requires local antigen encounter. Nat. Immunol. 2019;20:97–108. doi: 10.1038/s41590-018-0260-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McHeik S, et al. Coexpression of CCR7 and CXCR4 during B cell development controls CXCR4 responsiveness and bone marrow homing. Front Immunol. 2019;10:2970. doi: 10.3389/fimmu.2019.02970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suan D, et al. CCR6 defines memory B cell precursors in mouse and human germinal centers, revealing light-zone location and predominant low antigen affinity. Immunity. 2017;47:1142–1153.e1144. doi: 10.1016/j.immuni.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 73.Kunsch C, Rosen CA. NF-kappa B subunit-specific regulation of the interleukin-8 promoter. Mol. Cell Biol. 1993;13:6137–6146. doi: 10.1128/mcb.13.10.6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sims-Mourtada JC, et al. In vivo expression of interleukin-8, and regulated on activation, normal, T-cell expressed, and secreted, by human germinal centre B lymphocytes. Immunology. 2003;110:296–303. doi: 10.1046/j.1365-2567.2003.01745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Phalke S, et al. Age-associated B cells appear in patients with granulomatous lung diseases. Am. J. Respir. Crit. Care Med. 2020;202:1013–1023. doi: 10.1164/rccm.201911-2151OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mouat IC, Shanina I, Horwitz MS. Age-associated B cells are long-lasting effectors that impede latent gammaHV68 reactivation. Sci. Rep. 2022;12:21189. doi: 10.1038/s41598-022-25543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Elkington P, Tebruegge M, Mansour S. Tuberculosis: an infection-initiated autoimmune disease? Trends Immunol. 2016;37:815–818. doi: 10.1016/j.it.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belyaeva IV, Kosova AN, Vasiliev AG. Tuberculosis and autoimmunity. Pathophysiology. 2022;29:298–318. doi: 10.3390/pathophysiology29020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Joosten SA, et al. Mycobacterial growth inhibition is associated with trained innate immunity. J. Clin. Invest. 2018;128:1837–1851. doi: 10.1172/JCI97508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singhania A, et al. A modular transcriptional signature identifies phenotypic heterogeneity of human tuberculosis infection. Nat. Commun. 2018;9:2308. doi: 10.1038/s41467-018-04579-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lubbers R, van Essen MF, van Kooten C, Trouw LA. Production of complement components by cells of the immune system. Clin. Exp. Immunol. 2017;188:183–194. doi: 10.1111/cei.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weill JC, Reynaud CA. IgM memory B cells: specific effectors of innate-like and adaptive responses. Curr. Opin. Immunol. 2020;63:1–6. doi: 10.1016/j.coi.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schorey JS, Carroll MC, Brown EJ. A macrophage invasion mechanism of pathogenic mycobacteria. Science. 1997;277:1091–1093. doi: 10.1126/science.277.5329.1091. [DOI] [PubMed] [Google Scholar]
- 84.Irvine EB, et al. Robust IgM responses following intravenous vaccination with Bacille Calmette-Guerin associate with prevention of Mycobacterium tuberculosis infection in macaques. Nat. Immunol. 2021;22:1515–1523. doi: 10.1038/s41590-021-01066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, et al. Latently and uninfected healthcare workers exposed to TB make protective antibodies against Mycobacterium tuberculosis. Proc. Natl Acad. Sci. USA. 2017;114:5023–5028. doi: 10.1073/pnas.1611776114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ogongo P, et al. Differential skewing of donor-unrestricted and gammadelta T cell repertoires in tuberculosis-infected human lungs. J. Clin. Invest. 2020;130:214–230. doi: 10.1172/JCI130711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 88.Andreu N, et al. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS ONE. 2010;5:e10777. doi: 10.1371/journal.pone.0010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tezera, L. B., Bielecka, M. K. & Elkington, P. T. Bioelectrospray methodology for dissection of the host-pathogen interaction in human tuberculosis. Bio Protoc.7, e2418 (2017). [DOI] [PMC free article] [PubMed]
- 90.Reyfman PA, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The source data behind the graphs in the paper can be found in Supplementary Data 1. Further inquiries can be directed to the corresponding author. The raw single-cell RNA sequencing data have not been shared publicly due to ethical restrictions, given the at-risk nature of people with HIV, but are available on request through the corresponding authors and their contact to the IRB, Prof. Shenuka Singh (Singhshen@ukzn.ac.za) or the UKZN ethics board (BREC@ukzn.ac.za).