Abstract
The whiA sporulation gene of Streptomyces coelicolor A3(2), which plays a key role in switching aerial hyphae away from continued extension growth and toward sporulation septation, was cloned by complementation of whiA mutants. DNA sequencing of the wild-type allele and five whiA mutations verified that whiA is a gene encoding a protein with homologues in all gram-positive bacteria whose genome sequence is known, whether of high or low G+C content. No function has been attributed to any of these WhiA-like proteins. In most cases, as in S. coelicolor, the whiA-like gene is downstream of other conserved genes in an operon-like cluster. Phenotypic analysis of a constructed disruption mutant confirmed that whiA is essential for sporulation. whiA is transcribed from at least two promoters, the most downstream of which is located within the preceding gene and is strongly up-regulated when colonies are undergoing sporulation. The up-regulation depends on a functional whiA gene, suggesting positive autoregulation, although it is not known whether this is direct or indirect. Unlike the promoters of some other sporulation-regulatory genes, the whiA promoter does not depend on the sporulation-specific ς factor encoded by whiG.
Dispersal of the mycelial organism Streptomyces coelicolor A3(2) occurs by the formation of long chains of spores from aerial hyphae. A critical stage in this process is the subdivision of a multigenomic apical aerial hyphal compartment into many unigenomic prespore compartments by the synchronous formation of regularly spaced sporulation septa (26, 38). At least six genetic loci (whiA, whiB, whiG, whiH, whiI, and whiJ) are needed for sporulation septation (8, 10), in addition to cell division genes that are also involved in vegetative growth (24, 25). These six early whi loci appear to play no role in vegetative growth. Mutations in them have pleiotropic effects on the later stages of sporulation, including weak or undetectable transcription of the genes responsible for the production of grey spore pigment (20), hence the white colony phenotype after which they were named (18). Transcription of sigF, which encodes a late sporulation sigma factor, is also dependent on these early whi genes (21, 28). The complex early whi mutant phenotypes suggest that some or all of the six “early” whi genes encode regulatory elements. Indeed, whiG encodes a sigma factor (ςWhiG [11, 37]), whiH encodes a repressor-like protein (31), and whiI encodes a response regulator-like protein (1); the product of whiB has features that resemble those of some transcription factors (13, 34). Mutations in some other more recently discovered genes also affect sporulation septation but in a more allele-specific manner (30).
This paper is concerned with whiA. Like whiB mutants, whiA and whiA whiB double mutants have tightly coiled aerial hyphae that are markedly longer than normal spore chains, contain uncondensed DNA, and lack sporulation septa and readily detectable FtsZ (9, 15, 33). whiA and whiB are therefore believed to play a role in the cessation of aerial growth. Flärdh et al. (15) and Aínsa et al. (1) proposed that this is part of a hypothetical sequentially dependent series of developmental decisions. In this model, newly emerged aerial hyphae first switch to a sporulation-specific mode of elongation through the action of ςWhiG, eventually undergoing a WhiA- and WhiB-dependent orderly cessation of elongation and DNA replication, presumably in response to some signal(s); it is suggested that growth cessation then sets off further signals, which activate the WhiH and WhiI proteins, and the activated forms of these proteins switch on sporulation septation.
The available evidence points to a close interplay between WhiA and WhiB. whiB is one of at least six similar genes in S. coelicolor, all encoding small proteins whose conserved features include four cysteine residues (“Wbl” proteins [27, 34]). Likely orthologues of several of these six proteins are present in most actinomycetes, but no similar proteins are known in any other organisms (34). Here we characterize the whiA gene and show that it is the S. coelicolor version of a gene that is conserved and ubiquitous among gram-positive bacteria but for which there had been no functional attribution.
MATERIALS AND METHODS
Strains and media.
S. coelicolor A3(2)-derived strains used in this study were M145 (prototrophic, SCP1− SCP2−) (17), J1501 (hisA1 uraA1 strA1 Pgl− SCP1− SCP2−) (7), and the whiA strains C13, C72, C73, C85, C170, and C213 (all prototrophic SCP1+ SCP2+) (8). Escherichia coli strains were the general host DH5α (32) and the Dam− Dcm− HsdM− strain ET12567 (23). Media, culture conditions, and transformation procedures for S. coelicolor were as previously described (17), and those for E. coli strains were as described by Sambrook et al. (32).
Library screening.
The library of M145 chromosomal DNA used in this study was cloned in the low-copy-number SCP2*-based vector pIJ698 and had previously been used to clone whiH (31). In brief, the library was represented by a regular array of 2,400 patches of transformants of J1501 (auxotroph). The library was replicated onto a densely inoculated lawn of mycelial fragments of the prototrophic whiA mutant C72 on plates of nonselective minimal medium (MM) supplemented with mannitol as the carbon source and histidine and uracil to permit the growth of J1501. After 4 days of incubation at 30°C, the plates were replicated onto selective MM lacking histidine and uracil and containing thiostrepton at 50 μg/ml to select for C72 recipients of the pIJ698 derivatives. After incubation for at least 4 days, the transconjugants were screened for restoration of the wild-type phenotype (grey, sporulating aerial mycelium) instead of the white, nonsporulating aerial mycelium of mutant C72.
Subcloning and sequencing.
The 7-kb insert from pIJ6204 that complemented whiA (see Results) was cloned in both orientations into pDH5 (a vector, based on pUC119, that is unable to replicate in S. coelicolor and contains tsr, a thiostrepton resistance marker [16]), giving pIJ6217 and pIJ6218. The restriction map of the insert was established, and subclones with defined deletions were created by digesting pIJ6217 and pIJ6218 with restriction enzymes and then ligating to induce recircularization. Each subclone was passaged through the nonmethylating E. coli strain ET12567 and used to transform the whiA mutant C13, selecting for resistance to thiostrepton.
The 3.4-kb insert of the subclone that retained the ability to complement whiA mutants (see Results) was sequenced using dye termination chemistry and a model 373A DNA-sequencing system (Applied Biosystems) and then extended to an adjacent SacI site by using conventional sequencing as described by Bruton and Chater (6). The FRAME program (4) was used to detect open reading frames (ORFs). The sequencing of mutant alleles of whiA was as described by Ryding et al. (31). Sequences were processed using the GCG Inc. package (14), and databases were searched using BLAST (2). Preliminary sequence from unfinished genomes was obtained from The Institute of Genomic Research database (http://www.tigr.org). The sequence of S. coelicolor cosmid C54 (accession number AL035591) was obtained from the S. coelicolor Genome Project at The Sanger Centre (http://www.sanger.ac.uk/Projects/S_coelicolor). Predictions of secondary structure were done at the PSA server (36) of the BioMolecular Engineering Research Centre (http://bmerc-www.bu.edu/psa).
Disruption of ORF4.
Plasmid pIJ6413 is a pDH5 derivative containing the 7-kb insert from pIJ6204, in which the only PstI site was within the insert, internal to ORF4. A copy of the hygromycin resistance marker hyg (40) was cloned into the PstI site to give pIJ6414. S. coelicolor M145 was transformed with pIJ6414 prepared from E. coli ET12567, selecting for hygromycin resistance. Chromosomal DNA from three transformants that were sensitive to thiostrepton (and hence were presumed to be the products of recombination events on either side of the hyg marker) was examined by Southern hybridization and found to show the correct restriction pattern.
Transcription analysis.
To investigate transcription during development, RNA samples were isolated from cultures grown for different times on MM overlaid with cellophane disks (1). Indeed, the RNA samples for the wild-type strain M145 time course experiment used to study whiA transcription were the same as those reported in that study. S1 protection assays were performed using the hrdB probe as a control (21), with a probe generated by PCR (31). To provide a template for PCR to generate the whiA probe, a 0.47-kb PstI-SalI fragment from pIJ6221, containing the 5′ end of whiA and the presumptive promoter region, was cloned into pIJ2925 (19) to give pIJ6412. For amplification, the primers were the radiolabeled oligonucleotide 5′-CTGCAGCAGGTCCGGGTGAC-3′ (complement of nucleotides 2365 to 2384 of the deposited sequence) and the unlabeled oligonucleotide 5′-AATACCGCATCAGGCGCCATTCG-3′, which anneals in the vector pIJ2925, generating a probe with a nonhomologous tail of 220 nucleotides. For high-resolution S1 mapping, the radiolabeled oligonucleotide was 5′-GCCAGCAGCTCCGGGTCGTG-3′ (complement of nucleotides 2255 to 2274).
In vitro runoff transcription using purified ςWhiG and E. coli core RNA polymerase (37) was performed on DNA generated by PCR with the oligonucleotides 5′-TGCTGGACGCGCTGGTCGAG-3′ (nucleotides 1998 to 2017) and 5′-CCGCGTTCCCGGTGTCCAGC-3′ (complement of nucleotides 2463 to 2482) and with pIJ6221 as the template. The PCR product was digested with RsaI or AluI to produce templates of 285 and 221 nucleotides, respectively, in which the whiA ends of the fragment are located 149 and 85 nucleotides downstream of the putative transcription start site.
Reverse transcriptase-PCR was performed using the Titan One Tube RT-PCR System (Boehringer Mannheim), under the conditions specified by the manufacturer and with the primers mentioned above.
Microscopy.
For phase-contrast microscopy of Streptomyces, strains were cultivated on MM for up to 5 days at 30°C. Then, coverslips were touched against the top of the colonies, and the impression preparations obtained were observed in a Zeiss Axiophot microscope. Photographs were taken using Kodak Technical pan film. For scanning electron microscopy, colonies were processed and examined as described by Flärdh et al. (15).
Nucleotide sequence accession number.
The nucleotide sequence referred in the text has been deposited in the GenBank/EMBL/DDBJ databases under accession number AF106003.
RESULTS
The morphological phenotypes of whiA mutants.
Six whiA mutants had previously been identified on the basis of morphological phenotype combined with genetic mapping data (8). In broad agreement with earlier studies (8, 26), examination of the aerial hyphae of five of these mutants (C13, C72, C73, C85, and C213) by phase-contrast microscopy revealed aerial hyphae that were long and tightly coiled (like those of the constructed whiA null mutant shown in Fig. 1) and indistinguishable from those of whiB mutants. (Some fragmentation of C13 aerial hyphae was sometimes seen, although less than had previously been observed on MM with glucose as a carbon source [8].) However, C170 produced only straight aerial hyphae (similar to those of whiG mutants), in contrast to the previous description (8) (see below for a brief discussion).
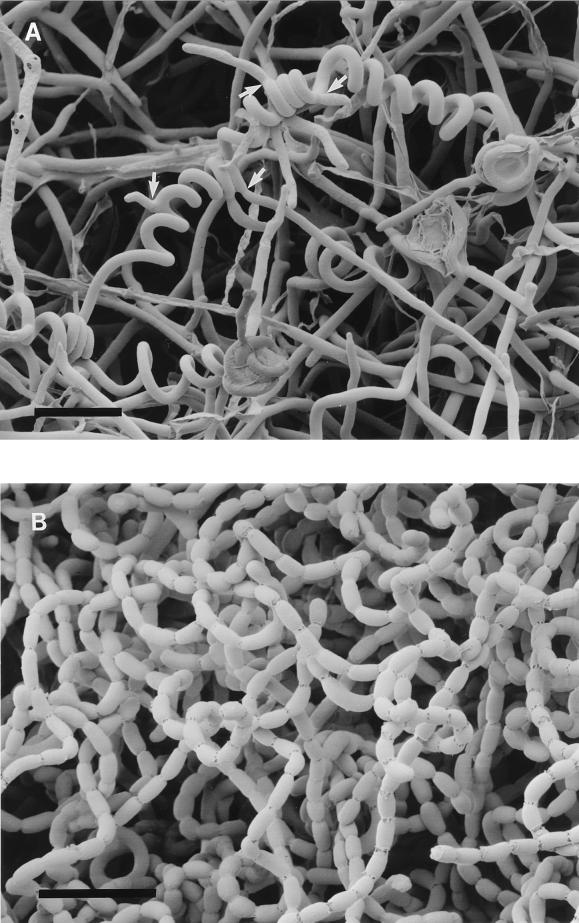
FIG. 1.
Scanning electron micrograph of aerial hyphae of the whiA disruption mutant J2401 (A) and the wild-type strain M145 (B). In contrast to the wild-type chains of unicellular spores, the whiA mutant exhibits slightly coiled “stalks” surmounted by coiled aerial hyphae that are often 100 μm or more long (the coiled segment shown here is ca. 150 μm long) and no indentations that might indicate sporulation septa. In this example, a number of branches (arrows) have emerged from the coiled hypha. We have not observed this previously in whiA mutants studied by phase-contrast microscopy, and we suspect that the branches are the equivalent of germ tubes, which are sometimes seen to emerge from spores in spore chains. Bar, 5 μm.
Cloning of DNA complementing whiA mutants.
A library of S. coelicolor DNA (31) was transferred by conjugation into the whiA mutant C72, and plasmids pIJ6203 and pIJ6204 caused sporulation and grey-pigment biosynthesis uniformly across the corresponding exconjugant patches. The same technique was used to transfer these plasmids into five other strains previously classified as whiA mutants (C13, C73, C85, C170, and C213 [8]). These plasmids fully restored grey-pigment biosynthesis and spore formation in C13 and C73. In C85, the plasmids only altered the degree of coiling and fragmentation of aerial hyphae, giving loosely coiled fragments, resembling those seen with whiH mutants (8, 31), and suggesting that C85 may contain both a whiA mutation and a mutation in another whi gene. Alternatively, whiA85 could be partially dominant. Neither pIJ6203 nor pIJ6204 affected the white-colony phenotype of C170, which was consistent with other evidence that it had acquired a second whi mutation (see below). Introducing either pIJ6203 or pIJ6204 into C213 restored spore pigment to at least wild-type levels, but the spacing of the sporulation septa was unusually variable, resulting in long and short spore compartments in the same spore chain. This effect was explained because C213 had been created by mutagenesis of an existing mutant (C58) that showed excessive production of spore pigment and also aberrant sporulation (H. M. Kieser, personal communication). Therefore, pIJ6203 and pIJ6204 complemented the whiA mutation and allowed the effect of the initial mutation to become apparent.
Plasmids with various parts of the 7-kb insert of pIJ6204 were used in attempts to complement C13. pIJ6221, containing a 3.4-kb XbaI-Asp718I fragment, restored sporulation in 94% of the transformants (the 6% of the transformants that were white may have arisen through homogenotization), while pIJ6223, whose insert (the 2.5-kb XbaI-PstI fragment) was just 0.9 kb shorter than that in pIJ6221, did not complement C13 (Fig. 2). This indicated that pIJ6221 contained most or all of the whiA gene, with at least part of it being located in the 0.9-kb PstI-Asp718I fragment.
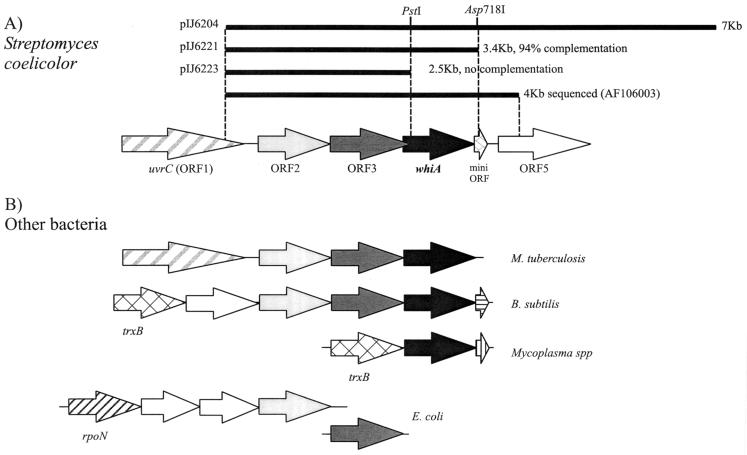
FIG. 2.
The whiA region and its comparison with similar regions of other bacterial chromosomes. (A) Subcloning analysis by complementation of strain C13 and schematic representation of the arrangement of the genes. Although one possible start codon for ORF2 overlaps the stop codon for ORF1, the codon usage for the region between this and the next possible start codon 114 bp downstream would not be typical for Streptomyces coding sequences, with a G+C content in the third position of only 73% compared to >90% for most genes (39). The end of ORF2 and the beginning of ORF3 are separated by only 7 bp. There are two likely ATG start codons for whiA, separated by 3 bp: the more upstream of them is 7 bp from a predicted ribosome-binding site and overlaps by 10 bp the end of ORF3; the other ATG codon (13 bp from the ribosome-binding site) overlaps the stop codon for ORF3. The stop codon of whiA and the putative start codon of the mini-ORF are separated by 11 bp, and the stop codon of the mini-ORF and the start codon for ORF5 are separated by 87 bp. In this noncoding intergenic region, there are some inverted repeats that might act as transcription terminators. (B) Arrangement of related genes from other bacteria. The genes are shaded to indicate familial relationships.
The pIJ6204 insert hybridized to cosmid C54 of the minimal ordered library of S. coelicolor constructed by Redenbach et al. (29), consistent with previous genetic mapping of whiA (8, 22). Southern blotting revealed that the BamHI and SalI digestion patterns of DNA from five of the whiA mutants (C13, C72, C85, C170, and C213) were the same as for the wild type (M145) when the 7-kb probe was used. However, C73 gave no signal, indicating that it contains a deletion of at least the 7-kb fragment (data not shown).
whiA-like genes and the accompanying ORFs are widespread among gram-positive bacteria.
DNA sequence analysis included the 3.4-kb XbaI-Asp718I fragment that complemented whiA13 and the sequence beyond the Asp718I site. This revealed the 3′ end of one ORF (designated ORF1), three complete ORFs (ORF2, ORF3, and ORF4), and the 5′ end of one ORF (ORF5), all orientated in the same direction and with little or no noncoding DNA between them. Subsequently, the sequence of cosmid C54 from the S. coelicolor genome project confirmed our primary sequence and also revealed a “mini-ORF” located immediately downstream of ORF4. The main features of this gene cluster are summarized in Fig. 2.
Most of ORF4 (termed SCC54.10c in the cosmid C54 annotation) was located between the PstI and Asp718I restriction sites mentioned in the section above, and so it was presumed to be responsible for the complementation of C13. This was confirmed in subsequent experiments (see below). Genes encoding WhiA-like products are present in the genomes of all sequenced gram-positive bacteria, showing amino acid identities ranging from 70% (mycobacteria) to 20–27% (low-G+C gram-positive bacteria). Moreover, a whiA-like gene is present only once in any one genome, and in all cases except mycoplasmas it is downstream of genes resembling ORF2 and ORF3.
WhiA does not resemble any protein of known function. Of the adjacent and possibly cotranscribed genes, only ORF1 encodes a protein of predictable function (the ATP-binding cassette excision nuclease subunit C, widely conserved in many bacteria). ORF2 shows strong similarity to a gene of unknown function that is located three genes downstream of rpoN, the gene which encodes an atypical sigma factor (ς54), in several gram-negative species. However, neither the sequence near ORF2 in S. coelicolor nor the complete sequence of Mycobacterium tuberculosis (12) contains any rpoN-like genes. Like ORF2, ORF3 homologues also occur in phylogenetically diverse bacteria that do not possess whiA-like genes. There are mini-ORFs unrelated to the one in S. coelicolor downstream of the whiA-like gene in Bacillus subtilis and the mycoplasmas (3) but not in M. tuberculosis. Finally, ORF5 is related to a number of carboxypeptidases from both prokaryotic and eukaryotic organisms.
Analysis of a constructed whiA null mutant.
We constructed an S. coelicolor M145 derivative strain, J2401, carrying a whiA::hyg disruption in the chromosome. This mutant developed a white aerial mycelium, and no spores could be detected under phase-contrast microscopy or SEM. The aerial hyphae bore long and tightly coiled tip regions on slightly coiled “stalks,” showing a phenotype indistinguishable from that of the original whiA point mutants. In the example shown in Fig. 1, the coiled part of the hypha is about 150 μm long whereas the wild-type spore chains seldom exceed 50 μm. Plasmid pIJ6204 (Fig. 2) restored full sporulation to J2401 (data not shown).
Sequencing the whiA mutant alleles.
For each of five whiA mutants, a 1.5-kb region containing the promoter region of whiA and the coding sequences of whiA and the mini-ORF was sequenced. Two mutants had single-amino-acid substitutions in whiA: in C85 the mutation changed leucine 162 into proline, and in C213 the mutation changed leucine 196 into proline. In the alignment of WhiA-like proteins (Fig. 3), these two leucine residues are highly conserved and are located inside regions with a high probability of folding into a helix (following a prediction of the secondary structure of WhiA), where the presence of the prolines would considerably distort the structure. Two mutants had deletions causing frameshifts: in C170 the loss of a G nucleotide caused a frameshift from arginine 99 onward, whereas in C72 the deletion of seven nucleotides (GCCTCGG) produced a frameshift from arginine 306 onwards, adding 61 aberrant amino acids to the C terminus of WhiA.
FIG. 3.
Alignment of WhiA-like proteins from diverse gram-positive bacteria. The sequences compared are from Mycoplasma pneumoniae (mycpne; accession number AAB96238), Mycoplasma genitalium (mycgen; accession number AAC71321), Mycobacterium tuberculosis (myctbc; accession number CAB02171), S. coelicolor (scwhia; this paper), and Bacillus subtilis (bacsub; accession number CAB08059). The M. tuberculosis protein is the most similar to WhiA (ca. 70% amino acid identity); the others, all from low-G+C bacteria, ranged from 20 to 27% identity. The consensus sequence is based on the occurrence of identical residues in at least three of the five sequences (black boxes). Grey boxes indicate residues similar to consensus residues. The positions of point mutations, frameshifts (fs), and the insertion of the hyg cassette in J2401 discussed in the text are marked underneath the consensus sequence. Regions predicted to form α-helices in the C-terminal part of the S. coelicolor WhiA are overlined.
C13 had a mutation just upstream of the putative ribosome-binding site and translation start codon of whiA, within the coding sequence of the preceding gene ORF3: TG was changed to CT, changing the wild-type sequence TGGAAGGA to CTGAAGGA (Fig. 4). This mutation introduces an in-frame premature stop codon in ORF3 that removes its last 6 residues, and it also reduces the complementarity of this region to the 3′ end of the 16S rRNA. Both effects are predicted to affect the translation of whiA, tending to uncouple the translation of ORF3 and whiA and reducing the translation of whiA independently initiated from its own ribosome-binding site.
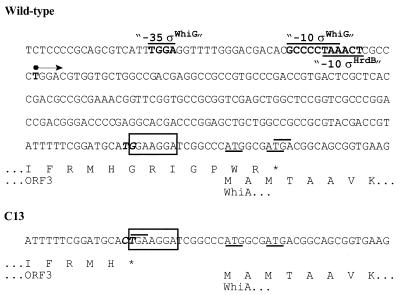
FIG. 4.
Sequence (nucleotides 2079 to 2343 from AF106003) at the overlap of ORF3 and whiA, showing the promoter regions and the transcription start point of whiA (bold), the stop codon of ORF3 (overlined), the putative ribosome-binding site for whiA (boxed), the two putative ATG start codons for whiA (underlined), and the proposed translational overlap between ORF3 and whiA (only the translation of the last line of the nucleotide sequence is shown). At the bottom, the sequence corresponding to bp 2291 to 2343 (and its translation) is reproduced, showing the mutation found in mutant C13 (TG→CT shown in bold italics in both sequences). The stop codon of ORF3 in C13 (overlined) is located 11 and 17 bp, respectively, upstream of the two proposed start codons for whiA, whereas in the wild type, ORF3 overlaps whiA. Thus, the mutation in C13 could affect the translational coupling of whiA and ORF3, as well as the efficiency of the ribosome-binding site located upstream of the whiA start codon.
As described in sections above, C85 and C170 were the only whiA mutants that were not complemented by pIJ6204. In C85, the mutation found (Leu162→Pro) could therefore represent a dominant allele, although the presence of a secondary mutation (perhaps in whiH, to judge from the phenotype of C85/pIJ6204, although this has not been further studied) is an alternative explanation. IN C170, the presence of a frameshift mutation in whiA confirms that this mutant was originally isolated and correctly classified as a whiA mutant. The “abnormal” phenotype currently detected in C170 (remarkably similar to whiG mutants) could be explained by the presence of a secondary mutation. The likely presence of a second whi mutation in two of the six whiA mutants was surprising and was not consistent with the results of an earlier genetic analysis (8). Possibly, there may be a selective advantage in such additional mutations: the aerial hyphae of whiA mutants, which are normally used for subculture and storage in the laboratory, have lower viability than the aerial hyphae of other whi mutants (our unpublished observations), and the accumulation of other mutations during culture preservation or subculture may alleviate this effect.
Transcriptional analysis.
whiA appears to be downstream of other genes in an operon, raising the possibility that at least part of its expression might arise from cotranscription with the upstream genes. Low-resolution S1 mapping and reverse transcription-PCR analysis of S. coelicolor M145 RNA (data not shown) showed that whiA could be transcribed both from a specific promoter within the gene immediately upstream, and from a promoter at least 200 nucleotides (perhaps much more) further upstream.
To characterize both transcripts further, we carried out S1 mapping with a radiolabeled probe of 690 nucleotides. Of this, 470 nucleotides corresponded to a region covering the 3′ end of ORF3 and the 5′ end of whiA (where the whiA-specific promoter had been located) while the remaining 220 nucleotides corresponded to vector sequences, allowing us to differentiate between probe-probe reannealing artifacts, readthrough transcription, and transcription produced from the whiA-specific promoter. Using RNA samples isolated from a time course of surface cultures of the whiA+ strain M145, two signals were detected (Fig. 5), the larger one (ca. 470 nucleotides) being due to readthrough from a promoter located upstream of the region included in the probe and the smaller one (ca. 250 nucleotides) corresponding to the mRNA produced from the whiA-specific promoter. The temporal regulation of these two signals was quite different; the readthrough transcription was rather constant throughout the time course, while the signal corresponding to the whiA-specific promoter was strongly induced when spore formation was taking place and then decreased (Fig. 5). (This decrease might reflect a real decrease in in vivo abundance or might be an artifact of possibly less efficient extraction of RNA from spores than from mycelium.) The transcription of the control gene hrdB was essentially constant during the time course. Even though their exposure time was five times longer, the intensity of the bands corresponding to the longer whiA transcript at all time points was considerably lower than for hrdB. (Although quantitative comparisons between different RNA-DNA hybrids are fallible, we note that similar low band intensity was also obtained with a probe prepared with a different oligonucleotide as the labeled reverse primer, and in all cases the apparent efficiency of incorporation of the 32P end label into the primer had appeared to be comparable.) However, during its developmental up-regulation, the apparent strength of the whiA-specific promoter approached that of hrdB. Since it is likely that the promoter is strongly expressed only at a particular developmental stage in any one hypha and since aerial hyphal development is not synchronized in colonies, the whiA-specific promoter may well be very strong during its transient period of maximum expression.
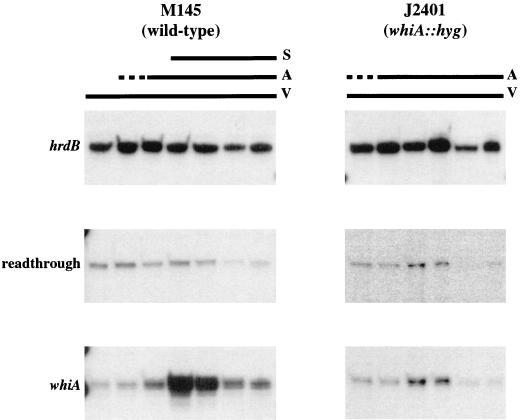
FIG. 5.
The developmental up-regulation of the whiA-specific mRNA is greatly reduced in a whiA mutant. RNA samples were extracted from surface-grown cultures at (left to right) 18, 24, 36, 48, 72, 96, and 120 h (M145) and 24, 36, 48, 72, 96, and 120 h (J2401) and subjected to S1 nuclease protection analysis using radiolabeled probes for the indicated transcripts. The developmental progress of the culture is indicated above the panels (V, vegetative growth; A, aerial mycelium; S, sporulation). Note that the autoradiographs obtained with the whiA probe were exposed for five times as long as the hrdB autoradiographs.
To study the extent to which the presence of WhiA affected the expression of whiA itself, we also analyzed the transcription of whiA in RNA samples from J2401, the M145 derivative with the whiA::hyg disruption. The readthrough transcription detected in the J2401 time course was essentially the same as in the wild-type M145 (Fig. 5). However, the strong induction of whiA mRNA from the whiA-specific promoter detected in the wild type did not occur. Only a weak increase in the level of that transcript was detected in the samples corresponding to 48 and 72 h, suggesting an apparent positive autoregulation, although it is not clear whether it is directly or indirectly mediated by WhiA itself.
Using high-resolution S1 mapping, the transcription start point of whiA was localized to the nucleotide T2133, 187 nucleotides upstream of the more upstream of the putative start codons for whiA (data not shown). The putative −35 and −10 regions contained TGGA and GCCCCTAA sequences separated by 16 nucleotides. These sequences partly resemble those of S. coelicolor promoters dependent on the sporulation-specific sigma factor ςWhiG (the main difference being in the −10 box, which in ςWhiG-dependent promoters has the consensus GCCGATAA, in which the central GA is completely conserved). To further examine this possible dependence, we used purified ςWhiG and E. coli RNA polymerase in an in vitro runoff transcription assay on DNA templates containing the whiA promoter. However, no transcript could be detected (data not shown), although the ςWhiG holoenzyme was active in a positive-control experiment with the whiI promoter (1). Using S1 mapping, we also detected the whiA transcript in RNA samples isolated from a whiG deletion mutant (data not shown), thus ruling out the dependence of whiA on ςWhiG.
An alternative candidate −10 sequence, TAAACT, separated by 5 bp from the transcription start point, is a potential recognition site for RNA polymerase holoenzyme containing the major sigma factor ςHrdB (5). However, no appropriately spaced classical −35-like sequence is present (the sequence TTGGAG, similar to the proposed consensus for the −35 sequence of the ςHrdB-dependent promoters, is, however, located 20 nucleotides upstream). In other bacteria, the absence of a −35-like sequence is often correlated with a requirement for transcriptional activation. If this were the case, the up-regulation of the transcription of whiA from its own promoter could be explained by the action of a transcription factor (perhaps WhiA itself) whose activity and/or concentration could increase specifically during sporulation.
DISCUSSION
Characterization of the whiA sporulation locus in S. coelicolor.
When the S. coelicolor whi mutants defective in sporulation were first isolated and characterized (8, 18), six of the mutations were mapped to the whiA locus, located between the hisC and argA markers, around the “12 o'clock” position in the chromosome. Here we have described the molecular nature of the whiA gene on the basis of a study of a fragment of DNA that complements the whiA mutations in four of the original mutants. (Two other mutants appeared to have accumulated second mutations in other whi genes.) The assignment of whiA to a particular ORF (ORF4) was based on the complementation of whiA mutants by subclones, the finding of different mutations affecting ORF4 in each of the whiA mutants, and the fact that the disruption of ORF4 produced a phenotype similar to that of the original whiA mutants.
In the mycobacterial species (distantly related actinomycetes), ORF1, ORF2, ORF3, and the whiA-like gene are conserved in the same order as in S. coelicolor, and only ORF2, ORF3, and the whiA-like gene are conserved in the other gram-positive organisms except Mycoplasma spp. The conservation of these genes indicates that this arrangement is ancient and significant and suggests that they may be coregulated. The readthrough transcription of whiA from an unknown start point(s)—possibly upstream of the whole gene cluster—may well be important to whiA activity, since the whiA13 mutation that probably uncouples translation with the upstream gene produces a phenotype like that of the whiA disruption mutant (although the whiA13 phenotype probably also arises in part from the altered ribosome-binding site). Neither of the genes immediately upstream of whiA are essential for viability, since they are both deleted in the C73 mutant, which is still able to undergo normal mycelial growth.
Is WhiA a transcriptional regulator?
In the constructed whiA disruption mutant, transcription of whiA from its own promoter has little of the sporulation-specific induction that can be detected in the wild type. At the molecular level, it is not clear whether this induction would involve a direct interaction between WhiA and the whiA-specific promoter or whether WhiA contributes to the activation of an unidentified transcriptional activator or the inactivation of a repressor. The up-regulation of whiA transcription seems to be associated with the morphological checkpoints of sporulation septation or controlled cessation of aerial growth, developmental stages that are not reached in a whiA mutant. The transcriptional arrangement within the operon would indicate that the WhiA protein is present throughout growth, but perhaps in vegetative cells it is in an inactive form that requires a signal before it is able to stimulate its own transcription. The apparent positive autoregulation of whiA is in interesting contrast to the apparent autorepression of whiH and whiI, which make up a ςWhiG-dependent part of the regulatory network for sporulation septation (1, 31). We note that both whiA and whiB (which are expressed independently of ςWhiG) have a low-level constitutive promoter and a stronger developmentally regulated promoter (35).
Although the simplest interpretation of the apparent autoregulation of whiA would cast WhiA as a transcription factor, predictions of secondary structure for WhiA have failed to reveal any similarity to transcriptional regulators or to detect any obvious DNA-binding motifs. These predictions indicate that in the N-terminal part of the protein (residues 1 to 156) there are short regions showing moderate preference for forming α-helices or β-strands, arranged without any apparent periodicity or pattern, some of them being connected by turns containing proline residues. However, the rest of the protein (residues 157 to the end) shows a very high probability of folding into several α-helixes of variable length (10 to 26 residues), some of them also connected by proline-containing loops (Fig. 3).
WhiA may act by controlling the cessation of aerial hyphal growth.
Among the whi mutants, one striking feature of both whiA and whiB mutants is that they produce long and tightly coiled aerial hyphae. The study of the phenotypes of different whi mutants and the analysis of double mutants has given rise to the idea that WhiA and WhiB are responsible for the decision to stop the growth of the aerial hyphae in a coordinated manner prior to sporulation septation (15). Although this might be the consequence simply of WhiA and WhiB accumulation to some critical level, it is also possible that one or both proteins may respond to some aspect of cellular physiology; for example, they could sense the gradual extinction of a compound that is present in limited quantities in the closed-off aerial hypha. (A variant of this would involve WhiA and WhiB detecting a signal that increases in strength when continued growth becomes limited.) The observation that whiA mRNA is maximally abundant when aerial hyphae are beginning to form spores leads us to suggest that the interaction of WhiA with some such signal is necessary for its positive autoregulation.
An additional perspective is suggested by looking at the morphological development of other actinomycetes. Members of the genera Microbispora and Microtetraspora produce short chains of only two and four spores, respectively, suggesting that they have evolved a mechanism by which they are able to strictly limit the number of chromosomal replications in the cell that is destined to form spores. Perhaps Streptomyces has a similar mechanism that is much less obvious because of the difficulty in precisely counting the relatively large number of spores in individual chains. One might imagine a model in which WhiA and WhiB combine to detect a changed level of a signal and then coordinate the completion of a fixed number of further cycles of DNA replication with the associated cessation of cell growth. It might be interesting to examine carefully the DNA phenotypes of stationary-phase cells in single-celled bacteria containing mutations in their whiA-like genes.
ACKNOWLEDGMENTS
N.J.R. and J.A.A. contributed equally to this work.
This work was supported by the Commitment and Development programme of the BBSRC (grant CAD 04380) and by a John Innes Foundation studentship to N.J.R.
We are grateful to Helen M. Kieser for providing filters containing the S. coelicolor cosmids and for providing lyophilized samples of the whi mutants. We acknowledge The Sanger Centre S. coelicolor genome-sequencing team for providing the sequence of cosmid C54 (and all the other cosmids, of course!) and The Institute of Genomic Research for making available the database of the unfinished genomes. We thank Sebastien Mouz, Tobias Kieser, Mervyn Bibb, Mark Buttner, and Gabriella Kelemen for critical reading of the manuscript.
REFERENCES
- 1.Aínsa J A, Parry H D, Chater K F. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolorA3(2) Mol Microbiol. 1999;34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellgard M I, Gojobori T. Identification of a ribonuclease H gene in both Mycoplasma genitalium and Mycoplasma pneumoniaeby a new method for exhaustive identification of ORFs in the complete genome sequences. FEBS Lett. 1999;445:6–8. doi: 10.1016/s0014-5793(99)00075-7. [DOI] [PubMed] [Google Scholar]
- 4.Bibb M J, Findlay P R, Johnson M W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984;30:157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- 5.Bourn W R, Babb B. Computer assisted identification and classification of streptomycete promoters. Nucleic Acids Res. 1995;23:3696–3703. doi: 10.1093/nar/23.18.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruton C J, Chater K F. Nucleotide sequence of IS110, an insertion sequence of Streptomyces coelicolorA3(2) Nucleic Acids Res. 1987;15:7053–7065. doi: 10.1093/nar/15.17.7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chater K F, Bruton C J, King A A, Suárez J E. The expression of Streptomyces and Escherichia coli drug resistance determinants cloned into the Streptomycesphage ΦC31. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 8.Chater K F. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1972;72:9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- 9.Chater K F. Construction and phenotypes of double sporulation deficient mutants in Streptomyces coelicolorA3(2) J Gen Microbiol. 1975;87:312–325. doi: 10.1099/00221287-87-2-312. [DOI] [PubMed] [Google Scholar]
- 10.Chater K F. Taking a genetic scalpel to the Streptomycescolony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- 11.Chater K F, Bruton C J, Plaskitt K A, Buttner M J, Méndez C, Helmann J. The developmental fate of S. coelicolor hyphae depends crucially on a gene product homologous with the motility sigma factor of B. subtilis. Cell. 1989;59:133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- 12.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry III C E, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail M A, Rajandream M-A, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston J E, Taylor K, Whitehead S, Barrell B G. Deciphering the biology of Mycobacterium tuberculosisfrom the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 13.Davis N K, Chater K F. The Streptomyces coelicolor whiBgene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol Gen Genet. 1992;232:351–358. doi: 10.1007/BF00266237. [DOI] [PubMed] [Google Scholar]
- 14.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flärdh K, Findlay K C, Chater K F. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolorA3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- 16.Hilleman D, Puhler A, Wohlleben W. Gene disruption and gene replacement in Streptomycesvia single stranded DNA transformation of integration vectors. Nucleic Acids Res. 1991;19:727–731. doi: 10.1093/nar/19.4.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: The John Innes Foundation; 1985. [Google Scholar]
- 18.Hopwood D A, Wildermuth H, Palmer H M. Mutants of Streptomyces coelicolordefective in sporulation. J Gen Microbiol. 1970;61:397–408. doi: 10.1099/00221287-61-3-397. [DOI] [PubMed] [Google Scholar]
- 19.Janssen G R, Bibb M J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia colicolonies. Gene. 1993;124:133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- 20.Kelemen G H, Brian P, Flärdh K, Chamberlin L, Chater K F, Buttner M J. Developmental regulation of the transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolorA3(2) J Bacteriol. 1998;180:2515–2521. doi: 10.1128/jb.180.9.2515-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelemen G H, Brown G L, Kormanec J, Potúčková L, Chater K F, Buttner M J. The positions of the sigma factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolorA3(2) Mol Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 22.Kieser H M, Kieser T, Hopwood D A. A combined genetic and physical map of the chromosome of Streptomyces coelicolorA3(2) J Bacteriol. 1992;174:5496–5507. doi: 10.1128/jb.174.17.5496-5507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Analysis of Streptomyces avermitilisgenes required for avermectin biosynthesis utilising a novel integrative vector. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 24.McCormick J R, Su E P, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol Microbiol. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 25.McCormick J R, Losick R. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolorA3(2) J Bacteriol. 1996;178:5295–5301. doi: 10.1128/jb.178.17.5295-5301.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McVittie A M. Ultrastructural studies on sporulation in wild-type and white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1974;81:291–302. doi: 10.1099/00221287-81-2-291. [DOI] [PubMed] [Google Scholar]
- 27.Molle V, Palframan W J, Findlay K C, Buttner M J. WhiD and WhiB: homologous proteins required for different stages of sporulation in Streptomyces coelicolorA3(2) J Bacteriol. 2000;182:1286–1295. doi: 10.1128/jb.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Potúčková L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. A new RNA polymerase sigma factor, ςF, is required for the late stages of morphological differentiation in Streptomycesspp. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 29.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolorA3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 30.Ryding N J, Bibb M J, Molle V, Findlay K C, Chater K F, Buttner M J. New sporulation loci in Streptomyces coelicolorA3(2) J Bacteriol. 1999;181:5419–5425. doi: 10.1128/jb.181.17.5419-5425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ryding N J, Kelemen G H, Whatling C A, Flärdh K, Buttner M J, Chater K F. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolorA3(2) Mol Microbiol. 1998;29:343–357. doi: 10.1046/j.1365-2958.1998.00939.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schwedock J, McCormick J R, Angert E R, Nodwell J R, Losick R. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol Microbiol. 1997;25:847–858. doi: 10.1111/j.1365-2958.1997.mmi507.x. [DOI] [PubMed] [Google Scholar]
- 34.Soliveri J A, Gomez J, Bishai W R, Chater K F. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomycesand other actinomycetes. Microbiology. 2000;146:333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- 35.Soliveri J, Brown K L, Buttner M J, Chater K F. Two promoters for the whiB sporulation gene of Streptomyces coelicolorA3(2) and their activities in relation to development. J Bacteriol. 1992;174:6215–6220. doi: 10.1128/jb.174.19.6215-6220.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stultz C M, Nambudripad R, Lathrop R H, White J V. Predicting protein structure with probabilistic models. In: Allewell N, Woodward C, editors. Protein structural biology in bio-medical research. Advances in molecular and cell biology. Vol. 22. Greenwich, Conn: JAI Press; 1997. [Google Scholar]
- 37.Tan H, Yang H, Tian Y, Wu W, Whatling C A, Chamberlin L C, Buttner M J, Nodwell J, Chater K F. The Streptomyces coelicolor sporulation-specific ςWhiGform of RNA polymerase transcribes a gene encoding a ProX-like protein that is dispensable for sporulation. Gene. 1998;212:137–146. doi: 10.1016/s0378-1119(98)00152-8. [DOI] [PubMed] [Google Scholar]
- 38.Wildermuth H, Hopwood D A. Septation during sporulation in Streptomyces coelicolor. J Gen Microbiol. 1970;60:57–59. doi: 10.1099/00221287-60-1-51. [DOI] [PubMed] [Google Scholar]
- 39.Wright F, Bibb M J. Codon usage in the G+C-rich Streptomycesgenome. Gene. 1992;113:55–65. doi: 10.1016/0378-1119(92)90669-g. [DOI] [PubMed] [Google Scholar]
- 40.Zalacaín M, González A, Guerrero M C, Mattaliano R J, Malpartida F, Jiménez A. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 1986;14:1565–1581. doi: 10.1093/nar/14.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]