Abstract
A pyrazinamidase (PZase)-deficient pncA mutant of Mycobacterium tuberculosis, constructed by allelic exchange, was used to investigate the effects of heterologous amidase gene expression on the susceptibility of this organism to pyrazinamide (PZA) and related amides. The mutant was highly resistant to PZA (MIC, >2,000 μg/ml), in accordance with the well-established role of pncA in the PZA susceptibility of M. tuberculosis (A. Scorpio and Y. Zhang, Nat. Med. 2:662–667, 1996). Integration of the pzaA gene encoding the major PZase/nicotinamidase from Mycobacterium smegmatis (H. I. M. Boshoff and V. Mizrahi, J. Bacteriol. 180:5809–5814, 1998) or the M. tuberculosis pncA gene into the pncA mutant complemented its PZase/nicotinamidase defect. In both pzaA- and pncA-complemented mutant strains, the PZase activity was detected exclusively in the cytoplasm, suggesting an intracellular localization for PzaA and PncA. The pzaA-complemented strain was hypersensitive to PZA (MIC, ≤10 μg/ml) and nicotinamide (MIC, ≥20 μg/ml) and was also sensitive to benzamide (MIC, 20 μg/ml), unlike the wild-type and pncA-complemented mutant strains, which were highly resistant to this amide (MIC, >500 μg/ml). This finding was consistent with the observation that benzamide is hydrolyzed by PzaA but not by PncA. Overexpression of PzaA also conferred sensitivity to PZA, nicotinamide, and benzamide on M. smegmatis (MIC, 150 μg/ml in all cases) and rendered Escherichia coli hypersensitive for growth at low pH.
The major human pathogen Mycobacterium tuberculosis is responsible for approximately 2 million deaths and 8 million new cases of tuberculosis per annum (7). Pyrazinamide (PZA) is one of the first-line drugs used in the recommended treatment of tuberculosis (36). This drug plays a key role in shortening the duration of chemotherapy by virtue of its ability to kill semidormant tubercle bacilli located in an acidic environment which are resistant to the bactericidal effects of other drugs (13). In a seminal study, Scorpio and Zhang demonstrated that PZA itself is a prodrug, which requires hydrolysis by the pncA-encoded pyrazinamidase (PZase) to its active metabolite, pyrazinoic acid (30). The critical role played by pncA in the mode of action of PZA is underscored by the fact that the overwhelming majority of PZA-resistant clinical isolates of M. tuberculosis contain mutations in this gene (4, 23). In subsequent studies on the inhibitory mechanisms of pyrazinoic acid, Zhang et al. showed that pyrazinoic acid accumulates under conditions of acidic external pH (39), accounting for why PZA is active against M. tuberculosis in vitro only under conditions of acidic pH (20). This group also showed that mycobacterial species differed considerably in their ability to expel pyrazinoic acid by active efflux (39). Therefore, although M. avium has an active PZase, its resistance to PZA is probably due to effective pyrazinoic acid efflux (34). Similarly, the resistance of M. kansasii to PZA could be ascribed to a combination of its low PZase and weak pyrazinoic acid efflux activities (35), whereas that of M. smegmatis, which possesses two PZases, the highly active PzaA (2) and PncA (11), is due to a highly active efflux mechanism (39). In summary, the susceptibility of a mycobacterium to PZA under acidic conditions thus appears to be determined by the relative contributions of its PZase and pyrazinoic acid efflux activities.
Pyrazinoic acid has been postulated to have both a specific activity against M. tuberculosis and a nonspecific activity due to the effect that accumulation of this membrane-permeant weak acid would have on intracellular pH (12). We previously characterized the major PZase/nicotinamidase of M. smegmatis and showed that overexpression of this amidase, which is structurally unrelated to PncA, rendered M. smegmatis sensitive to PZA and nicotinamide (2). To further explore the relationship between PZase/nicotinamidase activity and PZA susceptibility, we investigated the effects of PzaA expression on the susceptibility of M. tuberculosis, M. smegmatis, and Escherichia coli to PZA and related compounds, and in this paper we report that the PzaA protein confers hypersensitivity to PZA, nicotinamide, and benzamide on these organisms under acidic conditions. The results presented herein suggest that the weakly acidic hydrolysis products of these amides confer similar inhibitory effects on these organisms.
MATERIALS AND METHODS
Materials, bacterial strains, media, and growth conditions.
Restriction enzymes were from Roche Molecular Biochemicals or New England Biolabs, [14C]carbonyl nicotinamide (53 mCi/mmol) was from Sigma, [α-32P]dCTP was from Amersham, and PZA, pyrazinoic acid, nicotinamide, and benzamide were from Sigma. The BM Chemiluminescence Western blotting kit was from Roche Molecular Biochemicals. The bacterial strains and plasmids used in this study are shown in Table 1. M. smegmatis was grown in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glycerol, 0.085% NaCl, 0.2% glucose, and 0.05% Tween 80 and on Middlebrook 7H10 medium supplemented with 0.085% NaCl and 0.2% glucose as the solid medium. M. tuberculosis was grown in Middlebrook 7H9 broth supplemented with 0.2% glycerol, 0.05% Tween 80, and OADC supplement (Merck) and on Middlebrook 7H10 supplemented with OADC as the solid medium (16). E. coli strains were grown in Luria-Bertani broth or agar. E. coli DH5α and GM161 were used for plasmid manipulations (29), and E. coli BL21(DE3)/pLysS and E. coli TB-1 were used for protein expression. Kanamycin and hygromycin were used at 50 and 200 μg/ml, respectively, for E. coli strains and at 20 and 50 μg/ml, respectively, for mycobacterial strains. Ampicillin and chloramphenicol were used at 100 and 34 μg/ml, respectively, for E. coli.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| M. smegmatis | ||
| mc2155 | High-frequency transformation mutant of M. smegmatis ATCC 607 | 32 |
| pzaA::aph | pzaA knockout mutant of mc2155; Kmr | 2 |
| M. tuberculosis | ||
| H37Rv | Laboratory strain (ATCC 27294) | Laboratory collection |
| pncA::hyg | pncA knockout mutant of H37Rv; Hygr | This work |
| Plasmids | ||
| pGEM3Z(+)f | E. coli cloning vector; Apr | Promega |
| pET15b | E. coli expression vector; Apr | Novagen |
| pMAL-c2 | E. coli expression vector; Apr | New England Biolabs |
| pHINT | E. coli-Mycobacterium integrating shuttle vector; Hygr Apr | 21 |
| pAINT | pHINT derivative with hyg replaced by aph; Kmr Apr | This work |
| pOLYG | E. coli-mycobacterium replicating shuttle vector; Hygr oriM | 21 |
| Pac4 | Cosmid from pYUB328::H37Rv library carrying the pncA gene | This work |
| pGncA | pGEM3Z(+)f derivative carrying pncA on a 4.67-kb Asp718 fragment | This work |
| pGncAh | pGncA derivative carrying pncA insertionally inactivated by hyg (pncA::hyg) | This work |
| pGncAKO | pGncAh derivative carrying hsp60-sacB | This work |
| pHncA | pHINT derivative carrying pncA on a PstI fragment | This work |
| pOncA | pOLYG derivative carrying pncA on a PstI fragment | This work |
| pOppncA | pOLYG derivative carrying pncA with consensus ribosome binding site under control of the pzaA promoter | This work |
| pTncA | pET15b derivative carrying pncA | 2 |
| pAIncA | pAINT derivative carrying pncA on a 4.67-kb Asp718 fragment | This work |
| pMncA | pMAL-c2 derivative carrying malE-pncA translational fusion | This work |
| pGam | pGEM3Z(+)f derivative carrying pzaA with upstream sequence | 2 |
| pOam | pOLYG derivative carrying pzaA with upstream sequence | 2 |
| pAIam | pAINT derivative carrying pzaA with upstream sequence | This work |
| pTam | pET15b derivative carrying pzaA | 2 |
Amide susceptibility testing.
The MIC of PZA is pH dependent (27). To maximize the sensitivity of the susceptibility tests, the lowest pH value at which visible growth of the various strains occurred was therefore used as the test pH. Since a pH of 5.5 proved to be too low to support the growth of M. tuberculosis H37Rv, susceptibility testing was carried out at a pH of 5.8. For M. smegmatis, a pH of 5.1 was used, under which conditions visible colonies were formed in 5 to 6 days on solid medium (compared with 2 to 3 days for colony formation at pH 6.6). By analogy, a pH of 5.1 was used for susceptibility testing of E. coli strains. PZA, nicotinamide, and benzamide susceptibility tests for M. tuberculosis were performed in Middlebrook 7H9 broth supplemented with 0.3 g of citric acid per liter, with the pH adjusted to 5.8, and subsequently supplemented with glycerol, OADC, and 0.05% Tween 80. Amide stocks (0.1 M) were added to give the required final concentration. Log-phase cultures of M. tuberculosis (optical density at 600 nm ≈ 0.9) were diluted 10-fold in this medium. A 50-μl aliquot of diluted cells was then inoculated in 3 ml of drug-containing medium, and the culture was incubated at 37°C for 2 weeks. The MIC was defined as the lowest drug concentration at which no turbidity was visible after 2 weeks. Susceptibility tests were also performed in unmodified Middlebrook 7H9 broth (pH 6.6) supplemented with OADC and Tween 80, as above. Susceptibility to PZA and benzamide was confirmed radiometrically (5) using the BACTEC PZA test kit (Becton Dickinson). M. tuberculosis strains, subcultured in BACTEC 12B medium as specified by the manufacturer, were grown to a growth index of 150 for use in the inoculation of the BACTEC susceptibility test media. Susceptibility was determined as specified by the manufacturer but with the following minor modification: PZA and benzamide were dissolved in reconstituting fluid (Becton Dickinson) and filter sterilized, and 0.2 ml of each stock was added to the modified Middlebrook 12B medium to give final concentrations of 0, 50, 100, 200, 400, and 600 μg of PZA per ml and 0, 50, 100, and 200 μg of benzamide per ml. M. smegmatis susceptibility tests were performed as previously described (2). The drug susceptibility of E. coli BL21(DE3)/pLysS carrying pET15b-derived plasmids was determined by plating on M9 minimal medium, adjusted to pH 5.1, containing chloramphenicol, ampicillin, isopropyl-β-d-galactopyranoside (IPTG; 1 mM), and the appropriate amide.
Construction of a pncA::hyg knockout mutant of M. tuberculosis.
A cosmid (Pac4) containing pncA was identified from a library of M. tuberculosis (pYUB328::H37Rv) (1) using a PCR-amplified M. tuberculosis pncA probe (2). A 4.67-kb Asp718 fragment containing pncA was cloned in pGEM3Z(+)f to yield pGncA. The hyg resistance cassette was excised from pIJ963 (19) as a BamHI-BglII fragment and was cloned in the BclI site contained within the pncA gene to create pGncAh. A cassette carrying the Bacillus subtilis sacB gene under the control of the mycobacterial hsp60 (groEL) promoter (B. G. Gordhan, S. Quan, and V. Mizrahi, unpublished data) was cloned into the XbaI site of pGncAh to create pGncAKO. M. tuberculosis was electroporated with 1 μg of UV-treated pGncAKO as described by Gordhan and Parish (10), and the cells were plated on 7H10 medium containing hygromycin and 5% sucrose. Chromosomal DNA was extracted from Hygr Sucr clones and analyzed by Southern blotting as previously described (2, 9), using 32P-labeled probes generated using a random-prime labeling kit (Roche Molecular Biochemicals).
Construction of PncA and PzaA expression vectors.
A 3.39-kb PstI fragment from Pac4 containing the M. tuberculosis pncA gene and its promoter was cloned into pHINT to create pHncA and into pOLYG to create pOncA. The pncA gene was also cloned under the control of the stronger, M. smegmatis pzaA promoter and a consensus ribosome binding site, as follows. The pzaA promoter was excised as a 340-bp PstI fragment from pGam (2), cloned in pOLYG, and excised as a HindIII-XbaI fragment. This fragment was then ligated to the XbaI-BamHI fragment containing the pncA gene with its ribosome binding site (9, 22) from the pET15b derivative, pTncA (2), and cloned in HindIII-BamHI-digested pOLYG to form pOppncA. The integrating vector, pAINT, was derived from pHINT by excising the hyg gene by digestion with SmaI and SacI and replacing it with the Tn903-derived aph gene (15). The pncA-containing 4.67-kb Asp718 fragment from Pac4 was cloned in pAINT to form pAIncA, and the pzaA-containing HindIII-XbaI fragment from pGam was also cloned in this vector to produce pAIam. Replicating and integrating plasmids (100 ng) were electroporated into M. smegmatis and M. tuberculosis as previously described (9, 22). The vector pMncA, which directs the overexpression of a recombinant form of PncA fused to the maltose binding protein (MBP), was constructed as follows. The vector pTncA was digested with NcoI (which cuts at the start codon of the pncA gene in the plasmid) and was blunt ended before the pncA gene was excised on a BamHI fragment. This fragment was cloned in pMAL-c2 that had been digested with EcoRI, blunt ended, and subsequently restricted with BamHI, to yield pMncA, which contains an in-frame translational fusion between the MBP-encoding malE gene and pncA. The MBP::PncA fusion protein was induced by IPTG treatment of E. coli TB-1 cells carrying pMncA and was purified on amylose resin as specified by the manufacturer (New England Biolabs).
Enzyme assays.
Lysates of M. smegmatis were prepared as previously described (6). Lysates of M. tuberculosis were prepared by resuspending cell pellets in buffer A (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM dithiothreitol) and homogenizing them with glass beads in a Mini bead beater (Biospec Products, Bartlesville, Okla.) three times for 1 min each at maximum speed, with cooling on ice between pulses. Lysates were clarified by centrifugation at 13,000 × g for 5 min. Secreted M. tuberculosis proteins were prepared from supernatants of mid-log cultures by harvesting cells at 5,000 × g for 10 min and sterilization of the spent growth medium by filtration through 0.2-μm-pore-size filters. Proteins contained in the sterilized filtrate were concentrated either by centrifugation through a 30-kDa Ultrafree MC filter unit (Millipore) or by precipitation with 6 volumes of ice-cold ethanol, followed by centrifugation at 17,000 × g for 1 h and resuspension in buffer A. Amidase activities were determined using the following assays. (i) The presence of PZase activity in bacterial strains was determined using a spectrophotometric assay following ferrous ammonium sulfate addition, as previously described (2). (ii) PZase and nicotinamidase specific activities were assayed by high-pressure liquid chromatography (HPLC) using the following method, which is based on that described by Yan and Sloan (37). Enzyme reaction mixtures were incubated at 37°C in 20 mM PZA or nicotinamide in 100 mM sodium phosphate buffer at pH 7.5 in a total volume of 200 μl containing 60 to 240 μg of protein from cell lysates prepared from log-phase cultures. After incubation of the mixture for a sufficient period to result in 0 to 10% substrate conversion, the reaction was terminated by the addition of 20 μl of 80% (wt/vol) trichloroacetic acid. Precipitates were removed by centrifugation (13,000 × g for 10 min), and 120 μl of the reaction mixture was diluted in 400 μl of 100 mM sodium phosphate buffer. Samples were filtered through 0.45-μm-pore-size filters, and 20-μl aliquots were separated on a Phenomenex Luna C18 column (150 by 4.6 mm) with 10% methanol elution buffer using a SpectraSystem P4000 pump (Thermo Separation Products). Substrates and products were detected using a SpectraSystem UV3000 detector set at 254 and 280 nm. At a flow rate of 1 ml/min, pyrazinoic acid eluted at 3.31 min, PZA eluted at 4.15 min, nicotinic acid eluted at 2.68 min, and nicotinamide eluted at 2.95 min. Peaks were integrated using the PC1000 system software for LC (version 3.5; Thermo Separation Products). Each mycobacterial strain was tested in at least three independent experiments, using a minimum of five time points during the enzyme reaction. (iii) Nicotinamidase activity was additionally determined using the following radiochemical assay. Either 10 μl of cell lysate (containing 4 mg of protein per ml) or 20 μl of concentrated culture filtrate in buffer A was added to 2 μl of a solution containing 5 mM [14C]nicotinamide at a specific activity of 53 mCi/mmol. The reaction mixtures were incubated at 37°C for 0, 10, 75, or 140 min before the reactions were quenched by the addition of 2 μl of acetic acid (2 M). The total assay mixture was then spotted onto a silica gel thin-layer chromatography (TLC) plate (Aldrich) containing unlabeled nicotinic acid and nicotinamide as internal standards. The plates were developed in a mixture of 1-butanol, NH3, and water (8:1:1), and spots corresponding to the internal standards were visualized under UV light, excised, and counted in toluene-based liquid scintillation fluid in a Beckman LS6000IS scintillation counter. (iv) An analogous, nonradioactive assay was used to monitor the benzamidase activities of whole-cell suspensions or cell lysates in 10- to 20-μl assay mixtures containing 10 mM benzamide. The reaction products were spotted on TLC plates, which were developed and visualized for benzamide or benzoic acid under UV light, as described above. Protein concentrations were determined by a Bradford assay (Bio-Rad assay kit II).
Intracellular localization assays.
PzaA-specific antiserum was prepared by purifying PzaA from inclusion bodies in IPTG-induced E. coli BL21(DE3)/pLysS carrying pTam (2) by a combination of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the inclusion bodies and electroelution of the protein from gel slices using a BioTrap BT1000 device (Schleicher & Schuell). A New Zealand White rabbit was immunized subcutaneously with 200 μg of protein in 1 ml of phosphate-buffered saline at weekly intervals over a 6-week period before antiserum was prepared. Purified MBP::PncA was used for the production of PncA antiserum by immunization of a New Zealand White rabbit, as described above. M. tuberculosis expresses very low levels of PncA, in contrast to the higher levels detected in M. smegmatis recombinants carrying the various PncA expression vectors. To enrich for PncA-specific antibodies, serum was incubated with strips of polyvinylidene difluoride membrane with bound PncA [obtained from IPTG-induced BL21(DE3)/pLysS(pTncA) cell lysates that had been fractionated by SDS-PAGE and transferred to polyvinylidene difluoride], which had previously been treated with blocking solution (Western blotting kit). The strips were washed extensively in Tris-buffered saline prior to elution of antibody by incubation in 0.2 M glycine (pH 2.8)–1 mM EDTA for 20 min. Eluted antibody was neutralized with 1 M Tris-HCl and stabilized in 1% blocking solution, and precipitates were removed by centrifugation. Cell lysates and culture supernatants were prepared as described above, precipitated in 80% acetone, and denatured in SDS-PAGE loading buffer. For cell lysates, approximately 20 μg of total protein was loaded per lane of the gel. Western blotting was performed with a Western blotting chemiluminescence kit as specified by the manufacturer.
RESULTS
Construction and characterization of a pncA::hyg knockout mutant of M. tuberculosis.
To investigate the effects of M. smegmatis pzaA expression on the PZA susceptibility of M. tuberculosis, we constructed a PZase-deficient mutant of M. tuberculosis by targeted knockout of its PZase/nicotinamidase-encoding pncA gene (30), for use as a host strain. The insertionally inactivated pncA::hyg allele, in which the hyg marker was flanked on either side by 1,733 bp (5′) and 2,493 bp (3′) of homologous DNA, was cloned in a suicide plasmid carrying a sacB counterselection marker. Plasmid DNA, pretreated by UV irradiation (10, 14), was electroporated into M. tuberculosis, which was directly plated on medium containing hygromycin and sucrose. Of 19 colonies genotypically screened by Southern blot analysis, 8 corresponded to allelic exchange mutants, 8 were site-specific single-crossover recombinants that were spontaneously Sucr, and 3 were spontaneous Hygr mutants (Fig. 1). As predicted from the well-established role of the pncA gene in M. tuberculosis (30), the pncA::hyg mutant was highly resistant to PZA (MIC, >2,000 μg/ml [Table 2]), and it lacked PncA protein (Fig. 2A, lane 2, and data not shown) and PZase activity in both cell lysate and culture supernatant fractions (Table 2 and data not shown).
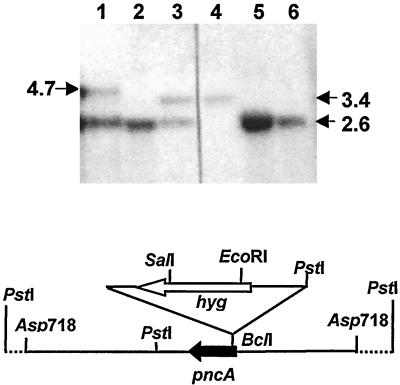
FIG. 1.
Construction and genotypic characterization of the M. tuberculosis pncA::hyg mutant and its pncA-complemented derivative. The restriction map of the pncA locus, illustrating the pncA::hyg allele carried on the Asp718 fragment of the allelic exchange substrate, pGncAKO, is shown beneath the Southern blot. Genomic DNA was digested with PstI, and Southern blots were probed with a PCR-generated pncA probe (2). The expected sizes of the wild-type and inactivated PstI alleles are 3.39 and 2.65 kb, respectively. Lanes: 1, pncA::hyg mutant complemented with pAIncA (showing the mutant allele and the additional 4.7-kb allele arising from the integrated copy of pncA); 2, 5, and 6, independent isolates of the pncA::hyg mutant; 3, product of homologous recombination by single crossover (showing both wild-type and mutant alleles); 4, wild-type H37Rv.
TABLE 2.
Correlation between amidase activity and amide drug susceptibility of strains of M. smegmatis, M. tuberculosis, and E. coli
| Strain | Amidase activity
|
MIC (μg/ml)a
|
||||
|---|---|---|---|---|---|---|
| PZA (U/mg)b | Nicotinamide (U/mg)b | Bentamide (+/−)c | PZA | Nicotinamide | Benzamide | |
| M. smegmatis | ||||||
| mc2155 | 0.210 ± 0.016 | 0.530 ± 0.040 | + | ≥2,200 | ≥2,200 | ≥500 |
| mc2155(pOam) | 6.47 ± 0.30 | 15.65 ± 0.51 | +++ | 150 | 150 | 150 |
| mc2155(pOppncA) | ≥2,200 | ≥2,200 | ≥500 | |||
| pzaA::aph | 0.0201 ± 0.0006 | 0.0220 ± 0.009 | − | ≥2,200 | ≥2,200 | ≥500 |
| pzaA::aph(pOncA) | 0.0690 ± 0.008 | 0.0961 ± 0.0154 | ≥2,200 | ≥2,200 | ≥500 | |
| pzaA::aph(pOppncA) | 0.551 ± 0.025 | 0.702 ± 0.033 | ≥2,200 | ≥2,200 | ≥500 | |
| M. tuberculosis | ||||||
| H37Rv | 0.0021 ± 0.0004 | 0.0042 ± 0.0005 | − | 20 | 20 | ≥500 |
| pncA::hyg | 0 | 0 | − | ≥2,000 | ≥2,000 | ≥500 |
| [pncA::hyg]::pAIncA | 0.0022 ± 0.0001 | 0.0056 ± 0.0002 | − | 20 | 20 | ≥500 |
| [pncA::hyg]::pAIam | 0.112 ± 0.0060 | 0.289 ± 0.010 | + | ≤10 | ≥20 | 20 |
| H37Rv(pOam) | 1.84 ± 0.16 | 7.1 ± 0.2 | NDe | |||
| E. colid | ||||||
| BL21(DE3)/pLysS | + | + | ≥2,000 | ≥2,000 | ≥500 | |
| BL21(DE3)/pLysS(pTncA) | +++ | +++ | − | ≤200 | ≤200 | ≥500 |
| BL21(DE3)/pLysS(pTam) | +++ | +++ | +++ | ND | ||
For M. smegmatis strains, the MIC is defined as the concentration of drug that causes ≥90% growth inhibition (measured by colony counts), since a defined cutoff value was obtained at this point. A 100% growth inhibition of M. smegmatis strains was not obtained, since at higher amide concentrations, the decrease in colony counts was dependent on the initial number of cells plated. For M. tuberculosis strains, the MIC is defined as the concentration of drug at which no visible growth occurs in tests performed at pH 5.8 and/or the concentration that causes >90% reduction in growth index relative to the drug-free control in the BACTEC test. For E. coli strains, the MIC is defined as the drug concentration causing a >99% growth inhibition (measured by colony counts).
Specific activity calculated from activity data obtained using the HPLC amidase assay and using the previously defined unit (2). The results represent the mean and standard deviation from at least three independent experiments.
+++, high activity; +, detectable activity; −, undetectable activity (as determined by the TLC benzamidase assay and the previously described PZase and nicotinamidase assays) (2).
Since the specific amidase activity in E. coli recombinants carrying pTncA and pTam is dependent on the duration of IPTG induction of gene expression, only a semiquantitative assessment of activity is shown in these cases (see footnote c for a key).
The pH sensitivity for growth of this strain precluded determination of the MIC of PZA.
FIG. 2.
Determination of the cellular localization of PncA and PzaA in mycobacteria by Western blot analysis. Western blotting was carried out as described in Materials and Methods. The locations of PncA and PzaA are indicated by arrowheads, and the sizes are given in kilodaltons. (A) Detection of PncA in M. tuberculosis strains. Lanes: 1, cell lysate from wild-type H37Rv; 2, cell lysate from the pncA::hyg mutant; 3, cell lysate from the pncA-complemented mutant, pncA::hyg::pAIncA; 4, culture filtrate protein precipitate from wild-type H37Rv; 5, culture filtrate protein precipitate from pncA::hyg::pAIncA. (B) Detection of PncA in M. smegmatis strains. Lanes: 1, cell lysate from mc2155(pOncA); 2, cell lysate from wild-type mc2155; 3, cell lysate from mc2155::pHncA; 4, cell lysate from mc2155(pOppncA); 5, culture filtrate protein precipitate from mc2155::pHncA. (C) Detection of PzaA in M. tuberculosis and M. smegmatis strains. Lanes: 1, cell lysate from wild-type mc2155; 2, cell lysate from the pzaA::aph mutant; 3, cell lysate from wild-type H37Rv; 4 and 5, cell lysate from the pzaA-complemented mutant, pncA::hyg::pAIam; 6, culture filtrate protein precipitate from the pncA::hyg::pAIam mutant; 7, culture filtrate protein precipitate from wild-type mc2155.
PZase and nicotinamidase specific activities were determined by HPLC analysis of enzyme reaction products. The PZase specific activities determined by the HPLC-based method were quantitatively different from those determined by the previously described spectrophotometric assay (2). The difference was most pronounced for M. tuberculosis, which expresses a very low PZase activity, whose absolute value appears to have been overestimated using the spectrophotometric assay (0.02 U/mg [2]). The sensitivity of pyrazinoic acid detection by this assay is limited by the high background caused by the gradual time- and buffer-dependent oxidation of the ferrous metal ion, which occurs following addition of the ferrous ammonium sulfate. In contrast, the HPLC assay provided a sensitive alternative method for specific amidase activity determination because it allowed the direct quantitation of separated substrate and product. HPLC analysis revealed very low but detectable PZase and nicotinamidase activities in the parental M. tuberculosis H37Rv strain, whereas no pyrazinoic or nicotinic acid conversion could be detected in cell lysates prepared from the pncA::hyg mutant strain over a 120-min incubation with 240 μg of total protein. These results were independently corroborated by determination of nicotinamidase activity analysis using [14C]nicotinamide as a substrate, which revealed that there was no detectable conversion to nicotinic acid by a cell lysate prepared from the mutant strain under conditions resulting in ≥90% hydrolysis by a lysate prepared from the wild-type strain (75-min incubation with 40 μg of total protein). As expected, integration of a functional copy of pncA at the attB locus of the mutant (Fig. 1, lane 1) restored the phenotype of the mutant to that of the wild type in terms of PZA susceptibility, level and cellular localization of PncA protein (Fig. 2A, lanes 3 and 5, and data not shown), and PZase and nicotinamidase activity (Table 2).
Expression of M. smegmatis pzaA in M. tuberculosis confers hypersensitivity to PZA and related aromatic amides.
Integration of a single copy of the M. smegmatis pzaA gene in the pncA::hyg mutant resulted in an increase in both the specific activity of PZase and the PZA sensitivity to levels higher than those conferred by integration of the M. tuberculosis pncA gene at the same (attB) locus (Table 2). This result is in agreement with the recently demonstrated ability of PzaA to confer PZA susceptibility on M. bovis BCG (11). An inverse correlation was observed between the PZase activity and the MIC of PZA. Furthermore, analysis of the cellular localization of the PzaA protein in M. tuberculosis by Western blotting confirmed that, like PncA, this protein localized exclusively in the cytoplasm (Fig. 2C, lanes 4 to 6). The pzaA-complemented mutant was also highly sensitive to both nicotinamide and benzamide in a pH-dependent manner: under acidic (pH 5.8) conditions, the MICs of PZA and benzamide for the pzaA-complemented mutant were ≤10 and 20 μg/ml, respectively, whereas at the normal pH of the growth medium (pH 6.6), the MIC of both amides was 100 μg/ml. In contrast, although pncA complementation restored nicotinamide sensitivity to the pncA::hyg mutant, the complemented strain, like the parental wild type, was completely insensitive to benzamide.
The biochemical basis for this observation was investigated by performing an amidase activity analysis on cell lysates from various strains with benzamide as a substrate (Fig. 3). Whereas strains of E. coli and M. smegmatis expressing PzaA hydrolyzed all three amides (Fig. 3A, lanes 2 to 4 and 10 to 12), those overexpressing PncA did not (lanes 13 to 15), suggesting that PzaA has a broader substrate range than PncA. The pncA-complemented pncA::hyg mutant of M. tuberculosis and the M. smegmatis pzaA::aph mutant did not hydrolyze benzamide (Fig. 3A, lanes 5 to 7; Fig. 3B, lanes 1 and 2). In contrast, wild-type M. smegmatis and the pzaA-complemented pncA::hyg mutant of M. tuberculosis were proficient in benzamidase activity (Fig. 3A, lanes 2 to 4; Fig. 3B, lanes 4 and 5). Although the M. smegmatis pzaA::aph mutant retained residual PZase activity (Fig. 3A, lane 18), which was recently attributed to the presence of a functional PncA in this organism (11), this mutant exhibited no benzamidase activity (lanes 5 to 7). In conjunction, these observations confirmed the restricted substrate specificity of PncA relative to PzaA.
FIG. 3.
Benzamidase and PZase activities of cell lysates expressing PncA or PzaA. (A) Cell lysates from M. smegmatis (lanes 2 to 9, 17 and 18) or IPTG-induced E. coli carrying either pTam or pTncA (lanes 10 to 15) were incubated with the respective amides, and the reaction products were analyzed by TLC, as described in Materials and Methods. Lanes: 1, benzamide (BZA) and benzoic acid (BOA) standards; 2, mc2155 incubated with benzamide for 0 min; 3, mc2155 incubated with benzamide for 10 min; 4, mc2155 incubated with benzamide for 40 min; 5, pzaA::aph incubated with benzamide for 0 min; 6, pzaA::aph incubated with benzamide for 10 min; 7, pzaA::aph incubated with benzamide for 40 min; 8, boiled mc2155 lysate incubated with benzamide for 0 min; 9, boiled mc2155 lysate incubated with benzamide for 40 min; 10, E. coli(pTam) incubated with benzamide for 0 min; 11, E. coli(pTam) incubated with benzamide for 7 min; 12, E. coli(pTam) incubated with benzamide for 20 min; 13, E. coli(pTncA) incubated with benzamide for 0 min; 14, E. coli(pTncA) incubated with benzamide for 7 min; 15, E. coli(pTncA) incubated with benzamide for 20 min; 16, benzoic acid standard; 17, mc2155 incubated with PZA for 20 min; 18, pzaA::aph incubated with PZA for 20 min; 19, pyrazinoic acid (POA) standard; 20, PZA plus pyrazinoic acid standards. (B) Benzamidase activity in cell lysates of M. tuberculosis strains. Lanes: 1, pncA::hyg::pAIncA incubated with benzamide for 30 min; 2, pncA::hyg::pAIncA incubated with benzamide for 150 min; 3, benzamide plus benzoic acid standard; 4, pncA::hyg::pAIam incubated with benzamide for 30 min; 5, pncA::hyg::pAIam incubated with benzamide for 150 min.
The phenotypic effects of heterologous expression of PzaA in M. tuberculosis were significantly exacerbated by overexpression of this amidase from the multicopy plasmid pOam. A transformant carrying this plasmid displayed marked pH sensitivity, as evidenced by its poor growth at pH 5.8 and by the fact that at pH 6.6, the MIC of PZA was 5 μg/ml whereas that of BZA was <20 μg/ml. This observation is consistent with the notion that the PZA susceptibility of M. tuberculosis is limited by its relatively weak PZase and pyrazinoic acid efflux activities.
Overexpression of M. smegmatis PzaA also confers aromatic amide sensitivity on M. smegmatis and E. coli.
We have previously shown that the natural resistance of M. smegmatis to PZA and nicotinamide can be overcome by overexpression of PzaA (2). This initial observation was extended by analyzing the effects of PzaA overexpression from pOam and PncA overexpression from pOncA and pOppncA on the susceptibility of M. smegmatis to these drugs. Western blot analysis was used to monitor the heterologous expression of M. tuberculosis PncA from these various pncA-based plasmids in M. smegmatis (Fig. 2B). This analysis confirmed the intracellular localization of PncA, expressed heterologously from the single-copy vector pHncA (Fig. 2B, lane 3) and from the multicopy vectors pOncA and pOppncA (Fig. 2B, lanes 1 and 4). However, in spite of the obvious overexpression of PncA revealed by Western blot analysis, neither multicopy vector conferred amide susceptibility on M. smegmatis, in accordance with the relatively low specific activity of PZase in strains carrying these plasmids (Table 2). In contrast, the PzaA-overexpressing plasmid pOam conferred a 30-fold increase in the specific activities of PZase and nicotinamidase in M. smegmatis, which resulted in a concomitant increase in the sensitivity of this organism to benzamide, in addition to its previously observed hypersensitivity to PZA and nicotinamide (2). The amide susceptibility study also revealed that M. smegmatis is naturally more susceptible to benzamide than to PZA (Table 2). However, comparison of the benzamide susceptibilities of the wild type and the PZase-deficient pzaA::aph mutant suggested that the susceptibility to benzamide at concentrations above 500 μg/ml was hydrolysis independent.
High-level overexpression of PncA resulted in a marked increase in the susceptibility of E. coli to PZA and nicotinamide but not to benzamide. The sensitivity of E. coli to overproduction of PZase was such that BL21(DE3)/pLysS cells carrying the PzaA-overexpressing plasmid pTam were unable to form colonies at low pH on minimal medium, even in the absence of exogenously added amides. The few colonies that did grow all carried rearranged plasmids, confirming the toxicity at low pH of PzaA overproduction. In conjunction, these observations underscored the close link between aromatic amide susceptibility at an acidic pH and the ability of the organism to hydrolyze the amide to its corresponding acid.
DISCUSSION
In this study, we constructed a PZase-defective pncA mutant of M. tuberculosis as a tool to investigate the effect of heterologous expression of a PZase/nicotinamidase enzyme of higher specific activity and broader substrate specificity than PncA on the sensitivity of M. tuberculosis to aromatic amides, which are structurally related to PZA. The observation of an exclusively intracellular localization for the PncA protein in M. tuberculosis is in agreement with the observations of others (25, 39), but our observation of the lack of nicotinamidase activity in both the intracellular and extracellular fractions of the pncA::hyg mutant differs from that of Raynaud et al. (24, 25), who have reported the existence of extracellular nicotinamidase activity in M. tuberculosis. However, this discrepancy might be due to differences in growth medium, since this factor could affect the expression and secretion of enzymes (38).
M. tuberculosis, M. smegmatis, and E. coli are all rendered hypersensitive to inhibition by benzamide by the heterologous expression of PzaA, which was shown to catalyze the hydrolysis of this amide. The MIC of benzamide for the pzaA-complemented pncA::hyg mutant (20 μg/ml) was comparable to that of PZA (≤10 μg/ml). As neutral molecules, PZA and benzamide are expected to equilibrate rapidly across the membrane of M. tuberculosis. Intracellular PzaA would then hydrolyze the amides to their corresponding, weak acids. Since the pKa values of benzoic acid and pyrazinoic acid are 4.1 and 2.9, respectively, the Henderson-Hasselbach equation predicts that both acids would be ≥99.9% deprotonated in the intracellular environment, which has a pH of 7 (39). The similarity in the MICs of PZA and benzamide implies a similar inhibitory mechanism for these two drugs and suggests that, as previously observed for pyrazinoic acid (39), benzoic acid accumulates in M. tuberculosis under conditions of an acidic external pH.
Inhibition by the intracellular accumulation of pyrazinoic acid, nicotinic acid, and benzoic acid, three aromatic acids with very different structures, makes it unlikely that pyrazinamide (or, more correctly, pyrazinoic acid) acts by binding and inhibiting a specific macromolecular target. The simplest assumption compatible with our results would be that the accumulation of these aromatic anions exerts a nonspecific inhibitory effect on cellular metabolism (27, 29). However, the bacteriostatic effect of these compounds may also involve an additional mechanism. Thus, when PzaA and PncA hydrolyze the parent amides in the cell, ammonia will be produced in addition to the aromatic acids. The ammonia can be assimilated in anabolic metabolism or can diffuse out of the mycobacterial cell, which will result in acidification of the cytoplasm. Most bacterial cells have a mechanism to reverse such cytosolic acidification; for example, in E. coli, intracellular pH homeostasis is maintained at an acidic external pH by its potassium transport systems (31), and in Streptococcus faecalis, the activity of H+-ATPase is critical in the maintenance of neutral cytosolic pH (17). During adaptation to acidic external pH, which tends to acidify the cytoplasm, several proteins are induced, some of which assist in regulating intracellular pH homeostasis (18). These mechanisms may be sufficient to keep the cytosolic pH neutral, even when ammonia is rapidly depleted and/or lost from the cytoplasm. However, there are two possible complications that could hinder the pH homeostasis in M. tuberculosis treated with PZA and other aromatic acid amides. First, even in the absence of these amides, the machinery that reverses cytosolic acidification may already be working close to its capacity in the acidic medium used, because under these conditions the efflux of protons will become difficult and a spontaneous influx of protons from the medium could possibly occur. Second, M. tuberculosis displays a pronounced sensitivity to acidic pH compared to that of other mycobacteria such as M. avium, both in vitro (3, 13, 33) and within the environment provided by the host cell vacuole (8), suggesting that M. tuberculosis may not be very competent in correcting the acidification of the cytoplasm. We therefore think that in addition to the accumulation of aromatic anions, acidification of the cytoplasm, or at least the metabolic consequences of reversing such acidification, may contribute to the inhibitory effects of these aromatic amides. The intracellular pH of M. tuberculosis was shown to remain neutral following a 2-h incubation at pH 5.0 with PZA at 50 μg/ml (39), but the acid response machinery of this organism may be functioning at maximum capacity under such conditions. It will therefore be of interest to examine the intracellular pH of M. tuberculosis treated with higher concentrations of PZA or that of M. tuberculosis expressing the pzaA gene under such conditions.
ACKNOWLEDGMENTS
This work was supported by the Glaxo Wellcome Action TB Initiative. V.M. was also supported by the South African Medical Research Council, the National Research Foundation and the South African Institute for Medical Research.
We thank Jo Michael and Chris van der Westhuizen for assistance with the HPLC assays, Bhavna Gordhan for assistance with targeted gene knockout, André Trollip for assistance with the BACTEC tests, Bill Jacobs for providing the pYUB328::H37Rv library, Ken Duncan and Ruth McAdam for advice and encouragement, and anonymous reviewers for constructive comments.
REFERENCES
- 1.Balasubramanian V, Pavelka M S, Jr, Bardarov S S, Martin J, Weisbrod T R, McAdam R A, Bloom B R, Jacobs W R., Jr Allelic exchange in Mycobacterium tuberculosis with long linear recombination substrates. J Bacteriol. 1996;178:273–279. doi: 10.1128/jb.178.1.273-279.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boshoff H I M, Mizrahi V. Purification, gene cloning, targeted knockout, overexpression and biochemical characterization of the major pyrazinamidase from Mycobacterium smegmatis. J Bacteriol. 1998;180:5809–5814. doi: 10.1128/jb.180.22.5809-5814.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman J S, Bernard J S. The pH tolerance of unclassified mycobacteria. Am Rev Respir Dis. 1962;86:582–583. doi: 10.1164/arrd.1962.86.4.582. [DOI] [PubMed] [Google Scholar]
- 4.Cheng S J, Thibert L, Sanchez T, Heifets L, Zhang Y. pncA mutations as a major mechanism of pyrazinmide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob Agents Chemother. 2000;44:528–532. doi: 10.1128/aac.44.3.528-532.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutler R R, Wilson P, Villaroel J, Clarke V. Evaluating current methods for determination of the susceptibility of mycobacteria to pyrazinmide, conventional, radiometric Bactec and two methods of pyrazinamidase testing. Lett Appl Microbiol. 1997;24:127–132. doi: 10.1046/j.1472-765x.1997.00367.x. [DOI] [PubMed] [Google Scholar]
- 6.Durbach S I, Andersen S J, Mizrahi V. SOS induction in mycobacteria: analysis of the DNA-binding activity of a LexA-like repressor and its role in DNA damage induction of the recA gene from Mycobacterium smegmatis. Mol Microbiol. 1997;26:643–653. doi: 10.1046/j.1365-2958.1997.5731934.x. [DOI] [PubMed] [Google Scholar]
- 7.Dye C, Scheele S, Dolin P, Pathania V, Raviglione M C. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA. 1999;282:677–686. doi: 10.1001/jama.282.7.677. [DOI] [PubMed] [Google Scholar]
- 8.Gomes M S, Paul S, Moreira A L, Appelberg R, Rabinovitch M, Kaplan G. Survival of Mycobacterium avium and Mycobacterium tuberculosis in acidified vacuoles of murine macrophages. Infect Immun. 1999;67:3199–3206. doi: 10.1128/iai.67.7.3199-3206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordhan B G, Andersen S J, De Meyer A R, Mizrahi V. Construction by homologous recombination and phenotypic characterization of a polA mutant of Mycobacterium smegmatis. Gene. 1996;178:125–130. doi: 10.1016/0378-1119(96)00350-2. [DOI] [PubMed] [Google Scholar]
- 10.Gordhan, B. G., and T. Parish. Gene replacement by UV pretreatment. Methods Mol. Biol., in press. [DOI] [PubMed]
- 11.Guo M, Sun Z, Zhang Y. Mycobacterium smegmatis has two pyrazinamidase enzymes, PncA and PzaA. J Bacteriol. 2000;182:3881–3884. doi: 10.1128/jb.182.13.3881-3884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heifets L B, Flory M A, Lindholm-Levy P J. Does pyrazinoic acid as an active moiety of pyrazinamide have specific activity against Mycobacterium tuberculosis? Antimicrob Agents Chemother. 1989;33:1252–1254. doi: 10.1128/aac.33.8.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heifets L, Lindholm-Levy P. Pyrazinamide sterilizing activity in vitro against semidormant Mycobacterium tuberculosis bacterial populations. Am Rev Respir Dis. 1992;145:1223–1225. doi: 10.1164/ajrccm/145.5.1223. [DOI] [PubMed] [Google Scholar]
- 14.Hinds J, Mahenthiralingam E, Kempsell K E, Duncan K, Stokes R W, Parish T, Stoker N G. Enhanced gene replacement in mycobacteria. Microbiology. 1999;145:519–527. doi: 10.1099/13500872-145-3-519. [DOI] [PubMed] [Google Scholar]
- 15.Husson R N, James B E, Young R A. Gene replacement and expression of foreign DNA in mycobacteria. J Bacteriol. 1990;172:519–524. doi: 10.1128/jb.172.2.519-524.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs W R, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems for mycobacteria. Methods Enzymol. 1991;381:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 17.Kobayashi H. A proton-translocating ATPase regulates pH of the bacterial cytoplasm. J Biol Chem. 1985;260:72–76. [PubMed] [Google Scholar]
- 18.Lambert L A, Abshire K, Blackenhorn D, Slonczewski J L. Proteins induced in Escherichia coli by benzoic acid. J Bacteriol. 1997;179:7595–7599. doi: 10.1128/jb.179.23.7595-7599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lydiate D J, Ashby A M, Henderson D J, Kieser H M, Hopwood D A. Physical and genetic characterization of chromosomal copies of the Streptomyces coelicolor mini-circle. J Gen Microbiol. 1989;135:941–955. [Google Scholar]
- 20.McDermott W, Tompsett R. Activation of pyrazinamide and nicotinamide in acidic environment in vitro. Am Rev Tuberc. 1954;70:748–754. doi: 10.1164/art.1954.70.4.748. [DOI] [PubMed] [Google Scholar]
- 21.O'Gaora P, Barnini S, Hayward C, Filley E, Rook G, Young D, Thole J. Mycobacteria as immunogens: development of expression vectors for use in multiple mycobacterial species. Med Princ Prac. 1997;6:91–96. [Google Scholar]
- 22.Parish T, Gordhan B G, McAdam R A, Duncan K, Mizrahi V, Stoker N G. Production of mutants in amino acid biosynthetic genes of Mycobacterium tuberculosis by homologous recombination. Microbiology. 1999;145:3497–3503. doi: 10.1099/00221287-145-12-3497. [DOI] [PubMed] [Google Scholar]
- 23.Ramaswamy S, Musser J M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuberc Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 24.Raynaud C, Etienne G, Peyron P, Lanéelle M A, Daffé M. Extracellular enzyme activities potentially involved in the pathogenicity of Mycobacterium tuberculosis. Microbiology. 1998;144:577–587. doi: 10.1099/00221287-144-2-577. [DOI] [PubMed] [Google Scholar]
- 25.Raynaud C, Lanéelle M A, Senaratne R H, Draper P, Lanéelle G, Daffé M. Mechanisms of pyrazinamide resistance in mycobacteria: importance of lack of uptake in addition to lack of pyrazinamidase activity. Microbiology. 1999;154:1359–1367. doi: 10.1099/13500872-145-6-1359. [DOI] [PubMed] [Google Scholar]
- 26.Roe A J, McLaggan D, Davidson I, O'Byrne C, Booth I R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salfinger M, Heifets L B. Determination of pyrazinamide MICs for Mycobacterium tuberculosis at different pHs by the radiometric assay. Antimicrob Agents Chemother. 1988;32:1002–1004. doi: 10.1128/aac.32.7.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Salmond C V, Kroll R G, Booth I R. The effect of food preservatives on pH homeostasis in Escherichia coli. J Gen Microbiol. 1984;130:2845–2850. doi: 10.1099/00221287-130-11-2845. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 30.Scorpio A, Zhang Y. Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug, pyrazinamide in tubercle bacillus. Nat Med. 1996;2:662–667. doi: 10.1038/nm0696-662. [DOI] [PubMed] [Google Scholar]
- 31.Slonczewski J L, Foster J W. pH-regulated genes and survival at extreme pH. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 2. Washington, D.C.: American Society for Microbiology; 1996. pp. 1539–1549. [Google Scholar]
- 32.Snapper S B, Melton R E, Kieser T, Mustafa S, Jacobs W R., Jr Isolation and characterization of high efficiency plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 33.Stottmeier K D, Beam R E, Kubica G P. Determination of drug susceptibility of mycobacteria to pyrazinamide in 7H10 agar. Am Rev Respir Dis. 1967;96:1072–1075. doi: 10.1164/arrd.1967.96.5.1072. [DOI] [PubMed] [Google Scholar]
- 34.Sun Z, Scorpio A, Zhang Y. The pncA gene from naturally pyrazinamide-resistant Mycobacterium avium encodes pyrazinamidase and confers pyrazinamide susceptibility to resistant M. tuberculosis complex organisms. Microbiology. 1997;143:3367–3373. doi: 10.1099/00221287-143-10-3367. [DOI] [PubMed] [Google Scholar]
- 35.Sun Z, Zhang Y. Reduced pyrazinamidase activity and the natural resistance of Mycobacterium kansasii to the antituberculosis drug pyrazinamide. Antimicrob Agents Chemother. 1999;43:537–542. doi: 10.1128/aac.43.3.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. WHO report on the tuberculosis epidemic: stop TB at the source. Geneva, Switzerland: Tuberculosis Control Programme, World Health Organization; 1995. [Google Scholar]
- 37.Yan C, Sloan D L. Purification and characterization of nicotinamide deamidase from yeast. J Biol Chem. 1987;262:9082–9087. [PubMed] [Google Scholar]
- 38.Zaborina O, Li X, Cheng G, Kapatral V, Chakrabarty A M. Secretion of ATP-utilizing enzymes, nucleoside diphosphate kinase and ATPase, by Mycobacterium bovis BCG: sequestration of ATP from macrophage P2Z receptors? Mol Microbiol. 1999;31:1333–1334. doi: 10.1046/j.1365-2958.1999.01240.x. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Scorpio A, Nikaido H, Sun Z. Role of acid pH and deficient efflux of pyrazinoic acid in unique susceptibility of Mycobacterium tuberculosis to pyrazinamide. J Bacteriol. 1999;181:2044–2049. doi: 10.1128/jb.181.7.2044-2049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]