Abstract
Vibrio parahaemolyticus is a ubiquitous, gram-negative marine bacterium that undergoes phase variation between opaque and translucent colony morphologies. The purpose of this study was to determine the factor(s) responsible for the opaque and translucent phenotypes and to examine cell organization within both colony types. Examination of thin sections of ruthenium red-stained bacterial cells by electron microscopy revealed a thick, electron-dense layer surrounding the opaque cells that was absent in preparations from translucent strains. Extracellular polysaccharide (EPS) material was extracted from both opaque and translucent strains, and the opaque strain was shown to produce abundant levels of polysaccharide, in contrast to the translucent strain. Compositional analysis of the EPS identified four major sugars: glucose, galactose, fucose, and N-acetylglucosamine. Confocal scanning laser microscopy was used to investigate cell organization within opaque and translucent colonies. Cells within both types of colonies exhibited striking organization; rod-shaped cells were aligned parallel to one another and perpendicular to the agar surface throughout the depth of the colony. Cells within translucent colonies appeared more tightly packed than cells in opaque colonies. In addition, a dramatic difference in the structural integrity of these two colony types was observed. When colonies were perturbed, the cell organization of the translucent colonies was completely disrupted while the organization of the opaque colonies was maintained. To our knowledge, this study represents the first description of how cells are organized in the interior of a viable bacterial colony. We propose that the copious amount of EPS produced by the opaque strain fills the intercellular space within the colony, resulting in increased structural integrity and the opaque phenotype.
Bacteria have evolved numerous adaptive mechanisms to facilitate their survival under changing environmental conditions. One such mechanism is phase variation, which occurs when the expression of a given factor is periodically altered such that it is either “on” or “off.” These variations are generally spontaneous and reversible and occur at relatively high frequency (>10−5 per generation) (13). Factors that are controlled by phase variation are often associated with the cell surface and include structures such as flagella, fimbriae, outer membrane proteins, and exo- and lipopolysaccharide (13). Variations in the expression of these structures often result in an observable phenotypic change, such as altered colony morphology.
Vibrio parahaemolyticus is a ubiquitous, gram-negative marine bacterium that undergoes phase variation that results in different colony morphologies. This organism “switches” between a large, flat, translucent colony type (TR) and a smaller, mounded, opaque colony type (OP) (19, 20). It is not clear whether environmental conditions influence OP-TR switching in V. parahaemolyticus or whether this variation is random. The factor(s) responsible for the OP and TR phenotypes in V. parahaemolyticus has not been identified. Although the colony morphologies are not identical to those of V. parahaemolyticus, there are other bacteria that exhibit OP-TR phase variation. For example, Neisseria gonorrhoeae undergoes phase variation, resulting in colonies with different degrees of opacity; these differences have been associated with differential expression of specific outer membrane proteins (Opa proteins) (17, 24, 38). Vibrio vulnificus also switches between OP and TR colony morphologies (36, 40, 43). For this organism, both opacity and virulence have been correlated with the production of capsular polysaccharide (29, 40).
Recently, a gene product that controlled opacity in V. parahaemolyticus was identified (20). The gene, designated opaR, was expressed in OP strains but not in TR strains. In addition, it was found that the OP and TR phenotypes could be controlled by artificially controlling the expression of opaR. These results suggested that OpaR was a positive regulatory protein required for the OP phenotype. Homologues of opaR include Vibrio harveyi luxR (25, 26, 34), a regulatory gene that controls bioluminescence and polyhydroxybutyrate production; Vibrio cholerae hapR (15), a positive regulatory gene that controls expression of the hemagglutinin protease; and Vibrio anguillarum vanS, a gene that positively regulates the expression of empA, a metalloprotease (D. Milton, V. Hope, M. Camara, and P. Williams, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. H-193, p. 366, 1999). The gene products that are responsible for the V. parahaemolyticus OP and TR phenotypes, and that presumably are regulated by OpaR, have not been identified.
Although cell surface structures that cause the OP and TR phenotypes have been identified in several bacteria, the physical basis for why these colony types appear either OP or TR is unclear. One possibility is that cell surface components alter cell packing or organization within a colony, ultimately resulting in differential transmission of light. Swanson has proposed this scenario as a possible cause of opacity in gonococci (37). However, further evidence to support this or other models for opacity is lacking. There is little information in general about how cells are organized within bacterial colonies. Such knowledge is important for understanding the colony microenvironment and the way in which bacteria function as multicellular entities, e.g., colony development and pattern formation (5, 32). A number of studies have used scanning and transmission electron microscopy to investigate colony organization in various bacteria (1, 3, 8, 9, 11, 30, 31, 37). These studies suggested that cells within colonies were ordered; however, they were limited to observations of cells at the colony surface or those contained in a single section of a colony that had been subjected to a fixation process.
The purpose of this study was to identify the factor(s) responsible for opacity in V. parahaemolyticus and investigate the physical basis of the OP and TR colony morphologies. Results strongly suggested that differences in the production of capsular polysaccharide (CPS) were responsible for the OP and TR phenotypes. CPS was isolated from the OP variant and initially characterized. Confocal scanning laser microscopy (CSLM) was used to investigate cell organization within OP and TR colonies. This method allowed the visualization of sequential sections through viable colonies. Results revealed striking cell organization within both colony types and significant differences between OP and TR strains. To our knowledge, these results represent the first description of how cells are organized in the interior of a viable colony. A model for the physical basis of opacity is presented.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains used in this study are listed in Table 1. The wild-type OP strain, BB22OP, has been previously described (20). Three TR strains were examined: (i) the wild-type TR strain, BB22TR (4); (ii) a newly isolated spontaneous TR derivative of BB22OP, LM5093; and (iii) BB22OP containing a Tn5 mutation in the opaR region (which converted the strain from OP to TR), LM4463. BB22TR exhibits a genetic rearrangement in the opaR region with respect to BB22OP (20); the opaR region in LM5093 has not been characterized. These independent strains were used to ensure that any differences observed were consistently correlated with the OP and TR colony morphologies. Plasmid pLM1876 was constructed by cloning a kanamycin cassette (2) into the HindIII site of pMMB66EH (12) (HindIII site destroyed). Plasmid pLM2384 was constructed by cloning a PCR-amplified gfpmut2 gene (7) into the SmaI site of pLM1876. Procedures for the transfer of plasmids from Escherichia coli to V. parahaemolyticus via conjugation have been previously described (35). Heart infusion agar (25 g of heart infusion broth [Difco], 20 g of NaCl, and 20 g of Bacto Agar per liter) was used for propagating V. parahaemolyticus strains. Kanamycin and isopropyl-β-d-thiogalactopyranoside (IPTG) were used at final concentrations of 50 μg/ml and 0.5 mM, respectively. Strains were grown overnight at 30°C. Variation between OP and TR strains is sufficiently slow that it is possible to obtain essentially “pure” cultures of either OP or TR strains having fewer than 1 alternate form per 1,000 colonies.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| BB22TR | Wild type; TR variant | R. Belas (4) |
| BB22OP | Wild type; OP variant | 20 |
| LM4463 | TR phenotype; Tn5 in opaR region | BB22OP (this work) |
| LM4892 | OP phenotype, BB22OP/pLM2384 | BB22OP (this work) |
| LM4893 | TR phenotype, BB22TR/pLM2384 | BB22TR (this work) |
| LM4894 | TR phenotype, LM5093/pLM2384 | LM5093 (this work) |
| LM5093 | TR phenotype; spontaneous derivative of BB22OP | BB22OP (this work) |
| Plasmids | ||
| pLM1876 | Kanr Apr broad-host-range expression vector carrying lacIq and Ptac | This work |
| pLM2384 | Kanr Apr broad-host-range expression vector carrying gfpmut2 under IPTG control | This work |
Electron microscopy.
Bacteria were grown on heart infusion agar plates overnight at 30°C, suspended in 0.3 M sucrose, and gently centrifuged in microcentrifuge tubes. Staining and fixation of the bacterial pellets was performed by the method described by Luft (18). The cells were fixed and stained for 1 h in a solution containing the following in equal amounts: 2.5% glutaraldehyde in 0.1 M cacodylate buffer, 0.2 M cacodylate buffer (pH 7.3), and 0.15% (wt/vol) ruthenium red. Bacteria were washed for 1 h with 0.1 M cacodylate buffer (pH 7.3) and then fixed for 2 h with an osmium solution containing equal parts of 5% OsO4, 0.2 M cacodylate buffer (pH 7.3), and 0.15% (wt/vol) ruthenium red. Fixed cells were then washed again with cacodylate buffer and dehydrated through a series of washes with 25% (15 min), 50% (15 min), 75% (15 min), 95% (30 min), and 100% (1 h) ethanol. Samples were then washed with solutions of ethanol-Epon (2:1), ethanol-Epon (1:2), and Epon, for 2, 2, and 4 h, respectively. The samples were baked overnight at 70°C, cut into thin sections, placed on a copper grid, double stained with uranyl acetate and lead citrate, and examined under a Hitachi H-7000 transmission electron microscope operating at an accelerating voltage of 75 kV.
Isolation and extraction of EPS.
A single bacterial colony was used to inoculate an entire petri plate by being streaked onto the plate and incubated for 20 h at 30°C. Bacteria were scraped from the plates and suspended in 4 to 5 ml of phosphate-buffered saline (20 mM sodium phosphate [pH 7.3], 100 mM NaCl) per plate. Samples were vortexed for 1 min and then shaken at 200 rpm on a rotary shaker for 1.5 h at 30°C; the vortexing and shaking process was then repeated. At this point, samples from OP strains were significantly more viscous than those from TR strains, and so the samples from the OP strains were diluted fivefold with phosphate-buffered saline. Cells and debris were subsequently removed by centrifugation at 10,000 × g for 15 min. Samples being prepared for monosaccharide analysis were also ultracentrifuged at 154,000 × g for 16 h. Supernatant containing the extracellular polysaccharide (EPS) was removed; RNase A (Sigma), DNase I (Sigma), and MgCl2 were added at final concentrations of 50 μg/ml, 50 μg/ml, and 10 mM, respectively; and the mixture was incubated for 8 h at 37°C. Proteinase K (Amresco) was subsequently added at a final concentration of 200 μg/ml. Following incubation for 17 h at 37°C, the sample was extracted twice with phenol-chloroform, precipitated with 2.5 volumes of ethanol, washed with 70% ethanol, and suspended in water. Samples being prepared for monosaccharide analysis were extensively dialyzed against nanopure H2O to remove salts. Multiple samples being prepared for EPS quantitation experiments were not routinely treated with RNase and DNase, because results indicated this treatment does not significantly affect EPS quantitation.
Analysis of EPS by SDS-PAGE.
Samples containing extracted EPS were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described for proteins (21) with the following modifications. The gels were 8.5 cm long, with the lower (12.5% polyacrylamide) gel being approximately 2 cm and the upper (5% polyacrylamide), stacking gel being approximately 6.5 cm. The gels were made in this manner because the EPS migrated only through the stacking gel. The gels were stained with the cationic carbocyanine dye 1-ethyl-2-[3-(1-ethylnaphtho(1,2d)-thiazolin-2-ylidene)-2-methylpropenyl]-naphtho(1,2d)thiazolium bromide (Stains-All; United States Biochemicals, Cleveland, Ohio) by the method described by Kelley and Parker (16).
Quantitative analysis of EPS.
Suspensions of plate-grown cells were prepared as described above. Half of the sample was used for quantitating EPS. EPS was extracted from these samples as described above, and the amount of carbohydrate was quantitated using the phenol-sulfuric acid method of Dubois et al. (10) with glucose as the standard. The other half of the sample was used to quantitate cellular protein using the method of Bradford (6). The carbohydrate concentration was expressed as micrograms of carbohydrate per milligram of protein. At least four independent polysaccharide preparations were quantitated.
Carbohydrate analysis.
Glycosyl composition analysis was performed at the University of Georgia Complex Carbohydrate Research Center. The monosaccharide composition was determined by combined gas chromatography-mass spectrometry (GC-MS) analysis of trimethylsilyl (TMS) derivatives of the component methyl glycosides released by methanolysis (23, 42). Briefly, methyl glycosides were prepared from the sample by methanolysis with 1 N HCl in 100% methanol followed by re-N-acetylation of amino sugars with acetic anhydride-pyridine. The samples were then treated with Tri-Sil (Pierce Chemical Co., Rockford, Ill.) to produce per-O-trimethylsily derivatives for GC-MS. GC-MS analysis of the TMS methyl glycosides was performed on an HP 5890 GC interfaced to a 5970 MSD, using a Supelco DB-5 or equivalent fused-silica capillary column.
Outer membrane protein and lipopolysaccharide analysis.
Outer membrane proteins and lipopolysaccharide from BB22OP and BB22TR were isolated and analyzed as described by McCarter and Silverman (21) and Hitchcock and Brown (14), respectively.
CSLM.
Colonies of green fluorescent protein (GFP)-expressing strains were obtained by streaking for isolated colonies on agar-coated microscope slides containing kanamycin and IPTG. After a 20-h incubation at 30°C, a coverslip was gently placed over the bacterial colonies on the slide. For some experiments, colonies were fixed with 2.5% glutaraldehyde for 30 min at room temperature before placement of the coverslip. A Zeiss (Jena, Germany) inverted LSM 510 confocal microscope (40× objective) was used to collect single optical sections (x-y plane) and z sections (x-z plane) through the colony of interest by using a 488-nm excitation wavelength for visualization of the fluorescent bacteria. The images were processed using Zeiss LSM510 software version 2.02.
RESULTS
Opacity is associated with an extracellular layer.
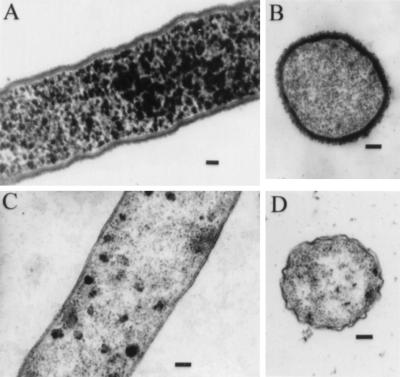
Electron microscopy was used to investigate the nature of the morphological differences between OP and TR strains of V. parahaemolyticus. Plate-grown bacteria were suspended in buffer and gently pelleted. Pellets were stained with ruthenium red, and thin sections were observed by transmission electron microscopy. Representative results are shown in Fig. 1. The wild-type OP strain, BB22OP, showed a heavy, fibrous, electron-dense layer that completely surrounded the bacterial cells (Fig. 1A and B). This layer was absent in preparations made from the wild-type TR strain, BB22TR (Fig. 1D). In addition, the layer was not observed around cells of a strain that had been converted from an OP to a TR phenotype by introduction of a Tn5 insertion in the opaR region (Fig. 1C). The OpaR protein was shown to be a positive regulator of opacity, and previous results suggested a genetic rearrangement of the opaR locus in BB22OP and BB22TR (20).
FIG. 1.
Electron micrographs of BB22OP (A and B), LM4463 (C), and BB22TR (D) stained with ruthenium red. Bars, 100 nm. BB22OP exhibits an OP phenotype, whereas LM4463 and BB22TR exhibit TR phenotypes.
Additional tests were performed with OP and TR strains to identify other potential differences in cell surface structures that might contribute to the OP and TR phenotypes. Outer membrane proteins and lipopolysaccharide were isolated from BB22TR and BB22OP and analyzed by SDS-PAGE. No profound differences in outer membrane protein or lipopolysaccharide profiles were observed (data not shown). Thus, the major cell surface characteristic that appeared to differ between OP and TR strains was an extracellular layer that stained with ruthenium red. This staining property suggested the presence of acidic polysaccharide (18, 28).
Correlation of opacity with EPS production.
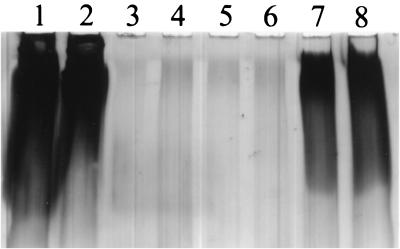
The above results suggested that the OP strain was producing increased levels of polysaccharide compared to the TR strain. To further test this possibility, we extracted EPS material from BB22OP, BB22TR, and a newly isolated spontaneous TR derivative of BB22OP, LM5093. Samples were analyzed by SDS-PAGE and stained with Stains-All, a cationic carbocyanine dye that typically stains proteins red, nucleic acids blue, and acidic polysaccharides variable colors. The results are shown in Fig. 2. The OP strain (lanes 1 and 2 [undiluted sample] and lanes 7 and 8 [fivefold dilution]) produced copious amounts of a material that was essentially absent in the two TR strain preparations (BB22TR [lanes 3 and 4] and LM5093 [lanes 5 and 6]). This material stained purple, consistent with its being a polysaccharide, and migrated through the 5% polyacrylamide stacking gel but not the 12.5% polyacrylamide lower gel. The presence of the material was not affected by the addition of RNase or DNase to the sample.
FIG. 2.
SDS-PAGE analysis of EPS produced by OP and TR strains. EPS samples were diluted twofold in electrophoresis sample buffer. Equal volumes were analyzed by SDS-PAGE and stained with Stains-All. The 5% polyacrylamide or “stacking” portion of the gel is shown. Two independent preparations from each strain were examined. Lanes: 1 and 2, BB22OP; 3 and 4, BB22TR; 5 and 6, LM5093; 7 and 8, fivefold dilution of BB22OP.
To confirm that the isolated material was polysaccharide and to estimate the carbohydrate levels in the samples, we measured the carbohydrate content of the extracted material using the phenol-sulfuric acid method of Dubois et al. (10). Glucose was used as a carbohydrate standard. A positive reaction of the extracellular material in this assay confirmed the presence of sugars. By using this assay and normalizing samples to total cellular protein, we detected 15.6 ± 1.1 μg of carbohydrate/mg of protein from BB22OP and 1.3 ± 0.2 μg of carbohydrate/mg of protein from BB22TR. Thus, these results demonstrated a significant difference in polysaccharide production between OP and TR strains. These measurements suggested that the levels of EPS extracted from BB22OP were at least 12-fold higher than those extracted from BB22TR. However, the SDS-PAGE data shown in Fig. 2 suggested a much larger difference and are likely to be more representative, given the quantitative limitations of the phenol-sulfuric acid method (see Discussion).
Carbohydrate analysis of V. parahaemolyticus EPS.
To initially characterize the EPS extracted from the OP strain, the monosaccharide composition was determined. Purified EPS was hydrolyzed by methanolysis, treated to form trimethylsilyl derivatives, and analyzed by GC-MS. Monosaccharide derivatives were identified by their retention times and mass spectra in comparison to those of standards derivatized and analyzed in parallel. The results are shown in Table 2. Four major sugars were identified: fucose, galactose, glucose, and N-acetylglucosamine. The molar proportions of fucose, galactose, and glucose most probably represent a 1:1:1 ratio of these residues in the polysaccharide. The slightly higher level of glucose in the sample may be because it is a commonly observed impurity. The molar ratio of N-acetylglucosamine appeared to be approximately 0.5. However, it is possible that this value is an underestimate since yields of acetamido sugars can vary somewhat depending on the primary structure of the original carbohydrate (S. Levery, personal communication). Very low levels (≤1%) of three other sugars, arabinose, mannose, and N-acetylgalactosamine, were present in the sample.
TABLE 2.
Glycosyl composition analysis of EPS
| Glycosyl residue | Percentagea |
|---|---|
| Glucose | 34.6 |
| Galactose | 27.8 |
| Fucose | 21.3 |
| N-Acetylglucosamine | 13.9 |
| Arabinose | 1.0 |
| Mannose | 0.7 |
| N-Acetylgalactosamine | 0.6 |
Values are expressed as the weight percentage of total carbohydrate detected.
Investigating colonial cell organization by CSLM.
The physical basis for why bacterial colonies appear opaque or translucent is not well understood. It is possible that due to different cell surface components, cells within OP and TR colonies are organized or packed differently, resulting in differential transmission of light. To address whether this idea would explain the phenotypic differences in V. parahaemolyticus strains, we investigated cell organization within colonies by CSLM. Since colonial cell organization has not been extensively studied in any bacteria, this investigation was also expected to be informative in a general sense. To enable the visualization of bacterial cells by CSLM, a kanamycin-resistant plasmid containing a gene encoding GFP under IPTG control was introduced into BB22OP, BB22TR, and LM5093 (TR). The resulting strains, LM4892, LM4893, and LM4894, respectively, were streaked onto agar-coated microscope slides containing kanamycin and IPTG. Coverslips were gently placed over individual colonies, which were examined using CSLM.
(i) Examination of cell organization in the x-y plane.
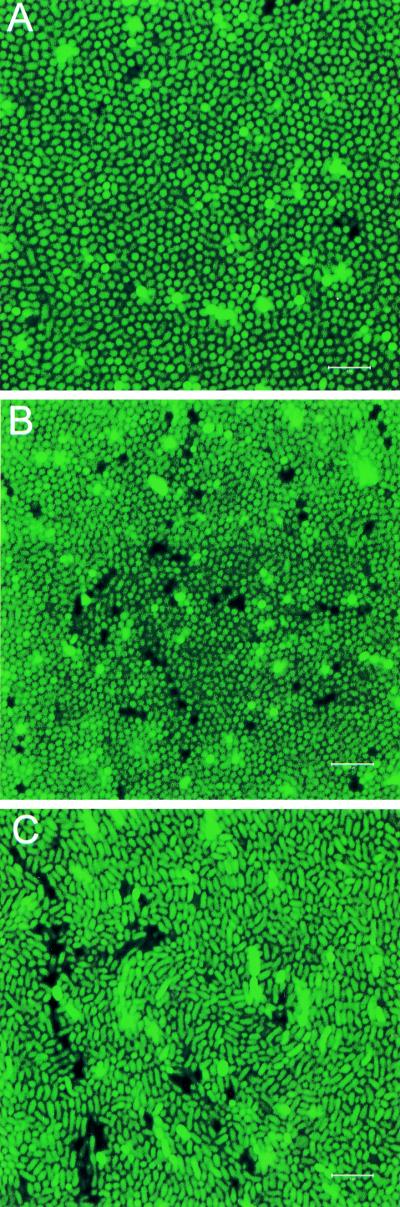
Representative images of single optical sections (x-y plane) from LM4892 (OP) and LM4894 (TR) colonies are shown in Fig. 3A and B, respectively. These images represent a horizontal slice through the bacterial colony. It was clear from these images that both colony types exhibited striking cellular organization within the colony. Circular cells were packed into an orderly array. This highly organized pattern was observed in x-y sections from the top of the colony to a depth of approximately 40 μm. At this point, resolution of individual cells became limiting, possibly due to the requirement of GFP activity for oxygen. Single optical sections were obtained from multiple colonies and from multiple locations within each colony. Images such as those shown in Fig. 3A and B were consistently observed at all locations with two exceptions: (i) the very first layer of cells at the top of the colony, which exhibited a slightly more random organization, possibly due to placement of the coverslip, and (ii) the periphery of the colony, where cell organization varied considerably (data not shown). Since V. parahaemolyticus is a rod-shaped organism, the consistent circular shapes of the bacterial cells throughout the depth of the colonies suggested that the rod-shaped cells were arranged perpendicular to the agar surface. Suspension of the colonies in liquid medium and subsequent examination using CSLM confirmed that the cells within the colonies were rod shaped (data not shown).
FIG. 3.
Confocal scanning laser micrographs through the x-y plane of V. parahaemolyticus colonies. The V. parahaemolyticus strains contained a plasmid expressing GFP. A coverslip was gently placed over single colonies prior to examination. The single optical sections shown (x-y plane) were obtained at a depth of approximately 10 μm from the top of the colony. (A) Unperturbed BB22OP colony (OP phenotype); (B) unperturbed LM5093 colony (TR phenotype); (C) perturbed LM5093 colony (TR phenotype). Dark spots and extra-bright spots in the image are probably due to alterations in GFP expression or activity. Bars, 5 μm.
Although OP and TR colonies showed similar organization, results suggested a significant difference in cell packing between these two colony types. Cells in TR colonies (LM4894 [Fig. 3B]) appeared more tightly packed than cells in OP colonies (LM4892 [Fig. 3A]). Images from the TR strain LM4893 were similar to those obtained from TR strain LM4894 (data not shown). To determine if these differences were quantitative, the average number of cells within a 100-μm2 area was determined for LM4892 (OP) and LM4894 (TR). Multiple images from at least six independent colonies of each strain were used for cell quantitation. This analysis determined that OP colonies had an average cell density of 91.6 ± 8.1 cells/100 μm2 while TR colonies had an average cell density of 128.9 ± 9.5 cells/100 μm2. Comparison of the two data sets using a two-sample t test indicated that the differences were statistically significant (P < 0.0001).
During the course of these experiments, it became clear that slight perturbations of TR colonies (for example, aggressive or quick movements of the microscope objective or pressure on the coverslip) resulted in very different images from those previously observed. As shown in Fig. 3C, cells of a “perturbed” TR colony (LM4894) appeared randomly organized compared to the high degree of organization of unperturbed colonies (Fig. 3B). This result was observed with multiple colonies of both TR strains, LM4893 and LM4894. In contrast, cells from OP colonies maintained a high degree of organization, similar to that shown in Fig. 3A, after similar, aggressive treatment of the colonies (data not shown). These results indicated that the structure and organization of the TR colonies were much more fragile and sensitive to physical force than were those of the OP colonies.
(ii) Examination of cell organization in the x-z plane.
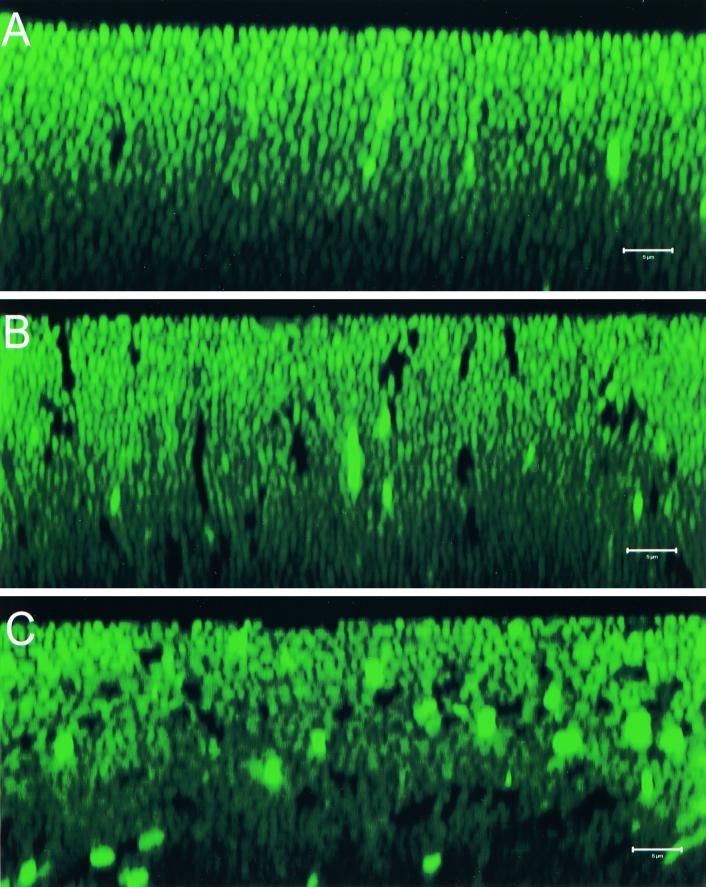
The data presented above suggested that the V. parahaemolyticus rod-shaped cells were uniformly arranged perpendicular to the surface in both the OP and TR colonies and, further, that the high degree of cellular organization of the TR colony could be easily disrupted. To further address these results, we performed z-series scans (x-z series), which provided a side view of the cells within the bacterial colony. Representative images from an OP colony (LM4892), a TR colony (LM4894), and a perturbed TR colony (LM4894) are shown in Fig. 4A, B, and C, respectively. The images of the nonperturbed colonies shown in Fig. 4A and B provided further evidence that the cells within these colonies were oriented uniformly upright. Rod-shaped cells could be seen that were aligned in a horizontal and highly organized fashion. As in the x-y scans, cells in the TR colony appeared to be more tightly packed (compare Fig. 4A and B). The x-z image obtained from the perturbed TR colony (Fig. 4C) showed sharp contrast to the images obtained from the nonperturbed colonies. The x-z image of the perturbed colony appeared blurred compared to the other x-z images; individual cells were difficult to distinguish with regard to their shape and/or orientation. This image was thus consistent with random cell organization within this colony.
FIG. 4.
Confocal scanning laser micrographs through the x-z plane of V. parahaemolyticus colonies. V. parahaemolyticus strains contained a plasmid that expressed GFP. A coverslip was gently placed over single colonies prior to examination. z sections (x-z plane) were obtained from the top of the colony to a depth of approximately 40 μm. Representative images are shown. The top of the colony is at the top of each image. (A) unperturbed BB22OP colony (OP phenotype); (B) unperturbed LM5093 colony (TR phenotype); (C) perturbed LM5093 colony (TR phenotype). Dark spots and extra-bright spots in the image are probably due to alterations in GFP expression or activity. Bars, 5 μm.
DISCUSSION
The purpose of this study was to determine the factor(s) responsible for the OP and TR phenotypes in V. parahaemolyticus and to examine colonial cell organization in this organism. Results presented here strongly suggest that opacity in V. parahaemolyticus correlates with increased production of EPS. Examination of thin sections of cells from OP and TR strains that had been stained with ruthenium red, which stains acidic polysaccharides, revealed a thick layer of material surrounding only the opaque cells. Based on the clear association of this layer with the cell surface, we suggest that this material be referred to as capsular polysaccharide (CPS). Abundant extracellular material was isolated from OP but not TR strains by using an EPS extraction procedure. Results obtained using the phenol-sulfuric acid assay confirmed that the extracted material was carbohydrate and suggested that the carbohydrate levels were at least 12-fold higher in samples prepared from the OP strain than in those from the TR strain. Analysis of the CPS material by SDS-PAGE suggested that the ratio between the two strains was significantly higher. The SDS-PAGE data are likely to be a more accurate reflection of the difference in CPS production between these two strains. The phenol-sulfuric acid assay has limitations for detecting different types of sugars, particularly when quantitating unknown saccharides with glucose as a standard. We conclude from the phenol-sulfuric acid and SDS-PAGE results that there is a minimum 12-fold difference in CPS production between the OP and TR strains. Based on the correlation between opacity and CPS production and our failure to detect differences in other cell surface structures, we suggest that CPS production plays a major role in the OP phenotype.
To our knowledge, the results presented here represent the first example of a polysaccharide produced by V. parahaemolyticus. Glycosyl composition analysis identified four major sugars present in CPS extracted from an opaque strain: glucose, galactose, fucose, and N-acetylglucosamine, in an approximate molar ratio of 1:1:1:0.5. This distinguishes V. parahaemolyticus CPS from EPS produced by other marine vibrios, including the CPS produced by the opaque variant of V. vulnificus (29), which contains 2-acetoamido-2,6-dideoxyhexopyranose and 2-acetamido hexouronate, the capsule-like material produced by V. cholerae O1 (39), and the amorphous exopolysaccharide produced by V. cholerae O1 El Tor (41).
The staining properties of V. parahaemolyticus CPS with ruthenium red and Stains-All, which are both cationic, suggested that the polysaccharide was acidic. Polysaccharides can be rendered acidic in a variety of ways. Constituent monosaccharides can be oxidized, resulting in uronic acids, in which the C-6 hydroxyl is oxidized to carboxylic acid, or in neuraminic acids, in which the C-1 aldehyde is oxidized to carboxylic acid. The monosaccharide composition analysis, which would have detected both of these constituents, failed to detect either in V. parahaemolyticus CPS. Another possibility is that one or more of the constituent monosaccharides is modified by derivatization with an acidic compound, e.g., pyruvate, succinate, or acetate, which are common substitutions of bacterial exopolysaccharides, or by esterification with sulfuric or phosphoric acid. Of these modifications, only pyruvate would have been detected by the composition analysis used. Further work will permit the identification of the complete structure of V. parahaemolyticus CPS. Interestingly, the amorphous exopolysaccharide produced by V. cholerae O1 El Tor (41) also exhibits acidic properties but does not contain acidic monosaccharide residues.
The results presented here predict that OpaR, a positive regulator of opacity (20), may be positively regulating the expression of genes responsible for CPS production. This possibility would be an addition to the list of functions regulated by this family of proteins, which so far include luminescence and polyhydroxybutyrate in V. harveyi (26, 34), the hemagglutinin/protease in V. cholerae (15), and a metalloprotease in V. anguillarium (Milton et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol.). Interestingly, hapR mutants of V. cholerae El Tor strains have a rugose colony morphology, which has been associated with polysaccharide production (15, 41). Therefore, HapR might be directly or indirectly repressing polysaccharide synthesis. V. parahaemolyticus is not luminescent, but it does produce extracellular protease activity, as assayed by the presence of clearing zones on plates containing casein (J. L. Enos-Berlage, unpublished results). However, similar proteolytic activity is observed with both a wild-type strain and an opaR mutant (Enos-Berlage, unpublished). Thus, a common theme for the function of this family of regulators has not emerged.
This study has provided insight into the physical basis of the OP phenotype. Examination of OP and TR colonies by CSLM revealed that cells in TR colonies were “packed” at significantly higher densities than were cells in OP colonies. Thus, it does not appear that increased cell density is responsible for opacity in V. parahaemolyticus. This is unlike the OP phenotype described for Neisseria gonorrhoeae, where a correlation between opacity and more highly aggregated cells has been observed (37). It is possible that multiple mechanisms can result in the OP phenotype. In V. parahaemolyticus OP colonies appeared to have an increased intercellular space. Based on the results presented here, we propose that this space contains and is the result of CPS production by the OP strain. We further propose that this “filling” in the intercellular space is the material that alters light transmission through the colony and results in the OP phenotype. This model is consistent with the increased structural integrity of OP colonies that was observed during the CSLM studies. Cells in the OP colonies remained in a highly organized pattern despite perturbation of the colony by physical force. The highly ordered pattern was completely abolished in TR colonies after undergoing the same treatment. We suggest that the polysaccharide material between cells in the OP colonies acts as an adhesive, holding the cells in these colonies together in a highly organized manner and making the colony structure resistant to physical force.
V. parahaemolyticus has the ability to sense and respond to surfaces (19, 22). A distinct motility system, which encompasses over 50 genes, is surface induced. It should be noted that all of the experiments described here were performed with surface-grown cells. Preliminary data suggest that the difference in CPS production between OP and TR strains grown in liquid medium is not as large. V. parahaemolyticus forms thick biofilms on the sides of culture vessels and at the air-liquid interface. The possibility of surface-regulated polysaccharide production and the role this synthesis might play in biofilm formation will be interesting to explore.
The results presented here have revealed important clues to the way bacterial cells are organized within colonies. To our knowledge, this is the first study that has examined cell organization in sequential sections on the interior of viable, nonfixed colonies. The results obtained strongly suggest that V. parahaemolyticus cells are uniformly aligned parallel to one another and perpendicular to the surface throughout the depth of the colony. These results are thus consistent with previous work with a number of bacteria suggesting that cells in colonies are highly ordered (1, 3, 8, 9, 11, 30, 31, 37). Some of the scanning electron micrographs from the elegant microscopy of Pseudomonas putida and Escherichia coli colonies by Shapiro bear some resemblance to images that we obtained of the colony interior (30, 31). Large, ordered, multicellular arrays were observed in the scanning electron microscopy studies, and some of these arrays exhibited uniformly arranged circular cells. It was unclear whether these cells were short, coccoid cells or whether they were bacilli standing on end. We believe that the latter is likely based on the results of the present study. The fact that the cell pattern we observed has been seen on the surface of colonies from different bacteria suggests that this pattern may be a general phenomenon and not specific to V. parahaemolyticus.
The basis for the striking colonial cell organization will be interesting to determine. The organization could be a consequence of the way cells within a colony grow and divide, or it could be the result of active cell movement through the colony. It is possible that the organization is “social” in nature and is dependent on physical cell-cell interactions or intercellular communication. These types of interactions are important in other cases where bacterial cells act in a coordinated fashion to form a multicellular entity, e.g., biofilm formation (27) or fruiting-body formation in Myxococcus xanthus (33). It will be interesting to determine if the highly organized cell structure we observed in V. parahaemolyticus colonies is dependent on specific cell surface components, motility systems, or cell-cell interactions.
ACKNOWLEDGMENTS
We especially thank Todd Ontl, who performed the electron microscopy; Adam Howe, who assisted with the confocal microscopy studies; Rex Montgomery and Byung Yang, who performed the initial monosaccharide composition analysis; and Michael Apicella, who provided constant expertise and methods for polysaccharide analyses. We thank The University of Georgia Complex Carbohydrate Research Center, in particular Steven Levery, and the Medical Photography and Graphics Facility and the Central Microscopy Research Facility at the University of Iowa, in particular Randy Nessler, for excellent support. We also thank Bonnie Stewart and Sanford Jaques for helpful discussions.
This material is based on work supported by the National Science Foundation under a fellowship awarded in 1999 to J.L.E.-B., Public Health Service grant GM43196 from the National Institutes of Health (NIH) to L.L.M., and in part by the NIH-funded Resource Center for Biomedical Complex Carbohydrates (NIH grant 2-P41-RR05351-06) to the Complex Carbohydrate Research Center.
REFERENCES
- 1.Afrikian E G, Julian G S, Bulla L A. Scanning electron microscopy of bacterial colonies. Appl Microbiol. 1973;26:934–937. doi: 10.1128/am.26.6.934-937.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–55. [PubMed] [Google Scholar]
- 3.Bauer H, Sigarlakie E, Faure J C. Scanning and transmission electron microscopy of three strains of Bifidobacterium. Can J Microbiol. 1975;21:1305–1316. doi: 10.1139/m75-197. [DOI] [PubMed] [Google Scholar]
- 4.Belas R, Simon M, Silverman M. Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Jacob E, Cohen I, Gutnick D L. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu Rev Microbiol. 1998;52:779–806. doi: 10.1146/annurev.micro.52.1.779. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 8.Dekker N P, Lammel C J, Mandrell R E, Brooks G F. Opa (protein II) influences gonococcal organization in colonies, surface appearance, size, and attachment to human fallopian tube tissues. Microb Pathog. 1990;9:19–31. doi: 10.1016/0882-4010(90)90037-q. [DOI] [PubMed] [Google Scholar]
- 9.Drucker D B, Whittaker D K. Examination of certain bacterial colonies by scanning electron microscopy. Microbios. 1971;4:109–113. [PubMed] [Google Scholar]
- 10.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith R. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–356. [Google Scholar]
- 11.Fass R J. Morphology and ultrastructure of staphylococcal L colonies: light, scanning, and transmission electron microscopy. J Bacteriol. 1973;113:1049–1053. doi: 10.1128/jb.113.2.1049-1053.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerste J P, Pansegrau W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 13.Henderson I R, Owen P, Nataro J P. Molecular switches—the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 14.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jobling M G, Holmes R K. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol Microbiol. 1997;26:1023–1034. doi: 10.1046/j.1365-2958.1997.6402011.x. [DOI] [PubMed] [Google Scholar]
- 16.Kelley J T, Parker C D. Identification and preliminary characterization of Vibrio cholerae outer membrane proteins. J Bacteriol. 1981;145:1018–1024. doi: 10.1128/jb.145.2.1018-1024.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambden P R, Heckels J E, James L T, Watt P J. Variations in surface protein composition associated with virulence properties in opacity types of Neisseria gonorrhoeae. J Gen Microbiol. 1979;114:305–312. doi: 10.1099/00221287-114-2-305. [DOI] [PubMed] [Google Scholar]
- 18.Luft J H. Ruthenium red and violet. I. Chemistry, purification, methods of use for electron microscopy and mechanism of action. Anat Rec. 1971;171:347–368. doi: 10.1002/ar.1091710302. [DOI] [PubMed] [Google Scholar]
- 19.McCarter L L. The multiple identities of Vibrio parahaemolyticus. J Mol Microbiol Biotechnol. 1999;1:51–57. [PubMed] [Google Scholar]
- 20.McCarter L L. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J Bacteriol. 1998;180:3166–3173. doi: 10.1128/jb.180.12.3166-3173.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarter L L, Silverman M. Phosphate regulation of gene expression in Vibrio parahaemolyticus. J Bacteriol. 1987;169:3441–3449. doi: 10.1128/jb.169.8.3441-3449.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarter L L, Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990;4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 23.Merkle R K, Poppe I. Carbohydrate composition analysis of glycoconjugates by gas-liquid chromatography/mass spectrometry. Methods Enzymol. 1994;230:1–15. doi: 10.1016/0076-6879(94)30003-8. [DOI] [PubMed] [Google Scholar]
- 24.Meyer T F, Gibbs C P, Haas R. Variation and control of protein expression in Neisseria. Annu Rev Microbiol. 1990;44:451–477. doi: 10.1146/annurev.mi.44.100190.002315. [DOI] [PubMed] [Google Scholar]
- 25.Miyamoto C M, Chatterjee J, Swartzman E, Szittner R, Meighen E A. The role of the lux autoinducer in regulating luminescence in Vibrio harveyi: control of luxR expression. Mol Microbiol. 1996;19:767–775. doi: 10.1046/j.1365-2958.1996.417948.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto C M, Sun W, Meighen E A. The LuxR regulator protein controls synthesis of polyhydroxybutyrate in Vibrio harveyi. Biochim Biophys Acta. 1998;1384:356–364. doi: 10.1016/s0167-4838(98)00028-4. [DOI] [PubMed] [Google Scholar]
- 27.Parsek M R, Greenberg E P. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Methods Enzymol. 1999;310:43–55. doi: 10.1016/s0076-6879(99)10005-3. [DOI] [PubMed] [Google Scholar]
- 28.Patterson H, Irvin R, Costerton J W, Cheng K J. Ultrastructure and adhesion properties of Ruminococcus albus. J Bacteriol. 1975;122:278–287. doi: 10.1128/jb.122.1.278-287.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy G P, Hayat U, Abeygunawardana C, Fox C, Wright A C, Maneval J C R, Bush C A, Morris J J G. Purification and determination of the structure of capsular polysaccharide of Vibrio vulnificus M06-24. J Bacteriol. 1992;174:2620–2630. doi: 10.1128/jb.174.8.2620-2630.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapiro J. Scanning electron microscope study of Pseudomonas putida colonies. J Bacteriol. 1985;164:1171–1181. doi: 10.1128/jb.164.3.1171-1181.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro J A. Organization of developing Escherichia coli colonies viewed by scanning electron microscopy. J Bacteriol. 1987;169:142–156. doi: 10.1128/jb.169.1.142-156.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro J A. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- 33.Shimkets L J. Intercellular signaling during fruiting-body development of Myxococcus xanthus. Annu Rev Microbiol. 1999;53:525–549. doi: 10.1146/annurev.micro.53.1.525. [DOI] [PubMed] [Google Scholar]
- 34.Showalter R, Martin M O, Silverman M R. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J Bacteriol. 1990;172:2946–2954. doi: 10.1128/jb.172.6.2946-2954.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman M, Showalter R, McCarter L. Genetic analysis in Vibrio. Methods Enzymol. 1991;204:515–536. doi: 10.1016/0076-6879(91)04026-k. [DOI] [PubMed] [Google Scholar]
- 36.Simpson L M, White V K, Zane S F, Oliver J D. Correlation between virulence and colony morphology in Vibrio vulnificus. Infect Immun. 1987;55:269–272. doi: 10.1128/iai.55.1.269-272.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanson J. Studies on gonococcus infection XII. Colony color and opacity variants of gonococci. Infect Immun. 1978;19:320–331. doi: 10.1128/iai.19.1.320-331.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanson J. Studies on gonococcus infection. XIV. Cell wall protein differences among color/opacity colony variants of Neisseria gonorrhoeae. Infect Immun. 1978;21:292–302. doi: 10.1128/iai.21.1.292-302.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wai S N, Mizunoe Y, Takade A, Kawabata S-I, Yoshida S-I. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–3655. doi: 10.1128/aem.64.10.3648-3655.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright A C, Simpson L M, Oliver J D, Morris J J G. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect Immun. 1990;58:1769–1773. doi: 10.1128/iai.58.6.1769-1773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yildiz F H, Schoolnik G K. Vibrio cholerae O1 El Tor: Identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci USA. 1999;96:4028–4033. doi: 10.1073/pnas.96.7.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.York W S, Darvill A G, McNeil M, Stevenson T T, Albersheim P. Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol. 1985;118:3–40. [Google Scholar]
- 43.Yoshida S-I, Ogawa M, Mizuguchi Y. Relation of capsular materials and colony opacity to virulence of Vibrio vulnificus. Infect Immun. 1985;47:446–451. doi: 10.1128/iai.47.2.446-451.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]