Abstract
The genes encoding cholera toxin (ctxA and ctxB) are encoded in the genome of CTXφ, a filamentous phage that infects Vibrio cholerae. To study the evolutionary history of CTXφ, we examined genome diversity in CTXφs derived from a variety of epidemic and nonepidemic Vibrio sp. natural isolates. Among these were three V. cholerae strains that contained CTX prophage sequences but not the ctxA and ctxB genes. These prophages each gave rise to a plasmid form whose genomic organization was very similar to that of the CTXφ replicative form, with the exception of missing ctxAB. Sequence analysis of these three plasmids revealed that they lacked the upstream control region normally found 5′ of ctxA, as well as the ctxAB promoter region and coding sequences. These findings are consistent with the hypothesis that a CTXφ precursor that lacked ctxAB simultaneously acquired the toxin genes and their regulatory sequences. To assess the evolutionary relationships among additional CTXφs, two CTXφ-encoded genes, orfU and zot, were sequenced from 13 V. cholerae and 4 V. mimicus isolates. Comparative nucleotide sequence analyses revealed that the CTXφs derived from classical and El Tor V. cholerae isolates comprise two distinct lineages within otherwise nearly identical chromosomal backgrounds (based on mdh sequences). These findings suggest that nontoxigenic precursors of the two V. cholerae O1 biotypes independently acquired distinct CTXφs.
Vibrio cholerae is the etiologic agent of the diarrheal disease cholera. Humans become infected with V. cholerae after ingestion of contaminated food or water. Of the nearly 200 recognized serogroups of V. cholerae, only the O1 and O139 serogroups are associated with epidemics of cholera (27). The V. cholerae O1 serogroup is further divided into the classical and El Tor biotypes on the basis of several phenotypic differences. Since 1817, seven cholera pandemics have been described. The classical biotype is believed to have given rise to the first six cholera pandemics (2). The ongoing seventh pandemic of cholera, which began in 1961, is caused by the El Tor biotype. In 1992, a newly recognized serogroup, O139, emerged and resulted in cholera epidemics in Southeast Asia (14). The emergence of this novel V. cholerae serogroup, along with the re-emergence in 1991 of cholera in South America after a nearly 100-year absence, has renewed interest in the origins and evolution of this pathogen.
Pathogenic V. cholerae isolates colonize the small intestine and secrete cholera toxin (CT), an A-B-type toxin, to cause the profuse secretory diarrhea characteristic of cholera (44). CT is encoded by ctxA and ctxB, which are not integral components of the V. cholerae genome but, instead, reside in the genome of CTXφ, a filamentous bacteriophage that infects V. cholerae, as well as its close relative V. mimicus (8, 18, 48). CTXφ utilizes the V. cholerae type IV pilus TCP, an essential intestinal colonization factor (46), as its receptor (48). In contrast to the well-characterized filamentous bacteriophages derived from Escherichia coli, such as f1, the CTXφ genome integrates into the genome of V. cholerae to form a prophage (48). Integration of CTXφ is site specific (39, 48). However, following infection of classical strains or El Tor strains lacking a CTXφ integration site, the El Tor-derived CTXφ remains extrachromosomal, replicating as a plasmid (48, 49). This plasmid form of CTXφ was designated the phage replicative form (RF), since cells harboring this plasmid produce relatively large amounts of viral particles (48, 49).
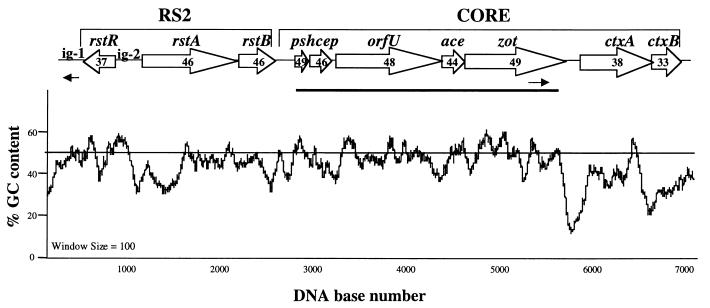
The 6.9-kb CTXφ genome has a modular structure composed of two functionally distinct domains, the core and RS2 regions (Fig. 1) (48). The core region encodes CT and the genes involved in phage morphogenesis, including genes that are thought to encode the major and minor phage coat proteins (Psh, Cep, OrfU, and Ace) and a protein required for CTXφ assembly (Zot) (48). The RS2 region encodes genes required for replication (rstA), integration (rstB), and regulation (rstR) of CTXφ (49). RS2 also contains two intergenic regions, ig-1 and ig-2 (Fig. 1). The RS2 region genes only show sequence similarity to two recently described Vibrio sp.-derived filamentous phages (13, 23). In contrast, the genes of the core region (with the exceptions of ctxA and ctxB) show sequence similarity to the morphogenesis genes of filamentous phages from a range of bacterial species (48). Interestingly, the percent GC contents of the ctxA and ctxB genes, 38 and 33%, respectively (Fig. 1), are significantly different from those of the rest of the core region genes. The distinct GC content of ctxAB, compared to the remainder of the CTXφ genome, suggests that these genes evolved separately from the remainder of the phage genome and that they were acquired after the emergence of a precursor form of CTXφ that lacked ctxAB. In fact, there have been a number of reports in the literature of zot+ V. cholerae isolates lacking ctxAB (15, 31).
FIG. 1.
(Top) Schematic representation of the organization of the El Tor-derived CTXφ genome. Open arrows represent CTXφ ORFs and the direction of transcription of each gene. Numbers within the arrows indicate the genes' percent GC contents. The horizontal bar below the CTXφ map indicates the position of the core probe used in DNA hybridization analysis. The two arrows below the CTXφ map indicate the positions of the PCR primers used to amplify the region between zot and ig-1 from pre-CTXφ RF DNA. (Bottom) Regional variation in the mean proportion of GC content based on a sliding window of 100 nucleotides.
The diversity among CTXφ genomes has only been examined with reference to the RS2 region genes. DNA sequence analysis has revealed that rstA and rstB from CTXETφ CTXclassφ, and CTXcalcφ are highly similar (99% amino acid identity) (16, 29). In contrast, comparisons of rstR sequences among CTXETφ, CTXclassφ, and CTXcalcφ revealed that they were extremely diverse, with less than 30% amino acid sequence similarity (16, 29). Furthermore, the three known rstR genes have a percent GC content (34 to 37%) that is distinct from those of most of the other CTXφ genes (Fig. 1). The hypervariability of rstR and its distinct GC content suggest that CTXφ variants obtained diverse rstR cassettes via horizontal gene transfer, followed by recombination. A similar mechanism is believed to account for the diversity of repressors in lambdoid phages (12). Since recombination, rather than mutation, probably accounts for the distinct CTXφ rstR genes, the diversity of rstR sequences does not provide an insight into the relatedness of distinct CTXφs.
We set out to study the evolution and relatedness of CTXφ variants and to examine whether distinct CTXφ genotypes are associated with each of the V. cholerae O1 biotypes, with different V. cholerae serogroups, or with V. mimicus. During the course of this work, three strains harboring CTX-like prophages that lacked the ctxA and ctxB genes were identified. We hypothesize that these CTX-like prophages lacking ctxAB represent derivatives of the ancestral precursor of CTXφ. Comparative nucleotide sequence analyses of two CTXφ core region genes, orfU and zot, from 13 V. cholerae strains revealed that there are distinct phage lineages in classical and El Tor V. cholerae isolates. These analyses suggest that acquisition of CTXφ by Vibrio spp. has occurred multiple times and has involved several CTXφ genotypes.
MATERIALS AND METHODS
Bacterial strains.
All of the bacterial strains used were cultured in Luria-Bertani (34) broth at 37°C. These strains included 13 V. cholerae and 4 V. mimicus isolates that encompassed seven serogroups (Table 1). These Vibrio sp. isolates were derived from both human and environmental sources with a wide geographic distribution between 1930 and 1993 (Table 1).
TABLE 1.
Strains used for comparative sequence analyses
| Strain | Serogroup (biotype) | Source | Yr | Locality | Reference |
|---|---|---|---|---|---|
| V. cholerae | |||||
| C1 | O1 (classical) | Unknown | 1955 | Unknown | 5 |
| CA401 | O1 (classical) | Clinical | 1953 | India | 20 |
| 569B | O1 (classical) | Clinical | 1948 | India | 33 |
| N16961 | O1 (El Tor) | Clinical | 1975 | Bangladesh | 26 |
| E7946 | O1 (El Tor) | Clinical | 1978 | Bahrain | 33 |
| C5 | O1 (El Tor) | Unknown | 1957 | Indonesia | This study |
| RV79 | O1 (El Tor) | Clinical | 1930 | Vietnam | 33 |
| SG20 | O139 | Clinical | 1993 | India | This study |
| CO130 | O37 | Environmental | 1993 | India | This study |
| V52 | O37 | Clinical | 1968 | Sudan | 50 |
| 151 | O37 | Environmental | 1993 | Mexico | 38 |
| V46 | O141 | Clinical | 1978 | United States | This study |
| 208 | O11 | Clinical | 1993 | Thailand | 38 |
| C325 | O1 | Clinical | 1995 | India | 31 |
| V. mimicus | |||||
| PT5 | O115 | Clinical | 1985 | Bangladesh | 45 |
| PT48 | O115 | Clinical | 1985 | Bangladesh | 45 |
| 523-80 | O115 | Clinical | 1980 | United States | 45 |
| 9583 | O115 | Clinical | 1980 | United States | 45 |
Molecular analyses.
V. cholerae DNA was extracted and purified using the G-nome DNA isolation kit from Bio 101, Inc., Vista, Calif. To assay for the presence of the CTXφ genome among Vibrio isolates, two pairs of primers were used for PCR assays, orfU1 plus zot2 and ctxA1 plus ctxB2, which were designed from published DNA sequences as previously described (8). For Southern hybridization analyses, a CTXφ core region DNA probe spanning the region between psh and zot was used (Fig. 1). Southern hybridization was carried out using horseradish peroxidase-labeled DNA probes, which were prepared and hybridized using the ECL direct labeling and detection system (Amersham Pharmacia, Little Chalfont, Buckinghamshire, England) in accordance with the manufacturer's instructions.
To test for the presence of extrachromosomal DNA corresponding to a precursor CTXφ (pre-CTXφ) lacking ctxAB in strains 151, 208, and C325, Qiaprep Spin kits (Qiagen, Valencia, Calif.) were used to first isolate plasmid DNA from mid-log-phase cultures. Then, Southern blot analysis with a CTXφ core region probe was used to detect CTXφ-related sequences in these plasmid preparations. O395, which contains a defective CTX prophage, and Bah-2 (pCTX-Kn) were used as negative and positive controls, respectively. To test for transduction of the pre-CTXφs, filtered cell-free supernatants from the same mid-log cultures were mixed with agglutinated (TCP+) O395 and grown overnight at 30°C. Then, plasmid DNA was prepared from these cells and the presence of CTXφ-related sequences was assayed for by Southern hybridization as described above.
Nucleotide sequencing.
Sequencing was performed with an Applied Biosystems 373A automated DNA sequencing system using the DyeDeoxy terminator cycle sequencing kit at the Tufts Medical School sequencing facility. For all of the genes analyzed, both DNA strands were sequenced. The BLAST programs (1) were used to compare sequences to those in the GenBank databases. A 715-bp region of orfU and a 708-bp segment of zot were amplified by PCR and sequenced. PCR primers to amplify the chromosomal mdh locus were designed from the mdh sequence of V. cholerae strain N16961. With the forward (5′ atgaaagtcgctgttatt 3′) and reverse (5′ gtatctaacatgccatcc 3′) primers, an 892-bp region of the 939-bp mdh sequence was amplified. PCR products were purified using the Qiaquick (Qiagen) PCR purification kit and subsequently sequenced on both strands. To generate sequencing templates from the three strains that were found to contain CTX-like prophage genomes that lacked ctxAB, the DNA sequences between zot and ig-1 in the RF were amplified with primers zot5 and rig1 (Fig. 1) using plasmid DNA derived from these strains as templates. The forward primer zot5 (5′ gcagtagcctttgactgag 3′) lies within the zot gene, and the reverse primer rig1 (5′ cacgctacgtcgcttatgt 3′) is located within the conserved part of the first intergenic region of CTXφ. The PCR products were cloned into pCR2.1 (Invitrogen, Carlsbad, Calif.), and the resulting plasmids were subsequently used as templates for sequencing.
Phylogenetic analyses.
Regional variation in GC content over the entire length of the CTXφ genome was calculated using a sliding window of 100 nucleotides. The GC content of each CTXφ gene was also calculated using the MacVector program. DNA sequence data were assembled and edited with Eyeball Sequence Editor (11). Gene trees were constructed with MEGA (30). Rates (per site) of synonymous (kS) and nonsynonymous (kN) substitutions were calculated by the methods of Nei and Gojobori (36) and Nei and Jin (37). The proportions of synonymous (pS) and nonsynonymous (pN) substitutions in the orfU and zot genes between pairs of strains were tabulated in a sliding-window analysis of 30 codons along each gene by the program PSWIN (T. S. Whittam, Pennsylvania State University).
Nucleotide sequence accession numbers.
The nucleotide sequences obtained during this study for orfU, zot, and mdh have been deposited in GenBank under accession numbers AF238329 to AF238373.
RESULTS
Identification and analysis of potential CTXφ precursors.
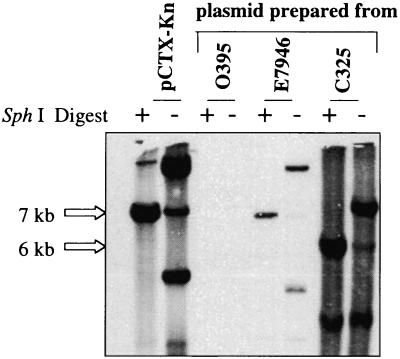
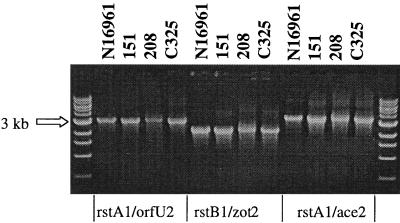
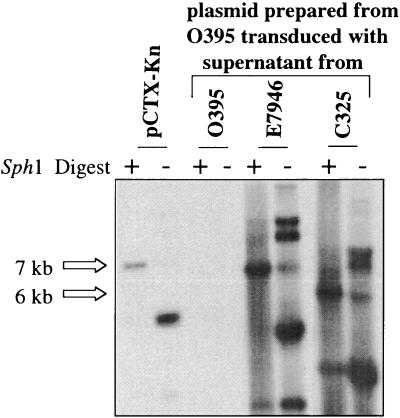
To begin to assess the diversity of CTXφ, we examined our laboratory collection of natural V. cholerae isolates for the presence of CTXφ-related sequences using PCR tests for the presence of orfU, ace, zot, and ctxAB. We identified two isolates, 151 (38) and 208 (38), that contained the CTXφ sequences orfU, ace, and zot but did not encode ctxAB. In addition to these two isolates, we obtained a third V. cholerae strain, C325, that was previously reported to contain CTXφ core region sequences without ctxAB (31). V. cholerae strains 151, 208, and C325 belonged to three serogroups, O11, O37, and O1, respectively, and were isolated in Mexico, Thailand, and India, respectively. Southern blots revealed that the phage structural genes were present within the chromosomal DNAs of these strains (data not shown). We hypothesized that the phage genes might be components of a prophage that could represent a derivative of a precursor of CTXφ (pre-CTXφ) that lacked ctxAB. The distinct GC content of ctxAB relative to the remainder of the CTXφ genome strongly suggests that the toxin genes were acquired subsequent to the development of a pre-CTXφ. To test whether the phage genes are part of functional prophages in these strains, we first examined whether these three strains contained extrachromosomal DNA that corresponded to the RF of the pre-CTXφ DNA. Southern blot analyses of plasmid DNAs prepared from these strains showed the presence of extrachromosomal DNA that hybridized with a core region probe in all three cases (Fig. 2 and data not shown). For C325 and 151, this plasmid was ∼6.0 kb, a size consistent with a CTXφ-like RF that is missing ctxAB. In strain 208, this plasmid was ∼7.0 kb. Furthermore, PCR analyses strongly suggested that the gene content and gene order of core region and RS2 region genes in these three plasmids are identical to those of CTXφ, with the exception of the missing ctxAB genes (Fig. 3). Finally, supernatants derived from all three strains could transmit these pre-CTXφ genomes to a classical recipient strain (Fig. 4 and data not shown). These results indicate that these putative pre-CTXφ prophages give rise to infectious particles. These experiments demonstrate that these three CTX-like prophages lacking ctxAB are functional and are consistent with the hypothesis that CTXφ evolved from a pre-CTXφ lacking ctxAB.
FIG. 2.
Detection of an extrachromosomal form of the pre-CTX prophage from ctxAB strain C325. Plasmid DNA was prepared from C325, classical strain O395 (a negative control), El Tor strain E7946 (a positive control), and Bah-2(pCTX-Kn). Southern hybridization with a core probe was then used to detect either SphI-digested or undigested plasmid DNA.
FIG. 3.
PCR analyses of the genomic organizations of the plasmid form of CTXφ from V. cholerae strain N16961 (El Tor) or from putative pre-CTXφs from V. cholerae strains 151, 208, and C325. The primer pairs used are indicated below the lanes. The left- and rightmost lanes contained molecular size markers.
FIG. 4.
Detection of an infectious form of pre-CTXφ derived from strain C325. Cell-free supernatants from C325, O395 (a negative control), and E7946 (a positive control) were mixed with agglutinated O395. Twenty-four hours later, plasmid DNA was prepared from these three cultures and the presence of CTXφ core genes was determined by Southern blot analysis.
To further characterize these potential pre-CTXφ derivatives and to determine if these three genomes contain another toxin-encoding gene in place of ctxAB, we sequenced the region 3′ of zot in these plasmids. To accomplish this, PCR primers within the 3′ end of zot and a conserved region of ig-1 (Fig. 1) were used to amplify this region from these three plasmids. The three nucleotide sequences of this region were highly similar. A comparison of these sequences with each other and with the corresponding sequence from El Tor strain N16961 is shown in Fig. 5. There is almost complete nucleotide sequence identity at the 5′ end of these four sequences; however, the terminal 60 bp of N16961 zot was highly divergent from the new sequences (28 polymorphic sites).
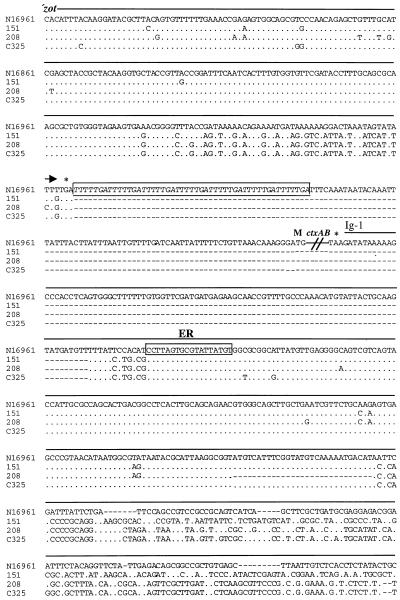
FIG. 5.
Alignment of the nucleotide sequences of zot to ig-1 from V. cholerae strain N16961 (El Tor) and from V. cholerae strains 151, 208, and C325. The latter three sequences were determined from PCR-amplified pre-CTXφ RF DNAs derived from these strains. The arrow above the sequences depicts the 3′ end of the zot ORFs. Dots indicate nucleotide identity, and dashes represent gaps introduced to allow alignment of the CTXφ sequences with the pre-CTXφ sequences. Boxed and italicized nucleotides indicate the repeat sequences that play a role in ToxR binding. The boxed nucleotides designated ER represent the end repeat sequence thought to constitute the core of the CTXφ attachment site. The ctxAB genes of strain N16961 are not shown. M represents the ctxA start codon, and the asterisk designates the stop codons of ctxB and zot. The line above the N16961 sequence beginning after the ctxB stop codon represents the El Tor ig-1 sequence.
The sequences downstream of zot in N16961 also were dissimilar to the new sequences. The heptad direct repeat sequences that facilitate regulation of ctxAB expression by the transcriptional activator ToxR (32, 35, 40) were absent from the 151-, 208-, and C325-derived plasmids, as were the ctxAB promoter and coding sequences (Fig. 5). No open reading frames replaced ctxAB in these plasmids; rather, 3′ of the presumed zot stop codon in these three sequences was a sequence that aligned nearly perfectly with a part of ig-1 from strain N16961. This region of identity began near the 18-bp end repeat sequence that is thought to constitute part of the CTXφ attachment site and extended for 215 bp (49). 3′ of this conserved region, which probably constitutes a critical part of CTXφ attP, these three sequences diverged from the N16961 sequence. However, the N16961 sequence for this region of ig-1 also differs from the ig-1 of classical CTX prophages (29). These data are consistent with a model of CTXφ evolution in which a pre-CTXφ simultaneously obtained ctxAB, cis-acting sequences required for activation of toxin gene expression, and a new carboxyl terminus for Zot. The presence of the 18-bp end repeat sequence that likely corresponds to the core region of the CTXφ attachment site, as well as other parts of the CTXφ attP sequence in the putative pre-CTXφ genomes, suggests that these phages have integration sites and integration mechanisms similar to those of CTXφ.
Diversity of CTX core region sequences.
Since our understanding of the diversity of CTXφs was limited and rested entirely on RS2 region sequences, we compared the sequences of core region genes from a set of Vibrio sp. isolates that we found to harbor CTXφ. This set included 13 V. cholerae strains and 4 V. mimicus strains (Table 1). These strains were chosen to represent the diversity of CTXφs in our laboratory collection based on the dates and sites of their isolation. The sample of V. cholerae included seven O1 serogroup strains (three classical and four El Tor biotype strains). Although classical and El Tor strains are thought to be essentially clonal (3, 10, 28), the similarity of the CTXφs in these strains has not been assessed. The six non-O1 serogroup strains included four serotypes (O139, O37, O141, and O11) and two strains that contained the putative pre-CTXφ derivatives (151 and 208).
To enable comparisons of the diversity of CTXφ sequences with the diversity of a V. cholerae chromosomal sequence, we also performed sequence analyses of mdh, which encodes the metabolic enzyme malate dehydrogenase. We selected mdh because this locus was used previously in several evolutionary studies of a number of enteric bacteria, including V. cholerae, E. coli, and Salmonella enterica (6, 7, 10, 41). These studies demonstrated that phylogenetic trees based on mdh are congruent with analyses of evolutionary relatedness based on other methods and that such trees give a reliable estimate of genotypic divergence. For this study, we analyzed 693 bp of the 936-bp coding region of mdh. Among the 13 V. cholerae and 4 V. mimicus isolates we compared, there were 82 polymorphic sites resulting in nine amino acid replacement substitutions among the V. cholerae and V. mimicus isolates examined (Table 2). The mdh sequence from V. cholerae isolates in pairwise comparisons revealed an average difference of four polymorphic nucleotide sites. Consistent with a recent report (10), the mdh sequences in epidemic V. cholerae isolates were essentially identical. Among V. mimicus strains, the mdh sequence differed, on average, by two polymorphic nucleotide sites. Most of the genetic variation observed at the mdh locus was between the two Vibrio species; thus, pairwise comparisons between V. cholerae and V. mimicus strains yielded an average difference of 68 polymorphic nucleotide sites.
TABLE 2.
Nucleotide sequence variation in the orfU and zot genes of CTXφs derived from V. cholerae and V. mimicus strains
| Gene | No. of sites
|
kS | kN | |||
|---|---|---|---|---|---|---|
| Total | Total polya | Silent polyb | AA polyc | |||
| CTXφ | ||||||
| orfU (V. cholerae only) | 715 | 71 | 44 | 27 | 0.0898 ± 0.0159d | 0.0272 ± 0.0047 |
| orfU (V. mimicus only) | 715 | 58 | 37 | 21 | 0.1040 ± 0.0199 | 0.0279 ± 0.0052 |
| zot (V. cholerae only) | 708 | 32 | 23 | 9 | 0.0407 ± 0.0096 | 0.0051 ± 0.0018 |
| zot (V. mimicus only) | 708 | 12 | 7 | 5 | 0.0244 ± 0.0093 | 0.0052 ± 0.0023 |
| V. cholerae vs V. mimicus | ||||||
| orfU (all strains) | 715 | 72 | 45 | 27 | 0.0993 ± 0.0176 | 0.0289 ± 0.0051 |
| zot (all strains) | 708 | 32 | 23 | 9 | 0.0385 ± 0.0088 | 0.0053 ± 0.0018 |
| Chromosomal | ||||||
| mdh (V. cholerae) | 693 | 13 | 4 | 7 | 0.0229 ± 0.0072 | 0.0013 ± 0.0008 |
| mdh (V. mimicus) | 693 | 4 | 2 | 2 | 0.0071 ± 0.005 | 0.0027 ± 0.0019 |
| V. cholerae vs V. mimicus | ||||||
| mdh (all strains) | 693 | 82 | 73 | 9 | 0.1618 ± 0.0216 | 0.0054 ± 0.0018 |
Total poly, total number of polymorphic sites.
Silent poly, silent polymorphic sites.
AA poly, amino acid polymorphic sites.
kS and kN values are means ± standard deviations.
The nucleotide sequences of 715 bp (codons 46 to 396) of the 1,188-bp coding region of orfU for the 13 V. cholerae strains and 4 V. mimicus strains were similarly compared. In total, there were 72 polymorphic sites within the orfU sequences, which resulted in 27 sites of amino acid replacements (Table 2). Among the 17 orfU sequences determined, there were 8 variant sequences. Interestingly, the orfU sequences were highly variable between the classical and El Tor biotypes but identical within biotypes, with the exception of El Tor strain RV79, a 1930 clinical isolate from Vietnam. The O139 orfU sequence (strain SG20) was identical to the sequences derived from El Tor strains N16961, C5, and E7946. V. mimicus strain PT5 shared orfU sequence identity with these three El Tor strains as well. Comparison of the orfU nucleotide sequences between classical and El Tor V. cholerae isolates revealed 61 polymorphic nucleotide sites resulting in 23 amino acid replacements. The orfU sequences from nonepidemic V. cholerae isolates CO130, V52, V46, 151, and 208 were more similar to the orfU sequences derived from the El Tor isolates, whereas V. mimicus isolates PT48, 523-80, and 9583 shared sequence similarity with orfU from classical V. cholerae isolates.
The nucleotide sequences of a 708-bp segment of zot (codons 54 to 290) of the 1,200-bp coding region of zot were also compared. From the 17 strains examined, there were eight variant zot sequences. Among the eight variant zot sequences, there were a total of 32 polymorphic sites of which 9 resulted in an amino acid replacement (Table 2). Similar to the findings for the orfU gene, the classical biotype V. cholerae strains all had the same zot sequence that was distinct from the El Tor strains' zot sequence. Also, again with the exception of RV79, the El Tor sequences and the O139 sequences were identical. The level of sequence divergence observed at the zot locus was less than that observed at the orfU locus. There were a total of 18 polymorphic sites between the classical and El Tor biotype strains.
For the two CTXφ genes orfU and zot, we estimated the genetic diversity in all pairwise comparisons using the methods of Nei and Gojobori (36) and Nei and Jin (37). The results are summarized in Table 2 along with those from analysis of the chromosomal gene mdh for comparison. The values kS and kN are the average numbers of nucleotide differences per synonymous (silent) site and per nonsynonymous (replacement) site, respectively, among all pairwise comparisons. Table 2 shows that the estimates of CTXφ gene diversity within either V. cholerae or V. mimicus are significantly greater than those calculated for the chromosomal locus mdh. Also, as noted above, orfU sequence diversity was greater than zot sequence diversity.
Spatial distribution of polymorphic sites.
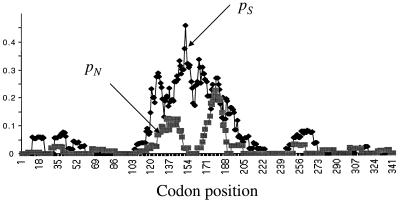
The OrfU protein is thought to be functionally equivalent to pIII of filamentous phages derived from E. coli (22, 48). pIII is a phage coat protein that mediates phage attachment to a host cell. We hypothesized that the significant differences in the OrfU sequences between classical strain- and El Tor strain-derived CTXφs reflect the functional constraints that these two OrfU proteins face in binding to the CTXφ receptor, TCP. The sequence of TcpA, the major subunit of TCP, is known to vary considerably between the two V. cholerae O1 biotypes (24, 42). To begin to address this possibility, we analyzed whether the synonymous and nonsynonymous substitutions within orfU are clustered in the central domain of the genes, which encodes the putative TCP-binding domain of OrfU (21). The results of this analysis, shown in Fig. 6, revealed a striking clustering of synonymous and nonsynonymous site variation in the central portion of orfU. Two distinct peaks of nonsynonymous site variation were present in the central region. We predict that these two orfU hypervariable regions constitute parts of OrfU that are important for OrfU-TcpA interaction. In contrast, similar analysis of the zot sequences did not reveal any clustering of the polymorphic sites.
FIG. 6.
Regional variation in the mean proportion of synonymous (pS) differences between pairs of strains and the mean proportion of nonsynonymous (pN) differences between pairs of strains based on a sliding window of 90 nucleotides in orfU. Squares represent pN values, and diamonds indicate pS values.
Evolutionary relationships among CTXφs and their Vibrio sp. host strains.
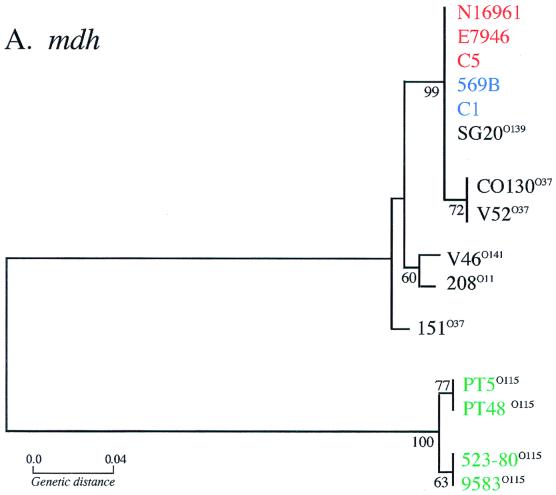
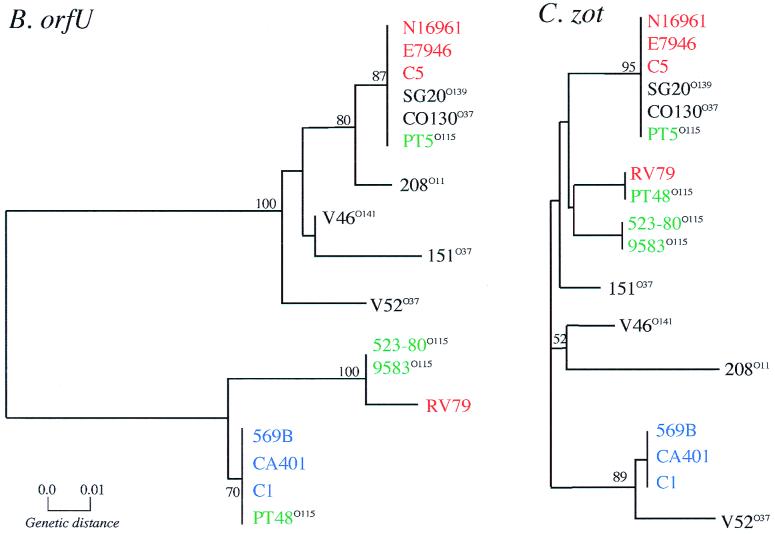
To compare the evolutionary relationships between CTXφs and their Vibrio sp. host strains, the 17 zot, orfU, and mdh sequences were used to construct three phylogenetic trees. These three trees, shown in Fig. 7, were constructed by the neighbor-joining method from a matrix of pairwise genetic distances based on all polymorphic nucleotide sites, with correction for multiple substitutions by the Jukes-Cantor method (25, 43). The most notable feature of this analysis is the divergent clustering of CTXφ genes from classical and El Tor biotype V. cholerae isolates (with the exception of RV79). The orfU and zot sequences derived from strains of the same biotype invariably clustered together (Fig. 7). For the most part, orfU sequences derived from nonepidemic V. cholerae isolates clustered with El Tor orfU sequences. The relationships among the V. mimicus strains based on CTXφ gene trees are very different. V. mimicus strain PT5 clusters with the El Tor strains on both the orfU and zot gene trees. However, V. mimicus strains PT48, 523-80, and 9583 grouped with the classical strains in the orfU tree but with the El Tor strains in the zot gene tree (Fig. 7). These discrepancies are probably due to the low number of polymorphic sites analyzed at the zot locus, as indicated by the low bootstrap values obtained for these nodes in the zot gene tree. Similarly, the limited number of polymorphic sites among the zot sequences probably explains the discrepancy between the orfU and zot gene trees for non-O1 serogroup strains V52, 208, and V46. Another notable feature of Fig. 7 is the lack of congruence of the phage gene trees with the gene tree derived from mdh sequences. The classical and El Tor epidemic V. cholerae isolates are all identical based on mdh sequence analysis, yet they have very divergent CTXφ sequences. This lack of similarity between the phage and chromosomal gene trees is indicative of horizontal transmission of CTXφ, as expected for a mobile genetic element like CTXφ. The clustering of V. mimicus strain PT5-derived CTXφ sequences with El Tor V. cholerae CTXφ sequences and the V. mimicus strain PT48-derived orfU sequence with the classical orfU sequence (Fig. 7) provides additional evidence for the proposition (8) that horizontal transfer of CTXφ between V. cholerae and V. mimicus occurred relatively recently. Similarly, these gene trees suggest that there was a relatively recent transfer of CTXφ between O1 and non-O1 strains of V. cholerae.
FIG. 7.
Evolutionary relationships based on synonymous site variation in the orfU, zot, and mdh genes. The neighbor-joining method was used to construct the trees. The V. cholerae El Tor strains are in red, classical strains are in blue, non-O1 serogroup strains are in black with the particular serogroup in the superscript, and V. mimicus strains are in green. Bootstrap values based on 1,000 computer-generated trees are indicated at the nodes, and only values greater than 50 are shown.
DISCUSSION
CTXφ evolution.
A number of toxins and other virulence factors are encoded within the genomes of bacteriophages (4, 47). In most, if not all, cases, phage-encoded virulence genes are not thought to directly influence the biological properties of the phage. Rather, these genes are believed to be accessories to the bacteriophage genome that can affect the properties of the host cell and thereby potentially indirectly influence the viability of the phage genome. Since neither CT nor its structural genes seem to affect CTXφ functions and since the GC content of ctxAB differs from that of most of the other CTXφ genes, we hypothesized that ctxAB was acquired after the evolution of a pre-CTXφ that lacked these genes.
We identified and analyzed three V. cholerae isolates (151, 208, and C325) that contained several CTXφ core region genes but lacked ctxAB, under the assumption that such strains may contain a derivative of a pre-CTXφ. These three isolates were all found to harbor an extrachromosomal circular DNA molecule whose genomic organizations were very similar to that of the CTXφ RF, with the exception of missing ctxAB, the ToxR binding sites found 5′ of ctxAB, and 173 bp of the ig-1 region 3′ of ctxAB. Since the GC content of ctxAB is distinct from the remainder of the core region, and since the 3′ end of zot in these three plasmids is significantly different than the zot sequence in CTXφ, we believe that these three plasmids are more likely to be derivatives of a pre-CTXφ that never contained ctxAB rather than of CTXφs that have lost ctxAB. If these plasmids (or their prophage forms) do represent derivatives of pre-CTXφs, then it seems probable that the sequences downstream from zot and 5′ of ctxAB were acquired along with ctxAB by a pre-CTXφ. Since ToxR and ToxT are required for ctxAB expression, this suggests that the strain that donated ctxAB to a pre-CTXφ contained these transcriptional activators. The origin of ctxAB and the mechanism of its acquisition by a pre-CTXφ remain matters for speculation. As is the case for several toxin-encoding phages (4), ctxAB is adjacent to the CTXφ attachment site, raising the possibility that an imprecise excision of the pre-CTX prophage generated a new phage that included adjacent chromosomal ctxAB sequences.
We determined the nucleotide sequence of large portions of two CTXφ core region genes, zot and orfU, from 13 V. cholerae and 4 V. mimicus strains in order to study the relatedness of different CTXφs. There were significant differences in these two sequences between the V. cholerae O1 biotypes. However, except for El Tor strain RV79, no differences in these sequences were found in strains of the same biotype that were isolated at different times and locations, reflecting the clonality of the sixth (classical)- and seventh (El Tor)-pandemic strains of V. cholerae. The similarity of RV79-derived CTXφ sequences to CTXφ sequences from classical V. cholerae (Fig. 7) most likely reflects the fact that this El Tor strain was isolated 31 years prior to the onset of the current seventh pandemic of cholera, during a period when the classical biotype was predominant.
The orfU sequences were significantly more diverse than the zot sequences. Also, the polymorphic sites in orfU were clustered in the central region of this gene whereas no clustering was evident in the zot polymorphic sites. We suspect that the clustering of the polymorphisms in orfU developed in response to a high level of selective pressure upon this domain of OrfU, which is predicted to bind CTXφ particles to TCP, the CTXφ receptor on V. cholerae. Interestingly, the amino acid sequence of TcpA, the major subunit of TCP, is known to be significantly different in classical and El Tor V. cholerae isolates (80% amino acid similarity). We therefore predict that the two hypervariable regions (Fig. 6) within OrfU interact directly with TcpA and that the abundant interbiotype polymorphisms in OrfU reflect selective pressures to change in parallel with TcpA, its ligand.
Diverse CTXφs in the evolution of toxigenic Vibrio spp.
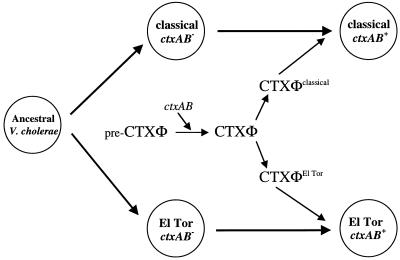
Comparative sequence analyses of the chromosomal mdh locus in CTXφ host strains yields an inferred phylogeny that differs significantly from the inferred phylogeny based on either zot or orfU sequences (compare Fig. 7A with B or C). It was recently reported (10) that epidemic isolates of V. cholerae are very closely related, and our comparisons of mdh sequences confirm this finding. All of the mdh sequences for V. cholerae serogroup O1 epidemic isolates that we analyzed were identical and thus cluster together on the mdh gene tree. However, this was not the case for the two CTXφ gene trees. The phage sequences derived from classical and El Tor strains formed two divergent branches on both the orfU and zot gene trees. This finding of distinct CTXφ lineages within essentially identical V. cholerae chromosomal backgrounds is not consistent with a simple evolutionary scenario of clonal descent, in which a single CTXφ progenitor infected an ancestral V. cholerae isolate and evolved within V. cholerae to the present level of diversity. Rather, it suggests the hypothesis that distinct CTXφs independently infected ctxAB progenitors of the classical and El Tor V. cholerae pandemic strains (Fig. 8).
FIG. 8.
Model for acquisition of ctxAB by the two V. cholerae O1 biotypes. An ancestral V. cholerae isolate gave rise to the classical and El Tor biotypes, which were subsequently independently infected with divergent CTXφs. CTXφ probably arose from a precursor CTXφ that acquired the CT genes by imprecise excision from a unknown donor strain.
We also found that mdh sequences were more variable between V. mimicus and V. cholerae isolates than either the orfU or zot sequences derived from these two Vibrio species (Table 2). V. mimicus mdh sequences and V. cholerae mdh sequences are located on distinct branches of the mdh gene tree, whereas there is no consistent clustering of V. mimicus-derived CTXφ sequences on the zot and orfU gene trees (Fig. 7). This indicates that CTXφ(s) infected V. cholerae and V. mimicus after these two Vibrio species diverged from their most recent common ancestor. As recently suggested (8), the identity of orfU and zot sequences in V. mimicus strain PT5 with the El Tor sequences suggests recent horizontal transfer of the El Tor CTXφ between these two Vibrio species. Likewise, the similarity of the orfU sequences from the other three V. mimicus isolates we studied (PT48, 523-80, and 9583) to the classical orfU sequence suggests that these isolates were infected by a CTXφ closely related to classical CTXφ. Although the classical CTX prophage is thought to be defective in classical V. cholerae isolates, the similarity of orfU in V. mimicus isolates to the classical orfU gene suggests that classical CTXφ was not always defective and that at some time in the past, classical CTXφ infected V. mimicus. However, since the zot sequences from these three V. mimicus isolates do not cluster with the classical zot sequences, it is possible that a distinct CTXφ, perhaps derived from recombination, infected the ctxAB progenitors of these V. mimicus strains.
The orfU and zot sequences analyzed from the six non-O1 V. cholerae strains examined were diverse. In O139 strain SG20, these sequences were identical to the El Tor zot and orfU sequences. Since serogroup O139 V. cholerae is believed to be derived from El Tor V. cholerae via recombination of the locus encoding the serogroup antigen, this sequence identity likely represents vertical inheritance of this phage genome from the El Tor precursor of this newly emerged epidemic serogroup given the known sequence identity of El Tor and O139 V. cholerae at several loci (42). CTXφ sequence identity was also found between serogroup O37 strain CO130 and El Tor isolates. This CTXφ sequence identity probably reflects horizontal transfer of CTXφ between these isolates, given the differences in chromosomal background among these isolates. Since strain CO130 is an environmental rather than a clinical isolate, such horizontal transfer of CTXφ may have occurred outside of human hosts in the aquatic ecosystems that are the natural habitats of V. cholerae. Faruque et al. recently proposed that the natural habitats of V. cholerae may be an important site for the emergence of new toxigenic strains (17). Although the site where CTXφ-mediated transfer of ctxAB occurred is not known, the identity of CTXφ sequences from otherwise diverse O1 and non-O1 V. cholerae strains and V. mimicus isolates strongly suggests that horizontal transmission of CTXφ has occurred relatively recently and that such transmission is an ongoing process that contributes to the emergence of new toxigenic Vibrio species.
ACKNOWLEDGMENTS
We thank our colleagues Andrew Camilli, Brigid Davis, and Bianca Hochhut for critically reading the manuscript. We are grateful to F. Mooi, G. B. Nair, L. Shi, Y. Takeda, and L. Campos for generous donations of bacterial isolates. We thank Anne Kane and the NEMC GRASP Center (grant P30DK-34928) for providing culture media.
This work was supported by the Howard Hughes Medical Institute and NIH grant AI-42347 to M.K.W. E.F.B. was supported by NIH training grant T32 AI-07329 and an Enterprise Ireland basic research grant. M.K.W. is a PEW Scholar of Biomedical Research and a Tupper Research Fellow.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barua B. History of cholera. In: Barua D, Greenough III W B, editors. Cholera. New York, N.Y: Plenum Publishing Corporation; 1992. pp. 1–36. [Google Scholar]
- 3.Beltran P, Delgado G, Navarro A, Trujillo F, Selander R K, Cravioto A. Genetic diversity and population structure of Vibrio cholerae. J Clin Microbiol. 1999;37:581–590. doi: 10.1128/jcm.37.3.581-590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishai W R, Murphy J R. Bacteriophage gene products that cause human disease. In: Calendar R, editor. The bacteriophages. New York, N.Y: Plenum Press; 1988. pp. 683–724. [Google Scholar]
- 5.Bockemuhl J, Meinicke D. Value of phage typing of Vibrio cholerae biotype El Tor in West Africa. Bull W H O. 1976;54:187–192. [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd E F, Wang F-S, Whittam T S, Selander R K. Molecular genetic relationships of the salmonellae. Appl Environ Microbiol. 1996;62:804–808. doi: 10.1128/aem.62.3.804-808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd E F, Nelson K, Wang F-S, Whittam T S, Selander R K. Molecular genetic basis of allelic polymorphism in malate dehydrogenase (mdh) in natural populations of Escherichia coli and Salmonella enterica. Proc Natl Acad Sci USA. 1994;91:1280–1284. doi: 10.1073/pnas.91.4.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyd E F, Moyer K E, Shi L, Waldor M K. Infectious CTXφ and the vibrio pathogenicity island prophage in Vibrio mimicus evidence for recent horizontal transfer between V. mimicus and V. cholerae. Infect Immun. 2000;68:1507–1513. doi: 10.1128/iai.68.3.1507-1513.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyd E F, Waldor M K. Alternative mechanism of cholera toxin acquisition by Vibrio cholerae: generalized transduction of CTXφ by bacteriophage CP-T1. Infect Immun. 1999;67:5898–5905. doi: 10.1128/iai.67.11.5898-5905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byun R, Elbourne L D, Lan R, Reeves P R. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect Immun. 1999;67:1116–1124. doi: 10.1128/iai.67.3.1116-1124.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cabot E L, Beckenbach A T. Simultaneous editing of multiple nucleic acid and protein sequences with ESEE. Comput Appl Biosci. 1989;5:233–234. doi: 10.1093/bioinformatics/5.3.233. [DOI] [PubMed] [Google Scholar]
- 12.Campbell A, Botstein D. Evolution of the lambdoid phages. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 365–380. [Google Scholar]
- 13.Chang B, Taniguchi H, Miyamoto H, Yoshida S. Filamentous bacteriophages of Vibrio parahaemolyticus as a possible clue to genetic transmission. J Bacteriol. 1998;180:5094–5101. doi: 10.1128/jb.180.19.5094-5101.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cholera Working Group. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet. 1993;342:387–390. [PubMed] [Google Scholar]
- 15.Chowdury M A R, Hill R T, Colwell R R. A gene for the enterotoxin zonula occludens toxin is present in Vibrio mimicus and Vibrio cholerae O139. FEMS Microbiol Lett. 1994;119:377–380. doi: 10.1111/j.1574-6968.1994.tb06916.x. [DOI] [PubMed] [Google Scholar]
- 16.Davis B M, Kimsey H H, Chang W, Waldor M K. The Vibrio cholerae O139 Calcutta bacteriophage CTXφ is infectious and encodes a novel repressor. J Bacteriol. 1999;181:6779–6787. doi: 10.1128/jb.181.21.6779-6787.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruque S M, Asadulghani A, Alim A R, Albert M J, Islam K M, Mekalanos J J. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect Immun. 1998;66:3752–3757. doi: 10.1128/iai.66.8.3752-3757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruque S M, Rahman M M, Asadulghani A, Islam K M N, Mekalanos J J. Lysogenic conversion of environmental Vibrio mimicus strains by CTXφ. Infect Immun. 1999;67:5723–5729. doi: 10.1128/iai.67.11.5723-5729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldberg I, Mekalanos J J. Effect of a recA mutation on cholera toxin gene amplification and deletion events. J Bacteriol. 1986;165:723–731. doi: 10.1128/jb.165.3.723-731.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanne L, Finkelstein R A. Characterization and distribution of the hemagglutinins produced by Vibrio cholerae. Infect Immun. 1982;36:209–214. doi: 10.1128/iai.36.1.209-214.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heilpren A J, Waldor M K. CTXφ infection of Vibrio cholerae requires the tolQRA gene products. J Bacteriol. 2000;182:1739–1747. doi: 10.1128/jb.182.6.1739-1747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holliger P, Riechmann L. A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure. 1997;5:265–275. doi: 10.1016/s0969-2126(97)00184-6. [DOI] [PubMed] [Google Scholar]
- 23.Honma Y, Ikema M, Toma C, Ehara M, Iwanaga M. Molecular analysis of a filamentous phage (fs1) of Vibrio cholerae O139. Biochim Biophys Acta. 1997;1362:109–115. doi: 10.1016/s0925-4439(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 24.Iredell J R, Manning P A. Biotype-specific tcpA genes in Vibrio cholerae. FEMS Microbiol Lett. 1994;121:47–54. doi: 10.1111/j.1574-6968.1994.tb07074.x. [DOI] [PubMed] [Google Scholar]
- 25.Jukes T H, Cantor C R. Evolution of protein molecules. New York, N.Y: Academic Press, Inc.; 1969. [Google Scholar]
- 26.Kaper J B, Lockman H, Balini M, Levine M M. Recombinant nontoxinogenic Vibrio cholerae strains as attenuated cholera vaccine candidates. Nature. 1984;308:655–658. doi: 10.1038/308655a0. [DOI] [PubMed] [Google Scholar]
- 27.Kaper J B, Morris J G, Levine M M. Cholera. Clin Microbiol Rev. 1995;8:48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karaolis D K, Lan R, Reeves P R. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J Bacteriol. 1995;177:3191–3198. doi: 10.1128/jb.177.11.3191-3198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimsey H, Waldor M K. CTXφ immunity: application in the development of cholera vaccines. Proc Natl Acad Sci USA. 1998;95:7035–7039. doi: 10.1073/pnas.95.12.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetics analysis, version 1.0. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 31.Kurazano H, Pal A, Bag P K, Nair G B, Karasawa T, Mihara T, Takeda Y. Distribution of genes encoding cholera toxin, zonula occludens toxin, accessory cholera toxin, and El Tor hemolysin in Vibrio cholerae of diverse origins. Microb Pathol. 1995;18:231–235. doi: 10.1016/s0882-4010(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 32.Li C C, Crawford J A, DiRita V J, Kaper J B. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol Microbiol. 2000;35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 33.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 34.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 35.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 36.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 37.Nei M, Jin L. Variances of the average numbers of nucleotide substitutions within and between populations. Mol Biol Evol. 1989;6:290–300. doi: 10.1093/oxfordjournals.molbev.a040547. [DOI] [PubMed] [Google Scholar]
- 38.Novais R C, Coelho A, Salles C A, Vicente A C. Toxin-co-regulated pilus cluster in non-O1, non-toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol Lett. 1999;171:49–55. doi: 10.1111/j.1574-6968.1999.tb13411.x. [DOI] [PubMed] [Google Scholar]
- 39.Pearson G D N, Woods A, Chiang S L, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfau J D, Taylor R K. Genetic footprint on the ToxR-binding site in the promoter for cholera toxin. Mol Microbiol. 1996;20:213–222. doi: 10.1111/j.1365-2958.1996.tb02502.x. [DOI] [PubMed] [Google Scholar]
- 41.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhine J A, Taylor R K. TcpA pilin sequences and colonization requirements for O1 and O139 Vibrio cholerae. Mol Microbiol. 1994;13:1013–1020. doi: 10.1111/j.1365-2958.1994.tb00492.x. [DOI] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Sears C L, Kaper J L. Enteric bacterial toxins: mechanism of action and linkage to intestinal secretion. Microbiol Rev. 1996;60:167–215. doi: 10.1128/mr.60.1.167-215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi L, Miyoshi S, Hiura M, Tomochika K, Shimada T, Shinoda S. Detection of genes encoding cholera toxin (CT), zonula occludens toxin (ZOT), accessory cholera enterotoxin (ACE) and heat-stable enterotoxin (ST) in Vibrio mimicus clinical strains. Microbiol Immunol. 1998;42:823–828. doi: 10.1111/j.1348-0421.1998.tb02357.x. [DOI] [PubMed] [Google Scholar]
- 46.Taylor R K, Miller V L, Furlong D B, Mekalanos J J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci USA. 1987;84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waldor M K. Bacteriophage biology and bacterial virulence. Trends Microbiol. 1998;6:295–297. doi: 10.1016/s0966-842x(98)01320-1. [DOI] [PubMed] [Google Scholar]
- 48.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 49.Waldor M K, Rubin E J, Pearson G D N, Kimsey H, Mekalanos J J. Regulation, replication, and integration functions of the Vibrio cholerae CTXφ are encoded by region RS2. Mol Microbiol. 1997;24:917–926. doi: 10.1046/j.1365-2958.1997.3911758.x. [DOI] [PubMed] [Google Scholar]
- 50.Zinnaka Y, Carpenter C C. An enterotoxin produced by noncholera vibrios. Johns Hopkins Med J. 1972;131:403–411. [PubMed] [Google Scholar]