Abstract
Background
Results regarding whether it is essential to incorporate genetic variants into risk prediction models for esophageal cancer (EC) are inconsistent due to the different genetic backgrounds of the populations studied. We aimed to identify single-nucleotide polymorphisms (SNPs) associated with EC among the Chinese population and to evaluate the performance of genetic and non-genetic factors in a risk model for developing EC.
Methods
A meta-analysis was performed to systematically identify potential SNPs, which were further verified by a case-control study. Three risk models were developed: a genetic model with weighted genetic risk score (wGRS) based on promising SNPs, a non-genetic model with environmental risk factors, and a combined model including both genetic and non-genetic factors. The discrimination ability of the models was compared using the area under the receiver operating characteristic curve (AUC) and the net reclassification index (NRI). The Akaike information criterion (AIC) and Bayesian information criterion (BIC) were used to assess the goodness-of-fit of the models.
Results
Five promising SNPs were ultimately utilized to calculate the wGRS. Individuals in the highest quartile of the wGRS had a 4.93-fold (95% confidence interval [CI]: 2.59 to 9.38) increased risk of EC compared with those in the lowest quartile. The genetic or non-genetic model identified EC patients with AUCs ranging from 0.618 to 0.650. The combined model had an AUC of 0.707 (95% CI: 0.669 to 0.743) and was the best-fitting model (AIC = 750.55, BIC = 759.34). The NRI improved when the wGRS was added to the risk model with non-genetic factors only (NRI = 0.082, P = 0.037).
Conclusions
Among the three risk models for EC, the combined model showed optimal predictive performance and can help to identify individuals at risk of EC for tailored preventive measures.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12885-024-12370-y.
Keywords: Esophageal cancer, Meta-analysis, Single-nucleotide polymorphism, Weighted genetic risk score, Risk prediction model
Background
Esophageal cancer (EC) remains a public health issue globally. EC was the seventh most common cancer in incidence and ranked as the sixth leading cause of cancer-related mortality worldwide in 2020 [1]. In China, new cases of EC and related deaths account for 53.70% and 55.35% of the world’s totals, respectively [1, 2]. Moreover, the overall 5-year survival rate for patients with EC in China remains dismal at only 15–25% [3]. Like other cancers, early diagnosis can contribute to a dramatically improved 5-year survival rate for patients with EC [4]. Epidemiological studies have shown that relevant variables, such as smoking and alcohol consumption, are risk factors for EC, and striking sex and age disparities also exist [5, 6]. In addition, the existence of various genetic variants is closely associated with susceptibility to EC [7, 8].
To improve early detection of EC, a promising approach is to establish a risk prediction model that incorporates well-recognized risk factors to identify high-risk individuals in advance. Furthermore, ethnic differences in either genetic factors or histologic subtypes deserve full consideration. EC includes esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). In China, ESCC is predominant.
As an effective tool to improve risk stratification, risk prediction models have been developed based on a combination of genetic and non-genetic factors for various malignancies, such as breast cancer [9] and colorectal cancer [10]. In 2008, Yokoyama et al. [11] constructed a prediction model for EC by incorporating a single-nucleotide polymorphism (SNP) and four individual risk factors. The results showed that compared with conventional screening protocols, the positive predictive value of endoscopy for the top 10% of risk in the model was increased by approximately 1.7%. However, one SNP cannot adequately represent the genetic variants related to EC, and the study was conducted only in the Japanese male population. In addition, Chang et al. [12] developed a prediction model for ESCC in Chinese population by including 25 SNPs and 4 non-genetic factors. However, inclusion of a large number of SNPs hampers cost-effectiveness. In 2018, Dong et al. [13] developed a risk model for EAC among people of European ancestry by including 23 genetic variants and several epidemiologic factors. The conclusions of these studies regarding whether it is essential to incorporate genetic factors into risk models for EC were inconsistent due to the different genetic backgrounds of the populations included. To the best of our knowledge, studies including genetic variants in risk prediction models for EC are still limited for the Chinese population to date. Genetic predisposition, as a well-established risk indicator of EC, warrants further research to clarify its value in predicting the risk of developing EC [14].
In this study, a meta-analysis was performed to comprehensively identify potential SNPs that may predispose individuals to EC in Chinese population. A case-control study was carried out to verify the associations of these SNPs with EC, followed by construction of risk prediction models based on a panel of well-established risk factors and promising SNPs to provide an effective tool for identifying individuals at high risk.
Methods
Meta-analysis for selecting candidate SNPs
The meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement.
Search strategy
To identify SNPs related to EC, a comprehensive literature search was performed using the following online databases up to July 1, 2020: PubMed, EMBASE, Web of Science, Cochrane Library, CNKI (Chinese), WanFang (Chinese), and CBM (Chinese). The following search terms were used: (risk factors) AND (esophageal OR esophagus) AND (neoplasm OR cancer OR tumor OR neoplastic OR carcinoma OR adenocarcinomas OR malignancy OR malignancies OR neoplasia) AND (single nucleotide polymorphism OR SNP OR variant OR variation OR polymorphism) AND (Chinese OR China).
Inclusion and exclusion criteria
The eligibility criteria were as follows: (1) studies on associations between SNPs and EC risk; (2) studies for which odds ratios (ORs) and 95% confidence intervals (CIs) were available; (3) studies for which the genotype distribution in the controls was in accordance with Hardy-Weinberg equilibrium (HWE); and (4) case-control or cohort-designed study. The exclusion criteria were as follows: (1) not original studies (reviews, meta-analyses, letters, and abstracts); (2) fewer than three studies for one SNP; (3) studies for which the sample size of cases or controls was less than 10; and (4) studies for which the minor allele frequency was less than 1% in the control group. For studies based on the same population, we selected only the study with the most informative data.
Data extraction and quality assessment
The following data were extracted independently by two authors: the first author, year of publication, study region, cancer type, gene, SNP, distribution of genotypes in case and control groups, type of controls, genotyping method, and quality control. Any discrepancies were resolved through discussion with a third investigator. The Newcastle-Ottawa Scale (NOS) was used to evaluate the quality of the studies. We rated the quality as 0–9, with scores of 5–6 and 7–9 being judged to represent moderate and high quality, respectively.
A case-control study for verifying candidate SNPs
Subjects
In total, 500 EC patients and 500 controls were enrolled for the current study. All cases were obtained from a third-level grade A hospital in Henan Province, China, in 2018 and confirmed by pathology reports. Controls were randomly selected from participants in a cardiovascular disease epidemiological survey simultaneously conducted in Henan Province and were frequency-matched to cases by sex. The exclusion criteria for patients and controls were as follows: (1) patients with EC who had a history of another tumor; (2) controls who experienced health problems, including tumors and esophagus-related diseases.
Basic information of the subjects with EC was retrieved from clinical records, and the controls were administered a professionally designed questionnaire that assessed information regarding non-genetic factors. Individuals who had smoked at least one cigarette every 1–3 days for more than six months were considered smokers. Individuals who had drunk alcohol at least once a week for more than six months were considered drinkers. This study was approved by the Institutional Review Board of Zhengzhou University, and all participants provided informed consent.
Genotyping and quality control
A GeneJET Whole Blood Genomic DNA Purification Mini Kit was used to extract DNA. Improved multiplex ligation detection reaction (iMLDR™) was used to genotype SNPs in the case group. ABI3730XL sequencer (AppliedBiosystems, U.S.A) and GeneMapper 4.0 were used for sequencing and identification of genotypes, respectively. Genotyping in the control group were performed via DNA sequencing. All DNA samples were successfully genotyped.
For quality control, agarose gel electrophoresis was applied for each sample before genotyping. The quality of genotyping was assessed by using negative quality control and repeated genotyping of 3% of the samples randomly selected. Moreover,10% of the samples in the case group were further genotyped by using DNA sequencing to verify the concordance of the two methods.
Construction of risk prediction models for esophageal cancer
Data were randomly split into a training set (60%, 301 cases and 299 controls) for developing risk prediction models and a verification set (40%, 199 cases and 201 controls) for evaluating the resulting models.
Three models containing different variables were developed: a genetic model with genetic markers only; a non-genetic model fit with environmental risk factors, including smoking, alcohol consumption, and family history of esophageal cancer; and a combined model including both genetic and non-genetic predictors.
Promising SNPs verified in the case-control study were utilized to calculate the weighted genetic risk score (wGRS). The genetic model was then constructed using this wGRS [15]. Logistic regression was employed to develop non-genetic and combined models.
The wGRS is estimated as follows:
The genetic score of single SNP was calculated based on the OR of the risk allele and the frequency of genotype in Chinese population (Chinese Han in Beijing, CHB).
Genetic score (W) = (1-p)2+2p(1-p)OR + p2OR2 (p is the risk allele frequency).
AA = 1/W; AB = OR/W; BB = OR2/W (A is the non-risk allele; B is the risk allele; AA, AB, and BB refer to the SNP genotype).
wGRS = SNP1×SNP2×SNP3×SNP4……SNPn (Missing value set to 1).
Statistical analysis
In the meta-analysis, ORs with 95% CIs were used for assessment of associations between genetic variants and EC risk. Statistical heterogeneity was evaluated by means of the Cochran Q-test and I2 statistic. A fixed-effects model (Mantel-Haenszel) was applied if the P value was ≥ 0.10 or I2 was ≤ 50%; otherwise, a random-effects model (DerSimonian-Laird) was applied. Begg’s test and Egger’s test were conducted to examine publication bias.
Unconditional logistic regression was performed to evaluate associations between genetic variants and EC risk in this case-control study. The chi-square test of goodness of fit was employed to analyze whether the distribution of genotypes in the control group matched HWE. For significant SNPs, the false-positive report probability (FPRP) was calculated to verify the authenticity of the summary results [16, 17]. The default value of the FPRP critical value was 0.5, and the prior probabilities were set to 0.25, 0.1, and 0.01. The attributable risk percentage (ARP) and population attributable risk percentage (PARP) were calculated to evaluate the epidemiological effect of each SNP.
Receiver operating characteristic (ROC) curves and the net reclassification index (NRI) were utilized to evaluate the discrimination of the different models with the area under the ROC curve (AUC), sensitivity, specificity, positive likelihood ratio, negative likelihood ratio, and accuracy rate. Comparison of AUCs was further performed by using DeLong’ test [18]. The Akaike information criterion (AIC) and Bayesian information criterion (BIC) were adopted to determine the goodness-of-fit of the models.
R software (version 4.2.2), MedCalc (version 20.027), SPSS (version 26.0), and Stata statistical software (version 15.1) were used in this study. Statistical significance was determined at α = 0.05, and all P values for statistical significance were two-sided.
Results
Main findings from the meta-analysis
The screening procedure is summarized in online Additional file 1: Figure S1. After duplicate exclusion (n = 2 865), title or abstract screening (n = 3 336), and full-text review (n = 336), a total of 100 articles (149 SNP-related studies) were ultimately included in the subsequent analysis (online Additional file 3: Supplementary References). If two populations or SNPs were present in one article, we considered it to be two independent studies. The studies included 48 654 cases and 58 373 controls, involving 29 SNPs located in 22 genes. The number of datasets for each SNP ranged from 3 to 11, with the most widely studied SNP being ALDH2 rs671. More details of the SNPs are provided in online Additional file 2: Table S1 and Additional file 1: Figure S2.
Twelve SNPs significantly decreased or increased the risk of EC (P53 rs1042522, CYP1A1 rs1048943, ADH1B rs1229984, ERCC2 rs13181, NQO1 rs1800566, MMP13 rs2252070, PLCE1 rs2274223, CDKN1A rs2395655, CYP2E1 rs3813867, TERT rs401681, CYP1A1 rs4646903, and IL23R rs6682925) (online Additional file 2: Table S2). Specifically, six SNPs (CYP1A1 rs1048943, ADH1B rs1229984, ERRCC2 rs13181, MMP13 rs2252070, PLCE1 rs2274223, and CYP2E1 rs3813867) were significant under all 5 genetic models. The most significant association with EC risk was observed for CYP1A1 rs1048943 under the homozygous model (OR = 2.44, 95% CI: 1.79 to 3.33). For the 12 significant SNPs indicated above, FPRP was the best for 12/12, 12/12 and 9/12 at the 0.25, 0.1 and 0.01 levels, respectively (online Additional file 2: Table S3), which suggests that the findings are relatively reliable. The top three SNPs for ARP were CYP2E rs3813867 (65.87%), CYP1A1 rs1048943 (59.02%), and ADH1B rs1229984 (55.16%). Moreover, the top three SNPs for PARP in the controls and CHB were the same as those for ARP. PARP for each SNP between the control group and CHB was similar, suggesting the controls to be representative (online Additional file 2: Table S4). Additionally, the findings from publication bias assessments provided little indication of publication bias except for ERCC2 rs13181 and NQO1 rs1800566.
Characteristics of the population
The detailed characteristics of the study subjects are shown in Table 1. There was no significant difference in sex between the patients and control subjects because of the frequency-matched design. The mean age was significantly older in the case group (63.00 ± 8.33) than in the control group (46.80 ± 11.55). As expected, compared with the controls, the EC patients were more likely to smoke, drink alcohol, and have a family history of esophageal cancer.
Table 1.
Baseline characteristics of participants in the case-control study
| Variables | Cases | Controls | t/χ2 | P value |
|---|---|---|---|---|
| (n = 500) | (n = 500) | |||
| Age, y, (Mean ± SD) | 63.00 ± 8.33 | 46.80 ± 11.55 | 25.418 | < 0.001 |
| Sex, n (%) | ||||
| Male | 363(72.6) | 363(72.6) | < 0.001 | 1.000 |
| Female | 137(27.4) | 137(27.4) | ||
| Smoking status, n (%) | ||||
| Yes | 222(44.4) | 173(34.6) | 10.047 | 0.002 |
| No | 278(55.6) | 327(65.4) | ||
| Drinking status, n (%) | ||||
| Yes | 194(38.8) | 148(29.6) | 9.403 | 0.002 |
| No | 306(61.2) | 352(70.4) | ||
| Family history of esophageal cancer, n (%) | ||||
| Yes | 78(15.6) | 10(2.0) | 57.616 | <0.001 |
| No | 422(84.4) | 490(98.0) |
NOTE: χ2 test was performed for categorical variables and Student t test was for continuous variables. SD: standard deviation
Evaluation and verification of SNPs in the case-control study
A total of 14 SNPs were evaluated, including 12 SNPs identified in the previous meta-analysis and another two SNPs, namely, MTHFR rs1801133 and ALDH2 rs671, included in a large number of studies in the above meta-analysis and considered to be significant in reviews [19, 20]. Finally, five promising SNPs (P53 rs1042522, MTHFR rs1801133, PLCE1 rs2274223, ALDH2 rs671, and ADH1B rs1229984) were validated as EC susceptibility loci (online Additional file 2: Table S5 and Table S6). For P53 rs1042522 and ADH1B rs1229984, the best-fitting genetic model was recessive and the ORs were 0.69 (95% CI: 0.48 to 1.00) and 1.78 (95% CI: 1.08 to 2.94), respectively. For MTHFR rs1801133, PLCE1 rs2274223, and ALDH2 rs671, the best-fitting genetic model was the dominant model, with ORs of 0.41 (95% CI: 0.26 to 0.65), 1.93 (95% CI: 1.37 to 2.71), and 2.42 (95% CI: 1.65 to 3.56), respectively. For these 5 promising SNPs, FPRP was the best for 5/5, 5/5 and 3/5 at the 0.25, 0.10 and 0.01 levels, respectively (online Additional file 2: Table S7).
Weighted genetic risk score (wGRS)
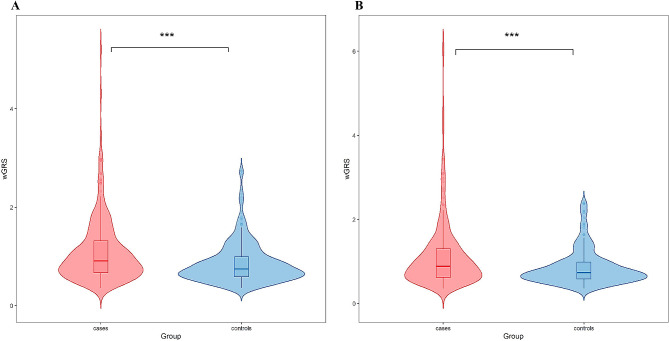
Details regarding the calculation of wGRS are described in online Additional file 2: Table S8. The wGRS was significantly greater in the patients than in the controls (Fig. 1). Next, we assessed the association between wGRS and EC risk (based on the quartile distribution in the controls), and found increased ORs across quartiles of wGRS (P for trend < 0.001 in the training set; P for trend = 0.005 in the validation set). The results showed that in the training set, individuals in only the highest quartile of wGRS had a 4.93-fold (95% CI: 2.59 to 9.38) increased risk of EC compared with those in the lowest quartile. In the validation set, a significantly increased risk was also observed only for the highest quartile (OR = 3.12, 95% CI: 1.53 to 6.36) (Table 2).
Fig. 1.
The distribution of wGRS in the case and control groups. (A) in the training set; (B) in the validation set. wGRS, weighted genetic risk score. ***P < 0.001
Table 2.
Association of wGRS with the risk of esophageal cancer
| wGRS | Training set | Validation set | ||||
|---|---|---|---|---|---|---|
| OR (95%CI) | P value | P for trend | OR (95%CI) | P value | P for trend | |
| Lowest | 1.00(Reference) | - | 1.00(Reference) | - | ||
| Second | 1.59(0.79,3.19) | 0.194 | 1.01(0.46,2.23) | 0.974 | ||
| Third | 1.50(0.77,2.91) | 0.235 | 1.45(0.68,3.11) | 0.335 | ||
| Highest | 4.93(2.59,9.38) | < 0.001 | < 0.001 | 3.12(1.53,6.36) | 0.002 | 0.005 |
NOTE: Based on the quartile distribution in the controls. Adjusted for age, smoking, alcohol consumption, and family history of esophageal cancer. wGRS, weighted genetic risk score. Training set: 301 cases and 299 controls; Validation set: 199 cases and 201 controls
Construction and evaluation of risk prediction models
In the training set, the genetic model was constructed based on wGRS. The equation of the non-genetic model was as follows: Y1 = 1/(1 + EXP(-(-0.236-0.584 × 1 + 2.038 × 2 + 1.392 × 3))) (X1, smoking; X2, family history of esophageal cancer; X3, the interaction of smoking and alcohol consumption). The combined model was expressed as follows: Y2 = 1/(1 + EXP(-(-1.110 + 0.908 × 1-0.558 × 2 + 1.976 × 3 + 1.393 × 4))) (X1, wGRS; X2, smoking; X3, family history of esophageal cancer; X4, the interaction of smoking and alcohol consumption) (online Additional file 2: Table S9).
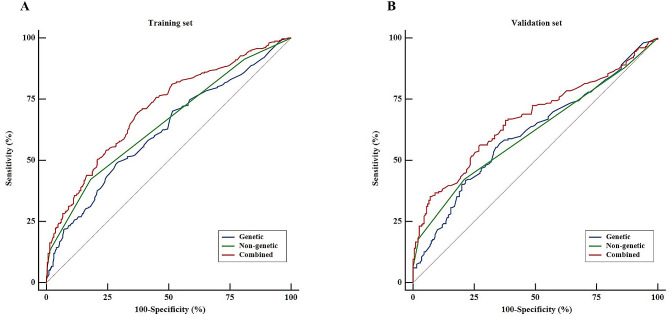
We evaluated the discriminative ability of the models. The non-genetic model achieved moderate accuracy in distinguishing EC patients from controls, with an AUC of 0.650 (95% CI: 0.610 to 0.688). The model containing the wGRS alone had a relatively lower AUC of 0.618 (95% CI: 0.578 to 0.657). When comparing the two AUCs, no statistical significance was found (Delong’s test, P = 0.301). However, with the addition of wGRS, the AUC for the non-genetic model significantly increased from 0.650 to 0.707 (Delong’s test, P < 0.001). Overall, the combined model was superior to the other models with genetic or non-genetic parameters alone (Fig. 2; Tables 3 and 4). As shown in Table 3, the combined model had a sensitivity of 69.44%, a specificity of 62.88%, and an accuracy of 66.17%.
Fig. 2.
Receiver operating characteristic curves for risk prediction models of esophageal cancer. (A) the three models were constructed in the training set; (B) these models were verified in the validation set
Table 3.
Evaluation of predictive performance and goodness of fit of risk prediction models
| Indicators | Training set | Validation set | ||||
|---|---|---|---|---|---|---|
| Genetic | Non-genetic | Combined | Genetic | Non-genetic | Combined | |
| AUC | 0.618 | 0.650 | 0.707 | 0.609 | 0.612 | 0.669 |
| AUC 95%CI | (0.578,0.657) | (0.610,0.688) | (0.669,0.743) | (0.559,0.657) | (0.563,0.660) | (0.620,0.715) |
| Youden index | 0.201 | 0.241 | 0.323 | 0.215 | 0.213 | 0.289 |
| Sensitivity (%) | 48.50 | 42.19 | 69.44 | 56.78 | 42.21 | 56.28 |
| Specificity (%) | 71.57 | 81.94 | 62.88 | 64.68 | 79.10 | 72.64 |
| Accuracy (%) | 60.00 | 62.00 | 66.17 | 60.75 | 60.75 | 64.50 |
| Positive likelihood ratio | 1.71 | 2.34 | 1.87 | 1.61 | 2.02 | 2.06 |
| Negative likelihood ratio | 0.72 | 0.71 | 0.49 | 0.67 | 0.73 | 0.60 |
| AIC | 805.41 | 776.78 | 750.55 | 538.84 | 530.50 | 511.80 |
| BIC | 814.21 | 785.58 | 759.34 | 546.82 | 538.49 | 519.78 |
NOTE: The genetic model was based on wGRS; the non-genetic model included non-genetic factors which were seen in the text; the combined model included both wGRS and non-genetic factors. AUC: area under the curve; AIC: Akaike information criterion; BIC: Bayesian information criterion; wGRS: weighted genetic risk score
Table 4.
Comparison of different esophageal cancer risk prediction models
| Model comparison | Difference of AUC | Za | P valuea | NRI | Zb | P valueb |
|---|---|---|---|---|---|---|
| Training set | ||||||
| Genetic vs. non-genetic | 0.032(-0.028,0.091) | 1.034 | 0.301 | 0.041 | 0.755 | 0.450 |
| Genetic vs. combined | 0.089(0.048,0.130) | 4.256 | < 0.001 | 0.122 | 3.234 | 0.001 |
| Non-genetic vs. combined | 0.058(0.030,0.086) | 4.067 | < 0.001 | 0.082 | 2.082 | 0.037 |
| Validation set | ||||||
| Genetic vs. non-genetic | 0.004(-0.072,0.079) | 0.091 | 0.927 | -0.002 | 0.020 | 0.984 |
| Genetic vs. combined | 0.060(0.007,0.113) | 2.223 | 0.026 | 0.075 | 1.214 | 0.225 |
| Non-genetic vs. combined | 0.057(0.021,0.092) | 3.084 | 0.002 | 0.076 | 2.128 | 0.033 |
NOTE: The difference of AUC was analyzed using Delong’s test. NRI, net reclassification improvement.
a represents the z-statistic and P value from Delong’s test;
b represents the z-statistic and P value from NRI analysis
Based on the NRI, the prediction effect of the combined model was significantly greater than that of the model with non-genetic parameters alone in both the training and validation sets (training set: NRI = 0.082, P = 0.037; validation set: NRI = 0.076, P = 0.033). When comparing the combined and genetic models, the NRI significantly improved only in the training set (training set: NRI = 0.122, P = 0.001; validation set: NRI = 0.075, P = 0.225) (Table 4). According to the AIC and BIC, the combined model was selected as the best fitting model (AIC = 750.55, BIC = 759.34) (Table 3). Overall, the model incorporating both genetic and non-genetic factors showed optimal predictive performance.
The predictive performance of these models was then evaluated by using another independent validation set. A similar discrimination ability was observed, which indicated that the models had rosy stability.
Discussion
In this study, a meta-analysis approach was used to identify potential SNPs related to EC risk in Chinese population, and a case-control study was designed to verify the associations of these SNPs with EC risk. A total of three models were effectively constructed and evaluated. The results suggested that the combined model was preferable to the other models, which further supports that the addition of multiple genetic variants may provide reliable value in EC risk prediction.
In the meta-analysis, although the results of some SNPs were consistent with those of previous meta-analyses or a genome-wide association study (GWAS) [21, 22], there were some inconsistencies [23, 24]. For instance, a previous meta-analysis [24] revealed that CYP1A1 rs4646903, which was significant in our meta-analysis, may not affect susceptibility to EC in Asian populations, while another meta-analysis [22] revealed that this statistically increasing risk was observed in the population from North China. These discrepant findings may be partly explained by differences in genetic susceptibility and environmental risk factors among diverse populations. Thus, to lessen the influence of different genetic backgrounds, our meta-analysis was conducted only in Chinese population.
Among the five promising SNPs used in the models, ADH1B rs1229984 and ALDH2 rs671 are involved in ethanol metabolism [25, 26]. The rs1229984 C allele and rs671 A allele can result in accumulation of acetaldehyde [27, 28]. Among individuals with a combination of the two risk alleles, the level of N2-ethylidene-dG in the DNA of leukocytes from alcoholics was significantly increased, which enhanced DNA damage, leading to an elevated risk of EC [29]. The rs2274223 polymorphism in PLCE1 affects esophageal carcinogenesis by enhancing the inflammatory response and upregulating phospholipase C epsilon mRNA, protein, and enzyme activity [30]. In addition, for P53 rs1042522 and MTHFR rs1801133, associations with EC risk may vary among different populations. The vital polymorphism P53 rs1042522, encoding proline or arginine, is located at codon 72 of exon 4 [31]. Several studies have reported an approximately twofold increase in the risk of EC in individuals with the rs1042522 CC genotype [32, 33], while other studies have shown that the GG genotype was a risk marker for human papillomavirus-associated EC [34, 35]. In our case-control study, the rs1042522 CC genotype reduced the risk of EC. Peng et al. also provided evidence that the CC genotype might be a risk factor for EC susceptibility in southern China but not in northern China [36]. Moreover, some studies have shown that the MTHFR rs1801133 TT genotype can increase the risk of EC [37, 38], while the rs1801133 T allele was showed to decrease EC risk in another study conducted in Henan Province, China [39]. There are several possible explanations for these different findings. The gene product of MTHFR is a central enzyme involved in folate metabolism, and the level of folate intake may influence the risk of EC associated with this polymorphism [40, 41]. In another study [42], the rs1801133 polymorphism increased EC risk, but the association disappeared after stratification by folate consumption. Additionally, the frequency of rs1801133 also differs by ethnicity [40].
Previous risk prediction models for EC were mostly based on non-genetic factors [14, 43–47], and easy-to-obtain variables were included in a standardized manner without any extra costs. However, for such a complex etiological disease, the actual predictive efficacy of environmental factors alone has not been completely established. In terms of numerous genetic variants, many studies [48, 49] on other cancers have reported that the predictive ability improved after adding genetic information to a model developed with non-genetic factors. For EC risk, Chang et al. [12] calculated the wGRS through the use of 25 SNPs and added the wGRS to the model with 4 non-genetic factors (sex, age, smoking status, and drinking status), with an elevated AUC ranging from 0.639 to 0.709. In another study, Dong et al. [13] used 23 GWAS-based SNPs to generate polygenic risk score (PRS) and found that individuals in the highest quartile had a more than 2-fold greater risk of developing EAC than those in the lowest quartile. However, Dong et al. noted that adding the PRS to a risk prediction model with non-genetic factors did not greatly improve its clinical use. Given that genetic predisposition is widely recognized as a well-established risk factor for EC, we constructed and evaluated risk prediction models with various combinations of genetic or non-genetic factors. Our findings provide supporting evidence that the addition of genetic predisposition significantly enhances performance in predicting EC risk.
There are several strengths of this study. First, meta-analysis was applied to comprehensively screen SNPs only in the Chinese population, avoiding the influence of different ethnicities. Second, the number of SNPs included in our risk models was relatively less than that in previous studies incorporating genetic variants [12, 13], which can improve cost-effectiveness. Third, when assessing whether there was improvement in discrimination after adding a new promising maker, the NRI was used to evaluate the degree of prediction increment in addition to AUC. The NRI especially focuses on the change in the number of individuals correctly discriminated by the new model compared to the old model, which can help to optimize limited resources. The goodness-of-fit of the model was evaluated using the AIC and BIC. After examining both the goodness-of-fit and predictive ability, the combined model was ultimately considered the optimal model in our study. Furthermore, the environmental risk predictors included in our models, such as smoking and alcohol consumption, were modifiable, which could enhance the awareness of adherence to healthy lifestyles.
Nevertheless, several limitations merit consideration. First, as mentioned previously, different genotyping methods were used in the case and control groups, which may bias the results. However, to minimize this bias, 10% of the samples from the cases were further genotyped by DNA sequencing, which was used for the controls, with consistent results. Second, external validation of our models was not conducted given the limited data availability, which included genetic data available outside of the present study. Third, genetic variants display regional and population differences, and our study constructed the wGRS associated with the risk of EC in the Chinese population through a case-control study, which may weaken the generalization of this wGRS to other racial or ethnic groups. Moreover, we must note that other related effect modifiers, such as the consumption of hot food and preserved vegetables, were not taken into consideration in our models; as such, they were not available for the study population. To address these issues, more comprehensive investigations should be performed when data are available.
Conclusions
In summary, three risk prediction models were developed based on various combinations of the wGRS or environmental risk factors. The results indicated that the combined model including both genetic and non-genetic factors showed the optimal predictive performance for EC risk, which can help to identify individuals with an increased risk of EC for individualized prevention from early stages in life. Further studies on external validation and cost effectiveness are needed to verify the practical feasibility of the model.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Additional file 1: Figure S1. Flow chart of literature selection in the meta-analysis. Figure S2. Distribution of studies included in mete-analysis by province in China. (docx)
Additional file 2: Table S1. SNPs identified from the meta-analysis. Table S2. Associations of genetic variants with esophageal cancer risk in the meta-analysis. Table S3. Heterogeneity test and evaluation of reliability for genetic variants significantly associated with esophageal cancer risk. Table S4. Epidemiological effect estimation for the relationship between genetic variants and esophageal cancer. Table S5. Associations of 14 candidate SNPs with risk of esophageal cancer in the case-control study. Table S6. Associations of genotypes of 14 candidate SNPs with esophageal cancer risk. Table S7. False positive report probability of 5 promising SNPs. Table S8. Risk score for each promising SNP. Table S9. Construction of non-genetic and combined models. (docx)
Additional file 3: Supplementary References. (docx).
Acknowledgements
Authors sincerely thank all the participants in our study. Authors thank Dr. Jianying Zhang for his support and helpful comments for this study.
Abbreviations
- EC
Esophageal cancer
- ESCC
Esophageal squamous cell carcinoma
- EAC
Esophageal adenocarcinoma
- SNP
Single-nucleotide polymorphism
- HWE
Hardy-Weinberg equilibrium
- OR
Odds ratio
- CI
Confidence interval
- wGRS
Weighted genetic risk score
- FPRP
False-positive report probability
- ARP
Attributable risk percentage
- PARP
Population attributable risk percentage
- ROC
Receiver operating characteristic
- NRI
Net reclassification index
- AUC
Area under ROC curve
- AIC
Akaike information criterion
- BIC
Bayesian information criterion
- CHB
Chinese Han in Beijing
- GWAS
Genome-wide association study
- PRS
Polygenic risk score
Author contributions
PW provided the design of the study. HL and KL performed the literature search and data extraction. PW, JX and HY confirmed the authenticity of all the raw data. HL and KL conducted experiments and analyzed data. HL wrote the first draft of the manuscript. KL, JX, JZ, YC, XZ, PW and HY revised the manuscript and assisted in analyzing the data in this study. All authors read and approved the final manuscript.
Funding
This work was supported by the Zhengzhou Major Project for Collaborative Innovation (18XTZX12007).
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participant
This study was approved by the Institutional Review Board of Zhengzhou University and all the participants signed informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and Mortality Worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin Med J (Engl) 2021;134(7):783–91. doi: 10.1097/CM9.0000000000001474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Huang B, Xu MC, Pennathur A, Li Z, Liu Z, Wu Q, Wang J, Luo K, Bai J, Wei Z, et al. Endoscopic resection with adjuvant treatment versus esophagectomy for early-stage esophageal cancer. Surg Endosc. 2022;36(3):1868–75. doi: 10.1007/s00464-021-08466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islami F, Fedirko V, Tramacere I, Bagnardi V, Jenab M, Scotti L, Rota M, Corrao G, Garavello W, Schüz J, et al. Alcohol drinking and esophageal squamous cell carcinoma with focus on light-drinkers and never-smokers: a systematic review and meta-analysis. Int J Cancer. 2011;129(10):2473–84. doi: 10.1002/ijc.25885. [DOI] [PubMed] [Google Scholar]
- 6.Thrift AP. Global burden and epidemiology of Barrett oesophagus and oesophageal cancer. Nat Rev Gastroenterol Hepatol. 2021;18(6):432–43. doi: 10.1038/s41575-021-00419-3. [DOI] [PubMed] [Google Scholar]
- 7.Wang K, Li J, Guo H, Xu X, Xiong G, Guan X, Liu B, Li J, Chen X, Yang K, et al. MiR-196a binding-site SNP regulates RAP1A expression contributing to esophageal squamous cell carcinoma risk and metastasis. Carcinogenesis. 2012;33(11):2147–54. doi: 10.1093/carcin/bgs259. [DOI] [PubMed] [Google Scholar]
- 8.Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T, Tsunoda T, Kamatani N, Kubo M, Nakamura Y, et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology. 2009;137(5):1768–75. doi: 10.1053/j.gastro.2009.07.070. [DOI] [PubMed] [Google Scholar]
- 9.Zheng W, Wen W, Gao YT, Shyr Y, Zheng Y, Long J, Li G, Li C, Gu K, Cai Q, et al. Genetic and clinical predictors for breast cancer risk assessment and stratification among Chinese women. J Natl Cancer Inst. 2010;102(13):972–81. doi: 10.1093/jnci/djq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jung KJ, Won D, Jeon C, Kim S, Kim TI, Jee SH, Beaty TH. A colorectal cancer prediction model using traditional and genetic risk scores in koreans. BMC Genet. 2015;16:49. doi: 10.1186/s12863-015-0207-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokoyama T, Yokoyama A, Kumagai Y, Omori T, Kato H, Igaki H, Tsujinaka T, Muto M, Yokoyama M, Watanabe H. Health risk appraisal models for mass screening of esophageal cancer in Japanese men. Cancer Epidemiol Biomarkers Prev. 2008;17(10):2846–54. doi: 10.1158/1055-9965.EPI-08-0397. [DOI] [PubMed] [Google Scholar]
- 12.Chang J, Huang Y, Wei L, Ma B, Miao X, Li Y, Hu Z, Yu D, Jia W, Liu Y, et al. Risk prediction of esophageal squamous-cell carcinoma with common genetic variants and lifestyle factors in Chinese population. Carcinogenesis. 2013;34(8):1782–6. doi: 10.1093/carcin/bgt106. [DOI] [PubMed] [Google Scholar]
- 13.Dong J, Buas MF, Gharahkhani P, Kendall BJ, Onstad L, Zhao S, Anderson LA, Wu AH, Ye W, Bird NC, et al. Determining risk of Barrett’s Esophagus and Esophageal Adenocarcinoma based on epidemiologic factors and genetic variants. Gastroenterology. 2018;154(5):1273–e12811273. doi: 10.1053/j.gastro.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han Y, Zhu X, Hu Y, Yu C, Guo Y, Hang D, Pang Y, Pei P, Ma H, Sun D, et al. Electronic Health Record-based Absolute Risk Prediction Model for Esophageal Cancer in the Chinese Population: Model Development and External Validation. JMIR Public Health Surveill. 2023;9:e43725. doi: 10.2196/43725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan F, Liu L, Chen X, Yang Q, Wang Y, Zhang Y, Wang K. Genetic risk and gastric cancer: polygenic risk scores in population-based case-control study. Expert Rev Mol Diagn 2023:1–10. [DOI] [PubMed]
- 16.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96(6):434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Canqing LL. Appliaction of false positive report probability in molecular epidemiological study. Clin J Prev Med. 2009;43(12):1141–2. [Google Scholar]
- 18.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–45. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 19.Li G, Song Q, Jiang Y, Cai A, Tang Y, Tang N, Yi D, Zhang R, Wei Z, Liu D, et al. Cumulative evidence for associations between genetic variants and risk of Esophageal Cancer. Cancer Epidemiol Biomarkers Prev. 2020;29(4):838–49. doi: 10.1158/1055-9965.EPI-19-1281. [DOI] [PubMed] [Google Scholar]
- 20.Matejcic M, Gunter MJ, Ferrari P. Alcohol metabolism and oesophageal cancer: a systematic review of the evidence. Carcinogenesis. 2017;38(9):859–72. doi: 10.1093/carcin/bgx067. [DOI] [PubMed] [Google Scholar]
- 21.Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, Li JM, Kong GQ, Qi H, Cui J, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42(9):759–63. doi: 10.1038/ng.648. [DOI] [PubMed] [Google Scholar]
- 22.Zheng H, Zhao Y. Association of CYP1A1 MspI polymorphism in the esophageal cancer risk: a meta-analysis in the Chinese population. Eur J Med Res. 2015;20(1):46. doi: 10.1186/s40001-015-0135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav U, Kumar P, Rai V. NQO1 gene C609T polymorphism (dbSNP: rs1800566) and Digestive Tract Cancer risk: a Meta-analysis. Nutr Cancer. 2018;70(4):557–68. doi: 10.1080/01635581.2018.1460674. [DOI] [PubMed] [Google Scholar]
- 24.Gong FF, Lu SS, Hu CY, Qian ZZ, Feng F, Wu YL, Yang HY, Sun YH. Cytochrome P450 1A1 (CYP1A1) polymorphism and susceptibility to esophageal cancer: an updated meta-analysis of 27 studies. Tumour Biol. 2014;35(10):10351–61. doi: 10.1007/s13277-014-2341-y. [DOI] [PubMed] [Google Scholar]
- 25.Edenberg HJ, Gelernter J, Agrawal A. Genetics of Alcoholism. Curr Psychiatry Rep. 2019;21(4):26. doi: 10.1007/s11920-019-1008-1. [DOI] [PubMed] [Google Scholar]
- 26.Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4(6):697–720. doi: 10.1517/17425255.4.6.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polimanti R, Gelernter J. ADH1B: from alcoholism, natural selection, and cancer to the human phenome. Am J Med Genet B Neuropsychiatr Genet. 2018;177(2):113–25. doi: 10.1002/ajmg.b.32523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li R, Zhao Z, Sun M, Luo J, Xiao Y. ALDH2 gene polymorphism in different types of cancers and its clinical significance. Life Sci. 2016;147:59–66. doi: 10.1016/j.lfs.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 29.Yukawa Y, Muto M, Hori K, Nagayoshi H, Yokoyama A, Chiba T, Matsuda T. Combination of ADH1B*2/ALDH2*2 polymorphisms alters acetaldehyde-derived DNA damage in the blood of Japanese alcoholics. Cancer Sci. 2012;103(9):1651–5. doi: 10.1111/j.1349-7006.2012.02360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang LD, Bi X, Song X, Pohl NM, Cheng Y, Zhou Y, Shears S, Ansong E, Xing M, Wang S, et al. A sequence variant in the phospholipase C epsilon C2 domain is associated with esophageal carcinoma and esophagitis. Mol Carcinog. 2013;52(Suppl 1):E80–86. doi: 10.1002/mc.22016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dumont P, Leu JI, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33(3):357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 32.Li M, Wang D, Wang Y, Sun G, Song W, Zhang B, Borjigin B. Association of TP53 codon 72 genotype polymorphism and environmental factors with esophageal squamous cell carcinoma in the Mongolian population of the Chinese region of Inner Mongolia. Oncol Lett. 2017;14(2):1484–90. doi: 10.3892/ol.2017.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma J, Zhang J, Ning T, Chen Z, Xu C. Association of genetic polymorphisms in MDM2, PTEN and P53 with risk of esophageal squamous cell carcinoma. J Hum Genet. 2012;57(4):261–4. doi: 10.1038/jhg.2012.15. [DOI] [PubMed] [Google Scholar]
- 34.Li T, Lu ZM, Guo M, Wu QJ, Chen KN, Xing HP, Mei Q, Ke Y. p53 codon 72 polymorphism (C/G) and the risk of human papillomavirus-associated carcinomas in China. Cancer. 2002;95(12):2571–6. doi: 10.1002/cncr.11008. [DOI] [PubMed] [Google Scholar]
- 35.Kawaguchi H, Ohno S, Araki K, Miyazaki M, Saeki H, Watanabe M, Tanaka S, Sugimachi K. p53 polymorphism in human papillomavirus-associated esophageal cancer. Cancer Res. 2000;60(11):2753–5. [PubMed] [Google Scholar]
- 36.Peng JZ, Xue L, Liu DG, Lin YH. Association of the p53 Arg72Pro polymorphism with esophageal cancer in Chinese populations: a meta-analysis. Genet Mol Res. 2015;14(3):9024–33. doi: 10.4238/2015.August.7.11. [DOI] [PubMed] [Google Scholar]
- 37.Tang W, Zhang S, Qiu H, Wang L, Sun B, Yin J, Gu H. Genetic variations in MTHFR and esophageal squamous cell carcinoma susceptibility in Chinese Han population. Med Oncol. 2014;31(5):915. doi: 10.1007/s12032-014-0915-6. [DOI] [PubMed] [Google Scholar]
- 38.Li D, Diao Y, Li H, Fang X, Li H. Association of the polymorphisms of MTHFR C677T, VDR C352T, and MPO G463A with risk for esophageal squamous cell dysplasia and carcinoma. Arch Med Res. 2008;39(6):594–600. doi: 10.1016/j.arcmed.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Qu HH, Cui LH, Wang K, Wang P, Song CH, Wang KJ, Zhang JY, Dai LP. The methylenetetrahydrofolate reductase C677T polymorphism influences risk of esophageal cancer in Chinese. Asian Pac J Cancer Prev. 2013;14(5):3163–8. doi: 10.7314/APJCP.2013.14.5.3163. [DOI] [PubMed] [Google Scholar]
- 40.Langevin SM, Lin D, Matsuo K, Gao CM, Takezaki T, Stolzenberg-Solomon RZ, Vasavi M, Hasan Q, Taioli E. Review and pooled analysis of studies on MTHFR C677T polymorphism and esophageal cancer. Toxicol Lett. 2009;184(2):73–80. doi: 10.1016/j.toxlet.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson SC, Giovannucci E, Wolk A. Folate intake, MTHFR polymorphisms, and risk of esophageal, gastric, and pancreatic cancer: a meta-analysis. Gastroenterology. 2006;131(4):1271–83. doi: 10.1053/j.gastro.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 42.Zhao P, Lin F, Li Z, Lin B, Lin J, Luo R. Folate intake, methylenetetrahydrofolate reductase polymorphisms, and risk of esophageal cancer. Asian Pac J Cancer Prev. 2011;12(8):2019–23. [PubMed] [Google Scholar]
- 43.Li H, Ding C, Zeng H, Zheng R, Cao M, Ren J, Shi J, Sun D, He S, Yang Z, et al. Improved esophageal squamous cell carcinoma screening effectiveness by risk-stratified endoscopic screening: evidence from high-risk areas in China. Cancer Commun (Lond) 2021;41(8):715–25. doi: 10.1002/cac2.12186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Han J, Wang L, Zhang H, Ma S, Li Y, Wang Z, Zhu G, Zhao D, Wang J, Xue F. Development and validation of an esophageal squamous cell Carcinoma Risk Prediction Model for Rural Chinese: Multicenter Cohort Study. Front Oncol. 2021;11:729471. doi: 10.3389/fonc.2021.729471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen W, Li H, Ren J, Zheng R, Shi J, Li J, Cao M, Sun D, He S, Sun X, et al. Selection of high-risk individuals for esophageal cancer screening: a prediction model of esophageal squamous cell carcinoma based on a multicenter screening cohort in rural China. Int J Cancer. 2021;148(2):329–39. doi: 10.1002/ijc.33208. [DOI] [PubMed] [Google Scholar]
- 46.Liu M, Liu Z, Cai H, Guo C, Li X, Zhang C, Wang H, Hang D, Liu F, Deng Q, et al. A model to identify individuals at high risk for Esophageal Squamous Cell Carcinoma and precancerous lesions in regions of high prevalence in China. Clin Gastroenterol Hepatol. 2017;15(10):1538–46. doi: 10.1016/j.cgh.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 47.Kunzmann AT, Thrift AP, Cardwell CR, Lagergren J, Xie S, Johnston BT, Anderson LA, Busby J, McMenamin UC, Spence AD, et al. Model for identifying individuals at risk for Esophageal Adenocarcinoma. Clin Gastroenterol Hepatol. 2018;16(8):1229–e12361224. doi: 10.1016/j.cgh.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 48.Iwasaki M, Tanaka-Mizuno S, Kuchiba A, Yamaji T, Sawada N, Goto A, Shimazu T, Sasazuki S, Wang H, Marchand LL, et al. Inclusion of a genetic risk score into a validated risk prediction model for Colorectal Cancer in Japanese men improves performance. Cancer Prev Res (Phila) 2017;10(9):535–41. doi: 10.1158/1940-6207.CAPR-17-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Yang L, Zhao X, Wang J, Qian J, Chen H, Fan W, Liu H, Jin L, Wang W, et al. Prediction of lung cancer risk in a Chinese population using a multifactorial genetic model. BMC Med Genet. 2012;13:118. doi: 10.1186/1471-2350-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Flow chart of literature selection in the meta-analysis. Figure S2. Distribution of studies included in mete-analysis by province in China. (docx)
Additional file 2: Table S1. SNPs identified from the meta-analysis. Table S2. Associations of genetic variants with esophageal cancer risk in the meta-analysis. Table S3. Heterogeneity test and evaluation of reliability for genetic variants significantly associated with esophageal cancer risk. Table S4. Epidemiological effect estimation for the relationship between genetic variants and esophageal cancer. Table S5. Associations of 14 candidate SNPs with risk of esophageal cancer in the case-control study. Table S6. Associations of genotypes of 14 candidate SNPs with esophageal cancer risk. Table S7. False positive report probability of 5 promising SNPs. Table S8. Risk score for each promising SNP. Table S9. Construction of non-genetic and combined models. (docx)
Additional file 3: Supplementary References. (docx).
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.