Abstract
Expression of the Yersinia enterocolitica inv gene is dependent on growth phase and temperature. inv is maximally expressed at 23°C in late-exponential- to early-stationary-phase cultures. We previously reported the isolation of a Y. enterocolitica mutant (JB1A8v) that shows a decrease in invasin levels yet is hypermotile when grown at 23°C. JB1A8v has a transposon insertion within uvrC. Described here is the isolation and characterization of a clone that suppresses these mutant phenotypes of the uvrC mutant JB1A8v. This suppressing clone encodes ClpB (a Clp ATPase homologue). The Y. enterocolitica ClpB homologue is 30 to 40% identical to the ClpB proteins from various bacteria but is 80% identical to one of the two ClpB homologues of Yersinia pestis. A clpB::TnMax2 insertion mutant (JB69Qv) was constructed and determined to be deficient in invasin production and nonmotile when grown at 23°C. Analysis of inv and fleB (flagellin gene) transcript levels in JB69Qv suggested that ClpB has both transcriptional and posttranscriptional effects. In contrast, a clpB null mutant, BY1v, had no effect on invasin levels or motility. A model accounting for these observations is presented.
Yersinia enterocolitica is a food-borne pathogen that causes a variety of intestinal syndromes (6, 8). One of the key features of the disease process is the ability of the bacteria to translocate through the intestinal epithelium and proliferate within the Peyer's patches (13, 37). Invasin, an outer membrane protein of Y. enterocolitica, is required for efficient translocation of the bacteria from the intestinal lumen to the Peyer's patches (37). Expression of the inv gene, which encodes invasin, is influenced by temperature and growth phase. inv expression is maximal when Y. enterocolitica is in late exponential to early stationary phase at 23°C (35, 38). In addition to temperature and growth phase, other environmental factors affect inv expression. Notable expression of inv is observed when the bacteria are grown at 37°C in L broth (LB) buffered at pH 5.5. Furthermore, an increase in Na+ ion concentration elicits a slight increase in inv expression at 37°C (35).
Expression of other identified Y. enterocolitica virulence factors is also thermally regulated in vitro. For example, ail encodes a protein that mediates Y. enterocolitica adherence, invasion, and serum resistance (5, 28, 39, 49). ail is highly expressed when bacteria are grown at 37°C, while at lower temperatures ail expression is repressed (39). Recently it was demonstrated that ClpP plays a role in repression of Ail synthesis at lower temperatures (34). In Escherichia coli, ClpP is the proteolytic subunit of an ATP-dependent serine protease (Clp) (11, 47).
Motility is also optimally expressed at 23°C (17, 20, 43). Y. enterocolitica possesses three flagellin structural genes (fleA, fleB, and fleC) (20). The fleA and fleB genes are transcribed at 25°C, while no flagellin transcripts were detected at 37°C (20). As in other Enterobacteriaceae, expression of the Y. enterocolitica flagellin genes has been shown to be dependent on the master regulators FlhDC (57) and alternative sigma factor FliA (ςF) (20, 21). Y. enterocolitica also possesses the anti-sigma factor FlgM, which negatively regulates FliA activity (21). fliA and flgM expression is coordinately regulated by temperature; expression of fliA and flgM is observed principally at lower temperatures (21). The mechanism for activation or repression of these genes in response to temperature has not been elucidated. However, FlhDC and FliA are required for the expression and secretion of at least one virulence determinant, the phospholipase YplA (56).
Little is known about the molecular basis for the environmental regulation of inv expression. However, we recently reported the isolation and characterization of two Y. enterocolitica invasin regulatory mutants. Both of the mutants, JB1A8v and JB16v, show a significant decrease in invasin expression but are hypermotile compared to the wild type when grown at 23°C. The first inv regulatory mutant, JB1A8v, has an mTn5Km transposon insertion that disrupts uvrC. It is not known at this time how the mTn5Km insertion mutation results in the observed phenotypes of JB1A8v (1). The second regulatory mutant, JB16v, was generated from a targeted disruption of the sspA locus in wild-type Y. enterocolitica (1). It was determined that the uvrC and sspA genes are separated by approximately 16 kb of DNA. In E. coli, sspA encodes the stringent starvation protein SspA, which has been shown to bind RNA polymerase (19, 54). In addition, E. coli SspA has been shown to negatively and positively regulate the expression of several proteins during the exponential- and post-exponential-growth phases (54). The expression of sspA itself is induced by entry into stationary phase as well as by starvation for phosphate, glucose, nitrogen, and amino acids (54). The exact mechanism for E. coli SspA regulation is presently unknown, but it is thought to be related to the ability of SspA to bind RNA polymerase.
Concurrently with the above studies, attempts were made to isolate a clone that could complement the phenotype of the Y. enterocolitica mutant JB1A8v. Thus, a clone was isolated that suppressed the phenotypes of decreased invasin and increased flagellin levels in JB1A8v. We report here the isolation and characterization of this suppressing clone, and we show evidence that ClpB (encoded on the suppressing clone) can play a role in the expression of invasin and the motility regulon.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
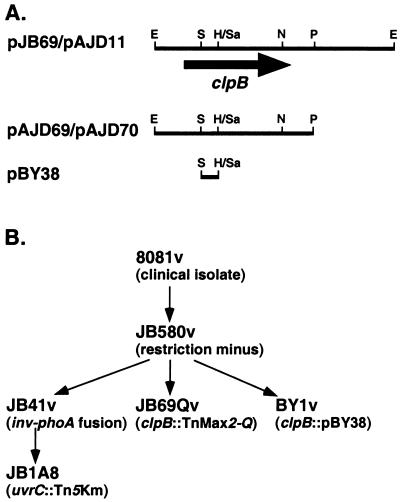
Strains and plasmids used in this study are listed in Table 1. Y. enterocolitica strains 8081v and JB580v (nalidixic acid resistant) are referred to as wild type. The latter is an R− M+ derivative of 8081v (22) and retains full virulence in BALB/c mice (unpublished data). A schematic of strain lineages for strains used or constructed in this study is shown in Fig. 1. The “v” designation refers to Y. enterocolitica strains harboring the virulence plasmid pYV8081. Expression of the inv gene was examined in Y. enterocolitica strains grown aerobically for 16 to 18 h at 23 or 37°C in LB. E. coli SM10λpir (46) was used to deliver mobilizable plasmids into Y. enterocolitica, and when necessary pRK2013 was used as as a conjugation helper plasmid (10). Antibiotics were used as necessary at the following concentrations (in micrograms per milliliter): nalidixic acid, 25; chloramphenicol, 25 for multicopy and 10 for single copy; tetracycline, 7.5; kanamycin, 50; ampicillin, 100; gentamicin, 100; and erythromycin, 150 for multicopy and 50 for single copy.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Y. enterocolitica | ||
| 8081v | Serogroup O:8; pYV8081 Nalr | R. Martinez |
| JB580v | Derivative of 8081v; R− M+ Nalr | 22 |
| JB41v | JB580v with wild-type inv; inv::phoA; Nalr Cmr | 1 |
| JB1A8v | JB580v uvrC::mTn5Km; regulatory mutant; Nalr Cmr Kmr | 1 |
| JB69Qv | JB580v clpB::TnMax2; Nalr Ermr | This work |
| BY1v | JB580v clpB::pEP185.2; Nalr Cmr | This work |
| E. coli | ||
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-TC::Mu; Kmr | 46 |
| DH5α | supE44 [lacU169(φ80dlacZΔM15)] hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 15 |
| E131 | F− φ(80dlacZΔM15)Δ(lacZYA argF)U169 deoR recA1 endA1 hsdR17 supE44 λ-thi-1 gyrA96 relA1λ-CH616 | 14 |
| LE392 | F−hsdR514 supE44 supF58 | 25 |
| Plasmids | ||
| pLAFR2 | Mobilizable cosmid vector; Tetr | S. Libby |
| pJQ200KS | Mobilizable suicide vector; Gmr | 41 |
| pTM100 | Mobilizable derivative of pACYC184; Tetr Cmr | 27 |
| pCR2.1 | PCR product cloning vector; Ampr Kmr | Invitrogen |
| pEP185.2 | Mobilizable suicide vector; Cmr | 22 |
| pRK2013 | ColE1 Tra (RK2)+; Kmr | 10 |
| pUC19 | Cloning vector; Apr | New England Biolabs |
| pTnMax2 | TnMax2 delivery plasmid; Ermr | 14 |
| pWSK129 and pWSK130 | Low-copy-number cloning vectors with pSC101 ori; Kmr | 51 |
| pJB32 | pLAFR2 containing an ∼25-kb insert; Tetr | This work |
| pJB69 | pTM100 containing a 5.4-kb EcoRI fragment from pJB32 carrying clpB; Tetr | This work |
| pJB69-Q | pJB69 clpB::TnMax2; Tetr Ermr | This work |
| pCRII-Q | pCR2.1 containing clpB::TnMax2 PCR product; Apr Kmr Ermr | This work |
| pEP185.2-Q | pEP185.2 containing clpB::TnMax2 PCR product; Cmr Ermr | This work |
| pJB302 | pCR2.1 containing clpB PCR product; Apr Kmr | This work |
| pJB305 | pTM100 containing clpB PCR product; Tetr | This work |
| pAVKF | pACY184 containing a 2.2-kb EcoRV fragment carrying fliA and upstream sequences; Cmr | 21 |
| pVM112 | pMT11HC containing inv; Apr | 36 |
| pKVS2 | pBluescript containing a 2.2-kb SacII fragment of the 3′ end of fleA, all of fleB, and the 5′ end of fleC; Apr | 20 |
| pBY38 | pEP185.2 with a 744-bp SalI-SacII fragment from pAJD70 | This work |
| pAJD69 | pWSK129 containing a 3.6-kb EcoRI-PstI fragment of pAJD11 carrying clpB | This work |
| pAJD70 | pWSK130 containing a 3.6-kb EcoRI-PstI fragment of pAJD11 carrying clpB | This work |
| pAJD11 | pWSK129 containing a 5.4-kb EcoRI fragment carrying clpB; Kmr | This work |
FIG. 1.
Physical maps of various clones used this study. (A) Restriction endonuclease sites: H, HindIII; R, EcoRI; S, SalI; Sa, SacII; P, PstI; N, NarI. (B) Schematic of strain lineages for strains used and/or constructed in this study.
DNA manipulations.
The chromosomal DNA library of Y. enterocolitica strain 8081v was constructed as follows. Chromosomal DNA was partially digested with the restriction enzyme Sau3AI and ligated into a BamHI site in the cosmid cloning vector pLAFR2 (obtained from S. Libby), a derivative of pLAFR1 (3). The ligation mixture was packaged into λ phage by use of a λ packaging mix (Promega, Madison, Wis.); the packaged DNA was transfected into E. coli strain LE392, (25). The complementing clone obtained from the library was designated pJB32.
To generate pJB69, pJB32 was partially digested with EcoRI and the fragments were ligated into pTM100 digested with EcoRI. The resulting recombinant plasmids were screened for the ability to complement JB1A8v. A plasmid with a 5.4-kb EcoRI insert was found to suppress both the invasin and motility phenotypes of mutant JB1A8v; this plasmid was designated pJB69.
pJB305 was constructed as follows. The Y. enterocolitica clpB locus was amplified by PCR using pJB32 as a template. PCR amplification was performed using oligonucleotide primers 691 (5′ GCTGATATACACATGTAG 3′) and 69Nr (5′ CCAATGAATGCTGCTG 3′), which anneal 5′ and 3′ of Y. enterocolitica clpB, respectively. The resulting ca. 3.5-kb PCR product was cloned into pCR2.1, generating a plasmid designated pJB302. pJB302 was subsequently digested with EcoRI, and insert DNA was isolated and ligated into pTM100 digested with EcoRI to yield pJB305. The insert in pJB305 does not carry any open reading frames other than clpB.
Construction of the clpB mutants JB69Qv and BY1.
JB69Qv was constructed as follows. Oligonucleotide primers used for PCR amplification of clpB::TnMax2-Q were 69GG (5′ CCTCAGGTCGATGATGAACCTG 3′) and 69Nr (5′ CCAATGAATGCTGCTG 3′), which anneal within and 3′ of the clpB gene, respectively. Template DNA used for PCR amplification was plasmid pJB69-Q, which contains clpB with a TnMax2 insertion in the 3′ end of the clpB gene. The resulting PCR product, ca. 3.8 kb, was cloned into pCR2.1, generating plasmid pCRII-Q. pCRII-Q was subsequently digested with AvaI and SpeI, and insert DNA was isolated and ligated into pEP185.2 digested with XhoI and XbaI. The resulting plasmid, pEP185.2-Q, was conjugated into wild-type Y. enterocolitica JB580v and recombined onto the chromosome by allelic exchange as described elsewhere (35). Double-crossover events at the homologous chromosomal locus were confirmed by Southern analysis using clpB-specific sequences as a probe (data not shown). BY1 was constructed as follows. A SalI/SacII internal fragment (744 bp) of pAJD70 was subcloned into pEP185.2 digested with SalI and SacII. This plasmid, pBY38, was conjugated into JB580v, and Nalr Cmr exconjugants were selected. The integration of the plasmid into the chromosome was confirmed by Southern analysis using clpB-specific sequences as a probe (data not shown). This mutation (clpB::pBY38) should create a truncated ClpB (364 amino acids of ClpB).
Cloning the clpB gene from the Y. enterocolitica chromosome.
Chromosomal DNA from Y. enterocolitica strain JB580v was digested with EcoRI. Fragments of approximately 5 to 6 kb were purified from a 0.8% (wt/vol) agarose gel and ligated into plasmid pWSK129. This library was then used to transform E. coli strain DH5α to kanamycin resistance. One hundred ninety-two transformants were grown directly on nylon filters on the surfaces of LB agar plates containing kanamycin (75 μg/ml) at 37°C overnight. The filters were then removed, dried for 10 min at room temperature, and placed on Whatman 3MM paper soaked with 0.4 M NaOH for 8 min. The filters were rinsed in 0.5 M Tris-HCl (pH 7.0) for 5 min and then in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for a further 5 min. The DNA was then cross-linked to the filters by UV fixation. The approximately 3.5-kb EcoRI insert fragment of plasmid pJB305 was labeled as described elsewhere (9) and used to probe the filters. A single clone that hybridized with the probe was identified. The plasmid had a single EcoRI insert fragment of approximately 5.4 kb and was designated pAJD11.
Western (immunoblot) analysis.
All the strains analyzed in this study had the same growth rates, and comparable culture densities were reached after overnight growth at 23°C. Whole-cell lysates were prepared from bacteria grown to early stationary phase in LB at either 23 or 37°C as described previously (35). Equal amounts of whole-cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Blots were then prepared for Western (immunoblot) analysis with a polyclonal anti-invasin antibody and visualized as described previously (2, 35). Coomassie-stained gels run in parallel confirmed that equivalent amounts of protein were loaded in each lane. Invasin levels as detected by immunoblot analysis were estimated by densitometry with the GDS2000 gel documentation system (UVP International). Cell surface-associated flagellin was obtained from the supernatants of vigorously vortexed cultures as described previously (1) and was detected by immunoblotting with monoclonal antibody 15DB (anti-E. coli flagellin) (Igen, Inc.) used at a dilution of 1:2,000 as described previously (1).
Phenotypic assays.
Motility assays were conducted in LB with 0.3% agar and appropriate antibiotics as described previously (1). Invasion assays were performed with bacterial cultures grown aerobically for 16 to 18 h in LB at 23 or 37°C as indicated. Bacteria were added to subconfluent human laryngeal epithelial (HEp-2) cells at a multiplicity of infection of ca. 100, and the invasion assay was performed as described elsewhere (28). Results were calculated by the following equation: % invasion = 100 × (number of bacteria recovered/number of bacteria added). Alkaline phosphatase (AP) activity was measured in permeablized cells, and results are expressed in enzyme units per OD600 (optical density at 600 nm) unit as described elsewhere (26). Assays were performed on duplicate cultures grown 16 to 18 h aerobically in LB. Y. enterocolitica JB580v was always assayed and had low but detectable background levels of AP activity (∼10 U) that were subtracted from the values presented.
mRNA slot blot analysis.
Total cellular RNA was purified from late-log- to early-stationary-phase cultures using the TRIzol reagent (Gibco BRL). RNA samples were treated with RNase-free DNase. In addition, RNase controls were always run for detection of possible DNA contamination. Purified RNA (2.5 μg) was applied to nylon membranes (Pro-Nytran; Schleicher and Schuell) using a slot blot apparatus (Bio-Rad). Membranes were UV cross-linked and subjected to prehybridization for 2 h at 42°C in 50% formamide–5× SSC–0.5% SDS–1× Denhardt's solution–10% dextran sulfate–150 μg of salmon sperm DNA/ml. Radiolabeled probes were added directly to membranes in prehybridization buffer and incubated for 12 to 24 h at 42°C. Membranes were washed twice in preheated 65°C 0.1% SDS–5× SSC for 15 min. A final wash in 2× SSC was performed at room temperature for 5 min. mRNA slot blots were visualized and analyzed with a Molecular Dynamics PhosphorImager and the ImageQuant program. The density of each slot was determined by volume analysis. Wild-type transcript levels were set at 100%, and relative amounts of transcripts from other strains were determined. Each result presented represents the mean ± the range from duplicate samples of an individual representative assay. Probes used in mRNA slot blot analysis were as follows: a 1.2-kb NdeI-BglII internal fragment of inv (36), an 840-bp StuI-XbaI fragment encompassing fliA (21), and a 460-bp AccI-HpaI fragment encompassing the 5′ and internal portions of fleB (20).
Nucleic acid purification and probe preparation.
Plasmid DNA was isolated by the alkaline lysis method (25) or with Wizard Minipreps (Promega). DNA fragments used in plasmid construction and probe preparation were prepared by digestion with the appropriate restriction endonuclease. After digestion the resulting fragments were gel purified using Gene Clean (Bio 101, La Jolla, Calif.). The purified fragments used as probes were labeled with [32P]dATP by the random primer method as previously described (9). DNA restriction enzymes, T4 DNA ligase, T4 DNA polymerase, and Klenow fragment were purchased from New England Biolabs (Beverly, Mass.) and used according to the manufacturer's instructions.
DNA sequence analysis.
Sequencing across TnMax2 transposon-plasmid junctions was facilitated by two primers, RES1 (5′-CCTGACAGAAATGGGC-3′) and TnMax2 (5′-CCTAAAGGGATCCAAAAGCT-3′), which anneal at the 5′ and 3′ ends of TnMax2, respectively. For each TnMax2 insertion, the DNA sequence of 200 to 400 bp in each direction from the site of insertion was determined. The nucleotide sequence was obtained by the dideoxynucleotide chain termination method (44) with the Sequenase sequencing kit (U.S. Biochemical Corp.). The DNA sequence of the insert in pAJD69 (complete clpB clone), from the EcoRI site (356 bp upstream of the putative clpB start codon) to the end of the clpB open reading frame, was determined. DNA sequencing was carried out using an Applied Biosystems DNA sequencing system and the BigDye terminator cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Some of the sequencing was performed by the Murdock Molecular Biology automated sequencing facility (University of Montana). Sequences were analyzed using the BLAST X program available in the NCBI database (www.ncbi.nlm.nih.gov).
Nucleotide sequence accession number.
The clpB sequence (3,025 bp) was deposited in GenBank under accession no. AF285784.
RESULTS
Isolation and characterization of a clone that suppresses the phenotypes of the JB1A8v mutant.
Y. enterocolitica strain JB41v carries wild-type inv and an inv::phoA translational fusion on the chromosome and is genotypically and phenotypically Inv+ (Table 1). Thus, modulation of inv expression can be monitored by AP activity, Western analysis, primer extension, or tissue culture invasion assays. Analysis of inv expression in JB41v by any of these assays showed the same pattern: expression of the inv gene and invasin levels were elevated at 23°C, while expression was reduced at 37°C (1). An mTn5Km insertion mutant of JB41v, JB1A8v, that demonstrated a significant reduction in inv expression and an increase in fleABC (flagellin) expression (1) was isolated. These phenotypes could be detected by both protein and RNA analyses (1). Sequencing of the insertion junction demonstrated that the disrupted gene was uvrC, but several different approaches taken to clone the wild-type locus disrupted by the mTn5Km were unsuccessful (1). However, we were able to isolate a low-copy-number cosmid clone that complemented the phenotypes of JB1A8v. This cosmid, pJB32, contains ca. 25 kb of Y. enterocolitica chromosomal DNA in pLAFR2. When bacteria were grown at 23°C, wild-type JB41v and mutant JB1A8v containing the cloning vector pLAFR2 showed 19.7 and 7.2 AP units, respectively. JB1A8v harboring pJB32 restored AP levels to 23.0 AP units. Western analysis and tissue culture invasion assays showed similar suppression of the JB1A8v invasin phenotype by pJB32 (data not shown). In addition, pJB32 restored motility and flagellin expression to wild-type levels (data not shown).
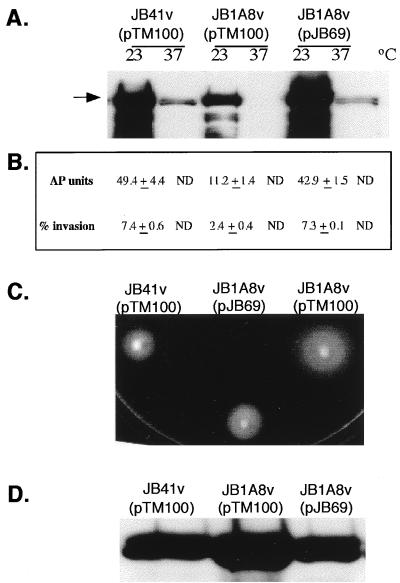
To determine the minimal region responsible for suppression of the JB1A8v mutant phenotypes, subclones from the insert of pJB32 were constructed and tested. A resulting subclone, pJB69, had a 5.4-kb EcoRI insert within the cloning vector pTM100 (Fig. 1). pJB69 restored normal invasin expression as verified via AP assays, Western analysis, and tissue culture invasion assays (Fig. 2A and B). In addition, the suppressing clone pJB69 restored flagellin synthesis and the motility phenotype to wild-type levels (Fig. 2C and D). JB41v harboring pJB69 showed a motility pattern similar to that of JB41v containing the cloning vector pTM100 (data not shown).
FIG. 2.
Suppression of JB1A8v invasin motility phenotypes by pJB69. (A) Western analysis of invasin expression by JB41v(pTM100), JB1A8v(pTM100), and JB1A8v(pJB69). Whole-cell extracts were fractionated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed with a rabbit polyclonal anti-invasin antibody as described in Materials and Methods. The arrow points to the band representing full-length invasin. It is typical to see full-length invasin along with several breakdown products. (B) Invasion phenotype and AP activity exhibited by JB41v(pTM100), JB1A8v(pTM100), and JB1A8v(pJB69). AP activities were assayed in duplicate and are represented as means ± ranges. Invasion assays with HEp-2 cells were performed in duplicate, and data represent means ± ranges as described in Materials and Methods. (C) Motility assays were performed with JB41v(pTM100), JB1A8v(pTM100), and JB1A8v(pJB69) in 0.3% motility soft agar plates incubated at 23°C for 16 h. (D) Western analysis of flagellin preparations probed with monoclonal antibody 15D8 as described in Materials and Methods. Data shown are from a single experiment and are representative of several experiments performed with similar results.
Sequence analysis of the suppressing clone pJB69.
To identify the genetic determinant encoded on the suppressing clone pJB69, the nucleotide sequences of portions of the insert DNA of pJB69 were determined. To this end, TnMax2 insertions into pJB69 were isolated and mapped. Primers complementary to the ends of TnMax2 were used to obtain the nucleotide sequences upstream and downstream of the site of insertion. Nucleotide sequence analysis revealed that plasmid pJB69 encodes the Y. enterocolitica homologue ClpB from E. coli (33 to 50% amino acid identity depending on the section of sequence). Subsequently the DNA sequence of the insert from the EcoRI site (356 bp upstream of the putative clpB start codon) to the end of the clpB open reading frame was determined and deposited in GenBank (3,025 bp; accession no. AF285784). Note that the clpB gene apparently has a GTG translation initiation codon (the first ATG codon is more than 350 bp after the start of the open reading frame).
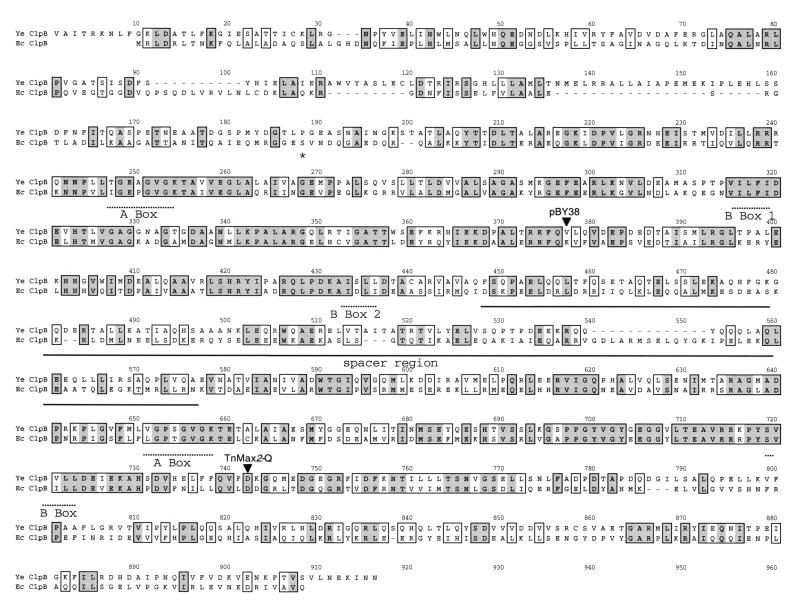
The predicted amino acid sequence of the Y. enterocolitica ClpB protein is 30 to 40% identical to those of the ClpB proteins from a variety of bacteria (data not shown) but is more than 80% identical to an open reading frame in Yersinia pestis (The Sanger Center, www.sanger.ac.uk/Projects/Microbes/). Homologues of the Y. enterocolitica ClpB include the ClpB protein of E. coli (Fig. 3). The Y. enterocolitica ClpB protein has several features in common with the Clp family of proteins (Fig. 3). There are apparently two nucleotide binding domains consisting of the conserved A and B consensus sequences (50). The first nucleotide binding region contains a second B segment, as is found in other Clp proteins (12, 47). The two nucleotide binding regions are separated by a large spacer region, a characteristic found only in the ClpB proteins (12, 47). The clpB gene of E. coli has an internal GTG initiation codon that leads to the synthesis of a truncated ClpB protein, the function of which is not known (33). This GTG codon is not conserved in the Y. enterocolitica gene (Fig. 3).
FIG. 3.
Comparison of the ClpB proteins of E. coli and Y. enterocolitica. The predicted amino acid sequence of the Y. enterocolitica (Ye) ClpB protein was aligned to that of the E. coli (Ec) ClpB protein (GenBank accession no. PO3815) using the ClustalW alignment software (MacVector) and the identity alignment matrix. Similar residues are boxed; identical residues are boxed and shaded. The Walker A and B boxes are underlined with dotted lines, and the spacer region is solid underlined. The asterisk indicates the position of an amino acid translated from a GTG codon that initiates the synthesis of a truncated ClpB protein in E. coli. Arrowheads point to the positions of the pBY38 and TnMax2-Q insertions (giving mutants BY1v and JB69Qv, respectively).
Construction and characterization of a Y. enterocolitica clpB mutant.
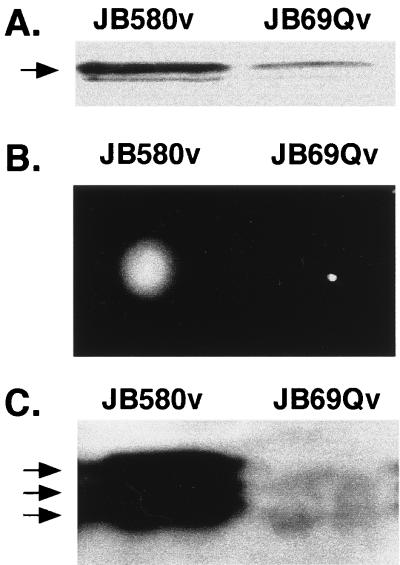
We constructed a clpB mutant of Y. enterocolitica to more clearly determine if ClpB plays a role in invasin expression and motility (flagellin expression). For this purpose, the plasmid with a TnMax2 insertion within the last one-third of clpB, pJB69-Q, was used as a template for a PCR to amplify clpB::TnMax2-Q; pJB69-Q does not suppress the phenotypes of JB1A8v (data not shown). The resulting PCR product, containing approximately 1.3 kb upstream and 1.0 kb downstream of the TnMax2 insertion site, was subcloned into the suicide vector pEP185.2. This clone was then recombined onto the chromosome of JB580v by allelic exchange. The subsequent homologous recombination leading to the replacement of clpB by clpB::TnMax2-Q was confirmed by Southern hybridization analysis with Y. enterocolitica clpB as a probe (data not shown). The resulting clpB::TnMax2-Q mutant, designated JB69Qv, has the same growth rate as JB580v at either 26 or 37°C in LB (data not shown). However, JB69Qv exhibited a significant decrease in invasin expression at 23°C compared to the wild-type strain JB580v, as analyzed by immunoblot analysis (Fig. 4A). In contrast to the mutant strain JB1A8v, the clpB::TnMax2-Q mutant JB69Qv was nonmotile at 23°C (Fig. 4B). Western analysis with a monoclonal anti-flagellin antibody showed that isolated cell surface flagellin protein levels were significantly decreased in the clpB::TnMax2-Q mutant JB69Qv compared to the wild-type strain JB580v (Fig. 4C).
FIG. 4.
Invasin and motility phenotypes of the clpB mutant JB69Qv. (A) Whole-cell extracts of indicated strains grown at 23°C were fractionated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were probed with a rabbit polyclonal anti-invasin antibody as described in Materials and Methods. The arrow points to the band representing full-length invasin. (B) Motility assays were performed with JB580v and JB69Qv in 0.3% motility soft agar plates incubated at 23°C for 16 h. (C) Western analysis of flagellin preparations probed with monoclonal antibody 15D8 as described in Materials and Methods. Arrows point to the three flagellin subunits in Y. enterocolitica. Data shown are from a single experiment and are representative of several experiments performed with similar results.
Complementation of the clpB mutant JB69Qv.
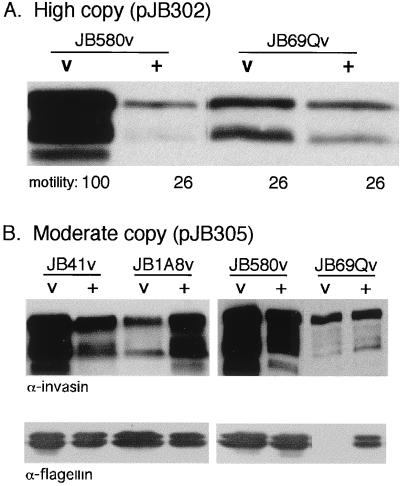
To more definitively assess complementation of the clpB mutant JB69Qv and suppression of mutant JB1A8v by clpB, a smaller subclone containing only clpB was generated. For this purpose Y. enterocolitica clpB was PCR amplified using pJB32 as a template, and the resulting PCR product was cloned into pCR2.1 and pTM100. The plasmids obtained were designated pJB302 and pJB305, respectively. When the high-copy-number clpB plasmid, pJB302, was introduced into the wild-type strain JB580v and the clpB::TnMax2-Q mutant JB69Qv, we observed decreased motility and decreased invasin levels (Fig. 5A), suggesting that overexpression of clpB negatively affects expression of inv and motility. Therefore the moderate-copy-number clpB plasmid, pJB305, was introduced into the mutant strains and assayed for invasin expression and motility. In contrast to the high-copy-number clpB plasmid, pJB305 suppressed the JB1A8v phenotype of decreased invasin levels, as analyzed by immunoblot analysis (Fig. 5B). In addition, JB1A8v harboring the moderate-copy-number complementing clone (pJB305) produced normal amounts of flagellin and showed a wild-type motility pattern (Fig. 5B; also data not shown). However, the clpB mutant JB69Qv containing pJB305 demonstrated only partial complementation of invasin expression and motility (Fig. 5B). Two other clpB complementing clones, pAJD69 and pAJD70, were independently constructed, but these also failed to fully complement the invasin and motility phenotypes of the clpB mutant JB69Qv (data not shown). This could potentially be due to a dominant negative effect of the clpB::TnMax2 mutation. In E. coli, clpB encodes two proteins, a full-length ClpB and a truncated form. The ClpB proteins have been shown to form the tetrameric ATPase in addition to hetero-oligomeric complexes (33). The clpB mutant JB69Qv was generated by a TnMax2 insertion in the carboxyl terminus of Y. enterocolitica ClpB; thus, if a truncated ClpB is synthesized, the clpB::TnMax2-Q mutation could have a trans-dominant effect. This possibility is supported by the observation that the expression of inv-phoA in JB41v carrying the clpB clone pJB69 is more than twice the expression of inv-phoA in JB41v carrying the clpB::TnMax2-Q mutant clone pJB69-Q (23.2 versus 9.8 U, respectively). It is also possible that ClpB is not expressed at sufficient levels from pJB305 for full complementation or that genes downstream of clpB are affected. To address this issue, a new clpB mutant, BY1v, was generated by plasmid insertion early in the gene; this mutation should create a truncated ClpB (364 amino acids). However, BY1v showed the same invasin and motility phenotypes as the wild-type strain JB580v (data not shown). Thus, the invasin and motility defects of JB69Qv may be specific to the clpB::TnMax2-Q allele.
FIG. 5.
Complementation of the clpB mutant JB69Qv and the uvrC mutant JB1A8v. (A) Western analysis of whole-cell extracts of indicated strains grown at 23°C using an antibody to invasin. The numbers listed at the bottom (motility) indicate the percentage of the size of the motility zone in soft agar seen with the wild type (i.e., zone for JB580v pCR2.1 = 100). The “v” indicates the presence of vector pCR2.1, and “+” indicates the presence of the clpB plasmid pJB302. (B) (Top) Western analysis of whole-cell extracts of indicated strains grown at 23°C using an antibody to invasin. (Bottom) Western analysis of flagellin preparations from the same strains probed with monoclonal antibody 15D8 as described in Materials and Methods. The “v” indicates the presence of vector pTM100, and “+” indicates the presence of the clpB plasmid pJB305. Data shown are from a single experiment and are representative of several experiments performed with similar results.
mRNA transcript levels in various mutants.
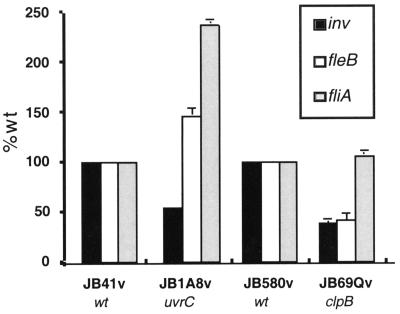
To determine if the changes in invasin and flagellin levels in the clpB::TnMax2-Q mutant JB69Qv were due to changes in the level of transcript, we performed RNA slot blot analysis as described in Materials and Methods. We first analyzed mRNA transcripts for fliA levels; fliA encodes a sigma factor required for expression of the flagellin genes (18, 21). fliA transcript levels in the clpB::TnMax2-Q mutant JB69Qv were comparable to wild-type JB580v levels. In contrast, the other inv regulatory mutant, JB1A8v, showed a significant increase in fliA transcript levels (Fig. 6). RNA transcripts were then analyzed using a fleB-specific probe; fleB encodes one of the flagellins. JB1A8v showed increased fleB transcription compared to that in the wild type, which correlates with the observed increase in motility and flagellin expression in this strain. In contrast, the clpB::TnMax2-Q mutant JB69Qv showed significantly lower fleB transcript levels compared to those in the wild type, consistent with the nonmotile phenotype of this mutant (Fig. 6). The increase in fliA transcript levels for JB1A8v suggests that the hypermotility phenotype of JB1A8v is due to increased expression of fliA and subsequent increased expression of fleABC. In contrast, the effect on motility observed for the clpB::TnMax2-Q mutant JB69Qv does not appear to work through alterations in fliA mRNA levels.
FIG. 6.
Analysis of transcript levels and summary of phenotypes of the various mutants. Transcription of inv, fleB, and fliA was measured by RNA slot blot analysis. RNA was prepared from late-log- to early-stationary-phase cells grown at 23°C. Wild-type transcript levels were set at 100%, and relative amounts of specific mRNA from other strains were determined. For mutant JB1A8v, transcript levels are shown relative to the parental strain JB41v (wild-type inv and inv::phoA). Transcript levels for JB69Qv are shown relative to the parental strain JB580v. Data presented represent the mean ± the range from an individual representative experiment containing duplicate samples and reflect the results from several experiments using independently prepared RNA samples.
We next analyzed inv transcript levels in these mutants. The clpB::TnMax2-Q mutant JB69Qv and the inv regulatory mutant JB1A8v demonstrated significant decreases in inv transcript levels. The decrease in inv transcript levels for JB1A8v correlates with inv-specific primer extension analysis reported previously (1). Although RNA analysis may suggest that an effect is at the transcriptional level, we cannot at this time distinguish between transcription initiation and stability of the transcript.
DISCUSSION
In a previous report we described the isolation and characterization of an invasin regulatory mutant, JB1A8v, that demonstrates decreased invasin levels and increased flagellin levels. JB1A8v has an mTn5Km insertion into uvrC. The uvrC mutant JB1A8v demonstrated a significant decrease in inv-specific transcript levels (1) and an increase in fleB-specific transcripts consistent with the observed expression levels of invasin and flagellin, respectively. It is not known at this time how the mTn5Km insertion mutation results in the observed phenotypes of JB1A8v, due to the inability to clone the wild-type locus (1). Interestingly, a clone encoding the Y. enterocolitica homologue of ClpB suppresses the mutant phenotypes of this mutant.
ClpB belongs to a recently identified family of proteins (Clp ATPases). Clp ATPases (ClpA, ClpX, and ClpY) were originally identified as regulatory components for ATP-dependent Clp proteases and have been shown to exist in organisms from all three kingdoms (12, 47). ClpB was initially identified as a heat shock protein (48) and has been shown to have ATPase activity (33, 55). Unlike other members of the Clp family (i.e., ClpA and ClpX), ClpB has yet to be shown to associate with the ATP-dependent protease subunit ClpP. In E. coli, clpB encodes two proteins, a full-length ClpB and a truncated form. The ClpB proteins form a tetrameric ATPase in addition to hetero-oligomeric complexes (33, 55). It has been suggested that the two forms of ClpB may provide the ClpB protein with separate functions (47). Recent data also suggest that Clp ATPases may act as chaperones independently of ClpP or other catalytic subunits (53).
The ClpB identified in this study as a suppressor of the mutant phenotypes of the inv regulatory mutant JB1A8 shows a relatively low level of identity to the ClpB of E. coli. This low level of identity with the E. coli protein is surprising given the close relationship of the two organisms. Conserved proteins of Y. enterocolitica and E. coli are often more than 80% identical. For example, the ClpX and ClpP proteins of E. coli and Y. enterocolitica are 94 and 90% identical, respectively (34). One possible explanation for the low identity of the ClpB proteins is that the clpB gene of Y. enterocolitica identified in this study was acquired by horizontal transfer. This explanation is supported by the observation that the Y. pestis chromosome encodes a close homologue of the Y. enterocolitica ClpB protein (more than 80% identical) in addition to a separate close homologue of the E. coli ClpB protein (more than 80% identical) (The Sanger Center, www.sanger.ac.uk/Projects/Microbes/). It is possible that the same is true for Y. enterocolitica.
What is the role of ClpB in Y. enterocolitica inv and fleABC (flagellin) expression? Two clpB mutants were constructed; one of these mutants, JB69Qv, shows decreased levels of invasin and is nonmotile. From densitometry analysis, the clpB::TnMax2-Q mutant JB69Qv shows 40% of wild-type inv message levels yet only 6% of wild-type protein levels. In addition, JB69Q shows 50% of wild-type fleB (flagellin) message levels, yet flagellin is virtually undetectable (Fig. 4C and 5B). These results suggest that the clpB mutation may affect inv expression both transcriptionally and posttranscriptionally. This mechanism of a Clp ATPase regulator working at several levels in the same genetic pathway is not uncommon. In Bacillus subtilis, MecB (a ClpC ATPase homologue) is a pleiotropic regulator of competence genes (29). In addition, it was recently determined that B. subtilis MecB/ClpC positively regulates autolysin gene expression and differentially regulates SigD (alternative sigma factor)-dependent genes, including flagellin and motility genes. The MecB/ClpC regulation of SigD-dependent genes has been demonstrated to act in both a SigD-dependent and a SigD-independent manner (42).
In addition, RNA analysis suggests that the clpB::TnMax2-Q mutation in JB69Qv affects motility at some point beyond transcription of fliA. By analogy to the ability of ClpA to activate and degrade the DNA binding protein RepA (53), one could postulate that ClpB may be necessary for the stability/ activation and degradation of FliA. In addition, ClpB might be required for activation/stability of a yet-to-be-identified regulator of inv expression. Complicating the interpretation of these results are the observations that a clpB mutation early in the gene (probably a null mutation, yielding mutant BY1v) shows the same inv expression pattern and motility as the wild type.
ClpB has two different ATP binding domains, and there is evidence in the literature that Clp proteins can have both positive and negative effects (52). Thus, one model to explain these results would be that each ATP binding domain of ClpB has a different function. In this model, ATP binding domain 1 functions to downregulate expression of inv and motility, and ATP binding domain 2 functions to upregulate expression of inv and motility; these effects could be direct or indirect. Therefore, under standard lab conditions, the functions of domain 1 and domain 2 cancel each other out. This would explain why the null mutant BY1v (ClpB1) phenotypically resembles the wild type under these conditions. Similarly, expression of inv and motility would be downregulated in JB69Qv because ClpB69 contains only a functional domain 1. This also would explain the partial complementation of JB69Qv by wild-type ClpB; the ratio of domain 1 to domain 2 would be greater than 1, and therefore expression of inv and motility would still be reduced relative to those in the wild type. Any condition that differentially affected the activity of domain 1 relative to that of domain 2 would alter the expression pattern of inv and motility.
In addition, the copy number of clpB also could affect expression of inv and motility. Consistent with this hypothesis, when Y. enterocolitica clpB is supplied in trans on a high-copy-number plasmid (pJB302), invasin levels are dramatically decreased. This phenomenon is seen in all strains tested, including the wild-type strain JB580v (J. L. Badger and V. L. Miller, unpublished data). In contrast, providing ClpB in trans on a moderate-copy-number plasmid (pJB305) restores invasin production and motility to wild-type levels in the uvrC mutant JB1A8v, partially complements the clpB mutant JB69Qv, and has a moderate effect on invasin levels and motility in the wild-type strains. This apparent requirement for Y. enterocolitica to maintain ClpB at appropriate levels in order to preserve proper expression is reminiscent of what has been observed for Saccharomyces cerevisiae. In S. cerevisiae a yeast non-Mendelian factor, [psi+], is suggested to be a self-modified protein analogous to mammalian prions, and recently it has been shown that intermediate amounts of the chaperone protein ClpB (Hsp104) are required for propagation of the [psi+] factor. It has also been observed that either overexpression or inactivation of ClpB (Hsp104) in S. cerevisiae leads to the same phenotype of being cured of the prion-like factor (7).
Interestingly, Clp ATPase homologues have been emerging as important factors for virulence. For example, Pederson et al. recently demonstrated that the repression of Y. enterocolitica ail expression at low temperatures is dependent on ClpP (34). In Salmonella enterica serovar Typhimurium, the alternative sigma factor ςS (RpoS) regulates genes involved in the environmental stress response in addition to regulating many virulence properties (16, 23, 24, 31, 32). In E. coli, ςS is negatively regulated by ClpXP-mediated degradation (45). MviA, a two-component response regulator homologue (also known as SprE in E. coli), plays an essential role in RpoS turnover (4, 30, 40). It has been demonstrated that MviA/SprE influences the susceptibility of RpoS to ClpXP-mediated degradation (40, 45). More recently, the Listeria monocytogenes MecB/ClpC homologue was determined to be necessary for macrophage survival and virulence in mice (44).
From evidence presented in this communication, it is apparent that ClpB plays a role in the regulation of the virulence factor invasin and of motility in Y. enterocolitica. Furthermore, it appears that the effects of ClpB take place at multiple levels. Based on evidence presented here, there are several levels at which expression of invasin and motility are cocontrolled, as follows. (i) A locus of JB1A8v (possibly uvrC) is necessary for proper expression of inv and fliA, and this effect occurs at the level of the inv and fliA transcripts. Overexpression of ClpB can compensate for this defect. (ii) ClpB-dependent effects on the inv and fleB transcripts are observed when only the N-terminal two-thirds of ClpB is expressed. (iii) ClpB-dependent regulation of FliA and invasin protein levels may take place. The complexity of the adaptive process by which Y. enterocolitica regulates invasin and motility may reflect the need to coordinate multiple functions associated with the pathogenic life cycle. It remains to be determined whether other factors associated with the pathogenicity or virulence of Y. enterocolitica are affected by ClpB.
ACKNOWLEDGMENTS
We are indebted to Jeff Pepe for construction of the cosmid library. We thank S. Minnich for providing pAVKF and pKVS2, S. Libby for providing pLAFR2, and P. Cotter and W. Goldman for critical reading of the manuscript. We especially thank Jeff F. Miller for support and advice as well as for the use of his lab during part of this project.
This work was supported in part by National Institutes of Health grants AI-27342 and AI-01230 to V.L.M. J.L.B. is a recipient of the UCPF award.
REFERENCES
- 1.Badger J L, Miller V L. Expression of invasin and motility are coordinately regulated in Yersinia enterocolitica. J Bacteriol. 1998;180:793–800. doi: 10.1128/jb.180.4.793-800.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger J L, Miller V L. Role of RpoS in survival of Yersinia enterocolitica to a variety of environmental stresses. J Bacteriol. 1995;177:5370–5373. doi: 10.1128/jb.177.18.5370-5373.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates P F, Swift R A. Double cos site vectors: simplified cosmid cloning. Gene. 1983;26:137–146. doi: 10.1016/0378-1119(83)90183-x. [DOI] [PubMed] [Google Scholar]
- 4.Bearson S M, Benjamin W H, Jr, Swords W E, Foster J W. Acid shock induction of RpoS is mediated by the mouse virulence gene mviA of Salmonella typhimurium. J Bacteriol. 1996;178:2572–2579. doi: 10.1128/jb.178.9.2572-2579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bliska J, Falkow S. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc Natl Acad Sci USA. 1992;89:3561–3565. doi: 10.1073/pnas.89.8.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottone E J. Yersinia enterocolitica: a panoramic view of a charismatic microorganism. Crit Rev Microbiol. 1977;5:211–241. doi: 10.3109/10408417709102312. [DOI] [PubMed] [Google Scholar]
- 7.Chernoff Y O, Lindquist S L, Ono B, Inge-Vechtomov S G, Liebman S W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 8.Cover T L, Aber R C. Yersinia enterocolitica. N Engl J Med. 1989;321:16–24. doi: 10.1056/NEJM198907063210104. [DOI] [PubMed] [Google Scholar]
- 9.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 10.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman S, Maurizi M R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman S, Squires C, Pichersky E, Carrington M, Hobbs M, Mattick J S, Dalrymple B, Kuramitsu H, Shiroza T, Foster T, et al. Conservation of the regulatory subunit for the Clp ATP-dependent protease in prokaryotes and eukaryotes. Proc Natl Acad Sci USA. 1990;87:3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grutzkau A, Hanski C, Hahn H, Riecken E O. Involvement of M cells in the bacterial invasion of Peyer's patches: a common mechanism shared by Yersinia enterocolitica and other enteroinvasive bacteria. Gut. 1990;31:1011–1015. doi: 10.1136/gut.31.9.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas R, Kahrs A F, Facius D, Allmeier H, Schmitt R, Meyer T F. TnMax—a versatile mini-transposon for the analysis of cloned genes and shuttle mutagenesis. Gene. 1993;130:23–31. doi: 10.1016/0378-1119(93)90342-z. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 16.Hengge-Aronis R. Back to log phase: sigma S as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 17.Iriarte M, Stainier I, Cornelis G R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995;63:1840–1847. doi: 10.1128/iai.63.5.1840-1847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iriarte M, Stainier I, Mikulskis A V, Cornelis G R. The fliA gene encoding ς28 in Yersinia enterocolitica. J Bacteriol. 1995;177:2299–2304. doi: 10.1128/jb.177.9.2299-2304.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishihama A, Saitoh T. Subunits of RNA polymerase in function and structure. IX. Regulation of RNA polymerase activity by stringent starvation protein (SSP) J Mol Biol. 1979;129:517–530. doi: 10.1016/0022-2836(79)90466-2. [DOI] [PubMed] [Google Scholar]
- 20.Kapatral V, Minnich S A. Co-ordinate, temperature-sensitive regulation of the three Yersinia enterocolitica flagellin genes. Mol Microbiol. 1995;17:49–56. doi: 10.1111/j.1365-2958.1995.mmi_17010049.x. [DOI] [PubMed] [Google Scholar]
- 21.Kapatral V, Olson J W, Pepe J C, Miller V L, Minnich S A. Temperature-dependent regulation of Yersinia enterocolitica class III flagellar genes. Mol Microbiol. 1996;19:1061–1071. doi: 10.1046/j.1365-2958.1996.452978.x. [DOI] [PubMed] [Google Scholar]
- 22.Kinder S A, Badger J L, Bryant G O, Pepe J C, Miller V L. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O:8 and construction of a transformable R− M+ mutant. Gene. 1993;136:271–275. doi: 10.1016/0378-1119(93)90478-l. [DOI] [PubMed] [Google Scholar]
- 23.Kowarz L, Coynault C, Robbe-Saule V, Norel F. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J Bacteriol. 1994;176:6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewen P C, Hengge-Aronis R. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 25.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 26.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michiels T, Cornelis G R. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller V L, Falkow S. Evidence for two genetic loci in Yersinia enterocolitica that can promote invasion of epithelial cells. Infect Immun. 1988;56:1242–1248. doi: 10.1128/iai.56.5.1242-1248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Msadek T, Kunst F, Rapoport G. MecB of Bacillus subtilis, a member of the ClpC ATPase family, is a pleiotropic regulator controlling competence gene expression and growth at high temperature. Proc Natl Acad Sci USA. 1994;91:5788–5792. doi: 10.1073/pnas.91.13.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muffler A, Fischer D, Altuvia S, Storz G, Hengge-Aronis R. The response regulator RssB controls stability of the ςS subunit of RNA polymerase in Escherichia coli. EMBO J. 1996;15:1333–1339. [PMC free article] [PubMed] [Google Scholar]
- 31.Nickerson C A, Curtiss R., III Role of sigma factor RpoS in initial stages of Salmonella typhimurium infection. Infect Immun. 1997;65:1814–1823. doi: 10.1128/iai.65.5.1814-1823.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norel F, Robbe-Saule V, Popoff M Y, Coynault C. The putative sigma factor KatF (RpoS) is required for the transcription of Salmonella typhimurium virulence gene spvB in Escherichia coli. FEMS Microbiol Lett. 1992;78:271–276. doi: 10.1016/0378-1097(92)90039-q. [DOI] [PubMed] [Google Scholar]
- 33.Park S K, Kim K I, Woo K M, Seol J H, Tanaka K, Ichihara A, Ha D B, Chung C H. Site-directed mutagenesis of the dual translational initiation sites of the clpB gene of Escherichia coli and characterization of its gene products. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 34.Pederson K J, Carlson S, Pierson D E. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol Microbiol. 1997;26:99–107. doi: 10.1046/j.1365-2958.1997.5551916.x. [DOI] [PubMed] [Google Scholar]
- 35.Pepe J C, Badger J L, Miller V L. Growth phase and low pH affect the thermal regulation of the Yersinia enterocolitica inv gene. Mol Microbiol. 1994;11:123–135. doi: 10.1111/j.1365-2958.1994.tb00295.x. [DOI] [PubMed] [Google Scholar]
- 36.Pepe J C, Miller V L. The Yersinia enterocolitica inv gene product is an outer membrane protein that shares epitopes with Yersinia pseudotuberculosis invasin. J Bacteriol. 1990;172:3780–3789. doi: 10.1128/jb.172.7.3780-3789.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe J C, Miller V L. Yersinia enterocolitica invasin: a primary role in the initiation of infection. Proc Natl Acad Sci USA. 1993;90:6473–6477. doi: 10.1073/pnas.90.14.6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pierson D, Falkow S. Nonpathogenic isolates of Yersinia enterocolitica do not contain functional inv-homologous sequences. Infect Immun. 1990;58:1059–1064. doi: 10.1128/iai.58.4.1059-1064.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pierson D E, Falkow S. The ail gene of Yersinia enterocolitica has a role in the ability of the organism to survive serum killing. Infect Immun. 1993;61:1846–1852. doi: 10.1128/iai.61.5.1846-1852.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pratt L A, Silhavy T J. The response regulator SprE controls the stability of RpoS. Proc Natl Acad Sci USA. 1996;93:2488–2492. doi: 10.1073/pnas.93.6.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;12:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 42.Rashid M H, Tamakoshi A, Sekiguchi J. Effects of mecA and mecB (clpC) mutations on expression of sigD, which encodes an alternative sigma factor, and autolysin operons and on flagellin synthesis in Bacillus subtilis. J Bacteriol. 1996;178:4861–4869. doi: 10.1128/jb.178.16.4861-4869.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohde J R, Fox J M, Minnich S A. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing using chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweder T, Lee K-H, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor (ςs) by ClpXP protease. J Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 47.Squires C, Squires C L. The Clp proteins: proteolysis regulators or molecular chaperones? J Bacteriol. 1992;174:1081–1085. doi: 10.1128/jb.174.4.1081-1085.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Squires C L, Pederson S, Ross B M, Squires C. ClpB is the Escherichia coli heat shock protein F84. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wachtel M R, Miller V L. In vitro and in vivo characterization of an ail mutant of Yersinia enterocolitica. Infect Immun. 1995;63:2541–2548. doi: 10.1128/iai.63.7.2541-2548.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 52.Wawrzynow A, Banecki B, Zylicz M. The Clp ATPases define a novel class of molecular chaperones. Mol Microbiol. 1996;21:895–899. doi: 10.1046/j.1365-2958.1996.421404.x. [DOI] [PubMed] [Google Scholar]
- 53.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams M D, Ouyang T X, Flickinger M C. Starvation-induced expression of SspA and SspB: the effects of a null mutation in sspA on Escherichia coli protein synthesis and survival during growth and prolonged starvation. Mol Microbiol. 1994;11:1029–1043. doi: 10.1111/j.1365-2958.1994.tb00381.x. [DOI] [PubMed] [Google Scholar]
- 55.Woo K M, Kim K I, Goldberg A L, Ha D B, Chung C H. The heat-shock protein ClpB in Escherichia coli is a protein-activated ATPase. J Biol Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- 56.Young G M, Schmiel D H, Miller V L. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc Natl Acad Sci USA. 1999;96:6456–6461. doi: 10.1073/pnas.96.11.6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young G M, Smith M J, Minnich S A, Miller V L. The Yersinia enterocolitica motility master regulatory operon, flhDC, is required for flagellin production, swimming motility, and swarming motility. J Bacteriol. 1999;181:2823–2833. doi: 10.1128/jb.181.9.2823-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]