Abstract
Background:
Co-morbid substance use is very common. Despite a historical focus using genetic epidemiology to investigate comorbid substance use and misuse, few studies have examined substance-substance associations using polygenic risk score (PRS) methods.
Methods:
Using summary statistics from the largest substance use GWAS to date (258,797– 632,802 subjects), GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN), we constructed PRSs for smoking initiation (PRS-SI), age of initiation of regular smoking (PRS-AI), cigarettes per day (PRS-CPD), smoking cessation (PRS-SC), and drinks per week (PRS-DPW). We then estimated the fixed effect of individual PRSs on 22 lifetime substance use and substance use disorder phenotypes collected in an independent sample of 2463 young Australian adults using genetic restricted maximal likelihood (GREML) in Genome-wide Complex Trait Analysis (GCTA), separately in females, males and both sexes together.
Results:
After accounting for multiple testing, PRS-SI significantly explained variation in the risk of cocaine (0.67%), amphetamine (1.54%), hallucinogens (0.72%), ecstasy (1.66%) and cannabis initiation (0.97%), as well as DSM-5 alcohol use disorder (0.72%). PRS-DPW explained 0.75%, 0.59% and 0.90% of the variation of cocaine, amphetamine and ecstasy initiation respectively. None of the 22 phenotypes including emergent classes of substance use were significantly predicted by PRS-AI, PRS-CPD, and PRS-SC.
Conclusions:
To our knowledge, this is the first study to report significant genetic overlap between the polygenic risks for smoking initiation and alcohol consumption and the risk of initiating major classes of illicit substances. PRSs constructed from large discovery GWASs allows the detection of novel genetic associations.
Keywords: Polygenic risk, Addiction, Genetics, Twins
1. Introduction
1.1. Genetic Influence on Substance Use
Based on non-molecular but genetically informative family and twin studies, it is widely accepted that licit and illicit substance use (SU) and substance use disorders (SUDs) are polygenic with 30–80% heritability estimated across different substances as summarised by a comprehensive review (Kendler et al., 2012b). Twin studies have also shown that the genetic covariance between major licit and illicit substances use disorders can be explained by common genetic risks (Kendler et al., 2003, 2012a), with some evidence suggesting that highly correlated () genetic risks underpinning legal (nicotine, alcohol and caffeine) and illegal (cannabis and cocaine) SUDs (Kendler et al., 2007). Recent meta-analyses of genome-wide association studies (GWAS) provide evidence that SU and SUDs support the conclusion these are polygenic genetic behaviours (Minica et al., 2018; Pasman et al., 2018; Prom-Wormley et al., 2017; Walters et al., 2018) with multiple genes of small effect contributing to the variation in the risk. Despite their complex polygenicity (Pan et al., 2013; Robinson et al., 2014) the direct effects of individual SU and SUD loci and genes are becoming increasingly clear. Multiple loci have been associated with liability to consuming nicotine (Stephens et al., 2013; Thorgeirsson et al., 2010; Tobacco and Genetics Consortium, 2010), alcohol (Agarwal, 1997; Agrawal et al., 2011; Bierut et al., 2012, 2010; Chen et al., 1999; Edenberg, 2007; Edenberg et al., 2010; Edenberg and Foroud, 2006; Ehlers et al., 2004; Frank et al., 2012; Guindalini et al., 2005; Hasin et al., 2002a, 2002b; Heath et al., 2011; Kendler et al., 2011; Lind et al., 2010, 2008; Luo et al., 2006; Macgregor et al., 2009; Moore et al., 2007; Schumann et al., 2011; Strat et al., 2008; Treutlein et al., 2009; Treutlein and Rietschel, 2011; Walters et al., 2018; Wang et al., 2012; Zuo et al., 2012, 2011), and cannabis (Minica et al., 2018; Pasman et al., 2018; Sherva et al., 2016; Stringer et al., 2016), and SNPs related to multi-substance use (Sherva et al., 2010) have now been identified. Given the availability of very large, informative GWAS summary statistics there now exists the opportunity to examine the degree to which the variation of SU and SUD risk can be explained by emerging allelic risks as opposed to statistical inference based on twin and family studies.
1.2. Polygenic Risk of Substance Use
A key implication of the recent molecular findings is that a complete list of true replicable signals is not required for GWAS to demonstrate significant concurrent or predictive criterion validity. The upper tails of well-powered GWAS summary test distributions are expected to be highly enriched with many true signals (The International Schizophrenia Consortium, 2009) that can be used to estimate individualized polygenic risk score for complex behaviours including SU and SUDs.
Polygenic risk score (PRS) analysis aggregates the effects of thousands of genetic variants that are associated with a trait using a spectrum of significance levels. A PRS is calculated as a weighted sum of the number of risk alleles at the selected SNPs that are carried by an individual. The weight is obtained from the effect size (e.g. beta for a continuous trait; log transformation of the odds ratio for a binary trait) associated with the SNPs. These scores allow us to compare and correlate PRSs between different individuals or PRSs for different phenotypes.
A common problem in PRS methodology is that a PRS typically explains a small proportion, in comparison to twin estimates (Kendler et al., 2012b) of the variation of a target phenotype, which may be improved with larger-sized discovery sample from which the PRS is calculated. The criterion validity of PRS to predict a variety of important behavioural outcomes has clearly been demonstrated. For example, PRS based on much smaller discovery samples already predict complex traits (Agerbo et al., 2015; Clarke et al., 2015; Jervis et al., 2015; Moor et al., 2015) including cannabis use and cannabis use frequency (Power et al., 2014). A number of reports have also linked PRS for psychiatric disorders to SU and SUDs outcomes (Carey et al., 2016; Du Rietz et al., 2017; Hartz et al., 2017; Reginsson et al., 2017; Verweij et al., 2017). However, very few studies (Verweij et al., 2017; Vink et al., 2014) have investigated the associations between PRSs for SU and SUDs, and none of these studies have leveraged the most recent and largest SU GWAS to date. With PRSs derived from a very large discovery sample GWAS and Sequencing Consortium of Alcohol and Nicotine use (GSCAN), the current study is likely to maximise the power to detect an association between PRSs and SU and SUDs.
1.3. Aims
The aim of this study was to establish the association between polygenic risks for licit substance use and the risk of self-report illicit substance use, alcohol use disorders (AUD) as well as measures of cannabis misuse in an Australian sample of young adults.
2. Materials and Methods
Our overall aim was to calculate the PRSs for five measures of tobacco and alcohol consumption, and then test their associations with self-reported illicit SU phenotypes while appropriately adjusting for multiple testing. PRSs are commonly calculated using independent SNPs that meet different association p-value thresholds, which allows us calculating PRS from SNPs that did not reach genome-wide significance.
2.1. Target Sample and Measures
Self-report measures of SU and SUDs were obtained from a sample of young adult Australian twins and their families who participated in the Brisbane Longitudinal Twin Study 19Up Project between 2009 and 2016 (Couvy-Duchesne et al., 2018; Gillespie et al., 2013) in Brisbane, Australia. Twins and their families were invited to participate in this project when they turned 19 or older. The 19Up study was approved by the QIMR Human Research Ethics Committee. Data were stored in compliance with national regulations regarding personal data protection. Informed consent was obtained from all the participants.
Our target sample comprised 2463 individuals who were genotyped (mean age: 26.1 years; age range: 18.7–38.6) at QIMR Berghofer. This included 1977 twin individuals (835 twin pairs), and 486 siblings from 1163 families. The Composite International Diagnostic Interview (CIDI; (Kessler and Üstün, 2004)) was used to identify Diagnostic and Statistical Manual of Mental Disorders (Fourth Edition, DSM-IV) diagnoses of lifetime use of illicit substance (cocaine, amphetamine, inhalants, sedatives, hallucinogens, opioids, ecstasy, prescription painkillers, prescription stimulants, and cannabis), alcohol abuse, alcohol dependence, cannabis abuse, and cannabis dependence. The prevalence of the ten binary initiation phenotypes ranged from 5% for lifetime inhalants to 47% for lifetime cannabis use (Table 1). For the four abuse and dependence phenotypes, the prevalence was 7% to 33%. Significantly different prevalence was seen between males and females in these 14 phenotypes, except for sedative, opioid and painkiller initiation (Table 1). The DSM-5 AUD and CUD outcomes (0 = absent, 1 =mild, 2 = moderate, and 3 =severe) were constructed based on the number of symptoms identified, with two or three symptoms indicating a mild SUD, four or five symptoms indicating a moderate SUD, and six or more symptoms indicating a severe SUD. In addition to these two four-point scales, we derived binary phenotypes by dichotomising those who scored 0 or 1 and those who score 2 or 3.
Table 1.
Number of female and male participants surveyed for lifetime use of illicit substance and substance use disorders and result for Chi-Sqaure test of association between prevalence and sex.
| Females | Males | Assoc: prevalence and sex (DF=1) | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Target phenotype | N (%) | Cases (%) | N (%) | Cases (%) | χ2 | p-value |
|
| ||||||
| Ever used cocaine | 1336 (58%) | 184 (14%) | 976 (42%) | 238 (24%) | 41.86 | < .0001 |
| Ever used amphetamine | 1335 (58%) | 235 (18%) | 975 (42%) | 223 (23%) | 9.51 | 0.0020 |
| Ever used inhalants | 1331 (58%) | 45 (3%) | 972 (42%) | 72 (7%) | 18.06 | < .0001 |
| Ever used sedatives | 1334 (58%) | 177 (13%) | 975 (42%) | 153 (16%) | 2.51 | 0.1133 |
| Ever used hallucinogens | 1334 (58%) | 139 (10%) | 974 (42%) | 201 (21%) | 45.97 | < .0001 |
| Ever used opioids | 1335 (58%) | 72 (5%) | 974 (42%) | 64 (7%) | 1.20 | 0.2725 |
| Ever used ecstasy | 1337 (58%) | 318 (24%) | 975 (42%) | 333 (34%) | 29.46 | < .0001 |
| Ever used prescription pain killers | 1335 (58%) | 246 (18%) | 974 (42%) | 179 (18%) | 0.00 | 1.0000 |
| Ever used prescription stimulants | 1334 (58%) | 151 (11%) | 973 (42%) | 162 (17%) | 13.18 | 0.0003 |
| Ever used cannabis | 1106 (59%) | 450 (41%) | 784 (41%) | 443 (57%) | 45.42 | < .0001 |
| Alcohol abuse | 1368 (59%) | 386 (28%) | 959 (41%) | 390 (41%) | 38.76 | < .0001 |
| Alcohol dependence | 1368 (59%) | 305 (22%) | 959 (41%) | 343 (36%) | 50.25 | < .0001 |
| DSM-5 AUD (ctrl mild vs moderate severe) | 1368 (59%) | 310 (23%) | 959 (41%) | 352 (37%) | 53.94 | < .0001 |
| Cannabis abuse | 1368 (59%) | 105 (8%) | 959 (41%) | 165 (17%) | 49.00 | < .0001 |
| Cannabis dependence | 1368 (59%) | 61 (4%) | 959 (41%) | 91 (9%) | 22.55 | < .0001 |
| DSM-5 CUD (ctrl mild vs moderate severe) | 1368 (59%) | 75 (5%) | 959 (41%) | 122 (13%) | 37.20 | < .0001 |
2.2. Discovery Sample
We used GWAS summary statistics from the global ‘GSCAN’ consortium (Liu et al., 2019) with the QIMR sample removed prior to constructing PRSs for four smoking and one alcohol-related discovery phenotypes: smoking initiation (SI) (N = 631,564, 52% smokers, 53.6% females); age of initiation of regular smoking (AI) (N = 258,251, 50.0% females); cigarettes per day (CPD) (N = 258,999, 55.1% females); smoking cessation (SC) (N = 312,273, 40% current smokers, 50.6% females); and drinks per week (DPW) (N = 527,402, 53.2% females). GWAS summary statistics for these five phenotypes were generated with sex as a covariate when possible.
2.3. Genetic Correlations Between Discovery Phenotypes
Genetic correlations () between discovery phenotypes were estimated using cross-trait LD score correlation (LDSC; Bulik-Sullivan et al., 2015). This method is suitable for our study as it requires only GWAS summary statistics, is not biased by sample overlap and estimates of rG between a binary and continuous trait without having to specify a scale. We merged each of the five GWAS files with the HapMap3 SNP list, converted the summary statistics into the LDSC format, and calculated the genetic correlation. The independent variables and weights for LD score regression were read from an LD score computed from the 1000 Genomes European data.
2.4. Calculation of Polygenic Risk Scores
Before estimating the PRS for the 19UP subjects we first ensured quality control (QC) of the discovery GWAS summary statistics SNPs by removing SNPs with more than one occurrence, keeping SNPs that began with “rs” in their variant IDs, removing SNPs from ambiguous strands (i.e. SNPs that had “A T”, “T A”, “C G” or “G C” in their reference alleles and alternative alleles), and excluding insertions or deletions. Next, we ensured QC for SNPs in the target BLTS cohort sample by retaining SNPs with minor allele frequencies between 0.01 and 0.99 and with genotype imputation R2 ≥ 0.6 (an indication of imputation quality across different platforms), and applying the same criteria as the QC used for the discovery sample SNPs. Thirdly, an inner join was then performed to obtain SNPs that were common to the discovery and target SNPs using chromosome number and base pair position (CHR: BP) as the merging key. Fourthly, we accounted for linkage disequilibrium (LD) by selecting one representative SNP per haplotype, a process known as LD-based SNP clumping. Specifically, we set the Rsquared threshold (plink –clump-r2) to 0.1, and the distance threshold (plink –clump-kb) to 10,000 kb. The number of SNPs that met our eight p-value thresholds of 5e-08, 1e-05, 1e-03, 1e-02, 5e-02, 0.1, 0.5 and 1 was compared before (see QC in Table S11) and after (see LD in Table S11) clumping based on their linkage disequilibrium (LD) in individual autosomes. Across all these autosomes (see Total in Table S11), less than 1% of SNPs were selected through LD-based clumping. The difference in the number of clumped SNPs tended to be small between the five discovery phenotypes at higher p-value thresholds, with 400,877– 403,341 clumped SNPs at p-value < 1, 280,074–281,387 clumped SNPs at p < 0.5, 86,672– 91,667 clumped SNPs at p < 0.1, 49,797–55,505 clumped SNPs at p < 5e-02, 13,235– 18,428 clumped SNPs at p < 1e02. The PRS-SI at a p-value threshold lower than 1e-03 had nearly twice the number of clumped SNPs compared to the four other discovery phenotypes constructed with the same p-value. Fifthly, we constructed PRSs from imputed genotype dosage by summing up allelic scores of the clumped SNPs across all the chromosomal blocks of 22 autosomes, resulting in one PRS per p-value threshold and individual in the target sample:
where is an individual, is the effect size of an independent for a discovery phenotype, and is the number of risk alleles at the in the individual . The number of independent SNPs, denoted as , varies with the discovery phenotypes and p-value thresholds. Prior to the PRS calculation, the genotypes had been imputed on the Michigan Imputation Server (Das et al., 2016) using the Haplotype Reference Consortium (McCarthy et al., 2016) version r1.1 as the reference panel and then converted to plink dosage format at QIMR. Lastly, we standardised the PRSs and merged them back into the QIMR cohort who were assessed with SU and SUDs.
2.5. Univariate Mixed Model That Models Familial and Cryptic Relatedness
To estimate the proportion of variance in each target phenotype that was explained by the PRS, we performed linear mixed modeling using genetic restricted maximum likelihood (GREML) by the software GCTA (Yang et al., 2011) :
where Y is a vector of either a binary phenotype or continuous phenotype with being the number of individuals in the input data, X is a vector of fixed effect covariates, and is the effect estimate of the X. Considering the predictive power of our PRSs may be confounded by sex, we conducted the mixed modelling separately for males (M), females (F) and both sexes combined (F + M). The covariates used in the both-sex model included categorical covariates of study wave, sex, GWAS array, and quantitative covariates, such as a single PRS, age, age2, age x sex, age2 x sex, and the first ten principal components derived from the SNP data. The two single-sex models used the same covariates except for sex, age x sex, and age2 x sex.
The and denote the random genetic effect and error term respectively. The genetic effect has a known variance-covariance structure that is defined by the genetic relationship matrix (GRM), which estimates the genetic relatedness between individuals using SNP data. Although the GCTA-GREML is developed mainly for estimating SNP heritability, this method provides additional functionality of estimating fixed effect parameters in a sample of related individuals. Our fixed effects were estimated using the GCTA –reml-est-fix option. We then derived an estimate of the association expressed as a proportion of the target phenotype (both binary and continuous) variance explained by PRS, R2 as
where is the fixed effect estimate of a PRS, is the standard deviation of a binary or continuous target phenotype Y, and is the standard deviation of the . Here, the R2 is defined as the square of the correlation between Y and PRS. We calculated two-sided p-values from at distribution in R.
2.6. Multiple Testing
We tested the association between each of the 22 target phenotypes and each of the 40 PRSs, giving a total of 880 association tests. To account for multiple testing (Table S2), we presented p-values adjusted for the effective number of independent target and discovery phenotypes (threshold T4) and Bonferroni-corrected p-values (threshold T5). Bonferroni correction is known to be extremely conservative and increase the likelihood of a type II error. We, therefore, interpreted our results based on the associations that remained significant after accounting for the T4 threshold, which was calculated as
Where is the nominal p-value, is the effective number of independent target phenotypes, and is the effective number of independent discovery phenotypes (Li and Ji, 2005; Nyholt, 2004). We estimated the as 14 and as five in females, males, and both sexes.
2.7. Statistical Software
The pipeline of PRS calculation was coded in BASH (Free Software Foundation, 2007) and R (R Core Team, 2017). LD-based SNP clumping and PRS calculation were performed using Plink (Chang et al., 2015). Univariate GREML was performed using GCTA (Yang et al., 2011). We used R and Base SAS 9.4 (SAS Institute Inc., 2017) for data cleaning, statistical analyses, graph and table generation.
3. Results
3.1. Genetic Correlations Between Discovery Phenotypes
Shown in Table 2, the five discovery phenotypes were significantly correlated with each other, with positive genetic correlation coefficients ranging between 0.08 and 0.43, and negative genetic correlation coefficients ranging between −0.69 and −0.15. AI negatively correlated with each of the other four phenotypes, suggesting that genetic liability to starting regular smoking at an early age could be related to the genetic liability to a higher chance of starting regular smoking (SI), smoking more cigarettes (CPD), or drinking more alcoholic beverage (DPW). There is also evidence for the genetic overlap between SI and CPD, SC and DPW.
Table 2.
Genetic correlations () between any two (trait1, trait2) of the five smoking and alcohol-related discovery phenotypes that were meta-analysed by genome-wide association studies.
| Trait1 | Trait2 | SE | p value | |
|---|---|---|---|---|
|
| ||||
| AI | CPD | −0.381 | 0.037 | < 0.001 |
| AI | DPW | −0.15 | 0.031 | < 0.001 |
| AI | SC | −0.294 | 0.041 | < 0.001 |
| AI | SI | −0.685 | 0.024 | < 0.001 |
| CPD | DPW | 0.084 | 0.028 | 0.0030 |
| CPD | SC | 0.426 | 0.033 | < 0.001 |
| CPD | SI | 0.28 | 0.033 | < 0.001 |
| DPW | SC | 0.104 | 0.033 | 0.0016 |
| DPW | SI | 0.403 | 0.018 | < 0.001 |
| SC | SI | 0.39 | 0.029 | < 0.001 |
The , standard errors (SE) and p values were estimated by cross-trait LD score regression. These five phenotypes are smoking initiation (SI, binary), age at starting regular smoking (AI, continuous), cigarettes per day (CPD, continuous), smoking cessation (SC, binary), and drinks per week (DPW, continuous).
3.2. Associations Between PRS and the Self-reported SU and SUD Measures
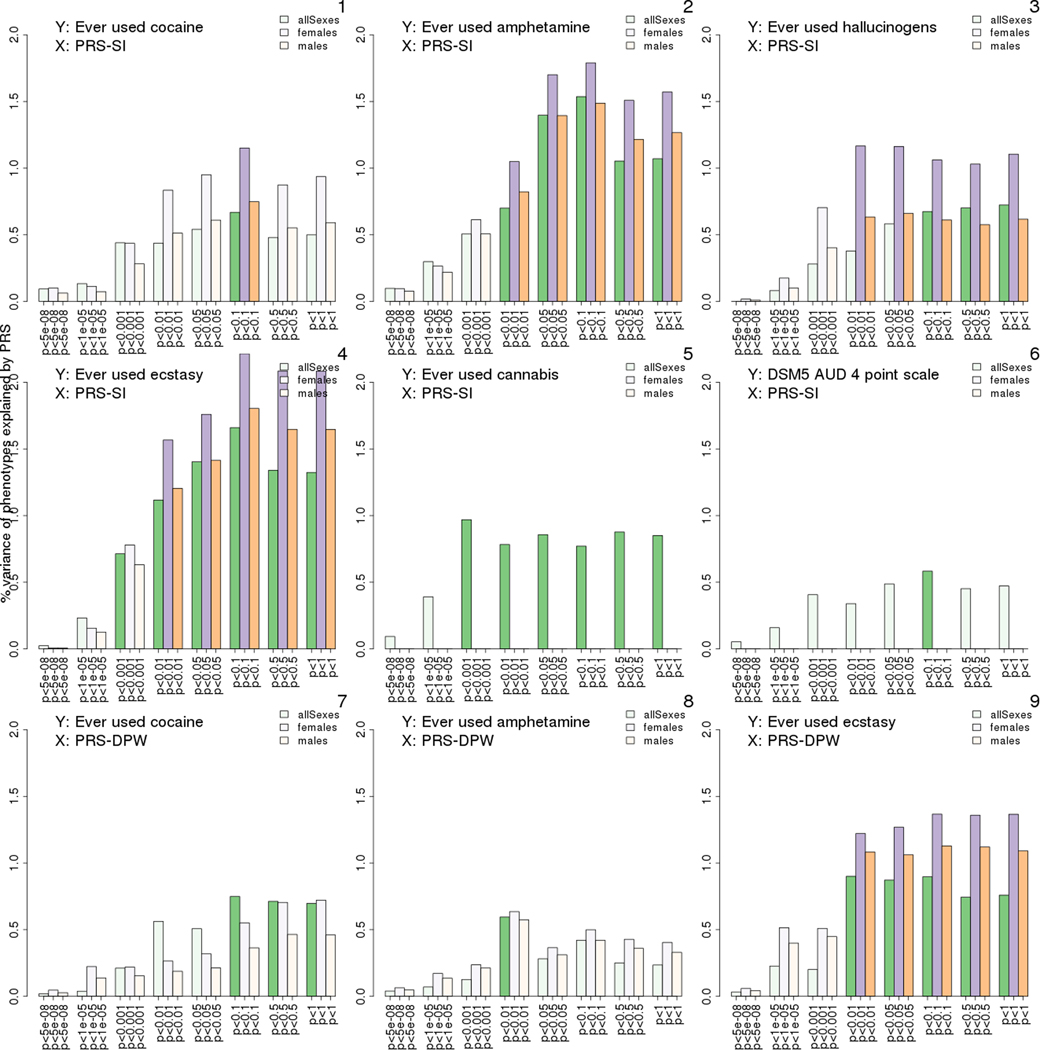
After correcting for multiple testing, significant predictors were only seen in PRS-SI, which explained the variance of six target phenotypes, and in PRS-DPW explaining the variance of three target phenotypes. Percentage of variation explained by the PRSs (prediction R2) in these significant associations are shown in Fig. 1. The associations that remained statistically significant after the Bonferroni-correction were found between PRS-SI and amphetamine initiation, ecstasy initiation, and alcohol abuse, as well as between PRS-DPW and ecstasy initiation. An overview of the results for the entire 880 substance-PRS associations is visualised for both sexes (Fig. S1), males (Fig. S2) and females (Fig. S3).1 Effect sizes of PRSs were all positive in these associations (estimates highlighted in blue and bold-face, Table S31). The effect size estimates were similar in males and females (Table S31). Phenotypic correlations between individual target phenotypes and PRSs were generally very small, ranging between −0.3 and 0.37 (Table S41), with the highest correlation found between age at onset of cannabis dependence and PRS-DPW calculated at p value < 5e-08.
Fig. 1.
Percentage variance of liability to illicit substance use (disorder) explained by polygenic risk scores (PRS) for smoking initiation (PRS-SI) or drinks per week (PRS-DPW) in males (M), females (F) and both sexes (F + M). Bar height represents the percent of the variation in a target phenotype explained by a PRS. Associations that remained significant after accounting for multiple testing (adjusted p-value threshold: 7.14e-04) are shown as green in both sexes combined, as orange in females and as purple in females. Bars from non-significant associations are shown in pale colour. Bar groups on the x-axis indicate the eight p-value thresholds at which the PRSs were calculated: 5e-08, 1e-05, 1e-03, 1e-02, 0.05, 0.1, 0.5, and 1 (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
4. Discussion
The aim of this study was to examine the association between molecular-based estimates of polygenic risk for alcohol and nicotine use and self-report measures of common and emergent types of illicit substance use and misuse. The pattern of genetic correlations (positive range: 0.08∼0.43, negative range: −0.68∼−0.15, see Table 2), as estimated by cross-trait LD score regression, between the five discovery PRS was very similar to those observed in the full GSCAN full sample ( range: −0.71∼0.42, Liu et al., 2019). The highest was between AI and SI, along with moderate to weak correlations in nine other pairwise correlations. Among the SU phenotypes predicted by PRS-SI or PRS-DPW were lifetime cocaine, amphetamine, hallucinogen, ecstasy, and cannabis use (Fig. 1). The DSM-5 AUD was the only clinical SUD diagnosis that was predicted by PRS-SI. Importantly, none of the PRSs that were based only on SNPs that reached genome-wide significant (p-value < 5e-08) significantly predicted any of the SU or SUD outcomes. However, significant associations were found when PRSs were based on less stringent p-values (5e-08 < p-value < 1). Despite the small-sized target sample, the use of PRSs based on a large discovery GWAS samples has the power to predict, to some extent, individuals at risk of comorbid SU and progression to AUDs.
4.1. Prediction by PRSs
Although the range in variances explained by our PRSs was small (0.67%–1.66% in both sexes, 0.00%∼2.21% in females, 0.58%∼1.80% in males, see Table S31), these estimates are commensurate with other studies that have used aggregated genetic risk based on well-powered GWAS discoveries to predict substance use and misuse. For example, GSCAN PRS for AI, CPD, SI and DPW explained between 1% and 4% of the variance in similar measures assessed by the Add Health and Health and Retirement Study datasets (Liu et al., 2019). Likewise, PRS based on a recent genome-wide meta-analysis of alcohol dependence explained between 0.3% and 1.7% of the variance in alcohol use and misuse phenotypes in the large Avon Longitudinal Study of Parents and Children (ALSPAC), Generation Scotland (GS), and Collaborative Study on the Genetics of Alcoholism (COGA) samples (Walters et al., 2018).
We found that cannabis initiation was significantly predicted by the PRS-SI, but not by PRS-CPD or PRS-SC. In contrast, Vink et al. (2014) reported an association between cannabis initiation and a PRS for CPD, based on the Tobacco and Genetics Consortium (TAG), but not with a TAG-based PRS for SI (N = 69,409), or PRS for SC. Apart from the obvious discrepancy in the two discovery sample sizes, the differences in the prediction by the smoking-related PRSs predicts may be attributable to the heterogeneity in terms of how the discovery GWAS consortia defined their phenotypes. For example, the GSCAN SI was a binary phenotype based on whether subjects had ever been a regular smoker, whereas the TAG-based SI used by Vink et al. was more strictly defined. The GSCAN CPD used the average number of cigarettes smoked per day scored on an ordinal scale with five response categories (1–5, 6–15, 16–25, 26–35, or 36 or more cigarettes smoked per day) whereas the CPD used by Vink et al. was the average or maximum number of cigarettes smoked per day from contributing studies. We cannot rule out the possibility that our study was underpowered to detect the effect of PRS-CPD on the initiation of cannabis, and even the other substances.
Given the sample sizes and success of recent GWSA meta-analyses investigating alcohol use and misuse (Liu et al., 2019; Walters et al., 2018) our a priori expectation was that the PRS- DPW should have predicted the self-reported measures of alcohol misuse. Instead, the PRS-DPW did not predict any alcohol-related disorders. In addition to the lack of power related to the BLTS sample size, this null finding could be attributable to the possibility that the PRS-DPW was assessing an unrelated genetic liability. Note that GSCAN DPW phenotype was based on either current or former drinkers, combined all types of liquor, or averaged across all assessments when based on longitudinal data (Liu et al., 2019). Kendler et al. (2012a) found that the DSM-IV syndrome of alcohol dependence does not reflect a single dimension but instead is best characterized by three distinct genetic factors: tolerance and heavy use; loss of control; and withdrawal symptoms (Kendler et al., 2012b). Therefore, it is plausible that GSCAN-based PRS for DPW may be indexing the genetics of tolerance and heavy use, which is in contrast to our measure of alcohol misuse that is more likely to be indexing genetic risks related to loss of control and withdrawal.
The pattern of non-significant associations between the alcohol and nicotine-based PRS and the non-medical use of prescribed or over the counter analgesics is commensurate with our recent twin modeling exploring the etiology of this emergent class of SU. Briefly, we have found that apart from opioids, broadly defined non-medical use of analgesics is genetically correlated () but mostly distinct from the genetic risks underpinning lifetime nicotine use (Gillespie et al., 2019). In terms of diagnostic outcomes, non-medical use of analgesics was only partially genetically correlated with either alcohol use disorder () and nicotine dependence () (Gillespie et al., 2019), which suggests that most of the genetic risks in non-medical use of analgesics are variable specific.
4.2. Limitations
Our results must be interpreted in the context of the following two limitations. First, the discovery sample was based predominately on Caucasian ancestral groups. Consequently, the significant PRS-phenotype correlations observed here may not apply to other ethnic groups especially those with different haplotype structures. The need to collect genetic data from non-European-ancestry populations has been recently discussed (Bentley et al., 2017; Kessler et al., 2016; Lewis and Vassos, 2017). Second, the significant associations between genetic risks for alcohol- and nicotine-related phenotypes and self-report measures of SU and SUDs do not imply causation. Third, our PRSs explained a relatively small proportion of the genetic variance, a common problem in addiction studies (Mies et al., 2017; Vink et al., 2014) that uses the PRS approach. Although it is premature to discuss the clinical utility of PRSs in genetic risk prediction for substance use, the PRS method has been shown to improve risk prediction in other diseases, such as prostate cancer (Helfand et al., 2016), when it is combined with a conventional risk factor family history. As discovery sample sizes increase and the PRS approach continues to refine, the PRS predictor can prove useful in discriminating patients in the top and bottom risk deciles (Lewis and Vassos, 2017).
4.3. Significance of this Study
The current results lend support for there being shared genetic risks between initiation of tobacco and various addictive substances (e.g. cocaine, amphetamine, hallucinogens, ecstasy, and cannabis), as well as between alcohol consumption and several of these substances. Twin studies show that common genetic factors tend to exert a varying degree of influence on the covariation of tobacco and cannabis use and misuse at different stages of the involvement of these two substances (Huizink et al., 2010; Neale et al., 2006), with the stronger influence at the earlier stages (e.g. initiation) than the later stages (e.g. progression) of the use. Taken together with our finding, this stage-dependent genetic relationship may be generalised to tobacco and other addictive substances (i.e. cocaine, amphetamine, hallucinogens, ecstasy).
Supplementary Material
Acknowledgements
We thank the twins and their families for participating in our studies. Special thanks to Jue-Sheng Ong, Jiyuan An, Xikun Han, MingChung Chiu and Chun-hui Yu for advice on better programming practices in BASH and R.
Role of Funding Source
Research in the current study was funded by National Health and Medical Research Council [APP10499110] in Australia and in part by National Institutes of Health [K99R00, R00DA023549, R21DA038852, R01DA037904]. LHC is supported by a QIMR International PhD scholarship. SEM is supported by an NHMRC fellowship [APP1103623].
Footnotes
Conflict of Interest
The authors certify that they have no commercial associations that might pose a conflict of interest in connection with this article
Appendix A. Supplementary data
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.drugalcdep.2019.01.015.
Supplementary material can be found by accessing the online version of this paper at https://doi.org/10.1016/j.drugalcdep.2019.01.015.
References
- Agarwal DP, 1997. Molecular genetic aspects of alcohol metabolism and alcoholism. Pharmacopsychiatry 30, 79–84. 10.1055/s-2007-979487. [DOI] [PubMed] [Google Scholar]
- Agerbo E, Sullivan PF, Vilhjalmsson BJ, Pedersen CB, Mors O, Borglum AD, Hougaard DM, Hollegaard MV, Meier S, Mattheisen M, Ripke S, Wray NR, Mortensen PB, 2015. Polygenic risk score, parental socioeconomic status, family history of psychiatric disorders, and the risk for schizophrenia: a Danish population-based study and meta-analysis. JAMA Psychiatry. 10.1001/jamapsychiatry.2015.0346. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Agrawal A, Freedman ND, Bierut LJ, 2011. Genome-wide association studies of alcohol intake–a promising cocktail? Am. J. Clin. Nutr 93, 681–683. 10.3945/ajcn.111.012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley AR, Callier S, Rotimi CN, 2017. Diversity and inclusion in genomic research: why the uneven progress? J. Community Genet 8, 255–266. 10.1007/s12687-017-0316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nothen MM, Nurnberger JI Jr., Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP, 2010. A genome-wide association study of alcohol dependence. Proc. Natl. Acad. Sci. U. S. A 107, 5082–5087. 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, Agrawal A, Bucholz KK, Grucza R, Hesselbrock V, Kramer J, Kuperman S, Nurnberger J, Porjesz B, Saccone NL, Schuckit M, Tischfield J, Wang JC, Foroud T, Rice JP, Edenberg HJ, 2012. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol. Psychiatry 17, 445–450. 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Consortium SWG of the P.G, Patterson N, Daly MJ, Price AL, Neale BM, 2015. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet 47, 291–295. 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey CE, Agrawal A, Bucholz KK, Hartz SM, Lynskey MT, Nelson EC, Bierut LJ, Bogdan R, 2016. Associations between polygenic risk for psychiatric disorders and substance involvement. Front. Genet 7, 149. 10.3389/fgene.2016.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ, 2015. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4. 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ, 1999. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am. J. Hum. Genet 65, 795–807. 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke TK, Smith AH, Gelernter J, Kranzler HR, Farrer LA, Hall LS, Fernandez-Pujals AM, MacIntyre DJ, Smith BH, Hocking LJ, Padmanabhan S, Hayward C, Thomson PA, Porteous DJ, Deary IJ, McIntosh AM, 2015. Polygenic risk for alcohol dependence associates with alcohol consumption, cognitive function and social deprivation in a population-based cohort. Addict. Biol 10.1111/adb.12245. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvy-Duchesne B, O’Callaghan V, Parker R, Mills N, Kirk KM, Scott Jan, Vinkhuyzen A, Hermens DF, Lind PA, Davenport TA, Burns JM, Connell M, Zietsch BP, Scott James, Wright MJ, Medland SE, McGrath J, Martin NG, Hickie IB, Gillespie NA, 2018. Nineteen and up study (19Up): understanding pathways to mental health disorders in young Australian twins. BMJ Open 8, e018959. 10.1136/bmjopen-2017-018959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh P-R, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C, 2016. Next-generation genotype imputation service and methods. Nat. Genet 48, 1284–1287. 10.1038/ng.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Rietz E, Coleman J, Glanville K, Choi SW, O’Reilly PF, Kuntsi J, 2017. Association of polygenic risk for attention-deficit/hyperactivity disorder with co-occurring traits and disorders. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 10.1016/j.bpsc.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, 2007. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health 30, 5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, 2006. The genetics of alcoholism: identifying specific genes through family studies. Addict. Biol 11, 386–396. 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI Jr., Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T, 2010. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol. Clin. Exp. Res 34, 840–852. 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Spence JP, Wall TL, Gilder DA, Carr LG, 2004. Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in southwest California indians. Alcohol. Clin. Exp. Res 28, 1481–1486. [DOI] [PubMed] [Google Scholar]
- Frank J, Cichon S, Treutlein J, Ridinger M, Mattheisen M, Hoffmann P, Herms S, Wodarz N, Soyka M, Zill P, Maier W, Mossner R, Gaebel W, Dahmen N, Scherbaum N, Schmal C, Steffens M, Lucae S, Ising M, Muller-Myhsok B, Nothen MM, Mann K, Kiefer F, Rietschel M, 2012. Genome-wide significant association between alcohol dependence and a variant in the ADH gene cluster. Addict. Biol 17, 171–180. 10.1111/j.1369-1600.2011.00395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Free Software Foundation, 2007. Bash (3.2.48) [Unix Shell Program] Retrieved from. http://ftp.gnu.org/gnu/bash/bash-3.2.48.tar.gz. [Google Scholar]
- Gillespie NA, Henders AK, Davenport TA, Hermens DF, Wright MJ, Martin NG, Hickie IB, 2013. The Brisbane Longitudinal Twin Study: pathways to cannabis use, abuse, and dependence project—current status, preliminary results, and future directions. Twin Res. Hum. Genet 16, 21–33. 10.1017/thg.2012.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie NA, Bates TC, Hickie IB, Medland SE, Verhulst B, Kirkpatrick RM, Kendler KS, Martin NG, Benotsch EG, 2019. Genetic and environmental risk factors in non-medically prescribed and over-the-counter analgesics, and their relationship to major classes of licit and illicit substance use and misuse in a population-based sample of young adult twins (submitted). Drug Alcohol Depend. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindalini C, Scivoletto S, Ferreira RG, Breen G, Zilberman M, Peluso MA, Zatz M, 2005. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am. J. Psychiatry 162, 1005–1007. 10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- Hartz SM, Horton AC, Oehlert M, Carey CE, Agrawal A, Bogdan R, Chen L-S, Hancock DB, Johnson EO, Pato CN, Pato MT, Rice JP, Bierut LJ, 2017. Association between substance use disorder and polygenic liability to schizophrenia. Biol. Psychiatry 82, 709–715. 10.1016/j.biopsych.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D, Aharonovich E, Liu X, Mamman Z, Matseoane K, Carr L, Li TK, 2002a. Alcohol and ADH2 in Israel: Ashkenazis, Sephardics, and recent Russian immigrants. Am. J. Psychiatry 159, 1432–1434. [DOI] [PubMed] [Google Scholar]
- Hasin D, Aharonovich E, Liu X, Mamman Z, Matseoane K, Carr Lg, Li TK, 2002b. Alcohol dependence symptoms and alcohol dehydrogenase 2 polymorphism: Israeli Ashkenazis, Sephardics, and recent Russian immigrants. Alcohol. Clin. Exp. Res 26, 1315–1321. 10.1097/01.alc.0000029597.07916.a9. [DOI] [PubMed] [Google Scholar]
- Heath AC, Whitfield JB, Martin NG, Pergadia ML, Goate AM, Lind PA, McEvoy BP, Schrage AJ, Grant JD, Chou YL, Zhu R, Henders AK, Medland SE, Gordon SD, Nelson EC, Agrawal A, Nyholt DR, Bucholz KK, Madden PA, Montgomery GW, 2011. A quantitative-trait genome-wide association study of alcoholism risk in the community: findings and implications. Biol. Psychiatry 70, 513–518. 10.1016/j.biopsych.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Kearns J, Conran C, Xu J, 2016. Clinical validity and utility of genetic risk scores in prostate cancer. Asian J. Androl 18, 509–514. 10.4103/1008-682X.182981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizink AC, Levälahti E, Korhonen T, Dick DM, Pulkkinen L, Rose RJ, Kaprio J, 2010. Tobacco, cannabis, and other illicit drug use among Finnish adolescent twins: Causal relationship or correlated liabilities? J. Stud. Alcohol Drugs 71, 5–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jervis S, Song H, Lee A, Dicks E, Harrington P, Baynes C, Manchanda R, Easton DF, Jacobs I, Pharoah PP, Antoniou AC, 2015. A risk prediction algorithm for ovarian cancer incorporating BRCA1, BRCA2, common alleles and other familial effects. J. Med. Genet 10.1136/jmedgenet-2015-103077. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Jacobson KC, Prescott CA, Neale MC, 2003. Specificity of genetic and environmental risk factors for use and abuse/dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am. J. Psychiatry 160 687–495. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Myers J, Prescott CA, 2007. Specificity of genetic and environmental risk factors for symptoms of cannabis, cocaine, alcohol, caffeine, and nicotine dependence. Arch. Gen. Psychiatry 64, 1313–1320. 10.1001/archpsyc.64.11.1313. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Kalsi G, Holmans PA, Sanders AR, Aggen SH, Dick DM, Aliev F, Shi J, Levinson DF, Gejman PV, 2011. Genomewide association analysis of symptoms of alcohol dependence in the molecular genetics of schizophrenia (MGS2) control sample. Alcohol. Clin. Exp. Res 35, 963–975. 10.1111/j.1530-0277.2010.01427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Aggen SH, Prescott CA, Crabbe J, Neale MC, 2012a. Evidence for multiple genetic factors underlying the DSM-IV criteria for alcohol dependence. Mol. Psychiatry 17, 1306–1315. 10.1038/mp.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Chen X, Dick D, Maes H, Gillespie N, Neale MC, Riley B, 2012b. Recent advances in the genetic epidemiology and molecular genetics of substance use disorders. Nat. Neurosci 15, 181–189. 10.1038/nn.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Üstün TB, 2004. The world mental health (WMH) survey initiative version of the World Health Organization (WHO) composite international diagnostic interview (CIDI). Int. J. Methods Psychiatr. Res 13, 93–121. 10.1002/mpr.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MD, Yerges-Armstrong L, Taub MA, Shetty AC, Maloney K, Jeng LJB, Ruczinski I, Levin AM, Williams LK, Beaty TH, Mathias RA, Barnes KC, Americas (CAAPA), C. on A. among A.P. in the, Boorgula MP, Campbell M, Chavan S, Ford JG, Foster C, Gao L, Hansel NN, Horowitz E, Huang L, Ortiz R, Potee J, Rafaels N, Scott AF, Vergara C, Gao J, Hu Y, Johnston HR, Qin ZS, Padhukasahasram B, Dunston GM, Faruque MU, Kenny EE, Gietzen K, Hansen M, Genuario R, Bullis D, Lawley C, Deshpande A, Grus WE, Locke DP, Foreman MG, Avila PC, Grammer L, Kim K-Y, Kumar R, Schleimer R, Bustamante C, Vega FMDL, Gignoux CR, Shringarpure SS, Musharoff S, Wojcik G, Burchard EG, Eng C, Gourraud P-A, Hernandez RD, Lizee A, Pino-Yanes M, Torgerson DG, Szpiech ZA, Torres R, Nicolae DL, Ober C, Olopade CO, Olopade O, Oluwole O, Arinola G, Song W, Abecasis G, Correa A, Musani S, Wilson JG, Lange LA, Akey J, Bamshad M, Chong J, Fu W, Nickerson D, Reiner A, Hartert T, Ware LB, Bleecker E, Meyers D, Ortega VE, Pissamai MRN, Trevor MRN, Watson H, Araujo MI, Oliveira RR, Caraballo L, Marrugo J, Martinez B, Meza C, Ayestas G, Herrera-Paz EF, Landaverde-Torres P, Erazo SOL, Martinez R, Mayorga A, Mayorga LF, Mejia-Mejia D-A, Ramos H, Saenz A, Varela G, Vasquez OM, Ferguson T, Knight-Madden J, Samms-Vaughan M, Wilks RJ, Adegnika A, Ateba-Ngoa U, Yazdanbakhsh M, O’Connor TD, 2016. Challenges and disparities in the application of personalized genomic medicine to populations with African ancestry. Nat. Commun 7, 12521. 10.1038/ncomms12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Vassos E, 2017. Prospects for using risk scores in polygenic medicine. Genome Med. 9. 10.1186/s13073-017-0489-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ji L, 2005. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity (Edinb) 95, 221–227. 10.1038/sj.hdy.6800717. [DOI] [PubMed] [Google Scholar]
- Lind PA, Eriksson CJ, Wilhelmsen KC, 2008. The role of aldehyde dehydrogenase-1 (ALDH1A1) polymorphisms in harmful alcohol consumption in a Finnish population. Hum. Genomics 3, 24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind PA, Macgregor S, Vink JM, Pergadia ML, Hansell NK, de Moor MH, Smit AB, Hottenga JJ, Richter MM, Heath AC, Martin NG, Willemsen G, de Geus EJ, Vogelzangs N, Penninx BW, Whitfield JB, Montgomery GW, Boomsma DI, Madden PA, 2010. A genomewide association study of nicotine and alcohol dependence in Australian and Dutch populations. Twin Res. Hum. Genet 13, 10–29. 10.1375/twin.13.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, … Vrieze S, 2019. Association studies of up to 1.2 million individuals yield new insights in the genetic etiology of tobacco and alcohol use (in press). Nat. Genet [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang BZ, Gelernter J, 2006. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology 31, 1085–1095. 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB, 2009. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum. Mol. Genet 18, 580–593. 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett J, Boomsma DI, Branham K, Breen G, Brummet C, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin L, Davey Smith G, Dedoussis G, Dorr M, Farmaki A-E, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee J, McGue M, Meitinger T, Melzer D, Min J, Mohlke KL, Vincent J, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards B, Sala C, Salomaa V, Schlessinger D, Schoenheer S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg L, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker P, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson C, Boehnke M, McCarthy MI, Durbin R, Abecasis G, Marchini J, 2016. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet 48, 1279–1283. 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mies GW, Verweij KJH, Treur JL, Ligthart L, Fedko IO, Hottenga JJ, Willemsen G, Bartels M, Boomsma DI, Vink JM, 2017. Polygenic risk for alcohol consumption and its association with alcohol-related phenotypes: Do stress and life satisfaction moderate these relationships? Drug Alcohol Depend. 183, 7–12. 10.1016/j.drugalcdep.2017.10.018. [DOI] [PubMed] [Google Scholar]
- Minica CC, Verweij KJH, van der Most PJ, Mbarek H, Bernard M, van Eijk KR, Lind PA, Liu MZ, Maciejewski DF, Palviainen T, Sanchez-Mora C, Sherva R, Taylor M, Walters RK, Abdellaoui A, Bigdeli TB, Branje SJT, Brown SA, Casas M, Corley RP, Smith GD, Davies GE, Ehli EA, Farrer L, Fedko IO, Garcia-Martinez I, Gordon SD, Hartman CA, Heath AC, Hickie IB, Hickman M, Hopfer CJ, Hottenga JJ, Kahn RS, Kaprio J, Korhonen T, Kranzler HR, Krauter K, van Lier PAC, Madden PAF, Medland SE, Neale MC, Meeus WHJ, Montgomery GW, Nolte IM, Oldehinkel AJ, Pausova Z, Ramos-Quiroga JA, Richarte V, Rose RJ, Shin J, Stallings MC, Wall TL, Ware JJ, Wright MJ, Zhao H, Koot HM, Paus T, Hewitt JK, Ribases M, Loukola A, Boks MP, Snieder H, Munafo MR, Gelernter J, Boomsma DI, Martin NG, Gillespie NA, Vink JM, Derks EM, 2018. Genome-wide association meta-analysis of age at first cannabis use. Addiction (Abingdon, England). 10.1111/add.14368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor MHM, de van den Berg SM, Verweij KJH, Krueger RF, Luciano M, Vasquez AA, Matteson LK, Derringer J, Esko T, Amin N, Gordon SD, Hansell NK, Hart AB, Seppälä I, Huffman JE, Konte B, Lahti J, Lee M, Miller M, Nutile T, Tanaka T, Teumer A, Viktorin A, We denoja J, Abecasis GR, Adkins DE, Agrawal A, Allik J, Appel K, Big deli TB, Busonero F, Campbell H, Costa PT, Smith GD, Davies G, Wit Hde, Ding J, Engelhardt BE, Eriksson JG, Fedko IO, Ferrucci L, Franke B, Giegling I, Grucza R, Hartmann AM, Heath AC, Heinonen K, Hen ders AK, Homuth G, Hottenga J-J, Iacono WG, Janzing J, Jokela M, Karlsson R, Kemp JP, Kirkpatrick MG, Latvala A, Lehtimäki T, Liewald DC, Mad den PAF, Magri C, Magnusson PKE, Marten J, Maschio A, Medland SE, Mihailov E, Milaneschi Y, Montgomery GW, Nauck M, Ouwens KG, Palotie A, Pettersson E, Polasek O, Qian Y, Pulkki-Råback L, Raitakari OT, Realo A, Rose RJ, Ruggiero D, Schmidt CO, Slutske WS, Sorice R, Starr JM, Pourcain BS, Sutin AR, Timpson NJ, Trochet H, Vermeulen S, Vuoksimaa E, Wi den E, Wouda J, Wright MJ, Zgaga L, Porteous D, Minelli A, Palmer AA, Rujescu D, Ciullo M, Hayward C, Rudan I, Metspalu A, Kaprio J, Deary IJ, Räikkönen K, Wilson JF, Keltikangas-Järvinen L, Bierut LJ, Hettema JM, Grabe HJ, van Duijn CM, E vans DM, Schlessinger D, Pe dersen NL, Terracciano A, McGue M, Penninx BWJH, Martin NG, Boomsma DI, 2015. Meta-analysis of genome-wi de association studies for neuroticism, and the polygenic association with major depressive disor der. JAMA Psychiatry 72, 642–650. 10.1001/jamapsychiatry.2015.0554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore S, Montane-Jaime K, Shafe S, Joseph R, Crooks H, Carr LG, Ehlers CL, 2007. Association of ALDH1 promoter polymorphisms with alcohol-related phenotypes in Trinidad and Tobago. J. Stud. Alcohol Drugs 68, 192–196. [DOI] [PubMed] [Google Scholar]
- Neale MC, Harvey E, Maes HHM, Sullivan PF, Kendler KS, 2006. Extensions to the modeling of initiation and progression: applications to substance use and abuse. Behav. Genet 36, 507–524. 10.1007/s10519-006-9063-x. [DOI] [PubMed] [Google Scholar]
- Nyholt DR, 2004. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am. J. Hum. Genet 74, 765–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Luo X, Liu X, Wu LY, Zhang Q, Wang L, Wang W, Zuo L, Wang KS, 2013. Genome-wide association studies of maximum number of drinks. J. Psychiatr. Res 47, 1717–1724. 10.1016/j.jpsychires.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, Abdellaoui A, Nivard MG, Baselmans BML, Ong J-S, Ip HF, van der Zee MD, Bartels M, Day FR, Fontanillas P, Elson SL, de Wit H, Davis LK, MacKillop J, Derringer JL, Branje SJT, Hartman CA, Heath AC, van Lier PAC, Madden PAF, Magi R, Meeus W, Montgomery GW, Oldehinkel AJ, Pausova Z, Ramos-Quiroga JA, Paus T, Ribases M, Kaprio J, Boks MPM, Bell JT, Spector TD, Gelernter J, Boomsma DI, Martin NG, MacGregor S, Perry JRB, Palmer AA, Posthuma D, Munafo MR, Gillespie NA, Derks EM, Vink JM, 2018. Genome-wide association analysis of lifetime cannabis use (N=184,765) identifies new risk loci, genetic overlap with mental health, and a causal influence of schizophrenia on cannabis use. Nat. Neurosci [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power RA, Verweij KJ, Zuhair M, Montgomery GW, Henders AK, Heath AC, Madden PA, Medland SE, Wray NR, Martin NG, 2014. Genetic predisposition to schizophrenia associated with increased use of cannabis. Mol. Psychiatry 19, 1201–1204. 10.1038/mp.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prom-Wormley EC, Ebejer J, Dick DM, Bowers MS, 2017. The genetic epidemiology of substance use disorder: a review. Drug Alcohol Depend. 180, 241–259. 10.1016/j.drugalcdep.2017.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2017. R: A Language and Environment for Statistical Computing (RRID:SCR_001905). Vienna, Austria. . [Google Scholar]
- Reginsson GW, Ingason A, Euesden J, Bjornsdottir G, Olafsson S, Sigurdsson E, Oskarsson H, Tyrfingsson T, Runarsdottir V, Hansdottir I, Steinberg S, Stefansson H, Gudbjartsson DF, Thorgeirsson TE, Stefansson K, 2017. Polygenic risk scores for schizophrenia and bipolar disorder associate with addiction. Addict. Biol 10.1111/adb.12496. n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MR, Wray NR, Visscher PM, 2014. Explaining additional genetic variation in complex traits. Trends Genet. 30, 124–132. 10.1016/j.tig.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAS Institute Inc, 2017. Base SAS 9.4 Procedures Guide, seventh edition SAS Institute, Cary, NC. [Google Scholar]
- Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, Desrivieres S, Aliev FA, Khan AA, Amin N, Aulchenko YS, Bakalkin G, Bakker SJ, Balkau B, Beulens JW, Bilbao A, de Boer RA, Beury D, Bots ML, Breetvelt EJ, Cauchi S, Cavalcanti-Proenca C, Chambers JC, Clarke TK, Dahmen N, de Geus EJ, Dick D, Ducci F, Easton A, Edenberg HJ, Esko T, Fernandez-Medarde A, Foroud T, Freimer NB, Girault JA, Grobbee DE, Guarrera S, Gudbjartsson DF, Hartikainen AL, Heath AC, Hesselbrock V, Hofman A, Hottenga JJ, Isohanni MK, Kaprio J, Khaw KT, Kuehnel B, Laitinen J, Lobbens S, Luan J, Mangino M, Maroteaux M, Matullo G, McCarthy MI, Mueller C, Navis G, Numans ME, Nunez A, Nyholt DR, Onland-Moret CN, Oostra BA, O’Reilly PF, Palkovits M, Penninx BW, Polidoro S, Pouta A, Prokopenko I, Ricceri F, Santos E, Smit JH, Soranzo N, Song K, Sovio U, Stumvoll M, Surakk I, Thorgeirsson TE, Thorsteinsdottir U, Troakes C, Tyrfingsson T, Tonjes A, Uiterwaal CS, Uitterlinden AG, van der Harst P, van der Schouw YT, Staehlin O, Vogelzangs N, Vollenweider P, Waeber G, Wareham NJ, Waterworth DM, Whitfield JB, Wichmann EH, Willemsen G, Witteman JC, Yuan X, Zhai G, Zhao JH, Zhang W, Martin NG, Metspalu A, Doering A, Scott J, Spector TD, Loos RJ, Boomsma DI, Mooser V, Peltonen L, Stefansson K, van Duijn CM, Vineis P, Sommer WH, Kooner JS, Spanagel R, Heberlein UA, Jarvelin MR, Elliott P, 2011. Genomewide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. U. S. A 108, 7119–7124. 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Kranzler HR, Yu Y, Logue MW, Poling J, Arias AJ, Anton RF, Oslin D, Farrer LA, Gelernter J, 2010. Variation in nicotinic acetylcholine receptor genes is associated with multiple substance dependence phenotypes. Neuropsychopharmacology 35, 1921–1931. 10.1038/npp.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherva R, Wang Q, Kranzler H, Zhao H, Koesterer R, Herman A, Farrer LA, Gelernter J, 2016. Genome-wide association study of Cannabis dependence severity, novel risk variants, and shared genetic risks. JAMA Psychiatry 73, 472–480. 10.1001/jamapsychiatry.2016.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens SH, Hartz SM, Hoft NR, Saccone NL, Corley RC, Hewitt JK, Hopfer CJ, Breslau N, Coon H, Chen X, Ducci F, Dueker N, Franceschini N, Frank J, Han Y, Hansel NN, Jiang C, Korhonen T, Lind PA, Liu J, Lyytikainen LP, Michel M, Shaffer JR, Short SE, Sun J, Teumer A, Thompson JR, Vogelzangs N, Vink JM, Wenzlaff A, Wheeler W, Yang BZ, Aggen SH, Balmforth AJ, Baumeister SE, Beaty TH, Benjamin DJ, Bergen AW, Broms U, Cesarini D, Chatterjee N, Chen J, Cheng YC, Cichon S, Couper D, Cucca F, Dick D, Foroud T, Furberg H, Giegling I, Gillespie NA, Gu F, Hall AS, Hallfors J, Han S, Hartmann AM, Heikkila K, Hickie IB, Hottenga JJ, Jousilahti P, Kaakinen M, Kahonen M, Koellinger PD, Kittner S, Konte B, Landi MT, Laatikainen T, Leppert M, Levy SM, Mathias RA, McNeil DW, Medland SE, Montgomery GW, Murray T, Nauck M, North KE, Pare PD, Pergadia M, Ruczinski I, Salomaa V, Viikari J, Willemsen G, Barnes KC, Boerwinkle E, Boomsma DI, Caporaso N, Edenberg HJ, Francks C, Gelernter J, Grabe HJ, Hops H, Jarvelin MR, Johannesson M, Kendler KS, Lehtimaki T, Magnusson PK, Marazita ML, Marchini J, Mitchell BD, Nothen MM, Penninx BW, Raitakari O, Rietschel M, Rujescu D, Samani NJ, Schwartz AG, Shete S, Spitz M, Swan GE, Volzke H, Veijola J, Wei Q, Amos C, Cannon DS, Grucza R, Hatsukami D, Heath A, Johnson EO, Kaprio J, Madden P, Martin NG, Stevens VL, Weiss RB, Kraft P, Bierut LJ, Ehringer MA, 2013. Distinct loci in the CHRNA5/CHRNA3/CHRNB4 gene cluster are associated with onset of regular smoking. Genet. Epidemiol 37, 846–859. 10.1002/gepi.21760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strat YL, Ramoz N, Schumann G, Gorwood P, 2008. Molecular genetics of alcohol dependence and related endophenotypes. Curr. Genomics 9, 444–451. 10.2174/138920208786241252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer S, Minică CC, Verweij KJH, Mbarek H, Bernard M, Derringer J, van Eijk KR, Isen JD, Loukola A, Maciejewski DF, Mihailov E, van der Most PJ, Sánchez-Mora C, Roos L, Sherva R, Walters R, Ware JJ, Abdellaoui A, Bigdeli TB, Branje SJT, Brown SA, Bruinenberg M, Casas M, Esko T, Garcia-Martinez I, Gordon SD, Harris JM, Hartman CA, Henders AK, Heath AC, Hickie IB, Hickman M, Hopfer CJ, Hottenga JJ, Huizink AC, Irons DE, Kahn RS, Korhonen T, Kranzler HR, Krauter K, van Lier PAC, Lubke GH, Madden PAF, Mägi R, McGue MK, Medland SE, Meeus WHJ, Miller MB, Montgomery GW, Nivard MG, Nolte IM, Oldehinkel AJ, Pausova Z, Qaiser B, Quaye L, Ramos-Quiroga JA, Richarte V, Rose RJ, Shin J, Stallings MC, Stiby AI, Wall TL, Wright MJ, Koot HM, Paus T, Hewitt JK, Ribasés M, Kaprio J, Boks MP, Snieder H, Spector T, Munafò MR, Metspalu A, Gelernter J, Boomsma DI, Iacono WG, Martin NG, Gillespie NA, Derks EM, Vink JM, 2016. Genome-wide association study of lifetime cannabis use based on a large meta-analytic sample of 32 330 subjects from the International Cannabis consortium. Transl. Psychiatry 6, e769. 10.1038/tp.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Schizophrenia Consortium, 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, van Duijn CM, Kaprio J, Gulcher JR, McCarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K, 2010. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet 42, 448–453. 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium, 2010. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet 42, 441–447. 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Rietschel M, 2011. Genome-wide association studies of alcohol dependence and substance use disorders. Curr. Psychiatry Rep 13, 147–155. 10.1007/s11920-011-0176-4. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nothen MM, Kiefer F, Spanagel R, Mann K, Rietschel M, 2009. Genome-wide association study of alcohol dependence. Arch. Gen. Psychiatry 66, 773–784. 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij KJH, Abdellaoui A, Nivard MG, Cort AS, Ligthart L, Draisma HHM, Minică C, International Cannabis Consortium, Gillespie NA, Willemsen G, Hottenga J-J, Boomsma DI, Vink JM, 2017. Genetic association between schizophrenia and cannabis use. Drug Alcohol Depend. 171, 117–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Hottenga JJ, de Geus EJC, Willemsen G, Neale MC, Furberg H, Boomsma DI, 2014. Polygenic risk scores for smoking: predictors for alcohol and cannabis use? Addiction 109, 1141–1151. 10.1111/add.12491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, Aliev F, Bacanu S-A, Batzler A, Bertelsen S, Biernacka JM, Bigdeli TB, Chen L-S, Clarke T-K, Chou Y-L, Degenhardt F, Docherty AR, Edwards AC, Fontanillas P, Foo JC, Fox L, Frank J, Giegling I, Gordon S, Hack LM, Hartmann AM, Hartz SM, Heilmann-Heimbach S, Herms S, Hodgkinson C, Hoffmann P, Jan Hottenga J, Kennedy MA, Alanne-Kinnunen M, Konte B, Lahti J, Lahti-Pulkkinen M, Lai D, Ligthart L, Loukola A, Maher BS, Mbarek H, McIntosh AM, McQueen MB, Meyers JL, Milaneschi Y, Palviainen T, Pearson JF, Peterson RE, Ripatti S, Ryu E, Saccone NL, Salvatore JE, Sanchez-Roige S, Schwandt M, Sherva R, Streit F, Strohmaier J, Thomas N, Wang J-C, Webb BT, Wedow R, Wetherill L, Wills AG, 23andMe Research Team, Boardman JD, Chen D, Choi D-S, Copeland WE, Culverhouse RC, Dahmen N, Degenhardt L, Domingue BW, Elson SL, Frye MA, Gäbel W, Hayward C, Ising M, Keyes M, Kiefer F, Kramer J, Kuperman S, Lucae S, Lynskey MT, Maier W, Mann K, Männistö S, Müller-Myhsok B, Murray AD, Nurnberger JI, Palotie A, Preuss U, Räikkönen K, Reynolds MD, Ridinger M, Scherbaum N, Schuckit MA, Soyka M, Treutlein J, Witt S, Wodarz N, Zill P, Adkins DE, Boden JM, Boomsma DI, Bierut LJ, Brown SA, Bucholz KK, Cichon S, Costello EJ, de Wit H, Diazgranados N, Dick DM, Eriksson JG, Farrer LA, Foroud TM, Gillespie NA, Goate AM, Goldman D, Grucza RA, Hancock DB, Harris KM, Heath AC, Hesselbrock V, Hewitt JK, Hopfer CJ, Horwood J, Iacono W, Johnson EO, Kaprio JA, Karpyak VM, Kendler KS, Kranzler HR, Krauter K, Lichtenstein P, Lind PA, McGue M, MacKillop J, Madden PAF, Maes HH, Magnusson P, Martin NG, Medland SE, Montgomery GW, Nelson EC, Nöthen MM, Palmer AA, Pedersen NL, Penninx BWJH, Porjesz B, Rice JP, Rietschel M, Riley BP, Rose R, Rujescu D, Shen P-H, Silberg J, Stallings MC, Tarter RE, Vanyukov MM, Vrieze S, Wall TL, Whitfield JB, Zhao H, Neale BM, Gelernter J, Edenberg HJ, Agrawal A, 2018. Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci 21, 1656–1669. 10.1038/s41593-018-0275-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KS, Liu X, Zhang Q, Wu LY, Zeng M, 2012. Genome-wide association study identifies 5q21 and 9p24.1 (KDM4C) loci associated with alcohol withdrawal symptoms. J. Neural Transm (Vienna, Austria: 1996) 119, 425–433. 10.1007/s00702-011-0729-z. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM, 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet 88, 76–82. 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Zhang CK, Wang F, Li CS, Zhao H, Lu L, Zhang XY, Lu L, Zhang H, Zhang F, Krystal JH, Luo X, 2011. A novel, functional and replicable risk gene region for alcohol dependence identified by genome-wide association study. PLoS One 6, e26726. 10.1371/journal.pone.0026726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Gelernter J, Zhang CK, Zhao H, Lu L, Kranzler HR, Malison RT, Li CS, Wang F, Zhang XY, Deng HW, Krystal JH, Zhang F, Luo X, 2012. Genome-wide association study of alcohol dependence implicates KIAA0040 on chromosome 1q. Neuropsychopharmacology 37, 557–566. 10.1038/npp.2011.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.