Abstract
The xylT gene product, a component of the xylene catabolic pathway of Pseudomonas putida mt2, has been recently characterized as a novel [2Fe-2S] ferredoxin which specifically reactivates oxygen-inactivated catechol 2,3-dioxygenase (XylE). In this study, three XylT-like proteins potentially involved in the catabolism of naphthalene (NahT) or cresols (PhhQ and DmpQ) have been overexpressed in Escherichia coli, purified, and compared with respect to their biochemical properties and interaction with XylE. The three XylT analogues show general spectroscopic characteristics common to plant-type [2Fe-2S] ferredoxins as well as distinctive features that appear to be typical for the XylT subgroup of these proteins. The midpoint redox potentials of the PhhQ and DmpQ proteins were −286 mV and −323 mV, respectively. Interestingly, all purified XylT-like proteins promoted in vitro reactivation of XylE almost as efficiently as XylT. The interaction of XylE with XylT and its analogues was studied by cross-linking experiments using the 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide. A polypeptide band with an Mr of 46,000, which corresponded to the cross-linked product between one XylE subunit and one molecule of ferredoxin, was obtained in all cases. The formation of the complex was affected by ionic strength, indicating that electrostatic forces are involved in the dioxygenase-ferredoxin interaction. In complementation experiments, plasmids expressing xylT or its analogues were introduced into an XylT-null mutant of P. putida which is unable to grow on p-methylbenzoate. All transconjugants regained the wild-type phenotype, indicating that all analogues can substitute for XylT in the in vivo reactivation of XylE. Our results provide evidence for a subgroup of [2Fe-2S] ferredoxins with distinct biochemical properties whose specific function is to reactivate intrinsically labile extradiol ring cleavage dioxygenases involved in the catabolism of various aromatic hydrocarbons.
In bacteria that are able to perform oxidative catabolism of aromatic hydrocarbons, degradation pathways converge towards a few common intermediates such as protocatechuate, salicylate, hydroxyquinol, and catechol (7). These intermediates are further oxidized by dioxygenases, which cleave the aromatic ring. Enzymatic cleavage of catechol is catalyzed by two categories of catechol dioxygenases, referred to as intradiol and extradiol dioxygenases, depending on the position of the cleavage site relative to the diol. The two types of enzymes are biochemically and structurally different and have distinct phylogenetic origins (9). The catechol 2,3-dioxygenase from Pseudomonas putida mt2, an archetypical extradiol dioxygenase, has been extensively studied, and its crystal structure has recently been solved (20). The enzyme is a tetramer composed of four identical subunits, each of which contains a ferrous iron atom. It is a labile enzyme which is inactivated upon exposure to oxidizing agents such as oxygen (24). Inactivation also occurs during catalytic turnover when substrate analogues such as alkyl- or chlorocatechols are provided (1, 4). The primary cause of the inactivation is thought to be an oxidation of the ferrous iron in the active site of the enzyme. In P. putida, a natural repair system which prevents irreversible inactivation of catechol 2,3-dioxygenase has been discovered (25). This system is dependent on a functional xylT gene and allows bacteria to avoid suicide inactivation of the dioxygenase by 4-methylcatechol when growing on p-xylene. The xylT gene product has been characterized as a [2Fe-2S] ferredoxin that specifically reactivates catechol 2,3-dioxygenase (12). It has been demonstrated that reactivation proceeds through an XylT-dependent reduction of the iron atom present essentially in the ferric state in the inactive enzyme.
In a recent search of databases for XylT homologues, we found more than a dozen genes encoding products which exhibited between 29 and 65% sequence identity with XylT (reference 16 and references cited therein). All of these genes are present in bacteria that degrade aromatic hydrocarbons and map close to a gene encoding a catechol dioxygenase, suggesting that these putative XylT analogues have a similar functional role. In this study, three XylT-like proteins have been purified in recombinant form from Escherichia coli, and their biochemical and spectroscopic properties have been investigated. Our results suggest that the three XylT-like proteins are dedicated to reactivation of catechol dioxygenase and play a physiological role similar to that of XylT in P. putida.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli strains DH5α (Gibco BRL) and BL21(DE3) (Novagen) were used for cloning and protein expression, respectively. P. putida mt2 (31) and XylT2 (25) were previously described. Plasmids used in this study are listed in Table 1. E. coli was grown in rich Luria-Bertani or very rich (Terrific) culture broth (26) containing appropriate antibiotics. P. putida was grown aerobically in either Luria-Bertani medium or mineral salts medium (M9) supplemented with 5 mM benzoate or p-methylbenzoate as the sole carbon source (12).
TABLE 1.
Plasmids used in this study
| Plasmid | Characteristics | Source or reference |
|---|---|---|
| pBBR1MCS-2 | Kmr cloning and expression vector for Pseudomonas | 21 |
| pRK600 | Cmr, ori ColE1, RK2-Mob+ RK2-Tra+ | 19 |
| pCR-Script II | Apr cloning vector | New England Biolabs |
| pGEM-T | Apr T-cloning vector | Promega |
| pET9a | Kmr expression vector for E. coli | Novagen |
| pAW31 | AprxylT-containing vector | 30 |
| pVI300Δ | AprdmpQ-containing vector | 28 |
| pLCN4 | AprphhQ-containing vector | 23 |
| pRAF104.3 | AprnahT-containing vector | 2 |
| pAJ90 | xylT PCR fragment cloned into pGEM-T | 12 |
| pAJ91 | 0.33-kb NdeI/BamHI fragment from pAJ90 | 12 |
| pAJ92 | dmpQ PCR fragment cloned into pGEM-T | This study |
| pAJ93 | 0.33-kb NdeI/BamHI fragment from pAJ92 | This study |
| pAJ94 | phhQ PCR fragment cloned into pGEM-T | This study |
| pAJ95 | 0.33-kb NdeI/BamHI fragment from pAJ94 | This study |
| pAJ96 | nahT PCR fragment cloned into pCR-Script-II | This study |
| pAJ97 | 0.33-kb NdeI/BamHI fragment from pAJ96 | This study |
| pBBX52 | Derivative of pBBR1MCS-2 containing xylT | This study |
| pBBN52 | Derivative of pBBX52 containing nahT | This study |
| pBBD52 | Derivative of pBBX52 containing dmpQ | This study |
| pBBP52 | Derivative of pBBX52 containing phhQ | This study |
Cloning and overexpression of xylT-like genes in E. coli.
The dmpQ, phhQ, and nahT genes were amplified by PCR using plasmids pVI300Δ, pLCN4, and pRAF104.3 as templates, respectively. Primers included NdeI or BamHI restriction sites to facilitate subcloning in expression plasmid pET9a (Table 2). PCR fragments were cloned in pGEM-T, checked by DNA sequencing, and finally subcloned in pET9a to give pAJ93 (dmpQ), pAJ95 (phhQ), and pAJ99 (nahT). These plasmids were introduced into E. coli strain BL21(DE3)(pLysS). Recombinant strains were grown at 37°C in Terrific broth growth medium under aeration until the culture reached an optical density at 600 nm of 2.0. The temperature was then lowered to 18°C, and overexpression of the cloned gene was triggered by addition of 0.2 mM isopropyl-β-d-thiogalactoside. Bacteria were harvested after 16 h of incubation and stored frozen at −80°C.
TABLE 2.
Oligonucleotides used in this study for amplification of DNA fragments by PCR
| Name | Sequencea | Relevant characteristics |
|---|---|---|
| AJ017 | CGGGCATATGAACCGTGCCGGTTATG | NdeI introduced into 5′ end of dmpQ |
| AJ108 | GCGCGGATCCTCATGACGACACCTCTTG | BamHI introduced into 3′ end of dmpQ |
| AJ019 | CGGGCATATGAGCAGCCCCCCGTTTC | NdeI introduced into 5′ end of phhQ |
| AJ020 | GCGCGGATCCTCAGGGATTGCTTCGCAC | BamHI introduced into 3′ end of phhQ |
| AJ021 | CGGGCATATGAACAGTGCCGGCTACG | NdeI introduced into 5′ end of xylT |
| AJ022 | GCGCGGATCCTCATGACGTCACCTCTTC | BamHI introduced into 3′ end of xylT |
| AJ364 | CGGGCATATGTCAGAGGTCTTTGAAATC | NdeI introduced into 5′ end of nahT |
| AJ365 | GCGCGGATCCCTATTCTCTTTGTTGTTGG | BamHI introduced into 3′ end of nahT |
| XylT52 | TCTGGGATGCATATGAACAGT | NdeI and Shine-Dalgarno sequence introduced into 5′ end of xylT |
Boldface indicates the restriction enzyme sequences introduced. The Shine-Dalgarno sequence which precedes xylT is underlined.
Purification of XylT analogues.
A procedure adapted from that described previously for XylT purification (12) was used for the isolation of the XylT-like proteins. All steps were carried out at 4°C under argon, using buffers adjusted to pH 7.5 and containing 2 mM dithionite, unless otherwise indicated. A crude extract was prepared by ultrasonication from about 10 g of frozen cells. Ammonium sulfate was added to a final concentration of 1.7 M. The precipitate was removed by centrifugation, and the supernatant fluid was applied to a 10-ml phenyl-Sepharose column (Amersham Pharmacia Biotech), equilibrated in 50 mM Tris-HCl buffer, containing 1.7 M ammonium sulfate. After extensive washing of the column with this buffer, a fraction containing the XylT-like protein was eluted with Tris buffer lacking ammonium sulfate. This fraction was concentrated to a volume of ca. 5 ml by ultrafiltration through an 8-kDa-cutoff Omega membrane (Pall-Gelman Laboratory) and then applied to a Φ 2.5- by 110-cm gel filtration column (AcA54; Biosepra) equilibrated in 25 mM Tris-HCl buffer containing 0.2 M NaCl. The purified protein was concentrated to a few milliliters by ultrafiltration as described above and stored as pellets in liquid nitrogen. The NahT protein was purified at pH 7.75 instead of 7.5 for stability reasons and was subjected to an additional chromatographic step in order to eliminate a 20-kDa contaminant polypeptide: the NahT fraction obtained from gel filtration was diluted fivefold in 25 mM Tris-HCl buffer (pH 8.5) and loaded on a 25-ml anion-exchange column (Q-hyperD; Biosepra), which was developed with a 0 to 0.2 M linear gradient of NaCl. The purified protein was concentrated and stored as indicated above. When needed, dithionite-free preparations of the purified proteins were obtained by anaerobic gel filtration on a Sephadex G25 column (Amersham Pharmacia Biotech) preequilibrated in 10 mM MOPS (morpholinepropanesulfonic acid) buffer, pH 7.5 (pH 7.75 for NahT).
Redox titrations.
The midpoint redox potentials of PhhQ and DmpQ were determined by spectrophotometric titration, using 5-deazaflavin as a photoreductant and safranin O (Sigma-Aldrich; Em,7 = −289 mV) as a redox indicator. The titration experiments were performed in an anaerobic glove box as previously described (13), except for the following changes. The extents of safranin and ferredoxin reduction were calculated from the absorbance at 520 and 406 nm, respectively. For calculations, the following experimentally determined absorption coefficients (millimolar−1 · centimeter−1) were used: ɛ406(ox) = 9.21 and ɛ406(red) = 5.23 for the oxidized and reduced forms of PhhQ at 406 nm, respectively; ɛ406(ox) = 9.20 and ɛ406(red) = 5.20 for the oxidized and reduced forms of DmpQ at 406 nm, respectively; ɛ520(ox) = 5.48 and ɛ520(red) = 3.01 for the oxidized and reduced forms of PhhQ and DmpQ at 520 nm, respectively; and ɛ520(ox) = 40.4 for oxidized safranin at 520 nm. The absorption of the ferredoxins at 406 nm was corrected for the contributions of safranin and 5-deazaflavin at this wavelength. Experimental data were fitted to the Nernst equation using the least-squares method.
Cross-linking experiments.
Cross-linking reactions were carried out under anaerobic conditions in 10 mM MOPS buffer, pH 7.75. XylE (3.6 μM) was mixed with each ferredoxin (20 μM) in a 4-ml reaction vial. Then, N-hydroxysulfosuccinimide (5 mM) and 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (12.5 mM) were successively added, and the reaction was allowed to proceed for 30 min at room temperature. The reaction was stopped by addition of 25 mM 2-mercaptoethanol and 10 mM ethanolamine. Samples (7.5 μl) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12.5% polyacrylamide gel.
Purification, inactivation, and reactivation of XylE.
Catechol 2,3-dioxygenase (XylE) was purified and subsequently inactivated by 4-methylcatechol according to published procedures (12). The preparation used in reactivation trials exhibited a residual activity of 3.05 U/mg of protein, which is equivalent to 2.5% of the initial activity. Ferredoxin-mediated reactivation of catechol dioxygenase was performed at 20°C under anaerobic conditions as described earlier (12).
Analytical methods.
UV-visible absorption and electron paramagnetic resonance (EPR) spectra were recorded as previously described (17). SDS-PAGE was performed on mini-slab gels using either a Tris-glycine buffer system (18) or a Tris-Tricine buffer system for resolution of peptides in the 2- to 20-kDa range (27). N-terminal sequence determination was performed by the Laboratoire de Chimie des Protéines at the CEA-Grenoble, after electrotransfer of proteins onto a Problott membrane (Applied Biosystems). Proteins were assayed using the bicinchoninic acid reagent kit (Pierce).
Complementation experiments.
The plasmids used for complementation analysis of an xylT deletion mutant were constructed as follows. The xylT gene, including the Shine-Dalgarno sequence, was amplified by PCR using plasmid pAW31 as a template. An NdeI restriction site was introduced upstream of the xylT coding sequence and a BamHI site was introduced downstream of this sequence, using primers XylT52 and AJ022, respectively (Table 2). The amplified fragment was cloned into the EcoRV restriction site of plasmid pBBR1MCS-2 (21). A plasmid carrying the xylT gene with the correct orientation with respect to the lac promoter was selected by restriction analysis and checked by DNA sequencing. This plasmid was named pBBX52. Derivatives of plasmid pBBX52 carrying nahT (pBBN52), dmpQ (pBBD52), and phhQ (pBBP52) were obtained by replacing xylT by the NdeI-BamHI fragments from plasmids pAJ97, pAJ93, and pAJ95, respectively. The recombinant plasmids were mobilized into the xylT deletion strain XylT2 (25) by means of triparental matings, with an E. coli helper strain containing plasmid pRK600. Transconjugants were selected on kanamycin medium and subsequently tested for the ability to grow on minimal medium supplemented with 5 mM p-methylbenzoate.
RESULTS
Purification and properties of NahT, DmpQ, and PhhQ.
The deduced amino acid sequences of the three XylT-like polypeptides considered in this study are aligned in Fig. 1. The NahT polypeptide was identified as the product of a gene associated with naphthalene catabolic genes in the marine bacterium P. stutzeri AN10 (2). A gene potentially encoding an identical polypeptide was found in P. putida KF715, a strain which degrades salicylate (22). The other two XylT analogues were described as gene products associated with catabolic pathways for the degradation of cresols (10, 28). The three XylT-like polypeptides exhibit 50% or more sequence identity with XylT, and all have a basic character, with calculated isoelectric points of 7.75 (NahT), 8.40 (DmpQ), and 7.85 (PhhQ).
FIG. 1.
Sequence alignment of the four ferredoxins considered in the present study. Sequences were aligned using the GeneWorks software (version 2.5N) from IntelliGenetics. Identical residues are shaded. Cysteines presumed to serve as ligands for the [2Fe-2S] cluster are indicated in boldface. The sources, accession numbers, and references for each sequence are as follows: XylT-MT2, XylT from P. putida mt-2 (M64747) (8); DmpQ-CF600, DmpQ from Pseudomonas sp. strain CF600 (X60835) (28); PhhQ-P35X, PhhQ from P. putida P35X (X79063) (23); and NahT-KF715, NahT from P. putida sp. strain KF715 (S78585) (22). The XylT sequence was modified to take into account a correction of the gene sequence resulting in an alanine-to-glycine replacement of residue 76 (12).
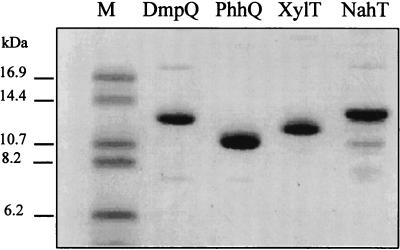
Overexpression of the nahT, dmpQ, and phhQ genes resulted in the accumulation in E. coli of polypeptides with Mrs of around 12,000. The nahT and dmpQ gene products tended to form insoluble inclusion bodies, but this was minimized by lowering the temperature of the cultures to 16 to 20°C during induction. The three proteins were purified anaerobically, yielding nearly homogeneous preparations as judged from SDS-PAGE (Fig. 2). The quantity of purified protein recovered from 1 liter of culture was about 1.0 mg for NahT, 3.0 mg for DmpQ, and 6.0 mg for PhhQ.
FIG. 2.
SDS-PAGE analysis of purified DmpQ, NahT, PhhQ, and XylT. The proteins were purified as described in Materials and Methods and analyzed (1.0 μg per well) by electrophoresis on a 16% polyacrylamide gel containing 10% glycerol in a Tris-Tricine system. Sizes of protein markers (lane M) are indicated.
The N-terminal sequence of each purified polypeptide was consistent with the deduced amino acid sequence for the corresponding gene. The analysis also revealed that the formylmethionines (fMets) of NahT and PhhQ were processed, whereas that of DmpQ was not. In the latter case, the fMet residue is followed by an asparagine, while the residue occupying the same position in PhhQ is a serine and that in NahT is a proline. In XylT, which like DmpQ has an fMet-Asn sequence at the N terminus, we found previously that the fMet residue was not processed (12). These results are consistent with the general finding that, in E. coli, the extent of fMet processing by methionyl-aminopeptidase is dependent on the length of the side chain of the next amino acid (11).
Analysis of the purified recombinant proteins by SDS-PAGE under conditions enhancing the resolution of small polypeptides revealed significant differences in their mobilities (Fig. 2). These differences correlated well with their molecular masses, which were calculated from the gene sequences to be 12,222 Da (DmpQ), 12,387 Da (NahT), and 10,834 Da (PhhQ). All three proteins showed similar gel filtration elution profiles, which indicated that they had monomeric structures with approximate molecular masses of 12 kDa.
Spectroscopic properties.
The UV-visible absorbance spectra of NahT, DmpQ, and PhhQ showed absorption bands centered near 336, 416, and 456 nm, indicative of the presence of a [2Fe-2S] cluster. Upon reduction, a general decrease of the absorbance occurred over the visible range, concomitant with the appearance of an absorption band near 540 nm, as previously observed for XylT (12) (data not shown).
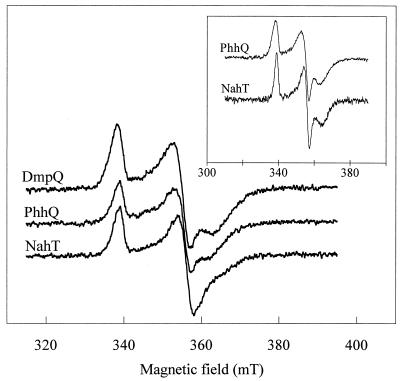
The EPR spectra of the XylT analogues in the reduced form showed similar rhombic signals, which are the signature of a [2Fe-2S] cluster with an S = 1/2 spin state (Fig. 3). The resonance lines were broad, and the resolution of the three components of the g tensor varied with the protein. The DmpQ signal showed the best rhombic symmetry with gx,y,z = 1.895, 1.94, and 2.04, while the gx component of the NahT signal was poorly resolved. The addition of glycerol to the protein samples prior to EPR analysis resulted in signals with a better resolution for NahT and PhhQ (Fig. 3, inset), while the spectrum of DmpQ was little affected (data not shown).
FIG. 3.
EPR spectra of the XylT-like ferredoxins. Purified samples of DmpQ (134 μM), PhhQ (133 μM), and NahT (96 μM) were reduced with 1 mM dithionite. Spectra were recorded at 10 K under the following conditions: microwave power, 0.01 mW at 9.65 GHz; modulation amplitude, 1 mT at 100 kHz. The inset shows spectra of PhhQ (110 μM) and NahT (70 μM) samples containing 16% glycerol.
Influence of oxygen and temperature on stability.
Our preliminary attempts to isolate NahT, DmpQ, and PhhQ indicated that these proteins were oxygen sensitive, although the instability varied widely among the proteins. Protein denaturation manifested itself in a progressive loss of the characteristic red color and also in precipitation of the protein. The stabilities of the three ferredoxins in terms of their half-lives were calculated from the kinetics of absorbance loss at 416 nm (Table 3). NahT was the most unstable of the three ferredoxins, reaching 50% denaturation within an hour at 37°C in an argon atmosphere, and was also highly sensitive to oxygen. PhhQ was also rather unstable but exhibited higher resistance to oxygen damage, particularly at pH 8.0. The DmpQ protein was the most stable, particularly in the absence of oxygen (Table 3).
TABLE 3.
Sensitivity of purified NahT, PhhQ, and DmpQ to oxygen and temperature
| Protein | Half-life under the following conditionsa:
|
||
|---|---|---|---|
| Air, 24°C | Argon, 24°C | Argon, 37°C | |
| NahT | 83 min | 13.5 h | 59 min |
| PhhQ | 6.7 h (11.6 h at pH 8.0) | 35.5 h | 2.4 h |
| DmpQ | 18.5 h | 107 h | >24 hb |
The pH was set at a predetermined optimum value of 7.5 (or 7.75 for NahT), and the ferredoxin concentration was adjusted within a limited range from 24 to 40 μM, as these two parameters were found to influence stability.
The A416 changed by less than 5% after the indicated time period.
Redox properties.
The three XylT-like ferredoxins were readily reducible by sodium dithionite or light-activated 5-deazaflavin. Spectrophotometric redox titration, performed as described in Materials and Methods, led to estimation of the midpoint redox potential of PhhQ to be −285 ± 5 mV and that of DmpQ to be −323 ± 4 mV, both at 25°C and pH 8.0. The instability of the NahT protein precluded reliable measurements of the extent of ferredoxin reduction during the titration.
In vitro reactivation of catechol 2,3-dioxygenase.
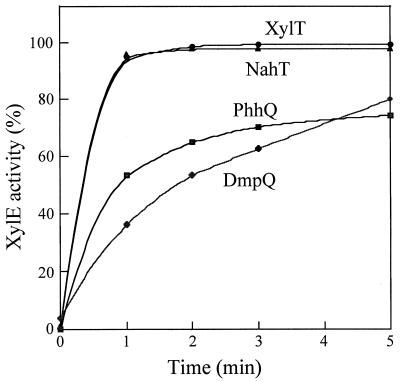
Since NahT, PhhQ, and DmpQ shared biochemical and redox properties similar to those of XylT, we determined whether these ferredoxins could reactivate 4-methylcatechol-inactivated XylE in in vitro reactivation assays performed as previously described (12). Kinetic data indicated that all three ferredoxins were able to reactivate XylE in a ferredoxin concentration-dependent manner. The XylE reactivation rates at constant ferredoxin concentrations varied according to the ferredoxin used: XylT and NahT promoted equally fast reactivation reactions, whereas PhhQ and DmpQ were less efficient, in terms of both the rates and extents of reactivation at the end of the 5-min reaction period (Fig. 4). These results could reflect differences in the interaction between the XylT analogues and catechol 2,3-dioxygenase. This possibility was investigated by performing cross-linking experiments.
FIG. 4.
Time course of ferredoxin-dependent XylE reactivation. A preparation of 4-methylcatechol-inactivated XylE (0.1 μM) was incubated in 0.2 ml of reactivating buffer (0.1 mM potassium phosphate [pH 7.5], 2.5 μM 5-deazaflavin, 10 mM glycine) containing the indicated ferredoxin at 0.5 μM. Reactivation was initiated by light. XylE activity was assayed on a 15-s time scale and expressed as a percentage of the maximal activity measured at 5 min in the XylT-mediated reactivation assay (100% = 120 U/mg).
Cross-linkage of XylT analogues and XylE.
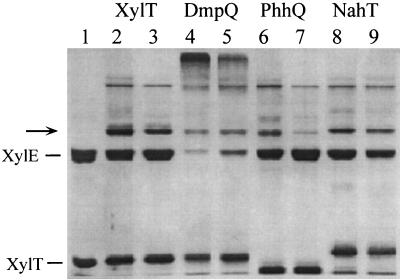
We treated the XylT analogues and XylE with the chemical reagent EDC under optimized conditions for the formation of a covalent complex between XylT and XylE, which will be described in detail elsewhere. When the ferredoxins were individually treated with EDC, the polypeptide mobility in SDS-PAGE was not affected and no additional band was detected, which is consistent with the monomeric nature of these proteins (data not shown). When XylE alone was subjected to EDC treatment, a band with an apparent Mr of around 100,000 was detected, in addition to a band corresponding to the 34-kDa subunit of the enzyme. This new band probably arose from the covalent coupling of three (or possibly four) subunits of the tetrameric enzyme (Fig. 5). When XylE was allowed to react with XylT or its analogues, a new band with an Mr of around 46,000 appeared on the polyacrylamide gel (Fig. 5). This product was formed with the same apparent yield irrespectively of whether the cross-linking was performed with active or inactive XylE. The size of this band is consistent with the formation of a covalent complex between a single XylE subunit and one ferredoxin molecule. N-terminal sequence analysis demonstrated that such a complex is indeed formed between XylE and XylT. The double sequence MN(KS)(GA)(VG)(MY)(RE)(PV) was found, which would be expected from the simultaneous sequencing of the N termini of the two proteins.
FIG. 5.
Cross-linking of XylE and XylT analogues. XylE (3.6 μM) was allowed to react with XylT, DmpQ, PhhQ, or NahT (20 μM) in the presence of EDC as described in Materials and Methods. Samples were analyzed by SDS-PAGE on a 12% polyacrylamide gel (lanes 2, 4, 6, and 8). The samples analyzed in lanes 3, 5, 7, and 9 were from cross-linking reactions performed in the presence of 0.1 M NaCl. Lane 1 was loaded with a mixture of unreacted XylE and XylT. The arrow indicates the cross-linked 46-kDa product.
When the cross-linking reactions were performed at a higher ionic strength (0.1 M NaCl), the intensity of the 46,000-Mr band decreased. The effect of ionic strength was particularly dramatic in the case of the PhhQ-XylE coupling product (Fig. 5, lane 7), suggesting that XylE interacted less strongly with PhhQ than with XylT or NahT. The case of DmpQ is atypical in that the cross-linking of this protein with XylE gave rise to high-molecular-weight aggregates, concurrent with an almost complete disappearance of the 34-kDa XylE subunit. While the reason for this different behavior is unknown, we noticed that the extent of protein aggregation increased with the DmpQ/XylE molar ratio and decreased when the ionic strength of the cross-linking reaction mixture was elevated.
Complementation of an XylT-deficient strain of P. putida.
The XylT2 mutant strain, which lacks the XylT protein, has lost the ability to grow on p-methylbenzoate because catechol dioxygenase becomes irreversibly inactivated by the intermediate metabolite 4-methylcatechol (25). Plasmid pBBX52, which carries an intact xylT gene, restored the ability of this strain to grow on p-methylbenzoate as the sole carbon source, suggesting that XylT was correctly synthesized. Plasmids carrying phhQ, dmpQ, or nahT also restored the wild-type phenotype, indicating that the corresponding proteins can functionally replace XylT in mediating XylE reactivation in vivo (data not shown).
DISCUSSION
The three XylT analogues considered in this study displayed absorbance and EPR spectra characteristic of [2Fe-2S] ferredoxins. Like XylT, however, they share little sequence homology with other [2Fe-2S] ferredoxins, including plant-type ferredoxins, except for four conserved cysteines that likely serve as ligands for the cluster (8). The three proteins are relatively unstable and oxygen sensitive, although DmpQ appears to be more resistant to denaturation (Table 3). Studies on cyanobacterial ferredoxins have recently shown that subtle structural changes, such as a double amino acid substitution in the sequence, can have a dramatic effect on oxygen sensitivity (29). Based on modelling experiments, it was proposed that oxygen sensitivity is dependent on the accessibility of the cavity surrounding the cluster. In this respect, we observed that the oxygen-sensitive NahT and PhhQ proteins yielded an EPR signal that changed upon glycerol addition, while the EPR signal of the DmpQ protein was unaffected by glycerol. These observations, which probably reflect differences in the accessibility of the cluster to the solvent, might also be interpreted in terms of oxygen stability of the relevant proteins.
The three XylT analogues described here were able to substitute for XylT in the in vitro reactivation of catechol 2,3-dioxygenase. Interestingly, kinetic experiments revealed that NahT was more efficient than DmpQ and PhhQ, although it showed greater instability and lower sequence similarity with XylT than the other two proteins. Perhaps the kinetics of reactivation of XylE is somehow related to the degree of exposure of the ferredoxin cluster to the solvent, which in turn might determine protein stability (see above). Alternatively, DmpQ and PhhQ might interact less efficiently with XylE because of a suboptimal molecular fit between the heterologous protein partners, as suggested by cross-linking experiments (see Fig. 5 and discussion below). Complementation experiments demonstrated that all XylT-like ferredoxins could replace XylT in vivo, providing evidence that they mediate XylE reactivation in P. putida.
All four ferredoxins specifically cross-reacted with XylE upon EDC treatment and formed a 46-kDa complex that likely consists of one molecule each of ferredoxin and XylE subunit, as shown in the case of the XylE-XylT coupling product. The formation of this complex was affected by ionic strength, indicating that electrostatic forces are implicated in the interaction between the two protein partners. This is in agreement with the general observation that ferredoxins interact with their physiological partners mainly through electrostatic forces (3, 5, 13, 14). Known ferredoxin structures have on their protein surfaces high local concentrations of negatively charged residues that contribute to the correct interaction with a corresponding positively charged domain of the partner protein (6, 15). In contrast to most ferredoxins, the proteins considered in the present study exhibit a net positive charge at neutral pH, and they probably interact with XylE through lysine or arginine residues. Assuming that all XylT-like proteins associate similarly with XylE, it is plausible that a conserved lysine residue participates in the cross-linked complexes. Sequence comparison reveals that only one lysine residue at position 34 is conserved in all four ferredoxin sequences (Fig. 1). When the comparison was extended to 12 additional putative XylT analogues available in the databases, it revealed that a lysine is present at the corresponding position in only six cases but that it is replaced by an arginine in six other sequences. In addition, a lysine at position 50 is conserved in 13 sequences but not in XylT, where it is replaced by an arginine. Hence, the occurrence of a basic residue in either or both of these positions might be important for the interaction of XylT-like ferredoxins with catechol dioxygenase. When XylE was cross-reacted with DmpQ, cross-linked products with apparent Mrs of greater than 200,000 were formed, in addition to the 46-kDa complex. This observation suggests that the interaction of DmpQ with XylE promotes multiple coupling reactions between the two proteins, perhaps because DmpQ offers more cross-linking possibilities to XylE. This is consistent with the fact that the DmpQ sequence contains twice as many lysines as the XylT sequence.
In this study, we described three new members of a subgroup of [2Fe-2S] ferredoxins that appear to be genetically and functionally associated with meta-cleaving catechol dioxygenases. Experimental data reported here constitute evidence that the physiological function of these proteins is to reactivate catechol dioxygenase, presumably through reduction of the iron atom at the enzyme active site, as has been demonstrated for XylT. Such a mechanism of enzyme regeneration probably occurs in all bacteria containing an xylT homologue and might contribute to the efficiency of degradation of many aromatic hydrocarbons.
ACKNOWLEDGMENTS
We thank V. Shingler, L. C. Ng, and R. Bosch for providing plasmids pVI300Δ, pLCN4, and pRAF104.3, respectively. We thank M. Louwagie for N-terminal sequence determinations.
This work was supported by grants from the Centre National de la Recherche Scientifique (UMR 5092), the Commissariat à l'Energie Atomique, and the Procope Franco-German exchange programme. K.N.T. expresses his gratitude to the Fonds des Chemischen Industrie for generous support. J.A. was supported by a grant from the FEBS.
REFERENCES
- 1.Bartels I, Knackmuss H J, Reineke W. Suicide inactivation of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-halocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch R, Garcia-Valdes E, Moore E R. Complete nucleotide sequence and evolutionary significance of a chromosomally encoded naphthalene-degradation lower pathway from Pseudomonas stutzeri AN10. Gene. 2000;245:65–74. doi: 10.1016/s0378-1119(00)00038-x. [DOI] [PubMed] [Google Scholar]
- 3.Brandt M E, Vickery L E. Charge pair interactions stabilizing ferredoxin-ferredoxin reductase complexes. Identification by complementary site-specific mutations. J Biol Chem. 1993;268:17126–17130. [PubMed] [Google Scholar]
- 4.Cerdan P, Wasserfallen A, Rekik M, Timmis K N, Harayama S. Substrate specificity of catechol 2,3-dioxygenase encoded by TOL plasmid pWW0 of Pseudomonas putida and its relationship to cell growth. J Bacteriol. 1994;176:6074–6081. doi: 10.1128/jb.176.19.6074-6081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coghlan V M, Vickery L E. Site-specific mutations in human ferredoxin that affect binding to ferredoxin reductase and cytochrome P450scc. J Biol Chem. 1991;266:18606–18612. [PubMed] [Google Scholar]
- 6.De Pascalis A R, Jelesarov I, Ackermann F, Koppenol W H, Hirasawa M, Knaff D B, Bosshard H R. Binding of ferredoxin to ferredoxin:NADP+ oxidoreductase: the role of carboxyl groups, electrostatic surface potential, and molecular dipole moment. Protein Sci. 1993;2:1126–1135. doi: 10.1002/pro.5560020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibson D T, Subramanian V. Microbial degradation of aromatic hydrocarbons. In: Gibson D T, editor. Microbial degradation of organic compounds. New York, N.Y: Marcel Dekker, Inc.; 1984. pp. 181–252. [Google Scholar]
- 8.Harayama S, Polissi A, Rekik M. Divergent evolution of chloroplast-type ferredoxins. FEBS Lett. 1991;285:85–88. doi: 10.1016/0014-5793(91)80730-q. [DOI] [PubMed] [Google Scholar]
- 9.Harayama S, Rekik M. Bacterial aromatic ring-cleavage enzymes are classified into two different gene families. J Biol Chem. 1989;264:15326–15333. [PubMed] [Google Scholar]
- 10.Herrmann H, Muller C, Schmidt I, Mahnke J, Petruschka L, Hahnke K. Localization and organization of phenol degradation genes of Pseudomonas putida strain H. Mol Gen Genet. 1995;247:240–246. doi: 10.1007/BF00705655. [DOI] [PubMed] [Google Scholar]
- 11.Hirel P H, Schmitter M J, Dessen P, Fayat G, Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc Natl Acad Sci USA. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hugo N, Armengaud J, Gaillard J, Timmis K N, Jouanneau Y. A novel [2Fe-2S] ferredoxin from Pseudomonas putida mt2 promotes the reductive reactivation of catechol 2,3-dioxygenase. J Biol Chem. 1998;273:9622–9629. doi: 10.1074/jbc.273.16.9622. [DOI] [PubMed] [Google Scholar]
- 13.Hurley J K, Hazzard J T, Martinez-Julvez M, Medina M, Gomez-Moreno C, Tollin G. Electrostatic forces involved in orienting Anabaena ferredoxin during binding to Anabaena ferredoxin:NADP(+) reductase: site-specific mutagenesis, transient kinetic measurements, and electrostatic surface potentials. Protein Sci. 1999;8:1614–1622. doi: 10.1110/ps.8.8.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurley J K, Salamon Z, Meyer T E, Fitch J C, Cusanovich M A, Markley J L, Cheng H, Xia B, Chae Y K, Medina M, Gomez-Moreno C, Tollin G. Amino acid residues in Anabaena ferredoxin crucial to interaction with ferredoxin-NADP+ reductase: site-directed mutagenesis and laser flash photolysis. Biochemistry. 1993;32:9346–9354. doi: 10.1021/bi00087a013. [DOI] [PubMed] [Google Scholar]
- 15.Jelesarov I, De Pascalis A R, Koppenol W H, Hirasawa M, Knaff D B, Bosshard H R. Ferredoxin binding site on ferredoxin:NADP+ reductase. Differential chemical modification of free and ferredoxin-bound enzyme. Eur J Biochem. 1993;216:57–66. doi: 10.1111/j.1432-1033.1993.tb18116.x. [DOI] [PubMed] [Google Scholar]
- 16.Jouanneau Y, Armengaud J, Hugo N, Meyer C, Timmis K N. Ferredoxin-mediated reactivation of catechol dioxygenase improves aromatic ring cleavage in pseudomonads. In: Fass R, Flashner Y, Reuveny S, editors. Novel approaches for bioremediation of organic pollution. New York, N.Y: Kluwer Academic/Plenum Publishers; 1999. pp. 117–126. [Google Scholar]
- 17.Jouanneau Y, Meyer C, Gaillard J, Forest E, Gagnon J. Purification and characterization of a novel dimeric ferredoxin (FdIII) from Rhodobacter capsulatus. J Biol Chem. 1993;268:10636–10644. [PubMed] [Google Scholar]
- 18.Jouanneau Y, Meyer C, Naud I, Klipp W. Characterization of an fdxN mutant of Rhodobacter capsulatus indicates that ferredoxin I serves as electron donor to nitrogenase. Biochim Biophys Acta. 1995;1232:33–42. doi: 10.1016/0005-2728(95)00106-x. [DOI] [PubMed] [Google Scholar]
- 19.Kessler B, de Lorenzo V, Timmis K N. A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria: regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol Gen Genet. 1992;233:293–301. doi: 10.1007/BF00587591. [DOI] [PubMed] [Google Scholar]
- 20.Kita A, Kita S, Fujisawa I, Inaka K, Ishida T, Horiike K, Nozaki M, Miki K. An archetypical extradiol-cleaving catecholic dioxygenase: the crystal structure of catechol 2,3-dioxygenase (metapyrocatechase) from Pseudomonas putida mt-2. Structure. 1999;7:25–34. doi: 10.1016/s0969-2126(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 21.Kovach M E, Phillips R W, Elzer P H, Roop R M, 2nd, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 22.Lee J, Min K R, Kim Y C, Kim C K, Lim J Y, Yoon H, Min K H, Lee K S, Kim Y. Cloning of salicylate hydroxylase gene and catechol 2,3-dioxygenase gene and sequencing of an intergenic sequence between the two genes of Pseudomonas putida KF715. Biochem Biophys Res Commun. 1995;211:382–388. doi: 10.1006/bbrc.1995.1825. [DOI] [PubMed] [Google Scholar]
- 23.Ng L, C, Shingler V, Sze C C, Poh C L. Cloning and sequences of the first eight genes of the chromosomally encoded (methyl)phenol degradation pathway from Pseudomonas putida P35X. Gene. 1994;151:29–36. doi: 10.1016/0378-1119(94)90629-7. [DOI] [PubMed] [Google Scholar]
- 24.Nozaki M, Ono K, Nakazawa T, Kotani S, Hayashi O. Metapyrocatechase. The role of iron and sulfhydryl groups. J Biol Chem. 1968;243:2682–2690. [PubMed] [Google Scholar]
- 25.Polissi A, Harayama S. In vivo reactivation of catechol 2,3-dioxygenase mediated by a chloroplast-type ferredoxin: a bacterial strategy to expand the substrate specificity of aromatic degradative pathways. EMBO J. 1993;12:3339–3347. doi: 10.1002/j.1460-2075.1993.tb06004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 28.Shingler V, Powlowski J, Marklund U. Nucleotide sequence and functional analysis of the complete phenol–3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992;174:711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh B B, Curdt I, Jakobs C, Schomburg D, Bisen P S, Bohme H. Identification of amino acids responsible for the oxygen sensitivity of ferredoxins from Anabaena variabilis using site-directed mutagenesis. Biochim Biophys Acta. 1999;1412:288–294. doi: 10.1016/s0005-2728(99)00069-9. [DOI] [PubMed] [Google Scholar]
- 30.Wasserfallen A. Ph.D. thesis. Geneva, Switzerland: University of Geneva; 1989. [Google Scholar]
- 31.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]