Abstract
Atherosclerosis differs across major arteries. Although the biological basis is not fully understood, limited evidence of genetic differences has been documented. This study, therefore, was aimed to identify differentially expressed genes between clinically relevant major arteries and investigate their enrichment in endothelial dysfunction-related gene sets. A bioinformatic analysis of publicly available gene-level read counts for coronary, aortic, and tibial arteries was performed. Differential gene expression was conducted with DeSeq2 at a false discovery rate of 0.05. Differentially expressed genes were then subjected to over-representation analysis and active-subnetwork-oriented enrichment analysis, both at a false discovery rate of 0.005. Enriched terms common to both analyses were categorized for each contrast into immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related terms, and the top differentially expressed genes validated against Swiss Institute of Bioinformatics’ Bgee database. There was mostly upregulation of differentially expressed genes for the coronary/tibial and aorta/tibial contrasts, but milder changes for the coronary/aorta contrast. Transcriptomic differences between coronary or aortic versus tibial samples largely involved immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related genes, suggesting potential to modulate endothelial dysfunction and atherosclerosis. These results imply atheroprone coronary and aortic environments compared with tibial artery tissue, which may explain observed relative inter-artery atherosclerosis risk.
Keywords: Differential gene expression, endothelium, coronary plaque, peripheral artery disease, aorta
Introduction
Globally, atherosclerosis remains cause for concern, especially among low- and middle-income countries.1,2 A 2020 estimate of global ischemic heart disease burden reported prevalence of 1.72%, projected to exceed 1.9% by 2030. 3 Similarly, a 2020 meta-analysis reported global prevalence of increased carotid intima-media thickness, carotid plaque, and carotid stenosis of 27.6%, 21.1%, and 1.5%, respectively, each increased since 2000. 4 In both studies, older age and male sex were associated with greater prevalence.3,4 These data highlight urgency to better understand atherogenesis that accounts for substantial mortality worldwide.5-7
Atherogenesis is a complex long-term process involving dyslipidemia, long-term inflammation, and autoimmunity.8-11 Key stages include low-density lipoprotein (LDL) uptake and oxidation, foam cell formation, 12 inflammasome activation producing interleukin (IL)-1, 10 IL-1-mediated smooth muscle cell (SMC) proliferation, 13 and subendothelial accumulation of foam cells, SMCs, and leukocytes.12,14 Endothelial dysfunction is an important intermediary and involves distorted vascular homeostasis.15,16 This contrasts the selectively permeable, anti-inflammatory, vasoregulatory, and anticoagulant normal endothelium. 17
However, despite interconnectivity of the entire vascular system, atherosclerosis differs clinically, structurally, and temporally across major arteries. Reiner et al 18 reported severe atherosclerosis more likely in coronary and abdominal aortic arteries in the presence of minimal or no mesenteric atherosclerosis, but not the reverse. Sawabe et al 19 later found differential atherosclerosis severity across intracranial, carotid, coronary, aortic, and femoral arteries, while a recent review identified atheroprotective (mostly upper limb) versus atheroprone (neck, most thoraco-abdominal, and lower limb) arteries. 20 Among atheroprone arteries, Dalager et al 21 reported primarily lipid-cored plaques in coronary arteries, contrasting foam cell and intermediate lesions within carotid arteries but normal or thickened femoral artery intimae. In this autopsy study, the presence of coronary and carotid atherosclerosis was observed from the third decade of life but not until the fourth decade, in femoral arteries. 21 This is consistent with observations that coronary plaque was always present when femoral plaque was found 22 and association of asymptomatic but advanced common iliac artery plaques with advanced coronary and carotid atherosclerosis. 23 Furthermore, when plaques occur in carotid and femoral arteries, histological analysis indicated more frequent fibrous cap atheromata and fibrocalcific plaques, respectively, consistent with higher calcium and lower cholesterol levels in femoral plaques. 24
The genetic basis for these differences was explored. Twenty-nine differentially expressed genes were reported between atheroprotective internal mammary arteries and atheroprone left anterior descending coronary arteries. 25 A comparison between 160 significant loci detected by a meta-analysis of coronary artery disease genome-wide association studies (GWAS) 26 and 5 loci detected by a meta-analysis of GWAS targeting single nucleotide polymorphisms (SNPs) associated with carotid intima-media thickness and carotid plaques 27 identified only one overlapping SNP. A comparison between the latter SNPs and coronary artery disease in the Coronary Artery Disease Genome-Wide Replication and Meta-Analysis consortium also identified only a handful of shared SNPs. 27 There is also evidence that cardiovascular risk factors modulate endothelial dysfunction/atherosclerosis-related gene expression. Previous work demonstrated down-regulation of endothelial cell (EC) inflammation-, proliferation-, and platelet adhesion-related genes, but up-regulation of EC survival-related genes in aortic ECs in response to shear stress. 28 On the contrary, high blood pressure reduces wall shear stress, 29 which is associated with enhanced atherogenesis. 30 A similar report identified largely nonoverlapping loci associated with aortic and carotid atherosclerotic lesions in a murine hyperlipidemic model. 31
The reports suggest that genetic differences between vascular beds may help explain the biology observed. However, differentially expressed genes among clinically relevant major arteries and their potential role in atherosclerosis are not well documented. In this study, therefore, differential gene expression between the coronary, aortic, and tibial arteries was investigated, with a view to identify atherosclerosis-relevant gene sets associated with known endothelial dysfunction processes.
Methods
Study design and samples
This was a bioinformatic analysis of publicly available data from the Genotype-Tissue Expression (GTEx) project, 32 comprising gene-level read counts derived from human postmortem aortic (n = 387), coronary (n = 206), and tibial artery (n = 602) samples. The samples were harvested as described in the GTEx Tissue Harvesting Work Instruction document version 03.05. 33 Briefly, aortic samples were from nonatherosclerotic ascending aorta or other thoracic regions, trimming away periaortic fat, and fibrous tissue, as well as trimming the adventitial wall inward to ensure thickness ⩽4 mm if required. Coronary artery samples were from noncalcific left and right coronary arteries, as well as noncalcific left and right descending and circumflex coronary arteries if required, after trimming away adjacent fat. Tibial artery samples were from the left tibial artery, after trimming away adjacent fat. The project datasets (GTEx Analysis V8: dbGaP Accession phs000424.v8.p2) were obtained from the GTEx Portal on April 18, 2023.
Raw data generation
RNA-seq was performed with Illumina TruSeq library construction protocol, as previously described.34,35 Briefly total RNA was quantified with Quant-iTTM RiboGreen®RNA Assay Kit and RNA Integrity Number (RIN) measured by Agilent Bioanalyzer. Messenger RNA was selected with oligo dT beads, with subsequent cDNA synthesis, end repair, addition of base “A” in preparation for adapter ligation and enrichment. Libraries were then quantified with KAPA Library Quantification Kit and sequencing performed with HiSeq 2000 or HiSeq 2500, according to manufacturer’s protocols, generating 76 bp paired-end reads.
Reads were aligned to the GRCh38/hg38 human reference genome with STAR v2.5.3a, using the GENCODE v26 annotation. 34 Gene-level read counts were generated with RNA-SeQC v1.1.9, 36 applying the “strictMode” flag. The latter filters for uniquely mapped reads contained within exon boundaries and aligned in proper pairs, with a read alignment distance of ⩽6. 34 The detailed methodology (V8) can be found at https://gtexportal.org/home/methods.
Bioinformatics analysis
Gene-level read count data for aortic, coronary, and tibial artery tissue (gct.gz files), as well as sample attribute and subject phenotype annotation data were downloaded from the GTEx project website at https://gtexportal.org/home/datasets (GTEx Analysis V8: dbGaP Accession phs000424.v8.p2). The raw count data comprised 54 592 rows (genetic features) and 1335 columns (samples), of which 432, 240, and 663 were aortic, coronary, and tibial artery samples, respectively. Sample attributes consisted of 63 variables, including the grouping variable (SMTSD: Tissue Type), site code, several batch-related variables, autolysis score, RIN, and several mapping and sequence-related variables. Subject phenotypes comprised 3 variables; sex, age, and the Hardy scale that classified cause of death into 5 categories, including “Fast death of natural causes” for which myocardial infarction was the model cause.
All files were merged, converted into a single GCT object (cmapR package, version 1.10.0 37 ), using annotation from the org.Hs.eg.db database (org.Hs.eg.db package, version 3.16.0 38 ), and then converted into a SummarizedExperiment object (SummarizedExperiment package, version 1.28.0 39 ). Initial exploratory analysis examined important variables, including demographics, Hardy category, quality-related variables, and site-, batch-, and test-related variables. Documented Hardy categories included 291 “Fast death of natural causes,” 61 “Intermediate death,” 120 “Slow death,” 793 “Ventilator Case,” and 57 “Violent and fast death.” RNA integrity number ranged between 3.2 and 10.0, total ischemic time ranged between 28 and 2076 minutes, and there were 15 samples with an autolysis score of “Moderate.’ Based on the preceding, the database was filtered to retain only samples with no or mild autolysis, RIN ⩾ 6, a recorded Hardy classification, and genes above a count threshold of ⩾10 in ⩾number of samples in the smallest group of the SMTSD variable (n = 206). This produced a dataset of 22 367 rows (features) and 1195 columns (samples), of which 387, 206, and 602 were aortic, coronary, and tibial artery samples.
Initial analysis also identified 2 RNA isolation processes used and 179 dates on which RNA was analyzed. Based on the potential impact of these test- and batch-related variables, associations between the grouping variable and other sample/subject variables, and consideration of other potentially relevant variables, a DeSeqDataSet class object was then constructed (DeSeq2 package, version 1.38.3 40 ), controlling for RIN, RNA isolation process, RNA analysis date, sex, age, and the Hardy scale category.
Differential gene expression was conducted with the DeSeq2 package, 40 with input from the DeSeqDataSet object, and specifying 3 contrasts: coronary versus tibial (CT), aorta versus tibial (AT), and coronary versus aorta (CA). The DeSeq function performed size factor estimation, dispersion estimation, and then negative binomial GLM fitting. 40 There were 339 nonconverging genetic features. As guided by the DESeq2 output, attempts were made to converge all genetic features. However, almost all these nonconverging features persisted after centering and scaling the sole numeric variable in the design formula, as well as increasing the maximum number of iterations allowed to 10 000 (default = 100). Further analysis revealed that the 339 nonconverging genetic features had significantly higher average number of zero (0) counts per feature (Welch 2 Sample t-test: 19.1% vs 1.7%, t = 20.813, P < .05) and significantly lower average mean counts (Welch 2 Sample t-test: 241.6 vs 2942.3, t = −24.321, P < .05), compared with converging features. These features were not removed at the initial filtering step to select features above a count threshold of ⩾10 in ⩾206 samples, and so were removed at this stage. The final filtered dataset, therefore, comprised 22 028 rows (features) and 1195 columns (samples), of which 387, 206, and 602 were aortic, coronary, and tibial artery samples. Differentially expressed genes (DEGs) were defined as those associated with an adjusted P value/false discovery rate (FDR) of 0.05, based on the Benjamini and Hochberg procedure, 41 and an absolute log 2 fold change ⩾1.0. Heatmaps of the differentially expressed genes for each contrast were visualized with the pheatmap package, version 1.0.12. 42
Over-representation analysis (ORA) was performed for the DEGs identified for each contrast, with the clusterProfiler package, version 4.6.2,43,44 targeting all 3 ontologies (Biological Processes, Molecular Functions, and Cellular Components) within the gene ontology (GO) knowledgebase.45,46 Enrichment analysis with clusterProfiler takes advantage of known functional conservation of genes across domains of life. 45 The enrichGO function was used to compute enrichment for GO terms based on the hypergeometric distribution. 43 Each contrast-related subset of DEGs was tested against a background gene list comprising genes identified by the assay (22 028 features). Enriched GO categories were defined as those associated with an FDR of .005, based on the Benjamini and Hochberg procedure, 41 and a q value of .005. For each contrast, the top 20 most significant over-represented GO terms were further visualized with the ggplot function from ggplot2 package, version 3.4.2. 47
Differentially expressed genes for each contrast were further examined by active-subnetwork-oriented enrichment analysis (ASOEA) using the pathfindR package, version 2.1.0. 48 Active subnetwork-based analysis exploits knowledge of protein-protein interaction networks (PINs) by identifying subgroups of correlated genes that share similar physical interactions, with the potential for improving the precision of functional enrichment analysis. 49 Differentially expressed genes for each contrast were mapped unto the PIN, BioGRID, to identify active subnetworks (active_snw_search function), using the “greedy search” algorithm at a score quantile threshold of 0.8 and minimum proportion of subnetwork significant genes criterion of 0.2. Enriched terms were identified with the enrichment_analyses function and defined as those associated with an FDR of 0.005, based on the Benjamini and Hochberg procedure. 41 Enriched terms were then clustered with the cluster_enriched_terms function, and the top clusters sorted by log2 fold change, visualized with the enrichment_chart function [pathfindR package. 48
Relevant “Biological Processes” enriched terms identified by both ORA and ASOEA were categorized into the following endothelial dysfunction-related processes for each contrast: immunity/inflammation-, lipid metabolism-, coagulation-, and membrane biology-related terms. Genes common to both GO and ASOEA terms for these 4 categories for each of the 3 contrasts were identified, their expression determined by filtering them from the lists of DEGs and were then visualized with the pheatmap package. 42
Validation
Three validation steps were performed. The categorization of enrichment terms into endothelial dysfunction-related processes was tested by matching the genes common to both GO and ASOEA terms in each category for all 3 contrasts, against compilations of relevant gene symbols and aliases, associated with each process (category). Symbols were obtained on July 18, 2023, from the Comparative Toxicogenomics Database that provides curated information regarding gene-disease relationships. 50 Datasets retrieved from searching the “Diseases” section, were genes associated with inflammation, edema (as a surrogate for loss of endothelial integrity), dyslipidemias, and blood coagulation disorders, each including their respective descendent conditions, which were therefore filtered for the primary disorder. Aliases were added using the humanSyno and alias2Symbol functions (geneSynonym version 1.7.23.7.9 and limma version 3.54.2 packages, respectively).51,52 Second, the differential expression of the top 5 up- and down-regulated genes common to GO and ASOEA terms (or less if the sample was <5) in each category for all 3 contrasts were compared with their previously reported relative tissue expression patterns, as documented in the Swiss Institute of Bioinformatics’ Bgee gene expression database. This resource curates healthy wild-type expression data and incorporates multiple sources, including the 2015 GTEx RNA-Seq data (release 6.p1). 53 Finally, to further evaluate the potential biological relevance of demonstrated changes, the genes common to GO and ASOEA terms in each category across contrasts were compared.
Statistical analysis
Associations between the grouping variable and other sample/subject variables were assessed by analysis of variance (ANOVA) or the Fisher exact test, and the relations between RIN and other sample/subject variables with the Pearson correlation, ANOVA, or t-test. Differential gene expression was performed using negative binomial GLM fitting with the Wald test. 40 All data preprocessing, statistical analysis, and data visualization were performed with R v4.2.2 (The R Foundation for Statistical Computing, 2022). A P value of <.05 was considered statistically significant. However, for identification of DEGs as well as enriched GO and ASOEA terms, P values were adjusted by the Benjamini and Hochberg procedure to control the FDR 41 at a threshold of 0.05 and 0.005, respectively.
Results
Descriptive analysis
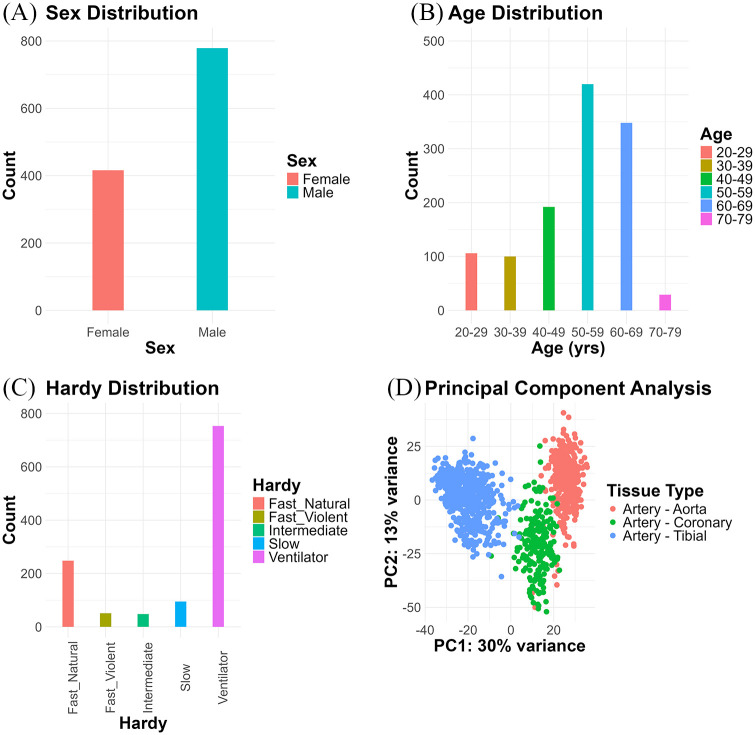
The final dataset of 22 028 rows (features) and 1195 columns (samples) comprised 416 females and 779 males (Figure 1A), with 138, 85, and 193 females, as well as 249, 121, and 409 males among the aortic, coronary, and tibial samples, respectively. There were 106, 100, 192, 420, 348, and 29 cases within the 20 to 29, 30 to 39, 40 to 49, 50 to 59, 60 to 69, and 70 to 79 age ranges (Figure 1B). Within each age range, aortic, coronary, and tibial samples accounted for the following number of cases: 35, 14, and 57; 34, 12, and 54; 59, 31, and 102; 139, 86, and 195; 113, 59, and 176; as well as 7, 4, and 18 cases, respectively. The cases fell into 5 Hardy categories: 248 “Fast death of natural causes,” 51 “Violent and fast death,” 48 “Intermediate death,” 95 “Slow death,” and 753 “Ventilator Case” (Figure 1C). Within each Hardy category, aortic, coronary, and tibial samples accounted for the following number of cases: 74, 30, and 144; 17, 9, and 25; 17, 5, and 26; 28, 15, and 52; as well as 251, 147, and 355 cases, respectively. The principal component analysis of the grouping variable (SMTSD: Tissue Type) revealed good sample separation (Figure 1D).
Figure 1.
Descriptive analysis: (A) Sex distribution. (B) Age distribution. (C) Hardy category distribution. (D) Principal component analysis of the grouping variable (SMTSD: tissue type).
Bivariate analysis
There were statistically significant associations between tissue type and several of the sample/subject variables, including RIN, autolysis score, total ischemic time, several test-related variables, as well as batch ID and date when RNA analyzed (Tables 1A and 2A). There were also statistically significant associations between RIN and several variables including autolysis score, total ischemic time, and several test- and batch-related variables including most of those significantly associated with tissue type (Tables 1B and 2B). RNA integrity number (was therefore controlled for in the design formula, in addition to the batch variables, RNA isolation process and RNA analysis date, known risk factors for atherosclerosis (sex and age), and the Hardy scale that includes the category “Fast death of natural causes” for which myocardial infarction was the model cause.
Table 1.
Association between SMTSD/SMRIN and categorical variables.
| A. SMTSD vs categorical variables (Fisher exact test) | B. SMRIN vs categorical variables (ANOVA or t-test) | ||||||
|---|---|---|---|---|---|---|---|
| P (unadjusted) | df | SumSq | MeanSq | F/t | p (unadjusted) | ||
| SMATSSCR | 0.000 | SMATSSCR | 1 | 16.43 | 16.43 | 32.13 | 0.000 |
| SMCENTER | 0.570 | SMCENTER | 3 | 0.63 | 0.21 | 0.40 | 0.760 |
| SMPTHNTS | 0.000 | SMPTHNTS | 1029 | 555.71 | 0.54 | 1.26 | 0.030 |
| SMUBRID | 0.000 | SMUBRID | 2 | 8.95 | 4.47 | 8.63 | 0.000 |
| SMNABTCH | 0.860 | SMNABTCH | 406 | 260.73 | 0.64 | 1.38 | 0.000 |
| SMNABTCHT | 0.130 | SMNABTCHT (t) | 1193 | –7.02 | 0.000 | ||
| SMNABTCHD | 1.000 | SMNABTCHD | 310 | 213.89 | 0.69 | 1.48 | 0.000 |
| SMGEBTCH | 0.000 | SMGEBTCH | 240 | 182.20 | 0.76 | 1.63 | 0.000 |
| SMGEBTCHD | 0.000 | SMGEBTCHD | 174 | 151.85 | 0.87 | 1.88 | 0.000 |
| SEX | 0.060 | SEX (wt) | 917.56 | 0.28 | 0.781 | ||
| AGE | 0.550 | AGE | 5 | 30.08 | 6.02 | 11.99 | 0.000 |
| DTHHRDY | 0.090 | DTHHRDY | 4 | 24.70 | 6.18 | 12.21 | 0.000 |
Abbreviations: DTHHRDY, Hardy Scale; SMATSSCR, Autolysis Score; SMCENTER, Code for collection site; SMGEBTCH, Genotype or Expression Batch ID; SMGEBTCHD, Date of Genotype or Expression Batch ID; SMNABTCH, Nucleic Acid Isolation Batch ID; SMNABTCHD, Date of nucleic acid isolation; SMNABTCHT, Type of nucleic acid isolation; SMPTHNTS, Pathology notes; SMRIN, RNA integrity number; SMTSD, tissue type; SMUBRID, Uberon ID, anatomical location as described by the Uber Anatomy Ontology (UBERON); t, 2 Sample t-test; wt, Welch 2 Sample t-test.
Bold = significant P-value.
Underline = t-test.
Table 2.
Association between SMTSD/SMRIN and numeric factors.
| A. SMTSD vs numeric factors (ANOVA) | B. SMRIN vs numeric factors (Pearson correlation) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| df | SumSq | MeanSq | F | p (unadjusted) | t | df | rho | lower_CI | upper_CI | p (unadjusted) | ||
| SMRIN | 2 | 8.95 | 4.47 | 8.63 | 0.000 | |||||||
| SMTSISCH | 2 | 6220739.21 | 3110369.60 | 24.31 | 0.000 | SMTSISCH | −7.45 | 1193 | −0.21 | −0.26 | −0.16 | 0.000 |
| SMTSPAX | 2 | 529555.21 | 264777.60 | 2.85 | 0.060 | SMTSPAX | 0.62 | 1190 | 0.02 | −0.04 | 0.07 | 0.530 |
| SME2MPRT | 2 | 0.19 | 0.09 | 21.80 | 0.000 | SME2MPRT | −2.49 | 1193 | −0.07 | −0.13 | −0.02 | 0.010 |
| SMCHMPRS | 2 | 70623032871.52 | 35311516435.76 | 0.61 | 0.550 | SMCHMPRS | 3.84 | 1193 | 0.11 | 0.05 | 0.17 | 0.000 |
| SMNTRART | 2 | 0.00 | 0.00 | 1.07 | 0.340 | SMNTRART | 4.61 | 1193 | 0.13 | 0.08 | 0.19 | 0.000 |
| SMMAPRT | 2 | 0.18 | 0.09 | 21.61 | 0.000 | SMMAPRT | −2.48 | 1193 | −0.07 | −0.13 | −0.01 | 0.010 |
| SMEXNCRT | 2 | 0.06 | 0.03 | 25.49 | 0.000 | SMEXNCRT | 12.05 | 1193 | 0.33 | 0.28 | 0.38 | 0.000 |
| SMGNSDTC | 2 | 193298544.40 | 96649272.20 | 75.00 | 0.000 | SMGNSDTC | −5.70 | 1193 | −0.16 | −0.22 | −0.11 | 0.000 |
| SMUNMPRT | 2 | 0.00 | 0.00 | 1.04 | 0.350 | |||||||
| SMRDLGTH | 2 | 0.00 | 0.00 | 1.04 | 0.350 | |||||||
| SME1MMRT | 2 | 0.00 | 0.00 | 0.74 | 0.480 | SME1MMRT | −3.07 | 1193 | −0.09 | −0.14 | −0.03 | 0.000 |
| SMSFLGTH | 2 | 15862.98 | 7931.49 | 1.49 | 0.230 | SMSFLGTH | 2.57 | 1193 | 0.07 | 0.02 | 0.13 | 0.010 |
| SMMPPD | 2 | 737121338684496.00 | 368560669342248.00 | 0.55 | 0.580 | SMMPPD | 1.04 | 1193 | 0.03 | −0.03 | 0.09 | 0.300 |
| SMNTERRT | 2 | 0.00 | 0.00 | 1.10 | 0.330 | SMNTERRT | −4.67 | 1193 | −0.13 | −0.19 | −0.08 | 0.000 |
| SMRRNANM | 2 | 5375538140210.74 | 2687769070105.37 | 84.17 | 0.000 | SMRRNANM | −4.51 | 1193 | −0.13 | −0.18 | −0.07 | 0.000 |
| SMRDTTL | 2 | 2706622286107600.00 | 1353311143053800.00 | 2.03 | 0.130 | SMRDTTL | 1.61 | 1193 | 0.05 | −0.01 | 0.10 | 0.110 |
| SMVQCFL | 2 | 473404280310761.00 | 236702140155380.00 | 10.71 | 0.000 | SMVQCFL | 1.31 | 1193 | 0.04 | −0.02 | 0.09 | 0.190 |
| SMTRSCPT | 2 | 194419485.16 | 97209742.58 | 74.33 | 0.000 | SMTRSCPT | −5.72 | 1193 | −0.16 | −0.22 | −0.11 | 0.000 |
| SMMPPDPR | 2 | 174940427959758.00 | 87470213979879.10 | 0.53 | 0.590 | SMMPPDPR | 1.04 | 1193 | 0.03 | −0.03 | 0.09 | 0.300 |
| SMNTRNRT | 2 | 0.06 | 0.03 | 29.00 | 0.000 | SMNTRNRT | −12.02 | 1193 | −0.33 | −0.38 | −0.28 | 0.000 |
| SMMPUNRT | 2 | 0.18 | 0.09 | 21.61 | 0.000 | SMMPUNRT | −2.48 | 1193 | −0.07 | −0.13 | −0.01 | 0.010 |
| SMEXPEFF | 2 | 0.01 | 0.01 | 1.64 | 0.200 | SMEXPEFF | 4.09 | 1193 | 0.12 | 0.06 | 0.17 | 0.000 |
| SMMPPDUN | 2 | 737121338684496.00 | 368560669342248.00 | 0.55 | 0.580 | SMMPPDUN | 1.04 | 1193 | 0.03 | −0.03 | 0.09 | 0.300 |
| SME2MMRT | 2 | 0.00 | 0.00 | 21.88 | 0.000 | SME2MMRT | 0.98 | 1193 | 0.03 | −0.03 | 0.08 | 0.330 |
| SME2ANTI | 2 | 30413560889060.40 | 15206780444530.20 | 0.48 | 0.620 | SME2ANTI | 0.96 | 1193 | 0.03 | −0.03 | 0.08 | 0.340 |
| SMALTALG | 2 | 43607887768855.60 | 21803943884427.80 | 0.42 | 0.660 | SMALTALG | −1.50 | 1193 | −0.04 | −0.10 | 0.01 | 0.130 |
| SME2SNSE | 2 | 32431275188696.10 | 16215637594348.00 | 0.51 | 0.600 | SME2SNSE | 0.97 | 1193 | 0.03 | −0.03 | 0.08 | 0.330 |
| SMMFLGTH | 2 | 10669.02 | 5334.51 | 1.18 | 0.310 | SMMFLGTH | 2.27 | 1193 | 0.07 | 0.01 | 0.12 | 0.020 |
| SME1ANTI | 2 | 31432236526126.70 | 15716118263063.30 | 0.49 | 0.610 | SME1ANTI | 1.05 | 1193 | 0.03 | −0.03 | 0.09 | 0.290 |
| SMSPLTRD | 2 | 131641586478760.00 | 65820793239380.30 | 2.64 | 0.070 | SMSPLTRD | 5.13 | 1193 | 0.15 | 0.09 | 0.20 | 0.000 |
| SMBSMMRT | 2 | 0.00 | 0.00 | 15.89 | 0.000 | SMBSMMRT | −0.26 | 1193 | −0.01 | −0.06 | 0.05 | 0.790 |
| SME1SNSE | 2 | 35984829389624.00 | 17992414694812.00 | 0.56 | 0.570 | SME1SNSE | 0.91 | 1193 | 0.03 | −0.03 | 0.08 | 0.370 |
| SME1PCTS | 2 | 1.43 | 0.72 | 9.75 | 0.000 | SME1PCTS | −4.11 | 1193 | −0.12 | −0.17 | −0.06 | 0.000 |
| SMRRNART | 2 | 0.00 | 0.00 | 110.85 | 0.000 | SMRRNART | −6.50 | 1193 | −0.19 | −0.24 | −0.13 | 0.000 |
| SME1MPRT | 2 | 0.18 | 0.09 | 21.36 | 0.000 | SME1MPRT | −2.46 | 1193 | −0.07 | −0.13 | −0.01 | 0.010 |
| SME2PCTS | 2 | 1.00 | 0.50 | 18.08 | 0.000 | SME2PCTS | 0.56 | 1193 | 0.02 | −0.04 | 0.07 | 0.580 |
Abbreviations: SMALTALG, Alternative Alignments: duplicate read entries providing alternative coordinates; SMBSMMRT, Base Mismatch Rate: Number of bases not matching the reference divided by the total number of bases aligned; SMCHMPRS, Chimeric Pairs: Pairs whose mates map to different genes; SME1ANTI, End 1 Antisense: Number of End 1 reads that were sequenced in the antisense direction; SME1MMRT, End 1 Mismatch Rate: The number of End 1 bases not matching the reference divided by the total number of End 1 bases; SME1MPRT, End 1 Mapping Rate: the number of End 1 mapped reads divided by the total number of End 1 reads; SME1PCTS, End 1 % Sense: Percentage of intragenic End 1 reads that were sequenced in the sense direction; SME1SNSE, End 1 Sense: Number of End 1 reads that were sequenced in the sense direction; SME2ANTI, End 2 Antisense: Number of reads that were sequenced in the antisense direction; SME2MMRT, End 2 Mismatch Rate: The number of End 2 bases not matching the reference divided by the total number of End 2 bases; SME2MPRT, End 2 Mapping Rate: the number of End 2 mapped reads divided by the total number of End 1 reads; SME2PCTS, End 2 % Sense: Percentage of intragenic End 2 reads that were sequenced in the sense direction; SME2SNSE, End 2 Sense: Number of End 2 reads that were sequenced in the sense direction; SMEXNCRT, Exonic Rate: The fraction of reads that map within exons; SMEXPEFF, Expression Profiling Efficiency: Ratio of exon reads to total reads; SMGNSDTC, Genes Detected: Total number of genes with at least 5 exon mapping reads; SMMAPRT, Mapping Rate: Ratio of total mapped reads to total reads; SMMFLGTH, Fragment Length M: The fragment length is the distance between the start of an upstream read and the end of the downstream pair mate; SMMPPD, Mapped: Total number of reads aligned/mapped; SMMPPDPR, Mapped Pairs: Total number of pairs for which both ends map; SMMPPDUN, Mapped Unique: Number of reads that were aligned and did not have duplicate flags; SMMPUNRT, Mapped Unique Rate of Total: Ratio of mapping of reads that were aligned and were not duplicates to total reads; SMNTERRT, Intergenic Rate: The fraction of reads that map to the genomic space between genes; SMNTRART, Intragenic Rate: The fraction of reads that map within genes (within introns or exons); SMNTRNRT, Intronic Rate: The fraction of reads that map within introns; SMRDLGTH, Read Length: maximum detected read length found; SMRDTTL, Total reads (filtered to exclude reads with vendor fail or alternative alignment flags); SMRIN, RNA integrity number; SMRRNANM, rRNA: Number of all reads (duplicate and non-duplicate) aligning to ribosomal RNA regions; SMRRNART, rRNA Rate: Ratio of all reads aligned to rRNA regions to total reads; SMSFLGTH, Fragment Length StdDev: The fragment length is the distance between the start of an upstream read and the end of the downstream pair mate; SMSPLTRD, Split Reads: The number of reads that span an exon-exon boundary; SMTRSCPT, Transcripts Detected: Total number of transcripts with at least 5 exon mapping reads; SMTSD, Tissue Type; SMTSISCH, Total Ischemic time for sample; SMTSPAX, Time sample spent in the PAXgene fixative; SMUNMPRT, Unique Rate of Mapped: Unique reads divided by all mapped reads; SMVQCFL, Failed Vendor QC Check: reads having been designated as failed by the sequencer.
Bold = significant p-value.
Underline = variables associated with SMTSD but not with SMRIN.
Overall summary of differential expression analysis
Among the 22 028 features, there were 7543 DEGs across the 3 contrasts, associated with 628 and 486 endothelial dysfunction-related GO and ASOEA terms, respectively. For the coronary x tibial (CT) contrast samples, there were 3006 DEGs, of which 2465 (82%) were up-regulated and 541 (18%) were down-regulated. Analysis of the aorta x tibial (AT) contrast, revealed 2547 DEGs, of which 1770 (69%) were up-regulated and 777 (31%) were down-regulated. For the coronary x aorta (CA) contrast, there were 1990 DEGs, of which 1301 (65%) were up-regulated and 689 (35%) were down-regulated. The full list of differentially expressed genes for the CT, AT, and CA contrasts are presented in Supplementary Tables 1–3. Functional enrichment analysis identified primarily “Biological Processes” terms. Over-representation analysis of the DEGs associated with the CT, AT, and CA contrasts identified 258, 224, and 146 GO terms, respectively (Supplementary Tables 4–6). Active-subnetwork-oriented enrichment analysis of the DEGs associated with the CT, AT, and CA contrasts identified 225, 147, and 114 ASOEA terms, respectively (Supplementary Tables 7–9). Characterization of GO and ASOEA terms into endothelial dysfunction-related processes revealed the following: immunity and inflammation-related terms included innate and adaptive responses, immune cell activation, proliferation, and migration, cytokine production and signaling, phagocytosis, and inflammatory responses; membrane biology terms included those related to cell adhesion and para-cellular transport; lipid metabolism terms involved enzyme activity, fatty acid transport, and lipid homeostasis; coagulation relevant terms included platelet activation, heparin binding and blood coagulation.
Coronary x tibial (CT) contrast
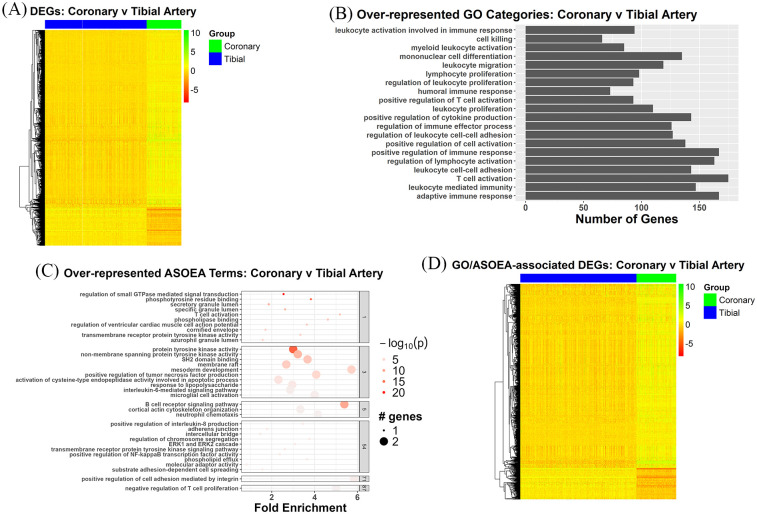
The heatmap of the 3006 CT contrast DEGs showed a mixed pattern of up- and down-regulated genes, with mostly up-regulation (Figure 2A). Examination of the top 20 GO terms for the CT contrast-revealed domination of immunity and inflammation-related terms (Figure 2B), while the top 20 ASOEA terms also included a few immunity and inflammation-related terms (Supplementary Table 7). However, the top clustered ASOEA terms included several immunity/inflammation-, lipid metabolism-, and membrane biology-related terms (Figure 2C). Among the 1556 unique CT contrast DEGs associated with enriched GO terms, 840 were shared with the 974 unique CT contrast DEGs associated with enriched ASOEA terms, with the common DEGs demonstrating mostly up-regulation (Figure 2D).
Figure 2.
CT contrast DEGs: Overview and enrichment terms. (A) Heatmap of 3006 DEGs. (B) Top 20 GO terms. (C) Top clustered ASOEA terms. (D) Heatmap of 840 DEGs associated with both enriched GO and ASOEA terms.
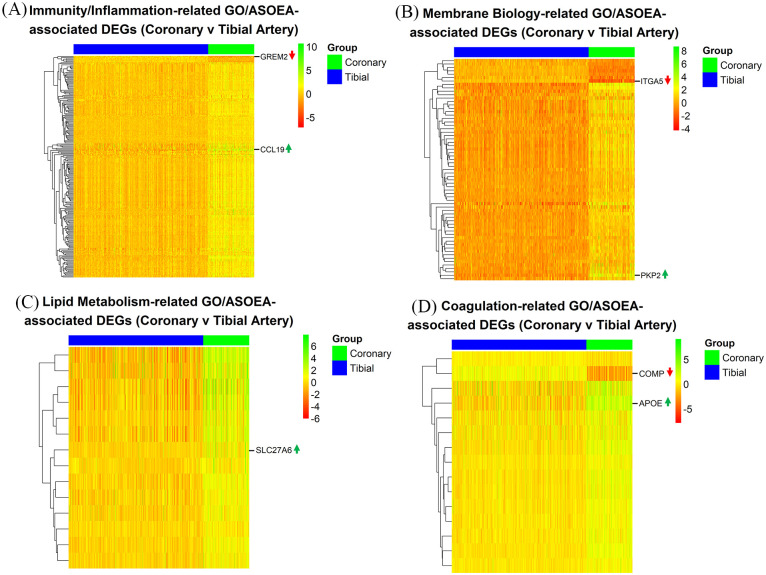
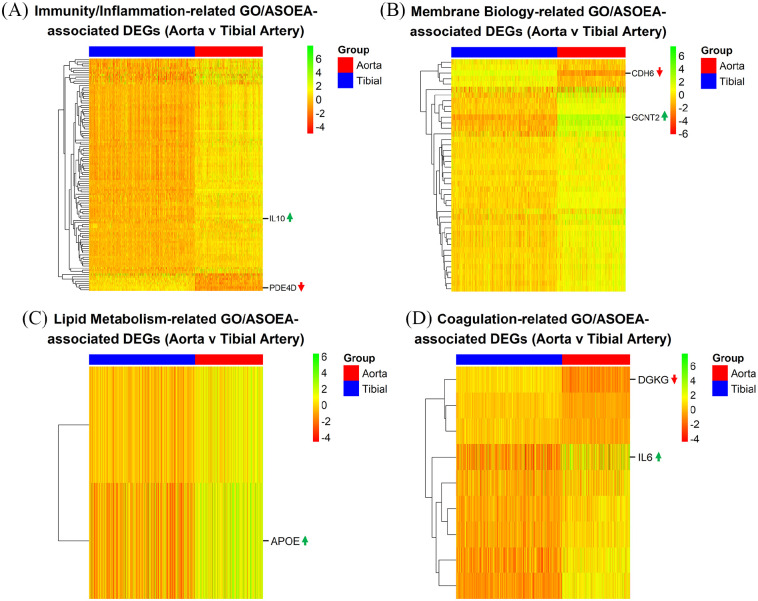
There were 65 (25.19%), 7 (2.71%), 7 (2.71%), and 1 (0.39%), as well as 40 (17.78%), 11 (4.89%), 5 (2.22%), and 2 (0.89%) immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related GO and ASOEA terms, respectively. Among these terms, there were 605, 270, 64, and 35 GO term-related genes, and 216, 115, 16, and 31 ASOEA term-related genes associated with immunity/inflammation, membrane biology, lipid metabolism, and coagulation, respectively. Of these, 210 (204 [97.14%] up-regulated), 65 (58 [89.23%] up-regulated), 14 (14 [100.00%] up-regulated), and 15 (13 [86.67%] up-regulated) genes were common to immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related GO and ASOEA terms, respectively (See Supplementary Tables 10–13 for complete lists). Visualization of common GO/ASOEA-related DEGs associated with each process demonstrated the general pattern of upregulation especially among the lipid metabolism- and immunity/inflammation-related genes (Figure 3A to D and Supplementary Figure 1a-1d).
Figure 3.
CT contrast: endothelial dysfunction-related DEGs. Heatmaps of common GO/ASOEA-related DEGs associated with (A) immunity/inflammation, (B) membrane biology, (C) lipid metabolism, and (D) coagulation, with the top up- and down-regulated genes labeled (up and down arrows respectively).
Aorta x tibial (AT) contrast
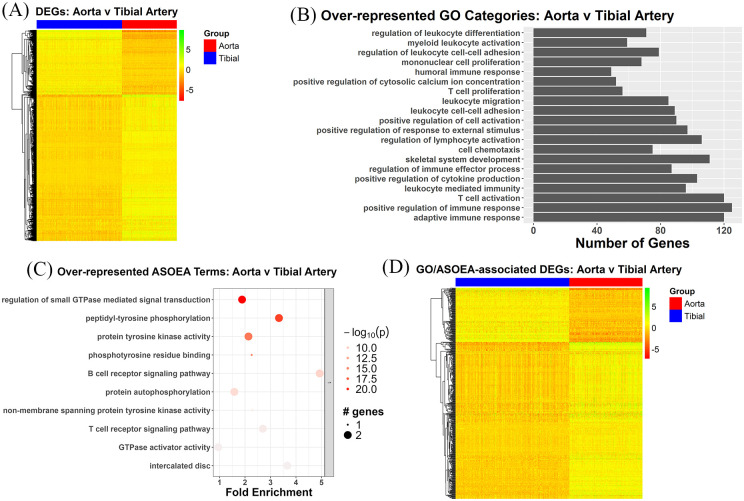
The 2547 AT contrast DEGs also demonstrated a mixture of up- and down-regulated genes, with mostly up-regulation (Figure 4A). The top 20 GO terms for the AT contrast revealed domination of immunity and inflammation-related terms (Figure 4B), while the top 20 ASOEA terms (Supplementary Table 8) and the top clustered ASOEA terms (Figure 4C) included a few immunity and inflammation-related terms. Among the 1248 unique AT contrast DEGs associated with enriched GO terms, 515 were shared with the 586 unique AT contrast DEGs associated with enriched ASOEA terms, with the common DEGs also demonstrating mostly up-regulation (Figure 4D).
Figure 4.
AT contrast DEGs: overview and enrichment terms. (A) Heatmap of 2547 DEGs. (B) Top 20 GO terms. (C) Top clustered ASOEA terms. (D) Heatmap of 515 DEGs associated with both enriched GO and ASOEA terms.
There were 55 (24.55%), 6 (2.68%), 3 (1.34%), and 2 (0.89%), as well as 26 (17.69%), 8 (5.44%), 1 (0.68%), and 1 (0.68%) immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related GO and ASOEA terms, respectively. Among these terms, there were 419, 188, 19, and 41 GO term-related genes, and 111, 62, 3, and 9 ASOEA term-related genes associated with immunity/inflammation, membrane biology, lipid metabolism, and coagulation, respectively. Of these, there were 107 (99 [92.52%] up-regulated), 42 (37 [88.10%] up-regulated), 2 (2 [100.00%] up-regulated), and 9 (6 [66.67%] up-regulated) genes common to immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related GO and ASOEA terms, respectively (See Supplementary Tables 14–17 for complete lists). Visualization of GO/ASOEA shared genes associated with each process also demonstrated a general pattern of upregulation, especially among the lipid metabolism- and immunity/inflammation-related genes (Figure 5A to D and Supplementary Figure 2a – 2d).
Figure 5.
AT contrast: endothelial dysfunction-related DEGs. Heatmaps of common GO/ASOEA-related DEGs associated with (A) immunity/inflammation, (B) membrane biology, (C) lipid metabolism, and (D) coagulation, with the top up- and down-regulated genes labeled (up and down arrows respectively).
Coronary x aorta (CA) contrast
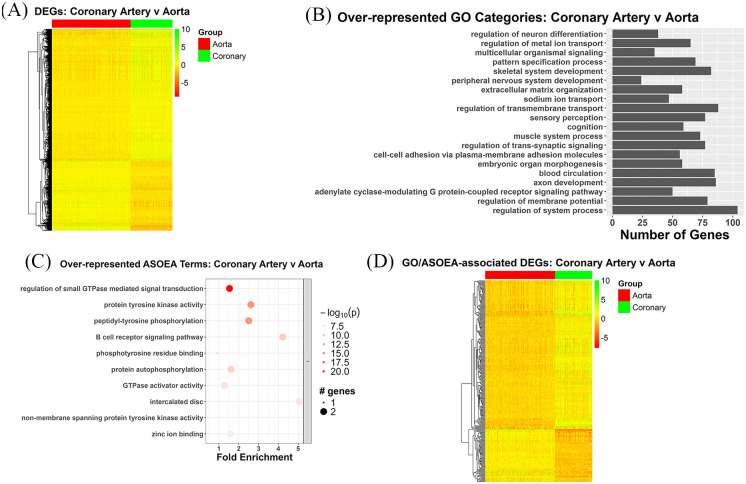
Similarly, the 1990 CA contrast DEGs demonstrated a mixed pattern of up- and down-regulated genes, with mostly up-regulation (Figure 6A). However, unlike the CT and AT contrasts, there was more diversity among the top 20 GO terms, which lacked specific immunity and inflammation-related terms (Figure 6B), as well as more diversity among top 20 ASOEA terms for the CA contrast with only 3 and 1 immunity and inflammation-related terms before (Supplementary Table 9) and after clustering (Figure 6 C), respectively. Among the 955 unique CA contrast DEGs associated with enriched GO terms, 314 were shared with the 375 unique CT contrast DEGs associated with enriched ASOEA terms, with the common DEGs also demonstrating a mixed pattern of expression, with mostly up-regulation (Figure 6D).
Figure 6.
CA contrast DEGs: overview and enrichment terms. (A) Heatmap of 1990 DEGs. (B) Top 20 GO terms. (C) Top clustered ASOEA terms. (D) Heatmap of 314 DEGs associated with both enriched GO and ASOEA terms.
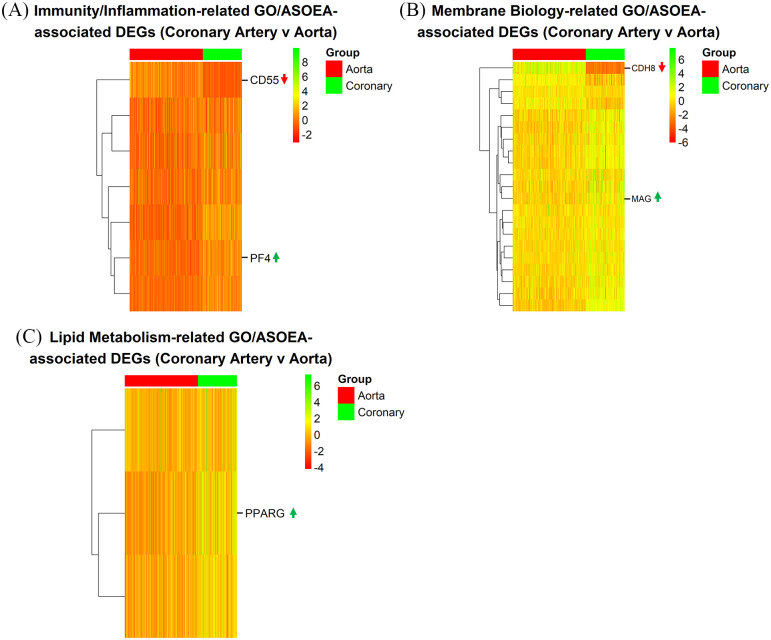
There were only 1 (0.68%), 3 (2.05%), 1 (0.68%), and 0 (0.00%), as well as 20 (17.54%), 7 (6.14%), 1 (0.88%), and 1 (0.88%) immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related GO and ASOEA terms, respectively. Among these terms, there were 30, 97, 20, and 0 GO term-related genes, and 34, 39, 3, and 4 ASOEA term-related genes associated with immunity/inflammation, membrane biology, lipid metabolism, and coagulation, respectively. Of these, there were 7 (6 [88.89%] up-regulated), 21 (17 [80.95%] up-regulated), 3 (3 [100.00%] up-regulated), and 0 (0 [0.00%] up-regulated) genes common to immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related GO and ASOEA terms, respectively (See Supplementary Tables 18–20 for complete lists). Visualization of the GO/ASOEA common genes associated with the first 3 processes demonstrated generally mild changes across arteries (Figure 7A to C and Supplementary Figure 3a – 3c).
Figure 7.
CA contrast: endothelial dysfunction-related DEGs. Heatmaps of common GO/ASOEA-related DEGs associated with (A) immunity/inflammation, (B) membrane biology, and (C) lipid metabolism, with the top up- and down-regulated genes labeled (up and down arrows respectively).
Validation
The categorization of enrichment terms into endothelial dysfunction-related processes was tested by matching genes common to GO and ASOEA terms in each category for all 3 contrasts against compilations of gene symbols and aliases associated with each process (category). Among lists of 104 154, 97 573, 92 014, and 81 024 genes/aliases associated with inflammation, edema (as a surrogate for impaired membrane biology), dyslipidemias, and blood coagulation disorders, all genes common to both GO and ASOEA terms in each category for all 3 contrasts were identified in their respective list. These results support the categorization of relevant GO and ASOEA terms into the 4 endothelial dysfunction-related processes.
The observed differential expressions of the top 5 up- and down-regulated genes (or less if the sample was <5) in each category for all 3 contrasts were compared with their previously reported relative tissue expression patterns, as documented in the Bgee gene expression database. 53 The top up-regulated DEGs identified for each category across all 3 contrasts are shown in Table 3. All top up-regulated genes in CT contrasts were found to be more highly expressed in left and right coronary arteries than tibial artery. Similarly, all top up-regulated AT contrast genes were found to be more highly expressed in ascending and descending thoracic aorta than tibial artery. All top up-regulated genes in CA contrasts were found to be more highly expressed in left and right coronary arteries than ascending and descending thoracic aorta, except for MS4A1 and PPARG. MS4A1 expression was the highest in the right coronary artery, followed by the ascending aorta, left coronary artery, and least expressed in the descending thoracic aorta, while PPARG expression was the highest in the left coronary artery, followed by descending thoracic aorta, the ascending aorta, and least expressed in the right coronary artery. 53 However, for both genes, average right, and left coronary artery expression was higher than average ascending aorta and descending thoracic aorta expression. 53
Table 3.
Top up-regulated DEGs.
| CT_ImIn | AT_ImIn | CA_ImIn | CT_Memb | AT_Memb | CA_Memb | CT_Lip | AT_Lip | CA_Lip | CT_Coag | AT_Coag |
|---|---|---|---|---|---|---|---|---|---|---|
| CCL19 | IL10 | PF4 | PKP2 | GCNT2 | MAG | SLC27A6 | APOE | PPARG a | APOE | IL6 |
| IGHA1 | IL6 | GATA3 | BMP7 | CDH8 | NRXN1 | APOC1 | PLTP | APOA1 | IL6 | DGKI |
| BLK | CCL19 | MS4A1 a | CDH2 | IL10 | PKP2 | APOE | ABCA8 | LCK | PDPN | |
| MS4A1 | TNF | IL7 | MAG | CDH2 | CDH6 | FABP4 | VAV1 | LCK | ||
| TNFSF14 | CCL3 | CXCL6 | TENM2 | TNF | NTNG1 | CD36 | TYRO3 | VAV1 |
Abbreviations: AT, Aorta vs Tibial; CA, Coronary vs Aorta; Coag, coagulation-related genes common to GO and ASOEA terms; CT, Coronary vs Tibial; ImIn, immunity/inflammation-related genes common to GO and ASOEA terms; Lip, lipid metabolism-related genes common to GO and ASOEA terms; Memb, membrane biology-related genes common to GO and ASOEA terms.
Genes for which average expression in Bgee database 53 used for validation.
The top down-regulated DEGs identified for each category across all 3 contrasts are shown in Table 4. Among the down-regulated genes, the converse pattern was observed. Genes in CT contrasts were found to be less expressed in left and right coronary arteries than tibial artery. Down-regulated AT contrast genes were found to be expressed at a lower level in ascending and descending thoracic aorta than tibial artery. Genes in CA contrasts were found to be less expressed in left and right coronary arteries than ascending and descending thoracic aorta, except for GRHL2. The GRHL2 expression was the highest in the right coronary artery, followed by the ascending aorta, descending thoracic aorta, and least expressed in the left coronary artery. 53 Also, average right and left coronary artery expression was slightly higher than average ascending aorta and descending thoracic aorta expression. 53
Table 4.
Top down-regulated DEGs.
| CT_ImIn | AT_ImIn | CA_ImIn | CT_Memb | AT_Memb | CA_Memb | CT_Coag | AT_Coag |
|---|---|---|---|---|---|---|---|
| GREM2 | PDE4D | CD55 | ITGA5 | CDH6 | CDH8 | COMP | DGKG |
| PDE4D | CHI3 L1 | CDH8 | NTNG1 | CNTN6 | F2 | F2 | |
| NCKAP1L | NCKAP1L | NLGN1 | NCKAP1L | ALCAM | DGKB | ||
| SDC4 | ADRA2A | NCKAP1L | ITGA5 | GRHL2 a | |||
| JAK2 | IL6R | CNTN6 | CTNNA3 |
Abbreviations: AT, Aorta vs Tibial; CA, Coronary vs Aorta; Coag, coagulation-related genes common to GO and ASOEA terms; CT, Coronary vs Tibial; ImIn, immunity/inflammation-related genes common to GO and ASOEA terms; Memb, membrane biology-related genes common to GO and ASOEA terms.
Gene with expression pattern not consistent with Bgee gene expression database. 53
To further test the findings and assess their potential biological relevance, comparisons were made of the genes common to GO and ASOEA terms in each category across contrasts. Among the immunity/inflammation-related up-regulated genes, there were 91, 5, and 3 genes in common between CT and AT, CT and CA, and the AT and CA contrast pairs, respectively. For the membrane biology-related genes, 30, 9, and 3 genes were common between these pairs, while there were 2 and 2 common lipid metabolism gene for the CT/AT and CT/CA contrast pairs, but none for the AT/CA pair. Finally, among coagulation-related genes, there were 5 genes in common between CT/AT, but none between CT/CA and AT/CA.
Among the immunity/inflammation-related down-regulated genes, there were 2 genes common between CT and AT contrast pairs, but none between the CT/CA and AT/CA pairs. For the membrane biology genes, 3 and 2 were common between CT/AT and CT/CA, respectively, but none between AT and CA. Only the coagulation-related gene F2 was common between CT/AT, with none between the CT/CA and AT/CA pairs. Finally, there were no down-regulated lipid metabolism-related genes. These findings support the prior evidence of greater similarity in the gene expression pattern between the CT and AT contrasts and, therefore, between the coronary and aortic tissues.
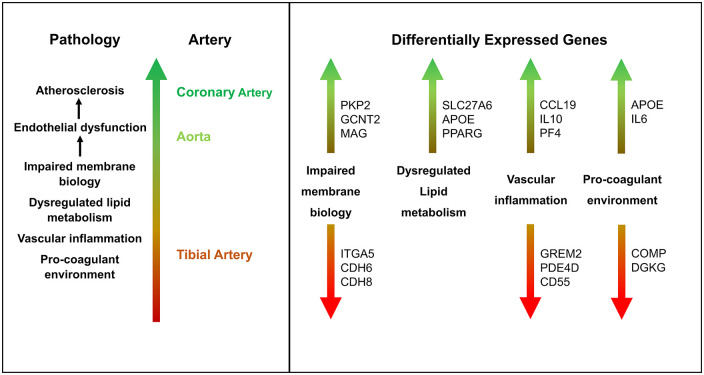
Taken together, the results imply that transcriptomic differences between coronary or aorta versus tibial samples largely involve immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related genes. Furthermore, these changes have the potential to modulate endothelial dysfunction and, therefore, atherosclerosis (Figure 8).
Figure 8.
Proposed model of genetic basis of relative inter-artery atherosclerosis risk.
Discussion
This study identified 7543 DEGs across 3 contrasts, associated with 628 and 486 endothelial dysfunction-related GO and ASOEA terms, respectively. Both CT and AT contrast DEGs and those common to GO and ASOEA terms showed primarily up-regulation, and were dominated by immunity/inflammation-related genes with contribution from membrane biology-, lipid metabolism-, and coagulation-related genes. However, although CA contrast DEGs and those common to GO and ASOEA terms also showed primarily up-regulation, there was no dominance of immunity/inflammation-, membrane biology-, and lipid metabolism-related genes, and there was a lack of coagulation-related genes.
Validity of the findings was tested by matching genes common to GO and ASOEA terms in each category and contrast against gene symbols and aliases known to be related to endothelial dysfunction-related processes.8-10, 15 This revealed full overlap between categorized genes and respective compiled lists, providing justification for categorization. The subsequent verification of expression patterns of DEGs by category and contrast was done by searching Bgee gene expression database, 53 which confirmed the relative expression pattern of almost all top up- and down-regulated genes. Furthermore, the comparisons of top DEGs for each category and contrast indicated greater overlap for both up- and down-regulated genes between CT and AT contrasts, compared with CT/CA and AT/CA contrast pairs, suggesting that gene expression patterns of coronary artery and aorta were similar, but distinct from tibial artery samples.
The biological relevance of the observed changes was further explored by reviewing the top DEG for each category and contrast, cognizant of key arterial cells with potential to modulate atherogenesis, including ECs, vascular smooth muscle cells (VSMCs), various immune cells, and platelets.12,14 Regarding CCL19, the top up-regulated immunity/inflammation-related CT contrast DEG, also among the top AT contrast genes, pro-inflammatory cytokines were found down-regulated in the thoracic aorta and serum of a CCL19-deficient murine model. 54 IL10, the top up-regulated AT contrast DEG, is widely considered an anti-inflammatory cytokine. However, although IL-10R1del macrophages showed pro-inflammatory phenotype in vitro, IL-10R1 deletion in hyperlipidemic mice reduced recruitment of macrophages and neutrophils, and attenuated plaque size, in the absence of systemic inflammation. These findings suggest complexity in IL10 atherogenesis signaling with possible interaction between vascular inflammatory and cholesterol homeostatic mechanisms. 55 With respect to the top CA contrast DEG, PF4 produced a VSMC inflammatory phenotype, with enhanced VSMC proliferation and cytokine production. 56 Among down-regulated DEGs, the top CT contrast gene GREM2, has been shown to block BMP2/TNFα activity within ECs including expression of inflammatory molecules and leukocyte adhesion, with excessive peri-infarct inflammation observed in a GREM2 knockdown murine model. 57 Similarly, inhibition of the top AT contrast gene PDE4D in a murine model was associated with vascular inflammation including coronary arteritis, 58 while the only CA contrast DEG CD55, among its pleotropic effects, functions as a complement system inhibitor, with deficiency associated with enhanced immune and inflammatory responses. 59 These findings imply potential for relatively pro-inflammatory coronary artery environment, in comparison with tibial artery tissue, with the aortic environment being intermediate, which could favor coronary and to a lesser extent aortic atherogenesis.15,16
Pro-inflammatory coronary and aortic environments would be facilitated by disruption of essential membrane functions, such as cell adhesion and membrane transport. VE-cadherin is a crucial endothelial adhesion molecule that, in concert with β-catenin, helps maintain endothelial integrity, including the control of leukocyte para-cellular transmigration. 60 Among the top up-regulated membrane biology-related DEGs, PKP2 has been shown to bind β-catenin, potentially reducing the latter’s availability,61,62 thereby destabilizing cadherin/β-catenin-mediated cell adhesion. 60 The impact of over-expressed GCNT2 may be more direct as seen by its role in TGF-β1–induced suppression of E-cadherin expression in the epithelial cell line NMuMG. 63 However, the significance of Myelin-associated glycoprotein (MAG), top gene in the CA contrast and among the top CT contrast DEGs, is less clear. Myelin-associated glycoprotein is a cell-adhesion molecule, highly expressed on myelinated nerve cells, and its dimerization was shown to maintain myelin-axon spacing and inhibit neurite outgrowth. 64 It is intriguing to consider whether MAG may play a similar role within the vascular endothelial layer, and whether this could impact endothelial integrity or transport. On the contrary, top down-regulated DEGs comprise the integrin ITGA5, and cadherins CDH6 and CDH8. Integrins and cadherins are known to be integral to cell-cell and cell-matrix adhesions.65,66 Their down-regulation could therefore be expected to compromise endothelial integrity. The expression of top membrane biology-related DEGs, therefore, implies relative endothelial junction disruption and enhanced membrane permeability in coronary artery samples, compared with tibial artery tissue, with the aortic samples being intermediate, again favoring coronary and to a lesser extent aortic atherogenesis.15,16
Relatively inflammatory coronary and aortic environments could be driven by dysregulated lipid metabolism. SLC27A6, the top up-regulated lipid metabolism-related CT contrast DEG, is a fatty acid transport protein that enhanced intracellular triglyceride and total cholesterol in nasopharyngeal carcinoma cells, 67 suggesting atherogenic potential. APOE was the top up-regulated AT contrast DEG, also among the top CT contrast genes. APOE is produced by several cell types including macrophages, and as a ligand constituent of various lipoproteins mediates the interaction between the latter and their receptors, thereby modulating blood and tissue lipid levels. Of the 3 major alleles, APOE4 is associated with higher LDL-cholesterol levels compared with APOE3, and although APOE2 generally lowers cholesterol levels, in the presence of certain genetic or environmental risk factors, can also increase cholesterol levels. 68 In addition, APOE4 was less efficient at cholesterol efflux from murine macrophages compared to APOE3, which could promote greater foam cell formation. 69 PPARG, the top up-regulated CA contrast DEG, is a nuclear receptor induced by oxidized-LDL in human monocytes and subsequently enhanced oxidized-LDL uptake and monocyte/macrophage differentiation. 70
Atherogenesis would be further promoted by a pro-coagulant environment. The top up-regulated coagulation-related CT and AT contrast DEGs were APOE and IL6, respectively. In addition to its role in lipid metabolism, APOE may play a role in coagulation, as revealed by its association with factor VIII and von Willebrand factor levels. 71 Similarly, during inflammation in humans, the pro-inflammatory cytokine IL-6 appeared to induce coagulation. 72 Vascular inflammation has been shown to enhance EC cytokine production including IL6, which can enhance platelet production and activation, stimulate monocyte tissue factor mRNA and surface expression, and shift the balance of hemostatic factors in favor of coagulation. 73 The top down-regulated CT and AT contrast DEGs were COMP and DGKG. Platelet-derived COMP was shown to inhibit coagulation, with COMP deficiency associated with shortened tail-bleeding and reduced clotting time in mice. 74 Although the role of DGKG in coagulation appears unclear, loss of its related gene isoform in a DGKζ-knockout murine model, upregulated platelet glycoprotein VI (GPVI) surface expression and enhanced GPVI-mediated platelet activation. 75
Taken together, these results are consistent with previous evidence of clinical, structural, and temporal differences in atherosclerosis across vascular beds, with a tendency toward increased central severity.18,19,21,22 They demonstrate a plausible mechanism: transcriptomic differences between coronary or aorta versus tibial samples largely involve immunity/inflammation-, membrane biology-, lipid metabolism-, and coagulation-related genes, with the potential to modulate endothelial dysfunction and therefore atherosclerosis. This finding further suggests the need for additional comparisons between other arteries to better understand the genetic basis of relative inter-artery atherosclerosis risk.
Possible triggers for the demonstrated transcriptomic differences are not fully understood. A 1964 report identified higher blood pressure and/or body build as correlated with cerebral, coronary, or aortic atherosclerosis severity. 76 Among an autopsy cohort, the comparison between right coronary and aortic arteries indicated increased lesion severity in the aorta among 30- to 34-year-old smokers and in the right coronary among >25-year-old subjects with hypertension. 77 Interconnectivity of the vascular system implies some potential systemic variables cannot fully account for the changes, for example, glucose levels between arteries do not differ significantly.78-80 On the contrary, systolic blood pressure does. 81 A progressive increase in arterial stiffness moving toward the periphery results in systolic pressure amplification. 82 Other local factors, for example, hemodynamics, vessel geometry and other wall properties, and peri-vascular mechanical forces may also contribute to the differences.20,31
This study had some important strengths. The samples exceeded 1000, with representation across a wide age range. The use of postmortem samples also allowed for the direct comparison of tissues not usually collected from the same individual. In addition, the bioinformatic analyses generated substantial amounts of data, which could be used for further investigation. There were also some limitations. There were nearly twice as many males as females. The samples were arterial tissue, which prevented an analysis of differential gene expression at the cellular level. Also, this study used data derived from donors enrolled at 4 sites, from RNA isolated using 2 different extraction processes on multiple dates, as well as from RNA analyzed on multiple dates. Owing to the heterogeneity in study design introduced by the meta-analytic approach, both RNA isolation process and RNA analysis date were controlled for in the analysis. Finally, postmortem samples may be subject to autolysis and other compromise to tissue quality. However, the present study screened out samples with more than mild autolysis and RIN < 6, to minimize the risk of poor sample quality.
In conclusion, in this study, it was obtained that differentially expressed genes across major arteries are enriched in endothelial dysfunction-related gene sets, including those with the potential to modulate endothelial integrity and permeability, lipid uptake, vascular inflammation, and coagulation. Given the role of endothelial dysfunction in atherosclerosis, this finding may help explain the observed relative inter-artery atherosclerosis risk. Understanding this risk could inform the development and timing of diagnostic and therapeutic strategies for the management of atherosclerotic disease. For example, unlike current systemic approaches, anti-inflammatory, anticoagulant, or other therapies targeted to relevant arteries in patients identified as being at risk, could potentially provide superior clinical efficacy with optimized risk-benefit ratios.
Supplemental Material
Supplemental material, sj-tif-21-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-tif-22-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-tif-23-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-1-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-10-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-11-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-12-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-13-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-14-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-15-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-16-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-17-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-18-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-19-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-2-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-20-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-3-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-4-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-5-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-6-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-7-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-8-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-9-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Acknowledgments
The project datasets (GTEx Analysis V8: dbGaP accession number phs000424.v8.p2) were obtained from the GTEx Portal on April 18, 2023.
Footnotes
The author declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Author Contributions: PAB: Conceptualization, Methodology, Software, Formal analysis, Writing—Original Draft, Writing—Review & Editing, Visualization, Final Approval.
Research Ethics and Patient Consent: Not applicable as this study involved bioinformatic analysis of publicly available data.
ORCID iD: Paul A Brown  https://orcid.org/0000-0003-3039-4752
https://orcid.org/0000-0003-3039-4752
Data Availability: The datasets used in this paper are publicly available from the project website at https://gtexportal.org/home/datasets, under “GTEx Analysis V8 (dbGaP Accession phs000424.v8.p2),” “Gene read counts by tissue.”
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Vaduganathan M, Mensah GA, Turco JV, Fuster V, Roth GA. The global burden of cardiovascular diseases and risk. J Am Coll Cardiol. 2022;80:2361-2371. doi: 10.1016/j.jacc.2022.11.005. [DOI] [PubMed] [Google Scholar]
- 2. Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of atherosclerosis and the potential to reduce the global burden of atherothrombotic disease. Circ Res. 2016;118:535-546. doi: 10.1161/CIRCRESAHA.115.307611. [DOI] [PubMed] [Google Scholar]
- 3. Khan MA, Hashim MJ, Mustafa H, et al. Global epidemiology of ischemic heart disease: results from the global burden of disease study. Cureus. 2020;12:e9349. doi: 10.7759/cureus.9349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song P, Fang Z, Wang H, et al. Global and regional prevalence, burden, and risk factors for carotid atherosclerosis: a systematic review, meta-analysis, and modelling study. Lancet Glob Health. 2020;8:e721-e729. doi: 10.1016/S2214-109X(20)30117. [DOI] [PubMed] [Google Scholar]
- 5. WHO. The top 10 causes of death. World Health Organization. Accessed August 10, 2023. https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death [Google Scholar]
- 6. Chen W, Li Z, Zhao Y, Chen Y, Huang R. Global and national burden of atherosclerosis from 1990 to 2019: trend analysis based on the global burden of disease study 2019. Chin Med J (England). 2023;136:2442-2450. doi: 10.1097/cm9.0000000000002839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vesnina A, Prosekov A, Atuchin V, Minina V, Ponasenko A. Tackling atherosclerosis via selected nutrition. Int J Mol Sci. 2022;23:8233. doi: 10.3390/ijms23158233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mannarino E, Pirro M. Molecular biology of atherosclerosis. Clin Cases Miner Bone Metab. 2008;5:57-62. [PMC free article] [PubMed] [Google Scholar]
- 9. Wolf D, Zirlik A, Ley K. Beyond vascular inflammation—recent advances in understanding atherosclerosis. Cell Mol Life Sci. 2015;72:3853-3869. doi: 10.1007/s00018-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wolf D, Ley K. Immunity and inflammation in atherosclerosis. Circ Res. 2019;124:315-327. doi: 10.1161/circresaha.118.313591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sobolevskaya EV, Shumkov OA, Smagin MA, et al. Markers of restenosis after percutaneous transluminal balloon angioplasty in patients with critical limb ischemia. Int J Mol Sci. 2023;24:9096. doi: 10.3390/ijms24109096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lusis AJ. Atherosclerosis. Nature. 2000;407:233-241. doi: 10.1038/35025203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of cantos and beyond. J Am Coll Cardiol. 2017;70:2278-2289. doi: 10.1016/j.jacc.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rafieian-Kopaei M, Setorki M, Doudi M, Baradaran A, Nasri H. Atherosclerosis: process, indicators, risk factors and new hopes. Int J Prev Med. 2014;5:927-946. [PMC free article] [PubMed] [Google Scholar]
- 15. Hadi HA, Carr CS, Al Suwaidi J. Endothelial dysfunction: cardiovascular risk factors, therapy, and outcome. Vasc Health Risk Manag. 2005;1:183-198. [PMC free article] [PubMed] [Google Scholar]
- 16. Peng Z, Shu B, Zhang Y, Wang M. Endothelial response to pathophysiological stress. Arterioscler Thromb Vasc Biol. 2019;39:e233-e243. doi: 10.1161/ATVBAHA.119.312580. [DOI] [PubMed] [Google Scholar]
- 17. Krüger-Genge A, Blocki A, Franke RP, Jung F. Vascular endothelial cell biology: an update. Int J Mol Sci. 2019;20:4411. doi: 10.3390/ijms20184411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reiner L, Jimenez FA, Rodriguez FL. Atherosclerosis in the mesenteric circulation. Observations and correlations with aortic and coronary atherosclerosis. Am Heart J. 1963;66:200-209. doi: 10.1016/0002-8703(63)90035. [DOI] [PubMed] [Google Scholar]
- 19. Sawabe M, Arai T, Kasahara I, et al. Sustained progression and loss of the gender-related difference in atherosclerosis in the very old: a pathological study of 1074 consecutive autopsy cases. Atherosclerosis. 2006;186:374-379. doi: 10.1016/j.atherosclerosis.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 20. Yamaguchi T, Morino K. Perivascular mechanical environment: a narrative review of the role of externally applied mechanical force in the pathogenesis of atherosclerosis. Front Cardiovasc Med. 2022;9:944356. doi: 10.3389/fcvm.2022.944356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dalager S, Paaske WP, Kristensen IB, Laurberg JM, Falk E. Artery-related differences in atherosclerosis expression. Stroke. 2007;38:2698-2705. doi: 10.1161/STROKEAHA.107.486480. [DOI] [PubMed] [Google Scholar]
- 22. Dalager S, Falk E, Kristensen IB, Paaske WP. Plaque in superficial femoral arteries indicates generalized atherosclerosis and vulnerability to coronary death: an autopsy study. J Vasc Surg. 2008;47:296-302. doi: 10.1016/j.jvs.2007.10.037. [DOI] [PubMed] [Google Scholar]
- 23. Nakamura E, Sato Y, Iwakiri T, et al. Asymptomatic plaques of lower peripheral arteries and their association with cardiovascular disease: an autopsy study. J Atheroscler Thromb. 2017;24:921-927. doi: 10.5551/jat.39669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Herisson F, Heymann MF, Chétiveaux M, et al. Carotid and femoral atherosclerotic plaques show different morphology. Atherosclerosis. 2011;216:348-354. doi: 10.1016/j.atherosclerosis.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 25. Archacki SR, Angheloiu G, Moravec CS, Liu H, Topol EJ, Wang QK. Comparative gene expression analysis between coronary arteries and internal mammary arteries identifies a role for the tes gene in endothelial cell functions relevant to coronary artery disease. Hum Mol Genet. 2011;21:1364-1373. doi: 10.1093/hmg/ddr574. [DOI] [PubMed] [Google Scholar]
- 26. Erdmann J, Kessler T, Munoz Venegas L, Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc Res. 2018;114:1241-1257. doi: 10.1093/cvr/cvy084. [DOI] [PubMed] [Google Scholar]
- 27. Bis JC, Kavousi M, Franceschini N, et al. Meta-analysis of genome-wide association studies from the charge consortium identifies common variants associated with carotid intima media thickness and plaque. Nat Genet. 2011;43:940-947. doi: 10.1038/ng.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen BP, Li YS, Zhao Y, et al. DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol Genomics. 2001;7:55-63. doi: 10.1152/physiolgenomics.2001.7.1.55. [DOI] [PubMed] [Google Scholar]
- 29. Kumar N, Pai R, Abdul Khader SM, Khan SH, Kyriacou PA. Influence of blood pressure and rheology on oscillatory shear index and wall shear stress in the carotid artery. J Braz Soc Mech Sci Eng. 2022;44:510. doi: 10.1007/s40430-022. [DOI] [Google Scholar]
- 30. Zhou M, Yu Y, Chen R, et al. Wall shear stress and its role in atherosclerosis. Front Cardiovasc Med. 2023;10:1083547. doi: 10.3389/fcvm.2023.1083547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jones MB, An A, Shi LJ, Shi W. Regional variation in genetic control of atherosclerosis in hyperlipidemic mice. G3 (Bethesda). 2020;10:4679-4689. doi: 10.1534/g3.120.401856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Carithers LJ, Ardlie K, Barcus M, et al. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank. 2015;13:311-319. doi: 10.1089/bio.2015.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. GTEx. GTEx tissue harvesting work instruction ver. 03.05. Accessed July 18, 2023. https://biospecimens.cancer.gov/resources/sops/docs/GTEx_SOPs/BBRB-PR-0004-W1%20GTEx%20Tissue%20Harvesting%20Work%20Instruction.pdf
- 34. GTEx. Laboratory methods. Published 2019. Accessed July 18, 2023. https://gtexportal.org/home/methods
- 35. Illumina. Truseq™ RNA and DNA library preparation kits v2. Published 2014. Accessed July 18, 2023. http://www.illumina.com/documents/products/datasheets/datasheet_truseq_sample_prep_kits.pdf
- 36. DeLuca DS, Levin JZ, Sivachenko A, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28:1530-1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Natoli T. CMap tools in R. Bioconductor. Published 2023. Accessed July 18, 2023. https://bioconductor.org/packages/release/bioc/html/cmapR.html
- 38. Carlson M. Genome wide annotation for human. Bioconductor. Published 2019. Accessed July 18, 2023. http://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html
- 39. Morgan M, Obenchain V, Hester J, Pagès H. SummarizedExperiment container. Bioconductor. Published 2023. Accessed July 18, 2023. https://bioconductor.org/packages/release/bioc/html/SummarizedExperiment.html
- 40. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol. 1995;57:289-300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 42. Kolde R. Pheatmap (version 1.0.12). RDocumentation. Published 2019. Accessed July 18, 2023. https://www.rdocumentation.org/packages/pheatmap/versions/1.0.12
- 43. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284-287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wu T, Hu E, Xu S, et al. ClusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reynolds KA, Rosa-Molinar E, Ward RE, Zhang H, Urbanowicz BR, Settles AM. Accelerating biological insight for understudied genes. Integr Comp Biol. 2021;61:2233-2243. doi: 10.1093/icb/icab029. [DOI] [PubMed] [Google Scholar]
- 46. The Gene Ontology Consortium. The gene ontology resource: enriching a gold mine. Nucleic Acids Res. 2021;49:D325-D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pedersen TL. ggplot2. RDocumentation. Published 2023. Accessed July 18, 2023. https://www.rdocumentation.org/packages/ggplot2/versions/3.4.2
- 48. Ulgen E, Ozisik O, Sezerman OU. Pathfindr: an R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front Genet. 2019;10:858. doi: 10.3389/fgene.2019.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chicco D, Agapito G. Nine quick tips for pathway enrichment analysis. PLoS Comput Biol. 2022;18:e1010348. doi: 10.1371/journal.pcbi.1010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. NC State University. Comparative toxicogenomics database. Published 2023. Accessed July 18, 2023. https://ctdbase.org/
- 51. Mancarcı O. geneSynonym. GitHub. Published 2023. Accessed July 18, 2023. https://github.com/oganm/geneSynonym
- 52. Smyth G. Limma (version 3.28.14). RDocumentation. Published June 2016. Accessed July 18, 2023. https://www.rdocumentation.org/packages/limma/versions/3.28.14
- 53. Swiss Institute of Bioinformatics. Gene expression data in animals. Published 2023. Accessed July 18, 2023. https://www.bgee.org/
- 54. Akhavanpoor M, Gleissner CA, Gorbatsch S, et al. CCL19 and CCL21 modulate the inflammatory milieu in atherosclerotic lesions. Drug Des Devel Ther. 2014;8:2359-2371. doi: 10.2147/dddt.S72394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lauran Stöger J, Boshuizen MCS, Brufau G, et al. Deleting myeloid IL-10 receptor signalling attenuates atherosclerosis in LDLR-/- mice by altering intestinal cholesterol fluxes. Thromb Haemost. 2016;116:565-577. doi: 10.1160/TH16-01. [DOI] [PubMed] [Google Scholar]
- 56. Shi G, Field DJ, Long X, et al. Platelet factor 4 mediates vascular smooth muscle cell injury responses. Blood. 2013;121:4417-4427. doi: 10.1182/blood-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sanders LN, Schoenhard JA, Saleh MA, et al. BMP antagonist Gremlin 2 limits inflammation after myocardial infarction. Circ Res. 2016;119:434-449. doi: 10.1161/circresaha.116.308700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Larson JL, Pino MV, Geiger LE, Simeone CR. The toxicity of repeated exposures to rolipram, a type iv phosphodiesterase inhibitor, in rats. Pharmacol Toxicol. 1996;78:44-49. doi: 10.1111/j.1600-0773.1996.tb00178.x. [DOI] [PubMed] [Google Scholar]
- 59. Dho SH, Lim JC, Kim LK. Beyond the role of CD55 as a complement component. Immune Netw. 2018;18:e11. doi: 10.4110/in.2018.18.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vestweber D. VE-cadherin. Arterioscler Thromb Vasc Biol. 2008;28:223-232. doi: 10.1161/ATVBAHA.107.158014. [DOI] [PubMed] [Google Scholar]
- 61. Neuber S, Mühmer M, Wratten D, Koch PJ, Moll R, Schmidt A. The desmosomal plaque proteins of the plakophilin family. Dermatol Res Pract. 2010;2010:101452. doi: 10.1155/2010/101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. van der Wal T, van Amerongen R. Walking the tight wire between cell adhesion and WNT signalling: a balancing act for β-catenin. Open Biol. 2020;10:200267. doi: 10.1098/rsob.200267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang H, Meng F, Wu S, et al. Engagement of I-branching {beta}-1, 6-N-acetylglucosaminyltransferase 2 in breast cancer metastasis and TGF-{beta} signaling. Cancer Res. 2011;71:4846-4856. doi: 10.1158/0008-5472.Can. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pronker MF, Lemstra S, Snijder J, et al. Structural basis of myelin-associated glycoprotein adhesion and signalling. Nat Commun. 2016;7:13584. doi: 10.1038/ncomms13584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Changede R, Sheetz M. Integrin and cadherin clusters: a robust way to organize adhesions for cell mechanics. Bioessays. 2017;39:1-12. doi: 10.1002/bies.201600123. [DOI] [PubMed] [Google Scholar]
- 66. Duong CN, Vestweber D. Mechanisms ensuring endothelial junction integrity beyond VE-Cadherin. Front Physiol. 2020;11:519. doi: 10.3389/fphys.2020.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhong X, Yang Y, Li B, et al. Downregulation of SLC27A6 by DNA hypermethylation promotes proliferation but suppresses metastasis of nasopharyngeal carcinoma through modulating lipid metabolism. Front Oncol. 2022;11:780410. doi: 10.3389/fonc.2021.780410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Khalil YA, Rabès J-P, Boileau C, Varret M. APOE gene variants in primary dyslipidemia. Atherosclerosis. 2021;328:11-22. doi: 10.1016/j.atherosclerosis.2021.05.007. [DOI] [PubMed] [Google Scholar]
- 69. Okoro EU, Zhao Y, Guo Z, Zhou L, Lin X, Yang H. Apolipoprotein E4 is deficient in inducing macrophage ABCA1 expression and stimulating the Sp1 signaling pathway. PLoS ONE. 2012;7:e44430. doi: 10.1371/journal.pone.0044430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tontonoz P, Nagy L, Alvarez JGA, Thomazy VA, Evans RM. PPARγ promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241-252. doi: 10.1016/S0092-8674(00)81575. [DOI] [PubMed] [Google Scholar]
- 71. Orsi FA, Lijfering WM, Van der Laarse A, et al. Association of apolipoproteins C-i, C-ii, C-iii and E with coagulation markers and venous thromboembolism risk. Clin Epidemiol. 2019;11:625-633. doi: 10.2147/clep.S196266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stouthard JML, Levi M, Hack CE, et al. Interleukin-6 stimulates coagulation, not fibrinolysis, in humans. Thromb Haemost. 1996;76:738-742. doi: 10.1055/s-0038. [DOI] [PubMed] [Google Scholar]
- 73. Kerr R, Stirling D, Ludlam CA. Interleukin 6 and haemostasis. Br J Haematol. 2001;115:3-12. doi: 10.1046/j.1365-2141.2001.03061.x. [DOI] [PubMed] [Google Scholar]
- 74. Liang Y, Fu Y, Qi R, et al. Cartilage oligomeric matrix protein is a natural inhibitor of thrombin. Blood. 2015;126:905-914. doi: 10.1182/blood-2015. [DOI] [PubMed] [Google Scholar]
- 75. Moroi AJ, Zwifelhofer NM, Riese MJ, Newman DK, Newman PJ. Diacylglycerol kinase ζ is a negative regulator of GPVI-mediated platelet activation. Blood Adv. 2019;3:1154-1166. doi: 10.1182/bloodadvances.2018026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bjurulf P. Atherosclerosis in different parts of the arterial system. Am Heart J. 1964;68:41-50. doi: 10.1016/0002-8703(64)90238. [DOI] [PubMed] [Google Scholar]
- 77. McGill HC, Jr, McMahan CA, Zieske AW, et al. Associations of coronary heart disease risk factors with the intermediate lesion of atherosclerosis in youth. Arterioscler Thromb Vasc Biol. 2000;20:1998-2004. doi: 10.1161/01.ATV.20.8.1998. [DOI] [PubMed] [Google Scholar]
- 78. Liappis N, Redel D, Kallfelz HC, Bantzer P. Verhalten der Glucose-Konzentration in fünf Gefässgebieten – Aorta, Arteria pulmonalis, Vena cava inferior, Vena cava superior, Vena hepatica – des kindlichen Organismus [Study of the glucose concentration in five vascular regions – aorta, arteria pulmonalis, vena cava inferior, vena cava superior, vena hepatica—in childhood (author’s transl)]. Klin Padiatr. 1979;191:385-389. [PubMed] [Google Scholar]
- 79. Gutierrez G, Venbrux A, Ignacio E, Reiner J, Chawla L, Desai A. The concentration of oxygen, lactate and glucose in the central veins, right heart, and pulmonary artery: a study in patients with pulmonary hypertension. Crit Care. 2007;11:R44. doi: 10.1186/cc5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hsieh PS, Moore MC, Neal DW, Cherrington AD. Importance of the hepatic arterial glucose level in generation of the portal signal in conscious dogs. Am J Physiol Endocrinol Metab. 2000;279:E284-E292. doi: 10.1152/ajpendo.2000.279.2.E284. [DOI] [PubMed] [Google Scholar]
- 81. Pauca AL, Wallenhaupt SL, Kon ND, Tucker WY. Does radial artery pressure accurately reflect aortic pressure? Chest. 1992;102:1193-1198. doi: 10.1378/chest.102.4.1193. [DOI] [PubMed] [Google Scholar]
- 82. McEniery CM, Cockcroft JR, Roman MJ, Franklin SS, Wilkinson IB. Central blood pressure: current evidence and clinical importance. Eur Heart J. 2014;35:1719-1725. doi: 10.1093/eurheartj/eht565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-tif-21-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-tif-22-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-tif-23-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-1-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-10-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-11-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-12-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-13-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-14-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-15-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-16-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-17-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-18-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-19-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-2-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-20-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-3-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-4-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-5-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-6-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-7-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-8-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights
Supplemental material, sj-xlsx-9-bbi-10.1177_11779322241251563 for Genes Differentially Expressed Across Major Arteries Are Enriched in Endothelial Dysfunction-Related Gene Sets: Implications for Relative Inter-artery Atherosclerosis Risk by Paul A Brown in Bioinformatics and Biology Insights