Abstract
Redundant TonB systems which function in iron transport from TonB-dependent ligands have recently been identified in several gram-negative bacteria. We demonstrate here that in addition to the previously described tonB locus, an alternative system exists for the utilization of iron from hemoglobin, transferrin, or lactoferrin in Neisseria meningitidis and Neisseria gonorrhoeae. Following incubation on media containing hemoglobin, N. meningitidis IR3436 (tonB exbB exbD deletion mutant) and N. gonorrhoeae PD3401 (tonB insertional mutant) give rise to colonies which can grow with hemoglobin. Transfer of Hb+ variants (PD3437 or PD3402) to media containing hemoglobin, transferrin, and/or lactoferrin as sole iron sources resulted in growth comparable to that observed for the wild-type strains. Transformation of N. meningitidis IR3436 or N. gonorrhoeae PD3401 with chromosomal DNA from the Hb+ variants yielded transformants capable of growth with hemoglobin. When we inactivated the TonB-dependent outer membrane hemoglobin receptors (HmbR or HpuB) in the Neisseria Hb+ variants, these strains could not grow with hemoglobin; however, growth was observed with transferrin and/or lactoferrin. These results demonstrate that accumulation of iron from hemoglobin, transferrin, and lactoferrin in the pathogenic neisseriae can occur via a system that is independent of the previously described tonB locus.
Acquisition of iron from transferrin, lactoferrin, heme, and heme-containing compounds in the pathogenic Neisseria spp. involves a family of distinct iron-regulated outer membrane receptors (1, 3, 5, 7–11, 14–16, 20, 21). The iron-repressible hemoglobin receptor HmbR of Neisseria meningitidis (20, 21) and HpuB of N. meningitidis and N. gonorrhoeae (7, 15, 16) are required for binding and utilization of hemoglobin or hemoglobin bound to haptoglobin, respectively. A second gene (hpuA), predicted to encode a lipoprotein, is found upstream of hpuB, and recent studies have demonstrated that both HpuA and HpuB are required for the utilization of hemoglobin in the pathogenic Neisseria spp. (7, 16). An N. gonorrhoeae hmbR homolog has been identified; however, the presence of a stop codon in the coding sequence in the gonococcal hmbR gene indicates that N. gonorrhoeae does not produce a functional HmbR (7, 21).
Energy for the transport of iron from host iron-binding proteins across the outer membrane into the periplasmic space in a number of gram-negative organisms is provided by TonB, in association with the ExbB and ExbD proteins (6, 12). The TonB system uses the proton motive force of the cytoplasmic membrane for the passage of ligands into the periplasm. Receptors which require energy supplied via the TonB system are termed TonB dependent and have amino acid homology in several regions termed TonB boxes. The TonB box represents the domain of the bacterial receptor which physically interacts with the energy-transducing protein TonB. The Neisseria HmbR and HpuB hemoglobin receptors have homology to TonB-dependent outer membrane receptors (7, 20), and N. meningitidis and N. gonorrhoeae tonB, exbB, and exbD homologs have been identified (2, 22). N. meningitidis and N. gonorrhoeae tonB, exbB, and exbD insertional mutants were observed to grow with hemin; however, these mutants were reported to be deficient in the ability to utilize hemoglobin, transferrin, and/or lactoferrin (2, 22).
It was recently reported that in Vibrio cholerae, a second TonB ExbB ExbD system is operative, and this system was demonstrated to functionally replace the TonB1 system for the acquisition of TonB-dependent ligands associated with the accumulation of iron (17). Likewise, in V. parahaemolyticus, V. alginolyticus, Serratia marcescens, Yersinia pestis, and Pseudomonas aeruginosa, two tonB genes have been identified (13, 18, 24). Based on these recent reports, we have examined a newly constructed N. meningitidis tonB exbB exbD deletion mutant and an N. gonorrhoeae tonB insertional mutant for the ability to grow with heme, hemoglobin, transferrin, and/or lactoferrin as sole iron sources. We have demonstrated that in addition to the previously described tonB locus, an alternative system is operative in N. meningitidis and N. gonorrhoeae for the utilization of iron from hemoglobin, transferrin, and/or lactoferrin. We have clearly demonstrated that this system can function with the TonB-dependent receptors HmbR in N. meningitidis and HpuB in N. gonorrhoeae for growth with hemoglobin.
Bacterial strains and growth conditions.
N. meningitidis strain IR1072 is a serogroup C clinical isolate which is hmbR hpuB. Attempts to PCR amplify the hpuB gene in IR1072 failed; however, we were able to amplify the hpuB gene from IR1075 using the same primers (data not shown). In addition, strain IR1072 is unable to grow with hemoglobin complexed to haptoglobin (20, 21). N. meningitidis IR3436 is a tonB exbB exbD deletion mutant constructed following transformation of strain IR1072 with a plasmid construct in which the tonB exbB exbD locus was deleted and replaced with the spectinomycin gene (I. Stojiljkovic, unpublished data). Genomic DNA isolated from IR3436 was used to transform N. meningitidis MC58, the serogroup B clinical isolate which has been fully sequenced, generating strain MC5801, an additional tonB exbB exbD deletion mutant. The N. gonorrhoeae tonB insertional mutant was constructed by transformation (4) of N. gonorrhoeae 340 with N. meningitidis IR2200 chromosomal DNA (Table 1). N. gonorrhoeae strain 340 does not utilize lactoferrin as an iron source. N. meningitidis or N. gonorrhoeae strains were typically maintained on gonococcal base (GCB) medium containing 1% Kellogg supplement and incubated aerobically (5% CO2) at 37°C for 16 to 18 h (9). When necessary, kanamycin (50 μg/ml), erythromycin (1.5 μg/ml), or spectinomycin (100 μg/ml) was used.
TABLE 1.
Bacterial strains used in this study
| Strain | Genotypic and phenotypic description | Reference or source |
|---|---|---|
| N. meningitidis | ||
| IR1072 | Clinical isolate, serogroup C (hmbR+ hpuB) | 20 |
| IR3436 | IR1072 but with tonB exbB exbD deletion | Stojiljkovic, unpublished |
| PD3437 | IR3436 variant which grows with hemoglobin, transferrin, and lactoferrin | This study |
| PD3438 | PD3437 but with hmbR::Kmr | This study |
| PD3439 | IR3436 transformed with genomic DNA from PD3437 | This study |
| IR1075 | Clinical isolate, serogroup C (hmbR+ hpuB+) | 21 |
| IR2200 | IR1075 but with tonB::Kmr | 22 |
| IR1098 | IR1072 but with hmbR::Kmr | 22 |
| MC58 | Clinical isolate, serogroup B (hmbR+ hpuB+) | Lab collection |
| MC5801 | MC58 but with tonB exbB exbD deletion | This study |
| N. gonorrhoeae | ||
| 340 | Wild type (hmbR+ hpuB+), but HmbR phenotypically not functional | Lab collection |
| PD3401 | 340 but with tonB::Kmr | This study |
| PD3402 | PD3401 variant which grows with hemoglobin and transferrin | This study |
| PD3403 | PD3402 but with hpuB::Ermr | This study |
| PD340E | 340 but with hpuB::Ermr | Lab collection |
Characterization of Neisseria tonB mutants.
The tonB mutation in N. meningitidis and N. gonorrhoeae strains was confirmed by either colony or genomic PCR analysis. The deletion of the tonB exbB exbD genes in N. meningitidis strains IR3436, PD3437, and PD3439, as well as in strain MC5801, was confirmed using primers del-A and del-D, designed to amplify the entire tonB exbB exbD locus and 400 bp of flanking DNA upstream of tonB gene and downstream of the exbD gene (Table 2). To confirm the disruption of the tonB gene in the N. gonorrhoeae insertional mutant, primers tonB-F2 and tonB-R1 were designed to amplify homologous regions between the 5′ end of the gene, upstream of the ATG codon, and the 3′ end of the gene, upstream of the stop codon. Briefly, a single colony was suspended in 100 μl of distilled H2O and incubated at 80 to 100°C for 5 min. A 10-μl volume of this crude preparation was then used for colony PCR analysis. Growth of N. meningitidis and N. gonorrhoeae strains was examined in a plate assay or in broth as described in the figure legends.
TABLE 2.
Oligonucleotide primers for PCR confirmation of N. meningitidis and N. gonorrhoeae tonB, hmbR, and hpuBmutations
| Primer | Sequence | Wild-type product | Mutant product |
|---|---|---|---|
| del-A del-D | 5′ GCGATCCGGTCTCAGTGGGTATAGC 3′ 5′ GGCTCGAGCGATATGGGGCAGGGGGTTCA 3′ | tonB, exbB, exbD genes and 400 nta of flanking DNA upstream of the tonB gene and downstream of the exbD gene (2.8-kb fragment) | 3.0-kb fragment corresponding to the omega cassette (2.2 kb) with 400 nt of flanking DNA |
| tonB-F2 tonB-R1 | 5′ TGCTATTATTATGCCCGAATT 3′ 5′ TTTTCACGATTTTAACGGAC 3′ | In N. meningitidis, F2 binds nt 101 upstream and R1 anneals to nt 724 downstream of the putative +1 initiation start codon (ATG) of the tonB gene, producing a 825-bp fragment | tonB gene with a kanamycin cassette insertion, producing a fragment of 2.03 kb |
| In N. gonorrhoeae, F2 anneals to nt 21 upstream and R2 anneals to nt 739 downstream of the putative +1 initiation start codon (ATG) of the tonB gene, producing a 760-bp fragment | tonB gene with a kanamycin cassette insertion, producing a fragment of 1.96 kb | ||
| hmbR-F1 hmbR-R1 | 5′ AACAAGAAATGATACGCGAC 3′ 5′ AGAGGTTGGTGTTACGCCGT 3′ | F1 anneals to nt 184 and R1 anneals to nt 928 downstream of the putative +1 initiation start codon (ATG) of the hmbR gene, producing a 744-bp fragment | hmbR gene with a kanamycin cassette insertion, producing a fragment of 1.94 kb |
| hpuB-F3 hpuB-R2 | 5′ TCAAACCCGTATTGGCTG 3′ 5′ GCACATCAGGAGCTTAGA 3′ | F3 anneals to nt 11 upstream of the putative +1 initiation start codon (ATG). R2 anneals to nt 2 upstream of the putative stop codon (TAA) of the hpuB gene, producing a 2.39-kb fragment | hpuB gene with a erythromycin cassette insertion, producing a fragment of 3.69 kb |
nt, nucleotides.
Construction of N. meningitidis hmbR and N. gonorrhoeae hpuB mutants.
N. meningitidis hmbR and N. gonorrhoeae hpuB insertional mutants were constructed in N. meningitidis strain PD3437 and N. gonorrhoeae strain PD3402, respectively, by transformation as previously described (4). To disrupt the hpuB gene, N. gonorrhoeae strain PD3402 was transformed using chromosomal DNA (1.0 μg) from N. gonorrhoeae strain PD340E (Table 1) and transformants were selected on GCB plates containing erythromycin (1.5 μg/ml) and kanamycin (2 μg/ml). To disrupt the hmbR gene, N. meningitidis strain PD3437 was transformed using chromosomal DNA (1.0 μg) from N. meningitidis strain IR1098 (Table 1) and transformants were selected on GCB plates containing kanamycin (25 μg/ml) and spectinomycin (50 μg/ml). The inactivation of hmbR and hpuB genes in strains PD3437 and PD3402 was confirmed by PCR (data not shown).
Utilization of hemoglobin, transferrin, and lactoferrin is not absolutely dependent on the tonB locus in the pathogenic Neisseria spp.
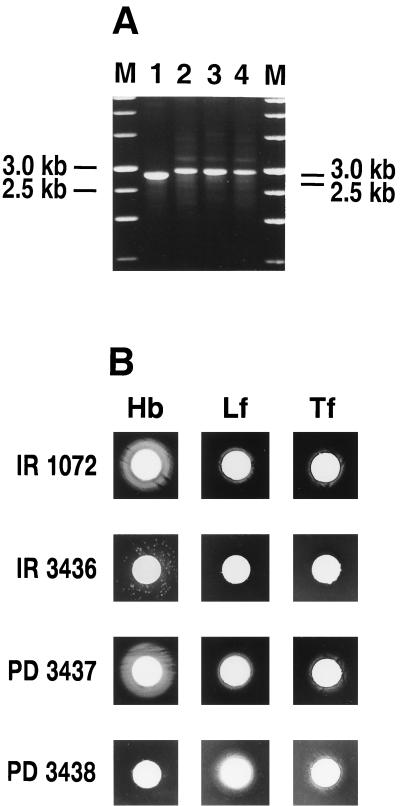
To determine if an alternate system independent of the previously described tonB locus could function in hemoglobin utilization in N. meningitidis, we examined the ability of an N. meningitidis tonB exbB exbD deletion (IR3436) mutant to grow with different iron sources. Both the wild-type and tonB N. meningitidis strains were capable of growth with Fe or heme as sole iron sources (data not shown). Discrete colonies around hemoglobin disks were observed with N. meningitidis strains IR3436 (tonB exbB exbD deletion) (Fig. 1B) and MC5801 (tonB exbB exbD deletion) (data not shown). Furthermore, when a single colony of N. meningitidis IR3436 isolated from around a hemoglobin disk was transferred to fresh GCB plates supplemented with desferal, spectinomycin, and hemoglobin and incubated for 18 h, confluent growth was observed (data not shown). We designated this hemoglobin-utilizing variant strain PD3437. When PD3437 was examined for the ability to use other iron sources in a plate assay, growth comparable to that of the wild-type strain IR1072 around disks containing hemoglobin, transferrin, or lactoferrin was observed (Fig. 1B). Furthermore, we also examined the growth of N. meningitidis strain PD3437 in liquid culture with hemoglobin or hemin as a sole iron source. For these studies, hemoglobin was supplemented with human serum albumin. In agreement with the results of the plate assays, we found that with hemoglobin as the sole iron source, N. meningitidis strain PD3437 grew at levels comparable to the original wild-type strain IR1072 (data not shown).
FIG. 1.
(A) The tonB exbB exbD deletion in N. meningitidis strains IR3436, PD3437, and PD3439 was confirmed by PCR. The tonB gene was amplified by colony PCR using N. meningitidis primers specific for the tonB gene (Table 2). Lanes: M, molecular size standard; 1, IR1072; 2, IR3436; 3, PD3437; 4, PD3438. Molecular sizes are indicated on the left; those on the right correspond to the approximate sizes of the expected amplified products. (B) Growth of N. meningitidis IR3436, PD3437, and PD3438 with various iron sources. A 10-μl volume of human hemoglobin (Hb) (10 mg/ml) or an iron-saturated human transferrin (Tf) (50 mg/ml) or lactoferrin (Lf) (25 mg/ml) was used. These solutions were applied to paper disks and placed onto the GCB-desferal (60 μM) plates seeded with bacteria (108 CFU). Strain IR1072 is the wild-type strain from which IR3436 and PD3437 were derived. Growth was recorded after 18 to 36 h; results are from one experiment and are representative of three separate experiments.
The tonB exbB exbD deletion in N. meningitidis strains IR3436 and PD3437 was confirmed by PCR analysis. As expected, we amplified a 2.8-kb fragment in the wild-type strain IR1072 and a 3.0-kb fragment in the tonB exbB exbD deletion mutants, strains IR3436 and PD3437 (Fig. 1A). Taken together, these results indicate that a stable change allowing the acquisition of iron from hemoglobin, transferrin, and lactoferrin had occurred and suggest that a locus independent of tonB exbB exbD can be utilized for growth with TonB-dependent ligands. Such a phenotype would be expected if a silent homolog of the tonB locus was present in N. meningitidis.
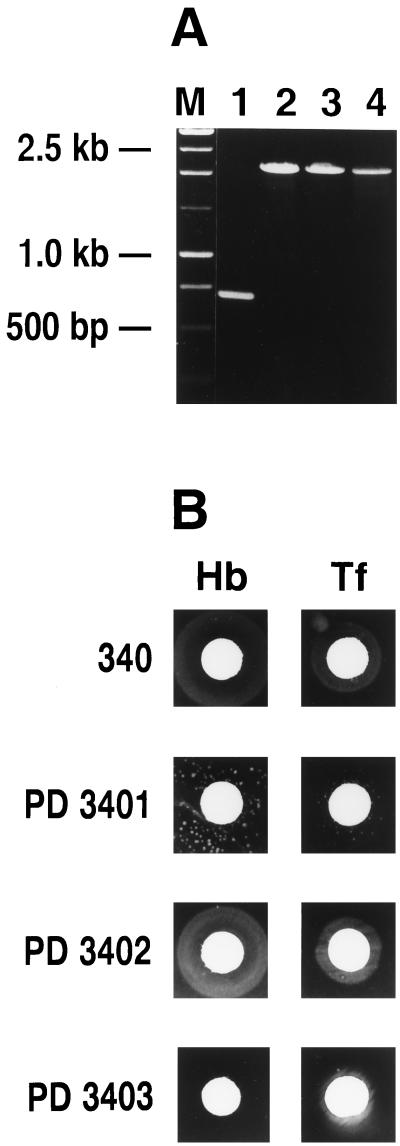
Using genomic DNA from the N. meningitidis tonB mutant (IR2200), we constructed a gonococcal tonB mutant in strain 340 and demonstrated that this strain (PD3401) grew with heme and ferric nitrate as sole iron sources (data not shown). As observed with the N. meningitidis tonB deletion mutant, we also observed colonies around disks containing hemoglobin and transferrin with strain PD3401 (Fig. 2B). We further characterized a hemoglobin-utilizing variant of strain PD3401 and designated this variant PD3402. When PD3402 colonies were transferred to GCB plates supplemented with desferal, kanamycin, and hemoglobin, confluent growth was observed (Fig. 2B). Furthermore, when PD3402 was transferred to media containing disks with various iron sources, growth was observed around the transferrin and hemoglobin disks, and this was comparable to the growth observed for the wild-type strain (strain 340) (Fig. 2B). To confirm that the hemoglobin utilization phenotype was not due to a reversion of the tonB mutation, we amplified the tonB gene from strains 340, PD3401, and PD3402 by PCR using primers specific for the tonB gene. As expected, the wild-type tonB gene was amplified as a ∼760-bp fragment from strain 340 whereas a ∼2.0-kb fragment was amplified in strains PD3401 and PD3402 (Fig. 2A). This was also confirmed by Southern blot hybridization analysis using the tonB gene as a probe; one intact copy of the tonB gene was observed in strain 340, and one copy of the disrupted tonB gene was observed in PD3401 and PD3402 (data not shown).
FIG. 2.
(A) Insertional inactivation of the tonB gene in N. gonorrhoeae strains PD3401, PD3402, and PD3404, was confirmed by PCR as described in the legend to Fig. 1. Lanes: M, molecular size standard; 1, 340; 2, PD3401; 3, PD3402; 4, PD3403. Molecular sizes are indicated on the left. (B) Growth of N. gonorrhoeae PD3401, PD3402, and PD3403 with various iron sources. Strains were examined for their ability to utilize various iron sources in a plate assay as described in the legend to Fig. 1. N. gonorrhoeae strain 340 is the wild-type strain from which strains PD3401 and PD3402 were derived. Growth was recorded after 18 to 36 h; results are from one experiment and are representative of three separate experiments. Hb, hemoglobin; Tf, transferrin.
Utilization of hemoglobin independently of the tonB locus is dependent on the N. meningitidis HmbR and N. gonorrhoeae HpuB hemoglobin receptors.
We next inactivated the hemoglobin receptors HmbR in N. meningitidis PD3437 and HpuB in N. gonorrhoeae PD3402. N. meningitidis strain IR3436, from which strain PD3437 was derived, is naturally hpuB, and thus inactivation of hmbR in this strain should result in a strain unable to grow with hemoglobin as a sole iron source. Likewise, since the hmbR locus is not expressed in N. gonorrhoeae (7, 21), the inactivation of the hpuB gene in N. gonorrhoeae strain PD3402 should result in a strain unable to grow with hemoglobin as the sole iron source. If the ability to utilize hemoglobin independently of the tonB locus was due to the removal of heme independent of HmbR or HpuB and subsequent transport into the cell via a heme acquisition pathway, we would expect that hmbR hpuB strains should still utilize heme from hemoglobin for growth, since hemin transport in N. meningitidis and N. gonorrhoeae occurs via an HmbR- and HpuB-independent mechanism (16, 20). The inactivation of hmbR or hpuB in these strains was confirmed by PCR. As expected, we observed a ∼744-bp PCR fragment using primers specific to an internal fragment of the hmbR gene in N. meningitidis strain PD3437 (data not shown). A PCR fragment of ∼1.9 kb was observed in the N. meningitidis hmbR insertional mutant strain PD3438, corresponding to the insertion of the kanamycin cassette. Likewise, using primers specific for the N. gonorrhoeae hpuB gene, we observed a ∼2.4-kb fragment in N. gonorrhoeae strain PD3402 (data not shown). A PCR fragment of ∼3.7 kb was observed in N. gonorrhoeae strain PD3403, corresponding to the insertion of the erythromycin cassette into this strain (data not shown).
N. meningitidis PD3438 and N. gonorrhoeae PD3403 exhibited normal growth with heme (data not shown), transferrin, and/or lactoferrin compared to the wild-type strains IR1072 and 340 (Fig. 1B and 2B). However, we did not observe any growth of strain PD3438 or PD3403 with hemoglobin as the sole iron source (Fig. 1B and 2B). These results clearly indicate that the ability to utilize hemoglobin independently of the above-described tonB locus in N. meningitidis and N. gonorrhoeae does not result from the removal of heme and subsequent transport into the cell via a heme acquisition pathway. These results also indicate that in N. meningitidis and N. gonorrhoeae a second TonB system must function with HmbR or HpuB for the transport of iron from hemoglobin. Furthermore, our observations suggest that the putative second tonB locus may be silent, and expression can be detected only when the first tonB locus is inactive.
The ability to utilize hemoglobin independently of the tonB locus can be genetically transformed into N. meningitidis IR3436 or N. gonorrhoeae PD3401.
The frequency of the appearance of N. meningitidis hemoglobin-utilizing (Hb+) variants for strain IR3436 was determined to be 1.37 × 10−7. Likewise, the frequency of the appearance of N. gonorrhoeae Hb+ variants for strain PD3401 was similar to that observed for N. meningitidis (2.9 × 10−7). To determine if the ability to utilize hemoglobin independently of the tonB locus was encoded by a gene which was now functional in the Hb+ variant (strain PD3437), we transformed N. meningitidis IR3436 with chromosomal DNA isolated from strain PD3437. Transformation of N. meningitidis strain IR3436 with chromosomal DNA from strain PD3437 yielded transformants capable of growth with hemoglobin at a frequency of 1.07 × 10−4. We randomly picked Hb+ transformants and transferred these colonies to media containing disks with various iron sources; for all Hb+ transformants tested, growth was observed around hemoglobin-, transferrin-, and lactoferrin-containing disks and was comparable to that observed for strain PD3437 (data not shown). The deletion of the tonB exbB exbD genes in N. meningitidis strain PD3439 was confirmed by PCR analysis (Fig. 1A).
Likewise, we obtained similar results when we transformed N. gonorrhoeae strain PD3401 with chromosomal DNA isolated from the Hb+ variant (strain PD3402). Transformation of N. gonorrhoeae strain PD3401 with chromosomal DNA isolated from strain PD3402 yielded transformants capable of growth with hemoglobin at a frequency of 4.9 × 10−3. We also randomly picked N. gonorrhoeae Hb+ transformants and transferred these colonies to media containing disks with various iron sources. As observed for N. meningitidis PD3439, we observed proficient growth of strain PD3404 around transferrin- and hemoglobin-containing disks. We also confirmed the tonB mutation in strain PD3404 by PCR analysis (Fig. 2A). These results indicate that the ability to utilize hemoglobin independently of the previously described tonB locus is encoded by a gene which is functional and stable in N. meningitidis strain PD3437 and N. gonorrhoeae strain PD3402.
Concluding remarks.
The results obtained in this study indicate that the previously described tonB locus is not solely responsible for the utilization of hemoglobin, transferrin, or lactoferrin in the pathogenic Neisseria spp. Based on these results, we postulate that N. meningitidis and N. gonorrhoeae possess a second, alternative TonB system for the utilization of iron from TonB-dependent ligands. Our results also indicate that in N. meningitidis and N. gonorrhoeae the second TonB system clearly functions with HmbR or HpuB for the acquisition of iron from hemoglobin. We found that the frequency of colonies which could grow with hemoglobin and transferrin was relatively low. Indeed, this may explain why, in previous studies (2, 22), these colonies either were not observed or appeared at a relatively low frequency (depending on the duration of incubation) and therefore were not reported. In addition, differences in the growth assays, the media used for plate assays (complex versus defined), the concentration of the various iron sources, or the concentration of organisms used may explain this discrepancy. In our studies we utilized plate assays with 100 μg of hemoglobin per disk (1.5 μM final concentration), whereas Stojiljkovic and Srinivasan (22) used plate assays with 50 μg of hemoglobin per disk (0.75 μM final concentration). In studies reported by Biswas et al. (2), the ability of gonococcal tonB exbB exbD mutants to utilize various iron sources was examined by streaking colonies on CDM plates containing various iron sources with a final hemoglobin concentration of 1 μM. Our observations suggest that the putative second tonB locus may be silent and that expression can be detected only when the first tonB locus is inactive. Furthermore, the ability to obtain stable Hb+ transformants following transformation with chromosomal DNA from the N. meningitidis or N. gonorrhoeae Hb+ variants (PD3437 or PD3402 respectively) indicates that, once this silent second tonB locus is expressed, its expression is stable.
In summary, we propose that the pathogenic Neisseria spp. possess a second TonB system which functions to transduce energy to TonB-dependent outer membrane receptors. Examination of the gonococcal and recently completed meningococcal database for Neisseria tonB homologs did not reveal any open reading frames with significant homology to any previously described tonB locus. This is not an unexpected finding, given that V. cholerae tonB2 has only 23% homology with Neisseria tonB. Similarly, P. aeruginosa tonB2 demonstrates 28% homology to P. aeruginosa tonB. Attempts to find conserved regions in both tonB proteins among these species were unsuccessful. We have constructed a genomic library from the hemoglobin-utilizing variant N. meningitidis PD3437, and studies are currently in progress to identify the second Neisseria tonB locus by complementation analysis.
Acknowledgments
This study was supported by Public Health Service grant AI30797 from the National Institute of Allergy and Infectious Diseases.
We thank Igor Stojiljkovic for providing N. meningitidis strains.
REFERENCES
- 1.Biswas G D, Anderson J E, Chen C-J, Cornelissen C N, Sparling P F. Identification and functional characterization of the Neisseria gonorrhoeae lbpB gene product. Infect Immun. 1999;67:455–459. doi: 10.1128/iai.67.1.455-459.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biswas G D, Anderson J E, Sparling P F. Cloning and characterization of Neisseria gonorrhoeae tonB, exbB and exbD genes. Mol Microbiol. 1997;24:169–179. doi: 10.1046/j.1365-2958.1997.3421692.x. [DOI] [PubMed] [Google Scholar]
- 3.Biswas G D, Sparling P F. Characterization of lbpA, the structural gene for a lactoferrin receptor in Neisseria gonorrhoeae. Infect Immun. 1995;63:2958–2967. doi: 10.1128/iai.63.8.2958-2967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas G D, Lacks S A, Sparling P F. Transformation-deficient mutants of piliated Neisseria gonorrhoeae. J Bacteriol. 1989;171:657–664. doi: 10.1128/jb.171.2.657-664.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnah R A, Schryvers A B. Preparation and characterization of Neisseria meningitidis mutants deficient in production of the human lactoferrin-binding proteins LbpA and LbpB. J Bacteriol. 1998;180:3080–3090. doi: 10.1128/jb.180.12.3080-3090.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun V. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD dependent receptor proteins. FEMS Microbiol Rev. 1995;16:295–307. doi: 10.1111/j.1574-6976.1995.tb00177.x. [DOI] [PubMed] [Google Scholar]
- 7.Chen C J, Sparling P F, Lewis L A, Dyer D W, Elkins C. Identification and purification of a hemoglobin-binding outer membrane protein from Neisseria gonorrhoeae. Infect Immun. 1996;64:5008–5014. doi: 10.1128/iai.64.12.5008-5014.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornelissen C N, Sparling P F. Binding and surface exposure characteristics of the gonococcal transferrin receptor are dependent on both transferrin-binding proteins. J Bacteriol. 1996;178:1437–1444. doi: 10.1128/jb.178.5.1437-1444.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai P J, Nzeribe R, Genco C A. Binding and accumulation of hemin in Neisseria gonorrhoeae. Infect Immun. 1995;63:4634–4641. doi: 10.1128/iai.63.12.4634-4641.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genco C A, Desai P J. Iron acquisition in the pathogenic neisseria. Trends Microbiol. 1996;4:179–184. doi: 10.1016/0966-842x(96)10029-9. [DOI] [PubMed] [Google Scholar]
- 11.Gray-Owen S D, Schryvers A B. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 1996;4:185–191. doi: 10.1016/0966-842x(96)10025-1. [DOI] [PubMed] [Google Scholar]
- 12.Larsen R A, Myers P S, Skare J T, Seachord C L, Darveau R P, Postle K. Identification of TonB homologs in the family Enterobacteriaceae and evidence for conservation of TonB-dependent energy transduction complexes. J Bacteriol. 1996;178:1363–1373. doi: 10.1128/jb.178.5.1363-1373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Letoffe S, Binet R, Paquelin A, Arnoux P, Szjzek M, Ghigo J M, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C. World Congress on Iron Metabolism: BIOIRON 99. 1999. Hemophore dependent heme acquisition systems in gram negative bacteria; p. 118. [Google Scholar]
- 14.Lewis L A, Rohde K, Gipson M, Behrens B, Gray E, Toth S I, Roe B A, Dyer D W. Identification and molecular analysis of lbpBA, which encodes the two-component meningococcal lactoferrin receptor. Infect Immun. 1998;66:3017. doi: 10.1128/iai.66.6.3017-3023.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lewis L A, Dyer D W. Identification of an iron-regulated outer membrane protein of Neisseria meningitidis involved in the utilization of hemoglobin complexed to haptoglobin. J Bacteriol. 1995;177:1299–1306. doi: 10.1128/jb.177.5.1299-1306.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis L A, Gray E, Wang Y-P, Roe B A, Dyer D W. Molecular characterization of hpuAB, the haemoglobin-haptoglobin utilization operon of Neisseria meningitidis. Mol Microbiol. 1997;23:737–749. doi: 10.1046/j.1365-2958.1997.2501619.x. [DOI] [PubMed] [Google Scholar]
- 17.Occhino D A, Wyckoff E E, Henderson D P, Wrona T J, Payne S M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 18.O'Malley S M, Mouton S L, Occhino D A, Deanda M T, Rashidi J R, Fuson K L, Rashidi C E, Mora M Y, Payne S M, Henderson D P. Comparison of the heme iron utilization systems of pathogenic vibrios. J Bacteriol. 1999;181:3594–3598. doi: 10.1128/jb.181.11.3594-3598.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto B R, Verweij-van Vught A M J J, MacLaren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 20.Stojiljkovic I, Hwa V, de Saint Martin L, O'Gaora P, Nassif X, Heffron F, So M. The Neisseria meningitidis haemoglobin receptor: its role in iron utilization and virulence. Mol Microbiol. 1995;15:531–541. doi: 10.1111/j.1365-2958.1995.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 21.Stojiljkovic I, Larson J, Hwa V, Anic S, So M. HmbR outer membrane receptors of pathogenic Neisseria spp.: iron-regulated, hemoglobin-binding proteins with a high level of primary structure conservation. J Bacteriol. 1996;178:4670–4678. doi: 10.1128/jb.178.15.4670-4678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stojiljkovic I, Shrinivasan N. Neisseria meningitidis tonB, exbB, and exbD genes: Ton-dependent utilization of protein bound iron in neisseriae. J Bacteriol. 1997;179:805–812. doi: 10.1128/jb.179.3.805-812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Q, Poole K. A second tonB gene in Pseudomonas aeruginosa is linked to the exbB and exbD genes. FEMS Microbiol Lett. 2000;184:127–132. doi: 10.1111/j.1574-6968.2000.tb09002.x. [DOI] [PubMed] [Google Scholar]