Abstract
Acetyl-CoA synthetase short-chain family member 1 (ACSS1) uses acetate to generate mitochondrial acetyl-CoA and is regulated by deacetylation by sirtuin 3. We generated an ACSS1-acetylation (Ac) mimic mouse, where lysine-635 was mutated to glutamine (K635Q). Male Acss1K635Q/K635Q mice were smaller with higher metabolic rate and blood acetate and decreased liver/serum ATP and lactate levels. After a 48-hour fast, Acss1K635Q/K635Q mice presented hypothermia and liver aberrations, including enlargement, discoloration, lipid droplet accumulation, and microsteatosis, consistent with nonalcoholic fatty liver disease (NAFLD). RNA sequencing analysis suggested dysregulation of fatty acid metabolism, cellular senescence, and hepatic steatosis networks, consistent with NAFLD. Fasted Acss1K635Q/K635Q mouse livers showed increased fatty acid synthase (FASN) and stearoyl-CoA desaturase 1 (SCD1), both associated with NAFLD, and increased carbohydrate response element–binding protein binding to Fasn and Scd1 enhancer regions. Last, liver lipidomics showed elevated ceramide, lysophosphatidylethanolamine, and lysophosphatidylcholine, all associated with NAFLD. Thus, we propose that ACSS1-K635-Ac dysregulation leads to aberrant lipid metabolism, cellular senescence, and NAFLD.
ACSS1–lysine-635 acetylation mimic knock-in mice exhibit disrupted lipid metabolism and acute NAFLD upon fasting.
INTRODUCTION
Acetyl–coenzyme A (acetyl-CoA) is essential for converting various fuel sources involved in cellular energy metabolism or bioenergetics, lipid synthesis, and epigenetic regulation (1, 2). Most acetyl-CoA in mammalian cells is generated through glycolysis, fatty acid oxidation (FAO), and, in specific organs, branched-chain amino acid oxidation (3, 4). Acetyl-CoA synthetase short-chain family member 1 (ACSS1; previously known as AceCS2) catalyzes the ligation of acetate and CoA to generate acetyl-CoA in the mitochondrial matrix, enabling the utilization of acetate for energy during conditions of nutrient deprivation (5). This metabolic flexibility or switch can ensure survival but, when dysregulated, can contribute to cellular dysfunction, including altered responses to metabolic and oxidative stress (6, 7).
Acetate is an important metabolic molecule that provides a key substrate for the generation of adenosine 5′-triphosphate (ATP) as well as lipogenesis in the liver (8). Acetate can also function as a small-molecule metabolic sensor that allows the cell and, importantly, the mitochondria to respond to nutrient and oxidative stress through both the regulation of gene expression and posttranslational modifications (PTMs) (1). In this regard, a seminal study showed that lysine-635–Ac (acetylation) can regulate the enzymatic activity of ACSS1 (9). Furthermore, it is well established that acetylation is a primary mitochondrial PTM that regulates multiple proteins, many of which are important for the maintenance of metabolic/bioenergetic balance (10, 11). These factors place acetate, ACSS1, and acetylation at the center of many mitochondrial processes that regulate metabolism and bioenergetics.
Sirtuin 3 (SIRT3) is the primary mitochondrial deacetylase that we, along with others, have shown to be activated by caloric restriction, fasting, and oxidative stress, as well as playing a role in oncogenicity (12–17). ACSS1 enzymatic activity is regulated, at least in part, through a single lysine amino acid at K635 in mice and K642 in humans (5, 18, 19). While acetylation has been shown to inhibit or decrease the ligase activity of ACSS1 in vitro (5), the physiological function of ACSS1 acetylation status in vivo and its role in cellular metabolism and mitochondrial bioenergetics have not been fully characterized.
RESULTS
Characterization of Acss1K635Q/K635Q mouse model
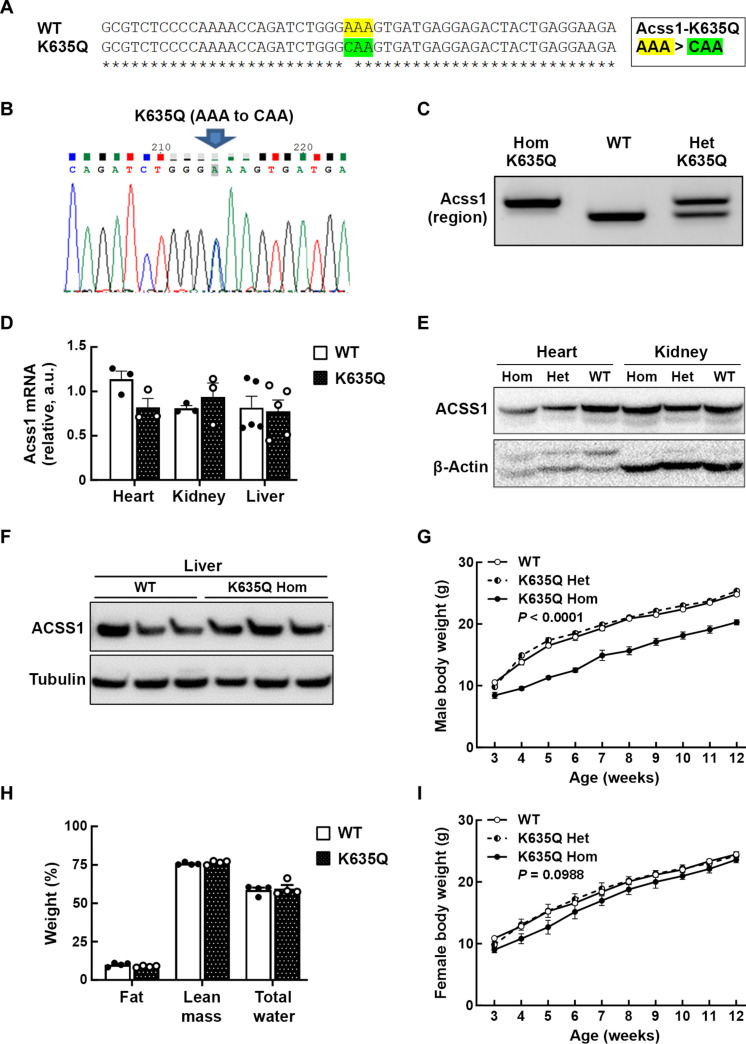
To investigate the role of ACSS1 acetylation in vivo, a knock-in mouse model was generated where the lysine (K) at amino acid position 635 was substituted with glutamine (Q) by changing the codon sequence from AAA to CAA (Fig. 1A) (see figs. S1 and S2, A to C, and Materials and Methods for details). This lysine-to-glutamine mutation has been shown to functionally and biochemically mimic lysine acetylation in ACSS1 (5). The Acss1K635Q mouse was first generated as a conditional knock-in model and sequenced to confirm the K635Q codon substitution (Fig. 1B). We crossed this conditional knock-in mouse with the Sox2Cre line to produce the full-body constitutive Acss1K635Q mouse used in this study. The line is maintained as heterozygous with wild-type Acss1 and followed by polymerase chain reaction (PCR) (Fig. 1C). We confirmed that Acss1K635Q/K635Q mice showed equivalent levels of Acss1 expression to the wild-type genotype, as measured by quantitative PCR (qPCR) (Fig. 1D), and equivalent ACSS1 protein levels by immunoblot in the heart, kidney (Fig. 1E), and liver (Fig. 1F).
Fig. 1. Characterization of ACSS1-K635Q mouse model.
(A) DNA sequence for the mice that were genetically altered to express the K635-Ac mimic mutant Acss1 gene by replacing lysine (AAA) at codon location 635 to glutamine (CAA). WT, wild type. (B) Genomic DNA was extracted from a hemizygous founder mouse and sequenced to confirm the AAA to CAA substitution at codon 635. (C) PCR product sizes of the target region of Acss1 confirm the genotypes of Acss1K635Q/K635Q, Acss1+/+ (WT), and heterozygous Acss1K635Q/+ mice. Hom, homozygous; Het, heterozygous. (D) RNA was isolated from Acss1+/+ (WT) and Acss1K635Q/K635Q (Hom) heart, kidney, and liver tissues, and Acss1 mRNA expression levels were quantified by qPCR. a.u., arbitrary units. (E) Immunoblot showing ACSS1 in heart and kidney extracts from Acss1+/+ (WT), Acss1K635Q/+ (Het), and Acss1K635Q/K635Q (Hom) mice. (F) Immunoblot showing ACSS1 in liver extracts from wild-type and Acss1K635Q/K635Q mice. (G) The body weights of male Acss1+/+, Acss1K635Q/+, and Acss1K635Q/K635Q mice were measured from 3 weeks of age until 12 weeks (N = 5 mice per group). Exact P value shown, calculated by two-way analysis of variance (ANOVA). (H) qMRI showing fat, lean mass, and total water as percentage of total body weight in 3-month-old male Acss1K635Q/K635Q mice versus wild-type mice (N = 4 mice per group). (I) The body weights of female Acss1+/+, Acss1K635Q/+, and Acss1K635Q/K635Q mice were measured from 3 weeks of age until 12 weeks (N = 5 mice per group). Exact P value shown, calculated by two-way ANOVA.
By 3 weeks after birth, it became apparent that male Acss1K635Q/K635Q mice were smaller than either heterozygous or wild-type mice, and they did not catch up over time (Fig. 1G). We confirmed that this size difference was not due to altered food consumption (fig. S2D) or absorption, as measured by fecal lipids (fig. S2E). Analysis of 3-month-old mice by quantitative magnetic resonance imaging (qMRI) showed that male Acss1K635Q/K635Q mice were smaller overall but had comparable body composition to wild-type mice, with similar percentages of fat, lean mass, and total water (Fig. 1H). While female Acss1K635Q/K635Q mice displayed reduced/delayed fertility, we observed no difference in weight among the genotypes (Fig. 1I). Since males seemed to exhibit greater sensitivity to Acss1 genotype, based on their smaller size (fig. S2F), we used male mice for the rest of this study.
Acss1K635Q alters metabolism
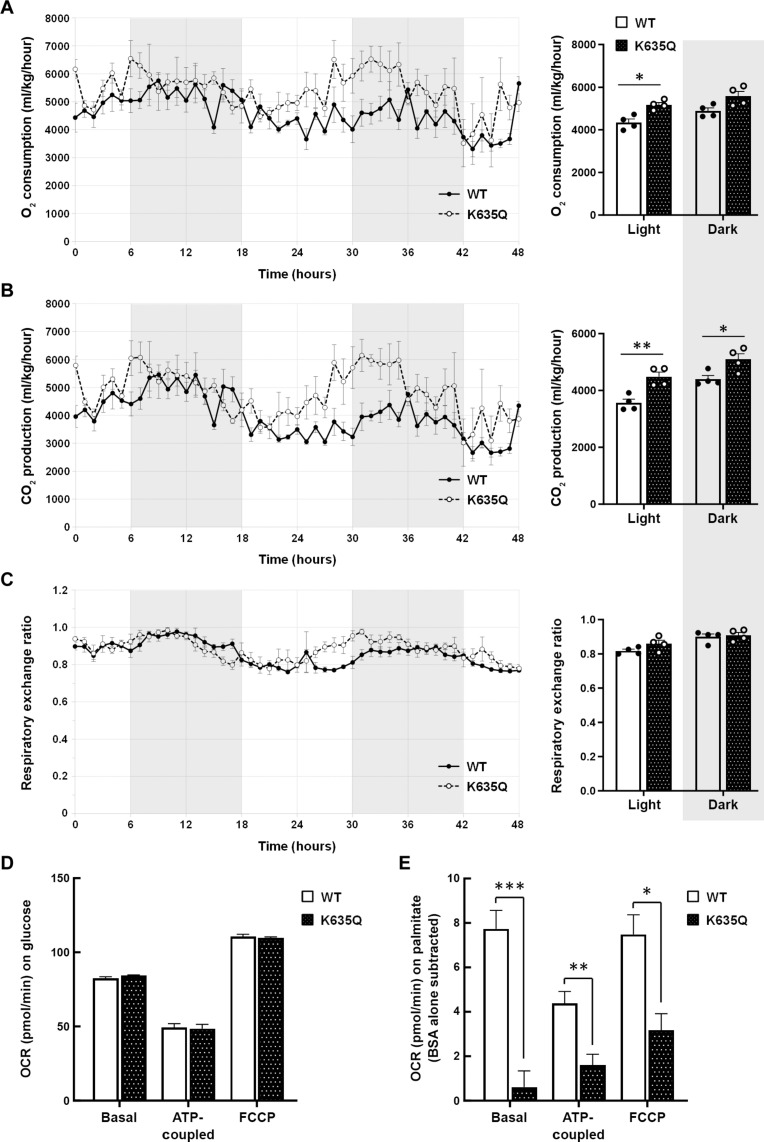
We performed indirect calorimetry studies on individually housed 3-month-old wild-type and Acss1K635Q/K635Q mice to assess whole-body metabolism. When normalized to lean body mass, the Acss1K635Q/K635Q mice exhibited higher oxygen consumption (Fig. 2A) and carbon dioxide production (Fig. 2B) than did wild-type mice. There was no difference between the respiratory exchange ratio (RER) of the genotypes, suggesting that there was not a preferential metabolic switch between fat and carbohydrate usage (Fig. 2C). We also put wild-type and Acss1K635Q/K635Q mice on a 3-week high-fat diet (HFD) and, again, found no difference in RER between the genotypes (fig. S3A).
Fig. 2. ACSS1-K635Q alters metabolism.
(A to C) Wild-type and Acss1K635Q/K635Q mice were individually housed to measure (A) oxygen consumption, (B) carbon dioxide production, and (C) RER during light and dark cycles (N = 4 mice per group). Data are normalized to lean body mass and represented as mean ± SEM. *P < 0.05, **P < 0.01 calculated by one-way ANOVA. (D) Basal, ATP-coupled, and FCCP-uncoupled OCRs in wild-type and Acss1K635Q/K635Q primary fibroblasts with 0.5 mM glucose (N = 2 mice per group, four to six technical replicates each). (E) Basal, ATP-coupled, and FCCP-uncoupled OCR in wild-type and Acss1K635Q/K635Q primary fibroblasts with 0.5 μM carnitine and 100 μM BSA-conjugated palmitate (N = 3 mice per group, four technical replicates each), as compared to BSA alone (OCR on BSA was subtracted from BSA-conjugated palmitate). Data are represented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, calculated by two-tailed unpaired Student’s t tests.
To further evaluate changes in metabolism, we examined mitochondrial function. We isolated primary hepatocytes and fibroblasts from wild-type and Acss1K635Q/K635Q mice and measured oxygen consumption rate (OCR) with Seahorse. Under standard glucose conditions, Acss1K635Q/K635Q hepatocytes had lower basal, ATP-coupled, and maximal (uncoupled) OCR than wild-type hepatocytes (Fig. 2D), suggesting a deficiency in mitochondrial respiration. However, primary fibroblasts in the same media showed no difference between the genotypes (fig. S3B) but did show a difference when palmitate was used as the primary substrate. Under these conditions, wild-type fibroblasts had a higher OCR than Acss1K635Q/K635Q as compared to bovine serum albumin (BSA) controls (Fig. 2E). Finally, we used qPCR to examine expression levels of genes involved in mitochondrial lipid uptake and metabolism in the primary hepatocytes (fig. S3C). These experiments suggest metabolic deficiencies in the Acss1K635Q/K635Q cells, including aberrant importation or utilization of palmitate to generate ATP.
Metabolic dysregulation in fasted Acss1K635Q/K635Q mice
It has been previously shown that, under conditions of fasting or other nutrient stress, acetate is an important energy source that is used to generate ATP (20, 21). Thus, we were interested in exploring how fasting would affect metabolism in mice expressing the ACSS1 constitutive acetylation mimic. Since 48 hours of fasting has previously been shown to ensure a switch to using stored triglycerides to fuel FAO and ketogenesis (22), experiments were done at this time point.
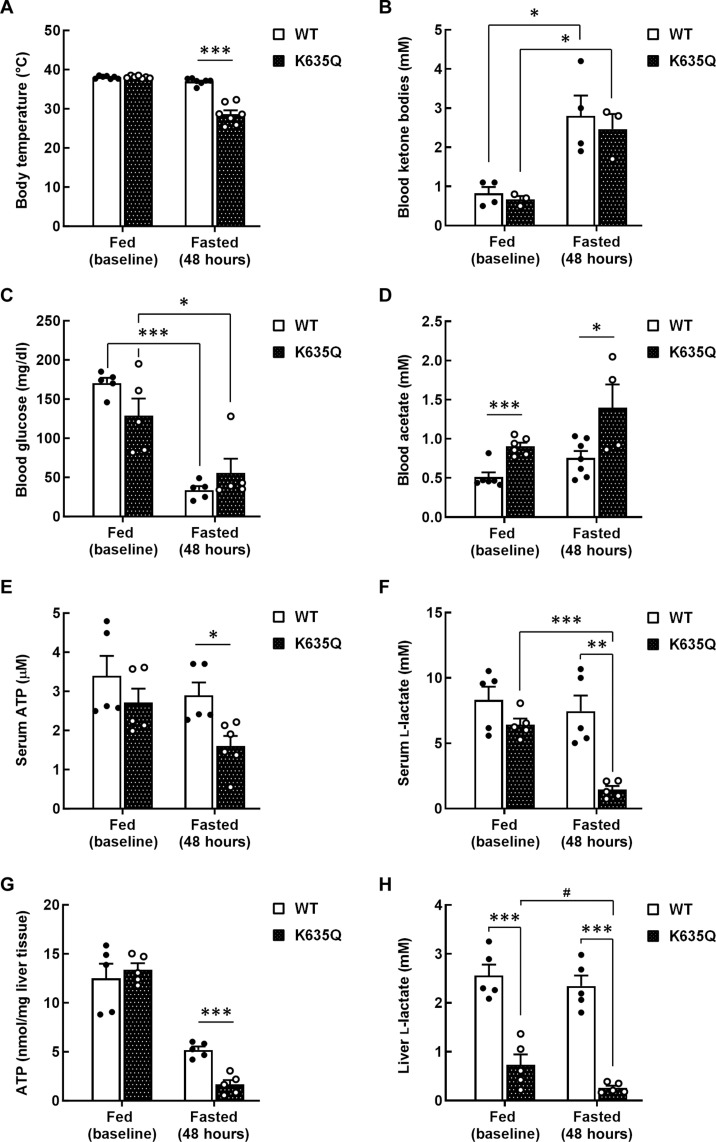
Both wild-type and Acss1K635Q/K635Q mice lost 20 to 25% percent of their body weight on a 48-hour fast (fig. S4A). However, Acss1K635Q/K635Q mice displayed hypothermia (Fig. 3A) and a general failure-to-thrive phenotype, many requiring euthanization. To identify any changes in metabolism, we initially examined blood/serum metabolites. In this regard, both wild-type and Acss1K635Q/K635Q mice exhibited a similar and significant increase in blood ketone body levels (Fig. 3B), as expected following a 48-hour fast. Both genotypes also exhibited similar baseline blood glucose levels and the expected decrease with fasting (Fig. 3C). While fasting increased blood acetate levels in wild-type mice, these remained significantly lower than those in Acss1K635Q/K635Q mice under both fed and fasted conditions (Fig. 3D). In the Acss1K635Q/K635Q mice, fasting also led to a significant reduction in both ATP (Fig. 3E) and lactate levels (Fig. 3F). While Acss1K635Q/K635Q mice displayed lower serum acylcarnitine levels than wild-type mice at baseline, there was no overall difference between the genotypes in the fasted state (fig. S4B). The increase in acetate, which was also observed in the Acss1 knockout mice (23), and the decrease in ATP seem consistent with dysregulation of ACSS1.
Fig. 3. Metabolic dysregulation in fasted Acss1K635Q/K635Q mice.
(A) Body temperature of fed and 48-hour–fasted wild-type and Acss1K635Q/K635Q mice. (B to F) Serum extracted from wild-type and Acss1K635Q/K635Q mice at baseline (fed) and after a 48-hour fast was measured for (B) ketones, (C) glucose, (D) acetate, (E) ATP, and (F) l-lactate. (G and H) Liver tissue isolated from fed and fasted wild-type and Acss1K635Q/K635Q mice was used to measure (G) ATP and (H) l-lactate levels. Data are represented as mean ± SEM. #P < 0.08, *P < 0.05, **P < 0.01, ***P < 0.001, calculated by two-tailed unpaired Student’s t tests.
We also examined metabolites in liver tissue from Acss1K635Q/K635Q and wild-type mice. While ATP levels were identical in both genotypes at baseline, a much greater decrease was observed in the Acss1K635Q/K635Q mice than in wild-type mice following the 48-hour fast (Fig. 3G). Lactate levels were unchanged in the wild-type mice with fasting, while the Acss1K635Q/K635Q mice exhibited decreased levels even at baseline and a significant reduction with fasting (Fig. 3H). Serum levels of both alanine and glutamine were significantly lower in Acss1K635Q/K635Q mice than in wild-type mice but were unchanged by fasting (fig. S4, C and D). In contrast to serum, baseline levels of liver acylcarnitine were similar in wild-type and Acss1K635Q/K635Q mice and did not show a significant increase upon fasting (fig. S4E). Finally, while acetyl-CoA was significantly reduced in the livers of wild-type mice after fasting, we found no change in Acss1K635Q/K635Q mice (fig. S4F). These results suggest aberrant mitochondrial bioenergetics in Acss1K635Q/K635Q mice that is exacerbated by fasting. Specifically, the decreased ATP and lactate levels along with increased acylcarnitine suggest dysregulation of fatty acid metabolism.
We ran high-resolution respirometry on fresh liver tissue dissected from fed and fasted wild-type and Acss1K636Q/K635Q mice to assess fatty acid–supported respiration (fig. 4SH). Specifically, we measured LEAK respiration [measure of inefficiency stimulated by palmitoylcarnitine but in the absence of adenosine 5′-diphosphate (ADP)], coupled respiration stimulated by palmitoylcarnitine alone or palmitoylcarnitine and octanoylcarnitine, and uncoupled respiration stimulated by palmitoylcarnitine and octanoylcarnitine, with and without succinate. Although we did not find any differences between the groups, the fasted Acss1K636Q/K635Q mice tended to have lower OCR on average, which might suggest that Acss1K636Q/K635Q mice are less able to adapt to conditions of food depravation.
Liver microsteatosis in fasted Acss1K635Q/K635Q mice
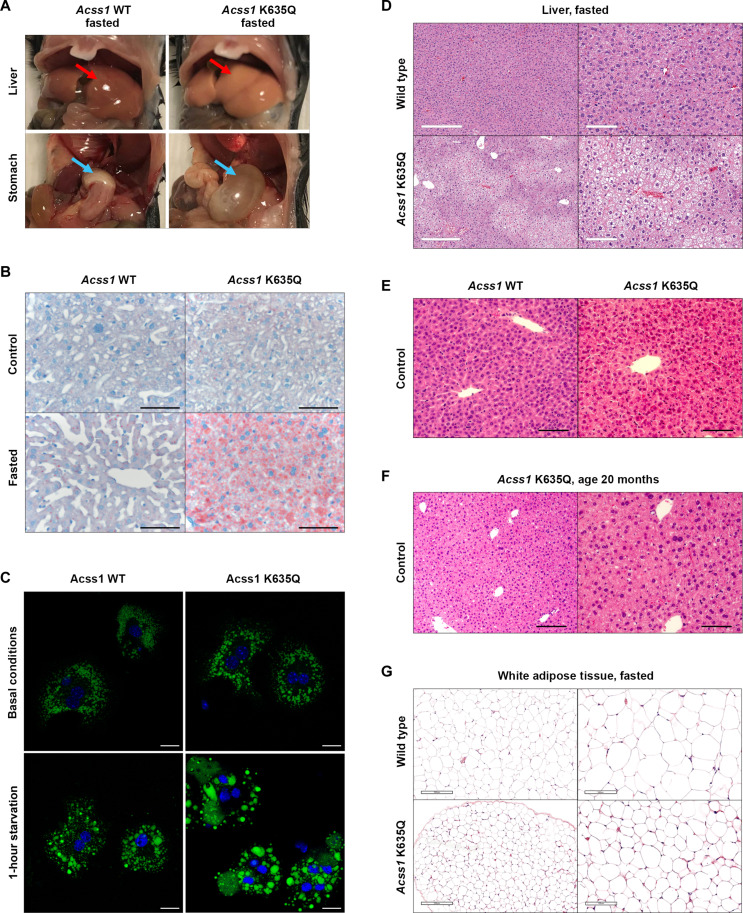
Examination of the abdominal cavity revealed that the livers of fasted Acss1K635Q/K635Q mice were noticeably larger and had numerous white and patchy areas suggestive of excess fat deposition, as commonly observed in a fatty liver phenotype (Fig. 4A, upper panels). In addition, the stomachs of the fasted Acss1K635Q/K635Q mice were enlarged and appeared bloated (Fig. 4A, lower panels). Histological examination of the livers of fasted Acss1K635Q/K635Q mice further revealed microsteatosis, the accumulation of fat-containing vacuoles within hepatocytes, which is a key histologic feature of nonalcoholic fatty liver disease (NAFLD) (Fig. 4B, upper panels versus lower panels). Oil Red O staining showed a notable increase in lipid droplets in the livers of fasted Acss1K635Q/K635Q compared to the fasted control mice, indicated by the areas of deep red staining (Fig. 4B, bottom right panel).
Fig. 4. Liver microsteatosis and fatty liver in fasted Acss1K635Q/K635Q mice.
(A) Images of liver and stomach from fasted wild-type and Acss1K635Q/K635Q mice (see arrows). (B) Oil Red O staining of liver from fed and fasted wild-type and Acss1K635Q/K635Q mice. Scale bars, 100 μm. (C) Lipid droplet staining in primary hepatocytes from wild-type and Acss1K635Q/K635Q mice under basal conditions (top) and after 1-hour starvation (bottom). Scale bars, 20 μm. (D to F) H&E staining of liver from (D) fasted wild-type and Acss1K635Q/K635Q mice [scale bars, 300 μm (left) and 100 μm (right)], (E) fed wild-type and Acss1K635Q/K635Q mice (scale bars, 50 μm), and (F) 20-month-old Acss1K635Q/K635Q mice [scale bars, 100 μm (left) and 50 μm (right)]. (G) H&E staining of white adipose tissue from fasted wild-type and Acss1K635Q/K635Q mice. Scale bars, 200 μm (left) and 100 μm (right).
We saw a similar phenotype in primary Acss1K635Q/K635Q hepatocytes. After 1-hour starvation, Acss1K635Q/K635Q hepatocytes showed a significant increase in the percentage of large lipid droplets (>1.5 μm2) and a decrease in small lipid droplets (<0.8 μm2) compared to either basal conditions (Fig. 4C, lower panels) or starved wild-type hepatocytes (Fig. 4C, right panels) (see fig. S4G for quantification). The increased lipid accumulation in fasted Acss1K635Q/K635Q mouse livers is also shown by hematoxylin and eosin (H&E) staining (Fig. 4D) and is not seen in Acss1K635Q/K635Q mice on normal diet (Fig. 4E), even when the mice are aged to 20 months (Fig. 4F). Notably, this acute NAFLD phenotype and the lipid droplet accumulation were not observed in the Acss1 knockout mice (23).
In addition, the white adipose tissue also appeared smaller with a more irregular border in the fasted Acss1K635Q/K635Q mice than in the fasted control mice (Fig. 4G, upper panels versus lower panels), while no differences were seen in brown adipose tissue, muscle, intestine, or testes (fig. S5). These biochemical and histological assays suggest that ACSS1 is critical to maintaining cellular metabolism and bioenergetics, leading to abnormal pathology in two critical bioenergetic organs in response to nutrient status, specifically in a fasting state.
Dysregulated cell senescence in Acss1K635Q/K635Q liver
To further investigate a potential mechanism for the development of acute NAFLD with fasting, we performed RNA sequencing (RNA-seq) analysis on liver tissue from fasted Acss1K635Q/K635Q and wild-type mice. Sequence reads were mapped to the UCSC/mm9 mouse genome using the HiSAT2 aligner, and quantification was carried out via StringTie. Differential expression analysis was done using the DESeq R package, and the following significance criteria were applied to the resulting gene list: Fragments Per Kilobase of transcript per Million mapped reads (FPKM) > 1, adjusted P value < 0.05, and fold change > 2. The differentially expressed genes were then processed using the Qiagen Ingenuity Pathway Analysis (IPA) software to determine enrichment and significance scores.
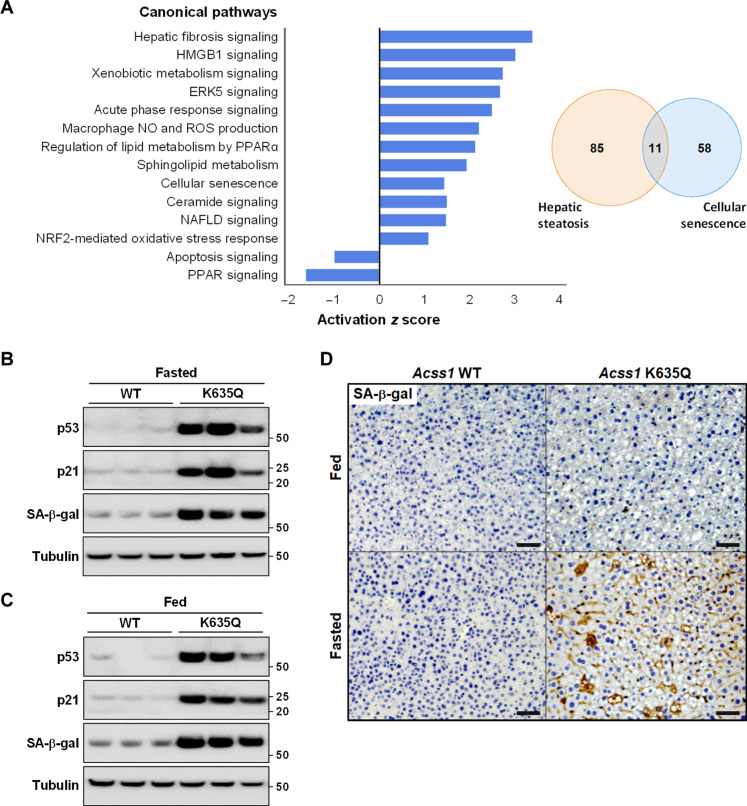
The RNA-seq analysis showed altered expression in multiple pathways involved in metabolism and stress response (Fig. 5A). This included altered expression of genes involved in (i) hepatic steatosis (fig. S6A), (ii) NAFLD (fig. S6B), (iii) cellular senescence (fig. S6C), and (iv) fatty acid metabolism (fig. S6D) in the fasted Acss1K635Q/K635Q mice, as compared to fasted controls. It has been established that, using both animal models and human samples, cellular senescence in the liver can be an early event in the development of a liver phenotype permissive to liver steatosis as well as subsequent progression to NAFLD (24). In this regard, a number of differentially regulated genes in our dataset are implicated in both cellular senescence and hepatic steatosis (Fig. 5A, Venn diagram).
Fig. 5. Dysregulated cell senescence and fatty acid metabolism in Acss1K635Q/K635Q liver.
(A) Total RNAs were isolated from the liver tissues of wild-type and Acss1K635Q/K635Q mice after a 48-hour fast and sent for RNA-seq (N = 3 mice per group). Differentially expressed genes were processed using the Qiagen Ingenuity Pathway Analysis (IPA) software to determine pathway enrichment and significance scores. Bar graph shows the top five differentially regulated Ingenuity Canonical Pathways by P value as well as select differentially regulated pathways involved in metabolism and stress response. Venn diagram shows overlap between differentially regulated genes involved in hepatic steatosis and cellular senescence. (B and C) Immunoblots showing relative p53, p21, and SA-β-gal levels in liver tissue from (B) fasted and (C) fed wild-type and Acss1K635Q/K635Q mice. (D) IHC staining for SA-β-gal in liver tissue from fed and fasted wild-type and Acss1K635Q/K635Q mice. Scale bars, 100 μm.
We examined liver tissue from fed and fasted wild-type and Acss1K635Q/K635Q mice for markers of cell senescence, including the senescence-associated isoform of β-galactosidase (SA-β-gal). Unexpectedly, we found elevated SA-β-gal not only in fasted Acss1K635Q/K635Q mice as compared to wild-type mice (Fig. 5B) but also in fed Acss1K635Q/K635Q mice (Fig. 5C and fig. S7A). We also observed a substantial increase in SA-β-gal by immunohistochemistry (IHC) staining in the livers of fasted Acss1K635Q/K635Q mice (Fig. 5D). In addition to SA-β-gal, we found increased levels of p21 (which can induce cell senescence) and p53 (transcriptional regulator of p21) (Fig. 5, B and C, and fig. S7A). These results suggest that Acss1K635Q/K635Q hepatocytes may exhibit preexisting cellular stress, priming them for metabolic dysfunction such as when further stressed by fasting (24).
In light of the evidence of metabolic dysregulation, microsteatosis, and increased cellular senescence in livers of fasted Acss1K635Q/K635Q mice, we expected to find elevated biomarkers of liver damage, specifically, aspartate transaminase (AST) or alanine transaminase (ALT). Unexpectedly, we did not find increased levels of either enzyme or any difference in the AST/ALT ratio in the sera of fasted Acss1K635Q/K635Q mice compared to fasted controls (fig. S7, B to D).
Dysregulated fatty acid synthesis in Acss1K635Q/K635Q liver
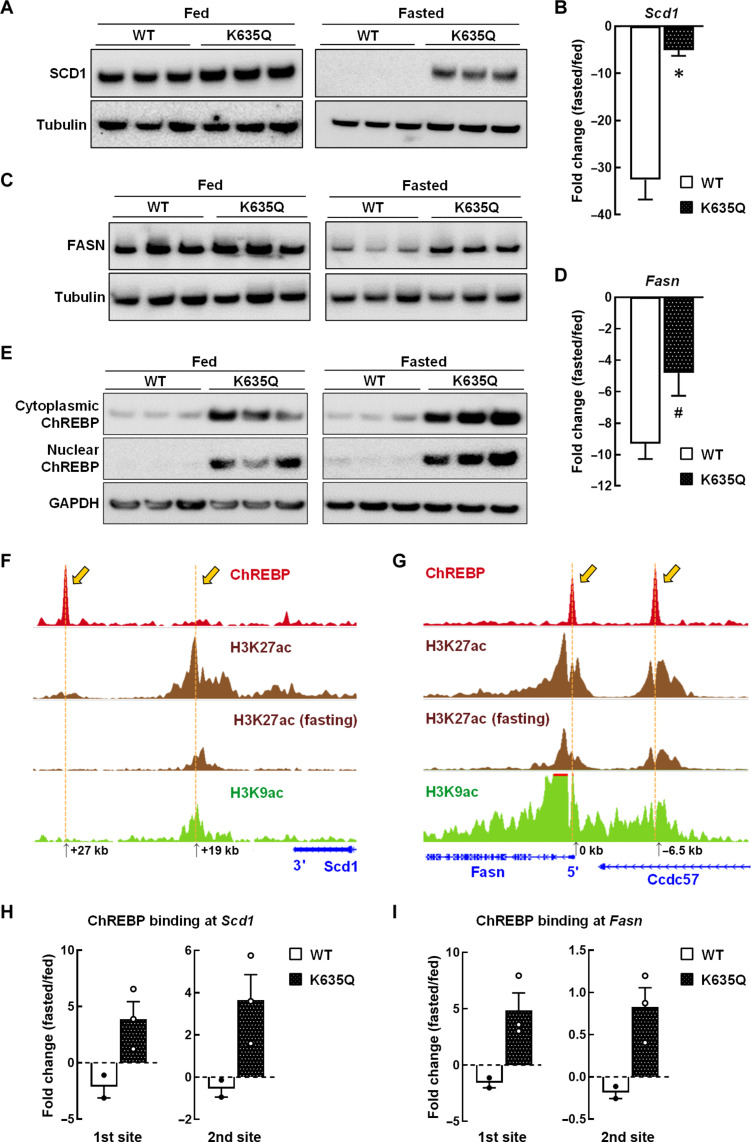
Next, we sought to further delineate the altered fatty acid metabolism gene expression identified in fasted Acss1K635Q/K635Q mice. Fatty acid synthesis pathway enzymes, including stearoyl-CoA desaturase (SCD1) (25, 26) and fatty acid synthase (FASN) (27, 28), are normally down-regulated during fasting as the liver switches to breaking down stored fat for the body to use as fuel. However, these enzymes are elevated in NAFLD murine models and patient samples (29). As expected, analysis of livers from fasted control mice showed nearly undetectable levels of SCD1 (Fig. 6A and fig. S7E) and significantly reduced transcription of Scd1 (Fig. 6B and fig. S7F). We found similar reductions in FASN at both protein (Fig. 6C and fig. S7G) and transcription (Fig. 6D and fig. S7H) levels. In contrast, fasted Acss1K635Q/K635Q mice maintained significantly higher levels of both SCD1 (Fig. 6A and fig. S7E) and FASN (Fig. 6C and fig. S7G). These aberrant protein levels were due, in part, to altered transcription as qPCR analysis showed the expected decrease in Scd1 and Fasn mRNA in fasted control mice, while fasted Acss1K635Q/K635Q mice showed a minimal decrease in either Scd1 (Fig. 6B and fig. S7F) or Fasn (Fig. 6D and fig. S7H). These data are consistent with previous publications showing that liver-specific inhibition of SCD1 and FASN attenuates the development of hepatic steatosis (30). By extension, the aberrant expression of these enzymes can lead to a liver phenotype permissive for lipid droplet accumulation and the development of steatosis and NAFLD.
Fig. 6. Dysregulated fatty acid synthesis in Acss1K635Q/K635Q liver.
(A) Immunoblots showing relative SCD1 in liver extracts from fed (left panel) and fasted (right panel) wild-type and Acss1K635Q/K635Q mice. (B) Scd1 was quantified by qPCR in liver from fed and fasted wild-type and Acss1K635Q/K635Q mice (N = 3 mice per group). Data are presented as mean fold change ± SEM. *P < 0.05, calculated by two-tailed unpaired Student’s t tests. (C) Immunoblots showing relative FASN in liver extracts from fed (left panel) and fasted (right panel) wild-type and Acss1K635Q/K635Q mice. (D) Fasn was quantified by qPCR in liver from fed and fasted wild-type and Acss1K635Q/K635Q mice (N = 3 mice per group). Data are presented as mean fold change ± SEM. #P < 0.08, calculated by two-tailed unpaired Student’s t tests. (E) Immunoblots showing relative ChREBP in cytoplasmic and nuclear subcellular fractions from livers of fed (left panel) and fasted (right panel) wild-type and Acss1K635Q/K635Q mice. (F and G) GEO datasets identify ChREBP DNA binding sites in (F) Scd1 and (G) Fasn in liver tissue (accession ID GSM6730577). Sites of H3K27ac, with and without fasting, and H3K9ac are shown. The Scd1 and Fasn gene locations are in blue. (H and I) ChIP assay using an anti-ChREBP antibody and primers overlapping the DNA binding sites in (H) Scd1 and (I) Fasn with liver extracts from fed and fasted wild-type and Acss1K635Q/K635Q mice. Data are represented as mean fold change ± SEM.
ChREBP binding to Scd1 and Fasn enhancer regions increases with fasting
Carbohydrate response element–binding protein (ChREBP) is expressed in lipogenic organs, such as the liver, and when dysregulated can contribute to the development of dyslipidemia and NAFLD, including through the transcriptional regulation of Scd1 and Fasn expression (31, 32). Immunoblot analysis showed significantly higher levels of ChREBP in both the cytoplasmic and nuclear subcellular fractions from livers of Acss1K635Q/K635Q mice, as compared to wild-type controls (Fig. 6E, left panel, and fig. S8, A and B). In addition, a further increase in ChREBP immunoreactive levels was observed in Acss1K635Q/K635Q mice after 48 hours of fasting, as compared to baseline levels in fed Acss1K635Q/K635Q mice (Fig. 6E, right panel, and fig. S8, A and B).
Since an increase in ChREBP was observed, we used the Gene Expression Omnibus (GEO) datasets to identify two potential DNA binding sites in the 3′ and 5′ regulatory regions of Scd1 (Fig. 6F) and Fasn (Fig. 6G), respectively (accession ID GSM6730577). This analysis identified strong ChREBP enrichment peaks at the upstream promoter region of Fasn and a downstream enhancer region of Scd1. The identification of these potential DNA binding sites was subsequently used to make primers (see Materials and Methods for primer sequence) that covered these two regulatory/binding site regions of ChREBP for each gene.
Next, we performed chromatin immunoprecipitation (ChIP) assay with ChREBP with these primers from liver extracts of wild-type and Acss1K635Q/K635Q mice on both normal diet and after fasting. The ChIP data showed a decrease in ChREBP enrichment at both sites in fasted wild-type mice for Scd1 (Fig. 6H and fig. S8, C and D) and Fasn (Fig. 6I and fig. S8, E and F). In contrast, an increase in ChREBP enrichment was observed in fasted Acss1K635Q/K635Q mice at both Scd1 (Fig. 6H and fig. S8, C and D) and Fasn (Fig. 6I and fig. S8, E and F). Regulation is likely through altered metabolites, as pulldown experiments showed no interaction between ACSS1 and ChREBP (fig. S8G), as expected. These experiments match the immunoblot data showing that the protein levels of both SCD1 and FASN remain elevated, instead of the expected decrease in the liver that is normally observed during fasting. The dysregulation of these two key proteins is consistent with publications showing that aberrant expression of these lipid enzymes can strongly contribute to NAFLD (33–35).
Liver lipidomics
On the basis of the observations of lipid accumulation and dysregulated fatty acid synthesis in the livers of fasted Acss1K635Q/K635Q mice, we measured free fatty acid (FFA) levels in the serum (fig. S9A) and triacylglycerols (TAGs) in the serum (fig. S9B) and liver (fig. S9C) in both fed and fasted wild-type and Acss1K635Q/K635Q mice. While we did see increased serum FFA and liver TAGs in Acss1K635Q/K635Q at baseline, unexpectedly, we did not find any differences between genotypes in the fasted state.
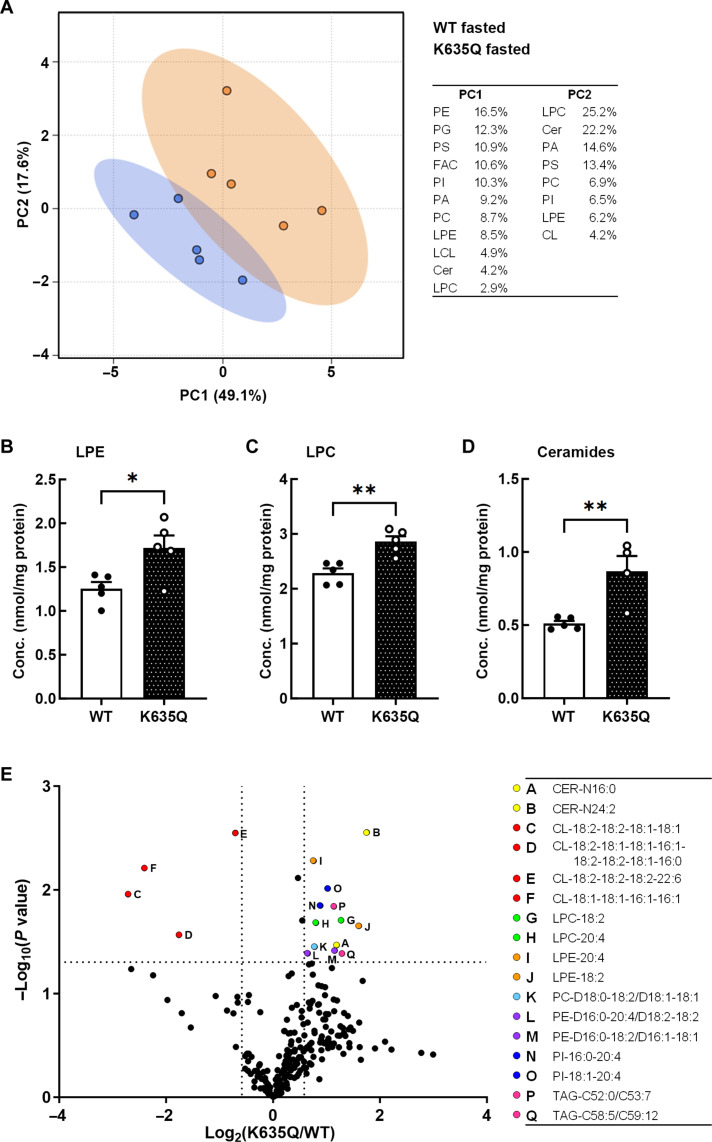
To find alterations in specific lipid classes or species, we performed a lipidomics analysis on liver samples collected from wild-type and Acss1K635Q/K635Q mice, both fed and after a 48-hour fast (see the Supplementary Materials for complete results). Principal components analysis showed that the greatest difference between fasted wild-type and Acss1K635Q/K635Q mice was in lysophosphatidylcholine (LPC), ceramide, and several classes of phospholipids (Fig. 7A and fig. S9, D and E). Three lipid classes were found to be significantly more abundant in fasted Acss1K635Q/K635Q mice, as compared to wild-type, lysophosphatidylethanolamine (LPE), LPC, and ceramides (Fig. 7, B to D). All three have been shown to play a role in aberrant liver lipid metabolism and NAFLD (36).
Fig. 7. Liver lipidomics comparing fasted wild-type and Acss1K635Q/K635Q mice.
(A) Principal components analysis (N = 5 mice per group). (B to D) LPE (B), LPC (C), and ceramides (D) are up-regulated 1.4-fold, 1.3-fold, and 1.7-fold, respectively, in fasted Acss1K635Q/K635Q mice compared to fasted wild-type mice. Data are represented as mean ± SEM. *P < 0.05 and **P < 0.01, calculated by two-tailed unpaired Student’s t tests. (E) Volcano plot showing the degree of differential regulation of specific lipid species. Cer, ceramide; CL, cardiolipin; FAC, fatty acyl chains; LCL, lysocardiolipin; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PS, phosphatidylserine; TAG, triacylglycerol.
Comparing the concentrations of total lipid class content in the fasted state, LPE was elevated 1.4-fold (Fig. 7B), LPC 1.3-fold (Fig. 7C), and ceramide 1.7-fold (Fig. 7D) in fasted Acss1K635Q/K635Q mice compared to controls. Analysis of specific lipid species is presented in a more granular volcano scatterplot, showing the significant enrichment of specific lipid metabolites in the liver tissue from fasted Acss1K635Q/K635Q mice (Fig. 7E). Again, differentially abundant lipids included a number of LPE, LPC, and ceramide species known to contribute to liver toxicity, including fatty liver and NAFLD. In addition, it has been previously shown that the increase in these lipids in NAFLD is due, at least in part, to the dysregulation of SCD1 and FASN (36, 37).
The compound myriocin is a potent inhibitor of serine palmitoyl transferase and an immunosuppressant (38) that has shown benefits in rat models of NAFLD (39, 40). To test the potential causative role of elevated ceramides in the fatty liver phenotype of fasted Acss1K635Q/K635Q mice, we administered myriocin (0.5 mg/kg) to wild-type and Acss1K635Q/K635Q mice before fasting them for 48 hours. While myriocin did lower ceramide levels in both genotypes (fig. S10A) and appeared to ameliorate fatty liver in fasted wild-type mice (fig. S10B), unexpectedly, there was no visible change in the livers of the fasted Acss1K635Q/K635Q mice (fig. S10C), suggesting that ceramide may be a consequence rather than a primary driver of the acute fatty liver phenotype of these mice.
Fasting increases lysine acetylation
Last, because it has been previously shown that fasting or calorie restriction can alter mitochondrial acetylation (41, 42), we measured pan-lysine acetylation in the livers of wild-type and Acss1K635Q/K635Q mice, both fed and after 48 hours of fasting. The results showed that, while fasting increased lysine acetylation as expected, the Acss1K635Q/K635Q mice had significantly greater acetylation than wild-type mice under both fed and fasting conditions (fig. S11, A and B).
DISCUSSION
Under conditions of limited nutrient availability, cells undergo metabolic reprogramming, which includes multiple compensatory cellular processes and alterations in lipid metabolism. One such enzyme involved in mitochondrial reprogramming to adapt to reduced nutrient availability is ACSS1, a mitochondrial ligase required for acetate metabolism. Used under fasting conditions, as well as in multiple disease models (43), acetate is converted to acetyl-CoA by ACSS2 in the cytoplasm and by ACSS1 in the mitochondria to generate ATP. In addition, acetate appears to function as more than a metabolic substrate and has been shown to be a regulatory molecule with diverse roles throughout the cell (44, 45). While most enzymes are regulated by more than one process, it has been demonstrated that ACSS1-K635-Ac is a key PTM that directs ACSS1 generation of acetyl-CoA from acetate and CoA in tissue culture and in vitro experiments (5, 9, 18). Thus, we proposed to determine the in vivo physiological effects of ACSS1-K635-Ac by generating a knock-in mouse that mimics the constitutive acetylation of K635-Ac (i.e., K535Q).
In our Acss1K635Q knock-in model, we observed that male mice were smaller than wild-type and female mice had dramatically reduced/delayed fecundity. This is in contrast to the previously published Acss1 null mice, which were reported overall indistinguishable from wild-type mice and had normal fecundity (23). However, both Acss1K635Q/K635Q and Acss1−/− male mice exhibited hypothermia when fasted and elevated blood acetate under both fed and fasted conditions (23), as expected given the enzymatic function of ACSS1.
In other regards, our results for the Acss1K635Q/K635Q mouse are quite different than those observed for the Acss1 knockout mice, despite both having the same C57B background (23). Specifically, while Sakakibara et al. (23) conducted multiple studies using a 48-hour fast, they did not report any changes in the appearance, weight, or histology of the liver. In contrast, we observed substantial differences in the liver that included enlargement and discoloration, with numerous white and patchy areas suggestive of excess fat deposition and microsteatosis, consistent with an NAFLD phenotype. On the basis of these data, while speculative, it seems reasonable to further investigate the idea that ACSS1-K635-Ac may have a different biological function than nonacetylated ACSS1, as has been shown for other acetylated enzymes such as manganese superoxide dismutase (MnSOD or SOD2) (46, 47).
Reduced lactate levels in both the serum and liver of Acss1K635Q/K635Q mice under normal fed conditions (Fig. 3, F and H) indicate the increased conversion of lactate to pyruvate to recover reduced ATP production, which may also contribute to elevated blood acetate levels (Fig. 3D). Serum alanine and glutamine levels were also lower in Acss1K635Q/K635Q mice (fig. S4, C and D), suggesting that these may also be used to fuel ATP production. That lactate levels in Acss1K635Q/K635Q mice were further reduced by fasting suggests extremely high conversion of lactate to pyruvate in the body. The reduced activity observed in fasting Acss1K635Q/K635Q mice may have contributed to the reduction in lactate since muscle is a major source of lactate production.
The NAFLD phenotype developed in Acss1K635Q/K635Q mice upon a 48-hour fast was accompanied by impaired ATP production (Fig. 3, E and G) and comparative up-regulation of the fatty acid synthesis enzymes SCD1 and FASN (Fig. 6, A to D), which have also been shown to play a role in developing fatty liver (30). This was unexpected since fatty acid synthesis is normally inhibited during fasting as the liver is instead directed to break down fatty acids and synthesize ketones to fuel the rest of the body. A possible cause for this maladaptive response is the elevated acetate resulting from reduced ability to ligate it with CoA in the mitochondria to dive the tricarboxylic acid (TCA) cycle. Excess acetate transferred out of the mitochondria can directly fuel fatty acid synthesis in the cytoplasm and, importantly, has been shown to increase histone acetylation and increase transcription of Scd1 and Fasn (fig. S11C) (48). Specifically, elevated acetate can increase promoter H3K9, H3K27, and H3K56 acetylation and activate transcription of lipogenic genes, including Scd1 and Fasn (48).
Another unexpected finding in the Acss1K635Q/K635Q mice was evidence of increased hepatic cell senescence, even in the fed state (Fig. 5C), suggesting that elevated acetate or ACSS1-K635Q itself, representing constitutive ACSS1-K635-Ac, causes aberrant signaling and cellular stress. In addition, chronic up-regulation of p53 in the Acss1K635Q/K635Q mice may have contributed to their development of acute hepatic steatosis upon fasting (49).
The lipid metabolites LPE, LPC, and ceramide have previously been shown to contribute to the progression of NAFLD. In this regard, elevated LPE participates in aberrant lipid droplet formation by suppressing lipolysis and fatty acid biosynthesis and thus plays a pathogenic role in inducing fatty liver (50, 51). LPC has been shown to promote endoplasmic reticulum (ER) stress and lipoapoptosis (52) and is also increased in the livers of patients with nonalcoholic steatohepatitis (8). Moreover, LPE and LPC are both cleavage products of phospholipase A2, a major driver of inflammatory responses associated with chronic liver disease (53, 54). Last, ceramides have been shown to induce steatosis and promote ER stress, thereby contributing to hepatocyte injury and a cellular liver environment permissive for NAFLD (36, 52, 55–57).
On the basis of the relative overabundance of SCD1 and FASN and lipidomics data, our results imply that ACSS1-K635Q, representing constitutive ACSS1-K635-Ac, disrupts liver acetate biology, lipid metabolism, mitochondrial bioenergetics, and ATP generation in vivo. Our results suggest that ACSS1-K635-Ac, when dysregulated, leads to several metabolic phenotypes consistent with the role of acetylation in the regulation of gene transcription and mitochondrial metabolism.
MATERIALS AND METHODS
Mouse model
The C57BL/6 (JAX strain no. 000664) and Sox2Cre B6.Cg-Edil3Tg(Sox2-cre)1Amc/J (JAX strain no. 008454) (58) were obtained from the Jackson Laboratory. The ACSS1-K635Q mouse was constructed (see fig. S1 for the schematic) using the bacterial artificial chromosome (BAC) clones RP23-226L10 and RP23-63M16 from the C57BL/6J mouse genome library, introducing the Acss1 K635Q mutation sequence (AAA > CAA), and constructing plasmids targeting genomic DNA. Mouse genomic fragments containing Acss1 exon 14 with the point mutation, homology arms, and a conditional knockout region were amplified using high-fidelity Taq DNA polymerase and sequentially assembled into a targeting vector together with recombination sites and selection markers. The Acss1 targeting construct was linearized and transfected into C57BL/6 embryonic stem (ES) cells via electroporation. Transfected ES cells were G418-selected, resistant clones were amplified and PCR-screened, and six were expanded and characterized by Southern blot analysis. Five of these were confirmed to be correctly targeted.
The targeted ES cell clone 1H5 was injected into C57BL/6 albino embryos, which were then re-implanted into CD-1 pseudo-pregnant females. Founder animals were identified by their coat color, and their germline transmission was confirmed by breeding with C57BL/6 females and subsequent genotyping of the offspring. The presence of the loxP sites (fig. S2A), deletion of the neomycin resistance cassette (fig. S2B), and presence of the knock-in (fig. S2C) were all confirmed by PCR. Three male and three female heterozygous conditional knock-in mice were generated from clone 1H5 as final deliverables (Cyagen).
These mice were intercrossed to produce homozygous conditional knock-in mice. These mice did not display any noticeable phenotype and had normal fertility. We crossed the homozygous conditional Acss1 knock-in mice with the Sox2Cre transgenic line. Since Sox2Cre expresses Cre recombinase in the female germline (59) as well as epiblast cells, this allowed for recombination of our conditional allele in the germline, replacing the endogenous exon 14 with the knock-in, to generate full-body constitutive Acss1K635Q. We maintain the line heterozygous with the wild-type Acss1 allele, followed by PCR (see table S1 for primers). The PCR product sizes from wild-type and constitutive knock-in Acss1 are 169 and 217 base pairs (bp), respectively.
Mice were maintained under pathogen-free conditions at 21 ± 2°C with 12-hour:12-hour light:dark cycle in the Animal Facility at the University of Texas Health Science Center at San Antonio (UTHSCSA). Mice had free access to water and, except those undergoing a 48-hour fast, pelleted Envigo Teklad Rodent Diet traditional formula LM-485, with 25% calories from protein, 17% from fat, and 58% from carbohydrates. For the 3-week HFD, mice were instead given Research Diets Inc. Rodent Diet with 60% calories from fat. For all experiments, mice were matched for age and sex. All animal experiments were conducted in compliance with the National Institutes of Health (NIH) Guidelines for Humane Care and the Use of Laboratory Animals, and all murine studies were approved by the UTHSCSA Institutional Animal Care and Use Committee.
The 48-hour fast
Four-week-old wild-type and Acss1K635Q/K635Q mice were either fasted for 48 hours or given normal chow ad libitum (N = 3 mice per genotype per feeding regimen). All mice had free access to water. Body weight, rectal temperature, and blood glucose were recorded before and at the end of the experiment. Mice were euthanized after being anesthetized with isoflurane by cervical dissociation. Blood was collected from the right cardiac ventricle and placed in EDTA precoated tube. Plasma was obtained after centrifugation at 1000g for 10 min at 4°C and stored at −80°C. Liver, heart, kidney, subcutaneous fat, testes, and muscle were harvested. Tissues were fixed in 4% formalin for histology and also snap-frozen in liquid nitrogen and kept at −80°C freezer for later use.
Cell culture
Primary hepatocytes were isolated from anesthetized mice by portal vein perfusion and collagenase digestion and purified by differential centrifugation as described previously (60). Hepatocytes were cultured in William’s E medium with 5% fetal bovine serum (basal conditions). For starved conditions, cells were incubated for 1 hour in Dulbecco’s modified Eagle’s medium with 1 mM glutamine but without glucose or pyruvate. Cells were fixed in 4% paraformaldehyde and washed three times with phosphate-buffered saline, and lipid droplets were stained with LipidTOX in a 1:1000 dilution for 30 min at room temperature as suggested by the manufacturer (Invitrogen). Cells were mounted with Vectashield and 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired with a Leica Stellaris 8 confocal microscope at ×63 magnification. Lipid droplets were analyzed by size and number with LASX software and classified by size.
qPCR analysis of mRNA expression
Total RNA was extracted and purified from mouse organ tissues/primary hepatocytes using TRIzol reagent (Invitrogen) and RNeasy Mini Plus Kit (Qiagen). cDNA was synthesized with High-Capacity cDNA Reverse Transcription Kit plus RNase inhibitor (Applied Biosystems) as per the manufacturer’s instructions. The primers used are listed in table S1. We used 2× PowerUp SYBR Green Master Mix (Applied Biosystems) and MicroAmp Optical 384-Well Reaction Plate with Barcode using ABI QuantStudio 5 Real-Time PCR system (Applied Biosystems). The qPCR thermal cycling conditions were 2 min at 50°C, 10 min at 95°C, 15 s at 95°C, and 1 min at 60°C for 40 cycles. The ΔΔCT data were normalized and analyzed by QuantStudio Design & Analysis Software v1.5.1 (Applied Biosystems).
Western blotting
Total protein was extracted from organ tissue samples using radioimmunoprecipitation assay buffer (Cell Signaling Technology). Protein concentrations were determined by bicinchoninic acid (BCA) protein assay (Pierce), and equal amounts from each sample were separated by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) in NuPAGE Bis-Tris 4 to 12% gradient gel (Invitrogen) and then transferred onto a polyvinylidene difluoride membrane (Bio-Rad). The membrane was probed with the primary antibodies, which were then detected using horseradish peroxidase (HRP)–conjugated mouse or rabbit secondary antibodies (Cell Signaling Technology) and enhanced chemiluminescence (SuperSignal West Femto Maximum Sensitivity Substrate, Thermo Fisher Scientific).
qMRI and indirect calorimetry
Whole-body composition was obtained with EchoMRI-100H and 130 (EchoMRI). Measures of total body fat, lean mass, and free water as grams of body weight were obtained in individual awake animals (anesthesia was not required). We ensured that mice were immobile and located at the bottom of the measurement tube. For calorimetry, mice were individually housed and allowed to acclimate in the room for 2 days. Mice were maintained under a 12-hour:12-hour light:dark cycle and had free access to food and water throughout the experiment. We used the MARS indirect calorimetry “pull mode” system (Sable Systems) to determine whole-body metabolic parameters such as O2 consumption and CO2 production for a period of 48 hours. Data from the third hour are examined to detect potential equipment-related problems so that animals can be quickly retested. To measure RER after the 3-week HFD, mice were singly housed in Sable Promethion metabolic cages, with all other conditions and procedures the same as above.
Histopathology
Formalin-fixed tissues were paraffin-embedded and processed for H&E staining by the UTHSCSA Histology/Immunohistochemistry Laboratory. Whole slides were scanned with Aperio microscopy. For Oil Red O staining, the liver tissue samples were embedded in an optical coherence tomography compound and frozen at −80°C before being sectioned and stained with Oil Red O staining. The lipid droplets in the liver were then examined under a microscope.
For IHC, formalin-fixed, paraffin-embedded mouse liver tissues were processed by the UTHSCSA Laboratory Medicine core using standard methods. Tissue sections were fixed in 3% hydrogen peroxide, followed by proteinaceous blocking solution (Avidin/Biotin Blocking Kit). Tissue sections were incubated with the primary antibody, followed by addition of the secondary antibody, with EnVision+ System, HRP polymer and diaminobenzidine (DAB) substrate (Dako), plus DAB sparkle (Biocare). When adequate color development was seen, slides were washed in water to stop the reaction, counterstained with Meyer’s hematoxylin (Dako), and covered with a Permount mounting medium (Richard-Allan Scientific). The micrographs were taken under a light microscope (Zeiss).
OCR with Seahorse
Mouse primary fibroblasts and primary hepatocytes were seeded in the XF96 plate for Seahorse at 100,000 cells per well. The day after, they were incubated for 1 hour in Seahorse recording media with 5 mM glucose, 1 mM sodium pyruvate, and 2 mM glutamine. Additional experiments were run with mouse primary fibroblasts incubated in Seahorse recording media with 0.5 mM glucose, 0.5 μM carnitine, and 100 μM palmitate conjugated to BSA or BSA alone as a control. Sequential injection of 1.5 μM oligomycin, 10 μM carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), and 0.5 μM rotenone and antimycin final concentrations was used to infer the respiratory parameters in the Seahorse XF96. Because of varied growth rates, hepatocyte OCR was additionally normalized to protein concentration per well.
Metabolite assays
All samples were measured in duplicates. Acetate levels were measured by colorimetric assay (Abcam), following the manufacturer’s instructions, using 5 μl of plasma per sample in a 96-well plate. Acetyl-CoA was measured with a fluorometric assay kit (Abcam) with a sensitivity of 0.01 nmol/well. ATP levels were measured via a colorimetric method (BioVision) with a detection limit of 50 pmol (1 μM). Blood glucose levels before and after fasting were measured with a glucose meter in tail venous blood. To measure ketone levels, 3-hydroxybutyric acid (BOH) and acetoacetic acid (AcAc) were measured with the Ketone body assay kit (Sigma-Aldrich) and total ketone body concentration (mM) was calculated as BOH + AcAc.
To measure lactate, collected sera or 10-mg samples of liver tissue homogenized in lactate assay buffer (BioVision) were filtered through 10-kDa Spin Column (Millipore), and 2 μl per sample was assayed in a 96-well plate following the manufacturer’s directions (BioVision) and measured at an absorbance of OD570 in a microplate reader. AST was measured by colorimetric assay (Cayman Chemical), following the manufacturer’s instructions, using 20 μl of plasma per sample. The reaction was performed at 37°C for 15 min, and the absorbance was measured at 340 nm. ALT was measured by colorimetric assay (BioVision), following the manufacturer’s instructions, using 10 μl of plasma per sample in a 96-well plate. Plasma glutamine (BioAssay systems) and alanine (Abcam) levels were measured by colorimetric assays, following the manufacturer’s instructions, using 10 μl of plasma per sample in 96-well plates.
High-resolution respirometry with Oroboros
Liver tissue was minced in ice-cold MiR05 buffer (110 mM sucrose, 60 mM potassium lactobionate, 2 mM MgCl2, 20 mM taurine, 10 mM KH2PO4, 0.5 mM EGTA, 20 mM Hepes, and 1 g/liter BSA) and placed inside the Oroboros Oxygraph-2k (O2k) at 37°C with the stir bars spinning at 750 rpm. Supplemental oxygen was added to each chamber to maintain O2 concentrations between 450 and 200 μM throughout the experiment. Substrates were added using the following titration protocol: malate (0.1 mM) and palmitoylcarnitine (0.5 mM) to stimulate LEAK respiration, ADP (2.5 mM) to stimulate palmitate-driven coupled respiration, octanoylcarnitine (0.5 mM) for palmitate- and octanoate-driven coupled respiration, cytochrome c (10 μM; quality control step), FCCP (0.5 μM steps) to uncouple respiration, succinate (10 mM), and antimycin A (2.5 μM) to inhibit respiration and quantify background oxygen consumption.
RNA sequencing
Total liver RNA was extracted using a NucleoSpin RNA kit (Macherey-Nagel) and submitted to the Genome Sequencing Facility for processing and library preparation. Paired-end sequencing was performed using an Illumina HiSeq 3000 system, and 75-bp length reads were determined. The reads were aligned to the UCSC/mm9 genome using the Tophat aligner. Read counts were generated via the HTseq Python package, and the DEseq R package was used for normalization and pairwise differential expression analysis. The criteria set for differential expression included P < 0.01, FPKM > 1, and fold change > 1.5. The resulting gene lists were then analyzed using the Qiagen IPA software to determine gene set and pathway enrichment scores.
Immunoprecipitation
Tracks for ChIP and sequencing (ChIP-seq) were generated from previously published ChIP-seq data in mouse liver tissues available in GEO, with the following accession numbers: ChREBP (GSM6730577), H3K9ac (GSM5404620), and H3K27ac (GSM3534485 and GSM3347654).
Liver tissue was harvested according to the Encode project protocol for tissue isolation (www.encodeproject.org). The resulting nuclei were fixed in 1% formaldehyde for 20 min and sonicated at low power for 15 s at 5 to 10 on/off cycles (Diagenode). The chromatin was incubated with ChREBP antibody (10 μg) overnight followed by Protein A magnetic beads (Invitrogen) for 4 hours. The IP fraction was washed, reverse cross-linked, and purified with Minelute columns (Qiagen). The resulting DNA was quantified with Fasn- and Scd1-specific primers by ABI Real Time qPCR (QuantStudio 5).
Protein immunoprecipitation was performed by incubating the liver lysate with 2 μg of ChREBP antibody (Cell Signaling Technology) overnight at 4°C followed by incubation for 4 hours with 40 μl of Protein A Dynabeads (Thermo Fisher Scientific). The complex was acid-eluted (0.1 M glycine HCl, pH 3.5) and concentrated using Amicon filters (10k, Millipore) and loaded to SDS-PAGE gels for Western blot.
Lipidomics
Lipid species were analyzed using multidimensional mass spectrometry–based shotgun lipidomic analysis (61). In brief, liver tissue homogenate containing 0.4 mg of protein that was determined with a Pierce BCA assay was accurately transferred to a disposable glass culture test tube. A premixture of lipid internal standards species, including D14:1 choline glycerophospholipid, D16:1 ethanolamine glycerophospholipid, D15:0 phosphatidylglycerol (Na), D14:0 phosphatidylserine (Na), D14:0 phosphatidic acid (Na), D4-16:0 FFA, N12:0 sphingomyelin, 14:0 lysoethanolamine glycerophospholipid, 17:0 lysocholine glycerophospholipid, T14:0 cardiolipin, N17:0 ceramide, 13C4 16:0 acylcarnitine, and T17:1 triglyceride (14.77, 14.83, 2.06, 0.64, 0.39, 2.91, 0.82, 0.42, 1.03, 3.65, 0.72, 0.006, and 15.01 nmol/mg protein, respectively), was added before conducting lipid extraction for quantification of the targeted lipid species. Lipid internal standard species of D4-16:0 FFA and N17:0 ceramide (310.292 and 1.185 nmol/ml plasma, respectively) were added before conducting lipid extraction for 50 μl of plasma. Lipid extraction was performed using a modified Bligh and Dyer procedure (61), and each lipid extract was reconstituted in chloroform:methanol (1:1, v:v) at a volume of 400 μl/mg protein or 500 μl/ml plasma. Phosphoethanolamine and FFA were derivatized as described previously (62, 63) before lipidomic analysis. All internal standards were purchased from Avanti Polar Lipids (USA).
For shotgun lipidomics, lipid extract was further diluted to a final concentration of ~500 fmol total lipids per microliter. Mass spectrometric analysis was performed on a triple quadrupole mass spectrometer (TSQ Altis, Thermo Fisher Scientific, San Jose, CA) and a Q Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA), both of which were equipped with an automated nanospray device (TriVersa NanoMate, Advion Bioscience Ltd., Ithaca, NY) as described previously (64). Identification and quantification of lipid species were performed using an automated software program (65, 66). Data processing (e.g., ion peak selection, baseline correction, data transfer, peak intensity comparison, and quantitation) was performed as described previously (66). The result was normalized to the protein content (nmol lipid/mg protein).
Statistical analysis
Except where otherwise indicated, data were processed and analyzed with GraphPad Prism v10.1.0 or Microsoft Excel v2210. Analysis of variance (ANOVA) or two-tailed Student’s unpaired t test was used to analyze the differences between groups as indicated in figure legends. P < 0.05 was considered statistically significant.
Acknowledgments
Funding: This work was supported by the National Cancer Institute grants R01CA152601 (D.G.) and R01CA214025 (D.G.); National Institute on Aging RL5 Career Development award (J.P.P.); National Institutes of Health grants R00HL148504 (X.S.), R00HL143277 (P.Z.), and R01DK133304 (P.Z.); National Institute on Aging training grant T32AG021890-20 (J.S.); American Cancer Society Research Scholar grant RSG-22-016-01-CCB (M.M.); American Federation of Aging Research (J.P.P.); American Heart Association Postdoctoral Fellowship (S.K.); Japan Science and Technology Agency Fusion Oriented Research for disruptive Science and Technology (FOREST) program award JPMJFR216D (M.M.); Cancer Prevention and Research Institute of Texas grants RR20012 (D.G.), RR200089 (P.Z.), RR210005 (X.S.), and RP220267 (M.M.); Voelker Fund Young Investigator Award (M.M.); and San Antonio Nathan Shock Center of Excellence in the Basic Biology of Aging P30AG013319 (E.D.M., J.P.P, and B.B.R.).
Author contributions: Conceptualization: D.G. and N.H. Methodology: G.X., S.Q., J.S., E.S.C., P.S., M.V., and M.P. Investigation: G.X., S.Q., J.S., E.S.C., P.S., D.C., M.V., J.P.P., X.H., M.P., P.Z., X.S., E.D.M., B.B.R., G.L., S.K., and M.M. Analysis: G.X., S.Q., J.S., E.S.C., J.P.P., X.S., and E.D.M. Visualization: G.X., S.Q., J.S., E.S.C., N.H., and E.M. Supervision: D.G., N.H., E.S.C., X.H., and M.M. Writing—original draft: S.Q., D.G., and E.M. Writing—review and editing: D.G. and E.M. All authors read and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The ACSS1-K635Q mouse can be provided by D.G. pending scientific review and a completed material transfer agreement. Requests for the ACSS1-K635Q mouse should be submitted to gius@UTHSCSA.edu. The RNA-seq data have been uploaded into GEO (accession number GSE260582). All other data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Table S1
REFERENCES AND NOTES
- 1.Schug Z. T., Peck B., Jones D. T., Zhang Q., Grosskurth S., Alam I. S., Goodwin L. M., Smethurst E., Mason S., Blyth K., McGarry L., James D., Shanks E., Kalna G., Saunders R. E., Jiang M., Howell M., Lassailly F., Thin M. Z., Spencer-Dene B., Stamp G., van den Broek N. J., Mackay G., Bulusu V., Kamphorst J. J., Tardito S., Strachan D., Harris A. L., Aboagye E. O., Critchlow S. E., Wakelam M. J., Schulze A., Gottlieb E., Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell 27, 57–71 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castro L. F., Lopes-Marques M., Wilson J. M., Rocha E., Reis-Henriques M. A., Santos M. M., Cunha I., A novel acetyl-CoA synthetase short-chain subfamily member 1 (Acss1) gene indicates a dynamic history of paralogue retention and loss in vertebrates. Gene 497, 249–255 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Gerhart-Hines Z., Rodgers J. T., Bare O., Lerin C., Kim S. H., Mostoslavsky R., Alt F. W., Wu Z., Puigserver P., Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J. 26, 1913–1923 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morise A., Mourot J., Boue C., Combe N., Amsler G., Gripois D., Quignard-Boulange A., Yvan-Charvet L., Fenart E., Weill P., Hermier D., Gender-related response of lipid metabolism to dietary fatty acids in the hamster. Br. J. Nutr. 95, 709–720 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Schwer B., Bunkenborg J., Verdin R. O., Andersen J. S., Verdin E., Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc. Natl. Acad. Sci. U.S.A. 103, 10224–10229 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yun M., Bang S. H., Kim J. W., Park J. Y., Kim K. S., Lee J. D., The importance of acetyl coenzyme A synthetase for 11C-acetate uptake and cell survival in hepatocellular carcinoma. J. Nucl. Med. 50, 1222–1228 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Calhoun S., Duan L., Maki C. G., Acetyl-CoA synthetases ACSS1 and ACSS2 are 4-hydroxytamoxifen responsive factors that promote survival in tamoxifen treated and estrogen deprived cells. Trans. Oncol. 19, 101386 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bissig-Choisat B., Alves-Bezerra M., Zorman B., Ochsner S. A., Barzi M., Legras X., Yang D., Borowiak M., Dean A. M., York R. B., Galvan N. T. N., Goss J., Lagor W. R., Moore D. D., Cohen D. E., McKenna N. J., Sumazin P., Bissig K. D., A human liver chimeric mouse model for non-alcoholic fatty liver disease. JHEP Rep. 3, 100281 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirschey M. D., Shimazu T., Capra J. A., Pollard K. S., Verdin E., SIRT1 and SIRT3 deacetylate homologous substrates: AceCS1,2 and HMGCS1,2. Aging 3, 635–642 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu Z., Scott I., Webster B. R., Sack M. N., The emerging characterization of lysine residue deacetylation on the modulation of mitochondrial function and cardiovascular biology. Circ. Res. 105, 830–841 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirschey M. D., Shimazu T., Huang J. Y., Verdin E., Acetylation of mitochondrial proteins. Methods Enzymol. 457, 137–147 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Hebert A. S., Dittenhafer-Reed K. E., Yu W., Bailey D. J., Selen E. S., Boersma M. D., Carson J. J., Tonelli M., Balloon A. J., Higbee A. J., Westphall M. S., Pagliarini D. J., Prolla T. A., Assadi-Porter F., Roy S., Denu J. M., Coon J. J., Calorie restriction and SIRT3 trigger global reprogramming of the mitochondrial protein acetylome. Mol. Cell 49, 186–199 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim H. S., Patel K., Muldoon-Jacobs K., Bisht K. S., Aykin-Burns N., Pennington J. D., van der Meer R., Nguyen P., Savage J., Owens K. M., Vassilopoulos A., Ozden O., Park S. H., Singh K. K., Abdulkadir S. A., Spitz D. R., Deng C. X., Gius D., SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 17, 41–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finley L. W., Carracedo A., Lee J., Souza A., Egia A., Zhang J., Teruya-Feldstein J., Moreira P. I., Cardoso S. M., Clish C. B., Pandolfi P. P., Haigis M. C., SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19, 416–428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torrens-Mas M., Pons D. G., Sastre-Serra J., Oliver J., Roca P., SIRT3 silencing sensitizes breast cancer cells to cytotoxic treatments through an increment in ROS production. J. Cell. Biochem. 118, 397–406 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Haigis M. C., Deng C. X., Finley L. W. S., Kim H. S., Gius D., SIRT3 is a mitochondrial tumor suppressor: A scientific tale that connects aberrant cellular ROS, the Warburg effect, and carcinogenesis. Cancer Res. 72, 2468–2472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vassilopoulos A., Pennington J. D., Andresson T., Rees D. M., Bosley A. D., Fearnley I. M., Ham A., Flynn C. R., Hill S., Rose K. L., Kim H. S., Deng C. X., Walker J. E., Gius D., SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid. Redox Signal. 21, 551–564 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallows W. C., Lee S., Denu J. M., Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc. Natl. Acad. Sci. U.S.A. 103, 10230–10235 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimura Y., Araki A., Maruta H., Takahashi Y., Yamashita H., Molecular cloning of rat acss3 and characterization of mammalian propionyl-CoA synthetase in the liver mitochondrial matrix. J. Biochem. 161, 279–289 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Bose S., Ramesh V., Locasale J. W., Acetate metabolism in physiology, cancer, and beyond. Trends Cell Biol. 29, 695–703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coniglio J. G., Anderson C. E., Robinson C. S., Acetate utilization in normal, fasting, and pyruvate-treated rats. J. Biol. Chem. 198, 525–532 (1952). [PubMed] [Google Scholar]
- 22.Jensen T. L., Kiersgaard M. K., Sorensen D. B., Mikkelsen L. F., Fasting of mice: A review. Lab. Anim. 47, 225–240 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Sakakibara I., Fujino T., Ishii M., Tanaka T., Shimosawa T., Miura S., Zhang W., Tokutake Y., Yamamoto J., Awano M., Iwasaki S., Motoike T., Okamura M., Inagaki T., Kita K., Ezaki O., Naito M., Kuwaki T., Chohnan S., Yamamoto T. T., Hammer R. E., Kodama T., Yanagisawa M., Sakai J., Fasting-induced hypothermia and reduced energy production in mice lacking acetyl-CoA synthetase 2. Cell Metab. 9, 191–202 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Papatheodoridi A. M., Chrysavgis L., Koutsilieris M., Chatzigeorgiou A., The role of senescence in the development of nonalcoholic fatty liver disease and progression to nonalcoholic steatohepatitis. Hepatology 71, 363–374 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Zhang M., Sun W., Zhou M., Tang Y., MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci. Rep. 7, 14493 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y. J., Yang Q. H., Han L., Zhang Y. P., Liu Y. Z., Jin L., Yan H. Z., Effects of soothing liver and invigorating spleen recipes on SREBP-1c, SCD-1 mRNA and proteins expression in hepatocytes of NAFLD rats. Zhong Yao Cai 37, 80–86 (2014). [PubMed] [Google Scholar]
- 27.Singh S., Karthikeyan C., Moorthy N., Fatty acid synthase (FASN): A patent review since 2016-present. Recent Pat. Anticancer Drug Discov. 19, 37–56 (2023). [DOI] [PubMed] [Google Scholar]
- 28.Liu B., Jiang S., Li M., Xiong X., Zhu M., Li D., Zhao L., Qian L., Zhai L., Li J., Lu H., Sun S., Lin J., Lu Y., Li X., Tan M., Proteome-wide analysis of USP14 substrates revealed its role in hepatosteatosis via stabilization of FASN. Nat. Commun. 9, 4770 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiappini F., Coilly A., Kadar H., Gual P., Tran A., Desterke C., Samuel D., Duclos-Vallee J. C., Touboul D., Bertrand-Michel J., Brunelle A., Guettier C., Le Naour F., Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci. Rep. 7, 46658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iizuka K., Takao K., Yabe D., ChREBP-mediated regulation of lipid metabolism: Involvement of the gut microbiota, liver, and adipose tissue. Front. Endocrinol. 11, 587189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Regnier M., Carbinatti T., Parlati L., Benhamed F., Postic C., The role of ChREBP in carbohydrate sensing and NAFLD development. Nat. Rev. Endocrinol. 19, 336–349 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Benhamed F., Denechaud P. D., Lemoine M., Robichon C., Moldes M., Bertrand-Michel J., Ratziu V., Serfaty L., Housset C., Capeau J., Girard J., Guillou H., Postic C., The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J. Clin. Invest. 122, 2176–2194 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang M., Tang Y., Tang E., Lu W., MicroRNA-103 represses hepatic de novo lipogenesis and alleviates NAFLD via targeting FASN and SCD1. Biochem. Biophys. Res. Commun. 524, 716–722 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Guo Y., Yu J., Wang C., Li K., Liu B., Du Y., Xiao F., Chen S., Guo F., miR-212-5p suppresses lipid accumulation by targeting FAS and SCD1. J. Mol. Endocrinol. 59, 205–217 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Kohjima M., Enjoji M., Higuchi N., Kato M., Kotoh K., Yoshimoto T., Fujino T., Yada M., Yada R., Harada N., Takayanagi R., Nakamuta M., Re-evaluation of fatty acid metabolism-related gene expression in nonalcoholic fatty liver disease. Int. J. Mol. Med. 20, 351–358 (2007). [PubMed] [Google Scholar]
- 36.Kartsoli S., Kostara C. E., Tsimihodimos V., Bairaktari E. T., Christodoulou D. K., Lipidomics in non-alcoholic fatty liver disease. World J. Hepatol. 12, 436–450 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hliwa A., Ramos-Molina B., Laski D., Mika A., Sledzinski T., The role of fatty acids in non-alcoholic fatty liver disease progression: An update. Int. J. Mol. Sci. 22, 6900 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyake Y., Kozutsumi Y., Nakamura S., Fujita T., Kawasaki T., Serine palmitoyltransferase is the primary target of a sphingosine-like immunosuppressant, ISP-1/myriocin. Biochem. Biophys. Res. Commun. 211, 396–403 (1995). [DOI] [PubMed] [Google Scholar]
- 39.Kurek K., Piotrowska D. M., Wiesiolek-Kurek P., Lukaszuk B., Chabowski A., Gorski J., Zendzian-Piotrowska M., Inhibition of ceramide de novo synthesis reduces liver lipid accumulation in rats with nonalcoholic fatty liver disease. Liver Int. 34, 1074–1083 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Jiang M., Li C., Liu Q., Wang A., Lei M., Inhibiting ceramide synthesis attenuates hepatic steatosis and fibrosis in rats with non-alcoholic fatty liver disease. Front. Endocrinol. 10, 665 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwer B., Eckersdorff M., Li Y., Silva J. C., Fermin D., Kurtev M. V., Giallourakis C., Comb M. J., Alt F. W., Lombard D. B., Calorie restriction alters mitochondrial protein acetylation. Aging Cell 8, 604–606 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anton S. D., Moehl K., Donahoo W. T., Marosi K., Lee S. A., Mainous A. G. III, Leeuwenburgh C., Mattson M. P., Flipping the metabolic switch: Understanding and applying the health benefits of fasting. Obesity 26, 254–268 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moffett J. R., Puthillathu N., Vengilote R., Jaworski D. M., Namboodiri A. M., Acetate revisited: A key biomolecule at the nexus of metabolism, epigenetics and oncogenesis—Part 1: Acetyl-CoA, acetogenesis and Acyl-CoA short-chain synthetases. Front. Physiol. 11, 580167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaworski D. M., Namboodiri A. M., Moffett J. R., Acetate as a metabolic and epigenetic modifier of cancer therapy. J. Cell. Biochem. 117, 574–588 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Moffett J. R., Puthillathu N., Vengilote R., Jaworski D. M., Namboodiri A. M., Acetate revisited: A key biomolecule at the nexus of metabolism, epigenetics, and oncogenesis—Part 2: Acetate and ACSS2 in health and disease. Front. Physiol. 11, 580171 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Y., Zou X., Dean A. E., Brien J. O., Gao Y., Tran E. L., Park S. H., Liu G., Kieffer M. B., Jiang H., Stauffer M. E., Hart R., Quan S., Satchell K. J. F., Horikoshi N., Bonini M., Gius D., Lysine 68 acetylation directs MnSOD as a tetrameric detoxification complex versus a monomeric tumor promoter. Nat. Commun. 10, 2399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He C., Danes J. M., Hart P. C., Zhu Y., Huang Y., de Abreu A. L., O'Brien J., Mathison A. J., Tang B., Frasor J. M., Wakefield L. M., Ganini D., Stauder E., Zielonka J., Gantner B. N., Urrutia R. A., Gius D., Bonini M. G., SOD2 acetylation on lysine 68 promotes stem cell reprogramming in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 116, 23534–23541 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao X., Lin S. H., Ren F., Li J. T., Chen J. J., Yao C. B., Yang H. B., Jiang S. X., Yan G. Q., Wang D., Wang Y., Liu Y., Cai Z., Xu Y. Y., Chen J., Yu W., Yang P. Y., Lei Q. Y., Acetate functions as an epigenetic metabolite to promote lipid synthesis under hypoxia. Nat. Commun. 7, 11960 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krstic J., Galhuber M., Schulz T. J., Schupp M., Prokesch A., p53 as a dichotomous regulator of liver disease: The dose makes the medicine. Int. J. Mol. Sci. 19, 921 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto Y., Sakurai T., Chen Z., Inoue N., Chiba H., Hui S.-P., Lysophosphatidylethanolamine affects lipid accumulation and metabolism in a human liver-derived cell line. Nutrients 14, 579 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto Y., Sakurai T., Chen Z., Furukawa T., Gowda S. G. B., Wu Y., Nouso K., Fujii Y., Yoshikawa Y., Chiba H., Hui S. P., Analysis of serum lysophosphatidylethanolamine levels in patients with non-alcoholic fatty liver disease by liquid chromatography-tandem mass spectrometry. Anal. Bioanal. Chem. 413, 245–254 (2021). [DOI] [PubMed] [Google Scholar]
- 52.Kakisaka K., Cazanave S. C., Fingas C. D., Guicciardi M. E., Bronk S. F., Werneburg N. W., Mott J. L., Gores G. J., Mechanisms of lysophosphatidylcholine-induced hepatocyte lipoapoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 302, G77–G84 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ishihara K., Miyazaki A., Nabe T., Fushimi H., Iriyama N., Kanai S., Sato T., Uozumi N., Shimizu T., Akiba S., Group IVA phospholipase A2 participates in the progression of hepatic fibrosis. FASEB J. 26, 4111–4121 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Pirisi M., Fabris C., Panozzo M. P., Soardo G., Toniutto P., Bartoli E., Increased serum phospholipase A2 activity in advanced chronic liver disease as an expression of the acute phase response. Dis. Markers 11, 103–111 (1993). [DOI] [PubMed] [Google Scholar]
- 55.Papandreou C., Bullo M., Tinahones F. J., Martinez-Gonzalez M. A., Corella D., Fragkiadakis G. A., Lopez-Miranda J., Estruch R., Fito M., Salas-Salvado J., Serum metabolites in non-alcoholic fatty-liver disease development or reversion; a targeted metabolomic approach within the PREDIMED trial. Nutr. Metab. 14, 58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Preuss C., Jelenik T., Bodis K., Mussig K., Burkart V., Szendroedi J., Roden M., Markgraf D. F., A new targeted lipidomics approach reveals lipid droplets in liver, muscle and heart as a repository for diacylglycerol and ceramide species in non-alcoholic fatty liver. Cells 8, 277 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hajduch E., Lachkar F., Ferre P., Foufelle F., Roles of ceramides in non-alcoholic fatty liver disease. J. Clin. Med. 10, 792 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hayashi S., Lewis P., Pevny L., McMahon A. P., Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119, S97–S101 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Hayashi S., Tenzen T., McMahon A. P., Maternal inheritance of Cre activity in a Sox2Cre deleter strain. Genesis 37, 51–53 (2003). [DOI] [PubMed] [Google Scholar]
- 60.Katsumura S., Siddiqui N., Goldsmith M. R., Cheah J. H., Fujikawa T., Minegishi G., Yamagata A., Yabuki Y., Kobayashi K., Shirouzu M., Inagaki T., Huang T. H., Musi N., Topisirovic I., Larsson O., Morita M., Deadenylase-dependent mRNA decay of GDF15 and FGF21 orchestrates food intake and energy expenditure. Cell Metab. 34, 564–580 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang M., Han X., Multidimensional mass spectrometry-based shotgun lipidomics. Methods Mol. Biol. 1198, 203–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Han X., Yang K., Cheng H., Fikes K. N., Gross R. W., Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J. Lipid Res. 46, 1548–1560 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang M., Han R. H., Han X., Fatty acidomics: Global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Anal. Chem. 85, 9312–9320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han X., Yang K., Gross R. W., Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: Development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom. 22, 2115–2124 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang M., Wang C., Han R. H., Han X., Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 61, 83–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang K., Cheng H., Gross R. W., Han X., Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 81, 4356–4368 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Table S1