Abstract
The secretion-responsive regulation of Escherichia coli secA occurs by coupling its translation to the translation and secretion of an upstream regulator, secM (formerly geneX). We revise the translational start site for secM, defining a new signal peptide sequence with an extended amino-terminal region. Mutational studies indicate that certain atypical amino acyl residues within this extended region are critical for proper secA regulation.
The eubacterial protein secretion machinery consists of a number of soluble and membrane-associated components (5). One critical element is SecA ATPase, which acts as a molecular motor to promote protein secretion at translocation sites that consist of SecYE, the SecA receptor, and SecG and SecDFyajC proteins, which regulate SecA membrane cycling (6, 7, 9, 15, 24). SecA appears to directly recognize the preprotein and guide its entry into the translocon by utilizing its membrane insertion and retraction activity (1, 8, 10, 12). Since SecA appears to initiate the first committed step in the protein translocation cycle, its level and activity are likely to be carefully regulated.
secA has been shown previously to be regulated at the translational level by the protein secretion-proficient state of the Escherichia coli cell, with derepression occurring when protein secretion becomes rate limiting (17, 20). Repression occurs by an autogenous mechanism whereby SecA binds to its translational operator site on geneX secA mRNA to block or dislodge ribosomes that initiate at the secA ribosome-binding site (21, 22). secA translation initiation requires a translational coupling mechanism that employs the upstream gene, geneX (11). Recently we have shown that the basis for the observed secretion-responsive regulation of secA relies on the secretability of geneX preprotein, since signal sequence mutations in geneX rendered secA expression constitutive and prlA signal sequence suppressor alleles restored secA regulation in this context (16). These results demonstrated that geneX is an important regulator of secA, and accordingly we suggest that it should be renamed secM (for “secretion monitor”).
In the original sequencing of the envA secM secA region, it was suggested that secM begins with an AUG codon to encode a 147-amino-acid-residue protein (2). Although it was subsequently pointed out that there are also two potential upstream GUG start sites for secM (23), the AUG initiator has been generally accepted as the secM start site. However, we show here that this is not the case.
No homologs of secM were found in GenBank, but two homologs were identified in the unfinished Salmonella enterica serovar typhi and Yersina pestis genome sequences kindly made available by the Sanger Centre (the S. typhi and Y. pestis secM sequence data were produced by the S. typhi and Y. pestis Sequencing Groups at the Sanger Centre and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens), and one homolog was found in the Klebsiella pneumoniae genome sequence produced by the Genome Sequencing Center at Washington University, St. Louis, Mo. (personal communication). Examination of the homologous sequences strongly suggested that the GUG codon 69 nucleotides upstream of the presumed AUG initiator is the correct translation start for several reasons. First, the protein sequence similarity extends upstream of the current AUG start and stops at the proposed GUG start, which is present in all four organisms (Fig. 1A). Second, the proposed GUG start sites are preceded by Shine-Dalgarno sequences that are able to promote translation initiation, whereas the downstream AUG codons are not (Fig. 1B and data not shown). Third, although it was demonstrated previously that E. coli secM possesses a signal sequence (18), the AUG-based signal peptide lacks a basic amino-terminal region (N region) and contains a charged residue (Glu) within its hydrophobic core region (H region); such features impair signal peptide function. In addition, the previously proposed cleavage site, Ala-Lys-Ala, is not conserved in the homologs. By contrast, the newly proposed secM signal peptides contain several basic amino acyl residues within their atypically long N region as well as a more typical H region. Furthermore, a new cleavage site nine residues before the old site is predicted by the SignalP program for all four secM sequences (14). Since the newly proposed secM signal peptide appeared to be more plausible, we performed a genetic test to verify this prediction.
FIG. 1.
(A) Predicted amino acid sequence of the 5′ portion of secM of E. coli (ec), S. typhi (st), K. pneumoniae (kp), and Y. pestis (yp) utilizing the proposed GUG initiator. The translation start site from the previously proposed AUG initiator is in bold. The predicted new and old signal peptide processing sites are indicated by the slash and double slash, respectively. (B) DNA sequences upstream of the proposed GTG initiator (in bold) of secM. The most conserved portion of the Shine-Dalgarno sequence is aligned above the corresponding region upstream of secM.
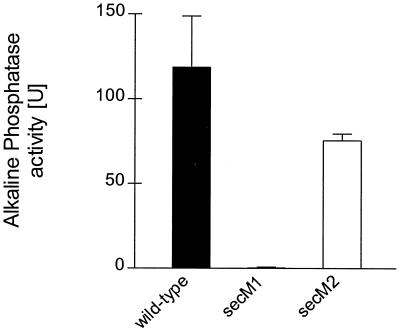
Since we have been previously unsuccessful in raising antibody to SecM for its detection and quantification, we constructed a secM-phoA fusion for measurement of SecM expression levels. PCR methods were utilized to amplify a 1.89-kb secM-phoA fragment from pCB9 (18) by using a forward primer upstream of secM and a reverse primer at the end of phoA that also contained a BamHI recognition sequence. This fragment was cut with BstBI and BamHI to generate a 1.6-kb fragment that was cloned into the analogous sites of pPhIF (16), thereby replacing the secM secA-lacZ region of pPhIF with the secM-phoA fusion to generate pSS1. Two mutations, GTG to GTA (Val) (secM1) and ATG to TGC (Cys) (secM2), were constructed to test the importance of the potential GUG and AUG initiation codons, respectively (Fig. 2). These mutations were chosen to abolish the potential of the respective start codons to initiate translation while preserving a chemically similar amino acid residue if the codon was noninitiating. The mutations were made in pPhIF using the Quik Change procedure as described by the manufacturer (Stratagene) and confirmed by DNA sequence analysis. This resulted in pPhIF derivatives pSS2 and pSS3 that contained secM1 and secM2, respectively. In order to generate secM-phoA fusions containing the secM1 and secM2 alleles, pSS2 and pSS3 were cut with BstBI and BamHI and the vector fragment was isolated and ligated to the 1.6-kb BstBI-BamHI DNA fragment containing the secM-phoA fusion to generate pSS4 and pSS5, respectively. These latter two plasmids are allelic with pSS1. CC118 containing each of these plasmids was assayed for alkaline phosphatase activity (Fig. 3). The results suggest strongly that the GUG codon and not the downstream AUG codon is the correct translation start site of secM, since the secM1 mutant essentially lacked alkaline phosphatase activity, while the secM2 mutant had alkaline phosphatase activity that was similar to that of the wild-type strain containing the secM-phoA fusion.
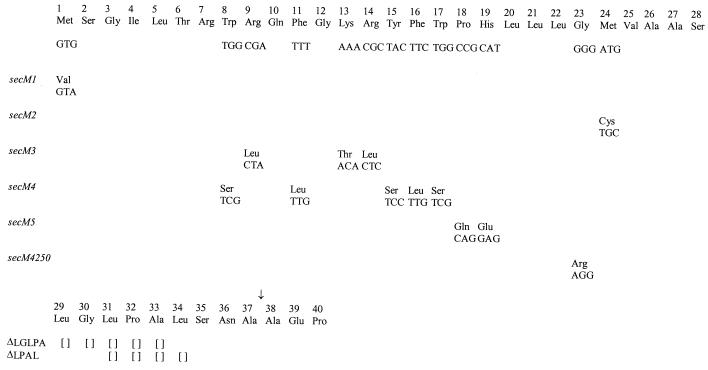
FIG. 2.
secM signal sequence alleles constructed in this study. The codon and amino acid substitutions for each allele are depicted. Empty brackets denote deletions. The presumed signal peptide processing site is indicated by an arrow.
FIG. 3.
CC118 [MC1000 phoA20 rpsE rpoB argE(Am) recA1] containing pSS1 (wild type) or the indicated allelic derivative was grown in Luria broth containing 100 μg of ampicillin per ml at 37°C to mid-logarithmic phase. Alkaline phosphatase assays were performed in duplicate for each of two duplicate cultures as described previously (18). The average result is given, with the error bar indicating the standard deviation.
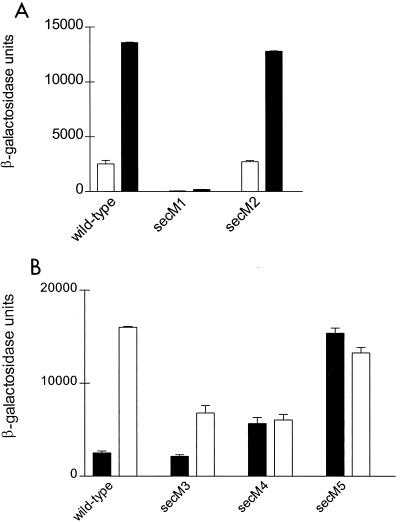
We next examined the effects of the secM1 and secM2 mutations on secA regulation using pPhIF that contains secM and a secA-lacZ translational fusion (11) or derivatives containing the secM1 and secM2 alleles. Secretion-proficient and secretion-defective conditions for examining secA regulation were created by utilizing a wild-type strain (CG155) or its isogenic secD1(Cs) derivative (CG29) as hosts. The secM1 alteration essentially abolished secA expression under both secretion-proficient and secretion-defective conditions, while secA expression and its regulation were normal with the secM2 mutation (Fig. 4A). These results are also consistent with the GUG start of secM as well as the documented translational coupling between secM and secA (11).
FIG. 4.
CG155 (MC1000 recA) (□) and CG29 [MC1000 recA1 secD1(Cs) phoR srl::Tn10] (■) containing pPhIF (wild type) or the indicated allelic derivative was grown in Luria broth containing 100 μg of ampicillin per ml at 39°C to mid-logarithmic phase when the culture was shifted to 23°C for 4 h. β-Galactosidase assays were performed in duplicate for each of two duplicate cultures as described previously (13). The average result is given with the error bar indicating the standard deviation.
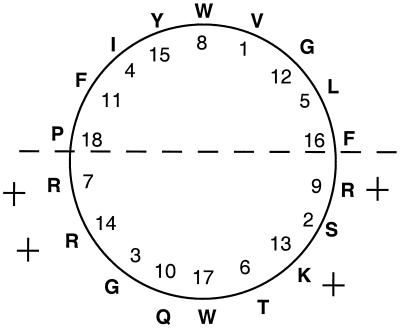
The newly proposed secM signal peptide (Fig. 1) is unusually long for a gram-negative organism due to its extended N region. Interestingly, this region can be modeled as an amphipathic helix (Fig. 5). Since the magnitude of the positive charge in the N region has been shown to be important for the affinity of SecA for signal peptides (1), the secM signal peptide may have a unique interaction with SecA that is related to its regulatory function. The Sec-independent twin arginine (TAT) signal peptides also possess a long N region, accounting for their larger size (4). Although the proposed secM signal peptides do not contain a twin arginine motif per se, we note that they do possess Arg-Arg or Lys-Arg pairs (Fig. 1). Furthermore, a comparison of the positional preferences of TAT- and Sec-dependent signal peptides from gram-negative bacteria for particular amino acid residues identified a striking preference for proline in the −6 position relative to the cleavage site for the former but not latter system (4). E. coli and Salmonella serovar typhi secM signal peptides do contain a proline at this position. As noted previously, a Pro-to-Arg substitution at this position causes derepression of secA expression (16).
FIG. 5.
Helical wheel diagram of the first 18 amino acyl residues of the E. coli secM signal peptide. A plus sign indicates the positively charged amino acid residues, and the dashed line depicts the boundary between the hydrophilic and hydrophobic faces of the α helix.
In light of these intriguing observations and the fact that the TAT and Sec pathways may be in competition with one another (4), we investigated the effect of tat gene deletions on secA regulation by utilizing ΔtatA, ΔtatE, ΔtatAE, and ΔtatC mutants (4) containing pPhIF. secA expression was normal in all of these strains as indicated by β-galactosidase activities that were similar to wild type (data not shown). We next examined the importance of the positively charged and aromatic amino acid residues within the N region of the E. coli secM signal peptide on secA regulation. For this purpose the secM3 and secM4 alleles were made employing the Quik Change procedure on pPhIF to generate pSS6 and pSS7, respectively (Fig. 2). Examination of secA regulation showed that the secM3 alteration caused a modest decrease in secA expression during a protein secretion block (Fig. 4B), suggesting that the highly positively charged N region and the Lys Arg motif were not essential for some level of secA regulation. By contrast, the secM4 alteration resulted in an increased basal level of secA expression and an inability to derepress secA expression further. Collectively these results suggest that the secM signal peptide is recognized by the Sec pathway and that certain aromatic amino acyl residues within its N region may be important for proper secA regulation.
Reassignment of the translation start site of secM is consistent with and illuminates a number of previous observations. First, the mutations that we assigned previously to the secM signal sequence are still within this structure (ΔLGLPA and ΔLPAL) (Fig. 2), although they are located in the distal rather than the proximal end of the H region (16). Second, in an effort to overexpress secM, we previously engineered a better Shine-Dalgarno sequence immediately preceding the presumed AUG start site of secM (secM5) (Fig. 2). Based on our results here, this mutation causes polar and charged amino acid residues (Gln and Glu) to be substituted into the H region of the secM signal peptide, and it should lead to derepression of secA expression similarly to other secM signal sequence defects (16). Indeed, this prediction is precisely what was observed (Fig. 4B). Third, Cook and Kumamoto (3) reported recently that secA overexpression can compensate for certain types of secB defects. The secM4250 allele (referred to as secA4250) that overproduces SecA protein 12-fold was suggested to affect secM translation, since it mapped three nucleotides upstream of the previously proposed secM translation start site (Fig. 2). However, consistent with its phenotype, this mutation is predicted to cause a severe disruption in the function of the reassigned secM signal peptide, resulting in a Gly-to-Arg substitution within the middle of the H region. Of note, all of the secM signal sequence mutations weaken the SignalP prediction relative to the wild type, but in most cases the various scored parameters do not drop below threshold values. This may indicate that secM-mediated regulation of secA is particularly sensitive to signal sequence variations. Finally, Riggs et al. (19) utilized a genetic selection based on up-regulation of a secA-lacZ fusion to select for sec mutants. Surprisingly, the vast majority of Lac+ mutants mapped to the secA-lacZ fusion. This result is readily understood since loss-of-function mutations can be obtained within the secM signal sequence as well as within novel sec genes that would have this phenotype. Additional studies will be required to investigate the unique features of the secM signal sequence and their relevance to promoting proper secA regulation.
Acknowledgments
We thank the Genome Sequencing Center, Washington University, St. Louis, Mo., for communication of the K. pneumoniae sequence data prior to publication. We thank Gunnar von Heijne for the kind gift of the tat mutant strains and Koreaki Ito for the suggestion of the secM nomenclature.
This work was supported by grants GM42033 and GM58560 from the National Institutes of Health to D.O. and K.R., respectively.
REFERENCES
- 1.Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990;265:8164–8169. [PubMed] [Google Scholar]
- 2.Beall B, Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987;169:5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cook H A, Kumamoto C A. Overproduction of SecA suppresses the export defect caused by a mutation in the gene encoding the Escherichia coli export chaperone SecB. J Bacteriol. 1999;181:3010–3017. doi: 10.1128/jb.181.10.3010-3017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cristobal S, de Gier J-W, Nielsen H, von Heijne G. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Driessen A J M, Fekkes P, van der Wolk J. The Sec system. Curr Opin Microbiol. 1998;1:216–222. doi: 10.1016/s1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 6.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG, and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–4879. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 9.Hartl F-U, Lecker S, Schiebel E, Hendrick J P, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 10.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 11.McNicholas P, Salavati R, Oliver D. Dual regulation of Escherichia coli secA translation by distinct upstream elements. J Mol Biol. 1997;265:128–141. doi: 10.1006/jmbi.1996.0723. [DOI] [PubMed] [Google Scholar]
- 12.Miller A, Wang L, Kendall D. Synthetic signal peptides specifically recognize SecA and stimulate ATPase activity in the absence of preprotein. J Biol Chem. 1998;273:11409–11412. doi: 10.1074/jbc.273.19.11409. [DOI] [PubMed] [Google Scholar]
- 13.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 14.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Nishiyama K-I, Suzuki T, Tokuda H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 16.Oliver D, Norman J, Sarker S. Regulation of Escherichia coli secA by cellular protein secretion proficiency requires an intact gene X signal sequence and an active translocon. J Bacteriol. 1998;180:5240–5242. doi: 10.1128/jb.180.19.5240-5242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver D B, Beckwith J. Regulation of a membrane component required for protein secretion in Escherichia coli. Cell. 1982;30:311–319. doi: 10.1016/0092-8674(82)90037-x. [DOI] [PubMed] [Google Scholar]
- 18.Rajapandi T, Dolan K M, Oliver D B. The first gene in the Escherichia coli secA operon, gene X, encodes a nonessential secretory protein. J Bacteriol. 1991;173:7092–7097. doi: 10.1128/jb.173.22.7092-7097.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggs P D, Derman A I, Beckwith J. A mutation affecting the regulation of a secA-lacZ fusion defines a new sec gene. Genetics. 1988;118:571–579. doi: 10.1093/genetics/118.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollo E E, Oliver D B. Regulation of the Escherichia coli secA gene by protein secretion defects: analysis of secA, secB, secD, and secY mutants. J Bacteriol. 1988;170:3281–3282. doi: 10.1128/jb.170.7.3281-3282.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salavati R, Oliver D. Competition between ribosome and SecA binding promotes Escherichia coli secA translational regulation. RNA. 1995;1:745–753. [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt M G, Oliver D B. SecA protein autogenously represses its own translation during normal protein secretion in Escherichia coli. J Bacteriol. 1989;171:643–649. doi: 10.1128/jb.171.2.643-649.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt M G, Rollo E E, Grodberg J, Oliver D B. Nucleotide sequence of the secA gene and secA(Ts) mutations preventing protein export in Escherichia coli. J Bacteriol. 1988;170:3404–3414. doi: 10.1128/jb.170.8.3404-3414.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Wolk J P W, de Wit J G, Driessen A J M. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]