Abstract
We have shown that the Escherichia coli phosphate-starvation-inducible psiE gene is regulated by both phosphate and the carbon source by using both lacZ and chloramphenicol acetyltransferase gene (cat) fusions. Yet, under all conditions tested, a single transcriptional start site lying 7 bp downstream of a predicted −10 region was revealed by primer extension analysis. DNase I footprinting showed that the PhoB transcriptional-activator protein protects two predicted pho boxes lying upstream of and near the −35 promoter region. Similar analysis showed that the cyclic AMP (cAMP)-cAMP receptor protein (cAMP-CRP) complex binds a region that overlaps with the downstream pho box. These results, together with measurements of the in vivo psiE promoter activity under various conditions, show that expression of the psiE gene is under direct positive and negative control by PhoB and cAMP-CRP, respectively.
The phosphate regulon (pho) of Escherichia coli includes at least 31 genes whose expression is induced under conditions of phosphate starvation and whose products have roles in the transport or degradation of various phosphorus sources. These genes are under the transcriptional control of the PhoB and PhoR proteins, which belong to a large family of two-component regulatory systems that respond to a variety of environmental stimuli (15, 26). PhoB is the transcriptional activator, which acts by binding pho box sites immediately upstream of all known pho regulon promoters (16, 17). These pho boxes comprise two well-conserved 7-bp direct repeats, which are separated by poorly conserved 4-bp spacer sequences, suggesting that PhoB molecules always bind on the same face of the DNA helix (20). PhoR is the histidine protein kinase that activates PhoB by phosphorylation and (presumably) inactivates phosphorylated PhoB (phospho-PhoB) by dephosphorylation in response to the extracellular phosphate concentration (14, 17, 22). Transcriptional activation by phospho-PhoB involves protein-protein interaction with the ς70 subunit of RNA polymerase holoenzyme (13).
By using λplacMu53 phage (3), we had previously identified several pho regulon genes, including ones in the phn (psiD) operon involved in phosphonate utilization (18); the phoH (psiH) gene, encoding an ATP binding protein with a helicase motif (9, 12); and a gene called psiE, the subject of this study. These phosphate-starvation-inducible (psi) genes had also been previously found using the Mu d1 transposon (24, 25, 27, 28). More recently, it was shown that the psiE gene of Salmonella enterica serovar Typhimurium is induced during infection of macrophages (23).
Wanner and coworkers (24, 28) had previously reported that expression of the psiE′-′lacZ gene in a cya or crp mutant was greater than that in the wild-type strain, indicating that psiE expression is negatively regulated by the cyclic AMP (cAMP)-cAMP receptor protein (cAMP-CRP) complex. They had also indicated that transcription from the promoter was also positively regulated by PhoB. However, these studies do not answer the following questions. How many promoters are there? How many transcription initiation sites are there? Where do PhoB and cAMP-CRP bind? How does RNA polymerase interact with the promoter(s) in the presence or absence of PhoB and/or cAMP-CRP?
Here we report more-detailed molecular studies on the regulation of the psiE gene, including identification of its transcription start site and DNA binding sites for PhoB and cAMP-CRP at the promoter region. Our results show that these proteins directly regulate transcription from only one mRNA initiation site of the psiE promoter. Based on the results, we discuss the possible mechanism of transcriptional regulation of the psiE gene.
Cloning of the psiE promoter region.
The psiE fusion strain SE5031 used in this study was found among a collection of E. coli SE5000 mutants made with λplacMu53 in an earlier study (9, 18). The strain displayed a strong Lac+ (dark blue) color on Tris-glucose low-phosphate (LP) agar and a weak Lac+ (pale blue) color on Tris-glucose high-phosphate (HP) agar containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and kanamycin. β-Galactosidase activities were induced by phosphate starvation of strain SE5031 grown in both Tris-glucose (667 U for LP medium; 90 U for HP medium) and Tris-glycerol (245 U for LP medium; 47 U for HP medium) media. However, the basal level (HP) and the phosphate-starvation-induced level (LP) were 1.9-fold and 2.7-fold higher, respectively, in Tris-glucose medium than in Tris-glycerol medium. These results suggest that expression of the psiE gene is regulated by both phosphate and the carbon source. Since it is known that the amount of the cAMP-CRP complex increases under glucose starvation conditions such as Tris-glycerol medium, these results suggested that expression of psiE is negatively regulated by cAMP-CRP independent of the phosphate concentration.
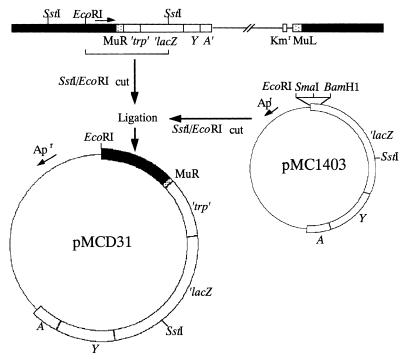
In order to further study the psiE promoter, we cloned the chromosomal psiE′-′lacZ fusion. Southern hybridization using the BamHI-SstI lacZ fragment of pMC1403 (5) as a probe revealed a single 12-kb EcoRI-SstI chromosomal fragment in SE5031 (Fig. 1). This fragment was gel purified, ligated with similarly digested pMC1403, and transformed into SE5000. A Lac+ transformant showed higher β-galactosidase activity on Tris-glucose LP medium than on Tris-glucose HP medium. It carried recombinant plasmid pMCD31, which contains the 12-kb insert, suggesting that pMCD31 carries a functional psiE promoter.
FIG. 1.
Strategy for cloning the chromosomal psiE′-′lacZ fusion. Black boxes, E. coli chromosomal DNA; thin lines, lambda or vector plasmid DNA; dotted boxes, Mu sequences; open boxes, ′trp′-′lacZYA′; arrow above the map, orientation of the psiE gene with respect to the λplacMu53 insertion. Apr, ampicillin resistance; Kmr, kanamycin resistance.
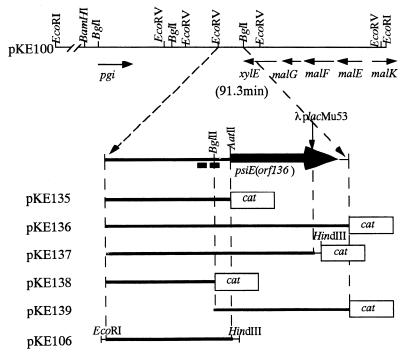
These results indicated that λplacMu53 had integrated into the psiE gene about 8.6 kb from a chromosomal EcoRI site (Fig. 1). Restriction analysis revealed that the psiE::λplacMu53 fusion in SE5031 lies near the carboxyl terminus-encoding region of psiE. By using the 12-kb EcoRI-SstI fragment in pMCD31 as a probe, two phages in an E. coli genomic λ library (11) that carry chromosomal DNA near 91.3 min (λ632 and λ633) were found to hybridize. As expected, hybridization was also detected in phages containing lacZ or trp sequences as these are present within the fusion sequences. The 15-kb EcoRI fragment of λ633 was subcloned into pUC9 (9), yielding pKE100 (Fig. 2).
FIG. 2.
Plasmids carrying the psiE regions. The restriction map and genes near the psiE gene in pKE100 are shown above. Vertical arrow, site of the λplacMu53 insertion; thick arrow and black boxes, psiE gene and pho boxes, respectively. Only relevant restriction enzyme sites are indicated. Boxes labeled cat show the gene encoding CAT.
Two other psiE′-′lacZ fusions were previously shown by DNA sequence analysis to lie within the same gene (19), which corresponds to an open reading frame (orf136) of unknown function adjacent to the xylE gene (6). The psiE gene corresponds to the yjbA locus at 91.3 min (2). However, it was unknown whether the psiE gene was translated since the earlier study employed only transcriptional fusions (24, 28). Because the λplacMu53 phage generates lacZ-protein fusions, the above results show that the psiE gene is also translated. Based on its hydrophobic characteristics, the psiE gene product may be a membrane protein.
Deletion analysis of the psiE promoter.
To define the functional promoter in vivo, we constructed five plasmids carrying different amounts of the psiE chromosomal region transcriptionally fused to cat (Fig. 2). These plasmids were introduced into ANCK10 (wild-type) and ANCH1 [Δ(phoB-phoR)] strains (8). The promoter activities were assessed by measuring chloramphenicol acetyltransferase (CAT) activities in transformants after growth in Tris-glucose LP and HP media (Table 1). Both wild-type and ΔphoB mutant cells carrying pKE135, pKE136, and pKE137 had high basal levels of CAT activity in comparison with cells carrying the vector control, pKK232-8 (4). The CAT activities of these wild-type transformants were induced 10-fold or more by phosphate starvation. This induction was not seen in ΔphoB mutant transformants. In contrast, transformants carrying pKE138 or pKE139, like transformants carrying the vector produced barely detectable levels of CAT. These data show that expression of psiE by phosphate starvation requires PhoB; however a substantial basal level is expressed independently of the phosphate concentration. Moreover, these data show that sequences flanking the BglII site are required for both PhoB-dependent and -independent expression (Fig. 2; see Fig. 5).
TABLE 1.
psiE promoter activity
| Plasmid | CAT activity in indicated medium for straina:
|
|||
|---|---|---|---|---|
| ANCK10
|
ANCH1 [Δ(phoB-phoR)]
|
|||
| LP | HP | LP | HP | |
| pKE135 | 310 | 25 | 24 (7) | 28 (8) |
| pKE136 | 240 | 22 | 21 (9) | 19 (7) |
| pKE137 | 320 | 23 | 25 (7) | 24 (8) |
| pKE138 | <1 | <1 | <1 | <1 |
| pKE139 | <1 | <1 | <1 | <1 |
| pKK232-8 | <1 | <1 | <1 | <1 |
Cells were grown overnight in Tris-glucose medium and were assayed. LP and HP, 0.1 and 2 mM phosphate concentrations, respectively. The activities of CAT are expressed in nanomoles of 5-thio-2-nitrobenzoate liberated per minute per optical density unit of the cell culture at 450 nm. CAT activities measured in cells grown in the presence of 2 mM cAMP are shown in parentheses.
FIG. 5.
Schematic presentation of the PhoB and cAMP-CRP binding sites in the psiE promoter. Boxes, pho boxes and the −10 region of psiE; brackets, regions protected from DNase I digestion in the presence of PhoB and cAMP-CRP; asterisks, positions identical with the consensus sequence for the cAMP-CRP binding site (7); thick line above pho box 1, putative −35 sequence. The BglII restriction enzyme site, the transcription initiation site, and a translation initiation codon (Met) are indicated.
As described in the previous section, expression of the chromosomal psiE′-′lacZ fusion gene occurred at higher levels in Tris-glucose medium than in Tris-glycerol medium, independent of the phosphate concentration. We therefore tested for involvement of the cAMP-CRP complex in the control of psiE expression. Expression of the psiE′-′cat fusions (pKE135, pKE136, and pKE137) was reduced in ΔphoB strain transformants when transformants were grown in the presence of cAMP in both Tris-glucose LP and HP media (Table 1), suggesting that expression of these constructs is also negatively regulated by cAMP-CRP. Previously, it was shown that expression of the psiE gene was negatively regulated by the crp and cya genes (24, 28).
Determination of the psiE transcription initiation site.
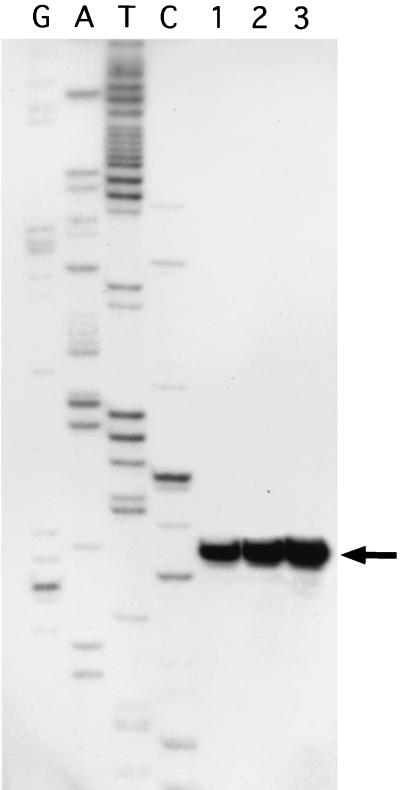
Primer extension analysis revealed a single band of the same size in all cells tested (Fig. 3). Each position of the gel corresponding to the bands was cut out, and the radioactivities were measured. When the radioactivity of the band derived from cells grown in Tris-glycerol HP medium was defined as 1 (Fig. 3, lane 1), the radioactivities of those derived from the cells grown in Tris-glucose HP and LP media were approximately 3 and 8, respectively (Fig. 3, lanes 2 and 3). These results are consistent with the promoter activities of the chromosomal psiE′-′lacZ gene as shown from in vivo data and indicate that transcription of the psiE gene initiates from the same start site under the various conditions examined. The data also suggest that transcription from this start site is positively regulated by PhoB and negatively regulated by cAMP-CRP. Seven base pairs upstream from the transcription initiation site, there is a DNA sequence (TATAca) similar to the consensus sequence for the −10 regions of E. coli promoters (see Fig. 5). Furthermore, 18 bp upstream from the −10 region, a putative −35 sequence (TaGAtc) exists. In general, PhoB-regulated promoters lack a consensus −35 sequence (15, 26).
FIG. 3.
Transcription initiation site of psiE. SE5000 carrying pKE106 (Fig. 2) was grown in Tris-glycerol HP medium or Tris-glucose HP or LP medium to early stationary phase, and RNA was isolated as described previously (1). The 23-base M13 forward sequencing primer (Gibco-BRL, Bethesda, Md.) was labeled with [γ-32P]ATP and T4 polynucleotide kinase. The end-labeled primer was hybridized to RNA extracted from SE5000 cells carrying pKE106 grown in Tris-glycerol HP (lane 1), Tris-glucose HP (lane 2), or Tris-glucose LP medium (lane 3). Primer extension was done using rTth reverse transcriptase for 10 min at 60°C. Arrow, position of primer extension products. Lanes G, A, T, and C, sequence ladders of plasmid pKE106 obtained by using the same primer by the dideoxy chain termination method (21). The transcription start site on the DNA sequence is shown in Fig. 5.
Determination of PhoB and cAMP-CRP binding sites within the psiE regulatory region.
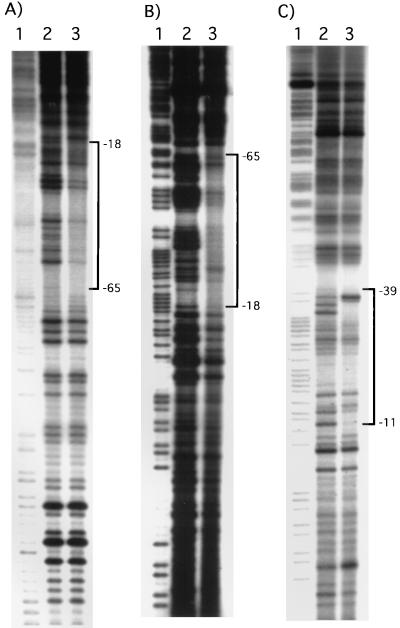
To determine whether PhoB or cAMP-CRP directly binds the psiE promoter, we performed DNase I footprinting experiments as described previously (17). The PhoB protein protected the DNA segment spanning nucleotides −18 to −65 from DNase I digestion on both strands (Fig. 4A and B). This region includes two pho boxes (Fig. 5). The upstream one, pho box 2, agrees poorly with the consensus pho box, CTGTCATAA(T)AT(A)CTGTCAC(T). Tandemly arranged pho boxes in the pstS and ugp promoter regions have also been reported (8, 10). In the ugp promoter, the pho boxes agree poorly with the consensus pho box. These data suggest that multiple pho boxes might be required for the binding of PhoB to regions containing weakly conserved pho boxes such as the psiE promoter.
FIG. 4.
DNase I footprinting of the psiE promoter region. The top and bottom strands were labeled at EcoRI and HindIII sites of pKE106 (Fig. 2) by [γ-32P]ATP using T4 polynucleotide kinase. These fragments were then gel purified. (A) Footprinting on the top strand with the PhoB protein. PhoB and PhoR1084 proteins were purified and used to prepare phospho-PhoB protein as described previously (17). PhoR1084 (4 pmol) and PhoB (4 pmol) were used. (B) Footprinting on the bottom strand with the PhoB protein. PhoR1084 (4 pmol) and PhoB (4 pmol) were used. (C) Footprinting on the bottom strand with CRP (1 pmol) and cAMP (10 μM). Lanes 1, G>A ladders from Maxam-Gilbert sequencing of the probe DNA; lanes 2, products of DNase I digestion in the absence of proteins; lanes 3, products of DNase I digestion in the presence of proteins. The regions protected from DNase I digestion are indicated by brackets, with nucleotide positions labeled from the transcription initiation site (+1).
The binding of the cAMP-CRP complex to the bottom strand was assayed. The cAMP-CRP complex bound to the region from −11 to −39, which includes a sequence similar to the consensus for the cAMP-CRP binding site (Fig. 4C and 5). cAMP-CRP binding enhanced the DNase I digestion at positions −36A and −37T from the transcription initiation site. Interestingly, the cAMP-CRP binding site covers the putative −35 region and overlaps the downstream pho box (Fig. 5).
Based on these results, we propose that the amount of the psiE transcript, which is initiated from only one start site, is versatilely regulated by culture conditions. Under high-glucose and LP conditions, where the amount of PhoB is large and that of cAMP-CRP is small, PhoB activates transcription. Under high-glucose and HP conditions, transcription from the typical −35 and −10 promoter (Fig. 5) by RNA polymerase alone would be the predominant mode of expression, because of low concentrations of cAMP-CRP and PhoB. Transcription from the promoter would be blocked by cAMP-CRP under low-glucose and HP conditions, because of a high concentration of cAMP-CRP and low concentration of PhoB. Under low-glucose and LP conditions, where both proteins are abundant, PhoB and cAMP-CRP would compete for binding to the psiE promoter region, which contains overlapping sequences recognized by these proteins. The in vivo data using the chromosomal psiE′-′lacZ gene suggested that, under these conditions, PhoB binding to the pho boxes predominates over cAMP-CRP binding to its site, which overlaps the pho box (Fig. 5). There may be a possibility that RNA polymerase interacts synergistically with PhoB but not with cAMP-CRP. Our data strongly suggest that transcription initiation from the psiE gene is regulated by dual regulatory systems that respond to environmental stimuli.
Acknowledgments
We thank H. Aiba for providing CRP protein. We also thank B.-S. Shin for helpful discussion.
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture of Japan and by grant 981-0503-019-2 from the Korea Science and Engineering Foundation. B.L.W. was supported by NIH GM57695.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1987. [Google Scholar]
- 2.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 3.Bremer E, Silhavy T J, Weisemann J M, Weinstock G M. λplacMu: a transposable derivative of bacteriophage lambda for creating lacZ protein fusions in a single step. J Bacteriol. 1984;158:1084–1093. doi: 10.1128/jb.158.3.1084-1093.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984;27:151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- 5.Casadaban M J, Chou J, Cohen S N. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis E O, Henderson P J F. The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J Biol Chem. 1987;262:13928–13932. [PubMed] [Google Scholar]
- 7.Ebright R H, Ebright Y W, Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989;17:10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasahara M, Makino K, Amemura M, Nakata A, Shinagawa H. Dual regulation of the ugp operon by phosphate and carbon starvation at two interspaced promoters. J Bacteriol. 1991;173:549–558. doi: 10.1128/jb.173.2.549-558.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim S-K, Makino K, Amemura M, Shinagawa H, Nakata A. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J Bacteriol. 1993;175:1316–1324. doi: 10.1128/jb.175.5.1316-1324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura S, Makino K, Shinagawa H, Amemura M, Nakata A. Regulation of the phosphate regulon of Escherichia coli: characterization of the promoter of the pstS gene. Mol Gen Genet. 1989;215:374–380. doi: 10.1007/BF00427032. [DOI] [PubMed] [Google Scholar]
- 11.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 12.Koonin E V, Rudd K E. Two domains of superfamily I helicases may exist as separate proteins. Protein Sci. 1996;5:178–180. doi: 10.1002/pro.5560050124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makino K, Amemura M, Kim S-K, Nakata A, Shinagawa H. Role of the ς70 subunit of RNA polymerase in transcriptional activation by activator protein PhoB in Escherichia coli. Genes Dev. 1993;7:149–160. doi: 10.1101/gad.7.1.149. [DOI] [PubMed] [Google Scholar]
- 14.Makino K, Amemura M, Kim S-K, Shinagawa H, Nakata A. Signal transduction of the phosphate regulon in Escherichia coli mediated by phosphorylation. In: Papa S, Azzi A, Tager J M, editors. Adenine nucleotides in cellular energy transfer and signal transduction. Basel, Switzerland: Birkhauser Verlag; 1992. pp. 191–200. [Google Scholar]
- 15.Makino K, Amemura M, Kim S-K, Yokoyama K, Kimura S. Mechanism of transcriptional activation of the phosphate regulon in Escherichia coli. Korean J Microbiol. 1998;36:231–238. [Google Scholar]
- 16.Makino K, Shinagawa H, Amemura M, Kimura S, Nakata A, Ishihama A. Regulation of the phosphate regulon of Escherichia coli: activation of pstS transcription by PhoB protein in vitro. J Mol Biol. 1988;203:85–95. doi: 10.1016/0022-2836(88)90093-9. [DOI] [PubMed] [Google Scholar]
- 17.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 18.Makino K, Kim S-K, Shinagawa H, Amemura M, Nakata A. Molecular analysis of the cryptic and functional phn operons for phosphonate use in Escherichia coli K-12. J Bacteriol. 1991;173:2665–2672. doi: 10.1128/jb.173.8.2665-2672.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metcalf W W, Steed P M, Wanner B L. Identification of phosphate-starvation-inducible genes in Escherichia coli K-12 by DNA sequence analysis of psi::lacZ(Mu d1) transcriptional fusions. J Bacteriol. 1990;172:3191–3200. doi: 10.1128/jb.172.6.3191-3200.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamura H, Hanaoka S, Nagadoi A, Makino K, Nishimura Y. Structural comparison of the PhoB and OmpR DNA-binding/transactivation domains and the arrangement of PhoB molecules on the phosphate box. J Mol Biol. 2000;295:1225–1236. doi: 10.1006/jmbi.1999.3379. [DOI] [PubMed] [Google Scholar]
- 21.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shinagawa H, Makino K, Yamada M, Amemura M, Sato T, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli: dual function of PhoR as a protein kinase and a protein phosphatase. In: Torriani-Gorini A, Yagil E, Silver S, editors. Phosphate in microorganisms. Washington, D.C.: American Society for Microbiology; 1993. pp. 20–25. [Google Scholar]
- 23.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 24.Wanner B L. Overlapping and separate controls on the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1983;166:283–308. doi: 10.1016/s0022-2836(83)80086-2. [DOI] [PubMed] [Google Scholar]
- 25.Wanner B L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986;191:39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- 26.Wanner B L. Phosphorus assimilation and control of the phosphate regulon. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C.: ASM Press; 1996. pp. 1357–1381. [Google Scholar]
- 27.Wanner B L, McSharry R. Phosphate-controlled gene expression in Escherichia coli using Mu d1-directed lacZ fusions. J Mol Biol. 1982;158:347–363. doi: 10.1016/0022-2836(82)90202-9. [DOI] [PubMed] [Google Scholar]
- 28.Wanner B L, Wieder S, McSharry R. Use of bacteriophage transposon Mu d1 to determine the orientation for three proC-linked phosphate-starvation-inducible (psi) genes in Escherichia coli K-12. J Bacteriol. 1981;146:93–101. doi: 10.1128/jb.146.1.93-101.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]