Abstract
Background:
Despite monogenic and polygenic contributions to cardiovascular disease (CVD), genetic testing is not widely adopted, and current tests are limited by the breadth of surveyed conditions and interpretation burden.
Methods:
We developed a comprehensive clinical genome CVD test with semi-automated interpretation. Monogenic conditions and risk alleles were selected based on the strength of disease association and evidence for increased disease risk, respectively. Non-CVD secondary findings genes, pharmacogenomic (PGx) variants and CVD polygenic risk scores (PRS) were assessed for inclusion. Test performance was modeled using 2,594 genomes from the 1000 Genomes Project, and further investigated in 20 previously tested individuals.
Results:
The CVD genome test is composed of a panel of 215 CVD gene-disease pairs, 35 non-CVD secondary findings genes, 4 risk alleles or genotypes, 10 PGx genes and a PRS for coronary artery disease. Modeling of test performance using samples from the 1000 Genomes Project revealed ~6% of individuals with a monogenic finding in a CVD-associated gene, 6% with a risk allele finding, ~1% with a non-CVD secondary finding, and 93% with CVD-associated PGx variants. Assessment of blinded clinical samples showed complete concordance with prior testing. An average of 4 variants were reviewed per case, with interpretation and reporting time ranging from 9–96 min.
Conclusions:
A genome sequencing based CVD genetic risk assessment can provide comprehensive genetic disease and genetic risk information to patients with CVD. The semi-automated and limited interpretation burden suggest that this testing approach could be scaled to support population-level initiatives.
Introduction
Identifying the genetic underpinnings of cardiovascular disease (CVD), including both monogenic and polygenic factors, can improve patient risk stratification, refine clinical diagnoses, and inform targeted treatment opportunities1–4. Guidelines recommend genetic testing for patients presenting with cardiomyopathy, arrhythmias, dyslipidemias, and aortopathies, but such testing is substantially underutilized. For example, a recent retrospective cohort study found that only 0.8% to 1.6% of patients newly diagnosed with CVD, and for whom guidelines recommended testing, received any kind of genetic investigation5. For those patients who do receive genetic testing, the results may be affected by inter-laboratory variability in assay differences (e.g. panel vs exome), the genes selected for interrogation, variant interpretation and variant classification6. Inconsistent testing practices in the CVD population can exaggerate inequities in access to a genetic diagnosis, undermine the value of genetic testing, and reduce the likelihood of reimbursement.
A growing body of evidence supports the use of genome sequencing (GS) as a first-line test, particularly in the neonatal acute care population7 and in children with signs and symptoms of a genetic disease 8. We reasoned that a GS-based CVD-focused genetic risk assessment test may simplify CVD genetic testing practices by providing a single, multifaceted, high-throughput testing platform for patients with a CVD phenotype for which genetic testing is recommended. Here we describe the development of the TruGenome™ CVD Test including monogenic CVD and risk allele evaluations, the selection of evidence-supported pharmacogenetic (PGx) alleles, identification of a population-sensitive polygenic risk score (PRS), and approaches to reduce test interpretation and reporting time. We also report the frequency of findings in unrelated individuals in the 1000 Genomes Project cohort, providing an estimate of CVD risk and enabling an assessment of test outcomes across multiple genetic ancestries. Finally, we explore test interpretation burden and compared findings from the TruGenome™ CVD Test to findings in a set of clinical samples from 20 previously tested individuals with CVD. The approach and outcomes reported here may inform future integration of GS applications in the diagnosis and management of CVD and other disease areas.
Methods
Full methods are available in the Supplemental Materials. The data that support the findings of this study are available from the corresponding author upon request. Institutional review board approval for activities involving human samples was received from WIRB-Copernicus Group (Protocol 20241866). Informed consent was not required.
Results
Design of the TruGenome™ CVD Test
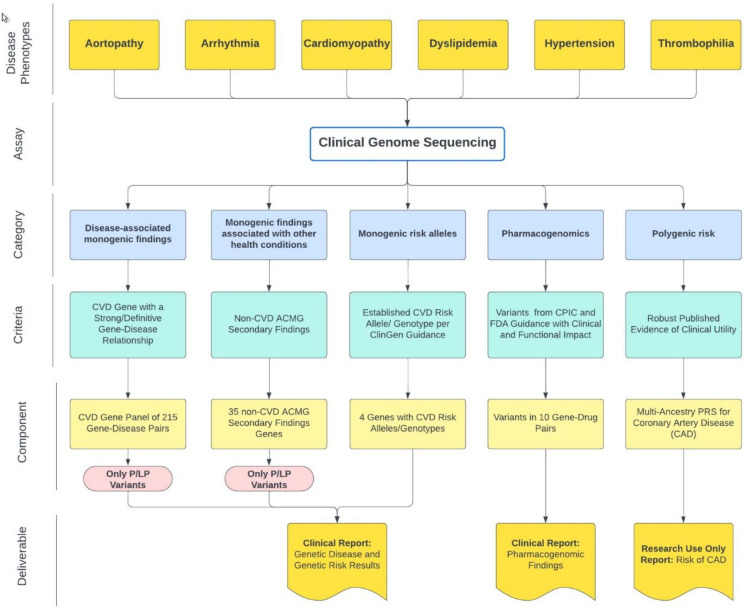
Test development focused on CVD phenotypes with a high prior probability of a genetic etiology and relevance in an adult cardiac clinic population, particularly aortopathy, arrhythmia, cardiomyopathy, dyslipidemia, coronary artery disease (CAD), hypertension and thrombophilia. The final test design includes five components: a CVD gene panel, select CVD risk alleles, non-CVD secondary findings, a PRS for CAD, and PGx findings. Each component integrates reporting thresholds set to support the return of high confidence, medically actionable genetic information (Figure 1).
Figure 1. TruGenome™ CVD test design and reportables.
Test design focused on CVD phenotypes relevant to cardiac clinic patients. Monogenic conditions and risk alleles were evaluated for the strength of evidence for increased disease risk. CVD PRSs and PGx findings were also explored for inclusion. The test includes five components: a 215 gene-disease pair CVD panel, the 35 remaining non-CVD ACMG secondary findings genes, 4 risk alleles or genotypes, PGx findings across 10 gene-drug pairs and a PRS for CAD. Test deliverables include a clinical report for genetic disease and genetic risk findings, an RUO CAD PRS report and a clinical report of pharmacogenomic findings.
Cardiovascular Disease Gene Panel
Three hundred and thirty-nine gene-disease pairs related to the relevant CVD phenotypes were identified from the literature, commercial gene panels, and publicly available gene-disease relationship (GDR) databases. One hundred and thirty nine of these 339 gene-disease pairs were included without further assessment based on a definitive or strong (D/S) GDR classification from ClinGen, Illumina Clinical Services Laboratory (ICSL) and groups that use the ClinGen Gene-Disease Validity framework. Forty-five gene-disease pairs were excluded based on a refuted or disputed GDR from ClinGen, ICSL or a group that uses the ClinGen framework. Using an expedited curation methodology (see Supplemental Methods) the remaining 155 gene-disease pairs were assessed and an additional 76 gene-disease pairs were included, yielding a final panel of 215 gene-disease pairs (Supplemental Figure 2, Supplemental Table 2).
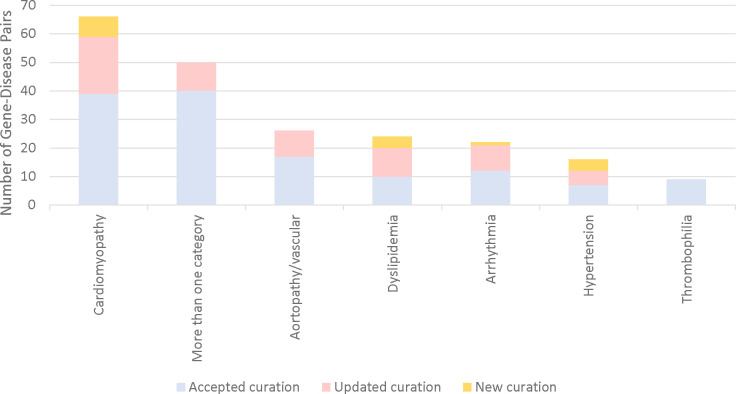
Cardiomyopathy was the most common CVD phenotype amongst the gene-disease pairs (66/215, 30%). Fifty GDRs (23%) were associated with more than one CVD phenotype, the majority of which included cardiomyopathy and arrhythmia in the phenotypic spectrum of disease (36/50). Curation of gene-disease pairs without a publicly available or ICSL GDR classification led to the inclusion of 16 gene-disease pairs, associated with cardiomyopathy (7 genes), dyslipidemia (4 genes), hypertension (4 genes), and arrhythmia (1 genes). Additionally, internal review and recuration of publicly available curations led to the inclusion of 63 gene-disease pairs, representing 29% of the gene-disease pairs on the final panel (Figure 2).
Figure 2. Number of Included Gene-Disease Pairs in Each CVD Phenotype by Curation Source.
All included gene-disease pairs had a GDR classification of strong or definitive based on the ClinGen Gene-Disease Validity Framework. Included gene-disease pairs came from one of three sources: 1) an accepted publicly available or internal curation, 2) an updated publicly available or internal curation or 3) a new internal curation. Updated curations and curations of gene-disease pairs with no current publicly available or internal curation data led to the inclusion of 79 gene-disease pairs on the total 215 gene-disease pair CVD panel.
To support test scalability and to prioritize the return of medically actionable information, variants identified in genes on the CVD gene panel were required to meet a pathogenic or likely pathogenic (P/LP) variant classification threshold for return, consistent with ClinGen guidance supporting return of P/LP variants in gene-disease pairs with a D/S classification31.
Secondary Findings Gene Panel
The American College of Medical Genetics and Genomics (ACMG) recommends testing for a minimum list of gene-disease pairs deemed to be medically actionable in the context of genomic sequencing-based diagnostic tests32. These non-diagnostic results, or ‘secondary findings’, include genes associated with inherited cancer risk, CVD, and metabolic conditions. In alignment with ACMG recommendations, analysis of secondary findings genes on the current version of the ACMG list is included as a component of the TruGenome™ CVD Test. Of the 81 current genes recommended for evaluation, 46 genes were included in the CVD panel, and the remaining 35 non-CVD genes were incorporated for testing as secondary findings. Variants identified in these 35 genes required a classification of P/LP in alignment with reporting of the CVD panel and ACMG recommendations.
Cardiovascular Disease Risk Alleles
A risk allele is defined as a genetic variant, which may or may not be common in the population, that is associated with an increased probability of an individual developing a disease when compared to the baseline risk observed in a control population without the allele19. The evaluation of CVD-associated risk alleles was framed using recommendations from the ClinGen Low Penetrance/Risk Allele Working group19. The risk allele reporting threshold included classification as an Established Risk Allele or an Established Risk Genotype. Relevant risk alleles or genotypes in six genes were identified through a comprehensive literature review, assessment of risk alleles previously evaluated in ICSL in support of a rare undiagnosed disease genome test, and through consultation with medical genetics experts. Two risk alleles and two risk genotypes met criteria for reporting: the Factor V Leiden variant in F5, the prothrombin variant in F2, the E2/E2 genotype in APOE and homozygosity or compound heterozygosity for the G1 and G2 alleles in APOL1. (Table 1). Heterozygosity for the E2 or E4 allele in APOE and heterozygosity of the c.253G>A p.(Asp85Asn) variant in KCNE1 were evaluated and did not meet the threshold for reporting based on the absence of replicated case-control studies supporting an associated >2-fold increased risk of CVD and long-QT syndrome, respectively.
Table 1.
Cardiovascular Disease Risk Alleles
| Gene | Allele/Genotype | Associated Risk | Classification | Outcome | Supporting Evidence |
|---|---|---|---|---|---|
| F5 | Heterozygosity or homozygosity for c.1601G>A (p.Arg534Gln) | Thrombophilia susceptibility | Established Risk Allele | Yes | Odds ratio >2 for increased risk for thromboembolism Replication across studies |
| F2 | Heterozygosity or homozygosity for c.*97G>A variant | Thrombophilia susceptibility | Established Risk Allele | Yes | Odds ratio >2 for increased risk for thromboembolism Replication across studies |
| APOE | Homozygosity for E2 allele [rs429358 (T:T) and rs7412 (T:T)] | Familial dysbetalipoproteinemia | Established Risk Genotype | Yes | Odds ratio >2 for increased risk for cardiovascular disease Replication across studies |
| APOL1 | Homozygosity or compound heterozygosity for G1 and G2 alleles | Chronic kidney disease | Established Risk Genotype | Yes | Odds ratio >2 for increased risk for chronic kidney disease with associated hypertension Replication across studies |
Clinical Pharmacogenomic Report
PGx variants endorsed by the FDA to have a potential impact on therapeutic management recommendations, safety or response, or pharmacokinetic properties20 and with published Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines21 as of May 2022, were reviewed for evidence of clinical and functional impact. Guidance from the Association for Medical Pathology (AMP) on allele selection was also reviewed22. Variants in ten genes associated with a total of 38 drugs were validated for inclusion (Table 2). Variants in CYP2C19, CYP2C9, CYP4F2, SLCO1B1, and VKORC1 are associated with drugs with implications for the management of CVD conditions. The remaining genes are associated with drugs that may be prescribed to individuals with cardiovascular events, for example, immunosuppressants following organ transplantation, antidepressants, and pain medications. Chemotherapy, HIV treatment, and proton pump inhibitors were also included. PGx reporting follows ACMG recommendations33.
Table 2.
Gene-Drug Pairs Included in Pharmacogenetic Testing
| Gene | Associated Drug(s) | Variant/Star Allele(s) Included |
|---|---|---|
| CYP2C19 | Citalopram, Clopidogrel, Doxepin, Escitalopram, Lansoprazole, Omeprazole, Pantoprazole, Voriconazole | *2, *3, *4.001, *4.002, *5, *6, *7, *8, *9, *10, *17, *35 |
| CYP2D6 | Amitriptyline, Atomoxetine, Clomipramine, Codeine, Desipramine, Doxepin, Imipramine, Fluvoxamine, Nortriptyline, Paroxetine, Tamoxifen, Tramadol, Trimipramine | *2 ,*3, *4.013, *4, *5, *6, *7, *8, *9, *10, *12, *13, *14, *15, *17, *21, *29, *31, *36, *40, *41, *42, *49, *56, *59, *68, copy number variants, tandem and hybrid variants |
| CYP3A5 | Tacrolimus | *3, *6, *7 |
| CYP2C9 | Celecoxib, Flurbiprofen, Fosphenytoin, Ibuprofen, Meloxicam, Phenytoin, Piroxicam, Warfarin | *2, *3, *5, *6, *8, *11, *12, *13, *14, *15 |
| VKORC1 | Warfarin | c.-1639G>A (rs9923231) |
| CYP4F2 | Warfarin | *3 |
| DPYD | Capecitabine, Fluorouracil | c.[1236G>A;1129–5923C>G] (rs56038477; rs75017182), c.1679T>G (rs55886062), c.1905+1G>A (rs3918290), c2846A>T (rs67376798), c.557A>G (rs115232898) |
| HLA-B (*57:01 proxy: HCP5) | Abacavir | n.852T>G (rs2395029) |
| SLCO1B1 | Simvastatin, Atorvastatin, Rosuvastatin | *5 |
| TPMT | Azathioprine, Mercaptopurine, Thioguanine | *2, *3A, *3B, *3C, *4 |
Polygenic Risk Score for Coronary Artery Disease
Comprehensive review of available PRSs, and the evidence supporting their validity and potential clinical utility, supported inclusion of a CAD PRS into the test design (see Supplemental Methods). After comparing the performance metrics and integration feasibility, an ancestry specific CAD PRS developed by Allelica, Inc. was selected for use35. A bespoke CAD PRS test report was developed which reports a binary ‘elevated’ or ‘not elevated’ outcome. The elevated threshold was defined as an estimate that the tested individual is at 2-fold increased risk of developing CAD compared to the remainder of the individuals in their ancestry group. The upper and lower bounds of the confidence interval are also reported to highlight uncertainty in the threshold estimate. Language regarding uncertainty of PRS predictions were included and the importance of genetic variants which may not be included in the PRS calculation, medical history, family history, and lifestyle factors was highlighted (Supplemental Figure 3). Given the uncertainty of the clinical utility of PRS scores the TruGenome™ CVD Test CAD PRS is provided as Research Use Only (RUO) finding, and the associated report highlights that the result is not validated for use for the diagnosis, prevention, or treatment of CAD.
Evaluation of test performance in the 1000 Genomes Project cohort
To ascertain TruGenome™ CVD Test performance, the frequency of findings across the monogenic CVD genes, risk allele, and PGx components was evaluated in 2,594 unrelated individuals from the 1,000 Genomes Project (1KGP) cohort. PRS performance was not assessed as the 1000 Genomes Project data was used in the development of the Allelica multi-ancestry CAD score25. For this analysis, likely reportable variants were defined as rare variants (MAF <1%) in genes on the CVD and/or secondary findings gene panels that have a P/LP ClinVar 2* classification or were predicted to have the functional effect of protein truncation (and thus have a high probability of being classified as LP/P) for which this variant type is an established disease-causing mechanism.
One hundred and thirty-five genomes (6%) had a single likely reportable variant (n= 131 genomes) or two likely reportable variants (n=4 genomes) identified in the nuclear genes associated with autosomal dominant or semi-dominant conditions on the CVD gene panel. No genomes had two likely reportable variants/alleles in a gene associated with autosomal recessive disease. The common TTR c.239C>T p.(Thr80Ile) variant associated with hereditary transthyretin amyloidosis was identified in 28 genomes (1%). Thirty-seven (35%) of the 105 unique variants were P/LP ClinVar 2* variants and 68 (65%) were predicted truncating variants. Likely reportable variants were identified most often in African populations, which was driven by TTR c.239C>T, p.(424G>A) (n=20) and predicted truncating variants (n=17).
Twenty-four genomes (0.9%) had a single likely reportable variant in one of the non-CVD secondary findings genes associated with autosomal dominant or semi-dominant conditions. These findings included 22 unique variants, 83% of which were P/LP ClinVar 2*. Secondary findings were identified in 17 of the 27 1000 Genomes Project population groups, with Northern and Western European (n=6) and African (n=7) populations accounting for nearly all these findings.
A risk allele was identified in 159 genomes (6%) (Supplemental Table 3). The most frequently identified risk alleles were homozygosity or compound heterozygosity for the G1 and G2 alleles in APOL1 (4% of genomes) and heterozygosity for the factor V Leiden variant in F5 (1% of genomes).
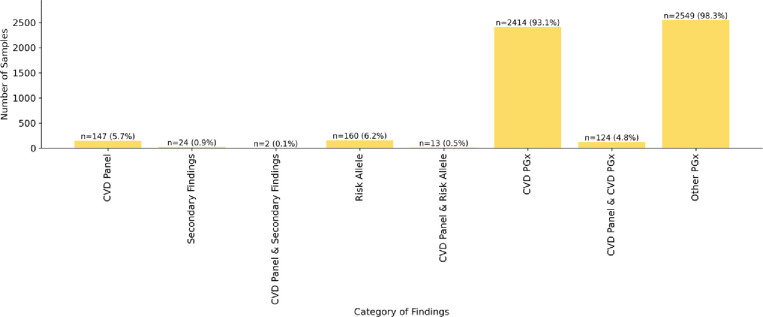
Almost all genomes (2,586, >99%) had at least one PGx finding reported. Each genome had on average of two PGx findings (range 0–5) in genes associated with a drug that has direct CVD implications, and two PGx findings (range 0–5) in the remaining genes on the pharmacogenomic test. Overall, 93% of genomes had one or more PGx finding in a gene associated with a drug with direct CVD implications (Figure 3 and Supplemental Table 3).
Figure 3. Percent of Genomes from an Unselected Population with Genetic Disease, Genetic Risk and PGx Findings in the TruGenome™ CVD Test.
To ascertain TruGenome™ CVD Test performance, the frequency of findings across the monogenic disease, risk allele, and PGx components was evaluated in 2,594 unrelated individuals in the 1,000 Genomes Project cohort. Six percent of genomes had a finding in a CVD-associated gene, 6% had a risk allele finding, 0.9% had a non-CVD-associated ACMG secondary finding, and 93% with at least one CVD-associated PGx variant.
TruGenome™ CVD test performance compared to prior genetic testing
TruGenome™ CVD performance was assessed in a blinded cohort of 20 individuals with a clinical diagnosis of a CVD who had previously received genetic testing (Table 3). Most individuals (16/20, 80%) were between the ages of 40–60 years old and approximately half had cardiomyopathy as a component of their CVD phenotype (11/20, 55%).
Table 3.
Age, Phenotypes, and Clinical Testing Details for Samples from Individuals at Risk for Genetic CVD
| Variable | Number of Individuals |
|---|---|
|
| |
| Age (years) | |
| 20–40 | 2 |
| 40–60 | 16 |
| >60 | 2 |
|
| |
| Phenotype(s) | |
| Aortopathy/aneurysms | 5 |
| Arrhythmia only | 1 |
| Cardiomyopathy and arrhythmia | 3 |
| Cardiomyopathy and/or heart failure | 8 |
| Hyperlipidemia | 3 |
|
| |
| Clinical genetic testing | |
| Aortic dysfunction/dilation panel | 5 |
| Cardiomyopathy panel | 7 |
| Comprehensive cardiac panel | 5 |
| Familial hypercholesterolemia panel | 2 |
| Long QT syndrome panel | 1 |
The average number of variants triaged in genes on the CVD panel and in non-CVD secondary findings genes per case was four (range 1 to 10 variants). Of the 77 variants triaged across the 20 cases, most were single nucleotide variants (SNVs; 67/77, 87%) and in genes on the CVD gene panel (59/77, 77%) vs. in non-CVD secondary finding genes.
Time to triage, interpret and report each case, including laboratory director review and sign out, ranged from 9 to 96 minutes. For cases that did not require a curation and without a P/LP variant reported (n=12) the average time was 20 minutes, ranging from 9 to 48 minutes. For those with a P/LP variant that had been previously curated, the range was 19 to 43 minutes (n=3). The range was 58 to 96 minutes for cases with one or more reportable P/LP variants requiring curation (n=5).
Seven P/LP variants were identified in genes on the CVD panel (FBN1, MYBPC3 ×3, SERPINC1, TTN x2) and one P/LP variant was identified in a non-CVD secondary findings gene (RYR1) (Table 4). One or more pharmacogenomic variant in genes associated with drugs determined to have direct CVD implications were identified in all 20 individuals.
Table 4.
Findings in Individuals with a Suspected Diagnosis of Genetic CVD
| Category of Result | Individuals with a Reportable Finding N, (% of Total) |
|---|---|
|
| |
| CVD gene panel | 7, (35) |
|
| |
| CVD risk allele | |
| F5 | 0, (0) |
| F2 | 1, (5)* |
| APOE | 0, (0) |
| APOL1 | 1, (5)** |
|
| |
| Non-CVD ACMG secondary findings | 1, (5) |
|
| |
| Elevated CAD PRS | 0, (0) |
|
| |
| Pharmacogenomic finding(s) | |
| CVD-related | 20, (100) |
| Non-CVD related | 16, (80) |
Heterozygosity for the c.*97G>A risk allele variant in F2 was identified
The APOL1 genotype G1/G2 was identified
All P/LP variants in the CVD gene panel identified on the TruGenome™ CVD test had been previously identified and reported by the clinical genetic testing performed by Greenwood Genetic Center, except a P variant in the SERPINC1 gene which was only detected by TruGenome™ CVD test. Variants in SERPINC1 are associated with antithrombin III deficiency. This individual had a Comprehensive Cardiac NGS Panel clinical test at Greenwood Genetic Center based on a diagnosis of obstructive cardiomyopathy and SERPINC1 was not part of the panel of genes tested.
Discussion
Genetic testing for patients with CVD is substantially underutilized, despite guidelines and established evidence of benefit. While many factors impact access to testing, the development of a comprehensive assay that enables concurrent assessment of multiple CVD-associated genetic factors on a genome backbone may support both laboratory and clinician adoption. This approach reduces the risk of false negative results that can occur when using focused NGS gene panel testing, enables the evaluation of complex CVD presentations which may have multiple genetic drivers, and supports the detection of variants that may not be assessed with other approaches. A broad testing approach also provides the option to identify ancillary genetic risk information including medically actionable secondary findings and pharmacogenomic information. A GS-based CVD genetic risk assessment may address genetic testing inconsistencies, underutilization and underdiagnosis in CVD by providing a single, scalable, multifaceted molecular diagnostic platform.
Gene curation is a necessary, and sometimes overlooked, step in robust clinical test development. The level of evidence supporting the association between a gene-disease pair impacts whether an identified variant in a gene would be expected to increase risk for the associated disease. The gene curation efforts described here expanded the CVD gene panel from 139 gene-disease pairs with previously curated D/S classifications to a total of 215 gene-disease pairs. Clinical laboratories, population genomic studies, and other institutions performing genomic analysis for individuals with CVD may consider utilizing this panel as a resource. Additions and subtractions are expected to be necessary as new information becomes available about these and other genes.
Polygenic risk scores have shown utility for population-based risk stratification, but their applicability to clinical medicine as individual level risk estimates across diverse ancestry groups is under investigation34. Data from a recent studies supports benefits and changes in patient management with the return of combined CAD PRS and monogenic disease risk information35.
However, more evidence is required to understand the clinical utility of the application of PRSs across ancestry groups and to inform best practices for PRS reporting to patients and providers. This ambiguity was a factor in our decision to return the CAD PRS finding as an RUO finding. Evidence generated from clinical trials integrating CAD PRSs as a component of a comprehensive genome-based test may help to fill these gaps and inform future integration of PRSs into clinical practice.
While the intended use of the TruGenome™ CVD Test is for adult patients with a diagnosis of CVD that may be genetic, the test development approach described here can inform the development and implementation of GS-based CVD risk assessment tests for broader populations. The rate of 6% of genomes in a large, unselected population having one or more likely reportable variant in the CVD gene panel aligns with expectations based on disease prevalence and penetrance36. Based on our test performance evaluation in a small cohort of individuals with CVD, the average number of variants requiring triage per case was 4, and the average time for interpretation and reporting for cases without a P/LP variant was 20 minutes. These data suggest that the TruGenome™ CVD Test and similar broad GS based CVD-risk assessment tests are likely amenable for population scale implementation, given that most individuals would not have a P/LP variant identified and therefore would not require time intensive interpretation and reporting.
This work has several limitations. The CVD gene panel content reported here may not represent all relevant gene-disease pairs with a D/S association, as available evidence to evaluate GDRs is continually generated. Similarly, there are likely other CVD risk alleles that could meet the threshold for inclusion that were either not evaluated or did not have sufficient evidence to support inclusion at the time of test development. The rate of genomes with a PGx finding in a gene-drug pair with direct CVD implications was not interpreted in the context of the dosing algorithm for warfarin, which incorporates the alleles tested in CYP2C9, VKORC1 and CYP4F2 in combination to modify warfarin dosing 37. The data from our evaluation of test performance in a cohort of individuals suspected to have genetic CVD was based on a small number of cases and may not be generalizable to other laboratory workflows. Finally, time estimates in the test performance evaluation were based on the use of internally developed software for the reporting of risk alleles, the CAD PRS and PGx findings without manual data review.
GS as a platform for CVD genetic risk assessment in adult patients with CVD phenotypes for which genetic testing is recommended may increase access to relevant and actionable genetic risk information. This approach is scalable, transferrable to other disease areas, and supports the use of genetic data across the lifespan of an individual.
Supplementary Material
Acknowledgments
We thank the ICSL Interpretation and Reporting Team for their support and feedback during test development work.
Sources of Funding
This work was supported by Illumina Inc. David E. Lanfear’s effort is supported in part by grants from the NIH (R01HL132154; P50MD017351).
Non-Standard Abbreviations and Acronyms
- ACMG
American College of Medical Genetics and Genomics
- AMP
Association for Molecular Pathology
- CVD
cardiovascular disease
- GDR
gene-disease relationship
- ICSL
Illumina Clinical Services Laboratory
- MAF
minor allele frequency
- GS
genome sequencing
- P
pathogenic
- LP
likely pathogenic
Footnotes
Disclosures
LMA, AJC, JL, JA, AM, AC, JPT, RR, CMB, KGG, RH, PM, BM, FM, AW, AH-V, ML, DL, DH, SSA, AK, SPS, EM and RJT were employees and/or shareholders of Illumina, Inc. when this work was completed. DEL is a consultant for Abbott Laboratories, ARMGO, Astra Zeneca, Illumina, Janssen, and Martin Pharmaceuticals, and has participated in clinical research from Akros, AstraZeneca, Bayer, Illumina, Janssen, Lilly, and Pfizer, and has a patent (held by Henry Ford Health) for a beta blocker polygenic score.
References
- 1.Cirino AL, Harris S, Lakdawala NK, Michels M, Olivotto I, Day SM, Abrams DJ, Charron P, Caleshu C, Semsarian C, et al. Role of Genetic Testing in Inherited Cardiovascular Disease: A Review. JAMA Cardiol. 2017;2:1153–1160. [DOI] [PubMed] [Google Scholar]
- 2.Gal DB, Morales A, Rojahn S, Callis T, Garcia J, Priest JR, Truty R, Vatta M, Nussbaum RL, Esplin ED, et al. Comprehensive Genetic Testing for Pediatric Hypertrophic Cardiomyopathy Reveals Clinical Management Opportunities and Syndromic Conditions. Pediatr. Cardiol. 2022;43:616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murdock DR, Venner E, Muzny DM, Metcalf GA, Murugan M, Hadley TD, Chander V, de Vries PS, Jia X, Hussain A, et al. Genetic testing in ambulatory cardiology clinics reveals high rate of findings with clinical management implications. Genet. Med. Off. J. Am. Coll. Med. Genet. 2021;23:2404–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reza N, Alford RL, Belmont JW, Marston N. The Expansion of Genetic Testing in Cardiovascular Medicine: Preparing the Cardiology Community for the Changing Landscape. Curr. Cardiol. Rep. 2024;26:135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longoni M, Bhasin K, Ward A, Lee D, Nisson M, Bhatt S, Rodriguez F, Dash R. Real-world utilization of guideline-directed genetic testing in inherited cardiovascular diseases. Front. Cardiovasc. Med. 2023;10:1272433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosseini SM, Kim R, Udupa S, Costain G, Jobling R, Liston E, Jamal SM, Szybowska M, Morel CF, Bowdin S, et al. Reappraisal of Reported Genes for Sudden Arrhythmic Death: Evidence-Based Evaluation of Gene Validity for Brugada Syndrome. Circulation. 2018;138:1195–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kingsmore SF, Cole FS. The Role of Genome Sequencing in Neonatal Intensive Care Units. Annu. Rev. Genomics Hum. Genet. 2022;23:427–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turro E, Astle WJ, Megy K, Gräf S, Greene D, Shamardina O, Allen HL, Sanchis-Juan A, Frontini M, Thys C, et al. Whole-genome sequencing of patients with rare diseases in a national health system. Nature. 2020;583:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musunuru K, Hershberger RE, Day SM, Klinedinst NJ, Landstrom AP, Parikh VN, Prakash S, Semsarian C, Sturm AC, American Heart Association Council on Genomic and Precision Medicine; Council on Arteriosclerosis, Thrombosis and Vascular Biology; Council on Cardiovascular and Stroke Nursing; and Council on Clinical Cardiology. Genetic Testing for Inherited Cardiovascular Diseases: A Scientific Statement From the American Heart Association. Circ. Genomic Precis. Med. 2020;13:e000067. [DOI] [PubMed] [Google Scholar]
- 10.Wilde AAM, Semsarian C, Márquez MF, Shamloo AS, Ackerman MJ, Ashley EA, Sternick EB, Barajas-Martinez H, Behr ER, Bezzina CR, et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the state of genetic testing for cardiac diseases. Eur. Eur. Pacing Arrhythm. Card. Electrophysiol. J. Work. Groups Card. Pacing Arrhythm. Card. Cell. Electrophysiol. Eur. Soc. Cardiol. 2022;24:1307–1367. [Google Scholar]
- 11.Mitochondrial Disease Genes Compendium - 1st Edition | Elsevier Shop [Internet]. [cited 2024 Apr 23];Available from: https://shop.elsevier.com/books/mitochondrial-disease-genes-compendium/falk/978-0-12-820029-2 [Google Scholar]
- 12.Strande NT, Riggs ER, Buchanan AH, Ceyhan-Birsoy O, DiStefano M, Dwight SS, Goldstein J, Ghosh R, Seifert BA, Sneddon TP, et al. Evaluating the Clinical Validity of Gene-Disease Associations: An Evidence-Based Framework Developed by the Clinical Genome Resource. Am. J. Hum. Genet. 2017;100:895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJ, Gripp KW, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993. [cited 2024 Apr 16]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1116/ [Google Scholar]
- 14.Orphanet [Internet]. [cited 2024 Apr 23];Available from: https://www.orpha.net/
- 15.Whiffin N, Minikel E, Walsh R, O’Donnell-Luria AH, Karczewski K, Ing AY, Barton PJR, Funke B, Cook SA, MacArthur D, et al. Using high-resolution variant frequencies to empower clinical genome interpretation. Genet. Med. Off. J. Am. Coll. Med. Genet. 2017;19:1151–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.PLAW-107publ280.pdf [Internet]. [cited 2024 Apr 23];Available from: https://www.congress.gov/107/plaws/publ280/PLAW-107publ280.pdf
- 17.Baldovino S, Moliner AM, Taruscio D, Daina E, Roccatello D. Rare Diseases in Europe: from a Wide to a Local Perspective. Isr. Med. Assoc. J. IMAJ. 2016;18:359–363. [PubMed] [Google Scholar]
- 18.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt RJ, Steeves M, Bayrak-Toydemir P, Benson KA, Coe BP, Conlin LK, Ganapathi M, Garcia J, Gollob MH, Jobanputra V, et al. Recommendations for risk allele evidence curation, classification, and reporting from the ClinGen Low Penetrance/Risk Allele Working Group. Genet. Med. Off. J. Am. Coll. Med. Genet. 2024;26:101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Health C for D and R. Table of Pharmacogenetic Associations. FDA [Internet]. 2022. [cited 2024 Apr 23];Available from: https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations
- 21.Guidelines – CPIC [Internet]. [cited 2024 Apr 23];Available from: https://cpicpgx.org/guidelines/
- 22.Practice Guidelines [Internet]. Assoc. Mol. Pathol. [cited 2024 Apr 23];Available from: https://www.amp.org/clinical-practice/practice-guidelines/
- 23.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50:1219–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J. Am. Coll. Cardiol. 2018;72:1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busby GB, Kulm S, Bolli A, Kintzle J, DiDomenico P, Bottà G. Development and Validation of Multi-Ancestry Polygenic Risk Scores for Coronary Artery Disease. [Google Scholar]
- 26.Lewis ACF, Perez EF, Prince AER, Flaxman HR, Gomez L, Brockman DG, Chandler PD, Kerman BJ, Lebo MS, Smoller JW, et al. Patient and provider perspectives on polygenic risk scores: implications for clinical reporting and utilization. Genome Med. 2022;14:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Index of /vol1/ftp/data_collections/1000G_2504_high_coverage/working/20201028_3202_raw_GT_with_annot [Internet]. [cited 2024 Apr 23];Available from: https://ftp.1000genomes.ebi.ac.uk/vol1/ftp/data_collections/1000G_2504_high_coverage/working/20201028_3202_raw_GT_with_annot/ [Google Scholar]
- 28. https://registry.opendata.aws/ilmn-dragen-1kgp . [Google Scholar]
- 29.Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, Harrison SM, ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI). Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum. Mutat. 2018;39:1517–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts AM, Ware JS, Herman DS, Schafer S, Baksi J, Bick AG, Buchan RJ, Walsh R, John S, Wilkinson S, et al. Integrated allelic, transcriptional, and phenomic dissection of the cardiac effects of titin truncations in health and disease. Sci. Transl. Med. 2015;7:270ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bean LJH, Funke B, Carlston CM, Gannon JL, Kantarci S, Krock BL, Zhang S, Bayrak-Toydemir P, ACMG Laboratory Quality AssuranceCommittee. Diagnostic gene sequencing panels: from design to report-a technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. Off. J. Am. Coll. Med. Genet. 2020;22:453–461. [DOI] [PubMed] [Google Scholar]
- 32.Miller DT, Lee K, Abul-Husn NS, Amendola LM, Brothers K, Chung WK, Gollob MH, Gordon AS, Harrison SM, Hershberger RE, et al. ACMG SF v3.2 list for reporting of secondary findings in clinical exome and genome sequencing: A policy statement of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. Off. J. Am. Coll. Med. Genet. 2023;25:100866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tayeh MK, Gaedigk A, Goetz MP, Klein TE, Lyon E, McMillin GA, Rentas S, Shinawi M, Pratt VM, Scott SA, et al. Clinical pharmacogenomic testing and reporting: A technical standard of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. Off. J. Am. Coll. Med. Genet. 2022;24:759–768. [DOI] [PubMed] [Google Scholar]
- 34.Lennon NJ, Kottyan LC, Kachulis C, Abul-Husn N, Arias J, Belbin G, Below JE, Berndt S, Chung W, Cimino JJ, et al. Selection, optimization, and validation of ten chronic disease polygenic risk scores for clinical implementation in diverse populations. MedRxiv Prepr. Serv. Health Sci. 2023;2023.05.25.23290535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maamari DJ, Brockman DG, Aragam K, Pelletier RC, Folkerts E, Neben CL, Okumura S, Hull LE, Philippakis AA, Natarajan P, et al. Clinical Implementation of Combined Monogenic and Polygenic Risk Disclosure for Coronary Artery Disease. JACC Adv. 2022;1:100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bourfiss M, van Vugt M, Alasiri AI, Ruijsink B, van Setten J, Schmidt AF, Dooijes D, Puyol-Antón E, Velthuis BK, van Tintelen JP, et al. Prevalence and Disease Expression of Pathogenic and Likely Pathogenic Variants Associated With Inherited Cardiomyopathies in the General Population. Circ. Genomic Precis. Med. 2022;15:e003704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JA, Caudle KE, Gong L, Whirl-Carrillo M, Stein CM, Scott SA, Lee MT, Gage BF, Kimmel SE, Perera MA, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin. Pharmacol. Ther. 2017;102:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.