Abstract
Endothelial cells play critical roles in the regulation of vascular tone, immunity, coagulation, and permeability. Endothelial dysfunction occurs in medical conditions including diabetes, atherosclerosis, sepsis, and acute lung injury. A reliable and reproducible method to isolate pure endothelial cells from mice is needed to investigate the role of endothelial cells in the pathogenesis of these and other conditions. In this protocol, lung microvascular endothelial cells were prepared from 5–7 day old neonatal mouse pups. Lungs are harvested, minced, and enzymatically digested with collagenase I, and released cells are cultured overnight. Endothelial cells are then selected using anti-PECAM1 (CD-31) IgG conjugated to magnetic beads, and cells again are cultured to confluence. A secondary cell selection then occurs with anti-ICAM2 (CD-102) IgG conjugated to magnetic beads to increase the purity of the endothelial cells, and the cells again are cultured to confluence. The entire process takes approximately 7–10 days before the cells can be used for experimentation. This simple protocol yields highly pure (purity >92%) endothelial cells that can be immediately used for in vitro studies, including the studies focused on endothelial cytokine and chemokine production, leukocyte-endothelial interactions, endothelial coagulation pathways, and endothelial permeability. With many knockouts and transgenic mouse lines available, this procedure also lends itself to understanding the function of specific genes expressed by endothelial cells in healthy and pathologic responses to injury, infection, and inflammation.
Introduction
Interest in studying the vascular endothelium has grown recently, as dysfunction of microvascular endothelial cells occurs in multiple human diseases, such as stroke, cardiovascular disease, diabetes, acute lung injury, sepsis, and non-infectious injuries1 , 2 , 3 , 4 , 5 . To define the pathogenesis of these conditions and understand the roles of specific genes in protective and dysregulated endothelial cell responses, there is a need for reliable methods to isolate and culture high purity microvascular endothelial cells from mice. While many studies utilize human endothelial cells such as human umbilical vein endothelial cells (HUVEC) or human lung microvascular endothelial cells (HMVEC), there are multiple reasons to use mouse endothelial cells to study endothelial function and dysfunction. First, there is considerable heterogeneity in responses of endothelial cells from different human donors, which leads to variability in results between individual experiments6 , 7 , 8 . Second, silencing gene targets in primary human cell lines can also lead to variable protein expression levels9 , 10 . In contrast, there is minimal heterogeneity in the responses of mouse endothelial cells11, assuming the genetic background is the same between study groups. Furthermore, large numbers of mouse endothelial cells can be prepared by pooling lungs from several neonatal mice. These factors allow for more consistent and reproducible results between experiments. Finally, the availability of knockout or transgenic mice also provides for the isolation of cell types lacking proteins of interest.
The considerable challenges with isolating healthy and pure populations of mouse lung endothelial cells using published methods12 , 13 , 14 , 15 , 16 led to developing a more reliable and straightforward method to isolate high-quality mouse lung microvascular endothelial cells. Advantages of the current protocol include using neonatal mice, which are readily available, limiting animal husbandry and housing costs. Additionally, in our experience, the viability of endothelial cells from neonatal mice is substantially higher than endothelial cells prepared from adult mice. Because neonatal mice have small lung tissue, endothelial cells can be harvested by digesting excised lungs into collagenase rather than using tracheal instillation of collagenase, which is recommended for collection from adult mice16. This also reduces the time between euthanizing mice and getting their endothelial cells into cell culture media and the incubator without affecting the purity or yield or the reproducibility of endothelial responses. Lastly, pure cell populations were isolated without flow cytometry, which can damage or lead to lower yields of the endothelial cells14 , 15 , 17 .
The current modified protocol consistently leads to high purity and viable endothelial cells in 7–10 days that can be used immediately for in vitro experiments. Isolating cells directly from knockout animals also minimizes the manipulation of cells before experimentation. These methods could be used to investigate the role of various proteins in endothelial function to discover endothelial-based therapeutic targets for multiple diseases.
Protocol
All animal procedures were approved by the Institutional Animal Care and Use Committee of the University of California San Francisco. Male C57BL/6 neonatal mouse pups (age 5–7 days) were used for the study. The entire protocol takes 7–10 days (Figure 1).
Figure 1: Schematic for isolation of murine lung endothelial cells.

Lungs from 5–7 day old neonatal pups are harvested and minced. Minced tissue was enzymatically digested with collagenase I. Cellular suspension is plated and cultured for 2–3 days. Cells then undergo primary immunobead isolation with CD-31-coated beads and are cultured for another 3–4 days. Second immunobead isolation with CD-102-coated beads before culturing another 2–3 days. Cells are now ready for functional experimentation. The image was created by a web-based illustration tool, Biorender.
1. Preparation of endothelial cell medium
Prepare the basal medium: Dulbecco’s Modified Eagle Medium with 4,500 mg/L of glucose and 4 mM of L-Glutamine (DMEM) supplemented with 20% (v/v) heat-inactivated fetal bovine serum, 100 U/100 μg/mL of penicillin/streptomycin, and 25 mM of HEPES (see Table of Materials). Sterile filter with 22 μM filter. Store basal medium at 4 °C and use it within a month of preparation.
Prepare the complete medium: Basal medium supplemented with 100 μg/mL of heparin, 100 μg/mL of endothelial cell growth supplement, 1x non-essential amino acids, and 1 mM sodium pyruvate (see Table of Materials). Sterile filter with 22 μM filter. Store at 4 °C and use within a month of preparation.
Materials
| Name | Company | Catalog Number | Comments |
|---|---|---|---|
| 50 mL conicals | Corning Life Sciences | 430829 | |
| Accutase Cell Detachment Solution | Innovative Cell Technologies, Inc | AT-104 | |
| BD PrecisionGlide Needle 25 G x 5/8” | Becton Disckinson | 305122 | |
| BioRender.com | BioRender | ||
| Blunt Cannula 15 G x 1-1/2” | Covidien | 8881202314 | |
| CD-102 Rat anti-mouse (3C4 (MIC2/4)) | BD Biosciences | 553326 | |
| CD-31 Rat anti-mouse (MEC 13.3) | BD Biosciences | 557355 | |
| Collagenase, Type 1 | Worthington Biochemical Corp | LS004194 | |
| DMEM, high glucose | Gibco | 11965092 | |
| Dynabeads Sheep Anti-Rat IgG | Invitrogen | 11035 | |
| Endothelial Cell Growth Supplment | EMD Millipore Corp | 02-102 | |
| Falcon cell strainers (70 µm) | Corning Life Sciences | 352350 | |
| Fetal Bovine Serum, heat inactivated | Gibco | 10082-147 | |
| Fine Scissors | Fine Science Tools | 14060-09 | |
| Fine Scissors | Fine Science Tools | 14060-10 | |
| Fisherbrand Disposable Borosilicate Glass Pasteur Pipets | Fisher Scientific | 13-678-20B | |
| FITC Rat Anti-mouse CD102 (3C4) | BD Biosciences | 557444 | |
| FITC Rat IgG2a, κ Isotype Control (R35-95) | BD Biosciences | 553929 | |
| Graefe Forceps | Fine Science Tools | 11050-10 | |
| HBSS, no calcium, no magnesium, no phenol red | Gibco | 14175095 | |
| Heparin Sodium Injections (1,000 Units/mL) | Medline | 0409-2720-02 | |
| Heparin sodium salt from porcine intestinal mucosa | Sigma | H3393 | |
| HEPES (1 M) | Gibco | 15630106 | |
| Nonessential amino acids (100x) | Gibco | 11140050 | |
| PE Rat Anti-mouse CD31 (MEC 13.3) | BD Biosciences | 553373 | |
| PE Rat IgG2a, κ Isotype Control R35-95) | BD Biosciences | 553930 | |
| Penicillin Streptomycin (10,000 U/mL, 10,000 µg/mL) | Gibco | 15140122 | |
| Recombinant Murine IFN-γ | Peprotech | 315-05 | |
| Recombinant Murine IL-1β | Peprotech | 211-11B | |
| Recombinant Murine TNF-α | Peprotech | 315-01A | |
| Round bottom polystyrene test tubes | Corning Life Sciences | 352058 | |
| Semken Forceps | Fine Science Tools | 11008-13 | |
| Sodium pyruvate (100 mM) | Gibco | 11360070 | |
| Stericup Quick Release-GP Sterile Vacuum Filtration System, 250 mL | Millipore | S2GPU02RE | |
| Steriflip-GP Sterile Centrifuge Tube Top Filter Unit | Milipore | SCGP00525 | |
| Sterile syringe (10 mL) | Fisher Scientific | 14-955-459 | |
| Sterile syringe (20 mL) | Fisher Scientific | 14-955-460 | |
| T-75 cell culture flask with vent cap, CellBIND treated | Corning Life Sciences | 3290 |
2. Precoating of anti-rat magnetic beads13
Make 50 mL of bead wash buffer: Phosphate-buffered solution (PBS) supplemented with 0.1% (w/v) bovine serum albumin (BSA). Sterile filter with 22 μM filter and store at 4 °C and use within a month of preparation.
-
Vortex the magnetic beads for a few seconds to resuspend the beads and pipette 50 μL of the beads (2 × 107 beads) into two 1.5 mL microcentrifuge tubes, one for CD-31 and one for CD-102 (see Table of Materials). Add 1 mL of bead wash buffer and mix well.
NOTE: All the mentioned volumes were used to isolate endothelial cells from 3–4 neonatal mouse pups (5–7 days old). CD-31 and CD-102 are the endothelial cell surface markers used for magnetic immunobead selection.
Place the tubes on a magnetic separator and remove the supernatant with a glass Pasteur pipet. Repeat wash 3 times with 1 mL of bead wash buffer each time.
Resuspend beads in original bead volume, 50 μL, with bead wash buffer. Then add 5 μL (2.5 μg) of antibody (CD-31/CD-102) to every 50 μL of beads.
Incubate bead suspension on gentle rotation on an end-over-end rotator overnight at 4 °C or for 3 h at room temperature. Repeat wash 4 times.
Resuspend the immunobeads in the original volume, 50 μL, with bead wash buffer and store at 4 °C and use within 1–2 weeks.
3. Isolation and plating of the lung cells
Prepare Type 1 collagenase solution on the day of isolation: Add 25 mg of Type 1 collagenase to 25 mL of HBSS (see Table of Materials). Incubate at 37 °C for 1 h with gentle rotation and sterile filter using 22 μM filter. Keep solution warm at 37 °C.
Inject 5–7 day old mouse pups intramuscularly with heparin (25 μL/mouse, 1,000 U/mL) 10 min before euthanasia to minimize endothelial cell activation and clot formation14 , 15 .
Euthanize the mouse pup with decapitation using sharp scissors in accordance with the 2020 AVMA Guidelines for Euthanasia. Then, pin the mouse to a dissection board with ventral side up.
-
Spray the carcass with 70% ethanol, then expose the thoracic cavity by first cutting the diaphragm and then cutting superiorly upwards along the entire lateral walls of the chest. Reflecting the rib cage back then exposes the heart and lungs.
NOTE: Use smaller forceps and scissors for this step given the small size of neonatal mice, and ensure not to pierce the heart or lungs as this will lead to inadequate blood removal from the lungs.
Attach a 25 G needle to a 10 mL syringe, and inject 5 mL of cold DMEM into the heart’s right ventricle to remove blood from the lungs. The lungs will turn white.
Remove the lungs from the thoracic cavity, one lobe at a time, by cutting the lobes distal to the corresponding bronchi.
Pool all the lung lobes into a 50 mL conical tube prefilled with 20 mL of ice-cold basal medium (from step 1.1).
Repeat steps 3.2–3.7 with the other mouse pups, pooling all lung lobes into the same 50-mL conical tube.
-
Gently agitate the tube by hand for 10–15 s to wash any excess red blood cells visually appreciated on the surface of the lungs.
NOTE: It is not necessary to remove all blood from within or around the lungs; however, this improves purity. Any further tissue handling needs to be done in a sterile hood.
Use a cell strainer to remove the lungs from the media, and transfer the tissue to a sterile, disposable 100 mm diameter tissue culture dish and mince with sterilized scissors.
-
Transfer the minced tissue to the 50 mL conical tube with 25 mL of pre-warmed collagenase solution (step 3.1). Gently agitate on a rotating mixer for 45 min at 37 °C.
NOTE: Overdigestion for even 60 min can lead to lower yields.
-
Attach a 20 mL syringe to a 15 G blunt cannula (see Table of Materials) and triturate the suspension 10–15 times to break up the tissue to a cellular suspension. Be vigorous, but avoid over-frothing.
NOTE: There will no longer be visible pieces of the lung, and instead, the suspension will be cloudy upon completion of this step.
Filter the suspension through a 70 μm cell strainer into a fresh 50 mL conical tube. Further, rinse the conical tube used for the digestion and the cell strainer with an additional 15 mL of basal medium.
Centrifuge the cellular suspension at 400 x g for 8 min at 4 °C.
Remove the supernatant with a glass Pasteur pipet and resuspend the pellet in 2 mL of complete medium. Transfer to a T-75 flask and add 8 mL of complete medium-culture at 37 °C in a humidified, 5% CO2 incubator.
-
The following day, wash flasks twice with 10 mL of Hank’s Balanced Salt Solution without calcium and magnesium (HBSS/CMF) (see Table of Materials) and add 10 mL complete medium.
NOTE: After 2–3 days, the culture will be 90–95% confluent and ready for primary immunobead isolation.
4. Primary immunobead isolation and culturing of endothelial cells
-
Wash adherent endothelial cells twice with 10 mL HBSS/CMF. Add 2 mL of trypsin-free cell detachment solution (see Table of Materials) and incubate at 37 °C for ~5 min. Ensure all the cells have been lifted from the flask.
NOTE: A trypsin-free detachment solution needs to be used instead of trypsin as trypsin can disrupt cell surface adhesion marker CD-31.
Add 8 mL of basal medium to neutralize the cell detachment solution and transfer contents to a 15 mL conical tube. Spin down at 400 x g for 4 min at room temperature.
Remove the supernatant using a glass Pasteur pipet and resuspend cells in 2 mL of basal medium. Transfer the 2 mL cell suspension to a 5 mL round bottom polystyrene tube (see Table of Materials).
Vortex anti-CD-31 coated beads for a few seconds to resuspend the beads and add 30 μL of beads for every 2 mL of cell suspension. Secure the lid.
Incubate tube for 10 min at room temperature on an end-over-end rotator. Then place the tubes on a magnetic separator and leave for 2 min.
Gently using a glass Pasteur pipet, aspirate the supernatant. Remove the tube from the magnet, resuspend the bead-cell pellet by adding 3 mL of basal medium to the tube, and pipette up and down several times.
Replace the tube on the magnetic separator for 2 min, and then carefully aspirate the supernatant using a glass Pasteur pipet.
Repeat washes (step 4.6–4.7) at least 4 times until the supernatant appears clear.
After the final wash, resuspend the bead-cell pellet in 3 mL complete growth medium and transfer it to a T-75 flask. Add 7 mL of the complete growth medium, and incubate at 37 °C in a humidified 5% CO2 incubator.
-
The next day wash cells with HBSS/CMF, then add 10 mL of fresh complete medium. Replace media with 10 mL of fresh complete medium every 2–3 days until 90–95% confluent.
NOTE: Once cells are ~90–95% confluent, which takes ~3–4 days, the culture is ready for secondary immunobead isolation.
5. Secondary immunobead isolation and culturing of endothelial cells
Wash adherent endothelial cells twice with 10 mL of HBSS/CMF. Add 2 mL of trypsin-free cell detachment solution and incubate at 37 °C for ~5 min. Ensure all the cells have been lifted from the flask.
Add 8 mL of basal medium to neutralize cell detachment solution and transfer to a 15 mL conical tube. Spin down at 400 x g for 4 min at room temperature.
Remove the supernatant using a glass Pasteur pipet and resuspend the cells in 2 mL of basal medium. Transfer the 2 mL cell suspension to a 5 mL round bottom polystyrene tube.
Vortex anti-CD-102 coated beads for a few seconds to resuspend the beads and add 30 μL of beads for every 2 mL of cell suspension. Secure the lid.
Incubate the tube for 10 min at room temperature on an end-over-end rotator. Place the tubes on a magnetic separator and leave for 2 min. Then gently aspirate the supernatant using a glass Pasteur pipet.
Remove the tube from the magnet, resuspend the bead-cell pellet by adding 3 mL of basal medium to the tube, and pipette up and down several times.
Replace the tube on the magnetic separator for 2 min before carefully aspirating the supernatant using a glass Pasteur pipet.
Repeat washes (step 5.6–5.7) 4 times or more until the supernatant appears clear.
After the final wash, resuspend the bead-cell pellet in 3 mL complete growth medium and transfer it to a T-75 flask. Add 7 mL complete growth medium, and incubate at 37 °C in a humidified 5% CO2 incubator.
The next day wash cells with HBSS/CMF, then add 10 mL of fresh complete medium. Replace media with 10 mL of fresh complete medium every 2–3 days until 90–95% confluent. Once the endothelial cells are fully confluent, they are ready for experimentation. The cells should not be used beyond passage six as senescence may occur, and other cell types may take over.
6. Confirmation of the endothelial cell surface markers by flow cytometry
After using trypsin-free cell detachment solution to detach the cells, resuspend cell pellet to 1 × 105 cells/mL and aliquot 100 μL of the cell suspension into three 1.5 mL microcentrifuge tubes, labeled 1, 2, and 3.
Wash cells with 1 mL of 1x PBS. Centrifuge at 500 x g for 2 min at 4 °C. Remove wash buffer and repeat twice.
Resuspend the cell pellet in 100 μL of 1x PBS. To tube 1, add 1 μL of conjugated anti-mouse CD-31 (PECAM-1) antibody (ex: phycoerythrin (PE) anti-mouse CD-31 antibody) and 1 μL of conjugated anti-mouse CD-102 (ICAM-2) (ex: fluorescein isothiocyanate (FITC) anti-mouse CD102 antibody) (see Table of Materials). To tube 2, add the appropriate isotype controls. Tube no. 3 will remain unstained.
Incubate the tubes on ice and in the dark for 30–45 min.
Add 1 mL of PBS to each tube, vortex gently, and centrifuge at 500 x g for 2 min at 4 °C. Remove wash and repeat three times. Resuspend the final pellet in 500 μL of PBS.
Keep the tubes covered with aluminum foil at 4 °C until analysis. Analysis needs to be completed within 3–4 h.
-
Acquire flow data using a cytometer. Use an unstained sample as a negative control to properly set laser voltage according to cell autofluorescence. Gate total cell population with a forward scattered plot and side scatter plot.
NOTE: After excluding doublets, CD31+ CD102+ double-positive cells are defined as endothelial cells.
Representative Results
This simple protocol allows one to isolate, culture, and characterize mouse microvascular endothelial cells over 7–10 days (Figure 1). Briefly, lungs are excised and enzymatically digested before plating. Immediately after plating, endothelial cells and other cells will float in cell culture media. By the following day, endothelial cells and the contaminating cells will be adhered to the plate, and a vigorous washing technique is needed to remove debris and the floating cells. Once the cells reach confluence, the first immunobead selection is performed. The CD-31-positive cells then begin to grow in a cobblestone formation (Figure 2). Cells that do not have cobblestone morphology or overgrow are contaminants and not endothelial cells13 , 14 , 15 , 16 .
Figure 2: Morphology of the murine lung endothelial cells.

Cultured murine lung endothelial cells display a cobblestone morphology.
A second immunobead selection is then performed using anti-CD-102 coated beads to increase the purity of endothelial cells, similar to other protocols13 , 16 . Cells that have grown to confluence after second immunobead selection can then be used for experimentation. Fluorescence-activated cell sorting (FACS) is used to confirm that the cell population is CD-31- and CD-102-positive (Figure 3).
Figure 3: Analysis of endothelial cells by flow cytometry.
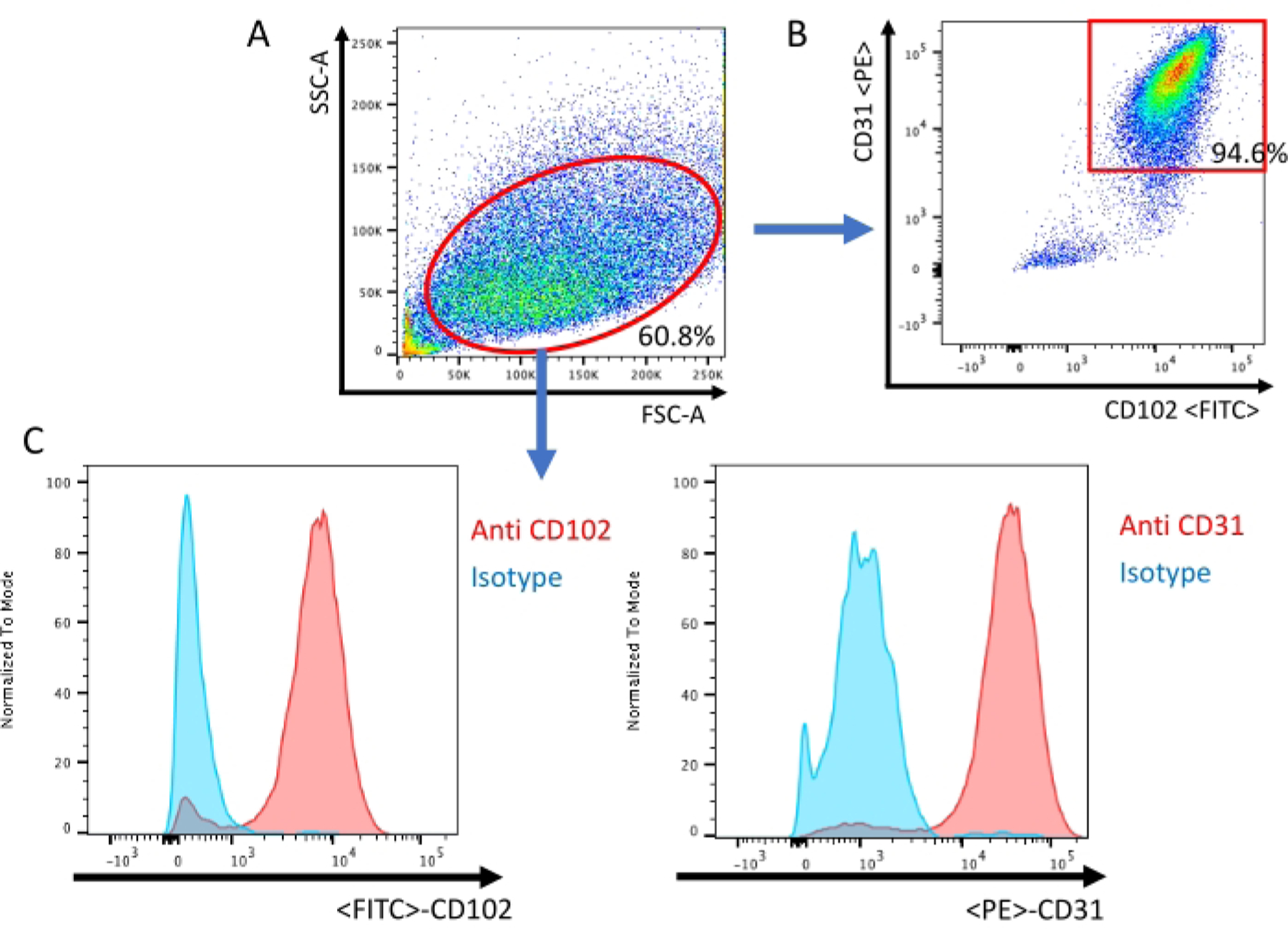
(A) All events are visualized on a dot plot of the Forward Scatter (FSC) and Side Scatter (SSC). (B) Scatter plot confirmation of murine lung endothelial cells being positive for both endothelial-specific cell surface antigens CD-31 and CD-102. (C) Histogram of the isotype sample and sample stained with CD-31/CD-102 specific antibody.
These endothelial cells can then be used for many applications, such as studying endothelial inflammatory pathways and endothelial barrier function. For example, it was observed that treatment with murine cytomix (IFNg, TNFa, and IL1b, each at 10 ng/mL) induced IL-6 production by endothelial cells (Figure 4A). Then Electric Cell-substrate Impedance Sensing (ECIS)18 was used to measure transendothelial resistance (TER) to the flow of electrical current across murine lung endothelial cell monolayers. It was observed that the cells treated with cytomix led to a decrease in TER (Figure 4B), which is consistent with increased endothelial permeability. These examples illustrate how the endothelial cells prepared using this protocol can study various endothelial processes. This allows investigators to more easily study genes of interest in endothelial cells, given the ever-growing numbers of available knockout and transgenic mice.
Figure 4: Cytomix induces murine lung endothelial cell IL-6 production and endothelial cell permeability.
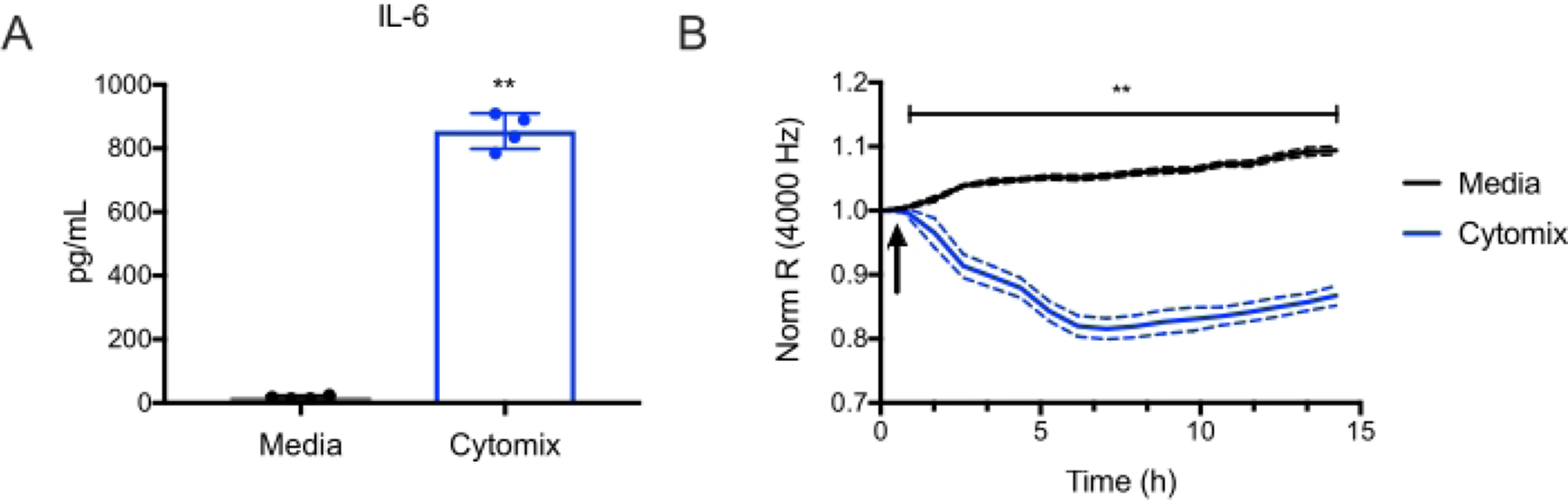
Murine lung microvascular endothelial cells from C57BL/6 mice were treated with cytomix (IFNg, TNFa, and IL1b, each at 10 ng/mL). (A) IL-6 levels were then quantified in culture supernatants at 15 h (**p<0.01, n = 4 wells/condition). (B) Electric Cell-Substrate Impedance Sensing was used to measure the transendothelial resistance (TER) repeatedly over 15 h. Data were normalized to the resistance immediately before the addition of the cytomix as designated and was analyzed by two-way analysis of variance (ANOVA) followed by the Tukey post-test19 (**p<0.01, n = 4 wells/condition).
Discussion
This simple protocol lends itself to high-purity endothelial cells from murine lungs. Although the total time from start to finish is 7–10 days, the hands-on time is around 3–4 h. Compared to other methods13 , 14 , 15 , 16 , the current protocol is streamlined, keeping essential steps without compromising yield and purity.
It is worth noting some critical aspects of this protocol. First, it is important to use neonatal pups rather than adult mice. In our experience, endothelial cells from adult mice do not proliferate as easily as cells from neonatal pups, and they are easily overrun by cells with spindle-like morphology consistent with fibroblasts or mesenchymal stem cells15 , 16 . One possible explanation for the difference is that lung development is in the alveolar stage in neonatal mice, characterized by the more rapid proliferation of endothelial cells20. There may also be some degree of senescence with aging21. Enzymatic digestion time also needs to be precise, as underdigestion can lead to a low yield single-cell suspension. In contrast, over digestion can lead to a substantial amount of cell death. We have determined that the optimal time for collagenase digestion is determined to be 45 minutes. This is then followed by mechanical dissociation with a blunt cannula. Instilling intratracheal collagenase during enzymatic digestion has also been reported14 , 15 ; however, this can be time-consuming and leads to incomplete digestion during the current work. Lastly, some protocols proceed directly to immunobead isolation immediately following enzymatic digestion13 , 16 . We have found that culturing the cells from the dissociated lung in the medium for a day or two before making the immunobead selection substantially increases the yield of viable and highly pure endothelial cells. This may be related to the speed of getting isolated cells into the tissue culture medium, which leads to less cell death in the early stages after removal from the mice, higher initial seeding density, and time for the endothelial cells to adhere and proliferate before the first immunobead isolation.
Limitations of the preparation and analyses of mouse endothelial cells are as follows. Each mouse only provides a limited number of cells that must be used within a few passages. This issue can be overcome by pooling lung digest from several mice. Unfortunately, we are unsuccessful in our efforts to cryopreserve and thaw the cells for future use. Despite these limitations, mouse endothelial cells have some advantages. They can be helpful, particularly in situations where human endothelial cells either cannot be used (e.g., gene knockout), when complementary studies are required to corroborate results with human cells, or when the heterogeneity of responses of endothelial cells from different human donors makes reproducibility between experiments problematic6. The homogenous endothelial cell population from genetically identical mice allows for reproducibility between experiments and the study of specific genes and pathways under stable background conditions.
Mouse endothelial cells can be used for multiple applications to provide insights into endothelial activation and dysfunction in different disease processes. For example, endothelial cells play an integral role in sepsis, contributing to both beneficial and pathological responses through their roles in activating localized and systemic inflammation, promoting leukocyte trafficking and activation in tissues, regulating coagulation, and modulating endothelial permeability2 , 22 , 23. This simple protocol provides viable, functional endothelial cells that can be used in assays for each of these endothelial processes.
Acknowledgments
This work was supported by NIH T32GM008440 (JH/EW) and NIH R01AI058106 (JH). We acknowledge the UCSF Parnassus Flow Cytometry Core (RRID:SCR_018206) supported in part by Grant NIH P30 DK063720 and by the NIH S10 Instrumentation Grant S10 1S10OD021822-01.
Footnotes
A complete version of this article that includes the video component is available at http://dx.doi.org/10.3791/63253.
Disclosures
The authors have nothing to disclose.
References
- 1.Maniatis NA, Orfanos SE The endothelium in acute lung injury/acute respiratory distress syndrome. Current Opinion Critical Care. 14 (1), 22–30 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Khakpour S, Wilhelmsen K, Hellman J Vascular endothelial cell Toll-like receptor pathways in sepsis. Innate Immunity. 21 (8), 827–846 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Tuttolomondo A, Daidone M, Pinto A Endothelial dysfunction and inflammation in ischemic stroke pathogenesis. Current Pharmaceutical Design. 26 (34), 4209–4219 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Roberts AC, Porter KE Cellular and molecular mechanisms of endothelial dysfunction in diabetes. Diabetes and Vascular Disease Research. 10 (6), 472–482 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Widmer RJ, Lerman A Endothelial dysfunction and cardiovascular disease. Global Cardiology Science and Practice. 2014 (3), 291–308 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joffre J et al. Catecholaminergic vasopressors reduce toll-like receptor agonist-induced microvascular endothelial cell permeability but not cytokine production. Critical Care Medicine. 49 (3), e315–e326 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan J, Nunn AD, Thomas R Selective induction of cell adhesion molecules by proinflammatory mediators in human cardiac microvascular endothelial cells in culture. International Journal of Clinical and Experimental Medicine. 3 (4), 315–331 (2010). [PMC free article] [PubMed] [Google Scholar]
- 8.Bouïs D, Hospers GA, Meijer C, Molema G, Mulder NH Endothelium in vitro: A review of human vascular endothelial cell lines for blood vessel-related research. Angiogenesis. 4 (2), 91–102 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Nolte A et al. Optimized basic conditions are essential for successful siRNA transfection into primary endothelial cells. Oligonucleotides. 19 (2), 141–150 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Auguste DT et al. Triggered release of siRNA from poly(ethylene glycol)-protected, pH-dependent liposomes. Journal of Controlled Release. 130 (3), 266–274 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gumkowski F, Kaminska G, Kaminski M, Morrissey LW, Auerbach R Heterogeneity of mouse vascular endothelium. In vitro studies of lymphatic, large blood vessel and microvascular endothelial cells. Blood Vessels. 24 (1–2), 11–23 (1987). [PubMed] [Google Scholar]
- 12.Alphonse RS et al. The isolation and culture of endothelial colony-forming cells from human and rat lungs. Nature Protocols. 10 (11), 1697–1708 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Lim Y-C, Luscinskas FW Isolation and culture of murine heart and lung endothelial cells for in vitro model systems. Methods in Molecular Biology. 341, 141–154 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Cao G, Abraham V, DeLisser HM Isolation of endothelial cells from mouse lung. Current Protocols in Toxicoly. 61 (24.2) 1–9 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Fehrenbach ML, Cao G, Williams JT, Finklestein JM, Delisser HM Isolation of murine lung endothelial cells. American Journal of Physiology - Lung Cellular and Molecular Physiology. 296 (6), L1096–1103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Niu N, Xu S, Jin ZG A simple protocol for isolating mouse lung endothelial cells. Scientific Reports. 9 (1), 1458 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sutermaster BA, Darling EM Considerations for high-yield, high-throughput cell enrichment: fluorescence versus magnetic sorting. Scientific Reports. 9 (1), 227 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szulcek R, Bogaard HJ, van Nieuw Amerongen GP Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. Journal of Visualized Experiments. 85, e51300 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong E et al. ERK1/2 has divergent roles in lps-induced microvascular endothelial cell cytokine production and permeability. Shock. 55 (3), 349–356 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domm W, Misra RS, O’Reilly MA Affect of early life oxygen exposure on proper lung development and response to respiratory viral infections. Frontiers in Medicine (Lausanne). 2, 55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia G, Aroor AR, Jia C, Sowers JR Endothelial cell senescence in aging-related vascular dysfunction. Biochimica et Biophysica Acta - Molecular Basis of Disease. 1865 (7), 1802–1809 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Joffre J, Hellman J, Ince C, Ait-Oufella H Endothelial responses in sepsis. American Journal of Respiratory and Critical Care Medicine. 202 (3), 361–370 (2020). [DOI] [PubMed] [Google Scholar]
- 23.Aird WC The role of the endothelium in severe sepsis and multiple organ dysfunction syndrome. Blood. 101 (10), 3765–3777 (2003). [DOI] [PubMed] [Google Scholar]


