Abstract
In Escherichia coli, gene products of the glp regulon mediate utilization of glycerol and sn-glycerol 3-phosphate. The glpFKX operon encodes glycerol diffusion facilitator, glycerol kinase, and as shown here, a fructose 1,6-bisphosphatase that is distinct from the previously described fbp-encoded enzyme. The purified enzyme was dimeric, dependent on Mn2+ for activity, and exhibited an apparent Km of 35 μM for fructose 1,6-bisphosphate. The enzyme was inhibited by ADP and phosphate and activated by phosphoenolpyruvate.
Growth of Escherichia coli on glycerol or sn-glycerol 3-phosphate (glycerol-P) as the sole carbon source is mediated primarily by the glp regulon (15). The glpFKX operon, one of the five operons of the regulon, encodes glycerol facilitator (glpF), glycerol kinase (glpK), and a protein of unknown function (glpX) (31, 32). It was initially reported that GlpX displays limited sequence similarity to the Synechococcus leopoliensis fructose 1,6-bisphosphatase (FBPase) (31). In our work, a more recent BLAST search revealed that GlpX manifests 39% identity to an FBPase of Synechococcus sp. strain PCC7492 (29). Until now, the only recognized E. coli FBPase was encoded by fbp (25). This FBPase (FBPase I) has only 10% identity to the amino acid sequence of glpX-encoded FBPase (FBPase II). E. coli FBPase I is dependent on Mg2+, is inhibited by low levels of AMP, is tetrameric (1), and is necessary for growth of E. coli on gluconeogenic substrates such as glycerol or succinate (10).
It is not clear why E. coli would maintain two distinct FBPases. FBPases can modulate the concentration of fructose 1,6-bisphosphate [Fru(1,6)P2] and fructose 6-phosphate. These two regulatory hexoses affect glycolysis enzymes 6-phosphofructokinases I and II, pyruvate kinase I, and phosphoenolpyruvate (PEP) carboxylase (2, 4, 13, 20); glycogen synthesis enzyme ADP-glucose pyrophosphorylase (12); and carbon-source import pathway enzymes glycerol kinase and 1-phosphofructokinase (6, 15). Flux through the Embden-Meyerhof pathway in the direction of glycolysis or gluconeogenesis can be allosterically controlled at the enzyme level by other metabolites as well: PEP, ATP, ADP or AMP (9). The potential “futile cycle” of phosphofructokinases and FBPases is also alleviated by this regulation. Therefore, regulation of FBPases is important.
In this communication, the FBPase activity of the glpX gene product is documented. The glpX-encoded enzyme, FBPase II, was purified and characterized, enabling comparison of the attributes of E. coli FBPases in vitro. Further, a chromosomal insertion mutation in glpX was constructed to test the physiological effects of the glpX mutation on carbohydrate metabolism.
E. coli strains and cloning of glpX.
E. coli strains used in this study are listed in Table 1. Strains were grown in Luria broth (LB) supplemented with antibiotics as needed or in minimal medium (7) containing 0.4% glycerol or 0.2% glucose.
TABLE 1.
E. coli strains used in this study
| Strain | Relevant genotype | Description and/or reference |
|---|---|---|
| MG1655 | Wild-type isolate (F−rph-1 λ−) | 3 |
| TL524 | MG1655 Δ(lacZYA–argF)U169 | TcS derivative of TL504 (22) |
| CAG12185 | argE86::Tn10 | 26 |
| CAG12019 | F−zjg-920::Tn10 rph-1 λ− | 26 |
| AM1 | Hfr pfkA2 fhuA22 ompF627 (T2r) fadL701 (T2r) relA1 pit-10 spoT1 rrnB2 mcrB1 creC510 | 21 |
| R6 | F−edd-1 gnd-1 gnt pps-1 ΔargH1 iclR15(Con) | 23 |
| DF657 | HfrC Δfbp287 fhuA22 spoT1 pit-10 relA1 λ−ompF627(T2r) | 25 |
| JB100 | DF657 Δfbp287a zjg-920::Tn10 | P1(CAG12019) into DF657 |
| JB108 | BL21(DE3) zjg920::Tn10 Δfbp287 | P1(JB100) into BL21(DE3) (27) |
| JLD1001b | TL524 glpX::Spcr | This work |
| JLD1101 | TL524 glpX::SpcrargE86::Tn10 | P1(CAG12185) into JLD1001, select Tcr, score Spr |
| JLD1301 | TL524 glpX::SpcrargE86::Tn10 | P1(JLD1101) into TL524 |
| JLD2401 | TL524 glpX::Spcr | P1(JLD1301) into TL524, select Spr, score Tcr |
| JLD2402 | TL524 glpX::Spcr Δfbp287 zjg-920::Tn10 | P1(JB100) into JLD2401 |
| JLD2403 | TL524 glpX::Spcrfbp+zjg-920::Tn10 | P1(JB100) into JLD2401 |
| JLD2404 | TL524 glpX+ Δfbp287 zjg-920::Tn10 | P1(JB100) into TL524 |
| JLD2405 | TL524 glpX+fbp+zjg-920::Tn10 | P1(JB100) into TL524 |
The Δfbp287 deletion affects not only fbp but also the flanking genes yjfF, yjfG, ytfT, and ytfS.
Construction of the glpX insertion mutation: strain JLD1001 was formed by insertion of the Spcr gene into the BsiWI site of glpX, using a plasmid or chromosomal integration selection system (16). pBS-glpX was constructed from SacI/BamHI-digested pBluescript KS(+) (Stratagene) ligated to the SacI/BamHI ∼3,400-bp fragment of pDW23 (32) encompassing the glpK, glpX, and fpr genes. Plasmid pBS-glpX::Spcr was constructed by ligation of SmaI-excised Spcr gene, from pHP45Ω (8), into the filled-in (now blunt) BsiWI site of glpX in pBS-glpX. pKO3-glpX::Spcr was formed by ligation of the SacI (blunted)/SalI piece of pBS-glpX::Spcr with SmaI/SalI-digested pKO3 (16). Plasmid pKO3-glpX::Spcr contains genes glpK, glpX::Spcr, and fpr. From pKO3-glpX::Spcr, glpX::Spcr was integrated into strain TL524 to form strain JLD1001. The integration site was verified by P1 transduction of JLD1001 to Tcr, with strain CAG12185 as the P1 donor, to form strain JLD1101. Comparison of PCR and Southern analyses of DNA of strains JLD2402 and JLD2403, containing the insertional mutation, with JLD2404 and JLD2405 (glpX+), also confirmed integration of the Spcr gene in the glpX gene (data not shown).
The glpX gene was PCR amplified from chromosomal DNA of strain MG1655 using the primer pair acgtgaaTTCCCCTGTGCTACACTTCG (hybridizes to a region 48 bp upstream of the initiation codon) and acgttctagaTTGCCTGTTACCCAATCAGC (hybridizes to a region 102 bp downstream of the termination codon), containing sequence mismatches (lowercase) and restriction sites EcoRI and XbaI (underlined). The PCR product was ligated into XbaI/EcoRI-cut pZE14 (18) forming pJB100. This plasmid was introduced into strain DF657 (Δfbp) to test for complementation of the Fbp− (glycerol-negative) phenotype. DF657(pJB100) was capable of slow growth on glycerol. Therefore, increased expression of glpX from the high-copy-number plasmid pJB100 complements the Fbp− phenotype.
Overexpression and purification of FBPase II.
To facilitate overexpression and purification of FBPase II, a pT7-7 (28) derivative containing glpX was constructed (pJB300B). The NdeI-SalI fragment that was inserted into the same sites of pT7-7 to form pJB300B was obtained by PCR amplification using primers acgtaccaattgaggagatatacaTATGAGACGAGAACTTGCCATC and acgtgtcgacTGCCTTATCTTCGTTCTCCG, with DNA from strain MG1655 as the template (sequence mismatches are lowercase and restriction sites are underlined). The expected nucleotide sequence of glpX in pJB300B was confirmed. Overexpression of glpX was induced in JB108(pJB300B) with 200 μM isopropyl-β-d-thiogalactopyranoside (IPTG) for 2 h in a 200-ml LB culture with an optical density at 600 nm of 0.6. Cells were harvested by centrifugation, resuspended in 2.5 ml of 20 mM Tricine (pH 7.7)–50 mM KCl–1 mM MgCl2–1 mM dithiothreitol (DTT)–0.5 mM EDTA, lysed by sonication, and centrifuged at 140,000 × g for 30 min. Nucleic acids were precipitated by addition of polyethylenimine (0.05%, vol/vol) and centrifugation at 10,000 × g for 20 min. FBPase II activity was precipitated by addition of 0.4 g of (NH4)2SO4 ml−1. The (NH4)2SO4 pellet was dissolved in 2.3 ml of 50 mM Tris-HCl (pH 7.7)–0.2 mM MgCl2–0.1 mM EDTA and fractionated by anion exchange chromatography on a Q HR15 column (Waters) at room temperature using a gradient of 0.075 to 0.5 M NaCl in 50 mM Tris-HCl (pH 7.7)–0.2 mM MgCl2–0.1 mM EDTA. Peak fractions were pooled and precipitated with 0.5 g of (NH4)2SO4 ml−1, redissolved, and then dialyzed against 20 mM Tricine (pH 7.7)–1 mM MgCl2–0.1 mM DTT–15% glycerol. All steps of the purification procedure were monitored by coupled spectrophotometric assay. A total of 2.8 mg of protein was purified 4.4-fold, with a 55% recovery and specific activity of 4.2 U mg−1. Analysis of purified product by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed that the enzyme was >95% pure. Protein concentrations were determined as described by Bradford (5) with bovine serum albumin as the standard.
Optimal conditions for enzyme activity.
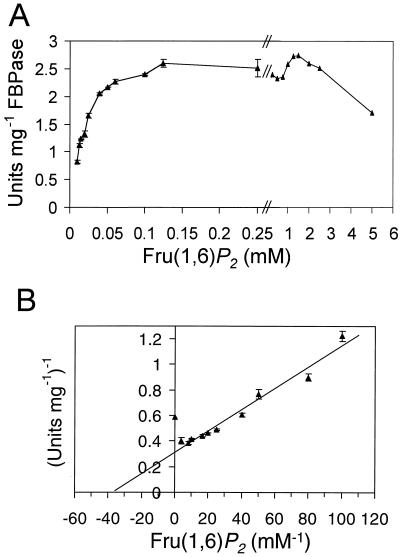
Using a coupled spectrophotometric assay (Fig. 1), the highest activity was obtained with 8 μg of FBPase II in a 1-ml reaction mixture containing 0.5 to 2 mM MnCl2, 50 to 125 mM KCl, 0.02 M Tricine (pH 7.7), and 1.5 mM Fru(1,6)P2. Omission of Mn2+ resulted in >90% loss of activity. Replacing Mn2+ with 1 mM ZnCl2, CaCl2, FeSO4, or CuSO4 resulted in almost complete loss of activity. Replacement with 3 to 10 mM MgCl2 resulted in <10% of the activity present with Mn2+. The presence of 1 mM DTT had no effect on FBPase II activity.
FIG. 1.
(A) Dependence of FBPase II activity on concentration of Fru(1,6)P2. Each data point is presented as an average of two to four determinations, with standard errors (error bars). Coupled spectrophotometric assay was used with a solution containing 8 μg of FBPase II, 0.2 U of phosphoglucose isomerase, 0.4 units of glucose-6-phosphate dehydrogenase, 20 mM Tricine (pH 7.7), 50 mM KCl, 1 mM MnCl2, and 0.25 mM NADP in a final volume of 1 ml at room temperature (24). The rate of reduction of NADP to NADPH was determined by monitoring absorbance at 340 nm. (B) Lineweaver-Burk replot of selected data points from panel A.
Molecular mass determination.
The molecular mass of the GlpX subunit estimated by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis was 40 kDa, which is near the expected size based on the deduced amino acid sequence (36 kDa). Gel filtration chromatography was performed using a Waters Protein Pak Glass 300SW column in 20 mM Tricine (pH 7.7)–100 mM KCl with 1 mM MnCl2 or 1 mM MgCl2. Standards included ribonuclease A (17 kDa), carbonic anhydrase (29 kDa), ovalbumin (45 and 90 kDa), bovine serum albumin (66 and 132 kDa) and phosphorylase b (97.4 kDa). Times of protein elution were determined by monitoring the absorbance at 280 nm. At high, medium, and low concentrations of FBPase II (320, 160, and 40 μg of FBPase II, respectively, in 200 μl of loading volume), the protein eluted at 90, 80, and 47 kDa, respectively. FBPase II is most likely a dimer at higher concentrations, in contrast to the tetrameric fbp-encoded FBPase I (1).
Catalytic properties.
A Km of ∼35 μM and Vmax of ∼3.3 U mg−1 were determined for FBPase II using Fru(1,6)P2 levels below 0.25 mM (Fig. 1). Substrate inhibition was observed at high Fru(1,6)P2 concentrations. FBPase II has lower affinity for Fru(1,6)P2 compared to FBPase I (apparent Km of 5 μM [1]). Based on specific activities of the purified enzymes, the deduced turnover number for FBPase II is about seven times less than that of FBPase I (19).
Substrate specificity of FBPase II.
Substrate specificity was determined by measuring the rate of enzyme-catalyzed production of inorganic phosphate from various potential substrates (see footnote d to Table 2). Fru(1,6)P2 was the best substrate found, while fructose 1-phosphate and ribulose 1,5-bisphosphate served less well as substrates. The apparent Km and Vmax of FBPase II with fructose 1-phosphate were 1 mM and 1.4 U mg−1, respectively, giving a Vmax/Km almost 70 times lower than that obtained using Fru(1,6)P2. Ribulose 1,5-bisphosphate at 1 mM produced 15% of the phosphatase activity obtained with 1.5 mM Fru(1,6)P2. Glucose 6-phosphate, fructose 6-phosphate, mannose 6-phosphate, glucose 1,6-bisphosphate, sedoheptulose 1,7-bisphosphate, sorbitol 6-phosphate, and glycerol-P were not significant substrates (all present at 1 mM [data not shown]).
TABLE 2.
Effectors of FBPase II with Fru(1,6)P2 substrate
| Effector (concn [mM]) | Reaction rate (%) |
|---|---|
| None | 100a |
| KH2PO4 (0.5) | 68a |
| Mn-AMP (1) | 100a |
| Mn-ADP (1.25) | 50b |
| Mn-ADP (1.9) | 38b |
| Mn-ATP (1.25) | 88b |
| Fructose 1-phosphate (2) | 63c |
| PEP (1) | 170d |
Enzyme activity was determined by coupled spectrophotometric assay containing 8 μg of FBPase, 0.05 mM Fru(1,6)P2, and effector. Reaction rates were compared to the rate with no effector added, which was 2.3 U mg−1 for this preparation.
Enzyme activity was determined by coupled spectrophotometric assay containing 8 μg of FBPase, 0.05 mM Fru(1,6)P2, effector, and Mn2+ in 1 mM excess of effector concentration. Reaction rates were compared to the rate with no effector added, which was 1.5 U mg−1 for this preparation.
Enzyme activity was determined as explained in footnote b but with 1 mM Mn2+.
Enzyme activity was determined by inorganic phosphate quantitation assay, containing 2 μg of FBPase, 0.6 mM Fru(1,6)P2, and PEP. Reaction rates were compared to the rate with no effector added, which was 2.3 U mg−1. Reaction mixtures (0.5 ml) containing 20 mM Tricine (pH 7.7), 50 mM KCl, 1 mM MnCl2, Fru(1,6)P2, and enzyme were incubated at room temperature. Aliquots were removed for phosphate determination using the malachite green-ammonium molybdate reagent (14). One unit of enzyme activity is defined as the amount of enzyme catalyzing the formation of 1 μmol of product in 1 min.
Effectors of FBPase II.
Fructose 1-phosphate, inorganic phosphate, ADP, and ATP inhibited FBPase II activity (Table 2). Fructose 1-phosphate and inorganic phosphate inhibited activity competitively, with apparent Kis of 1 and 0.35 mM, respectively. Fructose, fructose 6-phosphate, and glucose 6-phosphate were not inhibitors (data not shown). AMP had no effect on enzyme activity, but ATP and especially ADP inhibited activity at low Fru(1,6)P2 (≤0.1 mM). PEP almost doubled enzyme activity (Table 2), while addition of dihydroxyacetone phosphate, glycerol, or glycerol-P (all at 1 mM) did not affect activity (not shown). The response of FBPase II to effector molecules PEP, ADP, and ATP associates it with many enzymes of the Embden-Meyerhof pathway that are regulated by PEP, Fru(1,6)P2, ATP, ADP, or AMP (9). The properties of FBPase II are distinct from those of FBPase I, an enzyme exquisitely sensitive to inhibition by AMP (50% inhibition by 15 μM AMP [1]). AMP inhibition of FBPase I is alleviated by PEP, although higher concentrations of PEP are inhibitory (>1 mM PEP).
Physiological role of glpX.
Genetic studies were undertaken in an attempt to discern the physiological role of glpX. First, the roles of the two FBPases were investigated using congenic strains with the four possible combinations of wild-type and defective fbp and glpX alleles: JLD2402 (Δfbp glpX::Spcr), JLD2403 (fbp+ glpX::Spcr), JLD2404 (Δfbp glpX+), and JLD2405 (fbp+ glpX+) (Table 1). The fbp+ glpX::Spcr strain grew as well as the wild type on LB or glucose, fructose, succinate, or glycerol minimal medium, aerobically or anaerobically. Apparently, FBPase II is not crucial to cell growth under these conditions when FBPase I is present. Strains lacking FBPase I (e.g., Δfbp glpX+ strains DF657 and JLD2404) grew normally on glucose or fructose medium but were unable to grow on minimal medium supplemented with glycerol or other gluconeogenic substrates, indicating that chromosomal glpX+ does not compensate for the loss of fbp expression. (However, increased expression of glpX from multicopy plasmid pJB100 complemented the Fbp− phenotype.)
The Δfbp glpX+ strain (JLD2404) was able to revert to slow growth on glycerol minimal medium enriched with 0.03% Casamino acids and 2.5 mM KNO3 (with or without O2). These revertants lost the ability to grow on glycerol when transduced to glpX::Spcr. The Δfbp glpX::Spcr strain JLD2402 did not show any reversion under the same conditions. A possible explanation for reversion of Δfbp glpX+ strains to a glycerol-positive phenotype is a mutation yielding elevated glpX expression.
The effect of the glpX and fbp mutations on glycogen accumulation was tested by exposure of the four congenic strains to iodine vapor following growth on Kornberg medium containing 0.4% glycerol instead of glucose (17). Glycogen accumulated to higher levels in both fbp+ strains (maroon colony color) compared to levels found in the Δfbp mutants (orange colony color). Therefore, accumulation of glycogen to wild-type levels requires FBPase I but is not affected by the glpX mutation.
We hypothesized that the inability of Δfbp glpX+ strains to grow on gluconeogenic substrates may be due to low-level expression of glpX combined with competing hexose catabolic pathways that would prevent accumulation of hexose sufficient for cell growth. To test this possibility, the Δfbp allele was introduced by cotransduction with zjg920::Tn10 into a pfkA2 strain deficient in glycolysis (phosphofructokinase I; strain AM1) and into an edd gnd strain deficient in the Entner-Douderoff pathway (6-phosphogluconate dehydrase and 6-phosphogluconate dehydrogenase; strain R6). In both cases, transductants unable to grow on glycerol were obtained at the expected frequency, demonstrating that glpX function is insufficient to provide hexose for growth of these strains where presumed competing pathways are impaired. We did observe that a pfkA glpX::Spcr strain grew more slowly with glucose or glycerol than did the pfkA glpX+ parent strain AM1, indicating a functional importance of FBPase II in this strain.
Three classes of FBPases.
Bacterial FBPases are members of three different orthologous groups (30), including a group of glpX-encoded FBPases (class II). Some bacteria that contain orthologs of FBPase II include Klebsiella aerogenes (91% identity), Yersinia pestis (81%), Haemophilus influenzae (67%), Bacillus subtilis (49%), Clostridium acetobutylicum (45%), Mycobacterium tuberculosis (43%) and Synechococcus sp. strain PCC7942 (39% [29]). There is little similarity of these class II FBPases to the other two orthologous groups of FBPases, those of FBPase I (E. coli) (class I), and those of a very divergent FBPase of B. subtilis (class III [11]). From the sequence data available, no bacterial genome has a combination of class I and class III FBPases, although combinations of classes I and II and of classes II and III are seen.
In conclusion, glpX-encoded FBPase II is still somewhat of an unknown entity in the overall framework of E. coli metabolism. FBPase II is conserved in many other bacteria and is the only known FBPase in some organisms (e.g., M. tuberculosis). The enzyme is distinct in many aspects from FBPase I. However, the specificity and high affinity of FBPase II for substrate and its regulation by PEP and ADP suggest that the enzyme functions with FBPase I in the central pathways of carbohydrate metabolism.
Acknowledgments
We thank G. Church for providing the pKO3 plasmid system and Ali T. van Loo-Bhattacharya for technical support.
This work was supported in part by NSF grant MCB-9118757.
REFERENCES
- 1.Babul J, Guixe V. Fructose bisphosphatase from Escherichia coli. Purification and characterization. Arch Biochem Biophys. 1983;225:944–949. doi: 10.1016/0003-9861(83)90109-1. [DOI] [PubMed] [Google Scholar]
- 2.Blangy D, Buc H, Monod J. Kinetics of the allosteric interactions of phosphofructokinase from Escherichia coli. J Mol Biol. 1968;31:13–35. doi: 10.1016/0022-2836(68)90051-x. [DOI] [PubMed] [Google Scholar]
- 3.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 4.Boiteux A, Markus M, Plesser T, Hess B, Malcovati M. Analysis of progress curves. Interaction of pyruvate kinase from Escherichia coli with fructose 1,6-bisphosphate and calcium ions. Biochem J. 1983;211:631–640. doi: 10.1042/bj2110631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Buschmeier B, Hengstenberg W, Deutscher J. Purification and properties of 1-phosphofructokinase from Escherichia coli. FEMS Microbiol Lett. 1985;29:231–235. [Google Scholar]
- 7.Clark D J, Maaløe O. DNA replication and the division cycle in Escherichia coli. J Mol Biol. 1976;23:99–112. [Google Scholar]
- 8.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 9.Fraenkel D G. Glycolysis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 189–198. [Google Scholar]
- 10.Fraenkel D G, Horecker B L. Fructose-1,6-diphosphatase and acid hexose phosphatase of Escherichia coli. J Bacteriol. 1965;90:837–842. doi: 10.1128/jb.90.4.837-842.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita Y, Yoshida K I, Miwa Y, Yanai N, Nagakawa E, Kasahara Y. Identification and expression of the Bacillus subtilis fructose-1,6-bisphosphatase gene (fbp) J Bacteriol. 1998;180:4309–4313. doi: 10.1128/jb.180.16.4309-4313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Govons S, Gentner N, Greenberg E, Preiss J. Biosynthesis of bacterial glycogen. XI. Kinetic characterization of an altered adenosine diphosphate-glucose synthase from a “glycogen-excess” mutant of Escherichia coli B. J Biol Chem. 1973;248:1731–1740. [PubMed] [Google Scholar]
- 13.Kotlarz D, Buc H. Regulatory properties of phosphofructokinase 2 from Escherichia coli. Eur J Biochem. 1981;117:569–574. doi: 10.1111/j.1432-1033.1981.tb06375.x. [DOI] [PubMed] [Google Scholar]
- 14.Lanzetta P A, Alvarez L J, Reinach P S, Candia O A. An improved assay for nanomole amounts of inorganic phosphate. Anal Biochem. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- 15.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 307–342. [Google Scholar]
- 16.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu M Y, Gui G, Wei B, Preston J F, Oakford L, Yuksel U, Giedroc D P, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 18.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcus F, Edelstein I, Rittenhouse J. Inhibition of Escherichia coli fructose-1,6-bisphosphatase by fructose 2,6-bisphosphate. Biochem Biophys Res Commun. 1984;119:1103–1108. doi: 10.1016/0006-291x(84)90888-x. [DOI] [PubMed] [Google Scholar]
- 20.Morikawa M, Izui K, Taguchi M, Katsuki H. Regulation of Escherichia coli phosphoenolpyruvate carboxylase by multiple effectors in vivo. Estimation of the activities in the cells grown on various compounds. J Biochem (Tokyo) 1980;87:441–449. doi: 10.1093/oxfordjournals.jbchem.a132764. [DOI] [PubMed] [Google Scholar]
- 21.Morrissey A T, Fraenkel D G. Selection of fructose 6-phosphate kinase mutants in Escherichia coli. Biochem Biophys Res Commun. 1968;32:467–473. doi: 10.1016/0006-291x(68)90685-2. [DOI] [PubMed] [Google Scholar]
- 22.Podkovyrov S M, Larson T J. Identification of promoter and stringent regulation of transcription of the fabH, fabD and fabG genes encoding fatty acid biosynthetic enzymes of Escherichia coli. Nucleic Acids Res. 1996;24:1747–1752. doi: 10.1093/nar/24.9.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pouyssegur J M, Faik P, Kornberg H L. Utilization of gluconate by Escherichia coli. Uptake of D-gluconate by a mutant impaired in gluconate kinase activity and by membrane vesicles derived therefrom. Biochem J. 1974;140:193–203. doi: 10.1042/bj1400193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosen O M, Rosen S M, Horecker B L. Fructose-1,6-diphosphatase. Methods Enzymol. 1966;9:632–636. [Google Scholar]
- 25.Sedivy J M, Daldal F, Fraenkel D G. Fructose bisphosphatase of Escherichia coli: cloning of the structural gene (fbp) and preparation of a chromosomal deletion. J Bacteriol. 1984;158:1048–1053. doi: 10.1128/jb.158.3.1048-1053.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer M, Baker T A, Schnitzler G, Deischel S M, Goel M, Dove W, Jaacks K J, Grossman A D, Erickson J W, Gross C A. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev. 1989;53:1–24. doi: 10.1128/mr.53.1.1-24.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 28.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamoi M, Ishikawa T, Takeda T, Shigeoka S. Molecular characterization and resistance to hydrogen peroxide of two fructose-1,6-bisphosphatases from Synechococcus PCC 7942. Arch Biochem Biophys. 1996;334:27–36. doi: 10.1006/abbi.1996.0425. [DOI] [PubMed] [Google Scholar]
- 30.Tatusov R L, Galperin M Y, Natale D A, Koonin E V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Truniger V, Boos W, Sweet G. Molecular analysis of the glpFKX regions of Escherichia coli and Shigella flexneri. J Bacteriol. 1992;174:6981–6991. doi: 10.1128/jb.174.21.6981-6991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weissenborn D L, Wittekindt N, Larson T J. Structure and regulation of the glpFK operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J Biol Chem. 1992;267:6122–6131. [PubMed] [Google Scholar]