Abstract
BTB and TAZ domain proteins (BTs) function as specialized adaptors facilitating substrate recognition of the CUL3–RING ubiquitin ligase (CRL3) complex that targets proteins for ubiquitination in reaction to diverse pressures. Nonetheless, knowledge of the molecular mechanisms by which the apple scaffold protein MdBT2 responds to external and internal signals is limited. Here we demonstrate that a putative Ca 2+ sensor, calmodulin-like 15 (MdCML15), acts as an upstream regulator of MdBT2 to negatively modulate its functions in plasma membrane H+-ATPase regulation and iron deficiency tolerance. MdCML15 was identified to be substantially linked to MdBT2, and to result in the ubiquitination and degradation of the MdBT2 target protein MdbHLH104. Consequently, MdCML15 repressed the MdbHLH104 target, MdAHA8’s expression, reducing levels of a specific membrane H+-ATPase. Finally, the phenotype of transgenic apple plantlets and calli demonstrated that MdCML15 modulates membrane H+-ATPase-produced rhizosphere pH lowering alongside iron homeostasis through an MdCML15–MdBT2–MdbHLH104–MdAHA8 pathway. Our results provide new insights into the relationship between Ca2+ signaling and iron homeostasis.
Introduction
Iron (Fe), an indispensable micronutrient necessary for photosynthetic, respiratory, and hormone synthesis processes and for nitrogen fixation and hemoglobin synthesis [1]. Although Fe is abundant in the soil, it usually exists in the form of insoluble ferric hydroxides, especially in calcareous soil, resulting in the low Fe availability. This restricted Fe availability seriously harms crop yields and human health [2]. To adapt to Fe limitation, plants have been selected to generate two primary strategies for the acquisition of soil-localized Fe. The strategy I response is a reduction-based strategy in non-graminaceous plants, in which Fe3+ is solubilized via H+-ATPase, which acidifies the rhizosphere, and the iron is then reduced to Fe2+ through the action of the ferric chelate reductase known as FERRIC REDUCTION OXIDASE2 (FRO2). Subsequently, Fe2+ is introduced into roots via the metal transporter IRON REGULATED TRANSPORTER1 (IRT1) [3, 4]. The strategy II response is a chelation-based strategy in graminaceous plants, in which Fe3+ is chelated by phytosiderophores (mugineic acids, MAs) to form Fe3+–MA complexes. The complexes are then carried into the root through the transporters known as YELLOW STRIPE1/YELLOW STRIPE1-LIKE (YS1/YSL) [5, 6].
Transcriptional regulation plays an important role in the Fe-deficient responses, and a number of transcription factors (TFs) that modulate a set of Fe-deficient response genes have been identified, particularly the basic helix–loop–helix (bHLH) TFs [7, 8]. In Arabidopsis, the bHLH TF IIIa subgroup FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) is able to form a heterodimer with diverse Ib bHLHs (including bHLH38, bHLH39, bHLH100, and bHLH101), to co-regulate the expression of IRT1 and FRO2 to control Fe uptake and responses [9, 10]. POPEYE (PYE/bHLH47), a IVb subgroup bHLH TF, is a major player in maintaining Fe homeostasis through the inhibition of genes linked to Fe transport as well as homeostasis, such as NICOTIANAMINE SYNTHASE 4 (NAS4) and FRO3 [11]. Four bHLH TFs of the IVc subgroup, including IAA-LEUCINE RESISTANT3 (ILR3, also called bHLH105), bHLH34, bHLH104, and bHLH115, as well as the IVb subgroup bHLH121 are characterized as directly activating the transcription of PYE and bHLH38/39/100/101 involved in regulating the Fe deficiency response [12–17]. In apple (Malus domestica) and citrus, MdbHLH104 and MYB308 regulate rhizosphere acidification and Fe absorption by mediating the expression of plasma membrane Autoinhibited H+-ATPase (AHA) genes [18, 19].
In addition to transcriptional regulation, post-translational modification acting upstream of these TFs in response to Fe deficiency has also been extensively investigated. For instance, the RING E3 ubiquitin ligase named BRUTUS (BTS) and its paralogs, including BTS LIKE1 (BTSL1) and BTSL2 in Arabidopsis, as well as its orthologs HEMERYTHRIN MOTIF-CONTAINING RING AND ZINC-FINGER PROTEIN1 (OsHRZ1) in rice are found to ubiquitinate and degrade bHLH TFs (ILR3 and bHLH115, FIT, and POSITIVE REGULATOR OF IRON HOMEOSTASIS1 (OsPRI1), respectively), thereby negatively regulating Fe deficiency responses [11, 15, 20–23]. In apple, MdbHLH104 was recruited by the BTB/TAZ protein MdBT2 for degradation and sumoylated by the SUMO E3 ligase MdSIZ1, to co-control its stability to regulate PM H+-ATPase-caused rhizosphere pH lowering, as well as Fe absorption in response to Fe deficiency stress [24, 25]. Interestingly, recent studies found that in Malus xiaojinensis, an Fe-efficient apple rootstock, ROS-induced MxMPK6 could phosphorylate MxbHLH104 to promote the Fe deficiency tolerance [26, 27].
BTs belong to the BTB/TAZ subfamily of BTB/POZ proteins, which usually function as substrate-specific adaptors to bind Cullin 3 (CUL3), forming CUL3–RING ubiquitin ligase (CRL3) complexes, and target proteins for ubiquitination [28, 29]. In Arabidopsis, there are five members of the BTB/TAZ subfamily, from BT1 to BT5 [30]. Each member contains a conserved N-terminal BTB domain for interacting with the CUL3 complex, as well as a central TAZ domain for recruiting target proteins, and a C-terminal calmodulin-binding domain (CaMBD), which may be responsible for binding CaMs in a Ca2+-dependent manner [31, 32]. BT2 is able to react to several hormones and stresses, including the introduction of auxin, abscisic acid (ABA), nutrients, and circadian stress [33, 34]. Ca2+ is thought to regulate 35S enhancer-mediated transcription through the CaMBD domain of BT2, suggesting that Ca2+ may be sensed via CaMs in response to diverse signals [35]. When BT2 acts as a downstream target of TELOMERASE ACTIVATOR 1 (TAC1) to regulate telomerase activity, it is activated by increased Ca2+ concentrations [36]. BT2 also interacts with Ca2+-dependent protein kinases (CPKs) such as CPK4 and CPK11 [37], but the effects of these interactions have not been identified. These findings suggest an association between BT2 and Ca2+ signaling; however, the regulatory mechanisms involved are still poorly understood.
In apple, MdBT2 is a key component involved in various activities, such as Fe homeostasis, nitrogen utilization, anthocyanin biosynthesis, malate accumulation, and abiotic stress [38]. Here we searched for the MdBT2 interacting protein in apple through yeast two-hybrid (Y2H) screening, and identified a CaM-like (CML) protein, MdCML15. CaM/CML families contain EF-hand domains that trap Ca2+ and thus transduce calcium signaling, and are widely involved in regulating plant growth and development and coping with environmental stress [39, 40]. Recent studies have shown that MdCaM4 mediates phosphorylation of MdCYTOKININ RESPONSE FACTOR4 (MdCRF4), leading to ubiquitination degradation of MdCRF4, ultimately inhibiting ethylene biosynthesis during apple fruit ripening [41]. In the Fe-efficient rootstock Malus xiaojinensis, the MxMPK4-1 cascade is activated by Ca2+/MxCaM7 signaling, which phosphorylates MxIQM3, thereby promoting the separation of MxIQM3 from MxAHA2 and enhancing H+ secretion under Fe deficiency stress [42]. In this study, we found that MdCML15 negatively regulated Fe deficiency tolerance by promoting MdBT2-mediated MdbHLH104 degradation. Our results provide evidence for the relationship between Ca2+/CaM, BT2, and Fe homeostasis.
Results
CaM-like protein MdCML15 is identified as a putative MdBT2 interaction partner
To determine whether there is an association between BT2 and Ca2+ signaling, we screened the apple cDNA library with Y2H using MdBT2 as bait. A cDNA fragment of MDP0000227098 encoding part of the CaM-like protein was screened out. According to the chromosomal localization nomenclature of MdCMLs [43], the gene was named MdCML15. To know the evolutionary relationships of CML15s in plants, we made a phylogenetic tree and found that MdCML15 is most closely related to CML15s from other Rosaceae plants, followed by PbCML15, PpCML15, and FvCIP8 (Fig. 1A). Reverse transcription–quantitative PCR (RT–qPCR) examination demonstrated that MdCML15 was expressed throughout roots, stems, leaves, flowers, and fruits, with the highest transcript level in roots (Fig. 1B). Further, GUS staining of pMdCML15::GUS transgenic Arabidopsis thaliana showed different expression levels of MdCML15 in various tissues (Supplementary Data Fig. S1). We also detected the subcellular localization of MdCML15, and found that the green fluorescence signal of pMdCML15::GFP-MdCML15 mainly accumulated in the nucleus of apple leaf protoplasts (Fig. 1C).
Figure 1.
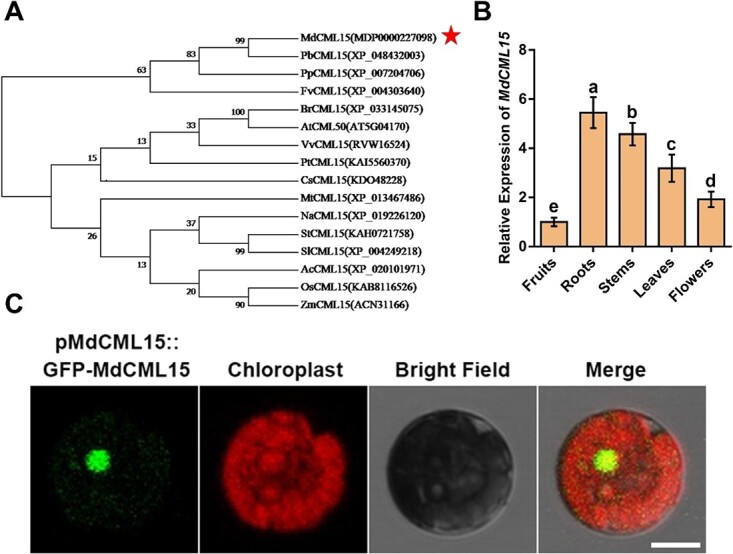
Expression pattern and subcellular localization of MdCML15. A Phylogenetic tree analysis of MdCML15 and 15 other plants’ CML15 protein sequences obtained from the NCBI database. MdCML15 is denoted by the red asterisk. PbCML15, Pyrus bretschneideri; PpCML15, Prunus persica; FvCML15, Fragaria vesca; CsCML15, Citrus spp.; MtCML15, Medicago truncatula; NaCML15, Nicotiana attenuata; SlCML15, Solanum lycopersicum; StCML15, Solanum tuberosum; AcCML15, Ananas comosus; VvCML15, Vitis vinifera; AtCML15, Arabidopsis thaliana; BrCML15, Brassica rapa; PtCML15, Populus trichocarpa; OsCML15, Oryza sativa; ZmCML15, Zea mays. B Expression analysis of MdCML15 in apple roots, stems, leaves, flowers, and fruits. Data are represented as relative to fruits. Results shown are mean ± standard deviation based on three independent biological replicates. Different letters represent significant differences (Tukey’s test, P < 0.05). C Subcellular localization of MdCML15 in GL-3 apple leaf protoplasts. Scale bar = 10 μm.
MdCML15 interacts with MdBT2
MdCML15, a protein that interacts with MdBT2, was selected through Y2H screening. Subsequently, various experiments were performed to verify the contact taking place between MdCML15 and MdBT2. First, we performed a Y2H assay to detect the MdCML15–MdBT2 interaction and its key domains. MdCML15 contains two EF-hand domains (Fig. 2A). Y2H results showed that full-length MdCML15 interacts with MdBT2, but the EF-hand domain alone cannot (Fig. 2B). MdBT2 consists of the N-terminal BTB domain, the middle TAZ domain, and the C-terminal CaMBD domain (Fig. 2C). MdBT2 fragments containing the TAZ and CaMBD domains interact with MdCML15, while MdBT2 fragments lacking the CaMBD domain or the TAZ domain do not (Fig. 2D), suggesting that these two domains were essential for the MdCML15–MdBT2 interaction. In addition to MdBT2, MdCML15 also interacts with MdBT1; however, it does not network with any of MdBT3.1, MdBT3.2, or MdBT4 (Supplementary Data Fig. S2). Moreover, MdBT2 interacts specifically with MdCML subfamily VIII members such as MdCML15, MdCML16, MdCML58 and MdCML60; it does not interact with other MdCMLs (Supplementary Data Fig. S3).
Figure 2.
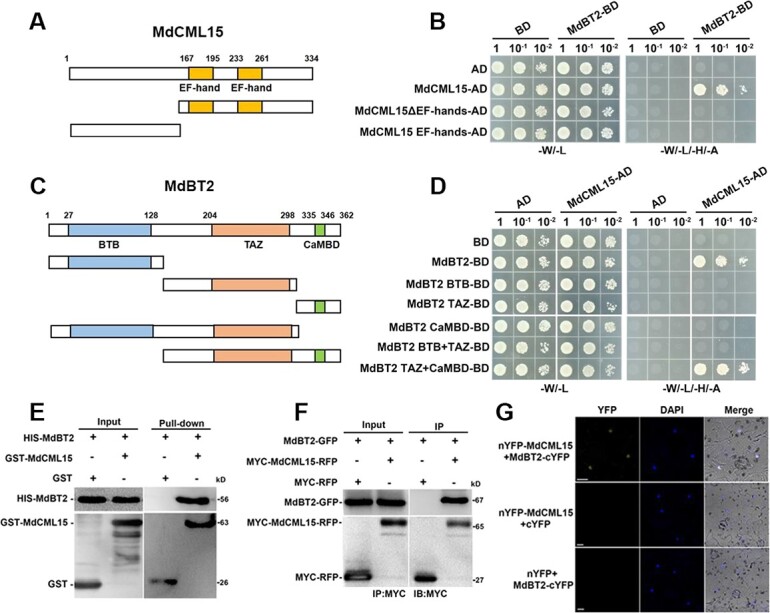
MdCML15 interacts with MdBT2. A Diagram of EF-hand domains in MdCML15 protein. B Y2H analysis between MdCML15 segments and full-length MdBT2. C Diagram of BTB, TAZ, and CaMBD domains in MdBT2 protein. D Y2H analysis between MdBT2 segments and full-length MdCML15. E Pull-down assay of the interaction between MdCML15 and MdBT2. HIS-MdBT2 was incubated with GST-MdCML15 or GST. GST was used as the negative control. F Co-IP assay of the interaction between MdCML15 and MdBT2. Total protein was extracted from 35S::MdBT2-GFP + 35S::MYC-MdCML15-RFP and 35S::MdBT2-GFP + 35S::MYC-RFP transgenic calli. G BiFC assay of the interaction between MdCML15 and MdBT2 in epidermal cells of tobacco leaves. nYFP–MdCML15 + cYFP and nYFP + MdBT2–cYFP served as negative controls. Nuclei are marked with DAPI. Scale bars = 10 μm.
Next, we conducted a pull-down experiment to determine whether MdCML15 physically interacts with MdBT2 in vitro. Prokaryotic-produced GST proteins or GST-MdCML15 recombinant proteins were incubated with HIS–MdBT2 proteins. HIS–MdBT2 was obtained using GST–MdCML15 but not when using GST alone (Fig. 2E), suggesting that MdCML15 is linked to MdBT2 in vitro. For the Co-IP assay, the immunoprecipitated proteins were analyzed with anti-MYC and anti-GFP antibodies. MdBT2–GFP was detected in the 35S::MdBT2-GFP + 35S::MYC-MdCML15-RFP calli but not in the 35S::MdBT2-GFP + 35S::MYC-RFP calli (Fig. 2F), indicating that MdCML15 interacts with MdBT2 in vivo. Then a bimolecular fluorescence complementation (BiFC) assay was performed to confirm the MdCML15–MdBT2 interaction in vivo. As a result, nYFP–MdCML15 + MdBT2–cYFP produced strong yellow fluorescence signal in the nucleus, but no signal was observed in the negative control nYFP–MdCML15 + cYFP or nYFP + MdBT2–cYFP (Fig. 2G), indicating that MdCML15–MdBT2 interaction occurs in the nucleus.
MdCML15 negatively modulates PM H+-ATPase-linked rhizosphere pH lowering and Fe insufficiency tolerance
MdBT2 is involved in regulating PM H+-ATPase activity and Fe deficiency response through its interaction with MdbHLH104, which directly activates the transcription of MdAHA8 [18, 24]. Since MdCML15 interacts with MdBT2, we investigated the role of MdCML15 in response to Fe deficiency. First, we tested whether MdCML15 responds to different iron treatments at transcript and protein levels. RT–qPCR analysis showed that the transcription level of MdCML15 was significantly downregulated after −Fe + Frz treatment (Supplementary Data Fig. S4A). Moreover, we used a cell-free protein degradation assay to detect the abundance of MdCML15 protein. Western blot results showed that, compared with +Fe treatment, −Fe + Frz treatment accelerated the degradation of HIS–MdCML15 protein (Supplementary Data Fig. S4B). In addition, the degradation of MdCML15 was significantly inhibited by the proteasome inhibitor MG132 (Supplementary Data Fig. S4B). Thus, these findings suggest that Fe deficiency inhibits MdCML15 expression at the transcriptional and post-translational levels.
Subsequently, we generated transgenic plants with overexpression of MdCML15 (35S::MdCML15). The expression of MdCML15 in 35S::MdCML15-1#/2#/3# was 7–10 times that of the wild type (WT; Supplementary Data Fig. S5). We treated the 35S::MdCML15-1#/2#/3# transgenic plants with Fe sufficiency or Fe deficiency for 30 days. The pH indicator bromocresol purple visualizes PM H+-ATPase-mediated rhizosphere acidification. In contrast to the WT, the 35S::MdCML15-1#/2#/3# lines exhibited reduced rhizosphere acidification when exposed to Fe-deficient conditions, as evidenced by the less intense yellow hue of the medium surrounding the roots (Fig. 3A). Additionally, all three 35S::MdCML15 lines demonstrated increased rhizosphere pH and decreased activity of the PM H+-ATPase compared with the WT when subjected to Fe deficiency (Fig. 3B and C). We also observed that the young leaves of the 35S::MdCML15-1#/2#/3# lines exhibited more severe chlorosis and lower chlorophyll and Fe contents than those of WT when exposed to Fe deficiency (Fig. 3D–F). Nonetheless, no significant variance was observed between the three 35S::MdCML15 transgenic lines and WT (Fig. 3). These results suggested that MdCML15 negatively regulates Fe deficiency tolerance.
Figure 3.
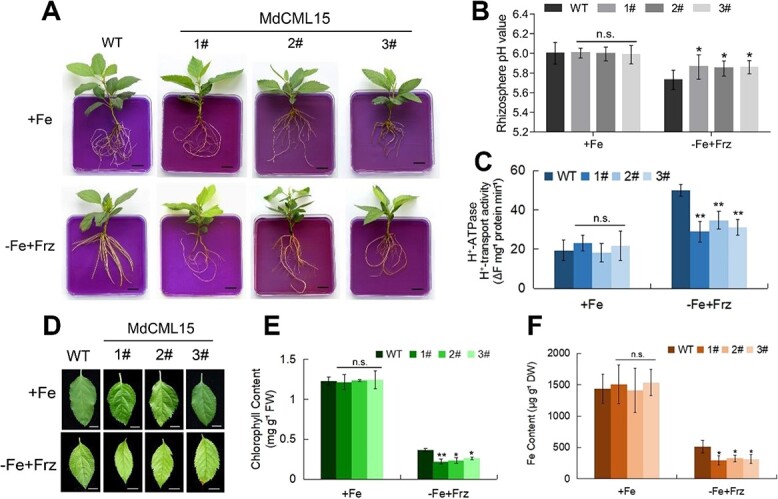
Transgenic apple seedlings overexpressing MdCML15 are more sensitive to Fe deficiency. A Rhizosphere acidification of 30-day-old WT and 35S::MdCML15 transgenic lines given +Fe (Fe-sufficient) or −Fe + Frz (Fe-deficient, without Fe and supplemented with 100 μM ferrozine) treatment for 10 days. The plants were then transferred to a flat containing bromocresol purple for 1 day; the yellow color represents rhizosphere acidification. Scale bars = 1 cm. B Root rhizosphere pH values of WT and 35S::MdCML15 transgenic plants under +Fe or −Fe + Frz treatment for 2 days. C The PM H+-ATPase activity of 30-day-old WT and 35S::MdCML15 transgenic lines treated with +Fe or −Fe + Frz for 10 days. D Young leaves of 30-day-old WT and 35S::MdCML15 transgenic lines after exposure to +Fe or −Fe + Frz medium for 3 weeks. Scale bars = 5 mm. E Total chlorophyll content of the young leaves in (D). FW, fresh weight. F Fe content of the young leaves in (D). DW, dry weight. In (B), (C), (E) and (F), error bars indicate standard deviation of three independent biological replicates. n.s., P > 0.05, *P < 0.05, **P < 0.01.
We also obtained transgenic plants with antisense expression of MdCML15 (35S::asMdCML15). The expression of MdCML15 in 35S::asMdCML15-A1#/A2#/A3# was 40% that of WT (Supplementary Data Fig. S5). Moreover, the transcripts of MdCML16, MdCML60, and MdCML58 homologous genes of MdCML15 did not change significantly in 35S::asMdCML15-A1#/A2#/A3#, indicating that the three 35S::asMdCML15 transgenic lines were specific to MdCML15 (Supplementary Data Fig. S6). The results showed that compared with WT, the 35S::asMdCML15-A1#/A2#/A3# lines displayed more rhizosphere acidification, lower rhizosphere pH and higher PM H+-ATPase activity under Fe deficiency conditions (Fig. 4A–C). Moreover, the three 35S::asMdCML15 lines had less chlorosis and higher chlorophyll and Fe contents than WT when subjected to Fe-deficient conditions (Fig. 4D–F). Nevertheless, no significant variability was observed between the three 35S::asMdCML15 lines and WT under Fe-sufficient conditions (Fig. 4). These results suggested that MdCML15 plays negative roles in Fe deficiency tolerance.
Figure 4.
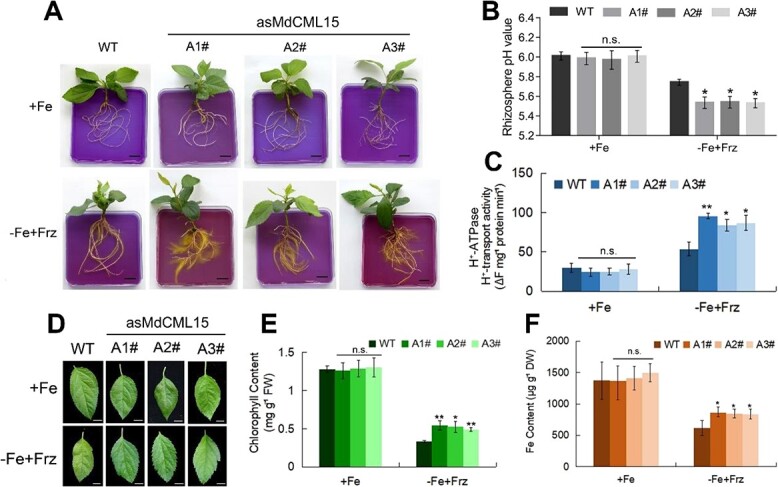
Transgenic apple seedlings antisense-expressing MdCML15 are tolerant to Fe deficiency. A Rhizosphere acidification of 30-day-old WT and 35S::asMdCML15 transgenic lines treated with +Fe or −Fe + Frz for 10 days. The plants were then transferred to a flat containing bromocresol purple for 1 day; the yellow color represents rhizosphere acidification. Scale bars = 1 cm. B Root rhizosphere pH values of WT and 35S::asMdCML15 transgenic plants under +Fe or −Fe + Frz treatment for 2 days. C PM H+-ATPase activity of 30-day-old WT and 35S::asMdCML15 transgenic lines treated with +Fe or −Fe + Frz for 10 days. D Young leaves of 30-day-old WT and 35S::asMdCML15 transgenic lines after exposure to +Fe or −Fe + Frz medium for 3 weeks. Scale bars = 5 mm. E Total chlorophyll content of the young leaves in (D). FW, fresh weight. F Fe content of the young leaves in (D). DW, dry weight. In (B), (C), (E) and (F), error bars indicate standard deviation of three independent biological replicates. n.s., P > 0.05, *P < 0.05, **P < 0.01.
In addition, 35S::MdCML15 and 35S::asMdCML15 transgenic calli were used to further examine the function of MdCML15 (Supplementary Data Fig. S7A). Compared with WT, 35S::MdCML15 overexpression calli exhibited less rhizosphere acidification, lower PM H+-ATPase activity, and lower Fe content under Fe deficiency conditions, while 35S::asMdCML15 suppression calli exhibited the opposite phenotype (Supplementary Data Fig. S7B–E), indicating that MdCML15 negatively modulates the PM H+-ATPase-caused rhizosphere pH lowering as well as Fe insufficiency tolerance.
MdCML15 is involved in MdBT2-mediated degradation of MdbHLH104
In our previous studies, MdBT2 negatively modulated MdbHLH104 protein abundance by mediating its ubiquitination and degradation [24]. We therefore examined whether MdCML15 affects the abundance of MdbHLH104 protein in vitro and in vivo. First, a cell-free degradation assay was carried out. Recombinant HIS−MdbHLH104 protein was incubated with extract from WT, 35S::MdCML15 and 35S::asMdCML15 calli. The findings demonstrated that the HIS−MdbHLH104 protein was broken down more rapidly in 35S::MdCML15 extract, while this process took a longer time in 35S::asMdCML15 extract compared with WT (Fig. 5A and B), indicating that MdCML15 promotes the degradation of MdbHLH104 protein. Further, the degradation of HIS−MdbHLH104 in 35S::MdCML15 extract was inhibited by the proteasome inhibitor MG132 (Fig. 5A and B). Moreover, compared with WT, HIS−MdbHLH104 degraded slower in 35S::asMdBT2 extract (Fig. 5A and B). When MdCML15 was overexpressed in 35S::asMdBT2 calli, the degradation of HIS−MdbHLH104 was slower than that in 35S::MdCML15 calli, similar to that in 35S::asMdBT2 calli (Fig. 5A and B; Supplementary Data Fig. S8A), suggesting that MdCML15-induced MdbHLH104 degradation requires MdBT2 in vitro.
Figure 5.
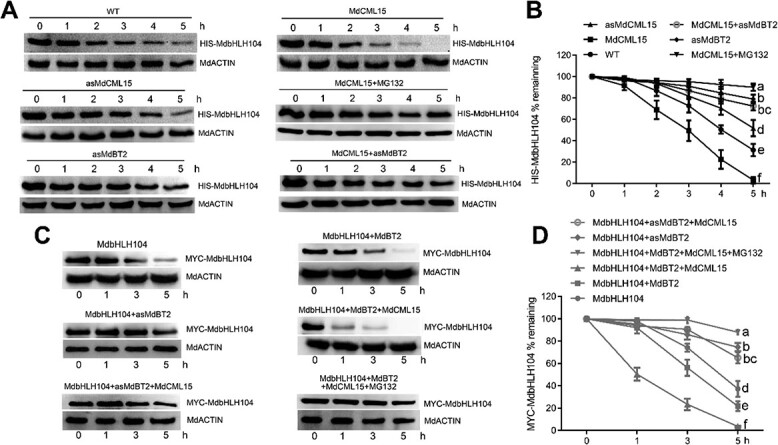
MdBT2 is required for MdCML15-induced MdbHLH104 degradation. A MdBT2 was required for MdCML15-induced MdbHLH104 degradation in vitro. Total proteins from WT, 35S::MdCML15 (MdCML15), 35S::asMdCML15 (asMdCML15), 35S::asMdBT2 (asMdBT2), and 35S::MYC-MdCML15 + 35S::asMdBT2 (MdCML15 + asMdBT2) transgenic calli were incubated with HIS−MdbHLH104 protein. The mixture was collected at the indicated times and detected with anti-HIS antibody. B The abundance of HIS−MdbHLH104 protein in (A) was quantified with Bio-Rad Quantity One software. C MdBT2 was required for MdCML15-induced MdbHLH104 degradation in vivo. MdCML15 was transiently overexpressed in 35S::MYC-MdbHLH104 + 35S::MdBT2 (MdbHLH104 + MdBT2) and 35S::MYC-MdbHLH104 + 35S::asMdBT2 (MdbHLH104 + asMdBT2) transgenic calli. Calli were collected at the indicated times and detected with anti-MYC antibody. D Abundance of MYC−MdbHLH104 protein in (C) was quantified with Bio-Rad Quantity One software. MdACTIN was used as a loading control. In (B) and (D) different letters represent significant differences (Tukey’s test, P < 0.05) and error bars indicate standard deviation of three biological replicates.
To assess the impact of MdCML15 on the levels of MdbHLH104 protein in vivo, we employed a viral vector, pIR, to induce transient overexpression of MdCML15 in calli containing 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP and 35S::MYC-MdbHLH104 + 35S::asMdBT2 (Supplementary Data Fig. S8B). The quantification of MdbHLH104 protein abundance was conducted using an anti-MYC antibody. The findings revealed that MYC–MdbHLH104 protein degradation occurred at a swifter pace in 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP calli compared with 35S::MYC-MdbHLH104 calli. Conversely, the degradation rate was slower in 35S::MYC-MdbHLH104 + 35S::asMdBT2 calli (Fig. 5C and D), signifying that MdBT2 facilitates the in vivo degradation of MdbHLH104 protein. Moreover, MYC−MdbHLH104 degradation was accelerated in 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP + pIR-MdCML15 calli when compared with 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP + pIR calli (Fig. 5C and D), suggesting that MdCML15 enhances the MdBT2-mediated degradation of MdbHLH104 protein. Notably, the degradation of MYC−MdbHLH104 in 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP + pIR-MdCML15 calli was nearly completely inhibited by MG132 (Fig. 5C and D). Additionally, no differences were observed in the degradation of MYC−MdbHLH104 between 35S::MYC-MdbHLH104 + 35S::asMdBT2 + pIR-MdCML15 and 35S::MYC-MdbHLH104 + 35S::asMdBT2 + pIR calli (Fig. 5C and D), indicating that MdBT2 is required for MdCML15-mediated MdbHLH104 degradation in vivo.
MdCML15 promotes MdBT2-mediated MdbHLH104 ubiquitination
To examine whether MdCML15 affects MdBT2-mediated ubiquitination of MdbHLH104 protein, we carried out ubiquitination assays in vitro and in vivo. In a semi-in vitro ubiquitination assay, the purified HIS−MdbHLH104 protein was used as substrate. MdBT2−GFP and MYC−MdCML15 proteins were immunoprecipitated using anti-GFP and anti-MYC antibodies, targeting the proteins in 35S::MdBT2-GFP and 35S::MYC-MdCML15 calli, respectively (Fig. 6A). Subsequently, the ubiquitination of the MdbHLH104 protein was assessed using anti-ubiquitin and anti-HIS antibodies. Western blotting results showed that HIS−MdbHLH104 was ubiquitinated when MdBT2−GFP was present, and strengthened by MYC−MdCML15 (Fig. 6A), indicating that MdCML15 promotes MdBT2-mediated ubiquitination of MdbHLH104 in vitro.
Figure 6.
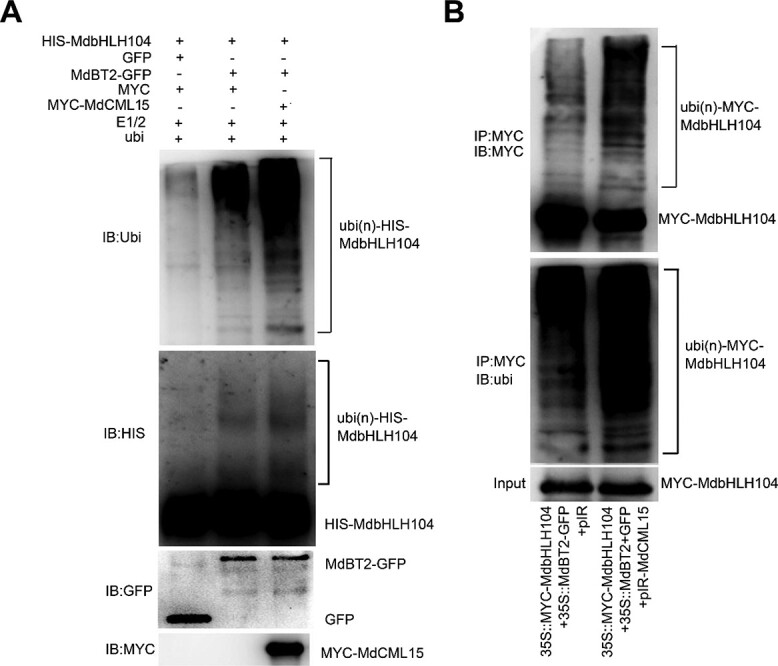
MdCML15 promotes MdBT2-mediated MdbHLH104 ubiquitination. A MdCML15 promoted MdBT2-mediated MdbHLH104 ubiquitination in vitro. HIS−MdbHLH104 protein in a mixture prior to reaction was detected with anti-HIS antibody and used as input. The indicated reaction products were detected with anti-HIS and anti-ubiquitin antibodies, respectively. Ubi(n)-HIS−MdbHLH104 refers to polyubiquitinated HIS−MdbHLH104 protein. B MdCML15 promoted MdBT2-mediated MdbHLH104 ubiquitination in vivo. The viral vector pIR-MdCML15 was transformed into 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP transgenic calli. The MYC−MdbHLH104 protein was immunoprecipitated with anti-MYC antibody and analyzed with anti-MYC and anti-ubiquitin antibodies. The empty vector pIR served as a negative control. Input refers to the MYC−MdbHLH104 protein prior to reaction detection with anti-MYC antibody.
To further detect the effect of MdCML15 on MdBT2-mediated MdbHLH104 ubiquitination in vivo, the viral vector pIR-MdCML15 was rapidly transformed into 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP calli (Supplementary Data Fig. S8B). Subsequently, the MYC−MdbHLH104 protein was subjected to immunoprecipitation using an anti-MYC antibody, with samples collected from both 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP + pIR and 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP + pIR-MdCML15 calli. The ubiquitination of the MdbHLH104 protein was assessed utilizing both anti-MYC and anti-ubiquitin antibodies. The results clearly demonstrated that the ubiquitination level of MYC−MdbHLH104 protein in 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP + pIR-MdCML15 calli was significantly higher compared with that in 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP + pIR calli (Fig. 6B). This observation underscores the role of MdCML15 in enhancing the ubiquitination of MdbHLH104 protein mediated by MdBT2 in vivo.
MdCML15 negatively regulates PM H+-ATPase activity and Fe deficiency tolerance in an MdBT2-dependent manner
MdbHLH104 directly activates MdAHA8 expression by binding to its promoter [18]. Since MdCML15 negatively regulates MdbHLH104 protein stability, we then investigated whether MdCML15 affects the expression of MdAHA8. The results showed that, compared with WT, the transcript level of MdAHA8 was lower in the roots of 35S::MdCML15-1#/2#/3# overexpression plants but higher in 35S::asMdCML15-A1#/A2#/A3# suppression plants in response to Fe deprivation (Fig. 7A). Moreover, we also determined the expression levels of MdIRT1 and MdFRO2, which play key roles in Fe3+ reduction and Fe2+ absorption. RT–qPCR results showed that after iron deficiency treatment MdCML15 could partially inhibit the expression of MdIRT1 and MdFRO2, but the degree of inhibition was smaller than that of MdAHA8 (Supplementary Data Fig. S9). To explore the role of MdBT2 in MdCML15-inhibited MdAHA8 expression, pIR-MdCML15 was transiently transformed into pMdAHA8::GUS and pMdAHA8::GUS + 35S::asMdBT2 calli under Fe deficiency conditions (Supplementary Data Fig. S10A). GUS staining and activity analysis results showed that the MdAHA8 promoter activity was higher in pMdAHA8::GUS + 35S::asMdBT2 calli than in pMdAHA8::GUS calli (Supplementary Data Fig. 7B). Meanwhile, overexpression of MdCML15 inhibited the MdAHA8 promoter activity in pMdAHA8::GUS + pIR-MdCML15 calli, while inhibition of MdBT2 in pMdAHA8::GUS + 35S::asMdBT2 + pIR-MdCML15 calli significantly eliminated this effect (Fig. 7B). These results indicated that MdCML15 negatively regulates MdAHA8 expression in an MdBT2-dependent manner.
Figure 7.
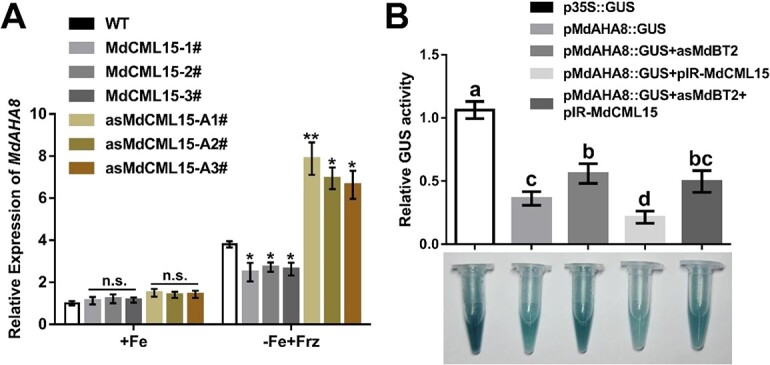
MdCML15-inhibited MdAHA8 expression depends on MdBT2. A RT–qPCR analysis of MdAHA8 in the roots of WT, 35S::MdCML15, and 35S::asMdCML15 transgenic plants. Error bars indicate standard deviation of three biological replicates. n.s., P > 0.05, *P < 0.05, **P < 0.01. B GUS staining and GUS activity analysis of MdAHA8 promoter using pMdAHA8::GUS in calli. The viral vector pIR-MdCML15 was transformed into pMdAHA8::GUS and pMdAHA8::GUS + 35S::asMdBT2 transgenic calli. GUS activity was measured using a 4-methylumbelliferyl-d-glucuronide method. The GUS activity of p35S::GUS was set to 1. Different letters represent significant differences (Tukey’s test, P < 0.05) and error bars indicate standard deviation of three biological replicates.
To investigate the function of MdBT2 in MdCML15 inhibition of the action of PM H+-ATPase and Fe insufficiency tolerance, MdBT2 was repressed within the roots of WT and 35S::MdCML15 transgenic apple plantlets by an Agrobacterium rhizogenes-mediated method to generate 35S::asMdBT2root and 35S::MdCML15shoot/(35S::MdCML15 + 35S::asMdBT2)root plantlets (Supplementary Data Fig. S10B). In response to Fe deprivation, the 35S::asMdBT2root plantlets exhibited lower rhizosphere pH, higher PM H+-ATPase activity, more obvious rhizosphere acidification, higher Fe content, and greener young leaves compared with WT (Fig. 8). Meanwhile, the 35S::MdCML15 plantlets showed the opposite phenotype, such as lower PM H+-ATPase activity, weaker rhizosphere acidification, lower Fe content, and more leaf chlorosis (Figs 3 and 8). However, MdBT2 suppression in 35S::MdCML15shoot/(35S::MdCML15 + 35S::asMdBT2)root transgenic roots exhibited the phenotype of 35S::asMdBT2root plantlets, largely abolishing the effects of MdCML15 (Fig. 8). These results illustrated that MdBT2 is necessary for MdCML15-mediated responses to Fe deficiency.
Figure 8.
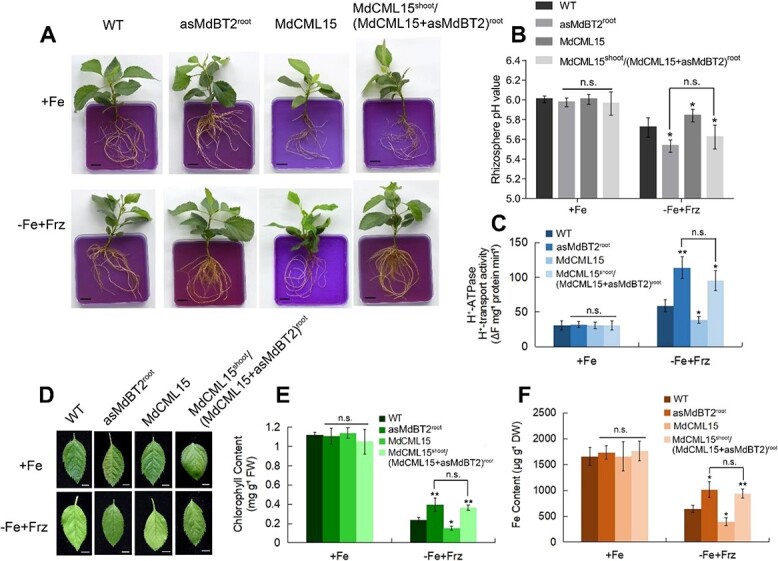
MdCML15-modulated PM H+-ATPase activity and Fe homeostasis requires MdBT2. A Rhizosphere acidification of 1-month-old plants under +Fe or −Fe + Frz conditions for 10 days. Acidification was visualized with bromocresol purple. Scale bars = 1 cm. B Root rhizosphere pH values of plants in (A) under +Fe or −Fe + Frz treatments for 2 days. (C) PM H+-ATPase activity of plants in (A). D Young leaves of 1-month-old plants exposed to +Fe or −Fe + Frz conditions for 20 days. Scale bars = 5 mm. E Total chlorophyll content of the young leaves in (D). FW, fresh weight. F Fe content of the young leaves in (D). DW, dry weight. In (B), (C), (E) and (F), error bars indicate standard deviation of three biological replicates. n.s., P > 0.05, *P < 0.05, **P < 0.01.
Finally, transgenic calli 35S::asMdBT2, 35S::MdCML15, and 35S::MdCML15 + 35S::asMdBT2 were used to further examine the role of MdBT2 in MdCML15 function (Supplementary Data Fig. S11). MdBT2 suppression partially abolished MdCML15 effects related to PM H+-ATPase-governed pH lowering and Fe content in 35S::MdCML15 + 35S::asMdBT2 calli in response to Fe-deficient conditions (Supplementary Data Fig. S11), suggesting that MdBT2 is required for the function of MdCML15.
Discussion
Ca2+ represents a ubiquitous second messenger with roles in various stages of growth and development in plants. Ca2+ signaling is recognized and decoded by Ca2+ sensors and then transmitted to downstream effectors to induce a series of physiological and biochemical processes [39]. Three primary categories of Ca2+ sensors exist, namely (i) calmodulins (CaMs) and calmodulin-like proteins (CMLs), (ii) calcineurin B-like proteins (CBLs), and (iii) calcium-dependent protein kinases (CPKs) [40]. In recent years, there have been a few reports on the mechanism of Ca2+ signaling regulating Fe homeostasis. In Arabidopsis, CPK21 and CPK23 phosphorylate and activate the iron transporter IRT1 to promote Fe absorption [44]. The Ser/Thr protein kinase CBL-INTERACTING PROTEIN KINASE11 (CIPK11), which is activated by Ca2+-CBL1/9, targets and phosphorylates FIT to adjust Fe acquisition [45]. In Malus xiaojinensis, an Fe-efficient apple rootstock, MxCaM7, decodes Ca2+ signaling to interact with MxIQM3, and simultaneously Ca2+ signaling activates the MxMPK4-1 cascade to phosphorylate MxIQM3 at the Ser393 site, thus causing MxIQM3 to dissociate from MxAHA2 and enhancing H+ secretion under Fe deficiency stress [42]. Interestingly, our results revealed that MdCML15 negatively regulates PM H+-ATPase-governed rhizosphere pH lowering and Fe insufficiency tolerance by interacting with MdBT2 and promoting MdBT2-mediated degradation of MdbHLH104 (Figs 2–6). Thus, plants maintain Fe homeostasis through different Ca2+ sensor-mediated regulatory pathways.
The CML proteins typically contain one to six EF-hand domains that bind Ca2+, and participate in diverse life processes, including polar cell growth, flowering time, and tolerance to drought, cold, and nutrient stresses [46, 47]. For example, CML23 and CML24 regulate flowering process, and CML38 regulates root growth in A. thaliana [48, 49], while MtCML10 and MtCML42 modulate cold tolerance in Medicago truncatula [50, 51]. Here we found that MdCML15 maintains Fe homeostasis in apple (Fig. 3), indicating that the functions of CMLs are extensive and diverse in plants. MdCML15 belongs to the VII subfamily of MdCMLs, and other members of the MdCML VIII subfamily, including MdCML16, MdCML58, and MdCML60, also interact with MdBT2 (Supplementary Data Fig. S3), indicating that MdCML15 may exhibit functional redundancy with MdCML16, MdCML58, or MdCML60. However, 35S::asMdCML15 suppression plants showed stronger resistance to Fe deficiency than WT (Fig. 4), suggesting that MdCML15 plays a major role, or that MdCML16, MdCML58, or MdCML60 may regulate other MdBT2-mediated activities.
As a key component of substrate recognition in the CRL3 ubiquitin complex, BTB/TAZ proteins regulate multiple plant growth and development and stress response processes [29]. There are five MdBT members in the apple genome, all of which contain the CaMBD domain [52]. In addition to MdBT2, MdCML15 also interacts with MdBT1 (Supplementary Data Fig. S2). Among them, MdBT2 has been the most widely studied and plays a central role in mediating various environmental and hormonal signals to regulate apple growth and development [38]. For example, MdBT2 regulates Fe homeostasis, anthocyanin biosynthesis, malate accumulation, leaf senescence, plant growth, and nitrogen usage by interacting with MdbHLH104, MdMYB1, MdCIbHLH1, MdMYB73, MdZAT10, MdRGL3a, and MdMYB88/124 proteins, and promoting their ubiquitination and degradation [24, 52–57]. However, the function of the CaMBD domain was not mentioned. In this study, we found that MdCML15 interacts with the CaMBD domain of MdBT2, albeit with the help of the TAZ domain (Fig. 2D), which may be due to the fact that the CaMBD domain is too small in the spatial conformation, containing 12 amino acids, to support the interaction with the entire protein of MdCML15, and therefore requires the assistance of the neighboring domain TAZ. Meanwhile, MdCML15 negatively regulates Fe deficiency tolerance by promoting MdBT2-mediated degradation of MdbHLH104 protein (Figs 2–6). CMLs are Ca2+ sensors and are capable of transducing calcium signaling [39, 40]. Our study not only clarified the role of the CaMBD domain, but also established the relationship between calcium signaling and MdBT2-mediated Fe homeostasis, and even other functions.
Apple stands as a pivotal economic fruit crop worldwide and ranks as one of the most susceptible fruit trees to Fe deficiency. Consequently, investigating the regulatory mechanisms governing Fe homeostasis in apple holds paramount significance. It was found that MdbHLH104 significantly enhanced Fe uptake by transcriptionally activating MdAHA8 expression and promoting H+ efflux [18]. Subsequently, three forms of protein modification for MdbHLH104 were uncovered. MdSIZ1 and MxMPK6 induced by Fe deficiency can sumoylate and phosphorylate MdbHLH104, respectively, and enhance the transcriptional activity of MdbHLH104, thereby promoting Fe absorption and coping with Fe deficiency stress [25, 26]. However, the ubiquitination modification of MdbHLH104 by MdBT2 occurs under Fe-sufficient conditions, where MdBT2 was induced to ubiquitinate and degrade MdbHLH104, preventing excessive Fe from being toxic to plants [24]. Our results suggest that MdCML15 negatively modulates Fe deficiency tolerance by interacting with MdBT2 and promoting MdBT2-mediated degradation of MdbHLH104 (Figs 2–6). Based on the above, we propose a simple model (Fig. 9). When apple roots are exposed to Fe-sufficient conditions, MdCML15 is induced to interact with MdBT2 in the nucleus to accelerate MdBT2-mediated ubiquitination and degradation of MdbHLH104, relieve transcriptional activation of MdAHA8, reduce H+ efflux, and thereby inhibit Fe absorption and avoid Fe toxicity. Our study not only enriched the regulatory network of Fe absorption, but also provided evidence for the link between calcium signaling and Fe homeostasis.
Figure 9.
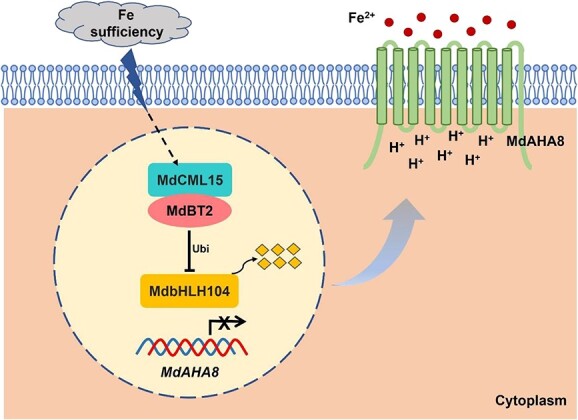
A model of Fe homeostasis regulation by MdCML15–MdBT2–MdbHLH104–MdAHA8 in apple trees. When apple roots are exposed to Fe-sufficient conditions, MdCML15 is induced to interact with MdBT2 in the nucleus to accelerate MdBT2-mediated ubiquitination and degradation of MdbHLH104, relieve transcriptional activation of MdAHA8, reduce H+ efflux, and thereby inhibit Fe absorption and prevent Fe toxicity.
Materials and methods
Plant resources and growth environments
Apple ‘Gala’ tissue cultures were employed as the control or WT plants in this study. These apple plants were cultivated at a temperature of 25°C under long-day conditions (16 h of light and 8 h of darkness) for 1 month. For subculturing, the growth medium employed was Murashige and Skoog (MS) supplemented with 1 mg l−1 of 6-benzylaminopurine (6-BA), 0.2 mg l−1 of naphthylacetic acid (NAA), and 0.5 mg l−1 of gibberellic acid (GA). Once rooted, a 1/2 MS medium supplemented with 1 mg l−1 of indole-3-acetic acid (IAA) was used. ‘Orin’ apple calli were cultured on MS medium supplemented with 1.5 mg l−1 of 2,4-dichlorophenoxyacetic acid (2,4-D) and 0.4 mg l−1 of 6-BA under darkness at 25°C and were subcultured at 3-week intervals.
For Fe starvation treatments, apple plants were transferred after rooting to Fe-sufficient (+Fe) liquid 1/2 MS medium or Fe-deficient (−Fe + Frz) liquid medium (1/2 MS without Fe supplemented with 100 μM ferrozine) for 10 or 20 days. The 3-week-old calli were transferred to +Fe medium (normal calli medium) or −Fe + Frz medium (normal callus medium without Fe supplemented with 100 μM ferrozine) for 15 days.
Vector construction and genetic transformation
To construct MdCML15 and MdBT2 overexpression vectors, the MdCML15 coding sequence (CDS) was linked with PXSN-MYC and PXSN vectors to form 35S::MYC-MdCML15 and 35S::MdCML15, and the MdBT2 CDS was linked with PRI–GFP vector to construct 35S::MdBT2-GFP. To construct the MdCML15 and MdBT2 silencing vectors, 200–300 bp specific antisense fragments of MdCML15 and MdBT2 were linked with PXSN or PRI to generate 35S::asMdCML15 and 35S::asMdBT2, respectively. The promoter fragment (2000 bp) of MdCML15 was connected with 1300-GUS to generate pMdCML15::GUS. Subsequently, the genetic transformation of apple tissue cultures and calli was performed using Agrobacterium tumefaciens containing the recombinant plasmids [58]. Transgenic 35S::MYC-MdbHLH104 and 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP calli were described in Zhao et al. [24].
For transient transformation, MdCML15 CDS was connected with the virus vector pIR [59]. IL-60-BS was used as auxiliary constructor to transform apple callus for 1 week together with pIR-MdCML15 or pIR.
For genetic analysis, the 200–300 bp specific antisense fragment of MdBT2 was cloned into pK7GWIWG2 to generate 35S::asMdBT2. The 35S::asMdBT2 vector was introduced into A. rhizogenes MSU440 and then genetically transformed into transgenic apple plantlets using an MSU440-mediated method [60]. The infected roots were cultivated using MS medium supplemented with 300 mg l−1 cephalosporin for 20 days. Then the transgenic roots were assessed by fluorescence screening, and rooting seedlings were chosen for phenotypic analysis.
Quantitative real-time PCR
Total RNA was extracted using the RNA Plant Plus Reagent (Tiangen, China). Subsequently, 1 μg of RNA was used to synthesize cDNA with the PrimeScript™ first-strand cDNA synthesis kit (Takara, Japan). A qRT–PCR assay was conducted using SYBR Green (Takara, Japan) and an ABI7500 system. The specific primers are detailed in Supplementary Data Table S1.
Phylogenetic tree analysis
The protein sequences from other species were blasted using MdCML15 as bait in the NCBI database (https://www.ncbi.nlm.nih.gov/). The AtCML50 (AT5G04170) protein sequence was downloaded from the Arabidopsis website (http://www.arabidopsis.org/). All protein sequences were multiple sequence aligned and used to construct a phylogenetic tree via the MEGA7 software.
Yeast two-hybrid screening and assays
Y2H screening and Y2H assays were carried out following the manufacturer’s guidelines (Clontech). The complete cDNA sequence of MdBT2, along with different segments of the gene, was cloned into the bait vector pGBT9. Additionally, the full-length cDNAs of MdBT1, MdBT3.1, MdBT3.2, and MdBT4 were also inserted into the bait vector. The complete cDNA alongside the domain deletion of MdCML15, as well as the full-length cDNAs of MdCaM4, MdCML12, MdCML19, MdCML24, MdCML30, MdCML58, MdCML16, MdCML60, MdCML65, MdCML71, and MdCML76, were inserted into the pGAD424 vector.
The Y2H screening experiment was carried out using MdBT2 as bait, and single colonies on −Trp−Leu−His−Ade (−W−L−H−A) medium were sequenced. Potential interacting proteins were identified by blasting the sequencing results. For Y2H assays, the resultant constructs were transformed into yeast, and yeast were grown on −W−L medium and screened using a −W−L−H−A medium.
Pull-down assay
To obtain recombinant protein, the MdCML15 cDNA was introduced into the pET32a vector, while the MdBT2 cDNA was integrated into the pGEX-4 T-1 vector. These constructed plasmids, along with the pGEX-4 T-1 vector itself, were introduced into Escherichia coli BL21 (DE3) for the expression of HIS–MdCML15, GST–MdBT2, and GST proteins, respectively. In the pull-down assay, 6 μg of HIS–MdCML15 was incubated with 10 μg of either GST–MdBT2 or GST. The pull-down procedure was performed using the Pierce GST Spin Purification Kit (Thermo).
Co-immunoprecipitation assay
Co-IP method was according to Kang et al. [61]. 35S::MYC-RFP and 35S::MYC-MdCML15-RFP were transiently transformed into 35S::MdBT2-GFP transgenic calli and cultured for 3 days for the Co-IP experiment. The calli were pretreated with MG132 for 1 h. Total proteins directly detected by anti-MYC and anti-GFP antibodies were used as input. The proteins were immunoprecipitated with an anti-MYC antibody, and then the immunoprecipitated proteins were detected with anti-GFP and anti-MYC antibodies.
Bimolecular fluorescence complementation assay
Full-length CDS sequences of MdCML15 and MdBT2 were inserted into the pSITE–nYFP and pSITE–cYFP vectors to create the nYFP–MdCML15 and MdBT2–cYFP fusions, respectively. The plasmids were then transformed into A. tumefaciens GV3101, and the indicated combinations were transiently co-transformed into Nicotiana benthamiana leaves [61]. The YFP fluorescence was observed after 2-day infiltration with a Zeiss LSM880 high-resolution laser confocal microscope.
Subcellular localization in apple leaf protoplasts
The transient transformation of apple leaf protoplasts was performed by a PEG-mediated method. pMdCML15::GFP-MdCML15 plasmids were transformed into protoplasts of 3-week-old apple tissue culture seedlings. After 16 h of light culture, the GFP fluorescence was observed with a Zeiss LSM880 high-resolution laser confocal microscope.
Rhizosphere pH measurements
The method of rhizosphere pH measurement was according to Sun et al. [62]. Apple seedlings were transferred after rooting to −Fe or −Fe + Frz liquid medium (pH = 6.0) for 2 days. The pH meter was slowly moved closer to the rhizosphere and the pH value was recorded when the value was stable. At least three plants were selected for rhizosphere pH determination.
GUS assay
For GUS histochemical staining, the pMdAHA8::GUS associated calli were submerged in staining buffer (0.075 M sodium phosphate buffer at a pH of 7.0, containing 0.05 mM ferricyanide, 0.05 mM ferrocyanide, 10 mM EDTA, 0.1% Triton X-100, 20% methanol, and 1 mM X-Gal) at a temperature of 37°C for 6 h. The GUS activity of calli was assessed as outlined by Liu et al. [58].
In vitro and in vivo degradation examination
Twenty-day-old transgenic calli grown in normal medium were subjected to extraction using a degradation buffer composed of 25 mM Tris–HCl (pH 7.5), 5 mM DTT, 4 mM PMSF, 10 mM ATP, 10 mM MgCl2, and 10 mM NaCl [63]. These samples were employed for cell-free protein degradation assays. Equal amounts of total proteins were co-incubated with prokaryotic-induced protein at 22°C, and samples were subsequently collected in loading buffer at specified time points.
To conduct degradation assays in vivo, total proteins were extracted from 20-day-old calli cultivated in normal medium and treated with cycloheximide, a protein synthesis inhibitor. Equal protein amounts were incubated at 22°C at the indicated time intervals. For the 26S proteasome inhibition assay, the calli were pretreated with 50 μM MG132, a proteasome inhibitor, for 1 h. Quantification of target proteins was carried out with the corresponding antibodies.
Ubiquitination assays
In order to conduct ubiquitination assays in vivo, 35S::MdBT2-GFP transgenic calli cultured under normal medium were subjected to a 1-h treatment with MG132. The reaction system (30 μl) comprised 200 ng HIS-MdbHLH104, 400 ng MdBT2–GFP or GFP, 500 ng MYC–MdCML15 or MYC, 25 ng UBE1 (E1; Boston Biochem), 50 ng E2 UbcH5b (E2; Boston Biochem), and 1 μg ubiquitin in a ubiquitination buffer (composed of 2 mM DTT, 50 mM Tris–HCl (pH 7.5), 5 mM MgCl2, and 2 mM ATP). This mixture was incubated for 16 h at 30°C. The samples taken before the reaction were assessed using an anti-HIS antibody (input), while the reaction products were subsequently analyzed using anti-HIS and anti-ubiquitin antibodies.
For the ubiquitination assay in vivo, 35S::MYC-MdbHLH104 + 35S::MdBT2-GFP or 35S::MYC-MdbHLH104 + 35S::asMdBT2 calli, which transiently co-expressed pIR or pIR-MdCML15 for 1 week under normal callus medium, were initially pretreated with 50 μM MG132 for 1 h. They were then incubated for 6 h. The MYC–MdbHLH104 proteins were immunoprecipitated using an anti-MYC antibody. Detection of the immunoprecipitated proteins was carried out using anti-MYC and anti-ubiquitin antibodies.
PM H+-ATPase activity and rhizosphere acidification assays
The PM H+-ATPase activity assay was conducted as previously outlined [25]. Calli that were treated with +Fe or −Fe + Frz for 15 days, as well as apple plantlets subjected to with +Fe or −Fe + Frz for 10 days, were utilized to isolate PM. PM proteins were combined with the enzyme reaction buffer (containing 25 mM 1,3-bis [tris (hydroxylmethyl) methylamino] propane-HEPES (pH 6.5), 3 mM MgSO4, 100 mM KCl, 10 μM quinacrine, and 250 mM mannitol) for 5 min in darkness. The reaction was initiated by adding 3 mM ATP, and fluorescence was measured at 430 nm excitation and 500 nm emission. At the conclusion of the reaction, 10 μM m-chlorophenylhydrazone (CCCP) was introduced. Finally, GUS activity was computed as the change in fluorescence per minute per unit mass of PM protein.
For the rhizosphere acidification assay, calli treated with +Fe or −Fe + Frz for 15 days, and apple plantlets subjected to +Fe or -Fe + Frz for 10 days, were transferred to plates containing 0.006% bromocresol purple, 0.2 mM CaSO4, and 1% agar adjusted to pH 6.5 for 1 day.
Accession numbers
Sequence data utilized in this article can be accessed from the Genome Database for Rosaceae (GDR; https://www.rosaceae.org) data libraries under the accession numbers MdBT2 (MDP0000643281), MdCML15 (MDP0000227098), MdbHLH104 (MDP0000825749), MdAHA8 (MDP0000181085), MdBT1 (MDP0000151000), MdBT4 (MDP0000215415), MdCaM4 (MDP0000865414), MdCML12 (MDP0000380330), MdCML19 (MDP0000859814), MdCML24 (MDP0000817646), MdCML30 (MDP0000164511), MdCML58 (MDP0000664492), MdCML16 (MDP0000120294), MdCML60 (MDP0000178485), MdCML65 (MDP0000602146), MdCML71 (MDP0000176317), MdBT3.1 (MDP0000296225), MdBT3.2 (MDP0000187156), and MdCML76 (MDP0000583167).
Acknowledgements
We would like to sincerely thank our team leader, Dr Yu-Jin Hao. He will be remembered for his great achievement and for his support and assistance in this work. This work was supported by grants from National Key Research and Development Program (2022YFD1201701), the National Natural Science Foundation of China (32001336 and 32272683), the China Agriculture Research System of MOF and MARA (CARS-27), and the Natural Science Foundation of Shandong Province (ZR2022QC093).
Author contributions
C.X.Y and X.F.W. planned and designed the research. X.J.L., H.K., X.L., Q.Z., and Y.H.D. performed experiments and analyzed the data. Q.L., Y.X., and Y.X.Y. provided technical assistance. X.J.L., H.K., and X.F.W. wrote the article.
Data availability
The data underlying this article are available in the manuscript and in its online supplementary material.
Conflict of interest
The authors declare that no competing interests exist.
Supplementary data
Supplementary data are available at Horticulture Research online.
Supplementary Material
Contributor Information
Xiao-Juan Liu, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China; State Key Laboratory of Tree Genetics and Breeding, Chinese Academy of Forestry, Beijing 100091, China.
Xin Liu, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China; Institute of Forestry and Pomology, Academy of Agriculture and Forestry Sciences, Beijing 100093, China.
Qiang Zhao, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China; College of Horticulture, Qingdao Agricultural University, Qingdao 266109, China.
Yuan-Hua Dong, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Qiangbo Liu, National Key Laboratory of Wheat Improvement, College of Life Sciences, Shandong Agricultural University, Tai-An, 271018, China.
Yuan Xue, State Key Laboratory of Tree Genetics and Breeding, Chinese Academy of Forestry, Beijing 100091, China.
Yu-Xin Yao, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Chun-Xiang You, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Hui Kang, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
Xiao-Fei Wang, National Key Laboratory of Wheat Improvement, Apple Technology Innovation Center of Shandong Province, Shandong Green Fertilizer Technology Innovation Center, College of Horticulture Science and Engineering, Shandong Agricultural University, Tai-An, 271018, Shandong, China.
References
- 1. Liang G. Iron uptake, signaling, and sensing in plants. Plant Commun. 2022;3:100349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gao F, Dubos C. Transcriptional integration of plant responses to iron availability. J Exp Bot. 2021;72:2056–70 [DOI] [PubMed] [Google Scholar]
- 3. Santi S, Schmidt W. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 2009;183:1072–84 [DOI] [PubMed] [Google Scholar]
- 4. Walker EL, Connolly EL. Time to pump iron: iron-deficiency-signaling mechanisms of higher plants. Curr Opin Plant Biol. 2008;11:530–5 [DOI] [PubMed] [Google Scholar]
- 5. Hindt MN, Guerinot ML. Getting a sense for signals: regulation of the plant iron deficiency response. BBA Mol Cell Res. 2012;1823:1521–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang X, Zhang D, Sun W. et al. The adaptive mechanism of plants to iron deficiency via iron uptake, transport, and homeostasis. Int J Mol Sci. 2019;20:2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015;20:124–33 [DOI] [PubMed] [Google Scholar]
- 8. Gao F, Robe K, Gaymard F. et al. The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors? Front. Plant Sci. 2019;10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuan Y, Wu H, Wang N. et al. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008;18:385–97 [DOI] [PubMed] [Google Scholar]
- 10. Wang N, Cui Y, Liu Y. et al. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol Plant. 2013;6:503–13 [DOI] [PubMed] [Google Scholar]
- 11. Long TA, Tsukagoshi H, Busch W. et al. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 2010;22:2219–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J, Liu B, Li M. et al. The bHLH transcription factor bHLH104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell. 2015;27:787–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li X, Zhang H, Ai Q. et al. Two bHLH transcription factors, bHLH34 and bHLH104, regulate iron homeostasis in Arabidopsis thaliana. Plant Physiol. 2016;170:2478–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liang G, Zhang H, Li X. et al. bHLH transcription factor bHLH115 regulates iron homeostasis in Arabidopsis thaliana. J Exp Bot. 2017;68:1743–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim SA, LaCroix IS, Gerber SA. et al. The iron deficiency response in Arabidopsis thaliana requires the phosphorylated transcription factor URI. Proc Natl Acad Sci USA. 2019;116:24933–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gao F, Robe K, Bettembourg M. et al. The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell. 2020;32:508–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lei R, Li Y, Cai Y. et al. bHLH121 functions as a direct link that facilitates the activation of FIT by bHLH IVc transcription factors for maintaining Fe homeostasis in Arabidopsis. Mol Plant. 2020;13:634–49 [DOI] [PubMed] [Google Scholar]
- 18. Zhao Q, Ren YR, Wang QJ. et al. Overexpression of MdbHLH104 gene enhances the tolerance to iron deficiency in apple. Plant Biotechnol J. 2016;14:1633–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fan Z, Wu Y, Zhao L. et al. MYB308-mediated transcriptional activation of plasma membrane H+-ATPase 6 promotes iron uptake in citrus. Hortic Res. 2022;9:uhac088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Selote D, Samira R, Matthiadis A. et al. Iron-binding E3 ligase mediates iron response in plants by targeting basic helix-loop-helix transcription factors. Plant Physiol. 2015;167:273–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang HM, Li Y, Yao XN. et al. POSITIVE REGULATOR OF IRON HOMEOSTASIS 1 (OsPRI1) positively regulates iron homeostasis in rice. Plant Physiol. 2017;175:543–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gratz R, Brumbarova T, Ivanov R. et al. Phospho-mutant activity assays provide evidence for alternative phospho-regulation pathways of the transcription factor FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR. New Phytol. 2020;225:250–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rodriguez-Celma J, Green RT, Connorton JM. et al. Arabidopsis BRUTUS-LIKE E3 ligases negatively regulate iron uptake by targeting transcription factor FIT for recycling. Proc Natl Acad Sci USA. 2019;116:17584–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao Q, Ren YR, Wang QJ. et al. Ubiquitination-related MdBT scaffold proteins target a bHLH transcription factor for iron homeostasis. Plant Physiol. 2016;172:1973–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou LJ, Zhang CL, Zhang RF. et al. The SUMO E3 ligase MdSIZ1 targets MdbHLH104 to regulate plasma membrane H+-ATPase activity and iron homeostasis. Plant Physiol. 2019;179:88–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li D, Sun Q, Zhang G. et al. MxMPK6-2-bHLH104 interaction is involved in reactive oxygen species signaling in response to iron deficiency in apple rootstock. J Exp Bot. 2021;72:1919–32 [DOI] [PubMed] [Google Scholar]
- 27. Zhai L, Sun Q, Gao M. et al. MxMPK4-1 phosphorylates NADPH oxidase to trigger the MxMPK6-2-MxbHLH104 pathway mediated Fe deficiency responses in apple. Plant Cell Environ. 2022;45:2810–26 [DOI] [PubMed] [Google Scholar]
- 28. Xu L, Wei Y, Reboul J. et al. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature. 2003;425:316–21 [DOI] [PubMed] [Google Scholar]
- 29. Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334 [DOI] [PubMed] [Google Scholar]
- 30. Robert HS, Quint A, Brand D. et al. BTB and TAZ domain scaffold proteins perform a crucial function in Arabidopsis development. Plant J. 2009;58:109–21 [DOI] [PubMed] [Google Scholar]
- 31. Du L, Poovaiah BW. A novel family of Ca2+/calmodulin-binding proteins involved in transcriptional regulation: interaction with fsh/Ring3 class transcription activators. Plant Mol Biol. 2004;54:549–69 [DOI] [PubMed] [Google Scholar]
- 32. Figueroa P, Gusmaroli G, Serino G. et al. Arabidopsis has two redundant Cullin3 proteins that are essential for embryo development and that interact with RBX1 and BTB proteins to form multisubunit E3 ubiquitin ligase complexes in vivo. Plant Cell. 2005;17:1180–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mandadi KK, Misra A, Ren S. et al. BT2, a BTB protein, mediates multiple responses to nutrients, stresses, and hormones in Arabidopsis. Plant Physiol. 2009;150:1930–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Araus V, Vidal EA, Puelma T. et al. Members of BTB gene family of scaffold proteins suppress nitrate uptake and nitrogen use efficiency. Plant Physiol. 2016;171:1523–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Irigoyen S, Ramasamy M, Misra A. et al. A BTB-TAZ protein is required for gene activation by cauliflower mosaic virus 35S multimerized enhancers. Plant Physiol. 2022;188:397–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ren S, Mandadi KK, Boedeker AL. et al. Regulation of telomerase in Arabidopsis by BT2, an apparent target of TELOMERASE ACTIVATOR1. Plant Cell. 2007;19:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Uno Y, Milla MAR, Maher E. et al. Identification of proteins that interact with catalytically active calcium-dependent protein kinases from Arabidopsis. Mol Gen Genomics. 2009;281:375–90 [DOI] [PubMed] [Google Scholar]
- 38. An JP, Wang XF, Hao YJ. BTB/TAZ protein MdBT2 integrates multiple hormonal and environmental signals to regulate anthocyanin biosynthesis in apple. J Integr Plant Biol. 2020;62:1643–6 [DOI] [PubMed] [Google Scholar]
- 39. Kudla J, Becker D, Grill E. et al. Advances and current challenges in calcium signaling. New Phytol. 2018;218:414–31 [DOI] [PubMed] [Google Scholar]
- 40. Tong T, Li Q, Jiang W. et al. Molecular evolution of calcium signaling and transport in plant adaptation to abiotic stress. Int J Mol Sci. 2021;22:12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li T, Liu Z, Lv T. et al. Phosphorylation of MdCYTOKININ RESPONSE FACTOR4 suppresses ethylene biosynthesis during apple fruit ripening. Plant Physiol. 2023;191:694–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun Q, Zhai L, Zhao D. et al. Kinase MxMPK4-1 and calmodulin-binding protein MxIQM3 enhance apple root acidification during Fe deficiency. Plant Physiol. 2023;191:1968–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Q, Liu X, Liu X. et al. Genome-wide identification, characterization, and expression analysis of calmodulin-like proteins (CMLs) in apple. Hortic Plant J. 2017;3:219–31 [Google Scholar]
- 44. Wang Z, Zhang Y, Liu Y. et al. Calcium-dependent protein kinases CPK21 and CPK23 phosphorylate and activate the iron-regulated transporter IRT1 to regulate iron deficiency in Arabidopsis. Sci China Life Sci. 2023;66:2646–62 [DOI] [PubMed] [Google Scholar]
- 45. Gratz R, Manishankar P, Ivanov R. et al. CIPK11-dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling. Dev Cell. 2019;48:726–740.e10 [DOI] [PubMed] [Google Scholar]
- 46. Dobney S, Chiasson D, Lam P. et al. The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J Biol Chem. 2009;284:31647–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeng H, Xu L, Singh A. et al. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015;6:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsai YC, Koo Y, Delk NA. et al. Calmodulin-related CML24 interacts with ATG4b and affects autophagy progression in Arabidopsis. Plant J. 2013;73:325–35 [DOI] [PubMed] [Google Scholar]
- 49. Song X, Li J, Lyu M. et al. CALMODULIN-LIKE-38 and PEP1 RECEPTOR 2 integrate nitrate and brassinosteroid signals to regulate root growth. Plant Physiol. 2021;187:1779–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun Q, Huang R, Zhu H. et al. A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J. 2021;108:1069–82 [DOI] [PubMed] [Google Scholar]
- 51. Yu S, Wu J, Sun Y. et al. A calmodulin-like protein (CML10) interacts with cytosolic enzymes GSTU8 and FBA6 to regulate cold tolerance. Plant Physiol. 2022;190:1321–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang XF, An JP, Liu X. et al. The nitrate-responsive protein MdBT2 regulates anthocyanin biosynthesis by interacting with the MdMYB1 transcription factor. Plant Physiol. 2018;178:890–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang K, An JP, Li CY. et al. The apple C2H2-type zinc finger transcription factor MdZAT10 positively regulates JA-induced leaf senescence by interacting with MdBT2. Hortic Res. 2021;8:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ren YR, Zhao Q, Yang YY. et al. Interaction of BTB-TAZ protein MdBT2 and DELLA protein MdRGL3a regulates nitrate-mediated plant growth. Plant Physiol. 2021;186:750–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang QY, Gu KD, Cheng LL. et al. BTB-TAZ domain protein MdBT2 modulates malate accumulation and vacuolar acidification in response to nitrate. Plant Physiol. 2020;183:750–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang D, Yang K, Kan Z. et al. The regulatory module MdBT2-MdMYB88/MdMYB124-MdNRTs regulates nitrogen usage in apple. Plant Physiol. 2021;185:1924–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang QY, Gu KD, Wang JH. et al. BTB-BACK-TAZ domain protein MdBT2-mediated MdMYB73 ubiquitination negatively regulates malate accumulation and vacuolar acidification in apple. Hortic Res. 2020;7:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Liu XJ, An XH, Liu X. et al. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. J Exp Bot. 2017;68:2977–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Peretz Y, Mozes-Koch R, Akad F. et al. A universal expression/silencing vector in plants. Plant Physiol. 2007;145:1251–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Xiao X, Ma F, Chen CL. et al. High efficient transformation of auxin reporter gene into trifoliate orange via Agrobacterium rhizogenes-mediated co-transformation. Plant Cell Tissue Organ Cult. 2014;118:137–46 [Google Scholar]
- 61. Kang H, Zhang TT, Li YY. et al. The apple BTB protein MdBT2 positively regulates MdCOP1 abundance to repress anthocyanin biosynthesis. Plant Physiol. 2022;190:305–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sun WJ, Zhang JC, Ji XL. et al. Low nitrate alleviates iron deficiency by regulating iron homeostasis in apple. Plant Cell Environ. 2021;44:1869–84 [DOI] [PubMed] [Google Scholar]
- 63. Ren YR, Zhao Q, Yang YY. et al. The apple 14-3-3 protein MdGRF11 interacts with the BTB protein MdBT2 to regulate nitrate deficiency-induced anthocyanin accumulation. Hortic Res. 2021;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the manuscript and in its online supplementary material.


