Abstract
A novel gene, FSY1, encoding a permease involved in active fructose uptake by a proton symport mechanism in the type strain of Saccharomyces carlsbergensis has been isolated. Fsy1p is only distantly related to the Hxt proteins that mediate facilitated diffusion of glucose and fructose in Saccharomyces cerevisiae and related species.
Hexose transport in Saccharomyces cerevisiae has been intensively studied at the molecular level over the last decade (2, 6, 11, 16). The facilitated diffusion transport system seems to be based on a large number of permeases (possibly 17), the so-called Hxt proteins (8, 16). Deletion of HXT1 to -7 (11) and the GAL2 gene, which encodes the galactose permease, leads to the virtual incapacity of the yeast to grow on glucose.
All Hxt proteins studied so far mediate facilitated diffusion of glucose and fructose. S. cerevisiae differs from other yeasts, which may exhibit, in addition to diffusion of glucose and fructose, proton symport systems for glucose and/or fructose (10, 15). However, previous work in our laboratory has shown that both Saccharomyces bayanus and Saccharomyces pastorianus (carlsbergensis), forming together with S. cerevisiae and Saccharomyces paradoxus the so-called Saccharomyces sensu stricto group, display fructose/H+ symport activities (1, 13). When further investigated, fructose/H+ symport was found to coexist with the well-characterized facilitated diffusion system for glucose and fructose, present in all members of the same Saccharomyces complex. The characterization of active fructose transport in S. pastorianus PYCC 4457 revealed that it mediates high-affinity fructose transport (Km, ∼0.2 mM) and that it does not accept d-glucose as a substrate. Besides d-fructose, only l-sorbose seems to be a substrate of the same carrier (1, 12), though the yeast is unable to metabolize this sugar.
To characterize the molecular composition of the active transport system for fructose, we set out to isolate the gene encoding the fructose/H+ transporter protein in S. pastorianus PYCC 4457, the type strain of S. carlsbergensis. Our strategy consisted of isolating the genes involved in active fructose transport by functional complementation of the absence of growth on fructose of an S. cerevisiae strain (RE800A) devoid of both the Hxt transporters 1 to 7 (11) and the galactose permease encoded by the GAL2 gene.
S. pastorianus genomic library.
A genomic library of S. pastorianus PYCC 4457 was constructed as follows. Genomic DNA of strain PYCC 4457 was partially digested with Sau3A, and the resulting DNA fragments were separated in 10 to 40% (wt/vol) discontinuous sucrose gradients. Fractions containing fragments in the ranges of 3.5 to 5 and 6 to 10 kb were pooled and used in separate ligations. The yeast shuttle vector YEplac195 (3) was digested with BamHI, dephosphorylated, and ligated to the purified genomic DNA fragments. Escherichia coli XL-GOLD supercompetent cells (Stratagene) were transformed by the ligations and plated onto Luria-Bertani medium supplemented with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml) and IPTG (isopropyl-β-d-thiogalactopyranoside) (40 μg/ml), yielding approximately 30,000 colonies, 90% of which were white. Restriction analysis of plasmid DNAs isolated from 12 white colonies revealed an insert length range of 3.5 to 8 kb. Library transformants were washed from the plates with Luria-Bertani medium, glycerol stocks were prepared, and the remaining cell suspension was left to grow for two generations. The cells were then harvested, and plasmid DNA was isolated.
Screening of the library.
We selected a spontaneous ura3 mutant of S. cerevisiae RE800A on medium containing 5-fluoroorotic acid to use it as a receptor strain for library screening. This auxotrophic strain was designated PYCC 5623.
Strain PYCC 5623 was transformed with library plasmid DNA by using the lithium acetate method (4). The transformation mixture was first plated onto solidified yeast nitrogen base (YNB) medium with 2% (wt/vol) maltose as the sole carbon source. After 5 days of incubation at 30°C, approximately 10,000 transformants were obtained. These colonies were then replica plated onto YNB medium containing 0.5% (wt/vol) fructose as the sole carbon source. After 5 days of incubation at 30°C, 15 colonies appeared on fructose plates (FRU+) and were further characterized. All FRU+ transformants were able to grow on YNB medium with either 0.5% (wt/vol) fructose or 0.5% (wt/vol) glucose as the sole carbon source. For five of the transformants, it could be readily shown that growth on fructose was dependent on the presence of a library plasmid. Each of these transformants was grown on YNB medium with 0.5% (wt/vol) fructose until an optical density at 640 nm of 0.5 was reached and tested for the presence of fructose/H+ symport activity. Symport activity was assessed by the proton influx elicited by the addition of fructose to an aqueous cell suspension (15). Only one of the transformants (T5) exhibited very strong sugar-proton symport activity when either fructose or sorbose was used as the substrate. Two different plasmids, pT5 I and pT5 II, were rescued after transformation of E. coli SURE competent cells with total DNA isolated from transformant T5. The results obtained upon transformation of strains PYCC 5623 (hxt-null ura3) and CEN.PK113-5D (wild type with respect to hexose transport) with these plasmids are summarized in Table 1. Surprisingly, both plasmids were able to complement the growth defect on fructose of the hxt-null strain. However, only transformants of both strains carrying the single plasmid pT5 II displayed fructose/H+ symport activity (Fig. 1). In addition, transformants of strain PYCC 5623 carrying plasmid pT5 II were unable to grow on glucose, which agrees with the previous observation that glucose is not a substrate of the active fructose transport system (1, 12). Thus, we concluded that plasmid pT5 II is likely to carry a gene involved in fructose/H+ symport in S. pastorianus PYCC 4457. As mentioned before, transformants carrying plasmid pT5 I lacked fructose/H+ symport activity, irrespective of the receptor strain. Partial sequencing of the insert in plasmid pT5 I indicated that it carries the homolog, in S. pastorianus, of GAL2 from S. cerevisiae (∼80% homology). This finding is corroborated by the fact that PYCC 5623/pT5 I transformants grow well on galactose, unlike PYCC 5623/pT5 II transformants. Apparently, the putative galactose permease from S. pastorianus is, similarly to S. cerevisiae Gal2p (7), able to support growth on glucose and fructose media (Table 1). This finding also explains the ability to grow on glucose of library transformant T5 containing pT5 I and pT5 II.
TABLE 1.
Phenotypes conferred to S. cerevisiae strains by plasmids pT5 I and pT5 II
| Plasmid | Strain | Growth on 0.5% glucose | Growth on 0.5% fructose | Fructose/H+ symport |
|---|---|---|---|---|
| pT5 I | CEN.PK113-5Da | + | + | − |
| PYCC 5623b | + | + | − | |
| pT5 II | CEN.PK113-5D | + | + | + |
| PYCC 5623 | − | + | + |
ura3 (wild type with respect to sugar transport).
hxt-null ura3.
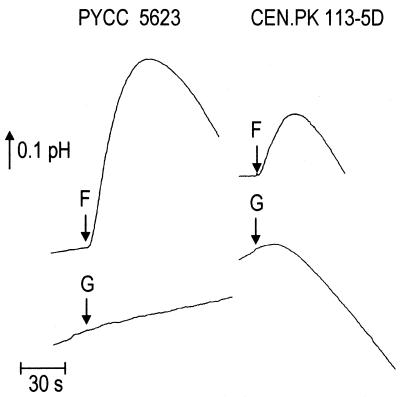
FIG. 1.
Illustration of the effect on extracellular pH of the addition of either 5 mM fructose (F) or 5 mM glucose (G) to aqueous fructose-grown cell suspensions (pH 5.0) of both the hxt-null strain (PYCC 5623) and the wild-type CEN.PK113-5D strain transformed with plasmid pT5 II. Increase in pH indicates proton uptake by the cells, and acidification indicates proton efflux, reflecting membrane ATPase activity.
In one of the PYCC 5623/pT5 II transformants, designated PYCC 5624, initial d-[14C]fructose (Amersham Pharmacia Biotech) uptake rates were measured (15). The results of a typical experiment are shown in Fig. 2. The affinity (Km = 0.16 mM) was similar to that estimated for the original strain, S. pastorianus PYCC 4457. In addition, measurements of d-[14C]glucose (Amersham Pharmacia Biotech) uptake, under the same conditions, indicated that strain PYCC 5624 cannot transport glucose (results not shown), which is in line with its inability to grow on glucose.
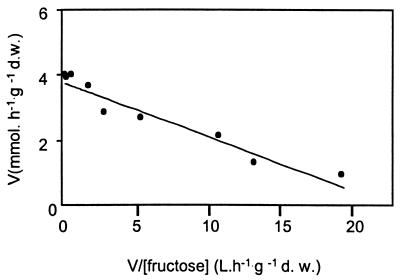
FIG. 2.
Eadie-Hofstee plot of the initial uptake rates of d-[14C]fructose measured in strain PYCC 5624, a S. cerevisiae hxt-null transformant carrying plasmid pT5 II. Cells were grown in YNB medium with 0.5% (wt/vol) fructose. Measurements for each concentration were made in triplicate. Kinetic parameters {Vmax = 3.8 ± 0.2 mmol · h−1 · g−1 (dry weight [d.w.]); Km = 0.16 ± 0.02 mM; 25°C, pH 5} were calculated using GraphPad Prism software, and the data were fitted to a one-component model according to Michaelis-Menten kinetics.
The FSY1 gene.
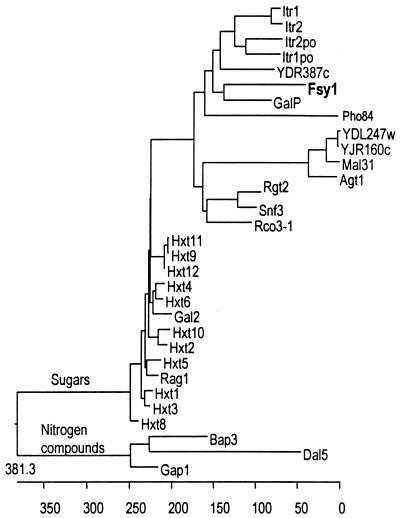
Plasmid pT5 II carries an insert of approximately 4 kb, of which a 2.2-kb region on both DNA strands was sequenced using an ALF express automated sequencer apparatus (Amersham Pharmacia Biotech) and 5′-Cy5-labeled sequence-specific primers. This region was found to contain an open reading frame of 570 amino acids, which we designated FSY1 (stands for fructose symport). Fsy1p contains two signature sequences (amino acids 199 to 224 and 402 to 417) characteristic of sugar transporters (the PredictProtein server, http://dodo.cpmc.columbia.edu/predictprotein/). It is predicted to have 12 membrane-spanning domains, with both ends of the protein residing in the cytoplasm (14). No significant homology was found between the FSY1 gene and DNA sequences present in public databases. However, searches using the Fsy1p complete amino acid sequence revealed a low, though significant, level of homology with transporter proteins belonging to the major facilitator superfamily (9). The GalP protein (galactose/H+ symporter) from E. coli exhibits the highest homology with Fsy1p (30% identity, 464 aligned residues). This novel permease seems to be only distantly related to the Hxt proteins responsible for facilitated diffusion of glucose and fructose in S. cerevisiae (Fig. 3).
FIG. 3.
Relationships between Fsy1p and other permeases belonging to the major facilitator superfamily. The dendrogram was obtained using Megalign software version 3.14 (Clustal method). All the permeases shown are S. cerevisiae proteins except the following: Fsy1, S. pastorianus; ltr1po and ltr2po, Schizosaccharomyces pombe; GalP, E. coli; Rco1, Neurospora crassa; and Rag1, Kluyveromyces lactis.
Fsy1p is, to our knowledge, the first specific permease to be identified in yeast that mediates the active transport of fructose but does not accept glucose as a substrate. Information about regions of hexose carriers that determine substrate specificity is scarce, although aromatic residues within transmembrane region 10 seem to be important (5). Interestingly, Fsy1p shares some features of transmembrane domain 10 with the Hxt proteins (where the positions corresponding to Tyr461 and to Trp466 in Fsy1p are also invariably occupied by aromatic residues). However, the region surrounding Phe431 in Hxt2p, which seems to be crucial for glucose recognition by this carrier, is considerably different in Fsy1p. Further characterization of the active fructose transport system in S. pastorianus (carlsbergensis) will address the specificity of Fsy1p, as well as the regulation of symport activity by substrate concentration.
Nucleotide sequence accession number.
The DNA sequence of the FSY1 open reading frame can be retrieved from the EMBL database under accession no. AJ250992.
Acknowledgments
We thank E. Boles (University of Düsseldorf, Düsseldorf, Germany) for the generous supply of strain RE800A.
P.G. and H.R.D.S. were recipients of PRAXIS XXI grants (BPD11825/97 and BPD18883/98, respectively) from the National Foundation for Science and Technology, Lisbon, Portugal. This work was in part supported by the EC project BIO4-CT95-0107.
REFERENCES
- 1.Cason D T, Spencer-Martins I, van Uden N. Transport of fructose by a proton symport in a brewing yeast. FEMS Microbiol Lett. 1986;36:307–309. [Google Scholar]
- 2.Diderich J A, Schepper M, van Hoek P, Luttik M A, van Dijken J P, Pronk J T, Klaassen P, Boelens H F, de Mattos M J, van Dam K, Kruckeberg A L. Glucose uptake kinetics and transcription of HXT genes in chemostat cultures of Saccharomyces cerevisiae. J Biol Chem. 1999;274:15350–15359. doi: 10.1074/jbc.274.22.15350. [DOI] [PubMed] [Google Scholar]
- 3.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 4.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/ss-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 5.Kasahara M, Maeda M. Contribution to substrate recognition of two aromatic amino acid residues in putative transmembrane segment 10 of the yeast sugar transporters Gal2 and Hxt2. J Biol Chem. 1998;273:29106–29112. doi: 10.1074/jbc.273.44.29106. [DOI] [PubMed] [Google Scholar]
- 6.Kruckeberg A L. The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol. 1996;166:283–292. doi: 10.1007/s002030050385. [DOI] [PubMed] [Google Scholar]
- 7.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Özcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paulsen I T, Sliwinski M K, Nelissen B, Goffeau A, Saier M H. Unified inventory of established and putative transporters encoded within the complete genome of Saccharomyces cerevisiae. FEBS Lett. 1998;430:116–125. doi: 10.1016/s0014-5793(98)00629-2. [DOI] [PubMed] [Google Scholar]
- 10.Postma E, van den Broek P J. Continuous-culture study of the regulation of glucose and fructose transport in Kluyveromyces marxianus CBS 6556. J Bacteriol. 1990;172:2871–2876. doi: 10.1128/jb.172.6.2871-2876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol. 1995;16:157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x. [DOI] [PubMed] [Google Scholar]
- 12.Rodrigues de Sousa H. Active fructose transport in Saccharomyces sensu stricto: taxonomic and industrial implications. Ph.D. thesis. Lisbon, Portugal: Universidade Nova de Lisboa; 1998. . (In portuguese.) [Google Scholar]
- 13.Rodrigues de Sousa H, Madeira-Lopes A, Spencer-Martins I. The significance of active fructose transport and maximum temperature for growth in the taxonomy of Saccharomyces sensu stricto. Syst Appl Microbiol. 1995;18:44–51. [Google Scholar]
- 14.Sonnhammer E L L, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. In: Glasgow J, Littlejohn T, Major F, Lathrop R, Sankoff D, Sensen C, editors. Proceedings of the 6th international Conference on Intelligent Systems for Molecular Biology. Menlo Park, Calif: AAAI Press; 1998. pp. 175–182. [PubMed] [Google Scholar]
- 15.Spencer-Martins I, van Uden N. Catabolite interconversion of glucose transport systems in the yeast Candida wickerhamii. Biochim Biophys Acta. 1985;812:168–172. doi: 10.1016/0005-2736(87)90285-9. [DOI] [PubMed] [Google Scholar]
- 16.Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg C P, Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464:123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]