Abstract
Cellular reprogramming technologies have been developed with different physicochemical factors to improve the reprogramming efficiencies of induced pluripotent stem cells (iPSCs). Ultrasound is a clinically applied noncontact biophysical factor known for regulating various cellular behaviors but remains uninvestigated for cellular reprogramming. Here, we present a new reprogramming strategy using low-intensity ultrasound (LIUS) to improve cellular reprogramming of iPSCs in vitro and in vivo. Under 3D microenvironment conditions, increased LIUS stimulation shows enhanced cellular reprogramming of the iPSCs. The cellular reprogramming process facilitated by LIUS is accompanied by increased mesenchymal to epithelial transition and histone modification. LIUS stimulation transiently modulates the cytoskeletal rearrangement, along with increased membrane fluidity and mobility to increase HA/CD44 interactions. Furthermore, LIUS stimulation with HA hydrogel can be utilized in application of both human cells and in vivo environment, for enhanced reprogrammed cells into iPSCs. Thus, LIUS stimulation with a combinatorial 3D microenvironment system can improve cellular reprogramming in vitro and in vivo environments, which can be applied in various biomedical fields.
Keywords: Induced pluripotent stem cell, Cellular reprogramming, Low-intensity ultrasound, Biophysical stimulation, Cytoskeletal rearrangement
Graphical abstract
Highlights
-
•
LIUS integrated with 3D hyaluronic acid hydrogel promotes cellular reprogramming into induced pluripotent stem cells.
-
•
LIUS modulates cytoskeletal changes, boosting cell-ECM interactions and CD44 expression to enhance cellular reprogramming.
-
•
LIUS stimulation enhances the reprogramming efficiencies of induced pluripotent stem cells both in vitro and in vivo.
1. Introduction
Cellular reprogramming technology has expanded several aspects of stem cell research as somatic cells with different origins can be induced to produce different cells with desired features, and these reprogramming technologies have developed from in vitro culture method to in vivo [1,2]. In particular, cellular reprogramming into a pluripotent state for induced pluripotent stem cells (iPSCs) is highlighted for its functional characteristics of self-renewal and differentiation into multiple cell lineages [[3], [4], [5], [6], [7], [8]]. Compared to other stem cells, iPSCs are favorable sources in that they can bypass ethical issues and immune rejections. Therefore, iPSCs have been utilized in various biomedical fields for disease screening, gene editing, and cell therapeutics [[9], [10], [11], [12]]. Although there still remains a necessity to increase iPSCs in number and purity, reprogramming of iPSCs is challenging as its efficiency is low (1–5%); this leads to incomplete reprogramming that lacks reproducibility and has a lengthy culture time [13]. Several defined sets of transcription factors, small-molecule cocktails, and cell culture supplements have been suggested to improve reprogramming efficiency [14,15], but these biochemical methods have limitations for increasing the reprogramming efficiency while neglecting other considerable adverse effects caused by biochemical factors [16]. Moreover, cellular reprogramming technology in vitro is often differently regulated in vivo due to differences in local microenvironment conditions. These circumstances cause more reduced reprogramming efficiencies in vivo. Therefore, development of alternative methods to enhance the reprogramming efficiencies are necessary for the increased production of iPSCs in therapeutic applications.
Biophysical changes have garnered attention for regulating cell fates during differentiation and reprogramming [17]. These biophysical changes include stiffness, topography, ligand patterning, stretch, compression, shear stress, and external stimulation [18,19]. Cells are continuously exposed to biophysical stimuli as they are perturbed by various surfaces, stresses, and mechanical forces. These biophysical changes can be controlled to influence a myriad of cellular processes by switching the mechanical cues to biological responses during cell migration, differentiation, proliferation, and apoptosis [[20], [21], [22], [23], [24]]. A few reprogramming studies have shown that biophysical changes to stiffness [25,26], topography [27,28], shear stress [29], mechanical stretching [30], and electromagnetic fields [31] can affect the reprogramming efficiencies of iPSCs. Especially, biophysical changes from 2D to 3D culture have shown increased cellular responses in migration, adhesion, interaction, and cytoskeletal organization [32,33]. As physiological microenvironment of cell is cultured in 3D, efforts to understand how biophysical factors synergistically activate cell behaviors in dynamic 3D microenvironment form [34,35]. In cell reprogramming studies, 3D culture system also showed improvement on producing more iPSCs [36,37]. These changes modulate cellular behaviors during the reprogramming process by varying the cytoskeletal changes, extracellular matrix (ECM)–receptor interactions, mesenchymal to epithelial transitions (MET), and epigenetic modulations [38].
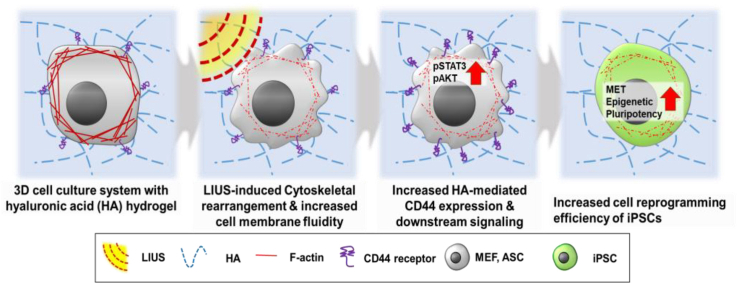
Ultrasound as a noninvasive acoustic wave force is another biophysical change that can apply mechanical stress in a noncontact manner. Among the various intensities and frequencies of ultrasound available, low-intensity ultrasound (LIUS) has been widely investigated as it is not lethal to cells yet provides active biophysical stimulus [39]. Unlike other inaccessible, expensive, invasive stimuli, LIUS has deep tissue-penetrating properties that can efficiently target sites with easily accessible apparatus [40]. Since the FDA has indicated that LIUS is generally safe with low risks, its safety profile is highly promising in various cell types [41]. LIUS has been commonly used for diagnostic and therapeutic purposes [42], as well as for modulating physiological changes in the proliferation, migration, and differentiation potentials (chondrocytes [43,44], neurons [45,46], and osteoblasts [47,48]) at the cellular level. Number of studies presented the effect of ultrasound stimulation on directed differentiation of various cell types, but limited number of studies support the reprogramming potential of ultrasound stimulation and its mechanotransduction [49]. Furthermore, mechanism of LIUS stimulation on cells and microenvironments are still unclear. A few studies have associated biophysical changes from LIUS with cytoskeletal rearrangement and fluidization of the cell membrane as well as transmembrane protein of the target cells [50]. Also, regulatory effects of LIUS on cell and tissue are associated with the interaction of cell with adjacent substrates, such as extracellular matrices [51,52]. Therefore, further investigations of dynamic culture with LIUS during cell reprogramming are necessary to examine cellular mechanisms of biophysical modulation and ECM interaction.
Previous studies have reported that 3D hydrogel system can improve the cell reprogramming process [53], but how cells in 3D microenvironment regulate in response to dynamic biophysical cues during cell reprogramming process has not been clearly elucidated. Here, we present the LIUS stimulation of 3D microenvironment system for improved cell reprogramming of iPSCs and identify mechanical action of LIUS on cell interaction with 3D microenvironment. A previous study reported enhanced reprogramming efficiency of iPSCs under a hyaluronic-acid (HA) based 3D hydrogel niche with controlled stiffness, initially upregulating the HA mediated CD44 expression [53]. In the present study, we apply the different duration of LIUS during cellular reprogramming of iPSCs within a HA based 3D hydrogel, and the reprogramming efficiency of the iPSCs is assessed. LIUS stimulation under 3D microenvironment culture is evaluated for pluripotency, mesenchymal to epithelial transition (MET), and histone modification. As LIUS induces acoustic wave force to modulate cytoskeletal rearrangement and cell surface change, cellular interaction with microenvironment may regulate increased reprogramming process in HA hydrogel. LIUS stimulated 3D microenvironment system is further confirmed its efficacy in human cell and in vivo environment. Our results indicate that LIUS stimulated microenvironment system induces more cell reprogramming process into iPSCs.
2. Results
2.1. LIUS enhances the reprogramming efficiency of iPSCs
In this study, LIUS stimulation was carried out by modulating the duration of application on the 3D hydrogel. The frequency and intensity of the LIUS were set constant at 40 kHz and 300 mW cm−2, respectively, as these values ranged within the viable condition [42]. Different durations of LIUS were induced on alternate days and analyzed throughout the study. Cell viability of the LIUS was first analyzed after stimulation for 7 days, and the fibroblasts stimulated with up to 20 min of LIUS showed high viability without dead cells (Supplementary Figs. 2a and b). However, 30 min of LIUS stimulation caused cell death, reducing the viability of the fibroblasts to ∼71 % on day 7. The results show that up to 20 min of LIUS can maintain cell viability for 14 days; thus, LIUS stimulations were limited to 20 min for further studies.
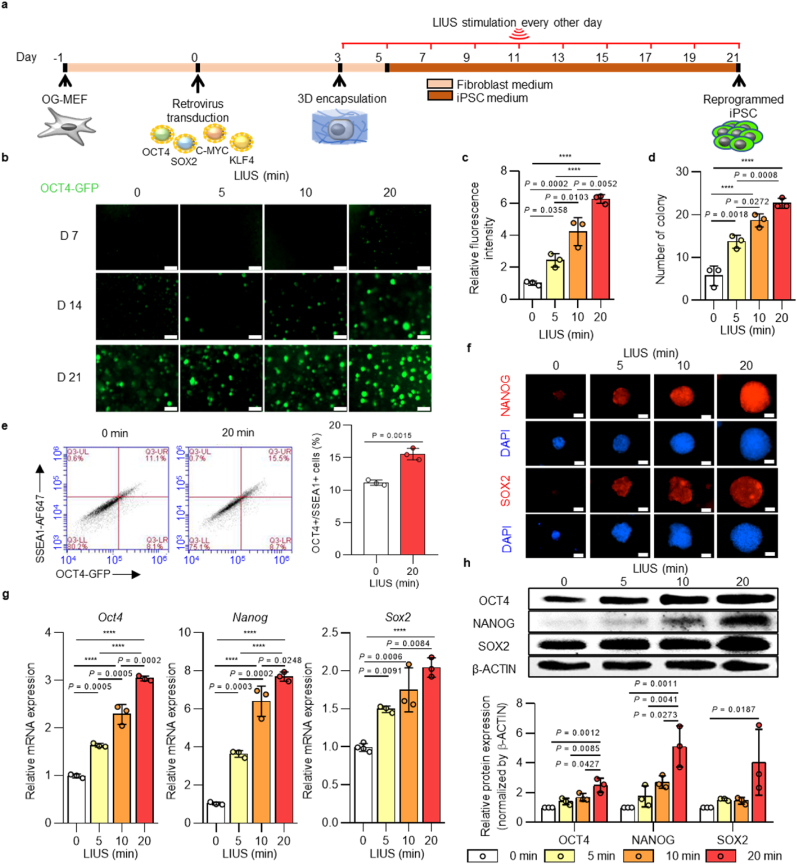
Next, LIUS stimulation was evaluated for reprogramming iPSCs. For the reprogramming process, OCT4-GFP expressing mouse embryonic fibroblasts (OG-MEFs) were induced with Yamanaka's four factors (OCT4, SOX2, c-MYC, and KLF4) using retroviral transduction. The transduced cells were then encapsulated in HA hydrogel using photo-crosslinking methods, and the hydrogels were stimulated with LIUS for a given time until analysis (Fig. 1a). Expression of OCT4-GFP at each time point showed that longer exposure to LIUS stimulation induced more OCT4-GFP-positive cells (Fig. 1b). While the unstimulated group started expressing OCT4-GFP on day 14 from transduction, 20 min of LIUS application accelerated GFP expression as early as 7 days after transduction. This observation is remarkable in that the reprogramming process of iPSCs can be further shortened to one week through LIUS stimulation. The quantification of OCT4-GFP-positive colonies on day 21 was also compared, and LIUS stimulation generated more GFP-expressing colonies in terms of numbers and intensities [20 min of LIUS produced ∼3.2-fold increase in colony numbers and ∼2.3-fold increase in GFP intensities compared with the unstimulated group] (Fig. 1c and d). These results correlated with those of flow cytometry analysis in that the ratio of OCT4+/SSEA1+ cells improved to 15.56 % when compared with the conventional 2D condition (∼1 %) and the 3D condition (∼11 %) (Fig. 1e and Supplementary Fig. 3). The fluorescent intensities of other pluripotent markers, NANOG and SOX2, were also observed, and higher intensities were detected after LIUS exposure, confirming the increased reprogramming efficiency (Fig. 1f). As the expressions of pluripotency proteins and genes demonstrate fully reprogrammed iPSCs [54], total expression of the pluripotent markers were examined. The gene expressions of pluripotent markers Oct4, Nanog, and Sox2 showed increased expression with longer LIUS stimulation (Fig. 1g). Compared to the unstimulated cells, 20 min of LIUS stimulation expressed Oct4, Nanog, and Sox2 markers by ∼2.7, ∼6.4, and ∼2.0 fold growth, respectively. Indeed, the protein expressions were also correlated with gene expressions, where longer LIUS stimulation upregulated the pluripotent markers of OCT4, NANOG, and SOX2 (up to 2.5, 5.1, and 4.0 times higher intensities of OCT4, NANOG, and SOX2 bands were measured in LIUS, respectively) (Fig. 1h).
Fig. 1.
| Ultrasound stimulation enhances reprogramming efficiency in 3D hydrogel. a, Schematic representation of the reprogramming of OCT4-GFP (OG)-MEFs into iPSCs under LIUS stimulation. b, OCT4-GFP expression of iPSCs derived from OG-MEFs under various LIUS stimulation times in HA hydrogel (scale bar, 300 μm). c, Quantitative analysis of OCT4 GFP intensity and d, number of iPSC colonies derived from OG-MEFs within 3D hydrogels with various ultrasound stimulation times (D21; n = 3). e, representative flow cytometry profile of the OCT4+/SSEA1+ cells under LIUS stimulation and its quantified results (D21; n = 3). Immunofluorescence images of f, NANOG and SOX2 stained colonies under LIUS-stimulated OG-MEFs after reprogramming (D21; scale bar, 300 μm). g, qPCR analysis of the pluripotent markers, (Oct4, Nanog, and Sox2) and h, western blot analysis of the pluripotency markers (OCT4, NANOG, and SOX2) in iPSCs derived from OG-MEFs under LIUS exposure in 3D hydrogels (D21; n = 3). All data are expressed as the mean ± s.d. The statistical significance was determined with one-way ANOVA followed by Tukey's multiple comparison post test. ****P < 0.0001.
2.2. LIUS accelerates initial changes in the iPSC reprogramming process
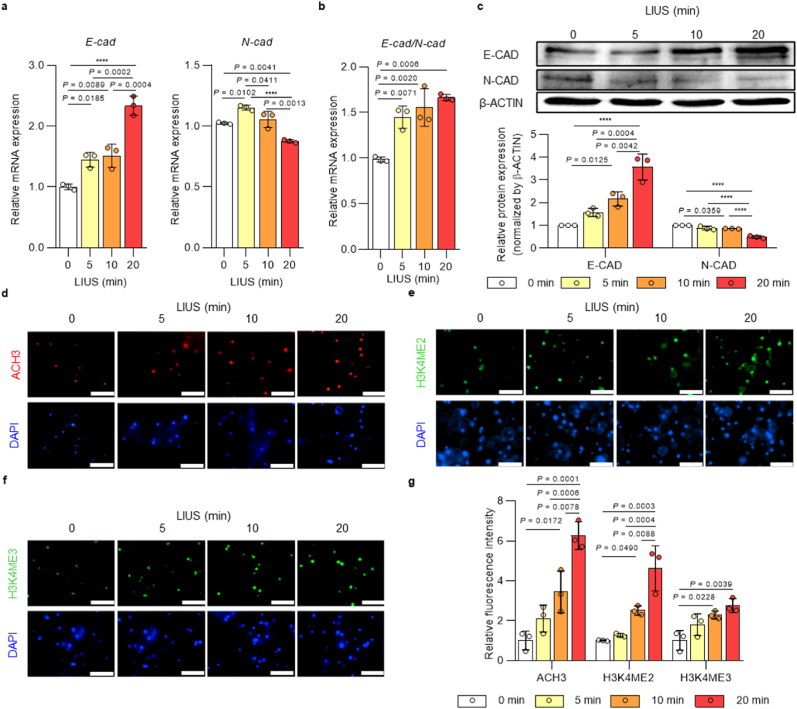
During the reprogramming process, cells undergo initial changes into fully reprogrammed iPSCs, such as METs and epigenetic modifications [55]. First, MET changes were analyzed by the gene expressions of epithelial marker E-cadherin and mesenchymal marker N-cadherin after LIUS stimulation (Fig. 2a). The results showed that LIUS stimulation significantly increased the expression of E-cad. On the contrary, N-cad expression was slightly reduced as LIUS was induced for 20 min. The ratio of E-cad to N-cad expression was more apparent such that LIUS stimulation increased the MET during iPSC reprogramming (Fig. 2b). Consistently, protein expression of E-cadherin was increased and expression of N-cadherin was decreased as LIUS stimulation was induced (Fig. 2c). Epigenetic modification is another aspect of iPSC reprogramming that activates or silences genes. Specifically, histone modifications are extensively reported transitions that occur in the early reprogramming process, which are key checkpoint molecules for iPSC reprogramming [56]. Here, we observed expressions of active H3 acetylation, H3K4 dimethylation, and H3K4 trimethylation during the early reprogramming process, and LIUS stimulation upregulated the expressions of all histone markers analyzed (Fig. 2d–g). Therefore, LIUS exposure during reprogramming enhanced the initial procedures by upregulating the METs and histone modifications.
Fig. 2.
| LIUS accelerates cell transition and epigenetic changes during iPSC reprogramming in 3D hydrogel. a, qPCR analysis of MET markers, E-cad and N-cad, and b, ratio of Cdh1/Cdh2 expression and c, western blot analysis of the MET markers (E-cadherin and N-cadherin) of OG-MEFs under LIUS (D1; n = 3). Immunofluorescence staining of d, Acetyl H3 (ACH3), e, dimethyl H3K4 (H3K4ME2), and f, trimethyl H3K4 (H3K4ME3) of OG-MEFs stimulated with LIUS (D1; scale bar, 100 μm). g, Quantification of fluorescence intensity in each immunofluorescence stained cell sample (n = 3). All data are expressed as the mean ± s.d. The statistical significance was determined with one-way ANOVA followed by Tukey's multiple comparison post test. ****P < 0.0001.
2.3. Cytoskeletal rearrangement by LIUS alters membrane fluidity and promotes CD44 signaling pathway
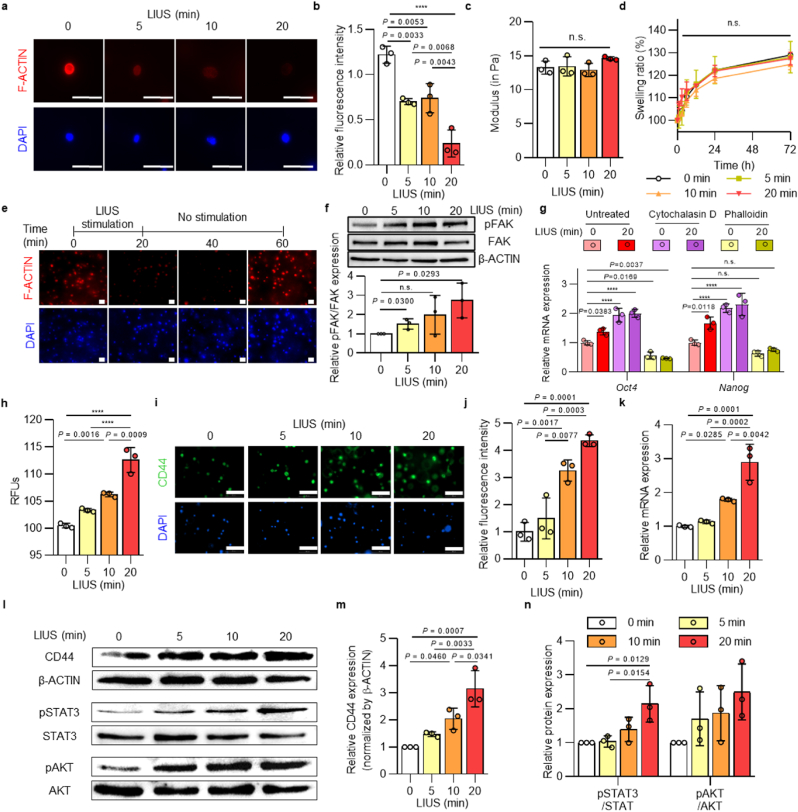
LIUS stimulation could potentially affect the mechanical forces on cells within the hydrogel system and adjust their physical properties. Thus, we examined the mechanisms for LIUS to modulate reprogramming efficiencies by regulating the cytoskeletal structures. F-actin staining was used to observe different expressions of actin stress fibers from LIUS stimulation. Interestingly, F-actin filaments start to lose their expression as LIUS was stimulated extensively (Fig. 3a and b, and Supplementary Figs. 4a and b). The measured intensities of F-actin staining showed ∼37 %, ∼43 %, and ∼75 % reductions for 5, 10, and 20 min of LIUS stimulations, respectively, compared to the unstimulated group. Aside from that, we did not observe any physical differences (modulus, swelling ratio) in the 3D hydrogels from LIUS stimulation (Fig. 3c and d). Actin stress fiber dynamics were further analyzed if LIUS stimulations permanently degraded F-actin expressions (Fig. 3e). As 20 min of LIUS application reduced F-actin expression, the fluorescence intensities remained at their reduced expressions until 40 min from initial LIUS stimulation. Interestingly, the expression of F-actin gradually reappeared and rapidly rearranged the cytoskeletal structure by 60 min. These results imply that the mechanical forces from LIUS only alter the temporal changes to the cytoskeletal rearrangements of the reprogrammed cells. The electron microscope images also show the changes to cell morphology after LIUS stimulation (Supplementary Fig. 5). Consequently, F-actin remodeling induced by LIUS regulates focal adhesion signaling associated with mechanical changes. Following LIUS-induced remodeling, focal adhesion kinase phosphorylation increased up to 2.7-fold (Fig. 3f), indicating its regulatory role in focal adhesion signaling. To further elucidate the involvement of cytoskeletal structures in cell reprogramming, we modulated F-actin structures using actin-associated small molecules known to transiently alter cytoskeletal organization [57], and assessed their effects on cells. In this experiment, cytochalasin D, which depolymerizes F-actin [58], and phalloidin, which stabilizes the F-actin structure [59], were employed. The results showed that each small molecule regulated the expression of F-actin in fibroblasts within a 3D hydrogel, even under the LIUS stimulation (Supplementary Fig. 6). Subsequently, these cells were evaluated its molecular changes in pluripotency, and observed significant alterations in the relative mRNA expression of pluripotent markers (Oct4 and Nanog) in response to each small molecule (Fig. 3g). F-actin depolymerization induced by cytochalasin D increased the Oct4 and Nanog expression by about 2-fold compared to the untreated group. Conversely, phalloidin, which stabilized F-actin polymerization, reduced Oct4 and Nanog expression to about 60 % of the untreated group. These results emphasize the contribution of LIUS-induced cytoskeletal remodeling to the cell reprogramming process.
Fig. 3.
| LIUS modulates cytoskeletal rearrangement and CD44 interaction. a, Immunofluorescence staining of F-actin after LIUS stimulation (scale bar, 50 μm) and b, its graphically quantified intensity (n = 3). Mechanical properties of hydrogel by LIUS application were analyzed by c, shear modulus and d, swelling ratio (n = 3). e, Time-dependent expressions of F-actin from LIUS were analyzed by the fluorescence intensities of F-actin (scale bar, 50 μm). f, Protein expression of phosphorylated FAK and total FAK under LIUS (D1; n = 3). g, mRNA expression of Oct4 and Nanog after F-actin modulation by cytochalasin D and phalloidin treatment (D1; n = 3). h, Cell membrane fluidity of LIUS was measured by the relative fluorescence intensity of excimer to monomer ratio (n = 3). i, Immunofluorescence staining of CD44 after LIUS stimulation (D1; scale bar, 100 μm) and j, its intensities (n = 3). k, Expressions of CD44 under LIUS were analyzed by qPCR (D1; n = 3). l, Protein expression of CD44 and signaling molecules were analyzed with western blotting. Relative protein expressions of m, CD44 and n, signaling molecules, STAT3 and AKT, were measured (D1; n = 3). All data are expressed as the mean ± s.d. The statistical significance was determined with one-way ANOVA followed by Tukey's multiple comparison post test. n.s., not significant, ****P < 0.0001.
The cytoskeletal change of LIUS may adjust the fluidity and mobility of the cell membrane composition. Using the diffusion rate of the lipid analog probes in the fluidized cell membranes, LIUS-stimulated cells were analyzed for their cell membrane fluidity (Fig. 3h). The relative fluorescence units (RFU) showed that more lipid probes were diffused in LIUS treatment, indicating highly fluidized cell membranes. Changes in the cell membrane fluidity from LIUS influence cell surface receptor mediated signaling to ECM interactions. A previous study reported the importance of HA/CD44 interactions facilitating the initial reprogramming processes [53], and enhanced membrane fluidity by LIUS could further improve CD44 expression. Fluorescence staining of CD44 showed up to 4 fold increased CD44-expressing cells with greater LIUS stimulation (Fig. 3i and j, and Supplementary Fig. 7). Likewise, up to 3 times high CD44 expressions from LIUS stimulation were found in the qRT-PCR (Fig. 3k) and western blot (Fig. 3l,m) results, confirming that LIUS stimulation increases CD44 expression. With increased expression of CD44, phosphorylation of downstream signaling molecules (STAT3 and AKT) was observed (Fig. 3l,n). Although all three signaling molecules show higher phosphorylation as LIUS was induced, significant increase in phosphorylation was observed from STAT3. Taken together, LIUS stimulation sequentially rearrange cytoskeletal fibers, modulate membrane fluidity, and regulate cell-ECM interaction to increase expression of CD44 and its downstream signals.
2.4. Presence of HA/CD44 is necessary for the increased reprogramming process during LIUS stimulation
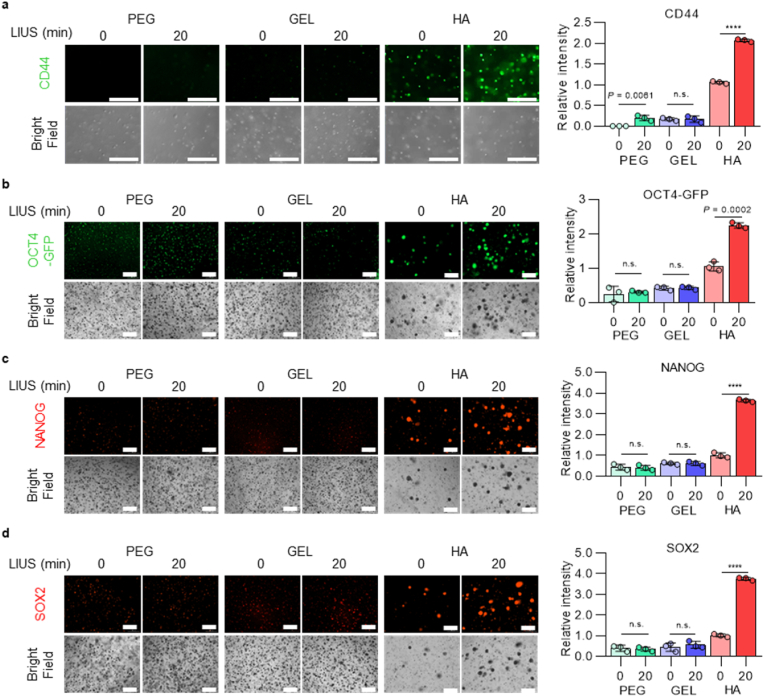
LIUS stimulation has shown increased reprogramming efficiencies from interaction of HA/CD44, and reprogramming efficiencies were further analyzed under LIUS stimulation when accessibility of HA or CD44 is limited. First, cells were exposed to different ECM-based hydrogels (PEG, Gel, and HA) before and after the LIUS stimulation. PEG and Gel were selected, as PEG is a biologically inert material, and Gel is the most widely used natural material for cell culture. While all hydrogels showed decreased F-actin expressions after LIUS stimulation (Supplementary Fig. 8), immunofluorescence stained CD44 showed high expression in HA hydrogel only, and LIUS stimulation further increased the CD44 expression to two folds (Fig. 4a). On the contrary, notable increased expression of CD44 were not observed from PEG and GEL hydrogels before and after the LIUS stimulation, as the CD44 expressions were ∼16 % from the HA hydrogel group. PEG hydrogel showed significant CD44 change after LIUS stimulation, but increased CD44 expression from PEG hydrogel with LIUS stimulus is too minimal (∼20 % of HA). These different hydrogels were further analyzed for the reprogramming efficiency into iPSCs by protein expressions of pluripotency. Fluorescence intensity of OCT4-GFP showed that LIUS stimulation only increased from HA hydrogel, while PEG and Gel did not change (Fig. 4b). Similarly, immunofluorescence stained SOX2 and NANOG also showed that LIUS stimulation regulated almost 3.6 fold increase from HA hydrogel, while other hydrogels remained its expressions (Fig. 4c and d). These results imply that HA ECM is necessary during LIUS stimulation to regulate CD44 expression and ultimately increase reprogramming efficiencies into iPSCs.
Fig. 4.
| LIUS stimulation under HA hydrogel mediates initial expression of CD44 and affects reprogramming efficiency. a, Immunofluorescence staining and its intensities of CD44 after LIUS stimulation under different microenvironment system (scale bar, 300 μm; n = 3). The reprogramming efficiency of LIUS system under different hydrogels were analyzed by immunofluorescence expression and its intensities of pluripotent markers, b, OCT4-GFP, c, NANOG, and d, SOX2 (scale bar, 300 μm; n = 3). All data are expressed as the mean ± s.d. The statistical significance was determined with two-sided t-tests for comparisons between two experimental groups. n.s., not significant, ****P < 0.0001.
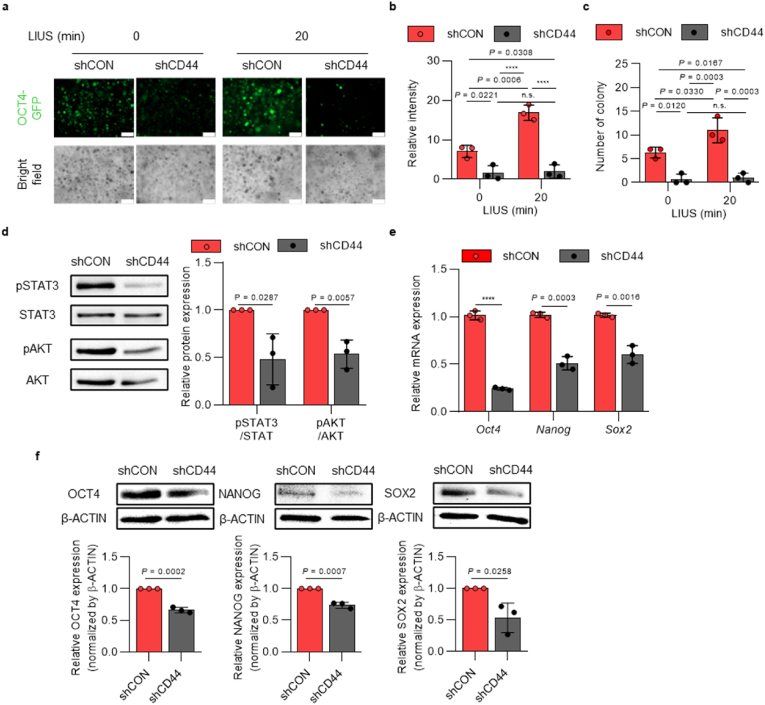
To further examine if LIUS solely improves the reprogramming efficiency of iPSCs without any CD44 modulations, we used downregulation of CD44 and performed the reprogramming with 20 min of LIUS application. In this experiment, shRNA was used to establish stably downregulated CD44-deficient OG-MEFs (shCD44) (Supplementary Figs. 9a–c). After the cells were characterized with CD44 downregulation, they were reprogrammed into iPSCs under 20 min of LIUS stimulation. Compared to the control group (shCon), the shCD44 group showed a dramatic reduction in OCT4-GFP expressions throughout reprogramming under LIUS stimulation (Fig. 5a). At day 21, the shCD44 cells demonstrated reduced number of colonies and fluorescence intensities of OCT4-GFP (Fig. 5b and c). LIUS mediated phosphorylation of downstream signaling molecules (STAT3 and AKT) was observed to be downregulated as CD44 was downregulated (Fig. 5d). The analyzed pluripotent markers also showed reduced mRNA and protein expressions when CD44 was downregulated (Fig. 5e and f). These results suggest that LIUS stimulation regulates the reprogramming efficiency of iPSCs mediated by the expression of CD44 interacting with the HA microenvironment.
Fig. 5.
| LIUS stimulation does not affect reprogramming efficiency into iPSCs in the absence of CD44 expression. a, OCT4-GFP expression of shCD44-induced cells under LIUS stimulation throughout the reprogramming process (scale bar, 300 μm). b, Relative fluorescence intensity and c, number of OCT4-GFP-expressing colonies of LIUS stimulated reprogrammed shCD44 cells (n = 3). d, Phosphorylation of signaling proteins (STAT3, AKT), analyzed with western blotting (n = 3). e, qRT-PCR results of pluripotent markers, along with f, western blotted images and intensities (n = 3). All data are expressed as the mean ± s.d. The statistical significance was determined with one-way ANOVA (b,c) followed by Tukey's multiple comparison post test or two-sided t-tests (d–f). n.s., not significant, ****P < 0.0001.
2.5. iPSC reprogramming applications of LIUS stimulation in HA microenvironment in human cell and in vivo condition
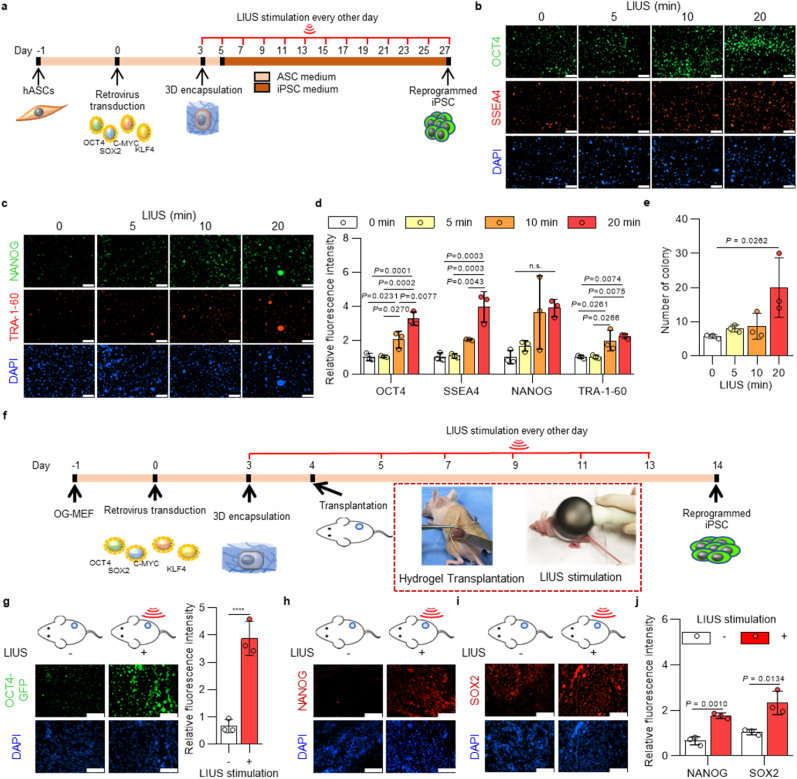
Results have shown that LIUS stimulation in HA microenvironment system enhances cellular reprogramming efficiencies in mouse cell, and we applied this system to human primary cells for increased iPSC production. For the reprogramming process of human primary cells, ASCs were isolated from patients and introduced retrovirally with Yamanaka's four factors. After encapsulation of transduced cells in HA hydrogel, LIUS was introduced every other day for a given time period (Fig. 6a). After the observation of viable LIUS conditions in human ASCs, up to 20 minutes of LIUS were stimulated for the analysis (Supplementary Fig. 10). Reprogrammed cells were stained with pluripotent protein markers to examine the effect of LIUS stimulation. Immunofluorescence images show that more pluripotent expressions were observed from the human ASCs as the LIUS stimulations increased (Fig. 6b and c). Along with the OCT4 and NANOG, pluripotent markers for both mouse and human iPSCs, SSEA4 and TRA-1-60 proteins, which solely present in human iPSCs [60], were also increased from LIUS stimulation. Protein expressions were quantified its intensities and showed that all the analyzed pluripotent markers are significantly increased at 20 min of LIUS stimulation (up to 3.27, 3.96, 3.89, and 2.22 times higher intensities of OCT4, SSEA4, NANOG, and TRA-1-60 proteins were quantified in LIUS, respectively) (Fig. 6d). Number of colonies formed were also indicated that 20 min of LIUS significantly increased in number (Fig. 6e), presenting that LIUS/HA hydrogel system can be applied to the reprogramming process in human cell.
Fig. 6.
| LIUS stimulation enhances reprogramming efficiencies into iPSCs in different applications. a, Schematic representation of the reprogramming of human adipose derived stem cells (hASCs) into iPSCs under ultrasound stimulation. Immunofluorescence staining of b, OCT4 (Green), SSEA4 (Red), c, NANOG (Green), and TRA-1-60 (Red) of hASCs under LIUS stimulation after reprogramming (scale bar, 300 μm). d, Quantification of fluorescence intensity in each immunofluorescence stained cell sample, and e, number of iPSC colonies derived from hASCs (n = 3). f, Schematic representation of the reprogramming of OG-MEFs into iPSCs under ultrasound stimulation in vivo. g, Fluorescence images of OCT4-GFP and its quantitative intensities from iPSCs reprogramming of OG-MEFs in vivo (scale bar, 100 μm; n = 3). Immunofluorescence images of pluripotent markers, h, NANOG and i. SOX2 from iPSCs derived from OG-MEFs under in vivo LIUS stimulation in HA hydrogel (scale bar, 100 μm), and j, quantified its fluorescence intensities (n = 3). All data are expressed as the mean ± s.d. The statistical significance was determined with one-way ANOVA (d,e) followed by Tukey's multiple comparison post test or two-sided t-tests (f,i). n.s., not significant, ****P < 0.0001.
Common procedures for cellular reprogramming are performed in vitro, understanding the cell fate specification and plasticity under controlled environment. To further utilize the LIUS/HA hydrogel technique in regenerative medicine, we investigated the reprogramming efficiency of LIUS in vivo environment. For LIUS stimulation in vivo, ultrasound therapy device was used to deliver sound waves in vivo. First, the ultrasound therapy device showed significant cytotoxicity of OG-MEFs from 20 min (Supplementary Fig. 11). Therefore, LIUS stimulation of the device was set to 10 min throughout the process. After the encapsulation of OG-MEFs under reprogramming process, HA hydrogel was transplanted to a mouse and further processed with LIUS stimulation until analysis (Fig. 6f). The immunofluorescent staining of pluripotent markers showed the elevated expression from LIUS stimulation (Fig. 6g–i). The presented result is meaningful that the LIUS/HA system can solely enhance the reprogramming process within in vivo condition. The relative fluorescence intensities also verify that expression of OCT4, NANOG, and SOX2 increased to 3.87, 1.77, and 2.33 times, respectively (Fig. 6j). Therefore, LIUS stimulation under HA hydrogel can be utilized in vivo reprogramming process to enhance reprogramming efficiency.
3. Discussion
Cell reprogramming technique has been highlighted as a promising tool for cell therapy and biomedical applications, yet key challenges still remain unsolved for its clinical use. Cell reprogramming is a complex process comprising some key events to convert somatic cells to iPSCs, and these key hallmarks include metabolic changes, inhibition of senescence, epigenetic modifications, morphological transitions, and pluripotency [55,61,62]. Bioengineering technologies have offered multiple approaches to enhance cell reprogramming process into desired cell types and numbers. In specific, engineering biomaterials to modulate biochemical and biophysical cues in a microenvironment have expanded the possibility to further control cell fate and function. Importance of microenvironment is a key regulating factor for cell behavior that it mimics the in vivo-like extracellular environment producing optimal biochemical and biophysical cues. Previous finding has shown that composition of ECM can impact reprogramming process, and it can be further modulated with biophysical cues [53,63]. Biophysical factors directly influence the cell surfaces, changing the phenotypic transitions by cytoskeletal rearrangements. Furthermore, biophysical changes can modulate the chromatin structure by regulating the size, shape, and stiffness of the nucleus [[64], [65], [66]]. These biophysical modulations can lead to regulation of transmembrane localization, signaling pathway, and nuclear localization that ultimately impact expression of transcription factors. LIUS, another stimulus to regulate biophysical cues, is a noninvasive acoustic wave that is well known for its safety for various cells, including fibroblast [67,68] and adipose-derived stem cells [[69], [70], [71]]. Although biophysical regulation impacting on cells or extracellular matrices are well-researched in tissue engineering and cell differentiation, there has been no studies exploring the reprogram cells into iPSCs clarifying the relationship of LIUS as biophysical regulation to microenvironments.
Here, we have presented LIUS as a new biophysical factor that can stimulates cell to promote reprogramming process of somatic cells into iPSCs throughout the MET, epigenetic modulation, and pluripotency expressions. LIUS directly alters cells to temporarily modulate cellular plasticity in cytoskeletal structure and membrane fluidity to facilitate actin remodeling and provide higher interaction with ECM components around the 3D microenvironment. 3D cell culture system is used in this study because it is advantageous over conventional 2D culture to observe the precise efficacy of LIUS stimulation, as the 3D system resembles cellular phenotypes, structures, and adhesion kinetics observed in vivo. Previous reports have indicated different regulatory mechanisms of mechanotransduction and cell stimulation between 2D and 3D [72,73]. Specifically, 2D culture generates forced polarity of cells, inducing varied biophysical modulations of LIUS or ECM interactions [74]. Through the LIUS stimulation, cytoskeletal remodeling and interaction between HA and CD44 has indicated as key factors to increase reprogramming efficiency of iPSCs, verifying that the effect of LIUS is highly influenced under optimal 3D microenvironment to improve cell reprogramming. As a result, biophysical modulation of LIUS stimulation within 3D microenvironment can substantially increase iPSC reprogramming efficiency up to 15-fold from traditional methods. The application potentials of LIUS are further confirmed with human primary cells isolated from human patients and in vivo condition that reprogramming efficiencies of iPSCs can be upregulated.
We suggest that LIUS directly initiates the cellular plasticity of a cell during reprogramming process. While LIUS induces insufficient forces to affect the mechanical properties of microenvironments [75,76], LIUS stimulates the intracellular cytoskeletal structure as a biophysical change, which initiates cell transition and cell-matrix interactions surrounding its microenvironments [77,78]. The report indicated that ultrasound-induced strains break down the intracellular cytoskeletal filaments under viable conditions [50], and the softening of the actin stress filaments affects cell mechanosensing and cell properties [79]. Mechanical modulations alter cytoskeletal remodeling of cells and biochemically shift signaling pathways to regulate stem cell fate [80,81]. Studies indicate that the stiffness of iPSCs is soft compared to those of fibroblasts and human-adipose-derived stromal cells that are not pluripotent [82]. Our results showed that LIUS reduces cytoskeletal structures of stimulated somatic cells, followed by increased membrane fluidity to soften the cell rigidity. Cytoskeletal structures were then remodeled to signal the focal adhesion pathway, which is responsible for the activation of STAT3 [83,84] and highly influenced cell reprogramming process in cell transition and epigenetic modification. Choi et al. validated the relationship between F-actin stress fibers and MET during cell reprogramming in that F-actin stress fibers reorganize as cells form the epithelium in the morphology [25]. Other studies have also indicated the role of mechanical modulation in epigenetic changes during cell reprogramming, and biophysical cues can induce actin depolymerization and reorganization to remodel cytoskeletal structure and transmit focal adhesion signaling for iPSC reprogramming [85,86]. The presented results correlate that cytoskeletal rearrangement and cellular rigidity from LIUS promotes cell transitions towards epithelial-like cells and epigenetic modification towards the reprogramming acceleration into iPSCs.
In addition to cell transitions by cytoskeletal disruption of LIUS, it is also known to influence the membrane fluidity of cells and mobility of the membrane receptor proteins (CD44) that interact with the surrounding biomaterials (HA). The acoustic force exerted by LIUS induces membrane fluidity of the cells, and the transmembrane receptors increase its mobility within the cell membrane. LIUS is already known to induce membrane fluidization by dynamic cortical F-actin cytoskeleton, allowing high cellular plasticity [87]. Different lineages from pluripotent to differentiated cells show variable cell membrane fluidities. As pluripotent stem cells lose their pluripotency towards the differentiated state, membrane fluidization decreases and the lipid structure gets ordered [88]. When cell membranes are disordered, the membrane receptors are less confined and have increased mobility, such that they encounter targets more easily [89]. Our results showed the cells surrounded by the HA substratum further upregulated the CD44 expression as a result from the LIUS stimulation, and it enhances the reprogramming efficiency of iPSCs. Interestingly, LIUS stimulation under removed HA substratum or inhibited CD44 expression did not increase pluripotency after cell reprogramming. Thus, the mobility of CD44 in the cell membrane is affected by LIUS stimulation in that more HA engages with the less confined CD44. A related study also mentioned that HA/CD44 binding is dependent on the spatial alignments of HA and CD44, and increased flexibility could facilitate the binding process [90]. Our investigations of LIUS uncovered the enhancement of cellular expression of CD44 under HA microenvironments that accelerates reprogramming efficiency, and it is possible to further modify LIUS stimulation for increased cellular interaction with other ECM microenvironments.
In our system, the HA hydrogel showed reprogramming efficiency of iPSCs by the interaction of HA with the CD44 transmembrane receptor that initiates reprogramming via enhanced MET, epigenetic, and pluripotent expressions [53]. Reports have indicated that HA acts as an adhesive bridging the molecules between cells that express CD44 [91], and CD44 expression is associated with the several signaling key molecules, such as STAT3, and AKT that are associated with transcription factors [[92], [93], [94]]. Our results also indicated high expression of CD44 induced phosphorylation of STAT3 and AKT proteins for activation. Each signaling molecule is highly important for pluripotency transcription markers, as STAT3 and AKT undergoes transcriptional activation of the pluripotent markers, KLF4, NANOG, SOX2, and OCT4 [[95], [96], [97]]. Phosphorylation of STAT3 at ser727 are found to regulate MET change, which our result also indicates CD44 induced STAT3 phosphorylation at ser727 increased epithelial marker over mesenchymal marker. Therefore, increased CD44 expression by LIUS also upregulated MET, epigenetic, and pluripotent markers. Additional studies on other intracellular signaling pathways are necessary to clarify the understanding of LIUS stimulation from 3D microenvironment during cell reprogramming, but the presented results imply that HA-CD44 signaling activates downstream molecules, STAT3 and AKT, for regulation of transcription factors facilitating reprogramming process.
Although LIUS/HA hydrogel system showed enhanced cell reprogramming efficiencies in mouse somatic cells, reproducible effects in human primary cells and in vivo condition are not validated. Since the cell reprogramming is intended to convert patient derived cells into iPSCs, investigating the reprogramming efficiency of LIUS stimulation under HA hydrogel should be verified from human cell. Human primary cells, isolated from patient's tissue, also showed high reprogrammed cells into iPSCs. More number of reprogrammed cells in human tissue demonstrates the potential for utilizing this method in clinical therapy and tissue regeneration. Moreover, LIUS stimulation in HA hydrogel is applied in in vivo environment. Recently, in vivo reprogramming has gained much attention for utilizing the reprogramming technique directly to the regenerative medicine applications [98]. However, generating reprogramming cells in vivo still raises challenges related to safety considerations and the constraint of limited conversion efficiency. HA hydrogels and LIUS are highly recognized materials for safety as these are widely utilized in clinical applications. These biocompatible materials are further validated its functional ability to increase reprogramming efficiency by transplanting HA encapsulated mouse cells and stimulating with LIUS. Results on pluripotency expression from mouse tissues showed more expressed pluripotent proteins from LIUS stimulation. The results are meaningful in that the previous cell reprogramming methods often encounter reduced reprogramming efficiencies when translated in vivo [99]. However, the LIUS/HA system show increased reprogramming efficiencies in both in vitro and in vivo condition. The result can be best explained by the HA hydrogel forming the surrounding microenvironment structure that mimics in vivo of reprogrammed cells, which the equivalent regulatory effects were induced from any condition when stimulated with LIUS. Other regulatory factors may be involved in LIUS during the reprogramming process in vivo, given the complexity of physiological responses. However, the LIUS stimulation presented here can be proposed as a potential tool for enhancing cell reprogramming systems. LIUS stimulation can also be extended to other in vivo cell reprogramming studies to demonstrate its applicability to different cell types, such as neuronal and musculoskeletal cells, as LIUS has shown potential for regenerating musculoskeletal and nerve systems [100,101]. Therefore, the presented LIUS/HA cell reprogramming technique can be suggested as a significant platform for various reprogramming applications in both human primary cell and in vivo condition.
In this study, we have suggested methods to increase the reprogramming efficiency of iPSCs via modulation of the biophysical factor LIUS in a 3D hydrogel system. While initial exposure to ultrasound has been reported to show pluripotent potential [49], no clear understanding of the mechanisms underlying the relationship between ultrasound and reprogramming is elucidated. The present study uses LIUS to modulate the cellular responses within a 3D hydrogel system. Within the viable conditions, increasing the LIUS time facilitates reprogramming efficiency of the iPSCs by upregulating pluripotent expressions. Moreover, LIUS stimulation enhances the MET and epigenetic markers, which indicate the initial changes during reprogramming. These changes were caused by LIUS activating the cytoskeletal rearrangements and mobility of the cell membrane and transmembrane proteins that easily interact with surrounding molecules. Utilization of LIUS as a biophysical stimulus holds many advantages for cellular reprogramming studies, as the reprogramming efficiency of LIUS is as effective as those of other biophysical cues. Furthermore, LIUS can be applied in other reprogramming processes (direct reprogramming or in vivo reprogramming) under modified microenvironments as other mechanical stimulants have already presented such potential in in vivo reprogramming [102,103]. This noninvasive biophysical LIUS stimulation represents a novel research area in bioengineering and mechanotransduction that can be expanded in the tissue engineering and in vivo reprogramming.
4. Conclusions
We demonstrated a new reprogramming strategy involving noncontact ultrasonic stimulation in a 3D hydrogel system, whereby Yamanaka's four-factor-induced reprogramming efficacy was improved. Integration of LIUS stimulation with the established 3D hydrogel system enhances the reprogramming efficiency, as more ultrasound stimulation leads to higher cellular reprogramming of iPSCs. LIUS stimulation also degrades the cytoskeletal structure, followed by increased fluidity and mobility of the cell membrane. These changes lead to increased expressions of CD44 with HA microenvironment only, and downstream signaling pathways of STAT3 and AKT led to the regulations of METs, epigenetics, and pluripotency during reprogramming. The established 3D microenvironment system integrated with a combinatorial LIUS is thus suggested as a new platform for cell reprogramming studies, and its capability for cell reprogramming can be applied in both human primary cells and in vivo tissue as well. Accordingly, the strategy presented herein is expected to be beneficial for various biological and biomedical applications.
5. Materials and methods
5.1. Preparation of photo-crosslinkable polymers
Photo-crosslinkable polymers were synthesized as previously reported [53]. In brief, methacrylated hyaluronic acid (MAHA) was synthesized by adding 1.5 % v/v of methacrylic anhydride to 1 % w/v hyaluronic acid (MW = 500 kDa, Bioland, South Korea) in aqueous solution (Supplementary Fig. 1a). While mixture was stirred for 24 h in the dark at 4 °C, 1.5 % v/v of 5 N NaOH was added to adjust the pH. Mixture was dialyzed for 3 days against deionized water using a dialysis membrane with a cutoff molecular weight of 100 kDa. After removing the remaining impurities, the mixture was lyophilized before use. The synthesized MAHA was analyzed for its relative peak of the methacrylate protons (peaks at ∼6.1, 5.6, and 1.85 ppm) using proton nuclear magnetic resonance spectroscopy (1H-NMR; 500 MHz FT-NMR Spectrometry, Bruker, USA) (Supplementary Fig. 1b). Methacrylated gelatin was synthesized from 10 % w/v Gelatin type B (Sigma Aldrich, St. Loius, USA) dissolved in phosphate buffer (25 mM potassium phosphate monobasic and 170 mM sodium phosphate dibasic) under 50 °C. While stirring, 10 % v/v methacrylic anhydride solution was added dropwise and reacted for 1 h, at pH of 7.5 in the dark. The reaction solution was then filtered with 100 μm cell strainer, dialyzed against distilled water for 3 days in 10 kDa membrane tube, and lyophilized for the final product.
5.2. Preparation of hydrogels
To prepare the HA, Gelatin (Gel), and Poly(ethylene glycol) (PEG) hydrogels, polymer solutions of MAHA (0.5 % w/v), methacrylated gelatin (7 % w/v), and PEG diacrylate (5 % w/v, Alfa Aesar, USA) in Dulbecco's phosphate-buffered saline (DPBS) with Irgacure 2959 (final concentration of 0.2 % w/v) were prepared. After adding 40 μL of the polymer solution to a polydimethylsiloxane (PDMS) mold of depth 2 mm and diameter 5 mm, the sample was exposed to UV light (365 nm, 60 mW cm−2, Sei Myung Vactron Co., Ltd., Korea) for 10 s (HA) and 50 s (Gel and PEG).
5.3. Hydrogel characterization
Swelling properties: The swelling ratio of the HA hydrogel was measured by immersing a sample in DPBS under LIUS at 37 °C for 3 days. Before weighing the swollen sample, the remaining water on the sample surface was blotted with a paper. The swelling ratio at each time point was defined by the weight ratio of the net liquid uptake to the dried hydrogel.
Rheological characterization: To characterize the mechanical properties of the HA hydrogel, the sample was measured using a HAAKE Rheostress1 (Thermo Scientific, USA). The volume of each sample was 500 μL. The oscillatory frequency sweep was applied at a constant oscillatory stress of 0.1 Pa for frequencies from 0.05 to 20 Hz. For all tests, the temperature was maintained at 37 °C.
5.4. Cell culture
Mouse embryonic fibroblasts (MEFs) were modified for OCT4-GFP expression (OG-MEF). The OG-MEFs were obtained from Dr. TaeHee Lee (Sejong University, South Korea) and incubated in a growth medium composed of Dulbecco's modified Eagle's medium (DMEM, Hyclone, USA) with 10 % v/v fetal bovine serum (FBS, Hyclone) and 1 % v/v penicillin/streptomycin (P/S, Hyclone). The cells were maintained at 37 °C in a humidified 5 % CO2 incubator and passaged with 0.25 % trypsin/EDTA (Hyclone). For F-actin modulation test, 2 μM of cytochalasin D (Sigma Aldrich) or 0.05 μM of phalloidin (Sigma Aldrich) was treated for 30 minutes. Human primary adipose-derived stem cells (ASCs) were obtained from the human infrapatellar fat pad of patients' knee, with approval of the Ethics Committee at Dongguk University (IRB No. DUIRB-202210-18). Briefly, human infrapatellar fat pad was washed with DPBS consisting 2 % P/S for three times. Washed tissue was digested with 0.5 mg mL−1 collagenase (Sigma Aldrich) and 1 % P/S diluted in DMEM with low glucose at 37 °C for 45 min. Then, digested tissue was processed to isolate cell pellets by filtration (45 μm strainer) and centrifugation (1000×g for 5 min). ASC cell pellet was resuspended and cultured in a growth medium composed of low glucose DMEM with 10 % fetal bovine serum and 1 % P/S at 37 °C in a humidified 5 % CO2 incubator.
For induction of the OG-MEFs into the mouse iPSCs, a mouse iPSC medium, consisted of DMEM with 15 % v/v FBS, 1 % v/v P/S, 1 % v/v non-essential amino acids (NEAA, Thermo Scientific), 1 % v/v N-2-hydroxyehtylpiperazine-N-2-ethane sulfonic acid (HEPES, Thermo Scientific), 1 % v/v Glutamax (Thermo Scientific), 1 % v/v EmbryoMax Nucleosides (Sigma Aldrich), 0.1 % v/v beta-mercaptoethanol (Thermo Scientific), and 1,000 units mL−1 leukemia inhibitory factor (LIF; Sigma Aldrich), was used. Human ASCs were induced into the human iPSCs using human iPSC medium, consisting of DMEM/F12 (Thermo Scientific) with 20 % KnockOut™ serum replacement (Thermo Scientific), 1 % P/S, 1 % NEAA, 0.1 % beta-mercaptoethanol, and 4 ng mL−1 recombinant human FGF-basic (rhFGF-b; Thermo Scientific).
5.5. Generation of iPSCs
On the day before transfection, GP2-293 packaging cells (Clontech, Germany) were seeded at a density of 4 × 106 cells per 100 mm dish; pMXs-hOCT4, pMXs-hSOX2, pMXs-hKLF4, and pMXs-hc-MYC (Addgene, USA) were transfected using retroviral packaging vector VSV-G (Thermo Scientific) and lipofectamine 2000 reagent (Thermo Scientific). The collected medium was centrifuged at 1,300 rpm for 3 min to remove debris, and the supernatant was filtered using a 0.45 μm syringe filter. Then, the filtered supernatant was loaded on an Amicon® Ultra-15 10 kDa Centrifugal Filter (Merck, USA) and centrifuged at 4,000×g and 4 °C for 20 min. The resulting solution was resuspended in fresh growth medium containing 8 μg mL−1 polybrene (Sigma Aldrich). Each OG-MEFs and ASCs were pre-seeded at a density of 1 × 106 and 2 × 105 cells, respectively, in 100 mm dishes before transduction. About 10 mL of the growth medium containing the retrovirus and polybrene was added to each dish, and the medium was replaced with a new growth medium after 24 h. After 48 h for complete expression, the transduction efficiencies of the OG-MEFs (>90 %) and ASCs (>50 %) were confirmed, and the detached cells were suspended in each hydrogel solution at a density of 2 × 106 cells mL−1. The transduced cells encapsulated hydrogel was replaced with the iPSC medium every day.
5.6. Ultrasound stimulation
Ultrasonic stimulation was applied using a Digital Ultrasonic Set (Daehan, Korea). Each OG-MEF-encapsulated HA hydrogel was collected in a 50 mL tube with 2 mL of the medium and exposed to LIUS for 0, 5, 10, and 20 min at a frequency of 40 kHz and an intensity of 300 mW cm−2 on alternate days during the culture period.
5.7. Live & dead staining
Live/dead fluorescence staining was performed to estimate the cytotoxicity of ultrasonic stimulation. First, the HA hydrogel was washed with DPBS and stained with 2 μM calcein AM (Thermo Scientific) as well as 4 μM ethidium homodimer-1 (Thermo Scientific) in DPBS solution for 30 min. Fluorescence images of the live (green) and dead (red) cells were then observed using a Cytation3 (Biotek, USA).
5.8. Gene expression analysis
For quantitative real-time polymerase chain reaction (qRT-PCR) analysis, the HA hydrogels were frozen in liquid nitrogen and disrupted with a homogenizer in 200 μL of TRIzol™ (Thermo Scientific). After complete homogenization, an additional 800 μL of TRIzol and 200 μL of chloroform were added. The samples were then vortexed and centrifuged at 13,000 rpm and 4 °C for 20 min. The supernatant was mixed with an equal amount of isopropanol, and the mixture was centrifuged at 13,000 rpm and 4 °C for 20 min. The supernatant was removed from the pellet, and washed with 75 % ethyl alcohol, followed by another round of centrifugation at 13,000 rpm and 4 °C for 10 min. The pellets were completely dried and resuspended in RNase-free water (Thermo Scientific). RNA quantification was then performed using the Cytation3. For cDNA synthesis, complementary DNA was synthesized from 1 μg of total RNA using the PrimeScript™ RT reagent kit (Takara, Japan). Then, qRT-PCR was performed using the Power SYBR® Green PCR Master Mix (Applied Biosystems, UK); the PCR conditions were 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s, 60 °C for 30 s, and melting curve stage of 95 °C for 15 s and 60 °C for 60 s. The primer sequences for the PCR are shown in Supplementary Table 1.
5.9. Western blot
Before western blotting, all hydrogel samples were washed three times with DPBS and frozen with liquid nitrogen. Then, the samples were disrupted with a homogenizer using 50 μL 5X RIPA buffer (Sigma Aldrich) supplemented with a protease inhibitor (Merck) and phosphatase inhibitor (Sigma Aldrich). The extract was centrifuged at 13,000 rpm and 4 °C for 20 min, and the supernatant was collected. The total protein concentration was quantified by bicinchoninic acid protein assay using the Pierce BCA Protein Assay Kit (Thermo Scientific). About 20 μg of each protein sample was separated by denaturing 10 % polyacrylamide gel electrophoresis. The separated proteins were transferred to nitrocellulose membranes and blocked with 5 % skim milk in tris-buffered saline and 0.05 % tween-20 (TBS-T) for an hour. The membranes were incubated overnight at 4 °C with primary antibodies in TBS-T with 5 % bovine serum albumin (BSA). The membranes were then washed three times with TBS-T and incubated for 2 h at room temperature with appropriate horseradish-peroxidase-conjugated secondary antibodies diluted 1:5000 in 5 % skim milk TBS-T. After washing with TBS-T thrice, the protein bands were detected using a Chemi-doc detection system (Bio-Rad, USA), and the images were visualized using Image Lab (Bio-Rad) software. Details of the primary and secondary antibodies are shown in Supplementary Table 2.
5.10. Immunofluorescence staining
The HA hydrogels were washed three times with DPBS, and the washed gels were fixed for an hour at room temperature with 4 % paraformaldehyde (Biosesang, Korea). The fixed hydrogels were permeabilized with 0.3 % (v/v) Triton X-100 in DPBS (PBS-T) at room temperature for 30 min. After blocking with 1 % (w/v) BSA in PBS-T for an hour at room temperature, the hydrogels were incubated in the primary antibody solution (1:200) overnight at 4 °C. The samples were then washed with DPBS and incubated in fluorescein-conjugated secondary antibodies (Thermo Scientific) diluted 1:200 in 1 % BSA in PBS-T for 2 h at room temperature under dark conditions. For F-actin staining, Texas red-X phalloidin (Thermo Scientific) was used. The unbound antibodies were washed with DPBS, and samples were counterstained with 4′6-diamidino-2-phenylindole (DAPI; Thermo Scientific) to observe the cellular nuclei. The fluorescence images were observed using Cytation3.
5.11. Flow cytometry
The surface antigens on the cells were analyzed via flow cytometry. Cells encapsulated in the hydrogels were washed twice with DPBS and dissociated using hyaluronidase from bovine testes Type I-s (Sigma Aldrich) before collection by centrifugation at 1,300 rpm for 3 min. The cell pellets were washed twice with DPBS and blocked with 2 % FBS in DPBS solution (FACS buffer). Specific antibodies (diluted 1:100 in FACS buffer) were incubated at 4 °C for 30 min and washed thrice with FACS buffer. Finally, fluorescence was detected using BD Accuri C6 (BD Bioscience, Japan). The percentage of expressed cell surface antigens was calculated for 10,000 gated cell events. The antibodies used in these experiments were anti-CD44 (103011, Biolegend, UK) and anti-SSEA1 (125608, Biolegend).
5.12. Cell membrane fluidity
The cells were analyzed for membrane fluidity via the membrane fluidity assay (Abcam, UK) according to manufacturer instructions. In brief, cells stimulated with LIUS were labeled with pyrenedecanoic acid (PDA) for an hour at 25 °C under dark conditions. The unincorporated PDA was washed, and the ratio of monomer (Em 400 nm) to excimer (Em 470 nm) fluorescence was normalized to that of the unstimulated cells.
5.13. CD44 knockdown
CD44 RNA was downregulated in the OG-MEFs by retroviral transduction to introduce a predesigned CD44 mouse shRNA into the retroviral vector (Origene, USA) targeting CD44. The CD44 mouse shRNA of the retroviral vector was purchased in annealed and purified form ready to be transfected into the GP2-293 packaging cells for retroviral synthesis. For the transduction, 1 × 106 cells were plated in 100 mm dish, and the adhered cells were transduced with retrovirus synthesized using polybrene. The OG-MEFs were transduced with CD44 mouse shRNA for 24 h and then replaced with new growth medium.
5.14. Electron microscopy
Cells cultured on the HA hydrogel with LIUS stimulation were harvested and washed in PBS, fixed with 2.5 % glutaraldehyde solution, and washed with distilled water. The hydrogel samples were lyophilized and coated with a 10-nm-thick layer of platinum/palladium using a sputter (E−1010, Hitachi, Japan). The morphology of the adhered cells on the hydrogels was observed by scanning electron microscopy (SEM; S-3000 N, Hitachi).
5.15. Ultrasound stimulation in in-vivo
Generation of iPSCs with LIUS stimulation in vivo condition was also analyzed, approved by the Institutional Animal Care and Use Committee of Dongguk University (IACUC-2021-018-3), in accordance with ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines). The transduced OG-MEFs encapsulated hydrogel was transplanted into a single subcutaneous space of randomly selected 6-8 weeks-old male BALB/c nude mice (Orient Bio, Inc., Korea). Each hydrogel is stimulated with ultrasound therapy (ST-10A, StraTek Co., Ltd., Korea). ultrasound was induced on top of hydrogel transplanted skin, at a frequency of 1 MHz and an intensity of 300 mW cm−2 on alternative days. On the day of analysis, each hydrogel was isolated from skin and analyzed.
5.16. Statistical analysis
In all figures, exact n values represent independent experiments performed for each condition. Statistical analysis of the results was then performed using GraphPad prism ver. 8.1.0 (GraphPad Software, USA). All data shown in this study are presented as means ± standard deviation (SD). Two-tailed Student's t-tests were used for comparisons between two experimental groups. One-way ANOVA using Tukey's multiple comparison post-test was implemented to compare the samples. Two-way ANOVA with Bonferroni posthoc testing was used for comparisons among more than two experimental groups with two varying parameters. The statistical significance values were set at n.s., not significant, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Data availability
The main data supporting the results in this study are available within the paper and its supplementary information. The raw and analyzed datasets generated during the study are available for research purposes from the corresponding author on reasonable request.
Ethics approval
Human primary adipose-derived stem cells (ASCs) were obtained from the human infrapatellar fat pad of patients' knee, with approval of the Ethics Committee at Dongguk University (IRB No. DUIRB-202210-18). All animals were bred according to the National Institutes of Health's Guide for Care and Use of Laboratory Animals. All animal studies were carried out, approved by the Institutional Animal Care and Use Committee of Dongguk University (IACUC-2021-018-3), in accordance with ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines).
CRediT authorship contribution statement
Deogil Kim: Writing – review & editing, Writing – original draft, Visualization, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization. Min-Ju Lee: Writing – original draft, Visualization, Validation, Methodology, Investigation, Data curation, Conceptualization. Yoshie Arai: Visualization, Methodology, Investigation, Funding acquisition, Data curation, Conceptualization. Jinsung Ahn: Methodology, Investigation, Data curation. Gun Woo Lee: Validation, Investigation. Soo-Hong Lee: Writing – review & editing, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
The authors gratefully acknowledge funding from the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT, MOE) (NRF-2019M3A9H1032376, NRF-2022R1A2C3004850, RS-2023-00214410, RS-2023-00257290, RS-2023-00246418, and RS-2023-00275407).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2024.05.011.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shi Y., Inoue H., Wu J.C., Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat. Rev. Drug Discov. 2017;16(2):115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Senís E., Mosteiro L., Wilkening S., Wiedtke E., Nowrouzi A., Afzal S., Fronza R., Landerer H., Abad M., Niopek D., Schmidt M., Serrano M., Grimm D. AAV vector-mediated in vivo reprogramming into pluripotency. Nat. Commun. 2018;9(1):2651. doi: 10.1038/s41467-018-05059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo J., Duan L., He X., Li S., Wu Y., Xiang G., Bao F., Yang L., Shi H., Gao M., Zheng L., Hu H., Liu X. A combined Model of human iPSC-derived liver organoids and hepatocytes reveals ferroptosis in DGUOK mutant mtDNA depletion syndrome. Adv. Sci. 2021;8(10) doi: 10.1002/advs.202004680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma A., Garcia G., Wang Y., Plummer J.T., Morizono K., Arumugaswami V., Svendsen C.N. Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Reports Medicine. 2020;1(4) doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burridge P.W., Matsa E., Shukla P., Lin Z.C., Churko J.M., Ebert A.D., Lan F., Diecke S., Huber B., Mordwinkin N.M., Plews J.R., Abilez O.J., Cui B., Gold J.D., Wu J.C. Chemically defined generation of human cardiomyocytes. Nat. Methods. 2014;11(8):855–860. doi: 10.1038/nmeth.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engle S.J., Blaha L., Kleiman R.J. Best practices for translational disease modeling using human iPSC-derived neurons. Neuron. 2018;100(4):783–797. doi: 10.1016/j.neuron.2018.10.033. [DOI] [PubMed] [Google Scholar]
- 7.Bloor A.J.C., Patel A., Griffin J.E., Gilleece M.H., Radia R., Yeung D.T., Drier D., Larson L.S., Uenishi G.I., Hei D., Kelly K., Slukvin I., Rasko J.E.J. Production, safety and efficacy of iPSC-derived mesenchymal stromal cells in acute steroid-resistant graft versus host disease: a phase I, multicenter, open-label, dose-escalation study. Nat. Med. 2020;26(11):1720–1725. doi: 10.1038/s41591-020-1050-x. [DOI] [PubMed] [Google Scholar]
- 8.Monkley S., Krishnaswamy J.K., Göransson M., Clausen M., Meuller J., Thörn K., Hicks R., Delaney S., Stjernborg L. Optimised generation of iPSC-derived macrophages and dendritic cells that are functionally and transcriptionally similar to their primary counterparts. PLoS One. 2020;15(12) doi: 10.1371/journal.pone.0243807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deinsberger J., Reisinger D., Weber B. Global trends in clinical trials involving pluripotent stem cells: a systematic multi-database analysis. npj Regenerative Medicine. 2020;5(1):15. doi: 10.1038/s41536-020-00100-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guhr A., Kobold S., Seltmann S., Seiler Wulczyn A.E.M., Kurtz A., Löser P. Recent trends in research with human pluripotent stem cells: impact of research and use of cell lines in experimental research and clinical trials. Stem Cell Rep. 2018;11(2):485–496. doi: 10.1016/j.stemcr.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J.Y., Nam Y., Rim Y.A., Ju J.H. Review of the current trends in clinical trials involving induced pluripotent stem cells. Stem Cell Reviews and Reports. 2022;18(1):142–154. doi: 10.1007/s12015-021-10262-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corbett J.L., Duncan S.A. iPSC-derived hepatocytes as a platform for disease modeling and drug discovery. Front. Med. 2019;6 doi: 10.3389/fmed.2019.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadtfeld M., Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes & development. 2010;24(20):2239–2263. doi: 10.1101/gad.1963910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al Abbar A., Ngai S.C., Nograles N., Alhaji S.Y., Abdullah S. Induced pluripotent stem cells: reprogramming platforms and applications in cell replacement therapy. Biores Open Access. 2020;9(1):121–136. doi: 10.1089/biores.2019.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G., Guo Y.e., Li C., Li S., Wan X. Small molecules that promote self-renewal of stem cells and somatic cell reprogramming. Stem Cell Reviews and Reports. 2020;16(3):511–523. doi: 10.1007/s12015-020-09965-w. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y., Li W., Laurent T., Ding S. Small molecules, big roles -- the chemical manipulation of stem cell fate and somatic cell reprogramming. J. Cell Sci. 2012;125(Pt 23):5609–5620. doi: 10.1242/jcs.096032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong S.Y., Soto J., Li S. Biophysical regulation of cell reprogramming. Current opinion in chemical engineering. 2017;15:95–101. doi: 10.1016/j.coche.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y., Wang K., Gu X., Leong K.W. Biophysical regulation of cell behavior—cross talk between substrate stiffness and nanotopography. Engineering. 2017;3(1):36–54. doi: 10.1016/J.ENG.2017.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Y., Wan B., Wang R., Zhang B., Luo P., Wang D., Nie J.-J., Chen D., Wu X. Mechanical stimulation on mesenchymal stem cells and surrounding microenvironments in bone regeneration: regulations and applications. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.808303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Love M.R., Palee S., Chattipakorn S.C., Chattipakorn N. Effects of electrical stimulation on cell proliferation and apoptosis. J. Cell. Physiol. 2018;233(3):1860–1876. doi: 10.1002/jcp.25975. [DOI] [PubMed] [Google Scholar]
- 21.Choi B., Kim D., Han I., Lee S.H. Microenvironmental regulation of stem cell behavior through biochemical and biophysical stimulation. Adv. Exp. Med. Biol. 2018;1064:147–160. doi: 10.1007/978-981-13-0445-3_9. [DOI] [PubMed] [Google Scholar]
- 22.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 23.Flanagan L.A., Ju Y.-E., Marg B., Osterfield M., Janmey P.A. Neurite branching on deformable substrates. Neuroreport. 2002;13(18):2411. doi: 10.1097/01.wnr.0000048003.96487.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang H.-B., Dembo M., Wang Y.-L. Substrate flexibility regulates growth and apoptosis of normal but not transformed cells. American Journal of Physiology-Cell Physiology. 2000;279(5):C1345–C1350. doi: 10.1152/ajpcell.2000.279.5.C1345. [DOI] [PubMed] [Google Scholar]
- 25.Choi B., Park K.S., Kim J.H., Ko K.W., Kim J.S., Han D.K., Lee S.H. Stiffness of hydrogels regulates cellular reprogramming efficiency through mesenchymal‐to‐epithelial transition and stemness markers. Macromol. Biosci. 2016;16(2):199–206. doi: 10.1002/mabi.201500273. [DOI] [PubMed] [Google Scholar]
- 26.Guo J., Wang Y., Sachs F., Meng F. Actin stress in cell reprogramming. Proc. Natl. Acad. Sci. USA. 2014;111(49):E5252–E5261. doi: 10.1073/pnas.1411683111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cordie T., Harkness T., Jing X., Carlson-Stevermer J., Mi H.-Y., Turng L.-S., Saha K. Nanofibrous electrospun polymers for reprogramming human cells. Cell. Mol. Bioeng. 2014;7(3):379–393. [Google Scholar]
- 28.Downing T.L., Soto J., Morez C., Houssin T., Fritz A., Yuan F., Chu J., Patel S., Schaffer D.V., Li S. Biophysical regulation of epigenetic state and cell reprogramming. Nat. Mater. 2013;12(12):1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sia J., Sun R., Chu J., Li S. Dynamic culture improves cell reprogramming efficiency. Biomaterials. 2016;92:36–45. doi: 10.1016/j.biomaterials.2016.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y.M., Kang Y.G., Park S.H., Han M.-K., Kim J.H., Shin J.W., Shin J.-W. Effects of mechanical stimulation on the reprogramming of somatic cells into human-induced pluripotent stem cells. Stem Cell Res. Ther. 2017;8(1):1–12. doi: 10.1186/s13287-017-0594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baek S., Quan X., Kim S., Lengner C., Park J.K., Kim J. Electromagnetic fields mediate efficient cell reprogramming into a pluripotent state. ACS Nano. 2014;8(10):10125–10138. doi: 10.1021/nn502923s. [DOI] [PubMed] [Google Scholar]
- 32.Chaicharoenaudomrung N., Kunhorm P., Noisa P. Three-dimensional cell culture systems as an in vitro platform for cancer and stem cell modeling. World J. Stem Cell. 2019;11(12):1065–1083. doi: 10.4252/wjsc.v11.i12.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu X., Su J., Wei J., Jiang N., Ge X. Recent advances in three-dimensional stem cell culture systems and applications. Stem Cell. Int. 2021;2021 doi: 10.1155/2021/9477332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huh D., Hamilton G.A., Ingber D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21(12):745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun M., Liu A., Yang X., Gong J., Yu M., Yao X., Wang H., He Y. 3D cell culture—can it Be as popular as 2D cell culture? Advanced NanoBiomed Research. 2021;1(5) [Google Scholar]
- 36.Caiazzo M., Okawa Y., Ranga A., Piersigilli A., Tabata Y., Lutolf M.P. Defined three-dimensional microenvironments boost induction of pluripotency. Nat. Mater. 2016;15(3):344–352. doi: 10.1038/nmat4536. [DOI] [PubMed] [Google Scholar]
- 37.Sun W., Zhang S., Zhou T., Shan Y., Gao F., Zhang Y., Zhang D., Xiong Y., Mai Y., Fan K., Davidson A.J., Pan G., Zhang X. Human urinal cell reprogramming: synthetic 3D peptide hydrogels enhance induced pluripotent stem cell population homogeneity. ACS Biomater. Sci. Eng. 2020;6(11):6263–6275. doi: 10.1021/acsbiomaterials.0c00667. [DOI] [PubMed] [Google Scholar]
- 38.Fang J., Hsueh Y.-Y., Soto J., Sun W., Wang J., Gu Z., Khademhosseini A., Li S. Engineering biomaterials with micro/nanotechnologies for cell reprogramming. ACS Nano. 2020;14(2):1296–1318. doi: 10.1021/acsnano.9b04837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker K.G., Robertson V.J., Duck F.A. A review of therapeutic ultrasound: biophysical effects. Phys. Ther. 2001;81(7):1351–1358. [PubMed] [Google Scholar]
- 40.Bakhshandeh B., Ranjbar N., Abbasi A., Amiri E., Abedi A., Mehrabi M.-R., Dehghani Z., Pennisi C.P. Recent progress in the manipulation of biochemical and biophysical cues for engineering functional tissues. Bioengineering & Translational Medicine. 2023;8(2) doi: 10.1002/btm2.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia P., Shi Y., Wang X., Li X. Advances in the application of low-intensity pulsed ultrasound to mesenchymal stem cells. Stem Cell Res. Ther. 2022;13(1):214. doi: 10.1186/s13287-022-02887-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xin Z., Lin G., Lei H., Lue T.F., Guo Y. Clinical applications of low-intensity pulsed ultrasound and its potential role in urology. Transl. Androl. Urol. 2016;5(2):255–266. doi: 10.21037/tau.2016.02.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Yang H., Wang Z., Zhu R., Cheng L., Cheng Q. 2021. Low-intensity Pulsed Ultrasound Promotes Mesenchymal Stem Cell Transplantation-Based Articular Cartilage Regeneration by Inhibiting the TNF Signaling Pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang T., Liang C., Chen L., Li J., Geng W. Low-intensity pulsed ultrasound alleviates hypoxia-induced chondrocyte damage in temporomandibular disorders by modulating the hypoxia-inducible factor pathway. Front. Pharmacol. 2020;11:689. doi: 10.3389/fphar.2020.00689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee I.C., Wu H.-J., Liu H.-L. Dual-frequency ultrasound induces neural stem/progenitor cell differentiation and growth factor utilization by enhancing stable cavitation. ACS Chem. Neurosci. 2019;10(3):1452–1461. doi: 10.1021/acschemneuro.8b00483. [DOI] [PubMed] [Google Scholar]
- 46.Ning G.-Z., Song W.-Y., Xu H., Zhu R.-S., Wu Q.-L., Wu Y., Zhu S.-B., Li J.-Q., Wang M., Qu Z.-G., Feng S.-Q. Bone marrow mesenchymal stem cells stimulated with low-intensity pulsed ultrasound: better choice of transplantation treatment for spinal cord injury. CNS Neurosci. Ther. 2019;25(4):496–508. doi: 10.1111/cns.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.An Y., Song Y., Wang Z., Wang J., Wu G., Zhu G., Chen L. Effect of low-intensity pulsed ultrasound on the biological behaviors of bone marrow mesenchymal stem cells on titanium with different surface topographies. Am. J. Tourism Res. 2018;10(1):67–76. [PMC free article] [PubMed] [Google Scholar]
- 48.Maung W.M., Nakata H., Miura M., Miyasaka M., Kim Y.K., Kasugai S., Kuroda S. Low-intensity pulsed ultrasound stimulates osteogenic differentiation of periosteal cells in vitro, tissue engineering. Part. Accel. 2021;27(1–2):63–73. doi: 10.1089/ten.TEA.2019.0331. [DOI] [PubMed] [Google Scholar]
- 49.Lee Y.S., Heo H., Lee J., Moon S.U., Jung W.Y., Park Y.K., Park M.G., Oh S.-H., Kim S. An ultra-effective method of generating extramultipotent cells from human fibroblasts by ultrasound. Biomaterials. 2017;143:65–78. doi: 10.1016/j.biomaterials.2017.07.033. [DOI] [PubMed] [Google Scholar]
- 50.Samandari M., Abrinia K., Mokhtari-Dizaji M., Tamayol A. Ultrasound induced strain cytoskeleton rearrangement: an experimental and simulation study. J. Biomech. 2017;60:39–47. doi: 10.1016/j.jbiomech.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 51.Atherton P., Lausecker F., Harrison A., Ballestrem C. Low-intensity pulsed ultrasound promotes cell motility through vinculin-controlled Rac1 GTPase activity. J. Cell Sci. 2017;130(14):2277–2291. doi: 10.1242/jcs.192781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim J., Chu Y.-S., Chu Y.-C., Lo C.-M., Wang J.-L. Low intensity ultrasound induces epithelial cell adhesion responses. J. Biomech. Eng. 2020;142(9) doi: 10.1115/1.4046883. [DOI] [PubMed] [Google Scholar]
- 53.Kim D., Cha B.-H., Ahn J., Arai Y., Choi B., Lee S.-H. Physicochemical properties in 3D hydrogel modulate cellular reprogramming into induced pluripotent stem cells. Adv. Funct. Mater. 2021;31(7) [Google Scholar]
- 54.Schmidt R., Plath K. The roles of the reprogramming factors Oct4, Sox2 and Klf4 in resetting the somatic cell epigenome during induced pluripotent stem cell generation. Genome Biol. 2012;13(10):251. doi: 10.1186/gb-2012-13-10-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buganim Y., Faddah D.A., Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat. Rev. Genet. 2013;14(6):427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin H., Zhao A., Zhang C., Fu X. Epigenetic control of reprogramming and transdifferentiation by histone modifications. Stem Cell Reviews and Reports. 2016;12(6):708–720. doi: 10.1007/s12015-016-9682-4. [DOI] [PubMed] [Google Scholar]
- 57.Gao J., Nakamura F. Actin-associated proteins and small molecules targeting the actin cytoskeleton. Int. J. Mol. Sci. 2022;23(4):2118. doi: 10.3390/ijms23042118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wakatsuki T., Schwab B., Thompson N.C., Elson E.L. Effects of cytochalasin D and latrunculin B on mechanical properties of cells. J. Cell Sci. 2001;114(Pt 5):1025–1036. doi: 10.1242/jcs.114.5.1025. [DOI] [PubMed] [Google Scholar]
- 59.Oliferenko S., Paiha K., Harder T., Gerke V., Schwärzler C., Schwarz H., Beug H., Günthert U., Huber L.A. Analysis of Cd44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. JCB (J. Cell Biol.) 1999;146(4):843–854. doi: 10.1083/jcb.146.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ginis I., Luo Y., Miura T., Thies S., Brandenberger R., Gerecht-Nir S., Amit M., Hoke A., Carpenter M.K., Itskovitz-Eldor J., Rao M.S. Differences between human and mouse embryonic stem cells. Dev. Biol. 2004;269(2):360–380. doi: 10.1016/j.ydbio.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 61.David L., Polo J.M. Phases of reprogramming. Stem Cell Res. 2014;12(3):754–761. doi: 10.1016/j.scr.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 62.Apostolou E., Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502(7472):462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin Y., Lee J.S., Kim J., Min S., Wi S., Yu J.H., Chang G.-E., Cho A.-N., Choi Y., Ahn D.-H., Cho S.-R., Cheong E., Kim Y.-G., Kim H.-P., Kim Y., Kim D.S., Kim H.W., Quan Z., Kang H.-C., Cho S.-W. Three-dimensional brain-like microenvironments facilitate the direct reprogramming of fibroblasts into therapeutic neurons. Nat. Biomed. Eng. 2018;2(7):522–539. doi: 10.1038/s41551-018-0260-8. [DOI] [PubMed] [Google Scholar]
- 64.Hernandez M., Patzig J., Mayoral S.R., Costa K.D., Chan J.R., Casaccia P. Mechanostimulation promotes nuclear and epigenetic changes in oligodendrocytes. J. Neurosci. 2016;36(3):806–813. doi: 10.1523/JNEUROSCI.2873-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iyer K.V., Pulford S., Mogilner A., Shivashankar G. Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophys. J. 2012;103(7):1416–1428. doi: 10.1016/j.bpj.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lovett D.B., Shekhar N., Nickerson J.A., Roux K.J., Lele T.P. Modulation of nuclear shape by substrate rigidity. Cell. Mol. Bioeng. 2013;6(2):230–238. doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ikeda K., Takayama T., Suzuki N., Shimada K., Otsuka K., Ito K. Effects of low-intensity pulsed ultrasound on the differentiation of C2C12 cells. Life Sci. 2006;79(20):1936–1943. doi: 10.1016/j.lfs.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 68.Zhou S., Schmelz A., Seufferlein T., Li Y., Zhao J., Bachem M.G. Molecular mechanisms of low intensity pulsed ultrasound in human skin fibroblasts. J. Biol. Chem. 2004;279(52):54463–54469. doi: 10.1074/jbc.M404786200. [DOI] [PubMed] [Google Scholar]
- 69.Song B.-W., Park J.-H., Kim B., Lee S., Lim S., Kim S.W., Choi J.-W., Lee J., Kang M., Hwang K.-C., Chae D.-S., Kim I.-K. A combinational therapy of articular cartilage defects: rapid and effective regeneration by using low-intensity focused ultrasound after adipose tissue-derived stem cell transplantation. Tissue Engineering and Regenerative Medicine. 2020;17(3):313–322. doi: 10.1007/s13770-020-00256-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang D., Gao Y., Wang S., Zhang W., Cao H., Zheng L., Chen Y., Zhang S., Chen J. Impact of low-intensity pulsed ultrasound on transcription and metabolite compositions in proliferation and functionalization of human adipose-derived mesenchymal stromal cells. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-69430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ahn J., Arai Y., Kim B.J., Seo Y.-K., Moon J.J., Shin D.A., Choi B., Lee S.-H. Combinatorial physicochemical stimuli in the three-dimensional environment of a hyaluronic acid hydrogel amplify chondrogenesis by stimulating phosphorylation of the Smad and MAPK signaling pathways. NPG Asia Mater. 2022;14(1):46. [Google Scholar]
- 72.Cukierman E., Pankov R., Stevens D.R., Yamada K.M. Taking cell-matrix adhesions to the third dimension. Science. 2001;294(5547):1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 73.Saraswathibhatla A., Indana D., Chaudhuri O. Cell-extracellular matrix mechanotransduction in 3D. Nat. Rev. Mol. Cell Biol. 2023;24(7):495–516. doi: 10.1038/s41580-023-00583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li Y., Kilian K.A. Bridging the gap: from 2D cell culture to 3D microengineered extracellular matrices. Adv. Healthcare Mater. 2015;4(18):2780–2796. doi: 10.1002/adhm.201500427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Handolin L., Pohjonen T., Partio E.K., Arnala I., Törmälä P., Rokkanen P. The effects of low-intensity pulsed ultrasound on bioabsorbable self-reinforced poly l-lactide screws. Biomaterials. 2002;23(13):2733–2736. doi: 10.1016/s0142-9612(02)00006-6. [DOI] [PubMed] [Google Scholar]
- 76.Jia W., Zhou Z., Zhan W. Musculoskeletal biomaterials: stimulated and synergized with low intensity pulsed ultrasound. J. Funct. Biomater. 2023;14(10):504. doi: 10.3390/jfb14100504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shao X., Li Q., Mogilner A., Bershadsky A.D., Shivashankar G.V. Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc. Natl. Acad. Sci. USA. 2015;112(20):E2595–E2601. doi: 10.1073/pnas.1504837112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yao H., Zhang L., Yan S., He Y., Zhu H., Li Y., Wang D., Yang K. Low-intensity pulsed ultrasound/nanomechanical force generators enhance osteogenesis of BMSCs through microfilaments and TRPM7. J. Nanobiotechnol. 2022;20(1):378. doi: 10.1186/s12951-022-01587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andreu I., Falcones B., Hurst S., Chahare N., Quiroga X., Le Roux A.-L., Kechagia Z., Beedle A.E.M., Elosegui-Artola A., Trepat X., Farré R., Betz T., Almendros I., Roca-Cusachs P. The force loading rate drives cell mechanosensing through both reinforcement and cytoskeletal softening. Nat. Commun. 2021;12(1):4229. doi: 10.1038/s41467-021-24383-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Putra V.D.L., Kilian K.A., Knothe Tate M.L. Biomechanical, biophysical and biochemical modulators of cytoskeletal remodelling and emergent stem cell lineage commitment. Commun. Biol. 2023;6(1):75. doi: 10.1038/s42003-022-04320-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McBeath R., Pirone D.M., Nelson C.M., Bhadriraju K., Chen C.S., Shape Cell, Tension Cytoskeletal. RhoA regulate stem cell lineage commitment. Dev. Cell. 2004;6(4):483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 82.Hammerick K.E., Huang Z., Sun N., Lam M.T., Prinz F.B., Wu J.C., Commons G.W., Longaker M.T. Elastic properties of induced pluripotent stem cells. Tissue Eng. 2010;17(3–4):495–502. doi: 10.1089/ten.tea.2010.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Visavadiya N.P., Keasey M.P., Razskazovskiy V., Banerjee K., Jia C., Lovins C., Wright G.L., Hagg T. Integrin-FAK signaling rapidly and potently promotes mitochondrial function through STAT3. Cell Commun. Signal. 2016;14(1):32. doi: 10.1186/s12964-016-0157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarkar Bhattacharya S., Thirusangu P., Jin L., Staub J., Shridhar V., Molina J.R. PFKFB3 works on the FAK-STAT3-SOX2 axis to regulate the stemness in MPM. Br. J. Cancer. 2022;127(7):1352–1364. doi: 10.1038/s41416-022-01867-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soto J., Song Y., Wu Y., Chen B., Park H., Akhtar N., Wang P.Y., Hoffman T., Ly C., Sia J., Wong S., Kelkhoff D.O., Chu J., Poo M.M., Downing T.L., Rowat A.C., Li S. Reduction of intracellular tension and cell adhesion promotes open chromatin structure and enhances cell reprogramming. Adv. Sci. 2023;10(24) doi: 10.1002/advs.202300152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Park S.-M., Lee J.-H., Ahn K.S., Shim H.W., Yoon J.-Y., Hyun J., Lee J.H., Jang S., Yoo K.H., Jang Y.-K., Kim T.-J., Kim H.K., Lee M.R., Jang J.-H., Shim H., Kim H.-W. Cyclic stretch promotes cellular reprogramming process through cytoskeletal-nuclear mechano-coupling and epigenetic modification. Adv. Sci. 2023;10(32) doi: 10.1002/advs.202303395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mizrahi N., Zhou E.H., Lenormand G., Krishnan R., Weihs D., Butler J.P., Weitz D.A., Fredberg J.J., Kimmel E. Low intensity ultrasound perturbs cytoskeleton dynamics. Soft Matter. 2012;8(8):2438–2443. doi: 10.1039/C2SM07246G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsuzaki T., Matsumoto S., Kasai T., Yoshizawa E., Okamoto S., Yoshikawa H.Y., Taniguchi H., Takebe T. Defining lineage-specific membrane fluidity signatures that regulate adhesion kinetics. Stem Cell Rep. 2018;11(4):852–860. doi: 10.1016/j.stemcr.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Freeman S.A., Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol. Rev. 2014;262(1):193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- 90.Wolny P.M., Banerji S., Gounou C., Brisson A.R., Day A.J., Jackson D.G., Richter R.P. Analysis of CD44-hyaluronan interactions in an artificial membrane system: insights into the distinct binding properties of high and low molecular weight hyaluronan. J. Biol. Chem. 2010;285(39):30170–30180. doi: 10.1074/jbc.M110.137562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ponta H., Sherman L., Herrlich P.A. CD44: from adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003;4(1):33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- 92.Yang C., Sheng Y., Shi X., Liu Y., He Y., Du Y., Zhang G., Gao F. CD44/HA signaling mediates acquired resistance to a PI3Kα inhibitor. Cell Death Dis. 2020;11(10):831. doi: 10.1038/s41419-020-03037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.So J.Y., Smolarek A.K., Salerno D.M., Maehr H., Uskokovic M., Liu F., Suh N. Targeting CD44-STAT3 signaling by gemini vitamin D analog leads to inhibition of invasion in basal-like breast cancer. PLoS One. 2013;8(1) doi: 10.1371/journal.pone.0054020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khurana S.S., Riehl T.E., Moore B.D., Fassan M., Rugge M., Romero-Gallo J., Noto J., Peek R.M., Stenson W.F., Mills J.C. The hyaluronic acid receptor CD44 coordinates normal and metaplastic gastric epithelial progenitor cell proliferation. J. Biol. Chem. 2013;288(22):16085–16097. doi: 10.1074/jbc.M112.445551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guofeng H., Hongtao W., Jijun H. In: Pluripotent Stem Cells. Deepa B., Nibedita L., editors. IntechOpen; Rijeka: 2013. Molecular mechanisms of embryonic stem cell pluripotency. Ch. 13. [Google Scholar]
- 96.Nakao S., Tsukamoto T., Ueyama T., Kawamura T. STAT3 for cardiac regenerative medicine: involvement in stem cell biology, pathophysiology, and bioengineering. Int. J. Mol. Sci. 2020;21(6):1937. doi: 10.3390/ijms21061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bourguignon L.Y.W., Earle C., Wong G., Spevak C.C., Krueger K. Stem cell marker (Nanog) and Stat-3 signaling promote MicroRNA-21 expression and chemoresistance in hyaluronan/CD44-activated head and neck squamous cell carcinoma cells. Oncogene. 2012;31(2):149–160. doi: 10.1038/onc.2011.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang H., Yang Y., Liu J., Qian L. Direct cell reprogramming: approaches, mechanisms and progress. Nat. Rev. Mol. Cell Biol. 2021;22(6):410–424. doi: 10.1038/s41580-021-00335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ofenbauer A., Tursun B. Strategies for in vivo reprogramming. Curr. Opin. Cell Biol. 2019;61:9–15. doi: 10.1016/j.ceb.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 100.Harrison A., Alt V. Low-intensity pulsed ultrasound (LIPUS) for stimulation of bone healing – a narrative review. Injury. 2021;52:S91–S96. doi: 10.1016/j.injury.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 101.Kim T., Park C., Chhatbar P.Y., Feld J., Mac Grory B., Nam C.S., Wang P., Chen M., Jiang X., Feng W. Effect of low intensity transcranial ultrasound stimulation on neuromodulation in animals and humans: an updated systematic review. Front. Neurosci. 2021;15 doi: 10.3389/fnins.2021.620863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jin Y., Seo J., Lee J.S., Shin S., Park H.-J., Min S., Cheong E., Lee T., Cho S.-W. Triboelectric nanogenerator accelerates highly efficient nonviral direct conversion and in vivo reprogramming of fibroblasts to functional neuronal cells. Adv. Mater. 2016;28(34):7365–7374. doi: 10.1002/adma.201601900. [DOI] [PubMed] [Google Scholar]
- 103.Chang Y., Yoo J., Kim J., Hwang Y., Shim G., Oh Y.-K., Kim J. Electromagnetized graphene facilitates direct lineage reprogramming into dopaminergic neurons. Adv. Funct. Mater. 2021;31(46) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The main data supporting the results in this study are available within the paper and its supplementary information. The raw and analyzed datasets generated during the study are available for research purposes from the corresponding author on reasonable request.