Abstract
The fields of Metagenomics and Metatranscriptomics involve the examination of complete nucleotide sequences, gene identification, and analysis of potential biological functions within diverse organisms or environmental samples. Despite the vast opportunities for discovery in metagenomics, the sheer volume and complexity of sequence data often present challenges in processing analysis and visualization. This article highlights the critical role of advanced visualization tools in enabling effective exploration, querying, and analysis of these complex datasets. Emphasizing the importance of accessibility, the article categorizes various visualizers based on their intended applications and highlights their utility in empowering bioinformaticians and non-bioinformaticians to interpret and derive insights from meta-omics data effectively.
Keywords: Metagenomics, Biodiversity, Ecosystems, Phylogeny, Databases, Visualization tools
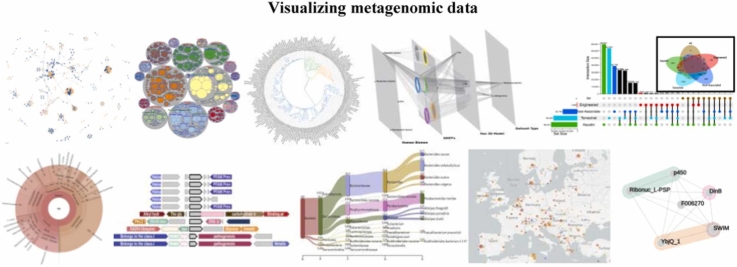
Graphical Abstract
A. Minimum-Evolution tree - Adh sequence data from eleven fruit fly species B. iTol circular tree - Alignment of temporally sampled data for using RelTime with Dated Tips (RTDT) to estimate times of divergence C. iTOL unrooted tree - Alignment of temporally sampled data for using RelTime with Dated Tips (RTDT) to estimate times of divergence. D. Pavian E. Krona sunburst chart - Taxonomic abundance of skin microbiome samples for 4 consecutive days F. iTOL rectangular tree - Adh sequence data from eleven fruit fly species.
1. Introduction
The total number of microbial cells on Earth is estimated to be 1030 [1], [2], outnumbering the stars of our Milky Way Galaxy (∼100 billion stars). Microorganisms, ubiquitous in nature, wield significant influence over Earth's biosphere. Every organism, spanning from humans to plants, interacts with the microorganisms in their environment. Nevertheless, a staggering percentage of > 98% remains largely unexplored due to the challenges of culturing them [3], [4]. In the human gut alone, microbial populations are estimated to range from 1013 to 1014 microbial cells, outnumbering human cells [5]. The study of the genomic material in metagenomes/metatranscriptomes allows researchers to gain insights into the genomic characteristics, functional potential, and ecological roles of specific microorganisms within complex microbial communities. It contributes to our understanding of microbial diversity, interactions, and the overall functioning of ecosystems.
Metagenomics and Metatranscriptomics [6], [7] are critical approaches in studying microbial communities and uncultured organisms. A metagenome encompasses the collective genomic content of a microbial community in a particular environment and includes the total genetic information from all the microbes present, including bacteria, archaea, viruses, and eukaryotic microorganisms such as protozoa or unicellular algae and fungi. Metagenomic analysis [8], [9], [10], [11] entails sequencing and analysis of DNA extracted directly from an environmental sample without the need for isolating and cultivating individual organisms. This approach allows researchers to explore the genetic diversity and functional potential of entire microbial communities.
Similarly, Metatranscriptomics [12], [13], [14] is a field of study that delves into the complex world of gene expression within microbial communities present in environmental samples. Unlike traditional transcriptomics, which focuses on the gene expression of individual organisms, metatranscriptomics examines the collective gene expression of all microbes within a given sample. A metatranscriptome represents the collection of all RNA transcripts (e.g., mRNA, rRNA, tRNA) produced by the microorganisms in a particular environment at a given point in time, and provides insights into the gene expression patterns and activities of the microbial community. A typical metatranscriptomic analysis involves the sequencing and analysis of the RNA transcripts, revealing which genes are actively being transcribed. One of the primary goals of metatranscriptomics is to elucidate the functional activities and metabolic processes occurring within microbial communities in their natural habitats. By analyzing the transcriptome, researchers can gain valuable insights into which genes are actively expressed, how they are regulated, and how microbial communities respond to changes in their environment. Overall, metatranscriptomics provides a powerful tool for exploring the functional potential and activities of microbial communities in diverse environments, offering valuable insights into their roles and interactions within ecosystems and their implications for human health and biotechnology.
Metagenome-assembled Genomes (MAGs) refer to the process of reconstructing individual genomes (at various levels of completion and possible contamination) of specific microorganisms from a metagenomic dataset. The process of extracting genomes from metagenomes is challenging due to the complex and diverse nature of metagenomic samples. However, advances in sequencing technology and computational methods have made it possible to extract and characterize genomes from metagenomes with increasing accuracy. These extracted genomes can provide valuable insights into the diversity and function of microbial communities, which can aid in the discovery of new organisms, metabolic pathways, and potential biotechnological applications.
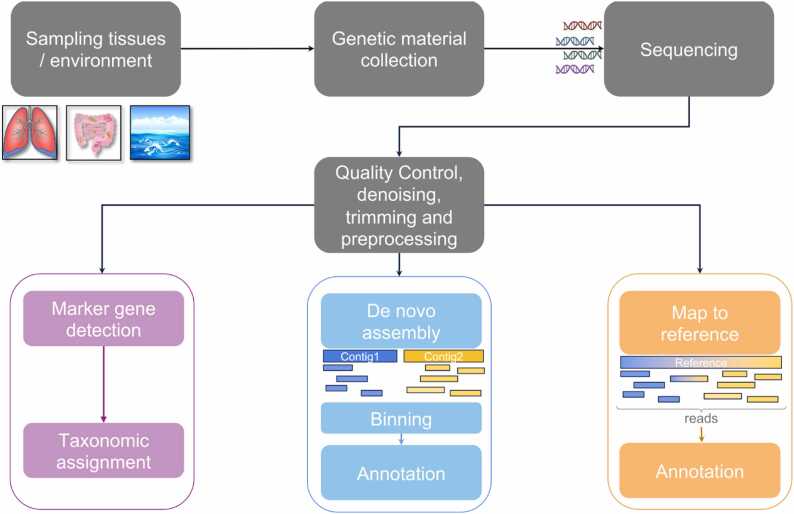
A typical Shotgun Metagenomic analysis involves steps (Fig. 1) such as:
-
•
Sequencing: Initially, researchers perform metagenomic sequencing on a sample, generating a dataset that contains DNA fragments from various microorganisms present in the environment.
-
•
Quality control: Raw metagenomic sequences are checked for quality and cleaned of contaminants such as adapters and primers.
-
•
Assembly/Read Mapping: In this step, short DNA fragments (reads) are aligned to reconstruct longer genomic sequences. The cleaned sequences are assembled into contigs and scaffolds using various assembly methods such as de novo assembly (no existence of reference genome), reference-based assembly (if a reference genome exists), or hybrid assembly (reference-guided and partially de novo).
-
•
Binning and Genome Reconstruction: Assembled contigs (contiguous DNA sequences) are grouped into similar operational taxonomic units based on similarities in nucleotide composition, coverage, and other features. The genomes that are reconstructed through binning are typically referred to as Metagenome Assembled Genomes (MAGs)
-
•
Annotation: MAGs are annotated with functional and taxonomic information similar to the isolate genomes.
Fig. 1.
Different steps of a typical metagenomic analysis: (i) Marker gene detection and taxonomic assignment, (ii) De novo assembly towards the generation of larger contigs, and (iii) Map to reference genome (if it exists).
Similarly, a typical Metatranscriptomics analysis involves steps such as:
-
•
Sample collection and RNA extraction: Samples are collected from the environment of interest, such as soil, water, or the human gut. Then, the extraction of total RNA from the collected samples follows to capture the actively transcribed genes.
-
•
cDNA synthesis: In this step, the extracted RNA is converted into complementary DNA (cDNA) using reverse transcription.
-
•
Sequencing library preparation: In this step, sequencing libraries from the cDNA samples are prepared, often using methods such as fragmentation and adapter ligation.
-
•
Sequencing: High-throughput sequencing of the prepared libraries using platforms like Illumina or PacBio is performed.
-
•
Data preprocessing: Like in metagenomics, data preprocessing such as adapter sequence trimming, low-quality read removal, and filtering out ribosomal RNA (rRNA) sequences is required.
-
•
Read mapping: The sequenced reads are mapped to a reference genome or transcriptome to identify the expressed genes and quantify their abundance.
-
•
Differential expression analysis: In this step, genes that are differentially expressed under different conditions or between different samples are identified.
-
•
Functional annotation and pathway analysis: In this step, the differentially expressed genes are annotated to assign putative functions based on databases like NCBI's RefSeq [15] or UniProt [16] as well as functional pathways enriched in the differentially expressed. The aim is to understand the biological processes at play.
In this review, we focus on the metagenomic visualization tools that aim at analyzing and displaying metagenomic data, including DNA sequences, functional information, and metadata. Visualization is crucial in the field of metagenomics as it allows researchers to understand complex microbial community structures, taxonomic compositions, and functional potentials. Although several visualization tools have been developed to aid researchers in exploring and interpreting metagenomic data, the field of metagenomic visualization is still in its infancy and the challenges regarding complexity, functionality, scalability, and interoperability remain open. Nonetheless, metagenomic visualization allows the automation of several important tasks:
-
•
Interactive and intuitive exploration and visualization of extensive datasets aid in the identification of patterns and trends within the data.
-
•
Comparison of multiple samples facilitates the recognition of similarities and differences, thereby enhancing comprehension of the diversity and complexity inherent in metagenomic data.
-
•
Integration of various data types, including functional, taxonomic, and metadata, contributes to a comprehensive understanding of metagenomic dataset(s).
-
•
Sharing of data and results among researchers fosters stronger collaboration and promotes improved reproducibility in research endeavors.
2. Databases and repositories
Currently, available metagenomes and metatranscriptome datasets, including raw reads, sequencing scaffolds, predicted genes and annotations, and associated metadata, are hosted in a wide range of publicly available repositories and databases [17] (Table 1). These include both standard sequence archives such as GenBank [18], the DNA Database of Japan (DDBJ) [19] and the European Nucleotide Archive (ENA) [20], (all three of which are members of the International Nucleotide Sequence Database Collaboration), the Sequence Read Archive (SRA) [21], or the Genomes OnLine Database (GOLD) [22], and specialized resources focusing exclusively on metagenomes. The most prominent databases in the latter category include IMG/M [23], [24], MGnify [25], SPIRE [26], and MG-RAST [27].
Table 1.
Databases and Repositories.
| Database Name | Description | Data Types | Accessibility | User Submission |
|---|---|---|---|---|
| GenBank | Archive for sequencing data | Genomes, Metagenomes, Metatranscriptomes, Amplicons | Publicly accessible | Yes |
| Sequence Read Archive (SRA) | Archive for sequencing data | Raw sequencing data | Publicly accessible | Yes |
| European Nucleotide Archive (ENA) | Archive for all publicly available nucleotide sequences | Genomes, Metagenomes, Metatranscriptomes, Amplicons | Publicly accessible | Yes |
| DOE Systems Biology Knowledgebase (KBase) | A platform for sharing, integrating, and analyzing microbial, plant, and community data | Genomes, Metagenomes, Metatranscriptomes, Amplicons | Publicly accessible | Yes |
| Genomes OnLine Database (GOLD) | Repository for genome projects and metadata (ecosystems) | Ecosystems | Publicly accessible | Yes |
| Integrated Microbial Genomes & Microbiomes (IMG/M) | Community-driven repository hosting genomes of cultivated and uncultivated microbial taxa, metagenomes, metatranscriptomes, amplicons, plasmids, and genome fragments | Metagenomes, Metatranscriptomes, Amplicons, Genomes | Publicly accessible | Yes |
| MGnify | Archive for exploration, and analysis, of microbiome sequencing datasets | Metagenomes, Metatranscriptomes, Amplicons, MAGs | Publicly accessible | Yes |
| Metagenome RAST (MG-RAST) | Microbiome repository with a unified pipeline for automated analysis of metagenomic samples | Metagenomes | Registered users | Yes |
| Integrated Microbial Viral Genomes (IMG/VR) |
Viral genomes and metagenomes | Viral Genomes, Viral Metagenomes | Publicly accessible | Yes |
| NMPFamsDB | Novel protein families from IMG’s metagenomes and metatranscriptomes | Protein Families | Publicly accessible | No |
| FESnov catalog | Catalog reporting functionally unannotated proteins derived from MAGs | Proteins | Publicly accessible | No |
| NIH Human Microbiome Project | Metagenomes from human host-associated systems, such as the gut microbiome | Human Microbiome Metagenomes | Publicly accessible | No |
| TerrestrialMetagenomeDB | Annotation of metagenomes obtained from soil samples | Soil Metagenomes | Publicly accessible | Yes |
| MarineMetagenomeDB | Annotation of metagenomes obtained from marine samples | Marine Metagenomes | Publicly accessible | Yes |
| HumanMetagenomeDB | Annotation of metagenomes obtained from human microbiome samples | Human Microbiome Metagenomes | Publicly accessible | Yes |
| SPIRE | Searchable resource of ecosystem metadata obtained from MAGs | Ecosystem Metadata | Publicly accessible | No |
| Marine Metagenomics Portal (MMP) | Collection of databases annotating marine-oriented metagenomic datasets | Marine Metagenomes | Publicly accessible | No |
| National Microbiome Data Collaborative (NMDC) | A platform for collaboration and data sharing among researchers studying microbiomes across diverse ecosystems | Microbiome Data | Publicly accessible | Yes |
The Integrated Microbial Genomes & Microbiomes (IMG/M) database is a community-driven repository that hosts genomes of cultivated and uncultivated microbial taxa from all domains of life, metagenomes and metatranscriptomes, amplicons, plasmids and genome fragments of interest, generated by targeted sequencing [23], [24]. IMG/M [23], [24] features a well-established, continuously updated metagenome analysis pipeline (DOE JGI Metagenome workflow), allowing researchers to submit their own genome or metagenome datasets, and automatically perform several types of analyses, including gene calling, taxonomic assignment, and functional annotation [28]. As a result, while a portion of the database’s content comes from other established sequence repositories, such as GenBank [18] or the SRA [21], the majority of its content is derived from user-submitted projects. Similar to IMG/M, MGnify [25] is a freely available database aimed at archiving, exploring, and analyzing microbiome sequencing datasets. The database accepts user-submitted data and provides a versatile annotation pipeline to cover the analysis of a wide range of dataset types, from studies targeting taxonomic markers (e.g., amplicon studies) to shotgun sequencing of metagenomes and metatranscriptomes, as well as metagenome-assembled genomes (MAGs). Furthermore, MGnify offers the option to provide assembly for user-submitted raw reads upon request [25]. Finally, the Metagenomes RAST service (MG-RAST) is another major microbiome repository and one of the earliest approaches to provide a unified pipeline for the automated analysis of metagenomic samples [27]. In contrast to other databases, MG-RAST imposes access limitations to its contents, with its database being restricted to its registered users. It focuses on the analysis of metagenome reads and mapping of the latter to reference genomes, rather than also analyzing other dataset categories (amplicons, assembled contigs/scaffolds, or MAGs).
In addition to IMG/M, MGnify, and MG-RAST, several, more specialized metagenomic databases are also available, focusing on specific microbiome types. For example, IMG/VR [29], [30] is a subset of IMG/M focusing exclusively on viral genomes and metagenomes [31], which has utilized specialized predictors to reanalyze IMG/M datasets and identify samples based on viral gene structure and virus-specific marker regions [32]. The DOE Systems Biology Knowledgebase (KBase) [33] is a freely accessible software and data platform facilitating the sharing, integration, and analysis of microbial, plant, and community data. NMPFamsDB [34], [35] hosts novel protein families [36] from IMG metagenomes and metatranscriptomes which do not have any hit to any known Pfam domain or similarity to any known reference genomes. Similarly, the FESnov catalog reports functionally unannotated proteins derived from MAGs [37]. Both databases offer several tools for the visualization of their data. Another similar, but more focused example is the Ocean Microbiomics Database [38], which hosts biosynthetic gene clusters formed by integrating isolate genomes from marine ecosystems with reconstructed draft genomes coming from seawater samples. The NIH Human Microbiome Project focuses on metagenomes from human host-associated systems, such as the gut microbiome [39], TerrestrialMetagenomeDB [40], MarineMetagenomeDB [41], and HumanMetagenomeDB [42] annotate metagenomes obtained from soil, marine, and human microbiome samples, respectively, originally deposited to GenBank [18], SRA [21], and MG-RAST [27]. SPIRE, hosted by EMBL, provides a searchable, planetary-scale resource of ecosystem metadata, obtained from MAGs [26]. Finally, the Marine Metagenomics Portal (MMP) [43] is a collection of databases annotating marine-oriented metagenomic datasets, retrieved from MGnify as well as super studies conducted by large microbiome initiatives, such as AtlantECO or the Tara Oceans expedition [44].
Finally, the National Microbiome Data Collaborative (NMDC) [45] is an innovative initiative designed to foster collaboration and data sharing among researchers studying microbiomes across diverse ecosystems. It serves as a centralized platform where scientists can access, analyze, and contribute to microbiome data, advancing our understanding of microbial communities and their impact on various environments and organisms. Through its collaborative framework, NMDC aims to accelerate discoveries and facilitate the development of novel solutions in fields ranging from healthcare to environmental science.
3. Sequence space
In this section, we describe today’s sequence metagenomic/metatransciptomic space across the aforementioned repositories (snapshot April 2024). IMG/M currently hosts 207,655 datasets, encompassing 54,030 metagenomic and 14,411 metatranscriptomic datasets (65,987,169,755 scaffolds). Similarly, the IMG/VR database, known for its comprehensive collection of uncultivated virus genomes contains a total of 14,203,973 viral genomes from metagenomes and 8023,647 viral OTUs. MGnify hosts 573,344 datasets derived from 2932 studies. Among these datasets, 459,617 are amplicons, 39,605 metagenomes, and 2581 metatranscriptomes. Additionally, MGnify features 429,448 genomes cataloged within 11 Metagenome-Assembled Genome (MAG) catalogs. The MGnify protein database hosts over 2.4 billion unique sequences predicted from metagenomic assemblies. SPIRE includes 99,146 metagenomic samples from 739 studies. With a total metagenomic assembly size of 16 terabase pairs (Tbp), SPIRE contains 35 billion predicted protein sequences and 1.16 million newly generated metagenome-assembled genomes (MAGs) of medium to high quality.
4. Pipelines
Metagenome annotation refers to the identification and functional characterization of genes and other genomic structure features in a metagenomic sample. The process can be performed using any number of sequence analysis tools [46]. However, due to the intricate nature of metagenome datasets, characterized by their complexity and diverse composition, dedicated pipelines are commonly used for effective analysis. Notable web-based examples include the DOE JGI Metagenome workflow [24], [28], EBI Metagenomics [30], and Metagenome RAST pipelines [27], integrated into the IMG/M, MGnify and MG-RAST databases, respectively. In addition, several standalone solutions also exist, including MetaErg [47], Prokka [48], MetaGOflow [49] (marine samples), PEMA [50] (metabarcoding analysis), PGAP [51], DFAST [52], and nf-core/mag [53].
While each pipeline may adopt different approaches and integrate different analysis methods, all currently available workflows focus on three main procedures: i) the identification of non-coding RNA genes (ncRNAs) and other marker regions, ii) the prediction of protein-coding genes, and iii) functional and taxonomic annotation of the sample. ncRNAs (e.g., rRNAs, tRNAs.) and marker regions (e.g., CRISPR elements) are detected by running searches against dedicated databases (e.g., Rfam [54]) with tools such as INFERNAL [55], or detecting sequence features with specialized tools (e.g., tRNAscan-SE [56] for tRNAs, CRISPRCasTyper [57] for spacer detection, CRT-CLI [58] for CRISPR sequences, geNomad for the identification of viruses and plasmids [59]). Protein gene calling can be performed using a wide array of gene prediction tools, most notable of which are Prodigal [60], GeneMark [61], its various implementations (GeneMarkS-2 for prokaryotic genes and GeneMark-ES/ET for eukaryotes) as well as FragGeneScan [62].
Following gene calling, functional annotation can be performed by searching the predicted genes against reference databases (e.g., RefSeq [15], UniRef90 [63], UniProtKB [16], Pfam [64], InterPro [65]) with pairwise alignments (e.g., BLAST [66], DIAMOND [67], MMseqs2 [68]) or Hidden Markov Model (HMM) - based methods (e.g., HMMER [69], HH-suite [70]). Finally, the taxonomic characterization of the dataset is based on the identified ncRNA genes, combined with the top most significant results of homology searches for the protein genes. In addition, detailed phylogenetic analysis can be performed using specialized tools such as Kraken 2 [71], PhymmBL [72] or MetaPhlAn [73].
5. Central visualization layouts used in metagenomics
Even though metagenomes are heterogeneous and complex to visualize, common visualization concepts can always be used for certain purposes (Fig. 2).
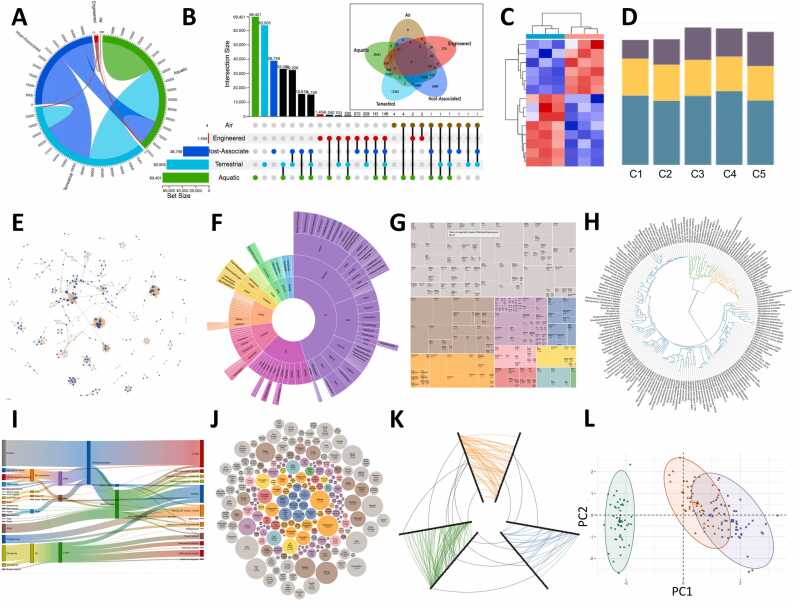
Fig. 2.
Different visualization concepts. (A) Circos diagram. (B) Upset plot & its corresponding Venn diagram. (C) HeatMap. (D) Bar chart (species). (E) Network. (F) Sunburst chart (Krona). (G) Treemap. (H) Phylogenetic tree. (I) Sankey plot. (J) Bubble chart. (K) Hive plot. (L) PCA map. All plots have been created using simulated data.
5.1. Circos
It is a circular data visualization tool that displays relationships between different entities arranged along the circumference of a circle (Fig. 2A). It was originally developed for genomics and bioinformatics applications but has since been used in various fields for visualizing complex relationships and patterns. In a Circos diagram, data is represented by ribbons or arcs connecting points on the circle. The position of each point along the circle represents a specific entity or category, and the ribbons indicate connections or relationships between them. The thickness or color of the ribbons can be used to encode quantitative information, making it effective for illustrating genomic data, such as genomic rearrangements, interactions between elements, or correlations in large datasets. Circos diagrams provide a unique and visually engaging way to represent intricate relationships and patterns in complex datasets. For example, NMPFamsDB [34], [35] is a database for novel protein families from metagenomes and offers the Ecosystem & Phylogeny option to allow users to visualize the association of a family with its organism categories or ecosystems at various levels via Circos plots.
5.2. Upset plots
An UpSet plot is a data visualization tool used to represent the intersections and cardinalities of sets in a more detailed and informative way than traditional Venn diagrams (Fig. 2B). UpSet plots are particularly useful when dealing with larger sets or multiple intersections between sets. They were designed to address some limitations of Venn diagrams, such as difficulties in scaling to a large number of sets and presenting the size of intersections. Key features of an UpSet plot include: (i) Matrix Display - Instead of using overlapping circles, UpSet plots use a matrix to represent the intersections of sets. Each row in the matrix corresponds to a unique combination of sets, and the cells indicate whether that particular combination is present or absent. (ii) Bars for Set Sizes - The plot typically includes bars or histograms that show the sizes of individual sets, providing a clear understanding of the distribution of elements across sets. (iii) Intersection Size Bars - The plot also includes bars that represent the size of each intersection, allowing for a quick comparison of the magnitudes of different intersections. (iv) Annotations - UpSet plots may include additional annotations or labels to provide context or highlight specific features of the data. For example, FLAME [74], [75], a web dedicated to functional enrichment, uses interactive UpSet plots to show overlapping annotations or enriched terms for various gene lists as well as the unions and intersections of the imported gene/protein lists.
5.3. Venn diagrams
This is a graphical representation that shows the relationships (unions and intersections) between sets or groups of elements (Fig. 2B). It consists of overlapping circles, each representing a set, and the overlaps or intersections between the circles represent the elements shared between those sets. The primary purpose of a Venn diagram is to visually depict the commonalities and differences between different groups or categories. Key components of a Venn diagram include (i) Circles or Ellipses - Each circle or ellipse in the diagram represents a set or category. The elements belonging to that set are enclosed within the circle. (ii) Overlap - The overlapping areas between circles indicate elements that are common to both sets. The size of the overlap reflects the extent of the shared elements. (iii) Non-overlapping Regions - The non-overlapping parts of each circle represent elements unique to that specific set. Venn diagrams are widely used in various fields, including metagenomics, to visualize the relationships and overlaps between different sets of elements such as taxonomic composition, functional gene annotations, comparing conditions or environments, and community structure. For instance, NMPFamsDB, a database housing novel protein families derived from microbial metagenomes and metatranscriptomes, utilized a Venn diagram in its graphical abstract. The diagram illustrates the distribution and coverage of novel protein families across the various domains of life. This visual representation effectively conveys that numerous novel protein families encompass members from multiple taxonomic groups, highlighting an intriguing discovery regarding the conservation and significance of these proteins.
5.4. Heatmap
It is a graphical representation that uses colors to visualize the intensity of a variable across a matrix or grid of data (Fig. 2C). It illustrates the values of a primary variable by arranging them in a grid of colored squares, with two axis variables divided into ranges similar to a bar chart or histogram. The color of each cell signifies the value of the main variable within the corresponding range of the axis variables. In the context of metagenome analysis, a heatmap can be employed to display the abundance or presence of specific microbial taxa or functional genes across different samples or conditions. Rows and columns in the heatmap may correspond to individual microbial taxa or genes and different samples, respectively, with colors indicating the relative abundance or occurrence of each element. This visualization type is valuable for identifying patterns, clustering related taxa or genes, and gaining insights into the composition and dynamics of microbial communities in metagenomic datasets. For instance, in [76], a heatmap is employed for the characterization of novel tissue microbiota using an optimized 16 S metagenomic sequencing pipeline. It visualizes the relative abundance of each bacterial family from sequencing of different mouse tissue samples performed in triplicate (three different mice for each tissue). Each line corresponds to a bacterial family. Each of the three columns for a tissue corresponds to a different mouse.
5.5. Bar Graphs
They represent data based on statistics and numerical figures. A bar graph uses the two axes to plot rectangular bars (Fig. 2D). One of the axes represents the observation/category which is usually a fixed variable, while the other axis represents the numerical magnitude that the observation carries. Typical types of bar graphs include horizontal bar charts, vertical bar charts, double bar graphs, multiple bar graphs, and bar lines. In the field of metagenomics, bar plots provide a useful visualization for representing the abundance or distribution of different taxonomic groups (e.g., species, genera, phyla) within a biological sample. Examples of such bar charts are the: (i) Stacked Bar Chart, (ii) Grouped Bar Chart, and (iii) Relative Abundance Bar Chart. In a stacked bar chart, each bar is divided into segments, with each segment representing a different taxonomic group. The height of each segment corresponds to the abundance of that group within the sample. A grouped bar chart can be used to compare the abundance of different taxonomic groups across multiple samples. Each group of bars represents a different sample, and within each group, bars represent the abundance of different taxonomic groups. A relative abundance bar chart displays the relative abundance of each taxonomic group rather than absolute counts. It can be useful for comparing the proportions of different taxa within a sample. For instance, in [77], a stacked barplot is employed to depict the distribution of symbiotic bacteria among species categorized as core or non-core. This study investigates honey collected across three harvesting seasons from a stable apiary to elucidate the diversity of species constituting the core and non-core bacterial communities. Through the use of a stacked barplot, the visualization effectively highlights differences in the characterization of core honeybee microbiota stability and the seasonal dynamics of five non-core bacterial strains.
5.6. Networks
In a general sense, a network visualization represents the connections and relationships between elements within a system, where these elements are nodes and the connections between them are edges. By using graphical representations, network visualization provides a clear and intuitive means to understand the structure, dependencies, and interactions within complex networks (Fig. 2D). Networks can be used to visualize data from several scientific fields. In Biology networks are often used to provide information about connectivity or other relationships between biological systems, samples, or entities [78], [79]. Typical cases of Biological Network Visualization are: (i) Biological Pathway Maps - These visualizations illustrate the sequences of biochemical reactions and molecular interactions involved in specific biological pathways. They provide a holistic view of how different molecules, such as proteins and metabolites, collaborate to perform essential cellular functions. (ii) Protein-Protein Interaction Networks - They are graphical representations of interactions between proteins that elucidate the intricate web of connections within cellular systems. Nodes represent proteins, and edges indicate interactions, allowing researchers to analyze the functional relationships critical for cellular processes. (iii) Gene Regulatory Networks - Visualization of gene regulatory networks demonstrates how genes control each other's expression. Nodes represent genes, and edges signify regulatory interactions, shedding light on the complex regulatory mechanisms governing cellular functions. (iv) Metabolic Networks - They depict the interconnected metabolic pathways within cells. Nodes represent metabolites, and edges indicate enzymatic reactions, providing insights into how cells process nutrients and energy. (v) Signaling Networks - They illustrate the pathways through which cells communicate with each other. (vi) Disease Networks - They capture the relationships between genes, proteins, and other biomolecules associated with specific diseases. (vii) Phylogenetic Networks - They represent the evolutionary relationships among different species. (viii) Ecological Networks - They Describe the interactions between different species in an ecosystem. This includes food webs, where species are connected by predator-prey relationships. For instance, in [35], networks were used to represent the distribution and association of novel protein clusters reported in NMPFamDB and their ecosystems. Eight ecosystem types were applied according to the GOLD ecosystem classification, represented by central, colored nodes (hubs). Gray peripheral nodes represent the protein clusters whereas edges represent the protein cluster–ecosystem associations.
5.7. Sunburst chart (Krona)
It is known by multiple names such as ring chart and radial treemap, and is used to visualize a hierarchical dataset (Fig. 2F). It demonstrates hierarchy by employing a series of concentric rings, where each ring corresponds to a specific level in the hierarchy. The segments within each ring are proportionally divided to represent the details at that level. By focusing on a segment within a ring, one can understand the relationship of that segment to the entire hierarchy and its parent ring segment. The Sunburst chart utilizes a radial layout, providing an immersive visualization experience for categorized datasets. Unlike a rectangular layout used in a Treemap, the Sunburst chart is space-filling and showcases how each ring is subdivided into sequential segments, effectively illustrating the hierarchical breakdown of the data. This visual representation of taxonomy in the chart proves to be valuable for metagenome analysis. Its radial layout allows for an intuitive exploration of the relationships between different taxonomic levels, offering insights into the composition and distribution of microbial communities. For example, a KRONA plot is employed in [80]. The plot provides insights into the major microbial taxonomies and functions within biogas plants (BGPs). It offers a comprehensive overview of the microbial community structure and metabolic functionality by summarizing identified microbial families and biological processes. The KRONA plot depicts the distribution of identified bacteria, archaea, and viruses across various taxonomic levels, from superkingdom to family, with abundances represented based on the number of identified spectra summed over all BGPs.
5.8. Treemap
It is a visualization that represents hierarchical data through nested rectangles (Fig. 2G). Each rectangle in the treemap corresponds to a specific category or sub-category, and the size of the rectangles reflects the quantitative value of the data they represent. The hierarchy is depicted by the nesting of rectangles within one another, with the top-level rectangle representing the overall dataset and subdividing it into smaller rectangles for each subsequent level. Treemaps are effective for displaying hierarchical structures and facilitating the intuitive exploration of complex datasets, making them particularly useful in areas such as information visualization, financial analysis, and project management. In metagenome analysis, treemaps can be applied as a visualization tool to represent hierarchical structures within microbial taxonomic or functional data. For instance, in [81], a treemap was utilized to visualize T-Cell Epitope Repertoire frequency patterns (TCEMs) within pathogen proteomes. Each rectangle within the treemap represents a distinct TCEM-sharing relationship among bacterial species and is sized proportionally to the number of motifs within that particular combination.
5.9. Phylogenetic trees
They are a specific type of tree diagram (dendrogram), useful for representing taxonomic relationships (Fig. 2H). These diagrams constructed from metagenomic data help illustrate the evolutionary relationships among these microorganisms by depicting the branching patterns based on genetic similarities, providing insights into the biodiversity and evolutionary history of entire microbial communities in a given ecosystem. For example, [82] presents a phylogenetic tree, showing the bacterial and archaeal tree of life, and presenting an updated view of domain-level relationships.
5.10. Sankey plots
A Sankey plot, also known as a Sankey diagram or flow diagram, is a visual representation that illustrates the flow of resources or information between multiple entities [83], [84] (Fig. 2I). The diagram consists of nodes (representing entities or categories) and direct links (weighted lines or arrows) that show the direction and quantity of the flow between the nodes. The width of the links is proportional to the quantity of the flow, allowing viewers to easily grasp the relative magnitudes of different pathways within the system. In metagenomic analysis, Sankey plots find application in illustrating the distribution and transitions of taxonomic or functional categories across different biological samples or conditions. These plots can represent the flow of microbial taxa or functional gene abundances, showcasing how these entities shift or remain consistent between various environmental samples, experimental treatments, or time points. The width of the links in the Sankey plot corresponds to the relative abundance of taxa or functional categories, providing a visual insight into the dynamics of microbial communities. For instance, BioSankey [85], facilitates the visualization of microbial communities over time. This tool assists in gaining a comprehensive understanding of experimental data and harnessing the full potential of a dataset by creating intuitive and interactive Sankey diagrams to depict changes in microbial species in microbiome studies across different time points.
5.11. Bubble charts
It is a visual representation that displays three-dimensional data using circles of varying sizes on a two-dimensional plane (Fig. 2J). Each circle, or "bubble", represents a data point and is positioned based on its values along two axes. The position on the chart conveys the relationship between two variables, while the size of the bubble indicates the magnitude of a third variable. In biology, a bubble chart can be applied to represent multivariate data, such as comparing species abundance across different habitats. The position of each bubble on the chart might signify environmental parameters, while the size of the bubbles could represent the population size of a particular species. This visualization method is powerful for identifying patterns, correlations, and potential ecological trends within diverse datasets. For example, [86] includes a bubble plot illustrating the relative taxonomic abundance of the samples. The size of each bubble indicates the taxon's abundance relative to its maximum abundance, with larger bubble sizes indicating higher abundance. Additionally, the size of each circle is scaled logarithmically to represent the number of Open Reading Frames (ORFs) assigned directly to the taxon. This visualization aids in comprehending the taxonomic composition of the microbial community and their potential roles in biogeochemical manganese cycling.
5.12. Hive Plots
The basic concept behind a hive plot is to visualize relationships or connections between multiple variables or categories in a structured and intuitive manner (Fig. 2K). It's often used to represent complex networks or datasets with multiple dimensions [87]. Overall, the key strength of hive plots lies in their ability to visualize multidimensional data in a concise and interpretable format, making them a valuable tool for exploratory data analysis, network visualization, and pattern recognition across diverse domains. They can be a useful tool for visualizing microbiome data, which often involves complex relationships between various microbial taxa and environmental factors. Microbiome data typically consists of abundance or presence/absence information for different microbial species or taxa across multiple samples. For instance, in [88], a three-axis hive plot was used to assess the characteristics of microbial networks associated with apparently healthy and diseased corals.
5.13. Dimensionality reduction methods
Dimensionality reduction methods [89], [90], [91], [92], [93], [94], [95] play a crucial role in analyzing high-dimensional datasets by transforming them into lower-dimensional representations while preserving important information. Principal Component Analysis (PCA) (Fig. 2L) is a widely used linear technique that identifies the axes of maximal variance in the data. It projects the data onto these axes to reduce dimensionality while retaining the most significant features. A PCA map is a visual representation employed to explore and understand the relationships among samples based on their overall composition. For instance, in [96], a 3D PCA plot is utilized to show the clustering result of four metagenomes from oil samples and 948 environmental metagenomes from the IMG database using the KO abundance. Such visualization can aid in examining the relationship among the functional compositions of metagenomes across diverse environments.
Other well-known dimensionality reduction methods include Uniform Manifold Approximation and Projection (UMAP), t-distributed Stochastic Neighbor Embedding (t-SNE), and Latent Dirichlet Allocation (LDA). Uniform Manifold Approximation and Projection (UMAP) is a nonlinear dimensionality reduction method that preserves both global and local structure in the data, making it effective for visualizing complex datasets. UMAP finds frequent application in the realm of Metagenomes, where its utilization is prevalent. The integration of such nonlinear machine learning methods is anticipated to significantly enhance our comprehension of the metagenome. t-distributed Stochastic Neighbor Embedding (t-SNE) is another popular nonlinear method focusing on preserving local relationships between data points, often used for visualizing high-dimensional data in two or three dimensions. Latent Dirichlet Allocation (LDA) is a probabilistic generative model commonly used for topic modeling in natural language processing. It reduces dimensionality by representing documents as distributions over topics, allowing for the exploration of underlying themes in large text corpora. Overall, these dimensionality reduction methods provide powerful tools for visualizing and exploring complex datasets across various domains (e.g., scRNA-seq, see SCALA application [97]).
5.14. Rarefaction curves
It is a method that adjusts for the variations in metagenomic clone library sizes across samples to aid comparisons of alpha diversity. The concept of rarefaction involves selecting a specified number of samples that are equal to or less than the number of samples in the smallest sample and then randomly eliminating reads from larger samples until the number of remaining samples reaches the threshold. Based on these subsamples of equal size, diversity metrics can be calculated to contradict ecosystems and are independent of disparity in sample sizes. Calculated rarefaction is represented by a line graph. The rarefaction curve not only copes with the sample coverage but also depicts whether the sampling depth was sufficient or not to estimate the diversity. A curve indicates sufficient sampling depth, while an ascending graph implies insufficient sampling depth. A rarefaction curve is commonly used in ecology and biodiversity studies to assess the sampling effort's adequacy in capturing the diversity of a biological community [98], [99], [100]. This curve plots the number of observed species or unique entities against the number of samples taken. Initially, as more samples are collected, the curve steeply rises, reflecting the discovery of new species. However, it eventually plateaus, indicating that the majority of the community's diversity has been sampled. Rarefaction curves assist researchers in estimating species richness, evaluating the effectiveness of sampling efforts, and making informed decisions about the comprehensiveness of their data collection in ecological studies. Rarefaction analysis is used to standardize diversity measures across different sample sizes, enabling fair comparisons between ecosystems or study sites. In [35], rarefaction curves have been used to show that while protein families from reference genomes seem to increase linearly, the equivalent families from metagenomes reveal exponential growth, thus never plateauing. Consequently, the study focuses on larger clusters for further analysis, yet highlights the vast unexplored protein sequence space.
5.15. Gene Map
Often referred to as a genetic map or genome map, it is a visual representation of the arrangement and location of genes on a particular chromosome or across an entire genome. Like Circos, it provides a graphical overview of the genetic structure, indicating the relative positions of genes, markers, and other genetic features. Gene maps are crucial tools in genomics and metagenomics research, aiding in the understanding of gene linkage, genetic distances, and the organization of genetic material. High-resolution gene maps are particularly important for studies involving gene identification, marker-assisted breeding, and investigations into the genetic basis of various traits or diseases. Advances in technology, such as next-generation sequencing, have significantly improved the accuracy and precision of gene mapping, contributing to our understanding of the genetic landscapes of various organisms, including humans. For instance, in [101], a gene map is used to show the extrication of the microbial interactions of activated sludge used in the textile effluent treatment of an anaerobic reactor through metagenomic profiling. This circular gene map illustrates the location and size of genes encoding for aldehyde dehydrogenase and numerous hypothetical proteins. Such visualization aids in comprehending the microbial organisms participating in degradation pathways and their interactions within the microbial community.
5.16. Tree Diagram
It is a graphical representation that depicts a hierarchical structure or relationship between different elements or components. It is called a "tree" because it often resembles an inverted tree with a single root or starting point, branching out into various branches and sub-branches. The structure of a tree diagram consists of nodes connected by edges, where each node represents a specific entity or concept, and the edges indicate the relationships or connections between them. Tree diagrams are commonly used in various fields such as computer science, linguistics, probability theory, and organizational charts to visually organize and illustrate hierarchical structures.
5.17. Space-filling maps
Space-filling curves like the one of Hilbert are intricate geometric patterns that traverse and cover a two-dimensional space in a continuous and non-overlapping manner. The Hilbert curve (or any other in this category) manifests as a continuous fractal structure, its formation rooted in the recursive subdivision of a square into four smaller subsquares, followed by the connection of their centers in a specific sequence. This intricate curve systematically traverses all points within a designated region, maintaining proximity between points on the original curve and their spatial arrangement on the plane. Historically, Hilbert curves have been used to produce genomic maps for large scaffolds (e.g., human chromosomes) and whole genome alignments for bacterial genomes [102]. Expanding the scope, this concept can be adapted for metagenomics in the configuration of a space-filling map. In this representation, each position or pixel corresponds to a genome within the reference collection. The intensity color value at a given position reflects the relative abundance of a particular genome in the metagenomic sample. These microbiome maps offer a versatile tool for exploration, enabling the investigation of taxonomy, ecosystem abundance, simultaneous comparison of multiple samples, and the analysis of microbial community dynamics through time-series analysis. In contrast to conventional visualization methods that often prioritize elements with the highest abundances in a population, Hilbert curve-based maps provide a more nuanced perspective. They offer enhanced resolution for taxa with smaller abundances, addressing a limitation commonly encountered in traditional visualization techniques. For example, the Meander application [102] has been used to compare chromosome 1 between strain ICE153 from central Asia and strain ICE97 from southern Italy, showing a deletion and a tandem duplication supported by both pair-end and read-depth information at higher resolution with the help of Hilber curves.
In the realm of metagenomic analysis, navigating through complex datasets and understanding intricate relationships among microbial communities pose significant challenges. To address these challenges, a diverse array of visualization concepts that are presented can be useful. In this table (Table 2), we focus on the major challenges encountered in metagenomic visualization, ranging from representing complex relationships to handling large datasets and understanding taxonomic hierarchies. Each visualization concept listed in the table offers unique functionalities tailored to address specific metagenomic challenges, providing researchers with invaluable tools to explore, analyze, and interpret complex biological data.
Table 2.
Visualization concepts organized by their relevance to metagenomic visualization challenges.
| Visualization Challenge | Visualization Concept |
|---|---|
| Representing complex relationships | Circos, Networks |
| Handling large sets or intersections | Upset plots, Venn diagrams |
| Visualizing abundance across samples | Heatmap, Bar graphs |
| Displaying hierarchical data structures | Treemap, Trees, Sunburst charts (Krona) |
| Understanding taxonomic relationships | Trees, Sunburst charts (Krona) |
| Illustrating flow or transitions | Sankey plots, Networks, Hive plots |
| Visualizing multidimensional data | Hive plots, 3D networks, Dimensionality Reduction methods |
| Standardizing diversity measures | Rarefaction curves |
| Visualizing genetic arrangements | Gene Map, Genome viewers |
| Linear representations at higher resolutions | Space-filling maps/curves |
6. Main applications of metagenomic visualization tools
Within this segment, we present an assortment of visualization tools, organizing them according to their primary functions. Although our compilation may not be exhaustive, we focus on spotlighting well-established tools, to illuminate a range of options available for visualizing metagenomic data in the ever-evolving landscape of data visualization. The tools are categorized into primary groups, including quality control, binning, assembly, genomic content viewers, taxonomy, community, and networks (Table 3).
Table 3.
Representative tools are organized by their main functionality.
| TOOL | CATEGORY BY MAIN FUNCTION | INPUT DATA TYPE | LICENSE TYPE | IMPLEMENTATION | LAST UPDATE |
|---|---|---|---|---|---|
| FastQC | Quality Control | Raw sequence data (before any alignment or assembly steps) | Open source | Stand-alone | 2023 |
| LongQC[103] | Quality Control | Raw Long-Read Sequencing Data (before any alignment or assembly steps - PacBio Sequencing, Oxford Nanopore Sequencing) | Open source | Stand-alone | 2023 |
| MinIONQC[104] | Quality Control | Raw sequence data (before any alignment or assembly steps - FASTQ, FAST5 format) | Open source | Stand-alone | 2020 |
| NanoPack[105] | Quality Control | Raw sequence data (before any alignment or assembly steps - FASTQ, FAST5 format) | Open source | Suite of tools | 2023 |
| SOAPnuke[106] | Quality Control | Raw sequence data (before any alignment or assembly steps - FASTQ format) | Open source | Stand-alone | 2024 |
| SequelTools[107] | Quality Control | Raw Long-Read Sequencing Data (before any alignment or assembly steps - PacBio Sequencing, Oxford Nanopore Sequencing) | Open source | Stand-alone | 2020 |
| ABySS-Explorer[108] | Assembly | ABySS Assemblies (scaffolds or contigs in FASTA format), Raw sequence data | Open source | Stand-alone | 2018 |
| Assembly Graph Browser (AGB)[109] | Assembly | Assembly Graph Files (GFA (Graphical Fragment Assembly)) | Open source | Stand-alone | 2019 |
| GfaViz[110] | Assembly | Assembly Graph Files (GFA (Graphical Fragment Assembly)) | Open source | Stand-alone | 2019 |
| SGTK[111] | Assembly | Assembly Graph Files (GFA (Graphical Fragment Assembly)) | Open source | Toolkit | Archived in 2023 |
| PanGraphViewer[112] | Assembly/Pangenome | Pangenome graphs (rGFA, GFA_v1, VCF), Annotation Files (BED, GTF / GFF) | Open source | Stand-alone | 2022 |
| MetagenomeScope | Assembly | GFA, FASTG, GML, LastGraph | Open source | Web-based tool | 2020 |
| BinaRena[113] | Binning | (Human) Assembled Data (FASTA) | BSD 3-Clause License | Web application | 2023 |
| CONCOCT[114] | Binning | Metagenomic Sequencing data, Contig Sequence | Open source | Stand-alone | 2019 |
| MetaWRAP[115] | Binning | Metagenomic sequencing data (FASTQ format), Assembled contigs (FASTA), | Open source | Pipeline | 2020 |
| VizBin[116] | Binning | Metagenomic Fragments (Contigs / reads)(FASTA) | BSD License (4-clause) | Stand-alone | 2019 |
| Anvio[117] | Contig & Genome Viewer / Communities / Taxonomy | DNA sequence (FASTA), Contigs (FASTA), Short reads (FASTA), External / Internal genome database | Open source | Stand-alone | 2023 |
| CGViewer.js[118] | Contig & Genome Viewer | JSON files | Open source | Web-based tool | 2019 |
| CRAMER[119] | Contig, Genome & MSA Viewer | Metagenomic sequence data (Raw DNA sequence / FASTA files) | Open source | Stand-alone | 2019 |
| Elviz[120] | Contig & Genome Viewer | Metagenomic sequence data (Raw DNA sequence / FASTA files) | Open source | Web-based application | 2024 |
| GDV[121] | Contig, Genome & MSA Viewer | RNA-seq data, ChIP-seq data, Genome Sequence Data, Proteomic Data & Epigenomic Data | Open source | Web-based application | 2021 |
| Gosling[122] | Contig, Genome & MSA Viewer | Metagenomic sequence data (Raw DNA sequence / FASTA files) | Open source | Toolkit | 2021 |
| IMG/M[23], IMG/VR[30] | Contig and Genome Viewer | Visualization of IMG/M and IMG/VR contig annotations | Open source | Web-based platforms | 2023 |
| IGV[123] | Genome Viewer | Metagenome sequence data (FASTA), Alignment Data, Variant Calls, Gene Annotations (GFF) | Open source | Stand-alone | 2023 |
| JBrowse[124] | Genome Viewer | Metagenome sequence data (FASTA), Alignment Data, Variant Calls, Gene Annotations (GFF) | Open source | Stand-alone | 2024 |
| MetaErg[47] | Contig Viewer | Metagenomic Contig, Gene Prediction File, Taxonomic Information File | Open source | Stand-alone pipeline | 2020 |
| Tablet[125] | Genome Viewer | SAM (Sequence Alignment/Map) and BAM (Binary Alignment/Map), Variant Call Format (VCF), Metagenome Sequence, Genome Assembly Files, Sequence Read Files | BSD-2-Clause license | Stand-alone | 2021 |
| UCSC Genome Browser[126] | Genome & MSA Viewer | Genome Sequence Data, Annotation Data (GFF), ChIP-Seq Data, RNA-seq Data, Multiple Sequence Alignments (MSA) | Open source | Online portal | 2022 |
| ENSEMBL[127] | Genome Viewer | Genome Sequence Data, Annotation Data (GFF), ChIP-Seq Data, RNA-seq Data, Multiple Sequence Alignments (MSA) | Open source | Suite of tools | 2024 |
| Artemis[128] | Genome Viewer | Genome Sequence Data, Annotation Data (Genebank, EMBL format) | Open source | Stand-alone | 2011 |
| UGENE[129] | Genome Viewer | Genome Sequence Data (FASTA, GFF, SAM/BAM, BED), Annotation Data (Genebank, EMBL format, BED, GFF), Multiple Sequence Alignments (MAF), Expression Data Files | Open source | Stand-alone | 2023 |
| Geneious[130] | Genome Viewer | Genome Sequence Data (FASTA, GFF, SAM/BAM, BED), Annotation Data (Genebank, EMBL format, BED, GFF), Multiple Sequence Alignments (MAF), Expression Data Files | Free trial - Requires subscription | Part of a software suite | 2023 |
| BV-BRC[131] | MSA Viewer | Multiple Sequence Alignments (MSA) | Portal | Web-based resource | 2022 |
| MSAViewer[132] | MSA Viewer | Multiple Sequence Alignments (MSA) | Open source | Web-based application | 2023 |
| Strudel[133] | MSA Viewer | Metadata (CSV,TSV), Aligned Sequence Data, Phylogenetic Tree Data, Annotation Data (GFF) | Open source | Stand alone | 2015 |
| SuiteMSA[134] | MSA Viewer | Multiple Sequence Alignments (MSA) | Open source | Stand alone | 2013 |
| JalView[135] | MSA Viewer | Multiple Sequence Alignments (ex FASTA, Clustal, Stockholm) | Open source | Stand alone | 2023 |
| MSABrowser[136] | MSA Viewer | Multiple Sequence Alignments (MSA) | Open source | Stand-alone web-based application | 2021 |
| Seaview[137] | MSA Viewer | Multiple Sequence Alignments (ex FASTA, Clustal, Stockholm, PHYLIP) | Open source | Stand-alone or helper application | 2024 |
| Panache[138] | Pangenome Viewer | Graphical Fragment Assembly (GFA) | Open source | Web-based interface | 2022 |
| Pan-Tetris[139] | Pangenome Viewer | Pangenome map files (ex PanGee), meta-information (TIGRFAM) | Open source | Software tool | 2015 |
| PanViz[140] | Pangenome Viewer | Pangenome Matrix (pattern of each gene group) and functional annotation files (GeneOntology) | Open source | Pipeline | 2017 |
| PanX[141] | Pangenome Viewer | Set of annotated bacterial strains (NCBI RefSeq, users input in GeneBank format) | Open source | Pipeline | 2018 |
| Pantools[142] | Pangenome & Panproteome Viewer | Annotation Files (GTF / GFF), Multiple Sequence Alignment File (FASTA), Genomic Sequence Files (FASTA), Variations adding (VCF files and a PAV table) | Open source | Stand-alone | 2024 |
| Bifrost[143] | Pangenome Viewer | Annotation Files (GTF / GFF), Multiple Sequence Alignment File (FASTA), Genomic Sequence Files (FASTA), | Open source | Stand-alone | 2024 |
| PanGenome Graph Builder[144] | Pangenome Viewer | Annotation Files (GTF / GFF), Multiple Sequence Alignment File (FASTA), Genomic Sequence Files (FASTA) | Open source | Stand-alone | 2024 |
| TwoPaCo[145] | Pangenome Viewer | Annotation Files (GTF / GFF), Multiple Sequence Alignment File (FASTA), Genomic Sequence Files (FASTA) | Open source | Stand-alone | 2022 |
| Minigraph-Cactus[146] | Pangenome Viewer | Annotation Files (GTF / GFF), Multiple Sequence Alignment File (FASTA), Genomic Sequence Files (FASTA) | Open source | Pipeline | 2024 |
| Jasper/Microbiome Maps[147] | Abundance analysis / Taxonomy / Ecosystem visualization | Abundance profiles / OTU table | Not open source | Stand-alone | 2023 |
| QIIME / QIIME 2[148] | Communities/ Taxonomy | raw DNA sequence reads | Open source | Analysis package | 2024 |
| Phyloseq[149] | Communities/ Taxonomy | OTU table (operational taxonomic units), phylogenetic tree | Open source | R package | 2013 |
| MicrobiomeAnalyet[150] | Communities/ Taxonomy/PCA visualization | OTU table (operational taxonomic units), taxon list, gene list, Gene abundance table, BIOM file | Open source | Web-based platform | 2024 |
| MetagenomeSeq[151] | Communities/ Taxonomy/PCA visualization | Taxonomic or Functional Annotations, Count Data Table | Open source | R package | 2019 |
| MEGA[152] | Taxonomy | Metagenome sequence data (FASTA), Phylogenetic Data (NEXUS, NEWICK) | Open source | Can be used as stand-alone and as part of a pipeline | 2022 |
| PAUP[153] | Taxonomy | Metagenome sequence data (FASTA), Phylogenetic Data (NEXUS, NEWICK) | Proprietary, and thus commercial | Stand-alone | 2007 |
| FigTree | Taxonomy | Phylogenetic Data (NEXUS, NEWICK) | Open source | Stand-alone | 2018 |
| iTOL[154], [155] | Taxonomy | Phylogenetic Data (NEXUS, NEWICK) | Open source | Web-based platform | 2023 |
| PhyD3[156] | Taxonomy | Phylogenetic Data (NEXUS, NEWICK) | Open source | Web-based tool | 2017 |
| Dendroscope[157] | Taxonomy (viewer) | Phylogenetic Data (NEXUS, NEWICK) | Open source | Stand-alone | 2023 |
| Cytoscape[158], [159] | Network visualization | Graphs - Lists (source - destination) | Open source | Stand-alone | 2023 |
| Gephi[160] | Network visualization | Graphs - Lists (source - destination) | Open source | Stand-alone | 2023 |
| Pajek[161] | Network visualization Large Networks |
Has its file format | Open source | Stand-alone | 2023 |
| Arena3Dweb[162], [163] | Network visualization 3D Multilayered Networks |
Network lists (source - destination but by defining their layers) | Open source | Web server and stand-alone | 2023 |
| NORMA[164], [165] | Network and group visualization | Network lists (source - destination) and annotation files (nodes and the annotation group they belong to) | Open source | Web server and stand-alone | 2022 |
6.1. Quality control
In metagenomic analysis, a common practice involves generating scaffolds or metagenome-assembled genomes (MAGs) from raw sequence data. A crucial initial phase in this procedure is conducting quality control (QC) on the raw data. This encompasses assessing read and base quality, trimming adapters, analyzing GC distribution, eliminating contaminated reads, addressing enrichment bias, generating quality metrics, and various other steps. Numerous tools have been created for this objective, generating visual representations of the aforementioned statistics, such as FastQC, LongQC [103], MinIONQC [104], and NanoPack [105] which, as implied by its name, is a package of a specific sub-category of tools consisting of NanoPlot, NanoComp, NanoQC, PHASIUS, Kyber, SOAPnuke, and SequelTools.
6.2. Assembly
Genome assembly is a complex process that involves piecing together the DNA sequences, essentially constructing extended DNA sequences (contigs) of an organism’s genomic data, in an attempt to reconstruct its complete genome. The genome of an organism is its entire DNA content, including genes and non-coding regions. If a reference genome is available, reads are aligned to that genome, while in the absence of a reference genome, de novo assembly is employed. De novo assembly is particularly important for studying non-model organisms, genomes with significant structural variations, or populations with diverse genomes.
Assembly visualization refers to the graphical representation of the results of genome assembly processes and aids researchers in understanding the structure and characteristics of assembled genomes. Visualizing genome assemblies is essential for quality assessment, identifying potential issues, and gaining insights into the overall genomic architecture. To this end, a plethora of tools can be used for the de novo metagenome assembly [166], [167], [168], [169], [170] (Fig. 3). Omega [171] assembler uses overlap graphs and has been specifically developed for metagenome assembly. Velvet [172] is designed for short-read sequencing data and an extension of it, MetaVelvet [173], is available aiming at the assembling of, specifically, metagenomic data using de-Bruijn graphs. MEGAHIT [174] uses succinct de Bruijn graphs for assembling large and complex metagenomic data while, BCALM 2 [175] aims to improve the scalability of the process by implementing the compaction of de Bruijn graphs. Another tool that uses de Bruijn graphs is metaSPAdes [176] which constitutes an extension of SPAdes adapted to the intricacies of metagenomic data. MetaCarvel [177] performs metagenome assembly, while at the same time, it can detect genomic variants. Some notable visualizers include ABySS-Explorer [108], AGB [109], Bandage [178], GfaViz [110], MetagenomeScope, PanGraphviewer [112], and SGTK [111].
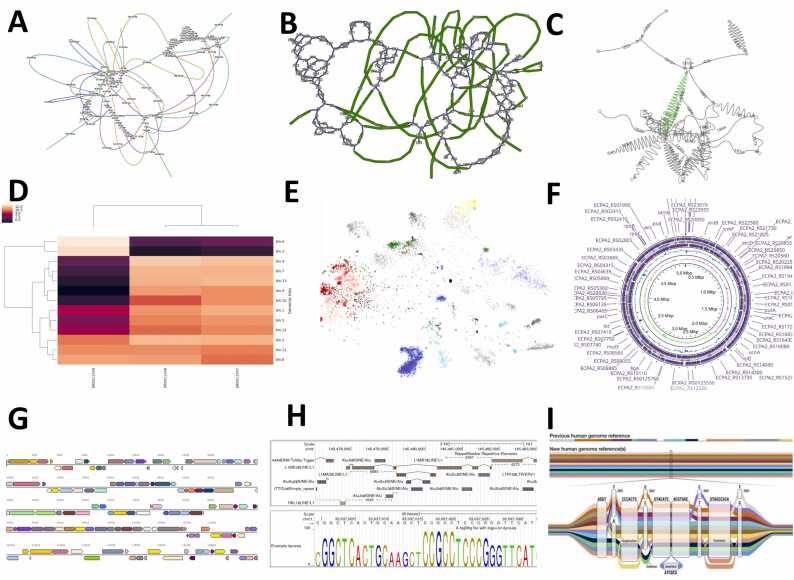
Fig. 3.
(A-C) Graph-based visualization of sequence assembly of Escherichia coli str. K-12 substrate MG1655 with (A) Bandage, (B) GFAviz, and (C) AbyssExplorer (NCBI:txid511145). (D) Heatmap visualizing the bin abundances of draft genomes using MetaWrap (Bioproject Accession: PRJEB2054, ID: 203783). (E) Binning of MAGs highlighting 214 bins of E.coli using BinaRena (BioProject: PRJNA382010). (F) CGView: Genome Contigs Viewer of Escherichia coli PA2 (NCBI RefSeq assembly GCF_000335355.2) in a circular format. (G-H) Scaffold visualization of E.coli K-12 with (G) IMG and (H) UCSC genome viewers. (I) Example of a pangenome graph.
6.3. Binning
Βinning is a crucial step in metagenomic analysis, which involves grouping genomic fragments (contigs) to reconstruct draft microbial genomes (MAGs) [179] (Fig. 3). Tools like MetaBAT [180], [181], BinaRena [113], ICoVeR [182], MyCC [183], gbtools [184], CONCOCT [114], VizBin [116], and MetaWRAP [115] aid in this process, employing different visualization methods and interactive interfaces to enable user-friendly exploration and refinement of bin assignments. BinaRena [113] offers a comprehensive interface, allowing scatter plot visualization of contigs and bin association editing. At the same time, ICoVeR [182] focuses on bin curation based on multiple binning algorithms using parallel coordinates and dimensionality reduction plots. MyCC [183] streamlines binning via a virtual machine, emphasizing marker gene-based clustering and genomic signature analysis. Gbtools [184] excels in visualizing coverage, GC content, and taxonomic annotations, aiding bin annotation and refinement. MetaWRAP [115], a modular pipeline, automates metagenomic data processing, extraction, and refinement of high-quality bins, offering taxonomy assignment, abundance estimation, functional annotation, and versatile visualization tools. These tools collectively address the need for accurate and efficient binning, catering to researchers' varying expertise levels and improving overall metagenomic analysis outcomes [179].
6.4. Community detection
Metagenomic analysis unfolds in several key steps, each contributing to a comprehensive understanding of the microbial communities. Clustering is a fundamental technique in bioinformatics and metagenomic analysis, allowing the uncovering of underlying patterns and relationships within complex datasets. Hierarchical clustering stands out as a significant non-graph-based methodology. It organizes sequences into a hierarchy of clusters, typically visualized as dendrograms, providing an insightful representation of the relationships between microbial entities. The agglomerative approach, where individual clusters progressively merge, and the divisive approach, where a single cluster iteratively divides, are two primary strategies. Widely used algorithms for agglomerative hierarchical clustering include single-linkage, complete-linkage, centroid-linkage, and average-linkage methods, as well as neighbor-joining [185] and the unweighted pair group method with arithmetic mean (UPGMA). Each iteration produces a new level in the dendrogram, and cutting thresholds, often user-defined or automated using methods like Dynamic Tree Cut or PAC Bayesian, delineate distinct clusters. While hierarchical clustering is powerful, its applicability to large-scale analyses is limited due to the requirement of a full-distance matrix and its high computational complexity.
Another approach is to apply graph-based clustering [186], [187] to detect communities on a constructed network (e.g., a Sequence Similarity Network [188], or an Average Nucleotide Identity (ANI) network [189]). Scalable graph-based clustering such as HipMCL [190], Louvain [191], or SPICi [192], can be directly applied to such networks. Notably, pairwise similarity comparisons can be made with scalable bioinformatic tools such as PASTIS [193], [194], last [195], or MMseqs [68]. ClusterMaker [168] is a Cytoscape plugin [136] that includes several network-based clustering algorithms.
Several tools facilitate clustering and visualization in metagenomic analyses like QIIME 2 [148], Anvi'o [196], and Phyloseq [149]. For example, the Quantitative Insights Into Microbial Ecology (QIIME, version 2) tool integrates hierarchical clustering methods for microbial community analysis and offers visualization through interactive plots [148], [197]. Additionally, Anvi'o [196], not only incorporates hierarchical clustering but also provides interactive interfaces for exploring and visualizing metagenomic data, enhancing the interpretability of complex microbial community structures. With its extensive interactive visualization capabilities, Anvi'o [196] is a comprehensive platform that integrates many aspects of the state-of-the-art computational strategies of data-enabled microbiology, such as phylogenomics, pangenomics, metagenomics, metatranscriptomics, genomics, and microbial population genetics, in a way that is user-friendly and seamless. Phyloseq [149] is an R package for analyzing and visualizing microbiome data. It offers a range of visualization options, including interactive plots and heatmaps, to explore the diversity and composition of microbial communities.
Principal Component Analysis (PCA) [198], aids in highlighting variations among microbial communities, providing a holistic view of the relationships between samples based on their compositional and abundance profiles. Tools that perform PCA analysis and visualization can be very useful. EMPeror [199] was one of the most useful tools for PCA analysis embedded into the QIIME suite.
The current version of QIIME2 [148] supports PCA visualization, enabling the interactive exploration of PCA results. QIIME2 offers dynamic and customizable plots that enhance the interpretability of metagenomic data. Additional tools for PCA analysis and visualization are MicrobiomeAnalyst [150], and MetagenomeSeq [151]. MicrobiomeAnalyst [150] is a web-based platform that integrates diverse statistical and bioinformatics tools. It includes PCA visualization as part of its multivariate statistical analysis suite, providing interactive visualizations for exploring the separation and clustering of microbial communities. MetagenomeSeq is an R package designed for the statistical analysis of metagenomic sequencing data. It incorporates PCA as a method for exploring variation across samples. Researchers can utilize the package to generate PCA plots and gain insights into the factors influencing the observed patterns in microbial community data.
6.5. Genome/Contig viewers
Genome viewers are tools used to visualize and analyze genomic data by providing researchers, scientists, and bioinformaticians with a graphical representation of genetic information, allowing them to explore, interpret, and understand the complexities of genomes [200]. Genome browser tools like CGViewer.js [118], Elviz [120], IMG/M [23], IMG/VR [30], Gosling [122], IGV [123], UCSC Genome Browser [126], GDV, JBrowse [124], Anvio [196], MetaErg [47], Tablet [125], Strudel [133], and CRAMER [119] offer diverse advantages and functionalities for the exploration of genomic data [201]. These tools enable multidimensional navigation through metagenome assemblies, plotting parameters such as GC content, relative abundance, phylogenetic affiliation, and contig length. They facilitate interactive exploration with real-time navigation, search, filtering, and drilling down from community profiles to individual gene annotations. Additionally, these browsers support flexible integration of various data types, including clinical data, aligned sequence reads, mutations, copy numbers, RNAi screens, gene expression, and genomic annotations. Users can benefit from the efficient exploration of large datasets across multiple resolution scales, resembling the seamless zoom and often pan functionality of Google Maps. These tools provide customizable track displays, metadata access, context menus for features, and diverse track selection methods, enhancing user interaction and data visualization.
Pangenome viewers are tools or software applications designed to visualize and analyze pangenomic data. These tools assist researchers in exploring the genetic diversity within a species or group of related organisms by providing interactive and informative visual representations of the pangenome [202], [203], [204]. Among others, popular pangenome viewers include Panache [138], Pan-Tetris [139], PanViz [140], and PanX [141] which were specifically created for gene-based pangenomes and struggle to handle extensive eukaryotic investigations. Other relevant tools include PanGP [205], Roary [206], Panseq [207], PanGraphViewer [112], Pantools [142], Bifrost [143], PanGenome Graph Builder [144], Minigraph-Cactus [146], and TwoPaCo [145].
Contig visualization tools are used to represent and analyze contiguous sequences of DNA or other biomolecules assembled from short DNA sequencing reads. Visualizing contigs is crucial for assessing the quality of a genome or transcriptome assembly, identifying structural variations, and gaining insights into the organization of genomic regions. Established tools are Bandage [178], Tablet [125], IGV (Integrative Genomics Viewer) [123], Artemis [128], UGENE [129], and Geneious [130]. Bandage is a graphical viewer to explore the connections between contigs, identify structural variations, and visualize the overall assembly graph. While IGV is primarily known as a genome browser, it also allows users to visualize contigs and their alignments. It is a versatile tool widely used for examining genomic data, including various types of sequencing data. Artemis is a genome browser and annotation tool that enables the visualization of contigs, genes, and other genomic features. It is particularly useful for annotating bacterial and archaeal genomes. Geneious is a comprehensive platform that includes tools for sequence analysis and assembly. It provides a user-friendly interface for visualizing contigs, exploring assemblies, and performing various molecular biology tasks.
Finally, Multiple Sequence Alignments (MSAs) are essential for comparing and understanding the similarities and differences between homologous sequences. Multiple Sequence Alignment (MSA) visualizers such as AlignmentViewer, BV-BRC [131], MSAViewer [132], Seaview [137], JalView [135], MSABrowser [136], NCBI MSA viewer, SuiteMSA [134], are used to display and analyze the alignment of multiple genomic sequences (DNA, RNA, or proteins).
6.6. Taxonomy
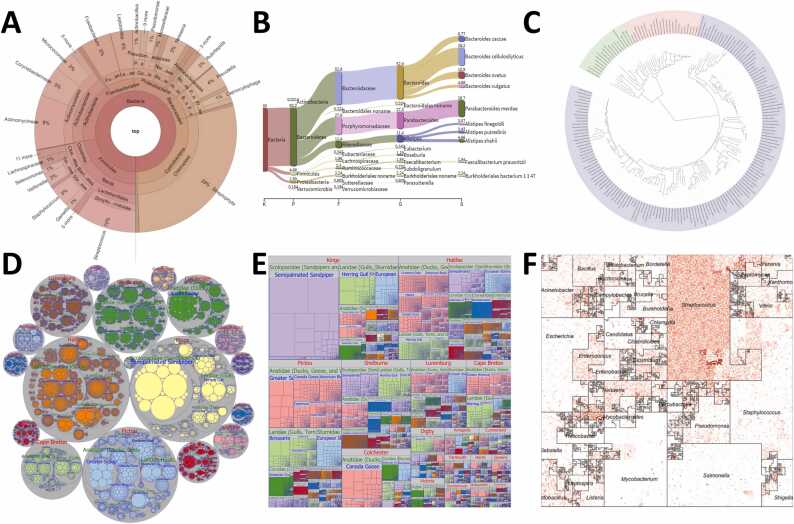
Taxonomy aims at the classification of organisms based on shared characteristics and evolutionary relationships. The classification system is presented in a hierarchical framework that ranges from broader to more specific categories. The Genome Taxonomy Database (GTDB; https://gtdb.ecogenomic.org) offers the most advanced genome-based taxonomy for prokaryotes that is both phylogenetically coherent and rank-normalized [208]. Various types of graphical representations are used to visualize the evolutionary ties between different organisms (Fig. 4). There are several tools and algorithms available for visualizing taxonomic connections [209]. Some phylogenetic tree visualization tools such as FigTree, iTOL [154], [155], MEGA [152], and Dendroscope [157] are designed with a user-friendly interface and also possess interactive capabilities. These tools offer a range of customization options, allowing the user to present, explore, and modify the appearance of phylogenetic trees. VAMPS (Visualization and Analysis of Microbial Population Structures) [210] is a repository that can provide visualization tools for the comparison of taxonomic distributions from different datasets. Additionally, Python toolkits such as ETE Toolkit (Environment for Tree Exploration) [211], DendroPy [212], and Bio.Phylo [213], which is all open-source, can be utilized for the analysis and visualization of phylogenetic trees. R packages such as Phyloseq [149], ampvis2 [214], and MetagenomeSeq [151] analyze and visualize metagenomic data using various statistical techniques. In addition to these, other visualization tools, including Treemap, Krona [215], and BioSankey [85] provide alternative representations for taxonomic data. Software tools like MEGA [152], and PAUP [153] are focused on molecular evolution and can be used for sequence alignment and phylogenetic tree construction. PhyD3 [156] is also utilized for DNA and amino acid sequence alignment. Anvi'o [196] provides tools for the visualization of taxonomic relationships within microbial communities.
Fig. 4.
(A) Sunburst chart (Krona) showing taxonomy. (B) Taxonomy with Sankey plot (Pavian). (C) Tree of Life visualized by iTOL. (D) Taxonomy visualized as a Bubble chart. (E) Taxonomy visualized as a Treemap. (F) Taxonomic Ordering with the use of Hilbert curves visualized by Jasper/Microbiome Maps. All the plots above have been created using example data provided with each tool.
6.7. Networks and associations
Leveraging networks within the realm of metagenomics offers valuable insights into the intricate interactions among microorganisms within a community. For instance, Taxonomic Networks aid in understanding relationships among diverse microbial taxa by employing taxonomic classifications. Nodes within these networks represent taxonomic units, while edges signify the extent of similarity or co-occurrence. Functional Networks enable the exploration of relationships among microbial genes or pathways, constructed based on functional annotations. Co-occurrence Networks illustrate patterns of co-existence among various microbial species or functional genes, shedding light on potential symbiotic or antagonistic relationships. Ecological Networks are employed to analyze community dynamics, identify keystone species, assess network stability, and gauge the influence of environmental factors on microbial interactions. Phylogenetic Networks display the evolutionary relationships among microbial species, unveiling patterns and aiding in the identification of closely related taxa with shared functions. A Host-Microbial Network represents the intricate interactions and relationships between a host organism and the microbial communities that inhabit various body sites. Humans, for example, are vulnerable to a vast array of microorganisms, including bacteria, viruses, fungi, and other microbes. Alternatively, a Disease Association Network plays a role in investigating the correlation between microbial communities and host health. Similarly to the previous category, these networks are constructed to encompass host-microbe and microbe-microbe interactions, providing pertinent insights into the microbiome's role in health and disease. Finally, a Microbiome Epidemiological Network denotes the interlinked associations among microorganisms within a population, concentrating on the epidemiological dimensions of microbial communities. This form of network analysis entails the examination of the dispersion, transmission, and elements impacting the prevalence of microorganisms in a population.
To this end, network visualizations, either individually or in combination, can contribute to the extraction of conclusions. For instance, examining temporal and spatial dynamics allows for the illustration of how microbial networks evolve over time or in diverse spatial locations, offering insights into the temporal and spatial shifts within microbial communities. Additionally, networks enable functional prediction. Utilizing network-based approaches facilitates the prediction of gene functions based on the functions of neighboring genes in the network, particularly beneficial in cases where functional annotations are incomplete.
All aforementioned relations can be captured and viewed in the form of networks [79], [216], [217], [218], [219], [220]. Network visualization tools are usually topic agnostic, in the sense that their functionalities are of general purpose, and the resulting network’s scientific field only depends on the type of input data. Widely used network visualization software in bioinformatics includes Cytoscape [158], [159], Graphia, Gephi [160], and Pajek [161]. However, for specific scenarios, dedicated network visualization tools can be employed. For instance, Arena3Dweb [162], [163], [221] facilitates interactive, multi-layered visualizations in 3D space to reveal intriguing data patterns. It allows network pseudo-alignment and excels in efficiently visualizing heterogeneous information, employing a multi-layered concept that proves particularly effective for time series analysis. Another specialized tool is NORMA [164], [165], which enables the highlighting of annotations over communities of nodes and supports layouts based on user-defined annotated groups. In the context of metagenomics, these nodes might for example represent bacterial species, whereas the overlaid annotations could indicate pathological functions, metabolic pathways, resistance to antibiotics, or symbiotic relationships.
Several examples involving the use of network visualization to describe metagenomic datasets are presented in Fig. 5a-c, created using data taken from the NMPFamsDB database. Fig. 5a presents a network visualization for the distribution of all available novel metagenome protein families (NMPFs) across eight major biome types (Freshwater, Marine, Soil, Plants, Human, Mammals, Other Host-associated and Engineered environments), rendered using Gephi. The biomes are represented by central, colored nodes (hubs), whereas the gray peripheral nodes represent the NMPFs, and edges represent NMPF-biome associations. Through this representation, NMPFs appearing in multiple biomes, as well as NMPFs confined to a specific biome can be visualized. Fig. 5b displays a three-dimensional (3D), multilayered network, featuring all the NMPFs connected with four major human microbiome systems (skin, respiratory, digestive, and reproductive systems), created with Arena3Dweb. In addition, each NMPF is annotated with annotation on the nature of its source microbiome sample (metagenome or metatranscriptome), and on whether it has a predicted protein structure model or not. This information is organized in multiple layers. The protein families themselves are depicted in the central layer, with nodes corresponding to NMPFs and intra-layer edges depicting the co-existence of NMPFs in the same metagenome sample. Inter-layer edges connect each NMPF with its corresponding annotation, including the association with a particular biome, the nature of the source dataset, and the availability of a 3D protein model. Finally, Fig. 5c displays a network representation of a gene neighborhood for a novel metagenome protein family (F006270) from NMPFamsDB, rendered using NORMA. The neighborhood of the family consists of proteins with hits to known Pfam domains (e.g., p450) and/or associations with COG functions (e.g., ‘Defense mechanisms’ or ‘metabolite biosynthesis’). Through these associations, a potential function for the unannotated genes of the protein family can be inferred. Overall, these examples demonstrate the capabilities of networks in providing advanced methods in the visualization, analysis, and annotation of metagenomic data and metadata.
Fig. 5.
(A-C) Various network visualization schemes for data retrieved from NMPFamsDB. (A) 2D Network visualization of NMPF distribution across different biomes, rendered using Gephi. (B) 3D, multi-layered network visualization of NMPFs associated with 4 human microbiomes, as well as additional annotation (sample type and availability of 3D model), created using Arena3Dweb. (C) A gene co-occurrence network describing the gene neighborhood of a novel metagenome protein family (F006270), constructed with data from NMPFamsDB and rendered using NORMA. The functional annotation of F006270’s neighboring genes is presented in the form of colored groups. (D) Gene neighborhood visualization for multiple MAGs through synteny conservation analysis, rendered using GeCoViz and the FESNov catalog. (E) Tree visualization of metagenome ecosystems, using the GOLD classification system. The number of metagenomic datasets associated with each ecosystem is given in parentheses. (F) Chronological progression of different SARS-Cov-2 strains in the form of a histogram, rendered using NextStrain. (G-H) Map visualizations of the geographical distribution across Europe (G) and global dispersion patterns of COVID-19 (H) rendered using NextStrain.
6.8. Gene neighborhood and synteny conservation analysis
In prokaryotic genomes, functionally related genes tend to be grouped, sharing common regulatory mechanisms and forming conserved gene neighborhoods. The study of these neighborhoods is usually performed in the form of a synteny conservation analysis, in which multiple genomes or, in the case of metagenomics, multiple metagenome scaffolds, are compared to investigate the existence of common coordinate patterns around one or multiple studied genes. Genome synteny refers to the conservation of the relative order of genes and other genomic elements in the chromosomes of different species and is often used to study evolutionary relationships between species and to identify orthologous genes, which are genes in different species that evolved from a common ancestral gene. Identifying common gene contexts among different scaffolds can be used to functionally annotate previously unknown metagenomic sequences (e.g., NMPFamsDB [34], [35], FESnov catalog [37]), predict protein-protein interactions, or discover novel functional roles. Synteny conservation can be explored through various means, ranging from simple MSAs to whole genome alignment visualization and, most notably through the use of synteny browsers. The latter are specialized genome browsers designed for the comparative analysis of multiple genomes/scaffolds, although standard browsers such as UCSC do offer limited synteny functionality. Examples include standalone tools such as JAX synteny browser [222], ALLMAPS [223] or GeneSpy [224] and web applications such as KEGG Synteny, WebFlaGs [225], and GeCoViz [226], including metagenome-oriented tools like FeGenie [227] and the EFI enzymology tools [228]. A complementary analysis to synteny conservation can be performed through the use of gene co-occurrence networks. In this approach, a gene neighborhood can be represented as an interaction network, in which the edges between genes represent their proximity to each other in multiple genomes or scaffolds. By annotating the neighbors (e.g., through association with Pfam, KEGG pathways, or COG functions), the potential function of an unannotated gene can also be inferred. Notable tools capable of providing this functionality include general-purpose network viewers such as Cytoscape, or specialized tools such as NORMA [164], [165]. Examples of gene neighborhood analysis, both through an association network and through a synteny browser, are shown in Fig. 5c-d.
6.9. Biome distribution / ecosystems / geographical distribution
Biome distribution, ecosystems, and geographical distribution are interconnected concepts that play a crucial role in understanding the diversity of life on Earth and the intricate relationships between living organisms and their environment. Biomes are large geographic regions characterized by distinct climates, vegetation, and animal life. The distribution of biomes across the planet is influenced by factors such as temperature, precipitation, and sunlight. Examples of biomes include tropical rainforests, deserts, tundras, and grasslands. Each biome has unique ecological features, and the global distribution of biomes contributes significantly to the Earth's biodiversity. Ecosystems are smaller, localized communities of living organisms interacting with their physical environment. These ecosystems, ranging from freshwater ponds to coral reefs, forests, or grasslands, experience distribution influenced by climate, topography, soil composition, and other environmental factors. Geographical distribution refers to the spatial arrangement of organisms on Earth, encompassing patterns of occurrence and abundance across regions. Factors such as climate, landforms, and human activities contribute to the geographical distribution of life forms. Understanding geographical distribution is essential for studying biodiversity, ecological patterns, and the impact of environmental changes on various species.
Biome distribution, ecosystems, and geographical distribution are intricately linked through complex ecological dynamics. The characteristics of a biome shape the types of ecosystems it harbors, and the geographical distribution of species is often associated with the specific biomes and ecosystems they inhabit. Environmental changes, whether natural or human-induced, exert profound effects on these interconnections, influencing the distribution of biomes and ecosystems over time.
Visualizing Biome Distribution, Ecosystems, and Geographical Distribution is instrumental in unraveling the intricate tapestry of Earth's biodiversity and ecological dynamics. Through advanced visualization techniques, researchers can map the global distribution of biomes, highlighting the distinct climates, vegetation, and animal life characterizing different geographic regions (See COVID-19 example in Fig. 5f-h). These visualizations not only provide a comprehensive understanding of the relationships between biomes, ecosystems, and geographical features but also serve as powerful tools for communicating complex ecological concepts to a broader audience, fostering environmental awareness and stewardship. While custom biome visualizations can be achieved using methods outlined in Section 4 (Visualization Concepts), various pre-built viewers are also accessible within metagenome resources. Databases such as IMG/M, MGnify, or SPIRE, use the GOLD ecosystem classification (Fig. 5e) and provide geolocation data visualization for each submitted dataset. GOLD also offers a specialized browser for exploring the geographical distribution of biomes based on microbiome metadata. NMPFamsDB provides visualization for the ecosystem and geographical distribution of each NMPF. In addition, the database offers dedicated tools for generating custom plots (bar charts, Venn diagrams, Circos plots, color-coded matrices, and Upset plots) measuring the ecosystem and phylogeny distribution of user-selected NMPFs, as well as the geographical spread of each NMPF. Finally, the Microbiome Maps resource uses Jasper [147] to visualize ecosystem distribution with Hilbert curves.
7. Discussion
Visualization tools represent indispensable assets in the analysis and interpretation of complex biological data in genomics and metagenomics. Genomics and metagenomics research have witnessed an exponential surge in data generation, necessitating robust visualization tools to unravel the intricacies encoded within these datasets. While advancements in visualization technologies have greatly enhanced researchers' ability to explore and interpret biological data, several challenges persist:
7.1. Conveying complexity
Despite advancements, visualization tools often struggle to effectively convey the complexity inherent in genomic and metagenomic datasets. For instance, the visualization of microbial community dynamics within ecological niches may oversimplify intricate interactions, leading to potential misinterpretation of ecological patterns.
7.2. Computational demands
Certain visualization tools impose significant computational demands, rendering them inaccessible to researchers with limited access to high-performance computing resources. For instance, tools that employ complex algorithms for three-dimensional visualization of genomic structures may require substantial computational power, limiting their utility in resource-constrained settings.
7.3. Compatibility issues
Compatibility issues between visualization tools, data formats, and operating systems pose substantial challenges. For example, the interoperability between bioinformatics pipelines and visualization platforms may necessitate complex data preprocessing steps, introducing potential errors and hindering seamless data analysis workflows.
7.4. Scalability limitations
The scalability of visualization tools is often tested when confronted with large-scale genomic and metagenomic datasets. For instance, tools designed for visualizing microbial community diversity may exhibit reduced performance or increased computational time when analyzing datasets encompassing diverse microbial populations or extensive sequencing depths.
7.5. Learning curve
Some visualization tools entail steep learning curves, requiring researchers to invest significant time and effort in mastering their functionalities.
7.6. Adjustment to future technologies
Visualization tools are poised to undergo a transformative evolution as they adapt to future technologies, such as virtual reality (VR). The integration of VR capabilities into visualization tools holds immense promise for revolutionizing how researchers explore and interact with biological data. By leveraging VR technology, visualization tools can offer immersive and interactive experiences that transcend the limitations of traditional 2D visualizations. For example, researchers could navigate through three-dimensional representations of genomic landscapes, manipulate molecular structures with hand gestures, or explore intricate biological networks in immersive virtual environments. Furthermore, the emergence of augmented reality (AR) technologies offers exciting possibilities for overlaying virtual data visualizations onto the physical world, enabling researchers to seamlessly integrate biological insights into their laboratory experiments or fieldwork. As VR and AR technologies continue to advance, visualization tools will play a pivotal role in harnessing the full potential of these immersive technologies to unlock new insights into the complexities of biological systems and accelerate scientific discovery.
Despite the challenges, advancements in visualization tools include a plethora of cutting-edge innovations. These advancements encompass a broad spectrum of transformative features such as:
7.7. Intuitive representations
Contemporary visualization tools offer intuitive representations that facilitate data exploration and interpretation. For instance, tools such as Krona utilize interactive sunburst visualizations to depict taxonomic hierarchies, enabling researchers to discern microbial community compositions with ease.
7.8. Interactive features and dynamic exploration
The incorporation of interactive features enables dynamic exploration of genomic and metagenomic data. Notable examples include Anvi'o which allows users to interactively visualize and annotate metagenomic assemblies, facilitating real-time exploration of genomic contexts.
7.9. Data integration
Bioinformatics visualization tools exhibit advanced data integration capabilities, revolutionizing researchers' ability to synthesize diverse omics datasets and unravel complex biological phenomena. These tools facilitate seamless integration of genomics/metagenomics, transcriptomics,/metatranscriptomics proteomics, and metabolomics data, enabling holistic analyses of biological systems.
7.10. Community engagement and continuous development
Popular visualization tools often boast active user communities, fostering collaborative development and continuous improvement. Galaxy for genomic analysis and Cytoscape for network analysis and visualization are two characteristic examples.
7.11. Customization flexibility
Tools that offer customization options empower researchers to tailor visualizations to their specific research questions and preferences. An exemplary tool in this regard is Circos which enables the creation of highly customizable circular plots for visualizing genomic data, allowing researchers to highlight genomic features of interest with precision.
7.12. Reproducibility
Genomic visualization tools play a crucial role in ensuring reproducibility by providing transparent and replicable means to visualize and analyze genomic data.
In conclusion, visualization tools represent indispensable assets for genomics and metagenomics research, offering valuable insights into complex biological phenomena. While recent advancements have significantly enhanced the utility and accessibility of visualization tools, several challenges persist, necessitating ongoing innovation and refinement. By addressing these challenges and capitalizing on emerging technologies, researchers can harness the full potential of visualization tools to advance our understanding of the intricacies of genomic and metagenomic landscapes.
Author contributions
All authors have tested and benchmarked various tools. All authors have read and approved the manuscript.
Funding
Fondation Santé; Onassis Foundation; ARISE program from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 945405; U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, supported by the Office of Science of the U.S. Department of Energy, operated under Contract No. DE-AC02–05CH11231; Startup funds from the Penn State College of Medicine and by the Huck Innovative and Transformational Seed Fund (HITS) award from the Huck Institutes of the Life Sciences at Penn State University; Hellenic Foundation for Research and Innovation (H.F.R.I) under the call ‘Greece 2.0 - Basic Research Financing Action (Horizontal support of all Sciences), Sub-action II’, Grant ID: 16718-PRPFOR; ‘Greece 2.0 - National Recovery and Resilience Plan’, Grant ID: TAEDR-0539180.
CRediT authorship contribution statement
Maria Chasapi: Data curation, Formal analysis, Investigation, Resources, Visualization, Writing – original draft, Writing – review & editing. Nikolaos Vergoulidis: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing. Maria Kokoli: Investigation. Nefeli K Venetsianou: Investigation. Evangelos Karatzas: Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. Ioannis Iliopoulos: Investigation, Project administration, Visualization, Writing – original draft, Writing – review & editing. Nikos C Kyrpides: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Evangelos Pafilis: Data curation, Investigation, Methodology, Writing – review & editing. Fotis A Baltoumas: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. Georgios A Pavlopoulos: Conceptualization, Investigation, Supervision, Visualization, Writing – original draft, Writing – review & editing. Eleni Panagiotopoulou: Investigation, Writing – review & editing. Eleni Aplakidou: Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Supervision, Visualization, Writing – original draft, Writing – review & editing. Ilias Georgakopoulos-Soares: Investigation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Glossary
- Accessory genome
A gene set commonly shared within only one or some strains
- Adapters
Short oligonucleotides ligated to the ends of DNA fragments of interest so that they can be combined with primers for amplification
- Amplicon
A piece of DNA or RNA that is the source and product of amplification or replication events. It can be formed naturally through gene duplication, or artificially with polymerase chain reactions
- Annotation
The process of deriving the structural and functional information of a protein or gene from a raw data set
- Assembly
The process of reconstructing a complete genome sequence from fragmented DNA sequences obtained through sequencing techniques.
- Average Nucleotide Identity (ANI)
A measure of nucleotide-level genomic similarity between the coding regions of two genomes
- Binning
The process of grouping reads or contigs into individual genomes and assigning each group to a specific taxon
- Biomes
Large geographic regions characterized by distinct climates, vegetation, and animal life
- Centroid-linkage method
A method of hierarchical clustering that defines the distance between clusters as being the distance between their centers/centroids
- Clustering
A data science technique that groups similar unlabeled objects
- Convolutional Neural Networks (CNN)
A network architecture for deep learning that learns directly from data
- COI marker genes
The mitochondrial cytochrome oxidase subunit 1 (COI) gene is one of the most popular markers used for molecular systematics
- Complete-linkage method
A method of Hierarchical clustering that defines the link between two clusters as a combination of all element’s pairs and the distance between those two clusters as the distance between two elements (one in each cluster) that are farthest away from each other.
- Contig
A set of DNA segments or sequences that overlap in a way that provides a contiguous representation of a genomic region
- Core genome
The core genome is defined as the set of genes that are ubiquitous—or nearly ubiquitous—to a set of genomes
- CRISPR elements
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) elements are specialized stretches of DNA found in the genomes of bacteria and other microorganisms. They are part of the microbial immune system, acting as a defense mechanism against foreign genetic material, such as viruses and plasmids. CRISPR elements consist of repeating sequences interspersed with unique spacer sequences derived from previous encounters with foreign genetic elements. They function in conjunction with CRISPR-associated (Cas) proteins to recognize and cleave specific sequences of foreign DNA or RNA, providing protection against future invasions. Additionally, CRISPR systems have been adapted for use in genetic engineering and gene editing technologies.
- De Bruijn graph (DBG)
A directed graph representing overlaps between sequences
- De novo assembly
De novo assembly is a method for constructing genomes from a large number of (short- or long-) DNA fragments, with no a priori knowledge of the correct sequence or order of those fragments.
- Ecosystem
A localized community of living organisms interacting with their physical environment
- Enrichment bias
A phenomenon where certain features are overrepresented or underrepresented in a sample or dataset due to experimental or analytical procedures
- Functional annotation
The process of attaching biological information to sequences of genes or proteins
- GC distribution
A measure that indicates the proportion of G and C bases out of an implied four total bases.
- Gene calling
The prediction of valid open reading frames (ORFs) for protein-coding genes in a sequence assembly
- Gene neighborhood
Segments of the genome with specific characteristics associated with them
- Gene prediction
The process of identifying the regions of genomic DNA that encode genes
- Genome synteny
The physical co-localization of genetic loci on the same chromosome within an individual or species
- Graph-based clustering
A method aims to partition a set of graphs into different groups that share some form of similarity.
- Hierarchical clustering
An unsupervised clustering technique which involves creating clusters in a predefined order
- Hybrid assembly
A technique that combines data from different sequencing technologies to create a more precise and complete genome sequence (reference-guided and partially de novo)
- ITS marker gene
The Internal transcribed spacer (ITS) is one of the most popular markers used for molecular systematics
- Metabarcoding analysis
The combined use of universal DNA barcodes and high-throughput sequencing (HTS) to characterize biological communities from genetic material collected from environmental samples
- Metadata
The descriptive data about the sample that a DNA/RNA sequence was obtained from
- Metagenome-assembled genome (MAGs)
A single-taxon assembly based on binned metagenomes that represents an entire individual genome
- Metagenome
The total amount of sequenced genetic material (DNA) from an environmental sample
- Metatranscriptome
Metatranscriptomics involves examining and analyzing the mRNA found within a metagenomic sample (known as the metatranscriptome). This approach reveals insights into the regulation and expression patterns of diverse microbial communities within the sampled environment.
- Multiple sequence alignment (MSA)
A bioinformatics technique used to align three or more biological sequences (such as DNA, RNA, or protein sequences) simultaneously. It aims to identify regions of similarity among the sequences, highlighting conserved motifs, domains, and functional elements
- NcRNAs
Functional RNA molecules that are not translated into proteins. Examples include rRNAs, tRNAs, micro-RNAs
- Neighbor-joining
A bottom-up (agglomerative) clustering method for the creation of phylogenetic trees
- NMPFams
Novel Metagenome Protein Families (no similarity to Pfam or reference genomes)
- Overlap-Layout Consensus (OLS)
A computational method used in bioinformatics to assemble DNA sequences by identifying overlapping regions between shorter DNA fragments and merging them to reconstruct longer contiguous sequences
- OTU
An operational taxonomic unit (OTU) is an operational definition used to classify groups of closely related individuals. A vOTU is a viral operation taxonomic unit.
- Pangenome
The entire set of genes from all strains within a clade
- Phylogenetic analysis
The study of the evolutionary relationship between the organisms
- Principal Component Analysis (PCA)
A versatile statistical method for reducing a cases-by-variables data table to its essential features, called principal components.
- Reference genomes
A digital nucleic acid sequence database, assembled by scientists as a representative example of the set of genes in one idealized individual organism of a species
- Reference-based assembly
A method for reconstruction of genomes or genetic sequences by aligning and assembling short DNA sequence reads against a known reference sequence
- Relative abundance
The evenness of distribution of individuals among species in a community.
- Scaffold
A portion of a genome sequence reconstructed from end-sequenced whole-genome shotgun clones. Scaffolds are composed of contigs and gaps
- Sequence alignment
A technique of arranging the sequences of DNA, RNA or protein to identify regions of similarity that may be a consequence of functional, structural or evolutionary relationships between the sequences
- Shotgun sequencing
A technique for determining the DNA sequence of an organism’s genome. The method involves randomly breaking up the genome into small DNA fragments that are sequenced individually. A computer program looks for overlaps in the DNA sequences, using them to reassemble the fragments in their correct order to reconstitute the genome
- Single-linkage method
A method of hierarchical clustering. It is based on grouping clusters in a bottom-up fashion (agglomerative clustering), at each step combining two clusters that contain the closest pair of elements not yet belonging to the same cluster as each other.
- Standalone tool
Any application or software that does not need to be bundled with other software or applications, nor does it require anything else to function
- Synteny conservation analysis
The analysis of the maintenance of the relative ordering of genes or genomic regions across different species, often used to infer evolutionary relationships and identify conserved genomic regions
- Taxonomic assignment
The process of classifying or assigning biological sequences (such as DNA sequences obtained from metagenomic or genomic data) to their respective taxonomic categories
- Trimming
The process of removing unwanted or low-quality regions from sequences, typically in the context of DNA or RNA sequences obtained from sequencing experiments
- Unweighted pair group method with arithmetic mean (UPGMA)
A hierarchical clustering algorithm used to construct dendrograms that illustrate the genetic or evolutionary relationships between biological sequences or taxa by sequentially merging the closest pairs of entities based on their pairwise distances.
Contributor Information
Georgios A. Pavlopoulos, Email: pavlopoulos@fleming.gr.
Fotis A. Baltoumas, Email: baltoumas@fleming.gr.
References
- 1.Turnbaugh P.J., Gordon J.I. An invitation to the marriage of metagenomics and metabolomics. Cell. 2008;134:708–713. doi: 10.1016/j.cell.2008.08.025. [DOI] [PubMed] [Google Scholar]
- 2.Rappuoli R., Young P., Ron E., Pecetta S., Pizza M. Save the microbes to save the planet. A call to action of the International Union of the Microbiological Societies (IUMS) One Health Outlook. 2023;5:5. doi: 10.1186/s42522-023-00077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steen A.D., Crits-Christoph A., Carini P., DeAngelis K.M., Fierer N., Lloyd K.G., et al. High proportions of bacteria and archaea across most biomes remain uncultured. ISME J. 2019;13:3126–3130. doi: 10.1038/s41396-019-0484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wade W. Unculturable bacteria--the uncharacterized organisms that cause oral infections. JRSM. 2002;95:81–83. doi: 10.1258/jrsm.95.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kho Z.Y., Lal S.K. The human gut microbiome – a potential controller of wellness and disease. Front Microbiol. 2018;9:1835. doi: 10.3389/fmicb.2018.01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Carlo P., Serra N., Alduina R., Guarino R., Craxì A., Giammanco A., et al. A systematic review on omics data (metagenomics, metatranscriptomics, and metabolomics) in the role of microbiome in gallbladder disease. Front Physiol. 2022;13 doi: 10.3389/fphys.2022.888233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguiar-Pulido V., Huang W., Suarez-Ulloa V., Cickovski T., Mathee K., Narasimhan G. Metagenomics, metatranscriptomics, and metabolomics approaches for microbiome analysis: supplementary issue: bioinformatics methods and applications for big metagenomics data. Evol Bioinform Online. 2016;12s1 doi: 10.4137/EBO.S36436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nam N., Do H., Loan Trinh K., Lee N. Metagenomics: an effective approach for exploring microbial diversity and functions. Foods. 2023;12:2140. doi: 10.3390/foods12112140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon C., Daniel R. Metagenomic analyses: past and future trends. Appl Environ Microbiol. 2011;77:1153–1161. doi: 10.1128/AEM.02345-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chistoserdova L. Recent progress and new challenges in metagenomics for biotechnology. Biotechnol Lett. 2010;32:1351–1359. doi: 10.1007/s10529-010-0306-9. [DOI] [PubMed] [Google Scholar]
- 11.Navgire G.S., Goel N., Sawhney G., Sharma M., Kaushik P., Mohanta Y.K., et al. Analysis and Interpretation of metagenomics data: an approach. Biol Proced Online. 2022;24:18. doi: 10.1186/s12575-022-00179-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shakya M., Lo C.-C., Chain P.S.G. Advances and challenges in metatranscriptomic analysis. Front Genet. 2019;10:904. doi: 10.3389/fgene.2019.00904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang, Thompson Y., Branck K.N., Yan Yan T., Nguyen L.H., Franzosa E.A., et al. Metatranscriptomics for the human microbiome and microbial community functional profiling. Annu Rev Biomed Data Sci. 2021;4:279–311. doi: 10.1146/annurev-biodatasci-031121-103035. [DOI] [PubMed] [Google Scholar]
- 14.Bashiardes S., Zilberman-Schapira G., Elinav E. Use of metatranscriptomics in microbiome research. Bioinform Biol Insights. 2016;10 doi: 10.4137/BBI.S34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haft D.H., Badretdin A., Coulouris G., DiCuccio M., Durkin A.S., Jovenitti E., et al. RefSeq and the prokaryotic genome annotation pipeline in the age of metagenomes. Nucleic Acids Res. 2024;52:D762–D769. doi: 10.1093/nar/gkad988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudhagara P., Bhavsar S., Bhagat C., Ghelani A., Bhatt S., Patel R. Web resources for metagenomics studies. Genom, Proteom Bioinforma. 2015;13:296–303. doi: 10.1016/j.gpb.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sayers E.W., Cavanaugh M., Clark K., Pruitt K.D., Schoch C.L., Sherry S.T., et al. GenBank. Nucleic Acids Res. 2022;50:D161–D164. doi: 10.1093/nar/gkab1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanizawa Y., Fujisawa T., Kodama Y., Kosuge T., Mashima J., Tanjo T., et al. DNA Data Bank of Japan (DDBJ) update report 2022. Nucleic Acids Res. 2023;51:D101–D105. doi: 10.1093/nar/gkac1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummins C., Ahamed A., Aslam R., Burgin J., Devraj R., Edbali O., et al. The European Nucleotide Archive in 2021. Nucleic Acids Res. 2022;50:D106–D110. doi: 10.1093/nar/gkab1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kodama Y., Shumway M., Leinonen R., on behalf of the International Nucleotide Sequence Database Collaboration The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012;40:D54–D56. doi: 10.1093/nar/gkr854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee S., Stamatis D., Li C.T., Ovchinnikova G., Bertsch J., Sundaramurthi J.C., et al. Twenty-five years of Genomes OnLine Database (GOLD): data updates and new features in v.9. Nucleic Acids Res. 2022 doi: 10.1093/nar/gkac974. gkac974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen I.-M.A., Chu K., Palaniappan K., Pillay M., Ratner A., Huang J., et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen I.-M.A., Chu K., Palaniappan K., Ratner A., Huang J., Huntemann M., et al. The IMG/M data management and analysis system v.7: content updates and new features. Nucleic Acids Res. 2022 doi: 10.1093/nar/gkac976. gkac976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell A.L., Almeida A., Beracochea M., Boland M., Burgin J., Cochrane G., et al. MGnify: the microbiome analysis resource in 2020. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt T.S.B., Fullam A., Ferretti P., Orakov A., Maistrenko O.M., Ruscheweyh H.-J., et al. SPIRE: a searchable, planetary-scale microbiome REsource. Nucleic Acids Res. 2024;52:D777–D783. doi: 10.1093/nar/gkad943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer F., Bagchi S., Chaterji S., Gerlach W., Grama A., Harrison T., et al. MG-RAST version 4-lessons learned from a decade of low-budget ultra-high-throughput metagenome analysis. Brief Bioinform. 2019;20:1151–1159. doi: 10.1093/bib/bbx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clum A., Huntemann M., Bushnell B., Foster B., Foster B., Roux S., et al. DOE JGI metagenome workflow. mSystems. 2021;6 doi: 10.1128/mSystems.00804-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux S., Páez-Espino D., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., et al. IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021;49:D764–D775. doi: 10.1093/nar/gkaa946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camargo A.P., Nayfach S., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., et al. IMG/VR v4: an expanded database of uncultivated virus genomes within a framework of extensive functional, taxonomic, and ecological metadata. Nucleic Acids Res. 2022 doi: 10.1093/nar/gkac1037. gkac1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paez-Espino D., Eloe-Fadrosh E.A., Pavlopoulos G.A., Thomas A.D., Huntemann M., Mikhailova N., et al. Uncovering Earth’s virome. Nature. 2016;536:425–430. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 32.Paez-Espino D., Chen I.-M.A., Palaniappan K., Ratner A., Chu K., Szeto E., et al. IMG/VR: a database of cultured and uncultured DNA Viruses and retroviruses. Nucleic Acids Res. 2017;45:D457–D465. doi: 10.1093/nar/gkw1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arkin A.P., Cottingham R.W., Henry C.S., Harris N.L., Stevens R.L., Maslov S., et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat Biotechnol. 2018;36:566–569. doi: 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baltoumas F.A., Karatzas E., Liu S., Ovchinnikov S., Sofianatos Y., Chen I.-M., et al. NMPFamsDB: a database of novel protein families from microbial metagenomes and metatranscriptomes. Nucleic Acids Res. 2024;52:D502–D512. doi: 10.1093/nar/gkad800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pavlopoulos G.A., Baltoumas F.A., Liu S., Selvitopi O., Camargo A.P., Nayfach S., et al. Unraveling the functional dark matter through global metagenomics. Nature. 2023;622:594–602. doi: 10.1038/s41586-023-06583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baltoumas F.A., Karatzas E., Paez-Espino D., Venetsianou N.K., Aplakidou E., Oulas A., et al. Exploring microbial functional biodiversity at the protein family level-From metagenomic sequence reads to annotated protein clusters. Front Bioinform. 2023;3 doi: 10.3389/fbinf.2023.1157956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodríguez Del Río Á., Giner-Lamia J., Cantalapiedra C.P., Botas J., Deng Z., Hernández-Plaza A., et al. Functional and evolutionary significance of unknown genes from uncultivated taxa. Nature. 2023 doi: 10.1038/s41586-023-06955-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paoli L., Ruscheweyh H.-J., Forneris C.C., Hubrich F., Kautsar S., Bhushan A., et al. Biosynthetic potential of the global ocean microbiome. Nature. 2022;607:111–118. doi: 10.1038/s41586-022-04862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lloyd-Price J., Mahurkar A., Rahnavard G., Crabtree J., Orvis J., Hall A.B., et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corrêa F.B., Saraiva J.P., Stadler P.F., da Rocha U.N. TerrestrialMetagenomeDB: a public repository of curated and standardized metadata for terrestrial metagenomes. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nata’ala M.K., Avila Santos A.P., Coelho Kasmanas J., Bartholomäus A., Saraiva J.P., Godinho Silva S., et al. MarineMetagenomeDB: a public repository for curated and standardized metadata for marine metagenomes. Environ Micro. 2022;17:57. doi: 10.1186/s40793-022-00449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasmanas J.C., Bartholomäus A., Corrêa F.B., Tal T., Jehmlich N., Herberth G., et al. HumanMetagenomeDB: a public repository of curated and standardized metadata for human metagenomes. Nucleic Acids Res. 2021;49:D743–D750. doi: 10.1093/nar/gkaa1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klemetsen T., Raknes I.A., Fu J., Agafonov A., Balasundaram S.V., Tartari G., et al. The MAR databases: development and implementation of databases specific for marine metagenomics. Nucleic Acids Res. 2018;46:D692–D699. doi: 10.1093/nar/gkx1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sunagawa S., Coelho L.P., Chaffron S., Kultima J.R., Labadie K., Salazar G., et al. Structure and function of the global ocean microbiome. Science. 2015;348 doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 45.The National Microbiome Data Collaborative Data Portal: an integrated multi-omics microbiome data resource. Nat. 2022 doi: 10.1093/nar/gkab990. [DOI] [Google Scholar]
- 46.Oulas A., Pavloudi C., Polymenakou P., Pavlopoulos G.A., Papanikolaou N., Kotoulas G., et al. Metagenomics: tools and insights for analyzing next-generation sequencing data derived from biodiversity studies. Bioinform Biol Insights. 2015;9:75–88. doi: 10.4137/BBI.S12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong X., Strous M. An integrated pipeline for annotation and visualization of metagenomic contigs. Front Genet. 2019;10:999. doi: 10.3389/fgene.2019.00999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Zafeiropoulos H., Beracochea M., Ninidakis S., Exter K., Potirakis A., De Moro G., et al. metaGOflow: a workflow for the analysis of marine Genomic Observatories shotgun metagenomics data. Gigascience. 2022;12 doi: 10.1093/gigascience/giad078. giad078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zafeiropoulos H., Viet H.Q., Vasileiadou K., Potirakis A., Arvanitidis C., Topalis P., et al. PEMA: a flexible Pipeline for Environmental DNA Metabarcoding Analysis of the 16S/18S ribosomal RNA, ITS, and COI marker genes. Gigascience. 2020;9 doi: 10.1093/gigascience/giaa022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanizawa Y., Fujisawa T., Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krakau S., Straub D., Gourlé H., Gabernet G., Nahnsen S. nf-core/mag: a best-practice pipeline for metagenome hybrid assembly and binning. NAR Genom Bioinform. 2022;4 doi: 10.1093/nargab/lqac007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalvari I., Nawrocki E.P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., et al. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021;49:D192–D200. doi: 10.1093/nar/gkaa1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chan P.P., Lin B.Y., Mak A.J., Lowe T.M. tRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Russel J., Pinilla-Redondo R., Mayo-Muñoz D., Shah S.A., Sørensen S.J. CRISPRCasTyper: An automated tool for the identification, annotation and classification of CRISPR-Cas loci. Bioinformatics. 2020 doi: 10.1101/2020.05.15.097824. [DOI] [PubMed] [Google Scholar]
- 58.Bland C., Ramsey T.L., Sabree F., Lowe M., Brown K., Kyrpides N.C., et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinforma. 2007;8:209. doi: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fast and accurate identification of plasmids and viruses in sequencing data using geNomad. Nat Biotechnol. 2023 doi: 10.1038/s41587-023-01982-7. [DOI] [PubMed] [Google Scholar]
- 60.Hyatt D., Chen G.-L., Locascio P.F., Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Borodovsky M., Lomsadze A. Gene identification in prokaryotic genomes, phages, metagenomes, and EST sequences with GeneMarkS suite. Curr Protoc Microbiol. 2014;32 doi: 10.1002/9780471729259.mc01e07s32. Unit 1E.7. [DOI] [PubMed] [Google Scholar]
- 62.Rho M., Tang H., Ye Y. FragGeneScan: predicting genes in short and error-prone reads. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suzek B.E., Wang Y., Huang H., McGarvey P.B., Wu C.H., UniProt Consortium UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics. 2015;31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021;49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paysan-Lafosse T., Blum M., Chuguransky S., Grego T., Pinto B.L., Salazar G.A., et al. InterPro in 2022. Nucleic Acids Res. 2023;51:D418–D427. doi: 10.1093/nar/gkac993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 67.Buchfink B., Xie C., Huson D.H. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 68.Steinegger M., Söding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol. 2017;35:1026–1028. doi: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 69.Potter S.C., Luciani A., Eddy S.R., Park Y., Lopez R., Finn R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–W204. doi: 10.1093/nar/gky448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steinegger M., Meier M., Mirdita M., Vöhringer H., Haunsberger S.J., Söding J. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinforma. 2019;20:473. doi: 10.1186/s12859-019-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brady A., Salzberg S.L. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manghi P., Blanco-Míguez A., Manara S., NabiNejad A., Cumbo F., Beghini F., et al. MetaPhlAn 4 profiling of unknown species-level genome bins improves the characterization of diet-associated microbiome changes in mice. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karatzas E., Baltoumas F.A., Aplakidou E., Kontou P.I., Stathopoulos P., Stefanis L., et al. Flame (v2.0): advanced integration and interpretation of functional enrichment results from multiple sources. Bioinformatics. 2023;39 doi: 10.1093/bioinformatics/btad490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thanati F., Karatzas E., Baltoumas F.A., Stravopodis D.J., Eliopoulos A.G., Pavlopoulos G.A. FLAME: a web tool for functional and literature enrichment analysis of multiple gene lists. Biol (Basel) 2021;10:665. doi: 10.3390/biology10070665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lluch J., Servant F., Païssé S., Valle C., Valière S., Kuchly C., et al. The characterization of novel tissue microbiota using an optimized 16S metagenomic sequencing pipeline. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0142334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galanis A., Vardakas P., Reczko M., Harokopos V., Hatzis P., Skoulakis E.M.C., et al. Bee foraging preferences, microbiota and pathogens revealed by direct shotgun metagenomics of honey. Mol Ecol Resour. 2022;22:2506–2523. doi: 10.1111/1755-0998.13626. [DOI] [PubMed] [Google Scholar]
- 78.Baltoumas F.A., Zafeiropoulou S., Karatzas E., Koutrouli M., Thanati F., Voutsadaki K., et al. Biomolecule and bioentity interaction databases in systems biology: a comprehensive review. Biomolecules. 2021;11:1245. doi: 10.3390/biom11081245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Koutrouli M., Karatzas E., Paez-Espino D., Pavlopoulos G.A. A guide to conquer the biological network era using graph theory. Front Bioeng Biotechnol. 2020;8:34. doi: 10.3389/fbioe.2020.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heyer R., Schallert K., Siewert C., Kohrs F., Greve J., Maus I., et al. Metaproteome analysis reveals that syntrophy, competition, and phage-host interaction shape microbial communities in biogas plants. Microbiome. 2019;7:69. doi: 10.1186/s40168-019-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bremel R.D., Homan E.J. Extensive T-Cell Epitope Repertoire Sharing among Human Proteome, Gastrointestinal Microbiome, and Pathogenic Bacteria: Implications for the Definition of Self. Front Immunol. 2015;6 doi: 10.3389/fimmu.2015.00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhu Q., Mai U., Pfeiffer W., Janssen S., Asnicar F., Sanders J.G., et al. Phylogenomics of 10,575 genomes reveals evolutionary proximity between domains Bacteria and Archaea. Nat Commun. 2019;10:5477. doi: 10.1038/s41467-019-13443-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Otto E., Culakova E., Meng S., Zhang Z., Xu H., Mohile S., et al. Overview of Sankey flow diagrams: Focusing on symptom trajectories in older adults with advanced cancer. J Geriatr Oncol. 2022;13:742–746. doi: 10.1016/j.jgo.2021.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kennedy A.B.W., Sankey H.R. The thermal efficiency of steam engines. report of the committee appointed to the council upon the subject of the definition of a standard or standards of thermal efficiency for steam engines: with an introductory note. (Including appendixes and plate at back of volume) Minutes Proc Inst Civ Eng. 1898;134:278–312. doi: 10.1680/imotp.1898.19100. [DOI] [Google Scholar]
- 85.Platzer A., Polzin J., Rembart K., Han P.P., Rauer D., Nussbaumer T. BioSankey: Visualization of Microbial Communities Over Time. J Integr Bioinforma. 2018;15 doi: 10.1515/jib-2017-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ghosh S., Das A.P. Metagenomic insights into the microbial diversity in manganese-contaminated mine tailings and their role in biogeochemical cycling of manganese. Sci Rep. 2018;8:8257. doi: 10.1038/s41598-018-26311-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krzywinski M., Birol I., Jones S.J., Marra M.A. Hive plots--rational approach to visualizing networks. Brief Bioinforma. 2012;13:627–644. doi: 10.1093/bib/bbr069. [DOI] [PubMed] [Google Scholar]
- 88.Sweet M., Burian A., Fifer J., Bulling M., Elliott D., Raymundo L. Compositional homogeneity in the pathobiome of a new, slow-spreading coral disease. Microbiome. 2019;7:139. doi: 10.1186/s40168-019-0759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Armstrong G., Rahman G., Martino C., McDonald D., Gonzalez A., Mishne G., et al. Applications and Comparison of Dimensionality Reduction Methods for Microbiome Data. Front Bioinform. 2022;2 doi: 10.3389/fbinf.2022.821861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nanga S., Bawah A.T., Acquaye B.A., Billa M.-I., Baeta F.D., Odai N.A., et al. Review of Dimension Reduction Methods. JDAIP. 2021;09:189–231. doi: 10.4236/jdaip.2021.93013. [DOI] [Google Scholar]
- 91.Ma Y., Zhu L. A Review on Dimension Reduction. Int Stat Rev. 2013;81:134–150. doi: 10.1111/j.1751-5823.2012.00182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang H., Wang Y., Rudin C., Browne E.P. Towards a comprehensive evaluation of dimension reduction methods for transcriptomic data visualization. Commun Biol. 2022;5:719. doi: 10.1038/s42003-022-03628-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. 2018 doi: 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 94.Velliangiri S., Alagumuthukrishnan S., Thankumar Joseph S.I. A Review of Dimensionality Reduction Techniques for Efficient Computation. Procedia Comput Sci. 2019;165:104–111. doi: 10.1016/j.procs.2020.01.079. [DOI] [Google Scholar]
- 95.Chari T., Pachter L. The specious art of single-cell genomics. PLoS Comput Biol. 2023;19 doi: 10.1371/journal.pcbi.1011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nie Y., Zhao J.-Y., Tang Y.-Q., Guo P., Yang Y., Wu X.-L., et al. Species Divergence vs. Functional Convergence Characterizes Crude Oil Microbial Community Assembly. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tzaferis C., Karatzas E., Baltoumas F.A., Pavlopoulos G.A., Kollias G., Konstantopoulos D. SCALA: A complete solution for multimodal analysis of single-cell Next Generation Sequencing data. Comput Struct Biotechnol J. 2023;21:5382–5393. doi: 10.1016/j.csbj.2023.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chakraborty J., Palit K., Das S. Microbial Biodegradation and Bioremediation. Elsevier,; 2022. Metagenomic approaches to study the culture-independent bacterial diversity of a polluted environment—a case study on north-eastern coast of Bay of Bengal, India; pp. 81–107. [DOI] [Google Scholar]
- 99.Wang L., Jin L., Xue B., Wang Z., Peng Q. Characterizing the bacterial community across the gastrointestinal tract of goats: Composition and potential function. MicrobiologyOpen. 2019;8 doi: 10.1002/mbo3.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weiss S., Xu Z.Z., Peddada S., Amir A., Bittinger K., Gonzalez A., et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Krishnaswamy V.G., Aishwarya S., Kathawala T.M. Extrication of the microbial interactions of activated sludge used in the textile effluent treatment of anaerobic reactor through metagenomic profiling. Curr Microbiol. 2020;77:2496–2509. doi: 10.1007/s00284-020-02020-4. [DOI] [PubMed] [Google Scholar]
- 102.Pavlopoulos G.A., Kumar P., Sifrim A., Sakai R., Lin M.L., Voet T., et al. Meander: visually exploring the structural variome using space-filling curves. Nucleic Acids Res. 2013;41 doi: 10.1093/nar/gkt254. e118–e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fukasawa Y., Ermini L., Wang H., Carty K., Cheung M.-S. LongQC: A Quality Control Tool for Third Generation Sequencing Long Read Data. G3 Genes|Genomes|Genet. 2020;10:1193–1196. doi: 10.1534/g3.119.400864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lanfear R., Schalamun M., Kainer D., Wang W., Schwessinger B. MinIONQC: fast and simple quality control for MinION sequencing data. Bioinformatics. 2019;35:523–525. doi: 10.1093/bioinformatics/bty654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.De Coster W., D’Hert S., Schultz D.T., Cruts M., Van Broeckhoven C. NanoPack: visualizing and processing long-read sequencing data. Bioinformatics. 2018;34:2666–2669. doi: 10.1093/bioinformatics/bty149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Y., Chen Y., Shi C., Huang Z., Zhang Y., Li S., et al. SOAPnuke: a MapReduce acceleration-supported software for integrated quality control and preprocessing of high-throughput sequencing data. GigaScience. 2018;7 doi: 10.1093/gigascience/gix120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hufnagel D.E., Hufford M.B., Seetharam A.S. SequelTools: a suite of tools for working with PacBio Sequel raw sequence data. BMC Bioinforma. 2020;21:429. doi: 10.1186/s12859-020-03751-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nielsen C.B., Jackman S.D., Birol I., Jones S.J.M. ABySS-Explorer: visualizing genome sequence assemblies. IEEE Trans Vis Comput Graph. 2009;15:881–888. doi: 10.1109/TVCG.2009.116. [DOI] [PubMed] [Google Scholar]
- 109.Mikheenko A., Kolmogorov M. Assembly Graph Browser: interactive visualization of assembly graphs. Bioinformatics. 2019;35:3476–3478. doi: 10.1093/bioinformatics/btz072. [DOI] [PubMed] [Google Scholar]
- 110.Gonnella G., Niehus N., Kurtz S. GfaViz: flexible and interactive visualization of GFA sequence graphs. Bioinformatics. 2019;35:2853–2855. doi: 10.1093/bioinformatics/bty1046. [DOI] [PubMed] [Google Scholar]
- 111.Kunyavskaya O., Prjibelski A.D. SGTK: a toolkit for visualization and assessment of scaffold graphs. Bioinformatics. 2019;35:2303–2305. doi: 10.1093/bioinformatics/bty956. [DOI] [PubMed] [Google Scholar]
- 112.Yuan Y., Ma RK-K, Chan T.-F. PanGraphViewer: a versatile tool to visualize pangenome graphs. Bioinformatics. 2023 doi: 10.1101/2023.03.30.534931. [DOI] [Google Scholar]
- 113.Pavia M.J., Chede A., Wu Z., Cadillo-Quiroz H., Zhu Q. BinaRena: a dedicated interactive platform for human-guided exploration and binning of metagenomes. Microbiome. 2023;11:186. doi: 10.1186/s40168-023-01625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alneberg J., Bjarnason B.S., de Bruijn I., Schirmer M., Quick J., Ijaz U.Z., et al. CONCOCT: Clust cONtigs Cover Compos. 2013 doi: 10.48550/ARXIV.1312.4038. [DOI] [Google Scholar]
- 115.Uritskiy G.V., DiRuggiero J., Taylor J. MetaWRAP—a flexible pipeline for genome-resolved metagenomic data analysis. Microbiome. 2018;6:158. doi: 10.1186/s40168-018-0541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Laczny C.C., Sternal T., Plugaru V., Gawron P., Atashpendar A., Margossian H.H., et al. VizBin - an application for reference-independent visualization and human-augmented binning of metagenomic data. Microbiome. 2015;3(1) doi: 10.1186/s40168-014-0066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eren A.M., Kiefl E., Shaiber A., Veseli I., Miller S.E., Schechter M.S., et al. Community-led, integrated, reproducible multi-omics with anvi’o. Nat Microbiol. 2020;6:3–6. doi: 10.1038/s41564-020-00834-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stothard P., Grant J.R., Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform. 2019;20:1576–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anastasiadi M., Bragin E., Biojoux P., Ahamed A., Burgin J., De Castro Cogle K., et al. CRAMER: a lightweight, highly customizable web-based genome browser supporting multiple visualization instances. Bioinformatics. 2020;36:3556–3557. doi: 10.1093/bioinformatics/btaa146. [DOI] [PubMed] [Google Scholar]
- 120.Cantor M., Nordberg H., Smirnova T., Hess M., Tringe S., Dubchak I. Elviz – exploration of metagenome assemblies with an interactive visualization tool. BMC Bioinforma. 2015;16:130. doi: 10.1186/s12859-015-0566-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rangwala S.H., Kuznetsov A., Ananiev V., Asztalos A., Borodin E., Evgeniev V., et al. Accessing NCBI data using the NCBI Sequence Viewer and Genome Data Viewer (GDV) Genome Res. 2021;31:159–169. doi: 10.1101/gr.266932.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.LYi S., Wang Q., Lekschas F., Gehlenborg N. Gosling: A Grammar-based Toolkit for Scalable and Interactive Genomics Data Visualization. IEEE Trans Vis Comput Graph. 2022;28:140–150. doi: 10.1109/TVCG.2021.3114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinforma. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., et al. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016;17:66. doi: 10.1186/s13059-016-0924-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Milne I., Bayer M., Cardle L., Shaw P., Stephen G., Wright F., et al. Tablet—next generation sequence assembly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nassar L.R., Barber G.P., Benet-Pagès A., Casper J., Clawson H., Diekhans M., et al. The UCSC Genome Browser database: 2023 update. Nucleic Acids Res. 2023;51:D1188–D1195. doi: 10.1093/nar/gkac1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cunningham F., Allen J.E., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., et al. Ensembl 2022. Nucleic Acids Res. 2022;50:D988–D995. doi: 10.1093/nar/gkab1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Carver T., Harris S.R., Berriman M., Parkhill J., McQuillan J.A. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Okonechnikov K., Golosova O., Fursov M., the UGENE team Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 130.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Olson R.D., Assaf R., Brettin T., Conrad N., Cucinell C., Davis J.J., et al. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res. 2023;51:D678–D689. doi: 10.1093/nar/gkac1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yachdav G., Wilzbach S., Rauscher B., Sheridan R., Sillitoe I., Procter J., et al. MSAViewer: interactive JavaScript visualization of multiple sequence alignments. Bioinformatics. 2016;32:3501–3503. doi: 10.1093/bioinformatics/btw474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bayer M., Milne I., Stephen G., Shaw P., Cardle L., Wright F., et al. Comparative visualization of genetic and physical maps with Strudel. Bioinformatics. 2011;27:1307–1308. doi: 10.1093/bioinformatics/btr111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Anderson C.L., Strope C.L., Moriyama E.N. SuiteMSA: visual tools for multiple sequence alignment comparison and molecular sequence simulation. BMC Bioinforma. 2011;12:184. doi: 10.1186/1471-2105-12-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Torun F.M., Bilgin H.I., Kaplan O.I. MSABrowser: dynamic and fast visualization of sequence alignments, variations and annotations. Bioinforma Adv. 2021;1 doi: 10.1093/bioadv/vbab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gouy M., Tannier E., Comte N., Parsons D.P. In: Multiple Sequence Alignment. Katoh K., editor. Springer US; New York, NY: 2021. Seaview Version 5: A Multiplatform Software for Multiple Sequence Alignment, Molecular Phylogenetic Analyses, and Tree Reconciliation; pp. 241–260. vol. 2231. vol. 2231. [DOI] [PubMed] [Google Scholar]
- 138.Durant É., Sabot F., Conte M., Rouard M. Panache: a web browser-based viewer for linearized pangenomes. Bioinformatics. 2021;37:4556–4558. doi: 10.1093/bioinformatics/btab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hennig A., Bernhardt J., Nieselt K. Pan-Tetris: an interactive visualisation for Pan-genomes. BMC Bioinforma. 2015;16 doi: 10.1186/1471-2105-16-S11-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pedersen T.L., Nookaew I., Wayne Ussery D., Månsson M. PanViz: interactive visualization of the structure of functionally annotated pangenomes. Bioinformatics. 2017;33:1081–1082. doi: 10.1093/bioinformatics/btw761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ding W., Baumdicker F., Neher R.A. panX: pan-genome analysis and exploration. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gkx977. e5–e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sheikhizadeh S., Schranz M.E., Akdel M., De Ridder D., Smit S. PanTools: representation, storage and exploration of pan-genomic data. Bioinformatics. 2016;32:i487–i493. doi: 10.1093/bioinformatics/btw455. [DOI] [PubMed] [Google Scholar]
- 143.Holley G., Melsted P. Bifrost: highly parallel construction and indexing of colored and compacted de Bruijn graphs. Genome Biol. 2020;21:249. doi: 10.1186/s13059-020-02135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li H., Feng X., Chu C. The design and construction of reference pangenome graphs with minigraph. Genome Biol. 2020;21:265. doi: 10.1186/s13059-020-02168-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Minkin I., Pham S., Medvedev P. TwoPaCo: an efficient algorithm to build the compacted de Bruijn graph from many complete genomes. Bioinformatics. 2017;33:4024–4032. doi: 10.1093/bioinformatics/btw609. [DOI] [PubMed] [Google Scholar]
- 146.Hickey G., Monlong J., Ebler J., Novak A.M., Eizenga J.M., Gao Y., et al. Pangenome graph construction from genome alignments with Minigraph-Cactus. Nat Biotechnol. 2023 doi: 10.1038/s41587-023-01793-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Valdes C., Stebliankin V., Ruiz-Perez D., Park J.I., Lee H., Narasimhan G. Microbiome maps: Hilbert curve visualizations of metagenomic profiles. Front Bioinform. 2023;3 doi: 10.3389/fbinf.2023.1154588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Estaki M., Jiang L., Bokulich N.A., McDonald D., González A., Kosciolek T., et al. QIIME 2 enables comprehensive end‐to‐end analysis of diverse microbiome data and comparative studies with publicly available data. CP Bioinforma. 2020;70 doi: 10.1002/cpbi.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McMurdie P.J., Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Dhariwal A., Chong J., Habib S., King I.L., Agellon L.B., Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–W188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Joseph Nathaniel Paulson HT. metagenomeSeq 2017. 10.18129/B9.BIOC.METAGENOMESEQ. [DOI]
- 152.Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wilgenbusch J.C., Swofford D. Inferring Evolutionary Trees with PAUP *. CP in Bioinformatics 2003;00. 10.1002/0471250953.bi0604s00. [DOI] [PubMed]
- 154.Letunic I., Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 155.Zhou T., Xu K., Zhao F., Liu W., Li L., Hua Z., et al. itol.toolkit accelerates working with iTOL (Interactive Tree of Life) by an automated generation of annotation files. Bioinformatics. 2023;39 doi: 10.1093/bioinformatics/btad339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kreft Ł., Botzki A., Coppens F., Vandepoele K., Van Bel M. PhyD3: a phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics. 2017;33:2946–2947. doi: 10.1093/bioinformatics/btx324. [DOI] [PubMed] [Google Scholar]
- 157.Huson D.H., Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 158.Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Lotia S., et al. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bastian M., Heymann S., Jacomy M. Gephi: An Open Source Software for Exploring and Manipulating Networks 2009. 10.13140/2.1.1341.1520. [DOI]
- 161.Mrvar A., Batagelj V. Analysis and visualization of large networks with program package Pajek. Complex Adapt Syst Model. 2016;4 doi: 10.1186/s40294-016-0017-8. [DOI] [Google Scholar]
- 162.Karatzas E., Baltoumas F.A., Panayiotou N.A., Schneider R., Pavlopoulos G.A. Arena3Dweb: interactive 3D visualization of multilayered networks. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kokoli M., Karatzas E., Baltoumas F.A., Schneider R., Pafilis E., Paragkamian S., et al. Arena3Dweb: interactive 3D visualization of multilayered networks supporting multiple directional information channels, clustering analysis and application integration. NAR Genom Bioinforma. 2023;5 doi: 10.1093/nargab/lqad053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Koutrouli M., Karatzas E., Papanikolopoulou K., Pavlopoulos G.A. NORMA: the network makeup artist — a web tool for network annotation visualization. Genom, Proteom Bioinforma. 2022;20:578–586. doi: 10.1016/j.gpb.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Karatzas E., Koutrouli M., Baltoumas F.A., Papanikolopoulou K., Bouyioukos C., Pavlopoulos G.A. The network makeup artist (NORMA-2.0): distinguishing annotated groups in a network using innovative layout strategies. Bioinforma Adv. 2022;2 doi: 10.1093/bioadv/vbac036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang C., Chowdhury D., Zhang Z., Cheung W.K., Lu A., Bian Z., et al. A review of computational tools for generating metagenome-assembled genomes from metagenomic sequencing data. Comput Struct Biotechnol J. 2021;19:6301–6314. doi: 10.1016/j.csbj.2021.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gupta S.K., Raza S., Unno T. Comparison of de-novo assembly tools for plasmid metagenome analysis. Genes Genom. 2019;41:1077–1083. doi: 10.1007/s13258-019-00839-1. [DOI] [PubMed] [Google Scholar]
- 168.Lapidus A.L., Korobeynikov A.I. Metagenomic data assembly – the way of decoding unknown microorganisms. Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.613791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Vollmers J., Wiegand S., Kaster A.-K. Comparing and evaluating metagenome assembly tools from a microbiologist’s perspective - not only size matters! PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0169662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Breitwieser F.P., Lu J., Salzberg S.L. A review of methods and databases for metagenomic classification and assembly. Brief Bioinforma. 2019;20:1125–1136. doi: 10.1093/bib/bbx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Haider B., Ahn T.-H., Bushnell B., Chai J., Copeland A., Pan C. Omega: an Overlap-graph de novo assembler for metagenomics. Bioinformatics. 2014;30:2717–2722. doi: 10.1093/bioinformatics/btu395. [DOI] [PubMed] [Google Scholar]
- 172.Zerbino D.R. Using the Velvet de novo assembler for short‐read sequencing technologies. CP Bioinforma. 2010;31 doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Namiki T., Hachiya T., Tanaka H., Sakakibara Y. MetaVelvet: an extension of Velvet assembler to de novo metagenome assembly from short sequence reads. Nucleic Acids Res. 2012;40 doi: 10.1093/nar/gks678. e155–e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li D., Liu C.-M., Luo R., Sadakane K., Lam T.-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 175.Chikhi R., Limasset A., Medvedev P. Compacting de Bruijn graphs from sequencing data quickly and in low memory. Bioinformatics. 2016;32:i201–i208. doi: 10.1093/bioinformatics/btw279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Nurk S., Meleshko D., Korobeynikov A., Pevzner P.A. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ghurye J., Treangen T., Fedarko M., Hervey W.J., Pop M. MetaCarvel: linking assembly graph motifs to biological variants. Genome Biol. 2019;20:174. doi: 10.1186/s13059-019-1791-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wick R.R., Schultz M.B., Zobel J., Holt K.E. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 2015;31:3350–3352. doi: 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yue Y., Huang H., Qi Z., Dou H.-M., Liu X.-Y., Han T.-F., et al. Evaluating metagenomics tools for genome binning with real metagenomic datasets and CAMI datasets. BMC Bioinforma. 2020;21:334. doi: 10.1186/s12859-020-03667-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kang D.D., Li F., Kirton E., Thomas A., Egan R., An H., et al. MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ. 2019;7 doi: 10.7717/peerj.7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kang D.D., Froula J., Egan R., Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3 doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Broeksema B., Calusinska M., McGee F., Winter K., Bongiovanni F., Goux X., et al. ICoVeR – an interactive visualization tool for verification and refinement of metagenomic bins. BMC Bioinforma. 2017;18:233. doi: 10.1186/s12859-017-1653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lin H.-H., Liao Y.-C. Accurate binning of metagenomic contigs via automated clustering sequences using information of genomic signatures and marker genes. Sci Rep. 2016;6 doi: 10.1038/srep24175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Seah B.K.B., Gruber-Vodicka H.R. gbtools: interactive visualization of metagenome bins in R. Front Microbiol. 2015;6 doi: 10.3389/fmicb.2015.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 186.Xu R., Wunsch D. Survey of clustering algorithms. IEEE Trans Neural Netw. 2005;16:645–678. doi: 10.1109/TNN.2005.845141. [DOI] [PubMed] [Google Scholar]
- 187.Brohée S., van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinforma. 2006;7:488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Atkinson H.J., Morris J.H., Ferrin T.E., Babbitt P.C. Using Sequence Similarity Networks for Visualization of Relationships Across Diverse Protein Superfamilies. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Yoon S.-H., Ha S.-M., Lim J., Kwon S., Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017;110:1281–1286. doi: 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 190.Azad A., Pavlopoulos G.A., Ouzounis C.A., Kyrpides N.C., Buluç A. HipMCL: a high-performance parallel implementation of the Markov clustering algorithm for large-scale networks. Nucleic Acids Res. 2018;46 doi: 10.1093/nar/gkx1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008;2008:P10008. doi: 10.1088/1742-5468/2008/10/P10008. [DOI] [Google Scholar]
- 192.Jiang P., Singh M. SPICi: a fast clustering algorithm for large biological networks. Bioinformatics. 2010;26:1105–1111. doi: 10.1093/bioinformatics/btq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Selvitopi O., Ekanayake S., Guidi G., Pavlopoulos G.A., Azad A., Buluc A. Distributed Many-to-Many Protein Sequence Alignment using Sparse Matrices. SC20: International Conference for High Performance Computing, Networking, Storage and Analysis, Atlanta, GA, USA: IEEE; 2020, p. 1–14. 10.1109/SC41405.2020.00079. [DOI]
- 194.Selvitopi O., Ekanayake S., Guidi G., Awan M.G., Pavlopoulos G.A., Azad A., et al. Extreme-Scale Many-against-Many Protein Similarity Search. SC22: International Conference for High Performance Computing, Networking, Storage and Analysis, Dallas, TX, USA: IEEE; 2022, p. 1–12. 10.1109/SC41404.2022.00006. [DOI]
- 195.Kiełbasa S.M., Wan R., Sato K., Horton P., Frith M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Eren A.M., Esen Ö.C., Quince C., Vineis J.H., Morrison H.G., Sogin M.L., et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3 doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jolliffe I.T., Cadima J. Principal component analysis: a review and recent developments. Philos Trans R Soc A. 2016;374:20150202. doi: 10.1098/rsta.2015.0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Vázquez-Baeza Y., Pirrung M., Gonzalez A., Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. GigaSci. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Pavlopoulos G.A., Malliarakis D., Papanikolaou N., Theodosiou T., Enright A.J., Iliopoulos I. Visualizing genome and systems biology: technologies, tools, implementation techniques and trends, past, present and future. Gigascience. 2015;4:38. doi: 10.1186/s13742-015-0077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Wang J., Kong L., Gao G., Luo J. A brief introduction to web-based genome browsers. Brief Bioinforma. 2013;14:131–143. doi: 10.1093/bib/bbs029. [DOI] [PubMed] [Google Scholar]
- 202.Eizenga J.M., Novak A.M., Sibbesen J.A., Heumos S., Ghaffaari A., Hickey G., et al. Pangenome Graphs. Annu Rev Genom Hum Genet. 2020;21:139–162. doi: 10.1146/annurev-genom-120219-080406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Andreace F., Lechat P., Dufresne Y., Chikhi R. Comparing methods for constructing and representing human pangenome graphs. Genome Biol. 2023;24:274. doi: 10.1186/s13059-023-03098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Vernikos G.S. In: The Pangenome. Tettelin H., Medini D., editors. Springer International Publishing; Cham: 2020. A Review of Pangenome Tools and Recent Studies; pp. 89–112. [DOI] [PubMed] [Google Scholar]
- 205.Zhao Y., Jia X., Yang J., Ling Y., Zhang Z., Yu J., et al. PanGP: A tool for quickly analyzing bacterial pan-genome profile. Bioinformatics. 2014;30(9):1297. doi: 10.1093/bioinformatics/btu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Page A.J., Cummins C.A., Hunt M., Wong V.K., Reuter S., Holden M.T.G., et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Laing C., Buchanan C., Taboada E.N., Zhang Y., Kropinski A., Villegas A., et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinforma. 2010;11:461. doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Parks D.H., Chuvochina M., Rinke C., Mussig A.J., Chaumeil P.-A., Hugenholtz P. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res. 2022;50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pavlopoulos G.A., Soldatos T.G., Barbosa-Silva A., Schneider R. A reference guide for tree analysis and visualization. BioData Min. 2010;3(1) doi: 10.1186/1756-0381-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Huse S.M., Mark Welch D.B., Voorhis A., Shipunova A., Morrison H.G., Eren A.M., et al. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinforma. 2014;15:41. doi: 10.1186/1471-2105-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Huerta-Cepas J., Serra F., Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Sukumaran J., Holder M.T. DendroPy: a Python library for phylogenetic computing. Bioinformatics. 2010;26:1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- 213.Talevich E., Invergo B.M., Cock P.J., Chapman B.A. Bio.Phylo: A unified toolkit for processing, analyzing and visualizing phylogenetic trees in Biopython. BMC Bioinforma. 2012;13:209. doi: 10.1186/1471-2105-13-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Andersen K.S., Kirkegaard R.H., Karst S.M., Albertsen M. ampvis2: an R package to analyse and visualise 16S rRNA amplicon data. Bioinformatics. 2018 doi: 10.1101/299537. [DOI] [Google Scholar]
- 215.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinforma. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Pavlopoulos G.A., Wegener A.-L., Schneider R. A survey of visualization tools for biological network analysis. BioData Min. 2008;1:12. doi: 10.1186/1756-0381-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Pavlopoulos G.A., Kontou P.I., Pavlopoulou A., Bouyioukos C., Markou E., Bagos P.G. Bipartite graphs in systems biology and medicine: a survey of methods and applications. Gigascience. 2018;7:1–31. doi: 10.1093/gigascience/giy014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.N. Moschopoulos C., A. Pavlopoulos G., Likothanassis S., Kossida S. Analyzing protein-protein interaction networks with web tools. CBIO. 2011;6:389–397. doi: 10.2174/157489311798072972. [DOI] [Google Scholar]
- 219.Papanikolaou N., Pavlopoulos G.A., Theodosiou T., Iliopoulos I. Protein-protein interaction predictions using text mining methods. Methods. 2015;74:47–53. doi: 10.1016/j.ymeth.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 220.Kontou P.I., Pavlopoulou A., Dimou N.L., Pavlopoulos G.A., Bagos P.G. Network analysis of genes and their association with diseases. Gene. 2016;590:68–78. doi: 10.1016/j.gene.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 221.Pavlopoulos G.A., O’Donoghue S.I., Satagopam V.P., Soldatos T.G., Pafilis E., Schneider R. Arena3D: visualization of biological networks in 3D. BMC Syst Biol. 2008;2:104. doi: 10.1186/1752-0509-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kolishovski G., Lamoureux A., Hale P., Richardson J.E., Recla J.M., Adesanya O., et al. The JAX Synteny Browser for mouse-human comparative genomics. Mamm Genome. 2019;30:353–361. doi: 10.1007/s00335-019-09821-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Tang H., Zhang X., Miao C., Zhang J., Ming R., Schnable J.C., et al. ALLMAPS: robust scaffold ordering based on multiple maps. Genome Biol. 2015;16:3. doi: 10.1186/s13059-014-0573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Garcia P.S., Jauffrit F., Grangeasse C., Brochier-Armanet C. GeneSpy, a user-friendly and flexible genomic context visualizer. Bioinformatics. 2019;35:329–331. doi: 10.1093/bioinformatics/bty459. [DOI] [PubMed] [Google Scholar]
- 225.Saha C.K., Sanches Pires R., Brolin H., Delannoy M., Atkinson G.C. FlaGs and webFlaGs: discovering novel biology through the analysis of gene neighbourhood conservation. Bioinformatics. 2021;37:1312–1314. doi: 10.1093/bioinformatics/btaa788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Botas J., Rodríguez Del Río Á., Giner-Lamia J., Huerta-Cepas J. GeCoViz: genomic context visualisation of prokaryotic genes from a functional and evolutionary perspective. Nucleic Acids Res. 2022;50:W352–W357. doi: 10.1093/nar/gkac367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Garber A.I., Nealson K.H., Okamoto A., McAllister S.M., Chan C.S., Barco R.A., et al. FeGenie: a comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol. 2020;11:37. doi: 10.3389/fmicb.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zallot R., Oberg N., Gerlt J.A. The EFI web resource for genomic enzymology tools: leveraging protein, genome, and metagenome databases to discover novel enzymes and metabolic pathways. Biochemistry. 2019;58:4169–4182. doi: 10.1021/acs.biochem.9b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]