Key Points
Transgenic chickens have been engineered to produce chicken VH sdAbs.
These chickens raise robust immune responses upon immunization.
sdAbs from the birds show favorable biophysical properties and antigen recognition.
Abstract
H chain–only Igs are naturally produced in camelids and sharks. Because these Abs lack the L chain, the Ag-binding domain is half the size of a traditional Ab, allowing this type of Ig to bind to targets in novel ways. Consequently, the H chain–only single-domain Ab (sdAb) structure has the potential to increase the repertoire and functional range of an active humoral immune system. The majority of vertebrates use the standard heterodimeric (both H and L chains) structure and do not produce sdAb format Igs. To investigate if other animals are able to support sdAb development and function, transgenic chickens (Gallus gallus) were designed to produce H chain–only Abs by omitting the L chain V region and maintaining only the LC region to serve as a chaperone for Ab secretion from the cell. These birds produced 30–50% normal B cell populations within PBMCs and readily expressed chicken sequence sdAbs. Interestingly, the H chains contained a spontaneous CH1 deletion. Although no isotype switching to IgY or IgA occurred, the IgM repertoire was diverse, and immunization with a variety of protein immunogens rapidly produced high and specific serum titers. mAbs of high affinity were efficiently recovered by single B cell screening. In in vitro functional assays, the sdAbs produced by birds immunized against SARS-CoV-2 were also able to strongly neutralize and prevent viral replication. These data suggest that the truncated L chain design successfully supported sdAb development and expression in chickens.
Introduction
The only species known to produce single-domain, H chain–only Abs belong to families of camelids and sharks (1, 2). In the case of camelid single-domain Abs (sdAbs), specific V regions (VHH) are used that contain framework changes that compensate for the lack of an L chain V region (VL) partner, notably in framework region 2 (FR2) to remove hydrophobic residues that would be solvent-exposed without the presence of a VL (3–5). In addition, the C region locus contains C region genes (IgG2 or IgG3) with a splice donor site mutation that causes skipping of the CH1-encoding exon to allow direct splicing of the VHH region to CH2 and the remainder of the C region (6). It is unclear whether there is an IgM stage for sdAbs in camelids (7, 8). Like other vertebrates, birds produce only heterodimeric “H plus L” Abs, which are similar to Abs found in other species but are mainly distinguished by the use of a single framework for both VL and VH (9–11), gene conversion to produce a diverse sequence repertoire (11), and noncanonical cysteines found in the VH regions (12, 13).
Previously, we reported that a small amount of H chain–only Ab is produced in homozygous IgL knockout (KO) chickens (14). We now report that normal levels of IgM are expressed when a truncated L chain (tLC) consisting of the L chain C region only (CL), with no VL, is introduced into the birds. sdAbs of chicken VH sequence are expressed, and high-affinity mAbs can be obtained. Sequence analysis shows that the frameworks (in particular, FR2) do not undergo “camelizing” mutations to alter residues that are normally involved in pairing with the VL. Interestingly, the conserved tryptophan in FR4 sometimes mutated to arginine, as observed in camelid species. Class switching to the IgY isotype, the major serum Ig normally found in chickens, is only found at low levels in the chickens expressing sdAbs.
These results indicate that the chicken sdAb platform is available to develop sdAbs for either research or therapeutic uses, with humanization of the frameworks a possible option (15, 16) if desired.
Materials and Methods
Transgenic design and development
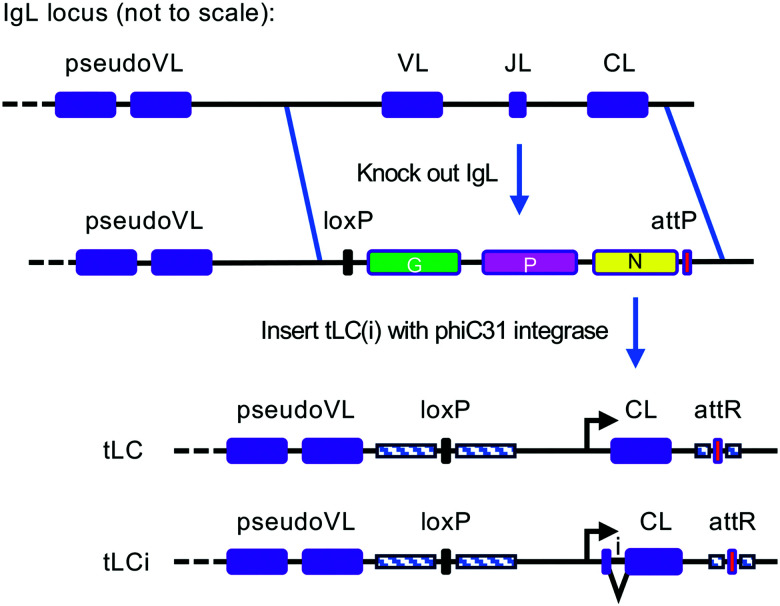
A tLC transgene consisting of the normal chicken L chain C region driven by the L chain promoter was built. The full 2.4-kb promoter region between the most proximal VL pseudogene and the functional VL was cloned upstream of the tLC, with the L chain leader sequence fused directly to the C region. Versions with the leader intron (tLCi) or without (tLC) were made (Fig. 1). A region comprising 283 bp of the 3′ untranslated region (UTR) was included in the transgene. The tLC insertion construct contained an attB site and was inserted into the L chain locus via phiC31 integrase recombination using an attP site previously integrated at the endogenous L chain locus (14), which had replaced the entire coding sequence of the L chain. (The 5845-bp deleted region spans from 1290 bp upstream of the start codon to 190 bp downstream of the stop codon; therefore, the tLC(i) insertion replaces the deleted 5′ and 3′ UTRs and inserts an extra copy of 1.2 kb of the upstream pseudogene-proximal region and 88 bp of the 3′ UTR.)
FIGURE 1.
Production of tLC(i) transgenic chickens. Previously, the functional chicken L chain locus (top line) was deleted by homologous recombination, replacing it with a selectable marker cassette containing a B-act-GFP gene (G), a CAG-puro gene (P), a promoterless neo gene (N), and an attP site (second line). IgL KO primordial germ cell line 229-92 was cotransfected with either the tLC or tLCi circular DNA construct and the pCMV-int phiC31 integrase expression plasmid pCMV-int (21) (not shown). The tLC(i) constructs were inserted into the attP site via an attB site on the incoming plasmids. Upon germline transmission of the tLC(i), selectable markers were removed by Cre recombination by breeding to Cre-expressing hens (51), leaving behind the tLC(i) insertion and single loxP and attR sites (bottom line). Because of the extent of the original deletion and the replacement by the tLC(i) insertion, 1.2 kb of the pseudogene-proximal region and 88 bp of the 3′ UTR are duplicated in the final locus (indicated by striped boxes).
Chicken primordial germ cells of line 229-92, carrying the attP-neo replacement of the L chain, were cultured as described previously (17–20) in KO-DMEM (Thermo Fisher, Waltham MA, 10829018) containing 40% Buffalo rat liver (BRL)-conditioned medium, 7.5% FCS (Hyclone, South Logan, UT, SH30088.03), 2.5% chicken serum (Thermo Fisher, 16110082), 2 mM GlutaMAX (Thermo Fisher, 35050-061), 1 mM pyruvate (Thermo Fisher, 11360-070), 1× nonessential amino acids (Thermo Fisher, 11140-050), 0.1 mM 2-ME (Thermo Fisher, 21985-023), 6 ng/ml recombinant murine stem cell factor (R&D Systems, Minneapolis, MN, 455-MC), and 4 ng/ml recombinant human FGF basic (R&D Systems, 234-FSE) on a feeder layer of irradiated BRL cells. A total of 5 × 106 cells were pelleted and resuspended in Nucleofector V buffer (Lonza, Walkersville, MD, VCA-1003) with 15 µg of circular tLC(i) insertion vector and the phiC31 integrase vector pCMV-int (21) in a total volume of 100 µl and transfected in a 2-cm cuvette with a BMX ECM830 square wave pulse electroporator (BTX, Holliston, MA) at 350 V, 100 µs, 8 pulses. Cells were resuspended in complete growth medium and distributed evenly over a 48-well plate with G418-resistant BRL feeder cells (4). G418 (Teknova, Hollister, CA, G5005) was added at 300 μg/ml 3 d after transfection, and the medium was changed every 2–3 d. G418-resistant clones were obtained at a frequency of ∼10 clones per 5 × 106 cells transfected and expanded until there were enough cells for injection. Single clones were injected into embryos to produce germline chimeras, and, in some cases, four clones were pooled and then injected. Chicks obtained from breeding chimeras were screened for the correct transgene insertion at the 5′ and 3′ insertion sites and by transgene-specific primers by PCR of comb biopsies.
Bird care and handling
All animal experiments were done in accordance with protocols approved by the OmniAb (previously Ligand Pharmaceuticals) Institutional Animal Care and Use Committee. The animal facility is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International.
Transgenic bird evaluations
Flow cytometry
At 11 wk of age, 14 transgenic birds were evaluated for B cell development, with 11 wild-type (WT) chickens serving as controls. These samples were also used for Western blot, ELISA, and RT-PCR evaluations. Whole-blood samples (2 ml per bird) were collected in EDTA anticoagulant, and PBMCs were isolated using Histopaque-1077 (Sigma-Aldrich, St. Louis, MO) following the manufacturer’s protocol. Collected PBMCs were washed once in PBS supplemented with 0.1% BSA, then aliquoted into 96-well U-bottomed plates for downstream immunostaining. PBMCs were incubated with primary Ab for 1 h on ice. Anti-chicken primary Abs were used as follows: monoclonal mouse anti-Bu1 at 5 μg/ml for the chicken B-cell marker Bu1 (SouthernBiotech, Birmingham, AL, 8395-01), monoclonal mouse anti-IgM at 5 μg/ml against IgM CH1 (SouthernBiotech, 8310-01), monoclonal mouse anti-IgL at 5 μg/ml (SouthernBiotech, 8340-01), monoclonal anti-TCR1 (TCRδγ) at 1:800 dilution (SouthernBiotech, 8230-01), combined monoclonal anti-TCR2 (TCRαβ/Vβ1) and anti-TCR3 (TCRαβ/Vβ2) at 1:200 dilution (SouthernBiotech, 8240-01 and 8250-01), and polyclonal goat anti-IgM at 4 μg/ml (Bethyl Laboratories, Montgomery, TX, A30-102A). After primary incubation, PBMCs were washed three times in PBS + 0.1% BSA, then incubated in secondary Ab for 1 h on ice, covered from light. Secondary Abs used were as follows: donkey anti-mouse Alexa Fluor 647 at 5 μg/ml (Thermo Fisher, A31571) or donkey anti-goat Alexa Fluor 647 at 4 μg/ml (Abcam, Cambridge, MA, AB150131). After secondary incubation, PBMCs were washed three times and loaded into Attune NxT flow cytometers for analysis of 10,000 cells per sample; resulting data were processed using FlowJo version 10 software. Data were analyzed in Excel by Student t test as compared with the WT/negative control and figures were created using Prism software.
Western blot analysis
Plasma samples were diluted to a final concentration of 1:100 in PBS. Samples were prepared to contain a final concentration of 1× sample loading buffer and 1.5 mM Tris carboxyethyl phosphene reducing agent and heated to 98°C for 10 min. Samples were loaded onto 4–12% gradient Bis-Tris NuPage SDS-PAGE gels (Thermo Fisher, NP0321) in MES SDS running buffer and run for 35 min at a constant 200 V. Gels were transferred to 0.2-μm nitrocellulose membranes using the iBlot2 7-min dry transfer system (Thermo Fisher). Membranes were immediately blocked in 3% skim milk in PBS for 1 h with oscillation, then incubated in directly conjugated primary Ab for 1 h with oscillation. Primary Abs used were as follows: polyclonal rabbit anti-IgY-HRP (Sigma-Aldrich, A90406) at 1:20,000 dilution in PBS + 0.05% Tween 20 (PBST) or polyclonal goat anti-IgM-HRP (Bethyl Laboratories, A30-102P) at 1 μg/ml in PBST. After incubation, membranes were washed five times for 5 min each with PBST and developed with West Pico PLUS chemiluminescent substrate (Thermo Fisher, 34580) for 15 min. Membranes were imaged for chemiluminescence using a Bio-Rad ChemiDoc XRS+ gel imager.
ELISA
High-binding ELISA plates (Greiner Bio-One, 655061) were coated with 50 μl/well of 2 μg/ml target protein diluted in PBS (for either 1 h at room temperature or overnight at 4°C). Plates were then blocked with 150 μl of 3% skim milk in PBS for 1 h at room temperature. Plasma samples were diluted in blocking buffer and applied to the plate at 50 μl/well and incubated at room temperature for 1 h. Plates were then washed four times with 300 μl/well PBST (0.05% Tween 20), and secondary Ab was applied at 1:5000 dilution in blocking buffer at 50 μl/well for 1 h incubation at room temperature. Secondary Abs were goat anti-chicken IgM-HRP (Bethyl, A30-120P) and rabbit anti-chicken IgY-HRP (Sigma-Aldrich, A90406). After secondary Abs, plates were washed five times. TMB substrate (Thermo Fisher, 002023) was applied at 50 μl/well for 10 min of color development, and the reaction was stopped with 50 μl/well 1 N HCl. Absorbance was read at 450 nm on a BioTek Synergy H1 plate reader.
RT-PCR
PBMC samples were pelleted and stored at –80°C until ready for processing. With use of the Qiagen RNeasy Plus kit (Qiagen, Germantown, MD, 74034), 107 cells were processed for mRNA extraction following the manufacturer’s procedure. Extracted mRNA was quantified using a Nano UV-Vis spectrometer for downstream normalization calculations. Extracted mRNA was used in Qiagen One-Step RT-PCRs (Qiagen, 210212) to detect IgM, IgY, and L chain (IgL) using the following primers: IgM, chVH-F9: 5′-CACCAGTCGGCTCCGCAACCATG-3′ and cIgM-CH2-R: 5′-GGGGTGCATGGTGACGAAAAG-3′; IgY, chVH-F9: 5′-CACCAGTCGGCTCCGCAACCATG-3′ and cIgY-CH2-R10: 5′-CTTCCCTTCCCTCCAATCGGTG-3′; IgL, cVL-5'UTR-F: 5′-GACACACAGCTGCTGGGATTC-3′ and chIgL-C-R: 5′-CCTGCAGGTGTAGGTCTCGT-3′. PCR parameters consisted of a 45-s reverse transcription step at 50°C followed by 35 cycles of amplification with 1-s extension time and 60°C annealing temperature. PCR products were run on 1% agarose ethidium bromide TAE gels, and bands were detected in a Bio-Rad ChemiDoc XRS+ gel imager.
Immunization program
Different cohorts of animals were immunized i.m. with protein immunogens. Three animals were immunized and screened in the progranulin (PGRN) program (bird numbers 48324, 48367, 48370) starting at 14 wk of age and with a prime followed by a total of five boosts. A quantity of 100 μg rhPGRN (Sino Biological, 10826-H08H) was used for each injection. A total of seven birds were immunized with SARS-CoV-2 proteins (cohort 1, birds 60376, 60396, 60413, starting at 6 wk of age and receiving prime plus four boosts; cohort 2, birds 63885 and 63938, starting at 10 wk of age and receiving prime plus five boosts; cohort 3, birds 66497 and 66499, starting at 8 wk of age and receiving prime plus four boosts), and all were screened except for 66499. The immunogens were as follows: Cohort 1 received recombinant SARS-CoV-2 His-tagged S1 spike protein monomer (n = 3 birds, Wuhan D614G, Acro Biosystems, S1N-C5256); cohort 2 received recombinant His-tagged spike trimers from Alpha, Beta, and Delta strains, alternating boosts (n = 2 birds, Acro Biosystems SPN-C52H6 [Alpha], SPN-C52Hk [Beta], SPN-C52He [Delta]); and cohort 3 received recombinant His-tagged Delta spike trimer (n = 2 birds, Acro Biosystems SPN-C52He) emulsified with 250 μl Freund’s complete adjuvant (Thermo Fisher, 77140) for primary immunizations. Two weeks later, animals began a biweekly i.m. boosting schedule of 100 μg protein with 250 μl Freund’s incomplete adjuvant (Thermo Fisher, 77145). Ag-specific titer was assessed by ELISA on the off week (plates directly coated with above Ags or with streptavidin followed by biotinylated Delta trimer, Acro Biosystems SPN-C82Ec, or biotinylated RBD, Acro Biosystems SPD-C82E4). Four days before euthanization, animals received a final boost of 100 μg protein i.v. with no adjuvant.
Next-generation sequencing
Splenocytes from human PGRN-immunized birds (two WT and three tLC) were prepared by Histopaque-1077 polysucrose gradient, RNA was extracted, and chicken VH was amplified. Seminested primers were used in the 5′ UTR (chVH-F9: 5′-CACCAGTCGGCTCCGCAACCATG-3′) and IgY C region (cIgY-CH2-P265: 5′-GTACGGGATCTACCCGGCAGA-3′ followed by another round of PCR with the same forward primer and cIgY-CH1-R26: 5′-GTCGGAACAACAGGCGGATA-3′). Amplicons were sequenced on a MiSeq at 2 × 300 (Illumina). Sequencing was performed by Abterra, Inc. (San Diego, CA).
Preprocessing of data was performed using the Immcantation (22, 23) suite of tools to assemble paired reads, filter good-quality sequences, mask the primers on 5′ and 3′ ends, collapse sequences to identify unique DNA sequences, and identify reliable sequences with two or more copies. Reliable sequences were then annotated using IgBlast and were clustered into lineages defined as 2-aa differences or less in the CDR3 region (24).
Ab candidates and characterization
mAb recovery
A single-cell screening method, the gel-encapsulated microenvironment (GEM) assay (U.S. patents 8030095 and 84151738), was employed to identify Ag-specific B cells (25). A quantity of 5 μm of aldehyde latex beads (Thermo Fisher, A37306) was coated with streptavidin followed by biotinylated Ag, overnight, blocked with 3% milk-PBS, and tested by labeling with plasma from immunized animals. GEMs were prepared (agarose droplets containing a single secreting B cell and Ag-coated beads) and incubated for 3 h at 37°C in RPMI/10% FCS containing 2 μg/ml Alexa Fluor 594 anti-chIgY (Thermo Fisher, A11042) as previously described (25) or with anti-IgM-FITC (Bethyl Laboratories, A30-102F). For the SARS-CoV-2 Ab discovery, cohort 1 birds (immunized with Wuhan D614G S1) were screened using Wuhan RBD; cohort 2 birds (immunized with Alpha, Beta, and Delta trimers) were screened using Alpha trimer; and cohort 3 birds (immunized with Delta trimer) were screened using B.1.1.529/Omicron trimer (Acro Biosystems SPN-C52Hz).
sdAb-hFc cloning
Single Ag-specific B cells were lysed, and mRNA was isolated. Chicken VH regions were amplified in a two-step seminested strategy previously described (25). In step 1, the primers used were in the 5′ UTR (chVH-F9; 5′-CACCAGTCGGCTCCGCAACCATG-3′) and in the IgM CH2 exon (cIgM-CH2-R; 5′-GGGGTGCATGGTGACGAAAAG-3′). In step 2, primers contained overhangs for cloning in addition to the VH-specific binding sites: forward in the VH leader (5′-TCGAACCCTTGCTAGCCGCCATGAGCCCACTCGTCTCCTCC-3′) and reverse in the JH region (5′-AGAAGATTTGGGATCCAGGCTGGAGGAGACGATGACTTCGGTCC-3′). The VH regions were inserted into a mammalian expression vector containing human Fc (IgG1 CH2-CH3) using InFusion (Takeda Bio, 638911).
Expression of chicken sdAb-hFc
Recombinant Abs were expressed in the Expi293 expression system (Thermo Fisher, A14525) as previously described (25).
Size exclusion chromatography
The rhPRGN (R&D Systems) and SARS-CoV-2 S1 (Acro Biosystems, S1N-C52H3) proteins used for kinetics and binning were buffer exchanged separately in size exclusion chromatography (SEC) buffer (10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.05% Tween 20) and further purified by Superdex 200 Increase 10/300 GL at 4°C. The peak elution fractions were collected and analyzed by SDS-PAGE.
Differential scanning fluorimetry and dynamic light scattering analysis
Samples were analyzed on an Unchained Labs Uncle instrument by monitoring full-spectrum fluorescence, static light scattering, and dynamic light scattering to determine Tm, Tagg, and monodispersity, respectively. Samples were heated from 25°C to 95°C with a ramping rate of 1°C/min, incubation time of 180 s, and plate hold of 60 s and analyzed by monitoring change in the 350/330-nm absorbance ratio. Dynamic Light Scattering data were collected after an incubation time of 180 s with a total of 10 acquisitions for 10 s per sample.
Surface plasmon resonance (SPR) epitope binning and kinetics assays were performed on the Carterra LSA platform. A “premixed” epitope binning assay was used to determine epitope diversity of the sdAbs as well as to compare with previously characterized anti-PGRN OmniChicken mAbs (26, 27). Briefly, an HC-200M sensor chip (Carterra) surface was activated for 5 min with EDC/sulfo-NHS at pH 5.5. The sdAb-hFc used for the amine-coupling reactions were affinity-purified from crude Expi293F supernatant, desalted, and diluted into 10 mM sodium acetate, pH 4.5, + 0.01% Tween 20 (Carterra) alongside the OmniChicken mAbs. The Ab ligand array was then printed on the activated surface by the 96-printhead, allowing 7-min contact time, followed by 5 min of quenching with 1 M ethanolamine, pH 8.5. The coupled HC-200M chip was then primed with a running buffer of 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, and 0.05% Tween 20 (HBSTE) supplemented with 0.5 mg/ml BSA. Binning was performed by iteratively injecting 100 nM rhPGRN (R&D Systems), which was preincubated for 1 h in the presence of >200 nM individual analyte Abs to deplete all Apo-PGRN molecules in solution. After every binning cycle, the arrayed Ab surface was regenerated with 10 mM glycine, pH 2.0, supplemented with 0.05% Tween 20 (2 × 30-s pulses). Binning and mapping data were analyzed in the Carterra Epitope software to generate heat maps and network/community plots as in Reference 28.
Binding affinities were determined in a “capture kinetic” assay format using an HC30M chip (Carterra) surface amine-crosslinked with a rabbit anti-human IgG Fc-specific polyclonal (Rockland) as the capture reagent. Using the 96-printhead, chicken sdAb-hFc (diluted up to 100-fold in HBSTE) in arrays of 96 along with the purified OmniChicken mAbs were captured onto the anti-human IgG Fc chip surface to enable a direct comparison. The Ab array surface was then primed with a running buffer of HBSTE + 0.5 mg/ml BSA before the kinetics assay. Recombinant Ags (PGRN or SARS-CoV-2 S1 monomer proteins) were prepared as a 3-fold dilution series spanning 76.2 pM–500 nM, and samples were injected for 5 min each in ascending concentration, allowing a 10-min dissociation phase following each associate phase, in a nonregenerative manner, as described (29). The Carterra kinetic software was used to process and analyze the data of each Ab globally by fitting the sensorgrams with a Langmuir 1:1 binding model. The affinity, or equilibrium KD, for Ag interacting with each captured mAb was determined by the ratio of the kinetic rate constants, KD = kd/ka.
Abs from cohort 1 (from immunization with Wuhan D614G S1 monomer) were tested in a pseudovirus neutralization assay against SARS-CoV-2 spike variants B.1.1.7 (Alpha), B.1.351 (Beta), and B.1.617.2 (Delta) at GenScript ProBio. Human ACE2-overexpressing cells were infected with a pseudotyped lentivirus carrying SARS-CoV-2 spike proteins. To measure infection levels, luciferase activity delivered by the lentivirus was detected. An ACE2-Fc protein was used as a positive control to bind virus and block infection. An 8-point dose response dilution series (range, 0.003–50 μg/ml) of the Abs was added to the cultures. The EC50 was calculated on the basis of mean values from triplicate assays for each dilution and converted to nanomolar values.
Results
Construct design: tLC
When we analyzed chickens with a homozygous deletion of the L chain locus, we observed that a small amount of H chain was still found in the serum, and a small population of B cells was found in the periphery (∼30-fold lower than normal) (14). This Ig was mainly of the IgM isotype, and very little IgY was produced. Because H chain that is not paired with L chain is bound to the ER protein BiP and thus sequestered inside the cell, it was perhaps unsurprising to find that the IgM found in serum contained a deletion of the CH1 domain, which occurred spontaneously by an unknown mechanism and allowed secretion of the H chain in the absence of L chain. (Chickens lack a surrogate L chain that could have enabled secretion in theory.) The CH1 domain of IgM is where BiP binds and blocks secretion. Ag-specific titer could be obtained upon immunization, although no mAbs were pursued.
To improve the efficiency of H chain–only expression in the birds, we decided to provide the H chain with a chaperone that would obviate the need for the spontaneous CH1 deletion and would allow H chain to be secreted without a full L chain. The L chain C region (CL) normally pairs with the CH1 domain, displacing BiP and enabling transit through the secretory pathway. Thus, we reasoned that expression of the CL alone as a tLC could serve to pair with the CH1 domain and enable full-length H chain expression of all isotypes, with the single Ag-binding modality determined by the VH domain.
We inserted a construct into the endogenous chicken L chain locus for expression of tLC driven by the L chain transcriptional regulatory elements so that it would be expressed in a tissue-specific manner at the appropriate level. We used the L chain KO allele that we previously made (14) and inserted two alternative tLC constructs: one without the leader intron (tLC) (Fig. 1) and one containing the leader intron (tLCi). The other intervening sequences (V to J and J to C) are absent from both constructs. Confirmation of the tLC(i) insertion into the L chain locus was performed by PCR on genomic DNA from the transfected primordial germ cells and hatched chicks and by reverse transcription PCR at the transcript level from PBMCs in the birds once they were hatched (Fig. 2).
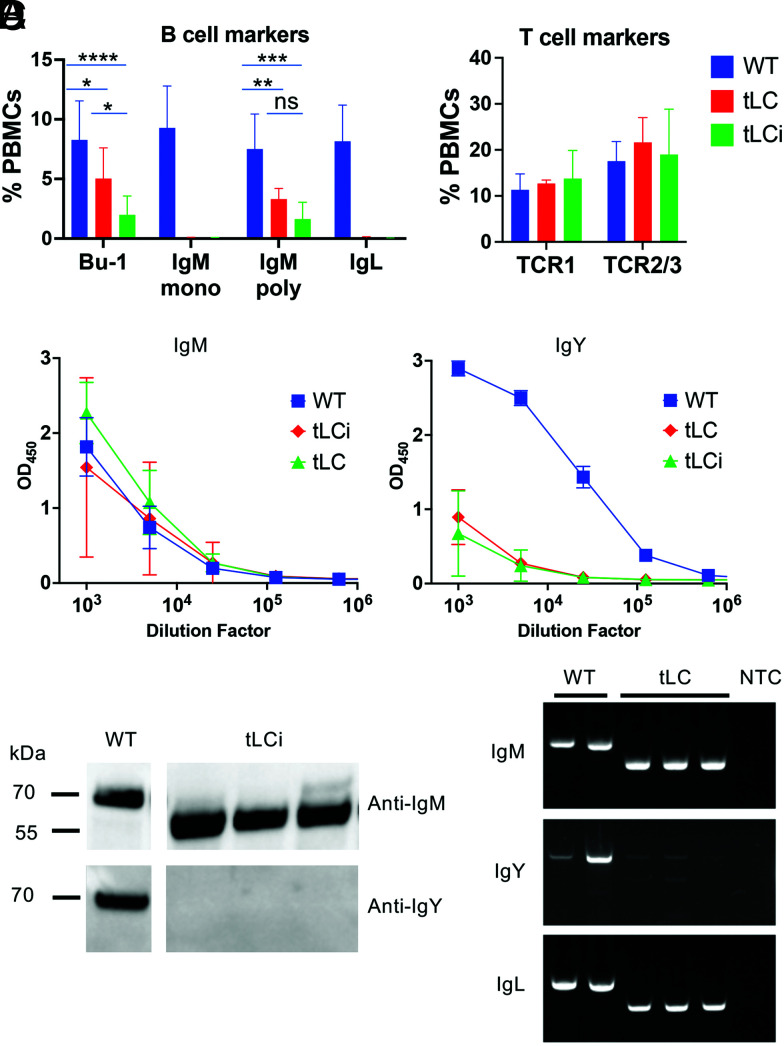
FIGURE 2.
B cell development and Ig expression in tLC(i) transgenic chickens. WT, tLC/IgL KO (tLC), and tLCi/IgL KO (tLCi) birds with WT H chains were analyzed. (A) PBMCs were analyzed by flow cytometry using B cell and T cell markers. mAbs specific to the Bu-1 protein (a chicken B cell marker), the CH1 domain of IgM, and a polyclonal serum to the IgM chain were used, as well as a mAb to the L chain. Data from 11 WT, 6 tLC, and 8 tLCi birds at 11 wk of age are shown. (B) Serum IgM and IgY levels were evaluated by ELISA. IgM levels appeared normal in tLC(i) birds, whereas IgY levels were ∼100-fold lower than normal. Data from four WT, three tLC, and eight tLCi birds at 11 wk of age are shown. (C) Western blot of serum Ig shows reduced size of IgM H chain. Data from three tLC birds at 11 wk of age are shown. (D) Reverse transcription PCR. RNA was obtained from PBMCs of three tLC birds at 16 wk of age, reverse transcribed, and amplified with primers specific for IgM, IgY, and IgL. The forward primer is in the 5′ UTR in each case, and, for the H chains, the reverse primers were in the CH2 domains. Products were cloned and sequenced to confirm that the IgM transcript consists of the VH region directly spliced inframe to the CH2 exon. A small amount of full-length IgY transcript is observed. *p = 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. NTC, no template control.
B cell development in tLC(i) birds
Heterozygous birds were produced with the genotype tLC(i)/IgL KO, with WT H chain. Birds carrying the tLC(i) construct produced a much more robust B cell population than the full IgL KO previously reported (14), with the proportion of Bu-1–positive (a B cell marker in chickens) cells ∼30–50% of normal (Fig. 2). Thus, the presence of the tLC transgene has an effect on B cell development, improving the B cell population compared with the full IgL KO. Staining with a polyclonal anti-IgM Ab confirmed that this population expressed surface IgM. However, staining with an anti-IgM mAb did not detect any cells in the tLC(i) birds, nor did staining with an anti-IgL mAb. This anti-IgM mAb is known to have its epitope in the CH1 domain of the IgM C region, which suggested that the CH1 domain might be missing from the surface IgM of the B cells. Further analysis by Western blotting and sequencing of RT-PCR products was consistent with CH1 deletion. The size of the IgM H chain by Western blot analysis was ∼15 kDa smaller than the WT, which would be expected if CH1 were missing. Similarly, the size of the RT-PCR product using a reverse primer in CH2 was reduced by ∼300 bp, and sequencing of RT-PCR products showed an inframe splice of the chicken VH directly to the IgM CH2 domain. The amount of IgM found in serum of tLC(i) birds was equivalent to WT chickens, as measured by ELISA (Fig. 2). However, very little IgY H chain was detected in serum, and the IgY transcript was only weakly detected by RT-PCR of PBMCs. IgA was also absent (Fig. 2). The main serum Ig in the tLC(i) birds is thus IgM. T cell numbers were unchanged compared with WT (Fig. 2).
Immunization of tLC(i) birds with PGRN and SARS-CoV-2 spike
tLC birds were immunized with hPGRN, a multidomain protein that we have used previously to compare immune responses in different OmniChicken strains, and the spike protein from SARS-CoV-2 (either S1 or full trimer, original strain with D614G mutation or Alpha, Beta, or Delta variants). Robust titers were obtained and the response was Ag-specific because no serum titer to nonrelated proteins was detected (Fig. 3).
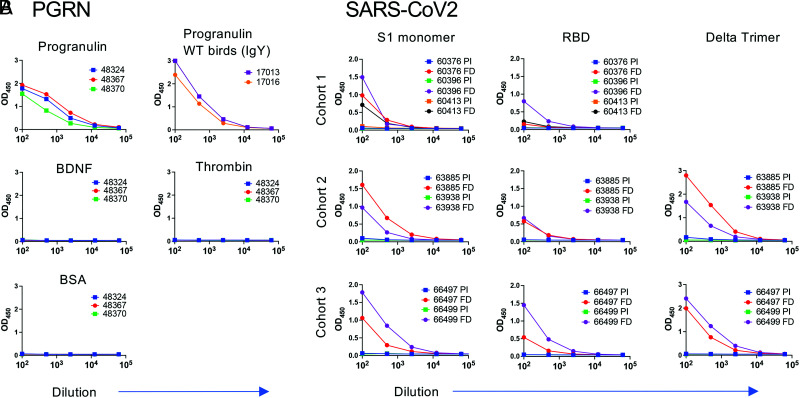
FIGURE 3.
tLC birds produce specific titer upon immunization. (A) First draw sera from three tLC/IgL KO birds immunized with hPGRN were titrated against PGRN, as well as human BDNF, thrombin, and BSA to test for cross-reactivity to unrelated proteins. Titers in the two WT birds immunized with PGRN are shown at the upper right, although they were measured with an anti-IgY secondary Ab. (B) Preimmune (PI) and final draw (FD) sera from three cohorts of SARS-CoV-2 spike immunized birds were titrated against S1 monomer (Wuhan D614G; left column), RBD WT strain (middle column), or Delta trimer (right column). Top row is cohort 1 (immunized with S1 Wuhan D614G), middle row is cohort 2 (immunized with alternating boosts of Alpha, Beta, and Delta trimers), bottom row is cohort 3 (immunized with Delta trimer). On the x-axis is serum dilution, and on the y-axis is OD450. All birds were tLC/IgL KO.
We performed next-generation sequencing of sdAb VH regions and WT VH regions from splenocytes of immunized birds to analyze the sdAb repertoire in the tLC birds (Fig. 4 and Table I). The single germline V gene in chickens simplifies the comparison of birds because the germline V sequence is the same in every case, obviating the possibility of skewed V gene usage. In the framework regions, the frequency of mutation at each position was essentially the same in VH from tLC and WT samples (Fig. 4A). The one exception was a position in framework 2, V42 (IMGT numbering), that was sometimes mutated to Met (∼30% of the sdAb sequences). Interestingly, W52, which in camelid VHH is changed to a G residue to increase solubility (5), is often mutated to Y or F in chickens, but both tLC and WT birds showed this change. The most notable mutation occurred in the canonical W118 (IMGT numbering) at the beginning of FR4. About 20% of the sdAb VH sequences contained Arg in this position, and a small proportion had other amino acids (Fig. 4B). Accordingly, the average hydrophobicity was similar in WT (−24.4 ± 11.2) and sdAb repertoires (slightly more hydrophilic at −26.3 ± 14.3; Kyte-Doolittle scale). Mean CDR-H3 lengths were slightly longer for sdAb repertoires (mean 19.2 ± 4.7 for sdAb VH; 18.3 ± 3.4 for WT VH), although frequencies of noncanonical cysteine residues in CDR-H3 were similar (68% of the sequences from sdAb had one or more Cys in CDR3 compared with 65% for WT). To assess levels of diversity in each bird, the percentages of sequences in each of the top 50 clonotype lineages were plotted (Fig. 4C). Two of the birds (one WT and one sdAb) showed a skewed pattern where a few lineages contained a significant proportion of the sequences, whereas the other three birds showed a more even distribution of similar-sized lineages across the repertoire.
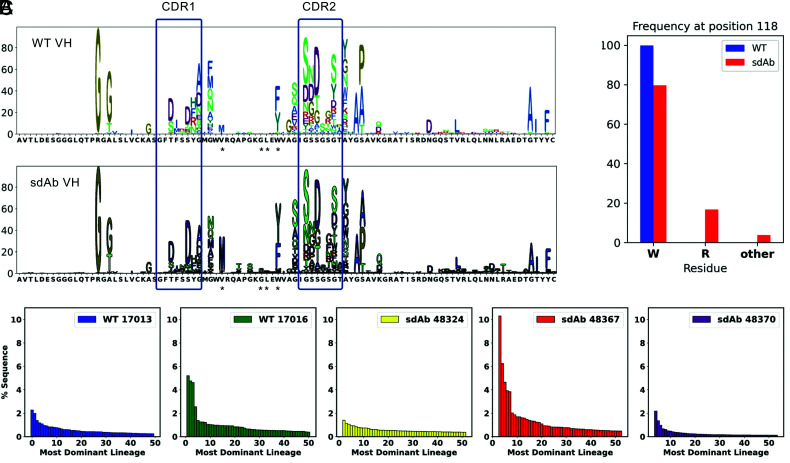
FIGURE 4.
tLC birds express sdAb VH repertoires similar to normal chicken VH repertoires. (A) H chain V regions were amplified from three tLC/IgL KO chickens and two WT chickens and sequenced using Illumina MiSeq 2 × 300 paired end reads. All amplicons were derived from splenocytes of PGRN-immunized birds. The frequency of mutation relative to the germline VH gene at each position, up to CDR3, was plotted for data pooled from all birds of each genotype. The sequence of the germline VH gene is shown on the x-axis. CDRs 1 and 2 (IMGT definitions) are indicated by boxes. The positions of V42 and W52 as well as the other two hallmark residues mutated in camelid VHH are indicated with asterisks. (B) The frequency of mutation at W118 in framework 4. WT chickens had 99.5% of the sequences with W at that position, but sdAb (tLC) birds showed ∼20% R and ∼5% other residues. (C) The percentages of the sequences present in each of the 50 most dominant lineages for each bird are shown. Birds 17016 and 48367 show marked skewing, with a few lineages containing a relatively large number of sequences. Bird 48367 had fewer total clonotypes than the other birds (approximately one-fourth the number), so the skewing is likely reflecting that fact.
Table I. Summary of next-generation sequencing preprocessing steps.
| Sample id | No. Raw Reads | AssemblePair | FilterSeq | MaskPrimer | Collapse | No. Reliable Seq | No. Filtered Seq |
|---|---|---|---|---|---|---|---|
| WT 17013 | 1,717,999 | 1,466,190 | 1,466,129 | 1,461,680 | 946,832 | 45,271 | 41,592 |
| WT 17016 | 1,704,597 | 1,493,714 | 1,493,652 | 1,486,506 | 976,539 | 44,035 | 40,537 |
| sdAb 48324 | 1,908,125 | 1,615,679 | 1,615,596 | 1,597,580 | 981,121 | 43,289 | 39,743 |
| sdAb 48367 | 1,673,983 | 1,412,722 | 1,412,648 | 1,392,903 | 817,010 | 44,831 | 41,984 |
| sdAb 48370 | 2,180,883 | 1,798,697 | 1,490,872 | 1,464,298 | 761,395 | 47,077 | 42,758 |
This table presents a summary of data at each stage of the next-generation sequencing preprocessing workflow for each sample. It begins with the total number of raw reads, followed by the count of sequences successfully assembled using the ImmunoSeq (Immcantation) pipeline in the “AssemblePair” column. The “FilterSeq” column indicates the number of high-quality sequences that passed the quality threshold (q > 20). The “MaskPrimer” column reports the count of clones for which the correct forward and reverse primers could be identified. “Collapse” details the number of sequences remaining after identical sequences have been merged. “Reliable Seq” lists the count of sequences with a duplicate count greater than 2, highlighting their reliability. Finally, “Filtered Seq” shows the sequences that underwent internal post-processing, ensuring the data’s quality and relevance. The internal filtering includes the following criteria: they must exhibit a productive, inframe VDJ; display a complete VDJ as determined by IgBlast; not contain any stop codons or ambiguous sites; they are required to be VH sequences (from the IGH locus); and all domains FR1, FR2, FR3, FR4, and CDR1, CDR2, and CDR3 must be present.
Ab discovery
Splenocytes from the three birds immunized with PGRN and the seven birds immunized with various forms of SARS-CoV-2 spike protein were screened for Ag-specific monoclonal chicken sdAbs. Screening was performed using GEM technology (25), and one bird was screened on our xPloration (30) platform. The chicken VH regions were amplified from single B cells identified in these platforms, cloned inframe with human Fc (IgG1), and expressed in 293 cells. Primers to amplify VH spliced to IgM constant regions were successful, whereas primers for IgY only rarely produced product (and none were cloned), consistent with the IgM expression observed in serum. Supernatant containing secreted sdAb-hFc was analyzed by ELISA for binding to Ag, and DNA sequences of the clones were determined. A total of 142 unique Abs binding to PGRN and 251 unique Abs binding to SARS-CoV-2 spike were obtained.
Sequence analysis of the sdAb regions revealed that few classical stabilizing mutations were observed (Supplemental Fig. 1), with one notable exception: The canonical W118 residue in FR4 was mutated to Arg in ∼20% of the Abs as seen in the bulk next-generation sequencing analysis. The levels of sequence diversity in the frameworks and CDRs appeared similar in the anti-PGRN sequences from the tLC(i) birds as compared with WT VH from WT birds previously studied (31). In camelid VHH, FR2 has less hydrophobic amino acid content than conventional VH (4) as a result of changes to four hallmark residues. The frequency of hydrophobic amino acids in FR2 was very similar in the chicken sdAbs compared with that of VH from WT birds (anti-PGRN Abs; 70% hydrophobic amino acids in sdAbs versus 71.7% in WT). The frequency of clonotypes (defined as 2-aa differences or less in CDR3) in the anti-PGRN sdAbs (29 clonotypes in 111 unique sequences) was lower than for WT clones (15 clonotypes in 31 sequences), whereas the frequency of singlets (clonotypes only represented by one clone) was about the same (20%) (Supplemental Fig. 1). The dominant clonotype in each case contained six sequences.
Biophysical characteristics
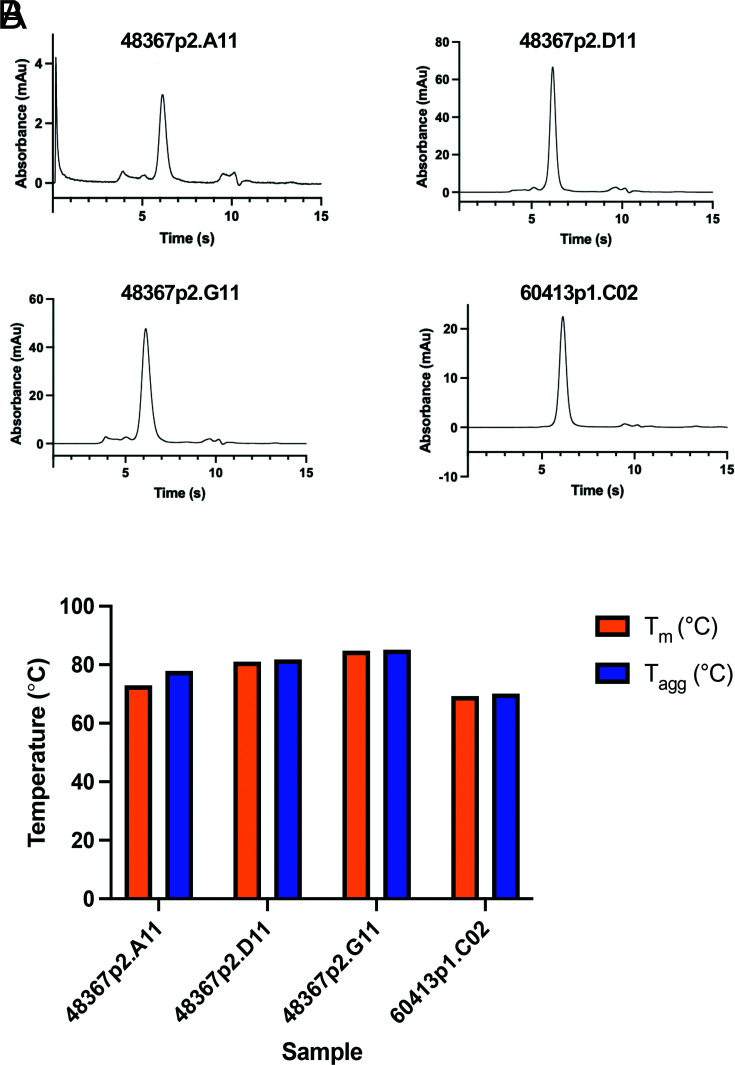
Chicken sdAb-hFc were initially characterized for biophysical properties such as monomericity and thermal stability (Fig. 5). The Abs were run on an analytical SEC, showing a pure Ab at the predicted molecular mass range of ∼85 kDa. Differential scanning fluorimetry was performed using Unchained Uncle to obtain the Tm and Tagg of the chicken sdAb-hFc, and the results showed that the Abs were very stable and required high temperatures to melt and aggregate.
FIGURE 5.
Analytical SEC and thermal stability of sdAbs. (A) Analytical SEC of four sdAb-hFc clones is shown, confirming mainly monomeric peak. Low amounts of protein were injected, so the minor peaks are more likely noise/impurities, not aggregates or degradation products. (B) Differential scanning fluorimetry analysis was run on the four sdAb-hFc that showed high Tm and Tagg.
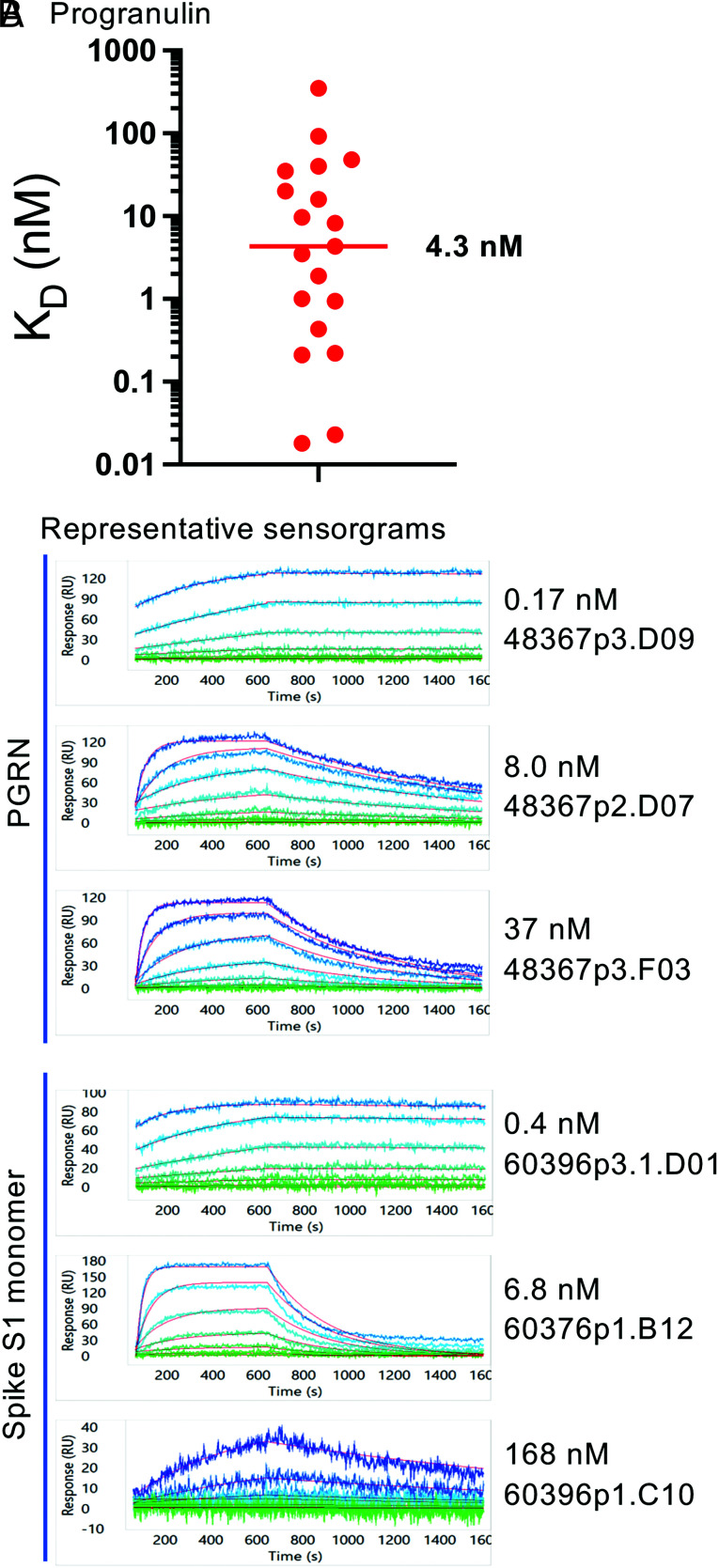
The kinetics of Ag binding were determined by SPR on the Carterra LSA instrument. Ab was captured on the chip, followed by binding of Ag in solution, either hPGRN or SARS-CoV-2 spike S1 monomer. The PGRN and spike S1 proteins were first confirmed to be monomeric for the purposes of monovalent Ag–Ab interactions by SEC (Supplemental Fig. 2). A range of affinities to PGRN were observed in the sdAb-hFc, including several subnanomolar clones (Fig. 6 and Supplemental Table I). For the anti–SARS-CoV-2 spike Abs, cohort 1 was immunized with the S1 monomer from the original Wuhan D614G strain and screened for mAb binders on the original receptor binding domain. Affinities to the Wuhan D614G S1 monomer were measured by SPR. Although all 18 of the clones in the cohort bound to S1 by ELISA, only 4 showed clear binding by SPR (Fig. 6B). Representative sensorgrams for a range of binding kinetics to PGRN or S1 are shown in Fig. 6B, illustrating accurate curve fits to the model, and kinetics statistics are listed in Supplemental Table I with a comparison with two benchmark IgG clones that showed effective viral neutralization early in the pandemic (32). To represent more faithfully how the sdAb-hFc would bind the native assembled spike trimer, we assessed “avidity-boosted binding” to spike trimers as analytes for cohorts 1, 2, and 3 Abs. Cohorts 2 and 3 birds were immunized with spike trimers and screened on Alpha and Omicron spike trimers, respectively. Highly avid binding to the Wuhan D614G and Delta strains was obtained, whereas binding to the Omicron variant was generally lower (Supplemental Fig. 3). Thus, despite weak or no binding to S1 monomer by SPR, the chicken sdAbs identified to the spike showed binding to the full trimer.
FIGURE 6.
Kinetics of Ag binding by chicken sdAbs. (A) Affinities (KD, in nM) to PGRN of a cohort of 19 unique sdAb-hFc clones are graphed. The line indicates the median KD (4.3 nM). (B) Representative sensorgrams for sdAb-hFc clones to PGRN (top three) and spike S1 monomer (lower three) for Abs with a range of affinities are shown.
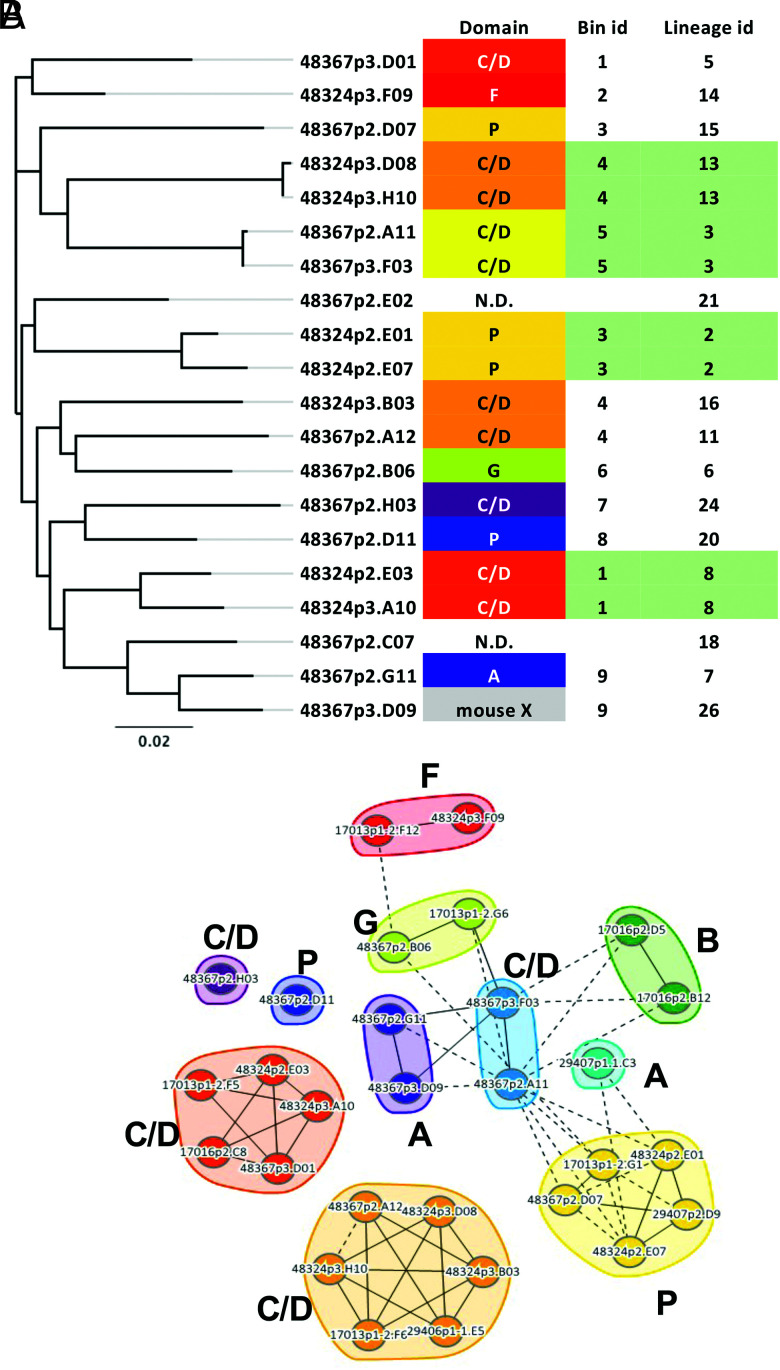
For the cohort of PGRN clones, the domain binding profiles were determined using a set of mouse-human chimeras in which specific domains of PGRN were swapped for the domain from the mouse PGRN (31). A given clone will bind to a chimera if its epitope is in a domain that is human, whereas if the domain containing its epitope is mouse, the clone will not bind. (This method only works for human-specific Abs.) Using this method, we found sdAb binders to domains P, G, F, A, and C/D among a small cohort of 20 Abs (Fig. 7). In addition, cross-blocking activity was measured on the Carterra instrument to place the Abs into epitope bins. A total of nine epitope bins across the five domains were targeted, indicating that some domains were bound at more than one site. For example, domain P was bound by Abs in bins 3 and 8 (Fig. 7). Clones in the same sequence lineage (as defined by two changes or fewer in CDR3) always fell into the same bin.
FIGURE 7.
Epitope binning. (A) The sequence dendrogram for a set of anti-PGRN Abs is aligned with the granulin domain and epitope bin for each clone. One Ab showed cross-reactivity to mouse (mouse X, and two Abs had weak binding that did not allow domain assignment. Bins were assigned identification numbers, and the bin each clone falls into is indicated. Lineage identification numbers for each clone (as determined by two changes or less in CDR-H3) are shown at right. Lineages populated by more than one clone are colored green, illustrating that both clones in each of those lineages are in the same epitope bin. (B) Nodal plot showing binning relationships, including control Abs from previous studies (31). Clones that block each other’s binding to the Ag are connected by lines; solid lines represent blocking relationships confirmed in both orders of addition, whereas dotted lines represent unidirectional blocking. Clones in the same bin are shown in a colored envelope.
Neutralization assay
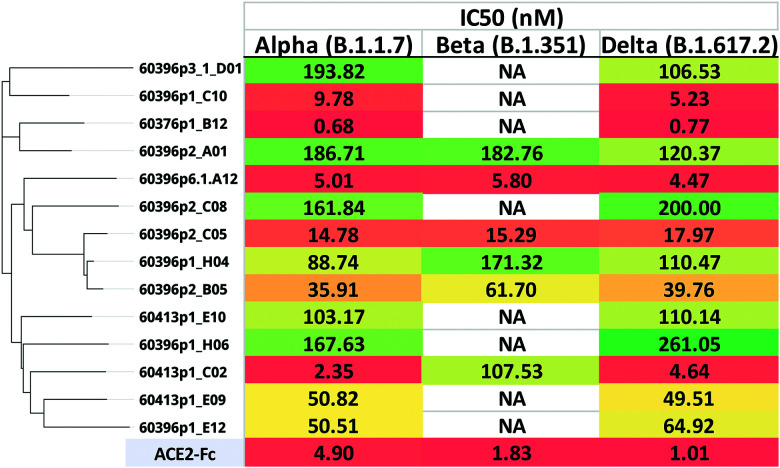
A subset of the anti–SARS-CoV-2 Abs (cohort 1) with diverse sequences (each representing a unique lineage) and known to bind to the receptor binding domain of S1 by ELISA was selected for testing in pseudovirus neutralization assays (GenScript ProBio). All of the selected Abs exhibited pseudoviral neutralization activity to varying degrees against the three subtypes of SARS-CoV-2 tested: Alpha, Beta, and Delta (Fig. 8). Consistent with other studies (33), the Beta variant with the E484K mutation allowed that variant to escape neutralization from some of the Abs. Three of the chicken sdAbs showed neutralization similar to or more potent than the ACE2-Fc control (Fig. 8).
FIGURE 8.
SARS-CoV-2 pseudovirus neutralization. sdAb-hFcs from cohort 1 immunized with the S1 monomer of the original SARS-CoV-2 strain (Wuhan D614G) were tested against Alpha, Beta, and Delta pseudoviruses in a neutralization assay. Each clone represented a unique sequence lineage. The IC50 values (nanomolar concentration) are shown, in order of the sequence dendrogram shown at left (NA, no activity). The ACE2-Fc positive control is also shown.
Discussion
Since their discovery in 1993 (34), camelid sdAbs (VHH Abs) have been a subject of interest for the development of therapeutics for their potential to bind antigenic epitopes that are inaccessible to normal Abs containing both L and H chains (35, 36). As of 2023, two VHH therapeutic Abs and one VHH-based chimeric Ag receptor T therapeutic have been clinically approved for use in patients (37).
Although WT chickens do not normally express H chain–only Abs, they can be easily induced to do so when a tLC transgene is introduced into the birds, leading to normal serum levels of IgM. Contrary to our prediction, the tLC does not support the full-length H chain by binding to the CH1 domain and chaperoning it through the secretory pathway; rather, it somehow enables the deletion of the CH1 domain, leading to expression of CH1-deleted IgM. Because the full L chain KO results in much lower levels of such CH1-deleted IgM (14), we reason that the tLC transgene and/or protein somehow promotes B cell development and CH1 deletion, although the mechanism remains unclear. It is unknown whether production of the tLC protein by the transgene is required, because we have not directly detected the tLC protein in birds, even though there is a clear effect on B cell development compared with a full KO. One possibility is that the tLC protein binds weakly to the CH1 domain of the full-length H chain, enabling enough surface IgM expression for the developing B cell to pass a developmental checkpoint, followed by CH1 deletion and increased levels of expression, independent of the tLC. Alternatively, there may be no tLC binding to the H chain at all, in which case the tLC expression could be providing an independent survival signal to the developing B cell, allowing time for the CH1 deletion to occur, leading to surface IgM expression. In the case of homozygous IgL KO, no such signal would be present, potentially the reason for much reduced B cell development.
The mechanism for CH1 deletion is unknown at this point, but in theory it could be either a genomic deletion of the CH1 coding sequence or a mutation that causes an alternative splice from the VH region directly to CH2. A genomic deletion that also disrupted the Cµ switch region could further explain the lack of class switching. The chicken C region locus is highly repetitive even in the areas not considered switch regions (Genome Reference Consortium GRCg6a) (38), so a DNA deletion between these repeat regions that removes CH1 seems plausible, especially given the fact that the switch region directly upstream of CH1 is normally the locus of a deletion breakpoint during class switching. The enzyme AID is required for gene conversion, somatic hypermutation, and class switching (39), so it is intriguing to speculate that it might be involved in CH1 deletion in the tLC birds. The repetitive and low-complexity nature of the sequences surrounding the CH1 exon poses a challenge to PCR approaches aimed at determining whether a genomic deletion is occurring in the B cells, and it thus remains unknown.
L chain KO mice were also found to produce low levels of H chain–only Abs also from spontaneous CH1 deletion, leading to the secretion of WT mouse VH as sdAbs (40, 41). In contrast to the chicken, the isotypes expressed in mouse were IgG and IgA, with no IgM detected. The sequences of the mouse sdAbs did not exhibit the stabilizing framework changes that would increase solubility, although no mAbs were produced that would have provided the materials for testing this idea through biophysical characterization. Transgenic mouse and rat strains have been developed as an alternative to camelid immunization for the discovery of therapeutic sdAb candidates. All of these engineered rodents (the UniRat [42], Harbor Biomed [43], and Crescendo mice [44]) contain fully human V region genes in the germline, and in the case of the UniRat, the JH gene contains the camelid mutation W118R (IMGT numbering) in FR4 (42). These transgenic rodents express human sequence V regions without stabilizing mutations, similar to the chicken sdAb in this report. Even though no somatically derived stabilizing mutations were found, the sdAbs derived from these rodents appear not to show signs of aggregation, at least in the limited studies shown. It remains to be seen how well these sdAbs perform in the clinic.
Consistent with the transgenic rodent-expressed human sdAbs, the chicken VH sdAbs presented in this study exhibit favorable biophysical characteristics despite the fact that they do not contain the hallmark solubilizing mutations found in camelid VHH sdAbs. The chicken sdAbs show high Tm/Tagg, good solubility, and no signs of aggregation, which may be surprising, considering that when humanizing a camelid VHH, it is not normally possible to fully humanize the FR2 mutations without compromising biophysical properties (45). Binding kinetics to PGRN of conventional Abs identified in our other transgenic chicken platforms were in a range similar to those observed in the present study: The median KD of PGRN clones from the common L chain bird OmniClic was ∼10 nM (46), and in OmniChickens, a median KD of ∼2.4 or 3.5 nM was observed (for κ- and λ-L chain birds, respectively) (26). The median KD of the chicken sdAbs to PGRN, 4.3 nM, is also in this range, indicating that the single-domain format can provide similar binding affinities despite having only one V region and three CDRs.
We have no structural information yet on the chicken sdAbs to investigate whether the paratopes formed by the chicken H chain–only Abs are adopting characteristics of camelid VHH Abs. Camelid VHH paratopes often take on a convex, protruding conformation compared with the more planar paratopes formed by the six CDRs of conventional VL+VH Abs (35). Moreover, camelid VHH Ag-binding contacts can include portions of the framework, such as FR2, FR3, or the N-terminus (36, 47, 48). On the basis of the similar frequency of mutations in the frameworks of chicken sdAb VH compared with normal chicken VH, there is no indication of increased frequency of mutations outside of the CDRs, which might reflect Ag binding in the frameworks for chicken sdAbs. With the possible exception of V42 in FR2, which had an increase in mutation rate in chicken sdAbs, the other framework residues that can contact Ag in camelid VHH Abs are not mutating in chicken sdAb VH sequences any more than in normal chicken VH. More generally, structural data on chicken Abs are extremely limited (49). However, we speculate that in the case of chicken sdAbs, the majority of Ag contacts are mediated by the CDRs, because only the CDRs are exhibiting a diversity of sequences. If true, this feature could simplify the humanization of any chicken sdAb because the sequence space involved in Ag contact, and thus necessary for grafting on a human framework, would be limited to the CDRs.
Camelid VHH often contains noncanonical disulfide bridges, usually between CDR1 and CDR3, which serve to stabilize the structure of the single domain (4, 50). Conventional chicken VH also often contains noncanonical cysteines, but more commonly they are found in intra-CDR3 disulfide bonds (13), which are thought to stabilize the longer CDR3 loops found in chickens. The chicken sdAb VH sequences found in the present study also contain these disulfide bridges, which raises the possibility that the chicken VH structure naturally lends itself to the single-domain format.
The chicken host expressing sdAbs retained the ability to recognize diverse epitopes and, in a small number of unique clones, was able to target many distinct sites with high affinity on our model Ag PGRN as well as produce anti–SARS-CoV-2 neutralizing Abs. The chicken sdAb platform retains the benefits of chicken host immunization and provides a route to unique sdAbs.
Supplementary Material
Footnotes
This work was supported by Ligand Pharmaceuticals and OmniAb.
The online version of this article contains supplemental material.
- BRL
- Buffalo rat liver
- FR2
- framework region 2
- GEM
- gel-encapsulated microenvironment
- KO
- knockout
- PGRN
- progranulin
- sdAb
- single-domain Ab
- SEC
- size exclusion chromatography
- SPR
- surface plasmon resonance
- tLC
- truncated L chain
- UTR
- untranslated region
- WT
- wild type
Disclosures
P.A.L., K.R., C.N.V., B.Z., Y.Z., G.S.R., D.B.S., D.P., E.C., S.I., M.-C.v.d.L., and W.H. are current employees of OmniAb. The other authors have no financial conflicts of interest.
References
- 1. Arbabi-Ghahroudi, M. 2017. Camelid single-domain antibodies: historical perspective and future outlook. Front. Immunol. 8: 1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flajnik, M. F., Deschacht N., Muyldermans S.. 2011. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 9: e1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nguyen, V. K., Muyldermans S., Hamers R.. 1998. The specific variable domain of camel heavy-chain antibodies is encoded in the germline. J. Mol. Biol. 275: 413–418. [DOI] [PubMed] [Google Scholar]
- 4. Vu, K. B., Ghahroudi M. A., Wyns L., Muyldermans S.. 1997. Comparison of llama VH sequences from conventional and heavy chain antibodies. Mol. Immunol. 34: 1121–1131. [DOI] [PubMed] [Google Scholar]
- 5. Riechmann, L., Muyldermans S.. 1999. Single domain antibodies: comparison of camel VH and camelised human VH domains. J. Immunol. Methods 231: 25–38. [DOI] [PubMed] [Google Scholar]
- 6. Nguyen, V. K., Hamers R., Wyns L., Muyldermans S.. 1999. Loss of splice consensus signal is responsible for the removal of the entire CH1 domain of the functional camel IGG2A heavy-chain antibodies. Mol. Immunol. 36: 515–524. [DOI] [PubMed] [Google Scholar]
- 7. Genst, E. D., Saerens D., Muyldermans S., Conrath K.. 2006. Antibody repertoire development in camelids. Dev. Comp. Immunol. 30: 187–198. [DOI] [PubMed] [Google Scholar]
- 8. Achour, I., Cavelier P., Tichit M., Bouchier C., Lafaye P., Rougeon F.. 2008. Tetrameric and homodimeric camelid IgGs originate from the same IgH locus. J. Immunol. 181: 2001–2009. [DOI] [PubMed] [Google Scholar]
- 9. Reynaud, C. A., Dahan A., Anquez V., Weill J. C.. 1989. Somatic hyperconversion diversifies the single Vh gene of the chicken with a high incidence in the D region. Cell 59: 171–183. [DOI] [PubMed] [Google Scholar]
- 10. Reynaud, C., Anquez V., Dahan A., Weill J.. 1985. A single rearrangement event generates most of the chicken immunoglobulin light chain diversity. Cell 40: 283–291. [DOI] [PubMed] [Google Scholar]
- 11. Reynaud, C. A., Anquez V., Grimal H., Weill J. C.. 1987. A hyperconversion mechanism generates the chicken light chain preimmune repertoire. Cell 48: 379–388. [DOI] [PubMed] [Google Scholar]
- 12. Ratcliffe, M. J. H. 2006. Antibodies, immunoglobulin genes and the bursa of Fabricius in chicken B cell development. Dev. Comp. Immunol. 30: 101–118. [DOI] [PubMed] [Google Scholar]
- 13. Wu, L., Oficjalska K., Lambert M., Fennell B. J., Darmanin-Sheehan A., Shúilleabháin D. N., Autin B., Cummins E., Tchistiakova L., Bloom L., et al. 2012. Fundamental characteristics of the immunoglobulin VH repertoire of chickens in comparison with those of humans, mice, and camelids. J. Immunol. 188: 322–333. [DOI] [PubMed] [Google Scholar]
- 14. Schusser, B., Collarini E. J., Pedersen D., Yi H., Ching K., Izquierdo S., Thoma T., Lettmann S., Kaspers B., Etches R. J., et al. 2016. Expression of heavy chain-only antibodies can support B-cell development in light chain knockout chickens. Eur. J. Immunol. 46: 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elter, A., Bogen J. P., Hinz S. C., Fiebig D., Palacios A. M., Grzeschik J., Hock B., Kolmar H.. 2021. Humanization of chicken-derived scFv using yeast surface display and NGS data mining. Biotechnol. J. 16: e2000231. [DOI] [PubMed] [Google Scholar]
- 16. Gjetting, T., Gad M., Fröhlich C., Lindsted T., Melander M. C., Bhatia V. K., Grandal M. M., Dietrich N., Uhlenbrock F., Galler G. R., et al. 2019. Sym021, a promising anti-PD1 clinical candidate antibody derived from a new chicken antibody discovery platform. MAbs 11: 666–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Song, Y., Duraisamy S., Ali J., Kizhakkayil J., Jacob V. D., Mohammed M. A., Eltigani M. A., Amisetty S., Shukla M. K., Etches R. J.. and M.-C. v. d. Lavoir. 2014. Characteristics of long-term cultures of avian primordial germ cells and gonocytes. Biol. Reprod. 90: 15. [DOI] [PubMed] [Google Scholar]
- 18. Lavoir, M.-C. V. D., Collarini E. J., Leighton P. A., Fesler J., Lu D. R., Harriman W. D., Thiyagasundaram T. S., Etches R. J.. 2012. Interspecific germline transmission of cultured primordial germ cells. PLoS One 7: e35664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Collarini, E. J., Leighton P. A., Lavoir M.-C. V. D.. 2019. Production of transgenic chickens using cultured primordial germ cells and gonocytes. Methods Mol. Biol. 1874: 403–430. [DOI] [PubMed] [Google Scholar]
- 20. Collarini, E., Leighton P., Pedersen D., Harriman B., Jacob R., Mettler-Izquierdo S., Yi H., Lavoir M.-C. V. D., Etches R. J.. 2014. Inserting random and site-specific changes into the genome of chickens. Poult. Sci. 94: 799–803. [DOI] [PubMed] [Google Scholar]
- 21. Thyagarajan, B., Olivares E. C., Hollis R. P., Ginsburg D. S., Calos M. P.. 2001. Site-specific genomic integration in mammalian cells mediated by phage C31 integrase. Mol. Cell. Biol. 21: 3926–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta, N. T., Heiden J. A. V., Uduman M., Gadala-Maria D., Yaari G., Kleinstein S. H.. 2015. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31: 3356–3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vander Heiden, J. A., Yaari G., Uduman M., Stern J. N. H., O’Connor K. C., Hafler D. A., Vigneault F., Kleinstein S. H.. 2014. pRESTO: a toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics 30: 1930–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rognes, T., Scheffer L., Greiff V., Sandve G. K.. 2022. CompAIRR: ultra-fast comparison of adaptive immune receptor repertoires by exact and approximate sequence matching. Bioinformatics 38: 4230–4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Izquierdo, S. M., Varela S., Park M., Collarini E. J., Lu D., Pramanick S., Rucker J., Lopalco L., Etches R., Harriman W.. 2016. High-efficiency antibody discovery achieved with multiplexed microscopy. Microscopy (Oxf.) 65: 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ching, K. H., Berg K., Morales J., Pedersen D., Harriman W. D., Abdiche Y. N., Leighton P. A.. 2020. Expression of human lambda expands the repertoire of OmniChickens. PLoS One 15: e0228164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ching, K. H., Collarini E. J., Abdiche Y. N., Bedinger D., Pedersen D., Izquierdo S., Harriman R., Zhu L., Etches R. J., Lavoir M.-C. V. D., et al. 2018. Chickens with humanized immunoglobulin genes generate antibodies with high affinity and broad epitope coverage to conserved targets. MAbs 10: 71–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Abdiche, Y. N., Yeung A. Y., Ni I., Stone D., Miles A., Morishige W., Rossi A., Strop P.. 2017. Antibodies targeting closely adjacent or minimally overlapping epitopes can displace one another. PLoS One 12: e0169535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Karlsson, R., Katsamba P. S., Nordin H., Pol E., Myszka D. G.. 2006. Analyzing a kinetic titration series using affinity biosensors. Anal. Biochem. 349: 136–147. [DOI] [PubMed] [Google Scholar]
- 30. Chen, B., Lim S., Kannan A., Alford S. C., Sunden F., Herschlag D., Dimov I. K., Baer T. M., Cochran J. R.. 2016. High-throughput analysis and protein engineering using microcapillary arrays. Nat. Chem. Biol. 12: 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Abdiche, Y. N., Harriman R., Deng X., Yeung Y. A., Miles A., Morishige W., Boustany L., Zhu L., Izquierdo S. M., Harriman W.. 2016. Assessing kinetic and epitopic diversity across orthogonal monoclonal antibody generation platforms. MAbs 8: 264–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baum, A., Fulton B. O., Wloga E., Copin R., Pascal K. E., Russo V., Giordano S., Lanza K., Negron N., Ni M., et al. 2020. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369: 1014–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang, P., Nair M. S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P. D., et al. 2021. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 593: 130–135. [DOI] [PubMed] [Google Scholar]
- 34. Hamers-Casterman, C., Atarhouch T., Muyldermans S., Robinson G., Hamers C., Songa E. B., Bendahman N., Hamers R.. 1993. Naturally occurring antibodies devoid of light chains. Nature 363: 446–448. [DOI] [PubMed] [Google Scholar]
- 35. Genst, E. D., Silence K., Decanniere K., Conrath K., Loris R., Kinne J., Muyldermans S., Wyns L.. 2006. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc. Natl. Acad. Sci. U. S. A. 103: 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zavrtanik, U., Lukan J., Loris R., Lah J., Hadži S.. 2018. Structural basis of epitope recognition by heavy-chain camelid antibodies. J. Mol. Biol. 430: 4369–4386. [DOI] [PubMed] [Google Scholar]
- 37. Jin, B-K., Odongo S., Radwanska M., Magez S.. 2023. Nanobodies: a review of generation, diagnostics and therapeutics. Int. J. Mol. Sci. 24: 5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Warren, W. C., Hillier L. W., Tomlinson C., Minx P., Kremitzki M., Graves T., Markovic C., Bouk N., Pruitt K. D., Thibaud-Nissen F., et al. 2017. A new chicken genome assembly provides insight into avian genome structure. G3 (Bethesda) 7: 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Papavasiliou, F. N., Schatz D. G.. 2002. Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell 109 Suppl: S35–S44. [DOI] [PubMed] [Google Scholar]
- 40. Zou, X., Osborn M. J., Bolland D. J., Smith J. A., Corcos D., Hamon M., Oxley D., Hutchings A., Morgan G., Santos F., et al. 2007. Heavy chain-only antibodies are spontaneously produced in light chain-deficient mice. J. Exp. Med. 204: 3271–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Geraldes, P., Rebrovich M., Herrmann K., Wong J., Jäck H.-M., Wabl M., Cascalho M.. 2007. Ig heavy chain promotes mature B cell survival in the absence of light chain. J. Immunol. 179: 1659–1668. [DOI] [PubMed] [Google Scholar]
- 42. Clarke, S. C., Ma B., Trinklein N. D., Schellenberger U., Osborn M. J., Ouisse L.-H., Boudreau A., Davison L. M., Harris K. E., Ugamraj H. S., et al. 2018. Multispecific antibody development platform based on human heavy chain antibodies. Front. Immunol. 9: 3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Drabek, D., Janssens R., Boer E. D, Rademaker R., Kloess J., Skehel J., Grosveld F.. 2016. Expression cloning and production of human heavy-chain-only antibodies from murine transgenic plasma cells. Front. Immunol. 7: 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teng, Y., Young J. L., Edwards B., Hayes P., Thompson L., Johnston C., Edwards C., Sanders Y., Writer M., Pinto D., et al. 2020. Diverse human VH antibody fragments with bio-therapeutic properties from the Crescendo Mouse. N. Biotechnol. 55: 65–76. [DOI] [PubMed] [Google Scholar]
- 45. Vincke, C., Loris R., Saerens D., Martinez-Rodriguez S., Muyldermans S., Conrath K.. 2009. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J. Biol. Chem. 284: 3273–3284. [DOI] [PubMed] [Google Scholar]
- 46. Ching, K. H., Berg K., Reynolds K., Pedersen D., Macias A., Abdiche Y. N., Harriman W. D., Leighton P. A.. 2021. Common light chain chickens produce human antibodies of high affinity and broad epitope coverage for the engineering of bispecifics. MAbs 13: 1862451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gordon, G. L., Capel H. L., Guloglu B., Richardson E., Stafford R. L., Deane C. M.. 2023. A comparison of the binding sites of antibodies and single-domain antibodies. Front. Immunol. 14: 1231623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murakami, T., Kumachi S., Matsunaga Y., Sato M., Wakabayashi-Nakao K., Masaki H., Yonehara R., Motohashi M., Nemoto N., Tsuchiya M.. 2022. Construction of a humanized artificial VHH library reproducing structural features of camelid VHHs for therapeutics. Antibodies 11: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Conroy, P. J., Law R. H. P., Gilgunn S., Hearty S., Caradoc-Davies T. T., Lloyd G., O’Kennedy R. J., Whisstock J. C.. 2014. Reconciling the structural attributes of avian antibodies. J. Biol. Chem. 289: 15384–15392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Govaert, J., Pellis M., Deschacht N., Vincke C., Conrath K., Muyldermans S., Saerens D.. 2012. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J. Biol. Chem. 287: 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leighton, P. A., Pedersen D., Ching K., Collarini E. J., Izquierdo S., Jacob R., Lavoir M.-C. V. D.. 2016. Generation of chickens expressing Cre recombinase. Transgenic Res. 25: 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.