Abstract
Background
The study aims were to evaluate the species distribution and antimicrobial resistance profile of Gram-negative pathogens isolated from specimens of intra-abdominal infections (IAI), urinary tract infections (UTI), respiratory tract infections (RTI), and blood stream infections (BSI) in emergency departments (EDs) in China.
Methods
From 2016 to 2019, 656 isolates were collected from 18 hospitals across China. Minimum inhibitory concentrations were determined by CLSI broth microdilution and interpreted according to CLSI M100 (2021) guidelines. In addition, organ-specific weighted incidence antibiograms (OSWIAs) were constructed.
Results
Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) were the most common pathogens isolated from BSI, IAI and UTI, accounting for 80% of the Gram-negative clinical isolates, while Pseudomonas aeruginosa (P. aeruginosa) was mainly isolated from RTI. E. coli showed < 10% resistance rates to amikacin, colistin, ertapenem, imipenem, meropenem and piperacillin/tazobactam. K. pneumoniae exhibited low resistance rates only to colistin (6.4%) and amikacin (17.5%) with resistance rates of 25–29% to carbapenems. P. aeruginosa exhibited low resistance rates only to amikacin (13.4%), colistin (11.6%), and tobramycin (10.8%) with over 30% resistance to all traditional antipseudomonal antimicrobials including ceftazidime, cefepime, carbapenems and levofloxacin. OSWIAs were different at different infection sites. Among them, the susceptibility of RTI to conventional antibiotics was lower than for IAI, UTI or BSI.
Conclusions
Gram-negative bacteria collected from Chinese EDs exhibited high resistance to commonly used antibiotics. Susceptibilities were organ specific for different infection sites, knowledge which will be useful for guiding empirical therapies in the clinic.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12879-024-09294-0.
Keywords: SMART, Carbapenem, OSWIA, Antibiogram, Cephalosporin, Antibiotic-Resistance
Background
Antimicrobials are frequently used in emergency departments (EDs) in China and a study noted that the proportion of emergency patients treated with antibiotics was as high as 39.31 to 43.45% from 2016 to 2019 [1]. Antibiotic stewardship in EDs should avoid administration of broad-spectrum antibiotics, shorten their use times, as well as minimizing their unnecessary use [2]. However, as patients presenting to EDs are often in an acute state, physicians have to make decisions in a very short time frame and they prescribe antibiotics empirically. For critically ill infected patients, guidelines recommend to start antibiotic treatment in the first hour of recognition [3], which means that it is frequently impossible to get microbiology results to guide the choice of antimicrobial therapy. In order to support the choice of empiric antibiotic treatments, consensus guidelines as well as local antibiotic drug susceptibility detection and various antimicrobial surveillance programs have been introduced in China [4, 5].
One approach to individualized empiric antibiotic therapy is the Weighted-Incidence Syndromic Combination Antibiogram, which is comprised of information about the likelihood a treatment regimen will be effective for all relevant organisms for a given infection based on existing large datasets [6–8]. A similar approach is the organ-specific weighted incidence antibiogram (OSWIA), which estimates probable susceptibilities of organ specific isolates to specific antibiotics [9]. The Study for Monitoring Antimicrobial Resistance Trends (SMART) global surveillance program monitors in vitro susceptibilities of clinical Gram-negative bacilli to antimicrobial agents obtained from blood stream infections (BSI), intra-abdominal infections (IAI), urinary tract infections (UTI) and respiratory tract infections (RTI). The purpose of the present study was to determine the prevalence and susceptibilities of various bacteria to conventional antibiotics in patients attending Chinese EDs through a retrospective analysis of the SMART data collected from 2016 to 2019 and to determine differences of organ distributions between the infecting bacterial strains.
Methods
Ethics
In this study, the patient informed consent was waived and authorized by the Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (Approval Number: 20210811-33).
Isolates
All bacterial isolates were collected from discarded clinical specimens of hospitalized patients with BSI, IAI, UTI and RTI between 2016 and 2019 who were admitted to the EDs of 18 hospitals across China (Supplementary Table 1). The IAI specimen is derived from tissues or organs within the abdominal cavity, including the stomach, intestines, liver, spleen, pancreas, kidneys that have been infected by pathogens, resulting in infectious diseases [10]. RTI refers to an infection of the tissues in the respiratory system by pathogens such as viruses, bacteria, or fungi. RTI specimens from the respiratory tract included nasal and throat swabs, sputum samples, bronchoalveolar lavage fluid, respiratory secretions, and others [11]. Identification of isolates was initially made by each hospital laboratory and then the specimens were sent for laboratory re-identification using MALDI-TOF/MS (Bruker Daltonics, USA). Any duplicate isolates collected from the same patient were excluded from the data analysis.
Antimicrobial susceptibility testing
Testing was carried out in the Peking Union Medical College Hospital clinical microbiology laboratory using the Trek Diagnostic System (Thermo Fisher Scientific). Clinical isolates and reference strains were detected using the microbroth dilution method. Minimum inhibitory concentrations (MICs) were determined with reference to the antimicrobial breakpoint of the CLSI M100 (2021) [12]. The antibiotics tested were amikacin (AMK), cefepime (FEP), ceftazidime (CAZ), aztreonam (ATM), ceftriaxone (CRO), colistin (COL), ertapenem (ETP), levofloxacin (LVX), cefoxitin (FOX), imipenem (IPM), tobramycin (TOB), meropenem (MEM) and piperacillin–tazobactam (TZP).
Definition of antimicrobial-resistant strains
Carbapenem resistance of Escherichia coli (E. coli) and Klebsiella pneumoniae (K. pneumoniae) refers to resistance to any of IPM, MEM or ETP. Carbapenem-resistant Pseudomonas aeruginosa (P. aeruginosa) was defined as resistance to IPM or MEM.
Quinolone resistance to E. coli, K. pneumoniae and P. aeruginosa was defined as resistance to LVX.
Third generation cephalosporin resistance to E. coli and K. pneumoniae was defined as resistance to any CRO or CAZ, and the strains susceptible or intermediate to carbapenems (IPM, MEM or ETP). P. aeruginosa resistant to third-generation cephalosporin was defined as resistance to CAZ (CAZ-resistant PA) and the strains susceptible or intermediate to carbapenems (IPM, MEM).
OSWIA calculation
Data were retrospectively analyzed to establish the distribution of bacteria in various organs for BSI, IAI, UTI and RTI. OSWIA values were determined as previously described [9].
Results
Patient characteristics and specimen source
Between January 01, 2016 and December 31, 2019, a total of 656 isolated were obtained from ED patients. The patient characteristics are detailed in Table 1. The patients average age was 60.6 years (range: 1–101), comprising 388 males and 268 females. Most infections were hospital-acquired (HA) (58.1%), while 249 (38.0%) were community-acquired (CA) and for 26 data were not applicable. The isolates included 210 strains from IAI collected during surgery from the peritoneal fluid, appendix, abscesses, pancreas, gall bladder, liver and stomach. A total of 122 strains from BSI, 112 strains from UTI mainly from the urine, and 208 strains from RTI taken from bronchoalveolar lavage, endotracheal aspirate, thoracentesis or sputum were identified, as well as 4 strains from unconfirmed organs.
Table 1.
Patient characteristics
| Variable | Patient (N = 656), n (%) |
|---|---|
| Gender | |
| Male | 388 (59.1) |
| Female | 268 (40.9) |
| Average age (range), years | 60.6 (1–101) |
| Age categories, years | |
| ≤ 39 | 104 (15.9) |
| 40-59 | 173 (26.4) |
| ≥ 60 | 379 (57.8) |
| Timing of infection onset | |
| Community-acquired | 249 (38.0) |
| Hospital-acquired | 381 (58.1) |
| Not applicable | 26 (4.0) |
| Specimen source | |
| BSI: Blood | 122 (18.6) |
| IAI | 210 (32.0) |
| Abscess | 26 (4.0) |
| Appendix | 100 (15.2) |
| Gall bladder | 29 (4.4) |
| Liver | 15 (2.3) |
| Pancreas | 2 (0.3) |
| Peritoneal fluid | 36 (5.5) |
| Stomach | 1 (0.2) |
| Other | 1 (0.2) |
| UTI | 112 (17.1) |
| Ureter | 3 (0.5) |
| Urine | 109 (16.6) |
| RTI | 208 (31.7) |
| Bronchoalveolar lavage | 8 (1.2) |
| Endotracheal aspirate | 2 (0.3) |
| Sputum | 196 (29.9) |
| Thoracentesis | 2 (0.3) |
| Unconfirmed infection | 4 (0.6) |
Abbreviations: BSI blood steam infection, IAI intra-abdominal infection, RTI respiratory tract infection, UTI urinary tract infection
Distribution of Gram-negative bacteria obtained from BSI, IAI, UTI and RTI
Enterobacterales were the most common Gram-negative bacilli isolated from emergency patients with BSI, IAI and UTI (Fig. 1). E. coli accounted for 48.4% in BSI, 58.6% in IAI and 72.3% in UTI, while K. pneumoniae accounted for 24.6%, 21.4% and 11.6%, respectively and other Enterobacterales were much less common than E. coli and K. pneumoniae. The pathogen distribution in RTI was distinctly different from the other three infection types, with P. aeruginosa and K. pneumoniae being the most common species each accounting for about 40% of the Gram-negative pathogens. Since the composition ratio of Gram-negative bacteria was different at different infection sites (Fig. 1, Supplementary Table 2) the varying patterns between infected organs should be considered when prescribing empirical treatments.
Fig. 1.
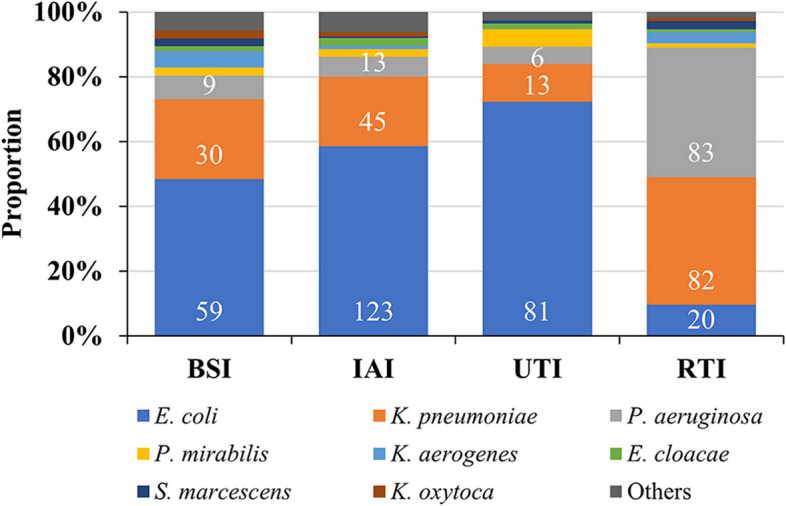
Distribution of Gram-negative bacilli in BSI, IAI, UTI and RTI. Abbreviations: BSI, blood steam infection; IAI, intra-abdominal infection; UTI, urinary tract infection
Distribution of Gram-negative bacteria from 2016 to 2019
The distribution of Gram-negative pathogens was stable between 2016 and 2019, with E. coli, K. pneumoniae and P. aeruginosa being the top 3 species, accounting for more than 80% of the clinical isolates (Fig. 2, Supplementary Table 3).
Fig. 2.
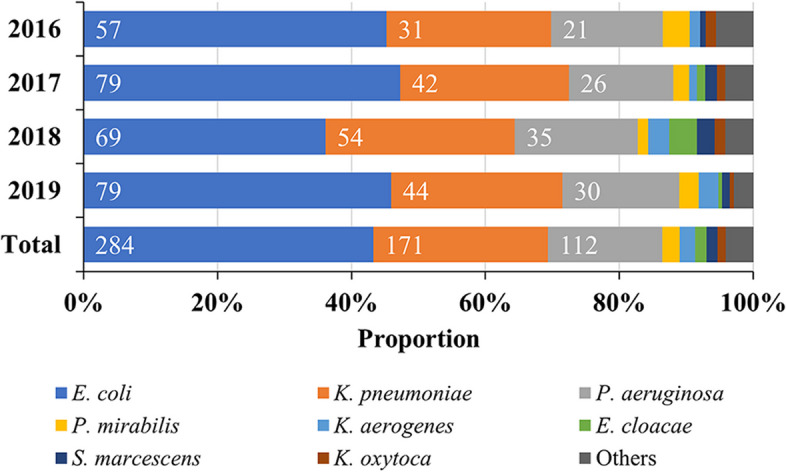
Composition ratio of Gram-negative bacteria in EDs from 2016 to 2019. Abbreviation: ED, emergency department.
Distribution of Gram-negative bacteria in different age groups of patients
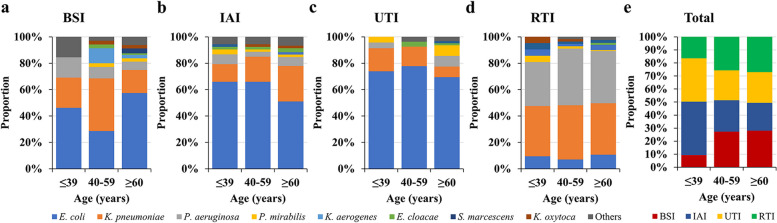
Among the strains collected, regardless of the organ of origin, the predominant species were P. aeruginosa, E. coli and K. pneumoniae. However, the composition ratio of the main bacterial groups were different in infection sites within age groups (Fig. 3a-d) and were generally different especially in the age group ≤ 39 years (Fig. 3e, Supplementary Table 4).
Fig. 3.
Comparison of the composition ratio of microbiota in different infected organs and age groups. Abbreviations: BSI, blood steam infection; IAI, intra-abdominal infection; RTI, respiratory tract infection; UTI, urinary tract infection.
Monitoring of drug susceptibility
Drug resistance rate monitoring of major Gram-negative bacteria from 2016 to 2019
E. coli exhibited < 10% resistance to AMK, COL, ETP, IPM, MEM and TZP, with the exception in 2016 to TZP, but generally TZP resistance rates were reduced between 2016 and 2019. Otherwise, resistance rates were more than 30%, with the exception of FOX (16.2%). K. pneumoniae exhibited < 20% resistance only to AMK and 6.4% to COL between 2016 and 2019. P. aeruginosa only exhibited low resistant rates of 13.4% to AMK, 11.6% to COL and 10.8% to TOB from 2016 to 2019 (Table 2).
Table 2.
In vitro antimicrobial resistance rates of major Gram-negative bacteria (E. coli, K. pneumoniae, P. aeruginosa) from 2016 to 2019 (%)
| AMK | ATM | CAZ | COL | CRO | ETP | FEP | FOX | IPM | LVX | MEM | TOB | TZP | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| E. coli (n = 284) | 7 (2.5%) | 51 (45.5%) | 88 (31.0%) | 16 (5.6%) | 162 (57.0%) | 12 (4.2%) | 127 (44.7%) | 46 (16.2%) | 8 (2.8%) | 161 (56.7%) | 9 (3.2%) | - | 26 (9.2%) |
| 2016 (n = 57) | 3 (5.3%) | 10 (47.6%) | 27 (47.4%) | 5 (8.8%) | 41 (71.9%) | 5 (8.8%) | 37 (64.9%) | 15 (26.3%) | 4 (7.0%) | 35 (61.4%) | 5 (8.8%) | - | 10 (17.5%) |
| 2017 (n = 79) | 2 (2.5%) | 13 (50.0%) | 16 (20.3%) | 5 (6.3%) | 39 (49.4%) | 4 (5.1%) | 33 (41.8%) | 16 (20.3%) | 3 (3.8%) | 43 (54.4%) | 3 (3.8%) | - | 7 (8.9%) |
| 2018 (n = 69) | 1 (1.4%) | 17 (48.6%) | 20 (29.0%) | 3 (4.3%) | 38 (55.1%) | 2 (2.9%) | 26 (37.7%) | 7 (10.1%) | 1 (1.4%) | 39 (56.5%) | 1 (1.4%) | - | 5 (7.2%) |
| 2019 (n = 79) | 1 (1.3%) | 11 (36.7%) | 25 (31.6%) | 3 (3.8%) | 44 (55.7%) | 1 (1.3%) | 31 (39.2%) | 8 (10.1%) | 0 (0.0%) | 44 (55.7%) | 0 (0.0%) | - | 4 (5.1%) |
| K. pneumoniae (n = 171) | 30 (17.5%) | 125 (44.0%) | 63 (36.8%) | 11 (6.4%) | 76 (44.4%) | 49 (28.7%) | 63 (36.8%) | 66 (38.6%) | 44 (25.7%) | 69 (40.4%) | 43 (25.1%) | - | 50 (29.2%) |
| 2016 (n = 31) | 7 (22.6%) | 35 (61.4%) | 11 (35.5%) | 3 (9.7%) | 14 (45.2%) | 11 (35.5%) | 11 (35.5%) | 13 (41.9%) | 8 (25.8%) | 14 (45.2%) | 8 (25.8%) | - | 8 (25.8%) |
| 2017 (n = 42) | 8 (19.0%) | 26 (32.9%) | 19 (45.2%) | 2 (4.8%) | 23 (54.8%) | 13 (31.0%) | 22 (52.4%) | 20 (47.6%) | 13 (31.0%) | 20 (47.6%) | 13 (31.0%) | - | 15 (35.7%) |
| 2018 (n = 54) | 7 (13.0%) | 29 (42.0%) | 16 (29.6%) | 3 (5.6%) | 23 (42.6%) | 10 (18.5%) | 16 (29.6%) | 14 (25.9%) | 10 (18.5%) | 19 (35.2%) | 9 (16.7%) | - | 10 (18.5%) |
| 2019 (n = 44) | 8 (18.2%) | 35 (44.3%) | 17 (38.6%) | 3 (6.8%) | 16 (36.4%) | 15 (34.1%) | 14 (31.8%) | 19 (43.2%) | 13 (29.5%) | 16 (36.4%) | 13 (29.5%) | - | 17 (38.6%) |
| P. aeruginosa (n = 112) | 15 (13.4%) | 51 (45.5%) | 39 (34.8%) | 13 (11.6%) | N | N | 37 (33.0%) | N | 44 (39.3%) | 41 (36.6%) | 40 (35.7%) | 7 (10.8%) | 42 (37.5%) |
| 2016 (n = 21) | 4 (19.0%) | 10 (47.6%) | 9 (42.9%) | 2 (9.5%) | N | N | 7 (33.3%) | N | 11 (52.4%) | 6 (28.6%) | 9 (42.9%) | - | 10 (47.6%) |
| 2017 (n = 26) | 5 (19.2%) | 13 (50.0%) | 11 (42.3%) | 1 (3.8%) | N | N | 11 (42.3%) | N | 9 (34.6%) | 12 (46.2%) | 9 (34.6%) | - | 9 (34.6%) |
| 2018 (n = 35) | 3 (8.6%) | 17 (48.6%) | 10 (28.6%) | 5 (14.3%) | N | N | 12 (34.3%) | N | 15 (42.9%) | 15 (42.9%) | 15 (42.9%) | 5 (14.3%) | 13 (37.1%) |
| 2019 (n = 30) | 3 (10.0%) | 11 (36.7%) | 9 (30.0%) | 5 (16.7%) | N | N | 7 (23.3%) | N | 9 (30.0%) | 8 (26.7%) | 7 (23.3%) | 2 (6.7%) | 10 (33.3%) |
N natural drug resistance, -, no detection
Abbreviations: AMK amikacin, ATM aztreonam, CAZ ceftazidime, COL colistin, CRO ceftriaxone, ETP ertapenem,FEP cefepime, FOX cefoxitin, IPM imipenem, LVX levofloxacin, MEM meropenem, TOB tobramycin, TZP piperacillin-tazobactam
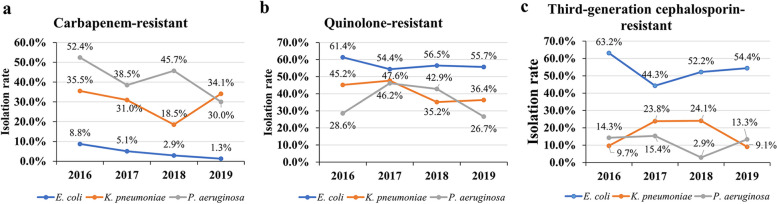
Detection rate and drug susceptibility of specific antibiotic-resistant bacteria from 2016 to 2019
Isolation of carbapenem-resistant, quinolone-resistant or third-generation cephalosporin-resistant E. coli, K. pneumoniae and P. aeruginosa
In isolates, the carbapenem-resistant E. coli and P. aeruginosa showed an overall downward trend. However, the rate of detection of carbapenem-resistant E. coli was relatively low, being only 1.3% in 2019, while for carbapenem-resistant P. aeruginosa in 2019 it was 30.0% and for carbapenem-resistant K. pneumoniae strains in 2019, 34.1%, the rate being higher than in 2018 (18.5%). The rate of detection of quinolone-resistant E. coli or K. pneumoniae showed a decreasing trend in the four years studied, being 55.7% in 2019 for the detection of quinolone-resistant E. coli and 36.4% for K. pneumoniae. The detection rate of quinolone-resistant P. aeruginosa was 26.7% in 2019 and lower than in 2018 (42.9%). The detection rates of third-generation cephalosporin-resistant E. coli and K. pneumoniae as well as P. aeruginosa showed an irregular trend from 2016 to 2019, being between 44.3%–63.2% and 9.1%–24.1% as well as 2.9%–15.4%, respectively throughout the years. Compared to E. coli, there were only few numbers of third-generation cephalosporin-resistant K. pneumoniae and P. aeruginosa isolates found between 2016 and 2019 (Fig. 4, Supplementary Table 5).
Fig. 4.
Isolation (detection) rate of carbapenem-resistant, quinolone-resistant, third-generation cephalosporin-resistant E. coli, K. pneumoniae and P. aeruginosa from 2016 to 2019
Specific drug resistance rates (%) of strains to antibiotics from 2016 to 2019
For E. coli, the resistance rate of carbapenem-resistant E. coli to AMK was 33.3%, and to the other antibiotics tested were > 60%, apart from COL (25.0%). Quinolone-resistant E. coli exhibited the lowest resistance rates to AMK (4.3%), ETP (5.6%), IPM (5.0%) and MEM (5.6%). The resistance rates of third-generation cephalosporin-resistant E. coli was 2.0% to AMK and 0% to ETP, IPM and MEM.
Carbapenem-resistant K. pneumoniae were 57.1% resistant to AMK and quinolone-resistant K. pneumoniae were 42.0% resistant to AMK, with low resistance rates only found to COL (8.2% and 10.1%), respectively. Third-generation cephalosporin-resistant K. pneumoniae was 6.7% resistant to AMK and 0% to ETP, IPM and MEM.
For carbapenem-resistant P. aeruginosa, the drug resistance to AMK was 30.4%, for TOB 28.0% and for COL (10.9%). For quinolone-resistant P. aeruginosa, only resistance rates to AMK (24.4%) and TOB (26.1%) as well as COL (9.8%) remained low. Third-generation cephalosporins resistant P. aeruginosa,showed low resistance rates of 0% to AMK, IPM, MEM and TOB (Table 3).
Table 3.
Drug-resistant rates (%) of carbapenem-resistant, quinolone-resistant, third-generation cephalosporin-resistant E. coli, K. pneumoniae and P. aeruginosa between 2016 and 2019 (n/%)
| E. coli | K. pneumoniae | P. aeruginosa | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Carbapenem - resistant (n = 12) | Quinolone - resistant (n = 161) | Third generation cephalosporin - resistant (n = 150) | Carbapenem - resistant (n = 49) | Quinolone - resistant (n = 69) | Third generation cephalosporin - resistant (n = 30) | Carbapenem - resistant (n = 46) | Quinolone - resistant (n = 41) | Third generation cephalosporin - resistant (n = 12) | |
| AMK | 4 (33.3%) | 7 (4.3%) | 3 (2.0%) | 28 (57.1%) | 29 (42.0%) | 2 (6.7%) | 14 (30.4%) | 10 (24.4%) | 0 (0.0%) |
| ATM | 11 (91.7%) | 98 (60.9%) | 113 (75.3%) | 47 (95.9%) | 61 (88.4%) | 19 (63.3%) | 31 (67.4%) | 33 (80.5%) | 11 (91.7%) |
| CAZ | 9 (75.0%) | 69 (42.9%) | 79 (52.7%) | 45 (91.8%) | 59 (85.5%) | 18 (60.0%) | 27 (58.7%) | 25 (61.0%) | 12 (100.0%) |
| COL | 3 (25.0%) | 10 (6.2%) | 7 (4.7%) | 4 (8.2%) | 7 (10.1%) | 4 (13.3%) | 5 (10.9%) | 4 (9.8%) | 1 (8.3%) |
| CRO | 12 (100.0%) | 117 (72.7%) | 150 (100.0%) | 49 (100.0%) | 65 (94.2%) | 27 (90.0%) | N | N | N |
| ETP | 12 (100.0%) | 9 (5.6%) | 0 (0.0%) | 49 (100.0%) | 45 (65.2%) | 0 (0.0%) | N | N | N |
| FEP | 9 (75.0%) | 94 (58.4%) | 118 (78.7%) | 45 (91.8%) | 58 (84.1%) | 18 (60.0%) | 30 (65.2%) | 25 (61.0%) | 7 (58.3%) |
| FOX | 11 (91.7%) | 31 (19.3%) | 21 (14.0%) | 49 (100.0%) | 58 (84.1%) | 14 (46.7%) | N | N | N |
| IPM | 8 (66.7%) | 8 (5.0%) | 0 (0.0%) | 44 (89.8%) | 43 (62.3%) | 0 (0.0%) | 44 (95.7%) | 25 (61.0%) | 0 (0.0%) |
| LVX | 9 (75.0%) | 161 (100.0%) | 108 (72.0%) | 45 (91.8%) | 69 (100.0%) | 21 (70.0%) | 27 (58.7%) | 41 (100.0%) | 6 (50.0%) |
| MEM | 9 (75.0%) | 9 (5.6%) | 0 (0.0%) | 43 (87.8%) | 43 (62.3%) | 0 (0.0%) | 40 (87.0%) | 27 (65.9%) | 0 (0.0%) |
| TOB | - | - | - | - | - | - | 7 (28.0%) | 6 (26.1%) | 0 (0.0%) |
| TZP | 9 (75.0%) | 22 (13.7%) | 15 (10.0%) | 45 (91.8%) | 48 (69.6%) | 4 (13.3%) | 32 (69.6%) | 26 (63.4%) | 9 (75.0%) |
N natural drug resistance; -, no detection
Abbreviations: AMK amikacin, ATM aztreonam, CAZ ceftazidime, COL colistin, CRO ceftriaxone, ETP ertapenem,FEP cefepime, FOX cefoxitin, IPM imipenem, LVX levofloxacin, MEM meropenem, TOB tobramycin, TZP piperacillin-tazobactam
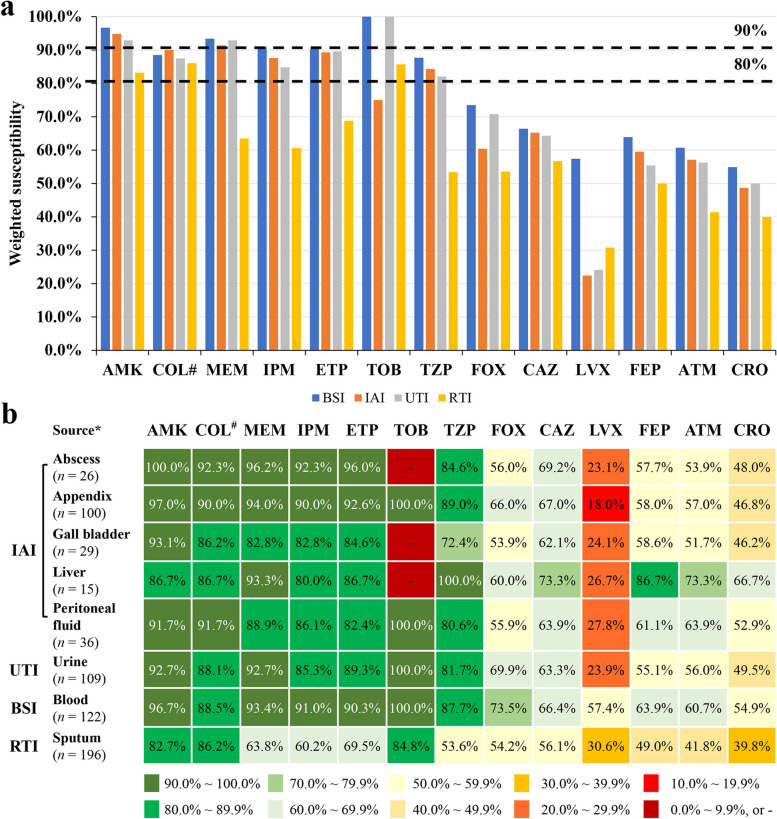
Antimicrobial susceptibility monitoring during empiric treatment of different infection sites and organs
In the weighted susceptibility assessment of different infection sites, it was found that the susceptibility of the same antibacterial drug at different organs and infection sites was different. For example, AMK, TOB, ETP, IPM and MEM were the antibiotics with > 90% susceptibility for BSI, but only AMK and MEM were > 90% effective antibiotics against IAI. High-susceptibility to antibiotics in UTI included AMK, TOB and MEM (all > 90%), and except for AMK, COL and TOB, the susceptibility to other antibiotics at the site of RTI infections was < 80% (Fig. 5, Supplementary Table 6).
Fig. 5.
Organ distribution related susceptibilities. a Differences in susceptibility of antibiotics at different infection sites. b Differences in weighted drug susceptibilities of antibiotics at different organs and infection sites. Note: *only sites from which more than 10 pathogenic bacteria were collected have been included; #, intermediate rate was shown for COL; -, no detection. Abbreviations: AMK, amikacin; ATM, aztreonam; BSI, blood steam infections; CAZ, ceftazidime; CRO, ceftriaxone; ETP, ertapenem; FEP, cefepime; FOX, cefoxitin; IAI, intra-abdominal infections; IPM, imipenem; LVX, levofloxacin; MEM, meropenem; RTI, respiratory tract infections; TOB, tobramycin; TZP, piperacillin/tazobactam; UTI, urinary tract infections
Discussion
This study analyzed Chinese data from the global SMART surveilance program and found that the most frequently isolated Gram-negative bacteria were Enterobacterales, a finding similar to previous results from SMART studies and the China Antimicrobial Surveillance Network (CHINET) [13, 14]. Enterobacterales are of particular concern given their ability to develop and spread resistance to penicillins, cephalosporins, carbapenems and quinolones [12, 15, 16]. As these are the most commonly used antibiotics in hospitals, such resistance would leave physicians with very limited treatment options. The vast majority of the pathogens were E. coli, K. pneumoniae and P. aeruginosa (86.4%), with resistance rates for cephalosporins in the range of 31.0%–57.0% and for ATM 44.0%–45.5%, indicating a high rate of ESBL-producing strains [17], which underlines the global health problem of cephalosporin resistance [18]. Previous SMART surveilence results found ESBL rates of 46.3%–49.1% for E. coli and 25.6%–26.8% for K. pneumoniae [13]. In the 2021 CHINET surveillance, resistance to third-generation cephalosporins was detected in 55.6% of E. coli and 43.8% of K. pneumoniae, also indicating the high ESBL prevalence in China [14]. The resistance rates for the fluoroquinolone LVX (36.6%–56.7%) were in a similar range to cephalosporins in this study, which might reflect the overuse of fluoroquinolones, especially since the development of cephalosporin-resistance [19, 20]. They were similar to the rates reported by the 2021 CHINET surveillance of LVX resistance detected as 53.6% for E. coli and 28.3% for K. pneumoniae isolates [14]. One approach to overcome cephalosporin resistance is the use of combination of a β-lactam and a β-lactamase inhibitor [21], such as tazobactam, which led to essentially reduced resistance rates of about 10% for otherwise cephalosporin-resistant E. coli and K. pneumoniae in the present study. Of concern is the rising resistance rate of K. pneumoniae to carbapenems, which reached25%–29% in the present study and was similar to the rate of about 25% found in the 2021 CHINET surveillance study [14].
Resistance rates for COL were generally low, but susceptibility breakpoints have been abolished in recent CLSI guidelines. Treatments with COL should be applied with maximum renally adjusted doses, since the previous MIC of 2 μg/mL could not be achieved in 50% of patients with normal renal functions and acute kidney injury occurs frequently with conventional doses. Recommendations include strongly preferred alternative drugs for active or combination treatments [22, 23]. Also, TOB is not commonly used in China, which might explain the low resistance rates found in the present study.
P. aeruginosa was the third most common pathogen detected and the most common Gram-negative pathogen found in RTI. It exhibited > 30% resistance to traditional antipesudomoas antibiotics, including CAZ, FEP, TZP, IPM, MEM and LVX. These results are similar to those reported in a recent SMART study that investigated P. seudomonas resistance to antibiotics in China [24].
This present survey included only isolates collected in EDs, since patients admitted to EDs are frequently candidates for urgent empiric antibiotic treatment, which should be administered according to the site of infection and the clinical severity of symptoms [25]. The characteristics of infected patients and physicians' routine treatments in the ED differed compared to other departments. This is because the infected site may initially not be clearly defined, patients cannot be observed for a long period of time, and etiological evidence is rarely available; doctors and nurses in EDs also have a very heavy workload. It is therefore convenient to use drugs which only need administration once a day to ED patients, including ETP once a day, AMK intramuscular injection, as well as LVX once a day, which can be sequenced simultaneously.
In order to offer guidelines for chosing an empiric treatment, organ specific therapy based on pathogen distribution differences and their antibiotic resistance variations have been developed [6, 8]. Of these, OSIWA was first applied for HA and CA infections of different intra-abdominal organs [9]. In the present study, we expanded the evaluation of OSIWA for the empirical treatment of IAI also to UTI, BSI and RTI. When considering the distribution of pathogens at a single infection site – for example E. coli caused more than 50% of all IAI and more than 60% of all UTI – whereas for RTI infections E. coli was the pathogen in < 15% of all cases (Fig. 3). The OSIWA shown in Fig 5 indicate the probabilities of successful empiric drug treatment for single infections sites. For RTI and IAI the choice of empirical antibiotics with expected efficacy was limited, but for UTI and BSI more antibiotics are available.
Limitations of the present survey were the relatively low sample numbers since only isolates from EDs have been included, which have had an influence on the resistance patterns reported and combination treatments with β-lactams, aminoglycoside and fluoroquinolone were not considered. In addition, since in the SMART data collection detailed data regarding resistance mechanisms are not included, susceptibility test results only can infer the mechanism of ESBL production in cephalosporin but not in carbapenem resistance.
Conclusions
The pathogens isolated from BSI, IAI, UTI and RTI between 2016 and 2019 in Chinese EDs were mainly E. coli, K. pneumoniae and P. aeruginosa,but therewere considerable resistance pattern differences as well as organ distributions between the bacterial strains. OSIWA determinations led to organ specific antibiotic drug effectiveness patterns, which should help to guide the choice of suitable empirical antibiotic treatments, especially for urgent infected cases in EDs.
Supplementary Information
Acknowledgements
The authors thank Shanghai BIOMED Science Technology Co., Ltd (Shanghai, China) for providing editorial assistance and MSD China for their financial support.
Abbreviations
- AMK
amikacin
- ATM
aztreonam
- BSIs
blood stream infections
- CA
community acquired
- CAZ
ceftazidime
- CHINET
China Antimicrobial Surveillance Network
- COL
colistin
- CRO
ceftriaxone
- EDs
emergency departments
- ETP
ertapenem
- FEP
cefepime
- FOX
cefoxitin
- HA
hospital acquired
- IAI
intra-abdominal infections
- IPM
imipenem
- LVX
levofloxacin
- MEM
meropenem
- MICs
minimum inhibitory concentrations
- OSWIAs
organ-specific weighted incidence antibiograms
- RTI
respiratory tract infections.
- SMART
Study for Monitoring Antimicrobial Resistance Trends
- TOB
tobramycin
- TZP
piperacillin–tazobactam
- UTI
urinary tract infections
Author contributions
All authors made substantial contributions to the conception and design of the study, acquisition of data, or analysis and interpretation of data, took part in drafting the manuscript or critically revising it for important intellectual content, agreed to submit it to the current journal, gave final approval of the version to be published and agreed to be accountable for all aspects of the work.
Funding
This study was sponsored by funding from Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, New Jersey, USA. The funding body was involved in the study design, analysis and interpretation of data, as well as the decision to submit the article for publication.
Availability of data and materials
The SMART database is not public and is only accessible to SMART investigators, but the data that support the findings of this study are directly available from MSD China or from the corresponding author Yunsong Yu upon reasonable request and with permission of MSD China.
Declarations
Ethical approval and consent to participate
In this study, the patient informed consent was waived and authorized by the Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (Approval Number: 20210811-33).
Consent for publication
Not applicable.
Competing interests
Pengcheng Li is an employee of MSD China. All other authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhu Y, Qiao Y, Dai R, Hu X, Li X. Trends and patterns of antibiotics use in China's Urban tertiary hospitals, 2016-19. Front Pharmacol. 2021;12:757309. doi: 10.3389/fphar.2021.757309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luyt C-E, Bréchot N, Trouillet J-L, Chastre J. Antibiotic stewardship in the intensive care unit. Crit Care. 2014;18(5):480. doi: 10.1186/s13054-014-0480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans L, Rhodes A, Alhazzani W, Antonelli M, Coopersmith CM, French C, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Crit Care Med. 2021;49(11):e1063–1143. doi: 10.1097/CCM.0000000000005337. [DOI] [PubMed] [Google Scholar]
- 4.Hay SI, Rao PC, Dolecek C, Day NPJ, Stergachis A, Lopez AD, et al. Measuring and mapping the global burden of antimicrobial resistance. BMC Med. 2018;16(1):78. doi: 10.1186/s12916-018-1073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnall J, Rajkhowa A, Ikuta K, Rao P, Moore CE. Surveillance and monitoring of antimicrobial resistance: limitations and lessons from the GRAM project. BMC Med. 2019;17(1):176. doi: 10.1186/s12916-019-1412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Randhawa V, Sarwar S, Walker S, Elligsen M, Palmay L, Daneman N. Weighted-incidence syndromic combination antibiograms to guide empiric treatment of critical care infections: a retrospective cohort study. Crit Care. 2014;18(3):R112. doi: 10.1186/cc13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook A, Sharland M, Yau Y, Group* P. Bielicki J. Improving empiric antibiotic prescribing in pediatric bloodstream infections: a potential application of weighted-incidence syndromic combination antibiograms (WISCA) Expert Rev Anti Infect Ther. 2022;20(3):445–456. doi: 10.1080/14787210.2021.1967145. [DOI] [PubMed] [Google Scholar]
- 8.Hebert C, Ridgway J, Vekhter B, Brown EC, Weber SG, Robicsek A. Demonstration of the weighted-incidence syndromic combination antibiogram: an empiric prescribing decision aid. Infect Control Hosp Epidemiol. 2012;33(4):381–388. doi: 10.1086/664768. [DOI] [PubMed] [Google Scholar]
- 9.Liu L, Ni Y. Antimicrobial susceptibilities of specific syndromes created with organ-specific weighted incidence antibiograms (OSWIA) in patients with intra-abdominal infections. BMC Infect Dis. 2018;18(1):584. doi: 10.1186/s12879-018-3494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinese Society of Surgical Infection and Intensive Care, Chinese Society of Surgery, Chinese Medical Association Chinese College of Gastrointestinal Fistula Surgeons, Chinese College of Surgeons, Chinese Medical Doctor Association. Chinese guideline for the diagnosis and management of intra-abdominal infection (2019 edition) Chin J Practical Surg. 2020;40(1):1–16. [Google Scholar]
- 11.Chinese Medical Association . Clinical Guidelines: Respiratory diseases. Beijing: People's Medical Publishing House; 2009. p. 174. [Google Scholar]
- 12.Nordmann P, Poirel L. Emergence of plasmid-mediated resistance to quinolones in enterobacteriaceae. J Antimicrob Chemother. 2005;56(3):463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Johnson A, Zhang G, Yang Y, Zhang J, Li D, et al. Susceptibilities of Gram-negative bacilli from hospital- and community-acquired intra-abdominal and urinary tract infections: a 2016–2017 update of the Chinese SMART study. Infect Drug Resist. 2019;12:905–914. doi: 10.2147/IDR.S203572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fupin HU, Yan GUO, Demei ZHU, Fu W, Xiaofei J, Yingchun XU, et al. CHINET surveillance of antimicrobial resistance among the bacterial isolates in 2021. Chin J Infect Chemother. 2022;22(5):521–530. [Google Scholar]
- 15.Ma J, Song X, Li M, Yu Z, Cheng W, Yu Z, et al. Global spread of carbapenem-resistant Enterobacteriaceae: Epidemiological features, resistance mechanisms, detection and therapy. Microbiol Res. 2023;266:127249. doi: 10.1016/j.micres.2022.127249. [DOI] [PubMed] [Google Scholar]
- 16.De Angelis G, Del Giacomo P, Posteraro B, Sanguinetti M, Tumbarello M. Molecular mechanisms, epidemiology, and clinical importance of beta-lactam resistance in enterobacteriaceae. Int J Mol Sci. 2020;21(14):5090. doi: 10.3390/ijms21145090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rawat D, Nair D. Extended-spectrum β-lactamases in gram negative bacteria. J Glob Infect Dis. 2010;2(3):263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pana ZD, Zaoutis T. Treatment of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBLs) infections: what have we learned until now? F1000Res.2018;7:F1000 Faculty Rev-1347. [DOI] [PMC free article] [PubMed]
- 19.Ezelarab HAA, Abbas SH, Hassan HA, Abuo-Rahma GEA. Recent updates of fluoroquinolones as antibacterial agents. Arch Pharm (Weinheim) 2018;351(9):e1800141. doi: 10.1002/ardp.201800141. [DOI] [PubMed] [Google Scholar]
- 20.Hou J, Long X, Wang X, Li L, Mao D, Luo Y, et al. Global trend of antimicrobial resistance in common bacterial pathogens in response to antibiotic consumption. J Hazard Mater. 2023;442:130042. doi: 10.1016/j.jhazmat.2022.130042. [DOI] [PubMed] [Google Scholar]
- 21.Yahav D, Giske CG, Grāmatniece A, Abodakpi H, Tam VH, Leibovici L. New β-Lactam-β-Lactamase Inhibitor Combinations. Clin Microbiol Rev. 2020;34(1):e00115. doi: 10.1128/CMR.00115-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satlin MJ, Lewis JS, Weinstein MP, Patel J, Humphries RM, Kahlmeter G, et al. Clinical and laboratory standards institute and European committee on antimicrobial susceptibility testing position statements on polymyxin b and colistin clinical breakpoints. Clin Infect Dis. 2020;71(9):e523–529. doi: 10.1093/cid/ciaa121. [DOI] [PubMed] [Google Scholar]
- 23.Tsuji BT, Pogue JM, Zavascki AP, Paul M, Daikos GL, Forrest A, et al. International consensus guidelines for the optimal use of the polymyxins: endorsed by the American College of Clinical Pharmacy (ACCP), European Society of Clinical Microbiology and Infectious Diseases (ESCMID), Infectious Diseases Society of America (IDSA), International Society for Anti-infective Pharmacology (ISAP), Society of Critical Care Medicine (SCCM), and Society of Infectious Diseases Pharmacists (SIDP) Pharmacotherapy. 2019;39(1):10–39. doi: 10.1002/phar.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Croche Santander B, Campos Alonso E, Sanchez Carrion A, Marcos Fuentes L, Diaz Flores I, Vargas JC, et al. [Appropriateness of antibiotic prescribing in paediatric patients in a hospital emergency department] Pediatr (Engl Ed) 2018;88(5):259–265. doi: 10.1016/j.anpedi.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Kollef MH, Shorr AF, Bassetti M, Timsit J-F, Micek ST, Michelson AP, et al. Timing of antibiotic therapy in the ICU. Crit Care. 2021;25(1):360. doi: 10.1186/s13054-021-03787-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The SMART database is not public and is only accessible to SMART investigators, but the data that support the findings of this study are directly available from MSD China or from the corresponding author Yunsong Yu upon reasonable request and with permission of MSD China.