Abstract
Growth rate maximization is an important fitness strategy for microbes. However, the wide distribution of slow-growing oligotrophic microbes in ecosystems suggests that rapid growth is often not favored across ecological environments. In many circumstances, there exist trade-offs between growth and other important traits (e.g., adaptability and survival) due to physiological and proteome constraints. Investments on alternative traits could compromise growth rate and microbes need to adopt bet-hedging strategies to improve fitness in fluctuating environments. Here we review the mechanistic role of trade-offs in controlling bacterial growth and further highlight its ecological implications in driving the emergences of many important ecological phenomena such as co-existence, population heterogeneity and oligotrophic/copiotrophic lifestyles.
Subject terms: Bacteriology, Microbial ecology, Bacterial physiology, Bacterial systems biology
Trade-offs play a key role in controlling bacterial growth and shaping microbial phenotypes, which further drives the emergence of ecologically relevant phenomena including co-existence, population heterogeneity and oligotrophic/copiotrophic lifestyles.
Introduction
Rapid growth is a fundamental property of microbes and is, in principle, advantageous for microbial fitness. Bacterial cells are capable of tightly coordinating macromolecule biosynthesis (mass accumulation) with cell cycle progression (number increase) to ensure rapid proliferation1–6. Protein synthesis lies at the core of bacterial growth as protein accounts for over half of the biomass and its synthesis consumes ~70% of the cellular energy budget3,5–8. Furthermore, mRNA transcription is globally coordinated with protein translation under various conditions9–11. For the model bacterium Escherichia coli, ribosome synthesis could be maximized in nutrient broth to support a rapid growth rate of ~20 min per doubling5,12–15. However, the nutrient status in ecological niches of bacteria is often highly fluctuating, leading to the so-called ‘feast and famine cycle’14,16–19. For enteric bacteria living in mammalian gut, feast-like conditions occur when abundant amino acids and carbohydrates are available after the meals of the host while famine-like conditions occur when bacteria are released into the sewer14,20,21. Similar scenarios also occur for soil bacteria living in plant rhizosphere, which feed on the root exudates of the host plant22–25 and also for marine bacteria living in either oligotrophic open sea or eutrophic coastal area16,26–29. Therefore, it is important to elucidate how microbes coordinate cell growth with gene expression to adapt to different nutrient environments.
Recent quantitative studies have revealed the fundamental role of proteome resource allocation in governing microbial growth control3,5,6,8,30–34. To maintain optimal growth statuses, bacteria attempt to balance their proteome investments in various functional sectors according to the nutrient conditions6,14,35–38. In a rich medium where nutrient sources are abundant, bacteria could save the proteome resource of anabolic proteins and further maximize ribosome synthesis to achieve fast growth8,13,15,39. When shifting to a minimal medium, bacteria employ (p)ppGpp-DksA to inhibit ribosome synthesis and devote more resources into anabolic proteins to fulfill the supply of in vivo amino acid flux5,8,19,40,41, which inevitably results in slower growth. Furthermore, cAMP-CRP allows bacteria to fine-tune the proteome allocation between catabolic and anabolic proteins to adapt to different carbon or nitrogen sources31,42. In this sense, bacteria employ sophisticated molecular strategies to efficiently modulate proteome allocation so that optimal growth statuses could be achieved in various nutrient conditions6. A three-sector proteome allocation model could quantitatively describe the tight relation between gene expression and growth rate with only a few phenomenological parameters6,30,31.
In order to further accelerate growth, microbes including bacteria and yeast attempt to balance energy biogenesis and biomass synthesis43–46. A long-standing puzzle associated with microbial energy biogenesis is the overflow metabolism, in which fast-growing cells (bacteria, yeasts and cancer cells) preferably use the seemingly low-efficient fermentation pathway instead of respiratory pathway to generate ATP during growth under aerobic conditions, thus secreting large amounts of fermentation products such as acetate and lactate47–49. Recent studies have shown that overflow metabolism results from the strategy of optimal proteome resource allocation for achieving rapid growth44,50. Although fermentation has a lower ATP production efficiency than respiration, it costs fewer enzymes and requires a lower proteome investment per ATP produced than respiratory pathway. In such case, employment of fermentation pathways for energy biogenesis allows microbes to save more proteome resources for ribosome synthesis to achieve faster growth under favorable conditions44,46,47. A similar logic applies to another puzzle of prokaryotic energy generation: the prevalence of Entner–Doudoroff (ED) pathway in many aerobes51 considering that ED pathway produces only one ATP per glucose—half of EMP pathway. This puzzle could again be rationalized by the fact that ED pathway costs much less enzymes to achieve the same glucose conversion rate than the EMP pathway, thus facilitating rapid growth43.
Although growth rate maximization is an important fitness strategy for bacterial cells, a seemingly paradox lies at the extremely large variations in the growth capacities of bacteria across phylogeny52,53. The maximal growth capacities of bacteria across species vary substantially with the shortest generation time ranging from 10 min (e.g. Vibrio natriegens)54 to days (e.g. Mycobacterium tuberculosis, Mtb and marine oligotrophic bacterium Pelagibacterales SAR11)55,56. In light of this fact, microbial ecologists conventionally divide bacteria into fast-growing copiotrophic bacteria (r-strategist) and slow-growing oligotrophic bacteria (k-strategist)56–58. Given that our ecosystems are often dominated by slow-growing oligotrophic bacteria25,27,28,58–61, e.g. the SAR11 bacterium that dominates the ocean ecosystem56, it is clear that rapid growth is often not the favorable trait to be selected across ecological environments. In addition, some clinically pathogens such as Mtb grow slowly and the slow growth phenotype is often associated with enhanced antibiotic tolerance, host adaptability and could further facilitate long-term latent infections and transmissions of pathogens62–67, posing the necessity of identifying the evolutionary driving force of slow growth phenotypes. Understanding the emergence of the diverse growth phenotypes of microbes is of fundamental importance for microbiology.
Recent studies have shown that a substantial fraction of proteome in bacteria could serve other important physiological objectives (e.g. adaptability and survival)14,68–71. These alternative objectives are often implemented at the cost of reduced growth rate as they drive proteome resources away from supporting biomass growth, leading to trade-offs70. Here we overview recent progress of trade-offs between growth and other physiological traits such as adaptability and survival, focusing on the mechanistic origins and physiological effects, and furthermore, highlighting their ecological implications in driving the emergence of phenotype diversity. We first introduce the trait of microbial adaptability to changing nutrient environments and its mechanistic link with proteome reserve. We then elaborate the trade-off mechanism between growth and adaptability across microbial species and its ecological implications in causing co-existence and phenotypic heterogeneity. We further discuss the trade-off relation between microbial growth and another core trait of fitness, survival and stress tolerance. We finally discuss how these fundamental trade-off principles could lead to the emergence of two major trophic phenotypes of microbes in our ecosystem: oligotrophic lifestyles versus copiotrophic lifestyles.
Proteome reserve promotes adaption to nutrient transition
Amino acid downshift
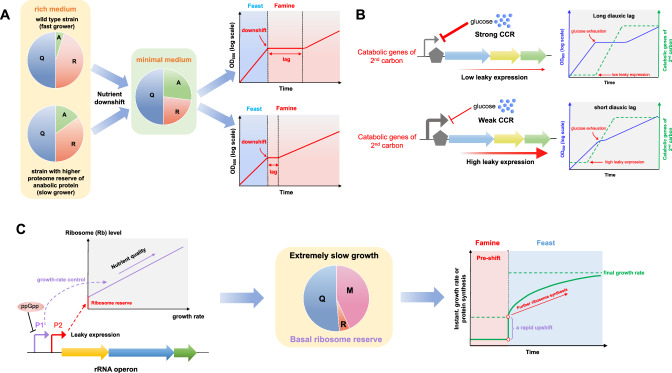
Since nutrient conditions are often highly fluctuating in nature, the adaptability to nutrient transition is crucial for microbial fitness14,19,68,72,73. The growth transition from good to poor nutrients is referred to as nutrient downshift while the opposite direction is referred to as nutrient upshift74. Upon a sudden nutrient downshift event, bacteria enter into a lag phase before fully adapting to the poor nutrient due to the time required for proteome re-allocation to meet the metabolic demand68,73,74. During the nutrient downshift from amino acid-supplemented rich medium to minimal medium, namely amino acid (AA) downshift, E. coli cells have to synthesize enough anabolic enzymes to supply the amino acid flux at the cost of downregulating ribosome synthesis19,35. A very recent study has shown that E. coli maintains a basal level of anabolic enzymes even when growing in nutrient broth14. The leaky expression of biosynthetic enzymes, namely proteome reserve, is actually crucial for the timely adaption of bacteria to sudden AA downshifts as it prevents the cells from being completely depleted of internal amino acid pools during downshift14. Slow-growing E. coli cells with increased basal levels of (p)ppGpp, harboring larger proteome reserve of anabolic proteins in rich medium, display much shorter lags during AA downshift and thus becomes a faster ‘switcher’ (Fig. 1A)19.
Fig. 1. Proteome reserve from leaky expressions promotes adaptability to nutrient fluctuation.
A During nutrient downshift from rich medium to minimal medium, bacterial cells have to re-allocate the proteome resource from ribosomes (R) to anabolic proteins (A). strains harboring a higher level of proteome reserve of anabolic proteins (A) could more quickly adapt to downshift with a shorter lag time than wild type strain. The Q sector denotes a growth-rate independent proteome sector for metabolic maintenance. B During carbon diauxic shift, strains of tight carbon catabolic repression (CCR) have low leaky expression for the catabolic genes of the 2nd carbon, and thus displaying a long diauxic lag. In contrast, strains of weak CCR have high leaky expression of related catabolic genes and could also trigger the induction of related catabolic genes before glucose exhaustion. As a result, strains of weak CCR display short or even no diauxic lag. C The control of rRNA synthesis contains two behaviors: a growth-rate dependent control mediated by (p)ppGpp on P1 promoter and a constitutive expression behavior from P2 promoter. The leaky expression of P2 promoter enables bacteria to maintain a basal ribosome reserve (R) in its proteome during extremely slow growth. The ribosome reserve allows bacteria to quickly initiate an upshift program during nutrient upshift from famine to feast conditions before making more ribosomes to further increase growth rate.
Carbon diauxie
Carbon diauxic shift is a special case of nutrient transition that first described by Jacques Monod, whereby bacteria consume all of a preferred carbon source (e.g. glucose) before utilizing the secondary carbon source, resulting in diauxic lags75,76. The diauxie phenomenon, also referred to as ‘glucose effect’, mainly results from the inhibitory effect of glucose on the transport and catabolism of the 2nd carbon via inducer exclusion77. Being similar to the case of AA downshift, the length of diauxic lag is tightly related to the leaky expression of the catabolic genes of the 2nd carbon source. For example, the glucose-lactose diauxic lag of E. coli could completely disappear if the leaky expression of lac operon is stimulated by addition of exogeneous cAMP78,79. In contrast, imposing proteome constraint by expressing non-required proteins could limit the expression of catabolic operons of the 2nd carbon and further prolong the diauxic lag69. A similar notion is also applicable to budding yeast. Compared with laboratory Saccharomyces cerevisiae, many natural isolates of S. cerevisiae and some other Saccharomyces species such as S. bayanus display much shorter or no diauxic lags during glucose-galactose shift due to a stronger leaky expression of the GAL gene (Fig. 1B)80–83. In addition, the induction of GAL genes could be bimodal for particular mixtures of glucose and galactose and for particular strains, leading to the emergence of two subpopulations including fast switcher (short diauxic lag) and slow switcher (long diauxic lag)80,82,84. Such a strategy of weakened carbon catabolic repression (CCR) is proposed to shift the yeast from is a ‘specialist’ to a ‘generalist’ in order to better adapt to changing environments80,84.
Mechanistically, both cAMP-CRP and (p)ppGpp regulate carbon diauxie of bacteria. cAMP-CRP promotes the emergence of carbon diauxic growth. CRP-cAMP activates the expression of glucose transporter (PtsG), enhancing the uptake of glucose and its conversion to glucose-6-phosphate, which then increases the dephosphorylated IIAGlc and further prevents lactose uptake by inhibiting LacY activity, namely inducer exclusion79. (p)ppGpp-mediated stringent response maintains transcription-translation coupling during carbon transitions, further ensuring the timely expression of the catabolic genes of the 2nd carbon source and facilitate the adaption of bacteria to the 2nd carbon19. As a result, the stringent-response deficient relA-null strain exhibits a longer diauxic lag.
Nutrient upshift
The strategy of proteome reserve is also crucial for bacteria during adaptation to nutrient upshift85,86. During nutrient upshift events, bacteria accelerate cell growth via upregulating ribosome synthesis and downregulating non-required metabolic proteins74. The P1 promoter of rrn operon is regulated by (p)ppGpp to achieve growth-rate dependent ribosome synthesis87. However, it has recently been found that E. coli and yeast cells maintain a ribosome reserve during slow growth conditions32,88–90, which comes from the leaky expression of rRNA genes (e.g. constitutive activity of rrn P2 promoter in E. coli)87. The ribosome reserve, being maintained in inactive states via either translation initiation inhibition or ribosome hibernation91–93, could be activated immediately during nutrient upshift to help microbes quickly resume protein synthesis and cell growth. In such case, microbes could significantly save the time required for de novo biosynthesis of more ribosomes (Fig. 1C)85,86.
Collectively, in both favorable and harsh nutrient environments, microbial cells manage to maintain a seemingly useless proteome reserve (either metabolic proteins or ribosomes). The proteome reserve, resulting from the leaky expression of related genes, is actually important for microbial cells to prepare for future changing environments and is thus advantageous for microbial fitness in fluctuating environments.
Trade-off between growth and adaptability
Although rapid growth and rapid adaption to nutrient fluctuations could in principle both promote microbial fitness, it is difficult to simultaneously optimize both traits by bacteria due to conflicts of resource allocation. For example, the proteome reserve of anabolic proteins in rich medium could limit the cellular ‘budget’ for ribosome synthesis, thus comprising the maximal growth capacity of bacteria14 (Fig. 1A). Artificial (p)ppGpp induction in rich medium, although favors faster adaption to AA downshift via increasing the proteome reserve of anabolic proteins, concurrently reduces cell growth by inhibiting ribosome synthesis19. In one word, the improvement of adaptability often occurs at the expense of reduced growth rate, resulting in trade-offs. Similarly, for those Saccharomyce variants with short or no diauxic lags, they exhibit lower growth rates in glucose medium than the laboratory S. cerevisiae strains with strong diauxic phenotype due to the cost of increased leaky expression of GAL genes81,83. A recent work has quantitatively established a negative relation between growth rate and diauxic lag for E. coli during transition from various glycolytic carbons to the gluconeogenic carbon, acetate68. They found that cells growing slowly on poor glycolytic carbons (e.g. mannose) require much shorter lags than cells growing rapidly on good carbons (e.g. glucose) during acetate downshift as slow growth enhances acetate catabolism such as glyoxylate shunt pathway. In addition to the trade-off between adaptability and growth, recent work has shown two other forms of trade-offs during glycolytic-gluconeogenic carbon transition that intrinsically rooted in metabolism and physiology94. Firstly, trade-off exists between lags to glycolysis and gluconeogenesis due to biochemical constraints of flux sensing. During shift between glycolytic and gluconeogenic carbons, metabolite levels could quickly collapse to their equilibrium so that cells could be unable to sense the proper direction of metabolic flux. Bacteria could alleviate this problem by selecting a preferred direction of regulation at the expense of increasing lag times in the opposite direction94. Species specialized in glycolysis (e.g. E. coli) have short lags during shift to glycolytic carbon but long lags during shift to gluconeogenic carbon while species specialized in gluconeogenesis (e.g. P. aeruginosa) follow the opposite behavior94. This notion of preferential directionality in central carbon metabolism (glycolysis or gluconeogenesis) has further been confirmed by the Cordero lab using 186 marine heterotrophic bacterial strains and 135 carbon sources95. Secondly, attempts of optimizing both directions (e.g. increasing the abundances of gluconeogenic enzymes in glycolytic growth or glycolytic enzymes in gluconeogenic growth) reduce both lag times simultaneously but at the expense of increasing futile cycling and energetic cost, resulting in trade-off between lags and futile cycling that could affect bacterial survival during stress94.
Co-existence of different phenotypes
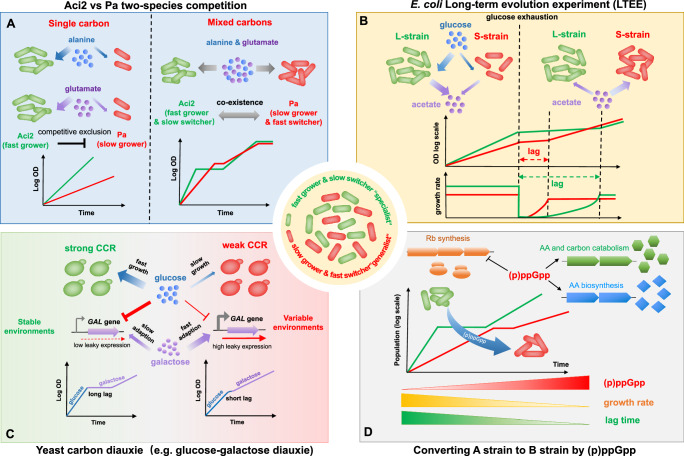
The trade-off between growth and adaptability is crucial in shaping microbial phenotypes and could often lead to the co-existence of different growth phenotypes (illustrated in Fig. 2). In a recent work by Bloxham et al., the authors investigated the coexistence of two species including Acinetobacter species (Aci2) and Pseudomonas aurantiaca (Pa)96. They found that Aci2 could competitively exclude Pa on either alanine or glutamate due to its faster growth on single carbon source. However, Pa has a much shorter lag than Aci2 during alanine-glutamate diauxic growth. Therefore, the ‘fast grower’ (Aci2) and the ‘fast switcher’ (Pa) could coexist when both alanine and glutamate are present. In this sense, the trade-off between growth and adaptability could allow slow-growing microbes to coexist with fast-growing microbes in multi-resource environments (Fig. 2A). Furthermore, Basan group has recently investigated the coexistence of different phenotypes in the E. coli long-term evolution experiment (LTEE)97. They identified two key phenotypes associated with LTEE: L-strain and S-strain (Fig. 2B). The L-strain is a fermentation specialist that grows more rapidly on glucose and could secret more fermentation products, acetate. In contrast, S-strain could switch more quickly to acetate utilization after glucose exhaustion, albeit at the cost of a lower growth rate on glucose. Therefore, co-existence again emerges between a ‘fast grower’ (L-strain) and a ‘fast switcher’ (S-strain) in the microecosystem of LTEE. It is fascinating that the co-existence in LTEE could robustly persist over thousands of generations of evolution without being replaced by fitter strains, which suggests that trade-off here is fundamental and at least is not easily overcome by minor tweaks in metabolism. Trade-off can thus result in evolutionarily stable coexistence of different phenotypes, which cannot be invaded and replaced by fitter strains, further resulting in evolutionarily stable ecosystems.
Fig. 2. Growth-adaptability trade-off leads to co-existence and phenotypic heterogeneity.
Two growth phenotypes could emerge in the population or microbial community: the ‘specialist’ population (fast grower & slow switcher) and the ‘generalist’ population (slow grower & fast switcher). A The co-existence of two bacterial species in the presence of multiple resources: Acinetobacter species (Aci2) and Pseudomonas aurantiaca (Pa). When alanine or glutamate is present as the sole carbon source, Aci2 could competitively exclude Pa due to its higher growth rates. However, these two strains could co-exist when both alanine and glutamate are available as Pa could more quickly adapt to the alanine-glutamate diauxic shift with a much shorter lag than Aci2. B Two subpopulations co-exist in the long-term evolution experiment (LTEE) of E. coli: the L-strain and S-strain. The L-strain grows faster than S-strain in glucose medium and could secret a higher amount of fermentation products, acetate, into the medium. However, S-strain could adapt more quickly to the growth transition to acetate at the cost of reduced growth rate in glucose medium. As a result, although growing more slowly than L-strain in glucose medium, S-strain could gain a fitness advantage during transition to acetate after the exhaustion of glucose. C During the carbon diauxic growth of budding yeast, stochastic gene expression generates two subpopulations: subpopulation A of strong carbon catabolite repression (CCR) could grow fast at glucose medium but requires a long lag phase to adapt to glucose-galactose diauxic shift; subpopulation B of weak CCR could grow slowly in glucose medium but adapt more quickly to the diauxic shift with a short or no lag. D (p)ppGpp simultaneously regulates bacterial growth and the adaptability to nutrient fluctuations. (p)ppGpp inhibits ribosome (Rb) synthesis but activates amino acid (AA) biosynthesis as well as some AA and carbon catabolism processes. As a result, (p)ppGpp reduces bacterial growth but meanwhile promotes the bacterial adaptation to various types of nutrient downshift. Therefore, (p)ppGpp acts as a key regulator that could convert the bacteria from fast grower & slow switcher to slow grower & fast switcher.
Bet-hedging via phenotypic heterogeneity
The trade-off between growth and adaptability poses challenges for microbes to adapt to an unpredictable environment. Phenotypic heterogeneity is an effective bet-hedging strategy for microbes to deal with the growth-adaptability trade-off. For example, during diauxic shift of S. cerevisiae, stochastic gene expression causes bimodality induction of the catabolic genes of the secondary carbon80,82, generating two subpopulations (Fig. 2C): a ‘fast grower’ (‘specialist’) subpopulation, with a stringent CCR, that displays a high growth rate in stable environments; a ‘fast switcher’ (‘generalist’) subpopulation, with a weak CCR, that could more quickly adapt to carbon transition at the cost of reduced growth rate in glucose due to high leaky gene expression. A similar phenomenon has been observed for Lactococcus lactis during glucose-cellobiose diauxie98. Therefore, phenotypic heterogeneity allows microbial population to balance growth and adaptability to maintain fitness in both stable and variable environments, achieving the objective of bet-hedging84. The emergence of phenotypic heterogeneity is found to be associated with metabolic heterogeneity during environmental changes99. The strategy of evolving phenotypic heterogeneity could have some specific fitness advantages over the simple strategy of adopting proteome re-allocation by a homogeneous population in highly fluctuating environments. In the former case, some subpopulations are capable of quickly adapting to the new environments with short lags, thus saving the time required for proteome re-allocation that could lead to long lags.
The effect of (p)ppGpp
In addition to CCR, (p)ppGpp signaling is also tightly related to the trade-off between growth and adaptability. In rich medium, (p)ppGpp induction inhibits bacterial growth but accelerate the bacterial adaption to AA downshift via triggering a global resource re-allocation from ribosome synthesis to anabolic biosynthesis19. Moreover, in minimal medium, (p)ppGpp induction could activate the catabolism of certain amino acids such as alanine and arginine, further shorten the lag of E. coli during carbon downshift (from glucose to alanine) and nitrogen downshift (from NH4Cl to alanine or arginine)100. Therefore, (p)ppGpp induction could convert the bacteria from a fast-grower & slow switcher to a fast-switcher & slow grower (Fig. 2D).
Trade-off between growth and survival
Growth-survival trade-off
To thrive in nature, bacterial cells must rapidly proliferate under favorable conditions but persist during stress. Therefore, survivability during stress is also a key trait that determines bacterial fitness, and thus bacteria adopt sophisticated molecular strategies to balance growth and survival101. The (p)ppGpp-mediated stringent response and RpoS-mediated general stress response, the two major signaling pathways involved in stress response in many bacterial species, are largely activated during stress such as nutrient deprivation while the levels of (p)ppGpp and RpoS are relatively low during exponential phase to avoid their inhibitory effects on cell growth102–104. Nevertheless, strong trade-offs between growth and survival do exist in bacteria in many circumstances. For example, clinically isolated pathogens often exhibit a slow growth phenotype which is proposed to facilitate the adaption and survival inside host62–66. Recent studies by Gerland group investigated the effect of growth rate on the survival of E. coli during nutrient starvation105,106. They find that the death rate of E. coli cells during nutrient starvation is negatively correlated with the growth rate of cells before entering into starvation as slow growth results in decreased maintenance rates of cells. Basan group further shows that a growth rate-dependent proteome response, especially on envelop proteins, contributes to the promotion of survival by slow growth107. A striking growth-survival trade-off, resulting from proteome resource competition, has recently been reported for the gram-positive model bacterium Bacillus subtilis70. The authors find that the resource allocation strategy of B. subtilis does not lead to growth maximization on various carbons sources. Knockout of a master regulator gene, spo0A, triggers a proteome re-allocation from survival & stress response pathway to biosynthesis pathways and further accelerates cell growth, however, at the cost of compromised bacterial survivability after nutrient deprivation due to a decreased stress adaptability.
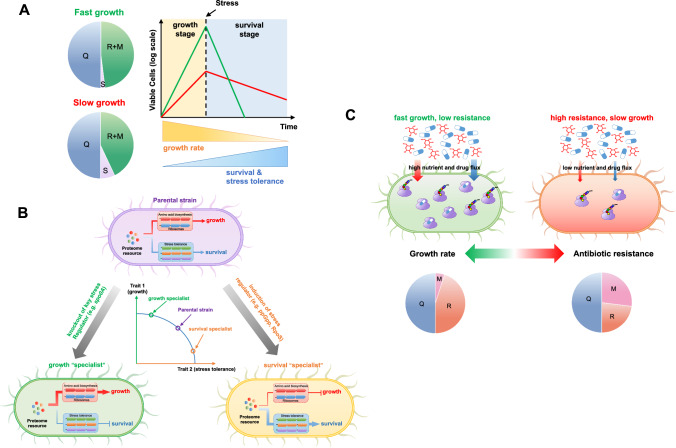
Mechanistically, the enhanced survivability during slow growth might attribute to an elevated stress response (Fig. 3A). The intracellular levels of key stress regulators, e.g. ppGpp and RpoS, are often growth rate dependent and increase significantly during slow growth102,108–111. Therefore, the leaky expressions of various stress responsive genes could also increase during slow growth and further facilitate bacterial survival under harsh environments112. In support of this picture, artificial induction of ppGpp or RpoS reduces cell growth but strongly enhances the stress tolerance of E. coli by triggering proteome re-allocations from biosynthesis to stress response100,109,113. In the case of B. subtilis, the positive effect of Spo0A on stress response could also be enlarged during slow growth due to an increased phosphorylation status of Spo0A under poor conditions114, imposing a stronger proteome burden on cell growth of B. subtilis in poor nutrients. As a result, the growth rate advantage of spo0A-null strain over wild type strain becomes even stronger on poor carbons70.
Fig. 3. Growth-survival trade-off.
A Slow growth could boost the survival and stress tolerance of bacteria. During slow growth conditions such as poor nutrients, bacteria could have a higher proteome investment on stress response (S), thus facilitating the long-term survival during sudden stress conditions. R+ M denotes the sum of growth rate dependent ribosomes (R) and metabolic sector (M). B Trade-off between growth and survival due to proteome allocation constraints. Growth and survival traits have proteome allocation conflicts and bacteria often need to balance the proteome investment on these two traits according to their environmental conditions. Knockout of some key stress regulators (e.g. spo0A in B. subtilis) favors growth at the cost of compromised survival, generating a growth ‘specialist’. In contrast, induction of certain stress regulators (e.g. ppGpp or RpoS protein) could favor survivability at the cost of reduced growth rate, generating a survival ‘specialist’. C An example of trade-off between growth and drug resistance against streptomycin for E. coli. Evolution favors the emergence of streptomycin resistant strains with compromised nutrient and drug uptake (right) so that the in vivo levels of both the drug and its ribosome targets decrease. As a result, resistance occurs at the cost of reduced drug-free growth rate due to decreased resource allocation towards ribosome synthesis (R sector) compared with native strain (left).
A similar example of growth-survival trade-off has been observed in the case of chemotaxis and motility, which are important for bacteria to survive in nutrient-limited ecological environments115. The chemotaxis and motility process of E. coli are positively regulated by CRP-cAMP and are thus upregulated on poor carbons116. Under carbon-limited conditions, a significant proteome resource is devoted to flagellar synthesis at the cost of reduced growth rates so that bacteria could become motile to search for more nutrient sources37,44. As a result, mutants devoid of flagellar synthesis gain a significant growth advantage over the parental strain in laboratory medium, especially on poor carbon sources116. Interestingly, Gude et al. has recently found that the growth-migration trade-off could also lead to co-existence between a fast-growing & slow-moving population and a fast-moving & slow-growing population117. They found that either of the two population could outcompete the other when low in relative abundance by active segregation and spatial exclusion within the patch. Such a type of inversion of the competitive hierarchy promotes the emergence of co-existence of the two populations and could, in principle, also promote phenotypic diversity in ecological niches.
The trade-off between growth and survival/stress response is not limited to bacteria but also found in eukaryotic microbes like yeast. A strikingly positive relation between division rate and death rate has been established in Schizosaccharomyces pombe118. Moreover, the expression of stress responsive genes is also negatively correlated with growth rate in S. cerevisiae, and thus slow growth also enhances the stress resistance of budding yeast119. Interestingly, trade-off between growth and mating also exists in S. cerevisiae due to the cost of gene expression120. Mutant devoid of mating gains a growth advantage during asexual stage while strains of increased mating efficiency mediated by GPA1 results in a growth disadvantage120.
Collectively, the fundamental growth-survival trade-off emerges as a result of conflicts of resource allocations. This principle could be taken advantage by microbial cells to convert their phenotype for surviving under various environmental conditions. For example, perturbing key signaling pathways (e.g. (p)ppGpp, RpoS, Spo0A) could favor either the growth trait or the survival trait by driving proteome resource towards either biosynthesis pathways or survival & stress response pathways (Fig. 3B)70,100,110,113,121.
Antibiotic tolerance and resistance
A widely concerned topic of bacterial survival is antibiotic tolerance and resistance, which are indicated by MDK (minimal duration for killing) and MIC (minimal inhibitory concentration) of bacteria during antibiotic treatment, respectively122. On one hand, it has long been proposed that slow growth could enhance drug tolerance. For example, slow-growing bacterial populations, associated with lower speeds of cell wall assembly and DNA replication, have higher tolerances (slower killing rate by antibiotics) than the fast-growing populations against β-lactams and fluoroquinolones treatment122,123. Tolerance could be enhanced by both inherited slow growth (e.g. slow growers such as Mtb) and non-inherited slow growth (e.g. due to poor nutrients)122. Increased drug tolerance could further favor the emergence of drug resistance124,125. Drug tolerance associated with slow growth phenotype has been observed for not only Mtb but also other pathogens such as Pseudomonas aeruginosa and Staphylococcus aureus62–64,66,125. A reversion of the slow growth phenotype, either by modulating key signaling pathways or adaptive evolution, could restore the antibiotic susceptibility and compromise the bacterial fitness inside host62,66. On the other hand, the development of antibiotic resistance is often associated with a fitness cost such as reduced drug-free growth rates126. The trade-off between resistance and growth could be due to several reasons: (1) because antibiotics target essential biological processes (e.g. translation, transcription and metabolism), resistance mutations could compromise the normal structures and functions of related macromolecules such as ribosome, RNA polymerase or metabolic enzymes and further reduce cell growth126–128; (2) resistance mediated by overexpression of resistance proteins such as efflux pumps or enzymes (e.g. beta-lactamase encoded in plasmids) imposes proteome burden and energy cost on bacteria126,129; (3) In a recent study focusing on evolution of antibiotic resistance against streptomycin130, the authors found that evolution favored the emergence of resistance mutants with attenuated antibiotic and nutrient uptake, which led to the reduction of both in vivo drug concentration and ribosome targets. Therefore, the cost of growth in such case is due to decreased nutrient uptake and ribosome synthesis (Fig. 3C).
Bet-hedging strategies to balance growth and survival
The fundamental growth-survival trade-off poses a challenge to microbial fitness as both of these two traits are crucial to sustain the bacterial population in natural niches, and therefore, bacteria sometimes manage to adopt bet-hedging strategies via generating phenotypic heterogeneity131. For example, B. subtilis initiates sporulation during unfavorable conditions such as starvation, however, only in a fraction of the population114,132. The rest cells could maintain growth with nutrients generated from cannibalism or autolysis70,133. Such a bet-hedging strategy allows the population to persist if the harsh environment continues or to quickly resume growth when the environments become favorable again. Starved E. coli population also adopts a bet-hedging strategy by evolving phenotypic heterogeneity when encountering new nutrient resources in order to break the fundamental trade-off between growth and survival134. The majority of cells could quickly resume growth after starvation with short lag times. Meanwhile, a small subpopulation with extreme long lags is retained and exhibits strong tolerance to environmental stressors such as antibiotics.
Oligotrophic versus copiotrophic lifestyles
Our current understanding of microbial physiology is primarily based on a few fast-growing model organisms such as E. coli and S. cerevisiae. According to the fundamental notion of r-/k- strategy, bacteria are conventionally divided into two classes53,57–59,135,136: the fast-growing copiotrophs (r-strategist) thriving in environments with high nutrient opportunities; slow-growing oligotrophs (k-strategist) living in nutrient-limited environments. The classification of copiotroph and oligotroph, although might be over-simplified, pinpoints to an important reality that our ecosystems are mainly dominated by slow-growing bacteria25,28,58–61. Perhaps the most well-known example lies at the oligotrophic bacterium SAR1156,137 and Prochlorococcus138,139, which are among the smallest but most abundant microorganisms on earth and constitute nearly half of all planktonic cells in ocean. Compared with common copiotrophic bacteria, the growth of oligotrophic bacteria such as SAR11 and Prochlorococcus are much slower (days per doubling). Furthermore, they have streamlined genomes (~1.5 Mb) and very small cell sizes (~0.1 μm3)56,137–139.
Growth efficiency and stress resistance
Direct comparisons between oligotrophs and copiotrophs highlight the intriguing trade-offs between growth rate and other traits such as growth efficiency26,52,60,140–142. It has been proposed that high-resource environments could favor faster growth but more energetically wasteful lifestyle (e.g. fermentation) due to increased competition while nutrient-limited environments favor relatively slow growth but more energy-efficient lifestyle (e.g. respiration)44,141. Although growing much more slowly, oligotrophic bacteria exhibit higher growth efficiency than copiotrophic bacteria26,52,60, especially in nutrient-limited environments as their high-affinity but low-specificity nutrient transport systems allow simultaneous uptake of mixed substrates26–28,140. The high growth efficiency of oligotrophic bacteria is associated with a low maintenance energy which could result from a minimization of the costs of various energy-consuming processes136,140,142. For example, the high-affinity, low-specificity transport systems enable oligotrophic bacteria to economically utilize multiple substrates under oligotrophic conditions with a relatively low amount of energy-intensive ABC transport systems27,136. Oligotrophic bacteria also tend to be non-motile136,143 and their genome streamlining properties53,144could further save the energy cost associated with macromolecular biosynthesis.
Strikingly, oligotrophic bacteria like Sphingopyxis alaskensis also display higher inherent stress resistance/tolerance than common copiotrophic bacteria against the treatment of hydrogen peroxide, high temperature, ethanol and UV during exponential growing26,27,136,145. Moreover, they could also sustain for a long time with no apparent loss of viability during nutrient starvation26,146. Many possibilities exist regarding the inherent stress resistance: bacteria have a constitutively high proteome investment on stress response; possession of some special stress protective mechanisms or/and the extremely slow growth itself simply results in stronger survivability.
Constitutive gene expression and fast adaption
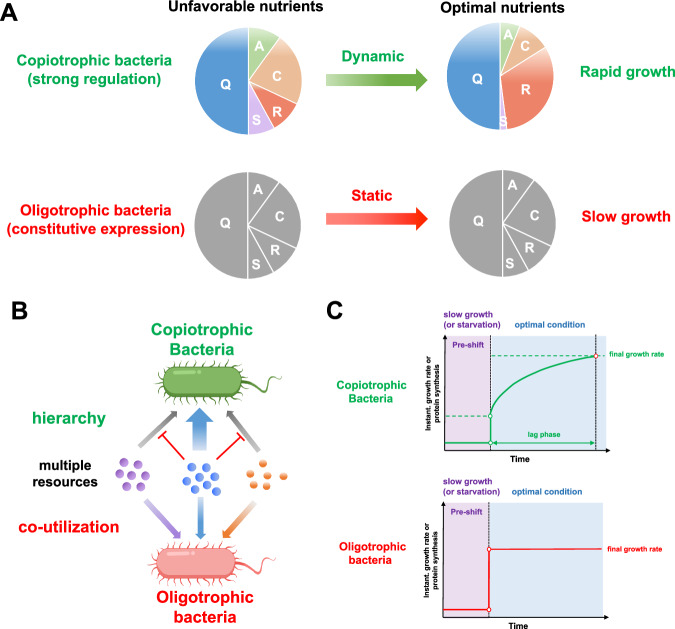
Compared with copiotrophic bacteria, oligotrophic bacteria exhibit a highly distinct feature of gene regulation: a significant reduction of transcription regulation with fewer sigma factors and two-component systems53,56,143. As a result, gene expressions in oligotrophic bacteria are highly constitutive and lack of growth-rate dependent control143. For example, oligotrophic bacteria, SAR11 and SAR92 are lack of growth control of ribosome synthesis147, a core mechanism employed by copiotrophic bacteria to achieve rapid growth in rich medium111. A series of global ‘omics’ studies have shown that oligotrophic bacteria exhibit weak alterations in transcriptome and proteome during changing growth conditions such as nitrogen limitation, sulfur limitation, glucose limitation as well as transition from exponential to stationary phase143,145,148–150. Transcriptome analysis suggests that only 0.1% of protein-encoding genes appear to be under transcriptional control in SAR11149. The constitutive gene expression pattern is not limit to bacteria and has also been found in methanogenic, oligotrophic archaeon Methanococcus maripaludis151. The abundances of various core proteome sectors such as translation machinery, catabolic proteins and anabolic proteins remain almost invariant across growth conditions for M. maripaludis. Instead, its modulation of the overall translation rate could be achieved by regulating ribosome activity rather than ribosome numbers151. Collectively, copiotrophic bacteria and oligotrophic bacteria follow distinct gene regulation strategies. A strong transcription regulation, e.g. mediated by (p)ppGpp and cAMP, enables copiotrophic bacteria to dynamically modulate the proteome allocation and achieve rapid growth via maximizing ribosome synthesis during favorable conditions5,6,13. In contrast, the constitutive gene expression pattern leads to a nearly static resource allocation strategy in oligotrophic bacteria and further limit the ribosome synthesis and maximal growth capacity (illustrated in Fig. 4A).
Fig. 4. The oligotrophic versus copiotrophic strategies.
A Proposed proteome allocation strategies for copiotrophic and oligotrophic bacteria. During transition from unfavorable nutrients to optimal nutrients, the strong transcription regulation in copiotrophic bacteria enables efficient dynamic proteome re-allocation among various sectors including catabolic proteins (C), anabolic proteins (A), ribosomes (R) and stress response proteins (S). As a result, ribosome synthesis could be maximized in optimal conditions to support fast growth. In contrast, the gene expressions of oligotrophic bacteria are largely constitutive, resulting in a static proteome allocation that limits ribosome synthesis and cell growth. B In the presence of multiple resources, the strong transcription regulation in copiotrophic bacteria such as catabolite repression leads to hierarchical utilization of substrates. In contrast, the constitutive expression strategy could enable oligotrophic bacteria to simultaneously utilize multiple substrates. C During nutrient upshift from unfavorable condition (poor nutrients or starvation) to optimal condition, copiotrophic bacteria need to re-allocate the proteome resource for maximizing ribosome synthesis to reach the final growth rate and thus require a lag time. Instead, oligotrophic bacteria adopt constitutive expression strategy and thus could reach the final growth rate immediately after a nutrient upshift process.
As described above for copiotrophic bacteria such as E. coli, the proteome reserve from the leaky expression (constitutive activity) of the promoters of metabolic proteins and ribosomes (Fig. 1), although limits growth rate, converts bacteria from ‘specialists’ to ‘generalists’ to efficiently utilize multiple resources with short or no delay. Due to the lack of transcription regulation, the constitutive expression strategy is much more prevalent in oligotrophic bacteria than copiotrophic bacteria143. In consistent with the constitutive expression strategy, oligotrophic bacteria could simultaneously utilize multiple substrates (Fig. 4B) and display no growth lags upon nutrient transition (Fig. 4C)26,136, allowing them to make full use of the limited nutrient resources available without any delays that could otherwise be associated with transcription reprograming process observed in copiotrophic bacteria143.
‘Oligotrophic state’ in copiotrophic bacteria
Although copiotrophic bacteria differ significantly from oligotrophic bacteria in the gene regulation strategy and lifestyle, it should be noted that sometimes an ‘oligotrophic state’ strategy is still retained in those fast-growing copiotrophic bacteria. For example, it has recently been found that a very small subpopulation of non-sporulating B. subtilis cells could enter into an extreme slow-growing ‘oligotrophic state’ during long-term starvation, which facilitates long-term survival and enables cells to become highly tolerant to stress and antibiotic treatment152. Moreover, the small fraction of slow-growing persister cells in the populations of E. coli and many clinical pathogens might also be viewed as a special case of oligotrophic lifestyle122,153,154. In this sense, copiotrophic bacteria still maintain a minimal set of oligotrophic strategies (e.g. proteome reserve strategy) as a bet-hedging approach to adapt to variable environments.
Future perspectives and conclusion
Understanding the emergence of diversity of microbial growth phenotypes across species and environments is a central topic of microbiology. Our review here highlights the importance of trade-off principles in shaping the growth phenotypes of microbial cells across species and environments. It has recently been suggested that trade-off lies at the core of determining the qualities of different nutrient substrates155. Rather than reflecting fundamental biochemical or biophysical limitations, nutrient quality is largely a self-determined, plastic property that reflects the safety, reliability, and profitability of different ecological environments for a particular species and could be quickly adapted by evolution if necessary155. Microbial cells could employ nutrients as a signal to infer information about their environments based on their experiences in natural niches and further implement a trade-off decision of fast growth versus preparation for changing environments. Accordingly, bacteria could sense low-quality carbon sources as a signal of environmental deterioration so they activate the expressions of many proteins related to adaptability and stress response by ppGpp and cAMP signaling as a strategy to prepare for the changing conditions155. Therefore, trade-off between growth and adaptability/survival in proteome allocation could thus set the qualities of different carbon sources and result in the simple bacterial growth laws31,156. From this view, trade-offs in combination with ecological conditions could be the root cause of different growth phenotypes. We propose here that achieving rapid growth is just one aspect of concerns of microbial cells that does not have a higher priority over other physiological traits like adaptation and survival. Those alternative physiological traits could affect growth rates at which bacteria have evolved to grow, as much as growth rates affect them. This point is vividly illustrated by coexistence in the LTEE, where evolution pushes S-strain to slow down its growth rate and the slow growth phenotype further facilitates this strain to occupy the ecological niche. Trade-offs can thus be selected for and be the root cause of slow growth rates. This notion is perhaps further reinforced by the fact of the wide distribution of slow-growing oligotrophic bacteria in our ecosystems, considering that oligotrophic bacteria often display better performances on alternative traits such as long-term survival than the fast-growing copiotrophic bacteria.
From a general perspective, the trade-off principles in growth control discussed here directly link three key levels of microbiological disciplines (Fig. 5). At molecular level, it is closely related to the fundamental design principle of gene regulation and resource allocation strategy in bacteria. At physiological level, it connects microbial growth with many other important physiological traits such as survival and adaption, stress and antibiotic tolerance. It is ultimately related to the ecological level with important implications in the emergences of various ecological phenomena such as co-existence, population heterogeneity and oligotrophic/copiotrophic lifestyles.
Fig. 5. Trade-off principles of microbial growth control links three levels of microbiological disciplines.
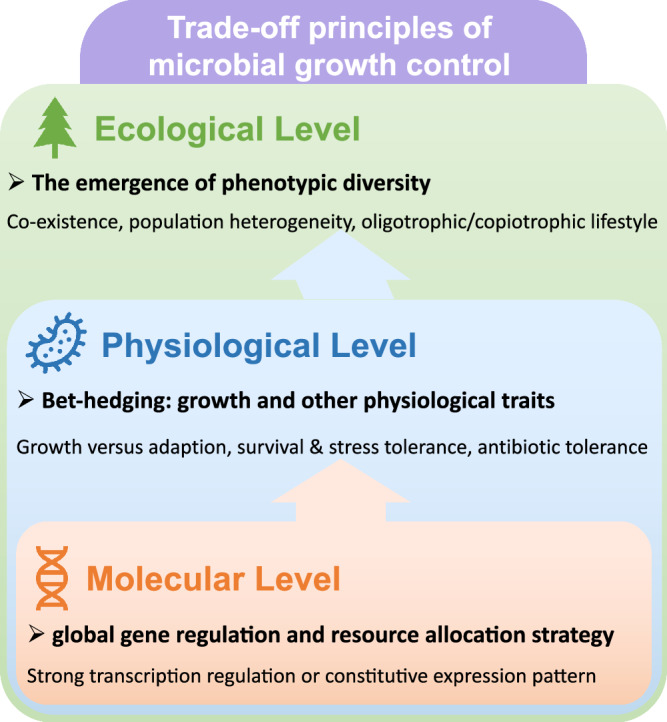
At molecular level, it is tightly related to the global gene regulation and resource allocation strategy of microbial cells. At physiological level, it is related to the bet-hedging strategy of microbial cells to balance growth and other key physiological traits such as adaptability, survival and stress tolerance as well as antibiotic tolerance. At ecology level, it is closely related to the emergency of phenotypic diversity that associated with many ecological phenomena such as co-existence of different phenotypes, population heterogeneity and the occurrence of oligotrophic/copiotrophic lifestyles.
Our current understanding of bacterial growth control is still largely limited to a few species. The proteome resource allocation strategy across conditions remains to be explored for those slow-growing oligotrophic species including extremophiles and archaea. It remains a grand challenge to identify key trade-off principles that drive the emergence of distinct growth phenotypes across species and across different ecological environments. To this end, it could be plausible to mimic certain natural environments of bacterial species in the laboratory and further interchange the living environments of different species to see how it could affect the evolution of growth and other related phenotypes. In addition, large interspecies variations in growth phenotypes emerge even among different natural isolates of the same species81,157. It is therefore important to investigate their distinct strategies of resource allocation and, furthermore, the underlying trade-off driving force. An ultimate challenge is whether we could quantitatively predict the evolutionary route of growth phenotypes from a long-term evolutionary scale. In another direction, trade-off principles shape the growth phenotype of many clinically related pathogens, further playing a key role in the evolvement of virulence and antibiotic tolerance. Mechanistic insights into the trade-off principles would guide the selection of proper antimicrobial drugs as well as development of novel antimicrobial therapies that further compromise the fitness of pathogens.
Acknowledgements
We thank Terry Hwa and various colleagues for useful discussions over these years. We thank Yiheng Wang for suggestions of this work. This work was supported by the National Key Research and Development Program of China (2022YFF1000400), National Natural Science Foundation of China (32270034, 32370044), Changjiang Young Scholar Program of Chinese Ministry of Education, Natural Science Funds for Distinguished Young Scholar of Hubei Province (2022CFA044), Wuhan Science and Technology Major Project (2023020302020708) and the Fundamental Research Funds for the Central Universities.
Author contributions
D.X. conceptualized this work. Z.M. and D.X. wrote and revised the manuscript together.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bremer, H. & Dennis, P. P. Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus3, 10.1128/ecosal.5.2.3 (2008). [DOI] [PubMed]
- 2.Reyes-Lamothe R, Sherratt DJ. The bacterial cell cycle, chromosome inheritance and cell growth. Nat. Rev. Microbiol. 2019;17:467–478. doi: 10.1038/s41579-019-0212-7. [DOI] [PubMed] [Google Scholar]
- 3.Dai X, Zhu M. Coupling of ribosome synthesis and translational capacity with cell growth. Trends Biochem. Sci. 2020;45:681–692. doi: 10.1016/j.tibs.2020.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Bruggeman FJ, Planqué R, Molenaar D, Teusink B. Searching for principles of microbial physiology. FEMS Microbiol. Rev. 2020;44:821–844. doi: 10.1093/femsre/fuaa034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belliveau NM, et al. Fundamental limits on the rate of bacterial growth and their influence on proteomic composition. Cell Syst. 2021;12:924–944.e922. doi: 10.1016/j.cels.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott M, Hwa T. Shaping bacterial gene expression by physiological and proteome allocation constraints. Nat. Rev. Microbiol. 2022;21:327–342. doi: 10.1038/s41579-022-00818-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klumpp S, Scott M, Pedersen S, Hwa T. Molecular crowding limits translation and cell growth. Proc. Natl Acad. Sci. USA. 2013;110:16754–16759. doi: 10.1073/pnas.1310377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott M, Klumpp S, Mateescu EM, Hwa T. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 2014;10:747. doi: 10.15252/msb.20145379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iyer S, Le D, Park BR, Kim M. Distinct mechanisms coordinate transcription and translation under carbon and nitrogen starvation in Escherichia coli. Nat. Microbiol. 2018;3:741–748. doi: 10.1038/s41564-018-0161-3. [DOI] [PubMed] [Google Scholar]
- 10.Zhu M, Mori M, Hwa T, Dai X. Disruption of transcription-translation coordination in Escherichia coli leads to premature transcriptional termination. Nat. Microbiol. 2019;4:2347–2356. doi: 10.1038/s41564-019-0543-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balakrishnan R, et al. Principles of gene regulation quantitatively connect DNA to RNA and proteins in bacteria. Science. 2022;378:eabk2066. doi: 10.1126/science.abk2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maaløe, O. Biological Regulation and Development (ed. Goldberger R. F.) (Plenum, 1979).
- 13.Kostinski S, Reuveni S. Ribosome composition maximizes cellular growth rates in E. coli. Phys. Rev. Lett. 2020;125:028103. doi: 10.1103/PhysRevLett.125.028103. [DOI] [PubMed] [Google Scholar]
- 14.Wu C, et al. Enzyme expression kinetics by Escherichia coli during transition from rich to minimal media depends on proteome reserves. Nat. Microbiol. 2023;8:347–359. doi: 10.1038/s41564-022-01310-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bosdriesz E, Molenaar D, Teusink B, Bruggeman FJ. How fast-growing bacteria robustly tune their ribosome concentration to approximate growth-rate maximization. FEBS J. 2015;282:2029–2044. doi: 10.1111/febs.13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorgensen BB, Boetius A. Feast and famine–microbial life in the deep-sea bed. Nat. Rev. Microbiol. 2007;5:770–781. doi: 10.1038/nrmicro1745. [DOI] [PubMed] [Google Scholar]
- 17.Navarro Llorens JM, Tormo A, Martínez-García E. Stationary phase in gram-negative bacteria. FEMS Microbiol. Rev. 2010;34:476–495. doi: 10.1111/j.1574-6976.2010.00213.x. [DOI] [PubMed] [Google Scholar]
- 18.Holscher HD. The gut microbiome in feast and famine. Nat. Rev. Gastroenterol. Hepatol. 2021;18:749–750. doi: 10.1038/s41575-021-00514-5. [DOI] [PubMed] [Google Scholar]
- 19.Zhu M, Dai X. Stringent response ensures the timely adaptation of bacterial growth to nutrient downshift. Nat. Commun. 2023;14:467. doi: 10.1038/s41467-023-36254-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conway, T., Krogfelt, K. A. & Cohen, P. S. The Life of Commensal Escherichia coli in the Mammalian Intestine. EcoSal Plus1, 10.1128/ecosalplus.8.3.1.2 (2004). [DOI] [PubMed]
- 21.Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: major fermentation by-products and their impact on host health. Microbiome. 2019;7:91. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 23.Cole BJ, et al. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 2017;15:e2002860. doi: 10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy A, et al. Genomic features of bacterial adaptation to plants. Nat. Genet. 2017;50:138–150. doi: 10.1038/s41588-017-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.López JL, et al. Growth rate is a dominant factor predicting the rhizosphere effect. ISME J. 2023;17:1396–1405. doi: 10.1038/s41396-023-01453-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavicchioli R, Ostrowski M, Fegatella F, Goodchild A, Guixa-Boixereu N. Life under nutrient limitation in oligotrophic marine environments: an eco/physiological perspective of Sphingopyxis alaskensis (formerly Sphingomonas alaskensis) Micro. Ecol. 2003;45:203–217. doi: 10.1007/s00248-002-3008-6. [DOI] [PubMed] [Google Scholar]
- 27.T. J. Williams, F. Joux, F. M. Lauro, S. Matallana-Surget, R. Cavicchioli, “Physiology of Marine Oligotrophic Ultramicrobacteria” in Extremophiles Handbook, K. Horikoshi, Ed. p. 1179-1199 (Springer Japan, 2011).
- 28.D. H. Hayakawa, M. J. Huggett, M. S. Rappé, “Ecology and Cultivation of Marine Oligotrophic Bacteria” in Extremophiles Handbook, K. Horikoshi, Ed. p. 1161-1178 (Springer Japan, 2011).
- 29.Dai T, et al. Nutrient supply controls the linkage between species abundance and ecological interactions in marine bacterial communities. Nat. Commun. 2022;13:175. doi: 10.1038/s41467-021-27857-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. Interdependence of cell growth and gene expression: origins and consequences. Science. 2010;330:1099–1102. doi: 10.1126/science.1192588. [DOI] [PubMed] [Google Scholar]
- 31.You C, et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metzl-Raz E, et al. Principles of cellular resource allocation revealed by condition-dependent proteome profiling. Elife. 2017;6:e28034. doi: 10.7554/eLife.28034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomáš, et al. Quantitative insights into the cyanobacterial cell economy. Elife. 2019;8:e42508. doi: 10.7554/eLife.42508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bjorkeroth J, et al. Proteome reallocation from amino acid biosynthesis to ribosomes enables yeast to grow faster in rich media. Proc. Natl Acad. Sci. USA. 2020;117:21804–21812. doi: 10.1073/pnas.1921890117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt A, et al. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 2016;34:104–110. doi: 10.1038/nbt.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori M, et al. From coarse to fine: the absolute Escherichia coli proteome under diverse growth conditions. Mol. Syst. Biol. 2021;17:e9536. doi: 10.15252/msb.20209536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xia J, et al. Proteome allocations change linearly with the specific growth rate of Saccharomyces cerevisiae under glucose limitation. Nat. Commun. 2022;13:2819. doi: 10.1038/s41467-022-30513-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu XP, Dourado H, Schubert P, Lercher MJ. The protein translation machinery is expressed for maximal efficiency in Escherichia coli. Nat. Commun. 2020;11:5260. doi: 10.1038/s41467-020-18948-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gourse RL, et al. Transcriptional responses to ppGpp and DksA. Annu Rev. Microbiol. 2018;72:163–184. doi: 10.1146/annurev-micro-090817-062444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu M, Dai X. Growth suppression by altered (p)ppGpp levels results from non-optimal resource allocation in Escherichia coli. Nucleic Acids Res. 2019;47:4684–4693. doi: 10.1093/nar/gkz211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kochanowski K, et al. Global coordination of metabolic pathways in Escherichia coli by active and passive regulation. Mol. Syst. Biol. 2021;17:e10064. doi: 10.15252/msb.202010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl Acad. Sci. USA. 2013;110:10039–10044. doi: 10.1073/pnas.1215283110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Basan M, et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature. 2015;528:99–104. doi: 10.1038/nature15765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Basan M. Resource allocation and metabolism: the search for governing principles. Curr. Opin. Microbiol. 2018;45:77–83. doi: 10.1016/j.mib.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 46.Malina C, Yu R, Björkeroth J, Kerkhoven EJ, Nielsen J. Adaptations in metabolism and protein translation give rise to the Crabtree effect in yeast. Proc. Natl Acad. Sci. USA. 2021;118:e2112836118. doi: 10.1073/pnas.2112836118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molenaar D, van Berlo R, de Ridder D, Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 2009;5:323. doi: 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dashko S, Zhou N, Compagno C, Piskur J. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 2014;14:826–832. doi: 10.1111/1567-1364.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Basan M, Hui S, Williamson JR. ArcA overexpression induces fermentation and results in enhanced growth rates of E. coli. Sci. Rep. 2017;7:11866. doi: 10.1038/s41598-017-12144-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conway T. The Entner-Doudoroff pathway: history, physiology and molecular biology. FEMS Microbiol. Rev. 1992;9:1–27. doi: 10.1111/j.1574-6968.1992.tb05822.x. [DOI] [PubMed] [Google Scholar]
- 52.Roller BRK, Stoddard SF, Schmidt TM. Exploiting rRNA operon copy number to investigate bacterial reproductive strategies. Nat. Microbiol. 2016;1:16160. doi: 10.1038/nmicrobiol.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissman JL, Hou S, Fuhrman JA. Estimating maximal microbial growth rates from cultures, metagenomes, and single cells via codon usage patterns. Proc. Natl Acad. Sci. USA. 2021;118:e2016810118. doi: 10.1073/pnas.2016810118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoff J, et al. Vibrio natriegens: an ultrafast-growing marine bacterium as emerging synthetic biology chassis. Environ. Microbiol. 2020;22:4394–4408. doi: 10.1111/1462-2920.15128. [DOI] [PubMed] [Google Scholar]
- 55.Cook GM, Berney M, Gebhard S, Heinemann M, Niederweis M. Physiology of mycobacteria. Adv. Micro. Physiol. 2009;55:81–182. doi: 10.1016/S0065-2911(09)05502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giovannoni SJ. SAR11 bacteria: the most abundant plankton in the oceans. Ann. Rev. Mar. Sci. 2017;9:231–255. doi: 10.1146/annurev-marine-010814-015934. [DOI] [PubMed] [Google Scholar]
- 57.Ho, A., Di Lonardo, D. P. & Bodelier, P. L. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol.93, fix006 (2017). [DOI] [PubMed]
- 58.Stone BWG, et al. Life history strategies among soil bacteria-dichotomy for few, continuum for many. ISME J. 2023;17:611–619. doi: 10.1038/s41396-022-01354-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Y, Neilson JW, Kushwaha P, Maier RM, Barberán A. Life-history strategies of soil microbial communities in an arid ecosystem. ISME J. 2021;15:649–657. doi: 10.1038/s41396-020-00803-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, et al. Substrate utilization and competitive interactions among soil bacteria vary with life-history strategies. Front. Microbiol. 2022;13:914472. doi: 10.3389/fmicb.2022.914472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caro TA, McFarlin J, Jech S, Fierer N, Kopf S. Hydrogen stable isotope probing of lipids demonstrates slow rates of microbial growth in soil. Proc. Natl Acad. Sci. USA. 2023;120:e2211625120. doi: 10.1073/pnas.2211625120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baek SH, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. PLoS Biol. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaiser P, et al. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol. 2014;12:e1001793. doi: 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kopf SH, et al. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc. Natl Acad. Sci. USA. 2016;113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pontes MH, Groisman EA. Slow growth determines nonheritable antibiotic resistance in Salmonella enterica. Sci. Signal. 2019;12:eaax3938. doi: 10.1126/scisignal.aax3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.La Rosa R, Rossi E, Feist AM, Johansen HK, Molin S. Compensatory evolution of Pseudomonas aeruginosa’s slow growth phenotype suggests mechanisms of adaptation in cystic fibrosis. Nat. Commun. 2021;12:3186. doi: 10.1038/s41467-021-23451-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dutta NK, Karakousis PC. Latent tuberculosis infection: myths, models, and molecular mechanisms. Microbiol. Mol. Biol. Rev. 2014;78:343–371. doi: 10.1128/MMBR.00010-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Basan M, et al. A universal trade-off between growth and lag in fluctuating environments. Nature. 2020;584:470–474. doi: 10.1038/s41586-020-2505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balakrishnan R, de Silva RT, Hwa T, Cremer J. Suboptimal resource allocation in changing environments constrains response and growth in bacteria. Mol. Syst. Biol. 2021;17:e10597. doi: 10.15252/msb.202110597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu M, et al. A fitness trade-off between growth and survival governed by Spo0A-mediated proteome allocation constraints in Bacillus subtilis. Sci. Adv. 2023;9:eadg9733. doi: 10.1126/sciadv.adg9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balakrishnan R, Cremer J. Conditionally unutilized proteins and their profound effects on growth and adaptation across microbial species. Curr. Opin. Microbiol. 2023;75:102366. doi: 10.1016/j.mib.2023.102366. [DOI] [PubMed] [Google Scholar]
- 72.Nguyen J, et al. A distinct growth physiology enhances bacterial growth under rapid nutrient fluctuations. Nat. Commun. 2021;12:3662. doi: 10.1038/s41467-021-23439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kratz JC, Banerjee S. Dynamic proteome trade-offs regulate bacterial cell size and growth in fluctuating nutrient environments. Commun. Biol. 2023;6:486. doi: 10.1038/s42003-023-04865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Erickson DW, et al. A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature. 2017;551:119–123. doi: 10.1038/nature24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Monod J. The growth of bacterial cultures. Annu. Rev. Microbiol. 1949;3:371–394. doi: 10.1146/annurev.mi.03.100149.002103. [DOI] [Google Scholar]
- 76.Blaiseau PL, Holmes AM. Diauxic Inhibition: Jacques Monod’s Ignored Work. J. Hist. Biol. 2021;54:175–196. doi: 10.1007/s10739-021-09639-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 78.Inada T, Kimata K, Aiba H. Mechanism responsible for glucose-lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 79.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc. Natl Acad. Sci. USA. 1997;94:12914–12919. doi: 10.1073/pnas.94.24.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.New AM, et al. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol. 2014;12:e1001764. doi: 10.1371/journal.pbio.1001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang J, et al. Natural variation in preparation for nutrient depletion reveals a cost-benefit tradeoff. PLoS Biol. 2015;13:e1002041. doi: 10.1371/journal.pbio.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Venturelli OS, Zuleta I, Murray RM, El-Samad H. Population diversification in a yeast metabolic program promotes anticipation of environmental shifts. PLoS Biol. 2015;13:e1002042. doi: 10.1371/journal.pbio.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Roop JI, Chang KC, Brem RB. Polygenic evolution of a sugar specialization trade-off in yeast. Nature. 2016;530:336–339. doi: 10.1038/nature16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siegal ML. Shifting sugars and shifting paradigms. PLoS Biol. 2015;13:e1002068. doi: 10.1371/journal.pbio.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mori M, Schink S, Erickson DW, Gerland U, Hwa T. Quantifying the benefit of a proteome reserve in fluctuating environments. Nat. Commun. 2017;8:1225. doi: 10.1038/s41467-017-01242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Korem Kohanim Y, et al. A bacterial growth law out of steady state. Cell Rep. 2018;23:2891–2900. doi: 10.1016/j.celrep.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 87.Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- 88.Dai X, et al. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol. 2016;2:16231. doi: 10.1038/nmicrobiol.2016.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li SH, et al. Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 2018;3:939–947. doi: 10.1038/s41564-018-0199-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yu R, et al. Nitrogen limitation reveals large reserves in metabolic and translational capacities of yeast. Nat. Commun. 2020;11:1881. doi: 10.1038/s41467-020-15749-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prossliner T, Skovbo Winther K, Sørensen MA, Gerdes K. Ribosome hibernation. Annu Rev. Genet. 2018;52:321–348. doi: 10.1146/annurev-genet-120215-035130. [DOI] [PubMed] [Google Scholar]
- 92.Diez S, Ryu J, Caban K, Gonzalez RL, Jr., Dworkin J. The alarmones (p)ppGpp directly regulate translation initiation during entry into quiescence. Proc. Natl Acad. Sci. USA. 2020;117:15565–15572. doi: 10.1073/pnas.1920013117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vinogradova DS, et al. How the initiating ribosome copes with ppGpp to translate mRNAs. PLoS Biol. 2020;18:e3000593. doi: 10.1371/journal.pbio.3000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schink SJ, et al. Glycolysis/gluconeogenesis specialization in microbes is driven by biochemical constraints of flux sensing. Mol. Syst. Biol. 2022;18:e10704. doi: 10.15252/msb.202110704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gralka M, Pollak S, Cordero OX. Genome content predicts the carbon catabolic preferences of heterotrophic bacteria. Nat. Microbiol. 2023;8:1799–1808. doi: 10.1038/s41564-023-01458-z. [DOI] [PubMed] [Google Scholar]
- 96.Bloxham B, Lee H, Gore J. Diauxic lags explain unexpected coexistence in multi-resource environments. Mol. Syst. Biol. 2022;18:e10630. doi: 10.15252/msb.202110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mukherjee A, et al. Coexisting ecotypes in long-term evolution emerged from interacting trade-offs. Nat. Commun. 2023;14:3805. doi: 10.1038/s41467-023-39471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Solopova A, et al. Bet-hedging during bacterial diauxic shift. Proc. Natl Acad. Sci. USA. 2014;111:7427–7432. doi: 10.1073/pnas.1320063111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Şimşek E, Kim M. The emergence of metabolic heterogeneity and diverse growth responses in isogenic bacterial cells. ISME J. 2018;12:1199–1209. doi: 10.1038/s41396-017-0036-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu M, Mu H, Dai X. Integrated control of bacterial growth and stress response by (p)ppGpp in Escherichia coli: a seesaw fashion. iScience. 2024;27:108818. doi: 10.1016/j.isci.2024.108818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Spira B, Ospino K. Diversity in E. coli (p)ppGpp levels and its consequences. Front. Microbiol. 2020;11:1759. doi: 10.3389/fmicb.2020.01759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hengge R. Stationary-phase gene regulation in Escherichia coli. EcoSal. 2011;4:1703. doi: 10.1128/ecosalplus.5.6.3. [DOI] [PubMed] [Google Scholar]
- 103.Battesti A, Majdalani N, Gottesman S. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Irving SE, Choudhury NR, Corrigan RM. The stringent response and physiological roles of (pp)pGpp in bacteria. Nat. Rev. Microbiol. 2021;19:256–271. doi: 10.1038/s41579-020-00470-y. [DOI] [PubMed] [Google Scholar]
- 105.Schink SJ, Biselli E, Ammar C, Gerland U. Death rate of E. coli during starvation is set by maintenance cost and biomass recycling. Cell Syst. 2019;9:64–73.e63. doi: 10.1016/j.cels.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 106.Biselli E, Schink SJ, Gerland U. Slower growth of Escherichia coli leads to longer survival in carbon starvation due to a decrease in the maintenance rate. Mol. Syst. Biol. 2020;16:e9478. doi: 10.15252/msb.20209478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schink S, Ammar C, Chang YF, Zimmer R, Basan M. Analysis of proteome adaptation reveals a key role of the bacterial envelope in starvation survival. Mol. Syst. Biol. 2022;18:e11160. doi: 10.15252/msb.202211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cunning C, Elliott T. RpoS synthesis is growth rate regulated in Salmonella typhimurium, but its turnover is not dependent on acetyl phosphate synthesis or PTS function. J. Bacteriol. 1999;181:4853–4862. doi: 10.1128/JB.181.16.4853-4862.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Phaiboun A, Zhang Y, Park B, Kim M. Survival kinetics of starving bacteria is biphasic and density-dependent. PLoS Comput. Biol. 2015;11:e1004198. doi: 10.1371/journal.pcbi.1004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Patange O, et al. Escherichia coli can survive stress by noisy growth modulation. Nat. Commun. 2018;9:5333. doi: 10.1038/s41467-018-07702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu C, et al. Cellular perception of growth rate and the mechanistic origin of bacterial growth law. Proc. Natl Acad. Sci. USA. 2022;119:e2201585119. doi: 10.1073/pnas.2201585119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fernández-Coll L, Cashel M. Possible roles for basal levels of (p)ppGpp: growth efficiency vs. surviving stress. Front. Microbiol. 2020;11:592718. doi: 10.3389/fmicb.2020.592718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Maharjan R, et al. The form of a trade-off determines the response to competition. Ecol. Lett. 2013;16:1267–1276. doi: 10.1111/ele.12159. [DOI] [PubMed] [Google Scholar]
- 114.Russell JR, Cabeen MT, Wiggins PA, Paulsson J, Losick R. Noise in a phosphorelay drives stochastic entry into sporulation in Bacillus subtilis. EMBO J. 2017;36:2856–2869. doi: 10.15252/embj.201796988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wadhwa N, Berg HC. Bacterial motility: machinery and mechanisms. Nat. Rev. Microbiol. 2022;20:161–173. doi: 10.1038/s41579-021-00626-4. [DOI] [PubMed] [Google Scholar]
- 116.Ni B, Colin R, Link H, Endres RG, Sourjik V. Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc. Natl Acad. Sci. USA. 2020;117:595–601. doi: 10.1073/pnas.1910849117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gude S, et al. Bacterial coexistence driven by motility and spatial competition. Nature. 2020;578:588–592. doi: 10.1038/s41586-020-2033-2. [DOI] [PubMed] [Google Scholar]
- 118.Nakaoka H. Live fast, die fast principle in a single cell of fission yeast. Micro. Cell. 2017;4:308–310. doi: 10.15698/mic2017.09.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zakrzewska A, et al. Genome-wide analysis of yeast stress survival and tolerance acquisition to analyze the central trade-off between growth rate and cellular robustness. Mol. Biol. Cell. 2011;22:4435–4446. doi: 10.1091/mbc.e10-08-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lang GI, Murray AW, Botstein D. The cost of gene expression underlies a fitness trade-off in yeast. Proc. Natl Acad. Sci. USA. 2009;106:5755–5760. doi: 10.1073/pnas.0901620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schellhorn HE. Function, evolution, and composition of the RpoS regulon in Escherichia coli. Front. Microbiol. 2020;11:560099. doi: 10.3389/fmicb.2020.560099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 123.Pontes MH, Groisman EA. A physiological basis for nonheritable antibiotic resistance. mBio. 2020;11:e00817–e00820. doi: 10.1128/mBio.00817-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Levin-Reisman I, et al. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- 125.Liu J, Gefen O, Ronin I, Bar-Meir M, Balaban NQ. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science. 2020;367:200–204. doi: 10.1126/science.aay3041. [DOI] [PubMed] [Google Scholar]
- 126.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 127.Rasouly A, et al. Analysing the fitness cost of antibiotic resistance to identify targets for combination antimicrobials. Nat. Microbiol. 2021;6:1410–1423. doi: 10.1038/s41564-021-00973-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Després PC, et al. Asymmetrical dose responses shape the evolutionary trade-off between antifungal resistance and nutrient use. Nat. Ecol. Evol. 2022;6:1501–1515. doi: 10.1038/s41559-022-01846-4. [DOI] [PubMed] [Google Scholar]
- 129.Olivares Pacheco J, Alvarez-Ortega C, Alcalde Rico M, Martínez JL. Metabolic compensation of fitness costs is a general outcome for antibiotic-resistant Pseudomonas aeruginosa mutants overexpressing efflux pumps. mBio. 2017;8:e00500–e00517. doi: 10.1128/mBio.00500-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pinheiro F, Warsi O, Andersson DI, Lässig M. Metabolic fitness landscapes predict the evolution of antibiotic resistance. Nat. Ecol. Evol. 2021;5:677–687. doi: 10.1038/s41559-021-01397-0. [DOI] [PubMed] [Google Scholar]
- 131.de Jong IG, Haccou P, Kuipers OP. Bet hedging or not? A guide to proper classification of microbial survival strategies. Bioessays. 2011;33:215–223. doi: 10.1002/bies.201000127. [DOI] [PubMed] [Google Scholar]
- 132.Veening JW, et al. Bet-hedging and epigenetic inheritance in bacterial cell development. Proc. Natl Acad. Sci. USA. 2008;105:4393–4398. doi: 10.1073/pnas.0700463105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.González-Pastor JE. Cannibalism: a social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 2011;35:415–424. doi: 10.1111/j.1574-6976.2010.00253.x. [DOI] [PubMed] [Google Scholar]
- 134.Moreno-Gámez S, et al. Wide lag time distributions break a trade-off between reproduction and survival in bacteria. Proc. Natl Acad. Sci. USA. 2020;117:18729–18736. doi: 10.1073/pnas.2003331117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Koch AL. Oligotrophs versus copiotrophs. Bioessays. 2001;23:657–661. doi: 10.1002/bies.1091. [DOI] [PubMed] [Google Scholar]
- 136.Lauro FM, et al. The genomic basis of trophic strategy in marine bacteria. Proc. Natl Acad. Sci. USA. 2009;106:15527–15533. doi: 10.1073/pnas.0903507106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Morris RM, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 138.Partensky F, Garczarek L. Prochlorococcus: advantages and limits of minimalism. Annu. Rev. Mar. Sci. 2010;2:305–331. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- 139.Biller SJ, Berube PM, Lindell D, Chisholm SW. Prochlorococcus: the structure and function of collective diversity. Nat. Rev. Microbiol. 2015;13:13–27. doi: 10.1038/nrmicro3378. [DOI] [PubMed] [Google Scholar]
- 140.Semenov AM. Physiological bases of oligotrophy of microorganisms and the concept of microbial community. Micro. Ecol. 1991;22:239–247. doi: 10.1007/BF02540226. [DOI] [PubMed] [Google Scholar]
- 141.Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- 142.Roller BR, Schmidt TM. The physiology and ecological implications of efficient growth. ISME J. 2015;9:1481–1487. doi: 10.1038/ismej.2014.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Noell SE, Hellweger FL, Temperton B, Giovannoni SJ. A reduction of transcriptional regulation in aquatic oligotrophic microorganisms enhances fitness in nutrient-poor environments. Microbiol. Mol. Biol. Rev. 2023;87:e0012422. doi: 10.1128/mmbr.00124-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ostrowski M, Cavicchioli R, Blaauw M, Gottschal JC. Specific growth rate plays a critical role in hydrogen peroxide resistance of the marine oligotrophic ultramicrobacterium Sphingomonas alaskensis strain RB2256. Appl. Environ. Microbiol. 2001;67:1292–1299. doi: 10.1128/AEM.67.3.1292-1299.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fegatella F, Cavicchioli R. Physiological responses to starvation in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 2000;66:2037–2044. doi: 10.1128/AEM.66.5.2037-2044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lankiewicz TS, Cottrell MT, Kirchman DL. Growth rates and rRNA content of four marine bacteria in pure cultures and in the Delaware estuary. ISME J. 2016;10:823–832. doi: 10.1038/ismej.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Smith DP, et al. Proteomic and transcriptomic analyses of “Candidatus Pelagibacter ubique” describe the first PII-independent response to nitrogen limitation in a free-living Alphaproteobacterium. mBio. 2013;4:e00133–00112. doi: 10.1128/mBio.00133-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Cottrell MT, Kirchman DL. Transcriptional control in marine copiotrophic and oligotrophic bacteria with streamlined genomes. Appl Environ. Microbiol. 2016;82:6010–6018. doi: 10.1128/AEM.01299-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Smith DP, et al. Proteome remodeling in response to sulfur limitation in “Candidatus Pelagibacter ubique”. mSystems. 2016;1:e00068–16. doi: 10.1128/mSystems.00068-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Müller AL, et al. An alternative resource allocation strategy in the chemolithoautotrophic archaeon Methanococcus maripaludis. Proc. Natl Acad. Sci. USA. 2021;118:e2025854118. doi: 10.1073/pnas.2025854118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gray DA, et al. Extreme slow growth as alternative strategy to survive deep starvation in bacteria. Nat. Commun. 2019;10:890. doi: 10.1038/s41467-019-08719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat. Rev. Microbiol. 2017;15:453–464. doi: 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 154.Gollan B, Grabe G, Michaux C, Helaine S. Bacterial persisters and infection: past, present, and progressing. Annu. Rev. Microbiol. 2019;73:359–385. doi: 10.1146/annurev-micro-020518-115650. [DOI] [PubMed] [Google Scholar]
- 155.Mukherjee A, et al. Plasticity of growth laws tunes resource allocation strategies in bacteria. PLoS Comput. Biol. 2024;20:e1011735. doi: 10.1371/journal.pcbi.1011735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Scott M, Hwa T. Bacterial growth laws and their applications. Curr. Opin. Biotechnol. 2011;22:559–565. doi: 10.1016/j.copbio.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mikkola R, Kurland CG. Selection of laboratory wild-type phenotype from natural isolates of Escherichia coli in chemostats. Mol. Biol. Evol. 1992;9:394–402. doi: 10.1093/oxfordjournals.molbev.a040731. [DOI] [PubMed] [Google Scholar]