Abstract
The ability to split one cell into two is fundamental to all life, and many bacteria can accomplish this feat several times per hour with high accuracy. Most bacteria call on an ancient homolog of tubulin, called FtsZ, to localize and organize the cell division machinery (divisome) into a ring-like structure at the cell midpoint. The divisome includes numerous other proteins, often including an actin homolog (FtsA), that interact with each other at the cytoplasmic membrane. Once assembled, the protein complexes that comprise the dynamic divisome coordinate membrane constriction with synthesis of a division septum, but only after overcoming checkpoints mediated by specialized protein-protein interactions. In this Review, we focus on the most recent evidence for how divisome proteins of Escherichia coli assemble at the cell midpoint, interact with each other, and regulate activation of septum synthesis. We also briefly discuss the potential of divisome proteins as novel antibiotic targets.
To survive and proliferate, a bacterial cell has to coordinate many metabolic and physical tasks. One of the most daunting challenges it faces is the need to split itself in two after accurately duplicating and partitioning its genome, all while maintaining a cell wall capable of withstanding high turgor pressure. In the case of fast-growing cells such as Escherichia coli, this act of binary fission needs to happen every ~20 minutes, and requires perfect coordination among the inner membrane, a cell wall, and the outer membrane all at the correct location at midcell between newly replicated daughter chromosomes. The complex protein nanomachine that drives bacterial cytokinesis is often called the divisome.
After several decades of study, we now know many of the proteins that assemble into the bacterial divisome. How the divisome is targeted to the cell midpoint by spatial regulatory systems is reasonably well understood in a number of species, although it is becoming clear that mechanisms for centering the divisome are diverse. On the other hand, the temporal regulation of divisome activity and the signaling between its cytoplasmic and periplasmic protein machines has been illuminated only recently. This Review covers some of the recent significant advances in our understanding of how the divisome is assembled, kept in check, and ultimately activated, emphasizing major spatial and temporal regulatory mechanisms. We will focus mainly on the E. coli model system, about which most is known, but compare and contrast to other species when relevant. In particular we will outline the key roles of the tubulin-like FtsZ and actin-like FtsA proteins in divisome organization and activation. We also discuss recent advances in divisome reconstitution in vitro, potential roles of biomolecular condensates, and targeting divisome proteins with small molecules.
Conserved divisome proteins
FtsZ is the master cytomotive protein organizer of the divisome in most bacteria. It assembles into polymers at the division plane to form the “Z ring” 1, which is required for the localization of all other divisome proteins. These FtsZ polymers move directionally around the inner membrane by treadmilling, which involves subunits joining at one end of a polymer while leaving the opposite end, ultimately acting as a molecular motor 2,3. However, FtsZ cannot attach to the membrane by itself, and needs to bind to membrane-associated proteins, through its conserved C-terminal tail, to organize into a Z ring. In E. coli, these membrane tethers include FtsA, one of several conserved bacterial actin homologs 4, and the less well-conserved ZipA, initially discovered as an FtsZ-Interacting Protein 5–7. Together, FtsZ, FtsA and ZipA comprise the “proto-ring”, which represents the initial cytoplasmic setup stage of the E. coli divisome 8 (FIG. 1A). The FtsE and FtsX proteins, which complex together, also arrive at this point 9.
Fig. 1. Centering and organizing the divisome.
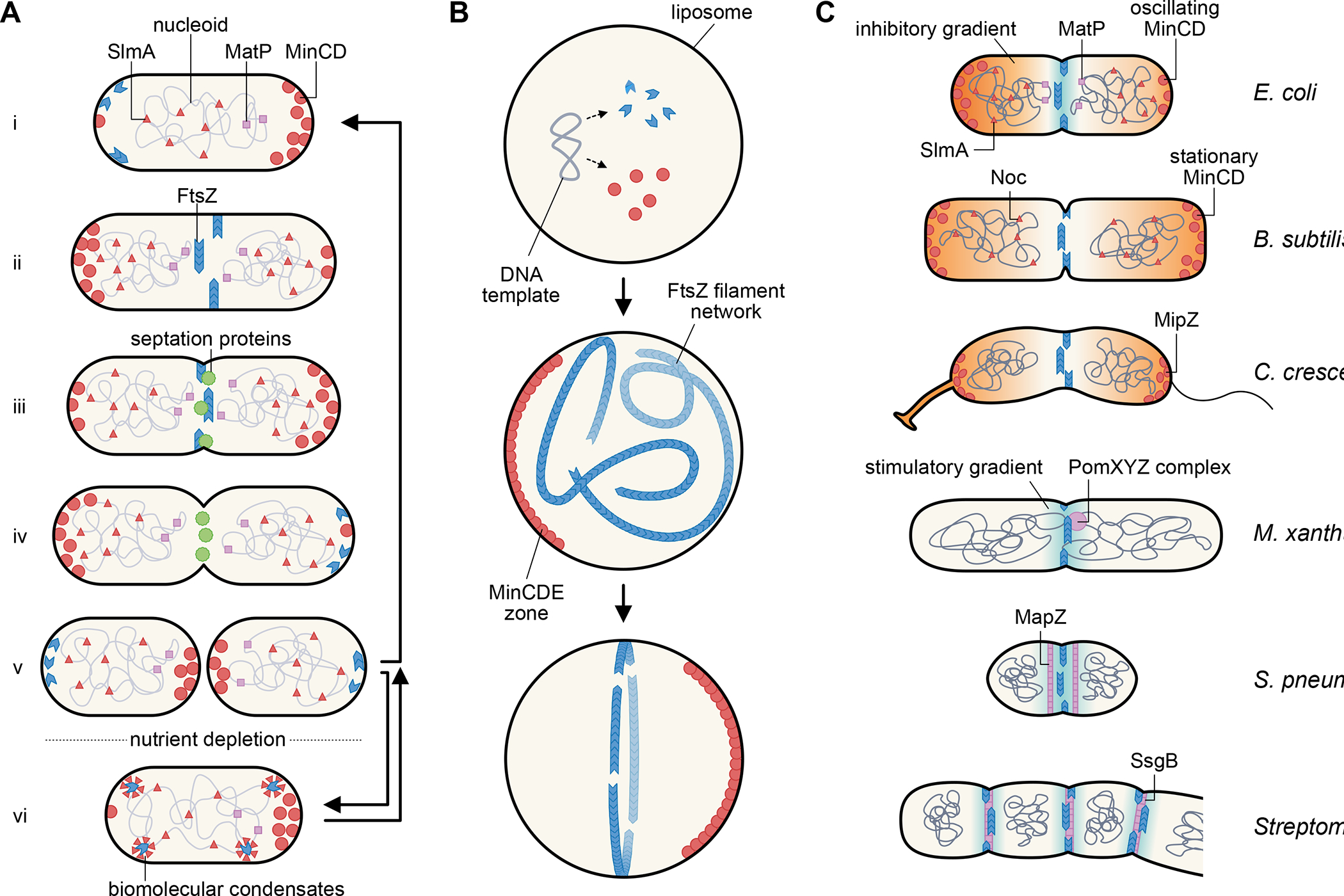
(A) Escherichia coli cell division cycle. Newborn cells (i) carry several spatial FtsZ positioning systems, including SlmA and MatP on the nucleoid and MinCD oscillating between the cell poles, that keep FtsZ from assembling into a Z ring at the cell poles or on top of the unsegregated nucleoid. As the replicated daughter nucleoids segregate, the Z ring assembles at midcell (ii) and recruits other septum-synthesis proteins (iii), which result in formation of the division septum. During septum synthesis, FtsZ leaves the division site prior to the other septation proteins (iv). The final step of cytokinesis results in two daughter cells (v), which begin the next cycle during growth conditions. Nutrient depletion or stress induces a quiescent state (vi) in which FtsZ localizes to putative biomolecular condensates that can revert back to polymer forms after growth restart. (B) Reconstitution of a centered proto-ring in lipid vesicles. After FtsZ, FtsA, and Min proteins are synthesized from a plasmid template or added as purified protein to lipid vesicles, they self-organize into membrane-associated polymers or gradients. Oscillation of MinCDE within the liposome helps to corral FtsA-tethered FtsZ polymers into a focused ring, mimicking in vivo behavior. (C) Theme and varations of Z ring positioning systems in diverse bacterial species, showing three examples of negative spatial regulators (orange gradients) and three examples of positive spatial regulators (blue gradients) and the proteins involved.
After a distinct delay 10,11, other E. coli divisome proteins are recruited to the proto-ring in a second stage in the order FtsK-(FtsQ/FtsL/FtsB)-FtsW-FtsI-FtsN, as shown by immunofluorescent or GFP-tagged localization of each of these proteins in mutants in which other divisome proteins are inactivated or depleted 12. Inactivation of any one of these proteins leads to a lethal cell division block, although division of cells lacking FtsE or FtsX is restored when grown in medium with high osmotic strength 13. Two of these, FtsW and FtsI, are essential septal wall synthesis enzymes conserved in most walled bacteria 14 and work together, so they will be referred to as FtsWI. There are numerous ancillary divisome proteins, such as the Zap proteins 15, cell wall amidases, and their regulators such as EnvC, whose absence individually does not result in major cell division defects but are important in combination with other members of their group in facilitating normal cell division. These have been covered in several recent reviews, including a comprehensive review of E. coli cell division 16.
Centering the divisome
A key task for a rapidly dividing rod-shaped bacterium such as E. coli is to identify the next site of cell division, which is identified very early in the cell division cycle 17. Several spatial regulatory systems contribute to Z-ring centering in E. coli, including the Min system and nucleoid occlusion (NO), both of which act as localized negative regulators of FtsZ polymerization 18,19. When the Min system is inactivated, cells divide either properly at the central site or asymmetrically near the cell poles to form DNA-less minicells, whereas mutants defective in NO display no defects unless other systems, such as the Min system itself, are disabled 20.
The E. coli Min system consists of three proteins, MinC, MinD, and MinE 21. MinD, which is a deviant Walker ATPase 22, uses its amphipathic helix to bind to the membrane when ATP-bound and leaves the membrane after ATP hydrolysis, which is stimulated by its membrane-associated partner, MinE 23,24. As a result, membrane-bound MinD-ATP is “chased” off the membrane by MinE as MinD-ADP until the regeneration of MinD-ATP allows it to rebind the membrane, repeating the cycle 25–27. This behavior is essentially a Turing reaction-diffusion system 28, which has been reconstituted in vitro as traveling waves of MinD and MinE on planar lipid bilayers 29. In the select group of rod-shaped species that harbor MinD and MinE, these waves organize into zones around cell poles that move periodically (and rapidly, within a few seconds) to the opposite cell pole, forming a bipolar concentration gradient (FIG. 1A).
Recent evidence suggests that this membrane-associated gradient may help gather polymerizing FtsZ toward the cell center 30,31. Nevertheless, MinC is clearly important for spatially restricting the Z ring to midcell, as it binds directly to two domains of FtsZ and inhibits FtsZ polymerization 32 and mutants lacking only MinC still divide aberrantly. The key to Z-ring centering is that MinC associates with MinD and oscillates with it 27,33, thus inhibiting Z-ring formation mostly at the cell poles where MinD is present most of the time. This inhibition is visible as an oscillation that runs counter to the Min oscillation 34,35. Cell geometry is crucial for the centering mechanism, as Min proteins seek the longer axis for their oscillations, even in spheroidal cells and compartments 36,37. During the septation process, Min oscillation transitions into a double oscillation in each daughter cell compartment to identify future division sites 38. There is evidence that MinC and MinD from E. coli and other species form copolymers in vitro 39–42, suggesting that copolymers of MinCD near cell poles may restrict FtsZ assembly there. However, so far there is no evidence that MinCD copolymers are important for Min function in E. coli cells 43.
The other major spatial regulator of the Z ring, NO, is mediated in E. coli by a DNA-binding protein called SlmA (originally isolated as a synthetic lethal allele of Min) 20. Specific chromosomal SlmA binding sites (SBS) are biased away from the replication terminus (Ter) near midcell and towards the chromosomal replication origin (OriC) near the cell poles, resulting in a bipolar gradient of SlmA in the cell 44,45. In addition, and crucial for its mechanism of action, SlmA also binds to FtsZ 46,47 and interferes with its assembly 48,49. Consequently, SlmA tends to inhibit FtsZ assembly in the vicinity of the nucleoid, particularly near OriC (FIG. 1A).
SlmA, however, is not well conserved in other species that use NO. In B. subtilis, NO is implemented by Noc, a ParB homolog that is structurally unrelated to SlmA. Like SlmA, Noc forms nucleoprotein complexes at several specific binding sites on the chromosome 50. Unlike SlmA, which binds to FtsZ and spreads on DNA flanking its binding sites 49, Noc uses an N-terminal amphipathic helix to spread on the membrane and recruit chromosomal DNA there, which indirectly discourages local Z ring formation 51, probably by inhibiting FtsZ subunit migration away from existing division septa 52.
Despite their different mechanisms of action, both Noc and SlmA have a strong tendency to form phase-separated biomolecular condensates at the membrane that recruit FtsZ, potentially sequestering FtsZ in a non-polymerized state 53,54 (FIG. 1A). These phase-separated states are reversible: addition of GTP to FtsZ-SlmA condensates triggers assembly of FtsZ polymers and dissolution of the condensates 53. Analogously, addition of CTP to Noc promotes its phase separation, and addition of high levels of KCl can dissolve both Noc and SlmA condensates 54,55.
It remains to be seen how these phase transitions function in bacterial cell division. A fluorescent derivative of FtsZ, along with associated divisome proteins, frequently forms polar foci in quiescent E. coli cells, which dissolve upon resumption of growth conditions 56. If these foci are biomolecular condensates, which has not yet been demonstrated conclusively but is supported by the ability of purified FtsZ to form condensates 57, they may sequester FtsZ monomers at low GTP concentrations during nutrient starvation and permit FtsZ to polymerize at higher GTP concentrations triggered by rapid growth. This has implications for the efficacy of antibacterials against bacterial persister cells, as a drug target inside a condensate may be more difficult for the drug to access 58. Very recently, it was reported that a protein called PomY forms biomolecular condensates that promote assembly of the Z ring at midcell in Myxococcus xanthus 59 (FIG. 1C). Fortunately, new tools are rapidly emerging to identify and confirm condensates in bacterial cells 60. For example, single particle tracking in combination with photobleaching recovery measurements can help determine whether a fluorescently tagged protein is forming condensates. In addition, fluorescently-tagged proteins with different tendencies to form condensates in bacteria can be used as key controls.
The Min system, FtsZ and a membrane tether have been used as a minimal system to reconstitute cell division in vitro, using lipid membranes as cell-like vessels. Following initial reports of FtsZ forming rings inside of liposomes and occasionally constricting them 61, and reconstitution of an oscillating Min system inside liposomes 62,63, two recent reconstitution studies expressed MinCDE along with FtsZ and a membrane tether inside cell-like liposomes using a cell-free transcription/translation system that optimizes protein folding and native stoichiometries 64,65. These studies were able to generate robust Min oscillations that organized FtsZ polymers around the middle of the liposome, ultimately producing isotropic constrictions. The most recent study reconstituted FtsZ, its FtsA membrane tether, and MinCDE inside liposomes to generate mid-liposome constrictions, and showed that Min oscillations and the condensation of FtsZ polymers at mid-liposome reinforce each other 66 (FIG. 1B). These reconstitutions have made impressive progress towards understanding how primitive wall-less cells may have duplicated. However, complete abscission of liposomes with FtsA and FtsZ has not yet been reported, possibly because additional proteins are needed to destabilize FtsZ polymers and/or fuse membranes 67. Future studies will also need to account for NO and other effects of the bacterial nucleoid, and should probably exploit spatial regulatory systems from other bacteria that lack Min or NO systems and may be simpler to implement.
Finally, several positively-acting spatial regulatory systems are known to center the Z ring in a number of species, including E. coli (FIG. 1C). For example, the MatP protein of E. coli binds to multiple DNA sites flanking the chromosomal Ter, which localizes near midcell prior to Z ring assembly 68. MatP also binds to ZapB, a protein that in turn binds to ZapA, which promotes bundling of FtsZ polymers 69–71. Along with its ability to bind to the membrane 53 and organize the chromosome, these properties demonstrate that MatP links the Z ring with Ter, providing spatial as well as temporal regulation with respect to chromosome replication 72,73. In other species, the Pom system in M. xanthus 74, mobile rings of MapZ in Streptococci 75, and SsgB protein in Streptomyces 76 act by positive regulation (FIG. 1C). PomXYZ constitute a MinD-MinE-like system that moves from the nucleoid to new division sites, whereas MapZ binds both peptidoglycan and FtsZ and helps to ferry FtsZ and proto-ring proteins towards new daughter division sites while the midcell septum is closing 77.
Stages of divisome assembly
Once the proto-ring is established, the remaining divisome proteins are recruited by multiple interactions. In E. coli, recruitment of FtsWI to midcell requires FtsK and the FtsQLB complex 78,79, which in turn require FtsA and ZipA 5,80,81 (FIG. 2). FtsK is a multi-pass transmembrane protein fused to a DNA motor that coordinates chromosome segregation with cell division 82–84. Another DNA translocase, SftA, is recruited to the Z ring in B. subtilis and may be involved in similar coordination 85. Only the N-terminal ~200 residues of FtsK (FtsKn), comprising all four transmembrane domains 86 are required for cell division 87–89. FtsQ, FtsL and FtsB are all conserved bitopic membrane proteins with short cytoplasmic tails and large periplasmic domains that interact with each other in a fairly stable FtsQLB complex 79,90–92. Recent genetic and structural studies, discussed more below, suggest that FtsB and FtsL interact directly with the periplasmic domains of FtsW and FtsI and trigger their activities 92–95.
Fig. 2. The E. coli divisome is built in stages.
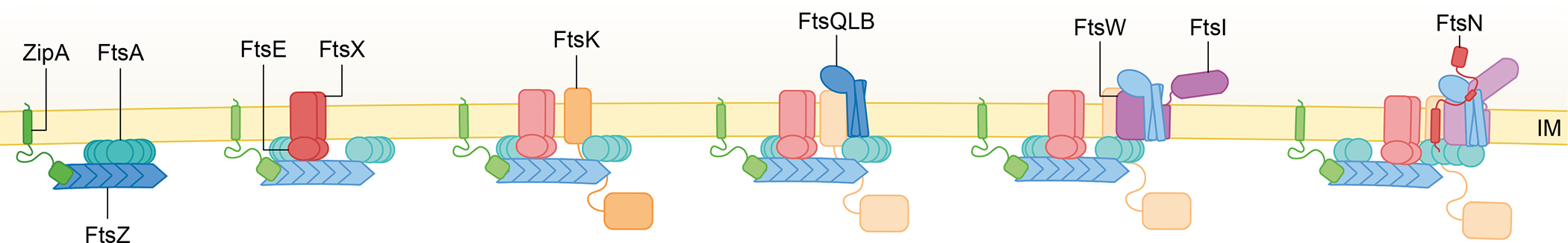
FtsZ is tethered to the membrane by FtsA and ZipA, forming the proto-ring. This is accompanied by the recruitment of the ABC transporter-like FtsEX complex and FtsK, which contains an N-terminal membrane bound domain required for cell division connected to a cytoplasmic DNA motor domain via a long linker. The next complex to be recruited is FtsQLB, which is important for recruitment and activation of the septum-specific transpeptidase (FtsI) and glycosyltransferase (FtsW). FtsN binds to and activates FtsWI enzymatic activity and is present at high concentrations at later stages. FtsN’s four distinct domains, the short cytoplasmic tail (FtsNcyto), transmembrane domain, essential periplasmic domain (FtsNE), and SPOR domain, are highlighted in the cartoon. The recruitment stage for each protein is highlighted by more intense coloring.
The mechanism by which FtsA recruits FtsK and FtsQLB is becoming clearer. Recent data from pull down and affinity copurification assays suggest that FtsA binds to FtsKn 96 and FtsKn forms a tight bimolecular complex with FtsQ 95, likely reflecting how this recruitment pathway occurs in vivo. Moreover, coevolutionary sequence analysis suggests that the first periplasmic loop of FtsKn interacts with a beta strand within the periplasmic domain of FtsQ between residues Q96 and K113 95. A better mechanistic understanding of how FtsQLB is recruited by FtsA and FtsK will necessitate structural dissection of the FtsQ-FtsK complex and a potential FtsA-FtsK complex.
FtsA-FtsN interactions have been characterized in greater detail. FtsA directly interacts with FtsN’s cytoplasmic tail, called FtsNcyto 97–99 (FIG. 2, 3A). However, FtsN’s recruitment to the divisome also depends on interactions with other divisome proteins including FtsI 100, making FtsN the last essential divisome protein to be recruited. FtsN’s periplasmic domain includes a short segment essential for FtsN function (FtsNE) and a nonessential C-terminal SPOR domain 101,102 (FIG. 2). The latter binds specifically to denuded cell wall glycans whose stem peptides have been cleaved off by cell wall amidases generated during septum synthesis 103. Consequently, increased septum synthesis by FtsWI, triggered by FtsN, results in more FtsN recruitment, which reinforces a positive feedback loop that drives septum synthesis forward 101. Divisome checkpoints, discussed in detail below, probably ensure that this feedback loop is not activated until the right conditions are met.
Fig. 3. FtsA oligomerization state regulates divisome activation.
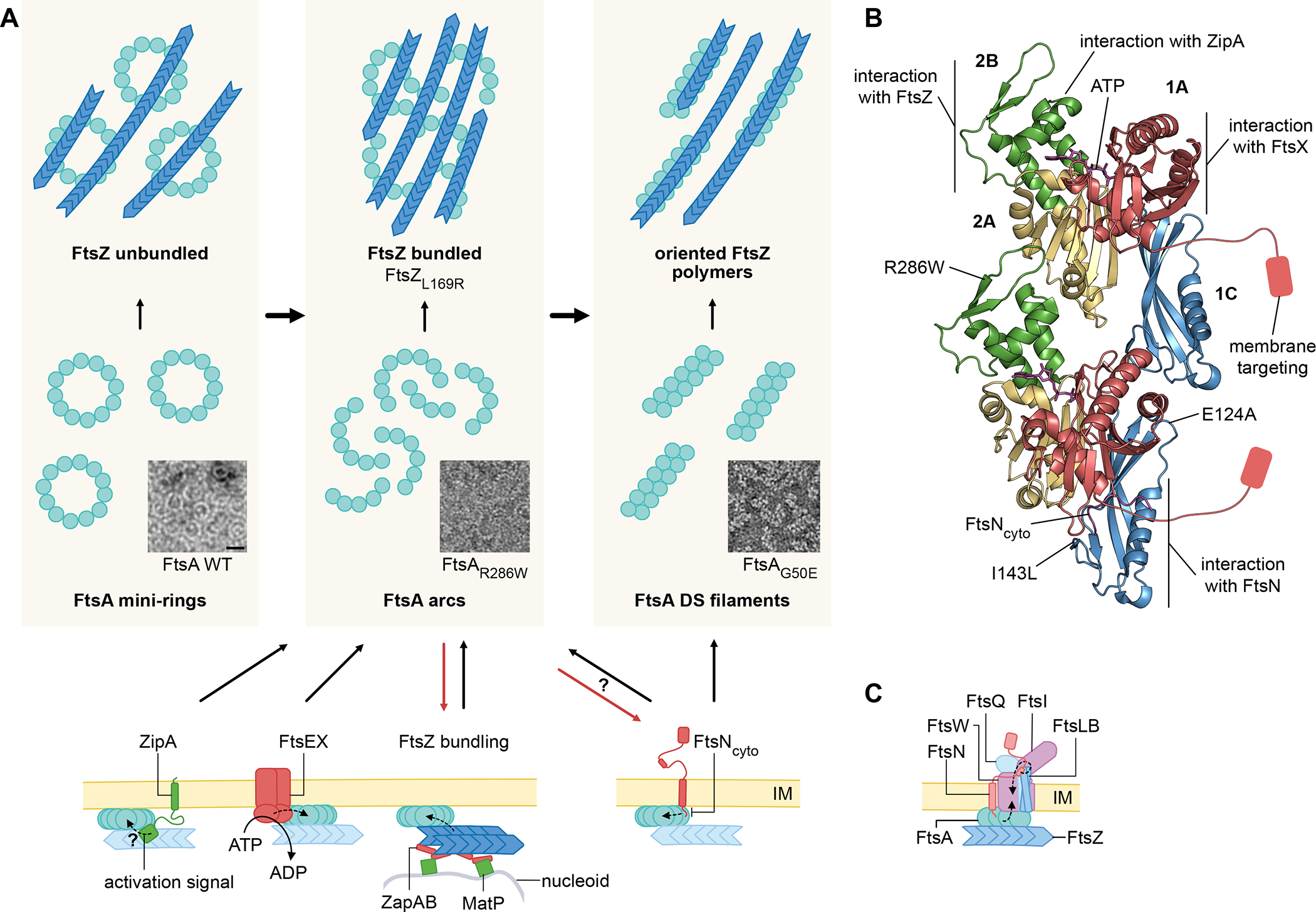
(A) Distinct oligomeric states of FtsA and their effects. WT FtsA initially forms mini-rings on lipid membranes that tether and align separated FtsZ polymers. Although mini-rings have not yet been visualized in vivo, unlocking of FtsA mini-rings by ZipA, FtsX, FtsN (arrows), or by mutations, would be expected to form open FtsA oligomers (arcs) or DS antiparallel FtsA filaments. In vitro, both of these unlocked FtsA states seem to induce tethered FtsZ polymers to laterally associate into bundles, possibly by increasing membrane packing density. FtsA*-like variants bypass several divisome defects in vivo and bypass the mini-ring form in vitro, instead assembling mostly into arcs (e.g. R286W) or DS filaments (G50E). FtsZ polymer bundling may be another divisome activation signal, as the self-bundling FtsZL169R (FtsZ*) variant bypasses several divisome defects in vivo and may convert FtsA into an unlocked, active form, suggesting a positive feedback loop (red and black arrows). In addition to FtsN’s ability to unlock FtsA mini-rings, unlocked mini-rings may facilitate binding of FtsN (red and black arrows). (B) FtsA interactions, variants and activities. FtsA interacts with FtsNcyto through subdomain 1C, FtsZ and ZipA through interactions in subdomain 2B, FtsX through subdomain 1A, another FtsA subunit through several interfaces, and the cytoplasmic membrane via a membrane-targeting amphipathic helix. FtsA*-like substitutions such as R286W likely weaken monomer-monomer interactions that result in unlocking of mini-rings in vitro. FtsN bypass alleles (I143L, E124A) map to subdomain 1C, potentially mimicking FtsNcyto binding. (C) The changes in FtsA oligomeric state signal the later divisome proteins such as FtsW to switch to the activated form (upward arrow). In addition to the cytoplasmic activation signal from FtsA, conformational changes in FtsQLB in the periplasm, along with FtsN’s essential domain, are also crucial for activation of FtsWI (downward arrow).
During early stages of cell division, treadmilling FtsZ polymers are needed to organize septum synthesis enzymes in diverse bacterial species 2,104–106 but are dispensable once septum synthesis is fully underway107,108. Consistent with this, a typical E. coli cell is ~ 1 μm in diameter, but as the septum closes and the membrane invaginates, FtsZ leaves first, starting at ~600 nm septal diameter (FIG. 1A), whereas later divisome proteins like FtsN start to depart after septal diameter decreases below 300–400 nm 72,109,110. In support of these findings, rapid inactivation of FtsZ at later stages of septation does not affect the completion of division 111,112. Later divisome proteins, whose domains mainly reside in the periplasm, also are spatially separated from FtsZ and other proto-ring proteins 113. Therefore, FtsZ is needed only to start the septum building process, and once the septal template has been built, FtsWI will ensure its faithful continuation in the proper position without the need for an FtsZ guide. What triggers FtsZ release? Under slow growth conditions, cellular levels of FtsZ increase during Z ring assembly and then decrease during disassembly due to proteolysis 114. However, the relatively rapid release of FtsZ from the constricting septum implicates other regulatory mechanisms. One may involve a recently discovered interaction between ZipA and the core domain of FtsZ 115, which is weaker than the well-known interaction between ZipA and FtsZ’s C-terminal tail. This weaker interaction with the polymerizing domain of FtsZ hints at a more regulatory role than simple membrane anchoring.
Divisome checkpoints and the key role of FtsA
The first hint of a divisome checkpoint came from isolation of a hypermorphic missense allele of FtsA, called FtsA* (FtsAR286W), that completely bypasses the requirement for ZipA and suppresses defects in other divisome proteins including FtsQ and FtsK 116,117. Cells expressing FtsA* are ~20% shorter than WT cells, consistent with circumventing a checkpoint 118. Subsequently, other single residue changes in FtsA were isolated that have FtsA*-like properties, and all map to the FtsA oligomer interaction interface (FIG. 3B) 119,120. These locations as well as genetic assays indicated that FtsA*like variants have altered oligomerization properties 119. Another hint of a checkpoint came from different variant, FtsAE124A, that is able to bypass FtsN 121. Unlike R286W, the E124A allele does not induce hyper-division, and maps to the 1C subdomain of FtsA instead of the 2B subdomain, farther from the longitudinal oligomer interface, suggesting a distinct regulatory mechanism (see below). In support of this idea, FtsAR286W poorly bypasses FtsN, FtsAE124A poorly bypasses ZipA, and neither can bypass the simultaneous loss of both ZipA and FtsN 121. Interestingly, ZipA and FtsN seem to have coevolved 122. This is consistent with their putative complementary roles in activating FtsA, and is further supported by the ability of either ZipA or FtsN to trigger preseptal peptidoglycan synthesis (independent of FtsWI) by activating the semi-redundant class A penicillin binding proteins PBP1A and PBP1B 123.
Recent biochemical studies have provided key new insights into the role of FtsA’s oligomeric state as a potential divisome checkpoint. When purified E. coli FtsA is added to lipid membranes and visualized by negative stain electron microscopy, it forms striking 20 nm-wide, roughly planar ring-shaped oligomers 124 (FIG. 3A). In contrast, purified FtsA* variants assemble into broken curved oligomers (e.g. FtsAR286W) and/or double stranded (DS) filaments (FtsAG50E) instead of mini-rings 125,126. Although the role of ATP in FtsA function is not yet understood, it is noteworthy that a well-known thermosensitive allele (FtsAS195P) that maps to the ATP site and is defective in ATP binding/hydrolysis can be intragenically suppressed by FtsA*-like mutants, which also have higher intrinsic ATPase activity and undergo more rapid dynamics than WT FtsA 118,120,127. Although it is not yet known what oligomeric structures FtsAS195P forms when inactive, and FtsA mini-rings have not yet been observed in cells, these data support the idea that “unlocking” the mini-rings switches FtsA to an active form that promotes septum synthesis. Furthermore, FtsA’s oligomerization activity can deform lipid membranes in vitro, which may help FtsZ to constrict the cytoplasmic membrane during septum synthesis 128.
As suggested by the ability of FtsAE124A (as well as another allele mapping to the 1C subdomain, FtsAI143L) to bypass it 129 (FIG. 3B), FtsN seems also to be involved in a checkpoint. Indeed, FtsN was originally discovered through its ability to suppress several divisome defects when overproduced 130. Although FtsN likely activates septum synthesis mainly through its periplasmic interactions with FtsWI, FtsN may also help to activate septum synthesis on the cytoplasmic side of the membrane by stabilizing the FtsA DS filament form. In support of this idea, adding a 5–10 fold molar excess of purified FtsNcyto (FtsN1–32) to FtsA on a lipid membrane is sufficient to switch most minirings to antiparallel DS filaments 126 (FIG. 3A). The unique lateral associations between FtsA oligomers that generate the DS filaments can be detected in vivo by crosslinking and are mediated by adjacent 1C subdomains, which is also where FtsNcyto binds 97,126 (FIG. 3B). Importantly, the FtsN bypass variant FtsAE124A forms mini-rings in vitro like wild-type FtsA but switches to DS filaments at lower concentrations of added FtsNcyto compared with WT FtsA 126. This suggests that the E124A residue change tips the balance towards DS filaments, perhaps by weakening interactions between mini-ring subunits, but may also rely on mini-rings to be unlocked by an earlier step. This is consistent with the aforementioned poor ability of FtsAE124A to bypass ZipA in vivo compared with FtsA* alleles.
Notably, FtsN-bound DS filaments of FtsA are curved in such a way that they can reinforce negative curvature on membranes to which they are attached 126. FtsA DS filaments would thus have a strong tendency to orient along the curved short axis of the cell instead of the straight long axis, which may help to gather treadmilling FtsZ polymers within a more concentrated band. It should be emphasized that although the periplasmic FtsNE segment is essential for E. coli cell division, FtsNcyto is dispensable 98,131, indicating that the FtsA-FtsN interaction is only one of several pathways to activate septum synthesis 99,129,132 (see also below). In addition to FtsN’s role in promoting or stabilizing FtsA DS filaments, FtsA oligomerization state seems to dictate whether FtsN can bind to it 99,119. For example, FtsA* interacts more efficiently with FtsN than WT FtsA and also earlier in the cell cycle 127,132,133. This implies that FtsNcyto may bind more efficiently to free ends of unlocked FtsA than to mini-rings, which may also be the case for other divisome proteins recruited by FtsA such as FtsK and/or FtsQLB.
The FtsA-FtsN interaction is also interesting in an evolutionary context. Bacterial Type IV pili are anchored to the membrane by a protein called PilM that has remarkable structural similarity to FtsA, with similar subdomain organization 97,134. Another similarity is that PilM also forms dodecameric rings on the inner surface of the cytoplasmic membrane 135,136. One of several pilus proteins that interact with the PilM ring, PilN, has a bitopic topology like FtsN. Even the binding sites for PilM and FtsN on FtsA are similar, as the cytoplasmic tails of both target the cleft between the 1A and 1C domains (near the FtsA E124 bypass residue), although at distinct specific sites 126,134.
In addition to FtsN, ZipA and FtsX are both implicated in switching FtsA to an activated state, perhaps by unlocking mini-rings. In contrast to FtsN, whose main function is activating an already assembled divisome, both ZipA and FtsX act early to recruit later divisome proteins 13,81,137 and can be bypassed by hypermorphic alleles or overproduction of some divisome proteins 138, again suggesting that free FtsA oligomer ends in unlocked mini-rings are important for recruitment of later divisome proteins. ZipA and FtsX interact directly with FtsA at sites in the 2B and 1A domains, respectively 137,139, suggesting that their mechanisms of activation are distinct (FIG. 3B). Although the mechanism of FtsA unlocking by ZipA is not yet known, unlocking of FtsA by FtsX is regulated by the ATPase activity of FtsX’s cytoplasmic binding partner FtsE (FIG. 3A). FtsX’s periplasmic binding partner EnvC, which stimulates cell wall amidases important for splitting septal peptidoglycan, is also regulated by FtsX’s ATPase activity 140,141. Interestingly, Z rings are less condensed in EnvC mutants, suggesting that signaling within the divisome proceeds not only outward from FtsEX to the amidases but also inward from EnvC through FtsEX to FtsZ 142.
How might these oligomeric switches in FtsA confer checkpoint-like properties? One way is through effects on FtsZ. On lipid membranes in vitro, protofilaments of FtsZ are held apart by FtsA mini-rings, and only bundle in the presence of FtsA* derivatives 124,125. Similar parallel but separated FtsZ filaments, which may represent the initial form of FtsZ prior to septum synthesis, have been observed in cells by cryo-EM tomography 143. These results are consistent with FtsA mini-rings, assuming that they are confirmed to exist in cells, being in an initial locked state and support the idea that FtsA oligomerization state regulates FtsZ assembly state 144.
Although the molecular mechanisms that trigger FtsA’s oligomeric switch or change FtsZ filament bundling state are not yet known, the switch to non-ring FtsA oligomers in vitro increases FtsA packing density on the membrane, which provides a denser array of membrane tethers for FtsZ to associate laterally and interact more dynamically 124,127 (FIG. 3A). As mentioned above, at least one part of the checkpoint likely involves increased bundling/condensation of FtsZ polymers. This bundling activity is enhanced by the binding of Zap proteins to FtsZ 15 and self-inhibited by FtsZ’s disordered C-terminal linker 145–147, but is crucial for promoting septum synthesis 148.
As is postulated for FtsA-FtsN interactions, FtsA-FtsZ oligomeric states may rely on a self-reinforcing mechanism. A variant of FtsZ (FtsZL169R), named FtsZ* 149 has a strong tendency to laterally associate into protofilament pairs and ribbons in vitro, and like FtsA* it can bypass ZipA and suppress some divisome defects in E. coli, including bundling defects caused by the lack of Zap proteins 149,150 (FIG. 3A). Importantly, these self-bundled FtsZ* protofilaments seem to promote unlocking of FtsA mini-rings in vitro, thus mimicking FtsA* 124. As FtsA mini-ring disassembly in vitro promotes FtsZ bundling and vice-versa, these two processes may reinforce a positive feedback loop in vivo that irreversibly drives septation forward, especially if bundled FtsZ can act as a direct template for FtsA DS filaments (FIG. 3A). However, hyper-bundled FtsZ also confers a fitness cost, as cells with extra FtsZ* or FtsZ* combined with FtsA* have septal abnormalities and are less viable 125,149. It should also be noted that other variants that increase FtsZ bundling in vitro such as FtsZE93R 151 do not bypass ZipA, so the degree and type of bundling is likely to be important for activation.
Another key part of the FtsA checkpoint may involve FtsA curved DS filaments themselves and their ability to activate septum synthesis. A recent model proposed that DS filaments of FtsA, triggered by FtsNcyto binding, act as rudders that guide septal wall synthesis powered by FtsWI 126. Such “surfing” cytoplasmic FtsA rudders would respond to the periplasmic activities of FtsWI directly through contacts with FtsN, which binds FtsA and FtsI, and by the FtsQLB complex, which binds and activates FtsW 93 and may also interact with FtsA 152 (FIG. 3C). This mechanism is roughly analogous to the membrane-associated rudders of the elongasome (cell wall elongation machinery) formed by DS filaments of MreB, which locally guide sidewall synthesis by PBP2 and RodA and are mediated by MreCD and RodZ 153,154. These potentially analogous mechanisms underscore the evolutionary kinship between divisome and elongasome 126,155 and their competition for proteins and substrates 142,156,157.
Other divisome checkpoints
The FtsA-FtsNcyto interaction is clearly an important regulator of E. coli divisome activity. However, the isolation of hypermorphic variants of FtsL, FtsB, FtsI and FtsW, named FtsL*, FtsB* FtsI* and FtsW*, which all map to periplasmic domains in these proteins, indicate that crucial checkpoints operate in the periplasm (FIG. 2, 3C). Like FtsA*, some periplasmic hypermorphs exhibit strong hyper-division phenotypes consistent with constitutive activation of septum synthesis. For example, ftsL* induces hyper-division and short cell length 158. In addition, ftsL* and ftsB* can partially bypass FtsK or FtsN. These data suggest that FtsB* and FtsL* proteins mimic the interaction between an activating protein such as FtsN and the FtsQLBWI complex, switching it to an active form. The absence of known ftsQ* alleles, along with evidence that FtsQ may interact directly with upstream divisome proteins such as FtsK and FtsA, suggest that it is the FtsBL portion of the complex that directly activates FtsWI. This idea is supported by genetic evidence pinpointing specific periplasmic domains in both FtsB and FtsL called CCD (constriction control domain), and a nearby domain in FtsL called AWI (activator of FtsWI) that harbor the hypermorphic alleles and activate the FtsWI septum synthases 159.
Moreover, recent structural studies combined with targeted mutations that decrease or increase function suggest that the activation signal travels from FtsB to FtsL to FtsI to FtsW 94, all via periplasmic interactions. The ultimate target of both checkpoints is probably the fourth extracellular loop (ECL4) of FtsW, as FtsW* mutants of E. coli such as FtsWE289G mapping to this loop can bypass FtsN and other divisome defects, including dominant negative mutations in ECL4 itself. Similarly, FtsW* alleles in Caulobacter crescentus can override upstream divisome defects, and the strongest of these alleles is in the same ECL4 161. In addition to periplasmic activation of FtsWI via FtsQLB, genetic evidence suggests that FtsA acts directly on the cytoplasmic side of the E. coli divisome to activate FtsW through the latter’s second cytoplasmic loop 93 (FIG. 3C). Consistent with a model of direct FtsA-FtsW signaling, FtsQ can be bypassed by coexpression of FtsA* along with a derivative of FtsW* that is artificially targeted to the divisome independently of FtsQ. Notably, ftsL and ftsB cannot be bypassed, at least so far, probably because conformational changes in FtsL and FtsB are essential for activating FtsWI.
In a different type of checkpoint, FtsK coordinates septum synthesis with chromosome segregation. This is particularly clear in C. crescentus, which, like other alpha-proteobacteria, lacks an obvious NO mechanism and instead uses a MinD-like multifunctional protein, MipZ, to couple chromosome segregation with Z ring placement 162. Moreover, it seems that a divisome protein called FzlA binds directly to FtsZ and the translocase domain of FtsK, thereby coupling activation of FtsWI with chromosome segregation 163. As it interacts with both proto-ring and late divisome proteins, FtsK probably acts similarly to coordinate divisome activity with chromosome segregation in E. coli 84, but the precise mechanism is not yet known. An interesting case of divisome-chromosome coordination operating in the opposite direction is the conserved CcrZ protein, which in S. pneumoniae uses its interaction with FtsZ at midcell to regulate the activity of DnaA, the key chromosome replication initiator protein 164.
Keeping up with treadmilling FtsZ
In addition to its checkpoint role, FtsA oligomers, along with ZipA, organize treadmilling polymers of FtsZ along the cell membrane 150,165 (FIG. 4A). B. subtilis FtsA oligomers seem to achieve this by transiently binding to treadmilling FtsZ subunits, which remain stationary in the middle of a treadmilling FtsZ polymer until they diffuse from the polymer end. This could achieve net translocation around the cell circumference (FIG. 4) by iterative diffusion and capture 104,166. Another possibility is that FtsA tracks the ends of FtsZ polymers by a Brownian ratchet mechanism 106,167. It is also possible that B. subtilis FtsA oligomers actually treadmill along with FtsZ polymers 3, but this requires that each FtsA polymer have a “minus” and “plus” end—i.e., polarity. This may be the case for B. subtilis FtsA, which forms polymers in parallel with FtsZ filaments 168. However, it is certainly not for E. coli FtsA, as its closed minirings and antiparallel filaments would have no plus or minus ends required for polarized subunit gain and loss. While E. coli FtsA arcs could theoretically treadmill, they would literally go around in circles and could not maintain the ~30 nm/sec directional velocity observed for FtsZ 2. Instead, FtsA oligomers—including mini-rings—probably keep up with FtsZ polymers in E. coli by disassembling and reassembling into new oligomeric structures that track the ends of FtsZ polymers. In support of this idea, turnover of fluorescently labeled FtsA in E. coli cells is nearly as rapid as FtsZ turnover 118 and the translocation velocities of fluorescent FtsA foci around the Z ring are similar to those of FtsZ 169 and of FtsA in other species 3,105, averaging ~30 nm/sec. Moreover, single molecules of FtsN that lack the periplasmic region, and thus mainly interact with FtsA, are either stationary or move fast at ~30 nm/sec (FIG. 4B), consistent with the idea that FtsA structures track treadmilling FtsZ 167.
Fig. 4. Divisome proteins build the septum in two tracks.
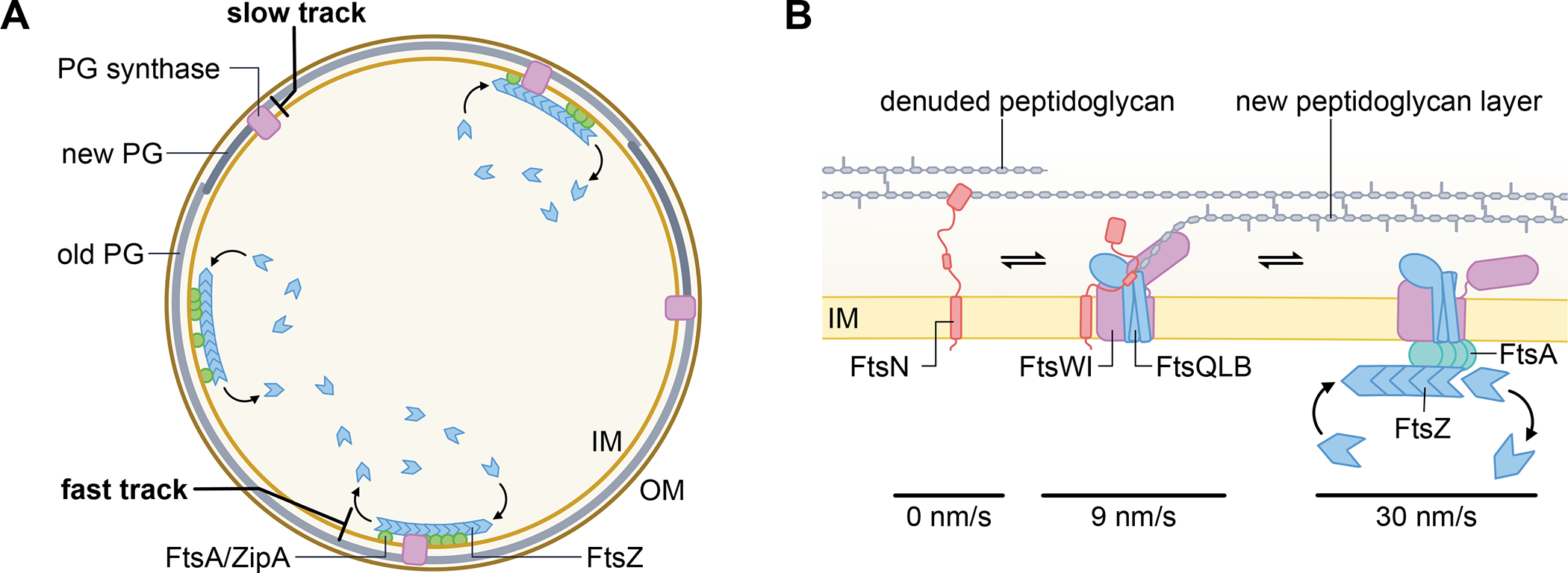
(A) Cross section of a dividing E. coli cell depicting several processive, FtsZ treadmilling complexes that move rapidly along the cytoplasmic face of the inner membrane to organize septum synthesis, in coordination with several independently moving, slower track of proteins that actively engage in septal peptidoglycan (PG) synthesis in the periplasm. (B) Detail of complexes comprising the different speed modes during septum formation. A subset of FtsN proteins are transiently stationary, possibly because they are bound to denuded septal PG resulting from amidase activity. Another subset of FtsN proteins is on the slow track (~9 nm/s) when in complex with FtsQLB and FtsWI as they synthesize septal PG, activated by FtsN. On the right is a putative fast moving (~30 nm/s) complex containing FtsZ and FtsA that spatially guides FtsQLB and FtsWI so the latter are properly placed to be handed off to the slow track for septum synthesis.
Although FtsA and ZipA are the primary FtsZ membrane tethers in E. coli, other proteins may provide this role in many other species. One of these is SepF, which is highly conserved in Gram-positive bacteria and archaea 170. SepF has an amphipathic helix for binding the membrane, and assembles into curved polymers at the membrane that bind directly to FtsZ and promote protofilament bundling 171,172. Moreover, SepF can functionally replace FtsA in B. subtilis 173 and regulates septum thickness 174, indicating that it shares FtsA’s ability to activate and guide septum synthesis activity. Archaeal SepF lacks a conserved glycine crucial for polymerization, suggesting that polymerization may not be important for SepF function in archaea 170. In addition to SepF, many Gram-positive bacteria express a protein with ZipA-like membrane topology called EzrA that binds to FtsZ and seems to act as a ZipA-like membrane tether. EzrA regulates FtsZ assembly, either negatively in B. subtilis or positively in Streptococcus pneumoniae, where it helps to condense Z rings 175,176. Finally, it is notable that the actinobacteria, which includes important pathogens such as Mycobacterium tuberculosis, completely lack FtsA (and ZipA). Perhaps to replace FtsA’s role, M. tuberculosis divisome proteins such as FtsW and FtsQ bind directly to FtsZ, at least in vitro 177,178, suggesting that FtsQ and FtsW act along with SepF as additional membrane tethers for FtsZ. However, it remains unclear why FtsZ needs multiple membrane tethers, unless they are involved in FtsZ’s early release (see above) or some other type of regulatory mechanism.
The Rings of Power
Consistent with the spatial separation of different divisome proteins mentioned earlier, it was recently discovered that the E. coli divisome consists of both a fast track that includes FtsZ and FtsA, which, as mentioned above, collectively translocate around midcell at an average speed of ~30 nm/sec, and a separate slow track that includes FtsN and the FtsWI processive septal synthase complex that move at ~9 nm/sec (FIG. 4A–B). The fast track uses the GTPase activity of FtsZ to rapidly sample cellular space and optimize positioning of FtsZ polymers to guide septal synthesis. The slow track, on the other hand, reflects direct engagement of proteins in septal wall synthesis, is independent of FtsZ’s GTPase activity, and persists throughout septum closure 167,179. Individual subunits of E. coli FtsA within mini-rings or arcs probably remain transiently stationary as they interact with stationary FtsZ subunits within the interior of treadmilling polymers. Initial FtsN binding to individual FtsA subunits might also be transiently stationary until the concentration of FtsN reaches sufficient levels to convert most of the FtsA into DS filaments, which then could move along the membrane as a unit in response to FtsWI activity. Single molecule tracking suggests that FtsN’s tracking of slow FtsWI movement occurs independently of FtsN-FtsA interactions 167,179, which is also consistent with FtsN interacting most strongly with the FtsWI complex. Whether FtsN remains bound to the FtsWI-directed FtsA DS filaments as septation continues is not clear. Overall, the transfer of proteins between tracks is reminiscent of a fast-moving ski gondola detaching from the main cable and transiently transferring to a slower-moving cable as it enters a lifthouse at the top or bottom of a ski slope to pick up or drop off passengers, followed by its reattachment. It remains to be determined how this protein handoff occurs at the molecular level, including how many FtsA subunits switch from fast to slow tracks after septation has started.
Divisome proteins as potential drug targets
Our improved understanding of the inner workings of the divisome can help develop new inhibitors of bacterial cell division as potential antimicrobials, and their mechanisms of action can shed new light on divisome function. The vast majority of naturally-occurring and synthetic peptide and small molecule inhibitors target FtsZ, and have contributed to our understanding of FtsZ function. For example, the B. subtilis protein MciZ, which inhibits Z ring formation in the mother cell during sporulation, binds to the bottom of an FtsZ subunit, capping the growing filament and increasing bulk GTP hydrolysis 180. This has provided important insights into the role of FtsZ’s GTPase activity and how FtsZ filaments grow 181. Similarly, the potent benzamide compound PC190723 binds to the interdomain cleft (IDC) of FtsZ (FIG. 5), freezing FtsZ monomers in a conformation that prevents treadmilling and thus inhibits cell division, at least in Gram-positive species 182. Newer benzamide derivatives optimized for docking in the IDC can inhibit Gram-positive FtsZ with sub-micromolar affinity and can also inhibit E. coli defective for efflux pumps 183, suggesting strategies that combine the small molecule with an efflux pump inhibitor or outer membrane disruptor 184.
Fig. 5. Binding of a small molecule inhibitor to the FtsZ interdomain cleft.
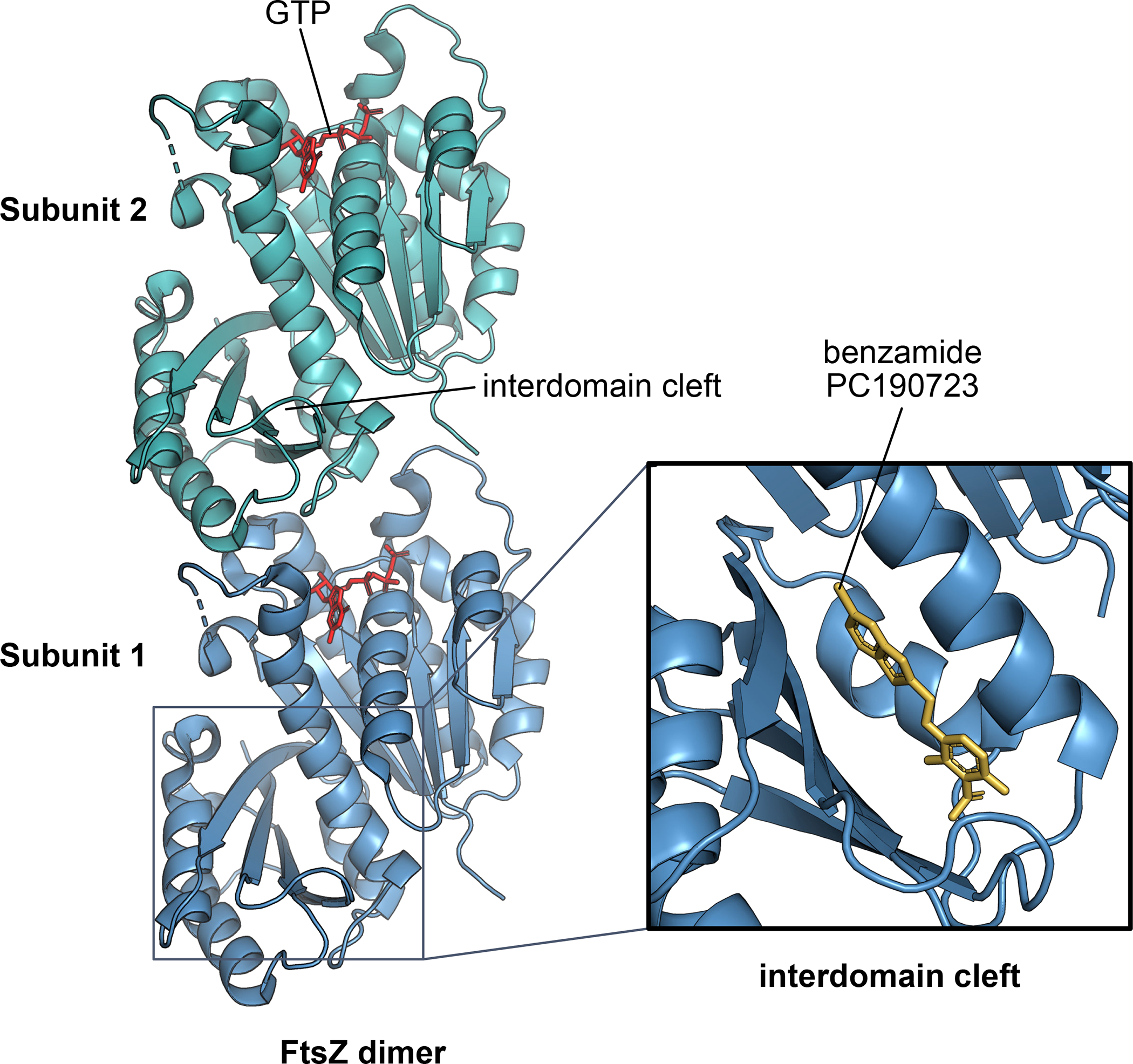
A dimer of the core polymerizing domain of FtsZ is shown, highlighting the interdomain cleft that connects the N-terminal GTP binding domain with the C-terminal domain. Many small molecule inhibitors of FtsZ, including the benzamide PC190723 and its derivatives, insert into the interdomain cleft (inset) and inhibit conformational changes within the FtsZ subunit required for its ability to dynamically treadmill.
Despite these advances, the targeting of other conserved divisome proteins has been limited. For example, no successful FtsA inhibitor has been reported, although there is potential for interfering with FtsA’s oligomeric state transitions. Another promising avenue is the targeting of protein-protein interactions within the divisome, which has been validated by isolation of an inhibitor of FtsQ-FtsB interactions 185. A macrocyclic peptide derived from FtsB was modified to make a covalent interaction, which inhibited division of E. coli cells with membrane permeability defects. In addition to optimizing known divisome inhibitors, it is clear that FtsW and the related RodA protein, now established as peptidoglycan polymerases 14, are particularly rich targets to exploit for antimicrobials because of their size, high conservation, and multiple interaction domains.
Future challenges and questions
Impressive progress has been made in the last few years towards understanding the mechanisms underlying regulation of divisome activity, including the existence of a fast track in the cytoplasm for septal guidance that signals to activate a slow track coordinated with septal wall synthesis. A very recent cryo-EM structure of Pseudomonas aeruginosa FtsQLB complexed with FtsWI reveals a large conformational change in FtsWI between inactive and active states 95. Moreover, another recent study based on structural predictions suggests that the essential role of FtsNE may be to directly tie FtsI and FtsL together in the periplasm 94.
In addition to learning more about how this switch is thrown in other model systems, there are several outstanding questions that remain. How do other parts of the divisome change structurally upon activation, and can alterations in FtsA oligomeric state be observed in dividing cells? What is the mechanism by which hypermorphic alleles such as FtsL*, FtsB*, FtsI* and FtsW* hyperactivate division? What switches divisome proteins from tracking FtsZ to tracking FtsWI and vice versa? More broadly, recent structural breakthroughs cited in this review suggest that a high resolution cryo-EM structure of an entire divisome complex, including the proto-ring, may be possible. Such a structure would help to address the above questions, facilitate the design of better small molecule inhibitors, and get closer to the goal of an autonomously replicating synthetic cell. In addition, improvements in high resolution fluorescence imaging of growing bacteria will be crucial for understanding how the proto-ring assembles at midcell and which cell cycle cues regulate it. Finally, understanding more about the variations on the cell division theme in diverse bacteria should provide the breadth of insight needed to fully understand how the simplest of cells duplicate, and how the first cells evolved.
Acknowledgements
The authors thank the National Institutes of Health (grant R35GM131705) for funding their research.
Footnotes
Competing Interests
The authors declare no competing interests.
REFERENCES
- 1.Bi E & Lutkenhaus J FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991). [DOI] [PubMed] [Google Scholar]
- 2.Yang X et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bisson-Filho AW et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Den Ent F & Löwe J Crystal structure of the cell division protein FtsA from Thermotoga maritima. EMBO J 19, 5300–5307 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pichoff S & Lutkenhaus J Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J 21, 685–693. (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pichoff S & Lutkenhaus J Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55, 1722–1734 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Hale CA & de Boer PA Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Rico AI, Krupka M & Vicente M In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J Biol Chem 288, 20830–6 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du S, Henke W, Pichoff S & Lutkenhaus J How FtsEX localizes to the Z ring and interacts with FtsA to regulate cell division. Mol. Microbiol. 112, 881–895 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM & Nanninga N Timing of FtsZ assembly in Escherichia coli. J Bacteriol 181, 5167–5175 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aarsman ME et al. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol 55, 1631–1645 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Attaibi M & den Blaauwen T An Updated Model of the Divisome: Regulation of the Septal Peptidoglycan Synthesis Machinery by the Divisome. Int. J. Mol. Sci. 23, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt KL et al. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J Bacteriol 186, 785–793 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taguchi A et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang KH, Durand-Heredia J & Janakiraman A FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J Bacteriol 195, 1859–1868 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin PA & Janakiraman A Localization, Assembly, and Activation of the Escherichia coli Cell Division Machinery. EcoSal Plus 9, eESP00222021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Q & Margolin W FtsZ dynamics during the cell division cycle of live Escherichia coli. J Bacteriol 180, 2050–2056 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowlett VW & Margolin W The Min system and other nucleoid-independent regulators of Z ring positioning. Front. Microbiol. 6, 478 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher MA Bacterial nucleoid occlusion: multiple mechanisms for preventing chromosome bisection during cell division. Subcell. Biochem. 84, 267–298 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Bernhardt TG & de Boer PA SlmA, a nucleoid-associated, FtsZ-binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18, 555–564 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Boer PAJ, Crossley RE & Rothfield LI A division inhibitor and a topological specificity factor coded for by the minicell locus determine the proper placement of the division site in Escherichia coli. Cell 56, 641–649 (1989). [DOI] [PubMed] [Google Scholar]
- 22.Lutkenhaus J & Sundaramoorthy M MinD and role of the deviant Walker A motif, dimerization and membrane binding in oscillation. Mol Microbiol 48, 295–303 (2003). [DOI] [PubMed] [Google Scholar]
- 23.de Boer PAJ, Crossley RE, Hand AR & Rothfield LI The MinD protein is a membrane ATPase required for the correct placement of the Escherichia coli division site. EMBO J 10, 4371–4380 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu Z & Lutkenhaus J Topological Regulation of Cell Division in E. coli. Spatiotemporal Oscillation of MinD Requires Stimulation of Its ATPase by MinE and Phospholipid. Mol Cell 7, 1337–1343. (2001). [DOI] [PubMed] [Google Scholar]
- 25.Raskin DM & de Boer PA The MinE ring: an FtsZ-independent cell structure required for selection of the correct division site in E. coli. Cell 91, 685–694 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Huang KC, Meir Y & Wingreen NS Dynamic structures in Escherichia coli: spontaneous formation of MinE rings and MinD polar zones. Proc Natl Acad Sci U A 100, 12724–8. (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raskin DM & de Boer PA MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181, 6419–6424 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glock P et al. Stationary Patterns in a Two-Protein Reaction-Diffusion System. ACS Synth. Biol. 8, 148–157 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Loose M, Fischer-Friedrich E, Ries J, Kruse K & Schwille P Spatial regulators for bacterial cell division self-organize into surface waves in vitro. Science 320, 789–792 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Ramm B et al. The MinDE system is a generic spatial cue for membrane protein distribution in vitro. Nat. Commun. 9, 3942 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Godino E, Doerr A & Danelon C Min waves without MinC can pattern FtsA-anchored FtsZ filaments on model membranes. Commun. Biol. 5, 675 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z & Lutkenhaus J Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ. J Bacteriol 182, 3965–3971 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raskin DM & de Boer PA Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96, 4971–4976 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanedar S & Margolin W FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr Biol 14, 1167–1173 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisicchia P, Arumugam S, Schwille P & Sherratt D MinC, MinD, and MinE drive counter-oscillation of early-cell-division proteins prior to Escherichia coli septum formation. mBio 4, e00856–00813 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbin BD, Yu X-C & Margolin W Exploring intracellular space: function of the Min system in round-shaped Escherichia coli. EMBO J 21, 1988–2008 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweizer J et al. Geometry sensing by self-organized protein patterns. Proc. Natl. Acad. Sci. U. S. A. 109, 15283–15288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juarez JR & Margolin W Changes in the Min oscillation pattern before and after cell birth. J Bacteriol 192, 4134–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosal D, Trambaiolo D, Amos LA & Lowe J MinCD cell division proteins form alternating copolymeric cytomotive filaments. Nat. Commun. 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conti J, Viola MG & Camberg JL The bacterial cell division regulators MinD and MinC form polymers in the presence of nucleotide. FEBS Lett. 589, 201–206 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Huang H et al. The cell division protein MinD from Pseudomonas aeruginosa dominates the assembly of the MinC-MinD copolymers. J. Biol. Chem. 293, 7786–7795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang N, Zhang T, Du S, Zhou Y & Chen Y How do MinC-D copolymers act on Z-ring localization regulation? A new model of Bacillus subtilis Min system. Front. Microbiol. 13, 841171 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park K-T, Du S & Lutkenhaus J MinC/MinD copolymers are not required for Min function. Mol. Microbiol. 98, 895–909 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernhardt TG & de Boer PA SlmA, a nucleoid-associated, FtsZ-binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18, 555–564 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tonthat NK et al. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J 30, 154–64 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du S & Lutkenhaus J SlmA antagonism of FtsZ assembly employs a two-pronged mechanism like MinCD. PLoS Genet. 10, e1004460 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumacher MA & Zeng W Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ. Proc. Natl. Acad. Sci. U. S. A. 113, 4988–4993 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cho H, McManus HR, Dove SL & Bernhardt TG Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci U A 108, 3773–8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tonthat NK et al. SlmA forms a higher-order structure on DNA that inhibits cytokinetic Z-ring formation over the nucleoid. Proc Natl Acad Sci U A 110, 10586–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu LJ & Errington J Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117, 915–925 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Adams DW, Wu LJ & Errington J Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J. 34, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu Y et al. The division defect of a Bacillus subtilis minD noc double mutant can be suppressed by Spx-dependent and Spx-independent mechanisms. J. Bacteriol. 203, e0024921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monterroso B et al. Bacterial FtsZ protein forms phase-separated condensates with its nucleoid-associated inhibitor SlmA. EMBO Rep. 20, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babl L, Merino-Salomón A, Kanwa N & Schwille P Membrane mediated phase separation of the bacterial nucleoid occlusion protein Noc. Sci. Rep. 12, 17949 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paccione G et al. Lipid surfaces and glutamate anions enhance formation of dynamic biomolecular condensates containing bacterial cell division protein FtsZ and its DNA-bound regulator SlmA. Biochemistry 15, 2482–2489 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu J, Liu Y, Yin H & Chang Z Regrowth-delay body as a bacterial subcellular structure marking multidrug-tolerant persisters. Cell Discov. 5, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robles-Ramos MÁ et al. Assembly of bacterial cell division protein FtsZ into dynamic biomolecular condensates. Biochim. Biophys. Acta Mol. Cell Res. 1868, 118986 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harms A, Maisonneuve E & Gerdes K Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354, aaf4268 (2016). [DOI] [PubMed] [Google Scholar]
- 59.Ramm B et al. A phase-separated biomolecular condensate nucleates polymerization of the tubulin homolog FtsZ to spatiotemporally regulate bacterial cell division. bioRxiv (2022) doi: 10.1101/2022.09.12.507586. [DOI] [Google Scholar]
- 60.Hoang Y, Azaldegui CA, Ghalmi M, Biteen JS & Vecchiarelli AG An experimental framework to assess biomolecular condensates in bacteria. bioRxiv 2023.03.22.533878 (2023) doi: 10.1101/2023.03.22.533878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osawa M & Erickson HP Liposome division by a simple bacterial division machinery. Proc Natl Acad Sci USA 110, 11000–11004 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Godino E et al. De novo synthesized Min proteins drive oscillatory liposome deformation and regulate FtsA-FtsZ cytoskeletal patterns. Nat. Commun. 10, 4969 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshida A, Kohyama S, Fujiwara K, Nishikawa S & Doi N Regulation of spatiotemporal patterning in artificial cells by a defined protein expression system. Chem. Sci. 10, 11064–11072 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furusato T et al. De Novo Synthesis of Basal Bacterial Cell Division Proteins FtsZ, FtsA, and ZipA Inside Giant Vesicles. ACS Synth. Biol. 7, 953–961 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Godino E et al. Cell-free biogenesis of bacterial division proto-rings that can constrict liposomes. Commun. Biol. 3, 539 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kohyama S, Merino-Salomón A & Schwille P In vitro assembly, positioning and contraction of a division ring in minimal cells. Nat. Commun. 13, 6098 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Godino E & Danelon C Gene-Directed FtsZ Ring Assembly Generates Constricted Liposomes with Stable Membrane Necks. Adv. Biol. 7, e2200172 (2023). [DOI] [PubMed] [Google Scholar]
- 68.Espeli O et al. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31, 3198–3211 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galli E & Gerdes K Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol 76, 1514–26 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Mannik J & Bailey MW Spatial coordination between chromosomes and cell division proteins in Escherichia coli. Front. Microbiol. 6, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buss J et al. A multi-layered protein network stabilizes the Escherichia coli FtsZ-ring and modulates constriction dynamics. PLoS Genet. 11, e1005128 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coltharp C, Buss J, Plumer TM & Xiao J Defining the rate-limiting processes of bacterial cytokinesis. Proc. Natl. Acad. Sci. U. S. A. 113, E1044–1053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Buss JA, Peters NT, Xiao J & Bernhardt TG ZapA and ZapB form an FtsZ-independent structure at midcell. Mol. Microbiol. 104, 652–663 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schumacher D et al. The PomXYZ proteins self-organize on the bacterial nucleoid to stimulate cell division. Dev. Cell 41, 299–314.e13 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Fleurie A et al. MapZ marks the division sites and positions FtsZ rings in Streptococcus pneumoniae. Nature 516, 259–262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Willemse J, Borst JW, de Waal E, Bisseling T & van Wezel GP Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev 25, 89–99 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Briggs NS, Bruce KE, Naskar S, Winkler ME & Roper DI The Pneumococcal Divisome: Dynamic Control of Streptococcus pneumoniae Cell Division. Front. Microbiol. 12, 737396 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Buddelmeijer N & Beckwith J A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol Microbiol 52, 1315–1327 (2004). [DOI] [PubMed] [Google Scholar]
- 79.Khadria AS & Senes A The transmembrane domains of the bacterial cell division proteins FtsB and FtsL form a stable high-order oligomer. Biochemistry 52, 7542–7550 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen JC, Weiss DS, Ghigo JM & Beckwith J Septal localization of FtsQ, an essential cell division protein in Escherichia coli. J Bacteriol 181, 521–530 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hale CA & de Boer PA Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J Bacteriol 181, 167–176 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu G, Draper GC & Donachie WD FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol Microbiol 29, 893–903 (1998). [DOI] [PubMed] [Google Scholar]
- 83.Aussel L et al. FtsK Is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108, 195–205. (2002). [DOI] [PubMed] [Google Scholar]
- 84.Dubarry N, Possoz C & Barre F-X Multiple regions along the Escherichia coli FtsK protein are implicated in cell division. Mol. Microbiol. 78, 1088–1100 (2010). [DOI] [PubMed] [Google Scholar]
- 85.Saaki TNV et al. SepF supports the recruitment of the DNA translocase SftA to the Z-ring. Mol. Microbiol. 117, 1263–1274 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Berezuk AM, Goodyear M & Khursigara CM Site-directed fluorescence labeling reveals a revised N-terminal membrane topology and functional periplasmic residues in the Escherichia coli cell division protein FtsK. J. Biol. Chem. 289, 23287–23301 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Draper GC, McLennan N, Begg K, Masters M & Donachie WD Only the N-terminal domain of FtsK functions in cell division. J Bacteriol 180, 4621–4627 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang L & Lutkenhaus J FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol 29, 731–740 (1998). [DOI] [PubMed] [Google Scholar]
- 89.Yu X-C, Tran AH, Sun Q & Margolin W Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol 180, 1296–1304 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kureisaite-Ciziene D et al. Structural Analysis of the Interaction between the Bacterial Cell Division Proteins FtsQ and FtsB. mBio 9, e01346–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Choi Y et al. Structural Insights into the FtsQ/FtsB/FtsL Complex, a Key Component of the Divisome. Sci. Rep. 8, 18061 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marmont LS & Bernhardt TG A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc. Natl. Acad. Sci. U. S. A. 117, 23879–23885 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Park K-T, Pichoff S, Du S & Lutkenhaus J FtsA acts through FtsW to promote cell wall synthesis during cell division in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 118, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Britton BM et al. Conformational changes in the essential E. coli septal cell wall synthesis complex suggest an activation mechanism. bioRxiv (2022) doi: 10.1101/2022.11.27.518129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kashammer L et al. Divisome core complex in bacterial cell division revealed by cryo-EM. BioRxiv (2022). [Google Scholar]
- 96.Berezuk AM, Roach EJ, Seidel L, Lo RY & Khursigara CM FtsA G50E mutant suppresses the essential requirement for FtsK during bacterial cell division in Escherichia coli. Can. J. Microbiol. 66, 313–327 (2020). [DOI] [PubMed] [Google Scholar]
- 97.Busiek KK, Eraso JM, Wang Y & Margolin W The early divisome protein FtsA interacts directly through Its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol 194, 1989–2000 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Busiek KK & Margolin W A role for FtsA in SPOR-independent localization of the essential Escherichia coli cell division protein FtsN. Mol Microbiol 92, 1212–26 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pichoff S, Du S & Lutkenhaus J The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol. Microbiol. 95, 971–987 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wissel MC & Weiss DS Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J Bacteriol 186, 490–502 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gerding MA et al. Self-enhanced accumulation of FtsN at Division Sites and Roles for Other Proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol 191, 7383–7401 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yahashiri A, Jorgenson MA & Weiss DS The SPOR domain, a widely conserved peptidoglycan binding domain that targets proteins to the site of cell division. J. Bacteriol. 199, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yahashiri A, Jorgenson MA & Weiss DS Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl. Acad. Sci. U. S. A. 112, 11347–11352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bisson-Filho AW et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Perez AJ et al. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A. 116, 3211–3220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McCausland JW et al. Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nat. Commun. 12, 609 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Monteiro JM et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Whitley KD et al. FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nat. Commun. 12, 2448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lyu Z, Coltharp C, Yang X & Xiao J Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional Superresolution imaging. Biopolymers 105, 725–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Soderstrom B et al. Coordinated disassembly of the divisome complex in Escherichia coli. Mol. Microbiol. 101, 425–438 (2016). [DOI] [PubMed] [Google Scholar]
- 111.Silber N, Mayer C, Matos de Opitz CL & Sass P Progression of the late-stage divisome is unaffected by the depletion of the cytoplasmic FtsZ pool. Commun. Biol. 4, 270 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Corbin Goodman LC & Erickson HP FtsZ at mid-cell is essential in Escherichia coli until the late stage of constriction. Microbiol. Read. Engl. 168, (2022). [DOI] [PubMed] [Google Scholar]
- 113.Soderstrom B, Chan H, Shilling PJ, Skoglund U & Daley DO Spatial separation of FtsZ and FtsN during cell division. Mol. Microbiol. (2017) doi: 10.1111/mmi.13888. [DOI] [PubMed] [Google Scholar]
- 114.Mannik J, Walker BE & Mannik J Cell cycle-dependent regulation of FtsZ in Escherichia coli in slow growth conditions. Mol. Microbiol. 110, 1030–1044 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cameron TA, Vega DE, Yu C, Xiao H & Margolin W ZipA Uses a Two-Pronged FtsZ-Binding Mechanism Necessary for Cell Division. mBio 12, e0252921 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Geissler B & Margolin W Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol 58, 596–612 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Geissler B, Elraheb D & Margolin W A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA 100, 4197–4202 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Geissler B, Shiomi D & Margolin W The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology 153, 814–825 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pichoff S, Shen B, Sullivan B & Lutkenhaus J FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA’s self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol 83, 151–67 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Herricks JR, Nguyen D & Margolin W A thermosensitive defect in the ATP binding pocket of FtsA can be suppressed by allosteric changes in the dimer interface. Mol. Microbiol. 94, 713–727 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bernard CS, Sadasivam M, Shiomi D & Margolin W An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol Microbiol 64, 1289–1305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mingorance J, Rico AI, & Gomez-Puertas P Bacterial morphogenes. in Molecules in Time and Space. Bacterial Shape, Division and Phylogeny (eds. Vicente M, Tamames J, Valencia A, & Mingorance J) (Springer, 2004). [Google Scholar]
- 123.Pazos M et al. Z-ring membrane anchors associate with cell wall synthases to initiate bacterial cell division. Nat. Commun. 9, 5090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Krupka M et al. Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat. Commun. 8, 15957 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schoenemann KM et al. Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol. Microbiol. 109, 676–693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nierhaus T et al. Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN. Nat. Microbiol. 7, 1686–1701 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Radler P et al. In vitro reconstitution of Escherichia coli divisome activation. Nat. Commun. 13, 2635 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Conti J, Viola MG & Camberg JL FtsA reshapes membrane architecture and remodels the Z-ring in Escherichia coli. Mol. Microbiol. 107, 558–576 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu B, Persons L, Lee L & de Boer PA Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 95, 945–970 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Dai K, Xu Y & Lutkenhaus J Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts). J Bacteriol 175, 3790–3797 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Goehring NW, Robichon C & Beckwith J A role for the non-essential N-terminus of FtsN in divisome assembly. J Bacteriol 189, 646–649 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pichoff S, Du S & Lutkenhaus J Disruption of divisome assembly rescued by FtsN-FtsA interaction in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 115, E6855–E6862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Männik J, Pichoff S, Lutkenhaus J & Männik J Cell Cycle-Dependent Recruitment of FtsN to the Divisome in Escherichia coli. mBio 13, e0201722 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Karuppiah V & Derrick JP Structure of the PilM-PilN inner membrane type IV pilus biogenesis complex from Thermus thermophilus. J Biol Chem 286, 24434–24442 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Llontop EE et al. The PilB-PilZ-FimX regulatory complex of the Type IV pilus from Xanthomonas citri. PLoS Pathog. 17, e1009808 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chang Y-W et al. Architecture of the type IVa pilus machine. Science 351, aad2001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Du S, Pichoff S & Lutkenhaus J FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl. Acad. Sci. U. S. A. 113, E5052–5061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Reddy M Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J Bacteriol 189, 98–108 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Vega DE & Margolin W Direct interaction between the two Z ring membrane anchors FtsA and ZipA. J. Bacteriol. 201, e00579–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang DC et al. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc Natl Acad Sci USA 108, E1052–1060 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Du S, Pichoff S & Lutkenhaus J Roles of ATP Hydrolysis by FtsEX and Interaction with FtsA in Regulation of Septal Peptidoglycan Synthesis and Hydrolysis. mBio 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Navarro PP et al. Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Nat. Microbiol. 7, 1621–1634 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Szwedziak P, Wang Q, Bharat TAM, Tsim M & Löwe J Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife 3, e04601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shiomi D & Margolin W Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol Microbiol 66, 1396–1415 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Buske PJ & Levin PA A flexible C-terminal linker is required for proper FtsZ assembly in vitro and cytokinetic ring formation in vivo. Mol Microbiol 89, 249–63 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Huecas S et al. Self-Organization of FtsZ Polymers in Solution Reveals Spacer Role of the Disordered C-Terminal Tail. Biophys. J. 113, 1831–1844 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Barrows JM, Sundararajan K, Bhargava A & Goley ED FtsA regulates Z-ring morphology and cell wall metabolism in an FtsZ C-terminal linker-dependent manner in Caulobacter crescentus. J. Bacteriol. 202, e00693–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Squyres GR et al. Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat. Microbiol. 6, 553–562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Haeusser DP, Rowlett VW & Margolin W A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol. Microbiol. 97, 988–1005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Krupka M, Sobrinos-Sanguino M, Jimenez M, Rivas G & Margolin W Escherichia coli ZipA organizes FtsZ polymers into dynamic ring-like protofilament structures. mBio 9, e1008–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jaiswal R et al. E93R substitution of Escherichia coli FtsZ induces bundling of protofilaments, reduces GTPase activity, and impairs bacterial cytokinesis. J Biol Chem 285, 31796–805 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Karimova G, Dautin N & Ladant D Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol 187, 2233–2243 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.van den Ent F, Izoré T, Bharat TA, Johnson CM & Löwe J Bacterial actin MreB forms antiparallel double filaments. eLife 3, e02634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hussain S et al. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Szwedziak P & Löwe J Do the divisome and elongasome share a common evolutionary past? Curr Opin Microbiol 16, 745–751 (2013). [DOI] [PubMed] [Google Scholar]
- 156.Begg KJ et al. The balance between different peptidoglycan precursors determines whether Escherichia coli cells will elongate or divide. J Bacteriol 172, 6697–6703 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lleo MM, Canepari P & Satta G Bacterial cell shape regulation: testing of additional predictions unique to the two-competing-sites model for peptidoglycan assembly and isolation of conditional rod-shaped mutants from some wild-type cocci. J Bacteriol 172, 3758–71 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tsang MJ & Bernhardt TG A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95, 925–944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Park K-T, Du S & Lutkenhaus J Essential Role for FtsL in Activation of Septal Peptidoglycan Synthesis. mBio 11, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Y et al. Genetic analysis of the septal peptidoglycan synthase FtsWI complex supports a conserved activation mechanism for SEDS-bPBP complexes. PLoS Genet. 17, e1009366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lariviere PJ et al. An Essential Regulator of Bacterial Division Links FtsZ to Cell Wall Synthase Activation. Curr. Biol. CB 29, 1460–1470.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kiekebusch D, Michie KA, Essen LO, Lowe J & Thanbichler M Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol Cell 46, 245–59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mahone CR et al. Integration of cell wall synthesis activation and chromosome segregation during cell division in Caulobacter. bioRxiv (2022) doi: 10.1101/2022.11.05.515301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gallay C et al. CcrZ is a pneumococcal spatiotemporal cell cycle regulator that interacts with FtsZ and controls DNA replication by modulating the activity of DnaA. Nat. Microbiol. 6, 1175–1187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Loose M & Mitchison TJ The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat Cell Biol 16, 38–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Baranova N et al. Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins. Nat. Microbiol. 5, 407–417 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Lyu Z et al. FtsN maintains active septal cell wall synthesis by forming a processive complex with the septum-specific peptidoglycan synthases in E. coli. Nat. Commun. 13, 5751 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Asymmetric localization of the cell division machinery during Bacillus subtilis sporulation. vol. 10 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cameron TA & Margolin W A fluorescent reporter for FtsA is functional as the sole FtsA in Escherichia coli and has hypermorphic properties. bioRxiv (2022) doi: 10.1101/2022.10.31.514644. [DOI] [Google Scholar]
- 170.Pende N et al. SepF is the FtsZ anchor in archaea, with features of an ancestral cell division system. Nat. Commun. 12, 3214 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Duman R et al. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc. Natl. Acad. Sci. 110, E4601–E4610 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dey S & Zhou H-X Membrane Tethering of SepF, a Membrane Anchor for the Mycobacterium tuberculosis Z-ring. J. Mol. Biol. 434, 167817 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M & Ogasawara N A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 60, 1364–1380 (2006). [DOI] [PubMed] [Google Scholar]
- 174.Wenzel M et al. Control of septum thickness by the curvature of SepF polymers. Proc. Natl. Acad. Sci. U. S. A. 118, e2002635118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Levin PA, Kurtser IG & Grossman AD Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc Natl Acad Sci USA 96, 9642–9647 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Perez AJ et al. FtsZ-Ring Regulation and Cell Division Are Mediated by Essential EzrA and Accessory Proteins ZapA and ZapJ in Streptococcus pneumoniae. Front. Microbiol. 12, 780864 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Datta P, Dasgupta A, Bhakta S & Basu J Interaction between FtsZ and FtsW of Mycobacterium tuberculosis. J Biol Chem 277, 24983–24987. (2002). [DOI] [PubMed] [Google Scholar]
- 178.Smrt ST, Escobar CA, Dey S, Cross TA, & Xhou HX An Arg/Ala-rich helix in the N-terminal region of M. tuberculosis FtsQ is a potential membrane anchor of the Z-ring. Commun Biol 6, 311 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yang X et al. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. 6, 584–593 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bisson-Filho AW et al. FtsZ filament capping by MciZ, a developmental regulator of bacterial division. Proc. Natl. Acad. Sci. U. S. A. 112, E2130–2138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Du S, Pichoff S, Kruse K & Lutkenhaus J FtsZ filaments have the opposite kinetic polarity of microtubules. Proc. Natl. Acad. Sci. U. S. A. 115, 10768–10773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Adams DW, Wu LJ, Czaplewski LG & Errington J Multiple effects of benzamide antibiotics on FtsZ function. Mol Microbiol 80, 68–84 (2011). [DOI] [PubMed] [Google Scholar]
- 183.Straniero V et al. Computational design and development of benzodioxane-benzamides as potent inhibitors of FtsZ by exploring the hydrophobic subpocket. Antibiotics 10, 442 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Du R-L et al. Gossypol acetate: A natural polyphenol derivative with antimicrobial activities against the essential cell division protein FtsZ. Front. Microbiol. 13, 1080308 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Paulussen FM et al. Covalent proteomimetic inhibitor of the bacterial FtsQB divisome complex. J. Am. Chem. Soc. 144, 15303–15313 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]


