Abstract
Objectives
This study aimed to explore the potential correlation between specific single nucleotide polymorphisms (TYK2, IFITM3, IFNAR2, and OAS3 variants) and the severity of COVID-19 in Moroccan patients.
Methods
A genetic analysis was conducted on 109 patients with PCR-confirmed SARS-CoV-2 infection in Morocco. Among these patients, 46% were hospitalized in the intensive care unit, while 59% were not hospitalized. Importantly, all patients lacked known risk factors associated with COVID-19 severity. Genotyping was performed to identify variations in TYK2 rs74956615, IFITM3 rs12252, IFNAR2 rs2236757, and OAS3 rs10735079. Statistical analysis was applied using codominant, dominant and recessive logistic regression models to assess correlations with COVID-19 severity.
Results
Our findings revealed no significant correlation between TYK2 rs74956615, IFITM3 rs12252, IFNAR2 rs2236757, and OAS3 rs10735079 with COVID-19 severity in Moroccan patients, as indicated in logistic regression models (p > .05). Interestingly, these results may offer insights into the mitigated impact of the COVID-19 pandemic and the reduced severity observed in SARS-CoV-2 infected patients in Morocco. Age, however, exhibited a significant correlation with severity (p < .001), with a trend towards increased likelihood of ICU admission with advancing age. Additionally, In the severe group, a higher proportion of patients were females (54%), indicating a statistically significant correlation with disease severity (p = .04). Nevertheless, female ICU patients aged above 60 years accounted for 37%, compared to 17% for males.
Conclusion
This study underscores the absence of a genetic association between the selected polymorphisms and COVID-19 severity in Moroccan patients. Advanced age emerges as the primary factor influencing the severity of COVID-19 patients without comorbidities. We recommend setting the threshold for advanced age at 60 years as a risk factor for severe forms of COVID-19.
Keywords: COVID-19, host genetics, genetic polymorphism, severity, TYK2, IFITM3, IFNAR2, OAS3
Introduction
Since the emergence of the COVID-19 pandemic, numerous scientists have been dedicated to unraveling the underlying mechanisms of SARS-CoV-2 infection. Beyond the study of the virus, host genetics play a crucial role in the clinical variability and severity of the response to this infection. 1 COVID-19, indeed, presents in a diverse array of symptoms among patients, spanning from asymptomatic form to multi-visceral failure requiring intensive care hospitalization or, in some cases, leads to fatal outcomes. 2 Various host-related risk factors have been linked with severe forms of COVID-19 including age, gender and other comorbidities such as hypertension, diabetes, obesity and more.3,4 Conversely, other studies suggest that host genetics and genetic polymorphisms in specific genes significantly influence the variation in COVID-19 disease phenotypes.1,5,6
Among the polymorphisms extensively studied for their involvement in the severity of SARS-CoV-2 infection, we identified the single nucleotide polymorphism (SNP) rs2236757 in the interferon alpha and beta receptor subunit 2 (IFNAR2) gene. This latter plays a crucial role in the antiviral immune response.7,8 Other interferon pathways implicated in severe forms of COVID-19 involve the Tyrosine Kinase 2 (TYK2) gene, which also plays a significant role in the antiviral immune response and whose TYK2 polymorphism rs74956615 has been correlated with severe forms. 9 In addition, the Interferon Induced Transmembrane Protein 3 (IFITM3) gene, responsible for encoding an immune effector protein crucial for viral restriction by limiting membrane fusion, harbors an IFITM3 rs12252 polymorphism. This variant has been associated to hospitalization and mortality in COVID-19 patients within a large Arab population.10,11 More recently, 2′-5′-oligoadenylate synthetase (OAS) genes have been found to be highly expressed in cardiomyocytes of SARS-CoV-2 infected patients and may serve as a therapeutic target for COVID-19 cardiac lesions. 12 Additionally, the OAS3 SNP rs10735079 has been associated with the risk of hospitalization. 13
Genome Wide Association Studies (GWAS) and several other In Silico and In vitro studies have affirmed the association of these genes with the severity of COVID-19.9,14–16 However, the associations of COVID-19 with the polymorphisms of these genes have not been the subject of study in Morocco.
The study at hand aims to investigate the correlation between severe forms of COVID-19 with TYK2, IFITM3, IFNAR2 and OAS3 polymorphisms. The purpose of this investigation is to identity the genetic variants associated with the hospitalization of Moroccan patients in intensive care unit (ICU). Consequently, the data derived from our work are valuable for illustrating the potential role of host genetic factors in the pathogenesis of SARS-CoV-2 infection.
Materials and methods
Study participants
The study recruited 109 Moroccan patients admitted to the virology laboratory of the Virology Center for Infectious and Tropical Diseases of the Mohammed V Military Training Hospital or to the Ibn Sina University Hospital Center between 2021 and 2022 for a COVID-19 diagnosis. All patients tested positive for SARS-CoV-2 virus through real-time Polymerase chain reaction (PCR) on a nasopharyngeal swab. Subsequently, socio-demographic data, medical and surgical history, as well as symptomatology were collected from the patients. Patients were then followed up and classified based on disease severity and clinical course. Following the clinical classification set by National Health Commission & National Administration of Traditional Chinese Medicine, severe cases were defined by the presence of significant respiratory distress (>30 breaths/min), oxygen saturation ≤93%, arterial partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) ≤300 mmHg, respiratory failure with mechanical ventilation, shock, or other organ failure requiring admission in the intensive care unit. 17
Inclusion criteria for this study encompassed SARS-CoV-2 positive patients aged between 18 and 65 years. Participants were required to have no identifiable risk factors or comorbidities associated with increased severity of COVID-19. This stringent selection aimed to isolate the impact of genetic factors on COVID-19 severity.
Our cohort consisted of 50 critically ill patients with severe forms of COVID-19 admitted to intensive care units, while 59 non-critically ill patients exhibited mild or asymptomatic forms of the disease, requiring neither respiratory assistance nor hospitalization.
For ethical considerations, written informed consent, including all necessary information about the study, was obtained from all patients or from an alternative legal representative acting on behalf of intensive care unit patients. The informed consent provided comprehensive details about the study, and participation was voluntary, allowing individuals to withdraw from the study at any time.
Sample size calculation
To ensure adequate statistical power for the detection of genetic associations in our study, we performed a sample size calculation using the pwr package in R. Our objective was to determine the number of participants required to identify significant associations between specific genetic loci and the disease under investigation with sufficient statistical power.
Parameters for sample size calculation
Significance Level (α): We set a significance level of 0.01 to minimize the probability of Type I error.
Power (1 - β): The desired power of the study was established at 0.8, indicating an 80% probability of detecting a true association.
Effect Size (Odds Ratio): An effect size with an odds ratio of 2 was assumed, based on the expected strength of association between the genotypes and the disease.
Control-to-Case Ratio: A 1:1 ratio of controls to cases was used, reflecting an equal distribution of participants in each group.
The odds ratio was first converted to log odds to facilitate the calculation of the effect size.
After which the effect size was computed using Cohen’s h, which is suitable for chi-squared tests in case-control studies. We assumed a 50% allele frequency in the control group for this calculation.
Using the pwr. chisq.test function from the pwr package, we calculated the total sample size required. This function is specifically designed for power calculations in studies utilizing chi-squared tests. In this case, the effect size happens to be 102.
Genotyping
Polymorphisms were selected via our upstream literature search on PubMed, for SNP analysis in the Moroccan population. For all studied patients, 2 mL EDTA tubes were collected and stored at −20°C. DNA was then extracted from 200 µl of blood using the High pure Genomic Nucleic Acid kit (Roche©). DNA concentration was then determined using a Qubit dsDNA BR Assay Kit (ThermoFisher Scientific©) and normalized to 10 ng/µl. The various polymorphisms, TYK2 rs74956615, IFITM3 rs12252, OAS3 rs10735079 and IFNAR2 rs2236757, were genotyped using specific Human TaqMan SNP Genotyping Assays 40x (ThermoFisher Scientific©), real-time PCR reactions were performed using the TaqPath ProAmp Multiplex Master mix (ThermoFisher Scientific©) and performed in the Quant studio 5 Real-Time PCR System 0.1 mL.
Hardy-weinberg equilibrium analysis
To evaluate the genetic equilibrium of the study population, we performed Hardy-Weinberg Equilibrium (HWE) tests for each of the genotyped loci using the MakeCounts function from the HardyWeinberg package, we converted the genotype data into allele counts suitable for HWE analysis. For each locus, we conducted an exact test of Hardy-Weinberg Equilibrium using the HWExact function from the HardyWeinberg package. 18
Statistical analysis
The application of the Shapiro-Wilk test and graphical representations aimed at testing the hypotheses of normality for numerical variables. Meanwhile, for clinical and demographic parameters with a non-Gaussian distribution, the Kruskal-Wallis test was carried to determine differences between groups. Regarding associations between categorical variables, the chi-squared test and Fischer’s exact test were employed. The correlation between ICU admission and SNPs was investigated using the SNPStats software. Logistic regression models were subsequently employed across all models, encompassing codominant, dominant, and recessive models. All analyses, including graphical representations, were conducted using Rstudio (version R 4.2.3). We considered a p value <.05 to be statistically significant.
Ethics committee
The protocol is designated as a non-interventional study in accordance with Moroccan law 28-13 on the Protection of Individuals Involved in Biomedical Research, and therefore does not require an assessment of the ethical committee or IRB approval. (https://www.sante.gov.ma/Reglementation/REGLEMENTATIONDESPRATIQUESMEDICALES/28-13.pdf). Written informed consent was diligently obtained from all participating patients, ensuring adherence to ethical standards.
Results
Demographic and phenotypic data are detailed in Table 1, where the mean age of all patients was 40 ± 13.8 years (minimum 22 and maximum 65). Our findings provide evidence of a statistically significant association between advance age and disease severity when comparing asymptomatic, mild and severe patients (p < .001). The median ages for asymptomatic, mild and severe cases were 26, 30 and 51 years respectively. Notably, Among patients with severe forms of the infection we observed 14 patients (28%) were aged >61 years (third quantile).
Table 1.
Patients demographics by clinical phenotype.
| Characteristic | N | Overall, N = 109 | Asymptomatic, N = 16 | Mild, N = 43 | Severe, N = 50 | p-value a |
|---|---|---|---|---|---|---|
| Age, median (IQR) | 109 | 36 (28, 51) | 26 (25, 32) | 30 (25,39) | 51 (39,61) | <0.001 |
| Sex, n (%) | 109 | 0.047 | ||||
| F | 51 (47%) | 10 (63%) | 14 (33%) | 27 (54%) | ||
| M | 58 (53%) | 6 (38%) | 29 (67%) | 23 (46%) | ||
| Hospitalization, n (%) | 109 | <0.001 | ||||
| Intensive care unit patients | 50 (46%) | 0 (0%) | 0 (0%) | 50 (100%) | ||
| Not hospitalized | 59 (54%) | 16 (100%) | 43 (100%) | 0 (0%) | ||
| Fever, n (%) | 109 | 61 (56%) | 0 (0%) | 28 (65%) | 33 (66%) | <0.001 |
| Asthenia, n (%) | 109 | 59 (54%) | 0 (0%) | 25 (58%) | 34 (68%) | <0.001 |
| Headaches, n (%) | 109 | 28 (26%) | 0 (0%) | 16 (37%) | 12 (24%) | 0.007 |
| ARDS b , n (%) | 109 | 50 (46%) | 0 (0%) | 0 (0%) | 50 (100%) | <0.001 |
| Pharyngitis, n (%) | 109 | 18 (17%) | 0 (0%) | 11 (26%) | 7 (14%) | 0.051 |
| Anosmia, n (%) | 109 | 33 (30%) | 0 (0%) | 24 (56%) | 9 (18%) | <0.001 |
| Agueusia, n (%) | 109 | 27 (25%) | 0 (0%) | 24 (56%) | 3 (6%) | <0.001 |
| Chill, n (%) | 109 | 27 (25%) | 0 (0%) | 12 (28%) | 15 (30%) | 0.025 |
| Myalgia, n (%) | 109 | 42 (39%) | 0 (0%) | 24 (56%) | 18 (36%) | <0.001 |
| Dyspnea, n (%) | 109 | 42 (39%) | 0 (0%) | 12 (28%) | 30 (60%) | <0.001 |
| Cough, n (%) | 109 | 55 (50%) | 0 (0%) | 29 (67%) | 26 (52%) | <0.001 |
| Digestive trouble, n (%) | 109 | 27 (25%) | 0 (0%) | 17 (40%) | 10 (20%) | 0.003 |
bARDS = Acute Respiratory Distress Syndrome.
aKruskal-Wailis rank sum test; Person’s Chi-squared test; Fisher’s exact test.
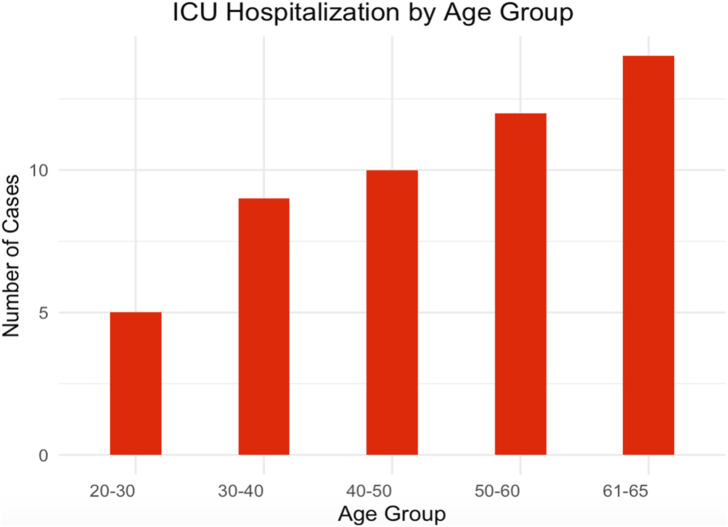
Our results indicate a correlation between age groups and ICU hospitalization, with a trend towards increased likelihood of ICU admission with advancing age, as observed in Figure 1.
Figure 1.
ICU Hospitalization by age group.
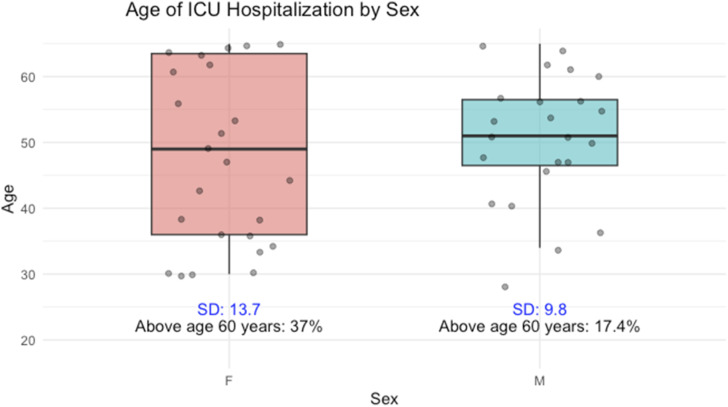
Additionally, in the severe group, a majority of patients were females (54%), and this difference was statically significant (p = .04), indicating a correlation between sex and disease severity. Regarding ICU patients, the standard deviation of ages for women is 13.7, while for men it is 9.8. Moreover, among patients aged over 60 years hospitalized in intensive care unit, 37% were women and 17.4% were men.
In order to determine the association between COVID-19 severity and the selected SNPs, genotyping was performed on the cohort of 109 patients. Results indicate no discernible correlation between the studied polymorphisms and the manifestation of severe SARS-CoV-2 infection. Logistic regression models indicated no statistically significant disparities detected between patients requiring intensive care and those not hospitalized concerning the SNPs IFNAR2 (rs2236757), TYK2 (rs74956615), OAS3 (rs10735079), and IFITM3 (rs12252) (p > .05) (Table 2). Furthermore, Hardy-Weinberg Equilibrium tests conducted on the genotyped loci consistently yielded p-values exceeding 0.05, affirming the adherence of all four loci to the principles of Hardy-Weinberg Equilibrium.
Table 2.
Association of genetic polymorphisms with COVID-19 ICU hospitalization.
| Model | Genotype | ICU Hospitalization | Not hospitalized | Odd Ratio (95% CI) | p-value |
|---|---|---|---|---|---|
| OAS rs10735079 | |||||
| Codominant | A/A | 24 (48%) | 34 (57.6%) | 1.00 | 0.3 |
| A/G | 23 (46%) | 19 (32.3%) | 0.58 (0.26-1.30) | ||
| G/G | 3 (6%) | 6 (10.2%) | 1.41 (0.32-6.21) | ||
| Dominant | A/A | 24 (48%) | 34 (57.6%) | 1.00 | 0.32 |
| A/G-G/G | 26 (52%) | 25 (42.4%) | 0.68 (0.32-1.45) | ||
| Recessive | A/A-A/G | 47 (94%) | 47 (94%) | 1.00 | 0.43 |
| G/G | 3 (6%) | 3 (6%) | 1.77 (0.42-7.49) | ||
| IFITM3 rs12252 | |||||
| Codominant | A/A | 31 (62%) | 41 (69.5%) | 1.00 | 0.36 |
| A/G | 18 (36%) | 18 (30.5%) | 0.76 (0.34-1.69) | ||
| G/G | 1 (2%) | 0 (0%) | 0.00 (0.00-NA) | ||
| Dominant | A/A | 31 (62%) | 41 (69.5%) | 1.00 | 0.41 |
| A/G-G/G | 19 (38%) | 18 (30.5%) | 0.72 (0.32-1.59) | ||
| Recessive | A/A-A/G | 49 (98%) | 59 (100%) | 1.00 | 0.21 |
| G/G | 1 (2%) | 0 (0%) | 0.00 (0.00-NA) | ||
| IFNAR2 rs2236757 | |||||
| Codominant | G/G | 29 (6%) | 29 (49.1%) | 1.00 | 0.44 |
| A/G | 18 (36%) | 28 (47.5) | 1.59 (0.71-3.41) | ||
| A/A | 3 (6%) | 2 (3.4%) | 0.67 (0.10-4.29) | ||
| Dominant | G/G | 29 (58%) | 29 (49.1%) | 1.00 | 0.36 |
| A/G-A/A | 21 (42%) | 30 (50.9%) | 1.43 (0.67-3.05) | ||
| Recessive | G/G-A/G | 47 (94%) | 57 (96.6) | 1.00 | 0.52 |
| A/A | 3 (6%) | 2 (3.4%) | 0.55 (0.09-3.43) | ||
| TYK2 rs74956615 | |||||
| T/T | 49 (98%) | 54 (91.5) | 1.00 | 0.12 | |
| T/A | 1 (2%) | 5 (8.5) | 4.54 (0.51-40.20) | ||
Discussion
The primary objective of our investigation was to determine whether genetic polymorphisms of the four appropriately selected genes are associated with severity or ICU hospitalization of Moroccan patients infected with SARS-CoV-2. Our findings indicate that the selected SNPs in the IFNAR2, TYK2, IFITM3 and OAS3 genes are not associated with susceptibility to severe forms in Moroccan patients.
The association of the IFNAR2 rs223675 locus was associated with critical forms of COVID-19 was identified in a GWAS study. 14 Yet, another study managed to associate the SNP rs2236757 A/A with the risk of ICU admission and death in non-white patients. 15 This contrast with our results may be explained by the inter-ethnic variability in frequencies of the IFNAR2 rs2236757 variant. 19 Similarly, GWAS reported that the TYK2 rs74956615 polymorphism was associated with critical forms of COVID-19. 14 However, a recent study demonstrated an association of this polymorphism with a reduced risk of COVID-19 complications and found no association with severe forms, aligning with our results. 9 The IFITM3 rs12252 SNP has been extensively studied in different populations due to its previous correlation with severe forms of influenza and other viral infections.20–22 Indeed, the SNP IFITM3 rs12252 was associated with hospitalization (p = .04) as well as death in COVID-19 patients (p < .0001).10,23 Conversely, our results align with those obtained in a German cohort since no correlation of the rs12252 polymorphism with susceptibility and severity was reported (p > .05). 24 Meanwhile, the role of IFITM3 gene variants in SARS-CoV-2 infections requires elucidation in ethnically diverse cohorts. Additionally, association studies has shown that the OAS gene locus represents a region associated with COVID-19 severity in individuals of European ancestry.14,25 However, our investigation of the OAS SNP rs10735079 reveals no relationship between the polymorphism and COVID-19 severity (p > .05). Similarly, Abdelhafez and his team observed no significant difference in the distribution of OAS3 SNP genotypes (p = .091).
It’s crucial to emphasize that our study specifically focuses on the impact of these genes on the Moroccan population, which had been previously recognized through GWAS as susceptibility genes to severe forms of the disease in diverse populations. Notably, these genes, extensively studied in the literature, have been implicated not only in the severity of COVID-19 but also in the severity of other viral infections across diverse populations. However, the associations of COVID-19 with the polymorphisms of these genes have not been the subject of study in Morocco. This gap in research has prompted us to investigate these associations among Moroccan patients, particularly considering, on one hand, that these SNPs have been implicated in severe forms of COVID-19 across various populations and large cohorts, and on the other hand, that these SNPs are present within genes crucial to antiviral immune response, suggesting a potential link to the severity of COVID-19 in patients without comorbidities. Therefore, our findings regarding the lack of association of these polymorphisms in our cohort could explain the low impact of the COVID-19 pandemic and the lower severity of SARS-CoV-2 infected patients in Morocco compared with Europe, America and Asia. 26
As presented, the relationship between polymorphisms and COVID-19 severity is inconsistent in previous studies conducted worldwide. This discrepancy may be attributed to ethnic variations between countries, and these variants indicate some population-based differences. However, it should be noted that other factors could explain the disparities between our results and those of previously cited studies, such as interactions with other polymorphisms or the influence of other risk and protective factors within each population.26,27 Indeed, our results showed a significant correlation between age and the severity of COVID-19 (p < .001), with severe cases having a median age of 51 years. Additionally, a peak was observed in the age group between 61 and 65 years in the ICU hospitalization group. This indicates that severity increases with advancing age as illustrated in Figure 2. These results align with existing literature. It is noteworthy that the number of patients hospitalized in ICU aged between 20 and 60 years, and without any comorbidities, is non-negligible. This underscores the necessity for exploring additional genetic polymorphisms that could be associated with the severity of COVID-19 in Morocco. Such investigations are crucial for a comprehensive understanding of the genetic factors influencing COVID-19 severity.
Figure 2.
Age of ICU hospitalization by sex.
In addition, our results revealed a significant correlation between gender and severity (p = .04), revealing a predominance of female patients within the ICU groups. These findings contradict existing literature. A recent meta-analysis comprising 229 studies was able to demonstrate that males have higher risk of hospitalization and admission to ICU following infection with SARS-CoV-2. 28 These conflicting findings may stem from the higher age of females in our cohort compared to males among ICU patients, with 37% of females ICU patients aged above 60 years compared to 17% for males as depicted in Figure 2. Notably, age emerges as the most influential factor affecting the severity of COVID-19.
The limitation of our study lies in the restricted sample size, which can be attributed to the stringent criteria applied for participant selection. Our study population consists of patients without comorbidities and risk factors, identifying participants meeting these criteria posed a significant challenge, especially among ICU patients. Furthermore, we attempted to recruit participants within a relatively short timeframe, primarily to mitigate the potential impact of the evolving landscape of SARS-CoV-2 variants on our study outcomes. This approach aimed to reduce the potential influence of comorbidities, risk factors and variant factors on our study outcomes. Nevertheless, this report contributes valuable insights into severe COVID-19 and provides information for the design of further studies.
Conclusion
Our study shows that there is no correlation between TYK2 rs74956615, IFITM3 rs12252, OAS3 rs10735079 and IFNAR2 rs2236757 with COVID-19 severity or ICU hospitalization in Morocco. Advanced age emerges as the primary factor influencing the severity of COVID-19 patients without comorbidities, and we suggest that setting the threshold for the age risk factor at 60 years may serve as a significant determinant for severe forms of the disease. Additionally, Further studies are recommended, including other genes involved in SARS-CoV-2 infection and a larger cohort, to confirm or refute the association of host genetics with COVID-19 in Moroccan patients.
Acknowledgements
The authors would like to thank the healthcare professionals of Mohamed V Military Teaching Hospital in Rabat, Morocco.
Footnotes
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
Ethical statement
Ethics approval
The research protocol received approval from the Ministry of Health for the purpose of conducting surveillance of respiratory diseases with epidemic potential, in accordance with Moroccan law (Law No. 28-13) concerning the protection of individuals participating in biomedical research (https://www.sante.gov.ma/Reglementation/REGLEMENTATIONDESPRATIQUESMEDICALES/28-13.pdf).
According to Article 26 of this law, non-interventional research must obtain the agreement of the head of the healthcare institution, whether public or private, where the research is planned. Consequently, the study did not necessitate formal assessment by an ethics committee. Nonetheless, informed consent was diligently obtained from all participating patients, ensuring adherence to ethical standards.
Informed consent
Written informed consent was obtained from all subjects before the study. And written informed consent was obtained from legally authorized representatives before the study.
ORCID iD
R. Benmansour https://orcid.org/0009-0002-7062-9536
References
- 1.Velavan TP, Pallerla SR, Rüter J, et al. (2021) Host genetic factors determining COVID-19 susceptibility and severity. EBioMedicine 72: 103629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feng Y, Ling Y, Bai T, et al. (2020) COVID-19 with different severities: a multicenter study of clinical features. Am J Respir Crit Care Med 201(11): 1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rahman A, Sathi NJ. (2021) Risk factors of the severity of COVID‐19: a meta‐analysis. Int J Clin Pract 75(7): e13916. https://onlinelibrary.wiley.com/doi/10.1111/ijcp.13916 [DOI] [PubMed] [Google Scholar]
- 4.Wang GQ, Zhao L, Wang X, et al. (2021) Diagnosis and treatment protocol for COVID-19 patients (tentative 8th edition): interpretation of updated key points. Infectious Diseases & Immunity 1(1): 17–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildirim Z, Sahin OS, Yazar S, et al. (2021) Genetic and epigenetic factors associated with increased severity of Covid-19. Cell Biol Int 45(6): 1158–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nhung VP, Ton ND, Ngoc TTB, et al. (2022) Host genetic risk factors associated with COVID-19 susceptibility and severity in Vietnamese. Genes 13(10): 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yaugel-Novoa M, Bourlet T, Longet S, et al. (2023) Association of IFNAR1 and IFNAR2 with COVID-19 severity. The Lancet Microbe 4(7): e487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takaoka A. Subchapter 39C - interferons. In: Ando H, Ukena K, Nagata S, éditeurs. Handbook of Hormones (2nd ed.). San Diego: Academic Press; 2021. p. 447–452. https://www.sciencedirect.com/science/article/pii/B9780128206492001157 [Google Scholar]
- 9.Regan JA, Abdulrahim JW, Bihlmeyer NA, et al. (2022) Phenome‐wide association study of severe COVID‐19 genetic risk variants. JAHA 11(5): e024004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alghamdi J, Alaamery M, Barhoumi T, et al. (2021) Interferon-induced transmembrane protein-3 genetic variant rs12252 is associated with COVID-19 mortality. Genomics 113(4): 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zani A, Yount JS. (2018) Antiviral protection by IFITM3 in vivo. Curr clin micro rpt. 5(4): 229–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao LJ, He ZM, Li YY, et al. (2023) Role of OAS gene family in COVID-19 induced heart failure. J Transl Med 21(1): 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beam TA, Klepser DG, Klepser ME, et al. (2023) COVID-19 host genetic risk study conducted at community pharmacies: implications for public health, research and pharmacists’ scope of practice. Res Soc Adm Pharm 2023: S155174112300298X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pairo-Castineira E, Clohisey S, Klaric L, et al. (2021) Genetic mechanisms of critical illness in COVID-19. Nature 591(7848): 92–98. [DOI] [PubMed] [Google Scholar]
- 15.Dieter C, de Almeida BL, Lemos NE, et al. (2022) Polymorphisms in ACE1, TMPRSS2, IFIH1, IFNAR2, and TYK2 genes are associated with worse clinical outcomes in COVID-19. Genes 14(1): 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steffen BT, Pankow JS, Lutsey PL, et al. (2022) Proteomic profiling identifies novel proteins for genetic risk of severe COVID-19: the Atherosclerosis Risk in Communities Study. Hum Mol Genet 31(14): 2452–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diagnosis and Treatment Protocol for Novel Coronavirus Pneumonia (Trial Version 7) (2020). Chinese Med J 133(9): 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graffelman J. (2015) Exploring diallelic genetic markers: the HardyWeinberg package. J Stat Software 64(3): 1–23. https://www.jstatsoft.org/v64/i03/ [Google Scholar]
- 19.Van Dyke AL, Cote ML, Wenzlaff AS, et al. (2009) Cytokine SNPs: comparison of allele frequencies by race and implications for future studies. Cytokine 46(2): 236–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang YH, Zhao Y, Li N, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat Commun 2013;4(1):1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.GenISIS Investigators T, MOSAIC Investigators T, Everitt AR, et al. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484(7395): 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang IC, Bailey CC, Weyer JL, et al. (2011) Distinct patterns of IFITM-mediated restriction of filoviruses, sars coronavirus, and influenza A virus. Baric rs, éditeur. PLoS Pathog 7(1): e1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahmadi I, Afifipour A, Sakhaee F, et al. (2022) Impact of interferon-induced transmembrane protein 3 gene rs12252 polymorphism on COVID-19 mortality. Cytokine 157: 155957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schönfelder K, Breuckmann K, Elsner C, et al. (2021) The influence of IFITM3 polymorphisms on susceptibility to SARS-CoV-2 infection and severity of COVID-19. Cytokine 142: 155492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niemi MEK, Karjalainen J, Liao RG, et al. (2021) Mapping the human genetic architecture of COVID-19. Nature 600(7889): 472–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udoakang A, Oboh M, Henry-Ajala A, et al. (2021) Low COVID-19 impact in africa: the multifactorial nexus. Open Res Africa 4: 47. [Google Scholar]
- 27.Weaver AK, Head JR, Gould CF, et al. (2022) Environmental factors influencing COVID-19 incidence and severity. Annu Rev Publ Health 43(1): 271–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pijls BG, Jolani S, Atherley A, et al. (2022) Temporal trends of sex differences for COVID-19 infection, hospitalisation, severe disease, intensive care unit (ICU) admission and death: a meta-analysis of 229 studies covering over 10M patients. F1000Res 11: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]