Abstract
Background:
Invasive lung adenocarcinoma with MPP/SOL components has a poor prognosis and often shows a tendency to recurrence and metastasis. This poor prognosis may require adjustment of treatment strategies. Preoperative identification is essential for decision-making for subsequent treatment.
Objective:
This study aimed to preoperatively predict the probability of MPP/SOL components in lung adenocarcinomas by a comprehensive model that includes radiomics features, clinical characteristics, and serum tumor biomarkers.
Design:
A retrospective case control, diagnostic accuracy study.
Methods:
This study retrospectively recruited 273 patients (males: females, 130: 143; mean age ± standard deviation, 63.29 ± 10.03 years; range 21–83 years) who underwent resection of invasive lung adenocarcinoma. Sixty-one patients (22.3%) were diagnosed with lung adenocarcinoma with MPP/SOL components. Radiomic features were extracted from CT before surgery. Clinical, radiomic, and combined models were developed using the logistic regression algorithm. The clinical and radiomic signatures were integrated into a nomogram. The diagnostic performance of the models was evaluated using the area under the curve (AUC). Studies were scored according to the Radiomics Quality Score and Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis guidelines.
Results:
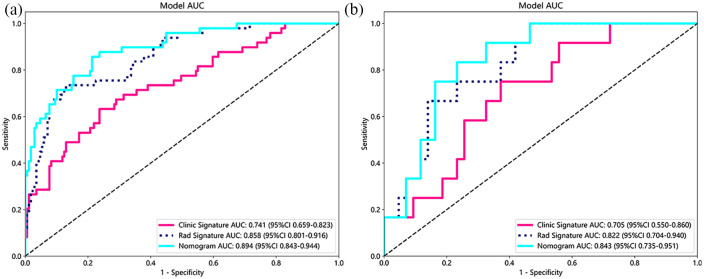
The radiomics model achieved the best AUC values of 0.858 and 0.822 in the training and test cohort, respectively. Tumor size (T_size), solid tumor size (ST_size), consolidation-to-tumor ratio (CTR), years of smoking, CYFRA 21-1, and squamous cell carcinoma antigen were used to construct the clinical model. The clinical model achieved AUC values of 0.741 and 0.705 in the training and test cohort, respectively. The nomogram showed higher AUCs of 0.894 and 0.843 in the training and test cohort, respectively.
Conclusion:
This study has developed and validated a combined nomogram, a visual tool that integrates CT radiomics features with clinical indicators and serum tumor biomarkers. This innovative model facilitates the differentiation of micropapillary or solid components within lung adenocarcinoma and achieves a higher AUC, indicating superior predictive accuracy.
Keywords: CT, lung adenocarcinoma, preoperative differential, radiomics
Plain language summary
A new tool to predict aggressive lung cancer types before surgery
We developed a tool to help doctors determine whether lung cancer is one of the more dangerous types, called micropapillary (MPP) or solid (SOL) patterns, before surgery. These patterns can be more harmful and spread quickly, so knowing they are there can help doctors plan the best treatment. We looked at the cases of 273 lung cancer patients who had surgery and found that 61 of them had these aggressive cancer types. To predict these patterns, we used a computer process known as logistic regression, analyzing CT scan details, health information, and blood tests for cancer markers. Based on CT scans, our tool was very good at predicting whether these patterns were present in two patient groups. However, predictions using only basic health information like the size of the tumor and whether the patient smoked needed to be more accurate. We found a way to make our predictions even better. Combining all information into one chart, known as a nomogram, significantly improved our ability to predict these dangerous cancer patterns. This combined chart could be a big help for doctors. It gives them a clearer picture of the cancer’s aggressiveness before surgery, which can guide them to choose the best treatment options. This approach aims to offer a better understanding of the tumor, leading to more tailored and effective treatments for patients facing lung cancer.
Introduction
Lung cancer is one of the major causes of cancer-related deaths in many countries around the world. Lung adenocarcinoma is the most common histological type of lung cancer. 1 Because lung adenocarcinoma has multiple histological subtypes, the subtypes show significant heterogeneity in molecular features, pathology, prognosis, and treatment strategies. The International Association for the Study of Lung Cancer, the American Thoracic Society (ATS), and the European Respiratory Society (ERS) have jointly published a new histological classification of lung adenocarcinomas. 2 Based on histological subtypes for prognostic studies, the classification system identifies several common subtypes of invasive lung adenocarcinoma, including lepidic, papillary, acinar, micropapillary (MPP), and solid (SOL). Typically, these subtypes appear in combination.3,4 The MPP/SOL-dominant subtype of invasive lung adenocarcinoma has a poor prognosis and often shows a tendency to recurrence and metastasis.5–7 Even if MPP/SOL accounts for <5% of lung adenocarcinoma tissue, its adverse prognostic impact on survival has been demonstrated.8,9 This poor prognosis may require adjustment of treatment strategies. In patients undergoing limited resection, MPP/SOL-containing subtypes have been identified as independent predictors of local recurrence.10,11 Therefore, preoperative identification of MPP/SOL components in lung adenocarcinomas is essential for surgical decision-making. However, presurgical lung biopsies can only demonstrate some of the features of heterogeneous tumors. 12 Thus, preoperative CT may help assess patient risk and decision-making for subsequent treatment or monitoring. Radiomics involves the extensive analysis of a vast quantity of medical images to extract numerous phenotypic features, also known as radiomic biomarkers, that reflect the characteristics of cancer. 13 These features contribute to clinical decision support and enhance diagnostic and prognostic predictions, especially in lung adenocarcinoma. 14 The development and validation of this quantitative imaging technique could introduce a new, noninvasive, and convenient method for improving therapeutic strategy management, leading to enhanced clinical and economic benefits for the patient.15,16
This study developed a comprehensive model that includes radiomics features, clinical characteristics, and serum tumor biomarkers. The objective of this model is to preoperatively predict the probability of MPP/SOL components based on chosen risk factors.
Materials and methods
This study was a retrospective case control, diagnostic accuracy study.
Quality system
The study followed the Radiomics Quality Score (RQS) system. 17 The model was developed and validated according to the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD) checklist for prediction model development and validation. 18 The RQS scoring criteria, the scores for this study, the rationale for the scores, and the TRIPOD statement for prediction model development were detailed in the Supplemental Material.
Patients
We retrospectively recruited patients who underwent resection of invasive lung adenocarcinoma at Zhejiang Hospital from March 2018 to May 2023. The inclusion criteria were: (1) patients who had undergone chest CT within 7 days before surgery; (2) CT images with thin sections (⩽1.5 mm) were adequate for analysis; (3) pathologically confirmed invasive lung adenocarcinoma with a complete pathology report describing pathological subtypes; (4) no prior history of chemotherapy or radiotherapy; (5) clinical data within 7 days before surgery; and (6) TNM staging does not exceed stage II. The exclusion criteria were: (1) patients with multifocal lesions; (2) tissue samples obtained by biopsy rather than surgery; (3) with other malignant tumors; (4) history of radiotherapy and chemotherapy prior to scanning; (5) microinvasive adenocarcinoma;(6) purely lepidic tumors; and (7) the concomitant presence of other malignancies.
Demographic information, laboratory tests, and clinical features were all obtained from electronic medical records. Clinical data include gender, age, height, weight, body mass index (BMI), body surface area (BSA), smoking status, years of smoking, daily smoking quantity, cytokeratin fragment 19 (CYFRA 21-1), thyroglobulin (TG), alpha-fetoprotein (AFP), carcinoembryonic antigen (CEA), neuron-specific enolase (NSE), carbohydrate antigen 125 (CA125), carbohydrate antigen 15-3 (CA15-3), carbohydrate antigen 19-9 (CA19-9), carbohydrate antigen 242 (CA242), carbohydrate antigen 50 (CA50), carbohydrate antigen 72-4 (CA72-4), and squamous cell carcinoma antigen (SCC).
Histological assessment
The surgically resected specimens were fixed in formalin for 48 h, embedded in paraffin, sectioned with a microtome, and stained with hematoxylin and eosin. Slides were evaluated by two pathologists blind to the patient’s medical history. Discrepancies in interpretation were resolved through discussion. Based on the histological classifications,19,20 all adenocarcinomas were divided into two categories: lung adenocarcinomas without MPP/SOL components and lung adenocarcinomas with MPP/SOL components.
Image acquisition and imaging evaluation
Chest CT images were obtained with multirow spiral CT scanners (SIEMENS SOMATOM Force; SIEMENS SOMATOM Definition; GE MEDICAL SYSTEMS Revolution; GE MEDICAL SYSTEMS CT 540; UNITED CT 510; and NeuCT Extra 2). Details regarding the acquisition parameters were set as follows: detector collimation, 1–1.5 mm; field of view: 20–38 cm; beam pitch, 0.800–1.5; beam width, 10–40 mm; gantry speed, 0.5 or 0.8 s per rotation;100–130 kV; 47–351 mA; reconstruction interval, 0.39–0.6 mm; matrix, 512 mm × 512 mm, and soft-tissue kernel. All CT data were acquired in the supine position at full inspiration. The scans ranged from the lung base to the level of the thoracic inlet. The patient’s CT data were downloaded from the Picture Archiving and Communication Systems.
Two independent radiologists review the thin-slice CT images. They are blind to the patient’s medical history. Tumor size (T_size) was the largest diameter of tumor measured on the axial plane in the lung window setting. In contrast, the solid tumor size (ST_size) was defined as the maximum diameter of the consolidation component. The consolidation-to-tumor ratio (CTR) was calculated as the ratio of the solid tumor size over the tumor size. 21 When the value of CTR is lower, it indicates that the lesion contains a higher proportion of ground-glass opacity components. The closer the value of CTR is to 1, the more the lesion tends toward consolidation. Discrepancies in interpretation were addressed and resolved through collaborative discussion.
Tumor segmentation and feature extraction
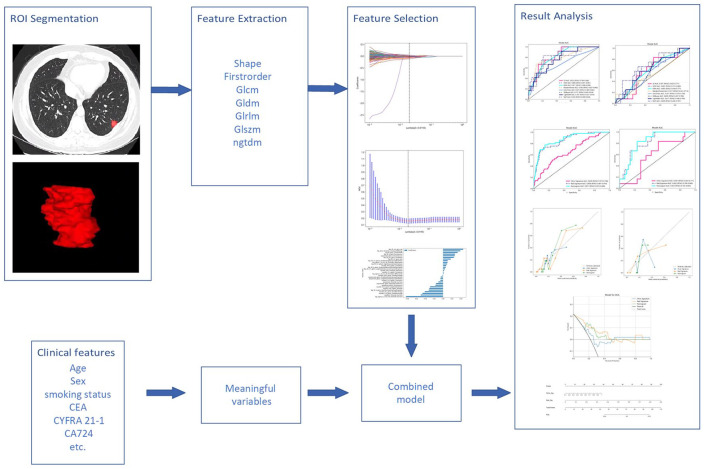
In order to improve the repeatability and reproducibility of radiomic features, data underwent a preprocessing stage. Z-score normalization was applied in the data; the resolution of all the images was resampled to 1 × 1 × 1 mm3; and the voxel intensity values were discretized with a fixed bin width of 25 units to reduce image noise, allowing for constant intensity resolution across all tumor images.22,23 Then, a radiologist with more than 6 years of work experience used the ITK-SNAP software (ITK-SNAP 3.8.0, www.itksnap.org) for 3D manual segmentation. When drawing the contours, necrosis, calcification, cavities, vascularity, and bronchial interference should be carefully avoided. Outlining all lesions was reviewed and confirmed by another senior radiologist with 26 years of work experience. They are blind to the patient’s medical history. Any deviation is effectively addressed and resolved through thorough discussion. The delineated regions of interest (ROI) is stored in Nifti(.NII) format for further analysis.
Subsequently, the radiomics features of each subject, including feature type and transformation type, were extracted from CT images using pyradiomics (version 2.2.0). 23 The feature types included first-order, shape, gray-level co-occurrence matrix (GLCM), gray-level size zone matrix (GLSZM), gray-level run length matrix (GLRLM), gray-level dependence matrix (GLDM), and neighboring gray tone difference matrix (NGTDM). The transform types included log, wavelet, local binary pattern, exponential, square, square root, and gradient. Except for shape, all radiomic features combine feature types and transform types. The framework is presented in Figure 1.
Figure 1.
The framework of model building.
Feature selection and radiomics model construction
The extracted radiomics features were screened by the Mann–Whitney U test. Only radiomic features with p-value <0.05 were retained. For features with high repetition, the Pearson correlation coefficient was used to calculate the correlation between features and retain the one feature with a correlation coefficient greater than 0.9 between any two features. To maximize the ability to depict features, we employed a greedy recursive deletion strategy in our feature filtering process: the feature with the highest redundancy within the current set is removed each time. The least absolute shrinkage and selection operator (LASSO) algorithm was used to reduce the dimension of the selected features. Based on the regulation weights λ, LASSO shrinks all regression coefficients toward zero and sets the coefficients of many irrelevant features exactly to zero. To find an optimal λ, 10-fold cross-validation with minimum criteria was employed, where the final value of λ yielded minimum cross-validation error. The retained features with nonzero coefficients were used in the regression model fitting and combined into a radiomics signature. Subsequently, a radiomics score (Rad-score) was calculated using a formula based on the radiomics features. The Python scikit-learn package was used for LASSO regression modeling.
After Lasso feature screening, we input the final features into machine learning models such as logistic regression (LR), support vector machines (SVM), k-nearest neighbor (KNN), random forest, extra trees, extreme gradient boosting (XGBoost), light gradient boosting machine (LightGBM), and multilayer perceptron (MLP) for risk model construction. Here, we used 10-fold cross-validation to obtain the final radiomics profile.
Clinical model construction
Establishing a clinical model is almost identical to that of radiomics model. First, the features used to build the clinical model were selected by baseline statistics with a p-value <0.05. We used the same machine learning model in the clinical model-building process. We set up a 10-fold cross-validation and a fixed test queue for a fair comparison.
Radiomics nomogram
We built a radiomics nomogram based on the radiomics and clinical model in the test cohort. The nomogram is based on LR analysis, combining radiomics signature and clinical risk factors.
In the test cohort, we evaluated the diagnostic efficacy of the radiomics nomogram. Receiver operating characteristic (ROC) curves were plotted to assess the diagnostic performance of the nomogram further. In order to assess the concordance between the predictions of the nomogram and the actual observations, we generated the calibration curves. The calibration capability of the nomogram was then evaluated using a Hosmer–Lemeshow analysis to check how well the predicted outcomes fit the observed outcomes. The clinical utility of the predictive model is evaluated using decision curve analysis (DCA). This method evaluates the model’s net benefits at different threshold probabilities to determine its overall usefulness in decision-making processes.
Statistical analysis
We performed statistical analysis using IBM SPSS software (version 22.0, http://www.ibm.com) and Python (3.7.6). Student’s t-test or Mann–Whitney U test (for continuous parameters, represented as mean ± standard deviation or median ± interquartile range) and χ2 test were used for feature comparison. During the training process, 10-fold cross-validation was used to evaluate the performance of radiomics, clinical, and combined models. In addition, these models were also validated in the test cohort. The AUC comparison was performed using DeLong’s test.
Results
Clinical data
This study retrospectively recruited 273 patients (males: females, 130: 143; mean age ± standard deviation, 63.29 ± 10.03 years; range 21–83 years) who underwent resection of invasive lung adenocarcinoma at Zhejiang Hospital from March 2018 to May 2023. In the cohort of 61 cases featuring MPP/SOL components, the average proportion of MPP/SOL was 18%, with a standard deviation of 21%, and proportions varied from a minimum of 2% to a maximum of 90%. Among the cases studied, 27 patients exhibited an MPP/SOL component that comprised less than 5% of the tumor, accounting for 44.3% of the cases with an identifiable MPP/SOL pattern. 273 patients were divided into two groups (0 for lung adenocarcinoma without MPP/SOL components and 1 for lung adenocarcinoma with MPP/SOL components). All lung adenocarcinoma patients were stratified and completely randomized at a ratio7:3 into training cohorts (n = 218) and validation cohorts (n = 55). We compared the clinical characteristics of the patients using independent samples t-tests, Mann–Whitney U-tests, or χ2 tests as appropriate. Table 1 shows the baseline clinical characteristics of the patients in our cohort. After one-way logistic regression analysis of the patient’s clinical data, the following factors were significantly associated with the presence or absence of MPP/SOL components within lung adenocarcinoma in the training cohort (p < 0.05, Table 1): T_size, ST_size, CTR, years of smoking, CYFRA 21-1, and SCC.
Table 1.
Baseline characteristics of patients in cohorts.
| Feature_name | Train-label = 0 | Train-label = 1 | p Value | Test-label = 0 | Test-label = 1 | p Value |
|---|---|---|---|---|---|---|
| T_size | 19.04 ± 7.81 | 24.16 ± 9.05 | <0.001 | 20.94 ± 8.68 | 20.82 ± 5.15 | 0.714 |
| ST_size | 13.62 ± 7.14 | 35.98 ± 115.53 | <0.001 | 14.58 ± 8.03 | 34.76 ± 60.75 | 0.048 |
| CTR | 0.71 ± 0.23 | 1.68 ± 6.13 | 0.007 | 0.70 ± 0.24 | 1.53 ± 2.34 | 0.017 |
| Height | 1.63 ± 0.08 | 1.66 ± 0.09 | 0.066 | 1.61 ± 0.09 | 1.64 ± 0.05 | 0.281 |
| Weight | 111.61 ± 634.11 | 66.21 ± 13.01 | 0.617 | 59.36 ± 10.34 | 62.38 ± 9.38 | 0.365 |
| BMI | 39.42 ± 206.82 | 23.99 ± 3.43 | 0.603 | 22.89 ± 3.46 | 23.13 ± 2.82 | 0.823 |
| BSA | 2.21 ± 8.13 | 1.66 ± 0.33 | 0.639 | 1.59 ± 0.17 | 1.65 ± 0.14 | 0.294 |
| Smoke_quantity | 6.11 ± 10.69 | 9.16 ± 13.42 | 0.099 | 7.37 ± 12.55 | 12.50 ± 12.15 | 0.213 |
| Smoking_years | 9.86 ± 16.48 | 15.51 ± 19.24 | 0.043 | 13.49 ± 19.87 | 20.67 ± 19.86 | 0.273 |
| Age | 63.73 ± 8.93 | 61.61 ± 12.29 | 0.183 | 63.40 ± 11.26 | 63.58 ± 10.69 | 0.959 |
| CYFRA21-1 | 2.70 ± 1.30 | 3.52 ± 4.47 | 0.036 | 2.98 ± 1.64 | 3.11 ± 1.34 | 0.803 |
| TG | 12.39 ± 12.55 | 11.99 ± 11.60 | 0.844 | 15.15 ± 22.99 | 15.63 ± 7.26 | 0.943 |
| AFP | 3.10 ± 1.29 | 3.22 ± 2.57 | 0.636 | 3.15 ± 1.39 | 3.13 ± 1.83 | 0.974 |
| CEA | 3.45 ± 5.40 | 3.76 ± 5.39 | 0.721 | 2.84 ± 2.18 | 4.84 ± 5.60 | 0.061 |
| NSE | 11.09 ± 2.15 | 10.89 ± 2.41 | 0.574 | 11.14 ± 2.11 | 11.85 ± 3.55 | 0.38 |
| CA125 | 11.95 ± 10.21 | 13.00 ± 16.98 | 0.59 | 11.09 ± 5.64 | 13.48 ± 7.26 | 0.228 |
| CA15-3 | 10.81 ± 5.67 | 12.92 ± 9.83 | 0.057 | 9.95 ± 4.83 | 12.20 ± 5.25 | 0.167 |
| CA19-9 | 13.17 ± 18.53 | 15.53 ± 13.14 | 0.406 | 11.55 ± 6.92 | 13.06 ± 12.25 | 0.579 |
| CA242 | 5.43 ± 10.12 | 6.55 ± 5.39 | 0.455 | 9.01 ± 28.28 | 4.52 ± 3.94 | 0.588 |
| CA50 | 7.17 ± 9.31 | 8.57 ± 8.28 | 0.342 | 6.71 ± 4.25 | 7.98 ± 7.28 | 0.445 |
| CA72-4 | 3.55 ± 7.34 | 3.56 ± 3.61 | 0.995 | 3.80 ± 5.70 | 23.56 ± 68.42 | 0.061 |
| SCC | 0.99 ± 0.50 | 1.46 ± 1.35 | <0.001 | 1.08 ± 0.59 | 1.67 ± 2.15 | 0.109 |
| Smoke | 0.08 | 0.1 | ||||
| 0 | 118(69.82) | 27(55.10) | 28(65.12) | 4(33.33) | ||
| 1 | 51(30.18) | 22(44.90) | 15(34.88) | 8(66.67) | ||
| Gender | 0.055 | 0.046 | ||||
| 0 | 94(55.62) | 19(38.78) | 27(62.79) | 3(25.00) | ||
| 1 | 75(44.38) | 30(61.22) | 16(37.21) | 9(75.00) |
Clinical data include: AFP, alpha-fetoprotein; BMI, gender, age, height, weight, body mass index; BSA, body surface area; CA125, carbohydrate antigen 125; CA15-3, carbohydrate antigen 15-3; CA19-9, carbohydrate antigen 19-9; CA242, carbohydrate antigen 242; CA50, carbohydrate antigen 50; CA72-4, carbohydrate antigen 72-4; CEA, carcinoembryonic antigen; CTR, consolidation-to-tumor ratio; CYFRA 21-1, cytokeratin fragment 19; NSE, neuron-specific enolase; SCC, squamous cell carcinoma antigen; smoke_quantity, smoking status, daily smoking quantity; smoking_years, years of smoking; ST_size, solid tumor size; T_size, tumor size; TG, thyroglobulin.
Radiomics model
A total of 1835 radiological features were extracted from the lesions on the lung window images, divided into 7 groups: 360 first-level, 14 shape, 440 GLCM, 320 GLSZM, 320 GLRLM, 280 GLDM, and 100 NGTDM. Figure 2 shows the number and proportion of all handcraft features. Figure 3 shows all the features and the corresponding p-value results.
Figure 2.
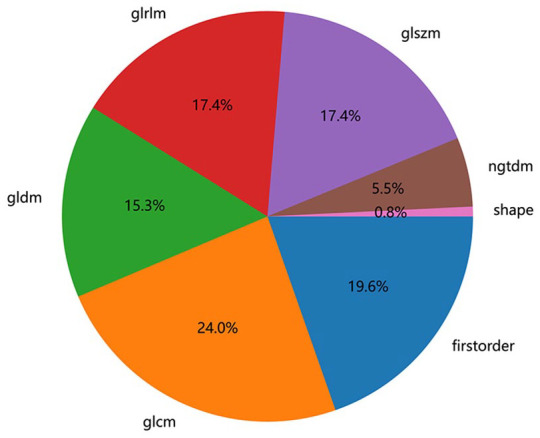
Number and proportion of handcrafted features.
Figure 3.
All the features and the corresponding p-value results.
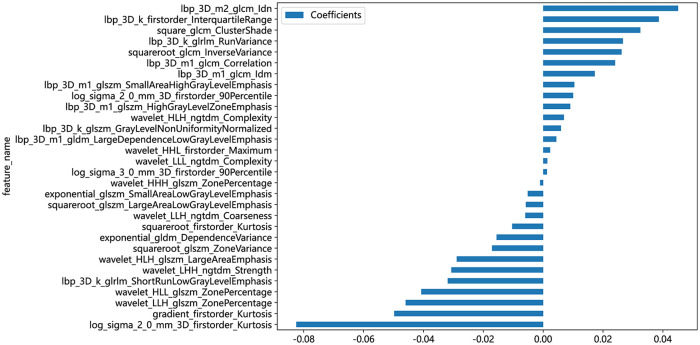
A radiomics signature was constructed based on the finally selected features. A Rad-score was generated for each patient using a linear combination of the values of the selected features weighted by their corresponding nonzero coefficients. The coefficients and mean standard error of ten-fold validation are shown in Figures 4 and 5. Figure 4 shows the coefficient values of the finally selected nonzero features. The Rad-score is calculated (Supplemental File).
Figure 4.
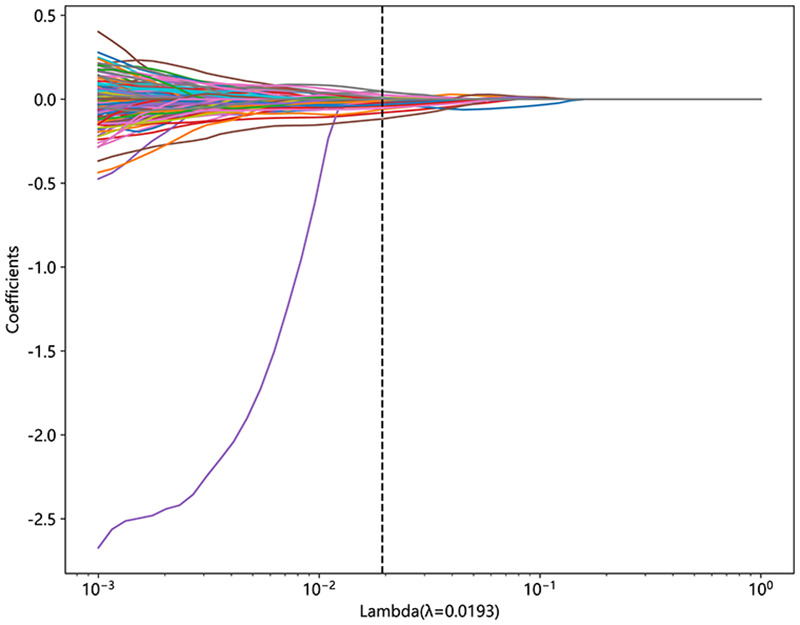
Coefficients of 10-fold cross-validation.
Figure 5.
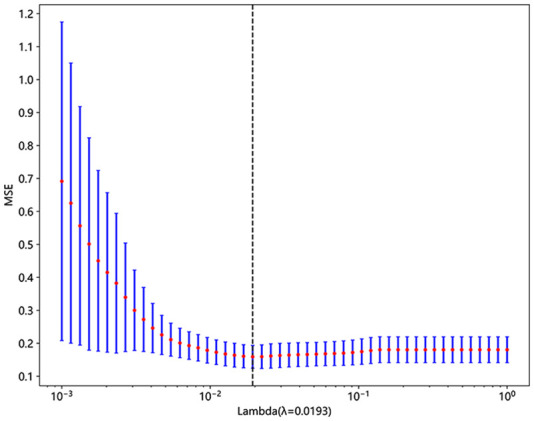
MSE of 10-fold cross-validation.
MSE, mean standard error.
The feature names and corresponding coefficients are displayed in Figure 6.
Figure 6.
The histogram of the Rad-score based on the selected features.
Model comparison
Radiomics features were used to construct models with classifiers such as LR, SVM, KNN, Random Forest, Extra Trees, XGBoost, LightGBM, and MLP (Table 2). Figure 7 shows the AUC of each radiomics model in the test cohort. LR achieved the best AUC values of 0.858 and 0.822 in the training and test cohort, respectively. Therefore, LR was the optimal classifier for constructing the radiomics model.
Table 2.
Table 2 is all model we used to predict micropapillary/solid component of lung adenocarcinoma, LR model preforms the best performance. So in the building of clinical signature, LR is selected as base model.
| Model_name | Accuracy | AUC | 95% CI | Sensitivity | Specificity | PPV | NPV | Task |
|---|---|---|---|---|---|---|---|---|
| LR | 0.839 | 0.858 | 0.8007–0.9158 | 0.870 | 0.735 | 0.919 | 0.621 | Label-train |
| LR | 0.636 | 0.822 | 0.7036–0.9398 | 0.535 | 1.000 | 1.000 | 0.375 | Label-test |
| SVM | 0.927 | 0.954 | 0.9122–0.9948 | 0.917 | 0.959 | 0.987 | 0.770 | Label-train |
| SVM | 0.800 | 0.806 | 0.6509–0.9615 | 0.791 | 0.833 | 0.944 | 0.526 | Label-test |
| KNN | 0.789 | 0.854 | 0.8052–0.9035 | 0.817 | 0.694 | 0.902 | 0.523 | Label-train |
| KNN | 0.764 | 0.811 | 0.6964–0.9257 | 0.791 | 0.667 | 0.895 | 0.471 | Label-test |
| RandomForest | 0.995 | 1.000 | 0.9996–1.0000 | 0.994 | 1.000 | 1.000 | 0.980 | Label-train |
| RandomForest | 0.836 | 0.785 | 0.6274–0.9424 | 0.884 | 0.667 | 0.905 | 0.615 | Label-test |
| ExtraTrees | 1.000 | 1.000 | 1.0000–1.0000 | 1.000 | 1.000 | 1.000 | 1.000 | Label-train |
| ExtraTrees | 0.800 | 0.673 | 0.4840–0.8629 | 0.907 | 0.455 | 0.848 | 0.556 | Label-test |
| XGBoost | 1.000 | 1.000 | 1.0000–1.0000 | 1.000 | 1.000 | 1.000 | 1.000 | Label-train |
| XGBoost | 0.764 | 0.771 | 0.6251–0.9176 | 0.791 | 0.667 | 0.895 | 0.471 | Label-test |
| LightGBM | 0.876 | 0.952 | 0.9267–0.9777 | 0.852 | 0.959 | 0.986 | 0.653 | Label-train |
| LightGBM | 0.618 | 0.767 | 0.6213–0.9136 | 0.535 | 0.917 | 0.958 | 0.355 | Label-test |
| MLP | 0.908 | 0.932 | 0.8940–0.9703 | 0.941 | 0.796 | 0.941 | 0.796 | Label-train |
| MLP | 0.782 | 0.818 | 0.6994–0.9363 | 0.791 | 0.750 | 0.919 | 0.500 | Label-test |
KNN, k-nearest neighbor; LightGBM, light gradient boosting machine; LR, logistic regression; MLP, multilayer perceptron; NPV negative predictive value; SVM, support vector machines; PPV, positive predictive value.
Figure 7.
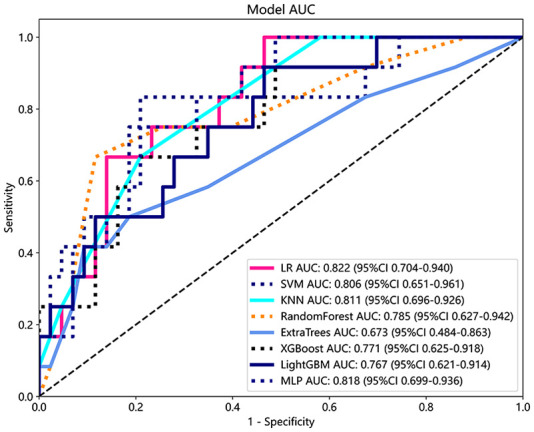
ROC analysis of different models on the rad signature.
Clinical model and combined model
T_size, ST_size, CTR, years of smoking, CYFRA 21-1, and SCC were used to construct the clinical model. Figure 8 shows the AUC of each clinical feature model in the test cohort. Given that LR demonstrated superior performance in the radiomics model, it was subsequently selected as the base model for constructing the clinical model. The clinical model achieved AUC values of 0.741 and 0.705 in the training and test cohorts, respectively.
Figure 8.
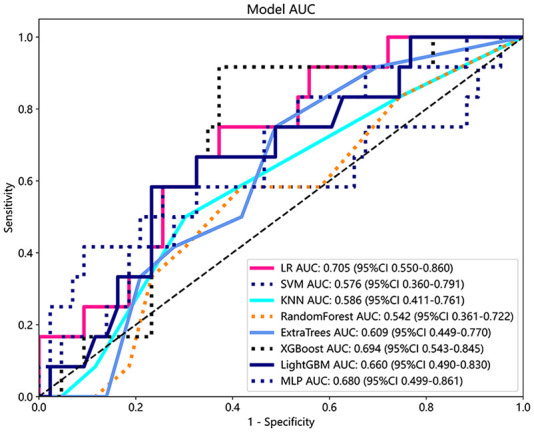
ROC analysis of different models on clinical signature.
A comprehensive model was formulated using the Rad-score and independent clinical risk factors. This model was then visually represented as a radiomics nomogram. The nomogram showed the higher AUCs of 0.894 and 0.843 in the training and test cohort, respectively (Table 3, Figure 9).
Table 3.
Main consequence of 3 models.
| Signature | Accuracy | AUC | 95% CI | Sensitivity | Specificity | PPV | NPV | Cohort |
|---|---|---|---|---|---|---|---|---|
| Clinic signature | 0.734 | 0.741 | 0.6592–0.8228 | 0.763 | 0.633 | 0.878 | 0.437 | Train |
| Rad signature | 0.839 | 0.858 | 0.8007–0.9158 | 0.870 | 0.735 | 0.919 | 0.621 | Train |
| Nomogram | 0.803 | 0.894 | 0.8434–0.9438 | 0.787 | 0.857 | 0.950 | 0.538 | Train |
| Clinic signature | 0.655 | 0.705 | 0.5505–0.8604 | 0.628 | 0.750 | 0.900 | 0.360 | Test |
| Rad signature | 0.636 | 0.822 | 0.7036–0.9398 | 0.535 | 1.000 | 1.000 | 0.375 | Test |
| Nomogram | 0.782 | 0.843 | 0.7354–0.9507 | 0.767 | 0.833 | 0.943 | 0.500 | Test |
AUC: In train and test cohorts, both clinical signature and rad signature get the prefect fitting. The Nomogram using the LR algorithm was performed to combine clinical signature and rad signature, which shows the best performance.
AUC, area under the curve; LR, logistic regression; NPV negative predictive value; PPV, positive predictive value.
Figure 9.
(a) The AUC of clinic signature, rad signature, and nomogram in train cohorts. (b) The AUC of clinic signature, rad signature, and nomogram in test cohorts. The combined model surpasses the clinical and radiomics models in both the train and test cohorts for the predictive performance regarding the micropapillary and solid components of lung adenocarcinoma.
AUC, area under the curve.
In order to compare the clinical model, the radiomics model, and the nomogram, we used the Delong test (Table 4). The AUCs of the nomogram-based models in the training and test cohorts were significantly different from those of the clinical model (p < 0.001). The nomogram calibration curve showed good consistency between the training and test cohorts (Supplemental Figure 1). The p-value of the Hosmer–Lemeshow test showed nonsignificant goodness of fit in the training and test cohorts (Table 5). In this study, we also evaluated each model through DCA. DCA of the clinical model, radiomics model, and the nomogram is presented in Supplemental Figure 2. DCA results showed a preferable clinical practicality potential for this nomogram.
Table 4.
Delong test.
| Cohort | Nomogram versus Clinic | Nomogram versus Rad |
|---|---|---|
| Train | 0.000002570 | 0.084 |
| Test | 0.00519 | 0.533 |
Table 5.
Hosmer–Lemeshow test.
| Cohort | Clinic signature | Rad signature | Nomogram |
|---|---|---|---|
| Train | 0.142 | 0.336 | 0.118 |
| Test | 0.067 | 0.402 | 0.083 |
Supplemental Figure 3 shows the nomogram used for clinical purposes. The Rad-score and clinical signature can be translated into numerical values based on the points axis. After calculating the individual points to reach the total sum displayed on the total point axis, a prediction of the MPP/SOL components was made.
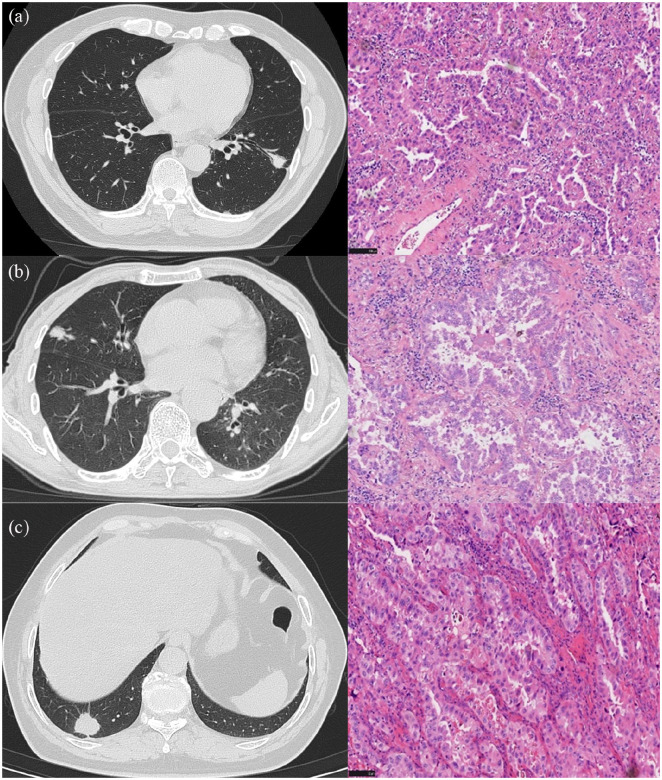
CT images and the corresponding pathology peritumoral lung tissue of three patients with lung adenocarcinoma confirmed by surgical resection are shown in Figure 10.
Figure 10.
CT images and corresponding pathology of peripheral invasive lung adenocarcinoma in three patients: (a) A 68-year-old man with a diagnosis of invasive lung adenocarcinoma. CT shows a solid nodule in the lower lobe of the left lung, with a diameter of 19.3 mm and a CTR = 1. High-power histologic image (hematoxylin and eosin, ×200) shows that the tumor exhibits irregular glandular-like structures with cells in a complex layered arrangement. The cytoplasm is abundant and eosinophilic, with nuclei of varying sizes, and significant pleomorphism is evident. The pathological classification is predominantly acinar with the presence of papillary patterns. The radiomics LR model predicts a probability of 0.21 for containing MPP/SOL components. (b) CT shows a solid nodule in the middle lobe of the right lung, with a diameter of 21.1 mm and a CTR = 1. High-power histologic image (hematoxylin and eosin, ×200) shows most of the tumor presents as acinar formations with irregular glandular lumens; locally, a sieve-like structure is observed. The cytoplasm is abundant, with some cells displaying clear cytoplasm. The nuclei are large and deeply stained, with prominent nucleoli. The final pathological classification is predominantly acinar, with lepidic parts and less than 5% solid. The radiomics LR model predicts a probability of 0.63 for containing MPP/SOL components. (c) A 66-year-old woman with a diagnosis of invasive lung adenocarcinoma. CT shows a solid nodule in the lower lobe of the right lung, with a diameter of 24 mm and a CTR = 1. High-power histologic image (hematoxylin and eosin, ×200) shows that the tumor appears as gland-like multilayered arrangements, with marked cellular atypia, abundant eosinophilic cytoplasm, and large, hyperchromatic nuclei. The final pathological classification is 50% micropapillary, 35% solid, and 15% lepidic. The radiomics LR model predicts a probability of 0.78 for containing MPP/SOL components.
CTR, consolidation-to-tumor ratio; LR, logistic regression; MPP, micropapillary; SOL, solid.
Discussion
The RQS analysis result of this study is 16, which demonstrates that this study’s quality is trustworthy and repeatable.
Micropapillary and solid patterns are essential in prognostic assessment when exploring the correlation between disease-free survival (DFS) and overall survival (OS) in patients with lung adenocarcinoma. Even if the MPP/SOL component represents only a tiny fraction of the tumor, it has been reported to significantly impact DFS and OS in patients with lung adenocarcinoma.8,9 Both are also considered essential markers for predicting the efficacy of adjuvant chemotherapy.24,25 Subsequent studies have revealed that the presence of MPP/SOL in lung adenocarcinoma may influence the selection of surgical methods. Patients who received sublobar resection demonstrated a higher likelihood of recurrence compared to comparable patients who were treated with lobectomy. This suggests that sublobar resection may not be the best procedure for patients with lung adenocarcinoma containing MPP/SOL. 10 However, for patients with significantly compromised cardiopulmonary function, elderly patients, or those necessitating multiple resections, the priority should be to preserve their lung function and avoid excessive resection. Indeed, determining the presence of MPP/SOL is crucial even for patients with lung adenocarcinoma who are not eligible for surgery. This is because biopsy samples may not fully capture the heterogeneity of the tumor and all of its subtype characteristics. 12 The complete histological subtype can only be accurately determined by evaluating the entirety of the tumor specimen. This provides a comprehensive view of the tumor’s characteristics and variability. Diagnosing the presence of MPP/SOL is integral to shaping personalized treatment strategies. It can influence the choice of therapies and interventions, ultimately impacting patient outcomes. Hence, preoperative awareness of the presence or absence of MPP/SOL components in lung adenocarcinomas is crucial for devising an optimal surgical strategy and determining early aggressive adjuvant therapy.
Different types of lung cancer may exhibit different clinical features. 26 Squamous cell carcinoma or small-cell carcinoma lesions are usually located in the central airways, and the external pressure of the tumor or mucosal invasion can cause coughing. Pleural effusion is most commonly seen in adenocarcinoma. The most common sites of metastasis for adenocarcinoma are the spine and ribs. A rapid disease progression characterizes small-cell lung cancer (SCLC) and is often associated with superior vena cava syndrome. 26 In the early stages, lung cancer often presents with nonspecific symptoms. 26
Smoking is the primary risk factor for lung cancer and is strongly related to the development of squamous cell carcinoma and SCLC. Smokers have a significantly increased risk of lung cancer compared to nonsmokers, which correlates directly with the quantity of cigarettes smoked and the duration of the smoking habit. 27 Additionally, smoking is linked to more aggressive forms of lung adenocarcinoma. Research has indicated that smoking is associated with more malignant subtypes of lung adenocarcinoma and specific patterns of gene mutations. 28 Long-term smokers may exhibit specific mutation patterns, such as TP53/KRAS mutations. These mutations are tied to lung adenocarcinoma’s malignancy level and the treatment response. 29 Among nonsmoking females, however, a family history of lung cancer is associated with an increased risk of developing lung adenocarcinoma. 30 These observations further underline the critical importance of smoking cessation as a preventative measure against the development of lung cancer. Occupational exposure, such as long-term exposure to asbestos, may increase the risk of developing non-SCLC and mesothelioma. 31
Serum tumor biomarkers serve as noninvasive diagnostic tools. Serum biomarkers can provide valuable information for diagnosing and prognosis of various malignant tumors. They are widely used in cancer screening and are indicators for prognosis and treatment effectiveness assessments. 32 Different types of lung cancer may be associated with specific changes in tumor markers. 32 For example, an elevated carcinoembryonic antigen (CEA) level is often associated with lung adenocarcinoma. In patients with SCLC, higher levels of neuron-specific enolase (NSE) and progastrin-releasing peptide are typically observed. CYFRA 21-1 is a fragment of cytokeratin 19, a protein crucial for preserving the structural integrity of epithelial cells. CYFRA 21-1 is overexpressed in the cytoplasm of various epithelial tumors, including lung cancer, and can be detected. CYFRA 21-1 can be released into the bloodstream due to cell lysis and tumor necrosis. CYFRA 21-1 levels in the serum have a positive correlation with the severity and stage of lung cancer, the presence of symptoms, and the specific type of pathological condition. CYFRA21-1 is frequently elevated in non-small cell lung cancer (NSCLC) and is currently considered the most robust biomarker for predicting postoperative survival in NSCLC patients. This suggests that when the severity or stage of lung cancer, the intensity of symptoms, or the prominence of the pathological condition increases or worsens, there is typically a corresponding rise in CYFRA 21-1 levels within the serum. 33 SCC was first identified as a tumor marker or biomarker for squamous cell carcinoma. It is a serum protein that is overexpressed in certain pathological states, especially in the presence of squamous cell carcinoma. It is instrumental in monitoring the effectiveness of treatment, detecting recurrence, and sometimes in the initial diagnosis of squamous cell carcinoma. However, it is not explicitly associated only with squamous cell carcinoma of the lung and may also be elevated in other types of lung cancer, such as lung adenocarcinoma. 34 Factors that influence the rate of SCC detection in serum include tumor size and volume, the aggressiveness of the primary or recurrent tumor, lymph node metastasis, distant metastasis, and impaired immune surveillance. All these factors could contribute to higher SCC levels even in patients with nonsquamous cell carcinomas, such as lung adenocarcinoma. This implies that the level of SCC could hold prognostic significance in patients with nonsquamous cell carcinoma. However, this correlation needs to be confirmed by further large-scale studies.35,36 In our study, SCC demonstrated statistical significance in the training cohort. By comprehensively analyzing these laboratory indicators, it is possible to more accurately determine the type of lung cancer in a patient, choose an appropriate treatment plan, and predict the course of the disease.
However, relying solely on clinical features or serum tumor biomarkers is not adequate for an accurate diagnosis of lung cancer. A comprehensive evaluation is typically needed. 37 Various imaging features have been proposed in the past to aid in identifying of distinct pathological subtypes and to classify patients accordingly. 38 However, features like the margins of a tumor or the presence of solid components can appear similar across different subtypes of lung cancer. The subtyping of lung cancer based on imaging features can be challenging. The performance of these features in correctly identifying the subtype of lung cancer has not been thoroughly evaluated. Many variables can affect the interpretation of these images, leading to a high degree of interobserver variability, meaning different healthcare professionals may interpret the same image differently.
The maximum diameter of the tumor is a critical parameter, with larger tumors typically correlating with poorer outcomes. 39 Takahiro et al. found a favorable prognosis for patients with solid components measuring less than or equal to 2 cm. In comparison, the behavior of solid components measuring greater than 2 cm is more aggressive. 40 Patients have different clinical and pathology with similar tumor size, as shown in Figure 10; but the same stage and differentiation degree cannot truly reflect the heterogeneity of the tumor and all of its subtype characteristics. The CTR serves as an insightful imaging biomarker to gauge the aggressiveness of lung adenocarcinomas, which, in turn, can inform prognosis and guide treatment decisions. 41 Lin et al. found that patients with a higher CTR presented more frequently with invasive adenocarcinomas and lymphovascular and visceral pleural invasion compared to those with lower CTR values. 42 This correlation is likely because a more prominent consolidation component often represents a denser, more solid mass suggestive of invasive carcinoma. CTR is also instrumental in distinguishing various adenocarcinoma subtypes. Xu and colleagues observed that lung adenocarcinomas with a CTR exceeding 0.5 were more prone to exhibiting micropapillary patterns. 43 Moreover, Chen et al. identified a CTR greater than 0.5, indicative of radiological invasiveness. They incorporated this threshold into a radiomic model designed to predict the presence of micropapillary and solid components. This model achieved a sensitivity of 90.00% and a specificity of 45.21%. 44 In our study, all three metrics demonstrated statistical significance in the training cohort and were subsequently utilized to construct the clinical model.
Radiomics is a revolutionary technology that extracts vast data from various medical images (such as CT, MRI, and PET scans) through high-throughput methods and uses advanced machine learning algorithms to select features with the highest clinical value. This technology plays a crucial role in deciphering the biological characteristics of tumors, particularly excelling in the early diagnosis of tumors and assessing treatment efficacy. Wang et al. developed a radiomic signature containing 15 radiological features that can distinguish between benign and malignant lung nodules with an accuracy of 86%. 45 The research report by Lu et al. indicates that the radiomic model excels in differentiating histopathological subtypes of lung cancer, with diagnostic performance (AUC) of 0.741 for SCLC versus non-SCLC, 0.822 for adenocarcinoma versus SCLC, 0.665 for squamous cell carcinoma versus SCLC, and 0.655 for adenocarcinoma versus squamous cell carcinoma. 46 Chen et al. integrated GPTV6 radiomic features with independent clinical predictive factors in patients with clinical stage IA non-SCLC, constructing a diagnostic model that performed well in predicting lymph node metastasis and prognosis, with AUCs of 0.85 in the training cohort, 0.80 in the internal validation cohort, and 0.74 in the external validation cohort. 47 Yang et al. reported that radiomic features based on pretreated CT scans can accurately predict tumor response in non-SCLC patients after first-line chemotherapy or targeted therapy, with an AUC of 0.746 (95% CI, 0.646–0.846). 48 Ding et al.’s research indicates that CT-based delta radiomic markers can improve the personalized prediction of OS and local recurrence in SCLC patients during early chemotherapy. The R32 marker showed the best performance, with C-indexes of 0.857 and 0.836, respectively, when clinical radiological features were integrated into the RRSOS model. 49 The radiomic model and the integrated model constructed by Chen et al. can noninvasively predict the EGFR mutation status and subtypes of lung adenocarcinoma, with AUCs in the test group reaching 0.759 and 0.554, respectively, which are of great value in saving clinical costs and guiding targeted therapy. 50
The microscale heterogeneity of tumor internal structures revealed by radiomics can be used to differentiate subtypes of SCLC and NSCLC, such as adenocarcinoma and squamous cell carcinoma. 51 In a previous radiomics study focusing on lung cancer subtypes, Yang et al. discovered that high-purity lung adenocarcinomas (70% pathological subtype purity) had an accuracy of 83% for five subtypes and 94% for three pathological gradings in the validation set. 52 Park et al. documented in their study that radiomics could distinguish between three pathological grades of the main subtypes of adenocarcinoma. The accuracy of this differentiation was commensurate with diagnostic evaluations made by professional radiologists. 53 Song et al. concluded in their research that the micropapillary component within lung adenocarcinomas can be predicted by evaluating the variance values of both the minimum and positive pixel values across the entire pixel value spectrum. 54 Bae et al. found that various subtypes of lung cancer could be distinguished using radiomics features derived from dual-energy CT data. They achieved AUC of 0.9307, 0.8610, and 0.8394 in cross-validation, suggesting a high degree of accuracy in their model. 55 By employing machine learning algorithms to analyze many radiomics features, it is possible to identify the most relevant features associated with specific types of lung cancer. This assists in distinguishing between benign and malignant lesions, as well as different subtypes of lung cancer. As a result, it enhances various aspects of cancer diagnosis and treatment, including early detection, auxiliary diagnosis, prognosis prediction, and immunotherapy practices. 56 Our study incorporated patients with lung adenocarcinoma, specifically those with micropapillary or solid components constituting <5% in the positive group. This approach more accurately reflects the real-world clinical application situation, enhancing the practical value and applicability of the study’s findings.
In this investigation, the authors used eight classifiers to compare their AUC in constructing radiomic models in this study. After a detailed comparison and analysis, it was determined that LR was the most effective classifier. This implies that the LR model demonstrated the highest performance level in predicting or classifying the outcome variable based on the given set of predictor variables. The final model incorporated 30 distinct radiomics features. The choice of these features could be attributed to the dynamic growth and conspicuous infiltration characteristic of micropapillary and solid components. These cells frequently intermingle with other tissue types, potentially leading to significant histological heterogeneity.38,57 Incorporating a larger number of radiomics features into the model enables a more detailed representation of the internal characteristics of lung cancer from various perspectives. This provides a more comprehensive understanding of its biological characteristics and gives insights into the behavior of the disease. It could enhance diagnosis accuracy and treatment strategies’ effectiveness. The Rad-score, or Radiomics score, is a calculated value typically obtained from multiple radiomic features using a specific computational model, such as linear regression. This quantified score aims to encapsulate the intricate information present in these features into one single, interpretable number. A Rad-score integrating multiple omics features could provide a more accurate representation of a tumor’s biological characteristics. This enhanced accuracy could improve the score’s predictive power, thereby aiding in the diagnosis and prognosis assessment of the disease. Furthermore, it could also provide valuable insights to guide therapeutic decision-making. This could explain why a higher degree of heterogeneity observed in CT images is often linked with increased aggressiveness and a less favorable prognosis. 58 These findings equip us with a powerful tool for predicting clinical outcomes in lung cancer more accurately. This can further assist in devising more effective treatment strategies.
By comprehensively analyzing these laboratory indicators, it is possible to more accurately determine the type of lung cancer in a patient, choose an appropriate treatment plan, and predict the course of the disease. Combining laboratory indicators with clinical assessment and imaging examinations helps obtain a comprehensive understanding of the patient’s condition. It provides crucial information for developing the optimal treatment plan. 59
At the same time, this study went beyond the use of radiomics methods and also considered serum tumor biomarkers and clinical features. These included factors such as T_size, ST_size, CTR, the number of years of smoking, and a range of serum tumor markers, including CYFRA 21-1. This approach offers a more holistic basis for diagnosing and treating lung adenocarcinoma, providing a more detailed and comprehensive understanding of the disease. Liu and colleagues present an innovative approach predicting high-grade components in lung adenocarcinoma. For this purpose, they masterfully unify radiomics, imaging features, and serum tumor markers. Their combined model has proven highly effective, achieving impressive AUCs of 0.88 and 0.94 in the training and validation cohorts. 16 The research reveals the significant potential of employing a multifaceted approach for predictive purposes. It opens up new avenues for precise classification and diagnosis of lung adenocarcinoma in the future. In this study, combining of radiomics features extracted from preoperative CT data with clinical features and serum tumor markers exhibits high sensitivity in diagnosing MPP/SOL components in lung adenocarcinoma. Our research provides a novel approach and an efficient tool for the medical community, fostering advancements in the precise diagnosis of lung adenocarcinoma. This study implies that prior to the resection of lung adenocarcinoma, physicians could alter their surgical plans based on the calculated presence of MPP/SOL components, thus opting for a more suitable resection range. 10 Furthermore, this study suggests that pathologists can heighten their vigilance during the frozen section analysis to identify evidence of MPP/SOL components better. This heightened awareness could lead to more accurate diagnoses and more effective treatment plans. However, in practical application, despite clear criteria for pathological subtyping, the inherent heterogeneity of tissue samples and potential sampling bias may still lead to misdiagnosis. This issue is particularly prevalent in the diagnostic accuracy of MPP/SOL.60,61 Our research introduces a novel diagnostic tool designed to aid doctors in more accurately identifying subtypes of lung adenocarcinoma. This innovative approach enhances diagnostic precision and provides a new reference point for formulating surgical plans.
To ensure the generalizability of the results, it is crucial to avoid selection bias. This can be achieved by refraining from cherry-picking patients/datasets. It is essential to establish explicit inclusion and exclusion criteria and adhere to them while including consecutive patients who meet these criteria. 62 Furthermore, incorporating data from multiple institutions or imaging devices can enhance the sample’s diversity, thereby improving the generalizability of the research findings. In this study, image data from six CT machines were included to accomplish this.
In all imaging modalities, the vendor, machine, and acquisition protocols are recognized as potential sources of bias. 63 Therefore, it is crucial to establish and implement standardized scanning protocols in the study. This includes defining image acquisition parameters, scanning sequences, and scanning parameters. Doing so ensures that the images collected from different devices and at other time points are comparable, thereby reducing biases resulting from device variations and changes in scanning parameters. Preprocessing plays a vital role in maximizing the reproducibility of radiomic features. 62 One of the initial preprocessing steps involves interpolating the imaging data to achieve isotropic voxel spacing. This enables improved comparison of heterogeneous, multi-institutional imaging data. 64 Additionally, discretization or quantization of image intensities is particularly important in facilitating comprehensible feature extraction. 65
The variability in segmentation can introduce bias in the subsequent stages of the analysis and should be taken into consideration. 63 One approach is to randomly select a subset of samples for reannotation and then calculate measures of inter-rater agreement, such as consistency or Kappa coefficients. Additionally, another experienced annotator or expert can review the annotations, and any discrepancies can be resolved through consistency analysis or expert consensus. 66 This procedure helps minimize the influence of individual annotators’ subjective preferences and ensures the accuracy and consistency of the annotations.
This study also has certain limitations. First, as this was a retrospective study, there is potential selection bias. In light of this, our future aim is to conduct a prospective study to control for confounding variables more effectively. Second, being a small-scale, single-center investigation, this study necessitates the external validation of our model’s stability and clinical applicability. The external validation of the stability and clinical applicability of the model needs to be added to a multicenter dataset in the future. Finally, it is essential to note that the manual segmentation of the ROI inherently carries the potential for inter- and intraobserver discrepancies. In the future, we plan to expand our patient base and implement advanced technical enhancements. This would include fully automated image segmentation, deep learning, and multiparametric modeling, all aimed at refining the precision of radiological diagnoses.
Conclusion
Our study has developed and validated a combined nomogram, a visual tool that integrates CT radiomics features with clinical indicators and serum tumor biomarkers. This innovative model facilitates the differentiation of micropapillary or solid components within lung adenocarcinoma. Importantly, our model achieves a higher AUC, indicating superior predictive accuracy.
Supplemental Material
Supplemental material, sj-docx-1-tar-10.1177_17534666241249168 for A combination of radiomic features, clinic characteristics, and serum tumor biomarkers to predict the possibility of the micropapillary/solid component of lung adenocarcinoma by Xiaowei Xing, Liangping Li, Mingxia Sun, Xinhai Zhu and Yue Feng in Therapeutic Advances in Respiratory Disease
Acknowledgments
Some of our experiments were carried out on the OnekeyAI platform. We thank OnekeyAI and their developers’ help in this scientific research work.
Footnotes
ORCID iD: Yue Feng  https://orcid.org/0000-0001-6251-9199
https://orcid.org/0000-0001-6251-9199
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Xiaowei Xing, Cancer Center, Department of Radiology, Zhejiang Provincial People’s Hospital, (Affiliated People’s Hospital), Hangzhou Medical College, Hangzhou, Zhejiang, China.
Liangping Li, Department of Radiology, Zhejiang Hospital, Hangzhou, Zhejiang, China.
Mingxia Sun, Department of Radiology, Zhejiang Hospital, Hangzhou, Zhejiang, China.
Xinhai Zhu, Department of Thoracic Surgery, Zhejiang Hospital, Hangzhou, Zhejiang, China.
Yue Feng, Cancer Center, Department of Radiology, Zhejiang Provincial People’s Hospital, (Affiliated People’s Hospital), Hangzhou Medical College, Hangzhou, Zhejiang, China.
Declarations
Ethics approval and consent to participate: This study protocol was approved by the Ethics Committee of the Zhejiang Hospital (approval number 2023-37K). This study adhered to the tenets of the Declaration of Helsinki. The authors are accountable for all aspects of the work and ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Considering the retrospective nature of this study, the requirement to obtain informed consent was waived.
Consent for publication: Not applicable.
Author contributions: Xiaowei Xing: Conceptualization; Writing – original draft; Writing – review & editing.
Liangping Li: Formal analysis; Methodology.
Mingxia Sun: Formal analysis; Validation.
Xinhai Zhu: Funding acquisition; Investigation.
Yue Feng: Conceptualization; Supervision; Writing – review & editing.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Zhejiang Medicine and Health Science and Technology Project (Grant/Award Number 2022PY035).
The authors declare that there is no conflict of interest.
Availability of data and materials: The experimental data used to support the findings of this study can be obtained from the corresponding author upon request.
References
- 1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. 20180912. [DOI] [PubMed] [Google Scholar]
- 2. Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J Thorac Oncol 2011; 6: 244-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Russell PA, Wainer Z, Wright GM, et al. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the New International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society International Multidisciplinary Lung Adenocarcinoma Classification. J Thorac Oncol 2011; 6: 1496–1504. [DOI] [PubMed] [Google Scholar]
- 4. Yoshizawa A, Motoi N, Riely GJ, et al. Impact of proposed IASLC/ATS/ERS classification of lung adenocarcinoma: prognostic subgroups and implications for further revision of staging based on analysis of 514 stage I cases. Mod Pathol 2011; 24: 653–664. [DOI] [PubMed] [Google Scholar]
- 5. Cao Y, Zhu LZ, Jiang MJ, et al. Clinical impacts of a micropapillary pattern in lung adenocarcinoma: a review. Onco Targets Ther 2016; 9: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leeman JE, Rimner A, Montecalvo J, et al. Histologic subtype in core lung biopsies of early-stage lung adenocarcinoma is a prognostic factor for treatment response and failure patterns after stereotactic body radiation therapy. Int J Radiat Oncol Biol Phys 2017; 97: 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qian F, Yang W, Wang R, et al. Prognostic significance and adjuvant chemotherapy survival benefits of a solid or micropapillary pattern in patients with resected stage IB lung adenocarcinoma. J Thorac Cardiovasc Surg 2018; 155: 1227–1235. e1222. [DOI] [PubMed] [Google Scholar]
- 8. Lee G, Lee HY, Jeong JY, et al. Clinical impact of minimal micropapillary pattern in invasive lung adenocarcinoma. Am J Surg Pathol 2015; 39: 660–666. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Zheng D, Zheng J, et al. Predictors of recurrence and survival of pathological T1N0M0 invasive adenocarcinoma following lobectomy. J Cancer Res Clin Oncol 2018; 144: 1015–1023. [DOI] [PubMed] [Google Scholar]
- 10. Nitadori J-i, Bograd AJ, Kadota K, et al. Impact of micropapillary histologic subtype in selecting limited resection vs lobectomy for lung adenocarcinoma of 2cm or Smaller. J Natl Cancer Inst 2013; 105: 1212–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hung J-J, Yeh Y-C, Jeng W-J, et al. Prognostic factors of survival after recurrence in patients with resected lung adenocarcinoma. J Thorac Oncol 2015; 10: 1328–1336. [DOI] [PubMed] [Google Scholar]
- 12. Huang K-Y, Ko P-Z, Yao C-W, et al. Inaccuracy of lung adenocarcinoma subtyping using preoperative biopsy specimens. J Thorac Cardiovasc Surg 2017; 154: 332–339.e331. [DOI] [PubMed] [Google Scholar]
- 13. Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012; 48: 441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magnet Reson Imaging 2012; 30: 1234–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weng Q, Hui J, Wang H, et al. Radiomic feature-based nomogram: a novel technique to predict EGFR-activating mutations for EGFR Tyrosin kinase inhibitor therapy. Front Oncol 2011; 11: 590937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu Y, Chang Y, Zha X, et al. A combination of radiomic features, imaging characteristics, and serum tumor biomarkers to predict the possibility of the high-grade subtypes of lung adenocarcinoma. Acad Radiol 2022; 29: 1792–1801. 20220326. [DOI] [PubMed] [Google Scholar]
- 17. Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14: 749–762. [DOI] [PubMed] [Google Scholar]
- 18. Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMJ 2015; 350: g7594. [DOI] [PubMed] [Google Scholar]
- 19. Austin JHM, Garg K, Aberle D, et al. Radiologic implications of the 2011 classification of adenocarcinoma of the lung. Radiology 2013; 266: 62–71. [DOI] [PubMed] [Google Scholar]
- 20. Butnor KJ. Controversies and challenges in the histologic subtyping of lung adenocarcinoma. Transl Lung Cancer Res 2020; 9: 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhai W, Liang D, Duan F, et al. Prognostic nomograms based on ground glass opacity and subtype of lung adenocarcinoma for patients with pathological stage IA lung adenocarcinoma. Front Cell Dev Biol 2021; 9: 769881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014; 5: 4006. 20140603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res 2017; 77: e104–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang C, Yang J, Lu M. Micropapillary predominant lung adenocarcinoma in stage IA benefits from adjuvant chemotherapy. Ann Surg Oncol 2020; 27: 2051–2060. [DOI] [PubMed] [Google Scholar]
- 25. Hung J-J, Wu Y-C, Chou T-Y, et al. Adjuvant chemotherapy improves the probability of freedom from recurrence in patients with resected stage IB lung adenocarcinoma. Ann Thorac Surg 2016; 101: 1346–1353. [DOI] [PubMed] [Google Scholar]
- 26. Kabalak PA, Yılmaz Ü. Clinical presentation of lung cancer. In: Cingi C, Yorgancıoğlu A, Bayar Muluk N, et al. (eds) Airway diseases. Cham: Springer International Publishing, 2023, pp.1–19. [Google Scholar]
- 27. Chen ZM, Peto R, Iona A, et al. Emerging tobacco-related cancer risks in China: a nationwide, prospective study of 0.5 million adults. Cancer 2015; 121 Suppl 17: 3097–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yanagawa N, Shiono S, Abiko M, et al. The clinical impact of solid and micropapillary patterns in resected lung adenocarcinoma. J Thorac Oncol 2016; 11: 1976–1983. [DOI] [PubMed] [Google Scholar]
- 29. Le Calvez F, Mukeria A, Hunt JD, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res 2005; 65: 5076–5083. [DOI] [PubMed] [Google Scholar]
- 30. Wu FZ, Huang YL, Wu CC, et al. Assessment of Selection criteria for low-dose lung screening CT among Asian Ethnic Groups in Taiwan: From mass screening to specific risk-based screening for non-smoker lung cancer. Clin Lung Cancer 2016; 17: e45–e56. [DOI] [PubMed] [Google Scholar]
- 31. Leiter A, Veluswamy RR, Wisnivesky JP. The global burden of lung cancer: current status and future trends. Nat Rev Clin Oncol 2023; 20: 624–639. [DOI] [PubMed] [Google Scholar]
- 32. Nakamura H, Nishimura T. History, molecular features, and clinical importance of conventional serum biomarkers in lung cancer. Surgery Today 2017; 47: 1037–1059. [DOI] [PubMed] [Google Scholar]
- 33. Muley T, Dienemann H, Ebert W. Increased CYFRA 21-1 and CEA levels are negative predictors of outcome in p-stage I NSCLC. Anticancer Res 2003; 23: 4085–4093. [PubMed] [Google Scholar]
- 34. Bian NN, Shi XY, Qi HY, et al. The relationship of plasma fibrinogen with clinicopathological stages and tumor markers in patients with non-small cell lung cancer. Medicine (Baltimore) 2019; 98: e16764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhu H. Squamous cell carcinoma antigen: clinical application and research status. Diagnostics (Basel) 2022; 12: 20220424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin WH, Chen IH, Wei FC, et al. Clinical significance of preoperative squamous cell carcinoma antigen in oral-cavity squamous cell carcinoma. Laryngoscope 2011; 121: 971–977. [DOI] [PubMed] [Google Scholar]
- 37. Nooreldeen R, Bach H. Current and future development in lung cancer diagnosis. Int J Mol Sci 2021; 22: 20210812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lederlin M, Puderbach M, Muley T, et al. Correlation of radio- and histomorphological pattern of pulmonary adenocarcinoma. Eur Respir J 2013; 41: 943–951. [DOI] [PubMed] [Google Scholar]
- 39. Xu S, Xi J, Jiang W, et al. Solid component and tumor size correlate with prognosis of stage IB lung adenocarcinoma. Ann Thorac Surg 2015; 99: 961–967. [DOI] [PubMed] [Google Scholar]
- 40. Mimae T, Tsutani Y, Miyata Y, et al. Solid tumor size of 2 cm divides outcomes of patients with mixed ground glass opacity lung tumors. Ann Thorac Surg 2020; 109: 1530–1536. [DOI] [PubMed] [Google Scholar]
- 41. Wu Y, Song W, Wang D, et al. Prognostic value of consolidation-to-tumor ratio on computed tomography in NSCLC: a meta-analysis. World J Surg Oncol 2023; 21: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin B, Wang R, Chen L, et al. Should resection extent be decided by total lesion size or solid component size in ground glass opacity-containing lung adenocarcinomas? Transl Lung Cancer Res 2021; 10: 2487–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu SJ, Tu JH, Chen H, et al. A multi-institutional analysis of the combined effect of micropapillary component and consolidation-to-tumor ratio >0.5 on the prognosis of pathological, Stage IA3, lung adenocarcinoma. Ann Surg Oncol 2023; 30: 5843–5853. [DOI] [PubMed] [Google Scholar]
- 44. Chen LW, Yang SM, Wang HJ, et al. Prediction of micropapillary and solid pattern in lung adenocarcinoma using radiomic values extracted from near-pure histopathological subtypes. Eur Radiol 2021; 31: 5127–5138. [DOI] [PubMed] [Google Scholar]
- 45. Jun W, Xia L, Di D, et al. Prediction of malignant and benign of lung tumor using a quantitative radiomic method. Annu Int Conf IEEE Eng Med Biol Soc 2016; 2016: 1272–1275. [DOI] [PubMed] [Google Scholar]
- 46. E L, Lu L, Li L, et al. Radiomics for classification of lung cancer histological subtypes based on nonenhanced computed tomography. Acad Radiol 2019; 26: 1245–1252. [DOI] [PubMed] [Google Scholar]
- 47. Chen QL, Li MM, Xue T, et al. Radiomics nomogram integrating intratumoural and peritumoural features to predict lymph node metastasis and prognosis in clinical stage IA non-small cell lung cancer: a two-centre study. Clin Radiol 2023; 78: e359–e367. [DOI] [PubMed] [Google Scholar]
- 48. Yang F, Zhang J, Zhou L, et al. CT-based radiomics signatures can predict the tumor response of non-small cell lung cancer patients treated with first-line chemotherapy and targeted therapy. Eur Radiol 2022; 32: 1538–1547. 20210926. [DOI] [PubMed] [Google Scholar]
- 49. Ding Z, Zhang C, Yao Q, et al. Delta radiomics model for the prediction of overall survival and local recurrence in small cell lung cancer patients after chemotherapy. Acad Radiol 2024; 31: 1168–1179. [DOI] [PubMed] [Google Scholar]
- 50. Chen Q, Li Y, Cheng Q, et al. EGFR mutation status and subtypes predicted by CT-based 3D radiomic features in lung adenocarcinoma. Onco Targets Ther 2022; 15: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tang X, Wu F, Chen X, et al. Current status and prospect of PET-related imaging radiomics in lung cancer. Front Oncol 2023; 13: 1297674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang SM, Chen LW, Wang HJ, et al. Extraction of radiomic values from lung adenocarcinoma with near-pure subtypes in the International Association for the Study of Lung Cancer/the American Thoracic Society/the European Respiratory Society (IASLC/ATS/ERS) classification. Lung Cancer 2018; 119: 56–63. [DOI] [PubMed] [Google Scholar]
- 53. Park S, Lee SM, Noh HN, et al. Differentiation of predominant subtypes of lung adenocarcinoma using a quantitative radiomics approach on CT. Eur Radiol 2020; 30: 4883–4892. [DOI] [PubMed] [Google Scholar]
- 54. Song SH, Park H, Lee G, et al. Imaging phenotyping using radiomics to predict micropapillary pattern within lung adenocarcinoma. J Thorac Oncol 2017; 12: 624–632. 20161205. [DOI] [PubMed] [Google Scholar]
- 55. Bae JM, Jeong JY, Lee HY, et al. Pathologic stratification of operable lung adenocarcinoma using radiomics features extracted from dual energy CT images. Oncotarget 2017; 8: 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y, Wu X, Yang P, et al. Machine learning for lung cancer diagnosis, treatment, and prognosis. Genomics Proteomics Bioinf 2022; 20: 850–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yanagawa N, Shiono S, Abiko M, et al. New IASLC/ATS/ERS classification and invasive tumor size are predictive of disease recurrence in Stage I lung adenocarcinoma. J Thorac Oncol 2013; 8: 612–618. [DOI] [PubMed] [Google Scholar]
- 58. Park SY, Lee JG, Kim J, et al. Imaging heterogeneity in lung cancer: techniques, applications, and challenges. Am J Roentgenol 2016; 207: 534–543. [DOI] [PubMed] [Google Scholar]
- 59. Vos D, Rao S, Pierce JD, et al. The past, present, and future (Liquid Biopsy) of serum tumor markers in lung cancer: a primer for the radiologist. J Comput Assist Tomogr 2021; 45: 950–958. [DOI] [PubMed] [Google Scholar]
- 60. Trejo Bittar HE, Incharoen P, Althouse AD, et al. Accuracy of the IASLC/ATS/ERS histological subtyping of stage I lung adenocarcinoma on intraoperative frozen sections. Mod Pathol 2015; 28: 1058–1063. [DOI] [PubMed] [Google Scholar]
- 61. Warth A, Stenzinger A, von Brünneck A-C, et al. Interobserver variability in the application of the novel IASLC/ATS/ERS classification for pulmonary adenocarcinomas. Eur Respir J 2012; 40: 1221–1227. [DOI] [PubMed] [Google Scholar]
- 62. Wagner MW, Namdar K, Biswas A, et al. Radiomics, machine learning, and artificial intelligence-what the neuroradiologist needs to know. Neuroradiology 2021; 63: 1957–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bibault JE, Xing L, Giraud P, et al. Radiomics: a primer for the radiation oncologist. Cancer Radiother 2020; 24: 403–410. [DOI] [PubMed] [Google Scholar]
- 64. Lohmann P, Bousabarah K, Hoevels M, et al. Radiomics in radiation oncology-basics, methods, and limitations. Strahlenther Onkol 2020; 196: 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zwanenburg A, Vallieres M, Abdalah MA, et al. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020; 295: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao B. Understanding sources of variation to improve the reproducibility of radiomics. Front Oncol 2021; 11: 633176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tar-10.1177_17534666241249168 for A combination of radiomic features, clinic characteristics, and serum tumor biomarkers to predict the possibility of the micropapillary/solid component of lung adenocarcinoma by Xiaowei Xing, Liangping Li, Mingxia Sun, Xinhai Zhu and Yue Feng in Therapeutic Advances in Respiratory Disease