Abstract
Neurodegenerative diseases, including Alzheimer’s disease (AD), are characterized by innate immune-mediated inflammation, but functional and mechanistic effects of adaptive immune system remain unclear. Here, we identify brain-resident CD8+ T cells that co-express CXCR6 and PD-1 and are in proximity to plaque-associated microglia in human and mouse AD brains. We also establish that CD8+ T cells restrict AD pathologies, including β-amyloid deposition and cognitive decline. Ligand–receptor interaction analysis identifies CXCL16–CXCR6 inter-cellular communication between microglia and CD8+ T cells. Further, Cxcr6 deficiency impairs accumulation, tissue residency programming, and clonal expansion of brain PD-1+CD8+ T cells. Ablation of Cxcr6 or CD8+ T cells ultimately increases pro-inflammatory cytokine production from microglia, with CXCR6 orchestrating brain CD8+ T cell–microglia co-localization. Collectively, our study reveals protective roles for brain CD8+ T cells and CXCR6 in mouse AD pathogenesis, and highlights that microenvironment-specific, inter-cellular communication orchestrates tissue homeostasis and protection from neuroinflammation.
Main text
Inflammation and aberrant immunity are hallmarks of Alzheimer’s disease (AD) and other neurodegenerative diseases1,2. Microglia contribute to AD by triggering neuroinflammation1,2, and adaptive immune cells also undergo phenotypic alterations in individuals with AD3,4. However, adaptive immune cells show both deleterious and protective effects in AD pathogenesis5-8, possibly attributed to temporal regulation or functional heterogeneity of the adaptive immune system. Further, inter-cellular communication is emerging as a crucial determinant of inflammation and disease outcomes in infections and cancer, yet our understanding of the regulation of such communication in the brain parenchyma and its contribution to neuroinflammation is lacking.
By integration of single-cell RNA-sequencing (scRNA-seq) and single-cell TCR-sequencing (scTCR-seq) profiling, we demonstrate disease-associated, age-dependent accumulation of CD8+ T cells in the brain parenchyma. Genetic disruption of total αβ+ or CD8+ T cells exacerbates β-amyloid (Aβ) aggregation and memory decline in a mouse model of AD. Through unbiased ligand–receptor network analysis, we reveal a CXCL16–CXCR6 communication axis between microglia and CD8+ T cells, associated with prevalent CXCL16 expression by microglia from mouse AD models and individuals with AD. CXCR6 deficiency reduces CD8+ T cell accumulation in the brain and leads to Aβ aggregation and impaired cognitive function. We identify clonally expanded CD8+ T cells with enhanced tissue residency programming in the brain parenchyma of AD mouse models, and establish the central role of CXCR6 in coordinating these processes. Loss of CXCR6 disrupts spatial localization of CD8+ T cells near Aβ plaque-associated microglia. We further show that CD8+ T cells and CXCR6 limit the pro-inflammatory activity of microglia. Collectively, our study reveals a role for inter-cellular crosstalk between brain-resident CXCR6+CD8+ T cells and microglia in restricting AD pathology.
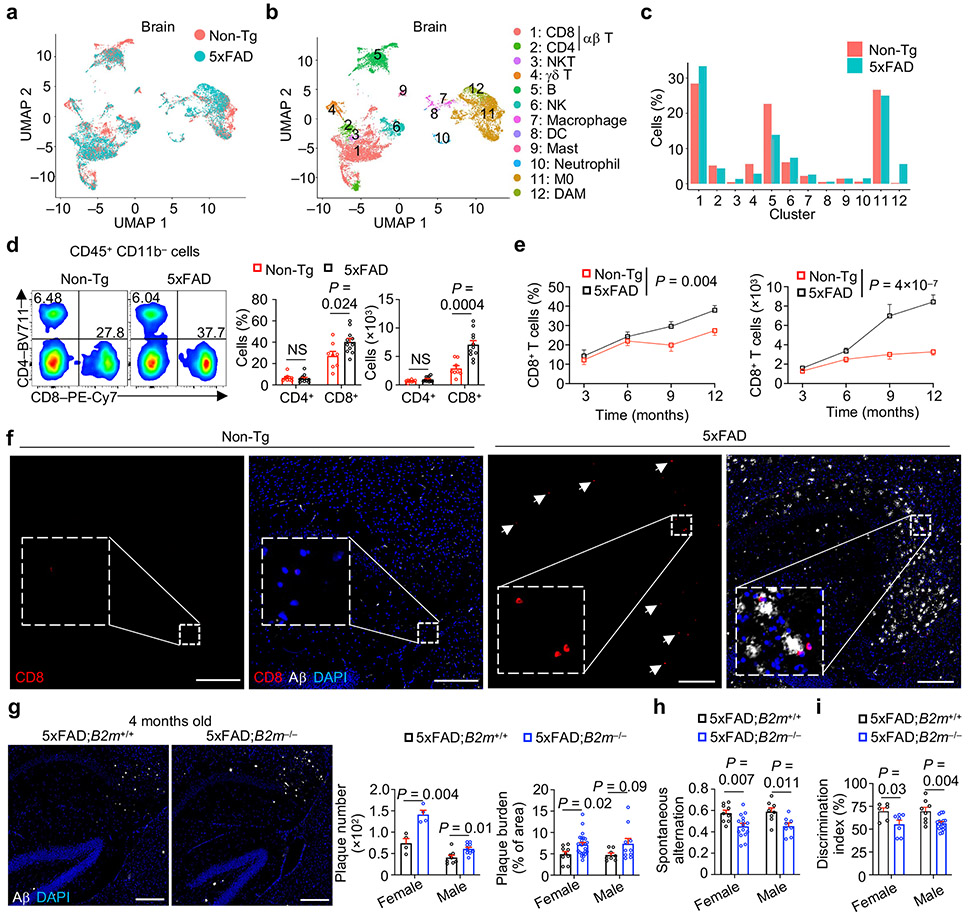
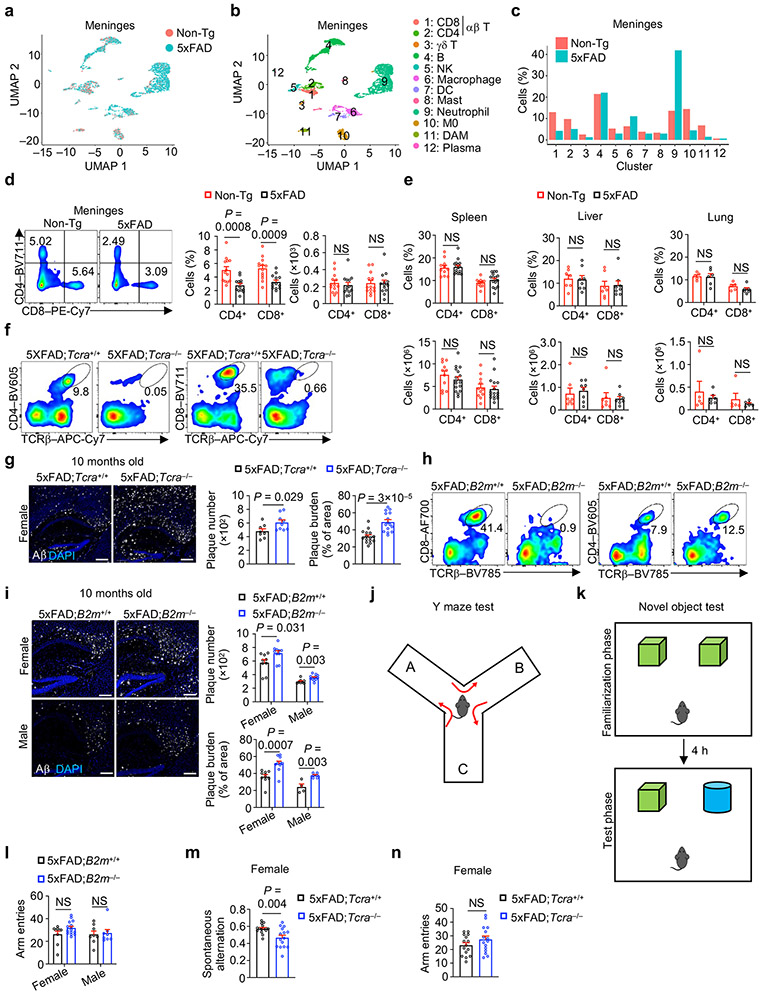
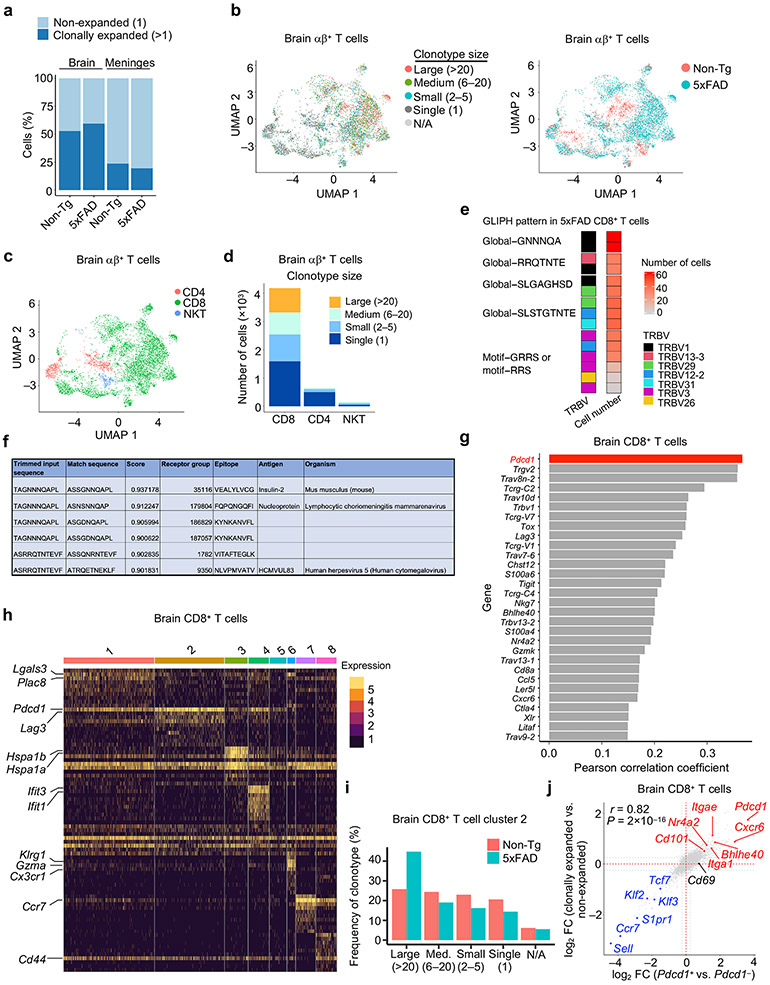
CD8+ T cells protect mice from AD-associated pathologies
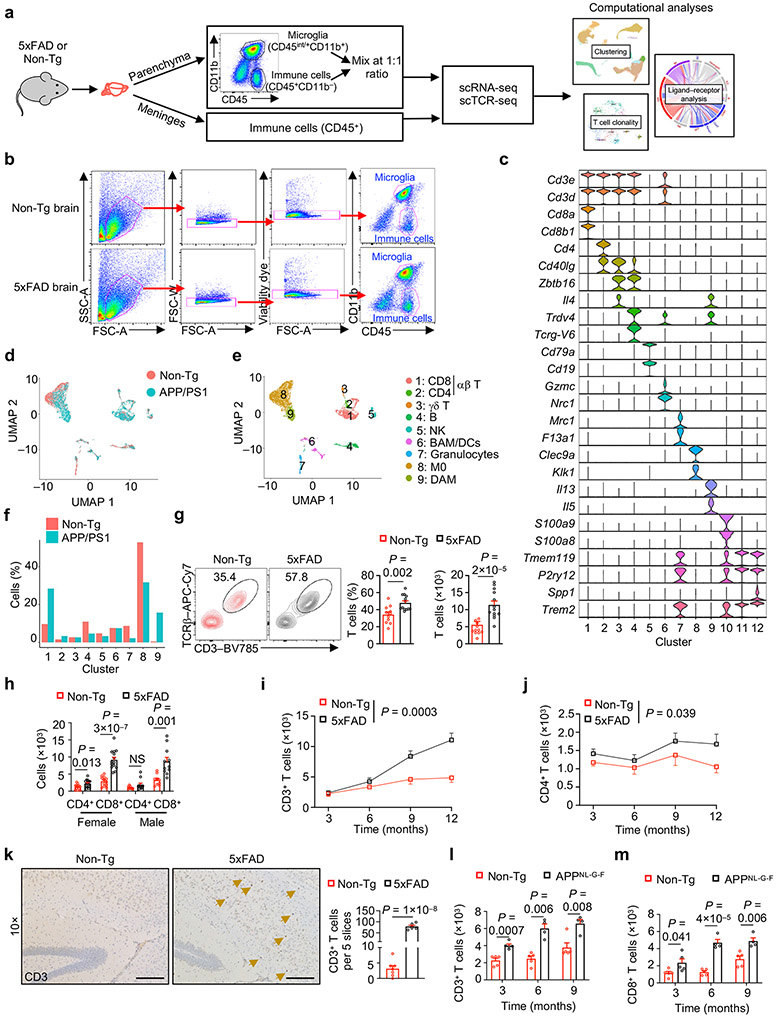
Immune cell alterations occur in the periphery and central nervous system (CNS) of individuals with AD3,4,9 and mouse models8,10,11, but less is known about changes in non-microglia immune cells in the brain parenchyma where disease-associated pathologies develop. Therefore, we performed scRNA-seq analysis on CD45int/+CD11b+ (primarily microglia) and CD45+CD11b− (non-microglia immune cells) cells from brain parenchyma (thereafter called brain) or meningeal dura and arachnoid tissues12 (thereafter called meninges) of 5xFAD13 and Non-transgenic (Non-Tg) control mice (Extended Data Fig. 1a, b). Uniform manifold approximation and projection (UMAP) and clustering analyses unveiled 12 distinct immune clusters, with disease-associated microglia (DAM) being restricted to 5xFAD mice as expected10 (Fig. 1a-c, Extended Data Fig. 1c). Notably, the frequency of CD8+ T cells, but not CD4+ T cells or γδ T cells, was elevated in 5xFAD mice (Fig. 1c), with such observations also extending to APP/PS111 mice (Extended Data Fig. 1d-f).
Figure 1. CD8+ T cells accumulate in AD and protect mice from AD-associated pathologies.
a–c, scRNA-seq analysis of CD45int/+CD11b+ (primarily microglia) and CD45+CD11b− non-microglia immune cells from the brains of 8-month-old Non-Tg or 5xFAD mice (n = 2 biological replicates, pooled from 3 mice per group). UMAP showing immune cell clusters colored by genotype (a) or cell types (b) (see Methods). M0: homeostatic microglia; DAM: disease associated microglia; DC: dendritic cells; NK: natural killer cells; NKT: natural killer T cells. c, Cell frequencies (normalized to total cells from each genotype) in each cluster in b. d, Brain CD4+ and CD8+ T cells from 10-month-old Non-Tg (n = 8) and 5xFAD (n = 11) mice. e, Brain CD8+ T cells at 3 (n = 6 per genotype), 6 (n = 13 for Non-Tg and 16 for 5xFAD), 9 (n = 8 for Non-Tg and 10 for 5xFAD), and 12 (n = 15 for Non-Tg and 18 for 5xFAD) months of age. f, Images of CD8+ T cells, Aβ, and DAPI in hippocampus of 12-month-old mice. Scale bars, 250 μm. White arrows indicate positive staining for CD8+ T cells. Boxed areas show the CD8 staining inset. g, Immunohistochemical analysis of Aβ plaques in 5xFAD;B2m−/− and 5xFAD;B2m+/+ mice at 4 months of age (representative images are from females). Number and burden of Aβ plaques in the brains of 5xFAD;B2m+/+ [n = 4 sections for female and 8 for male mice for plaque number; n = 9 sections for female and 8 for male mice for plaque burden] and 5xFAD;B2m−/− [n = 4 sections for female and 10 for male mice for plaque number; n = 24 sections for female and 10 for male mice for plaque burden] mice. Scale bars, 250 μm. A serial section from the same control mouse is shown in Fig. 3f. h, Spontaneous alternation from Y maze testing in 4-month-old 5xFAD;B2m+/+ (n = 9 for female and 8 for male) and 5xFAD;B2m−/− (n = 15 for female and 8 for male) mice. i, Quantified discrimination index from novel objection recognition testing in 4-month-old 5xFAD;B2m+/+ (n = 6 for female and 8 for male) and 5xFAD;B2m−/− (n = 7 for female and 15 for male) mice. Data were analyzed by two-tailed unpaired Student’s t-test (d, g for plaque burden, h, i), two-way analysis of variance (ANOVA; e), or two-tailed unpaired t-test with Welch’s correction for plaque number (g). Data are shown as mean ± s.e.m. in d, e and g–i; NS, not significant. Data were pooled from at least three (d, e, g, i), two (h), or are representative of three (f) independent experiments.
Flow cytometry analysis showed that 5xFAD mice had increased frequency and number of CD3+TCRβ+ T cells in the brain compared to Non-Tg mice , which was primarily attributed to the accumulation of CD8+ T cells (Fig. 1d, Extended Data Fig. 1g, h). CD3+ or CD8+ T cells, but not CD4+ T cells, showed more pronounced age-dependent accumulation in 5xFAD than in Non-Tg mice (Fig. 1e, Extended Data Fig. 1i, j), coinciding with the onset of Aβ plaque accumulation in 5xFAD mice10,13. Immunostaining experiments validated these observations in 5xFAD mice (Fig. 1f, Extended Data Fig. 1k). Further, CD3+ or CD8+ T cells were accumulated in the brains of APPNL-G-F mice14 (Extended Data Fig. 1l, m). As CD8+ T cells are increased in the brains of individuals with AD3,15, these results suggest that CD8+ T cell accumulation is a conserved feature in mouse AD models and individuals with AD.
Immune cells gain access to the brain through the meninges16, and meningeal lymphatic dysfunction exacerbates Aβ deposition and promotes inflammatory microglia responses in mouse models of AD12,17. In scRNA-seq profiling of immune cells from the meninges (Extended Data Fig. 1a), the frequency of CD8+ T cells was reduced in 5xFAD mice (Extended Data Fig. 2a-c), which was validated by flow cytometry analysis, although cellularity of CD8+ T cells was unaltered (Extended Data Fig. 2d). Analysis of lymphoid (spleen) and non-lymphoid (liver and lung) tissues showed no obvious alterations in T cells in 5xFAD mice (Extended Data Fig. 2e), indicating a selective accumulation of CD8+ T cells in the brains of 5xFAD mice.
Deposits of Aβ plaques and impaired cognitive memory function are hallmarks of AD13,18. To test the contribution of CD8+ T cells to the development of these AD hallmarks, we first generated TCRα-deficient 5xFAD mice that lacked both CD8+ and CD4+ T cells (5xFAD;Tcra−/− mice; Extended Data Fig. 2f). We observed increased Aβ deposition in 5xFAD;Tcra−/− mice (Extended Data Fig. 2g). To examine whether these effects extended to the selective loss of CD8+ T cells, we crossed 5xFAD mice with those deficient in the MHC class I molecule B2m (5xFAD;B2m−/− mice; Extended Data Fig. 2h), and also observed increased Aβ deposition at both 4 and 10 months of age (Fig. 1g, Extended Data Fig. 2i). Thus, deficiency of CD8+ T cells leads to increased Aβ plaque burden. To assess cognitive memory function, we used the spontaneous Y maze alternation19 and the novel object recognition19 tests (Extended Data Fig. 2j, k). Compared to 5xFAD;B2m+/+ mice, 4-month-old 5xFAD;B2m−/− mice had diminished ability to distinguish maze arms (Fig. 1h) and discriminate between the familiar and novel objects (Fig. 1i), indicating impaired cognitive memory function. These defects were not due to altered locomotive function, as evidenced by comparable numbers of total Y maze arm entries (Extended Data Fig. 2l). Such an impairment in cognitive memory function was also observed in 5xFAD;Tcra−/− mice (Extended Data Fig. 2m, n). These results collectively show that loss of CD8+ T cells results in increased Aβ plaque burden and cognitive impairment, thereby revealing a protective role of CD8+ T cells against mouse AD development.
CXCR6–CXCL16 coordinates CD8+ T cell–microglia crosstalk
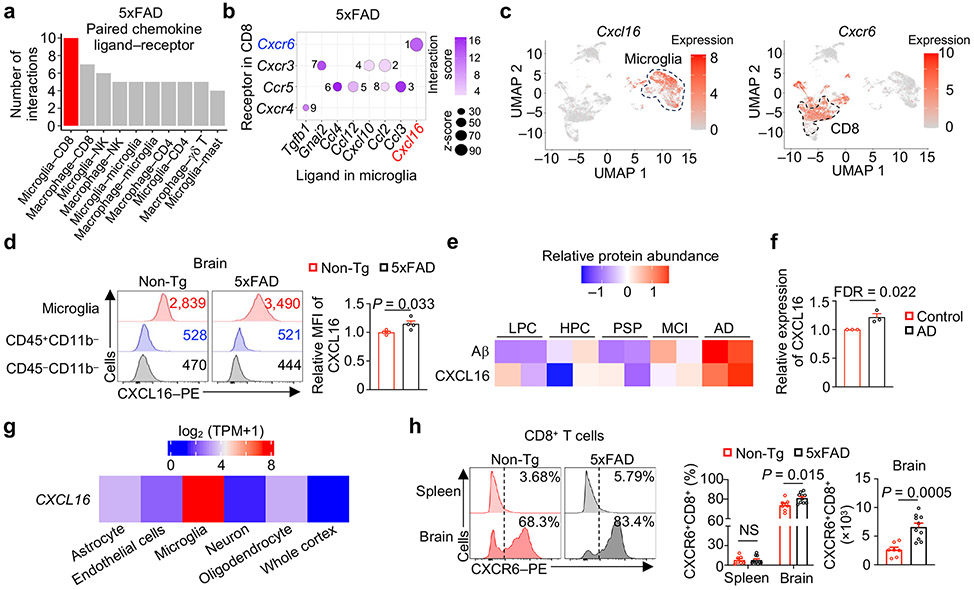
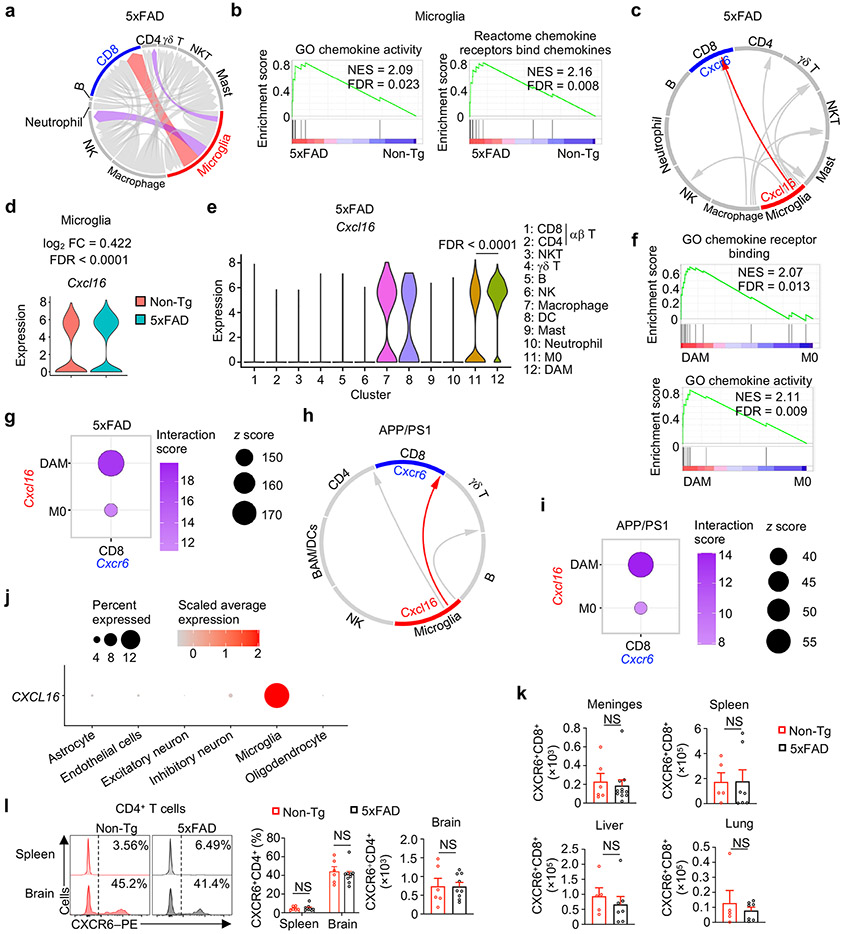
Inter-cellular communication networks influence tissue homeostasis and pathology in the CNS20. However, the specific signals, compartments, and contexts of inter-cellular communication in AD remain poorly understood, especially for non-microglia immune cells. Therefore, we inferred cellular crosstalk based on ligand–receptor interactions between immune cell types profiled by scRNA-seq21 and uncovered predicted interactions of microglia with CD8+ T cells (Extended Data Fig. 3a). Further, gene-set enrichment analysis (GSEA) showed that microglia from 5xFAD mice were enriched for chemokine- and chemokine receptor-associated signatures (Extended Data Fig. 3b). Accordingly, the number of chemokine–chemokine receptor interactions was predicted to be greater between microglia and CD8+ T cells as compared with other cell types (Fig. 2a), suggesting a potential inter-cellular communication between microglia and CD8+ T cells. Analysis of individual chemokine ligand–receptor pairs revealed CXCL16–CXCR6 as the top-ranked pair between microglia and CD8+ T cells in 5xFAD mice (Fig. 2b, Extended Data Fig. 3c). Accordingly, Cxcl16 and Cxcr6 were highly expressed in microglia and T cell clusters, respectively (Fig. 2c). We found that CXCL16 expression was moderately increased in microglia from 5xFAD mice (Fig. 2d, Extended Data Fig. 3d). Further, Cxcl16 expression was elevated in DAM compared with homeostatic microglia (microglia 0 or M0)10 (Extended Data Fig. 3e), in line with the enrichment of chemotactic pathways in DAM (Extended Data Fig. 3f). Additionally, we observed stronger inferred CXCR6–CXCL16 interaction between CD8+ T cells and DAM than between CD8+ T cells and M0 (Extended Data Fig. 3g). This inter-cellular communication axis was also evident in APP/PS1 mice11, including the stronger communication between DAM and CD8+ T cells than between M0 and CD8+ T cells (Extended Data Fig. 3h, i). Collectively, these results indicate that CXCL16–CXCR6-mediated communication between microglia and CD8+ T cells exists in multiple AD mouse models.
Figure 2. CXCL16–CXCR6 inter-cellular communication axis and CXCR6-dependent accumulation of CD8+ T cells in AD.
a, Predicted cell–cell interactions based on expression of chemokine ligand–receptor pairs in scRNA-seq from 5xFAD mice. CD8, CD8+ T cells; NK, NK cells (NK); CD4, CD4+ T cells; γδ Τ, γδ T cells; Mast, mast cells. b, Bubble plot depicting ranked chemokine ligand–receptor interactions between microglia and CD8+ T cells, respectively. The numbers indicate rankings of the interactions, in descending order based on z score. c, Cxcl16 and Cxcr6 expression within microglia and CD8+ T cell clusters, respectively (indicated by dashed circles). d, CXCL16 expression in microglia (CD45int/+CD11b+), non-microglia immune cells (CD45+CD11b−), and non-immune cells (CD45−CD11b−) from 10-month-old Non-Tg (n = 4) and 5xFAD (n = 4) mice. MFI: mean fluorescence intensity. e, Heatmap of Aβ and CXCL16 protein expression in individuals with different neurodegenerative conditions22. LPC, control individuals with low pathology of plaques; HPC, control individuals with Aβ pathology but no obvious cognitive defects; PSP, individuals with progressive supranuclear palsy who accumulate tau proteins; MCI, individuals with mild cognitive impairment with Aβ plaques; AD, diagnosed AD with cognitive defects, high pathology scores of plaques and tangles. f, Relative abundance of CXCL16 protein in individuals with AD versus healthy individuals23. g, CXCL16 expression across cell types or whole-brain cortex isolated from human brains24. TPM: transcripts per million. h, CXCR6 expression on CD8+ T cells from spleens and brains of 10-month-old Non-Tg (n = 7) and 5xFAD (n = 10) mice. Data were analyzed by two-tailed unpaired Student’s t-test (d, h) or one-tailed t-test followed by Benjamini–Hochberg false discovery rate correction (f). Data are shown as mean ± s.e.m. in d, f, and h. Data were pooled from at least three (d, h) independent experiments or three independent studies (f).
Microglia activation or inflammatory status coincide with progressive accumulation of Aβ and cognitive decline in individuals with AD2. Associated with these effects, CXCL16 expression was upregulated in postmortem brain tissue from individuals with late-stage AD22 (Fig. 2e), which was also evident in a meta-analysis of three independent proteomic datasets23 (Fig. 2f). Analysis of a bulk RNA-seq dataset24 and a single-nuclear RNA-seq (snRNA-seq) dataset25 revealed that CXCL16 gene expression was upregulated predominantly in microglia (Fig. 2g, Extended Data Fig. 3j). Thus, AD-associated alterations of CXCL16 expression occur in humans, indicating conserved regulation of CXCL16 between mouse AD models and humans.
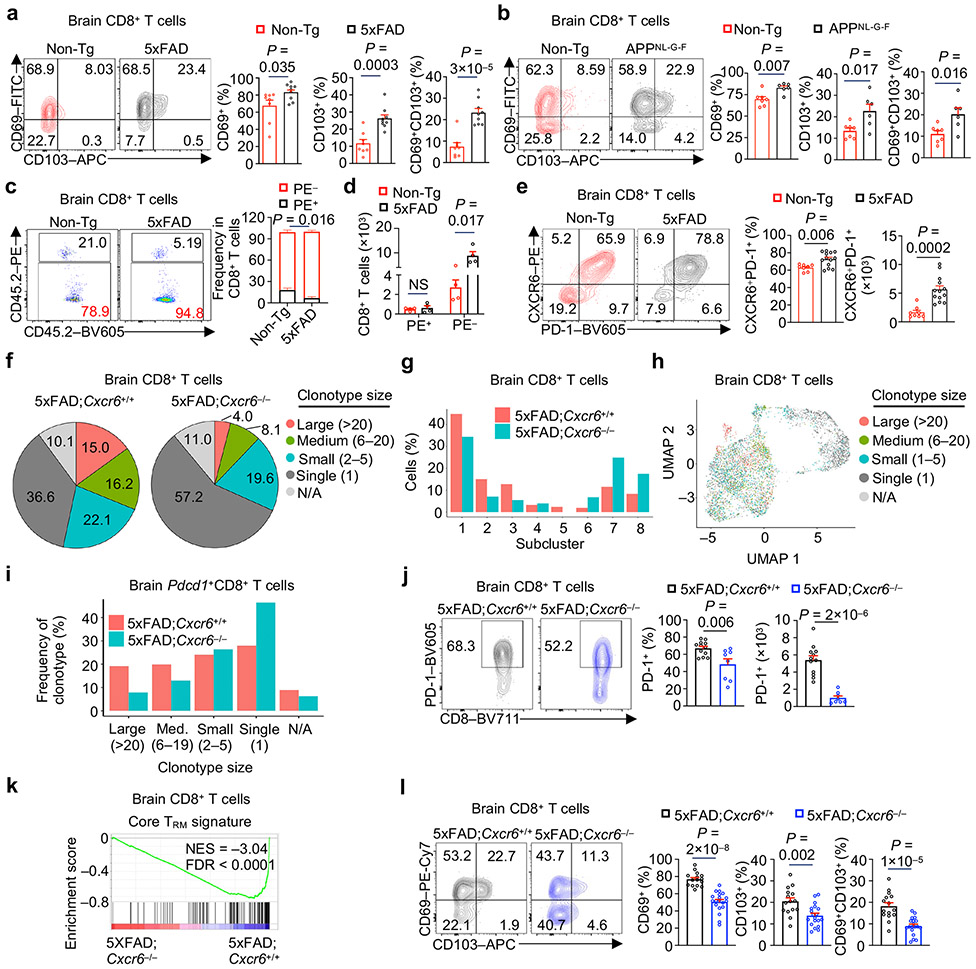
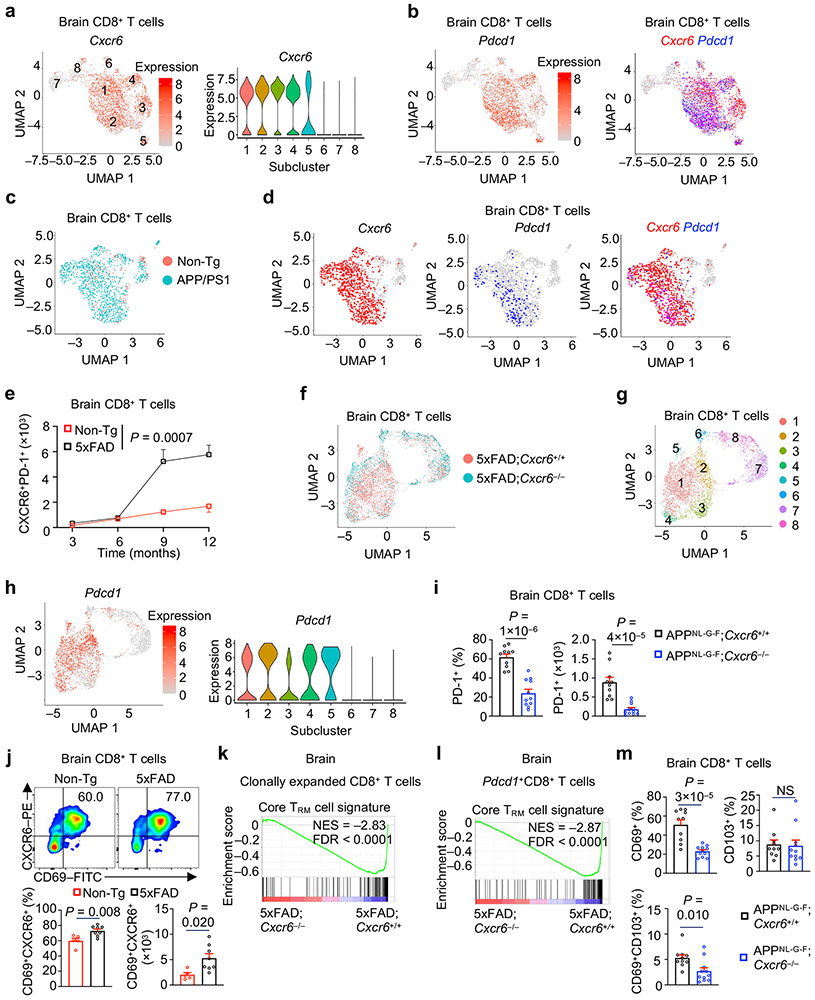
Next, we examined tissue-specific regulation and functional importance of CXCR6. A much larger proportion of CD8+ T cells expressed CXCR6 in the brain compared to the spleen, and the frequency and number of CXCR6+CD8+ T cells were increased in the brains (but not other tissues) of 5xFAD (Fig. 2h, Extended Data Fig. 3k). In contrast, an accumulation of CXCR6+CD4+ T cells was not observed (Extended Data Fig. 3l). Therefore, CXCR6 expression is upregulated in a tissue-specific manner on brain CD8+ T cells from 5xFAD mice.
Loss of CXCR6 exacerbates cognitive decline of 5xFAD mice
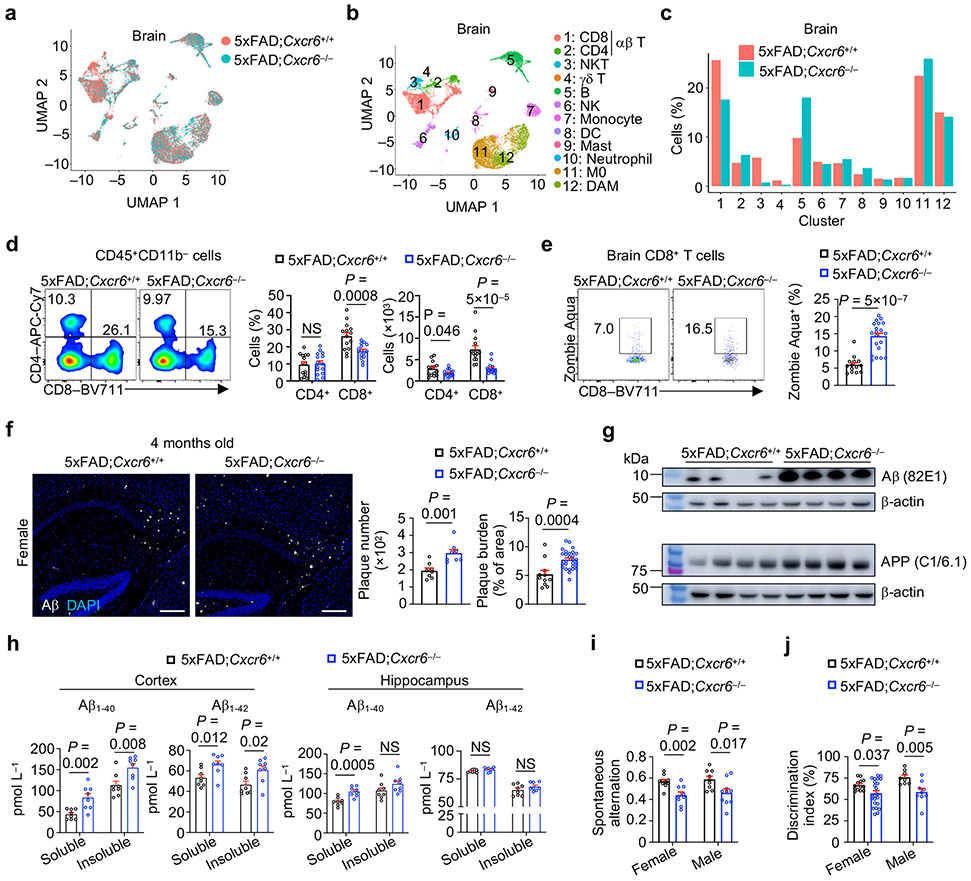
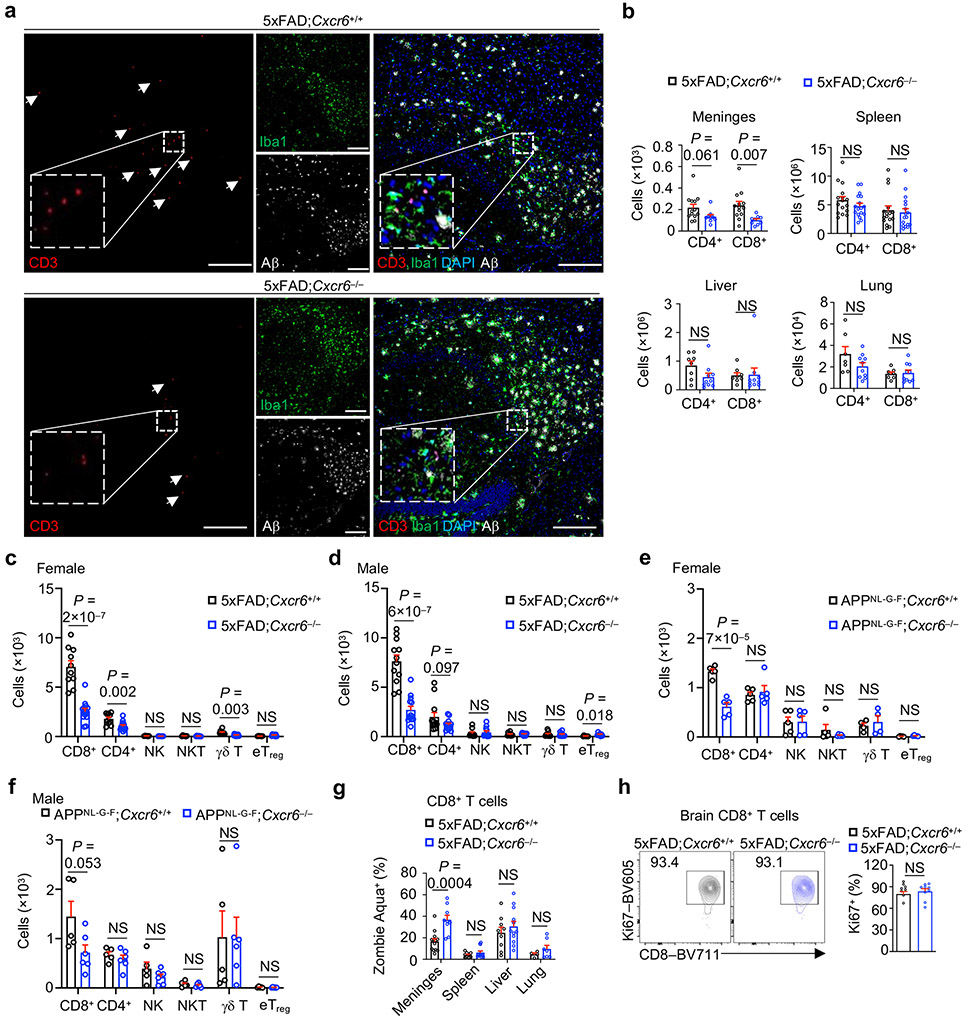
To test whether CXCR6 directs mouse AD-associated CD8+ T cell accumulation in the brain, we generated CXCR6-deficient 5xFAD mice (5xFAD;Cxcr6−/−) and compared their immune cell profiles in the brain with 5xFAD;Cxcr6+/+ mice (Fig. 3a, b). There was a reduction of brain CD8+ T cells or CD3+ T cells in 5xFAD;Cxcr6−/− mice (Fig. 3c, d, Extended Data Fig. 4a). This effect was specific to the CNS, as CD8+ T cells were reduced in the meninges but were largely unaltered in the spleen, liver, and lung from 5xFAD;Cxcr6−/− mice (Extended Data Fig. 4b). Further, compared to other CXCR6-expressing immune cells, CD8+ T cells had most pronounced reduction in the brains upon CXCR6 deletion in 5xFAD and APPNL-G-F mouse models (Extended Data Fig. 4c-f). Mechanistically, CD8+ T cells from the brain or meninges of 5xFAD;Cxcr6−/− mice displayed increased cell death but normal proliferative state (Fig. 3e, Extended Data Fig. 4g, h). Thus, CXCR6 is important for the accumulation and survival of CD8+ T cells in the brains of 5xFAD mice.
Figure 3. Loss of CXCR6 exacerbates cognitive decline of 5xFAD mice.
a, b, scRNA-seq analysis of microglia and non-microglia immune cells from the brains of 8-month-old 5xFAD;Cxcr6+/+ and 5xFAD;Cxcr6−/− mice (n = 2 biological replicates, pooled from 3 mice each per group). UMAP showing immune cell clusters colored by genotypes (a) or cell types (b). c, Cell frequencies (normalized to total cells from each genotype) in each cluster in b. d, Brain CD4+ and CD8+ T cells from 10-month-old 5xFAD;Cxcr6+/+ mice (n =14) and 5xFAD;Cxcr6−/− (n = 15) mice. e, Non-viable (Zombie-Aqua+) CD8+ T cells from the brains of 10-month-old 5xFAD;Cxcr6+/+ (n = 14) and 5xFAD;Cxcr6−/− (n = 20) mice. f, Immunohistochemical analysis of Aβ plaques in 4-month-old female 5xFAD;Cxcr6+/+ [n = 8 sections for plaque number; n = 11 sections for plaque burden] and 5xFAD;Cxcr6−/− [n = 8 sections for plaque number; n = 28 sections for plaque burden] mice. Number and burden of Aβ plaques were quantified. Scale bars, 250 μm. A serial section from the same control mouse is shown in Fig. 1g. g, Immunoblot analysis of Aβ and APP proteins in homogenized brain tissue from 4-month-old female 5xFAD;Cxcr6+/+ (n = 4) and 5xFAD;Cxcr6−/− (n = 5) mice. h, Soluble and insoluble fractions of Aβ1-40 and Aβ1-42 protein levels in hippocampus or cortex of 4-month-old female and male 5xFAD;Cxcr6+/+ (n = 8) and 5xFAD;Cxcr6−/− (n = 8) mice. i, Quantification of spontaneous alternation from Y maze testing in 4-month-old 5xFAD;Cxcr6+/+ (n = 10 for female and 9 for male) and 5xFAD;Cxcr6−/− (n = 9 for female and 10 for male) mice. j, Quantified discrimination index from novel objection recognition testing in 4-month-old 5xFAD;Cxcr6+/+ (n = 13 for female and 8 for male) and 5xFAD;Cxcr6−/− (n = 23 for female and 9 for male) mice. Data were analyzed by two-tailed unpaired Student’s t-test (d, e and f for plaque burden, h, i and j) or two-tailed unpaired t-test with Welch’s correction for plaque number (f). Data are shown as mean ± s.e.m. in d–f and h–j. Data were pooled from at least three (d, e, i, j), two (f), or one (h) independent experiments.
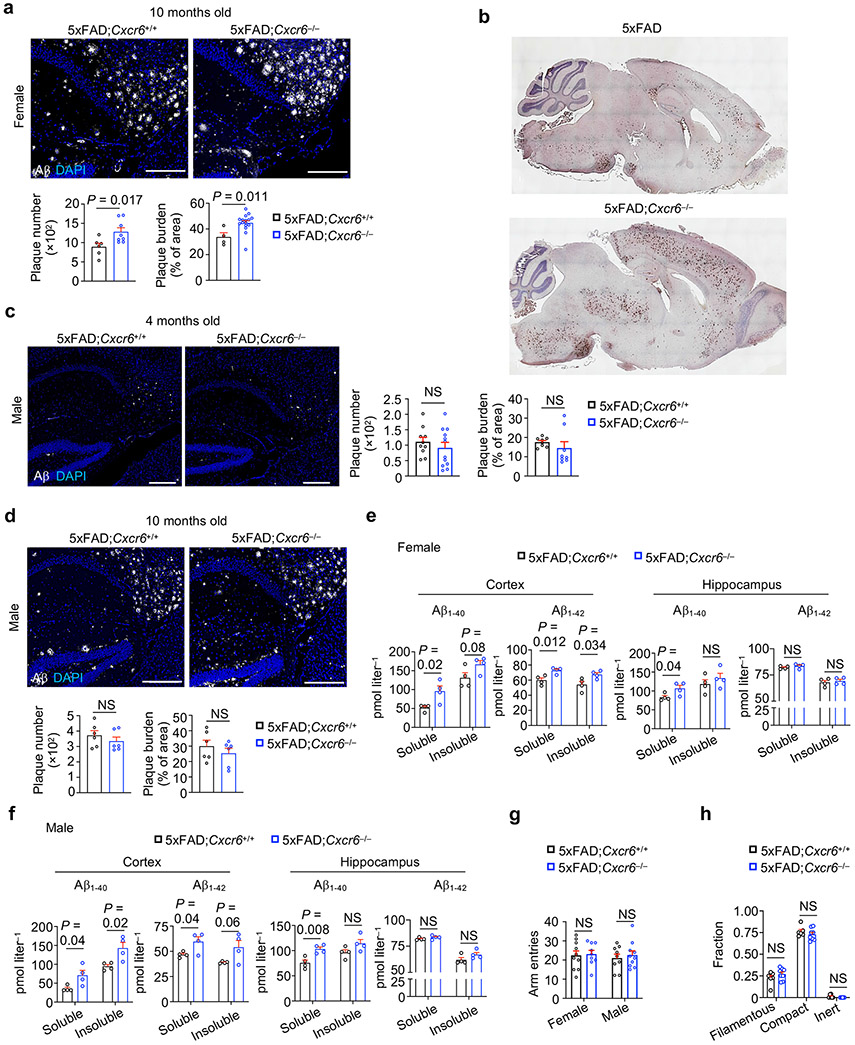
We next examined AD-associated pathology in 5xFAD;Cxcr6−/− mice. Female but not male 5xFAD;Cxcr6−/− mice had exacerbated Aβ deposits as analyzed by immunostaining (Fig. 3f, Extended Data Fig. 5a-d), which was also evident by immunoblot analysis of brain tissue homogenates (Fig. 3g). However, both female and male 5xFAD;Cxcr6−/− mice had increased concentrations of soluble and insoluble Aβ1-40 and Aβ1-42 levels in the cortex, and soluble Aβ1-40 in the hippocampus (Fig. 3h, Extended Data Fig. 5e, f). Moreover, both sexes of 5xFAD;Cxcr6−/− mice had impaired cognitive memory function (Fig. 3i, j), without alterations in locomotive abilities (Extended Data Fig. 5g). Finally, we examined whether CXCR6 deficiency impacts Aβ plaque phenotypes that are associated with differing degrees of neurotoxicity26, and found no differences in the frequencies of filamentous, compact, or inert Aβ plaque phenotypes (Extended Data Fig. 5h), suggesting that the enhanced AD pathology in these mice is associated with an increase in the overall quantities, but not phenotypes, of Aβ plaques. Together, CXCR6 deficiency is associated with the reduction in brain CD8+ T cells and exacerbated disease pathology in 5xFAD mice.
CXCR6 coordinates clonal expansion of brain CD8+ T cells
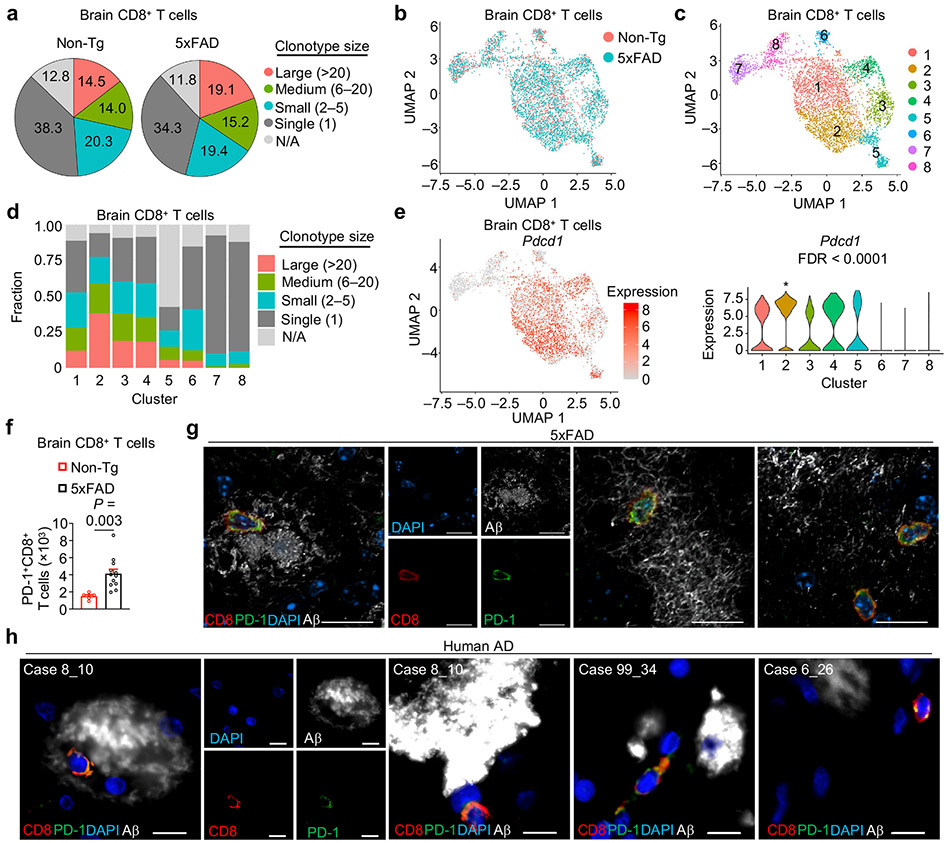
The phenotypic and functional properties of CD8+ T cells are impacted by immunological and local environmental signals. To dissect the molecular processes underlying CD8+ T cell accumulation in the brains of 5xFAD mice, we performed paired scRNA-seq with scTCR-seq analysis to identify clonally expanded T cell populations in the brain and meninges of 5xFAD and Non-Tg mice. Then, we integrated such TCR repertoire information with gene expression programs indicative of cellular phenotype and differentiation state. This analysis uncovered increased clonal expansion in the brain versus meninges, with T cells undergoing further clonal expansion in the brains of 5xFAD mice, especially among αβ+ T cells (Extended Data Fig. 6a, b). In particular, CD8+ T cells represented the majority of clonally expanded (defined as clonotype size > 1) T cells (Extended Data Fig. 6c, d). Moreover, highly expanded CD8+ T cell clones, especially those with clonotype size > 20, accounted for a larger fraction of total brain CD8+ T cells in 5xFAD mice (Fig. 4a). Using GLIPH27 to reveal the top 5 CDR3b patterns followed by TCRmatch28 analysis, we uncovered insulin-2, nucleoprotein of LCMV, and human herpes virus as possible epitopes that these clonally expanded cells may recognize (Extended Data Fig. 6e, f).
Figure 4. CXCR6 coordinates clonal expansion of brain CD8+ T cells.
a, Pie chart summarizing the percentages of brain CD8+ T cell clonotype sizes in age-matched Non-Tg and 5xFAD mice. N/A: not available. b, c, UMAP plots of brain CD8+ T cell subclusters colored by genotype (b) or subcluster (c). d, Bar plot showing the distribution of clonotype sizes for CD8+ T cell clusters shown in c [Clonotype size: Large (> 20); Medium (6–20); Small (2–5); and Single (1)]. e, Pdcd1 expression in brain CD8+ T cells shown on UMAP and violin plots in discrete subclusters. The asterisk labels significance in indicated subcluster (n = 1,199 cells) compared to all other subclusters (n = 3,442 cells). f, Number of PD-1+CD8+ T cells in the brains of 10-month-old Non-Tg (n = 6) and 5xFAD (n = 12) mice. g, Three separate imaging fields depicting brain CD8+ T cells (red) that co-express PD-1 (green) and are in proximity to β-amyloid (Aβ; white) in 12-month-old 5xFAD mice. Scale bars, 20 μm. h, Four separate imaging fields depicting brain CD8+ T cells (red) that co-express PD-1 (green) and are in proximity to Aβ (white). Case 8_10, Case 99_34, and Case 6_26 denote individual donors. Scale bars, 10 μm. Data were analyzed by two-tailed unpaired Student’s t-test (f) or two-tailed Wilcoxon rank-sum test (e). Data are shown as mean ± s.e.m. in f. Data were pooled from three (f) or are representative of three (g) independent experiments or are depicting images obtained from three separate donors (h).
To identify putative molecules that could mark clonally expanded brain CD8+ T cells, we calculated the Pearson correlation co-efficient between expression of individual genes and clonotype frequency29, leading to the nomination of Pdcd1 (encodes for PD-1) as the top-ranking gene to correlate with clonally expanded cells (Extended Data Fig. 6g). Subclustering of brain CD8+ T cells identified eight subclusters (Fig. 4b, c), distinguished by discrete gene signatures (Extended Data Fig. 6h). Many of these subclusters contained expanded clonotypes marked by high Pdcd1 expression, especially subcluster 2 (Fig. 4d, e). Also, an increased frequency of large clonotypes (clonotype size > 20) was observed in subcluster 2 from 5xFAD compared to Non-Tg mice (Extended Data Fig. 6i). Accordingly, we noted a strong positive correlation (r = 0.82) between the transcriptomes of clonally-expanded (versus non-expanded) and Pdcd1+ (versus Pdcd1−) cells (Extended Data Fig. 6j). Further, brain PD-1+CD8+ T cells were markedly increased in 5xFAD mice (Fig. 4f), further supporting disease-associated clonal expansion in the brain (Fig. 4a). Additionally, imaging analysis showed co-expression of CD8 and PD-1 in the brains of 5xFAD mice and individuals with AD (Fig. 4g, h). Together, these results indicate that PD-1 upregulation occurs on CD8+ T cells from the brains of 5xFAD mice and that PD-1 expression largely distinguishes clonally expanded versus non-expanded CD8+ T cells.
CXCR6 programs tissue residency in brain CD8+ T cells
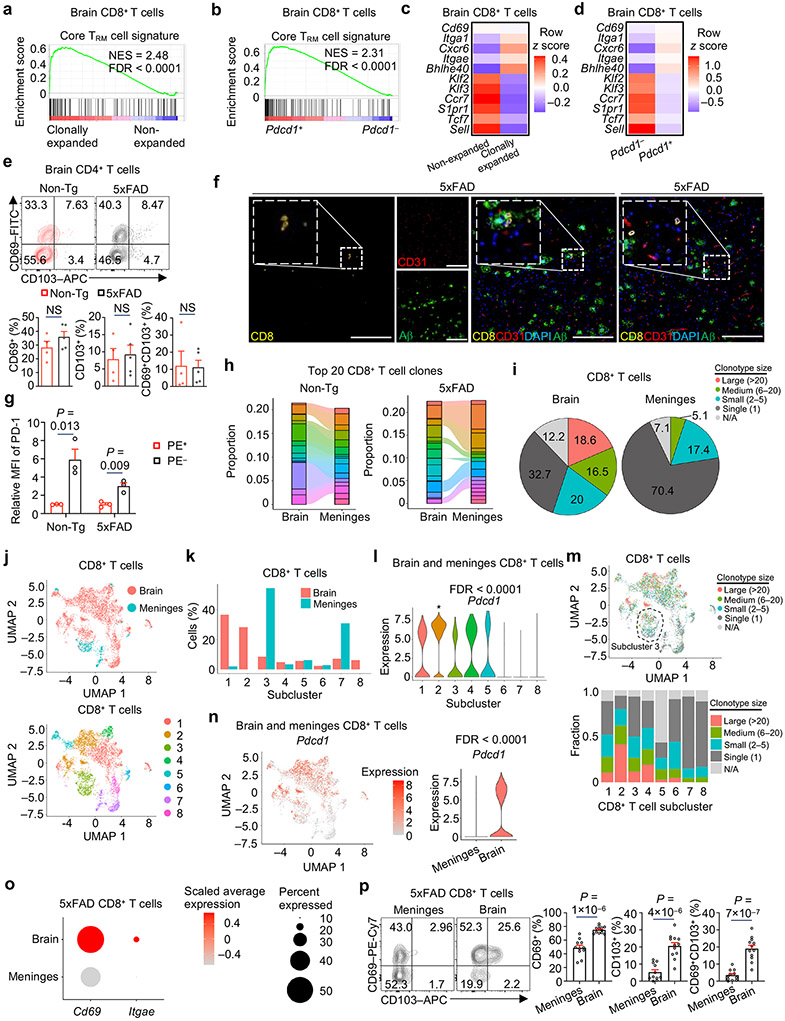
GSEA showed that a core tissue-resident memory (TRM) signature30 was enriched in clonally expanded CD8+ or Pdcd1+CD8+ T cells (Extended Data Fig. 7a, b), associated with an upregulation of TRM-associated marker genes (e.g., Cd69, Cxcr6 and Itgae, which encodes for CD103)30,31 (Extended Data Fig. 7c, d). Accordingly, brain CD8+ but not CD4+ T cells from 5xFAD or APPNL-G-F mice14 mice showed increased expression of TRM markers CD69 and CD103 compared to Non-Tg mice (Fig. 5a, b, Extended Data Fig. 7e), suggesting an enhanced TRM phenotype in brain CD8+ T cells under disease conditions. Consistent with this notion, immunostaining showed that the majority of brain CD8+ T cells were not within the vasculature (Extended Data Fig. 7f). Also, upon intravenous injection of mice with anti-CD45 PE-conjugated antibody32 to assess whether brain CD8+ T cells are derived from circulation (PE+) or resident to the brain (PE−), we found that the majority of brain CD8+ T cells were PE− cells (Fig. 5c), marked by elevated expression of PD-1 relative to PE+ cells (Extended Data Fig. 7g). Moreover, these PE−CD8+ T cells were increased in the brains of 5xFAD compared to Non-Tg mice (Fig. 5c, d). Therefore, the PD-1+CD8+ T cells that accumulate in 5xFAD mice are resident to the brain.
Figure 5. CXCR6 programs tissue residency of brain CD8+ T cells.
a, b, CD69+, CD103+, and CD69+CD103+ cells among brain CD8+ T cells from 10-month-old Non-Tg (n = 8) and 5xFAD (n = 9) mice (a) and 9-month-old Non-Tg (n = 7) and APPNL-G-F (n = 6) mice (b). CD69+ (%) graphs indicate CD69+CD103− plus CD69+CD103+ cells. CD103+ (%) graphs indicate CD69−CD103+ plus CD69+CD103+ cells. c, d, Frequencies (c) and numbers (d) of PE+ circulating and PE− tissue-resident CD8+ T cells in the brains of 10-month-old Non-Tg (n = 4) and 5xFAD (n = 4) mice. e, Brain CXCR6+PD-1+CD8+ T cells in 10-month-old Non-Tg (n = 8) and 5xFAD (n = 13) mice. f, Pie chart summarizing the percentages (normalized to total cells from each genotype) of brain CD8+ T cell clonotype sizes in 5xFAD;Cxcr6+/+ and 5xFAD;Cxcr6−/− mice. g, Frequencies (normalized to total cells from each genotype) of brain CD8+ T cells in each subcluster shown in Extended Data Fig. 8g. h, Clonotypes of brain CD8+ T cells colored by clonotype sizes (N/A: not available). i, Frequencies (normalized to total cells from each genotype) of clonotype sizes for Pdcd1+CD8+ T cells (subclusters 1–5 in Extended Data Fig. 8h). j, Brain PD-1+CD8+ T cells from 10-month-old 5xFAD;Cxcr6+/+ (n = 12) and 5xFAD;Cxcr6−/− (n = 8) mice. k, GSEA enrichment plot showing downregulation of core tissue-resident memory (TRM) signature30 in brain CD8+ T cells from 5xFAD;Cxcr6−/− mice versus 5xFAD;Cxcr6+/+ mice. NES, normalized enrichment score; FDR, false discovery rate. l, CD69+, CD103+, and CD69+CD103+ cells among brain CD8+ T cells isolated from 10-month-old 5xFAD;Cxcr6+/+ (n = 15) and 5xFAD;Cxcr6−/− (n = 19) mice. Data were analyzed by two-tailed unpaired Student’s t-test (a–e, j, l), two-tailed normalized weighted Kolmogorov–Smirnov test (NES for k), or two-tailed Benjamini–Hochberg adjusted P value test (FDR for k). Data are shown as mean ± s.e.m. in a–e, j and l. Data were pooled from at least three (a, b, e, j, l) or two (c, d) independent experiments.
Next, we performed pairwise comparison of the brain and meninges from individual mice, and found unique clonotypes of CD8+ T cells in the brain versus meninges of 5xFAD but not Non-Tg mice (Extended Data Fig. 7h). Further, the largest clonotype size (> 20), and to a lesser extent, medium clonotype size (6–20), were mainly responsible for the increased clonal expansion in the brain compared to the meninges (Extended Data Fig. 7i). To compare CD8+ T cell heterogeneity between the brain and meninges, we integrated cluster analysis of brain- and meninges-derived cells and identified preferential, or equal, contributions from each compartment (Extended Data Fig. 7j, k). Specifically, among subclusters 1–5 characterized by high expression of Pdcd1 (Extended Data Fig. 7l), the frequencies of subclusters 1, 2, and 4 were increased in the brain, while the frequency of subcluster 3 was elevated in the meninges (Extended Data Fig. 7k). Further, subclusters 1–4 contained higher frequencies of clonally expanded CD8+ T cells, followed by subclusters 5 and 6, while limited clonal expansion was observed in subclusters 7 and 8 (Extended Data Fig. 7m). Notably, subclusters (1, 2, and 4) with strong clonal expansion were composed mainly of brain-derived cells, and brain-derived cells were also mostly responsible for the clonal expansion in cluster 3 that contained a high frequency of meninges-derived cells (Extended Data Fig. 7j, k, m). Further, Pdcd1 expression was higher in brain than meningeal CD8+ T cells (Extended Data Fig. 7n). Moreover, expression of Cd69 and Itgae was increased in brain compared to meningeal CD8+ T cells from 5xFAD mice (Extended Data Fig. 7o). Accordingly, the proportion of CD8+ T cells expressing CD69 or CD103 alone or in combination was increased in the brain compared to the meninges in 5xFAD mice (Extended Data Fig. 7p). Thus, CD8+ T cell clonal expansion occurs more preferentially in the brain than meninges, thereby supporting a CNS compartment-specific and disease-associated clonal expansion of CD8+ T cells with a TRM phenotype.
We next examined the interplay between CXCR6 and PD-1 in brain CD8+ T cells, and the role of CXCR6 in tissue residency programming. Cxcr6 expression was evident in the clonally expanded, Pdcd1-expressing subclusters in the brains of 5xFAD or APP/PS1 mice11 (Extended Data Fig. 8a-d), supporting a conserved effect across different mouse models of AD. We validated the co-expression of these molecules on brain CD8+ T cells, with the frequency and number of CXCR6+PD-1+CD8+ T cells showing an increase in 5xFAD compared to Non-Tg mice (Fig. 5e), especially after 6 months of age (Extended Data Fig. 8e). We next applied scTCR-seq analysis to determine whether CXCR6 orchestrates the clonal expansion of the brain CD8+ T cells in 5xFAD mice, and found a marked reduction of clonally expanded CD8+ T cells upon CXCR6 deletion (Fig. 5f). CXCR6 deletion also reduced the proportions of Pdcd1-expressing subclusters (1–5) among total CD8+ T cells (Fig. 5g, Extended Data Fig. 8f-h). Further, clonally expanded CD8+ T cells (Fig. 5h), which largely overlapped with Pdcd1 expression (Extended Data Fig. 8f-h), showed a marked reduction of clonotypes with large- and medium-level expansion in the absence of CXCR6 (Fig. 5i). Accordingly, PD-1+CD8+ T cells were reduced in 5xFAD;Cxcr6−/− or APPNL-G-F;Cxcr6−/− mice (Fig. 5j, Extended Data Fig. 8i), thereby indicating an essential role of CXCR6 for the generation of clonally expanded PD-1+CD8+ T cells in the brains of mouse models of AD.
In addition to clonal expansion, we hypothesized that CXCR6 is important for TRM programming of brain CD8+ T cells in 5xFAD mice. Indeed, brain CD8+ T cells co-expressed the TRM-signature marker CD69 with CXCR6, and CD69+CXCR6+CD8+ T cells were accumulated in the brains of 5xFAD mice (Extended Data Fig. 8j). More importantly, the core TRM signature was downregulated in total, clonally-expanded, or Pdcd1+ CD8+ T cells from 5xFAD;Cxcr6−/− mice (Fig. 5k, Extended Data Fig. 8k, l). Further, 5xFAD;Cxcr6−/− and APPNL-G-F;Cxcr6−/− mice had decreased proportions of CD69+, CD103+, and CD69+CD103+ subpopulations of CD8+ T cells (Fig. 5l, Extended Data Fig. 8m). These results collectively indicate that CXCR6 is essential for TRM programming in brain CD8+ T cells in the context of mouse AD, and reveal a crucial role of CXCR6 in linking clonal expansion and tissue residency programs of brain CD8+ T cells.
CD8+ T cells restrain pro-inflammatory activity of microglia
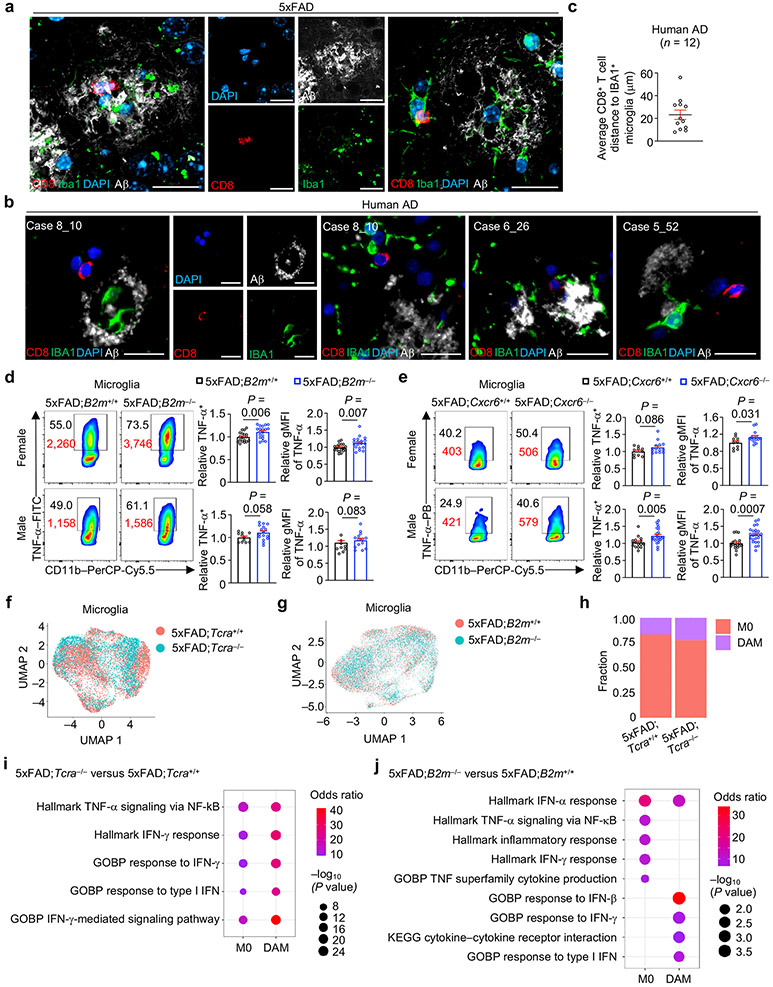
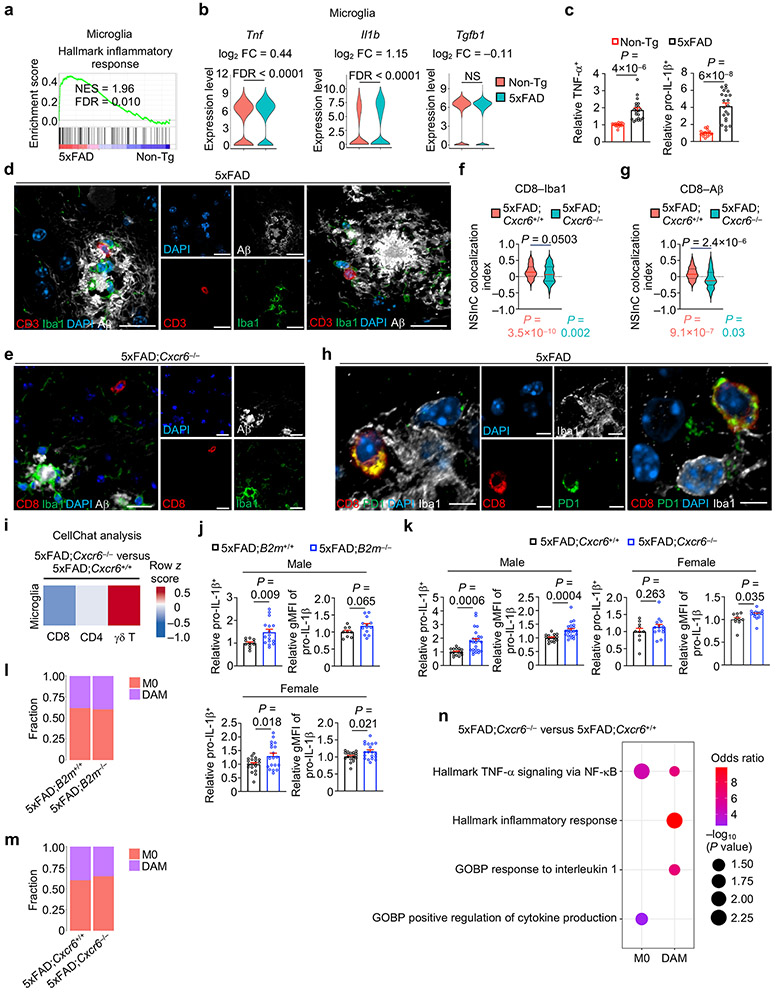
In accordance with their neuroinflammatory properties in AD1,2, we found that microglia from 5xFAD mice were enriched for inflammatory gene signatures and expression of pro-inflammatory cytokines Tnf (encodes for tumor necrosis factor-α (TNF-α)) and Il1b (encodes for interleukin-1β (IL-1β)) without alteration of anti-inflammatory Tgfb1 (Extended Data Fig. 9a-c). Consistent with our ligand–receptor analysis described above, high-resolution confocal imaging revealed co-localization of CD8+ T cells or CD3+ T cells with Iba1+ microglia near Aβ plaques in 5xFAD mice (Fig. 6a, Extended Data Fig. 9d). Such close proximity of CD8+ T cells with Aβ and IBA1+ microglia was also observed in the brains of individuals with AD (Fig. 6b, c). Thus, the co-localization of CD8+ T cells with microglia or Aβ is a conserved phenomenon in mouse models and humans3,33. To quantitatively examine spatial co-localization between CD8+ T cells and microglia or Aβ and the dependence on CXCR6, we performed normalized spatial intensity correlation (NSInC) analysis34. We observed a significant co-localization of CD8 with Iba1 or Aβ plaques in the brains of 5xFAD;Cxcr6+/+ mice, which was reduced in 5xFAD;Cxcr6−/− mice (Extended Data Fig. 9e-g). Similarly, CD8+ T cells expressing PD-1, which serves as a surrogate for CXCR6+CD8+ T cells (Fig. 5e), were also adjacent to microglia in brains of 5xFAD mice (Extended Data Fig. 9h). Moreover, 5xFAD;Cxcr6−/− mice showed reduced microglia and CD8+ T cell interaction strength than 5xFAD;Cxcr6+/+ mice based on CellChat analysis35 (Extended Data Fig. 9i). Altogether, our results reveal that CXCR6 contributes to spatial co-localization of CD8+ T cells with microglia and Aβ in the brain, thereby further supporting the inter-cellular communication between these cells and the CXCR6 dependence.
Figure 6. CD8+ T cells co-localize with microglia in AD and restrain pro-inflammatory activity of microglia in 5xFAD mice.
a, Three separate imaging fields depicting brain CD8+ T cells (red) in proximity to Aβ (white) and Iba1+ microglia (green) in 10-month-old 5xFAD mice. Scale bars, 20 μm. b, Four separate imaging fields depicting brain CD8+ T cells (red) in proximity to Aβ (white) and IBA1+ microglia (green) in individuals with AD. Case 8_10, Case 6_26, and Case 5_52 denote individual donors. Scale bars, 20 μm. c, Distance between an individual CD8+ T cell and microglia in the brains of individuals with AD (n = 12 individual CD8+ T cells from imaging analysis of four individuals). d, e, Relative percentage (normalized to sex-matched controls) of TNF-α+ microglia (shown in black color text) and geometric mean fluorescence intensity (gMFI; shown in red color text) from 10-month-old 5xFAD;B2m+/+ (percentage, n = 19 for female and 11 for male; gMFI, n = 19 for female and 9 for male) and 5xFAD;B2m−/− (percentage, n = 19 for female and 16 for male; gMFI, n = 19 for female and 13 for male) mice (d), or 5xFAD;Cxcr6+/+ (percentage, n = 10 for female and 16 for male; gMFI, n = 10 for female and 18 for male) and 5xFAD;Cxcr6−/− (n = 14 for female and 24 for male) mice (e). f–h, scRNA-seq analysis of CD45int/+CD11b+ microglia cells from the brains of indicated mice (see Methods). UMAPs shows cells separated by genotype (f, g), and the frequencies of microglia subclusters are presented (h). i, j, Functional enrichment analysis of upregulated genes in M0 and DAM from scRNA-seq analysis of indicated mice with the top enriched pathways shown. IFN, interferon; GOBP, Gene Ontology biological process. Data were analyzed by two-tailed unpaired Student’s t-test (d, e) or two-tailed Fisher’s exact test (i, j). Data are shown as mean ± s.e.m. in c–e. Data were pooled from at least three (d, e) or are representative of two (a) independent experiments. Human data are depicting images obtained from three separate donors (b) or quantified from imaging experiments of four separate donors (c).
Given this spatial proximity, we next tested the contribution of CD8+ T cell accumulation to microglial inflammatory status in vivo by analyzing microglia from 5xFAD;B2m−/− and 5xFAD;Cxcr6−/− mice. Microglia from 5xFAD;B2m−/− and 5xFAD;Cxcr6−/− mice showed an increase or a trending increase in TNF-α and pro-IL-1β expression (Fig. 6d, e, Extended Data Fig. 9j, k), indicative of an increased microglial inflammatory state in these mice. Additionally, we performed scRNA-seq analysis of microglia from 5xFAD;Tcra+/+ and 5xFAD;Tcra−/−and 5xFAD;B2m+/+ and 5xFAD;B2m−/− mice (Fig. 6f, g). Despite largely normal proportions of M0 and DAM cells (identified by DAM signature genes10; Fig. 6h, Extended Data Fig. 9l), we found enrichment of interferon signatures in both microglia subsets from 5xFAD;Tcra−/− and 5xFAD;B2m−/− mice (Fig. 6i, j). Further, TNF-α signaling signatures were enriched in both microglia subsets from 5xFAD;Tcra−/− mice and in M0 microglia from 5xFAD;B2m−/− mice (Fig. 6i, j). Finally, while the proportion of DAM was largely unaltered in 5xFAD;Cxcr6−/− mice (Extended Data Fig. 9m), inflammatory and TNF-α signaling signatures were modestly enriched in microglia subsets from 5xFAD;Cxcr6−/− mice (Extended Data Fig. 9n). Overall, these results suggest that the absence or reduction of brain CD8+ T cells results in an elevated inflammatory state of microglia.
Brain CD8+ T cells with regulatory features in 5xFAD mice
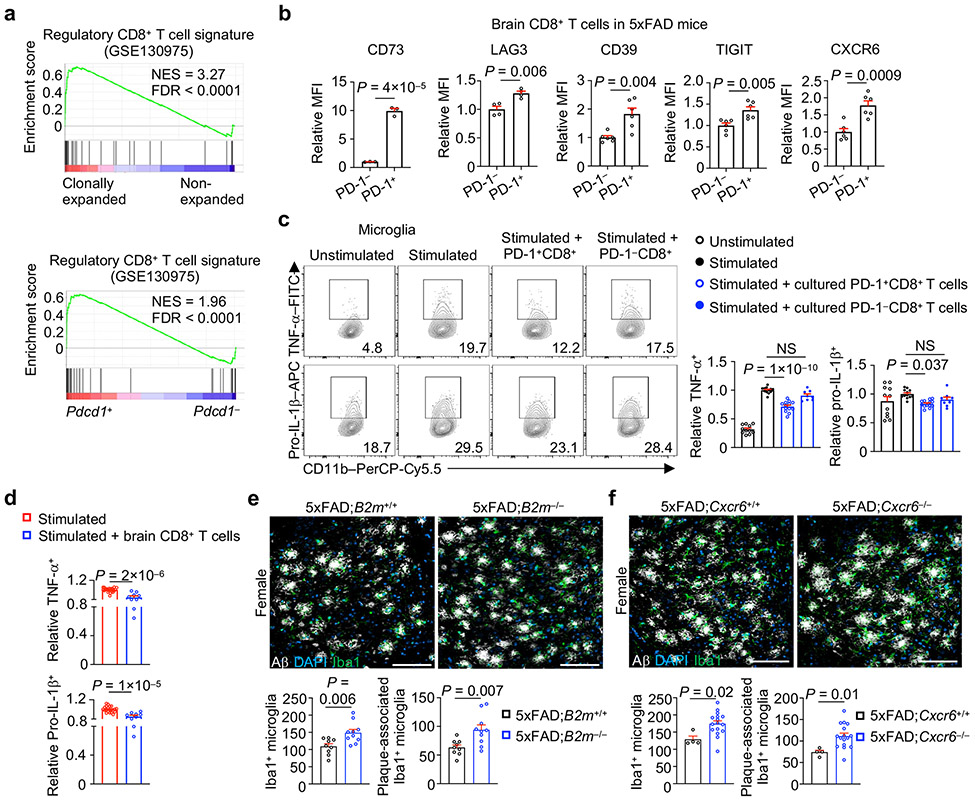
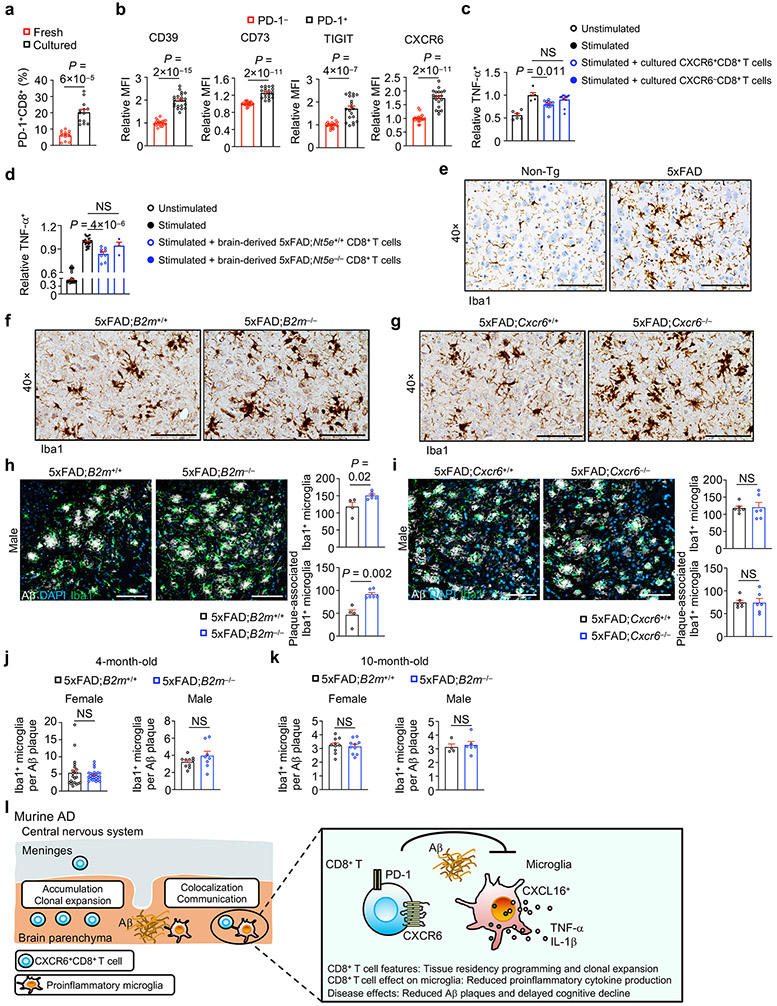
Regulatory CD8+ T cells with immunosuppressive functions exist in the diseased CNS36, and we found that clonally expanded and Pdcd1+ CD8+ T cells showed enrichment of this regulatory CD8+ T cell signature (Fig. 7a), associated with increased expression of suppression-associated molecules CD73, LAG3, CD39 and TIGIT together with CXCR6 (Fig. 7b). To directly test the whether CD8+ T cells exert suppressive effects on microglia, we adopted an in vitro system37,38 to generate PD-1+CD8+ T cells from splenic CD44hiCD8+ T cells (Extended Data Fig. 10a) and performed co-culture with primary microglia. Expression of TNF-α and pro-IL-1β in stimulated microglia was reduced in the presence of PD-1+CD44hiCD8+ T cells (Fig. 7c), in line with the increased expression of immunosuppressive markers (as well as CXCR6) on the cultured PD-1+CD44hiCD8+ T cells (Extended Data Fig. 10b). Similarly, CXCR6+CD8+ CD8+ T cells showed such suppressive effects (Extended Data Fig. 10c). Brain-derived CD8+ T cells from 5xFAD mice also inhibited pro-inflammatory cytokine production by microglia (Fig. 7d). However, brain CD8+ T cells from CD73-deficient 5xFAD (5xFAD;Nt5e−/−) mice were modestly defective for suppressing microglial pro-inflammatory cytokine production in vitro (Extended Data Fig. 10d), suggesting that CD73 may partially contribute to the suppressive mechanism of brain CD8+ T cells. Thus, PD-1+CD8+ T cells suppress production of pro-inflammatory cytokines in microglia.
Figure 7. Clonally expanded brain CD8+ T cells show regulatory gene expression and function on microglia in 5xFAD mice.
a, GSEA enrichment plots showing upregulation of a regulatory CD8+ T cell signature36 in clonally expanded versus non-expanded CD8+ T cells and Pdcd1+ versus Pdcd1− CD8+ T cells. NES, normalized enrichment score; FDR, false discovery rate. b, Relative expression (compared to PD-1−CD8+ T cells) of the indicated markers on PD-1+ versus PD-1− CD8+ T cells in the brains of 5xFAD mice (n = 3, 4, 6, 6, 6 for CD73, LAG3, CD39, TIGIT, CXCR6, respectively). MFI, mean fluorescence intensity. c, TNF-α+ and pro-IL-1β+ cells in microglia after in vitro co-culture with indicated in vitro-expanded, spleen-derived CD8+ T cell populations (see Methods for details). Stimulated condition indicates treatment with PMA and ionomycin plus 1 μM Aβ [Unstimulated (n = 11); Stimulated (n = 12); Stimulated + PD-1+CD8+ (n = 13); Stimulated + PD-1−CD8+ (n = 8)]. d, TNF-α+ and pro-IL-1β+ in microglia from 5xFAD mice upon their co-culture alone [Stimulated (n = 21)] or co-culture with freshly isolated brain CD8+ T cells from 5xFAD mice [Stimulated + brain CD8+ T cells (n = 9)]. e, f, Iba1+ microglia or Aβ-plaque-associated Iba1+ microglia from 10-month-old female 5xFAD;B2m+/+ (n = 9 sections) and 5xFAD;B2m−/− (n = 10 sections) mice (e), or female 5xFAD;Cxcr6+/+ (n = 4 sections) and 5xFAD;Cxcr6−/− (n = 16 sections) mice (f). One or two sections per mouse were quantified. Scale bars, 100 μm. Data were analyzed by two-tailed normalized weighted Kolmogorov–Smirnov test (NES for a), two-tailed Benjamini–Hochberg adjusted P value (FDR for a), or two-tailed unpaired Student’s t-test (b, d–f) or one-way ANOVA (c). Data are shown as mean ± s.e.m. in b–f. Data were pooled from at least three (c, d) or two (b, e, f) independent experiments.
Microgliosis occurs in mouse models and individuals with AD39,40. Accordingly, an activated amoeboid-like morphology of Iba1+ microglia was observed in 5xFAD mice (Extended Data Fig. 10e), which was further exacerbated in 5xFAD;B2m−/− and 5xFAD;Cxcr6−/− mice (Extended Data Fig. 10f, g). Further, we observed an accumulation of Iba1+ microglia and increased plaque-associated microglia in female and male 5xFAD;B2m−/− mice (Fig. 7e, Extended Data Fig. 10h), as well as female 5xFAD;Cxcr6−/− mice (Fig. 7f, Extended Data Fig. 10i), although there were comparable numbers of Iba1+ microglia per individual Aβ plaque (Extended Data Fig. 10j, k), indicative of progressive gliosis that coincides with plaque formation41,42. Collectively, these data suggest that CD8+ T cells act as a cellular brake to limit pro-inflammatory microgliosis (Extended Data Fig. 10l).
Discussion
Training of the innate immune system is an emerging concept in immunology43, with recent results showing that the adaptive immune system impacts microglial function under steady state32 and in neurodegenerative diseases5,6. However, the molecular basis of the inter-cellular communication in the brain microenvironment that shapes the homeostasis and functionality of microglia, especially the key signals that restrict their pro-inflammatory programs that can exacerbate neuroinflammatory pathologies, is largely unknown. Here, we reveal an accumulation of clonally expanded, PD-1+CD8+ T cells in the brains of mouse models of AD, which serve a key protective role to limit Aβ aggregation and cognitive memory impairment. This population is also essential to restrain the pro-inflammatory activity of microglia that is linked with neuroinflammation, increased Aβ burden, and AD pathogenesis2,44. We identify CXCR6 as a critical coordinator of brain CD8+ T cell accumulation, clonal expansion, tissue residency programming, and co-localization with microglia, associated with CD8+ T cell immunoregulatory function. Microglia cells in 5xFAD mice with a deficiency or impairment in brain CD8+ T cell accumulation show increased expression of pro-inflammatory cytokines and gene signatures, therefore suggesting a previously unrecognized inhibitory role of CD8+ T cells in impacting the localized immune training of microglia.
The composition of immune cell populations underlies tissue homeostasis or pathology in many diseases45, although the adaptive immune cell alterations in neurodegenerative diseases are underexplored. We found a disease- and age-associated accumulation of brain CD8+ T cells in 5xFAD mice, similar to other findings in the APP/PS1 amyloidosis15,46 and tauopathy8 mouse models of AD. Moreover, we ascribe this accumulation to two intertwined cellular mechanisms: clonal expansion (marked by expression of PD-1) and tissue residency programming (marked by expression of CD103 and CD69), with such observations largely consistent with analyses performed in other mouse models of AD and in individuals with AD3,8,46,47. More importantly, we demonstrate a tissue-specific dependence of both mechanisms on CXCR6, thereby providing key mechanistic insight governing the poorly understood regulation of TRM cells in neurodegenerative diseases. Associated with impaired clonal expansion and TRM programming, 5xFAD;Cxcr6−/− mice display enhanced disease pathologies, including increased Aβ burden, cognitive memory defects and pro-inflammatory microgliosis. Interestingly, γδ T cells are recruited to the meninges in a CXCR6-dependent manner shortly after birth and regulate anxiety-like behavior48, supporting the notion that CXCR6 is a central orchestrator of immune cell-mediated regulation of CNS homeostasis. Future studies are warranted to determine whether CXCR6 also affects CD8+ T cell accumulation in the CNS during other disease states, including tauopathy8.
Cell–cell interactions dictate functional immune responses under homeostasis and inflammation, yet how cells communicate in neurodegenerative diseases is poorly understood. We unbiasedly identify a CXCR6–CXCL16 interaction bridging CD8+ T cells and microglia in the brains of 5xFAD mice. Accordingly, CXCR6+CD8+ T cells and CXCL16+ microglia cells were increased in mouse AD brains, and CXCL16 expression was elevated in individuals with AD22-24. We also revealed a close proximity between PD-1+CD8+ T cells and IBA1+ activated microglia that were adjacent to Aβ plaques in the brains of 5xFAD mice and individuals with AD, consistent with recent reports3,8,33. Functionally, this spatial co-localization of CD8+ T cells and microglia was disrupted in 5xFAD;Cxcr6−/− mice, demonstrating the dependence on CXCR6 for CD8+ T cell–microglia interactions. Given the highly selective and conserved nature of the CXCL16–CXCR6 axis37,38,48, we propose that this bidirectional interaction between microglia and CD8+ T cells is clinically relevant, as it underlies a dynamic regulation of adaptive immunity and tissue homeostasis associated with AD pathogenesis.
Although innate immune cells, especially microglia, contribute to AD pathogenesis1,2, the role of the adaptive immune system in AD has remained controversial5-8. Indeed, 5xFAD mice lacking all T cells, B cells, NK cells, and innate lymphoid cells show enhanced disease burden5. This study also showed that B cell-derived antibodies to Aβ protect mice from cerebral amyloidosis by regulating the phagocytic activity of microglia5, although a recent study revealed a deleterious role for B cells7. By contrast, regulatory T cell depletion6 and anti-PD-1 treatment49 in 5xFAD mice cause activation of the immune system and decrease disease burden, suggesting that selective T cell populations are protective in AD, although the functional effects of discrete T cell subsets remain poorly defined. In this study, we establish a protective role for brain-resident CD8+ T cells with suppressive features, resembling those observed in viral and autoimmune contexts36,50, in restraining pro-inflammatory microgliosis as well as cerebral amyloidosis and cognitive decline. Of note, CD8+ T cells play a pathogenic role in disease development in a mouse model of AD tauopathy8. It is possible that Aβ and tau pathologies are associated with the accumulation of discrete CD8+ T cell populations with unique functions in the brain. Specifically, our data show that PD-1+CD8+ T cells with a suppressive phenotype accumulate in 5xFAD mice characterized by amyloidosis, with such an effect also observed in individuals with AD. In the tauopathy model, there is a decline in PD-1-expressing CD8+ T cells and an accumulation of activated CD8+ T cells that express pro-inflammatory factors like IFN-γ8. These complementary studies highlight the complex and context-dependent roles of the adaptive immune system in AD pathogenesis. Putative CD8+ T cell-targeted therapies may need to be fine-tuned to account for inflammatory versus regulatory effects of these discrete populations.
Collectively, our study highlights that the accumulation of tissue-resident CD8+ T cells acts to preserve microglial functional homeostasis in the context of increasing Aβ plaque burden. Our study suggests that, while Aβ plaque burden likely initiates pro-inflammatory microgliosis, CD8+ T cells accumulate at a later stage and play a homeostatic role in mitigating microglial inflammatory state to limit further Aβ plaque formation and cognitive decline. The bidirectional interaction between microglia and CD8+ T cells that is mediated by CXCL16–CXCR6, and the immunoregulatory actions of brain-resident CD8+ T cells, may be important immunotherapeutic targets for neurodegenerative diseases.
Methods
Mice
The research conducted in this study complies with all relevant ethical guidelines. Animal protocols were approved by and performed in accordance with the Institutional Animal Care and Use Committee (IACUC) of St. Jude Children’s Research Hospital. C57BL/6J, Tcra−/−, B2m−/−, Cxcr6−/−, and Nt5e−/− mice (all under the C57BL/6J background) were purchased from the Jackson Laboratory. 5xFAD mice13 were backcrossed to C57BL/6J background for more than 10 generations, and APPNL-G-F transgenic mice14 with knock-in of human APP mutated transgenes were obtained from Dr. Takaomi C. Saido (RIKEN Center for Brain Science, Saitama, Japan) and were backcrossed in C57BL/6J background for more than 10 generations. For intravenous labeling experiments, anti-CD45.2-PE was administrated to mice by tail vein injection for 5 min prior to euthanasia before harvesting the brain (see below)32. Age- and sex-matched (Non-Tg, 5xFAD, 5xFAD;Tcra−/−, 5xFAD;B2m−/−, 5xFAD;Cxcr6−/−, 5xFAD;Nt5e−/−, APPNL-G-F, and APPNL-G-F;Cxcr6−/−) mice with predetermined genotypes (not blinded to investigators) were randomly assigned to control and experimental groups. The phenotypes were stratified by sex as indicated in the figures and their legends. All mice were housed and bred under specific pathogen-free conditions in the Animal Resource Center at St. Jude Children’s Research Hospital. Mice were on 12-hour light/dark cycles that coincide with daylight in Memphis, TN, USA. The St. Jude Children’s Research Hospital Animal Resource Center housing facility was maintained at 20–25 °C and 30–70% humidity.
Preparation of single-cell suspensions
Splenocytes were released by mechanical force followed by red blood cell (RBC) lysis with ammonium chloride potassium buffer (Gibco). Cell suspensions were filtered through a 70 μm strainer and resuspended in Hank’s balanced salt solution (HBSS; Gibco) supplemented with 2% (vol/vol) fetal bovine serum (FBS). For brain immune cell isolation, euthanized mice underwent cardiovascular perfusion with PBS, followed by isolation of brain, meningeal dura, liver, and lung tissues. Of note, the meningeal dura was separated from brain tissue to avoid cellular contamination from this CNS compartment. Brain, liver, and lung were homogenized using an Octodissociator (Miltenyi) in HBSS containing 1 mg/ml collagenase IV (Worthington) and 0.5 mg/ml DNase I (Sigma) followed by digestion in a rotary shaker (at 240 r.p.m.) for 15 min (brain) or 30 min (liver and lung) at 37 °C, and filtration through a 70 μm strainer. Immune cells were purified by density centrifugation (1,500 ×g with low acceleration and low brake) in 38% Percoll (GE Healthcare) for 20 min. After centrifugation and RBC lysis (for lung and liver), pelleted immune cells were washed and filtered. Meninges (including meningeal dura and arachnoid tissues) was digested in medium with collagenase VIII (Sigma) and DNase I (Sigma) for 15 min at 37 °C and then pressed through 70 μm cell strainer12,51.
Flow cytometry
For analysis of surface markers, cells were stained in PBS containing 2% (wt/vol) FBS for 30 min on ice. Prior to surface staining, cells were incubated with fixable viability dye (Zombie Aqua Dye; 1:1,000, 423102, Biolegend) and anti-CD16/32 (for Fc receptor blocking; 1:1,000, 2.4G2, 70-0161-U500, Thermo Fisher Scientific) for 10 min at 24 °C. For intracellular staining of transcription factors and/or cytokines, cells were fixed with Foxp3 fixation and permeabilization buffer kits (Thermo Fisher Scientific) according to manufacturer’s instructions. For cytokine staining, cells were stimulated for 4 h with phorbol 12-myristate 13-acetate (PMA) and ionomycin in the presence of monensin (BD Biosciences). The following antibodies were used: anti-CD3 (1:300, 17A2, 100232), anti-TCRβ (1:300, H57-597, 109249 or 109220), anti-CD4 (1:300, RM4-5, 100550, 100526 or 100547), anti-CD8 (1:300, 53-6.7, 100759 or 100730), anti-CD11b (1:300, M1/70, 101259), anti-CXCR6 (1:300, SA051D1, 151104), anti-Ki67 (1:200, 16A8, 652413), anti-PD-1 (1:300, 29F.1A12, 135210 or 109116), anti-CD69 (1:300, H1.2F3, 104512), anti-TNF-α (1:200, MP6-XT22, 506304), anti-CD73 (1:300, TY/11.8, 127224) (all from Biolegend); anti-CD45.2 (1:300, 104, 75-0454-U100 or 50-0454-U100), anti-CD8 (1:300, 53-6.7, 60-0081-U100), anti-CD69 (1:300, H1.2F3, 35-0691-U100) (all from Tonbo); anti-CD11b (1:300, M1/70, 550993), anti-CXCL16 (1:300, 12-81, 566740), anti-TIGIT (1:300, GIGD7, 12-9501-82), anti-CD39 (1:300, 24DMS1, 50-0391-82) (all from BD Biosciences), anti-CD45 (1:300, 30-F11, 12-0451-83), anti-CD103 (1:300, 2E7, 17-1031-82), anti-pro-IL-1β (1:100, NJTEN3, 17-7114-80) (all from Thermo Fisher Scientific). Flow cytometry data were acquired using FACSymphony A3, LSRII, or Fortessa cytometers (BD Biosciences) and analyzed with FlowJo software v10 (TreeStar).
Cell purification and culture
For co-culture of CD8+ T cells and microglia, splenic CD44hiCD8+ T cells were sorted from 5xFAD mice and expanded in vitro for 2 d in the presence of rIL-2 (20 ng/ml, Tonbo 21-8021), rIL-12 (10 ng/ml, PeproTech 210-12), and rIL-15 (40 ng/ml, PeproTech 210-15)38. Expanded CD8+ T cells were re-sorted based on PD-1 or CXCR6 expression and co-cultured with CD45int/+CD11b+ microglia isolated from 5xFAD mice. For some co-culture experiments, brain CD8+ T cells were sorted from 5xFAD mice before co-culture with CD45int/+CD11b+ microglia. Co-cultured cells were stimulated with PMA52 and ionomycin in the presence of 1 μM Aβ (Anaspec, AS-20276; diluted in Click’s medium for 24 h at 4 °C for oligomerization before adding to cell culture) for 4 h.
Immunofluorescence and immunohistochemical staining
De-identified human postmortem brain tissues from individuals with AD (frontal gyrus) were obtained from the Brain and Body Donation Program at Banner Sun Health Research Institute. Individuals with clinical and pathological diagnoses53 were recruited from retirement communities in the greater Phoenix, Arizona, USA area. Informed consent was provided by all donors (females aged 61–87 at time of death) or their legal representatives, and the study was approved by the Banner Sun Health Research Institute Institutional Review Board as previously described53. Mouse brain was harvested, sagittally bisected, and fixed overnight in 4% paraformaldehyde (PFA) prior to processing and embedding in paraffin blocks, and 5 μm thick sections were cut and mounted on slides. Every tenth section was stained with H&E to match brain architecture across samples. Unstained, matched sections were deparaffinized, rehydrated and subjected to heat-mediated antigen retrieval in a citrate solution. Sections were then blocked with PBS containing 2% BSA and 0.05% Tween-20 and incubated with primary antibody overnight at 4 °C. Primary antibodies and dilutions were as follows: anti-amyloid beta (for mouse and human; 1:1,000; IBL 10323), anti-Iba1 (for mouse 1:500; Wako 019-01974), anti-IBA1 (for human 1:300; Biocare Medical CP290A), anti-CD3 (for mouse; 1:500; Cell Signaling Technologies 78588 or 1:1000; Santa Cruz sc-1127), anti-CD8 (for mouse; 1:500; Cell Signaling Technologies 98941), anti-CD8 (for human; 1:250, Abcam ab4055), anti-PD-1 (for mouse; 1:500; Cell Signaling Technologies 84651), anti-PD-1 (for human; 1:500; Cell Signaling Technologies 86163), and anti-CD31 (for mouse; 1:500; Cell Signaling Technologies 77699).
Immunofluorescence detection:
Sections were incubated with fluorescently tagged secondary antibodies (1:500; Thermo Fisher Scientific A32766, A32795) for 1 h at 24 °C, and coverslipped in a mounting media containing DAPI. For CD3 and CD8 staining, slides were incubated with a biotin blocking kit (Vector Labs, SP-2001), and signals were detected using a tyramide amplification kit (Thermo Fisher Scientific B40923), followed by fixation, secondary heat denaturation, and overnight incubation with antibodies against Aβ and directly-conjugated Iba1/IBA1, PD-1, or CD31 as indicated. CD8 and PD-1 co-detection was performed using sequential incubation and detection with a tyramide amplification kit (Thermo Fisher Scientific B40923) followed by secondary fixation and heat denaturation after each detection, and then overnight incubation with directly-conjugated anti-Iba1 antibody. Wide-field fluorescent images were acquired with an A1R HD25 Ti2 Eclipse inverted microscope (Nikon Instruments) equipped with a 20× S Fluor 0.75 NA objective, SOLA illuminator (Lumencor) and Orca-Fusion BT sCMOS camera (Hamamatsu) and analyzed using NIS-Elements Advanced Research software (v5.3.03; Nikon Instruments). Confocal imaging was performed using an A1R HD25 confocal with resonance scanning and a 40× Plan Fluor 1.3 NA oil objective.
Immunofluorescence quantification:
Fluorescence images were deconvolved using NIS-Elements Advanced Research software (v5.3.03; Nikon Instruments) and further analyzed in three dimensions using Imaris software (v9.5.1; Bitplane). To remove any batch effects between samples, Aβ plaque data from 5xFAD;Tcra−/−, 5xFAD;B2m−/−, and 5xFAD;Cxcr6−/− mice were normalized to respective control (i.e., 5xFAD) samples within a simultaneously-prepared batch. Aβ plaque numeration was performed on multi-field composite images (20×), wherein anatomical landmarks were used to compare similar regions of interest. Aβ plaque number and burden were determined following segmentation of fluorescent signals above local background and incorporation of a region growing algorithm, yielding total number and percent coverage of Aβ plaques per squared area of field. One or two sections per biological replicate were analyzed for each parameter. For quantification of Iba1+ microglia and plaque-associated Iba1+ microglia, a spot-fitting algorithm was used to identify individual Iba1+ cell bodies and Aβ plaques, with a maximum center-to-distance threshold of 25 μm used to designate co-localized objects. All parameters were fixed for each fluorescence channel across all samples for consistent measurement of both Aβ plaques and microglia. For normalized spatial intensity correlation (NCInC) analysis34 of CD8, Aβ, and Iba1, signals for each fluorescence channel were extracted from 87 images. NSInC indices and statistics were calculated pairwise (CD8–Aβ; CD8–Iba1) to determine the significance of marker co-localization compared to a simulated random distribution and also between genotype group. For Aβ plaque phenotype analysis, deparaffinized brain sections were incubated with anti-Aβ antibody (clone 82E1) and then stained with 1% Thioflavin S (Sigma T1892) in an aqueous buffer for 5 min prior to mounting and visualization by fluorescence microscopy. Aβ plaque phenotypes were manually scored as filamentous, compact, or inert based on published literature26.
Immunohistochemical detection:
Sections were exposed to 0.6% hydrogen peroxide for 30 min prior to primary antibody incubation. For detection, sections were incubated with biotinylated secondary antibodies followed by horseradish peroxidase-conjugated streptavidin (Elite ABC, Vector Labs PK-6101, PK-6102), visualized with ImmpactDAB substrate (Vector Labs SK-4105), counterstained with Hematoxylin QS (Vector Labs H-3404) and coverslipped with Permount (Fisher SP15-100). Images were acquired with an upright NI-Eclipse microscope (Nikon Instruments) equipped with a 10× Plan Apo 0.45 NA objective, 40× Plan Fluor 1.3 NA oil objective, DS-Ri2 16.25 megapixel digital camera. Composite wide-field images were generated and analyzed with NIS-Elements Advanced Research software (v5.3.03; Nikon Instruments).
ELISA and immunoblot
Mouse cortex and hippocampus brain regions were separated under a dissection microscope and flash frozen in liquid nitrogen. For soluble-fraction protein sample extraction, samples were homogenized in 500 μL Tris-buffered saline (TBS; pH 7.4) supplemented with protease inhibitors (Roche 04693132001) in a Dounce homogenizer followed by rinsing with 300 μL TBS to capture residual sample. Samples were incubated for 10 min on ice and then centrifuged for 1 h (20,000 ×g at 4 °C). Soluble-fraction sample supernatants were then flash frozen in liquid nitrogen and stored at −80 °C. For insoluble-fraction protein sample extraction, leftover sample pellets were washed with 200 μL TBS and then centrifuged for 5 min (20,000 ×g at 4 °C). Sample pellets were resuspended in 500 μL TBS plus 1% Triton X-100 and mixed by gentle rotation for 30 min followed by centrifugation for 1 h (20,000 ×g at 4 °C). Following centrifugation, insoluble-fraction sample supernatants were flash frozen in liquid nitrogen and stored at −80 °C until use. Quantitative analyses of soluble and insoluble Aβ1-40 and Aβ1-42 protein isoforms were conducted by ELISA (Fujifilm Wako Pure Chemical Corporation 294-62501, 292-64501) according to manufacturer’s instructions. For immunoblot analysis, soluble-fraction samples were resolved in Novex™ 4–20% Tris-Glycine plus Midi Gel (Invitrogen XP04205BOX) and transferred to PVDF membrane (Bio-Rad 1620177). Membranes were blocked using 5% BSA for 1 h and incubated overnight with mouse anti-Aβ (1:1,000; 82E1; 10323; IBL) or mouse anti-APP (1:1,000; C1/6.1; 802801; Biolegend). Rabbit anti-β actin (1:5,000; 13E5; 4970S; Cell Signaling Technologies) was used as a loading control. Membrane was washed with TBST and incubated with HRP-conjugated anti-mouse IgG (1:5,000; 7076S; Cell Signaling Technologies) or HRP-conjugated anti-rabbit IgG (1:5000; 7074S; Cell Signaling Technologies) for 2 h. After washing with TBST, the membrane was imaged using the Amersham Imager 600.
Behavior and short-term memory analyses
Mice were acclimated to the testing room for 30 min prior to behavioral experiments19. For spontaneous Y maze alternation testing, mice were placed in the center of the maze and allowed to freely explore for 5 min. Each session was recorded and scored by an observer. Spontaneous alternation was calculated by dividing the number of consecutive entries into 3 different maze arms by the number of total possible alternations (total arm entries – 2). Mice with less than 5 total arm entries were excluded from analysis19. For novel objection recognition, mice were habituated to an empty 40 cm × 40 cm open-field testing chamber (Noldus Information Technology) for 10 min. On the next day, mice were placed into the testing chamber containing two identical objects for 10 min, followed by a 4 h inter-session interval. For the test phase, the novel object was substituted, and mice were placed into the test chamber and allowed to explore both objects for 10 min. Test phase sessions were recorded and analyzed using EthoVision XT software (v16; Noldus Information Technology) to calculate novel object discrimination index (TNovel/(TNovel + TFamiliar) × 100)19.
scRNA-seq and data analysis
Library preparation:
For Non-Tg and 5xFAD or 5xFAD;Cxcr6+/+ and 5xFAD;Cxcr6−/− mice scRNA-seq datasets, brain or meninges were harvested from 8-month-old, sex-matched mice. After staining with fixable viability dye (1:1,000, 65-0865-18, eBioscience), anti-CD45.2 (1:200, 104, 17-0454-82, Thermo Fisher Scientific), anti-CD11b (1:200, M1/70, 48-0112-82, Thermo Fisher Scientific), cells (brain: CD45.2int/+CD11b+ and CD45.2+CD11b− cells or meninges: CD45+ cells) were sorted on a reflection cell sorter (iCyt), and cells from three mice per genotype were pooled as one biological replicate (n = 2 biological replicates for each genotype). For 5xFAD;Tcra+/+ and 5xFAD;Tcra−/− or 5xFAD;B2m+/+ and 5xFAD;B2m−/− mice scRNA-seq datasets, brains were harvested from 8-month-old, sex-matched mice. Cells were stained with fixable viability dye (1:1,000, 65-0865-18, eBioscience) and anti-CD45.2 (1:200, 104, 17-0454-82, Thermo Fisher Scientific) and sorted on a reflection cell sorter (iCyt). Cells (CD45.2int/+ and CD45.2− cells) from three mice per genotype were pooled as one biological replicate (n = 2 biological replicates for each genotype). The collected cells were counted and examined for viability by a Luna Dual Florescence Cell Counter (Logos Biosystems). The samples were centrifuged at 950 ×g for 5 min, and cells (was >98% viable) were resuspended in 1× PBS (100 mL; Thermo Fisher Scientific) plus 0.04% BSA (Amresco) with final a concentration of 1 × 106 cells/mL. Single-cell suspensions were loaded onto a Chromium Controller (10x Genomics) to generate 10,000 single-cell gel beads in emulsion per sample. Each sample was loaded into a separate channel. For Non-Tg and 5xFAD or 5xFAD;Cxcr6+/+ and 5xFAD;Cxcr6−/− mice scRNA-seq datasets, libraries were prepared using the Chromium Next GEM Single Cell 5’ v2 (Dual index) and Gel Bead Kit (10x Genomics). For 5xFAD;Tcra+/+ and 5xFAD;Tcra−/− mice scRNA-seq dataset, libraries were prepared using the Chromium Next GEM Single Cell 3’ v3.1 (Dual index) and Gel Bead Kit (10x Genomics). The cDNA was amplified (11 cycles), or after which cDNA was used for TCR enrichment/library preparation with the Chromium Single Cell V(D)J TCR kit, or for preparation of a gene expression library. The cDNA content of each sample and libraries were quality-checked using a high-sensitivity DNA chip with a 2100 Bioanalyzer (Agilent Technologies). The 5’ libraries were sequenced on NovaSeq (Illumina) with paired-end reads of 26 cycles for read 1 and 90 cycles for read 2. An average of 50,000,000 or 5,000,000 reads per samples were obtained for gene expression library (>70,000 reads per cell) and TCR library (>20,000 reads per cell), respectively. The 3’ libraries were sequenced on NovaSeq (Illumina) with paired-end reads of 28 cycles for read 1 and 90 cycles for read 2.
Data preprocessing:
The Cell Ranger v6.0 Single-Cell software suite (10x Genomics) was used to process the scRNA-seq FASTQ files. The ‘cellranger count’ command was performed to align the raw FASTQ files to the mm10 mouse reference genome and summarize the data into matrices that describe gene read counts per cell. For the datasets with matched TCR sequencing data, the ‘cellranger vdj’ command was used to generate a count matrix, which upon filtering, were used for downstream analyses.
For gene expression sequencing, the filtered count matrices were read into the R package Seurat (v4.0–v4.1)29,54,55. Samples were merged into a single Seurat object for consistent filtering, and features detected in fewer than 3 cells were removed from the dataset. Cells with abnormally low features or UMI (unique molecular identifier) counts or high mitochondrial read percentages (potentially dead or damaged cells) were removed. Cells with abnormally high UMI counts (potentially multiple cells in a single droplet) were also removed. Finally, any remaining multiplets expressing mutually-exclusive marker genes were removed. For cells from the brains of Non-Tg and 5xFAD mice, 15,923 cells were retained with an average of 1,828 genes per cell (UMI, median: 5,018; range: 504–29,991). For cells from the meninges of Non-Tg and 5xFAD mice, 6,337 cells were retained with an average of 1,747 genes per cell (UMI, median: 4,630; range: 502-29,955). For cells from the brains of 5xFAD; Cxcr6+/+ and 5xFAD;Cxcr6−/− mice, 20,760 cells were retained with an average of 2,022 genes per cell (UMI, median: 5,972; range: 500–49,890). For cells from the brains of 5xFAD;Tcra+/+ and 5xFAD;Tcra−/− mice, 19,432 CD45+ and CD45− cells were retained with an average of 3,242 genes per cell (UMI, median: 11,795; range: 2,002-34,998). For cells from the brains of 5xFAD;B2m+/+ and 5xFAD;B2m−/− mice, 20,075 CD45+ cells were retained with an average of 2,603 genes per cell (UMI, median: 8,367; range: 2,011–49,940). After quality control, libraries were normalized with NormalizeData function (scale.factor = 1 × 106) in Seurat R package.
For TCR sequencing, filtered contig annotation matrices from the Cell Ranger output were loaded into R. Annotation and quantification of TCR clonotypes were processed with the scRepertoire package (v1.3.5)56. Clonal frequencies were categorized as follows: Single (1), Small (2–5), Medium (6–20), and Large (> 20). Cells were considered clonally expanded if clonotype size was greater than 1 and non-expanded if clonotype size was equal to 1. Cells without clonal information were excluded from clonally expanded versus non-expanded analyses. GLIPH version 2 (GLIPH2) analysis was performed27,57 using CDR3b, Vb, Jb, and CDR3a sequence information from CD8+ T cells (generated from scTCR-seq analysis) isolated from brain and meninges of Non-Tg and 5xFAD mice. CD8+ T cells with their certain clonotypes were input into GLIPH2 (http://50.255.35.37:8080/; with parameters algorithm ‘GLIPH2’; reference version ‘mouse’; reference ‘CD8’; all_aa_interchangeable ‘yes’). After grouping the TCRs predicted to recognize the same epitope, 185 groups had Fisher score ≤ 0.05. The top 5 groups of CDR3b patterns in clonally expanded CD8+ T cells from the brains of 5xFAD mice were then visualized and input into TCRMatch tool28 (http://tools.iedb.org/tcrmatch/) to query annotated sequences in the Immune Epitope Database. The filtering level was selected as Medium (>0.90).
Cluster annotation and data visualization:
Normalized and filtered data were processed using the standard Seurat pipeline (v4.1). Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction was used for visualization, and Seurat’s FindClusters function (v4.1) was used to separate cells into unsupervised clusters. Cell types in clusters were defined using the following marker genes: CD8+ T cells (Cd3e+Cd8a+); CD4+ T cells (Cd3e+Cd4+); γδ T cells (Cd3e+Trdv4+Tcrg-V6+); NKT cells (Zbtb16+Cd4−Cd8a−Trdv4−Tcrg-V6−); B cells (Cd79a+Cd19+); NK cells (Ncr1+Cd3e−); monocytes (Cd14+); macrophages (Cd14+ Mrc1+); dendritic cells (Clec9a+); mast cells (Kit+Il13+); neutrophils (Hdc+); homeostatic microglia (M0; Tmem119+P2ry12+) and DAM (Tmem119+P2ry12+ApoehiSpp1hiTrem2hi).
As indicated, subclusters identified as Cd3e+ T cells were isolated, re-clustered, and re-labeled as CD4+ T cells, CD8+ T cells, γδ T cells, or NKT cells based on the above markers. To ensure accurate delineation between the microglia subpopulations, clusters identified as either DAM or M0 were isolated, re-clustered, and re-labeled as DAM or M0 based on the above markers. Curated violin plots or genes on UMAP that represent the expression level of genes were generated using the VlnPlot and FeaturePlot functions in Seurat.
For analysis of CD8+ T cells, Ptprc+Cd3e+Cd8a+Cd4− cells were isolated and re-clustered using Seurat workflow. To remove any batch effect between datasets, the package Harmony58 was used to integrate CD8+ T cells from different samples [with functions RunUMAP (dims = 1:30), FindNeighbors (dims = 1:30), and FindClusters (resolution = 0.5) resulting in 8 CD8+ T cell subclusters. To identify corresponding CD8+ T cell subclusters in the meninges compared with the brain, the CD8+ T cells in brain and meninges were integrated and re-clustered. The 8 subclusters in brain were used as a reference to determine the identity of subclusters in the meninges.
T cell receptor data analysis and visualization:
To identify genes correlated with clonal expansion, Seurat's GroupCorrelation function (v4.1) was used (with clonal frequency, output by the scRepertoire analysis). The top 20 shared clones in CD8+ T cells between the brain and meninges were plotted using the compareClonotypes function in scRepertoire (cloneCall = “aa”).
Public dataset analysis:
FASTQ files from 16-month-old APP/PS1 and Non-Tg mice11 (PRJNA529095) were downloaded. High-quality cells (gene number: 40–4,000; UMI: 200– 20,000; and mitochondrial reads <5.5%) were filtered and contaminants (CD45− cells and multiplets expressing mutually-exclusive marker genes) were removed, resulting in 6,203 cells (2,416 cells from Non-Tg and 3,787 cells from APP/PS1). The data were analyzed with the Seurat pipeline as described above, using the first 25 PCs for the KNN and UMAP dimensional reduction steps. Seurat’s FindClusters function (resolution 0.5) separated cells into 15 unsupervised clusters, which were annotated into 9 immune cell types (see above). Clusters expressing macrophage and/or DC markers were annotated “BAM/DCs”. For analysis of human snRNA-seq public dataset (GSE157827)25, Seurat RAW input files from single-nucleus transcriptome analysis of 169,496 nuclei from the prefrontal cortical samples of individuals with AD and normal control (NC) subject were downloaded. High-quality cells (gene number: < 7,500; UMI: 200–25,000; and mitochondrial reads <5%) were filtered, resulting in 153,687 cells remaining (74,148 cells from NC and 79,539 cells from AD). The data were analyzed using the Seurat pipeline as described above, using the first 20 PCs for the KNN and UMAP dimensional reduction steps. Seurat’s FindClusters function (resolution 0.1) separated cells into 15 unsupervised clusters, which were subsequently annotated into 6 cell types based on positive expression of the cell type markers as follows25: astrocytes (SLC1A2, ADGRV1, GPC5, RYR3, and GFAP); endothelial cells (CLDN5, FLT1, ABCB1, EBF1, and MT2A); excitatory neuron (RALYL, KCNIP4, CBLN2, LDB2, and KCNQ5); inhibitory neuron (NXPH1, LHFPL3, PCDH15, GRIK1, and ADARB2); microglia (LRMDA, DOCK8, ARHGAP24, ARHGAP15, and PLXDC2); and oligodendrocytes (ST18, PLP1, CTNNA3, MBP, and PIP4K2A). For analysis of human bulk proteomics dataset22, patient groups were stratified as follows: 1) control group with low pathology of plaques (LPC); 2) control group with Aβ pathology but no obvious cognitive defects (HPC); 3) control group with progressive supranuclear palsy who accumulate tau proteins (PSP); 4) control group with mild cognitive impairment with Aβ plaques (MCI); 5) physician-diagnosed AD with cognitive defects, high pathology scores of Aβ plaques and tau tangles (AD).
Identification of ligand–receptor pairs in scRNA-seq data
Ligand–receptor pairs were downloaded from published databases21,59,60. We defined a ligand or receptor as an expressed gene in a certain cell type if >10% cells had its expression level (cutoff:log2(normalized count + 1) > mean gene expression of the normalized count matrix). The expression value of unexpressed ligand or receptor was set to zero. A putative ligand–receptor pair interaction between cell type A and cell type B was scored as the product of average ligand expression across all cells in cell type A and the average receptor expression across all cells in cell type B using the following formula, where L is the ligand, R is the receptor, A is cell type A, and B is cell type B; Int and E stand for interactions and expression, respectively:
Statistical significance was assessed by randomly shuffling the cluster labels of all cells and repeating the above steps, which generated a null distribution for each ligand–receptor pair in each pairwise comparison between two cell types. After running 1,000 times permutations, Z-scores were calculated with the normal distribution curve generated from the permuted ligand–receptor pair interaction scores. Ligand–receptor interactions mediated by chemokine receptors and their ligands were identified by filtering with GO:0016493 (C–C chemokine receptor activity). Ligand–receptor interactions were considered significant if Z-score > 25.
To determine the difference of microglia and T cell interaction strength between 5xFAD;Cxcr6+/+ and 5xFAD;Cxcr6−/− mice, CellChat (v1.4.0)35 was used. CellChat objects were produced using the createCellChat function. The overall cell–cell interaction strength between microglia and CD4+, CD8+ or γδ T cells in 5xFAD;Cxcr6+/+ and 5xFAD;Cxcr6−/− mice was calculated by the default workflow based on ligand–receptor databases in the CellChat package. The CXCL16–CXCR6 pair was removed from ligand–receptor databases to avoid the contribution of the reduced CXCL16–CXCR6 interaction by CXCR6 deficiency to overall cell–cell communication. The workflow includes the following commands in sequential order, using default parameters unless otherwise specified: identifyOverExpressedGenes, identifyOverExpressedInteractions, projectData, computeCommunProb, filterCommunication (min.cells = 10), aggregateNet, subsetCommunication (thresh = 1), netAnalysis_computeCentrality. After running the pipeline on the two genotypes separately, the two CellChat objects were merged using the mergeCellChat function. Heatmaps of the differential cell–cell interaction strength between genotypes were generated using the function netVisual_heatmap (measure = "weight"), with row z-score to indicate the difference of the interaction strength on the heatmap.
Differential expression analysis
Differential expression (DE) gene analysis was performed on the log-normalized gene expression matrices with the Seurat FindMarkers function (v4.1) by using a two-tailed Wilcoxon rank-sum test and calculating adjusted P values via Bonferroni correction54.
Gene set enrichment and functional enrichment analyses
For gene set enrichment analysis (GSEA), genes were ranked in order of descending log-fold change values (derived from the DE analysis). Pre-ranked GSEA61 was performed using the Broad GSEA command-line software (v4.0.3). For the CD8+ T cells, a curated core TRM signature30 and the Hallmark signatures from the Molecular Signatures Database were used (MSigDB, v7.3) (https://www.broadinstitute.org/gsea/msigdb/). For the microglia, the Gene Ontology (C5), Hallmark, and C2 geneset collections from the MSigDB (v7.3) were used. For functional enrichment analysis, upregulated genes (log2 FC > 0.35; FDR < 0.05) in M0 and DAM subsets from 5xFAD;Tcra−/−, 5xFAD;B2m−/− or 5xFAD;Cxcr6−/− mice compared to their respective 5xFAD control subsets were used as inputs for funcEnrich.Fisher function in NetBID2 R package for pathway enrichment against MsigDB signatures (Hallmark, C2 and C5).
Statistical analysis
For biological (non-omics analysis), data were analyzed using Prism software (v7.0, GraphPad) by two-tailed unpaired Student’s t-test (with Welch’s correction for quantification of plaque number), one-way ANOVA, or two-way ANOVA. Two-tailed unpaired Student’s t-test, two-tailed Wilcoxon rank sum test, two-tailed Fisher’s exact test, two-tailed normalized weighted Kolmogorov–Smirnov test, two-tailed Benjamini–Hochberg adjusted P value (FDR), Pearson correlation coefficient, two-tailed F-test, and two-tailed Bonferroni correction were applied for analysis of scRNA-seq data. The sample sizes and statistical tests were mentioned in figure legends. Differences were considered statistically significant when P ≤ 0.05. No statistical method was used to predetermine the sample sizes, but our sample sizes are similar to those reported in other publications20,40. Data distribution was assumed to be normal but this was not formally tested. Age- and sex-matched mice with predetermined genotypes were randomly assigned to control and experimental groups. No other randomization was performed, and no data points were excluded for analysis. Data collection and analysis were not performed blind to the conditions of the experiments. Only human AD samples were used in this study, so randomization and blinding were not necessary.
Extended Data
Extended Data Fig. 1. T cells accumulate in mouse models of AD.
a, Schematic workflow of scRNA-seq and scTCR-seq experiments, showing tissue collection sites, cell sorting strategies, and computational analyses of Non-Tg and 5xFAD mice. b, Gating strategy for sorting microglia and non-microglia immune cells used for scRNA-seq and scTCR-seq analysis. Sorting gates for Non-Tg and 5xFAD samples were set according to the following parameters (in order): 1) cell size (to exclude debris); 2) singlet; 3) viable cells; 4) CD45int/+CD11b+ (for microglia) and CD45+CD11b− (for non-microglia immune cells). c, Violin plots showing expression of cell type-defining genes in clusters from Fig. 1b. d, e, CD45+ immune cell clusters colored by genotype (d) or cell types (e) from a public dataset of indicated mice11. f, Frequencies of cells described in d and e. g, Brain T cells (CD3+TCRβ+) from 10-month-old Non-Tg (n = 11) and 5xFAD (n = 13) mice. h, CD4+ and CD8+ T cells in the brains of female and male 10-month-old Non-Tg (n = 12 for female and 9 for male) and 5xFAD (n = 14 for female and 11 for male) mice. i, j, CD3+ [i, 3 months: Non-Tg (n = 4) and 5xFAD (n = 4); 6 months: Non-Tg (n = 8) and 5xFAD (n = 9); 9 months: Non-Tg (n = 8) and 5xFAD (n = 13); 12 months: Non-Tg (n = 11) and 5xFAD (n = 13)] or CD4+ [j, 3 months: Non-Tg (n = 6) and 5xFAD (n = 6); 6 months: Non-Tg (n = 13) and 5xFAD (n = 16); 9 months: Non-Tg (n = 8) and 5xFAD (n = 9); 12 months: Non-Tg (n = 14) and 5xFAD (n = 17)] T cells in the brains of Non-Tg and 5xFAD mice at indicated time points. k, Immunohistochemical analysis of CD3+ T cells in the brains of 10-month-old Non-Tg (n = 7) and 5xFAD (n = 6) mice. Scale bar, 200 μm. Arrows indicate positive staining for CD3+ T cells. l, m, CD3+ (l) or CD8+ (m) T cells in the brains of Non-Tg and APPNL-G-F mice [3 months: Non-Tg (n = 5) and APPNL-G-F (n = 5); 6 months: Non-Tg (n = 5) and APPNL-G-F (n = 4); 9 months: Non-Tg (n = 5) and APPNL-G-F (n = 4)]. Data were analyzed by two-tailed unpaired Student’s t-test (g, h, k–m) or two-way ANOVA (i, j). Data are shown as mean ± s.e.m. in g–m; NS, not significant. Data were pooled from at least three (g–j) or two (k–m) independent experiments.
Extended Data Fig. 2. Single-cell immune profiling of meninges and functional characterization of T cells by multiple genetic models.
a, b, scRNA-seq profiling of CD45+ immune cells from the meninges of male Non-Tg or 5xFAD mice (n = 2 biological replicates for each genotype) at 8 months of age. UMAP showing CD45+ immune cell clusters colored by genotype (a) or cell type (b). c, Frequencies (normalized to total cells from each genotype) of annotated clusters described in b. d, CD4+ and CD8+ T cells in the meninges of 10-month-old Non-Tg (n = 13) and 5xFAD (n = 13) mice. e, CD4+ and CD8+ T cells in the indicated tissues of 10-month-old Non-Tg and 5xFAD mice [spleen: Non-Tg (n = 11), 5xFAD (n = 16); liver: Non-Tg (n = 7), 5xFAD (n = 8); lung: Non-Tg (n = 5), 5xFAD (n = 6)]. f, Representative flow cytometry plots demonstrating a lack of CD8+TCRβ+ or CD4+TCRβ+ T cells in the brains of 5xFAD;Tcra−/− mice. g, Immunofluorescence analysis of the number and burden of Aβ plaques in 10-month-old female 5xFAD;Tcra+/+ (n = 8 sections for plaque number; n = 15 sections for plaque burden) and 5xFAD;Tcra−/− (n = 9 sections for plaque number; n = 15 sections for plaque burden) mice. Scale bar, 250 μm. h, Representative flow cytometry plots demonstrating a lack of CD8+ T cells (CD8+TCRβ+) but not CD4+ T cells (CD4+TCRβ+) in the brains of 5xFAD;B2m−/− mice. i, Immunofluorescence analysis of number and burden of Aβ plaques in 10-month-old male and female 5xFAD;B2m+/+ (n = 9 sections for female and 8 for male for plaque number; n = 10 sections for female and 4 for male for plaque burden) and 5xFAD;B2m−/− (n = 10 sections for female and 12 for male for plaque number; n = 10 sections for female and 6 for male for plaque burden) mice. Scale bar, 250 μm. j, k, Illustrations of Y maze (j) and novel objection recognition (k) behavioral tests. l, Total arm entries from spontaneous Y maze alternation testing in 4-month-old 5xFAD;B2m+/+ (n = 9 for female and 8 for male) and 5xFAD;B2m−/− (n = 15 for female and 8 for male) mice. m, n, Spontaneous alternation (m) and total arm entries (n) from spontaneous Y maze alternation testing in 4-month-old female 5xFAD;Tcra+/+ (n = 15) and 5xFAD;Tcra−/− (n = 15) mice. Data were analyzed by two-tailed unpaired Student’s t-test (d, e; g, i for plaque burden; l–n) or two-tailed unpaired t-test with Welch’s correction for plaque number (g, i). Data are shown as mean ± s.e.m. in d, e, g, i, and l–n; NS, not significant. Data were pooled from at least three (d, e, m, n) or two (g, i, l), or are representative of at least two (f, h) independent experiments.
Extended Data Fig. 3. CXCL16–CXCR6 inter-cellular communication axis between microglia and T cells.
a, Circle plot of predicted cell type interactions based on ligand–receptor pairs in brain immune cells in 5xFAD mice. Line thickness indicates number of ligand–receptor interactions; red color indicates CD8+ T cells and microglia as the cell-type pair with the highest number of interactions; purple color indicates comparatively weaker interactions present between CD4+ T or NK cells and microglia. b, GSEA enrichment plots showing upregulation of chemokine-associated signatures in microglia from 5xFAD mice compared to Non-Tg mice. c, Circle plot of predicted cell–cell interactions based on CXCL16 and CXCR6 pairing in brain immune cells of 5xFAD mice (red arrow indicates strongest interaction). d, Violin plot of Cxcl16 expression in microglia from Non-Tg (n = 1,782 cells) and 5xFAD (n = 2,870 cells) mice. e, Violin plot of Cxcl16 expression in immune cell clusters (n = 4,105 cells for M0 and 547 cells for DAM) in 5xFAD mice. f, GSEA enrichment plots showing upregulation of chemokine-associated signatures in DAM compared to M0. g, Bubble plot showing stronger predicted CXCL16–CXCR6 interaction between DAM and CD8+ T cells than M0 and CD8+ T cells in 5xFAD mice. h, Circle plot of predicted cell–cell interactions based on CXCL16 and CXCR6 pairing in brain immune cells of APP/PS1 mice (red arrow indicates the strongest interaction); BAM/DCs: border-associated macrophages/dendritic cells. i, Bubble plot showing stronger predicted CXCL16–CXCR6 interaction between DAM and CD8+ T cells than M0 and CD8+ T cells in APP/PS1 mice. j, Bubble plot of CXCL16 expression in brain cell types from a public human single-nuclear RNA-seq dataset25. k, CXCR6+CD8+ T cells in the meninges [Non-Tg (n = 13) and 5xFAD (n = 13)], spleen [Non-Tg (n = 5) and 5xFAD (n = 7)], liver [Non-Tg (n = 5) and 5xFAD (n = 7)], and lung [Non-Tg (n = 5) and 5xFAD (n = 7)] of 10-month-old Non-Tg and 5xFAD mice. l, CXCR6+CD4+ T cells from the spleens and brains of 10-month-old Non-Tg (n = 6) and 5xFAD (n = 9) mice. Data were analyzed by two-tailed normalized weighted Kolmogorov–Smirnov test (NES for b and f), two-tailed Benjamini–Hochberg adjusted P value (FDR for b and f), two-tailed Bonferroni correction (d), two-tailed Wilcoxon rank-sum test (e), or two-tailed unpaired Student’s t-test (k, l). Data are shown as mean ± s.e.m. in k and l; NS, not significant. Data were pooled from at least three (k, l) independent experiments.
Extended Data Fig. 4. CXCR6 promotes brain CD8+ T cell accumulation.
a, Widefield immunofluorescence images of CD3+ T cells, Aβ, and Iba1+ microglia in brains of indicated 10-month-old mice. Scale bar, 200 μm. White arrows indicate positive staining for CD3+ T cells. Boxed areas show CD3 staining insets. b, CD4+ and CD8+ T cells in the meninges [upper left, 5xFAD;Cxcr6+/+ (n = 13) and 5xFAD;Cxcr6−/− (n =9)], spleen [upper right, 5xFAD;Cxcr6+/+ (n = 16) and 5xFAD;Cxcr6−/− (n = 17)], liver [lower left, 5xFAD;Cxcr6+/+ (n = 8) and 5xFAD;Cxcr6−/− (n = 10)], or lung [lower right, 5xFAD;Cxcr6+/+ (n = 7) and 5xFAD;Cxcr6−/− (n = 10)] of indicated 10-month-old mice. c–f, Indicated immune cell populations in the brains of 10-month-old female and male 5xFAD;Cxcr6+/+ (c, d; n = 10, 10, 6, 6, 10, and 6 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg cells in female mice, respectively; and 12, 12, 9, 9, 9, and 7 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg cells in male mice, respectively) and 5xFAD;Cxcr6−/− mice (c, d; n = 14, 14, 7, 7, 13 and 7 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg cells in female mice, respectively; and 13, 13, 11, 11, 11, and 7 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg in male mice, respectively), or in 4-month-old APPNL-G-F;Cxcr6+/+ (e, f; n = 5, 5, 5, 5, 4 and 5 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg cells in female mice, respectively; and 5, 5, 5, 5, 5, and 5 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg cells in male mice, respectively) and APPNL-G-F;Cxcr6−/− (e, f; n = 5, 5, 5, 5, 4 and 5 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg cells in female mice, respectively; and 6, 6, 6, 6, 6, and 6 for CD8+ T cells, CD4+ T cells, NK1.1 cells, NKT cells, γδ T cells, and eTreg cells in male mice, respectively) mice. g, Non-viable (Zombie Aqua+) CD8+ T cells in indicated tissues of 10-month-old 5xFAD;Cxcr6+/+ (n = 13, 9, 9, 4 for meninges, spleen, liver, lung, respectively) and 5xFAD;Cxcr6−/− (n = 9, 12, 12, 7 for meninges, spleen, liver, lung, respectively) mice. h, Ki67+ cells among brain CD8+ T cells from 10-month-old 5xFAD;Cxcr6+/+ (n = 8) and 5xFAD;Cxcr6−/− (n = 9) mice. Data were analyzed by two-tailed unpaired Student’s t-test (b–h). Data are shown as mean ± s.e.m. in b–h; NS, not significant. Data were pooled from at least two (b–h) or are representative of two (a) independent experiments.
Extended Data Fig. 5. CXCR6 deletion increases Aβ plaques in 5xFAD mice.
a, Representative immunofluorescence images (upper) and quantification of number (lower left) and burden (% of area) (lower right) of Aβ plaques in 10-month-old female 5xFAD;Cxcr6+/+ (n = 6 sections for plaque number; n = 4 sections for plaque burden) and 5xFAD;Cxcr6−/− (n = 8 sections for plaque number; n = 16 sections for plaque burden) mice. Scale bar, 250 μm. b, Representative Aβ immunostaining in composite wide-field brain sections from indicated 10-month-old female mice. c, Immunofluorescence analysis of number and burden of Aβ plaques in 4-month-old male 5xFAD;Cxcr6+/+ (n = 10 sections for plaque number; n = 8 sections for plaque burden) and 5xFAD;Cxcr6−/− (n = 12 sections for plaque number; n = 8 sections for plaque burden) mice. Scale bar, 250 μm. d, Immunofluorescence analysis of number and burden of Aβ plaques in 10-month-old male 5xFAD;Cxcr6+/+ (n = 6 sections for plaque number; n = 6 sections for plaque burden) and 5xFAD;Cxcr6−/− (n = 6 sections for plaque number; n = 6 sections for plaque burden). Scale bar, 250 μm. e, f, Soluble and insoluble fractions of Aβ1-40 and Aβ1-42 protein levels in the hippocampus or cortex of 4-month-old female (e) and male (f) 5xFAD;Cxcr6+/+ (n = 4 for female and 4 for male) and 5xFAD;Cxcr6−/− (n = 4 for female and 4 for male) mice. g, Total arm entries from spontaneous Y maze alternation testing of 5xFAD;Cxcr6+/+ (n = 10 for female and 9 for male) and 5xFAD;Cxcr6−/− (n = 9 for female and 10 for male ) mice. h, Filamentous, compact, and inert Aβ plaque phenotypes in brain sections from 10-month-old female 5xFAD;Cxcr6+/+ (n = 6 sections) and 5xFAD;Cxcr6−/− (n = 8 sections) mice. Data were analyzed by two-tailed unpaired Student’s t-test (a, c, d for plaque burden, e–h) or two-tailed unpaired t-test with Welch’s correction for plaque number (a, c, d). Data are shown as mean ± s.e.m. in a and c–h; NS, not significant. Data were pooled from at least two (a, c, d, g) or one (b, e, f, h) independent experiment.
Extended Data Fig. 6. Clonal expansion of Pdcd1-expressing brain CD8+ T cells in 5xFAD mice.
a, Frequencies of clonally expanded (clonotype size > 1) versus non-expanded (clonotype size = 1) αβ+ T cells in brain and meninges from indicated mice. b, Subclustered brain and meninges αβ+ T cells colored by clonotype sizes and genotypes. c, Identification of CD4+ T cell, CD8+ T cell, and natural killer T (NKT) cell clusters among αβ+ T cells combined from both tissues and genotypes. d, TCR clones detected among CD8+ T cells, CD4+ T cells, and NKT cells, grouped by their clonotype sizes. e, Top 5 GLIPH patterns with TRBV sequence and number of cells in clonally expanded CD8+ T cells in the brains of 5xFAD mice from scTCR-seq analysis. f, TCRMatch analysis for predicted antigen epitopes that are recognized by clonally expanded brain CD8+ T cells from 5xFAD mice. g, Bar plot showing Pearson correlation of gene expression with clonotype sizes, with Pdcd1 identified as the top-ranked gene. h, Heatmap depicting relative expression values of the top 10 differentially expressed genes in each brain CD8+ T cell subcluster shown in Fig. 4c, with selected genes labeled. i, Frequencies of clonotype sizes for brain CD8+ T cells from Fig. 4c cluster 2 (representing the highest expression of Pdcd1) in Non-Tg versus 5xFAD mice. j, Fold change (FC)/FC plot analysis of transcriptomes from clonally expanded versus non-expanded CD8+ T cells compared with Pdcd1+ versus Pdcd1− CD8+ T cells. Red and blue dots indicate shared upregulated and downregulated genes, respectively. Data were analyzed by Pearson correlation coefficient (for r in j) and two-tailed F-test (for P value in j).
Extended Data Fig. 7. Clonal expansion and heterogeneity of CD8+ T cells in the brain and meninges.
a, b, GSEA enrichment plots showing increased core TRM signature30 in clonally expanded versus non-expanded (a) or Pdcd1+ versus Pdcd1− (b) brain CD8+ T cells. c, d, Heatmap of TRM signature genes30 in clonally expanded versus non-expanded (c) or Pdcd1+ versus Pdcd1− (d) brain CD8+ T cells. e, CD69+ (lower left), CD103+ (lower middle), and CD69+ CD103+ (lower right) among brain CD4+ T cells from 10-month-old Non-Tg (n = 4) and 5xFAD (n = 5) mice. f, Two separate imaging fields depicting brain CD8+ T cells (yellow) in proximity to Aβ (green) and CD31+ intravascular structures (red) in 10-month-old 5xFAD mice. Scale bar, 200 μm. Boxed areas show CD8 staining insets. g, PD-1 expression on PE+ and PE− CD8+ T cells from 10-month-old Non-Tg (n = 3) and 5xFAD (n = 3) mice. PE+ and PE− populations were identified after 5 min of labeling by intravenous injection of PE-conjugated anti-CD45 antibody; MFI, mean fluorescence intensity. h, Alluvial plots showing the top 20 CD8+ T cell clones (shared TCRα and TCRβ sequences) between brain and meninges from Non-Tg and 5xFAD mice. i, Pie chart summarizing the percentages (normalized to total cells in each compartment) of CD8+ T cell clonotype sizes in brain and meninges. j, Subclustered CD8+ T cells colored by CNS compartments or subclusters. k, Bar plot showing the frequencies (normalized to total cells from each compartment) of cells derived from the brain and meninges in subclusters as shown in j. l, Violin plots of Pdcd1 expression in CD8+ T cell subclusters as shown in j. The asterisk (*) labels significance in indicated subcluster (n = 1,274 cells) compared to all other subclusters (n = 3,900 cells). m, Clonotypes of CD8+ T cells and bar plot showing the distribution of clonotype sizes for CD8+ T cell subclusters shown in j. The encircled region indicates subcluster 3. Clonotype size: Large (>20); Medium (6–20); Small (2–5); and Single (1). n, UMAP and violin plots showing Pdcd1 expression from the brain (n = 4,641 cells) and meninges (n = 533 cells). o, Bubble plot showing relative Cd69 and Itgae expression in CD8+ T cells in the brain and meninges of 5xFAD mice. p, CD69+, CD103+, and CD69+CD103+ cells among CD8+ T cells in the meninges (n = 11) and brains (n = 12) of 10-month-old 5xFAD mice. Data were analyzed by two-tailed normalized weighted Kolmogorov–Smirnov test (NES for a and b), two-tailed Benjamini–Hochberg adjusted P value (FDR for a and b), two-tailed unpaired Student’s t-test (e, g, p), or two-tailed Wilcoxon rank-sum test (l, n). Data are shown as mean ± s.e.m. in e, g and p; NS, not significant. Data were pooled from at least three (p), two (g), or one (e) independent experiments.
Extended Data Fig. 8. CXCR6-dependent clonal expansion and tissue-residency programs in brain CD8+ T cells from 5xFAD mice.
a, UMAP and violin plots showing Cxcr6 expression in brain CD8+ T cells. b, UMAP of Pdcd1 expression (also shown in Fig. 4e) or Cxcr6 and Pdcd1 co-expression (purple color) in brain CD8+ T cell subclusters shown in a. c, UMAP of CD8+ T cells from a public dataset of Non-Tg and APP/PS1 mice11 colored by genotypes. d, Expression of Cxcr6, Pdcd1, or their overlap (purple color) on subclusters shown in c. e, CXCR6+PD-1+CD8+ T cells [3 months: Non-Tg (n = 3) and 5xFAD (n = 3); 6 months: Non-Tg (n = 4) and 5xFAD (n = 4); 9 months: Non-Tg (n = 4) and 5xFAD (n = 4); 12 months: Non-Tg (n = 5) and 5xFAD (n = 10)] in the brains of Non-Tg and 5xFAD mice at indicated time points. f, g, Subclustered brain CD8+ T cells from indicated mice on UMAP colored by genotype (f) or subclusters (g). h, UMAP and violin plot of Pdcd1 expression in brain CD8+ T cell subclusters in g. i, PD-1+CD8+ T cells in the brains of APPNL-G-F;Cxcr6+/+ (n = 10) and APPNL-G-F;Cxcr6−/− (n = 11) mice. j, CD69+CXCR6+CD8+ T cells from 10-month-old Non-Tg (n = 5) and 5xFAD (n = 8) mice. k, l, GSEA enrichment plots showing downregulated core TRM signature in clonally expanded (k) or Pdcd1+ brain CD8+ T cells (l) from 5xFAD;Cxcr6−/− versus 5xFAD;Cxcr6+/+ mice. m, CD69+, CD103+, and CD69+CD103+ among brain CD8+ T cells from 4-month-old APPNL-G-F;Cxcr6+/+ (n = 10) and APPNL-G-F;Cxcr6−/− (n = 11) mice. Data were analyzed by two-way ANOVA (e), two-tailed unpaired Student’s t-test (i, j, m), two-tailed normalized weighted Kolmogorov–Smirnov test (NES for k and l), or two-tailed Benjamini–Hochberg adjusted P value (FDR for k and l). Data are shown as mean ± s.e.m. in e, i, j and m; NS, not significant. Data were pooled from at least three (12 months in e; i,and j), two (6 and 9 months in e and m), or one (3-month-old mice in e) independent experiments.
Extended Data Fig. 9. Brain T cells co-localize with microglia and Aβ plaques in 5xFAD mice and inhibit microglia pro-inflammatory activity.
a, GSEA enrichment plot showing upregulation of inflammatory response signatures in microglia from 5xFAD mice versus Non-Tg mice. b, Violin plots showing expression of Tnf, Il1b, and Tgfb1 in microglia from Non-Tg (n = 1,782 cells) and 5xFAD (n = 2,870 cells) mice. c, Relative percentages (normalized to Non-Tg mice) of TNF-α+ and pro-IL-1β+ microglia from 10-month-old Non-Tg (n = 15) and 5xFAD (n = 21) mice. d, Immunofluorescence images of brain CD3+ T cells in proximity to Aβ or Iba1+ microglia in 10-month-old 5xFAD mice. Scale bar, 20 μm. e, Immunofluorescence image of brain CD8+ T cells (red), Aβ (white), and Iba1+ microglia (green) in 10-month-old 5xFAD;Cxcr6−/− mice. Scale bar, 20 μm. f, g, Spatial co-localization index between CD8+ T cells (CD8) and activated microglia (Iba1) (f) or CD8 and Aβ plaques (g) within the brains of 5xFAD;Cxcr6+/+ (n = 105 scored interactions) and 5xFAD;Cxcr6−/− (n = 71 scored interactions) mice. The index values for each target–pair were compared by genotype (P values shown in black color) and against simulated random distribution (P values shown in red or green color) (see Methods). h, Immunofluorescence images of PD-1 (green) and CD8 (red) co-expressing T cells in proximity to Iba1+ microglia (white) in 10-month-old 5xFAD mice. Scale bar, 5 μm. i, Heatmap showing the differential interaction strength, predicted by CellChat, between T cell subsets and microglia in indicated mice. Row z score indicates the differential interaction strengths. j, k, Relative percentages (normalized to sex-matched controls) of pro-IL-1β+ microglia (j, k) and geometric mean fluorescence intensity (gMFI; j, k) from 10-month-old 5xFAD;B2m+/+ (percentage, n = 19 for female and 11 for male; gMFI, n = 19 for female and 9 for male) and 5xFAD;B2m−/− (percentage, n = 19 for female and 16 for male; gMFI, n = 19 for female and 13 for male) mice (j), or 5xFAD;Cxcr6+/+ (n = 10 for female and 16 for male) and 5xFAD;Cxcr6−/− (n = 14 for female and 24 for male) mice (k). l, m Frequencies of M0 and DAM subclusters from scRNA-seq analysis of microglia from indicated mice. n, Functional enrichment analysis of upregulated genes in M0 and DAM from 5xFAD;Cxcr6−/− versus 5xFAD;Cxcr6+/+ mice. Data were analyzed by two-tailed normalized weighted Kolmogorov–Smirnov test (NES for a), two-tailed Benjamini–Hochberg adjusted P value (FDR for a), two-tailed unpaired Student’s t-test (b, c, f, g, j, k), or two-tailed Fisher’s exact test (n). Data are shown as mean ± s.e.m. (c, j, k) or medians with quartiles (f, g); NS, not significant. Data were pooled from at least three (c, j, k) or are representative of three (d, e) or two (f–h) independent experiments.
Extended Data Fig. 10. Brain CD8+ T cells mitigate pro-inflammatory features of microglia and limit microgliosis in 5xFAD mice.
a, PD-1+CD8+ T cells among freshly isolated splenic CD44hiCD8+ T cells from C57BL/6 mice [Fresh (n = 13)] and splenic CD44hiCD8+ T cells after cytokine culture [Cultured (n = 13)] (see Methods). b, Indicated marker expression on PD-1+ versus PD-1− CD44hiCD8+ T cells from 5xFAD mice (n = 20) after cytokine culture. c, TNF-α+ cells in microglia after in vitro co-culture with indicated in vitro-expanded, spleen-derived CD8+ T cell populations (see Methods for details). Stimulated condition indicates treatment with PMA and ionomycin plus 1 μM Aβ. [Unstimulated (n = 6); Stimulated (n = 5); Stimulated + cultured CXCR6+CD8+ T cells (n = 12); Stimulated + cultured CXCR6−CD8+ T cells (n = 12)]. d, TNF-α+ cells in microglia after in vitro co-culture with brain CD8+ T cells from the indicated mice [Unstimulated (n = 28); Stimulated (n = 28); Stimulated + brain-derived 5xFAD;Nt5e+/+ CD8+ T cells (n = 9); Stimulated + brain-derived 5xFAD;Nt5e−/− CD8+ T cells (n = 4)]. e–g, Immunohistochemical images of Iba1+ microglia in brains of indicated mice. Scale bar, 200 μm (e) or 100 μm (f, g). h, i, Immunofluorescence analysis of Iba1+ microglia or Aβ-plaque-associated Iba1+ microglia from 10-month-old male 5xFAD;B2m+/+ (n = 4 sections from 3 mice) and 5xFAD;B2m−/− (n = 6 sections from 3 mice) mice (h), or male 5xFAD;Cxcr6+/+ (n = 6 sections from 3 mice) and 5xFAD;Cxcr6−/− (n = 6 sections from 3 mice) mice (i). Scale bar, 100 μm. j, k, Immunofluorescence analysis of Iba1+ microglia cells associated with individual Aβ plaques in 4-month-old (j) or 10-month-old (k) female and male 5xFAD;B2m+/+ (n = 22 sections for 4-month-old female, 10 sections for 4-month-old male, 10 sections for 10-month-old female, 4 sections for 10-month-old male) and 5xFAD;B2m−/− (n = 25 sections for 4-month-old female, 8 sections for 4-month-old male, 10 sections for 10-month-old female, 6 sections for 10-month-old male) mice. l, Model for CD8+ T cell-dependent suppression of pro-inflammatory microglia. CXCR6+CD8+ T cells accumulate in the brains but not meninges of 5xFAD mice, where they co-localize with pro-inflammatory microglia near Aβ plaques. CXCL16-expressing microglia are present in the brains of individuals with AD and mouse models, and CXCR6 is required for CD8+ T cells to undergo enhanced tissue residency programing and clonal expansion (as marked by PD-1 expression) in the brain. These PD-1+CD8+ T cells limit pro-inflammatory microglia that contribute to Aβ plaque accumulation and cognitive impairments. Data were analyzed by two-tailed unpaired Student’s t-test (a, b, h–k) or one-way ANOVA (c, d). Data are shown as mean ± s.e.m. in a–d and h–k; NS, not significant. Data were pooled from at least four (d), three (b), two (a, c), or one (j, k), or are representative of two (e–i) independent experiments.
Supplementary Material
Acknowledgments
The authors acknowledge M. Hendren and R. Walton for animal colony management and animal behavior; P. Zhou and L. Long for performing tail vein injections; S. Yuan for tissue processing; J. Raynor and X. Sun for scientific insights; B. Heckmann for discussion of animal behavior experiments; Dr. Takaomi C. Saido (RIKEN Center for Brain Science, Saitama, Japan) for providing APPNL-G-F mice; and St. Jude Immunology flow cytometry core facility for cell sorting. This work was supported by ALSAC, the Arizona Department of Health Services, the Arizona Research Commission, the Michael J. Fox Foundation for Parkinson’s Research, and US National Institutes of Health grants AG072980, NS072026, and AG19610 (to T.G.B.), AG068581 (to J.P.) and AI131703 (to H.C.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Competing interests
H. Chi is a consultant for Kumquat Biosciences. The other authors have no competing interests to declare.
Data availability
The authors declare that data supporting the findings of this study are available within the paper. All scRNA-seq and scTCR-seq data described in the manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number GSE207702. Public data analyzed in this manuscript are also available under the GEO accession codes GSE73721 (ref. 24), GSE157827 (ref. 25), GSE98971 (for DAM cell signature; ref. 10), GSE107395 (for TRM signature; ref. 30), and GSE130975 (for regulatory CD8+ T cell signature; ref. 36), downloaded from SRA accession number PRJNA529095 (ref. 11), or the PRIDE database (http://www.proteomexchange.org) with accession number PXD007985 (ref. 22). Publicly available databases used in this study are available at the indicated locations: MSigDB Collections (for Hallmark, CP: Reactome, and C5: GO:BP), http://www.gsea-msigdb.org/gsea/msigdb/collections.jsp; GLIPH2 database (http://50.255.35.37:8080/). All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
- 1.Bettcher BM, Tansey MG, Dorothee G & Heneka MT Peripheral and central immune system crosstalk in Alzheimer disease - a research prospectus. Nat Rev Neurol 17, 689–701, doi: 10.1038/s41582-021-00549-x (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leng F & Edison P Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat Rev Neurol 17, 157–172, doi: 10.1038/s41582-020-00435-y (2021). [DOI] [PubMed] [Google Scholar]
- 3.Gate D. et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer's disease. Nature 577, 399–404, doi: 10.1038/s41586-019-1895-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phongpreecha T. et al. Single-cell peripheral immunoprofiling of Alzheimer's and Parkinson's diseases. Sci Adv 6, eabd5575, doi: 10.1126/sciadv.abd5575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh SE et al. The adaptive immune system restrains Alzheimer's disease pathogenesis by modulating microglial function. Proc Natl Acad Sci U S A 113, E1316–1325, doi: 10.1073/pnas.1525466113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baruch K. et al. Breaking immune tolerance by targeting Foxp3(+) regulatory T cells mitigates Alzheimer's disease pathology. Nat Commun 6, 7967, doi: 10.1038/ncomms8967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim K. et al. Therapeutic B-cell depletion reverses progression of Alzheimer's disease. Nat Commun 12, 2185, doi: 10.1038/s41467-021-22479-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X. et al. Microglia-mediated T cell infiltration drives neurodegeneration in tauopathy. Nature 615, 668–677, doi: 10.1038/s41586-023-05788-0 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olah M. et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer's disease. Nat Commun 11, 6129, doi: 10.1038/s41467-020-19737-2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Keren-Shaul H. et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer's Disease. Cell 169, 1276–1290 e1217, doi: 10.1016/j.cell.2017.05.018 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Van Hove H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci 22, 1021–1035, doi: 10.1038/s41593-019-0393-4 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Da Mesquita S. et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer's disease. Nature 560, 185–191, doi: 10.1038/s41586-018-0368-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oakley H. et al. Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer's disease mutations: potential factors in amyloid plaque formation. J Neurosci 26, 10129–10140, doi: 10.1523/JNEUROSCI.1202-06.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito T. et al. Single App knock-in mouse models of Alzheimer's disease. Nat Neurosci 17, 661–663, doi: 10.1038/nn.3697 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Unger MS et al. CD8(+) T-cells infiltrate Alzheimer's disease brains and regulate neuronal- and synapse-related gene expression in APP-PS1 transgenic mice. Brain Behav Immun 89, 67–86, doi: 10.1016/j.bbi.2020.05.070 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Alves de Lima K, Rustenhoven J & Kipnis J Meningeal Immunity and Its Function in Maintenance of the Central Nervous System in Health and Disease. Annu Rev Immunol 38, 597–620, doi: 10.1146/annurev-immunol-102319-103410 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Da Mesquita S. et al. Meningeal lymphatics affect microglia responses and anti-Abeta immunotherapy. Nature 593, 255–260, doi: 10.1038/s41586-021-03489-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheltens P. et al. Alzheimer's disease. Lancet 397, 1577–1590, doi: 10.1016/S0140-6736(20)32205-4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miedel CJ, Patton JM, Miedel AN, Miedel ES & Levenson JM Assessment of Spontaneous Alternation, Novel Object Recognition and Limb Clasping in Transgenic Mouse Models of Amyloid-beta and Tau Neuropathology. J Vis Exp 123, 55523, doi: 10.3791/55523 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAlpine CS et al. Astrocytic interleukin-3 programs microglia and limits Alzheimer's disease. Nature 595, 701–706, doi: 10.1038/s41586-021-03734-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L. et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 181, 442–459 e429, doi: 10.1016/j.cell.2020.03.048 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Bai B. et al. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer's Disease Progression. Neuron 105, 975–991 e977, doi: 10.1016/j.neuron.2019.12.015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bai B. et al. Proteomic landscape of Alzheimer's Disease: novel insights into pathogenesis and biomarker discovery. Mol Neurodegener 16, 55, doi: 10.1186/s13024-021-00474-z (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y. et al. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron 89, 37–53, doi: 10.1016/j.neuron.2015.11.013 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lau SF, Cao H, Fu AKY & Ip NY Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer's disease. Proc Natl Acad Sci U S A 117, 25800–25809, doi: 10.1073/pnas.2008762117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan P. et al. TREM2 Haplodeficiency in Mice and Humans Impairs the Microglia Barrier Function Leading to Decreased Amyloid Compaction and Severe Axonal Dystrophy. Neuron 90, 724–739, doi: 10.1016/j.neuron.2016.05.003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glanville J. et al. Identifying specificity groups in the T cell receptor repertoire. Nature 547, 94–98, doi: 10.1038/nature22976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chronister WD et al. TCRMatch: Predicting T-Cell Receptor Specificity Based on Sequence Similarity to Previously Characterized Receptors. Front Immunol 12, 640725, doi: 10.3389/fimmu.2021.640725 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529, doi: 10.1016/j.cell.2021.04.048 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milner JJ et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature 552, 253–257, doi: 10.1038/nature24993 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kok L, Masopust D & Schumacher TN The precursors of CD8(+) tissue resident memory T cells: from lymphoid organs to infected tissues. Nat Rev Immunol, doi: 10.1038/s41577-021-00590-3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pasciuto E. et al. Microglia Require CD4 T Cells to Complete the Fetal-to-Adult Transition. Cell 182, 625–640 e624, doi: 10.1016/j.cell.2020.06.026 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piehl N. et al. Cerebrospinal fluid immune dysregulation during healthy brain aging and cognitive impairment. Cell 185, 5028–5039 e5013, doi: 10.1016/j.cell.2022.11.019 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X. et al. Unbiased and robust analysis of co-localization in super-resolution images. Stat Methods Med Res 31, 1484–1499, doi: 10.1177/09622802221094133 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin S. et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 12, 1088, doi: 10.1038/s41467-021-21246-9 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saligrama N. et al. Opposing T cell responses in experimental autoimmune encephalomyelitis. Nature 572, 481–487, doi: 10.1038/s41586-019-1467-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dudek M. et al. Auto-aggressive CXCR6(+) CD8 T cells cause liver immune pathology in NASH. Nature 592, 444–449, doi: 10.1038/s41586-021-03233-8 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Di Pilato M. et al. CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 184, 4512–4530 e4522, doi: 10.1016/j.cell.2021.07.015 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hemonnot AL, Hua J, Ulmann L & Hirbec H Microglia in Alzheimer Disease: Well-Known Targets and New Opportunities. Front Aging Neurosci 11, 233, doi: 10.3389/fnagi.2019.00233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heckmann BL et al. LC3-Associated Endocytosis Facilitates beta-Amyloid Clearance and Mitigates Neurodegeneration in Murine Alzheimer's Disease. Cell 178, 536–551 e514, doi: 10.1016/j.cell.2019.05.056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen DV, Hanson JE & Sheng M Microglia in Alzheimer's disease. J Cell Biol 217, 459–472, doi: 10.1083/jcb.201709069 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarlus H & Heneka MT Microglia in Alzheimer's disease. J Clin Invest 127, 3240–3249, doi: 10.1172/JCI90606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wendeln AC et al. Innate immune memory in the brain shapes neurological disease hallmarks. Nature 556, 332–338, doi: 10.1038/s41586-018-0023-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heneka MT et al. NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678, doi: 10.1038/nature11729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chapman NM & Chi H Metabolic adaptation of lymphocytes in immunity and disease. Immunity 55, 14–30, doi: 10.1016/j.immuni.2021.12.012 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altendorfer B. et al. Transcriptomic Profiling Identifies CD8(+) T Cells in the Brain of Aged and Alzheimer's Disease Transgenic Mice as Tissue-Resident Memory T Cells. J Immunol 209, 1272–1285, doi: 10.4049/jimmunol.2100737 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smolders J. et al. Tissue-resident memory T cells populate the human brain. Nat Commun 9, 4593, doi: 10.1038/s41467-018-07053-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alves de Lima K. et al. Meningeal gammadelta T cells regulate anxiety-like behavior via IL-17a signaling in neurons. Nat Immunol 21, 1421–1429, doi: 10.1038/s41590-020-0776-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baruch K. et al. PD-1 immune checkpoint blockade reduces pathology and improves memory in mouse models of Alzheimer's disease. Nat Med 22, 135–137, doi: 10.1038/nm.4022 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Li J. et al. KIR(+)CD8(+) T cells suppress pathogenic T cells and are active in autoimmune diseases and COVID-19. Science 376, eabi9591, doi: 10.1126/science.abi9591 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 51.Derecki NC et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J Exp Med 207, 1067–1080, doi: 10.1084/jem.20091419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhat SA, Sood A, Shukla R & Hanif K AT2R Activation Prevents Microglia Pro-inflammatory Activation in a NOX-Dependent Manner: Inhibition of PKC Activation and p47(phox) Phosphorylation by PP2A. Mol Neurobiol 56, 3005–3023, doi: 10.1007/s12035-018-1272-9 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Beach TG et al. Arizona Study of Aging and Neurodegenerative Disorders and Brain and Body Donation Program. Neuropathology 35, 354–389, doi: 10.1111/neup.12189 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420, doi: 10.1038/nbt.4096 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502, doi: 10.1038/nbt.3192 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borcherding N, Bormann NL & Kraus G scRepertoire: An R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47, doi: 10.12688/f1000research.22139.2 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, Wang C, Rubelt F, Scriba TJ & Davis MM Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat Biotechnol 38, 1194–1202, doi: 10.1038/s41587-020-0505-4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korsunsky I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16, 1289–1296, doi: 10.1038/s41592-019-0619-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramilowski JA et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 6, 7866, doi: 10.1038/ncomms8866 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vento-Tormo R. et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 563, 347–353, doi: 10.1038/s41586-018-0698-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reimand J. et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 14, 482–517, doi: 10.1038/s41596-018-0103-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that data supporting the findings of this study are available within the paper. All scRNA-seq and scTCR-seq data described in the manuscript have been deposited in the NCBI Gene Expression Omnibus (GEO) database and are accessible through the GEO SuperSeries accession number GSE207702. Public data analyzed in this manuscript are also available under the GEO accession codes GSE73721 (ref. 24), GSE157827 (ref. 25), GSE98971 (for DAM cell signature; ref. 10), GSE107395 (for TRM signature; ref. 30), and GSE130975 (for regulatory CD8+ T cell signature; ref. 36), downloaded from SRA accession number PRJNA529095 (ref. 11), or the PRIDE database (http://www.proteomexchange.org) with accession number PXD007985 (ref. 22). Publicly available databases used in this study are available at the indicated locations: MSigDB Collections (for Hallmark, CP: Reactome, and C5: GO:BP), http://www.gsea-msigdb.org/gsea/msigdb/collections.jsp; GLIPH2 database (http://50.255.35.37:8080/). All other data supporting the findings of this study are available from the corresponding author on reasonable request. Source data are provided with this paper.