Abstract
Multiplexed imaging and spatial transcriptomics enable highly resolved spatial characterization of cellular phenotypes, but still largely depend on laborious manual annotation to understand higher-order patterns of tissue organization. As a result, higher-order patterns of tissue organization are poorly understood and not systematically connected to disease pathology or clinical outcomes. To address this gap, we developed an approach called UTAG to identify and quantify microanatomical tissue structures in multiplexed images without human intervention. Our method combines information on cellular phenotypes with the physical proximity of cells to accurately identify organ-specific microanatomical domains in healthy and diseased tissue. We apply our method to various types of images across healthy and disease states to show that it can consistently detect higher-level architectures in human tissues, quantify structural differences between healthy and diseased tissue, and reveal tissue organization patterns at the organ scale.
The recent development of technologies such as multiplexed imaging1–5 and spatial transcriptomics6–10 allows for both direct observation of cellular phenotypes and cellular interactions in intact tissues. Although these technologies provide a highly resolved view of cellular heterogeneity in tissues, they struggle to move beyond a cell-centric view of tissue, failing to uncover organizing principles of tissue architecture and tissue-specific physiology which are encoded at various scales of cellular and extracellular interactions. Understanding higher-level patterns of tissue and organ organization would be crucial to establishing a relationship between cellular phenotypes and organ-specific tissue physiology.
Visual inspection of histopathological images of biopsied or surgically removed tissue is a major component of disease diagnosis, but is a labor intensive job that requires manual annotations from specialized pathologists. Also, the process may require multiple specialists to reduce intra- and inter-observer variability. To assist and improve upon the inspection process, computational techniques have been developed for the automated detection and quantification of cells or tissue structures11–13, often in a supervised manner that requires manual annotations as training data. This approach is expensive and laborious, prone to learning biases from training data, and hard to employ with exceptionally abundant tissue features such as small capillaries or individual ducts in submucosal glands. Unsupervised methods try to accomplish similar tasks without the need for manual input. A popular method is the inference of cell neighborhoods based on multiplexed data by assembling a graph of cellular interactions based on physical proximity14,15. Clustering of cells based on these interactions yields cellular neighborhoods predictive of patient survival14–17. However, graph clustering per se does not make use of cell type identities or phenotypes and has only been applied to cancer tissue.
Recent studies applying unsupervised deep learning models to histopathological images such as hematoxylin eosin staining have shown that it is possible to extract morphological features that are, for example, predictive of gene expression18. Other studies have also employed deep learning of graphs of cellular proximity with cellular phenotypes for cell type prediction19, inference of cellular communication20 and data exploration21. These models are computationally expensive to train and their results heavily depend on training data, which may preclude joint analysis of expression and morphological features across studies and data types. There is thus a need for unsupervised, broadly applicable methods of tissue structure detection across organs and imaging modalities that incorporate cellular proximity, expression and morphological features. Here we present an accurate method to perform discovery and quantification of microanatomical tissue structures in multiplexed histopathological images without human intervention or prior knowledge. Our method, unsupervised discovery of tissue architecture with graphs (UTAG), combines information on cellular morphology and expression with the physical proximity of cells to discover domains of tissue architecture. We demonstrate that our approach is able to discover organ-specific microanatomical domains in the human lung with high accuracy in comparison with manual annotations and outperforms other methods. Furthermore, UTAG can be employed across various physiological states, such as infectious disease and cancer, and its results can reveal the high-level organization of tissues at the whole-organ scale.
Results
Unsupervised discovery of tissue architecture with graphs
To address the problem of discovery of microanatomical structure in tissue across data types and biological systems we developed a method called UTAG (Fig. 1a). Our method is generally applicable to images of cells in their native tissue context collected via highly multiplexed single-cell imaging data such as codetection by indexing (CODEX), cyclic immunofluorescence (CyCIF), imaging mass cytometry (IMC), multiplexed ion beam imaging (MIBI) and likewise multiplexed spatial platforms. The central aspect of UTAG is the combination of two matrices that represent phenotypic and positional information about each cell present in an image (Fig. 1a, gray areas), to generate a new feature space that encodes spatially aggregated phenotypic information. This matrix of new features can then be clustered into domains of cells that are both phenotypically and spatially related (Fig. 1a, orange area). The matrix of phenotypic information (feature matrix) is a numeric matrix of gene or protein abundance, or morphology for each cell, while the positional information of each cell is used to generate a graph of physical proximity between cells through binarization and optional normalization (adjacency matrix).
Fig. 1 |. Unsupervised discovery of tissue architecture with graphs.
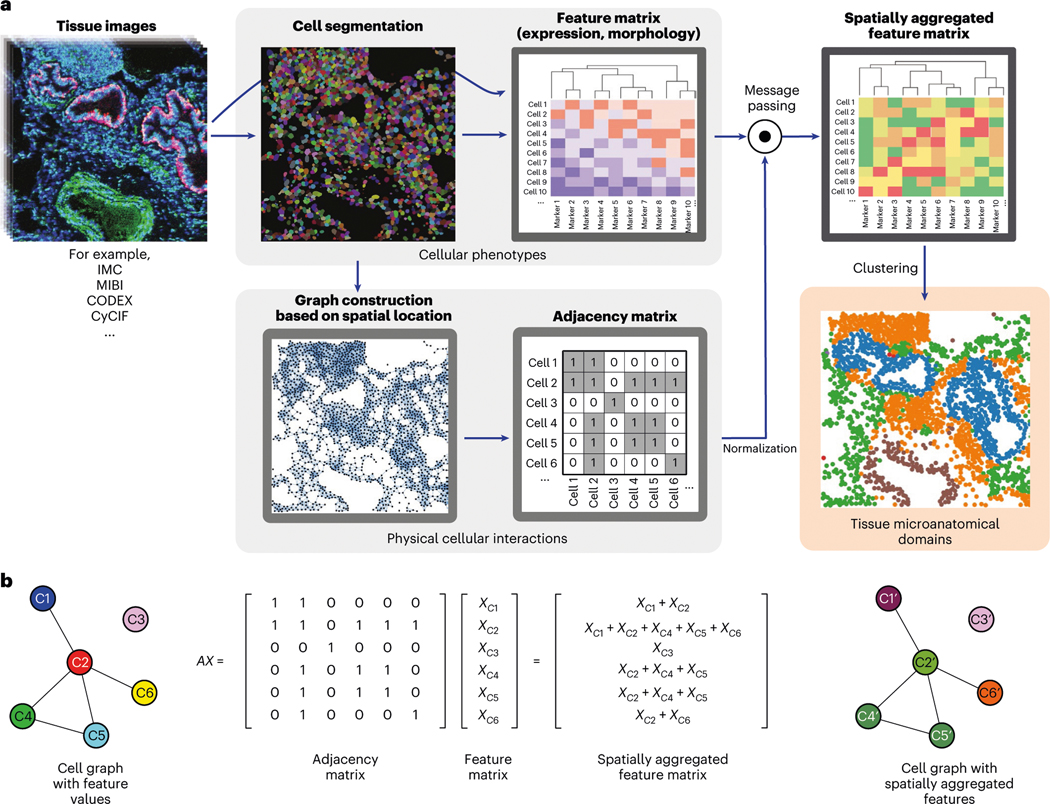
a, Schematic description of the methodology for the discovery of domains of tissue microanatomy and architecture using graphs of cellular interactions. Intensity values and cellular segmentation masks are used to derive an expression matrix containing the intensity of each marker in each cell and a graph of physical cellular interaction based on proximity, which can be represented as a binary adjacency matrix. Message passing (described in b) combines the expression and adjacency matrices into a new matrix of spatially aggregated expression values which serves as the input for clustering methods. The resulting clusters represent domains of tissue microanatomy underlying the tissue architecture. The procedure can be performed jointly across several images, yielding consistent microanatomical domains across images. b, Graphical description of the message passing procedure, in which the adjacency and expression matrices are combined with the dot product. Note how in the message-passed graph, the node colors are linear combinations of the colors of the nodes with which they share edges. Each element in the feature matrix in this example depicts a vector of features.
UTAG then leverages the properties of matrix multiplication through linear algebra to combine the matrices in a procedure known as message passing (Fig. 1b). In this, nodes of cells in physical proximity will receive a portion of the neighboring cell’s phenotypic information in a weighted manner, effectively diffusing the phenotypes into physically proximal cells determined by the adjacency matrix. The intermediate resulting spatially aggregated features therefore contain information on both cellular phenotypes and physical proximity between cells. This spatially aggregated feature matrix allows capture of microanatomical domains consisting of multiple cell types that are spatially homogeneously distributed. For example, arteries consist of a layer of endothelial cells surrounded by smooth muscle cells. Through message passing, endothelial cells become more like adjacent muscle cells and vice versa, effectively grouping cells with different phenotypic features based on their spatial distribution. Finally, this matrix is clustered using standard modern algorithms such as Leiden22 or Phenotyping by Accelerated Refined Community-Partitioning (PARC)23 clustering to derive domains of tissue structure in images (Fig. 1a, orange area). In this process, the number of captured domains is determined by a customizable resolution hyperparameter, which controls the coarseness in both Leiden and PARC clustering (Extended Data Fig. 1b). Biological interpretation of the discovered domains remains, however, dependent on the user by contextualization in terms of their cell type composition, frequency of cellular interactions or association with target variables such as clinically relevant outcomes. We provide a software package for the implementation of UTAG, including documentation and tutorials on its application to various datasets (https://github.com/ElementoLab/utag).
UTAG uncovers microanatomical principles in healthy lung
We first tested UTAG on healthy lung tissue images. The human lung is a highly compartmentalized tissue, with the organ physiology dictating an intricate interplay between cells and matrix to create functional structures such as the airway lumen, alveolar airspace and blood vessels. We applied UTAG to a dataset of 26 highly multiplexed IMC lung images from three donor lung specimens, consisting of 28 markers, with a particular focus on airways extending from proximal bronchi and succeeding divisions to terminal and respiratory bronchioles24 (Fig. 2a, first column). Importantly, in this dataset, each image has been manually annotated with organ-specific microanatomical domains such as airways, connective tissue, submucosal glands, vessels and alveolar space (Fig. 2a, fourth column). The annotated structures effectively serve as a reference for microanatomical annotation of the lung. In addition, the cells in these data had been phenotyped into seven broad clusters of cell identity (Fig. 2a, second column), which can be helpful when interpreting the composition of the domains, albeit not used by UTAG.
Fig. 2 |. Discovery of microanatomical domains and principles of tissue architecture in human lung.
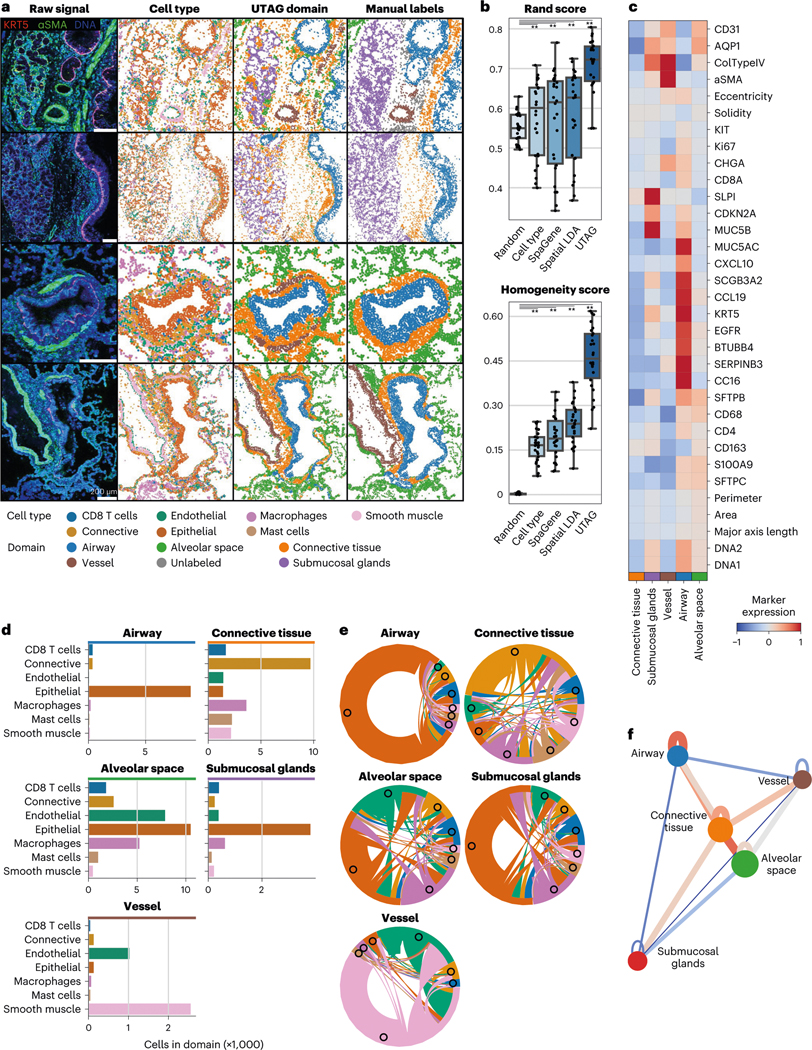
a, Microanatomical domains detected in IMC images of healthy human lung tissue. The first column illustrates the intensity of three selected channels in four representative images; the second column, the cell identity of the cells in those images; the third column displays the microanatomical domains discovered with UTAG; and the fourth column displays the microanatomical domains manually annotated by experts. Scale bars, 200 μm. b, Benchmarking of UTAG and competing methods against expert annotation. n = 26 highly multiplexed IMC lung images from three donor specimens. For a baseline comparison, we include randomized domain labels per cell and cell type identities. Each point represents one image and for both metrics values closer to 1 are optimal.** P < 0.0001, two-sided Mann-Whitney U-test after Benjamini-Hochberg P-value correction. Data in boxplots are presented by minimum, 25th percentile, median, 75th percentile and maximum. Values outside of 1.5 times interquartile range are classified as outliers and are denoted as fliers. c, Mean channel intensity for all channels aggregated by the discovered microanatomical domains. d, Cellular composition of microanatomical domains. e, Composition of microanatomical domains in terms of intercellular interactions derived from physical proximity. f, Model of physical proximity between microanatomical domains in the lung. The nodes of the graph represent the microanatomical domains, and the color of the edges between them show the strength and direction of their physical interactions. The node position is determined based on the edge weight using the Spring force-directed algorithm.
We applied UTAG to the IMC data by providing the position of the cells in the image and the intensity of each marker in each cell to the algorithm. We then labeled the resulting clusters with identities, splitting them into five groups depending on the intensity of markers and cellular composition (Extended Data Fig. 1c). The resulting microanatomical domains detected by UTAG largely recapitulated the microanatomy of manually labeled domains (Fig. 2a, third column, and Extended Data Fig. 2). To assess the performance of our method, we compared the discovered microanatomical domains with the labels applied by experts using Rand and Homogeneity score based on cell domain properties (Fig. 2b). Rand score measures label agreement and is a commonly used metric to benchmark unsupervised clustering. Homogeneity score assesses how uniquely each cluster maps to ground truth and does not penalize for detection of more granular subdomains. As a baseline comparison, we calculated the same metrics based on randomly shuffled domain labels and cell type identities. In addition, we also compare UTAG to other methods for inference of higher-level tissue structure in terms of their features and performance, such as SpaGene25 and SpatialLDA26 (Fig. 2b and Extended Data Fig. 3). UTAG is the only method that can infer microanatomical domains without cell type annotations jointly across images; most other methods focus on generating per-image results and may only be applicable to certain data types due to specific assumptions on the data (Supplementary Table 1). Furthermore, UTAG significantly outperformed all other methods both in terms of label Rand and Homogeneity score (Fig. 2b and Extended Data Fig. 3). UTAG outperformed SpatialLDA, the next best performing algorithm, by 2.42 fold (Homogeneity score), which shows that UTAG can discover accurate microanatomical domains in multiplexed imaging data.
The domains discovered by UTAG were enriched in protein expression specific to each domain, as evidenced by KRT5, CC16, SCGB3A2, MUC5B and MUC5AC expression in airways, or CD31, alpha smooth muscle actin (aSMA) and type IV collagen in vessels (Fig. 2c). Furthermore, we found the cell type composition to reflect the captured domains. Airways and submucosal glands consisted predominantly of epithelial cell types while being spatially distinct. Connective tissues were generally composed of sparse matrices of cells that had low expressions of all markers (Fig. 2c), but sometimes included supportive muscles and infiltrating immune cells (Fig. 2d). Other identified domains were well-balanced in terms of cell type composition. The alveolar space included a well-balanced proportion of epithelial and endothelial cells required for gas exchange (Fig. 2d). This reveals that UTAG, without specific training, is capable of effectively capturing both simple domains with a dominant cell type and more complex domains composed of multiple cell types. Beyond cell type composition, we identified distinct differences in the frequency of physical interactions between cells of different cell types across microanatomical domains (Fig. 2e). In airways, we observed a tight connection between epithelial cells and reciprocal proximity between epithelial and connective tissue. The connective tissue, as a transition tissue between airways and other functional domains in the lung, showed high diversity and balance in cellular interactions. The alveolar space domain has strong reciprocal interactions between epithelial and endothelial cells, which is a hallmark of alveolar type 1 cells closely connected to capillary endothelium. Taken together, the observed cell type abundance (Fig. 2d) and interaction relationships (Fig. 2e) within the microanatomical domains of the lung provide a signature of the architecture of the healthy human lung.
While the composition of an organ in microanatomical domains is an important part of its architecture, it is also important to understand the wider-scale architecture of an organ in relation to its physiology. To demonstrate how UTAG can be useful in uncovering organ-specific high-level architecture, we quantified physical interactions between microanatomical domains in IMC images and related domains based on the frequency of interactions (Fig. 2f). The resulting network, made by associating the strength of microanatomical domain interaction with attraction between nodes, summarizes the architecture of the lung—with a main anatomical axis of high-order tissue assembly from airway, connective tissue to alveolar space (Fig. 2f). Furthermore, we also found that both vessels and submucosal glands, while interacting with similar domains, are diametrically opposed to the main axis (Fig. 2f), which may suggest that segregation of vascular and secretory domains of the lung is a hallmark of healthy lung architecture. Overall, the microanatomical domains detected by UTAG in the lung, along with the inferred high-level structure of the organ, illustrate the accuracy and utility of UTAG in understanding tissue architecture at various scales with a completely unsupervised approach.
UTAG captures structural changes in diseased lung tissue
Having established the performance and usefulness of UTAG in multiplexed imaging of healthy tissue, we sought to determine whether UTAG is able to discover microanatomical domains in disease as well. To that end, we ran UTAG on a dataset of 239 IMC images with 37 markers from 27 deceased patients due to lung infection27 (Fig. 3a). Despite using a different set of markers from the healthy lung dataset (Fig. 2a), we were able to discover six largely similar microanatomical domains that were present in images of various disease groups: one domain representative of epithelial cells (predominantly airways), one domain of fibroblast-rich connective tissue, one domain for alveolar regions, one for vessels, one with clusters of various immune cells and a rare one of clustered neutrophils exclusively. Their relative abundance, however, reflects the changes in the morphology and cellular composition of the tissue after infection27, with, for example, an increased proportion of the epithelial domain following influenza and in late COVID-19, and an increase in the fraction of connective tissue in late COVID-19 that is indicative of fibrosis (Fig. 3b). Since topological domains aggregate spatially proximal cells of various cell types that contribute to tissue function, we hypothesized that the abundance of topological domains across images more easily explains the variance in the dataset than the abundance of cell types on their own. Indeed, in a principal component analysis (PCA) reduction of the data, we found that not only was the fraction of variance in the first component higher with topological domains, but they also more easily reconstructed the linear progression of healthy tissue in comparison with cell type identities alone (Fig. 3c).
Fig. 3 |. Microanatomical domains discovered by UTAG across data types and disease states.
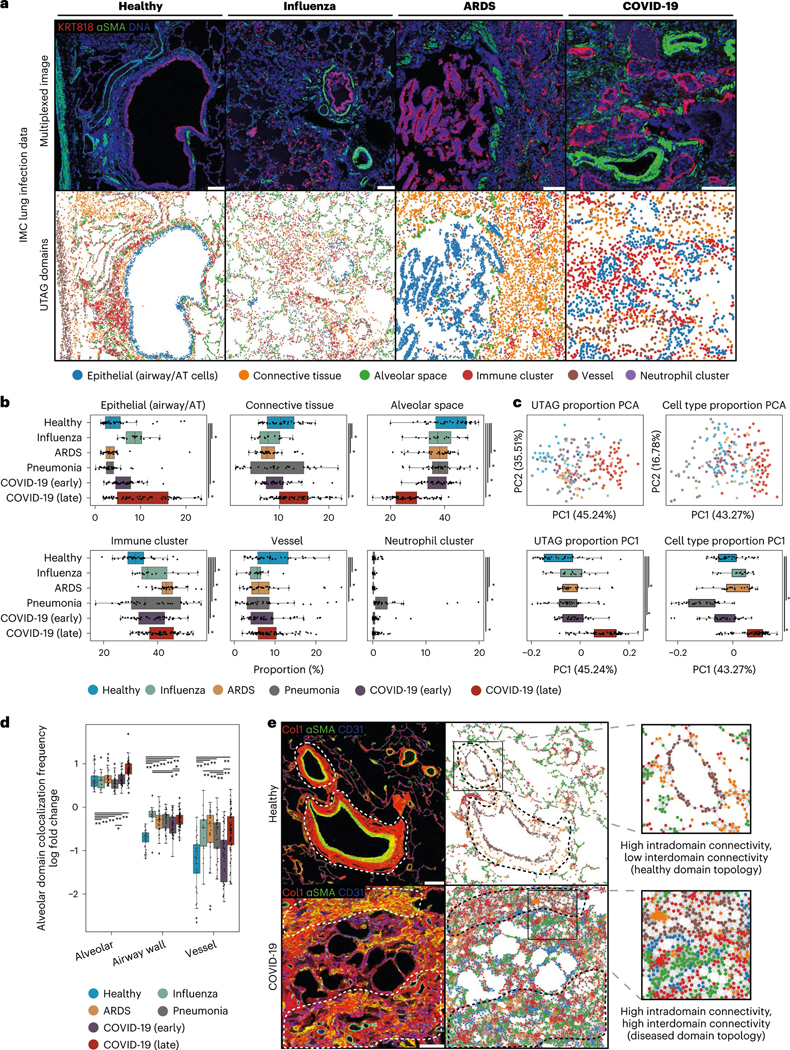
a, Discovery of microanatomical domains in IMC images of lung from patients of various pathologies. The top row illustrates the intensity of three selected channels and the bottom row displays the UTAG domains. Scale bars, 200 μm. b, Univariate analysis of microanatomical domain composition across lung infection disease. Microanatomical domain composition was percent normalized per slide. c, PCA for joint analysis of domain (left) or cell type (right) composition per image. The top two plots visualize the position of images in the first two principal components. The bottom two plots show the distribution of the first principal component aggregated by disease group. d, Log odds of domain colocalization frequencies across diseases in alveolar domains. Log odds indicates observed frequency over expected, as estimated empirically by random permutation. Positive values indicate high intradomain (alveolar–alveolar) colocalization compared to random mixtures and negative indicates low interdomain colocalization. **P < 0.01, *P < 0.05, two-sided Mann-Whitney U-test after Benjamini-Hochberg adjustment. e, IMC images of healthy and COVID-19 infected lung tissue. The image of healthy lung tissue shows highly compartmentalized domains, particularly in the vasculature, while the image of the diseased lung shows loss of compartmentalization. Scale bars, 200 μm. For b and c, n = 239 highly multiplexed IMC lung images from 27 deceased patients due to lung infection. *P < 0.05, two-sided Mann-Whitney U-test after Benjamini-Hochberg adjustment. Data in boxplots are presented by minimum, 25th percentile, median, 75th percentile and maximum. Values outside of 1.5 times interquartile range are classified as outliers and are denoted as fliers. ARDS, acute respiratory distress syndrome.
Since differences in cell type composition during lung infection have been reported27, we sought to investigate whether there are differences in the high-level composition of tissue, as quantified by the spatial proximity in topological domains across images (Extended Data Fig. 4). The most prominent differences in topological domain colocalization between disease states was observed between the alveolar space and vessel domains (Fig. 3d). In influenza, acute respiratory distress syndrome and late COVID-19, vessel domains interact with alveolar domains more tightly than in healthy lung or early COVID-19. In healthy lung sections, vessels often have high intradomain connectivity and are isolated from other domains, whereas in late COVID-19 lung sections we observed high connectivity of vessels with other domains, particularly the alveolar space (Fig. 3e). This likely reflects the previously described increase in vasculature due to pathology-induced angiogenesis28,29. The characterization of microanatomy across various disease states in the lung, along with the discovery of changes in the connectivity of tissue domains, demonstrate the versatility of unsupervised approaches such as UTAG to detect and quantify microanatomical structure in human tissue.
UTAG is applicable across imaging techniques and tissues
We have so far employed UTAG in the lung because we have annotated images allowing us to assess whether the discovered microanatomy aligns with current knowledge in the field. Given that UTAG is an unsupervised method, it is not guaranteed that its use across data types, organs and disease states will always discover microanatomical domains with physiological relevance or of pathologic interest.
To address whether UTAG generalizes to various types of multiplexed imaging data, we first applied it to a dataset of 19 CyCIF images with26markers from three lung cancerpatients30 (Extended Data Fig. 5a). We observed that the obtained domains largely reflected tumor or stromal microenvironments reflecting a complete departure from the tissue architecture seen in normal lung. This is likely due to proliferation of neoplastic cells, which is independent of the normal physiological function of the lung. In this setting, UTAG may be of use in cancer by detecting the interface between tumor and stromal, facilitating the investigation of cellular composition and interactions at this interface, without the need for manual annotation of images by an expert.
Second, to assess whether UTAG is capable of broadly discovering microanatomy across organs, we apply it to a set of 15 IMC images of COVID-19-infected intestine31 (Extended Data Fig. 5b). UTAG was able to clearly demarcate the intestinal epithelium from the remaining parenchyma and, within the epithelium, further differentiate between the E-cadherin-expressing enterocytes at the top of the villi and the proliferating(Ki67 + ) cellsofthecryptsand intestinal glands. Wealsoapplied UTAG to a dataset of 100 IMC images of pancreas32, where the predominant microanatomical division is between the endocrine Islets of Langerhans and the acinar cell-dominated exocrine regions. UTAG, in accordance, accurately identified the two major microanatomical subdivisions in a manner very comparable with supervised approaches (Extended Data Fig. 5c). Both the intestinal villi and the pancreatic islets constitute examples of specialized microanatomical structures with highly eccentric shapes and are therefore difficult to segment manually at scale.
Third, to benchmark UTAG on a different task, we employed a dataset of 58 IMC images with 28 markers from seven patients of upper tract urothelial carcinoma (UTUC)33 (Fig. 4a). In line with our observations in lung cancer (Fig. 3c), the five discovered domains largely reflected the division between tumor and stroma microenvironments. However, we did notice a gradient between the two, with domains with considerable immune infiltration for both tumor and stroma, and a domain present mostly at the interface between tumor and stroma (Fig. 4a). Of note, in this dataset, 16 images had been manually annotated with boundaries of tumor and stroma, which allowed us to assess the performance of UTAG in the delineation of these boundaries (Fig. 4b). We found that UTAG domains largely recapitulate these annotations and significantly outperformed both randomly shuffled labels and cell types as baseline, as well as other methods in overall agreement with cell type labels and purity of the domains, when compared with manual labels (Fig. 4b).
Fig. 4 |. Discovery of microanatomical domains associated in cancer.
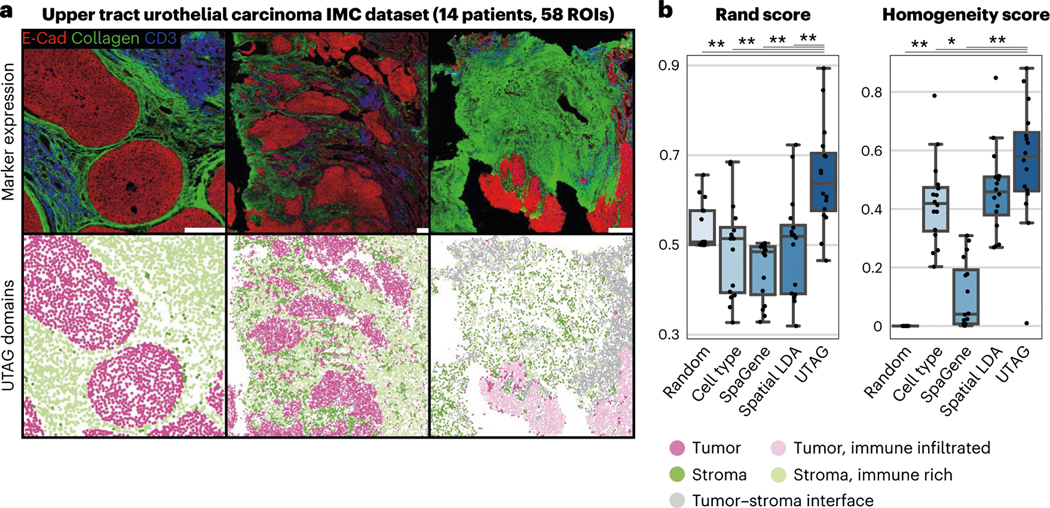
a, Discovery of tumor and stromal domains in IMC images of UTUC. The top row illustrates the intensity of three selected channels and the bottom row displays the UTAG domains. Scale bars, 200 μm. b, Benchmark of the UTAG domains against manual annotation of tumor and stromal domains. For comparison, we include randomized domain labels per cell and cell type identities. Each point represents one image and for both metrics values closer to 1 are optimal. n = 16 highly multiplexed IMC images with manually annotated microanatomical domains. Data in boxplots are presented by minimum, 25th percentile, median, 75th percentile and maximum. Values outside of 1.5 times interquartile range are classified as outliers and are denoted as fliers. **P < 0.01, *P < 0.05, two-sided Mann-Whitney U-test, Benjamini-Hochberg adjusted.
In summary, our analysis of tumor microenvironment domains in large cohorts of cancer patients revealed the accuracy of UTAG in detecting microenvironments reflecting tumor–stromal boundaries in agreement with manual annotations.
Discussion
UTAG performs discovery and quantification of microanatomical tissue structures in biological images with no prior knowledge. Our method leverages the combination of phenotypic and proximity information of cells to discover topology of tissues in various organs and various types of multiplexed imaging data. Given the lack of formal definition of microanatomical domains and healthy tissue datasets with such annotation that can be used as ground truth, benchmarking of our method relied on two datasets of lung microanatomy and tumor–stroma divisions in cancer.
UTAG performed significantly better than the baselines of random domain permutations and cell type identities, as well as SpaGene25 and SpatialLDA26 (Figs. 2 and 4b). We attribute this to the fact that 1) UTAG uses cell phenotypes as vectors of continuous variables of markers and 2) UTAG infers microanatomical domains for all images in a dataset jointly rather than on a per-image basis. The first is unique to UTAG and may explain the advantage against the SpatialLDA method, which uses counts of cell types neighboring each cell. The reliance on user-supplied cell type annotations can be a point of introduction of errors and does not fully leverage the quantitative information in multiplexing imaging data. The second is only common to UTAG and SpatialLDA and may explain why both perform better than SpaGene, which can only output microanatomical domain annotations on a single image basis—this means the burden of interpreting domains for each image separately is on the user, or that domains of various images have to be clustered a posteriori, which introduces one more step and does not guarantee discovery of domains present across images. Thus, UTAG being the only method capable of jointly inferring microanatomical domains across images without cell type information tailored for multiplexed imaging at single-cell resolution (Supplementary Table 1) not only likely contributes to its high performance but also requires less information and effort from the user before running (no cell type information is needed) and after (there is only one step of interpretation across all images).
Despite the good performance of UTAG in the discovery of tissue microanatomy, the ground truth set of manual annotations is inherently subjective to the observer and often incomplete by focusing on a subset of specific predefined structures. In fact, it is conceivable that a fully unsupervised method such as UTAG is able to capture gradients of mixtures between known domains or even new or poorly defined structure in tissue that is underappreciated.
On top of its ability to detect tissue architecture, UTAG can serve as a method to quantify biologically relevant processes such as angiogenesis in native tissue conformation. In this article, we presented ways to numerically quantify the loss of compartmentalization of vessels in alveolar space of COVID-19 infected lung (Fig. 3d). In a similar fashion, UTAG can be used to quantify the extent of various biological processes such as angiogenesis in individual samples—just as existing computational methods based on genomics and transcriptomics can, but with the advantage that the manifestation of biological processes are directly observable in the original physical context of the tissue.
While we believe our method provides a significant step toward the systematic discovery of tissue structure, one crucial aspect for its successful application is the interpretation of the discovered topological domains in terms of their identity and biological relevance. We demonstrated how on cases such as healthy tissue with well-defined structure related with organ-specific physiology, interpretation of domain identity based on cell type composition and interactions can be achieved (Fig. 2), while in tissues without strong structural patterning, or with undefined function such as cancer, interpretation of discovered domains can rely on the association with clinically relevant outcomes (Fig. 4). UTAG provides flexibility to the user to discover structures present in biological images, but we believe that its potential is maximized by the involvement of experts in the field, such as pathologists, in the discovery process and interpretation of results.
Beyond the conceptual limitation in the biological interpretation of UTAG results, a few technical issues must also be taken into account. UTAG relies on user-supplied cell segmentation to determine positional information from the cells and consequently infer physical interactions. Recent advances in cellular segmentation algorithms34–37 have greatly advanced the quality of segmentation masks for various types of images, but downstream results can only be as good as the segmentation. Furthermore, we greatly simplify the geometric complexity of two-dimensional tissue slices by assuming centroids capture most of the positional information of cells, which for eccentric cell types such as neurons, endothelial cells and various types of eccentric immune cells may not be the case.
The inference of cellular contacts and the scale at which microenvironmental signals diffuse across the local cellular context are fields of current study38–41 and of importance for the detection of tissue microanatomy. UTAG requires a user-provided parameter to discretize cellular contacts. In our experience, we found that changes in this parameter were most needed depending on the resolution of the images, since optical imaging typically has more resolution than, for example, laser-based tissue ablation in IMC. Nonetheless, this is something we purposefully designed to be tuned by the user so that UTAG is adaptable without making assumptions on the underlying structure of the tissue, such as has been done previously, for example, relying on the consistent shape of germinal centers42.
UTAG opens new possibilities in our ability to understand tissue architecture by detecting microanatomical domains, but also by quantifying how they interact at a higher level, to a point that we could infer the broad rules of human lung architecture. We envision that, in the future, UTAG could be applied to traditional histopathological images if an appropriate feature matrix can be extracted. That would open the possibility for the detection of microanatomical structures in large biobanks and association of these with clinical features at scale. Likewise, systematic application of UTAG in image data from various organs will undoubtedly accelerate projects such as spatial cell atlases43–45, by providing microanatomical context to the cells and enabling ground-up discovery of tissue architecture beyond cell type composition of tissues. Another exciting future application is the discovery of microanatomy in volumetric images of tissue13,46–48, since there is no conceptual limitation to using UTAG in three dimensions. This would enable robust morphometry of tissue structures, since a current challenge in two-dimensional analysis of tissue is the detection of structure independent of the cutting angle. Robust assessment of tissue microanatomy could enable the definition of tissue integrity ranges in human tissue across ages, detection of early precancer lesions and cancer invasion, and the study of age-associated diseases characterized by cellular degeneration, fibrosis and loss of tissue integrity.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41592-022-01657-2.
Methods
UTAG algorithm
The two inputs to the UTAG algorithm were the cell feature matrix and the location matrix. The cell feature matrix is designed to be as generalizable as possible to incorporate multiple imaging modalities and can contain any features ranging from generic cell properties such as cell area, perimeter and morphology to modality-specific attributes such as intensity of hematoxylin and eosin from H&E staining to marker expression quantification such as CD4, KRT8 or PD1 levels in IMC. From the location matrix, we build a graph using squidpy49 (v.1.1.0), where each node is a unique cell and each edge indicates whether two cells are within a threshold Euclidean distance. We then perform message passing, an inner product between the adjacency matrix of the graph and the feature matrix, so that each cell within the graph inherits features from its immediate neighbors. When aggregating spatial components with the feature matrix, we provide two possible ways to spatially aggregate the feature matrix. The default of the package is to aggregate by the mean, which sums all features from immediate neighbors and divides the resulting sum by the number of neighbors plus one to account for the cell itself. The second option is to aggregate by the sum, which skips normalizing by dividing the sum by the degree of the node’s connectivity. Reduction by mean is commonly used for a numerically more smooth aggregation. Sum aggregation, however, can be advantageous as it directly encodes cell density information, which can vary across structures, though the resulting sum values may be overly separated in cell-sparse regions where cells have only a few neighbors. While the spatial smoothing operation performed by UTAG may seem as if some of the details are diluted, the spatially smoothed matrix is only used for domain segmentation. Nuanced details such as rare cell types infiltrating specific domains can still be detected and used in downstream analysis, for example, on a per domain level. We denote the resulting matrix ‘spatially aggregated feature matrix’ that encodes information of both single cell features and cell locations. The cells in the spatially aggregated feature matrix are clustered into groups using the Leiden22 (v.0.8.7) and PARC23 (v.0.31) algorithm at multiple resolutions. Each cluster can then be annotated into microanatomical domains based on enrichment profiles or by inspecting user-provided cell type identities.
User guide on UTAG
UTAG greatly reduces the amount of manual labor involved in segmentation of microanatomical domains, but its successful application depends on three key user inputs. First is the max_dist parameter, which defines the threshold distance between cells for graph construction. Second is the clustering resolution to determine the coarsity of the clustering of cells. Last is user interpretation of the resulting clusters to label the microanatomical structures detected.
We intentionally leave the optimization of max_dist open to users to maximize the applicability of UTAG to unseen datasets. This is because this parameter is tightly related to the resolution or magnification of the data being used. In our manuscript, we apply the method on IMC data and optical imaging-based CyCIF, which have different per unit area pixel densities. In the case of IMC, we suggest that a suitable max_dist is between 10 and 20, as 1 pixel exactly maps to 1 micrometer. With an imaging-based technique like CyCIF, the optimal distance can vary with magnification, focal lengths, distance to tissue and other factors, which make it hard to suggest a one-fits-all rule. Also there might be nuanced differences for the exact tissue of interest that may vary across specimens under examination.
We believe that the optimal clustering resolution is a hyperparameter that should be explored to suit their biological question of interest. We therefore provide a list of resolutions as default to be explored by the user. A general rule is that increasing the resolution parameter will return more refined substructures, while decreasing it will return coarser, more broad structures. We also recommend users to use a higher resolution parameter when screening for a rare microanatomical domain, as higher resolution will capture more structures, and vice versa. In our benchmarking, we saw that, with the exception of extreme hyperparameter values, UTAG’s performance was fairly robust across various clustering resolutions (Extended Data Fig. 3).
Running UTAG on IMC data
To quantify cellular phenotypes, we used the cell masks and aggregated all pixels of a cell with the mean intensity for each IMC channel. We combined the per cells expression vector from all cells in all images into a single matrix. We then performed log transformation, z-score normalization truncated at positive and negative 3 standard deviations, followed by Combat50 (v.0.3.0) batch correction to phase out sample-specific biases. This was subsequently followed by a final z-score normalization truncated at 3 standard deviations.
For the healthy lung dataset, UTAG was run with a max_dist of 12, which, in physical dimensions, was 12 microns (Extended Data Fig. 1b). For lung infection and UTUC data, we ran UTAG with max_dist of 20; for COVID-19 intestine and diabetic pancreas data it was run with a max_dist of 15. Each dataset was clustered at resolutions of 0.05, 0.1, 0.3 and 0.5. The principle of selecting the optimal resolution was based on how diverse each dataset was, or in other words, how many patients and diseases each dataset contained. Higher resolutions, resulting in more clusters, were preferred in diverse datasets, whereas more homogenous datasets required only a few clusters. For the normal lung dataset, we used Leiden clustering at 0.3 resolution and annotated the resulting 11 clusters into 5 microanatomical domains (Extended Data Figs. 1c and d). For the infected lung, UTUC, intestine and pancreas dataset we used PARC clustering with resolution 0.3, 1.0, 0.5 and 0.1 respectively, which resulted in 20, 34, 12 and 5 clusters.
Running SpaGene and SpatialLDA
To benchmark UTAG against other methods for high-order tissue structure inference, we ran SpaGene25 and SpatialLDA26 on both datasets for which we have ground truth annotation of microanatomical domains. For this purpose, we also reran UTAG using a max_dist of 15 for both datasets, under Leiden clustering resolutions of 0.05, 0.07, 0.1, 0.3, 0.5, 0.8, 1.0 and 2.0, which resulted in 3, 5, 10, 11, 14, 17, 19, 25, 31 and 55 clusters for the healthy lung data, and 3, 4, 6, 22, 23, 27, 38 and 61 clusters for the UTUC data. We intentionally do not use the interpreted annotations in Fig. 2c and d and instead use the raw labels for consistent and fair comparison across methods. SpaGene was run using R v.4.1.3 on a per slide basis using the expression profile and cell location information, as designed by the authors. The number of nearest neighbors to build the graph was set to 24, and the number of latent topics was set to 10 to learn the various structures in the healthy lung dataset and 4 to learn the separation between tumor and stroma for the UTUC dataset. The number of resulting cell-to-topic and topic-to-marker matrices were imported back to Python. As there was no guarantee that topics learned for each slide was coherent across slides, we had to regroup the topics across slides. We used agglomerative clustering as implemented in the scikit-learn package to relabel the topics. The number of resulting clusters from agglomerative clustering was set to match the number of clusters from UTAG. Each cell was assigned the maximum probability relabeled topic to retrieve exactly one most likely topic per cell.
SpatialLDA was run using a working implementation from scimap51. For the cell type distribution in niche, the 7 cell type categories were used for the healthy lung data and 16 cell type categories were used for the UTUC data. Niche for each cell was defined by a radius of 15, matching the max_dist of 15 used for UTAG. The number of motifs was set to match the number of clusters from UTAG. Each cell was then assigned with the maximum probability topic to discretize the probability matrix.
Benchmarking against manual expert annotation
To show that the gain of information using the UTAG algorithm is statistically significant, we compare cell types, SpaGene results, SpatialLDA results and UTAG results against manual expert annotations (Extended Data Fig. 3a and b) across various resolutions. To objectively assess the performance of UTAG, we used Rand score and homogeneity score as an evaluation metric for the unsupervised segmentation task. Rand score, also known as Rand index, is a similarity measurement that is calculated by the ratio of agreeing pairs over all pairs between the predicted and true labels. The homogeneity score52 assesses how uniquely predicted labels associate with true labels (a measure of cluster purity). Ranges of both metrics are from 0.0 to 1.0 inclusive, with higher scores indicating better performance. To lay out a baseline for how the metrics work, we also show how random labels perform against the expert annotation. To test for differences in performance, we perform a two-tailed Mann-Whitney test between random labels scores, cell type scores and UTAG scores. The resulting performance was reported in Extended Data Fig. 3.
Quantification of cellular and microanatomical interactions
As UTAG achieves microanatomical domain annotation based on graphs leveraging spatial proximity, we can take advantage of the spatial neighborhood information for downstream analysis. Under the graph formalism, we can quantify cellular and domain interactions from edge counts connecting distinct nodes, identified by cell type and domain properties. Graphs were constructed with a threshold distance of 40 pixels for healthy lung IMC samples to allow a more lenient interaction threshold compared to the UTAG default. For cell-to-cell interactions, we quantify edges connecting a cell type to another and aggregate the connections into an adjacency matrix denoting the cell type colocalization. We present this cellular interaction matrix as a chord plot generated by holoviews python library. Microanatomical domains are similarly aggregated for each domain-to-domain interaction. These results are presented as a networkx53 (v.2.6.2) graph in a spring force layout, which visually demonstrates how each domain colocalizes with others. This was done on the logarithm of the counts of edge connections to ensure that the counts are on a comparable scale.
Lung infection univariate and principal component analysis
To quantify the difference in domain composition across disease types, each IMC slide was aggregated by the number of cells in each domain. Cell counts were subsequently percent normalized to take into account the difference in cell densities. We perform a univariate domain proportion comparison for each disease group with respect to healthy samples using a two-sided Mann-Whitney U-test. For a multivariate analysis, we reduce the dimensionality of domain proportion using PCA. We then perform a two-sided Mann-Whitney U-test on the first principal component, similar to the univariate analysis, to show how all domain distributions jointly vary across disease. To show that the first principal component of domain proportions better captures the difference in diseases, we perform the same analysis with cell type proportions. All Mann-Whitney U-tests were performed using pingouin54 (v.0.3.12) and were Bonferroni-Hochberg corrected.
Quantification of domain colocalization frequency
Quantifying domain-to-domain colocalization by counting the number of edges may not provide the most representative measurement because this value would be largely explained by the original domain abundance. For example, if there is one domain that is more abundant than every other domain, then that domain generally has the highest colocalization count with all other domains. To compensate for the original domain distribution, we repeatedly performed domain permutation, random shuffling of domains for cells in the graph, to establish an expected colocalization frequency given the domain distribution. We add one to both the observed colocalization frequency and expected frequency, computed by the mean of 100 random permutations, to avoid division by zero. Log-fold change for domain colocalization is then computed by taking the differences between two log-transformed values.
Running UTAG on CyCIF data
40X CyCIF lung cancer samples were downloaded from https://doi.org/10.7303/syn17865732. We used the provided cell segmentation probability maps generated with standard watershed algorithms in ImageJ or MATLAB to create cell masks using DeepCell, similar to the IMC data preprocessing. Cell fluorescence was mean aggregated, just as in the IMC data. All cells across images were combined together and the resulting matrix was log transformed, z-scaled, batch corrected with Combat and then z-scaled again.
Before running the UTAG algorithm, 11 DNA channels and 7 background channels were removed from the feature matrix, leaving 26 channels, to remove background noise and to ensure that the algorithm was not overly influenced by replicates of a single feature. The UTAG algorithm was run with a thresholding distance of 50 pixels because the per pixel distance was more than twice as high at this magnification. We ran both Leiden and PARC clustering at multiple resolutions of 0.05, 0.1, 0.3, 0.5 and 1.0. We annotated stromal and tumor regions based on 0.1 resolution, as the seven created clusters were more than enough for a small dataset with three patients and 16 slides.
Software used: squidpy49 v.1.1.0, Leiden22 v.0.8.7, PARC23 v.0.31, ilastik55 v.1.3.3, DeepCell37 v.0.10.0, Combat50 v.0.3.0, StarDist34, lifelines56 v.0.26.4, scikit-image57, scikit-learn58 v.0.24.2, scanpy59 v.1.8.0, pingouin54 v.0.3.12., scimap51 v.0.18.1 and R v.4.1.3.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. UTAG analysis of IMC images of healthy lung.
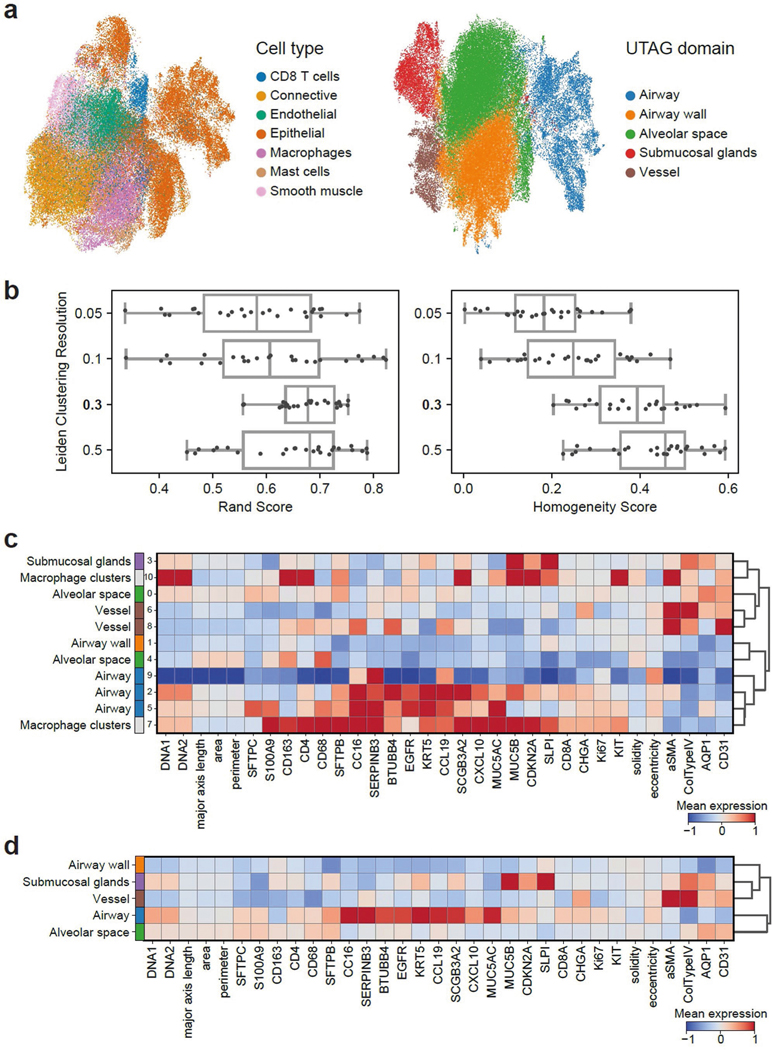
a) UMAP representation of all cells across all images based on cellular phenotypes only (left), or cellular phenotypes and positional information combined with UTAG (right). b) Labeling of domains from clustering indices. Leiden clustering at resolution 0.3 was mapped to domains based on expression profiles as it performed well on both Rand and Homogeneity score. Data in boxplots are presented by minimum, 25th percentile, median, 75th percentile, and maximum. **p < 0.01,,*p < 0.05, two-sided Mann-Whitney-U test Benjamini-Hochberg adjusted. c) Deciding optimal resolution for healthy lung IMC data. Leiden clustering for resolution of 0.1 was selected as the ideal resolution because it had the greatest median rand score across all slides.
Extended Data Fig. 2 |. Illustration of UTAG results on IMC images of healthy lung.
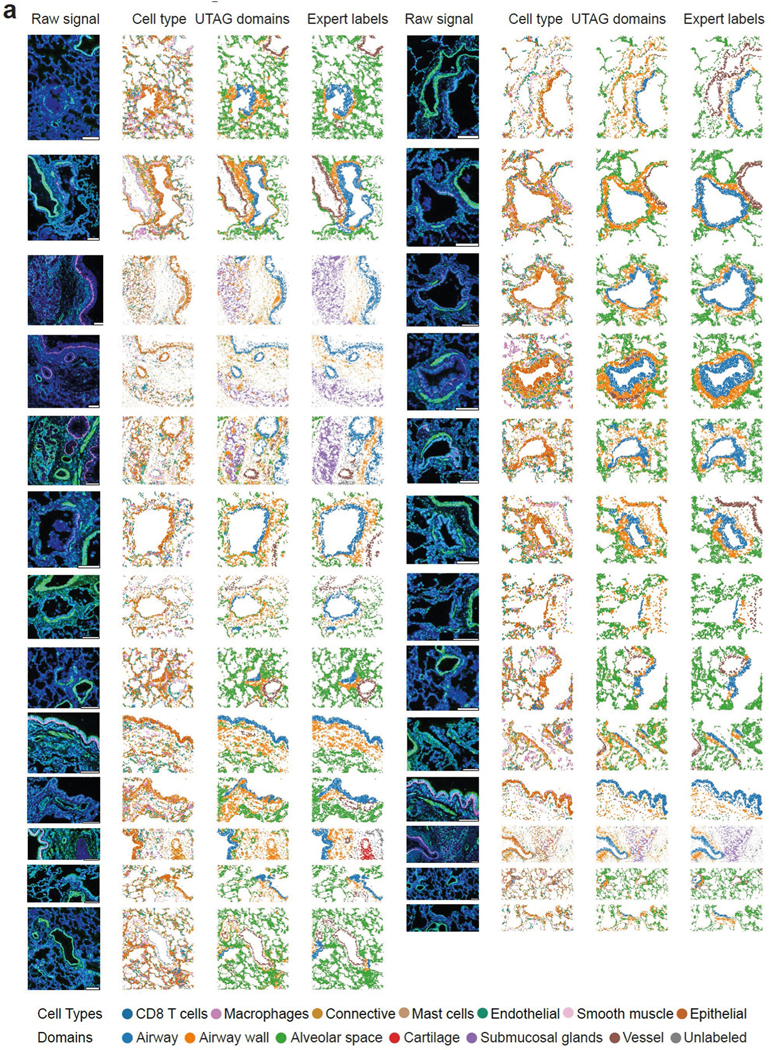
a) Illustration of lung IMC images where the first column illustrates three channels (KRT5, aαSMA, DNA), the second column cell type identities, the third column cells colored by manual annotation of microanatomical domains, and the fourth column cells colored by UTAG domains. Each channel on the raw signal is keratin 5 for red, alpha smooth muscle for green, and DNA for blue. Scale bars represent 200 μm.
Extended Data Fig. 3 |. Benchmarking UTAG and competing methods against expert labels.
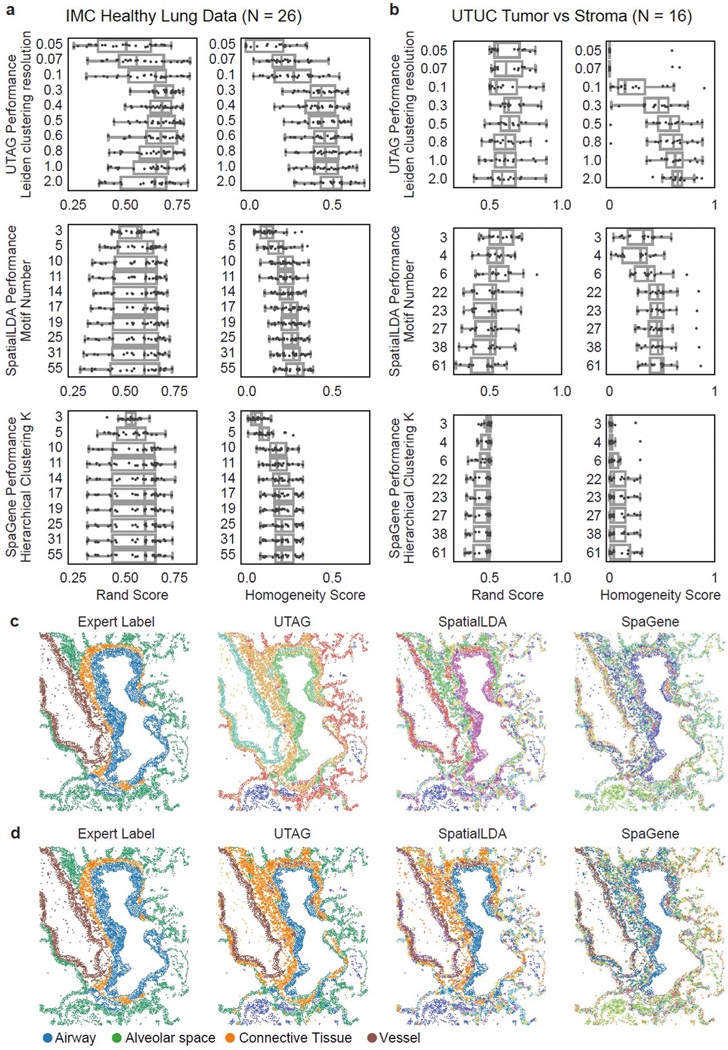
a) Results of each method on healthy lung data to segment microanatomical domains. Number of latent topics for SpaGene was set to 10 to capture the diverse target phenotypes. Due to supporting only single images, SpaGene topics were relabeled using agglomerative clustering to consistently label topics across slides. b) Results of each method on tumor vs. stroma on upper tract urothelial carcinoma. Number of latent topics for SpaGene was set to four to differentiate tumor versus stroma. c) Example of running UTAG, SpatialLDA, and SpaGene to demonstrate the difference in performance. The color mapping in this panel is different for each method as all three methods are unsupervised. d) Same as c) but with domain colors remapped to correspond to the ones from expert labels for ease of visual comparison. For a) and b), Data in boxplots are presented by minimum, 25th percentile, median, 75th percentile, and maximum. Values outside of 1.5 times interquartile range are classified as outliers and are denoted as fliers.
Extended Data Fig. 4 |. Application of UTAG to quantify domain colocalization frequency.
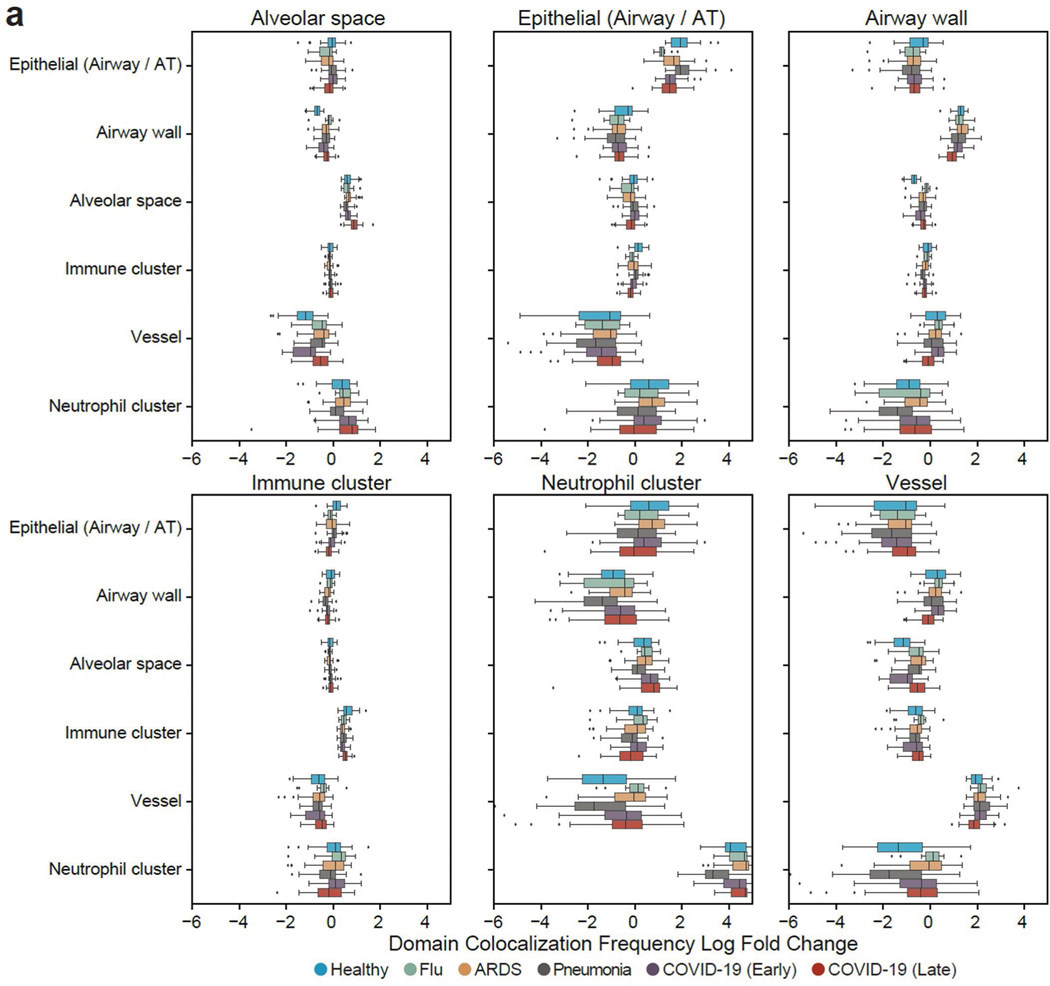
a) Full comparison of domain colocalization frequency for all pairwise microanatomical domains in lung infection data grouped by disease type. Data in boxplots are presented by minimum, 25th percentile, median, 75th percentile, and maximum. Values outside of 1.5 times interquartile range are classified as outliers and are denoted as fliers.
Extended Data Fig. 5 |. Application of UTAG to various data and tissue types.
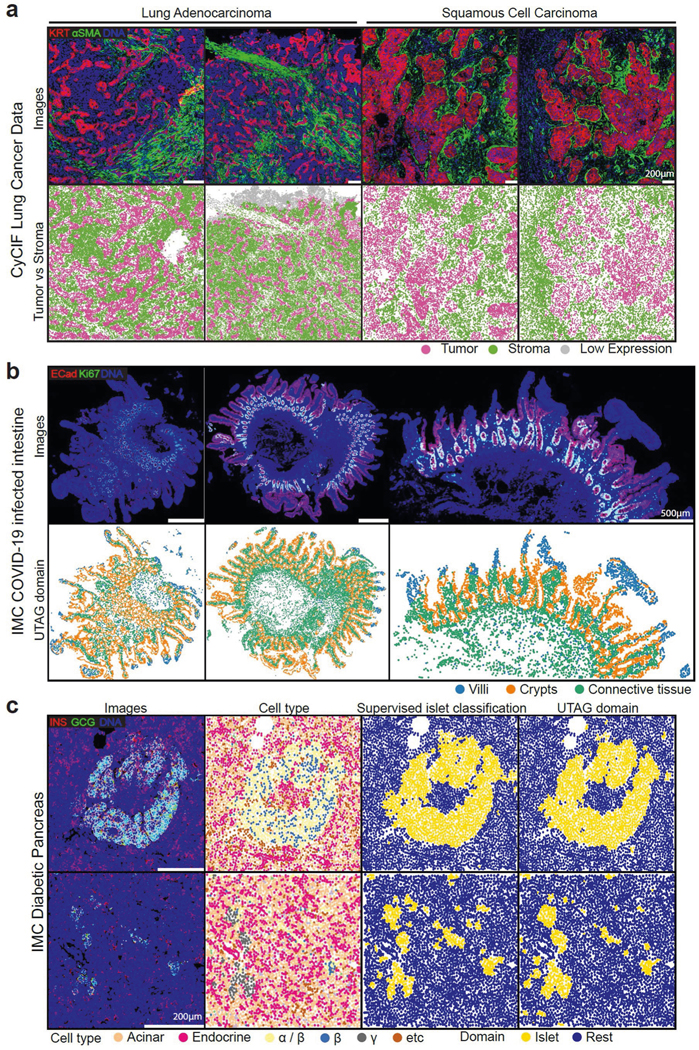
a)Discovery of tumor and stromal domains in CyCIF images of two types of lung cancer. The top row illustrates the intensity of three selected channels, while the bottom row displays the UTAG domains. Scale bars represent 200 μm. b) Discovery of structural domains in 15 intestine IMC images of COVID-19 infected patients30. The first row shows three channels of representative IMC images. The second row shows the corresponding segmented microanatomical domains. Scale bars represent 500 μm. c) Discovery of microanatomy in a dataset of 100 IMC images from pancreatic tissue of diabetes patients31. Each row represents a different region of interest. The first column shows three channels of IMC images. The second column shows identified cell types in the dataset. The third column shows supervised islet segmentation results from a trained random forest using manual labels available in the original publication. The fourth column shows unsupervised islet segmentation results from UTAG. Scale bars represent 200 μm.
Supplementary Material
Acknowledgements
A.F.R. is supported by an NCI T32CA203702 grant. O.E. is supported by NIH grants UL1TR002384 and R01CA194547, and Leukemia and Lymphoma Society SCOR 7012-16, SCOR 7021-20 and SCOR 180078-02 grants.
Footnotes
Code availability
Source code is publicly available at the following URL: https://github.com/ElementoLab/utag.
Competing interests
O.E. is scientific advisor and equity holder in Freenome, Owkin, Volastra Therapeutics and OneThree Biotech. The remaining authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41592-022-01657-2.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41592-022-01657-2.
Peer review information Nature Methods thanks Raza Ali and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team. Peer reviewer reports are available.
Reprints and permissions information is available at www.nature.com/reprints.
Data availability
Datasets used in this manuscript are publicly available at the repositories from the original publications: healthy lung IMC24: https://doi.org/10.5281/zenodo.6376766; COVID-19 lung IMC27: https://doi.org/10.5281/zenodo.4110559; lung cancer t-CyCIF30: https://doi.org/10.7303/syn17865732; upper tract urothelial carcinoma IMC33: https://doi.org/10.5281/zenodo.5719187. For convenience and reproducibility we make available a repository containing all processed datasets in h5ad format here: https://doi.org/10.5281/zenodo.6376766.
References
- 1.Giesen C. et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Angelo M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goltsev Y. et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Preprint at bioRxiv 10.1101/203166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J-R, Fallahi-Sichani M. & Sorger PK Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat. Commun. 6, 8390 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerdes MJ et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA 110, 11982–11987 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen KH, Boettiger AN, Moffitt JR, Wang S. & Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eng C-HL et al. Transcriptome-scale super-resolved imaging in tissues by RNA seqFISH. Nature 568, 235–239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stickels RR et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol. 10.1038/s41587-020-0739-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merritt CR et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Salmén F. et al. Barcoded solid-phase RNA capture for spatial transcriptomics profiling in mammalian tissue sections. Nat. Protoc. 13, 2501–2534 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Rizzardi AE et al. Quantitative comparison of immunohistochemical staining measured by digital image analysis versus pathologist visual scoring. Diagn. Pathol. 7, 42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rakhlin A, Shvets A, Iglovikov V. & Kalinin AA Deep convolutional neural networks for breast cancer histology image analysis. Preprint at bioRxiv 10.1101/259911 (2018). [DOI] [Google Scholar]
- 13.Kiemen A. et al. In situ characterization of the 3D microanatomy of the pancreas and pancreatic cancer at single cell resolution. Preprint at bioRxiv 10.1101/2020.12.08.416909 (2020). [DOI] [Google Scholar]
- 14.Keren L. et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell 174, 1373–1387.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson HW et al. The single-cell pathology landscape of breast cancer. Nature 10.1038/s41586-019-1876-x (2020). [DOI] [PubMed] [Google Scholar]
- 16.Raza Ali H. et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer 1, 163–175 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Schürch CM et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182, 1341–1359.e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ash JT, Darnell G, Munro D. & Engelhardt BE Joint analysis of expression levels and histological images identifies genes associated with tissue morphology. Nat. Commun. 12, 1609 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brbić M. et al. Annotation of spatially resolved single-cell data with STELLAR. Preprint at bioRxiv 10.1101/2021.11.24.469947 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Fischer DS, Schaar AC & Theis FJ Learning cell communication from spatial graphs of cells. Preprint at bioRxiv 10.1101/2021.07.11.451750 (2021). [DOI] [Google Scholar]
- 21.Innocenti C. et al. An unsupervised graph embeddings approach to multiplex immunofluorescence image exploration. Preprint at bioRxiv 10.1101/2021.06.09.447654 (2021). [DOI] [Google Scholar]
- 22.Traag VA, Waltman L. & van Eck NJ From Louvain to Leiden: guaranteeing well-connected communities. Scientific Reports 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stassen SV et al. PARC: ultrafast and accurate clustering of phenotypic data of millions of single cells. Preprint at bioRxiv 10.1101/765628 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rustam S. et al. A unique cellular organization of human distal airways and its disarray in chronic obstructive pulmonary disease. Preprint at bioRxiv 10.1101/2022.03.16.484543 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Hsu C-Y & Shyr Y. Scalable and model-free detection of spatial patterns and colocalization. Preprint at bioRxiv 10.1101/2022.04.20.488961 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen Z, Soifer I, Hilton H, Keren L. & Jojic V. Modeling multiplexed images with Spatial-LDA reveals novel tissue microenvironments. J. Comput. Biol. 27, 1204–1218 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rendeiro AF et al. The spatial landscape of lung pathology during COVID-19 progression. Nature 593, 564–569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Halawa S. et al. Potential long-term effects of SARS-CoV-2 infection on the pulmonary vasculature: a global perspective. Nat. Rev. Cardiol. 10.1038/s41569-021-00640-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ackermann M. et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N. Engl. J. Med. 10.1056/NEJMoa2015432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashid R. et al. Highly multiplexed immunofluorescence images and single-cell data of immune markers in tonsil and lung cancer. Sci. Data 6, 323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lehmann M. et al. Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8+ T cells. Mucosal Immunol. 14, 1381–1392 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damond N. et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 29, 755–768.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohara K. et al. The evolution of genomic, transcriptomic, and single-cell protein markers of metastatic upper tract urothelial carcinoma. Preprint at bioRxiv 10.1101/2021.11.16.468622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weigert M, Schmidt U, Haase R, Sugawara K. & Myers G. Star-convex polyhedra for 3D object detection and segmentation in microscopy. Preprint at arXiv 10.48550/arXiv.1908.03636 (2019). [DOI] [Google Scholar]
- 35.Mandal S. & Uhlmann V. SplineDist: automated cell segmentation with spline curves. Cold Spring Harb. Lab. 10.1101/2020.10.27.357640 (2020). [DOI] [Google Scholar]
- 36.Stringer C, Wang T, Michaelos M. & Pachitariu M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Greenwald NF et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 10.1038/s41587-021-01094-0 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CS, Tan J. & Tien J. Mechanotransduction at cell-matrix and cell-cell contacts. Annu. Rev. Biomed. Eng. 6, 275–302 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Snijder B. et al. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature 461, 520–523 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Imle A. et al. Experimental and computational analyses reveal that environmental restrictions shape HIV-1 spread in 3D cultures. Nat. Commun. 10, 2144 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zanotelli VRT et al. A quantitative analysis of the interplay of environment, neighborhood, and cell state in 3D spheroids. Mol. Syst. Biol. 16, e9798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bhate SS, Barlow GL, Schürch CM & Nolan GP Tissue schematics map the specialization of immune tissue motifs and their appropriation by tumors. Cell Syst. 10.1016/j.cels.2021.09.012 (2021). [DOI] [PubMed] [Google Scholar]
- 43.Regev A. et al. The Human Cell Atlas. eLife 6, e27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Consortium HuBMAP. The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ardini-Poleske ME et al. LungMAP: The Molecular Atlas of Lung Development Program. Am. J. Physiol. Lung Cell. Mol. Physiol. 313, L733–L740 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Currlin S. et al. 3D-mapping of human lymph node and spleen reveals integrated neuronal, vascular, and ductal cell networks. Preprint at bioRxiv 10.1101/2021.10.20.465151 (2021). [DOI] [Google Scholar]
- 47.Maric D. et al. Whole-brain tissue mapping toolkit using large-scale highly multiplexed immunofluorescence imaging and deep neural networks. Nat. Commun. 12, 1550 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuett L. et al. Three-dimensional imaging mass cytometry for highly multiplexed molecular and cellular mapping of tissues and the tumor microenvironment. Nature Cancer 10.1038/s43018-021-00301-w (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palla G. et al. Squidpy: a scalable framework for spatial single cell analysis. Preprint at bioRxiv 10.1101/2021.02.19.431994 (2021). [DOI] [Google Scholar]
- 50.Johnson WE, Li C. & Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8, 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Nirmal AJ, Chen Y-A & Sokolov A. labsyspharm/ scimap: Release v.0. 19. (2022); 10.5281/zenodo.6410307 [DOI] [Google Scholar]
- 52.Hirschberg JB & Rosenberg A. V-Measure: a conditional entropy-based external cluster evaluation. Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning,pp. 410–420 10.7916/D80V8N84 (2007). [DOI] [Google Scholar]
- 53.Hagberg Aric A., Schult Daniel A. and Swart Pieter J., “Exploring network structure, dynamics, and function using NetworkX”, in Proceedings of the 7th Python in Science Conference (SciPy2008), Varoquaux Gäel, Vaught Travis, and Millman Jarrod (Eds), (Pasadena, CA USA: ), pp. 11–15, Aug 2008. [Google Scholar]
- 54.Vallat R. Pingouin: statistics in Python. JOSS 3, 1026 (2018). [Google Scholar]
- 55.Berg S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 56.Davidson-Pilon C. lifelines: survival analysis in Python. J. Open Source Softw. 4, 1317 (2019). [Google Scholar]
- 57.van der Walt S. et al. scikit-image: image processing in Python. PeerJ 2, e453 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pedregosa F. & Varoquaux G. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011). [Google Scholar]
- 59.Wolf FA, Angerer P. & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets used in this manuscript are publicly available at the repositories from the original publications: healthy lung IMC24: https://doi.org/10.5281/zenodo.6376766; COVID-19 lung IMC27: https://doi.org/10.5281/zenodo.4110559; lung cancer t-CyCIF30: https://doi.org/10.7303/syn17865732; upper tract urothelial carcinoma IMC33: https://doi.org/10.5281/zenodo.5719187. For convenience and reproducibility we make available a repository containing all processed datasets in h5ad format here: https://doi.org/10.5281/zenodo.6376766.


