Abstract
Tumor biomarkers, the substances which are produced by tumors or the body’s responses to tumors during tumorigenesis and progression, have been demonstrated to possess critical and encouraging value in screening and early diagnosis, prognosis prediction, recurrence detection, and therapeutic efficacy monitoring of cancers. Over the past decades, continuous progress has been made in exploring and discovering novel, sensitive, specific, and accurate tumor biomarkers, which has significantly promoted personalized medicine and improved the outcomes of cancer patients, especially advances in molecular biology technologies developed for the detection of tumor biomarkers. Herein, we summarize the discovery and development of tumor biomarkers, including the history of tumor biomarkers, the conventional and innovative technologies used for biomarker discovery and detection, the classification of tumor biomarkers based on tissue origins, and the application of tumor biomarkers in clinical cancer management. In particular, we highlight the recent advancements in biomarker-based anticancer-targeted therapies which are emerging as breakthroughs and promising cancer therapeutic strategies. We also discuss limitations and challenges that need to be addressed and provide insights and perspectives to turn challenges into opportunities in this field. Collectively, the discovery and application of multiple tumor biomarkers emphasized in this review may provide guidance on improved precision medicine, broaden horizons in future research directions, and expedite the clinical classification of cancer patients according to their molecular biomarkers rather than organs of origin.
Subject terms: Cancer therapy, Tumour biomarkers
Introduction
Brief history of tumor biomarkers development
Biomarkers are designated as “a biological molecule found in blood, other body fluids, or tissues that is a sign of a normal or abnormal process, or a condition or disease. A biomarker may be used to see how well the body responds to a treatment for a disease or condition” according to the National Cancer Institute (http://www.cancer.gov/dictionary). Tumor biomarkers exist in tumor tissues or body fluids such as blood, urine, stool, saliva, and are produced by the tumor or the body’s response to the tumor.1 The goal of the tumor biomarker field is to develop sensitive, specific, reliable, cost-effective, reproducible, and powerful detection and monitoring strategies for tumor risk indication, tumor monitoring, and tumor classification so that patients can receive the most appropriate treatment and doctors can monitor the progress, regression, and recurrence of the tumors.2 Since the discovery of Bence-Jones protein (BJP), the first tumor biomarker, in 1846, this field has been through many stages and has made significant and substantial progress with the joint efforts of researchers, clinical staff, and patients.
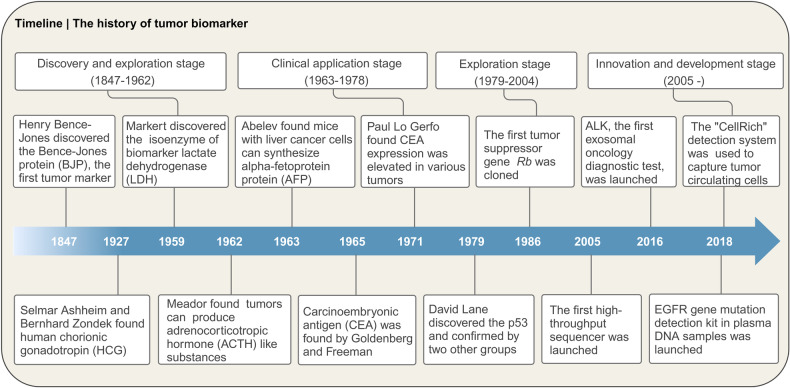
Discovery and exploration stage (1847–1962)
In 1847, Henry Bence-Jones described the findings of a large number of immunoglobulin light chains from the urine of a patient with multiple myeloma, and named it BJP, the first biochemical tumor biomarker described in diagnostic laboratory medicine3 (Fig. 1). The monitoring of BJP in urine has become one of the parameters related to the diagnosis and prognosis of multiple myeloma.4 The discovery of BJP marks the beginning of research on tumor biomarkers. Subsequently, hormones, isozymes, and other tumor biomarkers that displayed abnormalities during the occurrence and development of tumors were discovered. In 1927, Selmar Ashheim and Bernhard Zondek found a gonadal stimulating substance—human chorionic gonadotropin (HCG) from the blood and urine of pregnant women.5 Later on, HCG was identified as a tumor biomarker, which is frequently associated with gestational trophoblastic disease and testicular germ cell tumor.6 In 1959, lactate dehydrogenase (LDH), the first “isoenzyme”, was discovered in the bovine heart by Clement L Markert at Johns Hopkins University,7 and numerous clinical evidence subsequently demonstrated that LDH was an essential prognostic factor for different tumors.8 In 1962, Meador found that some tumors spontaneously produced adrenocorticotropic hormone-like substances, which hindered the normal secretion mechanism of adrenocorticotropic hormone and induced metabolic abnormalities dominated by hypokalemia.9 Despite the aforementioned breakthroughs in knowledge about tumor biomarkers, these biomarkers did not translate from bench to bedside for cancer diagnosis or monitoring.
Fig. 1.
Timeline of the history of tumor biomarker
Clinical application stage (1963–1978)
The next significant advances came from GI Abelev who is well known for his 1963 discovery that mice inoculated with liver cancer cells can synthesize alpha-fetoprotein (AFP)10 (Fig. 1). AFP has been used as a biomarker in clinical screening, diagnosis, prediction, and treatment evaluation of hepatocellular carcinoma (HCC).11 At around the same time (in 1965), Goldenberg and Freeman found that carcinoembryonic antigen (CEA) in fetal colon mucosa12 contributed a crucial part in tumor diagnosis and prognosis evaluation of lung cancer,13 breast cancer,14 ovarian cancer,15 colorectal cancer (CRC),16 etc. The discovery of AFP and CEA has promoted the clinical application of tumor biomarkers. However, the application of CEA as a tumor biomarker was later challenged by Paul Lo Gerfo and his colleagues in 1971. They measured the level of CEA in the serum or plasma of 674 hospitalized patients by radioimmunoassay (RIA). It was found that CEA expression level was elevated in the serum of patients with multitudinous diseases, but not cancer-specific, which hampered the potential absolute benefit of CEA assessment independent of other surveillance tools.
Exploration stage (1979–2004)
James Watson and Francis Crick’s discovery of the DNA double helix structure in 1953 ushered in a new era in tumor biomarker research.17 After this discovery, modern molecular biology significantly promoted the research on tumor biomarkers, with a large number of genes involved in tumor occurrence and progression being discovered. In 1979, p53 was found by David Lane and confirmed by other independent groups.18,19 Although being considered as a cell tumor antigen at the beginning, p53 was defined as a cancer suppressor gene in 198920 and more than 50% of p53 was mutant in cancer patients.21 In 1981, Robert Weinberg and Geoffrey Cooper discovered the small fragments of DNA in transgenic experiments in which the transformation of mouse NIH/3T3 fibroblast cells transfected with DNA extracted from human tumors cell lines was successfully induced and soon followed the isolation of homologous oncogenes HRAS and KRAS from human tumors.22,23 This discovery paved the way for the development of the first KRASG12C inhibitor, sotorasib, which was approved by the United States Food and Drug Administration (FDA) in 2021 for non-small cell lung cancer (NSCLC) treatment.24 The first human oncogene, retinoblastoma (Rb) gene, was successfully cloned in 198625 (Fig. 1). After that, large numbers of proto-oncogenes, oncogenes, tumor suppressors, receptors, and kinases were discovered, and some of them were successfully used as tumor diagnostic, prognostic, and therapeutic biomarkers.
Innovation and development stage (2005-)
The rapid development of science and technology is driving the field of tumor biomarkers to an innovation and development stage. An increasing number of methods and technologies have been developed and applied in tumor biomarker discovery and detection. The molecular biological technologies, such as genomics, transcriptomics, proteomics,26 metabolomics,27,28 the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) gene-editing technology,29,30 and high-throughput sequencing,31 make it possible to obtain large-scale information on the diversity of tumor biomarkers. In 2005, the first high-throughput sequencer was launched, which contributed to the sequencing technology. In recent years, nucleic acid-based liquid biopsy for monitoring cancer has attracted much attention.32 For example, cell-free DNA (cfDNA) in the plasma of cancer patients contains tumor-derived DNA sequences, which can be used as biomarkers for the early detection of cancer, guiding treatment, and monitoring drug resistance.33 In 2016, ExoDx Lung (ALK), the world’s first exosomal oncology diagnostic test, was launched. It can be used for the diagnosis of NSCLC and the screening of NSCLC patients for targeted therapy with anaplastic lymphoma kinase (ALK) inhibitors. Subsequently, the epidermal growth factor receptor (EGFR) gene mutation detection kit in plasma DNA samples was launched in 2018 and used to screen patients for EGFR-targeted drug therapy. China’s self-developed “CellRich” circulating tumor cell detection system was approved by the National Medical Products Administration in 2018 and used to capture tumor circulating cells. Furthermore, the emergence of “precision medicine” has pushed the field of tumor biomarkers to a new stage, which requires the discovery of more effective tumor biomarkers and the integration of multiple tumor biomarkers to support personalized medicine.34
Clinical application of tumor biomarkers
Early screening of tumors
Early screening of tumors is the most powerful public health tool that enables early detection, thus reducing annual cancer incidence, providing a higher chance of treatment, improving patient response to medical interventions, and prolonging patient survival, especially for those cancers with high mortality such as CRC.35 In addition to invasive or expensive screening methods such as endoscopy, low-dose computed tomography, and tissue biopsy, noninvasive and cost-effective screening based on biomarkers from body fluids including blood, stool, saliva, and urine has been gaining extensive attention. To date, thousands of these biomarkers including proteins, cytokines, metabolites, hormones, microRNA, and circulating DNA have been explored, and several of them have been successfully developed and used in the early screening of cancers.2,34 For example, AFP was the first blood biomarker used for the screening of HCC in the populations since 1964.36 After that, other biomarkers called “classical” tumor markers, such as CEA, carbohydrate antigen 19-9 (CA19-9),37 carbohydrate antigen 125 (CA125),38 prostate-specific antigen (PSA),39 and LDH,40 have been used in the clinical screening of various kinds of cancers. In addition to the “classical” tumor marker, a broad range of novel biomarkers have been explored in recent years, which include microRNA41 and other RNAs,42 microbial proteins,43 circulating nucleosomes,44 circulating tumor DNA,45 and circulating tumor cells.46 Albeit currently undergoing clinical trials or preclinical studies and unavailable in the market, they have great potential for clinical screening. As some biomarkers from body fluids may be difficult to detect because of fundamental biological barriers such as short circulation times and very low density, synthetic biomarkers including small-molecule, DNA-based, mammalian cell-based, and bacterial cell-based sensors have been developed to amplify tumor signals, thus enhancing the sensitivity and efficiency of early-stage tumor detection.47 New screening tests based on these novel techniques can be used in the clinic in the near future. Collectively, for the detection of early-stage cancer, the noninvasive or minimally invasive test is ideal, and developing such techniques is desirable in clinical applications.
Tumor auxiliary diagnosis
Due to the risk of false positive or negative results, relying solely on one biomarker level is not an accurate and reliable strategy for tumor diagnosis. Instead, the combination of tumor biomarkers with other methods such as tissue biopsy and endoscopy is a promising alternative to improve the effectiveness of screening.48,49 For example, the combined detection of AFP with cfDNA can improve the specificity of HCC diagnosis to 94.4%, which was superior to that of AFP alone in terms of higher sensitivity and better clinical correlation.50 The advantages of biomarker panels have been confirmed as compared with a single biomarker, especially a panel of biomarkers reflecting changes in independent pathways. The combination of periodin (POSTN) with CA15-3 and CEA for the diagnosis of breast cancer can improve the diagnostic performance of CA15-3 and CEA.14 For CRC, the detection of hemoglobin using fecal immunochemical testing in combination with transferrin in stool improves the diagnostic accuracy for CRC.51 The combination of various diagnosis strategies with biomarkers could result in an easier, faster, more accurate, and more specific diagnosis of cancer.
Prediction of tumor prognosis and curative effect
Precision stratification of cancer patients based on prognosis and therapeutic decision biomarkers has enabled the selection of treatment strategies and more effective treatments for individual cancer patients. One successful example is to distinguish the type of breast cancer by the expression of human epidermal growth factor receptor 2 (HER2), estrogen receptor (ER), and progesterone receptor (PR) in breast cancer tissues. These biomarkers help to identify the triple-negative breast cancer (TNBC) lacking the expression of ER, PR, and HER2 which is the most aggressive type of breast cancer associated with poor prognosis and limited treatment options, thus improving the management and treatment options with the ultimate goal of improving clinical outcomes.52 Moreover, identifying curative predictive biomarkers to distinguish patients who are most likely to respond to anticancer therapy from all cancer patients enhances therapeutic efficiency, decreases treatment costs, and avoids adverse events. For example, the implementation of patient selection prior to programmed cell death 1 (PD-1)/programmed cell death ligand 1 (PD-L1) inhibitors therapy by the combination of biomarkers reflecting tumor immune microenvironment and tumor cell-intrinsic features, such as PD-L1, tumor-infiltrating lymphocyte, tumor mutational burden, mismatch-repair deficiency, and gut microbiota, could enhance the treatment effect of anti-PD-1/anti-PD-L1 therapy in clinical practice.53
Tumor recurrence monitoring
The level of tumor biomarkers is valuable for indicating the disease recurrence of tumor patients. Some classic biomarkers for tumor diagnosis and prognosis, such as PSA, CEA, CA19-9, and CA72-4, are used for indicating the recurrence of cancers including prostate cancer, gastric cancer, breast cancer, and liver cancer.54,55 The CEA is increased in most liver recurrence cases of gastric cancer (90%), while the increase of CA19-9 after surgery in patients with gastric cancer could predict peritoneal recurrence more accurately (78.9%).56 In recent years, extensive molecular and genetic characterization of disseminated tumor cells and blood-based biomarkers have contributed significantly to monitoring cancer recurrence. Postoperative methylated septin 9 in plasma may represent a potential noninvasive biomarker for CRC recurrence monitoring in addition to CRC diagnosis and prognosis compared with CEA and CA19-9.57 The circulating tumor DNA (ctDNA) minimal residual disease (MRD) following treatment in solid tumors predicts relapse and highlights the application of this potentially transformative biomarker.58
Collectively, tumor biomarkers play an active role in all aspects of clinical application, such as early screening, diagnosis, prognosis, and relapse monitoring, and are of great value in helping patients prolong their survival and improve their quality of life. To date, excellent progress has been made in the discovery and application of biomarkers. Besides classical biomarkers used in clinical practice, recent advances in molecular biology technologies have significantly improved the discovery of new candidates for cancer management, but most of them are still in the early stage of development and validation. Great effort could be made to find new biomarkers with the right degree of specificity, sensitivity, and reliability, so as to provide evidence for individualized decision-making during the overall management of cancer patients. In this review, we summarize the current progress that has been made in cancer biomarker development and discuss the promise, limitations, and further challenges in biomarker development.
Technologies used in the detection of tumor biomarkers
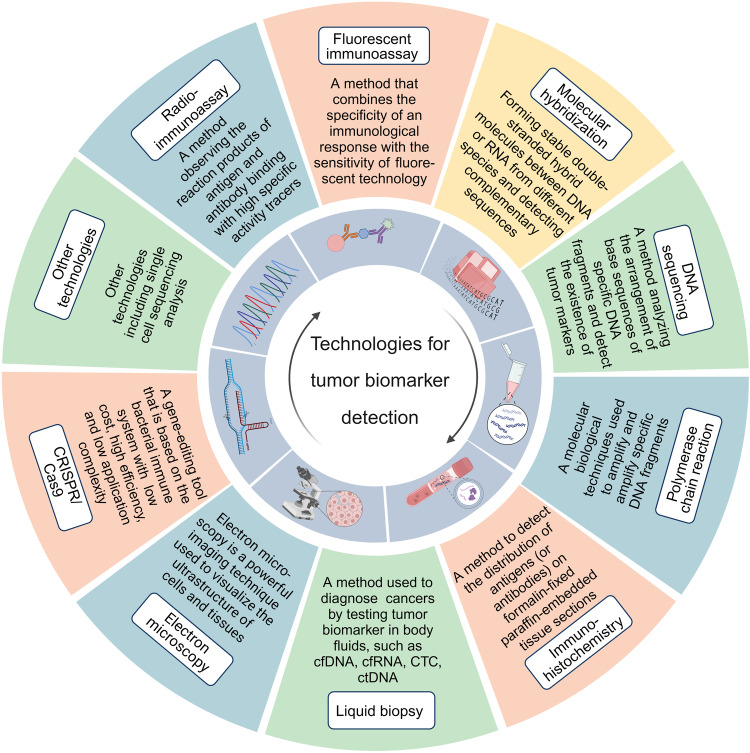
Multiple technologies have been developed for the detection of tumor biomarkers as follows (Fig. 2). In the past decades, various immunoassay methods have played crucial roles in the discovery of tumor biomarkers. Meanwhile, molecular hybridization technology and gene amplification detection technology further broaden the horizon of the application of tumor biomarkers in clinical practice. Immunohistochemistry (IHC) brings about the original distribution of biomarkers in fixed tissue. Furthermore, rapidly developed DNA sequencing and gene-editing technologies accelerate the speed and numbers of digging out prognostic and predictive tumor biomarkers. Other technologies, such as liquid biopsy and different microscopy technologies, as well as single-cell sequencing analysis,59 also provide tremendous convenience in cancer therapy.
Fig. 2.
Technologies for the detection of tumor biomarkers
RIA technology
RIA technology is an analytical method proposed in the late 1950s by the United States chemist Solomon A. Berson and Rosalyn S. Yalow.60 It integrates immunologic and radiolabeling techniques to quantitate minute amounts of biological substances based upon the competition between labeled and unlabeled antigens for specific antibody sites, forming antigen–antibody complexes.60,61 RIA usually uses radionuclide 125I as a tracer, which has been widely used for its advantages of highly radioactive.62–64 In addition, RIA is advantageous in measuring a variety of immunoreactive substances for its high sensitivity, and specificity.65,66 For example, RIA is utilized in the detection of early-stage tumors, and is an effective method in combination with clinical pathological assay to provide comprehensive evaluations of tumors.67 However, the shortcomings of RIA are also prominent, such as isotope contamination due to the radioactive wastes, the requirement of specific safety equipment, and the excessive radiation exposure of workers induced by the long incubation time. which limits its wide use.65,68 RIA tends to be eliminated with the rapid update of other immunoassay methods, such as enzyme-linked immunosorbent assay and fluorescent immunoassay (FIA), which use other substances such as fluorescent dye instead of radioactive isotopes to label antigens.
FIA technology
FIA, combining the specificity of the immunological response with the sensitivity of fluorescent technology, is a popular and fast-growing nonisotopic immunoassay technology. As a new immunoassay technology using fluorescein-labeled antibodies or antigens as tracers, the principle of FIA is similar to enzyme-linked immunosorbent assay. The fluorescein is chemically bound to the antibody (or antigen) molecule, and after that, the latter is combined with the matching antigen (or antibody). The fluorescence is observed, or the fluorescence intensity is measured by a fluorescence detector which determines the presence, distribution, and content of antigens (or antibodies) in samples. FIA has the advantages of high specificity, high sensitivity, and good practicability with cheap, stable, and safe reagents.69 Moreover, FIA avoids the risk of handling radioactive materials. Thus, FIA is widely used in the biomedical field in the measurement of drugs, hormones, and proteins; the identification of antibodies, and the quantification of antigens.70
The development of various fluorescent probes and instruments also contributes to the continuing evolution of FIA.2,71,72 Multiple FIA-related technologies with high detection sensitivities and various measurable properties have been developed, including fluorescent excitation transfer immunoassay, fluorescence polarization immunoassay, and time-resolved fluorescence immunoassays.73–77 For example, the multicolor quantum dots based on fluorescence polarization immunoassay have been applied in the detection of tumor biomarkers such as α-AFP and CEA.78
Molecular hybridization technology
Molecular hybridization technology is an important method for the investigation of gene expression and genome function by assessing chromosomal aberration using a fluorescent probe.79 The principle of molecular hybridization technology is to form stable double-stranded hybrid molecules between DNA or RNA from different species, thereby detecting complementary sequences or recognizing binding sites of transcription factors. Common molecular hybridization techniques include fluorescence in situ hybridization (FISH) and in situ hybridization.79,80 In situ hybridization uses labeled complementary DNA or RNA strands to localize specific DNA or RNA sequences on chromosomes or tissue sections fixed on slides (in situ), and the FISH technique helps to localize genes to different chromosomal locations. They are all molecular tools in cancer diagnosis, treatment, and prognosis.79,81 With the advantages of easy manipulation, fast hybridization process, possible automation of process and scoring, the FISH technique is wildly utilized to detect the tumor biomarkers in diagnosis and metastasis prognosis, such as the analysis of circulating tumor cells (CTCs) obtained from the patient blood sample82 in various of cancers, including lung cancer, glioma, breast cancer, ovarian cancer, and soft tissue sarcomas. Extensive prognostic and predictive biomarkers, such as ALK, mesenchymal-epithelial transition factor (c-MET), and ROS1, are identified by FISH.82 Significantly, the timeless and costless FISH remains a gold standard in ALK-rearrangements NSCLC.83 In 2011, a novel anticancer drug crizotinib and its companion, the ALK FISH probe detection kit, were simultaneously approved by the FDA, which highlights the crucial position of the FISH assay in guiding ALK-targeted therapy.84
In short, FISH is an increasingly demanded tool for biomarker research and personalized medicine despite the fact that the process of FISH may be time-consuming and costly when performed with standard chemicals and the retention of the fluorescence is limited.82
Gene amplification detection technology
Polymerase chain reaction (PCR), a molecular biological technology used to amplify specific DNA fragments, is an invaluable tool for the assessment of nucleic acids in tissues and body fluids. It can synthesize and amplify specific DNA into billions of copies in a few hours by separating the DNA into two strands and incubating it with oligonucleotide primers and DNA polymerase in vitro.85 PCR technology has developed to the third generation since the invention of Kary Mullis in 1985,86 and holds a pivotal position in biological research. The PCR technology includes three major steps: denaturation of double-stranded (ds) DNA template, annealing of primers (forward and reverse primers), and extension/elongation of dsDNA molecules.85 The quantitative real-time PCR (qPCR)-based assay is considered to be the gold standard for prognostic and predictive biomarker analysis for the quantitative advantage.85
The application of PCR in diagnostic gene mutation analysis, such as the B-raf proto-oncogene (BRAF), EGFR, Kirsten rat sarcoma viral oncogene homolog (KRAS), neuroblastoma RAS viral oncogene homolog (NRAS), and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) genes from the blood, is meaningful in initial cancer stratification and the monitoring of cancer progression. Moreover, several PCR assays approved by the FDA are used for the diagnosis of KRAS mutation status in formalin-fixed paraffin-embedded tissue, thereby guiding anti-EGFR antibody treatment for metastatic CRC.87 Similarly, qPCR assays are effective in the detection of MRD in leukemia, such as the quantification of BCR-ABL-positive cells post-induction chemotherapy/transplantation in acute lymphoblastic leukemia (ALL).85 PCR technology is also widely used to detect abnormal genes and abnormal mRNA amplification in tumors, such as MYCN amplification in neuroblastoma.88 Ligand-targeted PCR is essential for the detection of folate receptor-positive circulating tumor cells as a potential diagnostic biomarker in pancreatic cancer.89
PCR methods are of great advantages in the detection of nucleic acid biomarkers, including relatively simple manipulation, providing rapid inexpensive diagnosis with good sensitivity, valuable for clinical molecular pathology.87 Nevertheless, several intrinsic drawbacks of PCR that restrain its application have room for improvement, such as the requirement for instruments, experienced operators, laboratory setting, and sophisticated operations.90 Collectively, PCR is a valuable tool in tumor biomarker detection, while novel PCR-based methods remain to be explored to meet the needs of patient monitoring in clinic.87
DNA sequencing technology
DNA sequencing technology is a commonly used technology in molecular biology research, which is used to analyze the arrangement of the base sequence of specific DNA fragments. The world’s first method of DNA sequencing was invented by British biochemist Frederick Sanger, who performed the first complete DNA genome sequencing, bacteriophage ϕX174 in 1977.91,92 Since that, DNA sequencing technology has witnessed rapid development, which is now in its fourth generation of DNA sequencing technology.93 It not only opens up new perspectives in traditional biology, medical research, and other fields, but also promotes the further development of bioinformatics, molecular genetics,94 genomics,95 precision medicine,96 and other disciplines, which advances the progress of life science research.
DNA sequencing technology is not only the gold standard for microbial identification but can also be used to detect the existence of tumor biomarkers which indicate the occurrence and development of tumors. At present, next-generation sequencing technology (NGS) is the most widely used among four DNA sequencing technologies in clinical practice, which can detect multiple genomic alterations including nucleotide substitutions, small insertions, deletions, copy number variations, and chromosomal rearrangements. NGS promotes the identification of somatic mutations associated with acute myeloid leukemia (AML), melanoma, mesothelioma, small cell lung cancer (SCLC), breast cancer, and prostate cancer.97 For example, NGS was applied to detect mutations of many cancer-related genes, such as TP53,98 phosphatase and tensin homolog (PTEN),99 KRAS,100 and breast cancer type 1/2 susceptibility protein (BRCA1/2).101 These detections are valuable for assessing the family history and the risk of tumorigenesis, and improving clinical diagnostics.97 Except for providing a high sensitivity in gene mutations, NGS is dramatically cost-effective and less time cost compared with current PCR-based tests. For example, the cost of using PCR to detect RAS mutations is as high as several thousand dollars, while NGS only costs one-third for detecting the same mutations. Notably, NGS simultaneously sequences the remaining gene samples in the same pathway or multiple samples in a single sequencing run with high speed and accuracy, which avoids incurring additional operating costs.97
Moreover, RNA sequencing has been utilized in multifarious aspects of cancer management, including prognostic and predictive biomarker identification, the characterization of cancer heterogeneity, and the monitoring of drug resistance. Some special genomic biomarkers, including miRNA, lncRNA, and circRNA, have been discovered by RNA sequencing.102 For example, isocitrate dehydrogenase (IDH) mutation which is a good prognostic biomarker for glioma, and nuclear cyclooxygenase 2 combined with HER2 which serve as potential biomarkers for the diagnosis and prognosis of CRC, are identified by RNA sequencing.102
In conclusion, the advent of sequencing technology sequences individual cancer genomes, which opens a new chapter in precision cancer therapy. Novel sequencing technologies have the potential to decode massive amounts of cancer genomes rapidly and cheaply to benefit cancer precision therapy.
IHC technology
IHC, a technology used to detect the distribution of antigens (or antibodies) on formalin-fixed paraffin-embedded tissue sections,103 identifies targets through antigen–antibody interactions, and the antibody binding site is identified by direct labeling or secondary labeling method.
IHC is a gold standard and ubiquitously applied technology in cancer identification and diagnosis, especially in assessing biomarkers used for characterizing tumor subtypes, confirming tissue origin, distinguishing metastasis from primary tumor, providing prognosis information, stratifying patients for treatment selection, and predicting therapy response in various cancers.104–109 The American Society of Clinical Oncologists and College of American Pathologists (ASCO/CAP) provides the HER2 scoring guidelines to determine breast cancer pathological classification and clinical stage by using IHC-based staining intensity and the percentage of HER2+ cells in cancer tissues.110 IHC is used to detect p53 in cancer tissues.111,112 The detection of the excision repair cross-complementation group 1 protein by IHC has been approved for predicting the response of NSCLC patients to chemotherapy.105
As an indispensable technology, IHC holds the unique advantage of correlating the presence of protein with its location in tissues or cells compared with other protein detection methods, which is essential for illustrating protein function in normal and pathological tissues.113 Moreover, IHC can be operated by easy preparation and automated manipulation.82 However, several limitations still exist in IHC, especially a lack of reproducibility. Conflicting results often occur when different antibodies are used. The variables of the protocol affect the reliability of IHC, including the fixation time of tissues, the absolute level of the antigen, the affinity and concentration of antibody, and the sensitivity of the detection system.111 Thus, high-quality control of regents, standardized protocols, automated IHC, or combined IHC with transcriptomics will improve the accuracy, reproducibility, and reliability of IHC and accelerate its application in the discovery and validation of biomarkers.114
Liquid biopsy technology
Liquid biopsy, a minimally invasive methodology, is used to obtain tumor-derived information from body fluids so as to facilitate cancer diagnosis.115 Currently, liquid biopsy is used to detect cfDNA, cell-free RNA, CTCs, extracellular vesicles,116,117 ctDNA,118 circulating RNA, and exosomes119,120 in blood or other body fluids.116,121 Liquid biopsy can enhance patient overall survival (OS) by improving early cancer detection and monitoring treatment response continuously. Thus, liquid biopsies are widely used in the clinical biomarker screening of tumors, such as endometrial cancer,122 lung cancer,123 pancreatic cancer,124 CRC,125 melanoma,126 renal cell carcinoma (RCC),127 breast cancer, ovarian cancer, cervical cancer, and bladder cancer.128 In addition, liquid biopsy technology is also utilized to detect and monitor KRAS, BRAF, and EGFR mutations in patients with lung cancer, CRC, and breast cancer.128
Liquid biopsy can reduce the risk of biopsy by noninvasive sampling,128 and it has the advantages of convenient sampling and easy operation. Moreover, liquid biopsies have the potential to better detect heterogeneity across regions of the tumor.115 Although there are still some challenges to overcome in terms of assay sensitivity and specificity, liquid biopsy technology provides new opportunities for personalized cancer treatment and has the potential to revolutionize the field of oncology.
Electron microscopy technology
Electron microscopy (EM), a powerful imaging technique used to visualize the ultrastructure of cells and tissues with high resolution, is applied based on a special type of microscope, electron microscope.129,130 The first electron microscope was built in 1931 by a German engineer and academic professor Ernst Ruska.131 The electron microscope uses signals obtained from the interaction between an electron beam and the sample to achieve information about sample structure, morphology, and composition.132
EM has been widely used in investigating tumorigenesis-related cellular and subcellular change133 and observing the ultrastructure changes in cancer cells,134 as well as in clinical applications for cancer diagnosis and treatment.134–136 The ultrastructural features of tumor cells by EM can provide vital clues such as evidence or biomarkers of cytodifferentiation for correct diagnosis, which is difficult for diagnosis of light microscopy.137,138 Especially, the ultrastructural examination provided by EM is necessary for the precise categorization of biomarkers in apparently undifferentiated carcinoma.138 Thus, EM is useful in the differential diagnosis of tumors, particularly in small-cell “undifferentiated” tumors, such as neuroblastoma, rhabdomyosarcoma, Ewing’s sarcoma, undifferentiated squamous cell carcinoma (SCC) of the lymphoepithelioma type, and malignant lymphoma, amelanotic melanoma, and spindle-cell carcinoma.137 Scanning electron microscopy has been used as an alternative to examine the morphology of exosomes which is a diagnostic biomarker usually detected by liquid biopsy.139 In conclusion, EM is a valuable complementary tool for tumor diagnosis, especially providing valuable information on tumor differentiation which is difficult to define by light microscopies.134,140
CRISPR/Cas9 technology
The CRISPR/Cas9 technology is a gene-editing tool that is based on the bacterial immune system. The basic principle of CRISPR/Cas9 is to use a guide RNA molecule to direct a nuclease, Cas9, to a specific target gene. The nuclease then cleaves the DNA at the target site, allowing for precise modifications of genome.141,142
By using CRISPR/Cas9 technology to precisely edit cancer-related genes, researchers have created highly specific molecular probes for the detection of cancer biomarkers in body fluids, such as blood, urine, and saliva. CRISPR/Cas9 system is extensively used for different kinds of cancer biomarkers including virus nucleic acids, ctDNAs (i.e., EGFR mutation), miRNAs (i.e., miR-17, miR-31), proteins (i.e., TGF-β1, CEA, PSA, AFP), and extracellular vesicles.90 CRISPR/Cas9 can combine with other assays for tumor biomarker identification, such as qPCR, FISH, and nanotechnology, providing an efficient way for tumor biomarker discovery. Moreover, CRISPR/Cas9 has exerted significant effects in the treatment of cancers, such as pancreatic cancer, prostate cancer, breast cancer, ovarian cancer, liver cancer, and CRC.143
The CRISPR/Cas9 system enjoys some advantages, including low cost, high efficiency, low application complexity, easy-to-operate, and time-saving.30 The exquisite specificity is also a character of the CRISPR/Cas9 system which could distinguish single base mismatch in target nucleic acid.90 Moreover, CRISPR/Cas9-based nucleic acid amplification strategies exhibit high detection sensitivity comparable with PCR. However, several aspects of CRISPR/Cas9-based diagnosis still need to be improved. CRISPR/Cas9-based analysis requires the fluorescence spectrophotometer and electrochemical workstation which is inconvenient for detection. Thus, the portable and quantitative detection strategy should be further explored to monitor cancer biomarkers. Cancer progression is influenced by the level of multiple biomarkers such as various miRNAs, ctDNAs, and proteins, which makes the design of the high-throughput CRISPR/Cas9-based strategy for cancer biomarkers detection promising and significant.90 In conclusion, CRISPR/Cas9 technology is a powerful gene-editing tool that holds great promise and opportunities for the development of personalized cancer management.144
Classification of tumor biomarkers
Tumor biomarkers are diverse and can be classified by different standards. Here, we divide tumor biomarkers by tissue origin: tumor biomarkers derived from blood, tumor tissues, and other biofluids such as feces, urine, and saliva (Fig. 3).
Fig. 3.
Overview of human tumor biomarkers
Tumor biomarkers derived from blood
Tumor biomarkers in the blood are highly significant for tumor diagnosis and treatment. They have vital reference values for early tumor diagnosis, tumor stage assessment, anticancer strategy selection, treatment response monitoring, and prognosis.2,145 Here, we summarize the common tumor markers in blood and their roles in cancers.
Embryonic antigen tumor biomarkers
The 1960s saw the discovery of AFP and CEA, two tumor biomarkers that are still widely employed as tumor biomarkers. AFP and CEA are embryonic antigen substances which are proteins that only appear in the fetal period and gradually decline and disappear in adulthood.146–149 The reemergence of these embryonic antigens in cancer patients may be related to the activation of certain genes that have been turned off in adulthood when malignant cells transform, and these genes make embryonic antigens. While there aren’t many embryonic antigen tumor biomarkers, the ones that exist are crucial biomarkers for cancer care in clinical practice.
AFP
AFP, first discovered in 1956 by Bergstrand Czar,146 is a 3–5% carbohydrate-containing single-chain glycoprotein.150 Encoded by the AFP gene located in the q arm of chromosome 4 (4q25), AFP is a member of the albuminoid gene superfamily.151 As the amino acid sequences of AFP and albumin are very similar and highly homologous, AFP is considered as an analog of serum albumin in the fetal period and is the main protein in fetal circulation. At 18 months after birth, albumin synthesis gradually increases, and AFP concentration gradually decreases. The concentration of AFP in healthy adult serum is less than 10 μg/L.147 AFP is currently the most widely used tumor biomarker for HCC and has been used for more than 60 years. Elevated AFP can be seen in ~80% of HCC patients.152,153 Thus, AFP is currently applied for HCC screening, especially in China, Japan, Africa, and Alaska. The international academic community recommends limiting the reference value of AFP to 20 μg/L. Moreover, early-stage HCC is frequently detected by AFP detection combined with ultrasound.154 Tumor prognosis and treatment monitoring are additional applications for AFP. In patients with HCC, a sharp increase in AFP indicates tumor recurrence or metastasis. AFP >200 μg/L after surgery indicates incomplete removal or metastasis of HCC.155 Nonetheless, AFP levels are not the perfect diagnostic criteria for HCC. Approximately 40% of patients with early-stage HCC express normal or acceptable AFP levels. The elevation of AFP levels is observed in patients with chronic liver diseases, including ~20% of patients with hepatitis and 40% of patients with cirrhosis.156
CEA
CEA was first extracted from human CRC tissues and embryonic tissues in 1965, hence it was named for CEA.148 CEA belongs to a family of glycoproteins on the cell surface, and its gene is located on chromosome 19q.157 The production of CEA in the digestive tract starts at the early fetal stage (week 9–13). In addition to normal adult tissues such as the colon, stomach, cervix, sweat glands, and prostate, CEA is highly expressed in various tumors.149
As a broad-spectrum tumor biomarker, CEA is elevated in 70% of CRC, 55% of pancreatic cancer, 50% of gastric cancer, 45% of lung cancer, 40% of breast cancer, 40% of urethral carcinoma, and 25% of ovarian cancer patients.149,158,159 Serum CEA levels are proportional to tumor burden. Accordingly, CEA is applied to aid the diagnosis, determine the prognosis, monitor recurrence, and evaluate the therapeutic efficacy in cancer patients.160 In patients with breast cancer, CEA is one of the most frequently used biomarkers in the diagnosis, prognosis, and prediction of survival for different breast cancer molecular subtypes.161 In CRC patients, CEA level is a meaningful prognostic and diagnostic biomarker. The levels of CEA predict the 5-year survival rate of patients: 69% of patients have a CEA level below 5 ng/mL, 44% have a level of 5–200 ng/mL, and only 7% have a level equal or greater than 200 ng/mL.162 The elevated CEA level also has a bearing on poor prognosis and progression of lung adenocarcinoma patients with mutant EGFRs, and gastric cancer patients with lymph node metastasis.163 Additionally, CEA is also used for efficacy evaluation and recurrence detection after tumor treatment.164
Nevertheless, CEA lacks good sensitivity and specificity, which renders CEA inappropriate for tumor screening. A combination of CEA with other biomarkers could improve its actual significance in clinical practice.149,164
SCCA
Squamous cell carcinoma antigen (SCCA), a tumor-specific antigen, was first isolated from cervical SCC tissue by Kato and Torigoe in the 1970s.165 Initially, SCCA was used as a tumor biomarker for cervical cancer, and it has a high independent diagnostic value in cervical cancer.166 The serum level of SCCA correlates with the stage, the degree of invasion, recurrence, and the progression of cervical SCC.159 Cervical cancer patients with a high-level of pretreatment serum SCCA exhibit a higher risk for death than patients with low serum SCCA. Pretreatment SCCA cutoff ranging from 1.1 to 40.0 ng/mL is related to recurrence and death.166,167 Subsequent research has revealed that SCCA exists in tumors in the mouth, pharynx, esophagus, lung, and other tissues. In particular, high levels of SCCA have been found in multiple SCCs including lung cancer, esophageal cancer, and genitourinary system cancer in addition to cervical cancer, suggesting its essential role in the diagnosis and prognosis of the above cancers.168,169 Furthermore, elevated serum SCCA is associated with the therapeutic effect of postoperative chemotherapy in esophageal squamous cell carcinoma (ESCC),170 and with tumor-node-metastasis stage in head and neck squamous cell carcinoma (HNSCC).171 Peripheral SCCA has also been extensively utilized as one of the tumor biomarkers for monitoring NSCLC and predicting patients’ response to platinum combination chemotherapy, and serum SCCA level accurately reflects the survival status of patients.169 Despite its limited sensitivity in routine tests, SCCA is still a valuable diagnostic and prognostic biomarker in cancers.
TPS
Tissue polypeptide-specific antigen (TPS) is an M3 antigen determinant on the 18 fragments of cell keratin.172 TPS is synthesized in the S and G2 phases of the cell cycle, and the level of TPS in serum specifically indicates the proliferative activity of cells.173 The levels of TPS mostly depend on the number of cells in the proliferative phase instead of the total number of tumor cells, which is different from other tumor biomarkers.173 The serum levels of TPS are noticeably increased in multiple tumors, such as endometrial cancer, bladder cancer, NSCLC, skin cancer, carcinoma of male urethra, prostate cancer, pancreatic cancer, CRC, gastric cancer, esophageal cancer, neuroblastoma, and nephroblastoma.174–177 Thus, TPS has been employed as a serum tumor biomarker. Due to its lack of sensitivity and organ specificity, the prime application of TPS is monitoring treatment efficiency, and predicting tumor progression and recurrence, rather than diagnostic utility. In breast cancer patients, elevated serum levels of TPS could predict distant metastasis after treatment,178 and are recognized as an independent prognostic factor for disease-free survival (DFS) and OS of patients.179 In gastric cancer, TPS is applied in monitoring the palliative treatment response of patients with a 75% detection rate.180 The potential clinical role of TPS in RCC prognosis has also been demonstrated.181
Additionally, it is worth noting that TPS levels can alter in response to some pathological and physiological conditions, such as chronic pancreatitis, liver cirrhosis, ovulation, and menopausal status. Thereby, TPS in combination with other prognostic factors is necessary to improve the clinical use of serum TPS levels in predicting patient prognosis and facilitating the individualization of therapy for cancer patients.182 Further clinical studies are required to fully determine the utility of TPS alone or in combination.182,183 In conclusion, TPS has a unique value in the prediction of recurrence and metastasis, treatment monitoring, and prognostic evaluation in cancer patients.177
PSA
PSA, a serine protein kinase-releasing enzyme specifically secreted by the epithelial cells of prostate,184 is encoded by the prostate-specific gene kallikrein 3 which is a member of the tissue kallikrein family.185 PSA was first identified in the late 1970s.186 The elevated serum PSA levels represent prostate pathologies including prostatitis, benign prostatic hyperplasia, and prostate cancer.187,188 For the early diagnosis of prostate cancer, the positive cut-off value of serum PSA is greater than 10 ng/mL. In 1986, PSA was approved by the FDA as an adjunctive test for the detection of prostate cancer in men over the age of 50.185,189 Subsequently, in 1994, PSA was approved by the FDA as a diagnostic biomarker.189 Later on, PSA became popular in prostate cancer detection and patient management including screening, risk stratification for recurrence, surveillance following diagnosis, and monitoring therapy.186,188 Total PSA essentially consists of free PSA and bound PSA, and the higher percentage of the free PSA is connected to the lower the cancer risk. Studies have shown that a free PSA percentage >25% indicates the cancer risk is <10%, but a free PSA percentage <10% means the cancer risk is ~50%.187
However, PSA holds a poor specificity of 20–40% in prostate cancer diagnosis. Some noncancerous pathologies such as inflammation, trauma, or benign prostatic hyperplasia may also elevate the PSA level, which leads to a high rate of false positives. Besides, PSA is unable to differentiate between indolent and aggressive forms of prostate cancer, which may ignore aggressive prostate cancer with low initial serum PSA levels.187,190 All the aforementioned factors make prostate cancer now an “overdiagnosed” and “overtreated” cancer.185 To sum up, PSA level is a promising biomarker in prostate cancer diagnosis and prediction.
NSE
Neuron-specific enolase (NSE), a member of the enolase gene superfamily in glycolysis, was originally identified by Moore and McGregor in 1965 as an enzyme enriched in neurons and peripheral neuroendocrine cells.191,192 NSE consists of five dimeric isoenzymes with three different subunits, α, β, and γ, and is a sign of mature neural differentiation.193 Cell proliferation accelerates in response to oncogenic transformation in either central or peripheral neurons, accompanied by enhanced glycolysis and elevated NSE expression. Consequently, NSE plays pivotal roles in diagnosis, prognosis, and treatment efficacy evaluation in cancers originating from neural and neuroendocrine.194,195 Moreover, elevated NSE is also observed in SCLC which is with neuroendocrine properties. Serum NSE is currently believed to be a clinically potential biomarker for staging, monitoring treatment, and predicting relapse of SCLC.196,197 Interestingly, NSE also exerts a significant function in NSCLC. An analysis of 363 patients with advanced and metastatic NSCLC showed that patients with high NSE level (≥26.1 ng/mL) have significantly shorter progression-free survival (PFS) (5.69 vs 8.09 months) and OS than patients with low NSE level (11.41 vs 24.31 months).191 Besides, increased serum NSE levels are found in 30–69% of patients with NSCLC,198,199 which is in accordance with a study of 621 NSCLC patients which shows high NSE level (>12.5 ng/mL) is a prognosticate of poor outcome.200 Thus, serum NSE level is a predictive biomarker of cancer treatment response and an independent prognostic factor.191
AFU
α-l-Fucosidase (AFU), consisting of two isoforms, AFU1 and AFU2, which are encoded by FUCA1 and FUCA2 genes, respectively, is a lysosomal enzyme that clears the terminal α-l-fucose residues from glycoproteins.201 AFU is involved in the metabolism of glycoproteins, glycolipids, and oligosaccharides, and is widely distributed in human tissues and blood. The serum AFU level remains low under normal circumstances. While the serum AFU level increases rapidly as long as tumors attack the body, its level is closely related to the tumor stage and size.202 Multiple studies have shown that AFU is one of the most valuable biomarkers for HCC detection, with 85% sensitivity and 91% specificity.203,204 85% of patients with HCC can be diagnosed with AFU detection six months prior to the ultrasonography detection.205 Patients with a preoperative AFU >35 U/L have a lower recurrence-free survival (RFS) rate and OS rate than those with AFU ≤35 U/L, and they tend to form macrovascular invasion. Therefore, serum AFU is of great significance for judging the treatment effect, prognosis, and recurrence of HCC.205,206 Besides, the low AFU levels are significantly associated with longer OS in ESCC, which indicates that AFU is a potential prognostic biomarker for long-term survival in patients with early-stage ESCC.207 However, the serum levels of AFU are also mildly elevated in certain nonneoplastic conditions such as cirrhosis, chronic hepatitis, and gastrointestinal bleeding.203,208 Presently, the combination of AFU and AFP biomarkers is used in the diagnosis of HCC, which enhances the diagnostic specificity, and makes the diagnosis more stable and reliable for high-risk groups such as hepatitis and cirrhosis.155
LDH
LDH, an enzyme that catalyzes the reversible transfer of pyruvate to lactate and NADH to NAD + , consists of two different isoforms, lactate dehydrogenase A (LDHA) and LDHB.209,210 The two isoforms can form five homotetramers or heterotetramers with different functions.210 In the reverse reaction, LDHB is more effective at converting lactic acid back to pyruvate than LDHA is at converting pyruvate to lactic acid.211,212 Multiple factors, such as the oncogene c-Myc and hypoxia-inducible factor (HIF-1α), stimulate the transcription of LDHA,213,214 which results in the overexpression of LDHA in most tumor tissues.215
High expression of LDHA provides cancer cells with many benefits, and multiple studies have proved that high levels of serum LDH are associated with the proliferation of cancer-initiating cells, enhanced aggressiveness and metastasis, the poor prognosis of cancers, as well as radiation and chemotherapy resistance.216–218 The serum LDH level is considered to be a primary predictor of prognosis in patients with adverse prognosis and distant metastases in melanoma, RCC, and CRC.216 Accordingly, an analysis of 76 studies comprising 22,882 patients with solid tumors reveals that high serum LDH levels are linked to poor survival in patients with solid tumors, in particular in melanoma, prostate cancer, and RCC, and is a valuable and affordable prognostic biomarker in metastatic cancers.40 Serum LDH levels are closely correlated with OS in an analysis of 2507 cancer patients with brain metastasis216 and are a poor prognosticator for OS and DFS in nasopharyngeal carcinoma (NPC) patients. Furthermore, the elevated serum LDH levels could be used to develop individualized treatment strategies.219 A study of a total of 68 studies including 31,857 patients illustrates that LDH overexpression is a predictor to guide individual therapy in solid tumors,220 such as testicular cancer,221 SCLC,219 and gastrointestinal cancer.222,223 In conclusion, LDH is a valuable indicator of cancer diagnosis, efficacy evaluation, and recurrence and metastasis.
CA72-4
Carbohydrate antigen 72-4 (CA72-4) is a mucin carcinoid embryonic antigen found in liver metastases of breast cancer in 1981 and is highly expressed in human adenocarcinoma.224 Enhanced serum CA72-4 levels are effective indicators for the diagnosis of cancers, including gastric cancer, pancreatic cancer, CRC, breast cancer, ovarian cancer, lung cancer, cervical cancer, and endometrial cancer.225,226 Notably, CA72-4 exerts diagnostic value in patients with digestive system tumors, especially gastric cancer, with superior sensitivity and specificity.227 Studies have demonstrated that the sensitivity and specificity of CA72-4 applied in the diagnosis of gastric cancer alone are 49 and 96%, respectively, which outperforms other tumor biomarkers such as CEA (sensitivity 41%, specificity 93%), CA19-9 (sensitivity 44%, specificity 92%), and CA242 (sensitivity 38%, specificity 97%).228 The serum level of CA72-4 is also correlated with the malignant grade of gastric cancer. Thus, CA72-4 is used as the best serum marker for gastric cancer diagnosis in China.229 However, CA72-4 also has limitations. It has been uncovered that CA72-4 is highly expressed in normal tissues in addition to tumor tissues such as the endometrium and the colonic transitional mucosa, which results in false positives in patients with atrophic gastritis.230 The sensitivity of CA72-4 in the diagnosis of gastric cancer is far from satisfactory.231 The combination with other biomarkers may gain increased sensitivity and specificity of CA72-4 in tumor applications. In conclusion, serum CA72-4 is a unique biomarker of gastric cancer for screening, diagnosis, the prediction of metastasis and recurrence, and the evaluation of treatment efficiency.229
CA125
CA125, a highly glycosylated mucin, is originally discovered in a monoclonal antibody OC125 screening against the ovarian cancer cell line OVCA433.232,233 Thus, CA125 has become one of the most important biomarkers for monitoring epithelial ovarian cancer, and its sensitivity in the diagnosis of epithelial ovarian cancer reaches ~70%.234 The key role of CA125 in the prognosis of ovarian cancer patients has also been recognized. The Gynecologic Cancer Group (GCIG) has shown that the serum level of CA125 is associated with the progression and recurrence of ovarian cancer. According to the criteria of GCIG, patients with serum CA125 levels within the reference range (<35 U/mL) after surgery or chemotherapy are considered fully effective. While the CA125 level increased to twice of the minimum value (≥70 U/mL), the progression or recurrence is considered.235 Moreover, CA125 is also a diagnostic and prognostic biomarker for other nonovarian tumors, such as cervical cancer, endometrial carcinoma,236 and gastric cancer.237 Of note, ~1% of healthy people and 5% of patients with menstrual or benign diseases such as endometriosis and coronary artery disease have varying degrees of elevated serum CA125 levels.238–240
CA242
Carbohydrate antigen 242 (CA242), a sugar chain antigen containing sialic acid, is obtained after the immunization of mice with a human CRC cell line COLO 205.241 An analysis of serum CA242 levels from 34,680 patients with 27 clinically defined diseases suggests that patients with pancreatic cancer, cervical cancer, and lymphoma have the highest level of serum CA242, which are followed by esophagus cancer, CRC, ovarian cancer, and breast cancer.242 Hence, the primary application of CA242 is as a biomarker for CRC and pancreatic cancer.243 Serum CA242 has a normal reference value of less than 17 U/mL. The sensitivity for diagnosing pancreatic cancer and CRC is ~70 and 45%, respectively, and the specificity is ~95 and 83%, respectively.244–246 As CA242 exhibits a lower sensitivity for diagnosing pancreatic cancer, the combination of CA242 with CEA is a promising strategy for improving diagnosis sensitivity in pancreatic cancer.247 In addition, CA242 is also used as a clinical indicator of progression or recurrence during chemotherapy for pancreatic cancer.241,242
CA15-3
Carbohydrate antigen 15-3 (CA15-3, also known as mucin 1) is a large transmembrane glycoprotein derived from the MUC1 gene.248 It is expressed in normal tissues including the breast, esophagus, stomach, duodenum, pancreas, uterus, prostate, and lung.248,249 Notably, CA15-3 is overexpressed in the majority of human cancers, and is thought to be a key biomarker for cancers, especially for indicating cancer metastasis.250 The reference value for normal serum CA15-3 levels is less than 28 U/mL.243 In breast cancer, serum CA15-3 is used as an auxiliary diagnostic index with a diagnosis sensitivity of 61.5–70% which is higher than that of CEA.243 Thus, CA15-3 in combination with CEA is the most popular method for breast cancer diagnosis.251 Meanwhile, CA15-3 is also a crucial indicator for the evaluation of postoperative recovery, recurrence, and metastasis of breast cancer.248 It is noteworthy that serum CA15-3 is also elevated to varying degrees in benign diseases of the breast, liver, gastrointestinal tract, lung, and other organs, but the positive rate is low.250
CA27-29
Similar to CA15-3, carbohydrate antigen 27-29 (CA27-29) is a critical epitope for the MUC1 protein.243 With a sensitivity of 84% for breast cancer detection, CA27-29 is primarily utilized in breast cancer patients for diagnosis, and efficacy evaluation.243 Additionally, it also be used in combination with other markers to increase the specificity of breast cancer diagnosis.252 The elevated CA27-29 is also observed in other cancers including CRC, stomach cancer, pancreatic cancer, ovarian cancer, and benign diseases of the breast and liver.253
CA50
Carbohydrate antigen 50 (CA50) was initially identified as a cancer-specific antigen screened from monoclonal antibodies against CRC cell line COLO 205 in 1983.254 It is generally absent in normal tissues, but elevated in multifarious cancers. Patients suffering from pancreatic cancer, lung cancer, and colon cancer exhibit the highest levels of serum CA50. Serum CA50 is quite effective in the diagnosis of pancreatic cancer, with a sensitivity of more than 84%.255 Meanwhile, patients suffering from gastric cancer and rectum cancer reveal comparable serum CA50 levels.256,257 Similar to other carbohydrate antigens, serum CA50 is also increased in patients with non-neoplasm diseases such as chronic pancreatitis, colitis, cholecystitis, and pneumonia.256
CA19-9
CA19-9 is initially found in human CRC cell line SW1116 and belongs to the mucin glycoprotein antigen.258 It is extensively distributed on the cell membrane of Lewis antigen-positive epithelial cells such as the pancreatic duct, gallbladder, and gastrointestinal tract. CA19-9 is currently the most commonly used and gold-standard biomarker for pancreatic cancer,259,260 and holds a median sensitivity of 79% for diagnosis of pancreatic cancer.261,262 In addition, CA19-9 has also been used as a biomarker for other cancers, particularly digestive tract cancers.263 Other diseases such as liver damage, bile duct obstruction and inflammation, pancreatitis, acute diarrhea, stomach ulcer, and pulmonary fibrosis have also been linked to increased CA19-9 levels.264–266 Notably, CA 19-9 is not expressed in cells from patients with Lewis allele deficiencies, and it is necessary to ascertain the patient’s Lewis gene type information when applying CA19-9 as a diagnostic biomarker.267,268
HE4
Human epididymal protein 4 (HE4), an orotic acid protein, is first identified in distal epididymal epithelial cells.269 HE4 is widely expressed in the trachea, salivary gland, lung tissue, etc., and is highly expressed in ovarian cancer, endometrial cancer, and lung cancer. Meanwhile, age and menopausal status are also momentous factors affecting HE4 levels.270,271 At present, serum HE4 is primarily used for the diagnosis and recurrence monitoring of ovarian cancer with a sensitivity of 67%. HE4 is also used to evaluate the treatment effect of ovarian cancer.270,272 In addition, HE4 is also overexpressed in other non-gynecologic malignancies, including NSCLC, pancreatic cancer, and transitional cell carcinoma.273
Ferritin
Ferritin is the leading protein that is essential for iron storage and detoxification.274 Ferritin is present in numerous normal tissues such as liver, spleen, bone marrow, and body fluids.274 Serum ferritin levels are linked to a broad range of conditions. The low serum ferritin concentration indicates iron deficiency, e.g., anemia and diarrhea,275 and the high serum ferritin concentration indicates iron overload, e.g., hemochromatosis and hemolytic anemia, or infection and liver disease.276 Moreover, ferritin is overexpressed in various cancers, such as HCC, lung cancer, lymphoma, melanoma, and CRC.277,278 As indicated by its potential to promote tumor proliferation, angiogenesis, immunosuppression, and tumor drug resistance,276 ferritin is valuable in evaluating the progression and prognosis of cancer patients. Nevertheless, a number of factors influence ferritin levels, and ferritin’s limited specificity for tumor detection means that it is not an ideal diagnostic marker for cancers.279
p2PSA
Prostate-specific antigen precursor (p2PSA) is a precursor that is first secreted in the prostate gland ducts during the production of PSA.280 p2PSA is a relatively stable pro-PSA and has certain clinical value in the diagnosis of early prostate cancer. The prostate health index, which forecasts the diagnosis of prostate cancer, is calculated by PSA and p2PSA. Currently, the prostate health index is the strongest predictor of diagnosis at initial biopsy when total PSA levels are between 2.0 and 10 ng/mL in prostate cancer patients, and the prostate health index has been approved by the FDA for early diagnosis and risk grading of prostate cancer.280,281
HCG
HCG is a polypeptide hormone composed of two noncovalently linked subunits (α and β). The smaller α subunit is the part of follicle-stimulating hormone and luteinizing hormone, while the larger β subunit is unique to HCG.282,283 Serum levels of HCG in non-pregnant and menopause women maintain at a low level of 5–10 U/L, and increase dramatically during pregnancy.284 Increased serum HCG levels are observed in trophoblastic tumors, ovarian cancer, testicular cancer, breast cancer, lung cancer, HCC, CRC, and kidney cancer. Although HCG level could be employed for monitoring the disease progression, it is too low to be regarded as a diagnostic marker.282,285
CAM17.1
CAM17.1 is a mucin with high specificity for the digestive system, such as the pancreas, colon, small intestine, and biliary tract.286 Several studies revealed that CAM17.1 is particularly overexpressed in pancreatic cancer with a serum cut-off value is 39 U/L.287 CAM17.1 has a sensitivity of 86% for the diagnosis of pancreatic cancer, and a higher sensitivity of 89% in patients without jaundice.286 These findings suggested that CAM17.1 is a potential biomarker for pancreatic cancer diagnosis, which triggers the need for further study.
PIVKA-II
Protein induced by vitamin K absence or antagonist-II (PIVKA-II) is an abnormal prothrombin elevated in the conditions of vitamin K reduction or the presence of vitamin K antagonists.288 PIVKA-II is primarily used for the early detection of HCC, with a sensitivity and specificity of 97.5 and 90%, respectively.289,290 In other tumors such as gastric cancer and pancreatic cancer, PIVKA-II is also elevated at varying degrees.291 In addition to being able to differentiate between other non-malignant conditions such cirrhosis or chronic hepatitis, serum PIVKA-II is more accurate than AFP in the diagnosis of early-stage HCC.292,293 It is noteworthy that certain patients with vitamin K deficiency also exhibit elevated PIVKA-II levels.288
GRP
Gastrin-releasing peptide (GRP), first isolated from gastric nerve fibers by McDonald in 1978, is a gastrointestinal hormone that exits in the normal bronchial epithelial cells, pulmonary fibroblast, central nervous system cells, and neuroendocrine cells.294 Significantly, GRP is overexpressed in multiple cancers, including 62% of CRC patients, 59% of pancreatic cancer patients, 60% of prostate cancer patients, 39% of breast cancer patients, and 74% of SCLC patients.294 Since GRP has a short half-life and is unstable, it is more appropriate to detect its precursor, pro-GRP.294 With a sensitivity of 47 to 86%, serum pro-GRP detection is mainly utilized for the diagnosis, efficacy, and prognosis analysis of SCLC, outperforming NSE.294,295 The combined application of pro-GRP and NSE increases the sensitivity of SCLC detection.296 In addition, pro-GRP is also elevated in a few other diseases, such as gastritis and acute hepatitis, but the positive rate is generally low.297
Tumor biomarkers derived from tumor tissues
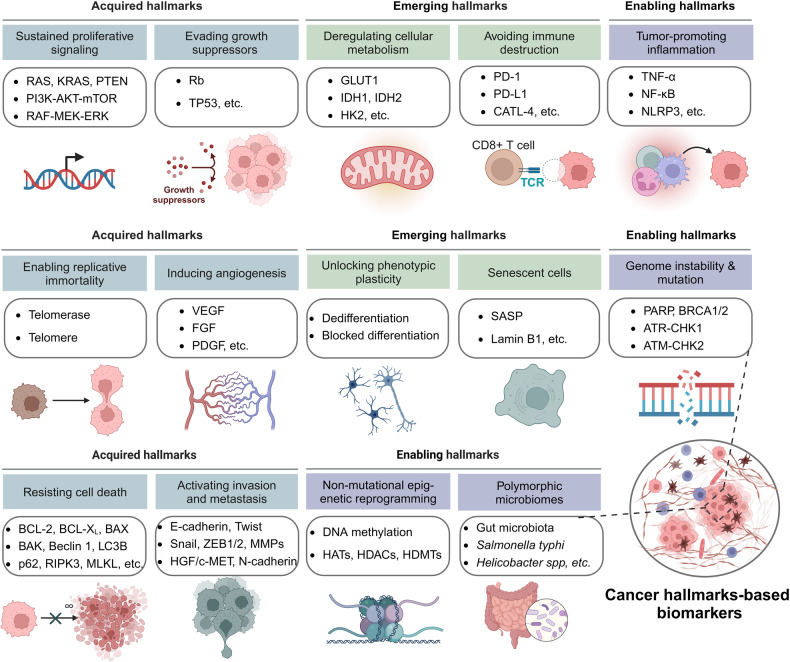
Since the six hallmarks of cancer were proposed in 2000, tumor characteristics are considered to be a set of functional capabilities acquired by human cells during the transition from a normal to a tumor growth state.298 To date, tumors have possessed fourteen major characteristics, including sustaining proliferative signaling, evading growth suppressors, enabling replicative immortality, inducing angiogenesis, resisting cell death, activating invasion and metastasis, genome instability, and mutation, tumor-promoting inflammation, deregulating cellular metabolism, avoiding immune destruction, unlocking phenotypic plasticity, nonmutational epigenetic reprogramming, and polymorphic microbiomes, and senescent cells.298–300 Herein, we summarize the tumor biomarkers from tumor tissues divided by cancer hallmarks (Fig. 4).
Fig. 4.
The 14 cancer hallmarks-based biomarkers. Fourteen major characteristics of tumor cells have been proven so far, which have been divided into acquired hallmarks including sustaining proliferative signaling, evading growth suppressors, resisting cell death, enabling replicative immortality, inducing angiogenesis, activating invasion and metastasis, enabling hallmarks including genome instability and mutation, tumor-promoting inflammation, nonmutational epigenetic reprogramming, and polymorphic microbiomes, and emerging hallmarks including deregulating cellular metabolism, avoiding immune destruction, unlocking phenotypic plasticity, and senescent cells. Each of the cancer hallmarks is involved in numerous essential biomarkers that play vital roles in tumor progression
Sustaining proliferative signaling
Cancer cells are capable of multiple approaches to acquire the ability to sustain proliferation. Stimulated by growth factors and other proliferative signals, proliferation-related signaling pathways, such as the RAS, the phosphoinositide 3-kinase (PI3K)-protein kinase B (AKT)-mammalian target of rapamycin (mTOR) pathway, and the RAF-mitogen-activated protein kinase (MAPK) kinase (MEK)-extracellular signal-related kinase (ERK) pathway, are activated in tumor cells, which subsequently regulate tumor cell proliferation, migration and invasion, gene transcription, cellular metabolic reprogramming, and tumor microenvironment (TME) remodeling.301–303
RAS
RAS genes, named after the rat sarcoma,304 were identified as the transformative factor in the Harvey and Kirsten strains of rat sarcoma viruses305 and were identified in the human genome in 1982.304,305 RAS proteins belong to a superfamily of GTPases, and three RAS genes (HRAS, NRAS, and KRAS) encode four highly homologous RAS proteins: HRAS, NRAS, KRAS4A, and KRAS4B, with the latter two KRAS isoforms arising from alternative splicing.306,307
RAS proteins couple cell surface receptors to intracellular effector pathways through binding to GTP or GDP, followed by a cycle between the GDP-bound inactive state (RAS-GDP) and the GTP-bound active state (RAS-GTP). Under physiological conditions, RAS proteins retain an inactive state, and are incapable of interacting with downstream effectors. When activated by upstream receptors, RAS is activated by guanine nucleotide exchange factors (GEFs) which promote GDP to GTP exchange, thereby recruiting diverse downstream effectors such as the RAF-MEK-ERK pathway and the PI3K-AKT-mTOR pathway.301,308 RAS activation has been linked to multiple tumor phenotypes, including cell cycle progression, proliferation, metastasis, and apoptosis resistance.301 Furthermore, RAS is involved in diverse metabolic processes such as aerobic glycolysis, glutaminolysis, redox homeostasis, and lipid metabolism in tumor cells to support tumor growth.309 Importantly, RAS activation remodels the TME,301 including the initiation and maintenance of proangiogenesis,310 the production of proinflammatory factors,311 and immune escape.301
RAS mutation is a prominent factor that plays a vital role in tumorigenesis and progression.312,313 Approximately 21% of all malignancies have RAS mutations,308 which include CRC,314 pancreatic ductal adenocarcinoma (PDAC),315 lung adenocarcinoma,316 and melanoma.317
Although the function of RAS in the physiological or pathological states has been thoroughly elucidated in the past decades, numerous unresolved concerns still need to be investigated. For instance, the regulatory relationship between RAS and downstream effectors other than PI3K and MAPK.318 To sum up, RAS is a crucial biomarker for tumor diagnosis, prognosis, and treatment.
KRAS
KRAS is by far the most frequently amplified and mutated RAS isoform among the three RAS genes, accounting for 85% of all RAS mutations.319 KRAS mutations were first identified in 1984 in patients with squamous cell lung cancer.320 Notably, KRAS mutations are present in 88% of pancreatic cancer, 50% of CRC, and 32% of lung cancer.319,321 The most common mutations in KRAS are G12D, G12V, G12C, G13D, and Q61R, which account for 70% of RAS mutations in cancer patients.321 KRASG12C mutation is the most frequent,321 and the G12C mutation alters KRAS conformation and shape by forming binding pockets, leading to increased affinity for GTP and sustained activation of KRAS, ultimately triggering the transduction of downstream oncogenic signaling.319,321
KRAS mutations have emerged as biomarkers for the prognosis, diagnosis, and treatment of some tumors, including PDAC,322 CRC,323 and lung cancer.324 A study in a pooled analysis has found that KRAS mutations are independently associated with shorter time to recurrence, survival after relapse, and OS in patients with microsatellite-stable resected stage III CRC.313 Patients with the KRASG12C mutation are related to inferior PFS and OS compared with patients with non-mutated tumors, according to a prognosis analysis in 1239 patients with metastatic CRC.323 Moreover, KRAS mutations link to the poor prognosis of patients with PDAC, and KRAS mutation assays provide significant predictive information on tumor progression and recurrence, which are of great value in the diagnosis, prognosis, and treatment of PDAC.325 Consistently, PDAC patients with KRASG12D mutation have shorter survival than all other PDAC patients.326 In lung adenocarcinoma patients, the patients with KRASG12C mutation have worse DFS than patients with nonG12C mutation KRAS or wild-type KRAS.324
Mechanistically, KRAS drives tumor development and progression through various signaling pathways. For example, the extensive metabolic reprogramming induced by KRAS mutations, such as glycolysis, glutamine metabolism, lipid metabolism, and nucleotide biosynthesis to facilitate tumorigenesis, has attracted much attention in recent years.327,328 KRAS-mutant cells exhibit the upregulation of glucose transporters329 and metabolic enzymes involved in the glycolysis,330,331 resulting in increased glucose flux in the glycolytic pathway.329 KRASG12D stimulates hexosamine biosynthesis and the pentose phosphate pathway to regulate glucose metabolism in PDAC.332 KRAS-mutant cells produce nicotinamide adenine dinucleotide phosphate (NADPH) by promoting glutamine catabolism,329 and intracellular fatty acid uptake and oxidation.333 Furthermore, KRAS leads to the transcriptional upregulation of MYC and the nonoxidative pentose phosphate pathway gene RPIA through activating MAPK, thereby enhancing nucleotide biosynthesis in PDAC cells.327
In summary, KRAS mutations are among the most prevalent drivers of tumorigenesis, and their activation is correlated with tumor progression and poor prognosis.334,335 The evidence presented above strongly suggests that KRAS is a crucial tumor biomarker.
PI3K-AKT-mTOR
The PI3K-AKT-mTOR pathway plays valuable roles in various cellular processes, such as cell proliferation, angiogenesis, protein translation, and metabolic reprogramming.302
In normal cells, growth factor-stimulated PI3K activation leads to the conversion of phosphatidylinositol-3,4-bisphosphate (PIP2) to phosphatidylinositol-3,4,5-trisphosphate (PIP3), followed by the recruitment of AKT and 3-phosphoinositide-dependent kinase 1 to the plasma membrane. Following that, 3-phosphoinositide-dependent kinase 1 phosphorylates and activates AKT, thus phosphorylating the downstream mTOR complex, contributing to cell survival and proliferation.302,336,337 The atypical serine/ threonine kinase mTOR consists of rapamycin-sensitive mTOR complex 1 (mTORC1) and rapamycin-insensitive mTORC2.338 AKT drives mTORC1 activation either directly by phosphorylating mTORC1 at Ser2448 or indirectly by inhibiting TSC1/TSC2.302 mTOR supports cell growth and proliferation by promoting cell cycle,302 sensing nutrient signaling339,340 by phosphorylating its downstream effectors such as S6K and 4EBP1.302,341 The tumor suppressor PTEN is a critical negative regulator of the PI3K signaling pathway.302 PETN rapidly metabolizes PIP3 by removing the 3’-phosphate of PIP3, which in turn terminates PI3K signaling.342
In cancer cells, the PI3K-AKT-mTOR signaling pathway is abnormally activated via the stimulation of tyrosine kinase growth factor receptors,343 the loss of PTEN functions, and the mutations of PIK3CA, thereby promoting tumorigenesis in a wide variety of human cancers.302,342 The PI3K-AKT-mTOR pathway exerts significant impacts on multiple cancers including lung cancer,344,345 ovarian cancer,302 breast cancer,346 and NPC.347,348 The PI3K-AKT-mTOR has been proven to be crucial in ovarian tumorigenesis and drug resistance.302 The level of pAKT is a diagnostic biomarker for the treatment of SCLC involving the combination of clinically approved inhibitors against isoform-specific PI3K and mTOR.345 In addition, the class I isoform of PI3K, the most well-known PI3K protein, contains four distinct isoforms of catalytic structural domain: p110α (PIK3CA), p110β (PIK3CB), p110γ (PIK3CG), and p110δ (PIK3CD).343 pIK3CA and PTEN aberrations lead to the activation of the PI3K-AKT-mTOR pathway.349,350 The TCGA database has shown that PIK3CA gene mutations occur in a variety of cancers, including 53% of endometrial cancer, 35% of breast cancer, 23% of cervical cancer, 21% of gastric cancer, 20% of head and neck cancer, 20% of CRC, 15% of lung cancer, and 10% of glioblastoma.343 PIK3CA mutation, PTEN loss, and pAKT activation are predictive biomarkers for the efficacy of tumor treatment.350,351 Moreover, PIK3CA mutations act as diagnostic biomarkers for HR+ and HER2- metastatic breast cancer.352 In summary, the PI3K-AKT-mTOR pathway is an essential biomarker pathway for tumor diagnosis, prognosis, and treatment.
RAF-MEK-ERK
The RAS-RAF-MEK-ERK pathway participates in the regulation of key processes such as cell proliferation, differentiation, migration, and apoptosis,303 which can be activated by growth factors, cytokines, integrins, and chemokine receptors.303,353 Active RAS binds to RAF kinase, which results in RAF dimerization and activation.354,355 RAF proteins possess three isoforms including BRAF, CRAF, and ARAF which share conserved regions of the regulatory domain and kinase domain,356 and among them, ARAF exhibits the lowest kinase activities.357 The active RAF dimer phosphorylates and activates MEK1/2 which subsequently phosphorylates and activates ERK1/2, followed by the phosphorylation and activation of downstream effectors and the proliferation of cells.336 Various proteins, such as Hsp90,358 p50CDC37,359 and KSR,360 are engaged in the regulation of RAF activation.
Abnormalities epically mutations in RAF-MEK-ERK signaling lead to the aberrations of cell proliferation.361 A mutation analysis of more than 3000 samples from 12 tumor types has shown that the mutations of RAF-MERK-ERK signaling occur in ~50% of cancers.362 In particular, BRAF mutations are widely investigated in cancers.362 Studies have revealed that the hyperactivity of the BRAF-MEK-ERK pathway is correlated with worse survival in patients with ER-negative or progesterone receptor-negative breast cancers,363 suggesting that the alterations of the RAS-RAF-MEK-ERK pathway could serve as predictive and prognostic biomarkers for breast cancer.303 Meanwhile, the aberrations of the RAS-RAF-MEK-ERK pathway can be predictive biomarkers of drug sensitivity in cancer therapies.303 In conclusion, the RAF-MEK-ERK signaling cascade functions as a significant biomarker in tumor progression.
PTEN
PTEN was discovered in 1997 as a tumor suppressor,364 and it was the first phosphatase proven to have tumor suppressive effects.365,366 As a phosphoinositide 3-phosphatase, PTEN negatively regulates the PI3K-AKT-mTOR pathway by converting PIP3 to PIP2, thereby hindering the proliferation and survival of tumor cells.365,367 Furthermore, PTEN exerts both enzymatic and nonenzymatic effects in cellular epithelial-mesenchymal transition (EMT), migration and invasion, glucose and lipid metabolism, cell cycle, DNA repair, genomic stability, and gene transcription.365,368
PTEN function and expression are frequently altered in a variety of cancers.369 Accordingly, PTEN acts as a prognostic and predictive biomarker in various cancers including prostate cancer, RCC, PDAC, CRC, breast cancer, endometrial cancer, brain cancers, skin cancers, and hematological malignancies.370 Aberration of PTEN is associated with the mutations, downregulation or deletion of the PTEN gene, and the abnormal subcellular localization of PTEN protein.371,372 PTEN deletion modulates the downstream effector of mTORC1 by regulating 4EBP1 and p70S6 kinase to increase protein synthesis.372 Significantly, PTEN deletion is strongly linked to a shorter OS and DFS of cancer patients.370 Taken together, PTEN is a significant biomarker for tumor prognosis. The mechanism studies of PTEN activation will be beneficial for the development of antitumor strategies based on the recovery of PTEN function.
Evading growth suppressors
In addition to inducing and maintaining growth stimulus signals, tumor cells also eschew powerful programs to evade growth restriction and blockade, which mainly rely on the action of tumor suppressor genes.
Rb
Rb, the first tumor suppressor gene to be identified, was originally discovered in retinoblastoma children.373,374 The alteration of the Rb gene or inactivation of the Rb protein is one of the most common events in cancers.375 Rb primarily restricts the transcription of cell cycle genes by regulating E2F transcription factors.376 Rb proteins are phosphorylated by cyclin-dependent kinases (CDKs),377 which lead to Rb functional inactivation, followed by E2F transcriptional activation and cell cycle progression.378 Inactivation of Rb causes abnormalities in cell division, defects in cell cycle withdrawal, impaired induction of cellular senescence, and impaired cell cycle checkpoint control.379 The function of Rb in tumor cells is disrupted in various ways including Rb gene mutation, Rb protein hydrolysis, Rb-E2F interaction elimination, and the overactivation of CDK.375 Consequently, Rb dysregulation acts as a prognostic biomarker in cancers.380
TP53
TP53, often referred to as the “guardian of the genome”, is a gene encoding the p53 tumor suppressor protein.381 Numerous studies have shown that p53 plays an integral role in biological processes such as cell cycle arrest, aging, DNA repair, and apoptosis.382–384
In human tumors, TP53 is the most commonly mutated gene with ~50% of tumors carrying the mutations or deletion of TP53.385,386 In addition to mutations or deletion, tumors may lose the function of p53 due to other mechanisms. For example, overexpression of viral oncoproteins or MDM2 leads to the degradation of p53 protein.387 The expression and function loss or gain function of TP53 are associated with poor prognosis, immune escape, and anticancer drug resistance. Thus, TP53 can serve as an effective predictive biomarker to evaluate prognosis and monitor therapeutic responses in various cancers.388 An analysis of over 29,000 cases from the International Agency for Research on Cancer database revealed that TP53 mutations are potential prognostic biomarkers, and can be used to bolster the predictive accuracy of the OS and DFS of cancer patients.389
Enabling replicative immortality
Unlimited proliferation is a critical characteristic of tumor cells.299 Normal cells undergo senescence due to their recurrent division cycle, whereas tumor cells are capable of unlimited replication, a phenomenon known as immortalization. The protection of chromosome ends by telomeres is crucial for tumor immortalization.299
Telomerase
Telomere is a repetitive DNA–protein complex located at the end of chromosome.390 Telomeres in normal cells gradually shorten with continuous cell division and eventually fail to protect the end of chromosomal DNA, thus triggering DNA damage, cellular senescence, and apoptosis. Therefore, the length of telomeres is closely related to the cellular lifespan.299,391
Telomerase is a DNA polymerase that maintains telomere length by adding telomeric repeat fragments to telomeric DNA ends, thus compensating for the attrition of chromosomal ends in continuous cell division.390,392 Telomerase is encoded by the human telomerase reverse transcriptase (hTERT) gene which is the catalytic subunit of telomerase holoenzyme.390,393 hTERT is silenced in almost all somatic cells and is significantly re-expressed in ~90% of human cancers by various approaches.390 Thus, the large majority of normal human somatic cells lack the telomerase-maintenance mechanism, while a tremendous proportion of cancer cells have a highly active telomerase-maintenance mechanism.392 The activation of the telomerase-maintenance mechanism is observed in numerous human cancers, such as breast cancer, CRC, kidney cancer, cervical cancer, liver cancer, lung cancer, pancreatic cancer, prostate cancer, thyroid cancer, and bladder cancer,394 which ensure the replicative immortality of cancer cells. In clinical practice, cancer patients with high hTERT levels are along with worse survival than those with low hTERT levels. Moreover, cancer patients with high hTERT levels have a higher risk of disease recurrence and death.395 Therefore, telomerase is an independent prognostic biomarker of OS in cancer patients.395 Besides, TERT promoter mutations increase the expression of telomerase directly, which contributes to tumorigenesis and is associated with poor OS of cancer patients, suggesting that TERT promoter mutations are prognostic biomarkers for cancers.396 Moreover, the nonenzymatic functions of telomeres promote cancer cell proliferation and the resistance of apoptosis,299 regulate chromatin structure,397 impair DNA damage repair,397 and increase antioxidant protein expression,393 although the detailed mechanism remains to be elucidated.299
Inducing angiogenesis
In tumor initiation and progression, the new vascular system can transport nutrients and oxygen, and excrete metabolic waste, which is critical for tumor growth.299,398 The transition from prevascular hyperplasia to highly vascularized and progressively outgrowing tumors is known as the “angiogenic switch”. In the early stage of tumor development, the angiogenic switch is highly activated, which in turn sustains the continuous generation of new blood vessels, and causes the transition from dormant hyperplasia to outgrowing vascularized tumor, ultimately promoting rapid proliferation of cancer cells.299,398,399 The angiogenic switch, which favors a proangiogenic outcome during tumor angiogenesis, is controlled by the balance between proangiogenic and antiangiogenic factors secreted by tumor cells or TME cells.398 Studies have ascertained that angiogenesis significantly contributes to the development of various cancers, including CRC, breast cancer, bladder cancer, RCC, and NSCLC.400,401 A large number of angiogenic factors such as vascular endothelial growth factors (VEGFs) have been found to induce the proliferation and differentiation of endothelial cells directly or indirectly.
VEGF
VEGF, originally known as vascular permeability factor, was discovered as a tumor secretory factor in 1983 by Senger et al.402 In 1989, Ferrara isolated VEGF and renamed it vascular endothelial growth factor.403 VEGFs are heparin-binding homodimeric glycoproteins whose family includes VEGF-A (commonly referred to as VEGF), VEGF-B, VEGF-C, VEGF-D, VEGF-E, VEGF-F, and placental growth factor (PlGF).401,404 VEGF has been demonstrated to be a potent inducer of angiogenesis,399 and is widely expressed in normal adult organs along with two other related endothelial growth factors VEGF-B and VEGF-C, suggesting their necessary roles in tissue angiogenesis and homeostasis.399 VEGFs in tumor tissues are extracted not only from tumor cells but also from host cells,405 and high levels of VEGFs are found in diverse tumor cells. Interestingly, tumor cells are able to produce VEGFs, but are unable to respond to them due to the absence of VEGF receptors (VEGFRs) on the cell surface.406 Multiple factors such as genetic and epigenetic regulation influence the VEGF levels in tumor cells. Among them, epigenetic factors include hypoxia-inducible transcription factors 1α and 2α, low pH, inflammatory cytokines, growth factors, androgens, estrogens, and chemokines.406,407 Genetic factors include the activation of oncogenes such as RAS, EGFR, HER2, and the deletion and mutational inactivation of oncogenes such as p53, PTEN, and VHL.406,408
VEGFs bind respectively to the three tyrosine kinase receptors (RTKs) VEGFR1–3 with different specificities and affinities and VEGFR2 mediates the main VEGFR signals.409 VEGFR-1, the first RTK to be identified as a VEGF receptor,410 binds VEGF with a binding affinity ten times higher than that of VEGFR-2, although its ability of signal transduction is quite weak.406 VEGFR-1 serves as a decoy receptor to chelate or trap VEGF under some circumstances, thus negatively regulating VEGF activity by preventing VEGF binding to VEGFR-2.411–414 The specific mechanism of VEGFR-1 in VEGF-mediated angiogenesis needs to be explored in further detail.406,415 In contrast, VEGFR-2 is expressed in almost all endothelial cells, and exerts function through the activation of VEGF, VEGF-B, C, or D.416 The binding of VEGF to VEGFR-2 causes receptor dimerization and subsequently activates the intracellular signaling cascades, such as the PI3K-AKT and the RAF-MEK-ERK pathways, which generates neovascular branches required for tumor growth, and ultimately promotes rapid tumor cell proliferation and migration.406 VEGFR-3 has similar functions to VEGFR-2, but its action site is mainly in the lymphatic blood vessels.401,415 VEGFR-3 is expressed in the lymphatic endothelial cells415 and mainly binds to VEGF-C and VEGF-D to induce lymphangiogenesis.417,418 In addition, VEGFs interact with the neuropilin receptor family.415
VEGF levels are associated with the aggressiveness of tumors.419 The plasma VEGF levels in various cancer patients are elevated and negatively correlated with tumor prognosis.420 Moreover, VEGF levels are used to predict the efficacy of oral tyrosine kinase inhibitors (TKIs) in cancer patients.401 For example, VEGFR inhibitor sorafenib has displayed better therapeutic efficacy against advanced clear cell renal cell carcinoma (ccRCC) patients with high levels of VEGF.401 In summary, circulating VEGF and VEGFR-2 have been used as crucial biomarkers for the prediction of prognosis and antiangiogenic drug efficacy.405,421
FGF
Fibroblast growth factor (FGF) is a secreted glycoprotein422 that engages in the regulation of organogenesis,423 angiogenesis, and wound repair.422,424 FGF binds to the transmembrane FGF receptor (FGFR) on the surface of target cells with high affinity.422,425 The mammalian FGFR family consists of four highly conserved transmembrane RTK FGFR1-4, and FGFR5 which has no intracellular tyrosine kinase structural domain but has FGF binding capacity.422 FGFR is widely expressed in a broad range of cells, especially endothelial cells.426
In tumors, FGF is essential for vascular endothelial integrity, angiogenesis, tumor proliferation, survival, and metastasis.425,426 Notably, abnormal FGF signaling accelerates tumor proliferation by promoting tumor angiogenesis.422 For example, the elevated level of FGF2 in prostate cancer induces neovascularization to boost tumor growth.427 The increased angiogenesis induced by FGF1 amplification in high-grade serous ovarian cancer leads to reduced OS in patients, suggesting that FGF1 is a prognostic biomarker for ovarian cancer.428
Furthermore, FGFR is strongly associated with the development of various tumors,422,429 such as prostate cancer,430,431 lung cancer,432 breast cancer,433 and pancreatic cancer.434 In particular, studies have revealed that FGFR with mutations or amplification functions as driving oncogene to aberrantly activate downstream pathways, resulting in mitogenic, mesenchymal, and antiapoptotic responses in cells.422 Somatic mutations of FGFR3 have been observed in more than 30% of bladder cancers.435,436 The somatic mutations of FGFR2 have been discovered in 12% of endometrial cancers, and mutant endometrial cancer cell lines are highly sensitive to FGFR TKIs.437 Besides, FGFR amplification is also tightly linked to the progression of numerous cancers.438,439 Approximately 10% of gastric cancers have shown FGFR2 amplification, which is associated with the poor prognosis of gastric cancer patients.440 Amplification of FGFR1 occurs in approximately 10% of breast cancers, especially ER+ type.441,442 In brief, FGF and FGFR are vital biomarkers of tumor prognosis and treatment.
PDGF
Platelet-derived growth factors (PDGFs), an α-granule component secreted in an autocrine manner during platelet activation,443 are critical proangiogenic factors for tumor angiogenesis.443,444 The PDGF family contains four different monomeric polypeptide chains: PDGF-A, -B, -C, and -D, which form four homodimers through disulfide bonds (PDGF-AA, -BB, -CC, -DD) and a PDGF-AB heterodimer.443,445 The PDGF receptor (PDGFR) consists of RTKs PDGFRα and PDGFRβ. PDGF isoforms trigger different receptor dimerization and phosphorylation by binding to the corresponding PDGFRs, thus activating multiple downstream growth signaling pathways, such as PI3K, MAPK, and JAK/STAT pathways, to promote cancer cell proliferation, migration and invasion, angiogenesis, and drug resistance.443,446,447
PDGFs and their receptors are extensively expressed in a number of cancers, such as oral squamous cell carcinoma (OSCC),448 skin SCC,449 soft tissue sarcomas,450 ccRCC,451 dermatofibrosarcoma protuberans, gastrointestinal stromal tumors (GIST),452 CRC,453 breast cancer,447 pancreatic cancer,454 gastric cancer,455 neuroendocrine tumors,456 NSCLC, ovarian cancer, and HCC.443 High PDGF-A levels correlated independently and inversely with the risk of metastatic relapse in cancer patients.450 The level of PDGF-D is associated with advanced tumor stages and the development of bone metastasis.457,458 High expression of PDGFR-β is independently linked to prostate cancer recurrence.445 In conclusion, PDGFs and PDGFRs are meaningful diagnostic biomarkers.
Resisting cell death
Resisting cell death is a significant tumor hallmark that contributes to tumor progression and therapeutic resistance.299 Apoptosis that leads to programmed cell death hinders tumorigenesis, and the apoptotic program is considerably reduced in highly aggressive and therapy-resistant tumor cells.299 Increasing autophagy activation might inhibit tumorigenesis in parallel with or in concert with apoptosis.459,460 Moreover, necrosis also significantly contributes to tumor cell death.461 The identification of biomarkers in these processes is useful for tumor diagnosis or prognosis.
Apoptosis
Sydney Brenner, Robert Horvitz, and John Sulston shared the 2002 Nobel Prize in Physiology or Medicine for their contributions to the discovery of apoptosis procedure.462 Cellular stress, DNA damage, and immune surveillance systems frequently cause apoptosis, a type of cell death that is initiated by the proteolytic cleavage of numerous proteins and the regulation of caspase protease activity.463 Apoptosis can be triggered through the intrinsic or mitochondrial pathway and the extrinsic pathway.464 The intrinsic pathway is controlled by the B-cell leukemia or lymphoma gene number 2 (BCL-2) family. BCL-2 induces mitochondrial outer membrane permeabilization and the release of multiple proapoptotic factors, followed by the release of cytochrome c from mitochondria to the cytoplasm. Subsequently, the apoptotic peptidase activating factor 1 interacts with cytochrome c, and form the apoptosome that induces the activation of the initiator caspase pro-caspase 9. Later on, the caspase 9 binds to the apoptosome and is cleaved and activated, which subsequently stimulates the activation of initiator caspase 3.465,466 This process in which cytochrome c is released from the mitochondria is negatively regulated by antiapoptotic BCL-2 family members such as BCL-2, B-cell lymphoma-extra large (BCL-XL), BCL-W, BCL-2-A1, and MCL1.463 The membrane permeabilization and the release of cytochrome c into cytoplasm are key processes for triggering apoptosis.463,467
The extrinsic apoptotic pathway is initiated through the proapoptotic death receptors which include Fas, the tumor necrosis factor receptor (TNFR) family such as TNFR1, TNFR2, and theTRAIL receptors DR4 and DR5. The proapoptotic death receptors bind to ligands and then trimerize and aggregate within the cell membrane, subsequently recruiting adapter proteins such as FADD, caspase 8 and/or caspase 10 to form the death-inducing signaling complex, which activates the initiator caspase 8, which in turn induces the activation of the effector caspases such as caspase 3, 6, and 7, and apoptosis.463,467 Consequently, the potential strategy for cancer therapy is targeting the proapoptotic and antiapoptotic proteins to induce apoptosis.468
BCL-2/BCL-XL
The BCL-2 family proteins have four conserved BCL-2 homology (BH) structural domains (BH1, 2, 3, and 4) which can be divided into three subfamilies based on the homology and function of proteins: the antiapoptotic BCL-2 family members (such as BCL-2 and BCL-XL), the multi-BH-domain proapoptotic members, such as the BCL-2-associated X protein (BAX) and the BCL-2 antagonist/killer (BAK), and the proapoptotic “BH3-only” proteins, such as the BCL-2 interacting mediator of cell death (BIM), and PUMA.469
BCL-2 was the first identified apoptosis regulator, which was activated by chromosome translocation in human follicular lymphoma oncoprotein.470 The BCL-X gene was cloned in 1993,471 and the BCL-XL protein, which is localized in the mitochondrion, is the first protein whose three-dimensional structure has been identified in the BCL-2 protein family.472,473 The “BH3-only” proteins can be divided into activators and sensitizers.474,475 Activators of BH3 proteins, such as BIM, BID, initiate apoptosis by directly inducing BAX and BAK oligomerization and cytochrome c release. However, sensitizer BH3 proteins, such as BAD, and BIK, exert proapoptotic functions by binding to antiapoptotic BCL-2 family members, rather than directly activating BAX or BAK.475–477 The interaction of one protein’s BH3 α-helix with a sizable hydrophobic pocket on binding partners regulates the activity of BH3-only proteins,475 which initiates apoptosis by activating proapoptotic proteins or by inhibiting antiapoptotic proteins.478
BCL-2 can drive oncogenic transformation, hinder apoptosis, and increase tumor cell survival.479,480 The high expression of BCL-XL is involved in tumor cell invasion, the maintenance of tumor stem cell phenotype, angiogenesis, and metastasis through inducing apoptosis resistance.480 The overexpression of BCL-2 and/or BCL-XL may contribute to tumor progression and the resistance of chemotherapeutic agents in various tumors,467,475 including pancreatic cancer,481 ovarian cancer,481 lung cancer,481 prostate cancer,481 breast cancer,482 neuroblastoma,483 CRC,484 gastric cancer,485 HCC,469 chronic lymphocytic leukemia (CLL),469 lymphoma,481 and multiple myeloma.481 Furthermore, BCL-XL can be used as an independent biomarker for the prognosis prediction of CRC patients.484 BCL-2 is a prognostic biomarker in TNBC patients. Lower BCL-2 expression level is associated with better outcomes of TNBC patients treated with both adjuvant and neoadjuvant chemotherapy.486 In summary, BCL-2 and BCL-XL are essential biomarkers in tumor prognosis and treatment.
BAX/BAK
BAX, a cytosolic membrane protein that works as a critical regulator of the apoptotic process, was identified by immunoprecipitation and yeast two-hybrid screening.480,487 BAX protein has BH1, BH2, and BH3 structural domains,480,488 which is highly homologous with BCL-2.480 BAX stimulates apoptosis either by inhibiting BCL-2 and BCL-XL or by directly triggering the apoptotic pathway.480 BAX moves from cytoplasm to mitochondria during apoptosis, followed by oligomerization and the formation of pores in the outer mitochondrial membrane, thus facilitating the release of cytochrome c which activates the downstream effector caspases and leads to cell death.469,489,490 Downregulation and mutations of BAX are essential for apoptosis resistance,491 and BAX acts as a potential prognostic and predictive biomarker in various cancers including gastric cancer, esophageal cancer, and CRC.492 The somatic frameshift mutations of the BAX gene highly occur in CRC with the microsatellite mutator phenotype.493 BAX mutations are found in ~21% of human hematopoietic malignancies such as ALL.494 Reduced BAX expression is a major factor in cisplatin resistance of ovarian cancer cells,495 5-FU resistance of CRC cells,496 and zoledronate resistance of lung cancer cells.497 The decreased BAX/BCL-2 ratio can be induced by BAX abnormalities, which affects the temozolomide-induced resistance in U87MG cells and paclitaxel-resistant breast cancer cells. Thus, the activation of BAX could be used to promote apoptotic cell death and overcome resistance.498
Furthermore, the high BAK expression is correlated with improved OS and PFS in patients with advanced gastric cancer. BAK is a predictive and prognostic biomarker for the therapeutic effect of docetaxel in patients with advanced gastric cancer.499 BAX-BAK heterodimer is also used as a pharmacodynamic biomarker of on-target drug action of MCL1 inhibitors.500
Autophagy
In 1955, Christian de Duve discovered the lysosome,501 a key organelle for intracellular degradation, and subsequently introduced the term “autophagy” at the CIBA Foundation Symposium on Lysosomes in 1963.502 In 2016, Yoshinori Ohsumi was awarded the 2016 Nobel Prize for Medicine or Physiology for elucidating the mechanism of autophagy, which led to increasing attention to autophagy in health and disease.460,503 To date, there are three main types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy. The general term “autophagy” usually means macroautophagy.504
Autophagy is a multistep, highly conserved degradation process: the initiation and nucleation of the autophagosome, the expansion, and elongation of the autophagosome membrane, the closure and fusion with the lysosome, and the degradation of products.460,505 Briefly, autophagy is triggered by a variety of factors, including nutrient or growth factor deprivation, energy status, hypoxia, ROS, and other stress inducers.506 Subsequently, a flat membrane named the phagophore or isolation membrane sequesters cytoplasmic constituents. The elongating phagophore results in complete sequestration and the formation of a double-membraned organelle autophagosome. Then, the autophagosome fuses with the lysosome, and the inner membrane of the autophagosome and the cytoplasm-derived materials it contains are subsequently degraded by the lysosome, resulting in the production of amino acids and lipids which are exported to the cytoplasm for recycling.504,507,508
Mechanistically, autophagy-associated (ATG) proteins, a group of evolutionarily conserved proteins, are responsible for this process.509 Autophagy begins with the activation of unc-51 like autophagy activating kinase 1 (ULK1) (also known as ATG1) complex which includes ULK1, ULK2, ATG13, FIP200, and ATG101. The ULK1 complex subsequently activates class III PI3K complex which includes VPS15, VPS34, ATG14, Beclin1, UVRAG, and AMBRA1, which mediates vesicle nucleation.460 Then, the ATG5-ATG12 complex binds to ATG16 to extend the autophagosomal membrane, and members of the LC3 and GABARAP protein families conjugate with lipid phosphatidylethanolamine (PE) and recruit PE to the membrane. ATG4B binds to ATG7 and then couples with LC3-I and PE to form LC3-II. Eventually, autophagosomes fuse with lysosomes to degrade macromolecules and reuse them. The adapter protein sequestosome-1 (also known as p62) targets autophagosome-specific substrates and LC3-II which are simultaneously degraded.460,510
Autophagy is a double-edged sword in cancer. The enhanced autophagic flow in tumor cells accelerates tumor cell growth, while the induction of autophagy can prevent the development of cancer.459,460 Therefore, autophagy inhibition and promotion are both promising strategies for cancer therapy, and their application depends on the actual situation.511
Beclin 1
Beclin 1 was identified as a BCL-2 interaction factor in the yeast two-hybrid screen in 1988.512,513 Human Beclin 1, the mammalian orthologue of yeast Atg6, consists of a BCL-2-homology 3 structural domain,514 a flexible helical domain,515 a coiled coil domain,516 and an evolutionarily conserved domain.514 Moreover, Beclin 1 contains a leucine-rich nuclear export signal that is essential for its autophagic and tumor suppressor functions.517 Beclin 1 is phosphorylated by ULK1 and acts as an integral component of the PI3K complex to localize autophagy proteins to the phagosome. Furthermore, Beclin 1 interacts with and is inhibited by BCL-2/BCL-XL in the BH3 structural domain, which blocks the formation of the Beclin 1-VPS34 complex and inhibits Beclin 1 interacting with UVRAG, thereby inhibiting autophagy. The bind of AMBRA1 to Beclin 1 stabilizes the Beclin 1-VPS34 complex, thus promoting autophagosome formation.518,519
Studies have found that Beclin 1 is a prognosis biomarker for various cancers. Reduced expression of Beclin 1 has been observed in brain tumors and cervical cell carcinomas.520 The absence of BECN1 has been found in 40 to 75% of sporadic breast cancer and ovarian cancer,521 and 40% of prostate cancer.522 Low expression of Beclin 1 is associated with the malignant phenotype and poor prognosis of gastric cancer.523 Beclin 1 inhibits the proliferation of human breast cancer cells MCF7 in vitro and in vivo through regulating autophagy.524 On the contrary, the elevated Beclin 1 expression is related to distant metastasis and poor prognosis in CRC patients, and reduced survival in CRC patients with 5-FU treatment.525 Taken together, Beclin 1 may serve as a valid prognostic indicator and therapeutic target for cancers although further research is needed to determine its specific mechanism in different cancers.526,527
LC3B
The microtubule-associated protein 1 light chain 3B (LC3B or MAP1LC3B) is a classical autophagy marker, is cleaved by protease at the C-terminus to form free LC3B-I, and LC3B-I binds to PE to form membrane LC3B-II in autophagy occurrence. The process in which LC3B-I converts to LC3B-II is essential for phagophore expansion and the formation of autophagosomes.528,529 Thus, LC3B is a marker for the detection of multiple autophagic fluxes.530 Accordingly, LC3B-II is one of the most commonly used biomarkers to detect the number of autophagosomes and autophagosome-related structures.518
The high LC3B expression is closely associated with the aggressive progression, and poor prognosis of multiple tumors, including gastric cancer,531 CRC,532 TNBC,533 melanoma,534 astrocytoma,535 esophageal cancer,536 and OSCC.537 Studies have found that LC3B has the highest expression in TNBC cells in different molecular subtypes of breast cancer,538 and its high expression is related to the progression and poor prognosis of TNBC patients.533 Moreover, LC3B is closely connected with the vascular invasion and lymph node metastasis of HCC, and is a potential therapeutic target for HCC.539 Collectively, LC3B is a meaningful prognostic biomarker in cancer management.527,534
ULK-1/2
ULK1, a conserved Ser/Thr kinase, plays a pivotal role in autophagy induction.540 High expression of ULK-1 is associated with poor prognosis in various tumors, including esophageal SCC,541 HCC,542 NPC,543 prostate cancer,544 and CRC.545 Studies have found that HCC patients with ULK1 and LC3B overexpression have larger tumors and a higher frequency of lymph node metastasis. The combination of ULK1 and LC3B is an independent predictor of OS and PFS in HCC patients.546 After androgen deprivation therapy, prostate cancer patients with high levels of ULK1 have shorter PFS and OS.544 In addition, elevated expression of ULK1 has been connected to lymph node metastasis547 and functions as a prognostic biomarker in patients with CRC.545 Interestingly, low expression of ULK1 is associated with operable breast cancer progression and is a poor prognostic biomarker for patient survival.548 In human NPC, ULK1 is also a promising biomarker for the prediction of poor prognosis and treatment response.543 Furthermore, ULK2 has been found to be expressed at higher levels in prostate cancer tissues compared with that in normal tissues.549 To better determine the prognostic value of ULK1 and ULK2 in different cancer types, comprehensive studies in prospective cohorts are necessary.530
p62
p62 (also known as sequestosome-1, SQSTM 1) was originally identified as an atypical protein kinase C (aPKC) interacting protein.550 p62 consists of several structural domains, including the N-terminal PB1 domain, the ZZ-type zinc finger (ZZ) domain, the tumor necrosis factor receptor-associated factor 6 (TRAF6) binding (TB) domain, the LIR domain, the Kelchlike ECH-associated protein 1 (Keap1)-interacting region (KIR), and the C-terminal UBA domain.551,552 Each structural domain of p62 has a different function. The PB1 domain is essential for the formation of homodimeric aggregates that regulate autophagic degradation. Moreover, p62 can interact with other proteins containing the PB1 domain, such as MAPK.551 ZZ structural domain is involved in the activation of NF-kB signaling pathway,553 and the TB structural domain can interact with TRAF6 which induces protein polyubiquitination.554 The LIR structural domain affects autophagosome formation and autophagic degradation by mediating LC3-p62 interactions,551,555 and the KIR structural domain activates Nrf2 by binding with Keap1.556,557 UBA structural domain is involved in autophagic lysosomal degradation558 and apoptosis signaling pathways.551
As a marker for autophagic flow detection, p62 accumulation usually represents the inhibition of autophagy.460,559 Upregulation or reduced degradation of p62 is associated with tumor progression and anticancer drug resistance.552 p62 expression is increased in 60% of lung adenocarcinomas and 90% of lung SCCs.550 Numerous studies have shown that high p62 expression is correlated with the aggressiveness and poor prognosis of cancers, including endometrial cancer,560 OSCC,537 epithelial ovarian cancer,561 and NSCLC.562 In addition, elevated p62 expression is also correlated with the high-grade, distant metastasis and reduced 5-year survival of breast cancer patients,563 especially in patients with TNBC cancer.564 In short, p62 is a meaningful prognostic biomarker and a potential target for cancer therapy.551,552
Necrosis
Necrosis is derived from the Greek “nekros” for corpse.461 Necroptosis is a programmed necrotic cell death type in a caspase-independent f manner, induced by TNFR superfamily and mediated by receptor-interacting protein kinase 1 (RIPK1, also known as RIP1), RIPK3 (also known as RIP3), and mixed lineage kinase domain-like (MLKL).565,566 Necrosis is caused by numerous stimuli such as cytokines, viral infection, pathogen-associated molecular, T-cell receptors, interferon receptors, Toll-like receptors, cellular metabolism, genotoxic stress, and various anticancer compounds.565,567 Common morphological features of necrotic cells include moderate chromatin condensation, cytoplasmic organelle swelling, and the rupture of plasma membrane.568 The biochemical characters of necrotic cells include a drop in ATP level, the activation of RIP1, RIP3, and MLKL, the release of damage-associated molecular pattern molecules (e.g., HMGB1), the hyperactivation of poly(ADP-ribose) polymerase 1 (PARP1).568 The basic feature that distinguishes necrosis from apoptosis is the rapid loss of cell membrane potential. Cellular energy depletion, membrane lipid damage, and the impairment of steady-state ion pump function lead to loss of membrane potential which in turn leads to cytoplasmic swelling, plasma membrane rupture, and cell lysis, thus promoting necrotic cell death.461
Studies have identified that necrosis is an essential predictor for prognosis and treatment response in various tumors, including pancreatic cancer,569 RCC,570 breast cancer, lung cancer, CRC,571 and soft tissue sarcoma.571,572 Tumor necrosis is closely associated with cancer-specific survival, OS, RFS, and PFS in patients with RCC, and it can be a prognostic biomarker of patients in clinical practice.570 Therefore, the discovery of biomarkers that identify necrosis and molecular mechanisms of necrosis enables the development of necrosis-based antitumor therapies.569
RIPK3
The serine/threonine kinase RIPK1 is a key regulator of necrosis, and RIPK3 is a downstream regulator of RIPK1.573,574 The RIPK1-RIPK3-MLKL complex, also known as the “necrosome”, mediates upstream cell death receptors and downstream signaling.565 Necrosome is a multiprotein complex that contributes to TNF-induced cell death.575,576 Necrotic cells trigger caspase 8 inactivation and activate RIPK1 and RIPK3, followed by autophosphorylation and cross-phosphorylation between RIPK1 and RIPK3 to form necrosome. Then, MLKL is phosphorylated, followed by oligomerizing and translocating to the plasma membrane and stimulating the necroptosis.577,578
The RIPK3 expression is significantly reduced in AML patients,579,580 which is consistent with the high methylation level near the transcriptional start site of RIPK3.581 RIPK3 deficiency promotes leukemogenesis by enhancing the accumulation of leukemia-initiating cells, and hinders myeloid differentiation through reducing cell death and IL-1β production.579,580 In addition, RIPK3 expression plays an important role in solid tumors. RIPK3 has been discovered to be downregulated in various cancer cells, including breast cancer,581 melanoma,582 lung cancer,583 and CRC.584 RIPK3 is downregulated in human CRC tissues compared with normal tissues,585,586 and the deletion of RIPK3 accelerates colorectal tumorigenesis in mice through sustained inflammation.577,585 Consistent with the above observations, low RIPK3 levels are strongly correlated with poor prognosis in patients with CRC586 and breast cancer.581 On the contrary, the expression of RIPK3 is elevated in several other tumors, such as serous ovarian cancer,587 pancreatic cancer,588 and colitis-associated cancer and colon cancer.589 RIPK3 promotes colitis-associated CRC through tumor cell proliferation and CXCL1-induced immunosuppression, and RIPK3 deficiency significantly reduces colitis-associated CRC development in mice.577,589 In conclusion, RIPK3 is a potential prognostic biomarker for tumors, although its role needs to be analyzed on a case-by-case basis.
MLKL
MLKL is a key factor in necroptosis execution,574,576,590 and a vital determinant of treatment response and poor prognosis in cancer patients.579,591 The low expression level of MLKL is significantly associated with lower OS in gastric cancer,592 ovarian cancer,593 cervical SCC,594 colon cancer,577,595 and pancreatic cancer.591 Moreover, in resected PADC patients receiving adjuvant chemotherapy, the low expression level of MLKL is related to decreased RFS. Thus, MLKL has become a prognostic biomarker for patients with early-stage resected PDAC.591 However, high levels of MLKL are tied to poor prognosis in patients with colon and esophageal cancers.596 The mRNA expression level of MLKL in gastric cancer tissues is significantly higher than that in normal tissues.592 The possible reason for this difference is that some cancer cells activate necrosis to modulate the immune system, and the exact mechanism needs to be further investigated.577 In short, MLKL is a potential prognostic biomarker for cancer patients.
Activating invasion and metastasis
Tumor metastasis is a process of transferring tumor cells from the primary lesion tumor to distant tissues and organ cascades.597,598 Tumor metastasis is divided into multiple steps: (1) tumor cells invade the extracellular matrix (ECM) and the surrounding stroma; (2) tumor cells enter into the bloodstream directly or the lymphatics; (3) tumor cells survive in the circulation; (4) tumor cells arrest in the circulation and arrive at distant organ sites; (5)tumor cells extravasate and invade into the parenchyma of distant tissues; (6) survival in the microenvironment and grow to form metastatic colonization599–601 (Fig. 5). In 1889, Stephen Paget vividly compared tumor metastasis to fertile “seeds” (tumor cells) falling on “congenial soil” (the metastatic microenvironment).602,603 Many changes occur in “seeds” during the metastasis process, including proteolytic degradation of basement membranes and ECM, changes in tumor cell adherence to cells and the ECM, and physical motility of tumor cells.599,604 Meanwhile, homeostasis of “soil” is also altered before tumor cells arrival by modulating the cellular composition, immune status, blood supply, and ECM of the metastatic site to create a microenvironment conducive to tumor cell colonization.605
Fig. 5.
The cancer invasion and metastasis and its targeted therapy. The tumor metastasis process consists of multiple steps. Initially, tumor cells invade the surrounding stroma and extracellular matrix from the primary tumor site, and then intravasate into the bloodstream or the lymphatics. Subsequently, tumor cells arrest in the circulation and arrive at distant organ sites, followed by extravasating and invading the parenchyma of distant tissues. Finally, tumor cells adapt to the new microenvironment and grow to form metastatic colonization. EMT is the basic embryonic developmental process that transforms polarized non-motile epithelial cells into motile and invasive mesenchymal cells. Multiple cellular stress conditions including hypoxia, inflammation, metabolic stress, and signaling cascades, can induce the expression of EMT transcription factors and prompt tumor metastasis. Meanwhile, MET amplification and mutation, the transcriptional dysregulation of c-MET, degradation deficiency, and abnormal HGF production result in the abnormal expression of HGF/c-MET and tumor progression. Various inhibitors including MMP inhibitors and HGF/c-MET inhibitors have been developed and emerging as promising tools in the suppression of tumor metastasis. c-MET mesenchymal-epithelial transition factor, EMT epithelial-mesenchymal transition, HGF hepatocyte growth factor, MMPs matrix metalloproteinases
Activated invasion and metastasis have been recognized as one of tumor hallmarks299,600 and a major cause of death in patients with solid tumors.606 Predicting tumor metastasis facilitates the implementation of personalized therapy in the clinical treatment of tumors, leading to better outcomes for cancer patients. Thus, identifying metastatic biomarkers helps to detect initial tumor metastasis or recurrence in clinical practice, thus improving the potential treatment and management strategy for cancer patients.
E-cadherin
Cadherins are a superfamily of at least 80 specific types of adhesion molecules characterized by the ability to form calcium-dependent intercellular homophilic bonds,607 which are involved in the regulation of tumor cell recognition, tumor suppression, and tissue morphogenesis.608 Common family members include Epithelial (E)-cadherin, Neuronal (N)-cadherin, and Placental (P)-cadherin.609 E-cadherin, a homophilic cell-cell adhesion molecule,610 is a type I cadherin expressed in epithelial cells. E-cadherin is the first member of the cadherin superfamily to be identified.607 The human E-cadherin gene (CDH1) is located on chromosome 16q22.1. The structure of mature E-cadherin consists of three parts: a highly conserved carboxyterminal cytodomain that is identical in all cadherin family members, a single-pass transmembrane domain, and an extracellular domain that consists of five cadherin-motif subdomains with putative calcium-binding sites.610,611 E-cadherin mediates cell adhesion through calcium-dependent trans-homodimeric interactions of its EC1 structure with the EC1 domain of adjacent cells, while the cytoplasmic part interacts with adherens junctions-related molecules such as β-catenin.612 E-cadherin plays a significant role in normal embryonic development, organ morphogenesis, and tissue formation by regulating proliferation, migration, or maintaining epithelial cell polarity.613 Thus, E-cadherin is a biomarker of the epithelial cell layer.613
The embryonic program EMT is of great importance in the progress of epithelial-derived tumors from benign lesions to invasive carcinomas and metastases.613,614 This process is accompanied by changes in cadherin expression599,615: from E-cadherin which promotes tumor adhesion and blocks invasion, to N-cadherin which is expressed in mesenchymal cells to promote tumor cell invasion,599 and E-cadherin dysfunction is an EMT landmark in this process.613 The causes of abnormal E-cadherin in tumor cells include reduced or absent E-cadherin expression, mutations or reduced transcription of E-cadherin genes, abnormal redistribution of E-cadherin within cells, the shedding of E-cadherin from the cell surface, and competition with other proteins for binding.610 In addition, E-cadherin is an important tumor growth suppressor,616 and inhibits tumor cell growth by upregulating p27-induced cell cycle arrest. Inhibition of E-cadherin leads to a decrease in cell adhesion, which promotes tumor metastasis.610
Several investigations have demonstrated the critical role that E-cadherin plays in tumor progression. E-cadherin is closely connected to pathological and clinical characteristics of tumor patients, such as the degree of differentiation, aggressiveness, venous permeation, peritoneal seeding, infiltrative growth, liver and bone metastasis, lymph node metastasis, tumor staging, and poor prognosis.609,617–619 The deletion or downregulation of E-cadherin promotes tumor invasion, infiltrative growth, and dedifferentiation.610,616 Thus, E-cadherin can be utilized as a prognostic biomarker of tumor metastasis for multiple tumors,609 including CRC,620 gastric cancer,621 pancreatic cancer,622 esophageal cancer,623 liver cancer,624 lung cancer,625 bladder cancer,626 prostate cancer,627,628 breast cancer,629 endometrial cancer,630 ovarian cancer,631 thyroid cancer,632 and HNSCC.633
EMT transcription factors
EMT is the basic embryonic developmental process that transforms polarized non-motile epithelial cells into motile and invasive mesenchymal cells.634,635 In tumor cells, EMT promotes tumor cell invasion and metastasis, induces cancer stem cell (CSC) stemness, chemoresistance, immune evasion, and cellular metabolic reprogramming,636,637 and inhibits senescence.638
EMT is regulated by EMT transcription factors which are classified according to their direct or indirect repression of E-cadherin.634 The direct repressors include zinc finger proteins of the Sail superfamily Snail1 (also known as Snail), Snai2 (Slug), and Snai3 (Smuc), zinc finger E-box binding protein (ZEB) family members ZEB1 and ZEB2. The indirect repressors including the basic helix-loop-helix proteins Twist1 and Twist2.634,639 Tumor cell stress conditions, such as hypoxia, inflammation, or metabolic stress, stimulate signaling cascades, such as Wnt, Notch, TGF-β, and RAS, and induce the expression of EMT transcription factors Snail, Slug, Twist, and ZEB.634,640 Then, EMT transcription factors induce downstream effects of EMT by a series of processes including regulating epithelial marker-related or mesenchymal marker genes, activating matrix metalloproteinases (MMPs) expression or interacting with epigenetic regulators to promote oncogenic transformation, modulating CSCs, generating chemoresistance, and increasing tumor angiogenesis, and ultimately promoting tumor cell motility and metastasis.634,638,641 In addition, EMT transcription factors also regulate tumor prosurvival phenotypes, such as participating in tumor cell DNA repair, the evasion of senescence and apoptosis, and immune evasion, providing survival advantages for tumor cells under various stress conditions.642
A significant enrichment analysis of 244 differentially expressed EMT-related genes in CRC has revealed that EMT-related signaling pathway genes are highly related to the prognosis prediction of CRC patients, where higher risk scores indicate poor prognosis.643 In conclusion, EMT transcription factors have been considered as prognostic biomarkers for tumor aggressiveness and metastasis in clinical practice.644–646
Twist
Twsit1 and Twsit2 are highly conserved basic helix-loop-helix transcription factors,638,647 which are pivotal regulators of embryonic morphogenesis.648 Twist is expressed in mesodermal and ectodermal-derived tissues, and it has been found that Twsit1 and Twsit2 which are structurally homologous are overexpressed in multiple human cancers.647,648 Twsit1 overexpression has been confirmed to be strongly associated with aggressiveness and metastasis in cancer patients, including sarcoma, glioma, melanoma, ESCC,649 neuroblastoma,650 cervical cancer,651 RCC,652 and hematological malignancies including AML, chronic myeloid leukemia (CML), ALL, CLL, lymphomas.653 In CML patients, the increased expression of Twist is related to tumor progression, tumor staging, and drug resistance, and Twist can be applied as a biomarker to assess MRD.653 Inhibition of Twist expression has been found to impair the high metastasis of breast cancer cells from the mammary gland to the lung.648 Collectively, Twist is a meaningful biomarker for tumor prognosis and metastasis.647
Snail
Snail, the first member of the snail superfamily, was first described in Drosophila melanogaster,654,655 and is essential for cellular mesoderm formation.654 The three members (Snail, Snai2, and Snai3) of the Snail family share a similar structure: a highly conserved C-terminal domain containing four to six C2H2-type zinc finger.656 In cancer cells, Snail functions as a transcriptional repressor by binding to the E-box motif (CAGGTG) of Snail-related genes with its C-terminal structural domain, thus inhibiting the transcription of target genes.654,656 For example, Snail downregulates E-cadherin expression and thereby induces EMT and basal-like phenotype conversion.636 The overexpression of Snail is associated with poor prognosis in patients with breast cancer,657 CRC,658 and liver cancer.659 Snail expression is significantly higher in the high-stage, high-grade, and significant lymphovascular invasion patients with upper urinary tract urothelial carcinoma.660 Slug, a member of the Snail family, also has a striking impact on EMT. Slug expression is an independent prognostic biomarker for poor survival in CRC661 and esophageal SCC patients.662
ZEB1/2
ZEB1 (also known as Zfhx1a and Zfhep) and ZEB2 (also known as SIP1 and Zfhx1b),663 members of the ZEB transcription factor family,638 which are encoded by the ZFHX1a and ZFHX1b genes.664 Both ZEB1 and ZEB2 possess two separated clusters of C2H2-type zinc fingers which bind to paired E-box promoter elements.664 ZEB1 is a key regulator of tumor cell plasticity and metastasis.665 Mechanically, ZEB1 binds directly to the E-box in the promoter of the CDH1 gene which encodes E-cadherin, blocking CDH1 transcription and inducing EMT.666 ZEB1 overexpression is strongly associated with highly aggressive precursor lesions and poor prognosis of pancreatic cancers.665,667,668 ZEB1 deficiency reduces stemness, tumorigenic, and colonization capacities in CSCs of pancreatic cancer, thereby inhibiting the formation of undifferentiated high-grade cancers, invasion, and metastasis.665 ZEB1 overexpression serves as a significantly independent adverse prognostic factor for RFS and OS in metaplastic breast cancer.669 The knockdown of ZEB1 in human breast cancer cells results in approximately 230 gene changes, most of which are related to epithelial differentiation and intercellular adhesion.666 Moreover, aberrant expression of ZEB1 is associated with multiple tumor progression and metastasis, including uterine cancer, osteosarcoma, lung cancer, liver cancer, and gastric cancer, which reveals the importance of ZEB1 in EMT induction and tumor development.666
ZEB2 is a DNA-binding transcriptional repressor consisting of multiple functional domains which interact with various transcriptional effectors.670 ZEB2 is proven to be highly expressed in human cancer cell lines lacking E-cadherin protein. Overexpression of ZEB2 blocks E-cadherin protein-mediated intercellular adhesion and promotes tumor cell metastasis.671 ZEB2 promotes the migration and invasion of breast cancer,672 bladder cancer, ovarian cancer, stomach cancer, CRC,673 OSCC,674 and pancreatic cancer.667
HGF/c-MET
c-MET, also known as RTK Met, was first identified as a proto-oncogene in the 1980s.675,676 c-MET is a disulfide-linked heterodimer composed of an extracellular α-subunit and a single-pass transmembrane β-subunit, which is translated and cleaved form pro-c-MET, a 170 kDa single-stranded precursor protein.677,678 The β-subunit of c-MET is involved in the regulation of kinase activity and effector signaling by forming extracellular and partially intracellular structural domains.677 Hepatocyte growth factor (HGF, also known as scatter factor) is the only known c-MET ligand,677 which is a 90 kDa heterodimer composed of an α chain and a β chain.677 HGF consists of six structural domain groups: amino-terminal domain (N), four kringle domains (K1–K4), and a serine proteinase homology (SPH) domain,675 of which the N-terminal and the first kringle region are c-MET high-affinity binding sites. HGF induces c-MET dimerization and phosphorylates c-MET residues Y1349/1356, subsequently activating various downstream signaling pathways including the ERK1/2, p38/MAPK, and PI3K-AKT, ultimately promoting cell proliferation and survival.675,677
Under normal physiological conditions, HGF/c-MET is involved in cellular processes such as embryogenesis, angiogenesis, wound healing, and organ regeneration. While abnormal expression of HGF/c-MET in tumor cells including MET amplification and mutation, the transcriptional dysregulation of c-MET, degradation deficiency, and abnormal HGF production are closely related to tumor progression.677,679 c-MET activation enhances tumorigenicity, invasion, and metastasis.680,681 High expression of HGF/c-MET is revealed in various cancers and is closely associated with the poor prognosis of cancer patients.675,682,683 For example, c-MET locus amplification occurs in patients with gastrointestinal cancers such as gastric cancer, metastatic CRC, gastroesophageal cancer, and esophageal adenocarcinoma.677,679 c-MET mRNA and protein levels are significantly higher in liver metastasis of CRC than in primary CRC, and its expression is positively correlated with tumor stage in CRC liver metastasis.684 Besides gastrointestinal cancers, c-MET mutations are found in papillary renal cancer,685 ovarian cancer,685 SCLC,686 HNSCC,687 and childhood HCC.679,685 Elevated HGF levels are found in various cancers including head and neck cancer,688 cervical cancer,689 HCC,690 and lung cancer,691 and are associated with poor prognosis. HGF promotes HCC migration and invasion, and is positively correlated with HCC metastasis.692 HGF has been observed to be an independent blood-based predictive biomarker and primary diagnostic marker in ovarian cancer patients.693 Moreover, HGF/c-MET can be used as a prognostic biomarker in various hematologic tumors, such as B-cell lymphoma, T and natural killer (NK) cell lymphoma, and Hodgkin lymphoma.694
Furthermore, c-MET activation mediates resistance to TKIs, chemotherapy, cetuximab, and radiotherapy in CRC patients.677 c-MET mediates radio-resistance by increasing cell motility and inhibiting apoptosis through autocrine and paracrine signaling.695 HGF co-amplification leads to clinical resistance in MET-amplified esophagogastric cancer.696 In conclusion, c-MET/HGF overexpression is an independent biomarker of poor prognosis and drug resistance in patients with various hematologic and solid tumors.
N-cadherin
N-cadherin, also known as cadherin 2 or CDH2,697 was identified in the 1980s.698 N-cadherin is a single-pass transmembrane calcium-binding glycoprotein that mediates intercellular adhesion,699,700 and consists of five extracellular substructural domains (EC1-EC5).701 In addition to expression in normal cells such as neuronal cells, osteoblasts, stromal cells, and endothelial cells.702 Studies have found that N-cadherin is highly expressed in various tumors including melanoma,703 neuroblastoma,704 breast cancer,705 urothelial cancer,702 ovarian cancer,706 and multiple myeloma.701 Abnormal expression of N-cadherin promotes tumor cell survival, proliferation, invasion, and metastasis by regulating signaling pathways, such as fibroblast growth factor receptor signaling, canonical Wnt signaling,701,702 and signalings involved in neovascularization and vascular stability regulation.701,707 In addition, N-cadherin exhibits great importance in hematological malignancies, such as leukemia and multiple myeloma,702 and is closely associated with poor prognosis in multiple myeloma.708 The N-cadherin antagonist ADH-1 induces cell apoptosis in various tumors including neuroblastoma,704 multiple myeloma,709 and pancreatic cancer,710 and improves the efficacy of tumor-infiltrating lymphocyte therapies.711 Blocking N-cadherin effectively inhibits prostate cancer invasion, metastasis, and castration resistance, which has become an important therapeutic target and biomarker for prostate cancer.712
MMPs
MMPs, also called matrixins, are highly conserved zinc-dependent endopeptidases belonging to the metzincin superfamily.713,714 MMP1, the first matrix metalloproteinase, was discovered in 1962 in the tadpole tail, which exerted the ability to degrade collagen.715 The members of the MMP family can be divided into six major groups: the astacins, the adamalysins (a proteinase with a disintegrin and metalloproteinases, ADAMs, the ADAMs with thrombospondin motif, the pappalysins, the serralysins, and the MMPs.714–716 Most catalytic domains of MMPs are highly homologous and basically consist of four structural domains. However, differences between each MMP still exit including substrate specificity, cellular and tissue localization, membrane binding, and regulation.713,717 MMPs consisting of 23 members with different structural domains in humans are widely expressed in various organs and tissues.713
The ECM is a fundamental component of body tissues and organs, which maintains tissue integrity by homeostatic balance between ECM production and its degradation.715 MMPs are proteolytic enzymes capable of degrading the basement membrane and the most of ECM components, thus remodeling the ECM.604 In addition, MMPs can also act as extracellular processing enzymes to regulate protein functions, as well as participate in various homeostatic regulations in tumor cells, such as immunity, angiogenesis, cell adhesion, cell proliferation, apoptosis, and EMT.713,718
The upregulation of MMPs has been observed in different tumors, such as breast cancer,719 CRC,720 gastric cancer,721 esophageal cancer,722 urinary bladder cancer,718 and lung cancer,723 which increases tumor metastasis and promotes cell invasion.604,724 In particular, MMP-9 is critical in cancer cell invasion and metastasis, and has been demonstrated to be a key biomarker in different cancers including NSCLC,725 cervical cancer,726 gastric cancer,727 ovarian cancer,728 breast cancer,729 osteosarcoma,730 and pancreatic cancer.731 The expression of MMP1, MMP2, and MMP16 are positively correlated with OS and DFS in patients with uveal melanoma.732 Collectively, MMPs can be potential biomarkers in various cancers.
Genome instability and mutation
DNA is a relatively stable organic molecule and genomic maintenance systems monitor and resolve damaged DNA, thus ensuring low mutation frequency within cells. During tumor development, cancer cells induce the accelerated accumulation of mutations by compromising genomic integrity or forcing genetically damaged cells to senescence or undergo apoptosis.299 DNA damage response (DDR) coordinates DNA repair by regulating cell cycle checkpoints and other global cellular responses. Genome instability and mutation caused by DDR defects are important hallmarks of cancer.733
PARP
DNA single-strand break (SSB) or single-strand nick are primarily recognized by PARP1 or PARP2, which catalyze the formation of poly (ADP-ribose) (PAR) chains on themselves and neighboring target proteins.733 PARP1 and its activity in poly(ADP-ribosyl)ation (PARylation) at SSBs recruit the scaffold protein XRCC1 which drives DNA ligase 3 (LIG3) and accessory repair factors to rejoin disruptions. The poly(ADP-ribose) polymerase (PARP) plays an important role in many cancer types, including ovarian cancer, breast cancer, pancreatic cancer, and prostate cancer.733 Furthermore, PAR chains are rapidly degraded by PAR glycohydrolase which restores PARP and PARylated proteins to a de-(ADP-ribosylated) state to promote SSB repair. As PARylation is a highly dynamic and transient process, the inhibition of both PAR glycohydrolase and PARP could reduce the repair efficiency of SSBs, exhibiting their anticancer efficiency.733 Especially, PARP inhibitors have been demonstrated to block the SSB repair pathway and trigger synthetic lethality in cancers with homologous recombination (HR) deficiency which results in impaired DNA double-strand breaks (DSB) repair.734,735
BRCA1/2
In addition to SSB, DSB exerts a vital role in genome integrity. There are two major DSB repair pathways in human cells: the nonhomologous end joining pathway and the HR pathway.733 The HR pathway uses the homologous DNA molecule (usually the sister chromatid) as the repair template. HR is initiated when nuclease digests double-stranded DNA ends at DSB sites to produce ssDNA overhangs. Immediately afterward, BRCA1 facilitates the recruitment of BRCA2 to DSB sites through interaction with PALB2, which loads RAD51 directly onto ssDNA ends. Nucleoprotein filaments are formed on ssDNA by RAD51, which subsequently promotes strand invasion and displacement loop (D-loop) ss formation. Finally, the invasion strand is replaced and strand annealing contributes to the HR completion.733
BRCA1/2 maintain genomic integrity after DNA damage by promoting accurate DNA repair via the HR pathway.733 BRCA1/2 regulate DNA replication by preventing nuclease degradation of nascent DNA and promoting the HR repair of broken replication forks to regulate DNA replication.733,736,737 The loss of BRCA1/2 function leads to the accumulation of DNA damage and genomic alterations including insertions, deletions, and chromosomal rearrangements, ultimately damaging genomic integrity and promoting tumorigenesis.733 The overexpression of BRCA1/2 is significantly associated with worse OS and clinicopathological characteristics in breast cancer.738 High expression of cytoplasmic BRCA1 and BRCA2 is significantly associated with favorable OS in digestive cancers, whereas BRCA1 nuclear expression usually predicts poor outcomes. Thus, BRCA1/2 could be used as clinicopathological biomarkers to evaluate the prognosis of digestive system cancers.739 Moreover, BRCA1/2 mutations are closely related to the progression of multiple cancers, including breast cancer, ovarian cancer, prostate cancer, and pancreatic cancer.733,740 BRCA1/2-deficient cells are highly sensitive to PARP inhibition,741 which is due to inhibition of PARP-dependent SSB repair resulting in the accumulation of DNA lesions (SSBs and DSBs) during replication.733 In conclusion, BRCA1/2 serves as a biomarker for prognosis and treatment response in cancer.
ATR-CHK1/ ATM-CHK2
Ataxia telangiectasia mutated (ATM) is a kinase responsible for orchestrating cellular responses to DSB and replication stress, including DNA repair, checkpoint activation, apoptosis, senescence, chromatin structural change, and transcription.742 Ataxia telangiectasia and Rad3-related protein (ATR), an essential regulator of the cellular replication stress response, is involved in cell-cycle arrest, inhibiting the beginning of replication origins, regulating global fork speed, and promoting fork stabilization.743 ATM and ATR respond to DNA damage by phosphorylating hundreds of substrates.744 The checkpoint kinase 1 (CHK1) and checkpoint kinase 2 (CHK2) are the major substrates downstream of ATR and ATM, respectively, and are responsible for downregulating the activity of CDKs, thereby preventing cell cycle progression under stress. ATM is recruited to DSB sites and promotes histone H2AX phosphorylation. Phosphorylated H2AX in turn recruits the mediator of DNA damage protein MDC1, and subsequent MDC1 phosphorylation by ATM leads to recruitment of DNA damage mediator proteins 53BP1 and BRCA1, thereby promoting DSB repair.733
ATM is frequently mutated or inactivated in a variety of tumors, including lung cancer, breast cancer, brain cancer,745 and pancreatic cancer.746 Endometrial cancer patients with ATM mutations exhibit a higher tumor mutational burden, a higher neoantigen load, and increased expression levels of immune checkpoints. Thus, ATM mutations can act as an independent prognostic factor and a potential biomarker for immune checkpoint therapy in endometrial cancer.747 Moreover, ATM mutations are independently associated with longer OS in patients with metastatic CRC.748 ATM deficiency also renders cancer cells sensitive to topoisomerase I inhibitors or PARP inhibitors. PARP and topoisomerase I inhibitors lead to single-ended DSB, while ATM inactivation delays DNA damage repair, leading to toxic chromosome fusions.733
Tumor-promoting inflammation
As one of the tumor hallmarks,299 persistent inflammation plays an essential role in a variety of human cancers by manipulating cancer development, angiogenesis, malignant transformation, invasion and migration, immune surveillance, and response to therapy.749,750 Inflammation-related regulators, including tumor necrosis factor-α (TNF-α), nuclear factor-κB (NF-κB), and Nod-like receptor protein 3 (NLRP3), are potential tumor prognostic biomarkers.
TNF-α
TNF-α, a vital member of the multifunctional TNF superfamily, is a 17 kDa type II transmembrane protein that was first isolated from the serum of mice infected with Bacillus Calmette-Guérin and endotoxin by E. A. Carswell in 1975.751,752 As a key molecule mediating the tumor-promoting inflammatory process, TNF-α drives inflammation directly by promoting inflammatory gene expression, or indirectly by triggering the inflammatory immune response and regulating cell death.753 Mechanistically, TNF-α binds as a homotrimer to two distinct homotrimeric receptors on the cell surface: TNFRI (p55 receptor) and TNFRII (p75 receptor),754 thus inducing downstream inflammatory mediators and growth factors, which further activates NF-κB and AP1.754 NF-κB signaling is a major mediator of protumor activity of inflammatory cytokines.755
TNF-α levels are abnormally elevated in various precancerous lesions, such as gastric lesions754,756 and inflammatory bowel disease, compared with normal tissues.754,757 In addition, TNF-α is overexpressed in the tumor and stroma of multiple malignancies, including breast cancer, ovarian cancer, CRC, prostate cancer, bladder cancer, esophageal cancer, renal cell cancer, melanoma, lymphoma, and leukemia.752 For example, ovarian cancer cells express 1000-fold more TNF-α mRNA than normal ovarian surface epithelial cells.758 The combination of upregulated TNF-α and C-reactive protein in the patient’s plasma is significantly related to shorter survival in HNSCC patients.759 In conclusion, TNF-α is a key regulator linking inflammation and tumorigenesis, and it may serve as a promising prognostic and therapeutic biomarker for tumor inflammation.755
NF-κB
NF-κB, first identified as a nuclear factor essential for immunoglobulin kappa light chain transcription in B cells in 1986,760 is a dimeric transcription factor. The mammalian NF-κB family consists of RELA (p65), NF-κB1 (p50; p105), NF-κB2 (p52; p100), c-REL, and RELB,761,762 all of which share a conserved amino-terminal region containing dimerization, nuclear localization, and DNA-binding domains. External stimuli, including infection factors, proteins, stress signals, and proinflammatory cytokines released by necrotic cells can activate NF-κB.762 The main activated form of NF-κB is a heterodimer of the p50 or p52 subunit associated with the p65 subunit.762 NF-κB proteins are present in the cytoplasm and are associated with inhibitory proteins of IκB. Activated IκB proteins are phosphorylated and ubiquitinated and then degraded by the proteasome, which induces NF-κB proteins to translocate to the nucleus.762 The nucleus NF-κB binds to cognate DNA-binding sites, promoting the transcription of various genes involved in cell cycle, proliferation, apoptosis resistance, and metastasis-promoting, ultimately enhancing cell growth, angiogenesis, stem cell formation, and cell metabolism.762–764
As a key regulator of inflammation,765 NF-κB is activated in various hematological and solid tumors and is closely associated with tumor development.766 A meta-analysis of 44 studies with a total of 4418 patients has revealed that NF-κB expression is connected to poor 3-year and 10-year OS in solid tumors.767 NF-κB level is significantly associated with large tumor size and high tumor grade in breast cancer patients.768 NF-κB also plays an important role in the TME. Activated NF-κB in cancer cells initiates and maintains the TME by upregulating chemokines that recruit immune response cells, inflammatory cells, and progenitors of cancer-associated fibroblasts.769 In addition, NF-κB regulates the EMT transition through the induction of EMT transcription factors.770,771 In conclusion, NF-κB is a prognostic biomarker of tumor inflammation in cancer.
NLRP3
NLRP3, belonging to the NLR protein family, is one of the most characterized inflammasomes.749 The NLR protein family has 22 members in humans.772 After the first inflammasome was discovered by Fabio Martinon in 2002,773 multiple PRRs have been identified and shown to be involved in inflammatory vesicle formation, such as NLRP1, NLRP2, NLRP3, and NLRC4.774
The inflammasome is a type of intracellular multiprotein hexamers or heptamers signaling complex that forms in cytoplasmic compartments, and the NLRP3 inflammasome has been intensively studied for its involvement in broad ranges of human diseases. Especially, dysregulation of the NLRP3 inflammasome is closely associated with the development of different cancers, including gastric cancer, CRC, HCC, head and neck cancer, lung cancer, breast cancer, prostate cancer, skin cancer, cervical cancer, and central nervous system tumors.749 A high NLRP3 level is correlated with the advanced tumor stage, distant metastasis, and the vascular invasion of cancers.775 It has been found that NLRP3 inflammasome promotes cancer cell differentiation by regulating cell cycle proteins and inducing the production of IL-1β which activates NF-κB by binding to its receptor, which ultimately leads to proliferation and invasion of gastric cancer.749 NLRP3 inflammasome activation in glioblastoma cells leads to IL-1β in aberrant expression.776 In addition, the NLRP3 inflammasome has been demonstrated to be elevated in HNSCC tissues, and its level is correlated with tumor prognosis.777 Activation of the NLRP3 inflammasome promotes the progression of prostate cancer,778 while reduced expression of NLRP3 inflammasome and IL-1β inhibits melanoma development.779 Furthermore, a high NLRP3 level is associated with a low 5-year and 10-year survival rate in CRC patients.780 Targeting NLRP3 inflammasome effectively inhibits HCC proliferation and metastasis.781 In conclusion, NLRP3 inflammasome activation leads to an inflammatory response that promotes cancer development and progression, and NLRP3 may serve as a prognostic and therapeutic biomarker for tumors.
Deregulating cellular metabolism
Otto Warburg first discovered the tendency of tumors to convert glucose to lactate in the presence of oxygen in 1924, known as “aerobic glycolysis“,782 which subsequently came to be termed the “Warburg effect“.783 Tumor cells reprogram glucose metabolism even in the presence of oxygen by restricting energy metabolism mainly to glycolysis, thus reprogramming energy production. Extensive alterations in energy metabolism in cancer cells are considered to be important hallmarks of cancer299 (Fig. 6).
Fig. 6.
The potential inhibitors that target cancer metabolic process. Glucose is taken up into the cell by glucose transporters GLUT1/4 and phosphorylated by hexokinases HK1 and HK2. Glucose 6-phosphate (P) and its downstream intermediates can either be converted to pyruvate or fuel biosynthesis through different pathways, such as the pentose phosphate pathway which provides ribose 5-P for nucleotide synthesis. Fructose-6-P is involved in the hexosamine biosynthesis pathway. Glycerol 3-P production contributes to the serine and glycine biosynthesis pathways which are regulated by the key enzymes PHGDH and SHMT1/2. Moreover, serine biosynthesis plays an essential role in amino acid metabolism and nucleotide metabolism by regulating one-carbon metabolism which is mediated by the methylenetetrahydrofolate dehydrogenase MTHFD1. Pyruvate can be converted to lactate by LDH and exported through the monocarboxylate transporter MCT-1. Besides, pyruvate can enter the TCA cycle as acetyl-CoA through the mitochondrial pyruvate carrier and pyruvate dehydrogenase. Various pathways influence the production of the mitochondrial acetyl-CoA, including fatty acid β-oxidation, glucose metabolism, and other sources that can condense with oxaloacetate to form citrate, which can then be exported from the mitochondrion. Citrate via ACLY is a vital source of cytoplasmic acetyl-CoA which forms malonyl-CoA by acetyl-CoA carboxylase ACC1 and ACC2. Subsequently, malonyl-CoA is cyclically extended by the addition of carbons from acetyl-CoA by FASN to make saturated fatty acids. Fatty acid catabolism is initiated with the formation of fatty acyl-CoA which is then converted by CPT1 to an acylcarnitine. Pyrimidine synthesis, a multistep process regulated by key enzymes such as CAD and DHODH, can produce pyrimidine nucleotides from glutamine, carbonate, and aspartate. Meanwhile, glutamine is taken up by transporters SLC1A5. Glutamate produced from glutamine by glutaminase enzymes can be used in glutathione synthesis. In addition, the complex V (ATP synthase) and the electron transport chain consisting of four complexes including complex I/II/III/IV (CI–IV), are promising targets for drug development. Inhibitors (red), key enzymes or transporters (blue), and key metabolites (purple) are shown. ACC acetyl-CoA carboxylase, ACLY ATP-citrate lyase, BP bisphosphate, CAD carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase, CoA coenzyme A, CI–IV, complex I/II/III/IV, CV complex V, CPT1 carnitine palmitoyltransferase 1, DHODH dihydroorotate dehydrogenase, FASN fatty acid synthase, GLUT1/GLUT4 glucose transporter 1/4, HK hexokinase, IDH1 isocitrate dehydrogenase 1, LDHA/B lactate dehydrogenase A/B, MCT-1 monocarboxylate transporter 1, MTHFD1 methylenetetrahydrofolate dehydrogenase 1, P phosphate, PHGDH phosphoglycerate dehydrogenase, PKM2 pyruvate kinase M2, SHMT serine hydroxymethyl transferase, SLC1A5 solute carrier family 1 member 5, TCA tricarboxylic acid
GLUT1
Tumor cells require the high uptake of glucose and glutamine to meet sustained proliferation.784 The polarity and hydrophilicity of glucose result in its inability to penetrate hydrophobic cell membranes. The transmembrane glucose transporter protein 1 (GLUT1, also known as SLC2A1) is the major glucose transporter protein, and GLUT1 expression is significantly upregulated in tumor cells.784 The crystal structure of human GLUT1 was first reported in 2014,785 and the expression of GLUT1 is regulated by various signaling pathways. The PI3K-AKT signaling pathway increases GLUT1 mRNA expression and drives GLUT1 protein transport from the inner membrane to the cell surface, thereby promoting glucose uptake.786 RAS upregulates GLUT1 mRNA expression and increases cellular glucose consumption.787 Tumor suppressor gene mutations, such as P53, block glycolysis by inhibiting GLUT1 expression.788 Additionally, the TME upregulates GLUT1 expression through HIF-1α.787
Overexpression of GLUT1 is an important biomarker for poor prognosis in multiple cancers, including breast cancer, ovarian cancer, prostate cancer, thyroid cancer, gastric cancer, HNSCC, glioblastomas, retinoblastomas, CRC, NSCLC, OSCC, esophageal cancer, urothelial papilloma, meningioma, brain cancer, diffuse large B-cell lymphoma, RCC, HCC, and cervical cancer.789–792 Studies have demonstrated that inhibitors targeting GLUT are an effective strategy for cancer treatment.787 In conclusion, GLUT1 is an essential target for tumor glucose metabolism, and it can be used as a diagnostic biomarker for tumors.
IDH1/2
Isocitrate dehydrogenase 1 (IDH1) is localized to peroxisomes and cytoplasmic lysosomes, whereas isocitrate dehydrogenase 2 (IDH2) is localized to mitochondria. Wild-type IDH1 and IDH2 metabolic enzymes catalyze the oxidative decarboxylation of isocitrate to generate α-ketoglutarate (α-KG). Cancer-associated IDH1 and IDH2 mutations occur almost exclusively at different arginine residues in the active site of the enzyme.793 IDH1 and IDH2 mutations occur in a wide variety of hematologic and solid tumors, including glioma, AML, intrahepatic cholangiocarcinoma, chondrosarcoma, thyroid cancer, and angioimmunoblastic T cell lymphoma.793–795 Mutant IDH1/2 catalyzes the conversion of α-KG to D-2-hydroxyglutarate (D2HG). D2HG is maintained at normal levels under physiological conditions, whereas mutant IDH leads to a large intracellular accumulation of D2HG in IDH mutant cancer. Elevated D2HG levels competitively inhibit α-KG-dependent lysine demethylases, leading to D2HG-induced dysregulation of histone and DNA methylation in cells, ultimately promoting tumor progression.793
IDH1/2 mutations have many advantages as easily detectable, reliable, and specific biomarkers. First, IDH1 and IDH2 mutations occur in highly restricted tumor types. Second, almost all tumor-derived mutant loci can be identified by simple PCR amplification and sequencing with a low volume of tumor samples. Third, IDH1 mutations can be identified by routine IHC.796 Fourth, techniques for the noninvasive detection of 2-hydroxyglutarate (2-HG) accumulation in glioma patients have been developed.797 Moreover, given that D2HG is upregulated in tumors with IDH mutations, elevated D2HG level in tumor tissues is used as a noninvasive detection biomarker for clinical IDH mutated tumors.793 In conclusion, IDH1/2 mutations are meaningful diagnostic biomarkers of tumor metabolism.
HK2
Hexokinases regulate the first step of glycolysis which produces and captures negatively charged glucose 6-phosphate ions within the cells. The hexokinases family has five isoforms in mammals including hexokinase 1–4 (HK1, HK2, HK3, HK4), and HKDC1.798 HK2 is the most active isozyme of the hexokinase family.799 In addition to being expressed in the muscle and heart,798 HK2 has been evaluated in various cancers, and is induced solely or synergistically by HIF-1 and MYC.800 An analysis of 21 studies with 2532 patients has revealed that HK2 overexpression is significantly associated with worse OS and PFS in solid tumors. For example, the negative effect of HK2 on OS is observed in HCC, gastric cancer, and CRC patients.801 HK2 expression is correlated with advanced-stage and high-grade ovarian cancer.798 HK2 downregulation inhibits tumor occurrence.802 Thus, HK2 is a meaningful prognostic tumor biomarker and a potential tumor treatment target.
Evading immune destruction
The immune system is responsible for monitoring and eliminating most early cancer cells, thereby inhibiting tumor formation. However, a significant increase in cancers due to low immune function is the evidence of defects in tumor immune surveillance. It has been found that cancer cells generate immune escape by disrupting the immune system, which ultimately promotes tumor progression, dissemination, and metastasis. Tumor cells suppress the action of cytotoxic lymphocytes by recruiting inflammatory cells with active immunosuppressive effects, such as regulatory T cells and myeloid-derived suppressor cells. Thus, immune evasion is another valuable hallmark of cancer.299
PD-1/PD-L1
PD-1 and PD-L1 participate in the evasion of the immune system by cancer cells.803 PD-1 (also called CD279), encoded by the PDCD1 gene,804 was cloned and identified from an apoptotic immune cell line in 1992.805 PD-1 is a type I transmembrane protein receptor consisting of 288 amino acids, whose structure consists of an IgV-like extracellular domain, a transmembrane domain, and a cytoplasmic (intracellular) domain.803 As a negative regulator of the immune response,806 PD-1 is mainly expressed in memory T cells in peripheral tissues, and less in B cells, activated monocytes, dendritic cells, and NK cells.803,804 Two ligands of PD-1, PD-L1 (also known as B7-H1 or CD274) and PD-L2 (also known as B7-DC or CD273),807 are type I transmembrane protein receptors. PD-L1 is a 290 amino acid protein receptor encoded by the Cd274 gene and includes two extracellular structural domains (IgV- and IgC-like domains), a transmembrane domain, and a cytoplasmic domain. Activated PD-1/PD-L1 signaling negatively regulates T cell-mediated immune responses in peripheral tissues, thereby limiting effector T cell responses and protecting tissues from damage.803,804
PD-1 signaling in the TME promotes tumor progression and survival by evading tumor immune surveillance. PD-1 is highly expressed in tumor-infiltrating lymphocytes in many types of cancers. PD-L1 is expressed on different types of tumor cells, including melanoma, ovarian cancer, lung cancer, and kidney cancer.808 The innate and adaptive immune resistance mechanisms contribute to the upregulated expression of PD-L1 in multifarious human cancers.809 A meta-analysis that analyzed 1251 patients from eight different microarray gene expression datasets has revealed that the expression levels of PD-1 and PD-L1 individually or jointly are potential prognostic factors for predicting the outcomes of patients with lung cancer.810 A study with 128 patients who are diagnosed with NSCLC, SCLC, melanoma, urothelial carcinoma, and other cancers, has proven that patients with high expression levels (>11.0 pg/μL) of soluble PD-L1 are more likely to exhibit progressive than those with low expression levels of PD-L1 (41.8 versus 20.7%). Moreover, a high expression level of soluble PD-L1 is also associated with a worse prognosis, the median PFS is 2.9 months versus 6.3 months, and the median OS is 7.4 months versus 13.3 months. Thus, high soluble PD-L1 is a predictive and prognostic biomarker for both decreased PFS and OS in advanced cancer patients who receive immune checkpoint blockade treatment.811 Moreover, a study of 293 HNSCC patients has concluded that strong PD-L1 expression is correlated with distant metastases, and dominates as the strongest prognostic factor of patient outcome.812 The PD-L1 expression level is a negative prognostic factor for patients with RCC813 and gastric cancer.814 Immune checkpoint inhibitors that block PD-1/PD-L1 interactions effectively prolong the survival of patients with various cancers and are promising cancer therapy.803 In conclusion, PD-/PD-L1 expression can be used as a predictive and prognostic biomarker for cancers.
CATL-4
Cytotoxic T lymphocyte antigen 4 (CTLA-4, also known as cluster of differentiation 152, CD152) is a receptor present on the surface of activated T cells, was discovered in 1987 by screening a cDNA library of mouse cytolytic T cell origin.815 CTLA-4 is normally expressed upon T cell activation,816 and activated CTLA-4 inhibits T cell proliferation and induces cell cycle arrest by cross-talk with PI3K and MAPK pathways that regulate cell proliferation.816 CTLA-4 is an inhibitory checkpoint commonly found in activated T cells and has been discovered to be the most reliable target for the treatment of cancer.817 CTLA-4 facilitates the tumor evasion of host immune surveillance, and participates in immune dysregulation in multifarious cancers, including lung cancer, cervical cancer, breast cancer, skin cancer, gastric cancer, CRC, B-cell CLL, and non-Hodgkin’s lymphoma.818 Moreover, targeting CTLA-4 significantly improves outcomes in multiple advanced cancers, including melanoma, lung cancer, breast cancer, head and neck cancer, bladder cancer, cervical cancer, liver cancer, gastric cancer, squamous cell skin cancer, classical Hodgkin’s lymphoma, and B-cell lymphoma.816
However, the correlation between CTLA-4 expression and patient prognosis in different cancers is controversial. Studies have found a significant correlation between the high expression of CTLA-4 and OS in single nucleotide polymorphisms subgroup cancers, including NPC, esophageal cancer, glioblastoma, and hematologic malignancy, in which CTLA-4 is a good prognostic biomarker.818 CTLA-4 overexpressed NSCLC is associated with a reduced death rate. Conversely, malignant pleural mesothelioma with high CTLA-4 exhibits poor prognosis.818 Higher CTLA-4 mRNA levels in breast cancer indicate higher clinical stage and axillary lymph node metastasis.819 A combined analysis of 844 ESCC patients has found that patients with both a low CTLA-4 and platelet lymphocyte ratio (PLR) level have longer OS.820 In conclusion, CTLA-4 is a prognostic biomarker in cancers and its positive or negative effects depend on specific cancer conditions.
Unlocking phenotypic plasticity
Cell development and organogenesis are accompanied by terminal differentiation that in most cases results in antiproliferative outcomes and suppresses tumor formation. It has been found that unlocking phenotypic plasticity to evade the state of terminal differentiation is a pivotal component of cancer development.300
Tumor cell differentiation is regulated by multiple factors. Liver enriched transcription factors are crucial regulators of hepatocyte differentiation and are essential for the maintenance of hepatocyte phenotype and function. Noncoding single-stranded RNA microRNAs are involved in the post-transcriptional regulation of gene expression, which is closely correlated with tumor dedifferentiation. The expression of miRNAs is negatively correlated with the degree of differentiation in HCC. Moreover, differentiation-related genes such as HMGCS2, BDH1, ALDH2, PIPOX, HAO1, AQP9, and PAH, have been identified to predict survival and poor prognosis in multifarious cancers.821
Differentiation and dedifferentiation are also essential for the developmental processes of many tumors. Melanocytes undergo dedifferentiation during tumorigenesis, and the malignant progression of pancreatic islet cell cancers to metastasis-prone carcinomas is associated with dedifferentiation. HDAC inhibitors induce the myeloid leukemia cell differentiation into mature myeloid morphology cells, thereby hindering the progression of leukemia.300 Furthermore, cellular plasticity in HCC is presented by the dynamic interconversion of cancer cell subpopulations in multiple developmental lineages and differentiation stages. Regardless, differentiation therapy unlocks phenotypic plasticity in HCC and induces terminal differentiation of CSCs, promoting their transformation into precursor cells that have lost self-renewal capacity, or converting them into non-CSCs that are sensitive to anticancer drugs.821 In conclusion, differentiation-related factors can be used as diagnostic and prognostic biomarkers for cancers.
Nonmutational epigenetic reprogramming
Since first described by Conrad Waddington in 1942,822 the epigenetic program of gene expression has become a hallmark of cancer that initiates and promotes tumorigenesis. The process of gene expression changes through pure epigenetic regulation is called “nonmutational epigenetic reprogramming”, which is different from genomic DNA instability and mutational mechanisms. Epigenetic alterations such as DNA methylation, histone modifications, chromatin remodeling, and noncoding RNA contribute to the signature ability during tumor progression.300
DNA methylation
DNA methylation is a chemical modification that plays a crucial role in chromatin-based transcriptional regulation, epigenetic gene expression, genomic stability, DNA repair, and replication. DNA methylation is mainly catalyzed by three DNA methyltransferases (DNMTs), DNMT1, DNMT3A, and DNMT3B.823 DNMTs are overexpressed in multiple cancers, including AML, CML, glioma, breast cancer, gastric cancer, CRC, HCC, pancreatic cancer, prostate cancer, and lung cancer.824 DNA methylation may lead to tumor suppressor gene silence, cell cycle dysregulation, DNA repair, and the misregulation of chromosomal stability genes, resulting in genomic instability in tumor cells.825 DNA methylation-based biomarkers have been hailed as an important event in cancer biomarker research.826 DNA methylation occurring mainly in centromeres, telomeres, inactive X-chromosomes, and repeat methylation is altered in 70% of mammalian promoter CpG islands, which are essential for gene transcriptional regulation and tumor malignant transformation.827,828 It has been found that 5–10% of CpG promoter islands are aberrantly methylated in various cancer genomes. DNMT1 is a CpG dinucleotides methyltransferase that recognizes hemimethylated DNA produced during DNA replication and methylates newly synthesized.828
The downregulation of tumor suppressor genes by hypermethylated CpG-rich regions of promoters is a typical example in tumor cells.826 Methylation of CpG dinucleotides (e.g., gene promoters) may serve as a clinically valuable biomarker. Moreover, CpG methylation is related to poor prognosis in patients with ccRCC.829 Methylation of GSTP1 has been discovered to be a promising diagnostic biomarker for HCC.826 The promoter methylations of NMDAR2B and PGP9.5 are linked to poor prognosis in patients with ESCC, and are meaningful clinical diagnostic and prognostic biomarkers for ESCC.830 The methylation status of single CpG dinucleotides affects the regulation of gene expression, and it can be utilized as a prognostic biomarker for CLL.831 O6-methylguanine-DNA methyltransferase (MGMT) methylation can also be used as a prognostic biomarker in glioblastoma patients.832
A study has searched the PubMed database for literature related to DNA methylation-based cancer biomarkers and retrieved a total of 14,743 research papers, which ultimately yields ~1800 tumor biomarkers through calculation and screening. However, only 13 DNA methylation-based biomarkers are currently commercially available and detectable, including GSTP1, APC, RASSF1, NDRG4, BMP3, SEPT9, SHOX2, TWIST1, OTX1, ONECUT2, MGMT, BCAT1, and IKZF1. Only nine of them (GSTP1, APC, RASSF1, NDRG4, BMP3, two SEPT9 biomarkers, SHOX2 and MGMT) have been included in the clinical guideline application.826 In addition, only two tests have been approved by the FDA: Cologuard (NDRG4 and BMP3), which analyzes stool DNA samples collected as part of a CRC screening protocol, and Epi proColon (SEPT9), which analyzes blood samples collected for the same purpose.826
As a promising biomarker for tumor diagnosis, prognosis, and prediction,833 DNA methylation has many advantages: frequent DNA methylation at the early stages of cancer, mature detection technology, good stability of DNA methylation in fixed samples, and presence in various body fluids and cell type specificity. Methylation at specific genomic sites can be a beneficial biomarker under the following conditions: clinically significant differences in methylation expression between the two groups, including diagnostic biomarkers in tumor versus nontumor tissues; prognostic biomarkers between tumor samples from patients with high-risk disease versus those with low-risk disease. In conclusion, DNA methylation in cancer is a clinically valuable biomarker for tumor management.826
Histone modification
Modification of histone proteins at amino-terminal tails such as acetylation, methylation, phosphorylation, and ubiquitination could alter the chromatin condensation, and DNA accessibility, subsequently interfering with gene expression. Histone modification is a dynamic process that is controlled by writers, such as histone acetyltransferases (HATs), histone methyltransferases (HMTs), readers, such as proteins containing bromodomains, and erasers, such as histone deacetylases (HDACs) and lysine demethylases. Histone modifications coregulate processes, such as DNA transcription, DNA replication, and DNA repair.834 Altered post-translational modifications of histones have been found in cancer cells, and changes in the overall level of histone modifications are found to predict clinical outcomes in various cancers.835
HATs
The Nε-acetylation of lysine residues is a major histone modification involved in the regulation of gene transcription, chromatin structure, and DNA repair. Acetylation neutralizes the positive charge of lysine, thereby weakening the electrostatic interaction between histones and negatively charged DNA. Thus, histone acetylation is associated with an open chromatin conformation. The HATs and HDACs family regulate the acetylation of histones.828 HATs are involved in a number of solid tumors and hematologic malignancies, and their expression levels are altered during tumor progression.836
HMTs
Histones are methylated on the side chains of arginine, lysine, and histidine residues and their methylation does not change the total charge of the molecule. The most characteristic sites of histone methylation are mono-, dimethyl- or trimethylation of lysine residues, including H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20. Among them, H3K4, H3K36, and H3K79 are correlated with active genes in euchromatin, while H3K9, H3K27, and H4K2 are associated with heterochromatic regions of the genome.828,837 In addition, different methylation states on the same residue have different functions. For example, H3K4me2/3 usually spans the transcriptional start site of the active gene,828 while H3K4me1 is linked to active enhancers.838 Trimethylation of H3K9 is involved in the repression of gene expression.828
KMTs are specific enzymes that target certain lysine residues, including the members of the EZH2 family. EZH2 is the catalytic subunit of polycomb repressive complex 2 and is primarily responsible for the methylation of H3K27. Studies have shown that EZH2 overexpression is strongly associated with poor prognosis in prostate and breast cancers.828 Loss-of-function mutations in the EZH2 gene in myeloid malignancies and T cell acute lymphoblastic leukemia (T-ALL) also lead to poor prognosis.839
HDACs
HDACs are enzymes that reverse lysine acetylation and restore the positive charge on the side chain. HDACs can be classified into four major groups based on sequence homology: class I (HDAC 1–3 and HDAC8), class II (HDAC 4–7 and HDAC 9-10), class III HDAC (sirtuin 1–7), and class IV only (HDAC11).828 HDACs promote leukemia development by mediating abnormal gene silencing in malignant tumors. Inhibition of HDACs induces growth arrest, differentiation, and apoptosis in tumor cells.840 In addition, studies have demonstrated that HDACs are usually connected to poor tumor prognosis.834
HDMTs
LSD1 (KDM1A) is a class of demethylases that demethylate lysine through an oxidation reaction with flavin adenine dinucleotide, which is restricted to demethylating mono- and dimethyl lysine. Jumonji is a class of demethylases with a conserved JMJC structural domain that demethylates all three methyl lysine states through an oxidative mechanism and radical attack.828 The most studied LSD1 is increased in a variety of cancers, and it is related to the differentiation of neuroblastoma cells. In addition, HDMTs are involved in the development of breast cancer, PDAC, and other tumorigenic processes.824
Polymorphic microbiomes
The microbiota, an increasingly hot topic in recent years, has been demonstrated to influence the microenvironment, tumorigenesis, and metastasis of various malignancies.841 An increasing number of studies have uncovered that the microbiota is critical for the development of various cancers,842 including gastric cancer,843 ovarian cancer,844 CRC,845 pancreatic cancer,846 prostate cancer,847 HCC,848 lung cancer,849 breast cancer,850 and cholangiocarcinoma.851
There are three major categories of regulatory mechanisms by which microbiota promote carcinogenesis: altering the balance of host cell proliferation and death such as DNA damage and DNA repair; regulating the tumorigenic inflammatory environment within the tissue and immune system function; and affecting host metabolism.852,853 Therefore, small molecule drugs targeting microbiota have become a hot research topic in antitumor therapy.854 Known oncogenic gut microbiota include Salmonella typhi855 and Helicobacter spp856 in biliary tract cancer, Helicobacter pylori857 in gastric cancer, etc. Helicobacter pylori has been identified as a class I carcinogen by the World Health Organization, and it is associated with gastric cancer and mucosa-associated lymphoid tissue.858 CRC is a classic case of dysregulation of the gut microbiota that promotes cancer development.859 Certain microbiota species can ultimately exert a pro-carcinogenic effect by stimulating inflammatory states, including the induction of proinflammatory toxins, the increase of ROS production,860 the aberration of signaling pathways,861 and the blockage of antitumor immune function.858 It has been confirmed that F. nucleatum is crucial in the progression of CRC,861 which was detected in lymph nodes and distant metastasis samples from patients.858 Peptostreptococcus anaerobius is more enriched in stool samples from CRC patients, and its ability to induce phosphorylation of adherent spot kinase in CRC cells activates NF-κB signaling, ultimately promoting the cause of chronic inflammation and tumor progression. In addition, microbiota in other sites still play carcinogenic roles. Oral micro-common pathogenic bacteria including Streptococcus anginosus, Veillonella, F. nucleatum, and P. gingivalis, are involved in several digestive cancers. P. gingivalis is enriched in ESCC at higher levels than normal tissues, and it utilizes the miR-194/GRHL3/PTEN/AKT signaling pathway to promote ESCC proliferation and migration. The concentration of F. nucleatum nucleic acid is significantly higher in esophageal cancer tissues than in normal esophageal tissues. P. gingivalis and F. nucleatum are associated with a high risk of pancreatic cancer, and P. gingivalis can promote the proliferation of pancreatic cancer cells.853 Collectively, certain microbiota can be carcinogenic by stimulating chronic inflammatory response. In addition, microbiota can alter key intracellular signaling pathways and attack gastric mucosa by utilizing various virulence factors.857 In conclusion, the microbiota is essential for cancer development, and the microbiota can be used as a potential biomarker for tumors.
Senescent cells
Cellular senescence is a classic form of irreversible proliferative arrest characterized by the shutdown of the cell division cycle, changes in cell morphology and metabolism, and the activation of the senescence-associated secretory phenotype (SASP), which is capable of transmitting signaling molecules in a paracrine manner to neighboring living cancer cells as well as to other cells in the TME. SASP involves the release of a large number of bioactive protein chemokines, cytokines, and proteases. A variety of conditions, such as microenvironmental stress, and telomere erosion, induce cell senescence.862–864 The cellular senescence is a significant biomarker of tumor cells.
SASP
SASP is mediated through the proinflammatory transcription factor NF-kB or through transcriptional processes that depend on epigenetic changes.863 The NF-κB, p38, mTOR and C/EBPβ signaling pathways induce the formation of SASP865 which include proinflammatory cytokines (e.g., IL-1α, IL-1β, IL-6, and IL-8), chemokines (e.g., CCL2, CCL5, and CXCL1), growth factors (e.g., HGF, EGF, and TGFα), MMPs, and various oxylipins SASP factors. These factors are the main paracrine messengers between senescent cells and their surrounding cells (including stromal bystander cells, immune cells, precancerous cells, and cancer cells).865
The SASP exerts a double-edged sword effect in tumorigenesis, which might be beneficial or detrimental to tumorigenesis.866 On the one hand, SASP exhibits tumor suppressive effects by maintaining the senescence program, permanently blocking tumor transformation of normal cells, and recruiting immune cells to remove damaged or oncogene-expressing cells from the organism.867 The SASP factor IL-6 inhibits osteosarcoma formation by inducing and enhancing senescence.868 Precancerous lesions in RAS-driven pancreatic cancer are accompanied by extensive senescence and SASP.869 On the other hand, some SASP factors are tumorigenic. Studies have revealed that SASP factors promote tumorigenesis due to paracrine mitogenic or metastatic effects on other premalignant cells, as well as interactions with surrounding endothelial cells, stromal cells, and tissues.867 Senescent cells and SASP factors favor the promotion of cell transformation, metastasis, and tumor growth. Specific PTEN deficiency in mouse prostate cancer tissues leads to precancerous lesion development with extensive senescence and SASP.867,870 In general, SASP factors regulated by NF-kB have tumor-suppressive, immunosurveillance effects, while SASP factors regulated by signal transducer and activator of transcription 3 (STAT3) have tumor-promoting and immunosuppressive effects.867
Lamin B1
Lamins are intermediate filament proteins that line the inner surface of the nuclear envelop which contribute to the size, shape, and stability of the nucleus.871 Nuclear lamins are type V intermediate filaments ranging in size from 60 to 80 kDa. Lamins are divided into A type (lamin A, C) or B type (lamin B1, B2) according to isoelectric points. Lamin regulates nuclear and cytoskeletal organization, mechanical stability, chromatin organization, gene regulation, genomic stability, differentiation, and tissue-specific functions by binding to a variety of nuclear protein complexes.871 Lamin B1 is essential for the regulation of normal organogenesis and organism survival.872 Lamin B1 knockdown triggers the formation of H3K27me3-enriched mesas and DNA hypomethylation regions overlapping with lamin B1-associated domains in cancer and accelerates the replicative and oncogene-induced senescence. Reduced lamin B1 expression has been discovered in multiple senescent cells, and its overexpression delays senescence.873,874 Silencing lamin B1 expression slows cell proliferation and induces premature senescence in WI-38 cells.873 Oncogenic Ras-induced premature senescence also reduces lamin B1 expression through a pRb-dependent mechanism. In addition, senescence is induced by DNA damage, replication failure, or oncogene expression when lamin B1 is lost in human and mouse primary cells. Lamin B1 loss is not dependent on p38-MAPK, NF-κB, or ROS signaling pathways which are positive regulators of senescence phenotypes.872
On the other hand, Lamin B1 upregulation is widely observed in tumor tissues of most cancer types. A high level of lamin B1 expression predicts poor OS and DFS for cancer patients.875 Lamin B1 is overexpressed and facilitates cell proliferation and metastasis in HCC, and increased lamin B1 expression indicates a dismal prognosis and immunotherapy response in HCC.876 Besides, lamin B1 has been proposed as a prognostic senescence biomarker in ccRCC877 and lung adenocarcinoma.878 To summarize, lamin B1 loss is a senescence-associated biomarker.
Liquid biopsy tumor biomarkers
Liquid biopsies have become a pivotal strategy for cancer diagnosis, real-time monitoring, and prognosis through minimally invasive detection of biofluids, such as blood, saliva, urine, pleural fluid, and ascites.879 Liquid biopsy tumor diagnostic biomarkers, including circulating tumor DNA, circulating tumor cells, and exosomes, are all effective monitoring tools for tumor diagnosis and treatment.
ctDNA
Cancer cells release naked DNA molecules into the circulation known as ctDNA which has become an essential biomarker for liquid biopsies to predict response to targeted therapies and immunotherapies to guide clinical anticancer treatment.880
ctDNA consists of small nucleic acid fragments that are not associated with cells or cellular fragments.881 Plasma ctDNA refers to tumor-derived DNA fragments and was detected in human plasma in 1948, and includes plasma cfDNA, circulating DNA derived from the death of hematopoietic cells.882,883 A study has revealed that ctDNA is detected in >75% of patients with advanced pancreatic cancer, ovarian cancer, CRC, bladder cancer, gastroesophageal cancer, breast cancer, melanoma, HCC, and head and neck cancer, and in 50% of patients with primary brain cancer, kidney cancer, prostate cancer, and thyroid cancer.881 As the plasma ctDNA content is less than 0.01%884 and its half-life is very short, plasma ctDNA levels allow for real-time dynamic assessment of tumor evolution through serial sampling, and represent intrapatient and interpatient variability to guide clinical drug use. In addition, plasma ctDNA captures tumor heterogeneity and effectively reflects DNA shed from multiple metastatic sites.884 The NCCN Guidelines for Breast Cancer (version 4.2020) recommend the use of ctDNA analysis for the evaluation of PIK3CA mutations in breast cancer.884,885
Moreover, ctDNA analysis is used for clinical real-time monitoring of treatment response in a variety of tumors, including breast cancer,886 NSCLC,887 prostate cancer, gastroesophageal cancer,884 HCC,50 and CRC.888 Plasma ctDNA analysis has been applied to monitor clinical cancer therapy resistance. Metastatic CRC is the first disease that utilizes liquid biopsy to evaluate treatment resistance. ctDNA analysis identifies CRC resistance to HER2-targeted therapy. The NCCN Guidelines for Gastric Cancer (version 2.2020) and the Esophageal and Esophagogastric Junction Cancer Guidelines (version 2.2020) recommend the use of plasma ctDNA analysis to detect drug sensitivity in patient treatment.884 Osimertinib resistance in NSCLC patients with EGFR mutation can be detected by plasma ctDNA analysis.889 Prostate cancer plasma ctDNA is used to detect BRCA reversion which mediates PARPi treatment resistance.890 In summary, ctDNA is widely used in patients with advanced solid tumors for the detection of MRD, the monitor of early recurrence, the prediction of treatment response, and drug resistance monitoring.58,888
CTCs
In 1869, Thomas Ashworth, an Australian physician, first identified CTCs, a type of cells shed into the bloodstream from primary or metastatic tumor sites.891 CTCs are cancer cells isolated from the primary tumor site and transported via the circulation to distant organs.892 CTC characteristics are defined as a nucleated circulating cell larger than 4 μm expressing the epithelial cell protein EpCAM and cytokeratin 8, 18, and 19, while not expressing the leukocyte-specific antigen CD45.893 This is the main basis and foundation of CTC testing.891
CTCs are clinically significant as biomarkers for the clinical management of patients with metastatic cancer and the implementation of precision medicine.894 More than 400 clinical trial studies have been conducted using CTCs as biomarkers. The key research contents and objectives are assessing prognostic information, the risk of recurrence and metastasis, stratifying and monitoring treatment in real-time, and identifying therapeutic targets and resistance mechanisms of cancer patients.895 CTC assays and molecular characterization have been applied to stratify patients with breast cancer, CRC, prostate cancer, lung cancer, pancreatic cancer, glioblastoma multiforme, and melanoma, and to monitor disease progression.892 Studies have discovered that CTC count is a prognostic indicator for multiple myeloma.896 The application of CTCs in the clinical assessment of gastric cancer has shown that CTCs are correlated with metastasis, poor prognosis, recurrence, and treatment response in gastric cancer patients.891 The comprehensive analyses of CTCs in metastatic breast cancer confirmed heterogeneous mechanisms of patient resistance to targeted therapies.897 CTCs can monitor therapeutic response and be utilized as a screening tool for brain micro-metastasis detection in breast cancer.898 In addition, CTCs have important clinical utility in the selection of therapeutic-specific biomarkers for the treatment of patients with prostate cancer. The response of prostate cancer to anticancer drugs is strongly correlated with the expression of AR-V7, a treatment-specific biomarker, in the CTC nuclei.899 The CTCs in the NSCLC pulmonary vein are independent predictors of NSCL recurrence after surgery.900 In addition, CTC analysis captures the tumor heterogeneity noninvasively and real-timely in cancer patients, which effectively explains the existence of heterogeneous drug resistance mechanisms in patients with refractory tumors901 and promotes the development of precise targeting strategies.901 In conclusion, CTC analysis is a clinically relevant noninvasive tool to monitor the progression and prognosis of cancers, and to deeply explore the biology of metastatic cancers, and has the potential to facilitate personalized precision treatment of cancers. Of course, the difficulties undoubtedly need to be solved in the future, including that the efficiency of CTC targeting approaches varies by cancer types, and the large patient cohorts are needed to flesh out the arguments in CTC application.894
Exosomes
Exosomes, first discovered in 1983 by Pan and Johnstone,902 are extracellular lipid bilayer vesicles of endosomal origin with an average size range of approximately 100 nm in diameter.903 The process of exosome biogenesis is well defined, starting with double invagination of the plasma membrane and the formation of intracellular multivesicular bodies containing intraluminal vesicles, followed by exocytosis of multivesicular bodies fused to the plasma membrane, and the intraluminal vesicles are finally secreted as exosomes with a diameter of ~40–150 nm.904 Exosomes released from the cell surface can fuse with the plasma membrane of recipient cells, thereby transporting their contents into the cytoplasm. In addition, proteins on the surface of exosomes can bind to the surface receptors of recipient cells to promote intracellular signaling.905 The exosomes are highly heterogeneous given their size, content, functional impact on recipient cells, and cellular complexity of origin.903 Among them, the content of exosomes is a key factor in the execution of functions. Recent advances have revealed that the contents of exosomes include proteins, DNA, mRNA, microRNA, long noncoding RNA, circular RNA, and other components.906
Exosomes have been demonstrated to be involved in cancer development, angiogenesis, metastasis, and therapeutic resistance, and can be used as diagnostic and prognostic biomarkers for tumor patients.906 For example, exosomes are useful in liquid biopsy to diagnose various cancers, including lung cancer,907 pancreatic cancer,908 gastric cancer,909 prostate cancer, breast cancer, ovarian cancer, glioblastoma, and melanoma.905 The exosomal DNA in serum exosomes has been proven to be of significant value in detecting cancer-related mutations, such as KRAS and TP53.910,911 The specific miRNAs that are differentially expressed in exosomes between cancer cells and normal cells have important diagnostic or prognostic value in the early detection of cancers.912 Studies have revealed that elevated serum-derived exosomal miR-21 is associated with multiple cancers, including pancreatic cancer, ovarian cancer, and lung cancer.913 The tumor suppressor miRNAs, such as miR-146a and miR-34a, are found in exosomes, and their low levels are correlated with poor prognosis of liver cancer, breast cancer, CRC, pancreatic cancer, and hematologic malignancies.903,912 The upregulation of other exosomal oncogenic microRNAs, such as miR-155, miR-17-92, and miR-1246, have also been shown to be connected to the progression of multiple cancers.903,912,914
Moreover, as the exosomal cargo exchange between cancer cells and stromal cells in the TME, exosomes from multiple cancer cells can effectively regulate TME angiogenesis and extracellular matrix remodeling at metastatic sites, thereby enhancing tumor growth and metastasis.903,904 Breast cancer-derived exosomes have been demonstrated to impair vascular integrity and enhance vascular permeability, thereby promoting breast cancer metastasis.915 Exosomes from glioblastoma cells promote tumor cell proliferation and induce angiogenesis.916 Ovarian cancer cell exosomes are involved in promoting their peritoneal dissemination.917 Moreover, cancer cell-derived exosomes also facilitate metastasis in melanoma,918 pancreatic cancer,919 gastric cancer, etc.920,921 Exosomes from different cellular sources, such as immune cells, cancer cells, epithelial cells, and mesenchymal cells, can influence the activity of recipient cells of both the innate and adaptive immune system. Exosomes stimulate or inhibit the function of CCD4+ and CD8+ T cells.903
In addition, exosomes secreted by cancer cells can benefit cell survival by interacting directly with drugs and reducing their antitumor efficacy, or by regulating cancer cell gene expression through TEM cell-derived exosomes. CAF-derived exosomes stimulate chemotherapy resistance in CRC922 and breast cancer,923 and promote resistance of ovarian cancer cells to paclitaxel.924 Moreover, macrophage-derived exosomes induce resistance to gemcitabine in pancreatic cancer cells.925 Thus, exosomes secreted by cancer cells can induce resistance to chemotherapeutic drugs.903
Exosomes have numerous advantages as liquid biopsy diagnostic biomarkers for cancer. First, exosomes can be secreted by all types of cells and are present in various biological fluids, such as blood, urine, semen, saliva, amniotic fluid, cerebrospinal fluid, ascites, tears, and breast milk.905 Therefore, it is easy and convenient to collect samples. Second, exosomes can reveal specific proteins from parental cells and target cells, which isolates the origin-specific exosomes and predict organ-specific metastasis.117 Third, the concentration of exosomes is higher than that of other liquid biopsy biomarkers, such as CTCs, thereby reducing the amount of sample collection. Fourth, exosomes are highly stable compared with other liquid biopsy biomarkers, such as ctDNA, which is rapidly degraded in the blood.926 In conclusion, exosomes are valuable biomarkers for the early diagnosis, prognosis prediction, and the therapeutic efficacy assessment of cancers. The possibility of combining protein, lipid, RNA, and miRNA exosome cargos in cancer may enhance the diagnostic and prognostic potential of exosomes, which is being considered.
Tumor biomarker-based cancer therapy
Optimizing the precise medical care of patients according to genetic and molecular characteristics can maximize the benefits of precision medicine. Therefore, discovering and developing tumor biomarkers and related cancer therapies contribute greatly to effective precision medicine. Compared with traditional chemotherapy drugs, targeted cancer therapy including small-molecule targeted drugs, monoclonal antibodies (mAbs), antibody-drug conjugates, and the proportion of biological drugs specifically targeting proteins or genes in cancer cells, results in high potency and low toxicity. During the past 30 years, remarkable progress has been achieved in this field. Targeted therapy has been the mainstream strategy for cancer therapy, and targeted drugs approved by the FDA, European Medicines Agency, Ministry of Health Labour and Welfare and National Medical Products Administration for cancer treatment have increased accordingly.927,928 Here, we summarize the pivotal tumor biomarker-based cancer therapies in preclinical and clinical studies in recent years and hope to provide comprehensive insights into cancer therapy.
Targeting tumor cell proliferative signaling
There is an increasing number of inhibitors related to tumor growth signaling pathways, including inhibitors of RAS, PI3K-AKT-mTOR, and RAF-MEK-ERK signaling pathways. In particular, multiple protein kinases have engaged in cell proliferation, and targeted drugs have rapidly developed and been applied for clinical use since the approval of the first small-molecule TKI, imatinib, by the FDA in 2001.929 Although their targets and mechanisms of action are different, all of them can effectively inhibit tumor cell proliferation.
RAS inhibitors
Since the discovery of RAS in the rat sarcoma virus, targeting RAS has attracted attention because of its vital role in tumorigenesis and progression. However, the development of RAS targeting therapy is extremely challenging.930 The RAS protein surface is very smooth, and the lack of drug-binding pockets makes targeting RAS problematic.931 The existence of high intracellular GTP concentrations, the activation of compensation pathway, and drug-resistant mutations in RAS-RAF-ERK pathway genes after RAS inhibitors administration, lead to poor inhibitory effects of RAS inhibitors.319,932 In recent years, due to the consecutive failure in the discovery of RAS inhibitors, RAS was once considered an undruggable target. Significantly, Shokat and colleagues opened a new chapter in RAS-targeted therapy by discovering a binding pocket containing the mutant cysteine residue in KRASG12C in 2013, which prompted the fast development of the first small-molecule KRASG12C inhibitor sotorasib.319 Subsequently, due to the importance of RAS in tumorigenesis and progression, alternative strategies such as targeting RAS mutations, and upstream and downstream effectors have been attempted. For example, drug exploitation in targeting RAS mutated malignancies can be accomplished using various strategies (Table 1), including interfering with RAS maturation and transport, promoting its localization to the plasma membrane, and inhibiting its downstream signaling.933 To date, inhibitors against KRASG12C have been approved for clinical use.308 Meanwhile, RAS inhibitor combination options are increasingly developed, i.e., combination with inhibitors that inhibit the RAS signaling pathway, maintain the GDP-binding status of KRAS proteins, and modulate the immune system.319
Table 1.
The FDA-approved and clinically developed RAS inhibitors in cancer therapies
| Target | Drug | Highest Phase | Indications | Company/Identifier | Status |
|---|---|---|---|---|---|
| Farnesyltr-ansferase | Tipifarnib | Approved | Head and neck squamous cell carcinoma harboring HRAS mutations who have progressed following platinum-containing chemotherapy | Kura Oncology | / |
| Lonafarnib (SCH66336) | Approved | Hutchinson-Gilford progeria syndrome and progeroid laminopathies | Merck & Co | / | |
| III | Carcinoma, non-small cell lung, metastases, neoplasm | NCT00050336 | Terminated | ||
| III | Myelodysplastic syndromes, leukemia, myelomonocytic, chronic, myelodysplasia, myelomonocytic | NCT00109538 | Terminated | ||
| Salirasib | II | Non-small cell lung cancer | NCT00531401 | Completed | |
| SOS | BI-1701963 | II | Advanced solid tumors, KRASG12C mutation | NCT04185883 | Recruiting |
| MRTX0902 | I/II | Solid tumor, advanced solid tumor, non-small cell lung cancer, colorectal cancer | NCT05578092 | Recruiting | |
| SHP2 | RMC-4630 | II | Non-small cell lung cancer | NCT05054725 | Active, not recruiting |
| I/II | Solid tumor | NCT03989115 | Completed | ||
| I/II | Advanced solid tumors, KRASG12C mutation | NCT04185883 | Recruiting | ||
| I/II | Metastatic neoplasm | NCT04418661 | Active, not recruiting | ||
| TNO155 | I/II | KRASG12C mutant solid tumors, non-small cell lung, carcinoma, colorectal, lung cancer, pulmonary cancer | NCT04699188 | Recruiting | |
| I/II | Advanced cancer, metastatic cancer, malignant neoplastic disease | NCT04330664 | Active, not recruiting | ||
| I/II | Advanced solid tumors, KRASG12C mutation | NCT04185883 | Recruiting | ||
| RLY-1971 | I | Advanced solid tumors, metastatic solid tumors | NCT05487235 | Recruiting | |
| I | Solid tumor, unspecified, adult | NCT04252339 | Completed | ||
| I | Colorectal cancer, non-small cell lung cancer | NCT05954871 | Recruiting | ||
| KRASG12C | Adagrasib (MRTX849) | Approved | Solid tumors harboring KRASG12C oncogenic driver mutation, including non-small cell lung cancer and colorectal cancer | Mirati Therapeutics | / |
| Sotorasib (AMG510) | Approved | KRASG12C-mutated locally advanced or metastatic non-small cell lung cancer | Amgen | / | |
| JNJ-74699157 (ARS-3248) | I | Neoplasms, advanced solid tumors, non-small cell lung cancer, colorectal cancer | NCT04006301 | Completed | |
| Divarasib (GDC-6036) | III | Non-small cell lung cancer | NCT03178552 | Recruiting | |
| D-1553 | II | Non-small cell lung cancer | NCT05383898 | Recruiting | |
| II | Non-small cell lung cancer | NCT05492045 | Recruiting | ||
| II | Solid tumor, non-small cell lung cancer, colorectal cancer | NCT04585035 | Recruiting | ||
| II | Solid tumor | NCT05379946 | Not yet recruiting | ||
| JDQ443 | III | Non-small cell lung cancer | NCT05132075 | Recruiting | |
| RMC-6291 | I | Non-small cell lung cancer, colorectal cancer, pancreatic ductal adenocarcinoma, advanced solid tumor | NCT05462717 | Recruiting | |
| Non-KRASG12C | MRTX1133 | I/II | Solid tumor, advanced solid tumor, non-small cell lung cancer, colorectal cancer, pancreatic adenocarcinoma | NCT05737706 | Recruiting |
| RMC-6236 | I | Non-small cell lung cancer, colorectal cancer, pancreatic ductal adenocarcinoma, advanced solid tumors | NCT05379985 | Recruiting | |
| siRNA strategies | KRASG12D iExosomes | I | KRASG12D metastatic pancreatic adenocarcinoma, pancreatic ductal adenocarcinoma | NCT03608631 | Recruiting |
| Antisense oligonucleotide | AZD4785 | I | Non-small cell lung cancer, advanced solid tumors | NCT03101839 | Completed |
| RAS analog | Rigosertib | III | Metastatic pancreatic adenocarcinoma | NCT01360853 | Completed |
| III | Myelodysplastic syndromes, chronic myelomonocytic leukemia | NCT01928537 | Completed | ||
| III | Myelodysplastic syndromes, chronic myelomonocytic leukemia | NCT01241500 | Completed | ||
| Cancer vaccines | mRNA-5671 | I |
Non-small cell lung cancer pancreatic cancer, colorectal cancer |
NCT03948763 | Completed |
| Antibody | NS1 | II |
Hematologic malignancy, acute leukemia, acute myeloid leukemia, acute lymphoblastic |
NCT05735717 | Recruiting |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov) and the United States Food and Drug Administration.gov (https://www.fda.gov/)
KRASG12C mutation inhibitors
Extensive studies on the structure and biofunction of KRAS mutations have shed light on the development of drugs targeting KRAS mutations. In particular, the development of KRAS mutation inhibitors has been encouraged by the approval of the first small-molecule KRASG12C inhibitor sotorasib (AMG510) for the treatment of KRASG12C mutant NSCLC patients by the FAD in 2021.934,935 Sotorasib is an FDA-approved KRASG12C-specific covalent inhibitor that irreversibly binds to the GDP-binding inactive conformation of KRASG12C, thus blocking its activity.931 Clinical trials have shown that sotorasib exerts anticancer activity in advanced solid tumor patients with KRASG12C mutations, including NSCLC,936 CRC, pancreatic cancer, endometrial carcinoma, appendiceal cancer, and melanoma.937 The first randomized phase III trial of targeting the KRASG12C inhibitor has revealed that sotorasib significantly increases PFS in NSCLC patients with KRASG12C mutation.938 Meanwhile, phase II clinical trials of sotorasib used for the treatment of previously treated locally advanced or metastatic NSCLC subjects with KRASG12C mutation or comorbidities are ongoing (NCT05631249 and NCT05311709). However, the objective response rate to sotorasib monotherapy is still far from satisfactory. A phase II clinical trial has shown that the objective response rate to sotorasib monotherapy in patients with advanced KRASG12C-mutated CRC is only 9.7%, indicating that its combination treatment strategy for KRASG12C-mutated CRC needs to be further evaluated.939 Following the successful development of sotorasib, adagrasib (MRTX849), an irreversible KRASG12C inhibitor received its first approval by the FDA for the treatment of advanced or metastatic NSCLC patients with KRASG12C mutation in December 2022. Adagrasib has a favorable pharmacokinetic profile, such as a long half-life (~24 h), broad tissue distribution, and dose-dependent pharmacokinetics.940 Notably, adagrasib penetrates the cerebrospinal fluid and causes the regression of lesions in patients with KRASG12C mutant NSCLC brain metastases.941 Clinical trials have demonstrated that adagrasib monotherapy is well tolerated with a disease control rate of 87% in 46 patients, and adagrasib in combination with cetuximab has shown clinical activity in patients with KRASG12C-mutated CRC. Moreover, adagrasib in combination with cetuximab is currently in a phase III clinical trial (NCT04793958) in patients with KRASG12C mutant CRC.942 The approval of sotorasib and adagrasib has opened the door to the possibility of developing more effective RASG12C inhibitors. ARS-853 is a highly cell-active, KRASG12C mutant-specific covalent inhibitor that targets the GDP-bound inactive state of KRAS and prevents its activation, resulting in the abrogation of KRAS mutation-induced signaling.930 ARS-853 is the first direct targeting KRAS inhibitor by covalently reacting with the RAS-GDP complex to trap it in its inactive state.930 Other KRASG12C mutation inhibitors such as JNJ-74699157 (ARS-3248), divarasib (GDC-6036), garsorasib (D-1553), JDQ443, and RMC-6291 are undergoing different phases of clinical trials in cancer patients (Table 1).Although extensive development of KRASG12C inhibitors is ongoing, they only work in a small fraction of patients with KRASG12C mutation, and the median PFS is fairly short, less than 1 year. Thus, an in-depth study of the resistance mechanism of KRASG12C mutation inhibitors is urgently needed. To date, KRAS inhibitor resistance mechanisms are distinguished into intrinsic resistance and acquired resistance.931,943 Intrinsic resistance is mainly influenced by the KRAS status in the cells. The cancer cells carrying KRAS mutations can be divided into two categories, KRAS-dependent and KRAS-independent,944 the latter of which may be less sensitive to KRAS inhibitors. Similarly, KRAS knockdown PDAC cells are able to maintain cell proliferation through PI3K alternative bypass pathway.931,945 The mechanisms of acquired resistance can be broadly classified into three categories: (1) KRAS alterations: mutations in the KRAS Y96, R68, and H95 residues. KRASY96D mutation can affect the binding pocket of inhibitors to KRAS protein switch-II.931,946 KRAS G12D/V/R, G13D, and Q61H mutations similarly cause resistance to KRAS G12C. Certain tumor cells produce new KRASG12C mutation after inhibitor treatment which maintains KRAS active and results in drug resistance.931 (2) Altered vertical signaling cascades: changes in upstream signaling pathways affecting KRAS-GTP binding affinity, downstream signaling pathways, such as MEK-ERK947 and EGFR,947 and other upregulation can all induce drug resistance.931 (3) Changes in the TME such as immune escape pathways produce resistance to KRASG12C inhibition931,948 Therefore, the combination of KRASG12C inhibitors with other drugs is considered an efficacious way to improve their antitumor effect. The results of clinical trials have proven that KRASG12C inhibitor combination strategies are well tolerated with no serious adverse effects in most patients. Of note, many clinical trials of the KRASG12C inhibitor combination are currently underway for the treatment of NSCLC and other solid tumors.931
In summary, KRASG12C inhibitors have promising therapeutic prospects for both monotherapy and combination use in cancer treatment. Given that KRAS inhibitor resistance is still a serious challenge in clinical practice, it remains to be elucidated in deeper detail to achieve precise therapies for cancer patients.
Non-KRASG12C mutation inhibitors
The promising advances in KRASG12C inhibitors have brought light to target other KRAS mutations. The KRASG12C mutation only represents a small part of the mutations, and other mutations include G12D, G12V, G12S, and G12R.931 MRTX1133 is a selective noncovalent KRASG12D inhibitor949 with a high affinity for KRASG12D and effectively inhibits the tumor growth of PDAC xenograft mouse models in vivo.950 RMC-9805 is a selective, orally bioavailable covalent KRASG12D inhibitor that effectively blocks the growth of KRASG12D mutant cancer cells in vitro and in vivo.931 In addition, RMC-6236 is a pan-RAS inhibitor that inhibits all RAS mutant subtypes,931 and is currently in phase I clinical trial (NCT5379985) in advanced solid tumors with KRAS mutations. However, due to the lack of active residues and intrinsic hydrolytic activity in these mutated proteins, the development of highly potent inhibitors against the above KRAS mutations remains a big challenge.931
The fast development of the above inhibitors targeting KRAS mutations may take advantage of the “addiction” of tumors to mutant RAS and create more effective regimens for more patients such as pancreatic cancer patients. The in-depth study of KRAS-specific mechanisms by which they enhance or hinder cancer proliferation will provide new insights for subsequent inhibitor development.
Other RAS targeting strategies
Existing KRASG12C inhibitors have narrow therapeutic windows and are only effective in a small proportion of cancer patients with KRAS mutation. Many studies are currently dedicated to discovering alternative strategies for targeting RAS. Pan-RAS inhibitors broaden the therapeutic windows by directly targeting multiple RAS-mutated cancers.951 The pan-RAS inhibitor RAS-IN-3144 can block the growth of KRASG13D mutant MDA-MB-231 cell-derived mouse xenograft tumors in vivo.952 A synthetic sos protein mimic has been found to suppress RAS activation as a pan-RAS inhibitor.953 Moreover, rigosertib is a RAS analog that inhibits RAS-mediated pancreatic cancer growth by interacting with the RAS-binding domain of RAF kinase, resulting in the inability of RAF.954 Additionally, oligonucleotides can inhibit protein synthesis by boosting mRNA degradation or interfering with translation.306 AZD4785 is an antisense oligonucleotide that targets KRAS mRNA with high affinity. It inhibits downstream signaling pathways by depleting cellular KRAS mRNA and proteins, thus exerting antitumor effects.955 A phase I clinical trial of AZD4785 (NCT03101839) in patients with KRAS mutated solid tumors has been completed, but no results have been posted. In addition, KRAS dimerization is essential for downstream signaling when KRAS is localized at the plasma membrane.951 NS1, a synthetic binding protein antibody, interferes with RAS dimerization and blocks CRAF-BRAF heterodimerization and activation.951,956 Recently, proteolysis-targeting chimeras (PROTACs) technology can directly degrade targeted proteins,308 which have been used to specifically degrade KRAS in cancer cells.957 In addition, CRISPR/Cas9 screening is also applied in the identification of RAS synthetic lethal gene.958 In addition, fibroblasts-derived exosomes loaded with G12D siRNA (iExosomes) can effectively restrain PDAC tumor growth.959,960 Mesenchymal stromal cells-derived exosomes with KRAS are currently in a phase I clinical trial (NCT03608631) evaluating efficacy in patients with pancreatic cancer. A phase I clinical trial (NCT03948763) of the KRAS mRNA vaccine V941 (mRNA-5671) in KRAS-mutated NSCLC, pancreatic cancer, and CRC has been completed, but no results have been posted.
Targeting upstream of RAS
Targeting RAS plasma membrane localization. Only plasma membrane localization of RAS proteins can stimulate downstream effectors and signaling pathways. If post-translational modification and membrane localization are blocked, RAS proteins will be inactive, suggesting that the inhibition of RAS membrane localization is an effective therapeutic strategy.951
Farnesyltransferase plays a vital role in RAS localization. The farnesyltransferase inhibitors (FTIs) can prevent RAS localization in the plasma membrane by suppressing farnesyltransferase, leading to the blockade of downstream signaling.931 Tipifarnib, an orally bioavailable, nonpeptidomimetic quinolinone FTI,961 was granted a fast track designation by the FDA in December 2019 for the treatment of patients with HNSCC harboring HRAS mutations who have progressed following platinum-containing chemotherapy. The objective response rate of tipifarnib in a phase II clinical study of HRAS-mutated recurrent and/or metastatic HNSCC was 55%, with common adverse effects of anemia and lymphocyte reduction.962 Subsequently, tipifarnib was given fast track status again by the FDA for the treatment of adult patients with different subtypes of peripheral T-cell lymphoma. Moreover, tipifarnib has been demonstrated to be effective in advanced refractory uroepithelial carcinoma with HRAS mutation,963 in recurrent and metastatic salivary gland carcinoma with HRAS mutation,964 and in HRAS-driven dedifferentiated thyroid cancers.965 Lonafarnib, originally discovered by Merck & Co as an investigational oncology drug966 and known as the world’s first drug approved by the FDA in November 2020 for the treatment of progeria and progeroid laminopathies, is also an orally active FTI that blockades RAS localization in the plasma membrane. Salirasib is a farnesylcysteine mimetic that blocks the function of all RAS isoforms (H-RAS, K-RAS, and N-RAS) by interfering with RAS binding to the plasma membrane.967 Numerous preclinical studies have shown the ability of salirasib to inhibit the proliferation of various human cancer cells,967 including breast cancer,968 glioblastoma,969,970 CRC,971 melanoma,972 ovarian cancer,973 HCC,974 and pancreatic cancer.975 However, a phase II clinical study revealed poor therapeutic activity of salirasib in KRAS-mutant lung cancer,976 and its development in subsequent clinical trials has been discontinued.931
Phosphodiesterase-δ phosphodiesterase-δ (PDEδ) is a prenyl-binding protein involved in regulating the membrane localization and signaling of farnesylated RAS. PDEδ binds and solubilizes farnesylated RAS proteins to enhance their diffusion in the cytoplasm and transfers RAS from the Golgi apparatus and endomembranes to the plasma membrane, thereby facilitating RAS enrichment at the plasma membrane and signaling transduction.977–979 Deltarasin inhibits KRAS-dependent proliferation of human PDAC cells by competitively binding the farnesyl-binding pocket of PDEδ and reducing RAS enrichment at the plasma membrane.978 Deltazinone has a similar mode of action to deltarasin, but possesses a higher selectivity and lower cytotoxicity.979 However, the fast release of KRAS-PDEδ inhibitors from PDEδ hindered drug-binding affinity, which resulted in poor antiproliferative activity of PDEδ inhibitors.980 Novel strategies, such as PDEδ degraders are underway.980
SOS1 inhibitors. Son of sevenless homolog 1 (SOS1), a guanine nucleotide exchange factor (GEF), catalytically promotes the activation of RAS which in turn consecutively enhances the GEF function of SOS1.306 Targeting SOS1 to disturb its interaction with KRAS in tumors is referred to as an effective way to inhibit a wide panel of KRAS-driven cancers.930,981,982 Multiple SOS1 inhibitors have been developed, mainly including quinazoline-based SOS1 inhibitors such as BAY-293 and BI-3406. BAY-293, first reported by Bayer and identified by combining high throughput screening and fragment screening in 2019, is a selective SOS1 inhibitor that suppresses the KRAS-SOS1 interaction at an IC50 value of 21 nM.983,984 BI-3406, an orally selective SOS1 inhibitor with quinazoline structure developed by Boehringer Ingelheim, can bind to the SOS1 catalytic domain, reduce GTP-RAS formation, and impair MEK inhibitor-induced feedback activation, eventually inhibiting KRAS-driven cancer cell proliferation.985 The most advanced SOS1 inhibitor is BI-1701963, an analog of BI-3406, which has demonstrated safety in clinical phase I trials and is under phase I clinical trial as monotherapy and in combination with trametinib in patients with KRAS mutated advanced or metastatic solid tumors (NCT04111458).931 MRTX0902 is a selective, orally bioavailable SOS1 inhibitor with antitumor effects in combination with MRTX849 in KRASG12C mutant NSCLC,931 and a phase I/II study of MRTX0902 in solid tumors with mutations in the KRAS MAPK pathway is ongoing (NCT05578092). In addition to the above representative small-molecule agonists, ZZ151, a potent, cooperative, and selective SOS1 PROTAC, has shown superior anticancer activities in KRASG12D and KRASG12V mutant xenografts in mice, which is worth further optimization.986 However, there is no marketable SOS1 compound, and only two cases are in clinical studies, with a lack of publicly disclosed clinical data. Exploiting highly selective and low-toxicity SOS1 inhibitors is a major research focus in the future.
SHP2 inhibitors. SHP2, a nonreceptor protein tyrosine phosphatase encoded by the PTPN11 gene, promotes GEF-mediated RAS-GTP interactions and activates the downstream RAS-RAF-ERK pathway.987,988 RMC-4630 is a selective orally bioavailable allosteric SHP2 inhibitor,988 which is undergoing a clinical phase I trial (NCT03634982) of monotherapy in participants with advanced relapsed or refractory solid tumors. A clinical phase I trial (NCT04916236) combining RMC-4630 and the ERK inhibitor LY3214996 for the treatment of KRAS-mutated cancer is ongoing. In addition, RMC-4630 and sotorasib are being used in combination in a clinical phase II trial (NCT05054725) to explore antitumor effects in patients with KRASG12C-mutated NSCLC. Similarly, TNO155, a selectively orally bioavailable SHP2 inhibitor,989 is being used alone or in combination with EGF816 (nazartinib) in phase I clinical trial (NCT03114319) for advanced solid tumor treatment. Other SHP2 inhibitors are also being tested in clinical trials. A clinical phase I trial of RLY-1971 in patients with advanced or metastatic solid tumors (NCT04252339) has been completed, but results have not yet been disclosed. SHP099 is a potent and selective small molecule SHP2 inhibitor that inhibits SHP2 activity by binding to the N-terminal and C-terminal ends of SH2 and the protein tyrosine phosphatase structural domain. SHP099 effectively suppressed tumor cell proliferation in vivo and in vitro by blocking RAS-ERK signaling.990 However, SHP2 inhibitor-based development has come a long way in the past few years. Earlier developed SHP2 inhibitors targeting the catalytic site failed in clinical practice due to poor selectivity and low bioavailability. Therefore, the development of SHP2 metathesis inhibitors has become an important research direction. Moreover, SHP2 inhibitors have a wide scope in drug combinations. In addition, due to the extensive expression of SHP2, a strategy that induces toxic effects and increases the safety of SHP2 inhibitors on normal cells is a primary problem to be solved.
Targeting RAS downstream effectors
The most classical downstream effector pathways of RAS are the RAF-MEK-ERK cascade pathway and the PI3K-AKT-mTOR pathway. A large number of inhibitors targeting the RAF-MEK-ERK and PI3K-AKT-mTOR pathways have been developed and are under clinical evaluation.958
PI3K-AKT-mTOR pathway inhibitors. Dysregulation of the PI3K-AKT-mTOR pathway is critical to the oncogenesis and progression of many human tumors, and their inhibitor development is of great significance991 (Table 2). However, the PI3K-AKT-mTOR pathway inhibitors have various resistance mechanisms. For example, the treatment of PI3K pathway inhibitors can result in the feedback activation of upstream signaling pathways, thus limiting their efficacy992
Table 2.
The FDA-approved and clinically developed PI3K-AKT-mTOR inhibitors
| Type | Target | Drug | Highest Phase | Indications | Company/Identifier | Status |
|---|---|---|---|---|---|---|
| PI3K | Pan-PI3K | Copanlisib (BAY80-6946) | Approved | Relapsed follicular lymphoma | Bayer | / |
| Buparlisib (BKM120) | III | Head and neck cancer | NCT04338399 | Recruiting | ||
| III | Metastatic breast cancer | NCT01633060 | Terminated | |||
| III | Breast cancer | NCT01610284 | Completed | |||
| III | Breast cancer | NCT01572727 | Completed | |||
| SF1126 (prodrug of LY294002) | I | Neuroblastoma | NCT02337309 | Terminated | ||
| p110α | Alpelisib (BYL719) | Approved | ER + /HER2-advanced metastatic breast cancer | Novartis Pharmaceuticals | / | |
| Inavolisib | III | Breast cancer | NCT05646862 | Recruiting | ||
| III | Breast cancer | NCT04191499 | Recruiting | |||
| III | Metastatic breast cancer | NCT05894239 | Recruiting | |||
| p110β | AZD8186 | I | Advanced castrate-resistant prostate cancer, squamous non-small cell lung cancer, triple-negative breast cancer | NCT01884285 | Completed | |
| GSK2636771 | II | Melanoma and other malignant neoplasms of skin, metastatic melanoma | NCT03131908 | Active, not recruiting | ||
| II |
Advanced lymphoma, advanced malignant solid neoplasm, hematopoietic and lymphoid cell neoplasm, refractory lymphoma, refractory malignant solid neoplasm, refractory multiple myeloma |
NCT04439188 | Active, not recruiting | |||
| II | Advanced lymphoma, advanced malignant solid neoplasm, hematopoietic and lymphoid cell neoplasm, refractory lymphoma, refractory malignant solid neoplasm, refractory multiple myeloma | NCT04439149 | Active, not recruiting | |||
| I/II | Advanced gastric adenocarcinoma | NCT02615730 | Completed | |||
| p110γ | IPI-549 | II | Head and neck squamous cell carcinoma, head and neck cancer, head and neck carcinoma, head and neck cancer stage IV, head and neck cancer stage III, HPV-Related carcinoma, HPV-Related malignancy, HPV-Related squamous cell carcinoma | NCT03795610 | Recruiting | |
| II | Bladder cancer, urothelial carcinoma, solid tumor, advanced cancer | NCT03980041 | Completed | |||
| II | Breast cancer, renal cell carcinoma | NCT03961698 | Active, not recruiting | |||
| p110δ | Idelalisib | Approved | Relapsed chronic lymphocytic leukemia | Gilead Sciences | / | |
| Duvelisib | Approved | Relapsed or refractory chronic lymphocytic leukemia, small lymphocytic lymphoma | Verastem | / | ||
| Umbralisib | Approved | Relapsed or refractory marginal zone lymphoma | TG Therapeutics | / | ||
| Dual PI3K/mTOR | Omipalisib (GSK2126458) | I | Solid tumors | NCT00972686 | Completed | |
| AKT | Capivasertib (AZD5363) | Approved | HR + , HER2- locally advanced or metastatic breast cancer with one or more PIK3CA/AKT1/PTEN-alterations | AstraZeneca | / | |
| Ipatasertib (GDC0068) | III | Breast cancer | NCT04060862 | Active, not recruiting | ||
| GSK2141795 | II | Cervical cancer | NCT01958112 | Terminated | ||
| II | Melanoma | NCT01941927 | Completed | |||
| II | Recurrent adult acute myeloid leukemia, untreated adult acute myeloid leukemia | NCT01907815 | Terminated | |||
| II | Estrogen receptor negative, HER2/Neu negative, invasive breast carcinoma, progesterone receptor negative, recurrent breast carcinoma, stage IV breast cancer, triple-negative breast carcinoma | NCT01964924 | Completed | |||
| MK-2206 | II | Colorectal neoplasms | NCT01333475 | Completed | ||
| II | Ovarian sarcoma, recurrent fallopian tube carcinoma, recurrent ovarian carcinoma, recurrent primary peritoneal carcinoma | NCT01283035 | Completed | |||
| II | Recurrent nasopharyngeal carcinoma | NCT01370070 | Completed | |||
| mTOR | mTORC1 | Everolimus | Approved | Advanced renal cell carcinoma following one prior antiangiogenic therapy | Novartis | / |
| Approved | HR+ and HER2- breast cancer | Novartis | / | |||
| Temsirolimus | Approved | Advanced renal cell carcinoma | Wyeth Pharmaceuticals | / | ||
| mTORC1/ mTORC2 | AZD2014 | II | Meningioma | NCT03071874 | Active, not recruiting | |
| II |
Neurofibromatosis 2 meningioma |
NCT02831257 | Completed | |||
| II | Diffuse large B-cell lymphoma | NCT02752204 | Completed | |||
| I/II |
Endometrial carcinoma, metastatic carcinoma, HR+ tumor |
NCT02730923 | Active, not recruiting | |||
| II | Metastatic breast cancer | NCT02299999 | Active, not recruiting | |||
| II | Non-small cell lung cancer metastatic | NCT02117167 | Active, not recruiting | |||
| II | Hepatocellular carcinoma | NCT03591965 | Terminated | |||
| INK128 | II | Metastatic castration-resistant prostate cancer | NCT02091531 | Completed | ||
| II | merkel cell carcinoma | NCT02514824 | Completed | |||
| II |
anaplastic thyroid cancer, thyroid cancer |
NCT02244463 | Active, not recruiting | |||
| OSI-027 | I | Any solid tumor or lymphoma | NCT00698243 | Completed |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov) and the United States Food and Drug Administration.gov (https://www.fda.gov/)
PI3K inhibitors. Numerous PI3K inhibitors have been developed and can be divided into three major classes: pan-PI3K inhibitors, isoform-specific PI3K inhibitors, and dual PI3K/mTOR inhibitors (Table 2). Most PI3K inhibitors are currently in clinical trials, and some of them (copanlisib, alpeilisib, idelalisib, duvelisib, and umbralisib) are approved by the FDA. However, the approvals/accelerated applications of some PI3K inhibitors, such as idelalisi, duvelisab, and umbrailisib, have been withdrawn due to frequent and severe adverse effects.993
Pan-PI3K inhibitors simultaneously inhibit the four catalytic subunits of class I PI3K p110α (PIK3CA), p110β (PIK3CB), p110γ (PIK3CG), and p110δ (PIK3CD). LY294002 and wortmannin are the first generation of pan-PI3K inhibitors, which build the foundation for the exploitation of novel high-efficiency and low-toxicity PI3K inhibitors. Unfortunately, the clinical applications of LY294002 and wortmannin are seriously limited by obvious adverse effects. LY294002 has poor aqueous solubility and adverse effects including severe respiratory depression and lethargy. Wortmannin has similar poor pharmacological properties, including a short half-life, chemical instability, and side effects, such as liver dysfunction and lymphocytopenia.993 Copanlisib (BAY80-6946) is an intravenous Pan-PI3K inhibitor developed by Bayer that inhibits α, β, γ, and δ isoforms with varying degrees of affinity,994 and received accelerated FDA approval in 2017 for the treatment of recurrent follicular lymphoma.993 Buparlisib (BKM120) is an oral ATP-competitive pan-PI3K inhibitor, being used for the treatment of stage II ESCC.995 The good brain penetration of buparlisib makes it a promising drug for the treatment of intracranial tumors.996 Buparlisib is currently being evaluated in a clinical phase III trial (NCT04338399) in combination with paclitaxel for the treatment of HNSCC. However, the lack of selectivity of pan-inhibitors results in nonselective inhibition of the PI3K pathway, leading to serious adverse side effects.
Researchers developed selective ATP-competitive inhibitors for each isoform of PI3K to limit the emergence of toxic effects. Isoform-specific PI3K inhibitors selectively inhibit p110α, p110β, p110δ, or p110γ subunits with greatly improved off-target effects. Alpelisib (BYL719), an oral inhibitor targeting PI3Kα developed by Novartis, was approved by the FDA for ER+/HER2-advanced metastatic breast cancer treatment in 2019.993 Inavolisib (GDC-0077) is also an oral inhibitor targeting PI3Kα with multiple ongoing clinical trials,997 including a phase III clinical trial (NCT05646862) for the treatment of HR+, HER2−, PIK3CA mutated breast cancer, and a phase II clinical trial (NCT05306041) for HR+, HER2+, PIK3CA mutated breast cancer. There are a few PI3Kβ-specific inhibitors, including GSK2636771,998,999 and AZD8186, developed by AstraZeneca.999 In addition, PI3Kδ inhibitors have mostly been approved by the FDA for clinical treatment. Idelalisib, developed by Gilead Sciences, was approved by the FDA in 2014 for the treatment of patients with relapsed CLL. Duvelisib, developed by Verastem, was approved by the FDA in 2018 for the treatment of relapsed or refractory CLL and small lymphocytic lymphoma, and subsequently received accelerated approval for adult patients with relapsed or refractory FL.1000 Umbralisib was developed by TG Therapeutics and received accelerated approval by the FDA in 2021 for the treatment of adult patients with relapsed or refractory marginal zone lymphoma.1001 Finally, no PI3Kγ inhibitors are currently available for clinical treatment. IPI-549, a potent and highly selective PI3Kγ inhibitor developed by Infinity Pharmaceuticals,1000 is being used in clinical phase II trials as a single agent for the treatment of HNSCC (NCT03795610) and in combination therapy for TNBC (NCT03961698). Furthermore, dual-targeted PI3K and mTOR inhibitors also exert potential roles in cancer therapy. Omipalisib (GSK2126458), a dual-targeted PI3K and mTOR inhibitor developed by GlaxoSmithKline,993 has completed a phase I clinical trial in solid tumors (NCT00972686), but no results have been disclosed yet.
In summary, although the side effects of PI3K inhibitors that can lead to serious and fatal immune-related adverse reactions remain an urgent issue,1002 PI3K inhibitors have great potential in clinical treatment. Exploring PI3K inhibitors in combination with other targeted therapies may be an effective strategy to reduce toxicity and improve clinical activity.346
AKT inhibitors. AKT belongs to the serine/threonine kinases family,343 which contains three isoforms with highly similar structures: AKT1, AKT2, and AKT3, making the development of their isoform-specific inhibitors challenging.346 Meaningfully, tumors with AKT1 mutations and AKT2 and AKT3 amplifications are highly sensitive to AKT inhibitors, while many PIK3CA mutant cancer cells were considerably less dependent on AKT,1003 which guides the rational use of AKT inhibitors. Currently, various selective ATP-competitive pan-AKT inhibitors (Table 2), including capivasertib by AstraZeneca Pharmaceuticals,1004 ipatasertib (GDC0068),1005 and GSK2141795,1006 have been developed which can inhibit all three AKT isoforms.1003 Significantly, on November 16, 2023, the FDA approved capivasertib (truqap) with fulvestrant for adult patients with HR+, HER2− locally advanced or metastatic breast cancer with one or more PIK3CA/AKT1/PTEN-alterations. In addition, AKT allosteric inhibitors such as the potent allosteric pan-AKT inhibitor MK-2206 possess better AKT specificity.1006 However, concerns such as the poor selectivity and toxicity of AKT inhibitors remain a burning challenge. Presently, PROTAC and AKT drug combination strategies may change the landscape of AKT drug development.
mTOR inhibitors. The mTOR pathway participates in multiple tumor cell processes, and hyperactivated mTOR signaling is observed in different types of cancers. Suppression of mTOR was approved by the FDA and the EMA as an effective strategy capable of inhibiting the PI3K-AKT-mTOR signaling.343 Currently, drug development against mTOR is a hot track, and mTOR inhibitors can be categorized into three generations: antibiotic allosteric inhibitors (first generation), ATP-competitive inhibitors (second generation), and novel mTOR inhibitors (third generation) (Table 2).1007 Rapamycin and its analogs (termed rapalogs) are members of the first generation of allosteric inhibitors of mTOR. Significantly, the rapamycin analogs everolimus (RAD001) has been approved for the treatment of RCC,1008 HR+ and HER2− breast cancer.1009 On May 30, 2007, temsirolimus (CCI-779) was approved for the treatment of advanced RCC.1010 Temsirolimus and metformin in combination with other drugs are currently in clinical phase I trials to evaluate their safety and dosage appropriateness in tumor patients. Researches on rapalogs have been relatively mature, but are further challenged by defects of large molecular weight, complex structure, difficulty in synthesis, and limited modification sites.1007 Therefore, the second-generation of small molecule-based mTOR inhibitors with simplified structures is a promising direction in the field of mTOR inhibitor development.1007 The second-generation mTOR inhibitors are structurally quite different from the first-generation mTOR inhibitors. They selectively target the active kinase site of mTOR and thus act as ATP-competitive inhibitors. A large number of ATP-competitive mTOR inhibitors are currently in clinical trials, including MLN01289 (INK128) in a phase II clinical trial (NCT02244463) for the treatment of anaplastic thyroid cancer. AZD2014 is in a phase I clinical trial (NCT02398747) for the treatment of advanced solid malignancies and a phase II clinical trial (NCT03071874) for the treatment of meningioma. OSI-027, an oral selective mTOR inhibitor, is being tested in a phase I clinical trial for advanced solid tumors or lymphoma (NCT02398747). A phase I clinical trial for AZD8055 in adults with recurrent gliomas (NCT01316809) has been completed, but no results have been disclosed. The third-generation mTOR inhibitor RapaLinks are important research breakthroughs that link rapamycin to ATP-competitive mTOR inhibitors via linkers to improve drug efficacy.1007 In addition, some natural products such as resveratrol can directly or indirectly regulate mTOR and mTOR signaling pathways.1007 Moreover, differences in mTOR inhibitor administration formula, doses, and oral bioavailability are found to result in differences in drug exposure and efficacy.1011 Improving the administration formula and combination with targeted drugs may be effective strategies to improve mTOR inhibitors.
RAF-MEK-ERK inhibitors
The RAF-MEK-ERK pathway regulates intracellular growth signaling1012 and is activated in more than 30% of human cancers.1013
BRAF is a commonly mutated protein kinase in human cancers, particularly frequent BRAF V600 point mutation in melanoma, and it has been thought to be an ideal target for cancer therapy.354 The first-generation selective RAF inhibitors, such as vemurafenib and dabrafenib, have been well-demonstrated to possess therapeutic effects in patients with BRAFV600E or BRAFV600K mutations although their efficacy was abrogated by quick-rising drug resistance.1014 To improve the efficiency and overcome drug resistance of first-generation RAF inhibitors, second-generation novel RAF inhibitors such as pan-RAF inhibitors and RAF dimer breakers have been developed.1015 Until now, DAY101 is the fastest progressing pan-RAF inhibitor in a phase II study to evaluate its safety and efficacy in patients with recurrent or progressive low-grade glioma or advanced solid tumors harboring a known BRAF alteration (NCT NCT04775485, NCT05760586). KIN-2787 is another pan-RAF inhibitor that is under a phase I clinical trial in adults with BRAF/NRAS-mutated advanced or metastatic solid tumors (NCT04913285). The phase I clinical trials of the pan-RAF inhibitor LY3009120 and LXH254 in advanced or metastatic cancers have been terminated because of their unfavorable efficiency.1016–1018 However, a phase I clinical trial of LXH254 in combination with LTT462 or trametinib or ribociclib for the treatment of KRAS or BRAF mutant NSCLC or NRAS mutant melanoma (NCT02974725) is ongoing.951 In addition, PLX8349 is an orally available, second-generation BRAF inhibitor with IC50 values of 3.8 nM, 14 nM, and 23 nM for BRAFV600E, BRAF, and CRAF, respectively. Importantly, PLX8349 effectively suppresses mutant BRAF cells without activating the MAPK pathway, thereby overcoming the resistance of first-generation RAF inhibitors.1019
Moreover, the inhibitors of MEK and ERK which are typical effectors of the RAS-RAF signaling pathway have been developed. Most MEK inhibitors are allosteric inhibitors rather than ATP-competitive inhibitors that block ERK phosphorylation via MEK.951 The oral ERK inhibitor MK-8353 is well tolerated and has good antitumor activity in BRAFV600 mutant melanoma patients.1020 The ATP-competitive ERK inhibitor SCH772984 effectively inhibited ERK1 and ERK2 activity and significantly suppressed the proliferation of BRAF, NRAS, or KRAS mutated tumor cells.1021 Clinical phase I trials have indicated that the ATP-competitive ERK1/2 kinase inhibitor ulixertinib (BVD-523) has antitumor effects in patients with advanced solid tumors.1013 In addition to monotherapy by the above inhibitors, the combination of MEK inhibitors or ERK inhibitors with RAF inhibitors effectively blocks the feedback activation pathway induced by RAF inhibitors or ERK inhibitors in BRAF-mutated or KRAS-mutated cancer cells.1022 In addition, the combination of the RAF-MEK-ERK pathway and PI3K-AKT-mTOR pathway for tumor-targeted therapy is a promising therapeutic strategy,951 which potently curbs the growth of various cancers such as KRAS-mutant lung cancer,1023 NRAS-mutated melanoma,1024 pancreatic cancer,1025 and many other cancers.1026
Taken together, inhibitors of the RAF-MEK-ERK pathway exhibit promising anticancer efficiency, although drug resistance impedes their further use. The in-depth investigation of the interaction of these inhibitors and growth regulatory mechanisms in specific tumor cell contexts and environments will improve the therapeutic effects and benefit future drug development. The combination of drug strategies based on RAS-targeted therapies and new technological approaches such as RNAi and CRISPR technologies will shed light on tumor therapy.992
Targeting evading growth suppressors
Cancer cells can maintain tumor progression by circumventing processes that negatively regulate cell proliferation which are supported by numerous tumor suppressor genes, such as RB and TP53. Therefore, targeting evading growth suppressors is a promising antitumor strategy.299 Indeed, CDK4/6 and MDM2 inhibitors are currently being used in clinical treatment or under development.1027,1028
CDK4/6 inhibitors
Relaxation of cell cycle mechanisms is associated with the dysregulation of CDKs, which ultimately promotes abnormal tumor proliferation and disease progression.1027 Over the past few decades, three generations of CDK inhibitors (CDKIs) have been developed. First- and second-generation CDKIs receive few clinical attention in the treatment of cancer patients due to their limited specificity and high toxicity. The development of third-generation CDKIs has made significant progress in clinical practice. Preclinical and clinical results suggest that these selective CDK4/6 inhibitors significantly reduce the progression of multiple malignancies.1029 Early in the cell cycle, mitotic signaling increases cyclin D expression, which binds to and activates CDK4/6. CDK4/6 phosphorylates the Rb protein which is further phosphorylated by CDK2, followed by releasing the E2F transcription factor which allows the cell to enter S phase.1030 Selective CDK4/6 inhibitors bind to ATP pockets, prevent CDK4/6 from binding to cyclin D, and prevent Rb phosphorylation, leading to G1 phase arrest and tumor cell death.1031 Currently, the typical CDK4/6 inhibitors palbociclib, ribociclib, and abemaciclib have been approved by the FDA for the treatment of different cancers alone or in combination with established therapies (Table 3).1032–1034
Table 3.
The FDA-approved and clinically developed CDK4/6 inhibitors
| Target | Drug | Highest Phase | Indications | Company/identifier | Status |
|---|---|---|---|---|---|
| CDK4/6 | Palbociclib (PD0332991) | Approved | HR+, HER2− advanced breast cancer in combination with hormonal therapy | Pfizer | / |
| Ribociclib (LEE-011) | Approved | HR+, HER2− advanced breast cancer in combination with hormonal therapy | Novartis | / | |
| Abemaciclib (LY2835219) | Approved | HR+, HER2− advanced breast cancer in combination with hormonal therapy, monotherapy for advanced HR+, HER2− breast cancer, adjuvant therapy for high-risk, early-stage HR+, HER2− breast cancer in combination with hormonal therapy | Eli Lilly | / | |
| Trilaciclib | Approved | Approved to reduce chemotherapy-induced bone marrow suppression in patients with extensive-stage small cell lung cancer | G1 Therapeutics | / | |
| TQB3616 | III | HR+, HER2− in advanced breast cancer | NCT05375461 | Recruiting | |
| III | Breast cancer | NCT05780567 | Recruiting | ||
| III | HR+, HER2− breast neoplasms | NCT05365178 | Not yet recruiting | ||
| SPH4336 | III | Locally advanced or metastatic breast cancer | NCT05860465 | Not yet recruiting | |
| III | Locally advanced or metastatic breast cancer | NCT05744687 | Recruiting | ||
| Dalpiciclib (SHR6390) | III | Advanced breast cancer | NCT05861830 | NCT05861830 | |
| III | Female breast cancer | NCT04842617 | NCT04842617 | ||
| III | Advanced breast cancer | NCT03966898 | NCT03966898 | ||
| Flavopiridol (L86-8275) | I/II | Lymphoma | NCT00445341 | NCT00445341 | |
| I/II | B-cell chronic lymphocytic leukemia, recurrent small lymphocytic lymphoma, refractory chronic lymphocytic leukemia, waldenström macroglobulinemia | NCT00058240 | NCT00058240 | ||
| II | Leukemia, lymphocytic, chronic | NCT00464633 | Recruiting | ||
| II | B-cell chronic lymphocytic leukemia, refractory chronic lymphocytic leukemia, stage I–IV chronic lymphocytic leukemia, | NCT00003620 | Completed | ||
| II | Refractory multiple myeloma, stage I–III multiple myeloma | NCT00047203 | Completed | ||
| II | Adenocarcinoma of the pancreas, recurrent pancreatic cancer, stage IV pancreatic cancer | NCT00331682 | Completed | ||
| II | Sarcoma | NCT00005974 | Completed | ||
| II | Prostate cancer | NCT00003256 | Completed | ||
| II | Adenocarcinoma of the gastroesophageal junction, diffuse adenocarcinoma of the stomach, intestinal adenocarcinoma of the stomach, mixed adenocarcinoma of the stomach, recurrent gastric cancer, stage IIIA gastric cancer, stage IIIB gastric cancer, stage IIIC gastric cancer, stage IV gastric cancer | NCT00991952 | Completed | ||
| II | Lymphoma | NCT00003039 | Completed | ||
| II | Kidney cancer | NCT00016939 | Completed | ||
| II | Melanoma (skin) | NCT00005971 | Completed | ||
| II | Endometrial cancer | NCT00023894 | Completed | ||
| II | Lymphoma | NCT00005074 | Completed | ||
| II | Liver cancer | NCT00087282 | Completed | ||
| I/II | Recurrent adult acute lymphoblastic leukemia, recurrent adult acute myeloid leukemia, secondary acute myeloid leukemia, untreated adult acute lymphoblastic leukemia, untreated adult acute myeloid leukemia | NCT00016016 | Completed | ||
| II | B-cell chronic lymphocytic leukemia, prolymphocytic leukemia, refractory chronic lymphocytic leukemia | NCT00098371 | Terminated | ||
| II | Head and neck cancer | NCT00020189 | Completed | ||
| II | Esophageal cancer | NCT00006245 | Completed | ||
| II | Breast cancer | NCT00020332 | Completed |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov) and the United States Food and Drug Administration.gov (https://www.fda.gov/)
Palbociclib, an orally reversible small molecule inhibitor developed by Pfizer, is the first selective CDK4/6 inhibitor. The chemical structure of palbociclib was identified in 2004,1030 and subsequently followed by 11 years of development. In February 2015, Palbociclib in combination with letrozole was accepted for accelerated approval by the FDA for the treatment of ER+, HER2− advanced breast cancer as initial endocrine-based therapy in postmenopausal women. Thereafter, in February 2016, the FDA approved palbociclib in combination with fulvestrant for the treatment of HR+, HER2− advanced or metastatic breast cancer in women with disease progression following endocrine therapy, followed by the approval of palbociclib in combination with an aromatase inhibitor for women with HR+, HER2− advanced or metastatic breast cancer and men in April 2019.1035 In preclinical studies, palbociclib exhibits significant antiproliferative effects on Rb-positive cells and leads to the selective arrest of the G1 phase in a series of tumor cells.1036 In a phase II clinical trial of palbociclib, the efficacy and safety of letrozole with or without palbociclib in the treatment of ER+, HER2− postmenopausal breast cancer patients were compared. The PFS of palbociclib plus letrozole versus letrozole alone was 20.2 months versus 10.2 months. The adverse effects especially hematologic aspects have been proven to be higher in the combination group compared with the letrozole monotherapy group.1037
Ribociclib is the second selective inhibitor of CDK4/6 developed by Novartis and approved by the FDA in 2017 for the treatment of patients with HR+, HER2− advanced or metastatic breast cancer.1030,1038 The IC50 values of ribociclib for CDK4 and CDK6 are much lower than those of other kinases, at 10 and 39 nM, respectively.1029 Ribociclib is used as a single agent or in combination with other drugs in preclinical studies. A phase III clinical trial of ribociclib has evaluated the efficacy and safety of ribociclib plus letrozole in patients with HR+, HER2− relapsed, or metastatic breast cancer. Results have shown PFS after 18 months in the ribociclib group versus placebo group 63.0 versus 42.2%. Moreover, adverse effects in the ribociclib group included nausea, infection (mainly upper respiratory tract infections and urinary tract infections), fatigue, diarrhea, neutropenia, leukopenia, hypertension, elevated alanine aminotransferases, lymphopenia, and QTc interval prolongation.1034 In the clinical trial MONALEESA-7, the efficacy and safety of ribociclib in combination with tamoxifen or nonsteroidal aromatase inhibitors have been observed in the treatment of advanced breast cancer with HR+ and HER2-. Moreover, ribociclib plus nonsteroidal aromatase inhibitors have been found to benefit more in advanced breast cancer without new adverse effects.1039
Abemaciclib is an oral CDK4/6 inhibitor developed by Eli Lilly with IC50 values of 2 and 10 nM for CDK4 and CDK6, respectively, and is approved by the FDA for the treatment of patients with HR+, HER2− advanced or metastatic breast cancer.1040 Abemaciclib is more active and less neutropenic than palbociclib and ribociclib.1041,1042 In addition, numerous studies have shown that abemaciclib can cross the blood-brain barrier into the central system, suggesting the possibility of treating primary or metastatic brain cancer.1043 In preclinical studies of abemaciclib, both monotherapy and in combination with endocrine therapy with abemaciclib have been found to have significant inhibitory effects on cancers.1043,1044 In the clinical study MONARCH-1, the safety and efficacy of abemaciclib monotherapy in patients with advanced breast cancer with refractory HR+, HER2− were evaluated. Results have shown that patients using abemaciclib have a significantly longer duration of response and PFS with milder adverse effects.1045 In a follow-up, a clinical study of abemaciclib in combination with fulvestrant or letrozole showed significant increases in PFS in the abemaciclib combination group compared with abemaciclib monotherapy group, and most patients were not severely neutropenic.1044,1046
Trilaciclib, a short-acting CDK4/6 inhibitor developed by G1 Therapeutics, has received approval from the FDA to decrease the incidence of chemotherapy-induced myelosuppression in adult patients when administered before a platinum/etoposide-containing regimen or topotecan-containing regimen for extensive-stage SCLC in 2021.1047 Other clinically CDK4/6 inhibitors have been developed including TQB3616, SPH4336, dalpiciclib (SHR6390), and flavopiridol (L86-8275). TQB3616, a novel CDK4/6 inhibitor, exhibits high selectivity and effectiveness in preclinical cancer models.1048 SPH3643, a highly selective CDK4/6 inhibitor, can efficiently and stably cross the blood‐brain barrier.1049 Dalpiciclib (SHR6390), a novel, highly selective CDK4/6 inhibitor, reveals high activity with IC50 of 12.4 nM and 9.9 nM against CDK4 and CDK6, respectively.1050 Flavopiridol, a pan-CDK inhibitor originally purified from Dysoxylum binectariferum,1051 is the first CDK inhibitor entering clinical trials and can target CDK1, CDK2, CDK4, CDK6, and CDK7.1052
MDM2 inhibitors
Since the first discovery of the co-crystal structure of MDM2-p53 complex in 1996,1053 an increasing number of researches have been dedicated to uncovering its functions in tumors.1054 Gene amplification, increased transcription, and accelerated translation cause the aberrant elevation of MDM2, which promotes p53 ubiquitination and increases p53 degradation. Thus, targeting MDM2-p53 interaction is a particularly attractive therapeutic strategy for p53 reactivation in tumors.1054,1055 Numerous small-molecule MDM2 inhibitors have been discovered so far, and nine of them, including RG7112, idasanutlin, AMG-232, SAR40583, APG-115, NVP-CGM097, siremadlin, and MK-8242, and milademetan, have undergone or been undergoing clinical trials for the treatment of cancers.1028
Nutlins, including Nutlin-1, Nutlin-2, and Nutlin-3, are the first selective and potent MDM2 inhibitors synthesized in 2004,1055 which lay the foundation for the following development of MDM2 inhibitors. RG7112, an MDM2 inhibitor developed by Roche based on the structure of Nutlins, can bind to the p53 pocket on MDM2 and suppress the p53-MDM2 interaction, and is the first MDM2 inhibitor to be assessed in clinical trials.387,1054 RG7112 has demonstrated clinical activity in a phase I trial for the treatment of patients with leukemia.1056 However, the poor tolerability and the adverse events, such as gastrointestinal toxicity, myelosuppression, sepsis, and hemorrhage, hinder its further developmen.387,1054,1056 Subsequently, idasanutlin (RG7388), another highly potent and selective MDM2 antagonist, exhibits superior potency, selectivity, and bioavailability compared with RG7112, although sharing the same action mechanism.1057 Idasanutlin is the second MDM2 inhibitor to be evaluated in clinical trials.387 The most common adverse events of idasanutlin include gastrointestinal toxicity (diarrhea and nausea) and hypokalemia.1054 A phase Ib study of idasanutlin in combination with XPO1 inhibitor selinexor for the treatment of children with progressive or recurrent atypical teratoid/rhabdoid tumors and malignant rhabdoid tumors is ongoing (NCT05952687). Meanwhile, a phase I/II study of idasanutlin monotherapy or in combination with chemotherapy or venetoclax for the treatment of acute leukemias or solid tumors is recruiting (NCT04029688). In addition, AMG-232 (navtemadlin, KRT-232) is an investigational oral, selective MDM2 inhibitor. Its most common adverse events include nausea, diarrhea, vomiting, decreased appetite, anemia, thrombocytopenia, and leukopenia.1058 Currently, five clinical trials of AMG-232 monotherapy or in combination with other drugs for the treatment of patients with cancers are recruiting (NCT03031730, NCT03041688, NCT03107780, NCT03217266, and NCT04190550).
Furthermore, SAR405838(MI-77301) and APG-115 are all spirooxindole-based MDM2 inhibitors developed by Wang Shaomeng’s research group.387,1059 APG-115 is designed to overcome stability-related issues observed in SAR405838.1054,1060 APG-115 can strongly bind with MDM2 protein, and has good chemical stability and excellent oral pharmacokinetic parameters.387 A phase I study of SAR405838 in patients with advanced cancer has been completed (NCT01636479), but no result has been posted. Significantly, seven clinical trials of APG-115 monotherapy or in combination with other drugs for the treatment of patients with tumors are recruiting. NVP-CGM097(CGM097), a highly potent and selective MDM2 inhibitor, has good cell activity, metabolic stability, and PK parameters.1061 A phase I study of CGM097 in patients with advanced solid tumors with p53 wild-type status has been completed with no results posted (NCT01760525). Siremadlin (HDM201) is an orally bioavailable and selective inhibitor of the p53-MDM2 interaction designed by Novartis.1062 Currently, three clinical trials of siremadlin monotherapy or in combination with other drugs for the treatment of patients with advanced soft-tissue sarcoma (NCT05180695), AML (NCT05447663, NCT05155709) are ongoing. MK-8242 (SCH900242) is a potent, orally bioavailable, small-molecule inhibitor of the MDM2-p53 interaction. Its common adverse events include anemia, leukopenia, pancytopenia, nausea, hyperbilirubinemia, hypophosphatemia, and anorexia.1063 A phase I trial of MK-8242 in patients with refractory/recurrent AML has been completed.1064 Furthermore, Milademetan (DS3032b) is an orally active MDM2 inhibitor by disrupting the MDM2-p53 interaction.1065 A phase I study of milademetan in combination with low-dose cytarabine with or without venetoclax for the treatment of AML has been completed, and has revealed modest therapeutic responses with recognizable gastrointestinal toxicity.1065
Several MDM2 inhibitors are currently being evaluated clinically for cancer therapy. However, there is no MDM2 inhibitor approved for clinical application.387 Challenges such as acquired resistance and toxicity remain. In addition, MDM2 PROTAC degraders have received heightened attention in recent years with higher cancer therapeutic efficacy, and their safety needs further determination1066.
Targeting enabling replicative immortality
Studies have illustrated that the protection of telomeres at the ends of chromosomes is essential for the replicative immortality ability of tumor cells. Tumor cells avoid senescence or apoptosis by upregulating telomerase expression to maintain telomeric DNA length. Therefore, targeting telomerase is of great importance. Various telomerase targeting strategies have been developed, such as vaccines, antisense oligonucleotides, and small molecule inhibitors.390,1067
Telomerase inhibitors
Given the critical role of telomere length in tumor proliferation, targeting telomerase is a promising antitumor treatment approach.392 It has been observed that the initial telomere length affects the therapeutic effect of telomerase inhibitors.1068 Therefore, it is valuable to detect telomerase length during clinical telomerase-targeted therapy.1069
The key goals of antitelomerase therapy are to induce tumor cell death and to reduce normal cytotoxicity. Numerous telomerase therapeutic options inhibit hTERT or hTR activity.390,1067 Imetelstat (GRN163L), developed by Geron, is a competitive inhibitor of telomerase that binds to the nucleotide region of the telomerase holoenzyme at a specific active site with high affinity, resulting in complete inhibition of telomerase activity.390 It has been identified that imetelstat has inhibitory effects on a series of cancer cells, including breast cancer,1070 lung cancer,1071 prostate cancer,1072 pancreatic cancer,1073 osteosarcoma,1074 glioblastoma,1075 HCC, and bladder cancer.390 In August 2023, Geron corporation announced the FDA acceptance of a new drug application for imetelstat for the treatment of transfusion-dependent anemia in patients with lower-risk myelodysplastic syndromes (MDS). Imetelstat is currently in a phase II clinical trial to evaluate its efficacy and safety in participants with high-risk MDS or AML that is relapsed/refractory to hypomethylating agent treatment (NCT05583552).
In addition, an increasing number of studies have identified telomerase as a promising anticancer immunotherapy target, and telomerase-based immunotherapy is a potential antitumor therapeutic strategy.1076 Antitelomerase immunotherapy exerts its antitumor effects mainly by enhancing the sensitivity of the immune system to tumor cells expressing telomerase-specific antigenic epitopes, thereby activating hTERT-specific CD8+ cells. The protein fragments or peptides formed by telomerase degradation in tumor cells are expressed on the surface of tumor cells as tumor-associated antigens via the human leukocyte antigen (HLA) class I pathway, which in turn triggers antitumor cytotoxic T lymphocyte responses.390 CD4+ or CD8+ cytotoxic T lymphocytes can target telomerase-specific antigenic epitopes to kill tumor cells.1076 Current telomerase-based cancer immunotherapy mainly includes the hTERT vaccine and dendritic cell strategy. Immunotherapy-based hTERT peptide (GV1001), cryptic peptides (Vx001), and dendritic cells (GRVAC1), three promising telomerase-targeted vaccines with low toxicity to normal cells and no autoimmunity,390 have been used for antitelomerase immune response therapy in cancer patients.1077 GV1001 is a 16 amino-acid hTER peptide vaccine containing the hTERT active site which significantly activates CD4+ or CD8+ cytotoxic T lymphocyte responses.1078 Vx001 is a vaccine containing the hTERT amino acid sequence with a high affinity for HLA class I. GRNVAC1, a dendritic cell-based vaccine with good activity and tolerance, exerts the antitumor effect by stimulating lysosomal degradation of hTERT into small peptide.390 Currently, GV1001, GRNVAC1, and Vx001 vaccines are in clinical trials. A phase II clinical trial (NCT00510133) of active immunotherapy with GRNVAC1 in patients with acute myelogenous leukemia has been completed, but no results have been posted.
Although targeting telomerase is an attractive antitumor strategy, some limitations still exist. First, as telomere shortening induced by cell division is a slow and long-term process, therapies that inhibit telomerase activity are hysteretic and are not suitable for first-line cancer therapy. Second, since telomerase is expressed in highly proliferative cells such as hematopoietic precursor cells and epidermal stem cells, telomerase targeting therapies are potentially toxic and cause side effects. Third, preclinical telomerase targeting therapy lacks a suitable model for evaluation.1068 It has been observed that mouse telomeres are 5–10 times longer than human,1068,1079 and their telomerases are widely expressed at low levels in adult tissue.1080 Thus, the dependence of mice on replicative immortality induced by telomerase activation is much lower than that of humans. Fourth, the structure characteristics of human telomerase holoenzymes need to be further resolved, which is crucial for telomerase inhibitor development.1068
Antiangiogenesis therapy
The concept of “antiangiogenesis” therapy was proposed by Dr. Judah Folkman in 1971, who found that tumor growth requires neovascularization for maintenance.1081 Antiangiogenic therapy has become an invaluable strategy to hinder tumor proliferation and metastasis. The current antiangiogenic inhibitors mainly include VEGF inhibitors, FGF inhibitors, and PDGF inhibitors.
VEGF inhibitors
Antiangiogenic therapies target angiogenesis via two major mechanisms: blocking intracellular receptor tyrosine kinases or neutralizing angiogenic factors such as VEGF or its receptor. The current drugs targeting VEGF include mAbs, VEGF decoy receptors, and small molecule TKIs. These drugs can be used as monotherapy or in combination with other chemotherapeutic agents for clinical treatment.401
In 2004, the anti-VEGF-A humanized mAb bevacizumab (avastin), the first antiangiogenic agent, was first approved by the FDA for the treatment of advanced CRC. In May 2009, bevacizumab was approved as a second-line cancer therapy for the treatment of glioblastoma. Subsequently, in July 2009, the FDA approved bevacizumab in combination with interferon-alpha for the treatment of RCC.401 Currently, combination chemotherapy is used for treating patients with advanced CRC, NSCLC, and breast cancer.398,1082 Ramucirumab (cyramza), a mAb that binds VEGFR2 to block the VEGF signaling pathway, has been approved for the treatment of many types of solid tumors, such as advanced or metastatic gastric cancer, gastroesophageal junction adenocarcinoma, uterine cancer, CRC, and ovarian cancer.425,1083,1084 Aflibercept, also known as VEGF-Trap, is a soluble recombinant fusion protein consisting of the extracellular binding domains of VEGFR-1 and VEGFR-2, which was approved by the FDA in 2012 for the treatment of metastatic CRC.1084 Aflibercept interacts with circulating VEGF, thus preventing its binding to receptors on endothelial cells.1085 There are clinical studies using aflibercept in combination with the chemotherapeutic agent for the treatment of various malignancies, such as patients with advanced CRC.401 In addition, ziv-aflibercept was approved for the treatment of patients with metastatic CRC in combination with FOLFIRI (folinic acid, fluorouracil, and irinotecan) in 2012.425,1086
Since angiogenesis contains multiple signaling pathways, such as the VEGFR family and FGFR family, selective antiangiogenic agents especially TKIs can minimize the induction of toxicities.1087 Compared with macromolecules, small molecule TKIs possess multitarget inhibitory efficiency, and penetrate into cells easily due to hydrophobic properties, thus blocking the activation of various signaling pathways, which ultimately increases treatment efficiency.1087 In addition, small molecule TKIs can be administered orally as inhibitor salt form.1087
Sorafenib (BAY43-9006) is a selective inhibitor targeting VEGFR-2 and VEGFR-3, PDGFR, FMS-like tyrosine kinase 3 (FLT-3), and c-Kit401 and is the first TKI approved by the FDA as a first-line treatment for advanced HCC.1088 A phase III clinical trial uncovered significant improvements in OS in patients with advanced HCC treated with sorafenib.1089 Common clinical adverse effects of sorafenib include diarrhea, fatigue, skin reactions on the hands and feet, rash or desquamation, and anorexia.416 Lenvatinib, an oral TKI that targets VEGFR, FGFR, PDGFR, KIT, and RET, exerts antiproliferative and immunomodulatory activity in preclinical cancer models, and has been approved as a first-line treatment for HCC. Compared with sorafenib, levatinib has improved OS and PFS in patients with HCC.1090 In addition, lenvatinib suppresses the development of various cancers by inhibiting VEGFR signaling, such as kidney cancer,1091 pancreatic cancer,1092 SCLC,1093 breast cancer,1094 differentiated thyroid cancer,1095 and anaplastic thyroid cancer.1090 The most common adverse effects of lenvatinib treatment include hypertension, diarrhea, loss of appetite, weight loss, and fatigue.416 Sunitinib, an oral TKI targeting VEGFR1-3, PDGFR, FLT-3, and c-Kit,401,1085 is approved by the FDA for the treatment of refractory GIST and metastatic RCC.401 There are clinical studies using sunitinib for the treatment of advanced or metastatic breast cancer and NSCLC.401 Pazopanib (GW786034) has been approved for treating advanced RCC. Its common adverse effects are diarrhea, hypertension, hair color change, nausea, anorexia, and a 2% occurrence of myocardial infarction or ischemia. Axitinib, a second-generation inhibitor of VEGF-1, 2, and 3, is more selective for VEGF without blocking PDGF, B-RAF, FLT-3, and KIT targets. In January 2012, axitinib was approved by the FDA for the treatment of RCC after failure of a prior systemic therapy.401
Despite the emerging number of antiangiogenic therapeutic agents, challenges remain to be solved in antiangiogenic drug development and application.398 First, the tumor vascular system in preclinical mouse models is more sensitive to antiangiogenic therapy than the human tumor vascular system, which enhances the inconsistency between preclinical and clinical practice.398 Second, the effect of antiangiogenic monotherapy is limited, and the appropriate strategy for the combination of antiangiogenic drugs with other drugs needs to be discovered. For example, immune checkpoint inhibitors and antiangiogenic agents have been proven to be a promising therapeutic strategy. Antiangiogenic agents can reshape the immune TME, while immune checkpoint inhibitors can downregulate vascular endothelial growth factor expression and alleviate hypoxic conditions to combat angiogenesis.1096 Moreover, withdrawal of antiangiogenic drugs may result in tumor growth and metastasis, and the strategy to prevent this rebound needs to be developed.1097 In conclusion, tumor angiogenesis is essential for tumor growth. The regulatory factors of tumor angiogenesis and targeted therapies are diverse and need to be further elucidated in the future.
FGF/FGFR inhibitors
Studies have found that abnormal FGF signaling contributes to tumor angiogenesis and subsequently promotes tumor growth.422 Researches on FGF-targeted therapies have flourished in recent years, but no FGF-targeted therapy has been approved for cancer treatment. Current FGF-targeted therapies include multitargeted small molecule TKIs, selective FGFR-targeted TKIs, mAbs against FGFR, and FGF ligand traps.1098,1099
Dovitinib (TKI258) is an orally active and nonselective TKI that targets VEGFR1-3, FGFR1-3, and PDGFR.1100 It has good antitumor activity against RCC1101 and breast cancer.1100 Clinical trials evaluating the efficacy and safety of dovitinib alone or in combination with other anticancer drugs in patients with solid tumors and hematologic malignancies are ongoing. Other nonselective multitargeted anti-FGFR TKIs include nintedanib (BIBF1120),1102 lucitanib (E3810),1103 and ponatinib (AP24534),1104 which reveal antitumor activity in advanced solid tumors. Given the toxicity of multitargeted TKIs, several FGFR-selective inhibitors have been developed that hinder FGFR1-3 kinase activity to varying degrees and are currently in clinical trials. Infigratinib (NVP-BGJ398), an oral ATP-competitive FGFR inhibitor developed by QED Therapeutics, was granted accelerated approval by the FDA on May 28, 2021, for the treatment of adults with previously treated, unresectable locally advanced or metastatic cholangiocarcinoma with an FGFR2 fusion or other rearrangement.1105 In addition, infigratinib is currently in a phase I clinical trial for renal pelvis and ureter urothelial carcinoma (NCT04228042), and phase II clinical trials for gastric cancer (NCT05019794), advanced solid tumors (NCT04233567), cholangiocarcinoma (NCT04233567), and metastatic and refractory malignant solid neoplasm (NCT04233567) treatment. Erdafitinib (JNJ-42756493), an FGFR inhibitor developed by Janssen Pharmaceutical, was granted accelerated approval by the FDA on April 12, 2019, for the treatment of patients with locally advanced or metastatic urothelial carcinoma with susceptible FGFR3 or FGFR2 genetic alterations.1106 Erdafitinib is currently in a phase II clinical trial for urinary bladder neoplasms (NCT04172675) and advanced solid tumor (NCT04083976) treatment. Moreover, futibatinib (TAS120), an oral, covalently binding, irreversible inhibitor of FGFR1-4, is being developed by Taiho Oncology and Taiho Pharmaceutical for the treatment of various cancers.1107 On September 30, 2022, the FDA granted accelerated approval to futibatinib for the treatment of adult patients with previously treated, unresectable, locally advanced or metastatic intrahepatic cholangiocarcinoma harboring FGFR2 gene fusions or other rearrangements.1108 Futibatinib is currently in a phase II clinical trial for the treatment of patients with metastatic breast cancer (NCT04024436), advanced and metastatic HCC (NCT04828486), and advanced and metastatic urothelial cancer (NCT04601857), phase I and II clinical trials for NSCLC (NCT04965818), and a phase III clinical trial for soft tissue sarcoma (NCT03784014). In addition to small molecules, there are few clinical anti-FGFR mAbs. MGFR1877S is an anti-FGFR3 mAb that is being studied in a phase I clinical trial in patients with solid tumors (NCT01363024). FP-1039 (GSK3052230) is a soluble fusion protein that belongs to an FGF ligand trap that isolates the FGF ligand and prevents its binding to the receptor.1099 FP-1039 was studied in a phase I trial in patients with advanced tumors (NCT00687505) and a phase I trial was withdrawn in endometrial cancers with FGFR2 mutations (NCT01244438) due to substandard recruitment of patients.
Although there are numerous inhibitors and antibodies targeting FGF/FGFR under investigation, the development of anti-FGF/FGFR agents remains a tough challenge. Tumors with higher FGFR amplification copy numbers are more sensitive to FGFR inhibitors, whereas the fraction of patients with high FGFR amplification is not frequent. Thus, monitoring the status of FGFR and choosing a subgroup of patients could be beneficial for FGRF-targeted therapy.438,1098 The precise treatment for patients with tumor heterogeneity and specific FGFR abnormalities remains an urgent task.1098 Moreover, the side effects and toxicity of nonselective FGFR inhibitors remain an unresolved challenge.1109
PDGF/PDGFR inhibitors
PDGF is an essential angiogenic factor and its expression is upregulated in a diverse range of tumor cells. Therefore, blocking PDGF/PDGFR signaling is a promising strategy for targeted therapy.
Targeting PDGFR
PDGFR-targeted therapies include specific inhibitors, nonspecific inhibitors, mAbs, and RNA aptamers, among which inhibitors have been extensively studied. PDGFR inhibitors are classified into two categories including specific and nonspecific inhibitors, based on their binding properties to the receptor, and many PDGFR inhibitors are currently used in the clinical treatment of different cancers.443 CP-673451, a specific ATP-competitive PDGFR inhibitor, effectively inhibits PDGFRβ activity.1110 CHMFL-PDGFR-159 is a highly selective inhibitor of PDGFRα, which significantly inhibits the proliferation of chronic eosinophilic leukemia cells.1111 In addition, there are a large number of nonspecific PDGFR inhibitors. As tyrosine kinases have a conserved ATP-binding pocket, most small molecule TKIs are in a multitarget binding mode and inhibit both PDGFRα and PDGFRβ activity, including imatinib, ponatinib, sorafenib, nilotinib, nintedanib, dasatinib, midostaurin, ripretinib, sitravatinib, masitinib, sunitinib, axitinib, pazopanib, crenolanib.1112 Imatinib, the milestone in the history of TKI development, is a multitarget TKI that targets PDGFR signaling1112 and is used for the first-line treatment of patients with BCR-ABL-positive leukemia.443 Sunitinib (SU11248) is a multitarget inhibitor that targets PDGFR, VEGFR, c-Kit, FLT-3, and Ret kinase,1113 which was approved by the FDA in 2006 for the first-line treatment of metastatic RCC.1114 Sorafenib is an inhibitor targeting PDGFR, c-Kit, FLT-3, and VEGFR for the treatment of patients with advanced HCC.443,1115 Ponatinib (AP24534) is a multitarget TKI targeting BCR-ABL, PDGFR, VEGFR, FGFR, and Src, which has been applied as a third-line agent for CML treatment.443 Regorafenib is a multitargeted TKI that suppresses PDGFR, VEGFR, c-Kit, BRAF, as well as EGFR, and ERK1116 for the treatment of advanced HCC, CRC, and GIST.443,1116,1117 Nilotinib is a TKI that blocks PDGFR α/β, c-Kit, and is primarily used in the treatment of patients with CML and ALL.443,1118 Nintedanib is an inhibitor that blocks PDGFRα/β, VEGFR1-3, and FGFR1-3.443 Dasatinib is a multitarget inhibitor that targets PDGFR α/β, BCR-ABL, YES, and c-Kit and is approved by the FDA for the treatment of chronic granulocytic leukemia.443,1119 Midostaurin (PKC412) blocks PDGFR, Kit, VEGFR2, and PKCα, and is primarily used in the treatment of AML with FLT-3 mutations.443,1120,1121 Ripretinib (DCC-2618) inhibits PDGFRα/β, VEGFR2, and Kit activity and is approved for the treatment of adult patients with advanced GIST.443,1122 Sitravatinib (MGCD516) and masitinib (AB1010) broadly target the PDGFR family.443 Axitinib is also a multitarget inhibitor targeting PDGFR, VEGFR1-3, and c-Kit, and is being used to treat patients with advanced RCC.443,1123 Pazopanib is an inhibitor that targets PDGFR, VEGFR, and c-Kit, and is useful in the treatment of RCC and soft tissue sarcoma.443,1124 Crenolanib (CP-868596) inhibits PDGFRα/β, FLT-3, and c-Kit and is a highly selective inhibitor of PDGFRβ. Moreover, lenvatinib and avapritinib are both oral inhibitors that target PDGFRα and c-Kit.443 Although a large number of TKIs targeting PDGFR have been approved for clinical practice, their low specificity and activity against PDGFR make accuracy evaluation of anti-PDGFR effect difficult. Moreover, the side effects and increased toxicity of PDGFR inhibitors induced by multitargets or in combination with other anticancer drugs are big issues and cannot be ignored. It is worthwhile to explore the structure-function relationship of PDGF inhibitors in order to discover PDGF/PDGFR inhibitors with high specificity and selectivity in the future. The nanotechnology may provide new perspectives for PDGF/PDGFR-targeted therapy by enhancing target specificity and drug delivery accuracy.443
Compared with extensive progress that has been made in TKIs targeting PDGFR, the progress on mAbs targeting PDGFR is limited. Olaratumab (IMC-3G3), a high-affinity human anti-PDGFRα mAb,1125 is currently in a phase I clinical trial for soft tissue sarcoma (NCT03126591). Tovetumab (MEDI-575) is also a human anti-PDGFRα mAb that is well tolerated and has a favorable pharmacokinetic profile in patients with advanced solid tumors.443 Gint4.T, a high-affinity RNA aptamer that specifically binds to PDGFRβ,1126 inhibits TNBC lung metastases1127 and glioma.443 The high specificity of mAbs offers a significant opportunity to selectively target PEGFR resulting in specific therapeutic effects, which warrant further investigation.
Targeting PDGF
Compared with the extensive studies targeting PDGFR, only a few studies are reporting PDGF targeting therapy. MOR8457 is a high-affinity PDGF mAb1128 that selectively binds PDGF-BB and masks its receptor PDGFRβ-binding epitope, thereby blocking receptor dimerization and tyrosine kinase activation, ultimately effectively preventing PDGF-BB-induced cell proliferation.443 6B3, a highly selective mAb, blocks PDGF-CC-induced PDGFRα phosphorylation and activation.1129 AX102 is a high-affinity single-stranded DNA aptamer that blocks PDGF-B and prevents activation of downstream proliferative signals.443,1130 E10030 is a PEG-modified aptamer that specifically binds to and restrains the function of PDGF-B.425
In summary, antiangiogenic drugs offer hope for antitumor therapy, but their resistance remains a pressing clinical issue. There are various reasons for antiangiogenic drug resistance. It has been revealed that lack of VEGF or VEGFR in certain metastatic tumors leads to poor efficacy in VEGF-targeted therapy, e.g., blocking the VEGFR2 pathway inhibits the growth of human RCC RBM1-IT4 cells implanted in the kidney but not in the bone of nude mice.1131 Tumor cells maintain sustained growth through existing blood vessels in organs with rich vascular systems (e.g., lungs), thus developing drug resistance.1132 Moreover, The abnormal induction of various proangiogenic factors such as bFGF,406 circulating PlGF, VEGF,1133 and FGF,1134 can cause acquired resistance to antiangiogenic therapy.406 In addition, the vascular dependence of tumor cells is heterogeneous and variable. Tumors with p53 mutation are less dependent on vascular supply and are more resistant to antiangiogenic drug treatment.1135 Therefore, an in-depth investigation into the mechanisms of antiangiogenic drug resistance and tumor heterogeneity is a promising strategy for tumor treatment.
Targeting resisting cell death
Autophagy inhibitors/inducers
The multiple steps of the autophagic pathway present many opportunities for the development of targeted autophagy inhibitors, which currently include small molecule autophagy inducers and autophagy inhibitors, the latter consisting of inhibitors of autophagy initiation, autophagosome maturation, and lysosomal activity, hijacking the autophagosome and lysosome for targeted protein degradation1136,1137 (Table 4).
Table 4.
The FDA-approved and clinically developed autophagy inhibitors/ inducers in cancer therapies
| Drug | Highest phase | Indications | Identifier | Status |
|---|---|---|---|---|
| Rapamycin | IV | Non-hodgkin’s lymphoma | NCT01180049 | Completed |
| IV | Angiomyolipoma | NCT01217125 | Completed | |
| IV | Refractory solid Tumors | NCT02688881 | Completed | |
| IV | Hemangioendothelioma of liver | NCT04406870 | Not yet recruiting | |
| Everolimus (RAD001) | Approved | Progressive, well-differentiated non-functional, neuroendocrine tumors of gastrointestinal or lung origin with unresectable, locally advanced or metastatic disease | Novartis | / |
| Approved | Advanced renal cell carcinoma following one prior antiangiogenic therapy | Novartis | / | |
| Approved | Advanced hormone receptor-positive, HER2-negative breast cancer in postmenopausal women | Novartis | ||
| AZD8055 | I | Glioblastoma multiforme, anaplastic astrocytoma, anaplastic oligodendroglioma, malignant glioma, brainstem glioma | NCT01316809 | Completed |
| I | Cancer, solid tumors, advanced solid malignancies | NCT00973076 | Completed | |
| I | Solid tumors | NCT00731263 | Completed | |
| I | Advanced hepatocellular carcinoma, cancer | NCT00999882 | Completed | |
| Chloroquine (CQ) | Approved | Prophylactic treatment of malaria | Bayer | / |
| II | Astrocytoma, grade IV, glioblastoma | NCT02432417 | Not yet recruiting | |
| Hydroxychloroquine (HCQ) | Approved | Chronic, discoid or systemic lupus erythematosus and acute or chronic rheumatoid arthritis | / | / |
| II | Advanced cancer, pancreatic cancer | NCT04386057 | Recruiting | |
| II | Melanoma | NCT04464759 | Recruiting | |
| II | Breast cancer | NCT04841148 | Recruiting | |
| II | Hepatocellular cancer | NCT03037437 | Recruiting | |
| II | Breast cancer | NCT04523857 | Recruiting | |
| II | Metastatic colorectal cancer | NCT05843188 | Recruiting |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov) and the United States Food and Drug Administration.gov (https://www.fda.gov/)
The first category of autophagy inducers is small molecule compounds which act primarily by directly inhibiting mTORC1 or activating AMPK. The cellular energy state sensor AMP can inhibit biosynthesis in response to energy stress by suppressing mTORC1. Thus, inhibition of mTORC1 blocks phosphorylation of ATG13, ULK1, and ULK2 in the ULK1 complex, and promotes AMPK activation or RAPTOR phosphorylation, thereby increasing autophagic flux.1136,1138 Moreover, rapalogs induce autophagy by forming a complex with FK506-binding protein (FKBP12), acting as a metamorphic inhibitor of mTORC1.1136,1139 Rapamycin, Torin-1, and AZD8055 can selectively inhibit mTORC1 kinase activity.1136,1140 In addition, the sodium voltage-gated channel blocker carbamazepine and the L-type calcium channel blocker felodipine are mTORC1 nondependent autophagy inducers and have been approved by the FDA.1136 The covalent acrylamide-based autophagy inducer EN6 indirectly induces mTORC1 inactivation and increases lysosomal acidification, leading to enhanced autophagic flow.1136 Additionally, disaccharide trehaloses SMER-28 and BRD5631 can induce autophagy in an mTORC1-independent manner.1141,1142 The natural product OSW-1 induces autophagy by blocking oxysterol-binding protein to inhibit cholesterol transport to lysosomes, thereby inhibiting mTORC1.1143,1144 Mucolipin 1 (TRPML1), an activator of the transient receptor potential cation channel, induces autophagosome biogenesis.1136,1145
The second category is the inhibitors of autophagy initiation. Autophagy inhibitors targeting PI3K and ULK1 can inhibit autophagy by preventing autophagosome formation. The pan-PI3K inhibitors 3-methyladenine and wortmannin are widely used in autophagy studies.1136 VPS34 is an important component of the class III PI3K complex. Autophinib and cinchona alkaloid-derived azaquindole-1 are both potent inhibitors of VPS34,1146,1147 and SAR405 and VPS34-IN1 are better selective VPS34 inhibitors.1136 ULK inhibitors include MRT68921 and SBI-0206965,1148,1149 which selectively inhibit ULK1 and ULK2. In addition, the inhibitor ULK-101 is a more selective and active ULK1 inhibitor, and is currently considered to be the most promising ULK1 inhibitory tool compound.1136 The cysteine protease autophagin-1 (ATG4B), a key protein for autophagosome formation and maturation, cleaves the C-terminus of LC3B and then binds to PE via ATG3. The development of ATG4B inhibitors is an effective way to inhibit autophagy. The styrrylquinoline-derived ATG4B inhibitor LV-320 has cysteine protease selectivity.1136,1150 The fluoromethylketone-based peptidomimetic FMK-9a, an irreversible covalent inhibitor of ATG4B, effectively inhibits ATG4B protein hydrolysis activity. In addition, altering the lipid composition of the nascent phagophore is also a new strategy for the development of autophagy inhibitors. The cholesterol transport protein GRAMD1A can effectively inhibit autophagosome biogenesis.1136
The third category is inhibitors of autophagosome maturation and lysosomal activity. The calcium channel TRPML1 is essential for autophagosome formation, and its small molecule inhibitor ML-SI3 can inhibit autophagic flow.1145,1151 In addition, inhibition of autophagosome-lysosome fusion by enhancing enhancement of 20S proteasome activity is an effective strategy to reduce autophagic flux.1152 The small molecule TCH-165 activates the 20S proteasome and specifically degrades important autophagosome-lysosome fusion regulators. Targeting autophagic terminal lysosomal activity is an effective method to inhibit autophagic flow.1136 These inhibitors inhibit lysosomal acidification by inhibiting v-ATPase or by directly increasing lysosomal pH and promoting lysosomal hydrolase inactivation. v-ATPase is a multisubunit proton pump responsible for maintaining low lysosomal pH. Natural products have been a rich source of highly potent v-ATPase inhibitors, including the macrolide antibiotics bafilomycin A1, concanamycin, benzolactone enamides salicylihalamide A, and lobatamide.1136,1153,1154 Lys01 is a tenfold more active autophagy inhibitor than hydroxychloroquine (HCQ), and its water-soluble salt Lys05 effectively promotes lysosomal deacidification and inhibits the proliferation of multiple tumor cell lines in vitro and the growth of tumor xenograft models in vivo.1155 Other lysosomal inhibitors, such as quinacrine, VATG-027, and VATG-032, also showed antitumor activity. VATG-027 is a potent inhibitor of autophagy with high cytotoxicity.1156 To date, the lysosomotropic agents chloroquine (CQ) and HCQ have been the main clinically applied autophagy inhibitors and are commonly used to alleviate acute and chronic inflammatory diseases, although a variety of autophagy inhibitors have been developed.1136,1157 They block the fusion of autophagosomes with lysosomal fusion to block organelle and protein degradation processes, thereby inhibiting nutrient recycling.460 CQ can enter the lysosome as a freely diffusing lysosomotropic agent and is deprotonated and trapped inside as a diacidic base in the lysosome.1156,1158 By sequestering the free hydrogen ions required to maintain an acidic pH, CQ increases the basicity of the lysosome, which renders pH-dependent lysosomal hydrolases and proteases, blocks lysosomal turnover, and inhibits the final stage of autophagy.1156 CQ and HCQ have been extensively studied as safety autophagy distributors for the treatment of various cancers, including breast cancer, melanoma, lung cancer, multiple myeloma, glioma, kidney cancer, prostate cancer, CRC, and other advanced solid tumors.1156,1159–1161
The fourth category is the degradation of the autophagosome and lysosome for targeted proteins. PROTACs have been applied to selectively target the degradation of autophagosomes or lysosomes and have promising applications.1136
In conclusion, there are various action mechanisms of autophagy inhibitors, which increase tumor chemotherapy drug sensitivity and inhibit tumor cell proliferation and metastasis.1159 Small molecule autophagy inhibitors remain excellent tools for autophagy-targeted therapy with the advantages of easy administration, rapid onset of action, and mostly reversible.1136 Given the nonautophagic targeted effects of most current autophagic targets1162 and the existence of mitochondrial autophagy pathways, the development of highly selective autophagy inhibitors is crucial.1136
Antiapoptotic therapy
Controlling cancer growth by promoting apoptosis is an effective antitumor strategy, and various apoptosis-based targeted therapies for tumors have been developed. Currently, the main focus on inhibitor development is targeting antiapoptotic family members. The inhibitors against BCL-2 are the most extensively studied, which include oligonucleotides that target BCL-2 expression, and proapoptotic BH3 mimetics that bind to antiapoptotic BCL-2 members (Table 5).463
Table 5.
The FDA-approved and clinically developed antiapoptotic inhibitors in cancer therapies
| Targets | Drug | Highest Phase | Indications | Company/Identifier | Status |
|---|---|---|---|---|---|
| BCL-2 | PNT-2258 | II | Non-hodgkin’s lymphoma | NCT01733238 | Completed |
| II | Diffuse large B-cell lymphoma | NCT02226965 | Completed | ||
| Dual BCL-2 and BCL-XL inhibitors | Navitoclax (ABT-263) | II |
Small cell lung cancer, Small cell lung carcinoma |
NCT00445198 | Completed |
| II | Chronic lymphocytic leukemia | NCT01557777 | Completed | ||
| II | Metastatic malignant solid neoplasm, recurrent lung small cell carcinoma | NCT03366103 | Terminated | ||
| II | Refractory acute lymphoblastic leukemia, relapsed acute lymphoblastic leukemia | NCT05192889 | Recruiting | ||
| II | Metastatic malignant solid neoplasm, refractory malignant solid neoplasm, unresectable malignant solid neoplasm | NCT02079740 | Active, not recruiting | ||
| II | Malignant solid neoplasm, melanoma | NCT01989585 | Active, not recruiting | ||
| II | Chronic lymphocytic leukemia | NCT01087151 | Completed | ||
| II | Platinum-resistant or Refractory ovarian cancer | NCT02591095 | Completed | ||
| I/II | Chronic lymphocytic leukemia | NCT00481091 | Completed | ||
| I/II | Chronic lymphoid leukemia, follicular lymphoma, lymphoid malignancies, mantle cell lymphoma, non-hodgkin’s lymphoma, peripheral T-cell lymphoma | NCT00406809 | Completed | ||
| II | Prostate cancer | NCT01828476 | Terminated | ||
| Palcitoclax (APG-1252) | II | Small cell lung cancer | NCT04210037 | Terminated | |
| ABT-737 | / | Ovarian cancer | NCT01440504 | Completed | |
| AZD0466 | II | Hematological malignancies | NCT04865419 | Recruiting | |
| Selective BCL-2 inhibitors | Venetoclax (ABT-199) | Approved | Chronic lymphocytic leukemia and acute myeloid leukemia | AbbVie Inc. and Genentech Inc | / |
| S55746 (S-055746, BCL201) | I | B-cell non-hodgkin lymphoma, chronic lymphocytic leukemia, multiple myeloma | NCT02920697 | Completed | |
| I | Follicular lymphoma, mantle cell lymphoma | NCT02603445 | Completed | ||
| I | Acute myeloid leukemia, myelodysplastic syndrome | NCT02920541 | Completed | ||
| Lisaftoclax (APG-2575) | II | Chronic lymphocytic leukemia, small lymphocytic lymphoma | NCT05147467 | Recruiting | |
| II | Multiple myeloma, amyloidosis | NCT04942067 | Recruiting | ||
| II | Breast cancer solid tumor, adult | NCT04946864 | Recruiting | ||
| II | Multiple myeloma | NCT04674514 | Recruiting | ||
| II | Chronic lymphocytic leukemia, small lymphocytic lymphoma | NCT04494503 | Recruiting | ||
| II | Relapsed/refractory acute myeloid leukemia, myeloid malignancy | NCT04501120 | Recruiting | ||
| I/II | Acute myeloid leukemia | NCT04964518 | Recruiting | ||
| BCL-XL inhibitors | ABBV-155 | I | Advanced solid tumors | NCT03595059 | Active, not recruiting |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov) and the United States Food and Drug Administration.gov (https://www.fda.gov/)
PNT-2258, a 24-base, single-stranded, chemically unmodified phosphodiester DNA oligonucleotide encapsulated in a specialized liposome, can target the regulatory region upstream of the BCL-2 gene.1163 Navitoclax (ABT-263) is the second-generation, potent, and orally bioavailable Bad-like BH3 mimetic with an oral bioavailability of 20 to 50% in preclinical models. ABT-263 disrupts the interaction of BCL-2/BC-XL with pro-death proteins and induces BAX translocation, cytochrome c release, and ultimately apoptosis.463,1164 Navitoclax inhibits growth in multiple preclinical tumor models1165 and is currently being evaluated in combination with the PARP inhibitor olaparib for the treatment of TNBC and ovarian cancer in a phase I clinical trial (NCT05358639). Palcitoclax (APG-1252) is a dual BCL-2 and BCL-XL inhibitor with safe and well-tolerated properties for treating patients with metastatic solid tumors.1166 ABT-737 is the first small molecule designed to selectively bind the hydrophobic pocket within BCL-2, BC-XL, and BCL-W, which is not bioavailable for oral administration. ABT-737 displaces BIM from BCL-2’s BH3-binding pocket, subsequently activates BAX and induces mitochondrial permeability, ultimately leading to apoptosis.463,475,1167 ABT-737 has antitumor activity against hematologic and solid tumors, including CLL, lymphoma, and SCLC.475,1168,1169 In addition, ABT-737 exerts synergistic cytotoxicity with chemotherapy and radiotherapy.475 AZD0466, a novel BH3-mimetic inhibitor targeting BCL-XL and BCL-2, has potent antitumor activity in preclinical models of malignant pleural mesothelioma.1170
Venetoclax (ABT-199), a selective inhibitor of BCL-2, has been approved by the FDA for the treatment of CLL and AML.1171 Venetoclax inhibits the proliferation of BCL-2 overexpressing small lymphocytic lymphoma.463 Venetoclax in combination with the FLT-3 TKIs gilteritinib or sorafenib synergistically inhibited FLT-3/ITD mutant AML proliferation and promoted apoptosis.1172 The toxic reactions of venetoclax include mild diarrhea (52%), upper respiratory tract infection (48%), nausea (47%), and grade 3 or 4 neutropenia (41%).1173 S55746 (S-055746, BCL201) is an orally selective BCL-2 inhibitor and can effectively impair hematological tumor growth.1174 Lisaftoclax (APG-2575), a selective oral BCL-2 inhibitor, demonstrates potent antitumor activity in preclinical models of hematologic malignancy.1175
ABBV-155 is a first-in-class selective BCL-XL inhibitor. ABBV-155 monotherapy or in combination with paclitaxel or docetaxel is currently in a phase I clinical trial in advanced solid tumors to assess its safety and preliminary activity (NCT03595059). In addition, as the BAX serine 184 regulatory site is responsible for subcellular localization and insertion into mitochondrial membranes, the agonists targeting BAX have been developed for cancer treatment. The small molecule BAX agonists SMBA1, SMBA2, and SMBA3 selectively bind to BAX and inhibit S184 phosphorylation, thereby promoting BAX insertion into mitochondrial membranes and the formation of BAX oligomers, and inducing conformational changes in BAX, ultimately leading to cytochrome c release and apoptosis.1176 Other BAX-activating compounds, such as BAM-7 and BTSA1, also have antitumor activity in glioblastoma and AML cells.463,1177 Meanwhile, some other preclinical inhibitors are also under investigation.463
Overall, apoptosis resistance is a hallmark of human cancers. The abnormal expression of antiapoptotic proteins and the downregulation or mutation of proapoptotic proteins promote the acquisition of apoptosis resistance in tumors.491 Tumor-targeted therapy inducing apoptosis is an effective approach to overcome apoptosis resistance and open up new directions for cancer treatment strategies.
Necroptosis-inducing anticancer agents
Various compounds and anticancer drugs with various mechanisms of action are capable of inducing necrosis in cancer cells, which include chemotherapeutic agents, natural compounds, and classical necrosis inducers.578,1178
Chemotherapeutic agents for necrosis have been widely developed in recent years. Necrostatin-1 is a small molecular alkaloid that was first considered an inhibitor of necrotic cell death in 20051179 and was subsequently identified as a specific inhibitor of RIPK1. Necrostatin-1 blocks RIPK1 kinase activity by interacting with an essential structure for the death domain receptor engagement T loop and blocking RIPK1 kinase activity.1180 In addition, necrostatin-1 analogs were also effective in inducing necrotic cell death.1181,1182 BI2536, a small molecule inhibitor of mitotic kinase polo-like kinase 1, induces necroptotic cell death in prostate cancer cells.1183
Moreover, natural compounds also hold an essential place in necrosis-based cancer therapy. Shikonin, a naturally occurring naphthoquinone, triggers necrotizing cell death, circumvents drug transporter proteins, and antiapoptotic BCL-2 protein-mediated apoptosis resistance.1184,1185 Shikonin inhibits glioma cells,1186 primary osteosarcoma, and pulmonary metastatic osteosarcoma1187 by inducing necrosis. Shikonin analogs such as deoxyshikonin, acetylshikonin, isobutyrylshikonin, beta-dimethylacrylshikonin, isovalerylshikonin, and alpha-methyl-n butylshikonin are able to induce necrosis, thus overcoming tumor resistances which are mediated by the resistance factors such as P-gp, BCL-2 and BCL-XL, MRP1, and BCRP1.1188 Obatoclax (GX15-070) is an indole bipyrrole compound antagonizing BCL-2, BCL-XL, BCL-W, and MCL1, which triggers necrotizing cell death by promoting necrosomes on autophagosomal membranes. Obatoclax induces nonapoptotic forms of cell death in rhabdomyosarcoma cells.1189 In addition, polyphenon E(R), a natural product of green tea extract, induces endoplasmic reticulum stress and causes necrotic death of prostate cancer cells.1190 Staurosporines, isolated from the bacterium Streptomyces staurosporeus in 1977, are protein kinase inducers of the intrinsic apoptotic pathway and trigger necrosis in the leukemia cell line U937 when cystatin proteases are impaired.1191,1192 FTY720, a sphingolipid analog that mimics ceramide, induces necroptotic cell death by modulating lipid signaling in glioblastoma cells.1193,1194 5′-Benzylglycinyl-amiloride (UCD38B) induces mitochondrial swelling, endoplasmic reticulum expansion, and nuclear condensation, and induces a nonapoptotic form of cell necrosis in glioma cells.1195
In addition to the above observations, other cell necrosis inducers are found to induce necrotic cell death via various mechanisms, such as selenosemicarbazone metal complexes,1196 protein disulfide isomerase PDIA6,1197 death receptor ligand TRAIL,1198 and mitochondrial activator of caspases mimetics (Smac).1184 Moreover, necrocytes can initiate adaptive immunity and recruit macrophages through activation of NF-κB, thereby activating immune cells and enhancing immunotherapy efficacy. The combined use of checkpoint inhibitors and necrocyte vaccines significantly improves the clinical outcomes of cancer patients.566,1184
Targeting invasion and metastasis
There are still limitations and tough challenges in the targeted treatment of tumor metastasis. Tumor cells have already spread to the blood, bone marrow, and distant organs by the time some cancer patients are first diagnosed with tumors.165,1199 Therefore, antimetastatic therapies need to take into account not only cells that metastasize from the primary tumor site, but also the inhibition of cancer cells that have already spread. Currently, strategies for preventing metastasis have been demonstrated preclinically (Fig. 5). However, drug development has been hindered due to poor trial design and therapeutic strategies.600 Encouragingly, potent and specific MMP inhibitors are being developed that may further improve efficacy and attenuate toxicity.1200 In addition, targeted HGF/c-MET inhibitors are bringing light to tumor treatment.677 In conclusion, more than 90% of cancer mortality is now attributed to metastasis, and the prospect of targeted tumor metastasis therapy is unlimited.165
MMP inhibitors
Since cancer cells require MMPs to degrade collagen to promote cell metastasis, the first generation of MMP inhibitors is structural analogs of collagen, among which the first compound belongs to the hydroxamic acid zinc-binding group.715,1201 Developed by British Biotech, batimastat (BB-94) is a potent pan-MMP inhibitor that acts by chelating zinc ions at the active site of each MMP enzyme. As the first anti-MMP drug to be tested in clinical trials,604 batimastat effectively inhibits breast cancer, ovarian cancer, and CRC tumor growth and metastasis in vivo, although its poor solubility in water hampers its further development.604,1202,1203 The subsequent development of marimastat (BB-2516) is an orally available second-generation synthetic MMP inhibitor.604 Marimasta has been tested in phase III clinical trials to evaluate its effectiveness in patients with SCLC (NCT00003011), stage III NSCLC (NCT00002911), and metastatic breast cancer (NCT00003010), but no results have been disclosed. In addition, the bryostatins are naturally occurring macrocyclic actone products that inhibit MMPs by modulating upstream regulators of MMPs.604 Tanomastat (BAY12-9566), a bryostatin compound developed by Bayer, is a nonpeptide biphenyl MMP inhibitor that is effective against a wide range of tumors.1204,1205 However, a phase III randomized trial of tanomastat as maintenance therapy in patients with advanced ovarian cancer responsive to primary surgery and paclitaxel/platinum-containing chemotherapy has shown that tanomastat is generally well tolerated but had no impact on PFS or OS.1206 Other MMPs inhibitors such as COL-3 (NSC-683551),1207 neovastat (AE-941),1208 prinomastat (AG3340),1209 BMS-275291,1210 and metastat(COL-3),1211 have been evaluated in clinical trials (Table 6).
Table 6.
The typical and clinically developed MMPs inhibitors in cancer therapy
| Drug | Highest phase | Indications | Identifier | Status |
|---|---|---|---|---|
| COL-3 (NSC-683551) | I | Lymphoma, melanoma, neoplasm metastasis, renal cell carcinoma | NCT00001683 | Completed |
| I | Unspecified adult solid tumor, protocol specific | NCT00003721 | Completed | |
| Neovastat (AE-941) | III | Kidney cancer | NCT00005995 | Completed |
| III | Adenocarcinoma of the lung, adenosquamous cell lung cancer, large-cell lung cancer, squamous cell lung cancer, stage IIIA non-small cell lung cancer, stage IIIB non-small cell lung cancer | NCT00005838 | Completed | |
| Prinomastat (AG3340) | III | Lung cancer | NCT00004199 | Completed |
| III | Prostate cancer | NCT00003343 | Completed | |
| II | Brain and central nervous system tumors | NCT00004200 | Completed | |
| Marimastat (BB-2516) | III | Lung cancer | NCT00003011 | Completed |
| III | Lung cancer | NCT00002911 | Completed | |
| III | Breast cancer | NCT00003010 | Completed | |
| BMS-275291 | II/III | Lung cancer | NCT00006229 | Completed |
| Metastat(COL-3) | I/II | Brain and central nervous system tumors | NCT00004147 | Completed |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov)
In conclusion, MMPs are a large family with different functions in tumor cells.1212 Depending on cell and tissue localization, disease type, etc., MMPs can be used as both drug targets and anti-targets.715 Therefore, given the cell or tumor specificity of MMPs, it is important to explore in-depth the contribution of MMPs in tumor progression and metastasis in various tumor types, which will facilitate the rational development of specific MMP inhibitors.604
HGF/c-Met inhibitors
There is an increasing number of studies on HGF/c-MET-targeted therapies, which mainly include anti-HGF/c-MET mAbs and small molecule inhibitors targeting the structural domain of c-MET kinase (Fig. 5 and Table 7). The mechanism of action of HGF/c-MET inhibitors is mainly neutralization or competition with HGF to inhibit receptor dimerization or induce c-MET degradation.677
Table 7.
The clinically developed HGF/c-MET inhibitors
| Type | Drug | Targets | Highest phase | Indications | Identifier | Status |
|---|---|---|---|---|---|---|
| Anti-c-MET mAbs | Emibetuzumab(LY2875358) | c-MET | II | Carcinoma, non-small-cell lung | NCT01900652 | Completed |
| II | Advanced cancer, gastric adenocarcinoma, gastroesophageal junction adenocarcinoma, hepatocellular cancer, renal cell carcinoma, non-small cell lung cancer | NCT02082210 | Completed | |||
| ABT-700 | c-MET | I | Advanced solid tumors | NCT01472016 | Completed | |
| Onartuzumab | c-MET | III | Solid tumor | NCT02488330 | Completed | |
| III | Non-squamous non-small cell lung cancer | NCT01456325 | Completed | |||
| III | Gastric cancer | NCT01662869 | Completed | |||
| III | Non-squamous non-small cell lung cancer | NCT01887886 | Completed | |||
| III | Non-small cell lung cancer | NCT02031744 | Completed | |||
| LY2875358 | c-MET | II | Gastric cancer | NCT01874938 | Completed | |
| II | Non-small cell lung cancer | NCT01897480 | Active, not recruiting | |||
| II | Non-small cell lung cancer | NCT01900652 | Completed | |||
| I/II | Advanced cancer, gastric adenocarcinoma, gastroesophageal junction adenocarcinoma, hepatocellular cancer, renal cell carcinoma, non-small cell lung cancer | NCT02082210 | Completed | |||
| Anti-HGF mAbs | Rilotumumab | HGF | III | Gastric cancer | NCT02137343 | Terminated |
| III | Gastric cancer | NCT01697072 | Terminated | |||
| III | Recurrent squamous cell lung carcinoma, stage IV squamous cell lung carcinoma | NCT02926638 | Terminated | |||
| II | Recurrent fallopian tube carcinoma, recurrent ovarian carcinoma, recurrent primary peritoneal carcinoma | NCT01039207 | Completed | |||
| Ficlatuzumab | HGF | II | Resistant, recurrent or metastatic head/neck squamous cell carcinoma | NCT03422536 | Completed | |
| II | Non-small cell lung cancer | NCT02318368 | Terminated | |||
| YYB-101 | HGF | Ib/IIa | Colorectal cancer | NCT04368507 | Completed | |
| Nonselective TKI (ATP-competitive) | Capmatinib | c-MET | IV | Non-small cell lung carcinoma | NCT05110196 | Recruiting |
| Tepotinib | c-MET | II | Non-small cell lung cancer | NCT03940703 | Active, not recruiting | |
| II | Solid tumor, MET exon 14 skipping mutation, MET amplification | NCT04647838 | Recruiting | |||
| II | Advanced (Stage IIIB/IV) non-small cell lung cancer with MET exon 14 skipping alterations or MET amplification lung adenocarcinoma stage IIIB/IV | NCT02864992 | Active, not recruiting | |||
| II | Colorectal neoplasms | NCT04515394 | Terminated | |||
| II | Recurrent lung non-small cell carcinoma, stage IV lung cancer | NCT06031688 | Not yet recruiting | |||
| II | Gastric cancer, gastroesophageal-junction cancer | NCT05439993 | Recruiting | |||
| I/II | Advanced non-small cell lung cancer with MET mutations | NCT04739358 | Recruiting | |||
| I/II | Non-small cell lung cancer | NCT01982955 | Completed | |||
| I/II | Hepatocellular carcinoma | NCT02115373 | Completed | |||
| I/II | Hepatocellular carcinoma | NCT01988493 | Completed | |||
| II |
Breast cancer, gastrointestinal cancer, non-small cell lung cancer, other cancer |
NCT04591431 | Active, not recruiting | |||
| AMG-337 | c-MET | II | Stomach neoplasms | NCT02016534 | Terminated | |
| II | Solid tumor | NCT03147976 | Withdrawn | |||
| II | Clear cell sarcoma | NCT03132155 | Terminated | |||
| I/II | Stomach neoplasms | NCT02096666 | Completed | |||
| Nonselective TKI (allosteric) | Tivantinib | c-MET | III | Hepatocellular carcinoma | NCT01755767 | Completed |
| III | Non-squamous, non-small cell lung cancer | NCT01244191 | Terminated | |||
| III | Non-small cell lung cancer | NCT01377376 | Terminated | |||
| III | Liver cancer | NCT02029157 | Completed | |||
| Nonselective TKI (ATP-competitive) | Crizotinib | c-MET, ALK, RON, AXL, TIE2, ROS1 | IV | Non-small cell lung cancer, anaplastic large-cell lymphoma, inflammatory myofibroblastic tumor | NCT05160922 | Recruiting |
| IV | Anaplastic lymphoma kinase or ROS1-positive non-small cell lung cancer | NCT03672643 | Active, not recruiting | |||
| IV | Systemic anaplastic large-cell lymphoma | NCT02487316 | Withdrawn | |||
| Cabozantinib | c-MET, c-RET, VEGFR1-3, c-Kit, FLT-3, TIE2, TRKB, AXL | IV | Hepatocellular carcinoma | NCT03963206 | Completed | |
| IV | Medullary thyroid cancer | NCT01896479 | Active, not recruiting | |||
| Foretinib | c-MET, VEGFR2, RON, ERK, AKT, PDGFRβ, c-Kit, TIE2 | II | Recurrent breast cancer | NCT01147484 | Completed | |
| II | Neoplasms, head and neck | NCT00725764 | Completed | |||
| II | Carcinoma, renal cell | NCT00726323 | Completed | |||
| II | Neoplasms, gastrointestinal tract | NCT00725712 | Completed | |||
| I/II | Breast cancer | NCT01138384 | Completed | |||
| I/II | Lung cancer | NCT01068587 | Completed | |||
| Glesatinib | c-MET, AXL | II | Non-small cell lung cancer | NCT02954991 | Completed | |
| Golvatinib | c-MET, VEGFR2, RON, Eph, c-Kit | I/II | Advanced solid tumors | NCT01433991 | Terminated | |
| I/II | Platinum-resistant squamous cell carcinoma of the head and neck | NCT01332266 | Completed | |||
| Merestinib | c-MET, MST1R, FLT-3, AXL, MERTK, TEK, ROS1, NTRK1/2/3, DDR1/2, MKNK1/2 | II | Carcinoma, non-small-cell lung, solid tumor | NCT02920996 | Active, not recruiting | |
| II | Biliary tract cancer, metastatic cancer, advanced cancer | NCT02711553 | Active, not recruiting |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov)
c-MET inhibitors
RTK is a cell surface receptor that binds to growth factor ligands such as HGF, VEGF, EGF, etc., and activates downstream signaling pathways. A number of small molecule TKI have been developed that selectively and nonselectively inhibit the catalytic activity of c-MET.1213 Significantly, two c-MET inhibitors, tepotinib developed by EMD Serono, and capmatinib developed by Novartis, have been approved by the FDA. Tepotinib inhibits MET kinase activity with an IC50 value of 1.7 nM and showed high selectivity for MET in screening against >400 kinases.1214 On February 3, 2021, the FDA granted accelerated approval to tepotinib for adult patients with metastatic NSCLC harboring MET exon 14 skipping alterations.1215 Capmatinib, an ATP-competitive, highly potent (IC50 value of 0.13 nM) and selective MET inhibitor, was granted regular approval by the FDA for the treatment of adult patients with metastatic NSCLC whose tumors have MET exon 14 skipping mutations in May 2020.1216 In addition, crizotinib (PF-02341066) is an orally bioavailable TKI that competitively inhibits the ATP-binding site of tyrosine kinases and inhibits c-MET, ALK, AXL and TIE2 activity.679 Crizotinib effectively inhibits c-MET phosphorylation and c-MET-dependent proliferation, migration, or invasion of tumor cells.679,1217 Notably, crizotinib has been approved for patients with ALK-positive and ROS1-positive metastatic NSCLC.677 Cabozantinib (XL184) is an orally available TKI that targets a variety of kinases including MET, VEGFR2, RET, FLT-3, and KIT.1218 It has shown efficacy in patients with prostate cancer1219 and advanced RCC.1220,1221 Foretinib (GSK1363089) is an oral and potent TKI targeting c-MET and VEGFR2. It binds to the ATP-binding pocket of the above kinases and makes kinase conformational changes. Foretinib has been tested in clinical trials in a variety of tumors including papillary renal cancer, gastric cancer, and head and neck cancer.1222 A phase II study of foretinib in adults with HNSCC revealed that 50.0% of participants have stable disease, and 21.4% have progressive disease (NCT00725764). Other phase II clinical trials of foretinib in solid tumors (NCT00742131) and breast cancer (NCT01138384) have been completed, but no results have been posted yet. Moreover, a phase II study of foretinib in adults with gastric cancer demonstrated that the serious adverse events of foretinib include abdominal pain (6.25%), dehydration (6.25%), malignant neoplasm progression (4.17%)(NCT00725712). Tivantinib (ARQ197) is a highly selective, non-ATP competitive inhibitor of c-MET.1223 Tivantinib inhibits the autophosphorylation of c-MET in many cancer cells and is highly selective for inactive or non-phosphorylated forms of c-MET, thus effectively blocking the activation of c-MET downstream effectors such as RAS, MAPK, and STAT3, ultimately inhibiting tumor proliferation, invasion and metastasis.1222,1224 Other nonselective c-MET inhibitors include glesatinib (MGCD-265), golvatinib (E-7050) and merestinib (LY-2801653), while selective c-MET inhibitors include tepatinib (EMD-1214063), AMG-337 and capatinib (INC-280), which are approved for clinical use or undergoing clinical study.
Anti-c-MET antibodies
Onartuzumab, an Escherichia coli-derived, humanized mAb against c-MET, can block the high-affinity binding of HGFα chain but not HGFβ chain to c-MET.1225 Preclinical studies found that onartuzumab effectively inhibits human glioblastoma and pancreatic cancer cell growth although further development of onartuzumab has been halted.1226 Emibetuzumab (LY2875358) is a humanized anti-c-MET bivalent antibody that effectively promotes internalization and degradation of c-MET, thereby blocking HGF-c-MET binding and HGF-induced c-MET phosphorylation.1227 Emibetuzumab combined with erlotinib is in a phase II clinical trial to evaluate their efficiency as a first-line treatment for patients with metastatic NSCLC with activated EGFR mutations (NCT01897480). LY3164530 is a bispecific anti-EGFR/c-MET antibody produced by fusing an anti-EGFR single-chain variable fragment to the N-terminal end of the emibetuzumab heavy chain.1226 LY3164530 disrupts signaling by binding and internalizing c-MET and EGFR.1226 Its phase I clinical trial in patients with advanced or metastatic cancer has been completed. However, significant toxicities associated with EGFR inhibition and the lack of a potential predictive biomarker limit its future development.1228 Amivantamab (JNJ-61186372) is a bispecific EGFR/c-MET antibody that binds the extracellular structural domain of each receptor, thereby avoiding resistance at the TKI binding site.1229 Amivantamab effectively inhibits tumors in various contexts, including tumors with T790M second-site resistance mutation in EGFR, c-MET pathway activation,1230 and EGFR exon 20 insertion driver mutations.1229 Amivantamab is currently being administered in monotherapy or combination with various drugs for cancer treatment in 15 clinical trials (NCT04077463, NCT02609776, NCT05845671, and NCT05653427). Encouragingly, amivantamab has been approved for marketing for the treatment of patients with metastatic NSCLC with EGFR exon 20 insertion mutations and platinum-based chemotherapy resistance (NCT04599712).
SAIT301 is a monoclonal humanized antibody developed by Samsung that promotes c-MET degradation.1226,1231 SAIT301 inhibits nasopharyngeal cell invasion and migration by downregulating EGR-1 via the degradation of c-MET.1232 Its clinical phase I trial in c-MET-overexpressed metastatic CRC has been completed, and the most common adverse effects were decreased appetite (50.0%), hypophosphatemia, fatigue, and dizziness (25.0%), diarrhea, and dyspnea (18.8%).1233 ABT-700 (h224G11) is a humanized bivalent mAb that inhibits c-MET dimerization and activation. A phase I clinical trial of ABT-700 in subjects with advanced solid tumors containing MET amplification or c-MET overexpression (NCT01472016) has been completed. ARGX-111 is an afucosylated IgG1 antibody that competitively binds c-MET, inhibits c-MET activity, and downregulates c-MET expression on the cell surface.1234 A clinical phase I trial of ARGX-111 in patients with advanced cancer overexpressing c-MET has been completed (NCT02055066). In addition. DN30 is a monovalent chimeric Fab that induces the cleavage of the extracellular portion of c-MET, leading to the shedding of its ectodomain.1235,1236 DN30 inhibits tumor growth in human gastric cancer, lung cancer, and glioblastoma.1226
Anti- HGF antibodies
Rilotumumab (AMG-102), an anti-HGF mAb binding to HGFβ chain structural domain, specifically blocks the activation of c-MET.1237 In particular, rilotumumab selectively alters the mature HGF, but shows no effect on the proteolytic activation process of pro-HGF.1238 To date, rilotumumab alone or in combination with other anticancer drugs, such as antiangiogenic agents, EGFR inhibitors, and chemotherapeutic agents, has been studied in clinical trials in patients with various solid tumors such as prostate cancer, kidney cancer, and advanced NSCLC.677,1239 Ficlatuzumab (AV-299), a humanized anti-HGF antibody, has been studied in clinical trials as a monotherapy or in combination with chemotherapeutic agents in patients with advanced pancreatic cancer (NCT03316599) and HNSCC (NCT02277197). YYB-101 is a humanized HGF antibody which inhibits c-MET activation by binding to the HGFα chain.1226 A clinical phase I trial of YYB-101 in patients with refractory advanced solid tumors (NCT02499224) has shown that YYB-101 exhibited favorable safety and efficacy in patients with refractory solid tumors. A clinical phase Ib/ IIa trial of YYB10 in combination with irinotecan in patients with metastatic or recurrent CRC (NCT04368507) has been completed, but no results are currently available.
Antibody mimetic engineered protein against HGF
MP0250, an ankyrin repeat protein capable of neutralizing VEGF and HGF, effectively inhibits multiple myeloma-mediated osteolysis and myeloma cell invasion.1226 Meanwhile, MP0250 can effectively improve bortezomib efficacy without increasing toxicity, suggesting that MP0250 combined with cytotoxic therapy may be a promising therapeutic approach.1240 A phase II clinical evaluation of MP0250 in combination with bortezomib and dexamethasone in patients with multiple myeloma (NCT03136653) has been completed, although the result has not yet been disclosed.
Competitive analogs of HGF
NK4 is a synthetic intramolecular fragment of HGF, originally purified as a fragment from elastase-digested HGF samples.1241 NK4 contains the HGF α-chain N-terminal hairpin domain and 4 kringle domains (K1–K4),1242 and lacks the 16 amino acids of the HGF C-terminus.1226 NK4 inhibits c-MET phosphorylation and activation by competing with HGF for binding to MET. NK4 effectively inhibits neovascularization, growth, invasion, and metastasis of many tumor cells.1243,1244
The HGF/c-MET pathway serves a critical role in cancer and is an attractive therapeutic target for cancer therapy. Over the past decade, great efforts have been devoted to the development of selectively c-MET inhibitors. Although small molecule c-MET inhibitors and antibody-based drugs have shown meaningful clinical efficacy, the challenges of resistance and side effects remain to be addressed. The c-MET amplification and overexpression, c-MET mutations, the activation of parallel signaling pathways, and the induction of HGF secretion are associated with acquired resistance after initial response to HGF/c-MET inhibitors. Therefore, how to overcome the acquired resistance as well as improve the safety of c-MET inhibitors needs to be solved urgently.1245 Moreover, stratifying patients appropriately based on the discovery of biomarkers may help identify the subgroups of patients who can benefit from anti- HGF/c-MET therapy.
Targeting DDR pathways
Multiple DDRs related small molecule inhibitors have been approved for clinical use or are under clinical investigation, including PARP inhibitors, ATM inhibitors, ATR inhibitors, and CHK1 inhibitors (Table 8). Herein we mainly focus on the inhibitors of PARP, ATM, ATR, and CHK1.
Table 8.
The clinically developed DDR inhibitors in cancer therapies
| Type | Drug | Highest phase | Indications | Identifier | Status |
|---|---|---|---|---|---|
| ATM inhibitor | AZD0156 | I | Advanced solid tumors | NCT02588105 | Completed |
| AZD1390 | I | Recurrent glioblastoma multiforme, primary glioblastoma multiforme, brain neoplasms, malignant, leptomeningeal disease | NCT03423628 | Recruiting | |
| I | Glioblastoma, glioma, glioblastoma multiforme, glioma, malignant | NCT05182905 | Recruiting | ||
| I | Solid tumor, metastatic solid tumor, solid carcinoma, solid tumor, adult, metastatic tumor, metastatic cancer | NCT05678010 | Recruiting | ||
| I | soft tissue sarcoma adult | NCT05116254 | Recruiting | ||
| I | non-small cell lung cancer | NCT04550104 | Recruiting | ||
| M4076 | I | Advanced solid tumors | NCT04882917 | Completed | |
| I | Metastatic or locally advanced unresectable solid tumors | NCT05396833 | Recruiting | ||
| ATM and DNA-PKcs inhibitor | XRD-0394 | I | Metastasis, locally advanced solid tumor, recurrent cancer | NCT05002140 | Active, not recruiting |
| ATR inhibitor | ART0380 | II | Advanced solid tumor, recurrent endometrial cancer, metastatic cancer | NCT05798611 | Recruiting |
| II | Advanced cancer, metastatic cancer, ovarian cancer, primary peritoneal cancer, fallopian tube cancer | NCT04657068 | Recruiting | ||
| ATRN-119 | II | Advanced solid tumor | NCT04905914 | Recruiting | |
| BAY1895344 | I | Advanced solid tumor | NCT04095273 | Completed | |
| I | Advanced solid tumor, non-hodgkin’s lymphoma, mantle cell lymphoma | NCT03188965 | Recruiting | ||
| I | Advanced solid tumors (excluding prostate cancer), ovarian cancer | NCT04267939 | Recruiting | ||
| Berzosertib (VX-970, M6620, VE-822) | II | Small cell lung cancer | NCT04768296 | Completed | |
| II | Small cell lung cancer, small cell cancer, advanced solid tumor, high grade neuroendocrine cancers | NCT04802174 | Recruiting | ||
| II | small cell lung cancer, advanced solid tumors | NCT04826341 | Recruiting | ||
| II | Lung non-small cell squamous carcinoma, stage IV lung cancer | NCT04216316 | Recruiting | ||
| II | Leiomyosarcoma, adult | NCT04807816 | Recruiting | ||
| I | Advanced solid tumor | NCT05246111 | Completed | ||
| II | Castration-resistant prostate carcinoma, metastatic prostate carcinoma, stage IV prostate cancer | NCT03517969 | Active, not recruiting | ||
| II | Ovarian serous tumor, recurrent fallopian tube carcinoma, recurrent ovarian carcinoma, recurrent primary peritoneal carcinoma | NCT02595892 | Active, not recruiting | ||
| II | Bladder small cell neuroendocrine carcinoma, extensive stage lung small cell carcinoma, extrapulmonary small cell neuroendocrine carcinoma, limited stage lung small cell carcinoma, platinum-resistant lung small cell carcinoma, platinum-sensitive lung small cell carcinoma, prostate small cell neuroendocrine carcinoma, recurrent lung small cell carcinoma | NCT03896503 | Active, not recruiting | ||
| II | Metastatic malignant solid neoplasm, refractory malignant solid neoplasm, unresectable malignant solid neoplasm | NCT04266912 | Active, not recruiting | ||
| II | Metastatic bladder urothelial carcinoma, metastatic renal pelvis and ureter urothelial carcinoma, metastatic ureter urothelial carcinoma, stage IV bladder urothelial carcinoma | NCT02567409 | Active, not recruiting | ||
| Ceralasertib (AZD6738) | III | Advanced or metastatic non-small cell lung cancer | NCT05450692 | Recruiting | |
| IMP9064 | I | Solid tumor, advanced solid tumor | NCT05269316 | Recruiting | |
| M4344 | II | Ovarian cancer recurrent | NCT04149145 | Withdrawn (never opened) | |
| I | Solid tumor, advanced solid tumor | NCT02278250 | Completed | ||
| RP-3500 | II | Advanced solid tumor, adult | NCT04972110 | Recruiting | |
| II | Solid tumor, metastatic cancer | NCT05566574 | Recruiting | ||
| I | Advanced solid tumor | NCT04855656 | Recruiting | ||
| II | Advanced solid tumor | NCT04497116 | Recruiting | ||
| CHK1 inhibitor | Prexasertib (LY2606368) | II | Small cell lung cancer | NCT02735980 | Completed |
| II | Ovarian cancer | NCT03414047 | Completed | ||
| II | Triple-negative breast cancer | NCT04032080 | Completed | ||
| II | Advanced cancers | NCT02873975 | Completed | ||
| II | Desmoplastic small round cell tumor, rhabdomyosarcoma | NCT04095221 | Active, not recruiting | ||
| II | Platinum-resistant ovarian cancer, endometrial adenocarcinoma, urothelial carcinoma | NCT05548296 | Recruiting | ||
| SRA737 | II | Advanced solid tumors or non-hodgkin’s lymphoma | NCT02797964 | Completed | |
| II | Advanced solid tumors | NCT02797977 | Completed | ||
| LY2880070 | II | Ewing sarcoma, Ewing-like sarcoma | NCT05275426 | Recruiting | |
| II | Solid tumors, colorectal cancer, breast cancer, ovarian cancer, colon cancer, rectal cancer, neoplasms, endometrial cancer, soft tissue sarcoma, triple-negative breast cancer, pancreatic cancer | NCT02632448 | Recruiting | ||
| II | Castration-resistant prostate carcinoma, metastatic malignant neoplasm in the lymph nodes, metastatic prostate carcinoma, stage IV prostate cancer | NCT04071236 | Recruiting | ||
| II | Locally advanced rectal cancer | NCT03770689 | Completed | ||
| II | Cholangiocarcinoma, gallbladder carcinoma, malignant solid neoplasm | NCT04068194 | Suspended |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov)
PARP inhibitors
PARP inhibitors are selective for targeting tumors deficient in the HR DNA repair factor BRCA1 or BRCA2 (BRCA1/2)741 or compromised HR.733 Six PARP inhibitors are currently approved for the clinical treatment of cancer patients including the specific subgroups with BRCA1/2 mutation: olaparib, rucaparib, niraparib, talazoparib, fuzuloparib, and pamiparib733 (Table 9). Olaparib is the first PARP inhibitor introduced into clinical practice.1246 In 2014, olaparib was approved for the treatment of patients with BRCA1/2-mutated metastatic ovarian cancer who had received three or more prior lines of chemotherapy. Subsequently, in 2016, rucaparib, a second PARP inhibitor, was authorized for the treatment of patients with advanced-stage ovarian cancers harboring deleterious BRCA1/2 mutations who had received two or more prior lines of chemotherapy. In 2019, niraparib was approved for the treatment of patients with HR-deficient advanced-stage ovarian cancers who had received three or more prior chemotherapy regimens.1246 Recently, in March 2022, olaparib was approved by the FDA for the adjuvant treatment of patients with hereditary BRCA1/2 mutations and HER2- high-risk early breast cancer1247 as well as for the maintenance treatment of patients with BRCA1/2-mutated ovarian cancers who are in complete or partial remission after platinum-based chemotherapy.1248 Moreover, PARP inhibitors are also used in patients with germline or somatic BRCA1/2-mutated ovarian cancer as maintenance therapy (olaparib)1249 or post-chemotherapy therapy (olaparib and rucaparib).1250 The FDA has approved olaparib and talazoparib for the treatment of advanced or metastatic HER2- breast cancer patients carrying deleterious germline BRCA1/2 mutations.1251,1252 Olaparib is also used for maintenance therapy in patients with germline BRCA1/2-mutated metastatic pancreatic cancer.1253 Meanwhile, rucaparib has been applied for second-line treatment of patients with metastatic castration-resistant prostate cancer with germline or somatic BRCA1/2 mutations.1254 Significantly, PARP inhibitors have been approved as first-line systemic therapies for patients with ovarian cancer.1246 However, acquired resistance to PARP inhibition is still an urgent question, which usually results from three types of mechanisms: drug target-related effects including the upregulation of drug efflux pumps or mutations of PARP or functionally related proteins; restoration of BRCA1/2 function leading to restoration of HR; or loss of DNA end-protection and/or restoration of replication fork stability.1246 Therefore, targeted strategies to overcome resistance to PARP inhibitors remain to be explored, and the identification of vulnerabilities of PARP inhibitor-resistant tumors is still challenging. Illustrating the properties of HR-deficient cancers will rationalize the treatment strategies to overcome resistance and improve the survival of patients.1246
Table 9.
The FDA-approved PARP inhibitors in cancer therapies
| Drug | Company | Indications | FDA approvals |
|---|---|---|---|
| Olaparib (Lynparza) | AstraZeneca | Ovarian (2014) | Olaparib capsules in patients with BRCA1/2 mutant advanced-stage ovarian cancers who have received ≥3 types of chemotherapies |
| Ovarian (2017) | Maintenance therapy for advanced -ovarian cancer patients with PR or CR to platinum-based chemotherapy | ||
| Ovarian (2018) | First-line maintenance therapy for patients with BRCA1/2 mutant advanced-stage ovarian cancers | ||
| Breast (2018) | Patients with BRCA1/2 mutant HER2-negative metastatic breast cancer who have been treated with chemotherapy | ||
| Breast (2022) | Patients with BRCA1/2 mutant HER2-negative high-risk early breast cancer who have been treated with adjuvant chemotherapy | ||
| Pancreatic (2019) | Adult patients with germline BRCA-mutated metastatic pancreatic adenocarcinoma | ||
| Prostate (2020) | Adult patients with HRR gene mutated metastatic castration-resistant prostate cancer | ||
| Rucaparib (Rubraca) | Clovis Oncology | Ovarian (2016) | Patients with BRCA1/2-mutant ovarian cancer refractory to ≥ prior lines of treatment |
| Ovarian (2018) | Maintenance treatment of patients with recurrent ovarian cancer | ||
| Prostate (2020) | BRCA-mutated metastatic castration-resistant prostate cancer | ||
| Niraparib | Tesaro | Ovarian (2019) | Patients with HR deficiency -positive, advanced ovarian cancer |
| Ovarian (2020) | First-line maintenance treatment of patients with advanced ovarian cancer | ||
| Talazoparib | Pfizer | Breast (2018) | Patients with germline BRCA-mutated, HER2-negative locally advanced or metastatic breast cancer |
Source: All the information is derived from the United States Food and Drug Administration.gov (https://www.fda.gov/)
ATM inhibitors
ATM is the apical DDR kinase that coordinates DSB repair, and a variety of compounds have been developed for selective inhibition of ATM.733 AZD0156 is a potent, selective, and orally active inhibitor of ATM,1255 and a phase I clinical trial of AZD0156 (NCT02588105) for safety and preliminary efficacy in advanced solid tumors has been completed, but no results have been posted. AZD1390, belonging to the same potent series as AZD0156, is an exquisitely potent, highly selective, and orally bioavailable ATM inhibitor.1256 AZD1390 effectively sensitizes the brain metastasis of breast cancers with DDR mutation to radiation therapy.1257 Multiple clinical trials of AZD1390 for cancer treatment are ongoing. M4076, an ATP-competitive ATM inhibitor with an IC50 value <1 nM, inhibits tumor cell growth by blocking DSB repair and enhances the sensitivity of tumor cells to radiation therapy both in vitro and in vivo.1258,1259 A phase I clinical trial of M4076 in advanced solid tumors is active (NCT04882917). The dual ATM and DNA-PK inhibitor XRD-0394 is a novel, potent, and orally active dual inhibitor.733 A phase I clinical trial of XRD-0394 for the treatment of metastatic locally advanced solid tumors and recurrent cancer is recruiting, but no data are yet publicly available (NCT05002140).
Given the important role of ATM in DSB signaling and repair, ATM inhibition combination therapy is currently an attractive strategy for cancer therapy in various clinical trials. ATM inhibitors enhance the anticancer activity of DNA damage agents such as topoisomerase inhibitors1260 and PARP inhibitor.1261 Collectively, ATM-targeted therapy has a promising potential in cancer therapy.
ATR inhibitors
ATR kinase maintains accurate DNA replication by regulating the DNA replication initiation and the process of replication forks, supporting that ATR is an important target for cancer therapy. To date, several ATR inhibitors have been developed.733,1262 Ceralasertib (AZD6738) is a selective and potent ATR inhibitor with good solubility, bioavailability, and pharmacokinetic properties.1263 Phase II clinical trials of ceralasertib in patients with osteosarcoma (NCT04417062) and advanced solid tumors (NCT04564027) are undergoing. Berzosertib (VX-970) is a highly potent, selective, and intravenous ATR inhibitor with an IC50 value of 19 nM.1264 A phase II clinical trial of berzosertib in patients with NSCLC (NCT04216316) is ongoing. BAY1895344, another novel potent and selective ATR inhibitor, exhibits strong monotherapy efficacy in cancers, and synergistic activity in combination with DNA damage therapies.1265 A phase II clinical trial of BAY1895344 in patients with recurrent solid tumors (NCT05071209) is in progress. Other ATR inhibitors, such as ART0380, ATRN-119, IMP9064, M4344, and RP-3500, are also in clinical trials. Studies also have found synergistic antitumor effects of ATR inhibitors with immunotherapy and other anticancer drugs.733 ATR inhibitors in combination with PARP inhibitors are used in the treatment of tumors with BRCA1/2 mutations.733,1266 Exploiting combination therapy based on ATR inhibitors may be a promising strategy for cancer therapy.
CHK1 inhibitors
CHK1, a downstream effector of ATR, is activated by DDR, and its inhibitors effectively suppress the proliferation of cancer cells with high levels of replication stress.1267 Some CHK1 inhibitors have been evaluated or are currently under evaluation in clinical trials, especially in combination with DNA damaging agents such as gemcitabine, cisplatin, and camptothecin.733,1268 LY2603618 is the first selective and potent CHK1 inhibitor.1269 However, the phase II evaluations of LY2603618 in combination with pemetrexed in patients with advanced NSCLC revealed no significant clinical activity and increased risk of thromboembolic events, which hindered its further development (NCT00988858, NCT01139775).1270,1271 MK-8776 (SCH900776) is a selective CHK1 inhibitor that induces cell death when it combines with antimetabolite drugs, such as hydroxyurea, gemcitabine, or pemetrexed in xenograft models.1269 However, clinical trials of MK-8776 combined with gemcitabine or cytarabine in patients with solid tumors or hematological malignancies have been completed and the results have shown no positive efficacy but some adverse events such as mucositis, nausea, and prolonged QT interval (NCT01870596, NCT00779584). LY2880070 is an oral, selective competitive CHK1 inhibitor,1272 and phase II studies in patients with solid tumors (NCT02632448) and Ewing sarcoma (NCT05275426) are in progress. In addition, LY2606368 (prexasertib) is a CHK1/2 dual inhibitor,1269,1273 and a phase I/II study to evaluate the efficacy and safety of LY2606368 in combination with irinotecan and temozolomide in participants with desmoplastic small round cell tumors and rhabdomyosarcoma (NCT04095221) is ongoing. SRA737, an orally bioavailable and selective CHK1 inhibitor, exhibits preclinical activity in MYC-amplified models of neuroblastoma and lymphoma.1274 Its phase II clinical trials in patients with advanced solid tumors or NHL (NCT02797964) and in patients with advanced solid tumors (NCT02797977) have been completed. The results have shown that SRA737 is well tolerated. However, further clinical development of SRA737 needs to be performed in combination therapy due to its poor monotherapy activity.1274 AZD7762, another ATP-competitive CHK1/2 dual inhibitor, suppresses the CHK1-mediated phosphorylation of CDC25C with an IC50 value of 5 nM. However, phase I clinical trials of AZD7762 have been terminated due to its cardiac toxicity and adverse effects.1269,1275
In summary, although CHK1 inhibitors are beneficial in preclinical studies, their clinical benefit remains to be confirmed. Studies have found that p53 mutation status may be a key factor in affecting the cell sensitivity to CHK1 inhibitors, suggesting that biomarkers affecting the efficacy of CHK1 inhibitors need to be identified.1269 Moreover, the development of novel CHK1 inhibitors with reduced toxicity is meaningful in the near future.
Targeting tumor inflammation pathways
Inflammation is considered to be one of the key characteristics of tumor initiation, progression, invasion, metastasis, and treatment resistance.299 The drugs modulating tumor inflammation pathways mainly include nonspecific agents, such as nonsteroidal anti-inflammatory drugs (NSAIDs), statins, and corticosteroids, and targeted drugs, such as neutralizing antibodies, small molecule inhibitors, and recombinant cytokines1276 (Table 10). NSAIDs, including aspirin, celecoxib, and ibuprofen, mainly exhibit anticancer efficiency by inhibiting COX activity and prostaglandin synthesis. NSAIDs have been discovered to reduce cancer mortality, and have shown good therapeutic and preventive effects in cancer patients with CRC, breast cancer, prostate cancer, and head and neck cancer.1277 Aspirin, one of the most widely used and typical anti-inflammatory drugs, has been applied as a broad-spectrum cancer-preventive agent. Phase III clinical trials of aspirin for the treatment of patients with gastric cancer (NCT04214990) and CRC (NCT02467582) are underway. Celecoxib, a COX-2 inhibitor, reveals anticancer activity in CRC, breast cancer, prostate cancer, and head and neck cancer.1278 A phase IV trial of celecoxib as adjuvant therapy to chemotherapy in subjects with metastasis CRC (NCT03645187) is ongoing. However, long-term treatment with NSAIDs can lead to side effects including mucosal lesions, bleeding, and peptic ulcers. Therefore, balancing the benefits of taking NSAIDs for the prevention and treatment of cancers is essential.1276 Statins, consisting of a series of compounds like rosuvastatin, can reduce blood cholesterol concentration by inhibiting the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase. Statins exert a significant role in antiangiogenic and anti-inflammatory therapy in preclinical studies. However, their clinical benefits remain to be further confirmed.1279 Corticosteroids, such as dexamethasone which is usually used as anti-inflammatory drugs for various chronic inflammatory diseases, are found to improve the efficacy of chemotherapy for glioma, breast cancer, lung cancer, and CRC in preclinical studies.1280 The strategies that specifically target inflammation pathways mainly include neutralizing antibodies, small molecule inhibitors, and recombinant cytokines.1276 As chronic inflammation cytokine IL-6 plays a pivotal role in cancer progression, the IL-6 antibody may exert therapeutic efficacy and benefit to cancer patients. Tocilizumab, a recombinant humanized mAb against the human IL-6 receptor, specifically binds to soluble and membrane-bound IL-6 receptor and inhibits signal transduction. Multiple clinical trials of tocilizumab for the treatment of patients with refractory AML (NCT04000698), NHL (NCT05171647), diffuse large B-cell lymphoma (NCT04408638), and relapsed or refractory follicular lymphoma (NCT04712097) are ongoing. In addition, siltuximab, a mAb against the human IL-6 receptor, was approved by the FDA for treating patients with multicentric Castleman disease on April 22, 2014. Its most common adverse events include pruritus, increased weight, rash, hyperuricemia, and upper respiratory tract infection.1281 To date, clinical trials of siltuximab are being conducted to evaluate the efficacy for the prevention of CAR-T cell related cytokine release syndrome in patients with NHL (NCT05665725), for the treatment of patients with metastatic pancreatic cancer (NCT04191421), large granular lymphocytic leukemia (NCT05316116), and multiple myeloma (NCT03315026).
Table 10.
The clinically anti-inflammatory inhibitors in cancer therapies
| Drug | Target | Highest phase | Indications | Identifier | Status |
|---|---|---|---|---|---|
| Aspirin | COX-1/2 | III | Gastric cancer | NCT04214990 | Recruiting |
| III | Colon cancer | NCT02467582 | Active, not recruiting | ||
| Celecoxib | COX-2 | IV | Colon cancer stage | NCT03645187 | Recruiting |
| IV | Hepatocellular carcinoma | NCT02961998 | Completed | ||
| IV | Bile duct cancer, pancreatic cancer | NCT01111591 | Unknown | ||
| IV | Colorectal cancer | NCT00473980 | Completed | ||
| Rosuvastatin | HMG-CoA | IV | Prostate cancer metastatic | NCT04776889 | Completed |
| Dexamethasone | Undefined | IV | Metastatic prostate cancer | NCT03432949 | Recruiting |
| IV | Cancer | NCT02815319 | Completed | ||
| IV | Early-stage breast cancer | NCT03348696 | Completed | ||
| IV | Pancreatic cancer | NCT04025840 | Recruiting | ||
| IV | Lung cancer | NCT02275702 | Completed | ||
| IV | Multiple myeloma | NCT01731886 | Completed | ||
| IV | Ovarian cancer | NCT00817479 | Completed | ||
| IV | Nasal and nasal-type NK/T-cell lymphoma | NCT01501149 | Unknown | ||
| IV | Relapsed refractory multiple myeloma | NCT03934684 | Active, not recruiting | ||
| IV | Peripheral T cell lymphoma | NCT03071822 | Unknown | ||
| IV | Hemophagocytic syndrome T/NK-cell lymphoma | NCT04999878 | Recruiting | ||
| Not Applicable | Mammary cancer | NCT05408676 | Completed | ||
| IV | Primary CNS lymphoma | NCT01960192 | Unknown | ||
| IV | PH+ acute lymphoblastic Leukemia | NCT02690922 | Unknown | ||
| Tocilizumab | IL-6R-specific antibody | III | Refractory acute myeloid leukemia Refractory acute lymphoblastic leukemia | NCT04000698 | Unknown |
| III | Non-hodgkin lymphoma | NCT05171647 | Recruiting | ||
| III | Diffuse large B-cell lymphoma | NCT04408638 | Recruiting | ||
| III | Relapsed or refractory follicular lymphoma | NCT04712097 | Recruiting | ||
| Siltuximab | anti-IL-6 antibody | II | Lymphoma, non-Hodgkin, multiple myeloma acute lymphoblastic leukemia | NCT04975555 | Recruiting |
| II | Multiple myeloma AL amyloidosis | NCT03315026 | Active, not recruiting | ||
| II | High-risk smoldering multiple myeloma | NCT01484275 | Completed | ||
| II | Multiple myeloma | NCT00402181 | Completed | ||
| II | Prostate cancer | NCT00433446 | Completed | ||
| II | Myeloma | NCT01531998 | Completed | ||
| II | Carcinoma, renal cell | NCT00265135 | Completed | ||
| II | Ovarian neoplasms, pancreatic neoplasms, colorectal neoplasms, head and neck neoplasms, lung neoplasms | NCT00841191 | Completed | ||
| II | Metastatic pancreatic adenocarcinoma, stage IV pancreatic cancer AJCC v8 | NCT04191421 | Completed | ||
| Itacitinib | CXCR4 | IV | Lymphoma | NCT05510544 | Recruiting |
| IV | Non-hodgkin’s lymphoma | NCT01164475 | Completed | ||
| Ruxolitinib | JAK1/2 | IV | Hemophagocytic syndrome, T/NK-cell lymphoma | NCT04999878 | Recruiting |
| Pacritinib | JAK2 | II | T-Cell neoplasm lymphoproliferative disorders | NCT04858256 | Recruiting |
| II | Prostate cancer | NCT04635059 | Recruiting | ||
| II | Breast cancer | NCT04520269 | Unknown | ||
| Bortezomib | NF-κB | IV | Multiple myeloma | NCT02268890 | Completed |
Source: All the information is derived from the United States Food and Drug Administration.gov (https://www.clinicaltrials.gov)
The significant role of the Janus kinase/signal transducers and activators of transcription (JAK/STAT) signaling pathway in cancer suggests targeting this pathway as a potential anticancer strategy.1276 Inhibition of the JAK/STAT signaling pathway has been demonstrated to downregulate cellular proliferation and survival, decrease stem cell properties and inflammatory response, suppress invasion and metastasis, ameliorate immunosuppress in malignant tumors.1282 Ruxolitinib is a first-in-class, potent, ATP-competitive, and small molecule JAK1/2 inhibitor (IC50 = 3.3 nM for JAK1, 2.8 nM for JAK2) developed by Incyte Corp.1283 Following its approval by the FDA for the treatment of diseases, such as multiple sclerosis and vitiligo, the potential of ruxolitinib in cancer therapy has garnered widespread interest. Several clinical trials of ruxolitinib in patients with pancreatic cancer, breast cancer, relapsed or refractory or post myeloproliferative AML, HNSCC, or NSCLC were either terminated or completed with results suggesting insufficient efficacy to justify further investigation. There are still multiple clinical trials undergoing to evaluate ruxolitinib monotherapy or in combination with decitabine for the treatment of accelerated/blast phase myeloproliferative neoplasms (NCT04282187), with trametinib for CRC and pancreatic adenocarcinoma (NCT04303403), with paclitaxel and carboplatin for stage III-IV epithelial ovarian and primary peritoneal cancer (NCT02713386), and with preoperative chemotherapy for triple-negative inflammatory breast cancer (NCT02876302), etc. Pacritinib, a potent inhibitor of JAK2 and FLT-3 with the IC50 values of 23 and 22 nM, respectively, was granted accelerated approval by the FDA on February for the treatment of adult patients with intermediate or high-risk primary or secondary myelofibrosis.1284 Although some clinical trials of pacritinib, such as those for the treatment of myeloproliferative neoplasmsrefractory CRC or AML, have been affected by increased side effects. Several clinical trials are still undergoing, including pacritinib in combination decitabine or the treatment of accelerated/blast phase myeloproliferative neoplasms (NCT04282187), prostate cancer (NCT04635059), relapsed/refractory T-cell lymphoproliferative neoplasms (NCT04858256). Moreover, other JAK inhibitors such as itacitinib, STAT3 inhibitors like OPB-31121, and TTI-101, and STAT3 antisense oligonucleotide danvatirsen (AZD9150) have been evaluated or are under investigation for their anticancer potential. It is important to note that the clinical studies targeting the JAK/STAT pathway have revealed the complexity of this approach and have underscored the necessity for in-depth investigation to combat cancer more effectively.
Inhibition of NF-κB is also an effective way to slow down tumor development and induce apoptosis in cancer cells. However, NF-κB deficiency can lead to severe immunodeficiency and long-term inhibition of NF-κB causes serious side effects, suggesting that an appropriate dosage regimen and administration time will facilitate NF-κB targeted therapy in the clinic.764 Moreover, multiple other inhibitors such as antibodies, cytokines and chemokines inhibitors, and inhibitors of inflammatory transcription factors, are in clinical trials and the results are eagerly anticipated.
Targeting tumor cell metabolism
The precedent of targeting the metabolism of cancer cells was created by Sydney Farber and colleagues in the 1940s, who successfully used antifolate agents such as aminopterin to induce remission in childhood ALL.1285 This discovery led to the development of chemotherapy drugs including methotrexate, 5-FU, gemcitabine, and pemetrexed which are widely used to treat various types of cancers by targeting one-carbon metabolic pathways and their downstream effectors, such as nucleotide metabolism. However, these drugs exhibit many deleterious side effects due to their nonspecific effect and the importance of one-carbon pathways in healthy cells.1286
Sixty years after Sidney Farber introduced antifolates for the treatment of childhood ALL, aberrations in cancer metabolism attracted much attention and have been extensively studied, although nearly one century has passed since Otto Warburg discovered aerobic glycolysis in cancer cells in the 1920s. However, therapeutic progress in targeting cancer metabolism remains limited and only a few metabolism-based drugs have been developed, and entered clinical trials for cancer therapy.1286
Targeting glycolysis
As glucose supplies the major source of energy, carbon intermediates and NADH for biosynthesis, targeted inhibition of glucose uptake and utilization in cancer cells is a promising therapeutic strategy1287 (Fig. 6). Multiple inhibitors against glycolytic enzymes and glycolytic product transporter proteins have been studied, such as GLUT1 inhibitors (STF-31, glutor, and BAY-876), HK2 inhibitors (2-deoxyglucose, benitrobenrazide), PKM2 inhibitors (TEPP-46 and mitapivat), LDHA inhibitors (GNE-140, NCI-006, and GSK28387808A), and MCT-1 inhibitors (AZD3965).1286–1288 Although these inhibitors have exhibited potent anticancer efficiency in various cancers both in vitro and in vivo preclinically, only a few of them have entered clinical trials, and no clinical success has been achieved thus far due to limited efficacy and toxicity. For example, the MCT-1 inhibitor AZD3965 blocks lactate-mediated tumor progression and has significant anticancer effects alone or combined with metformin. However, a phase I clinical trial in patients with advanced cancers including diffuse large B-cell lymphoma (DLBCL) and Burkitt’s lymphoma, has shown that a number of patients experienced dose-limiting AZD3965 related toxicities such as hematological, cardiac, and ophthalmic toxicities (NCT01791595).
Targeting amino acid metabolism
Similarly, amino acids, especially glutamine, participate in various cellular processes in cancer progression, which provide a major source of energy, cell component building blocks, and redox homeostasis, thereby providing a scientific rationale for targeting their metabolism for cancer treatment.1289 Anticancer drug candidates against glutamine metabolism and closely linking metabolic networks, such as glutamine transporter SLC1A5, glutaminase (GLS), and aminotransferase, have shown promising effects in cancer treatment (Fig. 6). The amino acid analog l-g-glutamyl-p-nitroanilide, the originally discovered compound V-9302, and specific synthetic mAbs (i.e., KM4008 and KM4012) were developed for the inhibition of SLC1A5. Although they suppressed glutamine-dependent growth of cancer cells to some extent, none of them entered clinical trials, for their specificity, efficiency, and safety profile need to be further evaluated and optimized. In addition to targeting glutamine transporters, targeting GLS, which transforms glutamine into glutamate in the mitochondria, is a notable drug development strategy. Among GLS1 inhibitors, telaglenastat (CB-839), a derivative of the allosteric inhibitor BPTES, has attracted much attention.1290,1291 CB-839 has been assessed in more than 10 completed clinical trials alone for the treatment of hematological and solid tumors, or in combination with everolimus for RCC (NCT03163667), talazoparib or palbociclib for solid tumors (NCT03875313, NCT03965845), paclitaxel for TNBC (NCT03057600), and azacitidine for myelodysplastic syndrome (NCT03047993). Disappointingly, CB-839 did not prove efficacious in the above clinical trials or some results were not disclosed. Moreover, some clinical trials of CB-839 alone or in combination with anticancer drugs are ongoing, such as with capecitabine for PIK3CA mutant CRC (NCT02861300), with temozolomide for IDH-mediated diffuse astrocytoma (NCT03528642), with carfilzomib and dexamethasone for recurrent or refractory multiple myeloma (NCT03798678), and with chemoradiation for advanced cervical cancer (NCT05521997). Sirpiglenastat (DRP-104), a glutamine analog that broadly targets glutamine metabolism, is under early-phase clinical trials for examining its efficacy as a single agent or in combination with immune checkpoint inhibitors for advanced cancer (NCT04471415 and NCT06027086).1292 The results from the above trials are still pending.
Targeting fatty acid metabolism
Cancer cells rely on de novo fatty acid synthesis for proliferation; thus, cancer cells are expected to be vulnerable to the inhibition of fatty acid synthetic enzymes. Inhibitors targeting fatty acid synthase, a key enzyme for de novo fatty acid synthesis, have been developed1293 (Fig. 6). Candidates such as TVB316 and TVB2640 have been demonstrated to be effective and less toxic than their predecessors, and more than 10 clinical trials of TVB2640 for the treatment of NSCLC, prostate cancer, and HER2-positive metastatic breast cancer are underway (NCT03808558, NCT03179904, and NCT05743621). The field is anxiously awaiting the results of these studies, decades after fatty acid synthase was identified as a potential cancer therapeutic target. ATP-citrate lyase (ACLY), a key enzyme for fatty acid chain elongation, converts citrate acetyl-CoA into the cytosol. The ACLY inhibitor bempedoic acid was approved by the FDA in 2020 as a lipid-lowering drug.1294 Furthermore, a series of allosteric ACLY inhibitors with low (nanomolar) competitive inhibitory activity were discovered, such as the allosteric inhibitor NDI-091143, which binds to homotetramer ACLY, shows potent inhibition and is competitive with citrate and noncompetitive with ATP.1295 PF-05221304, an orally administered inhibitor of acetyl-CoA carboxylases (ACC1 and ACC2), is currently undergoing clinical studies (NCT03248882) in nonalcoholic fatty liver disease with fibrosis. Its potential in cancer therapy needs to be further evaluated. ND-646, another allosteric inhibitor of ACC1 and ACC2, reduces tumor growth in NSCLC subcutaneous xenografts, suggesting potential avenues for therapeutic application.1291
Targeting mitochondria metabolism
The pivotal role of mitochondria as metabolic and biosynthetic organelles makes them attractive anticancer targets (Fig. 6). Although this approach thus far has been limited by toxicity due to difficulties in identifying specific compounds targeting metabolic enzymes, several compounds or candidates targeting the tricarboxylic acid (TCA) cycle and oxidative phosphorylation are currently in clinical trials for the treatment of both solid and hematological tumors.1296 The first successful antimetabolite drug came from targeting TCA. IDH catalyzes the oxidative carboxylation of isocitrate to produce α-KG, whereas its mutations result in the gain of function, converting α-KG to the oncometabolite 2-HG.1297 In 2017, the FDA approved enasidenib, a first-in-class IDH2 mutation inhibitor developed by Celegene, for the treatment of recurrent or refractory AML with IDH2 mutation. Subsequently, ivosidenib, developed by Agios Pharmaceuticals against IDH1 mutations, was approved by the FDA for the treatment of AML and cholangiocarcinoma with IDH1 mutation.1298 Moreover, AG-881 is undergoing clinical trials for the treatment of AML-carrying IDH2 or IDH1/2 mutations (NCT02492737). Similarly, CPI-613, targeting both the α-KG dehydrogenase complex and pyruvate dehydrogenase, is in phase I/II clinical trials for leukemias, lymphomas, and SCLC (NCT03699319 and NCT03793140). In addition to the TCA cycle, the electron transport chain, also known as the respiratory chain which consists of four complexes (CI–IV), is the main target for drug development. Metformin, the most well-known inhibitor of complex I, is approved for type-2 diabetes and has been found to exhibit anticancer effects against various cancers in preclinical studies and clinical trials. In contrast to metformin which is a nonspecific complex I inhibitor, IACS-010759 is a specific inhibitor of complex I that has undergone clinical trials for AML and advanced cancers. However, its toxicity has hindered its further development.1299 Rotenone and deguelin also inhibit complex I, while their neurotoxic effects are prominent. Antimycin A is an inhibitor of complex III commonly used in experimental research, while resveratrol has enrolled in clinical trials for different types of cancer. Complex IV can be inhibited by doxorubicin, a DNA intercalating chemotherapeutic drug, and the porphyrin photosensitizer photofrin, which is approved for esophageal cancer and NSCLC. No promising inhibitors have been reported to date for Complex V, except for oligomycin, which is only suitable for experimental use. Employing mitochondrial uncouplers is an alternative approach to impair the function of the electron transport chain. Niclosamide is in phase I/II clinical trials for prostate and colon cancer, while nitazoxanide is in phase II for different forms of advanced cancers.1296
Targeting one-carbon and nucleotide metabolism
In addition to the metabolism mentioned above, other metabolic processes also perform significant roles in tumors, such as one-carbon metabolism and nucleotide metabolism which have close connections. One-carbon metabolism provides one carbon unit in the form of methyl groups to several metabolic pathways and is responsible for the synthesis of methionine, serine/glycine, purine, and pyrimidine.1300 After a landmark study by Farber and colleagues revealed that the folate antagonist aminopterin induced remission in children with ALL, a series of classical inhibitors in this field, including methotrexate, pemetrexed, gemcitabine, and 5-FU, were used as frontline chemotherapy for a diverse range of cancers.1301 After that, multiple attempts were made to develop targeted molecules. MTHFD2 inhibitors LY345899 and DS18561882, have shown anticancer activity both in vitro and in vivo.1302 PHGDH catalyzes the transformation of the glycolytic intermediate 3-PG into 3-phosphohydroxy pyruvate, and its allosteric inhibitors, such as CBR-5884, NCT-503, α-ketothioamide derivatives, and compound b36, and orthosteric inhibitors which are indole derivatives such as BI-4916, inhibit PHGDH activity and moderately suppress cancer cell proliferation preclinically.1303 SHMT catalyzes the conversion of serine and tetrahydrofolate into glycine and 5,10-methylene- tetrahydrofolate, thus providing one carbon unit for nucleotide synthesis. Optimization has generated several experimental dual SHMT1/2 inhibitors, including SHIN1 and SHIN2, which have revealed some extent of anticancer effects preclinically.1304,1305 Human dihydroorotate dehydrogenase (hDHODH) is the fourth and rate-limiting enzyme of de novo pyrimidine synthesis, and inhibition of hDHODH is an effective strategy for the treatment of cancers. To date, classical DHODH inhibitors, such as leflunomide and teriflunomide, and several novel hDHODH inhibitors, such as brequinar, ASLAN003, BAY2402234, AG-636, PTC299, and JNJ-74856665 (NCT04609826), have been evaluated in clinical trials to investigate their safety and antitumor efficacy.1306
Targeting dietary interventions
Dietary interventions alone or in combination with various anticancer strategies have become promising tools for cancer therapy, including preventing tumorigenesis, delaying tumor growth, and improving the effectiveness of existing cancer treatments.1307 Dietary interventions potentially improve tumor therapy in several ways, such as eliminating specific nutrients that tumors use as fuel and building blocks, potentiating other forms of therapy including chemotherapy, radiotherapy, and targeted therapy by depriving tumors of nutrients, enhancing the antitumor immune response by modulating the growth factors or altering the systemic immune system.1308 Dietary interventions come in various forms, such as the restriction of energy or macronutrients, defined by timing such as intermittent fasting regimens.1308 Fasting mimicking diet and intermittent fasting sensitize anticancer medicines. For example, the combination of metformin and intermittent fasting is effective at targeting the metabolic plasticity of cancer. Understanding the interactions between cancer and diet is crucial for establishing diet as a line of treatment. Elucidating altered drug efficacy under a differential metabolic context will be important for future enhancing the dietary interventions in specific cancer therapies due to the heterogeneous nature of cancers and host metabolisms.1309,1310
Although extensive efforts have been made to develop targeted therapy against cancer metabolism, few have achieved clinical success, and metabolism-targeted therapy is still challenging for the following reasons. First, metabolism plays a crucial role in all kinds of cells including tumor cells, cells in the TME, such as immune cells, macrophages, cancer-related fibroblasts and other stromal cells, and normal cells. Moreover, extensive interactions of metabolites in these cells exist. These factors make strong antitumor activity and low toxicity by regulating metabolism extremely difficult. Second, both metabolite enzymes and metabolites possess unclassical functions such as acting as kinases or second messengers in addition to acting as enzymes and metabolites. Thus, inhibiting enzyme function may exhibit only moderate anticancer efficiency. Third, cancer cells reprogram their metabolism very quickly after various stimuli by increasing metabolic flexibility, uptake of extracellular metabolites via compensatory transporters and macropinocytosis, and upregulation of nutrient stress-response proteins. Blockage of one pathway by targeting a key enzyme could result in the activation of another metabolic hub. In this regard, targeting cancer metabolism must be based on a thorough understanding of how metabolic pathways affect the whole metabolic status of cancer hubs, which could promote the successful development of anticancer drugs targeting metabolism.
ICIs-based immunotherapy
Since Tasuku Honjo’s group at Kyoto University discovered PD-1 in 1992,806 and Allison’s team at MD Anderson Cancer Center reported that blocking CTLA-4 by its antibody could increase the antitumor activity of T cells and inhibit tumor growth in 1996,1311 immunotherapy has been considered as a breakthrough in clinical cancer treatment due to the promising efficacy.1312 Immune checkpoint inhibitor (ICI) therapies, including anti-CTLA-4, anti-PD-1, and anti-PD-L1 therapies, have revolutionized the systemic treatments for advanced hematological and solid tumors in the area of antitumor immunotherapy. Compared with chemotherapy and targeted therapies, ICIs induce unprecedented improvements in response rate and better survival rate in partial patients, even after cessation of treatment.1313,1314
Ipilimumab, the antibody against CTLA-4, was the first ICI approved by the FDA in 2011, which is a milestone in cancer immunotherapy. Ipilimumab successfully hindered cancer progression in patients with refractory metastatic melanoma.1315 Tremelimumab is another human IgG2 CTLA-4 antibody against HCC and was approved by the FDA in 2022.1316
PD-1/PD-L inhibitors
To date, PD-1/L1 inhibitors are the most widely applied ICIs, which undoubtedly changed the paradigm of cancer therapy. They have shown clinical efficacy against many different solid and hematologic malignancies. The binding of PD-L1 (initially identified as B7-H1) to its receptor PD-1 inhibits T-cell migration, proliferation, and secretion of cytotoxic mediators, thereby limiting tumor cell killing. Inhibitors of PD-1 and PD-L1 reverse T cell suppression by disrupting the PD-1 axis, thereby enhancing the endogenous antitumor immune response.1312 Until now, multiple PD-1/L1 inhibitors have been approved for commercialization in the US and China, which include pembrolizumab, nivolumab, dostarlimab, cemiplimab, sintilimab targeting PD-1, and atezolizumab, avelumab, and durvalumab targeting PD-L1 (Table 11). These inhibitors are now widely used for the treatment of various cancers, including NSCLC, melanoma, uroepithelial carcinoma, HNSCC, CRC, HCC, and Hodgkin’s lymphoma.1316 Nivolumab (Opdivo) and pembrolizumab (Keytruda) are particularly extensively used in clinical therapy. Nivolumab, developed by Bristol Myers Squibb, is the first clinical anti-PD-1 antibody approved in 2015 for the treatment of advanced SCLC and metastatic squamous NSCLC. After that, pembrolizumab, developed by Merck & Co, was approved by the FDA for the first-line treatment of patients with metastatic NSCLC in 2016.1312 Different from nivolumab, the prescription of pembrolizumab requires confirmed PD-L1 overexpression on tumors. At the same time, atezolizumab (Tecentriq) by Roche was approved by the FDA in 2016 to treat patients with advanced and metastatic urothelial carcinoma. Another two new PD-L1 antibodies, durvalumab (Imfinzi) and avelumab (Bavencio), were approved in 2017. Furthermore, Innovent Biologics in China developed a PD-1 antibody named sintilimab which achieved good efficiency after neoadjuvant administration.1317 Dostarlimab (Jemperli) developed by GSK was approved in 2021 and used in patients with mismatch repair-deficient advanced solid tumors.1318
Table 11.
The FDA-approved immune checkpoint inhibitors
| Target | Drugs | Indications | Company |
|---|---|---|---|
| CTLA-4 | Ipilimumab | Colorectal cancer, hepatocellular carcinoma, melanoma mesothelioma, non-small cell lung cancer, renal cell carcinoma | Bristol Myers Squibb |
| PD-1 | Cemiplimab | Basal cell carcinoma, cervical squamous cell cancer, non-small cell lung cancer | Regeneron Pharmaceuticals |
| Nivolumab | Colorectal cancer, esophageal squamous cell carcinoma, hepatocellular carcinoma, gastric cancer, hodgkin lymphoma, head and neck squamous cell carcinoma, melanoma, mesothelioma, non-small cell lung cancer, renal cell carcinoma, urothelial carcinoma | Bristol Myers Squibb | |
| Pembrolizumab | Breast cancer, cervical cancer, colorectal cancer, esophageal squamous cell cancer, endometrial carcinoma, esophageal carcinoma, gastric carcinoma, hepatocellular carcinoma, hodgkin lymphoma, non-small-cell lung cancer, melanoma, mesothelioma, Merkel cell carcinoma, non-small cell lung cancer, primary mediastinal large B-cell lymphoma, renal cell carcinoma, small-cell lung cancer, urothelial carcinoma, biliary tract cancer | Merck & Co | |
| Dostarlimab -gxly | Advanced or recurrent mismatch repair-deficient/microsatellite instability-high endometrial cancer, recurrent or advanced mismatch repair-deficient solid tumors | GlaxoSmithKline | |
| PD-L1 | Atezolizumab | Breast cancer, hepatocellular carcinoma, melanoma, non-small cell lung cancer, small-cell lung cancer, urothelial, alveolar soft part sarcoma, urothelial carcinoma | Genentech |
| Avelumab | Merkel cell carcinoma, renal cell carcinoma, urothelial carcinoma | EMD Serono | |
| Durvalumab | Non-small cell lung cancer, small-cell lung cancer, urothelial carcinoma, biliary tract cancer, hepatocellular carcinoma | AstraZeneca UK Limited |
Source: All the information is derived from the United States Food and Drug Administration.gov (https://www.fda.gov/)
There are currently three PD-L1 mAbs, atezolizumab, durvalumab, and avelumab, approved by the FDA for the treatment of NSCLC and merkel cell carcinoma. Atezolizumab, a humanized IgG1 mAb, abrogates antibody-dependent cytotoxicity and prevents depletion of PD-L1-expressing T cells.1316 Based on its favorable safety and efficacy profile, the FDA accelerated the approval of atezolizumab in May 2016 for the treatment of locally advanced or metastatic urothelial carcinoma treatment after the failure of cisplatin-containing chemotherapy, and subsequently approved for the treatment of advanced metastatic NSCLC during or following platinum-containing chemotherapy in October 2016.803 In addition, atezolizumab is the first ICI approved in combination with carboplatin and etoposide to treat advanced SCLC. Durvalumab is a fully human IgG1 mAb that binds PD-L1 with high affinity and specificity.803 Durvalumab obtained accelerated approval of the FDA in 2017 for the treatment of patients with advanced or metastatic urothelial carcinoma who have disease progression following platinum-containing chemotherapy, and for the treatment of patients with unresectable stage III NSCLC in 2018.1319 Avelumab is a fully human IgG1 mAb with a wild-type IgG1 crystallizable fragment (Fc) region, which enables avelumab to utilize both adaptive and innate immune mechanisms to suppress cancer cells.1320 Similarly, avelumab obtained accelerated approval for the treatment of patients with locally advanced or metastatic urothelial carcinoma in 2017, and subsequently approval for first-line treatment of patients with advanced RCC in combination with axitinib in 2019.1320 Compared with many oncology regimens, PD-1/PD-L1 blockade is associated with fewer adverse events including fatigue, diarrhea, and decreased appetite which are well tolerated. Moreover, there are still a large number of clinical trials undergoing to evaluate the therapeutic potential of the above inhibitors.
In addition to antibodies, novel strategies targeting PD-1/PD-L1 were developed. For example, AC-1, an antibody-based PROTAC termed AbTAC, simultaneously bound PD-L1 and E3 ligase RNF43 to degrade cell-surface PD-L1 via lysosomal degradation in different cell lines with high PD-L1 expression levels.1321,1322 Considering only a small fraction of cancer patients (lower than 50%) respond to PD-1/L1 inhibitors which are far from satisfactory, immune combination therapy which may improve the efficacy and expand the beneficiary population attracted much attention. The combinations of immune checkpoint blockade and costimulatory receptor activation, such as PD-L1 × 4-1BB (MCLA-145) and PD-1 × ICOS (XmAb23104), are under clinical investigation (NCT03922204, NCT03752398). Monovalent trispecific antibody NM21-1480 (αPD-L1, α4-1BB and αHSA) and GNC-038, a tetra-specific IgG-scFv conjugated antibody (αCD19/CD3/4-1BB/PD-L1) are in phase I clinical trials (NCT04442126 and NCT05192486). Checkpoint blockades incorporation with BsAbs achieved tumor-localized and TAA-dependent checkpoint blockage. For example, IBI315, an anti-PD-1 × HER2 developed by Innovent Bridge, is under phase I clinical study for patients with HER2-expressing advanced solid tumors (NCT04162327). Anti-PD-1 × CTLA-4 BsAbs, including AK104, MEDI5752, and MGD019, are expected to synergistically inhibit PD-1 and CTLA-4 double-positive lymphocytes, which are under clinical investigations (NCT06035224, NCT04522323, and NCT05293496).
Other ICIs
In addition to PD-1/PD-L1 and CTLA-4, novel immune checkpoints including lymphocyte activation gene-3 (LAG-3), T cell immunoglobulin and mucin-domain-containing 3 (TIM-3), and T cell immunoglobulin and ITIM domain (TIGIT) that mediate inhibitory signals through different mechanisms have been identified, and their inhibitors have been emerging for cancer immunotherapy.1312 The mAb drugs targeting these immune checkpoints transmissed inhibitory signals following ligand engagement, and their synergistic antitumor effect with PD-1/PD-L1 inhibitors were evaluated in preclinically and in multiple clinical trials (NCT03219268, NCT03708328, and NCT03440437).
Induced on CD4+ and CD8 + T cells under antigen stimulation, LAG-3 has become one of the most promising new targets of immune checkpoint blockage after PD-1 with great application prospects. Relatlimab is the most advanced mAb targeting LAG-3, which is under phase II/III clinical trial in unresectable or untreated metastatic melanoma in combination with nivolumab. The study resulted in a median PFS of 10.12 months in the combination group compared with 4.63 months in the monotherapy group, supporting its approval by the FDA for the treatment of metastatic melanoma combined with nivolumab. Relatlimab represents the third type of ICI to enter the market.1316 TM-3 is a T-cell surface inhibitor that is mainly expressed on CD4 + T helper cell 1 (Th1) and CD8 + CTL cells, and some innate immune cells including dendritic cells, NK cells, and macrophages. LY3321367, an anti-Tim-3 antibody, demonstrated good tolerability as monotherapy or in combination with an anti-PD-L1 antibody in phase I studies, and further clinical studies are needed to verify its efficacy and safety in larger cohorts of patients.1323,1324 TIGIT, a type I transmembrane protein which belongs to the immunoglobulin superfamily (IgSF), is expressed on T cells, regulatory T cells, memory T cells, and NK cells. Tiragolumab is the mAb targeting TIGIT which is currently under phase III clinical trial in extensive-stage SCLC in combination with atezolizumab (NCT04256421). In addition, mAbs targeting fibrinogen-like protein 1 (FGL1), a ligand for Lag-3 for NSCLC, nuclear receptor subfamily 2 group F member 6 (NR2F6), an intracellular IC molecule, and V-set immunoregulatory receptor (VISTA), an immunomodulatory protein expressed in lymphoid organs and bone marrow cells, are now being evaluated in phase I clinical studies for the treatment of solid tumors.1312
At present, approximately5000 registered clinical studies listed on the US trial registry site ClinicalTrials.gov are ongoing to evaluate the effectiveness of ICIs involving PD-1, PD-L1, or CTLA-4, both individually and in combinations against various hematological and solid tumors. Although ICIs have achieved great success in clinical treatment, some challenges still remain to be solved in this field. First, only parts of patients significantly benefit from ICI treatment. Thus, accurate prediction biomarkers by integrating multiple approaches to determine which patients are likely to benefit from ICIs is urgently needed. The combination and development of multiple functional approaches, including large-scale genomic sequencing, single-cell transcriptomic techniques, multi-omics, and computational immunogenomics, which integrate intratumour heterogeneity, tumor mutational burden, neoantigen expression, and immunogenicity, could improve the prediction of response to ICIs.1325 Second, although ICIs initially exhibited strong efficiency against tumor growth, patients still have relapse and/or develop acquired resistance. Combinational therapeutic strategies based on a deep understanding of the tumor and TME, and coordination of systemic and local intratumor immune responses enable to improve and maximize the potential benefit to more tumor patients.1326 Third, the development of novel cancer immunotherapy targets based on the mechanistic study can lead to the discovery of effective approaches, which in turn improve the efficacy of tumor immunotherapy. In summary, ICIs opened a new era of immunotherapy and changed the landscape of cancer treatment. They are promising treatment options although the response rate is far from satisfactory. Combination therapy and mechanism studies may improve efficacy, expand the beneficiary population, and further support immunotherapy as a mainstream cancer treatment alongside chemotherapy, radiotherapy, and surgery.
Differentiation therapy
The concept of differentiation therapy originates from the fact that hormones or cytokines can promote differentiation in vitro and thus irreversibly alter the phenotype of cancer cells. Certain signaling molecules and drugs, such as retinoic acid, cAMP, sodium butyrate, and cytokines, can induce terminal ex vivo differentiation in AML, embryonic carcinomas, or neuroblastoma.1327
Differentiation therapy is a meaningful tumor-targeting strategy, and many inhibitors have been developed to induce differentiation. The combination of small molecule inhibitors all-trans retinoic acid and arsenic trioxide for the treatment of acute promyelocytic leukemia is a watershed for differentiation therapy. In addition, retinoic acid is used to treat solid tumors. For example, retinoic acid induces the differentiation of tumor-initiating cells in HCC, suppresses the expression of stem cell markers, and induces the expression of liver-specific genes, ultimately increasing the sensitivity of cisplatin therapy. The small molecule drug arsenic trioxide, approved by the FDA for leukemia treatment, has shown effectiveness in various hematological malignancies and solid tumors. The natural product oroxylin A, a bioactive flavonoid in Scutellaria baicalensis with strong anticancer effects and safety, can induce tumor cell differentiation. Oncostatin M, a glycoprotein belonging to the IL-6 family of cytokines, is involved in cell growth and development and can induce differentiation and inhibit the proliferation of HCC cells.821
Since most solid tumor oncogenic signaling pathways are far more genetically complex than the genetic basis of leukemia, the efficacy of solid tumor differentiation inducers has not yet reached that of hematologic malignancies. Conventional cancer therapy aims to kill rapidly proliferating tumor cells, which can damage normal cells and lead to serious side effects. In contrast, differentiation therapies have low cytotoxicity and are effective in combination with classical tumor-killing cytotoxic compounds. Differentiation therapy reduces malignancy and inhibits the aggressiveness of tumors. Tumor differentiation therapy has many benefits, including reversing the malignant phenotype of tumors, restoring normal cellular functions, enhancing the immunogenicity of tumor cells, and enhancing the therapeutic sensitivity of tumor cells to conventional tumor therapy and ICIs.821 Therefore, the induction of cancer cell differentiation is a valuable tumor treatment strategy, which warrants further study.
Epigenetic reprogramming inhibitors
Epigenetic dysregulation in cancer has led to the exploration of epigenetic machinery as a promising target for drug development. Consequently, the field of developing epigenetic drugs, which target enzymes involved in regulating genome function through epigenetic mechanisms, has gained significant attention.1328 Currently, the focus of epigenetic drug development revolves around enzymes responsible for introducing (writers), recognizing (readers), and removing (erasers) epigenetic marks on DNA or core histones.1329 Inhibitors have been designed to target these enzymes, including DNMTs, and HMTs EZH2 and DOT1L as writers, HDM LSD1 and HDACs as erasers, and BET proteins as readers (Table 12).
Table 12.
The FDA-approved and clinically developed epigenetic reprogramming inhibitor in cancer therapies
| Type | Drug | Highest Phase | Indications | Company/Identifier | Status |
|---|---|---|---|---|---|
| DNMTi | Azacitidine (Vidaza) | Approved | Acute myeloid leukemia, chronic myelomonocytic leukemia, myelodysplastic syndromes | Celgene | / |
| Decitabine (Dacogen) | Approved | Acute myeloid leukemia, chronic myelomonocytic leukemia, myelodysplastic syndromes | MGI Pharma & SuperGen | / | |
| Guadecitabine | III | Acute myeloid leukemia | NCT02920008 | Completed | |
| III | Myelodysplastic syndromes, leukemia myelomonocytic chronic | NCT02907359 | Completed | ||
| III | Leukemia myeloid acute | NCT02348489 | Completed | ||
| NTX-301 | I | Acute myeloid leukemia, myelodysplastic syndromes, chronic myelomonocytic leukemia | NCT04167917 | Recruiting | |
| MG98 | I | Unspecified adult solid tumor | NCT00003890 | Completed | |
| HDACi | Belinostat (Beleodaq) | Approved | Peripheral T-cell lymphoma | Spectrum Pharma | / |
| Panobinostat (Farydak) | Approved | Multiple myeloma | Novartis | / | |
| Romidepsin (Istodax) | Approved | Cutaneous T cell lymphoma | Gloucester Pharma | / | |
| Vorinostat (Zolinza) | Approved | Cutaneous T cell lymphoma | Merk Sharp | ||
| Abexinostat | III | Renal cell carcinoma | NCT03592472 | Recruiting | |
| ACY-241 | I | Multiple myeloma | NCT02400242 | Active, not recruiting | |
| I | Malignant melanoma | NCT02935790 | Completed | ||
| I | Advanced solid tumors | NCT02551185 | Completed | ||
| I | Non-small cell lung cancer | NCT02635061 | Active, not recruiting | ||
| AR-42 | III | Neurofibromatosis type 2 | NCT05130866 | Recruiting | |
| CUDC-907 | II | Relapsed and/or refractory diffuse large B-cell lymphoma including myc alterations | NCT02674750 | Completed | |
| II | Thyroid neoplasms, poorly differentiated and undifferentiated thyroid cancer, differentiated thyroid cancer | NCT03002623 | Terminated | ||
| II | Prostate cancer | NCT02913131 | Terminated | ||
| CXD101 | II | Colorectal neoplasms malignant | NCT03993626 | Unknown | |
| II | Diffuse large B-cell lymphoma | NCT03873025 | Withdrawn | ||
| II | Hepatocellular carcinoma | NCT05873244 | Recruiting | ||
| Entinostat | III | Advanced breast cancer | NCT03538171 | Unknown | |
| III | Breast adenocarcinoma, HER2/Neu negative locally advanced breast carcinoma, metastatic breast carcinoma, recurrent breast carcinoma | NCT02115282 | Active, not recruiting | ||
| Givinostat (ITF2357) | II | Multiple myeloma | NCT00792506 | Terminated | |
| II | Chronic myeloproliferative neoplasms | NCT01761968 | Active, not recruiting | ||
| II | Hodgkin’s lymphoma | NCT00496431 | Terminated | ||
| II | Hodgkin’s lymphoma | NCT00792467 | Completed | ||
| Mocetinostat (MGCD0103) | II | Urothelial carcinoma | NCT02236195 | Completed | |
| II | Lymphocytic leukemia chronic | NCT00431873 | Completed | ||
| II | Hodgkin’s lymphoma | NCT00358982 | Terminated | ||
| II | Lymphoma | NCT00359086 | Completed | ||
| II | Myelogenous leukemia acute, myelodysplastic syndromes | NCT00374296 | Terminated | ||
| II | Lymphoma relapsed and refractory, diffuse large B-cell lymphoma and follicular lymphoma | NCT02282358 | Terminated | ||
| II | Myelodysplastic syndrome, acute myelogenous leukemia | NCT00324220 | Completed | ||
| Resminostat (4SC-201) | II | Hepatocellular carcinoma | NCT00943449 | Completed | |
| II | Advanced colorectal carcinoma | NCT01277406 | Completed | ||
| II | Hepatocellular carcinoma | NCT02400788 | Completed | ||
| II | Hodgkin’s lymphoma | NCT01037478 | Completed | ||
| II | Lymphoma | NCT02953301 | Active, not recruiting | ||
| Ricolinostat (ACY-1215) | II | Multiple myeloma | NCT01997840 | Active, not recruiting | |
| II | Lymphoma, Lymphoid malignancies | NCT02091063 | Completed | ||
| II | Multiple myeloma | NCT01323751 | Completed | ||
| EZH2i | Tazemetostat | Approved | Epithelioid sarcoma | Epizyme | / |
| GSK126 | I | Cancer, Neoplasms | NCT02082977 | Terminated | |
| CPI-1205 | II | Metastatic castration-resistant prostate cancer | NCT03480646 | Unknown | |
| CPI-1205 | I | Advanced solid tumors | NCT03525795 | Terminated | |
| CPI-1205 | I | B-cell lymphoma | NCT02395601 | Terminated | |
| DOT1Li | Pinometostat (EPZ-5676)- | II | Recurrent/refractory acute myeloid leukemia | NCT03701295 | Completed |
| II | Acute myeloid leukemia | NCT03724084 | Terminated | ||
| LSD1i | ORY-1001 | I | Small cell lung cancer | NCT02913443 | Completed |
| I | Acute myeloid leukemia | NCT05546580 | Recruiting | ||
| INCB059872 | II | Solid tumors and hematologic malignancy | NCT02712905 | Terminated | |
| II | Solid tumors | NCT02959437 | Terminated | ||
| II | Myeloproliferative neoplasms, myelodysplastic syndrome | NCT04061421 | Recruiting | ||
| IMG-7289 | II | Acute myeloid leukemia | NCT02842827 | Completed | |
| II | Myelofibrosis | NCT03136185 | Completed | ||
| II | Thrombocythemia | NCT04081220 | Recruiting | ||
| I | Acute myeloid leukemia | NCT05597306 | Recruiting | ||
| II | Extensive stage lung small cell carcinoma | NCT05191797 | Recruiting | ||
| GSK2879552 | I | Carcinoma small cell | NCT02034123 | Terminated | |
| I | Leukemia myelocytic acute | NCT02177812 | Terminated | ||
| CC-90011 | I | Lymphoma | NCT02875223 | Active, not recruiting | |
| I | Small cell lung carcinoma | NCT03850067 | Active, not recruiting | ||
| II | Neoplasms | NCT04350463 | Active, not recruiting | ||
| I | Prostatic neoplasms | NCT04628988 | Completed | ||
| I | Leukemia | NCT04748848 | Terminated | ||
| SP-2577 | II | Ewing sarcoma | NCT05266196 | Enrolling by invitation | |
| II | Recurrent chronic myelomonocytic leukemia | NCT04734990 | Active, not recruiting | ||
| BETi | TEN‐010 | I | Acute myeloid leukemia, myelodysplastic syndromes | NCT02308761 | Completed |
| I | Multiple myeloma | NCT03068351 | Completed | ||
| I | Solid tumors | NCT01987362 | Completed | ||
| GSK525762 | II | Neoplasms | NCT01943851 | Completed | |
| I | NUT midline carcinoma | NCT01587703 | Completed | ||
| OTX105 | I | Acute myeloid leukemia | NCT01713582 | Completed | |
| I | NUT midline carcinoma | NCT02259114 | Completed | ||
| CPI-0610 | I | Multiple myeloma | NCT02157636 | Completed | |
| I | Lymphoma | NCT01949883 | Completed |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov) and the United States Food and Drug Administration.gov (https://www.fda.gov/)
DNMT inhibitors
DNMT inhibitors, also known as hypomethylating agents, have become effective epigenetic therapies for cancer due to the crucial role of DNMTs in DNA methylation. There are two main classes of DNMT inhibitors: nucleoside analog inhibitors that are incorporated into newly synthesized DNA and recognized by DNMTs, and nonnucleoside inhibitors that interfere with DNMT binding.1330 Nucleoside inhibitors, such as 5-azacitidine (Vidaza), 5-aza-2′-deoxycytidine (decitabine, Dacogen), and SGI-110 (guadecitabine), belong to the first class. These cytidine analogs irreversibly sequester DNMT proteins into DNA, resulting in global DNA hypomethylation and reactivation of silenced genes in cancers. Currently, 5-azacitidine and decitabine have been approved for treating AML, chronic myelomonocytic leukemia, and MDS.1331 However, these drugs have notable side effects, including cellular and clinical toxicity, as well as chemical instability.1331 Next-generation nucleoside analog DNMT inhibitors, such as guadecitabine, exhibit improved pharmacokinetic profiles with longer plasma half-life and lower peak plasma concentrations, leading to reduced toxicity. Guadecitabine has shown promising outcomes in clinical studies for AML treatment.1332 Another novel hypomethylating agent called NTX-301 has demonstrated superiority over conventional agents such as 5-azacitidine and decitabine in preclinical studies, which supports ongoing clinical development efforts.1333 Clinical trials with NTX-301 are currently underway (NCT04167917, NCT03366116, and NCT04851834).
Nonnucleoside DNMT inhibitors directly target the catalytic site of specific DNMT enzymes, unlike nucleoside analogs that are incorporated into DNA. Researchers have discovered reversible and selective nonnucleoside DNMT inhibitors, including RG108, SGI-1027, GSK3685032, DC-05, and CM-272.1334 Additionally, MG98 is an antisense oligodeoxynucleotide that targets the 3′UTR of DNMT1 mRNA to achieve DNA demethylation.1335 However, further preclinical and clinical studies are necessary to determine if their outcomes are more favorable than those of nucleoside analogs.
HDAC inhibitors
Many HDAC inhibitors target Zn2+ in the active site of HDACs to inhibit their enzymatic activity, leading to changes in chromatin structure and gene expression. Five HDAC inhibitors have been globally approved, including vorinostat (SAHA), romodepsin, belinostat, panobinostat (approved by the FDA), and chidamide (also known as tucidinostat, approved by the NMPA) for specific hematologic malignancies.1336 In 2006, the FDA approved the first HDAC inhibitor, vorinostat, for the clinical treatment of cutaneous T-cell lymphoma.1337 Additionally, the FDA has successively approved romidepsin, belinostat, and panobinostat as HDAC inhibitors for the treatment of peripheral T-cell lymphoma, T-cell lymphoma, and multiple myeloma, respectively.1336 Since these drugs are all pan-inhibitors of HDAC and may exhibit potential toxic side effects, the first selective HDAC subtype inhibitor, tucidinostat, has been approved for the treatment of recurrent or refractory PTCL and breast cancer.1338
Currently, available HDAC inhibitors are nonselective or pan-HDAC inhibitors, which have drawbacks such as poor efficacy on solid tumors, limited therapeutic efficacy, drug resistance, and toxicity.1339 Thus, developing HDAC inhibitors with better activity and higher selectivity is an important area of research. Three schemes for designing HDAC inhibitors include drug design based on zinc-binding groups, selective inhibitors targeting different subtypes of HDAC, and dual mechanism or multitarget HDAC inhibitors.1340 Many HDAC inhibitors have been synthesized based on these schemes, with more than ten entering clinical trials.1341 For example, abexinostat, a potent oral pan-HDAC inhibitor designed based on ZBG, is currently undergoing phase II clinical trials for recurrent/refractory DLBCL and follicular lymphoma as a single-drug treatment (NCT03936153 and NCT03934567). Additionally, the combination of abexinostat and pazopanib for locally advanced or metastatic RCC has entered the phase III trial (NCT03592472). Another instance is entinostat, a synthetic benzamide derivative HDAC inhibitor that selectively inhibits class I and IV HDAC enzymes.1342 Entinostat has been evaluated in several phase II trials in patients with breast cancer and has improved patient median OS (NCT00676663). Furthermore, the combination of entinostat and exemestane for advanced breast cancer has entered phase III clinical trials (NCT03538171 and NCT02115282).
HMT inhibitors
In addition to broad epigenetic reprogrammers such as DNMT inhibitors and HDAC inhibitors, targeted therapy has been developed for specific mutations in epigenome-modifying enzymes.1343 For instance, tazemetostat (EPZ-6438, E7438) is used for patients with EZH2 mutations. Tazemetostat is the only approved EZH2 inhibitor granted FDA approval in 2020 for treating epithelioid sarcoma and follicular lymphoma based on clinical trials (NCT01897571 and NCT02601950).1344 Ongoing clinical trials are assessing its efficacy against various hematologic malignancies and solid tumors (NCT05567679, NCT04179864, NCT05023655, NCT04705818, and NCT04846478). Studies have shown that loss of Bap1 in mice can increase EZH2 expression and H3K27me3 levels. Moreover, mesothelioma cells with BAP1 mutations have shown sensitivity to EZH2 inhibition, leading to clinical trials investigating the use of tazemetostat in treating malignant mesothelioma with BAP1 inactivation (NCT02860286).1345
GSK126 is a potent and highly selective EZH2 inhibitor that targets both wild-type EZH2 and Y641 mutant EZH2.1346 However, a phase I clinical trial assessing its safety, pharmacokinetics, pharmacodynamics, and clinical activity in patients with relapsed or refractory DLBCL, other NHLs, multiple myeloma, and solid tumors was terminated. The results showed insufficient antitumor activity and a relatively short half-life, which limited effective exposure and did not support further clinical study (NCT02082977). On the other hand, CPI-1205 is an orally bioavailable EZH2 inhibitor with a unique indole-based structure different from pyridine-based compounds such as GSK126 and tazemetostat.1347 Preclinical studies have demonstrated that CPI-1205 significantly inhibits tumor growth in DLBCL xenograft models, with good oral bioavailability and an acceptable safety profile in rats and dogs. Currently, CPI-1205 is undergoing evaluation in a phase I clinical trial in patients with B-cell lymphoma (NCT02395601), advanced solid tumors (NCT03525795), and metastatic castration-resistant prostate cancer (CRPC) (NCT03480646).
DOT1L, the sole identified H3K79 methyltransferase, has been targeted for cancer treatment, especially in acute leukemias with MLL gene rearrangements.1348 Pinometostat, the first clinical inhibitor of DOT1L, exhibits improved potency, longer plasma half-life, enhanced selectivity, and efficacy in reducing leukemic cell proliferation. Phase I clinical trials have been conducted for MLL-rearranged leukemia using pinometostat. Although well tolerated, pinometostat requires continuous IV infusion due to rapid clearance and shows modest clinical effectiveness (NCT02141828 and NCT01684150).1349,1350
HDM inhibitors
Preclinical studies have shown LSD1 inhibitor-dependent differentiation and growth inhibition, which has led to the initiation of several clinical trials to assess its efficacy. These trials include iadademstat (ORY-1001), INCB059872, IMG-7289, GSK2879552, seclidemstat, and pulrodemstat (CC-90011).1351
Oryzon Genomics developed ORY-1001 in 2012, which uses TCP as the lead compound and is currently being studied in clinical trials for AML and SCLC (NCT05546580 and NCT02913443).1352 INCB059872, an irreversible LSD1 inhibitor reported by Lee et al. in 2016, has undergone five clinical trials to test its safety and efficacy in treating solid tumors and hematologic malignancies (NCT02712905, NCT02959437, NCT03132324, NCT03514407, and NCT04061421). While one phase I study was terminated due to sickle cell disease risks, other trials have shown potential in treating various cancers. IMG-7289 (Biomedestat), developed by Imago BioSciences in 2018, is currently undergoing clinical trials for treating AML and MDS (NCT02842827), bone marrow fibrosis (NCT03136185) and essential thrombocythemia (NCT04081220). GSK2879552, another irreversible LSD1 inhibitor, has shown antitumor activity in AML and SCLC but had to be discontinued in three clinical trials due to unfavorable risk-benefit profiles (NCT02034123, NCT02177812, and NCT02929498).
Two reversible LSD1 inhibitors, CC-90011 and SP-2577 (Seclidemstat), have undergone preclinical studies and clinical trials, showing promising results for treating different types of tumors. Celgene developed CC-90011, the first reversible LSD1 inhibitor, for clinical trials targeting relapsed or refractory solid tumors and NHL (NCT02875223), as well as advanced solid and hematological tumors (NCT03850067, NCT04350463, NCT04628988, and NCT04748848). Salarius Pharmaceuticals developed SP-2577, another LSD1 reversible inhibitor, which showed a manageable safety profile in a phase I clinical trial for advanced solid tumors (NCT03895684). Additionally, clinical trials have been initiated to investigate the combination of SP-2577 with topotecan, cyclophosphamide (NCT03600649), and azacytidine (NCT04734990) for cancer treatment.
BET inhibitors
In contrast to targeting the enzymatic domains responsible for modifying epigenetic marks, an alternative strategy focuses on inhibiting the proteins that recognize these modifications by disrupting protein–protein interactions. A prime example of this innovative approach is BET inhibitors, which disrupt the interaction between bromodomains and acetylated lysine residues. This disruption interferes with the recruitment of transcriptional machinery to specific gene loci.1353
Initial small molecule BET inhibitors such as JQ1 have revealed the oncogenic role of BET proteins and their impact on oncogene expression, leading to observed antitumorigenic effects in preclinical models. However, their clinical application has been hindered by poor pharmacokinetics, short half-life, and low oral bioavailability.1354 TEN-010 (RO6870810), a JQ1 derivative with improved pharmacological properties, has undergone clinical trials for AML, MDS, and solid tumors (NCT02308761 and NCT01987362). Other BET inhibitors have also advanced to clinical trials. For instance, GSK525762, evaluated in multiple studies for hematologic malignancies and solid tumors, has shown encouraging results, especially in NUT midline carcinoma, AML, and TNBC patients (NCT01943851 and NCT01587703). Further research is needed to establish its efficacy and safety. OTX015, another BET inhibitor, has demonstrated favorable antitumor activity, particularly in combination therapies, in clinical evaluations for various hematologic malignancies and solid tumors (NCT01713582 and NCT02259114). CPI-0610, a selective BET inhibitor, has entered the clinical trial for hematologic malignancies such as myelofibrosis and lymphoma to evaluate its safety, tolerability, and potential efficacy as monotherapy and combination therapy (NCT01949883). Early studies have shown promising activity, but some patients experienced adverse events such as thrombocytopenia and moderate diarrhea (NCT02157636).
In addition, efforts have been made to design PROTACs and molecular glues that can degrade BET proteins by utilizing the intracellular ubiquitin proteasome system.1343 A clinical trial is currently underway for the FHD-609 degrader in the treatment of synovial sarcoma (NCT04965753), and the CFT8634 degrader has recently entered phase I/II clinical trials for synovial sarcoma and SMARCB1-null solid tumors (NCT05355753).
IDH inhibitors
Mutations in isocitrate dehydrogenases IDH1 and IDH2, commonly found in lower-grade gliomas as well as in AML and other malignancies, result in neomorphic enzyme activity, leading to increased production of 2-HG from α-KG.1355 2-HG serves as a competitive inhibitor of various α-KG-dependent dioxygenases, such as the Jumonji-C domain family of histone demethylases and the TET family of DNA demethylases, disrupting the global methylation landscape and promoting cancer development by impairing cellular differentiation.1356 Consequently, mutant IDH has emerged as an appealing therapeutic target, leading to the development of several IDH inhibitors aimed at counteracting the effects of 2-HG.
Enasidenib, the first FDA-approved IDH2-mutant inhibitor, received approval in 2017 after positive results from a single-arm trial on relapsed or refractory AML patients with IDH2 mutations (NCT01915498). Similarly, based on favorable outcomes observed in a clinical trial (NCT02074839), the FDA approved ivosidenib, an IDH1-mutant inhibitor, for relapsed or refractory AML patients with IDH1 mutations.1357 More recently, the combination of ivosidenib and azacitidine received FDA approval for newly diagnosed, IDH1-mutated AML following a phase III trial (NCT03173248).1358 In 2021, ivosidenib was also approved by the FDA for advanced cholangiocarcinoma with IDH1 mutation after a phase III clinical trial (NCT02989857).1359 These examples highlight the success of biomarker-driven approaches in treating cancers with epigenetic alterations, emphasizing the importance of considering chromatin changes when evaluating drug targets that indirectly modulate chromatin.
Another strategy to target IDH1 genetic alterations in gliomas is the development of vaccines. The most common IDH1 mutation found in gliomas is the Arg132 mutation, resulting in the production of a tumor-specific neoantigen called IDH1(R132H). Several IDH1(R132H)-specific peptide vaccines are currently undergoing testing as monotherapy or in combination with other therapies (NCT02454634, NCT03893903, NCT02193347, and NCT02771301).1360
Despite the theoretical significance and rationale of epigenetic therapy, there are still several issues that need to be discussed and resolved. The first issue relates to selectivity, specifically how to selectively target widely expressed epigenetic regulators. Epigenetic events are generally present in both normal and cancer cells. However, certain cancers rely on specific epigenetic changes and are sensitive to their regulation. It is crucial to identify the most critical epigenetic changes in different types of cancers. The second issue is the differential susceptibility of hematologic malignancies and solid tumors to epigenetic intervention. While significant progress has been made in epigenetic therapy for hematologic malignancies, solid tumors have been less responsive. The complexity of the oncogenome and inherent cellular differences between hematologic malignancies and solid tumors may contribute to this variation in efficacy. Understanding these biological principles is essential for expanding the application of epigenetic therapy to solid tumors. Moreover, the heterogeneity and plasticity of human cancer highlight the importance of personalized and precise epigenetic therapies. Precision medicine approaches, such as using high-throughput epigenomics sequencing technology, can help create genome and epigenome maps of individual patient’s tumor cells. These maps can then be used for drug sensitivity testing and screening, enabling optimized treatments tailored to each patient.
Targeting tumor microbiomes
Targeted interference with gut and tissue-resident microbiota or microorganism-derived products-based therapies are effective ways to target tumors854 (Table 13). Several strategies are applied to target gut and tumor microorganisms, including fecal microbial transplantation (FMT), single strains or designer consortia-based targeted microbial strategies, diet-based and prebiotic, probiotic, and postbiotic-based interventions, as well as targeted antibiotic approaches.854 FMT, a promising way to modulate the gut microbiome, acts by transplanting the entire gut microbial complement from a donor such as a healthy individual into a recipient such as a patient with cancer. The early phase I study of FMT revealed that it is beneficial to treat steroid-refractory GI tract graft-versus-host disease, which is a complication of hematopoietic stem cell transplantation to treat leukemia.854,1361 While FMT methods transplant the entire donor microbiota, the specific transplantation of single microbial species or designer microbial consortia to improve treatment is needed in certain circumstances. For example, CBM588, a formulation that includes a strain of Clostridium butyricum, improves the PFS of patients with RCC in combination with immune checkpoint blockade therapy.854,1362 Moreover, numerous dietary strategies, such as long-term caloric restriction, short-term starvation, and ketogenic diets, have shown the potential to enhance the efficacy of immunotherapy.854 The dietary interventions have been widely applied in cancer therapy for target gut and tumor microorganism. However, lack of rigorous standard procedures and poor association between diet and clinical effects have hindered their clinical application.854
Table 13.
The typical and clinically developed microbiota inhibitor
| Drug | Highest Phase | Indications | Identifier | Status |
|---|---|---|---|---|
| Bismuth colloidal pectin granules quadruple therapy | IV | Gastric cancer, helicobacter pylori infection | NCT04660123 | Completed |
| IV | Gastric cancer, helicobacter Pylori infection | NCT04209933 | Completed | |
| Itraconazole | IV | Hematologic neoplasms | NCT02895529 | Terminated |
Source: All the information is derived from ClinicalTrials.gov (https://www.clinicaltrials.gov)
In addition, the probiotic and postbiotic-based interventions and targeted antibiotic approaches also play significant roles in targeting gut and tumor microorganisms.854 A study has found that oral probiotic candidate DTA81 is effective in preventing the development of early CRC.853,1363 In addition, H. pylori infection is an important oncogenic factor in gastric cancer and gastric mucosa-associated lymphoid tissue lymphoma.1364 Combination therapy of antisecretory proton pump inhibitors with the antibiotics amoxicillin, levofloxacin, clarithromycin, and metronidazole is the standard protocol for H. pylori eradication.853,1364 In addition, phytomedicines and probiotics are also used to treat H. pylori infections.1365 Bismuth collisional tuberculosis has been used in clinical trials to treat the eradication rate of H. pylori infection in cancer patients, as well as to evaluate the improvement of symptoms and the incidence of adverse reactions.853 Itraconazole is a broad-spectrum triazole antifungal agent with favorable pharmacodynamic and pharmacokinetic profiles and is used for the prevention or treatment of systemic fungal infections.1366,1367 Itraconazole inhibits cell proliferation, invasion, and migration of oral OSCC cells by suppressing the Hedgehog pathway-induced cell cycle arrest and apoptosis.1368 Moreover, itraconazole can inhibit the proliferation and growth of CRC cells by promoting autophagy and apoptosis, and it is an effective treatment for CRC.1369 In summary, the microbiota is an important driver of cancer, and targeting the microbiota in the gut is meaningful in precision cancer care. However, the field requires further elucidation of the specific mechanisms by which microorganisms impact cancer processes. Shortly, targeting microbiota in the gut may emerge as a promising tool for cancer care.
Therapeutic strategies for targeting cellular senescence
Induction of tumor cell senescence has been demonstrated as one of the underlying mechanisms by which cancer therapies such as radiation, chemotherapy, and targeted therapy exert their antitumor activity. Paradoxically, lingering senescent cells (SnCs) in tumor tissues fuel tumor progression, relapse, and metastasis partly through the expression of the SASP.865 Based on the above observations, targeted therapeutics inducing tumor cell senescence followed by senolytics to selectively clear newly induced SnCs, which is called “one-two punch” cancer therapy, represent an emerging and promising new strategy in cancer treatment. Moreover, the application of senomorphic drugs which reduce the production and secretion of SASP factors has attracted attention in cancer therapy.1370,1371
Therapy-induced senescence (TIS)
The currently available targeted therapeutics for inducing senescence include blocking the cell cycle, triggering DNA damage, manipulating epigenetic modulators, and regulating tyrosine kinases. As cell cycle arrest is a hallmark of senescent cells, drugs that inhibit CDK or enhance levels of CDK inhibitor proteins are currently being used in senescence-inducing cancer therapy.1372 In particular, CDK4/6 inhibitors such as palbociclib, abemaciclib and ribociclib, which are approved by the FDA for the treatment of advanced breast cancer, are able to induce senescence in various cancer cells.864 PF-06873600, a triple CDK2/4/6 inhibitor, is a potent senescence inducer in various cancer models, and is ongoing in breast cancer in combination with endocrine therapy.1373 AURKs and PLKs, which are serine/threonine kinases essential for cell mitosis, are potential targets for senescence-inducing therapy.1370 Multiple PLK1 inhibitors, such as BI-6727, and AURK inhibitors, such as alisertib, are currently undergoing clinical investigation (NCT02273388 and NCT06095505).
Triggering DNA damage is another strategy to induce senescence. For example, PARP inhibitors, including veliparib and olaparib, induce a reversible senescent phenotype caused by BCL-XL mediated resistance to apoptosis in ovarian cancer, breast cancer, and prostate cancer. Inhibition of DNA replication through small molecule inhibition of the kinase CDC7 using XL413 or TAK-931 leads to senescence induction in liver cancers.1374 Inhibition of the telomerase complex has been identified as inducing replicative senescence in anticancer therapy. Imetelstat, GX301, and BIBR1532, potent telomerase inhibitors, effectively induce senescence and suppress cancer cell proliferation in preclinical or clinical trials.1370,1375
Another approach to induce senescence is by modulating the epigenome of cancer cells. Decitabine, a DNMT inhibitor, and vorinostat, an HDAC inhibitor, upregulate the expression of multiple tumor suppressor genes, such as CDKN2A and TP53, thus inducing cellular senescence via these pathways in various cancer cells.865,1376
Moreover, numerous other drugs and antibodies can induce senescence in cancer cells. Tamoxifen, an estrogen receptor antagonist, and bicalutamide, an androgen receptor antagonist, can induce senescence in breast cancer or prostate cancer.1377 Trastuzumab and pertuzumab, which are antibodies targeting HER2, cause senescence in breast cancer. BRAF and MEK inhibitors, such as vemurafenib and trametinib, show great senescence-inducing effects in melanoma.1378
Senolytics
Although TIS contributes to antitumor effects and treatment outcomes, increasing evidence has demonstrated that the accumulation of SnCs can stimulate the relapse and metastasis of cancers. Thus, selective clearance of SnCs with senolytics will prevent tumor relapse and metastasis, overcome drug resistance, and minimize toxic side effects.
To date, drugs or compounds targeting the apoptosis modulator BCL-2/BCL-XL, PI3K-AKT-mTOR, BET, tyrosine kinases, and GLS have exhibited promising effects on the clearance of senescent cells.865,1370 Apoptosis resistance is a feature shared by both cancer and senescent cells; thus, blocking antiapoptotic proteins could selectively eliminate senescent cells. ABT-737 was one of the first senolytics selectively targeting BCL-2, BCL-XL, and BCL-W, thus removing SnCs by reactivating the apoptotic pathway.475 After that, Navitoclax (ABT-263), a Bcl-2/Bcl-xL inhibitor, and venetoclax (ABT-199), a BCL-2 inhibitor, were developed and used as adjuvant therapies with radiation to selectively eliminate TIS cells, and increase the survival of tumors, including glioblastoma, melanoma, and lung cancer cells.1379,1380 In addition, the PI3K/AKT inhibitors dasatinib and quercetin selectively kill senescent cells and reduce the secretion of proinflammatory cytokines.862 An mTOR inhibitor significantly reduced the tumor burden and increased survival in xenograft cancer models after treatment with a DNA-replication kinase CDC7 inhibitor, which induced senescent liver and lung cancer cells.1374 Another drug screen identified the BET family protein degrader as a senolytic drug and validated that ARV825, a PROTAC of BET, possesses strong senolytic activity.1381
Immune-targeted therapy may be an effective way to clear senescent cells.862 For example, chimeric antigen receptor T cells targeting the cell surface protein, urokinase-type plasminogen activator receptor were found to be effective in clearing senescent cells after mice with lung adenocarcinomas were exposed to MEK and CDK4/6 inhibitors to induce senescence.1382
Senomorphics
Preventing the development of SASP or improving SASP-related functions can reduce inflammation and cancer risk. Multiple signaling pathways are involved in regulating SASP function, including p38/MAPK, JAK/STAT, mTOR, NF-κB, and C/EBP-β.1383 The inhibitors that modulate the excretion of SASP are called senomorphics which potentially preserves the SASP-dependent protumor effects of senescent cells and exert a synergistic antitumor effect.865 For example, IL-6 mAb siltuximab, multiple signal inhibitor metformin, and the JAK inhibitor ruxolitinib reduced SASP, thereby reducing the protumor and damage induced by SASP.865,1383 In addition, the protein arginine methyltransferase (PRMT1) induces SASP at the promoter of proinflammatory genes. The PRMT1 inhibitor TC-E increases the apoptosis sensitivity of cancer cells by regulating the NF-κB pathway.862 Rapamycin, an inhibitor of mTOR, has been found to reduce SASP by inhibiting mTOR and to limit the growth-promoting effect of senescent bystander fibroblasts on prostate cancers.1384,1385
Although targets and several senolytics have been discovered and tested in preclinical or clinical settings, the development of senolytics is still a challenge for the following reasons. First, TIS is heterogeneous and context (e.g., tissue of origin, time after treatment) dependent. Therefore, a deeper understanding of tumor contexts is critical for the usage and development of novel drugs that induce senescence. Second, the different types of prosenescence drugs induce cellular senescence via different mechanisms, suggesting that different TIS cells may require different senolytics. Moreover, the characteristics of SnCs, and the physiological and pathogenic effects of SnCs have not been adequately identified. Thus, the discovery of novel senolytic targets, senolytics, and the optimization of prosenescence therapy and senolytic combinations need further investigation. Third, senescence-inducing drugs cause senescence not only in tumor tissues but also in the TME and normal tissues. Induction of senescence may induce immune escape which decreases the anticancer effect or causes unwanted side effects associated with lingering senescent cells. The selectivity of senescence-inducing should be improved. Last, as surveillance of SnCs is executed by cytokines and chemokines which are released in a time-dependent manner, the timing of senolytic intervention is crucial for the efficacy of the one-two punch strategy and to avoid side effects on normal tissues. Taken together, despite the various challenges, senotherapies is a promising strategy and is likely to be applied in the clinic in the future as our understanding of senescence improves.
Conclusion and perspectives
“It is far more important to know what sort of person the disease has than what sort of disease the person has”, which is put forward by Hippocrates and precisely interprets the trend of cancer management. Current cancer management aims to combine molecular data with traditional clinical information, such as symptoms, personal history, and histology, to tailor medical care with the most benefit and minimize risk. Meaningfully, multifarious biomarkers are conducive to monitoring and predicting the therapeutic response of precision and personalized medicine in clinical practice,1386 which highlights the pivotal position of tumor biomarkers in cancer therapy.
Tumor biomarkers, biologically indicating pathogenic processes or pharmaceutical responses to therapeutic interventions,145 consist of six different types of biomarkers that are biomarkers of early detection, diagnosis, prognosis, prediction, therapeutic target, and surrogate end point.145 The biomarkers of early detection are essential in screening patients with cancers at an early stage. The diagnostic biomarkers mainly contribute to the identification of the presence and characteristics of cancers. The prognostic biomarkers indicate the disease outcome of patients to achieve individualized management, and the predictive biomarkers are utilized to demonstrate the treatment response of patients, thereby identifying the best therapy. Thus, prognostic biomarkers can identify patients who are at high-risk of cancer. The predictive biomarkers suggest the patients that can benefit from a specific therapy.114 In addition, biomarkers of therapeutic targets can identify the molecular targets of novel therapies. Moreover, surrogate endpoint-related biomarkers are used as substitutes for clinical endpoints or to assess clinical benefits, such as posttherapy PSA changes to evaluate drugs in the clinic.145 Herein, we mainly summarize the development history, detection methods, and classification of tumor biomarkers, thereby illustrating the crucial roles of tumor biomarkers in cancer screening, diagnosis, treatment, prognosis, and targeted therapy.
Although tumor biomarkers are increasingly critical in cancer precision medicine, the biomarkers surviving from discovery to clinical trials are small in number.1386 Some challenges are still urgently being solved in the future. First, the characteristics and concentration of tumor biomarkers are influenced by different biologic factors, such as posttherapy, host heterogeneity, age and the presence of other diseases among different individuals, false positive biomarkers generated by other physiologic or pathologic processes, and exogenous interfering substances, i.e., foods, drugs, and natural alternative therapies. Second, it is necessary and urgent to explore novel tools or technologies that could discover novel and accurate biomarkers for the detection of preneoplastic neoplasia, micrometastatic spread, and states of early or aggressive cancer recurrence. Third, limitations in analytical sensitivity and specificity still exist. Clinical detection and measurement assays of biomarkers require sufficient sensitivity and improved specificity. Standard procedures, clear guidelines, and quality control schemes are essential to ensure accuracy and reproducibility for biomarker development.145 Fourth, the high risk of false positives arises when identifying a biomarker from thousands of molecules. Thus, the cost of massive screening multiple times and the increased risk of false positives of overdiagnosis need to be balanced. Consistent adherence to publishing guidelines of tumor biomarkers can improve transparency and better judge the quality of putative biomarker identification.1386 Artificial intelligence offers an intriguing opportunity in large-scale screens of available data and to develop novel tumor biomarkers.1387 Finally, a rational combination of various biomarkers could improve the efficiency and accuracy of the application of biomarkers. Extensive research along with the use of new technologies needs to be performed.
In conclusion, we see great enthusiasm in tumor biomarker discovery and application from the large number of studies in the late century. Subsequently, the continuous research and development of innovative tumor biomarkers and the continuous development of novel detection technologies will make it possible for sensitive and specific tumor biomarkers to be gradually applied in clinical practice, and make early screening, diagnosis, treatment, and prognosis assessment of tumors a reality.
Acknowledgements
This study was supported by 135 Project for Disciplines of Excellence of West China Hospital Sichuan University (ZYGD23011), National Natural Sciences Foundation of China (882272716), Natural Sciences Foundation of Sichuan (2022NSFSC0053). We also thank Biorender.com for creating all the figures which have obtained publication licensing right in the manuscript.
Author contributions
Yue Zhou and Yinglan Zhao designed the review and revised the manuscript. Lei Tao, Jiahao Qiu, Jing Xu, Xinyu Yang, Yu Zhang, Xinyu Tian, and Xinqi Guan revised the manuscript and the tables and figures. Yinglan Zhao and Xiaobo Cen revised the draft. All authors have read and approved the review.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Yue Zhou, Lei Tao, Jiahao Qiu
References
- 1.Schmitt CA, Wang B, Demaria M. Senescence and cancer - role and therapeutic opportunities. Nat. Rev. Clin. Oncol. 2022;19:619–636. doi: 10.1038/s41571-022-00668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu L, Qu X. Cancer biomarker detection: recent achievements and challenges. Chem. Soc. Rev. 2015;44:2963–2997. doi: 10.1039/C4CS00370E. [DOI] [PubMed] [Google Scholar]
- 3.Sewpersad S, Pillay TS. Historical perspectives in clinical pathology: Bence Jones protein - early urine chemistry and the impact on modern day diagnostics. J. Clin. Pathol. 2021;74:212–215. doi: 10.1136/jclinpath-2020-206675. [DOI] [PubMed] [Google Scholar]
- 4.Gupta N, Sharma A, Sharma A. Emerging biomarkers in multiple myeloma: a review. Clin. Chim. Acta. 2020;503:45–53. doi: 10.1016/j.cca.2019.12.026. [DOI] [PubMed] [Google Scholar]
- 5.Talwar GP, Gupta JC, Shankar NV. Immunological approaches against human chorionic gonadotropin for control of fertility and therapy of advanced-stage cancers expressing hCG/subunits. Am. J. Reprod. Immunol. 2011;66:26–39. doi: 10.1111/j.1600-0897.2011.01002.x. [DOI] [PubMed] [Google Scholar]
- 6.Grenache DG. Progress in understanding the use of human chorionic gonadotropin as a tumor marker. Clin. Chem. Lab Med. 2020;58:323–325. doi: 10.1515/cclm-2019-1288. [DOI] [PubMed] [Google Scholar]
- 7.Markert CL, Møller F. Multiple forms of enzymes: tissue, ontogenetic, and species specific patterns. Proc. Natl Acad. Sci. USA. 1959;45:753–763. doi: 10.1073/pnas.45.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurisic V, Radenkovic S, Konjevic G. The actual role of LDH as tumor marker, biochemical and clinical aspects. Adv. Exp. Med. Biol. 2015;867:115–124. doi: 10.1007/978-94-017-7215-0_8. [DOI] [PubMed] [Google Scholar]
- 9.Meador CK, et al. Cause of Cushing’s syndrome in patients with tumors arising from ‘nonendocrine’ tissue. J. Clin. Endocrinol. Metab. 1962;22:693–703. doi: 10.1210/jcem-22-7-693. [DOI] [PubMed] [Google Scholar]
- 10.Khramkova NI, Abelev GI. Antigenic structure of mouse hepatomas. II. Preparation of monospecific antibodies to the organospecific liver antigens. Neoplasma. 1963;10:121–126. [PubMed] [Google Scholar]
- 11.Hu X, Chen R, Wei Q, Xu X. The landscape of alpha fetoprotein in hepatocellular carcinoma: where are we? Int. J. Biol. Sci. 2022;18:536–551. doi: 10.7150/ijbs.64537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Primus FJ, Freeman JW, Goldenberg DM. Immunological heterogeneity of carcinoembryonic antigen: purification from meconium of an antigen related to carcinoembryonic antigen. Cancer Res. 1983;43:679–685. [PubMed] [Google Scholar]
- 13.Grunnet, M. & Sorensen, J. B. Carcinoembryonic antigen (CEA) as tumor marker in lung cancer. Lung Cancer. 76, 138–143 (2012). [DOI] [PubMed]
- 14.Jia L, et al. Soluble POSTN is a novel biomarker complementing CA153 and CEA for breast cancer diagnosis and metastasis prediction. BMC Cancer. 2022;22:760. doi: 10.1186/s12885-022-09864-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lertkhachonsuk AA, et al. Serum CA19-9, CA-125 and CEA as tumor markers for mucinous ovarian tumors. J. Obstet. Gynaecol. Res. 2020;46:2287–2291. doi: 10.1111/jog.14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das V, Kalita J, Pal M. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed. Pharmacother. 2017;87:8–19. doi: 10.1016/j.biopha.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 17.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 18.Oren M. p53: not just a tumor suppressor. J. Mol. Cell Biol. 2019;11:539–543. doi: 10.1093/jmcb/mjz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 20.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 21.Levine AJ. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer. 2020;20:471–480. doi: 10.1038/s41568-020-0262-1. [DOI] [PubMed] [Google Scholar]
- 22.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat. Rev. Cancer. 2003;3:459–465. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 23.Downward J. Signal transduction. prelude to an anniversary for the RAS oncogene. Science. 2006;314:433–434. doi: 10.1126/science.1134727. [DOI] [PubMed] [Google Scholar]
- 24.FDA approves first KRAS inhibitor: Sotorasib. Cancer Discov. 11, OF4 (2021). [DOI] [PubMed]
- 25.Slack RS, Miller FD. Retinoblastoma gene in mouse neural development. Dev. Genet. 1996;18:81–91. doi: 10.1002/(SICI)1520-6408(1996)18:1<81::AID-DVG9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 26.Belczacka I, et al. Proteomics biomarkers for solid tumors: current status and future prospects. Mass Spectrom. Rev. 2019;38:49–78. doi: 10.1002/mas.21572. [DOI] [PubMed] [Google Scholar]
- 27.Merrick, B. A., London, R. E., Bushel, P. R., Grissom, S. F. & Paules, R. S. Platforms for biomarker analysis using high-throughput approaches in genomics, transcriptomics, proteomics, metabolomics, and bioinformatics. IARC Sci. Publ. 121–142 (2011). [PubMed]
- 28.Qi S, et al. High-resolution metabolomic biomarkers for lung cancer diagnosis and prognosis. Sci. Rep. 2021;11:11805. doi: 10.1038/s41598-021-91276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabaan AA, et al. Application of CRISPR/Cas9 technology in cancer treatment: a future direction. Curr. Oncol. 2023;30:1954–1976. doi: 10.3390/curroncol30020152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosseini SA, et al. CRISPR/Cas9 as precision and high-throughput genetic engineering tools in gastrointestinal cancer research and therapy. Int. J. Biol. Macromol. 2022;223:732–754. doi: 10.1016/j.ijbiomac.2022.11.018. [DOI] [PubMed] [Google Scholar]
- 31.Parola, C., Neumeier, D. & Reddy, S. T. Integrating high-throughput screening and sequencing for monoclonal antibody discovery and engineering. Immunology. 153, 31–41 (2018). [DOI] [PMC free article] [PubMed]
- 32.Wurdinger T, In ‘t Veld SGJG, Best MG. Platelet RNA as pan-tumor biomarker for cancer detection. Cancer Res. 2020;80:1371–1373. doi: 10.1158/0008-5472.CAN-19-3684. [DOI] [PubMed] [Google Scholar]
- 33.Song P, et al. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 2022;6:232–245. doi: 10.1038/s41551-021-00837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou J, Wang E. Cancer biomarker discovery for precision medicine: new progress. Curr. Med. Chem. 2019;26:7655–7671. doi: 10.2174/0929867325666180718164712. [DOI] [PubMed] [Google Scholar]
- 35.Ladabaum U, Dominitz JA, Kahi C, Schoen RE. Strategies for colorectal cancer screening. Gastroenterology. 2020;158:418–432. doi: 10.1053/j.gastro.2019.06.043. [DOI] [PubMed] [Google Scholar]
- 36.Ayoub WS, et al. Current status of hepatocellular carcinoma detection: screening strategies and novel biomarkers. Ther. Adv. Med. Oncol. 2019;11:1758835919869120. doi: 10.1177/1758835919869120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita K, Watanabe M. Clinical significance of tumor markers and an emerging perspective on colorectal cancer. Cancer Sci. 2009;100:195–199. doi: 10.1111/j.1349-7006.2008.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma S. Tumor markers in clinical practice: general principles and guidelines. Indian J. Med. Paediatr. Oncol. 2009;30:1–8. doi: 10.4103/0971-5851.56328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma M, Patel P, Verma M. Biomarkers in prostate cancer epidemiology. Cancers. 2011;3:3773–3798. doi: 10.3390/cancers3043773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petrelli F, et al. Prognostic role of lactate dehydrogenase in solid tumors: a systematic review and meta-analysis of 76 studies. Acta Oncol. 2015;54:961–970. doi: 10.3109/0284186X.2015.1043026. [DOI] [PubMed] [Google Scholar]
- 41.Zhang C, et al. Overview of microRNAs as diagnostic and prognostic biomarkers for high-incidence cancers in 2021. Int. J. Mol. Sci. 2022;23:11389. doi: 10.3390/ijms231911389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghazimoradi MH, Karimpour-Fard N, Babashah S. The promising role of non-coding RNAs as biomarkers and therapeutic targets for leukemia. Genes. 2023;14:131. doi: 10.3390/genes14010131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dai, J.-H., Tan, X.-R., Qiao, H. & Liu, N. Emerging clinical relevance of microbiome in cancer: promising biomarkers and therapeutic targets. Protein Cell10.1093/procel/pwad052 (2023). [DOI] [PMC free article] [PubMed]
- 44.Wang H, Wang Y, Zhang D, Li P. Circulating nucleosomes as potential biomarkers for cancer diagnosis and treatment monitoring. Int. J. Biol. Macromol. 2024;262:130005. doi: 10.1016/j.ijbiomac.2024.130005. [DOI] [PubMed] [Google Scholar]
- 45.Wen X, Pu H, Liu Q, Guo Z, Luo D. Circulating tumor DNA-A novel biomarker of tumor progression and its favorable detection techniques. Cancers. 2022;14:6025. doi: 10.3390/cancers14246025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin D, et al. Circulating tumor cells: biology and clinical significance. Signal Transduct. Target. Ther. 2021;6:404. doi: 10.1038/s41392-021-00817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwong GA, et al. Synthetic biomarkers: a twenty-first century path to early cancer detection. Nat. Rev. Cancer. 2021;21:655–668. doi: 10.1038/s41568-021-00389-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calabrese F, et al. Are there new biomarkers in tissue and liquid biopsies for the early detection of non-small cell lung cancer? J. Clin. Med. 2019;8:414. doi: 10.3390/jcm8030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bresalier RS, et al. Biomarkers for early detection of colorectal cancer: the early detection research network, a framework for clinical translation. Cancer Epidemiol. Biomark. Prev. 2020;29:2431–2440. doi: 10.1158/1055-9965.EPI-20-0234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol. Cancer. 2019;18:114. doi: 10.1186/s12943-019-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirata I. Evaluation of the usefulness of the simultaneous assay of fecal hemoglobin (Hb) and transferrin (Tf) in colorectal cancer screening - for the establishment of the Hb and Tf two-step cutoff assay (HTTC assay) Diagnosis. 2020;7:133–139. doi: 10.1515/dx-2019-0049. [DOI] [PubMed] [Google Scholar]
- 52.Sukumar J, Gast K, Quiroga D, Lustberg M, Williams N. Triple-negative breast cancer: promising prognostic biomarkers currently in development. Expert Rev. Anticancer Ther. 2021;21:135–148. doi: 10.1080/14737140.2021.1840984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi M, et al. Biomarkers for predicting efficacy of PD-1/PD-L1 inhibitors. Mol. Cancer. 2018;17:129. doi: 10.1186/s12943-018-0864-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moradi A, Srinivasan S, Clements J, Batra J. Beyond the biomarker role: prostate-specific antigen (PSA) in the prostate cancer microenvironment. Cancer Metastasis Rev. 2019;38:333–346. doi: 10.1007/s10555-019-09815-3. [DOI] [PubMed] [Google Scholar]
- 55.Kim DH, et al. The relationships between perioperative CEA, CA 19-9, and CA 72-4 and recurrence in gastric cancer patients after curative radical gastrectomy. J. Surg. Oncol. 2011;104:585–591. doi: 10.1002/jso.21919. [DOI] [PubMed] [Google Scholar]
- 56.Choi SR, et al. Role of serum tumor markers in monitoring for recurrence of gastric cancer following radical gastrectomy. Dig. Dis. Sci. 2006;51:2081–2086. doi: 10.1007/s10620-006-9166-5. [DOI] [PubMed] [Google Scholar]
- 57.Lu P, et al. Methylated septin 9 as a promising biomarker in the diagnosis and recurrence monitoring of colorectal cancer. Dis. Markers. 2022;2022:7087885. doi: 10.1155/2022/7087885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moding EJ, Nabet BY, Alizadeh AA, Diehn M. Detecting liquid remnants of solid tumors: circulating tumor DNA minimal residual disease. Cancer Discov. 2021;11:2968–2986. doi: 10.1158/2159-8290.CD-21-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, et al. Single-cell transcriptome-based multilayer network biomarker for predicting prognosis and therapeutic response of gliomas. Brief. Bioinform. 2020;21:1080–1097. doi: 10.1093/bib/bbz040. [DOI] [PubMed] [Google Scholar]
- 60.Goldsmith SJ. Radioimmunoassay: review of basic principles. Semin. Nucl. Med. 1975;5:125–152. doi: 10.1016/S0001-2998(75)80028-6. [DOI] [PubMed] [Google Scholar]
- 61.Grange RD, Thompson JP, Lambert DG, Mahajan RP. Radioimmunoassay, enzyme and non-enzyme-based immunoassays. Br. J. Anaesth. 2014;112:213–216. doi: 10.1093/bja/aet293. [DOI] [PubMed] [Google Scholar]
- 62.Hosoki H, Mori M, Takahara J, Daito M. Measurement of serum cortisol with 125I-cortisol radioimmunoassay. Horumon Rinsho. 1975;23:721–729. [PubMed] [Google Scholar]
- 63.Hack CE, et al. A modified competitive inhibition radioimmunoassay for the detection of C3a. Use of 125I-C3 instead of 125I-C3a. J. Immunol. Methods. 1988;108:77–84. doi: 10.1016/0022-1759(88)90405-X. [DOI] [PubMed] [Google Scholar]
- 64.Langone JJ. 125I-Labeled protein A: reactivity with IgG and use as a tracer in radioimmunoassay. Methods Enzymol. 1980;70:356–375. doi: 10.1016/S0076-6879(80)70064-2. [DOI] [PubMed] [Google Scholar]
- 65.Kim J-H, Lee S-Y, Lee S-K. Development of novel lab-on-a-chip platform for high-throughput radioimmunoassay. Appl. Radiat. Isot. 2021;168:109526. doi: 10.1016/j.apradiso.2020.109526. [DOI] [PubMed] [Google Scholar]
- 66.Darwish IA. Immunoassay methods and their applications in pharmaceutical analysis: basic methodology and recent advances. Int. J. Biomed. Sci. 2006;2:217–235. doi: 10.59566/IJBS.2006.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chester SJ, et al. A new radioimmunoassay detecting early stages of colon cancer: a comparison with CEA, AFP, and Ca 19-9. Dis. Markers. 1991;9:265–271. [PubMed] [Google Scholar]
- 68.Booth JC, et al. Comparison of enzyme-linked immunosorbent assay, radioimmunoassay, complement fixation, anticomplement immunofluorescence and passive haemagglutination techniques for detecting cytomegalovirus IgG antibody. J. Clin. Pathol. 1982;35:1345–1348. doi: 10.1136/jcp.35.12.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hemmilä I. Fluoroimmunoassays and immunofluorometric assays. Clin. Chem. 1985;31:359–370. doi: 10.1093/clinchem/31.3.359. [DOI] [PubMed] [Google Scholar]
- 70.Hicks JM. Fluorescence immunoassay. Hum. Pathol. 1984;15:112–116. doi: 10.1016/S0046-8177(84)80049-0. [DOI] [PubMed] [Google Scholar]
- 71.Huang X, et al. Ratiometric optical nanoprobes enable accurate molecular detection and imaging. Chem. Soc. Rev. 2018;47:2873–2920. doi: 10.1039/C7CS00612H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang RR, et al. Beyond the margins: real-time detection of cancer using targeted fluorophores. Nat. Rev. Clin. Oncol. 2017;14:347–364. doi: 10.1038/nrclinonc.2016.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cobb M, Gotcher S. Fluorescence immunoassay in the clinical laboratory. Am. J. Med. Technol. 1982;48:671–677. [PubMed] [Google Scholar]
- 74.Nishiyama K, et al. One-step non-competitive fluorescence polarization immunoassay based on a Fab fragment for C-reactive protein quantification. Sens. Actuators B Chem. 2021;326:128982. doi: 10.1016/j.snb.2020.128982. [DOI] [Google Scholar]
- 75.Nielsen K, Lin M, Gall D, Jolley M. Fluorescence polarization immunoassay: detection of antibody to Brucella abortus. Methods. 2000;22:71–76. doi: 10.1006/meth.2000.1038. [DOI] [PubMed] [Google Scholar]
- 76.Ullman EF, Schwarzberg M, Rubenstein KE. Fluorescent excitation transfer immunoassay. A general method for determination of antigens. J. Biol. Chem. 1976;251:4172–4178. doi: 10.1016/S0021-9258(17)33277-5. [DOI] [PubMed] [Google Scholar]
- 77.Diamandis EP. Immunoassays with time-resolved fluorescence spectroscopy: principles and applications. Clin. Biochem. 1988;21:139–150. doi: 10.1016/S0009-9120(88)80104-8. [DOI] [PubMed] [Google Scholar]
- 78.Tian J, et al. Multiplexed detection of tumor markers with multicolor quantum dots based on fluorescence polarization immunoassay. Talanta. 2012;92:72–77. doi: 10.1016/j.talanta.2012.01.051. [DOI] [PubMed] [Google Scholar]
- 79.Jehan Z, Uddin S, Al-Kuraya KS. In-situ hybridization as a molecular tool in cancer diagnosis and treatment. Curr. Med Chem. 2012;19:3730–3738. doi: 10.2174/092986712801661031. [DOI] [PubMed] [Google Scholar]
- 80.Veselinyová D, et al. Selected in situ hybridization methods: principles and application. Molecules. 2021;26:3874. doi: 10.3390/molecules26133874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Fox JL, Hsu PH, Legator MS, Morrison LE, Seelig SA. Fluorescence in situ hybridization: powerful molecular tool for cancer prognosis. Clin. Chem. 1995;41:1554–1559. doi: 10.1093/clinchem/41.11.1554. [DOI] [PubMed] [Google Scholar]
- 82.Chrzanowska NM, Kowalewski J, Lewandowska MA. Use of fluorescence in situ hybridization (FISH) in diagnosis and tailored therapies in solid tumors. Molecules. 2020;25:1864. doi: 10.3390/molecules25081864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shackelford RE, Vora M, Mayhall K, Cotelingam J. ALK-rearrangements and testing methods in non-small cell lung cancer: a review. Genes Cancer. 2014;5:1–14. doi: 10.18632/genesandcancer.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu L, et al. Fluorescence in situ hybridization (FISH): an increasingly demanded tool for biomarker research and personalized medicine. Biomark. Res. 2014;2:3. doi: 10.1186/2050-7771-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gökmen-Polar, Y. In Predictive Biomarkers in Oncology (eds Badve, S. & Kumar, G. L.) Ch. 5 (Springer International Publishing, 2019).
- 86.Saiki RK, et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 87.Cree IA. Diagnostic RAS mutation analysis by polymerase chain reaction (PCR) Biomol. Detect. Quantif. 2016;8:29–32. doi: 10.1016/j.bdq.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maris JM. Recent advances in neuroblastoma. N. Engl. J. Med. 2010;362:2202–2211. doi: 10.1056/NEJMra0804577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cheng H, et al. Ligand-targeted polymerase chain reaction for the detection of folate receptor-positive circulating tumour cells as a potential diagnostic biomarker for pancreatic cancer. Cell Prolif. 2020;53:e12880. doi: 10.1111/cpr.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gong S, et al. CRISPR/Cas-based in vitro diagnostic platforms for cancer biomarker detection. Anal. Chem. 2021;93:11899–11909. doi: 10.1021/acs.analchem.1c02533. [DOI] [PubMed] [Google Scholar]
- 91.Sanger F, et al. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977;265:687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- 92.Heather JM, Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107:1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Feng Y, Zhang Y, Ying C, Wang D, Du C. Nanopore-based fourth-generation DNA sequencing technology. Genomics. Proteom. Bioinforma. 2015;13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lappalainen T, Scott AJ, Brandt M, Hall IM. Genomic analysis in the age of human genome sequencing. Cell. 2019;177:70–84. doi: 10.1016/j.cell.2019.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Foox J, et al. Performance assessment of DNA sequencing platforms in the ABRF next-generation sequencing study. Nat. Biotechnol. 2021;39:1129–1140. doi: 10.1038/s41587-021-01049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kumar KR, Cowley MJ, Davis RL. Next-generation sequencing and emerging technologies. Semin. Thromb. Hemost. 2019;45:661–673. doi: 10.1055/s-0039-1688446. [DOI] [PubMed] [Google Scholar]
- 97.Kim RY, Xu H, Myllykangas S, Ji H. Genetic-based biomarkers and next-generation sequencing: the future of personalized care in colorectal cancer. Per. Med. 2011;8:331–345. doi: 10.2217/pme.11.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wood AC, et al. Evaluation of tumor DNA sequencing results in patients with gastric and gastroesophageal junction adenocarcinoma stratified by TP53 mutation status. Oncologist. 2022;27:307–313. doi: 10.1093/oncolo/oyac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548:297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kris MG, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pilgrim SM, Pain SJ, Tischkowitz MD. Opportunities and challenges of next-generation DNA sequencing for breast units. Br. J. Surg. 2014;101:889–898. doi: 10.1002/bjs.9458. [DOI] [PubMed] [Google Scholar]
- 102.Hong M, et al. RNA sequencing: new technologies and applications in cancer research. J. Hematol. Oncol. 2020;13:166. doi: 10.1186/s13045-020-01005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hussaini HM, Seo B, Rich AM. Immunohistochemistry and Immunofluorescence. Methods Mol. Biol. 2023;2588:439–450. doi: 10.1007/978-1-0716-2780-8_26. [DOI] [PubMed] [Google Scholar]
- 104.Zaha DC. Significance of immunohistochemistry in breast cancer. World J. Clin. Oncol. 2014;5:382–392. doi: 10.5306/wjco.v5.i3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fitzgibbons, P. L. & Cooper, K. In Basic Concepts of Molecular Pathology (eds Allen, T. C. & Cagle, P. T.) Ch. 14 (Springer, 2009).
- 106.Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol. Cancer Ther. 2015;14:847–856. doi: 10.1158/1535-7163.MCT-14-0983. [DOI] [PubMed] [Google Scholar]
- 107.Zhao S, et al. Molecular subtyping of triple-negative breast cancers by immunohistochemistry: molecular basis and clinical relevance. Oncologist. 2020;25:e1481–e1491. doi: 10.1634/theoncologist.2019-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tay TKY, et al. Correlating SS18-SSX immunohistochemistry (IHC) with SS18 fluorescent in situ hybridization (FISH) in synovial sarcomas: a study of 36 cases. Virchows Arch. 2021;479:785–793. doi: 10.1007/s00428-021-03135-0. [DOI] [PubMed] [Google Scholar]
- 109.Luu TT. Review of immunohistochemistry biomarkers in pancreatic cancer diagnosis. Front. Oncol. 2021;11:799025. doi: 10.3389/fonc.2021.799025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kashyap A, et al. Quantitative microimmunohistochemistry for the grading of immunostains on tumour tissues. Nat. Biomed. Eng. 2019;3:478–490. doi: 10.1038/s41551-019-0386-3. [DOI] [PubMed] [Google Scholar]
- 111.Hall PA, Lane DP. p53 in tumour pathology: can we trust immunohistochemistry?–Revisited! J. Pathol. 1994;172:1–4. doi: 10.1002/path.1711720103. [DOI] [PubMed] [Google Scholar]
- 112.Rodrigues VDC, et al. Analysis of scanning electron microscopy images to investigate adsorption processes responsible for detection of cancer biomarkers. ACS Appl. Mater. Interfaces. 2017;9:5885–5890. doi: 10.1021/acsami.6b16105. [DOI] [PubMed] [Google Scholar]
- 113.Slaoui M, Fiette L. Histopathology procedures: from tissue sampling to histopathological evaluation. Methods Mol. Biol. 2011;691:69–82. doi: 10.1007/978-1-60761-849-2_4. [DOI] [PubMed] [Google Scholar]
- 114.Rizk EM, et al. Prognostic and predictive immunohistochemistry-based biomarkers in cancer and immunotherapy. Hematol. Oncol. Clin. North Am. 2019;33:291–299. doi: 10.1016/j.hoc.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J. Hematol. Oncol. 2022;15:131. doi: 10.1186/s13045-022-01351-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cescon DW, Bratman SV, Chan SM, Siu LL. Circulating tumor DNA and liquid biopsy in oncology. Nat. Cancer. 2020;1:276–290. doi: 10.1038/s43018-020-0043-5. [DOI] [PubMed] [Google Scholar]
- 117.Zhou B, et al. Application of exosomes as liquid biopsy in clinical diagnosis. Signal Transduct. Target Ther. 2020;5:144. doi: 10.1038/s41392-020-00258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat. Rev. Clin. Oncol. 2021;18:297–312. doi: 10.1038/s41571-020-00457-x. [DOI] [PubMed] [Google Scholar]
- 119.Li S, et al. The role of exosomes in liquid biopsy for cancer diagnosis and prognosis prediction. Int. J. Cancer. 2021;148:2640–2651. doi: 10.1002/ijc.33386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yu W, et al. Exosome-based liquid biopsies in cancer: opportunities and challenges. Ann. Oncol. 2021;32:466–477. doi: 10.1016/j.annonc.2021.01.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tatischeff I. Extracellular vesicle-DNA: the next liquid biopsy biomarker for early cancer diagnosis? Cancers. 2023;15:1456. doi: 10.3390/cancers15051456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Muinelo-Romay L, Casas-Arozamena C, Abal M. Liquid biopsy in endometrial cancer: new opportunities for personalized oncology. Int. J. Mol. Sci. 2018;19:2311. doi: 10.3390/ijms19082311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kemper M, et al. Liquid biopsies in lung cancer. Cancers. 2023;15:1430. doi: 10.3390/cancers15051430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhao Y, et al. Liquid biopsy in pancreatic cancer - Current perspective and future outlook. Biochim. Biophys. Acta Rev. Cancer. 2023;1878:188868. doi: 10.1016/j.bbcan.2023.188868. [DOI] [PubMed] [Google Scholar]
- 125.Zhou H, et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol. Cancer. 2022;21:86. doi: 10.1186/s12943-022-01556-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ricciardi E, et al. Metastatic melanoma: liquid biopsy as a new precision medicine approach. Int. J. Mol. Sci. 2023;24:4014. doi: 10.3390/ijms24044014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Li M, et al. Liquid biopsy at the frontier in renal cell carcinoma: recent analysis of techniques and clinical application. Mol. Cancer. 2023;22:37. doi: 10.1186/s12943-023-01745-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arneth B. Update on the types and usage of liquid biopsies in the clinical setting: a systematic review. BMC Cancer. 2018;18:527. doi: 10.1186/s12885-018-4433-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhao, J., Yu, X., Shentu, X. & Li, D. The application and development of electron microscopy for three-dimensional reconstruction in life science: a review. Cell Tissue Res. 10.1007/s00441-024-03878-7 (2024). [DOI] [PubMed]
- 130.Stahlberg H, Walz T. Molecular electron microscopy: state of the art and current challenges. ACS Chem. Biol. 2008;3:268–281. doi: 10.1021/cb800037d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lambert, L. & Mulvey, T. In Advances in Imaging and Electron Physics (ed. Hawkes, P. W.) Vol. 95, Ch.1 (Elsevier, 1996).
- 132.Dey, P. In Basic and Advanced Laboratory Techniques in Histopathology and Cytology. Ch.28 (Springer Nature, 2022).
- 133.Sobel HJ, Marquet E. Usefulness of electron microscopy in the diagnosis of tumors. Pathol. Res. Pract. 1980;167:22–44. doi: 10.1016/S0344-0338(80)80180-4. [DOI] [PubMed] [Google Scholar]
- 134.Ordóñez NG, Mackay B. Electron microscopy in tumor diagnosis: indications for its use in the immunohistochemical era. Hum. Pathol. 1998;29:1403–1411. doi: 10.1016/S0046-8177(98)90008-9. [DOI] [PubMed] [Google Scholar]
- 135.Cohen Hyams T, Mam K, Killingsworth MC. Scanning electron microscopy as a new tool for diagnostic pathology and cell biology. Micron. 2020;130:102797. doi: 10.1016/j.micron.2019.102797. [DOI] [PubMed] [Google Scholar]
- 136.Ferlosio A, Orlandi A. The use of electron microscopy for the diagnosis of malignant pleural mesothelioma. J. Thorac. Dis. 2016;8:E1487–E1489. doi: 10.21037/jtd.2016.11.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Battifora H, Applebaum EL. Electron microscopy in the diagnosis of head and neck tumors. Head Neck Surg. 1979;1:202–212. doi: 10.1002/hed.2890010303. [DOI] [PubMed] [Google Scholar]
- 138.Fisher C, Path FRC, Flood LM, Ramsey AD. The role of electron microscopy in the diagnosis of tumours of the head and neck. J. Laryngol. Otol. 1992;106:403–408. doi: 10.1017/S002221510011967X. [DOI] [PubMed] [Google Scholar]
- 139.Wu Y, Deng W, Klinke DJ. Exosomes: improved methods to characterize their morphology, RNA content, and surface protein biomarkers. Analyst. 2015;140:6631–6642. doi: 10.1039/C5AN00688K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Albero-González R, et al. Complementary value of electron microscopy and immunohistochemistry in the diagnosis of non-small cell lung cancer: a potential role for electron microscopy in the era of targeted therapy. Ultrastruct. Pathol. 2019;43:237–247. doi: 10.1080/01913123.2019.1692118. [DOI] [PubMed] [Google Scholar]
- 141.Tao J, Bauer DE, Chiarle R. Assessing and advancing the safety of CRISPR-Cas tools: from DNA to RNA editing. Nat. Commun. 2023;14:212. doi: 10.1038/s41467-023-35886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Malekshoar M, et al. CRISPR-Cas9 targeted enrichment and next-generation sequencing for mutation detection. J. Mol. Diagn. 2023;25:249–262. doi: 10.1016/j.jmoldx.2023.01.010. [DOI] [PubMed] [Google Scholar]
- 143.Allemailem KS, et al. Current updates of CRISPR/Cas9-mediated genome editing and targeting within tumor cells: an innovative strategy of cancer management. Cancer Commun. 2022;42:1257–1287. doi: 10.1002/cac2.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Almeida RS, Wisnieski F, Takao Real Karia B, Smith MAC. CRISPR/Cas9 genome-editing technology and potential clinical application in gastric cancer. Genes. 2022;13:2029. doi: 10.3390/genes13112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bensalah K, Montorsi F, Shariat SF. Challenges of cancer biomarker profiling. Eur. Urol. 2007;52:1601–1609. doi: 10.1016/j.eururo.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 146.Bergstrand CG, CZAR B. Demonstration of a new protein fraction in serum from the human fetus. Scand. J. Clin. Lab. Invest. 1956;8:174. doi: 10.3109/00365515609049266. [DOI] [PubMed] [Google Scholar]
- 147.Tomasi TBJ. Structure and function of alpha-fetoprotein. Annu. Rev. Med. 1977;28:453–465. doi: 10.1146/annurev.me.28.020177.002321. [DOI] [PubMed] [Google Scholar]
- 148.Gold, P. & Freedman, S. O. Specific carcinoembryonic antigens of the human digestive system. J. Exp. Med. 122, 467–481 (1965). [DOI] [PMC free article] [PubMed]
- 149.Hammarström S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- 150.Mizejewski GJ. Biological roles of alpha-fetoprotein during pregnancy and perinatal development. Exp. Biol. Med. 2004;229:439–463. doi: 10.1177/153537020422900602. [DOI] [PubMed] [Google Scholar]
- 151.Johnson P, Zhou Q, Dao DY, Lo YMD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2022;19:670–681. doi: 10.1038/s41575-022-00620-y. [DOI] [PubMed] [Google Scholar]
- 152.Daniele B, Bencivenga A, Megna AS, Tinessa V. Alpha-fetoprotein and ultrasonography screening for hepatocellular carcinoma. Gastroenterology. 2004;127:S108–S112. doi: 10.1053/j.gastro.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 153.Galle PR, et al. Biology and significance of alpha-fetoprotein in hepatocellular carcinoma. Liver Int. 2019;39:2214–2229. doi: 10.1111/liv.14223. [DOI] [PubMed] [Google Scholar]
- 154.Gupta S, Bent S, Kohlwes J. Test characteristics of alpha-fetoprotein for detecting hepatocellular carcinoma in patients with hepatitis C. A systematic review and critical analysis. Ann. Intern. Med. 2003;139:46–50. doi: 10.7326/0003-4819-139-1-200307010-00012. [DOI] [PubMed] [Google Scholar]
- 155.Waidely E, Al-Yuobi ARO, Bashammakh AS, El-Shahawi MS, Leblanc RM. Serum protein biomarkers relevant to hepatocellular carcinoma and their detection. Analyst. 2016;141:36–44. doi: 10.1039/C5AN01884F. [DOI] [PubMed] [Google Scholar]
- 156.Johnson PJ. The role of serum alpha-fetoprotein estimation in the diagnosis and management of hepatocellular carcinoma. Clin. Liver Dis. 2001;5:145–159. doi: 10.1016/S1089-3261(05)70158-6. [DOI] [PubMed] [Google Scholar]
- 157.Thomas P, Toth CA, Saini KS, Jessup JM, Steele GJ. The structure, metabolism and function of the carcinoembryonic antigen gene family. Biochim. Biophys. Acta. 1990;1032:177–189. doi: 10.1016/0304-419x(90)90003-j. [DOI] [PubMed] [Google Scholar]
- 158.Fiebiger W, Wiltschke C. [Tumor markers] Acta Med. Austriaca. 2001;28:33–37. doi: 10.1046/j.1563-2571.2001.01008.x. [DOI] [PubMed] [Google Scholar]
- 159.Li M, et al. Recent progress in biosensors for detection of tumor biomarkers. Molecules. 2022;27:7327. doi: 10.3390/molecules27217327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang Y, et al. Serum carcinoembryonic antigen elevation in benign lung diseases. Sci. Rep. 2021;11:19044. doi: 10.1038/s41598-021-98513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Fan Y, Chen X, Li H. Clinical value of serum biomarkers CA153, CEA, and white blood cells in predicting sentinel lymph node metastasis of breast cancer. Int. J. Clin. Exp. Pathol. 2020;13:2889–2894. [PMC free article] [PubMed] [Google Scholar]
- 162.Lakemeyer L, et al. Diagnostic and prognostic value of CEA and CA19-9 in colorectal cancer. Diseases. 2021;9:21. doi: 10.3390/diseases9010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Pal M, Muinao T, Boruah HPD, Mahindroo N. Current advances in prognostic and diagnostic biomarkers for solid cancers: detection techniques and future challenges. Biomed. Pharmacother. 2022;146:112488. doi: 10.1016/j.biopha.2021.112488. [DOI] [PubMed] [Google Scholar]
- 164.Sørensen CG, Karlsson WK, Pommergaard HC, Burcharth J, Rosenberg J. The diagnostic accuracy of carcinoembryonic antigen to detect colorectal cancer recurrence - a systematic review. Int. J. Surg. 2016;25:134–144. doi: 10.1016/j.ijsu.2015.11.065. [DOI] [PubMed] [Google Scholar]
- 165.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Charakorn C, et al. The association between serum squamous cell carcinoma antigen and recurrence and survival of patients with cervical squamous cell carcinoma: a systematic review and meta-analysis. Gynecol. Oncol. 2018;150:190–200. doi: 10.1016/j.ygyno.2018.03.056. [DOI] [PubMed] [Google Scholar]
- 167.Markovina S, et al. Serum squamous cell carcinoma antigen as an early indicator of response during therapy of cervical cancer. Br. J. Cancer. 2018;118:72–78. doi: 10.1038/bjc.2017.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kato H, Nagaya T, Torigoe T. Heterogeneity of a tumor antigen TA-4 of squamous cell carcinoma in relation to its appearance in the circulation. Gan. 1984;75:433–435. [PubMed] [Google Scholar]
- 169.Zhu H. Squamous cell carcinoma antigen: clinical application and research status. Diagnostics. 2022;12:1065. doi: 10.3390/diagnostics12051065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yang Y, et al. Clinical use of tumor biomarkers in prediction for prognosis and chemotherapeutic effect in esophageal squamous cell carcinoma. BMC Cancer. 2019;19:526. doi: 10.1186/s12885-019-5755-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Travassos DC, Fernandes D, Massucato EMS, Navarro CM, Bufalino A. Squamous cell carcinoma antigen as a prognostic marker and its correlation with clinicopathological features in head and neck squamous cell carcinoma: systematic review and meta-analysis. J. Oral Pathol. Med. 2018;47:3–10. doi: 10.1111/jop.12600. [DOI] [PubMed] [Google Scholar]
- 172.Polito M, et al. Serum markers for monitoring of prostatic carcinoma. Prostate. 1997;33:208–216. doi: 10.1002/(SICI)1097-0045(19971101)33:3<208::AID-PROS10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 173.Ahn SK, et al. Preoperative serum tissue polypeptide-specific antigen is a valuable prognostic marker in breast cancer. Int. J. Cancer. 2013;132:875–881. doi: 10.1002/ijc.27727. [DOI] [PubMed] [Google Scholar]
- 174.Berglund Å, Molin D, Larsson A, Einarsson R, Glimelius B. Tumour markers as early predictors of response to chemotherapy in advanced colorectal carcinoma. Ann. Oncol. 2002;13:1430–1437. doi: 10.1093/annonc/mdf220. [DOI] [PubMed] [Google Scholar]
- 175.Buccheri G, Ferrigno D. Lung tumor markers of cytokeratin origin: an overview. Lung Cancer. 2001;34:S65–S69. doi: 10.1016/S0169-5002(01)00347-6. [DOI] [PubMed] [Google Scholar]
- 176.van Dalen A. TPS in breast cancer–a comparative study with carcinoembryonic antigen and CA 15-3. Tumour Biol. 1992;13:10–17. doi: 10.1159/000217747. [DOI] [PubMed] [Google Scholar]
- 177.Valik D, Nekulova M. Serum tissue polypeptide-specific antigen (TPS): what is its diagnostic value? Br. J. Cancer. 2000;82:1756–175. doi: 10.1054/bjoc.1999.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zhang J, Wei Q, Dong D, Ren L. The role of TPS, CA125, CA15-3 and CEA in prediction of distant metastasis of breast cancer. Clin. Chim. Acta. 2021;523:19–25. doi: 10.1016/j.cca.2021.08.027. [DOI] [PubMed] [Google Scholar]
- 179.Xie S, Ding X, Mo W, Chen J. Serum tissue polypeptide-specific antigen is an independent predictor in breast cancer. Acta Histochem. 2014;116:372–376. doi: 10.1016/j.acthis.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 180.Kornek G, Schenk T, Raderer M, Djavarnmad M, Scheithauer W. Tissue polypeptide-specific antigen (TPS) in monitoring palliative treatment response of patients with gastrointestinal tumours. Br. J. Cancer. 1995;71:182–185. doi: 10.1038/bjc.1995.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chang C-H, et al. Tissue polypeptide specific antigen (TPS) as a tumor marker in renal cell carcinoma. Anticancer Res. 2002;22:2949–2950. [PubMed] [Google Scholar]
- 182.Noh, D. -Y., Ahn, S. K., Moon, H. -G., Han, W. & Kim, J. In Biomarkers in Cancer (eds Preedy, V. R. & Patel, V. B.) Ch.19 (Springer, 2015).
- 183.Inaba N, et al. Immunoradiometrical measurement of tissue polypeptide specific antigen (TPS) in normal, healthy, nonpregnant and pregnant Japanese women. Asia Ocean. J. Obstet. Gynaecol. 1993;19:459–466. doi: 10.1111/j.1447-0756.1993.tb00407.x. [DOI] [PubMed] [Google Scholar]
- 184.Wang MC, Valenzuela LA, Murphy GP, Chu TM. Purification of a human prostate specific antigen. J. Urol. 2017;197:S148–S152. doi: 10.1016/j.juro.2016.10.100. [DOI] [PubMed] [Google Scholar]
- 185.Prensner JR, Rubin MA, Wei JT, Chinnaiyan AM. Beyond PSA: the next generation of prostate cancer biomarkers. Sci. Transl. Med. 2012;4:127rv3. doi: 10.1126/scitranslmed.3003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Duffy MJ. Biomarkers for prostate cancer: prostate-specific antigen and beyond. Clin. Chem. Lab. Med. 2020;58:326–339. doi: 10.1515/cclm-2019-0693. [DOI] [PubMed] [Google Scholar]
- 187.Adamaki M, Zoumpourlis V. Prostate cancer biomarkers: from diagnosis to prognosis and precision-guided therapeutics. Pharm. Ther. 2021;228:107932. doi: 10.1016/j.pharmthera.2021.107932. [DOI] [PubMed] [Google Scholar]
- 188.Van Poppel H, et al. Serum PSA-based early detection of prostate cancer in Europe and globally: past, present and future. Nat. Rev. Urol. 2022;19:562–572. doi: 10.1038/s41585-022-00638-6. [DOI] [PubMed] [Google Scholar]
- 189.Kouriefs C, Sahoyl M, Grange P, Muir G. Prostate specific antigen through the years. Arch. Ital. di Urol. 2009;81:195–198. [PubMed] [Google Scholar]
- 190.Terada N, et al. Prognostic and predictive biomarkers in prostate cancer: latest evidence and clinical implications. Ther. Adv. Med. Oncol. 2017;9:565–573. doi: 10.1177/1758834017719215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yan P, et al. Prognostic value of neuron-specific enolase in patients with advanced and metastatic non-neuroendocrine non-small cell lung cancer. Biosci. Rep. 2021;41:BSR20210866. doi: 10.1042/BSR20210866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Kaiser E, Kuzmits R, Pregant P, Burghuber O, Worofka W. Clinical biochemistry of neuron specific enolase. Clin. Chim. Acta. 1989;183:13–31. doi: 10.1016/0009-8981(89)90268-4. [DOI] [PubMed] [Google Scholar]
- 193.Thelin EP, et al. Utility of neuron-specific enolase in traumatic brain injury; relations to S100B levels, outcome, and extracranial injury severity. Crit. Care. 2016;20:285. doi: 10.1186/s13054-016-1450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ferraro S, et al. Measurement of serum neuron-specific enolase in neuroblastoma: is there a clinical role? Clin. Chem. 2020;66:667–675. doi: 10.1093/clinchem/hvaa073. [DOI] [PubMed] [Google Scholar]
- 195.Reifenberger G, Szymas J, Wechsler W. Differential expression of glial- and neuronal-associated antigens in human tumors of the central and peripheral nervous system. Acta Neuropathol. 1987;74:105–123. doi: 10.1007/BF00692841. [DOI] [PubMed] [Google Scholar]
- 196.Yang G, et al. Recent advances in biosensor for detection of lung cancer biomarkers. Biosens. Bioelectron. 2019;141:111416. doi: 10.1016/j.bios.2019.111416. [DOI] [PubMed] [Google Scholar]
- 197.Schneider J. Tumor markers in detection of lung cancer. Adv. Clin. Chem. 2006;42:1–41. doi: 10.1016/S0065-2423(06)42001-1. [DOI] [PubMed] [Google Scholar]
- 198.Yu D, Du K, Liu T, Chen G. Prognostic value of tumor markers, NSE, CA125 and SCC, in operable NSCLC Patients. Int. J. Mol. Sci. 2013;14:11145–11156. doi: 10.3390/ijms140611145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ferrigno D, Buccheri G, Giordano C. Neuron-specific enolase is an effective tumour marker in non-small cell lung cancer (NSCLC) Lung Cancer. 2003;41:311–320. doi: 10.1016/S0169-5002(03)00232-0. [DOI] [PubMed] [Google Scholar]
- 200.Pujol JL, Boher JM, Grenier J, Quantin X. Cyfra 21-1, neuron specific enolase and prognosis of non-small cell lung cancer: prospective study in 621 patients. Lung Cancer. 2001;31:221–231. doi: 10.1016/S0169-5002(00)00186-0. [DOI] [PubMed] [Google Scholar]
- 201.Zhang C, et al. Alpha-L-fucosidase has diagnostic value in prostate cancer with ‘gray-zone PSA’ and inhibits cancer progression via regulating glycosylation. Front. Oncol. 2021;11:742354. doi: 10.3389/fonc.2021.742354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Giardina MG, et al. Serum alpha-L-fucosidase. A useful marker in the diagnosis of hepatocellular carcinoma. Cancer. 1992;70:1044–1048. doi: 10.1002/1097-0142(19920901)70:5<1044::AID-CNCR2820700506>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 203.Fawzy Montaser M, Amin Sakr M, Omar Khalifa M. Alpha-L-fucosidase as a tumour marker of hepatocellular carcinoma. Arab J. Gastroenterol. 2012;13:9–13. doi: 10.1016/j.ajg.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 204.Giardina MG, et al. Serum alpha-L-fucosidase activity and early detection of hepatocellular carcinoma: a prospective study of patients with cirrhosis. Cancer. 1998;83:2468–2474. doi: 10.1002/(SICI)1097-0142(19981215)83:12<2468::AID-CNCR9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 205.Wang K, et al. Alpha-1-fucosidase as a prognostic indicator for hepatocellular carcinoma following hepatectomy: a large-scale, long-term study. Br. J. Cancer. 2014;110:1811–1819. doi: 10.1038/bjc.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Xing H, et al. Clinical performance of α-L-fucosidase for early detection of hepatocellular carcinoma. Biomark. Med. 2019;13:545–555. doi: 10.2217/bmm-2018-0414. [DOI] [PubMed] [Google Scholar]
- 207.Yu X, et al. Alpha-l-fucosidase: a novel serum biomarker to predict prognosis in early stage esophageal squamous cell carcinoma. J. Thorac. Dis. 2019;11:3980–3990. doi: 10.21037/jtd.2019.08.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Liu D, et al. Diagnostic value of 5 serum biomarkers for hepatocellular carcinoma with different epidemiological backgrounds: a large-scale, retrospective study. Cancer Biol. Med. 2021;18:256–270. doi: 10.20892/j.issn.2095-3941.2020.0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Khan AA, Allemailem KS, Alhumaydhi FA, Gowder SJT, Rahmani AH. The biochemical and clinical perspectives of lactate dehydrogenase: an enzyme of active metabolism. Endocr. Metab. Immune Disord. Drug Targets. 2020;20:855–868. doi: 10.2174/1871530320666191230141110. [DOI] [PubMed] [Google Scholar]
- 210.Certo M, Tsai C-H, Pucino V, Ho P-C, Mauro C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2021;21:151–161. doi: 10.1038/s41577-020-0406-2. [DOI] [PubMed] [Google Scholar]
- 211.Akins NS, Nielson TC, Le HV. Inhibition of glycolysis and glutaminolysis: an emerging drug discovery approach to combat cancer. Curr. Top. Med. Chem. 2018;18:494–504. doi: 10.2174/1568026618666180523111351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Young A, Oldford C, Mailloux RJ. Lactate dehydrogenase supports lactate oxidation in mitochondria isolated from different mouse tissues. Redox Biol. 2020;28:101339. doi: 10.1016/j.redox.2019.101339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Abbaszadeh Z, Çeşmeli S, Biray Avcı Ç. Crucial players in glycolysis: cancer progress. Gene. 2020;726:144158. doi: 10.1016/j.gene.2019.144158. [DOI] [PubMed] [Google Scholar]
- 214.Mishra D, Banerjee D. Lactate dehydrogenases as metabolic links between tumor and stroma in the tumor microenvironment. Cancers. 2019;11:750. doi: 10.3390/cancers11060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Augoff K, Hryniewicz-Jankowska A, Tabola R. Lactate dehydrogenase 5: an old friend and a new hope in the war on cancer. Cancer Lett. 2015;358:1–7. doi: 10.1016/j.canlet.2014.12.035. [DOI] [PubMed] [Google Scholar]
- 216.Pei Y-Y, et al. Lactate dehydrogenase as promising marker for prognosis of brain metastasis. J. Neurooncol. 2022;159:359–368. doi: 10.1007/s11060-022-04070-z. [DOI] [PubMed] [Google Scholar]
- 217.Li J, et al. Suppression of lactate dehydrogenase A compromises tumor progression by downregulation of the Warburg effect in glioblastoma. Neuroreport. 2016;27:110–115. doi: 10.1097/WNR.0000000000000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wei Y, et al. Prognostic significance of serum lactic acid, lactate dehydrogenase, and albumin levels in patients with metastatic colorectal cancer. Biomed. Res. Int. 2018;2018:1804086. doi: 10.1155/2018/1804086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhou G-Q, et al. Baseline serum lactate dehydrogenase levels for patients treated with intensity-modulated radiotherapy for nasopharyngeal carcinoma: a predictor of poor prognosis and subsequent liver metastasis. Int. J. Radiat. Oncol. Biol. Phys. 2012;82:e359–e365. doi: 10.1016/j.ijrobp.2011.06.1967. [DOI] [PubMed] [Google Scholar]
- 220.Zhang J, et al. Prognostic value of pretreatment serum lactate dehydrogenase level in patients with solid tumors: a systematic review and meta-analysis. Sci. Rep. 2015;5:9800. doi: 10.1038/srep09800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Sartor O, et al. An exploratory analysis of alkaline phosphatase, lactate dehydrogenase, and prostate-specific antigen dynamics in the phase 3 ALSYMPCA trial with radium-223. Ann. Oncol. 2017;28:1090–1097. doi: 10.1093/annonc/mdx044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Colgan SM, Mukherjee S, Major P. Hypoxia-induced lactate dehydrogenase expression and tumor angiogenesis. Clin. Colorectal Cancer. 2007;6:442–446. doi: 10.3816/CCC.2007.n.014. [DOI] [PubMed] [Google Scholar]
- 223.Comandatore A, et al. Lactate dehydrogenase and its clinical significance in pancreatic and thoracic cancers. Semin. Cancer Biol. 2022;86:93–100. doi: 10.1016/j.semcancer.2022.09.001. [DOI] [PubMed] [Google Scholar]
- 224.Cho J, et al. The prognostic role of tumor associated glycoprotein 72 (TAG-72) in stage II and III colorectal adenocarcinoma. Pathol. Res. Pract. 2019;215:171–176. doi: 10.1016/j.prp.2018.10.024. [DOI] [PubMed] [Google Scholar]
- 225.Li M, Men X, Zhang X. Diagnostic value of carbohydrate antigen 72-4 combined with carbohydrate antigen 15.3 in ovarian cancer, cervical cancer and endometrial cancer. J. Buon. 2020;25:1918–1927. [PubMed] [Google Scholar]
- 226.Mariampillai AI, et al. Cancer antigen 72-4 for the monitoring of advanced tumors of the gastrointestinal tract, lung, breast and ovaries. Anticancer Res. 2017;37:3649–3656. doi: 10.21873/anticanres.11735. [DOI] [PubMed] [Google Scholar]
- 227.Guadagni F, et al. CA 72-4 measurement of tumor-associated glycoprotein 72 (TAG-72) as a serum marker in the management of gastric carcinoma. Cancer Res. 1992;52:1222–1227. [PubMed] [Google Scholar]
- 228.Chen X-Z, et al. Correlation between serum CA724 and gastric cancer: multiple analyses based on Chinese population. Mol. Biol. Rep. 2012;39:9031–9039. doi: 10.1007/s11033-012-1774-x. [DOI] [PubMed] [Google Scholar]
- 229.Xu Y, Zhang P, Zhang K, Huang C. The application of CA72-4 in the diagnosis, prognosis, and treatment of gastric cancer. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188634. doi: 10.1016/j.bbcan.2021.188634. [DOI] [PubMed] [Google Scholar]
- 230.Zhang Y, Zhang M, Bai X, Li C, Zhang L. Increased serum CA724 levels in patients suffering gout vs cancers. Prog. Mol. Biol. Transl. Sci. 2019;162:177–186. doi: 10.1016/bs.pmbts.2018.12.005. [DOI] [PubMed] [Google Scholar]
- 231.Yang A-P, et al. CA72-4 combined with CEA, CA125 and CAl9-9 improves the sensitivity for the early diagnosis of gastric cancer. Clin. Chim. Acta. 2014;437:183–186. doi: 10.1016/j.cca.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 232.Bast RC, et al. Reactivity of a monoclonal antibody with human ovarian carcinoma. J. Clin. Invest. 1981;68:1331–1337. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Diamandis EP, Bast RCJ, Gold P, Chu TM, Magnani JL. Reflection on the discovery of carcinoembryonic antigen, prostate-specific antigen, and cancer antigens CA125 and CA19-9. Clin. Chem. 2013;59:22–31. doi: 10.1373/clinchem.2012.187047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zhang M, Cheng S, Jin Y, Zhao Y, Wang Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim. Biophys. Acta Rev. Cancer. 2021;1875:188503. doi: 10.1016/j.bbcan.2021.188503. [DOI] [PubMed] [Google Scholar]
- 235.Rustin GJS, et al. Definitions for response and progression in ovarian cancer clinical trials incorporating recist 1.1 and CA 125 agreed by the gynecological cancer intergroup (GCIG) Int. J. Gynecol. Cancer. 2011;21:419–423. doi: 10.1097/IGC.0b013e3182070f17. [DOI] [PubMed] [Google Scholar]
- 236.Franier BDL, Thompson M. Early stage detection and screening of ovarian cancer: a research opportunity and significant challenge for biosensor technology. Biosens. Bioelectron. 2019;135:71–81. doi: 10.1016/j.bios.2019.03.041. [DOI] [PubMed] [Google Scholar]
- 237.Feng F, et al. Diagnostic and prognostic value of CEA, CA19–9, AFP and CA125 for early gastric cancer. BMC Cancer. 2017;17:737. doi: 10.1186/s12885-017-3738-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Bast RCJ, et al. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. N. Engl. J. Med. 1983;309:883–887. doi: 10.1056/NEJM198310133091503. [DOI] [PubMed] [Google Scholar]
- 239.Akinwunmi BO, et al. Chronic medical conditions and CA125 levels among women without ovarian cancer. Cancer Epidemiol. Biomark. Prev. 2018;27:1483–1490. doi: 10.1158/1055-9965.EPI-18-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Crosby DA, Glover LE, Martyn F, Wingfield M. CA125 measured during menstruation can be misleading. Ir. Med. J. 2018;111:738. [PubMed] [Google Scholar]
- 241.Haglund C, Lundin J, Kuusela P, Roberts PJ. CA 242, a new tumour marker for pancreatic cancer: a comparison with CA 19-9, CA 50 and CEA. Br. J. Cancer. 1994;70:487–492. doi: 10.1038/bjc.1994.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Dou H, Sun G, Zhang L. CA242 as a biomarker for pancreatic cancer and other diseases. Prog. Mol. Biol. Transl. Sci. 2019;162:229–239. doi: 10.1016/bs.pmbts.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 243.Tang Y, Cui Y, Zhang S, Zhang L. The sensitivity and specificity of serum glycan-based biomarkers for cancer detection. Prog. Mol. Biol. Transl. Sci. 2019;162:121–140. doi: 10.1016/bs.pmbts.2019.01.010. [DOI] [PubMed] [Google Scholar]
- 244.Ni XG, et al. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. Eur. J. Surg. Oncol. 2005;31:164–169. doi: 10.1016/j.ejso.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 245.Ozkan H, Kaya M, Cengiz A. Comparison of tumor marker CA 242 with CA 19-9 and carcinoembryonic antigen (CEA) in pancreatic cancer. Hepatogastroenterology. 2003;50:1669–1674. [PubMed] [Google Scholar]
- 246.Nilsson O, et al. Sensitivity and specificity of CA242 in gastro-intestinal cancer. A comparison with CEA, CA50 and CA 19-9. Br. J. Cancer. 1992;65:215–221. doi: 10.1038/bjc.1992.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Pasanen PA, et al. Clinical evaluation of a new serum tumour marker CA 242 in pancreatic carcinoma. Br. J. Cancer. 1992;65:731–734. doi: 10.1038/bjc.1992.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Nath S, Mukherjee P. MUC1: a multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014;20:332–342. doi: 10.1016/j.molmed.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Gendler SJ. MUC1, the renaissance molecule. J. Mammary Gland Biol. Neoplasia. 2001;6:339–353. doi: 10.1023/A:1011379725811. [DOI] [PubMed] [Google Scholar]
- 250.Yousefi M, et al. Aptasensors as a new sensing technology developed for the detection of MUC1 mucin: a review. Biosens. Bioelectron. 2019;130:1–19. doi: 10.1016/j.bios.2019.01.015. [DOI] [PubMed] [Google Scholar]
- 251.Price MR. High molecular weight epithelial mucins as markers in breast cancer. Eur. J. Cancer Clin. Oncol. 1988;24:1799–1804. doi: 10.1016/0277-5379(88)90088-0. [DOI] [PubMed] [Google Scholar]
- 252.Seale KN, Tkaczuk KHR. Circulating biomarkers in breast cancer. Clin. Breast Cancer. 2022;22:e319–e331. doi: 10.1016/j.clbc.2021.09.006. [DOI] [PubMed] [Google Scholar]
- 253.Duffy MJ. Serum tumor markers in breast cancer: are they of clinical value? Clin. Chem. 2006;52:345–351. doi: 10.1373/clinchem.2005.059832. [DOI] [PubMed] [Google Scholar]
- 254.Lindholm L, et al. Monoclonal antibodies against gastrointestinal tumour-associated antigens isolated as monosialogangliosides. Int. Arch. Allergy Appl. Immunol. 1983;71:178–181. doi: 10.1159/000233384. [DOI] [PubMed] [Google Scholar]
- 255.Kawa S, et al. Elevated serum levels of dupan-2 in pancreatic cancer patients negative for lewis blood group phenotype. Br. J. Cancer. 1991;64:899–902. doi: 10.1038/bjc.1991.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Shan M, Tian Q, Zhang L. Serum CA50 levels in patients with cancers and other diseases. Prog. Mol. Biol. Transl. Sci. 2019;162:187–198. doi: 10.1016/bs.pmbts.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 257.Bouhours JF, Bouhours D, Hansson GC. Developmental changes of gangliosides of the rat stomach. Appearance of a blood group B-active ganglioside. J. Biol. Chem. 1987;262:16370–16375. doi: 10.1016/S0021-9258(18)49265-4. [DOI] [PubMed] [Google Scholar]
- 258.Steinberg W. The clinical utility of the CA 19-9 tumor-associated antigen. Am. J. Gastroenterol. 1990;85:350–355. [PubMed] [Google Scholar]
- 259.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014;371:2140–2141. doi: 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 260.Kannagi R. Carbohydrate antigen sialyl Lewis a–its pathophysiological significance and induction mechanism in cancer progression. Chang Gung Med. J. 2007;30:189–209. [PubMed] [Google Scholar]
- 261.Goonetilleke KS, Siriwardena AK. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. 2007;33:266–270. doi: 10.1016/j.ejso.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 262.Luo G, et al. Roles of CA19-9 in pancreatic cancer: biomarker, predictor and promoter. Biochim. Biophys. Acta Rev. cancer. 2021;1875:188409. doi: 10.1016/j.bbcan.2020.188409. [DOI] [PubMed] [Google Scholar]
- 263.Scarà S, Bottoni P, Scatena R. CA 19-9: biochemical and clinical aspects. Adv. Exp. Med. Biol. 2015;867:247–260. doi: 10.1007/978-94-017-7215-0_15. [DOI] [PubMed] [Google Scholar]
- 264.Al-Janabi AAHS, Tawfeeq EF. Interfering effect of black tea consumption on diagnosis of pancreatic cancer by CA 19-9. J. Gastrointest. Cancer. 2017;48:148–150. doi: 10.1007/s12029-016-9855-z. [DOI] [PubMed] [Google Scholar]
- 265.Mujica VR, Barkin JS, Go VL. Acute pancreatitis secondary to pancreatic carcinoma. Study group participants. Pancreas. 2000;21:329–332. doi: 10.1097/00006676-200011000-00001. [DOI] [PubMed] [Google Scholar]
- 266.Zhang D, et al. Metformin reduces serum CA199 levels in type 2 diabetes Chinese patients with time-effect and gender difference. Diabetes Technol. Ther. 2015;17:72–79. doi: 10.1089/dia.2014.0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Luo G, et al. Optimize CA19-9 in detecting pancreatic cancer by Lewis and Secretor genotyping. Pancreatology. 2016;16:1057–1062. doi: 10.1016/j.pan.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 268.Wannhoff A, et al. FUT2 and FUT3 genotype determines CA19-9 cut-off values for detection of cholangiocarcinoma in patients with primary sclerosing cholangitis. J. Hepatol. 2013;59:1278–1284. doi: 10.1016/j.jhep.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 269.Kirchhoff C, Habben I, Ivell R, Krull N. A major human epididymis-specific cDNA encodes a protein with sequence homology to extracellular proteinase inhibitors. Biol. Reprod. 1991;45:350–357. doi: 10.1095/biolreprod45.2.350. [DOI] [PubMed] [Google Scholar]
- 270.Galgano MT, Hampton GM, Frierson HFJ. Comprehensive analysis of HE4 expression in normal and malignant human tissues. Mod. Pathol. 2006;19:847–853. doi: 10.1038/modpathol.3800612. [DOI] [PubMed] [Google Scholar]
- 271.Behrouzi R, Barr CE, Crosbie EJ. HE4 as a biomarker for endometrial cancer. Cancers. 2021;13:4764. doi: 10.3390/cancers13194764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Zhang R, Siu MKY, Ngan HYS, Chan KKL. Molecular biomarkers for the early detection of ovarian cancer. Int. J. Mol. Sci. 2022;23:12041. doi: 10.3390/ijms231912041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Qu W, et al. Physiopathological factors affecting the diagnostic value of serum HE4-test for gynecologic malignancies. Expert Rev. Mol. Diagn. 2016;16:1271–1282. doi: 10.1080/14737159.2016.1251317. [DOI] [PubMed] [Google Scholar]
- 274.Truman-Rosentsvit M, et al. Ferritin is secreted via 2 distinct nonclassical vesicular pathways. Blood. 2018;131:342–352. doi: 10.1182/blood-2017-02-768580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Mei Z, et al. Physiologically based serum ferritin thresholds for iron deficiency in children and non-pregnant women: a US National Health and Nutrition Examination Surveys (NHANES) serial cross-sectional study. Lancet Haematol. 2021;8:e572–e582. doi: 10.1016/S2352-3026(21)00168-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Koperdanova M, Cullis JO. Interpreting raised serum ferritin levels. BMJ. 2015;351:h3692. doi: 10.1136/bmj.h3692. [DOI] [PubMed] [Google Scholar]
- 277.Matzner Y, Konijn AM, Hershko C. Serum ferritin in hematologic malignancies. Am. J. Hematol. 1980;9:13–22. doi: 10.1002/ajh.2830090103. [DOI] [PubMed] [Google Scholar]
- 278.Gray CP, Arosio P, Hersey P. Association of increased levels of heavy-chain ferritin with increased CD4 + CD25+ regulatory T-cell levels in patients with melanoma. Clin. Cancer Res. 2003;9:2551–2559. [PubMed] [Google Scholar]
- 279.de Almeida SM, da Cunha DS, Yamada E, Doi EM, Ono M. Quantification of cerebrospinal fluid ferritin as a biomarker for CNS malignant infiltration. Arq. Neuropsiquiatr. 2008;66:720–724. doi: 10.1590/S0004-282X2008000500022. [DOI] [PubMed] [Google Scholar]
- 280.Lazzeri M, et al. Serum isoform [-2]proPSA derivatives significantly improve prediction of prostate cancer at initial biopsy in a total PSA range of 2-10 ng/ml: a multicentric european study. Eur. Urol. 2013;63:986–994. doi: 10.1016/j.eururo.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 281.Fossati N, et al. Preoperative prostate-specific antigen isoform p2PSA and its derivatives, %p2PSA and prostate health index, predict pathologic outcomes in patients undergoing radical prostatectomy for prostate cancer: Results from a multicentric european prospective stud. Eur. Urol. 2015;68:132–138. doi: 10.1016/j.eururo.2014.07.034. [DOI] [PubMed] [Google Scholar]
- 282.Gilligan TD, et al. American society of clinical oncology clinical practice guideline on uses of serum tumor markers in adult males with germ cell tumors. J. Clin. Oncol. 2010;28:3388–3404. doi: 10.1200/JCO.2009.26.4481. [DOI] [PubMed] [Google Scholar]
- 283.Gregory JJJ, Finlay JL. Alpha-fetoprotein and beta-human chorionic gonadotropin: their clinical significance as tumour markers. Drugs. 1999;57:463–467. doi: 10.2165/00003495-199957040-00001. [DOI] [PubMed] [Google Scholar]
- 284.Openshaw MR, et al. Circulating cell free DNA in the diagnosis of trophoblastic tumors. EBioMedicine. 2016;4:146–152. doi: 10.1016/j.ebiom.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Gallagher DJ, Riches J, Bajorin DF. False elevation of human chorionic gonadotropin in a patient with testicular cancer. Nat. Rev. Urol. 2010;7:230–233. doi: 10.1038/nrurol.2010.10. [DOI] [PubMed] [Google Scholar]
- 286.Gansauge F, et al. CAM 17.1–a new diagnostic marker in pancreatic cancer. Br. J. Cancer. 1996;74:1997–2002. doi: 10.1038/bjc.1996.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Rhodes JM. Usefulness of novel tumour markers. Ann. Oncol. 1999;10:118–121. doi: 10.1093/annonc/10.suppl_4.S118. [DOI] [PubMed] [Google Scholar]
- 288.Ryu MR, Kang E-S, Park H-D. Performance evaluation of serum PIVKA-II measurement using HISCL-5000 and a method comparison of HISCL-5000, LUMIPULSE G1200, and ARCHITECT i2000. J. Clin. Lab. Anal. 2019;33:e22921. doi: 10.1002/jcla.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Hu B, Tian X, Sun J, Meng X. Evaluation of individual and combined applications of serum biomarkers for diagnosis of hepatocellular carcinoma: a meta-analysis. Int. J. Mol. Sci. 2013;14:23559–23580. doi: 10.3390/ijms141223559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Xing, H. et al. Clinical application of protein induced by vitamin K antagonist-II as a biomarker in hepatocellular carcinoma. Tumour Biol. 10.1007/s13277-016-5443-x. (2016). [DOI] [PubMed]
- 291.Tartaglione S, et al. Protein induced by vitamin K absence II (PIVKA-II) as a potential serological biomarker in pancreatic cancer: a pilot study. Biochem. Med. 2019;29:20707. doi: 10.11613/BM.2019.020707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Marrero JA, et al. Des-gamma carboxyprothrombin can differentiate hepatocellular carcinoma from nonmalignant chronic liver disease in american patients. Hepatology. 2003;37:1114–1121. doi: 10.1053/jhep.2003.50195. [DOI] [PubMed] [Google Scholar]
- 293.Durazo FA, et al. Des-gamma-carboxyprothrombin, alpha-fetoprotein and AFP-L3 in patients with chronic hepatitis, cirrhosis and hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2008;23:1541–1548. doi: 10.1111/j.1440-1746.2008.05395.x. [DOI] [PubMed] [Google Scholar]
- 294.Wojcik E, Kulpa JK. Pro-gastrin-releasing peptide (ProGRP) as a biomarker in small-cell lung cancer diagnosis, monitoring and evaluation of treatment response. Lung Cancer. 2017;8:231–240. doi: 10.2147/LCTT.S149516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lv SP, et al. Meta-analysis of serum gastrin-releasing peptide precursor as a biomarker for diagnosis of small cell lung cancer. Asian Pac. J. Cancer Prev. 2017;18:391–397. doi: 10.22034/APJCP.2017.18.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Tutar N, et al. Clinical significance of progastrin-releasing peptide, neuron-specific enolase, chromogranin a, and squamous cell cancer antigen in pulmonary neuroendocrine tumors. Turkish J. Med. Sci. 2019;49:774–781. doi: 10.3906/sag-1810-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Busch RA, Heneghan AF, Pierre JF, Wang X, Kudsk KA. The enteric nervous system neuropeptide, bombesin, reverses innate immune impairments during parenteral nutrition. Ann. Surg. 2014;260:432–434. doi: 10.1097/SLA.0000000000000871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 299.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 300.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 301.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ediriweera MK, Tennekoon KH, Samarakoon SR. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: biological and therapeutic significance. Semin. Cancer Biol. 2019;59:147–160. doi: 10.1016/j.semcancer.2019.05.012. [DOI] [PubMed] [Google Scholar]
- 303.Rocca A, Braga L, Volpe MC, Maiocchi S, Generali D. The predictive and prognostic role of RAS-RAF-MEK-ERK pathway alterations in breast cancer: revision of the literature and comparison with the analysis of cancer genomic datasets. Cancers. 2022;14:5306. doi: 10.3390/cancers14215306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Barbacid M. ras genes. Annu. Rev. Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 305.Chang EH, Gonda MA, Ellis RW, Scolnick EM, Lowy DR. Human genome contains four genes homologous to transforming genes of Harvey and Kirsten murine sarcoma viruses. Proc. Natl Acad. Sci. USA. 1982;79:4848–4852. doi: 10.1073/pnas.79.16.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 307.Kwan AK, Piazza GA, Keeton AB, Leite CA. The path to the clinic: a comprehensive review on direct KRAS(G12C) inhibitors. J. Exp. Clin. Cancer Res. 2022;41:27. doi: 10.1186/s13046-021-02225-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Yang H, et al. Targeting RAS mutants in malignancies: successes, failures, and reasons for hope. Cancer Commun. 2023;43:42–74. doi: 10.1002/cac2.12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Mukhopadhyay S, Vander Heiden MG, McCormick F. The metabolic landscape of RAS-driven cancers from biology to therapy. Nat. Cancer. 2021;2:271–283. doi: 10.1038/s43018-021-00184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Buchanan FG, et al. Up-regulation of the enzymes involved in prostacyclin synthesis via Ras induces vascular endothelial growth factor. Gastroenterology. 2004;127:1391–1400. doi: 10.1053/j.gastro.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 311.Ancrile BB, O’Hayer KM, Counter CM. Oncogenic ras-induced expression of cytokines: a new target of anti-cancer therapeutics. Mol. Inter. 2008;8:22–27. doi: 10.1124/mi.8.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Murugan AK, Grieco M, Tsuchida N. RAS mutations in human cancers: roles in precision medicine. Semin. Cancer Biol. 2019;59:23–35. doi: 10.1016/j.semcancer.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 313.Taieb J, et al. Prognostic value of BRAF and KRAS mutations in MSI and MSS stage III colon cancer. J. Natl Cancer Inst. 2017;109:djw272. doi: 10.1093/jnci/djw272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature487, 330–337 (2012). [DOI] [PMC free article] [PubMed]
- 315.Cancer Genome Atlas Research Network. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell32, 185–203.e13 (2017). [DOI] [PMC free article] [PubMed]
- 316.Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature511, 543–550 (2014). [DOI] [PMC free article] [PubMed]
- 317.Cancer Genome Atlas Research Network. Genomic classification of cutaneous melanoma. Cell161, 1681–1696 (2015). [DOI] [PMC free article] [PubMed]
- 318.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Longo DL, Rosen N. Targeting oncogenic RAS protein. N. Engl. J. Med. 2022;387:184–186. doi: 10.1056/NEJMe2206831. [DOI] [PubMed] [Google Scholar]
- 320.Santos E, et al. Malignant activation of a K-ras oncogene in lung carcinoma but not in normal tissue of the same patient. Science. 1984;223:661–664. doi: 10.1126/science.6695174. [DOI] [PubMed] [Google Scholar]
- 321.Prior IA, Hood FE, Hartley JL. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Dai M, Chen S, Teng X, Chen K, Cheng W. KRAS as a key oncogene in the clinical precision diagnosis and treatment of pancreatic cancer. J. Cancer. 2022;13:3209–3220. doi: 10.7150/jca.76695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Modest DP, et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: Pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann. Oncol. 2016;27:1746–1753. doi: 10.1093/annonc/mdw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Gallina FT, et al. KRAS G12C mutation and risk of disease recurrence in stage I surgically resected lung adenocarcinoma. Lung Cancer. 2023;181:107254. doi: 10.1016/j.lungcan.2023.107254. [DOI] [PubMed] [Google Scholar]
- 325.Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020;17:153–168. doi: 10.1038/s41575-019-0245-4. [DOI] [PubMed] [Google Scholar]
- 326.Shen H, et al. KRAS G12D mutation subtype in pancreatic ductal adenocarcinoma: does it influence prognosis or stage of disease at presentation? Cells. 2022;11:3175. doi: 10.3390/cells11193175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Santana-Codina N, et al. Oncogenic KRAS supports pancreatic cancer through regulation of nucleotide synthesis. Nat. Commun. 2018;9:4945. doi: 10.1038/s41467-018-07472-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Weinberg F, et al. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc. Natl Acad. Sci. USA. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kerk SA, Papagiannakopoulos T, Shah YM, Lyssiotis CA. Metabolic networks in mutant KRAS-driven tumours: tissue specificities and the microenvironment. Nat. Rev. Cancer. 2021;21:510–525. doi: 10.1038/s41568-021-00375-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Amendola CR, et al. KRAS4A directly regulates hexokinase 1. Nature. 2019;576:482–486. doi: 10.1038/s41586-019-1832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Wang H, et al. Inhibition of glycolytic enzyme hexokinase II (HK2) suppresses lung tumor growth. Cancer Cell Int. 2016;16:9. doi: 10.1186/s12935-016-0280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ying H, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Padanad MS, et al. Fatty acid oxidation mediated by acyl-CoA synthetase long chain 3 is required for mutant KRAS lung tumorigenesis. Cell Rep. 2016;16:1614–1628. doi: 10.1016/j.celrep.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Akhave NS, Biter AB, Hong DS. Mechanisms of resistance to KRAS(G12C)-targeted therapy. Cancer Discov. 2021;11:1345–1352. doi: 10.1158/2159-8290.CD-20-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Zhu C, et al. Targeting KRAS mutant cancers: from druggable therapy to drug resistance. Mol. Cancer. 2022;21:159. doi: 10.1186/s12943-022-01629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Lu S, et al. Ras conformational ensembles, allostery, and signaling. Chem. Rev. 2016;116:6607–6665. doi: 10.1021/acs.chemrev.5b00542. [DOI] [PubMed] [Google Scholar]
- 337.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat. Rev. Cancer. 2018;18:744–757. doi: 10.1038/s41568-018-0074-8. [DOI] [PubMed] [Google Scholar]
- 339.Sawyers CL. Will mTOR inhibitors make it as cancer drugs? Cancer Cell. 2003;4:343–348. doi: 10.1016/S1535-6108(03)00275-7. [DOI] [PubMed] [Google Scholar]
- 340.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- 342.Fruman DA, et al. The PI3K pathway in human disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Polivka JJ, Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol. Ther. 2014;142:164–175. doi: 10.1016/j.pharmthera.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 344.Papadimitrakopoulou V, Adjei AA. The Akt/mTOR and mitogen-activated protein kinase pathways in lung cancer therapy. J. Thorac. Oncol. 2006;1:749–751. [PubMed] [Google Scholar]
- 345.Hung M-C, Wang W-P, Chi Y-H. AKT phosphorylation as a predictive biomarker for PI3K/mTOR dual inhibition-induced proteolytic cleavage of mTOR companion proteins in small cell lung cancer. Cell Biosci. 2022;12:122. doi: 10.1186/s13578-022-00862-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Nunnery SE, Mayer IA. Targeting the PI3K/AKT/mTOR pathway in hormone-positive breast cancer. Drugs. 2020;80:1685–1697. doi: 10.1007/s40265-020-01394-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Wang W, et al. Activation of Akt/mTOR pathway is associated with poor prognosis of nasopharyngeal carcinoma. PLoS ONE. 2014;9:e106098. doi: 10.1371/journal.pone.0106098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Li H-L, Deng N-H, He X-S, Li Y-H. Small biomarkers with massive impacts: PI3K/AKT/mTOR signalling and microRNA crosstalk regulate nasopharyngeal carcinoma. Biomark. Res. 2022;10:52. doi: 10.1186/s40364-022-00397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Janku F, et al. PI3K/AKT/mTOR inhibitors in patients with breast and gynecologic malignancies harboring PIK3CA mutations. J. Clin. Oncol. 2012;30:777–782. doi: 10.1200/JCO.2011.36.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Janku F, et al. PIK3CA mutation H1047R is associated with response to PI3K/AKT/mTOR signaling pathway inhibitors in early-phase clinical trials. Cancer Res. 2013;73:276–284. doi: 10.1158/0008-5472.CAN-12-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Owonikoko TK, Khuri FR. Targeting the PI3K/AKT/mTOR pathway: biomarkers of success and tribulation. Am. Soc. Clin. Oncol. Educ. Book. 2013;33:e395–e401. doi: 10.14694/EdBook_AM.2013.33.e395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Fusco N, et al. PIK3CA mutations as a molecular target for hormone receptor-positive, HER2-negative metastatic breast cancer. Front. Oncol. 2021;11:644737. doi: 10.3389/fonc.2021.644737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Punekar SR, Velcheti V, Neel BG, Wong KK. The current state of the art and future trends in RAS-targeted cancer therapies. Nat. Rev. Clin. Oncol. 2022;19:637–655. doi: 10.1038/s41571-022-00671-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat. Rev. Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Takács T, et al. The effects of mutant Ras proteins on the cell signalome. Cancer Metastasis Rev. 2020;39:1051–1065. doi: 10.1007/s10555-020-09912-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Bahar ME, Kim HJ, Kim DR. Targeting the RAS/RAF/MAPK pathway for cancer therapy: from mechanism to clinical studies. Signal Transduct. Target Ther. 2023;8:455. doi: 10.1038/s41392-023-01705-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Su W, et al. ARAF protein kinase activates RAS by antagonizing its binding to RASGAP NF1. Mol. Cell. 2022;82:2443–2457.e7. doi: 10.1016/j.molcel.2022.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.da Rocha Dias S, et al. Activated B-RAF is an Hsp90 client protein that is targeted by the anticancer drug 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2005;65:10686–10691. doi: 10.1158/0008-5472.CAN-05-2632. [DOI] [PubMed] [Google Scholar]
- 359.Grammatikakis N, Lin JH, Grammatikakis A, Tsichlis PN, Cochran BH. p50(cdc37) acting in concert with Hsp90 is required for Raf-1 function. Mol. Cell. Biol. 1999;19:1661–1672. doi: 10.1128/MCB.19.3.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Michaud NR, et al. KSR stimulates Raf-1 activity in a kinase-independent manner. Proc. Natl Acad. Sci. USA. 1997;94:12792–12796. doi: 10.1073/pnas.94.24.12792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Steelman LS, et al. Roles of the Ras/Raf/MEK/ERK pathway in leukemia therapy. Leukemia. 2011;25:1080–1094. doi: 10.1038/leu.2011.66. [DOI] [PubMed] [Google Scholar]
- 362.Imperial R, Toor OM, Hussain A, Subramanian J, Masood A. Comprehensive pancancer genomic analysis reveals (RTK)-RAS-RAF-MEK as a key dysregulated pathway in cancer: its clinical implications. Semin. Cancer Biol. 2019;54:14–28. doi: 10.1016/j.semcancer.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 363.Liu D, Zhou K. BRAF/MEK pathway is associated with breast cancer in ER-dependent mode and improves ER status-based cancer recurrence prediction. Clin. Breast Cancer. 2020;20:41–50.e8. doi: 10.1016/j.clbc.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 364.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat. Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 365.Worby CA, Dixon JE. PTEN. Annu. Rev. Biochem. 2014;83:641–669. doi: 10.1146/annurev-biochem-082411-113907. [DOI] [PubMed] [Google Scholar]
- 366.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J. Biol. Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 367.Hollander MC, Blumenthal GM, Dennis PA. PTEN loss in the continuum of common cancers, rare syndromes and mouse models. Nat. Rev. Cancer. 2011;11:289–301. doi: 10.1038/nrc3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Xie P, et al. Neddylation of PTEN regulates its nuclear import and promotes tumor development. Cell Res. 2021;31:291–311. doi: 10.1038/s41422-020-00443-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J. Clin. Oncol. 2004;22:2954–2963. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 370.Bazzichetto C, et al. Pten as a prognostic/predictive biomarker in cancer: an unfulfilled promise? Cancers. 2019;11:435. doi: 10.3390/cancers11040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Song MS, Salmena L, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor. Nat. Rev. Mol. Cell Biol. 2012;13:283–296. doi: 10.1038/nrm3330. [DOI] [PubMed] [Google Scholar]
- 372.Lee Y-R, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat. Rev. Mol. Cell Biol. 2018;19:547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 373.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 374.Benavente CA, Dyer MA. Genetics and epigenetics of human retinoblastoma. Annu. Rev. Pathol. 2015;10:547–562. doi: 10.1146/annurev-pathol-012414-040259. [DOI] [PubMed] [Google Scholar]
- 375.Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer. 2019;19:326–338. doi: 10.1038/s41568-019-0143-7. [DOI] [PubMed] [Google Scholar]
- 376.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Munro S, Carr SM, La Thangue NB. Diversity within the pRb pathway: is there a code of conduct. Oncogene. 2012;31:4343–4352. doi: 10.1038/onc.2011.603. [DOI] [PubMed] [Google Scholar]
- 378.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 379.Dyson NJ. RB1: a prototype tumor suppressor and an enigma. Genes Dev. 2016;30:1492–1502. doi: 10.1101/gad.282145.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Knudsen ES, et al. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun. Biol. 2020;3:158. doi: 10.1038/s42003-020-0873-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 382.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Bieging KT, Attardi LD. Cancer: a piece of the p53 puzzle. Nature. 2015;520:37–38. doi: 10.1038/nature14374. [DOI] [PubMed] [Google Scholar]
- 384.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 385.Wang M, Attardi LD. A balancing act: p53 activity from tumor suppression to pathology and therapeutic implications. Annu. Rev. Pathol. 2022;17:205–226. doi: 10.1146/annurev-pathol-042320-025840. [DOI] [PubMed] [Google Scholar]
- 386.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 387.Zhu H, et al. Targeting p53–MDM2 interaction by small-molecule inhibitors: learning from MDM2 inhibitors in clinical trials. J. Hematol. Oncol. 2022;15:91. doi: 10.1186/s13045-022-01314-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Shi C, Liu S, Tian X, Wang X, Gao P. A TP53 mutation model for the prediction of prognosis and therapeutic responses in head and neck squamous cell carcinoma. BMC Cancer. 2021;21:1035. doi: 10.1186/s12885-021-08765-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Li VD, Li KH, Li JT. TP53 mutations as potential prognostic markers for specific cancers: analysis of data from The Cancer Genome Atlas and the International Agency for Research on Cancer TP53 Database. J. Cancer Res. Clin. Oncol. 2019;145:625–636. doi: 10.1007/s00432-018-2817-z. [DOI] [PubMed] [Google Scholar]
- 390.Jafri MA, Ansari SA, Alqahtani MH, Shay JW. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016;8:69. doi: 10.1186/s13073-016-0324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc. Natl Acad. Sci. USA. 1989;86:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Gao J, Pickett HA. Targeting telomeres: advances in telomere maintenance mechanism-specific cancer therapies. Nat. Rev. Cancer. 2022;22:515–532. doi: 10.1038/s41568-022-00490-1. [DOI] [PubMed] [Google Scholar]
- 393.Marinaccio J, et al. TERT extra-telomeric roles: antioxidant activity and mitochondrial protection. Int. J. Mol. Sci. 2023;24:4450. doi: 10.3390/ijms24054450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Bajaj S, Kumar MS, Peters GJ, Mayur YC. Targeting telomerase for its advent in cancer therapeutics. Med. Res. Rev. 2020;40:1871–1919. doi: 10.1002/med.21674. [DOI] [PubMed] [Google Scholar]
- 395.Bertorelle R, et al. Telomerase is an independent prognostic marker of overall survival in pataients with colorectal cancer. Br. J. Cancer. 2013;108:278–284. doi: 10.1038/bjc.2012.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Powter B, et al. Human TERT promoter mutations as a prognostic biomarker in glioma. J. Cancer Res. Clin. Oncol. 2021;147:1007–1017. doi: 10.1007/s00432-021-03536-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Masutomi K, et al. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl Acad. Sci. USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Baeriswyl V, Christofori G. The angiogenic switch in carcinogenesis. Semin. Cancer Biol. 2009;19:329–337. doi: 10.1016/j.semcancer.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 399.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86:353–364. doi: 10.1016/S0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 400.Weidner N, et al. Tumor angiogenesis: a new significant and independent prognostic indicator in early-stage breast carcinoma. J. Natl Cancer Inst. 1992;84:1875–1887. doi: 10.1093/jnci/84.24.1875. [DOI] [PubMed] [Google Scholar]
- 401.Al-Husein B, Abdalla M, Trepte M, DeRemer DL, Somanath PR. Antiangiogenic therapy for cancer: an update. Pharmacotherapy. 2012;32:1095–1111. doi: 10.1002/phar.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Senger DR, et al. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science. 1983;219:983–985. doi: 10.1126/science.6823562. [DOI] [PubMed] [Google Scholar]
- 403.Ferrara N, Henzel WJ. Pituitary follicular cells secrete a novel heparin-binding growth factor specific for vascular endothelial cells. Biochem. Biophys. Res. Commun. 1989;161:851–858. doi: 10.1016/0006-291X(89)92678-8. [DOI] [PubMed] [Google Scholar]
- 404.Dabravolski SA, Khotina VA, Omelchenko AV, Kalmykov VA, Orekhov AN. The role of the VEGF family in atherosclerosis development and its potential as treatment targets. Int. J. Mol. Sci. 2022;23:931. doi: 10.3390/ijms23020931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Ebos JML, Lee CR, Christensen JG, Mutsaers AJ, Kerbel RS. Multiple circulating proangiogenic factors induced by sunitinib malate are tumor-independent and correlate with antitumor efficacy. Proc. Natl Acad. Sci. USA. 2007;104:17069–17074. doi: 10.1073/pnas.0708148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Kerbel RS. Tumor angiogenesis. N. Engl. J. Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Kerbel R, Folkman J. Clinical translation of angiogenesis inhibitors. Nat. Rev. Cancer. 2002;2:727–739. doi: 10.1038/nrc905. [DOI] [PubMed] [Google Scholar]
- 408.Lee SH, Jeong D, Han Y-S, Baek MJ. Pivotal role of vascular endothelial growth factor pathway in tumor angiogenesis. Ann. Surg. Treat. Res. 2015;89:1–8. doi: 10.4174/astr.2015.89.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Simons M, Gordon E, Claesson-Welsh L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016;17:611–625. doi: 10.1038/nrm.2016.87. [DOI] [PubMed] [Google Scholar]
- 410.de Vries C, et al. The fms-like tyrosine kinase, a receptor for vascular endothelial growth factor. Science. 1992;255:989–991. doi: 10.1126/science.1312256. [DOI] [PubMed] [Google Scholar]
- 411.Park JE, Chen HH, Winer J, Houck KA, Ferrara N. Placenta growth factor. Potentiation of vascular endothelial growth factor bioactivity, in vitro and in vivo, and high affinity binding to Flt-1 but not to Flk-1/KDR. J. Biol. Chem. 1994;269:25646–25654. doi: 10.1016/S0021-9258(18)47298-5. [DOI] [PubMed] [Google Scholar]
- 412.Ye X, Gaucher J-F, Vidal M, Broussy S. A structural overview of vascular endothelial growth factors pharmacological ligands: from macromolecules to designed peptidomimetics. Molecules. 2021;26:6759. doi: 10.3390/molecules26226759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Ceci C, Atzori MG, Lacal PM, Graziani G. Role of VEGFs/VEGFR-1 signaling and its inhibition in modulating tumor invasion: experimental evidence in different metastatic cancer models. Int. J. Mol. Sci. 2020;21:1388. doi: 10.3390/ijms21041388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Apte RS, Chen DS, Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176:1248–1264. doi: 10.1016/j.cell.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Ferrara N, Gerber H-P, LeCouter J. The biology of VEGF and its receptors. Nat. Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 416.Yao C, et al. Angiogenesis in hepatocellular carcinoma: mechanisms and anti-angiogenic therapies. Cancer Biol. Med. 2023;20:25–43. doi: 10.20892/j.issn.2095-3941.2022.0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Karkkainen MJ, Mäkinen T, Alitalo K. Lymphatic endothelium: a new frontier of metastasis research. Nat. Cell Biol. 2002;4:E2–E5. doi: 10.1038/ncb0102-e2. [DOI] [PubMed] [Google Scholar]
- 418.Alitalo K, Tammela T, Petrova TV. Lymphangiogenesis in development and human disease. Nature. 2005;438:946–953. doi: 10.1038/nature04480. [DOI] [PubMed] [Google Scholar]
- 419.Aguilar-Cazares D, et al. Contribution of angiogenesis to inflammation and cancer. Front. Oncol. 2019;9:1399. doi: 10.3389/fonc.2019.01399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Lacin S, Yalcin S. The prognostic value of circulating VEGF-A level in patients with hepatocellular cancer. Technol. Cancer Res. Treat. 2020;19:153303382097167. doi: 10.1177/1533033820971677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Bocci G, et al. Increased plasma vascular endothelial growth factor (VEGF) as a surrogate marker for optimal therapeutic dosing of VEGF receptor-2 monoclonal antibodies. Cancer Res. 2004;64:6616–6625. doi: 10.1158/0008-5472.CAN-04-0401. [DOI] [PubMed] [Google Scholar]
- 422.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 423.De Moerlooze L, et al. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 424.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 425.Mashreghi M, et al. Angiogenesis biomarkers and their targeting ligands as potential targets for tumor angiogenesis. J. Cell. Physiol. 2018;233:2949–2965. doi: 10.1002/jcp.26049. [DOI] [PubMed] [Google Scholar]
- 426.Presta M, et al. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 2005;16:159–178. doi: 10.1016/j.cytogfr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 427.Doll JA, et al. Thrombospondin-1, vascular endothelial growth factor and fibroblast growth factor-2 are key functional regulators of angiogenesis in the prostate. Prostate. 2001;49:293–305. doi: 10.1002/pros.10025. [DOI] [PubMed] [Google Scholar]
- 428.Birrer MJ, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J. Clin. Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 429.Dienstmann R, et al. Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors. Ann. Oncol. 2014;25:552–563. doi: 10.1093/annonc/mdt419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Zhang Y, et al. Role of epithelial cell fibroblast growth factor receptor substrate 2alpha in prostate development, regeneration and tumorigenesis. Development. 2008;135:775–784. doi: 10.1242/dev.009910. [DOI] [PubMed] [Google Scholar]
- 431.Schaeffer EM, et al. Androgen-induced programs for prostate epithelial growth and invasion arise in embryogenesis and are reactivated in cancer. Oncogene. 2008;27:7180–7191. doi: 10.1038/onc.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Desai A, Adjei AA. FGFR signaling as a target for lung cancer therapy. J. Thorac. Oncol. 2016;11:9–20. doi: 10.1016/j.jtho.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 433.Xian W, et al. Fibroblast growth factor receptor 1-transformed mammary epithelial cells are dependent on RSK activity for growth and survival. Cancer Res. 2009;69:2244–2251. doi: 10.1158/0008-5472.CAN-08-3398. [DOI] [PubMed] [Google Scholar]
- 434.Nomura S, et al. FGF10/FGFR2 signal induces cell migration and invasion in pancreatic cancer. Br. J. Cancer. 2008;99:305–313. doi: 10.1038/sj.bjc.6604473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Cappellen D, et al. Frequent activating mutations of FGFR3 in human bladder and cervix carcinomas. Nat. Genet. 1999;23:18–20. doi: 10.1038/12615. [DOI] [PubMed] [Google Scholar]
- 437.Dutt A, et al. Drug-sensitive FGFR2 mutations in endometrial carcinoma. Proc. Natl Acad. Sci. USA. 2008;105:8713–8717. doi: 10.1073/pnas.0803379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Pearson A, et al. High-level clonal FGFR amplification and response to FGFR inhibition in a translational clinical trial. Cancer Discov. 2016;6:838–851. doi: 10.1158/2159-8290.CD-15-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Weiss J, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci. Transl. Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Ahmad I, Iwata T, Leung HY. Mechanisms of FGFR-mediated carcinogenesis. Biochim. Biophys. Acta. 2012;1823:850–860. doi: 10.1016/j.bbamcr.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 441.Nakamura IT, et al. Comprehensive functional evaluation of variants of fibroblast growth factor receptor genes in cancer. NPJ Precis. Oncol. 2021;5:66. doi: 10.1038/s41698-021-00204-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Wang S, Ding Z. Fibroblast growth factor receptors in breast cancer. Tumour Biol. 2017;39:1010428317698370. doi: 10.1177/1010428317698370. [DOI] [PubMed] [Google Scholar]
- 443.Zou X, et al. Targeting the PDGF/PDGFR signaling pathway for cancer therapy: a review. Int. J. Biol. Macromol. 2022;202:539–557. doi: 10.1016/j.ijbiomac.2022.01.113. [DOI] [PubMed] [Google Scholar]
- 444.Kalra K, Eberhard J, Farbehi N, Chong JJ, Xaymardan M. Role of PDGF-A/B ligands in cardiac repair after myocardial infarction. Front. Cell Dev. Biol. 2021;9:669188. doi: 10.3389/fcell.2021.669188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Nordby Y, et al. High expression of PDGFR-β in prostate cancer stroma is independently associated with clinical and biochemical prostate cancer recurrence. Sci. Rep. 2017;7:43378. doi: 10.1038/srep43378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Demoulin J-B, Essaghir A. PDGF receptor signaling networks in normal and cancer cells. Cytokine Growth Factor Rev. 2014;25:273–283. doi: 10.1016/j.cytogfr.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 447.Thies KA, et al. Stromal platelet-derived growth factor receptor-β signaling promotes breast cancer metastasis in the brain. Cancer Res. 2021;81:606–618. doi: 10.1158/0008-5472.CAN-19-3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Lin L-H, et al. Overexpression of platelet-derived growth factor and its receptor are correlated with oral tumorigenesis and Poor Prognosis in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2020;21:2360. doi: 10.3390/ijms21072360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Bernat-Peguera A, et al. PDGFR-induced autocrine SDF-1 signaling in cancer cells promotes metastasis in advanced skin carcinoma. Oncogene. 2019;38:5021–5037. doi: 10.1038/s41388-019-0773-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Brahmi M, et al. Expression and prognostic significance of PDGF ligands and receptors across soft tissue sarcomas. ESMO Open. 2021;6:100037. doi: 10.1016/j.esmoop.2020.100037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Yang J, et al. A positive feedback loop between inactive VHL-triggered histone lactylation and PDGFRβ signaling drives clear cell renal cell carcinoma progression. Int. J. Biol. Sci. 2022;18:3470–3483. doi: 10.7150/ijbs.73398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Roskoski RJ. The role of small molecule platelet-derived growth factor receptor (PDGFR) inhibitors in the treatment of neoplastic disorders. Pharmacol. Res. 2018;129:65–83. doi: 10.1016/j.phrs.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 453.Manzat Saplacan RM, et al. The role of PDGFs and PDGFRs in colorectal cancer. Mediators Inflamm. 2017;2017:4708076. doi: 10.1155/2017/4708076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Taeger J, et al. Targeting FGFR/PDGFR/VEGFR impairs tumor growth, angiogenesis, and metastasis by effects on tumor cells, endothelial cells, and pericytes in pancreatic cancer. Mol. Cancer Ther. 2011;10:2157–2167. doi: 10.1158/1535-7163.MCT-11-0312. [DOI] [PubMed] [Google Scholar]
- 455.Qian H, et al. The clinical significance of platelet-derived growth factors (PDGFs) and their receptors (PDGFRs) in gastric cancer: A systematic review and meta-analysis. Crit. Rev. Oncol. Hematol. 2018;127:15–28. doi: 10.1016/j.critrevonc.2018.05.004. [DOI] [PubMed] [Google Scholar]
- 456.Cavalcanti E, Ignazzi A, De Michele F, Caruso ML. PDGFRα expression as a novel therapeutic marker in well-differentiated neuroendocrine tumors. Cancer Biol. Ther. 2019;20:423–430. doi: 10.1080/15384047.2018.1529114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Sethi S, Macoska J, Chen W, Sarkar FH. Molecular signature of epithelial-mesenchymal transition (EMT) in human prostate cancer bone metastasis. Am. J. Transl. Res. 2010;3:90–99. [PMC free article] [PubMed] [Google Scholar]
- 458.Ustach CV, et al. A novel signaling axis of matriptase/PDGF-D/ß-PDGFR in human prostate cancer. Cancer Res. 2010;70:9631–9640. doi: 10.1158/0008-5472.CAN-10-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Yang Y, et al. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol. Sin. 2013;34:625–635. doi: 10.1038/aps.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat. Rev. Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 462.Hata AN, Engelman JA, Faber AC. The BCL2 family: key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov. 2015;5:475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Pistritto G, Trisciuoglio D, Ceci C. Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging. 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Ledgerwood EC, Morison IM. Targeting the apoptosome for cancer therapy. Clin. cancer Res. 2009;15:420–424. doi: 10.1158/1078-0432.CCR-08-1172. [DOI] [PubMed] [Google Scholar]
- 466.Jin Z, El-Deiry WS. Overview of cell death signaling pathways. Cancer Biol. Ther. 2005;4:139–163. doi: 10.4161/cbt.4.2.1508. [DOI] [PubMed] [Google Scholar]
- 467.Letai AG. Diagnosing and exploiting cancer’s addiction to blocks in apoptosis. Nat. Rev. Cancer. 2008;8:121–132. doi: 10.1038/nrc2297. [DOI] [PubMed] [Google Scholar]
- 468.Hu W, Kavanagh JJ. Anticancer therapy targeting the apoptotic pathway. Lancet Oncol. 2003;4:721–729. doi: 10.1016/S1470-2045(03)01277-4. [DOI] [PubMed] [Google Scholar]
- 469.Delbridge ARD, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer. 2016;16:99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- 470.Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Boise LH, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-N. [DOI] [PubMed] [Google Scholar]
- 472.Lee, E. F. & Fairlie, W. D. The Structural Biology of Bcl-x(L). Int. J. Mol. Sci. 20, 2234 (2019). [DOI] [PMC free article] [PubMed]
- 473.Li M, Wang D, He J, Chen L, Li H. Bcl-X(L): a multifunctional anti-apoptotic protein. Pharmacol. Res. 2020;151:104547. doi: 10.1016/j.phrs.2019.104547. [DOI] [PubMed] [Google Scholar]
- 474.Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat. Rev. Mol. Cell Biol. 2019;20:175–193. doi: 10.1038/s41580-018-0089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Oltersdorf T, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 476.Vo T-T, Letai A. BH3-only proteins and their effects on cancer. Adv. Exp. Med. Biol. 2010;687:49–63. doi: 10.1007/978-1-4419-6706-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Letai A, et al. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell. 2002;2:183–192. doi: 10.1016/S1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 478.Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103:839–842. doi: 10.1016/S0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 479.Siddiqui WA, Ahad A, Ahsan H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: an update. Arch. Toxicol. 2015;89:289–317. doi: 10.1007/s00204-014-1448-7. [DOI] [PubMed] [Google Scholar]
- 480.Qian S, et al. The role of BCL-2 family proteins in regulating apoptosis and cancer therapy. Front. Oncol. 2022;12:985363. doi: 10.3389/fonc.2022.985363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Mohammad RM, et al. Broad targeting of resistance to apoptosis in cancer. Semin. Cancer Biol. 2015;35:S78–S103. doi: 10.1016/j.semcancer.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Schott AF, Apel IJ, Nuñez G, Clarke MF. Bcl-XL protects cancer cells from p53-mediated apoptosis. Oncogene. 1995;11:1389–1394. [PubMed] [Google Scholar]
- 483.Dole MG, et al. Bcl-xL is expressed in neuroblastoma cells and modulates chemotherapy-induced apoptosis. Cancer Res. 1995;55:2576–2582. [PubMed] [Google Scholar]
- 484.Jin-Song Y, et al. Prognostic significance of Bcl-xL gene expression in human colorectal cancer. Acta Histochem. 2011;113:810–814. doi: 10.1016/j.acthis.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 485.Kondo S, et al. Over-expression of bcl-xL gene in human gastric adenomas and carcinomas. Int. J. cancer. 1996;68:727–730. doi: 10.1002/(SICI)1097-0215(19961211)68:6<727::AID-IJC6>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 486.Bouchalova K, et al. Triple negative breast cancer - BCL2 in prognosis and prediction. Review. Curr. Drug Targets. 2014;15:1166–1175. doi: 10.2174/1389450115666141106151143. [DOI] [PubMed] [Google Scholar]
- 487.Cartron P-F, et al. The first alpha helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol. Cell. 2004;16:807–818. doi: 10.1016/j.molcel.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 488.Lalier L, et al. Bax activation and mitochondrial insertion during apoptosis. Apoptosis. 2007;12:887–896. doi: 10.1007/s10495-007-0749-1. [DOI] [PubMed] [Google Scholar]
- 489.Hantusch A, Das KK, García-Sáez AJ, Brunner T, Rehm M. Bax retrotranslocation potentiates Bcl-x(L)’s antiapoptotic activity and is essential for switch-like transitions between MOMP competency and resistance. Cell Death Dis. 2018;9:430. doi: 10.1038/s41419-018-0464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.McArthur K, et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science. 2018;359:eaao6047. doi: 10.1126/science.aao6047. [DOI] [PubMed] [Google Scholar]
- 491.Igney FH, Krammer PH. Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer. 2002;2:277–288. doi: 10.1038/nrc776. [DOI] [PubMed] [Google Scholar]
- 492.Pietrantonio F, et al. Role of BAX for outcome prediction in gastrointestinal malignancies. Med. Oncol. 2013;30:610. doi: 10.1007/s12032-013-0610-z. [DOI] [PubMed] [Google Scholar]
- 493.Rampino N, et al. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 494.Meijerink JP, et al. Hematopoietic malignancies demonstrate loss-of-function mutations of BAX. Blood. 1998;91:2991–2997. doi: 10.1182/blood.V91.8.2991.2991_2991_2997. [DOI] [PubMed] [Google Scholar]
- 495.Perego P, et al. Association between cisplatin resistance and mutation of p53 gene and reduced bax expression in ovarian carcinoma cell systems. Cancer Res. 1996;56:556–562. [PubMed] [Google Scholar]
- 496.Manoochehri M, Karbasi A, Bandehpour M, Kazemi B. Down-regulation of BAX gene during carcinogenesis and acquisition of resistance to 5-FU in colorectal cancer. Pathol. Oncol. Res. 2014;20:301–307. doi: 10.1007/s12253-013-9695-0. [DOI] [PubMed] [Google Scholar]
- 497.Aoyagi T, et al. Lung cancer cell line sensitivity to zoledronic acid is BAX-dependent. Anticancer Res. 2013;33:5357–5363. [PubMed] [Google Scholar]
- 498.Liu Z, et al. Direct activation of Bax protein for cancer therapy. Med. Res. Rev. 2016;36:313–341. doi: 10.1002/med.21379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Kubo T, et al. BAK is a predictive and prognostic biomarker for the therapeutic effect of docetaxel treatment in patients with advanced gastric cancer. Gastric Cancer. 2016;19:827–838. doi: 10.1007/s10120-015-0557-1. [DOI] [PubMed] [Google Scholar]
- 500.Srivastava A, et al. BAX-BAK heterodimer as a pharmacodynamic biomarker of on-target drug action of Mcl-1 inhibitors to evaluate in-vivo effectiveness. JCO. 2018;36:2582–2582. doi: 10.1200/JCO.2018.36.15_suppl.2582. [DOI] [Google Scholar]
- 501.de Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 1955;60:604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 503.Towers CG, Thorburn A. Therapeutic targeting of autophagy. EBioMedicine. 2016;14:15–23. doi: 10.1016/j.ebiom.2016.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 505.Barbeau LMO, Keulers TGH, Rouschop KMA. Tumors responsive to autophagy-inhibition: Identification and biomarkers. Cancers. 2020;12:1–24. doi: 10.3390/cancers12092463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 2019;9:1167–1181. doi: 10.1158/2159-8290.CD-19-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat. Rev. Cancer. 2007;7:961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 510.Gao W, Wang X, Zhou Y, Wang X, Yu Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct. Target Ther. 2022;7:196. doi: 10.1038/s41392-022-01046-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Long X, et al. Autophagy-targeted nanoparticles for effective cancer treatment: advances and outlook. NPG Asia Mater. 2022;14:71. doi: 10.1038/s41427-022-00422-3. [DOI] [Google Scholar]
- 512.Liang XH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998;72:8586–8596. doi: 10.1128/JVI.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 513.Wirawan E, et al. Beclin1: a role in membrane dynamics and beyond. Autophagy. 2012;8:6–17. doi: 10.4161/auto.8.1.16645. [DOI] [PubMed] [Google Scholar]
- 514.Fu LL, Cheng Y, Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int. J. Biochem. Cell Biol. 2013;45:921–924. doi: 10.1016/j.biocel.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 515.Mei Y, et al. Conformational flexibility enables the function of a BECN1 region essential for starvation-mediated Autophagy. Biochemistry. 2016;55:1945–1958. doi: 10.1021/acs.biochem.5b01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Li X, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat. Commun. 2012;3:662. doi: 10.1038/ncomms1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Liang XH, Yu J, Brown K, Levine B. Beclin 1 contains a leucine-rich nuclear export signal that is required for its autophagy and tumor suppressor function. Cancer Res. 2001;61:3443–3449. [PubMed] [Google Scholar]
- 518.Khan T, et al. Autophagy modulators for the treatment of oral and esophageal squamous cell carcinomas. Med. Res. Rev. 2020;40:1002–1060. doi: 10.1002/med.21646. [DOI] [PubMed] [Google Scholar]
- 519.Xiang H, et al. Targeting autophagy-related protein kinases for potential therapeutic purpose. Acta Pharm. Sin. B. 2020;10:569–581. doi: 10.1016/j.apsb.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Cao Y, Klionsky DJ. Physiological functions of Atg6/Beclin 1: a unique autophagy-related protein. Cell Res. 2007;17:839–849. doi: 10.1038/cr.2007.78. [DOI] [PubMed] [Google Scholar]
- 521.Tran S, Fairlie WD, Lee EF. Beclin1: protein structure, function and regulation. Cells. 2021;10:1522. doi: 10.3390/cells10061522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Vega-Rubín-de-Celis S. The role of Beclin 1-dependent autophagy in cancer. Biology. 2019;9:4. doi: 10.3390/biology9010004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Zhou W-H, et al. Low expression of Beclin 1, associated with high Bcl-xL, predicts a malignant phenotype and poor prognosis of gastric cancer. Autophagy. 2012;8:389–400. doi: 10.4161/auto.18641. [DOI] [PubMed] [Google Scholar]
- 524.Liang XH, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 525.Park JM, Huang S, Wu TT, Foster NR, Sinicrope FA. Prognostic impact of Beclin 1, p62/sequestosome 1 and LC3 protein expression in colon carcinomas from patients receiving 5-fluorouracil as adjuvant chemotherapy. Cancer Biol. Ther. 2013;14:100–107. doi: 10.4161/cbt.22954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Han Y, et al. Prognostic significance of Beclin-1 expression in colorectal cancer: a meta-analysis. Asian Pac. J. Cancer Prev. 2014;15:4583–4587. doi: 10.7314/APJCP.2014.15.11.4583. [DOI] [PubMed] [Google Scholar]
- 527.He Y, et al. The prognostic value of autophagy-related markers beclin-1 and microtubule-associated protein light chain 3B in cancers: a systematic review and meta-analysis. Tumour Biol. 2014;35:7317–7326. doi: 10.1007/s13277-014-2060-4. [DOI] [PubMed] [Google Scholar]
- 528.Hwang HJ, Kim YK. The role of LC3B in autophagy as an RNA-binding protein. Autophagy. 2023;19:1028–1030. doi: 10.1080/15548627.2022.2111083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Bhutia SK, et al. Monitoring and measuring mammalian autophagy. Methods Mol. Biol. 2019;1854:209–222. doi: 10.1007/7651_2018_159. [DOI] [PubMed] [Google Scholar]
- 530.Bortnik S, Gorski SM. Clinical applications of autophagy proteins in cancer: from potential targets to biomarkers. Int. J. Mol. Sci. 2017;18:1496. doi: 10.3390/ijms18071496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Masuda GO, et al. Clinicopathological correlations of autophagy-related proteins LC3, Beclin 1 and p62 in gastric cancer. Anticancer Res. 2016;36:129–136. [PubMed] [Google Scholar]
- 532.Guo G-F, et al. Predictive and prognostic implications of 4E-BP1, Beclin-1, and LC3 for cetuximab treatment combined with chemotherapy in advanced colorectal cancer with wild-type KRAS: analysis from real-world data. World J. Gastroenterol. 2019;25:1840–1853. doi: 10.3748/wjg.v25.i15.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Zhao H, et al. High expression of LC3B is associated with progression and poor outcome in triple-negative breast cancer. Med. Oncol. 2013;30:475. doi: 10.1007/s12032-013-0475-1. [DOI] [PubMed] [Google Scholar]
- 534.Lazova R, et al. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin. Cancer Res. 2012;18:370–379. doi: 10.1158/1078-0432.CCR-11-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Winardi D, et al. Correlation of altered expression of the autophagy marker LC3B with poor prognosis in astrocytoma. Biomed. Res. Int. 2014;2014:723176. doi: 10.1155/2014/723176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.El-Mashed S, et al. LC3B globular structures correlate with survival in esophageal adenocarcinoma. BMC Cancer. 2015;15:582. doi: 10.1186/s12885-015-1574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Liu JL, et al. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br. J. Cancer. 2014;111:944–954. doi: 10.1038/bjc.2014.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Choi J, Jung W, Koo JS. Expression of autophagy-related markers beclin-1, light chain 3 A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology. 2013;62:275–286. doi: 10.1111/his.12002. [DOI] [PubMed] [Google Scholar]
- 539.Wu D-H, et al. Autophagic LC3B overexpression correlates with malignant progression and predicts a poor prognosis in hepatocellular carcinoma. Tumour Biol. 2014;35:12225–12233. doi: 10.1007/s13277-014-2531-7. [DOI] [PubMed] [Google Scholar]
- 540.Papinski D, Kraft C. Regulation of autophagy by signaling through the Atg1/ULK1 complex. J. Mol. Biol. 2016;428:1725–1741. doi: 10.1016/j.jmb.2016.03.030. [DOI] [PubMed] [Google Scholar]
- 541.Jiang L, et al. Association of the expression of unc-51-Like kinase 1 with lymph node metastasis and survival in patients with esophageal squamous cell carcinoma. Int. J. Clin. Exp. Med. 2014;7:1349–1354. [PMC free article] [PubMed] [Google Scholar]
- 542.Xu H, et al. UNC51-like kinase 1 as a potential prognostic biomarker for hepatocellular carcinoma. Int. J. Clin. Exp. Pathol. 2013;6:711–717. [PMC free article] [PubMed] [Google Scholar]
- 543.Yun M, et al. ULK1: a promising biomarker in predicting poor prognosis and therapeutic response in human nasopharygeal carcinoma. PLoS ONE. 2015;10:e0117375. doi: 10.1371/journal.pone.0117375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Zhang H-Y, Ma Y-D, Zhang Y, Cui J, Wang Z-M. Elevated levels of autophagy-related marker ULK1 and mitochondrion-associated autophagy inhibitor LRPPRC are associated with biochemical progression and overall survival after androgen deprivation therapy in patients with metastatic prostate cancer. J. Clin. Pathol. 2017;70:383–389. doi: 10.1136/jclinpath-2016-203926. [DOI] [PubMed] [Google Scholar]
- 545.Zou Y, et al. High expression levels of unc-51-like kinase 1 as a predictor of poor prognosis in colorectal cancer. Oncol. Lett. 2015;10:1583–1588. doi: 10.3892/ol.2015.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Wu D-H, et al. Combination of ULK1 and LC3B improve prognosis assessment of hepatocellular carcinoma. Biomed. Pharmacother. 2018;97:195–202. doi: 10.1016/j.biopha.2017.10.025. [DOI] [PubMed] [Google Scholar]
- 547.Schmitz KJ, Ademi C, Bertram S, Schmid KW, Baba HA. Prognostic relevance of autophagy-related markers LC3, p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients with respect to KRAS mutational status. World J. Surg. Oncol. 2016;14:189. doi: 10.1186/s12957-016-0946-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Tang J, et al. Low expression of ULK1 is associated with operable breast cancer progression and is an adverse prognostic marker of survival for patients. Breast Cancer Res. Treat. 2012;134:549–560. doi: 10.1007/s10549-012-2080-y. [DOI] [PubMed] [Google Scholar]
- 549.John Clotaire DZ, et al. MiR-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem. Biophys. Res. Commun. 2016;472:194–200. doi: 10.1016/j.bbrc.2016.02.093. [DOI] [PubMed] [Google Scholar]
- 550.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Tao M, Liu T, You Q, Jiang Z. p62 as a therapeutic target for tumor. Eur. J. Med. Chem. 2020;193:112231. doi: 10.1016/j.ejmech.2020.112231. [DOI] [PubMed] [Google Scholar]
- 552.Islam MA, Sooro MA, Zhang P. Autophagic regulation of p62 is critical for cancer therapy. Int. J. Mol. Sci. 2018;19:1405. doi: 10.3390/ijms19051405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Sanz L, Sanchez P, Lallena MJ, Diaz-Meco MT, Moscat J. The interaction of p62 with RIP links the atypical PKCs to NF-kappaB activation. EMBO J. 1999;18:3044–3053. doi: 10.1093/emboj/18.11.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Jadhav T, Geetha T, Jiang J, Wooten MW. Identification of a consensus site for TRAF6/p62 polyubiquitination. Biochem. Biophys. Res. Commun. 2008;371:521–524. doi: 10.1016/j.bbrc.2008.04.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 555.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 556.Komatsu M, et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- 557.Moscat J, Karin M, Diaz-Meco MT. p62 in cancer: signaling adaptor beyond autophagy. Cell. 2016;167:606–609. doi: 10.1016/j.cell.2016.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Cohen-Kaplan V, et al. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26 S proteasome. Proc. Natl Acad. Sci. USA. 2016;113:E7490–E7499. doi: 10.1073/pnas.1615455113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 559.Pugsley, H. R. Assessing autophagic flux by measuring LC3, p62, and LAMP1 co-localization using multispectral imaging flow cytometry. J. Vis. Exp. 55637 (2017). [DOI] [PMC free article] [PubMed]
- 560.Iwadate R, et al. High expression of p62 protein Is associated with poor prognosis and aggressive phenotypes in endometrial cancer. Am. J. Pathol. 2015;185:2523–2533. doi: 10.1016/j.ajpath.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 561.Iwadate R, et al. High expression of SQSTM1/p62 protein is associated with poor prognosis in epithelial ovarian cancer. Acta Histochem. Cytochem. 2014;47:295–301. doi: 10.1267/ahc.14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Inoue D, et al. Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci. 2012;103:760–766. doi: 10.1111/j.1349-7006.2012.02216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Rolland P, et al. The ubiquitin-binding protein p62 is expressed in breast cancers showing features of aggressive disease. Endocr. Relat. Cancer. 2007;14:73–80. doi: 10.1677/erc.1.01312. [DOI] [PubMed] [Google Scholar]
- 564.Yuan Z-Y, et al. Accumulation of p62 is associated with poor prognosis in patients with triple-negative breast cancer. Onco. Targets Ther. 2013;6:883–888. doi: 10.2147/OTT.S46222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Su Z, Yang Z, Xie L, Dewitt JP, Chen Y. Cancer therapy in the necroptosis era. Cell Death Differ. 2016;23:748–756. doi: 10.1038/cdd.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Razaghi A, Heimann K, Schaeffer PM, Gibson SB. Negative regulators of cell death pathways in cancer: perspective on biomarkers and targeted therapies. Apoptosis. 2018;23:93–112. doi: 10.1007/s10495-018-1440-4. [DOI] [PubMed] [Google Scholar]
- 567.Yu X, Deng Q, Bode AM, Dong Z, Cao Y. The role of necroptosis, an alternative form of cell death, in cancer therapy. Expert Rev. Anticancer Ther. 2013;13:883–893. doi: 10.1586/14737140.2013.811180. [DOI] [PubMed] [Google Scholar]
- 568.Xie Y, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–379. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.He R, Wang Z, Dong S, Chen Z, Zhou W. Understanding necroptosis in pancreatic diseases. Biomolecules. 2022;12:828. doi: 10.3390/biom12060828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Zhang L, et al. Tumor necrosis as a prognostic variable for the clinical outcome in patients with renal cell carcinoma: a systematic review and meta-analysis. BMC Cancer. 2018;18:870. doi: 10.1186/s12885-018-4773-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Richards CH, Mohammed Z, Qayyum T, Horgan PG, McMillan DC. The prognostic value of histological tumor necrosis in solid organ malignant disease: a systematic review. Futur. Oncol. 2011;7:1223–1235. doi: 10.2217/fon.11.99. [DOI] [PubMed] [Google Scholar]
- 572.Salah S, Lewin J, Amir E, Abdul Razak A. Tumor necrosis and clinical outcomes following neoadjuvant therapy in soft tissue sarcoma: a systematic review and meta-analysis. Cancer Treat. Rev. 2018;69:1–10. doi: 10.1016/j.ctrv.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 573.Ofengeim D, Yuan J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat. Rev. Mol. Cell Biol. 2013;14:727–736. doi: 10.1038/nrm3683. [DOI] [PubMed] [Google Scholar]
- 574.Huang X, et al. Bypassing drug resistance by triggering necroptosis: Recent advances in mechanisms and its therapeutic exploitation in leukemia. J. Exp. Clin. Cancer Res. 2018;37:310. doi: 10.1186/s13046-018-0976-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Murphy JM, et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity. 2013;39:443–453. doi: 10.1016/j.immuni.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 577.Lalaoui N, Brumatti G. Relevance of necroptosis in cancer. Immunol. Cell Biol. 2017;95:137–145. doi: 10.1038/icb.2016.120. [DOI] [PubMed] [Google Scholar]
- 578.Qin X, Ma D, Tan Y-X, Wang H-Y, Cai Z. The role of necroptosis in cancer: a double-edged sword? Biochim. Biophys. Acta Rev. Cancer. 2019;1871:259–266. doi: 10.1016/j.bbcan.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 579.Chan FK-M. RIPK3 slams the brake on leukemogenesis. Cancer Cell. 2016;30:7–9. doi: 10.1016/j.ccell.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 580.Höckendorf U, et al. RIPK3 restricts myeloid leukemogenesis by promoting cell death and differentiation of leukemia initiating cells. Cancer Cell. 2016;30:75–91. doi: 10.1016/j.ccell.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 581.Koo GB, et al. Methylation-dependent loss of RIP3 expression in cancer represses programmed necrosis in response to chemotherapeutics. Cell Res. 2015;25:707–725. doi: 10.1038/cr.2015.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Geserick P, et al. Absence of RIPK3 predicts necroptosis resistance in malignant melanoma. Cell Death Dis. 2015;6:e1884. doi: 10.1038/cddis.2015.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Fukasawa M, et al. Microarray analysis of promoter methylation in lung cancers. J. Hum. Genet. 2006;51:368–374. doi: 10.1007/s10038-005-0355-4. [DOI] [PubMed] [Google Scholar]
- 584.Moriwaki K, Bertin J, Gough PJ, Orlowski GM, Chan FKM. Differential roles of RIPK1 and RIPK3 in TNF-induced necroptosis and chemotherapeutic agent-induced cell death. Cell Death Dis. 2015;6:e1636. doi: 10.1038/cddis.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Bozec D, Iuga AC, Roda G, Dahan S, Yeretssian G. Critical function of the necroptosis adaptor RIPK3 in protecting from intestinal tumorigenesis. Oncotarget. 2016;7:46384–46400. doi: 10.18632/oncotarget.10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Feng X, et al. Receptor-interacting protein kinase 3 is a predictor of survival and plays a tumor suppressive role in colorectal cancer. Neoplasma. 2015;62:592–601. doi: 10.4149/neo_2015_071. [DOI] [PubMed] [Google Scholar]
- 587.McCabe KE, et al. Triggering necroptosis in cisplatin and IAP antagonist-resistant ovarian carcinoma. Cell Death Dis. 2014;5:e1496. doi: 10.1038/cddis.2014.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 588.Seifert L, et al. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. doi: 10.1038/nature17403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Liu Z-Y, et al. RIP3 promotes colitis-associated colorectal cancer by controlling tumor cell proliferation and CXCL1-induced immune suppression. Theranostics. 2019;9:3659–3673. doi: 10.7150/thno.32126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Zhan C, Huang M, Yang X, Hou J. MLKL: functions beyond serving as the executioner of necroptosis. Theranostics. 2021;11:4759–4769. doi: 10.7150/thno.54072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.Colbert LE, et al. Pronecrotic mixed lineage kinase domain-like protein expression is a prognostic biomarker in patients with early-stage resected pancreatic adenocarcinoma. Cancer. 2013;119:3148–3155. doi: 10.1002/cncr.28144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 592.Ertao Z, et al. Prognostic value of mixed lineage kinase domain-like protein expression in the survival of patients with gastric caner. Tumour Biol. 2016;37:13679–13685. doi: 10.1007/s13277-016-5229-1. [DOI] [PubMed] [Google Scholar]
- 593.He L, Peng K, Liu Y, Xiong J, Zhu FF. Low expression of mixed lineage kinase domain-like protein is associated with poor prognosis in ovarian cancer patients. Onco. Targets Ther. 2013;6:1539–1543. doi: 10.2147/OTT.S52805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Ruan J, Mei L, Zhu Q, Shi G, Wang H. Mixed lineage kinase domain-like protein is a prognostic biomarker for cervical squamous cell cancer. Int. J. Clin. Exp. Pathol. 2015;8:15035–15038. [PMC free article] [PubMed] [Google Scholar]
- 595.Li X, et al. Association of mixed lineage kinase domain-like protein expression with prognosis in patients with colon cancer. Technol. Cancer Res. Treat. 2017;16:428–434. doi: 10.1177/1533034616655909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 596.Liu X, et al. Key roles of necroptotic factors in promoting tumor growth. Oncotarget. 2016;7:22219–22233. doi: 10.18632/oncotarget.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Suhail Y, et al. Systems biology of cancer metastasis. Cell Syst. 2019;9:109–127. doi: 10.1016/j.cels.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 599.Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- 600.Fares J, Fares MY, Khachfe HH, Salhab HA, Fares Y. Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target Ther. 2020;5:28. doi: 10.1038/s41392-020-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 601.Maishi N, Hida K. Tumor endothelial cells accelerate tumor metastasis. Cancer Sci. 2017;108:1921–1926. doi: 10.1111/cas.13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 603.Liu Q, et al. Factors involved in cancer metastasis: a better understanding to ‘seed and soil’ hypothesis. Mol. Cancer. 2017;16:176. doi: 10.1186/s12943-017-0742-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Curran S, Murray GI. Matrix metalloproteinases: molecular aspects of their roles in tumour invasion and metastasis. Eur. J. Cancer. 2000;36:1621–1630. doi: 10.1016/S0959-8049(00)00156-8. [DOI] [PubMed] [Google Scholar]
- 605.Steeg PS. Targeting metastasis. Nat. Rev. Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Ma B, Wells A, Clark AM. The pan-therapeutic resistance of disseminated tumor cells: role of phenotypic plasticity and the metastatic microenvironment. Semin. Cancer Biol. 2020;60:138–147. doi: 10.1016/j.semcancer.2019.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 607.Howard EW, Camm KD, Wong YC, Wang XH. E-cadherin upregulation as a therapeutic goal in cancer treatment. Mini Rev. Med. Chem. 2008;8:496–518. doi: 10.2174/138955708784223521. [DOI] [PubMed] [Google Scholar]
- 608.Hazan RB, Qiao R, Keren R, Badano I, Suyama K. Cadherin switch in tumor progression. Ann. N. Y. Acad. Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- 609.Wijnhoven BP, Dinjens WN, Pignatelli M. E-cadherin-catenin cell-cell adhesion complex and human cancer. Br. J. Surg. 2000;87:992–1005. doi: 10.1046/j.1365-2168.2000.01513.x. [DOI] [PubMed] [Google Scholar]
- 610.Beavon IR. The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation. Eur. J. Cancer. 2000;36:1607–1620. doi: 10.1016/S0959-8049(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 611.Steinberg MS, McNutt PM. Cadherins and their connections: adhesion junctions have broader functions. Curr. Opin. Cell Biol. 1999;11:554–560. doi: 10.1016/S0955-0674(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 612.Venhuizen J-H, Jacobs FJC, Span PN, Zegers MM. P120 and E-cadherin: double-edged swords in tumor metastasis. Semin. Cancer Biol. 2020;60:107–120. doi: 10.1016/j.semcancer.2019.07.020. [DOI] [PubMed] [Google Scholar]
- 613.Schmalhofer O, Brabletz S, Brabletz T. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 2009;28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 614.Yilmaz M, Christofori G, Lehembre F. Distinct mechanisms of tumor invasion and metastasis. Trends Mol. Med. 2007;13:535–541. doi: 10.1016/j.molmed.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 615.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 616.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem. Sci. 1999;24:73–76. doi: 10.1016/S0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 617.Fearon ER. Connecting estrogen receptor function, transcriptional repression, and E-cadherin expression in breast cancer. Cancer Cell. 2003;3:307–310. doi: 10.1016/S1535-6108(03)00087-4. [DOI] [PubMed] [Google Scholar]
- 618.Karayiannakis AJ, et al. Aberrant E-cadherin expression associated with loss of differentiation and advanced stage in human pancreatic cancer. Anticancer Res. 1998;18:4177–4180. [PubMed] [Google Scholar]
- 619.Pignatelli M, et al. Loss of membranous E-cadherin expression in pancreatic cancer: correlation with lymph node metastasis, high grade, and advanced stage. J. Pathol. 1994;174:243–248. doi: 10.1002/path.1711740403. [DOI] [PubMed] [Google Scholar]
- 620.Van Aken J, et al. Immunohistochemical analysis of E-cadherin expression in human colorectal tumours. Pathol. Res. Pract. 1993;189:975–978. doi: 10.1016/S0344-0338(11)80667-9. [DOI] [PubMed] [Google Scholar]
- 621.Mayer B, et al. E-cadherin expression in primary and metastatic gastric cancer: down-regulation correlates with cellular dedifferentiation and glandular disintegration. Cancer Res. 1993;53:1690–1695. [PubMed] [Google Scholar]
- 622.Chetty R, Serra S, Asa SL. Loss of membrane localization and aberrant nuclear E-cadherin expression correlates with invasion in pancreatic endocrine tumors. Am. J. Surg. Pathol. 2008;32:413–419. doi: 10.1097/PAS.0b013e31813547f8. [DOI] [PubMed] [Google Scholar]
- 623.Kadowaki T, et al. E-cadherin and alpha-catenin expression in human esophageal cancer. Cancer Res. 1994;54:291–296. [PubMed] [Google Scholar]
- 624.Slagle BL, Zhou YZ, Birchmeier W, Scorsone KA. Deletion of the E-cadherin gene in hepatitis B virus-positive Chinese hepatocellular carcinomas. Hepatology. 1993;18:757–762. doi: 10.1002/hep.1840180402. [DOI] [PubMed] [Google Scholar]
- 625.Bremnes RM, Veve R, Hirsch FR, Franklin WA. The E-cadherin cell-cell adhesion complex and lung cancer invasion, metastasis, and prognosis. Lung Cancer. 2002;36:115–124. doi: 10.1016/S0169-5002(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 626.Otto T, et al. Inverse relation of E-cadherin and autocrine motility factor receptor expression as a prognostic factor in patients with bladder carcinomas. Cancer Res. 1994;54:3120–3123. [PubMed] [Google Scholar]
- 627.Morton RA, Ewing CM, Nagafuchi A, Tsukita S, Isaacs WB. Reduction of E-cadherin levels and deletion of the alpha-catenin gene in human prostate cancer cells. Cancer Res. 1993;53:3585–3590. [PubMed] [Google Scholar]
- 628.Umbas R, et al. Expression of the cellular adhesion molecule E-cadherin is reduced or absent in high-grade prostate cancer. Cancer Res. 1992;52:5104–5109. [PubMed] [Google Scholar]
- 629.Rasbridge SA, Gillett CE, Sampson SA, Walsh FS, Millis RR. Epithelial (E-) and placental (P-) cadherin cell adhesion molecule expression in breast carcinoma. J. Pathol. 1993;169:245–250. doi: 10.1002/path.1711690211. [DOI] [PubMed] [Google Scholar]
- 630.Saito T, Nishimura M, Yamasaki H, Kudo R. Hypermethylation in promoter region of E-cadherin gene is associated with tumor dedifferention and myometrial invasion in endometrial carcinoma. Cancer. 2003;97:1002–1009. doi: 10.1002/cncr.11157. [DOI] [PubMed] [Google Scholar]
- 631.Veatch AL, Carson LF, Ramakrishnan S. Differential expression of the cell-cell adhesion molecule E-cadherin in ascites and solid human ovarian tumor cells. Int. J. Cancer. 1994;58:393–399. doi: 10.1002/ijc.2910580315. [DOI] [PubMed] [Google Scholar]
- 632.Brabant G, et al. E-cadherin: a differentiation marker in thyroid malignancies. Cancer Res. 1993;53:4987–4993. [PubMed] [Google Scholar]
- 633.Kurtz KA, Hoffman HT, Zimmerman MB, Robinson RA. Decreased E-cadherin but not beta-catenin expression is associated with vascular invasion and decreased survival in head and neck squamous carcinomas. Otolaryngol. Head Neck Surg. 2006;134:142–146. doi: 10.1016/j.otohns.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 634.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat. Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 635.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol. Cancer. 2016;15:18. doi: 10.1186/s12943-016-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Dong C, et al. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Debnath P, Huirem RS, Dutta P, Palchaudhuri S. Epithelial-mesenchymal transition and its transcription factors. Biosci. Rep. 2022;42:BSR20211754. doi: 10.1042/BSR20211754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 638.Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev. Cell. 2019;49:361–374. doi: 10.1016/j.devcel.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat. Cell Biol. 2019;21:102–112. doi: 10.1038/s41556-018-0196-y. [DOI] [PubMed] [Google Scholar]
- 640.Pearlman RL, Montes de Oca MK, Pal HC, Afaq F. Potential therapeutic targets of epithelial-mesenchymal transition in melanoma. Cancer Lett. 2017;391:125–140. doi: 10.1016/j.canlet.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 641.Sánchez-Tilló E, et al. EMT-activating transcription factors in cancer: Beyond EMT and tumor invasiveness. Cell Mol. Life Sci. 2012;69:3429–3456. doi: 10.1007/s00018-012-1122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 642.Brabletz S, Schuhwerk H, Brabletz T, Stemmler MP. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 2021;40:e108647. doi: 10.15252/embj.2021108647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 643.Zhang Z, et al. Genomics and prognosis analysis of epithelial-mesenchymal transition in colorectal cancer patients. BMC Cancer. 2020;20:1135. doi: 10.1186/s12885-020-07615-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Matysiak M, Kapka-Skrzypczak L, Jodłowska-Jędrych B, Kruszewski M. EMT promoting transcription factors as prognostic markers in human breast cancer. Arch. Gynecol. Obstet. 2017;295:817–825. doi: 10.1007/s00404-017-4304-1. [DOI] [PubMed] [Google Scholar]
- 645.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: Beyond EMT and MET. Biochim. Biophys. Acta Rev. Cancer. 2017;1868:584–591. doi: 10.1016/j.bbcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 646.Ng L, Poon RTP, Pang R. Biomarkers for predicting future metastasis of human gastrointestinal tumors. Cell Mol. Life Sci. 2013;70:3631–3656. doi: 10.1007/s00018-013-1266-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 647.Ansieau S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 648.Yang J, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 649.Khales SA, Mozaffari-Jovin S, Geerts D, Abbaszadegan MR. TWIST1 activates cancer stem cell marker genes to promote epithelial-mesenchymal transition and tumorigenesis in esophageal squamous cell carcinoma. BMC Cancer. 2022;22:1272. doi: 10.1186/s12885-022-10252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 650.Sepporta M-V, et al. TWIST1 expression is associated with high-risk neuroblastoma and promotes primary and metastatic tumor growth. Commun. Biol. 2022;5:42. doi: 10.1038/s42003-021-02958-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 651.Xiong H, et al. Twist1 enhances hypoxia induced radioresistance in cervical cancer cells by promoting nuclear EGFR localization. J. Cancer. 2017;8:345–353. doi: 10.7150/jca.16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 652.Ohba K, et al. High expression of twist is associated with tumor aggressiveness and poor prognosis in patients with renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2014;7:3158–3165. [PMC free article] [PubMed] [Google Scholar]
- 653.Norozi F, Ahmadzadeh A, Shahjahani M, Shahrabi S, Saki N. Twist as a new prognostic marker in hematological malignancies. Clin. Transl. Oncol. 2016;18:113–124. doi: 10.1007/s12094-015-1357-0. [DOI] [PubMed] [Google Scholar]
- 654.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 655.Nüsslein-Volhard C, Wieschaus E, Kluding H. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: I. Zygotic loci on the second chromosome. Wilhelm. Roux’s Arch. Dev. Biol. 1984;193:267–282. doi: 10.1007/BF00848156. [DOI] [PubMed] [Google Scholar]
- 656.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of Snail in EMT and tumorigenesis. Curr. Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Tran HD, et al. Transient SNAIL1 expression is necessary for metastatic competence in breast cancer. Cancer Res. 2014;74:6330–6340. doi: 10.1158/0008-5472.CAN-14-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.De Craene B, et al. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005;65:6237–6244. doi: 10.1158/0008-5472.CAN-04-3545. [DOI] [PubMed] [Google Scholar]
- 659.Zhang M, Dong X, Zhang D, Chen X, Zhu X. High expression of Snail and NF-κB predicts poor survival in Chinese hepatocellular carcinoma patients. Oncotarget. 2017;8:4543–4548. doi: 10.18632/oncotarget.13891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 660.Kosaka T, et al. Expression of snail in upper urinary tract urothelial carcinoma: prognostic significance and implications for tumor invasion. Clin. Cancer Res. 2010;16:5814–5823. doi: 10.1158/1078-0432.CCR-10-0230. [DOI] [PubMed] [Google Scholar]
- 661.Shioiri M, et al. Slug expression is an independent prognostic parameter for poor survival in colorectal carcinoma patients. Br. J. Cancer. 2006;94:1816–1822. doi: 10.1038/sj.bjc.6603193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 662.XU, S. et al. Expression of twist, slug and snail in esophageal squamous cell carcinoma and their prognostic significance. Oncol. Lett. 21, 184 (2021). [DOI] [PMC free article] [PubMed]
- 663.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell. Mol. Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 664.Brabletz S, Brabletz T. The ZEB/miR-200 feedback loop–a motor of cellular plasticity in development and cancer? EMBO Rep. 2010;11:670–677. doi: 10.1038/embor.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 665.Krebs AM, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017;19:518–529. doi: 10.1038/ncb3513. [DOI] [PubMed] [Google Scholar]
- 666.Zhang P, Sun Y, Ma L. ZEB1: at the crossroads of epithelial-mesenchymal transition, metastasis and therapy resistance. Cell Cycle. 2015;14:481–487. doi: 10.1080/15384101.2015.1006048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 667.Galván JA, et al. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br. J. Cancer. 2015;112:1944–1950. doi: 10.1038/bjc.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 668.Sangrador I, et al. Zeb1 in stromal myofibroblasts promotes Kras-driven development of pancreatic cancer. Cancer Res. 2018;78:2624–2637. doi: 10.1158/0008-5472.CAN-17-1882. [DOI] [PubMed] [Google Scholar]
- 669.Zawati I, et al. Association of ZEB1 and Vimentin with poor prognosis in metaplastic breast cancer. Ann. Diagn. Pathol. 2022;59:151954. doi: 10.1016/j.anndiagpath.2022.151954. [DOI] [PubMed] [Google Scholar]
- 670.Hegarty SV, Sullivan AM, O’Keeffe GW. Zeb2: a multifunctional regulator of nervous system development. Prog. Neurobiol. 2015;132:81–95. doi: 10.1016/j.pneurobio.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 671.Kang Y, Massagué J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277–279. doi: 10.1016/j.cell.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 672.Burks HE, et al. ZEB2 regulates endocrine therapy sensitivity and metastasis in luminal a breast cancer cells through a non-canonical mechanism. Breast Cancer Res. Treat. 2021;189:25–37. doi: 10.1007/s10549-021-06256-x. [DOI] [PubMed] [Google Scholar]
- 673.Li N, et al. An FBXW7-ZEB2 axis links EMT and tumour microenvironment to promote colorectal cancer stem cells and chemoresistance. Oncogenesis. 2019;8:13. doi: 10.1038/s41389-019-0125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 674.Maeda G, et al. Expression of SIP1 in oral squamous cell carcinomas: implications for E-cadherin expression and tumor progression. Int. J. Oncol. 2005;27:1535–1541. [PubMed] [Google Scholar]
- 675.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat. Rev. Mol. Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 676.Cooper CS, et al. Molecular cloning of a new transforming gene from a chemically transformed human cell line. Nature. 1984;311:29–33. doi: 10.1038/311029a0. [DOI] [PubMed] [Google Scholar]
- 677.Bradley CA, et al. Targeting c-MET in gastrointestinal tumours: rationale, opportunities and challenges. Nat. Rev. Clin. Oncol. 2017;14:562–576. doi: 10.1038/nrclinonc.2017.40. [DOI] [PubMed] [Google Scholar]
- 678.Faiella A, Riccardi F, Cartenì G, Chiurazzi M, Onofrio L. The emerging role of c-Met in carcinogenesis and clinical implications as a possible therapeutic target. J. Oncol. 2022;2022:5179182. doi: 10.1155/2022/5179182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 679.Zou HY, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–4417. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 680.Rong S, Segal S, Anver M, Resau JH, Vande Woude GF. Invasiveness and metastasis of NIH 3T3 cells induced by Met-hepatocyte growth factor/scatter factor autocrine stimulation. Proc. Natl Acad. Sci. USA. 1994;91:4731–4735. doi: 10.1073/pnas.91.11.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 681.Moshitch-Moshkovitz S, et al. In vivo direct molecular imaging of early tumorigenesis and malignant progression induced by transgenic expression of GFP-Met. Neoplasia. 2006;8:353–363. doi: 10.1593/neo.05634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 682.Otsuka T, et al. c-Met autocrine activation induces development of malignant melanoma and acquisition of the metastatic phenotype. Cancer Res. 1998;58:5157–5167. [PubMed] [Google Scholar]
- 683.Demkova L, Kucerova L. Role of the HGF/c-MET tyrosine kinase inhibitors in metastasic melanoma. Mol. Cancer. 2018;17:26. doi: 10.1186/s12943-018-0795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 684.Yao J-F, et al. Role of HGF/c-Met in the treatment of colorectal cancer with liver metastasis. J. Biochem. Mol. Toxicol. 2019;33:e22316. doi: 10.1002/jbt.22316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 685.Lee JH, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene. 2000;19:4947–4953. doi: 10.1038/sj.onc.1203874. [DOI] [PubMed] [Google Scholar]
- 686.Ma PC, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–6281. [PubMed] [Google Scholar]
- 687.Di Renzo MF, et al. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene. 2000;19:1547–1555. doi: 10.1038/sj.onc.1203455. [DOI] [PubMed] [Google Scholar]
- 688.Hartmann S, Bhola NE, Grandis JR. HGF/Met signaling in head and neck cancer: impact on the tumor microenvironment. Clin. Cancer Res. 2016;22:4005–4013. doi: 10.1158/1078-0432.CCR-16-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Boromand N, et al. Clinical and prognostic value of the C-Met/HGF signaling pathway in cervical cancer. J. Cell. Physiol. 2018;233:4490–4496. doi: 10.1002/jcp.26232. [DOI] [PubMed] [Google Scholar]
- 690.Wang H, et al. The function of the HGF/c-Met axis in hepatocellular carcinoma. Front. Cell Dev. Biol. 2020;8:55. doi: 10.3389/fcell.2020.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Miranda O, Farooqui M, Siegfried JM. Status of agents targeting the HGF/c-Met axis in lung cancer. Cancers. 2018;10:280. doi: 10.3390/cancers10090280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 692.Goyal L, Muzumdar MD, Zhu AX. Targeting the HGF/c-MET pathway in hepatocellular carcinoma. Clin. Cancer Res. 2013;19:2310–2318. doi: 10.1158/1078-0432.CCR-12-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 693.Kim HJ. Therapeutic strategies for ovarian cancer in point of HGF/c-MET targeting. Medicina. 2022;58:649. doi: 10.3390/medicina58050649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 694.Lam BQ, Dai L, Qin Z. The role of HGF/c-MET signaling pathway in lymphoma. J. Hematol. Oncol. 2016;9:135. doi: 10.1186/s13045-016-0366-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 695.Guryanova OA, Bao S. How scatter factor receptor c-MET contributes to tumor radioresistance: ready, set, scatter! J. Natl Cancer Inst. 2011;103:617–619. doi: 10.1093/jnci/djr103. [DOI] [PubMed] [Google Scholar]
- 696.Kwak EL, et al. Molecular heterogeneity and receptor coamplification drive resistance to targeted therapy in MET-Amplified esophagogastric cancer. Cancer Discov. 2015;5:1271–1281. doi: 10.1158/2159-8290.CD-15-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Mrozik KM, et al. Therapeutic targeting of N-cadherin is an effective treatment for multiple myeloma. Br. J. Haematol. 2015;171:387–399. doi: 10.1111/bjh.13596. [DOI] [PubMed] [Google Scholar]
- 698.Hatta K, Takeichi M. Expression of N-cadherin adhesion molecules associated with early morphogenetic events in chick development. Nature. 1986;320:447–449. doi: 10.1038/320447a0. [DOI] [PubMed] [Google Scholar]
- 699.Cao Z-Q, Wang Z, Leng P. Aberrant N-cadherin expression in cancer. Biomed. Pharmacother. 2019;118:109320. doi: 10.1016/j.biopha.2019.109320. [DOI] [PubMed] [Google Scholar]
- 700.Leckband DE, de Rooij J. Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 2014;30:291–315. doi: 10.1146/annurev-cellbio-100913-013212. [DOI] [PubMed] [Google Scholar]
- 701.Blaschuk OW. N-cadherin antagonists as oncology therapeutics. Philos. Trans. R. Soc. Lond. Ser. B, Biol. Sci. 2015;370:20140039. doi: 10.1098/rstb.2014.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Mrozik KM, Blaschuk OW, Cheong CM, Zannettino ACW, Vandyke K. N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer. BMC Cancer. 2018;18:939. doi: 10.1186/s12885-018-4845-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 703.Ciołczyk-Wierzbicka D, Laidler P. The inhibition of invasion of human melanoma cells through N-cadherin knock-down. Med. Oncol. 2018;35:42. doi: 10.1007/s12032-018-1104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 704.Lammens T, et al. N-Cadherin in neuroblastoma disease: expression and clinical significance. PLoS ONE. 2012;7:e31206. doi: 10.1371/journal.pone.0031206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Hulit J, et al. N-cadherin signaling potentiates mammary tumor metastasis via enhanced extracellular signal-regulated kinase activation. Cancer Res. 2007;67:3106–3116. doi: 10.1158/0008-5472.CAN-06-3401. [DOI] [PubMed] [Google Scholar]
- 706.Klymenko Y, et al. Cadherin composition and multicellular aggregate invasion in organotypic models of epithelial ovarian cancer intraperitoneal metastasis. Oncogene. 2017;36:5840–5851. doi: 10.1038/onc.2017.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 707.Mariotti A, Perotti A, Sessa C, Rüegg C. N-cadherin as a therapeutic target in cancer. Expert Opin. Investig. Drugs. 2007;16:451–465. doi: 10.1517/13543784.16.4.451. [DOI] [PubMed] [Google Scholar]
- 708.Groen RWJ, et al. N-cadherin-mediated interaction with multiple myeloma cells inhibits osteoblast differentiation. Haematologica. 2011;96:1653–1661. doi: 10.3324/haematol.2010.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 709.Sadler NM, Harris BR, Metzger BA, Kirshner J. N-cadherin impedes proliferation of the multiple myeloma cancer stem cells. Am. J. Blood Res. 2013;3:271–285. [PMC free article] [PubMed] [Google Scholar]
- 710.Shintani Y, et al. ADH-1 suppresses N-cadherin-dependent pancreatic cancer progression. Int. J. Cancer. 2008;122:71–77. doi: 10.1002/ijc.23027. [DOI] [PubMed] [Google Scholar]
- 711.Sun Y, et al. N-cadherin inhibitor creates a microenvironment that protect TILs from immune checkpoints and Treg cells. J. Immunother. Cancer. 2021;9:e002138. doi: 10.1136/jitc-2020-002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 712.Richards L. Genetics: N-cadherin—a target for prostate cancer therapy. Nat. Rev. Clin. Oncol. 2011;8:63. doi: 10.1038/nrclinonc.2010.212. [DOI] [PubMed] [Google Scholar]
- 713.Tokito A, Jougasaki M. Matrix metalloproteinases in non-neoplastic disorders. Int. J. Mol. Sci. 2016;17:1178. doi: 10.3390/ijms17071178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 714.Huang H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: recent advances. Sensors. 2018;18:3249. doi: 10.3390/s18103249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 715.de Almeida LGN, et al. Matrix metalloproteinases: from molecular mechanisms to physiology, pathophysiology, and pharmacology. Pharmacol. Rev. 2022;74:712–768. doi: 10.1124/pharmrev.121.000349. [DOI] [PubMed] [Google Scholar]
- 716.Roy R, Yang J, Moses MA. Matrix metalloproteinases as novel biomarkers and potential therapeutic targets in human cancer. J. Clin. Oncol. 2009;27:5287–5297. doi: 10.1200/JCO.2009.23.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 717.Klein T, Bischoff R. Physiology and pathophysiology of matrix metalloproteases. Amino Acids. 2011;41:271–290. doi: 10.1007/s00726-010-0689-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 718.Szarvas T, Vom Dorp F, Ergün S, Rübben H. Matrix metalloproteinases and their clinical relevance in urinary bladder cancer. Nat. Rev. Urol. 2011;8:241–254. doi: 10.1038/nrurol.2011.44. [DOI] [PubMed] [Google Scholar]
- 719.Talvensaari-Mattila A, Pääkkö P, Höyhtyä M, Blanco-Sequeiros G, Turpeenniemi-Hujanen T. Matrix metalloproteinase-2 immunoreactive protein: a marker of aggressiveness in breast carcinoma. Cancer. 1998;83:1153–1162. doi: 10.1002/(SICI)1097-0142(19980915)83:6<1153::AID-CNCR14>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 720.Murray GI, Duncan ME, O’Neil P, Melvin WT, Fothergill JE. Matrix metalloproteinase-1 is associated with poor prognosis in colorectal cancer. Nat. Med. 1996;2:461–462. doi: 10.1038/nm0496-461. [DOI] [PubMed] [Google Scholar]
- 721.Sier CF, et al. Tissue levels of matrix metalloproteinases MMP-2 and MMP-9 are related to the overall survival of patients with gastric carcinoma. Br. J. Cancer. 1996;74:413–417. doi: 10.1038/bjc.1996.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 722.Murray GI, et al. Matrix metalloproteinase-1 is associated with poor prognosis in oesophageal cancer. J. Pathol. 1998;185:256–261. doi: 10.1002/(SICI)1096-9896(199807)185:3<256::AID-PATH115>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 723.Vandenbroucke RE, Dejonckheere E, Libert C. A therapeutic role for matrix metalloproteinase inhibitors in lung diseases? Eur. Respir. J. 2011;38:1200–1214. doi: 10.1183/09031936.00027411. [DOI] [PubMed] [Google Scholar]
- 724.Isaacson KJ, Martin Jensen M, Subrahmanyam NB, Ghandehari H. Matrix-metalloproteinases as targets for controlled delivery in cancer: an analysis of upregulation and expression. J. Control. Release. 2017;259:62–75. doi: 10.1016/j.jconrel.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 725.Gong L, et al. Prognostic impact of serum and tissue MMP-9 in non-small cell lung cancer: a systematic review and meta-analysis. Oncotarget. 2016;7:18458–18468. doi: 10.18632/oncotarget.7607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 726.Li Y, Wu T, Zhang B, Yao Y, Yin G. Matrix metalloproteinase-9 is a prognostic marker for patients with cervical cancer. Med. Oncol. 2012;29:3394–3399. doi: 10.1007/s12032-012-0283-z. [DOI] [PubMed] [Google Scholar]
- 727.Lian P-L, et al. Integrin αvβ6 and matrix metalloproteinase 9 correlate with survival in gastric cancer. World J. Gastroenterol. 2016;22:3852–3859. doi: 10.3748/wjg.v22.i14.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 728.Li L-N, Zhou X, Gu Y, Yan J. Prognostic value of MMP-9 in ovarian cancer: a meta-analysis. Asian Pac. J. Cancer Prev. 2013;14:4107–4113. doi: 10.7314/APJCP.2013.14.7.4107. [DOI] [PubMed] [Google Scholar]
- 729.Yousef EM, Tahir MR, St-Pierre Y, Gaboury LA. MMP-9 expression varies according to molecular subtypes of breast cancer. BMC Cancer. 2014;14:609. doi: 10.1186/1471-2407-14-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 730.Wang J, et al. Matrix metalloproteinase 9 (MMP-9) in osteosarcoma: review and meta-analysis. Clin. Chim. Acta. 2014;433:225–231. doi: 10.1016/j.cca.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 731.Tian M, et al. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer. 2008;8:241. doi: 10.1186/1471-2407-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 732.Wang T, Zhang Y, Bai J, Xue Y, Peng Q. MMP1 and MMP9 are potential prognostic biomarkers and targets for uveal melanoma. BMC Cancer. 2021;21:1068. doi: 10.1186/s12885-021-08788-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Groelly FJ, Fawkes M, Dagg RA, Blackford AN, Tarsounas M. Targeting DNA damage response pathways in cancer. Nat. Rev. Cancer. 2023;23:78–94. doi: 10.1038/s41568-022-00535-5. [DOI] [PubMed] [Google Scholar]
- 734.Chu Y-Y, Yam C, Yamaguchi H, Hung M-C. Biomarkers beyond BRCA: promising combinatorial treatment strategies in overcoming resistance to PARP inhibitors. J. Biomed. Sci. 2022;29:86. doi: 10.1186/s12929-022-00870-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 735.Wang Y, et al. PARP inhibitors in gastric cancer: beacon of hope. J. Exp. Clin. Cancer Res. 2021;40:211. doi: 10.1186/s13046-021-02005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Quinet A, Lemaçon D, Vindigni A. Replication fork reversal: players and guardians. Mol. Cell. 2017;68:830–833. doi: 10.1016/j.molcel.2017.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 737.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2011;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 738.Jin TY, et al. BRCA1/2 serves as a biomarker for poor prognosis in breast carcinoma. Int. J. Mol. Sci. 2022;23:3754. doi: 10.3390/ijms23073754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 739.Wang G-H, et al. BRCA1 and BRCA2 expression patterns and prognostic significance in digestive system cancers. Hum. Pathol. 2018;71:135–144. doi: 10.1016/j.humpath.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 740.Breast Cancer Linkage Consortium. Cancer risks in BRCA2 mutation carriers. J. Natl Cancer Inst. 91, 1310–1316 (1999). [DOI] [PubMed]
- 741.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 742.Blackford AN, Jackson SP. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017;66:801–817. doi: 10.1016/j.molcel.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 743.Ngoi NYL, et al. Targeting ATR in patients with cancer. Nat. Rev. Clin. Oncol. 2024;21:278–293. doi: 10.1038/s41571-024-00863-5. [DOI] [PubMed] [Google Scholar]
- 744.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 745.Cremona CA, Behrens A. ATM signalling and cancer. Oncogene. 2014;33:3351–3360. doi: 10.1038/onc.2013.275. [DOI] [PubMed] [Google Scholar]
- 746.Bailey P, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 747.Sun L, Wang R-C, Zhang Q, Guo L-L. ATM mutations as an independent prognostic factor and potential biomarker for immune checkpoint therapy in endometrial cancer. Pathol. Res. Pract. 2020;216:153032. doi: 10.1016/j.prp.2020.153032. [DOI] [PubMed] [Google Scholar]
- 748.Randon G, et al. Prognostic impact of ATM mutations in patients with metastatic colorectal cancer. Sci. Rep. 2019;9:2858. doi: 10.1038/s41598-019-39525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 749.Moossavi M, Parsamanesh N, Bahrami A, Atkin SL, Sahebkar A. Role of the NLRP3 inflammasome in cancer. Mol. Cancer. 2018;17:158. doi: 10.1186/s12943-018-0900-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 750.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 751.Carswell EA, et al. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl Acad. Sci. USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 752.Szlosarek PW, Balkwill FR. Tumour necrosis factor alpha: a potential target for the therapy of solid tumours. Lancet Oncol. 2003;4:565–573. doi: 10.1016/S1470-2045(03)01196-3. [DOI] [PubMed] [Google Scholar]
- 753.van Loo G, Bertrand MJM. Death by TNF: a road to inflammation. Nat. Rev. Immunol. 2023;23:289–303. doi: 10.1038/s41577-022-00792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 754.Szlosarek P, Charles KA, Balkwill FR. Tumour necrosis factor-alpha as a tumour promoter. Eur. J. Cancer. 2006;42:745–750. doi: 10.1016/j.ejca.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 755.Balkwill F. Tumour necrosis factor and cancer. Nat. Rev. Cancer. 2009;9:361–371. doi: 10.1038/nrc2628. [DOI] [PubMed] [Google Scholar]
- 756.Bounder G, et al. Associations of the -238(G/A) and -308(G/A) TNF-α promoter polymorphisms and TNF-α serum levels with the susceptibility to gastric precancerous lesions and gastric cancer related to Helicobacter pylori infection in a Moroccan population. Asian Pac. J. Cancer Prev. 2020;21:1623–1629. doi: 10.31557/APJCP.2020.21.6.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 757.Noguchi M, Hiwatashi N, Liu Z, Toyota T. Secretion imbalance between tumour necrosis factor and its inhibitor in inflammatory bowel disease. Gut. 1998;43:203–209. doi: 10.1136/gut.43.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Szlosarek PW, et al. Expression and regulation of tumor necrosis factor alpha in normal and malignant ovarian epithelium. Mol. Cancer Ther. 2006;5:382–390. doi: 10.1158/1535-7163.MCT-05-0303. [DOI] [PubMed] [Google Scholar]
- 759.Andersson BÅ, et al. Plasma tumor necrosis factor-α and C-reactive protein as biomarker for survival in head and neck squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2014;140:515–519. doi: 10.1007/s00432-014-1592-8. [DOI] [PubMed] [Google Scholar]
- 760.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 761.Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 762.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat. Rev. Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 763.Taniguchi K, Karin M. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 2018;18:309–324. doi: 10.1038/nri.2017.142. [DOI] [PubMed] [Google Scholar]
- 764.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 765.Viatour P, Merville M-P, Bours V, Chariot A. Phosphorylation of NF-kappaB and IkappaB proteins: implications in cancer and inflammation. Trends Biochem. Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 766.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 767.Wu D, et al. NF-κB expression and outcomes in solid tumors: a systematic review and meta-analysis. Medicine. 2015;94:e1687. doi: 10.1097/MD.0000000000001687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 768.Sarkar DK, et al. Role of NF-κB as a prognostic marker in breast cancer: a pilot study in Indian patients. Indian J. Surg. Oncol. 2013;4:242–247. doi: 10.1007/s13193-013-0234-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 769.Ben-Neriah Y, Karin M. Inflammation meets cancer, with NF-κB as the matchmaker. Nat. Immunol. 2011;12:715–723. doi: 10.1038/ni.2060. [DOI] [PubMed] [Google Scholar]
- 770.Kumar M, et al. NF-κB regulates mesenchymal transition for the induction of non-small cell lung cancer initiating cells. PLoS ONE. 2013;8:e68597. doi: 10.1371/journal.pone.0068597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 771.Huber MA, et al. NF-kappaB is essential for epithelial-mesenchymal transition and metastasis in a model of breast cancer progression. J. Clin. Invest. 2004;114:569–581. doi: 10.1172/JCI200421358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 2002;10:417–426. doi: 10.1016/S1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- 774.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 775.Si Y, Liu L, Fan Z. Mechanisms and effects of NLRP3 in digestive cancers. Cell Death Discov. 2024;10:10. doi: 10.1038/s41420-023-01783-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 776.Tarassishin L, Casper D, Lee SC. Aberrant expression of interleukin-1β and inflammasome activation in human malignant gliomas. PLoS ONE. 2014;9:e103432. doi: 10.1371/journal.pone.0103432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 777.Bae JY, et al. P2X7 receptor and NLRP3 inflammasome activation in head and neck cancer. Oncotarget. 2017;8:48972–48982. doi: 10.18632/oncotarget.16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Veeranki S. Role of inflammasomes and their regulators in prostate cancer initiation, progression and metastasis. Cell. Mol. Biol. Lett. 2013;18:355–367. doi: 10.2478/s11658-013-0095-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 779.Dunn JH, Ellis LZ, Fujita M. Inflammasomes as molecular mediators of inflammation and cancer: potential role in melanoma. Cancer Lett. 2012;314:24–33. doi: 10.1016/j.canlet.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 780.Shi F, et al. Low NLRP3 expression predicts a better prognosis of colorectal cancer. Biosci. Rep. 2021;41:BSR20210280. doi: 10.1042/BSR20210280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 781.Fan S, et al. Luteoloside suppresses proliferation and metastasis of hepatocellular carcinoma cells by inhibition of NLRP3 inflammasome. PLoS ONE. 2014;9:e89961. doi: 10.1371/journal.pone.0089961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 782.Warburg O. Über den Stoffwechsel der Carcinomzelle. Naturwissenschaften. 1924;12:1131–1137. doi: 10.1007/BF01504608. [DOI] [Google Scholar]
- 783.Wang Y, Patti GJ. The Warburg effect: a signature of mitochondrial overload. Trends Cell Biol. 2023;33:1014–1020. doi: 10.1016/j.tcb.2023.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 784.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J. Cell. Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 785.Deng D, et al. Crystal structure of the human glucose transporter GLUT1. Nature. 2014;510:121–125. doi: 10.1038/nature13306. [DOI] [PubMed] [Google Scholar]
- 786.Wieman HL, Wofford JA, Rathmell JC. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol. Biol. Cell. 2007;18:1437–1446. doi: 10.1091/mbc.e06-07-0593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 787.Meng Y, et al. The progress and development of GLUT1 inhibitors targeting cancer energy metabolism. Future Med. Chem. 2019;11:2333–2352. doi: 10.4155/fmc-2019-0052. [DOI] [PubMed] [Google Scholar]
- 788.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The tumor suppressor p53 down-regulates glucose transporters GLUT1 and GLUT4 gene expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.CAN-03-0846. [DOI] [PubMed] [Google Scholar]
- 789.Zambrano A, Molt M, Uribe E, Salas M. Glut 1 in cancer cells and the inhibitory action of resveratrol as a potential therapeutic strategy. Int. J. Mol. Sci. 2019;20:3374. doi: 10.3390/ijms20133374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 790.Zhang B, Xie Z, Li B. The clinicopathologic impacts and prognostic significance of GLUT1 expression in patients with lung cancer: a meta-analysis. Gene. 2019;689:76–83. doi: 10.1016/j.gene.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 791.Amann T, Hellerbrand C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin. Ther. Targets. 2009;13:1411–1427. doi: 10.1517/14728220903307509. [DOI] [PubMed] [Google Scholar]
- 792.Wang J, et al. Glucose transporter GLUT1 expression and clinical outcome in solid tumors: a systematic review and meta-analysis. Oncotarget. 2017;8:16875–16886. doi: 10.18632/oncotarget.15171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 793.Waitkus MS, Diplas BH, Yan H. Biological role and therapeutic potential of IDH mutations in cancer. Cancer Cell. 2018;34:186–195. doi: 10.1016/j.ccell.2018.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 794.Turkalp Z, Karamchandani J, Das S. IDH mutation in glioma: new insights and promises for the future. JAMA Neurol. 2014;71:1319–1325. doi: 10.1001/jamaneurol.2014.1205. [DOI] [PubMed] [Google Scholar]
- 795.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann. Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 796.Capper D, et al. Characterization of R132H mutation-specific IDH1 antibody binding in brain tumors. Brain Pathol. 2010;20:245–254. doi: 10.1111/j.1750-3639.2009.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 797.Andronesi OC, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci. Transl. Med. 2012;4:116ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 798.Guo D, Meng Y, Jiang X, Lu Z. Hexokinases in cancer and other pathologies. Cell Insight. 2023;2:100077. doi: 10.1016/j.cellin.2023.100077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 799.Ciscato F, Ferrone L, Masgras I, Laquatra C, Rasola A. Hexokinase 2 in cancer: a prima donna playing multiple characters. Int. J. Mol. Sci. 2021;22:4716. doi: 10.3390/ijms22094716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 800.Kim J, Gao P, Liu Y-C, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 2007;27:7381–7393. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 801.Liu Y, et al. Prognostic significance of the metabolic marker hexokinase-2 in various solid tumors: a meta-analysis. PLoS ONE. 2016;11:e0166230. doi: 10.1371/journal.pone.0166230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 802.Patra KC, et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell. 2013;24:213–228. doi: 10.1016/j.ccr.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 803.Akinleye A, Rasool Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019;12:92. doi: 10.1186/s13045-019-0779-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 804.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev. Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 805.Ishida Y, Agata Y, Shibahara K, Honjo T. Induced expression of PD-1, a novel member of the immunoglobulin gene superfamily, upon programmed cell death. EMBO J. 1992;11:3887–3895. doi: 10.1002/j.1460-2075.1992.tb05481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 806.Nishimura H, Nose M, Hiai H, Minato N, Honjo T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity. 1999;11:141–151. doi: 10.1016/S1074-7613(00)80089-8. [DOI] [PubMed] [Google Scholar]
- 807.Freeman GJ, et al. Engagement of the PD-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J. Exp. Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 808.Dong H, et al. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 2002;8:793–800. doi: 10.1038/nm730. [DOI] [PubMed] [Google Scholar]
- 809.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat. Rev. Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 810.Chang C-H, et al. The prognostic significance of PD1 and PDL1 gene expression in lung cancer: a meta-analysis. Front. Oncol. 2021;11:759497. doi: 10.3389/fonc.2021.759497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 811.Oh SY, et al. Soluble PD-L1 is a predictive and prognostic biomarker in advanced cancer patients who receive immune checkpoint blockade treatment. Sci. Rep. 2021;11:19712. doi: 10.1038/s41598-021-99311-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 812.Müller T, et al. PD-L1: a novel prognostic biomarker in head and neck squamous cell carcinoma. Oncotarget. 2017;8:52889–52900. doi: 10.18632/oncotarget.17547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 813.Rosellini M, et al. Prognostic and predictive biomarkers for immunotherapy in advanced renal cell carcinoma. Nat. Rev. Urol. 2023;20:133–157. doi: 10.1038/s41585-022-00676-0. [DOI] [PubMed] [Google Scholar]
- 814.Gu L, et al. PD-L1 and gastric cancer prognosis: a systematic review and meta-analysis. PLoS ONE. 2017;12:e0182692. doi: 10.1371/journal.pone.0182692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 815.Brunet JF, et al. A new member of the immunoglobulin superfamily–CTLA-4. Nature. 1987;328:267–270. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 816.Rotte A. Combination of CTLA-4 and PD-1 blockers for treatment of cancer. J. Exp. Clin. Cancer Res. 2019;38:255. doi: 10.1186/s13046-019-1259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 817.Rowshanravan B, Halliday N, Sansom DM. CTLA-4: a moving target in immunotherapy. Blood. 2018;131:58–67. doi: 10.1182/blood-2017-06-741033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 818.Hu P, et al. The prognostic value of cytotoxic T-lymphocyte antigen 4 in cancers: a systematic review and meta-analysis. Sci. Rep. 2017;7:42913. doi: 10.1038/srep42913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 819.Mao H, et al. New Insights of CTLA-4 into its biological function in breast cancer. Curr. Cancer Drug Targets. 2010;10:728–736. doi: 10.2174/156800910793605811. [DOI] [PubMed] [Google Scholar]
- 820.Zhang C-Y, et al. Prognostic value of combined analysis of CTLA-4 and PLR in esophageal squamous cell carcinoma (ESCC) patients. Dis. Markers. 2019;2019:1601072. doi: 10.1155/2019/1601072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 821.Zheng S, et al. Differentiation therapy: unlocking phenotypic plasticity of hepatocellular carcinoma. Crit. Rev. Oncol. Hematol. 2022;180:103854. doi: 10.1016/j.critrevonc.2022.103854. [DOI] [PubMed] [Google Scholar]
- 822.Waddington CH. The epigenotype. 1942. Int. J. Epidemiol. 2012;41:10–13. doi: 10.1093/ije/dyr184. [DOI] [PubMed] [Google Scholar]
- 823.Nishiyama A, Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends Genet. 2021;37:1012–1027. doi: 10.1016/j.tig.2021.05.002. [DOI] [PubMed] [Google Scholar]
- 824.Cheng Y, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target Ther. 2019;4:62. doi: 10.1038/s41392-019-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 825.Meng H, et al. DNA methylation, its mediators and genome integrity. Int. J. Biol. Sci. 2015;11:604–617. doi: 10.7150/ijbs.11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 826.Koch A, et al. Analysis of DNA methylation in cancer: location revisited. Nat. Rev. Clin. Oncol. 2018;15:459–466. doi: 10.1038/s41571-018-0004-4. [DOI] [PubMed] [Google Scholar]
- 827.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 828.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 829.Van Vlodrop IJH, et al. Prognostic significance of Gremlin1 (GREM1) promoter CpG island hypermethylation in clear cell renal cell carcinoma. Am. J. Pathol. 2010;176:575–584. doi: 10.2353/ajpath.2010.090442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 830.Kim MS, et al. A promoter methylation pattern in the N-methyl-D-aspartate receptor 2B gene predicts poor prognosis in esophageal squamous cell carcinoma. Clin. Cancer Res. 2007;13:6658–6665. doi: 10.1158/1078-0432.CCR-07-1178. [DOI] [PubMed] [Google Scholar]
- 831.Claus R, et al. Quantitative DNA methylation analysis identifies a single CpG dinucleotide important for ZAP-70 expression and predictive of prognosis in chronic lymphocytic leukemia. J. Clin. Oncol. 2012;30:2483–2491. doi: 10.1200/JCO.2011.39.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 832.Yuan G, et al. Defining optimal cutoff value of MGMT promoter methylation by ROC analysis for clinical setting in glioblastoma patients. J. Neurooncol. 2017;133:193–201. doi: 10.1007/s11060-017-2433-9. [DOI] [PubMed] [Google Scholar]
- 833.Das PM, Singal R. DNA methylation and cancer. J. Clin. Oncol. 2004;22:4632–4642. doi: 10.1200/JCO.2004.07.151. [DOI] [PubMed] [Google Scholar]
- 834.Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 2014;13:673–691. doi: 10.1038/nrd4360. [DOI] [PubMed] [Google Scholar]
- 835.Seligson DB, et al. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- 836.Avvakumov N, Côté J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 837.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 838.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 839.Ntziachristos P, et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012;18:298–301. doi: 10.1038/nm.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 840.Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 2003;4:13–18. doi: 10.1016/S1535-6108(03)00165-X. [DOI] [PubMed] [Google Scholar]
- 841.Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014;15:317–328. doi: 10.1016/j.chom.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 842.de Martel C, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- 843.Ferreira RM, et al. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 844.Banerjee S, et al. The ovarian cancer oncobiome. Oncotarget. 2017;8:36225–36245. doi: 10.18632/oncotarget.16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 845.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014;12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 846.Pushalkar S, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8:403–416. doi: 10.1158/2159-8290.CD-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 847.Sfanos KS, et al. A molecular analysis of prokaryotic and viral DNA sequences in prostate tissue from patients with prostate cancer indicates the presence of multiple and diverse microorganisms. Prostate. 2008;68:306–320. doi: 10.1002/pros.20680. [DOI] [PubMed] [Google Scholar]
- 848.Mima K, et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 2017;402:9–15. doi: 10.1016/j.canlet.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 849.Mao Q, et al. Interplay between the lung microbiome and lung cancer. Cancer Lett. 2018;415:40–48. doi: 10.1016/j.canlet.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 850.Urbaniak C, et al. The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 2016;82:5039–5048. doi: 10.1128/AEM.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 851.Avilés-Jiménez F, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin. Microbiol. Infect. 2016;22:178.e11–178.e22. doi: 10.1016/j.cmi.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 852.Garrett WS. Cancer and the microbiota. Science. 2015;348:80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 853.Wang Y, et al. Crosstalk between autophagy and microbiota in cancer progression. Mol. Cancer. 2021;20:163. doi: 10.1186/s12943-021-01461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 854.Park EM, et al. Targeting the gut and tumor microbiota in cancer. Nat. Med. 2022;28:690–703. doi: 10.1038/s41591-022-01779-2. [DOI] [PubMed] [Google Scholar]
- 855.Di Domenico EG, Cavallo I, Pontone M, Toma L, Ensoli F. Biofilm producing Salmonella typhi: chronic colonization and development of gallbladder cancer. Int. J. Mol. Sci. 2017;18:1887. doi: 10.3390/ijms18091887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 856.Huang Y, et al. Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J. Clin. Pathol. 2004;57:1273–1277. doi: 10.1136/jcp.2004.018556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 857.Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196–202. doi: 10.1016/j.canlet.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 858.Helmink BA, Khan MAW, Hermann A, Gopalakrishnan V, Wargo JA. The microbiome, cancer, and cancer therapy. Nat. Med. 2019;25:377–388. doi: 10.1038/s41591-019-0377-7. [DOI] [PubMed] [Google Scholar]
- 859.Sun J, Kato I. Gut microbiota, inflammation and colorectal cancer. Genes Dis. 2016;3:130–143. doi: 10.1016/j.gendis.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 860.Mangerich A, et al. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc. Natl Acad. Sci. USA. 2012;109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 861.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 862.Hu D, et al. Cellular senescence and hematological malignancies: from pathogenesis to therapeutics. Pharm. Ther. 2021;223:107817. doi: 10.1016/j.pharmthera.2021.107817. [DOI] [PubMed] [Google Scholar]
- 863.Hernandez-Segura A, Nehme J, Demaria M. Hallmarks of cellular senescence. Trends Cell Biol. 2018;28:436–453. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 864.Wang B, Kohli J, Demaria M. Senescent cells in cancer therapy: friends or foes? Trends Cancer. 2020;6:838–857. doi: 10.1016/j.trecan.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 865.Schmitt CA, Wang B, Demaria M. Senescence and cancer — role and therapeutic opportunities. Nat. Rev. Clin. Oncol. 2022;19:619–636. doi: 10.1038/s41571-022-00668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 866.Birch J, Gil J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 2020;34:1565–1576. doi: 10.1101/gad.343129.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 867.Rao SG, Jackson JG. SASP: tumor suppressor or promoter? Yes! Trends Cancer. 2016;2:676–687. doi: 10.1016/j.trecan.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 868.Kansara M, et al. Immune response to RB1-regulated senescence limits radiation-induced osteosarcoma formation. J. Clin. Invest. 2013;123:5351–5360. doi: 10.1172/JCI70559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 869.Lesina M, et al. RelA regulates CXCL1/CXCR2-dependent oncogene-induced senescence in murine Kras-driven pancreatic carcinogenesis. J. Clin. Invest. 2016;126:2919–2932. doi: 10.1172/JCI86477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 870.Chen Z, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 871.Gruenbaum Y, Foisner R. Lamins: nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015;84:131–164. doi: 10.1146/annurev-biochem-060614-034115. [DOI] [PubMed] [Google Scholar]
- 872.Freund A, Laberge R-M, Demaria M, Campisi J. Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell. 2012;23:2066–2075. doi: 10.1091/mbc.e11-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 873.Shimi T, et al. The role of nuclear lamin B1 in cell proliferation and senescence. Genes Dev. 2011;25:2579–2593. doi: 10.1101/gad.179515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 874.Shah PP, et al. Lamin B1 depletion in senescent cells triggers large-scale changes in gene expression and the chromatin landscape. Genes Dev. 2013;27:1787–1799. doi: 10.1101/gad.223834.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 875.Qin H, et al. Pan-cancer analysis identifies LMNB1 as a target to redress Th1/Th2 imbalance and enhance PARP inhibitor response in human cancers. Cancer Cell Int. 2022;22:101. doi: 10.1186/s12935-022-02467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 876.Yang Y, et al. Lamin B1 is a potential therapeutic target and prognostic biomarker for hepatocellular carcinoma. Bioengineered. 2022;13:9211–9231. doi: 10.1080/21655979.2022.2057896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 877.Radspieler MM, et al. Lamin-B1 is a senescence-associated biomarker in clear-cell renal cell carcinoma. Oncol. Lett. 2019;18:2654–2660. doi: 10.3892/ol.2019.10593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 878.Li W, et al. Lamin B1 overexpresses in lung adenocarcinoma and promotes proliferation in lung cancer cells via AKT pathway. Onco. Targets Ther. 2020;13:3129–3139. doi: 10.2147/OTT.S229997. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 879.Yu D, et al. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer. 2022;21:56. doi: 10.1186/s12943-022-01509-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 880.Dang DK, Park BH. Circulating tumor DNA: current challenges for clinical utility. J. Clin. Invest. 2022;132:e154941. doi: 10.1172/JCI154941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 881.Bettegowda C, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra24. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 882.Aucamp J, Bronkhorst AJ, Badenhorst CPS, Pretorius PJ. The diverse origins of circulating cell-free DNA in the human body: a critical re-evaluation of the literature. Biol. Rev. Camb. Philos. Soc. 2018;93:1649–1683. doi: 10.1111/brv.12413. [DOI] [PubMed] [Google Scholar]
- 883.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 884.Cheng ML, et al. Circulating tumor DNA in advanced solid tumors: clinical relevance and future directions. CA Cancer J. Clin. 2021;71:176–190. doi: 10.3322/caac.21650. [DOI] [PubMed] [Google Scholar]
- 885.Higgins MJ, et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer Res. 2012;18:3462–3469. doi: 10.1158/1078-0432.CCR-11-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 886.Magbanua MJM, et al. Circulating tumor DNA and magnetic resonance imaging to predict neoadjuvant chemotherapy response and recurrence risk. NPJ Breast Cancer. 2021;7:32. doi: 10.1038/s41523-021-00239-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 887.Pellini B, Chaudhuri AA. Circulating tumor DNA minimal residual disease detection of non-small-cell lung cancer treated with curative intent. J. Clin. Oncol. 2022;40:567–575. doi: 10.1200/JCO.21.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 888.Malla M, Loree JM, Kasi PM, Parikh AR. Using circulating tumor DNA in colorectal cancer: current and evolving practices. J. Clin. Oncol. 2022;40:2846–2857. doi: 10.1200/JCO.21.02615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 889.Oxnard GR, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 890.Quigley D, et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 2017;7:999–1005. doi: 10.1158/2159-8290.CD-17-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 891.Ma S, et al. Clinical application and detection techniques of liquid biopsy in gastric cancer. Mol. Cancer. 2023;22:7. doi: 10.1186/s12943-023-01715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 892.Aceto N, Toner M, Maheswaran S, Haber DA. En route to metastasis: circulating tumor cell clusters and epithelial-to-mesenchymal transition. Trends Cancer. 2015;1:44–52. doi: 10.1016/j.trecan.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 893.van de Stolpe A, Pantel K, Sleijfer S, Terstappen LW, den Toonder JMJ. Circulating tumor cell isolation and diagnostics: toward routine clinical use. Cancer Res. 2011;71:5955–5960. doi: 10.1158/0008-5472.CAN-11-1254. [DOI] [PubMed] [Google Scholar]
- 894.Castro-Giner F, Aceto N. Tracking cancer progression: from circulating tumor cells to metastasis. Genome Med. 2020;12:31. doi: 10.1186/s13073-020-00728-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 895.Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin. Chem. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- 896.Kostopoulos IV, et al. Circulating plasma cells in newly diagnosed multiple myeloma: prognostic and more. J. Clin. Oncol. 2023;41:708–710. doi: 10.1200/JCO.22.01606. [DOI] [PubMed] [Google Scholar]
- 897.Paoletti C, et al. Comprehensive mutation and copy number profiling in archived circulating breast cancer tumor cells documents heterogeneous resistance mechanisms. Cancer Res. 2018;78:1110–1122. doi: 10.1158/0008-5472.CAN-17-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 898.Boral D, et al. Molecular characterization of breast cancer CTCs associated with brain metastasis. Nat. Commun. 2017;8:196. doi: 10.1038/s41467-017-00196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 899.Scher HI, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016;2:1441–1449. doi: 10.1001/jamaoncol.2016.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 900.Chemi F, et al. Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse. Nat. Med. 2019;25:1534–1539. doi: 10.1038/s41591-019-0593-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 901.Miyamoto DT, Ting DT, Toner M, Maheswaran S, Haber DA. Single-cell analysis of circulating tumor cells as a window into tumor heterogeneity. Cold Spring Harb. Symp. Quant. Biol. 2016;81:269–274. doi: 10.1101/sqb.2016.81.031120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 902.Pan BT, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 903.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 904.Paskeh MDA, et al. Emerging role of exosomes in cancer progression and tumor microenvironment remodeling. J. Hematol. Oncol. 2022;15:83. doi: 10.1186/s13045-022-01305-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 905.Kalluri R. The biology and function of exosomes in cancer. J. Clin. Invest. 2016;126:1208–1215. doi: 10.1172/JCI81135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 906.Dai J, et al. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct. Target Ther. 2020;5:145. doi: 10.1038/s41392-020-00261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 907.Cui S, Cheng Z, Qin W, Jiang L. Exosomes as a liquid biopsy for lung cancer. Lung Cancer. 2018;116:46–54. doi: 10.1016/j.lungcan.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 908.Guo W, et al. Liquid biopsy analysis of lipometabolic exosomes in pancreatic cancer. Cytokine Growth Factor Rev. 2023;73:69–77. doi: 10.1016/j.cytogfr.2023.07.006. [DOI] [PubMed] [Google Scholar]
- 909.Fu M, et al. Exosomes in gastric cancer: roles, mechanisms, and applications. Mol. Cancer. 2019;18:41. doi: 10.1186/s12943-019-1001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 910.Yang S, et al. Detection of mutant KRAS and TP53 DNA in circulating exosomes from healthy individuals and patients with pancreatic cancer. Cancer Biol. Ther. 2017;18:158–165. doi: 10.1080/15384047.2017.1281499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 911.Kahlert C, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 912.Salehi M, Sharifi M. Exosomal miRNAs as novel cancer biomarkers: challenges and opportunities. J. Cell. Physiol. 2018;233:6370–6380. doi: 10.1002/jcp.26481. [DOI] [PubMed] [Google Scholar]
- 913.Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. Extracell. Vesicles. 2016;5:31292. doi: 10.3402/jev.v5.31292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 914.Bhagirath D, et al. microRNA-1246 is an exosomal biomarker for aggressive prostate cancer. Cancer Res. 2018;78:1833–1844. doi: 10.1158/0008-5472.CAN-17-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 915.Zhou W, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 916.Kucharzewska P, et al. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl Acad. Sci. USA. 2013;110:7312–7317. doi: 10.1073/pnas.1220998110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 917.Yokoi A, et al. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat. Commun. 2017;8:14470. doi: 10.1038/ncomms14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 918.Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 919.Costa-Silva B, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 920.Zhang H, et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017;8:15016. doi: 10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 921.Le MTN, et al. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. J. Clin. Invest. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 922.Hu Y, et al. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS ONE. 2015;10:e0125625. doi: 10.1371/journal.pone.0125625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 923.Chen W, et al. Exosomes from drug-resistant breast cancer cells transmit chemoresistance by a horizontal transfer of microRNAs. PLoS ONE. 2014;9:e95240. doi: 10.1371/journal.pone.0095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 924.Au Yeung CL, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat. Commun. 2016;7:11150. doi: 10.1038/ncomms11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 925.Binenbaum Y, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018;78:5287–5299. doi: 10.1158/0008-5472.CAN-18-0124. [DOI] [PubMed] [Google Scholar]
- 926.Zhou Y, Zhang Y, Gong H, Luo S, Cui Y. The role of exosomes and their applications in cancer. Int. J. Mol. Sci. 2021;22:12204. doi: 10.3390/ijms222212204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 927.Wu Q, Qian W, Sun X, Jiang S. Small-molecule inhibitors, immune checkpoint inhibitors, and more: FDA-approved novel therapeutic drugs for solid tumors from 1991 to 2021. J. Hematol. Oncol. 2022;15:143. doi: 10.1186/s13045-022-01362-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 928.Li G, Qin Y, Xie C, Wu Y-L, Chen X. Trends in oncology drug innovation in China. Nat. Rev. Drug Discov. 2021;20:15–16. doi: 10.1038/d41573-020-00195-w. [DOI] [PubMed] [Google Scholar]
- 929.Cohen P, Cross D, Jänne PA. Kinase drug discovery 20 years after imatinib: progress and future directions. Nat. Rev. Drug Discov. 2021;20:551–569. doi: 10.1038/s41573-021-00195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 930.Patricelli MP, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 931.Parikh K, et al. Drugging KRAS: current perspectives and state-of-art review. J. Hematol. Oncol. 2022;15:152. doi: 10.1186/s13045-022-01375-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 932.LoRusso PM, Sebolt-Leopold JS. One step at a time — clinical evidence that KRAS is indeed druggable. N. Engl. J. Med. 2020;383:1277–1278. doi: 10.1056/NEJMe2026372. [DOI] [PubMed] [Google Scholar]
- 933.Keeton AB, Salter EA, Piazza GA. The RAS-effector interaction as a drug target. Cancer Res. 2017;77:221–226. doi: 10.1158/0008-5472.CAN-16-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 934.Canon J, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217–223. doi: 10.1038/s41586-019-1694-1. [DOI] [PubMed] [Google Scholar]
- 935.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 936.Skoulidis F, et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 2021;384:2371–2381. doi: 10.1056/NEJMoa2103695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 937.Hong DS, et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 2020;383:1207–1217. doi: 10.1056/NEJMoa1917239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 938.de Langen AJ, et al. Sotorasib versus docetaxel for previously treated non-small-cell lung cancer with KRAS(G12C) mutation: a randomised, open-label, phase 3 trial. Lancet. 2023;401:733–746. doi: 10.1016/S0140-6736(23)00221-0. [DOI] [PubMed] [Google Scholar]
- 939.Fakih MG, et al. Sotorasib for previously treated colorectal cancers with KRAS(G12C) mutation (CodeBreaK100): a prespecified analysis of a single-arm, phase 2 trial. Lancet Oncol. 2022;23:115–124. doi: 10.1016/S1470-2045(21)00605-7. [DOI] [PubMed] [Google Scholar]
- 940.Hallin J, et al. The KRAS(G12C) inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54–71. doi: 10.1158/2159-8290.CD-19-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 941.Sabari JK, et al. Activity of adagrasib (MRTX849) in brain metastases: preclinical models and clinical data from patients with KRASG12C-mutant non-small cell lung cancer. Clin. Cancer Res. 2022;28:3318–3328. doi: 10.1158/1078-0432.CCR-22-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 942.Weiss J, et al. LBA6 KRYSTAL-1: adagrasib (MRTX849) as monotherapy or combined with cetuximab (Cetux) in patients (Pts) with colorectal cancer (CRC) harboring a KRASG12C mutation. Ann. Oncol. 2021;32:S1294. doi: 10.1016/j.annonc.2021.08.2093. [DOI] [Google Scholar]
- 943.Yaeger R, Solit DB. Overcoming adaptive resistance to KRAS inhibitors through vertical pathway targeting. Clin. Cancer Res. 2020;26:1538–1540. doi: 10.1158/1078-0432.CCR-19-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 944.Singh A, et al. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 945.Muzumdar MD, et al. Survival of pancreatic cancer cells lacking KRAS function. Nat. Commun. 2017;8:1090. doi: 10.1038/s41467-017-00942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 946.Tanaka N, et al. Clinical acquired resistance to KRAS(G12C) inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021;11:1913–1922. doi: 10.1158/2159-8290.CD-21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 947.Amodio V, et al. EGFR blockade reverts resistance to KRAS(G12C) inhibition in colorectal cancer. Cancer Discov. 2020;10:1129–1139. doi: 10.1158/2159-8290.CD-20-0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 948.Tsai YS, et al. Rapid idiosyncratic mechanisms of clinical resistance to KRAS G12C inhibition. J. Clin. Invest. 2022;132:e155523. doi: 10.1172/JCI155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 949.Wang X, et al. Identification of MRTX1133, a noncovalent, potent, and selective KRAS(G12D) inhibitor. J. Med. Chem. 2022;65:3123–3133. doi: 10.1021/acs.jmedchem.1c01688. [DOI] [PubMed] [Google Scholar]
- 950.Hallin J, et al. Anti-tumor efficacy of a potent and selective non-covalent KRAS(G12D) inhibitor. Nat. Med. 2022;28:2171–2182. doi: 10.1038/s41591-022-02007-7. [DOI] [PubMed] [Google Scholar]
- 951.Ryan MB, Corcoran RB. Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 2018;15:709–720. doi: 10.1038/s41571-018-0105-0. [DOI] [PubMed] [Google Scholar]
- 952.Welsch ME, et al. Multivalent small-molecule Pan-RAS inhibitors. Cell. 2017;168:878–889.e29. doi: 10.1016/j.cell.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 953.Hong SH, et al. A Sos proteomimetic as a pan-Ras inhibitor. Proc. Natl Acad. Sci. USA. 2021;118:e2101027118. doi: 10.1073/pnas.2101027118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 954.Athuluri-Divakar SK, et al. A small molecule RAS-mimetic disrupts RAS association with effector proteins to block signaling. Cell. 2016;165:643–655. doi: 10.1016/j.cell.2016.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 955.Ross SJ, et al. Targeting KRAS-dependent tumors with AZD4785, a high-affinity therapeutic antisense oligonucleotide inhibitor of KRAS. Sci. Transl. Med. 2017;9:eaal5253. doi: 10.1126/scitranslmed.aal5253. [DOI] [PubMed] [Google Scholar]
- 956.Spencer-Smith R, et al. Inhibition of RAS function through targeting an allosteric regulatory site. Nat. Chem. Biol. 2017;13:62–68. doi: 10.1038/nchembio.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 957.Bond MJ, Chu L, Nalawansha DA, Li K, Crews CM. Targeted degradation of oncogenic KRAS(G12C) by VHL-recruiting PROTACs. ACS Cent. Sci. 2020;6:1367–1375. doi: 10.1021/acscentsci.0c00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 958.Papke B, Der CJ. Drugging RAS: know the enemy. Science. 2017;355:1158–1163. doi: 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 959.Kamerkar S, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 960.Hou P, Wang YA. Conquering oncogenic KRAS and its bypass mechanisms. Theranostics. 2022;12:5691–5709. doi: 10.7150/thno.71260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 961.Karp JE, Lancet JE. Tipifarnib in the treatment of newly diagnosed acute myelogenous leukemia. Biologics. 2008;2:491–500. doi: 10.2147/btt.s3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 962.Ho AL, et al. Tipifarnib in head and neck squamous cell carcinoma with HRAS mutations. J. Clin. Oncol. 2021;39:1856–1864. doi: 10.1200/JCO.20.02903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 963.Lee HW, et al. A phase II trial of tipifarnib for patients with previously treated, metastatic urothelial carcinoma harboring HRAS mutations. Clin. Cancer Res. 2020;26:5113–5119. doi: 10.1158/1078-0432.CCR-20-1246. [DOI] [PubMed] [Google Scholar]
- 964.Hanna GJ, et al. Tipifarnib in recurrent, metastatic HRAS-mutant salivary gland cancer. Cancer. 2020;126:3972–3981. doi: 10.1002/cncr.33036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 965.Untch BR, et al. Tipifarnib inhibits HRAS-driven dedifferentiated thyroid cancers. Cancer Res. 2018;78:4642–4657. doi: 10.1158/0008-5472.CAN-17-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 966.Dhillon S. Lonafarnib: first approval. Drugs. 2021;81:283–289. doi: 10.1007/s40265-020-01464-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 967.Bustinza-Linares E, Kurzrock R, Tsimberidou AM. Salirasib in the treatment of pancreatic cancer. Future Oncol. 2010;6:885–891. doi: 10.2217/fon.10.71. [DOI] [PubMed] [Google Scholar]
- 968.Yue W, Wang J, Li Y, Fan P, Santen RJ. Farnesylthiosalicylic acid blocks mammalian target of rapamycin signaling in breast cancer cells. Int. J. Cancer. 2005;117:746–754. doi: 10.1002/ijc.21222. [DOI] [PubMed] [Google Scholar]
- 969.Goldberg L, Kloog Y. A Ras inhibitor tilts the balance between Rac and Rho and blocks phosphatidylinositol 3-kinase-dependent glioblastoma cell migration. Cancer Res. 2006;66:11709–11717. doi: 10.1158/0008-5472.CAN-06-1878. [DOI] [PubMed] [Google Scholar]
- 970.Blum R, Jacob-Hirsch J, Amariglio N, Rechavi G, Kloog Y. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1alpha, causing glycolysis shutdown and cell death. Cancer Res. 2005;65:999–1006. doi: 10.1158/0008-5472.999.65.3. [DOI] [PubMed] [Google Scholar]
- 971.Halaschek-Wiener J, et al. A novel Ras antagonist regulates both oncogenic Ras and the tumor suppressor p53 in colon cancer cells. Mol. Med. 2000;6:693–704. doi: 10.1007/BF03402049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 972.Halaschek-Wiener J, Kloog Y, Wacheck V, Jansen B. Farnesyl thiosalicylic acid chemosensitizes human melanoma in vivo. J. Invest. Dermatol. 2003;120:1–7. doi: 10.1046/j.1523-1747.2003.12009.x. [DOI] [PubMed] [Google Scholar]
- 973.Beiner ME, et al. Ras antagonist inhibits growth and chemosensitizes human epithelial ovarian cancer cells. Int. J. Gynecol. Cancer. 2006;16:200–206. doi: 10.1136/ijgc-00009577-200602001-00032. [DOI] [PubMed] [Google Scholar]
- 974.Stärkel P, et al. Ras inhibition in hepatocarcinoma by S-trans-trans-farnesylthiosalicyclic acid: Association of its tumor preventive effect with cell proliferation, cell cycle events, and angiogenesis. Mol. Carcinog. 2012;51:816–825. doi: 10.1002/mc.20849. [DOI] [PubMed] [Google Scholar]
- 975.Gana-Weisz M, et al. The Ras inhibitor S-trans,trans-farnesylthiosalicylic acid chemosensitizes human tumor cells without causing resistance. Clin. Cancer Res. 2002;8:555–565. [PubMed] [Google Scholar]
- 976.Riely GJ, et al. A phase II trial of salirasib in patients with lung adenocarcinomas with KRAS mutations. J. Thorac. Oncol. 2011;6:1435–1437. doi: 10.1097/JTO.0b013e318223c099. [DOI] [PubMed] [Google Scholar]
- 977.Chandra A, et al. The GDI-like solubilizing factor PDEδ sustains the spatial organization and signalling of Ras family proteins. Nat. Cell Biol. 2011;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 978.Zimmermann G, et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638–642. doi: 10.1038/nature12205. [DOI] [PubMed] [Google Scholar]
- 979.Papke B, et al. Identification of pyrazolopyridazinones as PDEδ inhibitors. Nat. Commun. 2016;7:11360. doi: 10.1038/ncomms11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 980.Cheng J, Li Y, Wang X, Dong G, Sheng C. Discovery of novel PDEδ degraders for the treatment of KRAS mutant colorectal cancer. J. Med. Chem. 2020;63:7892–7905. doi: 10.1021/acs.jmedchem.0c00929. [DOI] [PubMed] [Google Scholar]
- 981.Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. Int. Ed. Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 982.Leshchiner ES, et al. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc. Natl Acad. Sci. USA. 2015;112:1761–1766. doi: 10.1073/pnas.1413185112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 983.Hillig RC, et al. Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS–SOS1 interaction. Proc. Natl Acad. Sci. USA. 2019;116:2551–2560. doi: 10.1073/pnas.1812963116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 984.Chen T, et al. Inhibition of son of sevenless homologue 1 (SOS1): promising therapeutic treatment for KRAS-mutant cancers. Eur. J. Med. Chem. 2023;261:115828. doi: 10.1016/j.ejmech.2023.115828. [DOI] [PubMed] [Google Scholar]
- 985.Hofmann MH, et al. Bi-3406, a potent and selective sos1–kras interaction inhibitor, is effective in kras-driven cancers through combined mek inhibition. Cancer Discov. 2021;11:142–157. doi: 10.1158/2159-8290.CD-20-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 986.Zhou Z, et al. Discovery of a potent, cooperative, and selective SOS1 PROTAC ZZ151 with in vivo antitumor efficacy in KRAS-mutant cancers. J. Med. Chem. 2023;66:4197–4214. doi: 10.1021/acs.jmedchem.3c00075. [DOI] [PubMed] [Google Scholar]
- 987.Ruess DA, et al. Mutant KRAS-driven cancers depend on PTPN11/SHP2 phosphatase. Nat. Med. 2018;24:954–960. doi: 10.1038/s41591-018-0024-8. [DOI] [PubMed] [Google Scholar]
- 988.Liu C, et al. Combinations with allosteric SHP2 inhibitor TNO155 to block receptor tyrosine kinase signaling. Clin. Cancer Res. 2021;27:342–354. doi: 10.1158/1078-0432.CCR-20-2718. [DOI] [PubMed] [Google Scholar]
- 989.Lamarche MJ, et al. Identification of TNO155, an allosteric SHP2 inhibitor for the treatment of cancer. J. Med. Chem. 2020;63:13578–13594. doi: 10.1021/acs.jmedchem.0c01170. [DOI] [PubMed] [Google Scholar]
- 990.Chen YNP, et al. Allosteric inhibition of SHP2 phosphatase inhibits cancers driven by receptor tyrosine kinases. Nature. 2016;535:148–152. doi: 10.1038/nature18621. [DOI] [PubMed] [Google Scholar]
- 991.De Santis MC, Gulluni F, Campa CC, Martini M, Hirsch E. Targeting PI3K signaling in cancer: Challenges and advances. Biochim. Biophys. Acta Rev. Cancer. 2019;1871:361–366. doi: 10.1016/j.bbcan.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 992.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 993.Meng D, et al. Development of PI3K inhibitors: advances in clinical trials and new strategies (Review) Pharmacol. Res. 2021;173:105900. doi: 10.1016/j.phrs.2021.105900. [DOI] [PubMed] [Google Scholar]
- 994.Liu N, et al. BAY 80-6946 is a highly selective intravenous pI3K inhibitor with potent p110α and p110δ activities in tumor cell lines and xenograft models. Mol. Cancer Ther. 2013;12:2319–2330. doi: 10.1158/1535-7163.MCT-12-0993-T. [DOI] [PubMed] [Google Scholar]
- 995.Kojima T, et al. Phase II study of BKM120 in patients with advanced esophageal squamous cell carcinoma (EPOC1303) Esophagus. 2022;19:702–710. doi: 10.1007/s10388-022-00928-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 996.De Gooijer, M. C. et al. Buparlisib is a brain penetrable pan-PI3K inhibitor. Sci. Rep. 8, 10784 (2018). [DOI] [PMC free article] [PubMed]
- 997.Song KW, et al. RTK-dependent inducible degradation of mutant PI3Kα drives GDC-0077 (Inavolisib) efficacy. Cancer Discov. 2022;12:204–219. doi: 10.1158/2159-8290.CD-21-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 998.Mateo J, et al. A first-time-in-human study of GSK2636771, a phosphoinositide 3 kinase beta-selective inhibitor, in patients with advanced solid tumors. Clin. Cancer Res. 2017;23:5981–5992. doi: 10.1158/1078-0432.CCR-17-0725. [DOI] [PubMed] [Google Scholar]
- 999.Barlaam B, et al. Discovery of (R)-8-(1-(3,5-difluorophenylamino)ethyl)-N,N-dimethyl-2-morpholino-4-oxo-4H-chromene-6-carboxamide (AZD8186): a potent and selective inhibitor of PI3Kβ and PI3Kδ for the treatment of PTEN-deficient cancers. J. Med. Chem. 2015;58:943–962. doi: 10.1021/jm501629p. [DOI] [PubMed] [Google Scholar]
- 1000.Yu M, et al. Development and safety of PI3K inhibitors in cancer. Arch. Toxicol. 2023;97:635–650. doi: 10.1007/s00204-023-03440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1001.Dhillon S, Keam SJ. Umbralisib: first approval. Drugs. 2021;81:857–866. doi: 10.1007/s40265-021-01504-2. [DOI] [PubMed] [Google Scholar]
- 1002.Roskoski RJ. Properties of FDA-approved small molecule phosphatidylinositol 3-kinase inhibitors prescribed for the treatment of malignancies. Pharmacol. Res. 2021;168:105579. doi: 10.1016/j.phrs.2021.105579. [DOI] [PubMed] [Google Scholar]
- 1003.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat. Rev. Clin. Oncol. 2013;10:143–153. doi: 10.1038/nrclinonc.2013.10. [DOI] [PubMed] [Google Scholar]
- 1004.Addie M, et al. Discovery of 4-amino-N-[(1 S)-1-(4-chlorophenyl)-3-hydroxypropyl]-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidine-4-carboxamide (AZD5363), an orally bioavailable, potent inhibitor of Akt kinases. J. Med. Chem. 2013;56:2059–2073. doi: 10.1021/jm301762v. [DOI] [PubMed] [Google Scholar]
- 1005.Lin J, et al. Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin. Cancer Res. 2013;19:1760–1772. doi: 10.1158/1078-0432.CCR-12-3072. [DOI] [PubMed] [Google Scholar]
- 1006.Toson B, Fortes IS, Roesler R, Andrade SF. Targeting Akt/PKB in pediatric tumors: a review from preclinical to clinical trials. Pharm. Res. 2022;183:106403. doi: 10.1016/j.phrs.2022.106403. [DOI] [PubMed] [Google Scholar]
- 1007.Chen Y, Zhou X. Research progress of mTOR inhibitors. Eur. J. Med. Chem. 2020;208:112820. doi: 10.1016/j.ejmech.2020.112820. [DOI] [PubMed] [Google Scholar]
- 1008.Coppin C. Everolimus: the first approved product for patients with advanced renal cell cancer after sunitinib and/or sorafenib. Biologics. 2010;4:91–101. doi: 10.2147/btt.s6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1009.Raphael J, et al. Everolimus in advanced breast cancer: a systematic review and meta-analysis. Target Oncol. 2020;15:723–732. doi: 10.1007/s11523-020-00770-6. [DOI] [PubMed] [Google Scholar]
- 1010.Kwitkowski VE, et al. FDA approval summary: temsirolimus as treatment for advanced renal cell carcinoma. Oncologist. 2010;15:428–435. doi: 10.1634/theoncologist.2009-0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1011.Dancey J. MTOR signaling and drug development in cancer. Nat. Rev. Clin. Oncol. 2010;7:209–219. doi: 10.1038/nrclinonc.2010.21. [DOI] [PubMed] [Google Scholar]
- 1012.Davies H, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 1013.Sullivan RJ, et al. First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: results of a phase I dose-escalation and expansion study. Cancer Discov. 2018;8:184–195. doi: 10.1158/2159-8290.CD-17-1119. [DOI] [PubMed] [Google Scholar]
- 1014.Callahan MK, et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N. Engl. J. Med. 2012;367:2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1015.Degirmenci U, Yap J, Sim YRM, Qin S, Hu J. Drug resistance in targeted cancer therapies with RAF inhibitors. Cancer Drug Resist. 2021;4:665–683. doi: 10.20517/cdr.2021.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1016.Peng SB, et al. Inhibition of RAF isoforms and active dimers by LY3009120 leads to anti-tumor activities in RAS or BRAF mutant cancers. Cancer Cell. 2015;28:384–398. doi: 10.1016/j.ccell.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 1017.Monaco KA, et al. LXH254, a potent and selective ARAF-Sparing inhibitor of BRAF and CRAF for the treatment of MAPK-Driven tumors. Clin. Cancer Res. 2021;27:2061–2073. doi: 10.1158/1078-0432.CCR-20-2563. [DOI] [PubMed] [Google Scholar]
- 1018.Sullivan RJ, et al. A phase I study of LY3009120, a pan-RAF inhibitor, in patients with advanced or metastatic cancer. Mol. Cancer Ther. 2020;19:460–467. doi: 10.1158/1535-7163.MCT-19-0681. [DOI] [PubMed] [Google Scholar]
- 1019.Zhang C, et al. RAF inhibitors that evade paradoxical MAPK pathway activation. Nature. 2015;526:583–586. doi: 10.1038/nature14982. [DOI] [PubMed] [Google Scholar]
- 1020.Moschos SJ, et al. Development of MK-8353, an orally administered ERK1/2 inhibitor, in patients with advanced solid tumors. JCI Insight. 2018;3:e92352. doi: 10.1172/jci.insight.92352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1021.Morris EJ, et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 1022.Poulikakos PI, Solit DB. Resistance to MEK inhibitors: should we co-target upstream? Sci. Signal. 2011;4:pe16. doi: 10.1126/scisignal.2001948. [DOI] [PubMed] [Google Scholar]
- 1023.Engelman JA, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat. Med. 2008;14:1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1024.Posch C, et al. Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proc. Natl Acad. Sci. USA. 2013;110:4015–4020. doi: 10.1073/pnas.1216013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1025.Junttila MR, et al. Modeling targeted inhibition of MEK and PI3 kinase in human pancreatic cancer. Mol. Cancer Ther. 2015;14:40–47. doi: 10.1158/1535-7163.MCT-14-0030. [DOI] [PubMed] [Google Scholar]
- 1026.She QB, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1027.Mughal MJ, Bhadresha K, Kwok HF. CDK inhibitors from past to present: a new wave of cancer therapy. Semin. Cancer Biol. 2023;88:106–122. doi: 10.1016/j.semcancer.2022.12.006. [DOI] [PubMed] [Google Scholar]
- 1028.Wang S, Chen F-E. Small-molecule MDM2 inhibitors in clinical trials for cancer therapy. Eur. J. Med. Chem. 2022;236:114334. doi: 10.1016/j.ejmech.2022.114334. [DOI] [PubMed] [Google Scholar]
- 1029.Ingham M, Schwartz GK. Cell-cycle therapeutics come of age. J. Clin. Oncol. 2017;35:2949–2959. doi: 10.1200/JCO.2016.69.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1030.Goel S, Bergholz JS, Zhao JJ. Targeting CDK4 and CDK6 in cancer. Nat. Rev. Cancer. 2022;22:356–372. doi: 10.1038/s41568-022-00456-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1031.Fassl A, Geng Y, Sicinski P. CDK4 and CDK6 kinases: from basic science to cancer therapy. Science. 2022;375:eabc1495. doi: 10.1126/science.abc1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1032.Dhillon S. Palbociclib: first global approval. Drugs. 2015;75:543–551. doi: 10.1007/s40265-015-0379-9. [DOI] [PubMed] [Google Scholar]
- 1033.Corona SP, Generali D. Abemaciclib: a CDK4/6 inhibitor for the treatment of HR + /HER2- advanced breast cancer. Drug Des. Devel. Ther. 2018;12:321–330. doi: 10.2147/DDDT.S137783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1034.Hortobagyi GN, et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N. Engl. J. Med. 2016;375:1738–1748. doi: 10.1056/NEJMoa1609709. [DOI] [PubMed] [Google Scholar]
- 1035.Wedam S, et al. FDA approval summary: palbociclib for male patients with metastatic breast cancer. Clin. Cancer Res. 2020;26:1208–1212. doi: 10.1158/1078-0432.CCR-19-2580. [DOI] [PubMed] [Google Scholar]
- 1036.Maurer C, Martel S, Zardavas D, Ignatiadis M. New agents for endocrine resistance in breast cancer. Breast. 2017;34:1–11. doi: 10.1016/j.breast.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 1037.Finn RS, et al. Overall survival results from the randomized phase 2 study of palbociclib in combination with letrozole versus letrozole alone for first-line treatment of ER + /HER2- advanced breast cancer (PALOMA-1, TRIO-18) Breast Cancer Res. Treat. 2020;183:419–428. doi: 10.1007/s10549-020-05755-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1038.Barroso-Sousa R, Shapiro GI, Tolaney SM. Clinical development of the CDK4/6 inhibitors ribociclib and abemaciclib in breast cancer. Breast Care. 2016;11:167–173. doi: 10.1159/000447284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1039.Tripathy D, et al. Ribociclib plus endocrine therapy for premenopausal women with hormone-receptor-positive, advanced breast cancer (MONALEESA-7): a randomised phase 3 trial. Lancet Oncol. 2018;19:904–915. doi: 10.1016/S1470-2045(18)30292-4. [DOI] [PubMed] [Google Scholar]
- 1040.Fujiwara Y, et al. Phase 1 study of abemaciclib, an inhibitor of CDK 4 and 6, as a single agent for Japanese patients with advanced cancer. Cancer Chemother. Pharmacol. 2016;78:281–288. doi: 10.1007/s00280-016-3085-8. [DOI] [PubMed] [Google Scholar]
- 1041.Torres-Guzmán R, et al. Preclinical characterization of abemaciclib in hormone receptor positive breast cancer. Oncotarget. 2017;8:69493–69507. doi: 10.18632/oncotarget.17778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1042.Palumbo A, Lau G, Saraceni M. Abemaciclib: the newest CDK4/6 inhibitor for the treatment of breast cancer. Ann. Pharmacother. 2019;53:178–185. doi: 10.1177/1060028018795146. [DOI] [PubMed] [Google Scholar]
- 1043.Patnaik A, et al. Efficacy and safety of abemaciclib, an inhibitor of CDK4 and CDK6, for patients with breast cancer, non-small cell lung cancer, and other solid tumors. Cancer Discov. 2016;6:740–753. doi: 10.1158/2159-8290.CD-16-0095. [DOI] [PubMed] [Google Scholar]
- 1044.Sledge GW, et al. MONARCH 2: abemaciclib in combination with fulvestrant in women with HR + /HER2-advanced breast cancer who had progressed while receiving endocrine therapy. J. Clin. Oncol. 2017;35:2875–2884. doi: 10.1200/JCO.2017.73.7585. [DOI] [PubMed] [Google Scholar]
- 1045.Dickler, M. N. et al. MONARCH 1, a phase II study of abemaciclib, a CDK4 and CDK6 inhibitor, as a single agent, in patients with refractory HR+/HER2− metastatic breast cancer. Clin. Cancer Res. 24, 5485–5485 (2018). [DOI] [PubMed]
- 1046.Goetz MP, et al. MONARCH 3: abemaciclib as initial therapy for advanced breast cancer. J. Clin. Oncol. 2017;35:3638–3646. doi: 10.1200/JCO.2017.75.6155. [DOI] [PubMed] [Google Scholar]
- 1047.Dhillon S. Trilaciclib: first approval. Drugs. 2021;81:867–874. doi: 10.1007/s40265-021-01508-y. [DOI] [PubMed] [Google Scholar]
- 1048.Hu W, Wang L, Luo J, Zhang J, Li N. The potent novel CDK4/6 inhibitor TQB3616 in hormone receptor positive breast cancer: preclinical characterization with in vitro and human tumor xenograft models. Breast Cancer. 2023;15:899–912. doi: 10.2147/BCTT.S434973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1049.Liao X, et al. SPH3643: a novel cyclin-dependent kinase 4/6 inhibitor with good anticancer efficacy and strong blood-brain barrier permeability. Cancer Sci. 2020;111:1761–1773. doi: 10.1111/cas.14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1050.Zhang P, et al. A phase 1 study of dalpiciclib, a cyclin-dependent kinase 4/6 inhibitor in Chinese patients with advanced breast cancer. Biomark. Res. 2021;9:24. doi: 10.1186/s40364-021-00271-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1051.Sedlacek H, et al. Flavopiridol (L86 8275; NSC 649890), a new kinase inhibitor for tumor therapy. Int. J. Oncol. 1996;9:1143–1168. doi: 10.3892/ijo.9.6.1143. [DOI] [PubMed] [Google Scholar]
- 1052.Murphy CG, Dickler MN. The role of CDK4/6 inhibition in breast cancer. Oncologist. 2015;20:483–490. doi: 10.1634/theoncologist.2014-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1053.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 1054.Konopleva M, et al. MDM2 inhibition: an important step forward in cancer therapy. Leukemia. 2020;34:2858–2874. doi: 10.1038/s41375-020-0949-z. [DOI] [PubMed] [Google Scholar]
- 1055.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 1056.Andreeff M, et al. Results of the phase I trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin. Cancer Res. 2016;22:868–876. doi: 10.1158/1078-0432.CCR-15-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1057.Ding Q, et al. Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J. Med. Chem. 2013;56:5979–5983. doi: 10.1021/jm400487c. [DOI] [PubMed] [Google Scholar]
- 1058.Erba HP, et al. Phase 1b study of the MDM2 inhibitor AMG 232 with or without trametinib in relapsed/refractory acute myeloid leukemia. Blood Adv. 2019;3:1939–1949. doi: 10.1182/bloodadvances.2019030916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1059.Wang S, et al. SAR405838: an optimized inhibitor of MDM2-p53 interaction that induces complete and durable tumor regression. Cancer Res. 2014;74:5855–5865. doi: 10.1158/0008-5472.CAN-14-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1060.Aguilar, A. et al. Discovery of 4-((3’R,4’S,5’R)-6″-chloro-4’-(3-chloro-2-fluorophenyl)-1’-ethyl-2″-oxodispiro[cyclohexane-1,2’-pyrrolidine-3’,3″-indoline]-5’-carboxamido)bicyclo[2.2.2]octane-1-carboxylic acid (AA-115/APG-115): a potent and orally active murine double minute 2 (MDM2) inhibitor in clinical development. J. Med. Chem. 60, 2819–2839 (2017). [DOI] [PMC free article] [PubMed]
- 1061.Holzer P, et al. Discovery of a dihydroisoquinolinone derivative (NVP-CGM097): a highly potent and selective MDM2 inhibitor undergoing phase 1 clinical trials in p53wt tumors. J. Med. Chem. 2015;58:6348–6358. doi: 10.1021/acs.jmedchem.5b00810. [DOI] [PubMed] [Google Scholar]
- 1062.Stein EM, et al. Results from a first-in-human phase I study of siremadlin (HDM201) in patients with advanced wild-type TP53 solid tumors and acute leukemia. Clin. Cancer Res. 2022;28:870–881. doi: 10.1158/1078-0432.CCR-21-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1063.Wagner AJ, et al. Phase I trial of the human double minute 2 inhibitor MK-8242 in patients with advanced solid tumors. J. Clin. Oncol. 2017;35:1304–1311. doi: 10.1200/JCO.2016.70.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1064.Ravandi F, et al. A phase I trial of the human double minute 2 inhibitor (MK-8242) in patients with refractory/recurrent acute myelogenous leukemia (AML) Leuk. Res. 2016;48:92–100. doi: 10.1016/j.leukres.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1065.Senapati J, et al. A phase I study of milademetan (DS3032b) in combination with low dose cytarabine with or without venetoclax in acute myeloid leukemia: clinical safety, efficacy, and correlative analysis. Blood Cancer J. 2023;13:101. doi: 10.1038/s41408-023-00871-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1066.Fang Y, Liao G, Yu B. Small-molecule MDM2/X inhibitors and PROTAC degraders for cancer therapy: advances and perspectives. Acta Pharm. Sin. B. 2020;10:1253–1278. doi: 10.1016/j.apsb.2020.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1067.Jackson MR, et al. Radiolabeled oligonucleotides targeting the RNA subunit of telomerase inhibit telomerase and induce DNA damage in telomerase-positive cancer cells. Cancer Res. 2019;79:4627–4637. doi: 10.1158/0008-5472.CAN-18-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1068.Guterres AN, Villanueva J. Targeting telomerase for cancer therapy. Oncogene. 2020;39:5811–5824. doi: 10.1038/s41388-020-01405-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1069.Lai TP, et al. A method for measuring the distribution of the shortest telomeres in cells and tissues. Nat. Commun. 2017;8:1356. doi: 10.1038/s41467-017-01291-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1070.Joseph I, et al. The telomerase inhibitor imetelstat depletes cancer stem cells in breast and pancreatic cancer cell lines. Cancer Res. 2010;70:9494–9504. doi: 10.1158/0008-5472.CAN-10-0233. [DOI] [PubMed] [Google Scholar]
- 1071.Dikmen ZG, et al. In vivo inhibition of lung cancer by GRN163L: a novel human telomerase inhibitor. Cancer Res. 2005;65:7866–7873. doi: 10.1158/0008-5472.CAN-05-1215. [DOI] [PubMed] [Google Scholar]
- 1072.Marian CO, Wright WE, Shay JW. The effects of telomerase inhibition on prostate tumor-initiating cells. Int. J. Cancer. 2010;127:321–331. doi: 10.1002/ijc.25043. [DOI] [PubMed] [Google Scholar]
- 1073.Burchett KM, Yan Y, Ouellette MM. Telomerase inhibitor imetelstat (GRN163L) limits the lifespan of human pancreatic cancer cells. PLoS ONE. 2014;9:e85155. doi: 10.1371/journal.pone.0085155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1074.Hu Y, Bobb D, He J, Hill DA, Dome JS. The HSP90 inhibitor alvespimycin enhances the potency of telomerase inhibition by imetelstat in human osteosarcoma. Cancer Biol. Ther. 2015;16:949–957. doi: 10.1080/15384047.2015.1040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1075.Marian CO, et al. The telomerase antagonist, imetelstat, efficiently targets glioblastoma tumor-initiating cells leading to decreased proliferation and tumor growth. Clin. Cancer Res. 2010;16:154–163. doi: 10.1158/1078-0432.CCR-09-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1076.Vonderheide RH. Telomerase as a universal tumor-associated antigen for cancer immunotherapy. Oncogene. 2002;21:674–679. doi: 10.1038/sj.onc.1205074. [DOI] [PubMed] [Google Scholar]
- 1077.Ruden M, Puri N. Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 2013;39:444–456. doi: 10.1016/j.ctrv.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 1078.Kyte JA. Cancer vaccination with telomerase peptide GV1001. Expert Opin. Investig. Drugs. 2009;18:687–694. doi: 10.1517/13543780902897631. [DOI] [PubMed] [Google Scholar]
- 1079.Hemann MT, Greider CW. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1080.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 1081.Sherwood, L. M., Parris, E. E. & Folkman, J. Tumor angiogenesis: therapeutic implications. N. Engl. J. Med. 285, 1182–1186 (1971). [DOI] [PubMed]
- 1082.Gasparini G, Longo R, Toi M, Ferrara N. Angiogenic inhibitors: a new therapeutic strategy in oncology. Nat. Clin. Pract. Oncol. 2005;2:562–577. doi: 10.1038/ncponc0342. [DOI] [PubMed] [Google Scholar]
- 1083.Wilke H, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15:1224–1235. doi: 10.1016/S1470-2045(14)70420-6. [DOI] [PubMed] [Google Scholar]
- 1084.Itatani Y, Kawada K, Yamamoto T, Sakai Y. Resistance to anti-angiogenic therapy in cancer-alterations to anti-VEGF pathway. Int. J. Mol. Sci. 2018;19:1–18. doi: 10.3390/ijms19041232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1085.Kelly RJ, Darnell C, Rixe O. Target inhibition in antiangiogenic therapy a wide spectrum of selectivity and specificity. Cancer J. 2010;16:635–642. doi: 10.1097/PPO.0b013e3181ff37cf. [DOI] [PubMed] [Google Scholar]
- 1086.de Oliveira Dias JR, de Andrade GC, Novais EA, Farah ME, Rodrigues EB. Fusion proteins for treatment of retinal diseases: aflibercept, ziv-aflibercept, and conbercept. Int. J. Retin. Vitr. 2016;2:3. doi: 10.1186/s40942-016-0026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1087.Gotink KJ, Verheul HMW. Anti-angiogenic tyrosine kinase inhibitors: what is their mechanism of action? Angiogenesis. 2010;13:1–14. doi: 10.1007/s10456-009-9160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1088.Akce M, El-Rayes BF, Bekaii-Saab TS. Frontline therapy for advanced hepatocellular carcinoma: an update. Ther. Adv. Gastroenterol. 2022;15:17562848221086126. doi: 10.1177/17562848221086126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1089.Cheng A-L, et al. Efficacy and safety of sorafenib in patients in the Asia-Pacific region with advanced hepatocellular carcinoma: a phase III randomised, double-blind, placebo-controlled trial. Lancet Oncol. 2009;10:25–34. doi: 10.1016/S1470-2045(08)70285-7. [DOI] [PubMed] [Google Scholar]
- 1090.Zhao Y, Zhang Y-N, Wang K-T, Chen L. Lenvatinib for hepatocellular carcinoma: from preclinical mechanisms to anti-cancer therapy. Biochim Biophys. Acta Rev. Cancer. 2020;1874:188391. doi: 10.1016/j.bbcan.2020.188391. [DOI] [PubMed] [Google Scholar]
- 1091.Matsuki M, et al. Targeting of tumor growth and angiogenesis underlies the enhanced antitumor activity of lenvatinib in combination with everolimus. Cancer Sci. 2017;108:763–771. doi: 10.1111/cas.13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1092.Yamamoto Y, et al. Lenvatinib, an angiogenesis inhibitor targeting VEGFR/FGFR, shows broad antitumor activity in human tumor xenograft models associated with microvessel density and pericyte coverage. Vasc. Cell. 2014;6:18. doi: 10.1186/2045-824X-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1093.Matsui J, et al. E7080, a novel inhibitor that targets multiple kinases, has potent antitumor activities against stem cell factor producing human small cell lung cancer H146, based on angiogenesis inhibition. Int. J. Cancer. 2008;122:664–671. doi: 10.1002/ijc.23131. [DOI] [PubMed] [Google Scholar]
- 1094.Matsui J, et al. Multi-kinase inhibitor E7080 suppresses lymph node and lung metastases of human mammary breast tumor MDA-MB-231 via inhibition of vascular endothelial growth factor-receptor (VEGF-R) 2 and VEGF-R3 kinase. Clin. Cancer Res. 2008;14:5459–5465. doi: 10.1158/1078-0432.CCR-07-5270. [DOI] [PubMed] [Google Scholar]
- 1095.Ogasawara S, et al. Antiproliferative effect of lenvatinib on human liver cancer cell lines in vitro and in vivo. Anticancer Res. 2019;39:5973–5982. doi: 10.21873/anticanres.13802. [DOI] [PubMed] [Google Scholar]
- 1096.Song Y, et al. Anti-angiogenic agents in combination with immune checkpoint inhibitors: a promising strategy for cancer treatment. Front. Immunol. 2020;11:1–17. doi: 10.3389/fimmu.2020.01956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1097.Zhang Y, et al. Maintenance of antiangiogenic and antitumor effects by orally active low-dose capecitabine for long-term cancer therapy. Proc. Natl Acad. Sci. USA. 2017;114:E5226–E5235. doi: 10.1073/pnas.1705066114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1098.Babina IS, Turner NC. Advances and challenges in targeting FGFR signalling in cancer. Nat. Rev. Cancer. 2017;17:318–332. doi: 10.1038/nrc.2017.8. [DOI] [PubMed] [Google Scholar]
- 1099.Hallinan N, et al. Targeting the fibroblast growth factor receptor family in cancer. Cancer Treat. Rev. 2016;46:51–62. doi: 10.1016/j.ctrv.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 1100.André F, et al. Targeting FGFR with dovitinib (TKI258): Preclinical and clinical data in breast cancer. Clin. Cancer Res. 2013;19:3693–3702. doi: 10.1158/1078-0432.CCR-13-0190. [DOI] [PubMed] [Google Scholar]
- 1101.Angevin E, et al. Phase I study of dovitinib (TKI258), an oral FGFR, VEGFR, and PDGFR inhibitor, in advanced or metastatic renal cell carcinoma. Clin. Cancer Res. 2013;19:1257–1268. doi: 10.1158/1078-0432.CCR-12-2885. [DOI] [PubMed] [Google Scholar]
- 1102.Okamoto I, et al. Phase I safety, pharmacokinetic, and biomarker study of BIBF 1120, an oral triple tyrosine kinase inhibitor in patients with advanced solid tumors. Mol. Cancer Ther. 2010;9:2825–2833. doi: 10.1158/1535-7163.MCT-10-0379. [DOI] [PubMed] [Google Scholar]
- 1103.Soria J-C, et al. Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann. Oncol. 2014;25:2244–2251. doi: 10.1093/annonc/mdu390. [DOI] [PubMed] [Google Scholar]
- 1104.Tan FH, Putoczki TL, Stylli SS, Luwor RB. Ponatinib: a novel multi-tyrosine kinase inhibitor against human malignancies. Onco. Targets Ther. 2019;12:635–645. doi: 10.2147/OTT.S189391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1105.Kang C. Infigratinib: first approval. Drugs. 2021;81:1355–1360. doi: 10.1007/s40265-021-01567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1106.Markham A. Erdafitinib: first global approval. Drugs. 2019;79:1017–1021. doi: 10.1007/s40265-019-01142-9. [DOI] [PubMed] [Google Scholar]
- 1107.Syed YY. Futibatinib: first approval. Drugs. 2022;82:1737–1743. doi: 10.1007/s40265-022-01806-z. [DOI] [PubMed] [Google Scholar]
- 1108.Gandhy SU, et al. FDA approval summary: futibatinib for unresectable advanced or metastatic, chemotherapy refractory intrahepatic cholangiocarcinoma with FGFR2 fusions or other rearrangements. Clin. Cancer Res. 2023;29:4027–4031. doi: 10.1158/1078-0432.CCR-23-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1109.Liang G, Chen G, Wei X, Zhao Y, Li X. Small molecule inhibition of fibroblast growth factor receptors in cancer. Cytokine Growth Factor Rev. 2013;24:467–475. doi: 10.1016/j.cytogfr.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 1110.Roberts WG, et al. Antiangiogenic and antitumor activity of a selective PDGFR tyrosine kinase inhibitor, CP-673,451. Cancer Res. 2005;65:957–966. doi: 10.1158/0008-5472.957.65.3. [DOI] [PubMed] [Google Scholar]
- 1111.Wang Q, et al. Discovery of 4-((N-(2-(dimethylamino)ethyl)acrylamido)methyl)-N-(4-methyl-3-((4-(pyridin-3-yl)pyrimidin-2-yl)amino)phenyl)benzamide (CHMFL-PDGFR-159) as a highly selective type II PDGFRα kinase inhibitor for PDGFRα driving chronic eosinophilic leukemia. Eur. J. Med. Chem. 2018;150:366–384. doi: 10.1016/j.ejmech.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 1112.Papadopoulos N, Lennartsson J. The PDGF/PDGFR pathway as a drug target. Mol. Asp. Med. 2018;62:75–88. doi: 10.1016/j.mam.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 1113.Ferrari SM, et al. Sunitinib in the treatment of thyroid cancer. Curr. Med. Chem. 2019;26:963–972. doi: 10.2174/0929867324666171006165942. [DOI] [PubMed] [Google Scholar]
- 1114.Schmid TA, Gore ME. Sunitinib in the treatment of metastatic renal cell carcinoma. Ther. Adv. Urol. 2016;8:348–371. doi: 10.1177/1756287216663979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1115.He W, et al. Artesunate regulates neurite outgrowth inhibitor protein B receptor to overcome resistance to sorafenib in hepatocellular carcinoma cells. Front. Pharmacol. 2021;12:615889. doi: 10.3389/fphar.2021.615889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1116.Mehta M, et al. Regorafenib sensitizes human breast cancer cells to radiation by inhibiting multiple kinases and inducing DNA damage. Int. J. Radiat. Biol. 2021;97:1109–1120. doi: 10.1080/09553002.2020.1730012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1117.Arai H, et al. Molecular insight of regorafenib treatment for colorectal cancer. Cancer Treat. Rev. 2019;81:101912. doi: 10.1016/j.ctrv.2019.101912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1118.Kantarjian HM, et al. Long-term outcomes with frontline nilotinib versus imatinib in newly diagnosed chronic myeloid leukemia in chronic phase: ENESTnd 10-year analysis. Leukemia. 2021;35:440–453. doi: 10.1038/s41375-020-01111-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1119.Naqvi K, et al. Long-term follow-up of lower dose dasatinib (50 mg daily) as frontline therapy in newly diagnosed chronic-phase chronic myeloid leukemia. Cancer. 2020;126:67–75. doi: 10.1002/cncr.32504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1120.Stone RM, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N. Engl. J. Med. 2017;377:454–464. doi: 10.1056/NEJMoa1614359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1121.Fischer T, et al. Phase IIB trial of oral midostaurin (PKC412), the FMS-like tyrosine kinase 3 receptor (FLT3) and multi-targeted kinase inhibitor, in patients with acute myeloid leukemia and high-risk myelodysplastic syndrome with either wild-type or mutated FLT3. J. Clin. Oncol. 2010;28:4339–4345. doi: 10.1200/JCO.2010.28.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1122.Roskoski RJ. Properties of FDA-approved small molecule protein kinase inhibitors: a 2022 update. Pharmacol. Res. 2022;175:106037. doi: 10.1016/j.phrs.2021.106037. [DOI] [PubMed] [Google Scholar]
- 1123.Soleimani M, Nappi L, Kollmannsberger C. Avelumab and axitinib combination therapy for the treatment of advanced renal cell carcinoma. Future Oncol. 2020;16:3021–3034. doi: 10.2217/fon-2020-0586. [DOI] [PubMed] [Google Scholar]
- 1124.Grünwald V, et al. Randomized comparison of pazopanib and doxorubicin as first-line treatment in patients with metastatic soft tissue sarcoma age 60 years or older: results of a German Intergroup Study. J. Clin. Oncol. 2020;38:3555–3564. doi: 10.1200/JCO.20.00714. [DOI] [PubMed] [Google Scholar]
- 1125.Lowery CD, et al. Olaratumab exerts antitumor activity in preclinical models of pediatric bone and soft tissue tumors through inhibition of platelet-derived growth factor receptor α. Clin. Cancer Res. 2018;24:847–857. doi: 10.1158/1078-0432.CCR-17-1258. [DOI] [PubMed] [Google Scholar]
- 1126.Wang F, et al. Gint4.T-modified DNA tetrahedrons loaded with doxorubicin inhibits glioma cell proliferation by targeting PDGFRβ. Nanoscale Res. Lett. 2020;15:150. doi: 10.1186/s11671-020-03377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1127.Camorani S, et al. Targeted imaging and inhibition of triple-negative breast cancer metastases by a PDGFRβ aptamer. Theranostics. 2018;8:5178–5199. doi: 10.7150/thno.27798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1128.Yoshida S, et al. Extrahepatic platelet-derived growth factor-β, delivered by platelets, promotes activation of hepatic stellate cells and biliary fibrosis in mice. Gastroenterology. 2014;147:1378–1392. doi: 10.1053/j.gastro.2014.08.038. [DOI] [PubMed] [Google Scholar]
- 1129.Zeitelhofer M, et al. Preclinical toxicological assessment of a novel monoclonal antibody targeting human platelet-derived growth factor CC (PDGF-CC) in PDGF-CChum mice. PLoS ONE. 2018;13:e0200649. doi: 10.1371/journal.pone.0200649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1130.Falcon BL, et al. Increased vascular delivery and efficacy of chemotherapy after inhibition of platelet-derived growth factor-B. Am. J. Pathol. 2011;178:2920–2930. doi: 10.1016/j.ajpath.2011.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1131.Karashima T, et al. Blockade of the vascular endothelial growth factor-receptor 2 pathway inhibits the growth of human renal cell carcinoma, RBM1-IT4, in the kidney but not in the bone of nude mice. Int. J. Oncol. 2007;30:937–945. [PubMed] [Google Scholar]
- 1132.Leenders WPJ, et al. Antiangiogenic therapy of cerebral melanoma metastases results in sustained tumor progression via vessel co-option. Clin. Cancer Res. 2004;10:6222–6230. doi: 10.1158/1078-0432.CCR-04-0823. [DOI] [PubMed] [Google Scholar]
- 1133.Motzer RJ, et al. Activity of SU11248, a multitargeted inhibitor of vascular endothelial growth factor receptor and platelet-derived growth factor receptor, in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 2006;24:16–24. doi: 10.1200/JCO.2005.02.2574. [DOI] [PubMed] [Google Scholar]
- 1134.Batchelor TT, et al. AZD2171, a pan-VEGF receptor tyrosine kinase inhibitor, normalizes tumor vasculature and alleviates edema in glioblastoma patients. Cancer Cell. 2007;11:83–95. doi: 10.1016/j.ccr.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1135.Yu JL, Rak JW, Coomber BL, Hicklin DJ, Kerbel RS. Effect of p53 status on tumor response to antiangiogenic therapy. Science. 2002;295:1526–1528. doi: 10.1126/science.1068327. [DOI] [PubMed] [Google Scholar]
- 1136.Whitmarsh-Everiss T, Laraia L. Small molecule probes for targeting autophagy. Nat. Chem. Biol. 2021;17:653–664. doi: 10.1038/s41589-021-00768-9. [DOI] [PubMed] [Google Scholar]
- 1137.Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: Therapeutic potential and persisting obstacles. Nat. Rev. Drug Discov. 2017;16:487–511. doi: 10.1038/nrd.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1138.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1139.Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. doi: 10.1016/0092-8674(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 1140.Chresta CM, et al. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70:288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 1141.Sarkar S, et al. Small molecules enhance autophagy and reduce toxicity in Huntington’s disease models. Nat. Chem. Biol. 2007;3:331–338. doi: 10.1038/nchembio883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1142.Kuo SY, et al. Small-molecule enhancers of autophagy modulate cellular disease phenotypes suggested by human genetics. Proc. Natl Acad. Sci. USA. 2015;112:E4281–E4287. doi: 10.1073/pnas.1512289112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1143.Lim C-Y, et al. ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat. Cell Biol. 2019;21:1206–1218. doi: 10.1038/s41556-019-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1144.Burgett AWG, et al. Natural products reveal cancer cell dependence on oxysterol-binding proteins. Nat. Chem. Biol. 2011;7:639–647. doi: 10.1038/nchembio.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1145.Scotto Rosato A, et al. TRPML1 links lysosomal calcium to autophagosome biogenesis through the activation of the CaMKKβ/VPS34 pathway. Nat. Commun. 2019;10:5630. doi: 10.1038/s41467-019-13572-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1146.Robke L, et al. Phenotypic identification of a novel autophagy inhibitor chemotype targeting lipid kinase VPS34. Angew. Chem. Int. Ed. Engl. 2017;56:8153–8157. doi: 10.1002/anie.201703738. [DOI] [PubMed] [Google Scholar]
- 1147.Foley DJ, et al. Phenotyping reveals targets of a pseudo-natural-product autophagy inhibitor. Angew. Chem. Int. Ed. Engl. 2020;59:12470–12476. doi: 10.1002/anie.202000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1148.Petherick KJ, et al. Pharmacological inhibition of ULK1 kinase blocks mammalian target of rapamycin (mTOR)-dependent autophagy. J. Biol. Chem. 2015;290:11376–11383. doi: 10.1074/jbc.C114.627778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1149.Egan DF, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol. Cell. 2015;59:285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1150.Bosc D, et al. A new quinoline-based chemical probe inhibits the autophagy-related cysteine protease ATG4B. Sci. Rep. 2018;8:11653. doi: 10.1038/s41598-018-29900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1151.Samie M, et al. A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis. Dev. Cell. 2013;26:511–524. doi: 10.1016/j.devcel.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1152.Njomen E, Tepe JJ. Regulation of autophagic flux by the 20 S proteasome. Cell Chem. Biol. 2019;26:1283–1294.e5. doi: 10.1016/j.chembiol.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1153.Xie X-S, et al. Salicylihalamide A inhibits the V0 sector of the V-ATPase through a mechanism distinct from bafilomycin A1. J. Biol. Chem. 2004;279:19755–19763. doi: 10.1074/jbc.M313796200. [DOI] [PubMed] [Google Scholar]
- 1154.Boyd MR, et al. Discovery of a novel antitumor benzolactone enamide class that selectively inhibits mammalian vacuolar-type (H + )-ATPases. J. Pharmacol. Exp. Ther. 2001;297:114–120. [PubMed] [Google Scholar]
- 1155.McAfee Q, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc. Natl Acad. Sci. USA. 2012;109:8253–8258. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1156.Goodall ML, et al. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy. 2014;10:1120–1136. doi: 10.4161/auto.28594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1157.Ferreira PMP, et al. Chloroquine and hydroxychloroquine in antitumor therapies based on autophagy-related mechanisms. Pharmacol. Res. 2021;168:105582. doi: 10.1016/j.phrs.2021.105582. [DOI] [PubMed] [Google Scholar]
- 1158.Solomon VR, Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur. J. Pharm. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 1159.Silva VR, et al. Challenges and therapeutic opportunities of autophagy in cancer therapy. Cancers. 2020;12:3461. doi: 10.3390/cancers12113461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1160.Ashrafizadeh M, et al. Targeting autophagy in prostate cancer: preclinical and clinical evidence for therapeutic response. J. Exp. Clin. Cancer Res. 2022;41:105. doi: 10.1186/s13046-022-02293-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1161.Morel E, et al. Autophagy: a druggable process. Annu. Rev. Pharmacol. Toxicol. 2017;57:375–398. doi: 10.1146/annurev-pharmtox-010716-104936. [DOI] [PubMed] [Google Scholar]
- 1162.Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177:1682–1699. doi: 10.1016/j.cell.2019.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1163.Ebrahim AS, et al. PNT2258, a novel deoxyribonucleic acid inhibitor, induces cell cycle arrest and apoptosis via a distinct mechanism of action: a new class of drug for non-Hodgkin’s lymphoma. Oncotarget. 2016;7:42374–42384. doi: 10.18632/oncotarget.9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1164.Tse C, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- 1165.Chen J, et al. The Bcl-2/Bcl-X L/Bcl-w inhibitor, navitoclax, enhances the activity of chemotherapeutic agents in vitro and in vivo. Mol. Cancer Ther. 2011;10:2340–2349. doi: 10.1158/1535-7163.MCT-11-0415. [DOI] [PubMed] [Google Scholar]
- 1166.Lakhani NJ, et al. First-in-human study of palcitoclax (APG-1252), a novel dual Bcl-2/Bcl-xL inhibitor, demonstrated advantages in platelet safety while maintaining anticancer effect in U.S. patients with metastatic solid tumors. J. Clin. Oncol. 2020;38:3509. doi: 10.1200/JCO.2020.38.15_suppl.3509. [DOI] [Google Scholar]
- 1167.Brinkmann K, Ng AP, de Graaf CA, Strasser A. What can we learn from mice lacking pro-survival BCL-2 proteins to advance BH3 mimetic drugs for cancer therapy? Cell Death Differ. 2022;29:1079–1093. doi: 10.1038/s41418-022-00987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1168.Del Gaizo Moore V, et al. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J. Clin. Invest. 2007;117:112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1169.Cory S, Adams JM. Killing cancer cells by flipping the Bcl-2/Bax switch. Cancer Cell. 2005;8:5–6. doi: 10.1016/j.ccr.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 1170.Arulananda S, et al. A novel BH3-mimetic, AZD0466, targeting BCL-XL and BCL-2 is effective in pre-clinical models of malignant pleural mesothelioma. Cell Death Discov. 2021;7:122. doi: 10.1038/s41420-021-00505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1171.Roberts AW, Wei AH, Huang DCS. BCL2 and MCL1 inhibitors for hematologic malignancies. Blood. 2021;138:1120–1136. doi: 10.1182/blood.2020006785. [DOI] [PubMed] [Google Scholar]
- 1172.Zhu R, et al. FLT3 tyrosine kinase inhibitors synergize with BCL-2 inhibition to eliminate FLT3/ITD acute leukemia cells through BIM activation. Signal Transduct. Target. Ther. 2021;6:186. doi: 10.1038/s41392-021-00578-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1173.Roberts AW, et al. Targeting BCL2 with venetoclax in relapsed chronic lymphocytic leukemia. N. Engl. J. Med. 2016;374:311–322. doi: 10.1056/NEJMoa1513257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1174.Casara P, et al. S55746 is a novel orally active BCL-2 selective and potent inhibitor that impairs hematological tumor growth. Oncotarget. 2018;9:20075–20088. doi: 10.18632/oncotarget.24744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1175.Deng J, et al. Lisaftoclax (APG-2575) is a novel BCL-2 inhibitor with robust antitumor activity in preclinical models of hematologic malignancy. Clin. Cancer Res. 2022;28:5455–5468. doi: 10.1158/1078-0432.CCR-21-4037. [DOI] [PubMed] [Google Scholar]
- 1176.Xin, M. et al. Small-molecule Bax agonists for cancer therapy. Nat. Commun. 5, 4935 (2014). [DOI] [PMC free article] [PubMed]
- 1177.Reyna DE, et al. Direct activation of BAX by BTSA1 overcomes apoptosis resistance in acute myeloid leukemia. Cancer Cell. 2017;32:490–505.e10. doi: 10.1016/j.ccell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1178.Zang X, Song J, Li Y, Han Y. Targeting necroptosis as an alternative strategy in tumor treatment: From drugs to nanoparticles. J. Control. Release. 2022;349:213–226. doi: 10.1016/j.jconrel.2022.06.060. [DOI] [PubMed] [Google Scholar]
- 1179.Degterev A, et al. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005;1:112–119. doi: 10.1038/nchembio711. [DOI] [PubMed] [Google Scholar]
- 1180.Degterev A, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1181.Takahashi N, et al. Necrostatin-1 analogues: critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis. 2012;3:e437. doi: 10.1038/cddis.2012.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1182.Cao L, Mu W. Necrostatin-1 and necroptosis inhibition: pathophysiology and therapeutic implications. Pharm. Res. 2021;163:105297. doi: 10.1016/j.phrs.2020.105297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1183.Deeraksa A, et al. Plk1 is upregulated in androgen-insensitive prostate cancer cells and its inhibition leads to necroptosis. Oncogene. 2013;32:2973–2983. doi: 10.1038/onc.2012.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1184.Fulda S. Therapeutic exploitation of necroptosis for cancer therapy. Semin. Cell Dev. Biol. 2014;35:51–56. doi: 10.1016/j.semcdb.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 1185.Han W, et al. Shikonin circumvents cancer drug resistance by induction of a necroptotic death. Mol. Cancer Ther. 2007;6:1641–1649. doi: 10.1158/1535-7163.MCT-06-0511. [DOI] [PubMed] [Google Scholar]
- 1186.Huang C, et al. Shikonin kills glioma cells through necroptosis mediated by RIP-1. PLoS ONE. 2013;8:e66326. doi: 10.1371/journal.pone.0066326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1187.Fu, Z. et al. The anti-tumor effect of shikonin on osteosarcoma by inducing RIP1 and RIP3 dependent necroptosis. BMC Cancer13, 580 (2013). [DOI] [PMC free article] [PubMed]
- 1188.Xuan Y, Hu X. Naturally-occurring shikonin analogues–a class of necroptotic inducers that circumvent cancer drug resistance. Cancer Lett. 2009;274:233–242. doi: 10.1016/j.canlet.2008.09.029. [DOI] [PubMed] [Google Scholar]
- 1189.Basit F, Cristofanon S, Fulda S. Obatoclax (GX15-070) triggers necroptosis by promoting the assembly of the necrosome on autophagosomal membranes. Cell Death Differ. 2013;20:1161–1173. doi: 10.1038/cdd.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1190.Rizzi F, et al. Polyphenon E®, a standardized green tea extract, induces endoplasmic reticulum stress, leading to death of immortalized PNT1a cells by anoikis and tumorigenic PC3 by necroptosis. Carcinogenesis. 2014;35:828–839. doi: 10.1093/carcin/bgt481. [DOI] [PubMed] [Google Scholar]
- 1191.Zhou B, et al. Bioactive staurosporine derivatives from the Streptomyces sp. NB-A13. Bioorg. Chem. 2019;82:33–40. doi: 10.1016/j.bioorg.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 1192.Dunai ZA, et al. Staurosporine induces necroptotic cell death under caspase-compromised conditions in U937 cells. PLoS ONE. 2012;7:e41945. doi: 10.1371/journal.pone.0041945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1193.Saddoughi SA, et al. Sphingosine analogue drug FTY720 targets I2PP2A/SET and mediates lung tumour suppression via activation of PP2A-RIPK1-dependent necroptosis. EMBO Mol. Med. 2013;5:105–121. doi: 10.1002/emmm.201201283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1194.Zhang L, Wang H, Ding K, Xu J. FTY720 induces autophagy-related apoptosis and necroptosis in human glioblastoma cells. Toxicol. Lett. 2015;236:43–59. doi: 10.1016/j.toxlet.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 1195.Pasupuleti N, Leon L, Carraway KL, Gorin F. 5-Benzylglycinyl-amiloride kills proliferating and nonproliferating malignant glioma cells through caspase- independent necroptosis mediated by apoptosis-inducing factor. J. Pharmacol. Exp. Ther. 2013;344:600–615. doi: 10.1124/jpet.112.200519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1196.Zec M, et al. Novel selenosemicarbazone metal complexes exert anti-tumor effect via alternative, caspase-independent necroptotic cell death. Med. Chem. 2014;10:759–771. doi: 10.2174/1573406410666140327122009. [DOI] [PubMed] [Google Scholar]
- 1197.Tufo G, et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ. 2014;21:685–695. doi: 10.1038/cdd.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1198.Jouan-Lanhouet S, et al. TRAIL induces necroptosis involving RIPK1/RIPK3-dependent PARP-1 activation. Cell Death Differ. 2012;19:2003–2014. doi: 10.1038/cdd.2012.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1199.Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nat. Rev. Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- 1200.Winer A, Adams S, Mignatti P. Matrix metalloproteinase inhibitors in cancer therapy: turning past failures into future successes. Mol. Cancer Ther. 2018;17:1147–1155. doi: 10.1158/1535-7163.MCT-17-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1201.Zucker S, Cao J, Chen WT. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene. 2000;19:6642–6650. doi: 10.1038/sj.onc.1204097. [DOI] [PubMed] [Google Scholar]
- 1202.Gautam J, et al. Down-regulation of cathepsin S and matrix metalloproteinase-9 via Src, a non-receptor tyrosine kinase, suppresses triple-negative breast cancer growth and metastasis. Exp. Mol. Med. 2018;50:1–14. doi: 10.1038/s12276-018-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1203.Macaulay VM, et al. Phase I study of intrapleural batimastat (BB-94), a matrix metalloproteinase inhibitor, in the treatment of malignant pleural effusions. Clin. Cancer Res. 1999;5:513–520. [PubMed] [Google Scholar]
- 1204.Gatto C, et al. BAY 12-9566, a novel inhibitor of matrix metalloproteinases with antiangiogenic activity. Clin. Cancer Res. 1999;5:3603–3607. [PubMed] [Google Scholar]
- 1205.Baidya SK, Amin SA, Jha T. Outline of gelatinase inhibitors as anti-cancer agents: a patent mini-review for 2010-present. Eur. J. Med. Chem. 2021;213:113044. doi: 10.1016/j.ejmech.2020.113044. [DOI] [PubMed] [Google Scholar]
- 1206.Hirte H, et al. A phase III randomized trial of BAY 12-9566 (tanomastat) as maintenance therapy in patients with advanced ovarian cancer responsive to primary surgery and paclitaxel/platinum containing chemotherapy: a National Cancer Institute of Canada Clinical Trials. Gynecol. Oncol. 2006;102:300–308. doi: 10.1016/j.ygyno.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 1207.Rudek MA, et al. Phase I clinical trial of oral COL-3, a matrix metalloproteinase inhibitor, in patients with refractory metastatic cancer. J. Clin. Oncol. 2001;19:584–592. doi: 10.1200/JCO.2001.19.2.584. [DOI] [PubMed] [Google Scholar]
- 1208.Mao J-W, He X-M, Tang H-Y, Wang Y-D. Protective role of metalloproteinase inhibitor (AE-941) on ulcerative colitis in rats. World J. Gastroenterol. 2012;18:7063–7069. doi: 10.3748/wjg.v18.i47.7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1209.Deryugina EI, Ratnikov BI, Strongin AY. Prinomastat, a hydroxamate inhibitor of matrix metalloproteinases, has a complex effect on migration of breast carcinoma cells. Int. J. cancer. 2003;104:533–541. doi: 10.1002/ijc.10977. [DOI] [PubMed] [Google Scholar]
- 1210.Rizvi NA, et al. A phase I study of oral BMS-275291, a novel nonhydroxamate sheddase-sparing matrix metalloproteinase inhibitor, in patients with advanced or metastatic cancer. Clin. Cancer Res. 2004;10:1963–1970. doi: 10.1158/1078-0432.CCR-1183-02. [DOI] [PubMed] [Google Scholar]
- 1211.Syed S, et al. A phase I and pharmacokinetic study of Col-3 (Metastat), an oral tetracycline derivative with potent matrix metalloproteinase and antitumor properties. Clin. Cancer Res. 2004;10:6512–6521. doi: 10.1158/1078-0432.CCR-04-0804. [DOI] [PubMed] [Google Scholar]
- 1212.Maurya, S. K., Poddar, N., Tandon, P. & Yadav, A. K. In Pathophysiological Aspects of Proteases (eds Chakraborti, S. & Dhalla, N. S.) Ch.10 (Springer, 2017).
- 1213.Parikh PK, Ghate MD. Recent advances in the discovery of small molecule c-Met Kinase inhibitors. Eur. J. Med Chem. 2018;143:1103–1138. doi: 10.1016/j.ejmech.2017.08.044. [DOI] [PubMed] [Google Scholar]
- 1214.Markham A. Tepotinib: first approval. Drugs. 2020;80:829–833. doi: 10.1007/s40265-020-01317-9. [DOI] [PubMed] [Google Scholar]
- 1215.Mathieu LN, et al. FDA approval summary: capmatinib and tepotinib for the treatment of metastatic NSCLC harboring MET exon 14 skipping mutations or alterations. Clin. Cancer Res. 2022;28:249–254. doi: 10.1158/1078-0432.CCR-21-1566. [DOI] [PubMed] [Google Scholar]
- 1216.Dhillon S. Capmatinib: first approval. Drugs. 2020;80:1125–1131. doi: 10.1007/s40265-020-01347-3. [DOI] [PubMed] [Google Scholar]
- 1217.Zou HY, et al. Sensitivity of selected human tumor models to PF-04217903, a novel selective c-Met kinase inhibitor. Mol. Cancer Ther. 2012;11:1036–1047. doi: 10.1158/1535-7163.MCT-11-0839. [DOI] [PubMed] [Google Scholar]
- 1218.Yakes FM, et al. Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor, simultaneously suppresses metastasis, angiogenesis, and tumor growth. Mol. Cancer Ther. 2011;10:2298–2308. doi: 10.1158/1535-7163.MCT-11-0264. [DOI] [PubMed] [Google Scholar]
- 1219.Smith DC, et al. Cabozantinib in patients with advanced prostate cancer: results of a phase II randomized discontinuation trial. J. Clin. Oncol. 2013;31:412–419. doi: 10.1200/JCO.2012.45.0494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1220.Choueiri TK, et al. Cabozantinib versus everolimus in advanced renal-cell carcinoma. N. Engl. J. Med. 2015;373:1814–1823. doi: 10.1056/NEJMoa1510016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1221.Choueiri TK, et al. Cabozantinib versus everolimus in advanced renal cell carcinoma (METEOR): final results from a randomised, open-label, phase 3 trial. Lancet Oncol. 2016;17:917–927. doi: 10.1016/S1470-2045(16)30107-3. [DOI] [PubMed] [Google Scholar]
- 1222.Eder JP, Vande Woude GF, Boerner SA, Lorusso PM. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin. Cancer Res. 2009;15:2207–2214. doi: 10.1158/1078-0432.CCR-08-1306. [DOI] [PubMed] [Google Scholar]
- 1223.Eathiraj S, et al. Discovery of a novel mode of protein kinase inhibition characterized by the mechanism of inhibition of human mesenchymal-epithelial transition factor (c-Met) protein autophosphorylation by ARQ 197. J. Biol. Chem. 2011;286:20666–20676. doi: 10.1074/jbc.M110.213801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1224.Zhao S, et al. Selective inhibitor of the c-Met receptor tyrosine kinase in advanced hepatocellular carcinoma: no beneficial effect with the use of tivantinib? Front. Immunol. 2021;12:731527. doi: 10.3389/fimmu.2021.731527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1225.Merchant M, et al. Monovalent antibody design and mechanism of action of onartuzumab, a MET antagonist with anti-tumor activity as a therapeutic agent. Proc. Natl Acad. Sci. USA. 2013;110:E2987–E2996. doi: 10.1073/pnas.1302725110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1226.Kim K-H, Kim H. Progress of antibody-based inhibitors of the HGF-cMET axis in cancer therapy. Exp. Mol. Med. 2017;49:e307. doi: 10.1038/emm.2017.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1227.Liu L, et al. LY2875358, a neutralizing and internalizing anti-MET bivalent antibody, inhibits HGF-dependent and HGF-independent MET activation and tumor growth. Clin. Cancer Res. 2014;20:6059–6070. doi: 10.1158/1078-0432.CCR-14-0543. [DOI] [PubMed] [Google Scholar]
- 1228.Patnaik A, et al. A phase I study of LY3164530, a bispecific antibody targeting MET and EGFR, in patients with advanced or metastatic cancer. Cancer Chemother. Pharmacol. 2018;82:407–418. doi: 10.1007/s00280-018-3623-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1229.Park K, et al. Amivantamab in EGFR exon 20 insertion-mutated non-small-cell lung cancer progressing on platinum chemotherapy: initial results from the CHRYSALIS phase I study. J. Clin. Oncol. 2021;39:3391–3402. doi: 10.1200/JCO.21.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1230.Moores SL, et al. A novel bispecific antibody targeting EGFR and cMet is effective against EGFR inhibitor-resistant lung tumors. Cancer Res. 2016;76:3942–3953. doi: 10.1158/0008-5472.CAN-15-2833. [DOI] [PubMed] [Google Scholar]
- 1231.Lee JM, et al. Cbl-independent degradation of Met: ways to avoid agonism of bivalent met-targeting antibody. Oncogene. 2014;33:34–43. doi: 10.1038/onc.2012.551. [DOI] [PubMed] [Google Scholar]
- 1232.Lee B-S, et al. Met degradation by SAIT301, a Met monoclonal antibody, reduces the invasion and migration of nasopharyngeal cancer cells via inhibition of EGR-1 expression. Cell Death Dis. 2014;5:e1159. doi: 10.1038/cddis.2014.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1233.Lee J, et al. Phase I trial of anti-MET monoclonal antibody in MET-overexpressed refractory cancer. Clin. Colorectal Cancer. 2018;17:140–146. doi: 10.1016/j.clcc.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 1234.Hultberg A, et al. Depleting MET-expressing tumor cells by ADCC provides a therapeutic advantage over inhibiting HGF/MET signaling. Cancer Res. 2015;75:3373–3383. doi: 10.1158/0008-5472.CAN-15-0356. [DOI] [PubMed] [Google Scholar]
- 1235.Petrelli A, et al. Ab-induced ectodomain shedding mediates hepatocyte growth factor receptor down-regulation and hampers biological activity. Proc. Natl Acad. Sci. USA. 2006;103:5090–5095. doi: 10.1073/pnas.0508156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1236.Pacchiana G, et al. Monovalency unleashes the full therapeutic potential of the DN-30 anti-Met antibody. J. Biol. Chem. 2010;285:36149–36157. doi: 10.1074/jbc.M110.134031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1237.Burgess TL, et al. Biochemical characterization of AMG 102: a neutralizing, fully human monoclonal antibody to human and nonhuman primate hepatocyte growth factor. Mol. Cancer Ther. 2010;9:400–409. doi: 10.1158/1535-7163.MCT-09-0824. [DOI] [PubMed] [Google Scholar]
- 1238.Yap TA, De Bono JS. Targeting the HGF/c-met axis: state of play. Mol. Cancer Ther. 2010;9:1077–1079. doi: 10.1158/1535-7163.MCT-10-0122. [DOI] [PubMed] [Google Scholar]
- 1239.Tarhini AA, et al. Phase 1/2 study of rilotumumab (AMG 102), a hepatocyte growth factor inhibitor, and erlotinib in patients with advanced non-small cell lung cancer. Cancer. 2017;123:2936–2944. doi: 10.1002/cncr.30717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1240.Fiedler U, et al. Potency of bortezomib in combination with MP0250, a bispecific VEGF- and HGF-targeting darpin, in a preclinical multiple myeloma model. J. Clin. Oncol. 2014;32:e19574–e19574. doi: 10.1200/jco.2014.32.15_suppl.e19574. [DOI] [Google Scholar]
- 1241.Date K, Matsumoto K, Shimura H, Tanaka M, Nakamura T. HGF/NK4 is a specific antagonist for pleiotrophic actions of hepatocyte growth factor. FEBS Lett. 1997;420:1–6. doi: 10.1016/S0014-5793(97)01475-0. [DOI] [PubMed] [Google Scholar]
- 1242.Mizuno S, Nakamura T. HGF-MET cascade, a key target for inhibiting cancer metastasis: the impact of NK4 discovery on cancer biology and therapeutics. Int. J. Mol. Sci. 2013;14:888–919. doi: 10.3390/ijms14010888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1243.Matsumoto K, Nakamura T. Mechanisms and significance of bifunctional NK4 in cancer treatment. Biochem. Biophys. Res. Commun. 2005;333:316–327. doi: 10.1016/j.bbrc.2005.05.131. [DOI] [PubMed] [Google Scholar]
- 1244.Matsumoto K, Nakamura T. NK4 (HGF-antagonist/angiogenesis inhibitor) in cancer biology and therapeutics. Cancer Sci. 2003;94:321–327. doi: 10.1111/j.1349-7006.2003.tb01440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1245.Rivas S, Marín A, Samtani S, González-Feliú E, Armisén R. MET signaling pathways, resistance mechanisms, and opportunities for target therapies. Int. J. Mol. Sci. 2022;23:13898. doi: 10.3390/ijms232213898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1246.Dias MP, Moser SC, Ganesan S, Jonkers J. Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 2021;18:773–791. doi: 10.1038/s41571-021-00532-x. [DOI] [PubMed] [Google Scholar]
- 1247.Tutt ANJ, et al. Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N. Engl. J. Med. 2021;384:2394–2405. doi: 10.1056/NEJMoa2105215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1248.Ledermann J, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N. Engl. J. Med. 2012;366:1382–1392. doi: 10.1056/NEJMoa1105535. [DOI] [PubMed] [Google Scholar]
- 1249.Moore K, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N. Engl. J. Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 1250.Kristeleit R, et al. Rucaparib versus standard-of-care chemotherapy in patients with relapsed ovarian cancer and a deleterious BRCA1 or BRCA2 mutation (ARIEL4): an international, open-label, randomised, phase 3 trial. Lancet Oncol. 2022;23:465–478. doi: 10.1016/S1470-2045(22)00122-X. [DOI] [PubMed] [Google Scholar]
- 1251.Robson M, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N. Engl. J. Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 1252.Litton JK, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N. Engl. J. Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1253.Golan T, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N. Engl. J. Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1254.Abida W, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J. Clin. Oncol. 2020;38:3763–3772. doi: 10.1200/JCO.20.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1255.Riches LC, et al. Pharmacology of the ATM inhibitor AZD0156: potentiation of irradiation and olaparib responses preclinically. Mol. Cancer Ther. 2020;19:13–25. doi: 10.1158/1535-7163.MCT-18-1394. [DOI] [PubMed] [Google Scholar]
- 1256.Durant ST, et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci. Adv. 2018;4:eaat1719. doi: 10.1126/sciadv.aat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1257.Tew BY, et al. ATM-inhibitor AZD1390 is a radiosensitizer for breast cancer CNS metastasis. Clin. Cancer Res. 2023;29:4492–4503. doi: 10.1158/1078-0432.CCR-23-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1258.Fuchss T, et al. Abstract 3500: highly potent and selective ATM kinase inhibitor M4076: a clinical candidate drug with strong anti-tumor activity in combination therapies. Cancer Res. 2019;79:3500. doi: 10.1158/1538-7445.AM2019-3500. [DOI] [Google Scholar]
- 1259.Zimmermann A, et al. A new class of selective ATM inhibitors as combination partners of DNA double-strand break inducing cancer therapies. Mol. Cancer Ther. 2022;21:859–870. doi: 10.1158/1535-7163.MCT-21-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1260.Hickson I, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 1261.Bryant HE, Helleday T. Inhibition of poly (ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 2006;34:1685–1691. doi: 10.1093/nar/gkl108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1262.Toledo LI, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 2011;18:721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1263.Boudny M, Trbusek M. ATR-CHK1 pathway as a therapeutic target for acute and chronic leukemias. Cancer Treat. Rev. 2020;88:102026. doi: 10.1016/j.ctrv.2020.102026. [DOI] [PubMed] [Google Scholar]
- 1264.Middleton MR, et al. Phase 1 study of the ATR inhibitor berzosertib (formerly M6620, VX-970) combined with gemcitabine ± cisplatin in patients with advanced solid tumours. Br. J. Cancer. 2021;125:510–519. doi: 10.1038/s41416-021-01405-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1265.Wengner AM, et al. The novel ATR inhibitor BAY 1895344 is efficacious as monotherapy and combined with DNA damage-inducing or repair-compromising therapies in preclinical cancer models. Mol. Cancer Ther. 2020;19:26–38. doi: 10.1158/1535-7163.MCT-19-0019. [DOI] [PubMed] [Google Scholar]
- 1266.Kim H, et al. Targeting the ATR/CHK1 axis with PARP inhibition results in tumor regression in BRCA-mutant ovarian cancer models. Clin. Cancer Res. 2017;23:3097–3108. doi: 10.1158/1078-0432.CCR-16-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1267.Brooks K, et al. A potent Chk1 inhibitor is selectively cytotoxic in melanomas with high levels of replicative stress. Oncogene. 2013;32:788–796. doi: 10.1038/onc.2012.72. [DOI] [PubMed] [Google Scholar]
- 1268.Massey AJ, et al. mTORC1 and DNA-PKcs as novel molecular determinants of sensitivity to Chk1 inhibition. Mol. Oncol. 2016;10:101–112. doi: 10.1016/j.molonc.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1269.Qiu Z, Oleinick NL, Zhang J. ATR/CHK1 inhibitors and cancer therapy. Radiol. Oncol. 2018;126:450–464. doi: 10.1016/j.radonc.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1270.Scagliotti G, et al. Phase II evaluation of LY2603618, a first-generation CHK1 inhibitor, in combination with pemetrexed in patients with advanced or metastatic non-small cell lung cancer. Invest. N. Drugs. 2016;34:625–635. doi: 10.1007/s10637-016-0368-1. [DOI] [PubMed] [Google Scholar]
- 1271.Wehler T, et al. A randomized, phase 2 evaluation of the CHK1 inhibitor, LY2603618, administered in combination with pemetrexed and cisplatin in patients with advanced nonsquamous non-small cell lung cancer. Lung Cancer. 2017;108:212–216. doi: 10.1016/j.lungcan.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 1272.Li Q, et al. A new wave of innovations within the DNA damage response. Signal Transduct. Target. Ther. 2023;8:338. doi: 10.1038/s41392-023-01548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1273.King C, et al. LY2606368 causes replication catastrophe and antitumor effects through CHK1-dependent mechanisms. Mol. Cancer Ther. 2015;14:2004–2013. doi: 10.1158/1535-7163.MCT-14-1037. [DOI] [PubMed] [Google Scholar]
- 1274.Kristeleit R, et al. A phase 1/2 trial of SRA737 (a Chk1 inhibitor) administered orally in patients with advanced cancer. Br. J. Cancer. 2023;129:38–45. doi: 10.1038/s41416-023-02279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1275.Sausville E, et al. Phase I dose-escalation study of AZD7762, a checkpoint kinase inhibitor, in combination with gemcitabine in US patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014;73:539–549. doi: 10.1007/s00280-014-2380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1276.Zhao H, et al. Inflammation and tumor progression: signaling pathways and targeted intervention. Signal Transduct. Target Ther. 2021;6:263. doi: 10.1038/s41392-021-00658-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1277.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat. Rev. Clin. Oncol. 2015;12:584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 1278.Jendrossek V. Targeting apoptosis pathways by celecoxib in cancer. Cancer Lett. 2013;332:313–324. doi: 10.1016/j.canlet.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 1279.Boudreau DM, Yu O, Johnson J. Statin use and cancer risk: a comprehensive review. Expert Opin. Drug Saf. 2010;9:603–621. doi: 10.1517/14740331003662620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1280.Dinarello CA. Anti-inflammatory agents: present and future. Cell. 2010;140:935–950. doi: 10.1016/j.cell.2010.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1281.Deisseroth A, et al. FDA approval: siltuximab for the treatment of patients with multicentric Castleman disease. Clin. Cancer Res. 2015;21:950–954. doi: 10.1158/1078-0432.CCR-14-1678. [DOI] [PubMed] [Google Scholar]
- 1282.Park H, Lee S, Lee J, Moon H, Ro SW. Exploring the JAK/STAT signaling pathway in hepatocellular carcinoma: unraveling signaling complexity and therapeutic implications. Int. J. Mol. Sci. 2023;24:13764. doi: 10.3390/ijms241813764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1283.Quintás-Cardama A, et al. Preclinical characterization of the selective JAK1/2 inhibitor INCB018424: therapeutic implications for the treatment of myeloproliferative neoplasms. Blood. 2010;115:3109–3117. doi: 10.1182/blood-2009-04-214957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1284.William AD, et al. Discovery of the macrocycle 11-(2-pyrrolidin-1-yl-ethoxy)-14,19-dioxa-5,7,26-triaza-tetracyclo[19.3.1.1(2,6).1(8,12)]heptacosa-1(25),2(26),3,5,8,10,12(27),16,21,23-decaene (SB1518), a potent Janus kinase 2/fms-like tyrosine kinase-3 (JAK2/FLT3) inhibitor. J. Med. Chem. 2011;54:4638–4658. doi: 10.1021/jm200326p. [DOI] [PubMed] [Google Scholar]
- 1285.FARBER S, DIAMOND LK. Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N. Engl. J. Med. 1948;238:787–793. doi: 10.1056/NEJM194806032382301. [DOI] [PubMed] [Google Scholar]
- 1286.Stine ZE, Schug ZT, Salvino JM, Dang CV. Targeting cancer metabolism in the era of precision oncology. Nat. Rev. Drug Discov. 2022;21:141–162. doi: 10.1038/s41573-021-00339-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1287.Martinez-Outschoorn UE, Peiris-Pagés M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 2017;14:11–31. doi: 10.1038/nrclinonc.2016.60. [DOI] [PubMed] [Google Scholar]
- 1288.Siebeneicher H, et al. Identification and optimization of the first highly selective GLUT1 inhibitor BAY-876. ChemMedChem. 2016;11:2261–2271. doi: 10.1002/cmdc.201600276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1289.Jin J, Byun J-K, Choi Y-K, Park K-G. Targeting glutamine metabolism as a therapeutic strategy for cancer. Exp. Mol. Med. 2023;55:706–715. doi: 10.1038/s12276-023-00971-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1290.Varghese S, et al. The glutaminase inhibitor CB-839 (Telaglenastat) enhances the antimelanoma activity of T-cell-mediated immunotherapies. Mol. Cancer Ther. 2021;20:500–511. doi: 10.1158/1535-7163.MCT-20-0430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1291.Kremer DM, Lyssiotis CA. Targeting allosteric regulation of cancer metabolism. Nat. Chem. Biol. 2022;18:441–450. doi: 10.1038/s41589-022-00997-6. [DOI] [PubMed] [Google Scholar]
- 1292.Leone RD, et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science. 2019;366:1013–1021. doi: 10.1126/science.aav2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1293.Camarda R, et al. Inhibition of fatty acid oxidation as a therapy for MYC-overexpressing triple-negative breast cancer. Nat. Med. 2016;22:427–432. doi: 10.1038/nm.4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1294.Ray KK, et al. Safety and efficacy of bempedoic acid to reduce LDL cholesterol. N. Engl. J. Med. 2019;380:1022–1032. doi: 10.1056/NEJMoa1803917. [DOI] [PubMed] [Google Scholar]
- 1295.Wei J, et al. An allosteric mechanism for potent inhibition of human ATP-citrate lyase. Nature. 2019;568:566–570. doi: 10.1038/s41586-019-1094-6. [DOI] [PubMed] [Google Scholar]
- 1296.Sainero-Alcolado L, Liaño-Pons J, Ruiz-Pérez MV, Arsenian-Henriksson M. Targeting mitochondrial metabolism for precision medicine in cancer. Cell Death Differ. 2022;29:1304–1317. doi: 10.1038/s41418-022-01022-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1297.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1298.Yen K, et al. AG-221, a first-in-class therapy targeting acute myeloid leukemia harboring oncogenic IDH2 mutations. Cancer Discov. 2017;7:478–493. doi: 10.1158/2159-8290.CD-16-1034. [DOI] [PubMed] [Google Scholar]
- 1299.Molina JR, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat. Med. 2018;24:1036–1046. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 1300.Ducker GS, Rabinowitz JD. One-carbon metabolism in health and disease. Cell Metab. 2017;25:27–42. doi: 10.1016/j.cmet.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1301.Locasale JW. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1302.Ju H-Q, et al. Modulation of redox homeostasis by inhibition of MTHFD2 in colorectal cancer: mechanisms and therapeutic implications. J. Natl Cancer Inst. 2019;111:584–596. doi: 10.1093/jnci/djy160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1303.Zhou X, et al. Discovery of novel inhibitors of human phosphoglycerate dehydrogenase by activity-directed combinatorial chemical synthesis strategy. Bioorg. Chem. 2021;115:105159. doi: 10.1016/j.bioorg.2021.105159. [DOI] [PubMed] [Google Scholar]
- 1304.García-Cañaveras JC, et al. SHMT inhibition is effective and synergizes with methotrexate in T-cell acute lymphoblastic leukemia. Leukemia. 2021;35:377–388. doi: 10.1038/s41375-020-0845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1305.Scaletti E, Jemth A-S, Helleday T, Stenmark P. Structural basis of inhibition of the human serine hydroxymethyltransferase SHMT2 by antifolate drugs. FEBS Lett. 2019;593:1863–1873. doi: 10.1002/1873-3468.13455. [DOI] [PubMed] [Google Scholar]
- 1306.Li C, et al. Design, synthesis, and biological evaluation of a novel series of teriflunomide derivatives as potent human dihydroorotate dehydrogenase inhibitors for malignancy treatment. J. Med. Chem. 2021;64:18175–18192. doi: 10.1021/acs.jmedchem.1c01711. [DOI] [PubMed] [Google Scholar]
- 1307.Kanarek N, Petrova B, Sabatini DM. Dietary modifications for enhanced cancer therapy. Nature. 2020;579:507–517. doi: 10.1038/s41586-020-2124-0. [DOI] [PubMed] [Google Scholar]
- 1308.Caffa I, et al. Fasting-mimicking diet and hormone therapy induce breast cancer regression. Nature. 2020;583:620–624. doi: 10.1038/s41586-020-2502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1309.Martínez-Garay C, Djouder N. Dietary interventions and precision nutrition in cancer therapy. Trends Mol. Med. 2023;29:489–511. doi: 10.1016/j.molmed.2023.04.004. [DOI] [PubMed] [Google Scholar]
- 1310.Taylor SR, Falcone JN, Cantley LC, Goncalves MD. Developing dietary interventions as therapy for cancer. Nat. Rev. Cancer. 2022;22:452–466. doi: 10.1038/s41568-022-00485-y. [DOI] [PubMed] [Google Scholar]
- 1311.Leach DR, Krummel MF, Allison JP. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 1996;271:1734–1736. doi: 10.1126/science.271.5256.1734. [DOI] [PubMed] [Google Scholar]
- 1312.Wang D-R, Wu X-L, Sun Y-L. Therapeutic targets and biomarkers of tumor immunotherapy: response versus non-response. Signal Transduct. Target. Ther. 2022;7:331. doi: 10.1038/s41392-022-01136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1313.Doroshow DB, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2021;18:345–362. doi: 10.1038/s41571-021-00473-5. [DOI] [PubMed] [Google Scholar]
- 1314.Pérez-Ruiz E, et al. Cancer immunotherapy resistance based on immune checkpoints inhibitors: Targets, biomarkers, and remedies. Drug Resist. Updat. 2020;53:100718. doi: 10.1016/j.drup.2020.100718. [DOI] [PubMed] [Google Scholar]
- 1315.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1316.Zinn S, et al. Advances in antibody-based therapy in oncology. Nat. cancer. 2023;4:165–180. doi: 10.1038/s43018-023-00516-z. [DOI] [PubMed] [Google Scholar]
- 1317.Wang K, et al. Overall survival of patients with hepatocellular carcinoma treated with sintilimab and disease outcome after treatment discontinuation. BMC Cancer. 2023;23:1017. doi: 10.1186/s12885-023-11485-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1318.André T, et al. Antitumor activity and safety of dostarlimab monotherapy in patients with mismatch repair deficient solid tumors: a nonrandomized controlled trial. JAMA Netw. Open. 2023;6:e2341165. doi: 10.1001/jamanetworkopen.2023.41165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1319.Antonia SJ, et al. Overall survival with durvalumab after chemoradiotherapy in stage III NSCLC. N. Engl. J. Med. 2018;379:2342–2350. doi: 10.1056/NEJMoa1809697. [DOI] [PubMed] [Google Scholar]
- 1320.Grivas P, et al. Avelumab first-line maintenance treatment for advanced urothelial carcinoma: review of evidence to guide clinical practice. ESMO Open. 2023;8:102050. doi: 10.1016/j.esmoop.2023.102050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1321.Cotton AD, Nguyen DP, Gramespacher JA, Seiple IB, Wells JA. Development of antibody-based PROTACs for the degradation of the cell-surface immune checkpoint protein PD-L1. J. Am. Chem. Soc. 2021;143:593–598. doi: 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1322.Li S, et al. PROTACs: novel tools for improving immunotherapy in cancer. Cancer Lett. 2023;560:216128. doi: 10.1016/j.canlet.2023.216128. [DOI] [PubMed] [Google Scholar]
- 1323.Girardi DM, et al. Cabozantinib plus nivolumab phase I expansion study in patients with metastatic urothelial carcinoma refractory to immune checkpoint inhibitor therapy. Clin. Cancer Res. 2022;28:1353–1362. doi: 10.1158/1078-0432.CCR-21-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1324.Harding JJ, et al. Blocking TIM-3 in treatment-refractory advanced solid tumors: a phase Ia/b study of LY3321367 with or without an anti-PD-L1 antibody. Clin. Cancer Res. 2021;27:2168–2178. doi: 10.1158/1078-0432.CCR-20-4405. [DOI] [PubMed] [Google Scholar]
- 1325.Addala V, et al. Computational immunogenomic approaches to predict response to cancer immunotherapies. Nat. Rev. Clin. Oncol. 2023;21:28–46. doi: 10.1038/s41571-023-00830-6. [DOI] [PubMed] [Google Scholar]
- 1326.Powles T, et al. Pembrolizumab plus axitinib versus sunitinib monotherapy as first-line treatment of advanced renal cell carcinoma (KEYNOTE-426): extended follow-up from a randomised, open-label, phase 3 trial. Lancet Oncol. 2020;21:1563–1573. doi: 10.1016/S1470-2045(20)30436-8. [DOI] [PubMed] [Google Scholar]
- 1327.De Thé H. Differentiation therapy revisited. Nat. Rev. Cancer. 2018;18:117–127. doi: 10.1038/nrc.2017.103. [DOI] [PubMed] [Google Scholar]
- 1328.Bates SE. Epigenetic therapies for cancer. N. Engl. J. Med. 2020;383:650–663. doi: 10.1056/NEJMra1805035. [DOI] [PubMed] [Google Scholar]
- 1329.Michalak EM, Burr ML, Bannister AJ, Dawson MA. The roles of DNA, RNA and histone methylation in ageing and cancer. Nat. Rev. Mol. Cell Biol. 2019;20:573–589. doi: 10.1038/s41580-019-0143-1. [DOI] [PubMed] [Google Scholar]
- 1330.Chen C, et al. DNA methylation: from cancer biology to clinical perspectives. Front. Biosci. 2022;27:326. doi: 10.31083/j.fbl2712326. [DOI] [PubMed] [Google Scholar]
- 1331.Ma J, Ge Z. Comparison between decitabine and azacitidine for patients with acute myeloid leukemia and higher-risk myelodysplastic syndrome: a systematic review and network meta-analysis. Front. Pharmacol. 2021;12:701690. doi: 10.3389/fphar.2021.701690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1332.Kantarjian HM, et al. Guadecitabine (SGI-110) in treatment-naive patients with acute myeloid leukaemia: phase 2 results from a multicentre, randomised, phase 1/2 trial. Lancet Oncol. 2017;18:1317–1326. doi: 10.1016/S1470-2045(17)30576-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1333.Lim B, et al. The preclinical efficacy of the novel hypomethylating agent NTX-301 as a monotherapy and in combination with venetoclax in acute myeloid leukemia. Blood Cancer J. 2022;12:57. doi: 10.1038/s41408-022-00664-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1334.Pappalardi MB, et al. Discovery of a first-in-class reversible DNMT1-selective inhibitor with improved tolerability and efficacy in acute myeloid leukemia. Nat. Cancer. 2021;2:1002–1017. doi: 10.1038/s43018-021-00249-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1335.Plummer R, et al. Phase I study of MG98, an oligonucleotide antisense inhibitor of human DNA methyltransferase 1, given as a 7-day infusion in patients with advanced solid tumors. Clin. Cancer Res. 2009;15:3177–3183. doi: 10.1158/1078-0432.CCR-08-2859. [DOI] [PubMed] [Google Scholar]
- 1336.Ramaiah MJ, Tangutur AD, Manyam RR. Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sci. 2021;277:119504. doi: 10.1016/j.lfs.2021.119504. [DOI] [PubMed] [Google Scholar]
- 1337.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 1338.Sun Y, et al. Therapeutic potential of tucidinostat, a subtype-selective HDAC inhibitor, in cancer treatment. Front. Pharmacol. 2022;13:932914. doi: 10.3389/fphar.2022.932914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1339.Roche J, Bertrand P. Inside HDACs with more selective HDAC inhibitors. Eur. J. Med. Chem. 2016;121:451–483. doi: 10.1016/j.ejmech.2016.05.047. [DOI] [PubMed] [Google Scholar]
- 1340.Yue K, et al. Comparison of three zinc binding groups for HDAC inhibitors - A potency, selectivity and enzymatic kinetics study. Bioorg. Med. Chem. Lett. 2022;70:128797. doi: 10.1016/j.bmcl.2022.128797. [DOI] [PubMed] [Google Scholar]
- 1341.Su M, Gong X, Liu F. An update on the emerging approaches for histone deacetylase (HDAC) inhibitor drug discovery and future perspectives. Expert Opin. Drug Discov. 2021;16:745–761. doi: 10.1080/17460441.2021.1877656. [DOI] [PubMed] [Google Scholar]
- 1342.Connolly RM, Rudek MA, Piekarz R. Entinostat: a promising treatment option for patients with advanced breast cancer. Future Oncol. 2017;13:1137–1148. doi: 10.2217/fon-2016-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1343.Davalos V, Esteller M. Cancer epigenetics in clinical practice. CA Cancer J. Clin. 2023;73:376–424. doi: 10.3322/caac.21765. [DOI] [PubMed] [Google Scholar]
- 1344.Hoy SM. Tazemetostat: first approval. Drugs. 2020;80:513–521. doi: 10.1007/s40265-020-01288-x. [DOI] [PubMed] [Google Scholar]
- 1345.Zauderer MG, et al. EZH2 inhibitor tazemetostat in patients with relapsed or refractory, BAP1-inactivated malignant pleural mesothelioma: a multicentre, open-label, phase 2 study. Lancet Oncol. 2022;23:758–767. doi: 10.1016/S1470-2045(22)00277-7. [DOI] [PubMed] [Google Scholar]
- 1346.McCabe MT, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 1347.Vaswani RG, et al. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J. Med. Chem. 2016;59:9928–9941. doi: 10.1021/acs.jmedchem.6b01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1348.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117:6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1349.Stein EM, et al. The DOT1L inhibitor pinometostat reduces H3K79 methylation and has modest clinical activity in adult acute leukemia. Blood. 2018;131:2661–2669. doi: 10.1182/blood-2017-12-818948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1350.Waters NJ, et al. Exploring drug delivery for the DOT1L inhibitor pinometostat (EPZ-5676): Subcutaneous administration as an alternative to continuous IV infusion, in the pursuit of an epigenetic target. J. Control. Release. 2015;220:758–765. doi: 10.1016/j.jconrel.2015.09.023. [DOI] [PubMed] [Google Scholar]
- 1351.Wang N, Ma T, Yu B. Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct. Target. Ther. 2023;8:69. doi: 10.1038/s41392-023-01341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1352.Salamero O, et al. First-in-human phase I study of Iadademstat (ORY-1001): a first-in-class lysine-specific histone demethylase 1 A inhibitor, in relapsed or refractory acute myeloid leukemia. J. Clin. Oncol. 2020;38:4260–4273. doi: 10.1200/JCO.19.03250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1353.Stathis A, Bertoni F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018;8:24–36. doi: 10.1158/2159-8290.CD-17-0605. [DOI] [PubMed] [Google Scholar]
- 1354.Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: a novel epigenetic approach. Ann. Oncol. 2017;28:1776–1787. doi: 10.1093/annonc/mdx157. [DOI] [PubMed] [Google Scholar]
- 1355.Pirozzi CJ, Yan H. The implications of IDH mutations for cancer development and therapy. Nat. Rev. Clin. Oncol. 2021;18:645–661. doi: 10.1038/s41571-021-00521-0. [DOI] [PubMed] [Google Scholar]
- 1356.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1357.DiNardo CD, et al. Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 2018;378:2386–2398. doi: 10.1056/NEJMoa1716984. [DOI] [PubMed] [Google Scholar]
- 1358.Montesinos P, et al. Ivosidenib and azacitidine in IDH1-mutated acute myeloid leukemia. N. Engl. J. Med. 2022;386:1519–1531. doi: 10.1056/NEJMoa2117344. [DOI] [PubMed] [Google Scholar]
- 1359.Zhu AX, et al. Final overall survival efficacy results of ivosidenib for patients with advanced cholangiocarcinoma with IDH1 mutation: the phase 3 randomized clinical ClarIDHy trial. JAMA Oncol. 2021;7:1669–1677. doi: 10.1001/jamaoncol.2021.3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1360.Platten M, et al. A vaccine targeting mutant IDH1 in newly diagnosed glioma. Nature. 2021;592:463–468. doi: 10.1038/s41586-021-03363-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1361.Danne C, Rolhion N, Sokol H. Recipient factors in faecal microbiota transplantation: one stool does not fit all. Nat. Rev. Gastroenterol. Hepatol. 2021;18:503–513. doi: 10.1038/s41575-021-00441-5. [DOI] [PubMed] [Google Scholar]
- 1362.Dizman N, et al. Nivolumab plus ipilimumab with or without live bacterial supplementation in metastatic renal cell carcinoma: a randomized phase 1 trial. Nat. Med. 2022;28:704–712. doi: 10.1038/s41591-022-01694-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1363.da Silva Duarte V, et al. Chemoprevention of DMH-induced early colon carcinogenesis in male BALB/c mice by administration of Lactobacillus paracasei DTA81. Microorganisms. 2020;8:1994. doi: 10.3390/microorganisms8121994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1364.Yang J-C, Lu C-W, Lin C-J. Treatment of Helicobacter pylori infection: current status and future concepts. World J. Gastroenterol. 2014;20:5283–5293. doi: 10.3748/wjg.v20.i18.5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1365.Vítor JMB, Vale FF. Alternative therapies for Helicobacter pylori: probiotics and phytomedicine. FEMS Immunol. Med Microbiol. 2011;63:153–164. doi: 10.1111/j.1574-695X.2011.00865.x. [DOI] [PubMed] [Google Scholar]
- 1366.Haria M, Bryson HM, Goa KL. Itraconazole. A reappraisal of its pharmacological properties and therapeutic use in the management of superficial fungal infections. Drugs. 1996;51:585–620. doi: 10.2165/00003495-199651040-00006. [DOI] [PubMed] [Google Scholar]
- 1367.Piérard GE, Arrese JE, Piérard-Franchimont C. Itraconazole. Expert Opin. Pharmacother. 2000;1:287–304. doi: 10.1517/14656566.1.2.287. [DOI] [PubMed] [Google Scholar]
- 1368.Ban, L. et al. Anti-fungal drug itraconazole exerts anti-cancer effects in oral squamous cell carcinoma via suppressing Hedgehog pathway. Life Sci. 254, 117695 (2020). [DOI] [PubMed]
- 1369.Deng H, et al. Itraconazole inhibits the Hedgehog signaling pathway thereby inducing autophagy-mediated apoptosis of colon cancer cells. Cell Death Dis. 2020;11:539. doi: 10.1038/s41419-020-02742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1370.Wang L, Lankhorst L, Bernards R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer. 2022;22:340–355. doi: 10.1038/s41568-022-00450-9. [DOI] [PubMed] [Google Scholar]
- 1371.Prasanna PG, et al. Therapy-induced senescence: opportunities to improve anticancer therapy. J. Natl Cancer Inst. 2021;113:1285–1298. doi: 10.1093/jnci/djab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1372.Gorgoulis V, et al. Cellular senescence: defining a path forward. Cell. 2019;179:813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 1373.Freeman-Cook KD, et al. Discovery of PF-06873600, a CDK2/4/6 inhibitor for the treatment of cancer. J. Med. Chem. 2021;64:9056–9077. doi: 10.1021/acs.jmedchem.1c00159. [DOI] [PubMed] [Google Scholar]
- 1374.Wang C, et al. Inducing and exploiting vulnerabilities for the treatment of liver cancer. Nature. 2019;574:268–272. doi: 10.1038/s41586-019-1607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1375.Waksal JA, Bruedigam C, Komrokji RS, Jamieson CHM, Mascarenhas JO. Telomerase-targeted therapies in myeloid malignancies. Blood Adv. 2023;7:4302–4314. doi: 10.1182/bloodadvances.2023009903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1376.Kaletsch A, et al. Effects of novel HDAC inhibitors on urothelial carcinoma cells. Clin. Epigenetics. 2018;10:100. doi: 10.1186/s13148-018-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1377.Tuttle R, et al. Novel senescence associated gene, YPEL3, is repressed by estrogen in ER+ mammary tumor cells and required for tamoxifen-induced cellular senescence. Int. J. Cancer. 2012;130:2291–2299. doi: 10.1002/ijc.26239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1378.Rosemblit C, et al. Oncodriver inhibition and CD4(+) Th1 cytokines cooperate through Stat1 activation to induce tumor senescence and apoptosis in HER2+ and triple negative breast cancer: implications for combining immune and targeted therapies. Oncotarget. 2018;9:23058–23077. doi: 10.18632/oncotarget.25208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1379.Zhu Y, et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl-2 family of anti-apoptotic factors. Aging Cell. 2016;15:428–435. doi: 10.1111/acel.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1380.Bousset L, Gil J. Targeting senescence as an anticancer therapy. Mol. Oncol. 2022;16:3855–3880. doi: 10.1002/1878-0261.13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1381.Wakita M, et al. A BET family protein degrader provokes senolysis by targeting NHEJ and autophagy in senescent cells. Nat. Commun. 2020;11:1935. doi: 10.1038/s41467-020-15719-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1382.Amor C, et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature. 2020;583:127–132. doi: 10.1038/s41586-020-2403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1383.Huang W, Hickson LJ, Eirin A, Kirkland JL, Lerman LO. Cellular senescence: the good, the bad and the unknown. Nat. Rev. Nephrol. 2022;18:611–627. doi: 10.1038/s41581-022-00601-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1384.Laberge R-M, et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 2015;17:1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1385.Wang R, et al. Rapamycin inhibits the secretory phenotype of senescent cells by a Nrf2-independent mechanism. Aging Cell. 2017;16:564–574. doi: 10.1111/acel.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1386.Vargas AJ, Harris CC. Biomarker development in the precision medicine era: lung cancer as a case study. Nat. Rev. Cancer. 2016;16:525–537. doi: 10.1038/nrc.2016.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1387.Sveen A, Kopetz S, Lothe RA. Biomarker-guided therapy for colorectal cancer: strength in complexity. Nat. Rev. Clin. Oncol. 2020;17:11–32. doi: 10.1038/s41571-019-0241-1. [DOI] [PMC free article] [PubMed] [Google Scholar]